-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGLD-4-Mediated Translational Activation Regulates the Size of the Proliferative Germ Cell Pool in the Adult Germ Line
Throughout adulthood, animal tissue homeostasis requires adult stem cell activities. A tight balance between self-renewal and differentiation protects against tissue overgrowth or loss. This balance is strongly influenced by niche-mediated signaling pathways that primarily trigger a transcriptional response in stem cells to promote self-renewal/proliferation. However, the cell-intrinsic mechanisms that modulate signaling pathways to promote proliferation or differentiation are poorly understood. Recently, post-transcriptional mRNA regulation emerged in diverse germline stem cell systems as an important gene expression mechanism, primarily preventing the protein synthesis of factors that promote the switch to differentiation. In the adult C. elegans germ line, this study finds that the evolutionarily conserved cytoplasmic poly(A) polymerase, GLD-4, plays an crucial role in maintaining a healthy balance between proliferation and differentiation forces. This is in part due to translational activation of the mRNA that encodes the germ cell-expressed Notch signaling receptor, an essential regulator of proliferation. Moreover, GLD-4 activity is part of a redundant genetic network downstream of Notch that, together with several other conserved mRNA regulators, promotes differentiation onset. Given the widespread expression of these conserved RNA regulators in metazoans, cell fate balances that are reinforced by translational activation and repression circuitries may therefore be a general mechanism of adult tissue maintenance.
Published in the journal: . PLoS Genet 10(9): e32767. doi:10.1371/journal.pgen.1004647
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004647Summary
Throughout adulthood, animal tissue homeostasis requires adult stem cell activities. A tight balance between self-renewal and differentiation protects against tissue overgrowth or loss. This balance is strongly influenced by niche-mediated signaling pathways that primarily trigger a transcriptional response in stem cells to promote self-renewal/proliferation. However, the cell-intrinsic mechanisms that modulate signaling pathways to promote proliferation or differentiation are poorly understood. Recently, post-transcriptional mRNA regulation emerged in diverse germline stem cell systems as an important gene expression mechanism, primarily preventing the protein synthesis of factors that promote the switch to differentiation. In the adult C. elegans germ line, this study finds that the evolutionarily conserved cytoplasmic poly(A) polymerase, GLD-4, plays an crucial role in maintaining a healthy balance between proliferation and differentiation forces. This is in part due to translational activation of the mRNA that encodes the germ cell-expressed Notch signaling receptor, an essential regulator of proliferation. Moreover, GLD-4 activity is part of a redundant genetic network downstream of Notch that, together with several other conserved mRNA regulators, promotes differentiation onset. Given the widespread expression of these conserved RNA regulators in metazoans, cell fate balances that are reinforced by translational activation and repression circuitries may therefore be a general mechanism of adult tissue maintenance.
Introduction
During development, tissues grow to form functional organs. In adulthood, animal tissues remain constant in size, in part, as a result of the dynamic balance between self-renewal/proliferation and differentiation. Perturbation of this balance affects tissue homeostasis and, consequently, compromises organ function. While excess proliferation contributes to tumorigenesis, a deficit in proliferation leads to tissue degeneration. Hence, tight regulatory mechanisms are in place to control the balance between self-renewal/proliferation and differentiation. One prevalent cell-extrinsic regulatory mechanism of stem cells to self-renew/proliferate is their dependency on supporting niche cells, which trigger established signal transduction pathways that primarily lead to changes at the transcriptional level. However, to fine-tune proper tissue homeostasis and to provide tight feedback controls, additional cell-intrinsic gene expression mechanisms are likely to exist.
In recent years, invertebrate germline tissues emerged as powerful in vivo models to investigate the balance between proliferation and differentiation. One influential paradigm is the adult “female” germ line of C. elegans, which depends on a single somatic niche cell and maintains a strict spatio-temporal organization of proliferating and differentiating cells [1]. Undifferentiated germ cells proliferate exclusively in the distal end of the germ line, termed the proliferative zone (PZ) [2], [3], [4]. The PZ is proposed to contain a distal pool of germline stem cell-like cells (GSCs) and a proximal pool of transit amplifying cells that gradually mature to start differentiation at a defined distance from the distal end [1], [5], termed the mitosis-to-meiosis boundary. Germ cells crossing this boundary enter meiotic prophase, which is here defined as differentiation onset [2], [6], [7]. Germline proliferation relies on the Notch signaling pathway that is instructed by the somatic distal tip cell (DTC) [8], [9], [10]. Consistent with its continuous requirement for germ cell proliferation in the adult, the inactivation of Notch signaling leads to progressive loss of GSCs, due to differentiation of all germ cells [9]. Conversely, constitutive activation of Notch results in the expansion of the proliferative GSC pool at the expense of differentiation [11], [12]. In agreement with this, germ cells in the PZ express the Notch receptor GLP-1, while differentiating cells lose GLP-1 expression [13]. Hence, Notch-mediated transcriptional regulation of mitotic fate-promoting genes is suggested to directly maintain the proliferative fate [14], [15], [16]. However, germ cell-intrinsic mechanisms that promote niche-mediated germ cell proliferation are still widely unknown.
In nematodes and flies, germ cells also utilize conserved translational repressors to promote the undifferentiated state [17]. In C. elegans, two nearly identical translational repressors of the PUF RNA-binding protein family, FBF-1 and FBF-2, jointly referred to as FBF, are essential for adult GSCs [18]. FBF recognizes specific sequence elements (FBEs) in its mRNA targets and, by translationally repressing numerous meiosis-promoting genes, FBF is critical for maintaining the undifferentiated, proliferative state [7], [18], [19], [20]. Moreover, the fbf-2 locus is a proposed target of Notch-mediated regulation [14], [21], thus linking transcriptional activation with translational repression, the two dominant mechanisms used for sustained germ cell proliferation in different organisms [1].
Across species, differentiation onset of germ cells depends on translational control [17]. In nematodes, the STAR-type RNA-binding protein, GLD-1, inhibits GLP-1 protein accumulation [22], [23] and recognizes glp-1 mRNA by three GLD-1-binding motifs (GBMs) present in its 3′UTR [24], [25]. Meiotic prophase entry also requires the Nanos protein family member, NOS-3, a presumed translational repressor of yet unknown mitosis-promoting genes [21]. However, in the absence of GLD-1, NOS-3, or both, germ cells enter meiotic prophase in the adult [7], [26]. The cytoplasmic poly(A) polymerase (cytoPAP) complex GLD-2/GLD-3 is a proposed translational activator of meiosis-promoting mRNAs, envisioned to extend their poly(A) tail lengths. GLD-2 is a non-canonical nucleotidyltransferase, stimulated by the Bicaudal-C family member, GLD-3 [27], [28]. However, in the absence of GLD-2, GLD-3, or both, the PZ is expanded but meiosis is still initiated [7]. This complexity highlights that differentiation onset is in general a multi-pathway-regulated process [17].
In the current model of the core genetic network underlying differentiation onset in C. elegans, the four meiosis-promoting RNA regulators act in two parallel pathways. The two translational repressors (gld-1 and nos-3) form the first pathway; the two translational activators (gld-2 and gld-3) form the second pathway. This genetic redundancy is most apparent in germ cells that lack GLD-3 and NOS-3, as they do not enter meiotic prophase and continue to proliferate [7]. Importantly, tumorous proliferation of gld-3 nos-3 double mutant germ cells is independent of Notch signaling and dependent on cyclin E activity [4], [7]. Intriguingly, germ cells lacking GLD-2 and NOS-3 are able to start meiotic prophase [6], [7]. This suggests that the current pathway assignments are too simplistic and emphasizes that more meiosis-promoting regulators must exist [4], [6], [7]. Especially, commitment to female meiotic progression provides precedence for redundant translational activation activities in C. elegans. Here, in addition to GLD-2 cytoPAP-mediated GLD-1 expression [29], the GLD-4/GLS-1 cytoPAP complex has been identified to translationally activate gld-1 mRNA [30]. As a non-canonical poly(A) polymerase, GLD-4 is most similar to the conserved group of Trf4/5-type RNA modifiers that regulate RNA stability in the nucleus [30], [31], [32]. However, GLD-4 poly(A) polymerase is cytoplasmic and requires for its functions the nematode-specific protein, GLS-1 [30], [33]. In the absence of GLD-2, the GLD-4/GLS-1 cytoPAP complex is essential for female meiotic progression into pachytene [30].
In this study, we report that the GLD-4/GLS-1 cytoPAP complex has a dual role in regulating the balance between proliferation and differentiation. We find that the GLD-4/GLS-1 cytoPAP complex is crucial to maintain germ cell proliferation in the adult, in part by promoting robust translation of glp-1 mRNA. Moreover, to ensure that meiosis-promoting factors are inefficiently translated, GLD-2 cytoPAP levels are kept low in the GSC pool by FBF-mediated translational repression. Lastly, we also find that GLD-4/GLS-1 cytoPAP promotes meiotic prophase entry, in parallel to GLD-2 cytoPAP and independently of Notch. Our data suggest that two translational feedback loops limit the size of the proliferative germ cell pool and maintain a healthy balance of germ cell proliferation and differentiation in the adult germ line.
Results
GLD-4/GLS-1 cytoPAP activity maintains the size of the proliferative zone
On average, the PZ of adult germ lines extends from the first germ cell row at the distal end further proximally to row 20, where germ cells start differentiation by entering meiotic prophase (Figure 1A,B). In wild type, the PZ is populated by about 225–250 germ cells (Figure 1C). Since there are no molecular markers for subpopulations of cells in the PZ, like stem cells, transit amplifying cells and cells in pre-meiotic S-phase, the start of meiotic prophase is commonly defined as the onset of differentiation [1], [34]. Differentiation is revealed by the germ cells' specific nuclear architecture and chromatin morphology, the combinatorial expression and localization of the meiotic cohesin REC-8, the synaptonemal protein HIM-3, and phosphorylated nuclear envelope protein pSUN-1 [34] (Figure 1A,B).
Fig. 1. gld-4 and gld-2 have opposing functions in regulating the balance between proliferation and differentiation. 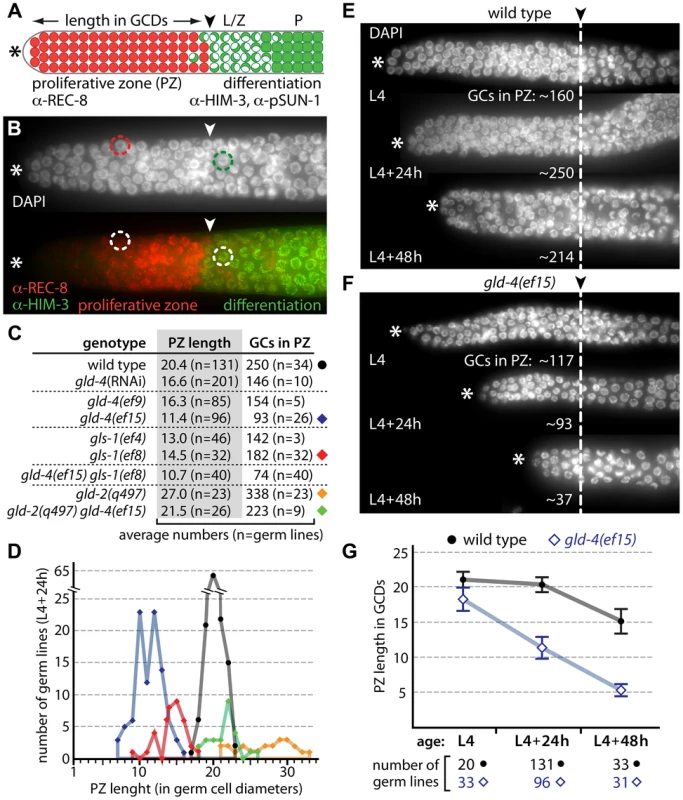
(A) Diagram of the distal end of an adult germ line (not to scale). Germ cells divide in the proliferative zone (PZ; red circles) and express REC-8 prior to meiotic prophase entry. Germ cells in meiotic prophase (green) express HIM-3 and pSUN-1; leptotene/zygotene (L/Z; half-filled circles) and pachytene (P; squares). In adulthood, a mitosis-to-meiosis boundary (arrowhead) is maintained at a defined distance from the distal tip (asterisk). GCDs, germ cell diameters. (B) Corresponding immunofluorescence micrograph of a distal gonad, illustrating a typical PZ nucleus (red circle) and a crescent-shaped meiotic prophase nucleus (green circle). The mitosis-to-meiosis boundary is apparent from REC-8 and HIM-3 expression. (C,D) Proliferative zone measurements of cytoPAP mutant gonads reveal germ cell (GC) number differences in young adults (L4+24 h). Circle and diamonds to the right side of the table in (C) correspond to the data shown in (D). (E,F) Age-dependent PZ changes are enhanced in gld-4 mutants. Dashed line, mitosis-to-meiosis boundary. (G) Quantification of PZ length changes of E and F over time. Error bars, standard deviations. Germ cells in single mutants of meiosis-promoting genes (i.e. gld-1, nos-3, gld-2, gld-3) initiate meiotic prophase [7]. However, shifts in the position of the mitosis-to-meiosis boundary suggest a role in proliferation or differentiation. For example, in gld-2 single mutants, the PZ is extended and contains more germ cells than wild type [7] (Figure 1C,D), consistent with gld-2's function in promoting meiotic entry [35]. We found that gld-4 and gls-1 single mutants have smaller PZs with fewer germ cells (Figure 1C,D). The strength of the reduction appears to correlate with the reported allelic strengths of the individual mutations [30], [33] (Figure 1C). As the PZ of gld-4 gls-1 double mutants is similarly reduced (Figure 1C), these results argue for a common role of gld-4 and gls-1 in promoting mitosis. The PZ of the gld-2 gld-4 double mutant is similar to wild type in size and germ cell number (Figure 1C,D). Together, these results suggest that gld-2 and gld-4 have independent and opposing roles to set the mitosis-to-meiosis boundary in adults.
