-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaReduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and Anti-pathogen Activities
The infectious agents that cause malaria and dengue are transmitted by Anopheles and Aedes mosquitoes, respectively. Bacteria found in the mosquito midgut have the potential to dramatically affect the susceptibility of the mosquito vector to the malaria parasite and dengue virus. In this work, we investigate one such microbe, Chromobacterium sp. (Csp_P), a bacterium isolated from a field-caught Aedes aegypti mosquito. We show that Csp_P can effectively colonize the midguts of Anopheles gambiae and Aedes aegypti mosquitoes and can, when ingested by the mosquito, significantly reduce the mosquito's susceptibility to infection with the malaria parasite and dengue virus. We also show that exposure to, and ingestion of, Csp_P can reduce the lifespan of larval and adult mosquitoes, respectively. We show that Csp_P has anti-Plasmodium and anti-dengue activity independent of the mosquito, suggesting that the bacterium secretes metabolites that could potentially be exploited to prevent disease transmission or to treat infection.
Published in the journal: . PLoS Pathog 10(10): e32767. doi:10.1371/journal.ppat.1004398
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004398Summary
The infectious agents that cause malaria and dengue are transmitted by Anopheles and Aedes mosquitoes, respectively. Bacteria found in the mosquito midgut have the potential to dramatically affect the susceptibility of the mosquito vector to the malaria parasite and dengue virus. In this work, we investigate one such microbe, Chromobacterium sp. (Csp_P), a bacterium isolated from a field-caught Aedes aegypti mosquito. We show that Csp_P can effectively colonize the midguts of Anopheles gambiae and Aedes aegypti mosquitoes and can, when ingested by the mosquito, significantly reduce the mosquito's susceptibility to infection with the malaria parasite and dengue virus. We also show that exposure to, and ingestion of, Csp_P can reduce the lifespan of larval and adult mosquitoes, respectively. We show that Csp_P has anti-Plasmodium and anti-dengue activity independent of the mosquito, suggesting that the bacterium secretes metabolites that could potentially be exploited to prevent disease transmission or to treat infection.
Introduction
The influence of the gut microbiota on the vector competence of disease vectors such as mosquitoes has gained increasing interest over the past decade [1]–[3]. Previous work has shown that co-infection of Anopheles mosquitoes with Plasmodium and with Serratia sp. or Enterobacter sp. bacteria leads to reduced Plasmodium infection [3], [4]. Additionally, the presence of certain bacterial species in Aedes mosquito midguts leads to a lower intensity of dengue virus infection [5]. Studies have also shown that Anopheles and Aedes mosquitoes that have had their gut microbiota experimentally reduced via antibiotic treatment show higher Plasmodium and dengue virus infection levels, respectively, than do their untreated counterparts [6]–[8]. The anti-pathogen activity of mosquito midgut bacteria has been attributed to the elicitation of the mosquito immune system in some instances, and to direct anti-pathogenic activity of bacteria-produced molecules in others [9]. Activation of the IMD pathway, the major anti-P. falciparum immune pathway, has been shown to be mediated through an interaction between the pattern recognition receptor PGRP-LC and the midgut microbiota [10]. In turn, microbe-derived anti-pathogen factors have been characterized in some microbe-host interaction systems and include cytotoxic metalloproteases, hemolysins, antibiotics, haemaglutinins, proteases, prodigiosin pigments, and iron chelators (siderophores) [9].
In nature, bacteria commonly grow attached to surfaces in complex matrices of cells, proteins, polysaccharides, and DNA (biofilm growth), rather than as single free-swimming cells (planktonic growth) [11], [12]. Biofilm formation allows the bacteria to survive exposure to host-derived antimicrobial factors and other environmental stressors [11], [12]. Furthermore, bacterial cells in a biofilm have quite different gene expression and metabolic profiles than do cells in a free-swimming planktonic state [11]. Studies of Pseudomonas aeruginosa colonization of the Drosophila melanogaster gut have shown that biofilm formation can dramatically affect dissemination in the hemolymph and fly mortality [13].
In this study, we show that a Chromobacterium sp. isolate, Csp_P, previously isolated from the midgut of field-collected Ae. aegypti mosquitoes [5], exerts in vitro anti-Plasmodium and anti-dengue activity when grown under biofilm conditions. Csp_P can effectively colonize the intestines of the two most important mosquito disease vectors, An. gambiae and Ae. aegypti, where it blocks Plasmodium and dengue infection. It also exerts entomopathogenic activity against both larval and adult stages and could therefore be used for the development of a biocontrol agent. Csp_P's anti-pathogen activities appear to be mediated by stable secondary metabolites, suggesting that Csp_P is a source of potentially interesting candidates for the development of therapeutic and transmission-blocking drugs.
Results/Discussion
In a previous study, we isolated a Gram-negative bacterium Chromobacterium sp. (Csp_P) from the midgut of field-collected Ae. aegypti mosquitoes in Panama [5]. The genus Chromobacterium spp. represents soil - and water-associated bacteria of tropical and subtropical regions [14], and members of this genus are known to produce a variety of bioactive compounds [14], [15] and to form biofilms. The most extensively studied member, Chromobacterium violaceum, has been found to produce violacein, a violet pigment compound with potent antimicrobial, antiparasitic, and tumoricidal activity [14], [16]. Csp_P can be cultured in Luria Bertani (LB) broth and on LB agar, on which it forms flat colonies with a tan color that become darker with time and are opaque when exposed to light. Csp_P does not produce violacein, but molecular characterization of its 16s rRNA gene sequence and phylogenetic analysis showed a 98% similarity to Chromobacterium haemolyticum and Chromobacterium aquaticum, probably its two closest relatives.
Csp_P colonization of the mosquito midgut
To assess the ability of Csp_P to colonize the mosquito midgut, we exposed antibiotic-treated mosquitoes to a sugar source containing106 colony forming units (CFU)/ml for Ae. aegypti or 108 CFU/ml for An. gambiae for 24 h and then dissected, homogenized and plated the midguts on LB agar plates at 3 days post-exposure. Treatment with antibiotics through the sugar meal was performed to remove the native microbial flora which can fluctuate in terms of load and species composition between individual mosquitoes of the same cage and generation, thereby complicating the interpretation of our data [7]. The presence of the native microbiota would also render it difficult to discriminate the Csp_P colonies from those of other species through visual inspection. Csp_P displayed an exceptional ability to rapidly colonize mosquito midguts, showing a prevalence of 80% in An. gambiae and 97% in Ae. aegypti cage populations at 3 days after exposure (Fig. 1A). Average bacterial loads at this time point were approximately 105 and 104 CFU per midgut in Ae. aegypti (Fig. 1B) and An. gambiae (Fig. 1C) females, respectively.
Fig. 1. Csp_P colonization of the mosquito midgut. 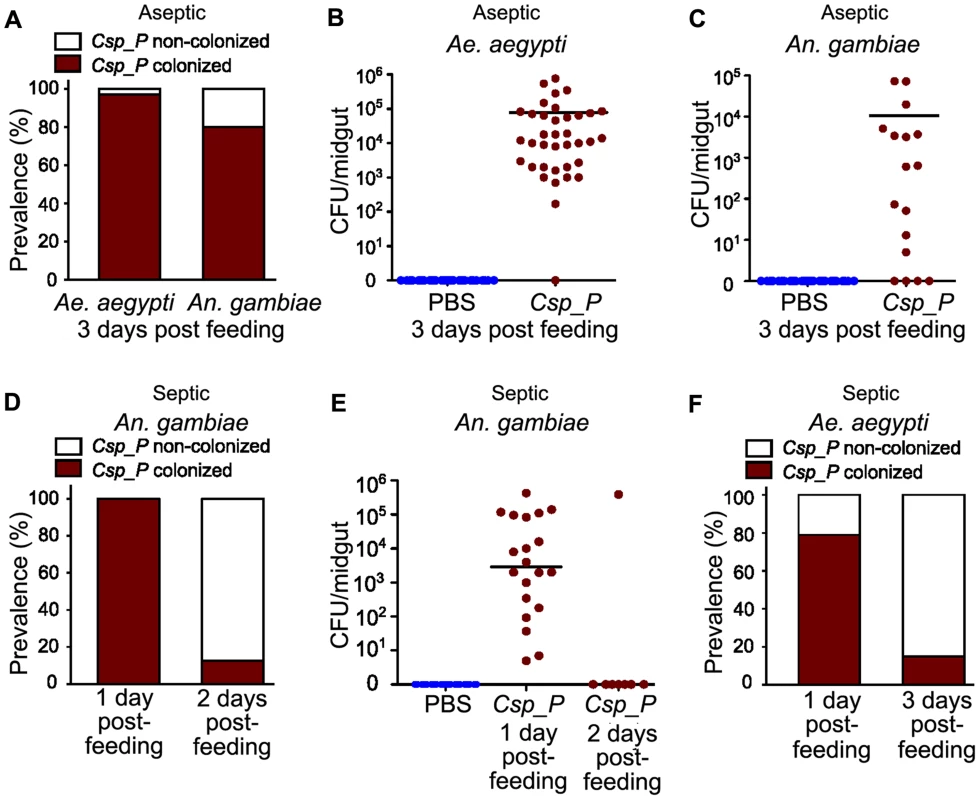
All mosquitoes were exposed to Csp_P via sugar meal. To introduce Csp_P via sugar meal, adults were allowed to feed for 24 h on 1.5% sucrose containing Csp_P liquid culture at a final concentration of ∼108 CFU/ml for An. gambiae and ∼106 (A, B) or 1010 (F) CFU/ml for Ae. aegypti. For antibiotic treated mosquitoes, the prevalence of Csp_P was measured in Ae. aegypti and An. gambiae midguts at 3 days post-exposure (A). The number of colony forming units (CFUs) of Csp_P was also measured in the midguts of (B) Ae. aegypti and (C) An. gambiae 3 days after exposure to Csp_P. Experiments for antibiotic treated Ae. aegypti and An. gambiae were replicated at least three times. Final sample sizes: nAe. aegypti/PBS = 37; nAe. aegypti/Csp_P = 37; nAn. gambiae/PBS = 30; nAn. gambiae/Csp_P = 17. For septic (i.e. non-antibiotic treated) mosquitoes, the prevalence and bacterial load of Csp_P was measured in An. gambiae midguts at 1 and 2 days post exposure (D,E). Experiments for septic An. gambiae were replicated twice. Final sample sizes: nAn. gambiae/PBS = 30; nAn. gambiae/Csp_P/Day 1 = 20; nAn. gambiae/Csp_P/Day 2 = 8. Prevalence of Csp_P was measured in Ae. aegypti midguts at 1 and 3 days post exposure (F). Experiments for septic Ae. aegypti were replicated twice. Final sample sizes: nAe. aegypti/Csp_P/Day 1 = 19; nAe. aegypti/Csp_P/Day 3 = 20. Horizontal lines indicate mean values. The following transformation was applied to all raw CFU data: y = log10(x+1), where x = original CFU count and y = plotted data values. We also tested the ability of Csp_P to colonize the midguts of non-antibiotic treated mosquitoes. Because nearly all septic (i.e. non-antibiotic treated) An. gambiae mosquitoes had died two days after Csp_P introduction through sugar-feeding at 108 CFU/ml (Fig. 2C), we were only able to assay prevalence and bacterial load of Csp_P at days one and two post feeding. At one day after Csp_P ingestion, we found that Csp_P was present in all sampled mosquitoes with an average bacterial load of 5.12×104 (Figure 1D, E). At two days after Csp_P exposure, only 5% of Csp_P-fed An. gambiae were still alive (Fig. 2C) and Csp_P was detected in only one (12.5%) of these remaining mosquitoes (Fig. 1D, E). In septic Ae. aegypti mosquitoes that had fed on a 1010 CFU/ml Csp_P-containing sugar solution, we identified Csp_P in 79% of mosquitoes sampled on day 1 post feeding (Fig. 1F). At three days after feeding on the Csp_P-containing sugar solution, approximately 30% of the Ae. aegypti were still alive (Fig. 2D) and Csp_P was detected in 15% of these mosquitoes (Fig. 1F). These data suggest that Csp_P colonized the vast majority of An. gambiae and Ae. aegypti mosquitoes by day 1 post exposure and that Csp_P caused rapid mortality in most individuals. The small percentage that survived up to day 2 or 3, post exposure, may have received a small dose of bacteria and succeeded in clearing it by the time they were dissected. It is difficult to compare the colonization efficiency between septic and antibiotic treated mosquitoes because the survival curves differ dramatically (Fig. 2). While it appears that Csp_P was better at colonizing the midgut of antibiotic treated An. gambiae (Fig. 1A vs. 1D) and Ae. aegypti (Fig. 1A vs. 1F), our measurement does not take into account that individuals died much more rapidly in the septic population. This rapid mortality likely selected for mostly Csp_P negative individuals by day 2 and 3 post-feeding.
