-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaBudding Yeast Greatwall and Endosulfines Control Activity and Spatial Regulation of PP2A for Timely Mitotic Progression
Entry into mitosis is triggered by cyclinB/Cdk1, whose activity is abruptly raised by a positive feedback loop. The Greatwall kinase phosphorylates proteins of the endosulfine family and allows them to bind and inhibit the main Cdk1-counteracting PP2A-B55 phosphatase, thereby promoting mitotic entry. In contrast to most eukaryotic systems, Cdc14 is the main Cdk1-antagonizing phosphatase in budding yeast, while the PP2ACdc55 phosphatase promotes, instead of preventing, mitotic entry by participating to the positive feedback loop of Cdk1 activation. Here we show that budding yeast endosulfines (Igo1 and Igo2) bind to PP2ACdc55 in a cell cycle-regulated manner upon Greatwall (Rim15)-dependent phosphorylation. Phosphorylated Igo1 inhibits PP2ACdc55 activity in vitro and induces mitotic entry in Xenopus egg extracts, indicating that it bears a conserved PP2A-binding and -inhibitory activity. Surprisingly, deletion of IGO1 and IGO2 in yeast cells leads to a decrease in PP2A phosphatase activity, suggesting that endosulfines act also as positive regulators of PP2A in yeast. Consistently, RIM15 and IGO1/2 promote, like PP2ACdc55, timely entry into mitosis under temperature-stress, owing to the accumulation of Tyr-phosphorylated Cdk1. In addition, they contribute to the nuclear export of PP2ACdc55, which has recently been proposed to promote mitotic entry. Altogether, our data indicate that Igo proteins participate in the positive feedback loop for Cdk1 activation. We conclude that Greatwall, endosulfines, and PP2A are part of a regulatory module that has been conserved during evolution irrespective of PP2A function in the control of mitosis. However, this conserved module is adapted to account for differences in the regulation of mitotic entry in different organisms.
Published in the journal: . PLoS Genet 9(7): e32767. doi:10.1371/journal.pgen.1003575
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003575Summary
Entry into mitosis is triggered by cyclinB/Cdk1, whose activity is abruptly raised by a positive feedback loop. The Greatwall kinase phosphorylates proteins of the endosulfine family and allows them to bind and inhibit the main Cdk1-counteracting PP2A-B55 phosphatase, thereby promoting mitotic entry. In contrast to most eukaryotic systems, Cdc14 is the main Cdk1-antagonizing phosphatase in budding yeast, while the PP2ACdc55 phosphatase promotes, instead of preventing, mitotic entry by participating to the positive feedback loop of Cdk1 activation. Here we show that budding yeast endosulfines (Igo1 and Igo2) bind to PP2ACdc55 in a cell cycle-regulated manner upon Greatwall (Rim15)-dependent phosphorylation. Phosphorylated Igo1 inhibits PP2ACdc55 activity in vitro and induces mitotic entry in Xenopus egg extracts, indicating that it bears a conserved PP2A-binding and -inhibitory activity. Surprisingly, deletion of IGO1 and IGO2 in yeast cells leads to a decrease in PP2A phosphatase activity, suggesting that endosulfines act also as positive regulators of PP2A in yeast. Consistently, RIM15 and IGO1/2 promote, like PP2ACdc55, timely entry into mitosis under temperature-stress, owing to the accumulation of Tyr-phosphorylated Cdk1. In addition, they contribute to the nuclear export of PP2ACdc55, which has recently been proposed to promote mitotic entry. Altogether, our data indicate that Igo proteins participate in the positive feedback loop for Cdk1 activation. We conclude that Greatwall, endosulfines, and PP2A are part of a regulatory module that has been conserved during evolution irrespective of PP2A function in the control of mitosis. However, this conserved module is adapted to account for differences in the regulation of mitotic entry in different organisms.
Introduction
Entry into mitosis in eukaryotic cells is driven by cyclin-dependent kinases (CDKs) bound to B-type cyclins. Many targets of cyclinB/CDKs have been identified in different organisms and include proteins involved in mitotic spindle formation/elongation, nuclear envelope breakdown, chromosome condensation and segregation.
Several mechanisms contribute to the rapid raise in cyclinB/CDKs activity at the onset of mitosis: i) cyclin accumulation, through transcriptional activation and inhibition of their proteolysis; ii) phosphorylation of their catalytic subunit Cdk1 by Cdk1-activating kinases (CAKs); iii) removal of Cdk1 inhibitory phosphorylations on Thr14 and Tyr15 (reviewed in [1]. Phosphorylation on Thr14 and Tyr15 of Cdk1 is carried out by Wee1 (Swe1 in budding yeast) and Myt1 protein kinases and is reversed by Cdc25-like phosphatases (Mih1 in budding yeast) that promote entry into mitosis. Polo kinase, which is activated in mitosis by cyclinB/Cdk1 [2], [3], activates in turn Cdc25 [4], [5]. In addition, the Wee1 kinase is phosphorylated and downregulated by cyclinB/Cdk1 [6], [7], [8], [9]. Together, these data have led to the idea of a positive autoregulatory loop for cyclinB/Cdk1 activation at the onset of M phase [10], [11]. In budding yeast a positive feedback loop for mitotic CDKs also exists, but differs from that of other organisms in many respects. The mitotic CDKs Clb1-4/Cdk1 drive a mitotic transcriptional program that leads to accumulation of several mitotic proteins, including the Clb1-4 cyclins themselves and the polo kinase Cdc5 (reviewed in [12]). Clb2-Cdk1 initially activates Swe1 through direct phosphorylation, thus leading to its own inhibition [6]. However, this phosphorylation primes further phosphorylations on Swe1 by multiple kinases, including Cdc5, that eventually target it to degradation [13].
Another key aspect of mitotic entry concerns the downregulation of phosphatases counteracting cyclinB/Cdk1 activity. In budding yeast, the Cdc14 phosphatase promotes inactivation of cyclinB/Cdk1 at the end of mitosis and reverses their phosphorylations [14], [15]. In interphase Cdc14 is kept inactive in the nucleolus [16], [17]. In all other eukaryotic cells analysed to date, phosphorylation events by mitotic CDKs are reversed mainly by PP2A and, to a lesser extent, by PP1 phosphatases (reviewed in [18]). Phosphatases are often protein complexes containing catalytic and regulatory subunits that confer substrate specificity. Core PP2A complexes are made of one catalytic (C subunit), one scaffold (A subunit) and one of many regulatory subunits (B subunit) [19]. Additional proteins, like the conserved Tap42, can also bind to PP2A and modulate its activity (reviewed in [20]). The B55 regulatory subunit, which exists in several isoforms, confers specificity of PP2A complexes towards Cdk1-dependent phosphorylation sites [21], suggesting that PP2A-B55 is the most relevant phosphatase in many eukaryotes for reversing Cdk1-dependent phosphorylations. Consistently, PP2A-B55δ prevents mitotic entry in Xenopus egg extracts [22], [23] and PP2A-B55α promotes mitotic exit in human cells [24].
In the past few years, the Greatwall protein kinase has emerged as a key factor for restraining PP2A-B55 activity in mitosis and allowing mitotic entry. In Xenopus egg extracts Greatwall is required for mitotic entry and to maintain the mitotic state [23], [25], [26], [27]. Depletion of Greatwall in Drosophila neuroblasts leads to mitotic defects that are compatible with a role of Greatwall in promoting entry into mitosis [28], [29]. Two closely related regulatory proteins, α-endosulfine (ENSA) and Arpp19, bind and inhibit PP2A-B55δ upon phosphorylation by Greatwall on a specific serine residue [30], [31]. Inactivation of Drosophila endosulfine leads to mitotic defects similar to those caused by Greatwall inactivation [32], while depletion of Arpp19 arrests Xenopus egg extracts in G2 [30], [31]. Indeed, Greatwall-dependent phosphorylation of α-endosulfine and Arpp19 promotes mitotic entry both by maintaining high levels of phosphorylated Cdk1 substrates and by feeding the Cdk1 autoregulatory loop (reviewed in [33], [34]).
In stark contrast to other organisms, budding yeast PP2ACdc55 promotes, rather than prevents, timely entry into mitosis by participating in the positive feedback loop for Cdk1 activation (reviewed in [20]). Indeed, PP2ACdc55 opposes the initial Swe1 phosphorylation by Cdk1, which stimulates Cdk1-Tyr19 inhibitory phosphorylation [35]. In addition, PP2ACdc55 dephosphorylates and activates Mih1 [36]. Consistently, mutants defective in PP2ACdc55 activity accumulate high levels of Tyr19-phosphorylated Cdk1 [35], [37], [38], [39].
Budding yeast possesses two redundant endosulfines, called Igo1 and Igo2 (Initiation of G zero), that are phosphorylated by the Greatwall-related kinase Rim15 on the same serine (Ser64 of Igo1 and Ser63 of Igo2) that in Xenopus Arpp19 and ENSA is phosphorylated by Greatwall [40]. Deletion of IGO1 and IGO2, as well as of RIM15, does not affect cell viability but compromises the establishment of the quiescence (G0) state. Indeed, Igo proteins phosphorylated by Rim15 bind to an mRNA decapping factor to shelter degradation of specific mRNAs during initiation of the quiescence program [40]. Furthermore, recent data indicate that Igo proteins help establishing the quiescence-specific transcriptional program through binding and inhibition of PP2A bound to its B subunit Cdc55 [41]. Similar to yeast, depletion of α-endosulfine by RNAi or deletion of its aminoacid sequence targeted by Greatwall has no obvious consequence on cell division and fertility in the worm C. elegans [42].
In this manuscript we report the roles of Rim15 and Igo proteins in the control of mitosis. In agreement with recently published data [41], we find that upon phosphorylation by Rim15 Igo1 binds to yeast PP2ACdc55 and can inhibit its activity in vitro. Consistently, phosphorylated Igo1 promotes mitotic entry in Xenopus egg extracts similarly to ENSA and Arpp19, suggesting that endosulfines from different species are interchangeable. However, phenotypic analyses of yeast mutants lacking Rim15 or Igo proteins, together with biochemical data, indicate that yeast endosulfines behave in vivo as activators, rather than inhibitors, of PP2ACdc55 and contribute to timely activation of mitotic CDKs. Our data indicate that Igo proteins help establishing the positive feedback loop for Cdk1 activation and retaining PP2ACdc55 in the cytoplasm [43], thereby promoting mitotic entry. We propose that endosulfines are accessory subunits of PP2A-B55 complexes that can contribute to their activation while inhibiting their phosphatase activity, to meet specific mitotic features in different organisms.
Results
Phosphorylation of Igo1 by Rim15 stimulates Igo1 interaction with PP2ACdc55 in late S/G2 phase
The budding yeast paralogous proteins Igo1 and Igo2 share significant homology with vertebrate endosulfines [44], especially around the serine that in endosulfines is phosphorylated by Greatwall (Gwl) (Fig. 1A). Indeed, Ser64 of Igo1 was shown to be phosphorylated by the yeast Greatwall-like kinase Rim15 [40]. To investigate if yeast endosulfines interact physically with PP2A in yeast cells, endogenous Igo1 was tagged at the C-terminus with 3 Pk epitopes and immunoprecipitated from yeast cells extracts. We then assessed the presence of various PP2A subunits in the immunoprecipitates. As shown in Fig. 1B, we found the catalytic subunit Pph21 and the Cdc55 regulatory subunit of PP2A to co-immunoprecipitate with Igo1-Pk3. In addition, Rts3 and Tap42, which associate with different PP2A phosphatases [45], [46], were also found in the immunoprecipitates. In contrast, we could not detect Rts1, the other major PP2A regulatory subunit, bound to Igo1-Pk3 (Fig. S1A). Based on several independent experiments, we estimated that about 1% of total Cdc55 is bound to Igo1 in asynchronous cycling cells. Interestingly, Igo1 association with PP2A subunits was dramatically impaired upon deletion of RIM15, suggesting that it is enhanced by Igo1 phosphorylation (Fig. 1B). Consistently, the interaction between HA-tagged Cdc55 and the mutant protein Igo1-S64A tagged with myc epitopes was markedly reduced compared to its wild type counterpart (Fig. 1C). Thus, efficient interaction between Igo1 and PP2A requires Rim15-dependent phosphorylation of Igo1 on Ser64. The residual binding between Igo1-S64A and Cdc55 appeared to be further impaired by RIM15 deletion (Fig. 1C), suggesting that Rim15 might phosphorylate additional Igo1 residues besides S64 to stabilize the Igo1-PP2ACdc55 complex. Indeed, recombinant Igo1-S64A could be still phosphorylated in vitro by human Greatwall, albeit inefficiently (Fig. S1B).