The PZ expands during larval development and is maintained during adulthood [8]. We measured the size of the PZ at the last larval stage (L4), and 24 hours (h), and 48 h later in young adults (Figure 1E–G). The difference between wild type and gld-4 is the smallest in L4 and greatest during adulthood, due to a large relative shrinkage of the PZ in gld-4 young adults (Figure 1E–G). Therefore, gld-4 activity is primarily important for the maintenance but not establishment of the PZ during early adulthood.
Expression of a glp-1 mRNA translational reporter depends on GLD-4/GLS-1, but not on GLD-2, cytoPAP
The documented presence of GLD-4 [30] and GLS-1 [33] in the distal end of the germ line and the single mutant phenotypes argue for a role of gld-4 and gls-1 in promoting germ cell proliferation. CytoPAPs are envisioned to regulate poly(A) tail metabolism of target mRNAs in a positive manner [31]. Biochemically, cytoPAPs elongate poly(A) tails, which in turn stabilize mRNAs and enhance their translation. We hypothesized that GLD-4 targets mRNAs encoding proteins important for proliferation in the PZ. An obvious, but not exclusive, candidate for this regulation is the Notch receptor-encoding glp-1 mRNA.
Notch expression is regulated at multiple levels in C. elegans [13], [23], [36]. To uncouple mRNA regulation from protein regulation, we used a translational reporter of GFP::H2B under the control of the glp-1 3′UTR [25] (Figure 2). The glp-1 3′UTR reporter is driven by a ubiquitous germ cell-specific promoter and encodes a translational fusion product of GFP and histone 2B (Figure 2A). This nuclear GFP signal reflects GLD-1-mediated regulation of the glp-1 mRNA [25].
Fig. 2. GLD-4 affects glp-1 3′UTR GFP reporter mRNA translation. 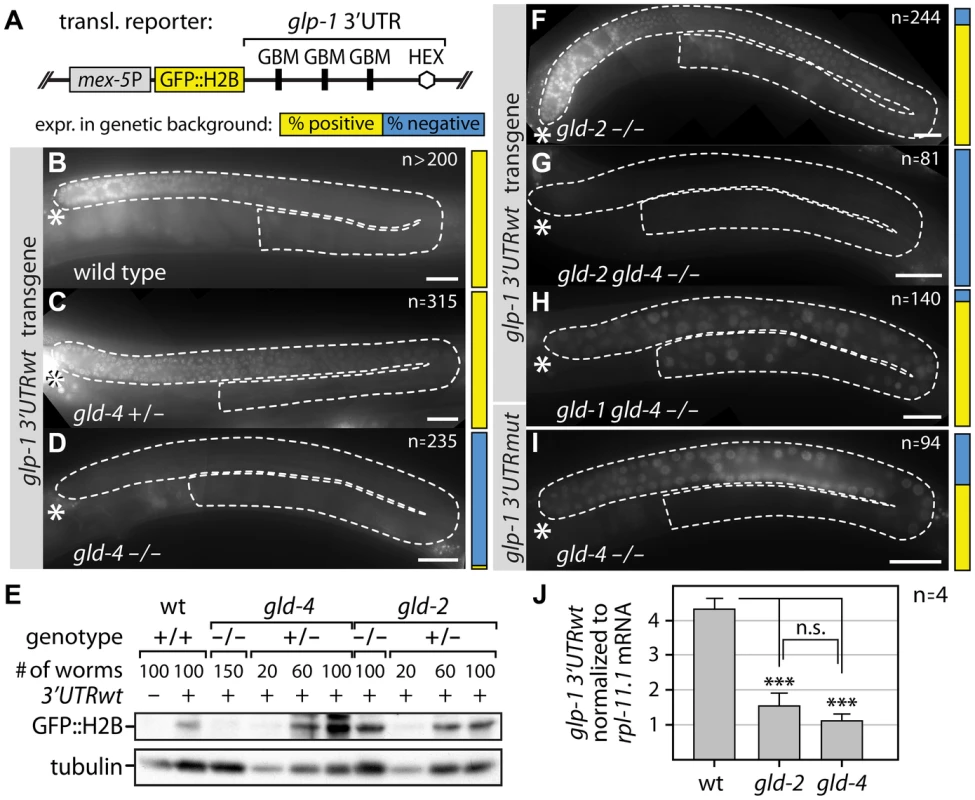
(A) Schematic representation of the glp-1 3′UTR reporter transgene. Germline expression of a GFP-Histone 2B-fusion product (GFP::H2B) is driven by the mex-5 promoter (mex-5P). GBM, GLD-1-binding motif; HEX, 3′end formation sequence. (B–D, F–I) Micrographs of adult gonads, expressing a GFP::H2B translational glp-1 3′UTR reporter in wild-type, heterozygous (+/−), or homozygous (−/−) mutant backgrounds, reveal gld-4-dependent reporter protein expression. Transgenic animals carry either a wild type (B–D, F–H) or a GBM-mutant (I) glp-1 3′UTR. The penetrance of analyzed germ lines (n) expressing the respective translational reporter is indicated by a color-coded bar (described in A) next to each micrograph. Dashed white lines mark gonads as assessed by DIC microscopy; asterisk, distal tip. Scale bars: 25 µm. (E) Quantification of GFP::H2B protein expression of the glp-1 3′UTRwt translational reporter in given genotypes by α-GFP immunoblotting; tubulin serves as loading control. #, number of animals per lane. (J) Quantification of glp-1 3′UTR reporter mRNA levels by RT-qPCR normalized to rpl-11.1 mRNA. Endogenous glp-1 mRNA levels were similar among all three genotypes. Error bars are standard error of the mean (SEM). ***, p<0.001; n.s., not significant (Student's t-test). In a wild-type background, reporter GFP expression is present in all animals analyzed and its pattern is similar to endogenous GLP-1 protein expression [13], [25] (Figure 2B). To assess whether reporter GFP expression is under the influence of GLD-4 cytoPAP activity, we crossed the glp-1 3′UTR reporter locus into the strong loss-of-function gld-4(ef15) mutant background (Figure 2C,D). To control for unexpected genetic background influences, we compared heterozygous and homozygous gld-4 siblings from the progeny of a heterozygous mother (see Materials and Methods). In the gld-4 heterozygous mutant, reporter GFP expression is similar to a wild-type background (compare Figure 2B with C). Strikingly, upon gld-4 removal, reporter expression was undetectable in almost all germ lines (Figure 2D). Consistent with a reduction in the GFP signal, we also observed lower GFP protein amounts by immunoblotting. When comparing gld-4 animals to wild-type background, we observed a reduction of >80% in protein abundance (Figure 2E). These results imply that the expression of the glp-1 3′UTR reporter depends on gld-4 cytoPAP activity.
GLS-1 and GLD-4 function together in meiotic progression [30], and in promoting differentiation onset (Figure 1C). Similar to the gld-4 mutant, reporter GFP expression is undetectable in most gls-1(ef8) mutant germ lines (∼87%, n = 220), suggesting that gls-1 promotes glp-1 3′UTR reporter expression similar to gld-4 activity.
To investigate whether gld-4 is the only known cytoPAP regulating reporter expression, we assessed GFP expression in gld-2 mutants and detected it in almost all germ lines (Figure 2F). Moreover, the amounts of GFP reach wild-type protein levels and are similar between gld-2 homozygous and heterozygous mutants (Figure 2E). Importantly, reporter GFP expression is still dependent on gld-4 activity in the gld-2 mutant background, as its expression is undetectable in all gld-2 gld-4 homozygous double mutants (Figure 2G). These results suggest that glp-1 3′UTR reporter expression is largely independent of gld-2 cytoPAP activity, and specifically dependent on gld-4 cytoPAP activity.
glp-1 translational reporter expression is post-transcriptionally controlled
To further investigate at which level GLD-4 cytoPAP may regulate glp-1 3′UTR reporter expression, we made use of GLD-1, a known translational repressor of glp-1 mRNA [23]. In gld-1 single mutants, reporter GFP is expressed in the PZ and in differentiating germ cells (100%, n = 140). To test, whether loss of GLD-1 would de-repress reporter GFP expression in the gld-4 mutant, we analyzed GFP::H2B expression in the gld-1 gld-4 double mutant background. Most germ lines weakly express GFP when compared to gld-4 mutants (compare Figure 2H with 2D). A similar weak de-repression is observed in gld-4 mutant germ lines that contain mutated GLD-1-binding site reporter mRNAs (glp-1 3′UTR mut) (Figure 2I). Taken together, these results confirm that the glp-1 3′UTR reporter can be translated in a gld-4 mutant background and that expression of the glp-1 3′UTR reporter is partly dependent on the GLD-4 cytoPAP even when GLD-1-mediated repression is removed.
Several mechanisms may account for reduced glp-1 3′UTR reporter expression in the absence of gld-4. To confirm that the effects on GFP::H2B expression are due to translational and not transcriptional regulation of the glp-1 3′UTR reporter, we examined the mRNA levels of the wild-type glp-1 3′UTR reporter by RT-qPCR (Figure 2J). Compared to wild type, we noticed a reduction of ∼4-fold in both gld-4 and gld-2 mutant backgrounds (Figure 2J), suggesting that glp-1 3′UTR reporter mRNA is less abundant in either cytoPAP mutant. Importantly, the glp-1 3′UTR reporter mRNA levels are similar to each other in both cytoPAP homozygous mutants, yet they give rise to different amounts of reporter protein (compare Figure 2D with 2F, and Figure 2E). Hence, we conclude that the major reduction in reporter GFP expression in gld-4 mutants is primarily at the translational and not at the transcriptional level.
Endogenous GLP-1 protein expression depends on GLD-4 cytoPAP activity
To further investigate whether endogenous GLP-1 protein expression is one likely candidate of gld-4-mediated regulation, we measured GLP-1 protein expression in gld-4 mutants and compared it to wild type (Figure 3). By quantifying GLP-1 intensities in distal germ lines of L4+24 h and L4+48 h animals, we observed a significant decrease in the gld-4 mutant background in the PZ over time (Figure 3A,B). When we measured endogenous glp-1 mRNA levels in L4+24 h animals we observed a mild increase in gld-4(ef15) mutants compared to wild type (Figure 3C). Together these observations suggest that gld-4 promotes GLP-1 expression post-transcriptionally.
