-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTargeted Sister Chromatid Cohesion by Sir2
The protein complex known as cohesin binds pericentric regions and other sites of eukaryotic genomes to mediate cohesion of sister chromatids. In budding yeast Saccharomyces cerevisiae, cohesin also binds silent chromatin, a repressive chromatin structure that functionally resembles heterochromatin of higher eukaryotes. We developed a protein-targeting assay to investigate the mechanistic basis for cohesion of silent chromatin domains. Individual silencing factors were tethered to sites where pairing of sister chromatids could be evaluated by fluorescence microscopy. We report that the evolutionarily conserved Sir2 histone deacetylase, an essential silent chromatin component, was both necessary and sufficient for cohesion. The cohesin genes were required, but the Sir2 deacetylase activity and other silencing factors were not. Binding of cohesin to silent chromatin was achieved with a small carboxyl terminal fragment of Sir2. Taken together, these data define a unique role for Sir2 in cohesion of silent chromatin that is distinct from the enzyme's role as a histone deacetylase.
Published in the journal: . PLoS Genet 7(2): e32767. doi:10.1371/journal.pgen.1002000
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002000Summary
The protein complex known as cohesin binds pericentric regions and other sites of eukaryotic genomes to mediate cohesion of sister chromatids. In budding yeast Saccharomyces cerevisiae, cohesin also binds silent chromatin, a repressive chromatin structure that functionally resembles heterochromatin of higher eukaryotes. We developed a protein-targeting assay to investigate the mechanistic basis for cohesion of silent chromatin domains. Individual silencing factors were tethered to sites where pairing of sister chromatids could be evaluated by fluorescence microscopy. We report that the evolutionarily conserved Sir2 histone deacetylase, an essential silent chromatin component, was both necessary and sufficient for cohesion. The cohesin genes were required, but the Sir2 deacetylase activity and other silencing factors were not. Binding of cohesin to silent chromatin was achieved with a small carboxyl terminal fragment of Sir2. Taken together, these data define a unique role for Sir2 in cohesion of silent chromatin that is distinct from the enzyme's role as a histone deacetylase.
Introduction
Proper segregation of chromosomes at mitosis and meiosis requires sister chromatid cohesion. The process ensures that newly replicated chromatids bi-orient on spindle microtubules such that a single copy of each chromosome transfers to progeny cells. Defects in the sister chromatid cohesion pathway lead to certain developmental diseases, and chromosome segregation defects like those seen in cancer [1]–[4].
Cohesion of sister chromatids is mediated by a protein complex known as cohesin [5], [6]. The core complex consists of a hetero-dimer of SMC proteins Smc1 and Smc3, as well as non-SMC proteins Scc3/Irr1 and Mcd1/Scc1/Rad21 (hereafter referred to as Scc3 and Mcd1, respectively). The subunits form a large protein ring with a striking central void. Thus, a prominently held view is that cohesin holds sister chromatids together by single complexes embracing both chromatids. Elegant protein-crosslinking studies showed that single cohesin rings can indeed hold together two partially purified minichromosomes [7]. Other data raises the possibility that cohesin might hold sister chromatids together by a different mechanism [8]–[10].
Cohesin binds discrete sites on chromosomal DNA. Most non-centromeric sites in budding and fission yeasts lie within the AT-rich regions between convergently transcribed genes [11]–[13]. Transcriptional elongation redistributes complexes from intragenic to intergenic regions, suggesting that cohesin enrichment is maintained dynamically. In contrast to the situation in these fungal systems, cohesin maps along the lengths of actively transcribed genes in Drosophila and to sites within transcribed genes in humans [14]–[16]. Thus, cohesin binding and transcription are not always mutually exclusive.
Cohesin is also found within pericentric heterochromatin regions where transcription is suppressed but not extinguished. In fission yeast, the complex is retained at these locations by Swi6, a homolog of heterochromatin protein HP1, which interacts with cohesin subunit Psc3 (Scc3 in budding yeast) [17], [18]. During meiosis, Swi6 also interacts with shugoshin, a protein that protects centromeric cohesin from being dismantled [19]. In heterochromatin mutants, cohesin does not bind pericentric domains and mitotic chromosomes fail to mount properly onto spindle microtubules.
Budding yeast lacks Swi6 and pericentric heterochromatin but it does contain transcriptionally silenced domains that nevertheless bind cohesin. Using the HMR locus as one representative example, we found that silencing mutations selectively disrupted cohesin binding and correspondingly abolished cohesion of sister chromatid DNA bearing the locus [9]. A search to understand why cohesin accumulates at HMR served as the impetus for this study.
Based on the chromatin-mediated mechanism of regional DNA inactivation, transcriptionally silenced domains in budding yeast are referred to as silent chromatin [20]. Like heterochromatin domains in other organisms, silent chromatin is packaged with histones that bear a distinct signature of post-translational modifications. Specifically, acetylation and methylation of lysines are absent. Silent chromatin domains associate with a complex of non-histone silencing factors known as the Sir proteins (Sir2, Sir3 and Sir4). Sir2 is a member of the evolutionarily conserved class of NAD+-dependent protein deacetylases known as sirtuins. The enzyme creates and maintains histone deacetylation within silent chromatin. Sir3 and Sir4 associate with the suitably deacetylated histones. The complex of Sir proteins is first recruited to sites of action by cis-acting elements known as silencers, which bind ORC, as well as Abf1 and Rap1 in various combinations. Following recruitment, cycles of histone deacetylation and histone binding allow the Sir proteins to spread over kilobases. A tRNA gene acts as a barrier element on the right side of HMR that blocks silent chromatin from spreading further downstream [21]. The element also augments HMR with sufficient cohesin for cohesion [22], probably through recruitment of the Scc2/4 cohesin loading complex [23], [24].
We considered two competing hypotheses to account for retention of cohesin at HMR. The first, based on a simple recruitment model, posits that a silent chromatin component interacts directly with cohesin or some factor associated with the complex. A second hypothesis stems from the ability of silent chromatin to impede a broad-range of DNA-based events, such as DNA replication, repair and transcription [20]. If silent chromatin also suppresses an activity that mobilizes cohesin, the complex would accumulate at silenced loci. To distinguish between these possibilities, we developed assays to determine whether silencing or silent chromatin components were required for cohesion of HMR. Our studies show that Sir2 is sufficient for cohesion, even in the absence of silencing.
Results
An assay for targeted sister chromatid cohesion
Our principle assay for cohesion at HMR utilizes a strain in which the locus is tagged with lac-GFP and flanked by target sites for a site-specific recombinase [9]. Inducible excision after arrest in M phase converts HMR loci on sister chromatids into a pair of extrachromosomal circles that produce one bright fluorescent focus if they are held together and two foci if they are not (Figure 1A and 1B).
