-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSLO-1-Channels of Parasitic Nematodes Reconstitute Locomotor Behaviour and Emodepside Sensitivity in Loss of Function Mutants
The calcium-gated potassium channel SLO-1 in Caenorhabditis elegans was recently identified as key component for action of emodepside, a new anthelmintic drug with broad spectrum activity. In this study we identified orthologues of slo-1 in Ancylostoma caninum, Cooperia oncophora, and Haemonchus contortus, all important parasitic nematodes in veterinary medicine. Furthermore, functional analyses of these slo-1 orthologues were performed using heterologous expression in C. elegans. We expressed A. caninum and C. oncophora slo-1 in the emodepside-resistant genetic background of the slo-1 loss-of-function mutant NM1968 slo-1(js379). Transformants expressing A. caninum slo-1 from C. elegans slo-1 promoter were highly susceptible (compared to the fully emodepside-resistant slo-1(js379)) and showed no significant difference in their emodepside susceptibility compared to wild-type C. elegans (p = 0.831). Therefore, the SLO-1 channels of A. caninum and C. elegans appear to be completely functionally interchangeable in terms of emodepside sensitivity. Furthermore, we tested the ability of the 5′ flanking regions of A. caninum and C. oncophora slo-1 to drive expression of SLO-1 in C. elegans and confirmed functionality of the putative promoters in this heterologous system. For all transgenic lines tested, expression of either native C. elegans slo-1 or the parasite-derived orthologue rescued emodepside sensitivity in slo-1(js379) and the locomotor phenotype of increased reversal frequency confirming the reconstitution of SLO-1 function in the locomotor circuits. A potent mammalian SLO-1 channel inhibitor, penitrem A, showed emodepside antagonising effects in A. caninum and C. elegans. The study combined the investigation of new anthelmintic targets from parasitic nematodes and experimental use of the respective target genes in C. elegans, therefore closing the gap between research approaches using model nematodes and those using target organisms. Considering the still scarcely advanced techniques for genetic engineering of parasitic nematodes, the presented method provides an excellent opportunity for examining the pharmacofunction of anthelmintic targets derived from parasitic nematodes.
Published in the journal: . PLoS Pathog 7(4): e32767. doi:10.1371/journal.ppat.1001330
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001330Summary
The calcium-gated potassium channel SLO-1 in Caenorhabditis elegans was recently identified as key component for action of emodepside, a new anthelmintic drug with broad spectrum activity. In this study we identified orthologues of slo-1 in Ancylostoma caninum, Cooperia oncophora, and Haemonchus contortus, all important parasitic nematodes in veterinary medicine. Furthermore, functional analyses of these slo-1 orthologues were performed using heterologous expression in C. elegans. We expressed A. caninum and C. oncophora slo-1 in the emodepside-resistant genetic background of the slo-1 loss-of-function mutant NM1968 slo-1(js379). Transformants expressing A. caninum slo-1 from C. elegans slo-1 promoter were highly susceptible (compared to the fully emodepside-resistant slo-1(js379)) and showed no significant difference in their emodepside susceptibility compared to wild-type C. elegans (p = 0.831). Therefore, the SLO-1 channels of A. caninum and C. elegans appear to be completely functionally interchangeable in terms of emodepside sensitivity. Furthermore, we tested the ability of the 5′ flanking regions of A. caninum and C. oncophora slo-1 to drive expression of SLO-1 in C. elegans and confirmed functionality of the putative promoters in this heterologous system. For all transgenic lines tested, expression of either native C. elegans slo-1 or the parasite-derived orthologue rescued emodepside sensitivity in slo-1(js379) and the locomotor phenotype of increased reversal frequency confirming the reconstitution of SLO-1 function in the locomotor circuits. A potent mammalian SLO-1 channel inhibitor, penitrem A, showed emodepside antagonising effects in A. caninum and C. elegans. The study combined the investigation of new anthelmintic targets from parasitic nematodes and experimental use of the respective target genes in C. elegans, therefore closing the gap between research approaches using model nematodes and those using target organisms. Considering the still scarcely advanced techniques for genetic engineering of parasitic nematodes, the presented method provides an excellent opportunity for examining the pharmacofunction of anthelmintic targets derived from parasitic nematodes.
Introduction
Infections with parasitic nematodes heavily affect the well-being, health, and productivity of humans and animals worldwide [1]. Since the 1960s several broad spectrum anthelmintic compounds have been available. During decades of frequent and sometimes inappropriate use of these anthelmintics, resistance to currently available drugs has developed and is an increasing problem in parasitic nematodes, especially in livestock [2]. In human medicine, where mass anthelmintic treatment programmes were employed during recent years in countries with endemic gastro-intestinal nematode infections, there is also growing concern regarding anthelmintic resistance, and several reports of treatment failure were published during recent years [3]-[6]. In livestock non-chemical worm control procedures such as pasture management, feeding, and breeding are being tested, but they are cost - and labour-intensive and often not practical [7]. In parasites of companion animals, resistance is less common. Nevertheless, populations of the canine hookworm Ancylostoma caninum were recently reported to show high degrees of resistance to pyrantel [8]. Therefore, the need for anthelmintic compounds with new modes of action is urgent.
Recently, three groups of anthelmintic compounds employing new mechanisms of action have been introduced. The oxindole alkaloid paraherquamide was described first in 1981 [9]. Paraherquamide and its derivative 2-deoxoparaherquamide (derquantel) are anthelmintically active by blocking acetylcholine receptors and therefore inhibiting neurotransmission [10], [11]. Derquantel has been launched in combination with abamectin as a drench for sheep in New Zealand in 2010. The combination showed high efficacies against field infections with strongyles in sheep [12]. The second group, comprising the amino-acetonitrile derivatives (AAD), was recently reported to act mainly through the nicotinic acetylcholine receptor ACR-23. This receptor is not present in mammals and is not involved in the action of levamisole, ivermectin, benomyl, dimethyl-4-phenylpiperazinium, and aldicarb. The derivative AAD 1470 was shown to have good efficacy against different species of gastrointestinal nematodes [13]. The first available AAD on the market was AAD 1566 (monepantel), which has been launched as a sheep drench. The third group are the cyclooctadepsipeptides. The parent compound of this class is PF1022A, which was discovered as a fermentation product of the fungus Mycelia sterilia [14]. The semi-synthetic derivative emodepside has a broad spectrum of anthelmintic activity [15], indicating that the mechanism of action might be conserved throughout nematode clades. Emodepside and PF1022A were also shown to be effective against anthelmintic-resistant populations of the sheep nematode Haemonchus contortus and the cattle nematode Cooperia oncophora [16]. Commercially, emodepside was first available as a spot-on preparation in combination with praziquantel for cats. Recently, emodepside has been launched as a tablet for dogs, also in combination with praziquantel.
In Caenorhabditis elegans, emodepside potently inhibits locomotion, egg-laying, and pharyngeal pumping [17]. Previous studies identified nematode latrophilin (LAT-1) as a target for emodepside [18], [19], but LAT-1 is not required for the inhibitory effects of emodepside on locomotion [19], [20]. Indeed, a mutagenesis screen revealed the large conductance calcium-gated potassium channel SLO-1 as a key component for the mechanisms of action of emodepside [20]. SLO-1 channels are regulated by voltage and by intracellular concentration of calcium ions [21]–[24]. They were first identified in experiments with the slowpoke mutant of Drosophila melanogaster, which exhibits abnormal locomotory behaviour and decreased flight ability [22], [24]. In C. elegans, SLO-1 was previously shown to control excitatory neurotransmitter release. It is expressed in the nerve ring and in the body wall muscle [21]. The slo-1 loss-of-function mutants show a characteristic locomotor phenotype consisting of an increase in locomotor reversal frequency [20], [21]. The mutagenesis screen for emodepside-resistant C. elegans mentioned above revealed nine independent lines that were able to move and to reproduce on agar plates with an emodepside concentration as high as 1 µM, a concentration that immobilises wild-type C. elegans. All nine lines fell into a single complementation group that mapped closely to the slo-1 locus on chromosome V. Four of them were sequenced and showed mutations in the slo-1 locus predicted to lead to a reduced or abolished function of the channel. In locomotion assays, the slo-1 mutants had different degrees of resistance to emodepside. Reduction-of-function mutants showed reduced susceptibility to emodepside whilst loss-of-function mutants were not at all inhibited after exposure to emodepside [20]. The putative slo-1 null allele reference strain NM1968 slo-1(js379)V [21] was also highly resistant to emodepside. The expression of slo-1 in slo-1(js379) animals from the pan-neuronal promoter snb-1 [21], [25] and the muscle cell-specific promoter myo-3 [21], [26], either in combination or separately, restored emodepside susceptibility to different degrees [20].
In this study, we identified slo-1 orthologues in H. contortus, A. caninum and C. oncophora. The slo-1 coding sequences of A. caninum and C. oncophora were subsequently expressed in the emodepside-resistant C. elegans strain slo-1(js379) to investigate their ability to rescue emodepside susceptibility of slo-1 loss-of-function mutants. Furthermore, we compared the ability of different C. elegans promoters as well as the slo-1 5′ flanking regions of A. caninum and C. oncophora to drive expression of slo-1 in slo-1 loss-of-function mutants and examined the locomotor phenotype as well as the degree of emodepside susceptibility in the transformants. Finally, we showed that penitrem A, an inhibitor of mammalian SLO-channels [27], is able to antagonise the paralysing effect of emodepside on infective A. caninum larvae as well as on the locomotion of young C. elegans adults in a dose-dependent manner.
Materials and Methods
Parasites
The animals used for the maintenance of the parasitic nematode strains were helminth-free prior to infection. All animals used in this study were handled in strict accordance with good animal practice as defined by the relevant national and local animal welfare bodies, and all animal work was approved by the appropriate committee. Calves were infected with approx. 30,000 C. oncophora third-stage larvae, and sheep were infected with 6,000-8,000 infective larvae of H. contortus. After 21 to 30 days, the animals were necropsied, and the small intestine or the abomasum, respectively, was removed. The worms were either washed off or picked directly from the mucosa. Dogs were infected with 400-500 infective A. caninum larvae. After reaching patency, the dogs were treated with 4 mg/kg arecoline. The subsequently deposited faeces were collected and sieved through a 100 µm mesh sieve. The adult A. caninum were picked directly from the sieve. The recovered parasites were sorted according to sex, washed in 0.9% NaCl solution and subsequently in DEPC-treated water. The worms were frozen at -80°C in sterile GIT buffer (4 M guanidine; 0.1 M Tris, pH 7.5; 1% β-mercapto-ethanol).
Ethics statement
All experiments with animals were performed in strict accordance to the German law for animal welfare (Tierschutzgesetz) and with the approval of the respective local authority, the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit (LAVES) under the reference numbers 01A38, 01A48 and 06A395. All efforts were made to avoid and minimize suffering of the animals.
