-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Activation of Effector Avr3b by Plant Cyclophilin is Required for the Nudix Hydrolase Activity of Avr3b
Phytophthora sojae, an oomycete pathogen that causes the Phytophthora root and stem rot disease of soybean, delivers variety of effectors into host cell to reprogram host immunity. Genome sequencing uncovers that P. sojae genome encode several hundreds of effector genes. However, the mode of action of most of the P. sojae effectors remains unknown. The investigation of effector-interacting proteins provides opportunities to better understand the pathogenesis mechanism of the pathogen and the defense mechanism of the host plants. Previously, we reported that P. sojae avirulence effector Avr3b modulates plant immunity through its Nudix hydrolase activity. Interestingly, the enzymatic activity is required for its virulence but not required for recognition by resistant gene. In this study, we identified soybean cyclophilin protein GmCYP1 as an Avr3b interactor. The enzymatic activity of GmCYP1 is required for the maturation Avr3b, which is directly related to both virulence and avirulence functions of Avr3b. This work provides a novel insight into how Phytophthora pathogens recruit host proteins to activate the enzymatic activity of effectors in order to gain successful infection.
Published in the journal: . PLoS Pathog 11(8): e32767. doi:10.1371/journal.ppat.1005139
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005139Summary
Phytophthora sojae, an oomycete pathogen that causes the Phytophthora root and stem rot disease of soybean, delivers variety of effectors into host cell to reprogram host immunity. Genome sequencing uncovers that P. sojae genome encode several hundreds of effector genes. However, the mode of action of most of the P. sojae effectors remains unknown. The investigation of effector-interacting proteins provides opportunities to better understand the pathogenesis mechanism of the pathogen and the defense mechanism of the host plants. Previously, we reported that P. sojae avirulence effector Avr3b modulates plant immunity through its Nudix hydrolase activity. Interestingly, the enzymatic activity is required for its virulence but not required for recognition by resistant gene. In this study, we identified soybean cyclophilin protein GmCYP1 as an Avr3b interactor. The enzymatic activity of GmCYP1 is required for the maturation Avr3b, which is directly related to both virulence and avirulence functions of Avr3b. This work provides a novel insight into how Phytophthora pathogens recruit host proteins to activate the enzymatic activity of effectors in order to gain successful infection.
Introduction
Plants have two layers of defense in response to microbial pathogen attacks [1]. Pattern-triggered immunity (PTI) is dependent on the recognition of microbe - or pathogen-associated molecular patterns (MAMPs or PAMPs) by surface-localized pattern recognition receptors (PRRs) [2]. The second defense system is effector-triggered immunity (ETI), which relies on the perception of pathogen effectors [1]. ETI produces a faster and stronger resistance response than PTI and often induces localized cell death in the infected area [1,3]. Effectors are usually rapidly evolving in pathogens due to dynamic and sometimes opposite evolutionary forces during the arms race with the hosts [4].
Plant pathogens secrete a diverse repertoire of effectors to overcome plant immunity and enable infection [5,6]; however, the biochemical functions of most effectors, especially those produced by eukaryotic pathogens, remain largely unknown. To our knowledge, only a few effectors produced by phytopathogenic oomycetes and fungi have clear biochemical functions and manipulate their host targets by enzymatic activity. For example, Cmu1 produced by the fungal pathogen Ustilago maydis has chorismate mutase activity and affects salicylic acid (SA) levels in maize cells [7]. Similarly, PsIsc1 from the oomycete pathogen Phytophthora sojae and VdIsc1 from the fungal pathogen Verticillium dahliae also suppress SA-mediated immunity through their isochorismatase activity [8].
Phytophthora is a group of notorious plant pathogens that infect a wide range of crop, vegetable, horticultural, pasture plants, and forest trees. Like many other plant pathogens, Phytophthora secretes effectors to gain virulence. The functions of an increasing number of Phytophthora effectors, particularly the RXLR (arginine, any, leucine, arginine) effectors that can be delivered into host cell, have recently been reported [5]. Many RxLR effectors can suppress PTI and ETI. For example, various effectors from the soybean (Glycine max) pathogen P. sojae, including Avr1b, Avr3b and Avr1d, are able to abolish cell death triggered by other effectors and/or PAMP in soybean or Nicotiana benthamiana [9–12]. Functional analysis of thirty-three RXLR effectors from the potato and tomato pathogen Phytophthora infestans revealed eight of them that could suppress PTI triggered by the bacterial flagellin peptide flg22 in tomato protoplasts [13].
Further investigations have revealed the molecular basis by which RXLR effectors suppress plant immunity. For example, PexRD2 perturbs plant immunity-related signaling by interacting with the kinase domain of MAPKKKε and reducing the accumulation of phosphorylated MAPK [14]. AVRblb2 interacts with host papain-like cysteine protease C14, which is a positive regulator of plant immunity, and prevents its secretion into apoplast [15]. Pi03192 targets NAC transcription factor NTP1 and NTP2 and prevents Phytophthora culture filtrate triggered re-localization of these proteins from the endoplasmic reticulum (ER) into the nucleus [16]. Another important discovery is that Phytophthora RXLR effectors could target RNA silencing pathway. Phytophthora suppressors of RNA silencing 1 and 2 (PSR1 and PSR2, respectively) of P. sojae are powerful suppressors of RNA silencing in plants and enhance plant susceptibility to Phytophthora by inhibiting the biogenesis of small RNAs [17,18].
We previously demonstrated that the P. sojae RXLR effector Avr3b possesses the Nudix hydrolase activity and contributes to full virulence of P. sojae [19]. Nudix hydrolases hydrolyze a wide range of organic pyrophosphates, including nucleoside di - and triphosphates, dinucleoside and diphosphoinositol polyphosphates, nucleotide sugars and RNA caps, which may be toxic or have regulatory roles [20]. As such, Nudix hydrolases play a key role in signaling, house-keeping processes and maintaining cellular homeostasis [21]. In particular, Arabidopsis thaliana Nudix protein AtNUDT7 was found to be a negative regulator of the basal defense response. Loss-of-function mutation of AtNUDT7 results in higher levels of salicylic acid (SA) and constitutive expression of defense-related genes, and thereby enhanced resistance to the bacterial pathogen Pseudomonas syringae and the oomycete pathogen Hyaloperonospora arabidopsidis (Hpa) [22–25]. Besides Avr3b, Nudix effector CtNUDIX from the fungal pathogen Colletotrichum truncatum is exclusively expressed during the late biotrophic phase and elicits a HR-like cell death in tobacco leaves, suggesting that CtNUDIX may signal the transition from biotrophy to necrotrophy [26]. In addition, Hpx26 produced by the bacterial pathogen Ralstonia solanacearum shares homology with known Nudix hydrolase [27]. So far, the biochemical functions of CtNUDIX and Hpx26 are unclear.
Cyclophilin (CYP) possesses peptidyl-prolyl cis-trans isomerase (PPIase) activity and catalyzes the isomerization of prolyl bonds in proteins [28,29]. CYP family members are involved in diverse aspects of cellular physiology including transcription, immune response, mitochondrial function, cell death, and chemotaxis [30–32]. Interestingly, CYP plays a critical role in the activation of bacterial effectors and the life cycle of viruses, especially hepatitis C virus (HCV) and human immunodeficiency virus (HIV) [30,33,34]. For example, AvrRpt2 from P. syringae is delivered into the plant cell as an inactive form and subsequently activated by host cyclophilin ROC1 (Rotamase CYP1) [33]. During HCV infection of animal cells, cyclophilin B interacts with the HCV RNA polymerase NS5B and functions as a stimulatory regulator of NS5B in HCV replication machinery [35].
The Nudix hydrolase activity of Avr3b is important for P. sojae infection. However, recombinant Avr3b proteins produced in Escherichia coli had no hydrolase activity unless it was incubated with plant extracts, suggesting that unknown plant factors is required for the activation of Avr3b in host cells. Here, we identified the soybean cyclophilin protein GmCYP1 as an Avr3b-interacting protein and showed that its PPIase activity is responsible for the activation of Avr3b enzymatic activity. We further identified the Pro132 residue in a potential GP motif of Avr3b as a key residue involved in Avr3b-GmCYP1 interaction. The mutant Avr3bP132A, no longer possessing the hydrolase activity, is also abolished for its virulence activity in N. benthamiana as well as HR-triggering activity in a resistant soybean cultivar that produces the cognate resistance (R) protein of Avr3b, Rps3b. Moreover, application of a PPIase inhibitor, cyclosporine A (CsA), or silencing of GmCYP1 greatly reduced the cell death triggered by Avr3b in soybean, suggesting that GmCYP1 is required for the maturation of Avr3b as a Nudix hydrolase and therefore playing an essential role in the virulence and avirulence activities of Avr3b in planta.
Results
Avr3b is activated in planta
Previous study demonstrated that Avr3b is a functional Nudix hydrolase, and this enzymatic activity is essential for the virulence function of Avr3b [19]. To further elucidate the enzymatic activity of Avr3b, we expressed the recombinant GST-Avr3b protein in N. benthamiana and E. coli, respectively. Interestingly, unlike Avr3b expressed in N. benthamiana, which exhibited clear Nudix hydrolase activity, purified Avr3b produced in E. coli could not hydrolyze the substrate NADH (Fig 1A). These results suggest that the enzymatic activity of Avr3b might need to be activated by some unknown factors from plants. To test this hypothesis, recombinant Avr3b produced by E. coli was incubated with dialyzed total protein extracts from P. sojae, soybean or N. benthamiana for 15 hours before the hydrolase activity was examined. Consistent with the previous results, addition of soybean or N. benthamiana extracts significantly promoted the hydrolase activity of Avr3b, while no change was observed after Avr3b was incubated with extracts from P. sojae mycelium (Fig 1B). These results suggest that Avr3b is likely in an inactive form in P. sojae and activated after it is delivered into plant cells by a plant factor(s).
