-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPervasive Adaptive Protein Evolution Apparent in Diversity Patterns around Amino Acid Substitutions in
In Drosophila, multiple lines of evidence converge in suggesting that beneficial substitutions to the genome may be common. All suffer from confounding factors, however, such that the interpretation of the evidence—in particular, conclusions about the rate and strength of beneficial substitutions—remains tentative. Here, we use genome-wide polymorphism data in D. simulans and sequenced genomes of its close relatives to construct a readily interpretable characterization of the effects of positive selection: the shape of average neutral diversity around amino acid substitutions. As expected under recurrent selective sweeps, we find a trough in diversity levels around amino acid but not around synonymous substitutions, a distinctive pattern that is not expected under alternative models. This characterization is richer than previous approaches, which relied on limited summaries of the data (e.g., the slope of a scatter plot), and relates to underlying selection parameters in a straightforward way, allowing us to make more reliable inferences about the prevalence and strength of adaptation. Specifically, we develop a coalescent-based model for the shape of the entire curve and use it to infer adaptive parameters by maximum likelihood. Our inference suggests that ∼13% of amino acid substitutions cause selective sweeps. Interestingly, it reveals two classes of beneficial fixations: a minority (approximately 3%) that appears to have had large selective effects and accounts for most of the reduction in diversity, and the remaining 10%, which seem to have had very weak selective effects. These estimates therefore help to reconcile the apparent conflict among previously published estimates of the strength of selection. More generally, our findings provide unequivocal evidence for strongly beneficial substitutions in Drosophila and illustrate how the rapidly accumulating genome-wide data can be leveraged to address enduring questions about the genetic basis of adaptation.
Published in the journal: . PLoS Genet 7(2): e32767. doi:10.1371/journal.pgen.1001302
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001302Summary
In Drosophila, multiple lines of evidence converge in suggesting that beneficial substitutions to the genome may be common. All suffer from confounding factors, however, such that the interpretation of the evidence—in particular, conclusions about the rate and strength of beneficial substitutions—remains tentative. Here, we use genome-wide polymorphism data in D. simulans and sequenced genomes of its close relatives to construct a readily interpretable characterization of the effects of positive selection: the shape of average neutral diversity around amino acid substitutions. As expected under recurrent selective sweeps, we find a trough in diversity levels around amino acid but not around synonymous substitutions, a distinctive pattern that is not expected under alternative models. This characterization is richer than previous approaches, which relied on limited summaries of the data (e.g., the slope of a scatter plot), and relates to underlying selection parameters in a straightforward way, allowing us to make more reliable inferences about the prevalence and strength of adaptation. Specifically, we develop a coalescent-based model for the shape of the entire curve and use it to infer adaptive parameters by maximum likelihood. Our inference suggests that ∼13% of amino acid substitutions cause selective sweeps. Interestingly, it reveals two classes of beneficial fixations: a minority (approximately 3%) that appears to have had large selective effects and accounts for most of the reduction in diversity, and the remaining 10%, which seem to have had very weak selective effects. These estimates therefore help to reconcile the apparent conflict among previously published estimates of the strength of selection. More generally, our findings provide unequivocal evidence for strongly beneficial substitutions in Drosophila and illustrate how the rapidly accumulating genome-wide data can be leveraged to address enduring questions about the genetic basis of adaptation.
Introduction
A central challenge of evolutionary biology is to elucidate the nature of adaptive changes to the genome: do they comprise a negligible or substantial fraction of differences among species? When they occur, are they driven by strong positive selection or are they fine-tunings of minor consequence to fitness? In Drosophila, perhaps the most studied taxon in these respects, there are conflicting accounts regarding the intensity of selection driving adaptations [1]–[4] but accumulating lines of evidence suggest that adaptation may be prevalent [5]–[7].
The evidence is based primarily on two kinds of signatures that beneficial substitutions leave in their wake. The first is an excess of divergence at functional sites compared to that expected under neutrality, detected using the approach introduced by McDonald and Kreitman [8]–[11]. Numerous studies based on extensions of this approach indicate that approximately one in two amino acid and one in five non-coding differences between Drosophila species may be adaptive [7], [11]–[14]. These findings remain tentative, however, because other factors, and notably plausible demographic scenarios, could cause a substantial overestimation of the fraction of beneficial substitutions [7], [8], [15]–[17]. Moreover, McDonald-Kreitman based approaches can provide only very limited information about the strength of positive selection.
The second footprint of adaptation is in diversity patterns. When a rare or new allele is favored and fixes in the population, it drags closely linked neutral alleles to loss or fixation. This “selective sweep” leads to a transient reduction in levels of neutral diversity around a beneficial substitution, where the size of the affected region decreases with the recombination rate and increases with the intensity of positive selection [18]–[20]. In accordance with a model of recurrent selective sweeps, levels of synonymous diversity across the genomes of a number of Drosophila species increase with rates of crossing over [21]–[23] and decrease with increasing numbers of amino acid substitutions [2], [3].
