-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPhotoreceptor Specificity in the Light-Induced and COP1-Mediated Rapid Degradation of the Repressor of Photomorphogenesis SPA2 in Arabidopsis
Plants have evolved photoreceptors that initiate a signaling cascade to adjust growth and development to the ambient light environment. The CUL4-dependent COP1/SPA E3 ubiquitin ligase is a key negative regulator of light signaling whose function is repressed by light. Recent research has identified mechanisms that are common to both phytochrome and cryptochrome photoreceptors. Here, we have identified a mechanism of light-induced COP1/SPA repression that is specific to phytochrome photoreceptors. We show that the SPA2 protein is very rapidly degraded in red, far-red and blue light in a phytochrome-dependent fashion. We further show that SPA2 degradation in the light depends on COP1 and on the interaction of SPA2 with COP1. Hence, our results suggest a light-induced degradation of SPA2, but not of COP1, by the COP1/SPA2 ubiquitin ligase. The human ortholog of COP1, which functions without the plant-specific SPA proteins, is known to be regulated by autodegradation following DNA damage. Hence, autodegradation of components of this E3 ligase is a regulatory mechanism used in both humans and plants.
Published in the journal: . PLoS Genet 11(9): e32767. doi:10.1371/journal.pgen.1005516
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005516Summary
Plants have evolved photoreceptors that initiate a signaling cascade to adjust growth and development to the ambient light environment. The CUL4-dependent COP1/SPA E3 ubiquitin ligase is a key negative regulator of light signaling whose function is repressed by light. Recent research has identified mechanisms that are common to both phytochrome and cryptochrome photoreceptors. Here, we have identified a mechanism of light-induced COP1/SPA repression that is specific to phytochrome photoreceptors. We show that the SPA2 protein is very rapidly degraded in red, far-red and blue light in a phytochrome-dependent fashion. We further show that SPA2 degradation in the light depends on COP1 and on the interaction of SPA2 with COP1. Hence, our results suggest a light-induced degradation of SPA2, but not of COP1, by the COP1/SPA2 ubiquitin ligase. The human ortholog of COP1, which functions without the plant-specific SPA proteins, is known to be regulated by autodegradation following DNA damage. Hence, autodegradation of components of this E3 ligase is a regulatory mechanism used in both humans and plants.
Introduction
As sessile organisms plants continuously monitor the ambient light conditions and adjust their growth and development with the aim to optimize growth and—ultimately—seed production in a competitive environment. Plants sense the intensity, color, direction and periodicity of light. Responses to these light parameters include seedling deetiolation (inhibition of hypocotyl elongation, opening of cotyledons and apical hook, greening), phototropism, shade avoidance, the accumulation of anthocyanins and the induction of flowering in particular day lengths [1].
To sense the light, plants have evolved several classes of photoreceptors [1,2]. The phytochrome photoreceptors sense red light (R) and far-red light (FR) and exist in two R/FR photointerconvertible conformations. Among the five phytochromes in Arabidopsis (phyA-phyE), the relatively light-stable phyB is the primary phytochrome controlling FR-reversible responses to R. These responses are also named low fluence responses (LFR). phyA is rapidly degraded in R and primarily mediates high-irradiance responses (HIR) to continuous FR (FRc) and very low fluence responses (VLFR) [3,4]. Blue light (B) is sensed by cryptochromes, phototropins and the ZEITLUPE family, but also by phyA. Cryptochromes are encoded by two genes in Arabidopsis, CRY1 and CRY2. Both mediate seedling deetiolation in B, while primarily cry2 is responsible for B-induced flowering in long days [5,6]. For both, phytochromes and cryptochromes, mutant photoreceptor variants have been identified that are constitutively active and thus signal also in darkness [7–10]. Recently, UVR8 was identified as the long-sought UV-B receptor [11,12].
In Arabidopsis, the phytochrome and cryptochrome photoreceptors act to inhibit a key repressor of light signaling that prevents light responses in darkness. This repressor, the CONSTITUTIVELY PHOTOMORPHOGENIC1/SUPPRESSOR OF PHYA-105 (COP1/SPA) complex, functions as an E3 ubiquitin ligase which ubiquitinates positively-acting light signaling intermediates, mainly transcription factors, thereby targeting them for proteolytic degradation in the 26S proteasome. In the light, photoreceptors directly interact with the COP1/SPA complex, leading to its inactivation which subsequently allows the target transcription factors to accumulate and to initiate vast reprogramming of gene expression [13,14]. The degradation of the light-labile photoreceptors phyA and cry2 is also in part dependent on COP1 and/or SPA genes [15–18]. The Arabidopsis COP1/SPA complex is likely a tetramer consisting of two COP1 and two SPA subunits [19]. COP1 is a single-copy gene in higher plants, while SPA proteins are encoded by a small gene family of four genes in Arabidopsis (SPA1-SPA4) and 2 genes in rice [13,20]. Mutations in either COP1 or all four SPA genes lead to constitutive photomorphogenesis in Arabidopsis, with seedlings showing the features of light-grown seedlings in complete darkness [21,22]. While cop1 null mutants arrest growth at the seedling stage, spa null mutants are viable. cop1 spa quintuple null mutants can complete embryogenesis, indicating that the COP1/SPA complex is not necessary for embryogenesis [23]. Apart from controlling seedling growth, the COP1/SPA complex also plays an important role during other light-induced responses, such as anthocyanin biosynthesis, elongation responses during shade avoidance, leaf expansion and the suppression of flowering under non-inductive short-day conditions. These responses are mediated through a number of COP1/SPA substrates including CO, HFR1, PAP1, PAP2 and BBX family proteins [24–32]. Moreover, COP1/SPA is a positive regulator in UV-B mediated photomorphogenesis [11,12]. The four SPA genes have overlapping but also distinct functions in controlling the various light responses during plant development [22,24–26,33].
The COP1/SPA complex acts as part of a CULLIN4 (CUL4)-based E3 ubiquitin ligase. CUL4-associated E3 ligases consist of CUL4, RBX1, DDB1 as well as a variable WD repeat protein which recognizes the substrate and binds DDB1 [34,35]. The WD repeat proteins COP1 and SPA are substrate adaptors in CUL4-DDB1COP1/SPA E3 ligase(s) [36]. Both COP1 and SPAs contain a central coiled-coil domain responsible for the formation of the COP1/SPA complex via homo - and heterodimerization [19,37,38]. In their C-termini, both COP1 and SPAs carry a WD-repeat domain which mediates interaction with substrates as well as with DDB1 [36,39]. The N-termini of COP1 and SPA are distinct, with COP1 harboring a RING finger domain and SPA proteins carrying a kinase-like domain [40,41].
Light is the key factor controlling COP1/SPA activity. Genetic studies showed that the SPA2 protein is particularly strongly inactivated by light when compared to the other three SPAs, making SPA2 a particularly interesting SPA when analyzing light-mediated inhibition of COP1/SPA activity [22,42]. How light inactivates the COP1/SPA complex is not fully understood. Evidence indicates that phytochrome and cryptochrome photoreceptors converge on COP1/SPA to promote light signaling in R, FR and B. Such light-induced inactivation of COP1/SPA occurs via multiple mechanisms. First, after light exposure, COP1 translocates from the nucleus into the cytoplasm [43,44]. Second, the B-dependent interaction of cry1 with SPA1 reduces the COP1/SPA1 interaction [45–47]. Similarly, an interaction of light-activated phytochromes A and B with members of the SPA family reduces the interaction between COP1 and SPA proteins [48,49]. For cry2, B acts to promote the interaction of cry2 with COP1 [50]. A third mechanism which reduces COP1/SPA activity in FRc-grown plants involves the degradation of SPA1 and SPA2 in the proteasome [42]. Here, we have analyzed the molecular mechanism of SPA2-degradation in different light qualities and uncover a photoreceptor-specific mechanism of light-induced COP1/SPA repression via COP1.
