-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDensovirus Is a Mutualistic Symbiont of a Global Crop Pest () and Protects against a Baculovirus and Bt Biopesticide
The old world cotton bollworm, Helicoverpa armigera, is one of the most significant pests of crops throughout Asia, Europe, Africa and Australia. Herein, we report a novel densovirus (HaDNV-1) which was widely distributed in wild populations of H. armigera and was beneficial to its host by increasing larval and pupal development rates, female lifespan and fecundity, suggesting a mutualistic interaction between the cotton bollworm and HaDNV-1. The cotton bollworm is currently widely controlled by the biopesticides Bacillus thuringiensis (Bt) toxin and the baculovirus HaNPV. It is therefore important to estimate the risk that the symbiotic virus will negatively impact on the efficiency of these biopesticides. Field and laboratory results suggest that HaDNV-1 infection significantly increases larval resistance to HaNPV and Bt toxin. These results have important implications for the selection of biopesticides for this species, and highlight the need for greater research into the elegant microbial interactions that may impact host individual and population dynamics.
Published in the journal: . PLoS Pathog 10(10): e32767. doi:10.1371/journal.ppat.1004490
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004490Summary
The old world cotton bollworm, Helicoverpa armigera, is one of the most significant pests of crops throughout Asia, Europe, Africa and Australia. Herein, we report a novel densovirus (HaDNV-1) which was widely distributed in wild populations of H. armigera and was beneficial to its host by increasing larval and pupal development rates, female lifespan and fecundity, suggesting a mutualistic interaction between the cotton bollworm and HaDNV-1. The cotton bollworm is currently widely controlled by the biopesticides Bacillus thuringiensis (Bt) toxin and the baculovirus HaNPV. It is therefore important to estimate the risk that the symbiotic virus will negatively impact on the efficiency of these biopesticides. Field and laboratory results suggest that HaDNV-1 infection significantly increases larval resistance to HaNPV and Bt toxin. These results have important implications for the selection of biopesticides for this species, and highlight the need for greater research into the elegant microbial interactions that may impact host individual and population dynamics.
Introduction
The interactions between symbiotic species and their hosts are becoming increasingly understood within insect systems [1], [2], [3]. Symbionts form diverse evolutionary relationships that influence the life history of their host, from mutualistic, by protecting them from natural enemies or increasing their host's fitness though a variety of means [1], [4], [5], [6], [7], [8], to parasitic, either by decreasing their resistance to harmful microorganisms or their tolerance to environmentally harmful factors, or by killing them directly [9], [10], [11]. There is a growing literature on the mutualistic interactions between intracellular bacterial symbionts, such as Wolbachia and their insect hosts, in which the symbionts spread through the host population by increasing the fitness of infected hosts [1], [6], [12], [13]. However, viral mutualistic symbioses have rarely been reported. This may be because, as obligate symbionts, viruses have long been considered harmful to their host and are usually isolated from cadavers killed by the virus. Moreover, until relatively recently, laboratory techniques only had the capacity to shed light on overtly pathogenic viruses, and not covert beneficial ones [14], [15], [16]. The development of molecular and sequencing technology facilitates the discovery and analysis of non-pathogenic virus species, using techniques such as suppression subtractive hybridization (SSH) and RNA-seq [17], [18]. Generally, viruses isolated from healthy individuals may be conditionally beneficial to their hosts. Recently, these ‘good viruses’ have attracted more attention, largely due to the prospect of using them in applications such as gene therapy and as tools for gene manipulation [2], [19]. As defined by Roossinck, there are few examples of viral mutualistic symbioses in insects (identified as conveying benefit to the host without any detectable fitness costs) [2].
The cotton bollworm moth, Helicoverpa armigera, is a major migratory pest of cotton and other economically-important crops throughout Asia, Africa, Europe and Australasia [20], [21], [22]. In China, the introduction of Bt-cotton in the 1990s has seen a dramatic decline in the H. armigera moth population. However, there are signs of Bt-resistance emerging [23], [24], fueling renewed interest in other forms of biological pest control, including the use of host-specific viral pesticides, derived from densoviruses [25], small RNA viruses [26] and baculoviruses [27], [28], [29], [30], [31]. Previously, we reported a novel densovirus (HaDNV-1, from the family Parvoviridae) in H. armigera moths that possesses a monosense genome that is 4926 nucleotides in length and clustered with the members of the genus Iteravirus in phylogenetic analysis [32]. This has allowed further investigation into the interactions between HaDNV-1 and its host H. armigera, which we report here. The main objective of this study was to establish the ecological significance of this virus within the migratory H. armigera system. Specifically, we undertook experiments to determine the transmission strategies of HaDNV-1, the impact of HaDNV-1 infection on host fitness, including its capacity to modulate resistance to potentially lethal biopesticides, and the prevalence of HaDNV-1 in field populations of H. armigera. Our results show that HaDNV-1 can be both horizontally - and vertically-transmitted in H. armigera; that HaDNV-1 infection increases host-fitness by increasing larval/pupal development rate, female lifespan and egg/offspring production; and that it also enhances larval resistance to H. armigera nucleopolyhedrovirus (HaNPV), a widely-used biopesticide. Resistance to Bt Cry1Ac protoxin was also enhanced, but only at relatively low toxin concentrations. Overall, we found no evidence for a negative effect of densovirus infection on H. armigera fitness-related traits, strongly suggesting a mutualistic interaction between the cotton bollworm and HaDNV-1.
Results
Transmission strategies of HaDNV-1 and host-tissue distribution
To establish the modes of transmission of the densovirus HaDNV-1, we first produced an uninfected laboratory colony from a single breeding pair of H. armigera (NONINF strain). An infected strain (INF strain) was subsequently produced using neonate larvae from the NONINF strain, dosing them with either purified HaDNV-1 (108/µl; method 1, see Materials and Methods) or filtered liquid from infected individuals (108/µl; method 2, see Materials and Methods). Thus, our results indicated that HaDNV-1 could efficiently infect larvae by oral ingestion. The efficiency of infection with filtered liquid was higher than that of the purified virus (Table 1, Fig. S1A, S1B), suggesting that the purification process might have inactivated the virus in some way. We also found that individuals artificially infected with HaDNV-1 via peroral infection could efficiently transmit the viral infection to their offspring (Fig. S1C), and the same was true for naturally infected individuals (Fig. S1D), suggesting vertical transmission of the virus.
Tab. 1. The transmission mode of HaDNV-1. 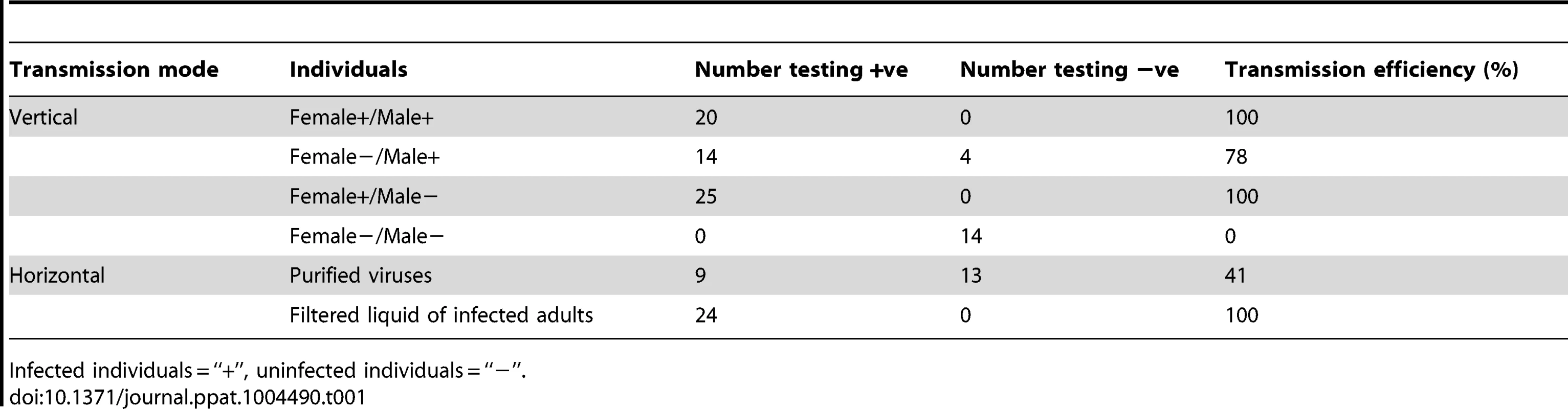
Infected individuals = “+”, uninfected individuals = “−”. HaDNV-1 was capable of being vertically-transmitted from both infected females and infected males, but transmission-efficiency was higher from infected females than males (Table 1, Fig. S1E, S1F, S1G). With qPCR, we tested whether vertical transmission of HaDNV-1 was due to virus contamination on the surface of the eggs (transovum), or whether the virus was transmitted within the egg itself (transovarial). HaDNV-1 titers were not significantly different between sodium hypochlorite-treated and non-treated eggs (t = 1.296, d.f. = 6, P = 0.24) (Fig. 1), suggesting that transovarial transmission was occurring.