Making reliable inferences about adaptation based on these relationships has been challenging, with two decades of effort focused on distinguishing the effects of positive selection from those of background (i.e., purifying) selection and from possible mutagenic effects of recombination [5], [24]–[29]. By necessity, previous studies relied on limited summaries of the data, thereby losing much of the information carried by the spatial signature of beneficial fixations. In particular, measurements of diversity, recombination, and functional divergence were taken in arbitrarily chosen window sizes, making it harder to distinguish the effects of adaptation from other evolutionary forces [29], [30], and likely biasing estimates of adaptive parameters of interest (e.g., the rate and intensity of selection) [7]. As an illustration, based on the relationship between diversity levels and amino acid divergence seen in 100 kb windows, Macpherson et al. [3] inferred few beneficial amino acid substitutions with a large selective coefficient of ∼1%; in contrast, focusing on the same relationship in individual genes, Andolfatto [2] inferred many beneficial amino substitutions with a selective coefficient of ∼10−3%; the two studies differed in other regards, but the disparate conclusions may reflect in part the choice of window size [7]. In summary, despite accumulating evidence that adaptation may be widespread in Drosophila, we still lack characterizations that capture genome-wide signatures that are specific to adaptive evolution and do not rely on an a priori choice of scale.
Results/Discussion
Here, we take advantage of genome-wide variation data from Drosophila in order to produce a readily interpretable characterization of the effects of positive selection that overcomes a number of limitations. To do so, we consider the average level of neutral diversity as a function of distance from amino acid substitutions. Our reasoning is as follows: Beneficial amino acids that fixed in the recent evolutionary past (∼Ne generations [20]) should create a trough in diversity levels around them, whereas amino acid substitutions that were selectively neutral or occurred farther in the past should have little effect on diversity patterns. If we consider the effects of all amino acid substitutions in the genome jointly, and a non-negligible fraction of amino acid fixations were favored – as McDonald-Kreitman based estimates suggest – then we should expect a trough in the average level of neutral diversity around amino acid substitutions. The depth of this trough is expected to increase with the fraction of beneficial amino acid substitutions, and its width will reflect the intensity of selection driving these substitutions. In contrast to previous approaches, this characterization does not depend on an a priori choice of window size, and captures much more of the footprint of adaptive substitutions.
To generate this plot, we use autosomal amino acid substitutions on the lineage leading from the common ancestor of Drosophila simulans and D. melanogaster to D. simulans, relying on the genomes of D. erecta and D. yakuba as outgroups [31]. As a measure of neutral diversity, we consider the number of synonymous polymorphisms divided by the overall number of codons at a given distance from an amino acid substitution. The polymorphism levels in D. simulans are measured using a recent dataset of six inbred lines [5], down-sampled to have a uniform sample size of 4 lines at ∼50% of the codons in the genome. Ideally, we would like to plot diversity levels as a function of genetic distance from amino acid substitutions, since the expected reduction in diversity depends on genetic rather than physical distance from the selected loci. Since there are no high-resolution estimates of recombination rates in D. simulans, we use physical distance instead, but consider only regions for which the homologous regions in D. melanogaster have an estimated recombination rate above 0.75cM/Mb. The collated plot in Figure 1A (red) thus obtained is averaged over n = 26,834 amino acid substitutions.
Fig. 1. The footprint of beneficial amino acid substitutions in neutral levels of diversity. 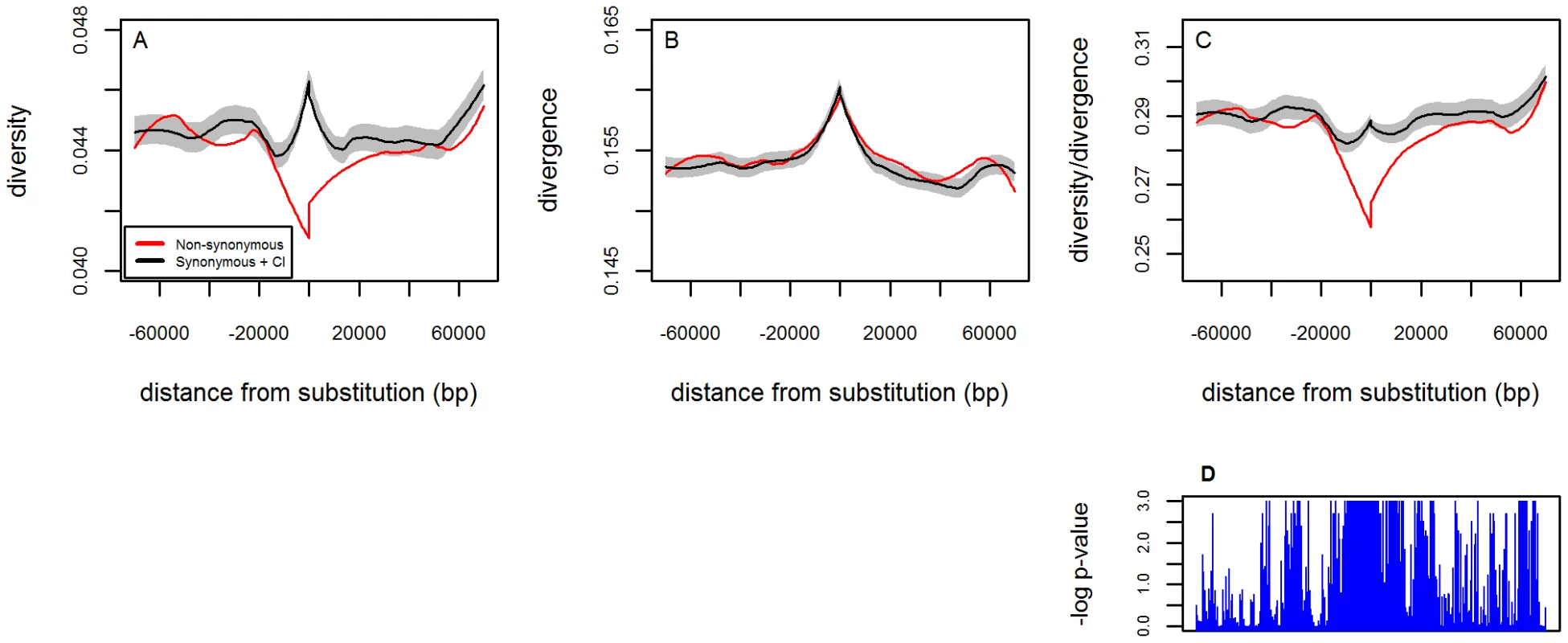