Fig. 2. Csp_P exposure causes high mortality in adults and larvae. 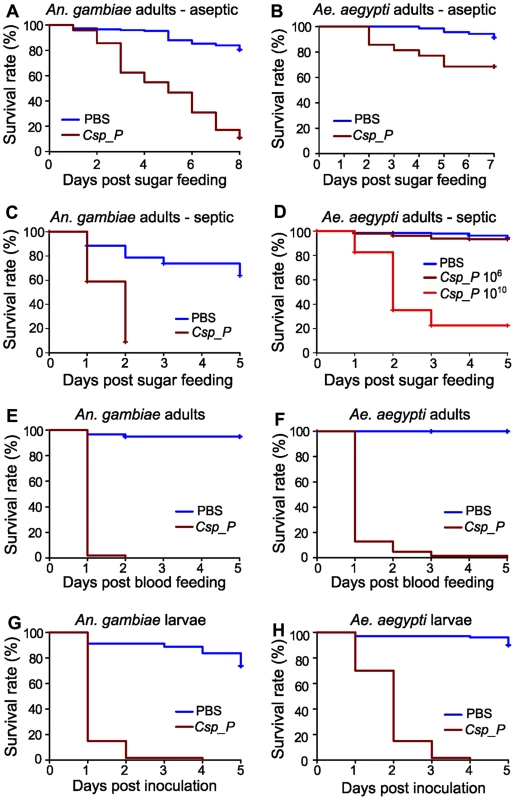
Csp_P was experimentally introduced into the adult midgut via either a sugar meal (A–D) or blood meal (E, F), and mortality was observed over 5–8 days. To introduce Csp_P via sugar meal, adults were allowed to feed for 24 h on 1.5% sucrose containing Csp_P liquid culture at a final concentration of ∼108 CFU/ml for An. gambiae and ∼106 or 1010 CFU/ml for Ae. aegypti. Csp_P ingestion significantly decreased survival in sugar-fed aseptic (i.e. pre-treated with antibiotics) An. gambiae (A, p<0.0001) and Ae. aegypti (B, p<0.0001). Each experiment was replicated three times. Total sample sizes: (A)PBS = 149; (A)Csp_P = 146; (B)PBS = 70; (B)Csp_P = 70. Ingestion of Csp_P significantly decreased survival in sugar-fed septic (i.e. not treated with antibiotics) An. gambiae (C, p<0.0001). In septic Ae. aegypti, survival was significantly decreased after feeding on a 1010 CFU/ml sugar meal (D, p<0.0001) but not after feeding on a 106 CFU/ml sugar meal (D, p = 0.08). Experiments in C and D were replicated twice. Total sample sizes: (C)PBS = 95; (C)Csp_P = 124; (D)PBS = 185; (D)Csp10∧6 = 223; (D)Csp10∧10 = 226. To introduce Csp_P via blood meal, Csp_P liquid culture (∼108 CFU/ml) was mixed 1∶1 with human blood/serum and fed to septic An. gambiae (E) and Ae. aegypti (F) adults. Experiments were replicated three times with total sample sizes: (E)PBS = 59; (E)Csp_P = 51; (F)PBS = 37; (F)Csp_P = 62. The effects of Csp_P on larval mortality were also tested by placing 2- to 4-day-old An. gambiae (G) and Ae. aegypti (H) larvae in water containing Csp_P at a starting concentration of 106 CFU/ml and monitoring survival over 5 days. Experiments were replicated 2–3 times with final sample sizes: (G)PBS = 80; (G)Csp_P = 60; (H)PBS = 100; (H)Csp_P = 60. P values reported above were obtained by performing pairwise Log-Rank Tests between PBS and Csp_P treatments. Survival curves were fitted using the Kaplan-Meier method. Vertical tick-marks indicate censored samples; in C and D multiple individuals were dissected on each day to measure Csp_P prevalence and bacterial load for Figure 1. Csp_P exerts entomopathogenic activity upon mosquito ingestion and larval exposure
We examined the influence of Csp_P midgut colonization on mosquito longevity by exposing antibiotic-treated An. gambiae and Ae. aegypti mosquitoes to a sugar source for 24 h containing Csp_P at a final concentration of 108 and 106 CFU/ml, respectively, and then monitoring survival. This treatment led to a decrease in the longevity of both species when compared to non-exposed control mosquitoes (Fig. 2A, B). We repeated this experiment with septic (i.e. not antibiotic treated) An. gambiae and Ae. aegypti. We found that feeding on a sugar source containing Csp_P at a concentration of 108 CFU/ml resulted in rapid mortality of An. gambiae adult females (Fig. 2C). Mortality of septic Ae. aegypti females was not increased after feeding on a sugar source containing Csp_P at a concentration of 106 CFU/ml but was dramatically increased when the sugar meal contained Csp_P at a concentration of 1010 CFU/ml (Fig. 2D). These data suggest that Csp_P has strong entomopathogenic activity regardless of whether other microbes are present in the mosquito gut. We observed lower survival in septic An. gambiae and Ae. aegytpi after feeding on a blood meal containing Csp_P at a final concentration of 108 CFU/ml (Fig. 2E, F). The stronger entomopathogenic effect upon Csp_P introduction through the blood meal was most likely because the mosquitoes received a large single bacterial dose upon bloodfeeding rather than the multiple low doses that would be expected during sugar feeding. It is also possible that Csp_P proliferated to high numbers in the nutritious blood.
To study the influence of Csp_P on larval viability, we placed 2 - to 4-day-old mosquito larvae in groups of 10 in pools containing 5 ml distilled water supplemented with 50 µl of a 1.0 OD600 liquid culture of Csp_P, and then monitored survival. This resulted in almost complete mortality of An. gambiae and Ae. aegypti larvae over a 3 - and 2-day period, respectively, when compared to the control larvae that were exposed to the normal breeding water microbiota (Fig. 2G, H). These studies suggest that Csp_P –mediated mortality may be the direct result of a mosquitocidal factor or systemic infection through dissemination into the hemolymph; alternatively, its colonization of the midgut (or other tissues) might cause mortality indirectly by interfering with vital functions of the mosquito. Studies of Pseudomonas aeruginosa colonization of the Drosophila melanogaster gut have shown that biofilm formation can dramatically affect both dissemination within the hemolymph and fly mortality [13]. Csp_P is capable of forming biofilms in vitro, though whether biofilm formation occurs within the mosquito midgut remains untested. C. violaceum produces cyanide at high cell density [17], [18] via the cyanide-producing hcnABC operon, a behavior that is reportedly regulated by quorum sensing [18], [19]. Cyanide production by bacteria has been shown to cause host mortality in both nematodes [20] and insects [21]. Chromobacterium subtsugae has previously been shown to exert oral toxicity in various insects of agricultural importance, but not in Culex mosquitoes [22].