Fig. 1. Viral load of HaDNV-1 in cotton bollworm eggs. 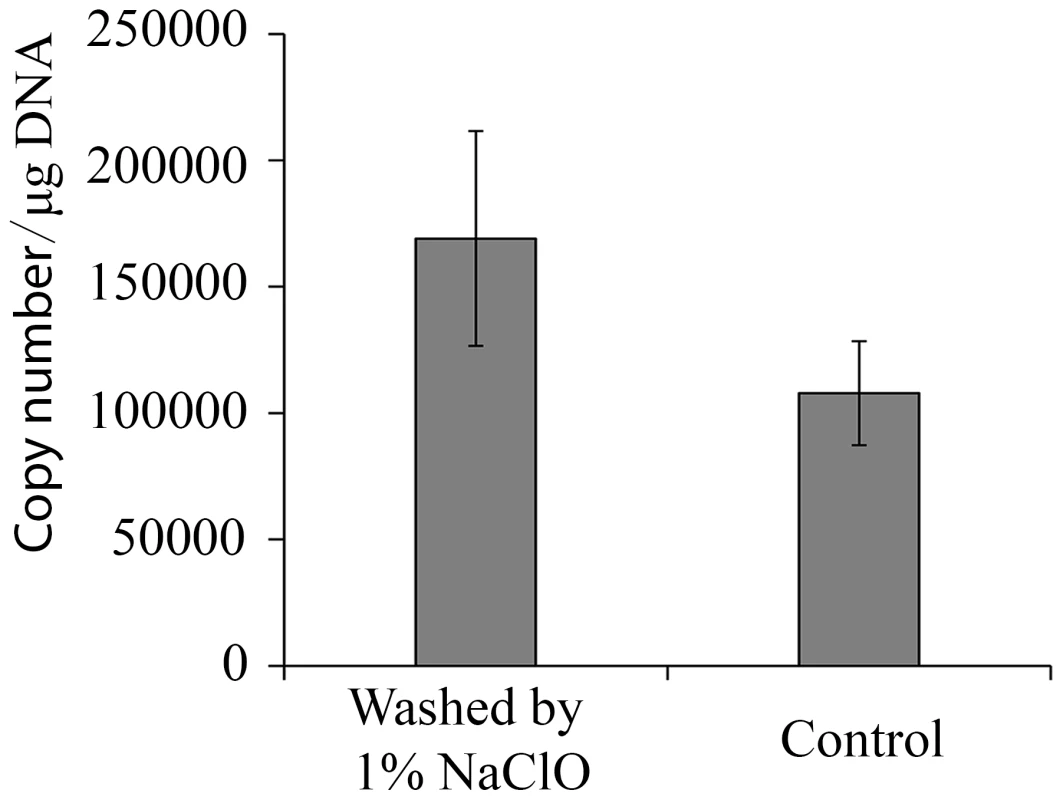
Absolute quantification of HaDNV-1 copy number per µg of host DNA in eggs washed or non-washed in 1% sodium hypochlorite (n = 4). To examine the possibility of horizontal transmission through ingestion of contaminated foodplant (as would be a possibility in wild populations), we placed uninfected neonate larvae in diet cells that had previously housed infected insects (n = 8). Our results indicated that horizontal virus transmission did not occur in this manner, despite our previous experimental evidence that larvae could be orally infected. To examine this further, we used a range of HaDNV-1 concentrations to infect larvae and subsequently examined virus intensity in host frass (faeces). As expected, larval infection rate was positively related to the magnitude of the HaDNV-1 oral challenge, with low infection rates at doses less than 106/µl (Table 2); but even for larvae challenged with large viral doses, their frass contained only very low levels of HaDNV-1, with only 3 out of 20 samples containing more than 1×105/mg and none with more than 5×105/mg. Therefore, while we cannot exclude the possibility that horizontal transmission of HaDNV-1 may occur via the oral-fecal route, the viral levels in frass were very low and may not be sufficient for oral infection.
Tab. 2. Detection of HaDNV-1 infecting larvae dosed at a range of concentrations. 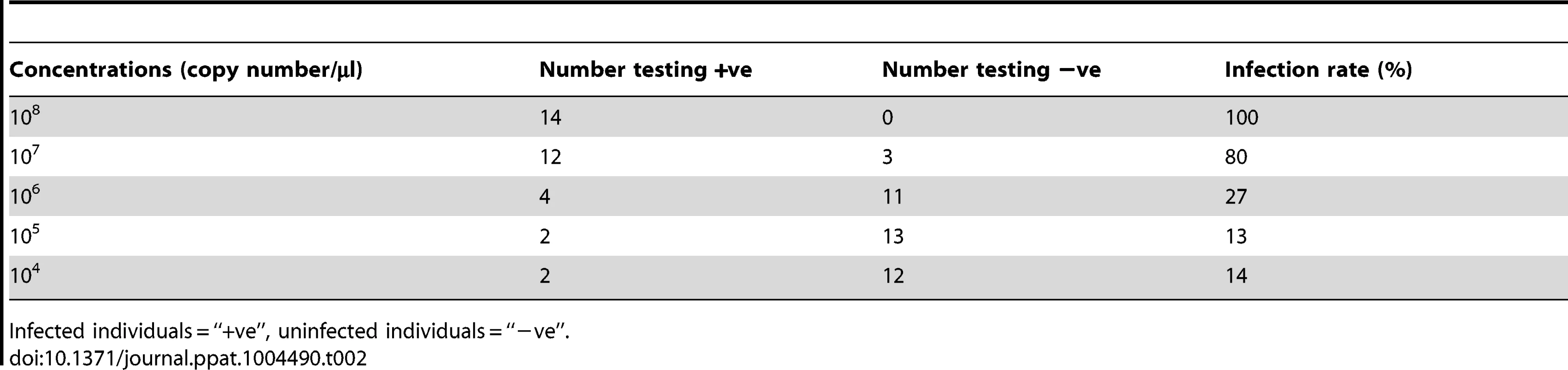
Infected individuals = “+ve”, uninfected individuals = “−ve”. HaDNV-1 distribution was quantified within different host body tissues using qPCR. In both larvae and adults, HaDNV-1 titers were significantly higher in the fat body than in all other tissues: larvae: F = 11.098, d.f. = 5,36, P<0.0001 (Fig. 2A); adult females: F = 26.601, d.f. = 5,30, P<0.0001 (Fig. 2B); adult males: F = 44.560, d.f. = 5,30, P<0.0001 (Fig. 2C).
Fig. 2. Tissue distribution of the HaDNV-1 in A) larvae, B) adult females and C) adult male cotton bollworms. 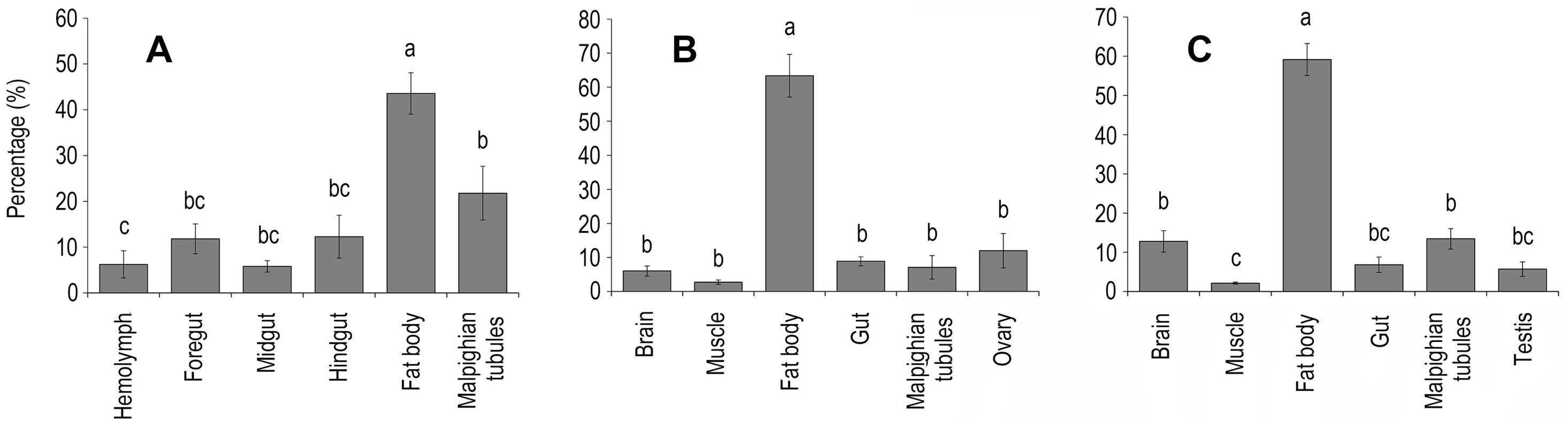
Within each figure, significant differences ascribed using Tukey tests are shown using different letters. Percentage (%) = the ratio of HaDNV-1 in different tissues (per mg), as described in Methods 2.3 (larvae: n = 7; adult males: n = 6; adult females: n = 6). Means ± SE. Host range of HaDNV-1 infection
Using H. armigera as a control, we tested four other species of lepidopterans for their potential to act as alternative hosts for HaDNV-1, by attempting oral inoculation in Spodoptera exigua, Spodoptera litura, Agrotis segetum and Agrotis ipsilon. Results indicated that while oral inoculation with HaDNV-1 could successfully infect H. armigera, none of the four other species tested positive (Fig. S2A). We also tested field-captured adults of the closely-related species H. assulta but failed to find any HaDNV-1 positive individuals (n = 9; Fig. S2B). Based on these available data, it appears that infection with HaDNV-1 is host-specific to H. armigera.
Effect of HaDNV-1 infection on the development, fecundity and adult longevity of H. armigera
To quantify the impact of HaDNV-1 infection on H. armigera development, a number of bioassays were performed using neonate larvae orally inoculated with filtered liquid from either HaDNV-1 infected (DNV+) or non-infected (DNV−) individuals (Fig. S3A, S3B). Both male and female DNV+ individuals developed significantly more quickly than the control individuals in both the larval (female: t = 2.732, d.f. = 312, P = 0.0067, male: t = 4.147, d.f. = 379, P<0.001) (Fig. 3A) and pupal stages (female: t = 5.100, d.f. = 312, P<0.001, male: t = 4.057, d.f. = 379, P<0.001) (Fig. 4A). Between 7–11 days post-hatch (approximately 3rd–5th instar) DNV+ larvae weighed significantly more than DNV - larvae by an average of ∼20% (GLMM with larval identity as a random term and log10-transformed larval weight as the dependent variable: Age (days): F = 2386.8, d.f. = 1,127, P<0.0001; HaDNV-1 infection status (+ve or −ve): F = 27.25, d.f. = 1,36, P<0.0001) (Fig. 3B, Fig. S4). However, their growth rates over this period did not differ, suggesting that densovirus effects on larval growth rate occurred prior to day 7 post-hatch (GLMM: interaction between infection status and age: F = 0.01, d.f. = 1,126, P = 0.91) (Fig. S4). A chloroform-wash assay indicated that at 9 days old, DNV+ larvae contained more lipid than DNV − individuals, measured as either lipid mass (t = 2.045, d.f. = 50, P = 0.046) or as a percentage of the whole body (t = 2.342, d.f. = 50, P = 0.023) (Fig. 3C, 4D). Larval mortality of DNV+ was significantly lower than DNV − (Table 3). However, there was no significant difference in pupal weight between DNV+ and DNV − insects (GLM: densovirus infection status: F = 0.99, d.f. = 1,692, P = 0.329; Sex: F = 41.08, d.f. = 1,693, P<0.0001; interaction term: F = 0.064, d.f. = 1,691, P = 0.80; female: t = 0.96, d.f. = 312, P = 0.34, male: t = 0.481, d.f. = 379, P = 0.63) (Fig. 4B), or pupation rate or eclosion rate between HaDNV-1 positive and HaDNV-1 negative insects (Table 3).
