-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSKN-1/Nrf, A New Unfolded Protein Response Factor?
article has not abstract
Published in the journal: . PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003827
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003827Summary
article has not abstract
Cell function requires simultaneous regulation of numerous processes, often under variable conditions. Several inducible pathways have been defined as being responsible for maintaining homeostasis under the threat of a particular stress such as heat, oxidizing conditions, exposure to pathogens, or loss of proteostasis. The challenge now is to understand how and why these pathways interact in basal, stress, and pathological states. The Caenorhabditis elegans inducible transcription factor SKN-1, a homolog of mammalian Nrf proteins, has been defined as the transcription factor that responds to oxidative stress. In this issue of PLOS Genetics, Glover-Cutter et al. [1] challenge this paradigm by showing that SKN-1 directly regulates the genes of core regulators and effectors of the endoplasmic reticulum (ER) unfolded protein response (UPR) and that the UPR plays a role in activation of the antioxidant/detoxification response.
ER homeostasis requires coordination of protein translation, folding, and covalent modification; availability of energy and substrates; and maintenance of a redox environment that is suitable for disulfide bond formation. Disruption of ER homeostasis can cause accumulation of misfolded proteins in the ER lumen, referred to as “ER stress,” which can impair cell function and eventually trigger cell death [2]–[4]. The UPR is a eukaryotic signaling program that responds to ER stress by inhibiting protein translation, inducing protein folding chaperones, and directing the degradation of misfolded proteins [2]–[4] (Figure 1). Redox homeostasis in animal cells is controlled in part by a family of transcription factors represented by SKN-1 in C. elegans and Nrf1, Nrf2, and Nrf3 in mammals (Figure 1). SKN-1 and Nrf2 have well-established roles in promoting redox homeostasis and small molecule detoxification, and SKN-1 has been shown to promote longevity [5]–[8].
Fig. 1. Summary of interactions between the endoplasmic reticulum (ER) unfolded protein response (UPR) and antioxidant/detoxification transcription factor SKN-1 in C. elegans. 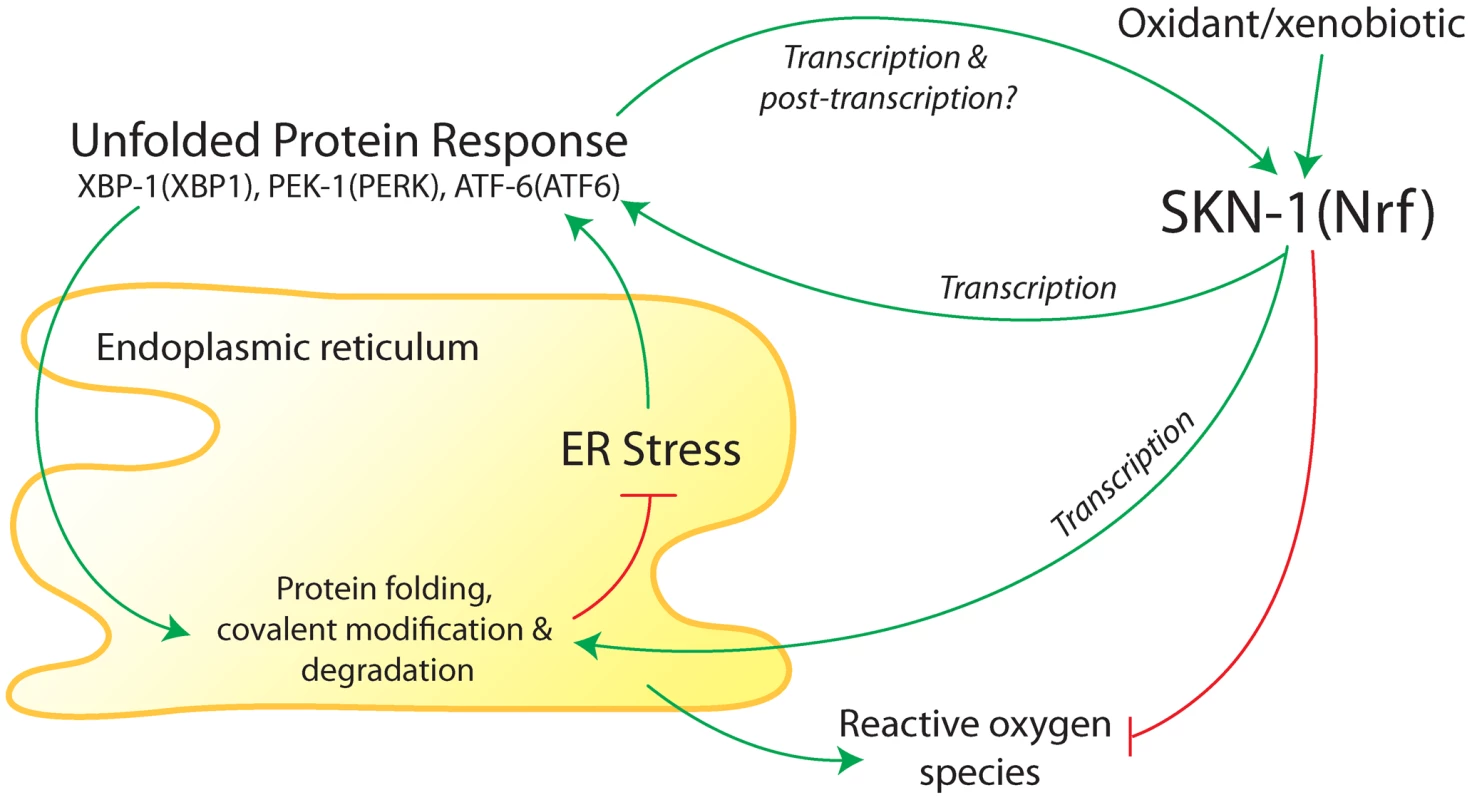
The ER folds and modifies newly synthesized membrane and secreted proteins. Accumulation of misfolded proteins in the ER activates three canonical branches of the UPR, which are mediated by XBP-1, PEK-1, and ATF-6 (mammalian homologs in parentheses). Transcriptional targets of the UPR function to promote protein folding and covalent modification of new proteins and degradation of misfolded proteins. SKN-1 (homolog of mammalian proteins Nrf1, Nrf2, and Nrf3) was found to transcriptionally regulate core UPR regulators and some downstream targets during ER stress [1]. Core UPR regulators also transcriptionally regulate SKN-1 during ER stress and oxidative/xenobiotic stress [1]. Based on studies in mammalian cells [12], [13], SKN-1 would also be expected to buffer reactive oxygen species that are produced in the ER. A long variant of SKN-1 may reside in the ER membrane (not shown). Regulation of SKN-1 during ER and oxidative/xenobiotic stress may include post-translational modifications. Glover-Cutter et al. [1] conducted an extensive series of genetic and molecular experiments to investigate regulatory interactions between SKN-1 and the UPR; many of their findings are summarized in Figure 1. They showed that SKN-1 regulates numerous genes involved in ER function during ER stress that are not typically activated by SKN-1 during oxidative stress. These include protein chaperones and homologs of the following core UPR components: BiP (an unfolded protein sensor), PERK (a protein kinase), IRE1 (a protein kinase and mRNA endonuclease), and the transcription factors XBP1, ATF4, and ATF6. During ER stress, SKN-1 protein was shown to associate with loci for homologs of ATF4, ATF6, XBP1, and IRE1, indicating that regulation of core UPR genes by SKN-1 is likely to be direct.
How is SKN-1 activated by ER stress? The authors observed elevated skn-1 mRNA and protein levels during ER stress [1]. Processing of protein disulfide bonds in the ER can elevate reactive oxygen species (ROS) [4], [9], a well-established stimulus for SKN-1 that could simply activate it secondarily. However, the authors demonstrated induction of skn-1 mRNA by a strong reducing agent and by silencing of the worm ER oxidoreductase, conditions that cause ER stress and decrease ROS [1]. Furthermore, the C. elegans homologs of XBP1 and ATF6, and SKN-1 itself, all associated with the skn-1 locus during ER stress and were found to play a role in induction of skn-1 mRNA [1]. Therefore, activation of SKN-1 during ER stress appears to be at least partly transcriptional via UPR transcription factors.
If SKN-1 is required for the UPR, then could the UPR also be required for the antioxidant/detoxification response? The answer may be yes. During oxidative stress, core components of the UPR were required for induction of skn-1 mRNA and some SKN-1 target genes. It is of additional interest that activation of p38 MAPK by phosphorylation, which activates SKN-1 under conditions of oxidative stress, also required components of the UPR [10].
Important regulatory and functional interactions between the UPR and Nrf2 were previously known for mammalian cells [4], [11]–[13]. Nrf2 is phosphorylated and activated by PERK during ER stress and promotes cell survival by maintaining redox homeostasis together with ATF4 [12]–[13]. Nrf2 also activates expression of proteasome subunits and may support degradation of misfolded proteins during ER stress [11]. So what is different about the current findings? Transcriptional regulation of core UPR transcription factors and downstream effectors by SKN-1 is a far more central function in the ER stress response than has previously been reported for SKN-1/Nrf family members. Details of the molecular interactions may not all be conserved, but the findings for SKN-1 raise the possibility that Nrf1, Nrf2, or Nrf3 may be centrally integrated into the mammalian UPR.
As with any new findings, important new questions follow. SKN-1 was shown to contribute to survival of ER stress in vivo [1]. Determining the relative importance of SKN-1–mediated UPR gene regulation versus redox homeostasis would be challenging, but is needed to understand the function of these newly identified regulatory interactions. Other than PERK phosphorylation of Nrf2 [12]–[13], little is known about post-translational regulation of SKN-1/Nrf proteins during ER stress. Evidence was provided for association of a long SKN-1 variant with the ER [1], and Nrf1 and Nrf3 each have a predicted transmembrane domain and have been reported to be associated with the ER membrane [14]–[16]. In unstressed cells, ATF6 is a membrane protein tethered to the ER by BiP [3]. During ER stress, ATF6 undergoes cleavage to its active transcription factor form in the golgi [3]. Further work is needed to determine if SKN-1/Nrf proteins at the ER have a similar fate.
Coordination between SKN-1/Nrf proteins and the UPR has been evolutionarily conserved and, therefore, may be fundamentally important to cell homeostasis. Direct regulation of the UPR implies that the ER may be able to prepare for protein damage under harsh conditions detected by SKN-1/Nrf family members. It may also ensure that redox homeostasis in the cytosol is compatible with redox-dependent protein processing in the ER. The activity of SKN-1 also responds to changes in the nucleolus [17], the proteasomes [18], nutrient signaling [5]–[6], and protein translation [19]. Therefore, an extremely complex network of signals likely converges on SKN-1 to ensure that redox status and detoxification activity are compatible with a number of cellular processes. Deciphering this signaling network will provide mechanistic insights into how a single transcription factor is influenced by multiple signals. As we continue to refine our understanding of cellular stress responses and their roles in disease and aging, it will be increasingly important to investigate how different pathways coordinate responses to optimize homeostasis, avoid incompatibilities, and mitigate competition for common substrates.
Zdroje
1. Glover-CutterKM, LinS, BlackwellTK (2013) Integration of the unfolded protein and oxidative stress responses through SNK-1/Nrf. PLoS Genet 9: e1003701 doi:10.1371/journal.pgen.1003701
2. HigaA, ChevetE (2012) Redox signaling loops in the unfolded protein response. Cell Signal 24 : 1548–1555.
3. CullinanSB, DiehlJA (2006) Coordination of ER and oxidative stress signaling: the PERK/Nrf2 signaling pathway. Int J Biochem Cell Biol 38 : 317–332.
4. BhandaryB, MarahattaA, KimHR, ChaeHJ (2012) An involvement of oxidative stress in endoplasmic reticulum stress and its associated diseases. Int J Mol Sci 14 : 434–456.
5. Robida-StubbsS, Glover-CutterK, LammingDW, MizunumaM, NarasimhanSD, et al. (2012) TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab 15 : 713–724.
6. TulletJMA, HertweckM, AnJH, BakerJ, HwangJY, et al. (2008) Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132 : 1025–1038.
7. SykiotisG, BohmannD (2010) Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal 3: re3.
8. SykiotisGP, BohmannD (2008) Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell 14 : 76–85.
9. MalhotraJD, KaufmanRJ (2007) Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal 9 : 2277–2293.
10. InoueH, HisamotoN, AnJH, OliveiraRP, NishidaE, et al. (2005) The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 19 : 2278–2283.
11. LeeCS, HoDV, ChanJY (2013) Nuclear factor-erythroid 2-related factor 1 regulates expression of proteasome genes in hepatocytes and protects against endoplasmic reticulum stress and steatosis in mice. FEBS J 280 : 3609–20 doi:10.1111/febs.12350
12. CullinanSB, ZhangD, HanninkM, ArvisaisE, KaufmanRJ, et al. (2003) Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol 23 : 7198–7209.
13. CullinanSB, DiehlJA (2004) PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem 279 : 20108–20117.
14. ZhangY, KobayashiA, YamamotoM, HayesJD (2009) The Nrf3 transcription factor is a membrane-bound glycoprotein targeted to the endoplasmic reticulum through its N-terminal homology box 1 sequence. J Biol Chem 284 : 3195–3210.
15. WangW, ChanJY (2006) Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J Biol Chem 281 : 19676–19687.
16. ZhangY, CrouchDH, YamamotoM, HayesJD (2006) Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem J 399 : 373–385.
17. LeungCK, EmpinadoH, ChoeKP (2012) Depletion of a nucleolar protein activates xenobiotic detoxification genes in Caenorhabditis elegans via Nrf/SKN-1 and p53/CEP-1. Free Radic Biol Med 52 : 937–950.
18. LiX, MatilainenO, JinC, Glover-CutterKM, HolmbergCI, et al. (2011) Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet 7: e1002119 doi:10.1371/journal.pgen.1002119
19. WangJ, Robida-StubbsS, TulletJM, RualJF, VidalM, et al. (2010) RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet 6: e1001048 doi:10.1371/journal.pgen.1001048
Štítky
Genetika Reprodukční medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 9
-
Všechny články tohoto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