Results
SPA1 and SPA2 are degraded in far-red, red and blue light
To investigate the dynamics and wave-length dependency of light-induced SPA2 degradation, we determined SPA2 protein levels in dark-grown seedlings that were briefly exposed to R, FR or B. These seedlings expressed HA-tagged SPA2 under the control of the 5′ and 3′ regulatory sequences of SPA2 (SPA2::SPA2-HA) [42]. The SPA2 promoter expresses at the same level in dark-grown and light-exposed seedlings [42,51]. Therefore, light-induced differences in SPA2-HA protein levels in these lines are due to changes in protein stability, as shown previously [42]. Exposure of dark-grown seedlings to a short, 200-second pulse of R (Rp) was sufficient to strongly reduce SPA2-HA protein levels within 5 min after subsequent transfer to darkness (Fig 1A). Ten minutes after the Rp, there was barely any SPA2-HA protein detectable. Similarly, when dark-grown seedlings were irradiated with a pulse of FR (FRp) or B (Bp), SPA2-HA protein abundance decreased to a very low level. The response time to FRp and Bp was also very rapid, but slightly longer when compared to Rp.
Fig. 1. Light exposure rapidly reduces SPA2 protein levels. 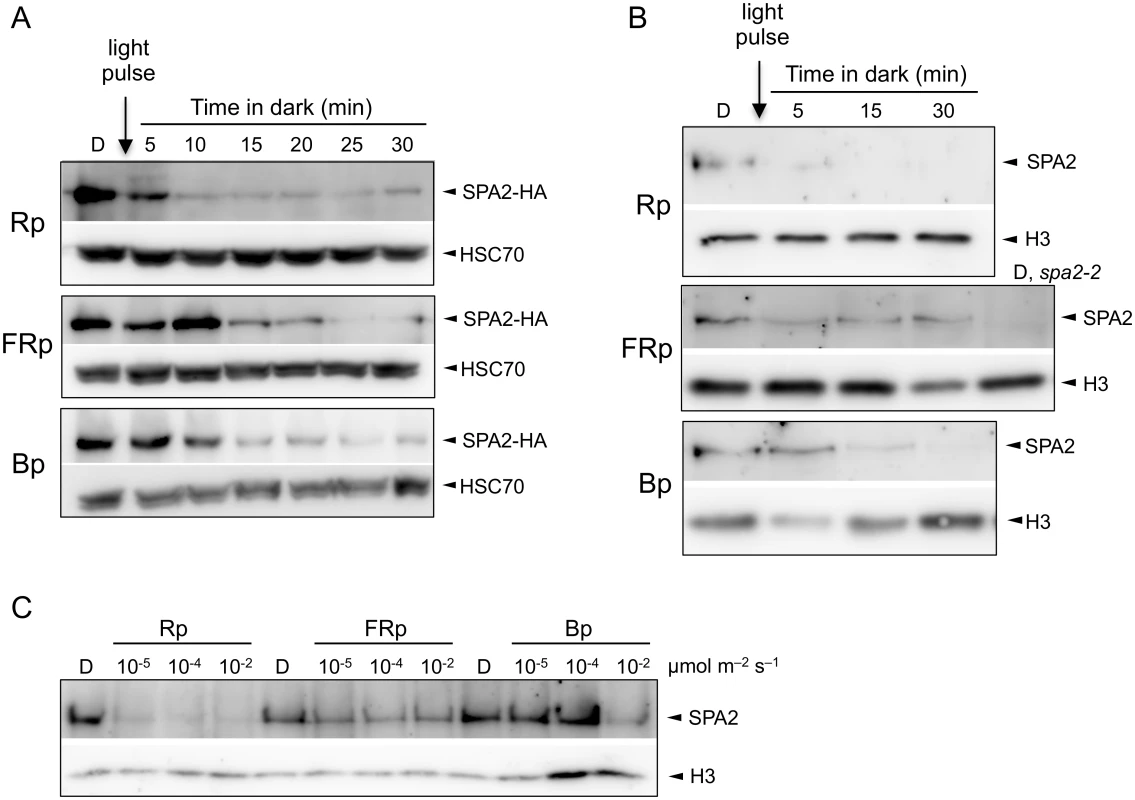
A. SPA2-HA protein levels in 4-day-old dark-grown (D) seedlings that were irradiated with a 200-s-pulse of 0.01 μmol m–2 s–1 FR (FRp), 0.01 μmol m–2 s–1 R (Rp) or 0.1 μmol m–2 s–1 B (Bp) and subsequently kept in the dark for the indicated time. Seedlings carried the SPA2::SPA2-HA transgene in a spa1 spa2 spa3 background. Proteins were detected using α-HA and α-HSC70 antibodies (loading control). B. SPA2 protein levels in 4-day-old dark-grown (D) wild-type seedlings that were irradiated with a 200-s-pulse of 0.01 μmol m–2 s–1 FR (FRp), 0.01 μmol m–2 s–1 R (Rp) or 0.2 μmol m–2 s–1 B (Bp) and subsequently kept in the dark for the indicated time. Proteins were detected in nuclear extracts using α-SPA2 antibodies. Histone H3 levels (H3) served as a loading control. C. SPA2 protein levels in wild-type seedlings that were transferred to a 200-s-pulse of R, FR or B of the indicated fluence rates and subsequently kept in darkness for 2 h. Proteins were detected in nuclear extracts using α-SPA2 antibodies. Histone H3 levels (H3) served as a loading control. To determine whether the native SPA2 protein behaves like the SPA2-HA protein, we analyzed SPA2 protein levels in wild-type seedlings using an α-SPA2 antibody. Because SPA2 levels are very low, we enriched the protein preparations through nuclear extracts to detect the constitutively nuclear-localized SPA2 protein [22,42]. Fig 1B shows that a pulse of FR, R or B rapidly and strongly reduced SPA2 protein abundance. Again, Rp was more effective in reducing SPA2 levels than FRp and Bp. We subsequently asked what fluences are necessary for the reduction of SPA2 protein levels. Fluences of 0.002 μmol m-2 of R, i.e. a 200-s-pulse of R with a fluence rate of 10−5 μmol m-2 s-1, was sufficient to reduce SPA2 protein levels to almost undetectable levels (Fig 1C), indicating that degradation in R is extremely sensitive to light and likely involves a VLFR. FRp and Bp were again less effective than Rp (Fig 1C).
We subsequently asked whether R, FR and B also cause a decrease in SPA1 levels. Here, the induction of SPA1 gene expression by light [40] precluded a specific analysis of protein stability using α-SPA1 antibodies. Therefore, we used transgenic lines expressing SPA1-HA under the control of the constitutive SPA2 promoter. These lines showed a strong reduction in SPA1-HA abundance in FR, R and B (Fig 2).
