-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaViral Infection Induces Expression of Novel Phased MicroRNAs from Conserved Cellular MicroRNA Precursors
RNA silencing, mediated by small RNAs including microRNAs (miRNAs) and small interfering RNAs (siRNAs), is a potent antiviral or antibacterial mechanism, besides regulating normal cellular gene expression critical for development and physiology. To gain insights into host small RNA metabolism under infections by different viruses, we used Solexa/Illumina deep sequencing to characterize the small RNA profiles of rice plants infected by two distinct viruses, Rice dwarf virus (RDV, dsRNA virus) and Rice stripe virus (RSV, a negative sense and ambisense RNA virus), respectively, as compared with those from non-infected plants. Our analyses showed that RSV infection enhanced the accumulation of some rice miRNA*s, but not their corresponding miRNAs, as well as accumulation of phased siRNAs from a particular precursor. Furthermore, RSV infection also induced the expression of novel miRNAs in a phased pattern from several conserved miRNA precursors. In comparison, no such changes in host small RNA expression was observed in RDV-infected rice plants. Significantly RSV infection elevated the expression levels of selective OsDCLs and OsAGOs, whereas RDV infection only affected the expression of certain OsRDRs. Our results provide a comparative analysis, via deep sequencing, of changes in the small RNA profiles and in the genes of RNA silencing machinery induced by different viruses in a natural and economically important crop host plant. They uncover new mechanisms and complexity of virus-host interactions that may have important implications for further studies on the evolution of cellular small RNA biogenesis that impact pathogen infection, pathogenesis, as well as organismal development.
Published in the journal: . PLoS Pathog 7(8): e32767. doi:10.1371/journal.ppat.1002176
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002176Summary
RNA silencing, mediated by small RNAs including microRNAs (miRNAs) and small interfering RNAs (siRNAs), is a potent antiviral or antibacterial mechanism, besides regulating normal cellular gene expression critical for development and physiology. To gain insights into host small RNA metabolism under infections by different viruses, we used Solexa/Illumina deep sequencing to characterize the small RNA profiles of rice plants infected by two distinct viruses, Rice dwarf virus (RDV, dsRNA virus) and Rice stripe virus (RSV, a negative sense and ambisense RNA virus), respectively, as compared with those from non-infected plants. Our analyses showed that RSV infection enhanced the accumulation of some rice miRNA*s, but not their corresponding miRNAs, as well as accumulation of phased siRNAs from a particular precursor. Furthermore, RSV infection also induced the expression of novel miRNAs in a phased pattern from several conserved miRNA precursors. In comparison, no such changes in host small RNA expression was observed in RDV-infected rice plants. Significantly RSV infection elevated the expression levels of selective OsDCLs and OsAGOs, whereas RDV infection only affected the expression of certain OsRDRs. Our results provide a comparative analysis, via deep sequencing, of changes in the small RNA profiles and in the genes of RNA silencing machinery induced by different viruses in a natural and economically important crop host plant. They uncover new mechanisms and complexity of virus-host interactions that may have important implications for further studies on the evolution of cellular small RNA biogenesis that impact pathogen infection, pathogenesis, as well as organismal development.
Introduction
RNA-mediated gene silencing is a widespread mechanism of host defense against viral [1]–[6] and bacterial [7] infections. The 21–24 nt small RNAs, produced from DICER processing of double-stranded RNAs (dsRNAs) or RNA transcripts with stem-loop structures, are broadly defined as small interfering RNAs (siRNAs) and microRNAs (miRNAs), respectively [8]–[10]. They are incorporated into ARGONAUTES (AGOs) to form an RNA-INDUCED SILENCING COMPLEX (RISC). The RISC then recognizes its target RNA/DNA sequences through specific base pairing, to activate RNA cleavage or translation repression or DNA methylation [9], [11]–[14]. In plants, the miRNA precursors are processed into miRNA/miRNA* duplexes mostly by DICER-LIKE 1 (DCL1) with 2-nt 3′overhangs [9], [15], [16]. After methylation at the 3′ end, the miRNA sequences are preferentially incorporated into RISC to regulate gene expression, whereas the miRNA* sequences are usually degraded [17], [18].
Viruses encode dedicated proteins that function as viral suppressors of RNA silencing (VSRs), or other multi-functional proteins, to defend against host RNA silencing by interfering with distinct steps of the silencing pathways [19]. Previous studies, based on RNA gel blots, showed that transgenic expression of VSRs from many different plant viruses often caused reduced accumulation of conserved miRNAs [20]–[26]. The Tobacco mosaic virus movement and coat protein interactions also alter accumulation of tobacco miRNAs [27]. The biological function of this down-regulated miRNA accumulation for viral infection or plant defense remains to be understood.
The siRNAs are produced via processing of dsRNAs derived from distinct sources and are classified into three types: trans-acting siRNAs (ta-siRNAs), natural antisense transcript-derived siRNAs (nat-siRNAs) and repeat-associated siRNAs (ra-siRNAs). The ta-siRNAs are generated in a phased pattern through DCL4-processing of dsRNA substrates formed via the activity of RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) [28]–[31]. The nat-sRNAs are produced from dsRNAs formed by natural antisense cis-transcript pairs [32], [33]. The ra-siRNAs are derived from transposons and repetitive elements [34], [35]. Transgenic expression of VSRs from some plant viruses also often led to reduced accumulation of siRNAs, likely as a means of dampening host silencing against viral infection [20], [23], [24].
There is evidence that some cellular miRNAs play anti-viral roles against animal viruses, although a particular miRNA is exploited to support viral infection [4], [5]. Many animal viruses cause generally down-regulated expression of host miRNAs as shown by microarrays [36]–[39] and quantitative real-time PCR [40]. Deep sequencing also revealed a similar pattern, and additionally identified a few new miRNAs induced only in virus-infected cells [41]–[43]. Some new miRNAs are induced in an organ-specific manner [41].
To gain further insights into viral interactions with the host RNA silencing pathways, we used deep sequencing to characterize the small RNA profiles of rice plants infected by two different RNA viruses together with microarray and quantitative RT-PCR analyses of the expression patterns of RNA silencing pathway genes. Rice is one of the most important crop plants and emerging models for RNA silencing studies [44]–[49]. Rice dwarf virus (RDV), which causes millions of dollars crop losses each year, is a member of Phytoreovirus whose genome consists of 12 dsRNAs that encode 12 proteins. The RDV non-structural protein Pns10 has been identified as a VSR, which has siRNA-duplex binding activities [50], [51]. Rice stripe virus (RSV), another devastating rice pathogen, is a member of Tenuivirus whose genome consists of four negative sense and ambisense single-stranded RNAs that encode seven proteins. The nonstructural protein NS3 functions as a VSR that also has siRNA-duplex binding activities [52]. Both viruses are transmitted via insect vector in a persistent manner and the eggs from viruliferous female adults also carry viruses and spread diseases. RSV and RDV are transmitted to rice plants by the small brown plant hopper (Laodelphax striatellus) and leafhopper (Nephotettix cincticeps or Resilia dorsalis), respectively.
Our analyses showed that RSV and RDV infections differentially affected rice small RNA profiles. RSV infection enhanced the accumulation of some rice miRNA*s, but not their corresponding miRNAs, as well as accumulation of phased siRNAs from a particular precursor. Significantly, RSV infection also induced the expression of novel miRNAs in a phased pattern from several conserved miRNA precursors. In comparison, no such changes in host small RNA expression was observed in RDV-infected rice plants. RSV infection significantly elevated the expression levels of selective OsDCLs and OsAGOs, whereas RDV infection only affected the expression of certain OsRDRs. Our results provide a comparative analysis, via deep sequencing, of changes in the small RNA profiles and in the genes of RNA silencing machinery induced by different viruses in a natural and economically important crop host plant. They uncover new mechanisms and complexity of virus-host interactions that may have important implications for further studies on the evolution of cellular small RNA biogenesis that impact pathogen infection, pathogenesis organismal development, and crop-protection technology development.
Results
Deep sequencing of small RNAs
We sequenced the small RNAs from rice plants infected with RDV and RSV, respectively, and from plants mock-inoculated controls by using the Solexa/Illumina deep sequencing method. The three-week-old rice seedlings were exposed, respectively, to viruliferous leafhopper, N. cincticeps (RDV), virus-free N. cincticeps (RDV, mock), viruliferous planthopper, L. Striatellus (RSV) and virus-free L. Striatellus (RSV, mock). After three weeks, when virus-induced symptoms appeared in the systemic leaves of virus-infected plants, the plants from each treatment (i.e., RDV-infected, RDV mock, RSV-infected and RSV mock) were pooled to prepare an RNA library for sequencing. We performed sequencing with three biological replicates for each treatment, with approximately 15 plants pooled in each replicate. (All the sequencing data can be available from the website: http://www.cbi.pku.edu.cn/download/rdsv/rdsv.html.) From each library, we obtained more than 50% of small RNA sequences that had at least one perfect match in the rice genome and no more than one mismatch in the virus genome (Table 1). The similar percentages of small RNA sequences matched to the rice and virus genomes in all replicates for a given treatment indicate similar quality of RNA preparation and sequencing. Because of the large volume of total sequence data and our primary focus in this study on learning about how the two viruses would affect small RNA profiles and the RNA silencing machinery in a common host, the virus-derived small RNA profiles and their biological implications will be analyzed and presented in a future report. For the rice small RNAs, we normalized the total sequence reads of each library to one million, and then used the average read value of unique sequences from all replicates in each treatment for further analysis.