Fig. 1. Igo1 interacts with PP2ACdc55 upon Ser64 phosphorylation in lateS/G2 phase of the cell cycle. 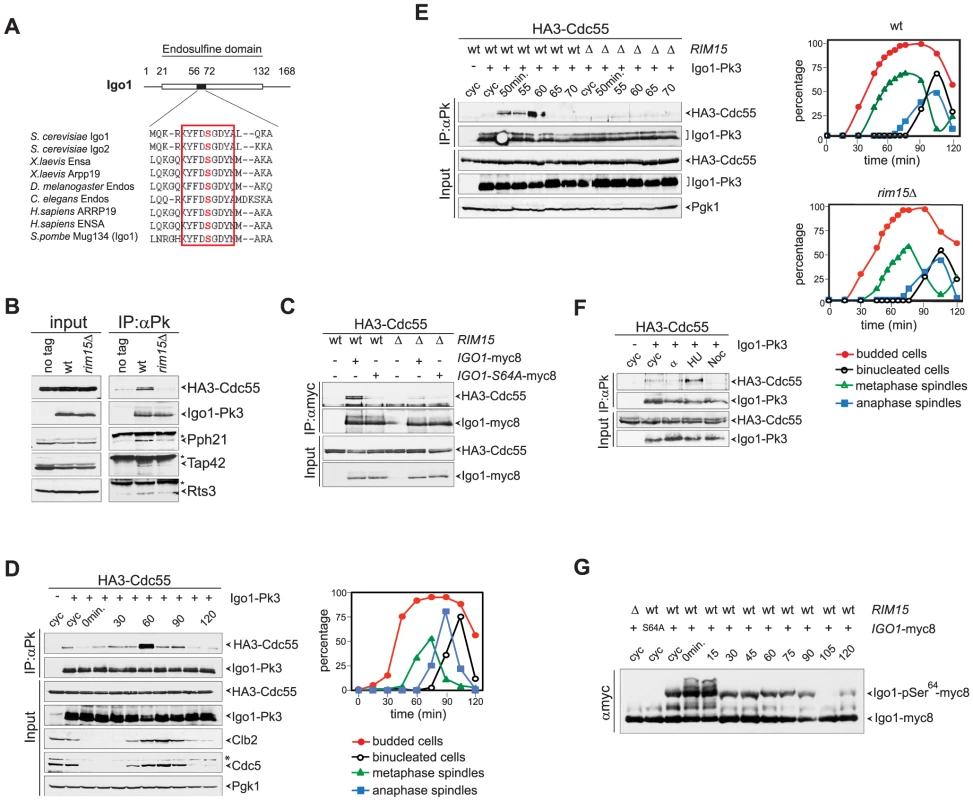
A. Primary sequence alignment of endosulfines from different species around the serine phosphorylated by Greatwall/Rim15. B. Cell extracts from wild type and rim15Δ cells expressing HA3-Cdc55 and Igo1-Pk3 as only sources of Cdc55 and Igo1 were subjected to immunoprecipitation with anti-Pk antibodies. The amount of HA3-Cdc55, Pph21, Tap42 and Rts3 co-immunoprecipitating with Igo1-Pk3 was assessed by western blot analysis. An extract from an Igo1-untagged strain was used as negative control (no tag). The asterisk marks the IgG in the IPs. C. Co-immunoprecipitations of HA3-Cdc55 with either wild type Igo1-myc8 or Igo1-S64A-myc8, in the presence or absence of RIM15. D–E. Cycling (cyc) cultures of cells expressing HA3-Cdc55 and Igo1-Pk3 was arrested in G1 by α-factor and released in fresh medium at 25°C. At different time points after release cell samples were collected to analyse the interaction between Igo1-Pk3 and HA3-Cdc55 after immunoprecipitation with anti-Pk antibodies (left, IP:αPk), the protein levels of HA3-Cdc55, Igo1-Pk3, Clb2, Cdc5 (left, Input), as well as the kinetics of budding, DNA replication (by FACS, data not shown), spindle formation/elongation and nuclear division (right graph). Pgk1 was used as loading control for the western blot. F. Cycling (cyc) cells were arrested in G1 by alpha factor or in S phase by hydroxyurea (HU) or in mitosis by nocodazole (Noc) to analyse the interaction between Igo1-Pk3 and HA3-Cdc55 after immunoprecipitation with anti-Pk antibodies. G. A cycling (cyc) culture of cells expressing Igo1-myc8 was arrested in G1 by alpha factor and released into the cell cycle at 25°C. At the indicated time points cell samples were collected for FACS analysis of DNA contents (not shown) and to prepare TCA protein extracts that were run on precast Phos-tag gels to visualize the phosphorylation of Igo1 by mobility shift. Extracts from rim15Δ and Igo1-S64A-myc8 cells were loaded as controls. Note that alpha factor induces an additional mobility shift likely corresponding to additional phosphorylations that are quickly lost upon cell cycle entry. We next asked if Igo1 interaction with PP2ACdc55 is regulated during the cell cycle. To this end, wild type cells co-expressing Pk-tagged Igo1 and HA-tagged Cdc55 were arrested in G1 by alpha-factor and released into fresh medium at 25°C. At different time points after release we analysed Igo1-Cdc55 interaction by co-immunoprecipitation, as well as other cell cycle parameters like budding, formation and elongation of bipolar spindles. As shown in Fig. 1D, a basal level of Cdc55 binding to Igo1 was detectable during most cell cycle stages but sharply peaked at 60 minutes from the G1 arrest. Based on the kinetics of bipolar spindle formation (starting at 60 minutes after release) and the appearance of the mitotic cyclin Clb2 and the polo kinase Cdc5 (peaking at 75 minutes after release), we conclude that Igo1-Cdc55 interaction is maximal in late S/G2 phase. The rise and fall in Igo1-Cdc55 interaction during the cell cycle was very sharp, as shown by a tighter time course, and depended on Rim15 (Fig. 1E). In addition, it was present at relatively high levels in cells treated with the replication inhibitor hydroxyurea, but not in cells arrested in G1 by alpha factor or arrested in mitosis upon spindle depolymerization by nocodazole (Fig. 1F). In spite of its cell cycle-regulated interaction with Cdc55, phosphorylation of Igo1 on Ser64 appeared to be constant throughout the cell cycle, as shown by Phos-tag phosphate affinity gel electrophoresis (Fig. 1G). Thus, Rim15-dependent phosphorylation of Ser64 is necessary for Igo1 binding to Cdc55, but other factors must be involved for this interaction to be maximal in late S or G2 phase.
Yeast endosulfines can be phosphorylated by Gwl and induce mitotic entry in Xenopus egg extracts
To test if yeast endosulfines inhibit PP2A catalytic activity like in other organisms, we affinity-purified PP2ACdc55 complexes from yeast cells expressing HA-tagged Cdc55 and measured its associated phosphatase activity on the P-Ser/P-Thr substrate phosphorylase a [47]. Recombinant GST-tagged Igo1, but not its S64A mutant variant, could be readily phosphorylated by a hyperactive version of the Gwl kinase (Gwl-K72M) purified from baculovirus-infected insect cells (Fig. 2A), suggesting that Gwl and Rim15 are interchangeable for Igo1 phosphorylation in vitro. Consistent with previous results [41], addition of phosphorylated Igo1 to PP2ACdc55 complexes inhibited their activity in vitro in a dose-dependent manner (Fig. 2B). The S64A mutation reduced, but did not abolish, the ability of Igo1 to inhibit PP2A activity (Fig. 2B).
Fig. 2. Yeast Igo1 induces mitotic entry in Xenopus egg extracts. 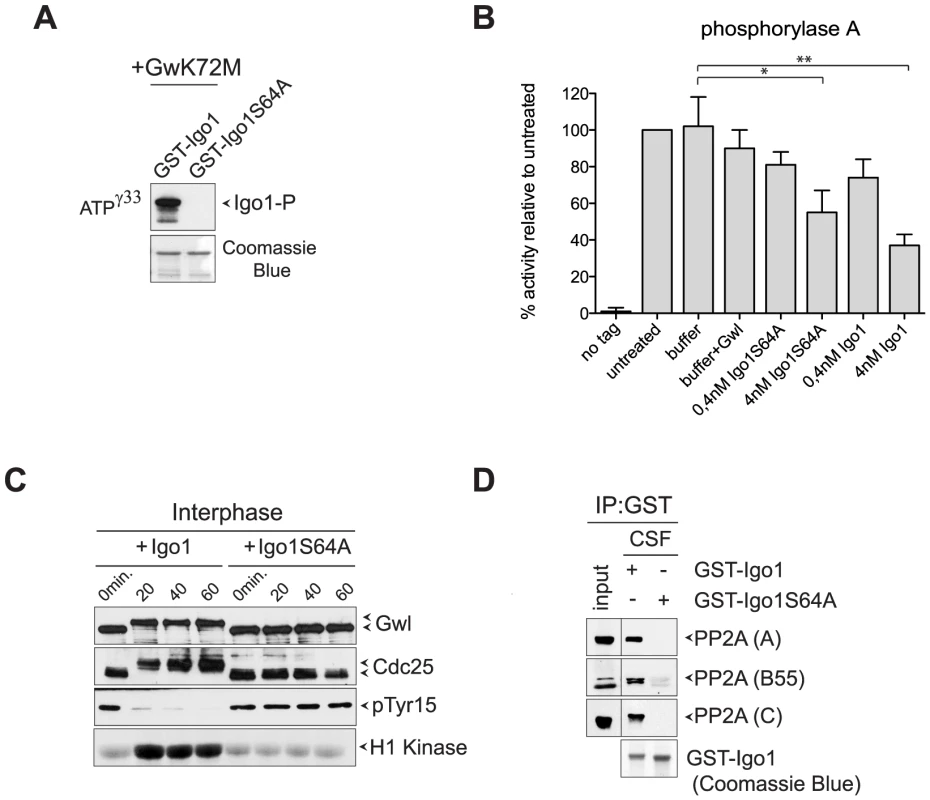
A. Human hyperactive Gwl (Gwl-K72M) was used to phosphorylate in vitro GST-Igo1 and GST-Igo1S64A purified from bacterial cells. B. Bacterially purified GST-Igo1 or GST-Igo1S64A were phosphorylated in vitro by human Gwl and added at the indicated concentrations to PP2ACdc55 complexes affinity-purified from yeast cells expressing HA-tagged Cdc55. The “no tag” control corresponds to the activity of an anti-HA affinity purification from cells expressing untagged Cdc55. C. Igo1 or Igo1S64A previously phosphorylated in vitro by Gwl were added to Xenopus egg interphase extracts at time 0. At the indicated time points aliquots of extracts were frozen in liquid nitrogen and subsequently analysed by western blot for Gwl and Cdc25 phosphorylation (causing a mobility shift in mitosis) and Cdk1 phosphorylation on Tyr15. Cdk1 kinase activity was measured using histone H1 as substrate. D. GST-Igo1 and GST-Igo1S64A were pulled down to assess their association with A, B55 and C subunit of PP2A by western analysis. To establish if Igo1 and Igo2 are functional orthologs of endosulfines, we asked if they could promote mitotic entry in Xenopus egg extracts. Addition of recombinant Igo1 phosphorylated in vitro by Gwl to Xenopus interphase extracts induced mitotic entry, as assessed by the appearance of phosphorylated forms of Gwl and Cdc25, the disappearance of Cdk1 inhibitory phosphorylation and raise in histone H1 kinase activity. In stark contrast, addition of Igo1-S64A had no effect (Fig. 2C). Since endosulfine-mediated mitotic entry under these conditions depends on inhibition of PP2A/B55, we asked if Igo1 could bind to PP2A in Xenopus egg extracts. Strikingly, wild type Igo1, but not Igo1-S64A, pulled down the catalytic (C), structural (A) and regulatory (B55) subunit of PP2A from CSF-induced mitotic extracts (Fig. 2D).
