-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNucleosomes Shape DNA Polymorphism and Divergence
In eukaryotic cells, the majority of DNA is packaged in nucleosomes comprised of ∼147 bp of DNA wound tightly around the highly conserved histone octamer. Nucleosomal DNA from diverse organisms shows an anti-correlated ∼10 bp periodicity of AT-rich and GC-rich dinucleotides. These sequence features influence DNA bending and shape, facilitating structural interactions. We asked whether natural selection mediated through the periodic sequence preferences of nucleosomes shapes the evolution of non-protein-coding regions of D. melanogaster by examining the inter - and intra-species genomic variation relative to these fundamental chromatin building blocks. The sequence changes across nucleosome-bound regions on the melanogaster lineage mirror the observed nucleosome dinucleotide periodicities. Importantly, we show that the frequencies of polymorphisms in natural populations vary across these regions, paralleling divergence, with higher frequencies of preferred alleles. These patterns are most evident for intronic regions and indicate that non-protein coding regions are evolving toward sequences that facilitate the canonical association with the histone core. This result is consistent with the hypothesis that interactions between DNA and the core have systematic impacts on function that are subject to natural selection and are not solely due to mutational bias. These ubiquitous interactions with the histone core partially account for the evolutionary constraint observed in unannotated genomic regions, and may drive broad changes in base composition.
Published in the journal: . PLoS Genet 10(7): e32767. doi:10.1371/journal.pgen.1004457
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004457Summary
In eukaryotic cells, the majority of DNA is packaged in nucleosomes comprised of ∼147 bp of DNA wound tightly around the highly conserved histone octamer. Nucleosomal DNA from diverse organisms shows an anti-correlated ∼10 bp periodicity of AT-rich and GC-rich dinucleotides. These sequence features influence DNA bending and shape, facilitating structural interactions. We asked whether natural selection mediated through the periodic sequence preferences of nucleosomes shapes the evolution of non-protein-coding regions of D. melanogaster by examining the inter - and intra-species genomic variation relative to these fundamental chromatin building blocks. The sequence changes across nucleosome-bound regions on the melanogaster lineage mirror the observed nucleosome dinucleotide periodicities. Importantly, we show that the frequencies of polymorphisms in natural populations vary across these regions, paralleling divergence, with higher frequencies of preferred alleles. These patterns are most evident for intronic regions and indicate that non-protein coding regions are evolving toward sequences that facilitate the canonical association with the histone core. This result is consistent with the hypothesis that interactions between DNA and the core have systematic impacts on function that are subject to natural selection and are not solely due to mutational bias. These ubiquitous interactions with the histone core partially account for the evolutionary constraint observed in unannotated genomic regions, and may drive broad changes in base composition.
Introduction
Sequence-dependent differences in the physical properties of DNA influence its associations with the histone core, as well as the kinetics of nucleosome assembly and stability [2]–[9]. One of the most generalizable sequence affinities of the histone octamer is the periodic variation of dinucleotide frequencies across nucleosomal DNA. Alignments of nucleosomal sequences from diverse eukaryotes display a prominent ∼10 bp periodic enrichment of AT-rich dinucleotides, along with an anti-correlated periodicity of GC-rich dinucleotides [8], [10]–[14]. The ∼10 bp spacing of AA/TT dinucleotides generates intrinsically curved DNA molecules with increased nucleosome binding affinity [5], [8], [14]–[17]. Peaks of AA/TT frequency are found specifically over positions where the minor groove bends interiorly, whereas GC dinucleotides peak where the major groove is facing the histone core. Structural data suggest that DNA shape, in particular the narrowing of the minor groove and the associated lowering of its electrostatic potential at AT-rich sequences facilitate contacts with key histone arginines [9], [18], [19]. GC dinucleotides contract the major groove, which also facilitates the tight winding of DNA around the core [9], [20].
Although these broadly conserved dinucleotide patterns have been cited as evidence for a genomic “code” for nucleosome positioning [10], the role of sequence in nucleosome function remains contested and unresolved [6], [8], [21], [22]. Correlation between in vitro and in vivo nucleosome maps in yeast may reflect the influence of the inherent sequence preferences of the histone core on nucleosome positioning [7], [23]. However, strong experimental evidence suggests that trans-acting factors (e.g. RNA polymerase II, transcription factors and ATP-dependent remodelers) are central to establishing nucleosome positions along genomic DNA (translational positions), particularly in genic regions, with sequence providing a weaker contribution [7], [8], [21], [24], [25]. In cases where DNA sequence does impact translational nucleosome positions, its influence is largely attributed to GC content and anti-nucleosomal sequences, such as poly-dA/dT tracts, rather than dinucleotide patterns [5], [8], [25]–[27].
Dinucleotides are instead thought to play a distinct but integrally connected role in directing and preserving the ‘rotational positioning’ of nucleosomal DNA [8], [9], [20], [28], which refers to the orientation of DNA relative to the core. Due to the structural constraints inherent to nucleosome formation, a given translational position in the genome will assemble with a particular rotational alignment. This determines which bases face the nucleosome interior and exterior, and also the positioning of the major and minor grooves relative to the core. Nucleosomes tend to occupy translational genomic positions which are offset by ∼10 bp increments [13], [28], [29]. Thus, due to the helical structure of DNA, with ∼10.4 bp per turn, the rotational orientation of DNA relative to the core is thought to be unchanged as nucleosomes assume new favored translational positions (Figure 1A). This 10 bp incremental movement leaves the exposure of sites at the surface unchanged [20], [28], and is in agreement with the reported step size of many chromatin remodelers [30]. By influencing the rotational positioning of DNA relative to the histone core, nucleotide changes at particular nucleosome positions (or in flanking regions) could have diverse functional impact, for example on nucleosome assembly, stability, remodeling efficiency, RNA and DNA polymerase processivities and transcription factor binding site access. However, despite considerable evidence that dinucleotide patterns impact nucleosome positioning and dynamics in vitro, in vivo evidence of function has remained elusive.
Fig. 1. Dinucleotide frequencies in the D. melanogaster genome show small (∼10 bp) and large (∼180) scale periodicities relative to isolated nucleosomal fragments. 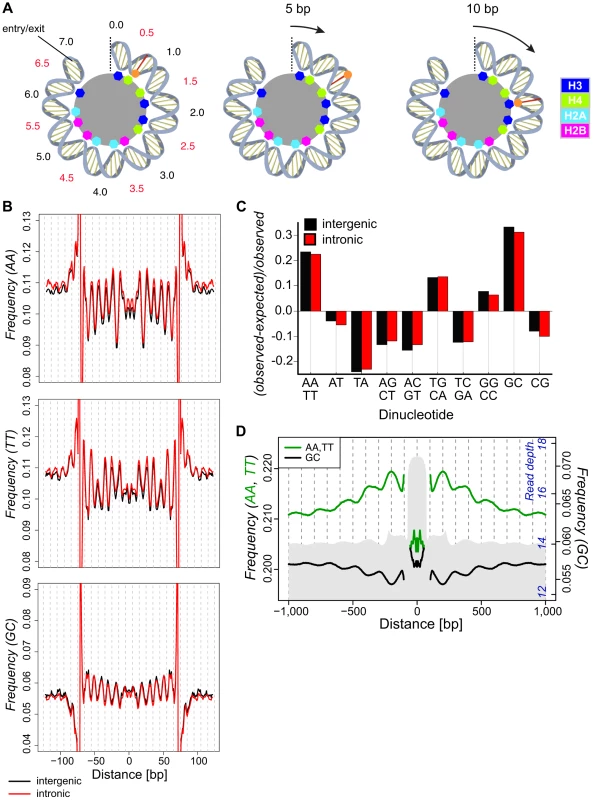
(A) A cartoon representation of half of the dyad symmetric nucleosome depicting DNA associations with the histone core. The dyad axis is marked with a dotted line, and SHL locations are noted in black and red. Colored octagons represent regions where the DNA minor groove contacts the histone core. A base in the minor groove is marked for reference (red line and orange dot). The results of a 5 (middle) or 10 (right) bp translational movement are depicted. A 5 bp movement rotates the marked base away from the core, moving it to a region where the major groove faces the interior. Translational movement of 10 bp preserves the rotational position of the base (interior) and, given a ∼10 bp periodic spacing of preferred dinucleotides, maintains favored sequence positions. (B) Frequencies of AA, TT and GC dinucleotides across intergenic (black) and intronic (red) nucleosomal n147 (±50 bp). Note that frequencies are outside the range of the plots near the edges of the n147 because of the inherent sequence bias of MNase. (C) Deviations in the observed genomic dinucleotide frequencies relative to expected values based on overall nucleotide frequencies in intergenic (black) and intronic (red) regions. (D) Average frequencies of AA/TT (green) and GC (black) dinucleotides surrounding intergenic n147 regions (±1 kb). Note that frequencies immediately flanking the edges (±5 bp) of the n147 are not plotted, since these largely reflect sequence bias of the MNase. Read depth of intergenic MNase-derived 142–152 bp (n142-152) nucleosomal DNA fragments in grey (axis in blue). One approach to discovering function is to look for evidence of natural selection in sequence polymorphism (variation within species) and divergence (variation between species). Individual mutations influencing histone-DNA interactions may have only slight, undetectable phenotypic effects in the laboratory; in contrast, the associated fitness consequences in large natural populations can strongly shape rates of divergence and levels of polymorphism over many generations [31]. Of course, strongly selected variants will go to fixation quickly and be maintained in very high frequency against the weaker force of mutational reversion. However, observations of extensive DNA sequence polymorphism and divergence throughout the genome, including nucleosomal sequences, indicate that such systematic selection is not dominating stochastic effects (mutation, genetic drift, and variation in selection coefficients) in the evolutionary dynamic.