Fig. 3. GLD-4 promotes endogenous GLP-1 expression. 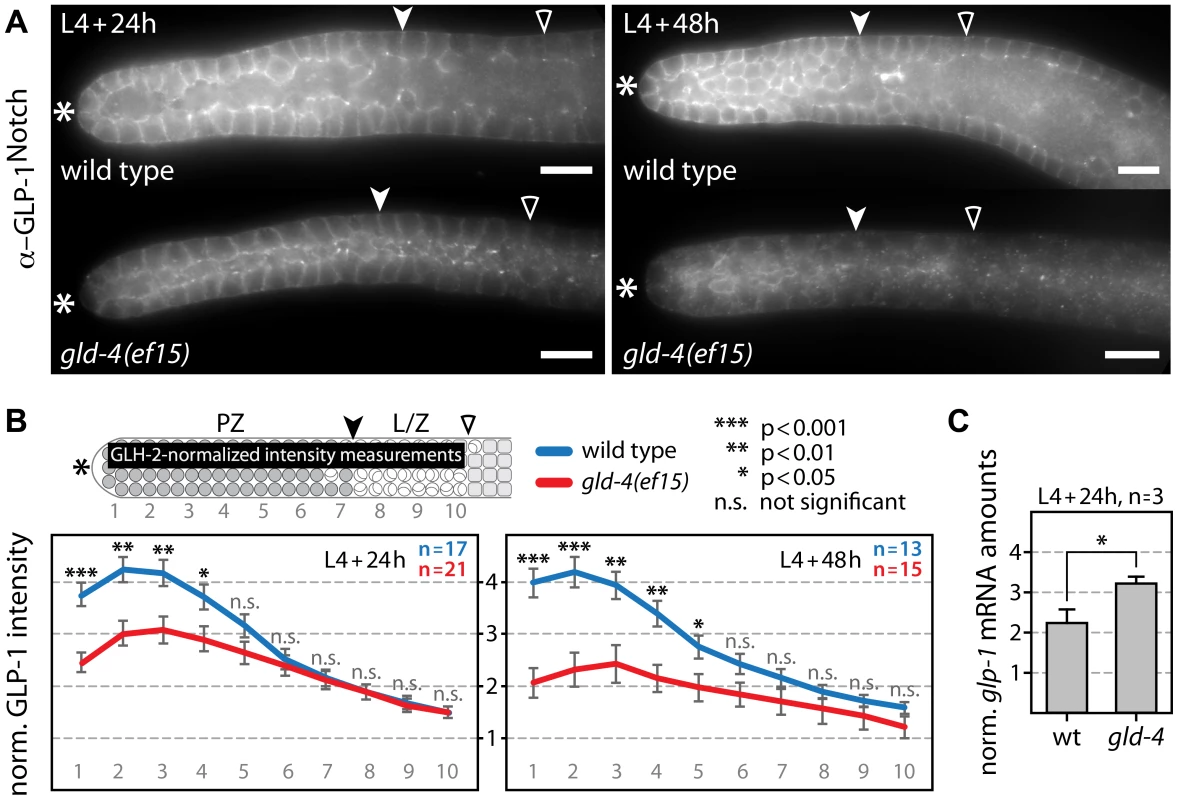
(A) Median images of extruded gonads stained with α-GLP-1 antibodies in given genetic backgrounds and at two developmental time points. Asterisk, distal tip; arrowhead, mitosis-to-meiosis boundary; empty carat, beginning of pachytene. Scale bars: 10 µm. (B) Quantification of the distal region of immunostained germ lines (n) from A. The α-GLP-1 fluorescent signal was normalized to α-GLH-2 signal. The values of the y-axis represent arbitrary units. Error bars are SEM; p values from Student's t-test. The top scheme indicates the area used for intensity measurements in the distal germ line (black bar). PZ, proliferative zone; L/Z, leptotene/zygotene; other label as in A. (C) Quantification of endogenous glp-1 mRNA levels by RT-qPCR normalized to rpl-11.1 mRNA. Error bars are SEM. *, p<0.05 (Student's t-test). A prerequisite for GLD-4/GLS-1-mediated glp-1 mRNA regulation is that they form an mRNP complex. To test for a possible association of GLD-4 and GLS-1 with glp-1 mRNA, we performed several RNA co-immunoprecipitation (RIP) experiments, using GLD-4-specific, GLS-1-specific, and non-specific antibodies. Subsequent RT-PCR (Figure 4A) and RT-qPCR (Figure 4B) analysis of different RIP experiments revealed a specific enrichment of endogenous glp-1 mRNA, which is similar to the positive control, gld-1 mRNA (Figure 4B). These results demonstrate an association of GLD-4/GLS-1 cytoPAP complex with endogenous glp-1 mRNA and establish a potential physical link for glp-1 mRNA translational regulation.
Fig. 4. glp-1 mRNA associates with GLD-4 and is a likely target of poly(A) tail extension and translational activation. 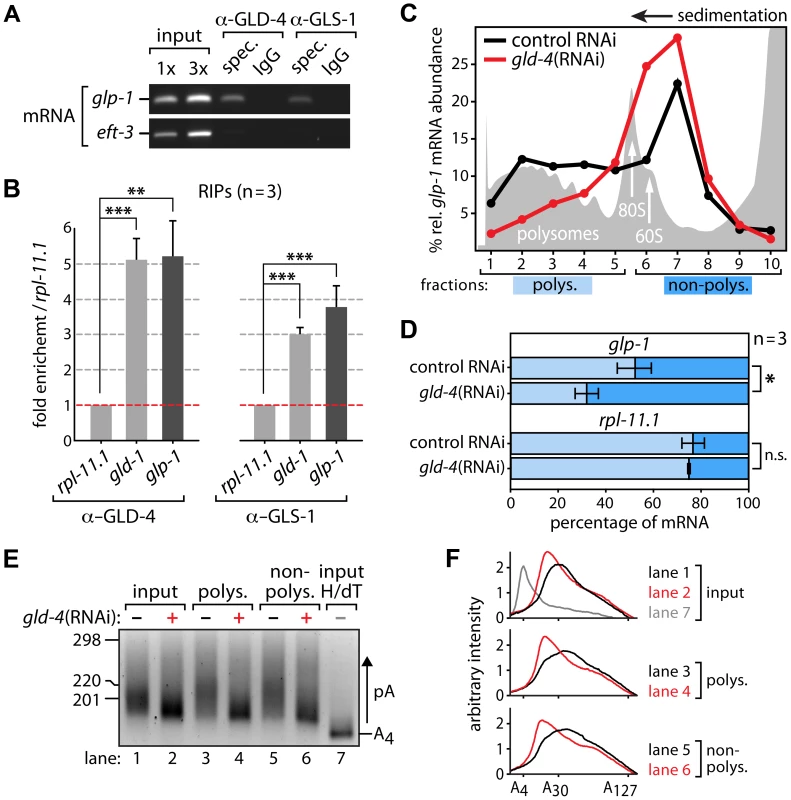
(A,B) RNA-coimmunoprecipitation experiments (RIPs) of GLD-4 and GLS-1 proteins specifically enrich glp-1 mRNA and the positive control gld-1 mRNA. eft-3 and rpl-11.1 mRNA served as negative controls. (A) A representative ethidium bromide-stained agarose gel of semiquantitative RT-PCR products from three independent biological replicates. (B) Quantitative RT-PCR measurements of three additional RIPs. Error bars are SEM. ***, p<0.001; **, p<0.01; n.s., not significant (Student's t-test). (C,D) Translational efficiency of glp-1 mRNA depends on gld-4 activity. The data are representative of three independent biological experiments. (C) Polysome gradient. Top is to the right; grey peaks represent optical density read of 258 nm; the peaks of the large ribosomal subunit (60S), monosomes (80S), and polysomes are indicated. Relative glp-1 mRNA levels are lower in polysome fractions of gld-4(RNAi) as measured by RT-qPCR. (D) Quantification and comparison of glp-1 mRNA in pooled polysomal (polys.) and non-polysomal (non-polys.) fractions. Each measurement was normalized to an internal spike-in control (see Materials and Methods). Error bars are SEM. *, p<0.05; n.s., not significant (Student's t-test). (E,F) poly(A) tails of glp-1 mRNA are reduced upon gld-4(RNAi). (E) Representative PAT assay (n = 2) of the glp-1 mRNA material from (C) and the gradient input material. Nucleotide size marker to the left. Lane 7 reflects a 3′UTR with a strongly reduced poly(A) tail (pA) after RNAase H and oligo dT treatment (H/dT). (F) Line scans of PAT assay from (E). Cytoplasmic polyadenylation affects RNA stability and translational efficiency [37]. To test whether ribosomal engagement of the endogenous glp-1 mRNA requires GLD-4 cytoPAP, we performed sucrose gradient sedimentation experiments. In theory, the more ribosomes are attached to an mRNA, the further the mRNA migrates into the gradient during ultra centrifugation. Therefore, efficiently translated mRNAs will be in the heavier, polysome fractions of the gradient, while poorly or non-translated mRNAs tend to sediment to lighter, non-polysomal fractions. Due to the large amounts of material needed, we compared control RNAi and gld-4(RNAi) knockdown worms (Figure 4C), knowing that gld-4(RNAi) efficacy is less robust than using mutants. In extracts of wild type and control RNAi (Figure 4C), about 50% of the endogenous glp-1 mRNA resides in the polysome fraction, suggesting that half of the glp-1 mRNA population is actively translated, consistent with the known germline and embryonic translational repression of glp-1 mRNA [23], [38]. Upon knockdown of gld-4, but not in control RNAi, we observed a shift of glp-1 mRNA into lighter fractions of the gradient (Figure 4D). This reflects a specific decrease in translational competence of endogenous glp-1 mRNA as rpl-11.1, a germ line-enriched mRNA that encodes a protein of the large ribosomal subunit [39], is unaffected (Figure 4D).
CytoPAPs modify the 3′ends of RNAs [31]. To investigate whether GLD-4 affects the length of the glp-1 mRNA poly(A) tail, we performed a poly(A) test (PAT) assay [40], and compared endogenous glp-1 mRNA poly(A) tails, using sucrose gradient fractioned mRNA and non-fractionated input as our starting material. To obtain enough RNA material for the PAT assay and to discriminate translationally active from inactive mRNA pools, we combined several samples of the non-polysomal and polysomal fractions. While all three samples show reduced glp-1 poly(A) tail lengths in gld-4(RNAi) compared to control RNAi knockdowns, we observe no clear difference between the respective non-polysomal and polysomal fractions (Figure 4E,F). The observed poly(A) tail differences are consistent with the contribution of gld-4 activity to gld-1 mRNA [40]. This data suggests that GLD-4 cytoPAP activity has an overall impact on glp-1 poly(A) tail status. Together, our combined results suggest that GLD-4 association with endogenous glp-1 mRNA may stimulate its efficient translation.
GLD-4 and GLD-2 cytoPAP expression differs in the PZ
GLD-4 and GLD-2 cytoPAP expression patterns are distinct in “female” germ lines. GLD-4 is expressed equally strong within the entire PZ and in meiosis [30] (Figure 5A). By contrast, GLD-2 is poorly expressed in the distal half of the PZ, becomes more abundant further proximal, and is most abundant in cells that have entered meiosis [28] (Figure 5A). Hence, the differential expression of the two proteins in the PZ may form the basis of GLD-4's unique role in mitosis and GLD-2's role in meiotic entry.