Thus, yeast endosulfines can inhibit the phosphatase activity of PP2A complexes and fulfill the role of their vertebrate counterparts in promoting mitotic entry.
Lack of Rim15 or Igo proteins delays mitotic entry under temperature stress conditions by maintaining the inhibitory phosphorylation of Cdk1
To further investigate the possible role of Rim15 and Igo1,2 in cell cycle progression, we analysed the latter upon deletion of RIM15 or IGO1 and IGO2. Deletion of RIM15 or IGO1 and IGO2 had no significant effect on the kinetics of cell division at physiological temperatures (25°C and 30°C, data not shown). However, rim15Δ and igo1Δ igo2Δ mutant cells where sensitive to thermal stress (14°C, 16°C and 38°C) and were hypersensitive to cell wall stressors, such as caffeine, calcofluor white and SDS (Fig. S2A).
We compared the cell cycle progression of rim15Δ and igo1Δ igo2Δ mutants to that of wild type cells at high and low temperatures. Cells were grown at 25°C, arrested in G1 by alpha-factor and then released in the cell cycle at 38°C. Although the kinetics of budding were similar in the three strains, mitotic events, such as spindle formation, spindle elongation and nuclear division, were delayed by 10–30 minutes in rim15Δ and igo1Δ igo2Δ cells relative to wild type. In addition, accumulation of the polo kinase Cdc5, as well as of the mitotic cyclin Clb2 and its associated kinase, were affected by RIM15 deletion and, even more pronouncedly, by IGO1 and IGO2 deletion (Fig. 3A). A similar experiment showed that deletion of RIM15 and IGO1 and IGO2 delayed mitosis also at 16°C, as shown by analysis of the same cell cycle markers above (Fig. 3B). Also at low temperature the mitotic defects of igo1Δ igo2Δ cells were more pronounced than those of rim15Δ cells. Taken together, these data indicate that Rim15 and its targets Igo1 and Igo2 are required for timely mitotic entry and mitotic progression under temperature stress, with Igo proteins having a more prominent role than Rim15 in this process. Consistent with a conserved function for Igo proteins and endosulfines in promoting mitotic entry, at least under certain conditions, expression of human Arpp19 or human ENSA from the IGO1 promoter partially rescued the temperature-sensitivity of igo1Δ igo2Δ cells at 37°C (Fig. S2B).
Fig. 3. Deletion of RIM15 or IGO1 and IGO2 delays mitotic entry under temperature stress. 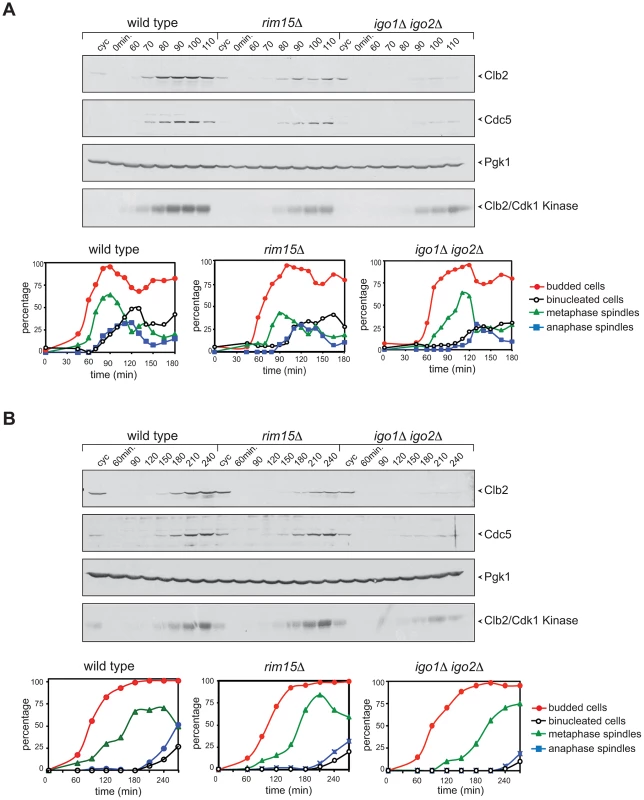
Cycling (cyc) cultures of wild type, rim15Δ and igo1Δ igo2Δ cells were arrested in G1 by α-factor and released in fresh medium at 38°C (A) or 16°C (B). At the indicated time points cells were collected for FACS analysis of DNA contents (not shown), kinetics of budding, spindle assembly/elongation and nuclear division (graphs), as well as to prepare protein extracts for western blot analysis of Clb2 and Cdc5 levels and for Clb2/Cdk1 kinase activity using histone H1 as substrate. Pgk1 was used as loading control. Rim15 and Igo1,2 promote mitotic entry by impinging on the positive feedback loop of Cdk1 activation
Since yeast PP2ACdc55 promotes timely entry into mitosis, the genetic data above raised the possibility that in vivo Igo proteins contribute to PP2ACdc55 activation. We therefore set out to measure PP2ACdc55 catalytic activity in wild type and igo1Δ igo2Δ cells. To this end, we immunoprecipitated HA-Cdc55 and tested the activity of the associated phosphatase using phosphorylase a and histone H1 as substrates. As shown in Fig. 4A, anti-HA immunoprecipitates contained the Pph21 catalytic and the Tpd3 scaffold subunits of PP2A. Remarkably, lack of Igo1 and Igo2 caused a small (15–20%) but significant decrease on PP2ACdc55 activity on both phosphorylase a and histone H1 (Fig. 4B). No further decrease in PP2ACdc55 activity was observed by incubating igo1Δ igo2Δ cells at the restrictive temperature (38°C, data not shown). Thus, although Igo proteins are PP2ACdc55 inhibitors, in vivo they sustain full PP2ACdc55 activity. However, they did not seem to affect the basic composition of the hetero-trimeric PP2ACdc55 complex, as shown by the similar levels of interaction between endogenous HA-tagged Cdc55 and the PPh21 and Tpd3 subunits in wild type and igo1Δ igo2Δ cells (Fig. 4C).
Fig. 4. Igo proteins are required for full PP2ACdc55 activity and timely Cdk1-Tyr19 and Mih1 dephosphorylation. 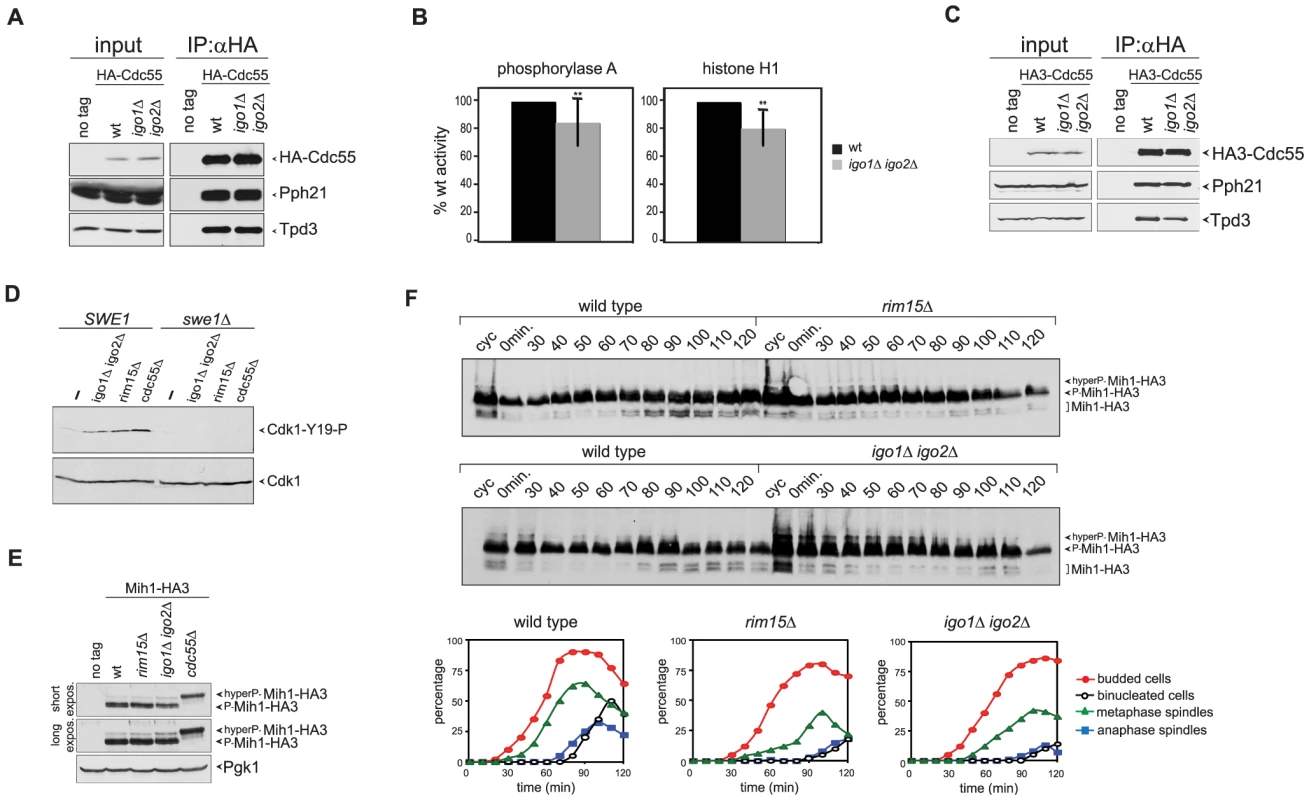
A–B. HA-Cdc55 expressed from the TPI1 promoter was immunoprecipitated from lysates of wild type (wt) or igo1Δ igo2Δ cells. Immunoprecipitates were analysed by western blot for the presence of HA-Cdc55, Pph21 and Tpd3 (A) and in parallel assayed for phosphatase activity using phosphorylase a (n = 12) and histone H1 (n = 5) as substrates (B). Statistical significance of differences was assesses by Student's t-test (** p<0.01). C. HA-Cdc55 expressed at endogenous levels from the CDC55 promoter was immunoprecipitated from lysates of wild type (wt) or igo1Δ igo2Δ cells. Immunoprecipitates were analysed by western blot for the presence of HA-Cdc55, Pph21 and Tpd3. D. Cells with the indicated genotypes were arrested in G1 by α-factor and released for 2 hours in the presence of nocodazole. The levels of Tyr19-phosphorylated Cdk1 (Cdk1-Y19-P) were measured by western blot analysis. E. Total lysates of wild type, rim15Δ, igo1Δ igo2Δ and cdc55Δ cells expressing HA-tagged Mih1 (Mih1-HA3) and logarithmically growing at 25°C were analysed by western blot with anti-HA antibodies. F. Logarithmically growing (cyc) cultures of the same strains as in (A) were arrested in G1 by alpha factor and released in the cell cycle at 38°C to analyse at the indicated time points the electrophoretic mobility of Mih1-HA3 by western blot. Cell samples were also collected at the indicated time points to measure DNA contents by FACS analysis (not shown), as well as kinetics of budding, spindle assembly/elongation and nuclear division (graphs). Deletion of CDC55 causes an accumulation of Tyr19-phoshorylated Cdk1 and delays mitotic entry [37], [39]. Our finding that Igo1 and Igo2 are required for full PP2ACdc55 activity and for timely mitotic entry under temperature stress conditions prompted us to test if Rim15 and Igo proteins regulate Cdk1 phosphorylation on Tyr19. To test if the levels of Tyr19 phosphorylation on Cdk1 were misregulated in the absence of Rim15 or Igo proteins, wild type, igo1Δ igo2Δ and rim15Δ mutant cells, as well as cdc55Δ cells used as control, were arrested in G1 and released in the presence of the microtubule-depolymerizer nocodazole at 25°C, i.e. at a temperature where RIM15 and IGO1/2 are not required for mitotic entry (data not shown). The phosphorylation status of Cdk1 was monitored by western analysis using a phospho-specific antibody that recognizes phosphorylated Cdk1-Y19. In agreement with previous reports [36], [39], [43], phosphorylation of Cdk1-Y19 was barely detectable in wild-type cells and high in cdc55Δ mutant cells under these conditions (Fig. 4D). Similarly, Cdk1-Y19 phosphorylation was increased in igo1Δ igo2Δor rim15Δ mutant cells relative to wild type cells and was fully abolished by SWE1 deletion (Fig. 4D). Similar results were obtained under conditions of thermal stress (data not shown). Thus, Rim15 and Igo proteins are required for timely Cdk1 dephosphorylation, even in conditions where they do not appear to regulate mitosis.