A. Average synonymous diversity level in D. simulans as a function of distance from amino acid (red) and synonymous (black) substitutions in the D. simulans lineage. B. Average synonymous divergence between D. melanogaster and D. yakuba (a proxy for the mutation rate) as a function of distance from amino acid (red) and synonymous (black) substitutions. C. Synonymous diversity levels divided by divergence as a function of distance from amino acid (red) and synonymous (black) substitutions (see Text S1). The curves in A–B were smoothed with LOESS on the left and right of substitutions separately, and C was calculated as a ratio of the value after smoothing (see Text S1). The gray sleeves represent the standard error of the mean of the synonymous control (black curve) estimated from 1000 bootstraps and smoothed by LOESS as above (see Text S1). D. A Manhattan plot of the one tailed p-value (on a logarithmic scale) testing the hypothesis that the average diversity divided by the average divergence around amino acid substitutions is the same as that around synonymous substitutions (shown in C). Results are shown as a function of distance from the substitution (based on 1000 bootstraps and calculated in bins of 0.5 kb; see Text S1 for details). Because the plot is constructed by conditioning on a substitution at the center, diversity patterns could be distorted even in the absence of adaptive evolution. Namely, if mutation rates vary across the genome then they might, on average, be elevated near substitutions. Considering the average synonymous divergence between D. melanogaster and D. yakuba as a proxy for the mutation rate confirms this expectation, as it reveals a small increase near substitutions (Figure 1B). To correct for this elevation in rates, we divide the average level of diversity around amino acid substitutions at a given distance by the average divergence (Figure 1C). Moreover, as a control, we compare the patterns around amino acid substitutions with plots that were constructed analogously but around synonymous substitutions instead (Figure 1A–1C: black) [28].
As predicted by a model of recurrent selective sweeps, we find a clear reduction in diversity levels around amino acid substitutions relative to the synonymous control. This reduction is statistically significant within a window of ∼15kb around amino acid substitutions (at the 1% level, as assessed by bootstrapping; see Text S1). Farther from substitutions, where sweeps are unlikely to have an effect on diversity, the curves for synonymous and amino acid substitutions are indistinguishable. This pattern is robust to the effects of synonymous codon usage bias (Figure 4 in Text S1), as well as to changes in the recombination rate threshold (Figure 5 in Text S1), and to the choice of outgroup used to correct for the mutation rate (not shown). In addition, we see similar patterns when we examine the substitutions that occur on any one of the autosomal chromosome arms (Figure 6 in Text S1).
This pattern is a distinctive signature of adaptive evolution. Demographic processes would not lead to systematically decreased diversity around amino acid substitutions. In turn, for background selection to generate the observed trough centered on amino acid substitutions, its effects in regions of the genome with moderate to high recombination rates would have to be strong enough to lead to both a substantial reduction in diversity and to the fixation of many weakly deleterious amino acid mutations. Modeling indicates that, given plausible parameters for Drosophila, this is highly unlikely [32].
Our analyses also reveal that amino acid substitutions are clustered near one another (Figure 2A: red). This clustering is greater and more localized than the clustering of synonymous substitutions around amino acid substitutions (Figure 2A: black), implying that it is caused by more than the spatial distribution of exons in the genome and an elevated mutation rate near amino acid substitutions. The difference between the clustering of amino acid and synonymous substitutions further suggests that variation in constraint and possibly in adaptability among and within genes contribute to the pattern for amino acid substitutions ([33]; also see Text S1).
Fig. 2. The reduction in diversity around amino acid substitutions, controlling for clustering. 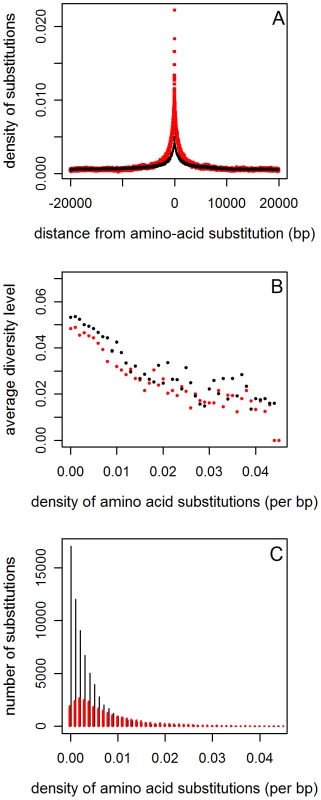
A. The density of amino acid (red) and synonymous (black) substitutions as a function of distance from an amino acid substitution. The synonymous density was multiplied by 0.4 (the ratio of the average amino acid to the average synonymous densities) in order to make the comparison of densities more transparent. B. A comparison between the average diversity levels around amino acid (red) and synonymous substitutions (black) as a function of the density of amino acid substitutions in their vicinity. Diversity levels and density of amino acid substitutions were measured in a window size of 1kb centered at the substitution under consideration. C. The numbers of amino acid (red) and synonymous substitutions (black) used to estimate the average diversity levels at each density. Aside from being an interesting finding in itself, this clustering could influence the observed reduction in diversity. If two amino acid substitutions occur in close proximity and one led to a recent selective sweep, the reduction in diversity that it caused will also be observed around the other substitution. This effect will reduce diversity around both non-synonymous and synonymous substitutions, but it will have a larger effect around amino acid substitutions because the density of amino acid substitutions nearby is on average greater (Figure 2A). Indeed, the level of synonymous diversity decreases strongly with the density of amino acid substitutions surrounding a substitution (Figure 2B; Figure 8 in Text S1; Spearman's ρ = −0.93 for amino acid substitutions and ρ = −0.88 for synonymous substitutions; p<10−15 for both), consistent with previous studies [2], [3]. We also find, however, that the average level of synonymous diversity around amino acid substitutions is consistently lower than that around synonymous substitutions when the two are matched for the density of amino acid substitutions in their vicinity (Figure 2B; Figure 8 in Text S1; signs test p<10−4). In other words, there is a substantial relative reduction in diversity around amino acid substitutions that is not explained by the amplifying effects of clustering.
In addition to providing compelling evidence for the prevalence of beneficial amino acid substitutions, the collated plot carries information about selection parameters, as the shape of the trough in diversity is indicative of the rate of adaptive protein evolution and of the distribution of selective effects of fixations. To learn about these parameters, we develop a coalescent-based model for average diversity levels as a function of distance from an amino acid substitution, accounting for their clustering (see Text S1). Using this model, we infer adaptive parameters by jointly maximizing the composite-likelihood of diversity patterns as a function of different distances from the focal substitution (i.e., the likelihood of points along the entire curve), thus mining a richer summary of the data than previous approaches. When we assume that a fraction α of beneficial substitutions were driven by a selection coefficient s and the rest were neutral, we estimate that ∼5% of the substitutions were beneficial with a relatively strong selection coefficient of ∼0.4% (Table 5 in Text S1). Using a Gamma distribution for the selection coefficients, α increases to ∼6.5% and the average selection coefficient remains similarly high; despite the additional parameter, the likelihood is barely higher (Table 5 in Text S1). These estimates are relatively insensitive to assumptions about other parameters (with the exception of the assumptions about recombination rates, as discussed below); in particular, simulations suggest that the estimated strength of selection is robust to demographic assumptions (see Text S1 for details).
A visual comparison suggests a reasonable fit of these models to the data (Figure 3A). However, the inference based on models with one selection coefficient, or even a Gamma distribution of coefficients, might be dominated by the broad features of the plot, such that any narrower trough caused by beneficial substitutions with weaker selection coefficients could be overlooked. A closer look around the focal substitutions supports this notion, revealing a small trough inside the main trough, on the scale of several hundred bps, which is not captured by either of the two models (Figure 3B). We therefore consider another model, with two beneficial selection coefficients. Using it, we estimate that ∼13% of the substitutions were beneficial, ∼3% with a large selective advantage of ∼0.5% and the rest with a much weaker effect, of approximately one hundredth of a percent (Table 5 in Text S1). A mixture model with two exponentials reveals a similar picture: ∼4% of substitutions are estimated to come from a distribution with a mean selective coefficient of ∼0.5% and 11% from a distribution with a mean of ∼4·10−5 (Table 5 in Text S1). Importantly, both models provide a substantially better fit to the data (Table 5 in Text S1) and they capture the smaller as well as the larger troughs in diversity (Figure 3A and 3B). In turn, estimates under a model with three beneficial selective coefficients are similar to those obtained in model with only two and offer no improvement to the fit (Table 5 in Text S1). Taken together, these findings indicate that selective sweeps are driven by two classes of beneficial fixations: a minority with large beneficial effects that account for most of the reduction in diversity and a majority with much weaker effects. Moreover, they help explain why previous inferences based on the signatures of sweeps in Drosophila yielded markedly different estimates (ranging over three orders of magnitudes) [1]–[4].
Fig. 3. The fit of recurrent selective sweep models to diversity patterns around amino acid substitutions. 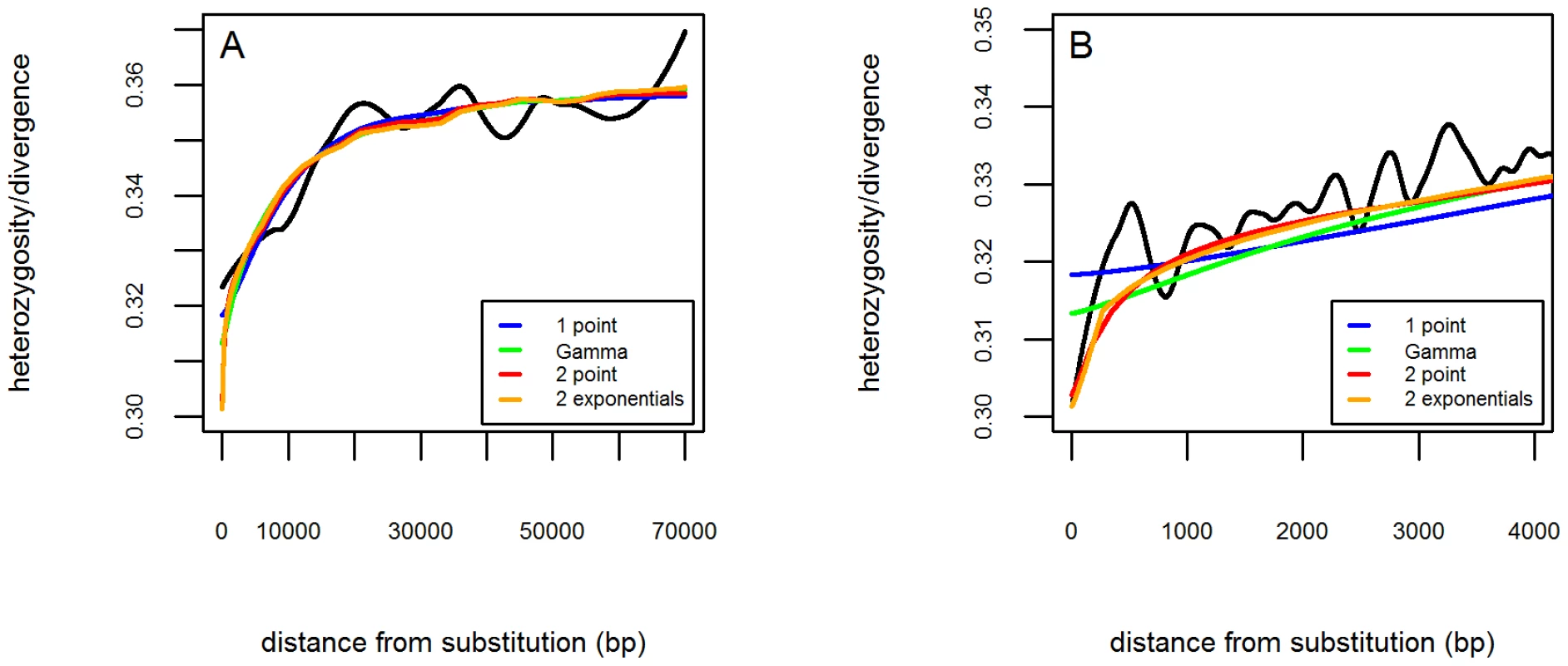
A. Observed and predicted curves for the average synonymous heterozygosity as a function of distance from amino acid substitutions. The curve based on the data (black) was smoothed using LOESS with a span of 0.5 and divided by divergence, as in Figure 1. The predicted curves correspond to maximum likelihood estimates based on different distributions of beneficial selection coefficients: “1 point” corresponds to a single selection coefficient (blue); “Gamma” to a Gamma distribution (green); “2 point” to two selection coefficients (red); “2 exponentials” to a mixture of two exponentials (orange). B. A close-up on distances up to 4 kb. To reveal more detail of the observed curve on this scale, we used LOESS smoothing with a smaller span of 0.002. See Text S1 for further details. Our estimates of the fraction of beneficial amino acid substitutions (∼13%) are on the same order of magnitude but lower than previous McDonald-Kreitman based estimates (∼50%; cf. [7]). Some of this difference might arise from violations of the assumptions on which the inferences rely; in particular, in our approach, that adaptive parameters have remained constant in the D. simulans lineage, or in McDonald-Krietman based inferences, that the efficacy of purifying selection has not changed markedly [8], [16], [34].
An intriguing alternative is that the two approaches are actually estimating parameters of somewhat different modes of adaptation. Our inference is based on the effects of beneficial substitutions that arise from new mutations and likely misses some contribution of adaptation from standing variation. Specifically, a subset of beneficial substitutions could stem from previously neutral or deleterious alleles that were segregating in the population before a change in the environment rendered them beneficial. If these alleles were young when the environment changed, they would still generate the signature of a selective sweep and contribute, at least partially, to our estimated fraction of beneficial substitutions. This is likely for alleles that were previously deleterious and at mutation-selection balance, but also possible for neutral alleles [35]–[37]. If, however, the segregating alleles were older when they became beneficial and at higher frequency in the population, they would lead to a negligible effect on diversity and would therefore not contribute to the signature on which our inference relies. These beneficial substitutions would nonetheless contribute to an excess of non-synonymous divergence compared to the neutral expectation, and should therefore be picked by the McDonald-Kreitman based inferences, leading to higher estimates of adaptive substitutions than obtained by our approach. Other modes of adaptation, such as polygenic selection, may also contribute differentially to the two inference methodologies [38].
We note that a current limitation of our inference is its reliance on rough estimates of the recombination rate, and its assumption of a constant rate per base. In the logistic approximation to the trajectory of a beneficial allele, the expected reduction in diversity as a function of distance from the beneficial substitution depends on s/r, where s is the selection coefficient and r is the genetic distance to the substitution (Equation 2 in Text S1). This implies, for example, that if our inference relies on a recombination rate consistently two-fold greater than the real rate, our estimated selection coefficient will be two-fold overestimated (see Table 3 in Text S1). We therefore consider our estimates of selection coefficients to be rough approximations. In addition, heterogeneity in the recombination rate, such as is known to exist in other taxa (e.g., [39], [40]), could also affect our inferences. The heterogeneity would have to be of a highly specific nature in order to account for our finding of two markedly different scales of selection coefficients, but at the moment, we cannot rule out the possibility. For these reasons, it would be important to revisit the inference once we possess high-resolution genetic maps in D. simulans.
In summary, our findings establish a distinctive, genome-wide signature of adaptation in D. simulans, suggesting that many amino acid substitutions are beneficial and are driven by two classes of selective effects. Enabled by a richer summary of diversity patterns that avoids an a priori choice of scale, these conclusions offer a coherent interpretation of the results of previous inferences. It will now be interesting to see whether similar findings emerge in other Drosophila species, which vary in their recombination rates, effective population sizes, and ecology.
Materials and Methods
Data
We reconstructed the sequence of the ancestor of D. melanogaster and D. simulans in order to identify substitutions along the D. simulans lineage. For that purpose, we use a four species alignement from the 12 Drosophila genomes project [31] consisting of D. simulans, D. melanogaster, D. yakuba and D. erecta, and removed codons containing gaps in either of them. We then inferred the ancestral sequences using PAML, with the CODEML model and the ((D. mel, D. sim), (D. yak, D. ere)) tree [41]. To measure polymorphism levels at coding regions of the D. simulans genome, we used resequencing data from six inbred lines of D. simulans and their alignment with D. melanogaster [5]. We applied quality control filters and randomly down-sampled the remaining codons to four, in order to maintain a uniform sample size in measuring polymorphism. In the end, we retained ∼50% of all protein-coding DNA. Unless otherwise noted, our analysis was performed on data from autosomal regions, for which the sex-averaged recombination rate in the homologous region of D. melanogaster was greater than 0.75cM/Mb (using the genetic map as in [3]). See Section 1 in Text S1 for more details.
Construction of the collated plot
We used synonymous polymorphisms to measure the average levels of diversity as a function of distance from amino acid and synonymous substitutions along the D. simulans lineage. To measure the average level of diversity at distance x, we divided the number of codons segregating for a synonymous polymorphism by the overall number of codons observed in the D. simulans polymorphism dataset at distance x from one of the amino acid (or synonymous) substitution. In order to control for variation in the neutral mutation rate around substitutions, we calculated the average synonymous divergence around both amino acid and synonymous substitutions. For that purpose, we identified synonymous substitutions between D. melanogaster and D. yakuba and measured the average level of divergence at distance x by dividing the number of codons exhibiting a synonymous substitution between D. melanogaster and D. yakuba by the overall number of codons observed in the alignment of these species at distance x from one of the amino acid (or synonymous) substitutions. For further details and the robustness analysis, see Sections 2–4 in Text S1.
Inference method
The shape of the collated plot around amino acid substitutions carries information about the rate of adaptive protein evolution and the intensity of selection driving it, two parameters of long-standing interest. To learn about these parameters, we developed a model describing the expected neutral diversity levels around substitutions, which relies on Gillespie's pseudohitchhiking coalescent model [42]. We then used a composite likelihood approach [43] to estimate the parameters. For a description of the approach and assessments of its reliability, see Section 6 in Text S1.
