-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Moonlighting Enzyme Links Cell Size with Central Metabolism
Growth rate and nutrient availability are the primary determinants of size in single-celled organisms: rapidly growing Escherichia coli cells are more than twice as large as their slow growing counterparts. Here we report the identification of the glucosyltransferase OpgH as a nutrient-dependent regulator of E. coli cell size. During growth under nutrient-rich conditions, OpgH localizes to the nascent septal site, where it antagonizes assembly of the tubulin-like cell division protein FtsZ, delaying division and increasing cell size. Biochemical analysis is consistent with OpgH sequestering FtsZ from growing polymers. OpgH is functionally analogous to UgtP, a Bacillus subtilis glucosyltransferase that inhibits cell division in a growth rate-dependent fashion. In a striking example of convergent evolution, OpgH and UgtP share no homology, have distinct enzymatic activities, and appear to inhibit FtsZ assembly through different mechanisms. Comparative analysis of E. coli and B. subtilis reveals conserved aspects of growth rate regulation and cell size control that are likely to be broadly applicable. These include the conservation of uridine diphosphate glucose as a proxy for nutrient status and the use of moonlighting enzymes to couple growth rate-dependent phenomena to central metabolism.
Published in the journal: . PLoS Genet 9(7): e32767. doi:10.1371/journal.pgen.1003663
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003663Summary
Growth rate and nutrient availability are the primary determinants of size in single-celled organisms: rapidly growing Escherichia coli cells are more than twice as large as their slow growing counterparts. Here we report the identification of the glucosyltransferase OpgH as a nutrient-dependent regulator of E. coli cell size. During growth under nutrient-rich conditions, OpgH localizes to the nascent septal site, where it antagonizes assembly of the tubulin-like cell division protein FtsZ, delaying division and increasing cell size. Biochemical analysis is consistent with OpgH sequestering FtsZ from growing polymers. OpgH is functionally analogous to UgtP, a Bacillus subtilis glucosyltransferase that inhibits cell division in a growth rate-dependent fashion. In a striking example of convergent evolution, OpgH and UgtP share no homology, have distinct enzymatic activities, and appear to inhibit FtsZ assembly through different mechanisms. Comparative analysis of E. coli and B. subtilis reveals conserved aspects of growth rate regulation and cell size control that are likely to be broadly applicable. These include the conservation of uridine diphosphate glucose as a proxy for nutrient status and the use of moonlighting enzymes to couple growth rate-dependent phenomena to central metabolism.
Introduction
Cell size control is a fundamental aspect of the cell cycle. Coordinating cell growth with division is essential to ensure that daughter cells have sufficient room for cytoplasmic and genetic material and are the correct size for a given condition or developmental fate. Despite the universal requirement for size control, how cells are able to detect the achievement of a particular size and communicate this information to the division apparatus remains an unresolved question in cell biology [1].
Nutrient availability is a primary determinant of cell size for single-celled organisms. In their seminal 1958 studies Schaechter, Maaløe, and Kjeldgaard determined that Salmonella cell size is coupled to growth rate, which is itself a function of nutrient availability [2]. Later work established that growth rate and nutrient availability are conserved determinants of cell size. Escherichia coli and Bacillus subtilis both coordinate cell size with nutrient availability, as do single-celled eukaryotes including the classic cell cycle model organism Schizosaccharomyces pombe [3], [4], [5].
To coordinate growth rate and nutrient availability with size, cells must have a mechanism to transmit information about growth rate and metabolic status to the division machinery. In B. subtilis, cell size is coordinated with central metabolism in part through uridine diphosphate glucose (UDP-glucose)-dependent changes in the oligomerization potential of the glucosyltransferase, UgtP [6], [7]. During growth in nutrient-rich medium UDP-glucose, synthesized in a two-step pathway from glucose-6-phosphate, stimulates interaction between UgtP and the highly conserved tubulin-like cell division protein FtsZ, delaying assembly of the division machinery and increasing cell size. Conversely, during growth in nutrient-poor medium and/or in the absence of UDP-glucose, UgtP favors self-interaction. This permits division to proceed unimpeded yielding a smaller size. UgtP is a moonlighting enzyme also required for synthesis of the diglucosyl-diacylglycerol (Di-glc-DAG) anchor for lipoteichoic acid (LTA), an anionic polymer that is a major component of the Gram-positive cell wall. Loss-of-function mutations in ugtP disrupt synthesis of the Di-glc-DAG moiety, but does not impact LTA synthesis [8].
Previous reports have indicated that inactivating UDP-glucose synthesis by inactivating the phosphoglucomutase, pgm, results in a ∼25% reduction in E. coli cell size under nutrient-rich conditions [9], [10]. This phenotype suggests that pgm mutant cells are unable to properly coordinate cell division with nutritional conditions and, furthermore, implicate UDP-glucose as a widely conserved intracellular proxy for nutrient-dependent size control. Interestingly, despite the apparent conservation of UDP-glucose as a signaling molecule, the identity of the effector is less obvious. As a Gram-negative bacterium, E. coli does not synthesize LTA and computational analysis does not reveal a ugtP homolog within its ∼4.6 MB genome.
Here we report the identification and characterization of the integral inner-membrane protein OpgH as a UDP-glucose-activated inhibitor of FtsZ ring formation in E. coli. Genetic and biochemical data indicate that OpgH interacts directly with FtsZ via its N-terminal domain to inhibit division in a UDP-glucose-dependent manner. OpgH is the functional homolog of UgtP, a sugar transferase in B. subtilis that modulates cell size in response to carbon availability. While both moonlighting enzymes serve as membrane-associated glucosyltransferases, they share no homology, have distinct enzymatic activities, and inhibit FtsZ assembly through different mechanisms thereby exposing a remarkable instance of convergent evolution. Moreover, this work significantly advances the understanding of nutrient-dependent cell size control in E. coli, the predominant model organism for studying bacterial physiology.
Results
E. coli utilizes UDP-glucose to couple cell size with nutrient availability
Previous work from our lab and others suggests that E. coli may employ UDP-glucose as an intracellular proxy for nutrient availability in the regulatory circuit responsible for coupling cell size with growth rate [10], [11]. To test this we examined the size of wild type cells and pgm null mutants that are defective in the first step of UDP-glucose biosynthesis (Figure 1A). If UDP-glucose is central to the nutrient-dependent control of E. coli cell size, the size differential between wild type and pgm mutant cells should be greatest under nutrient-rich conditions and least under nutrient-poor conditions.
Fig. 1. E. coli utilizes UDP-glucose to coordinate nutrient availability with cell size. 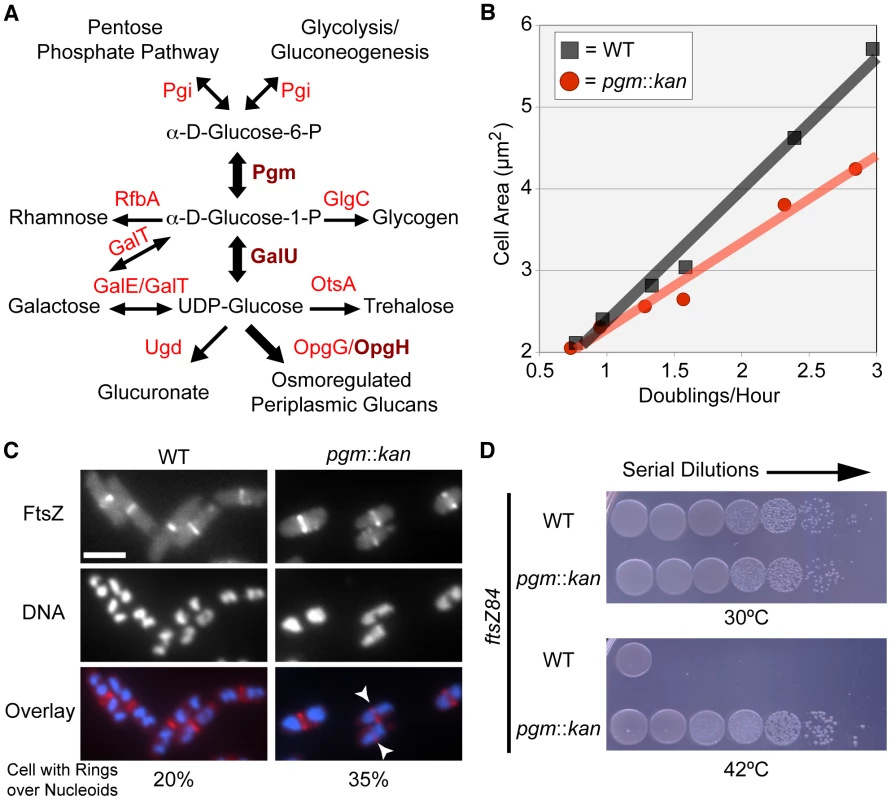
(A) A simplified schematic of UDP-glucose production and utilization in E. coli. Substrates are in black font, enzymes are in red font. Enzymes crucial to coordination of size with growth rate are in dark red and bold. Metabolic pathway information was derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) website [75], [76]. (B) Cell area measurements of wild type (black squares) versus pgm::kan (red circles) in various growth media (from left to right: AB-succinate, AB-glucose, AB-succinate + casamino acids (CAA), AB-glucose + CAA, LB, and LB-glucose). (C) Occurrence of FtsZ rings over incompletely segregated nucleoids in wild type or smaller pgm::kan cells. Both strains encode a chromosomal copy of Plac::gfp-ftsZ and were cultured in nutrient-rich conditions (LB-glucose with 1 mM IPTG). GFP-FtsZ (top), DAPI stained DNA (middle), and the overlay with FtsZ in red and DNA in blue (bottom). White arrows indicate cells with a division ring over partially segregated chromosomes. Bar = 2.5 µm. (See Table S1 for expanded analysis.) (D) Disrupting UDP-glucose production by disrupting pgm suppresses lethality of FtsZ84 in non-permissive conditions (42°C). Serial dilution plating of strains with ftsZ84 ± pgm::kan grown on nutrient-rich, low-salt media at 30°C (permissive) or 42°C (restrictive). See also Figure S1 (expression levels of FtsZ84). Measurements of the cross-sectional area of cells cultured under a range of nutrient conditions indicate that pgm mutants are indeed defective in the growth-rate-dependent control of cell size (Figure 1B). In LB+0.2% glucose (LB-glucose), the average cross-sectional area of wild-type E. coli was 5.66 µm2 while pgm mutants were 4.24 µm2, a difference of over 25%. However, the size inequity dissipated under nutrient-poor conditions. pgm mutants were only 4% smaller than wild type when cultured at a growth rate 4-times slower in a minimal growth medium supplemented with 0.4% succinate (AB-succinate).