Fig. 1. Tethered Sir2 mediates cohesion. 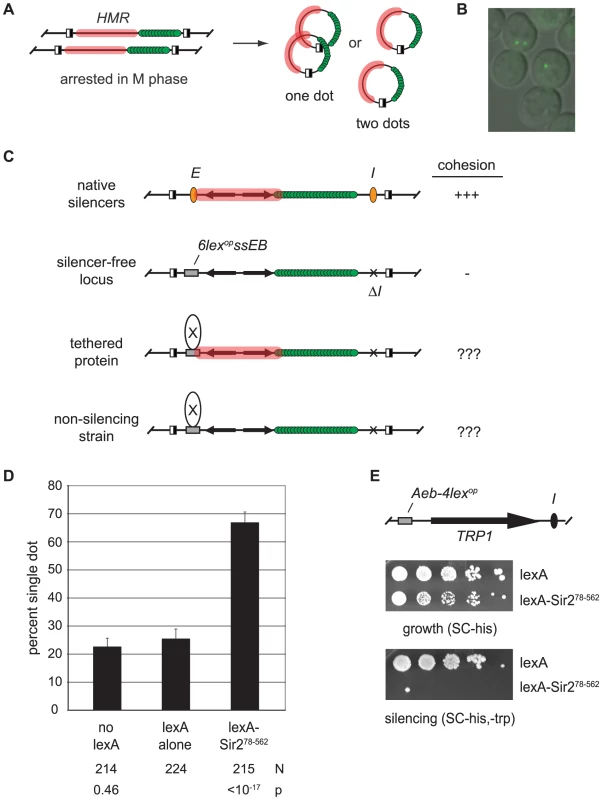
A) Evaluation of chromosomal cohesion by DNA circle formation. Site-specific recombination is initiated by galactose-induced expression of the R recombinase in cells arrested at M phase with microtubule inhibitors. Recombinase sites are depicted with half-filled boxes. Silent chromatin domains are highlighted with pink and lac-GFP is represented with green spheres. Cohesion of DNA circles yields a singe bright dot of fluorescence whereas lack of cohesion yields two dots. B) Representative image of M phase-arrested cells with circularized HMR loci. C) Protein-targeting assay for cohesion. Unexcised recombination cassettes are drawn for simplicity. The HMR silencers (orange ovals) were deleted and HMR-E is replaced with a fragment containing lexA sites (6lexopssEB). D) Tethered Sir2 mediates cohesion. HMR circles were counted in strain CSW19 that lacked lexA, and in strains CSW36 and CSW37 that contained integrated expression cassettes for lexA alone or lexA-Sir278–562, respectively. N represents the number of cells examined. The strain bearing lexA alone was used as point of comparison in significance tests. E) Tethered Sir2 mediates silencing. Strain GA-2050 was transformed with plasmids expressing either lexA alone (pBTM116H) or lexA-Sir278–562 (pCSW22) and spotted in 10-fold serial dilutions on SC-trp,-his to measure silencing of the TRP1 reporter and SC-his to measure growth. The strain contains an array of 4 lexA operators in place of the Rap1 and Abf1 binding sites at the HMR-E silencer (Aeb-4lexop) [76]. Parallel assays with the HMR-E silencer of the targeted cohesion assay (6lexopssEB) yielded similar results, albeit less dramatic ones (data not shown). To test whether silent chromatin components can mediate cohesion we tethered individual silencing factors directly to the DNA circles (Figure 1C). To this end, the E silencer of HMR was replaced with a synthetic construct (6lexopssEB) that includes binding sites for Rap1, Abf1 and the bacterial protein lexA. The I silencer was deleted. These modifications were previously shown to eliminate silencing of the locus [25]. Individual silencing factors were then targeted to HMR-6lexopssEB as lexA-linked fusion proteins. Cell cycle arrest in M phase, recombinase induction and fluorescence microscopy were performed as described previously [9].
Tethering silent chromatin components to DNA often nucleates silent chromatin assembly and restores transcriptional repression [25], [26]. In these situations, it would be impossible to determine whether the tethered protein, a co-recruited protein or the silenced state was responsible for cohesion. Therefore, tethered proteins were also examined under conditions that abolish silent chromatin assembly to evaluate their precise roles in cohesion.
Sir2 is sufficient for cohesion of HMR
Pilot experiments showed that excised circles bearing the HMR-6lexopssEB construct colocalized infrequently [9]. When lexA was expressed, only 22% of the nuclei contained the single bright fluorescent spot (Figure 1D). Strikingly, cohesion of the circles increased to 67% when lexA was fused to Sir2 (designated lexA-Sir278–562). Tethering Sir2 to DNA was essential. In a strain lacking lexA binding sites at HMR, the chimera failed to produce cohesion (Figure S1).
LexA-Sir278–562 lacks the first 77 amino acids of Sir2 that are dispensable for transcriptional repression [27]. We confirmed that lexA-Sir278–562 nucleates silencing at HMR using a strain that contains lexA binding sites and a TRP1 reporter gene at the locus (Figure 1E). Taken together, these initial findings demonstrate that tethered Sir2 confers both silencing and cohesion at HMR.
The Sir2 polypeptide consists of a conserved catalytic core, as well as N and C terminal domains that help target the deacetylase to sites of action [28]. An allele lacking the N-terminal 198 amino acids confers little transcriptional repression, even when tethered to DNA [29]. To generate a lexA chimera with similar characteristics, we eliminated the entire N-terminal domain (amino acids 1–242). Surprisingly, this construct (lexA-Sir2243–562) yielded a measurable degree of silencing in a strain with intact SIR genes (for comparisons, see lexA-Sir278–562 and lexA alone in figure 2A). Deletion of either the SIR2 or SIR4 genes eliminated silencing by lexA-Sir2243–562, indicating that 1) the chimera operates within the Sir pathway, and that 2) the chimera requires the endogenous full-length Sir2 for transcriptional repression. The reliance of lexA-Sir2243–562 on other SIR genes, including SIR2, for silencing made the chimera an ideal candidate for further study.
Fig. 2. Cohesion by tethered Sir2 does not require other Sir proteins. 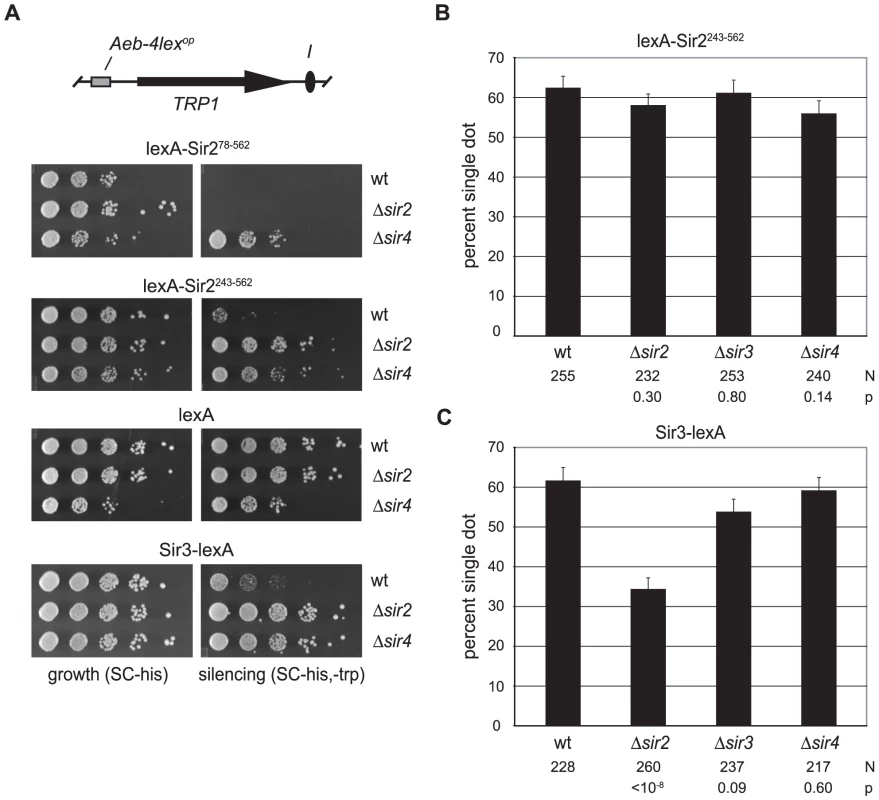
A) Silencing assays for tethered Sir2 constructs. Strains GA-2050 (wt), CSW98 (Δsir2) and CSW157 (Δsir4) expressing lexA-Sir278–562 (pCSW22), lexA-Sir2243–562 (pCSW21), lexA alone (pBTM116H) or Sir3-lexA (pCSW17) were spotted on SC-his (growth) and SC-trp,-his plates (silencing). In the Δsir4 strain, high expression of the lexA-Sir2 constructs caused a slight growth defect, as reported previously [77]. To compensate, the cell density of Δsir4 isolates bearing lexA-Sir2 constructs was concentrated 10-fold before plating. B) Cohesion of HMR circles by lexA-Sir2243–562 in silencing-deficient mutants. Strains CSW18 (wt), CSW42 (Δsir2), CSW85 (Δsir3) and CSW43 (Δsir4) were transformed with pCSW21. C) Cohesion of HMR circles by Sir3-lexA. Strains CSW19 (wt), CSW42 (Δsir2) and CSW85 (Δsir3) and CSW43 (Δsir4) were transformed with a plasmid expressing Sir3-lexA (pCSW14). Figure 2B shows that lexA-Sir2243–562 produced cohesion in over 60% of the nuclei examined. Importantly, cohesion of the excised HMR circles persisted in strains that lacked SIR2, SIR3 or SIR4. We conclude that tethered Sir2 can mediate cohesion in the absence of transcriptional silencing and without the aid of endogenous Sir proteins.
Sir3 was also examined directly with the targeted cohesion assay. When the protein was linked to lexA, HMR circles colocalized in over 60% of wild-type cells (Figure 2C). The tethered protein also conferred transcriptional repression in the wild-type reporter strain (Figure 2A). Both cohesion and silencing by Sir3-lexA were significantly diminished by deletion of Sir2. Elimination of Sir4, on the other hand, disrupted silencing but not cohesion. A simple explanation for the requirement of Sir2 but not Sir4 is that tethered Sir3 recruits Sir2, which in turn mediates cohesion of the locus.