Fig. 2. Light exposure reduces SPA1 protein levels. 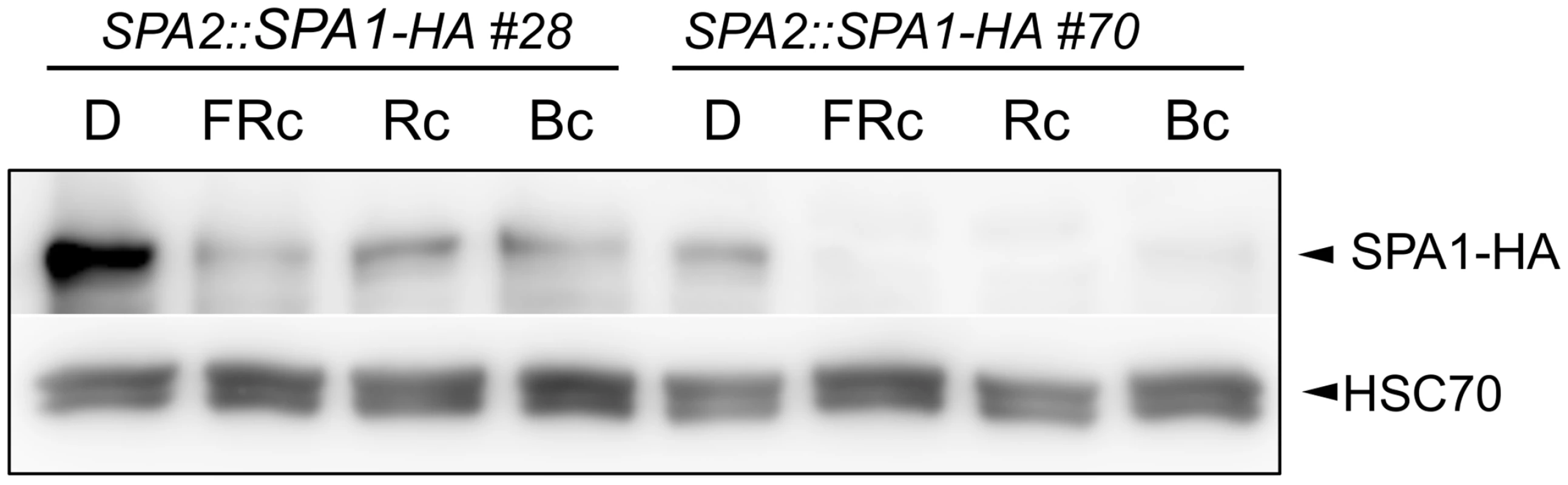
SPA1-HA protein levels in transgenic seedlings expressing SPA1-HA under the control of the native SPA2 promoter. Seedlings were grown in darkness (D) for 4 days and subsequently transferred to FRc (5 μmol m–2 s–1), Rc (10 μmol m–2 s–1) or Bc (50 μmol m–2 s–1) for 6 h. SPA1-HA was detected using an α–HA antibody. HSC70 levels served as a loading control. Rapid SPA2 degradation in R, FR and B is exclusively mediated by phytochromes
We asked which photoreceptor(s) are responsible for degradation of SPA2 in different light qualities and quantities. To this end we investigated SPA2 levels in various photoreceptor mutants. Degradation of SPA2 in response to FRc was fully abolished in a phyA mutant, in both Col and RLD accessions (Fig 3A). Similarly, a pulse of FR had no effect on SPA2 protein levels in a phyA mutant (Fig 3B). Hence, phyA is responsible for SPA2 degradation in FR.
Fig. 3. SPA2 degradation in FR and R requires phytochromes and is R/FR reversible. 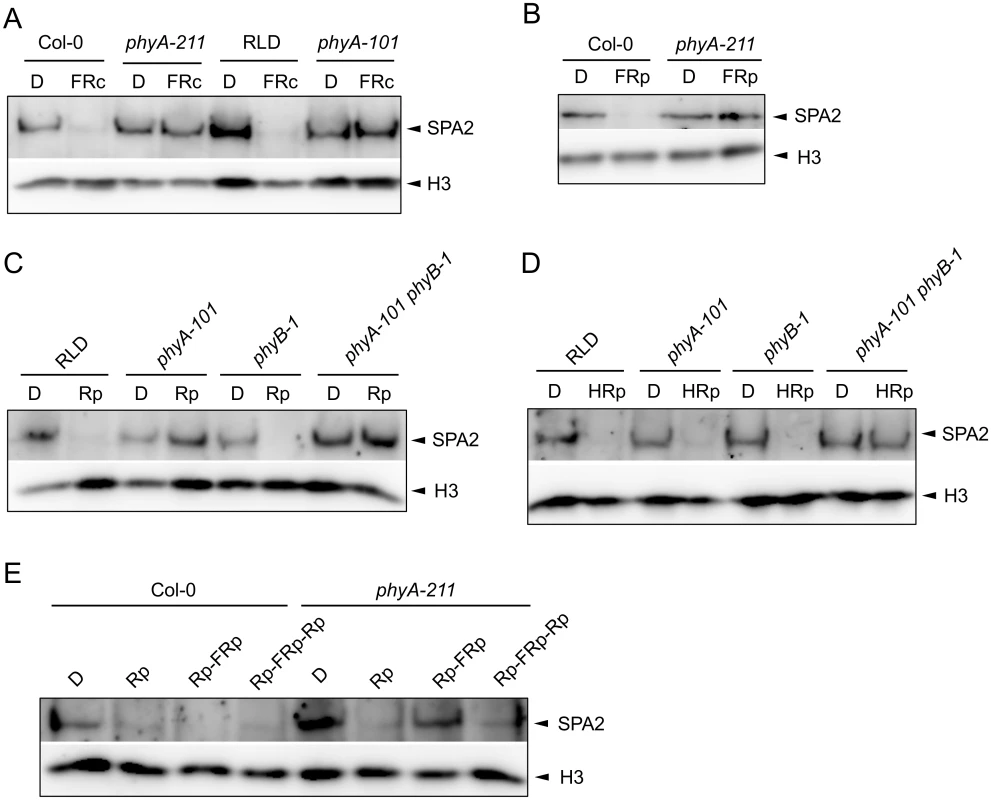
A. SPA2 protein levels in 4-day-old dark-grown wild-type and phyA mutant seedlings that were transferred to FRc (0.1 μmol m–2 s–1) for 30 min. phyA-211 is in Col and phyA-101 in RLD accession. B. SPA2 protein levels in 4-day-old dark-grown wild-type Col-0 and phyA-211 mutant seedlings that were irradiated with a 200-s-pulse of 5 μmol m–2 s–1 FR (FRp) and subsequently kept in the dark for 30 min. C, D. SPA2 protein levels in the 4-day-old dark-grown seedlings of wild-type RLD, phyA-101, phyB-1 and phyA-101 phyB-1 mutants that were irradiated with a 200-s-pulse of 0.01 μmol m–2 s–1 R (Rp)(C) or 10 μmol m–2 s–1 R (HRp)(D) and subsequently kept in the dark for 30 min. E. SPA2 protein levels in wild-type Col-0 and phyA-211 mutant seedlings that were grown in darkness for 4 days and subsequently irradiated with a 200-s-pulse of 10 μmol m–2 s–1 R (Rp), a 200-s-pulse of 10 μmol m–2 s–1 R followed by a 200-s-pulse of 5 μmol m–2 s–1 FR (Rp-FRp) or a 200-s-pulse of 10 μmol m–2 s–1 R in addition to Rp-FRp (Rp-FRp-Rp). After irradiation, seedlings were kept in the dark for 30 min. SPA2 levels were detected in nuclear extracts using an α-SPA2 antibody. Histone H3 levels (H3) served as a loading control. After a pulse of R with low fluence rates, SPA2 protein levels were not reduced in a phyA mutant nor in a phyA phyB double mutant. A phyB mutant, in contrast, showed a reduction in SPA2 levels after Rp and thus exhibited a similar response as the wild type (Fig 3C). The phyA-requirement for a response to low Rp confirms that this treatment initiates a phyA-perceived VLFR [3]. After a pulse with higher fluence rates of R, only the phyA phyB double mutant lacked a reduction in SPA2 protein abundance when compared to dark-grown seedlings (Fig 3D). Hence, phyA and phyB mediate the degradation of SPA2 after high Rp. This suggests that both VLFR and LFR responses trigger SPA2 degradation in red light. R/FR reversibility is a hall-mark of an LFR [4]. Indeed, SPA2 degradation after Rp was reversible by a pulse of FR in a phyA mutant background which would lack the VLFR (Fig 3E).