Tab. 1. Summary of deep sequencing results of small RNAs from virus-infected and mock-inoculated rice small RNA libraries. 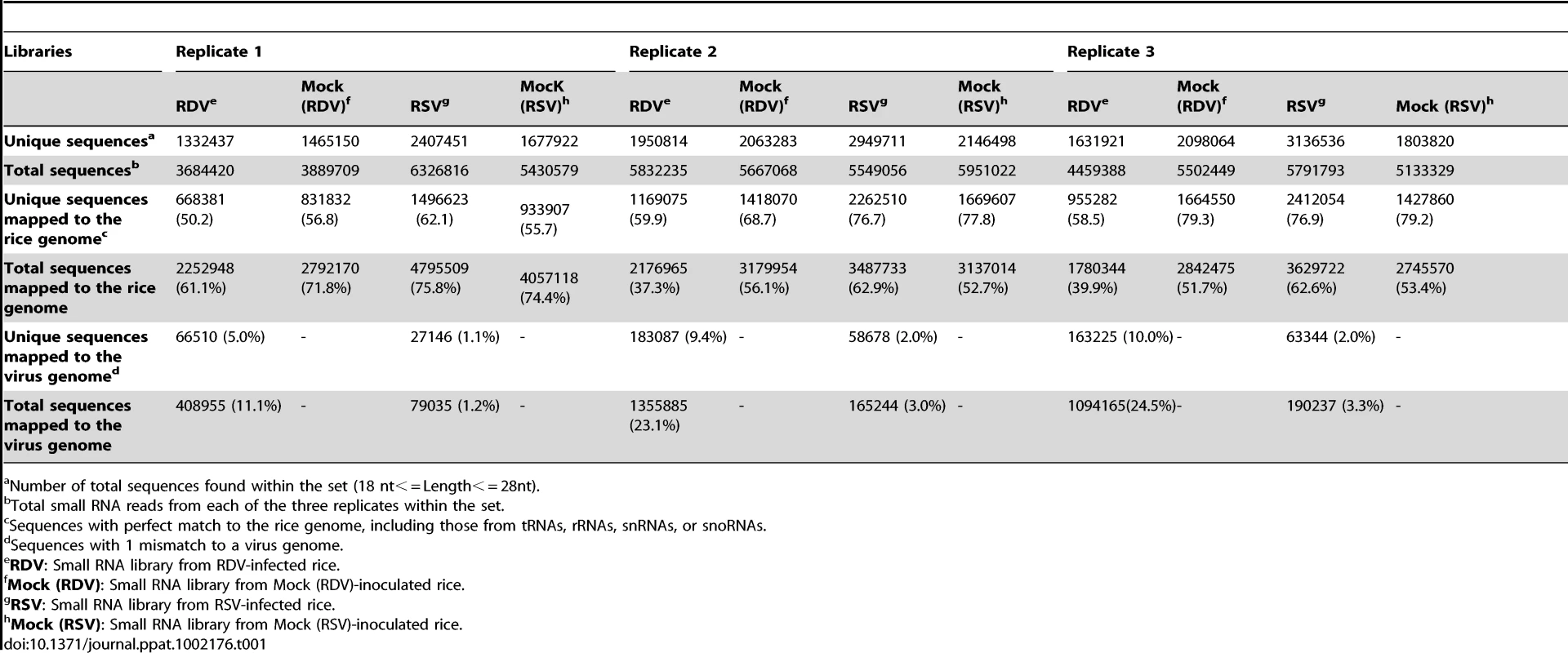
Number of total sequences found within the set (18 nt< = Length< = 28nt). Infection by RSV, but not by RDV, led to altered expression of selective rice miRNAs
We compared the total reads of rice miRNAs among the four libraries, using pooled data from the three independent biological replicates. There were 4060, 4852, 8449 and 5014 unique sequences, with 89036, 91409, 118975 and 90214 reads, that match perfectly to miRNA precursors from RDV-infected, mock (RDV)-inoculated, RSV-infected, and mock (RSV)-inoculated rice plants, respectively (Table 2). Notably, the number of unique sequences mapped to miRNA precursors from the RSV-infected rice plants was nearly twice of those from each of the other three types of plants.
Tab. 2. Summary of small RNAs mapped to known rice miRNA precursors in virus-infected and mock-inoculated rice plants. 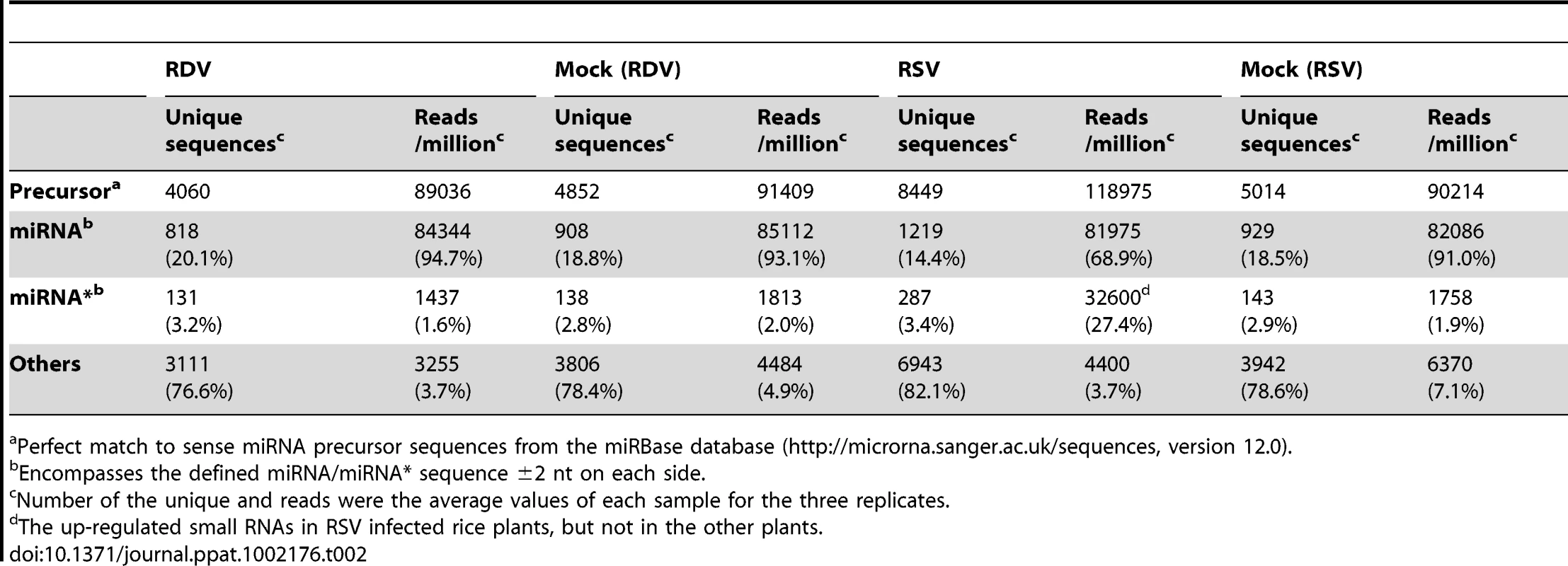
Perfect match to sense miRNA precursor sequences from the miRBase database (http://microrna.sanger.ac.uk/sequences, version 12.0). Of all the sequence reads mapped to miRNA precursors, 94.7% (84344), 93.1% (85112), 68.9% (81975) and 91.0% (82086) were mature miRNA sequences, and 1.6% (1437), 2.0% (1813), 27.4% (32600) and 1.9% (1758) were miRNA* sequences (Table 2). Interestingly, the reads of miRNA* sequences in the RSV-infected rice small RNA library were 15 times higher than those from the other three libraries, and was nearly half of the miRNA reads from the same library. The sequence data from the three biological replicates are presented in Supplemental Table S1, which reproducibly show more than about 10-times higher accumulation of miRNA* sequences in RSV-infected rice plants than in the other three groups of plants.
For many miRNAs there were no obvious differences in expression levels between RSV-infected and mock-inoculated rice plants. However, a number of miRNAs showed significant changes in expression in RSV-infected plants. These were defined as those having reads of 100 or more and showing at least two-fold changes in reads between RSV-infected and mock-inoculated plants. As shown in Table 3, expressions of miR156b, miR159a1, miR164a, miR166, miR167a, miR1884b, miR393b, miR396e and miR528 were down regulated, whereas miR535, miR390 and miR171 were up regulated in the infected plants. Although data from the three independent biological replicates were reproducible (Supplemental Table S1 and S2), we nonetheless used RNA gel blots to verify the altered expressions of some miRNAs with higher reads. As shown in Figure 1A, miR156, miR166 and miR167 were down regulated, whereas miR172 showed no obvious changes in accumulation. The expression of miR168 showed a two-fold increase on RNA gel blots, in near agreement with the sequence data (Table 3). By contrast, fewer miRNAs showed changes in accumulation levels in RDV-infected plant as compared to those in the mock-inoculated plants. Specifically, miR167a, miR171 and miR1863 were down regulated and only miR393 was induced in RDV-infected rice plants (Table 3). We confirmed the down-regulation of miR167 in RDV-infected rice in northern blots (Figure 1A). Thus, different viruses can differentially affect the expression of some miRNAs in a common host.