We note that in the absence of Sir2, Sir3-lexA yielded a slightly higher level of cohesion than lexA alone (Sir3-lexA = 34% vs lexA = 26.3%). This difference is sufficiently small (p = 0.03) that we cannot conclude equivocally whether Sir3 possesses a subtle intrinsic cohesion activity. Given the strong cohesion signals afforded by Sir2, we focused our attention on this Sir protein for the remainder of the study.
Tethered Sir2 can mediate cohesion at an ectopic locus
If Sir2 mediates cohesion at HMR then the protein ought to impart cohesion when tethered at other genomic positions. We explored this possibility by targeting the protein to the LYS2 locus. LYS2 is situated near the center of chromosome II, hundreds of kilobases away from silent chromatin domains at the chromosome ends [30]. The locus had previously been modified to contain lexA-binding sites, as well as lac repressor sites and recombinase sites for the DNA excision assay [31]. When lexA alone was expressed, LYS2 DNA circles colocalized in 37% of the cells (Figure 3). This value is higher than the baseline for HMR cohesion under similar conditions (Figure 1D). The value is sufficiently low, however, to detect increases in cohesion due to tethered Sir2 fragments. Indeed, cohesion of LYS2 circles increased to 60% when lexA-Sir2243–562 was expressed. Pairing persisted in a strain lacking SIR3 indicating that cohesion was due to the tethered protein and not due to formation of silent chromatin at LYS2. These findings indicate that Sir2 can impart cohesion at chromosomal locations other than HMR.
Fig. 3. Sir2 mediates cohesion when tethered to an ectopic site. 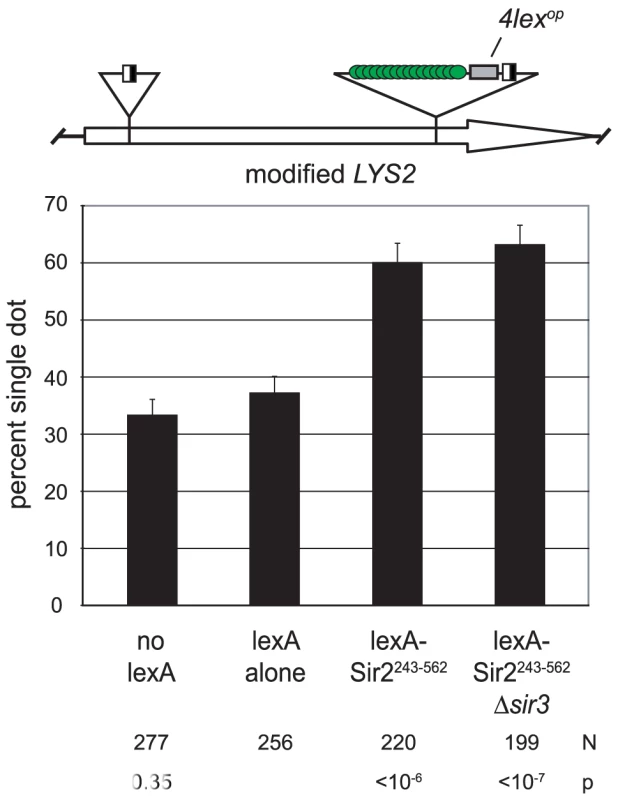
Colocalization of DNA circles excised from the LYS2 locus was measured in strains CSW91 (wt) or CSW95 (Δsir3) expressing either lexA-Sir2243–562 (pCSW1) or lexA alone (pCSW2). A non-catalytic domain of Sir2 mediates cohesion
We next asked whether the deacetylase activity of Sir2 was responsible for Sir2-mediated cohesion. To address this question, we introduced a well-characterized active site mutation (H364Y) into lexA-Sir2243–562. Previous studies showed that this mutation abolishes Sir2 deacetylase activity, silencing and silent chromatin formation [32], [33]. We found here that the mutated polypeptide conferred as much cohesion to HMR circles as the unaltered polypeptide (Figure 4A). This experiment was performed in a sir2 null strain to eliminate contributions of the endogenous gene (see figure 2A). Furthermore, acting on the remote possibility that tethered Sir2 mediates cohesion by recruiting one of the other yeast sirtuins (Hst1-4), we repeated the experiment in media supplemented with nicotinamide, a generic sirtuin inhibitor [28]. No decrease in cohesion was observed (64% vs. 61% with nicotinamide; p = 0.5). Collectively, these results show that the enzymatic activity of Sir2 or other sirtuins is not required for cohesion by tethered Sir2.
Fig. 4. A non-catalytic domain of Sir2 mediates cohesion. 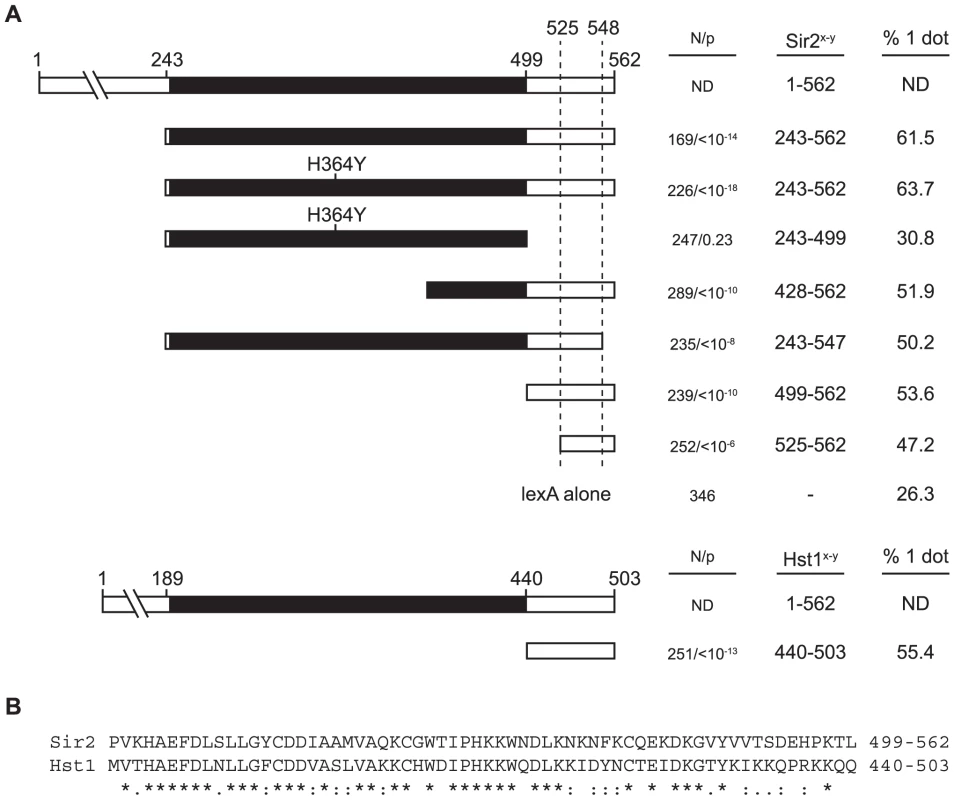
A) Cohesion assays with Sir2 fragments. A map Sir2 shows the conserved catalytic core domain in black. Strain CSW42 (Δsir2) was transformed with vectors expressing lexA-Sir2243–562 (pCSW1), lexA-Sir2243–562(H364Y) (pCSW3), lexA-Sir2243–499(H364Y) (pCSW56), lexA-Sir2428–562 (pCSW6), lexA243–547 (pCSW10), lexA-Sir2499–562 (pYFC1), lexA-Sir2525–562 (pYFC2), lexA-Hst1440–503 (pYFC6) or lexA alone (pCSW2). Immunoblotting with antibodies to lexA confirmed that the cohesion-deficient chimera lexA-Sir2243–499(H364Y) expressed appropriately (data not shown). ND = not determined. B) Alignment of the Sir2499–562 and Hst1440–503 with conservation noted according to CLUSTALW criteria [78]: identical residues (*); conservative substitutions (:); semi-conservative substitutions (.). To map the Sir2 domain responsible for cohesion we generated a set of truncation mutants. Figure 4A shows that all but one of the constructs yielded HMR cohesion levels significantly above background. The exception is a lexA chimera that bears just the conserved catalytic core of Sir2 (residues 243–499). All of the other cohesion-proficient chimeras share in common a small C-terminal domain of Sir2 spanning amino acids 525–547. The picture that emerges is that Sir2 contains a discrete motif within the non-catalytic, C-terminal region of the protein that mediates cohesion.
