-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaE3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3
IFITM3 is critical for limiting the severity of influenza virus infections in humans and mice. Optimal antiviral activity of IFITM3 is achieved when it is present at high levels within cells. Our results indicate that the E3 ubiquitin ligase NEDD4 decreases baseline IFITM3 levels by ubiquitinating IFITM3 and promoting its turnover. Depleting NEDD4 from cells results in IFITM3 accumulation and greater resistance to infection by influenza viruses. Therefore, we have identified NEDD4 as a regulator of IFITM3 levels and as a novel drug target for preventing influenza virus and other IFITM3-sensitive virus infections.
Published in the journal: . PLoS Pathog 11(8): e32767. doi:10.1371/journal.ppat.1005095
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005095Summary
IFITM3 is critical for limiting the severity of influenza virus infections in humans and mice. Optimal antiviral activity of IFITM3 is achieved when it is present at high levels within cells. Our results indicate that the E3 ubiquitin ligase NEDD4 decreases baseline IFITM3 levels by ubiquitinating IFITM3 and promoting its turnover. Depleting NEDD4 from cells results in IFITM3 accumulation and greater resistance to infection by influenza viruses. Therefore, we have identified NEDD4 as a regulator of IFITM3 levels and as a novel drug target for preventing influenza virus and other IFITM3-sensitive virus infections.
Introduction
Interferon (IFN)-induced transmembrane protein 3 (IFITM3) is a 15 kDa protein that restricts cellular infection by influenza virus [1,2,3]. IFITM3 is active against all strains of influenza virus that have been tested to date, regardless of serotype or species of origin [1,3,4,5,6], and it similarly inhibits many other medically important viruses such as HIV, SARS coronavirus, and Ebola virus [1,5,7,8,9]. Confirming its importance in vivo, IFITM3 knockout mice succumb to sublethal doses of influenza virus [10,11]. Likewise, IFITM3 is the only known protein for which a genetic polymorphism present in a significant percentage of the human population is associated with severe influenza virus infections [10,12,13,14]. In the cell, IFITM3 localizes to endosomes and lysosomes [15,16,17], and traps endocytosed virus particles within these degradative compartments by impeding the formation of the virus fusion pore [16,18,19]. Yet, even with this potent mechanism by which IFITM3 limits infections, influenza virus remains a significant health concern [20,21]. This may be explained by the fact that IFITM3 is present at low levels within most cells at steady state and is induced by IFNs only after infection has already been established [3,11,22]. The inability to up-regulate IFITM3 levels independently of infection or IFNs is a challenge preventing the field from harnessing the activity of IFITM3 for infection prevention.
We previously showed that ubiquitination increases the rate of IFITM3 turnover within the cell [15]. A non-ubiquitinated lysine-to-alanine mutant of IFITM3 possessed enhanced antiviral activity and a longer half-life as compared to WT IFITM3 [15]. These findings indicated that inhibition of IFITM3 ubiquitination could augment the activity and/or levels of endogenous IFITM3, thus offering a strategy for exploiting IFITM3 therapeutically or prophylactically against viral infections. The identification of the E3 ubiquitin ligase(s) capable of modifying IFITM3 among the more than 600 annotated E3 ligases in the human genome will be an important step toward validating this antiviral strategy.
Through our work studying tyrosine phosphorylation of IFITM3, we discovered that phosphorylation at tyrosine 20 (Y20) inhibited IFITM3 ubiquitination [23]. This led us to posit that phosphorylation of Y20 may block an E3 ubiquitin ligase recognition signal. Indeed, Y20 is part of a highly conserved PPxY motif (where P = proline, x = any amino acid, and Y = tyrosine, Fig 1A)[24]. PPxY motifs are commonly recognized by WW (characterized by two tryptophan residues spaced approximately 20 amino acids apart) domains of NEDD4-family E3 ubiquitin ligases, of which there are nine family members [25]. We chose to focus first on NEDD4, the prototypical member of this family, for several reasons: 1) NEDD4 and IFITM3 both have ubiquitous expression patterns while several other NEDD4-family members are tissue-specific (BioGPS.org [26]), 2) Like IFITM3, many of the known NEDD4 substrates are membrane proteins and are associated with endosomal and lysosomal pathways [25,27], 3) IFITM3 and NEDD4 are both S-palmitoylated, suggesting that they may localize to similar membrane subdomains [2,28], and 4) NEDD4 is reported to be inhibited by ISG15 [29,30,31], an IFN-inducible protein, thus providing an intriguing model whereby IFN might induce IFITM3 expression while also inhibiting its ubiquitination. Herein, we provide results demonstrating the ability of NEDD4 to ubiquitinate IFITM3 and identify a unique role for NEDD4 in decreasing steady state IFITM3 abundance, leading to increased cellular susceptibility to influenza virus infection.
Fig. 1. IFITM3 is ubiquitinated by NEDD4. 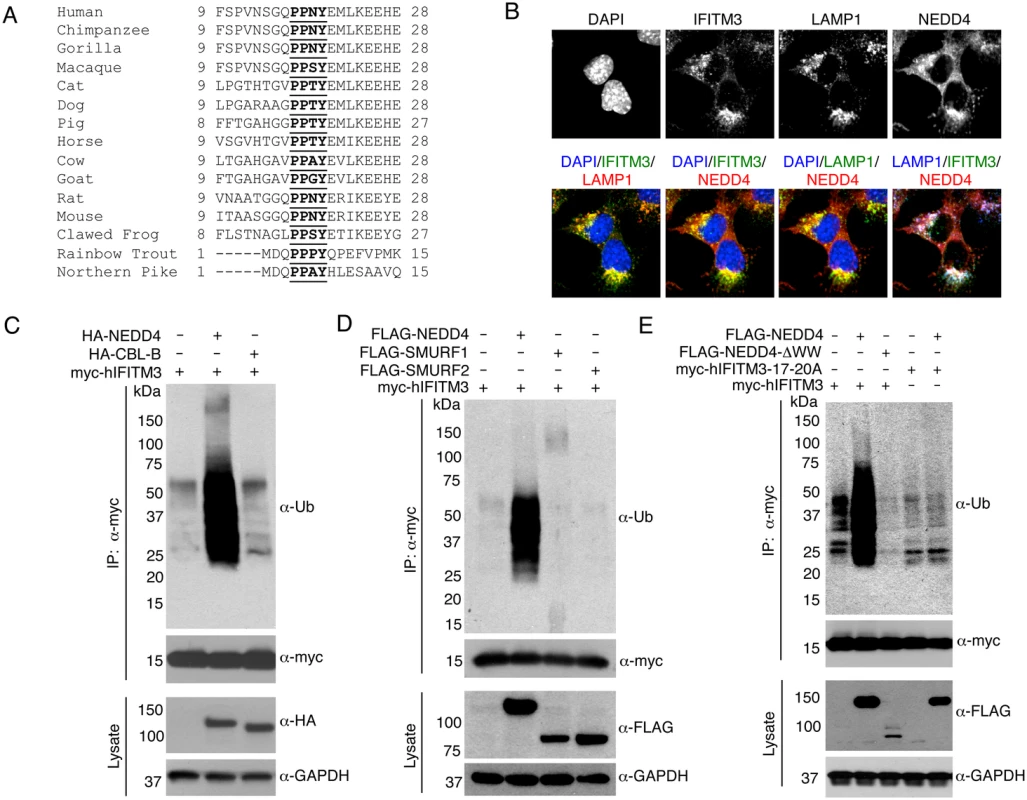
A) Alignment of IFITM3 N-terminal amino acids from various species. Bold and underlined text highlights the conserved PPxY motif. B) Mouse embryonic fibroblasts (MEFs) were stimulated overnight with IFN-α (160 units/mL) to ensure production of IFITM3, and imaged by fluorescent confocal microscopy with staining for endogenous IFITM3, NEDD4, LAMP1, and nuclei (DAPI). Images were taken with a 60x objective and 2.5x zoom. Pseudocolored merged images in different staining combinations are shown. C-E), HEK293T cells were co-transfected with plasmids expressing IFITM3 and epitope tagged ubiquitin ligases, NEDD4, CLB-B, SMURF1 and SMURF2, as indicated. Cell lysates were immunoprecipited with anti-myc resin, and examined by Western blotting with anti-myc and anti-ubiquitin (Ub) antibodies. Western blots of cell lysates with anti-HA (C) or anti-FLAG (D,E) antibodies were performed to confirm expression of the ubiquitin ligases. Anti-GAPDH Western blotting was performed to confirm comparable protein loading. Results
NEDD4 co-localizes with IFITM3 at lysosomes
To explore the possibility that NEDD4 ubiquitinates IFITM3, we first examined whether IFITM3 and NEDD4 are in proximity to one another within cells. We stimulated mouse embryonic fibroblasts (MEFs) with IFN-α to induce abundant expression of IFITM3. By performing immunofluorescence microscopy imaging of endogenous IFITM3 and NEDD4, we detected co-localization of these two proteins (Fig 1B). Further co-localization of these proteins with endogenous LAMP1, a lysosomal marker, indicates that NEDD4 and IFITM3 may interact at lysosomes (Fig 1B).
