-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPredicting Carriers of Ongoing Selective Sweeps without Knowledge of the Favored Allele
Methods for detecting the genomic signatures of natural selection have been heavily studied, and they have been successful in identifying genomic regions under positive selection. However, methods that detect positive selective sweeps do not typically identify the favored allele, or even the haplotypes carrying the favored allele. The main contribution of this paper is the development and analysis of a new statistic (the HAF score), assigned to individual haplotypes. Using both theoretical analyses and simulations, we describe how the HAF scores differ for carriers and non-carriers of the favored allele, and how they change dynamically during a selective sweep. We also develop an algorithm, PreCIOSS, for separating carriers and non-carriers. Our tool has broad applicability as carriers of the favored allele are likely to contain a future most recent common ancestor. Therefore, identifying them may prove useful in predicting the evolutionary trajectory—for example, in contexts involving drug-resistant pathogen strains or cancer subclones.
Published in the journal: . PLoS Genet 11(9): e32767. doi:10.1371/journal.pgen.1005527
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005527Summary
Methods for detecting the genomic signatures of natural selection have been heavily studied, and they have been successful in identifying genomic regions under positive selection. However, methods that detect positive selective sweeps do not typically identify the favored allele, or even the haplotypes carrying the favored allele. The main contribution of this paper is the development and analysis of a new statistic (the HAF score), assigned to individual haplotypes. Using both theoretical analyses and simulations, we describe how the HAF scores differ for carriers and non-carriers of the favored allele, and how they change dynamically during a selective sweep. We also develop an algorithm, PreCIOSS, for separating carriers and non-carriers. Our tool has broad applicability as carriers of the favored allele are likely to contain a future most recent common ancestor. Therefore, identifying them may prove useful in predicting the evolutionary trajectory—for example, in contexts involving drug-resistant pathogen strains or cancer subclones.
Introduction
With genome sequencing, we now have an opportunity to more completely sample genetic diversity in human populations, and probe deeper for signatures of adaptive evolution [1–3]. Genetic data from diverse human populations in recent years have revealed a multitude of genomic regions believed to be evolving under recent positive selection [4–16].
Methods for detecting selective sweeps from DNA sequences have examined a variety of signatures, including patterns represented in variant allele frequencies as well as in haplotype structure. Initially, the problem of detecting selective sweeps was approached primarily by considering variant allele frequencies, exploiting the shift in frequency at ‘hitchhiking’ sites linked to a favored allele relative to non-hitchhiking sites [17, 18]. The site frequency spectrum (SFS) within and across populations is often used as a basis for such inference [4, 6, 19–25]. More recently, methods based on haplotype structure have been developed using a variety of approaches, including the frequency of the most common haplotype [26], the number and diversity of distinct haplotypes [27], the haplotype frequency spectrum [28], and the popular approach of long-range haplotype homozygosity [29–32].
In general, haplotype-based methods seek to characterize the population with summary statistics that capture the frequency and length of different haplotypes. However, the haplotypes are related through a genealogy, and relationships among them are inherently lost in such analyses. In addition, data on the site frequency spectrum can be lost or hidden in analyses focused on haplotype spectra. In this paper, we connect related measures of haplotype frequencies and the site frequency spectrum by merging information describing haplotype relationships with variant allele frequencies. Our main contribution is a statistic that we term the haplotype allele frequency (HAF) score, which captures many of the properties shared by haplotypes carrying a favored allele.
