-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCo-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites
Viruses recruit cellular membranes and subvert cellular proteins involved in lipid biosynthesis to build viral replicase complexes and replication organelles. Among the lipids, sterols are important components of membranes, affecting the shape and curvature of membranes. In this paper, the tombusvirus replication protein is shown to co-opt cellular Oxysterol-binding protein related proteins (ORPs), whose deletion in yeast model host leads to decreased tombusvirus replication. In addition, tombusviruses also subvert Scs2p VAP protein to facilitate the formation of membrane contact sites (MCSs), where membranes are juxtaposed, likely channeling lipids to the replication sites. In all, these events result in redistribution and enrichment of sterols at the sites of viral replication in yeast and plant cells. Using in vitro viral replication assay with artificial vesicles, we show stimulation of tombusvirus replication by sterols. Thus, co-opting cellular ORP and VAP proteins to form MCSs serves the virus need to generate abundant sterol-rich membrane surfaces for tombusvirus replication.
Published in the journal: . PLoS Pathog 10(10): e32767. doi:10.1371/journal.ppat.1004388
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004388Summary
Viruses recruit cellular membranes and subvert cellular proteins involved in lipid biosynthesis to build viral replicase complexes and replication organelles. Among the lipids, sterols are important components of membranes, affecting the shape and curvature of membranes. In this paper, the tombusvirus replication protein is shown to co-opt cellular Oxysterol-binding protein related proteins (ORPs), whose deletion in yeast model host leads to decreased tombusvirus replication. In addition, tombusviruses also subvert Scs2p VAP protein to facilitate the formation of membrane contact sites (MCSs), where membranes are juxtaposed, likely channeling lipids to the replication sites. In all, these events result in redistribution and enrichment of sterols at the sites of viral replication in yeast and plant cells. Using in vitro viral replication assay with artificial vesicles, we show stimulation of tombusvirus replication by sterols. Thus, co-opting cellular ORP and VAP proteins to form MCSs serves the virus need to generate abundant sterol-rich membrane surfaces for tombusvirus replication.
Authors Summary
Cellular proteins and cellular membranes are usurped by positive-stranded RNA viruses to assemble viral replicase complexes required for their replication. Tombusviruses, which are small RNA viruses of plants, depend on sterol-rich membranes for replication. The authors show that the tombusviral replication protein binds to cellular oxysterol-binding ORP proteins. Moreover, the endoplasmic reticulum resident cellular VAP proteins also co-localize with viral replication proteins. These protein interactions likely facilitate the formation of membrane contact sites that are visible in cells replicating tombusvirus RNA. The authors also show that sterols are recruited and enriched to the sites of viral replication. In vitro replication assay was used to show that sterols indeed stimulate tombusvirus replication. In summary, tombusviruses use subverted cellular proteins to build sterol-rich membrane microdomain to promote the assembly of the viral replicase complex. The paper connects efficient virus replication with cellular lipid transport and membrane structures.
Introduction
Plus-stranded (+)RNA viruses subvert various intracellular and organellar membranes to assemble viral replicase complexes (VRCs) consisting of viral replication proteins and co-opted host proteins and the viral RNAs in the infected cells [1]–[6]. Although several of the co-opted host proteins affect the biochemical activities of the viral RNA-dependent RNA polymerases (RdRp), [7]–[11], other host proteins, such as the ESCRT proteins, reticulons and amphiphysins are proposed to function by facilitating membrane deformation occurring during VRC assembly [12]–[15], further highlighting the importance of membranes in viral replication.
RNA viruses also subvert cellular proteins involved in lipid biosynthesis or alter intracellular lipid metabolism and lipid transport [5], [6], [16], [17]. Viral RdRps of many (+)RNA viruses interact with membranes and build functional VRCs in single-membrane spherules and vesicle-like structures that have a narrow opening to the cytosol. Accordingly, virus-induced formation of spherules, double membrane vesicles or tubulovesicular cubic membranes is documented in a variety of cell organelles [6], [16], [18].
The virus-induced VRCs and membranous structures not only gather all the replication factors into confined cytosolic areas, but importantly, they also protect the fragile viral RNAs from degradation by host ribonucleases and help avoid recognition of viral components by the host antiviral surveillance system [6], [19]. Overall, assembly of the VRCs is an essential step during the replication of (+)RNA viruses that is absolutely dependent on cellular lipids and membranes in the infected cells.
Tomato bushy stunt virus (TBSV) and Carnation Italian ringspot virus (CIRV) are tombusviruses with small (+)RNA genome that serve as models to study virus - host interactions, virus replication and recombination using yeast (Saccharomyces cerevisiae) as a surrogate host [6], [20]–[22]. Tombusviruses code for two replication proteins involved in all the replication steps. The auxiliary TBSV p33 replication protein, which is an RNA chaperone, recruits the TBSV (+)RNA to the site of replication, which occurs at the cytosolic surface of peroxisomal membranes, while the auxiliary CIRV p36 replication protein recruits the viral (+)RNA to the outer mitochondrial membrane [23]–[25]. The interaction between the TBSV RdRp protein p92pol and the p33 replication protein is required for assembling the functional VRCs [26]. Multiple systematic genome-wide screens in combination with global proteomics approaches have led to the identification of ∼500 host proteins/genes that are implicated in TBSV replication and recombination [7], [9], [27]–[35]. The tombusvirus p33 and p92pol replication proteins, the viral (+)RNA and a ∼dozen co-opted host proteins are recruited to peroxisome membranes to assemble the tombusvirus VRCs [6], [15], [26], [34]–[45].
Previously, we have shown that TBSV replication depends on sterol and phospholipid biosynthesis in yeast and plant cells [46], [47]. Sterols are ubiquitous and essential membrane components in all eukaryotes, regulating many membrane functions, membrane rigidity, fluidity, curvature and permeability by interacting with other lipids and proteins within the membranes [48]–[50]. Sterols are also important for the organization of detergent-resistant membrane microdomains, called membrane rafts [51]. Most sterols are synthesized in the ER membrane and mainly accumulate in the plasma membrane in uninfected cells.
The lipid transfer proteins, including the sterol - and oxysterol-binding proteins [OSBP or Oxysterol-binding protein related proteins (ORPs)] are involved in many cellular processes, which includes nonvesicular sterol transfer, vesicular trafficking, lipid metabolism, and signaling [52]–[55]. The ORPs are important to increase the local concentration of sterols in membrane compartments that facilitate the bending of membranes [52]–[55]. Yeast has seven ORP genes (OSH1-7), while Arabidopsis has 12 predicted ORPs [56]. Because of these features, ORPs might facilitate the formation of viral replication organelles, including VRCs when subverted by viruses.
The ORPs are known to function at membrane contact sites (MCS or ER junctions), where the ER membrane is proximal to other intracellular organelles [57], [58]. MCSs are proposed to facilitate the non-vesicular trafficking of small molecules, including sterols and other lipids. The ORPs are recruited to MCSs by VAP (VAMP-associated protein) proteins [57], [58]. VAPs are present in all eukaryotes and implicated in the regulation of lipid metabolism and transport, membrane trafficking, microtubule organization and the unfolded protein response [59]. Interestingly, the global proteomics screens with TBSV identified the p33-interacting Scs2p protein [33], which is the major member of the VAP family in yeast. Arabidopsis has 10 VAP orthologs, which are grouped in the VAP33 subfamily and the best-characterized member, PVA12 is known to localize to the ER, similar to the yeast Scs2p VAP protein [56]. Scs2p is a tail-anchored, ER membrane-associated protein that anchors the ORPs to the ER membrane [58].
In this paper, we identify the yeast oxysterol-binding homology (Osh) proteins, which are co-opted to support tombusvirus replication. We also show the redistribution and enrichment of sterols to the site of tombusvirus replication, suggesting that the viral replication proteins interact with the yeast Osh proteins to create sterol-rich membrane microdomains. In addition, we present evidence that the viral replication proteins bind to the yeast Scs2p VAP protein in the ER membrane, likely facilitating the formation of MCSs and the recruitment of Osh proteins. Many of our findings are also observed in plants, suggesting that the subversion of ORPs and VAP proteins by tombusviruses also takes place in plants. Altogether, these virus-host interaction processes likely serve the virus need to generate abundant sterol-rich membrane surfaces for virus replication.
Results
Tombusvirus replication depends on recruited cellular oxysterol-binding proteins in yeast model host
Tombusvirus replication greatly depends on sterols [47], which are distributed in cells via vesicle transfer and sterol-binding ORP proteins in a vesicle independent pathway [53], [55]. Therefore, we have tested if TBSV replication proteins interact with the yeast ORPs, named Osh1-7p, to facilitate sterol transfer in virus-infected cells. Our co-purification experiments with the affinity-purified tombusvirus p33 replication protein from isolated membranous fraction of yeast model host showed that four of the seven yeast Osh proteins, which are lipid-transfer proteins involved in oxysterol/sterol-binding, were associated with the membrane-bound p33 (Fig. 1A and S1A). These yeast proteins, namely Osh3p, Osh5p, Osh6p and Osh7p, were not only efficiently co-purified with p33 replication protein, but they also bound directly to p33 in a pull-down assay (Fig. S1B) and interacted with p33 in a membrane (split-ubiquitin-based) yeast two-hybrid assay (Fig. S1C). Co-purification of Osh4p was less robust with p33 from membranous fraction of yeast, while the co-purified Osh1p and Osh2p were close to the detection limit (Fig. S1A).