Fig. 3. Larval life-history parameters of DNV- and DNV+ cotton bollworms. 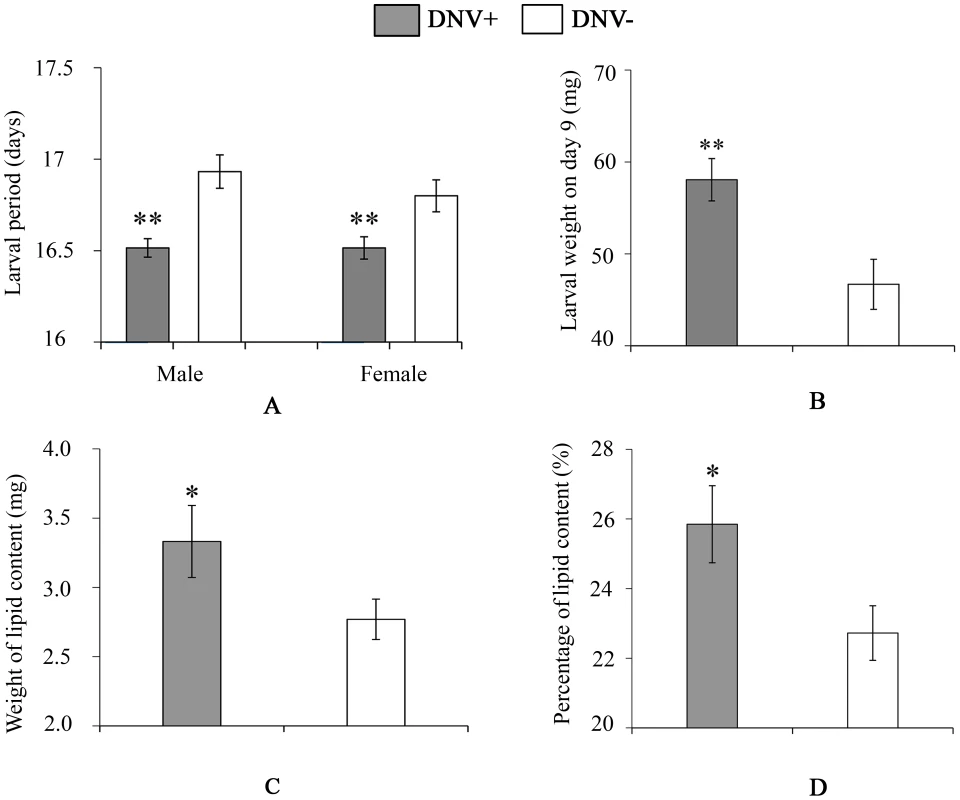
(A) Larval period (for male, n(DNV+) = 204, n(DNV−) = 177; for female, n(DNV+) = 169, n(DNV−) = 145). (B) Larval weight on day 9 (n = 19). (C) Lipid content in HaDNV-1 positive and negative larvae on day 9 (n(DNV+) = 19, n(DNV−) = 33). (D) Percentage of lipid content in HaDNV-1 positive and negative larvae on day 9 (n(DNV+) = 19, n(DNV−) = 33). DNV− = densovirus negative larvae, DNV+ = densovirus positive larvae. Means ± SE. * = P<0.05, ** = P<0.01. Fig. 4. Pupal and adult life-history parameters of DNV− and DNV+ cotton bollworms. 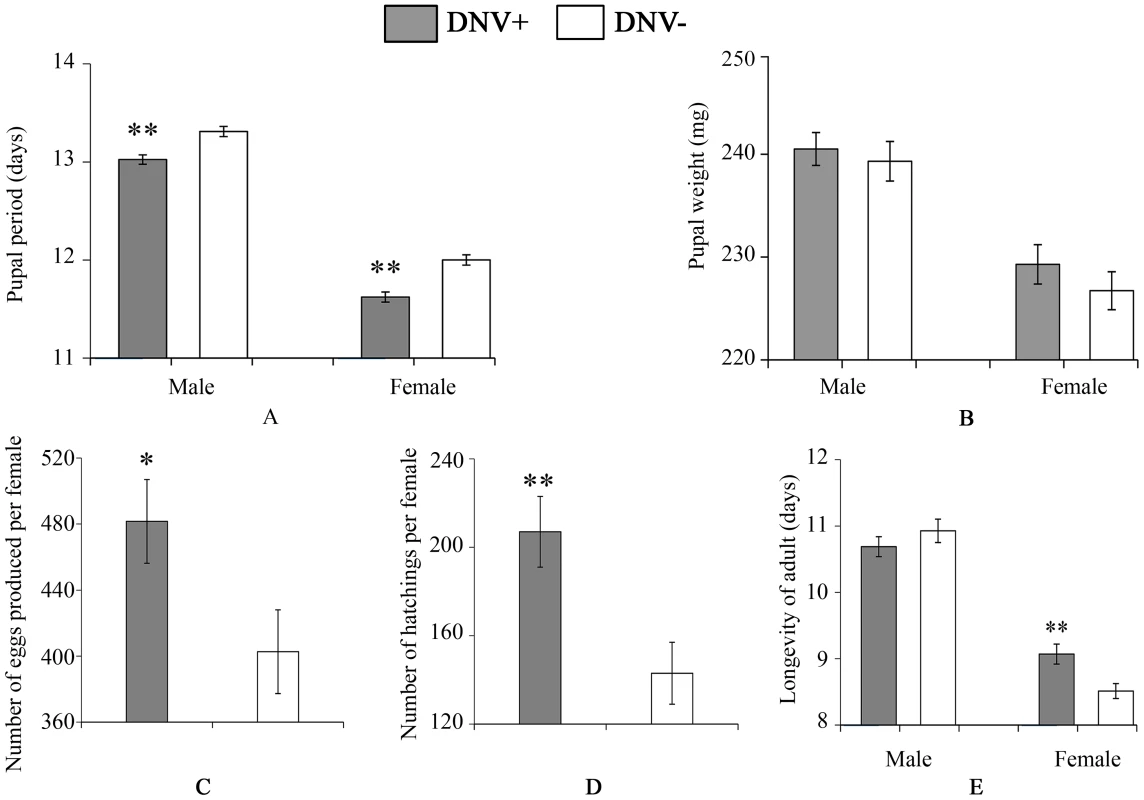
(A) Pupal period (for male, n(DNV+) = 204, n(DNV−) = 177; for female, n(DNV+) = 169, n(DNV−) = 145). (B) Pupal weight (for male, n(DNV+) = 204, n(DNV−) = 177; for female, n(DNV+) = 169, n(DNV−) = 145). (C) Number of eggs produced per female (n(DNV+) = 47, n(DNV−) = 48). (D) Egg hatch per female (n(DNV+) = 47, n(DNV−) = 48). (E) Longevity of adult (for male, n(DNV+) = 174, n(DNV−) = 195; for female, n(DNV+) = 174, n(DNV−) = 195). DNV− = densovirus negative larvae, DNV+ = densovirus positive larvae. Means ± SE. * = P<0.05, ** = P<0.01. Tab. 3. The influence of HaDNV-1 on survival rates of cotton bollworm. 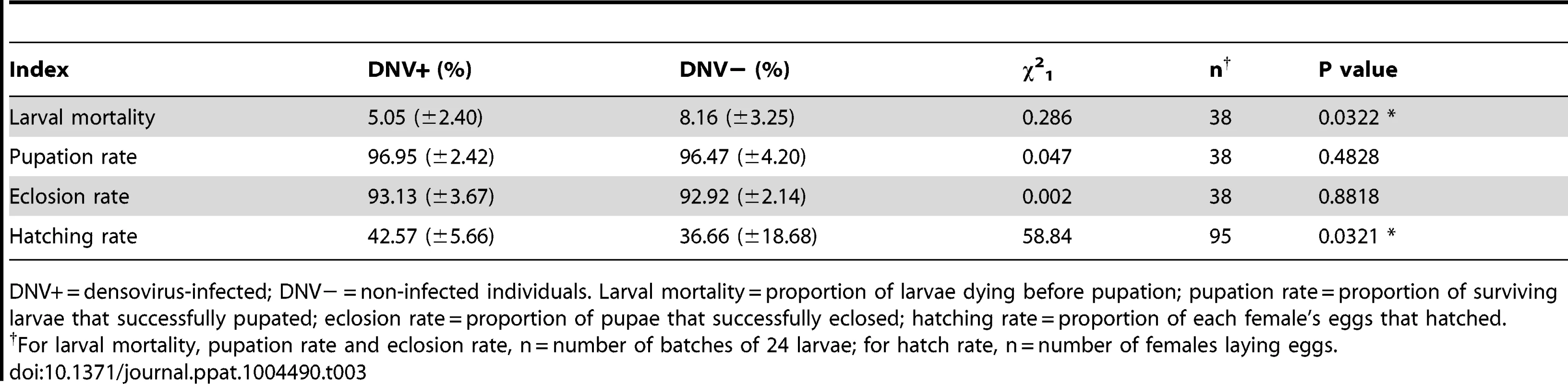
DNV+ = densovirus-infected; DNV− = non-infected individuals. Larval mortality = proportion of larvae dying before pupation; pupation rate = proportion of surviving larvae that successfully pupated; eclosion rate = proportion of pupae that successfully eclosed; hatching rate = proportion of each female's eggs that hatched. To determine the effect of HaDNV-1 infection on adult life-history traits, we used individuals from the non-infected (NONINF) and infected (INF) strains; and their infection status was confirmed by PCR (Fig. S3C, S3D). Infected INF strain moths produced significantly more eggs (t = 2.172, d.f. = 93, P = 0.032; Fig. 4C) and more neonates (t = 3.026, d.f. = 93, P = 0.0032; Fig. 4D) than individuals from the uninfected NONINF strain. Egg viability (hatch-rate) was significantly higher in the INF strain than in the NONINF strain (Table 3). The life-span of densovirus-infected females was significantly longer than that of females that were virus-free (χ21 = 13.5, d.f. = 1, P = 0.0002; Fig. 4E), but the longevity of males was not significantly different between the two strains (χ2 = 1.64, d.f. = 1, P = 0.2; Fig. 4E).