Deetiolation in blue light is mediated by the cryptochromes cry1 and cry2 as well as by phyA. We therefore investigated B-induced degradation of SPA2 in cry1 cry2 and in phyA mutants. After a pulse of B, the decrease in SPA2 levels was abolished in phyA mutant seedlings but was normal in the cry1 cry2 mutant (Fig 4A). These results indicate that a pulse of B only triggered phyA-mediated SPA2 degradation. Also after irradiation with continuous B of very high fluence rates (50 μmol m-2 s-1) for 30 min high SPA2 levels were retained in phyA mutant seedlings. In cry1 cry2 mutant seedlings, SPA2 levels were again strongly reduced similar to wild-type seedlings (Fig 4B). Only after prolonged irradiation with B of high fluence rates for 24 h, SPA2 levels decreased in a phyA-deficient mutant (Fig 4C). These results show that the rapid B-induced reduction in SPA2 levels is exclusively mediated by phyA. Only after very long irradiation with B of high fluence rates other photoreceptor(s) become active in reducing SPA2 levels.
Fig. 4. Degradation of SPA2 in B primarily requires phyA. 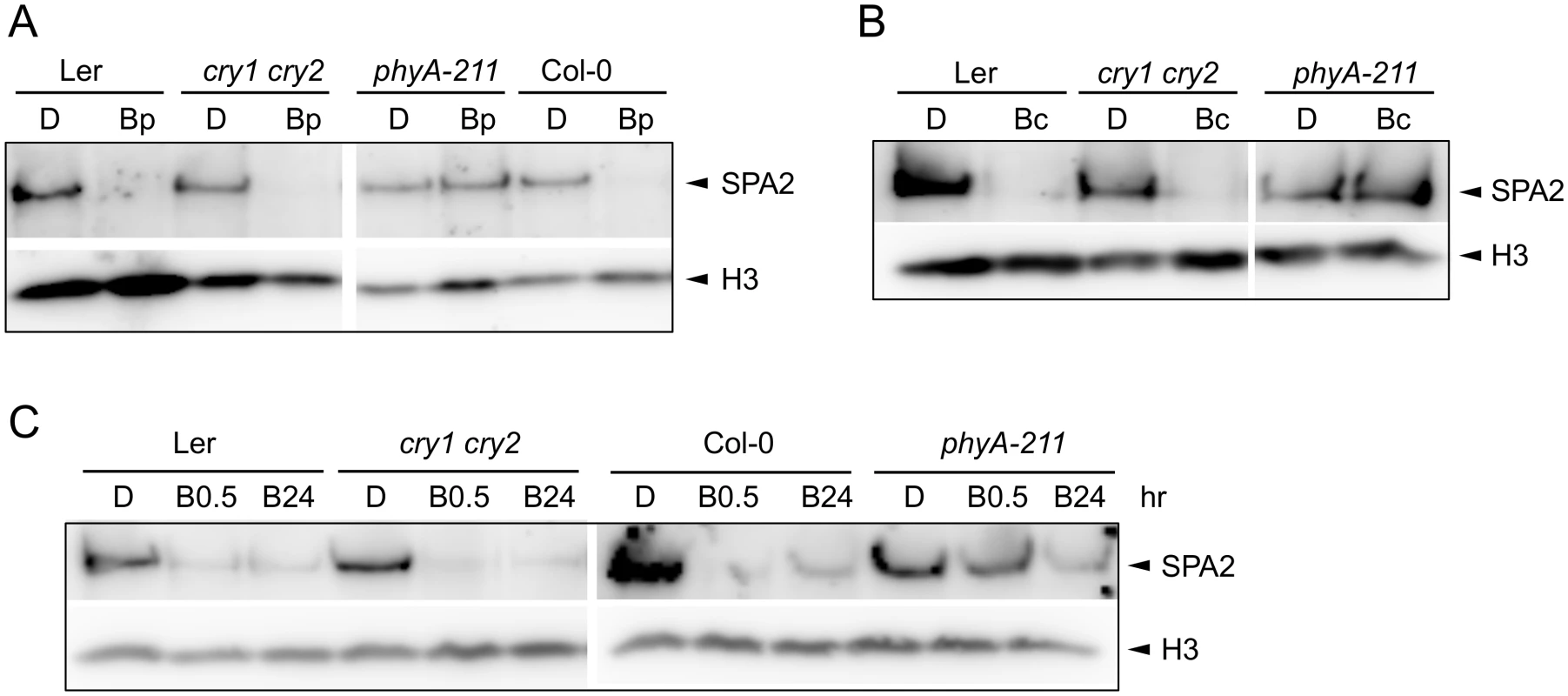
A-C. SPA2 protein levels in 4-day-old dark-grown (D) seedlings of the indicated genotypes that were (A) irradiated with a 200-s-pulse of 0.2 μmol m–2 s–1 B (Bp) and subsequently kept in the dark for 30 min, (B) transferred to Bc (50 μmol m–2 s–1) for 30 min or (C) transferred to 20 μmol m–2 s–1 Bc for 0.5 h (B0.5) or 24 h (B24). SPA2 levels were detected in nuclear extracts using an α-SPA2 antibody. Histone H3 levels (H3) were used as a loading control. In an attempt to uncover a possible role of cryptochromes in the response to long-term B irradiation, we analyzed SPA2 protein levels in a cry1 cry2 phyA-201 triple mutant background (Ler accession). SPA2 protein levels still decreased in this mutant after prolonged exposure to blue light (S1A Fig). However, SPA2 levels in phyA-201 also decreased after FRc (S1B Fig). Hence, degradation of SPA2 in the cry1 cry2 phyA-201 triple mutant may either be due to residual phyA activity or, alternatively, the regulation of SPA2 stability may be different in the Ler accession than in the Col and RLD accessions.
SPA2 levels in transgenic lines expressing constitutively active photoreceptors
Constitutively active photoreceptor variants have been described that initiate light signaling even in darkness. We therefore investigated whether these photoreceptor variants also cause a constitutive reduction in SPA2 protein abundance, i.e. also in dark-grown seedlings. To this end, we analyzed SPA2 protein levels in transgenic lines expressing the constitutively active phytochrome mutants phyBY276H and phyAY242H [7]. As reported previously, phyBY276H-expressing seedlings showed very strong constitutive photomorphogenesis, both in the PHYB wild-type and the phyB-5 mutant background [7] (Fig 5A). These phyBY276H lines showed very low SPA2 protein levels in dark-grown seedlings (Fig 5B), suggesting that the SPA2 protein is destabilized in darkness by the constitutively active phyB photoreceptor.