Fig. 1. Interaction between p33 replication protein and yeast oxysterol-binding Osh proteins. 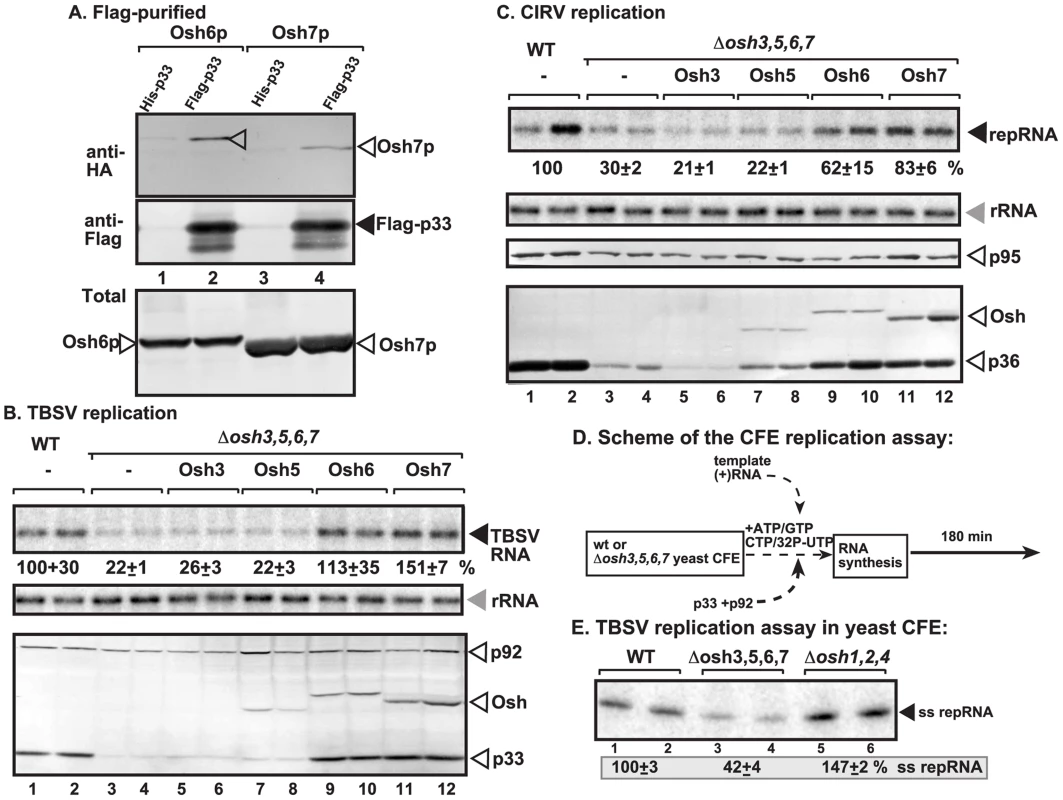
(A) Co-purification of the yeast Osh6p and Osh7p proteins with the p33 replication protein. Top panel: Western blot analysis of co-purified HA-tagged cellular proteins with Flag-affinity purified p33 from isolated membrane fraction of yeast cells. Osh6p and Osh7p were detected with anti-HA antibody. The negative control was His6-tagged p33 purified from yeast extracts using a FLAG-affinity column. Middle panel: Western blot of purified Flag-p33 detected with anti-FLAG antibody. Bottom panel: Western blot of HA-tagged Osh6p and Osh7p proteins in the total yeast extracts using anti-HA antibody. (B) Decreased TBSV repRNA accumulation in osh3,5,6,7Δ yeast. To launch TBSV repRNA replication, we expressed His6-p33 from the galactose-inducible GAL1 promoter, His6-p92 from the copper-inducible CUP1 promoter and DI-72(+) repRNA from the galactose-inducible GAL10 promoter in the parental (SEY6210) and in osh3,5,6,7Δ yeast strains. His6-tagged Osh3, 5, 6, 7 were expressed from GAL1 promoter. The yeast cells were pre-cultured for 24 hours at 23°C in 2% glucose SC minimal media, and then for 48 h at 23°C in 2% galactose SC minimal media supplemented with 50 µM CuSO4. Northern blot analysis was used to detect DI-72(+) repRNA accumulation. The accumulation level of DI-72(+) repRNA was normalized based on 18S rRNA levels. Bottom panel: Western blot analysis of the accumulation level of His6-tagged p33, His6-p92 and His6-Osh proteins using anti-His antibodies. Each experiment was performed three times. (C) Decreased accumulation of the mitochondrial CIRV in osh3,5,6,7Δ yeast. See further details in Panel B. (D) Scheme of the in vitro tombusvirus replicase assay based on yeast CFEs and purified recombinant TBSV replication proteins. (E) Reduced activity of the tombusvirus replicase assembled in CFE from osh3,5,6,7Δ yeast. Denaturing PAGE analysis of in vitro tombusvirus replicase activity in the CFEs. Note that this image shows the repRNAs made by a full cycle of replicase activity, producing both (−) and (+)-strands, in vitro. The CFEs contained the same amount of total yeast proteins (not shown). Each experiment was performed three times. To demonstrate that some members of the 12 ORP proteins from Arabidopsis thaliana plants [60] could also interact with the tombusvirus p33 replication protein, we expressed 6 small ORP (i.e., ORP3A-C and ORP4A-C) proteins from A. thaliana, which similar to the yeast Osh6p lack the FFAT motif, in yeast cells replicating TBSV repRNA. The FFAT motif in mammalian ORPs and several of the yeast ORPs is used for interaction with VAPs [53]. Affinity-purification of tombusvirus p33 from isolated membranous fraction of yeast co-expressing one of the AtORPs showed that all six AtORP proteins were co-purified with p33 (Fig. S2). Altogether, these data have established that a set of host ORPs directly interacted with the tombusvirus p33 replication protein and these cytosolic proteins are likely recruited to intracellular membranes by p33 replication protein.
To study if the recruited cellular Osh proteins affect TBSV replication in yeast, we used a yeast strain lacking the four p33-interacting OSH genes (osh3,5,6,7Δ) [61]. Interestingly, replication of TBSV RNA decreased by ∼5-fold in osh3,5,6,7Δ yeast in comparison with the wt yeast (compare lanes 3–4 with 1–2 in Fig. 1B). The amount of the p33 replication protein accumulating in osh3,5,6,7Δ yeast greatly decreased, suggesting that these cellular Osh proteins are needed for the stability of this viral replication protein, while their lesser effects on p92pol replication protein accumulation was also observed (Fig. 1B, lanes 3–4; also, Fig. S1D, lanes 3–4). Similarly, another tombusvirus, CIRV, which unlike TBSV that utilizes the peroxisome membrane, takes advantage of the mitochondrial outer membranes for replication [23]–[25], also accumulated at a ∼3-fold reduced level in osh3,5,6,7Δ yeast (compare lanes 3–4 with 1–2 in Fig. 1C). The accumulation of the CIRV p36 and p95pol replication proteins also decreased in osh3,5,6,7Δ yeast. Based on our findings that single deletion of each of the seven yeast OSH genes (not shown) or combined deletion of three genes in osh5,6,7Δ or osh1,2,4Δ yeasts (Fig. S1D) did not have a major effect on TBSV replication, while combined deletion of the four OSH genes in osh3,5,6,7Δ yeast decreased TBSV and CIRV accumulation, we suggest that Osh3/5/6/7 proteins likely play at least partially overlapping roles in tombusvirus replication.
Complementation studies with individually expressed Osh proteins in osh3,5,6,7Δ yeast revealed that Osh6p and Osh7p could fully complement TBSV replication, while also partially complementing CIRV replication (lanes 9–12, Fig. 1B–C). Complementation of tombusvirus replication by Osh3p and Osh5p was not detectable under these conditions (lanes 5–8, Fig. 1B–C).
To obtain evidence that Osh proteins can affect the assembly of the membrane-bound tombusvirus replicase complex, we took advantage of a cell-free TBSV replication assay based on yeast cell-free extracts (CFE) [26]. In this assay, the TBSV replicase is assembled in the test tube using CFE, purified recombinant p33 and p92pol and in vitro transcribed TBSV (+)repRNA (Fig. 1D) [26]. Under these experimental conditions, the CFE can support a single round of full tombusviral replication, producing 32P-labeled minus - (which is present in dsRNA form) and excess amount of viral plus-strand RNA progeny [26], [62]. We found that CFE prepared from osh3,5,6,7Δ yeast supported TBSV RNA replication only with ∼40% efficiency when compared with CFE from wt yeast (compare lanes 3–4 with 1–2 in Fig. 1E). In contrast, CFE prepared from osh1,2,4Δ yeast lacking the three Osh proteins not interacting with p33 (Fig. 1E), supported in vitro TBSV replication efficiently, suggesting that, unlike Osh3/5/6/7; the Osh1p, Osh2p and Osh4p ORP proteins are not required for the in vitro assembly of the TBSV replicase.
Overall, these data indicate that Osh3p, Osh5p, Osh6p and Osh7p are required for TBSV and CIRV RNA replication, as well as that they affect replicase assembly and the accumulation of tombusvirus replication proteins. However, these proteins likely play redundant roles since combined deletion of the four p33-interacting OSH genes was needed to have substantial effect on TBSV accumulation in yeast and yet, single expression of either Osh6p or Osh7p in osh3,5,6,7Δ yeast was satisfactory to complement the viral replication defect in osh3,5,6,7Δ yeast.
Co-localization of Osh6p with tombusvirus replication proteins in yeast
To test if Osh6p interacts and co-localizes with the tombusvirus p33 replication protein, we used bimolecular fluorescence complementation (BiFC) assay in yeast co-expressing the N-terminal half of Venus YFP fused to p33 and the C-terminal half of Venus YFP fused to Osh6p (Fig. 2A). The BiFC signal revealed interaction between p33 and Osh6p and these proteins were co-localized partially to the peroxisome membranes (Fig. 2A) and to the ER membrane (Fig. 2B). The BiFC signal with Osh6p in the absence of the p33 fusion part (i.e., expression of the N-terminal half of Venus YFP only) was close to background level (Fig. 2C) [63]. Using the BiFC approach, we also observed interaction between the CIRV p36 replication protein and Osh6p and partial co-localization to the mitochondrial membrane (Fig. 2D) and to the ER membrane (Fig. 2E). Thus, the mitochondria-localized CIRV p36 could also co-opt Osh6p, but the relocalization of Osh6p occurs at a different subcellular location than in the case of peroxisome-localized TBSV. Overall, these data support that Osh6p is recruited by tombusviruses to subcellular membranes likely via interaction between the replication proteins and Osh6p.
Fig. 2. The tombusvirus p33 replication protein interacts with the yeast Osh6p protein in the ER. 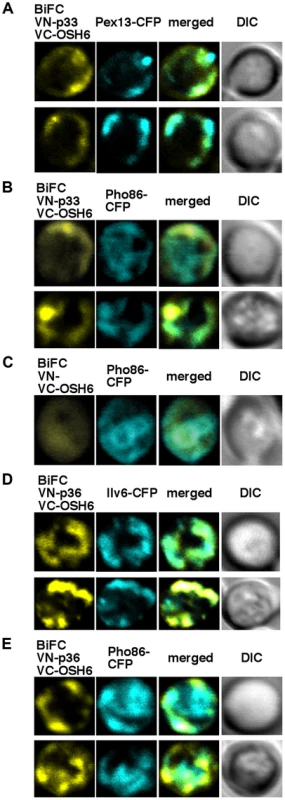
(A–B) Left panels: BiFC analysis of interaction between Osh6p and p33 in yeast cells. Confocal laser microscopy images also show the peroxisomal localization of Pex13-CFP or ER-localized Pho86-CFP. The merged images demonstrate the interaction between p33 and Osh6p (detected via BiFC) and their partial co-localization with Pex13-CFP and Pho86-CFP. DIC (differential interference contrast) images are shown on the right. (C) Control BiFC experiments with yeast lacking the p33 sequence. Yeast was grown under similar conditions and images were taken as in panel A–B. (D–E) BiFC analysis of interaction between Osh6p and the CIRV p36 in yeast cells. Confocal laser microscopy images show the mitochondrial localization of Ilv6-CFP or ER-localized Pho86-CFP. The merged images demonstrate the interaction between p36 and Osh6p and their partial co-localization with Ilv6-CFP and Pho86-CFP. Re-distribution of sterols to the sites of tombusvirus replication in yeast
Since the proposed major function of ORPs is to transfer sterols between organellar membranes inside cells [52], [64], we predicted that recruitment of Osh3/5/6/7 ORPs by tombusviruses to the sites of viral replication could result in enrichment of sterols in these viral subcompartments. Accordingly, testing the distribution of sterols in yeast using the fluorescent sterol probe filipin dye [61] revealed striking differences between yeast cells replicating or not replicating tombusviruses. Namely, the sterols, which are mostly enriched in the plasma membrane in yeast reaching ∼60% of total ergosterols (images without tombusviruses on the right, Fig. 3A) [53], were redistributed to internal punctate-like subcellular compartments in yeast replicating TBSV and CIRV tombusviruses (left and central images). The redistributed sterols were mostly present in punctate-like structures in yeast replicating the mitochondria-localized CIRV, while the punctate structures were also visible, but somewhat more diffused in yeast replicating the peroxisome-localized TBSV (Fig. 3A). The reduced localization of sterols in the plasma membrane was also noticeable in yeast replicating these tombusviruses.
Fig. 3. Enrichment of sterols at the sites of tombusvirus replication in yeast. 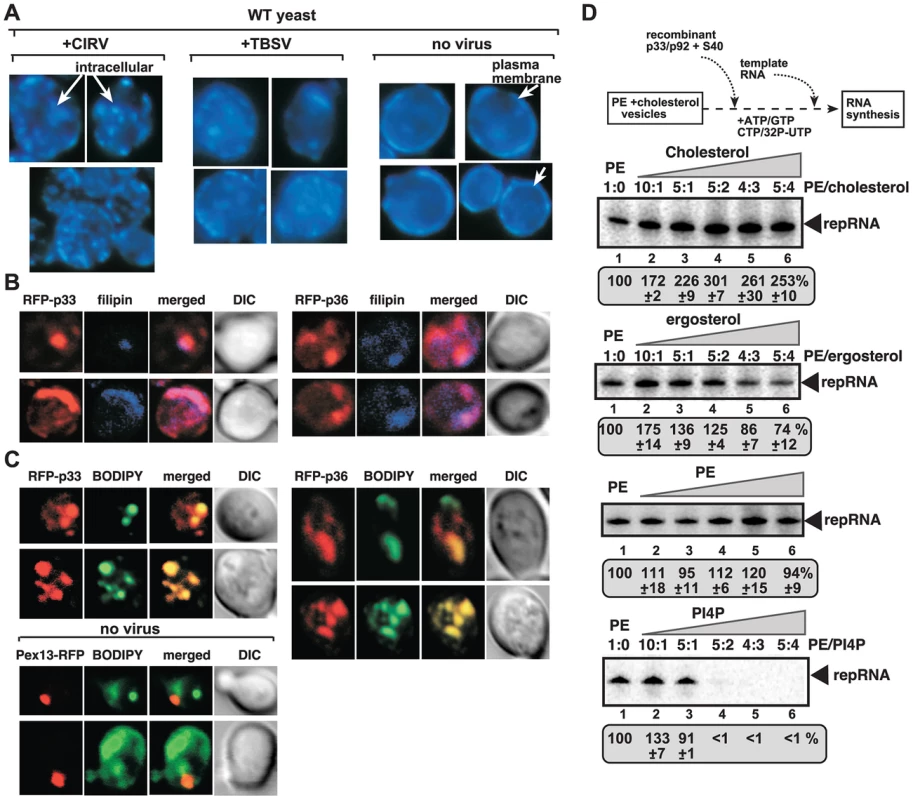
(A) Re-localization of ergosterols to internal punctate structures in yeast replicating CIRV or TBSV. Fluorescent microscopic images of yeast cells stained with filipin dye. Note that filipin stains ergosterols present mostly at the plasma membrane in virus-free cells. (B) Co-localization of ergosterols and CIRV RFP-p36 and CNV RFP-p33 is shown by confocal laser microscopy. Yeasts were stained with filipin dye. (C) Enrichment of fluorescently-labeled sterol at the sites of tombusvirus replication in yeast deficient in ergosterol synthesis (erg9Δ). The BODIPY-cholesterol was taken up by yeast from the culture media. The bottom two rows represent images from yeast not expressing tombusvirus proteins and serve as control. (D) Sterols stimulate in vitro TBSV replication in artificial vesicles. Artificial PE vesicles (liposomes) were made in the presence of increasing concentrations of cholesterol or ergosterol, followed by in vitro TBSV replication assay as shown schematically. The amount of TBSV repRNA synthesized in the in vitro assay is shown in PAGE images. PE and PI(4)P were used as controls. Note that addition of extra PE did not change TBSV replication (second panel from the bottom), while PI(4)P inhibited it when used at higher concentrations (bottom image). To test if the redistributed sterols ended up in the membranes supporting tombusvirus replication, we performed co-localization experiments with RFP-tagged replication proteins and filipin dye to monitor sterol distribution. These experiments showed the enrichment of sterols in the subcompartment containing either CIRV p36 or the peroxisomal p33 replication proteins (Fig. 3B). We also performed experiments with fluorescently-labeled sterols (BODIPY-cholesterol) added to the growth media of yeast lacking the capacity to synthesize sterols (due to the deletion of the ergosterol biosynthesis ERG9 gene) [65]. Interestingly, the fluorescently-labeled sterols were enriched in the same compartment as the tombusvirus p33 protein, or the CIRV p36, forming the characteristic punctate-like structures (Fig. 3C). Based on these data, we suggest that sterols (either synthesized internally in yeast or derived via intake from the culture media) are re-distributed and highly enriched at the sites of tombusvirus replication protein accumulation, which represent the sites of tombusvirus replication in yeast [12], [41].
Sterols stimulate tombusvirus replication in artificial vesicles
Artificial vesicles (liposomes) formed from phosphatidylethanolamine (PE) can support in vitro replication of TBSV repRNA in the presence of viral replication proteins and the soluble fraction of CFE (Xu and Nagy, unpublished). We have tested if presence of sterols in the artificial PE vesicles could promote TBSV replication. The in vitro replication assay revealed that TBSV replication was increased to the highest extent (by up to ∼3-fold) when either ∼30% cholesterol or 10% ergosterol was present in the PE vesicles (Fig. 3D). Thus, the artificial vesicles-based assay supports the stimulatory role of sterols in tombusvirus replication.
Tombusvirus p33 replication protein interacts with Scs2p VAP protein
The cellular ORPs are involved in non-vesicular lipid transfer and they are known to function at membrane contact sites (which are temporally formed areas where two membranes come to close vicinity) by facilitating the transfer of sterols and sterol derivatives from one membrane (the ER is usually the donor membrane) to the other membrane, serving as the acceptor membrane [54], [57], [58]. By hijacking the cellular ORPs, tombusviruses might be able to induce and/or stabilize the formation of MCSs to accelerate sterol transfer from the ER to peroxisomal membranes (or mitochondrial in case of CIRV). The MCS involving the ER membrane usually contains the ER-resident VAP proteins that facilitate the formation of MCSs [54], [57]. Indeed, we have identified the yeast Scs2p VAP protein in our previous genome-wide screens with tombusviruses [32], [35].
Therefore, to obtain evidence on the putative role of MCSs in tombusvirus replication, we studied the interaction between p33 replication protein and the yeast Scs2p VAP protein that is implicated in the regulation of lipid metabolism and transport. Scs2p is ER localized and binds to ORPs [53], [63]. Importantly, Scs2p is present at MCSs involving the ER [58]. We found that p33 replication protein strongly interacts with the yeast Scs2p based on membrane yeast two-hybrid assay (Fig. 4A) [33]. The interaction between the HA-tagged Scs2p and the FLAG-tagged p33 was confirmed using a co-purification assay based on FLAG-affinity purification of p33 replication protein from membrane fraction (Fig. 4B).
Fig. 4. The tombusvirus p33 replication protein binds to the yeast Scs2p VAP protein in the ER. 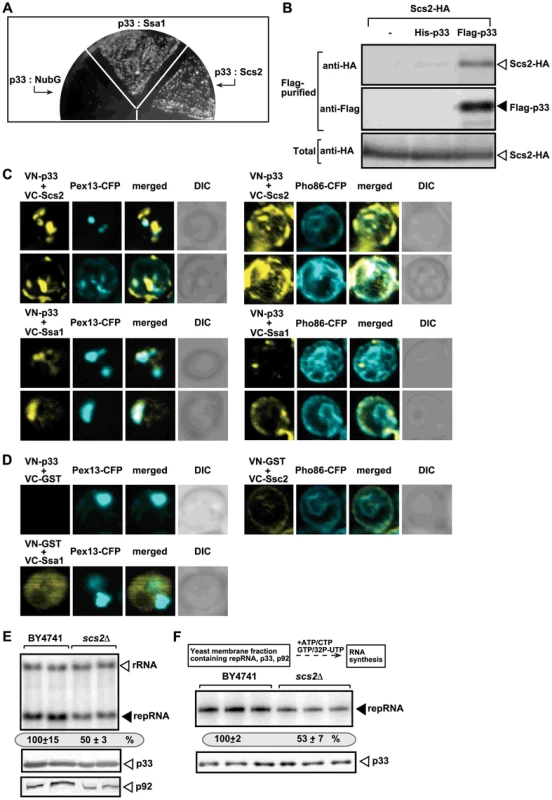
(A) The split ubiquitin assay was used to test binding between p33 and Scs2p in wt (NMY51) yeast. The bait p33 was co-expressed with the shown prey proteins. SSA1 (HSP70 chaperone), and the empty prey vector (NubG) were used as positive and negative controls, respectively. (B) Co-purification of the Scs2p protein with the tombusvirus p33 replication protein. The FLAG-tagged p33 was purified from the membrane fractions of yeast extracts using a FLAG-affinity column. Top panel: Western blot analysis of co-purified 6xHA-tagged Scs2p using anti-HA antibody. Middle panel: Western blot of purified p33 (either His6- or Flag-tagged, as shown) detected with anti-FLAG antibody. Bottom panel: Western blot of 6xHA-tagged Scs2p in the total yeast extract using anti-HA antibody. (C) BiFC analysis of interactions between Scs2p and p33 and between Hsp70 (Ssa1p) and p33. Confocal laser microscopy images also show the peroxisomal localization of Pex13p marker protein (left panels) or Pho86-CFP ER-marker protein (right panels). The merged images at the top show the interaction between p33 and Scs2p and their partial co-localization with Pex13p-CFP or Pho86-CFP ER-marker, while the merged images at the bottom demonstrate the interaction between p33 and Ssa1 and their co-localization with Pex13p. DIC (differential interference contrast) images are shown on the right. Each row represents a separate yeast cell. Note that the Venus-(N-terminal portion) tag was fused to the N-terminus of p33 and the Venus-C tag was fused to host proteins, which are all N-terminal tags. Both p33 and Scs2p have cytosolic N-terminal regions. (D) Control BiFC experiments. Yeast was grown under similar conditions and images were taken as in panel C. (E) Decreased TBSV repRNA accumulation in scs2Δ yeast. To launch TBSV repRNA replication, we expressed His6-p33 and FLAG-tagged p92 from the copper-inducible CUP1 promoter and DI-72(+) repRNA from the galactose-inducible GAL1 promoter in the parental (BY4741) and in scs2Δ yeast strains. The yeast cells were cultured for 16 hours at 23°C on 2% galactose SC minimal media, and then for 24 h at 23°C on 2% galactose SC minimal media supplemented with 50 µM CuSO4. Northern blot analysis was used to detect DI-72(+) repRNA accumulation. The accumulation level of DI-72(+) repRNA was normalized based on 18S rRNA levels. Bottom panels: Western blot analysis of the accumulation level of His6-tagged p33 and FLAG-tagged p92 proteins using anti-His or anti-FLAG antibodies. Each experiment was performed three times. (F) Reduced activity of the tombusvirus replicase assembled in scs2Δ yeast. Top: Scheme of the experimental design. Denaturing PAGE analysis of in vitro replicase activity in the membrane-enriched fraction from wt and scs2Δ yeasts using the co-purified repRNA. The yeast cells were harvested for analysis at 24 h time point after launching TBSV replication. Note that this image shows the repRNAs made by the replicase in vitro. Each experiment was performed three times. Using BiFC, we also show that the Scs2p: p33 interaction mostly takes place in the ER (Fig. 4C, top right panel), not in the peroxisome membrane (Fig. 4C, top left panel), where TBSV replication occurs, especially at the early stage of the replication process [41]. This is in contrast with the co-localization of Hsp70 (Ssa1p in yeast) and p33, which is mostly observed in the peroxisomal membrane (Fig. 4C, bottom panels), as demonstrated earlier [39]. The co-localization and interaction of p33 and Scs2p in the ER membrane are in agreement with the model that TBSV could interact with Scs2p and the Osh proteins to form/stabilize MCSs.
To study the relevance of p33: Scs2p interaction, we launched TBSV replication in scs2Δ yeast [59]. Interestingly, we found ∼50% reduction of TBSV replicon (rep)RNA level in scs2Δ yeast (Fig. 4E, top panel), suggesting that Scs2p is beneficial for TBSV replication. The p33 and p92 levels decreased in scs2Δ yeast (Fig. 4E, bottom panels), suggesting that Scs2p affects the stability of the tombusvirus replication proteins [46]. When we isolated the membrane-bound tombusvirus replicase from wt and scs2Δ yeast, we found ∼2-fold reduction in replicase activity in vitro after adjustment for comparable level of p33 replication protein in the replicase preparations (Fig. 4F). Overall, the inhibitory effect of SCS2 deletion on TBSV replication, replicase activity and replication protein levels in yeast are similar to the effects caused by deletion of Osh3/5/6/7 in yeast (see above).
Expression of Scs2p from the strong GAL1 promoter resulted in up to ∼8-fold increase in TBSV repRNA accumulation in scs2Δ yeast (Fig. S3), demonstrating that Scs2p is a positive host factor for TBSV. Moreover, complementation with the conserved VAP-motif containing MSP domain of Scs2p, which interacts with p33, also led to ∼5-fold stimulation of TBSV repRNA accumulation in scs2Δ yeast (Fig. S3), suggesting that the VAP region is critical to promote TBSV. These experiments established that two critical groups of cellular proteins, namely the ORP and VAP proteins, which are functional at MCSs, interact with the tombusvirus replication protein and stimulate tombusvirus replication.
Over-expression of plant VAP proteins increases tombusvirus replication in yeast and plants
Although over-expression of ORPs in yeast or plants had limited effect on tombusvirus replication (likely due to their abundance and/or redundant nature) (not shown), we attempted over-expression strategy with plant VAP proteins, which are conserved among eukaryotes [59]. First, we showed that six different Arabidopsis VAP proteins could interact with TBSV p33 protein in the yeast split-ubiquitin assay (Fig. 5A), similar to the interaction between p33 and the yeast Scs2p (Fig. 5A). Then, we demonstrated that expression of AtVAP27-1 and AtVAP27-2 proteins [66], [67] increased TBSV repRNA accumulation in scs2Δ yeast by up to ∼3-fold (Fig. 5B, lanes 7–12), suggesting that these plant VAP proteins can stimulate TBSV replication.
Fig. 5. Six Arabidopsis VAP proteins interact with the TBSV p33 replication protein in yeast. 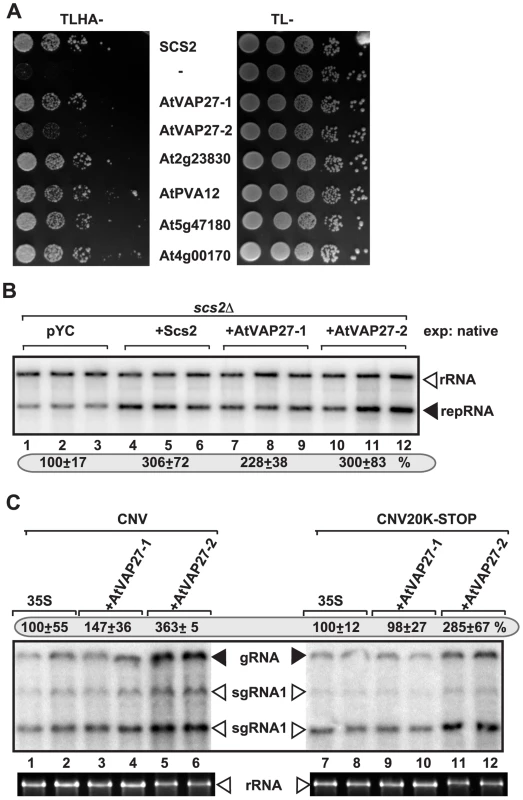
(A) The split ubiquitin assay was used to test binding between p33 and six Arabidopsis VAP proteins in scs2Δ yeast. The bait p33 was co-expressed with the shown prey proteins. SCS2 and the empty prey vector (NubG) were used as positive and negative controls, respectively. The left panel shows p33: VAP interactions, the right panel demonstrates that comparable amounts of yeasts were used for these experiments. (B) Expression of Arabidopsis VAP proteins can complement the defect in TBSV repRNA accumulation in scs2Δ yeast. Northern blot analysis of DI-72(+) repRNA accumulation in scs2Δ yeast expressing Arabidopsis VAP proteins from the native SCS2 promoter. Note that pYC expresses a short peptide and used as a negative control. (C) Stimulation of tombusvirus RNA accumulation in plants by expression of two Arabidopsis VAP proteins. Expression of the Arabidopsis VAP proteins was done in N. benthamiana leaves, which were co-infiltrated with Agrobacterium carrying a plasmid to launch CNV replication from the 35S promoter. The control samples were obtained from leaves expressing no VAP proteins (35S, lanes 1–2 and 7–8). Total RNA was extracted from leaves 3 days after agroinfiltration. The accumulation of CNV RNAs in N. benthamiana leaves was analyzed by Northern blot. The ribosomal RNA (rRNA), visualized by ethidium-bromide staining, was used as a loading control. Over-expression of AtVAP27-2, and to a lesser extent AtVAP27-1 proteins in Nicotiana benthamiana leaves also increased the accumulation of Cucumber necrosis virus (CNV) genomic RNA, which is very closely related to TBSV, by up to ∼3.5-fold (Fig. 5C, lanes 7–12). Based on these experiments, we suggest that the plant VAP proteins likely play similar roles in tombusvirus RNA replication to the yeast Scs2p VAP protein. The plant VAP proteins are important for the recruitment of plant ORP proteins to the ER membrane, suggesting similar functions for these cellular proteins to their counterparts in yeast and animals [56].
To further demonstrate the similarity between yeast and plant regarding the roles of ORPs and VAPs in TBSV replication, we used confocal laser microscopy to analyze the subcellular localization of BFP-tagged p33 and YFP-AtPVA12 in N. benthamiana leaves. Interestingly, a portion of BFP-p33 molecules is co-localized with YFP-AtPVA12 (Fig. 6A). In addition, BFP-p33 is also partially co-localized with AtOrp3A and AtPVA12 complexes (detected by BiFC, Fig. 6B). Altogether, the in planta data suggest comparable distribution and co-localization of viral p33 replication protein in relation with plant ORP and VAP proteins as observed in yeast.
Fig. 6. The tombusvirus p33 replication protein co-localizes with AtOrp3A and AtPVA12 proteins in Nicotiana benthamiana. 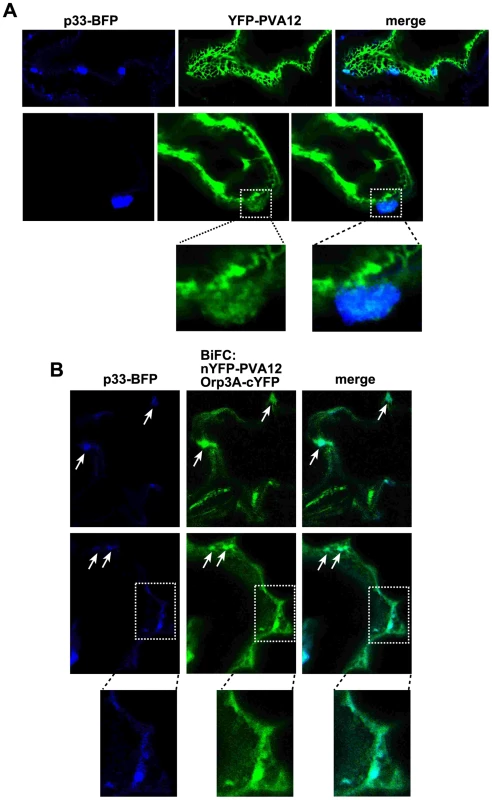
(A) Partial co-localization of TBSV BFP-tagged p33 replication protein with the YFP-AtPVA12 VAP protein in N. benthamiana cells. Expression of the above proteins from the 35S promoter was done after agro-infiltration into N. benthamiana leaves. Note the altered membrane structure in the subcellular area showing co-localization of BFP-p33 and YFP-AtPVA12 (portion of the image was enlarged at the bottom panel), which might be due to local membrane proliferation. (B) Co-localization of TBSV BFP-tagged p33 replication protein with the nYFP-AtPVA12 VAP and AtOrp3A-cYFP protein complexes in N. benthamiana cells. Expression of the above proteins from the 35S promoter was done after agro-infiltration into N. benthamiana leaves. Note that nYFP-AtPVA12 VAP and AtOrp3A-cYFP proteins were detected by BiFC. The subcellular areas (likely representing the ER membranes) where one viral and two cellular proteins are co-localized are marked with arrows. Control BiFC experiments were as in a previous paper (not shown) [56]. Formation of tombusvirus-induced spherules close to membrane contact sites in plants and yeast
By co-opting the cellular ORP and VAP proteins via interaction with the viral replication protein, tombusviruses likely induce and/or stabilize the formation of MCSs to accelerate sterol transfer from the ER to the sites of replication. To study the predicted formation of MCSs during viral replication, we used EM to visualize the tombusvirus-induced spherules (vesicle-like structures with narrow openings toward the cytosol), which represent the sites of viral RNA replication in tombusvirus-infected plants. Indeed, we frequently observed additional membranes in the close vicinity (separated by 10–30 nm distance) of the peroxisomal membranes containing the characteristic ∼50–70 nm tombusvirus-induced spherules (Fig. 7A). Higher magnification of these membranes indicates that ER sacks (with some ribosomes) are juxtaposed to the peroxisomal membrane (Fig. 7B). Additional EM images with either the genomic RNA or the DI-72 replicon RNA further supported the frequent occurrence of juxtaposed membranes, thus the formation of MCS-like structures, close to viral spherules in plants (Fig. S4). We also observed MCS-like structures in the vicinity of viral spherules in EM images of yeast cells expressing the viral replication proteins and the TBSV repRNA (Fig. 7C). Thus, these EM structures support the model that MCSs are formed between subcellular membranes in the vicinity of the tombusvirus-induced spherules in plant cells.
Fig. 7. The presence of MCS-like structures in the vicinity of tombusvirus-induced spherules in plant cells infected with CNV or in yeast cells replicating TBSV repRNA. 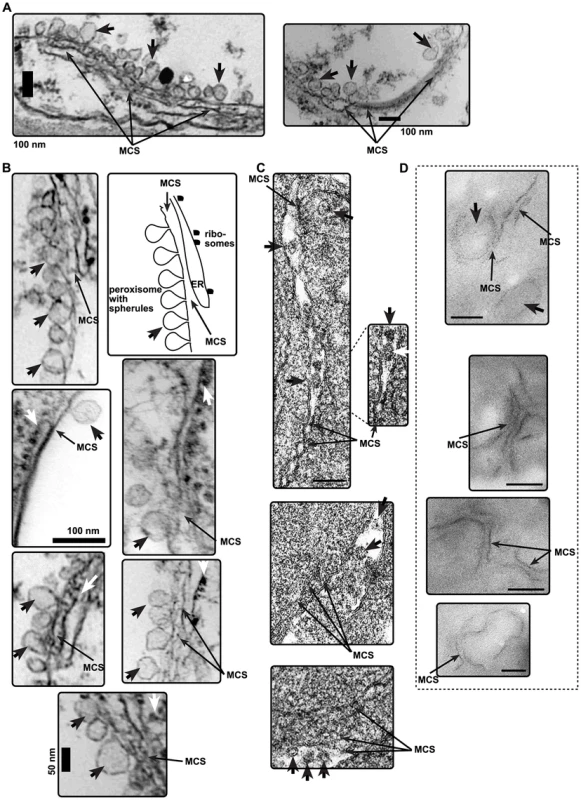
(A) Representative electron microscopic images of portions of N. benthamiana cells. Several characteristic virus-induced spherules are marked with arrowheads and the MCS-like structures are indicated by arrows. These spherules are formed via membrane invagination into peroxisome-derived membranes. (B) Close up view of spherules and MCS-like structures in plant cells infected with CNV. The ER and the associated ribosomes are indicated with white arrowheads. See further details in panel A. (C) TEM of stained ultra-thin sections in yeast cells replicating TBSV repRNA. Characteristic spherule-like vesicles are assembled in a peripheral membranous compartment (arrows) in the vicinity of MCS-like structures. (D) METTEM of ultra-thin sections of yeasts replicating TBSV repRNA. The MT-tagged p33 was visualized by ∼1 nm gold nano-clusters associated to MT. Black arrowhead points at a spherule-like structure, while black arrows show possible MCS with p33 present in opposing membranes. Bars in panels C-D represent 100 nm. To detect if p33 replication proteins are present in structures similar to MCSs, we used Metal-Tagging Transmission Electron Microscopy (METTEM) [68], [69]. The METTEM-based imaging of yeast cells expressing MT-tagged p33 revealed the presence of juxtaposed elongated p33 clusters that resembled MCSs (Fig. 7D and S5B). Distribution of MT-p33 in these METTEM images indicates that p33 is likely present both in the donor and acceptor membranes at MCSs (Fig. 7D). In several images, these MCS-like structures were in the vicinity of spherules (visible as more round-shaped structures). Gold-labeled antibody-based detection of dsRNA, which is present in the tombusvirus VRCs, revealed that the viral dsRNA is unlikely being made at the MCSs, but instead, in the VRCs that are detected as more globular structures by METTEM (Fig. S5A and C). Altogether, the obtained data with METTEM suggest that p33 replication protein could be present in both membranes (donor and acceptor) in the putative MCSs.
Discussion
Lipids and subcellular membranes are critical for (+)RNA viruses in order to assemble membrane-bound VRCs or form viral replication organelles during their replication in infected cells [5], [18], [70]. The VRC assembly depends on reshaping/deforming membranes to generate membranous structures, such as spherules and vesicles. In addition, many (+)RNA viruses also induce membrane proliferation by enhancing the synthesis of new lipids, or by redirecting lipids to the sites of viral replication [71]–[74]. Among the targeted lipids are phospholipids and sterols, which are major components of cellular membranes, affecting the size, shape, curvature and rigidity of membranes and intracellular organelles [75]. Therefore, not surprisingly, many genome-wide screens performed with (+)RNA viruses have led to the identification of a number of host genes affecting lipid biosynthesis or metabolism [8], [10], [76], [77].
In this paper, we show that members of lipid transfer proteins, namely the oxysterol-binding ORP proteins are recruited by tombusviruses to the sites of viral replication. The direct binding of Osh3/5/6/7 to the tombusvirus p33 replication protein leads to the enrichment of these ORPs in peroxisomal (or in mitochondrial in case of CIRV) and ER subcompartments. This also leads to enrichment of sterols at the site of viral replication. We propose that the subverted ORP proteins likely facilitate the re-distribution of sterols to viral sites (Fig. 8) at the expense of their natural plasma membrane localization. The enrichment of sterols in the membranes around the replicase complex likely promotes viral RNA replication as we observed more efficient TBSV repRNA replication in artificial PE vesicles containing cholesterol or ergosterol.
Fig. 8. Model on the role of ORP and VAP proteins and MCSs in tombusvirus replicase assembly. 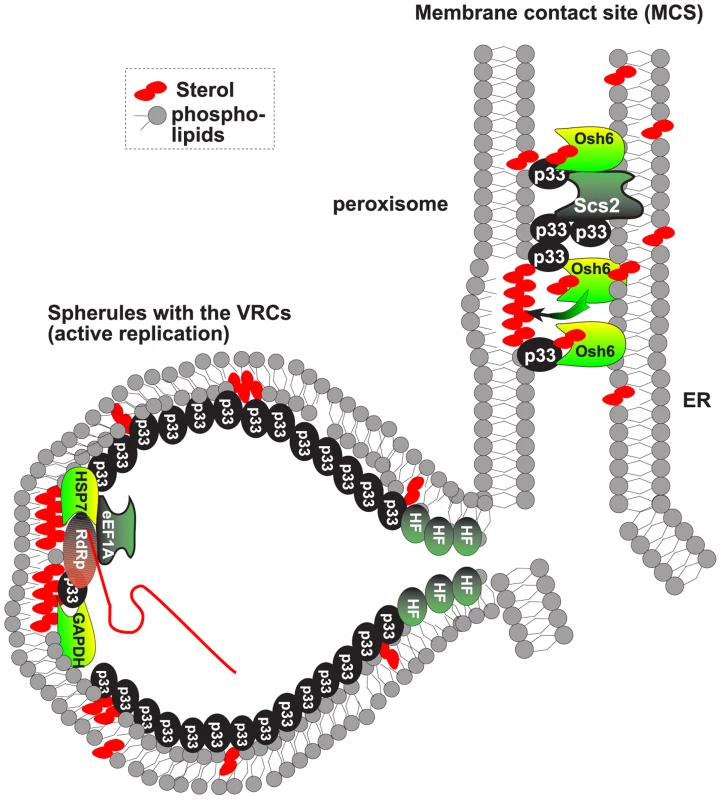
The ORP proteins, such as the yeast Osh6p, are recruited via binding to the tombusviral p33 replication protein and the ER-bound Scs2p VAP to MCS, formed between the ER and peroxisomal membranes. The ORPs then facilitate the enrichment of sterols in the peroxisomal membrane (or mitochondrial membrane in case of CIRV), forming sterol-rich microdomains needed for the formation of virus-induced spherules. These spherules contain the VRCs performing viral RNA synthesis. Note that the MCSs are not part of the VRCs based on the difference in BiFC localization of p33:Ssa1p Hsp70 complex and p33:Scs2p, which suggests that different subpopulations of p33 are involved in each interaction. We have previously shown that the Ssa1p is present in the active VRCs. The obtained data support the functional role of the cellular ORP proteins as vesicle-independent sterol transporters in tombusvirus replication. This is because combined deletion of OSH3, 5, 6 and 7 resulted in similar debilitating changes in viral replication to that observed by perturbing sterol metabolism via down regulation of ERG25 or deletion of ERG4 sterol biosynthesis genes in yeast model host [7], [47]. These changes included (i) major reduction in tombusvirus replication; (ii) instability of viral replication proteins; and (iii) the low efficiency of tombusvirus replicase assembly/activity. It is plausible that the enrichment of sterols around the viral replication proteins via recruitment of ORP proteins facilitates virus-induced membrane deformation and the generation of viral spherules that represent the sites of tombusvirus replication (Fig. 8) [12]. In the absence of ORP proteins, low concentration of sterols around the membrane-bound replication proteins could also hinder protein-protein interactions and lead to degradation of viral replication proteins not protected by the spherule-like structures.
Tombusviruses likely exploit ORP proteins at MCSs to enrich the sterols in subdomains of either peroxisomes (in case of TBSV) or mitochondria (CIRV). Indeed, the yeast Osh proteins and other eukaryotic ORPs are active in directional sterol transport when membranes of various organelles form temporal MCSs [54], [57], [58]. This model is supported by the observation that Osh6p : p33 replication protein complexes were co-localized to both peroxisomal and ER membranes, while the CIRV p36 : Osh6p complexes were localized to both mitochondrial and ER membranes (Fig. 2). Moreover, the ER-localized p33 : Scs2p complexes were observed in close vicinity of peroxisomal membranes (Fig. 4). Further evidence on the proposed role of MCSs in VRC/spherule formation during tombusvirus replication was obtained by EM images of tombusvirus-infected plant cells or yeast cells supporting TBSV replication (Fig. 7). The TBSV-induced spherules were frequently located in the vicinity of ER-like membrane sacks, suggesting the presence of MCSs at or in the vicinity of the site of tombusvirus replication.
Another supporting evidence for the role of MCSs is the interaction between the tombusvirus p33 replication protein and the yeast Scs2p VAP protein in the ER membrane. The interactions between Osh proteins and Scs2p are known to promote the formation of MCSs involving the ER membrane [54], [57], [58]. We found that the p33 replication protein interacts with both Scs2p VAP protein and ORPs (i.e., Osh3/5/6/7), indicating that tombusviruses might enhance sterol transfer to the site of replication via stabilizing MCSs to aid the formation of tombusvirus-induced spherules and VRCs (Fig. 8). Based on the observed interactions and intracellular localization data, it is likely that both peroxisome (in case of TBSV and CNV) and mitochondria-based (for CIRV) replication depends on MCSs in host cells.
ORPs and VAP sequence containing proteins similar to the yeast Scs2p also exist in plants, suggesting that comparable strategy involving cellular VAP proteins and ORPs could be used by tombusviruses in their native hosts. Accordingly, we show that the TBSV p33 replication protein interacts with six Arabidopsis ORPs and five different VAP proteins and expression of two of these VAP proteins in scs2Δ yeast increased TBSV repRNA accumulation. Moreover, over-expression of one of these Arabidopsis VAP proteins also increased tombusvirus genomic RNA accumulation in N. benthamiana leaves (Fig. 5C), indicating that the host VAP proteins are also beneficial to tombusvirus replication in plants. Moreover, partial co-localization of BFP-p33 molecules with YFP-AtPVA12 and p33 or peroxisomes (only in TBSV-infected cells) with AtOrp3A and AtPVA12 complexes (Fig. 6) suggests comparable roles of co-opted ORPs and VAPs in TBSV replication in yeast and plants.
Sterols not only affect tombusviruses, but replication of other RNA viruses, such as Dengue virus, Norwalk virus, hepatitis C virus (HCV) and picornaviruses [74], [78]–[81]. Interestingly, the HCV replicase complex is associated with cholesterol-rich lipid rafts that could promote high local concentration of viral proteins [82]. West Nile virus has been shown to induce the redistribution of cholesterol to the sites of virus replication, possibly from the plasma membrane [83]. The reduced sterol level in the plasma membrane likely inhibits antiviral responses, the entry process of animal viruses to cells, and the exit of virus particles from cells [84]–[87]. It will be interesting to see if lipid-transfer ORP proteins, VAP proteins and MCSs are involved in the infection process of other viruses and pathogens.
Summary
In this paper, we document that selected ORPs and VAP proteins are co-opted by tombusviruses. We propose that viral subversion of these host proteins leads to enrichment of sterols at the sites of tombusvirus replication likely via MCS formation. The increased amount of sterols in viral protein-containing microdomains is proposed to facilitate the formation of spherules and vesicle-like structures that are needed for tombusvirus replication.
Materials and Methods
Analysis of tombusvirus replication in yeast
Yeast strains SEY6210 and JRY6266 his3Δ (see plasmids used in Materials and Methods S1) were transformed with plasmids pESC/DI72/His33 (or pESC/DI72/His36), pGAD(Trp1)-His92-CUP1 (or pESC(Trp1)-Flag95-CUP1) and pYC2/NT-C plasmids expressing one of the His6-tagged Osh proteins. Yeast strains SEY6210, JRY6266, JRY62323 and JRY6207 were transformed with plasmids pESC(Ura3)/DI72/His33 and pGAD(Trp1)-His92-CUP1. SEY6210 and JRY6259 were transformed with pESC(Trp1)/DI72/His33 and pGAD-His92-CUP1. Transformed yeasts were pre-grown in SD minimal media supplemented with 2% glucose for 24 hours at 29°C [88]. Then, the yeast cultures were centrifuged, washed in 2% galactose minimal media and used to start cultures in 2% galactose minimal media plus 50 µM CuSO4 (starting with 0.25 OD600). Cultures were grown for 48 hours at 29°C.
Yeast strains BY4741 and scs2Δ were transformed with pGBK-His33-CUP1/DI72-GAL1, pGAD-flag92-CUP1 and pYC2/NT-C plasmids expressing one of the Scs2/VAP proteins. Transformed yeasts were grown in SD minimal media supplemented with 2% galactose for 20 hours at 29°C, then supplemented with 50 µM CuSO4 and grown for an additional 24 hours. Total RNA extractions and Northern blot analyses were performed as described previously [21], [88]. DI-72 (+)repRNA was detected with a 32p-labelled RNA probe and the relative accumulation determined using the 18s rRNA for normalization [89].
Accumulation of the viral replication proteins was determined by Western blot using anti-His or anti-Flag antibodies followed by anti-mouse conjugated to alkaline phosphatase and NBT/BCIP for detection [89].
Analysis of TBSV replication in cell-free extracts
Cell-free extracts (CFE) from yeast strains SEY6210, JRY6266 and JRY6259 were prepared as described previously [26]. MBP-tagged p33 and p92 proteins were purified from E. coli as described [90]. CFE-based reaction mixtures containing 0.1 µg of each purified protein, 0.5 µg in vitro-transcribed repRNA and CFE in a 20 µl final volume were incubated at 25°C for 3 hours. The amount of newly synthesized repRNA was analyzed in polyacrylamide/Urea gels as described [62].
Analysis of tombusvirus replication in plants
Nicotiana benthamiana leaves were co-infiltrated with Agrobacterium tumefaciens carrying the plasmid pGD-CNV or pGD-CNV20k-stop [12] and A. tumefaciens carrying pGD-L plasmids expressing A. thaliana VAP27-1 or VAP27-2 proteins. Total RNA was extracted from the infiltrated leaves 3 days after agro-infiltration and the accumulation of CNV genomic RNA was analyzed by agarose electrophoresis as described [12].
Analysis of protein interactions in vivo using yeast membrane two-hybrid assay
The yeast membrane two-hybrid assay, based on the split-ubiquitin system has been described before [35]. The plasmid pGAD-BT2-N-His33 was co-transformed with pPR-N-RE derived constructs into yeast strain NMY51 (Dualsystems). Transformed yeasts colonies were suspended in 100 µl of water and 5 µl of the suspension spread onto TL− plates, as loading control, or onto TLHA− plates to score protein interactions [35] and incubated at 30°C. Alternatively, colonies were suspended in 100 µl of water and serially diluted (by 10-fold) in water. 6 µl of each dilution were spotted onto TL− or TLHA− plates.