Fig. 1. The enzymatic activity of Avr3b is activated by plant factors. 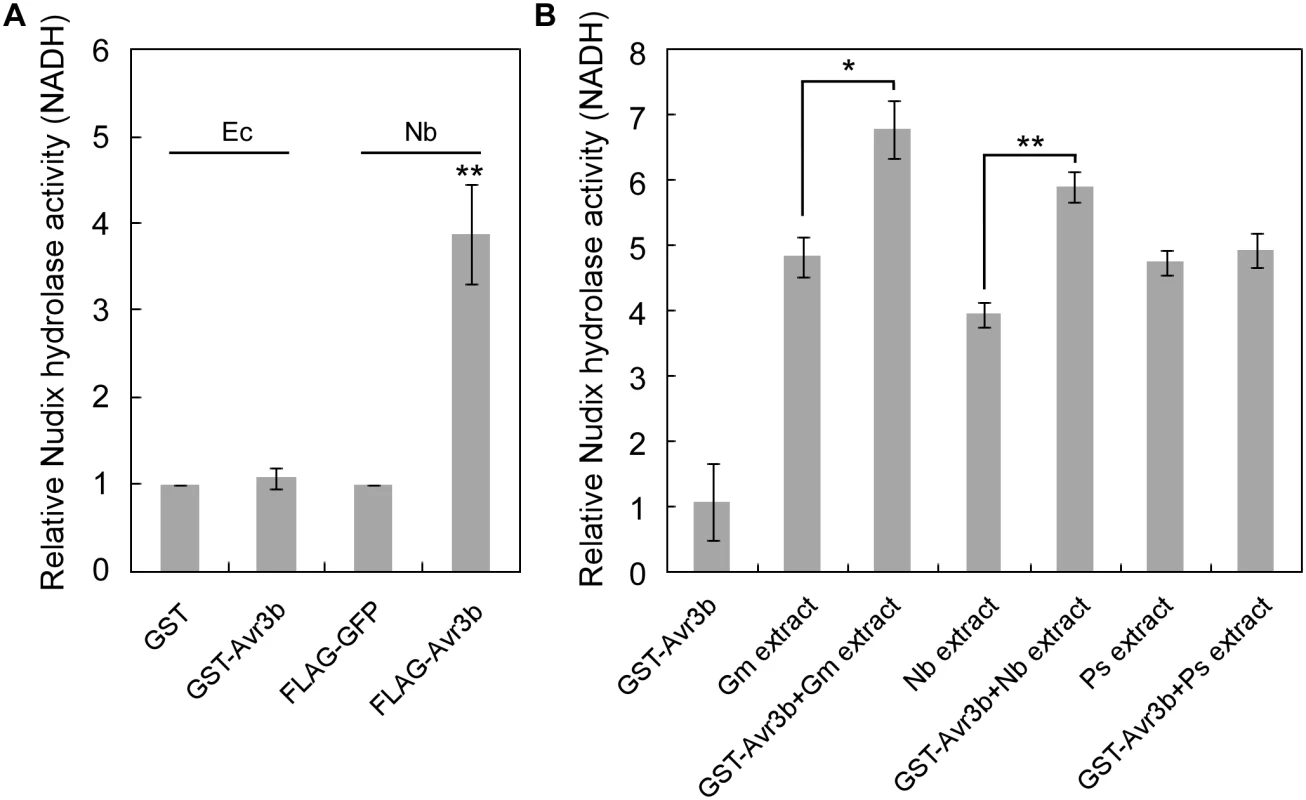
(A) Avr3b only has Nudix hydrolase activity when produced in plants. Avr3b was ectopically expressed in E. coli (Ec) or N. benthamiana (Nb). Relative Nudix hydrolase activity was calculated by comparing enzymatic activity of purified GST-Avr3b with GST produced in E. coli or purified FLAG-Avr3b with FLAG-GFP in N. benthamiana. NADH was applied as substrate in Nudix hydrolase activity assays. Means and standard errors from three replicates are shown. ** representing t test P < 0.01. (B) E. coli produced Avr3b protein can be activated by incubation with plant protein extracts. Recombinant GST-Avr3b protein was purified from E. coli. The recombinant protein (2 μg) was incubated with 100 μg dialyzed soybean (Glycine max, Gm) extract, 100 μg dialyzed N. benthamiana extract, or 100 μg dialyzed P. sojae (Ps) mycelium extract at 25°C for 15 hours. The Nudix hydrolase activity was then determined using FLAG-GFP as a control. Means and standard errors from three measurements are shown. ** or *, representing t test P < 0.01 or 0.05, respectively. Avr3b interacts with plant cyclophilins
To investigate the underlying mechanism of Avr3b activation in plant cells, we performed yeast two-hybrid (Y2H) screens and identified plant proteins that associate with Avr3b using a soybean cDNA library. One protein that was repeatedly identified from three independent screens is a soybean cyclophilin protein GmCYP1 (Gm11g10480). We then focused our research on GmCYP1 because cyclophilin was shown to activate the cysteine protease activity of a bacterial effector AvrRpt2 in Arabidopsis [33,34].
The Avr3b-GmCYP1 interaction was validated using three independent assays. First, we cloned the full length cDNA sequence of GmCYP1 into the Y2H vector pGADT7 and confirmed its association with Avr3b in yeast (Fig 2A). Secondly, we purified the recombinant GST-Avr3b, His-GmCYP1 proteins from E. coli and detected co-precipitation of His-GmCYP1 with GST-Avr3b in vitro, but not with GST, using glutathione resins (Fig 2B and S1 Fig). Finally, we confirmed the interaction between Avr3b and GmCYP1 in N. benthamiana using co-immunoprecipitation. FLAG-Avr3b was co-expressed with GFP-GmCYP1 or GFP in N. benthamiana leaves using Agrobacterium-mediated transient expression. Enrichment of GFP-GmCYP1 was detected in the FLAG-Avr3b precipitates using anti-FLAG affinity gel from total protein extracts (Fig 2C). Taken together, these experiments demonstrate that Avr3b directly interacts with GmCYP1 in vitro and in vivo.
Fig. 2. Avr3b interacts with plant cyclophilins. 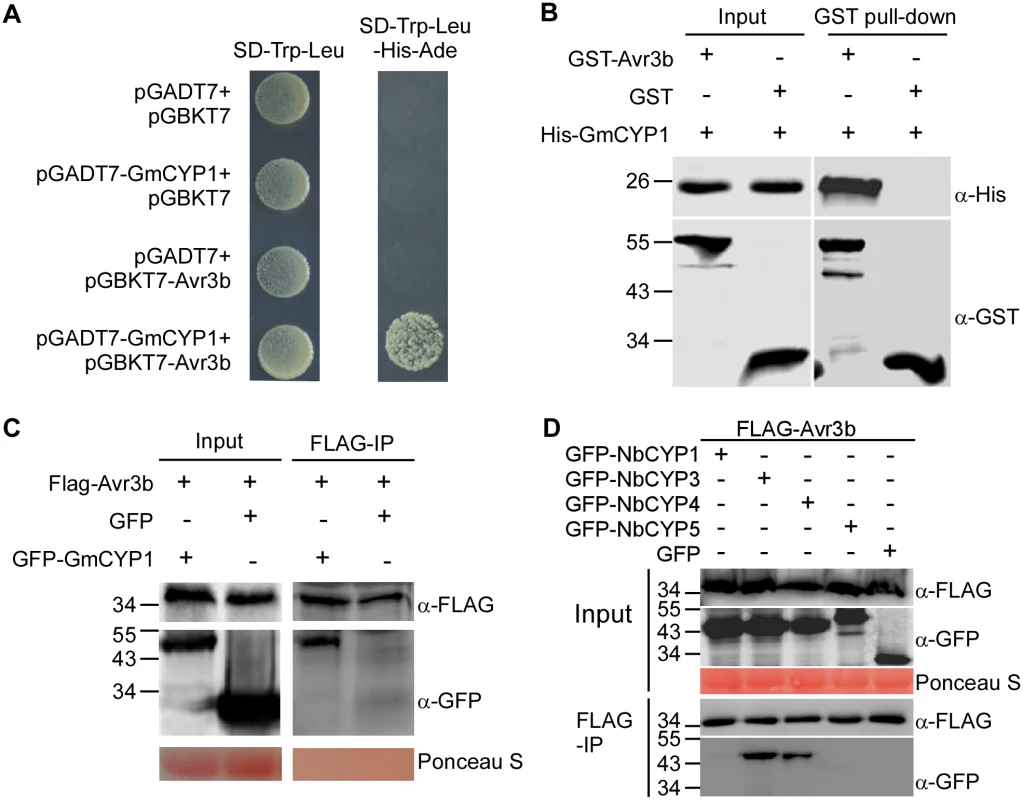
(A) Avr3b interacts with GmCYP1 in yeast. Avr3b and GmCYP1 were cloned into pGBKT7 and pGADT7 vectors, respectively. Yeast transformants were grown on SD/-Trp/-Leu (SD-2) or the selective medium SD/-Trp/-Leu/-His/-Ade (SD-4). The plates were photographed 2 days after inoculation. (B) Avr3b physically interacts with GmCYP1 in vitro. GST-Avr3b or GST bound resins were incubated with E. coli cell lysate containing His-GmCYP1. Co-precipitation of His-GmCYP1 with the GST-binding resins was examined by western blots using anti-His antibody before (Input) and after resins incubation (Pull-down). (C) Avr3b interacts with GmCYP1 in plant cells. FLAG-Avr3b was transiently expressed with either GFP-GmCYP1 or GFP in N. benthamiana leaves. FLAG-tagged Avr3b was immunoprecipitated by anti-FLAG M2 affinity gel from total plant extracts. Input controls (Input) and M2 gel binding proteins (IP) were analyzed by western blots using anti-FLAG or anti-GFP antibody. (D) Avr3b interacts with NbCYP3 and NbCYP4 in N. benthamiana. FLAG-Avr3b was transiently expressed with GFP-tagged protein fusions of NbCYPs or GFP in N. benthamiana. Immunoprecipitates pulled down using anti-FLAG M2 affinity gel from total protein extracts were immunoblotted with anti-FLAG or anti-GFP antibodies. IP, immunoprecipitate. These experiments were repeated three times with similar results. Because Avr3b could be activated by protein extract of N. benthamiana, we hypothesized that cyclophilin proteins in N. benthamiana could interact with Avr3b and activate its Nudix hydrolase activity. To test this hypothesis, we examined whether Avr3b interacts with GmCYP1 homologs in N. benthamiana. A phylogenetic tree was constructed based on the conserved cyclophilin domain of cyclophilin proteins from N. benthamiana, soybean and P. sojae (S2 Fig). Five N. benthamiana cyclophilins (NbCYP1—NbCYP5) were selected as they share the highest (74%-94%) similarity to GmCYP1 in amino acid sequences. Among them, NbCYP1 (NbC26100015g0001) and NbCYP2 (NbS00003953g0001) are identical in their full-length amino acid sequences; NbCYP3 (NbS00044621g0001) and NbCYP4 (NbS00014422g0001) also share high identity (98% identity). These NbCYPs were then examined for their interactions with Avr3b. Using in planta co-immunoprecipitation assay, we were able to detect enrichment of NbCYP3 and NbCYP4, but not NbCYP1 or NbCYP5 (NbS00058430g0005), in the FLAG-Avr3b precipitates (Fig 2D). These results demonstrate that NbCYP3 and NbCYP4 can interact with Avr3b in planta.