Fig. 5. SPA2 protein levels in transgenic seedlings expressing constitutively active photoreceptors. 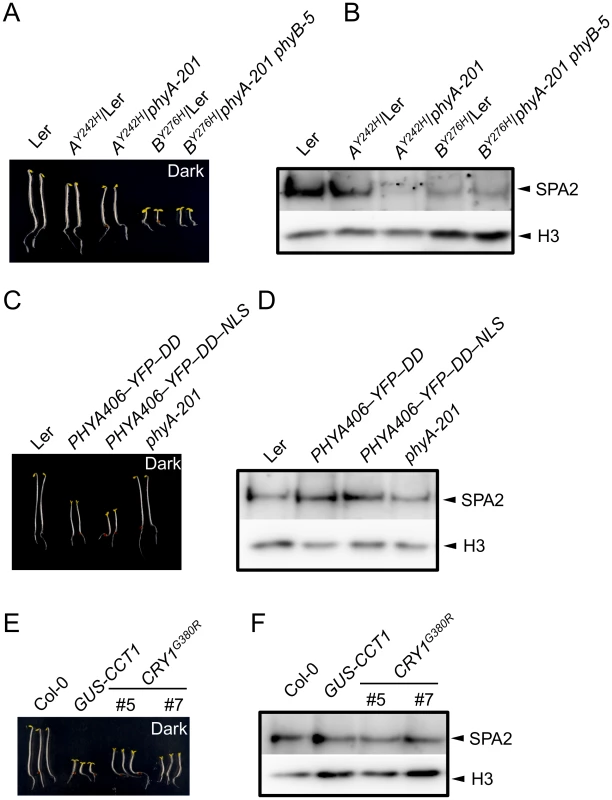
A, C, E. Phenotype of 4-day-old dark-grown seedlings of the indicated genotypes. Both PHYA406 constructs were in the phyA-201 background. B, D, F. Immunodetection of SPA2 protein levels in 4-day-old dark-grown seedlings. SPA2 was detected in nuclear extracts using α-SPA2 antibodies. Histone H3 levels (H3) served as a loading control. phyAY242H-expressing seedlings exhibit weaker constitutive photomorphogenesis than phyBY276H-expressing seedlings [7]. These seedlings had a shorter hypocotyl and a partially opened hook, especially in the phyA mutant background, when compared to the wild type (Fig 5A). This phenotype was somewhat weaker than reported previously which is likely due to the younger age of our seedlings and the absence of sucrose in the culture medium when compared to [7]. SPA2 abundance in dark-grown seedlings was strongly reduced in lines expressing phyAY242H in a phyA background, while it was similar to wild type in lines expressing phyAY242H in a PHYA wild-type background (phyAY242H/Ler) (Fig 5B). Hence, SPA2 levels were constitutively reduced in the presence of phyAY242H, but this effect is outcompeted by the presence of wild-type phyA. In summary, these mutations in both phyA and phyB cause constitutive degradation of SPA2 in darkness.
Expression of the N-terminal 406 amino acids of phyA fused to an artificial dimerization domain has also been shown to cause constitutive photomorphogenesis in darkness [10] (Fig 5C). However, this phyA variant did not alter SPA2 protein levels in darkness (Fig 5D), indicating that this constitutively active phyA variant was not capable of inducing SPA2 degradation in darkness.
Fusion of the cry1 C-terminal extension (CCT1) to an artificial dimerization domain (GUS) leads to a constitutively active cry1 photoreceptor. Similarly, a cry1G380R variant is constitutively active. Hence, seedlings expressing CCT1 or cry1G380R exhibit strong constitutive photomorphogenesis in darkness [8,9]. SPA2 protein levels were unaltered in dark-grown GUS-CCT1 - and cry1G380R-expressing seedlings when compared to the wild type, despite the constitutive photomorphogenesis displayed by these seedlings (Fig 5E and 5F). Hence, none of the constitutively active cry1 variants affected SPA2 protein levels in darkness. This is in agreement with the primary roles of phytochromes in SPA2 degradation.
B-induced reduction in SPA2 function in spa1 spa3 spa4 mutants strongly depends on phyA
Since rapid degradation of SPA2 in B was exclusively dependent on phyA, we predicted that phyA is of particular importance in inactivating SPA2 function in B. To test this hypothesis, we generated a phyA-deficient spa1 spa3 spa4 phyA mutant which only expresses functional SPA2 among the four SPA proteins. Hence, we can observe the effect of light on SPA2 activity in the absence of any other SPAs, and in the presence or absence of phyA. We had shown previously that spa1 spa3 spa4 mutant seedlings etiolate normally in darkness but are very hypersensitive to R, FR and B when compared to the wild type, thus resembling a spa quadruple mutant already at extremely low fluence rates of light [22,42] (Fig 6A–6C). Hence, SPA2 is sufficient for full repression of photomorphogenesis in darkness but is extremely effectively inactivated by light. In B, spa1 spa3 spa4 phyA mutant seedlings displayed much longer hypocotyls than spa1 spa3 spa4 mutant seedlings, indicating that the lack of phyA dramatically reduced the responsiveness of the spa1 spa3 spa4 mutant to B. The hypocotyl length of the spa1 spa3 spa4 phyA mutant in B was very similar to that of the phyA single mutant. Hence, in the absence of phyA, the mutations in SPA1, SPA3 and SPA4 had no detectable effect (Fig 6A). In Rc, the phyA mutation abolished the hypersensitivity of the spa1 spa3 spa4 mutant to lower fluence rates of Rc but not to higher fluence rates of Rc (Fig 6B). This is consistent with our finding that SPA2 degradation in lower fluence rates of R requires phyA, while in higher fluence rates of R phyB in addition to phyA mediates SPA2 degradation. As expected, the responsiveness of spa1 spa3 spa4 mutant seedlings to FRc was fully dependent on phyA (Fig 6C). Taken together, these results show that the hypersensitivity of the spa1 spa3 spa4 mutant to B fully depends on phyA. This agrees with our observation that rapid SPA2 degradation in B was exclusively dependent on phyA. Since the spa1 spa3 spa4 phyA mutant retained responsiveness to B, as indicated by the inhibition of hypocotyl elongation in B of higher fluence rates, additional phyA-independent mechanisms of SPA2 inactivation by B exist. These are likely mediated by the cryptochromes.
