-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNew Functions of Ctf18-RFC in Preserving Genome Stability outside Its Role in Sister Chromatid Cohesion
Expansion of DNA trinucleotide repeats causes at least 15 hereditary neurological diseases, and these repeats also undergo contraction and fragility. Current models to explain this genetic instability invoke erroneous DNA repair or aberrant replication. Here we show that CAG/CTG tracts are stabilized in Saccharomyces cerevisiae by the alternative clamp loader/unloader Ctf18-Dcc1-Ctf8-RFC complex (Ctf18-RFC). Mutants in Ctf18-RFC increased all three forms of triplet repeat instability—expansions, contractions, and fragility—with effect over a wide range of allele lengths from 20–155 repeats. Ctf18-RFC predominated among the three alternative clamp loaders, with mutants in Elg1-RFC or Rad24-RFC having less effect on trinucleotide repeats. Surprisingly, chl1, scc1-73, or scc2-4 mutants defective in sister chromatid cohesion (SCC) did not increase instability, suggesting that Ctf18-RFC protects triplet repeats independently of SCC. Instead, three results suggest novel roles for Ctf18-RFC in facilitating genomic stability. First, genetic instability in mutants of Ctf18-RFC was exacerbated by simultaneous deletion of the fork stabilizer Mrc1, but suppressed by deletion of the repair protein Rad52. Second, single-cell analysis showed that mutants in Ctf18-RFC had a slowed S phase and a striking G2/M accumulation, often with an abnormal multi-budded morphology. Third, ctf18 cells exhibit increased Rad52 foci in S phase, often persisting into G2, indicative of high levels of DNA damage. The presence of a repeat tract greatly magnified the ctf18 phenotypes. Together these results indicate that Ctf18-RFC has additional important functions in preserving genome stability, besides its role in SCC, which we propose include lesion bypass by replication forks and post-replication repair.
Published in the journal: . PLoS Genet 7(2): e32767. doi:10.1371/journal.pgen.1001298
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001298Summary
Expansion of DNA trinucleotide repeats causes at least 15 hereditary neurological diseases, and these repeats also undergo contraction and fragility. Current models to explain this genetic instability invoke erroneous DNA repair or aberrant replication. Here we show that CAG/CTG tracts are stabilized in Saccharomyces cerevisiae by the alternative clamp loader/unloader Ctf18-Dcc1-Ctf8-RFC complex (Ctf18-RFC). Mutants in Ctf18-RFC increased all three forms of triplet repeat instability—expansions, contractions, and fragility—with effect over a wide range of allele lengths from 20–155 repeats. Ctf18-RFC predominated among the three alternative clamp loaders, with mutants in Elg1-RFC or Rad24-RFC having less effect on trinucleotide repeats. Surprisingly, chl1, scc1-73, or scc2-4 mutants defective in sister chromatid cohesion (SCC) did not increase instability, suggesting that Ctf18-RFC protects triplet repeats independently of SCC. Instead, three results suggest novel roles for Ctf18-RFC in facilitating genomic stability. First, genetic instability in mutants of Ctf18-RFC was exacerbated by simultaneous deletion of the fork stabilizer Mrc1, but suppressed by deletion of the repair protein Rad52. Second, single-cell analysis showed that mutants in Ctf18-RFC had a slowed S phase and a striking G2/M accumulation, often with an abnormal multi-budded morphology. Third, ctf18 cells exhibit increased Rad52 foci in S phase, often persisting into G2, indicative of high levels of DNA damage. The presence of a repeat tract greatly magnified the ctf18 phenotypes. Together these results indicate that Ctf18-RFC has additional important functions in preserving genome stability, besides its role in SCC, which we propose include lesion bypass by replication forks and post-replication repair.
Introduction
DNA trinucleotide repeats are subject to frequent expansions and contractions in families affected by Huntington's disease (HD) and other inherited neurological disorders [1], [2]. Some expanded triplet repeats also cause chromosome fragility, as in fragile X syndrome [1]–[3]. The complexity of triplet repeat instability in humans makes it likely that multiple mechanisms contribute to the problem. Two major sources of instability have been identified [1], [2], [4]. The first is erroneous DNA repair, which can account for instability in both proliferating and non-proliferating cells. Evidence for erroneous repair of triplet repeats includes the finding of fewer expansions of long CAG/CTG alleles in knockout mice deficient in DNA repair factors Msh2, Msh3, Pms2, or Ogg1 (summarized in [4]. Less is known in mammals about a causative role of repair on contractions or fragility, although CAG repeat contractions in a human cell line depend on elements of mismatch and nucleotide-excision repair [5]. The second major source of instability is aberrant DNA replication in proliferating cells. Many DNA replication mutants show altered levels of triplet repeat instability [1], [2], and treatment of human cell lines with DNA replication inhibitors affects expansions, contractions, and fragility [3], [6]. Proliferating cells such as those in the male germ line are prone to expansions, although it is not known whether replication is causative in these cells. For example, expansions are present in pre-meiotic testicular germ cells from HD patients, with additional instability in meiotic and post-meiotic cells [7].
Sister chromatid cohesion is one important facet of DNA metabolism that has not been investigated for an effect on triplet repeat instability. A potential role of SCC in modulating triplet repeats is supported by the interplay of SCC with DNA repair [8], [9] and with replication (summarized in [10]. One protein complex that participates in SCC is the alternative clamp loader/unloader Ctf18-Dcc1-Ctf8-RFC (Ctf18-RFC). In the absence of the Ctf18-RFC, SCC is compromised [11], [12]. Biochemically, Ctf18-RFC can load and unload PCNA onto DNA [13]–[15]. It has been proposed that the PCNA unloading activity of Ctf18-RFC may be important to facilitate passage of the replication fork through the cohesin ring [15], [16]. Ctf18-RFC has also been proposed to play a more general role in fork stabilization [17]. The ability of Ctf18-RFC to recruit PCNA to hydroxyurea-stalled replication forks [16] and to act together with the checkpoint mediator protein Mrc1 in the DNA replication checkpoint [18] is consistent with a more general role at stalled forks.
This paper describes the discovery, through blind mutant screens, of yeast Ctf18-RFC mutants that destabilize triplet repeats. Genetic analysis indicates Ctf18-RFC likely acts through replication fork stabilization and/or post-replication repair (PRR), not SCC, to prevent triplet repeat instability, chromosome fragility and cell cycle delays in S and G2/M phases. Our data also support a general role for the Ctf18-RFC complex in preventing DNA damage, a role which becomes more crucial in the presence of an at-risk sequence such as an expanded trinucleotide repeat tract.
Results
Triplet repeat instability is increased in Ctf18-RFC but not SCC mutants
Two independent genetic screens identified mutants in Ctf18-RFC as defective in stabilization of trinucleotide repeats (Text S1). In screen one, the yeast strain contained a (CAG)20-URA3 reporter to monitor contractions (Figure S1). This strain was transformed with a gene disruption library, and transformants were screened for increased rate of contractions [19]. Following three rounds of testing with increasing stringency, vectorette PCR was used to identify a ctf18::LEU2 allele. The contraction phenotype was confirmed in a commercially obtained ctf18 strain. In screen two, mutants were sought that increased the rate of fragility for a (CAG)85 tract on a yeast artificial chromosome (YAC; Figure S2) [20]. Transfer of the YAC to the commercial haploid deletion strain set was followed by assays for increased fragility. Several rounds of screening showed that the dcc1 mutant reproducibly displayed the fragility phenotype. Thus, two different genetic screens for defects in regulating triplet repeat instability converged on the Ctf18 and Dcc1 components of the Ctf18-Dcc1-Ctf8-RFC complex.
