-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaBreak to Make a Connection
article has not abstract
Published in the journal: . PLoS Genet 7(2): e32767. doi:10.1371/journal.pgen.1002006
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1002006Summary
article has not abstract
Meiosis and the Regulation of Double-Strand Break Formation
Meiosis is the cell division program utilized by most sexually reproducing organisms as a strategy to produce haploid gametes (i.e., sperm and eggs) from diploid parental cells. As suggested by its name, which stems from the Greek word meaning “to diminish or reduce”, meiosis reduces the chromosome number by half. This is accomplished by following a single round of DNA replication with two consecutive rounds of cell division (meiosis I and meiosis II). At meiosis I, homologous chromosomes segregate away from each other (reductional division) and at meiosis II, sister chromatids segregate to opposite poles of the spindle (equational division). During prophase of meiosis I, chromosomes undergo a series of unique and well-orchestrated steps that promote accurate segregation. These steps include the formation of programmed DNA double-strand breaks (DSBs), homologous chromosome pairing, and synapsis. A subset of DSBs are repaired via recombination between homologous chromosomes such that there is a reciprocal exchange of genetic material between the homologs resulting in crossover formation. These crossover events, underpinned by flanking sister chromatid cohesion, generate physical attachments between the homologs (chiasmata), which are important for their proper alignment at the metaphase I plate. Either impaired DSB formation or a failure to form chiasmata during meiosis can result in the formation of eggs and sperm carrying an incorrect number of chromosomes, which in turn accounts for a large percentage of the miscarriages, birth defects, and infertility observed in humans [1]. Thus, DSB formation is an essential process for successful offspring production. Although it is known that DSB formation is catalyzed by Spo11, a conserved type II topoisomerase-like protein [2], [3], the regulation of DSB formation is not completely understood. In this issue of PLoS Genetics, Lake et al. [4] shed new light on this process by identifying Trade Embargo (Trem) as a critical protein for DSB production during Drosophila female meiosis.
Recent studies in yeast have started to uncover the molecular basis for the regulation of DSB induction. It is known that at least ten proteins (Spo11-Ski8, Mer2-Mei4-Rec114, Rec102-Rec104, Mre11-Rad50-Xrs2) are essential for DSB induction in Saccharomyces cerevisiae [5]. S phase cyclin-dependent kinase Cdc28–Clb5/Clb6 (CDK-S) and the Dbf4-dependent kinase Cdc7–Dbf4 (DDK) regulate the timing of DSB formation [6], [7]. Mer2 is an essential target of both CDK-S and DDK. Specifically, Mer2 is phosphorylated by CDK-S at Ser30. This phosphorylation primes Mer2 for subsequent phosphorylation by DDK on Ser29, creating a negatively charged “patch”. This coordinated phosphorylation triggers the interaction of Mer2 with Mei4 and Rec114 [6], [7]. CDK-S-mediated phosphorylation of Mer2 is also important for promoting the interaction between Mer2 and Xrs2 [8]. Thus, pS30 of Mer2 recruits the Mre11-Rad50-Xrs2 complex to DSB hotspots. Finally, Spo11-Ski8 and Rec102-Rec104 sub-complexes are recruited to the hotspots.
Efforts to identify DSB-inducing factors in other species have been hampered in part by the low level of sequence conservation shared with the factors first identified in S. cerevisiae. However, a sophisticated in silico analysis recently identified the orthologs of Mei4 and Rec114 in fission yeast, plants, and mammals [9]. Similar to yeast, mei4−/ − mice lack meiotic DSB induction [9]. In mammals, it has been reported that Prdm9/Meisetz, which is a multi zinc-finger protein that contains KRAB and methyl transferase domains, marks DSB hotspots [10]–[12]. Moreover, the polymorphism of the zinc fingers alters the binding activity to hotspot sequences [10], [11], [12], although Prdm9 is not essential for DSB formation [13]. In other model organisms, HIM-17 which is a six THAP (C2CH) repeat containing protein in Caenorhabditis elegans [14], MEI1 [15] in mice, and SWI1 in Arabidopsis thaliana [16] have been reported as factors required for DSB formation. However, how these proteins act to make DSBs remains unclear.