Hst1 is a yeast sirtuin that bears considerable amino acid similarity to Sir2 in the C-terminal region (Figure 4B). The deacetylase represses middle sporulation genes in vegetative cells, as well as genes involved in NAD+ and thiamine biosynthesis [34]–[36]. Hst1 differs from Sir2 in that the protein acts locally to repress specific promoters rather than by forming an extended repressive domain [37]. We tethered the C-terminal domain (amino acids 440–503) to HMR in the same Δsir2 strain used to evaluate the lexA-Sir2 constructs. Figure 4A shows that lexA-Hst1440–503 imparts a comparable degree of targeted cohesion. These results indicate that Sir2-mediated cohesion is not limited to just one member of the sirtuin family.
Sir2-mediated cohesion requires cohesin
Both Sir2 and its yeast paralog Hst2 can form homotrimers [38], [39]. Thus, one explanation for DNA colocalization is that tethered Sir2 fragments on different DNAs associate with one another directly. To explore this possibility we performed a two-hybrid analysis using lexA-Sir2243–562(H364Y) as a bait protein. The experiments utilized a HIS3 reporter strain that lacks the endogenous SIR2 gene. A weak positive interaction signal was obtained with a prey vector bearing full length Sir2 fused to the Gal4 activation domain (Figure 5A). Importantly, no interaction was seen with a prey vector bearing the shorter Sir2243–562 fragment. Given that all of our critical experiments were performed with this fragment in strains lacking full length Sir2, colocalization of HMR circles is not likely attributable to Sir2 self-association.
Fig. 5. Cohesion by Sir2 requires the cohesin genes. 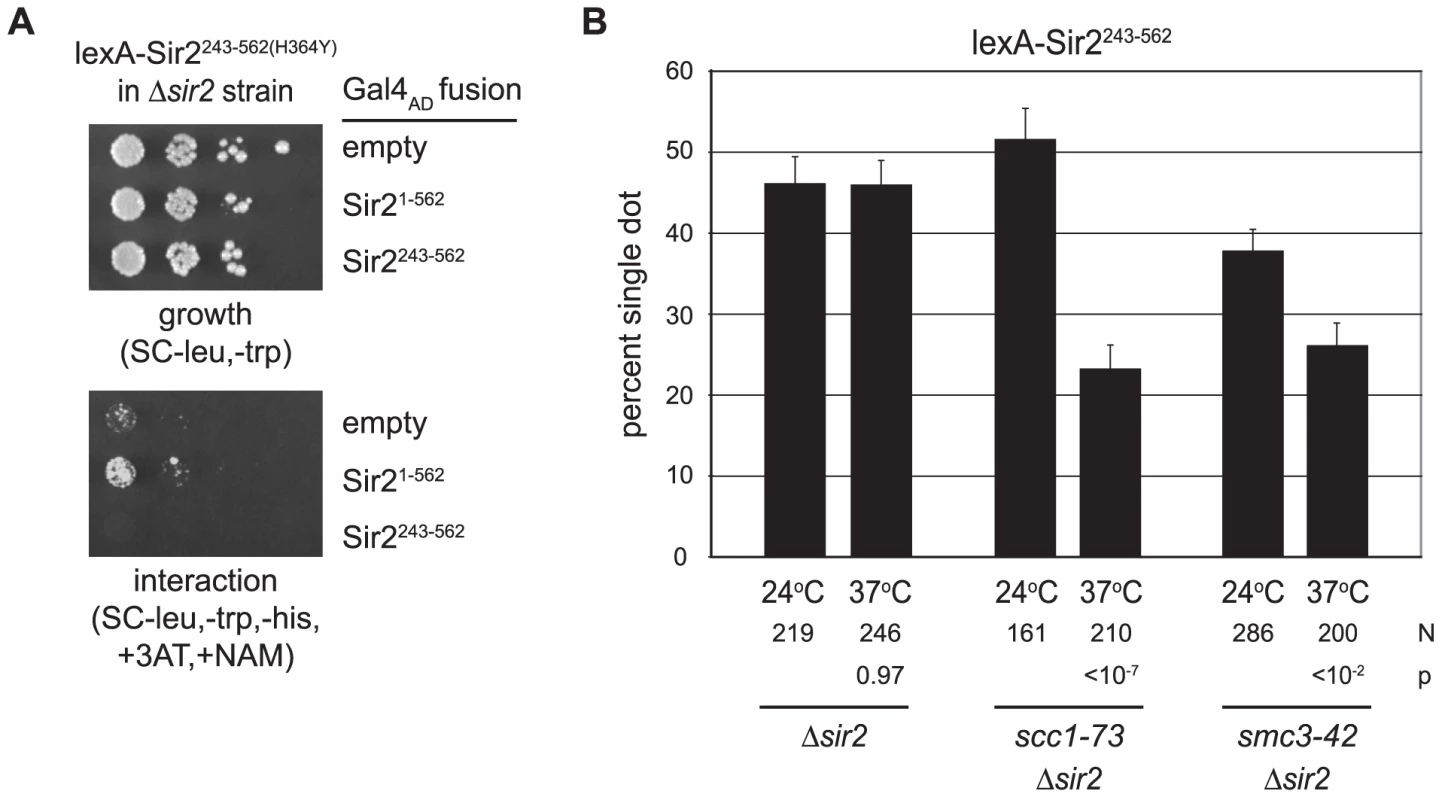
A) Two-hybrid assay for Sir2 self-association. Strain JCY13 (LYS2::(4xlexop)-HIS3 Δsir2) expressing lexA-Sir2243–562(H364Y) (pCSW55) in combination with Gal4AD (pGAD-C1), Gal4AD-Sir21–562 (pCTC85), or Gal4AD-Sir2243–562 (pCSW58) was plated in serial dilution on SC-trp,-leu to monitor growth and SC-trp,-leu,-his supplemented with 5 mM 3-aminotriazole (3AT) and 5 mM nicotinamide (NAM) to monitor interactions. 3-AT raises the level of HIS3 expression necessary for growth. NAM was included in case recruitment of functional Sir2 caused silencing of the reporter gene. B) Targeted cohesion assay in cohesin mutants. Cultures were pre-grown at 24°C according to the standard protocol (Materials and Methods). Two hours after the addition of galactose and benomyl, half of each culture was shifted to 37°C while the other half was maintained at 24°C. Two hours later, the cells were harvested and fixed. Strains CSW42 (Δsir2), CSW75 (scc1-73 Δsir2) and CSW152 (smc3-42 Δsir2) expressed lexA-Sir2243–562 (pCSW1). P values report the significance of the temperature shift on cohesion for each strain evaluated. Cohesin mediates cohesion of the native HMR locus [9]. We therefore anticipated that cohesin genes would also be required for cohesion by tethered Sir2. To test this possibility we crossed temperature sensitive alleles of MCD1/SCC1 and SMC3 into our DNA circle-producing strain. The scc1-73 and smc3-42 mutants and a wild-type counterpart were arrested in mitosis at permissive temperature (24°C). After recombining the HMR locus, the cultures were divided: half was maintained at the permissive temperature while the other half was shifted to 37°C, the non-permissive temperature for these mutants (see Figure 5 legend for details). In the wild-type strain, cohesion of the HMR circles by lexA-Sir2243–562 was unaffected by the temperature shift (Figure 5B). By contrast, both mutant strains displayed a significant reduction in HMR cohesion at the non-permissive temperature. This data indicates that cohesin is responsible for cohesion of DNA circles bound by Sir2.
Binding of cohesin at HMR requires Sir2 within silent chromatin
Chromatin immunoprecipitation (ChIP) was used previously to show that cohesin associates with HMR in a silencing-dependent manner [9]. We showed that Mcd1-TAP binding was lost when silent chromatin assembly was blocked by 1) deletion of SIR3, or 2) inhibition of the Sir2 deacetylase (see ChIPs of chromosomal templates in figures 5A and 7D of [9]). In the current study, a similar ChIP protocol was used to test whether lexA-Sir2243–562 retained cohesin at HMR-6lexopssEB. Unexpectedly, we could not obtain reproducible enrichment of the targeted locus. A variety of conditions and reagents were tested, and the procedure was validated with native silent chromatin (see below). We suspect that the level of cohesin necessary for colocalization in the targeted cohesion assay falls below the detection limit of this ChIP experiment.
We turned instead to a protein chimera approach we recently developed to study other aspects of transcriptionally silent chromatin [40]. In that study, Sir3 was fused to the Rpd3-family deacetylase Hos3 to show that the roles of Sir2 in silencing could be bypassed entirely. We demonstrated that the Sir3-Hos32–549 chimera 1) spread throughout HMR, 2) deacetylated histones across the locus and 3) required both silencers and Sir4 to mediate repression. Silencing could also be achieved by fusing Sir3 to a fragment of Sir2 that possessed enzyme activity but that lacked domains necessary for targeting. Here these Sir3 chimeras (Sir3-Sir2243–562 and Sir3-Hos32–549) were used to investigate the role for Sir2 in binding cohesin at a silenced domain. An additional chimera (Sir3-Hos32–549-Sir2499–562) was constructed to study the contribution of the 64 amino acid, cohesion-proficient fragment of Sir2.
Mating assays were used to evaluate the silencing potential of each chimera. In this assay, loss of HMR silencing in MATα cells creates a pseudo-diploid state that blocks mating and thus subsequent growth on SD indicator plates. Figure 6A confirms previous findings that Sir3-Sir2243–562 and Sir3-Hos32–549 mediate silencing of HMR in the absence of endogenous Sir2. The figure shows that Sir3-Hos32–549-Sir2499–562 also conferred silencing of HMR, albeit at a reproducibly reduced level. This functional assay indicates that Sir3-Hos32–549-Sir2499–562 delivers the Sir2499–562 fragment to the site where cohesion and cohesin binding were to be tested.
Fig. 6. Binding of cohesin at HMR requires Sir2. 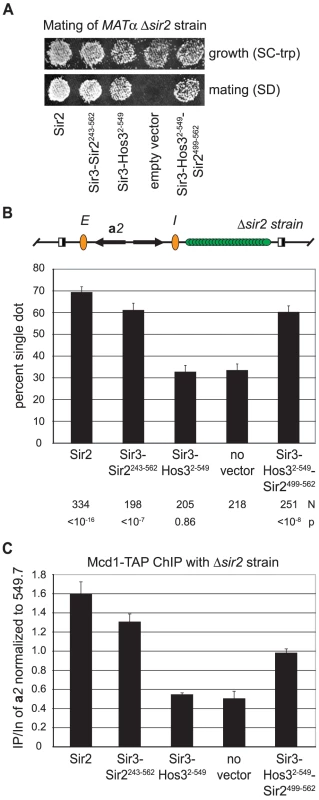
A) Silencing by Sir2 and the Sir3-deacetylase chimeras used as substitutes for Sir2. Strain YFC9 (MATα Δsir2) bearing plasmids expressing Sir2 (pCC7), Sir3-Hos32–549 (pCC10), Sir3-Sir2243–562 (pCC4), Sir3-Hos32–549-Sir2499–562 (pYFC14) or empty vector (pRS414) was mated with tester strain K125 (MATa hom3 ilv1). SC-trp plates monitored growth of the YFC9 transformants and SD plates monitored mating. B) Cohesion by Sir3-deacetylase chimeras. Colocalization of DNA circles bearing native HMR silencers was measured in strain CSW84 (Δsir2) bearing plasmids expressing Sir2 (pYFC16), Sir3-Sir2243–562 (pCC34), Sir3-Hos32–549 (pCC24) or Sir3-Hos32–549-Sir2499–562 (pYFC15) or no plasmid at all. C) Chromatin binding of Mcd1. Immunoprecipitation of Mcd1-TAP from strain CSW116 (SIR2) or strain YFC9 (Δsir2) bearing plasmids expressing Sir3-Hos32–549 (pCC10), Sir3-Sir2243–562 (pCC4), Sir3-Hos32–549-Sir2499–562 (pYFC14) or no plasmid at all. The ratio of the HMR a2 gene relative to the 549.7 site in the immunoprecipitated material (IP) was normalized to the same ratio of input material (In). Reported values represent the mean and standard deviation of at least three independent trials. See Figure S2 for representative gels. Cohesion by the Sir3 chimeras was evaluated in a sir2 null strain that produces GFP-tagged HMR circles with wild-type silencers (Figure 6B). In the absence of a chimera, HMR cohesion occurred in 33% of the cells. When Sir2 or the Sir3-Sir2243–562 was expressed, HMR cohesion levels rose to 69% and 61%, respectively. By contrast, expression of Sir3-Hos32–549 did not increase cohesion above background levels. Remarkably, addition of Sir2499–562 to the Sir3 - Hos32–549 chimera restored colocalization to the level obtained with Sir3 - Sir2243–562. This analysis indicates that the C-terminal fragment of Sir2 must be present within silenced chromatin for cohesion to occur.
ChIP of TAP-tagged Mcd1 was used to measure the ability of the chimeras to position cohesin at the HMR a2 gene. A cohesin-associated region of chromosome V (designated 549.7) that is not influenced by the SIR genes was used as a point of comparison [22]. Reference SIR2 and Δsir2 strains in figure 6C confirmed earlier findings: binding of Mcd1-TAP at HMR is hindered when silent chromatin is disrupted by loss of a single Sir protein, in this case Sir2. Expression of the Sir3-Sir2243–562 chimera restored cohesin binding at HMR to within 20% of the native level. By contrast, expression of the silencing-proficient Sir3-Hos32–549 chimera did not raise cohesin binding above the background sir2 null level. Importantly, the addition of 64 amino acids of Sir2 to the end of the Sir3-Hos32–549 chimera increased cohesin binding substantially. Taken together, these data indicate that Sir2 must be present within silent chromatin for cohesin to accumulate at silenced loci, and that a small C-terminal portion of Sir2 is sufficient for this activity.
Sir2 mediates HMR cohesion without proteins required for rDNA silencing and stability
Sir2 associates with the cluster of tandemly repeated ribosomal RNA genes known as the rDNA array. In this context the protein suppresses recombination between the repeated elements and suppresses RNA polymerase II transcription within each element [41]–[43]. Sir2 has been implicated in cohesin binding at the rDNA [44], [45]. It therefore seemed prudent to test whether rDNA-specific, protein partners of Sir2 modulate cohesion by the tethered protein. We first considered Net1, which along with Sir2 and Cdc14 forms the RENT complex [46], [47]. This protein is required for Sir2 binding at the rDNA and it has been found at HMR when over-expressed [46], [48]. A 15 amino acid C-terminal truncation of Sir2 disrupts the Net1-Sir2 interaction, abolishing rDNA silencing but not silencing of telomeres or the HM loci [49]. Figure 4A shows that deleting these 15 residues (lexA-Sir2243–547) did not interfere substantially with cohesion of HMR circles. We conclude that the RENT complex is not necessary for cohesion by tethered Sir2.
Transcriptional silencing by Sir2 at the rDNA recombinational enhancer requires a set of interacting proteins that includes Tof2 and a pair of bifunctional factors Csm1 and Lrs4. During meiosis I, Csm1 and Lrs4 form the monopolin complex that orients sister chromatid pairs towards the same spindle pole [50], [51]. Csm1 interacts with both Mcd1 and Smc1 prompting Huang and Moazed to hypothesize that these proteins link cohesin to the rDNA [52]. We tested whether these genes were required for cohesion by lexA-Sir2243–562. Figure 7 shows that neither TOF2, CSM1 nor LRS4 were required for colocalization of HMR circles. Collectively, the findings indicate that these rDNA silencing and stability proteins do not contribute to cohesion of HMR by tethered Sir2.