Consider a sample of haplotypes in a genomic region. We assume that all sites are biallelic, and at each site, we denote ancestral alleles by 0 and derived alleles by 1. We also assume that all sites are polymorphic in the sample. The HAF vector of a haplotype h, denoted c, is obtained by taking the binary haplotype vector and replacing non-zero entries (derived alleles carried by the haplotype) with their respective frequencies in the sample (Fig 1A). For parameter ℓ, we define the ℓ-HAF score of c as:
where the sum proceeds over all segregating sites j in the genomic region. The 1-HAF score of a haplotype amounts to the sum of frequencies of all derived alleles carried by the haplotype. The ℓ-HAF score is equivalent to the ℓ-norm of c raised to the ℓth power, or ( ‖ c ‖ ℓ ) ℓ. We will show that during a selective sweep, the HAF score of a haplotype serves as a proxy to its relative fitness.Fig. 1. The HAF score. 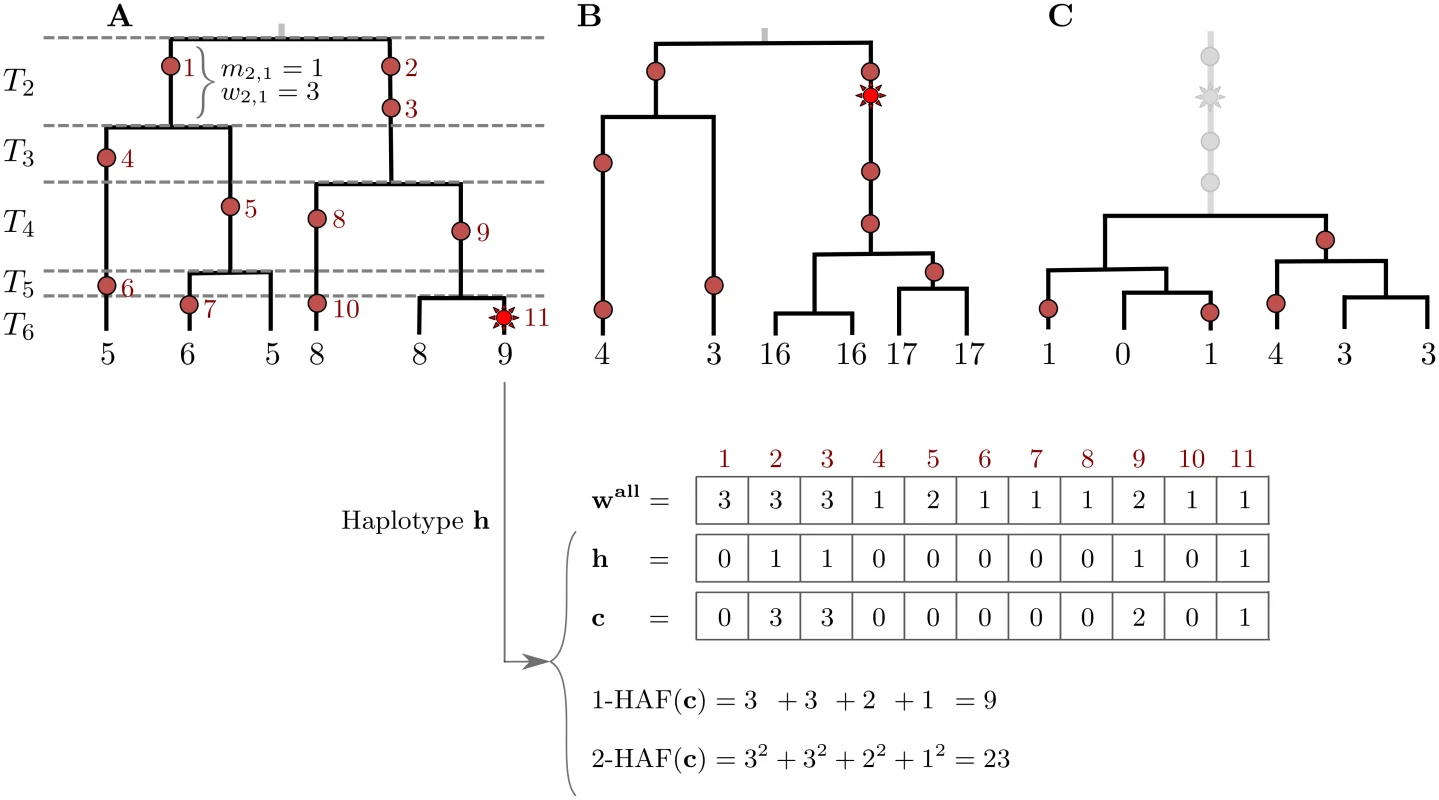
Genealogies of three samples (n = 6) progressing through a selective sweep, from left to right. Neutral mutations are shown as red circles, and are numbered in red; the favored allele is shown as a red star. The 1-HAF score of each haplotype is shown below its corresponding leaf, in black. For the rightmost haplotype in (A), the binary haplotype vector h is shown along with its HAF-vector c, and 1-HAF and 2-HAF scores. Vector wall lists the frequencies of all mutations. (A) The favored allele appears on a single haplotype. At this point in time, both the genealogy and the HAF score distribution are largely neutral. Coalescence times (T2, …, T6) are shown on the left, where Tk spans the epoch with exactly k lineages. (B) Carriers of the favored allele are distinguished by high HAF scores (in large part due to the long branch of high-frequency hitchhiking variation); non-carriers have low HAF scores. (C) After fixation, there is a sharp loss of diversity causing low HAF scores across the sample. Selective sweeps
The classical model for selection, and the one that has received most attention, is the “hard sweep” model, in which a single mutation conveys higher fitness immediately upon occurrence, and rapidly rises in frequency, eventually reaching fixation [17, 33]. Under this model, we can partition the haplotypes into carriers of the favored allele, and non-carriers. In the absence of recombination, the favored haplotypes form a single clade in the genealogy. As a sweep progresses, HAF scores in the favored clade will rise due to the increasing frequencies of alleles hitchhiking along with the favored allele. HAF scores of non-carrier haplotypes will decrease, as many of the derived alleles they carry become rare (Fig 1B). After fixation of the favored and hitchhiking alleles, HAF scores will decline sharply (Fig 1C), as the selected site and other linked sites are no longer polymorphic. Thus, this reduction in the HAF score results from the sudden loss of many high-frequency derived alleles from the pool of segregating sites [18, 20, 24]. Finally, as the site-frequency spectrum recovers to its neutral state due to new mutations and drift [23], so will the HAF scores.
