-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAccumulation of a Threonine Biosynthetic Intermediate Attenuates General Amino Acid Control by Accelerating Degradation of Gcn4 via Pho85 and Cdk8
Transcriptional activator Gcn4 maintains amino acid homeostasis in budding yeast by inducing multiple amino acid biosynthetic pathways in response to starvation for any amino acid—the general amino acid control. Gcn4 abundance is tightly regulated by the interplay between an intricate translational control mechanism, which induces Gcn4 synthesis in starved cells, and a pathway of phosphorylation and ubiquitylation that mediates its rapid degradation by the proteasome. Here, we discovered that accumulation of a threonine biosynthetic pathway intermediate, β-aspartate semialdehyde (ASA), in hom6Δ mutant cells impairs general amino acid control in cells starved for isoleucine and valine by accelerating the already rapid degradation of Gcn4, in a manner requiring its phosphorylation by cyclin-dependent kinases Cdk8/Srb10 and Pho85. Interestingly, our results unveil a division of labor between these two kinases wherein Srb10 primarily targets inactive Gcn4 molecules—presumably damaged under conditions of ASA excess—while Pho85 clears a greater proportion of functional Gcn4 species from the cell. The ability of ASA to inhibit transcriptional induction of threonine pathway enzymes by Gcn4, dampening ASA accumulation and its toxic effects on cell physiology, should be adaptive in the wild when yeast encounters natural antibiotics that target Hom6 enzymatic activity.
Published in the journal: . PLoS Genet 10(7): e32767. doi:10.1371/journal.pgen.1004534
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004534Summary
Transcriptional activator Gcn4 maintains amino acid homeostasis in budding yeast by inducing multiple amino acid biosynthetic pathways in response to starvation for any amino acid—the general amino acid control. Gcn4 abundance is tightly regulated by the interplay between an intricate translational control mechanism, which induces Gcn4 synthesis in starved cells, and a pathway of phosphorylation and ubiquitylation that mediates its rapid degradation by the proteasome. Here, we discovered that accumulation of a threonine biosynthetic pathway intermediate, β-aspartate semialdehyde (ASA), in hom6Δ mutant cells impairs general amino acid control in cells starved for isoleucine and valine by accelerating the already rapid degradation of Gcn4, in a manner requiring its phosphorylation by cyclin-dependent kinases Cdk8/Srb10 and Pho85. Interestingly, our results unveil a division of labor between these two kinases wherein Srb10 primarily targets inactive Gcn4 molecules—presumably damaged under conditions of ASA excess—while Pho85 clears a greater proportion of functional Gcn4 species from the cell. The ability of ASA to inhibit transcriptional induction of threonine pathway enzymes by Gcn4, dampening ASA accumulation and its toxic effects on cell physiology, should be adaptive in the wild when yeast encounters natural antibiotics that target Hom6 enzymatic activity.
Introduction
Cells undergo rapid transcriptional reprogramming in response to environmental changes by mobilizing transcriptional activators and repressors. Transcriptional activators function by binding to specific DNA sequences (UAS elements in yeast) and recruiting transcriptional cofactor proteins/complexes that remove repressive chromatin structure and directly recruit the transcriptional machinery to the promoters of genes under their control. Various mechanisms have been elucidated for stimulating activator function in response to environmental signals, including dissociation from a repressor, as in the case of yeast Gal4 [1], or increased entry into the nucleus as described for Pho4 and Gln3 [2]. The yeast activator Gcn4 is regulated by a unique translational control mechanism that rapidly increases the rate of Gcn4 synthesis in response to limitation for any amino acid—the conditions where increased transcription of amino acid biosynthetic genes under Gcn4 control is essential to maintaining cell growth. Gcn4 is also negatively regulated by a pathway that evokes its phosphorylation, ubiquitylation, and degradation by the proteasome, such that continued high-level translation of GCN4 mRNA is required to sustain induction of Gcn4 protein and its target genes. Together, these systems provide for reversible, short-lived induction of Gcn4, except under conditions of extreme starvation—in which protein synthesis is strongly impaired—where Gcn4 turnover is attenuated (reviewed in [3]). In addition to stimulating the transcription of genes encoding enzymes representing all of the amino acid biosynthetic pathways—the regulatory response dubbed general amino acid control (GAAC) — one-tenth or more of the yeast genome is induced by Gcn4, including genes involved in producing amino acid precursors, mitochondrial carrier proteins, vitamins and cofactors, amino acid transporters, autophagy, or the metabolism of purine, glycogen, and trehalose [4], [5].
The induction of Gcn4 expression at the translational level in amino acid-starved cells requires the protein kinase Gcn2, which is activated by uncharged tRNAs cognate to the limiting amino acid. Gcn2's sole substrate in yeast is the α subunit of general translation initiation factor 2 (eIF2). In its GTP-bound form, eIF2 delivers charged methionyl initiator tRNA (Met-tRNAiMet) to the small (40S) ribosomal subunit in the first step of translation initiation. The inactive eIF2-GDP complex is released at the end of the process and must be recycled to eIF2-GTP by the guanine nucleotide exchange factor eIF2B. Phosphorylation of eIF2α on serine-51 by Gcn2 converts eIF2-GDP from substrate to inhibitor of eIF2B, impeding the formation of the eIF2-GTP-Met-tRNAiMet ternary complex (TC). While this reduces the rate of bulk protein synthesis and limits amino acid consumption, it specifically induces translation of GCN4 mRNA owing to specialized regulatory sequences (upstream ORFs) present in the mRNA leader that couple reduced TC concentration to increased initiation at the GCN4 AUG start codon [3]. The newly synthesized Gcn4 enters the nucleus—a constitutive process for this activator [6], binds to the UAS elements of its target genes and recruits multiple cofactors to the promoter. The recruited cofactors include the nucleosome remodeling complexes SWI/SNF and RSC; and the SAGA and Mediator complexes, which carry out histone acetylation and/or function as adaptors to recruit general transcription factors and RNA polymerase II (PolII), culminating in increased assembly of transcription initiation complexes and elevated transcription of the coding sequences (CDS) [7]–[10].
In nutrient-replete yeast cells, and under conditions of moderate amino acid limitation, where Gcn2 is activated and translation of GCN4 mRNA induced, Gcn4 is a highly unstable protein owing to its ubiquitylation by ubiquitin ligase SCFCDC4 and attendant degradation by the proteasome [11]–[13]. This process helps to maintain Gcn4 at a low, basal level in nonstarved cells, and allows rapid restoration of the basal level when the translation rate of GCN4 mRNA is repressed by replenishing amino acids in starved cells. Rapid degradation of Gcn4 in sated or moderately starved cells requires its phosphorylation by the CDKs Cdk8/Srb10 and Pho85, with Pho85 making the greater contribution [12], [14]. In severely starved cells, Pho85's contribution to Gcn4 turnover is essentially eliminated, owing to the destabilization and consequent disappearance of its cyclin Pcl5 [15], [16], which accounts in large part for the stabilization of Gcn4 under these conditions. By contrast, Srb10 contributes to Gcn4 turnover under all conditions examined, making the minor contribution in sated or moderately starved cells but the major contribution in severely starved cells (where Pho85 is inactive) [14].
Despite its lesser importance in Gcn4 turnover, Srb10 appears to be responsible for clearing the fraction of Gcn4 that is sumoylated on Lys residues 50 and 58. It appears that sumoylation of Gcn4 on K50/K58 reduces its occupancy at the UAS elements of target genes in the early stages of GAAC induction during moderate starvation for Ile/Val imposed with the inhibitor sulfometuron (SM). However, the higher levels of UAS-bound unsumoylated Gcn4 that result from Arg substitutions of K50/K58 do not evoke increased PolII occupancy or higher transcription rates under these conditions in otherwise WT cells [17]. As sumoylation of transcription factors can inhibit transcriptional activation by impairing their ability to recruit RNA polymerase [18], sumoylation of Gcn4 might impair its activator function to dampen the general control response.
There is evidence that phosphorylation of Gcn4 by Srb10 or Pho85 reduces its activation function and that the phosphorylated species must be ubiquitylated and degraded by the proteasome to maintain WT basal expression of Gcn4 target genes under nonstarvation conditions. Thus, blocking proteasomal degradation of Gcn4 reduces target gene transcription in non-starved cells in a manner suppressed by eliminating CDK phosphorylation sites in Gcn4 or deleting both Srb10 and Pho85 [19]. It is unclear whether the accumulation of phosphorylated Gcn4 also impairs transcriptional activation under inducing conditions of amino acid starvation. The phosphorylation of Gcn4 by both kinases appears to be nucleus-localized, as Gcn4 mutants lacking nuclear localization signals are stabilized [6]. Srb10 is associated with the Mediator coactivator complex [20], which phosphorylates the CTD of the largest subunit of RNA polymerase II, Rpb1 [21]. The fact that Gcn4 recruits Mediator to the promoter [7], [8] is consistent with the possibility that Gcn4 participates in down-regulating its own function and stability by recruiting at least one of its inactivating CDKs [14].
Biosynthesis of threonine in yeast, as in other microorganisms and several plants, is a five-step pathway initiated with L-aspartic acid as the primary substrate [22],[23] (Fig. 1A). The absence of the threonine biosynthetic pathway in humans makes it a valuable target for drug development against fungal pathogens [24]. Transcription of at least 4 genes of the threonine pathway, HOM3, HOM2, THR1 and THR4 is under Gcn4 control (Fig. 1A) [4], [25]. Threonine biosynthesis is also subject to feedback inhibition by threonine, which inhibits the activity of the first enzyme in the pathway, aspartate kinase (Hom3), and also partially inhibits homoserine kinase (Thr1) (Fig. 1A) [26]. Peptide prolyl isomerase FKBP12 participates in the feedback inhibition of Hom3 by physical interaction between these two proteins [27], [28].
Fig. 1. hom6Δ impairs GAAC in cells starved for Ile/Val. 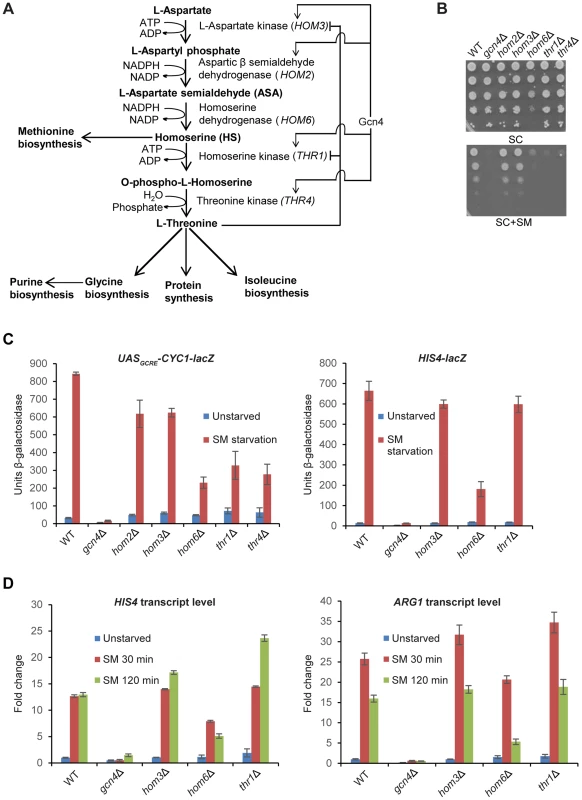
(A) Schematic diagram of the threonine biosynthesis pathway in Saccharomyces cerevisiae. Feedback inhibition by threonine of aspartate kinase (HOM3) and homoserine kinase (THR1) and transcriptional induction of certain pathway genes by Gcn4 under the GAAC are shown. (B) SM-sensitivity of hom6Δ, thr1Δ, and thr4Δ mutants. Parental WT strain BY4741/F729, gcn4Δ mutant F731, and deletion mutants lacking the indicated threonine biosynthetic gene (F2057, F1929, F941, F2056 and F926) were cultured overnight in SC-Ile/Val containing 2.5 mM threonine, washed and resuspended in sterile water at A600 = 1.0, and 10-fold serial dilations were spotted on agar plates of the same growth medium (SC) or medium also containing 0.5 µg/ml sulfomeutron methyl (SC+SM) and incubated at 30°C for 2d. (C) Yeast strains from (B) were transformed with pHYC2 (UASGCRE-CYC1-lacZ) or p367 (HIS4-lacZ) and cultured overnight in SC-Ura/Ile/Val containing 2.5 mM threonine. Duplicate cultures were diluted at A600 = 0.5 in SC-Ura/Ile/Val medium containing 1 mM threonine, and one set was harvested after 6 h at 30°C (Unstarved). For the duplicate set, SM was added to 0.5 µg/mL after 2.5 h and incubation continued another 6 h before harvesting. β-galactosidase activity (nmole of ONPG cleaved per min per mg) was measured in WCEs for three independent transformants of each strain, and mean and S.E.M. values are plotted. (D) WT (BY4741), gcn4Δ (F731), hom3Δ (F1929), hom6Δ (F941) and thr1Δ (F2056) strains were cultured as in (C) except that cultures were harvested at A600 = 0.4–0.6 after doubling at least twice (Unstarved) or treated with SM and cultured an additional 30 min or 120 min. Total RNA was purified and used for cDNA synthesis and using the appropriate fluorescently-labeled Taqman probes HIS4, ARG1, and ACT1 mRNAs were quantified by real-time qPCR. The levels of HIS4 or ARG1 mRNAs were normalized to those of ACT1 mRNA and expressed relative to the value determined for WT unstarved cells. Mean and S.D. values determined from two independent cultures are plotted. Besides threonine auxotrophy, thr1Δ and thr4Δ mutants exhibit myriad phenotypes that result from accumulation of the pathway intermediate homoserine (HS), as they are mitigated in thr1Δ hom3Δ double mutants that cannot produce HS (Fig. 1A) [29]. Accumulation of the substrate of Hom6, β-aspartate semialdehyde (ASA), also is toxic, as releasing feedback inhibition of Hom3 is lethal in hom6Δ cells (that accumulate ASA) in a manner rescued by simultaneously blocking ASA synthesis by eliminating Hom2 or Hom3 [27], [29]. However, hom6Δ mutants do not share all phenotypes of thr1Δ and thr4Δ mutants, and hom6Δ suppresses those unique to the latter mutants, implicating HS and ruling out a role for ASA in conferring many defects displayed by thr1Δ and thr4Δ cells. There is circumstantial evidence that HS toxicity results from its incorporation into proteins in place of threonine, which might evoke increased degradation of the HS-substituted proteins by the proteasome [29].