Sequences and constructs
Total RNA was isolated using Trizol reagent (Invitrogen, Karlsruhe, Germany), according to the manufacturer's recommendations. For cDNA synthesis and Rapid Amplification of cDNA Ends (RACE), the BD SMART RACE cDNA Amplification Kit (Clontech, St-Germain-en-Laye, France) was used following the manual. For isolation of genomic DNA, a standard phenol-chloroform method was used [28]. The GenomeWalker Universal Kit (Clontech) was used to amplify the putative slo-1 promoter regions of A. caninum and C. oncophora. Primers to amplify the putative C. elegans slo-1 promoter region were designed based on the sequence of YAC clone Y51A2D (GenBank Acc. No. AL021497). The first primers for fragments of the slo-1 coding sequence of H. contortus were designed based on EST (Expressed Sequence Tag) sequences revealed by the H. contortus EST Basic Local Alignment Search Tool (BLAST) of the Wellcome Trust Sanger Institute server, using the C. elegans slo-1 sequence (GenBank Acc. No. NM_001029089, accordant with slo-1 splice variant b) as template. The same primers were used to amplify a partial slo-1 coding sequence of C. oncophora. Primers for A. caninum slo-1 were designed based on a partial coding sequence detected in the whole genome shotgun library AIAAGSS 001 using the BLAST application of the Nematode Net [29]. Sequences of primers are given as supporting data, Table S1. PCR products were cloned into the pCR4-TOPO vector, using the TOPO TA Cloning Kit (Invitrogen) or into the pCR-Blunt vector, using the Zero Blunt PCR Cloning Kit (Invitrogen) and transformed into TOP10 Escherichia coli cells (Invitrogen). Vectors containing full-length slo-1 coding sequences were transformed into JM109 E. coli cells (Stratagene, La Jolla, CA, USA). Plasmid DNA preparation was performed using the NucleoSpin Plasmid Kit or the NucleoBond AX 100 Kit (Macherey and Nagel, Düren, Germany). To introduce the required restriction sites, PCR was performed using primers carrying the restriction sites (refer to supporting data, Table S1) with a plasmid, containing the respective full-length sequence, or with cDNA as template. The PCR products were cloned as described above and subcloned into the respective expression vector using T4 DNA ligase (Invitrogen). The basis of the expression plasmids was pBK3.1 [20], [21] (kindly provided by Lawrence Salkoff, Washington University School of Medicine, St. Louis), carrying the C. elegans slo-1 coding sequence downstream of the C. elegans snb-1 promoter, leading to neuron specific expression [21], [25]. The expression plasmids were propagated in XL10-Gold Ultracompetent Cells (Stratagene). The coding sequences of A. caninum and C. oncophora slo-1, respectively, were cloned between the XbaI and BamHI restriction sites within the pBK3.1, thus replacing the C. elegans slo-1 coding sequence. To test the functionality of the slo-1 coding sequences to be analysed in as natural an expression pattern as possible, constructs were built carrying the slo-1 coding sequences downstream of the C. elegans slo-1 promoter. To achieve a construct carrying the C. elegans slo-1 promoter and the C. elegans slo-1 coding sequence, a ligation was set up with three DNA fragments, since the coding sequence of C. elegans slo-1 contained an additional HindIII restriction site: 1) the vector backbone of pBK3.1 digested with BamHI/HindIII, 2) the promoter sequence (HindIII/partial XbaI digest), and 3) the coding sequence of pBK3.1 digested with XbaI/BamHI. The plasmids carrying the parasite slo-1 coding sequences downstream of the C. elegans slo-1 promoter were derived by modifying pBK3.1 constructs which already carried the slo-1 coding sequence of the parasitic nematodes. The snb-1 promoter was excised and replaced by the C. elegans slo-1 promoter sequence using the HindIII and XbaI restriction sites flanking the promoter region. For this purpose, the plasmid carrying the C. elegans slo-1 promoter sequence had to be digested completely with HindIII, but only partially with XbaI, since the promoter sequence had an additional XbaI restriction site. The plasmid carrying the C. oncophora slo-1 coding sequence downstream of the C. elegans slo-1 promoter was not used for functional analysis but as a starting point to construct a plasmid with the C. oncophora slo-1 coding sequence downstream of the C. oncophora slo-1 promoter region (see below). To test the functionality of the parasite promoter sequences, the parasite promoters were used to drive expression of the respective parasite slo-1 in C. elegans. For this purpose, the putative promoters were inserted between the HindIII and XbaI restriction sites in the modified pBK3.1 as described above, replacing the C. elegans slo-1 promoter. Due to additional HindIII and XbaI restriction sites in the C. oncophora slo-1 promoter sequence, the plasmid construction was done by blunt end ligation. All plasmids used for expression experiments in C. elegans were sequenced by custom sequencing (SeqLab Laboratories Goettingen, Germany), ensuring that the coding sequences and the ligation sites were intact. For an overview of constructs used for the transformation experiments refer to Table S2 (supporting data).
Sequences were analysed using the Sci Ed Central Align Plus 5 software, version 5.04 (Scientific and Educational Software; Cary, NC, USA), and the NCBI BLAST [30]. The predicted SLO-1 amino acid sequences and selected sequences of potassium channels of other species revealed by the BLAST search were aligned using the ClustalX2 [31] software package with default settings except that the alignment parameters were changed to BLOSUM. ClustalX2 calculates scores as percentages of the number of identities in the best alignment divided by the number of residues compared, excluding gap positions. The alignment constructed was manually edited and, after elimination of all positions containing gaps, a phylogenetic tree was built using bootstrap analysis (1000 replicates) and the Neighbour Joining method by the Mega4 software package [32] using the default Poisson correction model for multiple substitutions at the same site and assuming homogenous substitution rates for all sites. The analysis of the putative promoter regions was performed using the Genome2Promoter and MatInspector software packages (Genomatix, Munich, Germany). The putative slo-1 promoters of the three nematode species were compared by alignments using the BLAST bl2seq (filter inactivated for low complexity regions) [30].
Maintenance of C. elegans
The C. elegans strains were grown on nematode growth medium (NGM) agar plates containing 50 µl of E. coli (OP50) overnight culture as a food source at 20°C or room temperature. Strains employed were Bristol N2 and NM1968 slo-1(js379)V [21]. The latter contains a mutation within the transmembrane region of the SLO-1 channel which leads to the early termination of the protein and is therefore predicted to encode a non-functional ion-channel. Thus, slo-1(js379) animals show a slo-1 null phenotype due to a translational knock-out.
Preparation of assay plates
Emodepside was prepared as five different stock solutions (2 mM to 200 nM) in ethanol. 0.5 ml of stock solution was added to 100 ml NGM agar after autoclaving and at a temperature of 42°C. Accordingly, control plates were prepared containing 0.5 ml ethanol per 100 ml NGM agar, leading to a final concentration of 85 mM ethanol. This ethanol concentration does not significantly impair C. elegans locomotion [33], [34]. All plates were seeded with 50 µl E. coli OP50. In some of the experiments, agar plates also contained 1 µM penitrem A (Enzo Life Sciences, Lörrach, Germany) in 28 mM DMSO (final concentration) or only the DMSO vehicle as control. For the body bend counts, experiments were performed in the absence of E. coli, i.e. on plain un-seeded NGM plates.
Transformation of C. elegans
Hermaphrodite C. elegans were transformed by microinjection of plasmids into the gonads. Transformation with the differentially modified pBK3.1 plasmids (30 ng/µl) was accomplished by co-injecting the pPD118.33 (Addgene plasmid: 1596; 50 ng/µl) GFP-expressing marker. Successful transformation was determined by identification of the selection marker. For the behavioural and pharmacological analysis only worms carrying the selection marker were used as they were predicted to express the transgene of interest as well.
Confirmation of transcription
To confirm the transcription of the introduced slo-1 coding sequences in transgenic worms, RT-PCR was performed. Total RNA was isolated from a bulk of worms using the TriFast method (PeqLab), and contaminating DNA was removed by a DNase I treatment. 1 µg of total RNA was used for cDNA synthesis (RevertAid First Strand cDNA Synthesis Kit, Fermentas, St.Leon-Rot, Germany), and a -RT control (lacking the Reverse Transcriptase) was performed for each sample. PCR was performed using 1 µl of template in a 25 µl setup (High Fidelity PCR Enzyme Mix, Fermentas, St.Leon-Rot, Germany). Each cDNA was analyzed with all test primer pairs. For primer sequences refer to Table S3 (supporting data).
Behavioural analysis
The C. elegans slo-1 knockout strain NM1968 slo-1(js379)V shows an abnormal behaviour of locomotion in terms of increased reversals, i.e. to stop and reverse direction [21]. To analyse the impact of the heterologously expressed SLO-1 on this behaviour, the number of reversals was counted for all lines. Therefore, a total of 10 L4 stage larvae of each line were selected and placed on an OP50 seeded NGM agar plate. After 24 hours the young adult worms were transferred separately away from the bacterial lawn for one minute to allow removal of bacteria adherent to the worm. Then the worm was put on an un-seeded NGM-agar plate, and, after one minute of acclimatisation, the reversals were counted for 3 min. Numbers of body bends per minute and of reversals in different C. elegans lines were compared using One-Way-ANOVA and individual lines were then compared with Tukey's post hoc test implemented in GraphPad Prism. A p-value <0.05 was considered as statistically significant.
Locomotion assays
For locomotion assays L4 stage larvae of stable lines (at least F2 generation) were used. For each strain (transformed and control strains) ten worms were analysed for each concentration of emodepside (1 nM, 100 nM, 1 µM, and 10 µM, and in case of expression from the parasite promoters also 100 µM) and the ethanol control, respectively. The assays were repeated using two independent stable lines, so that in total 20 worms for each construct and concentration were analysed. The experiments were not repeated for the worms expressing the A. caninum slo-1 from the C. elegans slo-1 and snb-1 promoters due to the lack of sufficient numbers of transformants. The setup for the locomotion assay was as follows: L4 stage larvae of N2, slo-1(js379) and the transformed slo-1(js379) lines were transferred to NGM plates containing E. coli OP50 and either different concentrations of emodepside (10 µM to 1 nM) or ethanol vehicle. Worms were maintained on emodepside or control plates for 24 hours at 20°C and locomotion was examined afterwards. For that purpose, worms were transferred for one minute to plain un-seeded NGM plates to remove bacteria. Subsequently, the worms were transferred to a fresh un-seeded NGM plate and, after one minute, body bends were counted for each worm for another minute. A single body bend is defined as one full sinusoidal movement of the worm. For analysis of a transformant line at a certain concentration of emodepside, N2 and slo-1(js379) worms were tested on the same day as parallel controls.
For statistical comparisons, four-parameter logistic concentration-response-curves with variable slope were fitted using GraphPad Prism 5.0 after plotting the log10 of the emodepside concentration vs. the relative body bend activity at that concentration (percentage of maximum number of body bends in each data set). Bottom values were always constrained to greater than 0. Top values, Hill slopes and EC50 were not constrained. Calculation of means and 90% confidence intervals and statistical tests for differences in 1) EC50, 2) bottom or 3) all four parameters (top, bottom, Hill slope, and EC50) were also done using GraphPad Prism. For slo-1(js379), linear regression including testing for linearity and a significance test for a slope differing from 0 was performed with the same software. Statistical significance was assumed for p<0.05.
Larval migration inhibition assay
Infective larvae of A. caninum (non-exsheathed) were incubated for 24 h at room temperature in 1×PBS buffer containing either penitrem A or the vehicle dimethylsulfoxide (DMSO) in combination with different concentrations of emodepside or the respective vehicle ethanol. Penitrem A (500 µM stock solution in DMSO) was used in a final concentration of 1 µM penitrem A, resulting in a final DMSO concentration of 28 mM (0.2%). Emodepside (1 mM stock solution in ethanol) was used in final concentrations of 1 µM, 5 µM, and 10 µM, respectively. The final ethanol concentration was 170 mM (1%) in these experiments. The concentration of the vehicles was adjusted to the same final concentration in all setups by adding DMSO and/or ethanol. Furthermore, one control was performed without vehicles to estimate the impact of the vehicles. After 24 h, the larvae were used for a modified larval migration inhibition test (LMIT), similar to that described by Demeler et al. [35]. Briefly, 1800 µl containing approximately 100 larvae was pipetted onto precision sieves (mesh size 20 µm) in a 24 well plate. The volume of 1800 µl was sufficient that the sieves were hanging in the liquid and motile larvae were able to penetrate the meshes. After further incubation for 24 h at room temperature, the sieves were removed and the bottom side was carefully rinsed with approximately 300 µl 1×PBS to gather any adherent larvae. Thus, this well contained the migrated larvae. Then, the sieves were turned upside down, and each sieve was rinsed by carefully pipetting 1000 µl 1×PBS through the sieve meshes and collecting the buffer in a so far empty well to recover the non-migrated larvae. For each setup, migrated and non-migrated larvae were counted individually, and the percentage of migrated larvae was calculated as follows:
Each setup was performed in triplicate, and the whole experiment was performed three times in total. The results were compared to each other using a One-Way-ANOVA followed by a Tukey's post hoc test (GraphPad Prism) A p-value <0.05 was considered to be statistically significant.