Recombination is a source of ‘noise’ for the properties of the HAF score, predicted under the assumption of a hard sweep and no recombination, as it allows haplotypes to cross into and out of the favored clade. Recombination can lead to (i) haplotypes that carry the favored allele but little of the hitchhiking variation, thus having relatively low HAF scores despite their high fitness, or (ii) haplotypes that do not carry the favored allele but do carry much of the hitchhiking variation, thus having relatively high HAF scores despite their low fitness. By the same logic, recombination adds ‘noise’ after fixation by making the otherwise sharp decline in HAF scores more subtle and gradual. This more gradual decline occurs due to recombination weakening the linkage between the favored allele and hitchhiking variants.
Recently, “soft sweeps” have generated significant interest [34–36]. A soft sweep occurs when multiple sets of hitchhiking alleles in a given region increase in frequency, rather than a single favored haplotype. Soft sweeps may take place by one or more of the following mechanisms: (i) selection from standing variation: a neutral segregating mutation, which exists on several haplotypic backgrounds, becomes favored due to a change in the environment; (ii) recurrent mutation: the favored mutation arises several times on different haplotypic backgrounds; or, (iii) multiple adaptations: multiple favored mutations occur on multiple haplotypic backgrounds. Several methods have been developed for detecting soft sweeps [37, 38], as well as for distinguishing between soft and hard sweeps [39–41]. In soft sweeps, multiple sets of hitchhiking alleles rise to intermediate frequencies as the favored allele fixes. This could cause the pre-fixation peak and post-fixation trough in HAF scores to be less pronounced and to occur more gradually compared to a hard sweep.
We find (see Results) that the properties of the HAF score remain robust to many soft sweep scenarios. Moreover, the HAF score could potentially be used to detect soft sweeps. However, in this paper, we focus on the foundations, developing theoretical analysis and empirical work that predicts the dynamics of the HAF score. We also develop a single application. Recall that most existing methods for characterizing selective sweeps focus on identifying regions under selection. Here, given a region already identified to be undergoing a selective sweep, we ask if we can accurately predict which haplotypes carry the favored allele, without knowledge of the favored site. Successfully doing so may provide insight into the future evolutionary trajectory of a population. Haplotypes in future generations are more likely to be descended from, and therefore to resemble, extant carriers of a favored allele. This predictive perspective is of particular importance when a sweep is undesirable and measures may be taken to prevent it. For instance, consider a set of tumor haplotypes isolated from single cells, some of which are drug-resistant and therefore favored under drug exposure. Given a genetic sample of the tumor haplotypes, the HAF statistic may be applied to identify the resistant haplotypes—carriers of a favored allele—before they clonally expand and metastasize.
Below, we start with a theoretical explanation of the behavior of the HAF score under different evolutionary scenarios, validating our results using simulation. We then develop an algorithm (PreCIOSS: Predicting Carriers of Ongoing Selective Sweeps) to detect carriers of selective sweeps based on the HAF score. We demonstrate the power of PreCIOSS on simulations of both hard and soft sweeps, as well as on real genetic data from well-known sweeps in human populations. While our theoretical derivations make use of coalescent theory, and explicitly use tree-like genealogies, we note that HAF scores can be computed for any haplotype matrix including those with recombination events. Our results on simulated and real data imply that the utility of the HAF score extends to cases with recombination as well as other evolutionary scenarios.
Results
Theoretical and empirical modeling of HAF scores
We consider a sample of n haploid individuals chosen at random from a larger haploid population of size N. Let μ denote the mutation rate per generation per nucleotide, and let θ = 2NμL denote the population-scaled mutation rate in a region of length L bp. We consider both constant-sized and exponentially growing populations. For exponentially growing populations, let N0 denote the final population size, let r denote the growth rate per generation, and let α = 2 N0 r the population-scaled growth rate. Let ρ denote the population-scaled recombination rate. In our theoretical calculations, we assume no recombination (ρ = 0), and we derive expressions for the general ℓ-HAF score. We use simulations to demonstrate the concordance of theoretical and empirical values of the ℓ-HAF score, and show that the values are robust to the presence of recombination (see ‘Simulations’ in Methods for parameter choices). Although some of our theoretical calculations below derive expressions for the general ℓ-HAF score, we primarily use 1-HAF in the applied sections. Applications of ℓ-HAF with ℓ > 1 will be explored in future work.