Fig. 1. Confirmation of miRNA expression and analysis of miRNA target gene expression of in virus-infected plants. 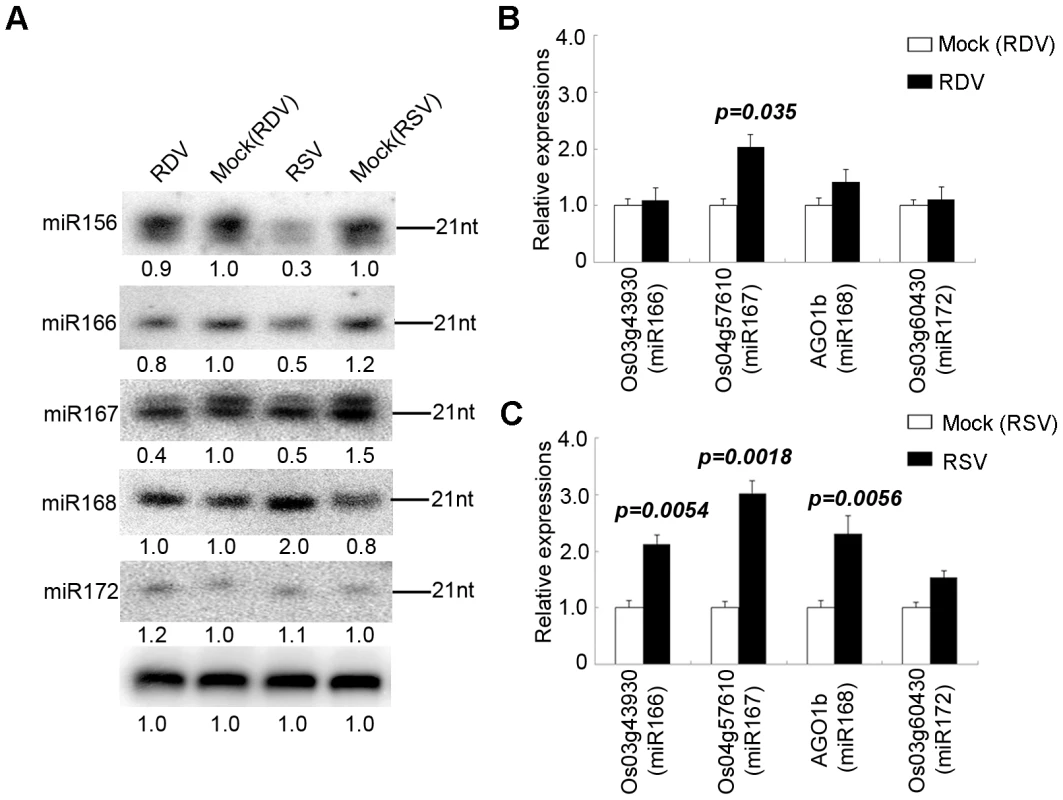
(A) RNA gel blots showing expression of miR156, miR164, miR166, miR167, miR168 and miR172 in virus infected rice plants. Rice U6 was reprobed as a control. The expression levels from mock (RDV)-inoculated plants are set to a value of 1.00 and the other three plants are expressed relative to this reference value. (B) Expression analysis of miRNA targets from RDV-infected plants and (C) from RSV-infected plants by quantitative real-time RT-PCR analysis. The expression levels of the assayed genes were normalized to the expression level of OsEF-1α.Os12g41680, Os03g43930, Os04g57610 and Os03g60430 are the targets of miR164, miR166, miR167 and miR172, respectively [54]. Tab. 3. Comparison of deep sequencing reads of known miRNAs and miRNA*s from virus-infected and mock-inoculated rice libraries. 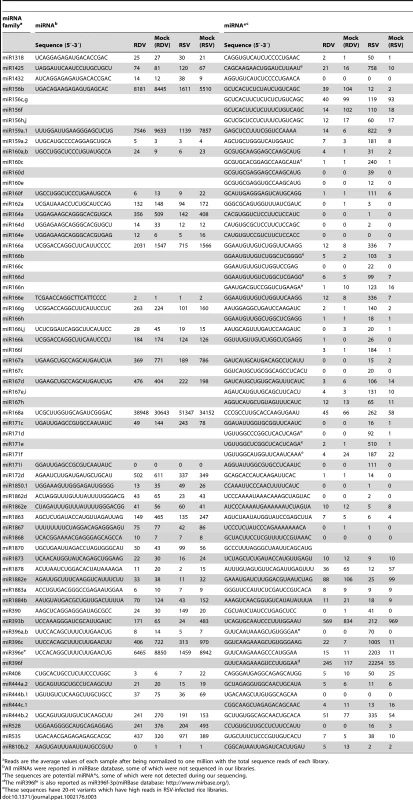
Reads are the average values of each sample after being normalized to one million with the total sequence reads of each library. Using quantitative real-time RT-PCR, we analyzed the expression of one target gene for each miRNA shown in Figure 1A. As shown in Figure 1B, the expression of AGO1b, a target of miR168, was induced during RSV infection (Figure 1C). This mimics the situation in Arabidopsis thaliana where the expressions of miR168 and AGO1 are transcriptionally co-regulated [53]. Os03g43930, a HD-Zip transcription factor and a target of miR166 [54], was up-regulated in agreement with the down-regulation of miR166 expression during RSV infection (Figure 1C). The expression of Os03g60430 (target of miR172) [55] showed no changes, whereas the expression of Os04g57610 (target of miR167) [54] increased in RSV infected rice (Figure 1C). These patterns correlated well with unaltered expression miR172 and down-regulated expression of miR167. Notably, none of these genes showed significant changes in expression levels, as did their cognitive miRNAs, in RDV-infected plants (Figure 1B). Using four different pairs of primers, we were unable to obtain conclusive data about the expression of potential miR156 targets (LOC_Os04g46580 and LOC_Os07g 32170).
RSV infection enhanced accumulation of specific rice miRNA*
In RSV-infected rice plants, many miRNA*s accumulated to high levels, whereas their corresponding miRNA sequences did not show any obvious changes compared to the mock control (Table 3). These include miRNA*s for some members of the miR160 family (miR160a–f), miR166 family (miR166a–e, g–l and n), miR167 family (miR167a, c–e, h and i), miR171 family (miR171c–f and miR171i), miR396 family (miR396a–c, e and f) and miRNA* of miR1318, miR1425, miR159a, miR168, miR172d, miR390, miR444b.2 and miR528.
These data were not attributed to sequencing bias. First, the vast majority of miRNA* in RSV-infected plants were present at low levels as in mock control plants. Second, the levels of both miRNA and miRNA* in RDV-infected plants were not different from those in the mock controls. Third, northern blots confirmed that miR1425*, miR160* and miR171* were significantly up regulated, whereas their corresponding miRNAs were not (Figure 2A). Thus, we concluded that RSV infection specifically enhanced the accumulation of certain miRNA* sequences, but not their corresponding miRNAs. We notice shorter sequences for miR160* and miR171* in RSV-infected rice (Figure 2A), which maybe 20-nt variants of miRNA* through sequencing data (Table 3; see also Table S2 for all sequencing data from three biological replicates).
Fig. 2. Predominant accumulation of selective miRNA*s in RSV-infected rice. 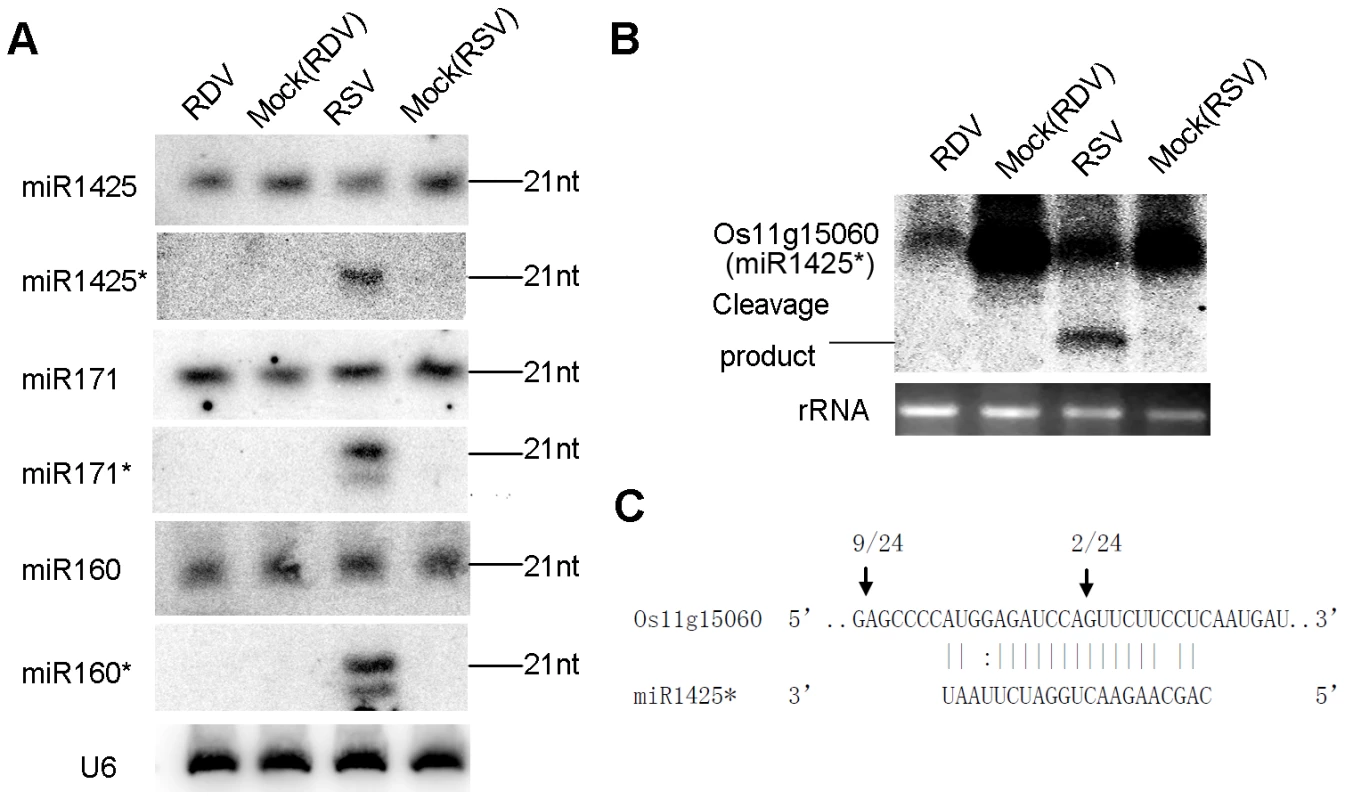
(A) Expression analysis of miR1425/miR1425*, miR171/miR171* and miR160/miR160* in RDV infected, mock (RDV)-inoculated, RSV-infected as well as mock (RSV)-inoculated rice plants by RNA gel blots. Rice U6 was reprobed as a control. (B) RNA gel blot analysis of the expression of Os11g15060, a predicted target of miR1425*. The 28S rRNA stained with ethidium bromide was used as a loading control. (C) Mapping of the cleavage site in Os11g15060 by 5′-RACE. The numbers above the arrows indicate the frequencies of sequenced RACE clones corresponding to each inferred cleavage site. We predicted the putative targets for some of these miRNAs*. Os11g15060, a SAM (S-adenosyl-L-Met)-dependent carboxyl methyltransferase, was predicted to be a target of miR1425*. Northern blots showed down-regulated expression of Os11g15060 in RSV-infected plants (Figure 2B). A cleavage product was detected (Figure 2B), and 5′-RACE (Rapid Amplification of 5′ Complementary DNA Ends) identified the cleavage site in the miR1425* binding region (Figure 2C), providing direct evidence that a plant miRNA* could direct the cleavage of its target mRNA. We also identified a second cleavage site outside the miR1425* binding region (Figure 2C). How this second site was derived is not clear, but many miRNA targets have more than one cleavage site as reported in other plants [56]–[58] and even in a green alga [59]. Whether this second cleavage results from the action of another yet-to-be identified small RNA induced by viral infection remains to be further investigated. Intriguingly, the expression of Os11g15060 was also down-regulated in RDV-infected plants (Figure 2B). However, absence of a cleavage product and failed 5′ RACE to identify a cleavage site (data not shown) suggests that this down-regulation in the RDV-infected plants was not caused by RNA silencing, or caused by partial silencing as well as another mechanism that remains to be identified. We also analyzed the expression of some potential targets of miR160* and miR171* including Os11g38140 and Os02g49240 (potential targets of miR160*), and Os03g38170 (potential target of miR171*). The expression of the three genes decreased in RSV-infected rice as compared with that in RDV-infected rice (Figure S1).