Accession numbers
Nucleotide sequences: C. elegans YAC clone Y51A2D containing the putative slo-1 promoter region (AL021497); C. elegans slo-1 splice variant b (NM_001029089); partial coding sequence of A. caninum slo-1 (CW974961); partial coding sequence of H. contortus slo-1 (genome version 20060127: contigs >004261, >0045106, >001213, and >057289); A. caninum slo-1 complete coding sequence (EU828635); C. oncophora slo-1 complete coding sequence (EF494185); H. contortus slo-1 complete coding sequence (EF494184);
Proteins sequences: C. elegans SLO-1a (AAL28102); C. elegans SLO-1b (AAL28103); C. elegans SLO-1c (AAL28104); C. briggsae hypothetical protein CBG12923 (XP_001675579.1); A. caninum SLO-1 (EU 828635); C. oncophora SLO-1 (EF494185); H. contortus SLO-1 (EF494184); Ixodes scapularis putative calcium-activated potassium channel (EEC10339.1); Cancer borealis calcium-activated potassium channel (AAZ80093.4); Manduca sexta calcium-activated potassium channel alpha subunit (AAT44358.1); Pediculus humanus corporis putative calcium-activated potassium channel alpha subunit (EEB13088.1); Drosophila melanogaster slowpoke, isoform P (NP_001014652.1); Tribolium castaneum predicted protein similar to slowpoke CG10693-PQ (XP_968651.2); Aplysia californica high conductance calcium-activated potassium channel (AAR27959.1); Xenopus laevis potassium large conductance calcium-activated channel, subfamily M, alpha member 1 (NP_001079159.1); Danio rerio novel calcium activated potassium channel (CAX13266.1); Trachemys scripta calcium-activated potassium channel (AAC41281.1); Gallus gallus calcium-activated potassium channel alpha subunit (AAC35370.1); Monodelphis domestica predicted protein similar to large conductance calcium-activated potassium channel subfamily M alpha member 1 (XP_001367795.1); Mus musculus mSlo (AAA39746.1); Homo sapiens potassium large conductance calcium-activated channel, subfamily M, alpha member 1, isoform CRA_d (EAW54600.1); Bos taurus BK potassium ion channel isoform C (AAK54354.1); Canis familiaris calcium-activated K+ channel, subfamily M subunit alpha-1 (Q28265.2); Strongylocentrotus. purpuratus predicted protein similar to calcium-activated potassium channel alpha subunit (XP_783726.2)
Results
Coding sequences
The search of the Wellcome Trust Sanger Institute H. contortus EST BLAST server using C. elegans slo-1 as template revealed four short fragments of 83 – 150 bp (from the contigs 004261 (two fragments) and 0045106 and 001213) within the coding sequence and a 599 bp fragment containing the last twenty codons of the coding sequence, the stop codon, and part of the 3′ untranslated region (UTR) (from contig 057289). Based on these sequences, primers were designed to amplify the partial coding sequence of H. contortus slo-1. The same primers were used to amplify the respective fragment of C. oncophora slo-1. A partial coding sequence of A. caninum slo-1 was detected in a whole genome shotgun library fragment (GenBank Acc. No.: CW974961) and primers were designed, according to that sequence. RACE-PCR completed the coding sequences and the 5′ and 3′ UTRs. The full-length coding sequences were 3309 bp (EU828635; 1103 predicted amino acids) for A. caninum slo-1, 3333 bp (EF494185; 1111 predicted amino acids) for C. oncophora slo-1, and 3315 bp (EF494184; 1105 predicted amino acids) for H. contortus. GC-contents of the coding sequences were 47.1 – 51.9%, molecular weight and isoelectric point of the proteins were predicted to be 125.02 - 125.88 kDa and 5.77-5.80, respectively. None of the 5′ UTR sequences contained a spliced leader 1 (SL1) sequence. Compared to the predicted sequences of A. caninum and H. contortus SLO-1, C. oncophora SLO-1 had six additional NH2-terminal amino acids. The identities of the nucleotide sequences within the coding region were 80% between A. caninum and C. oncophora, 79% between A. caninum and H. contortus, and 85% between C. oncophora and H. contortus. Based on the predicted amino acid sequences, the identities were 95% between A. caninum and C. oncophora, 95% between A. caninum and H. contortus, and 98% between C. oncophora and H. contortus. The splice variants slo-1a, b, and c of the C. elegans slo-1 cDNA coding sequence were all 73% identical with A. caninum, C. oncophora, and H. contortus slo-1, respectively. Based on predicted amino acid sequences, the identities were 87-88% between C. elegans SLO-1 (splice variants SLO-1 a, b, and c) and all three newly identified parasitic nematode SLO-1 sequences. A phylogenetic tree (Figure 1) shows the relationship of selected SLO channels on the protein level from several animal genera and species. All known nematode SLO-1 orthologues group together: however, within this nematode SLO-1 group, the predicted SLO-1 proteins of the parasitic nematodes cluster in a group distinct from the non-parasitic nematodes C. elegans and Caenorhabditis briggsae. Analysing EST and genome databases for putative SLO-1 orthologues in other nematodes, fragments of coding sequences were identified for a range of species, including Brugia malayi, Trichinella spiralis, Strongyloides ratti, and Trichuris muris (data not shown). As these sequences were either incomplete or of insufficient quality, they were not included in the phylogenetic analysis.
Fig. 1. Phylogenetic tree of SLO-1 amino acid sequences and related potassium channels. 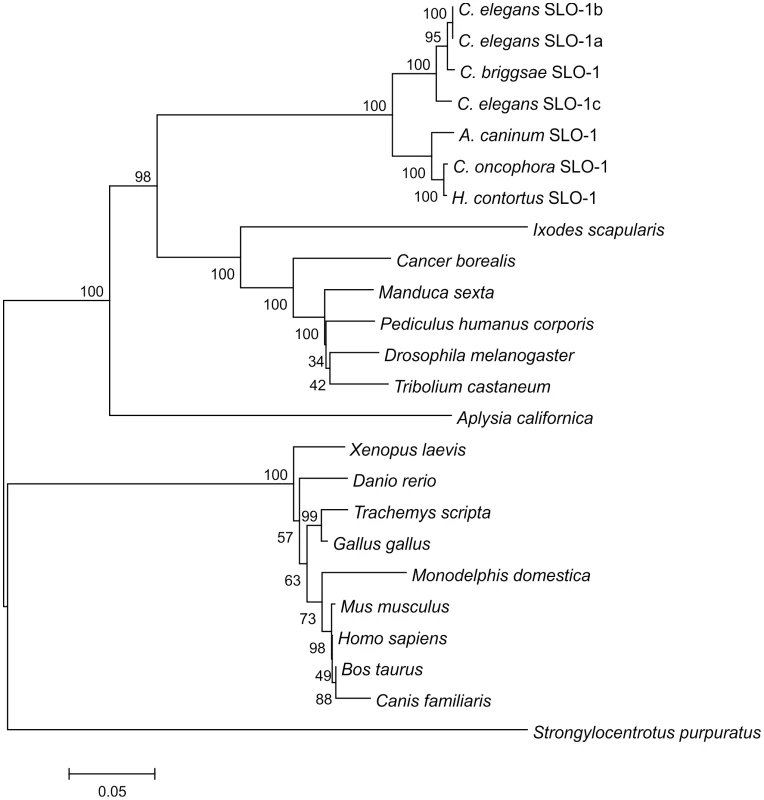
The tree was calculated using Neighbour Joining method. Numbers at the branches indicate bootstrap values (in %, 1000 replicates). The bar shows number of substitutions per mutation site. The selected sequences (GenBank accession numbers in brackets) are as follows: C. elegans SLO-1a (AAL28102); C. elegans SLO-1b (AAL28103); C. elegans SLO-1c (AAL28104); C. briggsae hypothetical protein CBG12923 (XP_001675579.1); A. caninum SLO-1 (EU828635); C. oncophora SLO-1 (EF494185); H. contortus SLO-1 (EF494184); Ixodes scapularis putative calcium-activated potassium channel (EEC10339.1); Cancer borealis calcium-activated potassium channel (AAZ80093.4); Manduca sexta calcium-activated potassium channel alpha subunit (AAT44358.1); Pediculus humanus corporis putative calcium-activated potassium channel alpha subunit (EEB13088.1); Drosophila melanogaster slowpoke, isoform P (NP_001014652.1); Tribolium castaneum predicted protein similar to slowpoke CG10693-PQ (XP_968651.2); Aplysia californica high conductance calcium-activated potassium channel (AAR27959.1); Xenopus laevis potassium large conductance calcium-activated channel, subfamily M, alpha member 1 (NP_001079159.1); Danio rerio novel calcium activated potassium channel (CAX13266.1); Trachemys scripta calcium-activated potassium channel (AAC41281.1); Gallus gallus calcium-activated potassium channel alpha subunit (AAC35370.1); Monodelphis domestica predicted protein similar to large conductance calcium-activated potassium channel subfamily M alpha member 1 (XP_001367795.1); Mus musculus mSlo (AAA39746.1); Homo sapiens potassium large conductance calcium-activated channel, subfamily M, alpha member 1, isoform CRA_d (EAW54600.1); Bos taurus BK potassium ion channel isoform C (AAK54354.1); Canis familiaris calcium-activated K+ channel, subfamily M subunit alpha-1 (Q28265.2); Strongylocentrotus. purpuratus predicted protein similar to calcium-activated potassium channel alpha subunit (XP_783726.2). Analysis of the putative slo-1 promoter sequences
The amplified putative promoter sequences covered approximately 3 kb upstream of the start codon (A. caninum slo-1 promoter 2997 bp, C. oncophora slo-1 promoter 3421 bp, C. elegans slo-1 promoter 3084 bp). The 5′ UTR of A. caninum slo-1 included an intron, which was not present in C. oncophora slo-1. The sequence analysis identified no known promoter elements or transcription factor binding sites in any of the slo-1 promoters employed. Just a few consensus sequences were detected, which might indicate RNA polymerase binding sites. No TATA or CAAT elements could be detected. Comparison of the putative slo-1 promoters of the three nematode species by alignments did not reveal any significant similarities. Comparing the sequences with the respective putative promoter regions of C. briggsae and Caenorhabditis remanei slo-1 (3000 bp upstream of the start codon) also revealed no significant similarities (data not shown).
Confirmation of transcription
In cDNA samples of all analysed transgenic lines, transcripts of the respective expression construct were detected. The primer pairs targeting the expression constructs containing slo-1 coding sequences of the other species gave no amplicon in PCR. In cDNA samples of the slo-1 null mutant strain slo-1(js379) – representing the genetic background of the transgenic strains – and in the Bristol N2 wild-type strain, no transcript of any expression construct could be detected, confirming the authenticity of the PCR results for the transgenic lines. To ensure that the absence of specific PCR products was not due to insufficient RNA-isolation or cDNA-synthesis, a control primer pair was used and gave a PCR product in all analysed cDNA samples (data not shown).
Behavioural phenotype of transgenic C. elegans
In all transgenic strains expressing functional slo-1 from one of the expression constructs, the phenotype of increased reversals exhibited by the slo-1 null mutant strain slo-1(js379) was completely rescued as the rate of reversals was statistically not significantly different (p = 0.87 in a one-way ANOVA) from that observed in Bristol N2 wild-type worms (Figure 2A) but significantly (p<0.001) lower than in mutant slo-1(js379).