Supporting Information
Zdroje
1. LiH
StephanW
2006 Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet 2 e166 doi:10.1371/journal.pgen.0020166
2. AndolfattoP
2007 Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res 17 1755 1762
3. MacphersonJM
SellaG
DavisJC
PetrovDA
2007 Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics 177 2083 2099
4. JensenJD
ThorntonKR
AndolfattoP
2008 An approximate bayesian estimator suggests strong, recurrent selective sweeps in Drosophila. PLoS Genet 4 e1000198 doi:10.1371/journal.pgen.1000198
5. BegunDJ
HollowayAK
StevensK
HillierLW
PohYP
2007 Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol 5 e310 doi:10.1371/journal.pbio.0050310
6. WrightSI
AndolfattoP
2008 The impact of natural selection on the genome: emerging patterns in Drosophila and Arabidopsis. Annual Review of Ecology and Systematics 39 193 213
7. SellaG
PetrovDA
PrzeworskiM
AndolfattoP
2009 Pervasive natural selection in the Drosophila genome? PLoS Genet 5 e1000495 doi:10.1371/journal.pgen.1000495
8. McDonaldJH
KreitmanM
1991 Adaptive protein evolution at the Adh locus in Drosophila. Nature 351 652 654
9. CharlesworthB
1994 The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet Res 63 213 227
10. SawyerSA
HartlDL
1992 Population genetics of polymorphism and divergence. Genetics 132 1161 1176
11. Eyre-WalkerA
2006 The genomic rate of adaptive evolution. Trends Ecol Evol 21 569 575
12. FayJC
WyckoffGJ
WuCI
2002 Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature 415 1024 1026
13. SmithNG
Eyre-WalkerA
2002 Adaptive protein evolution in Drosophila. Nature 415 1022 1024
14. AndolfattoP
2005 Adaptive evolution of non-coding DNA in Drosophila. Nature 437 1149 1152
15. OhtaT
1993 Amino acid substitution at the Adh locus of Drosophila is facilitated by small population size. Proc Natl Acad Sci U S A 90 4548 4551
16. Eyre-WalkerA
2002 Changing effective population size and the McDonald-Kreitman test. Genetics 162 2017 2024
17. NielsenR
2005 Molecular signatures of natural selection. Annu Rev Genet 39 197 218
18. Maynard SmithJM
HaighJ
1974 The hitch-hiking effect of a favourable gene. Genet Res 23 23 35
19. BravermanJM
HudsonRR
KaplanNL
LangleyCH
StephanW
1995 The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics 140 783 796
20. PrzeworskiM
2002 The signature of positive selection at randomly chosen loci. Genetics 160 1179 1189
21. AguadeM
MiyashitaN
LangleyCH
1989 Reduced Variation in the Yellow-Achaete-Scute Region in Natural Populations of Drosophila Melanogaster. Genetics 122 607 615
22. BerryAJ
AjiokaJW
KreitmanM
1991 Lack of polymorphism on the Drosophila fourth chromosome resulting from selection. Genetics 129 1111 1117
23. BegunDJ
AquadroCF
1992 Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356 519 520
24. CharlesworthB
MorganMT
CharlesworthD
1993 The effect of deleterious mutations on neutral molecular variation. Genetics 134 1289 1303
25. HudsonRR
1994 How can the low levels of DNA sequence variation in regions of the drosophila genome with low recombination rates be explained? Proc Natl Acad Sci U S A 91 6815 6818
26. StephanW
XingL
KirbyDA
BravermanJM
1998 A test of the background selection hypothesis based on nucleotide data from Drosophila ananassae. Proc Natl Acad Sci U S A 95 5649 5654
27. AndolfattoP
2001 Adaptive hitchhiking effects on genome variability. Curr Opin Genet Dev 11 635 641
28. KernAD
JonesCD
BegunDJ
2002 Genomic effects of nucleotide substitutions in Drosophila simulans. Genetics 162 1753 1761
29. KulathinalRJ
BennettSM
FitzpatrickCL
NoorMA
2008 Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc Natl Acad Sci U S A 105 10051 10056
30. InnanH
StephanW
2003 Distinguishing the hitchhiking and background selection models. Genetics 165 2307 2312
31. ClarkAG
EisenMB
SmithDR
BergmanCM
OliverB
2007 Evolution of genes and genomes on the Drosophila phylogeny. Nature 450 203 218
32. LoeweL
CharlesworthB
2007 Background selection in single genes may explain patterns of codon bias. Genetics 175 1381 1393
33. RidoutKE
DixonCJ
FilatovDA
2010 Positive selection differs between protein secondary structure elements in Drosophila. Genome Biology and Evolution doi:10.1093/gbe/evq008
34. OhtaT
1973 Slightly deleterious mutant substitutions in evolution. Nature 246 96 98
35. OrrHA
BetancourtAJ
2001 Haldane's sieve and adaptation from the standing genetic variation. Genetics 157 875 884
36. PrzeworskiM
CoopG
WallJD
2005 The signature of positive selection on standing genetic variation. Evolution Int J Org Evolution 59 2312 2323
37. HermissonJ
PenningsPS
2005 Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169 2335 2352
38. PritchardJK
PickrellJK
CoopG
2010 The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol 20 R208 215
39. MyersS
