-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaInhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
Toll-like receptor signaling requires functional Toll/interleukin-1 (IL-1) receptor (TIR) domains to activate innate immunity. By producing TIR homologous proteins, microbes inhibit host response induction and improve their own survival. The TIR homologous protein TcpC was recently identified as a virulence factor in uropathogenic Escherichia coli (E. coli), suppressing innate immunity by binding to MyD88. This study examined how the host MyD88 genotype modifies the in vivo effects of TcpC and whether additional, TIR-domain containing proteins might be targeted by TcpC. In wild type mice (wt), TcpC enhanced bacterial virulence, increased acute mortality, bacterial persistence and tissue damage after infection with E. coli CFT073 (TcpC+), compared to a ΔTcpC deletion mutant. These effects were attenuated in Myd88−/− and Tlr4−/− mice. Transcriptomic analysis confirmed that TcpC inhibits MYD88 dependent gene expression in CFT073 infected human uroepithelial cells but in addition the inhibitory effect included targets in the TRIF and IL-6/IL-1 signaling pathways, where MYD88 dependent and independent signaling may converge. The effects of TcpC on bacterial persistence were attenuated in Trif −/− or Il-1β −/− mice and innate immune responses to ΔTcpC were increased, confirming that Trif and Il-1β dependent targets might be involved in vivo, in addition to Myd88. Furthermore, soluble TcpC inhibited Myd88 and Trif dependent TLR signaling in murine macrophages. Our results suggest that TcpC may promote UTI-associated pathology broadly, through inhibition of TIR domain signaling and downstream pathways. Dysregulation of the host response by microbial TcpC thus appears to impair the protective effects of innate immunity, while promoting inflammation and tissue damage.
Published in the journal: . PLoS Pathog 6(9): e32767. doi:10.1371/journal.ppat.1001120
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001120Summary
Toll-like receptor signaling requires functional Toll/interleukin-1 (IL-1) receptor (TIR) domains to activate innate immunity. By producing TIR homologous proteins, microbes inhibit host response induction and improve their own survival. The TIR homologous protein TcpC was recently identified as a virulence factor in uropathogenic Escherichia coli (E. coli), suppressing innate immunity by binding to MyD88. This study examined how the host MyD88 genotype modifies the in vivo effects of TcpC and whether additional, TIR-domain containing proteins might be targeted by TcpC. In wild type mice (wt), TcpC enhanced bacterial virulence, increased acute mortality, bacterial persistence and tissue damage after infection with E. coli CFT073 (TcpC+), compared to a ΔTcpC deletion mutant. These effects were attenuated in Myd88−/− and Tlr4−/− mice. Transcriptomic analysis confirmed that TcpC inhibits MYD88 dependent gene expression in CFT073 infected human uroepithelial cells but in addition the inhibitory effect included targets in the TRIF and IL-6/IL-1 signaling pathways, where MYD88 dependent and independent signaling may converge. The effects of TcpC on bacterial persistence were attenuated in Trif −/− or Il-1β −/− mice and innate immune responses to ΔTcpC were increased, confirming that Trif and Il-1β dependent targets might be involved in vivo, in addition to Myd88. Furthermore, soluble TcpC inhibited Myd88 and Trif dependent TLR signaling in murine macrophages. Our results suggest that TcpC may promote UTI-associated pathology broadly, through inhibition of TIR domain signaling and downstream pathways. Dysregulation of the host response by microbial TcpC thus appears to impair the protective effects of innate immunity, while promoting inflammation and tissue damage.
Introduction
Toll-like receptors (TLRs) control innate host responses to mucosal and systemic infections and signaling involves the intracellular Toll/interleukin-1 receptor (TIR) domain [1]. Following ligand binding, signaling is initiated by the recruitment of adaptor proteins to the TIR domain [2], [3], [4], including myeloid differentiation factor-88 (MYD88), MYD88 adapter-like protein (Mal), TIR domain-containing adaptor protein inducing IFNβ (TRIF), TRIF-related adaptor molecule (TRAM) and the sterile α - and armadillo-motif-containing protein (SARM). Negative regulators of TLR signaling include SIGIRR, MyD88s and IRAK-M, which block MyD88 dependent activation, or Triad3A and SARM, which block the TRIF dependent pathway. The SIGIRR TIR domain resembles MyD88 but lacks two amino acids needed for signaling to occur [5], [6]. However, TIR-TIR interactions between SIGIRR and TLR4 prevent the recruitment of IRAK and TRAF6 to MyD88 [6]. MyD88s is a splice variant inhibiting MyD88 dependent TLR4 activation by allowing MyD88 to bind the intermediate IRAK-binding domain without inducing IRAK phosphorylation and NF-κB activation [7]. IRAK-M prevents IRAK and IRAK-4 dissociation from MyD88 and TRAF6 complex formation [8]; Triad3A interacts with TIR domains of TLRs, TRIF, TIRAP and RIP1 [9]; and SARM blocks gene induction downstream of TRIF [10]. Competition at the level of the TIR domain is thus used by host cells to modify TLR signaling in response to specific agonists [6], [7], [11], [12].
Pathogens have also evolved mechanisms to inhibit the TLR dependent host defense and to increase their fitness and virulence for a specific host niche [12]. The TIR domain plays a crucial role in the mammalian innate immune response and recently proteins containing TIR domains have been described in a wide variety of bacteria, fungi, archaea and viruses [13]. Whole genome sequencing and structural studies have revealed that several pathogens carry TIR-domain homologous sequences, including two proteins from Vaccinia virus A46R and A52R, which interfere with IL-1 and TLR4 mediated activation of NF-κB [14]. Similar proteins were identified in Salmonella, Brucella and uropathogenic E. coli (UPEC) [12], [15], [16]. On the other hand, Spear and co-workers suggested that most TIR domains in bacteria did not evolve to subvert the function of eukaryotic cells but simply to function as general purpose protein-protein interaction domains for diverse uses [13]. We recently showed that the TIR homologous protein TcpC in the UPEC strain E. coli CFT073 acts as a virulence factor by suppressing innate host responses in the kidney, enhancing bacterial persistence and tissue damage [12]. Epidemiologic studies of patient E. coli isolates showed that TcpC occurred more frequently in strains causing severe kidney infections than in E. coli causing other forms of urinary tract infection (UTI) [12]. The UTI model is therefore quite suitable to investigate the mechanisms of TcpC-modulation of the innate host response in vivo and the consequences for bacterial persistence and disease severity [11].
TcpC binds to MyD88 [12] but it remains unknown if other TIR-domain containing molecules of the host are influenced by TcpC in vivo. This study addressed this question with the aim to define the genetic control of TcpC mediated immune inhibition in vivo. We used the murine UTI model and mice lacking specific innate immune response genes to examine whether Myd88 controls the TcpC dependent response to UPEC infection and if additional Myd88 independent signaling pathways might modify the effects of TcpC in vivo. Furthermore, transcriptomic and proteomic analysis of infected human epithelial cells was used to define targets of TcpC and effects on innate immune activation by UPEC and inhibition of TLR signaling by soluble TcpC protein was defined in murine macrophages. The results suggest that TcpC partially inhibits TIR domain dependent signaling in vivo and in host cells including pathways downstream of MYD88, where MYD88 dependent and independent innate immune responses may converge. In this way, broad but incomplete suppression by microbial TcpC, may impair innate immune protection, while promoting inflammation and tissue damage.
Results
TcpC increases bacterial burden, abscess formation and tissue pathology
Experimental UTI was established in wild type (wt) C57BL/6 mice by intravesical infection with E. coli CFT073 (CFT073) or the CFT073tcpC::kan mutant (ΔTcpC) [12] and was monitored for seven days. A higher bacterial burden was present after CFT073 infection compared to ΔTcpC (p<0.001, Figure 1B) in urine samples obtained daily after infection. This difference was reproduced in kidneys (p<0.001) obtained on days four or seven after infection (Figure 1A). MIP-2 chemokine concentrations and neutrophil responses in urine were higher in mice infected with CFT073 compared to ΔTcpC (p<0.001; Figure 1C, D). Tissue damage was extensive in mice infected with CFT073; the kidneys were large, pale and soft and abscesses were present in 75% of the organs (Figure 1E, F). In contrast, kidneys of mice infected with ΔTcpC were normal in size, color and texture and lacked detectable abscesses (p<0.05, Figure 1E, F).