Subsequent analysis proved that ctf18 and dcc1 mutants exhibited increased levels of trinucleotide repeat contractions, expansions, and fragility. Contractions in these mutants were increased in every case, by up to 8-fold, for a wide range of repeat lengths (short (CAG)20, medium (CAG)70, and long (CAG)155 tracts; Figure 1A and 1B; Table S1). In fact, the contraction phenotype of dcc1 and ctf18 mutants for long tracts was so pronounced (Figure 1B) that the fraction of unaffected cells, only 20–30%, was too low for meaningful analysis of expansions in these strains. Expansions of medium length tracts were increased 10 - to 15-fold in ctf18 and dcc1 mutants (Figure 1C); however, these mutants did not increase expansion rates for very short (CAG)13 repeats. Fragility was increased 2 - to 3 fold for dcc1 and ctf18 mutants even in chromosomes without a repeat tract, but was further increased 3 - to 5 fold in the presence of an expanded repeat in a length-dependent manner (Figure 1D). In summary, inactivation of Ctf18-RFC substantially increased all three types of instability for CAG runs of 20–155 repeats and also increased general chromosome fragility.
Fig. 1. Contraction, expansion, and fragility phenotypes of SCC mutants. 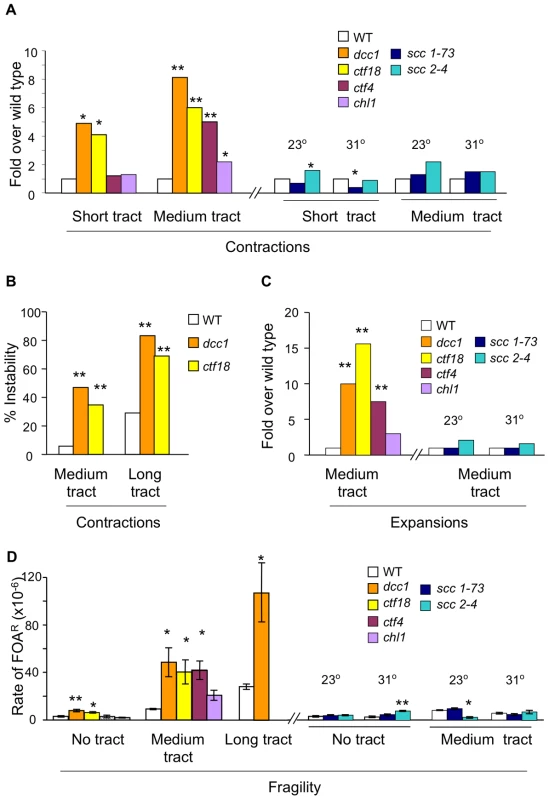
For all panels, * denotes p<0.05 and ** designates p<0.01 compared to the respective wild type strain. All assays are described in Table S1. (A) Contraction rates of (CAG)20 and contraction frequencies of (CAG)70, normalized to wild type. (B) Percentage of colonies showing contractions of medium (CAG)70 or long (CAG)155 tracts. (C) Expansions of (CAG)70 tracts, normalized to wild type. (D) Fragility rates with no repeat tract, or with medium (CAG)70 or long (CAG)155 tracts. Error bars denote ± one standard error of the mean (SEM). If these triplet repeat phenotypes are due to the sister chromatid cohesion (SCC) activity of Ctf18-RFC, then mutants in other SCC genes should show similar results. CTF4 and CHL1 were examined first, as these genes gave the closest match to Ctf18-RFC in a genetic interaction map of protein complexes involved in chromosome biology [21]. Mutants in CTF4 and CHL1 both show SCC defects [22]–[24]. Biochemically, Ctf4 couples polymerase a to Mcm and the replisome progression complex [25], [26], while Chl1 is a putative DNA helicase that associates with cohesion establishment factor Eco1 [24]. Inactivation of CTF4 did not affect short contractions, but increases were observed in contractions, expansions and fragility of the medium tract, with magnitudes similar to dcc1 and ctf18 mutants (Figure 1A, 1C, 1D). In contrast, the chl1 mutant gave virtually no phenotype in triplet repeat assays (Figure 1A, 1C, 1D). Since the ctf4 phenotype could be due to uncoupling of DNA polymerase α from the replication fork [25], [26] rather than SCC, the ctf4 and chl1 results suggested the possibility of an SCC-independent phenotype of mutants in Ctf18-RFC and Ctf4. Accordingly, assays were performed with scc1-73 and scc2-4 temperature-sensitive mutants defective in cohesion maintenance and establishment, respectively. At both permissive (23°) and at semi-permissive (31°) temperatures, scc1-73 and scc2-4 strains were indistinguishable from wild type in nearly every triplet repeat assay (Figure 1A, 1C, 1D). The lack of a triplet repeat instability phenotype in scc1-73, scc2-4, and chl1 strains is in contrast to the clear SCC defect seen in these mutants [22], [27]. While SCC cannot be rigorously excluded due to the essential nature of SCC1 and SCC2, the most likely explanation for the lack of phenotypes in chl1, scc1-73, and scc2-4 mutants is that Ctf18-RFC mitigates triplet repeat instability in an SCC-independent manner.
Other alternative RFC complexes do not stabilize CAG repeats to the same extent as Ctf18-RFC
In addition to the canonical clamp loader composed of Rfc1 and the core of Rfc2-5, there are three alternative clamp loaders; Ctf18-RFC, Elg1-RFC, and Rad24-RFC. The four clamp loaders have distinct biochemical properties, with Ctf18-RFC uniquely exhibiting efficient PCNA unloading [15]. Is Ctf18-RFC also distinct in regards to triplet repeat stabilization? The instability profiles of dcc1, elg1, and rad24 mutants show distinct patterns (Figure 2). Deficiency in DCC1 increased six of seven types of instability: all forms of contraction, expansion (save for short tracts), and fragility. In contrast, the elg1 and rad24 mutants showed elevated instability in only three or two assays, respectively (Figure 2). Aside from short tract expansions, the magnitude of elg1 or rad24 phenotypes was always weaker than for dcc1. Short tract expansions were previously shown [19] to be especially sensitive to defects in the DNA damage response, including rad24, consistent with the specificity for Rad24 seen in Figure 2. We conclude that while all three alternative RFC complexes help stabilize CAG/CTG repeats, Ctf18-RFC has the most potent and wide-ranging impact in our assays. Crabbe et al came to a similar conclusion regarding the predominance of Ctf18-RFC in the DNA replication checkpoint [18]. Therefore it remained the focus of this study.
Fig. 2. Triplet repeat phenotypes for mutants in three different alternative RFC complexes. 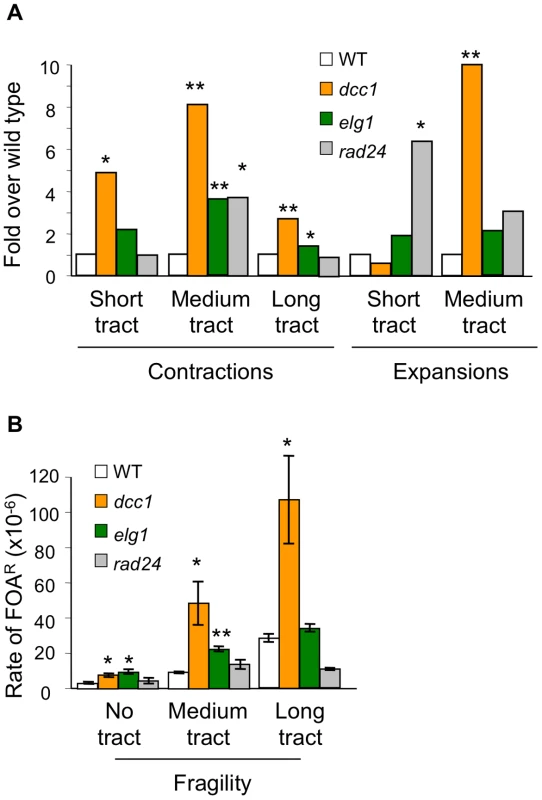
Assays, display, and symbols are as in legend to Figure 1. (A) Contractions and expansions normalized to wild type. (B) Rates of fragility for strains with no repeat tract, medium (CAG)70, or long (CAG)155 tracts. Error bars, ±1 SEM. Repeat instability in the absence of Ctf18-RFC is Rad52-dependent
Since SCC defects did not account for instability of triplet repeats, we tested the idea that strains deficient for Ctf18-RFC suffer enhanced DNA damage at the trinucleotide repeat, as suggested by the increased repeat fragility in mutants of the complex (Figure 1). If so, this damage might be susceptible to RAD52-dependent recombinational repair and therefore a rad52 background should alter the mutational spectrum in the absence of Ctf18-RFC. The results show that mutation of RAD52 suppressed, partially or completely, every dcc1 mutability phenotype–contractions of both short and medium CAG/CTG tracts and expansion of medium tracts (Figure 3A). We conclude that Rad52-dependent repair in the absence of Ctf18-RFC does not proceed with fidelity in the context of a CAG repeat, since it results in expansions and contractions. A similar result was also observed in srs2 and mre11 mutants, where increased levels of medium - and long-tract repeat expansions and contractions were dependent on Rad52 [28], [29]. Some medium tract contractions were Rad52-independent (Figure 3A); previous data indicated that an additional source of contractions could be processing of DSBs within the repeat tract followed by microhomology-mediated end joining [29]. Fragility was not suppressed or significantly increased in a dcc1 rad52 double mutant compared to the dcc1 single mutant (Figure 3B), indicating that Rad52 does not contribute to fragility resulting from DNA damage in Ctf18-RFC deficient cells. In summary, in the absence of Ctf18-RFC, a Rad52-dependent pathway is operative that is responsible for the majority of the observed contractions and expansions.