Co-purification of yeast Osh proteins with Flag-p33 from cellular membranes
For co-purification of chromosomally expressed 6xHA-tagged proteins with tombusvirus FLAG-p33 replication protein, yeast strains OSH6-6xHA, OSH7-6xHA and SCS2-6xHA were transformed with plasmids pGBK-His33-CUP1/DI72-GAL1 or pGBK-Flag33-CUP1/DI72-GAL1 and pGAD-His92-CUP1. For co-purification of His6-tagged proteins with CNV p33, yeast strain BY4741 was co-transformed with plasmids pESC/DI72/His33 or pESC/DI72/Flag33, pGAD-His92 and pYC2/NT-C plasmids expressing one of the His6-tagged Osh proteins. For co-purification of His6-tagged proteins with CIRV p36 replication protein, yeast BY4741 was co-transformed with plasmids pESC/DI72/His36 or pESC/DI72/Flag36, pESC(Leu2)-Flag95-CUP1 and pYC2/NT-C plasmids.
Transformed yeasts were pre-grown for 24 hours at 29°C in SD media containing 2% glucose. After centrifugation, the pre-grown yeast was used to inoculate 50 ml SD media supplemented with 2% galactose plus 50 µM CuSO4 and grown for another 24 hours at 29°C. The cultures were centrifuged, washed with phosphate buffer saline (PBS) and incubated in PBS plus 1% formaldehyde for 1 hour on ice to crosslink proteins [12]. Formaldehyde was quenched by addition of glycine (to 0.1 M) and the yeast was recovered by centrifugation. FLAG-p33 was purified from cellular membranes using anti-FLAG M2 agarose as described previously [35]. Purified FLAG-p33 was analyzed by Western blot using anti-FLAG antibody followed by anti-mouse antibody conjugated to alkaline phosphatase [35]. Co-purified His6-tagged Osh proteins were analyzed with anti-His antibody followed by anti-mouse antibody conjugated to alkaline phosphatase. Chromosomally expressed Osh6-6xHA, Osh7-6xHA, and Scs2-6xHA were analyzed with anti-HA antibody followed by anti-rabbit antibody conjugated to alkaline phosphatase [12]. Detection with NBT-BCIP was done as described [12], .
In vitro protein interaction assays
MBP-tagged TBSV p33 and its truncated derivatives were expressed in E. coli and purified in amylose columns as described [90]. Lysates of E. coli expressing recombinant GST-His6-Osh proteins were then passed through the amylose columns containing the captured MBP-p33 protein derivatives. After washing, MBP-p33 proteins were eluted with maltose. The amount of bound GST-His6-Osh proteins was analyzed by SDS-PAGE and Western blot using anti-His antibody as described [92].
Bimolecular fluorescence complementation (BiFC) assay in yeast
The ORF for the N-terminal portion of Venus fluorescent protein comprising amino acids 1 to 155 was PCR-amplified with primers #3907/#3762, digested with NcoI and BamHI and ligated into NcoI/BamHI-digested pESC/DI72/His33 [93]. The resulting plasmid, pESC-VenN-p33-DI72, expresses the N-terminal portion of Venus (aa 1–155) fused to the p33 protein. The NcoI/BamHI-digested Venus-N product was also ligated into NcoI/BamHI-digested pESC/DI72/His36 to generate pESC-VenN-p36-DI72. The sequence for the C-terminal portion of Venus (aa 155–236) was PCR-amplified with primers #3763/#3764, digested with HindIII and BamHI and ligated into HindIII/BamHI-digested pYC2/NT-C. OSH6, SCS2 and SSA1 sequences were PCR-amplified with primers #5569/#5132, #2990/#2991 and #2653/#2812, respectively, digested with BamHI and XhoI and ligated into BamHI/XhoI-digested pYC-VenC. pESC-VenN-p33-DI72 or pESC-VenN-p36-DI72 in combination with pYC-VenC-OSH6 and pGAD-pho86-CFP, pGAD-pex13-CFP or pGAD-ilv6-CFP [40] were co-transformed into Sc1 yeast strain. Transformed yeast colonies were grown in liquid SD media containing 2% galactose for 24 hours at 29°C and confocal laser microscopy was performed as previously described [40].
Co-localization and Bimolecular Fluorescence Complementation Assay in planta. pGD-35S-p33-BFP plasmid was created by PCR amplification of CNV p33 ORF with primers #424F and #3570R. Fluorescent protein TagBFP was amplified with primers #5877F and #5878R using TagBFP-AS-N plasmid (Evrogen) as a template. The PCR products were digested with NheI and XbaI respectively, ligated and a second PCR was performed with primers #424F and #5878R. The PCR product was digested with BamHI and SalI and then ligated into pGD-35S (the original was provided by M. Goodin) vector digested with the same enzymes. pGD-35S-BFPSKL plasmid was constructed by ligating the PCR product amplified with primers #5877F and #5879R into pGD-35S vector via BamHI and SalI restriction sites. The plasmids were transformed into Agrobacterium strain C58C1Rif. Plasmids, harboring YFP-PVA12, nYFP-PVA12 and ORP3a-cYFP were used as described earlier [56]. Four weeks old N. benthamiana leaves were agro-infiltrated for the transient expression of the tagged proteins. Transformed leaves were analyzed 48 h after agro-infiltration and confocal laser imaging was performed using an Olympus FV1000 microscope as described previously [94].
Filipin staining of sterols and confocal microscopy
Sc1 yeast strain was co-transformed with plasmids pYC-RFP-p33, pGAD-His92 and pGBK-DI72 or pYC-RFP-p36, pESC(Leu2)-Flag95-CUP1 and pGBK-DI72. Transformed yeasts were grown in SD minimal media supplemented with 2% galactose and plus 50 µM CuSO4 for 24 hours at 29°C. Cells were fixed and stained with filipin as described [95]. Briefly, yeast cultures were treated with formaldehyde (3%) for 15 minutes to fix cells. Then, cells were centrifuged, washed with water 2 times and finally re-suspended in 1 ml of water. 20 µl of filipin solution (5 mg/ml in ethanol) was added, followed by incubation in the dark for 15 minutes. Then 3 µl of the cell suspensions were directly spotted onto poly-lysine microscope slides and examined in a confocal microscope (405 nm laser for filipin and 543 laser for RFP) or in a UV light microscope (Zeiss) using a DAPI filter set.
Lipid vesicles preparation
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (PE, 18∶1), L-α-phosphatidylinositol-4-phosphate (PI4P, from brain), Cholesterol and Ergosterol were purchased from Avanti Polar lipids, Inc., dissolved and stored in chloroform. In each preparation, 269 nmol PE plus additional PE, PI4P or sterols were added into glass vial, mixed, and subsequently dried by a gentle stream of nitrogen for 1 hour, and further dried in speed-vacuum for 2–3 hours. 400 µl HEPES buffer (30 mM HEPES-KOH, pH 7.4) was added to each vial, and the preparations were subjected to sonication in a bath sonicator (Avanti Polar lipids, Inc.) filled with icy water for about 20 minutes, until the mixture become visually homogeneous. Lipid vesicles were used on the day of preparation for in vitro replication assay.
In vitro TBSV replication assay
The experimental procedure for in vitro replication assay using phospholipid vesicles was same as previously published procedure using purified yeast organelles [96], except that a 40,000 g supernatant (S40) from yeast cell-free extract was used, and lipid vesicles were used as membrane source instead of purified yeast organelles.
Fluorescence cholesterol labeling of yeast cells
Sterol auxotroph Saccharomyces cerevisiae erg9Δ strain [65] (MATa his3Δ, leu2Δ, met15Δ, ura3Δ, erg9Δ::hphNT1 derivative, kindly provided by Dr. Joe Chappell, UKY, USA) was transformed with plasmids expressing mRFP tagged CNV or CIRV replication proteins p33 or p36 [96]. Transformed yeast cells were pre-cultured overnight in synthetic complete dropout (SC) media containing 20 mg/L cholesterol, and diluted into SC media with galactose supplemented with 10 mg/L BODIPY-cholesterol (TopFluor Cholesterol, Avanti polar lipids, Inc.) at 0.5 OD600. The yeasts were incubated at 23°C for 16 hours and then subjected to confocal laser microscope analysis [40].
Electron microscopy
Methods have been previously described in details [12], [15]. Briefly, N. benthamiana leaves were agro-infiltrated with a construct expressing CNV 20 k stop or a combination of A. tumefaciens cultures expressing p33, p92, p19 proteins (viral suppressor of gene silencing), and DI-72 RNA. 2.5 days after agro-infiltration, leaf samples were incubated with a fixing buffer containing 0.1 M KPO4, pH 6.8, 3.5% glutaraldehyde and 1% paraformaldehyde. The leaves were injected with the fixing buffer using a syringe (without needle) and subsequently sectioned into 1×5 mm strips. The leaf sections were immersed in the fixing buffer and incubated overnight at 4°C. Leaf sections were washed three times for 10 minutes in 0.1 M KPO4 pH 6.8, plus 5% glucose, then treated with 1% OsO4 for 2 hours at room temperature (RT). Sections were washed in distilled water for 5 min and dehydrated sequentially in 50%, 70%, 80% and 90% ethanol for 10 min each at RT, followed by two incubations with 100% ethanol for 20 min and two with propylene oxide (PO) for 15 min. Samples were gradually infiltrated in 50/50 epon-araldite resin/PO overnight, 75/25 resin/PO for 4 hours and then 100% resin for 4 hours under vacuum. Samples were finally embedded in pure resin and incubated for 48 hours at 60°C for resin polymerization. After sectioning and mounting in copper grids, samples were stained with uranyl-acetate and lead-citrate and imaged in a Philips Biotwin12 transmision electron microscope. The images were cropped using Photoshop software.
For electron microscopy of yeast cells, the yeast BY4741 strain was pre-cultured from plated single colonies by inoculation in 2 ml of YPG (yeast extract peptone galactose). The culture was then incubated at 30°C and shaken overnight at 250 rpm. For inducing and maintaining viral replication, yeasts cells were grown in YPG at 23°C and shaken at 250 rpm for 24 h. When OD600 was around 2, cells were centrifuged for 5 min at 4000× g, re-suspended in TSD reduction buffer (Tris-sulfate DTT, pH 9.4) and treated for removing the cell wall to obtain spheroplasts. Compared to whole yeast cells, spheroplasts are well infiltrated with fixatives and resins allowing an optimal preservation and visualization of cell ultrastructure. For obtaining spheroplasts, yeast cells were maintained for 10 min at room temperature (RT) and then treated at 30°C with 0.1 µg/µl zymolyase 20T (AMS Biotechnology) in spheroplast medium A (1× yeast nitrogen base, 2% (w/v) glucose, 1× amino acids, 1 M sorbitol, 20 mM TrisCl, pH 7.5) for 5 or 15 min, depending on the yeast strain. After zymolyase treatment, cells were centrifuged for 5 min, at 1000 g and 23°C and washed once with spheroplast medium B (1× yeast nitrogen base, 2% (w/v) glucose, 1× amino acids, 1 M sorbitol) and twice with spheroplast medium A. Cells were chemically fixed in two steps: 1) 20 min in suspension at RT with 8% paraformaldehyde and 1% glutaraldehyde and 2) 1 h at RT with 4% paraformaldehyde and 0.5% glutaraldehyde in HEPES (pH 7.4); fixed cells were then processed by conventional embedding in the epoxy-resin EML-812 (Taab Laboratories). The embedding protocol, designed to provide an optimal preservation of cell membranes, was previously described in detail [97]–[99]. In brief, cells were post-fixed for 1 h at 4°C with 1% osmium tetroxide and 0.8% potassium ferricyanide in water, washed with HEPES, and incubated 40 min with 2% uranyl acetate at 4°C. Dehydration was done at 4°C with increasing concentrations of acetone (50, 70, 90, and twice in 100%, 10 min each). Samples were then incubated at room temperature with a mixture of acetone and resin (1∶1). Cells were infiltrated with 100% resin for 1 day and polymerized at 60°C for 3 days. Ultrathin (50–70 nm) sections were collected in formvar-coated 300 mesh cooper grids (G300-C3, Taab). Sections on grids were stained with saturated uranyl acetate and lead citrate following standard procedures and studied in a Jeol JEM 1011 transmission electron microscope operating at 100 kv.