Fig. 5. Differential GLD-4 and GLD-2 expression in the proliferative zone is FBF dependent. 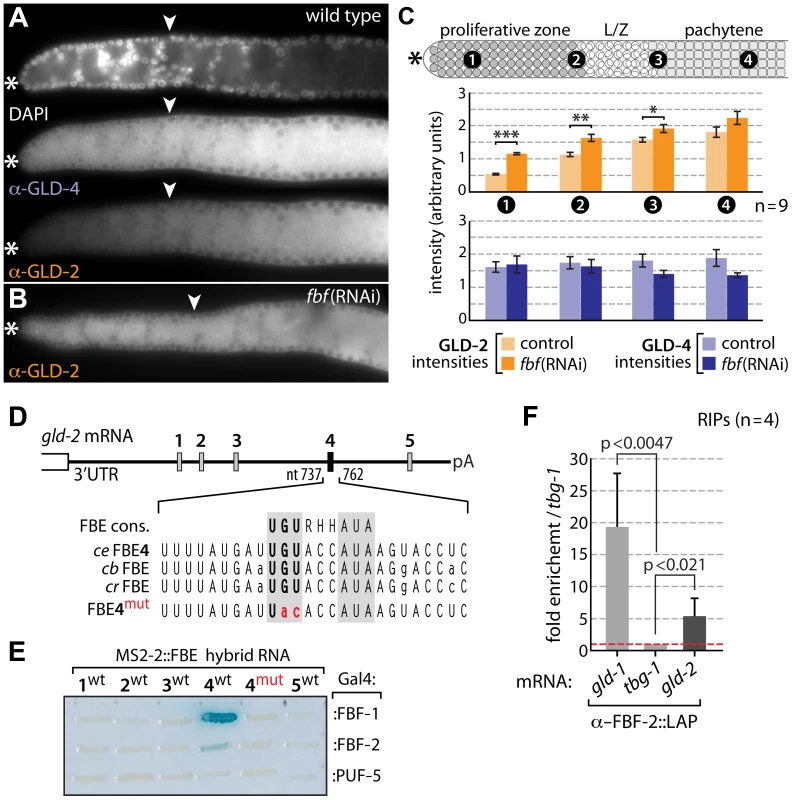
(A) GLD-4 expression is equal across the distal germ line. GLD-2 intensities increase from low-to-high in a distal-to-proximal manner. Extruded gonads of indicated genotype stained with DAPI, α-GLD-2, α-GLD-4, and α-GLH-2 as a positive tissue penetration control (not shown). Asterisk, distal tip; arrowhead, mitosis-to-meiosis boundary. (B,C) Distal GLD-2 expression is repressed by fbf activity. (B) Example of an fbf(RNAi) immunostained extruded gonad. For the complete RNAi experiment see Figure S1. (C) Quantification of the complete fbf(RNAi) experiment. Four different regions of nine germ lines per genotype were analyzed in their median, primarily cytoplasmic area. Error bars are SEM. ***, p<0.001; **, p<0.01; *, p<0.05; bars without indicated p value are statistically not significant (Student's t-test). (D, E) FBF binds specifically to at least one of the five predicted sequence elements in the gld-2 3′UTR. (D) Schematic drawing of the 1094 nt long gld-2 3′UTR. Sequence alignment of FBF-binding element consensus (FBE cons.) sequence [14] and the conserved FBE4 element in three Caenorhabditis species: ce, C. elegans; cb, C. briggsae; cr, C. remanei. pA indicates beginning of the poly(A) tail. (E) Yeast three-hybrid assay. RNA hybrid and Gal4-protein fusions are indicated. FBF-1, FBF-2 and PUF-5 belong to same RNA-binding protein family. Note, the wild-type (wt) and mutant (mut) sequence of FBE4 tested is larger than the given sequences (see Materials and Methods). A positive and negative control RNA was included (not shown) and protein expression was confirmed by western blotting (not shown). (F) LAP-tagged FBF-2 associates with endogenous gld-2 mRNA in RNA-coimmunoprecipitation experiments (RIPs) directed against the GFP portion of the fusion protein. Intriguingly, the protein expression pattern of GLD-2 does not match its ubiquitous mRNA expression pattern in the distal PZ [28]. This suggests translational regulation of GLD-2 expression. An obvious translational repressor in this region is FBF, which represses two mRNAs encoding meiosis-promoting regulators, GLD-1 [18] and GLD-3 [7]. To test for FBF-mediated gld-2 mRNA repression, we knocked down fbf by RNAi and assessed GLD-2 protein abundance in the distal-most germ line by indirect immunofluorescence, using GLD-4 as a reference, and quantified the amounts (Figure 5B,C). While GLD-4 levels are not significantly different between fbf(RNAi) and control RNAi germ lines, GLD-2 expression levels in the PZ are higher in fbf(RNAi) than in wild type (compare Figure 5A and 5B) or control RNAi experiments (Figure S1A–C). The GLD-2 protein increase is largely limited to the distal half of the PZ: ∼2.2-fold more in cells most distal (Figure 5C, area 1), compared to ∼1.5-fold more in cells most proximal to the PZ (Figure 5C, area 2). Such a restriction to the proliferative zone is consistent with previous reports on FBF activity [18], [20], [41] and suggests that GLD-2 but not GLD-4 is a specific target of FBF regulation.
FBF interacts with mRNAs through the conserved FBF-binding element (FBE) [18]. We identified five putative FBEs in the 3′UTR of gld-2 mRNA (Figure 5D) and tested each element for binding to FBF protein in a yeast 3-hybrid assay. Only FBE4 in its wild-type sequence was consistently and specifically bound by FBF (Figure 5E). Neither element was bound by PUF-5 (Figure 5E), a different C. elegans PUF protein that is abundantly expressed in differentiating female gametes [42]. Intriguingly, the bound FBE sequence is also present in two closely related Caenorhabditis species, suggesting that gld-2 mRNA translational repression may be conserved (Figure 5D). Moreover, RIP experiments of GFP-tagged FBF-2 confirmed a physical association of gld-2 mRNA with FBF in worm lysates, which appears to correlate with the number of active FBEs in the tested mRNAs (Figure 5F); the positive control, gld-1 mRNA, possesses two functional FBEs and was enriched strongest [18]. Taken together, we conclude that consistent with published FBF-1 RIP-Chip experiments [19], GLD-2 but not GLD-4 is most likely a direct target of the central mitosis-promoting translational repressor, FBF. Consistent with previous genetic findings [7], an evolutionary conserved translational repression of GLD-2 cytoPAP in undifferentiated cells might be pivotal for the robustness of the balance between proliferation and differentiation.
The GLD-4/GLS-1 cytoPAP has a second role in promoting meiosis
The current framework of the core regulatory network underlying meiotic entry appears incomplete and a third meiosis-promoting activity is likely to exist (Figure 6A) [4], [6], [7], [34]. Even though both meiosis-promoting pathways are inactive in the gld-2; nos-3 double mutant, germ cells enter meiosis [7], [26] (Figure 6B,D; Table 1). Intriguingly, GLD-2 and GLD-4 have a combined function during late meiosis when germ cells are past the onset of differentiation [30]. Hence, it seemed plausible that a further biological overlap of those two enzymes may exist at differentiation onset. Indeed, we find that the triple mutant gld-2 gld-4; nos-3 lacks any signs of differentiation and it is tumorous (Figure 6C,E; Table 1). This demonstrates that gld-4 activity promotes meiotic entry in the absence of gld-2 and nos-3.
Fig. 6. gld-4 and gls-1 promote onset of differentiation in parallel to gld-2 and nos-3. 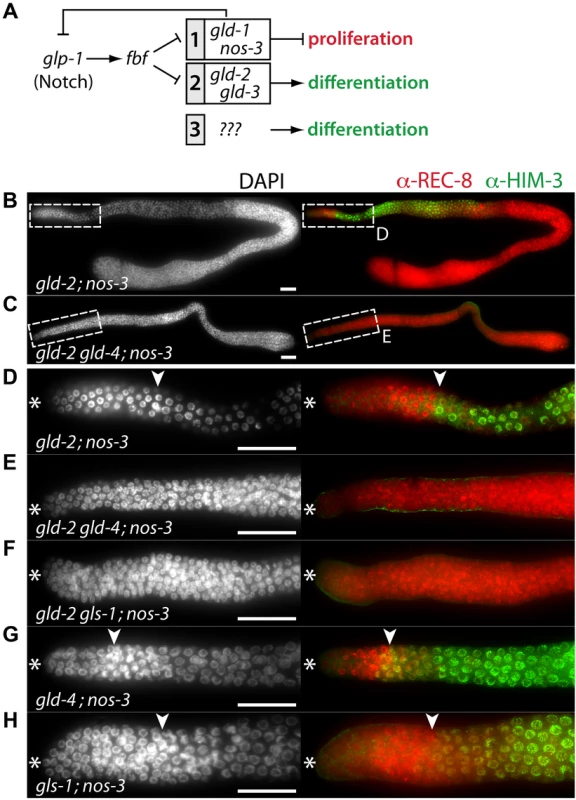
(A) The current genetic wiring of the core regulatory network that regulates the balance between proliferation and differentiation onset. Two genetic pathways of redundantly acting translational regulators operate downstream of the translational repressor, FBF. A third, yet undefined, pathway has been evoked [4], [6]. Note that not all genes are equivalent in the two pathways; only gld-3 nos-3 double mutants lack any signs of differentiation [7]. (B–H) Complete gonads stained with DAPI (left column), and with α-REC-8 and α-HIM-3 (right column) antibodies. Dashed boxes in B and C are close ups of D and E. See Table 1 for the total number of analyzed germ lines. (D–H) Distal region of extruded gonads. Asterisk, distal tip; arrowhead, mitosis-to-meiosis boundary. Scale bars: 50 µm. Tab. 1. Mitosis-to-meiosis decision phenotypes. 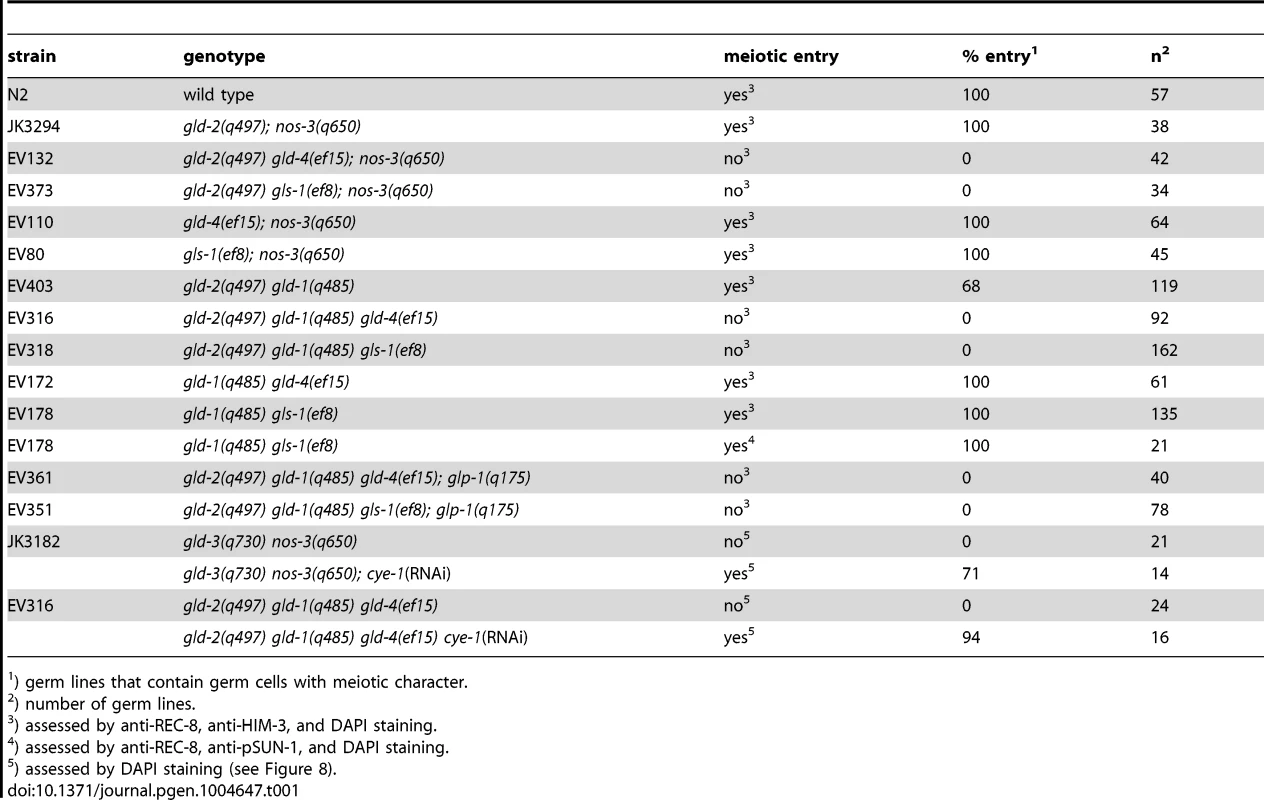
) germ lines that contain germ cells with meiotic character. GLS-1 stimulates GLD-4 enzymatic activity and the GLD-4/GLS-1 cytoPAP complex promotes late meiosis [30]. To test if gld-4 activity requires gls-1 function for promoting meiotic entry, we generated the gld-2 gls-1; nos-3 triple mutant. Similar to the gld-2 gld-4; nos-3 triple mutant, no meiotic entry was observed (Figure 6F; Table 1), indicating a shared function of gld-4 and gls-1. Together this suggests that in addition to a requirement for proliferation, the GLD-4/GLS-1 cytoPAP complex promotes the onset of differentiation in combination with GLD-2 cytoPAP.