Csp_P colonization of the mosquito midgut compromises pathogen infection
To investigate whether the presence of Csp_P in the mosquito midgut could influence the infection of An. gambiae with P. falciparum and of Ae. aegypti with the dengue virus DENV2, we assayed the infection of mosquitoes that had been exposed to Csp_P through sugar feeding 2 days prior to feeding on parasite - or virus-infected blood. Approximately one week after An. gambiae had fed on a P. falciparum gametocyte culture, parasite infection was assayed by counting oocyst-stage parasites on the basal side of the mosquito midgut. DENV2 infection of the midgut of Ae. aegypti was assayed through standard plaque assays 7 days after an infectious bloodmeal. All experiments were initiated using similar numbers of adult females for each treatment, but because Csp_P exposure causes high mortality in adults (Fig. 2), very few Csp_P-fed mosquitoes were still alive when the parasite and dengue infection assays were conducted. Nevertheless, we found that surviving mosquitoes exposed to Csp_P through sugar feeding prior to feeding on infectious blood displayed significantly increased resistance to P. falciparum infection and DENV infection (Fig. 3). The inhibition of P. falciparum infection was even greater when Csp_P was introduced through the gametocyte-containing blood meal at 106 CFU/ml (Fig. 3B), an effect most likely attributable to the larger number of ingested bacteria. Csp_P may inhibit pathogen infection directly through physical interaction with the pathogens or the production of anti-pathogen molecules. Alternatively, Csp_P may indirectly inhibit Plasmodium or dengue by (a) altering the long-term physiology or health of the mosquito such that pathogen infection is inhibited, (b) triggering a mosquito anti-pathogen response or (c) selecting for individuals that are more fit to resist Csp_P as well as DENV and Plasmodium infection. However, Csp_P's in vitro anti-pathogen activity (discussed below) suggests it has the potential to directly inhibit pathogen survival in the mosquito gut. Further studies are necessary to elucidate the mechanism by which Csp_P inhibits the pathogens in vitro and in vivo.
Fig. 3. Csp_P reduces mosquitoes' susceptibility to malaria and dengue infection. 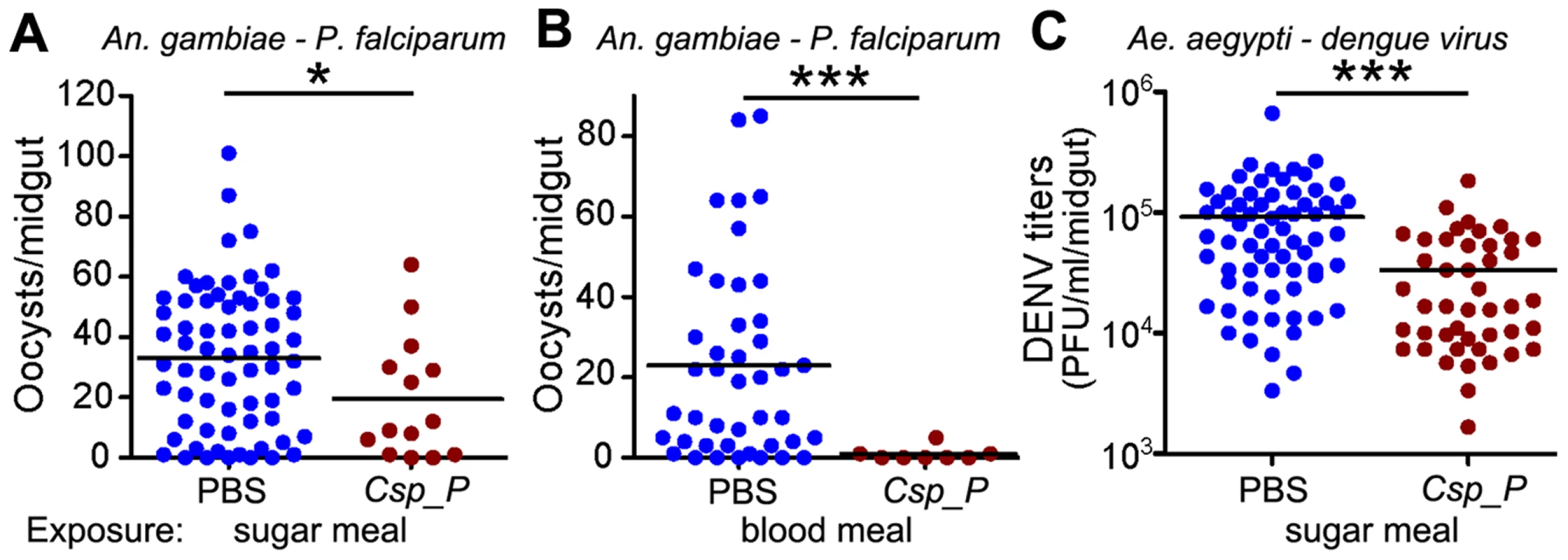
In (A) and (C), antibiotic-treated adults were allowed to feed for 24 h on 1.5% sucrose containing Csp_P liquid culture at a final concentration of ∼108 CFU/ml for An. gambiae (A) and ∼106 CFU/ml for Ae. aegypti (C). After introduction of Csp_P via the sugar meal, An. gambiae mosquitoes were given a blood meal that contained P. falciparum, and Ae. aegypti mosquitoes were given a blood meal that contained dengue virus. In (B), Csp_P (106 CFU/ml) was introduced concurrently with P. falciparum via blood meal through blood feeding of antibiotic-treated An. gambiae. In all experiments, PBS was used as the non-Csp_P-exposed control. At 7 days after infection, midguts were dissected. Oocysts were counted in P. falciparum-infected An. gambiae females, and dengue virus titers were assayed in dengue-infected Ae. aegypti females by conducting standard plaque assays. Experiments were initiated using similar numbers of adult females in each treatment (A, B starting numbers = 45–50/trtmt, C starting numbers = 30–40/treatment). All experiments were replicated at least three times with final samples sizes: (A)PBS = 67, (A)Csp_P = 14, (B)PBS = 43, (B)Csp_P = 8, (C)PBS = 68, (C)Csp_P = 45. Differences between treatments were assessed by Mann-Whitney test (*, p<0.05; ***, p<0.001). Csp_P induces mosquito innate immune system genes
We have previously shown that the An. gambiae and Ae. aegypti midgut microbiota elicit basal immune activity by elevating the expression of several immune factors, including antimicrobial peptides and antipathogen factors [5], [7], [8], [23], [24]. To determine Csp_P's potency in inducing the mosquito's innate immune system, we exposed mosquito SUA-5B cells to various concentrations of Csp_P and assayed for changes in the activity of a Cecropin1 promoter driving the expression of a luciferase reporter gene. We exposed these same cells to Pseudomonas putida, a Gram-negative bacterium that belongs to a bacterial genus commonly found in mosquito midguts [25]–[27]. This experiment showed that Cec1 expression increased with increasing Csp_P exposure, providing evidence that Csp_P is a potent immune elicitor (Fig. 4). We also compared the transcript abundance of mosquito immune genes in midguts from antibiotic-treated naïve mosqutioes to those from mosquitoes that had been provided a sugar source spiked with Csp_P (108 CFU/ml for An. gambiae and 106 CFU/ml for Ae. aegypti) 2 days earlier. We chose to assay gene expression at 2 days post exposure because this is the time at which increased mortality due to infection begins to occur. We hypothesized that infection levels and therefore any potential immune response would be high at this time. In Ae. aegypti, we found that cecropin E and G and defensin C displayed at least a 2-fold increase in transcript abundance in the midgut of Ae. aegypti colonized with Csp_P bacteria when compared to naïve controls (Fig. S1A). In An. gambiae, we found non-significant trends toward increased transcript abundance of the Rel2, FBN9 and cecropin genes and toward decreased transcript abundance of the defensin gene in the midgut tissue (Fig. S1B). These data represent a single time point post-infection, and while it is possible that additional time points may reveal dynamic patterns of Csp_P-induced changes in gene expression, our results generally agree with the cell culture data, and as a whole show that Csp_P has an immune-eliciting capacity in the mosquito gut.
