-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development
Plants have evolved sophisticated systems for adaptation to their natural habitat. In response to developmental and environmental cues, plants produce and perceive jasmonate (JA) signals, which induce degradation of JASMONATE-ZIM-Domain (JAZ) proteins and derepress the JAZ-repressed transcription factors to regulate diverse aspects of defense responses and developmental processes. Here, we identified the bHLH subgroup IIId transcription factors (bHLH3, bHLH13, bHLH14 and bHLH17) as novel targets of JAZs. These bHLH subgroup IIId transcription factors act as transcription repressors and function redundantly to negatively regulate JA responses. The quadruple mutant bhlh3 bhlh13 bhlh14 bhlh17 showed severe sensitivity to JA-inhibited root growth and JA-induced anthocyanin accumulation, and exhibited obvious increase in JA-regulated plant defense against pathogen infection and insect attack. Transgenic plants overexpressing bHLH13 or bHLH17 displayed reduced JA responses. Furthermore, these bHLH factors functioned as transcription repressors to antagonize the transcription activators, such as MYC2 and the WD-repeat/bHLH/MYB complex, through binding to their target sequences. Coordinated regulation of JA responses by transcription activators and repressors would benefit plants by allowing fine regulation of defense and development, and survival in their frequently changing environment.
Published in the journal: . PLoS Genet 9(7): e32767. doi:10.1371/journal.pgen.1003653
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003653Summary
Plants have evolved sophisticated systems for adaptation to their natural habitat. In response to developmental and environmental cues, plants produce and perceive jasmonate (JA) signals, which induce degradation of JASMONATE-ZIM-Domain (JAZ) proteins and derepress the JAZ-repressed transcription factors to regulate diverse aspects of defense responses and developmental processes. Here, we identified the bHLH subgroup IIId transcription factors (bHLH3, bHLH13, bHLH14 and bHLH17) as novel targets of JAZs. These bHLH subgroup IIId transcription factors act as transcription repressors and function redundantly to negatively regulate JA responses. The quadruple mutant bhlh3 bhlh13 bhlh14 bhlh17 showed severe sensitivity to JA-inhibited root growth and JA-induced anthocyanin accumulation, and exhibited obvious increase in JA-regulated plant defense against pathogen infection and insect attack. Transgenic plants overexpressing bHLH13 or bHLH17 displayed reduced JA responses. Furthermore, these bHLH factors functioned as transcription repressors to antagonize the transcription activators, such as MYC2 and the WD-repeat/bHLH/MYB complex, through binding to their target sequences. Coordinated regulation of JA responses by transcription activators and repressors would benefit plants by allowing fine regulation of defense and development, and survival in their frequently changing environment.
Introduction
Plant hormones are essential for the regulation of plant growth, differentiation, development, reproduction, and survival [1]–[3]. Jasmonates (JAs), a class of cyclic fatty acid-derived plant hormones originating from plastid membrane α-linolenic acid [4], [5], regulate diverse aspects of plant developmental processes [6], such as seedling growth [7], root development [8]–[10], plant fertility [11]–[13], trichome initiation [14], [15], pigment formation [14], [16], [17], and senescence [18]. It is also well established that jasmonates act as key defense signals in regulation of various abiotic and biotic stress such as mechanic wounding [19], [20], arthropod herbivores and necrotrophic pathogens [21]–[24] and drought [25], [26].
In response to external environmental signals and internal developmental cues, plants generate and perceive jasmonates (JA) signals to induce degradation of JASMONATE ZIM-Domain (JAZ) proteins [20], [27]–[33]. As consequence of JAZs degradation, JAZ-targeted transcription factors will be relieved to subsequently regulate their downstream signal cascades and modulate respective JA responses.
Current studies have identified several key transcription factors as direct targets of JAZ proteins [34]–[38]. The bHLH subgroup IIIe transcription factors (MYC2, MYC3 and MYC4) are targets of JAZs and play important roles in regulation of plant defense and developmental processes [39]–[41]. The R2R3-MYB transcription factors (MYB21, MYB24 and MYB57), and the transcription complexes WD-repeat/bHLH (TT8, GL3 or EGL3)/MYB (MYB75 or GL1) interact with JAZs to regulate JA-mediated male fertility, anthocyanin accumulation and trichome initiation, respectively [12], [14], [42]. Current research so far has not identified JAZ-targeted transcription repressors in JA pathway.
In this study, we identified the bHLH subgroup IIId transcription factors (bHLH3, bHLH13, bHLH14 and bHLH17) as new targets of JAZ proteins. These bHLH subgroup IIId transcription factors function redundantly to negatively regulate JA-mediated plant defense and development. Furthermore, these bHLH factors act as transcriptional repressors to suppress JA responses, which antagonize the previously reported transcription activators (such as MYC2 and the WD-repeat/TT8/MYB75 complex) through binding to their downstream target sequences. Coordinated regulation of JA responses by transcription repressors and activators would benefit plants for adaptation to their frequently changing environment.
Results
JAZ Proteins Interact with the bHLH Subgroup IIId Transcription Factors bHLH3, bHLH13, bHLH14 and bHLH17
To understand molecular basis of jasmonate action, we exhaustedly screened the Arabidopsis thaliana cDNA library using JAZ proteins (JAZ1 and JAZ8) as bait in the yeast two-hybrid (Y2H) system, and identified several transcription factors [12], [14]. We found that a bHLH transcription factor bHLH13 (AT1G01260) also interacted with various JAZ proteins (Figure 1A).
Fig. 1. JAZ proteins interact with bHLH3, bHLH13, bHLH14 and bHLH17. 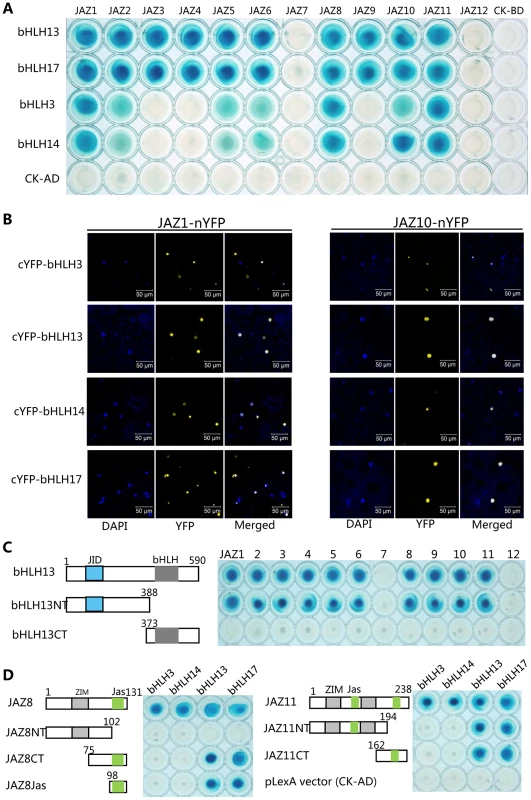
(A) Yeast two-hybrid (Y2H) assay to detect interactions of JAZs with bHLH3, bHLH13, bHLH14 and bHLH17 factors. Twelve Arabidopsis JAZs were fused with the LexA DNA binding domain (BD) in pLexA respectively. The bHLH3, bHLH13, bHLH14 and bHLH17 were fused with the activation domain (AD) in pB42AD respectively. Interactions (represented by blue color) were assessed on 2% Gal/1% raffinose/SD/-Ura/-His/-Trp/-Leu/X-β-Gal medium. (B) Bimolecular fluorescence complementation (BiFC) assay to detect interactions of JAZ1 and JAZ10 (fused with nYFP) with bHLH3, bHLH13, bHLH14 and bHLH17 (fused with cYFP). Construct pairs were coexpressed in leaves of N. benthamiana. YFP fluorescence was detected 50 hours after infiltration. The nuclei were indicated by DAPI (4,6-diamidino-2-phenylindole dihydrochloride) staining. (C) Y2H assay to test interactions of bHLH13 domain constructs with twelve JAZs. The schematic diagram shows the bHLH13 domain constructs. The conserved JID domain and bHLH domain were shown with blue and gray box respectively. The numbers indicate the positions of amino acid. Different bHLH13 domains were fused with AD in pB42AD, and JAZs were fused with BD in pLexA. (D) Y2H assay to test interactions between different domains of JAZ8 and JAZ11 with bHLH3, bHLH13, bHLH14 and bHLH17 respectively. The schematic diagram shows the JAZ8 and JAZ11 domain constructs. The conserved ZIM and Jas domains are indicated by gray and green boxes respectively. Different domains of JAZ8 and JAZ11 were fused with BD, and these bHLH factors were fused with AD individually. Phylogenetic analysis showed that bHLH13, together with bHLH3 (AT4G16430), bHLH14 (AT4G00870) and bHLH17 (AT2G46510/AtAIB), belongs to the subgroup IIId of the Arabidopsis bHLH family [43]–[45]. We further found that, in the yeast two-hybrid (Y2H) assays, all the bHLH subgroup IIId factors interacted with various JAZ proteins (Figure 1A).
To verify the interactions between JAZs and the bHLH3, bHLH13, bHLH14 or bHLH17 in planta, we performed bimolecular fluorescence complementation (BiFC) assays in leaves of Nicotiana benthamiana. As shown in Figure 1B, the strong signals of yellow fluorescent protein (YFP) in nucleus of N. benthamiana leaves were reconstructed by coexpression of JAZ1-nYFP (the JAZ1 fused with N-terminal fragment of YFP) with cYFP-bHLH3, cYFP-bHLH13, cYFP-bHLH14 and cYFP-bHLH17, respectively. Strong YFP signals were also detected when JAZ10 was coexpressed with these transcription factors in the BiFC assays (Figure 1B). These results suggested that these bHLH subgroup IIId factors interact with JAZs in planta.
To investigate which domain of these bHLH factors is responsible for the interaction with JAZ proteins, we representatively divided the bHLH13 into the N-terminal fragment (bHLH13NT) containing the JAZ-Interaction-Domain (JID) [40], and the C-terminal part (bHLH13CT) (Figure 1C). As shown in Figure 1C, bHLH13NT, but not bHLH13CT exhibited interactions with various JAZ proteins. Consistent with the previous speculation [40], these results suggest that the N-terminal fragment (JID domain) of the bHLH13 factor is responsible for the interactions with JAZ proteins.
To further examine which domain of JAZ proteins is critical for interactions with the bHLH subgroup IIId factors, we divided JAZ8 into the N-terminal fragment (JAZ8NT), the C-terminal part (JAZ8CT), and the Jas domain only (JAZ8Jas) (Figure 1D). Our results suggested that the Jas domain in JAZ8 is required for interactions with bHLH13 and bHLH17 (Figure 1D). Interestingly, bHLH3 and bHLH14 were able to interact with JAZ8, but not with the truncated fragments of JAZ8 (JAZ8NT, JAZ8CT and JAZ8Jas) (Figure 1D), suggesting that the entire JAZ8 is required for interactions with bHLH3 and bHLH14. Similarly, we found that the entire JAZ11 is required for interactions with bHLH3 and bHLH14, while either of Jas domains in JAZ11 is sufficient for interactions with bHLH13 and bHLH17 (Figure 1D).
The bHLH3, bHLH13, bHLH14 and bHLH17 Function Redundantly to Negatively Regulate JA Responses
To examine expression patterns of bHLH3, bHLH13, bHLH14 and bHLH17 in plant tissues, we generated Arabidopsis plants transgenic for the GUS reporter driven by endogenous promoter of each bHLH factor. Histochemical staining of the GUS activity demonstrated that all these bHLH factors are expressed in various plant tissues (Figure 2A), which is consistent with quantitative real-time PCR analysis (Figure 2B) and the public available data (www.bar.utoronto.ca) [43]. Interestingly, the COI1-dependent JA-induced gene expression was observed for bHLH13 and bHLH17, but not for bHLH3 and bHLH14 (Figure 2D). Further examination of subcellular localization indicated that bHLH3 and bHLH17 were nucleus-localized (Figure 2C), whereas bHLH13 and bHLH14 were localized in both nucleus and cytoplasm (Figure 2C).
Fig. 2. Expression patterns and subcellular localizations of bHLH3, bHLH13, bHLH14 and bHLH17. 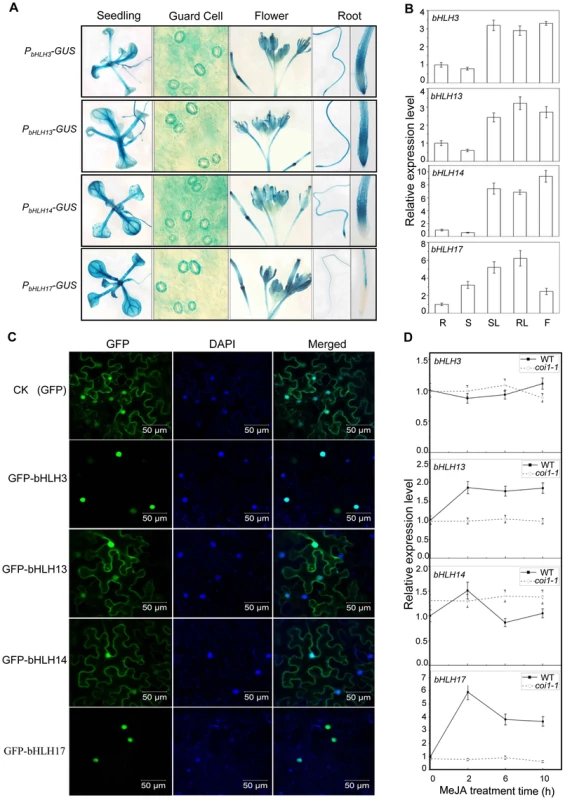
(A) GUS reporter gene was fused with the promoters of the four bHLH factors respectively to generate Arabidopsis transgenic plants (PbHLH3-GUS, PbHLH13-GUS, PbHLH14-GUS and PbHLH17-GUS). Histochemical GUS activity was detected in various tissues of transgenic seedlings. (B) Quantitative real-time PCR analysis of relative expression levels of bHLH3, bHLH13, bHLH14 and bHLH17 in root (R), stem (S), rosette leaf (RL), stem leaf (SL) and flower (F). ACTIN8 was used as the internal control. Error bars represent SE (n = 3). (C) Subcellular localization of bHLH3, bHLH13, bHLH14 and bHLH17 in epidermal cells of N. benthamiana leaves. Constructs indicated on the left were infiltrated in leaves of N. benthamiana. GFP fluorescence was detected 50 hours after infiltration. The nuclei were indicated by DAPI staining. (D) Quantitative real-time PCR analysis of bHLH3, bHLH13, bHLH14 and bHLH17 in 11-day-old WT and coi1-1 seedlings treated with 100 µM methyl-jasmonate (MeJA) for 0, 2, 6, and 10 hours. ACTIN8 was used as the internal control. Error bars represent SE (n = 3). To investigate the function of the bHLH subgroup IIId factors, we identified Arabidopsis mutants for bHLH3, bHLH13, bHLH14 and bHLH17 with the T-DNA insertion into the exon (for bHLH3, bHLH14 and bHLH17) or the 5′UTR (for bHLH13) (Figure 3A). Quantitative real-time PCR analysis showed that the expression of the full length bHLH subgroup IIId gene was abolished (for bHLH3, bHLH14 and bHLH17) or obviously reduced (for bHLH13) in their respective mutants (Figures 3A and 3B). Observation of typical JA-regulated responses, including plant fertility, JA-inhibitory root growth and JA-induced anthocyanin accumulation, showed that no obvious differences were detected among wild-type, bhlh3, bhlh13, bhlh14 and bhlh17 single mutants (Figure 3, and data not shown).
Fig. 3. JA responses were not obviously altered in the bhlh3, bhlh13, bhlh14 and bhlh17 single mutants. 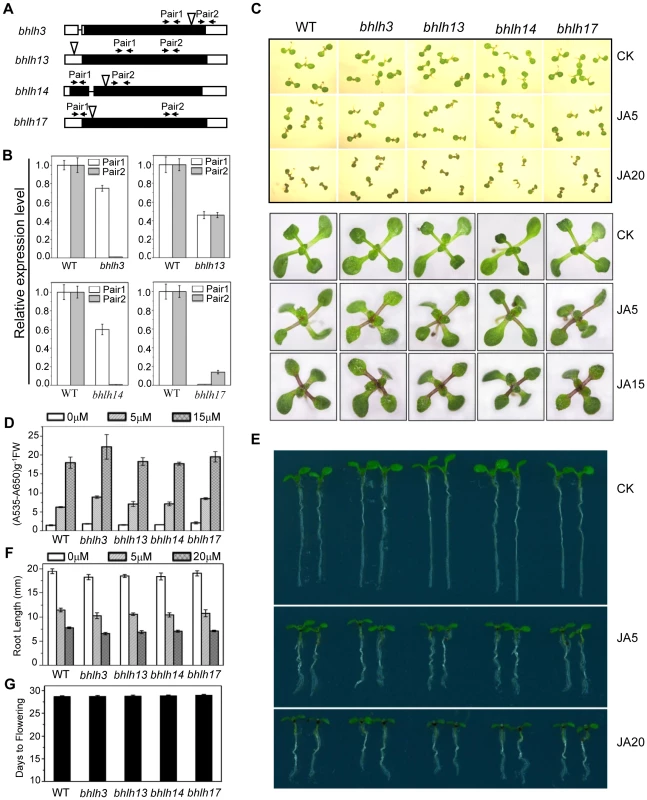
(A) Schematic diagrams of T-DNA insertion sites in bHLH3, bHLH13, bHLH14 and bHLH17. White box, UTR; black box, exon; black line, intron; white triangle, T-DNA insertion site. Pair1 and Pair2, indicated by arrows, are the primer pairs for analyzing gene expression in (B). (B) Quantitative real-time PCR analysis of bHLH3, bHLH13, bHLH14 and bHLH17 in the respective T-DNA insertion mutants using primer pairs indicated by arrow pairs in (A). ACTIN8 was used as the internal control. (C) Seedling phenotypes of 7-day-old (upper panel) and 11-day-old (bottom panel) Col-0 wild type (WT), bhlh3, bhlh13, bhlh14 and bhlh17 grown on MS medium supplied without (CK) or with indicated concentrations (µM) of MeJA. (D) Anthocyanin contents of the 11-day-old seedlings in WT, bhlh3, bhlh13, bhlh14 and bhlh17 single mutants grown on MS medium containing indicated concentrations of MeJA. FW, fresh weight. Error bars represent SE (n = 3). (E) Root phenotypes of 7-day-old seedlings of WT and single mutants of bhlh3, bhlh13, bhlh14 and bhlh17 grown on MS medium supplied without (CK) or with indicated concentrations (µM) of MeJA. (F) Root length of 11-day-old seedlings grown on MS medium containing indicated concentrations of MeJA. Error bars represent SE (n = 15). (G) Flowering time of WT, bhlh3, bhlh13, bhlh14 and bhlh17 single mutants. Data shown are the means from 24 plants. Error bars represent SE. As bHLH3, bHLH13, bHLH14 and bHLH17 display high similarity at the amino acid level and belong to the same subgroup IIId of the Arabidopsis bHLH family [44], we further generated double, triple and quadruple mutants for the bHLH subgroup IIId factors, through genetic cross among the bhlh3, bhlh13, bhlh14 and bhlh17 mutants, to investigate whether these factors function redundantly in regulation of JA responses. Interestingly, we found that the anthocyanin accumulation was gradually increased in the double mutant bhlh3 bhlh17, the triple mutant bhlh3 bhlh13 bhlh17, bhlh3 bhlh13 bhlh14, bhlh3 bhlh14 bhlh17, bhlh13 bhlh14 bhlh17, and the quadruple mutant bhlh3 bhlh13 bhlh14 bhlh17 in response to MeJA treatment (Figures 4A, 4C, and S1). Consistent with the tendency of JA-induced anthocyanin accumulation (Figures 4A and 4C), MeJA-induced expression of the anthocyanin biosynthetic genes, including DFR, LDOX and UF3GT [46], was gradually increased in the double, triple and quadruple mutants (Figure 4B).
Fig. 4. The bhlh3 bhlh13 bhlh14 bhlh17 quadruple mutant exhibited enhanced JA responses. 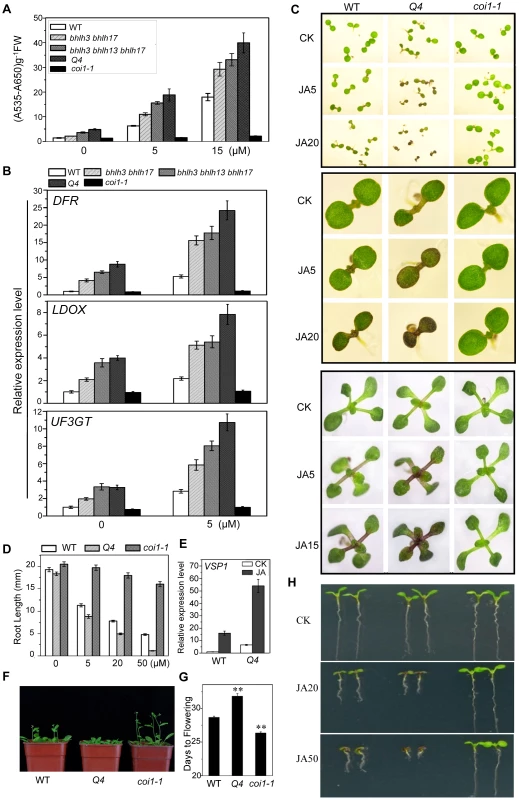
(A) Anthocyanin contents in the 11-day-old seedlings of WT, bhlh3 bhlh17, bhlh3 bhlh13 bhlh17, bhlh3 bhlh13 bhlh14 bhlh17 (Q4) and coi1-1 grown on MS medium containing indicated concentrations of MeJA. FW, fresh weight. Error bars represent SE (n = 3). (B) Quantitative real-time PCR analysis of DFR, LDOX and UF3GT expression levels in 11-day-old seedlings of indicated plants grown on MS medium supplied with 0 or 5 µM MeJA. ACTIN8 was used as the internal control. (C) Phenotypes of 7-day-old (the upper panel, the middle panel that showed enlarged seedlings) and 11-day-old seedlings (the bottom panel) of WT, the quadruple mutant (Q4) and coi1-1 grown on MS medium supplied without (CK) or with indicated concentrations (µM) of MeJA. (D) Root length of 11-day-old seedlings grown on MS medium containing indicated concentrations of MeJA. Error bars represent SE (n = 15). Asterisks denote Student's t-test significance compared with WT plants: **, P<0.01. (E) Quantitative real-time PCR analysis of VSP1 expression level in WT, Q4 and coi1-1 seedlings grown for 11 days on MS medium supplied without (CK) or with 5 µM MeJA (JA). ACTIN8 was used as the internal control. (F) Flowering phenotypes of five-week-old WT, Q4 and coi1-1 plants grown in a long-day growth chamber (16L/8D, 23°C). (G) Flowering time of WT, Q4 and coi1-1. Data shown are the means from 24 plants. Error bars represent SE. Asterisks denote Student's t-test significance compared with WT plants: **, P<0.01. (H) Root phenotypes of 7-day-old WT, Q4 and coi1-1 seedlings grown on MS medium supplied without (CK) or with indicated concentrations (µM) of MeJA. The quadruple mutant bhlh3 bhlh13 bhlh14 bhlh17 exhibited enhanced JA responses. They displayed obvious increase in JA-induced anthocyanin biosynthesis compared with wild-type control (Figures 4A and 4C). The JA-inhibitory root growth analysis also showed that the quadruple mutant was more sensitive to JA inhibition of root growth (Figures 4D and 4H). Observation of flowering time showed that the quadruple mutant also exhibited enhanced JA response: the quadruple mutant exhibited late flowering phenotype whereas the coi1-1 mutant plant flowered early (Figures 4F and 4G). Consistent with the enhanced JA-responses, expression of JA-inducible marker gene VSP1 [7] was significantly increased in the quadruple mutant (Figure 4E).
Taken together (Figures 3 and 4), mutations in bHLH3, bHLH13, bHLH14 and bHLH17 caused enhanced JA responses, demonstrating that these bHLH subgroup IIId factors function redundantly to negatively regulate JA responses.
The Quadruple Mutant bhlh3 bhlh13 bhlh14 bhlh17 Exhibited Enhanced JA-regulated Plant Defense
Botrytis cinerea, a necrotrophic fungus that causes gray mold disease in many plant species [47], induced severe wilting and high mortality in the Arabidopsis mutants coi1-1 [8], [48] or aos1[49], [50]. To investigate potential role for bHLH3, bHLH13, bHLH14 and bHLH17 in plant defense, we sprayed the B. cinerea spore suspension onto the quadruple mutant, wild-type and coi1-1 plants. As shown in Figure 5A and 5B, the quadruple mutant plants exhibited increased resistance against B. cinerea compared with wild type, whereas coi1-1 mutant plants displayed severe disease symptom, as revealed by disease severity and plant survival rate (Figures 5C and 5D). Consistent with the increased defense response, the PLANT-DEFENSIN gene PDF1.2 [51], antifungal gene THI2.1 [52], defense gene ERF1 [53] and wound-inducible gene LOX2 [54] were highly induced in the quadruple mutant compared with wild type when treated with MeJA (Figure 5E).
Fig. 5. The bHLH subgroup IIId factors negatively regulate JA-mediated plant defense. 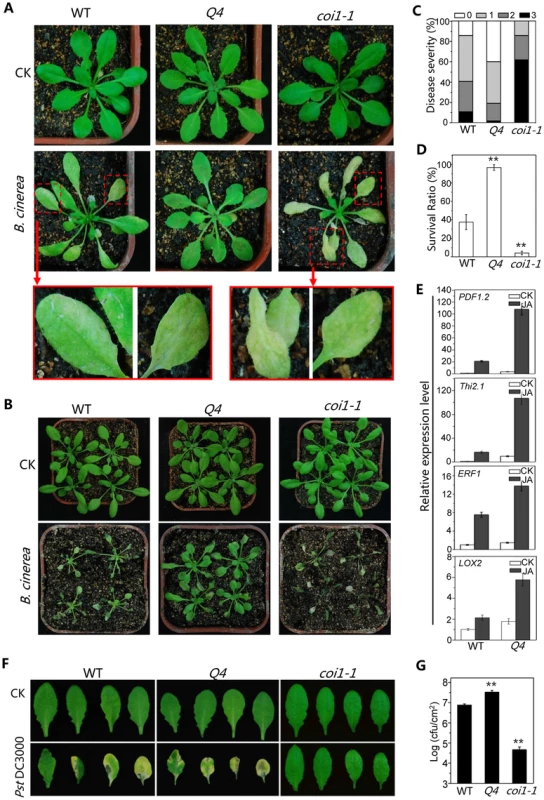
(A) Disease symptoms of WT, the quadruple mutant (Q4) and the coi1-1 mutant five days after spray inoculation with Botrytis cinerea or mock (CK) in the growth condition with ∼40% humidity. The enlarged leaves indicate obvious disease symptoms in WT, and severe disease symptoms in coi1-1. (B) Disease symptoms of WT, the quadruple mutant (Q4) and coi1-1 mutant seven days after spray inoculation with B. cinerea or mock (CK) in the growth condition with high humidity (>90%). (C) Disease severity of the plants in (A) seven days after spray inoculation with B. cinerea. Disease rating was represented as no visible (0, white), weak (1, light grey), severe symptoms (2, grey), and completely dead plants (3, dark). Experiments were repeated three times with similar results. (D) Survival ratio of the plants in (B) nine days after spray inoculation. Error bars represents SE. Asterisks denote Student's t-test significance compared with WT plants: **, P<0.01. (E) Quantitative real-time PCR analysis of expression levels of PDF1.2, Thi2.1, ERF1 and LOX2 in WT and Q4 germinated and grown on MS medium supplied without (CK) or with 5 µM MeJA (JA) for 11 days. ACTIN8 was used as the internal control. (F) Disease symptoms on leaves of WT, Q4 and coi1-1 mutant three days after spray inoculation with Pseudomonas syringae pv. tomato (Pst) DC3000. (G) Growth of Pst DC3000 on WT, Q4 and coi1-1 mutant plants three days after spray inoculation as in (F). Bacterial counts are expressed as log (cfu/cm2). Error bars represent SE. The results are representative of three independent biological experiments. Asterisks represent Student's t-test significance compared with WT plants: **, P<0.01. Previous studies showed that JA induces plant susceptibility to the bacterium strain Pst DC3000 of Pseudomonas syringae pv. Tomato. The coi1-1 mutant is more resistant to the Pst DC3000 inoculation [8], [55], [56] (Figures 5F and 5G). The quadruple mutant exhibited enhanced JA response in the Pst DC3000 inoculation assay: the quadruple mutant was more susceptible to the Pst DC3000 infection, whereas coi1-1 was resistant (Figures 5F and 5G).
To test whether the bHLH subgroup IIId factors regulate JA-mediated plant defense against insects, mature rosette leaves of wild-type, coi1-1, and the quadruple mutant were fed to Spodoptera exigua, a globally-significant agricultural pest with a broad host range [21]. We found that S. exigua larvae consumed the majority of the coi1-1 leaves (Figure 6A) and grew rapidly (Figures 6 B and C). However, the quadruple mutant leaves inhibited the growth of S. exigua larvae and showed reduced consumption by S. exigua (Figures 6 A–C). When insects were given the choice of selecting among wild type, coi1-1 and the quadruple mutant in the three-choice test, the quadruple mutant attracted fewer S. exigua larvae (∼6%), while wild type and coi1-1 accommodated the majority of S. exigua larvae (23% for wild type, 71% for coi1-1) (Figure 6D). In the two-choice test, the quadruple mutant attracted less larvae than wild type (∼28% for the quadruple mutant, ∼72% for wild type), while the wild-type attracted less larvae than coi1-1 (19% for wild type, ∼81% for coi1-1) (Figure 6E). These results suggested that the quadruple mutant displays enhanced JA-regulated plant defense against insect.
Fig. 6. The bhlh3 bhlh13 bhlh14 bhlh17 quadruple mutant exhibited enhanced JA-mediated plant defense against insect. 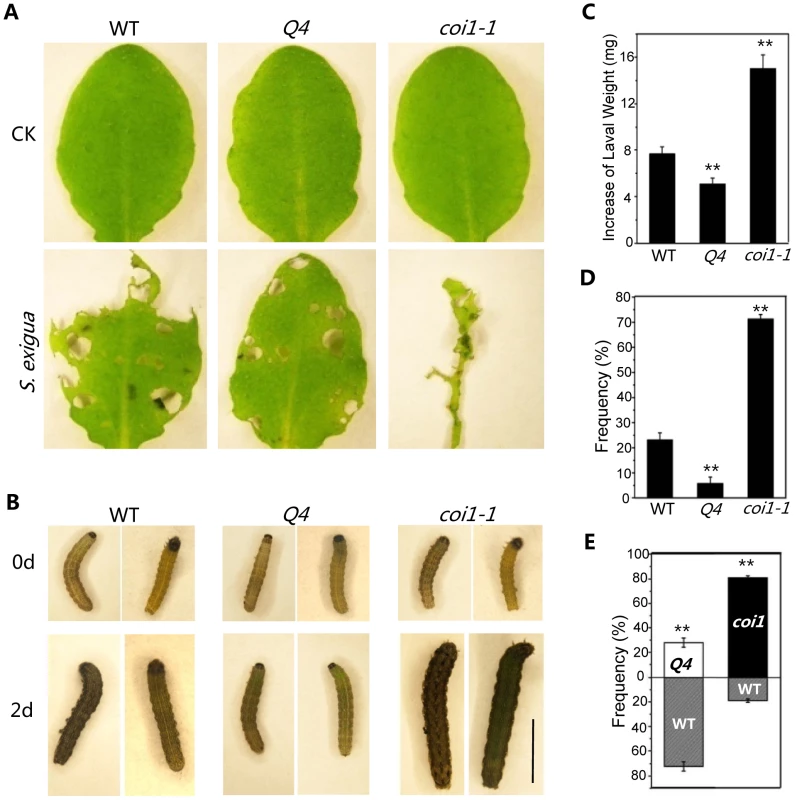
(A) Representative rosette leaves of wild-type (WT), the quadruple mutant bhlh3 bhlh13 bhlh14 bhlh17 (Q4), and coi1-1 after feeding without (CK) or with S. exigua. (B) Representative S. exigua larvae before feeding (0 d) or two days after feeding (2 d) with rosette leaves WT, Q4 and coi1-1. The scale bar represents 5 mm. (C) Increase of S. exigua larval weight two days after feeding with rosette leaves of WT, Q4 and coi1-1. Error bars represent SE. Asterisks represent Student's t-test significance compared with WT plants: **, P<0.01. (D) The preference of S. exigua larvae in the three-choice test among WT, Q4 and coi1-1. Error bars represent SE. Asterisks represent Student's t-test significance compared with WT plants: **, P<0.01. (E) The preference of S. exigua larvae in the two-choice test between WT and Q4, or between WT and coi1-1. Error bars represent SE. Asterisks represent Student's t-test significance compared with WT plants: **, P<0.01. In summary, we demonstrated that the bHLH subgroup IIId factors (bHLH3, bHLH13, bHLH14 and bHLH17) function as novel negative regulators of JA-mediated plant defense.
Overexpression of the Subgroup IIId bHLH Factor Caused Reduced JA Responses
Having demonstrated that the quadruple mutant exhibited enhanced JA responses (Figures 4–6), we further investigated whether overexpression of the bHLH subgroup IIId factors would lead to reduction in JA responses. As shown in Figure 7, the bHLH17 overexpression lines, 17OE-2 and 17OE-4 with highly expressed bHLH17 transcripts (Figure 7A), exhibited reduced JA responses compared with wild type, as indicated by reduction in JA-induced anthocyanin accumulation (Figures 7B and 7C), JA-inducible anthocyanin biosynthetic gene expression (Figure 7D), and JA-inhibitory root growth (Figures 7F and 7G). Similar to the bHLH17 overexpression plants, the bHLH13 overexpression lines (13OE-3 and 13OE-7) also exhibited reduced JA responses (Figure 7). These overexpression lines also exhibited the coi1-like phenotype to flower early (Figure 7H). Consistent with the decreased JA responses, JA-inducible gene expression of VSP1, LOX2, and PDF1.2 was reduced in the bHLH13 or bHLH17 overexpression lines (Figure 7E).
Fig. 7. Overexpression of bHLH 13 or bHLH 17 attenuated JA responses. 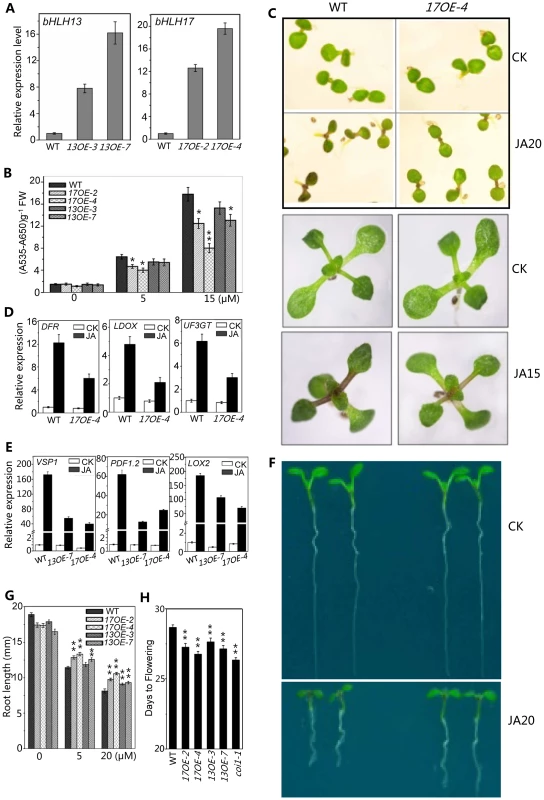
(A) Quantitative real-time PCR analysis of expression levels of bHLH13 and bHLH17 in 11-day-old seedlings of WT and transgenic plants overexpressing bHLH13 or bHLH17. Two individual transgenic lines for bHLH13 (13OE-3 and 13OE-7) and bHLH17 (17OE-2 and 17OE-4) were representatively used in analysis. ACTIN8 was used as the internal control. (B) Anthocyanin contents of the indicated 11-day-old seedlings grown on MS medium supplied with indicated concentrations of MeJA. FW, fresh weight. Error bars represent SE (n = 3). Asterisks represent Student's t-test significance compared with WT plants: *, P<0.05; **, P<0.01. (C) Phenotypes of 7-day-old (upper panel) and 11-day-old (bottom panel) seedlings of WT and 17OE-4 grown on MS medium supplied without (CK) or with indicated concentrations (µM) of MeJA. (D) Quantitative real-time PCR analysis of expression levels of DFR, LDOX and UF3GT in 11-day-old seedlings of WT and 17OE-4 grown on MS medium supplied without (CK) or with 15 µM MeJA (JA). ACTIN8 was used as the internal control. (E) Quantitative real-time PCR analysis of expression levels of VSP1, PDF1.2 and LOX2 in 11-day-old seedlings of WT, 13OE-7 and 17OE-4 treated with mock (CK) or with 100 µM MeJA (JA) for 6 hours. ACTIN8 was used as the internal control. (F) Root phenotypes of 7-day-old WT and 17OE-4 seedlings grown on MS medium supplied without (CK) or with 20 µM MeJA. (G) Root length of 11-day-old WT, bHLH13 and bHLH17 overexpression transgenic seedlings grown on MS medium containing indicated concentrations of MeJA. Error bars represent SE (n = 15). Asterisks represent Student's t-test significance compared with WT plants: **, P<0.01. (H) Flowering time of WT, bHLH13 and bHLH17 overexpression transgenic lines. Data shown are the means from 24 plants. Error bars represent SE. Asterisks represent Student's t-test significance compared with WT plants: **, P<0.01. Taken together with data of the genetic and physiological analysis on the quadruple mutant and the overexpression transgenic lines (Figures 4–7), we demonstrated that the bHLH3, bHLH13, bHLH14 and bHLH17 function as novel negative regulators of JA responses.
The bHLH Subgroup IIId Factors Act as Transcription Repressors
Having shown that the bHLH subgroup IIId factors (bHLH3, bHLH13, bHLH14 and bHLH17) negatively regulat JA responses, we examined whether these bHLH subgroup IIId factors function as transcriptional repressors using the GAL4-DNA - binding-domain (GAL4DB) and its binding sites (GAL4(4X)-D1-3(4X)-GUS)-based protoplast transient expression system [57], [58]. We found that the MYC2 functions as a transcription activator whereas JAZ1 acts as a negative regulator to inhibit the MYC2 activation activity in this transient expression system (Figure 8A and 8B). In contrast, bHLH17 functions as a transcription repressor whereas JAZ1 acts as a negative regulator to inhibit the bHLH17 repression function: expression of GAL4DB-fused bHLH17 obviously repressed the activity of the GUS reporter (Figure 8A and 8B); furthermore, coexpression of JAZ1 inhibited the bHLH17 repression function and rescued the bHLH17-repressed GUS activity in a dosage-dependent manner (Figure 8B).
Fig. 8. bHLH3, bHLH13, bHLH14 and bHLH17 act as transcription repressors. 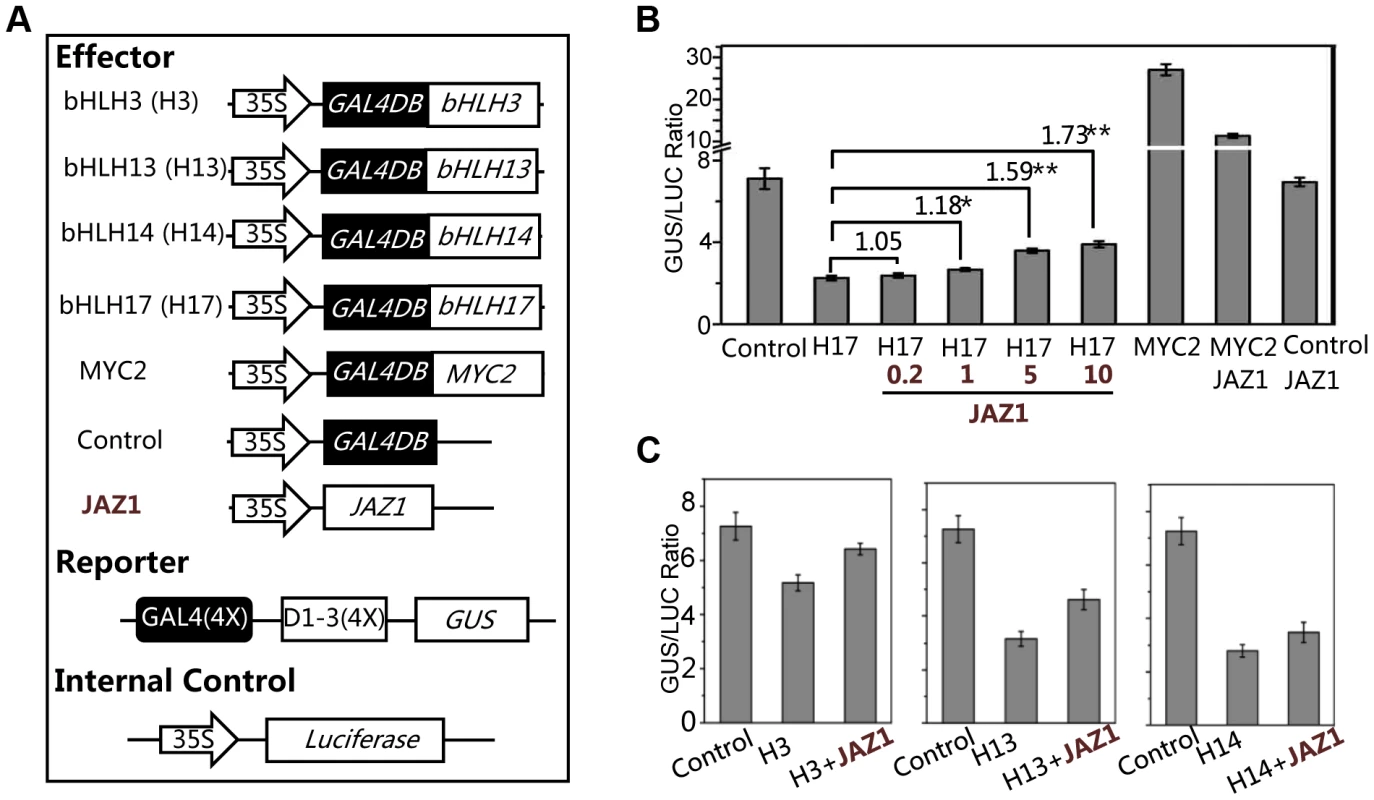
(A) The schematic diagram shows the constructs used in the transient expression assays. (B) Transient expression assays show that the bHLH17 (indicated by H17) acts as a transcription repressor, while JAZ1 attenuates the bHLH17 repression function in a dosage-dependent manner. The MYC2 functions as a transcription activator, while JAZ1 represses the MYC2 activation activity. The GUS reporter and the internal control luciferase were cotransformed with the Control, bHLH17 (H17) and MYC2 effectors. The 0.2, 1, 5 or 10 µg JAZ1 plasmid DNA were respectively used for co-transformation with bHLH17 (H17). 10 µg JAZ1 plasmid DNA was used for co-transformation with MYC2 and Control. When JAZ1 was used for co-transformation with the Control vector only, the GUS activity was not affected, which is consistent with the previous observations that the negative regulator JAZ1 is not a transcription factor and cannot repress the promoter sequences (JAZ1 interacts with and represses its targeted proteins/transcription factors). Numbers on the brackets indicate the relative values of the GUS/LUC ratio of bHLH17 with JAZ1 to that of bHLH17. The GUS/LUC ratio represents the GUS activity relative to the internal control LUC. Error bars represent SE (n = 3). Asterisks represent Student's t-test significance between pairs indicated with brackets (*, P<0.05; **, P<0.01). (C) Transient expression assays show that bHLH3, bHLH13 and bHLH14 act as transcription repressors and their repression function can be attenuated by JAZ1. The GUS reporter and the luciferase (LUC) internal control were cotransformed with the indicated constructs (10 µg for each construct). Error bars represent SE (n = 3). Similarly, we observed that GAL4DB-fused bHLH3, bHLH13 or bHLH14 repressed the activity of GUS reporter, and that their repression function could be partially inhibited by JAZ1 (Figure 8C). These results collectively suggest that these bHLH subgroup IIId factors act as transcriptional repressors, and that JAZ proteins interact with these transcription factors to attenuate their repression function.
The bHLH Subgroup IIId Factors Antagonize Transcription Activators in JA Pathway
We further generated a reporter construct PDFR-LUC in which the LUC reporter was driven by the promoter of DFR (Figure 9A), a direct target of the WD-repeat/TT8/MYB75 complex [46], [59], [60]. Consistent with previous data [61], [62], TT8 and MYB75 act as transcription activators to significantly induce expression of PDFR-LUC in the transient transcriptional activity assays (Figures 9A and 9B). We further found that bHLH17 repressed the TT8/MYB75-activated PDFR-LUC expression in a dosage-dependent manner (Figure 9B). Similarly, bHLH3 also repressed the TT8/MYB75-activated PDFR-LUC expression (Figure 9C).
Fig. 9. The bHLH3 and bHLH17 antagonize TT8/MYB75 and MYC2 to negatively regulate their downstream target genes. 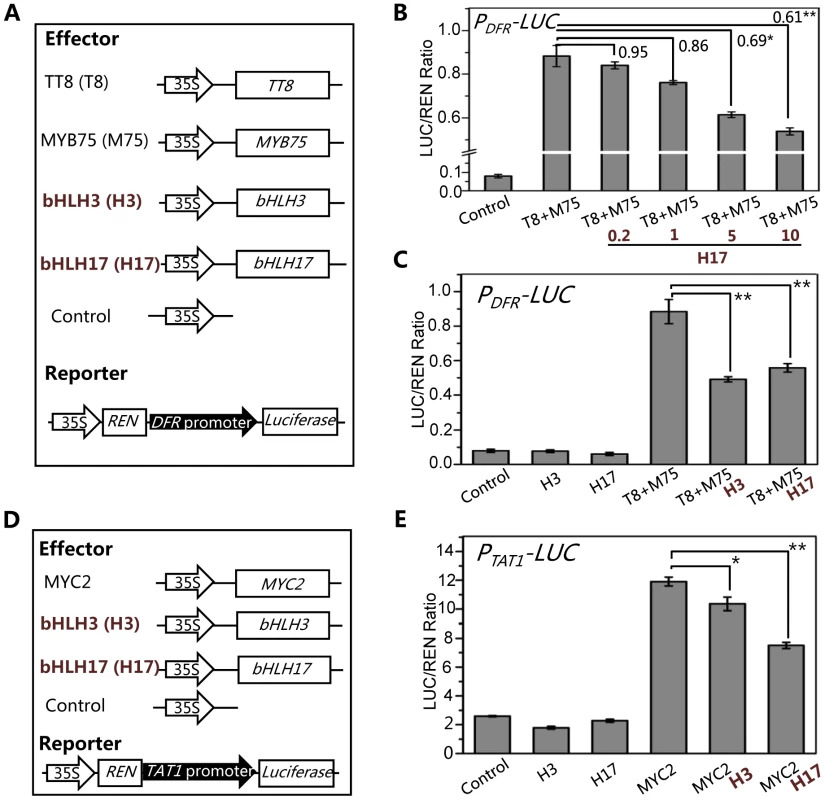
(A) The schematic diagram shows the constructs used in the transient transcriptional activity assays of (B) and (C). (B) Transient transcriptional activity assays show that activation of DFR promoter by TT8/MYB75 is repressed by bHLH17 in dosage-dependent manner. The PDFR-LUC reporter was cotransformed with the indicated constructs. The 0.2, 1, 5 or 10 µg bHLH17 (H17) plasmid DNA were respectively used for co-transformation with TT8 (T8) and MYB75 (M75). Numbers on the brackets indicate the relative values of the LUC/REN ratio of TT8/MYB75 with bHLH17 to that of TT8/MYB75. The LUC/REN ratio represents the PDFR-LUC activity relative to the internal control (REN driven by 35S promoter). Error bars represent SE (n = 3). Asterisks represent Student's t-test significance between pairs indicated with brackets (*, P<0.05; **, P<0.01). (C) Transient transcriptional activity assays show that activation of DFR promoter by TT8/MYB75 is repressed by bHLH3 and bHLH17. The PDFR-LUC reporter was cotransformed with the indicated constructs (10 µg for each construct). Error bars represent SE (n = 3). Asterisks represent Student's t-test significance (**, P<0.01). (D) The schematic diagrams show the constructs used in the transcriptional activity assays of (E). (E) Transient transcriptional activity assays show that activation of TAT1 promoter by MYC2 is repressed by bHLH3 and bHLH17. The PTAT1-LUC reporter was cotransformed with the indicated constructs (10 µg for each construct). Error bars represent SE (n = 3). Asterisks represent Student's t-test significance (*, P<0.05; **, P<0.01). Using similar approach, we generated a reporter construct PTAT1-LUC in which the LUC reporter was driven by the promoter of TAT1 (Figure 9D), a direct target of MYC2 [63]. Consistent with previous data [63], MYC2 acts as transcription activator to significantly induce expression of PTAT1-LUC in the transient transcriptional activity assays (Figure 9E). Furthermore, we found that bHLH3 and bHLH17 were able to repress the MYC2-activated TAT1 expression (Figures 9E). These results collectively suggest that the bHLH subgroup IIId factors antagonize transcription activators (such as MYC2, TT8 and MYB75) in JA pathway.
The Y2H and BiFC assays detected no direct interactions between the transcription repressor (bHLH3, bHLH13, bHLH14 or bHLH17) and transcription activator (such as MYC2 or TT8/MYB75) (Figures S2, S3, S4), which excluded the possibility that the bHLH subgroup IIId factors antagonize the previously reported transcription activators via direct interactions. Previous studies showed the transcription activators TT8/MYB75 and MYC2 bind to and activate their respective target sequences, such as promoters of DFR and TAT1 [61]–[63]. Here we used chromatin immunoprecipitation (ChIP)-PCR assays to investigate whether the bHLH subgroup IIId factors antagonize these transcription activators through binding to their target sequences. The ChIP-PCR assays showed that DFR promoter sequence, spanning two G-box motifs (CACGTG) at the position from −182 to −154, was highly enriched in the anti-myc-immunoprecipitated chromatin of the myc-bHLH3 transgenic plant, but not in the controls (the anti-myc-pulled wild-type chromatin, the empty beads-pulled chromatin of wild-type or the myc-bHLH3 transgenic plant) (Figure 10A), demonstrating a direct binding of myc-bHLH3 to the promoter sequence of DFR. Using the similar approach, we also detected the association of myc-bHLH3 with promoter of TAT1 (Figure 10B).
Fig. 10. bHLH3 directly binds to promoter sequences of TAT1 and DFR in ChIP-PCR assay. 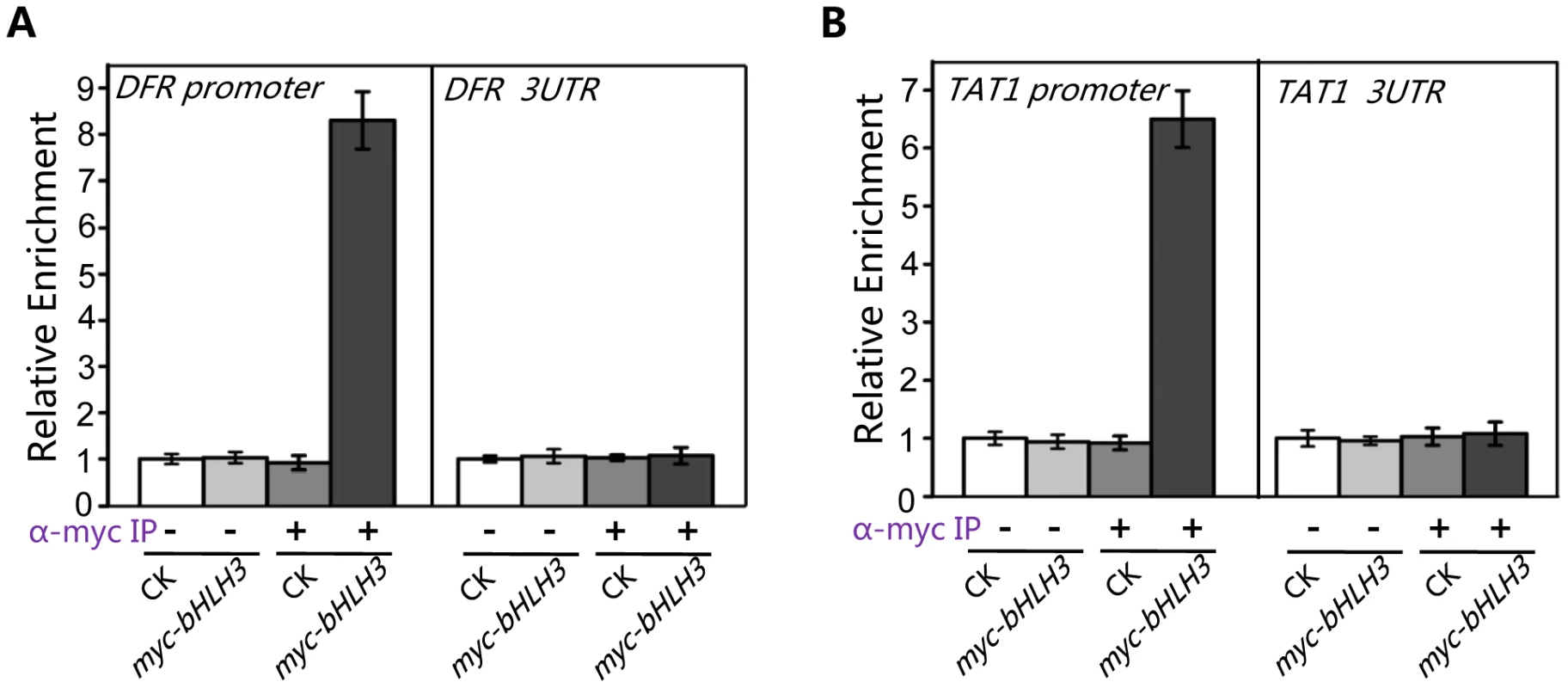
(A) ChIP-PCR analysis of the in vivo binding of myc-bHLH3 to the promoter of DFR. Chromatin from wild type plants (CK) and the transgenic plants constitutively expressing myc-bHLH3 (myc-bHLH3) was immunoprecipitated without (−) or with anti-myc antibody (+). Levels of the DFR promoter sequence (a 267-bp fragment containing two G-box motifs (CACGTG)) in the indicated chromatin were quantified by quantitative real-time PCR assay. PCR-amplification of a 77-bp 3′-UTR fragment of DFR was used as a negative control. The experiment was repeated three biological times with similar results. Error bars represent SE (n = 3). (B) ChIP-PCR analysis of the in vivo binding of myc-bHLH3 to the promoter of TAT1. Levels of the TAT1 promoter sequence (a 111-bp fragment containing one G-box motif) in the indicated chromatin were quantified by quantitative real-time PCR assay. PCR-amplification of a 82-bp 3′-UTR fragment of TAT1 was used as a negative control. The experiment was repeated three biological times with similar results. Error bars represent SE (n = 3). Taken together, our results suggest that bHLH3, bHLH13, bHLH14 and bHLH17 factors function as transcriptional repressors to repress the JA responses. These transcriptional repressors antagonize the previously reported transcription activators through binding to their downstream target sequences.
Discussion
The JAZ protéines [20], [29], [30], through formation of a core repression complex with TOPLESS and the NOVEL-INTERACTOR-OF-JAZ [57], are speculated to negatively regulate JA-mediated plant responses via interaction with and attenuation of their target transcription factors such as the bHLH subgroup IIIe transcription factors (MYC2, MYC3, MYC4) [29], [39]–[41], the R2R3-MYB transcription factors (MYB21, MYB24 and MYB57) [12], and the WD-repeat/bHLH (TT8, GL3 or EGL3)/MYB (MYB75 or GL1) complexes [14]. Here, we identified four bHLH subgroup IIId transcription factors (bHLH3, bHLH13, bHLH14 and bHLH17) as new targets of JAZ proteins (Figure 1). These transcription factors function as negative regulators to repress JA responses (Figures 4–7). Furthermore, our results showed that these negative regulators act as transcriptional repressors to antagonize the positive transcription factors through binding to their downstream target sequences (Figures 8–10). Coordinated regulation of JA responses by transcription repressors and transcription activators may benefit plants for adaptation to their frequently changing nature habitat.
Genetic and physiological analysis on the single, double, triple and quandruple mutants (bhlh3, bhlh13, bhlh14, bhlh17, bhlh3 bhlh17, bhlh3 bhlh13 bhlh17, bhlh3 bhlh13 bhlh14, bhlh3 bhlh14 bhlh17, bhlh13 bhlh14 bhlh17, bhlh3 bhlh13 bhlh14 bhlh17) showed that these bHLH subgroup IIId transcripiton factors function redundantly to repress JA responses (Figures 3–6 and S1). The quandruple mutant bhlh3 bhlh13 bhlh14 bhlh17 exhibited significant increase in JA responses, while the single mutant bhlh3, bhlh13, bhlh14 or bhlh17 displayed no obvious alterations in the tested JA responses (Figures 3–6 and S1), though we cannot exclude possibility that some JA responses may be mildly altered in single mutants. As shown in Figure 2 C and D, bhlh3 showed a mild increase in JA-inducible anthocyanin accumulation. Recent study also showed that the single mutant bhlh17/jam1 (another T-DNA insertion mutant of bHLH17/JAM1) displayed no obvious alteration in JA-inhibitory root growth, but exhibited the enhanced sensitivity in JA-inducible anthocyanin accumulation and defense against insect [64]. The functional redundancy among these bHLH3, bHLH13, bHLH14 and bHLH17 factors may result from their high similarity at amino acid level. It would be interesting to investigate whether and how these transcription factors exhibit homo - and heterodimerization to exert their redundant functions.
In addition to their redundant function, it is not clear whether the bHLH3, bHLH13, bHLH14 and bHLH17 factors have distinct roles in regulation of JA responses. The bHLH13 and bHLH17 exhibited similar interaction patterns with 10 JAZ proteins, and with the Jas domain of JAZ8/JAZ11 (Figure 1). However, for bHLH3 and bHLH14, they interacted with 7 JAZ proteins, and the full length of JAZ8/JAZ11 was required for interactions with bHLH3 and bHLH14 (Figure 1). Furthermore, the COI1-dependent and JA-induced gene expression was observed for bHLH13 and bHLH17, but not for bHLH3 and bHLH14 (Figure 2D). Interestingly, both bHLH3 and bHLH17 were nucleus-localized (Figure 2C), while bHLH13 and bHLH14 were localized in both nucleus and cytoplasm (Figure 2C). It remains to be elucidated whether these distinguished features would lead to distinct roles for these four transcription factors in JA pathway. Current studies so far have showed that bHLH17/AtAIB was positively involved in ABA signaling [43]. It is not clear whether the bHLH subgroup IIId factors play positive or negative roles in other signal pathways.
Previous studies showed that MYC2, and the WD-repeat/bHLH (TT8, GL3 or EGL3)/MYB (MYB75 or GL1) complexes act as transcription activators which bind to and activate promoter sequences of their respective target genes, such as TAT1 [63], and DFR [46], [60], [61]. We showed that the bHLH subgroup IIId factors act as transcription repressors (Figure 8), which bind to the promoter sequences of TAT1 and DFR (Figure 9 and 10), to antagonize the transcription function of MYC2 and MYB75/TT8 (Figure 9). It is interesting to investigate whether the bHLH subgroup IIId factors antagonize the activation function of MYC2 and MYB75/TT8 through competitive or concurrent binding to these target promoter sequences. Fernandez-Calvo et al. predicted that the activation domain is localized at the N-terminus of MYC2 [40]. It remains to experimentally investigate which domain in MYC2 (or in the bHLH subgroup IIId factors) is responsible for activation (or repression) of their target sequences.
The GAL4DB-based protoplast transient expression system is a well-established method for determination of transcription activators and repressors [57], [58], [65]–[68]. MYC2 acts as a transcription activator in this transient expression system (Figure 8A and 8B), which is consistent with previous observations [39], [57], [63], [69]. Consistent with its activation function, the MYC2 is a key gene that positively regulates a vast array of JA-responsive genes and diverse aspects of JA responses, including root growth [22], [70], anthocyanin accumulation [71], sesquiterpene synthase [72], nicotine biosynthesis [73], wound response [74], oxidative stress tolerance [75], and plant defense against both bacterial pathogen Pst DC3000 [40] and insects (Spodoptera littoralis and Helicoverpa armigera) [40], [75]. Interestingly, the MYC2 gene was shown to negatively regulate defense against necrotrophic fungi (B. cinerea and Plectosphaerella cucumerina) and expression of the related genes, such as PDF1.2 [22], [75], [76]. We showed that the bHLH subgroup IIId factors act as transcription repressors (Figures 8), which antagonize the activation function of MYC2 and TT8/MYB75 (Figure 9), to negatively regulate all the tested JA responses, including root growth, flowering, anthocyanin accumulation, and plant defense responses against bacterial pathogen Pst DC3000, necrotrophic pathogen B. cinerea, and insect S. exigua (Figures 4–7). Generation and characterization of the penta mutant myc2 bhlh3 bhlh13 bhlh14 bhlh17 would clarify whether mutations in the bHLH subgroup IIId factors are able to rescue the myc2-associated reduction of JA responses (such as root growth, anthocyanin accumulation, wound response, and defense against bacterial pathogen and insects), and to additively or synergistically affect the myc2-associated increase of defense response against necrotrophic pathogen B. cinerea.
It is speculated that, in response to JA signal, JAZ proteins are recruited by SCFCOI1 for ubiquitination and subsequent degradation. As a result, the JAZ-targeted transcription activators and repressors (bHLH3, bHLH13, bHLH14 and bHLH17) are released to antagonistically and coordinately regulate their target genes (such as TAT1 and DFR), which may further modulate expression of JA responsive genes essential for various JA responses (Figure 11). Plants live in fixed places and have to evolve sophisticated systems for adaptation to their frequently changing environment. The antagonistic and coordinated regulation of JA responses by transcription repressors and transcription activators may provide an important strategy for plant survival in their complicated nature habitat. It is possible that plants evolve this type of repressor-mediated negative regulation system to provide a fine feedback regulatory mechanism for avoiding exhausted and harmful excess JA responses.
Fig. 11. A simplified model for coordinated regulation of JA responses by JAZ-targeted transcription activators and transcription repressors. 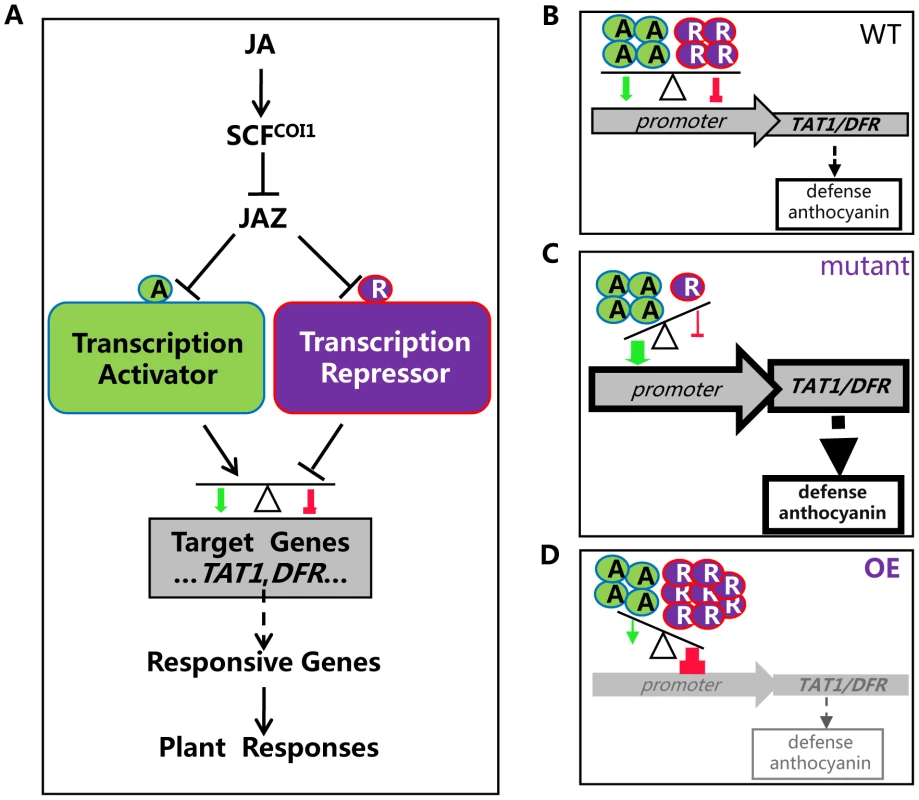
(A) JAZ proteins interact with transcription activators (such as MYC2, TT8 and MYB75) and transcription repressors (including bHLH3, bHLH13, bHLH14 and bHLH17) to attenuate their transcriptional function. Upon perception of JA, SCFCOI1 recruits JAZs for ubiquitination and degradation via 26S proteasome. The JAZ-targeted transcription activators and repressors are then released to antagonistically and coordinately regulate their target genes (such as TAT1 and DFR), which may further regulate JA responsive genes essential for various JA responses. (B) In wild type plant, both transcription activators and repressors, in response to JA signal, are released to regulate expression of their mutual target genes (such as TAT1 and DFR). The balance between repression and activation would lead to an appropriate output of JA-responsive genes, resulting in an appropriate level of JA responses such as plant defense and anthocyanin accumulation. A, transcription activator; R, transcription repressor. (C) In the bhlh3 bhlh13 bhlh14 bhlh17 quadruple mutant, the transcription repressors were severely abolished. The balance between repression and activation would lead to enhanced output of JA-responsive genes, resulting in increased JA responses. (D) In the plants overexpressing the bHLH subgroup IIId factors (OE), high level of the transcription repressors would exhibit strongest repression. The balance between repression and activation would lead to decreased output of JA-responsive genes, resulting in reduced JA responses. Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana mutant coi1-1 [77] was described as previously. bhlh3 (CS428877/GK-301G05), bhlh13 (CS466724/GK-696A04), bhlh14 (CS27164/GT1193.Ds3.05.24.99.b.288) and bhlh17 (CS874647/SAIL_536_F09) were obtained from the ABRC or NASC. The double, triple and quadruple mutants of the bHLH subgroup IIId factors were generated through genetic crossing among bhlh3, bhlh13, bhlh14 and bhlh17.
Arabidopsis thaliana seeds were sterilized with 20% bleach, plated on Murashige and Skoog medium (MS; Sigma-Aldrich), chilled at 4°C for 3 days, and transferred to a growth room under a 16-h (22–24°C)/8-h (16–19°C) light/dark photoperiod. Nicotiana benthamiana was grown in a growth room under a 16-h (28°C)/8-h (22°C) light/dark condition.
Y2H Screening and Y2H Assays
The yeast two-hybrid screening method was described as previously [12], [14].
For Y2H assay, all the CDS of JAZs, bHLH3, bHLH13, bHLH14, bHLH17, MYC2, MYC3, MYC4, TT8, GL3, EGL3, MYB75, GL1 and TTG1, and their domain derivatives were cloned into pLexA or pB42AD vectors. Primers used for the vector construction are presented in Table S1. The method details for yeast transformation and interaction detection using EGY48 were described as previously [12], [14]. Y2H images were taken 3 days after incubation at 30°C. Experiments were repeated three biological times.
BiFC Assays
For BiFC assays, full-length coding sequences of Arabidopsis JAZ1, JAZ10, bHLH3, bHLH13, bHLH14, bHLH17, MYC2, MYC3, MYC4, TT8, GL3, EGL3, MYB75 and GL1 were cloned into the binary nYFP or cYFP vector through enzyme digestion sites (KpnI/SalI) or Gateway reaction with pDONR207 vector system (Invitrogen) [12], [14]. Primer pairs for generation of constructs are listed in Table S1. Agrobacterium strains with indicated nYFP or cYFP vector were incubated, harvested, and resuspended in infiltration buffer (0.2 mM acetosyringone, 10 mM MgCl2, and 10 mM MES). Equal concentrations and volumes of Agrobacterium strains were mixed and coinfiltrated into N. benthamiana leaves by a needleless syringe. After infiltration, plants were placed at 24°C for 50 h before observation. The experiments were repeated three biological times.
Quantitative Real-time PCR
In Figure 2B, root, stem, rosette leaf, stem leaf and flowers were harvested for RNA extraction and subsequent reverse transcription. In Figures 2D and 7E, Arabidopsis seedlings were grown on MS medium for 11 days and then were treated with or without 100 µM MeJA for indicated time. In Figures 4B, 4E, 5E and 7D, Arabidopsis seedlings were grown on MS medium supplied with or without indicated concentration of MeJA for eleven days. These materials were harvested for RNA extraction and subsequent reverse transcription.
Real-time PCR analyses were performed with the RealMasterMix (SYBR Green I) (Takara) using the ABI7500 real-time PCR system as described previously [14]. ACTIN8 was used as the internal control. The primers used for real-time PCR analysis are presented in Table S2. All the experiments were repeated three biological times with similar results.
Protoplast Transfection Assays
For transient expression assay in Arabidopsis protoplast using the GUS reporter, the CDS of bHLH3, bHLH13, bHLH14, bHLH17 and MYC2 were fused with the GAL4DB under control of 35S promoter. The coding sequence of JAZ1 was cloned into the pGreenII 62-SK vector under control of 35S promoter [78]. Primers used for plasmid construction were shown in Table S1. Four copies of upstream GAL4 DNA binding sites (GAL4(4x)-D1-3(4x)) were used to drive the GUS gene generating the GUS reporter construct [58], [68]. The internal control contains a firefly luciferase gene (LUC) under control of 35S promoter. Arabidopsis mesophyll protoplasts preparation and subsequent transfection were performed as described previously [79]. Relative GUS activity was normalized against the LUC activity. In the JAZ1 dosage effect test of Figure 8B, 0.2, 1, 5 or 10 µg JAZ1 plasmid was respectively used, while for other reporter and effectors, 10 µg plasmid was used.
For transient transcriptional activity assay using the LUC reporter, the CDS sequences of MYC2, TT8, MYB75, bHLH3 and bHLH17 were cloned into pGreenII 62-SK vectors under control of 35S promoter respectively. The ∼519 bp and ∼980 bp promoter sequences of DFR and TAT1 were amplified from genomic DNA and cloned into pGreenII 0800-LUC respectively [78]. All primers used for making these constructs are listed in Table S1. After protoplasts preparation and subsequent transfection, firefly luciferase (LUC) and renillia luciferase (REN) activities were measured using the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer's instructions. Relative firefly luciferase (LUC) activity was calculated by normalizing against the renillia luciferase activity. In the experiment of bHLH17 dosage effect in Figure 9B, 0.2, 1, 5 or 10 µg bHLH17 plasmid was respectively used, while for other reporter and effectors, 10 µg plasmid was individually used. All the experiments were repeated three biological times with similar results.
ChIP Assays
The ChIP experiment was performed as described previously [80] using leaves of the 4-week-old myc-bHLH3 transgenic plants and the wild-type (Col-0) plants treated with 100 µM MeJA for 40 minutes. Immunoprecipitation was performed using Mouse anti-MYC antibody and protein G agarose beads. Enrichment of promoter DNA was confirmed by qRT-PCR using ACTIN2 as normazlization control. Primers for 3′UTR region were used as negative control. Primers for the ChIP assays are listed in Table S2. The experiments were repeated for three biological repeats with similar results.
Generation of Transgenic Plants
To generate Arabidopsis transgenic plants overexpressing bHLH13 and bHLH17, the full-length coding sequences of bHLH13 and bHLH17 were amplified and cloned into the modified pCAMBIA1300 vector under control of 35S promoter through the SalI and SpeI sites. To generate Arabidopsis myc-bHLH3 transgenic plants, coding sequence of bHLH3 was cloned into pROK2-myc vectors for fusion with six myc tags. These constructs were introduced into Arabidopsis plants using the Agrobacterium-mediated floral dip method. Two representative transgenic lines for bHLH13 and bHLH17 respectively were displayed in the article.
GUS Staining
∼2520 bp, ∼2500 bp, ∼1050 bp and ∼2570 bp promoter regions of bHLH3, bHLH13, bHLH14 and bHLH17 were respectively amplified and cloned into pCAMBIA1391Z vector to drive the GUS genes for generation of PbHLH3-GUS, PbHLH13-GUS, PbHLH14-GUS and PbHLH17-GUS. These constructs were transformed into Agrobacterium strains GV3101, and transferred into Arabidopsis by floral dip methods. Seedlings, inflorescences and roots from the transgenic plants harboring PbHLH3-GUS, PbHLH13-GUS, PbHLH14-GUS or PbHLH17-GUS were used for histochemical staining of GUS. Histochemical staining for GUS activity assay was performed as described previously [18].
Subcellular Localization
Coding sequences of bHLH3, bHLH13, bHLH14 and bHLH17 were respectively cloned into pEGAD vector for fusion with GFP under control of 35S promoter to generate the GFP-bHLH3, GFP-bHLH13, GFP-bHLH14 and GFP-bHLH17 constructs. The Agrobacterium containing the indicated constructs were resuspended in infiltration buffer (0.2 mM acetosyringone, 10 mM MgCl2, and 10 mM MES), and infiltrated into N. benthamiana leaves by a needleless syringe. After infiltration, plants were placed at 24°C for 50 h before GFP observation.
Anthocyanin Measurement
For anthocyanin measurement, 11-d-old Arabidopsis seedlings grown on MS medium with 0, 5, or 15 µM MeJA were measured as described previously [14] The anthocyanin content is presented as (A535–A650)/g fresh weight. The experiment was repeated three biological times.
Root Length Measurement
Seeds were grown on MS medium with 0, 5, 20 or 50 µM MeJA, chilled at 4°C for 3 days, and transferred to the growth room. Root lengths of fifteen 11-d-old seedlings for each genotype and treatment were measured and presented. The experiment was repeated three biological times.
Flowering Time
Flowering time of plants, grown in soil under long day condition with 16-h(22–25°C)/8-h(18–21°C) light/dark photoperiod, was recorded as the number of days from germination to the first appearance of buds in the rosette center. The experiment was repeated three biological times.
Infection with Pathogens
For Figures 5A and 5C, thirty-day-old plants were sprayed with Botrytis cinerea (106 spores/mL) solved in 0.025% tween or with 0.025% tween as control, placed in dark at the appropriate temperature (22°C) and high humidity (100%) for 36 hours, and transferred to a growth room under the growth conditions of a 16-h-light (21 to 23°C)/8-h-dark (16 to 19°C) photoperiod with ∼40% humidity. Infection symptoms were recorded at 7-day after infection. Infection ratings from 0 to 3 were assigned to the inoculated plants (0, no visible symptoms; 1, weak symptoms; 2, severe symptoms; 3, dead plants). For Figures 5B and 5D, four-week-old plants were sprayed with Botrytis cinerea (107 spores/mL) solved in 0.025% tween or with 0.025% tween as control, placed in dark at the appropriate temperature (22°C) and high humidity (100%) for 36 hours, transferred to a growth incubator under the growth conditions of a 16-h-light (21 to 23°C)/8-h-dark (16 to 19°C) photoperiod with high humidity (>90%). Plant survival ratio were recorded at 9-day after infection. At least thirty plants from each genotype were used in each experiment. The experiment was repeated three biological times.
Thirty-day-old plants were sprayed with Pseudomonas syringae pv tomato (Pst) DC3000 suspension containing 108 (colony-forming units)/mL bacteria (OD600 = 0.2) with 0.02% Silwet L-77 or 0.02% Silwet L-77 as control. Infection symptoms were recorded at 3-day after infection. At least thirty plants from each genotype were used in each experiment. The bacterial population counts in the plant was determined as previously described [81]. The experiment was repeated three biological times.
Insect Defense Assay with Spodoptera exigua
More than fifty mature rosette leaves with similar size from 4-week-old plants for each genotype were placed in one plastic Petri dishes (90 mm) containing wet filter paper. The 10 third-instar S. exigua larvae (∼8 mg each) were weighted, and reared on leaves in one Petri dish, for each genotype using five independent replicates. Two days after feeding, the weight of the 10 larvae were measured. The increase of average larval weight was recorded.
In the two-choice test, five rosette leaves from 4-week-old plants for each genotype were placed intervally in a circle in plastic Petri dishes (90 mm) containing wet filter paper. 40 newly hatched larvae were placed in the center of the Petri dishes for equal distance to the leaves. One day after incubation in the growth room, the numbers of larvae on leaves for each genotype were recorded. Four independent replicates were performed.
In the three-choice test, three rosette leaves from 4-week-old plants for each genotype were placed intervally in a circle in plastic Petri dishes (90 mm) containing wet filter paper. 40 newly hatched larvae were placed in the center of the Petri dishes for equal distance to the leaves. One day after incubation in the growth room, the numbers of larvae on leaves for each genotype were recorded. Six independent replicates were performed.
Accession Numbers
The Arabidopsis Genome Initiative numbers for genes mentioned in this article are as follows: JAZ1 (AT1G19180), JAZ2 (AT1G74950), JAZ3 (AT3G17860), JAZ4 (AT1G48500), JAZ5 (AT1G17380), JAZ6 (AT1G72450), JAZ7 (AT2G34600), JAZ8 (AT1G30135), JAZ9 (AT1G70700), JAZ10 (AT5G13220), JAZ11 (AT3G43440), JAZ12 (AT5G20900), bHLH3 (AT4G16430), bHLH13 (AT1G01260), bHLH14 (AT4G00870), bHLH17 (AT2G46510), MYC2 (AT1G32640), MYC3 (AT5G46760), MYC4 (AT4G17880), TT8 (AT4G09820), GL3 (AT5G41315), EGL3 (AT1G63650), MYB75 (AT1G56650), GL1 (AT3G27920), TTG1 (AT5G24520), DFR (AT5G42800), LDOX (AT4G22880), UF3GT (AT5G54060), VSP1 (AT5G24780), THI2.1 (AT1G72260), PDF1.2 (AT5G44420), LOX2 (AT3G45140), TAT1 (AT4G23600), ERF1 (AT3G23240) and ACTIN8 (AT1G49240).
Supporting Information
Zdroje
1. VanstraelenM, BenkovaE (2012) Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol 28 : 463–487.
2. JonesJD, DanglJL (2006) The plant immune system. Nature 444 : 323–329.
3. SpoelSH, DongX (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3 : 348–351.
4. SchallerA, StintziA (2009) Enzymes in jasmonate biosynthesis - structure, function, regulation. Phytochemistry 70 : 1532–1538.
5. WasternackC, HauseB (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot
6. CreelmanRA, MulletJE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48 : 355–381.
7. StaswickPE, SuWP, HowellSH (1992) Methyl jasmonate inhibition of root-growth and induction of a leaf protein are decreased in an Arabidopsis thaliana Mutant. Proc Natl Acad Sci USA 89 : 6837–6840.
8. FeysBJF, BenedettiCE, PenfoldCN, TurnerJG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male-sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 : 751–759.
9. SunJ, XuY, YeS, JiangH, ChenQ, et al. (2009) Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21 : 1495–1511.
10. ChenQ, SunJ, ZhaiQ, ZhouW, QiL, et al. (2011) The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23 : 3335–3352.
11. McConnM, BrowseJ (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8 : 403–416.
12. SongS, QiT, HuangH, RenQ, WuD, et al. (2011) The Jasmonate-ZIM Domain Proteins Interact with the R2R3-MYB Transcription Factors MYB21 and MYB24 to Affect Jasmonate-Regulated Stamen Development in Arabidopsis. Plant Cell 23 : 1000–1013.
13. LiL, ZhaoY, McCaigBC, WingerdBA, WangJ, et al. (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16 : 126–143.
14. QiT, SongS, RenQ, WuD, HuangH, et al. (2011) The Jasmonate-ZIM-Domain Proteins Interact with the WD-Repeat/bHLH/MYB Complexes to Regulate Jasmonate-Mediated Anthocyanin Accumulation and Trichome Initiation in Arabidopsis thaliana. Plant Cell 23 : 1795–1814.
15. YoshidaY, SanoR, WadaT, TakabayashiJ, OkadaK (2009) Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136 : 1039–1048.
16. LackmanP, Gonzalez-GuzmanM, TillemanS, CarqueijeiroI, PerezAC, et al. (2011) Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc Natl Acad Sci U S A 108 : 5891–5896.
17. YanJ, LiH, LiS, YaoR, DengH, et al. (2013) The Arabidopsis F-Box Protein CORONATINE INSENSITIVE1 Is Stabilized by SCFCOI1 and Degraded via the 26S Proteasome Pathway. Plant Cell doi:10.1105/tpc.112.105486.
18. ShanX, WangJ, ChuaL, JiangD, PengW, et al. (2011) A role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence. Plant Physiol 155 : 751–764.
19. RobsonF, OkamotoH, PatrickE, HarrisSR, WasternackC, et al. (2010) Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22 : 1143–1160.
20. YanYX, StolzS, ChetelatA, ReymondP, PagniM, et al. (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19 : 2470–2483.
21. HoweGA, JanderG (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59 : 41–66.
22. LorenzoO, ChicoJM, Sanchez-SerranoJJ, SolanoR (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 : 1938–1950.
23. ThalerJS, HumphreyPT, WhitemanNK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17 : 260–270.
24. HuP, ZhouW, ChengZ, FanM, WangL, et al. (2013) JAV1 Controls Jasmonate-Regulated Plant Defense. Mol Cell 50 : 504–515.
25. WathugalaDL, HemsleyPA, MoffatCS, CremelieP, KnightMR, et al. (2012) The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol 195 : 217–230.
26. SeoJS, JooJ, KimMJ, KimYK, NahmBH, et al. (2011) OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J 65 : 907–921.
27. YanJ, ZhangC, GuM, BaiZ, ZhangW, et al. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21 : 2220–2236.
28. FonsecaS, ChiniA, HambergM, AdieB, PorzelA, et al. (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nature Chemical Biology 5 : 344–350.
29. ChiniA, FonsecaS, FernandezG, AdieB, ChicoJM, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 : 666–671.
30. ThinesB, KatsirL, MelottoM, NiuY, MandaokarA, et al. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448 : 661–665.
31. KatsirL, SchilmillerAL, StaswickPE, HeSY, HoweGA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105 : 7100–7105.
32. SheardLB, TanX, MaoH, WithersJ, Ben-NissanG, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468 : 400–405.
33. BrowseJ (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60 : 183–205.
34. SantnerA, EstelleM (2007) The JAZ proteins link jasmonate perception with transcriptional changes. Plant Cell 19 : 3839–3842.
35. KazanK, MannersJM (2012) JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 17 : 22–31.
36. PauwelsL, GoossensA (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23 : 3089–3100.
37. De GeyterN, GholamiA, GoormachtigS, GoossensA (2012) Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci 17 : 349–359.
38. ShanX, YanJ, XieD (2012) Comparison of phytohormone signaling mechanisms. Curr Opin Plant Biol 15 : 84–91.
39. NiuY, FigueroaP, BrowseJ (2011) Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 62 : 2143–2154.
40. Fernandez-CalvoP, ChiniA, Fernandez-BarberoG, ChicoJM, Gimenez-IbanezS, et al. (2011) The Arabidopsis bHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses. Plant Cell 23 : 701–715.
41. ChengZ, SunL, QiT, ZhangB, PengW, et al. (2011) The bHLH Transcription Factor MYC3 Interacts with the Jasmonate ZIM-Domain Proteins to Mediate Jasmonate Response in Arabidopsis. Mol Plant 4 : 279–288.
42. ChengH, SongS, XiaoL, SooHM, ChengZ, et al. (2009) Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet 5: e1000440.
43. LiH, SunJ, XuY, JiangH, WuX, et al. (2007) The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol Biol 65 : 655–665.
44. HeimMA, JakobyM, WerberM, MartinC, WeisshaarB, et al. (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20 : 735–747.
45. Toledo-OrtizG, HuqE, QuailPH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 : 1749–1770.
46. GonzalezA, ZhaoM, LeavittJM, LloydAM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53 : 814–827.
47. GlazebrookJ (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 : 205–227.
48. ThommaBP, EggermontK, PenninckxIA, Mauch-ManiB, VogelsangR, et al. (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci U S A 95 : 15107–15111.
49. Mene-SaffraneL, DubugnonL, ChetelatA, StolzS, Gouhier-DarimontC, et al. (2009) Nonenzymatic oxidation of trienoic fatty acids contributes to reactive oxygen species management in Arabidopsis. J Biol Chem 284 : 1702–1708.
50. ParkJH, HalitschkeR, KimHB, BaldwinIT, FeldmannKA, et al. (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31 : 1–12.
51. PenninckxIA, EggermontK, TerrasFR, ThommaBP, De SamblanxGW, et al. (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8 : 2309–2323.
52. EppleP, ApelK, BohlmannH (1997) ESTs reveal a multigene family for plant defensins in Arabidopsis thaliana. FEBS Lett 400 : 168–172.
53. Berrocal-LoboM, MolinaA, SolanoR (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29 : 23–32.
54. MasonHS, DeWaldDB, MulletJE (1993) Identification of a methyl jasmonate-responsive domain in the soybean vspB promoter. Plant Cell 5 : 241–251.
55. MelottoM, UnderwoodW, KoczanJ, NomuraK, HeSY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126 : 969–980.
56. YangDL, YaoJ, MeiCS, TongXH, ZengLJ, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci U S A 109: E1192–1200.
57. PauwelsL, BarberoGF, GeerinckJ, TillemanS, GrunewaldW, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464 : 788–791.
58. TiwariSB, WangXJ, HagenG, GuilfoyleTJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13 : 2809–2822.
59. NesiN, DebeaujonI, JondC, PelletierG, CabocheM, et al. (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12 : 1863–1878.
60. ZhangF, GonzalezA, ZhaoM, PayneCT, LloydA (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130 : 4859–4869.
61. MatsuiK, UmemuraY, Ohme-TakagiM (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55 : 954–967.
62. ZimmermannIM, HeimMA, WeisshaarB, UhrigJF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40 : 22–34.
63. HouX, LeeLY, XiaK, YanY, YuH (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19 : 884–894.
64. NakataM, MitsudaN, HerdeM, KooAJ, MorenoJE, et al. (2013) A bHLH-Type Transcription Factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, Acts as a Repressor to Negatively Regulate Jasmonate Signaling in Arabidopsis. Plant Cell In Press: tpc.113.111112.
65. OhtaM, Ohme-TakagiM, ShinshiH (2000) Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J 22 : 29–38.
66. GuoS, XuY, LiuH, MaoZ, ZhangC, et al. (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4 : 1566.
67. TaoQ, GuoD, WeiB, ZhangF, PangC, et al. (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25 : 421–437.
68. ZhuJ, JeongJC, ZhuY, SokolchikI, MiyazakiS, et al. (2008) Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci U S A 105 : 4945–4950.
69. AbeH, Yamaguchi-ShinozakiK, UraoT, IwasakiT, HosokawaD, et al. (1997) Role of arabidopsis MYC and MYB homologs in drought - and abscisic acid-regulated gene expression. Plant Cell 9 : 1859–1868.
70. BoterM, Ruiz-RiveroO, AbdeenA, PratS (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18 : 1577–1591.
71. Laurie-BerryN, JoardarV, StreetIH, KunkelBN (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Molecular Plant-Microbe Interactions 19 : 789–800.
72. HongGJ, XueXY, MaoYB, WangLJ, ChenXY (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24 : 2635–2648.
73. ShojiT, HashimotoT (2011) Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol 52 : 1117–1130.
74. ZhangY, TurnerJG (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One 3: e3699.
75. DombrechtB, XueGP, SpragueSJ, KirkegaardJA, RossJJ, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19 : 2225–2245.
76. ZhaiQ, YanL, TanD, ChenR, SunJ, et al. (2013) Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity. PLoS Genet 9: e1003422.
77. XieDX, FeysBF, JamesS, Nieto-RostroM, TurnerJG (1998) COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 : 1091–1094.
78. HellensRP, AllanAC, FrielEN, BolithoK, GraftonK, et al. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1 : 13.
79. YooSD, ChoYH, SheenJ (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2 : 1565–1572.
80. SalehA, Alvarez-VenegasR, AvramovaZ (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3 : 1018–1025.
81. KatagiriF, ThilmonyR, HeSY (2002) The Arabidopsis thaliana-pseudomonas syringae interaction. Arabidopsis Book 1: e0039.
Štítky
Genetika Reprodukční medicína
Článek Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and MammalsČlánek Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative ElementsČlánek Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine ExposureČlánek Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 7
-
Všechny články tohoto čísla
- An Solution for Crossover Formation
- Genome-Wide Association Mapping in Dogs Enables Identification of the Homeobox Gene, , as a Genetic Component of Neural Tube Defects in Humans
- Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and Mammals
- Stepwise Activation of the ATR Signaling Pathway upon Increasing Replication Stress Impacts Fragile Site Integrity
- Genomic Analysis of Natural Selection and Phenotypic Variation in High-Altitude Mongolians
- Modification of tRNA by Elongator Is Essential for Efficient Translation of Stress mRNAs
- Role of CTCF Protein in Regulating Locus Transcription
- Gene Set Signature of Reversal Reaction Type I in Leprosy Patients
- Mapping of PARK2 and PACRG Overlapping Regulatory Region Reveals LD Structure and Functional Variants in Association with Leprosy in Unrelated Indian Population Groups
- Is Required for Formation of the Genital Ridge in Mice
- Monopolin Subunit Csm1 Associates with MIND Complex to Establish Monopolar Attachment of Sister Kinetochores at Meiosis I
- Recombination Dynamics of a Human Y-Chromosomal Palindrome: Rapid GC-Biased Gene Conversion, Multi-kilobase Conversion Tracts, and Rare Inversions
- Mechanisms of Protein Sequence Divergence and Incompatibility
- Histone Methyltransferase DOT1L Drives Recovery of Gene Expression after a Genotoxic Attack
- Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies
- Combinatorial Regulation of Meiotic Holliday Junction Resolution in by HIM-6 (BLM) Helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 Nucleases
- The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development
- The Role of Interruptions in polyQ in the Pathology of SCA1
- Dietary Restriction Induced Longevity Is Mediated by Nuclear Receptor NHR-62 in
- Fine Time Course Expression Analysis Identifies Cascades of Activation and Repression and Maps a Putative Regulator of Mammalian Sex Determination
- Genome-scale Co-evolutionary Inference Identifies Functions and Clients of Bacterial Hsp90
- Oxidative Stress and Replication-Independent DNA Breakage Induced by Arsenic in
- A Moonlighting Enzyme Links Cell Size with Central Metabolism
- Budding Yeast Greatwall and Endosulfines Control Activity and Spatial Regulation of PP2A for Timely Mitotic Progression
- The Conserved Intronic Cleavage and Polyadenylation Site of CstF-77 Gene Imparts Control of 3′ End Processing Activity through Feedback Autoregulation and by U1 snRNP
- The BTB-zinc Finger Transcription Factor Abrupt Acts as an Epithelial Oncogene in through Maintaining a Progenitor-like Cell State
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- The RNA-binding Proteins FMR1, Rasputin and Caprin Act Together with the UBA Protein Lingerer to Restrict Tissue Growth in
- Pattern Dynamics in Adaxial-Abaxial Specific Gene Expression Are Modulated by a Plastid Retrograde Signal during Leaf Development
- A Network of HMG-box Transcription Factors Regulates Sexual Cycle in the Fungus
- Bacterial Adaptation through Loss of Function
- ENU-induced Mutation in the DNA-binding Domain of KLF3 Reveals Important Roles for KLF3 in Cardiovascular Development and Function in Mice
- Interplay between Structure-Specific Endonucleases for Crossover Control during Meiosis
- FGF Signalling Regulates Chromatin Organisation during Neural Differentiation via Mechanisms that Can Be Uncoupled from Transcription
- The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression
- Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative Elements
- The Conserved ADAMTS-like Protein Lonely heart Mediates Matrix Formation and Cardiac Tissue Integrity
- The cGMP-Dependent Protein Kinase EGL-4 Regulates Nociceptive Behavioral Sensitivity
- RBM5 Is a Male Germ Cell Splicing Factor and Is Required for Spermatid Differentiation and Male Fertility
- Disease-Related Growth Factor and Embryonic Signaling Pathways Modulate an Enhancer of Expression at the 6q23.2 Coronary Heart Disease Locus
- Yeast Pol4 Promotes Tel1-Regulated Chromosomal Translocations
- A Dual Role for SOX10 in the Maintenance of the Postnatal Melanocyte Lineage and the Differentiation of Melanocyte Stem Cell Progenitors
- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Odoriferous Defensive Stink Gland Transcriptome to Identify Novel Genes Necessary for Quinone Synthesis in the Red Flour Beetle,
- Prediction of Complex Human Traits Using the Genomic Best Linear Unbiased Predictor
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
- Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine Exposure
- Exquisite Light Sensitivity of Cryptochrome
- miR-133a Regulates Adipocyte Browning In Vivo
- Strabismus Promotes Recruitment and Degradation of Farnesylated Prickle in Planar Polarity Specification
- Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
- Is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer
- Joint Molecule Resolution Requires the Redundant Activities of MUS-81 and XPF-1 during Meiosis
- The Mating Competence of Geographically Diverse Strains in Their Natural and Unnatural Sand Fly Vectors
- Defective Repair of Oxidative Base Lesions by the DNA Glycosylase Nth1 Associates with Multiple Telomere Defects
- Effective Blocking of the Enhancer Requires Cooperation between Two Main Mechanisms Suggested for the Insulator Function
- Trans-Ancestral Studies Fine Map the SLE-Susceptibility Locus
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Bacterial Adaptation through Loss of Function
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