Previously, we screened the entire library of viable haploid deletion mutants of Saccharomyces cerevisiae for sensitivity to SM (SMS phenotype) to identify genes required for a robust GAAC, which allowed us to implicate various cofactors in the mechanism of transcriptional activation by Gcn4 [7], [30], and certain vacuolar sorting proteins (Vps) in maintaining high-level Gcn4 activation function in cells starved for Ile/Val [13]. In the course of that work, we also discovered that hom6Δ, thr1Δ, and thr4Δ mutants are also SMS, and undertook here to elucidate the mechanisms underlying this phenotype. In fact, it had been shown previously that thr1Δ mutants are SMS, and that this phenotype is suppressed by deleting HOM3. As SM evokes derepression of threonine pathway enzymes by Gcn4 [4] (Fig. 1A), the SMS phenotype of thr1Δ mutants was attributed to Hom3-dependent accumulation of HS, and its attendant toxicity to cellular processes, when HOM3 and HOM2 transcription is induced by Gcn4 [29]. This explanation would not apply to hom6Δ cells, however, which cannot produce HS, leading us to examine whether the SMS phenotype in this instance results from ASA accumulation and impairment of the GAAC response. The results of our analysis indicate that ASA accumulation indeed attenuates GAAC, by accelerating further the already rapid degradation of Gcn4 triggered by the CDKs Pho85 and Srb10. They further suggest that Srb10 functions primarily in efficient clearance of inactive Gcn4 molecules, enriched for sumoylated species, whereas Pho85 clears unsumoylated, highly functional Gcn4 in addition to defective species.
Results
hom6Δ cells are defective in transcriptional activation by Gcn4
As noted above and displayed in Fig. 1B, yeast deletion mutants lacking HOM6, THR1, or THR4 are sensitive to sulfometuron methyl (SM), which evokes starvation for isoleucine and valine (Ile/Val) by inhibition of the ILV2-encoded biosynthetic enzyme [4]. At the SM concentration employed, growth of the hom6Δ, thr1Δ and thr4Δ strains is impaired to an extent similar to that of the gcn4Δ strain, lacking the activator of GAAC. Unlike these mutants, the hom3Δ and hom2Δ mutants grow like the wild-type (WT) strain on SM-containing medium (Fig. 1B, SC+SM). These findings indicate that thr1Δ, thr4Δ, and hom6Δ strains, but not hom3Δ and hom2Δ mutants, are sensitive to Ile/Val starvation imposed by SM. Moreover the hom6Δ mutant grows more slowly than WT (Slg- phenotype) even on medium lacking SM (Fig. 1B, SC).
To determine whether the SMS phenotypes of the thr1Δ, thr4Δ, and hom6Δ mutants reflect defective transcriptional activation by Gcn4, we measured induction of a UASGCRE-CYC1-lacZ reporter, driven by the CYC1 promoter and tandem Gcn4 binding sites from HIS4 (the UASGCRE) replacing the endogenous CYC1 UAS; and of a HIS4-lacZ reporter containing the native HIS4 5′-noncoding region. (HIS4 is a known Gcn4 target gene [5], [31].) As expected, treatment with SM for 6 h evokes a strong increase in UASGCRE-CYC1-lacZ reporter expression in WT, but not in gcn4Δ cells (Fig. 1C). Smaller induction ratios were observed for all five mutants of the threonine pathway, with the largest defect seen for the hom6Δ strain (∼75% reduction of induced UASGCRE-CYC1-lacZ expression) and the smallest defects observed for the hom3Δ and hom2Δ mutants (∼25% reductions) (Fig. 1C, left). In the case of the HIS4-lacZ reporter, the hom6Δ mutant, but not the hom3Δ or thr1Δ strains, displayed a marked (∼75%) reduction in induction by SM (Fig. 1C, right).
To confirm these findings, we measured induction of native mRNAs for HIS4 and ARG1 (another known Gcn4 target gene). Consistent with the HIS4-lacZ data, we observed induction defects for the hom6Δ mutant, but not the hom3Δ or thr1Δ strains, for both mRNAs (Fig. 1D). The magnitude of the induction defect in the hom6Δ mutant was considerably greater after 120 min versus 30 min of SM treatment, displaying ∼60% and ∼67% reductions for HIS4 and ARG1 mRNAs, respectively, at the longer incubation time, even though full induction of both mRNAs was achieved by 30 min of SM treatment in WT cells (Fig. 1D).
The foregoing results indicate that the SM-sensitivity of the hom6Δ mutant reflects a substantial defect in GAAC resulting from reduced transcriptional activation by Gcn4, which becomes more severe as starvation proceeds. By contrast, the other four threonine pathway mutants exhibit smaller defects in transcriptional activation, and the hom3Δ and thr1Δ strains actually display no detectable impairment of HIS4 and ARG1 induction by SM. The strong SMS phenotypes of the thr1Δ and thr4Δ mutants (Fig. 1B) can be reconciled with their moderate GAAC defects (Figs. 1C–D) by recalling that they accumulate the toxic intermediate HS, and that Gcn4-mediated induction of HOM2 and HOM3 under SM-induced starvation conditions is expected to elevate HS production in these strains (Fig. 1A), in the manner proposed previously for thr1Δ cells [29]. By contrast, induction of the threonine pathway during SM treatment in hom3Δ or hom2Δ mutants should have no effect on cell growth (as observed in Fig. 1B) because they cannot produce HS.
The catalytic activity of Hom6 is required for a robust GAAC response
The HOM6 product, homoserine dehydrogenase (HSD), converts β-aspartate semialdehyde (ASA) into HS. If the GAAC defect in hom6Δ cells results from the absence of this reaction, then hom6 mutants that produce catalytically defective HSD should display a strong GAAC defect. Based on a crystal structure of yeast HSD, 4 active site substitutions were generated that were previously characterized for their effects on HSD catalytic activity in vitro [32]. We introduced the corresponding mutations into plasmid-borne HOM6 and examined the ability of the mutant alleles to complement the transcriptional activation defects of hom6Δ cells. As expected, introduction of WT HOM6 complemented the Slg- and SMS phenotypes on media containing threonine, and the failure to grow on medium lacking threonine, of the hom6Δ strain (Fig. 2A, SC, SC+SM and SC-Thr, respectively). Except for the E208D allele, the plasmid-borne hom6 alleles encoding HSD active site substitutions abolished complementation of the threonine auxotrophy and SM-sensitivity of the hom6Δ strain (Fig. 2A). Consistent with this, the three defective alleles failed to restore SM-induction of the UASGCRE-CYC1-lacZ reporter, whereas E208D restored a WT level of induction (Fig. 2B). Interestingly, the previously determined kinetic parameters of the hom6-E208D product indicated a reduced substrate affinity, but high-level catalytic activity, in comparison to WT HSD [32]. Accordingly, our results demonstrate that HSD catalytic activity is required for a robust GAAC response. We presume that the diminished substrate affinity of the hom6-E208D mutant does not significantly reduce the rate of converting ASA to HS in living cells.
Fig. 2. Hom6 catalytic activity is required for robust GAAC in Ile/Val-starved cells. 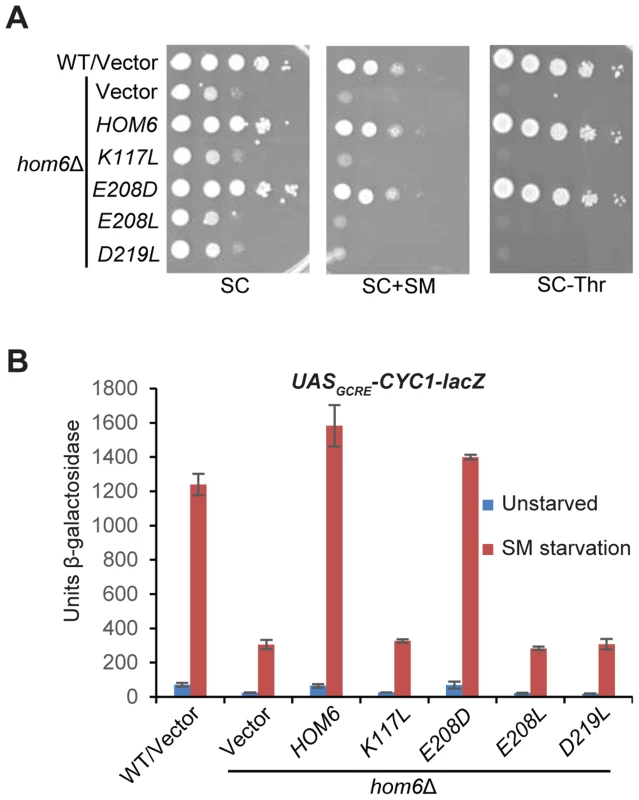
(A) The growth of WT (BY4741) and hom6Δ (F941) strains transformed with vector (YCplac111) and F941 (hom6Δ) transformed with single-copy plasmids carrying WT HOM6 (pYPR010) or active-site hom6 alleles K117L (pYPR018), E208D (pYPR020), E208L (pYPR022), or D219L (pYPR024) was analyzed essentially as in Fig. 1B except that growth on SC-Leu/Thr medium (SC-Thr) was also examined. (B) Transformants of the strains in (A) harboring pHYC2 were analyzed for β-galactosidase activity as in Fig. 1C. Means and S.E.Ms were calculated from three independent transformants of each strain. Accumulation of β-aspartate semialdehyde in hom6Δ cells impairs GAAC
We asked next whether the requirement for HSD activity for the GAAC response reflects a requirement for HS synthesis or, rather, the need to prevent accumulation of ASA. If the inability to produce HS is the salient defect, then supplementing hom6Δ cells with HS should restore their GAAC response. We found that a supplement of 1 mM HS restores growth on SC-Thr medium for the hom3Δ, hom2Δ and hom6Δ strains, but not for the thr1Δ or thr4Δ strains (Fig. 3A), consistent with the position of Thr1 and Thr4 downstream of HS production in the Thr pathway (Fig. 1A). A supplement of 5 mM HS was required to confer growth of the hom3Δ, hom2Δ and hom6Δ strains indistinguishable from that of WT; although this elevated HS concentration retards the growth of WT cells (Fig. 3A), presumably reflecting HS toxicity [29]. Importantly, HS supplementation did not rescue the defective SM-induction of the UASGCRE-CYC1-lacZ reporter in the hom6Δ mutant (Fig. 3B), indicating that its GAAC defect does not result from the inability to produce HS.
Fig. 3. Accumulation of aspartate semialdehyde in hom6Δ cells impairs GAAC. 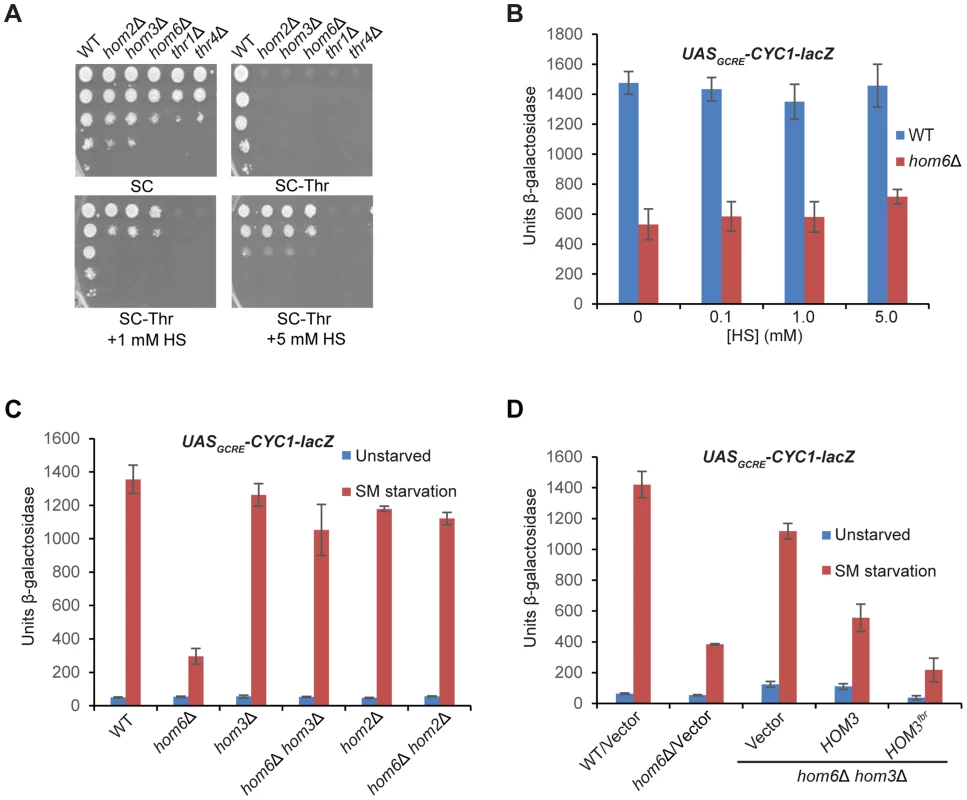
(A) Yeast strains described in Fig. 1B were analyzed for growth in spotting assays using SC medium containing Ile/Val and 1 mM Thr (SC), the same medium lacking Thr (SC-Thr) either with or without addition of homoserine (HS) at the indicated concentrations. (B–D) Transformants of the indicated relevant genotypes harboring pHYC2 were analyzed for UASGCRE-CYC1-lacZ expression as in Fig. 1C except for the addition of HS and the analysis of only SM-treated cultures in (B). pHYC2 transformants of the following strains were examined: (B) WT (BY4741) and hom6Δ (F941); (C) WT (BY4741), hom6Δ (YR001), hom3Δ (F1929), hom2Δ (F2057), hom6Δ hom2Δ (YR022), and hom6Δ hom3Δ (YR003); (D) WT (BY4741) and hom6Δ (YR001) strains transformed with vector pRS313, and hom6Δ hom3Δ strain YR003 transformed with pRS313, low copy (lc) HOM3 plasmid pYPR028, or lc HOM3fbr plasmid pYPR030. Means and S.E.Ms were calculated from three independent transformants of each strain. If accumulation of the Hom6/HSD substrate (ASA) in hom6Δ cells is responsible for the GAAC defect, then the GAAC response should be restored by preventing ASA production by eliminating Hom2 or Hom3; moreover, the GAAC defect should be exacerbated by eliminating feedback inhibition of the Hom3 product (Fig. 1A). Indeed, deleting HOM2 or HOM3 in the hom6Δ mutant restored SM-induction of the UASGCRE-CYC1-lacZ reporter essentially to the same levels observed in the hom2Δ or hom3Δ single mutants (Fig. 3C). Introducing WT HOM3 into the hom3Δ hom6Δ strain reinstated a defect in SM-induction of UASGCRE-CYC1-lacZ similar to that seen in the hom6Δ single mutant (Fig. 3D). Importantly, a relatively greater induction defect was observed when the feedback-resistant allele hom3-E282D (dubbed HOM3fbr) was introduced instead into the hom3Δ hom6Δ strain (Fig. 3D, cf. last two columns). As expected, introduction of HOM3fbr into the hom3Δ hom6Δ strain confers a strong Slg- phenotype (Fig. S1), owing to accumulation of ASA and its toxic effects on cell growth [27]. These findings demonstrate that the GAAC defect in hom6Δ cells results from ASA accumulation.