NEDD4 overexpression increases IFITM3 ubiquitination
We next examined the effect of overexpressing HA-tagged human NEDD4 (HA-NEDD4) on IFITM3 ubiquitination. We observed a significant increase in myc-tagged human IFITM3 (myc-hIFITM3) ubiquitination when HA-NEDD4 was expressed as compared to the transfection control (Fig 1C). On the contrary, no increase in IFITM3 ubiquitination was seen upon overexpression of HA-tagged human CBL-B, another E3 ubiquitin ligase that has been reported to interact with NEDD4 [32], and that is associated with regulation of immune responses [33] (Fig 1C). Additionally, we examined the effect of overexpressing FLAG-tagged human NEDD4 (FLAG-NEDD4) on IFITM3 ubiquitination in comparison to FLAG-tagged human SMURF1 and SMURF2 (FLAG-SMURF1 and FLAG-SMURF2), both of which are members of the NEDD4-family of ubiquitin ligases. FLAG-NEDD4 caused an increase in IFITM3 ubiquitination while FLAG-SMURF1 and FLAG-SMURF2 were unable to robustly modify IFITM3 (Fig 1D). These results demonstrate that NEDD4 possesses a degree of specificity for IFITM3 that is lacking for CBL-B and the NEDD4-family members, SMURF1 and SMURF2.
The IFITM3 PPxY motif is required for ubiquitination by NEDD4
As previously mentioned, NEDD4-family ubiquitin ligases possess two to four characteristic WW domains that interact with proline-rich motifs, including PPxY motifs, on substrate proteins [34]. NEDD4 has four WW domains and IFITM3 contains a highly conserved PPxY motif within its N-terminus (Fig 1A). To test whether these domains are required for IFITM3 ubiquitination, we generated an IFITM3 mutant in which each residue of the PPxY motif (17-PPNY-20 in IFITM3) was mutated to alanine (designated 17-20A) and utilized a FLAG-NEDD4 mutant in which its four WW domains were deleted (designated ΔWW). Upon co-overexpression of FLAG-NEDD4 with myc-hIFITM3, ubiquitination of IFITM3 was increased as expected, while the ΔWW mutant was unable to increase IFITM3 ubiquitination (Fig 1E). In fact, FLAG-NEDD4-ΔWW partially decreased steady state IFITM3 ubiquitination, perhaps indicating a dominant negative effect (Fig 1E). Moreover, the 17-20A mutant of IFITM3 showed less ubiquitination than WT IFITM3 and was unaffected by overexpression of NEDD4 (Fig 1E).
The IFITM3 PPxY motif shares its tyrosine with an overlapping YxxΦ motif known to be involved in the trafficking of IFITM3 from the plasma membrane to endosomes [23,35,36]. Thus, in order to be certain that the results we observed for the 17-20A mutant of IFITM3 was not because of interference with the YxxΦ motif, we tested additional PPxY mutants in which the two prolines were mutated to alanine (myc-hIFITM3-P17,18A) or in which the tyrosine was mutated to alanine (myc-hIFITM3-Y20A). Upon co-overexpression of FLAG-NEDD4, the ubiquitination of both of these mutants was only minimally increased as compared to the robust increase in ubiquitination of WT IFITM3 (Fig 2A). Interestingly, a truncated form of IFITM3 missing its first 21 amino acids, including the PPxY motif, is prevalent in certain human populations. This variant is associated with severe influenza virus infections [10,13,14] and more rapid progression of HIV-related disease [37]. A myc-hIFITM3 construct lacking these first 21 amino acids (Δ1–21) was, as expected, largely unaffected in terms of ubiquitination by overexpression of FLAG-NEDD4 (Fig 2B), identifying a potentially important difference between the truncated and full-length IFITM3 proteins.
Fig. 2. The IFITM3 PPxY motif is required for ubiquitination by NEDD4. 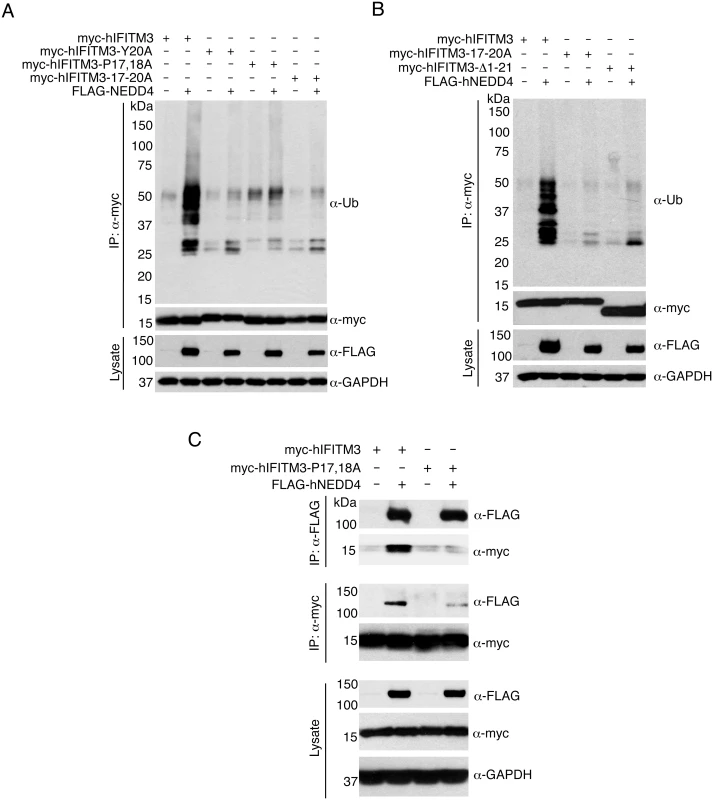
A-C) HEK293T cells were co-transfected with plasmids expressing myc-hIFITM3 or FLAG-NEDD4 as indicated. A-B) Cell lysates were immunoprecipitated with anti-myc resin, and examined by Western blotting with anti-myc and anti-ubiquitin (Ub). Western blotting of cell lysate with anti-FLAG antibodies was performed to confirm expression of NEDD4. Western blotting with anti-GAPDH antibodies was performed to confirm comparable protein loading. C) Cell lysates were immunoprecipitated with anti-myc or anti-FLAG resin, and co-immunoprecipitation was examined by Western blotting with both anti-myc and anti-FLAG antibodies for each immunoprecipitate. Western blots of cell lysates with anti-myc and anti-FLAG antibodies were performed to confirm expression of IFITM3 and NEDD4, respectively. Anti-GAPDH Western blotting was performed to confirm comparable protein loading. Next, since NEDD4 has been shown to physically interact with the PPxY motifs of its substrate proteins [25], we examined whether or not NEDD4 and IFITM3 co-immunoprecipitate with one another. We found that myc-hIFITM3 and FLAG-NEDD4 indeed co-immunoprecipitated with one another (Fig 2C), suggesting a physical interaction. Importantly, this interaction was greatly diminished between FLAG-NEDD4 and the P17,18A mutant of IFITM3 (Fig 2C). In sum, these results indicate that the IFITM3 PPxY motif is required for a strong interaction with and ubiquitination by NEDD4.
NEDD4-mediated IFITM3 ubiquitination is dependent upon NEDD4 catalytic activity
To determine whether a non-enzymatic activity of NEDD4 might be mediating its effect on IFITM3 ubiquitination, we tested a catalytically inactive NEDD4 point mutant. We found that this mutant was unable to increase IFITM3 ubiquitination, establishing that catalytic activity of NEDD4 is indeed required for its ability to increase IFITM3 ubiquitination (Fig 3). Since murine (m)IFITM3 also possesses a PPxY motif (Fig 1A), we tested the ability of NEDD4 to affect mIFITM3 modification. Like myc-hIFITM3, we observed an increase in myc-mIFITM3 ubiquitination when HA-NEDD4 was co-overexpressed and observed no effect of the catalytic mutant (Fig 3), suggesting a possible evolutionary conservation of NEDD4 modification of IFITM3 in mice and humans. These data further implicate NEDD4 as an E3 ubiquitin ligase capable of enzymatically modifying mouse and human IFITM3.
Fig. 3. NEDD4 catalytic activity is required for IFITM3 ubiquitination. 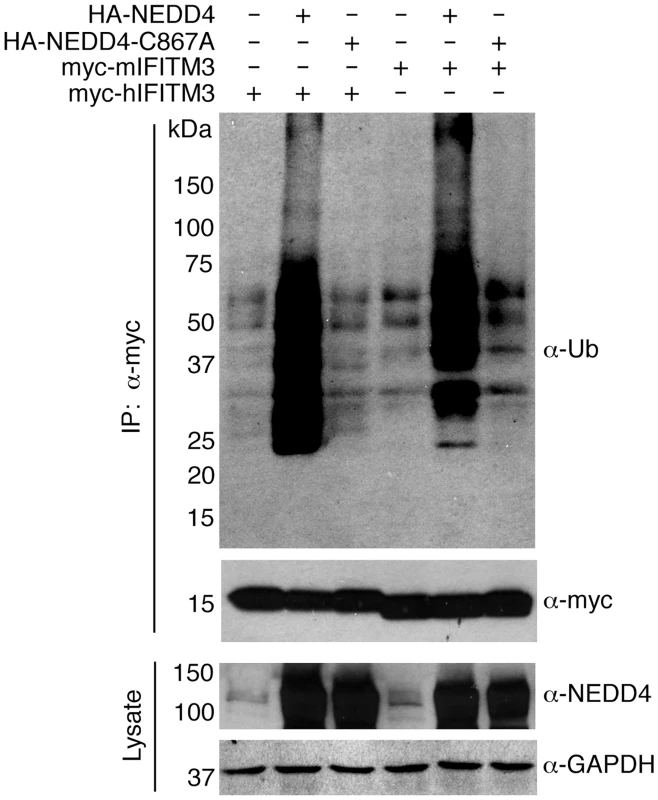
HEK293T cells were transfected with the indicated mouse or human IFITM3 constructs and were co-transfected with plasmids expressing HA-NEDD4 or a catalytically inactive HA-NEDD4-C867A mutant. IFITM3 was immunoprecipitated with anti-myc resin and subjected to anti-myc and anti-ubiquitin (Ub) Western blotting. Cell lysates were probed with anti-NEDD4 antibodies to confirm expression of NEDD4 constructs. Anti-GAPDH staining served as a protein loading control. NEDD4 can modify IFITM3 in vitro
While NEDD4 overexpression experiments suggest that NEDD4 directly ubiquitinates IFITM3 (Figs 1C, 1D and 1E, 2A and 2B and 3), this effect could be indirect. We therefore tested the ability of purified NEDD4 to ubiquitinate immunoprecipitated IFITM3 in vitro in order to confirm that NEDD4 can directly modify IFITM3. HA-hIFITM3 was incubated with purified NEDD4, enzymatic cofactors, and ubiquitin. We then re-immunoprecipitated IFITM3 and subjected it to anti-ubiquitin western blotting. Our results show that NEDD4 is capable of robustly ubiquitinating IFITM3 in vitro (Fig 4). Additionally, we employed ubiquitin mutants that could only be added via lysine 48 (K48) or lysine 63 (K63) linkages in order to examine whether NEDD4 preferentially utilizes one of these polyubiquitination linkages for modifying IFITM3. While both K48 and K63 linkages could be added to IFITM3 by NEDD4, we observed a preference for the K48 linkage in long polyubiquitin chains, which is traditionally associated with protein degradation (Fig 4). These results are consistent with our past results using linkage-specific anti-ubiquitin antibodies, which demonstrated that while both K48 and K63 ubiquitin linkages could be detected on IFITM3, K48 linkages are more prevalent [15]. These data are also consistent with our previous results indicating that ubiquitination of IFITM3 promotes its turnover [15].