Analysis of codon bias suggests that at equilibrium between selection, mutation and genetic drift, the ratio of the frequencies of two alternative synonymous codons throughout a single genome can be used to estimate the direction and magnitude of selection [32]. The action of natural selection can be inferred when synonymous codon pairs exhibit a strong “bias” towards one state relative to the other. This analysis extends to the distribution of polymorphic allele frequencies in genomes sampled from natural populations [33]–[35]. A similar approach can be applied to alternative nucleotides at particular positions within nucleosomal sequences. As the magnitude of selection increases, the expected frequency of preferred alleles increases. Consequently, the distribution of SNP frequencies (or “site frequency spectrum”) at a given nucleosome position (analogous to a synonymous SNP) is expected to shift towards relatively higher frequencies of preferred alleles.
If the observed dinucleotide patterns reflect selectively favored states, ancestrally unpreferred base pairs across nucleosomes should diverge towards the “preferred” state along species lineages. Further, if the “preferred” divergence patterns reflect the average impact of natural selection, then frequencies of polymorphisms in natural populations should be more skewed at sites experiencing stronger selection. “Unpreferred” variants, specifically substitutions or polymorphisms away from favored nucleotides, such as substitution of an ancestral A with a G at nucleosome positions which are systematically enriched for AA dinucleotides, should diverge more slowly and be rarer when polymorphic in the population. In contrast, “preferred” variants, such as substitution of an ancestral A with a G at positions of enriched for the GC dinucleotide, should diverge more rapidly and be more common when polymorphic.
At the lower resolution of an entire nucleosome and its nearby flanking regions, both divergence and polymorphism are observed to vary [36]–[39], but evidence of a role for natural selection in the underlying evolutionary dynamics remains sparse [40]–[43]. Studies of human SNPs [37], [38] and divergence in humans, yeast and medaka [36], [38], [39], [43] show that both expected heterozygosity and divergence between species are elevated near the central dyad and depressed in the adjacent linker regions, though these patterns appear to differ by substitutional pathway [38]. One possibility is that patterns of variation relative to nucleosomes derive from nucleosome-specific mutational biases. This could result from suppression of mutation by a protective aspect of nucleosome occupancy [44], or it could arise from an interaction between the histone core and DNA damage recognition or repair mechanisms [45]–[48]. Of course, natural selection mediated via DNA:nucleosome interactions may also strongly reshape the patterns of SNP variation and divergence between taxa [40]–[43]. Analysis of the site frequency spectrum promises to distinguish between these two alternatives.
The whole-nucleosome-resolution analyses considered above cannot leverage the specific structural predictions of dinucleotide interactions with the core and their strong mechanistic implications. Examination of polymorphism and divergence at each base pair position across the nucleosomal DNA opens a rich and precise view, as well as powerful tests of alternative mechanisms such as biased mutation and natural selection. We report the discovery of fine-scale periodicities in inter - or intra-species sequence variation relative to nucleosomes and discuss their implications for the role of natural selection mediated through nucleosome function. Our analysis of DNA sequence polymorphism and divergence across isolated nucleosomal fragments from D. melanogaster embryos reveals that nucleosomal sequences are diverging towards “preferred” nucleotides. Regions where the minor groove is interior are becoming more AT-rich, and regions where the major groove is interior are becoming more GC-rich along the melanogaster lineage. Using a new index for quantitating the frequency spectrum (Δπ), we identify clear signals associated with natural selection, which parallel the observed periodicities in divergence. This selection is strongest in intronic regions, where nucleosome assembly and positioning are expected to have greater functional impacts. These findings support the hypothesis that the widely observed sequence affinities of the core octamer have functional consequences that are subject to natural selection. Given the dominant role of nucleosomes in the packaging of the genome and their conserved sequence preferences, their interactions may broadly shape the sequence of melanogaster and other genomes.
Results
Periodicity in dinucleotide frequencies in D. melanogaster
To investigate the impact of nucleosomes on DNA sequence variation, we isolated nuclei from D. melanogaster embryos, performed Micrococcal nuclease (MNase) digestion, and used paired-end sequencing to position fragments on the genome (Figure S1). Previous studies in Drosophila identified a range of periodic dinucleotides in association with nucleosomes [10], [11]. Our collections of 276,614 intergenic and 270,998 intronic autosomal 147 bp nucleosomal fragments (hereafter n147, Tables S1 and S2) cover 68.5% of the unique intronic and intergenic euchromatic autosomal genome and display a ∼10 bp periodicity for many dinucleotide frequencies (Figures 1 and S2). In these and subsequent analyses, the 5′-3′ sequence from bases −73 to −1 were joined to the reverse complement of bases 1 to 73, to reflect the dyad symmetry of the nucleosome (see Materials and Methods). AA, TT and GC showed the strongest periodicity of WW and SS (where W = A|T, S = G|C) dinucleotide pairs, respectively (Figure 1B). These same dinucleotides show a distinct overrepresentation in the non-coding regions of the genome as a whole (Figure 1C). As noted in previous studies, AA and TT are similarly periodic and occur where the minor groove is interior (at superhelix locations, SHL, ±(i+0.5); where i is 0, 1,…6). However, noticeable differences between the distributions are apparent. For example, the frequency of TT displays a distinctly smaller peak at ∼SHL 4.5, and AA frequency displays a stronger drop at ∼SHL 2 (Figure 1B). GC frequency across n147 regions is anti-correlated with AA/TT and is characterized by a prominent upward concavity (Figure 1B). These dinucleotide periodicities extend well beyond n147 edges into linker regions, consistent with the proposed translational step size of 10 bp.
Upon examination of the dinucleotide frequencies flanking aligned n147 regions, we discovered an additional large-scale pattern in AA/TT and GC dinucleotide frequencies (Figure 1D). This ∼180 bp periodic variation in frequency tracks with overall nucleosome “occupancy” in the regions flanking the n147. Average AA/TT frequencies (Figure 1D) and overall A/T frequencies (Figure S3) are higher in regions of greater nucleosome “occupancy” and lower in putative “linker” regions. Thus, the AA/TT sequence features that facilitate nucleosome formation are enriched over regions with higher nucleosome “occupancy.” Conversely, GC frequency (and overall G/C frequencies, Figure S3) peaks at the periphery of more nucleosome-dense regions and in “linker” regions. These surprising “super-nucleosomal” periodicities extend the observed n147 patterns to flanking multi-nucleosomal arrays, and suggest a contribution of sequence to translational positioning. Consistent with chemical mapping of nucleosomes, this result suggests that the observed experimental correlation between MNase nucleosome “occupancy” and GC content [1], [8], [22], [26], [49], [50] reflects differential recovery, rather than positional preference [23], [51].
Divergence along the melanogaster lineage mirrors periodic nucleosomal base preferences
If variations in dinucleotide frequencies relative to nucleosomes result from accumulated sequence divergence, we expect substitution patterns to parallel the observed base preferences. However, the timescale(s) at which these patterns evolve is unknown. Lineage-specific or “polarized” divergence is the proportion of nucleotide sites that are different in melanogaster while identical in its sister taxa simulans (most recent common ancestor 2.5 MYA) and the proximate outgroup (yakuba or erecta; 6–7 MYA, see Materials and Methods). Overall genomic divergence on the melanogaster lineage shows a marked excess of G→A (inferred ancestral G, derived A in melanogaster) and C→T (ancestral C, derived T) substitutions compared to A→G and T→C (Figure 2A). This is in agreement with earlier estimates of divergence on the melanogaster lineage [52], [53] and with the observed two-fold greater mutation rate [54]. We next considered the average divergence at each site across n147 regions, normalized for underlying base frequencies. This analysis revealed a striking ∼10 bp periodicity in transitions (GC→AT and AT→GC) for two estimates of divergence; per-n147 in Figure 2B is weighted by the redundancy in the n147 set, while per-site in Figure S4 weights each site equally. Rates for GC→AT and AT→GC are anti-correlated and track with underlying dinucleotide frequencies. Thus, ancestral GC bases are more likely to become AT in nucleosomal regions where AA/TT dinucleotides are in higher frequency, and AT bases are more likely to become G or C at sites where GC is enriched. GC→AT divergence also shows a marked curvature, with a peak at the dyad axis.