Defects in UDP-glucose biosynthesis increase the frequency of FtsZ rings over incompletely segregated nucleoids
Rapidly growing cells are not only longer, but also have more total DNA, a consequence of multifork replication. Multifork replication is a phenomenon that allows E. coli and other bacteria to sustain mass doubling times shorter than the period required to complete chromosome replication and division. In previous work we determined that growth rate-dependent increases in cell size help prevent aberrant assembly of the division machinery over unsegregated bacterial chromosomes (nucleoids) in B. subtilis [6]. To determine the impact of a reduction in E. coli cell size on the frequency of division rings formed over nucleoids, we visualized FtsZ ring formation in rapidly growing pgm mutant cells with an inducible copy of ftsZ fused to gfp [12].
Consistent with growth rate-dependent increases in cell size ensuring that E. coli cells have sufficient room for DNA segregation during multifork replication, we observed a near two-fold increase in the frequency of division rings positioned over nucleoids in pgm mutants relative to wild type. In accordance with previous studies [13], approximately 20% (130/654) of wild type cells had an FtsZ ring over chromosomal material. The frequency increased to 35% (246/662) in diminutive pgm::kan cells (Figure 1C; Table S1). This suggests that the growth rate-dependent size increase is, in part, a mechanism to spatially coordinate the excess DNA generated by multifork replication.
Defects in UDP-glucose biosynthesis stabilize FtsZ assembly at midcell
To coordinate size with growth rate, cells must evaluate nutrient availability and subsequently transmit that information to the division machinery. To assess the impact of UDP-glucose on FtsZ assembly in E. coli, we determined if defects in UDP-glucose biosynthesis were sufficient to suppress the conditional lethality of the heat-sensitive ftsZ84 allele. FtsZ84 (G105S) supports assembly of the division ring at 30°C, but is unable at 42°C, leading to extensive filamentation and cell death [14], [15].
Consistent with increases in UDP-glucose activating an inhibitor of FtsZ assembly, the viability of pgm::kan ftsZ84 double mutants was ∼4.5-fold higher than ftsZ84 alone under restrictive conditions (Figure 1D). The plating efficiency (CFU restrictive/CFU permissive) of ftsZ84 cells grown at 30°C versus 42°C was just 0.013% (±0.001). In contrast, pgm::kan ftsZ84 double mutants exhibited essentially no reduction in viability under identical conditions 99.2% (±0.3). Notably, this is the first example of a loss-of-function mutation that suppresses the heat sensitivity of ftsZ84 without increasing the intracellular concentration of FtsZ84 (Figure S1). Together, the data presented in Figure 1 provides evidence for a UDP-glucose-activated factor coupling cell division directly to carbon metabolism.
Defects in the glucosyltransferase OpgH reduce E. coli cell size
To identify the UDP-glucose-dependent regulator of E. coli cell size, we systematically screened kanamycin resistance cassette insertions in genes predicted to be associated with UDP-glucose synthesis or utilization for defects in cell size, taking advantage of the Keio mutant collection (Figure 1A) [16]. Importantly, mass doubling rates in these mutants were indistinguishable from wild type in both nutrient-rich and nutrient-poor conditions (Table S2).
Of the 16 mutants we screened, knockouts in only three genes pgm, galU encoding a pyrophosphorylase required for synthesis of UDP-glucose from glucose-1-phosphate, and opgH (mdoH, b1049, EcoCyc:EG11886, UniProt:P62517) encoding a Family II glucosyltransferase, resulted in a statistically significant reduction in cell size (Figure 2A, 2B; Figure S3). Wild-type MG1655 cells had an average area of 5.66 µm2. The galU::kan and opgH::kan mutants were 18% (4.66 µm2) and 12% (5.01 µm2) smaller than wild type, respectively. As previously shown in Figure 1B, pgm null cells were 25% smaller at 4.24 µm2. Importantly, of the six known E. coli enzymes that utilize UDP-glucose (GalE, GalT, Ugd, OpgG, OpgH, and OtsA) mutations in only one, opgH::kan, led to a statistically significant size reduction (p>0.05). (Length and width data of the various mutants is presented in Table S2.) Why mutations in galU and opgH do not reduce cell size to the same extent as the pgm::kan mutation is not readily apparent, however it is reminiscent of what we have observed for mutations in the analogous genes in B. subtilis [6].
Fig. 2. OpgH acts as a nutrient-dependent division antagonist. 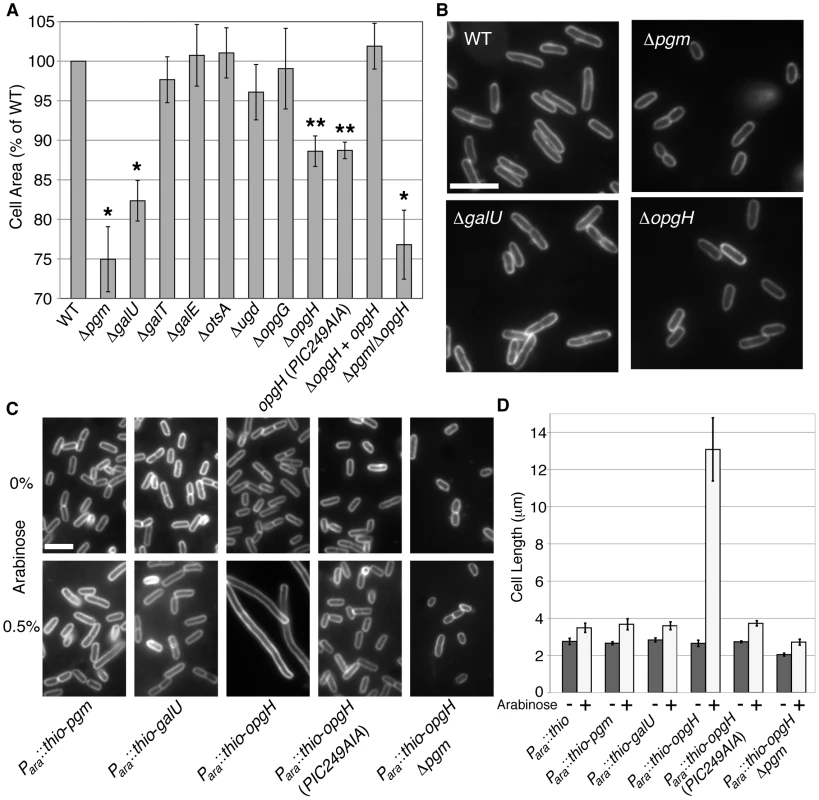
(A) Cell area measurements of mutants in UDP-glucose synthesis or utilization cultured in LB-glucose. WT is set to 100%. >250 cells were measured per sample, error bars are standard deviation (n>3). The opgH null is complemented with a plasmid encoding Plac::opgH-gfp and was cultured with 0.08 mM IPTG. This fusion is shown to be functional for glucosyltransferase ability (Figure S5). * denotes p>0.001, ** signifies p>0.05 as judged by chi2 analysis. (B) Micrographs of WT and knockouts of genes involved in UDP-glucose pathway grown in LB-glucose and stained with the membrane dye FM4-64. Bar = 5 µm. (C) Representative micrographs and (D) cell length measurements of strains cultured in LB overexpressing genes in the UDP-glucose pathway from an arabinose inducible promoter. Uninduced cells are on the upper panel. Induced constructs are on the bottom panel. Cells are stained with FM4-64. Bar = 5 µm. Error bar denotes standard deviation (n = 3). See also Table S2 (information of length, width, and growth rate), Figure S3 (measurements of cells defective in factors adjacent to the UDP-glucose biosynthesis pathway), and Figure S4 (Thio-OpgH-His overexpression levels). OpgH inhibits division in a UDP-glucose-dependent fashion
Based on our study of the parallel pathway in B. subtilis, we speculated that OpgH, rather than Pgm or GalU, was the UDP-glucose-dependent regulator of cell division in E. coli. To test this hypothesis, we measured the average size of cells expressing either pgm, galU, and opgH (fused with an N-terminal thioredoxin and a C-terminal polyhistidine) from a high-copy plasmid cultured in nutrient-rich media.
Consistent with our prediction, induction of thio-opgH-his, though not thio-pgm-his nor thio-galU-his, led to severe filamentation indicative of a block in cell division (Figure 2C, 2D). Cells expressing thio-opgH-his increased cell length over five-fold, from an average of ∼2.5 µm to ∼13.2 µm after 4 h of induction. Expression of either the thio-pgm-his or thio-galU-his fusion had a negligible impact on cell size.
UDP-glucose was absolutely required for OpgH-mediated division inhibition. Expression of wild-type thio-opgH-his in a pgm::kan background had no significant impact on cell size (Figure 2C, 2D; Figure S4). Cell size was similarly wild-type in cells expressing a mutant allele of opgH (PIC249AIA) defective in residues of the putative UDP-glucose binding site [17]. Strains with this mutation were also unable to complement normal glucosyltransferase function (Figure S5). Together, the data presented in Figure 2 suggests OpgH functions as a UDP-glucose-dependent antagonist of cell division.
OpgH regulates cell size independent from its role in OPG synthesis
OpgH is an inner-membrane glucosyltransferase that synthesizes osmoregulated periplasmic glucans (OPGs); branched glucans consisting of 5 to 13 glucose residues joined by β-1–2 linkages and branched by β-1–6 linkages [18], [19]. OpgH is responsible for the synthesis and subsequent periplasmic delivery of β-1–2 poly-glucose chains from UDP-glucose. opgH is co-transcribed with opgG, which encodes a periplasmic protein required for generating the β-1–6 branches [20].
