-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPathogenic Bacteria Target NEDD8-Conjugated Cullins to Hijack Host-Cell Signaling Pathways
The cycle inhibiting factors (Cif), produced by pathogenic bacteria isolated from vertebrates and invertebrates, belong to a family of molecules called cyclomodulins that interfere with the eukaryotic cell cycle. Cif blocks the cell cycle at both the G1/S and G2/M transitions by inducing the stabilization of cyclin-dependent kinase inhibitors p21waf1 and p27kip1. Using yeast two-hybrid screens, we identified the ubiquitin-like protein NEDD8 as a target of Cif. Cif co-compartmentalized with NEDD8 in the host cell nucleus and induced accumulation of NEDD8-conjugated cullins. This accumulation occurred early after cell infection and correlated with that of p21 and p27. Co-immunoprecipitation revealed that Cif interacted with cullin-RING ubiquitin ligase complexes (CRLs) through binding with the neddylated forms of cullins 1, 2, 3, 4A and 4B subunits of CRL. Using an in vitro ubiquitylation assay, we demonstrate that Cif directly inhibits the neddylated CUL1-associated ubiquitin ligase activity. Consistent with this inhibition and the interaction of Cif with several neddylated cullins, we further observed that Cif modulates the cellular half-lives of various CRL targets, which might contribute to the pathogenic potential of diverse bacteria.
Published in the journal: . PLoS Pathog 6(9): e32767. doi:10.1371/journal.ppat.1001128
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001128Summary
The cycle inhibiting factors (Cif), produced by pathogenic bacteria isolated from vertebrates and invertebrates, belong to a family of molecules called cyclomodulins that interfere with the eukaryotic cell cycle. Cif blocks the cell cycle at both the G1/S and G2/M transitions by inducing the stabilization of cyclin-dependent kinase inhibitors p21waf1 and p27kip1. Using yeast two-hybrid screens, we identified the ubiquitin-like protein NEDD8 as a target of Cif. Cif co-compartmentalized with NEDD8 in the host cell nucleus and induced accumulation of NEDD8-conjugated cullins. This accumulation occurred early after cell infection and correlated with that of p21 and p27. Co-immunoprecipitation revealed that Cif interacted with cullin-RING ubiquitin ligase complexes (CRLs) through binding with the neddylated forms of cullins 1, 2, 3, 4A and 4B subunits of CRL. Using an in vitro ubiquitylation assay, we demonstrate that Cif directly inhibits the neddylated CUL1-associated ubiquitin ligase activity. Consistent with this inhibition and the interaction of Cif with several neddylated cullins, we further observed that Cif modulates the cellular half-lives of various CRL targets, which might contribute to the pathogenic potential of diverse bacteria.
Introduction
Cycle inhibiting factors (Cif) constitute a family of bacterial cyclomodulins that inhibit eukaryotic cell proliferation by blocking the cell cycle [1], [2], [3]. First identified in enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC) [4], Cif homologues have been recently characterized in several other pathogenic proteobacteria including Burkholderia, Yersinia and Photorhabdus species [5], [6]. E. coli Cif is composed of a C-terminal active domain and a ∼20 amino acid N-terminal signal domain necessary for its injection into host cells by the type three secretion system (T3SS) [7], [8]. Crystal structures of Cif from E. coli, B. pseudomallei and P. luminescens revealed that Cif proteins possess a conserved putative catalytic triad composed of a cysteine, a histidine and a glutamine, and belong to the cysteine protease superfamily, a diverse group of enzymes with protease, acetyltransferase, deamidase, transglutaminase, and other biochemical activities [5], [9], [10]. Although the specific enzymatic activity of Cif has not been identified, the active site is essential for Cif cytostatic activity since mutation of the triad residues leads to the loss of phenotype [5], [6], [10].
Upon injection into host cells, Cif triggers a cytopathic effect characterized by cell cycle arrests both in G1/S and G2/M phase transitions and, in certain cell lines, the reorganization of the actin network [4], [11], [12]. The cell cycle arrest induced by Cif is irreversible and leads eventually to delayed cell death by apoptosis [13]. The inhibition of the cell cycle is independent of the DNA damage response and p53 pathway [14], but correlates with the accumulation of the cyclin-dependent kinase (CDK) inhibitors p21waf1/cip1 and p27kip1 (hereafter referred as p21 and p27 respectively) that inactivate CDKs whose activities are required for entry in both S - and M-phases [15], [16]. This accumulation of p21 and p27 results from protein stabilization, suggesting that Cif interferes with their 26S proteasome-mediated degradation [15].
In eukaryotic cells, degradation of intracellular proteins is processed mainly by ubiquitin-mediated proteolysis [17]. This highly regulated mechanism leads to conjugation of polyubiquitin chains to protein substrates, targeting them to the 26S proteasome for degradation [18]. Substrate specificity is determined by E3 ligase enzymes that recognize targets to be ubiquitinated. The most prominent class of E3s is the superfamily of cullin-RING ligases (CRLs) that are multi-subunit complexes based on cullin protein scaffolds. Among the different mechanisms regulating the activity of CRLs, conjugation of the cullin subunit with the ubiquitin-like protein NEDD8 (neddylation) stimulates the transfer of ubiquitin to target proteins, thereby enhancing their subsequent proteolysis by the 26S proteasome [19], [20], [21]. Degradation of many proteins involved in the control of cell cycle progression, including p21 and p27, is regulated by CRL activity [22].
To decipher the mode of action of the cyclomodulin Cif, we performed yeast two-hybrid screenings and identified NEDD8 as a specific Cif interaction partner. Cif co-compartmentalized with NEDD8 in the host cell nucleus and induced an accumulation of NEDD8-conjugated cullins that correlated with the accumulation of p21 and p27. Co-immunoprecipitation revealed that Cif interacted with the neddylated forms of cullin-RING complexes. Morevover, in vitro, Cif inhibited NEDD8-modified Cullin1/Fbw7-associated ubiquitin ligase activity. Thus, Cif manipulates the host ubiquitin-dependent proteasomal degradation by interfering with NEDD8-conjugated CRLs.
Results
Cif interacts with the ubiquitin-like protein NEDD8
To identify eukaryotic targets of Cif, yeast two-hybrid screenings were performed using the wild-type form of E. coli Cif protein, an inactive cysteine mutant [10] and two distinct cDNA libraries from human placenta and human colon. The neural precursor cell expressed, developmentally down-regulated 8 protein (NEDD8) was the sole common putative eukaryotic interacting partner detected in each screening. In the two-hybrid, the highest confidence score, which reflects the reliability and biological significance of each interaction [23], was obtained for NEDD8 interacting with the cysteine mutant, suggesting that inactivation of the Cif catalytic site could stabilize the interaction with NEDD8.
As a first step to validate the interaction between Cif and NEDD8, hexa-histidine tagged Cif (wild type or cysteine mutant) and NEDD8 were co-expressed in a laboratory E. coli strain and His-Cif was purified by nickel affinity chromatography. As a control, NEDD8 was also expressed in absence of His-Cif. Detection of NEDD8 in the eluted fractions from both wild type Cif and cysteine mutant purifications confirmed that Cif interacted with NEDD8 (Fig. 1A). Mirroring the yeast two-hybrid experiments, the cysteine mutant was more efficient to pull-down NEDD8 in the bacterial co-expression assay. In addition, Cif and NEDD8 co-eluted in the same fractions by gel filtration chromatography (Fig. 1B). Since NEDD8 is 60% identical and 80% homologous to ubiquitin, we also tested whether Cif could bind ubiquitin. In contrast to NEDD8, ubiquitin did not co-elute with His-Cif in the bacterial co-expression assay (Fig. 1A), demonstrating the specificity of Cif binding to NEDD8.
Fig. 1. Cif interacts in vitro with the ubiquitin-like protein NEDD8. 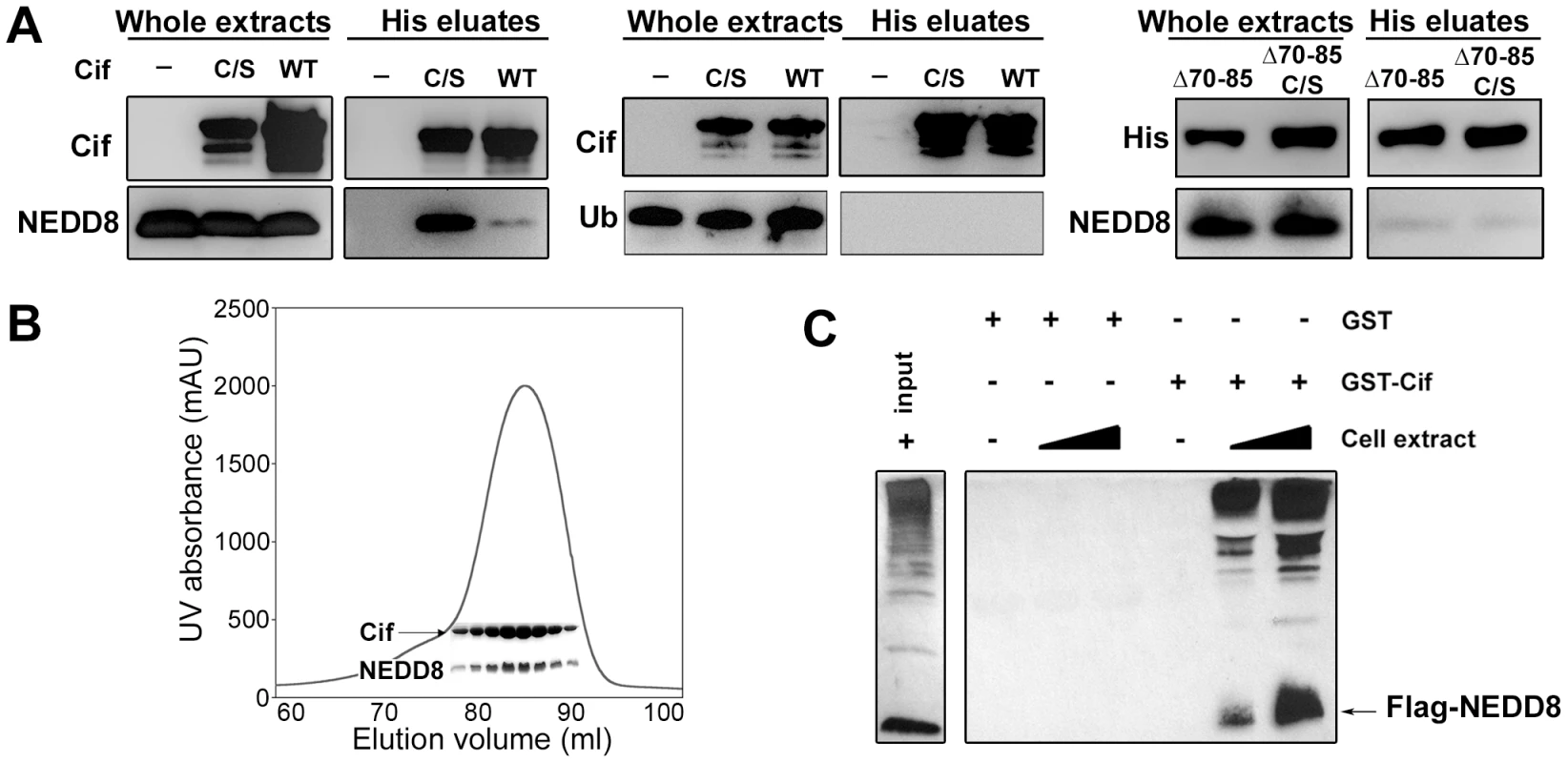
(A) Hexa-histidine tagged wild-type Cif (WT), Cif cysteine 109 mutant Cys-to-Ser (C/S), Cif deleted for amino-acids 70-85 mutant (Δ70-85) or Cif Δ70-85 Cys-to-Ser mutant(Δ70-65 C/S) and NEDD8 or ubiquitin (Ub) proteins were co-expressed in bacteria, and then His-Cif proteins were purified from bacterial lysates using nickel affinity. Whole extracts and eluted fractions were probed with indicated antibodies. (B) Wild-type His-Cif and NEDD8 were co-expressed in bacteria and His-Cif was purified using nickel affinity and gel filtration chromatography. The peak in the chromatograph corresponds to the His-Cif complex. NEDD8 and the corresponding fractions were visualized using SDS-PAGE stained with Coomassie blue. (C) Glutathione sepharose matrix was mixed with 100 µg of GST or GST-Cif Cys-to-Ser mutant purified proteins and then incubated with a lysate of HEK293T cells expressing a FLAG-NEDD8 construct. 100 µg or 250 µg of cells extract were used to obtain a GST constructs/cell extract ratio of 1∶1 and 1∶2.5 respectively. Cells extract (input) and eluted fractions were immunoblotted with anti-FLAG antibodies. FLAG-NEDD8 fusion protein is indicated with an arrow. As a second approach to confirm the interaction of Cif with NEDD8, lysates from HEK293T cells expressing FLAG-tagged NEDD8 were used to perform glutathione S-transferase (GST) pull-down experiments. The glutathione-sepharose matrix was first loaded with purified GST-Cif (cysteine mutant) fusion protein or GST alone as control, and then incubated with cell extracts. In contrast to the GST control, GST-Cif captured FLAG-NEDD8 and apparently FLAG-NEDD8-modified proteins present in the HEK293T extract (Fig. 1C). Together, these results demonstrate that Cif interacts specifically with the NEDD8 protein.
While members of the Cif family exhibit a low level of sequence similarity, crystal structure determination of Cif from E. coli and the xenologs from B. pseudomallei and P. luminescens revealed that all proteins are structurally well-conserved [5], [9], [10]. These studies have shown a head-and-tail domain arrangement. The tail corresponds to the N-terminal part of the protein. The C-terminal head-domain, which contains the catalytic triad, adopts an overall fold similar to cysteine proteases. Interestingly, deletion of the protruding α4 helix in the N-terminal part of Cif from B. pseudomalei, which is unlikely to affect the structural integrity of protein [5], abolishes the cell cycle arrest, suggesting that this domain could mediate substrate recognition [5]. Therefore, we constructed an E. coli Cif mutant deleted for residues 70 to 85 corresponding to the α4 helix (CifΔ70-85). The CifΔ70-85 mutant, like the Cif cysteine mutant, was translocated into host cells upon infection (Fig. 2A). However, removal of the α4 helix of Cif, even in combination with the cysteine mutation that increases in vitro NEDD8 binding efficiency, prevented association with NEDD8 (Fig. 1A). Furthermore, the CifΔ70-85 mutant did not induce actin stress fibers, nucleus enlargement, cell cycle arrest, or p21 and p27 accumulation (Fig 2B–C). Together these results suggest that interaction with NEDD8 via residues 70–85 is required for Cif-induced cell cycle arrest.
Fig. 2. Cytopathic activities of wild-type Cif, Cif C/A and CifΔ70-85 mutants. 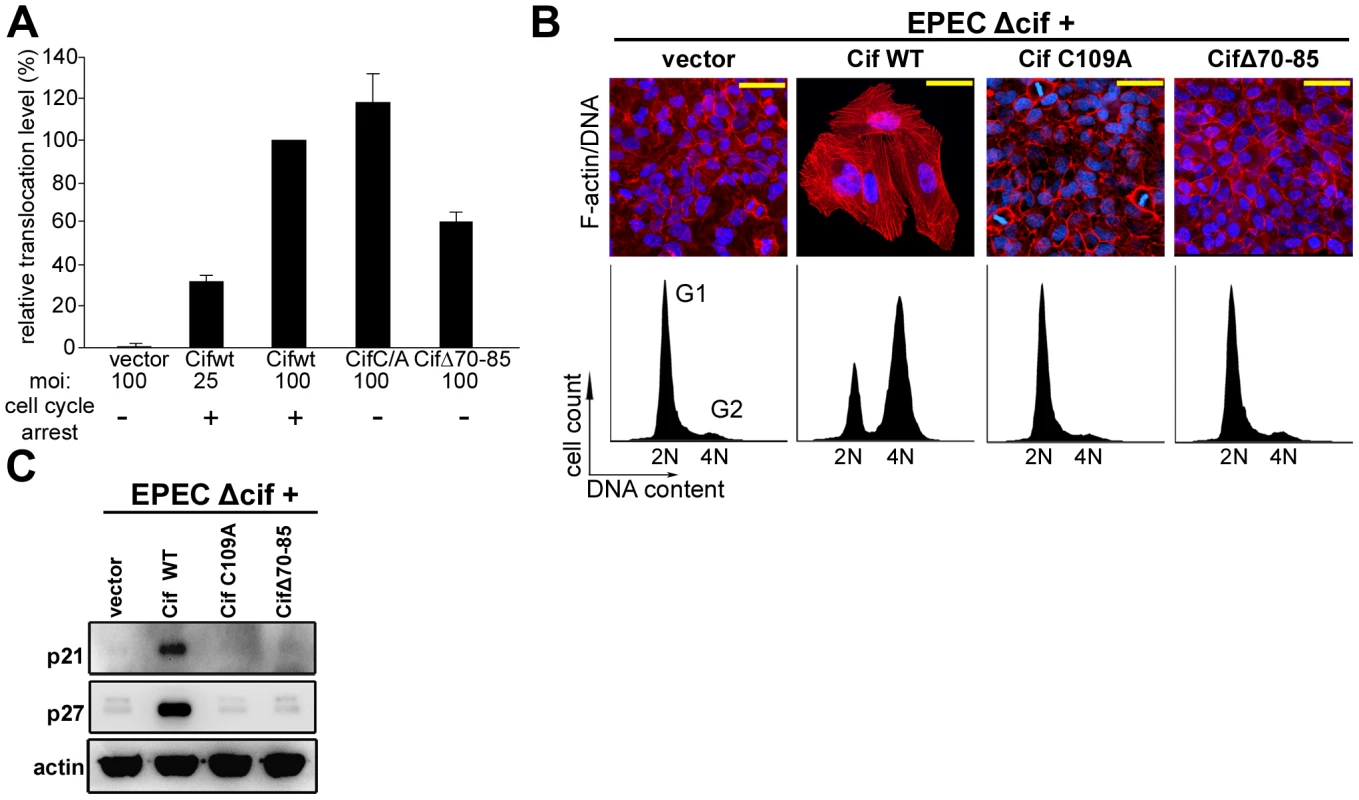
(A) Translocation efficiency of Cif wild-type and mutant proteins were assessed as follow: HeLa cells were loaded with CCF2/AM substrate and were infected 1 h 30 with E22Δcif hosting plasmids expressing the different Cif-TEM1 fusions and intracellular β-lactamase activity was detected by measuring cleavage of the CCF2/AM substrate. Translocation level is represented as percentages relative to translocation of the Cif WT-TEM protein (see materials and methods). Experiments were performed in triplicate and error bars represent standard errors of the mean. The multiplicity of infection (moi) and corresponding cell cycle arrest phenotype are indicated. Note that infection at moi of 25 with low level of translocation of wild-type Cif was sufficient to induce cell cycle arrest. (B) G1/S synchronized HeLa cells were infected 1.5 h with E22Δcif expressing the different Cif proteins, washed and incubated with antibiotic for 20-72 h. Upper panels: F-actin was labeled with phalloidin-rhodamine (red) and DNA with DAPI (blue) 72 h post-infection. Bars represent 50 µm. Lower panels: cell cycle distribution was analyzed by flow cytometry 20 h post-infection. 2N (G1) and 4N (G2) populations are indicated. (C) HeLa cells were infected as in B, washed and incubated with antibiotics for 24 h. Cell protein extracts were probed with anti-p21, anti-p27 and anti-actin antibodies. Cif co-compartmentalizes with NEDD8 in the host cell nucleus
Determination of Cif trafficking upon bacterial injection into host cells may provide clues to specify cellular functions hijacked by Cif. We therefore examined the subcellular localization of Cif into infected host cells by immunostaining and confocal microscopy. Optical sections revealed that Cif accumulated mostly in the host cell nucleus (Fig. 3A). A similar distribution of the inactive Cif cysteine mutant was observed. As a second step, we investigated whether Cif and NEDD8 co-localized in cells. Experiments were performed with stable HeLa Tet-on cells producing GFP-Cif (wild-type or cysteine mutant) fusion proteins. The distribution of the GFP signal showed that, as during infection experiments, wild-type Cif and the inactive cysteine mutant were mostly detected in the nucleus (Fig. 3B). In agreement with previous studies [24], immunodetection of NEDD8 showed its predominant localization also in the nucleus. Consistent with the nuclear localization of both proteins, dual GFP-Cif and NEDD8 labeling resulted in a partial coincident nuclear staining pattern (Fig. 3B). As expected, expression of GFP-Cif wild-type led to cell cycle arrest and to p21 and p27 accumulation whereas GFP-Cif cysteine mutant did not (Fig. 3C). These data demonstrate that Cif preferentially localizes to host cell nuclei, thus co-compartmentalizing with NEDD8.