Fig. 2. Divergence on the melanogaster lineage mirrors nucleosome sequence preferences. 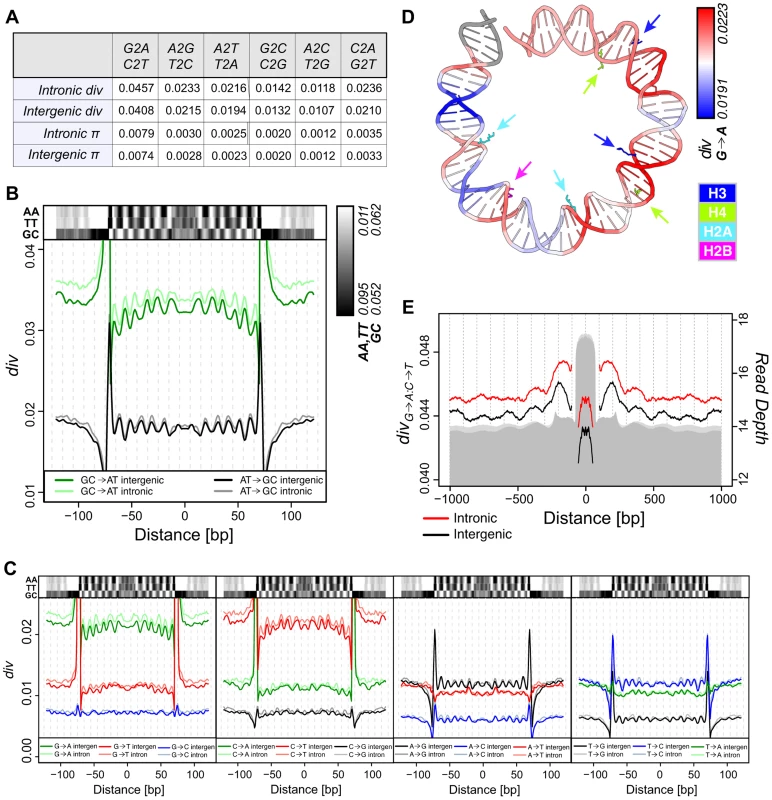
(A) Average polarized intergenic and intronic heterozygosity (π) and divergence on the melanogaster lineage for specific substitutions. (B) Smoothed average per-n147 polarized intergenic and intronic GC→AT (green = intergenic, light green = intronic) and AT→GC (black = intergenic, grey = intronic) divergence across aligned n147 regions (±50 bp). Intergenic dinucleotide frequencies are represented above. (C) Smoothed average per-n147 polarized divergence across intergenic and intronic n147 regions (±50 bp) for specific substitutions. Indicated dinucleotide frequencies plotted above for reference. Note that for both B and C, the estimated divergence rates are outside the range of the plots near the edges of the n147. (D) Smoothed intergenic per-n147 G→A divergence mapped onto bases +73 to −6 from the nucleosome structure [55]. Arginines that contact the minor groove are color coded by histone and highlighted by arrows. (E) Smoothed average combined G→A:C→T polarized divergence surrounding intergenic (black) and intronic (red) n147 regions (±1 kb). Note that divergences immediately flanking (±5 bp) the edges of the n147 are not plotted, since these largely reflect sequence bias of the MNase. Read depth of intronic (light grey) and intergenic (grey) n142-152 is represented below. Given the substantial variation in individual substitution rates, we next examined specific pathways to determine their relative contributions. Of all pathways, G→A, C→T, A→G and T→C exhibited the most obvious periodicities in divergence (Figure 2C; see Discussion). In some cases, divergence patterns reflect the subtleties observed for dinucleotide frequency patterns. For example, C→T rates are less peaked at SHL 4.5, the location of the lowest peak in TT frequency (Figure 2C). C→T also shows a greater difference in rates between the n147 periphery and linker regions (compared to G→A). Interestingly, for each ancestral base, the periodicities of substitutions that do not change GC content, appear weaker, perhaps due to both scaling and weaker signal to noise (Figure 2C). A subset of non-overlapping n147 regions showed similar patterns (Figure S5A).
When mapped onto the DNA from the nucleosome structure [55], peaks of intergenic G→A divergence clearly occur within regions where the minor groove is interior and in contact with key arginines of the histone core (Figure 2D). Note also the higher G→A divergence toward the central axis, as reflected by the downward concavity in Figure 2C. This is consistent with analyses of the impact of sequence variation on nucleosome structure, which identified this central region of H3/H4 interactions as most constrained [56]. Conversely, A→G substitution rates are highest in regions where the major groove is interior (Figure S5B). This pattern is consistent with established SS dinucleotide patterns [8], [10]–[14] and the observation that GC rich sequences are disfavored for minor groove compression and favor narrowing of the major groove [9], [18].
Divergence patterns should also reflect the observed nucleosome-scale periodicities in base and dinucleotide frequencies (Figures 1D and S3). To increase signal, we combined complementary substitutions, G→A and C→T (G→A:C→T) and A→G and T→C (A→G:T→C). Aligned n147 regions show substantially lower divergence rates than their immediate flanking sequences (Figure 2E). Rates drop to the local background within ∼500 bp, following the skew of AA/TT dinucleotides (and overall AT content; Figures 1D and S3). In spite of this local variation in rates, due at least in part to MNase preferences (Figure S6), we observe a large-scale (180 bp) periodicity in G→A:C→T divergence surrounding intergenic n147 nucleosomal regions (Figure 2E). Introns showed a similar but weaker pattern, potentially due to the influence of flanking coding regions (Figure 2E). Any periodicity of the A→G:T→C divergence in flanking regions is less obvious (Figure S5C), at least partially due to a 50% lower rate of divergence and thus inherently weaker signal.
These large-scale patterns allow us to resolve general trends in divergence relative to nucleosome occupancy. We find that, on average, G→A:C→T changes along the melanogaster lineage are fixed at higher rates across nucleosomes relative to linkers, mirroring underlying AA/TT dinucleotide frequencies. This is in apparent contrast to the report that the cytosine deamination mutational pathway (a major source of G→A:C→T transitions) and associated divergence is suppressed by nucleosome occupancy [44]. To clarify this discrepancy, we examined the interactions between divergence and “occupancy” of the n147 fragments, as estimated by depth of coverage by 142–152-bp nucleosomal fragments, n142-152. Indeed, we observe a negative correlation between this metric and all substitutional pathways (Table S3, Figure S7A). However, we note that n142-152 coverage is correlated with GC content of the n147 region (Figure S8D), as previously reported in other studies [1], [5], [8], [26], [49], and that correlations between nucleosome fragment GC and divergence are even more striking (Table S3, Figure S7B, S8C). This is also true for 500 bp intergenic windows, independent of nucleosome coverage (Table S3). When we parse n147 by n142-152 “occupancy,” we observe differences in AA/TT frequency, G→A:C→T divergence, and nucleosome phasing in flanking regions (Figure S8A). The periodicities of these features are most obvious surrounding highly occupied (GC-rich) n147 regions, but they do not appear to be unique to them. Thus, we conclude that nucleosome bound regions in D. melanogaster embryos are generally more AT-rich and have higher rates of G→A:C→T substitution than their adjacent “linker” regions, inconsistent with the fundamental claim in Chen, et al. [44] (mentioned above).
Periodicity in the site frequency spectra of natural populations: Evidence of natural selection
The divergence patterns we observe are consistent with known nucleosomal dinucleotide preferences [5], [8], [10]–[17]. This is analogous to observations for codons, where substitutions mirror genome-wide codon usage biases and are attributed to natural selection for preferred codons [34], [57], [58]. However, divergence patterns alone cannot exclude the hypothesis that substitutional patterns result from biased mutation relative to nucleosomes. Mutation rates may vary across nucleosome-bound regions and could lead to compositional variation and different rates of divergence. Nevertheless, once a new selectively neutral allele arises, its dynamics and thus its distribution of frequencies are independent of type (or rate) of mutation [59], [60]. While natural selection influences the probability of fixation (thus the rate of divergence), mild differences in fitness will also shift the site frequency spectra of polymorphic alleles [61]–[63]. Neutral and deleterious mutations tend to spend much of their typically short lives as rare alleles, while weakly favored alleles will be found at higher frequencies as many more drift towards fixation. Although the impacts of varying demographic histories [64] and of linked selection [48], [49] can lead to distributions of selectively neutral polymorphisms that mimic particular forms of selection, they should do so randomly across the genome and not show a positional relationship within nucleosomal sequences.
The hypothesis that nucleosome structure and function impose natural selection on genomic sequence variation predicts periodicities in the frequency spectra. Indeed, the average per-n147 frequencies of G-A and C-T SNPs in a sample of 36 D. melanogaster genomes from Raleigh (North Carolina) exhibit nucleosomal patterns paralleling those observed for dinucleotides and polarized divergence (Figures 3A and S9). Frequencies of A alleles at G-A SNPs show clear periodicity across intergenic and intronic n147 regions, extending into linker regions (Figure 3A). A alleles are relatively more common in SHL ±(i+0.5) regions, and G alleles are higher in regions where the major groove faces the histone core. Removal of singleton SNPs (cases where either allele is observed only once), which can mitigate the impact of possible sequencing errors, raises average A frequencies but does not eliminate the periodicity (Figure S9). Partitioning such SNPs by ancestral state can remove the impact of average mutation rate differences and reveal differences in the patterns of selection. Nucleosomal patterns of the average per-n147 frequencies of derived SNPs, such as G→GA (ancestral G and a derived, polymorphic A), exhibit clear periodicities that generally parallel divergence and nucleosomal dinucleotide frequencies (Figures S10, intergenic, and S11, intronic).