Fig. 4. NEDD4 ubiquitinates IFITM3 in vitro. 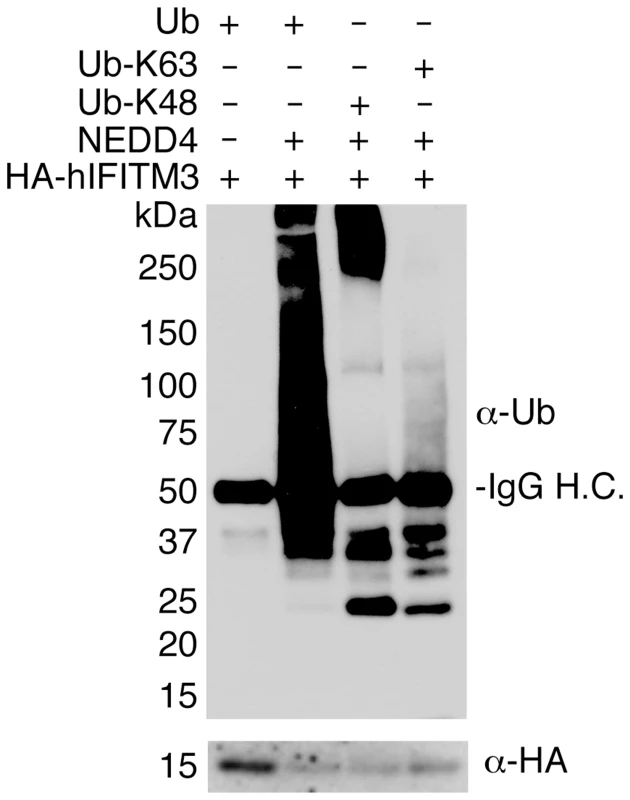
HA-hIFITM3 was added to reactions containing NEDD4-compatible E1 and E2 ubiquitin ligases, ubiquitin (WT or mutants in which only K48 or K63 were not mutated), and reaction buffer containing ATP in the presence or absence of NEDD4. The reaction was allowed to proceed for 1 h at 37°C, and IFITM3 was re-immunoprecipitated and subjected to Western blotting with anti-ubiquitin (Ub) and anti-HA antibodies. IgG H.C. indicates detection of the heavy chain of the immunoglobulin used for immunoprecipitation. NEDD4 knockout decreases IFITM3 ubiquitination and increases resistance to viral infection
In order to examine the effects of NEDD4 on endogenous IFITM3, we examined NEDD4 WT and knockout (KO) mouse embryonic fibroblasts (MEFs)[38]. We also utilized KO MEFs reconstituted with NEDD4 via retroviral transduction. Remarkably, Western blotting of lysates from NEDD4 KO cells showed an increase in steady state IFITM3 levels as compared to WT cells, while NEDD4 reconstitution decreased IFITM3 to WT levels (Fig 5A). To examine the requirement for NEDD4 in ubiquitinating IFITM3, we immunoprecipitated IFITM3 from large quantities of lysate from both WT and KO cells, expecting that the immunoprecipitation reagents would be saturated, thus providing us with comparable amounts of IFITM3 for examination of ubiquitination. Indeed, IFITM3 from NEDD4 KO cells was ubiquitinated much less than IFITM3 from WT cells (Fig 5B). These results demonstrate that NEDD4 is required for proper steady state ubiquitination of IFITM3, and that the absence of NEDD4 results in cellular accumulation of unmodified IFITM3.
Fig. 5. NEDD4 knockout decreases IFITM3 ubiquitination and protects cells from virus infection. 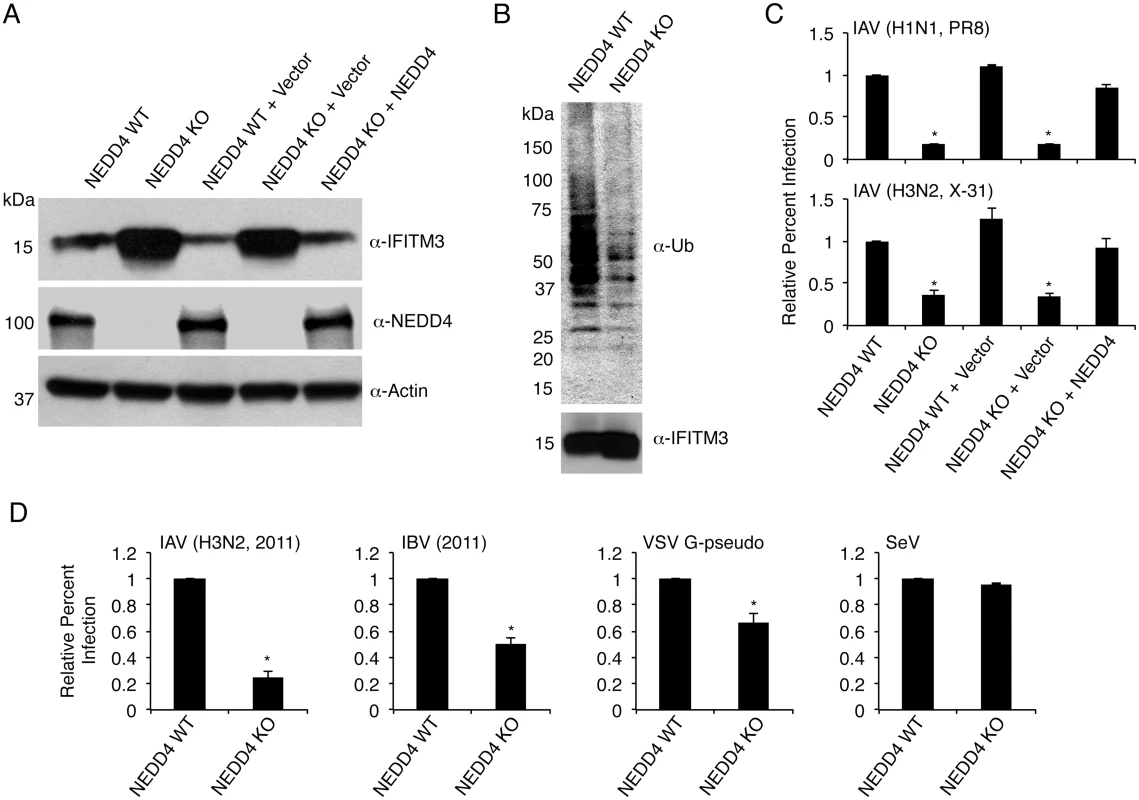
A) Cell lysates from NEDD4 WT and KO MEFs were subjected to anti-IFITM3, anti-NEDD4, and anti-actin immunoblotting to evaluate NEDD4 levels in each cell line and the effect of NEDD4 on endogenous IFITM3 levels. Retroviral reconstitution of the indicated cells with an empty retrovirus control or retrovirus expressing NEDD4 is denoted by + Vector and + NEDD4, respectively. B) 2 mg of protein from NEDD4 WT and KO cell lysates were immunoprecipitated for endogenous IFITM3 and examined by Western blotting with anti-ubiquitin and anti-IFITM3 antibodies. C) The indicated cell lines were infected for 24 h with influenza A virus (IAV) PR8 and X-31 strains at an MOI of 5. Cells were then fixed and stained with anti-influenza virus NP to measure the percentage of cells infected using flow cytometry. D) NEDD4 WT and KO MEFs were infected with influenza virus 2011 isolates (IAV or influenza B virus (IBV)) or Sendai virus (SeV) at an MOI of 5 for 24 h, or were infected with VSV G-pseudotyped retrovirus (VSV G-pseudo) for 48 h. Cells were then fixed and stained with anti-influenza NP in the case of influenza virus infections or were examined for GFP positivity in the case of VSV G-pseudo or SeV to measure the percentage of cells infected using flow cytometry. C,D) Non-infected samples were used as a baseline for gating of infected cells. Results shown are representative of at least three independent experiments, each performed with triplicate samples. The average percent infection of WT NEDD4 cells was set to 1 for the calculation of relative percent infection. Error bars represent standard deviation of triplicate samples. * Indicates a p-value less than 0.001 calculated by Student’s t-test in comparison to values for NEDD4 WT cells. Given the increase in baseline IFITM3 levels, we predicted that NEDD4 KO cells would be more resistant to influenza virus infection. We observed that NEDD4 KO MEFs were in fact significantly less susceptible to infections with influenza A virus (IAV) subtypes H1N1 and H3N2 (PR8 and X-31 strains, respectively) compared to WT control cells (Fig 5C). The decreased susceptibility of KO cells was returned to WT levels of infection upon NEDD4 reconstitution (Fig 5C). We also verified that the enhanced resistance of NEDD4 KO cells to influenza virus infection included resistance to recently circulating strains. NEDD4 KO cells were significantly less susceptible than WT cells to infection by both influenza B virus (IBV) and IAV H3N2 strains isolated in 2011 (Fig 5D). We also examined retrovirus pseudotyped with the vesicular stomatitis virus (VSV) G protein, which is also reported to be inhibited by IFITM3 [3,35,36,39,40]. As expected, the percent of NEDD4 KO cells infected with VSV G-pseudotyped virus was significantly less than WT cells (Fig 5D). Sendai virus (SeV), a parainfluenza virus that primarily fuses at the cell surface [41] and is thus only minimally affected by IFITM3 [4], was also tested. Unlike IAV, IBV, and VSV G-pseudotyped retrovirus, SeV was not appreciably affected by NEDD4 KO (Fig 5D). Thus, the pattern of virus restriction we observed is consistent with protection of NEDD4 KO cells by IFITM3.