ASA accumulation in hom6Δ cells reduces Gcn4 abundance
We sought next to determine whether ASA accumulation impairs the GAAC by reducing Gcn4 abundance. Starvation for Ile/Val by SM rapidly increases Gcn4 synthesis by inducing the translation of GCN4 mRNA [3]. Western analysis of WT cells reveals the expected rapid induction of Gcn4 after only 30 min of SM treatment, with a gradual decline in abundance as starvation continues up to 120 min [9] (Fig. 4A and B). Gcn4 abundance was decidedly reduced over much of the time course of SM treatment in vector transformants of the hom6Δ strain, again reaching its lowest level at 120 min of SM treatment. This reduction in Gcn4 abundance was mitigated by the absence of HOM3 in the hom6Δ hom3Δ double mutant, and exacerbated in transformants of the double mutant harboring feedback-resistant HOM3fbr, in which ASA accumulation is eliminated or exacerbated, respectively (Fig. 4A and B). Even after only 30 min of SM treatment, the hom6Δ HOM3fbr strain displayed low-level Gcn4 similar to that observed in hom6Δ/vector transformants after prolonged SM treatment for 120 min. The gradual decrease in Gcn4 abundance in hom6Δ cells (Fig. 4B) is consistent with the greater reduction in Gcn4 target gene transcription seen at 120 min versus 30 min of SM treatment (Fig. 1D). Moreover low-level HIS4 mRNA was observed in the hom6Δ HOM3fbr strain even after only 30 min of SM treatment (Fig. S2).
Fig. 4. Accumulation of ASA lowers Gcn4 abundance and stability. 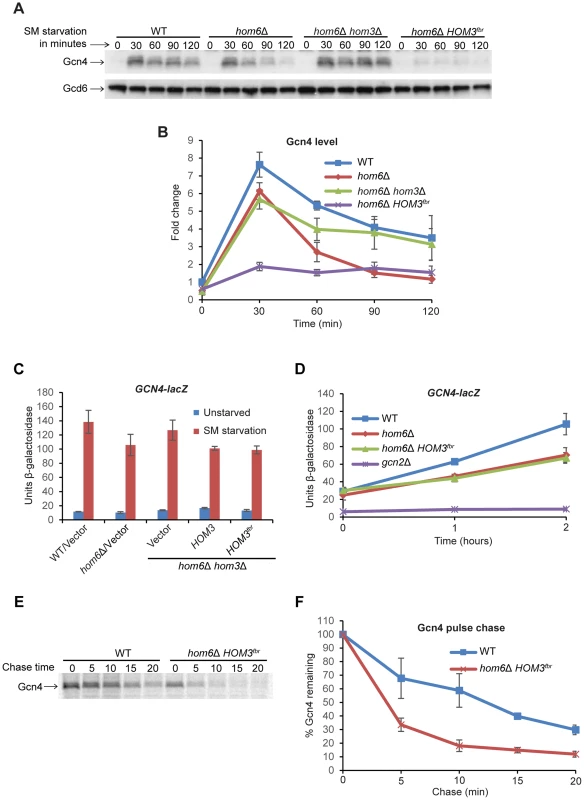
(A) WT (BY4741) and hom6Δ (YR001) strains transformed with vector (pRS313) and hom6Δ hom3Δ strain YR003 transformed with vector or lc HOM3fbr plasmid pYPR030 (indicated as hom6Δ hom3Δ and hom6Δ HOM3fbr respectively) were cultured in SC-His/Ile/Val for at least two doublings to A600 = 0.4–0.6 and subjected to SM treatment (0.5 µg/ml) for the indicated times. WCEs were extracted under denaturing conditions and subjected to Western analysis with α-Gcn4 and α-Gcd6 antibodies. (B) Western signals from (A) were quantified, Gcn4 signals were normalized to Gcd6 signals, and the Gcn4/Gcd6 ratios were expressed relative to that measured for the WT strain without SM treatment. Means and S.E.Ms were calculated from three independent transformants of each strain. (C) Yeast strains described in Fig. 3D transformed with p180 were analyzed for GCN4-lacZ expression as in Fig. 1C. Means and S.E.Ms were calculated from three independent transformants of each strain. (D) p180 transformants of strains from (C) and gcn2Δ strain H2931 were analyzed for GCN4-lacZ expression after SM treatment for the indicated times. Means and S.E.Ms were calculated from three independent transformants of each strain. (E) The WT and hom6Δ HOM3fbr strains from (A) were cultured in SC-His/Ile/Val, collected and resuspended in SC-His/Ile/Val/Met containing SM (1 µg/ml) for 15 min, and labeled with [35S] methionine/cysteine for 15 min. Cells were collected and resuspended in SC-His/Ile/Val containing Met and Cys (both at 10 mM) and aliquots were removed after the indicated times of chase. Aliquots of WCEs containing equal cpm were immunoprecipitated with α-Gcn4 antibodies, immunocomplexes were collected with protein A-agarose beads and resolved by SDS-PAGE, and the [35S]-Gcn4 signals were quantified by phosphorimaging. (F) Gcn4 signals from (E) are plotted relative to the value at 0 min of chase (100%). Means and S.D.s were calculated from two independent experiments. To determine whether the reduced Gcn4 abundance on ASA accumulation reflects decreased translation of GCN4 mRNA, we assayed a GCN4-lacZ fusion shown to be a faithful reporter of GCN4 transcription and the translational efficiency of GCN4 mRNA [33], [34]. Expression of this reporter shows the expected ∼10-fold induction in WT cells after 6 h of SM treatment, which is dampened somewhat both in the hom6Δ strain and in HOM3 transformants of the hom6Δ hom3Δ double mutant, but not in the vector transformants of the same strain (Fig. 4C). However, the HOM3fbr and HOM3 transformants of the double mutant exhibit indistinguishable levels of GCN4-lacZ expression. Similar results were obtained after only 1 h or 2 h of SM treatment (Fig. 4D), with the hom6Δ strain and HOM3fbr transformants of the hom6Δ hom3Δ double mutant both exhibiting similar reductions in GCN4-lacZ expression of ∼33% compared to the WT strain. As expected, a gcn2Δ mutant, lacking the key activator of GCN4 mRNA translation [3], is completely defective for GCN4-lacZ expression (Fig. 4D). While these findings suggest a reduction in Gcn4 synthesis on ASA accumulation in cells lacking HOM6, the ∼33% reductions in GCN4-lacZ expression observed in the hom6Δ and hom6Δ HOM3fbr strains do not account for the 60–70% reductions in Gcn4 abundance observed after 2 h of SM treatment in the same strains. These findings suggest that Gcn4 is also degraded more rapidly than usual in response to ASA accumulation.
To provide direct evidence supporting this last conclusion, we measured the turnover of newly synthesized Gcn4 by a pulse-chase experiment. Cells were cultured with SM for 30 min and pulse-labeled with [35S]-methionine/cysteine for the last 15 min of the starvation period, and then chased with excess nonradioactive methionine/cysteine. Consistent with previous reports, Gcn4 is normally a highly unstable protein and decays with a half-life of ∼10–12 min in SM-treated WT cells (Fig. 4E–F) [13]. Importantly, Gcn4 decay was markedly accelerated in the hom6Δ HOM3fbr strain, with the Gcn4 half-life dropping below 5 min, thus confirming that Gcn4 is degraded more rapidly in response to ASA accumulation (Fig. 4E–F).
Rapid degradation on ASA accumulation does not require UAS binding by Gcn4
As noted above, rapid degradation of Gcn4 is dependent on its phosphorylation by Pho85 and Srb10 in the nucleus, leading to its ubiquitylation and degradation by the proteasome [3]. It was also shown that the DNA-binding activity of Gcn4 is required for its sumoylation [17]. While it has been assumed that phosphorylation of Gcn4 by Srb10 likewise requires its binding to the UASGCRE [35], this has not been directly demonstrated. We hypothesized that the increased rate of Gcn4 turnover on ASA accumulation results from its increased phosphorylation by Pho85 or Srb10 and attendant degradation by the proteasome; and wished to determine whether, like sumoylation, the increased phosphorylation occurs when Gcn4 is bound to the UASGCRE. To this end, we asked whether inactivating the DNA-binding ability of Gcn4 would suppress the effect of ASA accumulation on its abundance by conducting Western analysis of Gcn4 variants described previously [36] lacking the C-terminal basic region or leucine zipper, which are both required for DNA binding by Gcn4 [37]. The variant lacking the DNA binding domain, gcn4-Δ235-250, also lacks one of two nuclear localization sequences (NLS2) identified in Gcn4, whereas the variant lacking the leucine zipper, gcn4-Δ251-281, retains both NLSs, and it was shown that the leucine zipper is dispensable for nuclear localization of GFP-tagged Gcn4 [6]. We verified that both gcn4 alleles are indistinguishable from deletion of the entire GCN4 coding sequence in the inability to permit growth on SM-containing medium (Fig. S3). Western analysis of SM-treated cells revealed that both variants differ dramatically from WT Gcn4 and display no detectable reduction in abundance in hom6Δ cells treated for 2 h with SM (Fig. 5A). (Note that both truncated variants are well expressed and show the expected increased electrophoretic mobility compared to WT Gcn4 (Fig. 5A)). Furthermore, introducing HOM3fbr into hom6Δ cells, which severely diminishes WT Gcn4 after only 30 min of SM treatment (Fig. 4A), has no effect on abundance of the gcn4-Δ235-250 and gcn4-Δ251-281 mutant proteins (Fig. S4A). While these results suggested that ASA evokes accelerated degradation only when Gcn4 is capable of UASGCRE-binding, it was possible that the greater stability of the mutant variants results from a failure to accumulate ASA on SM treatment owing to the absence of Gcn4-mediated derepression of threonine biosynthetic enzymes required for ASA production.