Fig. 2. Behavioural phenotype of transgenic C. elegans. 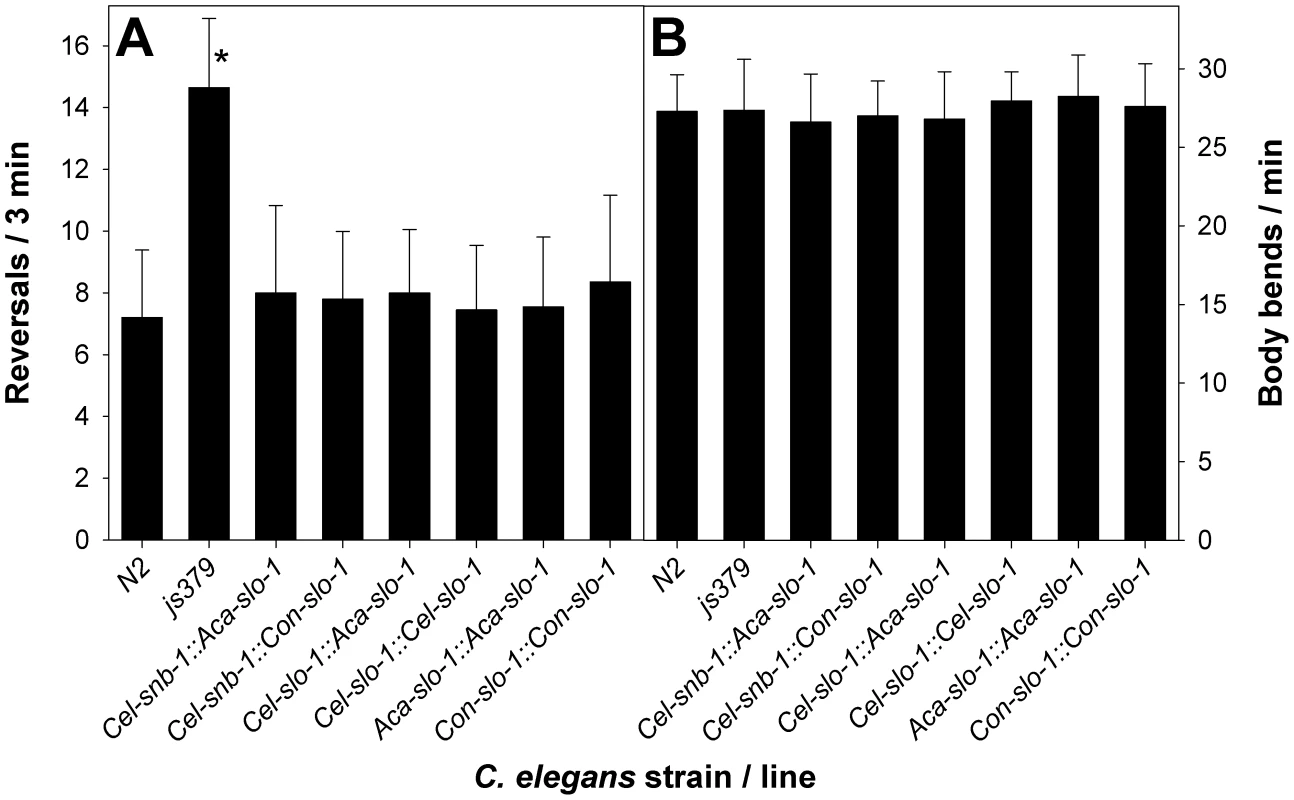
(A) number of reversals in 3 min were counted on NGM agar without bacteria for N2 Bristol, slo-1(js379) and the indicated transgenic lines derived from slo-1(js379). All values are means + SD. An asterisk (*) marks significant differences to all other lines (p<0.001) determined by One-Way-ANOVA followed by Tukey's test. (B) number of body bends per minute counted on NGM agar without bacteria. One-Way-ANOVA revealed no significant differences between different lines. N2, N2 Bristol wild-type strain; js379, slo-1(js379) mutant strain; Cel-snb-1::Aca-slo-1, line expressing A. caninum slo-1 from the C. elegans snb-1 promoter; Cel-snb-1::Con-slo-1, line expressing C. oncophora slo-1 from the C. elegans snb-1 promoter; Cel-slo-1::Aca-slo-1, line expressing A. caninum slo-1 from the C. elegans slo-1 promoter; Cel-slo-1::Cel-slo-1, line expressing C. elegans slo-1 from the C. elegans slo-1 promoter; Aca-slo-1::Aca-slo-1, line expressing A. caninum slo-1 from the A. caninum slo-1 promoter; Con-slo-1::Con-slo-1, line expressing C. oncophora slo-1 under control of the C. oncophora slo-1 promoter. Functional expression of parasitic nematode slo-1 in C. elegans
It was previously shown that C. elegans slo-1 loss-of-function mutants are highly resistant to the inhibition of locomotion behaviour by emodepside [20]. In our study, we expressed slo-1 orthologues of the parasitic nematodes A. caninum and C. oncophora in the emodepside-resistant slo-1(js379) genetic background in order to rescue sensitivity to emodepside and to investigate involvement of these proteins in the mode of action of emodepside. Locomotion was determined by measuring body bends of the worms in the absence of food. By transformation of C. elegans slo-1(js379), stable transgenic lines were obtained expressing 1) A. caninum slo-1 from the neuronal snb-1 promoter, 2) C. oncophora slo-1 from the snb-1 promoter, 3) A. caninum slo-1 from the C. elegans slo-1 promoter, 4) C. elegans slo-1 from the C. elegans slo-1 promoter 5) A. caninum slo-1 from the A. caninum slo-1 promoter, and 6) C. oncophora slo-1 from the C. oncophora slo-1 promoter (an overview is given in supporting data, Table S2). Transgenic lines were analysed for their susceptibility to emodepside. Their locomotion behaviour was compared to that of the wild-type strain Bristol N2 and to that of the loss-of-function mutant slo-1(js379) over a wide range of emodepside concentrations and concentration-response-curves were fitted to the data to allow statistical comparisons.
Animals of all analysed lines showed a comparable basic locomotion, measured as body bends per minute, on the control plates without emodepside (Figure 2B). Locomotion of the slo-1(js379) mutant strain was not at all affected by any of the emodepside concentrations tested (Figure 3) as revealed by concentration-response-curves that are not significantly different from a straight line with slope 0 (p = 0.91). In contrast, locomotion of the Bristol N2 wild-type strain was concentration-dependently inhibited by emodepside. The EC50 for this effect varied between 127.3 nM and 144.2 nM (Table 1) in this set of experiments. At the highest concentration used (10 µM), the Bristol N2 wild-type worms were nearly completely paralysed or dead. The transgenic worms expressing A. caninum (Figure 3A) or C. oncophora (Figure 3B) slo-1 from the snb-1 promoter showed significantly different concentration-response-curves (p<0.0001) with increased susceptibility to emodepside compared to the slo-1(js379) mutant but were not as susceptible as Bristol N2 wild-type animals. Although the EC50 values were not altered, the lines expressing parasitic nematode slo-1 from the snb-1 promoter showed significantly increased bottom values (refer to Table 1) indicating that even extremely high emodepside concentrations were not able to cause complete paralysis. At the highest concentration of 10 µM, worms of the transgenic lines were still able to show nearly half the body bend activity as the ethanol control, while the wild-type worms were almost completely immobilised. Expression of A. caninum slo-1 from the C. elegans slo-1 promoter (Figure 3C) showed a marked susceptibility to emodepside that was equivalent to N2 wild-type worms: worms expressing the parasite slo-1 from the C. elegans slo-1 promoter in slo-1(js379) animals fully restored susceptibility to emodepside as revealed by the absence of any significant differences in top and bottom values, Hill slope or EC50 (Table 1). A comparable effect was observed when the emodepside susceptibility of the slo-1(js379) mutant was rescued through the C. elegans slo-1 from the C. elegans slo-1 promoter (Figure 3D and Table 1).
Fig. 3. Parasite SLO-1 expressed from C. elegans promoters recover emodepside susceptibility in resistant slo-1 loss-of-function mutants. 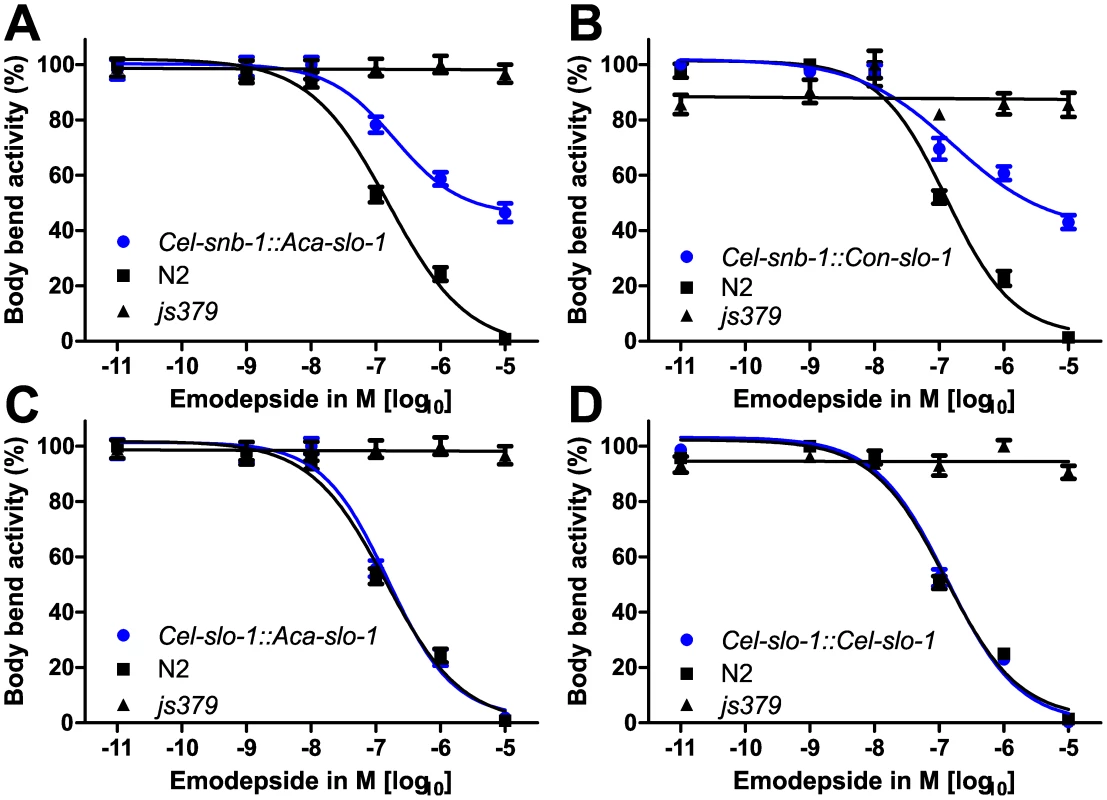