Fig. 6. The phyA-211 mutation abolishes the hypersensitivity of the spa1 spa3 spa4 mutant to B and FR but not to R. 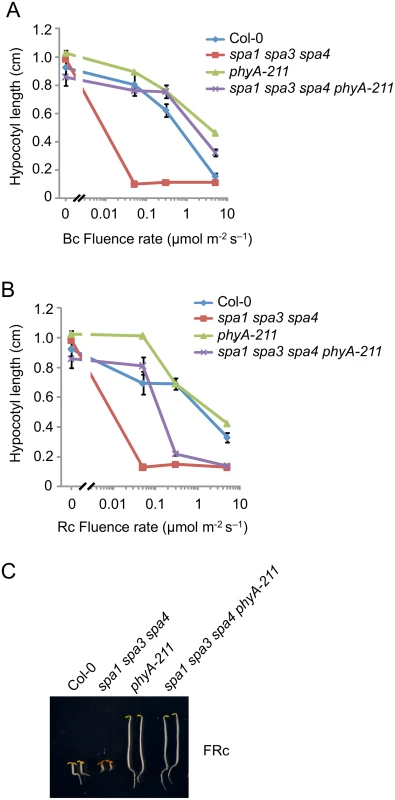
A, B. Hypocotyl length of seedlings of the indicated genotypes grown in various fluence rates of Bc (A) and Rc (B) for 4 days. Error bars represent the SEM. C. Visual phenotype of seedlings of the indicated genotypes grown in FRc (2 μmol m–2 s–1) for 4 days. SPA2 associates with cry1 but not with cry2 in B-treated seedlings
The lack of cryptochrome activity in B-induced SPA2 degradation might be caused by a failure of cryptochromes to rapidly interact with SPA2. Indeed, FRET/FLIM studies in transfected tobacco leaves failed to show an interaction between cry2 and SPA2. Similarly, recombinantly produced cry2 and SPA2 did not interact in in vitro pull-down assays [15]. On the contrary, SPA2 was shown to weakly interact with cry2 in B in the yeast two-hybrid system [50]. For cry1, no significant interaction with SPA2 was observed in the yeast two-hybrid assay [46]. To reinvestigate this question in planta, we conducted co-immunoprecipitation experiments using transgenic Arabidopsis seedlings expressing SPA2-HA and, as a positive control, SPA1-HA (Fig 7). To obtain similar protein levels of SPA1-HA and SPA2-HA in B, SPA1-HA was expressed under the control of the weaker SPA2 promoter (SPA2::SPA1-HA) and SPA2-HA from the stronger SPA1 promoter (SPA1::SPA2-HA). Moreover, seedlings were treated with proteasome inhibitor to reduce SPA degradation in B. Fig 7A shows that upon B-exposure both SPA1-HA and SPA2-HA co-immunoprecipitated higher-mobility cry1 isoforms which are formed in B. Hence, B induced the formation of a SPA2/cry1 complex, as it was previously reported for a SPA1/cry1 complex [46,47]. In addition, a lower-mobility cry1 which likely represents the non-phosphorylated isoform of cry1 showed weak constitutive interactions with SPA1-HA and SPA2-HA in B and darkness. The association of higher-mobility cry1 with SPA2 was very rapid. It occurred within 5 min of B-exposure (S2 Fig). cry2, in contrast, was not co-immunoprecipitated by SPA2-HA, neither in darkness nor in B. The positive control SPA1-HA showed the expected B-dependent association with cry2 (Fig 7B). In summary, in B, SPA2 associates with cry1 but not with cry2 in planta.
Fig. 7. SPA2 associates with cry1 in B but not with cry2. 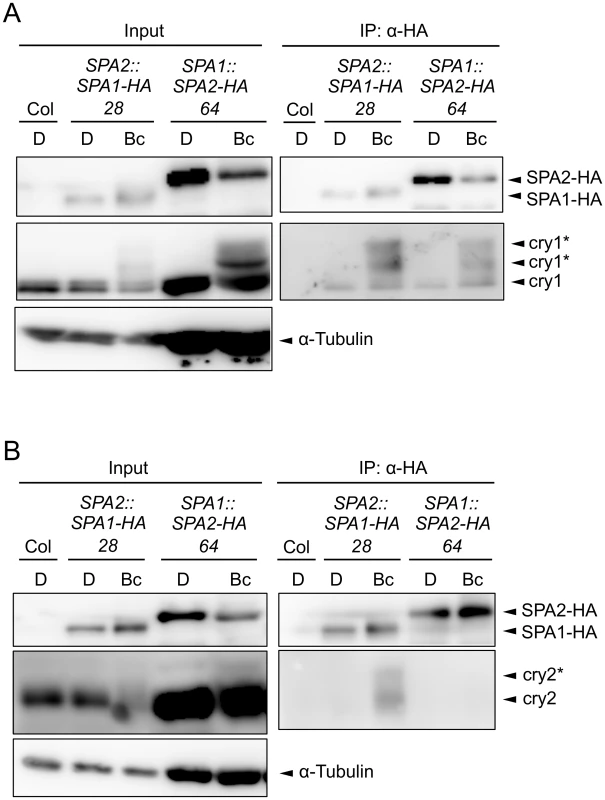
A, B. Co-immunoprecipitation of cry1 (A) and cry2 (B) by SPA1-HA and SPA2-HA. 4-day-old dark-grown seedlings (D) expressing SPA1-HA or SPA2-HA were transferred to 50 μmol m–2 s–1 Bc for 1 h (A) or 5 min (B). SPA1-HA and SPA2-HA proteins were immunoprecipitated using α-HA beads. To obtain similar protein levels of SPA1-HA and SPA2-HA in Bc, SPA1-HA was expressed under the control of the weaker SPA2 promotor (SPA2::SPA1-HA) and SPA2-HA from the stronger SPA1 promoter (SPA1::SPA2-HA). Moreover, seedlings were treated with proteasome inhibitor to prevent SPA degradation in Bc, and five times more protein extract was used for the SPA2-HA immunoprecipitation than for the SPA1-HA immunoprecipitation. α-HA antibody was used to detect SPA-HA proteins. α-cry1 and α-cry2 antibodies were used to detected cry1 and cry2, respectively. α-tubulin levels were used as loading control for the input. Asterisks likely indicate phosphorylated cry1 and cry2. COP1 is necessary for the light-induced degradation of SPA2
To identify the E3 ubiquitin ligase that mediates SPA2 degradation in the light, we asked whether the COP1/SPA E3 ligase itself may be responsible for ubiquitination of SPA2. We therefore investigated SPA2 protein levels in the hypomorphic cop1-4 mutant and in the cop1-5 null mutant, using light conditions that cause full degradation of SPA2. In a cop1-4 hypomorphic background, considerable SPA2 protein levels were retained in seedlings irradiated with FRc (Fig 8A and 8B). Hence, the FRc-induced reduction in SPA2 protein abundance was strongly attenuated, but not abolished, by the partial-loss-of-function cop1-4 mutation. We subsequently analyzed SPA2 protein levels in the cop1-5 null mutant. Because cop1 null mutants arrest growth at the very early seedling stage and, moreover, mostly fail to break the seed coat during germination, we could not obtain enough tissue for nuclear-enriched protein preparations which are necessary to detect the native SPA2 protein with α-SPA2 antibodies. We therefore crossed the SPA2::SPA2-HA transgene into a cop1-5 mutant background and detected the SPA2-HA protein using α-HA antibodies. This transgene-encoded SPA2-HA fully mimics function and behavior of the native SPA2 protein [42] (this study). As shown in Fig 8C and asreported above, SPA2-HA protein levels in the progenitor SPA2::SPA2-HA line decreased to almost undetectable levels upon irradiation with Rc. In a homozygous cop1-5 mutant background, in contrast, SPA2-HA levels were not reduced in Rc when compared to darkness. As an additional control, we also determined SPA2-HA protein levels in COP1 wild-type siblings that segregated in a progeny derived from the cross of cop1-5 with the SPA2::SPA2-HA line. In these siblings, SPA2-HA protein levels decreased upon Rc irradiation as in the progenitor SPA2::SPA2-HA line. Hence, the Rc-induced reduction in SPA2-HA protein abundance was fully dependent on COP1.