Fig. 7. Cohesion persists in the absence of rDNA-specific, protein partners of Sir2. 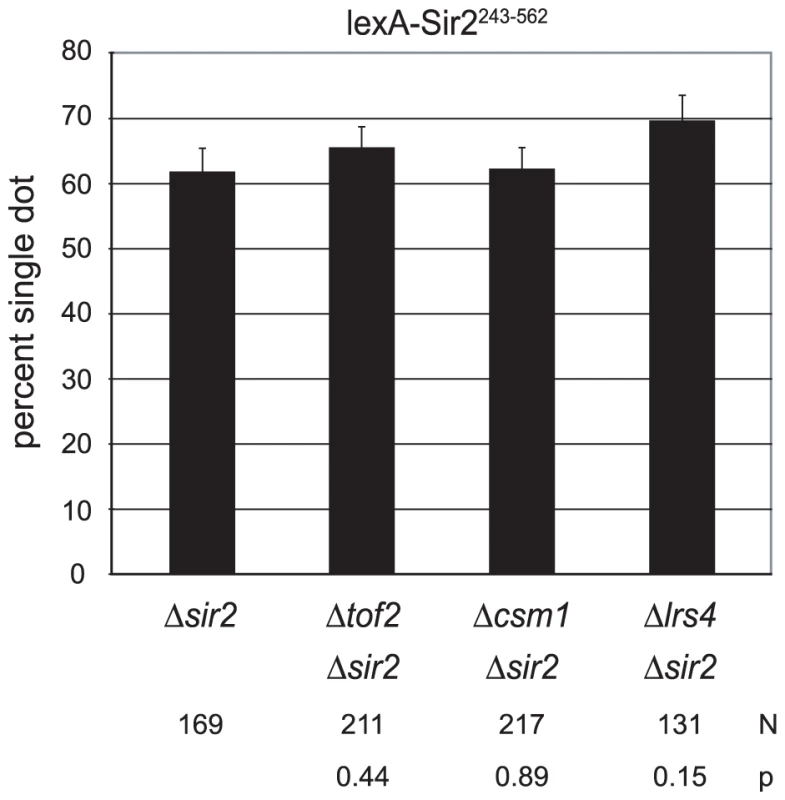
Colocalization of DNA circles bearing the 6lexopssEB silencer was measured in strains CSW42 (Δsir2), CSW65 (Δcsm1 Δsir2), CSW68 (Δtof2 Δsir2) and CSW66 (Δlrs4 Δsir2, only two trials) expressing lexA-Sir2243–562 (pCSW1). Discussion
In this study we examined the mechanistic basis for Sir-dependent cohesion of a silenced chromosomal domain in budding yeast. We developed a protein-targeting assay and found that the evolutionarily conserved Sir2 deacetylase was both necessary and sufficient for pairing DNA circles. Cohesin was required but other silencing factors like Sir3 and Sir4 were not. Through the use of mutants we showed that transcriptional silencing and cohesion are separable events: tethered Sir2 conferred cohesion in the absence of silencing and the Sir3-Hos3 chimera generated silencing in the absence of cohesion. Importantly, fusing a small fragment of Sir2 to Sir3-Hos3 was sufficient to restore cohesin binding and cohesion of the locus. We conclude that, in addition to deacetylating histones for silent chromatin assembly, Sir2 also orchestrates cohesin-dependent cohesion of silent chromatin domains on sister chromatids.
Sir2, cohesin and transcriptional silencing
Although our studies here focused on HMR we expect that the relationship between Sir2 and cohesion extends to other loci where Sir proteins assemble. Indeed, preliminary evidence indicates that the HML mating-type locus is also cohered in a silencing-dependent manner (Campor and Gartenberg, unpublished results). Why do silent chromatin and cohesion converge? Initial studies suggested a role in regulating transcriptional repression. Donze and Kamakaka first showed that silencing spread beyond HMR barrier elements in cohesin mutants [21]. Steve Bell and colleagues followed by showing that cohesin delayed establishment of silencing in cells that were walked step-wise through the cell cycle [53]. A parsimonious explanation for these observations is that cohesin impedes the conversion of active chromatin to silenced chromatin. Numerous studies in higher eukaryotes have further linked cohesin to gene regulatory phenomena (see [54] for a review). Intriguingly, cohesin was recently shown to form loops between enhancers and promoters by interacting with a transcriptional coactivation complex known as mediator [55]. Similarly, cohesin forms loops between distant sites by binding the mammalian CTCF, a protein that associates with insulators as well as other gene regulatory elements [56]–[59]. In yeast, silent chromatin domains fold-back upon themselves and interact with one another over great distances [60], [61]. Thus, one possibility is that cohesin facilitates long interactions to regulate silent chromatin domains.
Sir2, cohesin, and rDNA stability
The rationale for Sir2 mediating cohesion might alternatively be related to its role in genome stabilization at the rDNA. Binding of the deacetylase is necessary for binding of cohesin, which in turn is thought to block unequal sister chromatid exchange by maintaining the register between rDNA elements of opposing sister chromatids [44]. Exactly how Sir2 retains cohesin at the locus in not entirely clear. In one model, the deacetylase modulates cohesin levels indirectly by silencing a conserved RNA polymerase II promoter element near the rDNA recombinational enhancer [45]. According to the model, transcription by RNA polymerase II displaces cohesin when Sir2 is absent. A competing model by Huang and Moazed suggests that direct interaction between cohesin and one of the components of the rDNA silencing pathway, Csm1 specifically, could account for recruitment of the complex [52]. Our work with tethered fragments of Sir2 suggests an even more direct possibility: the polypeptide, not its capacity to silence, mediates cohesin recruitment directly. We note that a direct recruitment model for Sir2 need not be mutually exclusive with models based on transcriptional inhibition, or with other factors that contribute to rDNA stability [62].
Roles for protein deacetylation in sister chromatid cohesion
Acetylation and deacetylation of cohesin subunits plays a newly appreciated role in regulating cohesion during the cell cycle. Cohesion is established during S phase when the Eco1 protein acetyltransferase acetylates Smc3 [63]–[66]. This modification persists until cohesin complexes disassemble at anaphase onset. Deacetylation is a prerequisite for Smc3 to be re-used in the next cell cycle. Recently, the Rpd3-family member Hos1 was identified as the principle Smc3 deacetylase in yeast [67]–[69]. That residual deacetylation persists in the absence of Hos1 suggests that additional Smc3 deacetylation activities remain to be discovered [67]. Following DNA double strand breaks, Eco1 similarly acetylates Mcd1 to establish damage-induced cohesion [70]. Presumably, a parallel pathway exists for Mcd1 deacetylation. Whether Sir2 or a combination of sirtuins is involved in Mcd1 deacetylation or the residual deacetylation of Smc3 has not been determined.
Other sirtuins, other contexts, other genomes
The catalytic activity of Sir2 accounts for all other known functions of the enzyme. By contrast, cohesion by tethered Sir2 fragments does not even require the conserved catalytic core. Instead, we found that a small domain at the carboxyl-terminus was responsible. We anticipate that this domain retains cohesin at silenced loci by interacting directly with a cohesin subunit or with other proteins involved in cohesin utilization. Conversely, such an interaction could be important in situations where Sir2 might be recruited to sites where cohesin binds.
Significant homology exists between the C-terminal domain of Sir2 and Hst1. That this Hst1 domain also mediates cohesion when tethered to DNA suggests that cohesion occurs at the numerous promoters where Hst1 binds to regulate gene expression [34]–[36]. The lack of a homologous C-terminal domain in mammalian sirtuins thwarts a simple extrapolation to a cohesion connection in higher eukaryotes. However, the characteristics of two mammalian sirtuins, SirT1 and SirT6, warrant consideration (see [71], [72], and references therein). Like Sir2, these mammalian enzymes deacetylate histones (and other protein targets) to regulate gene expression. Additionally, SirT1 plays multiple roles in heterochromatic repression and SirT6 localizes to heterochromatin domains. Double strand breaks may represent sites of particular interest. Cohesin is recruited to these sites in yeast and in humans, as are Sir2, Hst1, SirT1 and SirT6 [73], [74]. The mammalian enzymes have been shown to suppress genomic instability, in part, by modifying DNA repair factors (see [75] for most recent example). Whether SirT1 and SirT6 also link sister chromatid cohesion to these chromosome-based events has not yet been tested.
Materials and Methods
Strain construction and application
Table S1 provides a complete list of strains used in this study. Cohesion of HMR by tethered proteins was measured with variants of strain CSW19 (RS::6lexopssEB-a2a1-256lacop-TRP1-ΔhmrI::RS (LEU2::GAL1-R)2::leu2-3,112 ADE2::HIS3p-lacGFP::ade2-1). Recombinase target sites are designated as RS. Cohesion of the LYS2 gene was measured with variants of strain CSW91 (RS::lys2-TRP1-4lexop-256lacop::RS). Cohesion of HMR by Sir3 chimeras was measured with native silencers in strain CSW84 (RS::HMRE-a2a1-HMRI-TRP1-256lacop::RS Δsir2). Silencing assays were performed with strains derived from GA-2050 (Aeb4lexop-TRP1-HMRI) and two hybrid assays were performed with strain JCY13 (LYS2::(4xlexop)-HIS3 Δsir2). ChIP assays were performed in variants of strain CSW116 (MCD1-TAP Δsir2).