We therefore analysed the levels and phosphorylation state of Swe1 during the cell cycle. Swe1 is phosphorylated in early mitosis by Clb2-Cdk1, which stimulates Swe1's ability to bind, phosphorylate and inhibit mitotic CDKs [6]. This initial Swe1 phosphorylation is opposed by PP2ACdc55 [35]. Later on during mitosis, Swe1 gets hyperphosphorylated and eventually degraded [13], thus feeding the positive feedback loop. We assayed Swe1 levels and phosphorylation in synchronized rim15Δ and igo1Δ igo2Δ cells released from G1 in the presence of nocodazole at 25°C. As shown in Fig. S3A, HA-tagged Swe1 (Swe1-HA3) accumulated at intermediate phosphorylation levels and got more slowly hyperphosphorylated in the absence than in the presence of Rim15 (Fig. S3A). Similar data were obtained in cells lacking Igo1 and Igo2 (data not shown). Delayed appearance of Swe1 hyperphosphorylated forms did not seem to be further affected by deletion of RIM15 or IGO1 and IGO2 upon release of G1 cells at the restrictive temperature of 38°C (Fig. S3B). Thus, Rim15 and Igo proteins contribute, like PP2ACdc55, to timely Swe1 phosphorylation even in conditions where these proteins are apparently not required for timely mitotic entry.
Cdk1-Y19 phosphorylation is reversed by the Mih1 phosphatase, which in turn undergoes a PP2ACdc55-dependent dephosphorylation that can be visualized by an increase in its electrophoretic mobility and correlates with mitotic entry [36]. We therefore asked if the levels of Mih1 phosphorylation are affected by deletion of RIM15 or IGO1 and IGO2. To this purpose, we expressed HA-tagged Mih1 (Mih1-HA3) in wild type, rim15Δ and igo1Δ igo2Δ mutant cells, as well as cdc55Δ cells used as control, and analysed Mih1 phosphorylation by western blot. Consistent with a previous report [36], most Mih1 was present in the cells in phosphorylated forms (Fig. 4E–F). Deletion of RIM15 or IGO1 and IGO2 led to accumulation of hyperphosphorylated forms already at 25°C, albeit not to the same levels as deletion of CDC55 (Fig. 4E). To analyse the transient appearance of the dephosphorylated, and presumably active, form of Mih1 during the cell cycle, wild type, rim15Δ and igo1Δ igo2Δ cells were arrested in G1 by alpha factor and released in the cell cycle at 38°C. Whereas dephosphorylated Mih1-HA3 started accumulating in wild type cells at 70–80 minutes after the release, coincident with the time of mitotic entry, its appearance was delayed in the absence of Rim15 or Igo proteins (Fig. 4F), in agreement with reduced PP2ACdc55 activity. Thus, the phosphorylation state of both Swe1 and Mih1 is affected by deletion of RIM15 or IGO1/2.
Consistent with a role of Rim15 and Igo proteins in the regulation of Cdk1 phosphorylation, genetic analyses showed that lack of Swe1 fully rescued the temperature-sensitive growth defect of igo1Δ igo2Δ and rim15Δ mutant cells at 38°C (Fig. 5A). Expression of non-phosphorylatable Cdc28-Y19F also rescued the cold - and temperature-sensitivity of igo1Δ igo2Δ and rim15Δ mutants, although somewhat less efficiently than SWE1 deletion (Fig. S4). Finally, MIH1 deletion enhanced the temperature-sensitivity of igo1Δ igo2Δ cells at 37°C and caused a cold-sensitive growth phenotype to rim15Δ cells at 16°C (it should be noted that igo1Δ igo2Δ cells are sensitive to this temperature ontheirown,Fig. 5B).
Fig. 5. Deletion of RIM15 or IGO1 and IGO2 affects the positive feedback loop for Cdk1 activation. 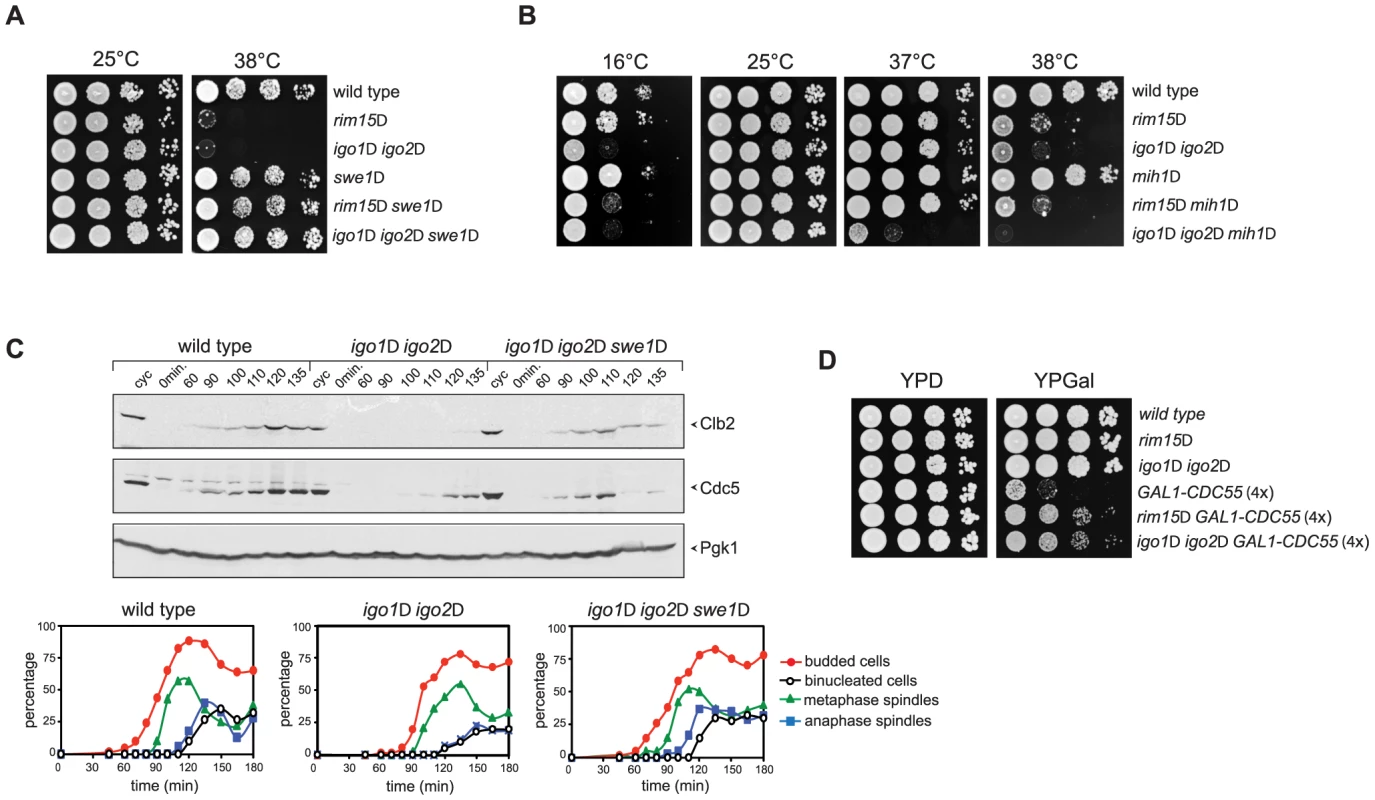
A–B. Serial dilutions of strains with the indicated genotypes were spotted on YEPD plates and incubated for 48 hours at the indicated temperatures. C. Cycling (cyc) cultures of wild type, igo1Δ igo2Δ and igo1Δ igo2Δ swe1Δ cells were arrested in G1 by α-factor and released in fresh medium at 38°C. At the indicated time points cells were collected for FACS analysis of DNA contents (not shown), kinetics of budding, spindle assembly/elongation and nuclear division (graphs), as well as to make protein extracts for western blot analysis of Clb2 and Cdc5 levels. Pgk1 was used as loading control. D. Serial dilutions of strains with the indicated genotypes were spotted on YPD and YPGal plates and incubated at 25°C for 2 days. We then asked if SWE1 deletion could rescue the mitotic entry delay of igo1Δ igo2Δ under temperature stress. To address this question, we arrested wild-type, igo1Δ igo2Δ and igo1Δ igo2Δswe1Δ cells in G1 and released them into fresh medium at 38°C. Under these conditions, igo1Δ igo2Δ cells showed a marked delay in the accumulation of Clb2 and Cdc5 and in spindle elongation relative to wild type cells (Fig. 3A and 5C). Strikingly, SWE1 deletion suppressed these mitotic defects and restored the normal kinetics of Clb2 and Cdc5 accumulation during the cell cycle (Fig. 5C). Altogether, these data suggest that Rim15 and Igo proteins regulate the phosphorylation state of Cdk1 for mitotic entry, presumably by affecting the PP2ACdc55–dependent regulation of Swe1 and Mih1, and indicate that excessive Cdk1 inhibitory phosphorylation is responsible for the mitotic delay of igo1Δ igo2Δ cells.
If budding yeast endosulfines were required for full PP2ACdc55 phosphatase activity, deletion of RIM15 or IGO1 and IGO2 should rescue the toxic effects caused by CDC55 overexpression, which prevents mitotic progression [48]. Consistent with our previous findings, cells carrying four integrated copies of a galactose-inducible GAL1-CDC55 construct were mostly unable to divide on galactose-containing plates (YPGal, Fig. 5D). Loss of Rim15 or Igo1/2 efficiently suppressed this lethality (Fig. 5D) without affecting the levels of CDC55 overexpression (data not shown).
Altogether, these data indicate that Rim15 and Igo proteins contribute to activation of PP2ACdc55, at least for what concerns some of its mitotic targets, thereby tuning the levels of Cdk1 phosphorylation to promote mitotic entry.
Igo proteins influence Cdc55 subcellular localization
The apparently opposite impact of Igo proteins on PP2A regulation in vitro and in vivo (i.e. inhibition versus activation, respectively), raised the possibility that in yeast Igo1/2 modulate PP2A activity at a different/additional level. For instance, the Zds1 and Zds2 proteins, which bind PP2ACdc55 in a stoichiometric complex and inhibit its activity in vitro [49], [50], were recently shown to regulate the subcellular localization of Cdc55. More specifically, Zds1/2 induce the nuclear export of Cdc55 into the cytoplasm, which in turn promotes mitotic entry [43]. We therefore asked if Rim15 and Igo proteins might play a similar role in controlling Cdc55 localization. HA-tagged Cdc55 was detected by indirect immunofluorescence in wild type and igo1Δ igo2Δ cells at various cell cycle stages, namely in G1 (unbudded cells), in S, G2 and early M phases (budded mononucleated cells) and in middle/late M phase (budded binucleated cells). As previously shown [43], Cdc55 was localized in both the nucleus and the cytoplasm. However, it was significantly more concentrated in the nucleus of rim15Δand igo1Δ igo2Δ cells than in the wild type in all cell cycle stages (Fig. 6A). Strikingly, deletion of SWE1 restored the normal nuclear/cytoplasmic ratio of Cdc55 (Fig. 6A), strongly indicating that the altered subcellular distribution of Cdc55 in mutants lacking Rim15 or Igo proteins is a consequence, rather than a cause, of misregulated positive feedback loop for Cdk1 activation.