Fig. 3. Periodicity in average SNP frequencies aligns with nucleosomal divergence and dinucleotide sequence preferences. 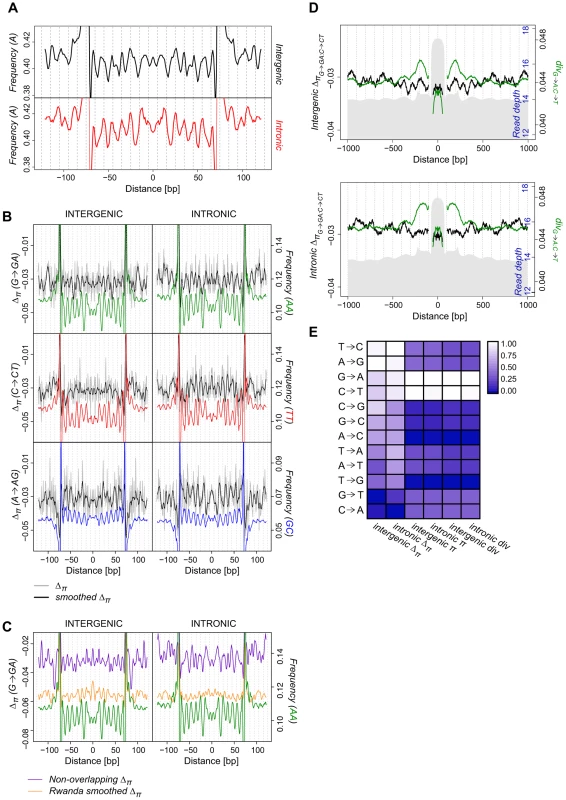
(A) Smoothed average intergenic (black) and intronic (red) per-n147 frequency of A alleles at G-A polymorphic sites in the Raleigh sample. (B) Average smoothed (black) and unsmoothed (grey) per-n147 Δπ across n147 regions (±50 bp) for G→GA, C→CT, and A→AG polymorphic sites. Color-coded dinucleotide frequencies are included for reference. (C) Average smoothed n147 G→GA Δπ for the Rwanda sample (orange) and across a non-overlapping subset of intergenic and intronic n147 regions (teal). Note that for both B and C the estimated divergence are outside the range of the plots near the edges of the n147. AA dinucleotide frequencies included for reference (green). (D) Combined intergenic and intronic (G→A:C→T; green) divergence and (G→GA:C→CT; black) Δπ surrounding intergenic n147 (±1 kb). n142-152 read depth in grey (axis in blue). Note that divergence and Δπ immediately flanking (±5 bp) the edges of the n147 are not plotted, since their true values are largely obscured by sequence bias of the MNase. (E) Average scaled intergenic and intronic per-n147 π, Δπ and divergence for specific substitutions (see Table S4). To systematically assess the periodicity in the frequency spectra we calculated a new index, Δπ, (closely related to Tajima's D [59]) across n147 regions for the Raleigh sample [65]. Where p is the frequency of a SNP in the sample and is the estimate of the heterozygosity, we define Δπ as the average (per SNP) deviation in from expectation under equilibrium between genetic drift and mutation to selectively equivalent alleles (see Materials and Methods). The “folding” of the frequency spectrum such that p is equivalent to (1−p) mitigates the impacts of errors in the inference of the ancestral state [66] and emphasizes variation in the midrange of p. Weak positive selection is predicted to skew toward higher values (more positive Δπ), while weak negative selection leads to more negative Δπ. Thus, systematic differences in selective forces at different positions across n147 regions should yield a pattern in Δπ that parallels that observed for divergence. These patterns of Δπ are superimposed on the observed genome-wide average negative skew [65] (Table S4) that can be attributed to strongly deleterious mutations [67], [68], varying demographic history [59], [64] or linked selection (background selection [69] and hitchhiking [70]).
Indeed, when we examined average Δπ for G→GA polymorphisms, we discovered a clear ∼10 bp periodic skew in frequency across nucleosomal regions, mirroring G→A divergence (per-n147 in Figure 3B and per-site in Figure S12). n147 G→GA Δπ are less negative in regions of higher AA dinucleotide frequency. Interestingly, intronic G→GA Δπ shows even more pronounced periodicity in the frequency spectrum, including the prominent drop at SHL 2 observed for AA frequency (Figures 3B and S12). Δπ for C→CT polymorphisms is also periodic in introns (both per-n147 and per-site), with peaks aligning with regions of high TT frequency; while intergenic n147 share a subset of these peaks (Figures 3B and S12), the overall patterns show much weaker periodicity (see below). Although peaks in intronic C→CT Δπ overlap roughly with those for G→GA sites, they show a more convex shape, similar to the C→T divergence. Substitutions in the complementary directions (e.g. A→AG) also show a periodic skew in allele frequencies. Introns display a striking periodicity in A→AG Δπ aligned with GC frequency (Figure 3B, while intergenic n147 A→AG sites show only two peripheral Δπ peaks and several peaks (valleys) that are discordant with the GC dinucleotide periodicity. Like underlying GC frequency, intronic A→AG Δπ has a concave upward shape. We observe weaker but interesting indications of continued periodicity in linker regions, consistent with selection for the preservation of rotational positioning in association with translational repositioning. The patterns of Δπ for 5 non-overlapping subsets of n147 regions were similar (Figure S13). We conclude that for several substitutional pathways there is strong evidence of selection maintaining the observed nucleosomal (di)nucleotide preferences.
The periodicity of nucleosomal Δπ is not limited to the Raleigh population. The strongest of these periodic patterns in Δπ are also apparent in a smaller, independent set of 21 sequenced genomes from a Rwandan (Africa) population [71] (Figures 3C and S14), which also exhibits more negative average Δπ values. This African sample is assumed to represent a larger, more stable population from the center of the species distribution, while the Raleigh sample represents the serial diasporas out-of-Africa and into North America. Notwithstanding differences in average Δπ, these strong and predicted periodicities in nucleosomal in Δπ support our hypothesis that direct interactions between the histone core and DNA sequence polymorphisms yield functional effects with fitness consequences.
An alternative hypothesis to explain these periodicities holds that the sequences evolve independently of natural selection and that the in vivo positions of our isolated nucleosomes reflect the innate preferred rotational positions of the particular genome used. Derived SNPs detected in a single strain are likely to be in high frequency, and thus we might observe periodicity in the frequency spectra at such SNPs in the absence of natural selection. To test for the impact of this hypothesized ascertainment bias on the periodicity of Δπ, we filtered the n147 for those in which the source genome bore the ancestral alleles. Despite the unavoidable thinning of the data, we observed clearly periodic polarized Δπ for those pathways with the strongest initial signals, e.g. intronic G→GA, C→CT and A→AG (Figure S15). These results indicate that the observed periodicities in the frequencies of preferred bases (parallel to the dinucleotide frequencies and the divergence) cannot be attributed to biases in the ascertainment associated with the genotype from which the nucleosomal sequences were prepared.
We next considered the values of Δπ surrounding n147 regions. The observed skew in intergenic G→GA:C→CT Δπ extends into adjacent sequence (Figure 3D), tracking with the periodicity of G→A:C→T divergence. Interestingly, in the ∼500 bp flanking n147 regions, there appear to be major and minor Δπ peaks associated with each divergence peak. Given the shoulder of C→CT Δπ values in linker regions adjacent to n147 (Figure 3A), this could represent a nucleosomal and a linker peak. Intronic regions show higher overall values of G→GA:C→CT Δπ and similar, but weaker, indications of increased G→GA:C→CT Δπ associated with nucleosome occupancy (Figure 3C). Among other interesting patterns in Δπ and contrasts to divergence in these flanking regions are those associated with the complementary set of substitutional paths, A→AG:T→TC, which exhibits peaks over apparent linker regions in Δπ but no parallel pattern in A→G:T→C divergence (Figure S16).
On average (per-n147), G→GA and C→CT are the most common polymorphisms and have among the most positive Δπ, indicating weak positive selection, in addition to being the most rapidly diverging bases (Figure 3D). Although rates of A→G and T→C divergence (and rates of associated polymorphisms) are much lower, these types of polymorphic sites also have high average Δπ (Figure 3E). Thus, substitutions with the most periodic divergence and Δπ also show the least overall negative skew in the frequency spectrum. Relative relationships of n147 average π, Δπ and divergence are quite similar to those of a non-overlapping subset and to the genome-wide averages (Table S4 and Figure S17). These broad genomic patterns appear inconsistent with equilibrium models and may reflect heterogeneity and/or recent (transient) shifts in selective forces [35], [52], [72].
Discussion
Histones are among the most ubiquitous and highly conserved eukaryotic proteins. Thus, it is not surprising that nucleosomal dinucleotide periodicities, which derive from key structural interactions between DNA sequence and the histone core, are shared widely across species. In spite of the near universality of these patterns among eukaryotes and decades of research, our understanding of their functional impact and evolutionary dynamics remain unsettled. In this work we examined genomic variation across regions defined by isolated nucleosomal DNA fragments. Our goal was to first determine if these regions showed interpretable variation in divergence between species, then to analyze population genomic variation for evidence of a role for natural selection in the generation and maintenance of nucleosome-associated sequence variation.
We find that divergence on the melanogaster lineage mirrors the sequence preferences of the histone core. This periodic variation in substitution rates across nucleosomal regions indicates that interior minor groove regions display more rapid substitution of AT for GC, and that AT base pairs in regions where the major groove faces inward are more likely to become GC rich. These striking patterns align directly with dinucleotide patterns that stabilize associations between DNA and the histone core, as documented in numerous biochemical and structural studies [5], [8], [15], [18], [19], [56]. If nucleosome-bound regions are evolving toward the observed nucleosome sequence preferences, a key question is whether this is the result of mutational bias relative to the positioning of chromatin proteins, or whether it is the consequence of natural selection based on functional differences. The available depth of population data and our new index Δπ allowed us to directly address this question. We find remarkable periodicities in Δπ that parallel the observed patterns of divergence. The spectra of SNP frequencies across n147 regions are variable, with higher Δπ when the inferred ancestral allele is unpreferred, and the derived allele is structurally favored. Therefore, we conclude that selection is, at least in part, driving the maintenance of nucleosome-associated sequence patterns on the melanogaster lineage.
If the fitness differences associated with such histone:DNA interactions are largely arising from nucleosomal dynamics (assembly, disassembly, movement and modification) and rotational positioning of functional elements, then we can further hypothesize that transcribed (intronic) nucleosomal sequences should exhibit stronger periodicity than untranscribed (intergenic) nucleosomal sequences. Consistent with this hypothesis, correlations of lineage specific divergence and Δπ with the relevant underlying dinucleotide frequencies are stronger for intronic sequences. Table 1 shows that in each case where a large difference between intergenic and intronic is apparent, it is the intronic that is larger. The two exceptions, G→A & AA and A→G & GC, are those where both correlations are among the highest. As might be expected given the longer timescale and greater number of variable sites, divergence correlates more strongly with dinucleotide frequencies than Δπ. Interestingly, Figures 2C and 3B show that these intronic vs. intergenic differences in correlation of divergence and Δπ with dinucleotide patterns may be attributable to large deviations from expectation in specific regions of the nucleosomal sequences, while other regions follow the expected periodic patterns.