Body bend activity in percent (relative to the highest number of body bends in that group) of young adults after 24 h exposure to emodepside. Comparison of wild-type N2, emodepside-resistant strain slo-1(js379), and transformed slo-1(js379) lines. Error bars represent standard errors of the mean. Dots (•) represent transformed lines, squares (▪) N2 Bristol wild-type strain, triangles (▴) js379(slo-1) mutant strain. (A) Cel-snb-1::Aca-slo-1, line expressing A. caninum slo-1 from the C. elegans snb-1 promoter. (B) Cel-snb-1::Con-slo-1, line expressing C. oncophora slo-1 from the C. elegans snb-1 promoter. (C) Cel-slo-1::Aca-slo-1, line expressing A. caninum slo-1 from the C. elegans slo-1. (D) Cel-slo-1::Cel-slo-1, C. elegans slo-1 expressed from C. elegans slo-1 promoter. Tab. 1. A summary of the pharmacological response to emodepside in transgenic lines expressing either C. elegans or parasite slo-1 under the control of C. elegans-derived promoters. 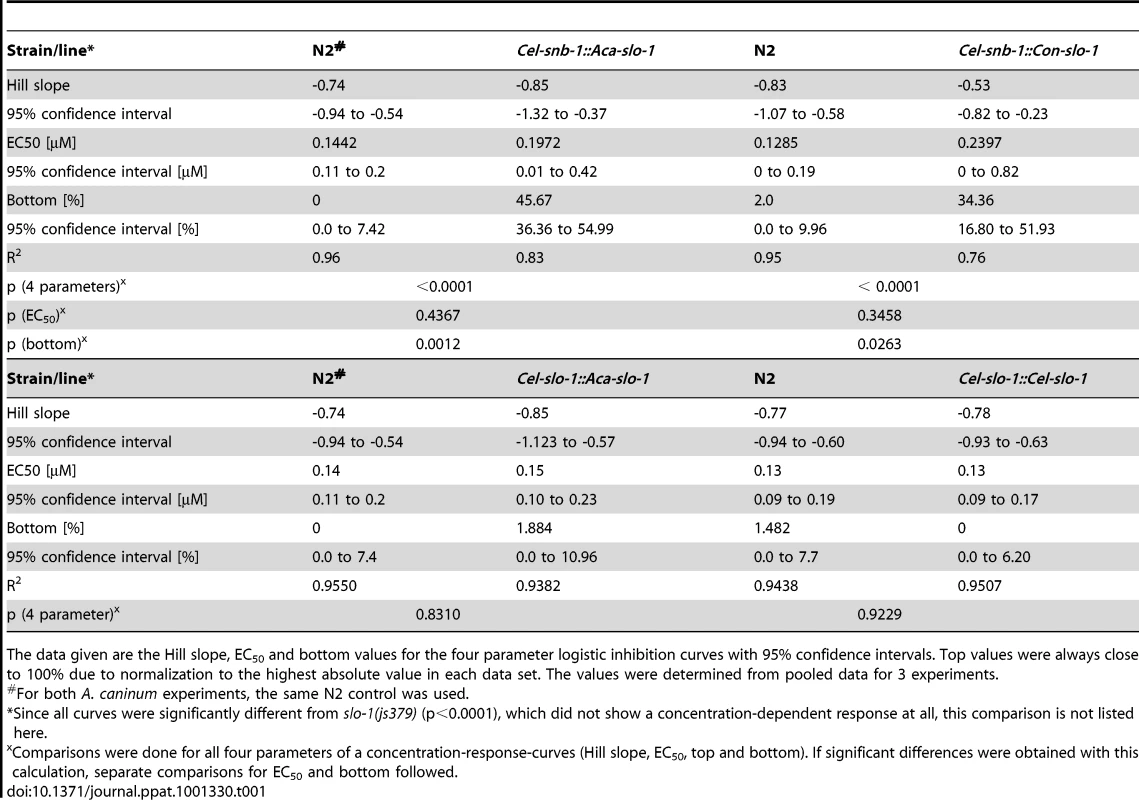
The data given are the Hill slope, EC50 and bottom values for the four parameter logistic inhibition curves with 95% confidence intervals. Top values were always close to 100% due to normalization to the highest absolute value in each data set. The values were determined from pooled data for 3 experiments. Transgenic worms expressing A. caninum or C. oncophora slo-1 from the respective A. caninum or C. oncophora slo-1 promoter showed increased susceptibility to emodepside compared to the slo-1(js379) mutant as well (Figure 4). However, the observed concentration-dependent effects were not as marked as seen for the transgenic worms expressing slo-1 from the C. elegans slo-1 promoter. The lines expressing A. caninum or C. oncophora slo-1 from the A. caninum or C. oncophora slo-1 promoter showed a 62 - and 72-fold higher EC50 than the wild type worms. EC50 and 95% confidence intervals and significance levels for comparisons are given in Table 2.
Fig. 4. Parasite SLO-1 expressed from parasite-derived slo-1 promoters partially recover emodepside susceptibility in resistant slo-1 loss-of-function mutants. 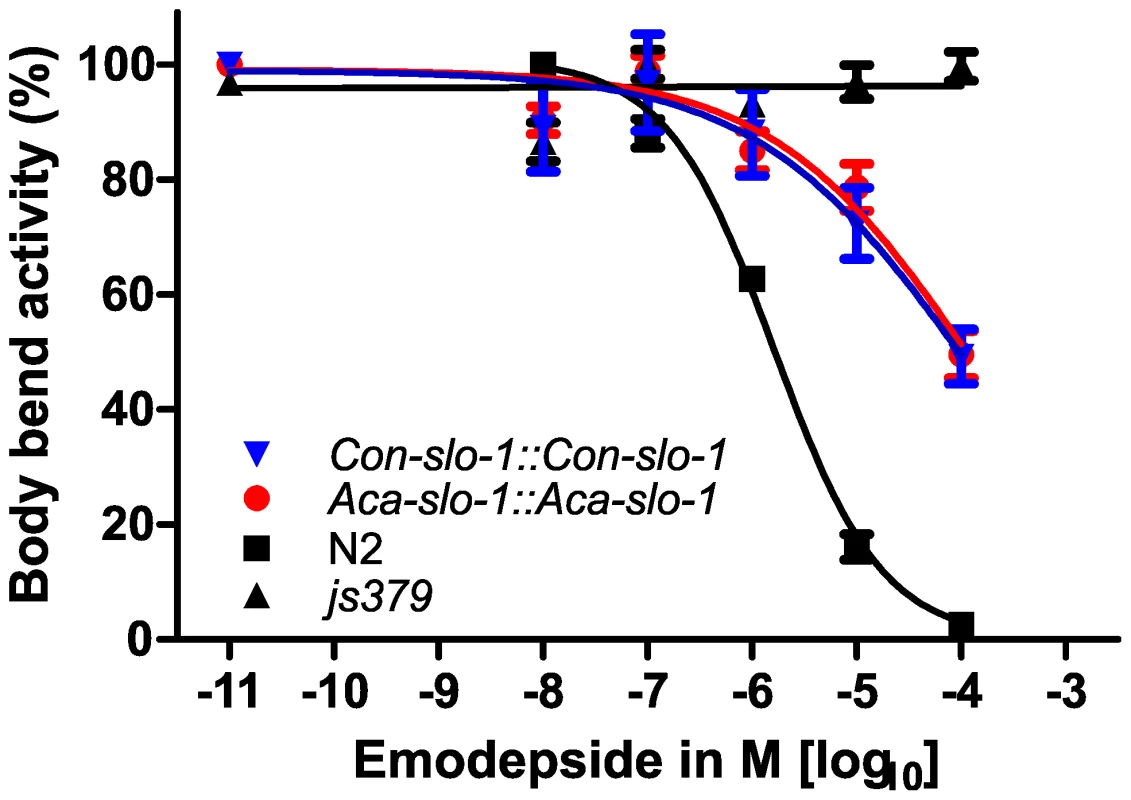
Body bend activity (relative to the highest number of body bends in each group) of young adults after 24 h exposure to emodepside. Comparison of wild-type N2, emodepside-resistant strain slo-1(js379), and transformed slo-1(js379) lines. Dots (•) represent Aca-slo-1::Aca-slo-1 lines (expressing A. caninum slo-1 from the putative A. caninum slo-1 promoter); inverted triangles (▾) Con-slo-1::Con-slo-1 lines (expressing C. oncophora slo-1 from the putative C. oncophora slo-1 promoter); squares (▪) N2 Bristol wild-type strain, triangles (▴) js379(slo-1) mutant strain. Tab. 2. Hill slope, EC50 and bottom value with 95% confidence intervals for transgenic lines expression slo-1 under control of a parasitic nematode-derived promoter. 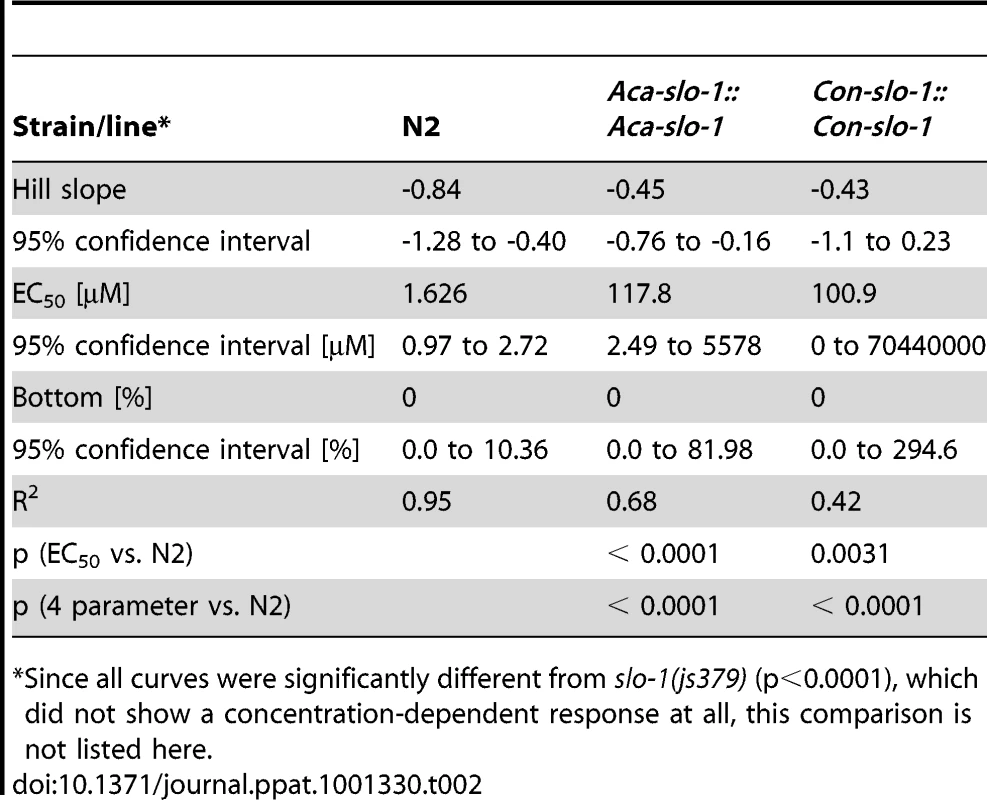
*Since all curves were significantly different from slo-1(js379) (p<0.0001), which did not show a concentration-dependent response at all, this comparison is not listed here. In all experiments, the susceptibility appeared not only as a simple reduction of the number of body bends, but also as an altered pattern of movement, since the worms seemed to be stiffened in the forepart of their body. None of the transformed strains showed coiling as was observed previously at 1 µM emodepside after transformation of slo-1(js379) with pBK3.1, the plasmid containing the C. elegans slo-1 coding sequence and the snb-1 promoter [20]. To conclude, a total functional rescue of the wild-type phenotype regarding the inhibitory effect of emodepside on locomotion was achieved with heterologous slo-1 genes expressed under the control of the C. elegans slo-1 promoter in C. elegans, as revealed by our statistical analysis showing no significant differences in the four parameters of the logistic concentration-response curve. These findings provide evidence that the slo-1 genes cloned from A. caninum and C. oncophora are functional, as well as structural, orthologues of C. elegans slo-1.
Inhibition of endogenous SLO-1 in A. caninum and C. elegans
The vehicles DMSO and ethanol in the concentrations used here did not have any statistically significant effect on the migration of A. caninum larvae through 20 µm meshes. In the presence of emodepside, a concentration-dependent inhibition of migration was observed (Figure 5A). The additional presence of 1 µM penitrem A clearly antagonized the effect of emodepside on migration. The difference in migration of larvae incubated with emodepside either with or without penitrem A was statistically highly significant with p-values of <0.001 for all emodepside concentrations tested. Body-bend assays with C. elegans worms produced highly similar results (Figure 5B).