RSV infection induced expression of new phased miRNAs from conserved precursors
The small RNA libraries of RSV-infected rice contained many unique sequences, absent from the other three libraries, which are mapped to several conserved miRNA precursors (Table 2). Some of these belong to the miR159 family, whose precursors are approximately 200 nt in length with a stem structure of 80–90 nt (Figure 3). As shown in Figure 3A–D (see also Supplemental Table S3 for all sequence data), besides the reported miRNA/miRNA* pair for each precursor of the family (i.e., miR159a.1/miR159a.1*, miR159a.2/miR159a.2*, miR159b.1/miR159b.1*, miR159c.1/miR159c.1* and miR159f.1/miR159f.1*, labeled red and blue respectively for each pair), new pairs of miRNA/miRNA* were produced from each precursor. These included miR159a.3/miR159a.3*, miR159b.2/miR159b.2*, miR159b.3/miR159b.3*, miR159c.2/miR159c.2*, miR159c.3/miR159c.3*, miR159f.2/miR159f.2* and miR159f.3/miR159f.3*. These pairs were generated in a phased pattern and often detected at higher levels than the reported pairs. The phased miRNA-miRNA*s have the characteristics of 2 nt overhangs at the 3′ end. These observations, together with the observation that such phased production of new miRNA/miRNA* were absent from RDV-infected or any mock-infected plants, ruled out the possibility that they were degradation products or sequencing errors.
Fig. 3. Phased miRNAs induced in RSV-infected rice. 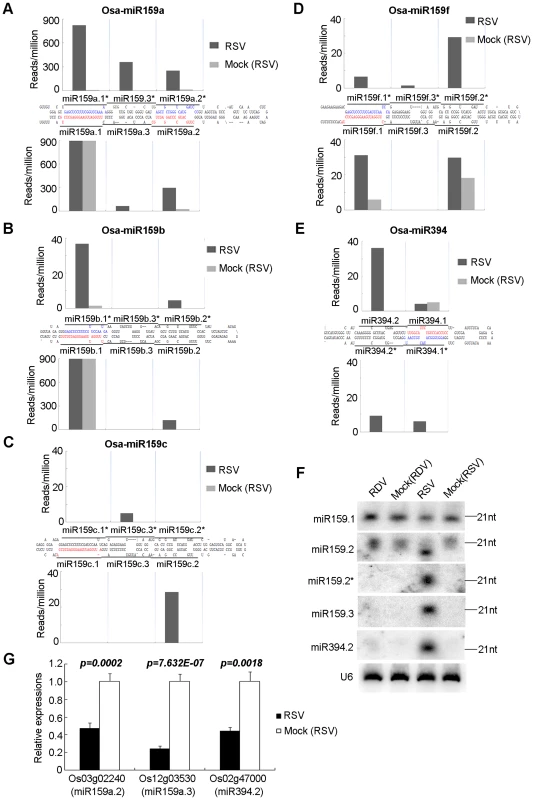
(A–E) Precursors of phased miRNAs. The sequences shown in red and blue are reported miRNAs and miRNA*s, respectively. The histograms indicate the reads of corresponding sequences on the precursors shown in black bars. (F) RNA gel blot showing the expression of phased miRNAs in RSV-infected rice. Rice U6 was reprobed as a control. (G) Expression analysis of the predicted targets of phased miRNAs by real time RT-PCR. Os03g02240 was previously validated as a target of miR159.2. [61]. Os12g03530 and Os02g4700 are predicted targets of miR159a.3 and miR394.2, respectively. The expression levels of the assayed genes were normalized to the expression level of OsEF-1α. In addition to the above three-duplex phase forms of miRNA/miRNA*s, we also found two-duplex phase forms derived from some other precursors. The precursor of miR394 in the miRBase database (http://microrna.sanger.ac.uk/sequences, version 12.0) is about 100 nt in length. However, we found that the actual precursor is longer and contains a 27-nt extension from the 5′ and 3′ ends of the reported precursor, respectively (Figure 3E and Figure S2). In this longer precursor, a new miR394.2/394.2* duplex appeared at the distal end of the stem structure, in phase with the reported 394.1/394.1* (Figure 3E and Figure S2).
Northern blots confirmed the expression of miR159.2/miR159.2*, miR159.3 and miR394.2 (Figure 3F). The resolution of northern blots did not permit distinction between family members, so that each band could contain multiple members of a family of miRNAs/miRNA*s.
Using clustalW [60], we analyzed the conservation of precursor sequences of miR159, miR319, and miR394 in different plants (Figure S3). We found that, compared with the reported mature miRNA/miRNA* sequences, the newly identified phased miRNA/miRNA* sequences are less well conserved.
The target of miR159a.2 is Os03g02240, a GT2 transcription factor, which has been verified by 5′ RACE [61]. Quantitative real-time RT-PCR analysis showed that RSV infection down-regulated the expression of Os03g02240 (Figure 3G). Os12g03530 and Os02g47000, the predicted targets of miR159a.3 and miR394.2, respectively, were down-regulated in RSV-infected rice plants (Figure 3G). However, in RDV-infected rice plants, the expression of these genes was unchanged (data not shown).
RSV infection enhanced the accumulation of distinct rice phased siRNA from a pre-existing RNA precursor
The transcripts encoded by the AK120922 locus of rice genome can fold into long inverted repeat structures, producing 12 21-nt phased small RNA duplexes [45], [62], [63]. From the stem region proximal to the terminal loop to the distal end of the RNA secondary structure, the 12 small RNA duplexes produced are named P1-12_5′ on the 5′ arm and P1-12_3′ on the 3′ arm (Figure 4A). One of these duplexes was initially characterized as miR436/miR436* duplex [62], but further studies demonstrated all small RNAs, including the so-called miR436, are DCL4-dependent siRNAs [45]. Analysis of our sequencing data showed that the reads of some AK120922-derived siRNAs in the RSV-infected rice plants were much higher than those in the mock-inoculated plants. In particular, the reads of P5_3′ increased by at least 100-fold. The higher expressions of these siRNAs were confirmed by northern blots (Figure 4B). No such changes were observed in RDV-infected plants (data not shown). Using realtime PCR, we analyzed the expression of the potential targets of P5_3′ and P10_3′. Surprisingly, we found that both of them were up-regulated in RSV-infected plants (Figure S4).