The loss of OPG synthesis has been implicated in a range of abnormal phenotypes including deficiencies in: envelope stability, flagellar synthesis and motility, phage infectivity, biofilm formation, and pathogenicity [21], [22], [23], [24], [25], [26], [27], [28], [29]. While some of these phenotypes can be attributed to an altered cell envelope, the majority are tied to aberrant activation of the Rcs phosphorelay [22], [30], [31]. To eliminate the possibility that the cell size defect was a secondary consequence of either lacking OPGs and/or Rcs-mediated gene expression, we examined the size of cells defective for OPG synthesis or Rcs activity.
Our data shows that OpgH's role in size modulation is independent of OPG synthesis or activation of the Rcs system. Inactivating opgG, thereby eliminating OPG production either by a non-polar mutation in opgG or by complementing opgH in trans in an opgGH double mutant, had no impact on cell size (Figure 2A; Figure S6A).
Based on previous reports indicating that one of the six promoters driving ftsZ is positively regulated by Rcs [32], [33], we were particularly concerned that induction of Rcs in the absence of UDP-glucose or OPGs might lead to overexpression of ftsZ, which in turn would reduce cell size. However, FtsZ levels are wild-type in Δpgm, ΔgalU, and ΔopgH mutant cells (Figure S6B). (FtsZ would have to be overexpressed by ∼50% to translate into a size reduction of 15–20% [10], [34].) Furthermore, a ΔopgH/ΔrcsB double mutant was ∼10% smaller than an ΔrcsB single mutant (Figure S6B). Deletion of rpoS, a transcriptional target of the Rcs system encoding the stationary phase transcription factor Sigma S [35], [36], had no impact on ΔopgH or Δpgm mutant size (Figure S6C). This data is not the first example of wild-type FtsZ levels in circumstances of Rcs induction [37], [38].
OpgH localizes to the division ring only in nutrient-rich conditions, yet independent of UDP-glucose
To determine OpgH's subcellular localization pattern, we used antiserum raised against OpgH's N-terminal cytoplasmic domain. During growth in LB-glucose (τ = 21), OpgH exhibited both peripheral and midcell localization. Of the 203/227 (89%) cells with an FtsZ ring, OpgH colocalized with FtsZ at midcell in 96% (195/203) of cells (Figure 3A). OpgH midcell localization was contingent on nutrient-rich conditions. At a slower growth rate (τ = 38), the covariance of OpgH and FtsZ medial localization was significantly reduced [FtsZ at midcell 69% (170/248); covariance 36% (61/170)]. When growth rates were further reduced by culturing cells in AB-glucose (τ = 60), OpgH was relegated to the cell periphery, rarely colocalizing with an FtsZ ring (3% 3/112) [55% (112/207) of cells had an FtsZ ring]. OpgH localization to midcell was dependent on FtsZ. We were unable to detect an instance in which OpgH localized to midcell in the absence of an FtsZ ring. Further, medial localization of OpgH was abolished when ftsZ was depleted (Figure 3B).
Fig. 3. OpgH localizes to midcell in a growth rate- and FtsZ-dependent manner. 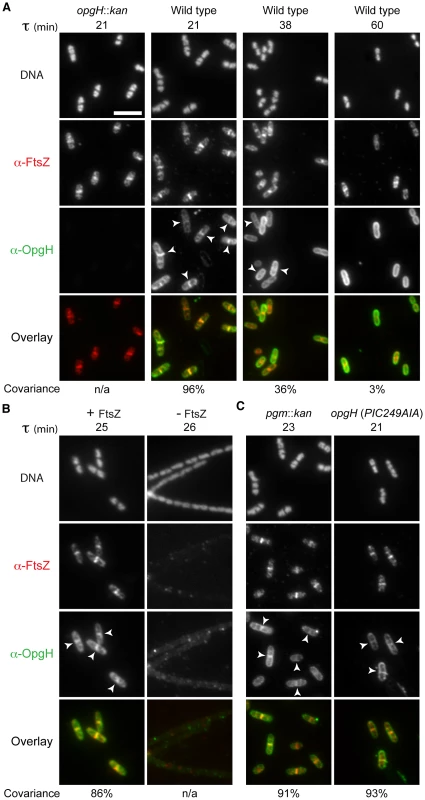
Immunofluorescence localization of FtsZ and OpgH in various growth conditions or genetic backgrounds. (A) OpgH localizes at midcell with FtsZ only at fast growth rates. Wild type cells were grown in either LB-glucose (τ = 21′), AB-glucose + casamino acids (τ = 38′), or AB-glucose (τ = 60′). (B) OpgH is unable to localize to midcell in the absence of FtsZ. A strain encoding a sodium salicylate inducible copy of ftsZ (PL3180) was grown to mid-log phase and back-diluted into LB broth ± inducer (2.5 µM sodium salicylate) for 2.5 h. (C) The frequency of OpgH at midcell is independent of UDP-glucose. Congenic strains either encoding a deletion of a key gene in UDP-glucose biosynthesis (pgm::kan) or a mutation in OpgH's putative UDP-glucose binding site. (A–C) DNA is stained by DAPI. Bar = 5 µm. White arrowheads indicate OpgH midcell localization. OpgH is in green and FtsZ is in red in the overlays. The percent covariance of FtsZ and OpgH at midcell is indicated below the micrographs. Based on our work the B. subtilis functional homolog UgtP, we speculated that changes in the intracellular levels of OpgH's substrate, UDP-glucose, might be responsible for its nutrient-dependent localization [6]. However, OpgH localization was unperturbed in either a background unable to synthesize UDP-glucose (pgm::kan) or a mutation in the putative UDP-glucose binding region (Figure 3C). Midcell covariance of OpgH and FtsZ localization at midcell in the pgm::kan cells was 91% (178/196) and 93% (212/227) in the nucleotide sugar-binding mutant when cultured in LB-glucose. Together, this data supports a model in which OpgH dynamically localizes to the division machinery in a growth rate-dependent, but UDP-glucose-independent manner.
The N-terminus of OpgH is both necessary and sufficient for division inhibition
OpgH is an 848 amino acid (97 kDa) integral inner-membrane protein. Reporter fusion analysis indicates that OpgH is composed of eight transmembrane domains with both N - and C-termini residing in the cytoplasm (Figure 4A) [39]. OpgH has three significant cytoplasmic domains referred to here as OpgHN (1–138), OpgHM (211–514), and OpgHC (702–848). OpgHM, the largest domain, contains the protein's putative UDP-glucose binding domain based upon sequence homology.
Fig. 4. The N-terminal cytoplasmic region of OpgH is necessary and sufficient to inhibit cell division. 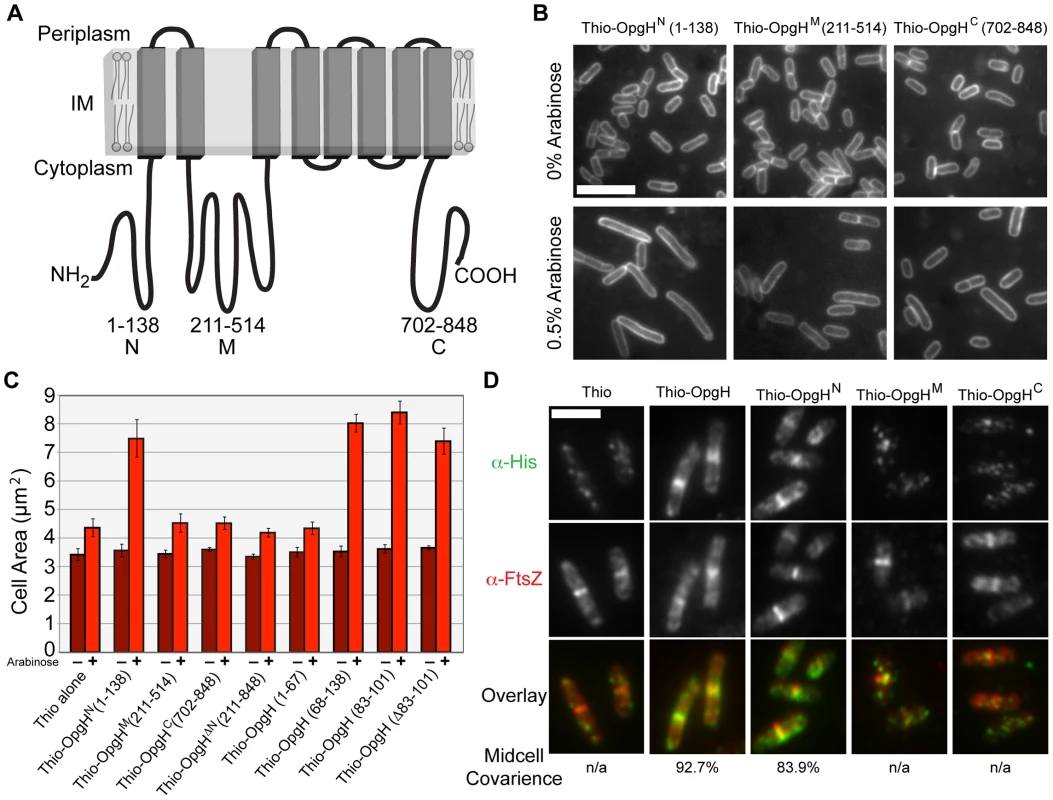
(A) A schematic representation of the inner-membrane glucosyltransferase OpgH [39]. (B) Overexpression of OpgHN increases cell size in the opgH::kan strain. Micrographs of cells encoding arabinose inducible N-terminal thioredoxin fusions to each of OpgH's cytoplasmic domains cultured in LB±0.5% arabinose. Cells are stained with FM4-64. Bar = 5 µm. (C) Cell area measurements of cells with the various thio-opgH-his constructs. Cells were cultured in LB with either 0% arabinose (dark red bars) or 0.5% arabinose (light red bars). Error bars equal standard deviation (n = 3). (D) Immunofluorescence micrographs of various Para::thio-opgH-his constructs following 2 h of induction in LB+0.5% arabinose. Overlays of OpgH (green) and FtsZ (red) localization are on the bottom row. Bar = 3 µm. See also Figure S7 (localization data on additional deletion constructions). To identify determinants within OpgH required for division inhibition, we expressed thioredoxin/polyhistidine fusions to opgHN, opgHM, or opgHC from an arabinose inducible promoter on multicopy vectors in an opgH::kan mutant background. Induction of thio-opgHN-his, but not thio-opgHM-his or thio-opgHC-his, resulted in a substantial increase in cell size relative to an uninduced control (Figure 4B, 4C). The area of cells expressing Thio-OpgHN-His increased more than two-fold ∼3 h post-induction (from 2.9 µm to 5.9 µm), indicative of a partial block in division. The differential in division inhibition efficacy between OpgH and OpgHN may be the consequence of the former being at the membrane where it can more readily access the division apparatus. Expression of an OpgHN deletion mutant (thio-opgH(Δ1-210)-his) had no impact on cell size (Figure 4C). These data are consistent with a model in which UDP-glucose binding to OpgHM leads to a conformational change that promotes OpgHN mediated division inhibition.
Expression of thioredoxin fusions to a set of nested deletions within the OpgHN domain identified an 18 amino acid peptide (residues 83–101) that was sufficient for division inhibition (Figure 4C; Figure S7). However, systematic alanine substitutions to this region or deletion of all 18 residues had no significant impact on the ability of OpgHN to mediate division inhibition (Figure 4C; data not shown). This finding suggests the presence of at least one additional determinant within OpgHN that is sufficient for its role in division inhibition.
Consistent with a role in cell division, OpgHN localized to midcell independent of the rest of the protein. Immunofluorescence microscopy using antibodies against the His tag, indicated that of the three cytoplasmic domains, only Thio-OpgHN-His exhibited medial localization on its own. During growth in LB-glucose, 84% (379/452) of cells expressing thio-opgHN-his displayed colocalization with FtsZ at midcell (Figure 4D). Strikingly, a Thioredoxin fusion to the 18 residue fragment of OpgHN (residues 83–101) that was sufficient for division inhibition, also colocalized with FtsZ in 82% (169/205) of cells (Figure S7B).
Although OpgHN was sufficient for medial localization, it was not necessary. A thioredoxin fusion to the OpgH(Δ1–210), the N-terminal deletion mutant, colocalized with FtsZ 67% of the time (148/221) during growth in LB-glucose, implying the N-terminal domain is not necessary for medial localization of OpgH (Figure S7B). Given the inability of the OpgHM or OpgHC domains to localize on their own, we hypothesize that the second localization determinant is situated in a transmembrane and/or periplasmic region of the protein (see Discussion).
OpgH inhibits division by blocking FtsZ ring formation
In light of our finding that cytoplasmic OpgHN was capable of blocking division in vivo (Figure 4B, 4C), as well as genetic data suggesting that the loss of UDP-glucose positively impacts FtsZ assembly at midcell (Figure 1D), we speculated that OpgH might modulate cell division through direct interactions with FtsZ. To test this possibility, we measured the proportion of cells with FtsZ rings following induction of either thio-his, thio-opgH-his or thio-opgHN-his. If OpgH inhibits division by antagonizing FtsZ polymerization dynamics, then induction of thio-opgH-his or thio-opgHN-his should reduce the number of FtsZ rings.
For this experiment, dilutions were calibrated to ensure cells would be in early exponential growth at each time point. Samples were taken at 25-minute intervals for ∼3 h, fixed, and the percentage of cells with an FtsZ ring were scored using immunofluorescence microscopy. Induction of both thio-opgH-his as well as thio-opgHN-his nearly abolished cells with FtsZ rings by ∼2.5 h post-induction (Figure 5A).
Fig. 5. OpgHN is an inhibitor of FtsZ assembly. 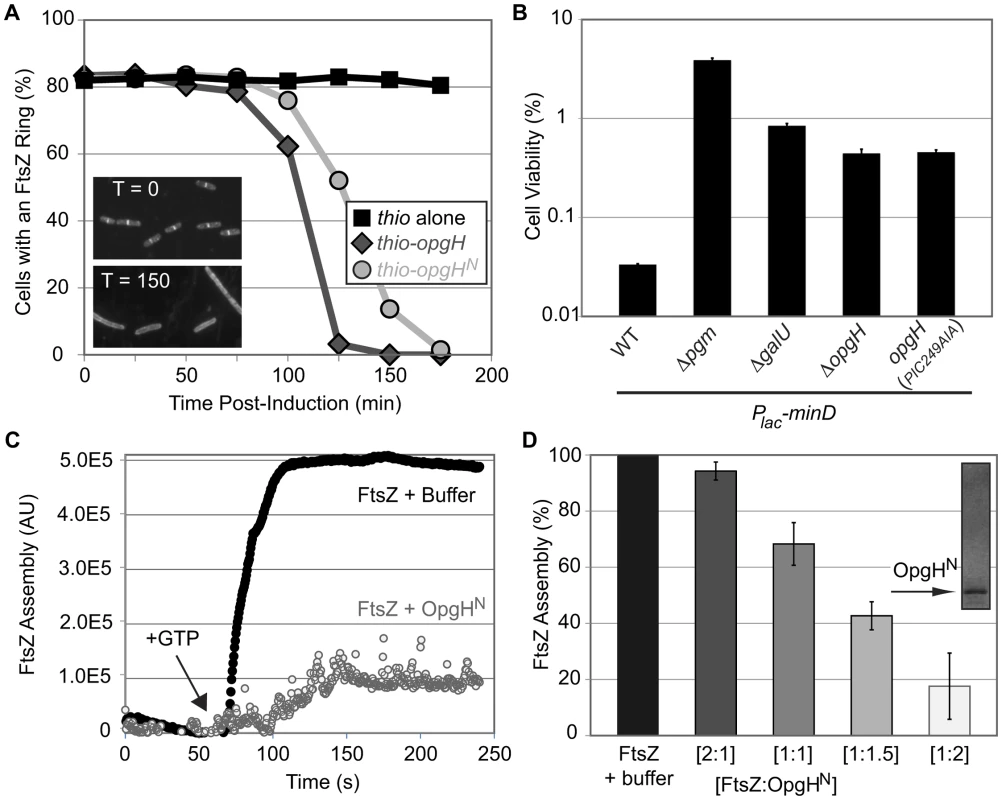
(A) Induction of OpgH inhibits FtsZ assembly in vivo. Cells were sampled and imaged for immunofluorescence microscopy at 25 minute intervals following the induction of either thio-his (black squares), thio-opgH-his (dark grey diamonds), or thio-opgHN-his (light grey circles). Cells were cultured in LB. >200 cells were evaluated per sample. Representative α-FtsZ immunofluorescence micrographs from time points 0′ and 150′ are shown in the lower left. (B) Mutations that disrupt synthesis of UDP-glucose or OpgH itself suppress the lethality of MinD overexpression. MinD is overexpressed by ∼2-fold, which is at the threshold of lethality in WT. Error bars equals standard deviation (n = 3). (See Figure S2 for relative MinD expression levels.) (C) A representative 90° angle light scattering plot of FtsZ assembly ± OpgHN. FtsZ is at 5 µM, OpgHN is at 10 µM. Arrow indicates addition of 1 mM GTP. (D) Concentration-dependent inhibition of FtsZ polymerization by OpgHN. The ratio of FtsZ to OpgHN is listed below. FtsZ is at 5 µM in all cases. (See Figure S9A, S9B for additional controls.) As an additional test of the impact of UDP-glucose and OpgH on FtsZ assembly dynamics in vivo, we evaluated the effects of overexpressing the spatial cell division regulator MinD. The precise temporal and spatial regulation of cell division is achieved through a concert of factors that modulate FtsZ assembly. Overexpression of an FtsZ inhibitor, and the associated lethality, can be balanced by deletion of another inhibitor [6], [40].
Consistent with defects in UDP-glucose synthesis or deletion of opgH enhancing FtsZ assembly, inactivating genes in the metabolic pathway suppressed lethality associated with minD overproduction (Figure 5B). Two-fold overexpression of MinD reduced the plating efficiency of wild-type E. coli to 0.03%. However, overexpression of MinD to identical levels in pgm null cells resulted in a plating efficiency of 3.9%, >100-fold higher than in the wild-type background (Figure 5B; Figure S2).
Consistent with OpgH interacting directly with FtsZ to delay division, an opgH::kan mutation increased the viability of cells overexpressing minD by ∼1.5 fold (Figure 5B; Figure S2). Cells expressing opgH(PIC249AIA) phenocopied the opgH null results, further corroborating that UDP-glucose binding is essential for OpgH mediated inhibition of FtsZ assembly. In contrast to a loss-of-function mutation in pgm, opgH::kan was unable to suppress the heat-sensitivity of ftsZ84 allele.
Genetic data suggests that OpgH reduces the pool of FtsZ available for assembly into the cytokinetic ring
To position OpgH within the regulatory hierarchy responsible for the temporal and spatial control of cell division, we examined the phenotypes of cells defective in either UDP-glucose synthesis (Δpgm) or opgH with defects in one of three previously characterized FtsZ antagonists: minCDE, slmA, or clpX. We reasoned that if OpgH shared a role in nucleoid occlusion, ring constriction, or preventing immediate division ring reassembly, then the double knockouts might exacerbate defects in division rings bisecting nucleoids, augmented cell size, or increased rate of minicells.
None of the double mutants had defects in growth rate, increased rates of division rings over unsegregated nucleoids, or a higher rate of inaccurately placed FtsZ rings during growth in LB-glucose (Table S1, data not shown). However, supporting a model in which OpgH reduces the effective concentration of FtsZ, a loss-of-function mutation in either opgH or pgm reduced the length of the abnormally long minCDE null mutants by approximately half (Figure S8; Table S1). This finding suggests that the pools of FtsZ available for assembly into the cytokinetic ring are elevated in the absence of either opgH or pgm.
OpgHN interacts directly with FtsZ to inhibit assembly
Based on the genetic evidence suggesting that OpgH antagonizes division through direct interactions with FtsZ, we next determined if purified OpgHN was sufficient to inhibit FtsZ assembly in vitro. For this experiment we employed 90° angle light scattering, a functional assay for FtsZ assembly [41], [42]. All experiments were performed with OpgHN (residues 1–138) and E. coli FtsZ in their native (untagged) form.
Consistent with our genetic data, OpgHN inhibited FtsZ assembly in a dose-dependent fashion (Figure 5C, 5D). At a 1∶1 ratio of FtsZ to OpgHN, FtsZ assembly was reduced by more than 30%. At higher ratios of FtsZ:OpgHN, 1∶1.5 and 1∶2, FtsZ assembly was reduced further by ∼60% and ∼80%, respectively. Heat-inactivating OpgH abolished its ability to inhibit FtsZ assembly (Figure S9A, S9B). This level of inhibition is slightly less potent than other E. coli FtsZ inhibitors [43], [44], [45], [46], however it is consistent with a model in which OpgH functions to delay division, not prevent it.
OpgH raises the apparent critical concentration of FtsZ required for GTP hydrolysis
Although 90° angle light scattering is valuable for measuring gross FtsZ assembly, it does not provide insight into the particular mechanism by which FtsZ assembly is obstructed. To elucidate the mechanism by which OpgH inhibits FtsZ assembly, we next determined the impact of OpgHN on FtsZ's intrinsic GTPase activity. FtsZ binds to GTP as a monomer, however dimerization is required for the formation of the GTPase active site and GTP hydrolysis (for reviews of FtsZ assembly dynamics see [7], [47], [48], [49], [50]). Factors that bind to FtsZ monomers and prevent them from being added to growing polymers (so-called sequesters) reduce GTP hydrolysis activity by interfering with dimerization and the formation of the GTPase active site [7], [51], [52], [53]. Conversely, factors that inhibit FtsZ assembly through other means (severing preformed polymers or interfering with lateral interactions between polymers) have little impact on GTP hydrolysis [43], [54], [55], [56].
Consistent with a mechanism in which OpgH sequesters FtsZ monomers and prevents them from assembling into single-stranded polymers, OpgHN reduced FtsZ's intrinsic GTP hydrolysis activity in a dose-dependent manner. Under our conditions, FtsZ hydrolyzed GTP at a rate of 5.1 GTP/minute/FtsZ (Figure 6A). GTP hydrolysis was reduced by 25% at a 1∶1 ratio (FtsZ:OpgHN), 55% at 1∶1.5, and 84% at 1∶2. OpgHN did not exhibit any significant GTPase activity on its own. Heat-treated OpgHN had no effect on FtsZ's GTPase activity (Figure S9C). Further, control experiments using a different GTPase, demonstrate that OpgHN does not interfere with the regenerative, NADH coupled GTPase assay (Figure S9D).
Fig. 6. OpgHN appears to function as an FtsZ monomer sequestering protein. 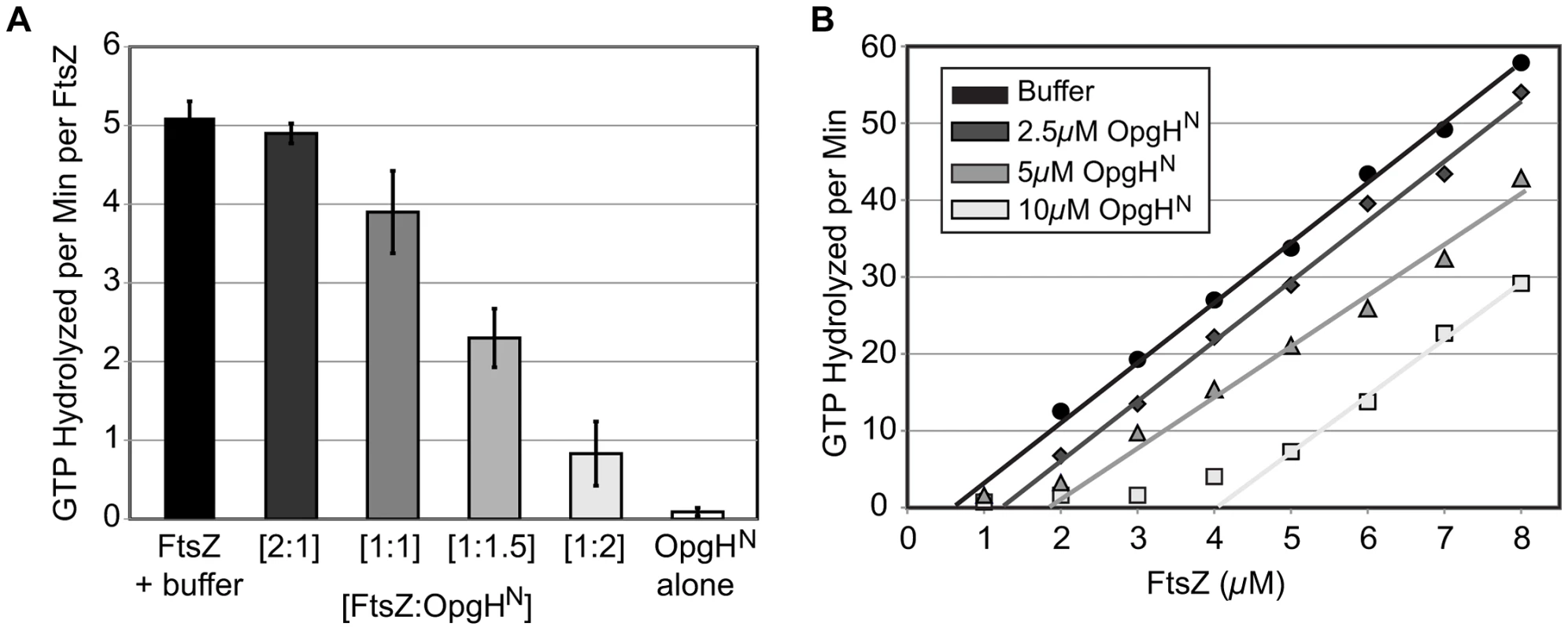
(A) Concentration-dependent inhibition of FtsZ's GTPase activity by OpgHN. The GTP hydrolysis rate of 5 µM FtsZ is shown at differing ratios of OpgHN. OpgHN alone is at 5 µM. Error bars equal standard deviation (n = 3). (B) FtsZ GTPase rates at increasing concentrations of OpgHN. The critical concentration for assembly of FtsZ was determined at OpgHN concentrations of 0 µM, 2.5 µM, 5 µM, or 10 µM. (See Figure S9C, S9D for additional controls.) Monomer binding reduces the pool of FtsZ subunits available for assembly. A hallmark of a sequestration-like mechanism is an increase in the apparent critical concentration (CcApp) of FtsZ required for GTP hydrolysis. To determine if OpgHN impacts FtsZ's CcApp, we measured GTP hydrolysis rates in reactions that kept levels of OpgHN constant, but increased the FtsZ concentration. Plotting GTP hydrolysis per minute versus FtsZ concentration identifies the X intercept for each OpgHN concentration. This intercept is equivalent to FtsZ's critical concentration under that condition [53].
In support of a model in which OpgH is a monomer binding protein, the addition of OpgHN increased FtsZ's CcApp for GTP hydrolysis. In the absence of OpgHN, E. coli FtsZ exhibited a CcApp of 0.69 µM. The CcApp was increased to 1.31 µM at a 2.5 µM OpgHN concentration, to 1.85 µM at 5 µM OpgHN, and finally to 4.08 µM in 10 µM OpgHN (Figure 6B).
Discussion
Classic work conducted over 50 years ago first identified nutritional content and growth rate as primary determinants of bacterial cell size [2]. Here we report that two factors, UDP-glucose and the glucosyltransferase OpgH, govern the regulatory circuit responsible for coordinating E. coli cell size with nutrient availability. Our genetic and biochemical data support a model in which binding of UDP-glucose by OpgH promotes a conformational change revealing an FtsZ interaction site on the OpgHN domain. Interactions between OpgHN and FtsZ obstruct assembly and/or maturation of the cytokinetic ring, delaying division and increasing cell size (Figure 7A). During slow growth in nutrient-poor conditions or defective for UDP-glucose production, OpgH assumes a conformation that obscures the interaction between OpgHN and FtsZ. FtsZ assembly is able to proceed unimpeded, reducing cell size (Figure 7B). OpgH, specifically OpgHN, is well conserved in the order Enterobacteriales, suggesting functional conservation in close relatives of E. coli (Figure S10).
Fig. 7. Glucosyltransferase OpgH couples cell size to nutritional availability and growth rate in E. coli. 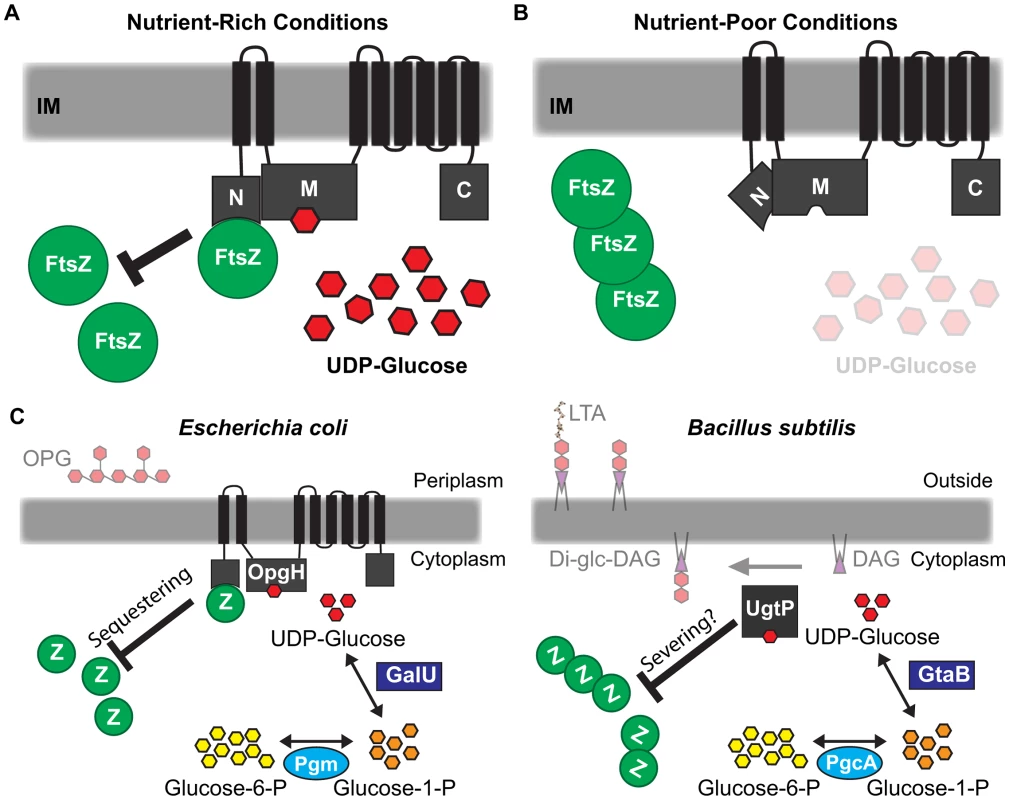
(A) The nucleotide sugar UDP-glucose acts as a proxy for nutritional status to ensure cells maintain an optimal size for a given growth rate. OpgH blocks division by inhibiting assembly of the essential bacterial cytoskeleton protein FtsZ. OpgH bound to UDP-glucose assumes a conformation where the N-terminus is able to interact with FtsZ. This interaction effectively reduces the pool of FtsZ able to participate in the formation and maturation of the FtsZ ring. (B) In nutrient-poor conditions, UDP-glucose is less available and no longer serves to promote interaction between OpgH and FtsZ. Consequently, division is unobstructed and cell size does not increase. (C) Evolutionarily divergent organisms E. coli and B. subtilis both utilize UDP-glucose and unrelated glucosyltransferases to coordinate growth rate-dependent size homeostasis. Both organisms have co-opted sugar transferases activated by UDP-glucose to antagonize assembly of FtsZ by different mechanisms. Based on our genetic and biochemical data, we propose that nutrient-dependent activation of OpgH leads to a reduction of FtsZ available for assembly into the cytokinetic ring. FtsZ levels are constant regardless of growth rate [57]. Nutrient-dependent increases in OpgH activity should translate into proportional reductions in the pool of available FtsZ subunits. Cells increase in size until they have accumulated sufficient FtsZ to overcome OpgH-mediated inhibition and support assembly of a mature cytokinetic ring [1]. This model is consistent with data from the Vicente lab demonstrating that modest reductions in FtsZ pools lead to a transient increases in cell size, but do not impact the timing of division under steady-state conditions [58].
Multiple determinants ensure OpgH localization to the division machinery
OpgH's localization to the division apparatus only occurs in nutrient-rich conditions (Figure 3). Intriguingly, this growth rate-dependent localization is not controlled by interaction with its substrate, UDP-glucose, unlike its functional B. subtilis homolog UgtP [6]. Thus, the mechanism behind OpgH's dynamic nutrient-dependent localization remains to be elucidated.
Analysis of OpgH deletion constructs suggests the presence of at least two determinants within the OpgH polypeptide are required for localization to the cytokinetic ring (Figure 4D; Figure S7B). One such determinant is within the soluble OpgHN domain. The other determinant resides somewhere in residues 211–848. Intriguingly, residues 651–713, encoding OpgH's 7th and 8th transmembrane domains, share homology to the SEDS (shape, elongation, division, and sporulation) family of proteins. The essential division protein FtsW and the shape determining protein RodA, two well-studied E. coli SEDS proteins, localize to the division apparatus through FtsZ-independent interactions [59], [60]. We hypothesize that OpgH employs its SEDS-like domain to bolster its medial localization by interacting with other components of the division machinery.
Specifically why OpgH employs more than one mechanism for localization to the cytokinetic ring is not immediately apparent. However, we speculate that concentrating OpgH at midcell may be essential to ensure that its effective concentration is high enough to meaningfully impact cell division. Although we were unable to obtain quantitative measurements of native OpgH levels, similar to other groups [39], qPCR data suggests opgH transcripts are ∼1000-fold less abundant than ftsZ transcripts (data not shown). A localization mechanism that depends on factors that localize to the division site downstream of FtsZ, would ensure that OpgH remains at midcell during the duration of the Z-period and is not displaced by competition with other FtsZ binding proteins.
A remarkable example of convergent evolution
When viewed in the context of the parallel pathway in B. subtilis, this study highlights an exceptional example of convergent evolution. E. coli and B. subtilis, organisms separated by a greater evolutionary distance than Homo sapiens and Saccharomyces cerevisiae, both utilize UDP-glucose and unrelated glucosyltransferases in the regulatory circuit coupling cell size with growth rate and nutrient availability (Figure 7C).
Why UDP-glucose-utilizing glucosyltransferase enzymes were chosen to coordinate this phenomenon is an intriguing question. In general, moonlighting enzymes tend to be more valuable if the multiple functions are both beneficial at the same time [61]. Thus, the question arises: Aside from growth in rich nutrient conditions, are there circumstances in which an increased need for their sugar transferase activity coincides with a need to prevent division?
Both effectors have previously described roles in envelope biogenesis: OpgH produces periplasmic glucans and UgtP synthesizes the Di-glc-DAG anchor for LTA. These products are particularly important during conditions that prompt the osmotic stress response [8], [62], [63]. Curiously, shifting the osmotic or turgor pressure has been reported to cause a temporary arrest of cell division that is alleviated only after cells equilibrate to the new conditions [64], [65]. Thus, while our work describes a role for OpgH and UgtP in nutrient-dependent size control, we speculate that both proteins may also function to inhibit division during osmotic stress or other cell envelope perturbations.
UDP-glucose is an ideal signaling molecule for transmitting information to the division apparatus under both conditions of rapid growth and cell envelope stress. Its accumulation is directly coupled to central carbon metabolism and is likely to accumulate primarily under nutrient-rich conditions, the same conditions that support multifork replication and necessitate an increase in cell size. Likewise, consistent with a role in the osmotic stress response pathway, genes involved in UDP-glucose synthesis are up-regulated during cell envelope stress and UDP-glucose is directly incorporated into the cell envelope [66], [67].
Curiously, synthesis of both OPG and LTA is the primary source of diacylglycerol in E. coli and B. subtilis [68], raising the possibility that diacylglycerol may serve as a secondary messenger in the regulatory circuit governing bacterial cell size. (FabH, an enzyme involved in key steps of fatty acid biosynthesis, has recently been implicated in the nutrient-dependent control of cell size, suggesting another potential role for lipids in cell size homeostasis [69].)
In closing, our findings represent a major advance in our understanding of cell size control in E. coli, the predominant model system for the study of bacterial physiology. Moreover, through comparison with a parallel pathway in B. subtilis, our work reveals conserved aspects of growth rate regulation and cell size control – including conservation of UDP-glucose, a molecule common to all domains of life, as a proxy for nutrient availability and the use of moonlighting enzymes to couple growth rate-dependent phenomena to central metabolism – that are likely to be broadly applicable.
Materials and Methods
Strains and media
E. coli strains and plasmids and their construction are described in Supporting Information (Text S1, Table S3, and Table S4). Cells were cultured in Luria-Bertani (LB) ±0.2% glucose or AB defined media [70] supplemented with 10 µg/ml thymine, ±0.5% casamino acids (CAA), and either 0.2% glucose or 0.4% succinate as a carbon source. Cells were grown at 37°C and used for experimentation at early exponential growth phase (an OD of 0.15–0.3) unless otherwise stated. Standard techniques were employed for cloning, P1vir transductions, and other genetic manipulations.
Quantitative immunoblotting
Cells were lysed at early to mid-log phase (an OD600 of 0.2–0.5) using physical or chemical techniques. Lysates were then normalized to either OD or total protein using a bicinchoninic acid (BCA) assay and subjected to SDS-PAGE. Immunoblots were performed using either rabbit α-MinD antibody (the gift of William Margolin), chicken α-His antibody (Millipore), rabbit α-FtsZ (the gift of David Weiss), or rabbit α-DnaA (the gift of Jon Kaguni) with cognate goat α-rabbit or donkey α-chicken secondary antibody conjugated to horseradish peroxidase (Jackson Immunoresearch). Band intensity was determined using ImageJ software and processed in Microsoft Excel [71].
Immunofluorescence microscopy
Microscopy performed was essentially similar to [10]. Cells were fixed in paraformaldehyde similar to as described in [72]. His-tagged proteins were detected using a chicken α-polyhistidine tag antibody (Millipore) with cognate goat α-chicken serum conjugated to Alexa488 (Invitrogen). FtsZ was detected using affinity-purified polyclonal rabbit α-FtsZ serum (the gift of William Margolin) in combination with goat α-rabbit serum conjugated to Alexa546 (Invitrogen). OpgH was detected using mouse α-OpgHN polyclonal serum with goat anti-mouse Alexa488 (Invitrogen). Genetic content was stained with DAPI.
Cell size measurements
Cells in early to mid-log phase were stained with FM-464 and adhered to 15-well glass slides using poly-L-lysine. The length and width of cells were calculated using the Openlab software. Only length was measured in cases of severe cell filamentation when width was irrelevant to the interpretation.
ftsZ84 and minD plating efficiency
Plating efficiency was done as previously described [73]. Briefly, strains encoding ftsZ84 or an inducible copy of minD were cultured in permissive conditions to early/mid-log phase (OD600 0.1–0.4), back-diluted to an OD600 of 0.01 and grown to an OD600 of 0.15–0.3. Cultures were normalized to optical density and then serially diluted from 10−1 to 10−8. Equal volumes were plated at permissive conditions (LB with 0.05% NaCl at 30°C for ftsZ84; LB-glucose + 0 mM IPTG for minD) and restrictive conditions (LB with 0.05% NaCl at 42°C for ftsZ84; LB-glucose + 0.15 mM IPTG for minD). Plating efficiency was calculated as the ratio of colony forming units at restrictive versus permissive conditions.
Protein purification
Untagged E. coli FtsZ was purified as described previously [42]. Native OpgHN was purified using the IMPACT Protein Purification System (New England Biolabs) as follows. BL21(DE3) cells harboring pBH616 (pTYB4 + opgHN) was grown in one-liter cultures with 100 µg/ml ampicillin at 37°C to mid-log, then induced with 1 mM IPTG for 4–6 h. (Inducting at temperatures under 37°C did not yield functional OpgHN.) Cells were pelleted, washed in 1×PBS (pH 7.4), repelleted, and frozen at −80°C for later use. On the day of purification, cell pellets were thawed and resuspended in 30 ml of ice-cold IMPACT lysis buffer (20 mM Tris-HCl, 500 mM NaCl, 1 mM EDTA, pH 8) containing 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (Sigma). Multiple passes in a prechilled French press cell at 1,000 lb/in2 lysed cells. All subsequent steps were performed at 4°C. Lysates were cleared by centrifugation, then loaded on to pre-equilibrated columns with 10–15 ml of chitin beads (New England Biolabs). The column was then washed with ∼10 column volumes of IMPACT lysis buffer. Subsequently, IMPACT lysis buffer containing 0.05 M dithiothreitol (DTT) was loaded to stimulate Intein auto-cleavage. The column was left incubating with DTT at 4°C overnight (16–20 h). The eluate yielded both the OpgHN-Intein fusion (71.9 kDa) and OpgHN (15.9 kDa). The fractions were pooled and transferred into dialysis tubing (7,000 MWCO) and concentrated from ∼25 ml to ∼1 ml using polyethylene glycol (PEG) 12,000. The 1 mL was immediately loaded onto a S-300 size exclusion column and equilibrated in OpgHN buffer (20 mM Tris-HCl, 100 mM NaCl, 10% glycerol, pH 8). OpgHN fractions were pooled and transferred to dialysis tubing (7,000 MWCO) and concentrated with PEG. Aliquots were flash frozen in liquid nitrogen and stored at −80°C. Protein concentrations were determined by a Coomassie Plus (Pierce) assay using a SPECTRAmax Plus spectrophotometer (Molecular Devices) using a BSA standard.
90° angle light scattering assay
Light scattering assays were performed as described previously using a DM-45 spectrofluorimeter (Olis) [40], [73]. Readings were taken every 0.5 s at 30°C. A baseline was established ∼60 s previous to the addition of 1 mM GTP. The reaction mixtures contained a final concentration of 5 µM FtsZ diluted in polymerization buffer (50 mM morpholineethanesulfonic acid (MES), 2.5 mM MgCl2, 1 mM EGTA, 50 mM KCl, pH 6.5) ± OpgHN or OpgHN buffer (20 mM Tris-HCl, 100 mM NaCl, 10% glycerol, pH 8). Data was collected by SpectralWorks (Olis) and exported into Microsoft Excel for processing. Baseline corrections were applied in Microsoft Excel to remove the background signal from unassembled FtsZ.
Regenerative coupled GTPase assay
FtsZ's GTPase activity was evaluated as previously described [42] using a continuous, regenerative coupled GTPase assay [74]. Briefly, experiments were done in the same buffer conditions as used for light scattering. A 150 µl reaction volume included: 1 mM phosphoenolpyruvate (PEP), 80 units/ml lactose dehydrogenase, 80 units/ml pyruvate kinase, 250 µM NADH, 1 mM GTP then varying amounts of FtsZ and OpgHN or equivalent volume of respective buffer. Absorbance at 340 nm was measured at 30°C for 3 min in a quartz cuvette (1 cm path length) using a SPECTRAmax Plus spectrophotometer (Molecular Devices). Raw data from 60–120 s was translated into activity with the extinction coefficient for NADH at 340 nm of 6220 M−1 cm−1.
Supporting Information
Zdroje
1. ChienAC, HillNS, LevinPA (2012) Cell size control in bacteria. Curr Biol 22: R340–349.
2. SchaechterM, MaaløeO, KjeldgaardNO (1958) Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol 19 : 592–606.
3. PierucciO, HelmstetterCE, RickertM, WeinbergerM, LeonardAC (1987) Overexpression of the dnaA gene in Escherichia coli B/r: chromosome and minichromosome replication in the presence of rifampin. J Bacteriol 169 : 1871–1877.
4. SargentMG (1975) Control of cell length in Bacillus subtilis. J Bacteriol 123 : 7–19.
5. FantesP, NurseP (1977) Control of cell size at division in fission yeast by a growth-modulated size control over nuclear division. Exp Cell Res 107 : 377–386.
6. WeartRB, LeeAH, ChienAC, HaeusserDP, HillNS, et al. (2007) A metabolic sensor governing cell size in bacteria. Cell 130 : 335–347.
7. ChienAC, ZarehSK, WangYM, LevinPA (2012) Changes in the oligomerization potential of the division inhibitor UgtP co-ordinate Bacillus subtilis cell size with nutrient availability. Mol Microbiol 86 : 594–610.
8. LazarevicV, SoldoB, MedicoN, PooleyH, BronS, et al. (2005) Bacillus subtilis alpha-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl Environ Microbiol 71 : 39–45.
9. LuM, KlecknerN (1994) Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J Bacteriol 176 : 5847–5851.
10. HillNS, KadoyaR, ChattorajDK, LevinPA (2012) Cell size and the initiation of DNA replication in bacteria. PLoS Genet 8: e1002549.
11. LuM, CampbellJ, BoyeE, KlecknerN (1994) SeqA: a negative modulator of replication initiation in E. coli. Cell 77 : 413–426.
12. HaleCA, de BoerPA (1999) Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol 181 : 167–176.
13. BernhardtTG, de BoerPA (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18 : 555–564.
14. RicardM, HirotaY (1973) Process of cellular division in Escherichia coli: physiological study on thermosensitive mutants defective in cell division. J Bacteriol 116 : 314–322.
15. LutkenhausJF, WuHC (1980) Determination of transcriptional units and gene products from the ftsA region of Escherichia coli. J Bacteriol 143 : 1281–1288.
16. BabaT, AraT, HasegawaM, TakaiY, OkumuraY, et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2 : 2006 0008.
17. BretonC, SnajdrovaL, JeanneauC, KocaJ, ImbertyA (2006) Structures and mechanisms of glycosyltransferases. Glycobiology 16 : 29R–37R.
18. BohinJP (2000) Osmoregulated periplasmic glucans in Proteobacteria. FEMS Microbiol Lett 186 : 11–19.
19. SchneiderJE, ReinholdV, RumleyMK, KennedyEP (1979) Structural studies of the membrane-derived oligosaccharides of Escherichia coli. J Biol Chem 254 : 10135–10138.
20. HanoulleX, RolletE, ClantinB, LandrieuI, Odberg-FerragutC, et al. (2004) Structural analysis of Escherichia coli OpgG, a protein required for the biosynthesis of osmoregulated periplasmic glucans. J Mol Biol 342 : 195–205.
21. RajagopalS, EisN, BhattacharyaM, NickersonKW (2003) Membrane-derived oligosaccharides (MDOs) are essential for sodium dodecyl sulfate resistance in Escherichia coli. FEMS Microbiol Lett 223 : 25–31.
22. EbelW, VaughnGJ, PetersHK3rd, TrempyJE (1997) Inactivation of mdoH leads to increased expression of colanic acid capsular polysaccharide in Escherichia coli. J Bacteriol 179 : 6858–6861.
23. FiedlerW, RoteringH (1988) Properties of Escherichia coli mutants lacking membrane-derived oligosaccharides. J Biol Chem 263 : 14684–14689.
24. GirgisHS, LiuY, RyuWS, TavazoieS (2007) A comprehensive genetic characterization of bacterial motility. PLoS Genet 3 : 1644–1660.
25. HoltjeJV, FiedlerW, RoteringH, WalderichB, van DuinJ (1988) Lysis induction of Escherichia coli by the cloned lysis protein of the phage MS2 depends on the presence of osmoregulatory membrane-derived oligosaccharides. J Biol Chem 263 : 3539–3541.
26. Penaloza-VazquezA, SreedharanA, BenderCL (2010) Transcriptional studies of the hrpM/opgH gene in Pseudomonas syringae during biofilm formation and in response to different environmental challenges. Environ Microbiol 12 : 1452–1467.
27. LoubensI, DebarbieuxL, BohinA, LacroixJM, BohinJP (1993) Homology between a genetic locus (mdoA) involved in the osmoregulated biosynthesis of periplasmic glucans in Escherichia coli and a genetic locus (hrpM) controlling pathogenicity of Pseudomonas syringae. Mol Microbiol 10 : 329–340.
28. PageF, AltabeS, Hugouvieux-Cotte-PattatN, LacroixJM, Robert-BaudouyJ, et al. (2001) Osmoregulated periplasmic glucan synthesis is required for Erwinia chrysanthemi pathogenicity. J Bacteriol 183 : 3134–3141.
29. BhagwatAA, JunW, LiuL, KannanP, DharneM, et al. (2009) Osmoregulated periplasmic glucans of Salmonella enterica serovar Typhimurium are required for optimal virulence in mice. Microbiology 155 : 229–237.
30. GottesmanS, McCullenCA, GuillierM, VanderpoolCK, MajdalaniN, et al. (2006) Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol 71 : 1–11.
31. ShibaY, MatsumotoK, HaraH (2006) DjlA negatively regulates the Rcs signal transduction system in Escherichia coli. Genes Genet Syst 81 : 51–56.
32. GervaisFG, PhoenixP, DrapeauGR (1992) The rcsB gene, a positive regulator of colanic acid biosynthesis in Escherichia coli, is also an activator of ftsZ expression. J Bacteriol 174 : 3964–3971.
33. CarballesF, BertrandC, BoucheJP, CamK (1999) Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol Microbiol 34 : 442–450.
34. WardJEJr, LutkenhausJ (1985) Overproduction of FtsZ induces minicell formation in E. coli. Cell 42 : 941–949.
35. HenggeR (2008) The two-component network and the general stress sigma factor RpoS (sigma S) in Escherichia coli. Adv Exp Med Biol 631 : 40–53.
36. BohringerJ, FischerD, MoslerG, Hengge-AronisR (1995) UDP-glucose is a potential intracellular signal molecule in the control of expression of sigma S and sigma S-dependent genes in Escherichia coli. J Bacteriol 177 : 413–422.
37. NagahamaH, SakamotoY, MatsumotoK, HaraH (2006) RcsA-dependent and -independent growth defects caused by the activated Rcs phosphorelay system in the Escherichia coli pgsA null mutant. J Gen Appl Microbiol 52 : 91–98.
38. CanoDA, Dominguez-BernalG, TierrezA, Garcia-Del PortilloF, CasadesusJ (2002) Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics 162 : 1513–1523.
39. DebarbieuxL, BohinA, BohinJP (1997) Topological analysis of the membrane-bound glucosyltransferase, MdoH, required for osmoregulated periplasmic glucan synthesis in Escherichia coli. J Bacteriol 179 : 6692–6698.
40. WeartRB, NakanoS, LaneBE, ZuberP, LevinPA (2005) The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol Microbiol 57 : 238–249.
41. MukherjeeA, LutkenhausJ (1999) Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacteriol 181 : 823–832.
42. BuskePJ, LevinPA (2012) Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J Biol Chem 287 : 10945–10957.
43. ChoH, McManusHR, DoveSL, BernhardtTG (2011) Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci U S A 108 : 3773–3778.
44. HuZ, MukherjeeA, PichoffS, LutkenhausJ (1999) The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci U S A 96 : 14819–14824.
45. SugimotoS, YamanakaK, NishikoriS, MiyagiA, AndoT, et al. (2010) AAA+ chaperone ClpX regulates dynamics of prokaryotic cytoskeletal protein FtsZ. J Biol Chem 285 : 6648–6657.
46. MukherjeeA, CaoC, LutkenhausJ (1998) Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci U S A 95 : 2885–2890.
47. EricksonHP, AndersonDE, OsawaM (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74 : 504–528.
48. LutkenhausJ, PichoffS, DuS (2012) Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton (Hoboken)
49. AdamsDW, ErringtonJ (2009) Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7 : 642–653.
50. RombergL, LevinPA (2003) Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu Rev Microbiol 57 : 125–154.
51. CordellSC, RobinsonEJ, LoweJ (2003) Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc Natl Acad Sci U S A 100 : 7889–7894.
52. MukherjeeA, LutkenhausJ (1998) Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J 17 : 462–469.
53. ChenY, MilamSL, EricksonHP (2012) SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 51 : 3100–3109.
54. ShenB, LutkenhausJ (2010) Examination of the interaction between FtsZ and MinCN in E. coli suggests how MinC disrupts Z rings. Mol Microbiol 75 : 1285–1298.
55. ThanbichlerM, ShapiroL (2006) MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126 : 147–162.
56. TonthatNK, AroldST, PickeringBF, Van DykeMW, LiangS, et al. (2011) Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J 30 : 154–164.
57. WeartRB, LevinPA (2003) Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol 185 : 2826–2834.
58. PalaciosP, VicenteM, SanchezM (1996) Dependency of Escherichia coli cell-division size, and independency of nucleoid segregation on the mode and level of ftsZ expression. Mol Microbiol 20 : 1093–1098.
59. BoyleDS, KhattarMM, AddinallSG, LutkenhausJ, DonachieWD (1997) ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol 24 : 1263–1273.
60. VatsP, ShihYL, RothfieldL (2009) Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol Microbiol 72 : 170–182.
61. HubertsDH, van der KleiIJ (2010) Moonlighting proteins: an intriguing mode of multitasking. Biochim Biophys Acta 1803 : 520–525.
62. KennedyEP (1982) Osmotic regulation and the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. Proc Natl Acad Sci U S A 79 : 1092–1095.
63. KennedyEP, RumleyMK (1988) Osmotic regulation of biosynthesis of membrane-derived oligosaccharides in Escherichia coli. J Bacteriol 170 : 2457–2461.
64. WoodJM (1999) Osmosensing by bacteria: signals and membrane-based sensors. Microbiol Mol Biol Rev 63 : 230–262.
65. MeuryJ (1988) Glycine betaine reverses the effects of osmotic stress on DNA replication and cellular division in Escherichia coli. Arch Microbiol 149 : 232–239.
66. Bury-MoneS, NomaneY, ReymondN, BarbetR, JacquetE, et al. (2009) Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet 5: e1000651.
67. VaronD, BoylanSA, OkamotoK, PriceCW (1993) Bacillus subtilis gtaB encodes UDP-glucose pyrophosphorylase and is controlled by stationary-phase transcription factor sigma B. J Bacteriol 175 : 3964–3971.
68. ZhangYM, RockCO (2008) Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6 : 222–233.
69. YaoZ, DavisRM, KishonyR, KahneD, RuizN (2012) Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc Natl Acad Sci U S A 109: E2561–2568.
70. ClarkDJ, MaaløeO (1967) DNA replication and the division cycle in Escherichia coli. J Mol Biol 23 : 99–112.
71. SchneiderCA, RasbandWS, EliceiriKW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9 : 671–675.
72. Levin PA (2002) Light Microscopy Techniques for Bacterial Cell Biology. In: Sansonetti PJ, Zychlinsky A, editors. Molecular Cellular Microbiology. London: Academic Press, Ltd. pp. 115–132.
73. HaeusserDP, SchwartzRL, SmithAM, OatesME, LevinPA (2004) EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol Microbiol 52 : 801–814.
74. IngermanE, NunnariJ (2005) A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol 404 : 611–619.
75. KanehisaM, GotoS (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28 : 27–30.
76. TanabeM, KanehisaM (2012) Using the KEGG database resource. Curr Protoc Bioinformatics Chapter 1: Unit1 12.
Štítky
Genetika Reprodukční medicína
Článek Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and MammalsČlánek The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and DevelopmentČlánek Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative ElementsČlánek Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine ExposureČlánek Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 7
-
Všechny články tohoto čísla
- An Solution for Crossover Formation
- Genome-Wide Association Mapping in Dogs Enables Identification of the Homeobox Gene, , as a Genetic Component of Neural Tube Defects in Humans
- Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and Mammals
- Stepwise Activation of the ATR Signaling Pathway upon Increasing Replication Stress Impacts Fragile Site Integrity
- Genomic Analysis of Natural Selection and Phenotypic Variation in High-Altitude Mongolians
- Modification of tRNA by Elongator Is Essential for Efficient Translation of Stress mRNAs
- Role of CTCF Protein in Regulating Locus Transcription
- Gene Set Signature of Reversal Reaction Type I in Leprosy Patients
- Mapping of PARK2 and PACRG Overlapping Regulatory Region Reveals LD Structure and Functional Variants in Association with Leprosy in Unrelated Indian Population Groups
- Is Required for Formation of the Genital Ridge in Mice
- Monopolin Subunit Csm1 Associates with MIND Complex to Establish Monopolar Attachment of Sister Kinetochores at Meiosis I
- Recombination Dynamics of a Human Y-Chromosomal Palindrome: Rapid GC-Biased Gene Conversion, Multi-kilobase Conversion Tracts, and Rare Inversions
- Mechanisms of Protein Sequence Divergence and Incompatibility
- Histone Methyltransferase DOT1L Drives Recovery of Gene Expression after a Genotoxic Attack
- Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies
- Combinatorial Regulation of Meiotic Holliday Junction Resolution in by HIM-6 (BLM) Helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 Nucleases
- The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development
- The Role of Interruptions in polyQ in the Pathology of SCA1
- Dietary Restriction Induced Longevity Is Mediated by Nuclear Receptor NHR-62 in
- Fine Time Course Expression Analysis Identifies Cascades of Activation and Repression and Maps a Putative Regulator of Mammalian Sex Determination
- Genome-scale Co-evolutionary Inference Identifies Functions and Clients of Bacterial Hsp90
- Oxidative Stress and Replication-Independent DNA Breakage Induced by Arsenic in
- A Moonlighting Enzyme Links Cell Size with Central Metabolism
- Budding Yeast Greatwall and Endosulfines Control Activity and Spatial Regulation of PP2A for Timely Mitotic Progression
- The Conserved Intronic Cleavage and Polyadenylation Site of CstF-77 Gene Imparts Control of 3′ End Processing Activity through Feedback Autoregulation and by U1 snRNP
- The BTB-zinc Finger Transcription Factor Abrupt Acts as an Epithelial Oncogene in through Maintaining a Progenitor-like Cell State
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- The RNA-binding Proteins FMR1, Rasputin and Caprin Act Together with the UBA Protein Lingerer to Restrict Tissue Growth in
- Pattern Dynamics in Adaxial-Abaxial Specific Gene Expression Are Modulated by a Plastid Retrograde Signal during Leaf Development
- A Network of HMG-box Transcription Factors Regulates Sexual Cycle in the Fungus
- Bacterial Adaptation through Loss of Function
- ENU-induced Mutation in the DNA-binding Domain of KLF3 Reveals Important Roles for KLF3 in Cardiovascular Development and Function in Mice
- Interplay between Structure-Specific Endonucleases for Crossover Control during Meiosis
- FGF Signalling Regulates Chromatin Organisation during Neural Differentiation via Mechanisms that Can Be Uncoupled from Transcription
- The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression
- Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative Elements
- The Conserved ADAMTS-like Protein Lonely heart Mediates Matrix Formation and Cardiac Tissue Integrity
- The cGMP-Dependent Protein Kinase EGL-4 Regulates Nociceptive Behavioral Sensitivity
- RBM5 Is a Male Germ Cell Splicing Factor and Is Required for Spermatid Differentiation and Male Fertility
- Disease-Related Growth Factor and Embryonic Signaling Pathways Modulate an Enhancer of Expression at the 6q23.2 Coronary Heart Disease Locus
- Yeast Pol4 Promotes Tel1-Regulated Chromosomal Translocations
- A Dual Role for SOX10 in the Maintenance of the Postnatal Melanocyte Lineage and the Differentiation of Melanocyte Stem Cell Progenitors
- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Odoriferous Defensive Stink Gland Transcriptome to Identify Novel Genes Necessary for Quinone Synthesis in the Red Flour Beetle,
- Prediction of Complex Human Traits Using the Genomic Best Linear Unbiased Predictor
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
- Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine Exposure
- Exquisite Light Sensitivity of Cryptochrome
- miR-133a Regulates Adipocyte Browning In Vivo
- Strabismus Promotes Recruitment and Degradation of Farnesylated Prickle in Planar Polarity Specification
- Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
- Is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer
- Joint Molecule Resolution Requires the Redundant Activities of MUS-81 and XPF-1 during Meiosis
- The Mating Competence of Geographically Diverse Strains in Their Natural and Unnatural Sand Fly Vectors
- Defective Repair of Oxidative Base Lesions by the DNA Glycosylase Nth1 Associates with Multiple Telomere Defects
- Effective Blocking of the Enhancer Requires Cooperation between Two Main Mechanisms Suggested for the Insulator Function
- Trans-Ancestral Studies Fine Map the SLE-Susceptibility Locus
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Bacterial Adaptation through Loss of Function
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