Tab. 1. Pearsons correlation of lineage specific (<i>per-site</i>) divergence or Δ<sub>π</sub> with dinucleotide frequencies. 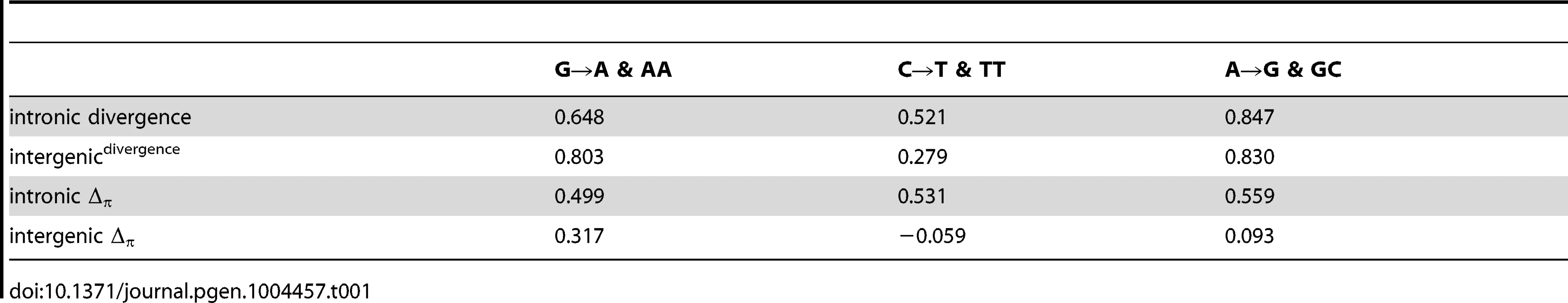
While natural selection is the most direct interpretation of these results, interactions between chromatin proteins and DNA damage and repair are well documented [44]–[48], [73]–[76]. Contextually biased mutation (substitution) pathways could underlie the observed periodicities in nucleosomal divergence. However, Drosophila does not have a significant level of 5-methylcytosine [77], the deamination of which is thought to drive the strong contextual biases (NpCpG) in vertebrates [78]. Indeed, a recent genomic sequencing study of Drosophila mutation accumulation lines yielded no evidence for contextual biases [54]. Most importantly, such sequence-contextual as well as nucleosome-mediated biases in mutation rates are excluded as an explanation for the observed periodicities in the skew of the SNP frequency spectrum (Δπ), since strictly neutral mutations should display the same frequency distribution across the genome [31], [33], [59]. Support for a role of natural selection maintaining these periodicities is bolstered by the stronger periodicities in intronic nucleosomal sequences, where transcription-associated remodeling and disruption of nucleosome-DNA interactions are more likely to have functional impacts.
There is, however, one potential “selectively neutral” mechanism to explain the observed periodic patterns in Δπ. Biased gene conversion (BGC), a process where heteroduplex regions formed between homologs are repaired in a direction favoring one base, can create SNP dynamics analogous to those of directional selection [79]. BGC systematically favoring GC over AT has been observed in a few species and indirectly implicated in others by associations of local GC content with estimated rates of crossing over [80]. However, evidence for such an association is not observed in Drosophila [81]. Given that the magnitudes of average Δπ and its periodicities for G→GA and C→CT SNPs are comparable to those of A→AG, any explanation of our results invoking BGC would have to involve multiple distinct gene conversional biases that depend on nucleosome position. While this is conceivable and worthy of further investigation, we conclude that the canonical GC-biased gene conversion is not a significant component of the evolutionary dynamics leading to these intricate nucleosomal patterns of polymorphism and divergence.
Whether these periodic patterns are the product of natural selection or BGC, the magnitude of the average force shaping the dynamics of nucleosomal SNPs must be small compared to that affecting the evolution of nonsynonymous variants. The shifts in G→A divergence between peaks and valleys in the n147 are ∼0.001 against a background average of ∼0.01, suggesting relatively weak constraint of 1 in 10 mutations. The non-synonymous rate of divergence on the melanogaster lineage, ∼0.006, is about one tenth of that for synonymous divergence corresponding to 9 out of 10 mutations being selected against [61]. Comparable conclusions could be drawn from the modest magnitudes of periodic fluctuation in expected heterozygosities and, indeed, in the widely observed periodicities in dinucleotide frequencies of nucleosomal sequences. Still, by virtue of its four-fold greater genomic footprint, the net selective impact of just the selection associated with such nucleosomal periodicities could approach the magnitude of non-synonymous variants. As is the case for coding sequence, differences in the relative (average) rates reflect the aggregate impact of selection that must vary substantially among nucleosomes, as well as among sites.
Evidence of natural selection supporting nucleosome-associated sequence periodicities and the implication of their biological impact casts the potential functions of non-protein-coding regions in a new light. Substantial portions of Drosophila, human and other genomes appear to be under evolutionary constraint, yet lack any functional annotation [67], [68]. Further, SNPs identified by genome-wide association studies (GWASs) of interesting human phenotypes often have mild attributable effects and map to unannotated intronic or intergenic regions, where mechanistic hypotheses concerning the impacts of such genomic variation are lacking. We demonstrate that at least part of the constraint in Drosophila arises from interactions between histone proteins and DNA sequence.
Our results suggest dinucleotide periodicities and the rotational positioning that they guide have significant biological consequences. Sequences affecting rotational positioning can influence the binding of transcriptional activators and participate in regulation of expression or gene splicing [25], [28], [82]–[84]. More generally, they impact nucleosome assembly and stability [2]–[7], [9], [17], properties that broadly impact chromatin dynamics and may influence higher order chromatin structures. Further, the observed large-scale periodicities in dinucleotide frequencies (and divergence and Δπ patterns supporting them) demonstrate that sequences that facilitate rotational positioning are specifically enriched relative to adjacent nucleosomes. So, while periodic sequence patterns are considered more relevant to rotational positioning, they clearly interact with the translational positioning of arrayed nucleosomes in Drosophila. Going forward, deeper and more detailed population genomic analyses should provide a unique window into the complex in vivo interactions between DNA sequence and nucleosome function.
The significance of these periodic patterns of polymorphism and divergence is amplified in light of the substantial proportion of the eukaryotic genome packaged in nucleosomes (four-fold greater than that of coding sequence in Drosophila) and the broad conservation of dinucleotide interactions with the histone core. Indeed, no other DNA-protein interaction remotely approaches the genomic density or structural impact of nucleosomes. The striking periodic variation we observe relative to nucleosomes fundamentally changes expectations about divergence and SNP frequency, particularly in non-protein-coding regions. Our results point to a layer of evolutionary forces across entire genomes, emanating from the interactions of DNA sequence variation with the structure and function of the histone core.
Materials and Methods
Nuclear isolation and MNase digestion
Embryos were collected from population cages [85] over a 1 hr period and aged at 25°C for 2–3 hr. Staged embryos were dechorionated in 50% bleach for 2 minutes, washed extensively, and then homogenized on ice in SEC buffer (10 mM HEPES, 150 mM NaCl, 10 mM EDTA 10% glycerol, 1 mM DTT) with Protease Inhibitors (PI) (0.1 mM PMSF and 2X Roche EDTA-free Protease Inhibitor tablets). After lysate filtering and centrifugation, pelleted nuclei were resuspended in CIB (15 mM Tris pH 7.5, 60 mM KCl, 15 mM NaCl. 0.34 M Sucrose, 0.15 mM Spermine, 0.5 mM Spermidine)+PI and then repelleted. Centrifugation of nuclei in CIB was repeated 3 times, and the resultant pellet was flash frozen and stored at −80°C.
Pelleted frozen nuclei were resuspended in CIB+PI, and chromatin was digested with 0.5 U/ml Micrococcal nuclease (Sigma) for 37°C for 15 min. MNase treated nuclei were pelleted, resuspended in 0.1% NP-40 PBS+PI, and incubated at 4°C for 3 hrs to release (primarily) mononucleosomes. Nuclei were re-pelleted, and chromatin from the supernatant was phenol-chloroform extracted. Digestion was analyzed on an agarose gel.
Library construction and sequencing
Approximately 882 ng of DNA was used as starting material for paired-end sequencing library construction following the Illumina protocol (PE-102-1001). 10 µl of paired end adapter oligos were ligated to the end-repaired, A-tailed fragments in a 50 µl reaction. The adapter-ligation product was gel-purified to select molecules approximately 150–700 bp in length and re-suspended in 30 µl total volume. 1 µl of size-selected ligation product was used as template for 12 cycles of library enrichment PCR in a 50 µl reaction volume. The enriched library was purified using QIAGEN MinElute columns and sequenced (2×36 cycles) on one lane of a flow cell (FC42JB8) with an Illumina GAIIx running the Illumina software SCS v2.4.135/Pipeline v1.4.0.
Subsequently, 8 µl of the same size-selected ligation product was used as a template for 10 cycles of library enrichment PCR in a 100 µl reaction volume. To enrich for 147 bp fragments, the library was purified using QIAGEN MinElute columns, then size selected on an agarose gel to recover fragments approximately 273 bp in length (as determined by an Agilent Bioanalyzer). This size-selected library was sequenced (2×36 cycles) on four lanes of a flow cell (FC61BGN) with an Illumina GAIIx running the Illumina software SCS v2.5.38/Pipeline v1.5.0.