BottoloL
FreemanC
McVeanG
DonnellyP
2005 A fine-scale map of recombination rates and hotspots across the human genome. Science 310 321 324
40. PetesTD
2001 Meiotic recombination hot spots and cold spots. Nat Rev Genet 2 360 369
41. YangZ
1997 PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13 555 556
42. GillespieJH
2000 Genetic drift in an infinite population. The pseudohitchhiking model. Genetics 155 909 919
43. HudsonRR
2001 Two-locus sampling distributions and their application. Genetics 159 1805 1817
Štítky
Genetika Reprodukční medicína
Článek Break to Make a ConnectionČlánek A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on DiseaseČlánek The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 2
-
Všechny články tohoto čísla
- Break to Make a Connection
- A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on Disease
- Single-Tissue and Cross-Tissue Heritability of Gene Expression Via Identity-by-Descent in Related or Unrelated Individuals
- Pervasive Adaptive Protein Evolution Apparent in Diversity Patterns around Amino Acid Substitutions in
- The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
- MiRNA Control of Vegetative Phase Change in Trees
- New Functions of Ctf18-RFC in Preserving Genome Stability outside Its Role in Sister Chromatid Cohesion
- Genome-Wide Association Studies of the PR Interval in African Americans
- Mapping of the Disease Locus and Identification of As a Candidate Gene in a Canine Model of Primary Open Angle Glaucoma
- Mapping a New Spontaneous Preterm Birth Susceptibility Gene, , Using Linkage, Haplotype Sharing, and Association Analysis
- A Population Genetic Approach to Mapping Neurological Disorder Genes Using Deep Resequencing
- and Genes Modulate the Switch between Attraction and Repulsion during Behavioral Phase Change in the Migratory Locust
- Targeted Sister Chromatid Cohesion by Sir2
- Correlated Evolution of Nearby Residues in Drosophilid Proteins
- Parallel Evolution of a Type IV Secretion System in Radiating Lineages of the Host-Restricted Bacterial Pathogen
- Lipophorin Receptors Mediate the Uptake of Neutral Lipids in Oocytes and Imaginal Disc Cells by an Endocytosis-Independent Mechanism
- Genome-Wide Association Study of Coronary Heart Disease and Its Risk Factors in 8,090 African Americans: The NHLBI CARe Project
- The Evolution of Host Specialization in the Vertebrate Gut Symbiont
- Genome-Wide Association of Familial Late-Onset Alzheimer's Disease Replicates and and Nominates in Interaction with
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- Association between Common Variation at the Locus and Changes in Body Mass Index from Infancy to Late Childhood: The Complex Nature of Genetic Association through Growth and Development
- AID Induces Double-Strand Breaks at Immunoglobulin Switch Regions and Causing Chromosomal Translocations in Yeast THO Mutants
- A Study of CNVs As Trait-Associated Polymorphisms and As Expression Quantitative Trait Loci
- Whole-Genome Comparison Reveals Novel Genetic Elements That Characterize the Genome of Industrial Strains of
- Prevalence of Epistasis in the Evolution of Influenza A Surface Proteins
- Srf1 Is a Novel Regulator of Phospholipase D Activity and Is Essential to Buffer the Toxic Effects of C16:0 Platelet Activating Factor
- Two Frizzled Planar Cell Polarity Signals in the Wing Are Differentially Organized by the Fat/Dachsous Pathway
- Phosphoinositide Regulation of Integrin Trafficking Required for Muscle Attachment and Maintenance
- Pathogenic VCP/TER94 Alleles Are Dominant Actives and Contribute to Neurodegeneration by Altering Cellular ATP Level in a IBMPFD Model
- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues
- Nucleosomes Containing Methylated DNA Stabilize DNA Methyltransferases 3A/3B and Ensure Faithful Epigenetic Inheritance
- Mutations in Zebrafish Result in Adult-Onset Ocular Pathogenesis That Models Myopia and Other Risk Factors for Glaucoma
- [], the Prion Formed by the Chromatin Remodeling Factor Swi1, Is Highly Sensitive to Alterations in Hsp70 Chaperone System Activity
- Characterization of Transcriptome Remodeling during Cambium Formation Identifies and As Opposing Regulators of Secondary Growth
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
- Epistatic Interaction Maps Relative to Multiple Metabolic Phenotypes
- Quantitative Models of the Mechanisms That Control Genome-Wide Patterns of Transcription Factor Binding during Early Development
- Genome-Wide Transcript Profiling of Endosperm without Paternal Contribution Identifies Parent-of-Origin–Dependent Regulation of
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- MiRNA Control of Vegetative Phase Change in Trees
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