Fig. 4. Csp_P elicits immune gene expression in the mosquito. 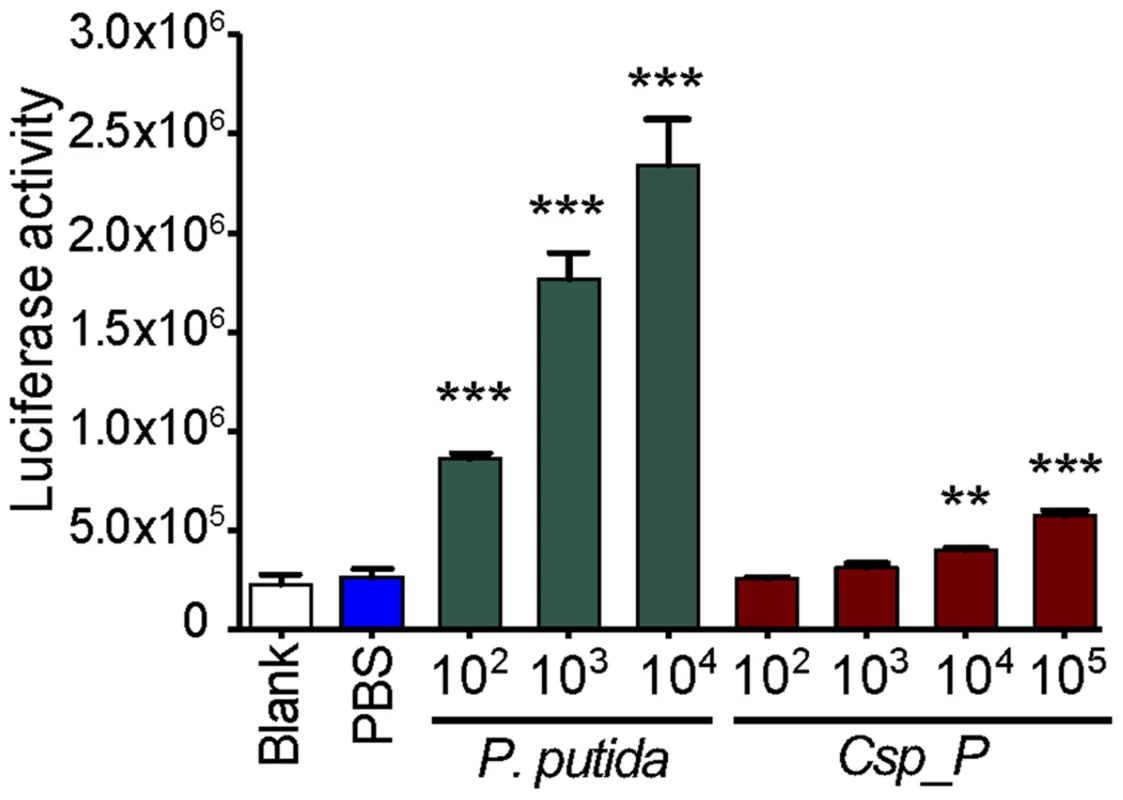
Induction of the Cec1 promoter in the SUA-5B cell-line exposed to P. putida and Chromobacterium sp. Csp_P. SUA-5B cells expressing a luciferase reporter gene driven by a Cec1 promoter were exposed to increasing concentrations of Csp_P and P. putida bacteria. Differences between bacteria-treated samples and PBS control samples were assessed by Dunnett's Multiple Comparison Test (**, p<0.01; ***, p<0.001). Csp_P inhibits Plasmodium development and abolishes dengue virus infectivity in vitro, independent of the mosquito
To test whether Csp_P could exert a direct anti-Plasmodium or anti-dengue effect in vitro that is independent of the mosquito, we performed experiments in which parasite development and virus infectivity were assayed after exposure to various preparations of either planktonic or biofilm cultures of Csp_P. Planktonic-state Csp_P was obtained by culturing Csp_P in liquid LB at 30°C overnight on a platform shaker. Biofilm was produced by culturing Csp_P in LB without agitation in a polystyrene 24-well plate at room temperature for 48 h, unless otherwise indicated. The anti-dengue and anti-Plasmodium activity of the following five different preparations of Csp_P was then tested: (a) 1 ml (108 CFU/ml) planktonic-state liquid culture, (b) 1 ml (109 CFU/ml) biofilm supernatant consisting of liquid LB drawn off freshly cultured biofilm, (c) 5 mg (109 CFU/ml) fresh biofilm resuspended in 1× PBS, (d) 5 mg dessicated biofilm prepared from biofilm collected 1–2 days prior to assay and allowed to completely dessicate at room temperature and then rehydrated in 1× PBS, (e) 5 mg heat-inactivated biofilm prepared by heating biofilm at 90°C for 24 h, collected 1 day prior to assay.
Our in vitro assays showed that Csp_P exerts potent anti-Plasmodium activity against both asexual and sexual parasite stages. We exposed P. falciparum 3D7 asexual stage parasites to all five bacterial preparations in vitro. Because bacterial growth can interfere with determining parasite number, we removed bacterial cells by filtering all preparations though a 0.2-µm filter. We found that all filtrates from 36-h biofilm preparations (fresh, supernatant, and dessicated) possessed strong anti-Plasmodium activity, resulting in inhibition of asexual stage parasites at a level comparable to the chloroquine-treated positive control (p<0.001, Fig. 5A). We also detected moderate anti-asexual stage activity in planktonic Csp_P preparations (p<0.001), while heat-inactivated Csp_P biofilm and biofilm from another bacterial species, Comamonas sp., had no inhibitory effect. We exposed an in vitro Plasmodium ookinete culture to all five filtered bacterial preparations to assess sexual-stage inhibition and found that the Csp_P 48-h biofilm (fresh, p<0.001; and dessicated, p<0.05) and biofilm supernatant (p<0.001) strongly blocked ookinete development (Fig. 5B). Exposure of the ookinete culture to the filtered planktonic Csp_P liquid culture resulted in a moderate but non-significant inhibition of ookinete development, and exposure to heat-inactivated Csp_P biofilm or Comomonas sp. biofilm filtrate had no effect on ookinete development (Fig. 5B). We also tested the effect of Csp_P bacterial preparations on P. falciparum gametocyte viability. Exposure to 42-h fresh biofilm filtrate resulted in 100% inhibition (p<0.001, Fig 5C) and exposure to 42-h dessicated biofilm resulted in approximately 60% inhibition (p<0.05, Fig. 5C) of P. falciparum gametocyte development. Gametocytemia could not be estimated for 42-h biofilm supernatant because this preparation caused hemolysis of RBCs (Fig. 5C). However, 36-h biofilm supernatant (which is not hemolytic) caused approximately 60% gametocyte inhibition when compared to the LB+PBS control (p = 0.06, Fig. S2).
Fig. 5. Csp_P has anti-Plasmodium and anti-dengue activity in vitro. 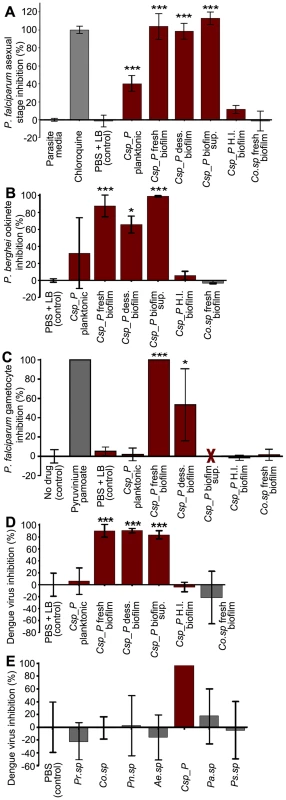
Csp_P was grown under planktonic and/or biofilm conditions and tested for anti-pathogen activity independent of the mosquito. Five different preparations of Csp_P were tested: (a) planktonic-state liquid culture, (b) biofilm supernatant, (c) fresh biofilm, (d) dessicated biofilm, and (e) heat-inactivated biofilm. A fresh Comamonas sp biofilm was also tested as control. (A) Csp_P 36-h biofilm has anti-parasite activity against asexual-stage P. falciparum. Csp_P cultures were filtered using a 0.2-µm filter and mixed with ring-stage P. falciparum parasite cultures. SYBR green I was then added to each sample, and inhibition of asexual-stage P. falciparum by Csp_P was measured by assaying fluorescence relative to the negative control (parasite medium, standardized to 0% inhibition). Chloroquine was used as a positive control and standardized to 100% inhibition. We performed a Tukey's test on the raw data to determine whether each bacterial treatment differed significantly from the PBS+LB control (*** p<0.001). (B) Csp_P has anti-parasite activity against ookinete-stage P. falciparum. Csp_P bacterial preparations were filtered using a 0.2-µm filter and mixed with blood taken from female Swiss Webster mice infected with Renilla luciferase-expressing transgenic P. berghei. Ookinete-stage P. berghei parasite counts were determined using the Renilla luciferase assay system, and percent inhibition by Csp_P was calculated relative to the negative control (PBS+LB control, standardized to 0% inhibition). We performed a Tukey's test to determine whether each bacterial treatment differed significantly from the control (*p<0.05, ***, p<0.001). (C) Csp_P 42-h biofilm has anti-parasite activity against gametocyte-stage P. falciparum. Csp_P cultures were filtered using a 0.2-µm filter and mixed with gametocyte-stage P. falciparum cultures. Erythrocytes were examined for gametocytes using Giemsa-stained blood films collected 3 days after Csp_P exposure. The red X indicates that the supernatant caused hemolysis and was therefore unusable. We determined gametocyte density per 1000 RBCs for each sample and performed a Tukey's test to determine whether each bacterial treatment significantly differed from the PBS+LB control (*p<0.05, *** p<0.001). (D) Csp_P has anti-dengue activity. Each Csp_P bacterial preparation (75 µl, unfiltered) was mixed with 75 µl MEM containing dengue virus serotype 2 and incubated at room temperature for 45 min. Samples were then filtered through a 0.2-µm filter and used to infect BHK21-15 cells. Percent inhibition was calculated as the percent decrease in PFU/ml relative to the negative control (PBS+LB, standardized to 0% inhibition). We analyzed the significance of pairwise comparisons between each treatment and the control using a Tukey's test (***, p<0.001). (E) Csp_P has anti-dengue activity when virus is suspended in human blood. Biofilms from multiple bacteria were tested for anti-dengue activity. All bacteria tested were isolated from field-caught Ae. aegypti mosquitoes. Ps.sp = Pseudomonas sp., Pr.sp = Proteus sp., Pn.sp = Paenobacillus sp., Co.sp = Comamonas sp., Pa.sp = Pantoea sp., Ae.sp = Aeromonas sp. [5]. The biofilm from each species was grown for 48 h at room temperature, and dengue virus mixed 1∶1 with human blood was added directly to the biofilm. After a 45-min incubation, the virus+blood/bilofilm solution was filtered and used to infect C6/36 cells. Biofilm sup = biofilm supernatant, H. I. biofilm = heat inactivated biofilm, dess. biofilm = dessicated biofilm re-suspended in 1× PBS. To test the inhibitory effect of Csp_P preparations on dengue virus infectivity in vitro, we mixed dengue virus (106 PFU/ml) in MEM 1∶1 with each of the five bacterial preparations of Csp_P for 45 min. Samples remained unfiltered during intial exposure to dengue and were filtered through a 0.2-µm filter before proceeding with standard plaque assays to avoid contamination of host cells. We found that exposure of dengue virus to a planktonic Csp_P culture did not affect its infectivity in BHK21-15 cells, whereas exposure to Csp_P biofilm, dessicated biofilm, or biofilm supernatant did abolish dengue virus infectivity (p<0.001, Fig. 5D). To better replicate the effect that Csp_P biofilm might have on dengue virus in human blood, we exposed dengue virus in human blood to Csp_P fresh biofilm for 45 min. We then filtered the blood+virus/biofilm mixture and assessed dengue virus infectivity by standard plaque assay. We found that fresh Csp_P biofilm displayed strong anti-dengue activity when the virus was suspended in human blood (Fig. 5E). Csp_P fresh biofilm was unique in its anti-dengue activity, since the biofilms of several other bacterial isolates from the guts of field-caught mosquitoes [5] did not exert any antiviral activity against dengue virus in human blood (Fig. 5E). The anti-dengue activity of Csp_P was apparently dependent on biofilm maturation, since biofilm grown for 24 h showed only weak inhibition when compared to 48-h biofilm (Fig. S3A). The Csp_P biofilm-associated anti-Plasmodium and antiviral activity was also heat-sensitive, since it could be inactivated through a 24-h incubation at 90°C (Fig. 5A–D).