A prediction of this model is that the function of a single cytoPAP is enough to promote entry into meiosis in the absence of nos-3. Hence, we generated the gld-4; nos-3 and the gls-1; nos-3 double mutants. In either double mutant, in comparison to the triple mutant with gld-2, we found robust entry into meiosis (Figure 6G,H; Table 1). In conclusion, gld-4 and gls-1 promote meiotic entry in parallel to gld-2 and nos-3, suggesting that gld-4 and gls-1 might be additional pathway components that promote differentiation onset. Moreover, the striking similarity between the gld-3 nos-3 double and gld-2 gld-4; nos-3 triple tumorous germ lines suggest that gld-2 and gld-4 or gls-1 activities are largely equivalent to gld-3 activity with regard to the meiotic entry process.
gld-4 and gls-1 promote meiosis in parallel to gld-1 and gld-2
NOS-3 and GLD-1 are assumed to act in a pathway parallel to the GLD-2/GLD-3 cytoPAP pathway (Figure 6A). To complete our analysis of the genetic interactions between the NOS-3/GLD-1 and the GLD-4/GLS-1 cytoPAP pathways, we generated triple mutant strains that had either one of the GLD-4/GLS-1 cytoPAP complex components removed in a gld-2 gld-1 double mutant background (Figure 7; Figure S2; Table 1).
Fig. 7. gld-4 and gls-1 promote onset of differentiation in parallel to gld-2 and gld-1. 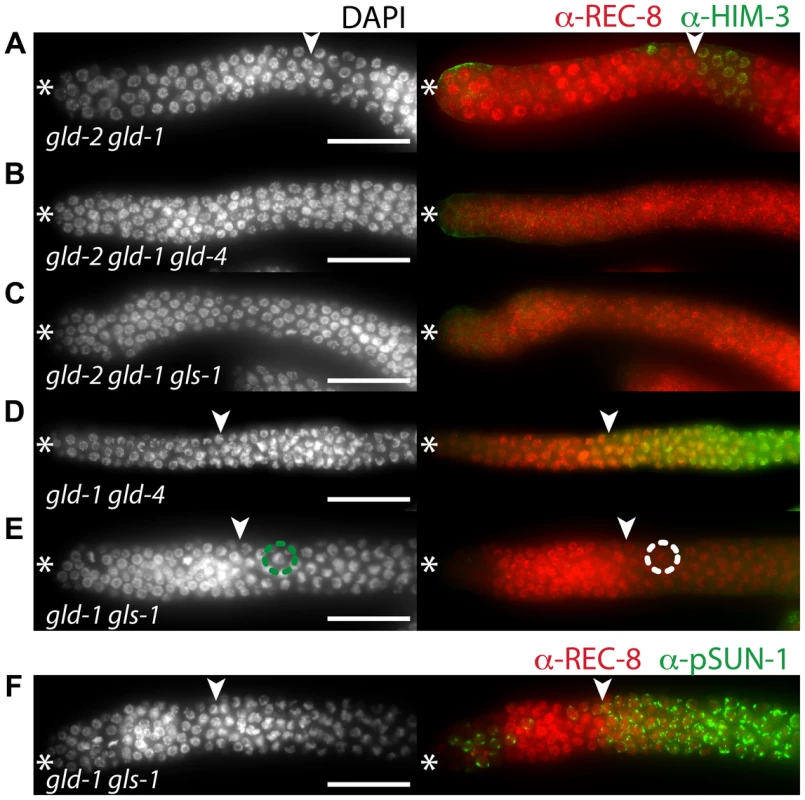
(A–F) Distal region of extruded gonads stained with DAPI, and with α-REC-8, α-HIM-3 (A–E), and α-pSUN-1 (F) antibodies. Asterisk, distal tip; arrowhead, mitosis-to-meiosis boundary. Scale bars: 50 µm. See Table 1 for the total number of analyzed germ lines. (E–F) gld-1 gls-1 double mutant germ lines possess a mitosis-to-meiosis boundary, albeit HIM-3 fails to be detected in E. A strong reduction of nucleoplasmic REC-8, the appearance of crescent-shaped nuclei in meiotic prophase (circles in E), and the abundant expression of pSUN-1 (F) reveal onset of differentiation. Germ cells, double mutant for gld-2 gld-1, enter meiosis in the majority of germ lines (Figure 7A; Figure S2; Table 1). Germ cells, triple mutant for gld-2 gld-1 gld-4 (Figure 7B) or gld-2 gld-1 gls-1 (Figure 7C), failed to enter meiosis and all germ lines are tumorous (Table 1). Importantly, germ cells in the gld-1 gld-4 (Figure 7D) and the gld-1 gls-1 (Figure 7E) double mutants enter meiosis (Table 1). Surprisingly, the gld-1 gls-1 double mutant did not stain for HIM-3 (Figure 7E). However, gld-1 gls-1 germ cells entered meiosis, as judged by their nuclear architecture, chromosome morphology, and the expression of pSUN-1 (Figure 7F). Our combined results are consistent with the previous triple mutant results, in which a nos-3 mutant gene replaced gld-1 (Figure 6), and establish a role of gld-4 and gls-1 in the onset of differentiation, suggesting that both genes operate in parallel to gld-2, gld-1 and nos-3.
Notch activity is dispensable for the proliferation of germ cells that cannot enter meiosis
Notch signaling promotes proliferation, upstream of the meiosis-promoting network [35]. To investigate whether proliferation of tumorous triple mutant gld-2 gld-1 gld-4 and gld-2 gld-1 gls-1 germ lines depends on Notch activity, we investigated GLP-1 protein expression and genetically ablated glp-1 function (Figure S3). In either triple mutant, GLP-1 remains expressed throughout the tumorous germ lines (Figure S3A,C). Consistent with their proliferative activity, dividing cells are scattered throughout the germ line and stain positively for phospho-histone-3 (PH-3) (Figure S3A,C), a marker for cells in prometaphase [43]. Loss of glp-1 in either triple mutant neither abolishes proliferation nor leads to meiotic entry and cells remain undifferentiated (Figure S3B,D). These results suggest that Notch is not required for proliferation in germ cells that are fully compromised in all meiosis-promoting pathways.
Proliferation in gld-2 gld-1 gld-4 tumorous germ lines depends on cyclin E activity
Germ cell proliferation in gld-3 nos-3 tumorous germ lines is independent of Notch signaling but depends on cyclin E [4] (Figure 8A; Table 1). In cye-1 RNAi knockdown experiments, we found that also gld-2 gld-1 gld-4 tumorous proliferation requires cyclin E activity (Figure 8B; Table 1). Moreover, consistent with gld-3 nos-3; glp-1 cye-1(RNAi) germ lines [4], an additional removal of Notch activity in gld-2 gld-1 gld-4; cye-1(RNAi) animals increases the ability of germ cells to start meiotic prophase more distally (Figure 8C). In either case, however, differentiation onset is aborted immediately after zygotene/very early pachytene and germ cells do not commit to meiosis. Together, these similarities among the gld-3 nos-3 and gld-2 gld-1 gld-4 tumorous germ lines suggest that gld-4 and, most likely gls-1, are components of a meiosis-promoting pathway that acts on the gld-2 side of both known meiosis-promoting pathways, rather than in a separate, third meiotic entry pathway (summarized in Figure 9) [4].
Fig. 8. Tumorous proliferation of gld-2 gld-1 gld-4 triple mutants depends on cye-1. 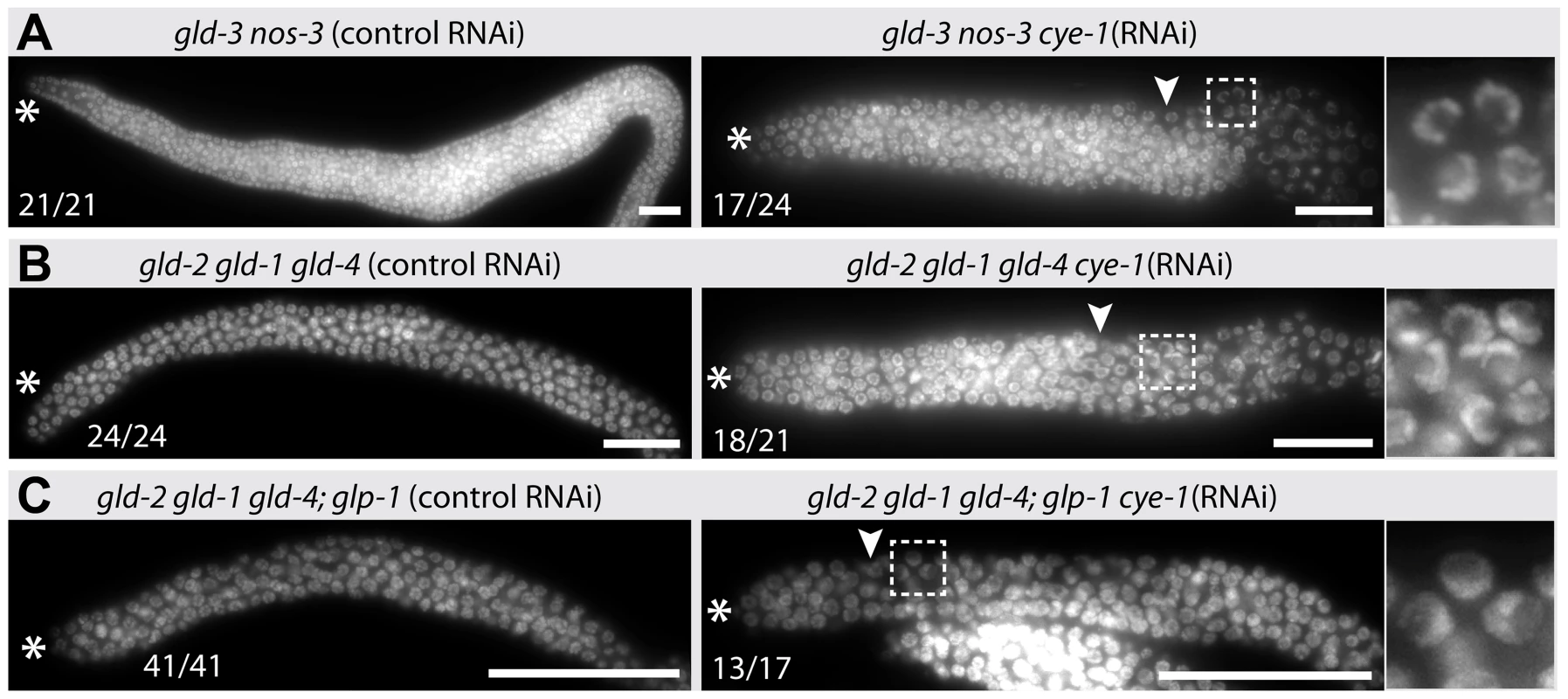
(A–C) DAPI staining of extruded distal gonads from indicated genotypes. Control RNAi and cye-1(RNAi) was conducted by feeding RNAi bacteria to heterozygote mothers and their homozygote progeny was analyzed 24 hrs past L4. Images of the prevailing phenotype and its occurrence out of all germ lines analyzed are given. The gld-3 nos-3 tumors served as a positive control for RNAi efficacy. Asterisk, distal tip; arrowhead, mitosis-to-meiosis boundary. Scale bars: 25 µm. Fig. 9. Translational regulators maintain a robust proliferative zone in the adult germ line. 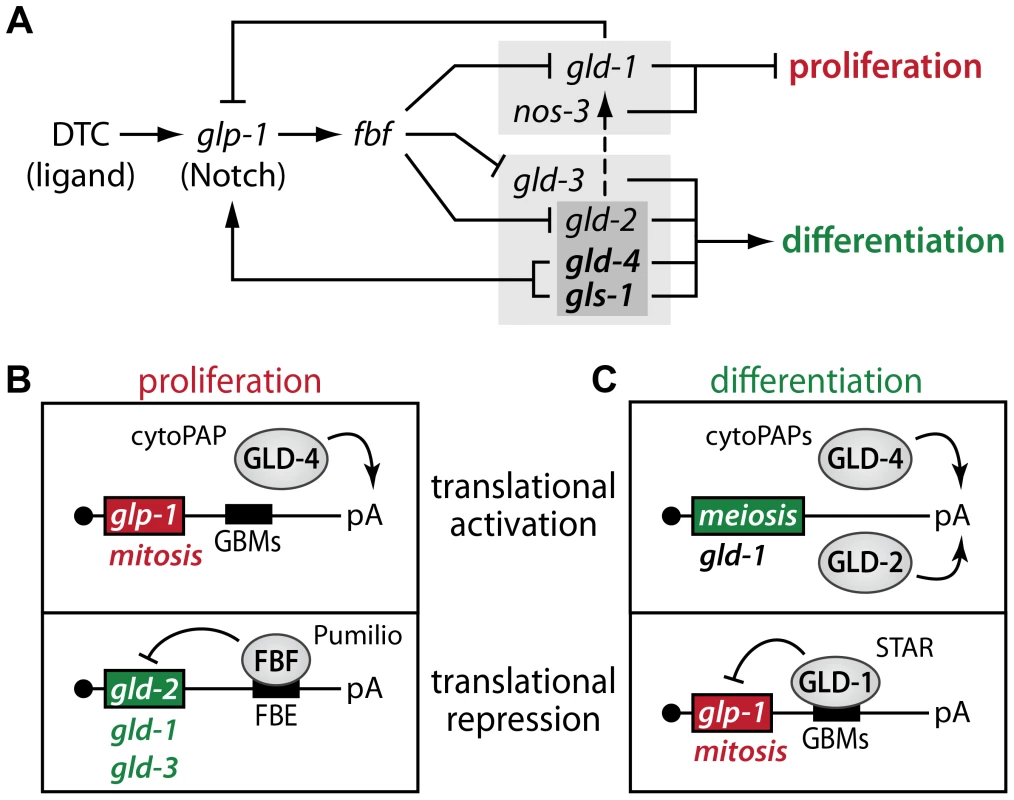
(A) Expanded genetic circuitry of primarily translation regulators that fine-tunes the balance between proliferation and differentiation. A light grey box highlights pathway members that regulate differentiation onset. A dark grey box highlights redundant activities that promote GLD-1 expression, primarily when cells commit into meiotic progression (dashed line). See text for details. Note, this simplified circuitry focuses on the RNA regulatory network downstream of GLP-1/Notch and does neither include other known downstream RNA targets nor potential upstream protein regulators. (B) Diagram of translational control examples in proliferative germ cells. Next to glp-1 mRNA, GLD-4 may also translationally activate additional mRNAs, encoding proliferation-promoting genes. Additional FBF-regulated mRNAs are known that promote the meiotic program. See text for details. (C) Diagram of translational control examples in differentiating germ cells. Additional GLD-regulated mRNAs are known that promote the proliferative fate. See text for details. Discussion
Our summed findings highlight that translational control, in the combined form of translational activation and repression, serves as a key regulatory mechanism to maintain adult tissue homeostasis in the C. elegans germ line (Figure 9). Central to our findings is the dual activity of the GLD-4/GLS-1 cytoPAP complex, which has a major role in promoting germ cell proliferation and a minor role in differentiation onset. By focusing on the activity of numerous key RNA regulators, this work expands the known core genetic circuitry downstream of niche-mediated Notch signaling that governs the balance between proliferation and differentiation (summarized in Figure 9A). At the molecular level, we propose a rheostat that consists of two translational control modules, one specific for proliferation (Figure 9B) and one specific for differentiation onset (Figure 9C). Both modules are interconnected via their mRNA targets, and this reciprocal translational activation and repression of either proliferation or differentiation factors may fine-tune the size of the proliferative zone.