Fig. 5. Effect of penitrem A and emodepside on nematode locomotion. 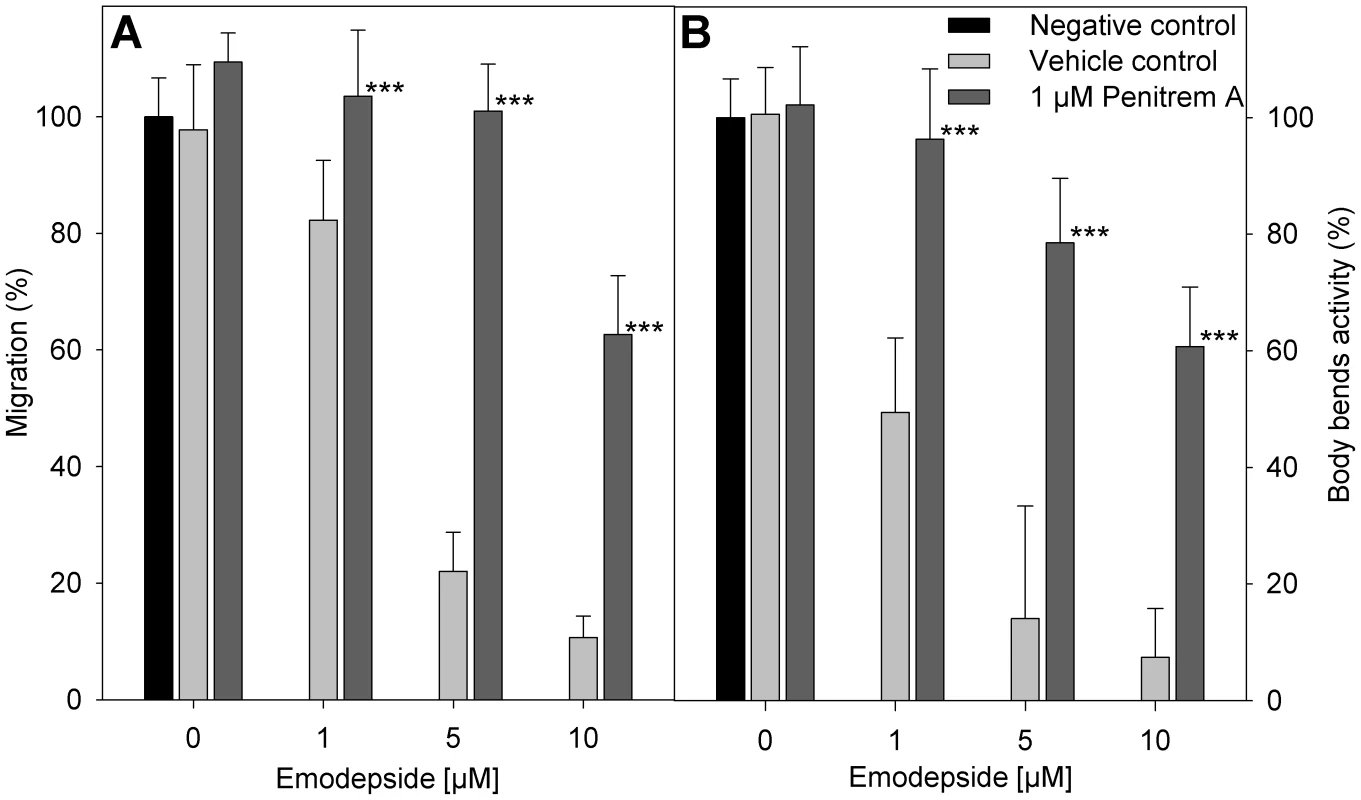
(A) Migration of infective A. caninum larvae (relative to negative control without vehicle) through a 20 µm precision sieve after incubation in different concentrations of emodepside in presence or absence of penitrem A. (B) Body bend activity of C. elegans (relative to negative control without vehicle). (A, B) Negative control (black bar), without vehicle or substance; vehicle control (light grey bars), with 28 mM DMSO, 170 mM ethanol, and the indicated emodepside concentrations; 1 µM penitrem A (dark grey bars), with 1 µM penitrem A, 170 mM ethanol and the indicated emodepside concentrations. Error bars represent standard deviations. Asterisks mark a significant difference between vehicle controls and the experiments with 1 µM penitrem A at the same emodepside concentration (*** p<0.001) determined by One-Way-ANOVA followed by a Tukey's pairwise comparison. Discussion
In the present study, we identified orthologues of the Ca2+-activated K+ (BK) channel C. elegans slo-1 in the parasitic nematodes H. contortus, C. oncophora, and A. caninum. Subsequently, we analysed the ability of A. caninum and C. oncophora slo-1 to functionally rescue emodepside susceptibility in slo-1 knockout mutant C. elegans. The examination of anthelmintic targets of parasitic nematodes is of great importance, since, in contrast to their orthologues in C. elegans, they are the direct targets for drugs used in veterinary and human medicine. Unfortunately, the parasitic stages of the nematodes, which mainly represent the target population for drugs, cannot be examined easily, and especially functional analysis of gene products in parasitic nematodes is usually not feasible. Up to now, parasitic nematodes cannot be maintained in in vitro cultures for their complete life cycle. Therefore, although it has been occasionally successful in some species such as filaria or Strongyloides spp. [36]-[38], genetic engineering, i.e. expression or knockout of genes, in parasitic nematodes is still an unsolved problem [39]. RNAi experiments in parasitic nematodes had very variable outcomes, depending on the target gene, the delivery method, and the species tested [40]–[44]. This might be due to the fact that parasitic nematodes seem to lack orthologues for a transporter responsible for the systemic spread of RNAi in C. elegans, facilitating the accessibility of cells for RNAi in the latter [45]. Therefore, the use of C. elegans as a model and expression system is currently one of the most powerful tools for the functional analysis of genes of parasitic nematodes, especially if the genes have close orthologues in C. elegans [39].
One approach is the overexpression of a parasitic nematode gene in C. elegans with a wild-type genetic background for the respective gene. This approach can be used if the knockout mutant phenotype for the gene to be examined is lethal or not evident. Couthier et al. [46] expressed the H. contortus transcription factor elt-2 ectopically in C. elegans and found that this expression had similar effects as ectopic expression of the endogenous elt-2.
Another experimental setup is exemplified by the experiments described here for slo-1, namely the rescue of the C. elegans loss-of-function mutant by expression of the homologous gene of a parasitic nematode. For that purpose, the mutant should have a clear phenotype and the effects of the rescue should be measurable. Similar experiments examining functionality of parasitic nematode genes in C. elegans have been performed previously. In the study of Kwa et al. [47], ß-tubulin (isotype 1) of H. contortus was expressed in benzimidazole-resistant mutants of C. elegans (TU1054 ben-1(u462)). The benzimidazole-resistance of the ben-1(u462) C. elegans mutants is due to a mutation disrupting the ß-tubulin gene ben-1 [47], [48]. The mutants showed a significantly higher EC50 with regard to the benzimidazole thiabendazole in a larval development inhibition assay compared to the wild-type Bristol N2. In contrast to the resistant ben-1 mutants, H. contortus ß-tubulin expressing ben-1(u462) mutants showed a lower EC50, though not as low as the wild-type larvae [47]. Thus, a total rescue of the wild-type phenotype regarding the effect of thiabendazole on egg-development was not achieved. The effect of expression of H. contortus ß-tubulin on susceptibility of adult ben-1(u462) worms to benzimidazoles has not been reported. Cook et al. [49] expressed the α-subunit of the glutamate-gated chloride channel (GluClα) of H. contortus in C. elegans GluClα mutants, which show a lower sensitivity to ivermectin and a decreased duration of forward movement. Here, a rescue of the wild-type phenotype in respect of the natural locomotion behaviour was observed. However, the effect of ivermectin was not described. Another study showed that expression of the transcription factor of the FOXO/FKH family of Strongyloides stercoralis in C. elegans daf-16 mutants was able to rescue the dauer-forming capability [50]. Very recently, the acetylcholinesterase of the plant-parasitic nematode Globodera pallida was expressed in C. elegans and was shown to functionally rescue the phenotype of the C. elegans double mutant ace-1;ace-2 [51]. In another recent study, Gillan et al. expressed the heat-shock protein 90 (hsp-90) of H. contortus and Brugia pahangi in C. elegans. While expression of H. contortus hsp-90 in C. elegans daf-21 heat shock protein 90 mutants (C. elegans daf-21(nr2081)) partially rescued the phenotype of the mutant, the B. pahangi hsp-90 failed to do so, although the construct was transcribed and translated [52].
The great advantage of C. elegans as an expression system for parasite genes is that posttranslational modifications of recombinantly expressed proteins, which can be necessary for the biological function of the protein, are more conserved between nematodes than between nematodes and standard expression systems [53]. In our experiments, we did not use the recombinantly expressed protein, but the whole transgenic organism to measure the influence of the heterologously expressed proteins on susceptibility to emodepside.
The expression of A. caninum slo-1 and C. oncophora slo-1 in the emodepside-resistant C. elegans slo-1(js379) mutant fully rescued the phenotype of worm locomotion: transgenic worms no longer showed increased reversal movement. These findings indicate a complete functional rescue and at least sufficient expression to restore SLO-1 dependent signalling to wild-type levels in the locomotor circuits. The subsequent pharmacological analysis showed that the transgenesis also rescued the phenotypic behaviour of the animals in terms of inhibited locomotion activity in the presence of emodepside. Animals expressing parasitic nematode slo-1 driven by the snb-1 promoter responded to emodepside in a manner qualitatively similar to wild-type animals, although the inhibition of locomotion was significantly weaker than that of the wild-type worms as determined by counting body bends. No complete paralysis was obtained even with an emodepside concentration that completely paralysed the wild-type animals. This phenotype might reflect the fact that expression of slo-1 was only reconstituted in one of its normal compartments, neuronal cells, whereas it was absent from another compartment, the muscle cells. The findings with parasite slo-1 under control of the snb-1 promoter are similar to previous experiments, in which C. elegans slo-1(js379) mutants were rescued by expression of endogenous slo-1 from the snb-1 promoter [20]. Interestingly, the coiled paralysis of the transgenic C. elegans upon exposure to emodepside observed in earlier experiments with the snb-1 promoter driven expression and also with the combination of snb-1 and myo-3 promoter driven expression of the endogenous slo-1 was not observed in our experiments. The coiling previously observed for slo-1(js379) animals expressing slo-1 from the snb-1 promoter in the presence of 1 µM emodepside was supposed to be due to overexpression or to ectopic expression in neurons usually not expressing slo-1 [20]. The most likely reason for the absence of this phenotype in the present study is the altered plasmid used for transformation. Although the linkage between the promoter and the slo-1 coding sequence was identical for the plasmids carrying the parasite slo-1 and the parental pBK3.1 plasmid used in the previous study, the downstream coding sequence may have influenced the level of expression. While the earlier study by Guest et al. [20] aimed to determine whether the mediation of the effects of emodepside is controlled via a neuronal or a muscular pathway, we were now interested in whether the parasitic nematode SLO-1 channels were also able to act as key components for emodepside action. Therefore, we chose to express the parasite slo-1 not only from the neuronal promoter snb-1, which showed a stronger effect in that former study than the muscle-specific promoter myo-3, but also from the putative endogenous C. elegans slo-1 promoter to achieve a pattern resembling the natural expression pattern, and from the putative parasite slo-1 promoters to test their ability to drive expression in C. elegans. The constructs were designed to be comparable to the pBK3.1 construct, which carries the snb-1 promoter sequence, 2987 bp in size.
The transgenic animals expressing parasitic nematode slo-1 driven by the C. elegans slo-1 promoter were highly susceptible to emodepside, and since their susceptibility was statistically not different from the susceptibility of the wild-type worms, we considered this phenotype as a full rescue. For some drug targets, such as β-tubulin, a single nucleotide polymorphism can abolish their functionality as a drug target [54]. Therefore, the overall sequence identity between parasite and C. elegans SLO-1 orthologues of 87-88% per se did not ensure a conserved function with regard to emodepside. In the study of Gillan et al. the H. contortus hsp-90 sequence showed 88% identity with the C. elegans orthologue, but its expression rescued the mutant phenotype only partially [52]. The finding that expression of slo-1 from different nematode species restored the susceptibility to emodepside in the slo-1(js379) mutants emphasises that the mode of action is most likely conserved between these species. Generally, SLO-1 channels belong to a relatively conserved ion channel family [23]. This was also confirmed by our BLAST search results, which identified channels in very distantly related genera.
The expression of parasite slo-1 under control of the putative slo-1 promoters from A. caninum and C. oncophora aimed to examine the capacity of the parasite-derived promoters to drive expression of the coding sequence of their natural gene within the heterologous background of C. elegans. The transformants showed only partial rescue of emodepside susceptibility. However, in contrast to the lines with snb-1 driven expression, the lines expressing slo-1 from the putative slo-1 promoters of A. caninum and C. oncophora, respectively, did not show increased bottom values. In these experiments the rescued lines clearly had a higher EC50, suggesting that the expression pattern might have been qualitatively restored but that expression levels in general were too low. Since, as was shown in our experiments using the C. elegans slo-1 promoter, the coding sequences of parasite slo-1 appeared to be able to rescue the resistant phenotype completely, the reason for the incomplete rescue is most likely the promoter.
The lack of TATA or CAAT elements which we observed for the slo-1 promoters from A. caninum, C. oncophora as well as from C. elegans is consistent with other studies on nematode promoters and strengthens the assumption that the absence of these elements is a characteristic feature of protein-coding genes of this phylum [26], [55]-[59]. Transcriptional regulatory elements can be located at large distances from the start codon, within intron sequences, and also within the 3′ UTR. Furthermore, expression can be influenced by post-transcriptional regulation, e.g. by microRNAs [60]. Nevertheless, most common reporter gene constructs only use upstream intergenic sequence, and it is recommended to include as much of the upstream sequence as possible. Even so, all phenotypes obtained with such reporter constructs must be interpreted with caution as they may not necessarily reflect the endogenous gene expression pattern [61].
We conclude from the present experiments that the parasite slo-1 promoters drive expression in a functionally appropriate pattern, as the parasite slo-1 expressed from the respective parasite slo-1 promoter qualitatively restored emodepside susceptibility in resistant slo-1(js379) C. elegans. The fact that the emodepside susceptibility of the transformants was significantly lower than in transformants expressing parasite slo-1 from the C. elegans snb-1 or slo-1 promoter, respectively, in turn indicates that the expression pattern obtained with the parasite promoters is not equivalent to that obtained with the C. elegans promoters used in this study. Interestingly, the phenotype of slo-1(js379) C. elegans concerning increased reversals was completely rescued by the parasite slo-1 expressed from the parasite slo-1 promoters. The fact that the rescue regarding emodepside susceptibility was less complete again strengthens the assumption that the spatial pattern or some other characteristics of expression such as expression levels in certain cell types might not have been sufficient to completely fill in the function of the wild-type slo-1 expression. An approach to use the slo-1 promoters of C. elegans, A. caninum, and C. oncophora to express GFP for localisation studies in C. elegans was only partially successful. Within the offspring of the microinjected hermaphrodites only single worms were found exhibiting GFP-expression. Fluorescence was detected as punctate structures in the pharynx region of the transformed animals, indicating expression in pharyngeal neurons, furthermore in the neuron-rich anal region of the worms and in locations consistent with expression in the nerve cords (data not shown). For the C. elegans slo-1 promoter reporter construct, GFP expression was observed in body wall muscle cells within the forepart of the body (data not shown). However, due to the restricted number of observations these investigations thus far do not allow to draw final conclusions and therefore need to be further pursued.
The hypothesis of the functional involvement of SLO-1 in the mechanism of action of emodepside in parasites was further supported by a series of experiments with emodepside and penitrem A. Penitrem A is a tremorgenic mycotoxin known to completely suppress bovine BK channel currents at a concentration of 10 nM [27]. It has also been used as a BK channel inhibitor in a study on muscle fibres of the liver fluke Fasciola hepatica [62]. The concentration in those experiments was 10 µM, but the authors do not report, whether they tested other concentrations. In our experiments, we used penitrem A in a concentration of 1 µM and showed its ability to antagonise the paralysing effect of up to 10 µM emodepside on A. caninum larvae and young C. elegans adults. While lower concentrations of penitrem A (10 nM and 100 nM, data not shown) did not impair the effect of 10 µM emodepside, 1 µM penitrem A antagonised emodepside at all emodepside concentrations analyzed. The need for higher penitrem A concentrations than in experiments with cultured mammalian cells might be explained by a lower accessibility of the target in the intact nematode larvae, e.g. due to the cuticula – at least for the non-feeding A. caninum third-stage larvae. Currently there are no data available on whether penitrem A is indeed also a specific BK channel inhibitor in nematodes and on what penitrem A concentrations are needed for this inhibition. However, the present data show antagonistic effects of emodepside and penitrem A, indicating that both drugs target the same pathway requiring SLO-1.
To conclude, the examination of the actual role of SLO-1 in the signalling of emodepside is still under way. The prevailing view is that emodepside directly or indirectly activates SLO-1 [20], [63]. In contrast to the effects of emodepside on pharyngeal pumping, the effects of emodepside on locomotion are not mediated by the previously described latrophilin-activating pathway [19]. The current model includes latrophilin and SLO-1 for the pharyngeal neurons and SLO-1 but not latrophilin for the body wall musculature [63].
The presented study aimed primarily to test the hypothesis that the mechanism of action of emodepside as far as currently known is conserved in nematodes. Our results are based on functional expression of A. caninum and C. oncophora slo-1 in C. elegans driven by different promoters and demonstrate the ability of the parasitic SLO-1 to act in the mode of action of emodepside. These results are further supported by the experiments with the BK channel inhibitor penitrem A antagonising emodepside. Therefore the current findings suggest that the mode of action is conserved across the three nematode species, providing an important example for functional analysis of the role of individual parasite genes as targets for anthelmintic drugs. Furthermore, these experiments emphasise the potency of C. elegans as an authentic functional model for expression of parasitic nematode genes – at least from clade V – and the subsequent physiological examination of drug/target interactions. Experiments of this type close the gap between research in model organisms and in parasitologically relevant target species. The results presented in this work open new perspectives on functional analysis of parasitic nematode genes in general and in particular allow further analysis of putative targets for emodepside and the elucidation of the mode of action in detail. Transgenic worms from the present study expressing C. elegans slo-1 driven by the C. elegans slo-1 promoter have already been used as a control in a parallel study regarding the expression of the human slo-1 orthologue kcnma1 in C. elegans (Crisford et al., submitted). Another possible application of the system is its use to analyse the impact of certain mutations on emodepside susceptibility, for instance single nucleotide polymorphisms (SNP), identified in resistant populations and suspected to contribute to resistance development. In the long-term, these methods might also enhance development of new anthelmintically active agents.
Supporting Information
Zdroje
1. JasmerDP
GoverseA
SmantG
2003
Parasitic nematode interactions with mammals and plants.
Annu Rev Phytopathol
41
245
270
2. WolstenholmeAJ
FairweatherI
PrichardR
Samson-HimmelstjernaG
SangsterNC
2004
Drug resistance in veterinary helminths.
Trends Parasitol
20
469
476
3. De ClercqD
SackoM
BehnkeJ
GilbertF
DornyP
1997
Failure of mebendazole in treatment of human hookworm infections in the southern region of Mali.
Am J Trop Med Hyg
57
25
30
4. AlbonicoM
BickleQ
RamsanM
MontresorA
SavioliL
2003
Efficacy of mebendazole and levamisole alone or in combination against intestinal nematode infections after repeated targeted mebendazole treatment in Zanzibar.
Bull World Health Organ
81
343
52
5. ReynoldsonJA
BehnkeJM
PallantLJ
MacnishMG
GilbertF
1997
Failure of pyrantel in treatment of human hookworm infections (Ancylostoma duodenale) in the Kimberley region of north west Australia.
Acta Trop
68
301
12
6. FlohrC
TuyenLN
LewisS
MinhTT
CampbellJ
2007
Low efficacy of mebendazole against hookworm in Vietnam: two randomized controlled trials.
Am J Trop Med Hyg
76
732
6
7. BesierB
2007
New anthelmintics for livestock: the time is right.
Trends Parasitol
23
21
24
8. KoppSR
KotzeAC
McCarthyJS
ColemanGT
2007
High-level pyrantel resistance in the hookworm Ancylostoma caninum.
Vet Parasitol
143
299
304
9. YamazakiM
OkuyamaE
KobayashiM
InoueH
1981
The Structure of Paraherquamide, A Toxic Metabolite from Penicillium paraherquei.
Tetrahedron Letters
22
135
136
10. ZinserEW
WolfML
Alexander-BowmanSJ
ThomasEM
DavisJP
2002
Anthelmintic paraherquamides are cholinergic antagonists in gastrointestinal nematodes and mammals.
J Vet Pharmacol Ther
25
241
250
11. LeeBH
ClothierMF
DuttonFE
NelsonSJ
JohnsonSS
2002
Marcfortine and paraherquamide class of anthelmintics: discovery of PNU-141962.
Curr Top Med Chem
2
779
793
12. LittlePR
HodgesA
WatsonTG
SeedJA
MaederSJ
2010
Field efficacy and safety of an oral formulation of the novel combination anthelmintic, derquantel-abamectin, in sheep in New Zealand.
N Z Vet J
58
121
129
13. KaminskyR
DucrayP
JungM
CloverR
RufenerL
2008
A new class of anthelmintics effective against drug-resistant nematodes.
Nature
452
176
180
14. SasakiT
TakagiM
YaguchiT
MiyadohS
OkadaT
1992
A new anthelmintic cyclodepsipeptide, PF1022A.
J Antibiot (Tokyo)
45
692
697
15. HarderA
Samson-HimmelstjernaG
2002
Cyclooctadepsipeptides–a new class of anthelmintically active compounds.
Parasitol Res
88
481
488
16. Samson-HimmelstjernaG
HarderA
SangsterNC
ColesGC
2005
Efficacy of two cyclooctadepsipeptides, PF1022A and emodepside, against anthelmintic-resistant nematodes in sheep and cattle.
Parasitology
130
343
347
17. BullK
CookA
HopperNA
HarderA
Holden-DyeL
2006
Effects of the novel anthelmintic emodepside on the locomotion, egg-laying behaviour and development of Caenorhabditis elegans.
Int J Parasitol
37
627
36
18. SaegerB
Schmitt-WredeHP
DehnhardtM
BentenWP
KruckenJ
2001
Latrophilin-like receptor from the parasitic nematode Haemonchus contortus as target for the anthelmintic depsipeptide PF1022A.
FASEB J
15
1332
1334
19. WillsonJ
AmliwalaK
DavisA
CookA
CuttleMF
2004
Latrotoxin receptor signaling engages the UNC-13-dependent vesicle-priming pathway in C. elegans.
Curr Biol
14
1374
1379
20. GuestM
BullK
WalkerRJ
AmliwalaK
O'ConnorV
2007
The calcium-activated potassium channel, SLO-1, is required for the action of the novel cyclo-octadepsipeptide anthelmintic, emodepside, in Caenorhabditis elegans.