Bacterial biofilms are composed of a matrix of extracellular polymeric substances containing polysaccharides, proteins, DNA, and secondary metabolites [12]. To investigate whether the anti-viral activity could simply be a result of virus particle sequestration by the biofilm, we mixed a dengue virus suspension with biofilm and incubated the mixture for a period of 45 min. Samples were then centrifuged, and viral RNA in the supernatants was quantified by RT-qPCR and compared between experimental (biofilm+DENV) and control (LB+DENV) treatments. Our results indicated that the dengue virus was not sequestered by Csp_P biofilm, since similar viral RNA copies were detected in the biofilm-exposed sample and the LB control sample (Fig. S3B). To investigate whether the loss of dengue virus infectivity was due to a biofilm-mediated change in the pH of the medium, we measured the pH of a dengue virus-Csp_P biofilm mixture at the end of a 45-min incubation period. The pH measurements showed an increase in the pH of the medium from 7.6 to 8.3 (Fig. S4A). A similar change in the pH was observed when we used the biofilms of other bacteria (Pantoea sp. Pasp_P and Proteus sp. Prsp_P) that do not affect dengue virus infectivity (Fig. S4A). To further investigate the effect of pH on dengue virus infectivity, we adjusted the pH of the MEM medium with NaOH and HCl to pH values of 5.0, 7.7, 8.5, and 10.0 prior to a 45-min incubation with the dengue virus. A decrease in virus infectivity was only observed after exposure to a pH of 5.0, suggesting that the moderate increase in pH did not mediate the Csp_P biofilm's inhibition of virus infectivity (Fig. S4B). We also showed that Csp_P biofilm does not exert a cytotoxic effect on insect or mammalian cells, as assessed by standard trypan blue cell staining (Invitrogen) (Fig. S5). We finally tested whether the Csp_P biofilm could influence the host cells' susceptibility to dengue virus infection by exposing C6/36 cells to Csp_P biofilm, then removing the biofilm through washes with PBS prior to dengue virus infection assays. This treatement did not influence the virus's ability to infect the host cells (Fig. S6), suggesting that the anti-DENV activity of Csp_P biofilm is not due to a reduced host cell susceptibility to the virus but is likely a direct anti-viral effect.
Csp_P produces broad-spectrum antibacterial activity(ies)
To provide baseline information on the potential production of antibacterial factors by Csp_P, we performed a basic growth inhibition assay by investigating the ability of a number of other mosquito midgut-derived bacterial isolates (Ae. aegypti-derived microbiota: Ps.sp = Pseudomonas sp., Pr.sp = Proteus sp., C.sp_P = Chromobacterium sp_P, C.viol = C. violaceum, Pn.sp = Paenobacillus sp.; An. gambiae-derived microbiota: Co.sp = Comamonas sp., Ac.sp = Acinetobacter sp., P.pu = Pseudomonas putida, E.sp = Enterobacter sp., Pa.sp = Pantoea sp., Ps.sp = Pseudomonas sp., S.sp = Serratia sp., Ch.sp = Chryseobacterium sp.) to grow in proximity to Csp_P on LB agar plates (Fig. 6). Csp_P was streaked on LB agar and allowed to grow for 48 hours. Midgut-derived bacterial isolates were then vertically streaked up to the Csp_P streak, and allowed to grow in the presence of Csp_P. This assay showed a prominent growth inhibition zone around the Csp_P streak, with inhibition of the growth of all the bacterial isolates that were derived from field-collected Ae. aegypti [5] and An. gambiae [3], including a close relative known for its production of a variety of bioactive factors, C. violaceum (Fig. 6A).
Fig. 6. Csp_P has anti-bacterial activity against many species commonly found in the midguts of Aedes and Anopheles mosquitoes. 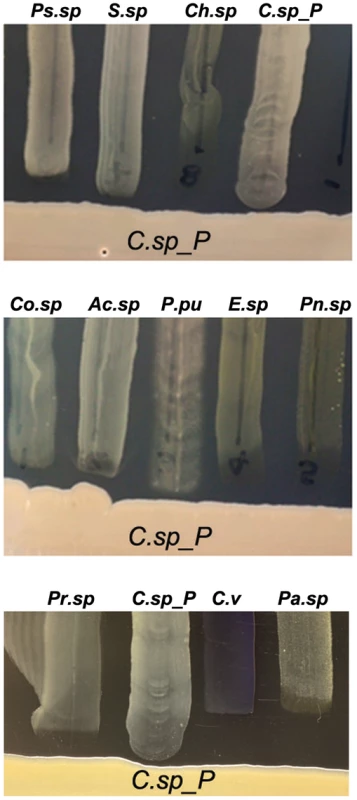
Csp_P was streaked on LB agar along with multiple bacterial species, and plates were observed for formation of zones of inhibition around Csp_P. Ps.sp = Presudomonas sp., Pr.sp = Proteus sp., C.sp_P = Chromobacterium sp_P, C.v = C. violaceum, Pn.sp = Paenobacillus sp., Co.sp = Comamonas sp., Ac.sp = Acinetobacter sp., P.pu = Pseudomonas putida, E.sp = Enterobacter sp., Pa.sp = Pantoea sp., S.sp = Serratia sp., Ch.sp = Chryseobacterium sp. [3], [5], [7]. Conclusions
Insect-bacteria associations can influence vector competence in multiple ways; these include shortening the insect's life span, blocking infection with human pathogens by the production of bioactive anti-pathogen factors, and eliciting the insect immune system. We have identified a Chromobacterium sp. (Csp_P) bacterium from the midgut of field-derived Aedes aegypti that exerts broad-spectrum anti-pathogen activity against Plasmodium and dengue virus. Specifically, Csp_P renders An. gambiae and Ae. aegypti more resistant to infection by the human malaria parasite Plasmodium falciparum and dengue virus, respectively. Csp_P inhibits the growth of a variety of other bacterial species found in the mosquito midgut and is capable of rapidly colonizing the mosquito midgut. Csp_P appears to exert entomopathogenic activity, since exposure of larvae to Csp_P in the breeding water and ingestion of Csp_P by adult mosquitoes result in high mosquito mortality. It is possible that Csp_P could be effectively used as a transmission blocking agent if it was delivered to mosquitoes through baited sugar traps [28]. Csp_P's ability to colonize the mosquito gut could be further enhanced through established selection procedures based on consecutive passages of the bacterium through the mosquito intestine [29]. Csp_P could be used alone in baited sugar traps or in combination with other microbes that have also been shown to either kill the mosquito or reduce pathogen infection, or both, when present in the mosquito gut [3], [24]. The larvicidal activity of Csp_P also renders it interesting for potential use in mosquito population suppression. The anti-pathogen activities of Csp_P appear to be mediated by bacteria-produced metabolites that also inhibit parasite and virus infection in vitro, making them interesting as possible lead compounds for transmission blocking and therapeutic drug development. The entomopathogenic, anti-bacterial, anti-viral, and anti-Plasmodium properties of Csp_P make this bacterium a particularly interesting candidate for the development of novel control strategies for the two most important vector-borne diseases, and they therefore warrant further in-depth study.
Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Mice were only used for mosquito rearing as a blood source according to approved protocol. The protocol was approved by the Animal Care and Use Committee of the Johns Hopkins University (Permit Number: M006H300). Commercial anonymous human blood, supplied from Interstate Blood Bank Inc., was used for Plasmodium and dengue virus infection assays in mosquitoes, and informed consent was therefore not applicable. The Johns Hopkins School of Public Health Ethics Committee has approved this protocol. Mosquito collections were performed in residences after owners/residents permission.
Mosquito rearing and antibiotic treatment
Aedes aegypti mosquitoes were from the Rockefeller strain, and Anopheles gambiae mosquitoes were from the Keele strain. Both were maintained on a 10% sugar solution at 27°C and 95% humidity with a 12-h light/dark cycle. Sterile cotton, filter paper, and sterilized nets were used to maintain the cages as sterilely as possible. For experiments utilizing aseptic mosquitoes, females were maintained on a 10% sucrose solution with 20 U penicillin and 20 µg streptomycin from the first day post-eclosion until 1–2 days prior to challenge. The effectiveness of the antimicrobial treatment was confirmed by colony forming unit assays prior to blood-feeding or bacterial challenge.
Introduction of bacteria via sugar meal
In cases where mosquitoes were antibiotic treated, reintroduction of bacteria through a sugar meal was done by first treating mosquitoes with antibiotics for 2–3 days after emergence, then providing them with 10% sucrose (for An. gambiae) or sterile water (for Ae. aegypti) for 24 h post-antibiotic treatment. When mosquitoes were not antibiotic treated, they were maintained on 10% sucrose for 2–5 days post emergence. Ae. aegypti were given sterile water during the final 24 hours of this period. In all cases, mosquitoes were then starved overnight and fed for 24 h on cotton strips moistened with a 1.5% sucrose solution containing Csp_P at a final concentration of approximately 108 CFU/ml for An. gambiae and 106 CFU/ml for Ae. aegypti. In some experiments (Figures 1 and 2), Ae. aegypti mosquitoes were also fed Csp_P at a final concentration of 1010 CFU/ml.
Assaying prevalence and bacterial load of Csp_P
In antibiotic treated mosquitoes, midguts were dissected three days post ingestion of Csp_P, homogenized in 1× PBS and plated on LB agar. Colonies were then counted to estimate colony forming units (CFUs) per midgut as well as prevalence of Csp_P. In mosquitoes not treated with antibiotics, prevalence and/or bacterial load was estimated in one of two ways. For An. gambiae, midguts were dissected at one and two days post Csp_P ingestion, homogenized in 1× PBS and serial dilutions of the homogenate were plated on LB agar supplemented with ampicillin (10,000 ug/ml). Csp_P is highly resistant to ampicillin and grows readily even at this high concentration. We verified that Csp_P was the only bacterium growing on antibiotic treated plates by first confirming that all colonies that grew were similar in color, growth rate and colony morphology. 16s rDNA was then sequenced from a subset of colonies and verified to match the sequence of Csp_P from pure freezer stock.
It was not possible to use this method for Ae. aegypti because their midguts commonly contained other highly ampicillin-resistant bacteria. These contaminants grew to very high numbers on the ampicillin-treated plates and interfered with the detection of Csp_P. DNA was therefore extracted using the ZR Soil Microbe DNA MicroPrep kit (Zymo Research) from samples dissected 1 and 3 days after feeding on a sugar meal containing either PBS or Csp_P (1010 CFU/ml).The manufacturer's protocol was altered in the following way: instead of using lysis buffer to disrupt cells, each midgut was put in 500 µl 1× PBS, 25 µl lysozyme (10 mg/ml) and 7.5 µl mutanolysin (10 KU/ml) were added and the samples were incubated at 37°C for 1.5 h. 15 µl proteinase K and 25 µl 10% SDS were then added, samples were incubated at 55°C for 1 h, and the standard protocol was then resumed. A diagnostic PCR was performed to assess the presence of Csp_P in each individual midgut. Primers were designed to amplify a 415 bp fragment of the Csp_P hydrogen cyanide synthase B gene and the primers were verified to be Csp_P-specific using Primer BLAST from NCBI (Forward primer: 5′AGGGCGTAACCCTGGACTAT 3′, Reverse primer: 5′ CCGAAGGAACTGGCTTCGTA 3′). PCR was performed with the above primers using 10 ng DNA as template and Phusion High-Fidelity DNA Polymerase according to the manufacturer's instructions, with the following exceptions: 0.5 µl of each primer (10 µm) was used, and 0.25 µl BSA was added to each reaction. Cycling conditions were as follows: 95°C for 30 seconds, [95°C for 30 s, 65°C for 30 s, 72°C for 45 s]×27 cycles, 72°C for 10 minutes. 8 µl of each sample was run on a 1% agarose gel and visualized at 400 ms exposure. A visible 415 bp band was considered positive evidence of Csp_P bacteria (see Fig. S7 for a representative example). A very faint band was detected in one of 40 PBS samples, suggesting a minor contamination event or the presence of another bacterium with high sequence identity to Csp_P. This was an isolated incident and was not seen in any other PBS samples. Two independent PCR products were sequenced from Csp_P fed samples and verified to be a perfect match to the sequence obtained from Csp_P sequenced directly from freezer stock. To serve as a positive control and to allow estimation of the sensitivity of the diagnostic PCR, a standard curve was run in which a range of 107–101 copies of the Csp_P hcn B PCR product was used as template. In this way, it was possible to estimate the minimum detection threshold of this assay. Using the above mentioned PCR conditions, a band was detectable in wells containing 103 initial copies of the hcn B product but not in wells containing 102 initial copies, suggesting that this assay is capable of detecting a minimum of 103 copies of Csp_P/midgut.
Introduction of bacteria via blood meal
At 2 days prior to blood feeding, sucrose was removed, and the mosquitoes were given sterile water. They were then starved for 12 h prior to blood feeding. Csp_P was grown overnight in liquid LB at 30°C. The overnight culture (1 ml) was then pelleted, washed with 1× PBS, and resuspended in 1× PBS to OD600 = 1.0, which equals a concentration of approximately 108 CFU/ml. Mosquitoes were then allowed to membrane-feed on blood containing bacteria or 1× PBS as a control (blood mixture: 50% 1.0 OD600 bacterial culture or 1× PBS, 40% blood, 10% human serum). Bacteria-fed adult females ingested approximately 105 CFU per mosquito.
Exposure of larvae to Csp_P
At 2–4 days post-hatching, larvae were placed in cell culture plates in groups of 10 per well. Each well contained 5 ml sterile water plus a small amount of larval food (liver powder, tropical fish flake food, and rabbit food pellets mixed in a 2∶1∶1 ratio). We then added 50 µl of an overnight culture of Csp_P diluted to OD600 = 1.0 (108 CFU/ml) to each well; 1× PBS was added to control wells, and mortality was monitored in all wells for a 5-day period.
Cell culture maintenance, mosquito infections with dengue virus, and titration of infected midguts
Dengue virus serotype 2 (New Guinea C strain, DENV-2) was propagated in the C6/36 mosquito cell line according to previously published methods [8]. In brief, cell line infection was allowed to proceed for 5–7 days, at which time the cells were harvested with a cell scraper and lysed by freezing and thawing in dry CO2 and a 37°C water bath, then centrifuged at 800 g for 10 min. Dengue virus serotype 2 was isolated and mixed 1∶1 with commercial human blood and used for infections as described in [8]. Mosquitoes that had previously fed on Csp_P bacteria-sucrose solution were starved overnight prior to dengue virus infection. Infected mosquitoes were collected at 7 days post-infection and surface-sterilized by dipping them in 70% ethanol for 1 min and then rinsing them twice in 1× PBS for 2 min each. Midgut dissection was done in one drop of 1× PBS under sterile conditions, and the midgut was transferred to a microcentrifuge tube containing 150 µl of MEM. Midguts were homogenized using a Kontes pellet pestle motor, filtered, and stored at −80°C until ready for virus titration.
Dengue virus titration of infected midguts was done as previously reported [8], [30]. In brief, the infected midgut homogenates were serially diluted and inoculated into C6/36 cells in 24-well plates. After an incubation of 5 days at 32°C and 5% CO2, the plates were fixed with 50%/50% methanol/acetone, and plaques were assayed by peroxidase immunostaining using mouse hyperimmune ascitic fluid specific for DENV-2 as the primary antibody and a goat anti-mouse HRP conjugate as the secondary antibody. In addition, where indicated, dengue virus plaque assays were conducted in BHK-21 cells. At 5 days post-infection, the 24-well plates were fixed and stained with crystal violet. Plaques (formed by cells with cytopathic effect) were counted and analyzed.
P. falciparum cultivation, mosquito infections, and oocyst counts
P. falciparum strain NF54 was maintained in continuous culture according to the method described by Tragger and Jensen [31]. In brief, P. falciparum was grown in O+ red blood cells (RBCs) at 2% hematocrit and RPMI 1640 medium supplemented with glutamine, HEPES, hypoxanthine, and 10% O+ human serum. To maintain a microaerophilic environment, parasites were maintained in a candle jar at 37°C. Use of human erythrocytes to support the growth of P. falciparum was approved by the internal review board of the Bloomberg School of Public Health. Gametocytemia and exflagellation events were assessed after 18 days of P. falciparum culture. The gametocyte culture was centrifuged and diluted in a mixture of RBCs supplemented with serum. Mosquitoes were rendered aseptic via antibiotic treatment and then fed on membrane feeders for 30 min with blood containing P. falciparum gametocytes. Csp_P was either added directly to the infectious blood meal (bacterial concentration = 106 CFU/mL) or introduced via sugar meal as described above 3 to 4 days prior to the infectious blood meal. On the same day as the blood meal, mosquitoes were sorted, and the unfed mosquitoes were removed. At 7 to 8 days after blood feeding, the fed mosquitoes were dissected, and their midguts were stained with 0.1% mercurochrome. The number of oocysts per midgut was determined with a light-contrast microscope, and the median was calculated for the control and each experimental condition. More than three independent replicates were used per group.
Csp_P culture preparations for in vitro anti-Plasmodium and anti-dengue activity assays
To grow bacteria in planktonic conditions, we spiked 5 ml sterile LB with 5 µl of bacterial freezer stock and allowed the culture to grow overnight at 30°C with shaking. We then diluted planktonic cultures to OD600 = 1.0 (±0.1) with additional sterile LB broth which, for Csp_P, results in a concentration of approximately 108 CFU/ml. To grow bacteria under biofilm conditions, we dispensed 1 ml of sterile LB into each well of a 24-well cell culture plate and spiked each well with 1 µl of bacterial freezer stock. We then allowed the culture to grow at room temperature without shaking for 48 h. Csp_P biofilm supernatant was harvested from single bacterial culture wells containing 48-h biofilm and was found to have an average bacterial concentration of approximately 109 CFU/ml. To harvest fresh biofilm, we removed the supernatant from five wells containing 48-h biofilm, resuspended the biofilm from each well in 100 µl 1× PBS and pooled the five wells. For Csp_P, this pooled biofilm solution contained approximately 109 CFU/ml and an average of 5 mg of biofilm (dry weight). To obtain desiccated biofilm, we collected the fresh biofilm from five wells as indicated, centrifuged the biofilm at 5000 rpm for 2.5 min, removed the PBS supernatant, and allowed the biofilm to dry at room temperature. On the day of the experiment, we resuspended the five wells of desiccated biofilm in 500 µl 1× PBS to mimic the fresh biofilm treatment. To heat-inactivate the fresh biofilm, we collected fresh biofilm as indicated and incubated samples at 90°C for 24 h prior to the experiment.
In vitro anti-Plasmodium activity assays
We prepared Csp_P bacterial cultures as described above and filtered all samples through a 0.2-µm filter.