Fig. 6. Igo1 and Igo2 control the subcellular localization of PP2ACdc55 independently of Zds proteins. 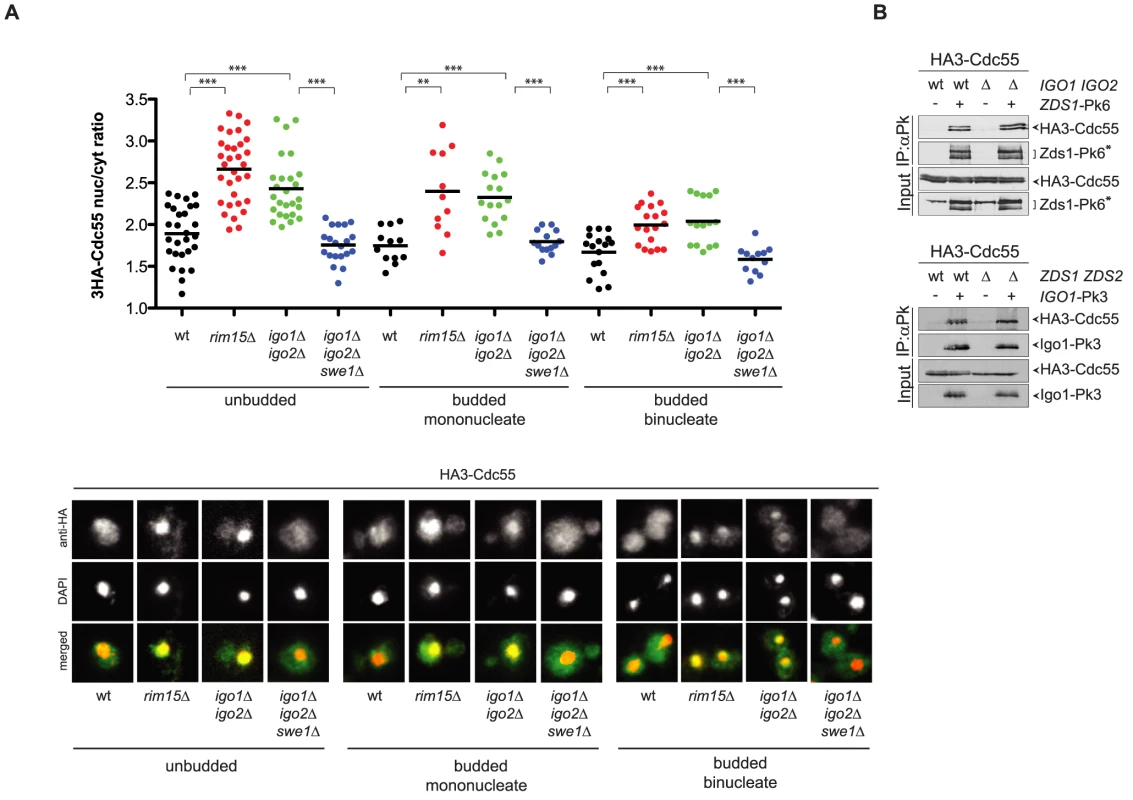
A. Localization of HA3-Cdc55 was analysed by indirect immunofluorescence with anti-HA antibodies after formaldehyde fixation of cycling cultures of wild type, rim15Δ igo1Δ igo2Δand igo1Δ igo2Δ swe1Δcells. Cells were scored according to their cell cycle stage: unbudded (G1), budded mononucleated (S, G2, early M) and budded binucleated (late M). At least 15 cells were scored for each class. B. Interaction between Zds1-Pk6 and HA3-Cdc55 in the presence or absence of Igo proteins (left panel) and interaction between Igo1-Pk3 and HA3-Cdc55 in the presence or absence of Zds proteins (right panel) was assessed by immunoprecipitation with anti-Pk antibodies followed by western blot analysis with anti-HA and anti-Pk antibodies. The asterisk marks a protein aspecifically recognized by the anti-Pk antibodies in the total extracts (input) and that co-migrates with Zds1-Pk6. The similar phenotype of igo1Δ igo2Δ and zds1Δ zds2Δ cells with respect to Cdc55 localization raised the possibility that Igo and Zds proteins work in concert and/or are part of the same complex. We therefore decided to analyse the possible interdependence between Igo and Zds proteins for their interaction with Cdc55. As previously shown, Pk-tagged Zds1 efficiently co-immunoprecipitated HA-tagged Cdc55 from extracts obtained from cycling cells. Interaction between Zds1 and Cdc55 was not affected by deletion of IGO1 and IGO2 (Fig. 6B). Similarly, co-immunoprecipitation of HA-tagged Cdc55 with Pk-tagged Igo1 was unaffected by deletion of ZDS1 and ZDS2. It is worth noting that in zds1Δ zds2Δ cells the mobility shift of Cdc55, which was previously shown to be due to phosphorylation [47], disappears, indicating another possible way for Zds proteins to regulate PP2ACdc55 activity besides its subcellular localization. Altogether, these data suggest that Igo and Zds proteins bind independently to PP2ACdc55 and might regulate its activity in an independent manner. Consistent with this conclusion, deletion of IGO1 and IGO2 caused synthetic sickness at high temperatures when combined with deletion of ZDS1 and ZDS2 (Fig. S5).
Discussion
Conservation of the Greatwall-Endosulfine-PP2A regulatory module
In several organisms endosulfine-like proteins bind to PP2A-B55 upon Greatwall-dependent phosphorylation of a conserved serine and inhibit it [33], [34]. Recent data showed that in quiescent yeast cells Rim15 phosphorylates the paralogous endosulfines Igo1 and Igo2, which in turn promote the transcription of specific nutrient-regulated genes by direct inhibition of the phosphatase PP2ACdc55 [41]. We show here that Igo1 phosphorylated on Ser64 by Rim15 interacts also during the unperturbed cell cycle with the catalytic subunit of PP2A (Pph21), the B regulatory subunit Cdc55, Tap42 and Rts3 and that phosphorylated Igo1 can bind PP2A complexes in yeast and Xenopus egg extracts. PP2A has multiple functions during the cell cycle (reviewed in [20]). In yeast it associates with two major and alternative B subunits, Cdc55 and Rts1. Rts1 does not appear to interact with Igo1 in our co-immunoprecipitations. Cdc55 has been involved in many cellular processes, such as mitotic entry and exit, morphogenesis and cytokinesis, mitotic checkpoints and stress response [35], [36], [38], [39], [48], [51], [52], [53], [54], [55], [56]. Of particular interest is the presence in our Igo1 immunoprecipitates of the essential Tap42 subunit, the orthologue of human PP2A-associated α4/IgBP1. Tap42 associates to the catalytic subunit of PP2A and PP2A-like phosphatases independently of the A and B subunit during logarithmic growth as opposed to stationary phase, suggesting that its interaction with PP2A phosphatases is regulated by nutrients [57]. Consistently, Tap42 phosphorylation, which mediates its interaction with PP2As, depends on the phosphatidylinositol-related kinases Tor1 and Tor2 (Target of rapamycin), which regulate cell growth in response to nutrient availability and cell stress [58]. The last PP2A subunit that we found in Igo1 immunoprecipitates is Rts3, a poorly characterized protein that was found to interact with different PP2A and PP2A-like complexes [45], [59], [60] and whose deletion causes sensitivity to caffeine [61], which in turn inhibits the Tor complex TORC1 [62].
The peak of interaction between Igo1 and Cdc55 is cell cycle-regulated and peaks in late S or G2 phase (note that it is not possible to discriminate between late S and G2 phase in budding yeast due to the lack of specific markers), i.e. when PP2ACdc55 participates to the positive feedback loop for Cdk1 activation (see below). In agreement with recently published data [41], we show that mutation of Igo1 Ser64, which is targeted by Rim15 [40], and deletion of RIM15 reduce significantly Igo1 interaction with PP2ACdc55, indicating that, like in higher organisms, endosulfine phosphorylation is required for its interaction with PP2A. In spite of its cell cycle-regulated interaction with PP2ACdc55, Igo1 phosphorylation on Ser64 appears to be constitutive during the cell cycle, raising the interesting possibility that cell cycle-controlled factors are involved to make the Igo1-Cdc55 binding periodic. Along the same line, it is also interesting to notice that phosphorylation of Ser64 of Igo1 and Ser63 of Igo2 is stimulated after inhibition of Cdk1 [63], suggesting that Cdk1 activation in early mitosis may cause the dissociation of Igo-PP2ACdc55 complexes.
The evolutionary conservation of the Greatwall-Endosulfine pathway is further strengthened by the finding that yeast Rim15 can phosphorylate in vitro human ENSA and Arpp19 [40], whereas human and Xenopus Greatwall can phosphorylate yeast Igo1 (this manuscript), indicating that Rim15 and Igo proteins are the true counterparts of vertebrate Greatwall and endosulfines, respectively. Thus, the Greatwall-Endosulfine-PP2A regulatory module appears to be conserved in all organisms analysed to date, with the notable exception of the nematode C. elegans where an obvious Greatwall-like kinase seems to be missing [42].
Are endosulfines positive or negative regulators of PP2A?
ENSA and Arpp19 inhibit PP2A activity in Xenopus egg extracts [30], [31]. Inhibition of PP2A by endosulfines is in turn essential for mitotic entry in Xenopus and human cells and for mitotic progression in Drosophila [23], [27], [29], [30], [31], [64]. Similarly, we and others [41] find that phosphorylated Igo1 inhibit PP2A activity in vitro and induces interphase Xenopus egg extracts to enter mitosis, indicating that budding yeast endosulfines can inhibit PP2A, like their vertebrate counterparts. PP2ACdc55 inhibition in rapamycin-treated yeast cells is important to establish a quiescence-specific transcriptional program [41]. We provide several lines of evidence indicating that Rim15 and Igo proteins also contribute to activate PP2ACdc55 for mitotic entry in vivo. First, lack of Igo1 and Igo2 causes a slight reduction, rather than an increase, in PP2ACdc55 phosphatase activity. Taking into account that only about 1% of Cdc55 is bound to Igo1 (and presumably a similar fraction of Cdc55 is bound to Igo2), the 15–20% reduction in PP2ACdc55 activity in cells lacking Igo proteins would imply a prominent role for this proteins in the full activation of PP2ACdc55 complexes. Second, deletion of RIM15 or IGO1 and IGO2 delays accumulation of active cyclinB/Cdk1 and dephosphorylation of Cdk1 Tyr19, similarly to inactivation of PP2ACdc55 [35], [39]. This is accompanied by a misregulation in the phosphorylation of the Swe1 kinase and the dephosphorylation of the Mih1 phosphatase during the cell cycle, which are both controlled by PP2ACdc55 [35], [36]. Third, the toxic effects caused by CDC55 overexpression [48] are rescued by RIM15 or IGO1 and IGO2 deletion. Yet, not only phosphorylated Igo1 can inhibit PP2A activity in vitro, but expression of human ENSA or Arpp19 rescues the temperature-sensitivity of igo1Δ igo2Δ cells, suggesting that vertebrate and yeast endosulfines are interchangeable for their mitotic function(s). Therefore, the apparently opposite modes of PP2A regulation by endosulfines in yeast versus vertebrates are unlikely linked to intrinsic differences in the structure of endosulfines and/or in their ability to bind and inhibit PP2A. Several other examples of proteins that behave as activators or inhibitors of their binding partner(s) depending on the organism and/or the conditions have been reported. Similarly to Igo1 and Igo2, budding yeast Zds proteins were shown to inhibit PP2ACdc55 phosphatase activity [49] and to promote efficient mitotic entry through nuclear export of Cdc55 and Cdc55-dependent regulation of the Cdk1 positive feedback loop [43], [50], [65]. Tap42 has been proposed to work as activator and inhibitor of PP2A [57], [66], [67], [68]. Finally, securin is both an inhibitor and a chaperone of separase for the regulation of sister chromatid splitting in anaphase. Depending on the organism, securin depletion/inactivation prevents sister chromatid separation or causes premature anaphase onset (reviewed in [69]). We speculate that Igo proteins might be chaperones for PP2ACdc55 and assist its proper folding while keeping the complex inhibited. The presence of phosphorylated substrates could competitively bind to PP2ACdc55 and overcome its inhibition. The exact biochemical mechanism of PP2A-B55 inhibition by endosulfines is currently unknown and will be likely clarified by structural and biochemical data.