Fig. 1. TcpC acts as a virulence factor by promoting bacterial persistence in the urinary tract and abscess formation in the kidneys of wild type mice. 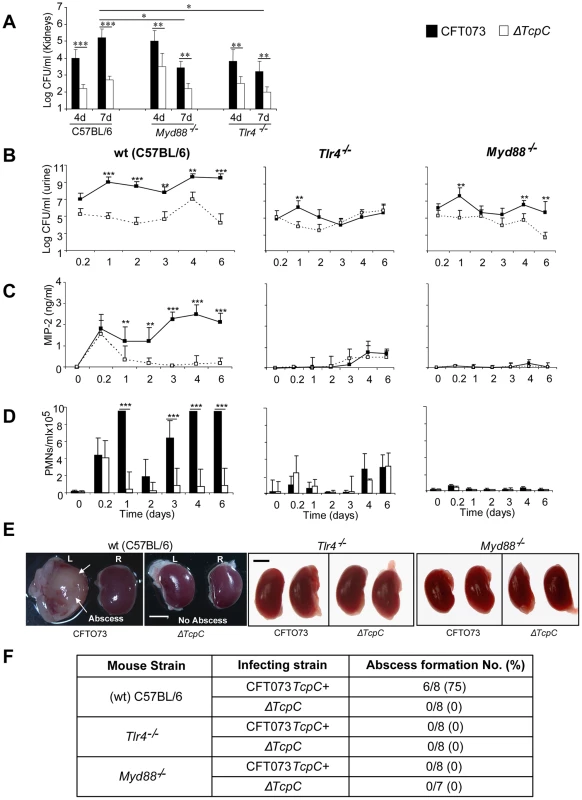
(A) Bacterial counts (CFUs) in kidneys of C57BL/6, Tlr4−/− and Myd88−/− on days 4 and 7 post-infection. (B) Bacterial counts (CFUs) in urine of C57BL/6, Tlr4−/− and Myd88−/− mice after infection with CFT073 or ΔTcpC. (C) MIP-2 response kinetics in urine samples of wt and mutant mice. (D) Neutrophil response kinetics in urine samples of wt and mutant mice. P values (*p<0.05, **p<0.01 and ***p<0.001 for CFT073 versus ΔTcpC mutant and for wt versus mutant mice; Fisher's exact test). Eight to ten mice were used per time point. (E) Abscess formation (arrows) in the kidneys of wt mice infected with CFT073 but not in Tlr4−/− and Myd88−/− mice infected with CFT073 or ΔTcpC (Scale bar, 2 mm).(F) Abscess formation after CFT073 or ΔTcpC infection, in percent of the total number of kidneys examined. The host Tlr4 and Myd88 genotypes control CFT073 and ΔTcpC infection and renal pathology
Experimental UTI with CFT073 or the ΔTcpC mutant was subsequently established in Tlr4 and Myd88 adaptor protein knockout mice in the C57BL/6 background (Tlr4−/− and Myd88−/− respectively). Total bacterial counts and the TcpC dependent difference in bacterial persistence were reduced in the mutant mice, compared to wt mice (Figure 1). Bacterial counts in urine were lower in Myd88−/− (p<0.05) and Tlr4−/− (p<0.05) mice as compared to wt mice (Figure 1). In addition, the renal bacterial counts were significantly lower in mutant mice on day seven than in wt animals which were infected with CFT073 (p<0.05, Figure 1A). However, there was no difference in bacterial counts in wt and mutant mice which were infected with ΔTcpC (Figure 1A). The MIP-2 and neutrophil responses were absent in infected Myd88−/− mice and drastically reduced in Tlr4−/− compared to C57BL/6 wt mice (p<0.0001, Figure 1C, D), confirming that these aspects of the early innate immune response require Myd88 and Tlr4 dependent signaling. Kidneys of Tlr4−/− and Myd88−/− mice infected with CFT073 or the ΔTcpC mutant were normal in size, color and texture and had no abscesses (Figure 1E, F).
TcpC-related differences in bacterial persistence were observed also in the mutant mice (Figure 1). Tlr4−/− and Myd88−/− mice developed significant bacteriuria (≥105 CFU/ml of urine) six hours after infection with CFT073 or ΔTcpC and bacteria persisted in urine until the experimental end point. Higher numbers of CFT073 than ΔTcpC were observed on day one in Tlr4−/− mice (p<0.01); and on days one, four and six in Myd88−/− mice (p<0.01, Figure 1B). In addition, CFT073 numbers were higher than ΔTcpC in kidneys of Myd88−/− and Tlr4−/− mice (p<0.01) on days four and seven (Figure 1A). The difference between CFT073 and ΔTcpC was reduced compared to wt mice in Tlr4−/− (p<0.01) and Myd88−/− mice (p<0.01) by more than two logs in urine (Figure 1A). TcpC dependent increases in MIP-2 and neutrophil responses observed in wt mice were lost in Tlr4−/− mice, the, confirming the essential role of TLR4 and its TIR domain for innate immune responses to UTI (Figure 1C and D). The Myd88−/− mice did not mount MIP-2 and neutrophil responses to CFT073 or ΔTcpC infection. These results suggest that TcpC affects bacterial persistence and pathology, in part, via Tlr4 and Myd88 dependent but also via Myd88 independent pathways.
Absence of kidney pathology in Tlr4 and Myd88 knockout mice
Kidney sections from CFT073 infected wt mice (htx-eosin, day 7) showed swollen collecting ducts and inflammatory cell infiltrates into the kidney parenchyma (Figure 2A). P-fimbriated bacteria were present in the tissues, from the pelvic region to the cortex, as shown by PapG specific antibody staining. Neutrophils were abundant in the abscesses and scattered throughout the tissue, as shown by a neutrophil specific antibody (areas AI-AIII in Figure 2A). By dual staining, P fimbriae and neutrophils were detected in the abscesses and collecting ducts (Figure 2A). In contrast, kidney sections from mice infected with ΔTcpC showed normal structure, no bacteria and few inflammatory cells (areas BI-BII in Figure 2B). Figure 2C shows htx-eosin stained sections from an uninfected control kidney and the inset shows a negative control section stained with the anti-neutrophil and anti-PapG antibodies. The results suggest that TcpC reduces host resistance and increases inflammation, resulting in a high bacterial burden and tissue damage as these sequels become attenuated in mice infected with the ΔTcpC mutant.
Fig. 2. Deletion of TcpC abrogates abscess formation in kidney sections from wild type mice infected with CFT073. 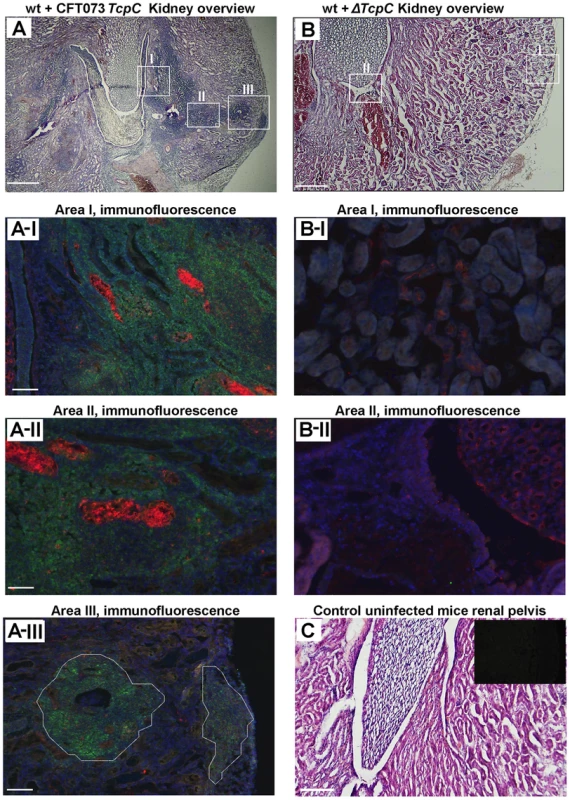
(A) Overview of htx-eosin stained kidney section of wt mice infected with CFT073, showing abscesses (scale bar, 500 µm). Magnified areas of section A shown as A-I, A-II and A-III are stained with specific antibodies to neutrophils (green, NIMP-R14) and the PapG adhesin (red, antiserum to a synthetic PapG peptide) (scale bar, 100 µm). Abscesses in wt mice are shown by dotted lines. (B) Overview of kidney section of wt mice infected with ΔTcpC (scale bar, 300 µm). Magnified areas of section B shown as areas B-I and B-II are stained with specific antibodies as described above. (C) Kidney section of uninfected control mice with htx-eosin staining (scale bar, 200 µm) and inset picture showing negative control for anti-neutrophil and anti-PapG antibodies. Htx-eosin stained sections from Myd88−/− and Tlr4−/− mice infected with CFT073 showed normal collecting ducts, few inflammatory cells and no bacteria in the medulla or cortex of kidneys from either host (Figure 3A). P fimbriae or neutrophils were not detected in infected kidneys by immunohistochemistry (Figure 3B). Similarly, there was no tissue pathology in Myd88−/− and Tlr4−/− mice infected with the ΔTcpC mutant. The results suggest that a host response involving Tlr4 and Myd88 leads to TcpC dependent kidney pathology after CFT073 infection and that hosts lacking Tlr4 or the adaptor are protected from such tissue damage.
Fig. 3. Morphology of intact kidney tissue in Tlr4 −/− and adaptor protein mutant mice infected with CFT073 and ΔTcpC. 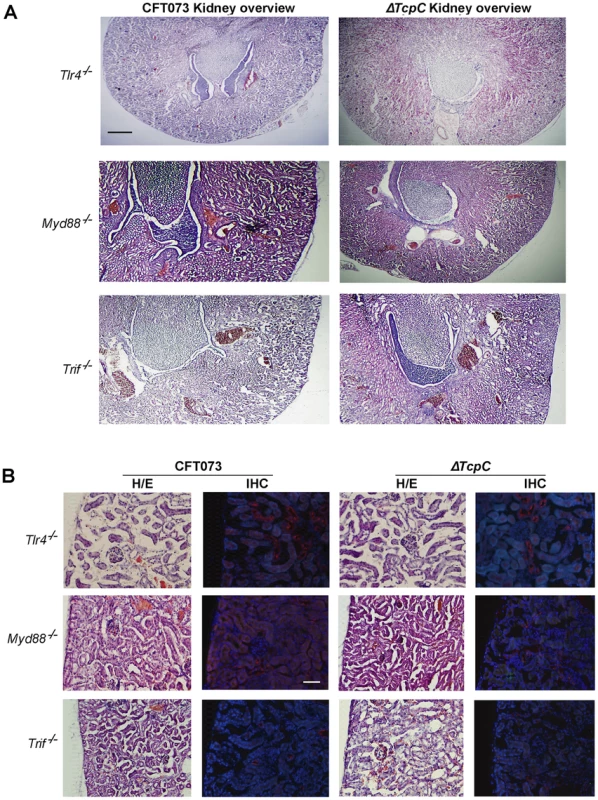
(A) Overview of htx-eosin stained kidney section of Tlr4−/−, Myd88−/− and Trif −/− mice. Scale bar, 500 µm. (B) Histology of renal cortex stained with specific antibodies to neutrophils (green, NIMP-R14) or the PapG adhesion (red, antiserum to a synthetic PapG peptide). Scale bar, 100 µm. Neutrophils or bacteria were not detected in the tissues. The Tlr4−/− and Myd88−/− mice were compared to C57BL/6 wt mice and Trif −/− mice to C57BL6/129 wt mice. Transcriptomic analysis of host responses to CFT073 and ΔTcpC in human uroepithelial cells
While the in vivo experiments confirmed that TcpC mediated virulence depends on pathways controlled by Myd88, they also suggested that TcpC modifies additional host response pathways. To identify such pathways, human A498 kidney epithelial cells were infected in vitro with CFT073 or ΔTcpC (4 hours, 108 CFU/ml) and complementary RNA was hybridized to Illumina whole genome microarrays. A heat map of significantly regulated genes is shown in Figure 4A (means of triplicate spots). In identification of CFT073 or ΔTcpC-specific genes, fold changes of ≥2 were used. For the comparison of CFT073 or ΔTcpC, a fold change in response to either strain >1.5 relative to the respective control was used.