Asexual-stage assay: Inhibition of asexual-stage P. falciparum was assessed using a SYBR green I-based fluorescence assay as described earlier [32]. Csp_P biofilm was grown for 36 h for this experiment because 48-h biofilm causes hemolysis of RBCs (Fig. S8), which interferes with the assay. Parasites were synchronized using 5% sorbitol [33]; 5 µl of each bacterial preparation was dispensed in triplicate wells of 96-well microplates, followed by addition of 95 µl of synchronous ring-stage P. falciparum cultures at 1% hematocrit and 1% parasitemia. Chloroquine (250 nM) was used as a positive control, and parasite growth medium was used as a negative control. After 72 h of incubation in a candle jar at 37°C, an equal volume of SYBR green-I solution in lysis buffer (Tris [20 mM; pH 7.5], EDTA [5 mM], saponin [0.008%; w/v], and Triton X-100 [0.08%; v/v]) was added to each well and mixed gently, then incubated 1–2 h in the dark at room temperature. Plates were read on a fluorescence plate reader (HTS 7000, Perkin Elmer), with excitation and emission wavelengths of 485 and 535 nm, respectively. Percent inhibition was calculated relative to negative (0% inhibition) and positive controls (100% inhibition). Three biological replicates were assayed.
Ookinete-stage assay: To assess inhibition of ookinete-stage P. berghei parasites, female Swiss Webster mice (6–8 weeks old) were infected with a transgenic strain of P. berghei that expresses Renilla luciferase. Starting at 3 days post-infection, exflagellation assays were performed until at least 20 exflagellation events were recorded in a 20× field. At this time, mice were bled by heart puncture using a heparinized needle, and the blood was diluted in 10 volumes of ookinete medium (RPMI 1640, 10% FBS, 50 mg/ml hypoxanthine, and 2 mg/ml NaHCO3, pH 8.3) with 4% mouse RBC lysate. Samples (50 µl) of each bacterial preparation were then mixed with the infected blood and incubated for 24 h at 19°C. Ookinete counts were determined using the Renilla luciferase assay system (Promega, USA) according to the manufacturer's instructions. The experiment was performed on two independent days, and each sample was assayed in triplicate on each day.
Gametocyte-stage assay: Inhibition of gametocyte-stage P. falciparum by Csp_P was assessed as described previously [34]. To prevent hemolysis of RBCs, Csp_P biofilm was grown for 36 and 42 hours for this experiment. In brief, NF54 P. falciparum cultures were started at 0.5% asexual parasitemia and 4% hematocrit. Csp_P bacterial preparations were added 15 days after Plasmodium cultures were initiated, and gametocytemia was determined 18 days after culture initiation. At least three biological replicates were tested for each culture preparation. More than 500 erythrocytes were examined for gametocytes across Giemsa-stained blood films from each sample.
In vitro anti-dengue activity assays
We prepared Csp_P bacterial cultures as described above (planktonic state, biofilm, biofilm supernatant, dessicated biofilm, and heat-inactivated biofilm), mixed 75 µl of each bacterial culture preparation with 75 µl of MEM containing dengue virus serotype 2 and incubated the mixture at room temperature for 45 min. Samples were then filtered through a 0.2-µm filter, serially diluted, and used to infect BHK21-15 cells. Plaque assays were conducted as described above to assess dengue virus infectivity. Percent inhibition was calculated as the percent decrease in PFU/ml relative to the PBS+LB control, which was standardized to 0% inhibition. The experiment was performed on two independent days, and each assay was performed in triplicate on each day. In experiments in which dengue was mixed with human blood before exposure to Csp_P, bacterial biofilms were not removed from the cell culture plate. Rather, dengue virus was mixed 1∶1 with human blood, and 150 µl of this mixture was added directly to each well containing Csp_P biofilm and incubated for 45 min at 30°C. Following this incubation period, the blood-dengue virus solution was mixed with the biofilm, and 50 µl of the mixture was then drawn from the well, diluted in MEM, and filtered through a 0.2-µm filter. The resulting filtrate solution was then serially diluted and used to infect C6/36 cells.
Assay for sequestration of viral particles by Csp_P biofilm
To assess whether the anti-dengue activity of Csp_P was due to sequestration of DENV by the Csp_P biofilm, we mixed a dengue virus suspension with Csp_P 48 hr biofilm or LB broth and incubated it for a period of 45 min. Samples were then centrifuged at 5,000 rpm for 5 min. The supernatants were collected, and RNA was extracted from equal volumes (50 µl) of experimental (biofilm+DENV) and control (LB+DENV) samples using the RNeasy kit (Qiagen). Comparison of viral RNA loads in the extracted supernatant was done via RT-qPCR relative quantification, using 2 µl of the viral RNA in a 20-µl reaction volume.
Assessing pH effects on dengue virus infectivity
The pH of bacterial biofilms and supernatants was assessed with a micro-pH electrode (Lazar Lab) at room temperature. Effects of pH changes on dengue virus infectivity were assessed by adjusting the pH of the MEM with NaOH and HCl until the desired range of pH values was obtained: 5.0, 7.7, 8.5, and 10.0. The pH-adjusted MEM was then mixed with dengue virus-laden blood and incubated for 45 min prior to serial dilution and infection of C6/36 cells.
Cell viability assays
Cell viability assays on the mosquito cell line C6/36 and the vertebrate cell line BHK-21 were performed via trypan blue staining (0.4%, Invitrogen) according to the manufacturer's instructions. In brief, 50 µl of suspended cells were placed in a microcentrifuge tube and mixed with 10 µl of Csp_P filtered fresh biofilm or PBS as a control. C6/36 cells were incubated at 32°C and BHK-21 cells were incubated at 37°C+5% CO2 for 45 min. Cells were then mixed with 12 µl of 0.4% trypan blue stain. The mixture was allowed to stand for 5 min at room temperature and then loaded into a hemocytometer for cell viability assessment and counting under a microscope.
Assay of the effects of Csp_P biofilm on host cell susceptibility to DENV
To assess whether exposure to Csp_P biofilm changes the susceptibility of the host cell to DENV, we conducted assays exposing C6/36 cells to Csp_P-filtered biofilm prior to dengue virus infection. Cells were grown to 80% confluency; the cell medium was then removed, washed once with 1× PBS, and then overlaid with 100 µl of Csp_P biofilm that had been filtered using a 2-µm filter or with 1× PBS (control) for about 10 min. Plates were then washed three times with 1× PBS and then infected with 100 µl of dengue virus for about 45 min. Cells were assessed for plaque formation at 6 days post-infection.
Hemolysis assay
Human erythrocytes were washed with RPMI 1640 medium until the supernatant was visually free of hemoglobin pigment. The washed erythrocytes were suspended in malaria complete medium to yield a 1% hematocrit. Filtered Csp_P biofilm was mixed with erythrocytes and incubated up to 24 h at 37°C. To separate lysed RBC cytosol from whole RBCs, the suspension was centrifuged at 2000 rpm for 5 min. The resulting supernatant was carefully aspirated and plated in new 96-well microplates. Control erythrocytes without any bacterial material were used as a negative control (blank), and freeze-thawed erythrocyte lysate was used as positive control (100% hemolysate). To determine the % lysis in test samples, plates were read at 405 nm in an ELISA plate reader (HTS 7000 Perkin Elmer), and the reading was expressed as a fraction of the positive control.
Real-time qPCR assays
To conduct real-time PCR assays, RNA samples were treated with Turbo DNase (Ambion, Austin, Texas, United States) and reverse-transcribed using M-MLV Reverse Transcriptase (Promega, USA). The real-time PCR assays were performed using the SYBR Green PCR Master Mix Kit (Applied Biosystems, Foster City, California, USA) in a 20-µl reaction volume; all samples were tested in duplicate. The ribosomal protein S7 gene was used for normalization of cDNA templates. Primer sequences used in these assays are given in Table S1.
Statistical analysis
The Mann-Whitney U test, one-way ANOVA with Dunnett's post-test and pairwise Log-Rank tests for survival analysis were conducted using the GraphPad Prism statistical software package (Prism 5.05; GraphPad Software, Inc., San Diego, CA). Data in Figure 5 were analyzed using an ANOVA, followed by a Tukey's test in R (R Foundation for Statistical Computing).
Supporting Information
Zdroje
1. CirimotichCM, RamirezJL, DimopoulosG (2011) Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe 10 : 307–310 doi:10.1016/j.chom.2011.09.006
2. CirimotichCM, ClaytonAM, DimopoulosG (2011) Low - and high-tech approaches to control Plasmodium parasite transmission by anopheles mosquitoes. J Trop Med 2011 : 891342 doi:10.1155/2011/891342
3. CirimotichCM, DongY, ClaytonAM, SandifordSL, Souza-Neto JA, et al. (2011) Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332 : 855–858 doi:10.1126/science.1201618
4. Gonzalez-CeronL, SantillanF, RodriguezMH, MendezD, Hernandez-AvilaJE (2003) Bacteria in Midguts of Field-Collected Anopheles albimanus Block Plasmodium vivax Sporogonic Development. J Med Entomol 40 : 371–374.
5. RamirezJL, Souza-NetoJ, Torres CosmeR, RoviraJ, OrtizA, et al. (2012) Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl Trop Dis 6: e1561 doi:10.1371/journal.pntd.0001561
6. BeierMS, PumpuniCB, BeierJC, DavisJR (1994) Effects of Para-Aminobenzoic Acid, Insulin, and Gentamicin on Plasmodium falciparum Development in Anopheline Mosquitoes (Diptera: Culicidae). J Med Entomol 31 : 561–565.
7. DongY, ManfrediniF, DimopoulosG (2009) Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog 5: e1000423 doi:10.1371/journal.ppat.1000423
8. XiZ, RamirezJL, DimopoulosG (2008) The Aedes aegypti Toll Pathway Controls Dengue Virus Infection. PLoS Pathog 4: e1000098.
9. AzambujaP, GarciaES, RatcliffeNA (2005) Gut microbiota and parasite transmission by insect vectors. Trends Parasitol 21 : 568–572.