GLD-4 cytoPAP and FBF function to maintain the proliferative zone
The GLD-4/GLS-1 cytoPAP complex has multiple roles in germ cell development [30], [33]. In this work, we demonstrate that both complex members contribute to the maintenance of the size of the proliferative zone by primarily influencing adult germline proliferation and secondarily differentiation onset. This dual role is consistent with the ubiquitous expression of both proteins in the respective regions of the adult germline tissue [30], [33]. GLD-4 is the enzymatic component of the GLD-4/GLS-1 cytoPAP complex and is evolutionarily most similar to nuclear Trf4/5-type polymerases, which add short poly(A) tails to nonproductive RNA molecules to initiate exosome-mediated RNA degradation [32]. By contrast, GLD-4 and its enzymatic activator GLS-1 are cytoplasmic proteins implicated in translational control [30], [33]. The notion that translational activation of mRNAs is coupled to cytoplasmic poly(A) tail extension or maintenance is primarily shaped by the work on poly(A) polymerases, such as members of the conserved GLD-2 family [31]. By analogy, GLD-4 cytoPAP's role in proliferative germ cells may therefore translationally activate mitotic-fate promoting mRNAs. We provided four pieces of evidence that the Notch receptor-encoding glp-1 mRNA is a likely mRNA target of GLD-4/GLS-1 cytoPAP activity: (1) GLD-4 associates with glp-1 mRNA, and (2) positively influences its poly(A) tail length. (3) Furthermore, we found that expression of a glp-1 3′UTR translational reporter and that of endogenous GLP-1 protein depends on GLD-4 presence. (4) Lastly, translational efficiency of endogenous glp-1 mRNA requires gld-4 activity. Therefore, these results combined support the idea that abundant GLP-1 expression is maintained by GLD-4-mediated translational activation. However, the partial reduction in glp-1 poly(A) tail length might either reflect an intrinsic enzymatic difference between Trf4-type PAPs and GLD-2, or suggests that other, yet undiscovered cytoPAPs may work redundantly to GLD-4. Alternatively, additional poly(A) tail-independent mechanisms for GLD-4-mediated translational activation may exist. Regardless of the precise molecular function of GLD-4, the translational repressor of glp-1 mRNA, GLD-1 protein, starts to accumulate in the proximal part of the PZ, prior to the mitosis-to-meiosis boundary [44], suggesting that glp-1 mRNA may already be subject to translational repression in the proximal PZ. Therefore, to ensure robust GLP-1 protein expression, GLD-4-mediated translational activation of glp-1 mRNA may help to counteract GLD-1-mediated translational repression to maintain the size of the PZ in the adult (Figure 9A,B). However, glp-1 mRNA is presumably not the only target of the GLD-4/GLS-1 cytoPAP complex, and others are likely to exist.
In the balance between proliferation and differentiation, the two translational activators, GLD-4 and GLD-2, seem to have antagonistic roles that may also constrain their regulation and function in the PZ. A loss of GLD-4 shrinks the PZ and a loss of GLD-2 expands the PZ. Therefore, GLD-2 may promote meiosis at the expense of mitosis in the gld-4 single mutant. Conversely, GLD-4 may be responsible for the expansion of the PZ in the gld-2 single mutant. Importantly, upon loss of both cytoPAP activities, the PZ re-adjusts to an intermediate size, arguing that they form an antagonistic pair. In particular, the distinct expression profile of either cytoPAP presumably reflects and affects their divergent roles in regulating mRNA-specific gene expression. The delay of GLD-2 protein expression in the PZ correlates with its genetic requirement for the onset of differentiation and a putatively required absence in undifferentiated cells. Moreover, its 2–3 fold lower abundance in the distal half of the PZ may selectively favor and functionally constrain GLD-4-mediated germ cell proliferation. Hence, a healthy balance between GLD-2 and GLD-4 functions appears to be perpetuated to maintain the size of the adult PZ.
To maintain adult germ cell proliferation and prevent progressive shrinkage of the PZ, gld-2 mRNA translation is delayed by FBF, a dominant translational repressor of several meiosis-promoting genes [7], [18], [19], [45]. We found that GLD-2 but not GLD-4 cytoPAP accumulation in the PZ appears to be inhibited by FBF, and that gld-2 mRNA associates with FBF most likely at least through one FBF-binding site in its 3′UTR. Therefore, a translational repressor (FBF) that turns off the activities of mRNAs encoding meiosis-promoting proteins (e.g. GLD-2) is combined with a translational activator (GLD-4) that turns on mRNA activities that encode mitosis-promoting proteins (e.g. GLP-1) to maintain germ cell proliferation (Figure 9A,B).
GLD-4 and GLD-2 cytoPAP stimulate entry into meiotic prophase
Conversely to germ cell proliferation, the onset of differentiation requires translational repressors (GLD-1 and NOS-3) that presumably turn off mRNA activities encoding mitosis-promoting proteins and translational activators (GLD-2, GLD-3, GLD-4, and GLS-1) that presumably turn on mRNA activities encoding meiosis-promoting proteins [6], [7], [26], [35] (Figure 9A,C). Previous genetic work established two parallel pathways, which either indirectly or directly promote differentiation onset (Figure 6A). However, not all components are equal in their potential to contribute to meiotic prophase entry. In this regard, the synergism of NOS-3 and GLD-3 is of equal strength, as is NOS-3 with both GLD-2 and GLD-4/GLS-1, or, GLD-1 with both GLD-2 and GLD-4/GLS-1. Hence, our findings of a dual role for GLD-4 cytoPAP strengthens the role of translational control even further, highlights the importance of translational activation for the balance of proliferation and differentiation, and clarifies the many levels of redundancy within the two, major, parallel pathways of the current genetic circuitry (Figure 9A).
Differentiation onset deploys two translational activators of presumed meiosis-promoting mRNAs (Figure 9C). In this regard, GLD-2 cytoPAP performs a more prevalent role in activating meiosis-promoting mRNAs as its combined loss with genes of the first, translational repressor pathway (i.e. gld-1 gld-2 or gld-2; nos-3 doubles) causes more germ cell overproliferation than is observed in the respective gld-4 double mutant germ lines. Importantly, germ cells of gld-2; nos-3 or gld-2 gld-1 double mutants enter meiosis in a gld-4- and gls-1-dependent manner, as triple mutant germ cells (e.g. gld-1 gld-2 gld-4 or gld-2 gld-4; nos-3) do not enter meiosis. Consistent with previous findings that germline proliferation in tumorous gld-2 gld-1 or gld-3 nos-3 double mutants is glp-1-independent [4], [35], tumorous triple mutant gld-2 nos-3 germ cells that lack in addition either gld-4 or gls-1 do not require GLP-1 activity to remain in mitosis either, arguing for their genetic position downstream of Notch and in parallel to each other for meiotic entry (Figure 9A). Intriguingly, the similarities between the gld-3 nos-3 double and gld-2 gld-4; nos-3 or gld-2 gls-1; nos-3 triple mutants suggest further that gld-3 activity equals the combined activities of gld-2 and gld-4/gls-1 with respect to the loss of nos-3, which positions gld-4/gls-1 within the second, translational activator pathway at the level of gld-2 (Figure 9A). These genetic behaviors appear to parallel the known molecular protein interactions. The multi-KH domain protein GLD-3 binds directly to GLD-2 cytoPAP and GLS-1 [30], [33], illustrating that GLD-3 may serve as an integral regulatory factor for both GLD-2 and GLD-4/GLS-1 cytoPAPs to promote differentiation onset.