Read mapping and filtering
Reads that passed the Illumina pipeline's quality filters were then aligned to the Berkeley Drosophila Genome Project's Release 5 reference sequence [86] using Version 0.7.0 of the MAQ program [85]. Read pairs that mapped more than 1,000 bp apart and those for which the combined sum of the quality scores of mismatches exceeded 300 were filtered using the maq map –a 1000 -e 300 command. Otherwise, default map parameters were used.
147 bp Fragments in intronic and intergenic regions
Mapped paired end clones of length 147 bp were then filtered based on Release 5.16 FlyBase Annotation of the D. melanogaster genome and classified as intronic or intergenic. For classification, all bases, including the flanking ±50 bp, were required to map entirely to a contiguous intronic or intergenic region. Heterochromatic reads were removed using cytogenomically-defined boundaries [87]. Downstream analysis was carried out on 276,614 intergenic and 270,998 intronic autosomal 147 bp nucleosomal fragments, referred to as intronic and intergenic n147. These cover 61% and 79% respectively of the target intergenic and intronic regions in the euchromatic autosomes (chr2 and chr3). The coordinates of the n147±50 bp flanking regions are in Tables S1 and S2. Average nucleosomal read depths, where represented, are a pileup representation of a set of similarly processed paired end clones with lengths ranging from 142–152 bp.
In calculating dinucleotide frequencies, divergence, π and Δπ across n147 regions (and, where relevant, ±50 bp flanking), both positional and average calculations took the dyad symmetry into account. Substitutional pathways were switched at the dyad axis, such that positions −73 to −1 were joined to the reverse complement of bases 1–73. Where included, flanking regions (±50 bp) were treated similarly. For larger scale positional divergence and genomic averages, data from complementary substitutional pathways were combined.
Dinucleotide frequencies
The n147 dinucleotide frequencies are the averages (in the reference sequence) over all n147 fragments for each position. Genomic over/underrepresentations of dinucleotides were calculated by dividing the difference between observed and expected frequencies by the expected frequency. Estimates of expected intergenic and intronic dinucleotide frequencies were calculated based on underlying base frequencies. Observed frequencies were computed directly from the reference sequence.
Population genomic data
The sequences of the euchromatic portions of 36 D. melanogaster genomes from Raleigh, North Carolina were released by DPGP (http://www.dpgp.org/1K_50genomes.html - Reference_Release_1.0). The sequencing, alignment and assignment of estimated quality scores are described in Langley. et al., 2012 [65]. The sequences of the 22 D. melanogaster genomes from Rwanda, Africa were released by DPGP (http://www.dpgp.org/dpgp2/update_20Jan2012/dpgp2_v2_rg.ID5.nohets.fastq.bz2). The sequencing, alignment and assignment of estimated quality scores are described in Pool et al., 2012 [71]. For both data sets, only bases with a minimum quality score of Q30 or greater were included in the analyses.
Divergence, frequency, π and Δπ
Calculations of divergence on the melanogaster lineage, frequency, expected heterozygosity (π) and the index of skew in the frequency spectrum (Δπ) were based solely on sites that could be polarized using a multiple alignment of D. melanogaster, D. simulans, D. yakuba and D. erecta genomes [65], i.e., simulans and yakuba and/or erecta have the identical base. For consideration of the potential impact of ascertainment bias association with the isolation of nucleosomes, sites were subjected to a more stringent polarization (simulans, yakuba and erecta have the identical base) and calculations were done only on sites where the experimental genome had the ancestral allele. These statistics (divergence, π and Δπ) were estimated with two alternative weightings. The first, per-n147, gave weight to genomic sites proportional to their occurrence at nucleosome positions among the n147. Thus, in instances where a particular site was found multiple times in the n147 set, the divergences or SNPs at that site were give proportionally more weight. This in effect weights the signal by nucleosome site “occupancy.” The second, per-site, counted normalized the weighting such that each site (conserved, divergence or SNP) contributed equally, independent of its recurrence in the n147. The per-n147 estimates reflect those nucleosomal sequences that were readily isolated, while the per-site method treats each population genomic variant equally.
A third representation of the mapping of divergence and Δπ consists of random non-overlapping regions sampled from the n147. These analyses are presented to simply address whether the periodicities arise solely from a ∼10 bp periodicity in the overlap of n147 fragments. For divergence, non-overlapping n147 sets (72,710 intronic and 72,859 intergenic sequences) were generated by random sampling of intergenic and intronic n147 without replacement. Newly drawn sequences were added to the non-overlapping subset only if they did not share any positions with prior sampled sequences. These non-overlapping subsets together cover 53% of the target intergenic and intronic regions in the euchromatic autosomes (chr2 and chr3) covered by the full n147. For non-overlapping Δπ, intergenic and intronic n147 (not including the flanking ±50 bp) were first filtered for only those nucleosomal regions containing the relevant SNP (taking dyad symmetry into account). This produced non-overlapping intergenic and intronic sets of ∼90,000 regions each for G→GA and C→CT and ∼55,000 regions for A→AG. These sets were then subjected to random sampling without replacement. A new n147 was added to the set if it did not overlap any already in the set. Non-overlapping G→GA and C→CT intronic and intergenic sets contained ∼46,000 regions each and A→AG sets contained ∼30,000.
All three of these methods for calculating divergence and Δπ yielded similarly periodic patterns reflecting the fact that while the genomic coverage of the n147 is not deep, it is also relatively uniform (cv 0.65) and the periodicities are not arising from a small subset of the n147 or interactions from overlapping n147 sequences.
Definition of divergence
Divergence was based on the species reference sequences (see above). Polarized divergence estimates were calculated as the number of specific substitutions (e.g. number of G→A) divided by the number of polarized sites inferred to have been the specific ancestral state (i.e., G in this case). For n147 average divergence, 5 bp at each end of the fragments were trimmed from the analysis to minimize the influence of sequence bias at the enzyme cleavage site (Figure S4).
Definition of frequency and π
All estimates of frequency and expected heterozygosity, π were calculated over sample sizes between nmin and nmax as
where L is the total number of sites (bp) with sample size between nmin and nmax. xj is the frequency of an allele at the jth of Si sites with sample size i. These SNPs can be categorized simply by state (A, C, G or T) or also as derived from an inferred ancestral state (e.g., A in a G→GA SNP). For n147 average , n147 regions were trimmed as described above for divergence.Definition of Δπ
We require a sample-based index of the skew in the site frequency spectrum. Tajima [59] proffered the test statistic, D, a normalization of d, the difference between two estimates of the same population parameter, 4Nμ, where μ is the mutation rate to selectively neutral alleles and N is the population size. These estimates are
and in a sample of size n, where xi is the frequency of one of the two alleles at each of the S segregating sites in an arbitrary genomic segment of length L base pairs. Thus (expected heterozygosity) and are per site (base pair) estimates of 4Nμ.But here we seek not a test statistic for a genomic segment, but an index of the same deviation, that can be aggregated across heterogeneously sampled data and compared across classes of genomic annotation.
To that end consider
a simple rescaling of Tajima's (little) d, Tajima [59] also presents the distribution of the proportion of S segregating sites with frequency i/n in the sample, Gn(i)/S. Gn(i) can not be used to compute properties of a sample unless one can argue that the sites evolved independently and are sampled independently. If we choose a set of S segregating sites (assumed to be independently sampled from a population, i.e. no linkage disequilibrium), rather than a genomic segment, we have the expected heterozygosity in a sample of size n, and so our index, This index is thus a measure of the deviation of the population (expected) heterozygosity per segregating site from its predicted value under the assumptions of equilibrium between selectively neutral mutation and genetic drift in a Wright-Fisher population.To estimate this deviation across sites with different sample sizes, we can calculate the weighted average, weighting by the reciprocal of the variance.
The variance of is
Notice that this is the theoretical variance in at a single segregating site in a sample of size n under the assumptions of the neutral model (above).If the frequencies at different sites are independent then we can estimate the sample variance of Δπ(n) for S sites with sampling depth n, simply as Var[Δπ(n)]/S.
Assuming again that these SNPs are sampled independently both within and over sample sizes (i.e., no linkage disequilibrium), the average Δπ can be estimated by the weighted average
where nmin>3 and nmax = largest sample size.Δπ for each position in the n147 regions (and its average across positions) was calculated over polarized sites with sampling sizes (n) between 32 and 34 in the Raleigh [65] and 18 to 21 for Rwandan data [71]. For n147 average Δπ the n147 regions were trimmed as described above for divergence. Data from complementary pathways were merged for larger scale positional Δπ.
Correlations of divergence with GC frequency and nucleosome “occupancy”
n142-152 coverage of intergenic n147 regions was defined as the sum of the coverage of the region by the larger set of 142–152 bp nucleosomal fragments. This corresponds to what some authors call “occupancy.” GC frequency of intergenic n147 regions was calculated based on the nucleotide frequencies in the D. melanogaster reference sequence from bases 6 to 141 of the n147 regions (to minimize the potential impact of MNase sequence bias). For density plots and Spearman's ρ, only n147 regions with at least 50 (out of 147) intergenic bases polarizable were included in the analyses. As above, the interior 5 bp on each end of n147 regions were trimmed from the analysis. For correlations between divergence and genomic intergenic GC frequencies, Spearman's ρ were reported for non-overlapping 500 bp windows in which at least 166 intergenic bases were polarizable and no more than 250 bp in the reference were “N”.