For visualization of MT-tagged p33, we processed the yeast strain DB-614 (BY4741 ADH1p-His-p92::kanMX4, GAL1p-His-MT-p33/GAL10p-DI72::hphNT1) expressing His6-p92 under control of the ADH1 promoter and ADH1 terminator [100], and the metallothionein 1 (MT-1)-tagged p33 (His6-MT-p33) under control of GAL1 promoter and CYC1 terminator, and DI-72 under control of GAL10 promoter and ADH1 terminator [15]. For building gold nano-clusters in MT-p33 molecules, these yeast cells were incubated with gold salts and processed for embedding in the acrylic resin LRWhite [15]. The embedding protocol allowed an adequate preservation of protein epitopes and the optimal visualization of the small gold nano-clusters [68]. Spheroplasts were incubated for 75 min with 0.2 mM HAuCl4 (SIGMA-ALDRICH) in spheroplast medium A. This treatment builds gold nano-clusters in MT-tagged proteins allowing detection of protein molecules in cells with high sensitivity and at molecular scale resolution [68], [69]. Cells were washed with spheroplast medium A and fixed 1 h at RT with 4% paraformaldehyde and 0.2% glutaraldehyde in PHEM (20 mM PIPES, 50 mM HEPES, 20 mM EGTA and 4 mM MgCl2, pH 6.9). After short dehydration steps of 10 min each in increasing concentrations of ethanol (30, 50, 70, 90 and twice with100%) at 4°C, spheroplasts were incubated in mixtures of ethanol and LR White (2∶1, 2∶2, 1∶2) and in 100% resin for 24 h. For resin polymerization samples were maintained at 60°C for 48 h. Ultra-thin sections were collected in 300 mesh Quantifoil holey carbon grids (R 3.5/1 Cu/Rh, Quantifoil Micro Tools) and studied without staining. For immunogold labeling of dsRNA, sections were incubated for 6 min with 1% bovine serum albumin (BSA) in PBS, with an anti-dsRNA primary antibody diluted 1∶200 in 1% BSA and with secondary antibodies conjugated with 5 or 10 nm colloidal gold particles and diluted 1∶40 in 1% BSA. The mouse anti-dsRNA MAb J2 was from English & Scientific Consulting and the secondary antibodies conjugated with colloidal gold particles were from BB International.
Supporting Information
Zdroje
1. NagyPD (2008) Yeast as a model host to explore plant virus-host interactions. Annu Rev Phytopathol 46 : 217–242.
2. HuangYW, HuCC, LinNS, HsuYH (2012) Unusual roles of host metabolic enzymes and housekeeping proteins in plant virus replication. Curr Opin Virol 2 : 676–682.
3. ShullaA, RandallG (2012) Hepatitis C virus-host interactions, replication, and viral assembly. Curr Opin Virol 2 : 725–732.
4. MineA, OkunoT (2012) Composition of plant virus RNA replicase complexes. Curr Opin Virol 2 : 669–675.
5. BelovGA, van KuppeveldFJ (2012) (+)RNA viruses rewire cellular pathways to build replication organelles. Curr Opin Virol 2 : 740–747.
6. NagyPD, PoganyJ (2012) The dependence of viral RNA replication on co-opted host factors. Nature Reviews Microbiology 10 : 137–149.
7. PanavasT, ServieneE, BrasherJ, NagyPD (2005) Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc Natl Acad Sci U S A 102 : 7326–7331.
8. CherryS, DoukasT, ArmknechtS, WhelanS, WangH, et al. (2005) Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev 19 : 445–452.
9. JiangY, ServieneE, GalJ, PanavasT, NagyPD (2006) Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J Virol 80 : 7394–7404.
10. KushnerDB, LindenbachBD, GrdzelishviliVZ, NoueiryAO, PaulSM, et al. (2003) Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A 100 : 15764–15769.
11. KrishnanMN, NgA, SukumaranB, GilfoyFD, UchilPD, et al. (2008) RNA interference screen for human genes associated with West Nile virus infection. Nature 455 doi:10.1038/nature07207
12. BarajasD, JiangY, NagyPD (2009) A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog 5: e1000705.
13. DiazA, WangX, AhlquistP (2010) Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc Natl Acad Sci U S A 107 : 16291–16296.
14. NeuvonenM, KazlauskasA, MartikainenM, HinkkanenA, AholaT, et al. (2011) SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog 7: e1002383.
15. BarajasD, MartinIF, PoganyJ, RiscoC, NagyPD (2014) Noncanonical Role for the Host Vps4 AAA+ ATPase ESCRT Protein in the Formation of Tomato Bushy Stunt Virus Replicase. PLoS Pathog 10: e1004087.
16. de CastroIF, VolonteL, RiscoC (2013) Virus factories: biogenesis and structural design. Cell Microbiol 15 : 24–34.
17. SchaeferEA, ChungRT (2013) HCV and host lipids: an intimate connection. Semin Liver Dis 33 : 358–368.
18. den BoonJA, DiazA, AhlquistP (2010) Cytoplasmic viral replication complexes. Cell Host Microbe 8 : 77–85.
19. PaulD, BartenschlagerR (2013) Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2 : 32–48.
20. NagyPD, PoganyJ (2006) Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 344 : 211–220.
21. PanavasT, NagyPD (2003) Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314 : 315–325.
22. NagyPD, PoganyJ, LinJY (2014) How yeast can be used as a genetic platform to explore virus-host interactions: from ‘omics’ to functional studies. Trends Microbiol 22 : 309–16 doi: 10.1016/j.tim.2014.02.003
23. PohMK, ShuiG, XieX, ShiPY, WenkMR, et al. (2012) U18666A, an intra-cellular cholesterol transport inhibitor, inhibits dengue virus entry and replication. Antiviral Res 93 : 191–198.
24. HwangYT, McCartneyAW, GiddaSK, MullenRT (2008) Localization of the Carnation Italian ringspot virus replication protein p36 to the mitochondrial outer membrane is mediated by an internal targeting signal and the TOM complex. BMC Cell Biol 9 : 54.
25. Weber-LotfiF, DietrichA, RussoM, RubinoL (2002) Mitochondrial targeting and membrane anchoring of a viral replicase in plant and yeast cells. J Virol 76 : 10485–10496.
26. PoganyJ, StorkJ, LiZ, NagyPD (2008) In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc Natl Acad Sci U S A 105 : 19956–19961.
27. Shah Nawaz-Ul-RehmanM, Reddisiva PrasanthK, BakerJ, NagyPD (2013) Yeast screens for host factors in positive-strand RNA virus replication based on a library of temperature-sensitive mutants. Methods 59 : 207–216.
28. ServieneE, ShapkaN, ChengCP, PanavasT, PhuangratB, et al. (2005) Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci U S A 102 : 10545–10550.
29. ServieneE, JiangY, ChengCP, BakerJ, NagyPD (2006) Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J Virol 80 : 1231–1241.
30. Shah Nawaz-Ul-RehmanM, Martinez-OchoaN, PascalH, SasvariZ, HerbstC, et al. (2012) Proteome-wide overexpression of host proteins for identification of factors affecting tombusvirus RNA replication: an inhibitory role of protein kinase C. J Virol 86 : 9384–9395.
31. NagyPD (2011) The roles of host factors in tombusvirus RNA recombination. Adv Virus Res 81 : 63–84.
32. NagyPD, PoganyJ (2010) Global genomics and proteomics approaches to identify host factors as targets to induce resistance against Tomato bushy stunt virus. Adv Virus Res 76 : 123–177.
33. MenduV, ChiuM, BarajasD, LiZ, NagyPD (2010) Cpr1 cyclophilin and Ess1 parvulin prolyl isomerases interact with the tombusvirus replication protein and inhibit viral replication in yeast model host. Virology 406 : 342–351.
34. LiZ, PoganyJ, PanavasT, XuK, EspositoAM, et al. (2009) Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 385 : 245–260.
35. LiZ, BarajasD, PanavasT, HerbstDA, NagyPD (2008) Cdc34p Ubiquitin-Conjugating Enzyme Is a Component of the Tombusvirus Replicase Complex and Ubiquitinates p33 Replication Protein. J Virol 82 : 6911–6926.
36. ServaS, NagyPD (2006) Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J Virol 80 : 2162–2169.
37. LiZ, NagyPD (2011) Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol 8 : 305–315.
38. LiZ, PoganyJ, TupmanS, EspositoAM, KinzyTG, et al. (2010) Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog 6: e1001175.
39. WangRY, StorkJ, NagyPD (2009) A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J Virol 83 : 3276–3287.
40. JonczykM, PathakKB, SharmaM, NagyPD (2007) Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362 : 320–330.
41. PanavasT, HawkinsCM, PanavieneZ, NagyPD (2005) The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 338 : 81–95.
42. NagyPD, BarajasD, PoganyJ (2012) Host factors with regulatory roles in tombusvirus replication. Curr Opin Virol 2 : 685–692.
43. McCartneyAW, GreenwoodJS, FabianMR, WhiteKA, MullenRT (2005) Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17 : 3513–3531.
44. KovalevN, NagyPD (2014) The Expanding Functions of Cellular Helicases: The Tombusvirus RNA Replication Enhancer Co-opts the Plant eIF4AIII-Like AtRH2 and the DDX5-Like AtRH5 DEAD-Box RNA Helicases to Promote Viral Asymmetric RNA Replication. PLoS Pathog 10: e1004051.
45. NagyPD, PoganyJ, LinJY (2014) How yeast can be used as a genetic platform to explore virus-host interactions: from ‘omics’ to functional studies. Trends Microbiol 22 : 309–316.
46. SharmaM, SasvariZ, NagyPD (2011) Inhibition of phospholipid biosynthesis decreases the activity of the tombusvirus replicase and alters the subcellular localization of replication proteins. Virology 415 : 141–152.
47. SharmaM, SasvariZ, NagyPD (2010) Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J Virol 84 : 2270–2281.
48. BlochK (1992) Sterol molecule: structure, biosynthesis, and function. Steroids 57 : 378–383.
49. BlochKE (1983) Sterol structure and membrane function. CRC Crit Rev Biochem 14 : 47–92.
50. SchekmanR (2007) How sterols regulate protein sorting and traffic. Proc Natl Acad Sci U S A 104 : 6496–6497.
51. RocheY, Gerbeau-PissotP, BuhotB, ThomasD, BonneauL, et al. (2008) Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. Faseb J 22 : 3980–3991.
52. SchulzTA, ChoiMG, RaychaudhuriS, MearsJA, GhirlandoR, et al. (2009) Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol 187 : 889–903.
53. RaychaudhuriS, PrinzWA (2010) The diverse functions of oxysterol-binding proteins. Annu Rev Cell Dev Biol 26 : 157–177.
54. Weber-BoyvatM, ZhongW, YanD, OlkkonenVM (2013) Oxysterol-binding proteins: functions in cell regulation beyond lipid metabolism. Biochem Pharmacol 86 : 89–95.
55. RidgwayND (2010) Oxysterol-binding proteins. Subcell Biochem 51 : 159–182.
56. SaravananRS, SlabaughE, SinghVR, LapidusLJ, HaasT, et al. (2009) The targeting of the oxysterol-binding protein ORP3a to the endoplasmic reticulum relies on the plant VAP33 homolog PVA12. Plant J 58 : 817–830.
57. MesminB, BigayJ, Moser von FilseckJ, Lacas-GervaisS, DrinG, et al. (2013) A Four-Step Cycle Driven by PI(4)P Hydrolysis Directs Sterol/PI(4)P Exchange by the ER-Golgi Tether OSBP. Cell 155 : 830–843.
58. StefanCJ, ManfordAG, BairdD, Yamada-HanffJ, MaoY, et al. (2011) Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144 : 389–401.