To confirm that the increased resistance of NEDD4 KO cells to influenza virus infection was due to increased levels of basal IFITM3, we knocked down IFITM3 in NEDD4 WT and KO cells for 24 hours prior to infection. Knockdown was verified through Western blotting of cell lysates prepared at the time of infection (Fig 6A). Importantly, knockdown of IFITM3 in both NEDD4 WT and KO MEFs resulted in an increase in influenza virus susceptibility, and largely eliminated the resistance of NEDD4 KO cells to infection (Fig 6B). Overall, these experiments demonstrate that NEDD4 promotes cellular susceptibility to influenza virus infection by decreasing levels of IFITM3.
Fig. 6. NEDD4 regulates cellular susceptibility to influenza virus infection by controlling IFITM3 levels. 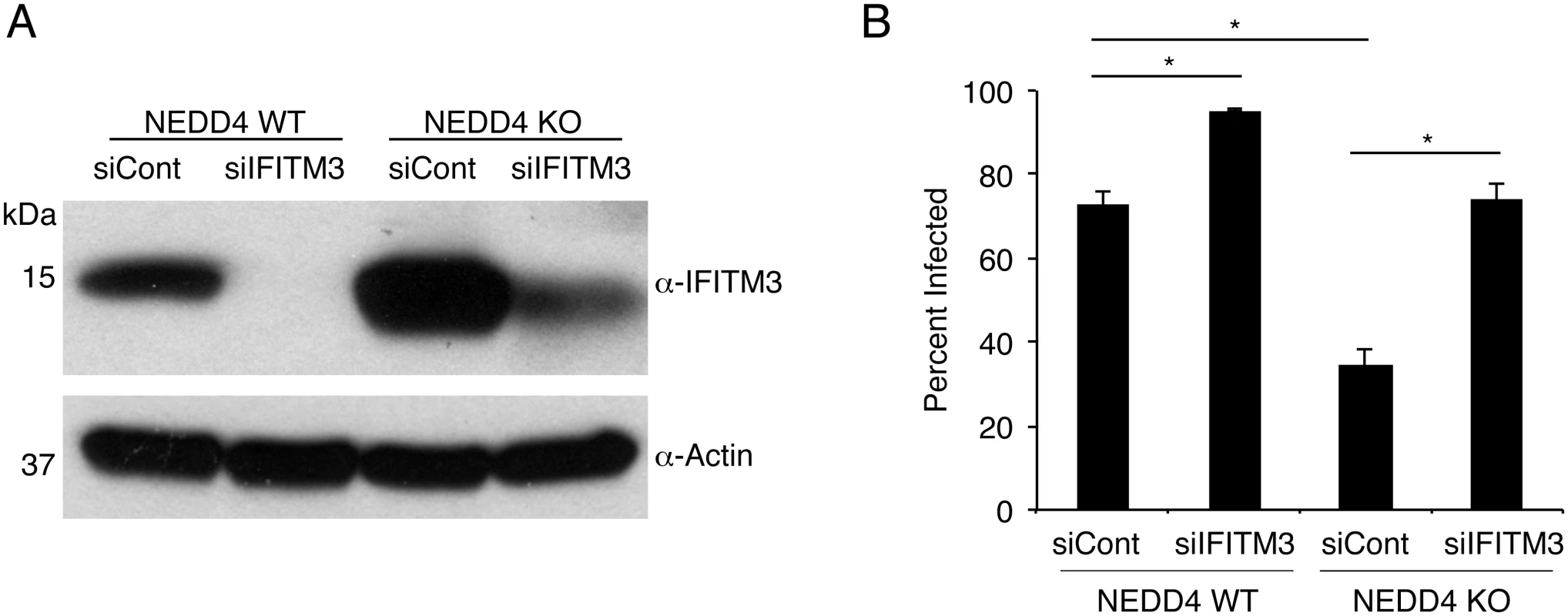
A,B) NEDD4 WT and KO MEFs were transfected for 24 h with control siRNA (siCont) or siRNA targeting IFITM3 (siIFITM3). A) Cells were collected just prior to infection for confirmation of IFITM3 knockdown by anti-IFITM3 Western blotting, with anti-actin blotting serving as a protein loading control. B) Following siRNA treatment, cells were infected with influenza virus strain PR8 at an MOI of 10 for 24 h. Cells were then fixed and stained with anti-influenza virus NP to measure the percentage of cells infected using flow cytometry. Results shown are representative of three independent experiments, each with samples run in triplicate. Error bars represent standard deviation. * Indicates p-value less than 0.001 calculated with Student’s t-test for comparison of samples denoted by horizontal lines. NEDD4 knockdown in human lung cells increases IFITM3 levels and resistance to influenza virus infection
To extend our results to more relevant human lung cells, we utilized the A549 human alveolar epithelial cell line to study the role of NEDD4 in the regulation of steady state IFITM3 levels. Knockdown of NEDD4 with siRNA in A549 cells led to a significant increase in endogenous IFITM3 compared to non-targeting control siRNA (Fig 7A). As expected, NEDD4 knockdown led to a significantly greater resistance to IAV infection (Fig 7B and 7C). Importantly, we found that the relationship between NEDD4 knockdown and increased IFITM3 levels was preserved in two additional human lung cell lines (Fig 7D). Taken together with experiments presented in Figs 3, 5 and 6, these data confirm an evolutionary conservation between mice and humans in the regulation of cellular IFITM3 levels by NEDD4. This work also identifies NEDD4 as a novel target in human cells for improving resistance to influenza virus infection independently of IFNs or adaptive immunity.
Fig. 7. NEDD4 knockdown in human lung cells increases IFITM3 levels and resistance to influenza virus infection. 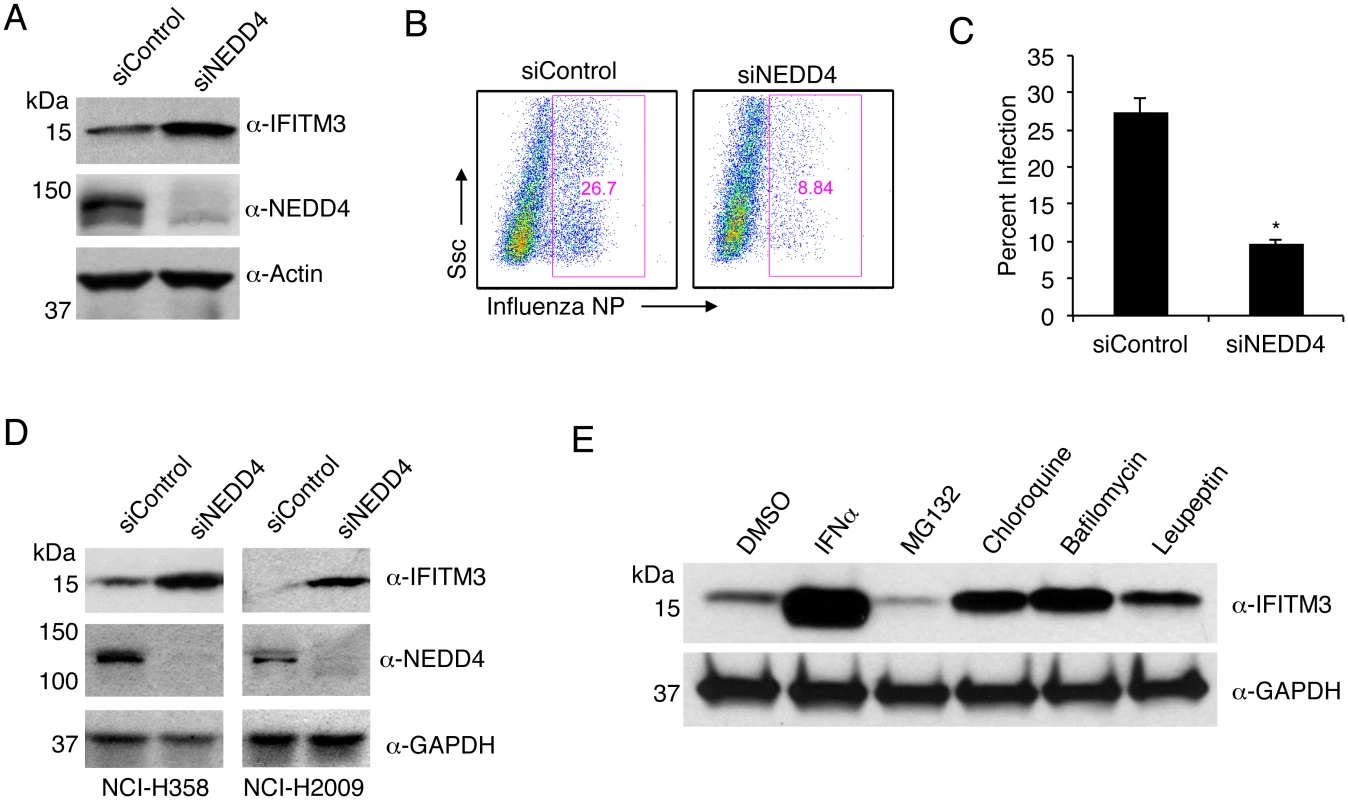
A-C) A549 cells were transfected for 48 h with control siRNA (siControl) or siRNA targeting human NEDD4 (siNEDD4). A) Cells were collected just prior to infection for confirmation of NEDD4 knockdown by anti-NEDD4 Western blotting, with anti-actin blotting serving as a loading control. Anti-IFITM3 blotting demonstrates an increase in IFITM3 upon NEDD4 knockdown. B,C) Following siRNA treatment, cells were infected with influenza virus strain PR8 at an MOI of 2.5 for 6 h. Cells were then fixed and stained with anti-influenza virus NP to measure the percentage of cells infected using flow cytometry. Results shown are representative of three independent experiments, with samples run in triplicate. Error bars represent standard deviation of triplicate samples. * Indicates a p-value less than 0.0001 calculated with Student’s t-test. D) NCI-H358 and NCI-H2009 cells were transfected for 48 h with siControl or siNEDD4. Cell lysates were subjected to immunoblotting with anti-NEDD4 to confirm NEDD4 knockdown, anti-IFITM3 to demonstrate increase in endogenous IFITM3 upon NEDD4 knockdown, and anti-GAPDH as a loading control. E) A549 cells were treated with equal volumes Dimethyl Sulfoxide (DMSO) as a control, MG132 (10 μM), Chloroquine (10 μM), Bafilomycin A1 (1 μM), or Leupeptin (100 μM) for 24 h. Cells were also treated with IFN-α (100 units/mL) for comparison. Cell lysates were subjected to anti-IFITM3 immunoblotting to evaluate endogenous IFITM3 levels with each treatment. Western blotting with anti-GAPDH served as a loading control. IFITM3 is turned over by the lysosomal degradation pathway
The degradative pathway involved in the turnover of steady state IFITM3 has not been previously investigated. Since NEDD4 is known to associate with the endosomal and lysosomal system and to target several of its substrates for lysosomal degradation [25], our results identifying NEDD4 as the primary ubiquitin ligase for IFITM3 would suggest that IFITM3 is degraded in lysosomes. To test this hypothesis, we utilized chloroquine and bafilomycin, which inhibit endosomal and lysosomal acidification and thus the activation of pH-dependent lysosomal proteases. We observed that treatment of A549 lung cells with these two inhibitors caused an accumulation of IFITM3 (Fig 7E). Similarly, treatment with leupeptin, an inhibitor of specific lysosomal proteases resulted in a similar increase in IFITM3 levels (Fig 7E). This is in contrast to the treatment of cells with the proteasomal inhibitor MG132, which consistently caused a modest decrease in IFITM3 levels, perhaps due to up-regulation of lysosomal degradation pathways when proteasome activity is inhibited. Overall, these experiments demonstrate that, consistent with the co-localization of IFITM3 and NEDD4 at lysosomes (Fig 1B), and the ubiquitination of IFITM3 by NEDD4 (Figs 1C, 1D and 1E, 2,3,4 and 5B), IFITM3 is turned over by the lysosomal degradation pathway.