Expected ℓ-HAF score under neutrality, constant population size
First, we assume that the genomic region of interest is evolving neutrally, the population size remains constant at N, and that the ancestral states are known or can be derived. In a sample of size n, let c(v) denote the HAF vector c for the vth haplotype (v ∈ {1, …, n}). Let ξw be the number of sites with derived allele frequency w. We only consider polymorphic sites in the sample, so the frequency is in the range w ∈ {1, …, n − 1}; a mutation present in all or none of the haplotypes in the sample would not be detectable. Each of the ξw sites of frequency w contributes wℓ to the ℓ-HAF score of each of the w haplotypes with the mutation, and contributes 0ℓ = 0 for each of the other n − w haplotypes. The mean of the ℓ-HAF scores of all n haplotypes in the sample is
Under the coalescent model, [42, Eq. (22)] shows that
Zdroje
1. Fu W, Akey JM. Selection and adaptation in the human genome. Annu Rev Genomics Hum Genet. 2013;14 : 467–489. doi: 10.1146/annurev-genom-091212-153509 23834317
2. Lachance J, Tishkoff SA. Population Genomics of Human Adaptation. Annu Rev Ecol Evol Syst. 2013 Nov;44 : 123–143. doi: 10.1146/annurev-ecolsys-110512-135833 25383060
3. Vitti JJ, Grossman SR, Sabeti PC. Detecting natural selection in genomic data. Annu Rev Genet. 2013;47 : 97–120. doi: 10.1146/annurev-genet-111212-133526 24274750
4. Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005 Nov;15(11):1566–1575. doi: 10.1101/gr.4252305 16251466
5. Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009 May;19(5):826–837. doi: 10.1101/gr.087577.108 19307593
6. Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010 Mar;20(3):393–402. doi: 10.1101/gr.100545.109 20086244
7. Berg JJ, Coop G. A population genetic signal of polygenic adaptation. PLoS Genet. 2014 Aug;10(8):e1004412. doi: 10.1371/journal.pgen.1004412 25102153
8. Jeong C, Di Rienzo A. Adaptations to local environments in modern human populations. Curr Opin Genet Dev. 2014 Dec;29C:1–8. doi: 10.1016/j.gde.2014.06.011
9. Tekola-Ayele F, Adeyemo A, Chen G, Hailu E, Aseffa A, Davey G, et al. Novel genomic signals of recent selection in an Ethiopian population. Eur J Hum Genet. 2014 Nov; advance online publication. doi: 10.1038/ejhg.2014.233 25370040
10. Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, et al. Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude. Science. 2010;329(5987):75–78. Available from: http://www.sciencemag.org/content/329/5987/75.abstract. doi: 10.1126/science.1190371 20595611
11. Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010 Jul;329(5987):72–75. doi: 10.1126/science.1189406 20466884
12. Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, Beggs W, et al. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 2012;13(1):R1. doi: 10.1186/gb-2012-13-1-r1 22264333
13. Alkorta-Aranburu G, Beall CM, Witonsky DB, Gebremedhin A, Pritchard JK, Di Rienzo A. The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 2012;8(12):e1003110. doi: 10.1371/journal.pgen.1003110 23236293
14. Huerta-Sanchez E, Degiorgio M, Pagani L, Tarekegn A, Ekong R, Antao T, et al. Genetic signatures reveal high-altitude adaptation in a set of ethiopian populations. Mol Biol Evol. 2013 Aug;30(8):1877–1888. doi: 10.1093/molbev/mst089 23666210
15. Udpa N, Ronen R, Zhou D, Liang J, Stobdan T, Appenzeller O, et al. Whole genome sequencing of Ethiopian highlanders reveals conserved hypoxia tolerance genes. Genome Biol. 2014 Feb;15(2):R36. doi: 10.1186/gb-2014-15-2-r36 24555826
16. Zhou D, Udpa N, Ronen R, Stobdan T, Liang J, Appenzeller O, et al. Whole-genome sequencing uncovers the genetic basis of chronic mountain sickness in Andean highlanders. Am J Hum Genet. 2013 Sep;93(3):452–462. doi: 10.1016/j.ajhg.2013.07.011 23954164
17. Kaplan NL, Hudson RR, Langley CH. The “hitchhiking effect” revisited. Genetics. 1989 Dec;123(4):887–899. 2612899
18. Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974 Feb;23(1):23–35. doi: 10.1017/S0016672300014634 4407212
19. Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989 Nov;123(3):585–595. 2513255
20. Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000 Jul;155 : 1405–1413. 10880498
21. Pavlidis P, Jensen JD, Stephan W. Searching for footprints of positive selection in whole-genome SNP data from nonequilibrium populations. Genetics. 2010 Jul;185(3):907–922. doi: 10.1534/genetics.110.116459 20407129
22. Lin K, Li H, Schlotterer C, Futschik A. Distinguishing positive selection from neutral evolution: boosting the performance of summary statistics. Genetics. 2011 Jan;187(1):229–244. doi: 10.1534/genetics.110.122614 21041556
23. Ronen R, Udpa N, Halperin E, Bafna V. Learning natural selection from the site frequency spectrum. Genetics. 2013 Sep;195(1):181–193. doi: 10.1534/genetics.113.152587 23770700
24. Simonsen KL, Churchill GA, Aquadro CF. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics. 1995 Sep;141(1):413–429. 8536987
25. Braverman JM, Hudson RR, Kaplan NL, Langley CH, Stephan W. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics. 1995 Jun;140(2):783–796. 7498754
26. Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ. Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics. 1994 Apr;136(4):1329–1340. 8013910
27. Depaulis F, Mousset S, Veuille M. Haplotype tests using coalescent simulations conditional on the number of segregating sites. Mol Biol Evol. 2001 Jun;18(6):1136–1138. doi: 10.1093/oxfordjournals.molbev.a003885 11371602
28. Innan H, Zhang K, Marjoram P, Tavare S, Rosenberg NA. Statistical tests of the coalescent model based on the haplotype frequency distribution and the number of segregating sites. Genetics. 2005 Mar;169(3):1763–1777. doi: 10.1534/genetics.104.032219 15654103
29. Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002 Oct;419(6909):832–837. doi: 10.1038/nature01140 12397357
30. Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006 Mar;4(3):e72. doi: 10.1371/journal.pbio.0040072 16494531
31. Toomajian C, Hu TT, Aranzana MJ, Lister C, Tang C, Zheng H, et al. A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 2006 May;4(5):e137. doi: 10.1371/journal.pbio.0040137 16623598
32. Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007 Oct;449(7164):913–918. doi: 10.1038/nature06250 17943131
33. Kim Y, Stephan W. Selective sweeps in the presence of interference among partially linked loci. Genetics. 2003 May;164(1):389–398. 12750349
34. Messer PW, Petrov DA. Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol Evol (Amst). 2013 Nov;28(11):659–669. doi: 10.1016/j.tree.2013.08.003
35. Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005 Apr;169(4):2335–2352. doi: 10.1534/genetics.104.036947 15716498
36. Pennings PS, Hermisson J. Soft sweeps II–molecular population genetics of adaptation from recurrent mutation or migration. Mol Biol Evol. 2006 May;23(5):1076–1084. doi: 10.1093/molbev/msj117 16520336
37. Ferrer-Admetlla A, Liang M, Korneliussen T, Nielsen R. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol Biol Evol. 2014 May;31(5):1275–1291. doi: 10.1093/molbev/msu077 24554778
38. Garud NR, Messer PW, Buzbas EO, Petrov DA. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 2015 Feb;11(2):e1005004. doi: 10.1371/journal.pgen.1005004 25706129
39. Peter BM, Huerta-Sanchez E, Nielsen R. Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet. 2012;8(10):e1003011. doi: 10.1371/journal.pgen.1003011 23071458
40. Schrider DR, Mendes FK, Hahn MW, Kern AD. Soft Shoulders Ahead: Spurious Signatures of Soft and Partial Selective Sweeps Result from Linked Hard Sweeps. Genetics. 2015 Feb; advance online publication.
41. Wilson BA, Petrov DA, Messer PW. Soft selective sweeps in complex demographic scenarios. Genetics. 2014 Oct;198(2):669–684. doi: 10.1534/genetics.114.165571 25060100
42. Fu YX. Statistical properties of segregating sites. Theor Popul Biol. 1995 Oct;48(2):172–197. doi: 10.1006/tpbi.1995.1025 7482370
43. Hudson RR. Gene genealogies and the coalescent process. In: Futuyma D, Antonovics J, editors. Oxford Surveys in Evolutionary Biology. Oxford: Oxford University Press; 1990. p. 1–44.
44. Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991 Oct;129(2):555–562. 1743491
45. Graham R, Knuth DE, Patashnik O. Concrete Mathematics: A Foundation for Computer Science. 2nd ed. Reading, Mass: Addison-Wesley; 1994.
46. Nordborg M. Coalescent Theory. In: Balding DJ, Bishop M, Cannings C, editors. Handbook of statistical genetics. 3rd ed. John Wiley & Sons, Ltd; 2008. p. 843–877.
47. Ewing G, Hermisson J. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics. 2010 Aug;26(16):2064–2065. doi: 10.1093/bioinformatics/btq322 20591904
48. Brodersen KH, Ong CS, Stephan KE, Buhmann JM. The Balanced Accuracy and Its Posterior Distribution. In: Pattern Recognition (ICPR), 2010 20th International Conference on; 2010. p. 3121–3124.
49. Grossman SR, Shlyakhter I, Shylakhter I, Karlsson EK, Byrne EH, Morales S, et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010 Feb;327(5967):883–886. doi: 10.1126/science.1183863 20056855
50. Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, et al. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci USA. 2011 Jul;108(29):11983–11988. doi: 10.1073/pnas.1019276108 21730125
51. Altshuler DM, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010 Sep;467(7311):52–58. doi: 10.1038/nature09298 20811451
52. Sequencing TC, Consortium A. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005 Sep;437(7055):69–87. doi: 10.1038/nature04072
53. Kuokkanen M, Enattah NS, Oksanen A, Savilahti E, Orpana A, Jarvela I. Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut. 2003 May;52(5):647–652. doi: 10.1136/gut.52.5.647 12692047
54. Olds LC, Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet. 2003 Sep;12(18):2333–2340. doi: 10.1093/hmg/ddg244 12915462
55. Troelsen JT, Olsen J, Møller J, Sjöström H. An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology. 2003 Dec;125(6):1686–1694. doi: 10.1053/j.gastro.2003.09.031 14724821
56. Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, et al. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004 Oct;2(10):e286. doi: 10.1371/journal.pbio.0020286 15361935
57. Stajich JE, Hahn MW. Disentangling the effects of demography and selection in human history. Mol Biol Evol. 2005 Jan;22(1):63–73. doi: 10.1093/molbev/msh252 15356276
58. Akey JM, Swanson WJ, Madeoy J, Eberle M, Shriver MD. TRPV6 exhibits unusual patterns of polymorphism and divergence in worldwide populations. Hum Mol Genet. 2006 Jul;15(13):2106–2113. doi: 10.1093/hmg/ddl134 16717058
59. Bhatia G, Patterson N, Pasaniuc B, Zaitlen N, Genovese G, Pollack S, et al. Genome-wide comparison of African-ancestry populations from CARe and other cohorts reveals signals of natural selection. Am J Hum Genet. 2011 Sep;89(3):368–381. doi: 10.1016/j.ajhg.2011.07.025 21907010
60. Sakamoto H, Yoshimura K, Saeki N, Katai H, Shimoda T, Matsuno Y, et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008 Jun;40(6):730–740. doi: 10.1038/ng.152 18488030
61. Wu X, Ye Y, Kiemeney LA, Sulem P, Rafnar T, Matullo G, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009 Sep;41(9):991–995. doi: 10.1038/ng.421 19648920
62. Whitfield JB. Alcohol dehydrogenase and alcohol dependence: variation in genotype-associated risk between populations. Am J Hum Genet. 2002 Nov;71(5):1247–1250. doi: 10.1086/344287 12452180
63. Peng Y, Shi H, Qi XB, Xiao CJ, Zhong H, Ma RL, et al. The ADH1B Arg47His polymorphism in east Asian populations and expansion of rice domestication in history. BMC Evol Biol. 2010;10 : 15. doi: 10.1186/1471-2148-10-15 20089146
64. Osier MV, Pakstis AJ, Soodyall H, Comas D, Goldman D, Odunsi A, et al. A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am J Hum Genet. 2002 Jul;71(1):84–99. doi: 10.1086/341290 12050823
65. Eng MY, Luczak SE, Wall TL. ALDH2, ADH1B, and ADH1C genotypes in Asians: a literature review. Alcohol Res Health. 2007;30(1):22–27. 17718397
66. Li H, Mukherjee N, Soundararajan U, Tarnok Z, Barta C, Khaliq S, et al. Geographically separate increases in the frequency of the derived ADH1B*47His allele in eastern and western Asia. Am J Hum Genet. 2007 Oct;81(4):842–846. doi: 10.1086/521201 17847010
67. McGovern PE, Zhang J, Tang J, Zhang Z, Hall GR, Moreau RA, et al. Fermented beverages of pre - and proto-historic China. Proc Natl Acad Sci USA. 2004 Dec;101(51):17593–17598. doi: 10.1073/pnas.0407921102 15590771
68. Fujimoto A, Ohashi J, Nishida N, Miyagawa T, Morishita Y, Tsunoda T, et al. A replication study confirmed the EDAR gene to be a major contributor to population differentiation regarding head hair thickness in Asia. Hum Genet. 2008 Sep;124(2):179–185. doi: 10.1007/s00439-008-0537-1 18704500
69. Kimura R, Yamaguchi T, Takeda M, Kondo O, Toma T, Haneji K, et al. A common variation in EDAR is a genetic determinant of shovel-shaped incisors. Am J Hum Genet. 2009 Oct;85(4):528–535. doi: 10.1016/j.ajhg.2009.09.006 19804850
70. Bryk J, Hardouin E, Pugach I, Hughes D, Strotmann R, Stoneking M, et al. Positive selection in East Asians for an EDAR allele that enhances NF-kappaB activation. PLoS ONE. 2008;3(5):e2209. doi: 10.1371/journal.pone.0002209 18493316
71. Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007 Oct;449(7164):913–918. doi: 10.1038/nature06250 17943131
72. Williamson SH, Hernandez R, Fledel-Alon A, Zhu L, Nielsen R, Bustamante CD. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc Natl Acad Sci USA. 2005 May;102(22):7882–7887. doi: 10.1073/pnas.0502300102 15905331
73. Luksza M, Lassig M. A predictive fitness model for influenza. Nature. 2014 Mar;507(7490):57–61. doi: 10.1038/nature13087 24572367
74. Lee MC, Lopez-Diaz FJ, Khan SY, Tariq MA, Dayn Y, Vaske CJ, et al. Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc Natl Acad Sci USA. 2014 Nov;111(44):E4726–4735. doi: 10.1073/pnas.1404656111 25339441
75. Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000 Sep;156(1):297–304. 10978293
76. Campbell CD, Chong JX, Malig M, Ko A, Dumont BL, Han L, et al. Estimating the human mutation rate using autozygosity in a founder population. Nat Genet. 2012 Nov;44(11):1277–1281. doi: 10.1038/ng.2418 23001126
77. Hey J, Wakeley J. A coalescent estimator of the population recombination rate. Genetics. 1997 Mar;145(3):833–846. 9055092
78. Szpiech ZA, Hernandez RD. selscan: An Efficient Multithreaded Program to Perform EHH-Based Scans for Positive Selection. Mol Biol Evol. 2014 Oct;31(10):2824–2827. doi: 10.1093/molbev/msu211 25015648
79. Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007 Oct;449(7164):851–861. doi: 10.1038/nature06258 17943122
Štítky
Genetika Reprodukční medicína
Článek The Chromatin Protein DUET/MMD1 Controls Expression of the Meiotic Gene during Male Meiosis inČlánek Tissue-Specific Gain of RTK Signalling Uncovers Selective Cell Vulnerability during Embryogenesis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 9
-
Všechny články tohoto čísla
- Retraction: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Signaling from Within: Endocytic Trafficking of the Robo Receptor Is Required for Midline Axon Repulsion
- A Splice Region Variant in Lowers Non-high Density Lipoprotein Cholesterol and Protects against Coronary Artery Disease
- The Chromatin Protein DUET/MMD1 Controls Expression of the Meiotic Gene during Male Meiosis in
- A NIMA-Related Kinase Suppresses the Flagellar Instability Associated with the Loss of Multiple Axonemal Structures
- Slit-Dependent Endocytic Trafficking of the Robo Receptor Is Required for Son of Sevenless Recruitment and Midline Axon Repulsion
- Expression of Concern: Protein Under-Wrapping Causes Dosage Sensitivity and Decreases Gene Duplicability
- Mutagenesis by AID: Being in the Right Place at the Right Time
- Identification of as a Genetic Modifier That Regulates the Global Orientation of Mammalian Hair Follicles
- Bridges Meristem and Organ Primordia Boundaries through , , and during Flower Development in
- Evaluating the Performance of Fine-Mapping Strategies at Common Variant GWAS Loci
- KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome
- Differential Expression of Ecdysone Receptor Leads to Variation in Phenotypic Plasticity across Serial Homologs
- Receptor Polymorphism and Genomic Structure Interact to Shape Bitter Taste Perception
- Cognitive Function Related to the Gene Acquired from an LTR Retrotransposon in Eutherians
- Critical Function of γH2A in S-Phase
- Arabidopsis AtPLC2 Is a Primary Phosphoinositide-Specific Phospholipase C in Phosphoinositide Metabolism and the Endoplasmic Reticulum Stress Response
- XBP1-Independent UPR Pathways Suppress C/EBP-β Mediated Chondrocyte Differentiation in ER-Stress Related Skeletal Disease