Complete ORF deletions were generated by PCR-mediated gene replacement using purified plasmids or extracted yeast DNA as PCR templates. All modifications were confirmed by combined gain and loss of diagnostic PCR products. Strains CSW18 and CSW19 are segregants of a cross between CSW10 and YCL49. Strains CSW47 and CSW48 are segregants of crosses between CSW36 and either K5832 or CRC85. Strain CSW91 is a segregant of a cross between CSW19 and GA-2627. Plasmids pRS403-lexA-Sir278–562 and pRS403-lexA were integrated in single copy at the HIS3 locus of CSW19 to yield strains CSW36 and CSW37, respectively.
Plasmid construction and confirmation
Tables S2–S3 provide detailed information about the plasmids used and how they were constructed. In addition to using traditional bacterial cloning techniques, plasmids were constructed in yeast by PCR-mediated plasmid gap repair (P-MPGR) or fragment-mediated plasmid gap repair (F-MGPR) using restriction digestion products. SIR2 truncations were generated by oligonucleotide-mediated plasmid gap repair (O-MPGR). All modifications within the gene chimeras were confirmed by sequencing. Plasmid sequences are available upon request.
Cohesion assay
Colocalization of excised DNA circles in M phase was measured as described in Chang et al. [9], unless specified otherwise. To retain plasmids, selective media was used for pregrowth on dextrose and subsequent growth on raffinose overnight. When the cultures reached mid-log phase the following morning, an equal volume of YPA media plus raffinose was added. Twenty minutes later, nocodazole (stock 1 mg/ml in DMSO, Cf = 10 µg/ml) was added to initiate M phase arrest. After three hours, galactose (Cf = 2%) and benomyl (stock 1 mg/ml, Cf = 10 µg/ml) were added. Two hours later, cells were harvested, fixed and mounted on microscope slides with agar pads. Serial sections were obtained by fluorescence microscopy and GFP-foci/nucleus were counted manually. All measurements (reported as the percentage of cells with single dots) are based on at least three independent trials, which were pooled because they satisfied χ2 tests of homogeneity of proportions. Error bars represent the standard error of proportion. In each data panel, values were compared to an appropriate control by χ2 tests and judged as significant using a 95% confidence interval.
Assays for two-hybrid interactions and silencing of reporter genes
To measure silencing of TRP1 inserted at HMR or two-hybrid interactions with the HIS3 reporter construct, plasmid-bearing strains were grown to saturation in selective media to retain plasmids and spotted in 10-fold serial dilutions. One set of selective plates was used to measure reporter gene expression and a second set was used as a loading control.
Mating assays for HMR silencing
Strain YFC9 (MATα Δsir2) bearing Sir2-substitution plasmids was grown to saturation in selective medium, diluted 10-fold and then spotted on a lawn of mating tester strain K125 (MATa) on YPDA plates. After at least 5 hr at 30°C, the cells were replica plated to SD agar to measure mating and SC-trp as a loading control.
Chromatin immunoprecipitation
Nocodazole was added to mid-log cultures that were either grown in YPDA overnight or that were sub-cultured in YPDA for 3 hours after overnight growth in selective media to retain plasmids. Three hours later, cross-linking and subsequent ChIP procedures were performed according to [22] using anti-TAP antibody (Open Biosystems) and Protein A beads (Invitrogen). PCR reactions were run in multiplex using primer sets listed in Table S4. Simultaneous amplification of a cohesin-free site (534) was included as an internal negative control of the immunoprecipitation reaction (Figure S2). Gels were stained with EtBr and destained in water before digital photography (Alpha Innotech). All bands were found to be non-saturating and within the linear range. Reported values were calculated as (a2/549.7)IP/(a2/549.7)In.
Supporting Information
Zdroje
1. SchüleBOviedoAJohnstonKPaiSFranckeU 2005 Inactivating mutations in ESCO2 cause SC phocomelia and Roberts syndrome: no phenotype-genotype correlation. Am J Hum Genet 77 1117 1128
2. VegaHWaisfiszQGordilloMSakaiNYanagiharaI 2005 Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet 37 468 470
3. BarberTDMcManusKYuenKWReisMParmigianiG 2008 Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A 105 3443 3448
4. ZhangNGeGMeyerRSethiSBasuD 2008 Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci U S A 105 13033 13038
5. OnnIHeidinger-PauliJMGuacciVUnalEKoshlandDE 2008 Sister chromatid cohesion: a simple concept with a complex reality. Ann Rev Cell Dev Biol 24 105 129
6. NasmythKHaeringCH 2009 Cohesin: its roles and mechanisms. Annu Rev Genet 43 525 558
7. HaeringCHFarcasAMArumugamPMetsonJNasmythK 2008 The cohesin ring concatenates sister DNA molecules. Nature 454 297 301
8. MilutinovichMKoshlandDE 2003 SMC complexes-wrapped up in controversy. Science 300 1101 1102
9. ChangCRWuCSHomYGartenbergMR 2005 Targeting of cohesin by transcriptionally silent chromatin. Genes Dev 19 3031 3042
10. ZhangNKuznetsovSGSharanSKLiKRaoPH 2008 A handcuff model for the cohesin complex. J Cell Biol 183 1019 1031
11. LengronneAKatouYMoriSYokobayashiSKellyGP 2004 Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430 573 578
12. GlynnEFMegeePCYuHGMistrotCUnalE 2004 Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2 e259 doi:10.1371/journal.pbio.0020259
13. BauschCNooneSHenryJMGaudenzKSandersonB 2007 Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol Cell Biol 27 8522 8532
14. MisulovinZSchwartzYBLiXYKahnTGGauseM 2008 Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117 89 102
15. ParelhoVHadjurSSpivakovMLeleuMSauerS 2008 Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132 422 433
16. WendtKSYoshidaKItohTBandoMKochB 2008 Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451 796 801
17. BernardPMaureJFPartridgeJFGenierSJaverzatJP 2001 Requirement of heterochromatin for cohesion at centromeres. Science 294 2539 2542
18. NonakaNKitajimaTYokobayashiSXiaoGYamamotoM 2002 Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4 89 93
19. YamagishiYSakunoTShimuraMWatanabeY 2008 Heterochromatin links to centromeric protection by recruiting shugoshin. Nature 455 251 255
20. RuschéLNKirchmaierALRineJ 2003 The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Ann Rev Biochem 72 481 516
21. DonzeDAdamsCRRineJKamakakaRT 1999 The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes & Dev 13 698 708
22. DubeyRNGartenbergMR 2007 A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev 21 2150 2160
23. D'AmbrosioCSchmidtCKKatouYKellyGItohT 2008 Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22 2215 2227
24. KogutIWangJGuacciVMistryRKMegeePC 2009 The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev 23 2345 2357
25. LiY-CChengT-HGartenbergMR 2001 Establishment of transcriptional silencing in the absence of DNA replication. Science 291 650 653
26. ChienC-TBuckSSternglanzRShoreD 1993 Targeting of Sir1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75 531 541
27. ShermanJMStoneEMFreeman-CookLLBrachmannCBBoekeJD 1999 The conserved core of a human SIR2 homologue functions in yeast silencing. Mol Biol Cell 10 3045 3059
28. SauveAAWolbergerCSchrammVLBoekeJD 2006 The biochemistry of sirtuins. Annu Rev Biochem 75 435 465
29. CockellMMPerrodSGasserSM 2000 Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics 154 1069 1083
30. LiebJDLiuXBotsteinDBrownPO 2001 Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28 327 334
31. GartenbergMRNeumannFNLarocheTBlaszczykMGasserSM 2004 Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119 955 967
32. ArmstrongCMKaeberleinMImaiSIGuarenteL 2002 Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity. Mol Biol Cell 13 1427 1438
33. HoppeGJTannyJCRudnerADGerberSADanaieS 2002 Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22 4167 4180
34. XieJPierceMGailus-DurnerVWagnerMWinterE 1999 Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO 18 6448 6454
35. BedalovAHiraoMPosakonyJNelsonMSimonJA 2003 NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol Cell Biol 23 7044 7054
36. LiMPetteysBJMcClureJMValsakumarVBekiranovS 2010 Thiamine biosynthesis in Saccharomyces cerevisiae is regulated by the NAD+-dependent histone deacetylase Hst1. Mol Cell Biol 30 3329 3341
37. RuschéLNRineJ 2001 Conversion of a gene-specific repressor to a regional silencer. Genes Dev 15 955 967
38. ZhaoKChaiXClementsAMarmorsteinR 2003 Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat Struct Biol 10 864 871
39. CubizollesFMartinoFPerrodSGasserSM 2006 A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol Cell 21 825 836