Covariation of densovirus and baculovirus within field populations
In larval field-collections, there was a non-random association between the two viruses (Chi-square test with Yates' correction: χ2 = 35.63, d.f. = 1, P<0.0001). Thus, there were relatively fewer larvae infected with both HaDNV-1 and HaNPV than would be expected by chance alone (14% versus 20%). When split by year, this effect was significant in 2012, when the overall HaNPV prevalence was 61% (χ2 = 19.75, d.f. = 1, P<0.0001; proportion infected with both viruses = 20% observed versus 26% expected), but not in 2013, when HaNPV prevalence was just 4% (χ2 = 0.82, d.f. = 1, P = 0.36; 2% observed versus 2% expected) (Table S1).
In adult field-collections, the prevalence of HaDNV-1 infection was uniformly high each year between 2008 and 2012 - 87%, 81%, 77%, 68% and 67%, respectively (Fig. S5). However, there was evidence for a significant decline in densovirus prevalence over the five years (GLMM with location as a random effect: χ21 = 39.06, P<0.0001). Despite high levels of baculovirus being observed in the larval field populations, we failed to detect any HaNPV-positive individuals in a random selection adult moths collected from four geographically diverse sites (n = 361 samples).
Interaction between densovirus and microbial biopesticides
To determine the interaction between the densovirus HaDNV-1 and the baculovirus HaNPV, we first confirmed individuals from NONINF strain were NPV-free using PCR with specific primers. Then, NONINF strain neonates were inoculated with either HaDNV-1 (DNV+) or water (DNV − controls), and infections verified using PCR. Survival to pupation in larvae not exposed to HaNPV did not differ between DNV+ (95%) and DNV − (92%) larvae (χ2 = 0.27, d.f. = 1, P = 0.60). However, for those larvae exposed to the baculovirus, there was a significant difference between DNV+ and DNV − larvae in their susceptibility to HaNPV (GLM: HaDNV-1 infection-status: χ2 = 4.04, d.f. = 1, P = 0.044, parameter estimate ± standard error = 0.4645±0.2319), with densovirus-infected larvae suffering lower mortality rates for a given virus dose (GLM: log10 virus dose: χ21 = 98.56, P<0.0001; LC50s = 3.13×107 versus 9.10×107 OB per ml, for DNV − and DNV+ larvae, respectively; Fig. 5A); the interaction between viral dose and infection status was marginally non-significant (dose*status: χ2 = 3.72, d.f. = 1, P = 0.054).
Fig. 5. Relationship between the baculovirus HaNPV and the densovirus HaDNV-1 in cotton bollworm larvae. 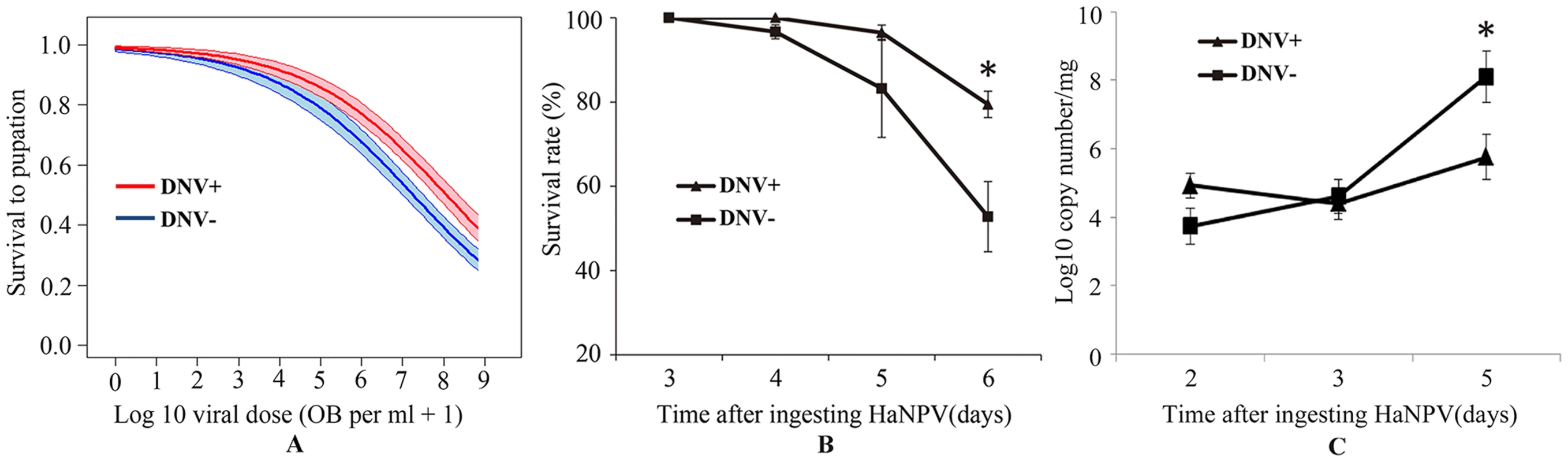
(A) Effect of HaNPV dose (log10-transformed number of occlusion bodies per ml) on larval survival to pupation. The thick lines are the fitted values and the shaded zones are the standard errors around these fitted values; blue line and shading = DNV− (control) larvae; red line and shading = DNV+ (densovirus-infected) larvae. The numbers of larvae that survived or died at differet concentrations (0, 106, 107, 108, 109 OB/ml) were 46/4, 38/8, 32/14, 20/32, 9/64 for DNV− individuals and 39/2, 31/10, 32/13, 31/17, 10/36 for DNV+ individuals. Temporal variation in (B) survival rate (%) (n = 216) and HaNPV copy numbers (log10-transformed) (C) (for day 2, n = 16; for day 3, n = 24; for day 5, n = 18) at different times after ingesting viruses. The concentrations of HaDNV-1 and HaNPV were 108/µl and 108 OBs/ml, respectively DNV− = densovirus negative larvae, DNV+ = densovirus positive larvae. Means ± SE. * = P<0.05, ** = P<0.01, based on t-tests at each time-point. We tested the differences of HaNPV replication between HaDNV-1 positive and negative individuals by repeating the HaNPV bioassay with 108 OBs/ml. The baculovirus bioassay indicated that there was no HaNPV-induced mortality in the control larvae that were exposed to water only, and that most mortality in the HaNPV-challenged larvae started at day 5 (120 h post-inoculation) (Fig. 5B). In NPV-challenged larvae, those carrying HaDNV-1 suffered significantly lower mortality overall than HaDNV-1 negative insects (Likelihood-ratio test: χ2 = 23.24, d.f. = 1, P<0.0001; linear coefficient (95% confidence interval) = 0.248 (0.134, 0.457)). Therefore, we collected samples before day 5 post-challenge to estimate HaNPV viral loads using qPCR. As would be expected, HaNPV titers (log-transformed) increased over time post-challenge and the rate of HaNPV titer increase was lower for HaDNV-1 positive larvae than in larvae lacking HaDNV-1, as indicated by a significant interaction term (linear model: Time post-challenge: F = 27.02, d.f. = 1,112, P<0.0001; DNV infection status: F = 5.69, d.f. = 1,112, P = 0.019; Time* DNV status interaction: F = 8.69, d.f. = 1,112, P = 0.0038; Fig. 5C). However, HaNPV titers were not directly correlated with HaDNV-1 titers in HaDNV-1 positive individuals (r = 0.066, n = 58, P = 0.623). These results suggest that HaDNV-1 protected H. armigera from HaNPV, possibly by slowing the accumulation of HaNPV.
A similar bioassay using the Bt toxin Cry1Ac instead of the baculovirus generated consistent results. As expected, larval development score increased over time and declined with increasing Bt dose (linear mixed-effects model with larval identity as a random term: Day: F = 18147.38, d.f. = 1,4172, P<0.0001; Log2Btdose: F = 1335.48, d.f. = 1,4172, P<0.0001). However, development was also influenced by the interaction between DNV infection status and the dose of Bt administered (DNV status: F = 120.21, d.f. = 1,4172, P<0.0001; DNV status * Bt dose interaction: F = 111.81, d.f. = 1,4172, P<0.0001), with the enhanced development of HaDNV-1 positive larvae at low Bt concentrations declining as Bt dose increased, such that mean development rate was independent of DNV infection status as Bt concentrations above 1.6 µg/g (Fig. 6). We also performed the bioassay with Bt cotton. As expected, there was a significant effect of Bt cotton on larval development rate, with development being significantly stunted in larvae exposed to the Bt plants (linear model: Diet: F = 63.74, d.f. = 1,476, P<0.001; mean score ± s.e.: Bt cotton = 1.717±0.153; non-Bt cotton = 3.529±0.167). However, whilst DNV positive larvae tended to have slightly higher development scores than DNV negative larvae (2.754±0.176 versus 2.492±0.164), this difference was non-significant and the interaction between DNV status and Bt exposure was also non-significant (DNV status: F = 1.336, d.f. = 1,476, P = 0.24; DNV status * Diet interaction: F = 0.0084, d.f. = 1,476, P = 0.93).