Fig. 3. Analysis of rad52 effects on triplet repeat instability phenotype of dcc1. 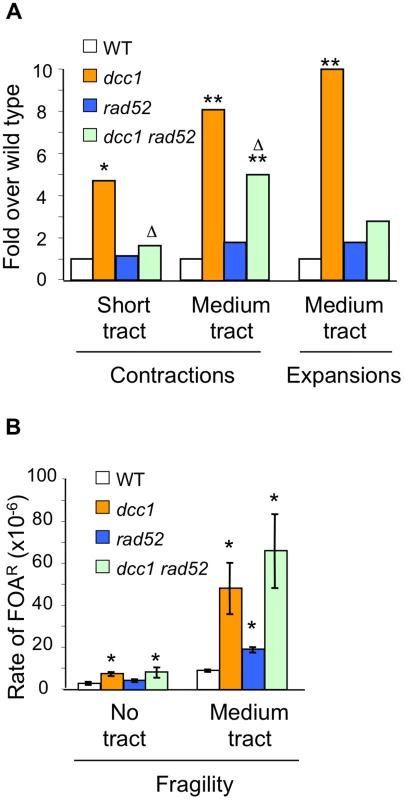
Contractions, expansions, and fragility were measured as described in Table S1. (A) Contraction and expansion phenotypes normalized to wild type. *, p<0.05, **, p<0.01 compared to wild type; Δ, p<0.05 compared to dcc1. (B) Fragility measurements as in Figure 1; the dcc1 rad52 mutant had a (CAG)65 repeat. Error bars, ±1 SEM. Ctf18-RFC and Mrc1 function in separate pathways
We considered the possibility that stabilization of replication forks by Ctf18-RFC explains its effects on triplet repeat mutations and fragility. This model is supported by studies showing Ctf18 localization to hydroxyurea-stalled forks in S. cerevisiae [16], its association with replication origins in unperturbed S. pombe cells [17], and its physical association with DNA polymerase ε [30]. If true, the fork stabilization model predicts that uncoupling DNA pol ε from the replicative helicase with an mrc1 mutation [31] should exacerbate the triplet repeat phenotype of Ctf18-RFC mutants. Mrc1 is important both for coupling the helicase and polymerase functions at the replication fork, and in signalling during the replication checkpoint and the DNA damage response [31]–[33]. We showed that triplet repeat expansions, contractions, and fragility are elevated in mrc1 mutants [19], [34], [35]. Also, mrc1 interacts genetically with ctf18, ctf8, and dcc1 [36].
To test mrc1 effects on triplet repeat instability in the absence of Ctf18-RFC, contraction rates were compared for short CAG tracts in single and double mutants of mrc1, dcc1, and ctf18. The results in Table 1 show 4.1 - to 7.1-fold increased contraction rates for single mutants of mrc1, dcc1, or ctf18. The two double mutants gave effects that were significantly greater than additive: 20-fold for mrc1 dcc1 (p = 0.05) and 16-fold for mrc1 ctf18 (p = 0.04). This result suggests that Ctf18-RFC and Mrc1 work in parallel to stabilize short repeats. Forward mutation rates at CAN1 were also significantly greater than additive in the mrc1 ctf18 double mutants: 23.4-fold over wt compared to 6.5-fold and 2.6-fold for the respective single mutants (Table 1; p = 0.002), indicating that these proteins act in different pathways in the context of a non-trinucleotide repeat sequence as well. In contrast, Mrc1, Ctf18, Ctf8, and Dcc1 have been proposed to function in the same SCC pathway [22]. This difference is consistent with our earlier conclusion that Ctf18-RFC stabilizes triplet repeats independently of SCC. The mrc1 dcc1 and mrc1 ctf18 double mutants could not be assayed with medium and long tracts due to cell lethality.
Tab. 1. Double mutant analysis of mrc1 with dcc1 and ctf18. 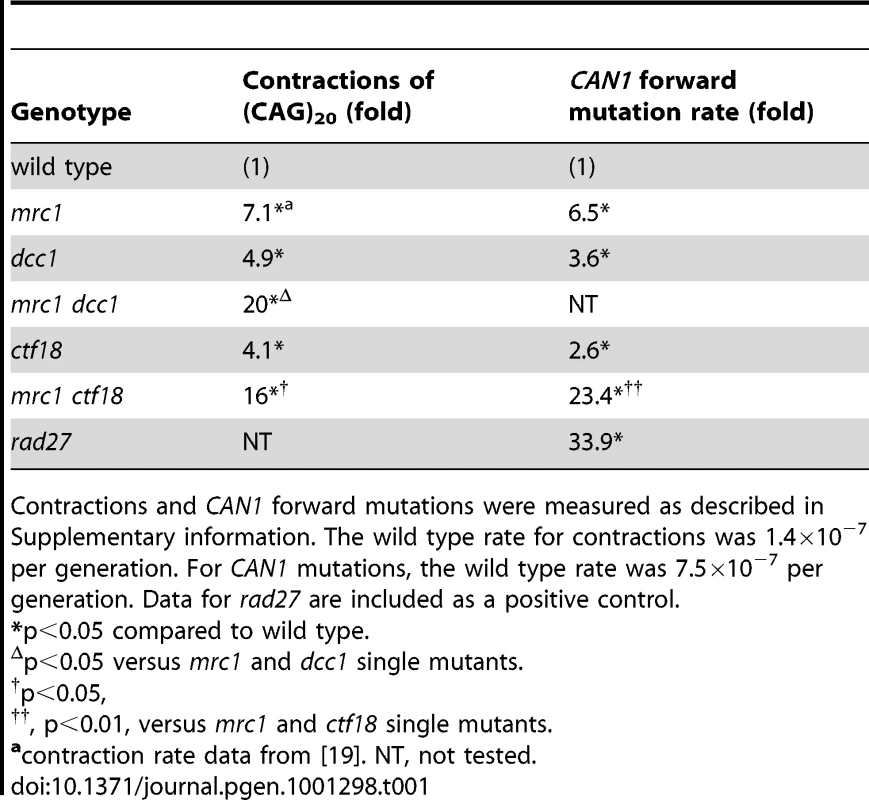
Contractions and CAN1 forward mutations were measured as described in Supplementary information. The wild type rate for contractions was 1.4×10−7 per generation. For CAN1 mutations, the wild type rate was 7.5×10−7 per generation. Data for rad27 are included as a positive control. Ctf18-RFC–deficient cells exhibit S phase delays and G2/M arrests with a multi-budded morphology, phenotypes exacerbated by the presence of a repeat tract
The results above suggested that Ctf18-RFC helps cope with triplet repeat-associated damage and in stabilizing replication forks, so we tested directly whether the Ctf18-RFC complex has a role in progression through the cell cycle. Cells from a log phase liquid culture were plated on solid media, and microscopic analysis was used to monitor the proportion of cells in each phase of the cell cycle: unbudded (G1), small budded (bud size one-third or less the size of the mother cell (S), and large budded (G2/M). The results are quantified in Figure 4A, and representative micrographs are shown in Figure 4B. In wild-type cells with no CAG/CTG tract, there was a distribution of 30% unbudded, 12% small budded, and 55% large budded (Figure 4A). The presence of a CAG/CTG tract changed this distribution in two ways. First, there were more small budded (S phase) cells, consistent with replication stress. Second, a new category of cells was observed that were either swollen with large buds or contained multiple buds (Figure 4B), a phenotype that is indicative of unresolved damage in G2/M [37]. The proportion of the multi-budded/swollen cells rose with increasing repeat tract length to as much as 20% of the wild type population (Figure 4A). In general the swelling was modest and most multi-budded clusters contained only one extra bud in wild-type cells (Figure 4B). In dcc1 and ctf18 cells, even without a repeat, multi-budded/swollen cells comprised 18–30% of the population, a level significantly greater than wild-type cells with no tract (Figure 4A). This indicates that the absence of the Ctf18-RFC complex leads to some level of repeat-independent damage that causes accumulation of cells in G2/M. Even more strikingly, the combination of an expanded repeat plus the lack of a functional Ctf18-RFC led to an increase in the multi-budded category to 42–54% of cells (Figure 4B). In addition, the morphological defects in dcc1 and ctf18 mutants with a repeat tract often showed a more severe phenotype, with extreme swelling and many connected buds (Figure 4B). Staining of nuclei revealed that some of the cells within multibudded clusters, often the more swollen ones, had fragmented or missing DNA (example in Figure 6A). We conclude that Ctf18-RFC has an important function in helping resolve repeat-independent DNA damage, and that damage is persisting into the G2 or M phase. Since this phenotype is enhanced by an expanded triplet repeat, and since the expanded repeat causes replication stress, we also infer that Ctf18-RFC helps cope with repeat-induced replication stress during S phase.