The function of Rim15 and Igo proteins in the control of mitosis
Consistent with a positive role of Igo proteins in PP2ACdc55 regulation for mitosis, we find that the phosphatase activity of PP2ACdc55 complexes in igo1Δ igo2Δ cells is reduced compared to wild type cells. This results in delayed phosphorylation of Swe1 and Mih1, which are known targets of PP2ACdc55 [35], [36], thus preventing the sharp rise in CDK activity during mitotic entry. Indeed, rim15Δ and igo1Δ igo2Δ cells are unable to timely dephosphorylate Tyr19 of Cdk1 and to efficiently raise cyclinB/Cdk1 activity in mitosis, resulting in a delayed spindle elongation, which requires high cyclinB/Cdk1 levels [70]. The observation that rim15Δ and igo1Δ igo2Δ mutants display lower activity of PP2ACdc55 complexes, nuclear retention of Cdc55 and accumulation of Tyr19-phosphorylated Cdk1 at physiological temperatures (e.g. 25°C), while progressing through mitosis with normal timing, is somewhat puzzling. Since mitotic defects of rim15Δ and igo1Δ igo2Δ mutants become apparent only under stress conditions, it is possible that the physiological state of cells can make them differentially responsive to increased levels of Tyr19-phosphorylated Cdk1. Deletion of SWE1 or expression of the non-phosphorylatable Cdc28-Y19F mutant protein rescues the temperature-sensitive growth and mitotic defects of rim15Δ and igo1Δ igo2Δ cells, whereas deletion of MIH1 aggravates them. The role of PP2ACdc55 in the nucleolar retention and inhibition of Cdc14 [52], which is the major CDK-counteracting phosphatase in yeast [15], might also be affected by lack of endosulfines and contribute to timely mitotic entry.
The activity of PP2ACdc55 has been recently proposed to be spatially regulated by nuclear export of Cdc55, which is regulated by Zds1 and Zds2 and promotes mitotic entry [43]. In this respect it is important to notice that a key determinant in Swe1 downregulation is its proteolysis, which requires Swe1 recruitment to the bud neck (i.e. out of the nucleus) [71], [72]. Thus, the cytoplasmic pool of PP2ACdc55 might be instrumental to trigger efficient Swe1 degradation [39]. We show that the nuclear/cytoplasmic ratio of Cdc55 is increased throughout the cell cycle upon deletion of RIM15 or IGO1 and IGO2. The increased concentration of Cdc55 in the nuclei of rim15Δ and igo1Δ igo2Δ cells might be a consequence of decreased PP2ACdc55 activity or be caused by a more direct function of yeast endosulfines in the control of Cdc55 nuclear-cytoplasmic shuttling. Our finding that SWE1 deletion restores proper subcellular localization of Cdc55 in the absence of Rim15 and Igo proteins argues against the latter possibility. We thus favor the idea that yeast endosulfines only impact on PP2ACdc55 activity, participating in the positive feedback loop for Cdk1 activation like their Xenopus counterparts [27]. CDK activity could in turn drive, directly or indirectly, Cdc55 export from the nucleus to the cytoplasm (Fig. 7). It is nevertheless possible that nuclear retention of Cdc55 in cells lacking Rim15 or endosulfines has a greater impact than the slight decrease in PP2A activity by itself on dephosphorylation of PP2ACdc55 substrates, including Swe1 and Mih1. In fact, Cdc55 mislocalization could keep PP2ACdc55 spatially segregated from its substrates. Further experiments will be required to understand the links between PP2A activity and its subcellular localization.
Fig. 7. Model. 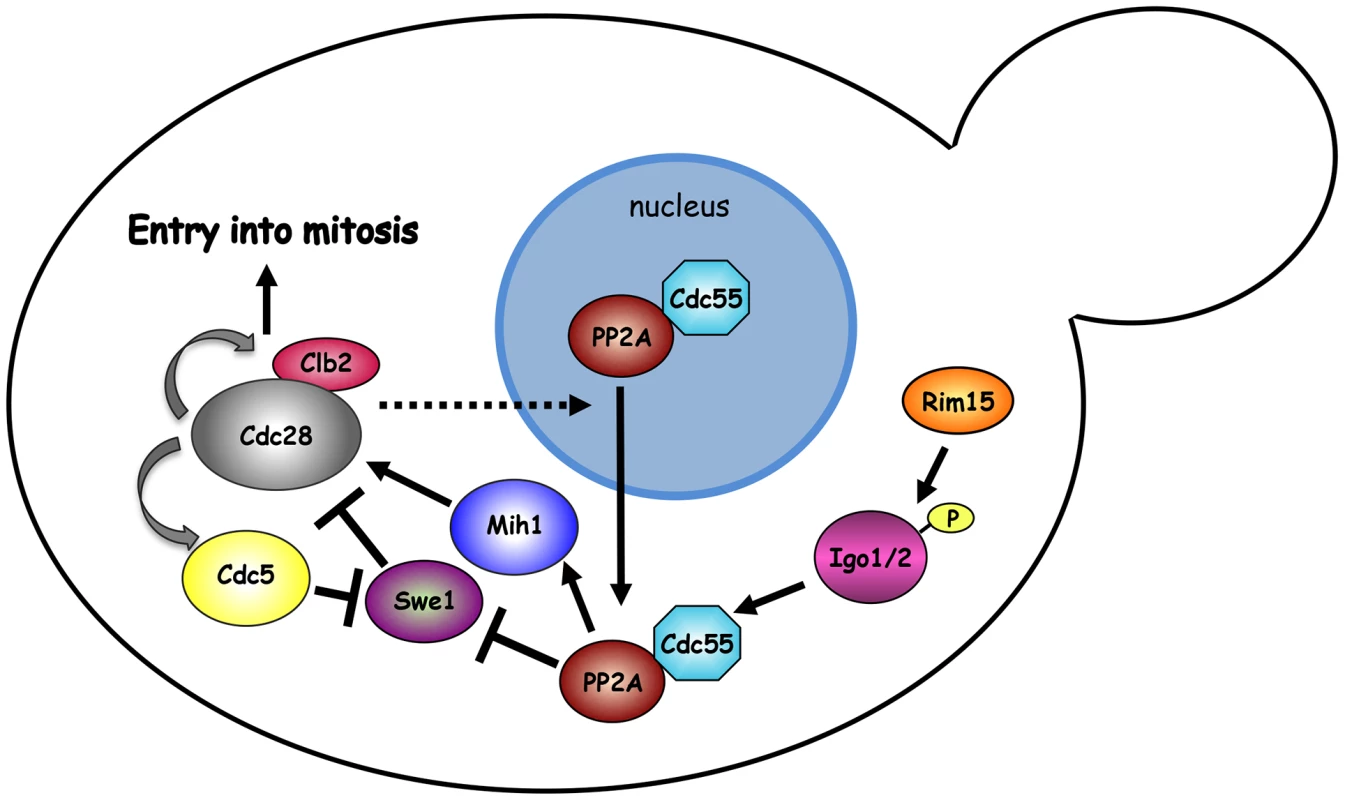
The mitotic CDK Clb2-Cdk1 is part of a positive feedback loop where it contributes to its own activation by promoting a transcriptional mitotic program for its own expression and that of the polo kinase Cdc5. Although Swe1 phosphorylation by Clb2-Cdk1 promotes Tyr19 phosphorylation, and thereby inhibition, of Cdk1 (not depicted), it primes further Swe1 phosphorylations by several other kinases, including Cdc5, that finally target Swe1 to degradation. Initial Swe1 phosphorylation by Cdk1 is counteracted by the PP2ACdc55 phosphatase, which also dephosphorylates and activates Mih1, thereby participating in the positive feedback loop for Cdk1 activation. The endosulfines Igo1 and Igo2, upon phosphorylation by Rim15, bind to PP2ACdc55 and contribute to their activation and nuclear export. The observation that SWE1 deletion restores normal localization of Cdc55 in igo1Δ igo2Δ mutant cells suggests that Swe1 and/or Clb2-Cdk1 might regulate the nuclear export of Cdc55 through an unknown mechanism. See text for further details. Similar to endosulfines, the Zds1 and Zds2 proteins bind tightly to PP2ACdc55 [49], [50], affect its subcellular localization [43] and target PP2ACdc55 activity to Mih1 [65], thus promoting timely mitotic entry [43], [50], [65]. In spite of the similarity, Igo and Zds proteins seem to work independently. Indeed, binding of Igo1 to Cdc55 does not require Zds1/2, whereas Zds1 interaction with Cdc55 does not require Igo proteins. In addition, deletion of IGO1 and IGO2 causes synthetic growth defects when combined to deletion of ZDS1 and ZDS2.
In conclusion, our data emphasize the plasticity of the Greatwall-Endosulfine-PP2A module. This module is conserved, but it has adapted to account for the differences in mitotic regulation depending on the cellular context. In cells where PP2A antagonizes Cdk1 activity the Greatwall-endosulfine module only inhibits PP2A, whereas in budding yeast where PP2A promotes timely mitotic entry the same module also stimulates PP2A activity, resulting in both cases in a sharp mitotic switch [73].
Materials and Methods
Strains, media and reagents, genetic manipulations
All yeast strains (Table S1) were derivatives of W303 (ade2-1, trp1-1, leu2-3,112, his3-11,15, ura3, ssd1), except for strains used for phosphatase assays that were derivatives of S288C. Cells were grown in either synthetic minimal medium (SD) supplemented with the appropriate nutrients or YEP (1% yeast extract, 2% bactopeptone, 50 mg/l adenine) medium supplemented with 2% glucose (YEPD) or 2% galactose (YEPG). Unless differently stated, alpha factor was used at 4 µg/ml, hydroxyurea (HU) at 200 mM and nocodazole at 15 µg/ml. Standard techniques were used for genetic manipulations [74], [75]. Gene deletions were generated by one-step gene replacement [76]. One-step tagging techniques [77], [78] were used to tag at their C-terminus Igo1 (Igo1-Pk3), Swe1 (Swe1-HA3) and Mih1 (Mih1-HA3).
Fluorescence microscopy
In situ immunofluorescence was performed on formaldehyde-fixed cells expressing HA-tagged Cdc55 using anti-HA monoclonal antibody (16B12, Covance Research Products), followed by indirect immunofluorescence using Cy3-conjugated goat anti-mouse antibody (GE Healthcare). To detect spindle formation and elongation, anti-tubulin immunostaining was performed with the YOL34 monoclonal antibody (Serotec) followed by indirect immunofluorescence using rhodamine-conjugated anti-rat antibody (Pierce Chemical Co). Digital images were taken with an oil 63X 1,4-0,6 HCX Plan-Apochromat objective (Zeiss) with a Coolsnap HQ2-1 charge-coupled device camera (Photometrics) mounted on a Zeiss AxioimagerZ1/Apotome fluorescence microscope controlled by the MetaMorph imaging system software. Fluorescence intensity of HA3-Cdc55 in the nucleus and the cytoplasm was quantified with ImageJ on a single focal plane. Significance of the differences between fluorescence intensities was statistically tested by means of a two-tailed t-test, assuming unequal variances.