40. ChouCCLiYCGartenbergMR 2008 Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing. Mol Cell 31 650 659
41. GottliebSEspositoRE 1989 A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56 771 776
42. SmithJSBoekeJD 1997 An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev 11 241 254
43. BrykMBanerjeeMMurphyMKnudsonKEGarfinkelDJ 1997 Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev 11 255 269
44. KobayashiTHoriuchiTTongaonkarPVuLNomuraM 2004 SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117 441 453
45. KobayashiTGanleyAR 2005 Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309 1581 1584
46. StraightAFShouWDowdGJTurckCWDeshaiesRJ 1999 Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97 245 256
47. HuangJMoazedD 2003 Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev 17 2162 2176
48. KasulkeDSeitzSEhrenhofer-MurrayAE 2002 A role for the Saccharomyces cerevisiae RENT complex protein Net1 in HMR silencing. Genetics 161 1411 1423
49. CuperusGShafaatianRShoreD 2000 Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO 19 2641 2651
50. TothARabitschKPGalovaMSchleifferABuonomoSB 2000 Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 103 1155 1168
51. RabitschKPPetronczkiMJaverzatJPGenierSChwallaB 2003 Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell 4 535 548
52. HuangJBritoILVillenJGygiSPAmonA 2006 Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev 20 2887 2901
53. LauABlitzblauHBellSP 2002 Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes & Dev 16 2935 2945
54. BoseTGertonJL 2010 Cohesinopathies, gene expression, and chromatin organization. J Cell Biol 189 201 210
55. KageyMHNewmanJJBilodeauSZhanYOrlandoDA 2010 Mediator and cohesin connect gene expression and chromatin architecture. Nature 467 430 435
56. HadjurSWilliamsLMRyanNKCobbBSSextonT 2009 Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460 410 413
57. NativioRWendtKSItoYHuddlestonJEUribe-LewisS 2009 Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 5 e1000739 doi:10.1371/journal.pgen.1000739
58. MishiroTIshiharaKHinoSTsutsumiSAburataniH 2009 Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J 28 1234 1245
59. HouCDaleRDeanA 2010 Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A 107 3651 3656
60. ValenzuelaLDhillonNDubeyRNGartenbergMRKamakakaRT 2008 Long-range communication between the silencers of HMR. Mol Cell Biol 28 1924 1935
61. MieleABystrickyKDekkerJ 2009 Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet 5 e1000478 doi:10.1371/journal.pgen.1000478
62. IdeSMiyazakiTMakiHKobayashiT 2010 Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327 693 696
63. Ben-ShaharTRHeegerSLehaneCEastPFlynnH 2008 Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321 563 566
64. UnalEHeidinger-PauliJMKimWGuacciVOnnI 2008 A molecular determinant for the establishment of sister chromatid cohesion. Science 321 566 569
65. ZhangJShiXLiYKimBJJiaJ 2008 Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 31 143 151
66. RowlandBDRoigMBNishinoTKurzeAUluocakP 2009 Building sister chromatid cohesion: Smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33 763 774
67. BorgesVLehaneCLopez-SerraLFlynnHSkehelM 2010 Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol Cell 39 677 688
68. BeckouetFHuBRoigMBSutaniTKomataM 2010 An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol Cell 39 689 699
69. XiongBLuSGertonJL 2010 Hos1 is a lysine deacetylase for the Smc3 subunit of cohesin. Curr Biol 20 1660 1665
70. Heidinger-PauliJMUnalEKoshlandD 2009 Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol Cell 34 311 321
71. HaigisMCSinclairDA 2010 Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 5 253 295
72. TennenRIChuaKF 2010 Chromatin regulation and genome maintenance by mammalian SIRT6. Trends Biochem Sci
73. UnalEArbel-EdenASattlerUShroffRLichtenM 2004 DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 16 991 1002
74. StromLLindroosHBShirahigeKSjogrenC 2004 Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16 1003 1015
75. KaidiAWeinertBTChoudharyCJacksonSP 2010 Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 329 1348 1353
76. TaddeiAHedigerFNeumannFRBauerCGasserSM 2004 Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J 23 1301 1312
77. HolmesSGRoseABSteuerleKSaezESayeghS 1997 Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics 145 605 614
78. LarkinMABlackshieldsGBrownNPChennaRMcGettiganPA 2007 Clustal W and Clustal X version 2.0. Bioinformatics 23 2947 2948
Štítky
Genetika Reprodukční medicína
Článek Break to Make a ConnectionČlánek A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on DiseaseČlánek The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 2
-
Všechny články tohoto čísla
- Break to Make a Connection
- A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on Disease
- Single-Tissue and Cross-Tissue Heritability of Gene Expression Via Identity-by-Descent in Related or Unrelated Individuals
- Pervasive Adaptive Protein Evolution Apparent in Diversity Patterns around Amino Acid Substitutions in
- The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
- MiRNA Control of Vegetative Phase Change in Trees
- New Functions of Ctf18-RFC in Preserving Genome Stability outside Its Role in Sister Chromatid Cohesion
- Genome-Wide Association Studies of the PR Interval in African Americans
- Mapping of the Disease Locus and Identification of As a Candidate Gene in a Canine Model of Primary Open Angle Glaucoma
- Mapping a New Spontaneous Preterm Birth Susceptibility Gene, , Using Linkage, Haplotype Sharing, and Association Analysis
- A Population Genetic Approach to Mapping Neurological Disorder Genes Using Deep Resequencing
- and Genes Modulate the Switch between Attraction and Repulsion during Behavioral Phase Change in the Migratory Locust
- Targeted Sister Chromatid Cohesion by Sir2
- Correlated Evolution of Nearby Residues in Drosophilid Proteins
- Parallel Evolution of a Type IV Secretion System in Radiating Lineages of the Host-Restricted Bacterial Pathogen
- Lipophorin Receptors Mediate the Uptake of Neutral Lipids in Oocytes and Imaginal Disc Cells by an Endocytosis-Independent Mechanism
- Genome-Wide Association Study of Coronary Heart Disease and Its Risk Factors in 8,090 African Americans: The NHLBI CARe Project
- The Evolution of Host Specialization in the Vertebrate Gut Symbiont
- Genome-Wide Association of Familial Late-Onset Alzheimer's Disease Replicates and and Nominates in Interaction with
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- Association between Common Variation at the Locus and Changes in Body Mass Index from Infancy to Late Childhood: The Complex Nature of Genetic Association through Growth and Development
- AID Induces Double-Strand Breaks at Immunoglobulin Switch Regions and Causing Chromosomal Translocations in Yeast THO Mutants
- A Study of CNVs As Trait-Associated Polymorphisms and As Expression Quantitative Trait Loci
- Whole-Genome Comparison Reveals Novel Genetic Elements That Characterize the Genome of Industrial Strains of
- Prevalence of Epistasis in the Evolution of Influenza A Surface Proteins
- Srf1 Is a Novel Regulator of Phospholipase D Activity and Is Essential to Buffer the Toxic Effects of C16:0 Platelet Activating Factor
- Two Frizzled Planar Cell Polarity Signals in the Wing Are Differentially Organized by the Fat/Dachsous Pathway
- Phosphoinositide Regulation of Integrin Trafficking Required for Muscle Attachment and Maintenance
- Pathogenic VCP/TER94 Alleles Are Dominant Actives and Contribute to Neurodegeneration by Altering Cellular ATP Level in a IBMPFD Model
- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues
- Nucleosomes Containing Methylated DNA Stabilize DNA Methyltransferases 3A/3B and Ensure Faithful Epigenetic Inheritance
- Mutations in Zebrafish Result in Adult-Onset Ocular Pathogenesis That Models Myopia and Other Risk Factors for Glaucoma
- [], the Prion Formed by the Chromatin Remodeling Factor Swi1, Is Highly Sensitive to Alterations in Hsp70 Chaperone System Activity
- Characterization of Transcriptome Remodeling during Cambium Formation Identifies and As Opposing Regulators of Secondary Growth
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
- Epistatic Interaction Maps Relative to Multiple Metabolic Phenotypes
- Quantitative Models of the Mechanisms That Control Genome-Wide Patterns of Transcription Factor Binding during Early Development
- Genome-Wide Transcript Profiling of Endosperm without Paternal Contribution Identifies Parent-of-Origin–Dependent Regulation of
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- MiRNA Control of Vegetative Phase Change in Trees
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