Fig. 3. Cif is sorted to the host cell nucleus and co-compartmentalizes with NEDD8. 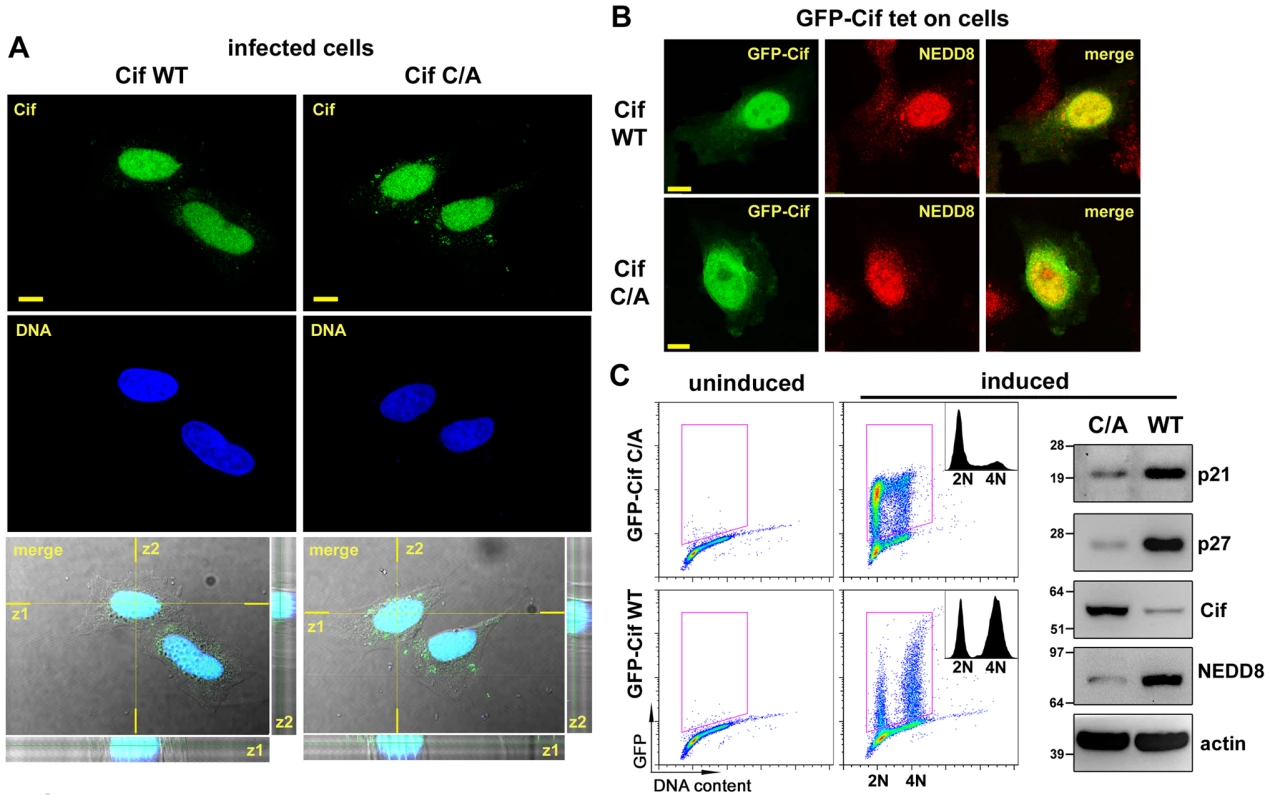
(A) HeLa cells were infected 2 h with EPECΔcif expressing wild-type Cif (WT) or the CifC109A (C/A) mutant, washed and further incubated 3 h with antibiotics. Cif was then visualized by indirect immunofluorescence and confocal imaging, with anti-Cif antibodies (green) and TO-PRO-3 to stain DNA (dark blue). Single optical XY slices and the corresponding Z slices of the image stacks are shown. In the merged images, coincident fluorescent sources appear cyan. Bars represent 10 µm. (B) Stable HeLa TetOn cells expressing GFP-Cif (WT) or GFP-CifC109A (C/A) were induced for 24 h with doxycycline. GFP-Cif was visualized by GFP fluorescence acquisition (green) and NEDD8 was detected by indirect immunofluorescence with anti-NEDD8 antibodies (red). In the merged images, coincident fluorescent sources appear orange-yellow. Bars represent 10 µm. (C) HeLa TetOn cells were induced or not with doxycycline, then DNA was stained with propidium iodide and DNA/GFP content was analyzed by flow cytometry. GFP-positive cells were gated as shown in the DNA content/GFP fluorescence 2D plots to generate the cell cycle histograms (insets). 2N and 4N DNA content are indicated. Induced cell protein lysates were probed with anti-p21, anti-p27, anti-Cif, anti-NEDD8 and anti-actin antibodies. Molecular weights in kDa are indicated. Cif induces accumulation of neddylated proteins in host cells
To investigate the biological significance of the Cif/NEDD8 interaction, we monitored the level of neddylated proteins in cells infected with wild type EPEC or strains deleted for the Cif gene (Δcif). Immunoblotting with anti-NEDD8 antibodies revealed that the level of neddylated protein(s) with an apparent size of ∼80 kDa increased in cells infected with wild-type EPEC, but not with the Δcif mutant strain (Fig. 4A). Accumulation of these neddylated proteins was detectable as soon as the end of infection (0 h) and was still observable 24 h post-infection. The increased level of neddylated proteins correlated with accumulation of p21 and p27 (Fig. 4A). The rapid accumulation of NEDD8-conjugated proteins was also observed in non-transformed rat small intestine epithelial IEC-6 cells and in human colon HCT116 cells (Fig. 4B). This accumulation correlated with their cell cycle arrest upon infection with EPEC expressing Cif [15]. Consistent with Cif and NEDD8 cellular localization (Fig. 3B), cell fractionation revealed that the 80 kDa neddylated proteins were mostly detected in the nuclear compartment (Fig. 4C). Cif alone was sufficient to modulate the level of neddylated proteins since eukaryotic expression of the wild-type GFP-Cif in the Tet-on cells also led to accumulation of the 80 kDa NEDD8-conjugated proteins (Fig. 3C). The Cif from B. pseudomallei, the most divergent xenolog of Cif, also induced accumulation of the 80 kDa neddylated proteins (Fig. 4D). In contrast, the level of NEDD8-conjugated proteins did not increase in cells infected with bacteria expressing Cif mutated on the catalytic cysteine, or the Cif mutant deleted for the α4helix (CifΔ70-85). Thus, a functional Cif was required for neddylated protein accumulation (Fig. 4D). Together, these data demonstrate that Cif alters the neddylation pattern in host cells and provokes an early accumulation of NEDD8-conjugated protein(s) in the nucleus of infected cells.
Fig. 4. Infection with Cif-producing EPEC induces the accumulation of neddylated proteins. 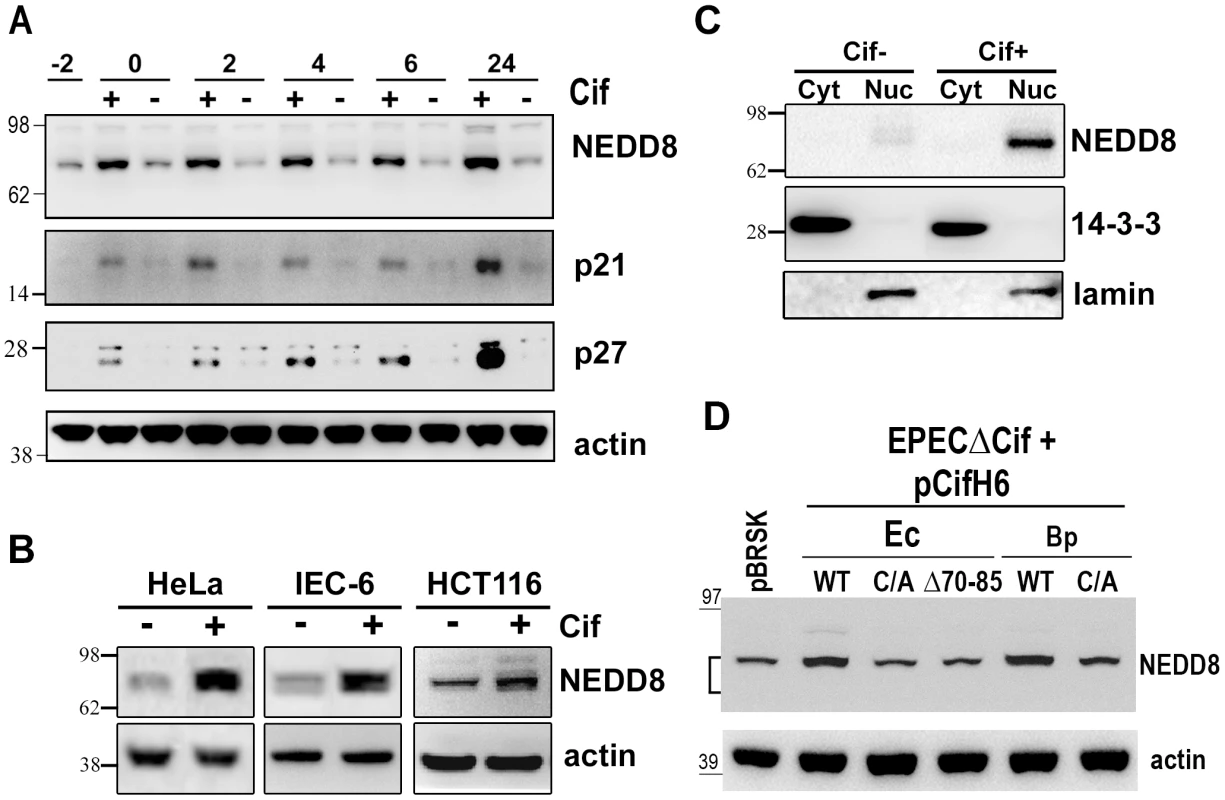
(A) HeLa cells were infected (starting at -2 h) 2 h with wild-type EPEC (+) or a Cif-deleted mutant EPEC (-), washed and further incubated with antibiotics up to 24 h. Cell protein extracts were probed with anti-NEDD8, anti-p21, anti-p27 and anti-actin antibodies. Molecular weights in kDa are indicated. (B) HeLa, IEC-6 or HCT116 cells were infected as in A and further incubated 6 h with antibiotics. Cell protein extracts were probed with anti-NEDD8 and anti-actin antibodies. (C) HeLa cells were infected as in A, washed and further incubated 6 h with antibiotics. Cytoplasmic (Cyt) and nuclear (Nuc) extracts were probed with anti-NEDD8, anti-14-3-3 (cytoplasmic control) and anti-lamin (nuclear control) antibodies. (D) HeLa cells were infected 3 h with EPECΔcif strain carrying the empty vector (pBRSK) or plasmids coding for His-tagged wild-type (WT), cysteine mutant (C/A) or Δ70-85 mutant of Cif from E. coli (Ec) or from B. pseudomallei (Bp) and further incubated 3 h with antibiotics. Cell protein extracts were probed with anti-NEDD8 and anti-actin antibodies. The brackets encompass the 73-81 kDa region. Neddylated cullins 1, 2, 3, 4A and 4B accumulate in the presence of Cif
Among known NEDD8-conjugated proteins, members of the cullin protein family are major substrates of the neddylation machinery [25]. Most eukaryotic genomes encode at least six cullins (CUL1, 2, 3, 4A, 4B and 5) that are scaffold proteins of cullin-RING E3 ubiquitin ligases. Since most of the cullin proteins are known to compartmentalize in the nucleus and have an approximate molecular weight of 80 kDa, we examined whether the Cif-induced neddylated proteins corresponded to several of the cullins (CUL1, 2, 3, 4A and 4B) [26]. The neddylation status of these cullins was investigated by immunoblotting following infection with EPEC Δcif exogenously expressing Cif proteins (wild-type or cysteine mutants) from E. coli or B. pseudomallei. As expected, two bands were detected for the cullin proteins, with estimated molecular weights of ∼75 and a slower migrating form of ∼80 kDa (Fig. 5A). The slower migrating species corresponded to the signal detected with the anti-NEDD8 antibody, and treatment of cell extracts with the NEDD8 protease 1 (NEDP1) induced disappearance of both the higher molecular form of cullins and the corresponding NEDD8 signal (Fig. 5B), indicating that these slower migrating proteins were neddylated cullins. Following infection, the levels of neddylated forms of CUL1, 2, 3, 4A and 4B increased in presence of the wild-type Cif proteins from E. coli or from B. pseudomallei (Fig. 5A). These findings indicated that the 80 kDa neddylated proteins accumulating in presence of Cif were proteins of the cullin family.
Fig. 5. Cif induces the accumulation of NEDD8-conjugated cullins in host cells. 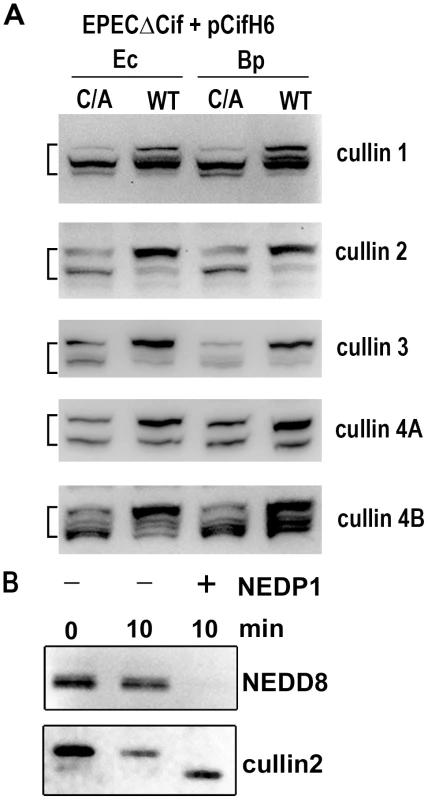
(A) HeLa cells were infected 3 h with EPECΔcif strain carrying plasmids coding for His-tagged wild-type (WT) or cysteine mutant (C/A) from E. coli (Ec) or from B. pseudomallei (Bp) and further incubated 3 h with antibiotics. Cell protein extracts were probed with anti-cullins (cullin 1 to cullin 4B) antibodies. The brackets encompass the 73-81 kDa regions. (B) Protein extracts from HeLa cells infected with EPEC expressing wild-type Cif were incubated for 10 min in presence (+) or absence (-) of NEDP1 and probed with anti-NEDD8 and anti-cullin2 antibodies. Cif interacts with neddylated cullin-RING complexes
We next examined whether Cif interacted with the neddylated forms of cullins. After infection of HeLa cells with EPEC expressing His-tagged Cif protein from E. coli (wild-type, Δ70-85 or C/A mutants), His-Cif proteins were immunoprecipited with anti-His antibodies coupled to magnetic beads and the different fractions were analyzed by western-blot (Fig. 6A). High molecular weight proteins detected with the anti-NEDD8 antibody co-eluted with the wild-type Cif protein. These data raised the possibility that Cif interacts with NEDD8-conjugated proteins. We next verified whether these neddylated proteins targeted by Cif were members of the cullin family. Figure 6B shows that many cullins (CUL1 to 4B) were found in the eluted fraction from the wild-type Cif precipitate. Interestingly, only the slowly-migrating neddylated forms of the cullins were recovered in the wild-type Cif eluates. These data suggest that Cif interacts with cullins through their NEDD8 modification. In contrast to our previous result showing that the Cif cysteine mutant interacted with NEDD8 in vitro (Fig. 1A–B), the Cif mutant did not co-elute with neddylated cullins in HeLa cells (Fig. 6B). However, after loading more concentrated amounts of the histidine-eluted fractions, western blot assays showed a weak (compared to wild-type His-Cif) but specific (compared to empty vector) signal using anti-cullin 2 and 4B antibodies in the His-Cif cysteine mutant immunoprecipitated fraction (Fig. 6C). Most likely, this weak signal reflects the low level of neddylated cullins observed in absence of active Cif, although it is also possible that the cysteine mutation partially impairs Cif ability to interact with neddylated cullins. These findings demonstrate that Cif induces the accumulation of neddylated cullins and also interacts with the NEDD8-conjugated form of these cullins.
Fig. 6. Cif interacts with NEDD8-conjugated cullins. 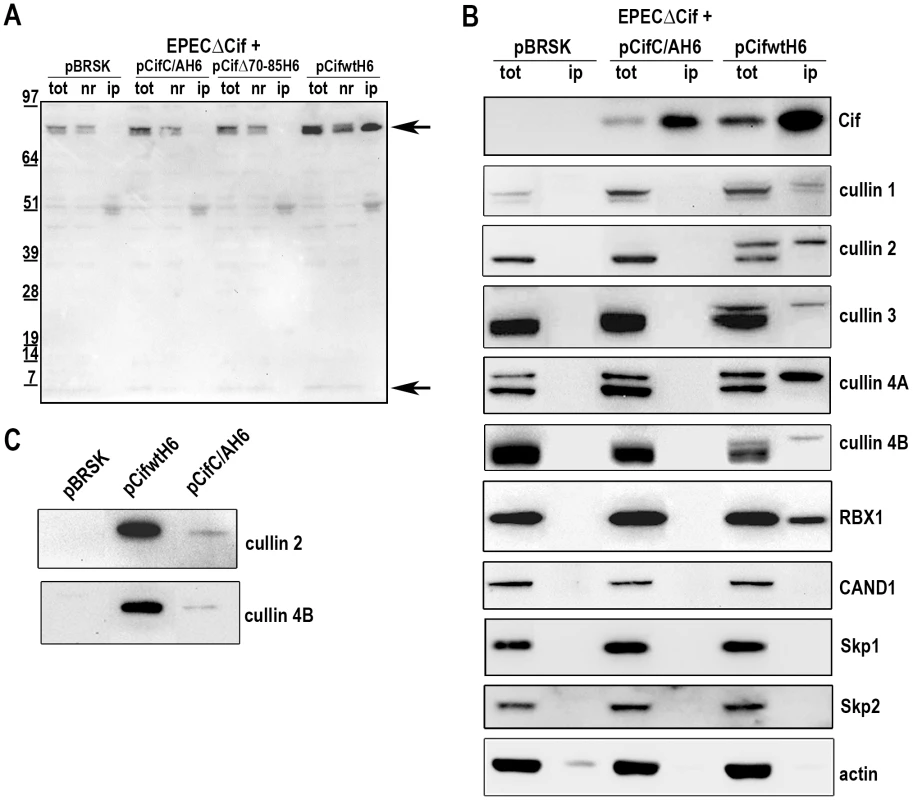
HeLa cells were infected 3 h with EPECΔcif carrying the empty vector (pBRSK) or plasmids expressing His-tagged E. coli Cif cysteine mutant (C/A), 70-85 deleted mutant (Δ70-85) or wild-type (WT) and further incubated 3 h with antibiotics. Cell extracts were immunoprecipitated with anti-His antibodies. (A) Total (tot), non-retained (nr) and immunoprecipitated (ip) protein fractions were probed with anti-NEDD8 antibodies. Arrows indicate neddylated cullins (top) and monomeric NEDD8 (bottom). Molecular weights in kDa are indicated. (B) Total and immunoprecipitated protein fractions were probed with anti-Cif, anti-cullins (CUL1 to CUL4B), anti-RBX1, anti-CAND1, anti-Skp1, anti-Skp2 and anti-actin antibodies. (C) High loads of immunoprecipitated fractions were probed with anti-cullins (CUL2 and CUL4B) antibodies. Since cullins nucleate CRLs by recruiting a RING protein and substrate recognition module [27], we investigated which cullin-associated proteins are also pulled out of lysates by Cif. Analysis of immunoprecipited fractions showed that the RING protein subunit, RBX1, co-eluted with Cif (Fig. 6B). However, neither substrate adaptators for the CUL1-associated CRL, Skp1 and Skp2, nor the CRL inhibitor CAND1 (cullin-associated and neddylation-dissociated 1) were recovered in these fractions (Fig. 6B). Lack of CAND1 association is consistent with the fact that CAND1 only binds non-neddylated cullins [27], which are absent in the Cif eluate. The data also indicate that while Cif is able to interact with cullin-RING complexes via neddylated cullin subunits, these complexes may not contain substrate adaptor modules.
Cif inhibits in vitro ubiquitylation activity of a NEDD8-modified CRL
Given that Cif associates with NEDD8-modified cullin-RING complexes, we wished to test the effects of Cif in an in vitro assay monitoring ubiquitylation activity of a neddylated CRL. We examined ubiquitylation of a biotin-conjugated phosphopeptide corresponding to a well-characterized degron from Cyclin E [28], [29], [30], [31]. When Cif is preinubated with a neddylated SCFFbw7 (NEDD8∼CUL1-Rbx1-Skp1-Fbw7ΔD), ubiquitylation of the cyclin E phosphopeptide was inhibited in a manner that depended on the putative Cif active site cysteine (Fig. 7A). This inhibition also required preincubation of Cif with the neddylated CRL complex, prior to initiating the ubiquitylation reaction (Fig. 7A), indicating that Cif probably acts through a progressive process rather than an immediate effect. A titration experiment showed a concentration-dependent inhibition of in vitro ubiquitylation activity, with some inhibition observed even when the concentration of wild-type Cif was lower than that of the neddylated CRL (Fig. 7B). Thus, it seems likely that inhibition relies on Cif enzymatic activity toward the neddylated CRL, and not toward other components of the ubiquitylation reaction. Together, these data indicate that Cif acts directly to inhibit the neddylated CUL1-associated ubiquitin ligase activity.
Fig. 7. Cif decreases neddylated SCF-mediated substrate polyubiquitylation in vitro. 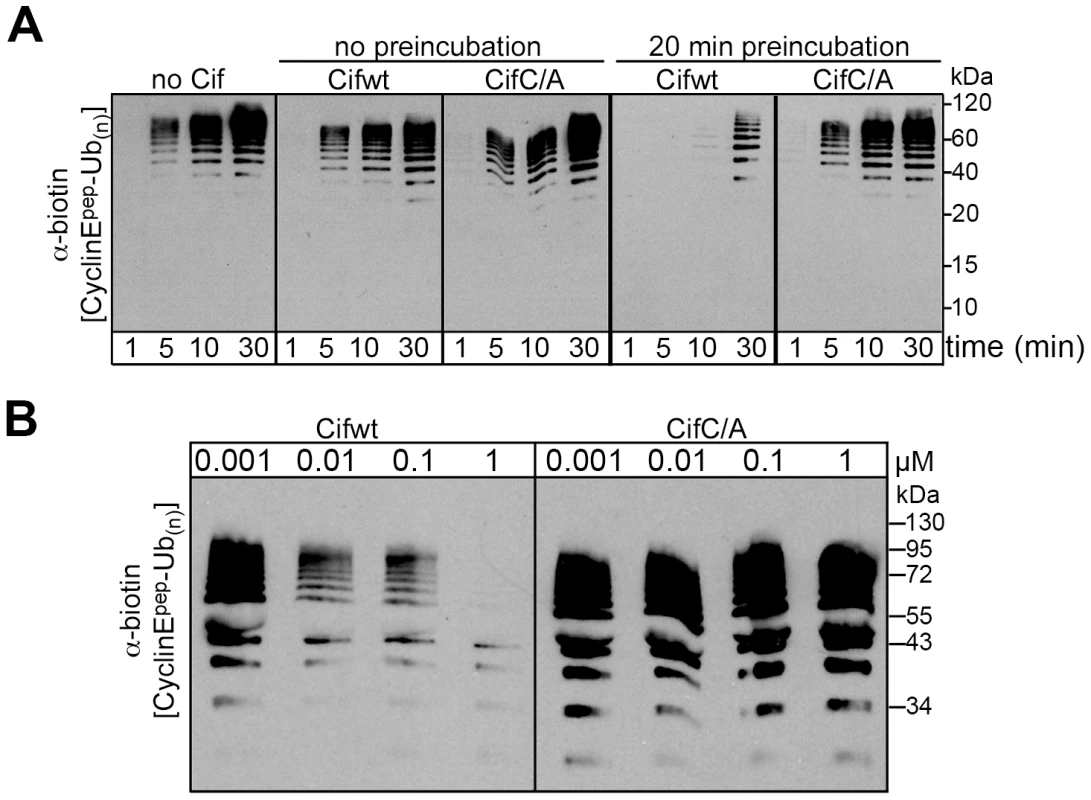
(A) Anti-biotin western blot showing time courses of polyubiquitylation of a biotin-labeled cyclin E phosphopeptide by NEDD8-modified SCFFbw7ΔD in the presence or absence of purified His-tagged wild type Cif (Cifwt) or Cif catalytic Cys-to-Ala (CifC/A) mutant. Reactions were performed by adding Cif straight to the reaction (no preincubation), or after 20 min room temperature incubation of NEDD8-modified SCFFbw7(263-C) with Cif prior to initiating the reaction by adding all other proteins for polyubiquitylation activity (20 min preincubation). (B) Anti-biotin western blot showing titration of the indicated amount of Cifwt or CifC/A with 200 nM SCFFbw7ΔD, prior to 10-min long polyubiquitylation reactions for a biotin-labeled cyclin E phosphopeptide. In all these reactions, the indicated version of Cif was preincubated with NEDD8-modified SCFFbw7ΔD for 1 h prior to reaction initiation. Cif blocks the degradation of various CRL substrates in cells
Given Cif association with cullin-RING complexes, and inhibition of in vitro ubiquitylation activity for a CUL1-based CRL, we wondered whether Cif might exert such inhibition in vivo to block the degradation of various CRL substrates. To address this, the total levels of several known CRL substrates were monitored during a time course following HeLa cell infection with EPEC expressing the wild-type Cif or the Cif cysteine mutant. Western blot analyses showed that, p21, p27 (CUL1-Skp2 substrates), phospho-IκB (a CUL1-βTrCP substrate) and RhoA (a CUL3-BACURD substrate) accumulated in the presence of Cif (Fig. 8A). The total levels of other potential targets (Cdt1 and β-catenin) were only slightly modified in presence of Cif (data not shown). To evaluate the effect of Cif on these CRL substrates in a more sensitive assay, the half-lives of Cdt1 (a CUL4-DDB1-CDT2 substrate) and β-catenin (a CUL1-βTrCP substrate) were analyzed in the presence of the protein synthesis inhibitor cycloheximide. Both Cdt1 and β-catenin appeared to be stabilized by wild-type Cif, but not the cysteine mutant (Fig. 8B). We also examined the half-lives of substrate adaptors of CUL1, Skp1 and Skp2. In contrast to Skp1, whose level remained stable irrespective of the Cif status, the half-life of Skp2 increased in the presence of wild-type Cif (Fig. 8B). Altogether, these results demonstrate that Cif is able to control the stability of several key host cell regulators by targeting their cullin-associated ubiquitin ligases.
Fig. 8. Cif induces stabilization of various targets of cullin-RING ubiquitin ligases. 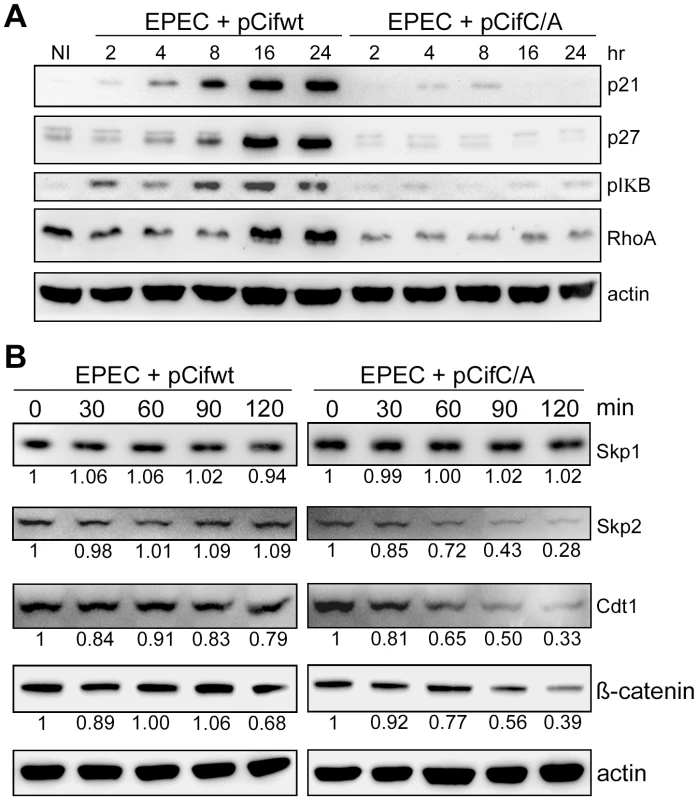
(A) HeLa cells were infected (starting at 0 h) for 2 h with EPECΔcif carrying a plasmid encoding wild-type (WT) or cysteine mutant (C/A) Cif and further incubated for indicated times with antibiotics. Cell protein extracts were probed with anti-p21, p27, phospho-IκB, RhoA or actin antibodies. NI: non-infected. (B) HeLa cells were infected for 2 h with EPECΔcif carrying the plasmid expressing cysteine mutant (pCifC/A) or wild-type Cif (pCifwt) and then (starting at 0 min) incubated for the indicated time with cycloheximide. Cell protein extracts were probed with anti-Skp1, Skp2, Cdt1, β-catenin or actin antibodies. The relative amount of protein (relative to 0 min and normalized to actin level) is indicated below each western blot. Discussion
Cif proteins from vertebrate and invertebrate bacterial pathogens induce accumulation of p21 and p27 resulting in cell cycle arrest [6], [15]. Here, we have shown that Cif binds to NEDD8 and neddylated cullins, inducing their accumulation early after infection. Cullins are scaffold proteins nucleating Cullin-RING E3 ubiquitin-Ligases (CRLs), and neddylation activates cullins to modulate numerous cell functions, including cell cycle progression by targeting p21 and p27 for ubiquitin-dependent degradation [22], [32]. In this study, we show that Cif can directly inhibit the ubiquitin ligase activity of a neddylated CUL1-associated complex, which correlates with the Cif-mediated accumulation of p21 and p27. Consistent with Cif's association with different neddylated cullins, we observed a stabilization of substrates from a range of CRLs. The mode of action of Cif represents a novel strategy for modulation of a broad range of host cell functions by bacterial pathogens.
Cif inhibits the ubiquitin ligase function of CRLs
NEDD8 conjugation of cullins is a major form of CRL regulation. Neddylation activates CRL-mediated substrate ubiquitylation by inducing a drastic conformational change of the C-terminal part of the cullin that frees the RBX1 RING, and thereby imparts catalytic geometries to an associated E2 enzyme [19], [27], [33]. Here, we report that Cif associates with the neddylated form of CUL1 to 4B, and that pre-incubation of Cif with a neddylated SCFFbw7 complex inhibits its ligase activity. This latter result suggests that Cif acts on one or several components present in this reconstituted ubiquitylation assay. Whether Cif uses an enzymatic activity to usurp the CRL functions, and/or poisons the complex by binding to neddylated cullins is still elusive. However, the fact that preincubation of wild-type Cif with the neddylated SCF-Fbw7 is necessary to inhibit the ubiquitin ligase activity, and that Cif mutated on the cysteine residue of the catalytic triad is not inhibitory, support a predominant role for the putative enzymatic activity of Cif, which is (based on structure data) a member of the superfamily of cysteine protease, acetyl transferases, deamidases or transglutaminases. Interestingly, co-immunoprecipitation assays revealed an absence of association of Cif-bound CUL1 with Skp1-Skp2 adaptators. This Cif-induced loss of substrate recognition subunits would further lead to CRL inhibition in vivo. However, how Cif could induce dissociation/prevent association of Skp1-Skp2 from cullins, or how this effect is related to CRL inhibition by Cif remains to be elucidated.
Cif induces cellular accumulation of neddylated cullins
CRLs transit through different stages of assembly, sequestration, neddylation, and deneddylation. To be fully active, CRLs undergo an activation cycle in which cullins oscillate between neddylated and non-neddylated (potentially sequestered by CAND1) states [27], [33]. Based on our results, we propose a model in which Cif interferes with this cycle and thus locks CRLs in a neddylated but inactive state. Cif appears to block many steps in the CRL activation cycle. First, we observe that CAND1, Skp1 and Skp2 are not associated with Cif-associated CRLs. Cullin neddylation may correlate with lack of CAND1 binding. Second, the levels of CRL substrates, and substrate-binding adaptors, may also influence cullin neddylation. A regulatory negative feed-back loop via cullin de-neddylation was reported to modulate CRL activity once substrate level decreases under a critical threshold [34]. Furthermore, Bornstein and collaborators reported that supplementation of Skp1-Skp2 and substrate to extracts of HeLa cells synergistically increased the level of neddylated CUL1 [35]. We found Cif-inhibition of CRLs also to correlate with the accumulation of numerous substrates and increased half-life of Skp2, which might secondarily result in accumulation of neddylated cullins, even though Cif-associated neddylated cullins are depleted of Skp1-Skp2 and presumably also inactivated. Finally, it is also possible that Cif interferes with cullin deneddylation. Since Cif treatment also inactivates neddylated cullins, their accumulation may essentially shut down both CRL assembly cycles as well as CRL ubiquitylation activities.
Cif stabilizes numerous targets of CRLs
CRLs represent the largest subfamily of E3s and therefore play regulatory roles in numerous and diverse cellular functions. Since Cif binds to various neddylated cullins, we anticipated that, in addition to p21 and p27, Cif could modulate the stability of substrates of multiple different CRLs. Indeed, Cif induces not only cell cycle arrest but also stress fibers formation in certain cell lines [4], [6], [11], [12], [14]. A recent study showed that dysfunction of the CUL3-BACURD complex results in abnormal actin stress fibers and distorted cell morphology, owing to impaired ubiquitylation and degradation of the small GTPase RhoA [36]. Our results show that Cif inhibition/interaction with neddylated CUL3 correlates with accumulation of RhoA, thus providing a likely explanation for Cif-dependent stress fiber formation. We also investigated the stability of Cdt1 whose level is controlled by CUL4-DDB1-Cdt2 ligase complex [37], [38], [39]. Cdt1 is a licensing factor that plays a major role in the control of the S-phase [40]. We showed that Cif increases the half-life of Cdt1, thus providing a mechanistic link for the DNA re-replication that occurs in a fraction of HeLa cells 2 or 3 days post-infection [4], [14]. Cif also increases the stability of β-catenin whose level is important in the Wnt signaling pathway that is involved in homeostatic self-renewal in a number of adult tissues including intestinal epithelium, and in colon carcinogenesis [41]. Finally, we have also shown in this study that Cif inhibits the degradation of phospho-IκB, an inhibitor of the NF-κB pro-inflammatory response. Further experiments will be necessary to investigate thoroughly the role of Cif in these pathways and to evaluate whether this effector could participate to escape the host immune response and plays an important role in bacterial pathogenesis.
Cif, a unique tool to study CRL regulation
Many bacterial toxins or microbial products mimic or modulate the activity of E3 ubiquitin ligases [42], [43], [44], [45]. For instance, deneddylation of CRL and consequently down-regulation of the NF-κB pathway have been proposed to explain the ability of intestinal bacterial communities to regulate the inflammatory tolerance of the mammalian intestinal epithelia [46], [47]. Similarly, a deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases [48]. However, to our knowledge, inhibition of CRLs through a pathogen's protein binding to NEDD8 has never been reported in the pathogenesis of bacterial or viral infections. The requirement that CRLs be activated by NEDD8 conjugation on the cullin proteins offers an “achilles heel” for modulating this entire subfamily. Owing to this remarkable property, Cif appears as a unique tool to dissect the regulation of CRL-dependent ubiquitylation and neddylation processes and more generally, to investigate many host-signaling pathways including cell cycle regulation, DNA repair, immune and inflammatory responses, antigen processing or cytoskeleton dynamics.
Materials and Methods
Cell lines and bacterial strains
Homo Sapiens HeLa (ATCC CCL-2), and HEK 293T (ATCC CRL-11268) cell lines were cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Eurobio) and 50 µg mL−1 gentamicin at 37°C in a 5% CO2 atmosphere. Rattus Norvegicus IEC-6 (CRL-1592) cells were grown in the above medium supplemented with bovine insulin (0.1 units mL−1; Sigma). H. sapiens HCT116 (ATCC CCL-247) were maintained in McCoy's 5A Medium (Invitrogen) supplemented with FBS and gentamicin as above. Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were cultured in lysogeny broth (LB) or in interaction medium (DMEM with 25 mM Hepes and 5% FBS). Antibiotics were used at the following concentrations: carbenicillin 50 µg mL−1, streptomycin 60 µg mL−1, chloramphenicol 20 µg mL−1 and kanamycin 25 µg mL−1.
Tab. 1. <i>E. coli</i> strains and plasmids used in this study. 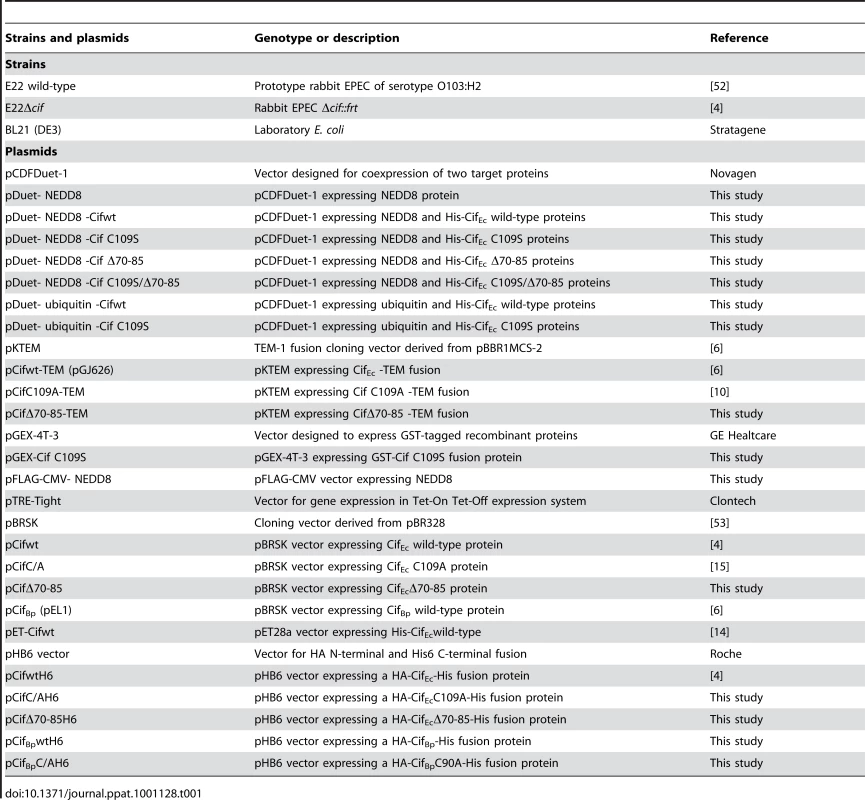
Generation of stable cell lines expressing GFP-Cif and GFP-Cif C109A
Fusion genes encoding GFP-Cif wild-type and C109A were amplified by PCR from plasmids pRN1 and pRN2 respectively [6] with primers pTRE-GFPCif-F (CGC CAC CAT GAG CGG GGG CGA) and pTagGFP-Cif-R (CGG GAT CCC TAA CTA CAT AGT GAT TTT ATT ATC TC). PCR fragments were digested with BamHI and ligated into the SmaI and BamHI sites of pTRE-Tight vector (Clontech). Both constructs were verified by DNA sequencing. After PvuI digestion, each plasmid was co-transfected with linear hygromycin selection marker into HeLa Tet-On Advanced cells (Clontech) using FuGENE6 transfection reagent (Roche) and cells were plated with the addition of 200 µg mL−1 of hygromycin B. Positive clones were identified by screening expression of GFP-Cif fusion proteins in presence of 2 µg mL−1 doxycycline inducer by fluorescence microscopy and western blot using anti-Cif and anti-Tag(CGY)FP antibodies (Evrogen).
Yeast two hybrid screenings
Yeast two hybrid (Y2H) screenings were performed and analyzed by Hybrigenics (Paris, France). Briefly, wild-type Cif and Cif C109S mutant were PCR amplified and cloned into a Y2H vector optimized by Hybrigenics, checked by sequencing, and used as bait for Y2H screenings of both a human colon and a placenta cDNA libraries. Candidate prey fragments were sequenced and compared to the GenBank database using BLASTN. A predicted biological score was used to assess the reliability of each interaction. This score takes into account (i) the redundancy and independency of prey fragments as well as the distributions of reading frames and stop codons in overlapping fragments, (ii) the interactions found in all the screens performed at Hybrigenics using the same library. This score has been shown to positively correlate with the biological significance of interactions [49].
NEDD8 binding assays
For bacterial co-expression experiments, the gene encoding NEDD8 was cloned into the second multiple cloning site of pCDFDuet-1 vector (Novagen) and genes encoding Cif proteins (wild-type or the mutants C109S, Δ70-85, C109S/Δ70-85) were cloned into the first multiple cloning site allowing expression of Cif tagged with a N-terminal hexahistidine motif. E. coli BL21(DE3) harboring the different constructs was grown at 37°C to OD 600 nm of 0.8, then induced with 1 mM IPTG and grown for an additional 3 h. The bacteria were lysed by sonication in presence of protease inhibitors (Complete, Roche). After centrifugation, bacterial extracts (supernatant) were incubated with Ni-NTA beads at 4°C for 16 h. Beads were washed four times with phosphate buffered saline (PBS) containing 0.1% NP40 and 20 mM imidazole. Bound proteins were eluted by resuspending beads with 60 µl of 3× SDS loading buffer and heating samples 10 min at 95°C for western blot analysis or with PBS buffer containing 250 mM imidazole for gel filtration chromatography analysis.
For GST pull-down, Cif C109S was cloned into the pGEX4T3 vector and the fusion protein GST-CifC109S was expressed in BL21(DE3) cells and purified using glutathione sepharose (GE Healthcare). HEK293T cells were transfected with pFLAG-CMV-NEDD8 using GenePORTER2 Transfection Reagent (Genlantis) following the manufacturer's protocol. 48 h post transfection, HEK 293T were lysed in RIPA buffer (50 mM Tris pH 8, 250 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS) in 3 freeze thaw cycles. Firstly, 100 µg of purified GST-CifC109S was bound to pre-equilibrated Glutathione Sepharose 4B in equilibrating buffer containing 20 mM Hepes pH 7.5, 5 mM MgCl2, 100 mM NaCl, and 2 mM DTT at 4°C for 1 h on a rotator. Either 100 µg or 250 µg of HEK 293T lysate was added to the GST-C109S bounded glutathione beads and further incubated at 4°C for 2 h on a rotator. The glutathione beads were washed once with equilibrating buffer, twice with wash buffer (20 mM Hepes pH 7.5, 300 mM NaCl, 5 mM MgCl2, 0.1% NP-40, 2 mM DTT), and lastly once with equilibrating buffer. 40 µl of 3xSDS loading buffer were added to the protein bound beads, and the beads were boiled at 95°C for 5 minutes. The samples containing only the supernatant were separated by SDS-PAGE and analyzed by immunoblotting membranes with anti-FLAG monoclonal antibody (Sigma).
Infection and cell treatments
Infection experiments were performed as previously described [6]. Briefly, cells were infected 2 h with bacteria grown in interaction medium, with a multiplicity of infection (moi) of 50 bacteria per cell. After the infection, cells were washed with Hank's balanced salt solution (HBSS; Invitrogen) and cultivated for the indicated times in DMEM medium supplemented with 10% FBS and 200 µg mL−1 gentamicin. When needed, cycloheximide was added to the medium at 50 µg mL−1. For synchronization in G1/S phase, HeLa cells were treated with 2 mM thymidine (Sigma) for 18 h, washed 3 times with HBSS, incubated in normal medium for 9 h and treated again with 2 mM thymidine for 16 h.
Actin stress fiber and cell cycle analyses
For cell morphology and actin cytoskeleton visualization, cells were fixed for 15 min in PBS 4% formaldehyde, permeabilized with 0.1% Triton X-100 and stained with rhodamine-phalloidin (Molecular Probes), DAPI (Sigma) or TO-PRO-3 (Invitrogen). Images were acquired with a DMRB fluorescence microscope equipped with a DFC300FX digital camera (Leica). For cell cycle distribution analyses, cells were trypsinized, washed, fixed with ethanol, stained with propidium iodide and analyzed using a FACScalibur flow cytometer (Becton Dickinson). For GFP quantification and cell cycle analyses of stable HeLa Tet On cells expressing GFP-Cif, cells were trypsinized, washed with ice-cold PBS, fixed for 3 h at 4°C in PBS with 1% formaldehyde, permeabilized overnight at 4°C in PBS ethanol 70%, stained with propidium iodide for 30 min at 37°C and analyzed by flow cytometry. Data from at least 20 000 cells were analyzed using FloJo software v8.5 (Tree Star).
Cif and NEDD8 localization
Cells were grown and treated on LabTek glass slides, fixed with PBS 3.7% formaldehyde 15 min at room temperature, permeabilized with PBS 0.1% Triton-X-100 for 5 min, and blocked with PBS 0.1% Tween-20 1% bovine serum albumine (BSA) 1% normal goat serum. Slides were incubated overnight at 4°C with rabbit anti-NEDD8 antibodies (CST) or affinity purified anti-Cif antibodies [10] diluted 1∶200 in blocking buffer. Cells were washed and incubated 30 min at room temperature with affinity purified FITC or TRITC-conjugated goat anti-rabbit antibodies (Zymed) diluted 1∶150. After washing DNA was stained 5 min with TO-PRO-3 (Invitrogen) and the slides were mounted with Vectashield (Vector Laboratories). Images were captured with an Olympus-IX70 laser scanning confocal microscope using a ×60 PLANAPO NA = 1.4 objective. The confocal aperture was set for a z-axis thickness of 0.23 µm and images stacks were acquired in sequential mode, and analyzed with the Olympus FV500 FluoView software.
Cell sample preparation and western blot analyses
For western blot analysis, 6×105 cells were lysed in 80 µl of SDS-PAGE sample buffer, sonicated for 2 s to shear DNA and then boiled for 5 min. Cytoplasmic and nuclear extracts were obtained as described elsewhere [50]. For deneddylation assays, 50 µL of samples were treated with 0.5 µg of recombinant NEDP1 (NEDD8 protease 1; Biomol). Cell extracts were heated for 5 min at 100°C after addition of 4× Laemli loading buffer. Protein samples were resolved on 4–12% NuPage gradient gels (Invitrogen) and blotted on PVDF membranes. Membranes were blocked in TBST (10 mM Tris pH 7.8, 150 mM NaCl, 0.1% Tween20) 5% non-fat dry milk, then probed with primary antibody (0.5 µg mL−1) in TBST 5% non-fat dry milk. Primary antibodies were: anti-FLAG (Sigma), anti-NEDD8, anti-Cdt1, anti-lamin, anti-Skp1 (Cell Signaling Technology), anti-actin (ICN), affinity-purified anti-Cif [10], anti-p21, anti-p27, anti-Skp2, anti-14-3-3, anti-RhoA, anti-ubiquitin (Santa Cruz Biotechnology), anti-RBX1 ( = ROC1), anti-CAND1 ( = TIP120A) and anti-cullins 1 to 4B (Abcam). Bound antibodies were visualized with horseradish peroxidase-conjugated secondary antibodies. Acquisitions were performed with a Molecular Imager ChemiDoc XRS system (Bio-Rad).
Immunoprecipitation assays
HeLa cells were infected with a moi of 100 bacteria per cell. After 3 h infection, cells were washed with HBSS and cultivated for 3 h in DMEM medium supplemented with 10% FBS and 200 µg mL−1 gentamicin and then were harvested in lysis buffer (50 mM Tris HCl pH 8, 150 mM NaCl, 1% Triton X-100, protease inhibitors). Extracts were processed according to manufacturer's protocol with µMACS anti-His microbeads (Miltenyi Biotec). Samples corresponding to total and non-retained (flow-through) proteins were collected during the immunoprecipitation protocol.
Translocation assays
Translocation levels of Cif-TEM fusion proteins were determined using CCF2/AM (Invitrogen) as a substrate for intracellular TEM enzyme as described previously [6], [8]. Briefly, HeLa cells seeded in black 96-well plates were loaded for 1 h at 37°C with 1.7 mM of CCF2/AM diluted in DMEM with 2 mM probenecid and then infected for 2 h with bacteria expressing TEM fusion proteins. Fluorescence was quantified in a microplate reader (TECAN Infinite M200) with excitation set at 410 nm (9 nm bandwidth) and emission at 450 nm for blue fluorescence and 520 nm for green fluorescence (20 nm bandwidth). Translocation was expressed as the emission ratio at 450/520 nm.
In vitro assays for neddylated SCFFbw7-mediated polyubiquitylation
NEDD8 modification of SCF was carried out as previously described [19]. UBA1 was expressed as described [51]. GST-Fbw7ΔD (i.e., the well-behaved monomeric Fbw7 containing residue 263 to the C-terminus) and Skp1 were coexpressed, and GST-Cdc34B, His-tagged Cif wild-type and Cif C/A were expressed in BL21 (DE3) Gold cells (Stratagene). GST-tagged proteins were purified by glutathione affinity, TEV or thrombin-cleaved and further purified by anion exchange and gel filtration chromatography. His-tagged Cif was purified by nickel-NTA affinity (Qiagen) and anion exchange chromatography. All proteins were desalted into 30 mM Tris-HCL, 20 mM NaCl pH 7.6. The neddylated SCFFbw7ΔD ubiquitylation substrate peptide was synthesized and purified by reversed-phase HPLC by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude (Memphis, TN, USA), and corresponds to the high-affinity phosphodegron sequence from Cyclin E, with the sequence KAMLSEQNRASPLPSGLLT*PPQS*GRRASY (* = phosphorylation) [30], [31]. The peptide N-terminal sequence is acetylated, and the C-terminus is linked to a biotin moiety. Polyubiquitylation without Cif preincubation was assayed at room temperature with 200 nM NEDD8 modified SCFFbw7ΔD, 0.5 µM Cdc34B, 50 µM ubiquitin, 1 nM–1 µM Cif as indicated (wild-type or catalytically-inactive Cys-to-Ala mutant), 2 mg mL−1 BSA and 5 µM biotin-conjugated cyclin E phosphopeptide in 30 mM Tris-HCL, 20 mM NaCl, 10 mM MgCl2, 5 mM ATP, pH 7.6, and started with 250 nM UBA1. Reactions were stopped with SDS sample buffer, and products were separated by SDS-PAGE, and visualized by western blot with anti-biotin antibodies (Rockland, 100-4198). For the preincubation time-courses, 1 µM Cif was mixed with 200 nM NEDD8 modified SCFFbw7ΔD for 20 min at room temperature prior to the addition of other reaction components, and reactions were stopped at the indicated times. For the titration experiments, a range of concentrations of Cif were incubated with 200 nM NEDD8 modified SCFFbw7ΔD for 1 h at room temperature prior to the addition of other reaction components, and reactions were stopped after 10 min.
Zdroje
1. Samba-LouakaA
TaiebF
NougayredeJP
OswaldE
2009 Cif type III effector protein: a smart hijacker of the host cell cycle. Future Microbiol 4 867 877
2. NougayredeJP
TaiebF
De RyckeJ
OswaldE
2005 Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol 13 103 110
3. OswaldE
NougayredeJP
TaiebF
SugaiM
2005 Bacterial toxins that modulate host cell-cycle progression. Curr Opin Microbiol 8 83 91
4. MarchesO
LedgerTN
BouryM
OharaM
TuX
2003 Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol Microbiol 50 1553 1567
5. YaoQ
CuiJ
ZhuY
WangG
HuL
2009 A bacterial type III effector family uses the papain-like hydrolytic activity to arrest the host cell cycle. Proc Natl Acad Sci U S A 106 3716 3721
6. JubelinG
ChavezCV
TaiebF
BanfieldMJ
Samba-LouakaA
2009 Cycle inhibiting factors (CIFs) are a growing family of functional cyclomodulins present in invertebrate and mammal bacterial pathogens. PLoS ONE 4 e4855
7. MillsE
BaruchK
CharpentierX
KobiS
RosenshineI
2008 Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe 3 104 113
8. CharpentierX
OswaldE
2004 Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J Bacteriol 186 5486 5495
9. CrowA
RacePR
JubelinG
Varela ChavezC
EscoubasJM
2009 Crystal structures of Cif from bacterial pathogens Photorhabdus luminescens and Burkholderia pseudomallei. PLoS One 4 e5582
10. HsuY
JubelinG
TaiebF
NougayredeJP
OswaldE
2008 Structure of the cyclomodulin Cif from pathogenic Escherichia coli. J Mol Biol 384 465 477
11. NougayredeJP
BouryM
TascaC
MarchesO
MilonA
2001 Type III secretion-dependent cell cycle block caused in HeLa cells by enteropathogenic Escherichia coli O103. Infect Immun 69 6785 6795
12. De RyckeJ
ComtetE
ChalarengC
BouryM
TascaC
1997 Enteropathogenic Escherichia coli O103 from rabbit elicits actin stress fibers and focal adhesions in HeLa epithelial cells, cytopathic effects that are linked to an analog of the locus of enterocyte effacement. Infect Immun 65 2555 2563
13. Samba-LouakaA
NougayredeJP
WatrinC
OswaldE
TaiebF
2009 The EPEC effector Cif induces delayed apoptosis in epithelial cells. Infect Immun 77 5471 5477
14. TaiebF
NougayredeJP
WatrinC
Samba-LouakaA
OswaldE
2006 Escherichia coli cyclomodulin Cif induces G2 arrest of the host cell cycle without activation of the DNA-damage checkpoint-signalling pathway. Cell Microbiol 8 1910 1921
15. Samba-LouakaA
NougayredeJP
WatrinC
JubelinG
OswaldE
2008 Bacterial cyclomodulin Cif blocks the host cell cycle by stabilizing the cyclin-dependent kinase inhibitors p21 and p27. Cell Microbiol 10 2496 2508
16. CayrolC
KnibiehlerM
DucommunB
1998 p21 binding to PCNA causes G1 and G2 cell cycle arrest in p53-deficient cells. Oncogene 16 311 320
17. CiechanoverA
FinleyD
VarshavskyA
1984 The ubiquitin-mediated proteolytic pathway and mechanisms of energy-dependent intracellular protein degradation. J Cell Biochem 24 27 53
18. PickartCM
CohenRE
2004 Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol 5 177 187
19. DudaDM
BorgLA
ScottDC
HuntHW
HammelM
2008 Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134 995 1006
20. SahaA
DeshaiesRJ
2008 Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 32 21 31
21. RabutG
PeterM
2008 Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 9 969 976
22. NakayamaKI
NakayamaK
2005 Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol 16 323 333
23. YuJ
FinleyRLJr
2009 Combining multiple positive training sets to generate confidence scores for protein-protein interactions. Bioinformatics 25 105 111
24. KamitaniT
KitoK
NguyenHP
YehET
1997 Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem 272 28557 28562
25. XirodimasDP
2008 Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans 36 802 806
26. JonesJ
WuK
YangY
GuerreroC
NillegodaN
2008 A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J Proteome Res 7 1274 1287
27. BosuDR
KipreosET
2008 Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div 3 7
28. WelckerM
ClurmanBE
2008 FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 8 83 93
29. YeX
NalepaG
WelckerM
KesslerBM
SpoonerE
2004 Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J Biol Chem 279 50110 50119
30. KleigerG
SahaA
LewisS
KuhlmanB
DeshaiesRJ
2009 Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139 957 968
31. PierceNW
KleigerG
ShanSO
DeshaiesRJ
2009 Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462 615 619
32. PetroskiMD
DeshaiesRJ
2005 Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6 9 20
33. MerletJ
BurgerJ
GomesJE
PintardL
2009 Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci 66 1924 1938
34. ChewEH
HagenT
2007 Substrate-mediated regulation of cullin neddylation. J Biol Chem 282 17032 17040
35. BornsteinG
GanothD
HershkoA
2006 Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc Natl Acad Sci U S A 103 11515 11520
36. ChenY
YangZ
MengM
ZhaoY
DongN
2009 Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell 35 841 855
37. JinJ
AriasEE
ChenJ
HarperJW
WalterJC
2006 A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 23 709 721
38. NishitaniH
SugimotoN
RoukosV
NakanishiY
SaijoM
2006 Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J 25 1126 1136
39. SengaT
SivaprasadU
ZhuW
ParkJH
AriasEE
2006 PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem 281 6246 6252
40. XouriG
DimakiM
BastiaensPI
LygerouZ
2007 Cdt1 interactions in the licensing process: a model for dynamic spatiotemporal control of licensing. Cell Cycle 6 1549 1552
41. CleversH
2006 Wnt/beta-catenin signaling in development and disease. Cell 127 469 480
42. HicksSW
GalanJE
2010 Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr Opin Microbiol 13 41 46
43. SpallekT
RobatzekS
GohreV
2009 How microbes utilize host ubiquitination. Cell Microbiol 11 1425 1434
44. AngotA
VergunstA
GeninS
PeetersN
2007 Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog 3 e3
45. RytkonenA
HoldenDW
2007 Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe 1 13 22
46. Collier-HyamsLS
SloaneV
BattenBC
NeishAS
2005 Cutting edge: bacterial modulation of epithelial signaling via changes in neddylation of cullin-1. J Immunol 175 4194 4198
47. KumarA
WuH
Collier-HyamsLS
HansenJM
LiT
2007 Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J 26 4457 4466
48. GastaldelloS
HildebrandS
FaridaniO
CallegariS
PalmkvistM
A deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat Cell Biol 12 351 361
49. FormstecherE
ArestaS
ColluraV
HamburgerA
MeilA
2005 Protein interaction mapping: a Drosophila case study. Genome Res 15 376 384
50. PortalD
RosendorffA
KieffE
2006 Epstein-Barr nuclear antigen leader protein coactivates transcription through interaction with histone deacetylase 4. Proc Natl Acad Sci U S A 103 19278 19283
51. HuangDT
AyraultO
HuntHW
TaherbhoyAM
DudaDM
2009 E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell 33 483 495
52. NougayredeJP
MarchesO
BouryM
MainilJ
CharlierG
1999 The long-term cytoskeletal rearrangement induced by rabbit enteropathogenic Escherichia coli is Esp dependent but intimin independent. Mol Microbiol 31 19 30
53. MarchesO
NougayredeJP
BoullierS
MainilJ
CharlierG
2000 Role of tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect Immun 68 2171 2182
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek SRFR1 Negatively Regulates Plant NB-LRR Resistance Protein Accumulation to Prevent AutoimmunityČlánek Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on VirulenceČlánek Phylogenetic Approach Reveals That Virus Genotype Largely Determines HIV Set-Point Viral LoadČlánek A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 9- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Azole Drugs Are Imported By Facilitated Diffusion in and Other Pathogenic Fungi
- Two Genes on A/J Chromosome 18 Are Associated with Susceptibility to Infection by Combined Microarray and QTL Analyses
- Impact of Simian Immunodeficiency Virus Infection on Chimpanzee Population Dynamics
- Breaking the Stereotype: Virulence Factor–Mediated Protection of Host Cells in Bacterial Pathogenesis
- The Canine Papillomavirus and Gamma HPV E7 Proteins Use an Alternative Domain to Bind and Destabilize the Retinoblastoma Protein
- Rescue of HIV-1 Release by Targeting Widely Divergent NEDD4-Type Ubiquitin Ligases and Isolated Catalytic HECT Domains to Gag
- Steric Shielding of Surface Epitopes and Impaired Immune Recognition Induced by the Ebola Virus Glycoprotein
- Dynamics of the Multiplicity of Cellular Infection in a Plant Virus
- HLA Class I Binding of HBZ Determines Outcome in HTLV-1 Infection
- Pathogenic Bacteria Target NEDD8-Conjugated Cullins to Hijack Host-Cell Signaling Pathways
- The HA and NS Genes of Human H5N1 Influenza A Virus Contribute to High Virulence in Ferrets
- SRFR1 Negatively Regulates Plant NB-LRR Resistance Protein Accumulation to Prevent Autoimmunity
- Cyclin-Dependent Kinase Activity Controls the Onset of the HCMV Lytic Cycle
- The N-Terminal Domain of the Arenavirus L Protein Is an RNA Endonuclease Essential in mRNA Transcription
- Generation of Neutralizing Antibodies and Divergence of SIVmac239 in Cynomolgus Macaques Following Short-Term Early Antiretroviral Therapy
- Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
- Intracellular Proton Conductance of the Hepatitis C Virus p7 Protein and Its Contribution to Infectious Virus Production
- The Transcriptome of the Human Pathogen at Single-Nucleotide Resolution
- The Epidermal Growth Factor Receptor (EGFR) Promotes Uptake of Influenza A Viruses (IAV) into Host Cells
- Surface Co-Expression of Two Different PfEMP1 Antigens on Single -Infected Erythrocytes Facilitates Binding to ICAM1 and PECAM1
- Sequestration and Tissue Accumulation of Human Malaria Parasites: Can We Learn Anything from Rodent Models of Malaria?
- Phylogenomics of Ligand-Gated Ion Channels Predicts Monepantel Effect
- Generation of Covalently Closed Circular DNA of Hepatitis B Viruses via Intracellular Recycling Is Regulated in a Virus Specific Manner
- CpG-Methylation Regulates a Class of Epstein-Barr Virus Promoters
- Molecular and Evolutionary Bases of Within-Patient Genotypic and Phenotypic Diversity in Extraintestinal Infections
- A Bistable Switch and Anatomical Site Control Virulence Gene Expression in the Intestine
- Are Members of the Fungal Genus (a) Commensals; (b) Opportunists; (c) Pathogens; or (d) All of the Above?
- Structures of Receptor Complexes of a North American H7N2 Influenza Hemagglutinin with a Loop Deletion in the Receptor Binding Site
- Phylogenetic Approach Reveals That Virus Genotype Largely Determines HIV Set-Point Viral Load
- The Coevolution of Virulence: Tolerance in Perspective
- Involvement of the Cytokine MIF in the Snail Host Immune Response to the Parasite
- Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens
- A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins
- High Content Phenotypic Cell-Based Visual Screen Identifies Acyltrehalose-Containing Glycolipids Involved in Phagosome Remodeling
- A Novel Small Molecule Inhibitor of Hepatitis C Virus Entry
- The Microbiota Mediates Pathogen Clearance from the Gut Lumen after Non-Typhoidal Diarrhea
- RNA Polymerases (L-Protein) Have an N-Terminal, Influenza-Like Endonuclease Domain, Essential for Viral Cap-Dependent Transcription
- Pathogen Specific, IRF3-Dependent Signaling and Innate Resistance to Human Kidney Infection
- Cellular Entry of Ebola Virus Involves Uptake by a Macropinocytosis-Like Mechanism and Subsequent Trafficking through Early and Late Endosomes
- The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis
- Formation of Mobile Chromatin-Associated Nuclear Foci Containing HIV-1 Vpr and VPRBP Is Critical for the Induction of G2 Cell Cycle Arrest
- Association of Tat with Promoters of PTEN and PP2A Subunits Is Key to Transcriptional Activation of Apoptotic Pathways in HIV-Infected CD4+ T Cells
- Metal Hyperaccumulation Armors Plants against Disease
- Cyclin-Dependent Kinase-Like Function Is Shared by the Beta- and Gamma- Subset of the Conserved Herpesvirus Protein Kinases
- Role of Acetyl-Phosphate in Activation of the Rrp2-RpoN-RpoS Pathway in
- Ebolavirus Is Internalized into Host Cells Macropinocytosis in a Viral Glycoprotein-Dependent Manner
- A Novel Family of IMC Proteins Displays a Hierarchical Organization and Functions in Coordinating Parasite Division
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens
- The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis
- Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
- The Coevolution of Virulence: Tolerance in Perspective
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