59. LevS, Ben HalevyD, PerettiD, DahanN (2008) The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol 18 : 282–290.
60. UmateP (2011) Oxysterol binding proteins (OSBPs) and their encoding genes in Arabidopsis and rice. Steroids 76 : 524–529.
61. BehCT, CoolL, PhillipsJ, RineJ (2001) Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics 157 : 1117–1140.
62. PoganyJ, NagyPD (2008) Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J Virol 82 : 5967–5980.
63. BehCT, McMasterCR, KozminskiKG, MenonAK (2012) A detour for yeast oxysterol binding proteins. J Biol Chem 287 : 11481–11488.
64. VillasmilML, BankaitisVA, MousleyCJ (2012) The oxysterol-binding protein superfamily: new concepts and old proteins. Biochem Soc Trans 40 : 469–473.
65. TakahashiS, YeoY, GreenhagenBT, McMullinT, SongL, et al. (2007) Metabolic engineering of sesquiterpene metabolism in yeast. Biotechnol Bioeng 97 : 170–181.
66. CaretteJE, VerverJ, MartensJ, van KampenT, WellinkJ, et al. (2002) Characterization of plant proteins that interact with cowpea mosaic virus ‘60K’ protein in the yeast two-hybrid system. J Gen Virol 83 : 885–893.
67. PetersenNH, JoensenJ, McKinneyLV, BrodersenP, PetersenM, et al. (2009) Identification of proteins interacting with Arabidopsis ACD11. J Plant Physiol 166 : 661–666.
68. RiscoC, Sanmartin-ConesaE, TzengWP, FreyTK, SeyboldV, et al. (2012) Specific, sensitive, high-resolution detection of protein molecules in eukaryotic cells using metal-tagging transmission electron microscopy. Structure 20 : 759–766.
69. DiestraE, FontanaJ, GuichardP, MarcoS, RiscoC (2009) Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol 165 : 157–168.
70. MillerS, Krijnse-LockerJ (2008) Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6 : 363–374.
71. HeatonNS, RandallG (2010) Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8 : 422–432.
72. HsuNY, IlnytskaO, BelovG, SantianaM, ChenYH, et al. (2010) Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141 : 799–811.
73. ReissS, RebhanI, BackesP, Romero-BreyI, ErfleH, et al. (2011) Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9 : 32–45.
74. IlnytskaO, SantianaM, HsuNY, DuWL, ChenYH, et al. (2013) Enteroviruses harness the cellular endocytic machinery to remodel the host cell cholesterol landscape for effective viral replication. Cell Host Microbe 14 : 281–293.
75. NohturfftA, ZhangSC (2009) Coordination of lipid metabolism in membrane biogenesis. Annu Rev Cell Dev Biol 25 : 539–566.
76. KrishnanMN, NgA, SukumaranB, GilfoyFD, UchilPD, et al. (2008) RNA interference screen for human genes associated with West Nile virus infection. Nature 455 : 242–245.
77. LiQ, BrassAL, NgA, HuZ, XavierRJ, et al. (2009) A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A 106 : 16410–16415.
78. ChangKO (2009) Role of cholesterol pathways in norovirus replication. J Virol 83 : 8587–8595.
79. RothwellC, LebretonA, Young NgC, LimJY, LiuW, et al. (2009) Cholesterol biosynthesis modulation regulates dengue viral replication. Virology 389 : 8–19.
80. SaganSM, RouleauY, LeggiadroC, SupekovaL, SchultzPG, et al. (2006) The influence of cholesterol and lipid metabolism on host cell structure and hepatitis C virus replication. Biochem Cell Biol 84 : 67–79.
81. KapadiaSB, ChisariFV (2005) Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci U S A 102 : 2561–2566.
82. AizakiH, LeeKJ, SungVM, IshikoH, LaiMM (2004) Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324 : 450–461.
83. MackenzieJM, KhromykhAA, PartonRG (2007) Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2 : 229–239.
84. LeeCJ, LinHR, LiaoCL, LinYL (2008) Cholesterol effectively blocks entry of flavivirus. J Virol 82 : 6470–6480.
85. MarquardtMT, PhalenT, KielianM (1993) Cholesterol is required in the exit pathway of Semliki Forest virus. J Cell Biol 123 : 57–65.
86. SimonM, JohanssonC, MirazimiA (2009) Crimean-Congo hemorrhagic fever virus entry and replication is clathrin-, pH - and cholesterol-dependent. J Gen Virol 90 : 210–215.
87. Amini-Bavil-OlyaeeS, ChoiYJ, LeeJH, ShiM, HuangIC, et al. (2013) The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 13 : 452–464.
88. PoganyJ, PanavasT, ServieneE, Nawaz-Ul-RehmanMS, NagyPD (2010) A high-throughput approach for studying virus replication in yeast. Current Protocols in Microbiology 19 : 16J.11.11–16J.11.15.
89. PanavasT, ServieneE, PoganyJ, NagyPD (2008) Genome-wide screens for identification of host factors in viral replication. Methods Mol Biol 451 : 615–624.
90. RajendranKS, NagyPD (2003) Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J Virol 77 : 9244–9258.
91. PanavieneZ, PanavasT, ServaS, NagyPD (2004) Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J Virol 78 : 8254–8263.
92. BarajasD, LiZ, NagyPD (2009) The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J Virol 83 : 11751–11764.
93. PathakKB, SasvariZ, NagyPD (2008) The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology 379 : 294–305.
94. WangRY, NagyPD (2008) Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3 : 178–187.
95. BehCT, RineJ (2004) A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J Cell Sci 117 : 2983–2996.
96. XuK, HuangTS, NagyPD (2012) Authentic in vitro replication of two tombusviruses in isolated mitochondrial and endoplasmic reticulum membranes. J Virol 86 : 12779–12794.
97. FontanaJ, Lopez-MonteroN, ElliottRM, FernandezJJ, RiscoC (2008) The unique architecture of Bunyamwera virus factories around the Golgi complex. Cell Microbiol 10 : 2012–2028.
98. FontanaJ, TzengWP, CalderitaG, Fraile-RamosA, FreyTK, et al. (2007) Novel replication complex architecture in rubella replicon-transfected cells. Cell Microbiol 9 : 875–890.
99. RiscoC, RodriguezJR, Lopez-IglesiasC, CarrascosaJL, EstebanM, et al. (2002) Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J Virol 76 : 1839–1855.
100. KovalevN, PoganyJ, NagyPD (2012) A Co-Opted DEAD-Box RNA Helicase Enhances Tombusvirus Plus-Strand Synthesis. PLoS Pathog 8: e1002537.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance SystemČlánek Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer CellsČlánek APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse ModelČlánek Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 10- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Theory and Empiricism in Virulence Evolution
- -Related Fungi and Reptiles: A Fatal Attraction
- Adaptive Prediction As a Strategy in Microbial Infections
- Antimicrobials, Stress and Mutagenesis
- A Novel Function of Human Pumilio Proteins in Cytoplasmic Sensing of Viral Infection
- Social Motility of African Trypanosomes Is a Property of a Distinct Life-Cycle Stage That Occurs Early in Tsetse Fly Transmission
- Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer
- Identification of the Microsporidian as a New Target of the IFNγ-Inducible IRG Resistance System
- mRNA Structural Constraints on EBNA1 Synthesis Impact on Antigen Presentation and Early Priming of CD8 T Cells
- Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages
- Neutrophil Crawling in Capillaries; A Novel Immune Response to
- Live Attenuated Vaccine Protects against Pulmonary Challenge in Rats and Non-human Primates
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
- HIV Acquisition Is Associated with Increased Antimicrobial Peptides and Reduced HIV Neutralizing IgA in the Foreskin Prepuce of Uncircumcised Men
- Uses a Unique Ligand-Binding Mode for Trapping Opines and Acquiring A Competitive Advantage in the Niche Construction on Plant Host
- Involvement of a 1-Cys Peroxiredoxin in Bacterial Virulence
- Ethanol Stimulates WspR-Controlled Biofilm Formation as Part of a Cyclic Relationship Involving Phenazines
- Densovirus Is a Mutualistic Symbiont of a Global Crop Pest () and Protects against a Baculovirus and Bt Biopesticide
- Insights into Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria
- Mycobacterial Antigen Driven Activation of CD14CD16 Monocytes Is a Predictor of Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome
- Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion
- Dampens the DNA Damage Response
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- Vaginal Challenge with an SIV-Based Dual Reporter System Reveals That Infection Can Occur throughout the Upper and Lower Female Reproductive Tract
- Detecting Differential Transmissibilities That Affect the Size of Self-Limited Outbreaks
- One Small Step for a Yeast - Microevolution within Macrophages Renders Hypervirulent Due to a Single Point Mutation
- Expression Profiling during Arabidopsis/Downy Mildew Interaction Reveals a Highly-Expressed Effector That Attenuates Responses to Salicylic Acid
- Human Cytomegalovirus Drives Epigenetic Imprinting of the Locus in NKG2C Natural Killer Cells
- Interaction with Tsg101 Is Necessary for the Efficient Transport and Release of Nucleocapsids in Marburg Virus-Infected Cells
- The N-Terminus of Murine Leukaemia Virus p12 Protein Is Required for Mature Core Stability
- Sterol Biosynthesis Is Required for Heat Resistance but Not Extracellular Survival in
- Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility
- Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly
- APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse Model
- Structural Basis for the Recognition of Human Cytomegalovirus Glycoprotein B by a Neutralizing Human Antibody
- Systematic Analysis of ZnCys Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus
- Epstein-Barr Virus Nuclear Antigen 3A Promotes Cellular Proliferation by Repression of the Cyclin-Dependent Kinase Inhibitor p21WAF1/CIP1
- The Host Protein Calprotectin Modulates the Type IV Secretion System via Zinc Sequestration
- Cyclophilin A Associates with Enterovirus-71 Virus Capsid and Plays an Essential Role in Viral Infection as an Uncoating Regulator
- A Novel Alpha Kinase EhAK1 Phosphorylates Actin and Regulates Phagocytosis in
- The pH-Responsive PacC Transcription Factor of Governs Epithelial Entry and Tissue Invasion during Pulmonary Aspergillosis
- Sensing of Immature Particles Produced by Dengue Virus Infected Cells Induces an Antiviral Response by Plasmacytoid Dendritic Cells
- Co-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites
- Characteristics of Memory B Cells Elicited by a Highly Efficacious HPV Vaccine in Subjects with No Pre-existing Immunity
- HPV16-E7 Expression in Squamous Epithelium Creates a Local Immune Suppressive Environment via CCL2- and CCL5- Mediated Recruitment of Mast Cells
- Dengue Viruses Are Enhanced by Distinct Populations of Serotype Cross-Reactive Antibodies in Human Immune Sera
- CD4 Depletion in SIV-Infected Macaques Results in Macrophage and Microglia Infection with Rapid Turnover of Infected Cells
- A Sialic Acid Binding Site in a Human Picornavirus
- Contact Heterogeneity, Rather Than Transmission Efficiency, Limits the Emergence and Spread of Canine Influenza Virus
- Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of
- HTLV-1 Tax Stabilizes MCL-1 via TRAF6-Dependent K63-Linked Polyubiquitination to Promote Cell Survival and Transformation
- Species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence
- A Critical Role for IL-17RB Signaling in HTLV-1 Tax-Induced NF-κB Activation and T-Cell Transformation
- Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90
- Role of Non-conventional T Lymphocytes in Respiratory Infections: The Case of the Pneumococcus
- Kaposi's Sarcoma-Associated Herpesvirus Induces Nrf2 during Infection of Endothelial Cells to Create a Microenvironment Conducive to Infection
- A Relay Network of Extracellular Heme-Binding Proteins Drives Iron Acquisition from Hemoglobin
- Glutamate Secretion and Metabotropic Glutamate Receptor 1 Expression during Kaposi's Sarcoma-Associated Herpesvirus Infection Promotes Cell Proliferation
- Reduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and Anti-pathogen Activities
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence
- MicroRNAs Suppress NB Domain Genes in Tomato That Confer Resistance to
- The ESAT-6 Protein of Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage
- Characterization of Uncultivable Bat Influenza Virus Using a Replicative Synthetic Virus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