Plot details
Plots were generated using R [88]. Prior to plotting, all calculations were symmetrized around the dyad axis. n147 (±50 bp) divergence and Δπ plots (Figure 1D, Figure 2B,C, Figure 3A) were smoothed using running average in a window of 5 bp (weights: 0.125, 0.250, 0.250, 0.250, 0.125). For large scale plots of dinucleotide frequency, divergence and Δπ (Figure 1C, Figure 2D, Figure 3B), flanking regions were smoothed using running average smoothing with a window of 50 bp of equal weights. In those plots, the central n147 regions were smoothed separately using a 30 bp window of equal weights. Regions of 5 bp upstream and downstream of the n147 edges were trimmed prior to smoothing for large-scale plots.
Structural mapping of divergence rates
To elucidate the distribution of the smoothed (as above) divergence on DNA from the structure of the nucleosome we colored-coded values of each base pair in a schematic rendering of the DNA strands of pdb1kx5 [55] using PyMOL [89]. Only the “top” turn of the DNA (base pairs 73 to −6) is shown. Bases 73 to 70 were rendered as grey, due to extreme values induced by MNase sequence bias.
Supporting Information
Zdroje
1. LeeW, TilloD, BrayN, MorseRH, DavisRW, et al. (2007) A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39 : 1235–1244.
2. PhamCD, HeX, SchnitzlerGR (2010) Divergent human remodeling complexes remove nucleosomes from strong positioning sequences. Nucleic Acids Res 38 : 400–413.
3. MoshkinYM, ChalkleyGE, KanTW, ReddyBA, OzgurZ, et al. (2012) Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol Cell Biol 32 : 675–688.
4. LowaryPT, WidomJ (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276 : 19–42.
5. ThastromA, LowaryPT, WidlundHR, CaoH, KubistaM, et al. (1999) Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol 288 : 213–229.
6. KaplanN, MooreI, Fondufe-MittendorfY, GossettAJ, TilloD, et al. (2010) Nucleosome sequence preferences influence in vivo nucleosome organization. Nat Struct Mol Biol 17 : 918–920 author reply 920–912.
7. KaplanN, MooreIK, Fondufe-MittendorfY, GossettAJ, TilloD, et al. (2009) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458 : 362–366.
8. Radman-LivajaM, RandoOJ (2010) Nucleosome positioning: how is it established, and why does it matter? Dev Biol 339 : 258–266.
9. WidomJ (2001) Role of DNA sequence in nucleosome stability and dynamics. Q Rev Biophys 34 : 269–324.
10. SegalE, Fondufe-MittendorfY, ChenL, ThastromA, FieldY, et al. (2006) A genomic code for nucleosome positioning. Nature 442 : 772–778.
11. MavrichTN, JiangC, IoshikhesIP, LiX, VentersBJ, et al. (2008) Nucleosome organization in the Drosophila genome. Nature 453 : 358–362.
12. IoshikhesIP, AlbertI, ZantonSJ, PughBF (2006) Nucleosome positions predicted through comparative genomics. Nat Genet 38 : 1210–1215.
13. GaffneyDJ, McVickerG, PaiAA, Fondufe-MittendorfYN, LewellenN, et al. (2012) Controls of nucleosome positioning in the human genome. PLoS Genet 8: e1003036.
14. SatchwellSC, DrewHR, TraversAA (1986) Sequence periodicities in chicken nucleosome core DNA. J Mol Biol 191 : 659–675.
15. AnselmiC, BocchinfusoG, De SantisP, SavinoM, ScipioniA (1999) Dual role of DNA intrinsic curvature and flexibility in determining nucleosome stability. J Mol Biol 286 : 1293–1301.
16. DrewHR, TraversAA (1985) DNA bending and its relation to nucleosome positioning. J Mol Biol 186 : 773–790.
17. ShraderTE, CrothersDM (1989) Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A 86 : 7418–7422.
18. RohsR, WestSM, SosinskyA, LiuP, MannRS, et al. (2009) The role of DNA shape in protein-DNA recognition. Nature 461 : 1248–1253.
19. WestSM, RohsR, MannRS, HonigB (2010) Electrostatic interactions between arginines and the minor groove in the nucleosome. J Biomol Struct Dyn 27 : 861–866.
20. JiangC, PughBF (2009) Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10 : 161–172.
21. ZhangY, MoqtaderiZ, RattnerBP, EuskirchenG, SnyderM, et al. (2009) Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16 : 847–852.
22. ValouevA, JohnsonSM, BoydSD, SmithCL, FireAZ, et al. (2011) Determinants of nucleosome organization in primary human cells. Nature 474 : 516–520.
23. BrogaardK, XiL, WangJP, WidomJ (2012) A map of nucleosome positions in yeast at base-pair resolution. Nature 486 : 496–501.
24. ZhangZ, WippoCJ, WalM, WardE, KorberP, et al. (2011) A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332 : 977–980.
25. FieldY, KaplanN, Fondufe-MittendorfY, MooreIK, SharonE, et al. (2008) Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216.
26. TilloD, HughesTR (2009) G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10 : 442.
27. SekingerEA, MoqtaderiZ, StruhlK (2005) Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell 18 : 735–748.
28. AlbertI, MavrichTN, TomshoLP, QiJ, ZantonSJ, et al. (2007) Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446 : 572–576.
29. CuiF, ColeHA, ClarkDJ, ZhurkinVB (2012) Transcriptional activation of yeast genes disrupts intragenic nucleosome phasing. Nucleic Acids Res 40 : 10753–10764.
30. BowmanGD (2010) Mechanisms of ATP-dependent nucleosome sliding. Curr Opin Struct Biol 20 : 73–81.
31. WrightS (1931) Evolution in Mendelian Populations. Genetics 16 : 97–159.
32. BulmerM (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics 129 : 897–907.
33. HartlDL, MoriyamaEN, SawyerSA (1994) Selection intensity for codon bias. Genetics 138 : 227–234.
34. AkashiH (1994) Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136 : 927–935.
35. AkashiH (1996) Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics 144 : 1297–1307.
36. SasakiS, MelloCC, ShimadaA, NakataniY, HashimotoS-I, et al. (2009) Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science (New York, NY) 323 : 401–404.
37. TolstorukovMY, VolfovskyN, StephensRM, ParkPJ (2011) Impact of chromatin structure on sequence variability in the human genome. Nature Structural & Molecular Biology 18 : 510–515.
38. PrendergastJ, SempleC (2011) Widespread signatures of recent selection linked to nucleosome positioning in the human lineage. Genome Research 21 : 1777–1877.
39. WashietlS, MachneR, GoldmanN (2008) Evolutionary footprints of nucleosome positions in yeast. Trends Genet 24 : 583–587.
40. BabbittGA, CotterCR (2011) Functional conservation of nucleosome formation selectively biases presumably neutral molecular variation in yeast genomes. Genome Biol Evol 3 : 15–22.
41. BabbittGA, KimY (2008) Inferring Natural Selection on Fine-Scale Chromatin Organization in Yeast. Molecular Biology and Evolution 25 : 1714–1727.
42. BabbittGA, TolstorukovMY, KimY (2010) The molecular evolution of nucleosome positioning through sequence-dependent deformation of the DNA polymer. J Biomol Struct Dyn 27 : 765–780.
43. WarneckeT, BatadaNN, HurstLD (2008) The impact of the nucleosome code on protein-coding sequence evolution in yeast. PLoS Genet 4: e1000250.
44. ChenX, ChenZ, ChenH, SuZ, YangJ, et al. (2012) Nucleosomes suppress spontaneous mutations base-specifically in eukaryotes. Science 335 : 1235–1238.
45. JavaidS, ManoharM, PunjaN, MooneyA, OttesenJJ, et al. (2009) Nucleosome remodeling by hMSH2-hMSH6. Mol Cell 36 : 1086–1094.
46. LiF, TianL, GuL, LiGM (2009) Evidence that nucleosomes inhibit mismatch repair in eukaryotic cells. J Biol Chem 284 : 33056–33061.
47. HinzJM, RodriguezY, SmerdonMJ (2010) Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme. Proc Natl Acad Sci U S A 107 : 4646–4651.
48. RodriguezY, SmerdonMJ (2013) The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes. J Biol Chem 288 : 13863–13875.
49. WangJ, ZhuangJ, IyerS, LinX, WhitfieldTW, et al. (2012) Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22 : 1798–1812.
50. PeckhamHE, ThurmanRE, FuY, StamatoyannopoulosJA, NobleWS, et al. (2007) Nucleosome positioning signals in genomic DNA. Genome Res 17 : 1170–1177.
51. AllanJ, FraserRM, Owen-HughesT, Keszenman-PereyraD (2012) Micrococcal nuclease does not substantially bias nucleosome mapping. J Mol Biol 417 : 152–164.
52. PohYP, TingCT, FuHW, LangleyCH, BegunDJ (2012) Population Genomic Analysis of Base Composition Evolution in Drosophila melanogaster. Genome Biol Evol 4 : 1245–1255.
53. MoriyamaEN, PowellJR (1996) Intraspecific nuclear DNA variation in Drosophila. Mol Biol Evol 13 : 261–277.
54. KeightleyPD, TrivediU, ThomsonM, OliverF, KumarS, et al. (2009) Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res 19 : 1195–1201.
55. DaveyCA, SargentDF, LugerK, MaederAW, RichmondTJ (2002) Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 a Resolution. Journal of Molecular Biology 319 : 1097–1113.
56. ChuaEY, VasudevanD, DaveyGE, WuB, DaveyCA (2012) The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Res 40 : 6338–6352.
57. IkemuraT (1982) Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol 158 : 573–597.
58. DuretL (2002) Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev 12 : 640–649.
59. TajimaF (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 : 585–595.
60. WatersonGA (1974) The Sampling Theory of Selectively Neutral Alleles. Advances in Applied Probability 6 : 463–488.
61. McVeanGAT, CharlesworthB (1999) A Population Genetic Model for the Evolution of Synonymous Codon Usage: Patterns and Predictions. Genetics Research 74 : 145–158.