Fig. 4. Induction of OsDCL4-dependent phased 21-nt siRNAs produced from the long hairpin of RNA encoded by the AK120922 locus in RSV-infected rice. 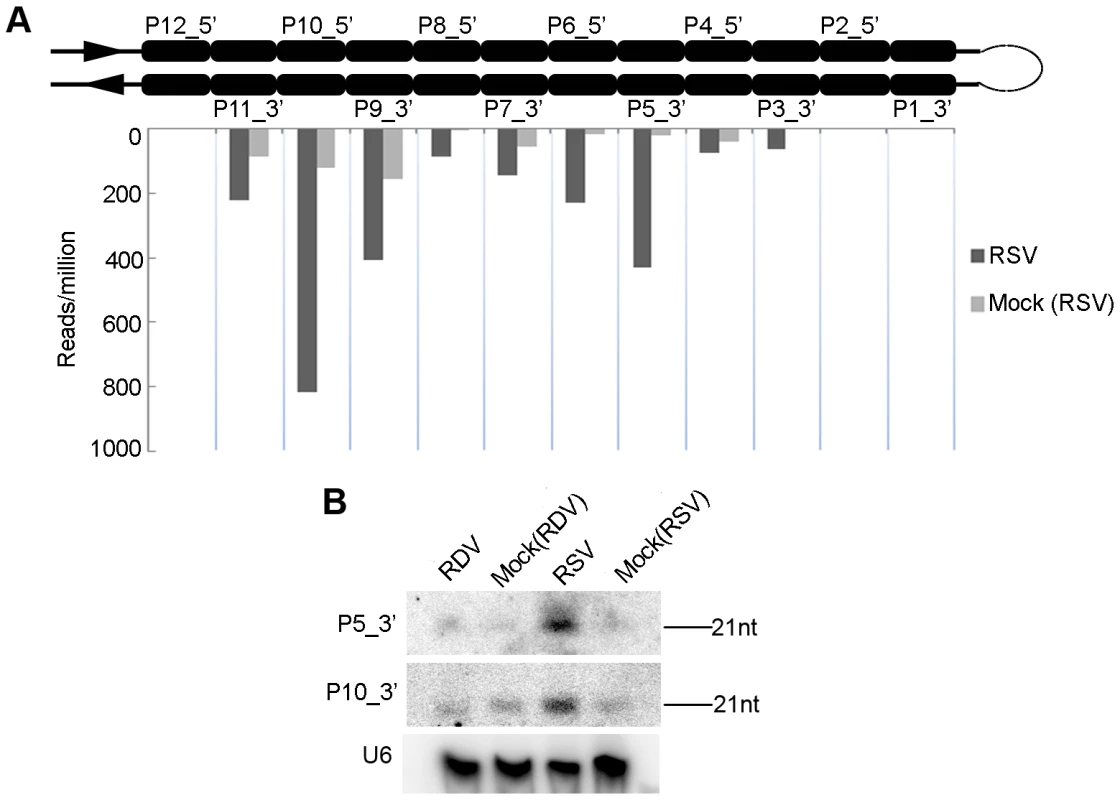
(A) Accumulation of siRNAs in RSV-infected rice compared that in mock-inoculated rice. The upper structure illustrates the long RNA hairpin. The read numbers below the hairpin, which were each normalized to one million with total reads of the corresponding library, indicate the expression levels of siRNAs at corresponding positions in the 3′ arm of the hairpin. (B) RNA gel blots confirm accumulation of the highly induced P10_3′ and P5_3′, hybridized with the complementary 32P-labeled probes. RDV and RSV infections differentially modified the expression of rice RNA silencing pathway genes
To gain additional insights into the effects of RDV and RSV infection on the RNA silencing pathways/machinery, we characterized the expression profiles of various genes involved in the biogenesis/function of miRNAs/siRNAs by using the same plant materials as used for small RNA deep sequencing. These genes include 8 homologs of OsDCLs, five OsRDRs and 19 AGOs that have been identified in rice [44]–[49].
We first analyzed the expression profiles of OsDCLs, OsRDRs and OsAGOs, according to the annotations of Kapoor et al [48]. Figure 5A presents microarray data showing that RDV and RSV infections affected the expression of different members of OsDCL, OsRDR and OsAGO families. The microarray data were further verified by quantitative real-time RT-PCR measurements (Figure 5B and C). Of the 8 DCL homologs in the rice genome [48], OsDCL3a (LOC_Os01g68120) and OsDCL3b (LOC_Os10g34430) were significantly down-regulated, and OsDCL2 (LOC_Os03g38740) significantly up-regulated, in RSV-infected plants (Figure 5C). In contrast, RDV infection had almost no influence on the expression of OsDCLs in rice plants (Figure 5B).
Fig. 5. Expression analysis of OsDCLs, OsAGOs and OsRDRs of rice plants infected with RDV and RSV as compared to mock controls. 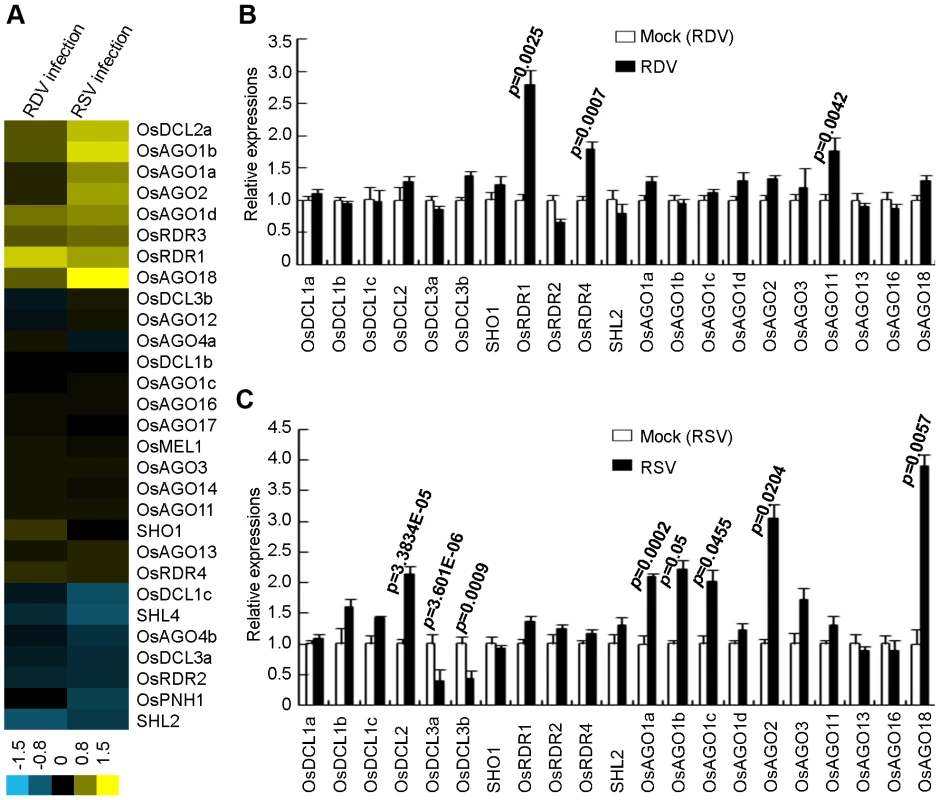
(A) Expression profiles from microarrays. The bar on the bottom represents the scale of relative expression levels of genes (Log2). (B) Expression profiles from RDV-infected plants and (C) Expression profiles from RSV-infected plant based on quantitative real-time RT-PCR analysis. The expression levels of the assayed genes were normalized to the expression level of OsEF-1α. The expression level in the corresponding mock rice was set to 1.0. The nomenclature of these genes is based on Kapoor et al. (2008). In rice, there are four AGO1 homologs: OsAGO1a (LOC_Os02g45070), OsAGO1b (LOC_Os04g47870), OsAGO1c (LOC_Os02g58490) and OsAGO1d (LOC_Os06g51310) [32], [33]. The expression of OsAGO1a-c as well as OsAGO2 (LOC_Os04g52540) and OsAGO18 (LOC_Os07g28850) increased significantly in RSV-infected rice plants (Figure 5C). In RDV-infected plants, only the expression of OsAGO11 increased significantly (Figure 5B).
The expression of OsRDR1 (LOC_Os02g50330) and OsRDR4 (LOC_Os01g10140) was significantly enhanced in RDV-infected plants (Figure 5B), but not in RSV-infected plants (Figure 5C). OsRDR2 did not change expression levels in plants infected by either virus.
These data demonstrate that the two viruses have distinct effects on the expression of different genes of the RNA silencing pathways in the common host rice.
Discussion
Recent studies used deep sequencing to obtain profiles of viral siRNAs [64]–[68] and viroid small RNAs [69], [70] in infected plants. Such data laid a foundation for further investigations on the biogenesis mechanisms and functions of viral and viroid small RNAs. Our present work provides the first deep sequencing analysis of plant small RNA profiles under viral infection conditions. This analysis not only filled a critical knowledge gap in RNA silencing-based virus-host interactions, but provided novel insights into the impact of viral infection on host small RNA biogenesis. Our results showed down-regulated as well as up-regulated accumulation of certain rice miRNAs, with up-regulation being more extensive. Most significantly, RSV infection, but not RDV infection, induced the expression of novel phased miRNAs from several families of conserved cellular miRNA precursors. Real-time RT-PCR experiments showed reduced accumulations of the predicted target mRNAs for these phased miRNAs (i.e., miR159.2 families, miR159.3 and miR394.2), indicating that the induced phased miRNAs have regulatory functions. How the reduced accumulation of these target mRNAs contributes to the establishment of viral infection and/or host defense will be an important focus of future research.
As compared to the short animal miRNA precursors, which are usually 70–80 nt, plant miRNA precursors are generally much longer with most known precursors to be 200–300 nt [15]. These long plant precursors make it possible to produce multiple miRNAs. Indeed, multiple, phased miRNAs are produced from some miRNA precursors in the single-cell alga, Chlamydomonas reinhardtii [59], [71] and in higher plants [72]. Some of these miRNAs are differentially regulated by bacterial infection in A. thaliana [72]. Our finding that new phased miRNAs are induced during the infection of a plant virus significantly broadens the landscape of phased miRNA biogenesis during pathogen infection. Further analyses of the host small RNA profiles involving different pathogens and hosts may lead to additional examples, and an understanding of the broad biological significance, of pathogen infection-induced expression of phased and other novel miRNAs.
The enhanced production of miRNA*s during RSV infection may be attributed to inhibition of RISC activity and miRNA/miRNA* unwinding by the RSV VSR, via direct or indirect interaction with the miRNA/miRNA* duplexes, as have been shown for VSRs of other viruses [22], [24]. If this were the case, one would expect elevated accumulations of miRNAs and miRNA*s at the same time. However, our deep sequencing showed that only the accumulations of miRNA*s, not the corresponding miRNAs from many miRNA precursors were enhanced. Such special accumulation of miRNA*, but not miRNAs, may be explained if RSV enhanced the activities of certain RISCs specially associated with miRNA*s or interfered with loading of some miRNAs into RISCs. Since we do not have data yet to support this postulation, we need to leave other alternative possibilities open for exploration. Schnettler et al. (2010) reported stabilization of miR171c/miR171c* complex by several tospoviruses in infected N. benthamiana, with elevated accumulation of miR171c* in the infected plants. This stabilization appears to be due to the activity of the viral NSs proteins [73].