Taken together, these results suggest that Avr3b associates with specific cyclophilin proteins in plant cells, such as GmCYP1 in soybean and NbCYP3 and NbCYP4 in N. benthamiana.
Cyclophilin activates the hydrolase activity of Avr3b
Cyclophilins serve as protein folding catalysts by regulating cis-trans isomerization of prolyl bonds [36]. We then confirmed that GmCYP1 possesses the peptidyl-prolyl cis-trans isomerase (PPIase) activity by the chymotrypsin-coupled assay using N-Succinyl-Alanine-Proline-Phenylalanine-P-Nitroanilide (Suc-AAPF-pNA) as the alpha-chymotrypsin substrate [37]. In the presence of functional PPIase, the Suc-AAPF-pNA prolyl bond is more rapidly converted to the trans conformation, which can be cleaved by chymotrypsin, leading to the formation of a colored product 4-nitroaniline [38]. As such, in the presence of active PPIase, we would observe a rapid increase in absorbance at 390 nm. As expected, Recombinant His-GmCYP1 protein purified from E. coli showed clear PPIase activity compared to GST, which served as a negative control (Fig 3A). Based on previous studies, Arg62 of GmCYP1 might be essential for the PPIase activity [34,39], we constructed the mutant GmCYP1R62A, which indeed lost the PPIase activity but still interacted with Avr3b (Fig 3A and S3 Fig). In addition, the PPIase activity of GmCYP1 can be significantly inhibited in the presence of 20 μM cyclosporine A (CsA), a chemical inhibitor of cyclophilin (Fig 3A). These results demonstrate that GmCYP1 is a canonical cyclophilin with the PPIase activity.
Fig. 3. Activation of Avr3b is dependent on the PPIase activity of GmCYP1. 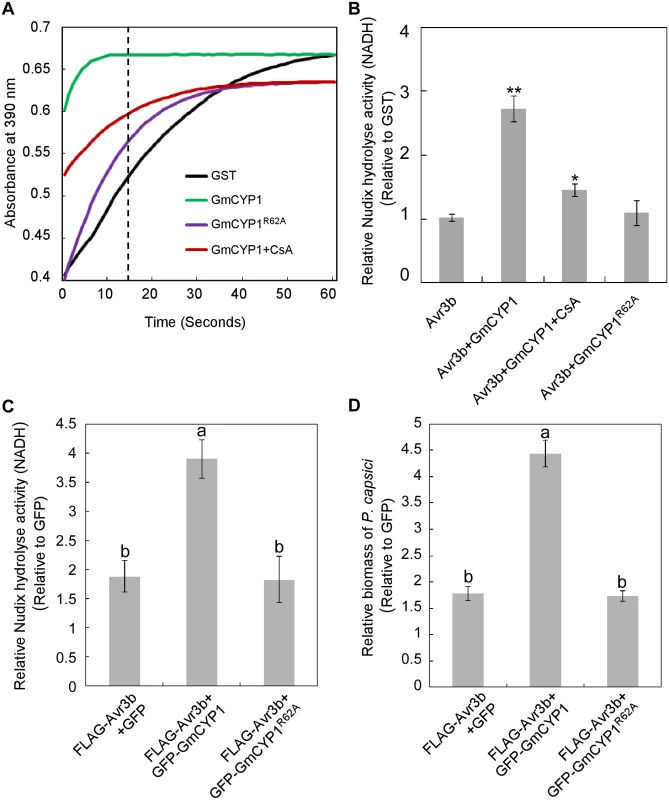
(A) GmCYP1 possesses peptidyl-prolyl cis-trans isomerase (PPlase) activity. GmCYP1R62A (PPIase-deficient mutant) was generated by site-directed mutagenesis. The PPIase enzymatic activity of GST, GmCYP1, and GmCYP1R62A produced in E. coli was analyzed using a chymotrypsin-coupled assay. Enzyme activity can be assessed by analyzing the peak of the curve 15 s (dotted line indicated) after the adding of α-chymotrypsin. A higher absorbance in 390 nm indicates increased PPIase activity. 20 μM cyclosporine A (CsA) was added to the reaction when appropriate as a chemical inhibitor of the PPIas activity. Three independent replicates were performed with similar results. (B) Avr3b can be activated by GmCYP1 in vitro. GST-Avr3b proteins produced from E. coli were incubated with purified GmCYP1or GmCYP1R62A. CsA was supplemented to the reactions when appropriate. The Nudix hydrolase activity was then determined using GST as a control. Means and standard errors from three measurements are shown. ** or *, representing significantly different than the Avr3b t test P < 0.01 or 0.05, respectively. (C) GmCYP1 enhances the Nudix hydrolase activity of Avr3b in planta. Avr3b was expressed with GFP-GmCYP1, GFP-GmCYP1R62A or GFP in N. benthamiana leaves using Agrobacterium-mediated transient expression and Nudix hydrolase activity was analyzed 48 hours post Agro-infiltration (hpi). Bars represent standard errors from four biological replicates. The same letter indicates no significant difference between values, and different letters indicate significant differences between values (P < 0.01, nonparametric Kruskal-Wallis test). (D) GmCYP1 enhances the virulence activity of Avr3b in N. benthamiana. Leaves expressing Avr3b with GFP-GmCYP1, GFP-GmCYP1R62A or GFP were inoculated with zoospore suspension of P. capsici 48 hpi. DNA from P. capsici infected regions was isolated at 36 hours post P. capsici inoculation, and the biomass of P. capsici in infected tissues was determined using quantitative PCR comparing with GFP-expressing tissues. Bars represent standard errors from three biological replicates. The letters represent statistical significance (P < 0.01 nonparametric Kruskal-Wallis test). Next, we examined whether Avr3b is activated by GmCYP1 via its PPIase activity. Purified GST-Avr3b proteins produced by E. coli were incubated with His-GmCYP1 or His-GmCYP1R62A (S4 Fig). The Nudix hydrolase activity of GST-Avr3b was significantly enhanced after incubation with His-GmCYP1, but not with His-GmCYP1R62A (Fig 3B and S1 Table). Consistently, activation of Avr3b Nudix hydrolase activity by GmCYP1 was significantly weakened in the presence of CsA (Fig 3B). These results suggest that Avr3b is activated by GmCYP1 in a manner that is dependent on PPIase activity.
To study the interaction between Avr3b and GmCYP1 in planta, Avr3b was co-expressed with GFP-GmCYP1, GFP-GmCYP1R62A or GFP in N. benthamiana leaves. N. benthamiana leaves transiently expressing Avr3b and GmCYP1 showed a higher level of Nudix hydrolase activity (Fig 3C and S1 Table) and increased susceptibility to P. capsici (Fig 3D). This difference was not due to different protein expression levels of Avr3b as shown by western blots (S5 Fig). These results suggest that GmCYP1 can promote Avr3b maturation in planta.
NbCYP3 and NbCYP4 activate Avr3b in N. benthamiana
Our previous experiments showed that NbCYP3 and NbCYP4 interact with Avr3b in planta, we next determined whether NbCYP3 and NbCYP4 can activate Avr3b processing. NbCYP3 and NbCYP4 were silenced in N. benthamiana by virus-induced gene silencing (VIGS) using Tobacco Rattle Virus (TRV)-based vectors. Because NbCYP3 and NbCYP4 are highly similar, our TRV:NbCYP3 construct effectively silenced both genes, but the transcript levels of the homologous genes, NbCYP1, NbCYP2, and NbCYP5, remained unchanged (S6A Fig). These results confirmed that the silencing construct specifically knocked down the expression of the Avr3b-interacting NbCYP3 and NbCYP4.
Plants expressing the TRV:NbCYP3/4 constructs did not show any marked phenotypic alterations compared to the control plants expressing TRV:GFP (S6B Fig). We then expressed Avr3b and RFP (as a control) in NbCYP3/4-silenced leaves through Agro-infiltration and analyzed the Nudix hydrolase activity. In contrast to the clear Nudix hydrolase activity detected in leaves expressing TRV:GFP, Avr3b did not exhibit any activity in NbCYP3/4 - silenced leaves (Fig 4A). This is not due to different protein expression levels of Avr3b in these leaves (S6C Fig). Next, we infected these leaves with the P. capsici. In leaves that transiently expressed Avr3b, we observed significantly enlarged lesions in the TRV:GFP plants; however, this virulence activity of Avr3b was completely abolished in NbCYP3/4-silenced leaves (Fig 4B). From these experiments, we conclude that silencing of NbCYP3 and NbCYP4 disrupted the activation of Avr3b in N. benthamiana. To verify whether NbCYP3 and NbCYP4 are responsible for activating the Nudix hydrolase activity of Avr3b, we co-expressed Avr3b with NbCYP3, NbCYP4 or GFP in N. benthamiana. Over-expression of NbCYP3 and NbCYP4 in N. benthamiana led to further enhancement on the Nudix hydrolase activity of Avr3b (Fig 4C), supporting a role of NbCYP3 or NbCYP4 in Avr3b activation in N. benthamiana.
Fig. 4. NbCYP3 and NbCYP4 are required for the Nudix hydrolase and the virulence activity of Avr3b in N. benthamiana. 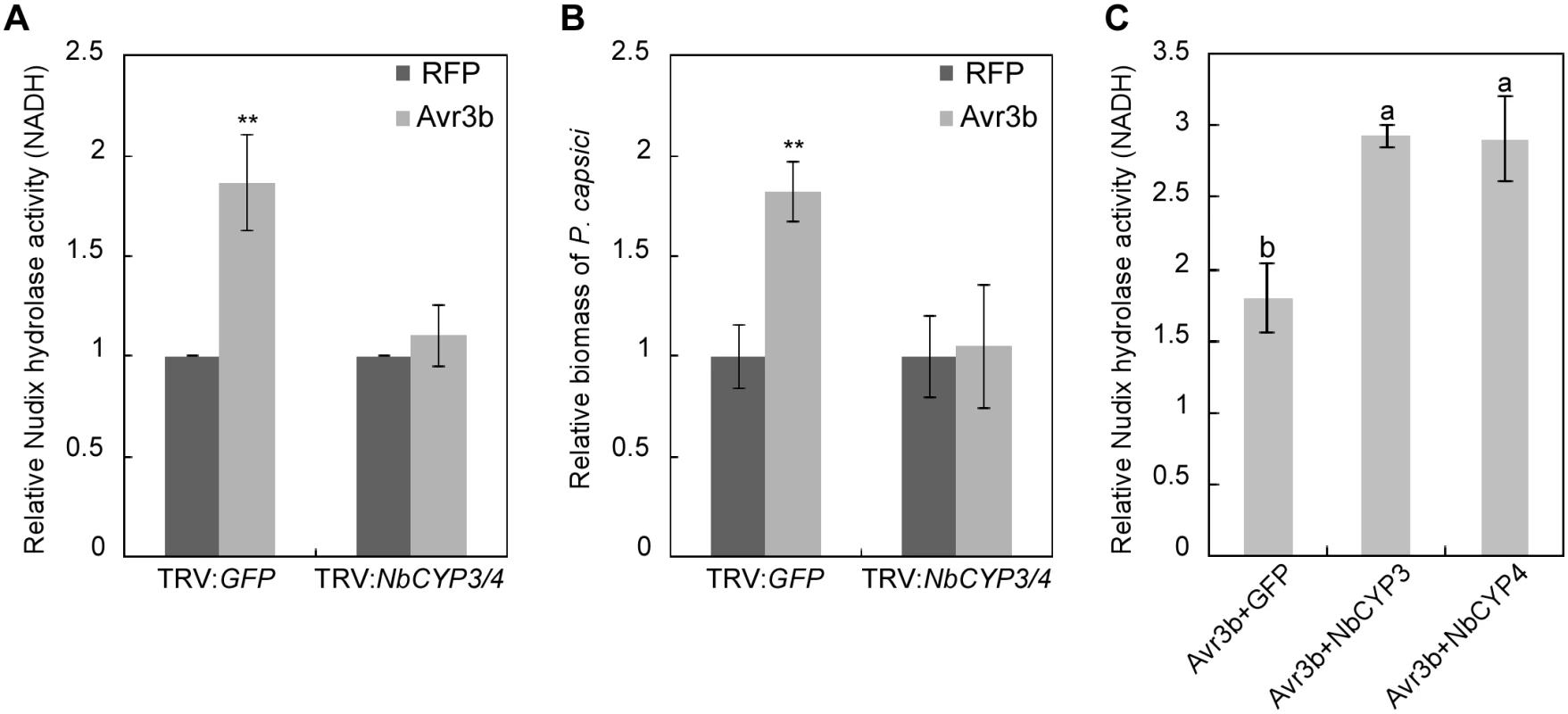
(A) NbCYP3/4-silenced N. benthamiana can not activate the Nudix hydrolase activity of Avr3b. Avr3b and RFP (as a negative control) were transiently expressed in NbCYP3/4-silenced N. benthamiana leaves. Total proteins were extracted at 48 hpi, and the Nudix hydrolase activity was measured using NADH as substrate. Means and standard errors from four measurements are shown. **, t test P<0.01. (B) NbCYP3 and NbCYP4 are required for the virulence activity of Avr3b in N. benthamiana. FLAG-RFP or FLAG-Avr3b proteins were transiently expressed in N. benthamiana leaves by Agro-infiltration. The leaves were inoculated with P. capsici at 48 hours post Agro-infiltration. Infection was determined using quantitative PCR to measure the ratios of P. capsici and N. benthamiana DNA at 36 hours post inoculation. Means and standard errors from four measurements are shown. ** representing t test P < 0.01. (C) NbCYP3 and NbCYP4 can enhance the Nudix hydrolase activity of Avr3b in N. benthamiana. FLAG-Avr3b was co-expressed in N. benthamiana leaves with GFP-NbCYP3 or GFP-NbCYP4 by Agro-infiltration method, and the Nudix hydrolase activity was measured using NADH as substrate comparing with GFP-expressing leaves. Bars represent standard errors from three biological replicates. The same letter indicates no significant difference between values, and different letters indicate significant differences between values (P < 0.01, nonparametric Kruskal-Wallis test). PPIase activity of GmCYP1 is required for Avr3b-induced cell death in soybean
In soybean, resistance to P. sojae strains expressing Avr3b is conferred by the resistance protein Rps3b [19]. To test the role of GmCYP1 in Avr3b-induced cell death in resistant cultivars of soybean, CsA was used to suppress the PPIase activity in soybean leaves, which were subsequently tested for Avr3b-triggered HR. Our results showed that transient expression of Avr3b in Rps3b-producing soybean could induce cell death, which was readily blocked by spraying 20 μM CsA on the leaves (Fig 5A and 5B). To rule out the possibility that application of CsA inhibits general cell death in soybean, another P. sojae effector Avr1b was tested using soybean carrying its cognate R protein Rps1b [40]. The same CsA treatment did not disturb Avr1b-triggered cell death in Rps1b-producing soybean, indicating that CsA specifically suppressed Avr3b-triggered cell death (Fig 5C and 5D). These results suggest that the PPIase activity is important for the recognition of Avr3b, but not a general factor of ETI, in soybean.
Fig. 5. Avr3b recognition in soybean is dependent on the PPlase activity of GmCYP1. 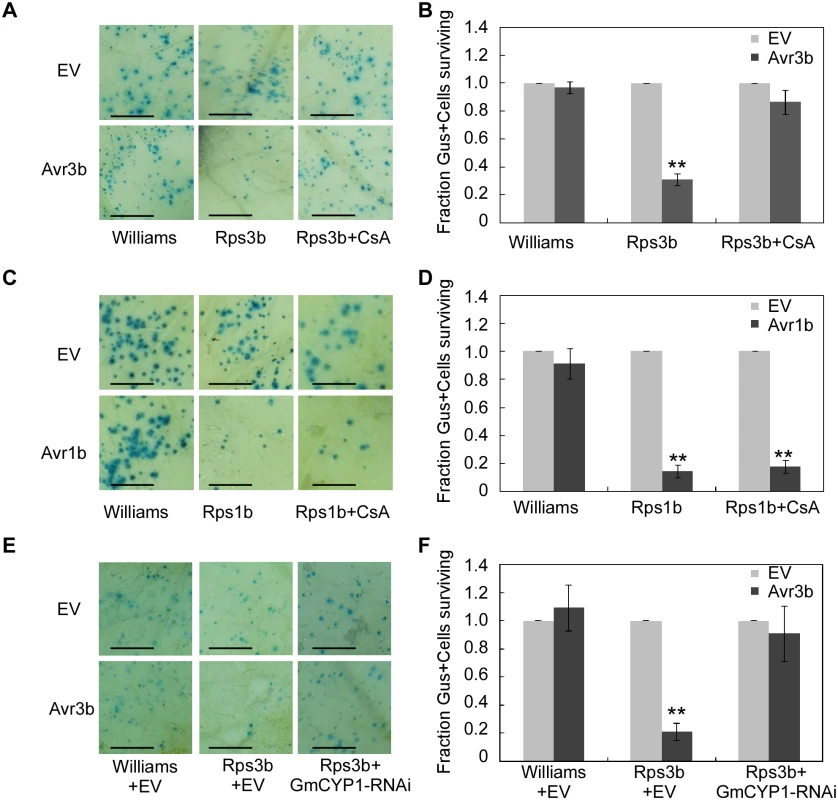
(A, B) CsA suppresses Avr3b-triggered cell death in soybean leaves. (A) Soybean cultivars Williams (rps3b) and PRX146-36 (Rps3b) were transformed by co-bombardment with a plasmid mixture consisting of a β-glucuronidase (GUS) expression vector and Avr3b or empty vector (EV). Soybean leaves were sprayed 20 μM CsA solution immediately after bombardment and incubated for 2 days in darkness at 28°C. The leaves were then stained using X-gluc. Scale bars, 3mm. (B) Percentage rate of GUS-positive blue spots following bombardment with Avr3b compared with EV. Bars represent standard errors from four independent replicates. **, t test P<0.01. (C, D) CsA could not suppress Avr1b-triggered cell death. (C) Avr1b was transiently expressed in soybean cultivars Williams (rps1b) and L77-1863 (Rps1b) by co-bombardment with GUS expression vector and Avr1b or EV. The leaves were incubated for 2 days in darkness at 28°C after bombardment, and then stained using X-gluc. Scale bars, 3mm. (D) Percentage rate of GUS-positive blue spots following bombardment with Avr1b compared with the empty vector. Bars represent standard errors from four independent replicates. **, t test P<0.01. (E, F) Avr3b failed to induce Rps3b-mediated cell death in GmCYP1-silenced soybean leaves. (E) Avr3b was delivered into leaves of soybean cultivars Williams (rps3b) and PRX146-36 (Rps3b) together with a GmCYP1-RNAi construct by co-bombardment. The leaves were incubated for 2 days in darkness at 28°C after bombardment, and then stained using X-gluc. Scale bars, 3mm. EV was used as a negative control. (F) Percentage rate of GUS-positive blue spots following bombardment with Avr3b compared with the EV in the presence of GmCYP1-RNAi. Bars represent standard errors from four independent replicates. **, t test P<0.01. To further investigate the role of GmCYP1 in Rps3b-mediated cell death in soybean, a hairpin RNAi construct targeting GmCYP1 was introduced into soybean leaves together with Avr3b by co-bombardment (S7A Fig). Quantitative RT-PCR data showed that GmCYP1 transcript level was significantly reduced in the delivery region two days after bombardment (S7B Fig). On the contrary, the transcript levels of another two soybean cyclophilin genes, Gm04g00700 and Gm06g00740, both of which share high sequence identity (83% and 84% in full-length nucleic acid sequences respectively) with GmCYP1, were not reduced in leaves expressing the GmCYP1-RNAi construct (S7B Fig). The cell death assay indicated that Avr3b-triggered cell death was weakened in soybean when GmCYP1 was silenced. (Fig 5E and 5F). Taken all the data together, our experiments demonstrate that GmCYP1 is required for the recognition of Avr3b by Rps3b, likely by modulating Avr3b protein structure via the PPIase activity.
Pro132 of Avr3b is required for association with GmCYP1
In general, cyclophilins preferentially bind to the Glycine-Proline (GP) motif in substrate proteins and catalyze prolyl bond isomerization [41,42]. Sequence analysis identified a potential GP motif (G131P132) in Avr3b (S8 Fig). To examine whether this putative GP motif is involved in the interaction between Avr3b and GmCYP1, we generated the mutant Avr3bP132A and examined its Nudix hydrolase activity when expressed in N. benthamiana. Western blot data showed that Avr3b and the mutant Avr3bP132A was expressed in a similar level (S9A and S9B Fig). Proteins extracted from tissues expressing Avr3bP132A showed the same low level of Nudix hydrolase activity as those extracted from GFP-expressing tissues, indicating that Avr3bP132A was no longer activated (Fig 6A). We then determined that the interaction between GmCYP1 and Avr3bP132A was significantly weakened in yeast (Fig 6B and S9C Fig), consistent with the notion that Pro132 plays a key role in mediating Avr3b-GmCYP1 interaction. Overall, these results demonstrated that Pro132 of Avr3b is required for Avr3b maturation, probably through direct interaction with GmCYP1.
Fig. 6. Proline132 of Avr3b plays a critical role in activation by cyclophilins. 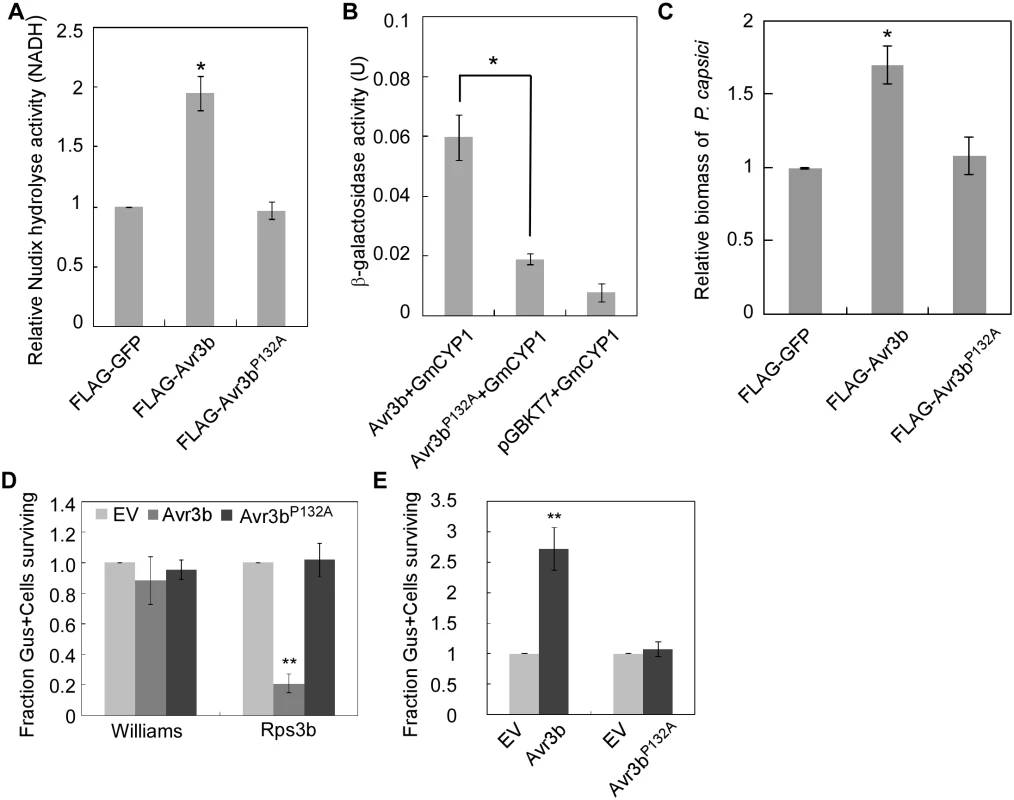
(A) Avr3bP132A mutant can not be activated in N. benthamiana. FLAG-GFP, FLAG-Avr3b or FLAG-Avr3bP132A were expressed in N. benthamiana leaves. Relative Nudix hydrolase activity was analyzed at 48 hpi. Bars represent standard errors from three independent replicates. * representing significantly different than FLAG-GFP (t test P<0.05). (B) Avr3bP132A is impaired in its interaction with GmCYP1. Yeast cells were co-transformed with pGADT7-GmCYP1 and pGBKT7 carrying Avr3b or Avr3bP132A. Transformants were grown on a selective medium.β-galactosidase activity in yeast cells was measured to determine the Avr3b-GmCYP1 interaction. Bars represent standard errors from three independent replicates. *, t test P<0.05. (C) The P132A mutation of Avr3b resulted in loss of virulence activity. FLAG-GFP, FLAG-Avr3b or FLAG-Avr3bP132A proteins were transiently expressed in N. benthamiana leaves by Agro-infiltration. The leaves were inoculated with P. capsici at 48 hours post Agro-infiltration. Biomass of P. capsici was determined at 36 hours post inoculation by quantitative PCR. Bars represent standard errors from three independent replicates. *, t test P<0.05. (D) Avr3bP132A failed to induce Rps3b-mediated cell death in soybean. Percentage rate of β-glucuronidase (GUS)-positive blue spots following bombardment with Avr3bP132A compared with the EV. Bars represent standard errors from four independent replicates. **, t test P<0.01. (E) Avr3bP132A could not suppress Avr1b-triggered HR in soybean producing Rps1b. Percentage rate of GUS-positive blue spots following co-bombardment of Avr1b with Avr3b or Avr3bP132A compared with EV. Data are the means of four independent experiments. Bars represent standard errors from four independent replicates. **, t test P<0.01. To further detect the effect of GmCYP1 on the biological functions of Avr3b in planta, Avr3b and Avr3bP132A were expressed in N. benthamiana, and the detached leaves were subsequently challenged with P. capsici. P. capsici produced larger lesions on Avr3b-expressing leaves than on Avr3bP132A-expressing leaves (S9D Fig). Quantitative PCR confirmed a higher biomass of P. capsici in Avr3b-expressing leaves, suggesting that Avr3bP132A was unable to promote P. capsici infection (Fig 6C).
Regarding to the avirulence role of Avr3b during soybean-P. sojae interaction, we tested whether Avr3bP132A could be recognized by soybean Rps3b plant. Transient expression of Avr3bP132A by bombardment could not induce Rps3b-mediated cell death on soybean leaves (Fig 6D and S9E Fig). Previous studies have shown that Avr3b suppresses cell death induced by interactions between Avr1b and Rps1b [19]; thus, we examined whether Avr3bP132A still retains this virulence function using bombardment. Our data showed that, unlike Avr3b, Avr3bP132A was unable to suppress Avr1b-triggered cell death in soybean (Fig 6E and S9F Fig). These results provide convincing evidence that Pro132 is essential for both avirulence and virulence functions of Avr3b, probably by mediating the interaction of Avr3b with GmCYP1.
Avr3b specifically interacts with plant cyclophilins, but not with Phytophthora CYP homologs
Cyclophilins belong to a large protein family that is present in all cell types of all the organisms studied so far [43]. Previous results showed that Avr3b could not be activated by P. sojae extracts (Fig 1B), suggesting that Avr3b may not interact with or be processed by P. sojae cyclophilins. By searching the genomes of soybean and P. sojae, we found 17 cyclophilin homologues in P. sojae and 72 cyclophilin homologues in soybean (S2 Table). To test whether P. sojae cyclophilins could interact with Avr3b, we cloned the genes Ps108795 and Ps108195, which are the most similar to GmCYP1 (79% and 60% identity in full-length amino acid sequences), for Y2H analysis. Our data showed that neither of these P. sojae cyclophilins could interact with Avr3b in yeast (Fig 7A and 7B). This result supports a hypothesis that Avr3b is likely present as an inactive form before entering the plant cells.
Fig. 7. Avr3b specifically interacts with GmCYP1 in soybean. 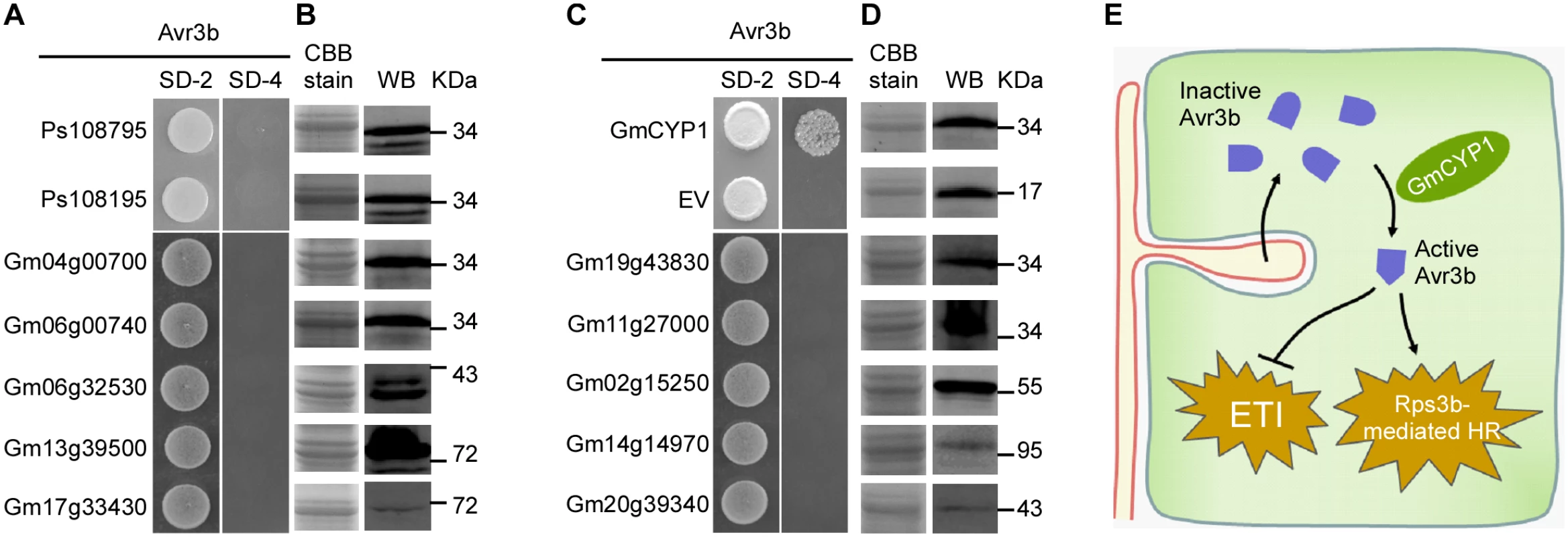
(A, C) Avr3b does not interact with other GmCYP1 homologs in soybean or two CYPs of P. sojae. Selected cyclophilin genes were cloned into pGADT7. Yeast cells were co-transformed with the empty prey vector, pGADT7 or pGADT7 containing one of homologue genes and the bait vector pGBKT7-Avr3b. Yeast transformants were grown on the selective minimal SD medium lacking tryptophan, and leucine (SD-2) or the selective SD medium lacking adenine, tryptophan, histidine, and leucine (SD-4). The plates were photographed 2 days after inoculation. (B, D) Western blots showing protein expression of cyclophilin homologs in yeast cells. Anti-HA antibody was used to detect the expression of cyclophilin homologous proteins of soybean and P. sojae. The same protein gel was stained with Coomassie brilliant blue (CBB) to show loading. WB, western blot. (E) A schematic summary illustrating maturation of Avr3b by plant cyclophilin proteins during Phytophthora infection. Inactive Avr3b enters plant cell and interacts with plant cyclophilin to gain the Nudix hydrolase activity. Active Avr3b suppresses ETI-associated cell death triggered by Avr1b. However, in the presence of Rps3b, active Avr3b triggers Rps3b-induced HR. GmCYPs have diverse gene structures and subcellular localizations, indicating that they involve in a large variety of cellular functions [44]. Since GmCYP1 is predicted as a cytosolic cyclophilin [44], we then selected ten soybean cyclophilins including other cytosolic cyclophilins that share high sequence similarity with GmCYP1 for interaction analysis with Avr3b. Interestingly, none of these tested GmCYPs could interact with Avr3b in yeast (Fig 7A–7D). These results suggest that the enzymatic activity of Avr3b is specifically activated by GmCYP1 in soybean.
Discussion
In this study, we report that a soybean cyclophilin protein GmCYP1 physically interacts with the Phytophthora effector Avr3b and activates the Nudix hydrolase activity of Avr3b, which is required for the full virulence of P. sojae. GmCYP1 possesses the PPIase activity that can be suppressed by the chemical inhibitor CsA. Application of CsA or mutation of catalytic residue Arg62 of GmCYP1 abolished its activating activity on Avr3b, suggesting that maturation of Avr3b in plant cells is dependent on PPIase activity of GmCYP1. Moreover, we identified Pro132 of Avr3b as a key residue to mediate the interaction with GmCYP1. In planta expression of Avr3bP132A lost its hydrolase activity and is no longer able to enhance Phytophthora infection when expressing in N. benthamiana. The mutation of Pro132 also abolished Avr3b-induced HR in Rps3b-producing soybean.
Interestingly, we found Avr3b Nudix hydrolase activity can be activated by both N. benthamiana and soybean extracts, but not P. sojae extracts. Since the activation of Avr3b by N. benthamiana extract could be largely impaired by CsA treatment, N. benthamiana likely provides “helper” molecule(s) possessing PPIase activity for Avr3b activation (S10 Fig). Our experiments further confirmed that two N. benthamiana cyclophilins, i.e. NbCYP3 and NbCYP4, and one soybean cyclophilin, i.e. GmCYP1, are likely the “helpers” that activate Avr3b in plant cells.
One outstanding question is whether other GmCYP1 homologs can process Avr3b. Our large scale yeast two hybrid assays testing interaction between Avr3b and GmCYP1 homologs suggest that Avr3b specifically interacts with GmCYP1, indicating substrate specificity in cyclophilin homologs in soybean (Fig 7A–7D). This is consistent with previous reports that cyclophilins interact and process different classes of proteins through sequence-specific binding and diverse localization [41,43,45,46].
Previous studies showed that cyclophilins can regulate the protease activity of effector AvrRpt2 from bacteria, conformational change of plant ETI receptor RPM1-interacting protein RIN4, and the binding activity of HCV RNA polymerase NS5B through their PPIase activity [31,33,35]. Our experiments showed that cyclophilins are also associated with the activation of Nudix hydrolase, and that GmCYP1 activates Avr3b in a PPIase-dependent manner. However, whether cis-trans isomerization of Avr3b protein actually occurs during Phytophthora infection requires further investigation.
To our surprise, the site direct Avr3bP132A mutation not only impaired Avr3b Nudix hydrolase activity but also abolished the Avr3b triggered immunity, suggesting Pro132 is a key residue associated with Rps3b recognition. Interestingly, the investigation of P. sojae natural populations uncovered only two Avr3b natural alleles, namely the avirulence allele Avr3bP6497 and the virulence allele Avr3bP7076. Sequence analysis of both alleles revealed a mutation on the Pro132 site in the avirulence sequence into an Alanine at the corresponding position in virulence allele Avr3bP7076 (S11 Fig). This natural mutant and the phenotype of the Avr3bP132A mutant constructed in this study suggest that Pro132 is a key residue in Avr3b recognition event. On the other hand, our previous study indicated that the Avr3bQQQQ mutant (Avr3bR220Q, E221Q, E225Q, E226Q, a non-functional Nudix hydrolase mutant) could still trigger cell death in soybean, suggesting that Avr3b recognition by the cognate Rps3b receptor is independent on the Nudix hydrolase activity [19]. A possible explanation of these results is that P132 is required for not only the Nudix hydrolyase activity, but also other feature of Avr3b, such as protein conformation changes, which may be required for Avr3b-Rps3b recognition.
In addition to P. sojae, Avr3b-like effectors are extensively present in other Phytophthora species including P. infestans, P. ramorum and P. capsici. Sequence alignment of Avr3b-like effectors indicated that the GP motif containing Pro132 is not conserved among these effectors, although other potential GP motifs are present in a few other effectors (S11 Fig). These data suggested that effector processing by plant cyclophilin proteins might not be required for all the Phytophthora Nudix effectors.
It has been widely accepted that pathogen effectors directly target host proteins and manipulate host targets [47]. However, host proteins also manipulate the activity of pathogen effectors. It has been proposed that some host proteins could act as “helpers” to facilitate effector functions, whereas others are “targets” [4]. Previous studies demonstrated that folding of bacterial effector facilitated by host factors might be important to regulate effector functions in host cells during pathogenesis. Previous studies showed that Arabidopsis cyclophilin ROC1 is required to activate the cysteine protease activity of the bacterial effector AvrRpt2 [33,34]. Our study demonstrated that a soybean cyclophilin GmCYP1 is responsible for the activation of a Phytophthora effector Avr3b as a Nudix hydrolase (Fig 7E). In another word, GmCYP1 is the host helper recruited by Avr3b to become an active virulence protein in plant cells. These results suggest that recruitments of host factors as “helpers” is a common pathogenesis mechanism shared by eukaryotic and prokaryotic pathogens.
Materials and Methods
Plant and microbe cultivation
Plants were grown in greenhouse at 22–25°C with a cycle of 16 and 8 hours of high light intensity and darkness, respectively. P. sojae isolate P6497 and P. capsici used in this study were grown in 10% vegetable (V8) juice agar medium at 25°C in the dark.
Yeast two-hybrid screens
Avr3b gene without the secretion signal was cloned into the yeast vector pGBKT7 (Clontech). The expression of the BD-Avr3b fusion proteins has been verified by western blots (S12 Fig). A soybean (Glycine max, William 82) cDNA library was constructed in the Y2H vector pGADT7 using total RNA extracted from soybean hypocotyl tissues collected 12 and 24 hours after the plants were inoculated with P. sojae zoospores (Clontech). Approximately 6×106 primary yeast clones (three times coverage of the library) were screened using Avr3b as the baits. Potential yeast transformants containing cDNA clones interacting with Avr3b were selected using the SD/-Trp/-Leu/-His/-Ade selective medium.
In vitro GST pull-down assays
To construct GST-fusion plasmids, Avr3b was inserted into the vector pGEX4T-2 (GE Healthcare Life Science). To construct His-fusion plasmid, GmCYP1 was inserted into the vector pET28a. Pull-down assay was performed using ProFound pull-down GST protein-protein interaction kit (Pierce) according to the manufacturer’s instructions. GST, GST-Avr3b, and His-GmCYP1 was expressed in E. coli strain BL21 (DE3) respectively. The soluble total proteins were incubated with 50 μl glutathione agarose beads (Invitrogen) for 2 hours at 4°C. The beads were washed five times and then incubated with equal amount of bacterial lysates containing His-GmCYP1 for another hour at 4°C. The beads were washed five times again, and the presence of His-GmCYP1 was detected by western blot using anti-His antibody.
Nudix hydrolase activity assays
To analysis Nudix hydrolase activity of Avr3b, a general approach was used as described [23]; Reaction mixture (50 μl for each sample) contained 50 mM Tris-HCl, pH 8.5, 1 mM dithiothreitol, 5 mM MgCl2, 2 mM substrate (NADH), 2 units of calf alkaline phosphatase, and 2 μg of protein samples. After incubation for 30 min at 37°C, the reaction was stopped by adding 150 μl of 0.5 M H2SO4, followed by addition of 100 μl of water. For color development, 700 μL of a freshly made mixture containing 600 μl of 0.42% (w/v) ammonium molybdate and 100 μL of 10% (w/v) ascorbic acid was used. The reaction tubes were incubated in 45°C water bath for 20 min for color development and then cooled down to room temperature. The solutions were measured using spectrophotometer (Beckman, USA) at A820. The reaction mixture without protein was used as a blank control. For Nudix hydrolase activity assay of total plant protein extract, one N. benthamiana leaf was infiltrated with FLAG-GFP bacteria in one side and with FLAG-Avr3b or FLAG-Avr3bP132A in another side. A total of 0.2 g of GFP or Avr3b infiltrated leaf tissue was collected at 2 days post infiltration (dpi). For both kinds of protein preparations, the relative hydrolase enzyme activity was calculated as the ratio of A820 reading value from sample mixture over control mixture.
Western blot and co-immunoprecipitation assays
A standard SDS-PAGE protocol was performed for protein separation. Proteins were transferred onto polyvinylidene difluoride (PVDF) membranes using a semi-wet apparatus (Bio-Rad, Hercules, CA,U.S.A.). Then, the membrane was blocked using phosphate-buffered saline (PBS; pH 7.4) with 3% nonfat dry milk for 1 hour at room temperature. Antibodies were added to PBST with 3% nonfat dry milk (PBSTM) at a ratio of 1 : 4,000 and incubated at 4°C overnight, followed by three washes (10 min each) with PBST. Then, the membrane was incubated with a goat anti-mouse IRDye 800CW (Odyssey,number 926–32210; Li-Cor, Lincoln, NE, U.S.A.) at a ratio of 1 : 10,000 in PBSTM at room temperature for 2 hours. The membrane was washed four times (10 min each) with PBST, then visualized using Odyssey scanner with excitation at 700 and 800 nm [11].
Avr3b with a N-terminal FLAG-tag was inserted into pGR107, and GmCYP1 was inserted into pBinGFP for expression in N. benthamiana. The Avr3bP132A and GmCYP1R62A mutants were generated by overlapping PCR using primer described in (S3 Table). FLAG-tagged Avr3b and GFP-tagged GmCYP1 or GFP were co-expressed in N. benthamiana leaves by agroinfiltration. FLAG-tagged proteins were immunoprecipitated by anti-FLAG M2 affinity gel (Sigma-Aldrich) from total extracts harvested from N. benthamiana leaves at 2 dpi. The interacting GFP fusion proteins were detected as described above. The expression of FLAG - or GFP-tagged protein in total extract was confirmed by western blot using either anti-FLAG or anti-GFP antibody.
Recombinant protein expression and purification
Recombinant Avr3b and mutants were overexpressed in E. coli strain BL21 (DE3). E. coli clones were grown in LB medium to a density of OD600 = 0.5, and then protein expression was induced overnight at 18°C with 0.4 mM isopropyl β-D-thiogalactopyranoside (IPTG). Cells were lysed in a buffer containing 100 mM Tris-HCl, 150 mM NaCl, 1mM imidazole, 1 mM DTT (pH 8.0). Recombinant GST-Avr3b were affinity purified by Glutathione Sepharose affinity chromatography (GE Healthcare) and eluted from the Glutathione Sepharose with 10 mM reduced glutathione in 100mM HEPES, 150 mM NaCl, 2 mM DTT, and 10% glycerol (pH 7.5). Recombinant CYP1 or mutant proteins were expressed E. coli BL21(DE3) by pET28a and isolated under native conditions. E. coli clones were grown in LB to a density of OD600 = 0.6. Protein expression was induced for 6 h at 28°C with 0.5 mM IPTG. Cells were lysed in a buffer containing 10 mM imidazole, 20 mM Tris pH 8.0, 150 mM NaCl. Proteins were purified by Ni+-sepharose affinity chromatography and washed with the same buffer containing 50 mM imidazole. The proteins were eluted with 250 mM imidazole in the same buffer.
PPIase assays
PPIase assays were conducted essentially as previously described [28]. The mixture of following components were incubated on ice for 10 min: 975 μl HEPES Buffer (35 mM HEPES, 0.015% TritonX-100, pH 8.0), 20 μl of 5 mM N-succinyl-ala-ala-pro-pNa (Baychem), and 8nM of His-GmCYP1 or the GST negative control. Each sample was placed in a spectrophotometer pre-cooled to 8°C. After the addition of 10 mM alpha-chymotrypsin (Sigma), the absorbance at 390 nm was recorded every second for one min at 8°C immediately.
Avr3b activation reactions
To test the activation of Avr3b proteins by soybean extract, the recombinant Avr3b protein (2 μg) produced and purified from E. coli was incubated with 100 μg of dialyzed soybean extract, 100 μg of dialyzed N. benthamiana extract or 100 μg of dialyzed P. sojae (P6497) mycelium extract respectively in a total volume of 50 μl in buffer containing 50 mM HEPES, 10 mM sodium bisulfite, 10 mM sodium metabisulfite and 1mM DTT (pH 7.5) at 25°C for 15 hours. To assess the activation of Avr3b by GmCYP1, purified recombinant Avr3b (2 μg) was incubated with 1 μg of E. coli expressed recombinant GmCYP1 protein in a total volume of 50 μl. For inhibition of PPIase activity, 20 μM of cyclosporine A (Sigma) was used in each reaction.
Virulence assay of Phytophthora on N. benthamiana
4 to 5 week old N. benthamiana were infiltrated with Agrobacterium carrying FLAG-GFP, FLAG-Avr3b or Avr3b mutant. The infiltrated leaves were inoculated with zoospore suspension of P. capsici 48 hours post Agro-infiltration. The total DNA isolated at 36 hours post inoculation. Then primers specific for P. capsici and N. benthamiana actin genes were used to quantify the relative biomass of pathogen by quantitative PCR (S3 Table). PCR reactions were performed on an ABI Prism 7500 Fast real-time PCR System (Applied Biosystems, Foster City, CA, U.S.A.).
Silencing of GmCYP1 in soybean
The GmCYP1 hairpin constructs were prepared in a two-step cloning process in pFF19::CHSA plasmid. The chalcone synthase (CHSA) intron was amplified from plasmid pFGC5941. The CHSA intron amplicon was inserted into plant transient expression plasmid pFF19::GUS (replacing GUS with CHSA intron, creating pFF19::CHSA). The PCR-amplified GmCYP1 cDNA fragments were first insert at outer restriction site (BamHI) and then the inner restriction site (AscI) on either side of the intron sequence. The construct were used by bombardment to induce transiently silencing GmCYP1 in soybean.
VIGS of cyclophilins in N. benthamiana
We used the Tobacco Rattle Virus (TRV)-based VIGS system, which uses bipartite sense RNA1 and RNA2 viruses [48], to silence cyclophilin genes in N. benthamiana. The NbCYP3 fragment was amplified by PCR and then cloned into pTRV2 vector. Primer pairs TRVNbCYP3F and TRVNbCYP3R (S3 Table) were used to amplify NbCYP3. At four-leaf stage, the N. benthamiana plants were selected for Agro-infiltration. Prior to Agro-infiltration, A. tumefaciens strain GV3101 cells carrying pTRV1 and pTRV2 constructs were collected and resuspended in infiltration buffer (10 mM MgCl2, 150 mM acetosyringone, and 10 mM MES, pH 5.6) and mixed in a 1 : 1 ratio. Plants were grown for 2 weeks after infiltration. The fully expanded six leaves of the silenced plants were then used for inoculation and quantitative PCR assays. TRV2:GFP vector was used as the negative control.
Particle bombardment assay
Avr3b, Avr3bP132A and Avr1b genes were amplified using specific primers without signal peptide sequences and inserted into plant transient expression plasmid pFF19-GUS (replacing GUS with the test genes). The double-barreled particle bombardment assays was performed on leaves to deliver a parallel control shot in every case that contained GUS DNA plus EV DNA as described [9]. For each paired shot, Specific cell death activity was calculated as the ratio of blue spot numbers for various test gene constructs compared to that of the control. After bombardment, the leaves were incubated for 2 days in darkness at 28°C. The leaves were then stained for 12 h at 37°C using 0.8 mg/mL X-gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid, cyclohexylammonium salt), 80 mM Na phosphate, pH 7.0, 0.4 mM K3Fe(CN)6, 0.4 mM K4Fe(CN)6, 8 mM Na2EDTA, and 0.1% (v/v) Triton X-100 and then de-stained in 100% ethanol.
Bioinformatics
Nudix hydrolase homologs used in this study are acquired as described [19]. The P. sojae and soybean homolog searches were performed at the Joint Genome Institute database (http://genome.jgi-psf.org) and soybean genome (http://www.phytozome.net/soybean.php). For N. benthamiana genome searching, the genome sequence was downloaded from the N. benthamiana database (http://bti.cornell.edu/research/projects/nicotiana-benthamiana/) and a local BLAST search was conducted. Protein domain and motif analyses were conducted using the NCBI conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) and Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan). Sequence alignment was performed using BioEdit2005.
Supporting Information
Zdroje
1. Jones JD, Dangl JL The plant immune system. Nature. 2006; 444 : 323–329. 17108957
2. Zipfel C Plant pattern-recognition receptors. Trends Immunol. 2014; 35 : 345–351. doi: 10.1016/j.it.2014.05.004 24946686
3. Dodds PN, Rathjen JP Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010; 11 : 539–548. doi: 10.1038/nrg2812 20585331
4. Win J, Chaparro-Garcia A, Belhaj K, Saunders DG, Yoshida K, Dong S, et al. Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb Symp Quant Biol. 2012; 77 : 235–247. doi: 10.1101/sqb.2012.77.015933 23223409
5. Bozkurt TO, Schornack S, Banfield MJ, Kamoun S Oomycetes, effectors, and all that jazz. Curr Opin Plant Biol. 2012; 15 : 483–492. doi: 10.1016/j.pbi.2012.03.008 22483402
6. Okmen B, Doehlemann G Inside plant: biotrophic strategies to modulate host immunity and metabolism. Curr Opin Plant Biol. 2014; 20 : 19–25. doi: 10.1016/j.pbi.2014.03.011 24780462
7. Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, et al. Metabolic priming by a secreted fungal effector. Nature. 2011; 478 : 395–398. doi: 10.1038/nature10454 21976020
8. Liu T, Song T, Zhang X, Yuan H, Su L, Li W, et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat Commun. 2014; 5 : 4686. doi: 10.1038/ncomms5686 25156390
9. Dou D, Kale SD, Wang X, Chen Y, Wang Q, Jiang RH, et al. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell. 2008; 20 : 1118–1133. doi: 10.1105/tpc.107.057067 18390593
10. Wang Q, Han C, Ferreira AO, Yu X, Ye W, Tripathy S, et al. Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell. 2011; 23 : 2064–2086. doi: 10.1105/tpc.111.086082 21653195
11. Yin W, Dong S, Zhai L, Lin Y, Zheng X, Wang Y The Phytophthora sojae Avr1d gene encodes an RxLR-dEER effector with presence and absence polymorphisms among pathogen strains. Mol Plant Microbe Interact. 2013; 26 : 958–968. doi: 10.1094/MPMI-02-13-0035-R 23594349
12. Yu X, Tang J, Wang Q, Ye W, Tao K, Duan S, et al. The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 2012; 196 : 247–260. doi: 10.1111/j.1469-8137.2012.04241.x 22816601
13. Zheng X, McLellan H, Fraiture M, Liu X, Boevink PC, Gilroy EM, et al. Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. Plos Pathogens. 2014; 10: e1004057. doi: 10.1371/journal.ppat.1004057 24763622
14. King SR, McLellan H, Boevink PC, Armstrong MR, Bukharova T, Sukarta O, et al. Phytophthora infestans RXLR Effector PexRD2 Interacts with Host MAPKKK{varepsilon} to Suppress Plant Immune Signaling. Plant Cell. 2014.
15. Bozkurt TO, Schornack S, Win J, Shindo T, Ilyas M, Oliva R, et al. Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc Natl Acad Sci U S A. 2011; 108 : 20832–20837. doi: 10.1073/pnas.1112708109 22143776
16. McLellan H, Boevink PC, Armstrong MR, Pritchard L, Gomez S, Morales J, et al. An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. Plos Pathogens. 2013; 9: e1003670. doi: 10.1371/journal.ppat.1003670 24130484
17. Qiao Y, Liu L, Xiong Q, Flores C, Wong J, Shi J, et al. Oomycete pathogens encode RNA silencing suppressors. Nat Genet. 2013; 45 : 330–333. doi: 10.1038/ng.2525 23377181
18. Xiong Q, Ye W, Choi D, Wong J, Qiao Y, Tao K, et al. Phytophthora Suppressor of RNA Silencing 2 Is a Conserved RxLR Effector that Promotes Infection in Soybean and Arabidopsis thaliana. Mol Plant Microbe Interact. 2014; 27 : 1379–1389. doi: 10.1094/MPMI-06-14-0190-R 25387135
19. Dong S, Yin W, Kong G, Yang X, Qutob D, Chen Q, et al. Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. Plos Pathogens. 2011; 7: e1002353. doi: 10.1371/journal.ppat.1002353 22102810
20. McLennan AG The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006; 63 : 123–143. 16378245
21. Jambunathan N, Penaganti A, Tang Y, Mahalingam R Modulation of redox homeostasis under suboptimal conditions by Arabidopsis nudix hydrolase 7. BMC Plant Biol. 2010; 10 : 173. doi: 10.1186/1471-2229-10-173 20704736
22. Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006; 18 : 1038–1051. 16531493
23. Ge X, Li GJ, Wang SB, Zhu H, Zhu T, Wang X, et al. AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol. 2007; 145 : 204–215. 17660350
24. Ge X, Xia Y The role of AtNUDT7, a Nudix hydrolase, in the plant defense response. Plant Signal Behav. 2008; 3 : 119–120. 19704728
25. Wang H, Lu Y, Liu P, Wen W, Zhang J, Ge X, et al. The ammonium/nitrate ratio is an input signal in the temperature-modulated, SNC1-mediated and EDS1-dependent autoimmunity of nudt6-2 nudt7. Plant J. 2012.
26. Bhadauria V, Banniza S, Vandenberg A, Selvaraj G, Wei Y Overexpression of a novel biotrophy-specific Colletotrichum truncatum effector, CtNUDIX, in hemibiotrophic fungal phytopathogens causes incompatibility with their host plants. Eukaryot Cell. 2013; 12 : 2–11. doi: 10.1128/EC.00192-12 22962277
27. Tamura N, Murata Y, Mukaihara T Isolation of Ralstonia solanacearum hrpB constitutive mutants and secretion analysis of hrpB-regulated gene products that share homology with known type III effectors and enzymes. Microbiology. 2005; 151 : 2873–2884. 16151200
28. Fischer G, Wittmann-Liebold B, Lang K, Kiefhaber T, Schmid FX Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature. 1989; 337 : 476–478. 2492638
29. Takahashi N, Hayano T, Suzuki M Peptidyl-Prolyl Cis-Trans Isomerase Is the Cyclosporin-a-Binding Protein Cyclophilin. Nature. 1989; 337 : 473–475. 2644542
30. Watashi K, Shimotohno K Cyclophilin and viruses: cyclophilin as a cofactor for viral infection and possible anti-viral target. Drug Target Insights. 2007; 2 : 9–18. 21901058
31. Li M, Ma X, Chiang YH, Yadeta KA, Ding P, Dong L, et al. Proline Isomerization of the Immune Receptor-Interacting Protein RIN4 by a Cyclophilin Inhibits Effector-Triggered Immunity in Arabidopsis. Cell Host Microbe. 2014; 16 : 473–483. doi: 10.1016/j.chom.2014.09.007 25299333
32. Zhang Y, Li B, Xu Y, Li H, Li S, Zhang D, et al. The cyclophilin CYP20-2 modulates the conformation of BRASSINAZOLE-RESISTANT1, which binds the promoter of FLOWERING LOCUS D to regulate flowering in Arabidopsis. Plant Cell. 2013; 25 : 2504–2521. doi: 10.1105/tpc.113.110296 23897924
33. Coaker G, Falick A, Staskawicz B Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science. 2005; 308 : 548–550. 15746386
34. Coaker G, Zhu G, Ding Z, Van Doren SR, Staskawicz B Eukaryotic cyclophilin as a molecular switch for effector activation. Mol Microbiol. 2006; 61 : 1485–1496. 16968222
35. Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, et al. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005; 19 : 111–122. 15989969
36. Matouschek A, Rospert S, Schmid K, Glick BS, Schatz G Cyclophilin catalyzes protein folding in yeast mitochondria. Proc Natl Acad Sci U S A. 1995; 92 : 6319–6323. 7603990
37. Bru R, Walde P Product inhibition of alpha-chymotrypsin in reverse micelles. Eur J Biochem. 1991; 199 : 95–103. 1712303
38. Kofron JL, Kuzmic P, Kishore V, Colon-Bonilla E, Rich DH Determination of kinetic constants for peptidyl prolyl cis-trans isomerases by an improved spectrophotometric assay. Biochemistry. 1991; 30 : 6127–6134. 2059621
39. Zydowsky LD, Etzkorn FA, Chang HY, Ferguson SB, Stolz LA, Ho SI, et al. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1992; 1 : 1092–1099. 1338979
40. Shan W, Cao M, Leung D, Tyler BM The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol Plant Microbe Interact. 2004; 17 : 394–403. 15077672
41. Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP Structural insights into the catalytic mechanism of cyclophilin A. Nat Struct Biol. 2003; 10 : 475–481. 12730686
42. Piotukh K, Gu W, Kofler M, Labudde D, Helms V, Freund C Cyclophilin A binds to linear peptide motifs containing a consensus that is present in many human proteins. J Biol Chem. 2005; 280 : 23668–23674. 15845542
43. Wang P, Heitman J The cyclophilins. Genome Biol. 2005; 6 : 226. 15998457
44. Mainali H, Chapman P, Dhaubhadel S Genome-wide analysis of Cyclophilin gene family in soybean (Glycine max). BMC Plant Biol. 2014; 14 : 282. doi: 10.1186/s12870-014-0282-7 25348509
45. Harrison RK, Stein RL Substrate specificities of the peptidyl prolyl cis-trans isomerase activities of cyclophilin and FK-506 binding protein: evidence for the existence of a family of distinct enzymes. Biochemistry. 1990; 29 : 3813–3816. 1693856
46. Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996; 87 : 1285–1294. 8980234
47. Rovenich H, Boshoven JC, Thomma BP Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr Opin Plant Biol. 2014; 20C: 96–103.
48. Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002; 30 : 415–429. 12028572
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type GenesČlánek Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion IsolatesČlánek -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated GlycolipidsČlánek Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 8- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- The Long and Winding Road (Apologies to the Beatles)
- The Ebola Virus: From Basic Research to a Global Health Crisis
- Riding the R Train into the Cell
- The Two-Phase Emergence of Non Pandemic HIV-1 Group O in Cameroon
- Tumor Progression Locus 2 Promotes Induction of IFNλ, Interferon Stimulated Genes and Antigen-Specific CD8 T Cell Responses and Protects against Influenza Virus
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies
- E3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3
- The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type Genes
- Hyperinvasive Meningococci Induce Intra-nuclear Cleavage of the NF-κB Protein p65/RelA by Meningococcal IgA Protease
- Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen
- HTLV-1 Tax Stimulates Ubiquitin E3 Ligase, Ring Finger Protein 8, to Assemble Lysine 63-Linked Polyubiquitin Chains for TAK1 and IKK Activation
- Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion Isolates
- Widespread Reassortment Shapes the Evolution and Epidemiology of Bluetongue Virus following European Invasion
- Inhibiting the Recruitment of PLCγ1 to Kaposi’s Sarcoma Herpesvirus K15 Protein Reduces the Invasiveness and Angiogenesis of Infected Endothelial Cells
- Goblet Cell Derived RELM-β Recruits CD4 T Cells during Infectious Colitis to Promote Protective Intestinal Epithelial Cell Proliferation
- HLA Class-II Associated HIV Polymorphisms Predict Escape from CD4+ T Cell Responses
- An siRNA Screen Identifies the U2 snRNP Spliceosome as a Host Restriction Factor for Recombinant Adeno-associated Viruses
- Extracellular Adenosine Protects against Lung Infection by Regulating Pulmonary Neutrophil Recruitment
- : Adaptations to the Dixenous Life Cycle Analyzed by Genome Sequencing, Transcriptome Profiling and Co-infection with
- Which Way In? The RalF Arf-GEF Orchestrates Host Cell Invasion
- Intracellular Uropathogenic . Exploits Host Rab35 for Iron Acquisition and Survival within Urinary Bladder Cells
- A Non-enveloped Virus Hijacks Host Disaggregation Machinery to Translocate across the Endoplasmic Reticulum Membrane
- Supporting Role for GTPase Rab27a in Hepatitis C Virus RNA Replication through a Novel miR-122-Mediated Effect
- -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated Glycolipids
- The Activation of Effector Avr3b by Plant Cyclophilin is Required for the Nudix Hydrolase Activity of Avr3b
- A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in
- Double-Edge Sword of Sustained ROCK Activation in Prion Diseases through Neuritogenesis Defects and Prion Accumulation
- The Rsb Phosphoregulatory Network Controls Availability of the Primary Sigma Factor in and Influences the Kinetics of Growth and Development
- Inhibits Virulence through Suppression of Pyochelin and Pyoverdine Biosynthesis
- Illuminating Targets of Bacterial Secretion
- Chemical Signals and Mechanosensing in Bacterial Responses to Their Environment
- Interdisciplinarity and Infectious Diseases: An Ebola Case Study
- Fungi That Infect Insects: Altering Host Behavior and Beyond
- Plasticity and Redundancy in Proteins Important for Invasion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
- A Novel Virus Causes Scale Drop Disease in
- STAT2 Knockout Syrian Hamsters Support Enhanced Replication and Pathogenicity of Human Adenovirus, Revealing an Important Role of Type I Interferon Response in Viral Control
- Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
- Twenty-Eight Years of Poliovirus Replication in an Immunodeficient Individual: Impact on the Global Polio Eradication Initiative
- AAV-Delivered Antibody Mediates Significant Protective Effects against SIVmac239 Challenge in the Absence of Neutralizing Activity
- Interferon-γ Promotes Inflammation and Development of T-Cell Lymphoma in HTLV-1 bZIP Factor Transgenic Mice
- Transgenic Rabbits Expressing Ovine PrP Are Susceptible to Scrapie
- Mitochondrial Activity and Cyr1 Are Key Regulators of Ras1 Activation of . Virulence Pathways
- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Serine Phosphorylation of HIV-1 Vpu and Its Binding to Tetherin Regulates Interaction with Clathrin Adaptors
- Inhibition of mTORC1 Enhances the Translation of Chikungunya Proteins the Activation of the MnK/eIF4E Pathway
- Nanoformulations of Rilpivirine for Topical Pericoital and Systemic Coitus-Independent Administration Efficiently Prevent HIV Transmission
- Arming of MAIT Cell Cytolytic Antimicrobial Activity Is Induced by IL-7 and Defective in HIV-1 Infection
- sRNA-Mediated Regulation of P-Fimbriae Phase Variation in Uropathogenic
- Evolutionary and Functional Analysis of Old World Primate TRIM5 Reveals the Ancient Emergence of Primate Lentiviruses and Convergent Evolution Targeting a Conserved Capsid Interface
- Hepcidin and Host Defense against Infectious Diseases
- Type I IFN Induction via Poly-ICLC Protects Mice against Cryptococcosis
- Mucosal B Cells Are Associated with Delayed SIV Acquisition in Vaccinated Female but Not Male Rhesus Macaques Following SIV Rectal Challenge
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Illuminating Targets of Bacterial Secretion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