Fig. 4. Uroepithelial gene expression in response to in vitro infection with CFT073 or ΔTcpC. 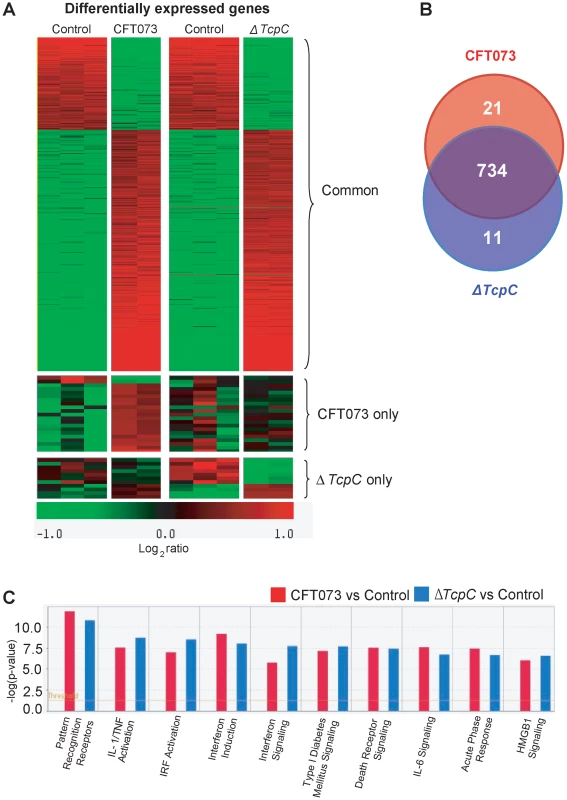
(A) A two-dimensional hierarchical heat map illustrating differentially transcribed genes in human A498 cells infected with CFT073 or ΔTcpC versus the control group. Analysis was performed on 766 genes with a fold change of at least two over untreated cells, and classified based on whether they are CFT073 or ΔTcpC specific, or common to both infections. Induced genes are represented by brighter shades of red, while down regulated genes are represented by brighter shades of green (see color scale). (B) Venn diagram showing numerical distribution of differentially expressed genes in CFT073 (red) and ΔTcpC infected (blue) cells compared to mock infected cells. (C) A comparison of the 10 highest scoring biological pathways between infected cells and controls as analyzed by Ingenuity Pathway Analysis with default settings. Infection stimulated marked changes in gene expression and three major groups of genes were altered; 734 regulated genes were shared, 21 genes responded only to CFT073, while 11 responded only to ΔTcpC (Figure 4B, Supplementary Table S1). Differentially expressed genes between unstimulated and CFT073 or ΔTcpC treated cells were then characterized using Ingenuity Pathway Analysis. Notably, signaling downstream of pattern recognition receptors, interferon induction, interferon response and IL-6/IL-1 signaling pathways were among the top-scoring pathways identified (Figure 4C). To further study the mechanisms of TcpC mediated innate immune inhibition, differentially transcribed genes were assigned to known response pathways (Figure 5, Supplementary Table S1). Consistent with the proposed role of TcpC as a MYD88 inhibitor [12], in vitro infection with ΔTcpC upregulated MYD88 dependent transcripts involved in pathogen pattern recognition, compared to CFT073 infected cells (p>0.01). These include the inflammatory cytokines IL-6, IL-8, TNF-α, IL-1α/β and the transcription factors IRF7 and NF-κB1, 2 (Figure 5A, Green). In addition, the expression of downstream pro-inflammatory genes including IL-1α/β, TNFAIP6 and TNFRSF11b were upregulated in ΔTcpC compared to CFT073 infected cells (Figure 5B, Green), while the transcription of negative regulators IL-1RA, NFkBIa and NFkBIb was reduced, consistent with activation of the MYD88 pathway (Figure 5B, Red). Interestingly, a corresponding reduction in the MAP kinase MAP2K3, JUN and FOS transcripts was observed, suggesting a partial rather than complete suppression of the MYD88 pathway by TcpC (Figure 5B, Red). NF-κB and IRF7 transcript levels, which are both MYD88 and TRIF dependent, were significantly increased after ΔTcpC infection compared to CFT073 (p<0.01), suggesting that also TRIF-dependent signaling, might be inhibited by TcpC. Other genes downstream of TRIF maintained their activity regardless of TcpC, however.
Fig. 5. Identification of pathway-specific genes by Ingenuity Pathway Analysis in response to in vitro infection. 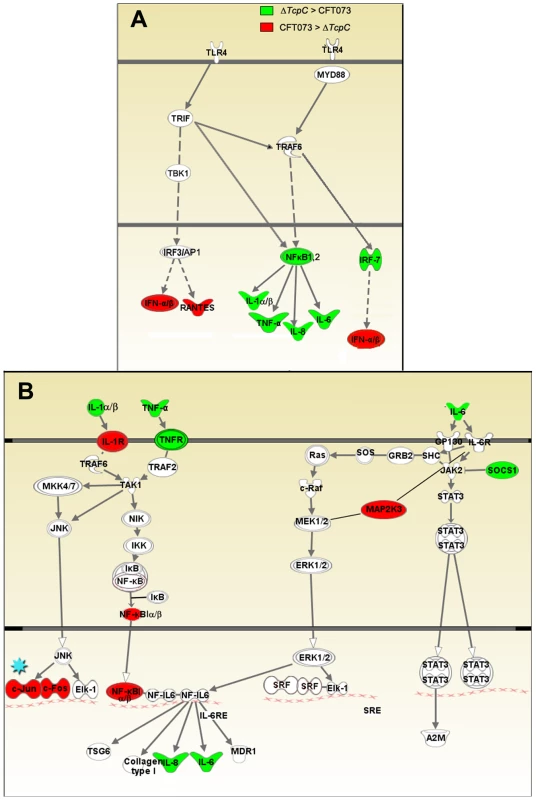
(A-I) Pattern recognition pathway. (A-II) IL-6/IL-1 pathway. Each node represents a protein whose functional class is represented by various shapes (ovals = transcription factors, spiral = kinases, dumbbell = transcription regulators, V shape = cytokine or growth factors, Y shape = transmembrane receptors, circles = others). Direct protein interactions are represented by solid lines, while dashed lines indicate indirect interaction. A red node indicates up regulation of a protein in response to CFT073, while a green node indicates down regulation by CFT073 relative to ΔTcpC. In order to validate the transcriptomic analysis, expression levels of selected genes were confirmed by RT-PCR in the cells infected with CFT073 or ΔTcpC (Figure 6A). Significant differences were observed for IL-8, IL-6, NFkB1, TNF - α and c-FOS. The pattern reflected a trend similar to that revealed by microarray analysis for the gene products tested (Figure 5B*, Red).
Fig. 6. Protein array analysis in response to in vitro infection with CFT073 or ΔTcpC. 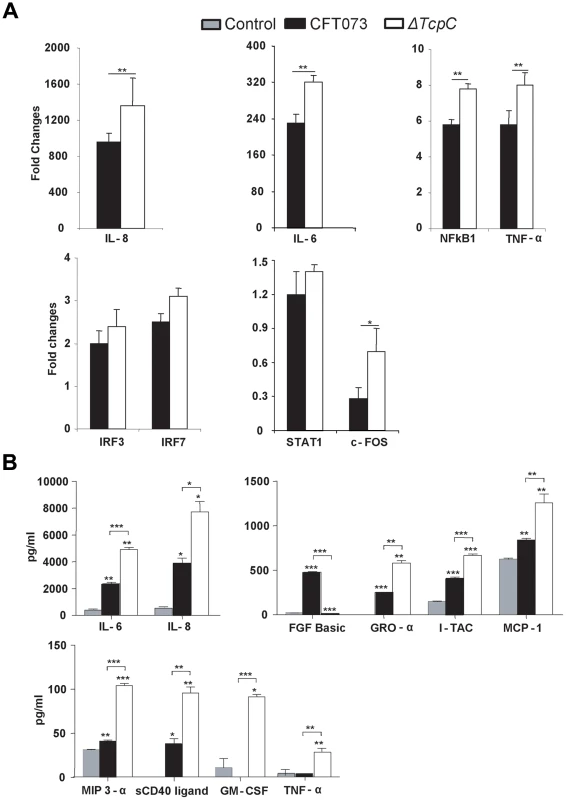
(A) RT-PCR analysis of human kidney cell lines infected with CFT073 or ΔTcpC for 4 hrs. All values represent means ± SEMs of three experiments. (B) Uroepithelial cytokine responses to infection with CFT073, ΔTcpC or mock-infected controls. A498 cells were stimulated for 4 hours with the indicated bacterial strains. Supernatant cytokine levels are in pg/ml and represent means ± SEMs of three experiments. Significant differences are indicated by single * (p<0.05), double ** (p<0.01) or triple asterisks *** (p<0.001, Student's t-test). To generate further insights into the effects of TcpC, cultured human kidney cells were stimulated with either CFT073 or ΔTcpC and culture medium was assayed using the Procarta human cytokine kit (Figure 6B). ΔTcpC stimulation resulted in significantly elevated levels of MYD88 dependent proinflammatory cytokines (IL-6, IL-8, TNF-α) (p<0.01) compared to the wild type strain, corroborating the transcriptomic analysis and suggesting that MyD88 and TRIF dependent pathways are modified by TcpC. In addition, ΔTcpC stimulation resulted in increased expression of the neutrophil chemoattractants MCP-1, GRO-α and MIP-3α (Figure 6B), while in vivo infection with ΔTcpC caused a lower MIP-2 response than CFT073.
The apparent discrepancies between the in vivo data and in vitro results were further examined, by establishing murine tubular kidney cell lines from wt and Myd88−/− or Trif Lps2/Lps2 mutant mice. The cells were infected with CFT073 or ΔTcpC and MIP-2 secretion was quantified by ELISA, four hours after infection (Supplementary Figure S1). MIP-2 secretion was reduced in ΔTcpC infected compared to CFT073 infected cells from wt mice, consistent with the increased response to CFT073 in wt mice. In Trif Lps2/Lps2 cells, MIP-2 secretion was also reduced in ΔTcpC compared to CFT073 infected cells (Supplementary Figure S1). Furthermore, the in vitro response of cells from Myd88−/− mice was very low, both to CFT073 and ΔTcpC, thus reproducing the lack of response in mutant mice. By RT-PCR, the MIP-2 response to ΔTcpC was reduced compared to CFT073 in cells from wt mice and also very low in cells from Myd88−/− mice but not different in cells from Trif Lps2/Lps2 mice (data not shown), Thus most but not all of the in vitro results were compatible with the in vivo data, either in human or murine kidney cells. Differences between in vivo data and in vitro assays are expected to occur, as the in vivo response of an entire organ system is unlikely to be reflected by a single cell type in vitro.
TcpC dependent virulence and innate immune responses act via the Trif signaling pathway
The cellular studies and findings in Myd88−/− and Tlr4−/− mice suggested, that bacterial TcpC might inhibit TLR4 dependent signaling, also through the Trif adaptor. To examine this hypothesis, Trif adaptor protein knockout mice (Trif −/−) were infected with CFT073 or the ΔTcpC mutant and compared to wt C57BL6/129 mice. The ΔTcpC mutant established higher bacterial counts in urine than CFT073 in Trif −/− mice (p<0.001), in contrast to C57BL6/129 wild type mice (Figure 7B, p<0.001). Furthermore, the difference in kidney counts between CFT073 and ΔTcpC in wt mice (Figure 7A, p<0.001) was not observed in Trif −/− mice, indicating that the effects of TcpC on bacterial persistence are neutralized in mice with defective Trif signaling. Trif −/− mice also exhibited a significant MIP-2 response to ΔTcpC compared to CFT073 (p<0.05, day 3 and 6) (Figure 7C) and the neutrophil response to ΔTcpC was significantly higher (p<0.001, Figure 7D).
Fig. 7. Effects of TcpC virulence in Trif −/− and Trif Lps2/Lps2 mice infected with CFT073 or ΔTcpC. 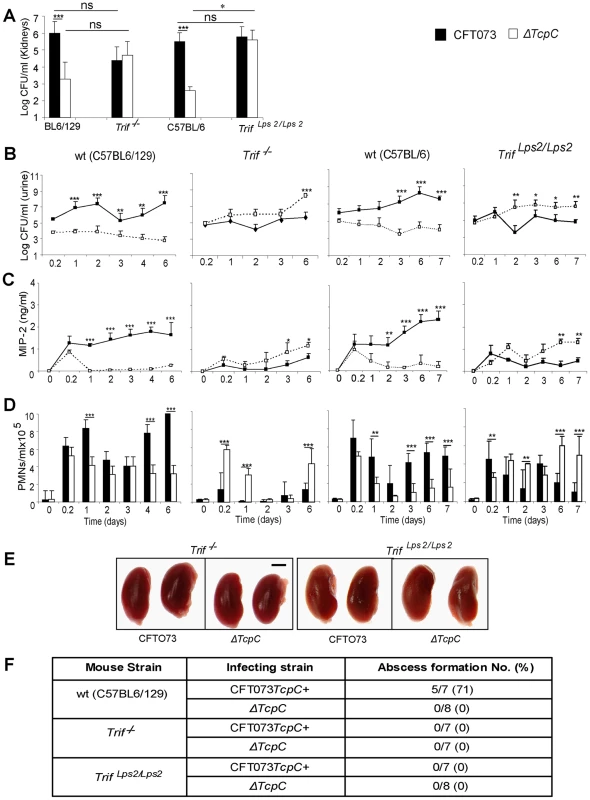
(A) Bacterial burden (CFU) in kidneys of wt (C57BL6/129), Trif −/−, wt (C57BL/6) and Trif Lps2/Lps2 mice on day seven after infection with CFT073 or ΔTcpC. (B) Bacterial numbers in urine of wt and mutant mice after infection. (C) Kinetics of the MIP-2 response, measured in urine samples. (D) Neutrophil response kinetics in urine of wt and mutant mice. P values (*p<0.05, **p<0.01 and ***p<0.001 for CFT073 versus ΔTcpC mutant and for wt versus mutant mice; Fisher's exact test; ns = not significant). (E) Abscess formation in mouse kidneys, seven days post-infection (Scale bar, 2 mm). (F) Abscess formation after CFT073 or ΔTcpC infection, in percent of the total number of kidneys examined. The effects of Trif on TcpC inhibition were confirmed in Trif Lps2/Lps2 mice, which carry a non-functional co-dominant Trif allele, induced by N-ethyl-N-nitrosourea mutagenesis on a pure C57BL/6 background [17] (Figure 7). As in Trif −/− mice, the ΔTcpC mutant established higher bacterial counts in urine than CFT073 in contrast to C57BL/6 wt mice (Figure 7B, p<0.001). The TcpC related difference in kidney counts between CFT073 and ΔTcpC in wt mice (Figure 7A, p<0.001) was not observed in Trif Lps2/Lps2 mice and the MIP-2 and neutrophil responses to ΔTcpC were significantly higher compared to CFT073 (Figure 7C, D; p<0.01). However, neither CFT073 nor ΔTcpC induced kidney abscesses in Trif −/− mice or in Trif Lps2/Lps2 mice (Figure 7E, F). The results suggest that the Trif adaptor protein is involved in the innate immune mechanisms controlling the persistence of TcpC expressing bacteria.
Inhibitory effects of TcpC on TLR signaling in murine macrophages
To confirm that the TIR domain of TcpC impaired MyD88-dependent TLR signaling, bone marrow derived macrophages (BMDMs) from wild type or Myd88−/− mice were stimulated with different TLR ligands in the presence of titrated amounts of TIR-TcpC, the c-terminal half of TcpC containing the TIR domain. TIR-TcpC impaired TNF secretion induced by the different TLR ligands with the exception of TLR3 mediated stimulation, as expected from the MyD88 independence of TLR3 (Figure 8A) [12]. Also as expected only poly(I:C) and LPS were able to induce TNF secretion in Myd88−/− BMDMs, presumably via Trif. Interestingly, TIR-TcpC impaired this pathway as well, consistent with the in vivo observation that the Trif pathway was influenced by TcpC (Figure 8B). In addition, control experiments showed that TcpC is secreted into the urine of infected mice (data not shown).
Fig. 8. Inhibitory effects of the soluble TcpC TIR-domain on TLR signaling in murine macrophages. 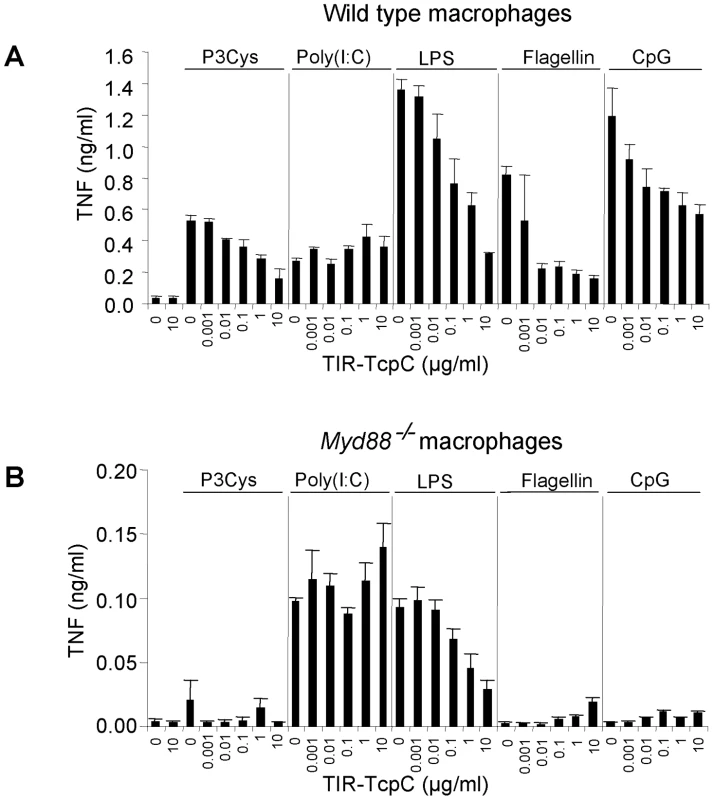
(A) Wt BMDM were stimulated with Pam3Cys (1 µg/ml), poly(I:C) (2.5 µg/ml), ultrapure LPS from E. coli K12 (100 ng/ml), flagellin from S. typhimurium (1 µg/ml) and CpG 1826 (2 µM) in the presence of titrated amounts of the purified TIR domain of TcpC (TIR-TcpC) as indicated. TNF secretion was analyzed 3 hr after stimulation. (B) The experiment in (A) was repeated at the same time, side by side with Myd88−/− BMDM. Error bars represent SD from three individual cultures. The experiment was repeated once with similar results. Influence of Il-1β and Irf3 on TcpC dependent virulence
The transcriptomic analysis suggested that TcpC strongly regulates the pro-inflammatory cytokines IL-6 and IL-1α/β, as well as downstream signaling pathways. Enhanced expression of IL-1α/β (p<0.03 for IL-1α and p<0.02 for IL-1β) in ΔTcpC infected human cells suggested that the inhibitory effect of TcpC includes IL-1 dependent inflammation. To address this question, Il-1β−/− mice were infected with CFT073 or ΔTcpC, as described. The TcpC dependent difference in bacterial persistence and host response induction was reduced in these mice (p<0.0001, Figure 9). Renal abscess formation or tissue pathology was not observed.
Fig. 9. Effects of TcpC virulence in Il-1β−/− and Irf3−/− mice infected with CFT073 or ΔTcpC. 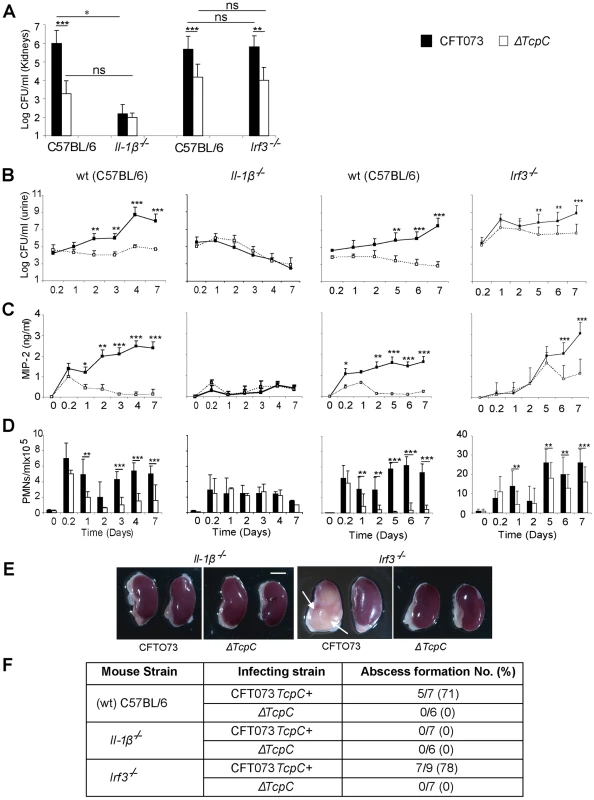
(A) Bacterial burden (CFU) in kidneys of wt (CF7BL/6), Il-1β−/− and Irf3−/− mice on day seven after infection with CFT073 or ΔTcpC. (B) Bacterial numbers in urine after infection. (C) Kinetics of the MIP-2 response, measured in urine samples. (D) Neutrophil response kinetics in urine of wt and mutant mice. P values (**p<0.01 and ***p<0.001 for CFT073 versus ΔTcpC mutant and for wt versus mutant mice; Fisher's exact test; ns = not significant). (E) Abscess formation in mouse kidneys, seven days post-infection (Scale bar, 2 mm). (F) Abscess formation after CFT073 or ΔTcpC infection, in percent of the total number of kidneys examined. IRF3 is a transcription factor controlling interferon responses to viral infection [18]. More recently, the involvement of IRFs in immunoregulation by TLRs has received more attention, since NF-kB, IRF3 and AP-1 form transcriptional complexes that regulate innate immune responses in monocytes [19]. We have recently identified IRF3 as an essential transcription factor in UTI, acting downstream of TLR4/TRAM (unpublished observation). To examine if Irf3 might be involved in TcpC mediated innate immune suppression, infection with CFT073 or ΔTcpC was established in Irf3 mutant mice (Irf3−/−), using wt C57BL/6 mice as controls. The difference in persistence between CFT073 and ΔTcpC in wild type mice was not observed during the early phase of infection in Irf3−/− mice (<day 5) (Figure 9A, B). Furthermore, in Irf3−/− mice, the early chemokine and neutrophil responses were delayed compared to responses in wt controls (Figure 9C, D). Significantly reduced responses to ΔTcpC were only observed from day five post-infection in Irf3−/− mice (Figure 9C, D), showing that the response kinetics differed from wt mice. Abscess formation in response to CFT073 was as frequent in the Irf3−/− as in wt mice (Figure 9E, F). The results suggest that the effect of TcpC on bacterial persistence and on the MIP-2 response are attenuated during the early phase of infection in IRF3-deficient mice and that IRF3 signaling is differentially activated depending on the TcpC status of the infecting strain (Please see figure 9B–D).
Taken together, the results show that bacterial TcpC modifies the innate host response broadly through inhibition of Tlr4, Myd88, Trif, IL-6/IL-1 and Irf3 dependent antibacterial effector functions.
Discussion
Bacterial TIR-like proteins are important virulence factors, which act by inhibiting innate immunity, thus facilitating the survival and persistence of several pathogens. The Salmonella enteritica TlpA protein enhances bacterial survival in macrophages and mice [15] and the Brucella TcpB protein inhibits TIRAP mediated signaling and reduces systemic spread of bacteria during the early stages of infection [16]. The E. coli TcpC protein increases virulence in the urinary tract and we have previously proposed that TLR signaling is impeded through the MYD88 adaptor via direct binding of TcpC to MYD88 [12]. This study addressed the mechanism through which TcpC modifies the innate immune response in the infected host, by varying the innate immune genetic repertoire. The results show that TcpC requires Myd88 to act as a virulence factor in vivo. Transcriptomic analysis identified additional targets for TcpC in human cells, including the TRIF and IL-6 pathways, as well as IL-1α/β. Tlr4−/−, Trif −/−, Lps2−/− and Il-1β−/− mice exhibited markedly different immune responses to TcpC stimulation and the TcpC dependent change in bacterial persistence and pathology was attenuated in these mice. The results thus suggest that pathways for host-defense can be fine-tuned by a bacterial virulence factor in order to paradoxically promote bacterial replication and pathology.
MyD88 was the first TLR adaptor to be identified and is shared by the TLRs as well as the interleukin-1 receptor [20], [21]. By targeting MyD88, TcpC would thus be expected to impair both TLR - and IL-1 dependent signaling pathways as well as related antibacterial effector functions. This interpretation was supported by the in vivo effects, which were not restricted to Myd88 but regulated by a group of genes encoding proteins with a TIR domain or regulated by such proteins. In Myd88−/− mice, essential chemokine and neutrophil responses to CFT073 infection were completely abrogated consistent with the importance of MyD88 for the overall innate immune response to these infections. The response to infection was strongly reduced also in Tlr4−/− mice confirming our previous findings that Tlr4 signaling is essential for the innate immune response to UTI and suggesting that that inhibition of TLR4 responses may be protective at the mucosal level [11], [22], [23], [24]. We have recently extended the analysis of potential eukaryotic interaction partners and have shown that the TIR-domain of TcpC binds to TLR4 in addition to MyD88 (unpublished observation). TcpC did not bind to TRIF or TLR2, however [12]. Thus, as expected by the differing aminoacid sequences, only selected TIR-domains bind TcpC. The difference in bacterial persistence, while reduced, was not abrogated in Tlr4−/− or MyD88−/− mice, however, possibly reflecting the involvement of pathways that remains intact in Myd88−/− mice. The interaction with TLR4 provides a molecular basis for inhibition of the TRIF-dependent arm of the TLR4 signaling cascade, thus possibly explaining, in part, the effects of TcpC in Trif −/− mice.
This mode of action of TcpC was also supported by the experiments in murine macrophages, where responses to most of the TLR specific ligands were inhibited by purified TcpC-TIR protein. TcpC inhibited LPS-driven, MyD88 dependent and LPS-driven MyD88-independent TNF secretion, including TLR4-TRIF signaling. However, TcpC did not influence poly (I:C)-induced TNF-secretion, whether MyD88 dependent or not. The fact that poly (I:C)-induced TNF-secretion in the absence of TcpC was lower in MyD88-deficient cells compared to wild type cells cannot be interpreted to imply that poly (I:C) stimulates MyD88-dependent TNF-secretion, since resting MyD88-deficient macrophages are impaired in their basal expression of several cytokines including TNF [25]. Cytokines like TNF are in general harder to induce in MyD88-deficient cells, as also reported by Sun et al. [26]. In preliminary experiments, we have also analyzed the secretion of the chemokine KC and have not found differences in wild type or MyD88-deficient cells after stimulation with poly (I:C). Thus, poly (I:C) stimulates cells in a MyD88-independent but TRIF-dependent manner and the results are compatible with the in vivo effect.
TRIF has a well conserved TIR domain and several TRAF6 binding regions within the N - and C-terminal RIP homotypic interaction motifs (RHIM) [27], [28], [29]. TRAM is TLR4 specific [30], [31] and the myristoylated N-terminus associates to the plasma membrane and protein kinase Cε phosphorylation of Serine 6 and 16 is essential for TRAM activation [32], [33]. In contrast to MYD88, TRIF dependent signaling activates both NF-κB and IRF3/7 [30]. After TRAM dependent TLR4 activation, TRIF forms a complex with TRAF3, IRAK1 and an IKK-like kinase. TRAF family member associated NF-κB activator (TANK)-binding kinase 1 (TBK1) and the IKK homolog, IKKε, phosphorylate IRF3 at its C terminus [30], [34], initiating IFN-α/β transcription [29]. IRF7 becomes activated in a similar manner [4]. In this study, TcpC suppressed transcription of NF-κB and IRF7 as well as IL-8, TNF-α, IL-1α/β and IL-6, which is consistent with the effects on MYD88 and possibly TRIF. The in vivo response to infection supports the conclusion that TcpC also suppresses Trif dependent effector functions, however, as Trif −/− mice have a functional Myd88 response, except for the cooperative TLR4 response to LPS [35]. The lack of pathology in infected Trif −/− mice further suggests that Trif signaling is essential for efficient innate immune-mediated clearance of UPEC infection.
The transcriptomic analysis of infected human kidney cells suggested that TcpC also modifies proinflammatory cytokine signaling downstream of MYD88 and TRIF. The IL-6 pathway was strongly regulated and IL-1α/β expression was reduced by TcpC. The involvement of IL-1 was confirmed by infection of Il-1β−/− mice and the phenotype of the Il-1β−/− mice was quite convincing, as the difference in bacterial persistence between CFT073 and the TcpC deficient mutant in wt mice was abrogated in Il-1β−/− mice and the innate immune response to the two isogenic strains was similar. Bacterial clearance was rapid, further suggesting that IL-1 may be a significant factor in the mucosal response to UPEC and in the establishment of tissue pathology. The mechanism of TcpC modulation of IL-1 is not clear, however and may either relate to a TIR domain dependent interaction of TcpC with the IL-1 receptor, effects on upstream signaling involving Myd88 and Trif and the resulting IL-1 response or to other mechanisms.
Recently, Trif dependent innate immune responses have been shown to activate IRFs that regulate the transcription of pro-inflammatory genes, including IL-8, IL-6 and TNF, in addition to interferon genes [18]. In a parallel study, we have characterized a new TLR4/TRAM dependent pathway that mediates innate responses to P-fimbriated, uropathogenic E. coli through Irf3 (unpublished observation). Using a combination of transcriptomics, phosphorylation arrays and imaging technology, we detected TRAM phosphorylation, activation of MAP kinases including p38, CREB phosphorylation and nuclear IRF3 translocation. Irf3−/− mice lacking this pathway, developed rapid lethal kidney infections with extensive tissue damage and patients prone to acute pyelonephritis were shown to carry IRF3 promoter polymorphisms that reduce transcription efficiency. In the present study, interferon dependent pathways were differentially regulated by TcpC in vitro (data not shown) and in Irf3−/− mice the effects of TcpC were significantly delayed compared to wt mice. While this effect was transcient, the results further support the results suggesting that TcpC may modify the effects of the TRIF/Irf3 pathway and the progression to disease and pathology.
Innate immune activation by uropathogenic E. coli is orchestrated by specific virulence factors and essential aspects of the mucosal response show pathogen specificity [36]. Such interactions are needed, as innate immune responses are not activated by asymptomatic carrier strains in the epithelial tissue, which forms a barrier against interactions with inflammatory cells, with broader reactivity. For example, epithelial cells lack surface expressed CD14 and do not respond to conserved bacterial PAMPS like LPS [11], [36]. In addition, asymptomatic carrier strains may actively suppress the mucosal immune response, but the mechanisms are not fully understood. We have previously shown that pathogen specific TLR4 activation favours TRIF or MyD88, depending on the surface fimbriae, which are expressed in a pathogen specific manner and serve as crucial virulence factors involved in attachment and host tissue perturbation [11]. P fimbriated bacteria preferentially activate TLR4/TRIF signalling while Type 1 fimbriae trigger TLR4 responses mainly involving MyD88. The adaptor protein usage in infected host cells can even be shifted from TRIF to MyD88 by a change in fimbrial expression [11]. E. coli CFT073, used in the present study, expresses both P and type 1 fimbriae, thus activating TLR4 signalling involving TRIF and MyD88 responses in infected cells, providing a basis for independent targeting of these pathways by TcpC.
In conclusion, our results suggest that TcpC may act as a broad microbial innate immune response modulator in vivo, by preventing bacterial clearance and dysregulating inflammation, with destructive effects on infected tissues. This adds TcpC to an increasing number of components that regulate TLR and MYD88 dependent responses. On the host side, MYD88s inhibits the recruitment of IRAK-4, thus acting as a negative regulator of TLR signaling [37]. SIGIRR, IRAK-M, SOCS1, Triad3A and SARM, and the cytosolic domain of SIGIRR block TLR4 signaling through TIR-TIR interactions preventing the recruitment of IRAK and TRAF6 to MYD88. TcpC binds to TLR4 (unpublished data) and MYD88 [12] through TIR domain interactions and inhibits TLR4 and MYD88 dependent signaling in vivo, as well as downstream effector functions. While many molecular interactions remain to be defined, it is clear that this microbially induced suppression of the host defense differs, in the classical sense, from mucosal tolerance, which may be triggered by microbial or other mucosal antigens, but is defined by the involvement of specific immunity, with T cells as the main effector cells. In the case of TcpC, the innate immune response is modified and the “tolerant” state appears to result from active microbial inhibition, rather than from a lasting change in the immune status of the host, and from a direct modification of host resistance rather than by inducing tolerance. Further insights into these mechanisms may be helpful to distinguish “bad” from “good” inflammation and to understand how partial inhibition of MYD88, TRIF and TLR4 by TcpC results in pathology while complete gene deletion is protective. These findings also illustrate the importance of the host response as a generator of pathology and suggest the possibility of intervention to support “good” host responses, promoting bacterial clearance and tissue integrity while inhibiting pathology.
Materials and Methods
Ethics statement
C57BL/6 (wt), C57BL6/129 (wt), C57BL/6 Tlr4−/−, C57BL/6 Myd88−/−, C57BL/6 Irf3−/−, C57BL/6 Il-1β−/−, C57BL/6 Trif Lps2/Lps2 and C57BL6/129 Trif −/− mice were bred and housed in the specific pathogen-free facilities of the MIG animal facilities (Lund, Sweden) with free access to food and water. All procedures were approved by the Animal Experimental Ethics Committee at the Lund District Court, Sweden (numbers M166-04 and M87-07), following Institutional, National, and European Union guidelines.
Cell lines and reagents
The human epithelial cell line A498 (ATCC HTB44, human kidney carcinoma, Manassas, VA, USA) was cultured in RPMI-1640 supplemented with 1 mM sodium pyruvate, 1 mM non-essential amino acids, gentamycin (50 µg mL−1) and 5% fetal calf serum (PAA Laboratories, Pasching, Austria).
Alexa-fluor 568-goat anti-rabbit IgG and Alexa-fluor 488-goat anti-rat were from Invitrogen (Eugen, Oregon, USA). Anti-rat NIMP-R14 (ab2557) were from Abcam (Cambridge, UK) and goat normal serum were from Dako (Denmark). The reagents paraformaldehyde (Merck KGaA, Darmstadt, Germany), triton X-100 (VWR International Ltd, England), isopentane (Sigma-Aldrich, Germany), VECTASHIELD mounting medium (Vector Laboratories, USA), poly-L-lysine-coated glass slides (Thermo Scientific, Waltham, USA), DAPI (Invitrogen, Oregon, USA), tryptic soy agar (Difco, Detroit, USA) and hematoxylin-eosin stain (Histolab Products AB, Gothenburg, Sweden) were used. Mouse MIP-2 quantification kit was from R&D systems (Abingdon, UK). TargetAmp Nano-g Biotin-aRNA Labelling kit was from Epicentre Biotechnologies (Madison, USA), RNeasy clean-up from Qiagen (Alabama, USA) and the Procarta Human Cytokine 50-plex kit from Panomics (Fremont, USA).
Mouse strains
Tlr4−/− [38], Myd88−/− [39], Irf3−/− [40] and Il-1β−/− [41] mice were generated in the C57BL/6 genetic background and Trif −/− [28] mice were generated in the C57BL6/129 genetic background. Lps2, a non-functional codominant allele of Trif, was induced by N-ethyl-N-nitrosourea mutagenesis on a pure C57BL/6 mouse background [17] in the Scripps Institute animal facilities (La Jolla CA). Il-1β−/− mice were provided by Max Planck (Institute for Infection Biology, Berlin, Germany). Wild type C57BL6/129 mice are the 50% backcross of C57BL/6 mice. All the knock out mice (Tlr4−/−, Myd88−/−, Irf3−/− and Il-1β−/−) are backcross in 100% C57BL/6 wt mice and are pure.
Bacterial strains
E. coli CFT073 was isolated from the blood and urine of a woman admitted to the University of Maryland Medical System for treatment of acute pyelonephritis [42]. This hly1+, pap1+, sfa1+ and pil1+ strain expresses P fimbriae, hemolysin, and Type 1 fimbriae and is highly virulent in the CBA mouse model of ascending UTI. It is cytotoxic for cultured human renal proximal tubular epithelial cells [43]. CFT073 expresses the TcpC protein and the ectcp sequences, which encode TcpC and were deleted in the ectcp::kan mutant (ΔTcpC), as described [12]. The strains were grown to stationary phase overnight on tryptic soy agar with appropriate selection and suspended in 10 ml of phosphate-buffered saline (PBS) (pH 7.2) to generate the bacterial suspension used for inoculation (109 CFU, colony forming units/ml). The bacteria were tested for virulence factor expression, including P and Type 1 fimbriae [44].
Experimental UTI mice model
Female C57BL/6, C57BL6/129, Tlr4−/−, Myd88−/−, Trif −/−, Trif Lps2/Lps2, Il-1β−/− and Irf3−/− mice were used at 8–12 weeks of age. E. coli urinary tract infection was established as described [45]. In brief, 0.1 ml of the bacterial suspension was administered into the bladders of anesthetized mice (109 CFU/ml) with the help of a soft polyethylene catheter (inner diameter 0.28 mm, outer diameter 0.61 mm; Clay Adams, New Jersey USA). Prior to inoculation, urine samples were collected and cultured on blood agar plates to ensure that the mice were uninfected. After infection, urine was collected into sterile tubes through gentle pressure on the mouse's abdomen (5 h, 24 h and up to 6 days) and neutrophils were quantified by light microscopy using a Burker chamber. The urine samples and serial dilutions were quantitatively cultured on tryptic soy agar (TSA) plates. Mice were sacrificed by cervical dislocation while anesthetized. Bladders and kidneys were removed under sterile conditions, placed into a plastic bag containing 5 ml PBS (pH 7.2), homogenized in a Stomacher 80 homogenizer (Seward medical, London, UK) and plated on TSA plates. Subsequently, blood agar and TSA plates were incubated at 37°C overnight and visually scored for bacterial colonies. Kidneys were also prepared for hematoxylin-eosin staining or immunohistochemistry.
Histology and immunohistochemistry
Kidneys were fixed in freshly prepared 4% paraformaldehyde soon after dissection and incubated overnight at 4°C. Further, the fixed tissues were incubated in 15% sucrose (4°C/24 hr) and washed in 25% ice cold sucrose (4°C/24 hr). Tissues were then frozen in isopentane at −40°C and stored at −80°C for further use. Cryostat sections were made with a steel knife (10 µm), mounted onto poly-L-lysine-coated glass slides and stained.
The kidney sections from all mouse groups were stained for dual immunohistochemistry as described [46]. Briefly, tissue sections were dried at 37°C for 15 min, washed in PBS-0.2% Triton X-100 (pH 7.2) (2×10 min/RT) and incubated (30 min/RT) with PBS-0.2% Triton X-100+ 5% goat normal serum (Dako, Denmark). Then the sections were incubated with primary anti-rat NIMP-R14 (ab2557, Abcam, Cambridge, UK; 1∶200) and antiserum to a synthetic peptide within the PapG adhesin (1∶200) for 2–3 hr at RT. Negative controls consisted of only normal goat serum (1∶200). The kidney sections were washed in PBS (3×5 min) and incubated (1 hr/RT) with secondary goat anti-rat immunoglobulins (1∶200), conjugated with Alexa 488 dye (A488; 495ext/519em nm), and secondary goat anti-rabbit immunoglobulins (1∶200), conjugated with Alexa 568 dye (A568; 578ext/603em nm), as fluorochrome (Invitrogen, USA). After washing in PBS (2×5 min), specimens were counterstained (3 min/RT) with DAPI (0.05 µM) for nuclei staining. Sections were washed again in PBS (10 min) and mounted with VECTASHIELD mounting medium (Vector Laboratories, USA) and kept in the dark. Sections were analyzed by fluorescence microscopy (AX60, Olympus Opticals, Hamburg, Germany) in the Department of Pathology, Lund University, Sweden.
Cytokine measurements
Urine samples were collected at 0.6 hr and after 1, 2, 3, 4, and 6 days and stored at −20°C. MIP-2 in urine was quantified by ELISA using the Mouse MIP-2 quantification kit (R&D systems, Abingdon, UK) according to the manufacturer's instructions and urine was diluted five fold in sample buffer. The ELISA plates were read at 450 nm in a Labsystems Multiskan PLUS reader (Analytical Instruments LLC, Golden Valley, USA).
DNA microarray analysis
For the microarray study, 350,000 A498 cells were seeded in 6-well plates and infected with CFT073 or ΔTcpC (108 CFU/ml) upon confluency. Total RNA was extracted 4 hr after stimulation by acid guanidinium thiocyanate-phenol-chloroform extraction (Trizol, Invitrogen, USA) followed by a Qiagen RNeasy clean-up procedure. RNA was reverse-transcribed to double stranded cDNA and converted to biotin-labelled cRNA using a TargetAmp Nano-g Biotin-aRNA Labelling kit (Epicentre Biotechnologies, Madison, USA). Labelled cRNAs were hybridized onto an Illumina Human WG6v3 Expression Beadchip for 16 hours at 58°C. The arrays were then washed and stained based on the Illumina Wash Protocol and then scanned using a BeadArray Scanner 500GX. The background subtracted data were pre-processed to correct negative and non significant intensities. Pre-processed data was normalized using the cross correlation [47] and genes with a fold change of 2 were identified as differentially expressed. Data was preprocessed using RMA implemented in the free software packages R and Bioconductor (http://www.r-project.org). In identification of CFT073 or ΔTcpC-specific genes, fold change of ≥2 was used for the comparison CFT073 or ΔTcpC with its control and fold change of ≤1.5 was used for the comparison ΔTcpC or CFT073 with its control, respectively. In addition to the above fold change criteria, statistical t-test with p≤0.05 was further used in selection of differentially expressed genes or CFT073/ΔTcpC specific genes. To further study signaling pathways altered by TcpC, the differentially expressed genes were analyzed using the Ingenuity Pathway Analysis software with default settings (Ingenuity Systems, Redwood City, CA). In parallel, cDNA was also quantified by RT-PCR using human primers IL-6 (Hs00174131_m1), IL-8 (Hs00174103_m1), NFKB1 (Hs00231653_m1), cFOS (Hs01119266_g1), IRF3 (Hs01547283_m1), STAT1 (Hs01014002_m1), IRF7 (Hs00185375_m1) and TNF-α (Hs01113624_g1) from Applied Biosystems.
Protein arrays
Kidney A498 cells were stimulated with CFT073 or ΔTcpC as described above. Culture supernatants were collected and cleared by centrifugation at 20,000×g before storage at −80°C. Cytokine profile analysis was performed in triplicate using the Procarta Human Cytokine 50-plex kit (Panomics, Fremont, USA) according to manufacturer's protocol. Cytokine levels were evaluated using a Luminex 100 system (Luminex, Austin, TX, USA).
RT-PCR and ELISA of murine tubular kidney cells
Primary murine renal tubular epithelial cells (MRTEC) were harvested from murine kidneys (C57BL/6, Trif Lps2/Lps2, Myd88−/−) following microdissection and brief collagenase digestion (2 mg/ml of Collagenase Type II). Cells were grown in hormonally defined RPMI supplemented with epidermal growth factor (Sigma, 50 pg/ml), insulin-transferrin-sodium selenite media supplement (Gibco, diluted 1∶100), Prostoglandin E1 (Sigma, 1.25 ng/ml), T3 (Sigma, 34 pg/ml), hydrocortisone (Sigma, 18 ng/ml), 10% heat-inactivated FCS, and 0.5 µg/ml gentamycin at 37°C. The cells were stimulated 5 days after the primary explantation at 100% confluence and infected with CFT073 or ΔTcpC (108 CFU/ml) for 4 hrs. Total extracted mRNAs were converted to cDNA using RT2 First Strand Kit (SABioscience Corporation, Fredrick, MA, USA). The mouse primers used for RT-PCR GAPDH (QT01658692) and MIP-2 (QT00113253) were from Qiagen. Gene expression levels were calculated by the ΔCt method and normalized to house-keeping genes. cDNA was quantified by RT-PCR using a Rotor gene 2000 instrument (Corbett Life Science, Sydney, Australia) and normalized against GAPDH for each gene. In parallel, MIP-2 in culture supernatants was also quantified by ELISA using the Mouse MIP-2 quantification kit (R&D systems, Abingdon, UK) according to the manufacturer's instructions.
BMDM and ligand stimulation assays
Wt or Myd88−/− BMDM (bone marrow derived macrophages) were cultured in 6-well plates at a concentration of 0.5×106/well in DMEM supplemented with 10% FCS, 1% penicillin-streptomycin and 0.1% 2-mercaptoethanol. Cells were stimulated with the TLR ligands Pam3Cys, poly(I:C), LPS, flagellin or CpG-ODN 1826 in the absence or presence of titrated amounts of the purified recombinant TIR domain of TcpC (TIR-TcpC) for 3 h. TNF was quantified in the culture supernatant using ELISA Duo sets (R&D Systems) as described by the manufacturer.
Statistical analysis
Bacterial counts and immune responses are presented as geomean ± SEM. Fisher's exact test and two-tailed T test was used for the analysis of bacterial counts and immune response. Student's t-test was used for protein array analysis. Wilcoxon's matched pairs test was used for paired comparisons. The level of significance was set at p<0.05 for all tests.
List of ID numbers for genes and proteins of mouse and humans
Gene ID number for human TLR4 is 7099, mouse Tlr4 is 21898, human MYD88 is 4615, mouse Myd88 is 17874, human TRIF is 148022, mouse Trif is 106759, human IRF3 is 3661, mouse Irf3 is 54131, human IL-1B is 3553 and mouse Il-1b is 16176.
Supporting Information
Zdroje
1. PoltorakA
HeX
SmirnovaI
LiuMY
Van HuffelC
1998 Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282 2085 2088
2. RodriguezN
WantiaN
FendF
DurrS
WagnerH
2006 Differential involvement of TLR2 and TLR4 in host survival during pulmonary infection with Chlamydia pneumoniae. Eur J Immunol 36 1145 1155
3. KawaiT
AkiraS
2006 TLR signaling. Cell Death Differ 13 816 825
4. KawaiT
SatoS
IshiiKJ
CobanC
HemmiH
2004 Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol 5 1061 1068
5. ThomassenE
RenshawBR
SimsJE
1999 Identification and characterization of SIGIRR, a molecule representing a novel subtype of the IL-1R superfamily. Cytokine 11 389 399
6. WaldD
QinJ
ZhaoZ
QianY
NaramuraM
2003 SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat Immunol 4 920 927
7. BurnsK
JanssensS
BrissoniB
OlivosN
BeyaertR
2003 Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med 197 263 268
8. KobayashiK
HernandezLD
GalanJE
JanewayCAJr
MedzhitovR
2002 IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 110 191 202
9. FearnsC
PanQ
MathisonJC
ChuangTH
2006 Triad3A regulates ubiquitination and proteasomal degradation of RIP1 following disruption of Hsp90 binding. J Biol Chem 281 34592 34600
10. CartyM
GoodbodyR
SchroderM
StackJ
MoynaghPN
2006 The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol 7 1074 1081
11. FischerH
YamamotoM
AkiraS
BeutlerB
SvanborgC
2006 Mechanism of pathogen-specific TLR4 activation in the mucosa: fimbriae, recognition receptors and adaptor protein selection. Eur J Immunol 36 267 277
12. CirlC
WieserA
YadavM
DuerrS
SchubertS
2008 Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med 14 399 406
13. SpearAM
LomanNJ
AtkinsHS
PallenMJ
2009 Microbial TIR domains: not necessarily agents of subversion? Trends Microbiol 17 393 398
14. BowieA
Kiss-TothE
SymonsJA
SmithGL
DowerSK
2000 A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc Natl Acad Sci U S A 97 10162 10167
15. NewmanRM
SalunkheP
GodzikA
ReedJC
2006 Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect Immun 74 594 601
16. RadhakrishnanGK
YuQ
HarmsJS
SplitterGA
2009 Brucella TIR Domain-containing Protein Mimics Properties of the Toll-like Receptor Adaptor Protein TIRAP. J Biol Chem 284 9892 9898
17. HoebeK
DuX
GeorgelP
JanssenE
TabetaK
2003 Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature 424 743 748
18. TaniguchiT
OgasawaraK
TakaokaA
TanakaN
2001 IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19 623 655
19. HondaK
TaniguchiT
2006 IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6 644 658
20. BurnsK
MartinonF
EsslingerC
PahlH
SchneiderP
1998 MyD88, an adapter protein involved in interleukin-1 signaling. J Biol Chem 273 12203 12209
21. KawaiT
AdachiO
OgawaT
TakedaK
AkiraS
1999 Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11 115 122
22. Svanborg EdenC
BrilesD
HagbergL
McGheeJ
MichalecS
1985 Genetic factors in host resistance to urinary tract infection. Infection 13 Suppl 2 S171 176
23. LundstedtAC
McCarthyS
GustafssonMC
GodalyG
JodalU
2007 A genetic basis of susceptibility to acute pyelonephritis. PLoS ONE 2 e825
24. RagnarsdottirB
SamuelssonM
GustafssonMC
LeijonhufvudI
KarpmanD
2007 Reduced toll-like receptor 4 expression in children with asymptomatic bacteriuria. J Infect Dis 196 475 484
25. ShiS
NathanC
SchnappingerD
DrenkowJ
FuortesM
2003 MyD88 primes macrophages for full-scale activation by interferon-gamma yet mediates few responses to Mycobacterium tuberculosis. J Exp Med 198 987 997
26. SunD
DingA
2006 MyD88-mediated stabilization of interferon-gamma-induced cytokine and chemokine mRNA. Nat Immunol 7 375 381
27. FitzgeraldKA
Palsson-McDermottEM
BowieAG
JefferiesCA
MansellAS
2001 Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature 413 78 83
28. YamamotoM
SatoS
HemmiH
HoshinoK
KaishoT
2003 Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301 640 643
29. SatoS
SugiyamaM
YamamotoM
WatanabeY
KawaiT
2003 Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol 171 4304 4310
30. FitzgeraldKA
RoweDC
BarnesBJ
CaffreyDR
VisintinA
2003 LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med 198 1043 1055
31. YamamotoM
SatoS
HemmiH
UematsuS
HoshinoK
2003 TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol 4 1144 1150
32. RoweDC
McGettrickAF
LatzE
MonksBG
GayNJ
2006 The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. Proc Natl Acad Sci U S A 103 6299 6304
33. McGettrickAF
BrintEK
Palsson-McDermottEM
RoweDC
GolenbockDT
2006 Trif-related adapter molecule is phosphorylated by PKC{epsilon} during Toll-like receptor 4 signaling. Proc Natl Acad Sci U S A 103 9196 9201
34. OganesyanG
SahaSK
GuoB
HeJQ
ShahangianA
2006 Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439 208 211
35. BeutlerB
HoebeK
GeorgelP
TabetaK
DuX
2005 Genetic analysis of innate immunity: identification and function of the TIR adapter proteins. Adv Exp Med Biol 560 29 39
36. BergstenG
SamuelssonM
WulltB
LeijonhufvudI
FischerH
2004 PapG-dependent adherence breaks mucosal inertia and triggers the innate host response. J Infect Dis 189 1734 1742
37. JanssensS
BurnsK
VercammenE
TschoppJ
BeyaertR
2003 MyD88S, a splice variant of MyD88, differentially modulates NF-kappaB - and AP-1-dependent gene expression. FEBS Lett 548 103 107
38. HoshinoK
TakeuchiO
KawaiT
SanjoH
OgawaT
1999 Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol 162 3749 3752
39. AdachiO
KawaiT
TakedaK
MatsumotoM
TsutsuiH
1998 Targeted disruption of the MyD88 gene results in loss of IL-1 - and IL-18-mediated function. Immunity 9 143 150
40. SatoM
SuemoriH
HataN
AsagiriM
OgasawaraK
2000 Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13 539 548
41. HoraiR
AsanoM
SudoK
KanukaH
SuzukiM
1998 Production of mice deficient in genes for interleukin (IL)-1alpha, IL-1beta, IL-1alpha/beta, and IL-1 receptor antagonist shows that IL-1beta is crucial in turpentine-induced fever development and glucocorticoid secretion. J Exp Med 187 1463 1475
42. KaoJS
StuckerDM
WarrenJW
MobleyHL
1997 Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect Immun 65 2812 2820
43. MobleyH
GreenD
TrifillisA
JohnsonD
ChippendaleG
1990 Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: Role of hemolysin in some strains. Infect Immun 58 1281 1289
44. JohansonIM
PlosK
MarklundBI
SvanborgC
1993 Pap, papG and prsG DNA sequences in Escherichia coli from the fecal flora and the urinary tract. Microb Pathog 15 121 129
45. HagbergL
EngbergI
FreterR
LamJ
OllingS
1983 Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun 40 273 283
46. MajewskaM
PanasiewiczG
MajewskiM
SzafranskaB
2006 Localization of chorionic pregnancy-associated glycoprotein family in the pig. Reprod Biol 6 205 230
47. ChuaSW
VijayakumarP
NissomPM
YamCY
WongVV
2006 A novel normalization method for effective removal of systematic variation in microarray data. Nucleic Acids Res 34 e38
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek SRFR1 Negatively Regulates Plant NB-LRR Resistance Protein Accumulation to Prevent AutoimmunityČlánek Phylogenetic Approach Reveals That Virus Genotype Largely Determines HIV Set-Point Viral LoadČlánek A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 9- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Azole Drugs Are Imported By Facilitated Diffusion in and Other Pathogenic Fungi
- Two Genes on A/J Chromosome 18 Are Associated with Susceptibility to Infection by Combined Microarray and QTL Analyses
- Impact of Simian Immunodeficiency Virus Infection on Chimpanzee Population Dynamics
- Breaking the Stereotype: Virulence Factor–Mediated Protection of Host Cells in Bacterial Pathogenesis
- The Canine Papillomavirus and Gamma HPV E7 Proteins Use an Alternative Domain to Bind and Destabilize the Retinoblastoma Protein
- Rescue of HIV-1 Release by Targeting Widely Divergent NEDD4-Type Ubiquitin Ligases and Isolated Catalytic HECT Domains to Gag
- Steric Shielding of Surface Epitopes and Impaired Immune Recognition Induced by the Ebola Virus Glycoprotein
- Dynamics of the Multiplicity of Cellular Infection in a Plant Virus
- HLA Class I Binding of HBZ Determines Outcome in HTLV-1 Infection
- Pathogenic Bacteria Target NEDD8-Conjugated Cullins to Hijack Host-Cell Signaling Pathways
- The HA and NS Genes of Human H5N1 Influenza A Virus Contribute to High Virulence in Ferrets
- SRFR1 Negatively Regulates Plant NB-LRR Resistance Protein Accumulation to Prevent Autoimmunity
- Cyclin-Dependent Kinase Activity Controls the Onset of the HCMV Lytic Cycle
- The N-Terminal Domain of the Arenavirus L Protein Is an RNA Endonuclease Essential in mRNA Transcription
- Generation of Neutralizing Antibodies and Divergence of SIVmac239 in Cynomolgus Macaques Following Short-Term Early Antiretroviral Therapy
- Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
- Intracellular Proton Conductance of the Hepatitis C Virus p7 Protein and Its Contribution to Infectious Virus Production
- The Transcriptome of the Human Pathogen at Single-Nucleotide Resolution
- The Epidermal Growth Factor Receptor (EGFR) Promotes Uptake of Influenza A Viruses (IAV) into Host Cells
- Surface Co-Expression of Two Different PfEMP1 Antigens on Single -Infected Erythrocytes Facilitates Binding to ICAM1 and PECAM1
- Sequestration and Tissue Accumulation of Human Malaria Parasites: Can We Learn Anything from Rodent Models of Malaria?
- Phylogenomics of Ligand-Gated Ion Channels Predicts Monepantel Effect
- Generation of Covalently Closed Circular DNA of Hepatitis B Viruses via Intracellular Recycling Is Regulated in a Virus Specific Manner
- CpG-Methylation Regulates a Class of Epstein-Barr Virus Promoters
- Molecular and Evolutionary Bases of Within-Patient Genotypic and Phenotypic Diversity in Extraintestinal Infections
- A Bistable Switch and Anatomical Site Control Virulence Gene Expression in the Intestine
- Are Members of the Fungal Genus (a) Commensals; (b) Opportunists; (c) Pathogens; or (d) All of the Above?
- Structures of Receptor Complexes of a North American H7N2 Influenza Hemagglutinin with a Loop Deletion in the Receptor Binding Site
- Phylogenetic Approach Reveals That Virus Genotype Largely Determines HIV Set-Point Viral Load
- The Coevolution of Virulence: Tolerance in Perspective
- Involvement of the Cytokine MIF in the Snail Host Immune Response to the Parasite
- Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens
- A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins
- High Content Phenotypic Cell-Based Visual Screen Identifies Acyltrehalose-Containing Glycolipids Involved in Phagosome Remodeling
- A Novel Small Molecule Inhibitor of Hepatitis C Virus Entry
- The Microbiota Mediates Pathogen Clearance from the Gut Lumen after Non-Typhoidal Diarrhea
- RNA Polymerases (L-Protein) Have an N-Terminal, Influenza-Like Endonuclease Domain, Essential for Viral Cap-Dependent Transcription
- Pathogen Specific, IRF3-Dependent Signaling and Innate Resistance to Human Kidney Infection
- Cellular Entry of Ebola Virus Involves Uptake by a Macropinocytosis-Like Mechanism and Subsequent Trafficking through Early and Late Endosomes
- The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis
- Formation of Mobile Chromatin-Associated Nuclear Foci Containing HIV-1 Vpr and VPRBP Is Critical for the Induction of G2 Cell Cycle Arrest
- Association of Tat with Promoters of PTEN and PP2A Subunits Is Key to Transcriptional Activation of Apoptotic Pathways in HIV-Infected CD4+ T Cells
- Metal Hyperaccumulation Armors Plants against Disease
- Cyclin-Dependent Kinase-Like Function Is Shared by the Beta- and Gamma- Subset of the Conserved Herpesvirus Protein Kinases
- Role of Acetyl-Phosphate in Activation of the Rrp2-RpoN-RpoS Pathway in
- Ebolavirus Is Internalized into Host Cells Macropinocytosis in a Viral Glycoprotein-Dependent Manner
- A Novel Family of IMC Proteins Displays a Hierarchical Organization and Functions in Coordinating Parasite Division
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens
- The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis
- Inhibition of TIR Domain Signaling by TcpC: MyD88-Dependent and Independent Effects on Virulence
- The Coevolution of Virulence: Tolerance in Perspective
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