Trem Is Required for DSB Formation in Drosophila
Trem was originally identified in a germline clone screen for meiotic mutants in Drosophila [17]. Trem encodes a 439 amino acid protein with an N-terminal Zinc finger Associated Domain (ZAD) and five C2H2-type zinc finger domains at the C-terminus. The original mutation alleles are tremF9 and tremf05981. tremF9 carries a C to T transition, corresponding to a proline to leucine change (P299L) in the second zinc finger domain. tremf05981 is a pBac-element insertion located in the gene's 5′UTR that results in a transcriptional null allele. The first analysis of tremF9 mutants revealed a 90-fold decrease in the frequency of crossovers [17]. However, the mechanistic function for Trem in crossover formation remained unknown. In the current study, the authors suggest that Trem functions in DSB formation. They observed that Trem is expressed throughout ovariole nuclei, but is enriched in cells in region 1 of the germarium, where mitotic proliferation and premeiotic S phase take place [18]. DSBs are marked by phosphorylated histone H2AX (γH2AX) in mammals and by phosphorylated His2Av (γHis2Av) in Drosophila. Similar to spo11/mei-W68 and mei-P22 [19] mutants, γHis2Av foci were not observed in tremF9 and tremf05981mutants. Mei-P22 forms discrete chromosome-associated foci during early prophase that are thought to mark the position of future DSBs [19], [20]. Intriguingly, Mei-P22 foci disappeared in trem mutants, suggesting that Trem is required to localize Mei-P22 to discrete foci during early prophase. Two strong lines of evidence are presented which support the idea that DSBs are not induced in trem mutants: 1) trem mutations suppress the defects in egg shell formation, karyosome formation, and oocyte selection detected in spn-B/rad57 and okr/rad54 DNA repair mutants; 2) X-ray exposure, which introduces exogenous DSBs, partially suppresses the defects in γHis2Av formation and meiotic spindle formation observed in trem mutants. These results suggest that Trem is required for DSB formation during meiotic prophase (Figure 1).
Fig. 1. A model for stepwise formation of DSBs during female meiosis in Drosophila. 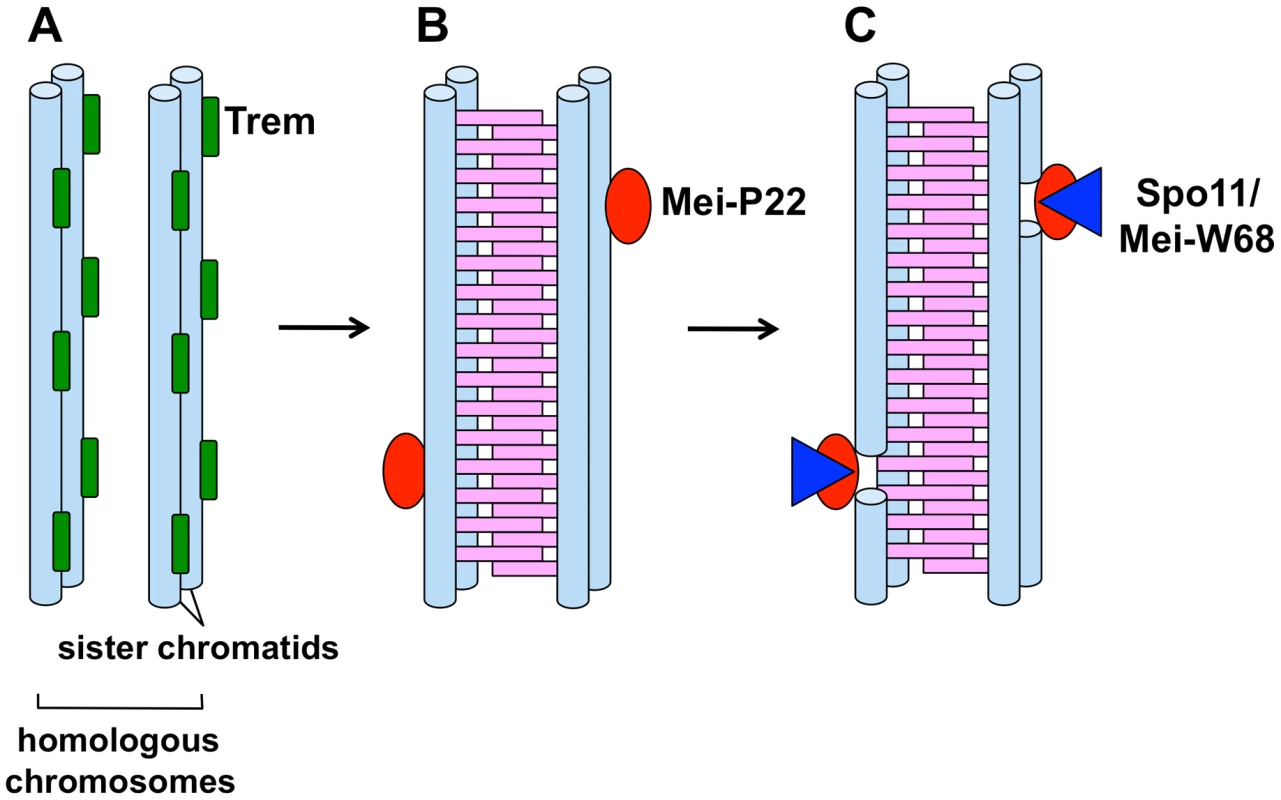
(A) Thread-like localization of Trem along sister chromatids in mitotically proliferating cells (not shown) and premeiotic S phase cells (premeiotic pairing occurs at this stage). (B) Synaptonemal complex formation (pink ladder) is completed by the pachytene stage where Trem-dependent chromosome-associated Mei-P22 foci are detected. (C) DSB formation by Mei-W68/Spo11 at the Mei-P22-marked sites. Setting the Stage for DSB Formation
How does Trem contribute to DSB formation? In Drosophila, homologous chromosome pairing occurs during the mitotic proliferation stage. This phenomenon, combined with the expression pattern of Trem, leads to the hypothesis that the preparation for DSB formation also occurs before meiotic prophase. One possibility is that Trem somehow changes the chromatin state via its DNA binding activity and an unknown interacting partner. After premeiotic replication, Mei-P22 is recruited to the DSB sites. Intriguingly, Mei-P22 has nine potential CDK target sites. Investigation into Mei-P22 regulation by CDK might therefore be very revealing. Although Trem does not harbor a methyl transferase domain and its localization pattern looks evenly distributed on the chromosomes, it will be important to assess whether Trem acts similarly to Prdm9 because Trem is also a multi zinc-finger protein. Moreover, specific posttranslational modifications of Trem may localize Trem to the DSB sites that are the future locations occupied by Mei-P22. Indeed, analysis through the Netphos, SUMOsp, and BDM-PUB programs identify several potential phosphorylation, ubiquitination, and sumoylation sites on Trem. Future studies will therefore reveal how Trem conditions or primes the chromatin state for recruitment of Mei-P22 to the DSB sites.
Zdroje
1. HassoldTHuntP 2001 To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2 280 291
2. KeeneySGirouxCNKlecknerN 1997 Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375 384
3. BergeratAde MassyBGadelleDVaroutasPCNicolasA 1997 An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386 414 417
4. LakeCMNielsenRJHawleyRS 2011 The Drosophila zinc finger protein Trade Embargo is required for double strand break formation in meiosis. PLoS Genet 7 e1002005 doi:10.1371/journal.pgen.1002005
5. KeeneyS 2007 Spo11 and the formation of DNA double-strand breaks in meiosis. EgelRLankenauDH Berlin Springer 81 123
6. SasanumaHHirotaKFukudaTKakushoNKugouK 2008 Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev 22 398 410
7. WanLNiuHFutcherBZhangCShokatKM 2008 Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev 22 386 397
8. HendersonKAKeeKMalekiSSantiniPAKeeneyS 2006 Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell 125 1321 1332
9. KumarRBourbonHMde MassyB 2010 Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev 24 1266 1280
10. BaudatFBuardJGreyCFledel-AlonAOberC 2010 PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327 836 840
11. MyersSBowdenRTumianABontropREFreemanC 2010 Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327 876 879
12. ParvanovEDPetkovPMPaigenK 2010 Prdm9 controls activation of mammalian recombination hotspots. Science 327 835
13. HayashiKYoshidaKMatsuiY 2005 A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438 374 378
14. ReddyKCVilleneuveAM 2004 C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118 439 452
15. LibbyBJReinholdtLGSchimentiJC 2003 Positional cloning and characterization of Mei1, a vertebrate-specific gene required fornormal meiotic chromosome synapsis in mice. Proc Natl Acad Sci U S A 100 15706 15711
16. MercierRArmstrongSJHorlowCJacksonNPMakaroffCA 2003 The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130 3309 3318
17. PageSLNielsenRJTeeterKLakeCMOngS 2007 A germline clone screen for meiotic mutants in Drosophila melanogaster. Fly (Austin) 1 172 181
18. CarpenterAT 1975 Recombination nodules and synaptonemal complex in recombination-defective females of Drosophila melanogaster. Chromosoma 75 259 292
19. LiuHJangJKKatoNMcKimKS 2002 mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162 245 258
20. MehrotraSMcKimKS 2006 Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet 2 e200 doi:10.1371/journal.pgen.0020200
Štítky
Genetika Reprodukční medicína
Článek A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on DiseaseČlánek The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 2
-
Všechny články tohoto čísla
- Break to Make a Connection
- A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on Disease
- Single-Tissue and Cross-Tissue Heritability of Gene Expression Via Identity-by-Descent in Related or Unrelated Individuals
- Pervasive Adaptive Protein Evolution Apparent in Diversity Patterns around Amino Acid Substitutions in
- The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
- MiRNA Control of Vegetative Phase Change in Trees
- New Functions of Ctf18-RFC in Preserving Genome Stability outside Its Role in Sister Chromatid Cohesion
- Genome-Wide Association Studies of the PR Interval in African Americans
- Mapping of the Disease Locus and Identification of As a Candidate Gene in a Canine Model of Primary Open Angle Glaucoma
- Mapping a New Spontaneous Preterm Birth Susceptibility Gene, , Using Linkage, Haplotype Sharing, and Association Analysis
- A Population Genetic Approach to Mapping Neurological Disorder Genes Using Deep Resequencing
- and Genes Modulate the Switch between Attraction and Repulsion during Behavioral Phase Change in the Migratory Locust
- Targeted Sister Chromatid Cohesion by Sir2
- Correlated Evolution of Nearby Residues in Drosophilid Proteins
- Parallel Evolution of a Type IV Secretion System in Radiating Lineages of the Host-Restricted Bacterial Pathogen
- Lipophorin Receptors Mediate the Uptake of Neutral Lipids in Oocytes and Imaginal Disc Cells by an Endocytosis-Independent Mechanism
- Genome-Wide Association Study of Coronary Heart Disease and Its Risk Factors in 8,090 African Americans: The NHLBI CARe Project
- The Evolution of Host Specialization in the Vertebrate Gut Symbiont
- Genome-Wide Association of Familial Late-Onset Alzheimer's Disease Replicates and and Nominates in Interaction with
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- Association between Common Variation at the Locus and Changes in Body Mass Index from Infancy to Late Childhood: The Complex Nature of Genetic Association through Growth and Development
- AID Induces Double-Strand Breaks at Immunoglobulin Switch Regions and Causing Chromosomal Translocations in Yeast THO Mutants
- A Study of CNVs As Trait-Associated Polymorphisms and As Expression Quantitative Trait Loci
- Whole-Genome Comparison Reveals Novel Genetic Elements That Characterize the Genome of Industrial Strains of
- Prevalence of Epistasis in the Evolution of Influenza A Surface Proteins
- Srf1 Is a Novel Regulator of Phospholipase D Activity and Is Essential to Buffer the Toxic Effects of C16:0 Platelet Activating Factor
- Two Frizzled Planar Cell Polarity Signals in the Wing Are Differentially Organized by the Fat/Dachsous Pathway
- Phosphoinositide Regulation of Integrin Trafficking Required for Muscle Attachment and Maintenance
- Pathogenic VCP/TER94 Alleles Are Dominant Actives and Contribute to Neurodegeneration by Altering Cellular ATP Level in a IBMPFD Model
- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues
- Nucleosomes Containing Methylated DNA Stabilize DNA Methyltransferases 3A/3B and Ensure Faithful Epigenetic Inheritance
- Mutations in Zebrafish Result in Adult-Onset Ocular Pathogenesis That Models Myopia and Other Risk Factors for Glaucoma
- [], the Prion Formed by the Chromatin Remodeling Factor Swi1, Is Highly Sensitive to Alterations in Hsp70 Chaperone System Activity
- Characterization of Transcriptome Remodeling during Cambium Formation Identifies and As Opposing Regulators of Secondary Growth
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
- Epistatic Interaction Maps Relative to Multiple Metabolic Phenotypes
- Quantitative Models of the Mechanisms That Control Genome-Wide Patterns of Transcription Factor Binding during Early Development
- Genome-Wide Transcript Profiling of Endosperm without Paternal Contribution Identifies Parent-of-Origin–Dependent Regulation of
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- MiRNA Control of Vegetative Phase Change in Trees
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