Discussion
Our previous work established that ubiquitination promotes the turnover of IFITM3 [15]. Thus, identification of the IFITM3 ubiquitin ligase would provide a potential target for increasing IFITM3 abundance and resistance to virus infections. In our previous work studying regulation of IFITM3 endocytosis by phosphorylation, we made the serendipitous discovery that the amino acid Y20 within IFITM3 is involved in regulating IFITM3 ubiquitination [23], which led us to identify the involvement of the IFITM3 PPxY motif in its ubiquitination by NEDD4 (Figs 1E and 2). NEDD4 knockdown or knockout in human or mouse cells, respectively, resulted in substantially greater levels of steady-state IFITM3 (Figs 5A, 6A and 7A and 7D). This accumulation of unmodified IFITM3 is consistent with the observed decrease in IFITM3 ubiquitination in NEDD4 KO cells (Fig 5B).
An additional intriguing aspect of our finding that IFITM3 steady state levels are regulated by NEDD4 is the previously described role of the IFN effector ISG15 in inhibiting NEDD4 [29,30]. ISG15 is a ubiquitin-like protein that specifically binds to NEDD4, blocking its productive interaction with Ubiquitin-E2 ligase complexes [29,30]. The importance of this pathway was highlighted by two independent studies demonstrating that ISG15 blocks NEDD4-mediated monoubiquitination of the VP40 matrix protein of Ebola virus, thereby inhibiting the budding of Ebola virus-like particles [29,30]. Importantly, several studies have implicated ISG15 as a critical antiviral effector against IAV and IBV [42,43,44]. Two studies have demonstrated conjugation of ISG15 onto the IAV NS1 protein by the E3 ligase HERC5, and found that ISGylation of IAV NS1 antagonizes virus replication [44,45]. Interestingly, IBV NS1 specifically blocks human ISG15 conjugation by preventing ISG15 interaction with the ISG15 activating enzyme UbE1L, effectively counteracting its antiviral effect [43,46,47,48]. We posit that high levels of IFITM3 attained after IFN stimulation result from both IFITM3 gene induction, as well as increased IFITM3 protein stability as a result of ISG15 inhibition of NEDD4. We are currently investigating this exciting potential synergistic link between ISG15 and IFITM3.
Our work demonstrates that NEDD4 is required for proper basal ubiquitination of IFITM3 (Fig 5B). However, our results would also suggest that additional ubiquitin ligases are also able to modify IFITM3, particularly when IFITM3 is present at high levels. This is supported by detection of partial ubiquitination of our various IFITM3-PPxY mutants (Figs 1E and 2A and 2B) and by detection of modest IFITM3 ubiquitination in NEDD4 KO cells (Fig 5B). The identities of secondary ubiquitin ligases for IFITM3 are still unknown. Of particular interest are the ubiquitin ligases capable of modifying the truncated Δ1–21 splice variant of human IFITM3, which was not significantly ubiquitinated by NEDD4 (Fig 2B). Identifying the ubiquitin ligases that modify this disease-associated variant may implicate crucial differences in the stability and degradative pathways potentially underlying the defect possessed by this protein. Nonetheless, our data clearly implicate NEDD4 as the primary E3 ubiquitin ligase for IFITM3, and demonstrate that NEDD4 is essential for maintaining low steady state IFITM3 levels.
This current work is in contrast to a prior study that concluded the IFITM3 PPxY motif was not involved in regulating the levels or antiviral activity of overexpressed IFITM3 [36]. However, this previous work did not directly assess ubiquitination of IFITM3 upon mutation of the PPxY motif. Additionally, our experiences studying IFITM3 ubiquitination here and in our prior work suggest that when examining overexpressed IFITM3 constructs, ubiquitination has only subtle effects on total protein levels detected by Western blotting despite significant effects on the IFITM3 half-life13,20. Thus, overexpression likely masked any effects of mutating the PPxY motif on the parameters previously tested [36].
Our study has uncovered a novel mechanism by which NEDD4 indirectly promotes cellular entry of influenza virus by decreasing IFITM3 levels (Figs 5, 6 and 7). Although this work is the first of its kind to identify NEDD4 as a negative regulator of IFITM3 levels, NEDD4 is well described to be necessary for the replication of several important RNA viruses. For example, NEDD4 interacts with proline-rich motifs in the viral late budding domains of Ebola virus [49], rabies virus [50], and HIV [51]. Mono-ubiquitination of these domains promotes efficient budding and viral egress necessary for productive viral spread. However, it remains to be determined whether inhibition of NEDD4 will serve as an effective in vivo antiviral strategy, particularly since NEDD4 is a developmentally essential molecule as demonstrated by the embryonic lethality of NEDD4 KO mice [52]. Likewise, NEDD4 has been implicated in regulation of insulin-like growth factor signaling [53], T-cell-mediated immunity [54,55], and tumor suppression [56]. On the other hand, neuron - and skeletal muscle-specific NEDD4 KO mice are viable [57,58], and NEDD4 is naturally inhibited by ISG15 during virus infections [29,30], perhaps suggesting that short-term inhibition of NEDD4 can occur without adverse effects. Additional experimentation will be needed to answer these vital questions, and this will be aided by the development of selective NEDD4 inhibitors, which is an area of active investigation [59,60]. Overall, our study identifies inhibition of NEDD4 as a novel strategy for preventing infection by influenza virus and other IFITM3-sensitive viruses through the increased accumulation of the antiviral restriction factor IFITM3.
Materials and Methods
Cell culture, transfections, and siRNA knockdowns
All cell lines used in these studies (HEK293T, A549, NCI-H358, NCI-H2009, and MEFs) were cultured in DMEM supplemented with 4.5 g/L D-glucose, L-glutamine, 110 mg/L sodium pyruvate, and 10% fetal bovine serum (Thermo Scientific) at 37°C and 5% CO2 in a humidified incubator. HEK293T and A549 cells were purchased from ATCC. NCI-H358 and NCI-H2009 cells were obtained from the ATCC and provided to us by Dr. Gustavo Leone (The Ohio State University). NEDD4 WT and KO MEFs used in this study were generated by Dr. Hiroshi Kawabe (Max Planck Institute)[38] and were kindly provided to us by Dr. Matthew Pratt (University of Southern California) who also generated the retrovirally reconstituted control cell lines. For Western blotting, cells were plated for 90% confluency in 6-well plates for 24 h prior to transfection with 2 μg/well of plasmids using Lipofectamine 2000 (Invitrogen). For microscopy, MEFs were plated for 50% confluency on glass coverslips in 12-well plates for 24 h prior to overnight treatment with IFN-α (BEI Resources). IFITM3 constructs were expressed from the pCMV-HA or pCMV-myc vectors (Clontech) as described previously [2,4,23]. IFITM3 mutants were made using the QuikChange Multi site-directed mutagenesis kit (Stratagene). Plasmids expressing HA-NEDD4 and HA-NEDD4-C867A were obtained from Addgene (plasmids 27002 and 26999, deposited by Dr. Joan Massagué, Memorial Sloan Kettering Cancer Center)[61], and plasmids expressing FLAG-NEDD4, FLAG-NEDD4-ΔWW, and HA-CBL-B were kindly provided by Dr. Jian Zhang (The Ohio State University). FLAG-SMURF1 and FLAG-SMURF2 were obtained from Addgene (plasmids 11752 and 11746, desposited by Jeff Wrana, University of Toronto)[62].
IFITM3 knockdown in MEFs was performed using Silencer Select Ifitm3 siRNA (Ambion, catalog no. 4390816) and negative control (Ambion, catalog no. 4390844). Human NEDD4 knockdown in A549, NCI H358, and NCI H2009 cells was performed using Dharmacon ON-TARGETplus SMARTpool Human NEDD4 (GE Healthcare, catalog no. L-007178-00) and Dharmacon ON-TARGETplus Control Pool Non-targeting control (GE Healthcare, catalog no. D-001810-10-20). siRNAs were transfected into cells using Lipofectamine RNAiMax transfection reagent (Invitrogen). Transfection of siRNA was performed for 24 h for mIFITM3 knockdown, and 48 h for NEDD4 knockdown. For Western blotting, cells were lysed with 1% Brij buffer (0.1 mM triethanolamine, 150 mM NaCl, 1% BrijO10 (Sigma), pH 7.4) containing EDTA-free protease inhibitor mixture (Roche) and 25 μM MG132 (Sigma). Immunoprecipitations were performed using EZview Red anti-c-myc or anti-HA affinity gel (Sigma), or with Protein G Plus Agarose Suspension (Calbiochem) in conjunction with anti-mIFITM3. Chloroquine, bafilomycin, and leupeptin were purchased from Sigma.
Co-immunoprecipitation (Co-IP) assays
Co-immunoprecipitation assays were adapted from a previously described protocol [63]. HEK293T cells were co-transfected overnight with plasmids expressing myc-hIFITM3 and FLAG-NEDD4. Cells were washed twice with PBS, lysed on ice in Triton X-100 lysis buffer (50 mM Hepes, pH 7.5, 150 mN NaCl, 1% Triton X-100, 10% glycerol, 1.5 mM MgCl2, 1.0 mM EGTA, 10 μg/mL leupeptin, 10 μg/mL aprotinin, 10 μg/mL pepstatin, and 1 mM PMSF) for 5 min, and centrifuged at 1,000 x g for 5 min at 4°C. 50 μg of cell lysate was set aside for each sample in order to evaluate, via Western blotting, expression of myc-hIFITM3, FLAG-NEDD4, and GAPDH as a loading control. Equal concentrations of cell lysate were immunoprecipitated using 15 μL EZview Red anti-c-myc or anti-FLAG affinity gel (Sigma) per sample for 1 h at 4°C with gentle nutation. Immunoprecipitations were washed three times with lysis buffer and examined by Western blotting with both anti-myc and anti-FLAG for each immunoprecipitate.
Western blotting and antibodies
Western blotting was performed with anti-myc (Developmental Studies Hybridoma Bank at the University of Iowa, deposited by Dr. J. Michael Bishop, catalog no. 9E 10), anti-HA (Clontech, catalog no. 631207), anti-hIFITM3 (Proteintech Group, catalog no. 11714-1-AP), anti-mIFITM3 (Abcam, catalog no. ab65183), anti-NEDD4 (Millipore, catalog no. 07–049), anti-FLAG (Sigma, catalog no. F7425), anti-actin (Abcam, catalog no. ab3280), or anti-GAPDH (Invitrogen, catalog no. 398600) antibodies. All primary antibodies were used at a 1 : 1000 dilution. Secondary antibodies, Goat Anti-Mouse IgG, HRP conjugate (Millipore catalog no. 12–349), Goat Anti-Rabbit IgG, HRP-linked (Cell Signaling, catalog no. 70745), and Goat Anti-Mouse, IgG1 Gamma 1 Heavy Chain Specific (SouthernBiotech, catalog no. 1070–05, specifically used for detecting immunoprecipitated protein ubiquitination) were all diluted at 1 : 20,000.
In vitro ubiquitination
HEK293T cells were transfected overnight with plasmid expressing HA-hIFITM3. Protein collected from all wells of one 6-well plate was immunoprecipitated using anti-HA affinity gel and was washed extensively. Immunoprecipitated protein on affinity gel was resuspended in PBS. 10% of the retrieved protein was used in each reaction containing 500 μM ubiquitin or ubiquitin mutants (Boston Biochem, catalogue nos. U-100H, UM-K630, or UM-K480), 0.5 μM UbcH5b E2 ligase (Boston Biochem, catalogue no. E2-622), 100 nM UBE1 E1 ligase (Boston Biochem, catalogue no. E305), and 1x Ubiquitin Conjugation Reaction Buffer containing ATP (Boston Biochem, catalogue no SK-10) in the presence or absence of 100 ng recombinant human NEDD4 (Sigma, catalogue no. SRP0226). Reactions were allowed to proceed at 37°C for 1 h and were stopped by boiling for 5 min. The reactions were then diluted 1 : 100 in ice cold 1% Brij buffer, and IFITM3 was re-immunoprecipitated at 4°C using newly added anti-HA affinity gel prior to Western blot analysis.
Fluorescence microscopy
Cells were fixed for 10 min with 3.7% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS for 10 min, and blocked for 10 min with 2% FBS in PBS. Primary antibodies, anti-mIFITM3 (Fragilis, Abcam, catalogue no. ab15592) (1 : 500), anti-NEDD4 (1 : 500), and anti-LAMP1 (Santa Cruz Biotechnology, catalogue no. sc-19992), and Alexa Fluor-labeled anti-mouse and anti-rabbit secondary antibodies (Life Technologies, 1 : 1000) or anti-rat DyLight 550-labeled secondary antibody (Abcam, catalogue no. ab96888, 1 : 1000) were diluted in 0.1% Triton X-100 in PBS. Cells were treated with antibodies sequentially for 20 min at room temperature and washed five times with 0.1% Triton X-100 in PBS after each antibody treatment. Glass slides were mounted in ProLong Gold antifade reagent containing DAPI (Life Technologies). Images were captured using a Fluoview FV10i confocal microscope (Olympus).
Infections and flow cytometry
Influenza viruses A/Puerto Rico/8/1934 (H1N1, PR8), a PR8 reassortant virus possessing the hemagglutinin and neuraminidase genes from A/Aichi/2/1968 (H3N2, X-31), A/Victoria/361/2011 (H3N2), and B/Texas/06/2011 were propagated in 10-day embryonated chicken eggs (purchased as day 0 eggs from Charles River Laboratories) for 48 h at 37°C as described previously [64]. PR8 and X-31 were provided to us by Drs. Bruno Moltedo and Thomas Moran (Mount Sinai School of Medicine) and the 2011 virus isolates were obtained from BEI Resources sponsored by the NIH/NIAID. SeV expressing green fluorescent protein (SeV-GFP)[65] was generated by Dr. Dominique Garcin (University de Geneve) and provided to us by Dr. Mark Peeples (Nationwide Children’s Hospital Research Institute). SeV-GFP was propagated in 10-day embryonated chicken eggs for 40 h at 37°C as described previously [66]. VSV G-pseudotyped retrovirus expressing green fluorescent protein was generated by transfection of the viral vector pLenti-CMV-GFP-puro (Addgene plasmid 17448, deposited by Dr. Eric Campeau)[67] and packaging plasmids (provided by Dr. Li Wu, The Ohio State University) along with plasmid expressing VSV G into HEK293T cells. Direct inhibition of GFP production by IFITM3 was not expected since this is driven by the CMV immediate early promoter and bypasses retrovirus-specific expression machinery [68]. Media was changed 18 h post-transfection, and media containing virus was then harvested 48 h post-transfection. Virus-containing media was centrifuged at 1200 x g for 5 min, filtered with 0.45 μm filters, frozen, stored at -80°C, and used for infection at a dose that provided approximately 70% infection of WT MEFs. MEFs were infected with IAV PR8, X-31, and H3N2 2011 strains, SeV and IBV at a multiplicity of infection of 5.0 or 10.0. MEFs were infected for 24 h, except in the case of VSV G-pseudotyped retrovirus infections, which were analyzed after 48 h of infection. A549 cells were infected with IAV strain PR8 at a multiplicity of infection of 2.5 for 6 h. Infected cells were washed with PBS and harvested in 0.25% trypsin EDTA. Cells were fixed in 3.7% paraformaldehyde for 10 min and permeabilized with 0.1% Triton X-100 for 10 min. IAV infected cells were stained with anti-influenza nucleoprotein (Abcam, catalog no. ab20343, 1 : 333) directly conjugated to Alexa Fluor 647 using a 100 μg antibody labeling kit (Life Technologies). IBV infected cells were stained with anti-IBV nucleoprotein (Thermo Scientific catalogue no. MA1-80712, 1 : 1000) followed by anti-mouse secondary antibodies conjugated directly to Alexa Fluor 488 (Life Technologies). Measurement of SeV and VSV G-pseudotyped retrovirus infection rates was done by detecting virus-encoded GFP. All antibodies were diluted in 0.1% Triton X-100 in PBS, and cells were stained for 20 min. Cells were washed three times with 0.1% Triton X-100 in PBS after each antibody treatment. PBS was used for final resuspension of cells for flow cytometric analysis using a FACSCanto II flow cytometer (BD Biosciences). Results were analyzed using FlowJo software.
Zdroje
1. Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, et al. (2011) Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS pathogens 7: e1001258. doi: 10.1371/journal.ppat.1001258 21253575
2. Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, et al. (2010) Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nature chemical biology 6 : 610–614. doi: 10.1038/nchembio.405 20601941
3. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, et al. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139 : 1243–1254. doi: 10.1016/j.cell.2009.12.017 20064371
4. Hach JC, McMichael T, Chesarino NM, Yount JS (2013) Palmitoylation on conserved and non-conserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. Journal of virology 87 : 9923–9927. doi: 10.1128/JVI.00621-13 23804635
5. Perreira JM, Chin CR, Feeley EM, Brass AL (2013) IFITMs restrict the replication of multiple pathogenic viruses. Journal of molecular biology 425 : 4937–4955. doi: 10.1016/j.jmb.2013.09.024 24076421
6. Diamond MS, Farzan M (2013) The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nature reviews Immunology 13 : 46–57. doi: 10.1038/nri3344 23237964
7. Lu J, Pan Q, Rong L, He W, Liu SL, et al. (2011) The IFITM proteins inhibit HIV-1 infection. Journal of virology 85 : 2126–2137. doi: 10.1128/JVI.01531-10 21177806
8. Compton AA, Bruel T, Porrot F, Mallet A, Sachse M, et al. (2014) IFITM Proteins Incorporated into HIV-1 Virions Impair Viral Fusion and Spread. Cell host & microbe 16 : 736–747.
9. Tartour K, Appourchaux R, Gaillard J, Nguyen XN, Durand S, et al. (2014) IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology 11 : 103. doi: 10.1186/s12977-014-0103-y 25422070
10. Everitt AR, Clare S, Pertel T, John SP, Wash RS, et al. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484 : 519–523. doi: 10.1038/nature10921 22446628
11. Bailey CC, Huang IC, Kam C, Farzan M (2012) Ifitm3 limits the severity of acute influenza in mice. PLoS pathogens 8: e1002909. doi: 10.1371/journal.ppat.1002909 22969429
12. Wang Z, Zhang A, Wan Y, Liu X, Qiu C, et al. (2013) Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proceedings of the National Academy of Sciences of the United States of America 111.
13. Zhang YH, Zhao Y, Li N, Peng YC, Giannoulatou E, et al. (2013) Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nature Communications 4.
14. Xuan Y, Wang LN, Li W, Zi HR, Guo Y, et al. (2015) IFITM3 rs12252 T>C polymorphism is associated with the risk of severe influenza: a meta-analysis. Epidemiology and infection: 1–10.
15. Yount JS, Karssemeijer RA, Hang HC (2012) S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. The Journal of biological chemistry 287 : 19631–19641. doi: 10.1074/jbc.M112.362095 22511783
16. Feeley EM, Sims JS, John SP, Chin CR, Pertel T, et al. (2011) IFITM3 Inhibits Influenza A Virus Infection by Preventing Cytosolic Entry. PLoS pathogens 7: e1002337. doi: 10.1371/journal.ppat.1002337 22046135
17. Melvin WJ, McMichael TM, Chesarino NM, Hach JC, Yount JS (2015) IFITMs from Mycobacteria Confer Resistance to Influenza Virus When Expressed in Human Cells. Viruses 7 : 3035–3052. doi: 10.3390/v7062759 26075508
18. Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, et al. (2013) IFITM Proteins Restrict Viral Membrane Hemifusion. PLoS pathogens 9: e1003124. doi: 10.1371/journal.ppat.1003124 23358889
19. Desai TM, Marin M, Chin CR, Savidis G, Brass AL, et al. (2014) IFITM3 Restricts Influenza A Virus Entry by Blocking the Formation of Fusion Pores following Virus-Endosome Hemifusion. PLoS pathogens 10: e1004048. doi: 10.1371/journal.ppat.1004048 24699674
20. Taubenberger JK, Kash JC (2010) Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7 : 440–451. doi: 10.1016/j.chom.2010.05.009 20542248
21. Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, et al. (2007) The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25 : 5086–5096. 17544181
22. Friedman RL, Manly SP, McMahon M, Kerr IM, Stark GR (1984) Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell 38 : 745–755. 6548414
23. Chesarino NM, McMichael TM, Hach JC, Yount JS (2014) Phosphorylation of the Antiviral Protein IFITM3 Dually Regulates its Endocytosis and Ubiquitination. The Journal of biological chemistry.
24. Chesarino NM, McMichael TM, Yount JS (2014) Regulation of the trafficking and antiviral activity of IFITM3 by post-translational modifications. Future microbiology 9 : 1151–1163. doi: 10.2217/fmb.14.65 25405885
25. Rotin D, Kumar S (2009) Physiological functions of the HECT family of ubiquitin ligases. Nature reviews Molecular cell biology 10 : 398–409. doi: 10.1038/nrm2690 19436320
26. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, et al. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology 10: R130. doi: 10.1186/gb-2009-10-11-r130 19919682
27. Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD (2005) HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. The Journal of cell biology 168 : 89–101. 15623582
28. Chesarino NM, Hach JC, Chen JL, Zaro BW, Rajaram MV, et al. (2014) Chemoproteomics reveals Toll-like receptor fatty acylation. BMC biology 12 : 91. doi: 10.1186/s12915-014-0091-3 25371237
29. Malakhova OA, Zhang DE (2008) ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. The Journal of biological chemistry 283 : 8783–8787. doi: 10.1074/jbc.C800030200 18287095
30. Okumura A, Pitha PM, Harty RN (2008) ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proceedings of the National Academy of Sciences of the United States of America 105 : 3974–3979. doi: 10.1073/pnas.0710629105 18305167
31. Pincetic A, Kuang Z, Seo EJ, Leis J (2010) The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. Journal of virology 84 : 4725–4736. doi: 10.1128/JVI.02478-09 20164219
32. Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, et al. (2003) WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. The Journal of biological chemistry 278 : 43169–43177. 12907674
33. Liu Q, Zhou H, Langdon WY, Zhang J (2014) E3 ubiquitin ligase Cbl-b in innate and adaptive immunity. Cell cycle 13 : 1875–1884. doi: 10.4161/cc.29213 24875217
34. Scheffner M, Kumar S (2014) Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochimica et biophysica acta 1843 : 61–74. doi: 10.1016/j.bbamcr.2013.03.024 23545411
35. Jia R, Xu F, Qian J, Yao Y, Miao C, et al. (2014) Identification of an endocytic signal essential for the antiviral action of IFITM3. Cellular Microbiology.
36. Jia R, Pan Q, Ding S, Rong L, Liu SL, et al. (2012) The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. Journal of virology 86 : 13697–13707. doi: 10.1128/JVI.01828-12 23055554
37. Zhang Y, Makvandi-Nejad S, Qin L, Zhao Y, Zhang T, et al. (2015) Interferon-induced transmembrane protein-3 rs12252-C is associated with rapid progression of acute HIV-1 infection in Chinese MSM cohort. AIDS 29 : 889–894. doi: 10.1097/QAD.0000000000000632 25784441
38. Fouladkou F, Landry T, Kawabe H, Neeb A, Lu C, et al. (2008) The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proceedings of the National Academy of Sciences of the United States of America 105 : 8585–8590. doi: 10.1073/pnas.0803233105 18562292
39. Weidner JM, Jiang D, Pan XB, Chang J, Block TM, et al. (2010) Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. Journal of virology 84 : 12646–12657. doi: 10.1128/JVI.01328-10 20943977
40. Amini-Bavil-Olyaee S, Choi YJ, Lee JH, Shi M, Huang IC, et al. (2013) The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell host & microbe 13 : 452–464.
41. Hoekstra D, Klappe K, de Boer T, Wilschut J (1985) Characterization of the fusogenic properties of Sendai virus: kinetics of fusion with erythrocyte membranes. Biochemistry 24 : 4739–4745. 3000417
42. Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, et al. (2007) IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proceedings of the National Academy of Sciences of the United States of America 104 : 1371–1376. 17227866
43. Lai C, Struckhoff JJ, Schneider J, Martinez-Sobrido L, Wolff T, et al. (2009) Mice Lacking the ISG15 E1 Enzyme UbE1L Demonstrate Increased Susceptibility to both Mouse-Adapted and Non-Mouse-Adapted Influenza B Virus Infection. Journal of virology 83 : 1147–1151. doi: 10.1128/JVI.00105-08 19004958
44. Zhao C, Hsiang TY, Kuo RL, Krug RM (2010) ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proceedings of the National Academy of Sciences of the United States of America 107 : 2253–2258. doi: 10.1073/pnas.0909144107 20133869
45. Tang YJ, Zhong GX, Zhu LH, Liu X, Shan YF, et al. (2010) Herc5 Attenuates Influenza A Virus by Catalyzing ISGylation of Viral NS1 Protein. Journal of immunology 184 : 5777–5790.