62. SawyerSA, HartlDL (1992) Population genetics of polymorphism and divergence. Genetics 132 : 1161–1176.
63. WrightS (1938) The Distribution of Gene Frequencies Under Irreversible Mutation. Proc Natl Acad Sci U S A 24 : 253–259.
64. WallJD, PrzeworskiM (2000) When did the human population size start increasing? Genetics 155 : 1865–1874.
65. LangleyCH, StevensK, CardenoC, LeeYC, SchriderDR, et al. (2012) Genomic Variation in Natural Populations of Drosophila melanogaster. Genetics 192 : 533–598.
66. HernandezRD, WilliamsonSH, BustamanteCD (2007) Context dependence, ancestral misidentification, and spurious signatures of natural selection. Mol Biol Evol 24 : 1792–1800.
67. HalliganDL, KeightleyPD (2006) Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res 16 : 875–884.
68. CasillasS, BarbadillaA, BergmanCM (2007) Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol Biol Evol 24 : 2222–2234.
69. CharlesworthB (2012) The effects of deleterious mutations on evolution at linked sites. Genetics 190 : 5–22.
70. BravermanJM, HudsonRR, KaplanNL, LangleyCH, StephanW (1995) The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics 140 : 783–796.
71. PoolJE, Corbett-DetigRB, SuginoRP, StevensKA, CardenoCM, et al. (2012) Population Genomics of Sub-Saharan Drosophila melanogaster: African Diversity and Non-African Admixture. PLoS Genet 8: e1003080.
72. BegunDJ, WhitleyP (2002) Molecular population genetics of Xdh and the evolution of base composition in Drosophila. Genetics 162 : 1725–1735.
73. SchopfB, BregenhornS, QuivyJP, KadyrovFA, AlmouzniG, et al. (2012) Interplay between mismatch repair and chromatin assembly. Proc Natl Acad Sci U S A 109 : 1895–1900.
74. GaillardH, FitzgeraldDJ, SmithCL, PetersonCL, RichmondTJ, et al. (2003) Chromatin remodeling activities act on UV-damaged nucleosomes and modulate DNA damage accessibility to photolyase. J Biol Chem 278 : 17655–17663.
75. JagannathanI, PepenellaS, HayesJJ (2011) Activity of FEN1 endonuclease on nucleosome substrates is dependent upon DNA sequence but not flap orientation. J Biol Chem 286 : 17521–17529.
76. YeY, StahleyMR, XuJ, FriedmanJI, SunY, et al. (2012) Enzymatic excision of uracil residues in nucleosomes depends on the local DNA structure and dynamics. Biochemistry 51 : 6028–6038.
77. LykoF, RamsahoyeBH, JaenischR (2000) DNA methylation in Drosophila melanogaster. Nature 408 : 538–540.
78. HwangDG, GreenP (2004) Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci U S A 101 : 13994–14001.
79. NagylakiT (1983) Evolution of a large population under gene conversion. Proc Natl Acad Sci U S A 80 : 5941–5945.
80. BirdsellJA (2002) Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol 19 : 1181–1197.
81. GaltierN, BazinE, BierneN (2006) GC-biased segregation of noncoding polymorphisms in Drosophila. Genetics 172 : 221–228.
82. LiQ, WrangeO (1995) Accessibility of a glucocorticoid response element in a nucleosome depends on its rotational positioning. Mol Cell Biol 15 : 4375–4384.
83. TilgnerH, NikolaouC, AlthammerS, SammethM, BeatoM, et al. (2009) Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16 : 996–1001.
84. KwakH, FudaNJ, CoreLJ, LisJT (2013) Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339 : 950–953.
85. LiXY, MacArthurS, BourgonR, NixD, PollardDA, et al. (2008) Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27.
86. CelnikerSE, WheelerDA, KronmillerB, CarlsonJW, HalpernA, et al. (2002) Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol 3: RESEARCH0079.
87. RiddleNC, MinodaA, KharchenkoPV, AlekseyenkoAA, SchwartzYB, et al. (2011) Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Research 21 : 147–163.
88. R Development Core Team (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing
89. Schrödinger L The PyMOL Molecular Graphics System, Version 1.5.0.4
Štítky
Genetika Reprodukční medicína
Článek Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome EvolutionČlánek Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder PopulationČlánek Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among PancrustaceansČlánek Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery DiseaseČlánek An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation inČlánek Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 7
-
Všechny články tohoto čísla
- Cuba: Exploring the History of Admixture and the Genetic Basis of Pigmentation Using Autosomal and Uniparental Markers
- Clonal Architecture of Secondary Acute Myeloid Leukemia Defined by Single-Cell Sequencing
- Mechanisms of Functional Variants That Impair Regulated Bicarbonate Permeation and Increase Risk for Pancreatitis but Not for Cystic Fibrosis
- Nucleosomes Shape DNA Polymorphism and Divergence
- Functional Diversification of Hsp40: Distinct J-Protein Functional Requirements for Two Prions Allow for Chaperone-Dependent Prion Selection
- Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome Evolution
- Activation of the Immune System by Combinations of Common Alleles
- Age-Associated Sperm DNA Methylation Alterations: Possible Implications in Offspring Disease Susceptibility
- Muscle-Specific SIRT1 Gain-of-Function Increases Slow-Twitch Fibers and Ameliorates Pathophysiology in a Mouse Model of Duchenne Muscular Dystrophy
- MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361
- Hypersensitivity of Primordial Germ Cells to Compromised Replication-Associated DNA Repair Involves ATM-p53-p21 Signaling
- Intrapopulation Genome Size Variation in Reflects Life History Variation and Plasticity
- SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD
- Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population
- Determinative Developmental Cell Lineages Are Robust to Cell Deaths
- DELLA Protein Degradation Is Controlled by a Type-One Protein Phosphatase, TOPP4
- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among Pancrustaceans
- UVB Induces a Genome-Wide Acting Negative Regulatory Mechanism That Operates at the Level of Transcription Initiation in Human Cells
- The Nesprin Family Member ANC-1 Regulates Synapse Formation and Axon Termination by Functioning in a Pathway with RPM-1 and β-Catenin
- Combinatorial Interactions Are Required for the Efficient Recruitment of Pho Repressive Complex (PhoRC) to Polycomb Response Elements
- Recombination in the Human Pseudoautosomal Region PAR1
- Microsatellite Interruptions Stabilize Primate Genomes and Exist as Population-Specific Single Nucleotide Polymorphisms within Individual Human Genomes
- An Intronic microRNA Links Rb/E2F and EGFR Signaling
- An Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding
- Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery Disease
- The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent Mutation
- Evolution and Genetic Architecture of Chromatin Accessibility and Function in Yeast
- An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation in
- Stage-Dependent and Locus-Specific Role of Histone Demethylase Jumonji D3 (JMJD3) in the Embryonic Stages of Lung Development
- Genome Wide Association Identifies Common Variants at the Locus Influencing Plasma Cortisol and Corticosteroid Binding Globulin
- Regulation of Feto-Maternal Barrier by Matriptase- and PAR-2-Mediated Signaling Is Required for Placental Morphogenesis and Mouse Embryonic Survival
- Apomictic and Sexual Germline Development Differ with Respect to Cell Cycle, Transcriptional, Hormonal and Epigenetic Regulation
- Functional EF-Hands in Neuronal Calcium Sensor GCAP2 Determine Its Phosphorylation State and Subcellular Distribution , and Are Essential for Photoreceptor Cell Integrity
- Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data
- Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
- Cis and Trans Effects of Human Genomic Variants on Gene Expression
- 8.2% of the Human Genome Is Constrained: Variation in Rates of Turnover across Functional Element Classes in the Human Lineage
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- A Loss of Function Screen of Identified Genome-Wide Association Study Loci Reveals New Genes Controlling Hematopoiesis
- Unraveling Genetic Modifiers in the Mouse Model of Absence Epilepsy
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
- The Coding and Noncoding Architecture of the Genome
- A Novel Locus Is Associated with Large Artery Atherosclerotic Stroke Using a Genome-Wide Age-at-Onset Informed Approach
- Brg1 Loss Attenuates Aberrant Wnt-Signalling and Prevents Wnt-Dependent Tumourigenesis in the Murine Small Intestine
- The PTK7-Related Transmembrane Proteins Off-track and Off-track 2 Are Co-receptors for Wnt2 Required for Male Fertility
- The Co-factor of LIM Domains (CLIM/LDB/NLI) Maintains Basal Mammary Epithelial Stem Cells and Promotes Breast Tumorigenesis
- Essential Genetic Interactors of Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates
- Meiosis-Specific Cohesin Component, Is Essential for Maintaining Centromere Chromatid Cohesion, and Required for DNA Repair and Synapsis between Homologous Chromosomes
- Silencing Is Noisy: Population and Cell Level Noise in Telomere-Adjacent Genes Is Dependent on Telomere Position and Sir2
- The Two Cis-Acting Sites, and , Contribute to the Longitudinal Organisation of Chromosome I
- A Broadly Conserved G-Protein-Coupled Receptor Kinase Phosphorylation Mechanism Controls Smoothened Activity
- Requirements for Acute Burn and Chronic Surgical Wound Infection
- LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription
- WAPL Is Essential for the Prophase Removal of Cohesin during Meiosis
- Expression in Planarian Neoblasts after Injury Controls Anterior Pole Regeneration
- Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
- Accumulation of a Threonine Biosynthetic Intermediate Attenuates General Amino Acid Control by Accelerating Degradation of Gcn4 via Pho85 and Cdk8
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání













