-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaBacterial and Host Determinants of MAL Activation upon EPEC Infection: The Roles of Tir, ABRA, and FLRT3
Infection of host cells by pathogenic microbes triggers signal transduction pathways leading to a multitude of host cell responses including actin cytoskeletal re-arrangements and transcriptional programs. The diarrheagenic pathogens Enteropathogenic E. coli (EPEC) and the related Enterohemorrhagic E. coli (EHEC) subvert the host-cell actin cytoskeleton to form attaching and effacing lesions on the surface of intestinal epithelial cells by injecting effector proteins via a type III secretion system. Here we use a MAL translocation assay to establish the effect of bacterial pathogens on host cell signaling to transcription factor activation. MAL is a cofactor of Serum response factor (SRF), a transcription factor with important roles in the regulation of the actin cytoskeleton. We show that EPEC induces nuclear accumulation of MAL-GFP. The translocated intimin receptor is essential for this process and phosphorylation of Tyrosine residues 454 and 474 is important. Using an expression screen we identify FLRT3, C22orf28 and TESK1 as novel activators of SRF. Importantly we demonstrate that ABRA (actin-binding Rho-activating protein, also known as STARS) is necessary for EPEC-induced nuclear accumulation of MAL and the novel SRF activator FLRT3, is a component of this pathway. We further demonstrate that ABRA is important for structural maintenance of EPEC pedestals. Our results uncover novel components in pathogen-activated cytoskeleton signalling to MAL activation.
Published in the journal: . PLoS Pathog 7(4): e32767. doi:10.1371/journal.ppat.1001332
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001332Summary
Infection of host cells by pathogenic microbes triggers signal transduction pathways leading to a multitude of host cell responses including actin cytoskeletal re-arrangements and transcriptional programs. The diarrheagenic pathogens Enteropathogenic E. coli (EPEC) and the related Enterohemorrhagic E. coli (EHEC) subvert the host-cell actin cytoskeleton to form attaching and effacing lesions on the surface of intestinal epithelial cells by injecting effector proteins via a type III secretion system. Here we use a MAL translocation assay to establish the effect of bacterial pathogens on host cell signaling to transcription factor activation. MAL is a cofactor of Serum response factor (SRF), a transcription factor with important roles in the regulation of the actin cytoskeleton. We show that EPEC induces nuclear accumulation of MAL-GFP. The translocated intimin receptor is essential for this process and phosphorylation of Tyrosine residues 454 and 474 is important. Using an expression screen we identify FLRT3, C22orf28 and TESK1 as novel activators of SRF. Importantly we demonstrate that ABRA (actin-binding Rho-activating protein, also known as STARS) is necessary for EPEC-induced nuclear accumulation of MAL and the novel SRF activator FLRT3, is a component of this pathway. We further demonstrate that ABRA is important for structural maintenance of EPEC pedestals. Our results uncover novel components in pathogen-activated cytoskeleton signalling to MAL activation.
Introduction
Infection of host cells by pathogenic microbes triggers signal transduction pathways leading to a multitude of host cell responses including actin cytoskeletal re-arrangements and transcriptional programs. This is achieved via the delivery of virulence factors directly into target cells [1]. Often structurally divergent, these effector proteins mimic eukaryotic functions [2] and are usually delivered into the host-cell cytosol by needle-like, type III (T3SS), type IV (T4SS) and type VI (T6SS) secretion systems [3]. These secretion systems are large multi-protein complexes that span the entire cell envelope. More than 25 species of Gram-negative bacteria have a Type III secretion system [4]. Many of the T3SS secreted bacterial virulence factors seem to fall into two general classes: 1) those that indirectly subvert actin dynamics by modulating the host-cell machinery involved in actin organization, or 2) those that directly bind actin [3]. Although the types of virulence factors introduced by various organisms differ, there is a shared theme of the subversion of nucleation promoting factors directly or indirectly via Rho, Rac or Cdc42.
Bacterial pathogens can manipulate a host-cell's cytoskeleton to attach, invade and/or move in the cell. A conserved strategy involves manipulating F-actin by modulating or mimicking G proteins in the host cell. Among transcription factors, Globular (G)-actin to Filamentous (F)-actin changes are sensed by serum response factor (SRF). SRF is a widely expressed transcription factor that controls the expression of many immediate early, muscle-specific and cytoskeletal genes [5], [6]. The activity of SRF is primarily controlled by its interaction with signal-regulated or tissue-specific regulatory cofactors. Two families of signal-regulated cofactors have been identified: the ternary complex factor (TCF) family, which are activated by mitogen activated protein (MAP) kinase phosphorylation [7], and the myocardin-related transcription factors (MRTFs). The MRTFs include Myocardin, MAL (also known as MRTF-A, BSAC or MKL1) and MRTF-B (also called MKL2 or MAL16). Rho-family GTPases and monomeric actin regulate the activity of MAL and MRTF-B [8], [9]. Rho family-mediated changes in actin dynamics are sensed by MAL, which contains G-actin-binding RPEL motifs at the N-terminus. Stimulation of Rho family-GTPases releases MAL from an inhibitory complex with G-actin and strongly activates SRF-regulated transcription [9], [10].
When overexpressed in heterologous systems, a number of wild type proteins involved in RhoGTPase signalling to actin dynamics, including Cdc42, Rac and VASP can activate SRF [8], [11]. However, these results have not been explored in the context of a potential link between bacterial pathogenesis and SRF mediated transcriptional programs. Furthermore understanding actin biology in the context of pathogen triggers also offers insight into regulation of the actin machinery in the host cell. This has previously proven to be a very successful avenue, especially for the study of bacterial factors targeting host cell GTPases. Both cellular and microbiological approaches have brought great insight into the bacterial infection process and host physiology [12], [13].
We developed a screen to identify both bacterial and host-cell factors important for pathogenesis. We use MAL-GFP translocation to establish the effect of bacterial pathogens on actin-mediated, host cell signalling to transcription factors and identify novel host cell factors involved in the maintenance of the EPEC pedestal. Here we report that EPEC infection induces nuclear accumulation of MAL-GFP and subsequent transcription of SRF target genes, in a manner dependent on pedestal formation. The translocated EPEC effector Tir is essential, as is phosphorylation of Tir by host cell kinases. We show that the host gene ABRA (also known as STARS), is necessary for MAL translocation and that FLRT3 is a novel SRF activator that functions as a signalling intermediary between the pedestal and nucleus.
Results
EPEC but not AIEC, S. Typhimurium, or E. coli K12, causes nuclear accumulation of MAL-GFP
SRF activation through the co-factor MAL requires Rho-mediated actin signalling [8]. G-actin binds directly to MAL [9]; extracellular stimuli activate cellular GTPases (Rho, Rac and CDC42) driving actin polymerization and altering the G-/F-actin ratio. This releases MAL, allowing it to accumulate in the nucleus, form a complex with SRF and drive transcription. In many cell types MAL is predominantly cytoplasmic and accumulates in the nucleus only upon stimulation to activate target genes [9], [14]–[16]. Using MAL nuclear accumulation as a readout we developed a microscopy-based screen for SRF activation in epithelial cells (Figure 1A).
Fig. 1. EPEC induces MAL-GFP nuclear accumulation. 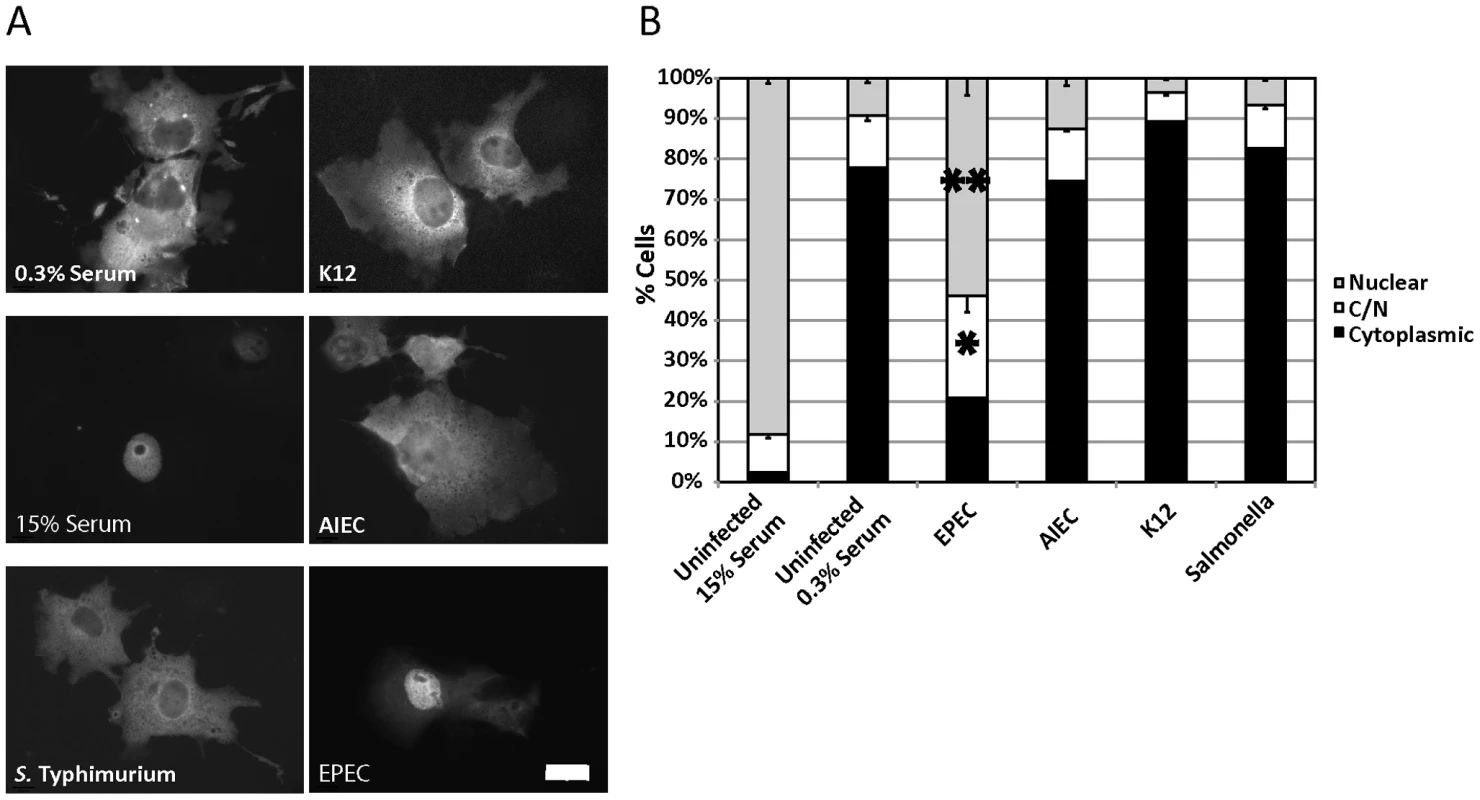
A. EPEC induces nuclear accumulation of MAL-GFP. COS-7 cells were transfected with MAL-GFP, serum starved then stimulated with 15% foetal calf serum (FCS) or infected with the indicated bacteria. B. The percentage of transfected cells from panel A that had MAL-GFP in the nucleus, cytoplasm or both nucleus and cytoplasm was determined. Data are the means of at least 3 experiments where a minimum of 150 transfected cells were counted for each condition of each experiment. Values are means ± SEM. **P = 7.31336−05, *P = 0.027. Scale Bar = 20 µm. To test the effects of bacterial infection on the regulation of the actin cytoskeleton, we screened a panel of gastrointestinal tract-associated bacterial pathogens including Enteropathogenic E. coli (EPEC), Adherent Invasive E. coli (AIEC), Salmonella enterica serovar Typhimurium (S. Typhimurium), and a non-pathogenic E. coli K12 for the ability to induce the nuclear accumulation of the SRF co-factor MAL. COS-7 cells were transfected with MAL-GFP. After 18 hours, the transfected cells were serum starved for a further 24 hours and then infected in DMEM containing 0.3% Foetal Calf Serum (FCS) with bacteria for a total of 5 hours. Following infection, cells were washed, fixed and stained for immunofluorescence. We found that EPEC, but not AIEC, K12 or S. Typhimurium could induce robust nuclear accumulation of MAL-GFP (Figure 1A). The percentage of cells exhibiting nuclear localization and both cytoplasmic and nuclear (C/N) localization of MAL-GFP increased significantly from 9.19%±1.09% to 53.94%±4.21%, and from 13%±1.36% to 25.23±4.02% respectively, when compared to the uninfected 0.3% serum control (Figure 1B). The nuclear localization induced by EPEC was less efficient than that of the 15% serum control (Figure 1A and B). These data suggest that nuclear localisation of MAL is specific to EPEC infection and not merely a general response to host/pathogen interaction or actin-mediated invasion events.
SRF is necessary for EPEC induced MAL-GFP accumulation in the nucleus
MAL (MRTF-A) is a well-described cofactor for Serum Response Factor (SRF). We wanted to confirm that the EPEC-induced nuclear localization of MAL-GFP was actually associated with SRF. To test this we transfected COS-7 cells with siRNA targeting SRF or a non-targeting control siRNA (Invitrogen), and determined the knockdown efficiency by quantitative RT-PCR (Figure 2A).
Fig. 2. SRF is important for EPEC induced MAL-GFP translocation. 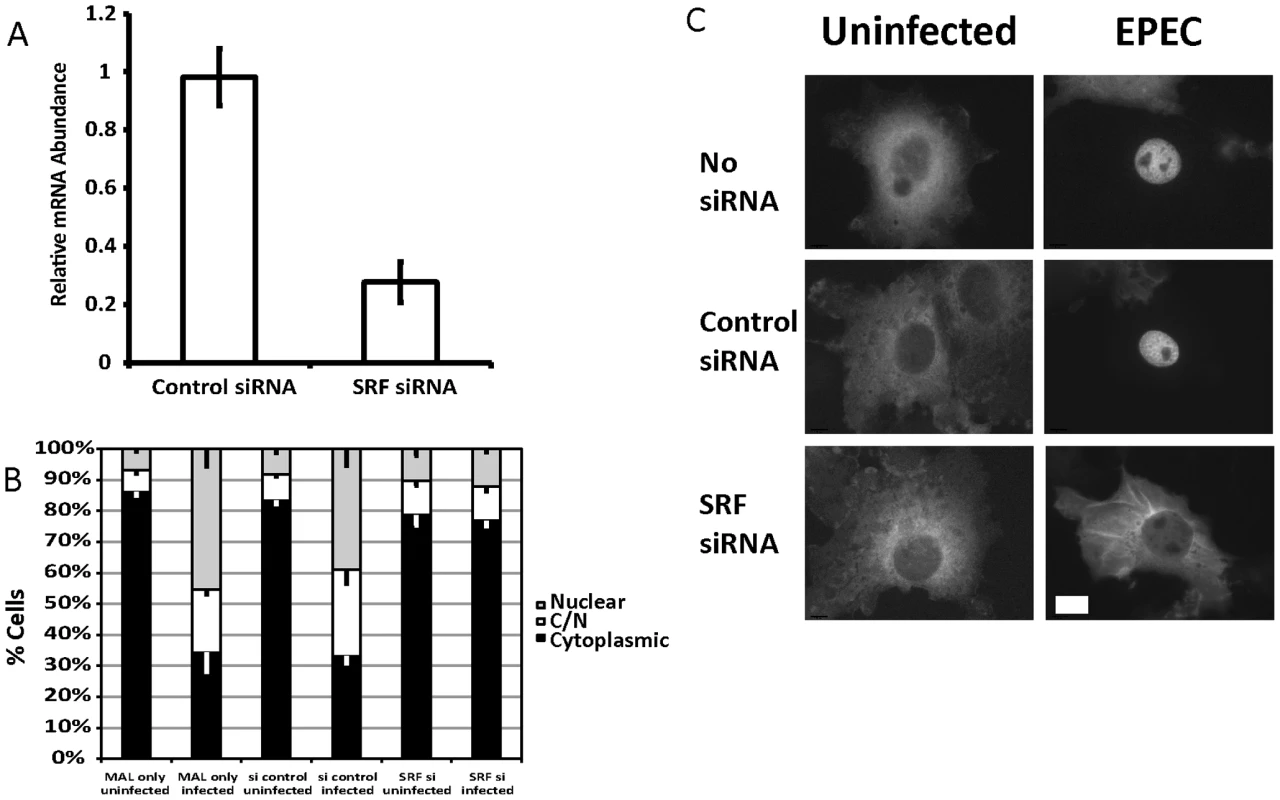
A. COS-7 cells transfected with siRNA targeting SRF or a non-targeting siRNA. After 72 hours knockdown efficiency was assessed using real-time quantitative RT-PCR with SRF specific primers and normalized to GAPDH. Shown are means of three experiments ± standard deviation. B. Infection of SRF-knockdown cells with EPEC results in a significant reduction in nuclear accumulation of MAL-GFP, P = 0.0081 relative to the no siRNA control. Data are the means of at least 3 experiments where a minimum of 150 transfected cells were counted for each condition of each experiment. Values are means ± SEM. C. Representative images of cells counted in panel B. Scale Bar = 20 µm. Infection of SRF-knockdown cells with EPEC resulted in a significant reduction in nuclear accumulation of MAL-GFP to 13.39%±1.61% compared to infection of wild type or non-targeting siRNA transfected COS-7 cells 45.38%±5.5% and 39.04%±5.39% respectively (Figure 2B and C). It is likely that SRF knockdown affects cytoskeletal gene expression, which in turn affects MAL localization.
SRF target genes are activated by EPEC infection
To confirm that MAL functions as a coactivator of SRF during EPEC infection we measured the expression levels of known SRF target genes at 3, 5 and 8 hours post infection (Figure S1). Of those tested Cdc42ep3 (CDC42 effector protein), ARHGDIB (Rho GDP dissociation inhibitor (GDI) beta), Acta2 (Alpha actin 2), Egr2 (Early growth response 2), IL-6 (Interleukin 6) and Vav3 (vav 1 guanine nucleotide exchange factor), were induced by EPEC infection but not by infection with EPEC Δtir (Figures 3 and S1). Fyn, Rsu1 and c-fos were not activated by EPEC infection during the timepoints measured. This data supports the hypothesis that nuclear MAL functions as an SRF cofactor during EPEC infection.
Fig. 3. Transcription of SRF target genes is activated by EPEC infection. 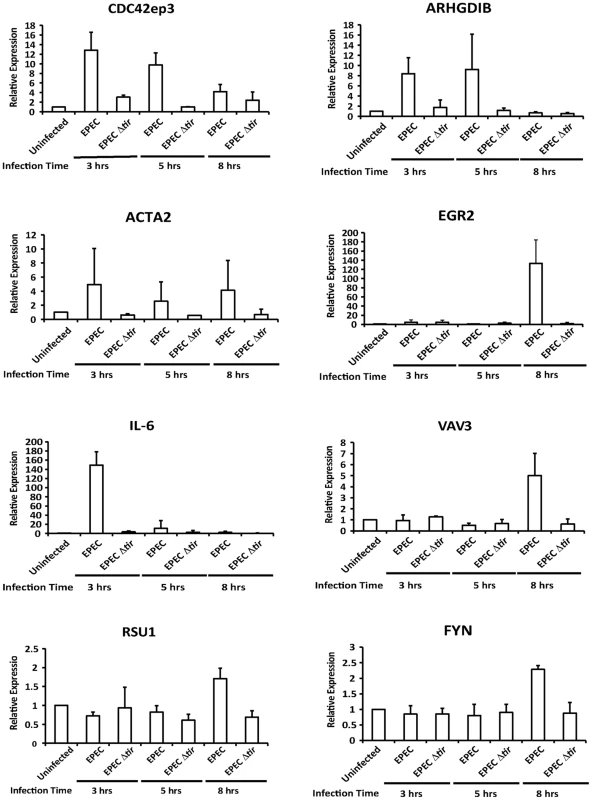
Transcription of SRF target genes measured by quantitative polymerase chain reaction (qRT-PCR). Data are the means of at least 3 experiments ± standard deviation. EPEC-induced MAL-GFP translocation requires Tir and is dependent on phosphorylation of Y454 and Y474
Given the relationship between SRF and actin, and the actin cytoskeleton rearrangements induced by pedestal formation, we hypothesized that pedestal formation would be necessary to induce the observed nuclear accumulation of MAL-GFP. To test this hypothesis we infected COS-7 cells with EPEC Δtir, which are unable to build actin pedestals [17]. In COS-7 cells infected with EPEC Δtir, MAL-GFP remained predominantly cytosolic, with 77.5% of cells±1.75%, displaying no significant difference to the 0.3% FCS control 75.7%±1.46% (Figure 4A and B). To confirm that this loss of phenotype was due solely to the lack of Tir, we infected COS-7 cells with EPEC Δtir rescued with a plasmid carrying Tir (pTir [17]). COS-7 infected with EPEC Δtir/pTIR efficiently rescued the MAL-GFP nuclear accumulation phenotype, with 57.38%±1.73% of cells exhibiting nuclear localization of MAL-GFP (Figure 4A and B). Similar to COS-7 cells infected with wild-type EPEC (60.08%±1.03% nuclear). Therefore the formation of the F-actin-rich pedestal is clearly necessary for EPEC induced MAL-GFP accumulation in the nucleus. This is supported by the fact that no SRF target genes were induced by infection with EPEC Δtir (Figure 3).
Fig. 4. Tir is essential for EPEC induced MAL-GFP translocation. 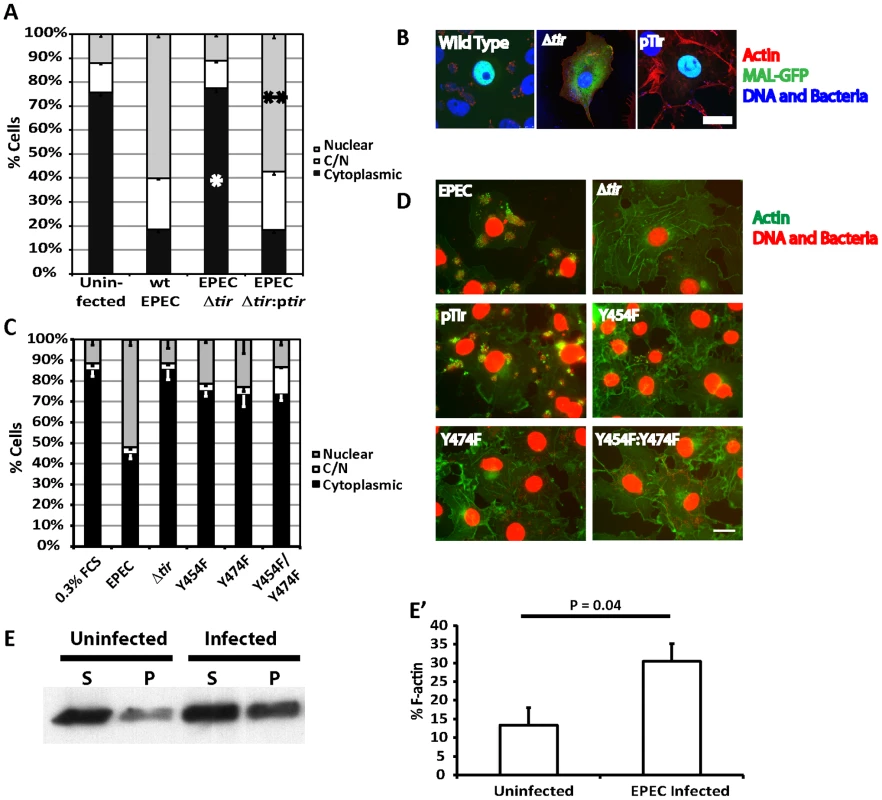
A. EPEC Δtir cannot induce the nuclear translocation of MAL-GFP. COS-7 cells were transfected with MAL-GFP, serum starved then infected with the indicated bacteria. The percentage of transfected cells that had MAL-GFP in the nucleus, cytoplasm or both nucleus and cytoplasm was determined. Data are the means of at least 3 experiments where a minimum of 150 transfected cells were counted for each condition of each experiment. Values are means ± standard deviation. Relative to the uninfected control *P = <0.45, ** P = 7.27−7. B. Representative images from A, MAL-GFP- (green), F-actin (red), DNA/bacteria (blue). C. Phosphorylation of Tir residues Y454 and Y474 is necessary for EPEC-induced MAL-GFP translocation. D. Representative images from C, F-actin (green), DNA/bacteria (Red). Scale Bar = 20 µm. E. Cells untreated or infected with EPEC for 3.5 hours were lysed and separated into 100,000 –g supernatant (S) or pellet (P) fractions. Equal amounts were separated by SDS-PAGE and actin in each fraction detected using an anti-actin antibody. E′ F-∶G-actin ratios from E were quantified as described in materials and methods. The mean % F-actin from at least 3 experiments is shown ± standard deviation. Among all the secreted EPEC effector proteins, only Tir is involved in signalling host cells to generate actin pedestals [18]. Phosphorylated Tir Y474 binds the adapter protein Nck to recruit N-WASP [17], while phosphorylated Y454 stimulates a lower efficiency Nck-independent pathway [19].
To determine if the activation of SRF by pedestal formation was dependent on a specific pathway, i.e. Nck dependent or independent, we infected MAL-GFP expressing COS-7 cells with EPEC Δtir strains rescued with pTIR Y474F, Y454F or Y474F/Y454F mutants and determined the percentage of cells displaying cytosolic or nuclear localization of MAL-GFP. Infecting cells with EPEC Δtir expressing either TirY454F or TirY474F significantly reduced the percentage of cells showing nuclear accumulation of MAL-GFP to 21.2%±0.6% (p = 3.96*10−5) and 23.04%±6.8% (p = 2.9*10−5) respectively, relative to those infected with wild-type EPEC (51.9%±2.9%, Figure 4C and D). The double mutant decreased the percentage of cells with nuclear MAL-GFP further, to a level not significantly different from the EPEC Δtir control, 14.86%±1.26% and 11.39%±1.7% respectively, which suggests that stimulation of actin assembly by Tir is crucial for MAL-GFP nuclear accumulation in response to EPEC infection.
To further understand the Tir requirements for EPEC induced MAL-GFP translocation we expressed a plasma membrane-targeted construct containing the intimin-binding extracellular loop and the COOH-terminal cytoplasmic domain of Tir (TirMC), or a similar plasma membrane-targeted Tir construct in which Tyr474 had been mutated to phenylalanine (TirMC (Y474F)) [18]. These constructs were clustered at the plasma membrane by infecting cells with EPEC Δtir, which cannot induce MAL-GFP translocation. Clustering the COOH terminus of Tir beneath the plasma membrane is sufficient to drive actin pedestal formation [18]. TirMC or TirMC (Y474F) expressing cells were identified by anti-HA fluorescence. 68.2%±6.17% of cells expressing TirMC displayed a nuclear localization of MAL-GFP after 5 hours of infection with EPEC Δtir (Figure S2). In contrast, nuclear localization of MAL-GFP was significantly reduced to 27.9%±4.03% (p = 0.000225) in cells expressing TirMC (Y474F) following infection with EPEC Δtir (Figure S2). These results suggest that the pathway of activation is unimportant, but rather the act of building and maintaining the pedestal is necessary to activate SRF.
EPEC infection in epithelial cells alters F∶G-actin ratios
To test the hypothesis that EPEC-induced nuclear accumulation of MAL is driven by infection-driven changes in G∶F-actin ratios within the host cell, we quantified the G - and F-actin in EPEC infected cells relative to uninfected cells. Cells were extracted with a Triton X-100 lysis buffer (see materials and methods) and separated into 100,000-g supernatant and pellet fractions. Under these conditions G-actin is found in the supernatant and F-actin in the pellet. As shown in figure 4E, at timepoints early in the infection, consistent with the kinetics of pedestal formation in tissue culture cells, we could detect an average 2.3-fold increase in F-actin in EPEC infected cells (Figure 4E).
Together these data demonstrate that pedestal formation can alter G∶F-actin ratios in infected cells and that pedestal formation is necessary for accumulation of MAL-GFP in the nucleus.
EPEC and EHEC Tir components are interchangeable for infection-induced MAL-GFP nuclear accumulation
EPEC and EHEC induce attaching and effacing lesions by different signalling mechanisms. Whereas EPEC Tir is the only translocated EPEC effector required to trigger pedestal formation, EHEC translocates two effectors, Tir(EHEC) and EspFu (also known as Tccp) to generate pedestals in an Nck-independent manner [18], [20]. We reasoned that if the act of building pedestals was enough to activate SRF, then TirEHEC+EspFU would be commensurable to TirEPEC in inducing MAL-GFP nuclear accumulation. As such, we tested to see if TirEHEC could rescue the EPEC Δtir phenotype. COS-7 cells expressing MAL-GFP were infected with EPEC Δtir exogenously expressing TirEHEC (KC12) or TirEHEC+EspFU (KC12/pEspFU) [21]. Post-infection, the cells were fixed, stained and MAL-GFP localization determined by fluorescence microscopy. Under these conditions 39.9%±4.56% of cells infected with KC12/pEspFU exhibited a nuclear localization of MAL-GFP compared to 26.9±2.06% of cells infected with KC12 (Figure 5A and B). The nuclear localization of MAL-GFP induced by KC12 was significantly reduced compared to cells infected with wild-type EPEC (48.4%±4.6%).
Fig. 5. TirEHEC can rescue the EPEC Δtir loss of MAL-GFP translocation. 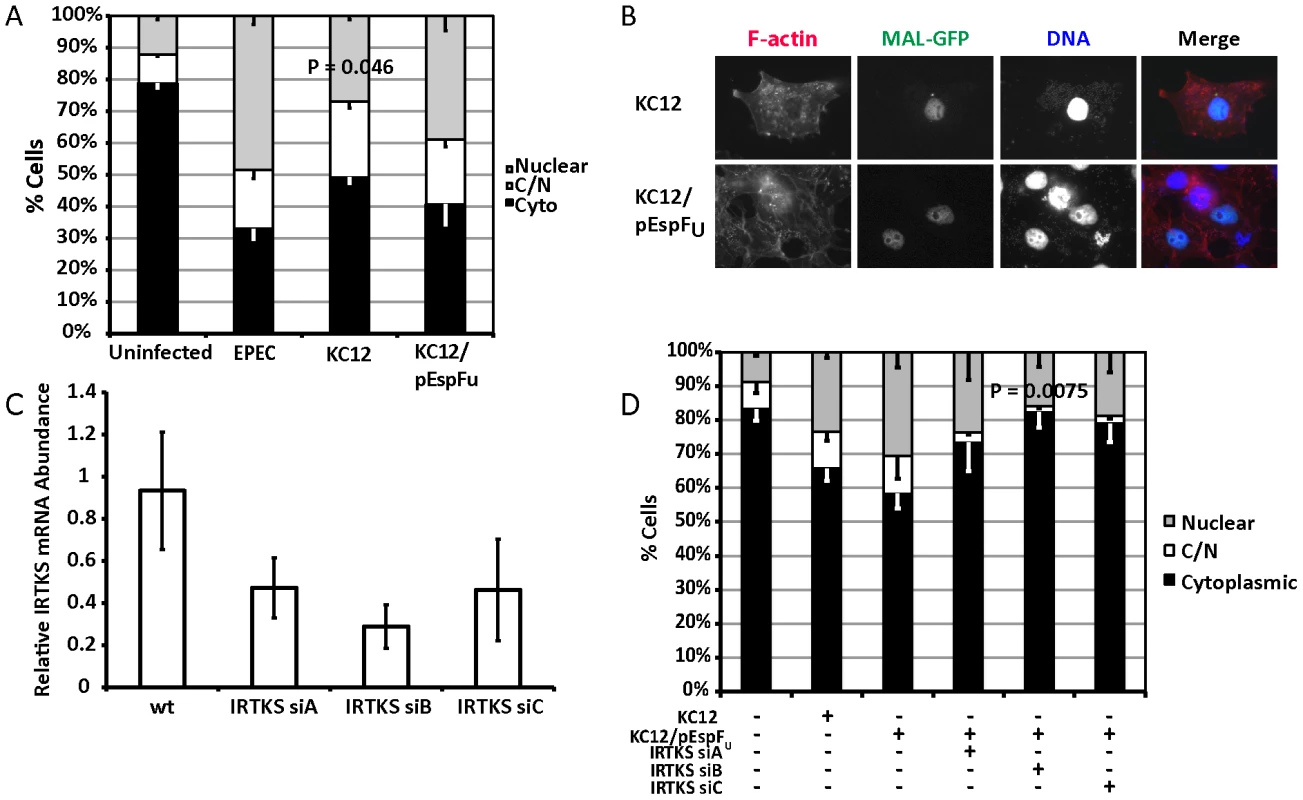
A. Cellular distribution of MAL-GFP in serum-starved COS-7 cells infected with bacteria as indicated. Data are the means of at least 3 experiments where a minimum of 150 transfected cells were counted for each condition of each experiment. Values are means ± standard deviation. B. Representative images of pedestal formation induced by TirEHEC rescue strains. C. COS-7 cells transfected with siRNA targeting IRTKS. After 72 hours knockdown efficiency was assessed using real-time quantitative RT-PCR with IRTKS specific primers and normalized to GAPDH. Shown are means of three experiments ± standard deviation. D. Cellular distribution of MAL-GFP in IRTKS knockdown cells infected with KC12 pEspFU. Data are means of three experiments ± standard deviation. Scale bar = 20 µm. Recent studies have demonstrated that the I-BAR family protein insulin receptor tyrosine kinase substrate (IRTKS) is central to EHEC pedestal formation, forming a ternary complex with TirEHEC, pEspFU and N-WASP necessary for pedestal formation [22], [23]. We tested whether IRTKS was necessary for MAL-GFP translocation in the TirEHEC rescue system. We first confirmed the ability of three siRNAs to knockdown IRTKS in COS-7 cells. siRNA B reproducibly gave the best knockdown (Figure 5C). We tested the ability of EPEC KC12/pEspFU to induce nuclear accumulation of MAL-GFP in the IRTKS knockdown cells. Knockdown of IRTKS significantly reduced the ability of KC12/pEspFU to induce nuclear accumulation of MAL-GFP from 30.5% of cells in the control to 15.9%±1.79% in knockdown cells (Figure 5D).
These data are consistent with significant actin-rearrangement induced by pedestal formation, being central to the nuclear accumulation of MAL-GFP. KC12 are inefficient builders of actin pedestals [21] and, under these conditions, cause very little nuclear translocation of MAL-GFP. However, the rescue expressing TirEHEC and EspFu, the two EHEC effectors required for robust pedestal formation, induces nuclear accumulation of MAL-GFP comparable to wild type EPEC. Secondly IRTKS has been shown to be necessary for efficient pedestal formation by EHEC [22], [23]. In the TirEHEC rescue system used here, knockdown of IRTKS led to reduced pedestal formation and a subsequent lack of MAL-GFP nuclear accumulation and activation of SRF.
ABRA and FLRT3 are SRF activators required for EPEC-induced MAL-GFP translocation
To identify host factors involved in the nuclear translocation of MAL-GFP we tested the ability of a number of known or putative actin binding proteins to induce nuclear translocation of MAL-GFP. Candidate expression plasmids were cotransfected with MAL-GFP into COS-7 cells and the cellular localization of MAL-GFP was determined by fluorescence microscopy. We defined the minimum cut-off point for activation as a 2-fold increase over the vector only control. Both ABRA and SRF were able to significantly induce the nuclear accumulation of MAL-GFP (Figures 6a and S3). 79.9%±6.04% of cells overexpressing ABRA exhibited nuclear localization of MAL-GFP and 87.05%±1.14% of cells overexpressing SRF displayed nuclear localization of MAL-GFP, an 8-fold increase over the vector only control. In addition we, identified three novel genes that could induce nuclear accumulation of MAL-GFP by overexpression. These genes are FLRT3 (32.2% nuclear), TESK1 (54.3% nuclear) and C22orf 28 (35.59% nuclear, Figures 6A and S3).
Fig. 6. ABRA and FLRT3 are SRF activators required for EPEC-induced MAL-GFP translocation. 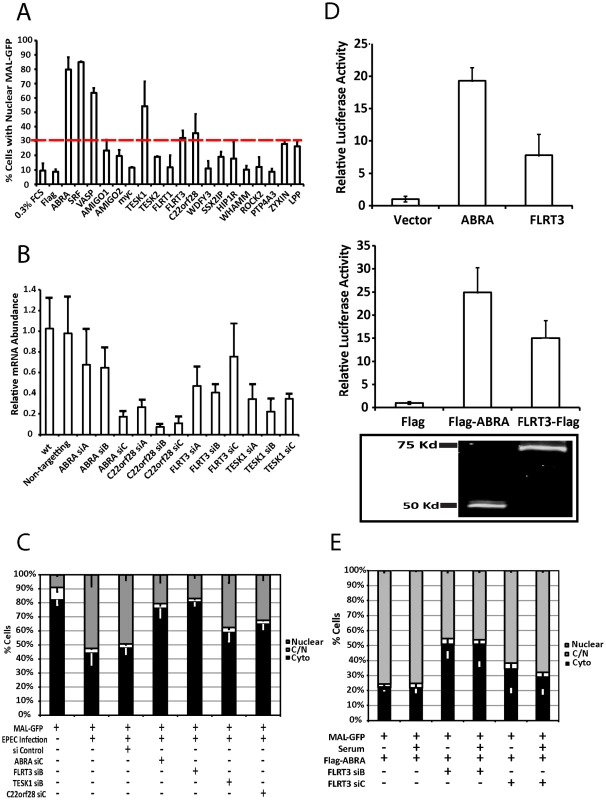
A. The percentage of transfected cells displaying nuclear localization of MAL-GFP after serum starvation. COS-7 cells were cotransfected with MAL-GFP and cDNA expression constructs as indicated. After 18 hours they were serum starved for 24 hours then fixed and stained. The dotted red line represents the cut-off threshold for the percentage of cells displaying nuclear MAL-GFP required for a gene to be declared a hit. The cut-off (30%) was defined as a 2-fold increase over background (10% for 0.3% FCS or empty vector controls). Data represents the mean of 500 cells from 3 individual experiments ± standard deviation. B. COS-7 cells transfected with siRNA targeting the indicated genes or a non-targeting siRNA. After 72 hours knockdown efficiency was assessed using real-time quantitative RT-PCR with FLRT3, TESK1 or C22orf28 specific primers and normalized to GAPDH. Shown are means of three experiments, each using independent cDNA samples, ± standard deviation. C. EPEC-induced nuclear translocation of MAL-GFP is significantly reduced in ABRA (P = 0.0021) and FLRT (P = 0.000363) knockdown cells. Localization of MAL-GFP in serum starved COS-7 cells with respective protein knockdown after infection with EPEC. Data are the means of three experiments ± standard deviation. D. ABRA and FLRT3 induce transcription of SRE-luciferase. E. FLRT3 knockdown significantly reduces ABRA-induced nuclear translocation of MAL-GFP, P = 0.0017 relative to the no siRNA control. Data are the means of three experiments ± standard deviation. ABRA is an actin binding protein that can induce nuclear accumulation of MAL-GFP and activate SRF [24]. C22orf28 (also known as HSPC117 or FAAP in mice) is a cell adhesion protein with Ankyrin repeats, that interacts with vinculin and talin [25]. TESK1 (testis-specific kinase 1) is a LIM kinase-related serine/threonine kinase that has been shown to influence actin organization via its ability to phosphorylate cofillin [26]. FLRT3 (Fibronectin leucine rich transmembrane protein 3) is a putative type I transmembrane protein containing 10 leucine-rich repeats, a fibronectin type III domain, and an intracellular tail. It has been implicated in neurite outgrowth [27] and cell adhesion [28] and has a predicted SRF binding site in its promoter. Furthermore we have recently demonstrated that Flrt3 is induced by bacterial infection [29].
To assess the significance of the overexpression screen hits for EPEC induced activation of SRF, we determined the ability of EPEC to induce MAL-GFP translocation in the absence of each protein individually. Knockdown efficiency was first established for the three candidate genes FLRT3, TESK1 and C22orf28 (Figure 6B). While C22orf28 and TESK1 had no effect, knockdown of ABRA or FLRT3 significantly reduced MAL-GFP nuclear translocation induced by EPEC to 20.6%±3.2% and 16.9±11.16% respectively (Figure 6C). This indicates that ABRA and FLRT3 are both required for EPEC-induced translocation of MAL-GFP to the nucleus. Furthermore we confirmed the ability of ABRA and FLRT3 to activate an SRE-luciferase reporter in the absence of serum (Figure 6D). Both epitope tagged and untagged constructs of ABRA and FLRT could induce transcriptional activity of a luciferase gene under the control of the serum response element. Together these results demonstrate that ABRA and FLRT3 are components of the pathway involved in EPEC induced signaling to SRF.
ABRA-induced GFP-MAL translocation is dependent on FLRT3
As both ABRA and FLRT3 can induce nuclear accumulation of MAL-GFP and are required for EPEC-induced nuclear accumulation of MAL-GFP (Figures 6), we sought to undertake an epistasis analysis of ABRA and FLRT3. We tested to see if FLRT3 knockdown would inhibit ABRA-induced translocation of MAL-GFP. We found that ABRA-induced nuclear accumulation of MAL-GFP was significantly reduced to 45.4%±7.6% of cells in the FLRT3 knockdown cells compared to 75.7%±3.3% in the wild type control (Figure 6E and S2C). In the reciprocal experiment, ABRA knockdown had no effect on FLRT3 induced nuclear accumulation of MAL-GFP (Figure S4). These findings are consistent with FLRT3 functioning downstream of ABRA but upstream of MAL.
Surprisingly FLRT3 siRNA reduces ABRA-induced MAL nuclear localization with or without serum induction under these conditions, whereas FLRT3 siRNA alone has no effect on serum induced MAL nuclear localization (Figure S2C). It therefore appears that the combination of ABRA overexpression and FLRT3 knockdown can block serum induction of MAL.
FRT3 and ABRA localize to the EPEC pedestal and ABRA is necessary for maintenance of discreet pedestals
We next sought to establish the cellular localization of each of the candidate proteins during EPEC infection to determine their involvement in pedestal formation (Figure 7 and S5). ABRA colocalized with F-actin and was enriched in the EPEC pedestal (Figure 7A, arrowheads), while SRF was always localized to the nucleus (Figure S3). FLRT3 localized to the plasma membrane and was enriched at pedestal sites (Figure 7A).
Fig. 7. ABRA is necessary for correct pedestal formation. 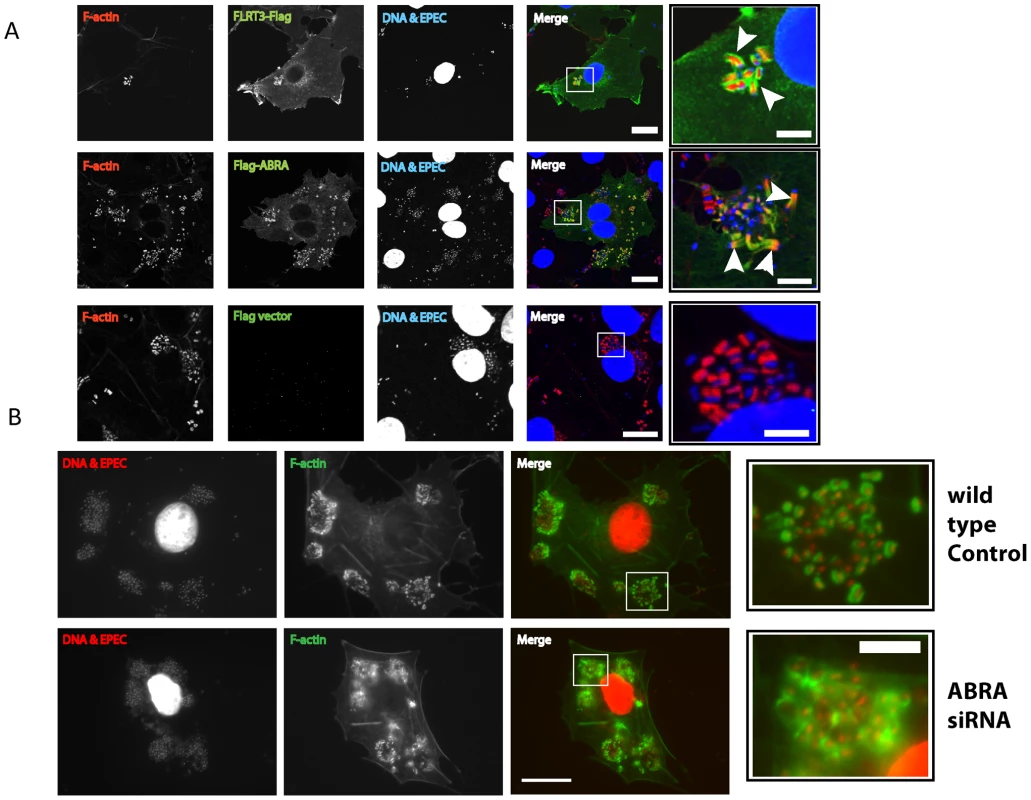
A. COS-7 cells transfected with empty pCMV-3xFlag vector, FLRT3-Flag or Flag-ABRA. Cells were infected with EPEC, fixed and stained with an anti-Flag antibody. FLRT3-Flag and Flag-ABRA are enriched at the EPEC pedestal (Arrowheads). B. Wild-type and ABRA knockdown cells were infected with EPEC for a total of 5 hours, fixed and stained with DAPI (red) and phalloidin (green). Pedestals in ABRA knockdown cells are disorganised (close-up). Scale bars = 20 µm and 5 µm in close-up panels. With ABRA localizing to the EPEC pedestal and knockdown inhibiting MAL-GFP nuclear accumulation, we questioned whether loss of ABRA would also affect EPEC pedestal morphology. Although pedestals associated with single bacteria appeared normal, pedestals associated with micro colonies of EPEC appeared unstructured (Figure 7B and S6), taking on an appearance akin to a ruffle. This suggests that maintenance of discreet pedestals is lost in the ABRA knockdown, with single pedestals merging into one large structure. We therefore suggest that ABRA is necessary for proper pedestal formation, which in turn, is necessary for SRF activation.
Discussion
Recent studies have suggested a connection between pathogen mediated actin re-organization and serum response factor (SRF) transcriptional programs [11]. We screened a panel of gastrointestinal tract-associated pathogens for the ability to induce nuclear accumulation of MAL-GFP. Surprisingly only EPEC caused a significant change in MAL-GFP localization under the infection conditions tested. We suspected S. Typhimurium would have some effect on MAL-GFP localization. It has been shown that S. Typhimurium induces actin ruffles during entry and activates host cell Rho-GTPases [30], [31]. However, unlike EPEC infection, Salmonellae rapidly return the host cell cytoskeleton to its resting state following engulfment, via the action of the effector protein SptP [32]. Perhaps this down-modulation of actin polymerization by S. Typhimurium is sufficient to stifle the activation of SRF, whereas the prolonged actin remodelling induced by EPEC infection is not.
We confirmed that MAL translocation correlates with upregulation of SRF target genes during EPEC infection (Figure 3 and S1). EPEC infection selectively activates SRF target genes, most significantly EGR2 and IL-6, but also CDC42EP3, ARHGDIB, ACTA2 and VAV3, relative to uninfected controls. None of these genes were activated by EPEC Δtir infection. CDC42EP3, ARHGDIB, and VAV3 are all involved in Rho, Rac or Cdc42-mediated signalling and are consistent with the Rho dependent pathway of MAL translocation and SRF activation [7]. Whether upregulation of these genes is required for pedestal formation or pathogen survival, or is a natural consequence of pedestal formation is unclear at this time, but warrants further study.
EGR2 is an immediate-early, zinc finger transcription factor with two serum response elements in its 5′ flanking sequence [33]. EGR2 can be activated by a number of infectious agents including viruses (Human T-cell Leukemia virus type 1), bacteria, and parasites (Toxoplasma gondii) [34]–[36]. Interestingly in T. gondii infection EGR2 induction was dependent on rhoptry secretion, a process analogous to secretion of proteins into a host cell by the bacterial type III secretion system [36]. Likewise, we find the secreted protein Tir to be essential for EPEC-induced activation of EGR2. In other infections EGR2 expression is often accompanied by EGR1 and c-FOS. Under our experimental conditions the expression of EGR1 and c-FOS was not induced. This may suggest that this is an EPEC-specific response rather than a general innate pathogen response.
It is clear that host signalling pathways are activated in response to many infectious agents, suggesting they are functioning in innate immunity. Although IL-6 is a well-known SRF target [10], [37] its expression can be induced by a number of bacteria [38], it is possible therefore, that IL-6 may function as an innate sentinel in this context. The fact that none of these genes were induced by infection with EPEC Δtir demonstrates that pedestal formation is fundamental to this signalling cascade.
Tir is an essential effector for the assembly of F-actin pedestals. Following secretion, Tir inserts into the host cell membrane, presenting an extracellular domain that binds the bacterial surface protein intimin [39]. The C-terminal region of TirEHEC is phosphorylated at Tyr474 by host-cell kinases [40] in a manner similar to host receptor phosphorylation [41], [42]. Phosphorylated Y474 and its flanking residues bind Nck via its SH2 domain [17], [43]. Nck subsequently recruits and activates N-WASP stimulating ARP2/3 driven F-actin assembly. In addition, TirEPEC can promote weak actin polymerization in an Nck-independent manner via phosphorylation of Tir residue Y454 [19]. In this report we show that Tir is essential for EPEC-induced MAL-GFP nuclear accumulation and subsequent transcriptional activation of selective SRF target genes. Infection of epithelial cells with EPEC Δtir does not induce MAL-GFP nuclear accumulation, but this phenotype is rescued by the exogenous expression of Tir (Figure 4A). This is consistent with actin rearrangement driven by pedestal formation being key for SRF activation rather than a translocated effector activating SRF directly. In further support of this idea TirEHEC+pEspFU could also rescue the EPEC Δtir phenotype (Figure 5). TirEHEC is functionally divergent from TirEPEC [17], [44], [45]. TirEHEC lacks a residue equivalent to Tyr474 [40], is not tyrosine phosphorylated in cells [46] and does not bind Nck [43]. To efficiently form actin pedestals EHEC requires a second translocated effector EspFU (TccP) [20], [21]. EspFU is recruited indirectly to Tir by IRTKS [22], [23], where it can than activate N-WASP which results in actin polymerization. Although the initial signalling methods used to recruit and activate host cell nucleation factors between the related pathogens are different, the net result is the same. Likewise, single mutations of either TirEPEC Y454 or Y474 to non-phosphorylatable phenylalanines drastically reduced the nuclear accumulation of MAL-GFP to similar levels (Figures 4C and S2), suggesting that Nck dependent or independent activation of N-WASP is irrelevant to EPEC-induced MAL-GFP nuclear accumulation. In addition, knockdown of SRF reduced EPEC-induced MAL-GFP accumulation in the nucleus to near uninfected levels (Figure 2). This is likely the result of altered cytoskeletal gene expression, resulting from the loss of SRF.
In order to identify the host signaling cascades that are co-opted by bacterial virulence factors to regulate the cytoskeleton, we sought a scheme to identify genes generally employed in mammalian cytoskeleton control. We picked known and putative actin-associated or regulatory genes and tested their ability to induce nuclear accumulation of MAL-GFP. Novel genes inducing MAL-GFP nuclear accumulation with probable involvement in actin-cytoskeletal rearrangement were then evaluated for involvement in host-pathogen interactions.
We identified FLRT3, TESK1 and C22orf28 as novel inducers of MAL nuclear accumulation and confirmed the involvement of FLRT3 in EPEC induced MAL translocation by siRNA (Figure 6). Overexpression of ABRA induced nuclear accumulation of MAL-GFP consistent with published data for the Murine homologue STARS [14], [24]. Knockdown of ABRA significantly decreased EPEC induced accumulation of MAL-GFP in the nucleus, suggesting that ABRA is a necessary component in the signaling pathway. In addition we found ABRA was enriched in EPEC pedestals and that ABRA knockdown adversely affected pedestal morphology. STARS has been shown to activate SRF and stabilize the F-actin cytoskeleton in a RhoA dependent manner, with the carboxy terminal being sufficient and necessary to activate SRF and bind actin [24]. The pedestal phenotype observed in ABRA knockdown cells is consistent with ABRA stabilizing the F-actin cytoskeleton in this context (Figure 7B). Loss of this stabilization function in microcolonies leads to the dissolution of discreet pedestals and results in a structure more similar to a ruffle.
Under our experimental conditions overexpression of SRF also resulted in the nuclear accumulation of MAL-GFP. The specific reason for this is currently unclear. Currently the prevailing hypothesis states that MAL continually shuttles between the cytoplasm and the nucleus. Perhaps MAL has a higher binding affinity for SRF than G-actin, and upon entering the nucleus, preferentially complexes with SRF and is retained in the nucleus.
Transcription of SRF is controlled by SRF its self [47], the overexpression of SRF may be interpreted by the cell as activation of the pathway, leading to an upregulation of SRF target genes and subsequent decrease in G-actin. These are just two potential hypotheses that may not be mutually exclusive, but warrant further study.
Of the proteins identified in this study ABRA and FLRT3 localized to the EPEC pedestal (Figure 7), and were both necessary for EPEC-induced translocation of MAL-GFP (Figure 6). Epistasis analysis showed that knockdown of FLRT3 could significantly reduce ABRA-induced nuclear accumulation of MAL-GFP, but ABRA knockdown had no effect on FLRT3-induced nuclear accumulation of MAL-GFP (Figure 6E and S4). This places FLRT3 downstream of ABRA and identifies it as an intermediary protein from pedestal to nucleus. Based on this data we hypothesize a new model (Figure 8), where EPEC-induced remodeling of the actin cytoskeleton, via Tir, activates SRF in an ABRA and FLRT3 dependent manner. Our findings therefore reveal a novel mechanism for pathogen-induced activation of a host transcription factor. They shed light on the relationship between ABRA and SRF and identify FLRT3 as a new component of this signalling pathway.
Fig. 8. A model for EPEC-induced activation of host-cell transcription factor SRF. 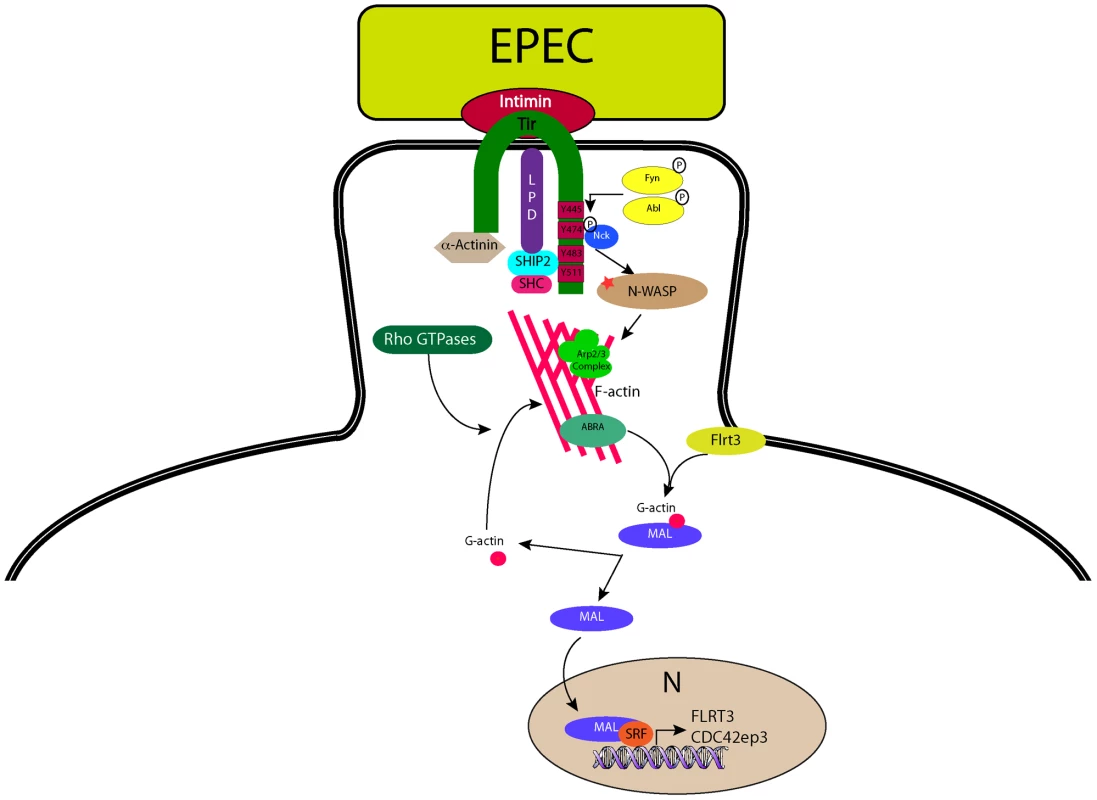
During infection with the extracellular pathogen EPEC, Tir translocates to the host cell and inserts into the plasma membrane where it interacts with the bacterial surface protein intimin and anchors it to the cell. The C-terminus of Tir is phosphorylated by host cell kinases leading to the recruitment and binding of Nck. N-WASP and the Arp2/3 complex are recruited leading to the generation of actin filaments beneath the bacterium. ABRA can bind actin in the newly formed (or forming) pedestal and stabilise the structure. The change in G-actin to F-actin ratio induced by pedestal formation is “sensed” by MAL via direct or indirect actions from ABRA and FLRT3, whereupon it is freed from its inhibitory complex with G-actin, to enter the nucleus and interact with SRF, driving transcription of target genes. Materials and Methods
Cell culture and bacterial strains
COS-7 cells were obtained from ATCC and routinely cultured in DMEM supplemented with 10% iron supplemented foetal calf serum (Hyclone, USA) and 40 µg/ml gentamycin sulphate. EPEC strains carrying tir deletions and complementation plasmids have been described previously [17]. S. Typhimurium SL1344 DsRed2 was given by Dr. H.C. Reinecker.
Constructs
MAL was amplified from a mouse cDNA template by PCR using a forward primer introducing a XhoI site: 5′ CTCGAGATGCCGCCTTTGAAAAGCCCC 3′; and a reverse primer introducing a SacII site: 5′ CCGCGGCAAGCAGGAATCCCAGTGGAG 3′. The resulting product was ligated into pEGFP-N1 (Clontech).
To generate constructs for mammalian expression of ABRA, SRF, FLRT3, TESK1and C22orf28 the coding sequences were amplified from cDNA clones in pCMV-SPORT6 obtained from Open Biosystems. Coding sequences were amplified using the primers in Table 1.
Tab. 1. Cloning primers. 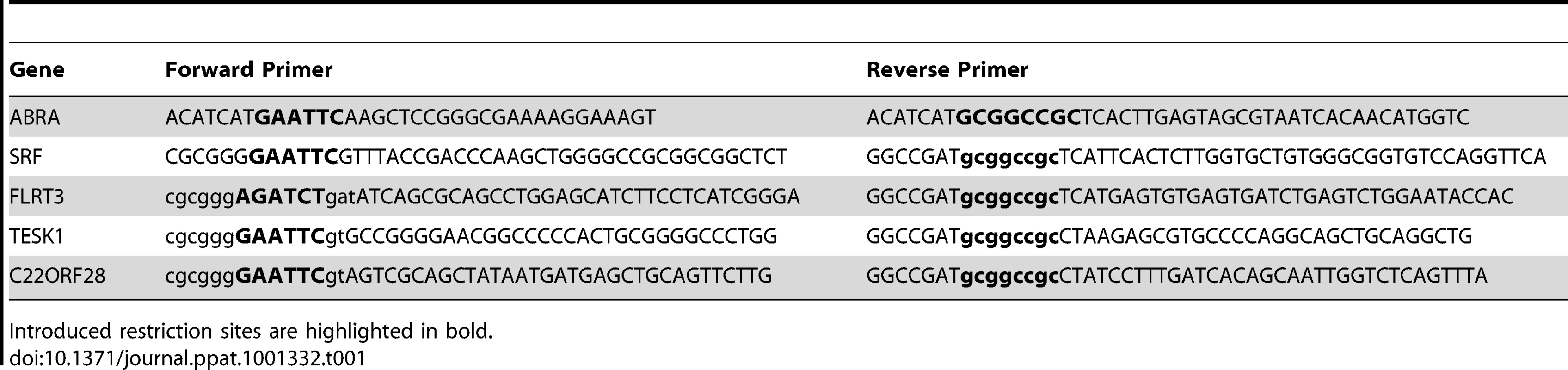
Introduced restriction sites are highlighted in bold. After digestion with the appropriate restriction enzymes, the coding sequence was subcloned into N-terminally tagged pCMV-3xFlag or -3xMyc vectors derived from the pCMV-Myc vector (Clontech, catalog no. 631604).
Transfections and knockdowns
COS-7 cells were plated onto 18 mm glass coverslips in 12-well plates at a density of 4×104 cells per well. After 24 h cells were transfected in antibiotic-free medium with MAL-GFP plus additional myc - or Flag - tagged constructs (in a modified pCMV vector, Clontech, USA), where noted, at a 1∶1 ratio, using GeneJuice (Novagen, UK), according to the manufacturers instructions. 18 h post-transfection cells were washed twice in PBS and incubated in DMEM 0.3% FCS for a further 18 h, prior to bacterial infections.
RNA interference
COS-7 cells were plated onto 18 mm glass coverslips in 12-well plates at a density of 4×104 cells per well. After 24 h, 20 pmol of modified RNA oligoduplexes (Stealth RNAi; Invitrogen), were transfected into each well using X-tremeGENE (Roche), according to the manufacturers instructions. siRNA silencing sequences are shown in Table 2.
Tab. 2. siRNA silencing sequences. 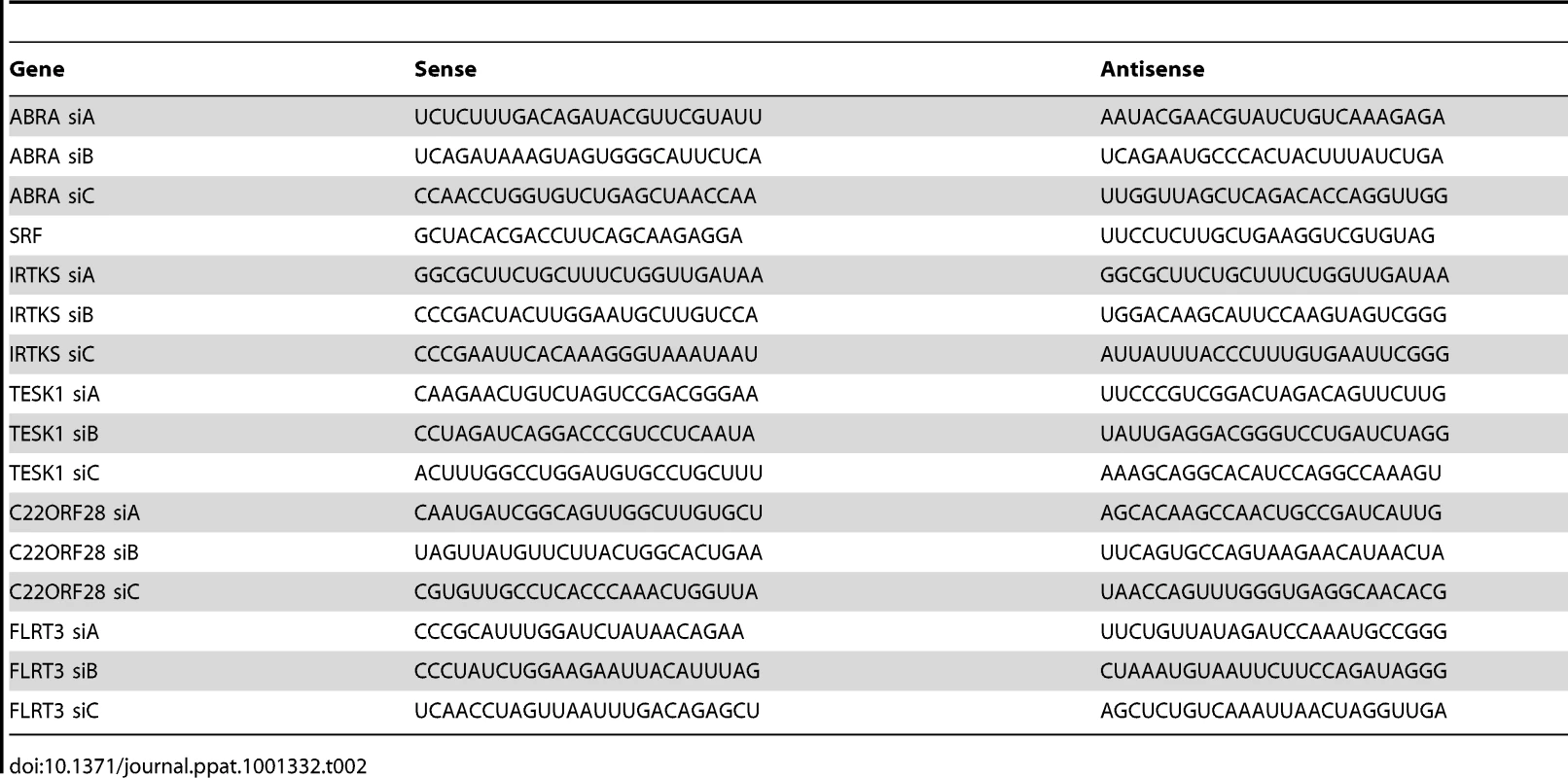
Cells were serum starved 48 h post-transfection as described above and infected, fixed and stained as described below.
Reverse transcription and real-time PCR
RNA extraction was performed by using an RNeasy kit (Qiagen) in accordance with the manufacturer's instructions. 500 ng of total RNA was reverse-transcribed using an iScript cDNA synthesis kit (Bio-Rad). The gene expression reported is representative of three independent experiments. Real-time quantitative PCR was performed in triplicate in a Bio-Rad iCycler thermal cycler equipped with an iQ5 optical module using the iQ SYBR Green Super Mix (Bio-Rad). In brief, 100 ng of reverse-transcribed cDNA was used for each PCR with forward and reverse primers at 250 nM. The thermal cycling conditions were 4 min at 95°C, followed by 40 cycles at 94°C for 15 s and 59°C for 1 min. Values were normalized to that of GAPDH. All PCR products were analyzed on a 2% agarose gel to verify the correct size of the amplicons. RT-PCR primer sequences are shown in Table 3.
Tab. 3. RT-PCR primer sequences. 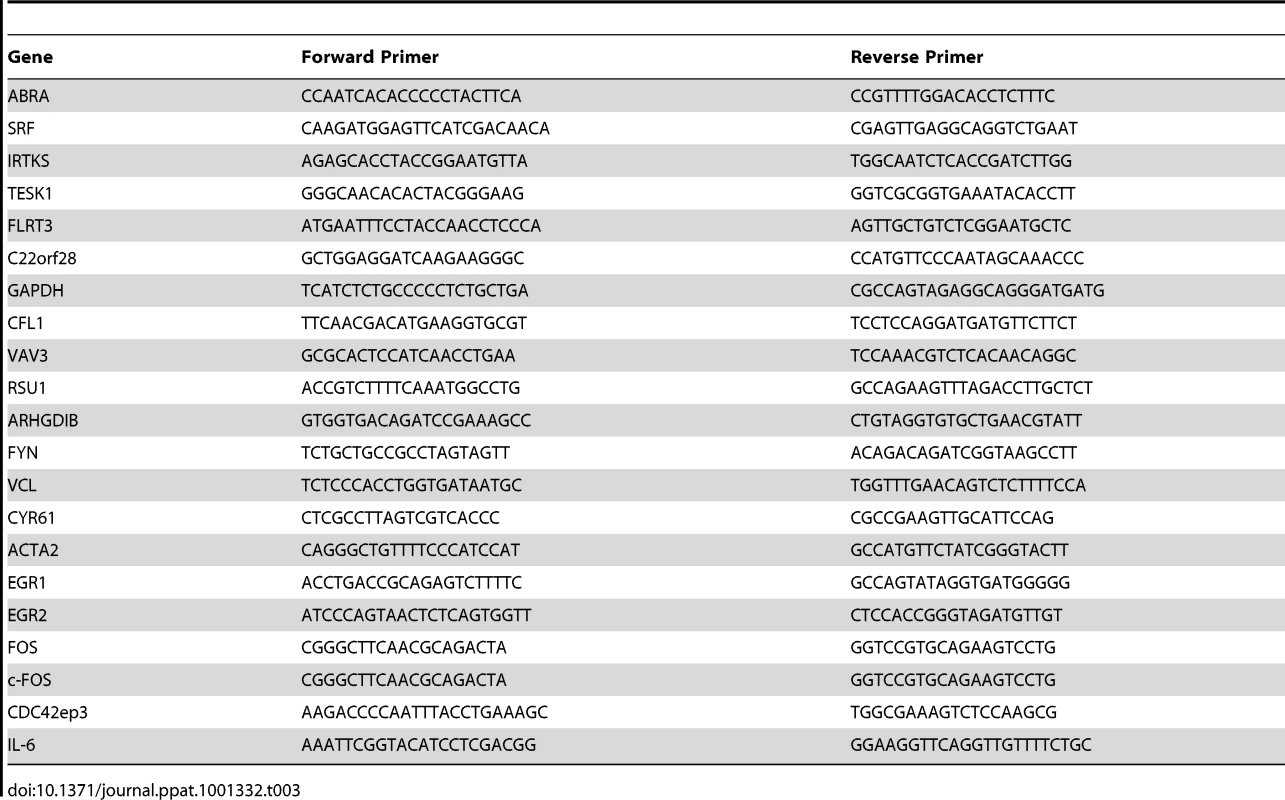
Bacterial infection conditions
For EPEC, AIEC and E.coli K12 infections were performed as previously described [21] with slight modifications to normalise infection conditions between the different strains. Briefly, colonies were seeded from fresh agar plates into 3 mls of LB broth with relevant antibiotics and grown with agitation at 37°C overnight. Cultures were then diluted 1∶1000 in DMEM containing 0.3% foetal calf serum and 1 ml added to each well of a 12-well plate. Plates were incubated at 37°C, 5% C02 for 5 hours.
S. Typhimurium infections were performed as described [48], with slight modifications to extend the infection time to the same duration as the EPEC infections. Briefly, SL1344 colonies from fresh agar plates were grown in LB broth plus 100 µg/ml ampicillin with agitation at 37°C overnight. Cultures were diluted 1∶33 and grown for a further 4 hours. Infections were performed using 1∶1000 dilutions of these sub-cultures, yielding a multiplicity of infection of 1∶10. Infections were allowed to proceed for 30–40 minutes at 37°C, 5% C02, then washed twice in DMEM+100 µg/ml Gentamycin to remove external bacteria and incubated for a further 4.5 hours at 37°C, 5% C02.
Immunofluorescence
Following infection, transfected cells were washed in PBS and fixed in 4% formaldehyde solution in PBS for 15 min. Cells were then permeabilised in 0.1% Triton-X 100 in PBS for 2 min, blocked with 10% donkey serum for 15 min and stained using appropriate antibodies for 1 h. Primary antibodies used were anti-Flag (Sigma Aldrich), anti-HA (Covance, USA) and anti-myc 9E10 (Covance, USA). The secondary antibody was Alexa488 or Alexa568-conjugated donkey anti-mouse (Jackson Immunoresearch). Actin was stained with Alexa568 or Alexa488-conjugated phalloidin (Invitrogen), DNA was labelled with DAPI (Invitrogen). Following staining coverslips were washed three times in PBS and mounted in ProLong Gold antifade reagent (Invitrogen). Cells were imaged using a Leica SP5 confocal microscope or a Ziess Axioplan widefield microscope.
Overexpression screen
Expression constructs in pCMV-SPORT6 were obtained from Open Biosystems. COS-7 cells were transfected with 250 ng of MAL-GFP and 250 ng of expression construct or an empty vector control. Eighteen hours post transfection the cells were washed twice with PBS and incubated in DMEM 0.3% FCS for a further 24 hours. Cells were washed in PBS and fixed in 4% formaldehyde solution in PBS for 15 min, and co-stained with DAPI (Invitrogen). Cellular localization of MAL-GFP was determined by epifluorescence microscopy. Data are the means ± standard deviation of 3 experiments. A minimum of 150 transfected cells were counted for each condition of each experiment.
Luciferase reporter assays
COS-7 cells were transfected with 50 ng of SRE-luciferase reporter plasmid [8], 1 ng of renilla luciferase (Promega) and either 500 ng of Flag-ABRA/untagged pCMV-ABRA or 150 ng FLRT3-Flag/pCMV-FLRT3. Controls were transfected with the appropriate empty vector. 8 hours post transfection cells were washed twice in PBS and resuspended in DMEM containing 0.3% FCS. 18 hours later cells were lysed in passive lysis buffer (Promega) and luciferase activities were measured with a Glomax 20/20 luminometer (Promega).
Quantification of F - and G-actin
G-∶F-actin ratios were quantified using a G-actin/F-actin In vivo assay kit (Cytoskeleton), in accordance with the manufacturers guidelines. Cells were infected as described above. 3.5 hours post infection samples were washed once in PBS, scraped and lysed with a bent 21 gauge needle in LAS2 lysis buffer. F-actin was then separated from G-actin by centrifugation at 100,000×g for 60 min at 37°C. The F-actin-containing pellet was resuspended in LAS2 buffer containing 2 µM cytochalasin D at a volume equivalent to the G-actin-containing supernatant volume. The resuspended F-actin pellet was kept on ice for 60 min with mixing by pipette every 15 min to dissociate F-actin. The F-actin and G-actin preparations were then assayed for protein. Equal amounts of protein were separated by 10% SDS-PAGE and detected by blotting with anti-actin. Band intensities were quantified with Odyssey application software (LI-COR).
Accession numbers
The following are the Entrez IDs (http://www.ncbi.nlm.nih.gov/) for the genes discussed in this article.
-
VCL, 7414
-
SRF, 6722
-
Cyr61, 3491
-
Acta2, 59
-
EGR1, 1958
-
EGR2, 1959
-
Fos, 2353
-
GAPDH, 2597
-
CFL1, 1072
-
VAV3, 10451
-
RSU1, 6251
-
ARHGDIB, 397
-
FYN, 2534
-
FLRT3, 23767
-
TESK1, 7016
-
MAL, 57591
-
VASP, 7408
-
AMIGO1, 57463
-
AMIGO2, 347902
-
TESK2, 10420
-
FLRT1, 23769
-
C22orf28, 51493
-
WDFY3, 23001
-
SSX2IP, 117178
-
HIP1R, 9026
-
WHAMM, 123720
-
PTP4A3, 11156
-
ZYXIN, 7791
-
LPP, 4026
Supporting Information
Zdroje
1. HaywardRD
LeongJM
KoronakisV
CampelloneKG
2006 Exploiting pathogenic Escherichia coli to model transmembrane receptor signalling. Nat Rev Microbiol 4 358 370
2. StebbinsCE
GalanJE
2001 Structural mimicry in bacterial virulence. Nature 412 701 705
3. BhavsarAP
GuttmanJA
FinlayBB
2007 Manipulation of host-cell pathways by bacterial pathogens. Nature 449 827 834
4. CornelisGR
2010 The type III secretion injectisome, a complex nanomachine for intracellular ‘toxin’ delivery. Biol Chem 391 745 51
5. TreismanR
1990 The SRE: a growth factor responsive transcriptional regulator. Semin Cancer Biol 1 47 58
6. ChaiJ
TarnawskiAS
2002 Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J Physiol Pharmacol 53 147 157
7. PosernG
TreismanR
2006 Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 16 588 596
8. HillCS
WynneJ
TreismanR
1995 The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81 1159 1170
9. MirallesF
PosernG
ZaromytidouAI
TreismanR
2003 Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113 329 342
10. VartiainenMK
GuettlerS
LarijaniB
TreismanR
2007 Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316 1749 1752
11. GrosseR
CopelandJW
NewsomeTP
WayM
TreismanR
2003 A role for VASP in RhoA-Diaphanous signalling to actin dynamics and SRF activity. EMBO J 22 3050 3061
12. BoquetP
LemichezE
2003 Bacterial virulence factors targeting Rho GTPases: parasitism or symbiosis? Trends Cell Biol 13 238 246
13. JaffeAB
HallA
2005 Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 21 247 269
14. KuwaharaK
BarrientosT
PipesGC
LiS
OlsonEN
2005 Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol 25 3173 3181
15. TabuchiA
EstevezM
HendersonJA
MarxR
ShiotaJ
2005 Nuclear translocation of the SRF co-activator MAL in cortical neurons: role of RhoA signalling. J Neurochem 94 169 180
16. DuKL
ChenM
LiJ
LeporeJJ
MerickoP
2004 Megakaryoblastic leukemia factor-1 transduces cytoskeletal signals and induces smooth muscle cell differentiation from undifferentiated embryonic stem cells. J Biol Chem 279 17578 17586
17. CampelloneKG
GieseA
TipperDJ
LeongJM
2002 A tyrosine-phosphorylated 12-amino-acid sequence of enteropathogenic Escherichia coli Tir binds the host adaptor protein Nck and is required for Nck localization to actin pedestals. Mol Microbiol 43 1227 1241
18. CampelloneKG
RankinS
PawsonT
KirschnerMW
TipperDJ
2004 Clustering of Nck by a 12-residue Tir phosphopeptide is sufficient to trigger localized actin assembly. J Cell Biol 164 407 416
19. CampelloneKG
LeongJM
2005 Nck-independent actin assembly is mediated by two phosphorylated tyrosines within enteropathogenic Escherichia coli Tir. Mol Microbiol 56 416 432
20. GarmendiaJ
PhillipsAD
CarlierMF
ChongY
SchullerS
2004 TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol 6 1167 1183
21. CampelloneKG
RobbinsD
LeongJM
2004 EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev Cell 7 217 228
22. VingadassalomD
KazlauskasA
SkehanB
ChengHC
MagounL
2009 Insulin receptor tyrosine kinase substrate links the E. coli O157:H7 actin assembly effectors Tir and EspF(U) during pedestal formation. Proc Natl Acad Sci U S A 106 6754 6759
23. WeissSM
LadweinM
SchmidtD
EhingerJ
LommelS
2009 IRSp53 links the enterohemorrhagic E. coli effectors Tir and EspFU for actin pedestal formation. Cell Host Microbe 5 244 258
24. AraiA
SpencerJA
OlsonEN
2002 STARS, a striated muscle activator of Rho signaling and serum response factor-dependent transcription. J Biol Chem 277 24453 24459
25. HuJ
TengJ
DingN
HeM
SunY
2008 FAAP, a novel murine protein, is involved in cell adhesion through regulating vinculin-paxillin association. Front Biosci 13 7123 7131
26. ToshimaJ
ToshimaJY
AmanoT
YangN
NarumiyaS
2001 Cofilin phosphorylation by protein kinase testicular protein kinase 1 and its role in integrin-mediated actin reorganization and focal adhesion formation. Mol Biol Cell 12 1131 1145
27. TsujiL
YamashitaT
KuboT
MaduraT
TanakaH
2004 FLRT3, a cell surface molecule containing LRR repeats and a FNIII domain, promotes neurite outgrowth. Biochem Biophys Res Commun 313 1086 1091
28. ChenX
KohE
YoderM
GumbinerBM
2009 A protocadherin-cadherin-FLRT3 complex controls cell adhesion and morphogenesis. PLoS One 4 e8411
29. NgAC
EisenbergJM
HeathRJ
HuettA
RobinsonCM
2010 Microbes and Health Sackler Colloquium: Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci U S A E-pub ahead of print. doi: 10.1073/pnas.1000093107
30. PatelJC
GalanJE
2006 Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J Cell Biol 175 453 463
31. McGhieEJ
BrawnLC
HumePJ
HumphreysD
KoronakisV
2009 Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol 12 117 124
32. FuY
GalanJE
1999 A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401 293 297
33. RangnekarVM
AplinAC
SukhatmeVP
1990 The serum and TPA responsive promoter and intron-exon structure of EGR2, a human early growth response gene encoding a zinc finger protein. Nucleic Acids Res 18 2749 2757
34. FujiiM
NikiT
MoriT
MatsudaT
MatsuiM
1991 HTLV-1 Tax induces expression of various immediate early serum responsive genes. Oncogene 6 1023 1029
35. KobayashiSD
BraughtonKR
WhitneyAR
VoyichJM
SchwanTG
2003 Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci U S A 100 10948 10953
36. PhelpsED
SweeneyKR
BladerIJ
2008 Toxoplasma gondii rhoptry discharge correlates with activation of the early growth response 2 host cell transcription factor. Infect Immun 76 4703 4712
37. LeeSM
VasishthaM
PrywesR
2010 Activation and repression of cellular immediate early genes by serum response factor cofactors. J Biol Chem 285 22036 22049
38. BoldrickJC
AlizadehAA
DiehnM
DudoitS
LiuCL
2002 Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci U S A 99 972 977
39. KennyB
DeVinneyR
SteinM
ReinscheidDJ
FreyEA
1997 Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91 511 520
40. KennyB
1999 Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol Microbiol 31 1229 1241
41. PhillipsN
HaywardRD
KoronakisV
2004 Phosphorylation of the enteropathogenic E. coli receptor by the Src-family kinase c-Fyn triggers actin pedestal formation. Nat Cell Biol 6 618 625
42. SwimmA
BommariusB
LiY
ChengD
ReevesP
2004 Enteropathogenic Escherichia coli use redundant tyrosine kinases to form actin pedestals. Mol Biol Cell 15 3520 3529
43. GruenheidS
DeVinneyR
BladtF
GoosneyD
GelkopS
2001 Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat Cell Biol 3 856 859
44. DeVinneyR
PuenteJL
GauthierA
GoosneyD
FinlayBB
2001 Enterohaemorrhagic and enteropathogenic Escherichia coli use a different Tir-based mechanism for pedestal formation. Mol Microbiol 41 1445 1458
45. KennyB
2001 The enterohaemorrhagic Escherichia coli (serotype O157:H7) Tir molecule is not functionally interchangeable for its enteropathogenic E. coli (serotype O127:H6) homologue. Cell Microbiol 3 499 510
46. DeVinneyR
SteinM
ReinscheidD
AbeA
RuschkowskiS
1999 Enterohemorrhagic Escherichia coli O157:H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect Immun 67 2389 2398
47. HerreraRE
ShawPE
NordheimA
1989 Occupation of the c-fos element in vivo is unaltered by growth factor induction. Nature 340 68 70
48. RiouxJD
XavierRJ
TaylorKD
SilverbergMS
GoyetteP
2007 Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 39 596 604
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Innate Immune Sensing of DNA
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 4- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Low Diversity Variety Multilocus Sequence Types from Thailand Are Consistent with an Ancestral African Origin
- Genetic Assignment Methods for Gaining Insight into the Management of Infectious Disease by Understanding Pathogen, Vector, and Host Movement
- Innate Immune Sensing of DNA
- Engineered Resistance to Development in Transgenic
- -Mediated Detoxification of Reactive Oxygen Species Is Required for Full Virulence in the Rice Blast Fungus
- SLO-1-Channels of Parasitic Nematodes Reconstitute Locomotor Behaviour and Emodepside Sensitivity in Loss of Function Mutants
- Structure-Function Analysis of the LRIM1/APL1C Complex and its Interaction with Complement C3-Like Protein TEP1
- A Effector with Enhanced Inhibitory Activity on the NF-κB Pathway Activates the NLRP3/ASC/Caspase-1 Inflammasome in Macrophages
- The MARCH Family E3 Ubiquitin Ligase K5 Alters Monocyte Metabolism and Proliferation through Receptor Tyrosine Kinase Modulation
- is an Unstable Pathogen Showing Evidence of Significant Genomic Flux
- The Cell Wall Protein CwpV is Antigenically Variable between Strains, but Exhibits Conserved Aggregation-Promoting Function
- Engineering HIV-Resistant Human CD4+ T Cells with CXCR4-Specific Zinc-Finger Nucleases
- SUMO-Interacting Motifs of Human TRIM5α are Important for Antiviral Activity
- Completion of Hepatitis C Virus Replication Cycle in Heterokaryons Excludes Dominant Restrictions in Human Non-liver and Mouse Liver Cell Lines
- : Reservoir Hosts and Tracking the Emergence in Humans and Macaques
- On Being the Right Size: The Impact of Population Size and Stochastic Effects on the Evolution of Drug Resistance in Hospitals and the Community
- Bacterial and Host Determinants of MAL Activation upon EPEC Infection: The Roles of Tir, ABRA, and FLRT3
- Respiratory Syncytial Virus Interferon Antagonist NS1 Protein Suppresses and Skews the Human T Lymphocyte Response
- NF-κB Hyper-Activation by HTLV-1 Tax Induces Cellular Senescence, but Can Be Alleviated by the Viral Anti-Sense Protein HBZ
- Human Cytomegalovirus IE1 Protein Elicits a Type II Interferon-Like Host Cell Response That Depends on Activated STAT1 but Not Interferon-γ
- A New Model to Produce Infectious Hepatitis C Virus without the Replication Requirement
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- NF-κB Hyper-Activation by HTLV-1 Tax Induces Cellular Senescence, but Can Be Alleviated by the Viral Anti-Sense Protein HBZ
- Bacterial and Host Determinants of MAL Activation upon EPEC Infection: The Roles of Tir, ABRA, and FLRT3
- : Reservoir Hosts and Tracking the Emergence in Humans and Macaques
- On Being the Right Size: The Impact of Population Size and Stochastic Effects on the Evolution of Drug Resistance in Hospitals and the Community
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