Fig. 6. Relationship between dose of Bt toxin (log2-transformed) and mean development score for DNV+ and DNV- cotton bollworm larvae (averaged over days 4 to 9 post-challenge). 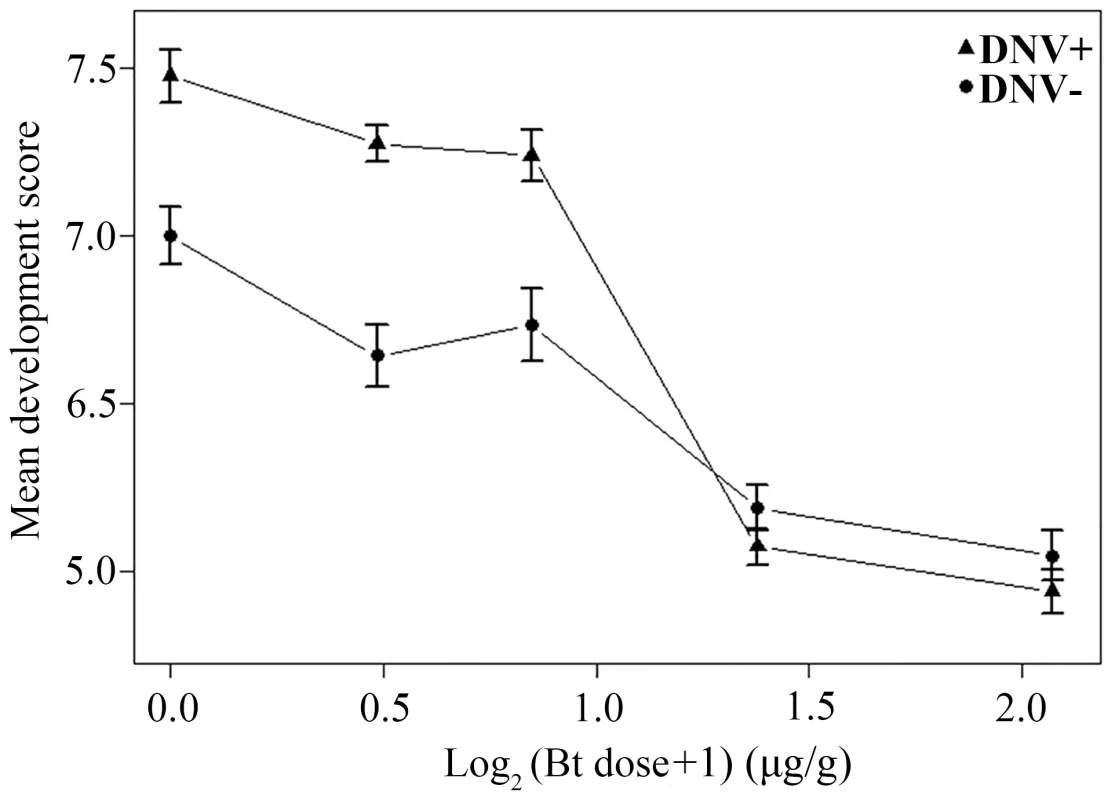
Development score = a qualitative measure of average development stage achieved on a scale from 0 (death) to 11 (mid-4th instar) (see Material and Methods for more details). DNV− = densovirus negative larvae, DNV+ = densovirus positive larvae. Discussion
To date, viral mutualistic symbioses have attracted little attention and are rarely reported, most likely due to a lack of obvious pathogenicity within their insect hosts. In our study system, SSH was previously used to detect and isolate a novel densovirus (HaDNV-1) from healthy migratory cotton bollworms, H. armigera [32]. To date, most reported DNVs have been pathogenic to their hosts, even resulting in mortality, and as a result DNVs have been considered as potential biological control agents of insect pests [33], [34], [35], [36]. However, in our present study, for the first time, we show a mutualistic relationship without any detectable negative interactions between a DNV and its host.
Although endosymbionts of insects do have the capacity for horizontal transmission, they are usually transmitted via maternal inheritance [1], [3]. However, viral symbionts can be efficiently transmitted both vertically and horizontally [37], [38], [39], [40], [41], [42], [43], [44]. We found that HaDNV-1 was efficiently vertically-transmitted via both the paternal and maternal lines. This was most likely via transovarial infection, with the efficiency of transmission being higher from infected females than males. The results presented here also suggest that HaDNV-1 can be horizontally-transmitted to H. armigera by peroral infection of larvae, in a dose-dependent manner. However, we failed to detect horizontal transmission by diet contamination, suggesting that although larvae can be infected orally, peroral infection may only be possible at very high HaDNV-1 concentrations. Indeed, infection rate and intensity were both positively correlated with the magnitude of the HaDNV-1 challenge, and the frass of larvae contained only very low levels of HaDNV-1. This suggests that in the field, HaDNV-1 is likely to be almost exclusively transmitted vertically from parents to offspring. Previous studies suggest that DNVs may vary in their host ranges, for example Junonia coenia densovirus (JcDNV), Mythimna loreyi densovirus (MlDNV) and Periplaneta fuliginosa densovirus (PfDNV), all infect several host species, whereas Galleria mellonella densovirus (GmDNV) infects only one species [33]. Our results suggest that HaDNV-1 is also strongly host-specific following oral exposure, only infecting H. armigera.
Certain bacterial beneficial symbionts have been reported to benefit their hosts by shortening host development time and increasing host fecundity [1], [45]. However, evidence of viruses increasing host fecundity has rarely been reported. One exception is in a vector-virus complex in the whitefly Bemisia tabaci: a plant virus transmitted by B. tabaci was found to accelerate the population growth rate of its insect host [46]. In our system, HaDNV-1 infection intensity was greatest in the host fat body, suggesting that the virus might play a role in the development of H. armigera. Indeed, the significantly shortened development time and faster growth rate of H. armigera infected with HaDNV-1 could be mediated by the virus promoting the accumulation of fat body by the host. Our results showed that at 9 days old, HaDNV-1-infected larvae contained more lipid than uninfected larvae. The positive effect of the HaDNV-1 on these life-history traits, including egg/offspring production, suggests a possible mutualistic relationship. Taken together with the results of the baculovirus bioassay, these results suggest that HaDNV-1 benefits H. armigera, but is not an obligate microbe required by the host to survive.
The baculovirus HaNPV is a large double-stranded DNA virus, which was first isolated in China in 1975 and has since become an important biopesticide for a number of agricultural pests [27], [28], [29], [30], [31]. To determine the interaction between HaDNV-1 and HaNPV in H. armigera, we collected samples of larvae and adults from the field to determine the natural infection rates of HaDNV-1 and HaNPV. Most significantly, we found that there was a clear negative interaction between the two viruses across larval populations, with there being more insects infected with one or other of the viruses than would be expected by chance alone, and fewer with both viruses or neither. One possible explanation for this observation is that there is a negative interaction between the two viruses: perhaps HaDNV-1 increases susceptibility to HaNPV disease, resulting in those individuals with both viruses being more likely to die, as seen in larvae of the African armyworm moth, Spodoptera exempta, co-infected with Wolbachia and the baculovirus SpexNPV [9]. However, our results from the HaNPV-HaDNV-1 bioassay suggest the opposite, with HaDNV-1 infected larvae being significantly more resistant to HaNPV than those not carrying the densovirus. Therefore, it is likely that fewer than expected HaNPV-HaDNV-1 co-infected individuals were detected in field populations because HaDNV-1 protects its host against HaNPV infection. Our qPCR assay supported this hypothesis: HaNPV was found to accumulate in HaDNV-1 infected larvae at a slower rate than in uninfected larvae. Another possibility to explain the dearth of co-infected individuals is that rather than there being a direct interaction between the viruses, the interaction is indirect. Baculoviruses only infect the larval stages of Lepidoptera and early larval instars are generally more susceptible to viral infection (via oral ingestion) than older larvae, possibly because they slough virus-infected midgut cells at a slower rate [47]. If a larva can grow more quickly than its peers in the same cohort, then it will be less susceptible to virus infection and potentially “escape” disease (via this developmental resistance mechanism). Consistent with this, we found that HaDNV-1-positive larvae developed faster than HaDNV-1-negative larvae (Fig. 3) and accumulated HaNPV at a slower rate (Fig. 5C). In field populations of adults, the infection rate of HaDNV-1 remained high from 2008 to 2012 (more than 67%). However, we failed to detect any HaNPV baculovirus in any of the 361 adults sampled. Only the larval stage is susceptible to baculovirus infection and so one possible explanation for this is that most of the baculovirus-infected individuals are lost from the system before adulthood due to increased larval mortality, abnormal pupation, or unsuccessful eclosion [48]. Alternatively, enhanced resistance to HaNPV in the adult stage may effectively clear all viral infections gained in the larval stage.
Theory suggests that the presence of a beneficial symbiont should result in a high frequency of infection, spreading rapidly through a population until reaching infection fixation [1]. However, our data from adult moths suggest that although there was a high frequency of HaDNV-1 infection, there was also, perhaps unexpectedly, a steady decline in prevalence from 2008 to 2012, which would suggest an unidentified cost of DNV infection. One possible explanation for this decline is that the prevalence of HaDNV-1 is related to the recent widespread introduction to China of genetically-modified Bacillus thuringiensis (Bt) cotton [49], [50], [51]. For example, it might be that selection for Bt-resistance has selected against densovirus infection. If this was the case, then we might expect to observe a negative association between HaDNV-1 infection and resistance to Bt. However, in our laboratory experiment with Bt protoxin and artificial diet, HaDNV-1-positive larvae showed significantly higher resistance to Bt than HaDNV-1-negative larvae at low Bt concentrations (≤0.8 µg/g), while no significant difference was observed at high Bt concentrations (≥1.6 µg/g). Interestingly, the bioassay with Bt cotton plants showed that although HaDNV-1 positive larvae developed faster than negative ones, the difference was not statistically significant, possibly because the leaves of the Bt cotton used (at the seedling stage) contained a high concentration of Bt protein (about 1 µg/g) [52]. A related possibility is that densovirus prevalence is positively associated with the size of the H. armigera population in the wild, which has markedly declined since Bt-cotton was introduced [51], perhaps because horizontal transmission of the densovirus is enhanced at high population densities. The possibility of unknown competitive factors, including other microorganisms, can also not be excluded. Therefore, despite some evidence suggesting that HaDNV-1 could impact the population dynamics of H. armigera, our data are currently not comprehensive enough to explain the long-term dynamics of HaDNV-1, and more monitoring of field populations will be required to answer some of these intriguing questions.