The specific accumulation of miRNA*s of several miRNA families and the conservation of the miRNA* sequences within the families suggest that there may be certain base preferences among the miRNA*s accumulated during RSV infection. Using WebLogo [74], we found that there was an ‘A’ bias in the 19th nucleotide from the 5′ terminus (Figure S5). Considering that the A. thaliana AGO2 preferentially produces miRNA* sequences [75] and the up-regulation of OsAGO2 and OsAGO18 during RSV infection of rice, we propose that the ‘A’ bias may direct certain miRNA* sequences into these OsAGOs. Still we cannot exclude the potential influence of OsAGO1, as the ‘A’ bias in the 19th nucleotide from the 5′ terminus of miRNA* corresponds to the 5′ terminal ‘U’ of miRNA. In Drosophila melanogaster, miRNA*s were reported to have regulatory functions [76]. Here, by northern blots and 5′ RACE, we demonstrated that Os11g15060, a predicted target of miR1425*, was specifically down regulated by cleavage during RSV infection (Figure 2C). This is the first direct experimental demonstration, to the best of our knowledge, that a plant miRNA* can regulate the stability of its predicted target mRNA.
Although many siRNAs were produced in rice [77], our current analysis showed that RSV infection enhanced mainly the accumulation of some phased siRNAs derived from the AK120922 transcripts. The specificity of this enhancement was supported by the observation that RDV infection did not have such an effect. Previous studies showed that accumulation of siRNAs decreased in transgenic plants expressing the VSRs of several plant viruses [20], [23], [24]. The inhibitory effect of VSRs on siRNA accumulation, via binding of VSRs with siRNAs, is one of the mechanisms of viral counter-defense [1], [2], [19]. The enhanced accumulation of phased siRNAs from AK120922 during RSV infection, in contrast to the generally observed decrease in siRNA accumulations, may be biologically significant. Whether the VSR or other proteins encoded by RSV play a role in this enhancement is an outstanding mechanistic question to be answered in the future.
The induced expression of new phased miRNAs and the enhanced production of selective miRNA*s and phased siRNAs in rice plants infected by RSV, but not by RDV, indicate that such changes were not a general response to viral infection. Rather, they were caused by distinct virus-host interactions. One of the primary consequences of such interactions was conceivably altered expression and/or function of the RNA silencing machinery, which further leads to altered small RNA profiles. Indeed, our microarray and quantitative real-time RT-PCR analyses demonstrated that the expression files of OsDCLs, OsAGOs and OsRDRs were differentially altered in rice infected with the two viruses. During RDV infection, with the exception of OsRDR1 being enhanced, there were no significant changes in the expression of RNA silencing pathway genes. In contrast, during RSV infection, the expression levels of OsDCL3a and OsDCL3b were down-regulated, whereas the expression of OsDCL2 was enhanced. Three of the four OsAGO1 homologs, OsAGO1a, OsAGO1b and OsAGO1c, were up-regulated in RSV-infected plants. Whether the altered expression patterns of the OsDCLs, OsRDRs and OsAGOs are responsible for the altered miRNA/siRNA biogenesis/accumulations in the RSV - and RDV-infected rice plants remains to be determined. Current data indicate that OsDCL1 participates in the production of 21-nt miRNAs and DCL4 mainly produces some siRNAs [28], [29]. DCL3 was recently reported to produce 24-nt miRNAs [14]. The four small RNA libraries we generated comprised mainly 24-nt sequences. However, in the RSV-infected rice small RNA libraries, the number of 24-nt small RNAs was reduced to about three quarters of the other three libraries (Figure S6). This correlated with a decreased level of DCL3 expression in RSV-infected rice. These results suggested that OsDCL3 may be primarily responsible for producing 24 nt small RNAs, just as the A. thaliana homolog does (Figure 5) [78], [79]. The production of 21 nt and 24 nt phased miRNAs suggests involvement of DCL1 and potentially also DCL3.
In summary, our data indicate that at least some viruses may have evolved mechanisms to induce expression of phased miRNAs from well-conserved cellular miRNA precursors. Whether such new miRNAs play a role in host defense or in viral infection will be an important question to be investigated in the future. While this manuscript was under review, Hu et al. (2011) reported that Oilseed rape mosaic tobamovirus infection of A. thaliana also led to elevated accumulation of host siRNAs and some miRNA-like small RNAs (ml-sRNAs). These ml-sRNAs are derived in phase with known miRNAs from miRNA-precursors [80]. They are different from the sequences we report here. Altogether, as for many previous discoveries, virus-induced production of phased miRNAs and ml-sRNAs may provide yet another useful model system to study the molecular mechanisms underlying the evolution and biogenesis of miRNAs. It remains to be seen whether virus-induced biogenesis of phased miRNAs is more widespread in plants and other organisms. Another important question is how different viruses affect the production of different small RNAs and how such differences impact specific mechanisms of viral infection and host responses.
Materials and Methods
Plant growth, virus infection and small RNA sequencing
RDV Fujian isolate and RSV Jiangsu isolate, China, were maintained in “Oryza sativa spp. japonica” rice plants grown in greenhouses at 25±3°C, 55±5% RH and under natural sunlight. Insects (Nephotettix cincticeps and Laodelphax striatellus) were maintained in cages that contained rice seedlings in greenhouses at 25±3°C. To obtain high viruliferous insects, nymphs were reared on virus-infected rice plants for 1 week, and insects were maintained up to the adult stage with occasional replacement of seedlings by healthy rice seedlings. Rice seedlings were grown in a greenhouse at 25–28°C and 70±5% relative humidity under natural sunlight. Three-week-old seedlings were placed individually in single tubes of 4-cm in diameter and 25-cm in height that each contained 15–20 ml of liquid nutrient medium at the bottom. The viruliferous insects of N. cincticeps (carrying RDV), L. Striatellus (carrying RSV) as well as virus-free N. cincticeps (mock for RDV) and L. Striatellus (mock for RSV) were added to each tube. Each tube was then sealed with a nylon net. After 2 days in growth chambers with a 14-h/10-h light/dark cycle, 70±5% relative humidity and a temperature regime of 28°C (day)/25°C (night), the insects were removed and the rice seedlings were returned to the greenhouse to grow under the above greenhouse conditions. After approximately three weeks of growth, when the newly developed leaves started to show viral symptoms, the whole seedlings were harvested. With each sample, at least 15 rice seedlings were pooled for RNA extractions. Total RNAs were extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) for RT-PCR, small RNA sequencing, microarray analysis, and northern blotting. RT-PCR was used to test individual rice seedlings for infection with RDV or RSV. All PCR primers are listed in Table S4. The small RNA library construction and Illumina 1G sequencing were carried out at BGI-Shenzhen (Shenzhen, Guangdong, China) using standard Solexa/Illumina protocols. Briefly, the total RNA was separated through 15% TBE urea denaturing PAGE gels and small RNAs of 15–30 nt were recovered. Then, 5′ and 3′ RNA adaptors were ligated to these small RNAs followed by reverse transcription into cDNAs. These cDNAs were finally amplified by PCR and subjected to Solexa/Illumina sequencing.
Bioinformatics
After removing the adaptor sequences, small RNA sequences with 18–28 nt in length were used for further analysis. BOAT provided by CBI (http://boat.cbi.pku.edu.cn/) was used for mapping small RNAs to the O. sativa genome sequences (TIGR Rice Annotation Release 5.0, ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/) as well as RDV (ftp://ftp.ncbi.nih.gov/genomes/Viruses/Rice_dwarf_virus_uid14797/) and RSV (ftp://ftp.ncbi.nih.gov/genomes/Viruses/Rice_stripe_virus_uid14795/) Rice stripe virus) genome sequences. Small RNAs with perfect genomic matches were used for further analysis. The small RNAs were annotated with reference to the following databases: miRBase (http://microrna.sanger.ac.uk/sequences, version 12.0) for miRNA sequences, Rfam (http://www.sanger.ac.uk/Software/Rfam/) for noncoding RNA (rRNAs, tRNAs, snoRNAs, and snRNAs) sequences and Repbase (http://www.girinst.org) for transposons and repeats.
WebLogo [74] was used for analyzing of relative frequencies of nucleotides at each position of small the sRNAs, and the mfold program [81] was used for predicting the stem-loop structures. Target genes were predicted by using the miRU web server (http://bioinfo3.noble.org/miRNA/miRU.htm) and our own Perl script [59]. For submission to the web server, we chose the default parameters (score for each 20 nt≤3, G: U pairs≤6, indels≤1 and other mismatches ≤3) and the TIGR Rice Genome mRNA dataset (OSA1 release 5, 01/23/2007). For our own Perl script, based on the score standard mentioned before [59], targets with score ≤5 were chosen. Identical search results from both methods were considered potential targets of newly identified miRNAs or phased miRNAs. For miRNA*s induced during RSV infection, their targets were validated based on the microarray data. ClustalW was used for the alignment of miRNA precursors and miRNA* sequences.
Microarray hybridization and data analysis
Hybridization of GeneChip rice genome array (Affymetrix), scanning and analysis were performed by the Affymetrix custom service (CapitalBio, Beijing, China) following standard protocols (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). Three biological replicates were conducted for RNA from each type of plant samples. Data analysis and comparison of the samples was finished using the standard Affymetrix protocol (CapitalBio). Cluster3.0 software was used for cluster analysis. The expression profile of some mentioned gene through microarray analysis was shown in Table S5.