Redundancy of cytoPAP-mediated translational activation has been previously reported in a later step of meiotic prophase of female germ cells that require abundant GLD-1 expression for meiotic commitment [30]. Intriguingly, gld-2 gld-4 double mutant germ cells enter meiosis [30], suggesting that the remaining low GLD-1 amounts might be sufficient to promote meiotic entry. Consistent with this idea, gld-2 gld-4 gld-1 triple mutant germ cells never enter meiosis, arguing that in the absence of cytoPAP activity, the remaining gld-1 activity/GLD-1 amount is indeed crucial for meiotic entry. In agreement with previous findings [4], [46], [47], our work suggests that for differentiation onset in gld-2 gld-4 double mutants, cyclin E represents an important target of GLD-1-mediated translational repression. However, we expect additional differentiation onset-promoting mRNA targets to be positively regulated by GLD-2 and GLD-4, either in a combinatorial manner or separately. Alternatively, other RNA-directed molecular functions, such as miRNA stability described for GLD-2 orthologs in mammals [48], might be relevant. Future research on the RNA-regulatory repertoire of GLD-2 and GLD-4 will be required to better resolve these issues.
We propose that two modules of translational activation and repression, interconnected via their mRNA targets, establish a molecular rheostat that leads to a reciprocal expression of either proliferation or differentiation factors. Together they maintain adult germline proliferation in adult C. elegans animals. Translational repression, in particular, is an established mechanism in Drosophila and C. elegans development. Translational control is also an essential mechanism of the transition from self-renewal/proliferation to differentiation in Drosophila germ cells [49], [50]. Our work suggests that the regulation of turning translation on is equally important for maintaining a healthy balance between proliferation and differentiation as turning translation off. With this work, we begin to fill this obvious gap in our understanding of adult tissue maintenance.
Materials and Methods
Strains and RNAi knockdown
C. elegans strains were handled according to standard procedures [51]. Worms were grown at 20°C and used for most experiments at an age of 24 hours (h) past mid-L4. Bristol N2 served as the wild-type strain.
Mutations used: LGI: gld-2(q497), gld-1(q485), fer-1(b232), gls-1(ef4), gls-1(ef8), gld-4(ef9), gld-4(ef15). LGII: gld-3(q730), nos-3(q650). LGIII: glp-1(q175). Transgenes used: rrrSi117[Pmex-5::GFP::H2B::glp-1(wt 3′UTR) unc-119(+)] II, rrrSi118[Pmex-5::GFP::H2B::glp-1(GBM1,2,3 mut 3′UTR) unc-119(+)] II; both are Mos1-mediated single copy gene insertions and their sequences are described in the supplemental text of [25]. JH2929 expresses the LAP-tagged FBF-2 [52].To generate the new gld-2(q497) gld-1(q485) double mutant, we crossed heterozygous gld-2 males with heterozygous gld-1 hermaphrodites. Next, we crossed F1 non-green worms, containing gld-1 and gld-2, and green siblings, containing the hT2[qIs48] I;III balancer. In the F2 progeny heterozygous balancer animals were screened for a recombination event between the gld-2 and the gld-1 locus by genomic PCR for the q485 deletion and sequencing for the q497 point mutation. Homozygote hT2[qIs48] I;III animals are embryonic lethal and cannot be analyzed as a wild-type sibling control.
All other double and triple mutants on LGI were generated in a similar manner and balanced by hT2[qIs48] I;III and are listed in Table 1. Quadruple mutants containing genes on LGI and LGIII were balanced by hT2[qIs48] I;III and validated by PCR for deletions and by sequencing for gld-2(q497) and glp1(q175). Double and triple mutant combinations on LGI and LGII were maintained by a closely linked GFP transgene (ccIs4251) to unc-15(e73) on LGI and mIn1[mIs14 dpy-10(e128)] on LGII. The presence of all mutations was validated by PCR for deletions or by sequencing. Primer sequences are available on request.
RNAi experiments were performed according to published feeding RNAi procedures [53]. The fbf RNAi construct corresponds to fbf-1 (nts 1040–1845), cye-1 is described elsewhere [54], and the empty pL4440 vector served as a control. L4-staged N2 animals were placed on RNAi plates and analyzed 24 h later. The efficiency of fbf knockdown was confirmed by a loss of anti-FBF immunoreactivity 24 h past L4, and after continued feeding at 48 h past the L4 stage by phenotypic changes of the germ lines, i.e. the shrinkage of the PZ and even later the appearance of male-fated germ cells [55].
Immunocytochemistry and immunoblotting
Antibodies against the following proteins were used as described: anti-HIM-3 1∶200 [56], anti-pSUN-1 1∶1000 [57], anti-GLD-4 1∶20 [30], anti-GLP-1 1∶10 [13], anti-PH-3 1∶500 (9706, Ser-10, 6G3, Cell Signaling), anti-FBF-1 1∶100 [55], anti-GLH-2 1∶200 [58], anti-GLS-1 1∶20 [33]. Monoclonal anti-REC-8 1∶20 (mo560-G25-1, at 10 ng/ul) and anti-GLD-2 1∶20 (A4-4, at 10 ng/ul) antibodies were generated against recombinant HIS-REC-8(aa330–525) and GST-GLD-2(aa959–1113) fusion peptides. The antibodies are specific to the respective proteins; no immunocytochemistry signal was observed in corresponding null mutants and the protein expression patterns in wild type match published ones [28], [59]. Secondary antibodies (1∶500) were coupled to FITC, CY3 and CY5 (Jackson Labs).
Extruded germ lines were prepared in solution as described [33]. The correct localization and comparable intensities of GLH-2 served as a tissue penetration control for all immunofluorescence experiments. Images were acquired with Axiovision Software (Zeiss) on a wide-filed Imager Z1 (Zeiss) microscope, equipped with an AxioCam MRm (Zeiss) camera. Raw images were processed in Photoshop CS5 (Adobe) and assembled in Illustrator CS5 (Adobe). For quantification of immunofluorescent intensities, all images for comparison were taken with identical settings. A median focal plane was chosen where the syncytium was at its maximum width. The pixel intensities were measured in Fiji (ImageJ). To compare GLP-1 intensities, a line scan was performed as is indicated in Figure 3B, ranging from the distal germline tip to the beginning of pachytene. Then all values were binned into the 10 fractions whose positions are displayed in Figure 3B. Averages of those fractions between all analyzed germ lines were calculated and normalized to GLH-2 intensities (measured in the same way). To compare cytoplasmic GLD-2 and GLD-4 intensities, four identical circles were placed over the rachis of the distal arm along the distal-to-proximal axis as indicated in Figure 5C (five germ cell diameters (GCD) proximal of the distal tip, at the end of the PZ, at the beginning of pachytene, and ten GCD into pachytene) and averaged for all germ lines per genotype. The GLD-2/GLD-4 intensities given are not normalized to the GLH-2 signals, which were in these cytoplasmic regions very low. To ensure equal penetration, we independently measured the peri-nuclear GLH-2 signal in neighboring nuclei and found it very similar among all analyzed germ lines.
Immunoblots were performed according to standard procedures with a mixture of two anti-GFP antibodies, at a final dilution of 1∶1000 (11814460001, clones 7.1 and 13.1, Roche) and 1∶200 (sc-9996, B-2, Santa Cruz), anti-tubulin 1∶100000 (T 5168, clone B-5-1-2, Sigma), and HRP-conjugated anti-mouse secondary antibodies (Jackson Labs).
Yeast three-hybrid
Three-hybrid experiments were performed as described [60]. gld-2 RNA sequences were cloned into the XmaI and SphI sites of the vector pIIIA/MS2-2, using either PCR-amplified fragment (FBE4) or annealed synthetic oligonucleotides (remaining FBE sites). Their nucleotide positions in relation to the first nucleotide of the gld-2 3′UTR are as follows: FBE1 (nts 298–335); FBE2 (nts 354–394); FBE3 (nts 460–493); FBF4 (nts 683–763); FBE5 (nts 952–986). For binding specificity, a mutation (UG to AC, see Figure 4D) was engineered by site-directed mutagenesis using Quikchange (Stratagene).
mRNA analysis
For the sucrose gradient experiments, whole-worm extracts of L1 synchronized adult animals (L4+24 h) grown in comparable feeding-RNAi conditions were prepared by pulverizing frozen worms and adding lysis buffer [50 mM HEPES pH 7.5, 125 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.005% NP-40, 2× Protease Inhibitor Cocktail without EDTA (Roche), 100 U/ml Ribolock (Fermentas), 2 mM PMSF, 4 mM Benzamidine, 2 µg/ml Leupeptin, 2 µg/ml Pepstatin, 0.1 µg/ml Pefabloc, 2 mM NaF, 2 mM Na3VO3 and 200 µg/ml Cycloheximide], followed by a low speed centrifugation to removed insoluble components. The clear supernatant of three biological replicates was layered onto a 17–50% w/v sucrose gradient and processed as previously described [61] with the only exception that the gradients were spun for 210 min. For the mRNA distributions analysis, 10 fmole of a polyadenylated in vitro transcribed luciferase mRNA was added to each fraction prior to RNA isolation as an internal RNA standard for extraction efficiency. The Trizol (Invitrogen) isolated RNA from individual fractions was resolved in equal volumes of water and further analyzed by qRT-PCR or pooled for splint-mediated poly(A) tests [40].
RIP experiments were performed from mixed-stage animals as described [62] using anti-GFP (MPI-CBG), anti-GLD-4 [30], or anti-GLS-1 [33] antibodies. Semiquantitative RT-PCR samples of three independent RIP experiments were resolved on ethidium bromide-stained agarose gels. The control samples without Reverse transcriptase were negative and are not shown. For the qRT-PCR experiments, equal volumes of gradient fractionated RNA, total RNA, or RIP material of three biological replicates was used as input material. cDNA synthesis was performed using Revert Aid Premium Transcriptase (Fermentas) in combination with oligo dT primers, following the manufactures protocol. qPCRs were performed on a Mx3000P qPCR system (Stratagene) using ABsolute qPCR SYBR Green mix (Thermo) under standard conditions.
For measuring poly(A) tail length, we pooled five non-polysome and polysome fractions as indicated in Figure 4. Together with 4 µg total RNA of the input material, we processed the pooled sucrose gradient experiments by ligating an RNA anchor to the 3′ends, preformed a gene-specific RT-PCR, and resolved the DNA samples on a high-resolution agarose gel, according to [40]. Lane quantifications were performed using Fiji (ImageJ), as described in [40].
Supporting Information
Zdroje
1. KimbleJ (2011) Molecular regulation of the mitosis/meiosis decision in multicellular organisms. Cold Spring Harbor perspectives in biology 3 : 1–16.
2. CrittendenSL, LeonhardKA, ByrdDT, KimbleJ (2006) Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Molecular biology of the cell 17 : 3051–3061.
3. MaciejowskiJ, UgelN, MishraB, IsopiM, HubbardEJ (2006) Quantitative analysis of germline mitosis in adult C. elegans. Developmental biology 292 : 142–151.
4. FoxPM, VoughtVE, HanazawaM, LeeMH, MaineEM, et al. (2011) Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 138 : 2223–2234.
5. CinquinO, CrittendenSL, MorganDE, KimbleJ (2010) Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proceedings of the National Academy of Sciences of the United States of America 107 : 2048–2053.
6. HansenD, HubbardEJ, SchedlT (2004) Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Developmental biology 268 : 342–357.
7. EckmannCR, CrittendenSL, SuhN, KimbleJ (2004) GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168 : 147–160.
8. KimbleJE, WhiteJG (1981) On the control of germ cell development in Caenorhabditis elegans. Developmental biology 81 : 208–219.
9. AustinJ, KimbleJ (1987) glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51 : 589–599.
10. ByrdDT, KimbleJ (2009) Scratching the niche that controls Caenorhabditis elegans germline stem cells. Seminars in cell & developmental biology 20 : 1107–1113.
11. PepperAS, KillianDJ, HubbardEJ (2003) Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163 : 115–132.
12. BerryLW, WestlundB, SchedlT (1997) Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124 : 925–936.
13. CrittendenSL, TroemelER, EvansTC, KimbleJ (1994) GLP-1 is localized to the mitotic region of the C. elegans germ line. Development 120 : 2901–2911.
14. LamontLB, CrittendenSL, BernsteinD, WickensM, KimbleJ (2004) FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Developmental cell 7 : 697–707.
15. LeeMH, HookB, PanG, KershnerAM, MerrittC, et al. (2007) Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS genetics 3: e233.