Fig. 4. Cell cycle distribution and morphological abnormalities of dcc1 and ctf18 mutants. 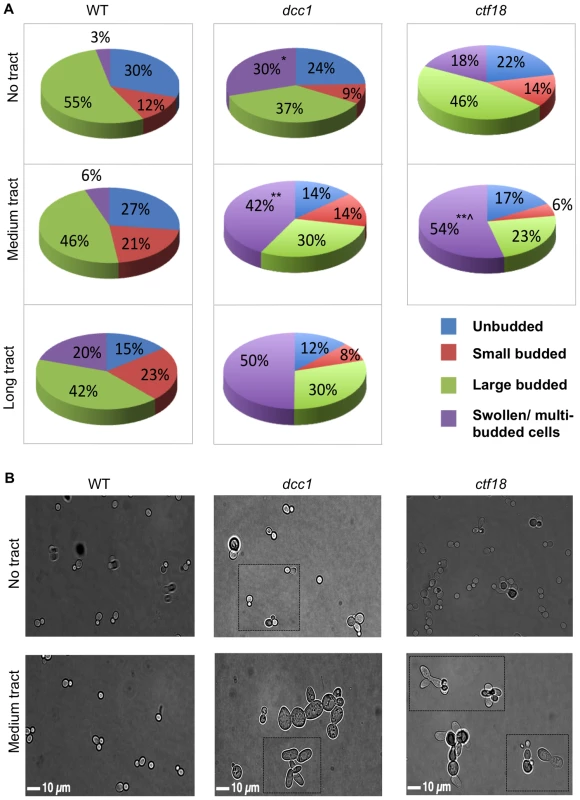
(A) Quantification of cell morphology in log phase cultures. Several hundred cells (range 227–723) were scored for each genotype. Note that the dcc1Δ long tract was a mixture of cells with 155 repeats and contracted tracts by the end of the experiment; we were not able to complete a ctf18 long tract experiment without substantial contractions. Differences in the percentage of multi-budded cells were analyzed by a pooled variance t test using the Systat program; *, p<0.05, **, p<0.01 compared to wild type of the same tract length; ∧, p<0.05 compared to the no tract control of the same strain (e.g. p = 0.054 for dcc1-70 compared to dcc1 no tract, and p = 0.013 for ctf18-70 compared to ctf18 no tract). (B) Microscopic images of cells; all images are at the same scale, dcc1 and ctf18 mutants are characterized by an increase in cell size and the formation of protruded and multiple buds. Dotted lines indicate an overlay of another image to provide additional examples of cells of that genotype. To measure cell cycle dynamics with more precision, we isolated unbudded G1 cells by micromanipulation and followed their progression through 2–3 cell cycles by microscopy. This single-cell approach measures the time spent in each phase of the cell cycle, and therefore it allows assignment of the cell cycle stage in which defects can first be detected. A schematic example of the approach and some representative data are shown in Figure 5A. The majority of wild type cells with no repeat spent ∼30 min in S phase, with a slight shift to longer S phases when a medium-length (CAG)70 repeat was present (Figure 5B). Cells containing a (CAG)70 tract and lacking DCC1 or CTF18 exhibited several cell cycle phenotypes. First, they divided much more slowly. Average division time was 5.8 h for dcc1 (range 2.5–8.5 h) and 3.5 h for ctf18 (range 2.0–6.0 h), compared to 2.0 h for wild type (CAG)70 strain. The presence of the repeats enhanced the delay as the dcc1 and ctf18 mutants with no repeat averaged 2.5 h and 2.0 h per division, respectively. Second, some ctf18 and dcc1 cells stayed small budded 1–2 h, consistent with an S-phase delay, a phenotype that was exacerbated by the presence of the repeat (Figure 5B). In contrast, all wild type cells completed S phase in 1 h or less, regardless of whether the repeat tract was present. Thus, single-cell analysis provides additional evidence for a role of Ctf18-RFC during S phase, as its absence leads to an extended S phase in some cells.
Fig. 5. Single cell analysis of cell cycle dynamics. 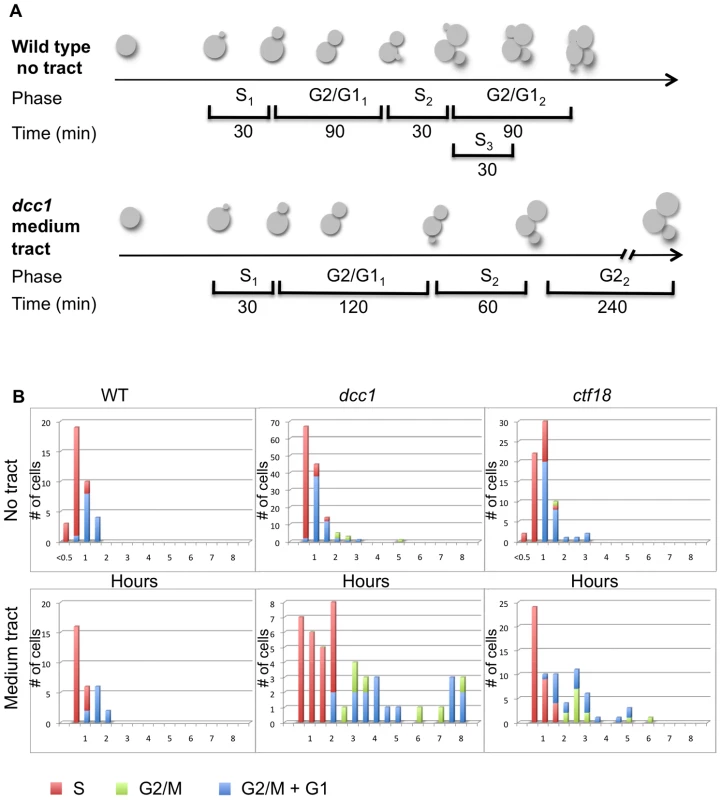
Aliquots from mid-logarithmic phase liquid cultures were plated onto solid media. Single unbudded cells were isolated by micromanipulation, and their progression was monitored by microscopy every 30 min for 6.0–8.5 h (1–4 cell divisions). (A) Examples of how cells were followed and scored. (B) Time spent in each phase of the cell cycle, as scored by budding index (see Materials and Methods). Red bars, S phase; green bars, G2 phase; blue bars, G2+G1 phases. Effects in G2/M were also evident from the single-cell analysis (Figure 5B). All wild type cells had a G2/M phase of 2 h or less, regardless of the presence of the trinucleotide repeat. In contrast, some dcc1 and ctf18 cells were detected with G2/M phases of 2 hours or more, even when no repeat was present. The length of the dcc1 and ctf18 strain G2/M phase was greatly increased in the presence of the (CAG)70 repeat, with some cells remaining in G2/M up to 6–8 h (after which time yeast cells are able to adapt to DNA damage and continue through M even without repair [38], [39]). This single-cell analysis also proved that arrested G2/M cells gave rise to the multi-budded cells described earlier. Finally, when the fate of the colony growth beyond 8 h was monitored, we observed that a majority of the dcc1 and ctf18 cells containing the (CAG)70 repeat tract only accomplished a few additional cell divisions and did not form colonies visible by eye. This observation indicates that we likely underestimated the fragility and instability phenotypes obtained for (CAG)70 repeats, and presumably for (CAG)155 repeats.