10. MeisterS, AgianianB, TurlureF, RelógioA, MorlaisI, et al. (2009) Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog 5: e1000542 doi:10.1371/journal.ppat.1000542
11. O'TooleG, KaplanHB, KolterR (2000) Biofilm formation as microbial development. Annu Rev Microbiol 54 : 49–79.
12. FlemmingH-C, WingenderJ (2010) The biofilm matrix. Nat Rev Microbiol 8 : 623–633 doi:10.1038/nrmicro2415
13. MulcahyH, SibleyCD, SuretteMG, LewenzaS (2011) Drosophila melanogaster as an animal model for the study of Pseudomonas aeruginosa biofilm infections in vivo. PLoS Pathog 7: e1002299 doi:10.1371/journal.ppat.1002299
14. DuránN, MenckCF (2001) Chromobacterium violaceum: a review of pharmacological and industiral perspectives. Crit Rev Microbiol 27 : 201–222.
15. Creczynski-PasaTB, Antônio RV (2004) Energetic metabolism of Chromobacterium violaceum. Genet Mol Res 3 : 162–166.
16. LopesSCP, BlancoYC, JustoGZ, NogueiraPA, RodriguesFLS, et al. (2009) Violacein extracted from Chromobacterium violaceum inhibits Plasmodium growth in vitro and in vivo. Antimicrob Agents Chemother 53 : 2149–2152 doi:10.1128/AAC.00693-08
17. MichaelsR, CorpeWA (1965) Cyanide formation by Chromobacterium violaceum. J Bacteriol 89 : 106–112.
18. BlomD, FabbriC, EberlL, WeisskopfL (2011) Volatile-mediated killing of Arabidopsis thaliana by bacteria is mainly due to hydrogen cyanide. Appl Environ Microbiol 77 : 1000–1008 doi:10.1128/AEM.01968-10
19. Ribeiro de VasconcelosAT, AlmeidaDF, HungriaM, et al. (2003) The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc Natl Acad Sci U S A 100 : 11660–11665 doi:10.1073/pnas.1832124100
20. GallagherLA, ManoilC (2001) Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol 183 : 6207–6214 doi:10.1128/JB.183.21.6207-6214.2001
21. BroderickKE, ChanA, BalasubramanianM, FealaJ, ReedSL, et al. (2008) Cyanide produced by human isolates of Pseudomonas aeruginosa contributes to lethality in Drosophila melanogaster. J Infect Dis 197 : 457–464 doi:10.1086/525282
22. MartinPAW, Gundersen-RindalD, BlackburnM, BuyerJ (2007) Chromobacterium subtsugae sp. nov., a betaproteobacterium toxic to Colorado potato beetle and other insect pests. Int J Syst Evol Microbiol 57 : 993–999 doi:10.1099/ijs.0.64611-0
23. ClaytonAM, CirimotichCM, DongY, DimopoulosG (2013) Caudal is a negative regulator of the Anopheles IMD pathway that controls resistance to Plasmodium falciparum infection. Dev Comp Immunol 39 : 323–332 doi:10.1016/j.dci.2012.10.009
24. BahiaAC, DongY, BlumbergBJ, MlamboG, TripathiA, et al. (2014) Exploring Anopheles gut bacteria for Plasmodium blocking activity. Environ Microbiol 16(9): 2980–94 doi:10.1111/1462-2920.12381
25. StraifSC, MbogoCN, ToureAM, WalkerED, KaufmanM, et al. (1998) Midgut bacteria in Anopheles gambiae and An. funestus (Diptera: Culicidae) from Kenya and Mali. J Med Entomol 35 : 222–226.
26. LindhJM, TereniusO, FayeI (2005) 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl Environ Microbiol 71 : 7217–7223 doi:10.1128/AEM.71.11.7217-7223.2005
27. RaniA, SharmaA, RajagopalR, AdakT, BhatnagarRK (2009) Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi-an Asian malarial vector. BMC Microbiol 9 : 96 doi:10.1186/1471-2180-9-96
28. MüllerGC, SchleinY (2008) Efficacy of toxic sugar baits against adult cistern-dwelling Anopheles claviger. Trans R Soc Trop Med Hyg 102 : 480–484 doi:10.1016/j.trstmh.2008.01.008
29. RiehleMA, MoreiraCK, LampeD, LauzonC, Jacobs-LorenaM (2007) Using bacteria to express and display anti-Plasmodium molecules in the mosquito midgut. Int J Parasitol 37 : 595–603.
30. DasS, GarverL, RamirezJR, XiZ, DimopoulosG (2007) Protocol for dengue infections in mosquitoes (A. aegypti) and infection phenotype determination. J Vis Exp 220.
31. TragerW, JensenJB (1976) Human malaria parasites in continuous culture. Science 193 : 673–675.
32. BennettTN, PaguioM, GligorijevicB, SeudieuC, KosarAD, et al. (2004) Novel, Rapid, and Inexpensive Cell-Based Quantification of Antimalarial Drug Efficacy. Antimicrob Agents Chemother 48 : 1807–1810.
33. LambrosC, VanderbergJ (1979) Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol 65 : 418–420.
34. FerrerP, TripathiAK, ClarkMA, HandCC, RienhoffHY, et al. (2012) Antimalarial iron chelator, FBS0701, shows asexual and gametocyte Plasmodium falciparum activity and single oral dose cure in a murine malaria model. PLoS One 7: e37171.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance SystemČlánek Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer CellsČlánek APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse ModelČlánek Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 10- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Theory and Empiricism in Virulence Evolution
- -Related Fungi and Reptiles: A Fatal Attraction
- Adaptive Prediction As a Strategy in Microbial Infections
- Antimicrobials, Stress and Mutagenesis
- A Novel Function of Human Pumilio Proteins in Cytoplasmic Sensing of Viral Infection
- Social Motility of African Trypanosomes Is a Property of a Distinct Life-Cycle Stage That Occurs Early in Tsetse Fly Transmission
- Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer
- Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance System
- mRNA Structural Constraints on EBNA1 Synthesis Impact on Antigen Presentation and Early Priming of CD8 T Cells
- Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages
- Neutrophil Crawling in Capillaries; A Novel Immune Response to
- Live Attenuated Vaccine Protects against Pulmonary Challenge in Rats and Non-human Primates
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
- HIV Acquisition Is Associated with Increased Antimicrobial Peptides and Reduced HIV Neutralizing IgA in the Foreskin Prepuce of Uncircumcised Men
- Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host
- Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence
- Ethanol Stimulates WspR-Controlled Biofilm Formation as Part of a Cyclic Relationship Involving Phenazines
- Densovirus Is a Mutualistic Symbiont of a Global Crop Pest () and Protects against a Baculovirus and Bt Biopesticide
- Insights into Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria
- Mycobacterial Antigen Driven Activation of CD14CD16 Monocytes Is a Predictor of Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome
- Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion
- Dampens the DNA Damage Response
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- Vaginal Challenge with an SIV-Based Dual Reporter System Reveals That Infection Can Occur throughout the Upper and Lower Female Reproductive Tract
- Detecting Differential Transmissibilities That Affect the Size of Self-Limited Outbreaks
- One Small Step for a Yeast - Microevolution within Macrophages Renders Hypervirulent Due to a Single Point Mutation
- Expression Profiling during Arabidopsis/Downy Mildew Interaction Reveals a Highly-Expressed Effector That Attenuates Responses to Salicylic Acid
- Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer Cells
- Interaction with Tsg101 Is Necessary for the Efficient Transport and Release of Nucleocapsids in Marburg Virus-Infected Cells
- The N-Terminus of Murine Leukaemia Virus p12 Protein Is Required for Mature Core Stability
- Sterol Biosynthesis Is Required for Heat Resistance but Not Extracellular Survival in
- Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility
- Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly
- APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse Model
- Structural Basis for the Recognition of Human Cytomegalovirus Glycoprotein B by a Neutralizing Human Antibody
- Systematic Analysis of ZnCys Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus
- Epstein-Barr Virus Nuclear Antigen 3A Promotes Cellular Proliferation by Repression of the Cyclin-Dependent Kinase Inhibitor p21WAF1/CIP1
- The Host Protein Calprotectin Modulates the Type IV Secretion System via Zinc Sequestration
- Cyclophilin A Associates with Enterovirus-71 Virus Capsid and Plays an Essential Role in Viral Infection as an Uncoating Regulator
- A Novel Alpha Kinase EhAK1 Phosphorylates Actin and Regulates Phagocytosis in
- The pH-Responsive PacC Transcription Factor of Governs Epithelial Entry and Tissue Invasion during Pulmonary Aspergillosis
- Sensing of Immature Particles Produced by Dengue Virus Infected Cells Induces an Antiviral Response by Plasmacytoid Dendritic Cells
- Co-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites
- Characteristics of Memory B Cells Elicited by a Highly Efficacious HPV Vaccine in Subjects with No Pre-existing Immunity
- HPV16-E7 Expression in Squamous Epithelium Creates a Local Immune Suppressive Environment via CCL2- and CCL5- Mediated Recruitment of Mast Cells
- Dengue Viruses Are Enhanced by Distinct Populations of Serotype Cross-Reactive Antibodies in Human Immune Sera
- CD4 Depletion in SIV-Infected Macaques Results in Macrophage and Microglia Infection with Rapid Turnover of Infected Cells
- A Sialic Acid Binding Site in a Human Picornavirus
- Contact Heterogeneity, Rather Than Transmission Efficiency, Limits the Emergence and Spread of Canine Influenza Virus
- Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of
- HTLV-1 Tax Stabilizes MCL-1 via TRAF6-Dependent K63-Linked Polyubiquitination to Promote Cell Survival and Transformation
- Species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence
- A Critical Role for IL-17RB Signaling in HTLV-1 Tax-Induced NF-κB Activation and T-Cell Transformation
- Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90
- Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
- Kaposi's Sarcoma-Associated Herpesvirus Induces Nrf2 during Infection of Endothelial Cells to Create a Microenvironment Conducive to Infection
- A Relay Network of Extracellular Heme-Binding Proteins Drives Iron Acquisition from Hemoglobin
- Glutamate Secretion and Metabotropic Glutamate Receptor 1 Expression during Kaposi's Sarcoma-Associated Herpesvirus Infection Promotes Cell Proliferation
- Reduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and Anti-pathogen Activities
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