46. Yuan W, Krug RM (2001) Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. The EMBO journal 20 : 362–371. 11157743
47. Zhao C, Collins MN, Hsiang TY, Krug RM (2013) Interferon-induced ISG15 pathway: an ongoing virus-host battle. Trends in microbiology 21 : 181–186. doi: 10.1016/j.tim.2013.01.005 23414970
48. Versteeg GA, Hale BG, van Boheemen S, Wolff T, Lenschow DJ, et al. (2010) Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. Journal of virology 84 : 5423–5430. doi: 10.1128/JVI.02395-09 20219937
49. Harty RN, Brown ME, Wang GL, Huibregtse J, Hayes FP (2000) A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: Implications for filovirus budding. Proceedings of the National Academy of Sciences of the United States of America 97 : 13871–13876. 11095724
50. Harty RN, Brown ME, McGettigan JP, Wang G, Jayakar HR, et al. (2001) Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. Journal of virology 75 : 10623–10629. 11602704
51. Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG (2000) A role for ubiquitin ligase recruitment in retrovirus release. Proceedings of the National Academy of Sciences of the United States of America 97 : 13063–13068. 11087860
52. Fouladkou F, Lu C, Jiang C, Zhou L, She Y, et al. (2010) The ubiquitin ligase Nedd4-1 is required for heart development and is a suppressor of thrombospondin-1. The Journal of biological chemistry 285 : 6770–6780. doi: 10.1074/jbc.M109.082347 20026598
53. Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, et al. (2008) Nedd4 controls animal growth by regulating IGF-1 signaling. Science signaling 1: ra5. doi: 10.1126/scisignal.1160940 18812566
54. Guo H, Qiao G, Ying H, Li Z, Zhao Y, et al. (2012) E3 ubiquitin ligase Cbl-b regulates Pten via Nedd4 in T cells independently of its ubiquitin ligase activity. Cell reports 1 : 472–482. 22763434
55. Yang B, Gay DL, MacLeod MK, Cao X, Hala T, et al. (2008) Nedd4 augments the adaptive immune response by promoting ubiquitin-mediated degradation of Cbl-b in activated T cells. Nature Immunology 9 : 1356–1363. doi: 10.1038/ni.1670 18931680
56. Xu C, Fan CD, Wang X (2015) Regulation of Mdm2 protein stability and the p53 response by NEDD4-1 E3 ligase. Oncogene 34 : 281–289. doi: 10.1038/onc.2013.557 24413081
57. Kawabe H, Neeb A, Dimova K, Young SM Jr., Takeda M, et al. (2010) Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron 65 : 358–372. doi: 10.1016/j.neuron.2010.01.007 20159449
58. Nagpal P, Plant PJ, Correa J, Bain A, Takeda M, et al. (2012) The ubiquitin ligase Nedd4-1 participates in denervation-induced skeletal muscle atrophy in mice. PloS one 7: e46427. doi: 10.1371/journal.pone.0046427 23110050
59. Han Z, Lu J, Liu Y, Davis B, Lee MS, et al. (2014) Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. Journal of virology 88 : 7294–7306. doi: 10.1128/JVI.00591-14 24741084
60. Mund T, Lewis MJ, Maslen S, Pelham HR (2014) Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proceedings of the National Academy of Sciences of the United States of America 111 : 16736–16741. doi: 10.1073/pnas.1412152111 25385595
61. Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, et al. (2009) Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Molecular cell 36 : 457–468. doi: 10.1016/j.molcel.2009.09.043 19917253
62. Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu HT, et al. (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Molecular cell 6 : 1365–1375. 11163210
63. Milkereit R, Rotin D (2011) A Role for the Ubiquitin Ligase Nedd4 in Membrane Sorting of LAPTM4 Proteins. PloS one 6.
64. Moltedo B, Li W, Yount JS, Moran TM (2011) Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. PLoS pathogens 7: e1002345. doi: 10.1371/journal.ppat.1002345 22072965
65. Strahle L, Marq JB, Brini A, Hausmann S, Kolakofsky D, et al. (2007) Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. Journal of virology 81 : 12227–12237. 17804509
66. Lopez CB, Yount JS, Hermesh T, Moran TM (2006) Sendai virus infection induces efficient adaptive immunity independently of type I interferons. Journal of virology 80 : 4538–4545. 16611914
67. Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, et al. (2009) A versatile viral system for expression and depletion of proteins in mammalian cells. PloS one 4: e6529. doi: 10.1371/journal.pone.0006529 19657394
68. Chutiwitoonchai N, Hiyoshi M, Hiyoshi-Yoshidomi Y, Hashimoto M, Tokunaga K, et al. (2013) Characteristics of IFITM, the newly identified IFN-inducible anti-HIV-1 family proteins. Microbes and infection / Institut Pasteur 15 : 280–290. doi: 10.1016/j.micinf.2012.12.003 23376165
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type GenesČlánek Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion IsolatesČlánek -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated GlycolipidsČlánek Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 8- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- The Long and Winding Road (Apologies to the Beatles)
- The Ebola Virus: From Basic Research to a Global Health Crisis
- Riding the R Train into the Cell
- The Two-Phase Emergence of Non Pandemic HIV-1 Group O in Cameroon
- Tumor Progression Locus 2 Promotes Induction of IFNλ, Interferon Stimulated Genes and Antigen-Specific CD8 T Cell Responses and Protects against Influenza Virus
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies
- E3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3
- The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type Genes
- Hyperinvasive Meningococci Induce Intra-nuclear Cleavage of the NF-κB Protein p65/RelA by Meningococcal IgA Protease
- Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen
- HTLV-1 Tax Stimulates Ubiquitin E3 Ligase, Ring Finger Protein 8, to Assemble Lysine 63-Linked Polyubiquitin Chains for TAK1 and IKK Activation
- Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion Isolates
- Widespread Reassortment Shapes the Evolution and Epidemiology of Bluetongue Virus following European Invasion
- Inhibiting the Recruitment of PLCγ1 to Kaposi’s Sarcoma Herpesvirus K15 Protein Reduces the Invasiveness and Angiogenesis of Infected Endothelial Cells
- Goblet Cell Derived RELM-β Recruits CD4 T Cells during Infectious Colitis to Promote Protective Intestinal Epithelial Cell Proliferation
- HLA Class-II Associated HIV Polymorphisms Predict Escape from CD4+ T Cell Responses
- An siRNA Screen Identifies the U2 snRNP Spliceosome as a Host Restriction Factor for Recombinant Adeno-associated Viruses
- Extracellular Adenosine Protects against Lung Infection by Regulating Pulmonary Neutrophil Recruitment
- : Adaptations to the Dixenous Life Cycle Analyzed by Genome Sequencing, Transcriptome Profiling and Co-infection with
- Which Way In? The RalF Arf-GEF Orchestrates Host Cell Invasion
- Intracellular Uropathogenic . Exploits Host Rab35 for Iron Acquisition and Survival within Urinary Bladder Cells
- A Non-enveloped Virus Hijacks Host Disaggregation Machinery to Translocate across the Endoplasmic Reticulum Membrane
- Supporting Role for GTPase Rab27a in Hepatitis C Virus RNA Replication through a Novel miR-122-Mediated Effect
- -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated Glycolipids
- The Activation of Effector Avr3b by Plant Cyclophilin is Required for the Nudix Hydrolase Activity of Avr3b
- A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in
- Double-Edge Sword of Sustained ROCK Activation in Prion Diseases through Neuritogenesis Defects and Prion Accumulation
- The Rsb Phosphoregulatory Network Controls Availability of the Primary Sigma Factor in and Influences the Kinetics of Growth and Development
- Inhibits Virulence through Suppression of Pyochelin and Pyoverdine Biosynthesis
- Illuminating Targets of Bacterial Secretion
- Chemical Signals and Mechanosensing in Bacterial Responses to Their Environment
- Interdisciplinarity and Infectious Diseases: An Ebola Case Study
- Fungi That Infect Insects: Altering Host Behavior and Beyond
- Plasticity and Redundancy in Proteins Important for Invasion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
- A Novel Virus Causes Scale Drop Disease in
- STAT2 Knockout Syrian Hamsters Support Enhanced Replication and Pathogenicity of Human Adenovirus, Revealing an Important Role of Type I Interferon Response in Viral Control
- Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
- Twenty-Eight Years of Poliovirus Replication in an Immunodeficient Individual: Impact on the Global Polio Eradication Initiative
- AAV-Delivered Antibody Mediates Significant Protective Effects against SIVmac239 Challenge in the Absence of Neutralizing Activity
- Interferon-γ Promotes Inflammation and Development of T-Cell Lymphoma in HTLV-1 bZIP Factor Transgenic Mice
- Transgenic Rabbits Expressing Ovine PrP Are Susceptible to Scrapie
- Mitochondrial Activity and Cyr1 Are Key Regulators of Ras1 Activation of . Virulence Pathways
- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Serine Phosphorylation of HIV-1 Vpu and Its Binding to Tetherin Regulates Interaction with Clathrin Adaptors
- Inhibition of mTORC1 Enhances the Translation of Chikungunya Proteins the Activation of the MnK/eIF4E Pathway
- Nanoformulations of Rilpivirine for Topical Pericoital and Systemic Coitus-Independent Administration Efficiently Prevent HIV Transmission
- Arming of MAIT Cell Cytolytic Antimicrobial Activity Is Induced by IL-7 and Defective in HIV-1 Infection
- sRNA-Mediated Regulation of P-Fimbriae Phase Variation in Uropathogenic
- Evolutionary and Functional Analysis of Old World Primate TRIM5 Reveals the Ancient Emergence of Primate Lentiviruses and Convergent Evolution Targeting a Conserved Capsid Interface
- Hepcidin and Host Defense against Infectious Diseases
- Type I IFN Induction via Poly-ICLC Protects Mice against Cryptococcosis
- Mucosal B Cells Are Associated with Delayed SIV Acquisition in Vaccinated Female but Not Male Rhesus Macaques Following SIV Rectal Challenge
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Illuminating Targets of Bacterial Secretion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