Protein extracts, immunoprecipitations and kinase assays
TCA protein extracts were prepared as previously described [79] for Phos-tag phosphate affinity gel electrophoresis (Wako, Fig. 1G) and to analyse the electrophoretic mobility of HA-tagged Swe1 and Mih1. For immunoprecipitations of Igo1-Pk3 or Zds1-Pk6, pellets from 50 ml yeast cultures (107 cells/ml) were lysed at 4°C with acid-washed glass beads in lysis buffer (50 mM Tris-Cl pH 7.5, NaCl 150 mM, 10% glycerol, 1 mM EDTA, 1% NP40, supplemented with protein inhibitors (Complete, Roche), 1 mM Na-orthovanadate and 60 mM ß-glycero-phosphate). Total extracts were cleared by spinning at 12000 rpm for 10 minutes and quantified by NanoDrop. Same amounts of protein extracts were subjected to immunoprecipitation with an anti-Pk antibody (from AbD serotec) pre-adsorbed to protein A-sepharose. Inputs represent 1/50th of the IPs final volumes.
For Clb2/Cdk1 kinase assays protein extracts were prepared as previously described [80]. 50 µg of extract were used for measuring kinase activity on histone H1 [81] and 30 µg for Western blot analysis.
Hyperactive human Gwl (Gwl-K72M) purified from baculovirus-infected cells or endogenous Gwl immunoprecipitated from CSF-treated Xenopus egg extracts [30] were used to phosphorylate in vitro GST-Igo1 purified from E.coli for the in vitro phosphorylation assay (Fig. 2A and Fig. S1B) and the mitotic entry experiment (Fig. 2C), respectively.
Yeast protein extracts were prepared according to [82] for western blot analysis. Proteins transferred to Protran membranes (Schleicher and Schuell) were probed with monoclonal anti-HA 12CA5, anti-Pgk1 (Molecular Probes), anti-Pk (AbD serotec), anti-Myc 9E10 or polyclonal Cdc5 antibodies (sc-6733 Santa Cruz), anti-Clb2 (sc-9071 Santa Cruz) or anti-phospho-cdc2 (pTyr15 Cell Signalling). Anti-Tap42, anti-Pph21 and anti-Rts3 antibodies were described [47]. Affinity-purified antibodies against Gwl and Cdc25 were also previously described [30]. Monoclonal anti-PP2A/C subunit (1D6) and anti-PP2A/A (6G3) antibodies were obtained from Upstate/Millipore and Cell Signalling, respectively. Polyclonal antibodies against PP2A/B55 were raised in rabbits and affinity-purified. Secondary antibodies were purchased from Amersham and proteins were detected by an enhanced chemiluminescence system according to the manufacturer.
Protein phosphatase assay and treatment of PP2A-complexes with phosphorylated Igo1
For immunoprecipitation assays, yeast whole-cell extracts were prepared as described previously [83] except that lysis was performed using a Fastprep (MP Biomedicals, 1×40 s, 6 m/s). HA-tagged Cdc55 containing a flexible glycine linker (GL) after the HA epitope (GGGSGGGGS) was expressed from the strong constitutive TPI1 promoter on an episomal centromeric plasmid [47]. HA-GL-Cdc55 was immunoprecipitated with anti-HA (clone 12CA5) antibodies cross-linked to BSA-coated protein A–Sepharose beads (GE Healthcare). Phosphatase activity of PP2A immunoprecipitates was assayed toward 32P-labeled phosphorylase a (Figs. 2B, 4A) or towards 32P-labeled histone H1 (Fig. 4B), as previously described [47], [83]. Immunoprecipitates were analyzed by 10% SDS-PAGE, immunoblotted and incubated with specific antibodies against Pph21 (rabbit pAB), Cdc55 (clone 9D3-H6) or Tpd3 (clone 5G2). The assay values (average of at least five independent experiments) are presented as a percentage of the wild-type strain activity, which was set at 100%. Values were normalized to the amount of Pph21 co-immunoprecipitated with HA-Cdc55 as determined by immunoblot and densitometer analysis using an Odyssey Infrared Imaging System (LI-COR, http://www.licor.com). For Figure 2B immunoprecipitates were split into 7 aliquots and in-vitro Greatwall-phosphorylated recombinant GST-Igo1 or GST-Igo1S64A (where kinase assays were carried out in 50 mM Tris pH7.2, 10 mM MgCl2, 1 mM ATP, 30°C for 30′) was added as indicated and phosphatase activity assays toward 32P-labeled phosphorylase a were conducted. The activities of the different samples are presented as percent of activity with respect to the activity of the untreated control, which was set to 100%. Data are presented as mean values ± standard deviation (SD), and were analyzed using two-tailed Student's t-test, assuming unequal variances. Differences with p-values lower than 0.05 were considered statistically significant (* p<0.05 ; ** p<0.01 ; *** p<0.001).
Other techniques
Nuclear division was scored with a fluorescence microscope on cells stained with propidium iodide (Sigma Aldrich). Flow cytometric DNA quantification was performed according to [84] on a Becton-Dickinson FACSCalibur.
Supporting Information
Zdroje
1. MorganDO (1995) Principles of CDK regulation. Nature 374 : 131–134.
2. AbrieuA, BrassacT, GalasS, FisherD, LabbeJC, et al. (1998) The Polo-like kinase Plx1 is a component of the MPF amplification loop at the G2/M-phase transition of the cell cycle in Xenopus eggs. J Cell Sci 111(Pt 12): 1751–1757.
3. MortensenEM, HaasW, GygiM, GygiSP, KelloggDR (2005) Cdc28-dependent regulation of the Cdc5/Polo kinase. Curr Biol 15 : 2033–2037.
4. KumagaiA, DunphyWG (1996) Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science 273 : 1377–1380.
5. QianYW, EriksonE, TaiebFE, MallerJL (2001) The polo-like kinase Plx1 is required for activation of the phosphatase Cdc25C and cyclin B-Cdc2 in Xenopus oocytes. Mol Biol Cell 12 : 1791–1799.
6. HarveySL, CharletA, HaasW, GygiSP, KelloggDR (2005) Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell 122 : 407–420.
7. MuellerPR, ColemanTR, DunphyWG (1995) Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell 6 : 119–134.
8. TangZ, ColemanTR, DunphyWG (1993) Two distinct mechanisms for negative regulation of the Wee1 protein kinase. EMBO J 12 : 3427–3436.
9. WatanabeN, AraiH, IwasakiJ, ShiinaM, OgataK, et al. (2005) Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc Natl Acad Sci U S A 102 : 11663–11668.
10. LindqvistA, Rodriguez-BravoV, MedemaRH (2009) The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol 185 : 193–202.
11. PomereningJR, SontagED, FerrellJEJr (2003) Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat Cell Biol 5 : 346–351.
12. BahlerJ (2005) Cell-cycle control of gene expression in budding and fission yeast. Annu Rev Genet 39 : 69–94.
13. AsanoS, ParkJE, SakchaisriK, YuLR, SongS, et al. (2005) Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J 24 : 2194–2204.
14. GrayCH, GoodVM, TonksNK, BarfordD (2003) The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. Embo J 22 : 3524–3535.
15. VisintinR, CraigK, HwangES, PrinzS, TyersM, et al. (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk - dependent phosphorylation. Mol Cell 2 : 709–718.
16. ShouW, SeolJH, ShevchenkoA, BaskervilleC, MoazedD, et al. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 : 233–244.
17. VisintinR, HwangES, AmonA (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus [see comments]. Nature 398 : 818–823.
18. WurzenbergerC, GerlichDW (2011) Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol 12 : 469–482.
19. Mayer-JaekelRE, HemmingsBA (1994) Protein phosphatase 2A–a ‘menage a trois’. Trends Cell Biol 4 : 287–291.
20. JiangY (2006) Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70 : 440–449.
21. Mayer-JaekelRE, OhkuraH, FerrignoP, AndjelkovicN, ShiomiK, et al. (1994) Drosophila mutants in the 55 kDa regulatory subunit of protein phosphatase 2A show strongly reduced ability to dephosphorylate substrates of p34cdc2. J Cell Sci 107(Pt 9): 2609–2616.
22. MochidaS, IkeoS, GannonJ, HuntT (2009) Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J 28 : 2777–2785.
23. CastilhoPV, WilliamsBC, MochidaS, ZhaoY, GoldbergML (2009) The M phase kinase Greatwall (Gwl) promotes inactivation of PP2A/B55delta, a phosphatase directed against CDK phosphosites. Mol Biol Cell 20 : 4777–4789.
24. SchmitzMH, HeldM, JanssensV, HutchinsJR, HudeczO, et al. (2010) Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol 12 : 886–893.
25. LorcaT, BernisC, VigneronS, BurgessA, BrioudesE, et al. (2010) Constant regulation of both the MPF amplification loop and the Greatwall-PP2A pathway is required for metaphase II arrest and correct entry into the first embryonic cell cycle. J Cell Sci 123 : 2281–2291.
26. VigneronS, BrioudesE, BurgessA, LabbeJC, LorcaT, et al. (2009) Greatwall maintains mitosis through regulation of PP2A. EMBO J 28 : 2786–2793.
27. YuJ, ZhaoY, LiZ, GalasS, GoldbergML (2006) Greatwall kinase participates in the Cdc2 autoregulatory loop in Xenopus egg extracts. Mol Cell 22 : 83–91.
28. Bettencourt-DiasM, GietR, SinkaR, MazumdarA, LockWG, et al. (2004) Genome-wide survey of protein kinases required for cell cycle progression. Nature 432 : 980–987.
29. YuJ, FlemingSL, WilliamsB, WilliamsEV, LiZ, et al. (2004) Greatwall kinase: a nuclear protein required for proper chromosome condensation and mitotic progression in Drosophila. J Cell Biol 164 : 487–492.
30. Gharbi-AyachiA, LabbeJC, BurgessA, VigneronS, StrubJM, et al. (2010) The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science 330 : 1673–1677.
31. MochidaS, MaslenSL, SkehelM, HuntT (2010) Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330 : 1670–1673.
32. RangoneH, WegelE, GattMK, YeungE, FlowersA, et al. (2011) Suppression of scant identifies Endos as a substrate of greatwall kinase and a negative regulator of protein phosphatase 2A in mitosis. PLoS Genet 7: e1002225.
33. GloverDM (2012) The overlooked greatwall: a new perspective on mitotic control. Open Biol 2 : 120023.
34. LorcaT, CastroA (2012) The Greatwall kinase: a new pathway in the control of the cell cycle. Oncogene 32 : 537–543.
35. HarveySL, EncisoG, DephoureN, GygiSP, GunawardenaJ, et al. (2011) A phosphatase threshold sets the level of Cdk1 activity in early mitosis in budding yeast. Mol Biol Cell 22 : 3595–3608.
36. PalG, ParazMT, KelloggDR (2008) Regulation of Mih1/Cdc25 by protein phosphatase 2A and casein kinase 1. J Cell Biol 180 : 931–945.
37. LinFC, ArndtKT (1995) The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. Embo J 14 : 2745–2759.
38. MinshullJ, StraightA, RudnerAD, DernburgAF, BelmontA, et al. (1996) Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr Biol 6 : 1609–1620.
39. YangH, JiangW, GentryM, HallbergRL (2000) Loss of a protein phosphatase 2A regulatory subunit (Cdc55p) elicits improper regulation of Swe1p degradation. Mol Cell Biol 20 : 8143–8156.
40. TalarekN, CameroniE, JaquenoudM, LuoX, BontronS, et al. (2010) Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5′-3′ mRNA decay pathway. Mol Cell 38 : 345–355.
41. BontronS, JaquenoudM, VagaS, TalarekN, BodenmillerB, et al. (2012) Yeast Endosulfines Control Entry into Quiescence and Chronological Life Span by Inhibiting Protein Phosphatase 2A. Cell Rep 3 : 16–22.
42. KimMY, BucciarelliE, MortonDG, WilliamsBC, Blake-HodekK, et al. (2012) Bypassing the Greatwall-Endosulfine Pathway: Plasticity of a Pivotal Cell-Cycle Regulatory Module in Drosophila melanogaster and Caenorhabditis elegans. Genetics 191 : 1181–1197.