Elevated levels of DNA damage are generated during S phase and persist into G2/M phase in ctf18 cells
The cell cycle delays, increased fragility, and Rad52-dependent instability all suggested that DNA damage may be occurring at CAG repeats in the absence of Ctf18-RFC. To directly test for damage, we measured the proportion of wild type and ctf18 cells with a Rad52 focus in the presence or absence of CAG repeats (Figure 6). Rad52 focus formation occurs at DSBs or at broken replication forks, but not at forks stalled by HU [40]. In the absence of any repeat, only 1.1% of wild type cells had a Rad52-YFP focus (Figure 6A and 6B). The presence of the repeats significantly increased the frequency of Rad52 foci to 3.3% and 3.8% for (CAG)70 and (CAG)155, respectively (Figure 6B). In ctf18 cells without a repeat tract, the incidence of Rad52 foci was elevated to 8% (Figure 6B), indicating that significant levels of DNA damage are occurring in this background, consistent with the increased fragility observed above. In addition, there was a further increase in cells with foci in ctf18 cells with an expanded repeat, to 17% for both (CAG)70 and (CAG)155 (Figure 6B). Considering that the repeat is in single copy, these data suggest a significant level of damage occurring at the repeat when the Ctf18-RFC complex is not functional.
Fig. 6. Cell cycle dependency of Rad52 focus formation in ctf18 cells. 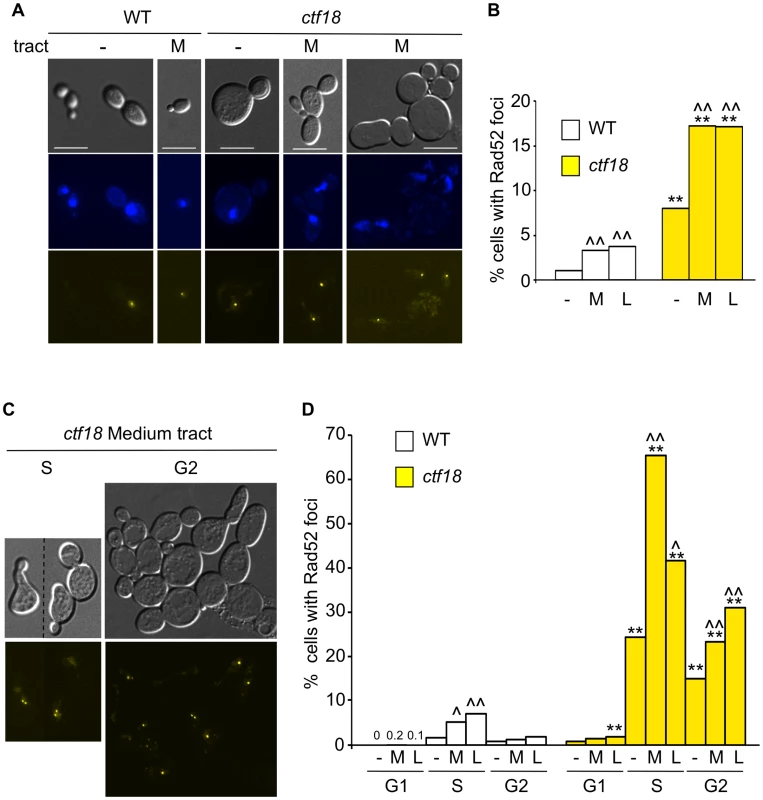
(A) Cultures of wild type and ctf18 cells were grown to mid-log phase before mounting on a microscope slide. The panel shows differential interference contrast (DIC), DAPI-stained DNA, and Rad52-yellow fluorescent protein (Rad52-YFP) images of selected cells among WT and ctf18 cells with no tract (−) or with a medium (CAG)70 tract (M). Scale bar is 10 µm. (B) Quantification of Rad52 foci formation in WT or ctf18 cells with no tract (−), medium (CAG)70 tract (M) or long (CAG)155 tract (L). *, p<0.05, **, p<0.01 compared to wild type of same tract length; ∧, p<0.05, ∧∧, p<0.01 compared to no tract of the same strain. (C) Cell cycle distribution of Rad52 foci. The occurrence of Rad52 foci in G1, S or G2 cells was determined after incubation with α-factor, 40 min after release from G1 or nocodazole, respectively (see Methods for details). Representative examples of ctf18 cells with medium (CAG)70 tract in S or G2 are shown as DIC images (top) or Rad52-YFP foci (bottom). (D) Quantification of Rad52 foci in G1, S, or G2 cell cycle stage. Labels and statistical analysis are as in (B). Percentages obtained for WT cells treated with α-factor are indicated. See Table S2 for complete set of data. To determine when in the cell cycle the damage occurs, we visualized foci in cells at different stages (Figure 6C and 6D). In wild type cells without a repeat, foci levels were very low, less than 2%, at all stages (Figure 6D). Interestingly, wild type cells containing an expanded CAG repeat had a detectable increase in Rad52 foci in S phase, 40 min after release from α-factor (Figure 6D). This timing coincides with replication through the repeat (assessed by 2D gel electrophoresis; R. Anand and C. Freudenreich, data not shown) indicating that Rad52-dependent events at the repeat may be replication-associated. The lower level of foci in cells arrested by nocodazole (Figure 6D) suggests that the DNA damage induced in S is usually repaired by G2/M in wild type cells.
In ctf18 cells, the percentage of Rad52 foci was very low in G1 cells arrested by α-factor, suggesting that the complex does not have a genome protective function in G1 (Figure 6D). In contrast, the proportion of cells with a Rad52 focus rose to 24% in S phase, and this number was dramatically increased in the presence of a repeat, to 65% for (CAG)70 or 42% for (CAG)155 (which was a mixture of 155 and contracted tracts). These data show that Ctf18-RFC has an important S phase role. We also observed ctf18 S phase cells with more than one Rad52 focus (Figure 6C). Contrary to the wild type situation, Rad52 foci frequently persisted into G2/M. 15–30% of the ctf18 cells still showed Rad52 foci when arrested in G2/early M by nocodazole (Figure 6D). DAPI staining revealed that these foci were detectable in both G2 cells with nuclei at the bud neck as well as in cells entering M phase. Thus the repeat-induced damage sometimes persisted into M phase.