Northern blot hybridization
Total RNA was used for mRNA and small RNA northern blot hybridization. For small RNA blots, 10–60 µg of total RNA (depending on the relative expression levels from deep sequencing) of each sample was separated on 15% polyacrylamide denaturing gels and then transferred to Hybond-N+ membranes (Amersham BioScience, Piscataway, NJ, USA). The membranes were cross-linked by UV transillumination and dried at 120°C for 30 min. DNA oligonucleotides complementary to small RNAs, which were labeled with γ-32P-ATP by T4 polynucleotide kinase (New England Biolabs, Beverly, MA, USA), were used as probes for hybridization. Membranes were prehybridized with buffer for 2 h followed by hybridization with the DNA probes overnight at 40°C in 5X SSC, 20 mM Na2HPO4 (PH 7.2), 7% SDS, 2X Denhardt's Solution, 100 µg/ml salmon sperm DNA. After washing twice at 40°C with 2X SSC and 0.1% SDS, the blots were imaged with a PhosphorImager (PerkinElmer, Shelton, CT, USA). The membranes were stripped with 0.1X SSC and 1% SDS at 100°C and reprobed. Probes complementary to U6 sequences were used as a loading control.
For mRNA blots, 15 µg of total RNA was separated by 1% formaldehyde agarose gel and transferred to Hybond-N+ membranes that were then cross-linked and dried as described above. Prehybridization and hybridization solution was 5X SSC, 1% SDS, 5X Denhardt's Solution, 100 µg/ml salmon sperm DNA and 50% formamide. The probes were labeled with α-32P-dCTP by DNA polymerase 1 large (Klenow) fragment (Promega, Madison, Wisconsin, USA) and were complementary to a 500-bp fragment corresponding to the 5′ partial sequence of Os11g15060 (target of miR1425*).
Quantitative real-time RT-PCR
Total RNA was treated with RNase-free DNase I (TAKARA Biotechnology, Dalian, China) at 37°C for 30 min. After phenol/chloroform extraction and isopropanol precipitation, the RNA was quantified with a UV/ visible spectrophotometer (Amersham). Two µ\g of total RNA was reverse-transcribed using poly (T) adapter with SuperScript Reverse Transcriptase (Invitrogen). qPCR was performed using SYBR Green Realtime PCR Master Mix (Toyobo) OsEF-1a gene was used as an internal control, with primers CX1597 (59-GCACGCTCTTCTTGCTTTCACTCT-39) and CX1598 (59-AAAGGTCACCACCATACCAGGCTT-39) [45]. Three independent biological replicates were conducted. These data were further normalized with the normalized expression values obtained from qRT-PCRs and bar charts plotted by using Microsoft Excel and SPSS (Statistical Product and Service Solutions) software (IBM; Version.10.0). All the other primers used are listed in Supplemental Table S4.
Target gene validation by 5′ RACE
Validation of target genes by mapping the cleavage sites was conducted with 5′ RACE by following the GeneRacer Kit (Invitrogen) protocols described previously [57]. Total RNA of RSV-infected rice was directly ligated to GeneRacer RNA Oligo adaptor without any modifications. RT-PCR was used to synthesize cDNAs using GeneRacer Oligo dT primer. GeneRacer 5′ Primer (5′CGACTGGAGCACGAGGACACTGA3′) and the target gene-specific outer primers (Table S4) were used for the first round of amplification. Then the GeneRacer 5′ Nested Primer (5′ GGACACTGACATGGACTGAAGGAGTA) and gene-specific inner primers (Table S4) were used for the second round of amplification. The PCR products were cloned and sequenced to identify the cleavage sites.
Supporting Information
Zdroje
1. LiFDingSW 2006 Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol 60 503 531
2. DingSWVoinnetO 2007 Antiviral immunity directed by small RNAs. Cell 130 413 426
3. MlotshwaSPrussGJVanceV 2008 Small RNAs in viral infection and host defense. Trends Plant Sci 13 375 382
4. UmbachJLCullenBR 2009 The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 23 1151 1164
5. CullenBR 2010 Five Questions about Viruses and MicroRNAs. PLoS Pathog 6 e1000787
6. DingSW 2010 RNA-based antiviral immunity. Nat Rev Immunol 10 632 644
7. PadmanabhanCZhangXMJinHL 2009 Host small RNAs are big contributors to plant innate immunity. Curr Opin Plant Biol12 465 472
8. RamachandranVChenXM 2008 Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321 1490 1492
9. VoinnetO 2009 Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 136 669 687
10. MaloneCDHannonGJ 2009 Small RNAs as Guardians of the Genome. Cell 136 656 668
11. BaulcombeD 2004 RNA silencing in plants. Nature 431 356 363
12. BrodersenPSakvarelidze-AchardLBruun-RasmussenMDunoyerPYamamotoYY 2008 Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 1185 1190
13. ZamorePDHaleyB 2005 Ribo-gnome: The big world of small RNAs. Science 309 1519 1524
14. WuLZhouHYZhangQQZhangJGNiFR 2010 DNA Methylation Mediated by a MicroRNA Pathway. Mol Cell 38 465 475
15. BartelDP 2004 MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281 297
16. KimVNNamJW 2006 Genomics of microRNA. Trends Genet 22 165 173
17. BaumbergerNBaulcombeDC 2005 Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits rnicroRNAs and short interfering RNAs. Proc Natl Acad Sci U S A 102 11928 11933
18. YuBYangZYLiJJMinakhinaSYangMC 2005 Methylation as a crucial step in plant microRNA biogenesis. Science 307 932 935
19. Diaz-PendonJADingSW 2008 Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu Rev Phytopathol 46 303 326
20. MalloryACReinhartBJBartelDVanceVBBowmanLH 2002 A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc Natl Acad Sci U S A 99 15228 15233
21. KasschauKDXieZAllenELlaveCChapmanEJ 2003 P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell 4 205 217
22. ChapmanEJProkhnevskyAIGopinathKDoljaVVCarringtonJC 2004 Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step (vol 18, pg 1179, 2004). Genes Dev 18 1510 1510
23. ChenJLiWXXieDPengJRDingSW 2004 Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression. Plant Cell 16 1302 1313
24. DunoyerPLecellierCHParizottoEAHimberCVoinnetO 2004 Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16 1235 1250
25. ChellappanPVanitharaniRFauquetCM 2005 MicroRNA-binding viral protein interferes with Arabidopsis development. Proc Natl Acad Sci U S A 102 10381 10386
26. MlotshwaSSchauerSESmithTHMalloryACHerrJMJr 2005 Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell 17 2873 2885
27. BazziniAAHoppHEBeachyRNAsurmendiS 2007 Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc Natl Acad Sci U S A 104 12157 12162
28. PeragineAYoshikawaMWuGAlbrechtHLPoethigRS 2004 SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18 2368 2379
29. VazquezFVaucheretHRajagopalanRLepersCGasciolliV 2004 Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16 69 79
30. YoshikawaMPeragineAParkMYPoethigRS 2005 A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19 2164 2175
31. AllenEXieZXGustafsonAMCarringtonJC 2005 microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207 221
32. BorsaniOZhuJHVersluesPESunkarRZhuJK 2005 Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279 1291
33. Katiyar-AgarwalSGaoSVivian-SmithAJinH 2007 A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21 3123 3134
34. VazquezF 2006 Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci 11 460 468
35. HendersonIRJacobsenSE 2007 Epigenetic inheritance in plants. Nature 447 418 424
36. GodshalkSEBhaduri-McIntoshSSlackFJ 2008 Epstein-Barr virus-mediated dysregulation of human microRNA expression. Cell Cycle 7 3595 3600
37. WangFZWeberFCroceCLiuCGLiaoXD 2008 Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol 82 9065 9074
38. LiYChanEYLiJNiCPengX 2010 MicroRNA expression and virulence in pandemic influenza virus-infected mice. J Virol 84 3023 3032
39. LiuXWangTWakitaTYangW 2010 Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology 398 57 67
40. BuckAHPerotJChisholmMAKumarDSTuddenhamL 2010 Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA 16 307 315
41. WangYBrahmakshatriyaVZhuHLupianiBReddySM 2009 Identification of differentially expressed miRNAs in chicken lung and trachea with avian influenza virus infection by a deep sequencing approach. BMC Genomics 10 512
42. ParameswaranPSklanEWilkinsCBurgonTSamuelMA 2010 Six RNA Viruses and Forty-One Hosts: Viral Small RNAs and Modulation of Small RNA Repertoires in Vertebrate and Invertebrate Systems. PLoS Pathog 6 e1000764
43. CuiLGuoXQiYQiXGeY 2010 Identification of microRNAs involved in the host response to enterovirus 71 infection by a deep sequencing approach. J Biomed Biotechnol 425939
44. LiuBLiPCLiXLiuCYCaoSY 2005 Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol 139 296 305
45. LiuBChenZYSongXWLiuCYCuiX 2007 Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell 19 2705 2718
46. NagasakiHItohJIHayashiKHibaraKISatoh-NagasawaN 2007 The small interfering RNA production pathway is required for shoot meristern initiation in rice. Proc Natl Acad Sci U S A 104 14867 14871
47. NonomuraKIMorohoshiANakanoMEiguchiMMiyaoA 2007 A germ cell-specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19 2583 2594
48. KapoorMAroraRLamaTNijhawanAKhuranaJP 2008 Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics 9 1471 2164
49. WuLZhangQQZhouHYNiFRWuXY 2009 Rice MicroRNA Effector Complexes and Targets. Plant Cell 21 3421 3435
50. CaoXSZhouPZhangXMZhuSFZhongXH 2005 Identification of an RNA silencing suppressor from a plant double-stranded RNA virus. J Virol 79 13018 13027