Conclusion
In conclusion, our studies to date suggest a mutualistic relationship between the cotton bollworm and HaDNV-1, in which the cotton bollworm appears to benefit from HaDNV-1 infection, with all host fitness parameters so far tested (larval growth rate, larval and pupal development rate, fertility, adult female lifespan, and resistance to baculovirus and low doses of Bt toxin) enhanced at no detectable cost. The study of beneficial viruses in both vertebrate and invertebrate systems has only relatively recently attracted researchers' attention [2], predominantly due to the explosion of new technologies that now make the detection of such organisms possible. It should be noted that the coevolution between viral mutualistic symbionts and their hosts could be an important factor to consider when studying the adaptability of insect host species. Illuminating the function of such viral symbionts may offer novel insights for future pest management strategies.
Materials and Methods
Insect culture and preparation of HaDNV-1 virus
Cotton bollworms (H. armigera) were reared using artificial diet [53] at 25±1°C with a 14 : 10, light:dark photoperiod. Adult moths were provided with 10% sugar and 2% vitamin complex. The colony was established from thirty breeding pairs captured at Langfang (Hebei province, China) in 2005. Individuals successfully producing offspring were tested for the presence of HaDNV-1, using the methods described below. Offspring from a single uninfected breeding pair were reared to produce the NONINF strain (uninfected) laboratory culture.
HaDNV-1 virus was isolated from migrating H. armigera adults captured in 2010 and 2011 using a vertical-pointing trap, and stored in liquid nitrogen [20]. Briefly, DNA was extracted from host tissues (except for the abdomens) of each individual, and PCR undertaken to detect the presence of HaDNV-1. Subsequently, the abdomens of positive individuals were divided into two groups: one group was used to purify the HaDNV-1 using the method described by La Fauce et al. (method 1) [54]; the other group was used to prepare a filtered liquid, containing an unpurified form of virus (method 2). Briefly, this second method involved grinding four abdomens under liquid nitrogen and transferring to 1 ml PBS buffer (0.01M, pH 7.4). The homogenate was centrifuged at 6500×g for 15 min at 4°C, and the liquid supernatant subsequently filtered with Sartorius Minisart 0.2 µm PES (Invitrogen, Grand Island, USA). The abdomens of negative individuals were filtered using the same method. Quantification of the viruses was performed using the qPCR method described below. All the samples were stored at −20°C.
Virus detection and quantification
To detect the existence of HaDNV-1 in H. armigera, specific primers amplifying a 496 bp fragment, DVVPF/DVVPR (Table S2) were designed according to the genomic sequence of HaDNV-1. The PCR program was as follows: 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C for 40 cycles. For detection of H. armigera nucleopolyhedrovirus (HaNPV), a pair of specific primers amplifying a fragment of 445 bp, NPVF/NPVR, were designed according to the open reading frame 14 (ORF14) of the genomic sequence of HaNPV. The PCR program was as follows: 30 s at 94°C, 30 s at 57°C, and 30 s at 72°C for 40 cycles.
For quantifying the copy numbers of HaDNV-1 and HaNPV, an absolute quantification qPCR methodology using a standard curve was performed [55]. Fragments containing the primers and probes of HaDNV-1 and HaNPV were amplified with our de novo primers (PF/PR for HaDNV-1, NPVF/NPVR for HaNPV) using the program: 30 s at 94°C, 30 s at 53°C, and 60 s at 72°C for 40 cycles, and cloned into the pEASY-T Cloning Vector (TransGen, Beijing, China). These plasmids were subsequently used for the quantification standard curve assay. qPCR was carried out with the TaqMan method in 20 µl reaction agent comprised of 1 µl of template DNA, 2×Premix Ex Taq (Takara, Japan), 0.2 µM of each primer and 0.4 µM probe, using a 7500 Fast Real-time PCR System (Applied Biosystems). Thermal cycling conditions were: 45 cycles of 95°C for 15 s, 60°C for 34 s. The DNA sample of each group was replicated three times. All primers used in this study were shown in Table S2. The equation of y = −1.052x+42.327 (y = the logarithm of plasmid copy number to base 2, x = Ct value, R2 = 0.9997) and y = −0.9861x+44.647 (y = the logarithm of plasmid copy number to base 2, x = Ct value, R2 = 0.9999) were used to calculate the copy number of HaDNV-1 and HaNPV, respectively.
HaDNV-1 transmission and host tissue distribution
We constructed an infected line (INF strain) of H. armigera by orally infecting NONINF strain larvae with HaDNV-1 (from filtered liquid, method 2 - see above) and maintained them by vertical transmission of the virus, using the primers DVVPF/DVVPR to confirm successful establishment of HaDNV-1 infection. Subsequently, individuals from both NONINF strain and INF strain were used to determine the transmission modes of HaDNV-1. For vertical transmission, ♀+/♂−, ♀−/♂+, ♀+/♂+ and ♀−/♂ − pairs were crossed and DNA from 3rd instar offspring larvae used to probe for HaDNV-1.
For the diet contamination assay, (to determine horizontal transmission efficiency), infected individuals from the INF strain were reared in diet cells until the start of the 3rd instar and then removed. Uninfected NONINF strain neonates were then placed in the vacated cells and reared to the pupal stage. DNA was extracted from the adults and probed for HaDNV-1 infection using PCR. Horizontal transmission of HaDNV-1 was determined using PCR with adult DNA as temples and different concentrations of the densovirus: 108, 107, 106, 105, 104/µl. The frass of larvae from HaDNV-1 positive individuals were also quantified by qPCR, as described above.
To examine virus infection in different body tissues, DNA was extracted from body parts of infected individuals (both larval and adult stages) and the copy numbers of HaDNV-1 were quantified by qPCR. To account for individual variation, we first calculated the copy numbers per milligram of tissue and then summed all the copy numbers from different tissues from the same individual and the percentage of each tissue was statistically analyzed (larvae: n = 7; adult males: n = 6; adult females: n = 6).
Quantification of HaDNV-1 in eggs
To further establish the role of vertical transmission in the life-cycle of the densovirus, we quantified HaDNV-1 infections in H. armigera eggs, primarily to distinguish between transovarial and transovum infection routes. Eggs from INF strain breeding pairs, which both of females and males were infected by HaDNV-1, were submerged in 1% sodium hypochlorite for 10 minutes. They were then filtered through a damp cloth, thoroughly rinsed, and allowed to dry. Four groups of hypochlorite-treated eggs (n = 50 eggs per group) were tested against non-treated eggs (control) and HaDNV-1 infections tested by qPCR.
Quantifying HaDNV-1 impact on host development and fecundity
To test the impact of HaDNV-1 infection on the life table parameters of its host, neonate NONINF strain larvae were first orally inoculated with either filtered-liquid containing HaDNV-1, or filtered-liquid from uninfected individuals (control). One hundred NONINF strain neonates were placed in each treatment Petri-dish for 2 days to ensure that larvae ingested the treated diet. They were then transferred to a 24-well plate (one individual per well: diameter = 1.5 cm; height = 2 cm) until the 5th larval instar; larvae were then individually reared in glass tubes until eclosion (diameter = 2 cm; height = 7.5 cm) (Fig. S6). The status of individuals was checked every day at 9 : 00 am. The weight of larvae from the 7th to 11th day post hatch, and the pupa on the 3rd day were recorded. Fifth-instar larvae were randomly selected to estimate the infection rate of HaDNV-1 during the experiment. This bioassay was replicated twice (n = 288 and n = 168 individuals, respectively). Individuals dying within 24 hours of the experimental set up were considered handling deaths, and excluded from the analysis.
In addition, newly eclosed adults from both the HaDNV-1 negative NONINF strain and HaDNV-1 positive INF strain were used to determine longevity, egg production and hatch rate. Three pairs of adults were put in each plastic cup (diameter = 8.5 cm; height = 10 cm) (Fig. S6). The experimental replicates were 3×77 for NONINF strain and 3×60 for INF strain, respectively. We recorded the number of eggs and newly hatched larvae every day. After death, individuals were used to detect HaDNV-1 via PCR. Data from failed matings were excluded.
Lipid quantification in HaDNV-1 positive and negative individuals
To quantify the impact of HaDNV-1 infection on host growth, we measured relative lipid mass within larvae of H. armigera. Larvae 9 days post-hatch were chosen to compare the lipid content between HaDNV-1 positive (n = 19) and HaDNV-1 negative (n = 33) individuals. The protocol was undertaken as Clissold et al. [56]. Briefly, the larval samples were freeze-dried, weighed, chloroform-extracted 3 times, dried again and weighed. The lipid mass was calculated by subtracting the post-chloroform-wash mass from the pre-chloroform-wash mass.
Baculovirus and Bt bioassays
To assess the capacity of HaDNV-1 to act as a beneficial symbiont, we quantified the interaction between HaDNV-1 and the common baculovirus pathogen HaNPV, via a series of laboratory bioassay studies. As previously described, neonate larvae were first treated with HaDNV-1 filtered liquid (either from HaDNV-1 infected or HaDNV-1 negative individuals). Two-day old larvae were then transferred to a 24-well plate and maintained on diet until the 9th day after hatching. Individuals weighing between 5–11 mg (early third-instar stage) were chosen for the HaNPV bioassay. Purified powder of HaNPV at a concentration of 5×1011 occlusion bodies (OBs) per g was generously provided by Dr. Qilian Qin in the Institute of Zoology, Chinese Academy of Science, Beijing, China. Larvae were orally dosed with 4 treatments of HaNPV (30 larvae per treatment at: 0 (control), 1×106, 1×107, 1×108, and 1×109 OBs/ml). Only larvae that ingested all the NPV within a 24 h period were used for the bioassay. Larvae were subsequently monitored daily for NPV mortality until pupation, and all viral deaths stored at −20°C. PCR with specific primers was used to test for NPV in dead larvae with non-obvious symptoms.
To assess HaNPV infection levels in HaDNV-1 positive and negative individuals, we performed a separate HaNPV bioassay with 108 OBs/ml. There were 24 individuals in each replicate and three replicates per treatment. Only larvae that ingested all the NPV within a 24 h period were used for the bioassay. The absolute quantification qPCR methodology was used to quantifying the copy numbers of HaNPV as described above. Survival analysis was conducted using Cox's proportional hazards model.
For the Bacillus thuringiensis bioassays, various concentrations of the Bt Cry1Ac protoxin were added and thoroughly mixed with standard artificial diet to obtain the desired concentrations (0 (control), 0.4 µg/g, 0.8 µg/g, 1.6 µg/g and 3.2 µg/g). After mixing, the diet solidified and solid 1 mg pieces were placed into each well of a 24-well plate and two-day old larvae infected or uninfected by HaDNV-1 were then transferred to each well (Fig. S6). There were 24 individuals in each replicate and three replicates per treatment. We graded the larvae from day 4 to day 9 after hatching according to the development rate: death = 0, early first instar stage = 1, middle first instar stage = 2, last first instar stage = 3, early second instar stage = 4, middle second instar stage = 5, last second instar stage = 6, early third instar stage = 7, middle third instar stage = 8, last third instar stage = 9, early fourth instar stage = 10, middle fourth instar stage = 11 [57].
At seedling stage with 5 leaves, we chose the new cotton 33B with Cry1Ac (Monsanto Company, Bt cotton) using Shi Yuan 321 (Shijiazhuang Acadamy of Agricultural Sciences, NonBt cotton) as control to perform the bioassay. Two-day old larvae infected or uninfected by HaDNV-1 were transferred to a 24-well plate with Bt-cotton or NonBt-cotton. There were 40 individuals in each replicate and three replicates per treatment. We graded the larvae after 7 days according to the development rate.
Detection of HaDNV-1 and HaNPV in wild populations of H. armigera
Samples of larvae were collected at 7 locations in 2012 (Jinan, Dezhou and Taian, Shandong province; Cangzhou, Heibei province; Tianmen and Qianjing, Hubei province; Maanshan, Anhui province) and 6 locations in 2013 (Luohe, Luoyang, Yuanyang and Nanyang, Henan province; Langfang and Cangzhou, Hebei province). The infection rate of HaDNV-1 and HaNPV was determined using the PCR method described as above.
Samples of adults were collected at fifteen locations from 2008 to 2012: A = Xinxiang, Henan province; B = Dezhou; C = Langfang; D = Yantai Shandong province; E = Yancheng, Jiangsu province; F = Handan, Shandong province; G = Changde, Hunan province; H = Tianmen, I = Qianjiang; J = Maanshan; K = Taian; L = Luohe; M = Weinan, Shanxi province; N = Shihezi, O = Kashi, Xinjiang province. We also randomly selected four places to detect HaNPV in the populations, including site 1 in 2010 (54 samples), site 2 in 2010 (103 samples), site 4 in 2012 (104 samples) and site 13 in 2011 (100 samples).
Host range of HaDNV-1
Using the same oral inoculation method as previously described (section 2.5), we chose four species of Lepidoptera (Spodoptera exigua, Spodoptera litura, Agrotis segetum, Agrotis ipsilon) to determine the host range of HaDNV-1 infection. We also collected nine adults of H. assulta from field populations, and PCR was used to detect HaDNV-1 infection.
Statistics
Statistical analyses were conducted using STATA v.9.0 and R v3.0.1 [58]. Student's t-test or ANOVA with Tukey were used to determine the level of significance in the relative levels of HaDNV-1. Egg hatch rates and larval/pupal mortality, pupation and eclosion rates were determined using generalized linear models (GLMs) with binomial errors. Analysis of the NPV and Bt bioassay data was also conducted using GLMs with binomial errors. A generalised linear mixed effects model (GLMM) with binomial errors was used to determine temporal variation in HaDNV-1 infection rates. A GLMM with Gaussian errors was used to quantify variation in larval growth rates with larval identity included as a random term. Development following exposure to Bt toxin in artificial diet was analyzed using linear mixed effects models using the lme function in R, with larval identity as a random term to account for the repeated measures data structure.
Accession number
The GenBank accession number of genomic sequence of HaDNV-1 and HaNPV were HQ613271 and AF303045, respectively.
Supporting Information
Zdroje
1. HimlerAG, Adachi-HagimoriT, BergenJE, KozuchA, KellySE, et al. (2011) Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332 : 254–256.
2. RoossinckMJ (2011) The good viruses: viral mutualistic symbioses. Nat Rev Microbiol 9 : 99–108.
3. SaridakiA, BourtzisK (2010) Wolbachia: more than just a bug in insects genitals. Curr Opin Microbiol 13 : 67–72.
4. VilcinskasA, StoeckerK, SchmidtbergH, RohrichCR, VogelH (2013) Invasive harlequin ladybird carries biological weapons against native competitors. Science 340 : 862–863.
5. WeeksAR, TurelliM, HarcombeWR, ReynoldsKT, HoffmannAA (2007) From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol 5: e114.
6. HedgesLM, BrownlieJC, O'NeillSL, JohnsonKN (2008) Wolbachia and virus protection in insects. Science 322 : 702.
7. BrownlieJC, JohnsonKN (2009) Symbiont-mediated protection in insect hosts. Trends Microbiol 17 : 348–354.
8. HaineER (2008) Symbiont-mediated protection. Proc Biol Sci 275 : 353–361.
9. GrahamRI, GrzywaczD, MushoboziWL, WilsonK (2012) Wolbachia in a major African crop pest increases susceptibility to viral disease rather than protects. Ecol Lett 15 : 993–1000.
10. LomerCJ, BatemanRP, JohnsonDL, LangewaldJ, ThomasM (2001) Biological control of locusts and grasshoppers. Annu Rev Entomol 46 : 667–702.
11. RogoffMH, YoustenAA (1969) Bacillus thuringiensis: microbiological considerations. Annu Rev Microbiol 23 : 357–386.
12. DouglasAE (1989) Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc 64 : 409–434.
13. OliverKM, DegnanPH, HunterMS, MoranNA (2009) Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325 : 992–994.
14. StoltzDB, WhitfieldJB (2009) Virology. Making nice with viruses. Science 323 : 884–885.
15. ShenHH (2009) The challenge of discovering beneficial viruses. J Med Microbiol 58 : 531–532.
16. CowlingBJ, JinL, LauEH, LiaoQ, WuP, et al. (2013) Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 382 : 129–137.
17. DiatchenkoL, LauYF, CampbellAP, ChenchikA, MoqadamF, et al. (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A 93 : 6025–6030.
18. MargueratS, BahlerJ (2010) RNA-seq: from technology to biology. Cell Mol Life Sci 67 : 569–579.
19. KootstraNA, VermaIM (2003) Gene therapy with viral vectors. Annu Rev Pharmacol Toxicol 43 : 413–439.
20. FengH, WuX, WuB, WuK (2009) Seasonal migration of Helicoverpa armigera (Lepidoptera: Noctuidae) over the Bohai Sea. J Econ Entomol 102 : 95–104.
21. FengH, WuK, NiY, ChengD, GuoY (2005) Return migration of Helicoverpa armigera (Lepidoptera: Noctuidae) during autumn in northern China. Bull Entomol Res 95 : 361–370.
22. FengH, WuK, ChengD, GuoY (2004) Northward migration of Helicoverpa armigera (Lepidoptera: Noctuidae) and other moths in early summer observed with radar in northern China. J Econ Entomol 97 : 1874–1883.
23. GunningRV, DangHT, KempFC, NicholsonIC, MooresGD (2005) New resistance mechanism in Helicoverpa armigera threatens transgenic crops expressing Bacillus thuringiensis Cry1Ac toxin. Appl Environ Microbiol 71 : 2558–2563.
24. ZhangH, TianW, ZhaoJ, JinL, YangJ, et al. (2012) Diverse genetic basis of field-evolved resistance to Bt cotton in cotton bollworm from China. Proc Natl Acad Sci U S A 109 : 10275–10280.
25. El-FarM, SzeleiJ, YuQ, FediereG, BergoinM, et al. (2012) Organization of the ambisense genome of the Helicoverpa armigera densovirus. J Virol 86 : 7024.
26. ChristianPD, MurrayD, PowellR, HopkinsonJ, GibbNN, et al. (2005) Effective control of a field population of Helicoverpa armigera by using the small RNA virus Helicoverp aarmigera stunt virus (Tetraviridae: Omegatetravirus). J Econ Entomol 98 : 1839–1847.
27. AllawayGP, PayneCC (1984) Host range and virulence of five baculoviruses from lepidopterous hosts. Ann Appl Biol 105 : 29–31.
28. FuxaJR (2004) Ecology of insect nucleopolyhedroviruse. Agric Ecosyst Environ 103 : 27–43.
29. ChenX, IJkelWF, TarchiniR, Sun X. SandbrinkH, et al. (2001) The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J Gen Virol 82 : 241–257.
30. SunX, ChenX, ZhangZ, WangH, BianchiFJ, et al. (2002) Bollworm responses to release of genetically modified Helicoverpa armigera nucleopolyhedroviruses in cotton. J Invertebr Pathol 81 : 63–69.
31. SunX, WangH, SunX, Chen X. PengC, et al. (2004) Biological activity and field efficacy of a genetically modified Helicoverpa armigera singlenucleocapsid nucleopolyhedrovirus expressing an insect-selective toxin from a chimeric promoter. Biol Control 29 : 124–137.
32. XuP, ChengP, LiuZ, LiY, MurphyRW, et al. (2012) Complete genome sequence of a monosense densovirus infecting the cotton bollworm, Helicoverpa armigera. J Virol 86 : 10909.