- Integration of Genome-Wide SNP Data and Gene-Expression Profiles Reveals Six Novel Loci and Regulatory Mechanisms for Amino Acids and Acylcarnitines in Whole Blood
- A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism
- Cell Cycle Regulates Nuclear Stability of AID and Determines the Cellular Response to AID
- A Genome-Wide Association Analysis Reveals Epistatic Cancellation of Additive Genetic Variance for Root Length in
- Tissue-Specific Gain of RTK Signalling Uncovers Selective Cell Vulnerability during Embryogenesis
- RAB-10-Dependent Membrane Transport Is Required for Dendrite Arborization
- Basolateral Endocytic Recycling Requires RAB-10 and AMPH-1 Mediated Recruitment of RAB-5 GAP TBC-2 to Endosomes
- Dynamic Contacts of U2, RES, Cwc25, Prp8 and Prp45 Proteins with the Pre-mRNA Branch-Site and 3' Splice Site during Catalytic Activation and Step 1 Catalysis in Yeast Spliceosomes
- ARID1A Is Essential for Endometrial Function during Early Pregnancy
- Predicting Carriers of Ongoing Selective Sweeps without Knowledge of the Favored Allele
- An Interaction between RRP6 and SU(VAR)3-9 Targets RRP6 to Heterochromatin and Contributes to Heterochromatin Maintenance in
- Photoreceptor Specificity in the Light-Induced and COP1-Mediated Rapid Degradation of the Repressor of Photomorphogenesis SPA2 in Arabidopsis
- Autophosphorylation of the Bacterial Tyrosine-Kinase CpsD Connects Capsule Synthesis with the Cell Cycle in
- Multimer Formation Explains Allelic Suppression of PRDM9 Recombination Hotspots
- Rescheduling Behavioral Subunits of a Fixed Action Pattern by Genetic Manipulation of Peptidergic Signaling
- A Gene Regulatory Program for Meiotic Prophase in the Fetal Ovary
- Cell-Autonomous Gβ Signaling Defines Neuron-Specific Steady State Serotonin Synthesis in
- Discovering Genetic Interactions in Large-Scale Association Studies by Stage-wise Likelihood Ratio Tests
- The RCC1 Family Protein TCF1 Regulates Freezing Tolerance and Cold Acclimation through Modulating Lignin Biosynthesis
- The AMPK, Snf1, Negatively Regulates the Hog1 MAPK Pathway in ER Stress Response
- The Parkinson’s Disease-Associated Protein Kinase LRRK2 Modulates Notch Signaling through the Endosomal Pathway
- Multicopy Single-Stranded DNA Directs Intestinal Colonization of Enteric Pathogens
- Recurrent Domestication by Lepidoptera of Genes from Their Parasites Mediated by Bracoviruses
- Three Different Pathways Prevent Chromosome Segregation in the Presence of DNA Damage or Replication Stress in Budding Yeast
- Identification of Four Mouse Diabetes Candidate Genes Altering β-Cell Proliferation
- The Intolerance of Regulatory Sequence to Genetic Variation Predicts Gene Dosage Sensitivity
- Synergistic and Dose-Controlled Regulation of Cellulase Gene Expression in
- Genome Sequence and Transcriptome Analyses of : Metabolic Tools for Enhanced Algal Fitness in the Prominent Order Prymnesiales (Haptophyceae)
- Ty3 Retrotransposon Hijacks Mating Yeast RNA Processing Bodies to Infect New Genomes
- FUS Interacts with HSP60 to Promote Mitochondrial Damage
- Point Mutations in Centromeric Histone Induce Post-zygotic Incompatibility and Uniparental Inheritance
- Genome-Wide Association Study with Targeted and Non-targeted NMR Metabolomics Identifies 15 Novel Loci of Urinary Human Metabolic Individuality
- Outer Hair Cell Lateral Wall Structure Constrains the Mobility of Plasma Membrane Proteins
- A Large-Scale Functional Analysis of Putative Target Genes of Mating-Type Loci Provides Insight into the Regulation of Sexual Development of the Cereal Pathogen
- A Genetic Selection for Mutants Reveals an Interaction between DNA Polymerase IV and the Replicative Polymerase That Is Required for Translesion Synthesis
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Arabidopsis AtPLC2 Is a Primary Phosphoinositide-Specific Phospholipase C in Phosphoinositide Metabolism and the Endoplasmic Reticulum Stress Response
- Bridges Meristem and Organ Primordia Boundaries through , , and during Flower Development in
- KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome
- XBP1-Independent UPR Pathways Suppress C/EBP-β Mediated Chondrocyte Differentiation in ER-Stress Related Skeletal Disease
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání





