-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe T-Box Factor MLS-1 Requires Groucho Co-Repressor Interaction for Uterine Muscle Specification
T-box proteins are conserved transcription factors that play crucial roles in development of all metazoans; and, in humans, mutations affecting T-box genes are associated with a variety of congenital diseases and cancers. Despite the importance of this transcription factor family, very little is known regarding how T-box factors regulate gene expression. The Caenorhabditis elegans genome contains 21 T-box genes, and their characterized functions include cell fate specification in a variety of tissues. The C. elegans Tbx1 sub-family member MLS-1 functions during larval development to specify the fate of non-striated uterine muscles; and, in mls-1 mutants, uterine muscles are transformed to a vulval muscle fate. Here we demonstrate that MLS-1 function depends on binding to the Groucho-family co-repressor UNC-37. MLS-1 interacts with UNC-37 via a conserved eh1 motif, and the MLS-1 eh1 motif is necessary for MLS-1 to specify uterine muscle fate. Moreover, unc-37 loss-of-function produces uterine muscle to vulval muscle fate transformation similar to those observed in mls-1 mutants. Based on these results, we conclude that MLS-1 specifies uterine muscle fate by repressing target gene expression, and this function depends on interaction with UNC-37. Moreover, we suggest that MLS-1 shares a common mechanism for transcriptional repression with related T-box factors in other animal phyla.
Published in the journal: . PLoS Genet 7(8): e32767. doi:10.1371/journal.pgen.1002210
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002210Summary
T-box proteins are conserved transcription factors that play crucial roles in development of all metazoans; and, in humans, mutations affecting T-box genes are associated with a variety of congenital diseases and cancers. Despite the importance of this transcription factor family, very little is known regarding how T-box factors regulate gene expression. The Caenorhabditis elegans genome contains 21 T-box genes, and their characterized functions include cell fate specification in a variety of tissues. The C. elegans Tbx1 sub-family member MLS-1 functions during larval development to specify the fate of non-striated uterine muscles; and, in mls-1 mutants, uterine muscles are transformed to a vulval muscle fate. Here we demonstrate that MLS-1 function depends on binding to the Groucho-family co-repressor UNC-37. MLS-1 interacts with UNC-37 via a conserved eh1 motif, and the MLS-1 eh1 motif is necessary for MLS-1 to specify uterine muscle fate. Moreover, unc-37 loss-of-function produces uterine muscle to vulval muscle fate transformation similar to those observed in mls-1 mutants. Based on these results, we conclude that MLS-1 specifies uterine muscle fate by repressing target gene expression, and this function depends on interaction with UNC-37. Moreover, we suggest that MLS-1 shares a common mechanism for transcriptional repression with related T-box factors in other animal phyla.
Introduction
T-box transcription factors play essential roles in the development of all multicellular organisms, where their functions include the specification of primary germ layers and the specification of cell fates during organogenesis [1], [2]. In humans, both decreased and increased activity of these factors are associated with congenital disease (Holt-Oram syndrome, Ulnar-Mammary syndrome, DiGeorge syndrome, etc) [3], auto immune disorders [4], and cancers [5], [6]. Despite this importance the mechanisms by which T-box factors regulate target gene expression are not well established.
Groucho family (Gro/TLE) proteins are conserved transcriptional co-repressors that interact with distinct Engrailed homology 1 (eh1) or WRPW/Y motifs in a wide variety of transcription factors and, in many cases, recruit histone deacetylases to target gene promoters [reviewed in [7]]. Gro/TLE factors have recently been implicated in the regulatory mechanism of several T-box factors. Xenopus Tbx6 and Tbx1, and zebrafish Tbx24 and Ntl/Brachyury interact indirectly with Gro/TLE factors through Ripply/Bowline family proteins, and this interaction can convert these proteins from transcriptional activators to repressors [8]–[10]. Two closely related members of the mouse Tbx1 subfamily Tbx15 and Tbx18 interact directly with the Gro/TLE protein TLE3 via eh1 motifs to repress reporter gene expression in mammalian cells [11]. Additional T-box factors likely function with Gro/TLE proteins, as T-box factors in several species contain eh1 motifs, including the Caenorhabditis elegans T-box factors MLS-1 and MAB-9 [12]. While this accumulating evidence suggests a variety of T-box factors interact with Gro/TLE factors, the significance of these interactions has not been examined in vivo.
In this report we investigate the interaction between MLS-1 and the C. elegans Gro/TLE protein UNC-37. MLS-1 is a member of the Tbx1 subfamily that includes mouse Gro/TLE-interacting proteins Tbx15 and Tbx18 [1], [13]. MLS-1 functions to specify uterine muscle fate in the mesodermal (M) lineage during hermaphrodite larval development [14]. In wild-type hermaphrodites, the M mesoblast produces all post-embryonic mesoderm cells, including two sex myoblasts (SMs) that divide during the late L3 and L4 stages to produce eight uterine muscles (four um1 and four um2 uterine muscles) and eight vulval muscles (four vm1 and four vm2 vulval muscles) [15]. mls-1 loss-of-function results in a transformation of uterine muscle precursors to a vulval muscle fate resulting in the loss of all um1 and um2 muscles and the formation of excess vm1 and vm2 muscles. In comparison, ectopic expression of mls-1 throughout the M lineage results in supernumerary uterine muscles [14].
Here we demonstrate that MLS-1 interacts with UNC-37 in both yeast two-hybrid and in C. elegans bimolecular fluorescence complementation (BiFC) assays. This interaction is mediated by an eh1 motif near the MLS-1 N-terminus, and mutation of this eh1 motif eliminates the ability of MLS-1 to specify uterine muscles. Furthermore, unc-37 loss-of-function results in a loss of uterine muscles and a corresponding gain of vulval muscles similar to mls-1 loss-of-function. Taken together, these results indicate MLS-1 functions as an UNC-37 dependent transcriptional repressor to specify uterine muscle fate, and they provide the first in vivo evidence that interaction with Gro/TLE factors is essential for T-box factor function.
Results
MLS-1 and UNC-37 interact in both yeast and worms via a conserved eh1 motif
C. elegans MLS-1 is a relatively small T-box protein (252 aa; Accession NP_498640) consisting of a 187 residue T-box DNA-binding domain flanked by short N-terminal and C-terminal amino acid stretches [14]. A bioinformatic screen of predicted transcription factors in C. elegans identified a high-scoring eh1 motif outside of the T-box near the MLS-1 N-terminus [12]. The MLS-1 eh1 motif is conserved in the N-terminus of MLS-1 proteins of several Caenorhabditis species (Figure 1A) suggesting it is biologically significant, and we hypothesize MLS-1 functions as a Groucho-dependent transcriptional repressor.
Fig. 1. MLS-1 and UNC-37 physically interact in an MLS-1 eh1 motif-dependent manner. 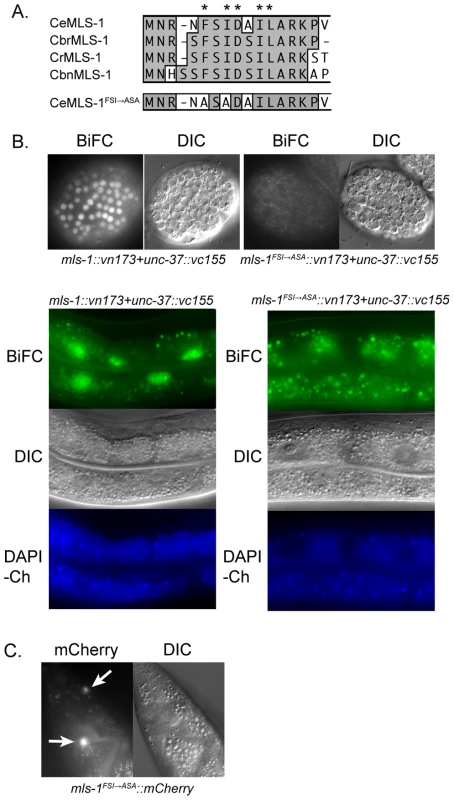
A) ClustalW alignment of the N-terminus of C. elegans (Ce), C. briggsae (Cbr), C. remanei (Cr), C. brenneri (Cbn), and mutated C. elegans MLS-1 sequences. Asterisks are above eh1 motif core residues. Two of these residues in C. elegans MLS-1 were mutated to alanines producing C. elegans MLS-1FSI→ASA. B) Transgenic animals (OK0691) co-expressing mls-1::vn173 and unc-37::vc155 (left column) show BiFC signal in embryo (top) and adult intestinal (bottom) nuclei. Animals (OK0710) co-expressing mls-1FSI→ASA::vn173 and unc-37::vc155 (right column) show no BiFC signal. Transgene expression is controlled by hsp-16.4 promoter. Gut granule autofluorescence is shown by images of DAPI channel (DAPI-Ch). C) MLS-1FSI→ASA::mCherry fusion proteins expressed using the mls-1 promoter are observed in enteric muscles (arrows). To test this hypothesis, we first asked if MLS-1 can interact with the C. elegans Groucho protein UNC-37 using yeast two-hybrid assays. A full-length MLS-1 bait was tested for interaction with a prey encoding UNC-37 residues 70–612, which consists of the entire WD repeat domain (Accession O02482). The WD repeat domain of Gro/TLE proteins mediates interaction with target proteins [7], [16], [17]. We found that MLS-1 interacts specifically with the UNC-37 prey, but does not interact with the empty prey vector. Mutating two consensus residues in the MLS-1 eh1 motif to alanines abolished this interaction (Figure 1A). In subsequent experiments we refer to this mutation as MLS-1FSI→ASA.
To further explore this physical interaction we asked if MLS-1 and UNC-37 are able to undergo bimolecular fluorescence complementation (BiFC) in C. elegans. In BiFC assays separate pieces of a fluorescent protein are attached to potential protein interactors, and if these proteins interact the fluorescent protein is able to reconstitute giving off a fluorescent signal [18], [19]. We generated three independent transgenic lines expressing mls-1 fused to a Venus N-terminal fragment (mls-1::vn173) and unc-37 fused to a Venus C-terminal fragment (unc-37::vc155) under control of the heat shock promoter hsp-16.41. Heat shocking each of these lines resulted in BiFC signal in the nuclei of several cell types in which the hsp-16.41 promoter is strongly active after heat shock, most noticeably in nuclei in embryos and the intestinal cells (Figure 1B) [20]. The nuclear BiFC signal in the intestine was distinct from autofluorescence from endogenous gut granules (Figure 1B), and transgenic lines expressing only unfused Venus protein halves had no BiFC signal as previously reported [18].
To determine whether the interaction between MLS-1 and UNC-37 requires the MLS-1 eh1 motif, we asked whether co-expression of mls-1FSI→ASA::vn173 and unc-37::vc155 would present a BiFC signal. After a one hour heat shock, transgenic animals co-expressing these proteins displayed no BiFC signal, whereas animals co-expressing wild-type mls-1::vn173 with unc-37::vc155 exhibited abundant BiFC signal (Figure 1B). Substantially longer heat shocks caused some nuclei co-expressing mls-1FSI→ASA::vn173 and unc-37::vn155 to exhibit low intensity BiFC signal, suggesting that the eh1 mutation reduced but did not completely eliminate interaction with UNC-37, and that the MLS-1FSI→ASA protein is expressed. To further demonstrate that the MLS-1FSI→ASA protein is expressed, we examined strains containing an mls-1FSI→ASA::mCherry transgene regulated by the mls-1 promoter, and found that this mutant fusion protein is expressed in enteric muscles at a level comparable to wild-type mls-1::mCherry (Figure 1C).
Taken together, these results indicate that MLS-1 can interact with UNC-37 in yeast and in C. elegans, and that these interactions depend on the MLS-1 eh1 motif. Furthermore, in C. elegans this interaction was detected only in cell nuclei, consistent with the hypothesis that these proteins interact to repress target gene expression.
MLS-1 and UNC-37 interact in biologically relevant cell types
MLS-1 and UNC-37 are capable of interacting in yeast and when expressed broadly in C. elegans using a heat shock promoter, but it is not clear from these results whether these proteins interact in any of the cells in which MLS-1 functions. UNC-37 is believed to be ubiquitously expressed throughout development [17], but MLS-1 is expressed in a very restricted pattern in uterine muscles, type 2 vulval muscles (after specification of these cells as vulval muscles), and three enteric muscles (left and right intestinal muscles and anal depressor muscle) [14]. Our analysis of mls-1 promoter activity confirms that mls-1 is expressed in the above mentioned enteric muscles throughout the life of animals post-hatching, but is only very transiently expressed in the uterine muscles and vm2s at L4 and young adult stages (Figure S1).
To determine if MLS-1 and UNC-37 interact in cells normally expressing MLS-1, we generated three independent strains co-expressing mls-1::vc155 and unc-37::vn155 under control of the mls-1 and unc-37 promoters, respectively, and found that each of these lines exhibited BiFC signal in most cells where mls-1 is normally expressed. BiFC signal was observed postembryonically in one or two cells in the tail, which we tentatively identify as enteric muscles (Figure 2C). BiFC signal was also observed near the vulva in young adults, in cells that we identify as the vm2 vulval muscles and the um1 and um2 uterine muscles based on position and morphology (Figure 2A, 2B). The um2 cells had BiFC signal localized in either nuclei or cytoplasm, and we believe these differences reflect the different morphology of type 1 and type 2 uterine muscles [21]. The signals in the vulval and uterine muscles were very transient and were not observed in L4s or in older adults.
Fig. 2. MLS-1 and UNC-37 are both co-expressed and interact in uterine, vulval, and enteric muscles. 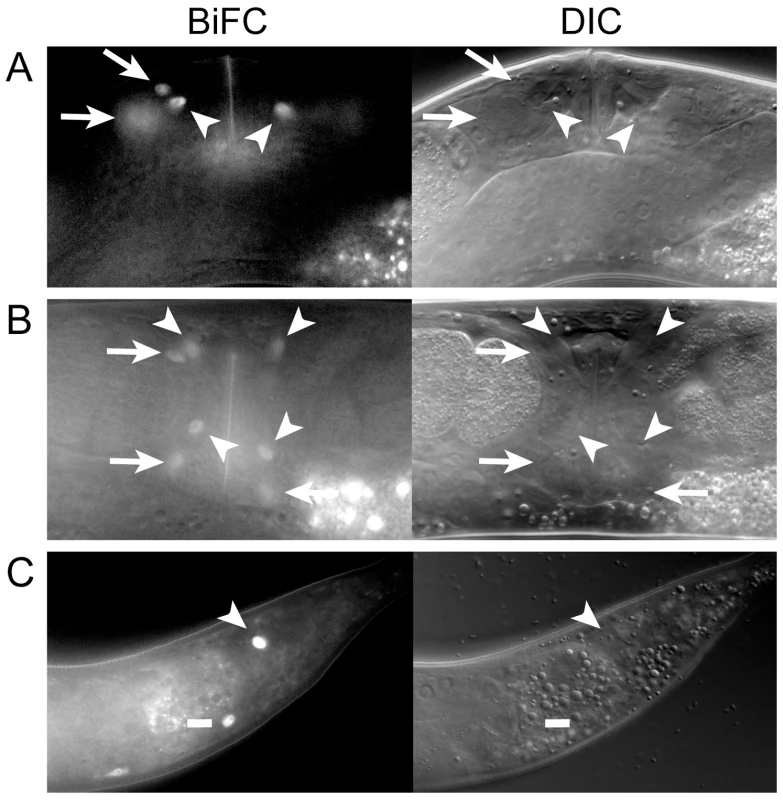
A, B) Lateral (A) and ventral (B) views of OK0742 young adult hermaphrodites co-expressing mls-1::vn173 and unc-37::vc155 under control of their own promoters exhibiting BiFC signal in uterine muscle (arrows) and vulval muscle (arrowheads) nuclei. Uterine muscle cytoplasm and nuclei displayed BiFC (compare arrows). C) Adult (OK0744) animal exhibiting BiFC in one intestinal muscle (dash) and the anal depressor (arrowhead). The MLS-1 eh1 motif is necessary for uterine muscle specification
MLS-1 promotes uterine muscle fate, and we wanted to determine if this activity depends on the MLS-1 eh1 motif. In mls-1 mutants uterine muscles are transformed into vulval muscles. This phenotype can be easily scored using an egl-15::gfp reporter, which in wild-type animals is expressed in four vm1 cells that form a characteristic X-shape near the vulva [22]–[24], but in mls-1 mutants egl-15::gfp expression can be seen in four additional cells [14] (Figure 3). mls-1 mutants can be efficiently rescued by transformation with a wild-type mls-1 genomic DNA fragment injected at 10 ng/µl [14]. We wanted to ask if MLS-1 requires an intact eh1 motif to specify uterine muscle fate, but we were unable to obtain transgenic lines containing the same genomic fragment bearing the mls-1FSI→ASA mutation when injected at this concentration. This result strongly suggests that the mls-1FSI→ASA is expressed, but is toxic to the animals.
Fig. 3. MLS-1 uterine muscle specification activity depends on its eh1 motif. 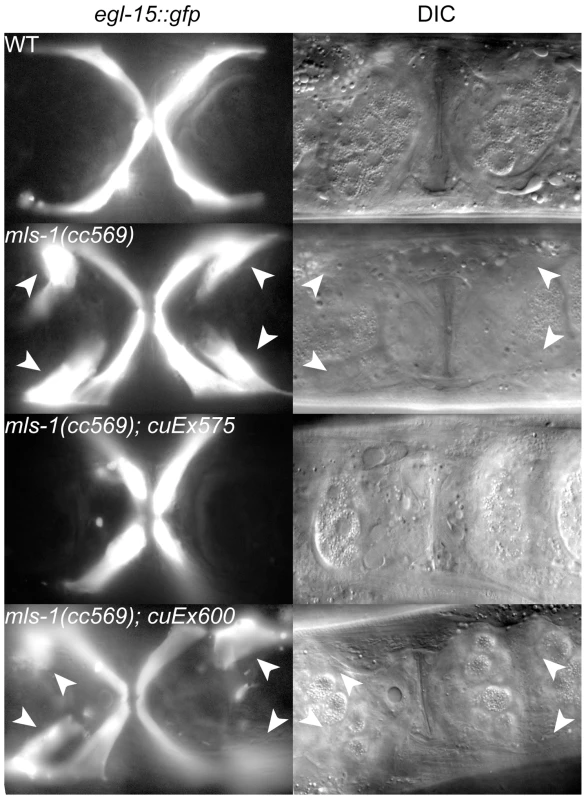
Ventral view of wild-type (WT) and mutant animals of the indicated genotypes expressing egl-15::gfp in vm1 cells. The transgene cuEx575 contains wild-type mls-1, while cuEx600 contains mls-1FSI→ASA. Supernumerary vm1 cells (arrowheads) are present in mls-1(cc569) and mls-1(cc569); cuEx600[mls-1FSI→ASA]. We were able to generate both wild-type mls-1 and mls-1FSI→ASA transgenic strains by reducing the DNA concentration to 2.5 ng/µl. Wild-type mls-1 DNA at this lower concentration was able to rescue mls-1(cc569) mutants in five independent lines, but at reduced percentages (Figure 3) (Table 1). In contrast the mls-1FSI→ASA transgene did not rescue mls-1(cc569) in seven of seven lines (Figure 3) (Table 1). These results indicate that the MLS-1 eh1 motif is required for MLS-1 to specify uterine muscle fate.
Tab. 1. Percentage of mls-1(cc569) animals rescued with mls-1 and mls-1FSI→ASA transgenes. 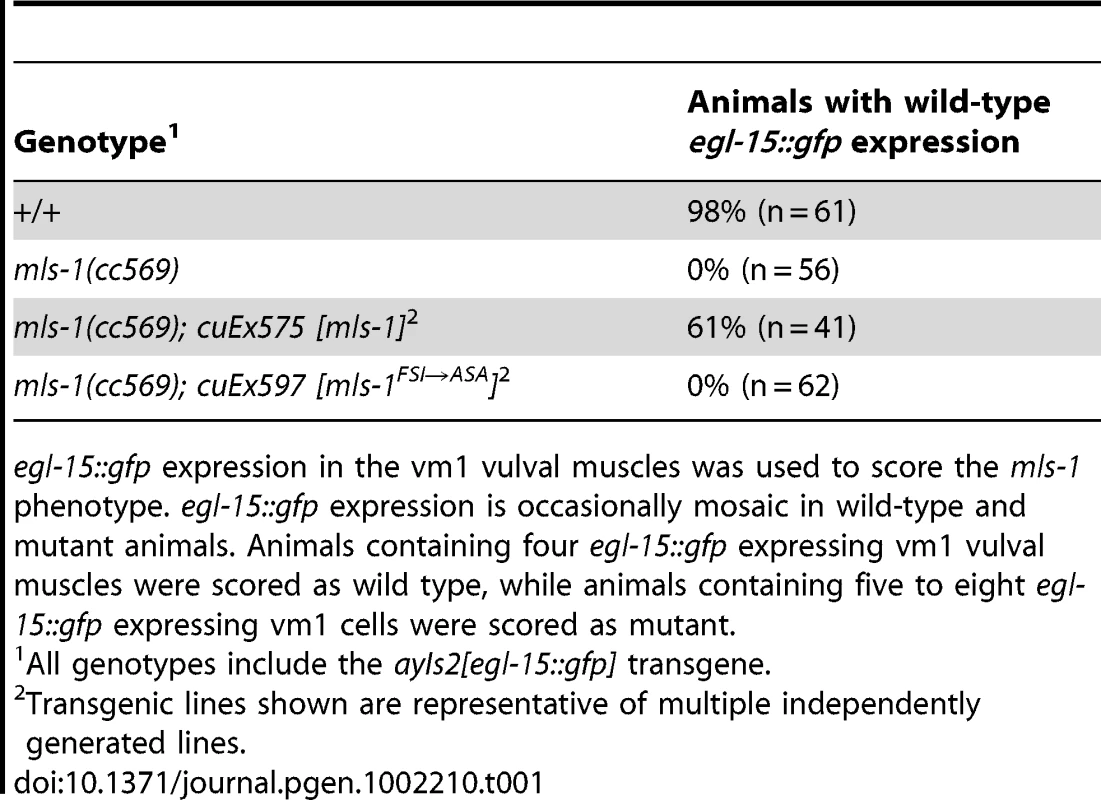
egl-15::gfp expression in the vm1 vulval muscles was used to score the mls-1 phenotype. egl-15::gfp expression is occasionally mosaic in wild-type and mutant animals. Animals containing four egl-15::gfp expressing vm1 vulval muscles were scored as wild type, while animals containing five to eight egl-15::gfp expressing vm1 cells were scored as mutant. unc-37 loss-of-function animals show sex muscle and mesoderm lineage defects
Because the MLS-1 eh1 motif mediates interaction with UNC-37 and is necessary for rescue of mls-1 mutants, we hypothesize MLS-1 requires UNC-37 to specify uterine muscle fate. Therefore unc-37 loss-of-function animals should phenocopy mls-1 mutants. To test this hypothesis, we performed unc-37(RNAi) on egl-15::gfp animals. These RNAi affected animals displayed egl-15::gfp expression in the four vm1 cells but also in up to four additional cells (Figure 4A, Table 2). To more directly examine the effect on uterine muscle specification we repeated unc-37(RNAi) experiments on animals with an integrated rgs-2::gfp transgene. In wild-type adults rgs-2::gfp is expressed in the pharynx, the ventral nerve cord, lumbar ganglia, and uterine muscles [14], [25]. unc-37(RNAi) greatly reduced the number of and intensity of rgs-2::gfp expressing uterine muscles, while having no effect on rgs-2::gfp expression in other tissues (Figure 4B, Table 2). Note that the morphology of uterine muscles made it much more difficult to count rgs-2::gfp expressing cells than egl-15::gfp expressing vulval muscles, but nearly half the unc-37(RNAi) animals we examined had no rgs-2::gfp expressing uterine muscles.
Fig. 4. unc-37 loss of function animals phenocopy mls-1(cc569). 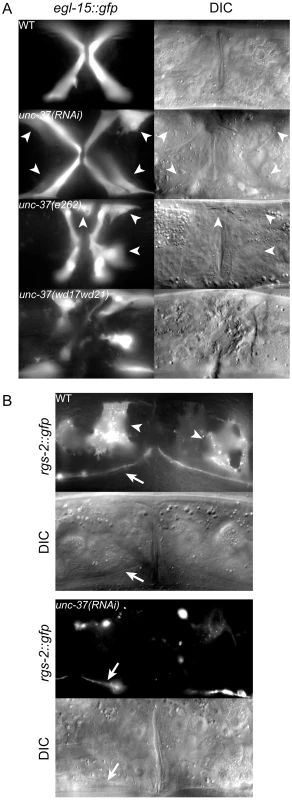
A) Ventral view of wild-type (WT) and mutant animals of the indicated genotypes expressing egl-15::gfp in vm1 cells. Supernumerary vm1 cells (arrowheads) are present in unc-37(RNAi), unc-37(e262), and unc-37(wd17wd21) animals. unc-37(wd17wd21) mutants exhibit a protruding vulva phenotype with mispositioned vulval muscles, so the supernumerary muscles are not marked. B) Ventral views of wild-type (WT) and mutant animals of the indicated genotypes expressing rgs-2::gfp. In wild type, rgs-2::gfp is detected in uterine muscles (arrowheads), which appear as sheet-like muscles and in the ventral nerve cord (arrow). unc-37(RNAi) causes a decrease in number of uterine muscles cells expressing rgs-2::gfp and decreased fluorescence intensity in these cells. Tab. 2. Vulval and uterine muscle pattern in unc-37(RNAi) animals. 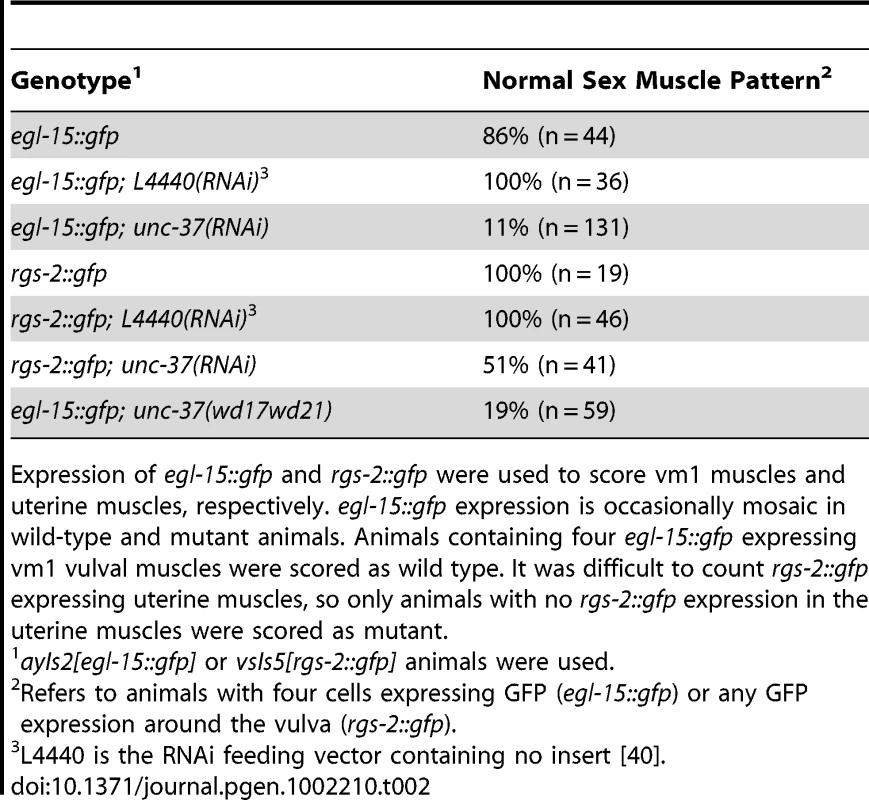
Expression of egl-15::gfp and rgs-2::gfp were used to score vm1 muscles and uterine muscles, respectively. egl-15::gfp expression is occasionally mosaic in wild-type and mutant animals. Animals containing four egl-15::gfp expressing vm1 vulval muscles were scored as wild type. It was difficult to count rgs-2::gfp expressing uterine muscles, so only animals with no rgs-2::gfp expression in the uterine muscles were scored as mutant. We next examined egl-15::gfp expression in two unc-37 mutants: unc-37(e262) and unc-37(wd17wd21). unc-37(e262) is a hypomorphic allele that can be maintained as a homozygous strain, and it encodes a missense mutation affecting a conserved residue in the WD repeat domain that specifically affects UNC-37 function in VA motor neurons [17]. In contrast unc-37(wd17wd21) is a putative null allele containing a splice acceptor site mutation upstream of the WD repeats [17]. unc-37(wd17wd21) mutants exhibit a maternal effect embryonic lethal phenotype, but unc-37(wd17wd21) homozygotes produced from heterozygous mutant hermaphrodites grow to adulthood and exhibit uncoordinated and protruding vulva phenotypes. Most unc-37(e262) mutants exhibited a wild-type pattern of egl-15::gfp expression, but approximately 5% of these animals expressed egl-15::gfp in extra cells. A much higher percentage of unc-37(wd17wd21) adult animals (81%, n = 59) contained supernumerary egl-15::gfp expressing vulval muscles. In both cases GFP-expressing cells were near the vulva similar to mls-1(cc569) animals (Figure 4A), but in other cases they were located in the posterior body wall muscle quadrants (Figure S2). A similar localization of egl-15::gfp expressing cells has previously been observed in mls-2 mutants [26]. This phenotype may be significant as MLS-2 is predicted to have a high scoring eh1 motif [12], and it suggests that unc-37(e262) may affect the activity of both MLS-1 and MLS-2 in specifying cell fates in the M lineage.
Discussion
MLS-1 represses transcription to specify uterine muscle fate
MLS-1 is a selector gene that is necessary and sufficient to specify uterine muscle development in the M lineage [14]. Here we show that MLS-1 interacts with the UNC-37/Groucho co-repressor, and that MLS-1 function in C. elegans depends on this interaction. These results indicate that MLS-1 specifies uterine muscle fate at least in part by repressing target gene transcription.
What types of genes does MLS-1 regulate? We suggest that MLS-1 represses expression of regulatory genes that themselves encode inhibitors of uterine muscle development and activators of vulval muscle development. This model is consistent with previous observations that mls-1 loss-of-function results in a transformation of uterine muscle to vulval muscle, whereas ectopic mls-1 expression results in production of ectopic uterine muscles [14]. Analogous negative regulatory pathways have been suggested for specification of body wall muscle and coelomocyte cell fates elsewhere in the M lineage [27], [28]. The MLS-1 targets must be expressed in the M lineage outside of the descendants of the SMs, because widespread expression of mls-1 in the M lineage can convert many M lineage cells to a uterine muscle fate [14]. We do not yet know of any direct targets of MLS-1. The egl-15 promoter is active in the supernumerary vulval muscles in mls-1 mutants, and we asked if it might be directly repressed by MLS-1. However mutation of predicted T-box binding sites in this promoter did not lead to expanded expression egl-15::gfp reporter (R. Miller and P. Okkema, unpublished), suggesting MLS-1 regulation of the egl-15 promoter is indirect.
MLS-1 may also function with UNC-37/Groucho in other MLS-1 expressing cells. When expressed with its own promoter, MLS-1 interacts with UNC-37 in BiFC assays in the vm2 vulval muscles and one to two intestinal/enteric muscles. The function of mls-1 in these cell types is unknown [14], so we cannot determine if interaction with UNC-37 is necessary for MLS-1, but we suggest that MLS-1 may similarly function as a Groucho dependent repressor in these cells.
In addition to MLS-1, UNC-37/Groucho likely interacts with other factors that are involved in vulval development or that are expressed in the M-lineage. unc-37(RNAi) and some mutant alleles produce a protruding vulva (Pvl) phenotype that is not observed in mls-1 mutants [29]. Likewise, we also found egl-15::gfp expressing cells that looked like body wall muscles in unc-37(e262) and more frequently in unc-37(wd17wd21) mutants, while we never observed this phenotype in mls-1 mutants. At least two other transcription factors expressed in the M lineage contain high scoring eh1 motifs (CEH-24 and MLS-2 see [12]), and we suggest that these and other factors expressed in the M lineage function as Groucho-dependent repressors.
mls-1FSI→ASA may be a gain-of-function mutation
Our results suggest that mutation of the MLS-1 eh1 motif creates a gain-of-function protein that is deleterious. It was much more difficult to generate transgenic lines expressing mls-1FSI→ASA than lines expressing wild-type mls-1 using either the mls-1 promoter or a heat-inducible promoter, and we were only able to generate transgenic lines expressing mls-1FSI→ASA with arrays containing a low concentration of the expression vector. MLS-1FSI→ASA contains an intact T-box, and, because many T-box factors bind similar sequences, MLS-1FSI→ASA could recognize binding sites for wild-type MLS-1 or for other T-box factors. It is unlikely that mls-1FSI→ASA is simply a dominant negative mutation that interferes with wild-type mls-1 function, because mls-1 null mutants are viable and healthy [14]. Instead, we suggest that MLS-1FSI→ASA interferes with function of other T-box factors that are required for viability. Alternatively, mutation of the eh1 motif may allow MLS-1 to function as a transcriptional activator and inappropriately activate T-box target genes. Groucho has been shown to convert a variety of transcriptional activators to repressors, including several T-box factors [7]–[10].
Relationship to T-box factors in other species
MLS-1 is a member of the Tbx1 sub-family, which includes the mammalian proteins Tbx1, Tbx15, Tbx18, Tbx20, and Tbx22. Like MLS-1, each of these proteins has been shown to repress transcription, and there is evidence that these proteins interact either directly or indirectly with Gro/TLE proteins [8]–[12]. Thus this T-box sub-family shares a common mechanism for transcriptional repression in different animal phyla.
Among the mammalian T-box genes, MLS-1 is most closely related to Tbx15, Tbx18 and Tbx22. These genes have diverse functions in mesoderm development, but recently described functions for Tbx18 in smooth muscle development may be most closely related to the function of MLS-1. Tbx18 is expressed in the developing urogenital ridge where it is necessary for development of the ureter smooth muscle [30], [31]. Likewise Tbx18 is also expressed in endocardial cells that contribute to the coronary smooth muscles, although its function in this tissue has not yet been characterized [32]. The C. elegans muscle types expressing MLS-1 share both structural and functional similarities with mammalian smooth muscles. C. elegans uterine muscles are non-striated with loosely organized myofilaments arranged circumferentially around the uterus, and contractions of these muscles help move embryos through the uterus toward the vulva [14], [15]. Ultrastructurally, the uterine muscles contain thin filaments attached to the uterine basal lamina at randomly arranged points, which is similar to the organization found in smooth muscles [33]. Together, these observations suggest the interesting possibility that Tbx18 and MLS-1 share conserved function in smooth muscle development.
Materials and Methods
Strains and plasmids
C. elegans were grown under standard conditions and were raised at 20°C unless otherwise noted [34]. The following strains were used: NH2447 ayIs2 [egl-15::gfp] IV [35]; PD4285 mls-1(cc569) I; ayIs2 [egl-15::gfp] IV [14]; LX354 lin-15(n765ts); vsIs5 [rgs-2::gfp; lin-15(+)] [25]; CB262 unc-37(e262) I [36], NC93 unc-37(wd17wd21)/dpy-14(e188) I [17], OK0675 unc-37(e262) I; ayIs2 [egl-15::gfp] IV, and OK0787 unc-37(wd17wd21)/dpy-14(e188) I; ayIs2 [egl-15::gfp] IV. The following transgenic strains were constructed for this work.
mls-1(cc569) rescue lines
Transgenic lines were constructed by injecting PD4825 with plasmids containing either an mls-1(+) genomic fragment (pSAK244.13, provided by A. Fire, Stanford) or mls-1FSI→ASA (pOK257.02) at 2.5 ng/µl and pRF4 at 100 ng/µl [37]: OK0720 mls-1(cc569); ayIs2; cuEx575[mls-1(+)], OK0721 mls-1(cc569); ayIs2; cuEx576[mls-1(+)], OK0722 mls-1(cc569); ayIs2; cuEx577[mls-1(+)], OK0723 mls-1(cc569); ayIs2; cuEx578[mls-1(+)], OK0724 mls-1(cc569); ayIs2; cuEx579[mls-1(+)], OK0725 mls-1(cc569); ayIs2; cuEx580[mls-1(+)], OK0683 mls-1(cc569); ayIs2; cuEx568[mls-1FSI→ASA], OK0745 mls-1(cc569); ayIs2; cuEx596[mls-1FSI→ASA], OK0746 mls-1(cc569); ayIs2; cuEx597[mls-1FSI→ASA], OK0747 mls-1(cc569); ayIs2; cuEx598[mls-1FSI→ASA], OK0748 mls-1(cc569); ayIs2; cuEx599[mls-1FSI→ASA], OK0749 mls-1(cc569); ayIs2; cuEx600[mls-1FSI→ASA], OK0750 mls-1(cc569); ayIs2; cuEx601[mls-1FSI→ASA].
BiFC lines
Transgenic lines were constructed by injecting N2 with pRF4 at 100 ng/µl and the BiFC plasmids pCE-BiFC-VN173, pCE-BiFC-VC155, pOK257.05 (mls-1::vn173), pOK257.06 (unc-37::vc155), pOK258.06 (mls-1FSI→ASA::VN173), pOK263.03 (genomic mls-1::VN173), or pOK266.02 (genomic unc-37::VC155) at 15 ng/µl: OK0708 cuEx566[VN173+VC155], OK0689 cuEx573[mls-1::vn173+unc-37::vc155], OK0690 cuEx574[mls-1::vn173+unc-37::vc155], OK0691 cuEx575[mls-1::vn173+unc-37::vc155], OK0709 cuEx567[mls-1FSI→ASA::VN173+unc-37::vc155], OK0710 cuEx568[mls-1FSI→ASA::VN173+unc-37::vc155], OK0742 cuEx593[genomic mls-1::VN173+genomic unc-37::VC155], OK0743 cuEx594[genomic mls-1::VN173+genomic unc-37::VC155], OK0744 cuEx595[genomic mls-1::VN173+genomic unc-37::VC155].
Bimolecular fluorescence complementation (BiFC)
mls-1 (Open Biosystems) and unc-37 (yk727f10 provided by Y. Kohara, National Institute of Genetics, Japan) cDNA were cloned into pCE-BiFC-VN173 and pCE-BiFC-VC155 plasmids (provided by Chang-Deng Hu, Purdue) to generate pOK257.05 (mls-1::vn173) and pOK257.06 (unc-37::vc155). mls-1FSI→ASA cDNA was generated by site directed mutagenesis of mls-1 cDNA (Quikchange II XL, Stratagene), and cloned into pCE-BiFC-VN173 producing pOK258.06 (mls-1FSI→ASA::vn173). Transgenic adults were picked to OP50 seeded plates, heat shocked for one hour at 33°C as described in Results, allowed to recover for one hour at 20°C, and examined for BiFC signal at 63× magnification.
Genomic mls-1::vn173 and unc-37::vc155 constructs, with gene expression under the control of their respective endogenous promoters, were produced by PCR of promoter+gene fragments using pSAK244.13 and N2 genomic DNA templates, respectively. hsp-16.41 promoter fragments were removed from pCE-BiFC-VN173 and pCE-BiFC-VC155 and replaced with these mls-1 and unc-37 promoter+gene cassettes generating pOK263.03 and pOK266.02. Transgenic animals from various stages (L1 to Adult) were examined at 63× for BiFC signal.
Yeast two-hybrid assays
Yeast two-hybrid assays were performed as previously described [38]. The mls-1 bait plasmid (pOK248.03) was constructed by inserting the full-length mls-1 cDNA in to pLexA-NLS, and the unc-37 prey plasmid was previously isolated from the pACT-RB1 cDNA library (provided by R. Barstead).
mls-1(cc569) rescue assay
mls-1(cc569); ayIs2 strains with mls-1(+) or mls-1FSI→ASA extrachromosomal arrays were generated as above. Adult transgenic animals from these lines and animals from NH2447 and PD4285 were examined and scored at 40× and 63× magnification for the number of cells expressing egl-15::gfp.
unc-37 loss of function
unc-37 cDNA was cloned into the L4440 (double T7 promoter) vector and the resulting plasmid (pOK247.03) was used to transform HT115(DE3) E. coli cells. unc-37 “feeding” RNAi was performed as before with some modifications [39], [40]. Plates with large numbers of ayIs2 [egl-15::gfp] IV (NH2447) or lin-15(n765ts) X; vsIs5 [rgs-2::gfp] (LX354) (provided by M. Koelle, Yale) adults and embryos were bleached and embryos were transferred to unseeded NGM plates and placed at 25°C overnight to allow embryos to hatch and synchronize as L1s. The next day synchronized L1s were transferred to unc-37(RNAi) seeded plates and placed at 20°C for 48 hours to 72 hours. Adult animals were examined and scored at 40× and 63× magnification for egl-15::gfp or rgs-2::gfp (mis)expression. Note that it is only possible to qualitatively evaluate rgs-2::gfp expression due to the unusual morphology of uterine muscles.
Supporting Information
Zdroje
1. NaicheLAHarrelsonZKellyRGPapaioannouVE 2005 T-box genes in vertebrate development. Annu Rev Genet 39 219 239
2. ShowellCBinderOConlonFL 2004 T-box genes in early embryogenesis. Dev Dyn 229 201 218
3. PackhamEABrookJD 2003 T-box genes in human disorders. Hum Mol Genet 12 R37 44
4. PengSL 2006 The T-box transcription factor T-bet in immunity and autoimmunity. Cell Mol Immunol 3 87 95
5. AbrahamsAParkerMIPrinceS 2010 The T-box transcription factor Tbx2: its role in development and possible implication in cancer. IUBMB Life 62 92 102
6. RowleyMGrotheyECouchFJ 2004 The role of Tbx2 and Tbx3 in mammary development and tumorigenesis. J Mammary Gland Biol Neoplasia 9 109 118
7. GasperowiczMOttoF 2005 Mammalian Groucho homologs: redundancy or specificity? J Cell Biochem 95 670 687
8. HitachiKDannoHTazumiSAiharaYUchiyamaH 2009 The Xenopus Bowline/Ripply family proteins negatively regulate the transcriptional activity of T-box transcription factors. Int J Dev Biol 53 631 639
9. KawamuraAKoshidaSTakadaS 2008 Activator-to-repressor conversion of T-box transcription factors by the Ripply family of Groucho/TLE-associated mediators. Mol Cell Biol 28 3236 3244
10. KondowAHitachiKOkabayashiKHayashiNAsashimaM 2007 Bowline mediates association of the transcriptional corepressor XGrg-4 with Tbx6 during somitogenesis in Xenopus. Biochem Biophys Res Commun 359 959 964
11. FarinHFBussenMSchmidtMKSinghMKSchuster-GosslerK 2007 Transcriptional repression by the T-box proteins Tbx18 and Tbx15 depends on Groucho corepressors. J Biol Chem 282 25748 25759
12. CopleyRR 2005 The EH1 motif in metazoan transcription factors. BMC Genomics 6 169
13. PocockRMioneMHussainSMaxwellSPontecorviM 2008 Neuronal function of Tbx20 conserved from nematodes to vertebrates. Dev Biol 317 671 685
14. KostasSAFireA 2002 The T-box factor MLS-1 acts as a molecular switch during specification of nonstriated muscle in C. elegans. Genes Dev 16 257 269
15. SulstonJEHorvitzHR 1977 Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56 110 156
16. JenningsBHPicklesLMWainwrightSMRoeSMPearlLH 2006 Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell 22 645 655
17. PflugradAMeirJYBarnesTMMillerDM 1997 The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development 124 1699 1709
18. HiattSMShyuYJDurenHMHuCD 2008 Bimolecular fluorescence complementation (BiFC) analysis of protein interactions in Caenorhabditis elegans. Methods 45 185 191
19. HuCDChinenovYKerppolaTK 2002 Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9 789 798
20. StringhamEGDixonDKJonesDCandidoEP 1992 Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol Biol Cell 3 221 233
21. SchaferW 2005 Egg-laying. CommunityTCeR La Jolla, CA WormBook 1 7 doi/10.1895/wormbook.1.38.1, http://www.wormbook.org
22. EimerSDonhauserRBaumeisterR 2002 The Caenorhabditis elegans presenilin sel-12 is required for mesodermal patterning and muscle function. Dev Biol 251 178 192
23. HarfeBDVaz GomesAKenyonCLiuJKrauseM 1998 Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev 12 2623 2635
24. LiuJFireA 2000 Overlapping roles of two Hox genes and the exd ortholog ceh-20 in diversification of the C. elegans postembryonic mesoderm. Development 127 5179 5190
25. DongMQChaseDPatikoglouGAKoelleMR 2000 Multiple RGS proteins alter neural G protein signaling to allow C. elegans to rapidly change behavior when fed. Genes Dev 14 2003 2014
26. JiangYHornerVLiuJ 2005 The HMX homeodomain protein MLS-2 regulates cleavage orientation, cell proliferation and cell fate specification in the C. elegans postembryonic mesoderm. Development 132 4119 4130
27. AminNMLimSEShiHChanTLLiuJ 2009 A conserved Six-Eya cassette acts downstream of Wnt signaling to direct non-myogenic versus myogenic fates in the C. elegans postembryonic mesoderm. Dev Biol 331 350 360
28. AminNMShiHLiuJ 2010 The FoxF/FoxC factor LET-381 directly regulates both cell fate specification and cell differentiation in C. elegans mesoderm development. Development 137 1451 1460
29. FlowersEBPooleRJTursunBBashllariEPe'erI 2010 The Groucho ortholog UNC-37 interacts with the short Groucho-like protein LSY-22 to control developmental decisions in C. elegans. Development 137 1799 1805
30. AirikRBussenMSinghMKPetryMKispertA 2006 Tbx18 regulates the development of the ureteral mesenchyme. J Clin Invest 116 663 674
31. KrausFHaenigBKispertA 2001 Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev 100 83 86
32. CaiCLMartinJCSunYCuiLWangL 2008 A myocardial lineage derives from Tbx18 epicardial cells. Nature 454 104 108
33. AltunZFHallDH 2009 Muscle system, nonstriated muscle. HerndonLA WormAtlas
34. LewisJAFlemingJT 1995 Basic culture methods. Methods Cell Biol 48 3 29