33. FediereG (2000) Epidemiology and pathology of Densovirinae. Contrib Microbiol 4 : 1–11.
34. JiangH, ZhangJ, WangJ, YangB, LiuC, et al. (2007) Genetic engineering of Periplaneta fuliginosa densovirus as an improved biopesticide. Arch Virol 152 : 383–394.
35. LedermannJP, SuchmanEL, BlackWCt, CarlsonJO (2004) Infection and pathogenicity of the mosquito densoviruses AeDNV, HeDNV, and APeDNV in Aedes aegypti mosquitoes (Diptera: Culicidae). J Econ Entomol 97 : 1828–1835.
36. MutuelD, RavallecM, ChabiB, MulteauC, SalmonJ, et al. (2010) Pathogenesis of Junonia coenia densovirus in Spodoptera frugiperda: A route of infection that leads to hypoxia.Virology. 403 : 137–144.
37. ChenY, EvansJ, FeldlauferM (2006) Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J Invertebr Pathol 92 : 152–159.
38. ZhouM, SunX, VlakJM, HuZ, van der WerfW (2005) Horizontal and vertical transmission of wild-type and recombinant Helicoverpa armigera single-nucleocapsid nucleopolyhedrovirus. J Invertebr Pathol 89 : 165–175.
39. LongdonB, JigginsFM (2012) Vertically transmitted viral endosymbionts of insects: do sigma viruses walk alone. Proc R Soc B 279 : 3889–3898.
40. FleurietA (1988) Maintenance of a hereditary virus - the sigma-virus in populations of its host, Drosophila melanogaster. Evol Biol 23 : 1–30.
41. FinePE (1975) Vectors and vertical transmission: an epidemiologic perspective. Ann N Y Acad Sci 266 : 173–194.
42. L'HeritierPH (1970) Drosophila viruses and their role as evolutionary factors. Evol Biol 4 : 185–209.
43. LongdonB, WilfertL, ObbardDJ, JigginsFM (2011) Rhabdoviruses in two species of Drosophila: vertical transmission and a recent sweep. Genetics 188 : 141–150.
44. VirtoC, ZarateCA, Lopez-FerberM, MurilloR, CaballeroP, WilliamsT (2013) Gender-mediated differences in vertical transmission of a nucleopolyhedrovirus. PLoS ONE 8: e70932.
45. WeeksAR, StouthamerR (2004) Increased fecundity associated with infection by a cytophaga-like intracellular bacterium in the predatory mite, Metaseiulus occidentalis. Proc Biol Sci 271: S193–195.
46. JiuM, ZhouX, TongL, XuJ, YangX, et al. (2007) Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS ONE 2: e182.
47. EngelhardEK, VolkmanLE (1995) Developmental resistance in fourth instar Trichoplusia ni orally inoculated with Autographa californica M nuclear polyhedrosis virus. Virology 209 : 384–389.
48. GeorgievskaL, HooverK, van der WerfW, MunozD, CaballeroP, et al. (2010) Dose dependency of time to death in single and mixed infections with a wildtype and egt deletion strain of Helicoverpa armigera nucleopolyhedrovirus. J Invertebr Pathol 104 : 44–50.
49. FengH, GouldF, HuangY, JiangY, WuK (2010) Modeling the population dynamics of cottonbollworm Helicoverpa armigera (Hübner) (Lepidoptera:Noctuidae) over a wide area in northern China. Ecol Model 221 : 1819–1830.
50. GaoY, FengH, WuK (2010) Regulation of the seasonal population patterns of Helicoverpa armigera moths by Bt cotton planting. Transgenic Res 19 : 557–562.
51. WuK, LuY, FengH, JiangY, ZhaoJ (2008) Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 321 : 1676–1678.
52. ZhangY, WuK, GuoY (2001) On the spatio-temporal expression of the contents of Bt insecticidal protein and the resistance of Bt transgenic cotton to cotton bollworm. Acta Phytophy Sin 28 : 1–6.
53. LiangG, WuK, YuH, LiK, FengX, et al. (2008) Changes of inheritance mode and fitness in Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae) along with its resistance evolution to Cry1Ac toxin. J Invertebr Pathol 97 : 142–149.
54. La FauceKA, EllimanJ, OwensL (2007) Molecular characterisation of hepatopancreatic parvovirus (PmergDNV) from Australian Penaeus merguiensis. Virology 362 : 397–403.
55. WongML, MedranoJF (2005) Real-time PCR for mRNA quantitation. Biotechniques 39 : 75–85.
56. ClissoldFJ, BrownZP, SimpsonSJ (2013) Protein-induced mass increase of the gastrointestinal tract of locusts improves net nutrient uptake via larger meals rather than more efficient nutrient absorption. J Exp Biol 216 : 329–337.
57. GaoY, WuK, GouldF (2009) Frequency of Bt resistance alleles in H. armigera during 2006–2008 in northern china. Environ Entomol 38 : 1336–1342.
58. R Development Core Team: R: A language and environment for statistical computing (2008) R Foundation for Statistical Computing, Vienna, Austria (ISBN 3-900051-07-0). Retrieved from http://www.R-project.org.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance SystemČlánek Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer CellsČlánek APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse ModelČlánek Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 10- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Theory and Empiricism in Virulence Evolution
- -Related Fungi and Reptiles: A Fatal Attraction
- Adaptive Prediction As a Strategy in Microbial Infections
- Antimicrobials, Stress and Mutagenesis
- A Novel Function of Human Pumilio Proteins in Cytoplasmic Sensing of Viral Infection
- Social Motility of African Trypanosomes Is a Property of a Distinct Life-Cycle Stage That Occurs Early in Tsetse Fly Transmission
- Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer
- Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance System
- mRNA Structural Constraints on EBNA1 Synthesis Impact on Antigen Presentation and Early Priming of CD8 T Cells
- Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages
- Neutrophil Crawling in Capillaries; A Novel Immune Response to
- Live Attenuated Vaccine Protects against Pulmonary Challenge in Rats and Non-human Primates
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
- HIV Acquisition Is Associated with Increased Antimicrobial Peptides and Reduced HIV Neutralizing IgA in the Foreskin Prepuce of Uncircumcised Men
- Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host
- Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence
- Ethanol Stimulates WspR-Controlled Biofilm Formation as Part of a Cyclic Relationship Involving Phenazines
- Densovirus Is a Mutualistic Symbiont of a Global Crop Pest () and Protects against a Baculovirus and Bt Biopesticide
- Insights into Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria
- Mycobacterial Antigen Driven Activation of CD14CD16 Monocytes Is a Predictor of Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome
- Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion
- Dampens the DNA Damage Response
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- Vaginal Challenge with an SIV-Based Dual Reporter System Reveals That Infection Can Occur throughout the Upper and Lower Female Reproductive Tract
- Detecting Differential Transmissibilities That Affect the Size of Self-Limited Outbreaks
- One Small Step for a Yeast - Microevolution within Macrophages Renders Hypervirulent Due to a Single Point Mutation
- Expression Profiling during Arabidopsis/Downy Mildew Interaction Reveals a Highly-Expressed Effector That Attenuates Responses to Salicylic Acid
- Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer Cells
- Interaction with Tsg101 Is Necessary for the Efficient Transport and Release of Nucleocapsids in Marburg Virus-Infected Cells
- The N-Terminus of Murine Leukaemia Virus p12 Protein Is Required for Mature Core Stability
- Sterol Biosynthesis Is Required for Heat Resistance but Not Extracellular Survival in
- Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility
- Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly
- APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse Model
- Structural Basis for the Recognition of Human Cytomegalovirus Glycoprotein B by a Neutralizing Human Antibody
- Systematic Analysis of ZnCys Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus
- Epstein-Barr Virus Nuclear Antigen 3A Promotes Cellular Proliferation by Repression of the Cyclin-Dependent Kinase Inhibitor p21WAF1/CIP1
- The Host Protein Calprotectin Modulates the Type IV Secretion System via Zinc Sequestration
- Cyclophilin A Associates with Enterovirus-71 Virus Capsid and Plays an Essential Role in Viral Infection as an Uncoating Regulator
- A Novel Alpha Kinase EhAK1 Phosphorylates Actin and Regulates Phagocytosis in
- The pH-Responsive PacC Transcription Factor of Governs Epithelial Entry and Tissue Invasion during Pulmonary Aspergillosis
- Sensing of Immature Particles Produced by Dengue Virus Infected Cells Induces an Antiviral Response by Plasmacytoid Dendritic Cells
- Co-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites
- Characteristics of Memory B Cells Elicited by a Highly Efficacious HPV Vaccine in Subjects with No Pre-existing Immunity
- HPV16-E7 Expression in Squamous Epithelium Creates a Local Immune Suppressive Environment via CCL2- and CCL5- Mediated Recruitment of Mast Cells
- Dengue Viruses Are Enhanced by Distinct Populations of Serotype Cross-Reactive Antibodies in Human Immune Sera
- CD4 Depletion in SIV-Infected Macaques Results in Macrophage and Microglia Infection with Rapid Turnover of Infected Cells
- A Sialic Acid Binding Site in a Human Picornavirus
- Contact Heterogeneity, Rather Than Transmission Efficiency, Limits the Emergence and Spread of Canine Influenza Virus
- Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of
- HTLV-1 Tax Stabilizes MCL-1 via TRAF6-Dependent K63-Linked Polyubiquitination to Promote Cell Survival and Transformation
- Species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence
- A Critical Role for IL-17RB Signaling in HTLV-1 Tax-Induced NF-κB Activation and T-Cell Transformation
- Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90
- Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
- Kaposi's Sarcoma-Associated Herpesvirus Induces Nrf2 during Infection of Endothelial Cells to Create a Microenvironment Conducive to Infection
- A Relay Network of Extracellular Heme-Binding Proteins Drives Iron Acquisition from Hemoglobin
- Glutamate Secretion and Metabotropic Glutamate Receptor 1 Expression during Kaposi's Sarcoma-Associated Herpesvirus Infection Promotes Cell Proliferation
- Reduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and Anti-pathogen Activities
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