Fig. 8. COP1 is required for SPA2 degradation in the light. 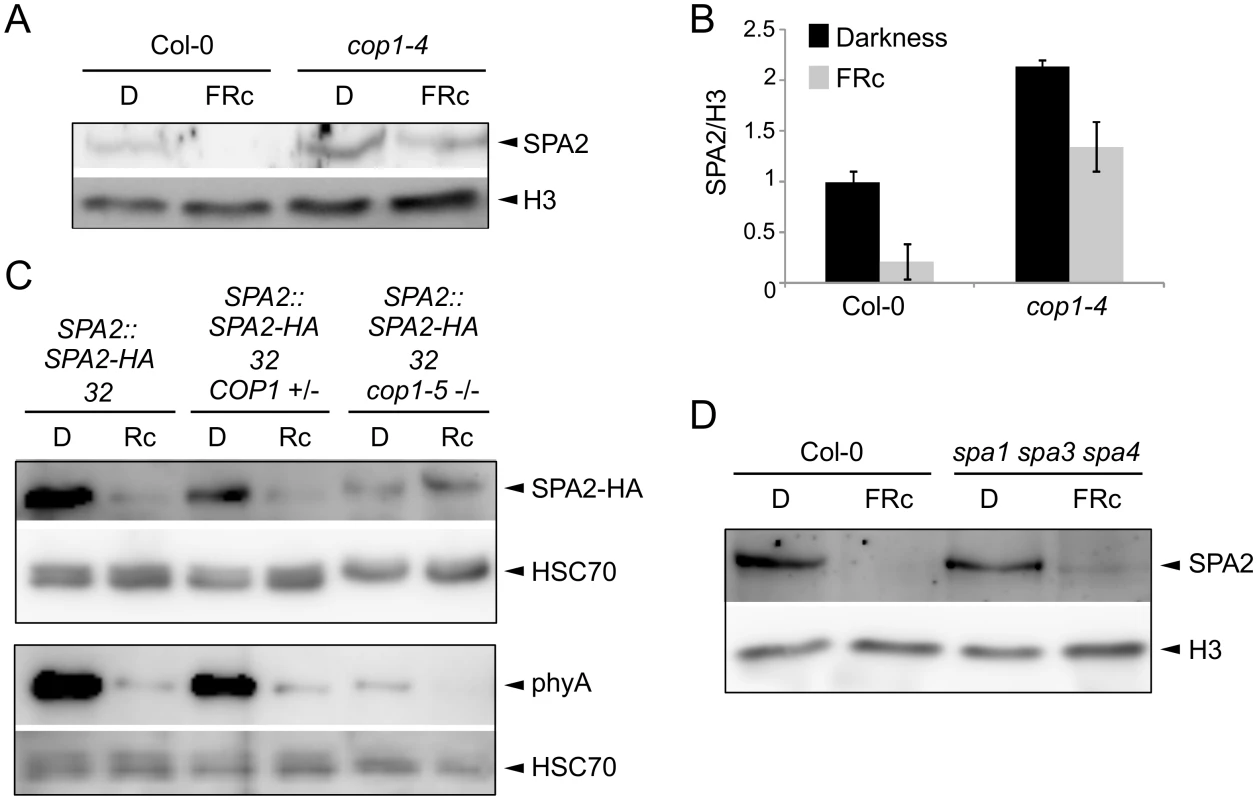
A, B. Immunodetection (A) and quantification (B) of SPA2 protein levels in 4-day-old wild-type Col-0 and cop1-4 mutant seedlings grown in darkness (D) or in FRc (0.35 μmol m–2 s–1). The error bars represent the SEM of two biological replicates. C. SPA2-HA protein levels in the cop1-5 null mutant background. The SPA2::SPA2-HA 32 transgene was crossed into a cop1-5 mutant background. SPA2::SPA2-HA 32 cop1-5 (-/-) represents homozygous cop1-5 mutants selected from a segregating population based on the black seed phenotype. SPA2::SPA2-HA 32 COP1 (+/-) represents homozygous COP1 wild-type or heterozygous COP1 cop1-5 siblings of the same segregating population. Seeds were incubated in darkness for 1 day and subsequently transferred to Rc (40 μmol m–2 s–1) for 6 h or kept in darkness. D. Immunodetection of SPA2 protein levels in 4-day-old wild-type Col-0 and spa1-100 spa3-1 spa4-3 mutant seedlings grown in darkness (D) or FRc (0.35 μmol m–2 s–1). Native SPA2 was detected in nuclear extracts using an α-SPA2 antibody. SPA2-HA and phyA were detected using an α-HA and an α-phyA antibody, respectively. Histone H3 levels (H3) or HSC70 levels were used as loading controls. Because the cop1 null mutations severely affect seedling growth and cause growth arrest, we wished to exclude the possibility that premature lethality is an indirect reason for the lack of SPA2 degradation in cop1-5. To do so, we made use of previous findings showing that degradation of phyA-Pfr is in part COP1-independent [16,18] and thus should occur in cop1-5. Indeed, phyA levels strongly decreased upon Rc irradiation (Fig 8C). Hence, the cop1-5 tissue used was clearly still capable of light perception and light response. The analysis of phyA abundance indicated that phyA levels were considerably lower in cop1-5 than in the wild type in both dark-grown and light-exposed tissues. The reasons for this are unknown. It may relate to the low efficiency of protein extraction using cop1-5 when compared to using the wild type. Hence, normalization to HSC70 levels may be unreliable. Consistent with this idea, SPA2-HA levels were also unexpectedly lower in cop1-5 than in the wild type.
Since the four SPA proteins heterodimerize in the tetrameric COP1/SPA complex [19], we asked whether the presence of SPA1, SPA3 and SPA4 affects SPA2 protein levels in the light. The light-induced reduction in SPA2 protein levels was also dramatic in the spa1 spa3 spa4 mutant, but slightly higher SPA2 protein levels consistently remained in FRc in spa1 spa3 spa4 when compared to the wild type (Fig 8D). Hence, the COP1/SPA2 complex which forms in the spa1 spa3 spa4 mutant is sufficient to allow SPA2 degradation in the light. Whether SPA2 is required for its own degradation cannot be determined from the presented experiments. The finding that the other three SPAs slightly increase SPA2 degradation in FRc hints at the possibility that SPA2 is involved in its own degradation.
The COP1-interacting coiled-coil domain of SPA2 is necessary for SPA2 degradation
Our finding that COP1 is required for SPA2 degradation in the light suggests that SPA2 is directly ubiquitinated by the COP1 or COP1/SPA2 ubiquitin ligase. If so, it is expected that interaction of SPA2 with COP1 is necessary for SPA2 degradation to occur. To test this hypothesis, we expressed a SPA2 deletion derivative that lacks the COP1-interacting coiled-coil domain under the control of the native SPA2 promoter (SPA2::ΔCC SPA2-HA; Fig 9A). Indeed, the ΔCC SPA2-HA protein failed to co-immunoprecipitate COP1 in extracts of transgenic plants, confirming that ΔCC SPA2-HA does not incorporate into a COP1/SPA complex (Fig 9C). Consistent with this finding, the ΔCC SPA2-HA transgene did not complement the spa1 spa2 spa3 mutant phenotype, whereas the full-length SPA2-HA transgene did (S3 Fig). ΔCC SPA2-HA protein abundance did not change in response to light (Fig 9B). The levels of full-length SPA2-HA, in contrast, decreased to undetectable levels in FRc. This difference in the behavior of the SPA2-HA and ΔCC SPA2-HA proteins is not due to any differences in SPA2-HA and ΔCC SPA2-HA transcript levels because transcript levels were not regulated by light, as expected for a gene expressed from the SPA2 promoter (S4 Fig). These results show that the COP1-interacting coiled-coil domain of SPA2 is necessary for SPA2 degradation in the light.