16. KershnerAM, ShinH, HansenTJ, KimbleJ (2014) Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proceedings of the National Academy of Sciences of the United States of America 111 : 3739–3744.
17. LehmannR (2012) Germline stem cells: origin and destiny. Cell stem cell 10 : 729–739.
18. CrittendenSL, BernsteinDS, BachorikJL, ThompsonBE, GallegosM, et al. (2002) A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417 : 660–663.
19. KershnerAM, KimbleJ (2010) Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proceedings of the National Academy of Sciences of the United States of America 107 : 3936–3941.
20. MerrittC, SeydouxG (2010) The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development 137 : 1787–1798.
21. CrittendenSL, EckmannCR, WangL, BernsteinDS, WickensM, et al. (2003) Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philosophical transactions of the Royal Society of London Series B, Biological sciences 358 : 1359–1362.
22. JonesAR, SchedlT (1995) Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes & development 9 : 1491–1504.
23. MarinVA, EvansTC (2003) Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130 : 2623–2632.
24. RyderSP, FraterLA, AbramovitzDL, GoodwinEB, WilliamsonJR (2004) RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nature structural & molecular biology 11 : 20–28.
25. WrightJE, GaidatzisD, SenftenM, FarleyBM, WesthofE, et al. (2011) A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. The EMBO journal 30 : 533–545.
26. HansenD, Wilson-BerryL, DangT, SchedlT (2004) Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131 : 93–104.
27. EckmannCR, KraemerB, WickensM, KimbleJ (2002) GLD-3, a Bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Developmental cell 3 : 697–710.
28. WangL, EckmannCR, KadykLC, WickensM, KimbleJ (2002) A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419 : 312–316.
29. SuhN, JedamzikB, EckmannCR, WickensM, KimbleJ (2006) The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proceedings of the National Academy of Sciences of the United States of America 103 : 15108–15112.
30. SchmidM, KuchlerB, EckmannCR (2009) Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes & development 23 : 824–836.
31. MinasakiR, EckmannCR (2012) Subcellular specialization of multifaceted 3′end modifying nucleotidyltransferases. Current opinion in cell biology 24 : 314–322.
32. SchmidtK, ButlerJS (2013) Nuclear RNA surveillance: role of TRAMP in controlling exosome specificity. Wiley interdisciplinary reviews RNA 4 : 217–231.
33. RybarskaA, HarterinkM, JedamzikB, KupinskiAP, SchmidM, et al. (2009) GLS-1, a novel P granule component, modulates a network of conserved RNA regulators to influence germ cell fate decisions. PLoS genetics 5: e1000494.
34. HansenD, SchedlT (2013) Stem cell proliferation versus meiotic fate decision in Caenorhabditis elegans. Advances in experimental medicine and biology 757 : 71–99.
35. KadykLC, KimbleJ (1998) Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125 : 1803–1813.
36. OguraK, KishimotoN, MitaniS, Gengyo-AndoK, KoharaY (2003) Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development 130 : 2495–2503.
37. NouschM, EckmannCR (2013) Translational control in the Caenorhabditis elegans germ line. Advances in experimental medicine and biology 757 : 205–247.
38. EvansTC, CrittendenSL, KodoyianniV, KimbleJ (1994) Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell 77 : 183–194.
39. MaciejowskiJ, AhnJH, CiprianiPG, KillianDJ, ChaudharyAL, et al. (2005) Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics 169 : 1997–2011.
40. MinasakiR, RudelD, EckmannCR (2014) Increased sensitivity and accuracy of a single-stranded DNA splint-mediated ligation assay (sPAT) reveals poly(A) tail length dynamics of developmentally regulated mRNAs. RNA biology 11.
41. SuhN, CrittendenSL, GoldstrohmA, HookB, ThompsonB, et al. (2009) FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics 181 : 1249–1260.
42. LublinAL, EvansTC (2007) The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Developmental biology 303 : 635–649.
43. HendzelMJ, WeiY, ManciniMA, Van HooserA, RanalliT, et al. (1997) Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106 : 348–360.
44. JonesAR, FrancisR, SchedlT (1996) GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage - and sex-specific expression during Caenorhabditis elegans germline development. Developmental biology 180 : 165–183.
45. KalchhauserI, FarleyBM, PauliS, RyderSP, CioskR (2011) FBF represses the Cip/Kip cell-cycle inhibitor CKI-2 to promote self-renewal of germline stem cells in C. elegans. The EMBO journal 30 : 3823–3829.
46. BiedermannB, WrightJ, SenftenM, KalchhauserI, SarathyG, et al. (2009) Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Developmental cell 17 : 355–364.
47. JeongJ, VerheydenJM, KimbleJ (2011) Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS genetics 7: e1001348.
48. KatohT, SakaguchiY, MiyauchiK, SuzukiT, KashiwabaraS, et al. (2009) Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & development 23 : 433–438.
49. HarrisRE, PargettM, SutcliffeC, UmulisD, AsheHL (2011) Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Developmental cell 20 : 72–83.
50. InscoML, BaileyAS, KimJ, OlivaresGH, WapinskiOL, et al. (2012) A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell stem cell 11 : 689–700.
51. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
52. VoroninaE, PaixA, SeydouxG (2012) The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development 139 : 3732–3740.
53. KamathRS, AhringerJ (2003) Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30 : 313–321.
54. CowanCR, HymanAA (2006) Cyclin E-Cdk2 temporally regulates centrosome assembly and establishment of polarity in Caenorhabditis elegans embryos. Nature cell biology 8 : 1441–1447.
55. ZhangB, GallegosM, PuotiA, DurkinE, FieldsS, et al. (1997) A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390 : 477–484.
56. ZetkaMC, KawasakiI, StromeS, MullerF (1999) Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & development 13 : 2258–2270.
57. PenknerAM, FridkinA, GloggnitzerJ, BaudrimontA, MachacekT, et al. (2009) Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139 : 920–933.
58. GruidlME, SmithPA, KuznickiKA, McCroneJS, KirchnerJ, et al. (1996) Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 93 : 13837–13842.
59. PasierbekP, JantschM, MelcherM, SchleifferA, SchweizerD, et al. (2001) A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes & development 15 : 1349–1360.
60. BernsteinDS, ButerN, StumpfC, WickensM (2002) Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods 26 : 123–141.
61. ClancyJL, NouschM, HumphreysDT, WestmanBJ, BeilharzTH, et al. (2007) Methods to analyze microRNA-mediated control of mRNA translation. Methods in enzymology 431 : 83–111.
62. JedamzikB, EckmannCR (2009) Analysis of RNA-protein complexes by RNA coimmunoprecipitation and RT-PCR analysis from Caenorhabditis elegans. Cold Spring Harbor protocols 2009: pdb prot5300.
Štítky
Genetika Reprodukční medicína
Článek An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of MovementČlánek Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced ActivityČlánek Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene NetworksČlánek Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS LocusČlánek tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa CellsČlánek Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced ApoptosisČlánek A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II FidelityČlánek The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites inČlánek Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell SenescenceČlánek BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct MechanismsČlánek Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 9
-
Všechny články tohoto čísla
- Translational Regulation of the Post-Translational Circadian Mechanism
- An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of Movement
- Eliminating Both Canonical and Short-Patch Mismatch Repair in Suggests a New Meiotic Recombination Model
- Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced Activity
- Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene Networks
- Targeted H3R26 Deimination Specifically Facilitates Estrogen Receptor Binding by Modifying Nucleosome Structure
- Role for Circadian Clock Genes in Seasonal Timing: Testing the Bünning Hypothesis
- The Tandem Repeats Enabling Reversible Switching between the Two Phases of β-Lactamase Substrate Spectrum
- The Association of the Vanin-1 N131S Variant with Blood Pressure Is Mediated by Endoplasmic Reticulum-Associated Degradation and Loss of Function
- Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS Locus
- Regulation of Flowering by the Histone Mark Readers MRG1/2 via Interaction with CONSTANS to Modulate Expression
- The Actomyosin Machinery Is Required for Retinal Lumen Formation
- Plays a Conserved Role in Assembly of the Ciliary Motile Apparatus
- Hidden Diversity in Honey Bee Gut Symbionts Detected by Single-Cell Genomics
- Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria
- tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa Cells
- Causal Variation in Yeast Sporulation Tends to Reside in a Pathway Bottleneck
- Tissue-Specific RNA Expression Marks Distant-Acting Developmental Enhancers
- WC-1 Recruits SWI/SNF to Remodel and Initiate a Circadian Cycle
- Clonal Expansion of Early to Mid-Life Mitochondrial DNA Point Mutations Drives Mitochondrial Dysfunction during Human Ageing
- Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels
- Differential Management of the Replication Terminus Regions of the Two Chromosomes during Cell Division
- Obesity-Linked Homologues and Establish Meal Frequency in
- Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced Apoptosis
- Stress-Induced Nuclear RNA Degradation Pathways Regulate Yeast Bromodomain Factor 2 to Promote Cell Survival
- The MAPK p38c Regulates Oxidative Stress and Lipid Homeostasis in the Intestine
- Widespread Genome Reorganization of an Obligate Virus Mutualist
- Trans-kingdom Cross-Talk: Small RNAs on the Move
- The Vip1 Inositol Polyphosphate Kinase Family Regulates Polarized Growth and Modulates the Microtubule Cytoskeleton in Fungi
- Myosin Vb Mediated Plasma Membrane Homeostasis Regulates Peridermal Cell Size and Maintains Tissue Homeostasis in the Zebrafish Epidermis
- GLD-4-Mediated Translational Activation Regulates the Size of the Proliferative Germ Cell Pool in the Adult Germ Line
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Translational Regulation of the DOUBLETIME/CKIδ/ε Kinase by LARK Contributes to Circadian Period Modulation
- Positive Selection and Multiple Losses of the LINE-1-Derived Gene in Mammals Suggest a Dual Role in Genome Defense and Pluripotency
- Out of Balance: R-loops in Human Disease
- A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II Fidelity
- Altered Behavioral Performance and Live Imaging of Circuit-Specific Neural Deficiencies in a Zebrafish Model for Psychomotor Retardation
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Meta-analysis of Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments
- The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites in
- Hydroxymethylated Cytosines Are Associated with Elevated C to G Transversion Rates
- Memory and Fitness Optimization of Bacteria under Fluctuating Environments
- Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell Senescence
- Interspecific Tests of Allelism Reveal the Evolutionary Timing and Pattern of Accumulation of Reproductive Isolation Mutations
- PRO40 Is a Scaffold Protein of the Cell Wall Integrity Pathway, Linking the MAP Kinase Module to the Upstream Activator Protein Kinase C
- Low Levels of p53 Protein and Chromatin Silencing of p53 Target Genes Repress Apoptosis in Endocycling Cells
- SPDEF Inhibits Prostate Carcinogenesis by Disrupting a Positive Feedback Loop in Regulation of the Foxm1 Oncogene
- RRP6L1 and RRP6L2 Function in Silencing Regulation of Antisense RNA Synthesis
- BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct Mechanisms
- Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
- Atkinesin-13A Modulates Cell-Wall Synthesis and Cell Expansion in via the THESEUS1 Pathway
- Dopamine Signaling Leads to Loss of Polycomb Repression and Aberrant Gene Activation in Experimental Parkinsonism
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
- Bipartite Recognition of DNA by TCF/Pangolin Is Remarkably Flexible and Contributes to Transcriptional Responsiveness and Tissue Specificity of Wingless Signaling
- The Olfactory Transcriptomes of Mice
- Muscular Dystrophy-Associated and Variants Disrupt Nuclear-Cytoskeletal Connections and Myonuclear Organization
- Interplay of dFOXO and Two ETS-Family Transcription Factors Determines Lifespan in
- Evidence for Widespread Positive and Negative Selection in Coding and Conserved Noncoding Regions of
- Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer's Disease and Related Dementias
- Rejuvenation of Meiotic Cohesion in Oocytes during Prophase I Is Required for Chiasma Maintenance and Accurate Chromosome Segregation
- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Local Effect of Enhancer of Zeste-Like Reveals Cooperation of Epigenetic and -Acting Determinants for Zygotic Genome Rearrangements
- Differential Responses to Wnt and PCP Disruption Predict Expression and Developmental Function of Conserved and Novel Genes in a Cnidarian
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