51. RenBGuoYYGaoFZhouP 2010 Multiple Functions of Rice Dwarf Phytoreovirus Pns10 in Suppressing Systemic RNA Silencing. J Virol 84 12914 12923
52. XiongRYWuJXZhouYJZhouXP 2009 Characterization and subcellular localization of an RNA silencing suppressor encoded by Rice stripe tenuivirus. Virology 387 29 40
53. VaucheretHMalloryACBartelDP 2006 AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 22 129 136
54. LiuQZhangYCWangCYLuoYCHuangQJ 2009 Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett 583 723 728
55. ZhouMGuLLPSongXWeiL 2010 Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L . ssp . indica). Front Biol 5 67 90
56. Jones-RhoadesMWBartelDP 2004 Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Mol Cell 14 787 799
57. LlaveCXieZXKasschauKDCarringtonJC 2002 Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 2053 2056
58. AllenEXieZXGustafsonAMSungGHSpataforaJW 2004 Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36 1282 1290
59. ZhaoTLiGLMiSJLiSHannonGJ 2007 A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev 21 1190 1203
60. ThompsonJDHigginsDGGibsonTJ 1994 Clustal-W - Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res 22 4673 4680
61. LacombeSNagasakiHSantiCDuvalDPieguB 2008 Identification of precursor transcripts for 6 novel miRNAs expands the diversity on the genomic organisation and expression of miRNA genes in rice. BMC Plant Bio 8 123
62. SunkarRGirkeTJainPKZhuJK 2005 Cloning and characterization of MicroRNAs from rice. Plant Cell 17 1397 1411
63. SunkarRGirkeTZhuJK 2005 Identification and characterization of endogenous small interfering RNAs from rice. Nucleic Acids Res 33 4443 4454
64. QiXPBaoFSXieZX 2009 Small RNA Deep Sequencing Reveals Role for Arabidopsis thaliana RNA-Dependent RNA Polymerases in Viral siRNA Biogenesis. PloS One 4 e4971
65. WangXBWuQFItoTCilloFLiWX 2010 RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci U S A 107 484 489
66. SzittyaGMoxonSPantaleoVTothGRusholme PilcherRL 2010 Structural and functional analysis of viral siRNAs. PLoS Pathog 6 e1000838
67. Garcia-RuizHTakedaAChapmanEJSullivanCMFahlgrenN 2010 Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22 481 496
68. LinKYChengCPChangBCWangWCHuangYW 2010 Global analyses of small interfering RNAs derived from Bamboo mosaic virus and its associated satellite RNAs in different plants. PloS One 5 e11928
69. NavarroBPantaleoVGiselAMoxonSDalmayT 2009 Deep sequencing of viroid-derived small RNAs from grapevine provides new insights on the role of RNA silencing in plant-viroid interaction. PloS One 4 e7686
70. BolducFHoareauCSt-PierrePPerreaultJP 2010 In-depth sequencing of the siRNAs associated with peach latent mosaic viroid infection. BMC Mol Biol 11 16
71. MolnarASchwachFStudholmeDJThuenemannECBaulcombeDC 2007 miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447 1126 1129
72. ZhangWGaoSZhouXXiaJChellappanP 2010 Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol 11 R81
73. SchnettlerEHemmesHHuismannRGoldbachRPrinsM 2010 Diverging affinity of tospovirus RNA silencing suppressor proteins, NSs, for various RNA duplex molecules. J Virol 84 11542 54
74. CrooksGEHonGChandoniaJMBrennerSE 2004 WebLogo: A sequence logo generator. Genome Res 14 1188 1190
75. MiSJCaiTHuYGChenYHodgesE 2008 Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133 116 127
76. OkamuraKPhillipsMDTylerDMDuanHChouYT 2008 The regulatory activity of microRNA star species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol 15 354 363
77. NobutaKVenuRCLuCBeloAVemarajuK 2007 An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol 25 473 477
78. BlevinsTRajeswaranRShivaprasadPVBeknazariantsDSi-AmmourA 2006 Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34 6233 6246
79. DelerisAGallego-BartolomeJBaoJSKasschauKDCarringtonJC 2006 Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313 68 71
80. HuQHollunderJNiehlAKørnerCJGereigeD 2006 Specific impact of tobamovirus infection on the Arabidopsis small RNA profile. PLoS One 6 e19549
81. ZukerM 2003 Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31 3406 3415
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Crystal Structure of Reovirus Attachment Protein σ1 in Complex with Sialylated OligosaccharidesČlánek A Protein Thermometer Controls Temperature-Dependent Transcription of Flagellar Motility Genes inČlánek Modulation of NKp30- and NKp46-Mediated Natural Killer Cell Responses by Poxviral Hemagglutinin
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 8- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Phenotypic Screens, Chemical Genomics, and Antimalarial Lead Discovery
- Characterisation of Regulatory T Cells in Nasal Associated Lymphoid Tissue in Children: Relationships with Pneumococcal Colonization
- Crystal Structure of Reovirus Attachment Protein σ1 in Complex with Sialylated Oligosaccharides
- Absence of Cross-Presenting Cells in the Salivary Gland and Viral Immune Evasion Confine Cytomegalovirus Immune Control to Effector CD4 T Cells
- Transcriptomic Analysis of Host Immune and Cell Death Responses Associated with the Influenza A Virus PB1-F2 Protein
- A Quorum Sensing Regulated Small Volatile Molecule Reduces Acute Virulence and Promotes Chronic Infection Phenotypes
- Autocrine Regulation of Pulmonary Inflammation by Effector T-Cell Derived IL-10 during Infection with Respiratory Syncytial Virus
- A Protein Thermometer Controls Temperature-Dependent Transcription of Flagellar Motility Genes in
- Association of Human TLR1 and TLR6 Deficiency with Altered Immune Responses to BCG Vaccination in South African Infants
- Histo-Blood Group Antigens Act as Attachment Factors of Rabbit Hemorrhagic Disease Virus Infection in a Virus Strain-Dependent Manner
- MrkH, a Novel c-di-GMP-Dependent Transcriptional Activator, Controls Biofilm Formation by Regulating Type 3 Fimbriae Expression
- Beta-HPV 5 and 8 E6 Promote p300 Degradation by Blocking AKT/p300 Association
- Modulation of NKp30- and NKp46-Mediated Natural Killer Cell Responses by Poxviral Hemagglutinin
- Transportin 3 Promotes a Nuclear Maturation Step Required for Efficient HIV-1 Integration
- Coordination of KSHV Latent and Lytic Gene Control by CTCF-Cohesin Mediated Chromosome Conformation
- A Novel Persistence Associated EBV miRNA Expression Profile Is Disrupted in Neoplasia
- The Plant Pathogen pv. Is Genetically Monomorphic and under Strong Selection to Evade Tomato Immunity
- IL-10 Blocks the Development of Resistance to Re-Infection with
- Anti-Apoptotic Machinery Protects the Necrotrophic Fungus from Host-Induced Apoptotic-Like Cell Death during Plant Infection
- Crystal Structure of PrgI-SipD: Insight into a Secretion Competent State of the Type Three Secretion System Needle Tip and its Interaction with Host Ligands
- Evades Immune Recognition of Flagellin in Both Mammals and Plants
- Tumor Cell Marker PVRL4 (Nectin 4) Is an Epithelial Cell Receptor for Measles Virus
- Provides Insights into the Evolution of the Salmonellae
- B Cell Repertoire Analysis Identifies New Antigenic Domains on Glycoprotein B of Human Cytomegalovirus which Are Target of Neutralizing Antibodies
- Thy1 Nk Cells from Vaccinia Virus-Primed Mice Confer Protection against Vaccinia Virus Challenge in the Absence of Adaptive Lymphocytes
- The Cytokine Network of Acute HIV Infection: A Promising Target for Vaccines and Therapy to Reduce Viral Set-Point?
- Dendritic Cell Status Modulates the Outcome of HIV-Related B Cell Disease Progression
- Differential Contribution of PB1-F2 to the Virulence of Highly Pathogenic H5N1 Influenza A Virus in Mammalian and Avian Species
- A Communal Bacterial Adhesin Anchors Biofilm and Bystander Cells to Surfaces
- Two Group A Streptococcal Peptide Pheromones Act through Opposing Rgg Regulators to Control Biofilm Development
- Activation of HIV Transcription by the Viral Tat Protein Requires a Demethylation Step Mediated by Lysine-specific Demethylase 1 (LSD1/KDM1)
- Unique Evolution of the UPR Pathway with a Novel bZIP Transcription Factor, Hxl1, for Controlling Pathogenicity of
- Disruption of PML Nuclear Bodies Is Mediated by ORF61 SUMO-Interacting Motifs and Required for Varicella-Zoster Virus Pathogenesis in Skin
- Flagellar Motility Is Not Directly Required to Maintain Attachment to Surfaces
- Viral Infection Induces Expression of Novel Phased MicroRNAs from Conserved Cellular MicroRNA Precursors
- Functional Cure of SIVagm Infection in Rhesus Macaques Results in Complete Recovery of CD4 T Cells and Is Reverted by CD8 Cell Depletion
- Recruitment of the Major Vault Protein by InlK: A Strategy to Avoid Autophagy
- The Steroid Catabolic Pathway of the Intracellular Pathogen Is Important for Pathogenesis and a Target for Vaccine Development
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tumor Cell Marker PVRL4 (Nectin 4) Is an Epithelial Cell Receptor for Measles Virus
- Two Group A Streptococcal Peptide Pheromones Act through Opposing Rgg Regulators to Control Biofilm Development
- Differential Contribution of PB1-F2 to the Virulence of Highly Pathogenic H5N1 Influenza A Virus in Mammalian and Avian Species
- Recruitment of the Major Vault Protein by InlK: A Strategy to Avoid Autophagy
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