Altogether, cell cycle analysis (Figure 4, Figure 5, Figure 6) indicates that damage at the repeat tract likely initiates in S phase, and that presence of Ctf18-RFC is important for completing S phase without delay and without accumulation of DNA damage. Judging by the accumulation of cells and presence of Rad52 foci in G2 and even into M phase, a significant portion of damage persists beyond S phase in Ctf18-RFC deficient cells.
Discussion
This work provides evidence for new functions of Ctf18-RFC in preserving genomic integrity, outside its role in sister chromatid cohesion (SCC). These discoveries stemmed from the application of sensitive and specific genetic assays that revealed Ctf18-RFC's role in protecting a broad range of CAG/CTG repeat lengths from expansion, contraction, and fragility (Figure 1). Ctf18-RFC mutant phenotypes at triplet repeats were distinct from those shown by mutants in other SCC factors, such as chl1, scc1-73, scc2-4, and mrc1 (Figure 1, Table 1). Ctf18-RFC also was more important at trinucleotide repeats compared to the alternative RFC complexes Elg1-RFC or Rad24-RFC (Figure 2). A novel role for Ctf18-RFC in replication fork bypass of lesions that arise from triplet repeats was suggested by analysis of double mutants between Ctf18-RFC and either Mrc1 (Table 1) or Rad52 (Figure 3). In agreement with the idea of a role at the replication fork, cells defective in Ctf18-RFC show an extended S phase and increased S-phase levels of Rad52 foci even in the absence of a triplet repeat (Figure 5, Figure 6). These mutants also accumulate in G2/M, often with altered morphology and persistence of Rad52 foci (Figure 4, Figure 5, Figure 6), a phenotype consistent with unresolved DNA damage in G2 [41]. These altered cell cycle phenotypes occur even in the absence of a triplet repeat, but the presence of an expanded repeat tract severely exacerbated the mutant defects. Taken together, the results of this study suggest that Ctf18-RFC helps avoid DNA damage arising during replication, that Ctf18-RFC may also be important in coping with damage when it persists into G2, and that triplet repeats make budding yeast especially dependent on Ctf18-RFC.
Our observations are consistent with action of Ctf18-RFC in S phase, during or soon after passage of the replication fork. Previous work localized Ctf18 at or near hydroxyurea-stalled forks by Chromatin IP in S. cerevisiae [16], and to replication origins in unperturbed S. pombe cells [17]. It was recently shown by DNA combing that fork speed is slowed 3-fold in ctf18 mutant cells [18]. Ctf18-RFC shows a physical association with DNA polymerase ε [30], [42], suggesting that it could act directly at the fork in a fork stabilization role. Although our results do not support a role for Ctf18-RFC in cohesion establishment, they are compatible with a role in facilitating replication through the cohesion ring, as proposed in [15], [16]. One way to explain the repeat-specific effects we see is if Ctf18-RFC promotes dis-assembling or re-assembling the replisome to facilitate bypass through replication barriers, such as a cohesin ring or a hairpin structure. Another possibility is Ctf18-RFC could have a more general fork stabilizing function to prevent formation of hairpins or other aberrant secondary structures associated with trinucleotide repeats. In either case, fork integrity would be affected in ctf18, dcc1, or ctf8 mutants, leading to fragility and increased recombination, and ultimately to triplet repeat mutations. This model would also explain the ctf4 phenotype on instability (Figure 1), based on the role of Ctf4 in coupling DNA polymerase α to the replication fork [25], [26]. Alternatively, the Ctf18-RFC S-phase role could be in a repair process that occurs behind the fork, but still in S phase, as described below.
The striking G2/M accumulation phenotype and persistence of Rad52 foci was unexpected in mutants of Ctf18-RFC, in part because this phenotype suggests the presence of unresolved DNA damage that persists beyond S phase. Thus, either the damage incurred during S phase in ctf18 cells is often not easily repaired, or Ctf18-RFC also plays a role in helping resolve or repair damage during G2. Recently, it was shown that post-replication gap repair can operate effectively when limited to the G2 phase, and the authors proposed that PRR occurs primarily on gaps left behind replication forks that have re-primed and continued [43], [44]. Interestingly, their data suggest that error-free PRR, which is dependent on Rad5-catalyzed polyubiquitylation of PCNA, usually commences in S-phase and continues into G2/M. Thus with regard to timing, our data would be consistent with a role for Ctf18-RFC in error-free PRR. The dependence on HR for instability is also consistent with a role in error-free PRR, since strand invasion is needed for the template-switching step. In contrast, it is unlikely that our results with Ctf-18 RFC relate to error-prone PRR, since HR does not occur during translesion synthesis. Notably, the triplet repeat defects associated with absence of Ctf18-RFC are different than results when the PRR pathway is abolished by deletion of Rad5 or abolishing PCNA modification, as those mutants specifically increased expansions, but not contractions, of short repeats [45]. Thus, our results would be most consistent with aberrant PRR, rather than ablation of the pathway.
How might Ctf18-RFC function biochemically to protect triplet repeats? Ctf18-RFC was shown to load and unload PCNA in vitro in a manner that is more efficient on single stranded DNA and inhibited by RPA [15]. Significant unloading was not seen for the other complexes, RFC, Rad24-RFC, and Elg1-RFC. This unique biochemical activity of Ctf18-RFC mirrors the distinct pattern in our genetic observations (Figure 2). Possibly the loading/unloading function is especially important at triplet repeats either to bypass a previously formed hairpin structure or to minimize exposed single strands during replication and thereby reduce secondary structure formation and instability. A second possibility is that a defect in PCNA unloading during a gap repair event could lead to a persistent HR structure that would be prone to breakage or cleavage. In this model, contractions could occur during DSB repair end processing as proposed in [29]. In contrast, timely unloading of PCNA could facilitate proper resolution of a recombination intermediate without breakage, for example by Sgs1 dissolution. Sgs1 has a role in resolution of X-shaped SCJs that form upon replication of damaged templates [46], and a recent study links Sgs1 to resolution of an intermediate that occurs during ubiquitylated PCNA-dependent gap repair [43]. Intriguingly, deletion of SGS1 also led to increased contractions and fragility, similar to (though not as dramatic as) ctf18 or dcc1 mutants, and in some contexts, the contractions were also Rad52-dependent [28]. Thus, for example, a defect in Ctf18-dependent unloading of ubiquitylated PCNA could lead to toxic recombination products, similar to a defect in Sgs1 activity. It will be interesting to learn how Ctf18-RFC functions on ubiquitylated PCNA, as we showed previously that expansion rates are elevated when PCNA ubiquitylation is blocked [45].
Does repeat stabilization by Ctf18-RFC extend to human cells? Human Ctf18-RFC was shown to control the velocity, spacing, and restart activity of replication forks via acetylation of the cohesin ring [47]. Also, human Ctf18-RFC has been recently shown to be necessary for accumulation of polymerase ε during repair of UV lesions induced outside of S phase [48]. A key future study is to see whether, and how, human Ctf18-RFC protects triplet repeats.
Methods
Most strains were derived from BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) or BY4705 (MATα his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0), isogenic derivatives of Saccharomyces cerevisiae strain S288C (Open Biosystems; [49]. Isogenic derivatives were obtained commercially (Open Biosystems) or were created by targeted deletion of BY4741 or BY4705. The scc1-73, scc2-4 and wild type parent strains (MATa ade2-1 can1-100 leu2-3,112 his3-11,15 ura3-1 trp1-1, except scc1-73 strain that was TRP1) were provided by Philippe Pasero, CNRS, Montpellier, France. Strains used for foci experiments were derived from W303 (MATa ADE2 his3-11,15 leu2-3,112 bar1::LEU2 trp1-1 RAD52-YFP), obtained from R. Rothstein, Columbia University, NYC, NY. The triplet repeat sequences reported here all have the CAG repeat on the lagging strand template, and CTG repeats on the Okazaki fragment. This CAG nomenclature is used throughout.