Fig. 5. The DNA binding domain is dispensable, but CDK phosphorylation site Thr-165 is required, for depletion of Gcn4 in hom6Δ cells treated with SM. 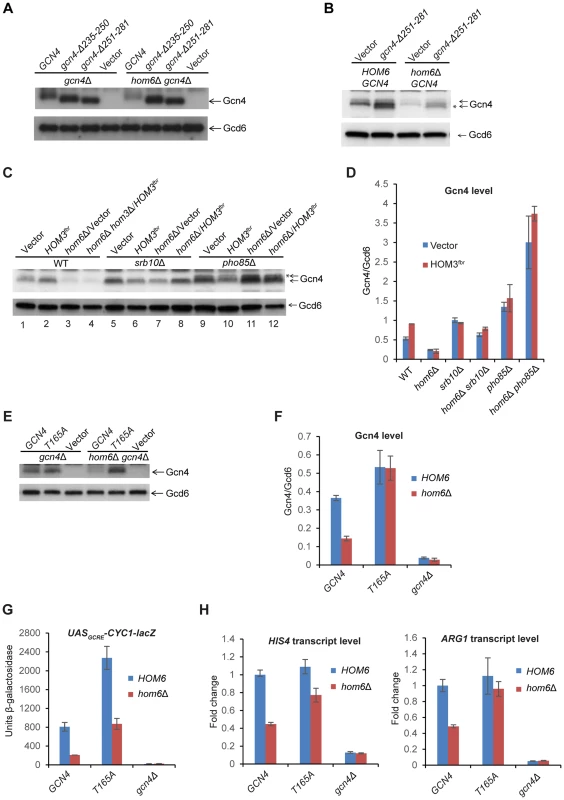
(A) gcn4Δ (F731) and hom6Δ gcn4Δ (YR009) strains transformed with sc plasmids with WT GCN4 (p164), mutant alleles gcn4-Δ235-250 (pCD114-1) or gcn4-Δ251-281 (pCD115-1), or empty vector (YCplac33), were subjected to Western analysis as in Fig. 4A, after SM treatment for 2 h. (B) WT (BY4741) and hom6Δ (YR001) strains transformed with vector YCplac33 or sc mutant allele gcn4-Δ251-281 (pCD115-1) were analyzed as in (A). * indicates mutant gcn4-Δ251-281 protein, displaying slightly greater electrophoretic mobility than WT Gcn4. (C) WT (BY4741), hom6Δ (YR001), hom6Δ hom3Δ (YR003), srb10Δ (F736), hom6Δ srb10Δ (YR004), pho85Δ (F947) and hom6Δ pho85Δ (YR006) strains transformed with vector (pRS313) or HOM3fbr plasmid pYPR030; were analyzed as in (A). * indicates putative phosphorylated isoforms of Gcn4. In Fig. S5, we verified that the level of Gcn4 measured by Western analysis is indistinguishable between the hom6Δ hom3Δ HOM3fbr strain examined here in lane 4 and a hom6Δ HOM3fbr strain that represents the most appropriate WT control for the mutants analyzed here in lanes 8 and 12, as would be expected from the dominance of HOM3fbr. (D) Western signals from (C) were quantified and the mean and S.E.M. Gcn4/Gcd6 ratios were calculated from 3 independent transformants. (E) gcn4Δ (F731) and hom6Δ gcn4Δ (YR009) strains transformed with sc plasmids harboring WT GCN4 (pYPR013), gcn4-T165A (pYPR047), or vector (YCplac111) were analyzed as in (A). (F) Western signals from (E) were analyzed as in (D). (G) Transformants of the strains in (E) harboring pHYC2 were analyzed for UASGCRE-CYC1-lacZ expression as in Fig. 1C. Means and S.E.Ms were calculated from three independent transformants of each strain. (H) Strains in (E) were analyzed for HIS4 and ARG1 mRNA levels as in Fig. 1D, after SM treatment for 2 h. Mean and S.D. values determined from two independent cultures were plotted relative to the value determined for WT cells. To address this last possibility, we repeated the experiment with the gcn4-Δ251-281 strains containing or lacking HOM6 after introducing WT GCN4 to reinstate the GAAC. Now we observed that the abundance of the truncated gcn4-Δ251-281 product was strongly reduced in hom6Δ cells, mirroring the behavior of full-length WT Gcn4 present in the same cells (Fig. 5B). The same results were observed in the corresponding strains also containing HOM3fbr (Fig. S4B). These results indicate that UAS-binding by Gcn4 is not required for its rapid degradation on ASA accumulation. Considering that Pho85 is responsible, whereas UAS-binding is dispensable, for the bulk of Gcn4 turnover under normal growth conditions [6], [12], [14], these findings are consistent with the possibility that Pho85 plays a prominent role in the accelerated degradation of Gcn4 evoked by excess ASA.
Srb10 and Pho85 are required for the reduced abundance of Gcn4 evoked by ASA accumulation in hom6Δ cells
We sought next to determine the contributions of Srb10 and Pho85 to the enhanced degradation of Gcn4 in response to excess ASA. Consistent with previous findings [12], [14], deletion of either SRB10 or PHO85 increases the abundance of Gcn4 in otherwise WT cells treated with SM, with pho85Δ evoking a somewhat greater increase than srb10Δ (Fig. 5C, lanes 1,5,9; Fig. 5D, vector transformants). As already shown above, Gcn4 abundance is severely diminished after 120 min of SM treatment in hom6Δ or hom6Δ HOM3fbr cells compared to the isogenic HOM6 cells (Fig. 5C, lanes 3–4 vs. 1–2). Importantly, eliminating SRB10 almost completely eliminates this reduction in Gcn4 abundance in both hom6Δ and hom6Δ HOM3fbr cells treated with SM (Fig. 5C, lanes 7–8 vs. 3–4; & Fig. 5D). The slower migrating Gcn4 species evident in WT cells is considerably reduced in srb10Δ cells, suggesting that it represents a phosphorylated form of Gcn4 that depends on Srb10, which is generally consistent with previous results [14].
Deletion of PHO85 also suppresses the reduction in Gcn4 abundance evoked by SM treatment of hom6Δ or hom6Δ HOM3fbr cells (Fig. 5C, cf. lanes 3–4 vs. 11–12). In fact in pho85Δ strains, Gcn4 abundance is higher in hom6Δ and hom6Δ HOM3fbr cells (where ASA accumulates) compared to the isogenic HOM6 cells (Fig. 5C, cf. lanes 11–12 vs. 9–10; and Fig. 5D). Interestingly, deleting PHO85 seems to increase the relative abundance of the slower migrating Gcn4 species, which presumably represent products of Srb10 phosphorylation that are not efficiently cleared in pho85Δ cells (Fig. 5C, cf. lanes 9–12 vs. 1–4). The results in Figs. 5C–D suggest that both Srb10 and Pho85 are required for the strong depletion of Gcn4 that occurs on ASA accumulation. The stronger effect of pho85Δ versus srb10Δ on Gcn4 abundance observed on ASA accumulation in these experiments is consistent with previous results indicating a relatively greater contribution of Pho85 to Gcn4 degradation under normal growth conditions [12], [14].
Gcn4 was found to be phosphorylated in vitro by Srb10 on multiple CDK consensus sites, including Ser17, Ser210, Thr61, Thr105 and possibly Thr165 [14], and by Pho85 both in vivo and in vitro on Thr165 [12]. Moreover, the T165A substitution alone was sufficient to confer marked stabilization of Gcn4 in vivo [12]. Importantly, we found that the Gcn4-T165A variant showed no reduction in abundance in SM-induced hom6Δ cells compared to HOM6 cells (Fig. 5E–F). Moreover, replacing WT GCN4 with the GCN4-T165A allele in hom6Δ cells restored UASGCRE-CYC1-lacZ reporter (Fig. 5G) and ARG1 mRNA expression (Fig. 5H) to levels essentially equivalent to those seen in HOM6 GCN4 cells. Expression of HIS4 mRNA also was boosted by GCN4-T165A in hom6Δ cells, although expression remained below that seen in HOM6 GCN4 cells (Fig. 5H), suggesting either that the Gcn4-T165A variant is not functionally equivalent to WT Gcn4 or that a fraction of Gcn4-T165A rescued in hom6Δ cells has a lower than WT specific activity. In any event, these findings provide strong evidence that phosphorylation of T165 by Pho85 and/or Srb10 is required for the pronounced depletion of Gcn4 evoked by ASA accumulation in hom6Δ cells.
Evidence that Srb10 and Pho85 eliminate functionally distinct pools of Gcn4 on ASA accumulation
Having found that removing either Srb10 or Pho85 restores high-level Gcn4 abundance during ASA accumulation in hom6Δ cells, we expected to find that transcriptional activation by Gcn4 would likewise be restored in both hom6Δ srb10Δ and hom6Δ pho85Δ strains, particularly since these CDKs have been implicated in reducing Gcn4 activation function via phosphorylation of Gcn4 [19]. However, we observed distinct differences in the activation function of Gcn4 in cells lacking Srb10 versus Pho85.
First, we found that eliminating PHO85 restores the ability of hom6Δ cells to grow on SM containing plates (Fig 6A). By contrast, hom6Δ srb10Δ cells cannot grow on SM medium, even though HOM6 srb10Δ cells grow at the WT rate on SM medium (Fig. 6A). These findings suggest that deletion of SRB10 does not rescue the defective GAAC response to SM treatment in hom6Δ cells, whereas deletion of PHO85 does. Consistent with the growth assays, we found that eliminating PHO85 fully restores transcriptional activation of HIS4 and ARG1 in hom6Δ cells, conferring even higher than WT levels of both transcripts in the hom6Δ pho85Δ double mutants (Fig. 6B). It is noteworthy that deleting HOM6 provokes no reduction in HIS4 or ARG1 mRNAs, and even seems to elevate HIS4 mRNA, in pho85Δ cells (Fig. 6B). By contrast, deleting SRB10 evokes little or no increase in HIS4 or ARG1 mRNA levels in SM-treated hom6Δ cells (Fig. 6B). The failure of srb10Δ to rescue activation of these genes in hom6Δ cells cannot be attributed simply to the loss of a coactivator function of Srb10 [10], as srb10Δ had little or no effect on levels of HIS4 or ARG1 mRNAs in otherwise WT HOM6 cells (Fig. 6B, srb10Δ vs. WT), nor on the ability to grow in SM medium (Fig. 6A, srb10Δ vs. WT). These findings suggest that the Gcn4 molecules rescued from accelerated degradation on ASA accumulation by elimination of Srb10 are relatively nonfunctional in transcriptional activation. By contrast, the Gcn4 molecules rescued from degradation by elimination of Pho85 from hom6Δ cells appear to include highly functional species capable of evoking a greater than WT level of transcriptional activation.
Fig. 6. Deleting SRB10 or PHO85 stabilizes functionally distinct populations of Gcn4 in hom6Δ cells treated with SM. 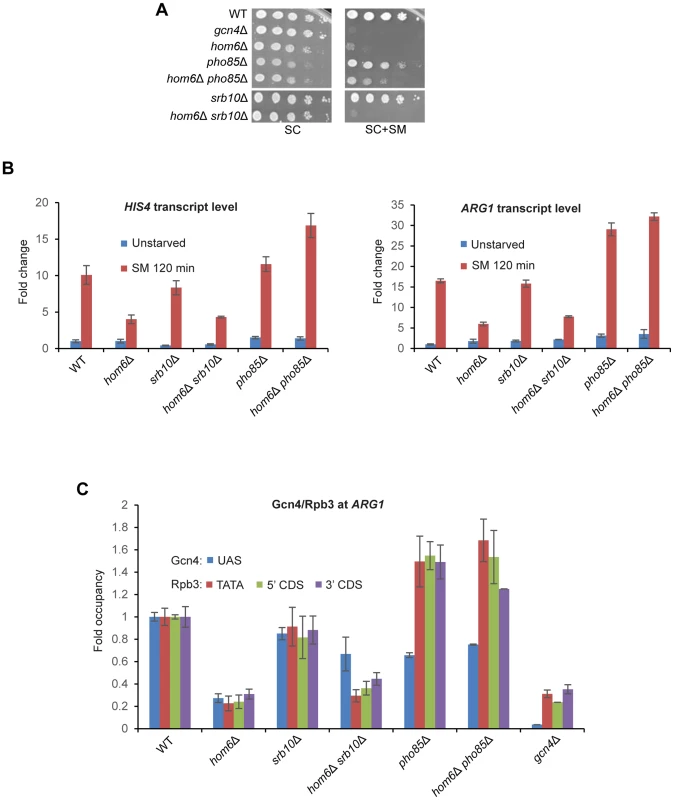
(A) WT (BY4741), gcn4Δ (F731), hom6Δ (YR001), pho85Δ (F947), hom6Δ pho85Δ (YR006), srb10Δ (F736) and hom6Δ srb10Δ (YR004) strains were analyzed as in Fig. 1B, except that the plates were incubated for 4d for SC+SM versus 2d for SC. Results for SC or SC+SM conditions derive from images of the same plate. (B) Yeast strains from (A) were analyzed for HIS4 and ARG1 mRNA levels in unstarved cells and after 120 min of SM starvation, as in Fig. 1D. Mean and S.D. values determined from two independent cultures are plotted. (C) Chromatin immunoprecipitation analysis of strains from (A) starved with SM for 120 min and crosslinked with formaldehyde. Sheared chromatin was immunoprecipitated with antibodies against Gcn4 or PolII subunit Rpb3, and DNA extracted from the immunoprecipitates and equivalent input samples was analyzed by quantitative PCR to determine levels of the ARG1 UAS for Gcn4; ARG1 promoter (TATA) and coding sequences (5′CDS/3′CDS) for Rpb3; and non-coding sequences on chromosome V (ChrV) as a negative control. Mean and S.D. occupancies of Gcn4/Rpb3 were calculated as (targetIP/ChrVIP)/(targetInput/ChrVInput) from duplicate PCR amplifications of DNA samples from immunoprecipitations of chromatin prepared from two independent cultures of each strain and expressed relative to those measured for the WT strain. Having found above that deleting PHO85 restores a higher level of Gcn4 in hom6Δ cells than does deleting SRB10 (Fig. 5D), it was important to determine whether the higher level of transcriptional activation seen in hom6Δ pho85Δ versus hom6Δ srb10Δ cells (Figs. 6A–B) arises simply from relatively greater UAS occupancy by Gcn4 in hom6Δ pho85Δ cells. To address this possibility, we conducted ChIP analysis to measure the occupancy of Gcn4 at the ARG1 UAS and the occupancies of Rpb3 (a PolII subunit) at the promoter (TATA element) and the 5′ or 3′ ends of the CDS at ARG1 after 2 h of SM treatment. It was shown previously that SM treatment of WT cells evokes large increases in occupancies of Gcn4 and Rpb3 at ARG1 that are completely absent in gcn4Δ cells [9], [10]. Importantly, these increases in occupancy are strongly diminished in SM-treated hom6Δ cells (Fig. 6C), providing direct evidence that the GAAC defect in the hom6Δ mutant results from low-level Gcn4 occupancy of the UAS with attendant reduced recruitment of PolII to the promoter.
As expected from the ability of srb10Δ to restore cellular Gcn4 abundance in hom6Δ cells (Fig. 5C–D), Gcn4 occupancy of the ARG1 UAS is substantially higher in hom6Δ srb10Δ versus hom6Δ SRB10 cells (Fig. 6C, blue bars). However, this increase in Gcn4 occupancy is associated with much smaller increases in Rpb3 occupancies at all three locations at ARG1 (Fig. 6C, orange, green, purple bars), consistent with the idea that the Gcn4 recovered in hom6Δ srb10Δ cells is relatively inactive.
Deletion of PHO85 had strikingly different consequences on Gcn4 activity. In HOM6 cells, the pho85Δ mutation evokes a reduction in UAS occupancy of Gcn4, but actually increases Rpb3 occupancies compared to the WT strain (Fig. 6C, pho85Δ vs. WT), which is consistent with the higher than WT levels of ARG1 mRNA in pho85Δ cells shown above (Fig. 6B). This effect of pho85Δ was noted previously [17], and is not understood mechanistically; however, it might indicate that Pho85 clears fully functional Gcn4 molecules as a homoeostatic mechanism to prevent hyperinduction of the GAAC response, such that UAS-bound Gcn4 has a greater than WT specific activity in pho85Δ cells.
Despite the much higher total cellular abundance of Gcn4 observed in hom6Δ pho85Δ versus hom6Δ srb10Δ cells (Fig. 5D), Gcn4 occupancy of the ARG1 UAS is comparable in these two strains (Fig. 6C, blue bars). In contrast, the Rpb3 occupancies at all three locations at ARG1 are substantially higher in the hom6Δ pho85Δ versus hom6Δ srb10Δ cells (Fig. 6C, orange, green, purple bars). In fact, the Rpb3 occupancies observed in hom6Δ pho85Δ cells exceed those in WT cells despite a lower than WT level of Gcn4 UAS occupancy in the mutant cells (Fig. 6C). These findings suggest that the Gcn4 molecules rescued in hom6Δ pho85Δ cells that are capable of UAS binding have a greater than WT specific activity.
As elaborated in the Discussion, the reduced ability of UAS-bound Gcn4 to activate transcription in the hom6Δ srb10Δ double mutant could be explained by proposing that Gcn4 is rendered less functional in response to ASA accumulation and that Srb10 is required to clear the inactive Gcn4 molecules from the promoter by targeting them for degradation. The apparent hyperactivity of UAS-bound Gcn4 in hom6Δ pho85Δ cells could be explained by proposing that Pho85 targets both fully functional Gcn4 and defective species rendered incapable of UAS-binding on ASA accumulation in hom6Δ cells.
Evidence that ASA accumulation increases sumoylation of Gcn4
It was shown recently that Gcn4 is sumoylated at target gene promoters, and Srb10 was implicated in clearing these sumoylated Gcn4 molecules [17]. We considered the possibility that sumoylation of Gcn4 bound to promoters increases on ASA accumulation and enhances the clearance of inactive Gcn4 by Srb10. If so, we would expect to find elevated sumoylation of Gcn4 in srb10Δ hom6Δ strains, but not in pho85Δ hom6Δ strains. To examine this possibility, we immunoprecipitated Gcn4 from whole cell extracts (WCEs) and probed the immune complexes with antibodies against Smt3 (yeast SUMO). After normalizing the Smt3 signal for Gcn4 abundance in the immune complexes, we observed that the Gcn4 present in hom6Δ srb10Δ cells after 2 h of SM treatment has an ∼2-fold higher level of sumoylation than observed in WT or hom6Δ cells under the same conditions, whereas sumoylation of Gcn4 is ∼2-fold lower in hom6Δ pho85Δ compared to WT or hom6Δ cells (Fig. 7A–B).
Fig. 7. Gcn4 is hypersumoylated in hom6Δ srb10Δ cells treated with SM. 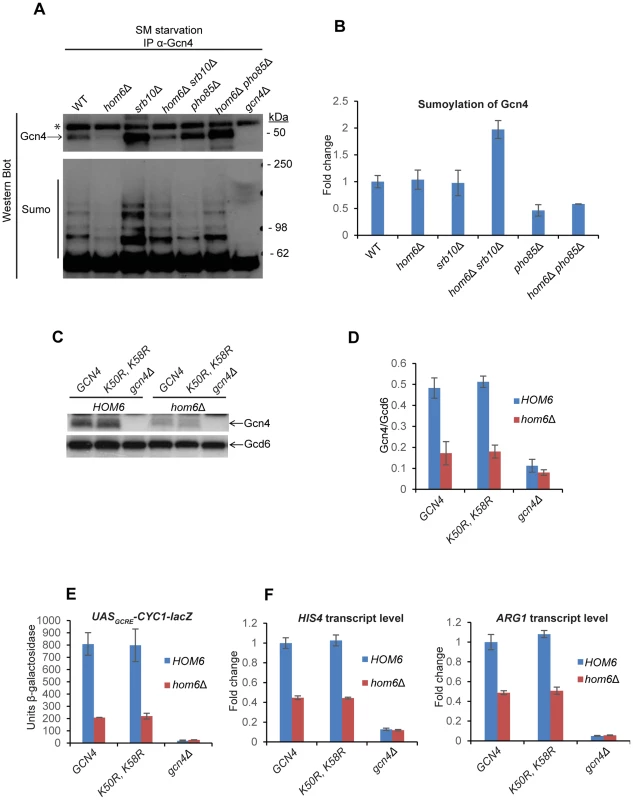
(A) Yeast strains from Fig. 6A were treated with 0.5 µg/ml SM for 120 min and aliquots of WCE containing 1 mg total protein were immunoprecipitated with α-Gcn4 antibodies. Immunocomplexes were subjected to Western analysis with α-Gcn4 and α-Smt3 (sumo) antibodies. Relevant molecular weight markers (SeeBlue Pre-Stained Standard) are indicated. * indicates a nonspecific band observed even in the gcn4Δ strain. Gcn4 Western signals do not necessarily reflect the relative Gcn4 levels in the starting WCEs presumably owing to variability in the efficiency of immunoprecipitating Gcn4. (B) Western signals for sumoylated Gcn4 from the lower panel of (A) were normalized to the Gcn4 signals from the upper panel of (A) and the Mean and S.D. ratios calculated from two independent cultures were expressed relative to that measured for WT cells. (C) gcn4Δ (F731) and hom6Δ gcn4Δ (YR009) strains transformed with sc plasmids harboring WT GCN4 (pYPR013), gcn4-K50R, K58R (pYPR038), or vector (YCplac111) were analyzed as in Fig. 5A. (D) Western signals from (C) were analyzed as in Fig. 5D. (E) Transformants of the strains in (C) harboring pHYC2 were analyzed for UASGCRE-CYC1-lacZ expression as in Fig. 1C. Means and S.E.Ms were calculated from three independent transformants of each strain. (F) Strains in (C) analyzed for HIS4 and ARG1 mRNA levels as in Fig. 5H. Mean and S.D. values determined from two independent cultures are plotted. The finding that sumoylated Gcn4 is elevated specifically in the srb10Δ hom6Δ strain supports the idea that sumoylation of Gcn4 increases during SM-starvation of hom6Δ cells and that Srb10 targets the sumoylated Gcn4 for clearance from target promoters. As a result of Srb10 function, the proportion of Gcn4 that is sumoylated should not increase in response to ASA accumulation in hom6Δ SRB10 cells, as we observed (Fig. 7B). By the same token, the fact that elimination of PHO85 from hom6Δ cells does not significantly alter the sumoylation of Gcn4 following SM treatment implies that Pho85 plays little role in clearing sumoylated Gcn4 and is therefore restricted primarily to clearing unsumoylated Gcn4, which would include Gcn4 molecules not bound to the UASGCRE.
It was shown that sumoylation of Gcn4 at Lys50 and Lys58 contributes to clearing Gcn4 from the UAS via Srb10 phosphorylation in the early stages of SM induction; and this process is eliminated by arginine substitutions at both Lys residues [17]. To determine whether sumoylation of Gcn4 stimulates the clearing of Gcn4 from promoters on ASA accumulation, we examined the effects of the K50R and K58R substitutions on Gcn4 abundance in SM-treated hom6Δ cells. We found that the Gcn4-K50R, K58R mutant displayed a reduction in abundance on SM-treatment of hom6Δ cells very similar to that observed for WT Gcn4 (Fig. 7C–D). The K50R, K58R substitution also had no effect on Gcn4 abundance in SM-treated pho85Δ and pho85Δ hom6Δ cells (Fig. S6), where turnover of Gcn4 is dependent on Srb10, thus suggesting that Srb10-dependent degradation of Gcn4 on ASA accumulation is not enhanced by sumoylation of Lys50/Lys58. We also found that, in otherwise WT cells, the Gcn4-K50R, K58R variant confers essentially WT SM-induction of the UASGCRE-CYC1-lacZ reporter and HIS4 and ARG1 mRNAs, in accordance with previous findings [17]; and that the Gcn4-K50R, K58R variant resembles WT Gcn4 in being unable to sustain efficient SM-activation of UASGCRE-CYC1-lacZ, HIS4 and ARG1 expression in hom6Δ cells (Fig. 7E–F). Thus, although our data suggest that sumoylation of promoter-bound Gcn4 increases on ASA accumulation, and that the sumoylated Gcn4 molecules are cleared from the promoter primarily by Srb10, as concluded previously [17], the sumoylation of Lys50/Lys58 is not critically required for the enhanced degradation of Gcn4 that occurs under conditions of ASA excess.
Discussion
In this report we have shown that accumulation of ASA in hom6 mutants lacking functional homoserine dehydrogenase (HSD) impairs the GAAC response to starvation for Ile/Val by accelerating degradation of the activator Gcn4. It is remarkable that ASA accumulation increases the rate of Gcn4 turnover considering that Gcn4 is already exceedingly short-lived under normal growth conditions [11], [12], [14]. The effect of ASA accumulation in reducing Gcn4 abundance and occupancy of the ARG1 UAS was mitigated in mutants lacking either of the CDKs, Srb10 and Pho85, known to phosphorylate Gcn4 and target it for ubiquitylation and rapid degradation by the proteasome. Deletion of SRB10 restored an essentially WT level of cellular Gcn4 in Ile/Val-starved hom6Δ cells, whereas deletion of PH085 conferred an even greater than WT level of cellular Gcn4 in starved hom6Δ cells. Similarly, mutating a key phosphorylation site of Pho85 and possibly Srb10, Thr-165, also rescued WT Gcn4 abundance in Ile/Val-starved hom6Δ cells. These findings are consistent with the model that ASA accumulation evokes an increased rate of phosphorylation-dependent degradation of Gcn4 by the proteasome. While both Srb10 and Pho85 are required for the accelerated Gcn4 turnover, it appears that Pho85 plays the larger role—just as observed under normal growth conditions [12]. This last conclusion is consistent with our finding that UASGCRE-binding by Gcn4 is dispensable for its rapid turnover on ASA accumulation, which is also the case under normal growth conditions [6].
Interestingly, the outcome on the GAAC response differed significantly depending on which of the two CDKs was eliminated in hom6Δ cells. On removal of Srb10 from hom6Δ cells, the recovery of UAS-bound Gcn4 was accompanied by only a small increase in transcriptional activation of ARG1, such that srb10Δ hom6Δ cells cannot grow on SM medium. By contrast, hom6Δ cells lacking Pho85 can grow on SM medium, and we observed an even greater than WT activation of ARG1 transcription conferred by essentially the same level of UAS-bound Gcn4 seen in hom6Δ srb10Δ cells, which is actually less than the UAS occupancy of Gcn4 found in fully WT cells. It could be argued that the low-level activation of ARG1 transcription seen in the srb10Δ hom6Δ double mutant reflects the requirement for Srb10 for efficient activation by Gcn4 observed previously [7], [10]. However, here we observed no effect of deleting SRB10 on cell growth, and little or no effect on the induction of ARG1 and HIS4 mRNAs or PolII occupancy of ARG1 CDS in otherwise WT SM-treated cells; and the small defects we observed seem inadequate to explain the nearly complete absence of increased ARG1 and HIS4 transcription and PolII occupancy at ARG1 occurring in SM-treated hom6Δ srb10Δ cells. Hence, we favor the alternative explanation that the specific activity of the UAS-bound Gcn4 rescued by eliminating Srb10 in hom6Δ cells is lower than that rescued by eliminating Pho85 in hom6Δ cells. This in turn suggests that these CDKs target different populations of Gcn4.
The notion that Srb10 and Pho85 recognize different populations of Gcn4 also fits with our demonstration that Gcn4 is more highly sumoylated in Ile/Val-starved srb10Δ hom6Δ cells than in starved WT or hom6Δ cells, whereas Gcn4 is hypo-sumoylated in Ile/Val-starved pho85Δ hom6Δ cells. This finding is consistent with the previous conclusion that Srb10 is required to clear sumoylated Gcn4 from promoters [17]. Hence, we suggest that the putative population of defective Gcn4 molecules that are phosphorylated by Srb10 and subsequently degraded also tend to be hyper-sumoylated. However, we found that sumoylation of the known sites of this modification in Gcn4, Lys50/Lys58, was unimportant for the accelerated degradation of Gcn4 in Ile/Val-starved hom6Δ cells. Thus, while sumoylation appears to be a characteristic of Gcn4 molecules that are phosphorylated by Srb10 and subsequently cleared from the promoter, we have no evidence that sumoylation enhances the unusually rapid degradation of these Gcn4 molecules that occurs during ASA accumulation.
To explain in greater detail our proposal that Srb10 and Pho85 target distinct populations of Gcn4, we begin by positing that phosphorylation of the Gcn4 activation domain (AD) by Srb10 and Pho85 occurs most rapidly when the AD is not engaged with coactivators at the promoter. Hence, both functional and non-functional Gcn4 molecules not bound to the UAS would be susceptible to rapid turnover, whereas UAS-bound Gcn4 would turn over more slowly unless it harbors a damaged or modified AD that cannot engage with coactivators. Pho85 is located in the nucleus [6]; however, we have observed only low-level recruitment of Pho85 to the ARG1 UASGCRE by ChIP analysis, at a level decidedly smaller than that seen for Srb10 (Fig. S7) or other Mediator subunits [38]. Moreover, Pho85 is responsible for the majority of Gcn4 degradation under both nonstarvation conditions and moderate-starvation conditions where the Pho85/Pcl5 complex is abundant [12], which includes our SM-induction conditions. This can explain the previous finding that Gcn4 DNA binding activity is dispensable for rapid Gcn4 turnover under such conditions [6]. Thus, we envision that Pho85 primarily targets Gcn4 molecules when they are not bound to a UASGCRE. In contrast, Srb10 is recruited by Gcn4 to the ARG1 promoter and, hence, likely plays a prominent role in the degradation of UAS-bound Gcn4 molecules that become disengaged from coactivators either stochastically or because of damage or modification of the AD (Fig. 8A). Again, this proposal is consistent with the previous finding that Srb10 is required to clear sumoylated Gcn4, as sumoylation is impaired by mutations that impair UAS-binding by Gcn4 [17].
Fig. 8. Model for the roles of Pho85 and Srb10 in accelerated turnover of Gcn4 in response to ASA accumulation in hom6Δ cells. 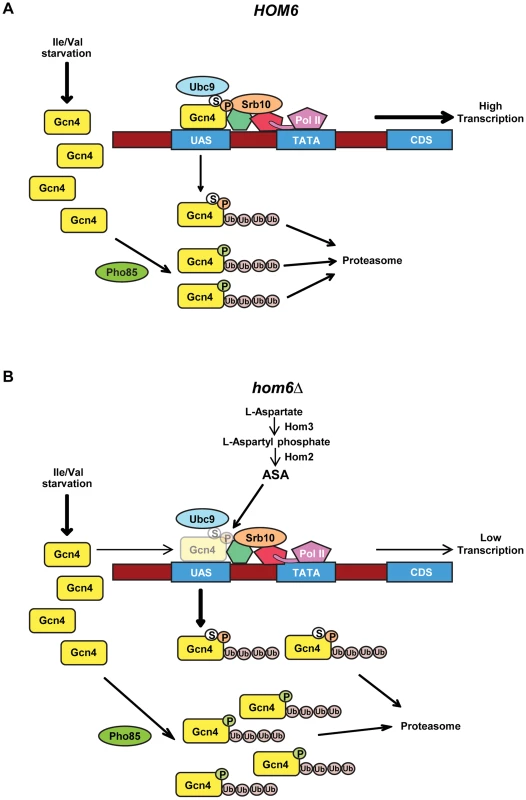
(A) Starvation of WT (HOM6) cells for Ile/Val evokes increased synthesis of Gcn4 at the translational level and subsequent increased binding of Gcn4 to UAS elements of genes subject to GAAC. Gcn4 recruits coactivators, including Mediator (green and red shapes connecting Gcn4 to Pol II), and Pol II to the promoter (TATA) for increased transcription of target gene coding sequences (CDS). As Srb10 is associated with Mediator and recruited by UAS-bound Gcn4, we propose that Srb10 phosphorylates the bulk of UAS-bound Gcn4 molecules (orange balls labeled with “P”), stimulating their ubiquitylation and attendant degradation by the proteasome. UAS-bound Gcn4 is also sumoylated by Ubc9 (white balls labeled with “S”) and the sumoylated molecules are targeted for degradation by Srb10. Pho85 is responsible for the majority of Gcn4 turnover, and we propose it phosphorylates (green balls labeled with “P”) and triggers degradation of both active and defective non-UAS bound Gcn4 species. (B) Starvation of hom6Δ cells for Ile/Val also evokes increased synthesis of Gcn4 and attendant increased binding of Gcn4 to UAS elements. However, we hypothesize that ASA accumulation evokes damage or inactivating modifications of Gcn4. Damage/modification restricted to the activation domain, disengages UAS-bound Gcn4 from coactivators and increases its rate of phosphorylation by Srb10 at the promoter, with attendant increased degradation by the proteasome. (Gcn4 opacity is reduced to depict its decreased occupancy of the UAS.) Damage/modification that extends to the DNA binding or dimerization domain disengages Gcn4 from the UAS and makes it susceptible to phosphorylation by Pho85 and subsequent proteasomal degradation. Pho85 also targets functional Gcn4 molecules when they disengage from the UAS, just as in HOM6 cells. Hence, the absence of Srb10 in srb10Δ cells spares from degradation defective Gcn4 molecules capable of UAS-binding and thereby reduces the specific activity of UAS-bound Gcn4. The absence of Pho85 in pho85Δ cells spares both fully functional Gcn4 molecules and inactive species incapable of stable UAS-binding and thereby increases the specific activity of UAS-bound Gcn4 while simultaneously decreasing the fraction of Gcn4 capable of UAS binding. There is evidence that Gcn4 is deactivated under normal growth conditions by Srb10 and Pho85, and that the phosphorylated, inactive protein must be degraded by the proteasome to prevent a reduction in the specific activity of UAS-bound Gcn4 [19]. We suggest that ASA accumulation in hom6Δ cells provokes damage or modification of Gcn4 that increases its rate of phosphorylation and subsequent turnover by the proteasome. The inability of the putative damaged or modified Gcn4 molecules to bind to the UAS or engage with coactivators could be responsible for their enhanced phosphorylation. Because eliminating the DNA binding activity of Gcn4 did not abolish its rapid turnover in hom6Δ cells harboring an intact GAAC, and DNA binding is not required for the Pho85-dominated turnover of Gcn4 under normal conditions [6], [12], we propose that Pho85 plays a predominant role in targeting the putative defective Gcn4 molecules generated under ASA excess, presumably when they are dissociated from the promoter; whereas Srb10 would make a lesser contribution and mediate the rapid degradation of defective, UAS-bound Gcn4 species (Fig. 8B). Accordingly, eliminating Srb10 will spare from degradation defective Gcn4 molecules that are capable of UAS binding, and because Pho85 will continue to target functional molecules when they become disengaged from the UAS, the specific activity of UAS-bound Gcn4 should decline in srb10Δ cells, as we observed. By contrast, eliminating Pho85 will rescue both damaged molecules incapable of UAS binding as well as functional Gcn4 molecules that are phosphorylated by Pho85 when they disengage from the UAS; and because Srb10 will continue to clear activation-defective Gcn4 species capable of UAS binding, the specific activity of UAS-bound Gcn4 should increase in pho85Δ cells, as we observed (Fig. 8B). Even in HOM6 pho85Δ cells, where no ASA accumulation occurs, the specific activity of UAS-bound Gcn4 exceeds that in WT cells, as seen both here and previously [17], and this phenomenon is also explained by our model (Fig. 8A). The proposal that Pho85 is responsible for clearing defective molecules incapable of binding to the UAS can also explain why a sizeable fraction of the Gcn4 spared from degradation in pho85Δ cells appears to be incapable of UAS binding, as indicated by the lower than WT UAS occupancy despite higher than WT cellular abundance of Gcn4 seen in pho85Δ and pho85Δ hom6Δ cells (Fig. 6C vs. Fig. 5C–D). As noted above, it is also possible that the lower than WT UAS-occupancy of Gcn4 in pho85Δ cells reflects an unknown feedback regulatory mechanism that limits Gcn4 binding to the UAS as a way to prevent hyperactivation of Gcn4 target genes beyond the elevated levels seen in pho85Δ cells.
An intriguing observation not anticipated by the model in Fig. 8 is that ASA accumulation provoked by SM-treatment of hom6Δ pho85Δ cells leads to a higher level of Gcn4 than occurs in HOM6 pho85Δ cells where ASA does not accumulate (Fig. 5C–D). We recently obtained evidence that most of this effect can be accounted for by an unexpected increase in GCN4 transcription or translation, as expression of the GCN4-lacZ reporter was found to be ∼2-fold higher in SM-treated hom6Δ pho85Δ versus HOM6 pho85Δ cells (Fig. S8).
A final interesting question is whether attenuation of the GAAC evoked by ASA accumulation is adaptive in WT yeast in the wild. Perhaps the enzyme HSD is frequently targeted for inhibition by plants, animals, or other microorganisms as a means inhibiting yeast growth. Indeed, the threonine pathway does not exist in mammals and has been identified as a valuable target for developing new antifungal therapeutics [24]. Moreover, it was shown that a strain of Streptomyces produces a natural antibiotic that targets HSD [39]. Reducing threonine biosynthesis by inhibiting HSD should activate eIF2α phosphorylation by Gcn2 and thereby reduce general protein synthesis, which is an appropriate response to limitation for threonine as a means of reducing the rate of threonine consumption. However, the concurrent transcriptional induction of Gcn4 target genes, including threonine biosynthetic pathway genes, evoked by translational upregulation of GCN4 mRNA might not be adaptive in this instance, owing to the toxic effects of ASA on cell physiology, including cytokinesis [27]. This toxicity of ASA provides a plausible rationale for the ability of this intermediate to suppress GAAC by accelerating Gcn4 turnover in the manner discovered here.
Materials and Methods
Media and growth conditions
Yeast strains were grown at 30°C in rich YPD medium (1% yeast extract, 2% peptone and 2% glucose) or defined synthetic complete (SC) medium (1.45g yeast nitrogen base, 5g ammonium sulfate, 2% glucose and 2g amino acid mix per liter) lacking leucine, uracil or histidine wherever appropriate for selection of plasmids; and lacking isoleucine and valine (Ile/Val) for treatment with sulfometuron (SM) at 0.5 µg/ml. Increasing threonine in SC medium from approximately 1 mM to 2.5 mM diminished the slow growth phenotype of hom6Δ cells, and eliminated that of thr1Δ and thr4Δ cells, in SC medium lacking Ile/Val. Therefore, overnight growth to saturation was achieved in SC medium supplemented with 2.5 mM threonine and thereafter yeast strains were cultured in SC with 1 mM threonine. Accordingly, a moderate threonine limitation was imposed in our experiments and, as threonine is a precursor in Ile/Val biosynthesis (Fig. 1A), this should intensify the limitation for Ile/Val provoked by SM treatment.
Yeast strain construction and verification
Yeast strains used in this study are listed in Table 1. Yeast strains purchased from Research Genetics or previously reported were verified for all auxotrophic requirements indicated in the genotype; and gene deletions were confirmed by PCR amplification of predicted deletion junctions using primers described in Table S1. To generate HOM6 deletion strains, the appropriate hphMX4 gene deletion cassette conferring hygromycin B resistance [40] was PCR-amplified from plasmid pAG32 using primers HOM6-MX4-F and HOM6-MX4-R, thus introducing homologous flanking sequences upstream and downstream of HOM6 coding sequences, and used to delete HOM6 by transforming the appropriate strains to hygromycin B resistance on YPD agar plates. HOM6 deletion was further confirmed by demonstrating acquisition of threonine auxotrophy, except when deleted in hom2Δ or hom3Δ strains F2057 and F1929, respectively; and by PCR-amplification of predicted junction fragments containing hphMX4 and sequences upstream or downstream of HOM6 coding sequences using primer pairs HOM6-A/HphMX-R1 and HOM6-DN-R/HphMX-F1 respectively. To generate SRB10-myc13 and PHO85-myc13 strains, a myc13::HIS3MX6 cassette was PCR-amplified from plasmid pFA6a-13myc-HIS3MX6 using primer pairs SRB10-MYC13-F/SRB10-MYC13-R or PHO85-MYC13-F/PHO85-MYC13-R, respectively, and used to transform strains BY4741 and YR001to His+. Cassette insertions were confirmed by PCR analysis of genomic DNA using the appropriate primers specific for the myc13::HIS3MX6 cassette and SRB10 or PHO85, and by Western analysis of whole cell extracts (WCEs) using anti-Myc antibodies (Roche). To generate GCN4 deletion strains, plasmid pHQ1240 containing a gcn4Δ::hisG::URA3::hisG cassette was digested with SspI and used to transform strains F947 and YR006 to Ura+. Deletion of GCN4 was indicated by acquisition of SM-sensitivity and verified by PCR amplification of gcn4Δ::hisG::URA3::hisG from chromosomal DNA using primer pairs specific for sequences upstream and downstream of the GCN4 CDS. The URA3 gene was subsequently evicted by selecting for growth on medium containing 5-fluoroorotic acid.
Tab. 1. Yeast strains used in this study. 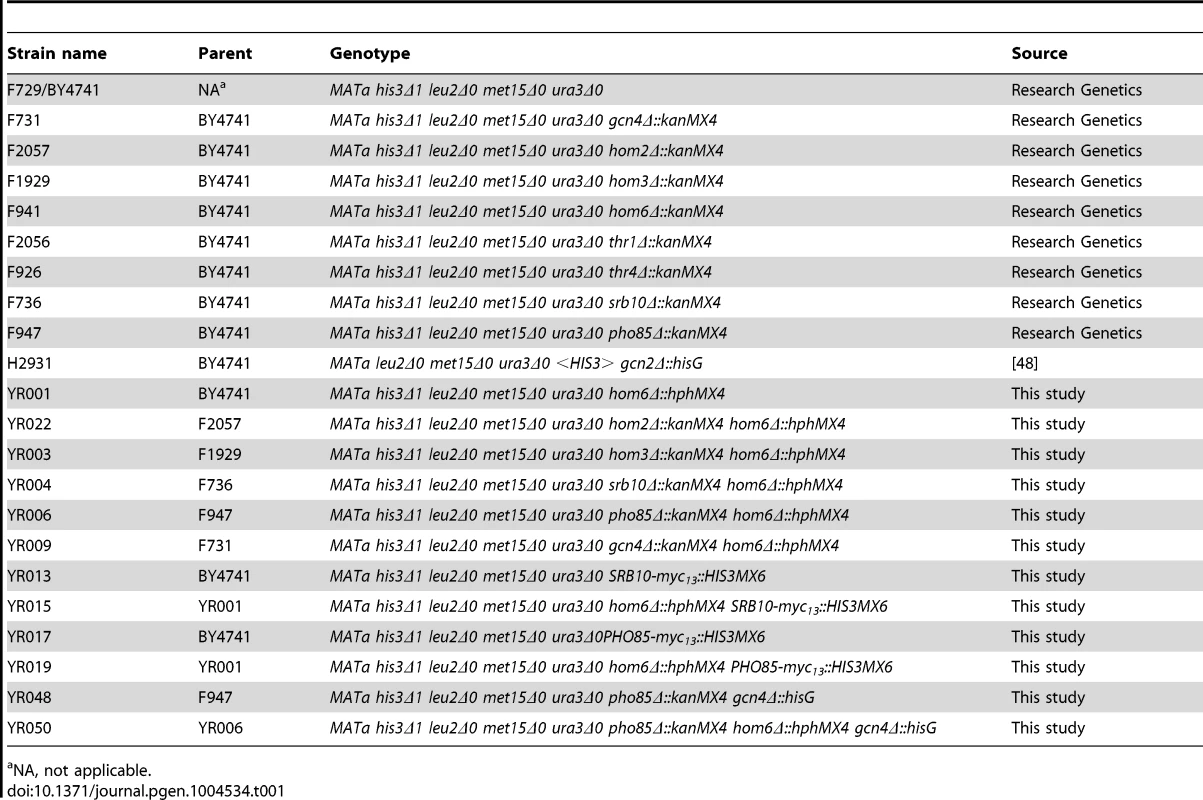
NA, not applicable. Plasmid constructions
All plasmids used in this study are listed in Table 2, and primers used in plasmid constructions are listed in Table S1. To construct pYPR010, HOM6 (chrX:689,322.690,749) was PCR-amplified from chromosomal yeast DNA of strain BY4741, using primers HOM6-HindIII-F and HOM6-BamHI-R, and inserted between the HindIII and BamHI sites in YCplac111. To construct pYPR018, pYPR020, pYPR022 and PYPR024, HOM6 in pYPR010 was mutagenized using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) to produce hom6 mutant alleles encoding the K117A, E208D, E208L and D219L substitutions, respectively, using sets of complementary primer pairs harboring the corresponding mutations (Table S1).
Tab. 2. Plasmids used in this study. 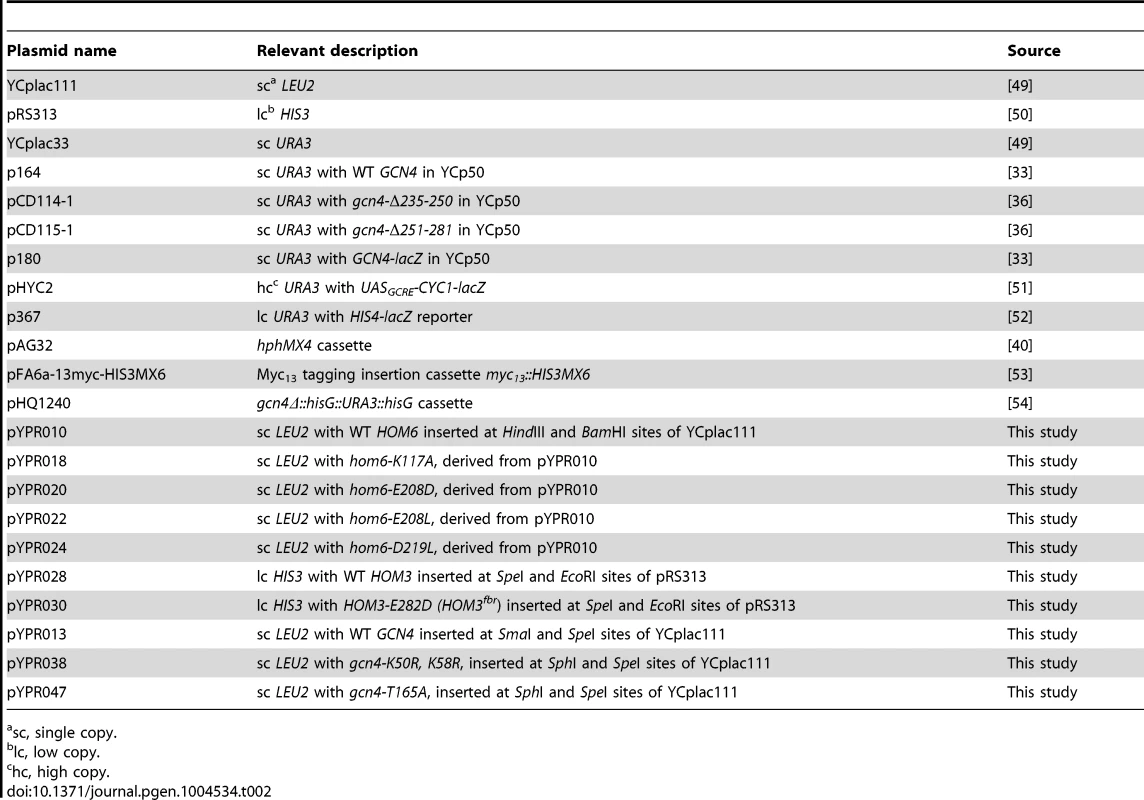
sc, single copy. To construct pYPR028, HOM3 (chrV:256,132.258,737) was PCR-amplified from chromosomal yeast DNA of BY4741 using primers HOM3-F1 and HOM3-R1 and inserted between the SpeI and EcoRI sites in pRS313. pYPR030, containing HOM3-E282D (HOM3fbr), was generated by fusion-PCR using HOM3-F1 and HOM3-R1 as outside primers and complementary primers HOM3-E282D-F and HOM3-E282D-R encoding the appropriate mutation, and inserted between the SpeI and EcoRI sites of pRS313. To construct pYPR013, the ApaI-SpeI fragment containing GCN4 was isolated from plasmid p164, polishing the ApaI end using Klenow polymerase exonuclease activity, and inserted between the SmaI and SpeI sites of YCplac111. pYPR038 and pYPR047 were constructed by fusion-PCR using Gcn4c-SphI-F and GCN4c-SpeI-R as outside primers in combination with primers GCN4-K50,58R-F and GCN4-K50,58R-R or primers GCN4-T165A-F and GCN4-T165A-R, respectively, and pYPR013 as PCR template. The PCR products were inserted between the SphI and SpeI sites in YCplac111.
Assaying lacZ reporters
Yeast strains transformed with plasmids pHYC2 (UASGCRE-CYC1-lacZ), p367 (HIS4-lacZ), or p180 (GCN4-lacZ) were grown to saturation and diluted in two identical cultures in SC-Ura/Ile/Val at A600 = 0.5, and after 2.5 h of growth, 0.5 µg/ml SM was added to one set of cultures. Cells were harvested from untreated (unstarved) cultures after a total 6 h of growth and SM-treated cultures grown for 6 h in the presence of SM [7]. Whole cell extracts (WCEs) were prepared and assayed for β-galactosidase activity as previously described [41]. Mean specific activities were calculated from results obtained from three independent transformants.
Quantification of mRNA abundance by real-time qRT-PCR
Yeast strains were cultured to an A600 of 0.4–0.6 in SC-Ile/Val, achieving at least two cell doublings, and treated with 0.5 µg/ml SM for the indicated times or left untreated. Total RNA was isolated by hot phenol extraction as previously described [42]. RNA concentration was quantified by Nanodrop spectroscopy and analyzed for integrity by agarose gel electrophoresis and ethidium bromide staining. An aliquot of 1 µg total RNA was used for cDNA synthesis using SuperScript III First-strand Synthesis Supermix for qRT-PCR (Invitrogen) and the resulting cDNA was diluted 10-fold. qRT-PCR was performed using Brilliant III Ultra-Fast qPCR Master Mix (Agilient Technologies) using the diluted cDNA in multiplex PCR and the appropriate TaqMan probes (Table S1) to quantify ACT1 (labelled with FAM), ARG1, or HIS4 (both labelled with HEX). qRT-PCR reactions were performed in triplicate using cDNA synthesized from RNA extracted from at least two independent cultures. ARG1 or HIS4 cDNA abundance was normalized to that of ACT1 by calculating 2(−ΔCt), where ΔCt is (Ct (Target) - Ct (ACT1)). Fold changes in mRNA abundance were normalized to those measured in uninduced WT cells, or as indicated, and plotted.
Chromatin immunoprecipitations (ChIP)
ChIP assays were conducted essentially as described previously [7], [43]. Yeast strains were cultured in 100 ml SC-Ile/Val as described above for RNA isolation, treated with 0.5 µg/ml SM for 2 h or as indicated, cross-linked for 15 min with 10 ml formaldehyde solution (50 mM HEPES KOH, pH 7.5, 1 mM EDTA, 100 mM NaCl and 11% formaldehyde) and quenched with 15 ml of 2.5 M glycine. WCEs were prepared by glass beads lysis in 400 µl FA lysis buffer (50 mM HEPES KOH, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% TritonX-100 and 0.1% Na-deoxycholate) with protease inhibitors for 45 min at 4°C and the supernatant collected after removing the beads was pooled with 600 µl FA lysis buffer used for washing the beads. The resulting lysate was sonicated to yield DNA fragments of 300–500 bp and cleared by centrifugation. 50 µl aliquots of lysates were immunoprecipitated for 2 h at 4°C with α-Gcn4, (Rabbit) [13] or α-Rpb3 antibodies (Mouse, Neoclone) coupled with α-rabbit IgG or α-mouse IgG conjugated magnetic beads (Dynabeads, Invitrogen), respectively, or with α-c-Myc (Rabbit, Roche) coupled with α-rabbit IgG conjugated magnetic beads. Recovered immune complexes were washed and eluted as described [43]. For matched input and IP samples, the crosslinks were reversed by incubation at 65°C overnight, treated with proteinase K, extracted twice with phenol:chloroform:isoamyl alcohol (25∶24∶1) and once with chloroform:isoamyl alcohol (24∶1), and ethanol precipitated, resuspending the resulting pellets in 30–40 µl TE containing RNAase as described earlier [43]. Quantitative PCRs were performed in the presence of [33P]-dATP with undiluted IP DNA and 500-fold diluted input DNA and further analyzed as previously described [7], [43]. The primers employed for ChIP analysis are listed in Table S1.
Western blot analysis
WCEs were prepared in denaturing conditions with trichloroacetic acid, as described previously [44] and analyzed by immunoblotting with α-Gcd6 [45] and affinity purified α-Gcn4 antibodies [13]. Western signals were quantified by ImageJ software.
Pulse-chase analysis of Gcn4 degradation
The analysis was performed essentially as previously described [13], [46]. Yeast cells collected from a 10 ml culture at A600 = 0.4–0.6 were washed with SC-Met/Ile/Val, inoculated into 0.5 ml SC-Met/Ile/Val containing 1 µg/ml SM and incubated for 15 min in a shaking water bath at 30°C; after which 1.0 mCi [35S]methionine/cysteine labelling mix was added and incubation continued for an additional 15 min. Cells were collected, transferred to 5 ml of pre-warmed SC-Ile/Val containing 10 mM methionine and 10 mM cysteine, and an 1 ml aliquots were removed immediately or after appropriate times of chase. Aliquots were denatured with 170 µl of 1.85 M NaOH, 7.4% 2-marcaptoethanol and precipitated with 70 µl 100% TCA on ice, washed with chilled acetone and dried under vacuum in a SpeedVac. The dried pellets were resuspended in 120 µl of 2.5% SDS, 5 mM EDTA, 1 mM PMSF by vortexing, boiled for 1 min, and cleared by centrifugation. Incorporation of label was measured by scintillation counting [13] and aliquots of extract containing equal amounts of radioactivity (5.7×105 cpm) were combined with 1 ml of immunoprecipitation (IP) buffer (50 mM Na-HEPES [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1 mM PMSF) containing 1 mg/ml BSA and 1 µl affinity-purified α-Gcn4 antibodies and mixed by rotating at 4°C for 2 h. Twenty µl of a 50% slurry of protein A-agarose beads pretreated with IP buffer containing BSA (1 mg/ml) was added, and mixing continued for 2 h. The beads were washed thrice with 500 µl cold IP buffer containing 0.1% SDS, resuspended in loading buffer, boiled, and resolved by SDS-PAGE using 4 to 20% gels. The gel was dried and subjected to autoradiography, and the [35S]-labeled Gcn4 was quantified by phosphorimaging analysis.
Analysis of Gcn4 sumoylation
A modification of a previously described protocol was employed [47], as follows. Yeast strains were cultured and treated with SM as described above for ChIP analysis. 40–60 A600 units of cells were lysed at 4°C with glass beads by 10 cycles of vortexing, 30s-on and 30s-off, in 500 µL of chilled lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.2% Triton ×100 and 1 mM PMSF) containing 10 mM sodium ethyl maleimide (NEM) and protease inhibitors. The resulting lysate was cleared by centrifugation at 13,000 rpm for 30 min at 4°C and soluble protein concentration was determined by the Bio-Rad protein assay. For each sample, a 40 µl suspension of magnetic beads conjugated with α-Rabbit IgG (Dynabeads, Invitrogen) was washed twice with lysis buffer containing 5 mg/mL BSA and rotated with 1 µl affinity purified α-Gcn4 antibody [13] in 200 µl lysis buffer/BSA for 3 h at 4°C. The magnetic beads coupled with α-Gcn4 antibody were washed twice with lysis buffer/BSA to remove unbound antibody and resuspended in 200 µl lysis buffer/BSA. Aliquots containing 1 mg of protein were added to the magnetic beads suspension, adjusting the final volume to 500 µl with lysis buffer/BSA, and further rotated for 2 h at 4°C. IP samples were washed thrice with lysis buffer containing 0.1% SDS and resuspended in 30 µl 1× Novex tris-glycine SDS sample buffer (Invitrogen) and boiled for 3 min. Aliquots of 5 µl and 25 µl were subjected to Western analysis with α-Gcn4 antibodies [13] and α-SUMO (α-Smt3) polyclonal antibodies [47].
Supporting Information
Zdroje
1. Johnston M, Carlson M (1992) Regulation of carbon and phosphate utilization. In: Jones EW, Pringle JR, Broach JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Cold Spring Harbor: Cold Spring Harbor Press. pp. 193–282.
2. KaffmanA, O'SheaEK (1999) Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol 15 : 291–339.
3. HinnebuschAG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59 : 407–450.
4. JiaMH, LarossaRA, LeeJM, RafalskiA, DeroseE, et al. (2000) Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol Genomics 3 : 83–92.
5. NatarajanK, MeyerMR, JacksonBM, SladeD, RobertsC, et al. (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21 : 4347–4368.
6. PriesR, BomekeK, IrnigerS, GrundmannO, BrausGH (2002) Amino acid-dependent Gcn4p stability regulation occurs exclusively in the yeast nucleus. Eukaryot Cell 1 : 663–672.
7. SwansonMJ, QiuH, SumibcayL, KruegerA, KimS-J, et al. (2003) A Multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. MolCellBiol 23 : 2800–2820.
8. QiuH, HuC, ZhangF, HwangGJ, SwansonMJ, et al. (2005) Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol 25 : 3461–3474.
9. GovindCK, YoonS, QiuH, GovindS, HinnebuschAG (2005) Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol Cell Biol 25 : 5626–5638.
10. QiuH, HuC, YoonS, NatarajanK, SwansonMJ, et al. (2004) An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol Cell Biol 24 : 4104–4117.
11. KornitzerD, RaboyB, KulkaRG, FinkGR (1994) Regulated degradation of the transcription factor Gcn4. EMBO J 13 : 6021–6030.
12. MeimounA, HoltzmanT, WeissmanZ, McBrideHJ, StillmanDJ, et al. (2000) Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCFCDC4 ubiquitin-ligase complex. Mol Biol Cell 11 : 915–927.
13. ZhangF, GaurNA, HasekJ, KimSJ, QiuH, et al. (2008) Disrupting vesicular trafficking at the endosome attenuates transcriptional activation by Gcn4. Mol Cell Biol 28 : 6796–6818.
14. ChiY, HuddlestonMJ, ZhangX, YoungRA, AnnanRS, et al. (2001) Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev 15 : 1078–1092.
15. ShemerR, MeimounA, HoltzmanT, KornitzerD (2002) Regulation of the transcription factor Gcn4 by Pho85 cyclin PCL5. Mol Cell Biol 22 : 5395–5404.
16. AviramS, SimonE, GildorT, GlaserF, KornitzerD (2008) Autophosphorylation-induced degradation of the Pho85 cyclin Pcl5 is essential for response to amino acid limitation. Mol Cell Biol 28 : 6858–6869.
17. RosoninaE, DuncanSM, ManleyJL (2012) Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes Dev 26 : 350–355.
18. GillG (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15 : 536–541.
19. LipfordJR, SmithGT, ChiY, DeshaiesRJ (2005) A putative stimulatory role for activator turnover in gene expression. Nature 438 : 113–116.
20. GuglielmiB, van BerkumNL, KlapholzB, BijmaT, BoubeM, et al. (2004) A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res 32 : 5379–5391.
21. LiaoS-M, ZhangJ, JefferyDA, KoleskeAJ, ThompsonCM, et al. (1995) A kinase-cyclin pair in the RNA in the polymerase II holoenzyme. Nature 374 : 193–196.
22. GaliliG (1995) Regulation of Lysine and Threonine Synthesis. Plant Cell 7 : 899–906.
23. Jones EW, Fink GR (1982) Regulation of amino acid and nucleotide biosynthesis in yeast. In: "The molecular biology of the yeast Saccharomyces Metabolism and gene expression" Strathern J N Jones E W and Broach J R eds. Cold Spring Harbor Laboratory Cold Spring Harbor NY: 181–300.
24. KingsburyJM, McCuskerJH (2010) Fungal homoserine kinase (thr1Delta) mutants are attenuated in virulence and die rapidly upon threonine starvation and serum incubation. Eukaryot Cell 9 : 729–737.
25. MountainHA, BystromAS, Tang LarsenJT, KorchC (1991) Four major transcriptional responses in the methionine/threonine biosynthetic pathway of }U}Saccharomyces cerevisiae}u}. Yeast 7 : 781–803.
26. RamosC, DelgadoMA, CalderonIL (1991) Inhibition by different amino acids of the aspartate kinase and the homoserine kinase of the yeast Saccharomyces cerevisiae. FEBS Lett 278 : 123–126.
27. Arevalo-RodriguezM, PanX, BoekeJD, HeitmanJ (2004) FKBP12 controls aspartate pathway flux in Saccharomyces cerevisiae to prevent toxic intermediate accumulation. Eukaryot Cell 3 : 1287–1296.
28. AlarconCM, HeitmanJ (1997) FKBP12 physically and functionally interacts with aspartokinase in Saccharomyces cerevisiae. Mol Cell Biol 17 : 5968–5975.
29. KingsburyJM, McCuskerJH (2010) Homoserine toxicity in Saccharomyces cerevisiae and Candida albicans homoserine kinase (thr1Delta) mutants. Eukaryot Cell 9 : 717–728.
30. KimSJ, SwansonMJ, QiuH, GovindCK, HinnebuschAG (2005) Activator Gcn4p and Cyc8p/Tup1p are interdependent for promoter occupancy at ARG1 in vivo. Mol Cell Biol 25 : 11171–11183.
31. HinnebuschAG (1988) Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev 52 : 248–273.
32. DeLaBarreB, ThompsonPR, WrightGD, BerghuisAM (2000) Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases. Nat Struct Biol 7 : 238–244.
33. HinnebuschAG (1985) A hierarchy of trans-acting factors modulate translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol 5 : 2349–2360.
34. MuellerPP, HinnebuschAG (1986) Multiple upstream AUG codons mediate translational control of GCN4. Cell 45 : 201–207.
35. TanseyWP (2001) Transcriptional activation: risky business. Genes Dev 15 : 1045–1050.
36. DrysdaleCM, DueñasE, JacksonBM, ReusserU, BrausGH, et al. (1995) The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol 15 : 1220–1233.
37. Hinnebusch AG (1992) General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In: Broach JR, Jones EW, Pringle JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. pp. 319–414.
38. ZhangF, SumibcayL, HinnebuschAG, SwansonMJ (2004) A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol Cell Biol 24 : 6871–6886.
39. YamakiH, YamaguchiM, TsuruoT, YamaguchiH (1992) Mechanism of action of an antifungal antibiotic, RI-331, (S) 2-amino-4-oxo-5-hydroxypentanoic acid; kinetics of inactivation of homoserine dehydrogenase from Saccharomyces cerevisiae. J Antibiot (Tokyo) 45 : 750–755.
40. GoldsteinAL, McCuskerJH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 : 1541–1553.
41. MoehleCM, HinnebuschAG (1991) Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol 11 : 2723–2735.
42. SchmittME, BrownTA, TrumpowerBL (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18 : 3091–3092.
43. GaurNA, HasekJ, BricknerDG, QiuH, ZhangF, et al. (2013) Vps factors are required for efficient transcription elongation in budding yeast. Genetics 193 : 829–851.
44. ReidGA, SchatzG (1982) Import of proteins into mitochondria. Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J Biol Chem 257 : 13056–13061.
45. CiganAM, FoianiM, HannigEM, HinnebuschAG (1991) Complex formation by positive and negative translational regulators of GCN4. Mol Cell Biol 11 : 3217–3228.
46. KornitzerD (2002) Monitoring protein degradation. Methods Enzymol 351 : 639–647.
47. MontpetitB, ThomsenND, HelmkeKJ, SeeligerMA, BergerJM, et al. (2011) A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472 : 238–242.
48. SattleggerE, SwansonMJ, AshcraftEA, JenningsJL, FeketeRA, et al. (2004) YIH1 is an actin-binding protein that inhibits protein kinase GCN2 and impairs general amino acid control when overexpressed. J Biol Chem 279 : 29952–29962.
49. GietzRD, SuginoA (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74 : 527–534.
50. SikorskiRS, HieterP (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 : 19–27.
51. HinnebuschAG, LucchiniG, FinkGR (1985) A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene CYC1 of yeast. Proc Natl Acad Sci U S A 82 : 498–502.
52. DonahueTF, CiganAM (1988) Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol 8 : 2955–2963.
53. LongtineMS, McKenzie IIIA, DemariniDJ, ShahNG, WachA, et al. (1998) Additonal modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 : 953–961.
54. YoonS, QiuH, SwansonMJ, HinnebuschAG (2003) Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol Cell Biol 23 : 8829–8845.
Štítky
Genetika Reprodukční medicína
Článek Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome EvolutionČlánek Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder PopulationČlánek Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among PancrustaceansČlánek Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery DiseaseČlánek An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation inČlánek Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 7
-
Všechny články tohoto čísla
- Cuba: Exploring the History of Admixture and the Genetic Basis of Pigmentation Using Autosomal and Uniparental Markers
- Clonal Architecture of Secondary Acute Myeloid Leukemia Defined by Single-Cell Sequencing
- Mechanisms of Functional Variants That Impair Regulated Bicarbonate Permeation and Increase Risk for Pancreatitis but Not for Cystic Fibrosis
- Nucleosomes Shape DNA Polymorphism and Divergence
- Functional Diversification of Hsp40: Distinct J-Protein Functional Requirements for Two Prions Allow for Chaperone-Dependent Prion Selection
- Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome Evolution
- Activation of the Immune System by Combinations of Common Alleles
- Age-Associated Sperm DNA Methylation Alterations: Possible Implications in Offspring Disease Susceptibility
- Muscle-Specific SIRT1 Gain-of-Function Increases Slow-Twitch Fibers and Ameliorates Pathophysiology in a Mouse Model of Duchenne Muscular Dystrophy
- MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361
- Hypersensitivity of Primordial Germ Cells to Compromised Replication-Associated DNA Repair Involves ATM-p53-p21 Signaling
- Intrapopulation Genome Size Variation in Reflects Life History Variation and Plasticity
- SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD
- Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population
- Determinative Developmental Cell Lineages Are Robust to Cell Deaths
- DELLA Protein Degradation Is Controlled by a Type-One Protein Phosphatase, TOPP4
- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among Pancrustaceans
- UVB Induces a Genome-Wide Acting Negative Regulatory Mechanism That Operates at the Level of Transcription Initiation in Human Cells
- The Nesprin Family Member ANC-1 Regulates Synapse Formation and Axon Termination by Functioning in a Pathway with RPM-1 and β-Catenin
- Combinatorial Interactions Are Required for the Efficient Recruitment of Pho Repressive Complex (PhoRC) to Polycomb Response Elements
- Recombination in the Human Pseudoautosomal Region PAR1
- Microsatellite Interruptions Stabilize Primate Genomes and Exist as Population-Specific Single Nucleotide Polymorphisms within Individual Human Genomes
- An Intronic microRNA Links Rb/E2F and EGFR Signaling
- An Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding
- Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery Disease
- The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent Mutation
- Evolution and Genetic Architecture of Chromatin Accessibility and Function in Yeast
- An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation in
- Stage-Dependent and Locus-Specific Role of Histone Demethylase Jumonji D3 (JMJD3) in the Embryonic Stages of Lung Development
- Genome Wide Association Identifies Common Variants at the Locus Influencing Plasma Cortisol and Corticosteroid Binding Globulin
- Regulation of Feto-Maternal Barrier by Matriptase- and PAR-2-Mediated Signaling Is Required for Placental Morphogenesis and Mouse Embryonic Survival
- Apomictic and Sexual Germline Development Differ with Respect to Cell Cycle, Transcriptional, Hormonal and Epigenetic Regulation
- Functional EF-Hands in Neuronal Calcium Sensor GCAP2 Determine Its Phosphorylation State and Subcellular Distribution , and Are Essential for Photoreceptor Cell Integrity
- Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data
- Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
- Cis and Trans Effects of Human Genomic Variants on Gene Expression
- 8.2% of the Human Genome Is Constrained: Variation in Rates of Turnover across Functional Element Classes in the Human Lineage
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- A Loss of Function Screen of Identified Genome-Wide Association Study Loci Reveals New Genes Controlling Hematopoiesis
- Unraveling Genetic Modifiers in the Mouse Model of Absence Epilepsy
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
- The Coding and Noncoding Architecture of the Genome
- A Novel Locus Is Associated with Large Artery Atherosclerotic Stroke Using a Genome-Wide Age-at-Onset Informed Approach
- Brg1 Loss Attenuates Aberrant Wnt-Signalling and Prevents Wnt-Dependent Tumourigenesis in the Murine Small Intestine
- The PTK7-Related Transmembrane Proteins Off-track and Off-track 2 Are Co-receptors for Wnt2 Required for Male Fertility
- The Co-factor of LIM Domains (CLIM/LDB/NLI) Maintains Basal Mammary Epithelial Stem Cells and Promotes Breast Tumorigenesis
- Essential Genetic Interactors of Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates
- Meiosis-Specific Cohesin Component, Is Essential for Maintaining Centromere Chromatid Cohesion, and Required for DNA Repair and Synapsis between Homologous Chromosomes
- Silencing Is Noisy: Population and Cell Level Noise in Telomere-Adjacent Genes Is Dependent on Telomere Position and Sir2
- The Two Cis-Acting Sites, and , Contribute to the Longitudinal Organisation of Chromosome I
- A Broadly Conserved G-Protein-Coupled Receptor Kinase Phosphorylation Mechanism Controls Smoothened Activity
- Requirements for Acute Burn and Chronic Surgical Wound Infection
- LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription
- WAPL Is Essential for the Prophase Removal of Cohesin during Meiosis
- Expression in Planarian Neoblasts after Injury Controls Anterior Pole Regeneration
- Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
- Accumulation of a Threonine Biosynthetic Intermediate Attenuates General Amino Acid Control by Accelerating Degradation of Gcn4 via Pho85 and Cdk8
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