35. HarfeBDBrandaCSKrauseMSternMJFireA 1998 MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development 125 2479 2488
36. BrennerS 1974 The genetics of Caenorhabditis elegans. Genetics 77 71 94
37. MelloCFireA 1995 DNA transformation. Methods Cell Biol 48 451 482
38. Roy ChowdhuriSCrumTWoollardAAslamSOkkemaPG 2006 The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev Biol 295 664 677
39. KamathRSMartinez-CamposMZipperlenPFraserAGAhringerJ 2001 Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2 research0002.0001 0002.0010
40. TimmonsLCourtDLFireA 2001 Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 103 112
Štítky
Genetika Reprodukční medicína
Článek B Chromosomes Have a Functional Effect on Female Sex Determination in Lake Victoria Cichlid FishesČlánek Distinct Cdk1 Requirements during Single-Strand Annealing, Noncrossover, and Crossover RecombinationČlánek Specification of Corpora Cardiaca Neuroendocrine Cells from Mesoderm Is Regulated by Notch SignalingČlánek Ongoing Phenotypic and Genomic Changes in Experimental Coevolution of RNA Bacteriophage Qβ and
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 8
-
Všechny články tohoto čísla
- Polo, Greatwall, and Protein Phosphatase PP2A Jostle for Pole Position
- Genome-Wide Association Analysis of Incident Coronary Heart Disease (CHD) in African Americans: A Short Report
- The T-Box Factor MLS-1 Requires Groucho Co-Repressor Interaction for Uterine Muscle Specification
- B Chromosomes Have a Functional Effect on Female Sex Determination in Lake Victoria Cichlid Fishes
- Analysis of DNA Methylation in a Three-Generation Family Reveals Widespread Genetic Influence on Epigenetic Regulation
- PP2A-Twins Is Antagonized by Greatwall and Collaborates with Polo for Cell Cycle Progression and Centrosome Attachment to Nuclei in Drosophila Embryos
- Discovery of Sexual Dimorphisms in Metabolic and Genetic Biomarkers
- Pervasive Sharing of Genetic Effects in Autoimmune Disease
- DNA Methylation and Histone Modifications Regulate Shoot Regeneration in by Modulating Expression and Auxin Signaling
- Mutations in and Reveal That Cartilage Matrix Controls Timing of Endochondral Ossification by Inhibiting Chondrocyte Maturation
- Variance of Gene Expression Identifies Altered Network Constraints in Neurological Disease
- Frequent Beneficial Mutations during Single-Colony Serial Transfer of
- Increased Gene Dosage Affects Genomic Stability Potentially Contributing to 17p13.3 Duplication Syndrome
- Distinct Cdk1 Requirements during Single-Strand Annealing, Noncrossover, and Crossover Recombination
- Hunger Artists: Yeast Adapted to Carbon Limitation Show Trade-Offs under Carbon Sufficiency
- Suppression of Scant Identifies Endos as a Substrate of Greatwall Kinase and a Negative Regulator of Protein Phosphatase 2A in Mitosis
- Temporal Dynamics of Host Molecular Responses Differentiate Symptomatic and Asymptomatic Influenza A Infection
- MK2-Dependent p38b Signalling Protects Hindgut Enterocytes against JNK-Induced Apoptosis under Chronic Stress
- Specification of Corpora Cardiaca Neuroendocrine Cells from Mesoderm Is Regulated by Notch Signaling
- Genome-Wide Gene-Environment Study Identifies Glutamate Receptor Gene as a Parkinson's Disease Modifier Gene via Interaction with Coffee
- Identification of Functional Toxin/Immunity Genes Linked to Contact-Dependent Growth Inhibition (CDI) and Rearrangement Hotspot (Rhs) Systems
- Genomic Analysis of the Necrotrophic Fungal Pathogens and
- Celsr3 Is Required for Normal Development of GABA Circuits in the Inner Retina
- Genetic Architecture of Aluminum Tolerance in Rice () Determined through Genome-Wide Association Analysis and QTL Mapping
- Predisposition to Cancer Caused by Genetic and Functional Defects of Mammalian
- Regulation of p53/CEP-1–Dependent Germ Cell Apoptosis by Ras/MAPK Signaling
- and but Not Interact in Genetic Models of Amyotrophic Lateral Sclerosis
- Gamma-Tubulin Is Required for Bipolar Spindle Assembly and for Proper Kinetochore Microtubule Attachments during Prometaphase I in Oocytes
- Ongoing Phenotypic and Genomic Changes in Experimental Coevolution of RNA Bacteriophage Qβ and
- Genetic Architecture of a Reinforced, Postmating, Reproductive Isolation Barrier between Species Indicates Evolution via Natural Selection
- -eQTLs Reveal That Independent Genetic Variants Associated with a Complex Phenotype Converge on Intermediate Genes, with a Major Role for the HLA
- The GATA Factor ELT-1 Works through the Cell Proliferation Regulator BRO-1 and the Fusogen EFF-1 to Maintain the Seam Stem-Like Fate
- and Control Optic Cup Regeneration in a Prototypic Eye
- A Comprehensive Map of Mobile Element Insertion Polymorphisms in Humans
- An EMT–Driven Alternative Splicing Program Occurs in Human Breast Cancer and Modulates Cellular Phenotype
- Evidence for Hitchhiking of Deleterious Mutations within the Human Genome
- A Broad Brush, Global Overview of Bacterial Sexuality
- Global Chromosomal Structural Instability in a Subpopulation of Starving Cells
- A Pre-mRNA–Associating Factor Links Endogenous siRNAs to Chromatin Regulation
- Glutamine Synthetase Is a Genetic Determinant of Cell Type–Specific Glutamine Independence in Breast Epithelia
- The Repertoire of ICE in Prokaryotes Underscores the Unity, Diversity, and Ubiquity of Conjugation
- Genome-Wide Association Analysis of Autoantibody Positivity in Type 1 Diabetes Cases
- Natural Polymorphism in BUL2 Links Cellular Amino Acid Availability with Chronological Aging and Telomere Maintenance in Yeast
- Chromosome Painting Reveals Asynaptic Full Alignment of Homologs and HIM-8–Dependent Remodeling of Chromosome Territories during Meiosis
- Ku Must Load Directly onto the Chromosome End in Order to Mediate Its Telomeric Functions
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- An EMT–Driven Alternative Splicing Program Occurs in Human Breast Cancer and Modulates Cellular Phenotype
- Chromosome Painting Reveals Asynaptic Full Alignment of Homologs and HIM-8–Dependent Remodeling of Chromosome Territories during Meiosis
- Discovery of Sexual Dimorphisms in Metabolic and Genetic Biomarkers
- Regulation of p53/CEP-1–Dependent Germ Cell Apoptosis by Ras/MAPK Signaling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