43. RossioV, YoshidaS (2011) Spatial regulation of Cdc55-PP2A by Zds1/Zds2 controls mitotic entry and mitotic exit in budding yeast. J Cell Biol 193 : 445–454.
44. DulubovaI, HoriuchiA, SnyderGL, GiraultJA, CzernikAJ, et al. (2001) ARPP-16/ARPP-19: a highly conserved family of cAMP-regulated phosphoproteins. J Neurochem 77 : 229–238.
45. BreitkreutzA, ChoiH, SharomJR, BoucherL, NeduvaV, et al. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328 : 1043–1046.
46. WangH, WangX, JiangY (2003) Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol Biol Cell 14 : 4342–4351.
47. HombauerH, WeismannD, MudrakI, StanzelC, FellnerT, et al. (2007) Generation of active protein phosphatase 2A is coupled to holoenzyme assembly. PLoS Biol 5: e155.
48. ChiroliE, RossioV, LucchiniG, PiattiS (2007) The budding yeast PP2ACdc55 protein phosphatase prevents the onset of anaphase in response to morphogenetic defects. J Cell Biol 177 : 599–611.
49. QueraltE, UhlmannF (2008) Separase cooperates with Zds1 and Zds2 to activate Cdc14 phosphatase in early anaphase. J Cell Biol 182 : 873–883.
50. YasutisK, VignaliM, RyderM, TameireF, DigheSA, et al. (2010) Zds2p regulates Swe1p-dependent polarized cell growth in Saccharomyces cerevisiae via a novel Cdc55p interaction domain. Mol Biol Cell 21 : 4373–4386.
51. HealyAM, ZolnierowiczS, StapletonAE, GoeblM, DePaoli-RoachAA, et al. (1991) CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol Cell Biol 11 : 5767–5780.
52. QueraltE, LehaneC, NovakB, UhlmannF (2006) Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125 : 719–732.
53. WangY, BurkeDJ (1997) Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol Cell Biol 17 : 620–626.
54. WangY, NgTY (2006) Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell 17 : 80–89.
55. YellmanCM, BurkeDJ (2006) The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell 17 : 658–666.
56. SanthanamA, HartleyA, DuvelK, BroachJR, GarrettS (2004) PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot Cell 3 : 1261–1271.
57. Di ComoCJ, ArndtKT (1996) Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev 10 : 1904–1916.
58. JiangY, BroachJR (1999) Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J 18 : 2782–2792.
59. GavinAC, AloyP, GrandiP, KrauseR, BoescheM, et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440 : 631–636.
60. HoY, GruhlerA, HeilbutA, BaderGD, MooreL, et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415 : 180–183.
61. Hood-DeGrenierJK (2011) Identification of phosphatase 2A-like Sit4-mediated signalling and ubiquitin-dependent protein sorting as modulators of caffeine sensitivity in S. cerevisiae. Yeast 28 : 189–204.
62. ReinkeA, ChenJC, AronovaS, PowersT (2006) Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem 281 : 31616–31626.
63. HoltLJ, TuchBB, VillenJ, JohnsonAD, GygiSP, et al. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325 : 1682–1686.
64. BurgessA, VigneronS, BrioudesE, LabbeJC, LorcaT, et al. (2010) Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci U S A 107 : 12564–12569.
65. WickyS, TjandraH, SchieltzD, YatesJ3rd, KelloggDR (2011) The Zds proteins control entry into mitosis and target protein phosphatase 2A to the Cdc25 phosphatase. Mol Biol Cell 22 : 20–32.
66. DuvelK, SanthanamA, GarrettS, SchneperL, BroachJR (2003) Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol Cell 11 : 1467–1478.
67. JacintoE, GuoB, ArndtKT, SchmelzleT, HallMN (2001) TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol Cell 8 : 1017–1026.
68. YanG, ShenX, JiangY (2006) Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J 25 : 3546–3555.
69. UhlmannF (2001) Secured cutting: controlling separase at the metaphase to anaphase transition. EMBO Rep 2 : 487–492.
70. RahalR, AmonA (2008) Mitotic CDKs control the metaphase-anaphase transition and trigger spindle elongation. Genes Dev 22 : 1534–1548.
71. LongtineMS, TheesfeldCL, McMillanJN, WeaverE, PringleJR, et al. (2000) Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol 20 : 4049–4061.
72. McMillanJN, LongtineMS, SiaRA, TheesfeldCL, BardesES, et al. (1999) The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol Cell Biol 19 : 6929–6939.
73. Domingo-SananesMR, KapuyO, HuntT, NovakB (2011) Switches and latches: a biochemical tug-of-war between the kinases and phosphatases that control mitosis. Philos Trans R Soc Lond B Biol Sci 366 : 3584–3594.
74. Maniatis T, Fritsch EF, Sambrook J (1992) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, NY.
75. ShermanF (1991) Getting started with yeast. Methods Enzymol 194 : 3–21.
76. WachA, BrachatA, PohlmannR, PhilippsenP (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 : 1793–1808.
77. JankeC, MagieraMM, RathfelderN, TaxisC, ReberS, et al. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21 : 947–962.
78. SheffMA, ThornKS (2004) Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21 : 661–670.
79. RancatiG, CrispoV, LucchiniG, PiattiS (2005) Mad3/BubR1 phosphorylation during spindle checkpoint activation depends on both Polo and Aurora kinases in budding yeast. Cell Cycle 4 : 972–980.
80. SchwobE, BohmT, MendenhallMD, NasmythK (1994) The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae [see comments] [published erratum appears in Cell 1996 Jan 12;84(1):following 174]. Cell 79 : 233–244.
81. SuranaU, AmonA, DowzerC, McGrewJ, ByersB, et al. (1993) Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. Embo J 12 : 1969–1978.
82. FraschiniR, D'AmbrosioC, VenturettiM, LucchiniG, PiattiS (2006) Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J Cell Biol 172 : 335–346.
83. FellnerT, LacknerDH, HombauerH, PiribauerP, MudrakI, et al. (2003) A novel and essential mechanism determining specificity and activity of protein phosphatase 2A (PP2A) in vivo. Genes Dev 17 : 2138–2150.
84. FraschiniR, FormentiE, LucchiniG, PiattiS (1999) Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J Cell Biol 145 : 979–991.
85. CalabriaI, BaroB, Rodriguez-RodriguezJA, RussinolN, QueraltE (2012) Zds1 regulates PP2ACdc55 activity and Cdc14 activation during mitotic exit through its Zds_C motif. J Cell Sci 125 : 2875–2884.
Štítky
Genetika Reprodukční medicína
Článek Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and MammalsČlánek The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and DevelopmentČlánek Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative ElementsČlánek Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine ExposureČlánek Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 7
-
Všechny články tohoto čísla
- An Solution for Crossover Formation
- Genome-Wide Association Mapping in Dogs Enables Identification of the Homeobox Gene, , as a Genetic Component of Neural Tube Defects in Humans
- Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and Mammals
- Stepwise Activation of the ATR Signaling Pathway upon Increasing Replication Stress Impacts Fragile Site Integrity
- Genomic Analysis of Natural Selection and Phenotypic Variation in High-Altitude Mongolians
- Modification of tRNA by Elongator Is Essential for Efficient Translation of Stress mRNAs
- Role of CTCF Protein in Regulating Locus Transcription
- Gene Set Signature of Reversal Reaction Type I in Leprosy Patients
- Mapping of PARK2 and PACRG Overlapping Regulatory Region Reveals LD Structure and Functional Variants in Association with Leprosy in Unrelated Indian Population Groups
- Is Required for Formation of the Genital Ridge in Mice
- Monopolin Subunit Csm1 Associates with MIND Complex to Establish Monopolar Attachment of Sister Kinetochores at Meiosis I
- Recombination Dynamics of a Human Y-Chromosomal Palindrome: Rapid GC-Biased Gene Conversion, Multi-kilobase Conversion Tracts, and Rare Inversions
- Mechanisms of Protein Sequence Divergence and Incompatibility
- Histone Methyltransferase DOT1L Drives Recovery of Gene Expression after a Genotoxic Attack
- Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies
- Combinatorial Regulation of Meiotic Holliday Junction Resolution in by HIM-6 (BLM) Helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 Nucleases
- The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development
- The Role of Interruptions in polyQ in the Pathology of SCA1
- Dietary Restriction Induced Longevity Is Mediated by Nuclear Receptor NHR-62 in
- Fine Time Course Expression Analysis Identifies Cascades of Activation and Repression and Maps a Putative Regulator of Mammalian Sex Determination
- Genome-scale Co-evolutionary Inference Identifies Functions and Clients of Bacterial Hsp90
- Oxidative Stress and Replication-Independent DNA Breakage Induced by Arsenic in
- A Moonlighting Enzyme Links Cell Size with Central Metabolism
- Budding Yeast Greatwall and Endosulfines Control Activity and Spatial Regulation of PP2A for Timely Mitotic Progression
- The Conserved Intronic Cleavage and Polyadenylation Site of CstF-77 Gene Imparts Control of 3′ End Processing Activity through Feedback Autoregulation and by U1 snRNP
- The BTB-zinc Finger Transcription Factor Abrupt Acts as an Epithelial Oncogene in through Maintaining a Progenitor-like Cell State
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- The RNA-binding Proteins FMR1, Rasputin and Caprin Act Together with the UBA Protein Lingerer to Restrict Tissue Growth in
- Pattern Dynamics in Adaxial-Abaxial Specific Gene Expression Are Modulated by a Plastid Retrograde Signal during Leaf Development
- A Network of HMG-box Transcription Factors Regulates Sexual Cycle in the Fungus
- Bacterial Adaptation through Loss of Function
- ENU-induced Mutation in the DNA-binding Domain of KLF3 Reveals Important Roles for KLF3 in Cardiovascular Development and Function in Mice
- Interplay between Structure-Specific Endonucleases for Crossover Control during Meiosis
- FGF Signalling Regulates Chromatin Organisation during Neural Differentiation via Mechanisms that Can Be Uncoupled from Transcription
- The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression
- Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative Elements
- The Conserved ADAMTS-like Protein Lonely heart Mediates Matrix Formation and Cardiac Tissue Integrity
- The cGMP-Dependent Protein Kinase EGL-4 Regulates Nociceptive Behavioral Sensitivity
- RBM5 Is a Male Germ Cell Splicing Factor and Is Required for Spermatid Differentiation and Male Fertility
- Disease-Related Growth Factor and Embryonic Signaling Pathways Modulate an Enhancer of Expression at the 6q23.2 Coronary Heart Disease Locus
- Yeast Pol4 Promotes Tel1-Regulated Chromosomal Translocations
- A Dual Role for SOX10 in the Maintenance of the Postnatal Melanocyte Lineage and the Differentiation of Melanocyte Stem Cell Progenitors
- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Odoriferous Defensive Stink Gland Transcriptome to Identify Novel Genes Necessary for Quinone Synthesis in the Red Flour Beetle,
- Prediction of Complex Human Traits Using the Genomic Best Linear Unbiased Predictor
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
- Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine Exposure
- Exquisite Light Sensitivity of Cryptochrome
- miR-133a Regulates Adipocyte Browning In Vivo
- Strabismus Promotes Recruitment and Degradation of Farnesylated Prickle in Planar Polarity Specification
- Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
- Is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer
- Joint Molecule Resolution Requires the Redundant Activities of MUS-81 and XPF-1 during Meiosis
- The Mating Competence of Geographically Diverse Strains in Their Natural and Unnatural Sand Fly Vectors
- Defective Repair of Oxidative Base Lesions by the DNA Glycosylase Nth1 Associates with Multiple Telomere Defects
- Effective Blocking of the Enhancer Requires Cooperation between Two Main Mechanisms Suggested for the Insulator Function
- Trans-Ancestral Studies Fine Map the SLE-Susceptibility Locus
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Bacterial Adaptation through Loss of Function
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