Int J Parasitol
37
1577
1588
21. WangZW
SaifeeO
NonetML
SalkoffL
2001
SLO-1 potassium channels control quantal content of neurotransmitter release at the C. elegans neuromuscular junction.
Neuron
32
867
881
22. AtkinsonNS
RobertsonGA
GanetzkyB
1991
A component of calcium-activated potassium channels encoded by the Drosophila slo locus.
Science
253
551
555
23. SalkoffL
ButlerA
FerreiraG
SantiC
WeiA
2006
High-conductance potassium channels of the SLO family.
Nat Rev Neurosci
7
921
931
24. ElkinsT
GanetzkyB
WuCF
1986
A Drosophila mutation that eliminates a calcium-dependent potassium current.
Proc Natl Acad Sci USA
83
8415
8419
25. NonetML
SaifeeO
ZhaoH
RandJB
WeiL
1998
Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants.
J Neurosci
18
70
80
26. OkkemaPG
HarrisonSW
PlungerV
AryanaA
FireA
1993
Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans.
Genetics
135
385
404
27. KnausHG
McManusOB
LeeSH
SchmalhoferWA
Garcia-CalvoM
1994
Tremorgenic indole alkaloids potently inhibit smooth muscle high-conductance calcium-activated potassium channels.
Biochemistry
33
5819
5828
28. SambrookJ
FritschEF
ManiatisT
1989
Purifications of Nucleic Acids.
Molecular Cloning – A Laboratory Manual, second edition
New York
Cold Spring Harbor Laboratory Press
E3
E4
29. WylieT
MartinJC
DanteM
MitrevaMD
CliftonSW
2004
Nematode.net: a tool for navigating sequences from parasitic and free-living nematodes.
Nucleic Acids Res
32
D423
D426
30. AltschulSF
MaddenTL
SchafferAA
ZhangJ
ZhangZ
1997
Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.
Nucleic Acids Res
25
3389
3402
31. LarkinMA
BlackshieldsG
BrownNP
ChennaR
McGettiganPA
2007
Clustal W and Clustal X version 2.0.
Bioinformatics
23
2947
2948
32. TamuraK
DudleyJ
NeiM
KumarS
2007
MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0.
Mol Biol Evol
24
1596
1599
33. MitchellPH
BullK
GlautierS
HopperNA
Holden-DyeL
2007
The concentration-dependent effects of ethanol on Caenorhabditis elegans behaviour.
Pharmacogenomics J
7
411
417
34. DaviesAG
Pierce-ShimomuraJT
KimH
VanHovenMK
ThieleTR
2003
A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans.
Cell
115
655
666
35. DemelerJ
KüttlerU
Samson-HimmelstjernaG
2010
Adaptation and evaluation of three different in vitro tests for the detection of resistance to anthelmintics in gastro intestinal nematodes of cattle.
Vet Parasitol
170
61
70
36. HigaziTB
MerriweatherA
ShuL
DavisR
UnnaschTR
2002
Brugia malayi: transient transfection by microinjection and particle bombardment.
Exp Parasitol
100
95
102
37. LokJB
MasseyHCJr
2002
Transgene expression in Strongyloides stercoralis following gonadal microinjection of DNA constructs.
Mol Biochem Parasitol
119
279
284
38. JackstadtP
WilmTP
ZahnerH
HobomG
1999
Transformation of nematodes via ballistic DNA transfer.
Mol Biochem Parasitol
103
261
266
39. BrittonC
MurrayL
2006
Using Caenorhabditis elegans for functional analysis of genes of parasitic nematodes.
Int J Parasitol
36
651
659
40. GeldhofP
MurrayL
CouthierA
GilleardJS
McLauchlanG
2006
Testing the efficacy of RNA interference in Haemonchus contortus.
Int J Parasitol
36
801
810
41. IssaZ
GrantWN
StasiukS
ShoemakerCB
2005
Development of methods for RNA interference in the sheep gastrointestinal parasite, Trichostrongylus colubriformis.
Int J Parasitol
35
935
940
42. VisserA
GeldhofP
de MaereV
KnoxDP
VercruysseJ
2006
Efficacy and specificity of RNA interference in larval life-stages of Ostertagia ostertagi.
Parasitology
133
777
783
43. KotzeAC
BagnallNH
2006
RNA interference in Haemonchus contortus: suppression of beta-tubulin gene expression in L3, L4 and adult worms in vitro.
Mol Biochem Parasitol
145
101
110
44. ZawadzkiJL
PresidentePJ
MeeusenEN
De VeerMJ
2006
RNAi in Haemonchus contortus: a potential method for target validation.
Trends Parasitol
22
495
499
45. VineyME
ThompsonFJ
2008
Two hypotheses to explain why RNA interference does not work in animal parasitic nematodes.
Int J Parasitol
38
43
47
46. CouthierA
SmithJ
McGarrP
CraigB
GilleardJS
2004
Ectopic expression of a Haemonchus contortus GATA transcription factor in Caenorhabditis elegans reveals conserved function in spite of extensive sequence divergence.
Mol Biochem Parasitol
133
241
253
47. KwaMS
VeenstraJG
Van DijkM
RoosMH
1995
Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans.
J Mol Biol
246
500
510
48. DriscollM
DeanE
ReillyE
BergholzE
ChalfieM
1989
Genetic and molecular analysis of a Caenorhabditis elegans beta-tubulin that conveys benzimidazole sensitivity.
J Cell Biol
109
2993
3003
49. CookA
AptelN
PortilloV
SineyE
SihotaR
2006
Caenorhabditis elegans ivermectin receptors regulate locomotor behaviour and are functional orthologues of Haemonchus contortus receptors.
Mol Biochem Parasitol
147
118
125
50. MasseyHCJr
BhopaleMK
LiX
CastellettoM
LokJB
2006
The fork head transcription factor FKTF-1b from Strongyloides stercoralis restores DAF-16 developmental function to mutant Caenorhabditis elegans.
Int J Parasitol
36
347
352
51. CostaJC
LilleyCJ
AtkinsonHJ
UrwinPE
2009
Functional characterisation of a cyst nematode acetylcholinesterase gene using Caenorhabditis elegans as a heterologous system.
Int J Parasitol
39
849
858
52. GillanV
MaitlandK
McCormackG
HimNA
DevaneyE
2009
Functional genomics of hsp-90 in parasitic and free-living nematodes.
Int J Parasitol
39
1071
1081
53. MurrayL
GeldhofP
ClarkD
KnoxDP
BrittonC
2007
Expression and purification of an active cysteine protease of Haemonchus contortus using Caenorhabditis elegans.
Int J Parasitol
37
1117
1125
54. KwaMS
VeenstraJG
RoosMH
1994
Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1.
Mol Biochem Parasitol
63
299
303
55. CulettoE
CombesD
FedonY
RoigA
ToutantJP
1999
Structure and promoter activity of the 5′ flanking region of ace-1, the gene encoding acetylcholinesterase of class A in Caenorhabditis elegans.
J Mol Biol
290
951
966
56. DibbNJ
MaruyamaIN
KrauseM
KarnJ
1989
Sequence analysis of the complete Caenorhabditis elegans myosin heavy chain gene family.
J Mol Biol
205
603
613
57. TaweW
WalterRD
Henkle-DuhrsenK
2000
Onchocerca volvulus superoxide dismutase genes: identification of functional promoters for pre-mRNA transcripts which undergo trans-splicing.
Exp Parasitol
94
172
179
58. KrauseS
SommerA
FischerP
BrophyPM
WalterRD
2001
Gene structure of the extracellular glutathione S-transferase from Onchocerca volvulus and its overexpression and promoter analysis in transgenic Caenorhabditis elegans.
Mol Biochem Parasitol
117
145
154
59. Gomez-EscobarN
GregoryWF
BrittonC
MurrayL
CortonC
2002
Abundant larval transcript-1 and -2 genes from Brugia malayi: diversity of genomic environments but conservation of 5′ promoter sequences functional in Caenorhabditis elegans.
Mol Biochem Parasitol
125
59
71
60. AmbrosV
2004
The functions of animal microRNAs.
Nature
431
350
355
61. BoulinT
EtchbergerJF
HobertO
2006
Reporter gene fusions, WormBook Apr
5
1
23
62. KumarD
WhiteC
FairweatherI
McGeownJG
2004
Electrophysiological and pharmacological characterization of K+-currents in muscle fibres isolated from the ventral sucker of Fasciola hepatica.
Parasitology
129
779
793
63. Holden-DyeL
O'ConnorV
HopperNA
WalkerRJ
HarderA
2007
SLO, SLO, quick, quick, slow: calcium-activated potassium channels as regulators of Caenorhabditis elegans behaviour and targets for anthelmintics.
Invert Neurosci
7
199
208
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Innate Immune Sensing of DNA
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 4- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Low Diversity Variety Multilocus Sequence Types from Thailand Are Consistent with an Ancestral African Origin
- Genetic Assignment Methods for Gaining Insight into the Management of Infectious Disease by Understanding Pathogen, Vector, and Host Movement
- Innate Immune Sensing of DNA
- Engineered Resistance to Development in Transgenic
- -Mediated Detoxification of Reactive Oxygen Species Is Required for Full Virulence in the Rice Blast Fungus
- SLO-1-Channels of Parasitic Nematodes Reconstitute Locomotor Behaviour and Emodepside Sensitivity in Loss of Function Mutants
- Structure-Function Analysis of the LRIM1/APL1C Complex and its Interaction with Complement C3-Like Protein TEP1
- A Effector with Enhanced Inhibitory Activity on the NF-κB Pathway Activates the NLRP3/ASC/Caspase-1 Inflammasome in Macrophages
- The MARCH Family E3 Ubiquitin Ligase K5 Alters Monocyte Metabolism and Proliferation through Receptor Tyrosine Kinase Modulation
- is an Unstable Pathogen Showing Evidence of Significant Genomic Flux
- The Cell Wall Protein CwpV is Antigenically Variable between Strains, but Exhibits Conserved Aggregation-Promoting Function
- Engineering HIV-Resistant Human CD4+ T Cells with CXCR4-Specific Zinc-Finger Nucleases
- SUMO-Interacting Motifs of Human TRIM5α are Important for Antiviral Activity
- Completion of Hepatitis C Virus Replication Cycle in Heterokaryons Excludes Dominant Restrictions in Human Non-liver and Mouse Liver Cell Lines
- : Reservoir Hosts and Tracking the Emergence in Humans and Macaques
- On Being the Right Size: The Impact of Population Size and Stochastic Effects on the Evolution of Drug Resistance in Hospitals and the Community
- Bacterial and Host Determinants of MAL Activation upon EPEC Infection: The Roles of Tir, ABRA, and FLRT3
- Respiratory Syncytial Virus Interferon Antagonist NS1 Protein Suppresses and Skews the Human T Lymphocyte Response
- NF-κB Hyper-Activation by HTLV-1 Tax Induces Cellular Senescence, but Can Be Alleviated by the Viral Anti-Sense Protein HBZ
- Human Cytomegalovirus IE1 Protein Elicits a Type II Interferon-Like Host Cell Response That Depends on Activated STAT1 but Not Interferon-γ
- A New Model to Produce Infectious Hepatitis C Virus without the Replication Requirement
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- NF-κB Hyper-Activation by HTLV-1 Tax Induces Cellular Senescence, but Can Be Alleviated by the Viral Anti-Sense Protein HBZ
- Bacterial and Host Determinants of MAL Activation upon EPEC Infection: The Roles of Tir, ABRA, and FLRT3
- : Reservoir Hosts and Tracking the Emergence in Humans and Macaques
- On Being the Right Size: The Impact of Population Size and Stochastic Effects on the Evolution of Drug Resistance in Hospitals and the Community
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