Contraction and expansion rates for short CAG tracts were measured by fluctuation analysis, and authenticated by PCR, as previously described [19]. Statistical analyses were performed using the Wilcoxon Mann Whitney test. P values of less than 0.05 were considered statistically significant. Forward mutation rates for the CAN1 gene were determined by fluctuation analysis using canavanine at 60 µg/ml, and statistical analysis was performed using Student's t-test for comparison with wt and a two-way ANOVA with interaction tests [50] for comparison between single and double mutants.
Expansions, contractions, and fragility of medium and long CAG tracts were measured using a YAC system, as described previously [20]. Diagrams of these assays are shown in Figure S2. Contraction and expansion frequencies for medium (CAG)70 and long (CAG)155 tracts were determined as previously described previously [29]. For each strain, approximately 150 colonies were analyzed for CAG repeat length by colony PCR in at least three separate experiments, using primers flanking the CAG repeat (P1 and P2 in Figure S2). PCR products were separated on a 2% Metaphor gel (Cambrex Bio Science Rockland, Inc.) and sized. The frequency of repeat expansions and contractions in each strain background was calculated and statistical significance determined by the Fisher's exact test. Repeat lengths from 0 to ∼200 CAG repeats with an accuracy of +/−3 repeats can be obtained by this method.
Fragility assays were performed as in [29]. Mutation rate was determined using the method of maximum likelihood [51] and data presented are an average of 3–5 experiments. Error bars indicate the standard error of the mean. Significance compared to the wild-type value for the same tract length was determined using a pooled variance t-test. Growth temperature was 30°C unless otherwise indicated. A summary of the whole instability and fragility data is represented in the Table S1.
Cell cycle distribution and cell morphologies (Figure 4) were obtained as follows: YC-Leu-Ura liquid cultures were grown to mid-logarithmic phase and analyzed microscopically for the presence of unbudded cells, small-budded cells (bud smaller than one-third of the mother cell), large-budded cells (bud equal to or larger than one-third of the mother cell), and other cells (malformed cells with protruded or multiple buds). The assays were repeated at least 3 times for each strain. Pictures of the cells were taken using a Zeiss AX10 microscope, under 63× magnification. Single cell assays (Figure 5) were initiated by plating mid-logarithmic phase liquid cultures onto solid YC-Leu-Ura media. Single unbudded cells were isolated by micromanipulation, and their progression was monitored by microscopy every 30 min for 6.0–8.5 h. (1–4 cell divisions). The numbers of single cell lineages monitored were: wild type, no repeat (12); wild type, medium tract (12); ctf18Δ, no repeat (24); ctf18Δ, medium tract (24); dcc1Δ, no repeat (53); and dcc1Δ, medium tract (33). Because we did not micromanipulate daughter cells away from each other, and because many of them in fact could not be separated (indicating incomplete cell division and leading to a multi-budded cell), it was not always possible to distinguish the end of G2/M and beginning of G1. In these cases, we grouped the two cell cycle phases (G2+G1). At the end of the experiment, cells still in G2 phase for a time less than the wild-type G2/M average time (1 h for no tract and 1.5 h for medium tract, respectively) were not considered to be informative for their G2 phase, and were not counted.
Cell imaging and fluorescent microscopy: examination of Rad52-YFP focus levels by microscopy was performed as previously described [52]. Briefly, cells were grown overnight in SC-Leu-Ura media at 23°C and exponentially growing cultures were prepared for microscopy. To visualize nuclear DNA by DAPI staining (50 ng/ml), cells were fixed in ethanol before mounting on the slide. Cell images were captured using a Zeiss AX10 microscope (Carl Zeiss, Thornwood, NY) equipped with a Retiga EXi camera (Qimaging), and acquired using SlideBook software (Intelligent Imaging Innovations, Denver, CO). All images were taken at 63-fold magnification. A single DIC image and 17 YFP images obtained at 0.3-µm intervals along the z-axis were captured for each frame, and Rad52-YFP foci were counted by inspecting all focal planes intersecting each cell. For each strain, ∼200–800 cells (range 172–1483; Table S2) were scored for Rad52-YFP foci. Fisher's exact t-test was used to calculate significance.
For cell synchronization, cells were arrested in G1 phase by treatment with α-mating factor (3.4 µg/ml; Sigma, St. Louis, MO) for 2–5 hrs (2 for wt, 5 for ctf18). S phase was evaluated by releasing cells from G1 arrest by washing three times in water, resuspension in SC-Leu-Ura medium, and 40 min incubation (consistent with replication timing of the CAG repeat on the yeast artificial chromosome, R. Anand and C. H. Freudenreich, unpublished results) prior to processing for fluorescence microscopy. Prolonged time in the presence of α-mating factor didn't eliminate the multi-budded category; ctf18 multi-budded clusters were excluded from the G1 and S phase Rad52 foci quantification. For G2/M arrest, cells were treated with 0.2 M nocodazole for 3–5 hrs. Because of the high frequency of multi-budded cells in the ctf18 strain, DNA DAPI staining was used as the reference to set the total number of cells.
Supporting Information
Zdroje
1. PearsonCE
EdamuraKN
ClearyJD
2005 Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 6 729 742
2. MirkinSM
2007 Expandable DNA repeats and human disease. Nature 447 932 940
3. FreudenreichCH
2007 Chromosome Fragility: Molecular mechanisms and cellular consequences. Frontiers in Bioscience 12 4911 4924
4. KovtunIV
McMurrayCT
2008 Features of trinucleotide repeat instability in vivo. Cell Res 18 198 213
5. LinY
DionV
WilsonJH
2006 Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol 13 179 180
6. YangZ
LauR
MarcadierJL
ChitayatD
PearsonCE
2003 Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type I disease locus in human cells. Am J Hum Genet 73 1092 1105
7. YoonS-R
DubeauL
de YoungM
WexlerNS
ArnheimN
2003 Huntington disease expansion mutations in humans can occur before meiosis is completed. Proc Natl Acad Sci USA 100 8834 8838
8. StrömL
KarlssonC
LindroosHB
WedahlS
KatouY
2007 Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317 242 245
9. ÜnalE
Heidinger-PauliJM
KoshlandD
2007 DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317 245 248
10. SkibbensRV
2005 Unzipped and loaded: the role of DNA helicases and RFC clamp-loading complexes in sister chromatid cohesion. J Cell Biol 169 841 846
11. HannaJS
KrollES
LundbladV
SpencerFA
2001 Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21 3144 3158
12. MayerML
GygiSP
AebersoldR
HieterP
2001 Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell 7 959 970
13. BermudezVP
ManiwaY
TappinI
OzatoK
HurwitzJ
2003 The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc Natl Acad Sci USA 100 10237 10242
14. ShiomiY
ShinozakiA
SugimotoK
UsukuraJ
ObuseC
2004 The reconstituted human Chl12-RFC complex functions as a second PCNA loader. Genes Cells 9 279 290
15. BylundGO
BurgersPMJ
2005 Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol Cell Biol 25 5445 5455
16. LengronneA
McIntyreJ
KatouY
KanohY
HopfnerK-P
2006 Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23 787 799
17. AnsbachAB
NoguchiC
KlansekIW
HeidlebaughM
NakamuraTM
2008 RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol Biol Cell 19 595 607
18. CrabbeL
ThomasA
PantescoV
De VosJ
PaseroP
2010 Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat Struct Mol Biol 17 1391 1398
19. RazidloDF
LahueRS
2008 Mrc1, Tof1 and Csm3 inhibit CAG·CTG repeat instability by at least two mechanisms. DNA Repair 7 633 640
20. CallahanJL
AndrewsKJ
ZakianVA
FreudenreichCH
2003 Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol Cell Biol 23 7849 7860
21. CollinsSR
MillerKM
MaasNL
RoguevA
FillinghamJ
2007 Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 806 810
22. XuH
BooneC
BrownGW
2007 Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics 176 1417 1429
23. PetronczkiM
ChwallaB
SiomosMF
YokobayashiS
HelmhartW
2004 Sister-chromatid cohesion mediated by the alternative RFC-Ctf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J Cell Sci 117 3547 3559
24. SkibbensRV
2004 Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics 166 33 42
25. GambusA
van DeursenF
PolychronopoulosD
FoltmanM
JonesRC
2009 A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J 28 2992 3004
26. ErricoA
CosentinoC
RiveraT
LosadaA
SchwobE
2009 Tipin/Tim1/And1 protein complex promotes Polα chromatin binding and sister chromatid cohesion. EMBO J 28 3681 3692
27. SuterB
TongAHY
ChangM
YuL
BrownGW
2004 The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics 167 579 591
28. KerrestA
AnandRP
SundararajanR
BermejoR
LiberiG
2009 SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat Struct Mol Biol 16 159 167
29. SundararajanR
GellonL
ZunderRM
FreudenreichCH
2010 Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics 184 65 77
30. GavinAC
AloyP
GrandiP
KrauseR
BoescheM
2006 Proteome survey reveals modularity of the yeast cell machinery. Nature 440 631 636
31. LouH
KomataM
KatouY
GuanZ
ReisC
2008 Mrc1 and DNA polymerase ε function together in linking DNA replication and the S phase checkpoint. Mol Cell 32 106 117
32. ZegermanP
DiffleyJFX
2003 Lessons in how to hold a fork. Nat Struct Biol 10 778
33. TourrièreH
PaseroP
2007 Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair 6 900 913
34. LahiriM
GustafsonTL
MajorsER
FreudenreichCH
2004 Expanded CAG repeats activate the DNA damage checkpoint pathway. Mol Cell 15 287 293
35. FreudenreichCH
LahiriM
2004 Structure-forming CAG/CTG repeat sequences are sensitive to breakage in the absence of Mrc1 checkpoint function and S-phase checkpoint signaling: implications for trinucleotide repeat expansion diseases. Cell Cycle 3 1370 1374
36. PanX
YeP
YuanDS
WangX
BaderJS
2006 A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124 1069 1081
37. EnserinkJM
SmolkaMB
ZhouH
KolodnerRD
2006 Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae. J Cell Biol 175 729 741
38. SandellLL
ZakianVA
1993 Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75 729 739
39. ToczyskiDP
HartwellLH
1997 CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90 1097 1106
40. LisbyM
BarlowJH
BurgessRC
RothsteinR
2004 Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118 699 713
41. SchmidtKH
KolodnerRD
2004 Requirement for Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol Cell Biol 24 3213 3226
42. MurakamiT
TakanoR
TakeoS
TaniguchiR
OgawaK
2010 Stable interaction between the human proliferating cell nuclear antigen loader complex Ctf18-replication factor C (RFC) and DNA polymerase epsilon is mediated by the cohesion-specific subunits, Ctf18, Dcc1, and Ctf8. J Biol Chem 285 34608 34615
43. KarrasGI
JentschS
2010 The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141 255 267
44. DaigakuY
DaviesAA
UlrichHD
2010 Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465 951 956
45. DaeeDL
MertzT
LahueRS
2007 Postreplication repair inhibits CAG·CTG repeat expansions in Saccharomyces cerevisiae. Mol Cell Biol 27 102 110
46. BranzeiD
VanoliF
FoianiM
2008 SUMOylation regulates Rad18-mediated template switch. Nature 456 915 920
47. TerretM-E
SherwoodR
RahmanS
QinJ
JallepalliPV
2009 Cohesin acetylation speeds the replication forks. Nature 462 231 234
48. OgiT
LimsirichaikulS
OvermeerRM
VolkerM
TakenakaK
2010 Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell 37 714 727
49. BrachmannCB
DaviesA
CostGJ
CaputoE
LiJ
1998 Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 115 132
50. SlinkerBK
1998 The statistics of synergism. J Mol Cell Cardiol 30 723 731
51. ZhengQ
2002 Statistical and algorithmic methods for fluctuation analysis with SALVADOR as an implementation. Math Biosci 176 237 252
52. LisbyM
RothsteinR
MortensenUH
2001 Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A 98 8276 8282
Štítky
Genetika Reprodukční medicína
Článek Break to Make a ConnectionČlánek A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on DiseaseČlánek The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 2
-
Všechny články tohoto čísla
- Break to Make a Connection
- A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on Disease
- Single-Tissue and Cross-Tissue Heritability of Gene Expression Via Identity-by-Descent in Related or Unrelated Individuals
- Pervasive Adaptive Protein Evolution Apparent in Diversity Patterns around Amino Acid Substitutions in
- The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
- MiRNA Control of Vegetative Phase Change in Trees
- New Functions of Ctf18-RFC in Preserving Genome Stability outside Its Role in Sister Chromatid Cohesion
- Genome-Wide Association Studies of the PR Interval in African Americans
- Mapping of the Disease Locus and Identification of As a Candidate Gene in a Canine Model of Primary Open Angle Glaucoma
- Mapping a New Spontaneous Preterm Birth Susceptibility Gene, , Using Linkage, Haplotype Sharing, and Association Analysis
- A Population Genetic Approach to Mapping Neurological Disorder Genes Using Deep Resequencing
- and Genes Modulate the Switch between Attraction and Repulsion during Behavioral Phase Change in the Migratory Locust
- Targeted Sister Chromatid Cohesion by Sir2
- Correlated Evolution of Nearby Residues in Drosophilid Proteins
- Parallel Evolution of a Type IV Secretion System in Radiating Lineages of the Host-Restricted Bacterial Pathogen
- Lipophorin Receptors Mediate the Uptake of Neutral Lipids in Oocytes and Imaginal Disc Cells by an Endocytosis-Independent Mechanism
- Genome-Wide Association Study of Coronary Heart Disease and Its Risk Factors in 8,090 African Americans: The NHLBI CARe Project
- The Evolution of Host Specialization in the Vertebrate Gut Symbiont
- Genome-Wide Association of Familial Late-Onset Alzheimer's Disease Replicates and and Nominates in Interaction with
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- Association between Common Variation at the Locus and Changes in Body Mass Index from Infancy to Late Childhood: The Complex Nature of Genetic Association through Growth and Development
- AID Induces Double-Strand Breaks at Immunoglobulin Switch Regions and Causing Chromosomal Translocations in Yeast THO Mutants
- A Study of CNVs As Trait-Associated Polymorphisms and As Expression Quantitative Trait Loci
- Whole-Genome Comparison Reveals Novel Genetic Elements That Characterize the Genome of Industrial Strains of
- Prevalence of Epistasis in the Evolution of Influenza A Surface Proteins
- Srf1 Is a Novel Regulator of Phospholipase D Activity and Is Essential to Buffer the Toxic Effects of C16:0 Platelet Activating Factor
- Two Frizzled Planar Cell Polarity Signals in the Wing Are Differentially Organized by the Fat/Dachsous Pathway
- Phosphoinositide Regulation of Integrin Trafficking Required for Muscle Attachment and Maintenance
- Pathogenic VCP/TER94 Alleles Are Dominant Actives and Contribute to Neurodegeneration by Altering Cellular ATP Level in a IBMPFD Model
- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues
- Nucleosomes Containing Methylated DNA Stabilize DNA Methyltransferases 3A/3B and Ensure Faithful Epigenetic Inheritance
- Mutations in Zebrafish Result in Adult-Onset Ocular Pathogenesis That Models Myopia and Other Risk Factors for Glaucoma
- [], the Prion Formed by the Chromatin Remodeling Factor Swi1, Is Highly Sensitive to Alterations in Hsp70 Chaperone System Activity
- Characterization of Transcriptome Remodeling during Cambium Formation Identifies and As Opposing Regulators of Secondary Growth
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
- Epistatic Interaction Maps Relative to Multiple Metabolic Phenotypes
- Quantitative Models of the Mechanisms That Control Genome-Wide Patterns of Transcription Factor Binding during Early Development
- Genome-Wide Transcript Profiling of Endosperm without Paternal Contribution Identifies Parent-of-Origin–Dependent Regulation of
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- MiRNA Control of Vegetative Phase Change in Trees
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