Fig. 9. The coiled-coil domain of SPA2 is required for the interaction of SPA2 with COP1 and for SPA2 degradation in the light. 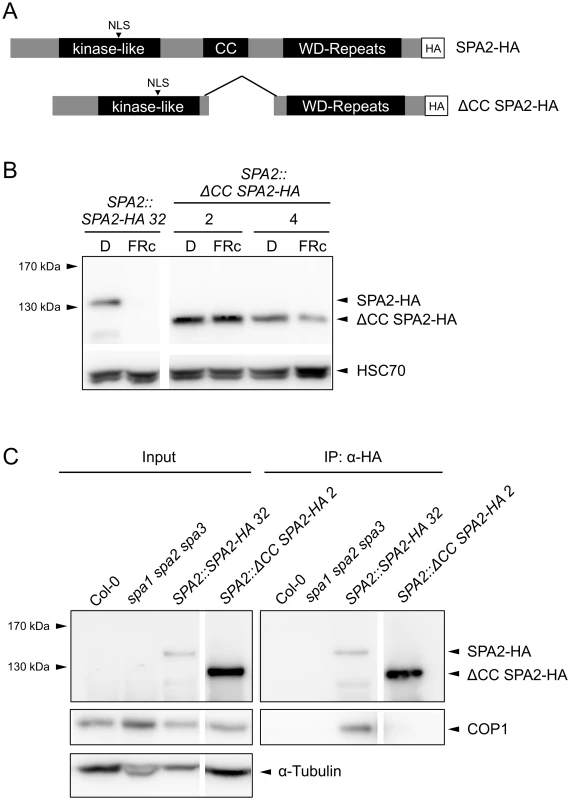
A. Schematic representation of the full-length SPA2-HA and the ΔCC SPA2-HA proteins. All proteins were expressed in transgenic spa1 spa2 spa3 mutant plants under the control of the native SPA2 promoter. B. SPA2-HA and ΔCC SPA2-HA protein levels in transgenic spa1 spa2 spa3 mutant seedlings grown in darkness or FRc (5 μmol m–2 s–1) for 4 days. SPA-HA proteins were detected using an α–HA antibody. HSC70 levels were used as a loading control. Line numbers represent independent transgenic lines. C. ΔCC SPA2-HA does not interact with COP1. SPA2-HA and ΔCC SPA2-HA were immunoprecipitated by α–HA beads. Wild-type Col-0 and the spa1 spa2 spa3 mutant were used as negative controls. SPA-HA proteins were detected by an α–HA antibody. COP1 proteins were detected by an α-COP1 antibody. α-tubulin levels were used as a loading control for the input fractions. Line numbers represent independent transgenic lines. Discussion
The four SPA proteins are components of the COP1/SPA E3 ubiquitin ligase and have redundant but also distinct functions in regulating plant growth and development in response to the light environment. The phenotypic analysis of spa mutants showed that SPA2, among the four SPA proteins, exhibits the greatest difference in activity between dark - and light-grown seedlings and is therefore a particularly interesting SPA protein when investigating light-induced inactivation of COP1/SPA activity [22,42]. Here, we have analyzed the molecular mechanism of SPA2 degradation in different light qualities and have uncovered a photoreceptor-specific mechanism of light-induced COP1/SPA repression via COP1.
Our results demonstrate that the SPA2 protein is degraded very rapidly, i.e. within 5–15 min after dark-grown seedlings were exposed to a brief pulse of R, FR or B. Since COP1 function depends on SPA proteins, this rapid, light-induced degradation of SPA2 provides a very effective mechanism to inactivate COP1/SPA2 activity in light-grown plants. We and others have shown previously that COP1 levels do not significantly change in response to R, FR or B [41,42]. Hence, light does not affect the stability of the whole COP1/SPA2 complex but only that of SPA2. This shows that the presence of SPA2 in the COP1/SPA2 E3 ubiquitin ligase provides a means for light-induced inactivation of the COP1/SPA2 complex. Though both phytochrome and cryptochrome photoreceptors inactivate COP1/SPA function in the respective light qualities [14], we found that the rapid degradation of SPA2 specifically required phytochromes not only in R and FR, but also in B. Thus, this mechanism of rapid COP1/SPA2 inactivation is specific to phytochrome action. In summary, our analysis shows that a photoreceptor-specific mechanism of COP1/SPA2 inactivation developed during evolution. Evidence indicates that multiple mechanisms have evolved that inactivate COP1/SPA function in the light. Another mechanism of inactivation was found to be common to phytochromes and cry1 since phyA, phyB and cry1 induce a dissociation of COP1 from SPA1 in R or B, respectively [46–49]. A third mechanism, the light-induced exclusion of COP1 from the nucleus also occurs in R, FR and B and is primarily mediated by phyA, phyB and cry1 in FR, R and B, respectively [52]. On the other hand, B-control of COP1 nuclear abundance was found to also require biosynthesis of the phytochrome chromophore [53], suggesting an essential role of phytochromes also in B. In total, evidence indicates that photoreceptor-specific mechanisms and common mechanisms induced by both phy and cry photoreceptors co-act to allow an appropriate response to a changing light environment.
Our results show that rapid SPA2 degradation in R involves a phyA-dependent VLFR and a phyB-dependent LFR which is also reversible by FR. In FR, SPA2 degradation was fully dependent on phyA. This demonstrates that the responsiveness of the SPA2 protein to R and FR directly correlates with our current knowledge on phyA and phyB activities in R and FR [3,4] and thus appears to be an immediate output of light-induced phytochrome action. Previous findings showing that SPA2 directly interacts with phyA and phyB [49] are in good agreement with this conclusion. phyA is also a well-known B-photoreceptor that together with cry1 and cry2 is responsible for seedling deetiolation in B [4]. The particular biological significance of phyA in B-induced repression of SPA2 function is supported by our finding that the extreme hypersensitivity to B in spa1 spa3 spa4 triple mutants which only have functional SPA2 was indeed fully dependent on phyA. We therefore suggest that light inactivates COP1/SPA2 function in B primarily through rapid, phyA-induced degradation of SPA2. Residual SPA2 protein that escapes degradation may be inactivated by additional mechanisms, such as cry1-mediated dissociation from COP1, as it has been described for SPA1 [46,47], and phyA-mediated dissociation from COP1 [49]. The latter, however, has not been analyzed in B so far.
Since the SPA1 protein is also degraded in R, FR and B, albeit with lower efficiency than SPA2, a SPA1-containing COP1 complex may also be inactivated through phytochrome-mediated degradation of SPA1, i.e. via the same or a very similar mechanism as the light-induced degradation of SPA2. Interestingly, the mutant phenotypes of spa single mutant seedlings defective in SPA1, SPA3 or SPA4 are also fully dependent on phyA, even in R. These single mutants etiolate normally in darkness, but exhibit hypersensitivity in the light in a PHYA wild-type background only [33,54,55]. The mechanistic reason for this observation has so far remained unknown but could be explained by a phyA-mediated de-stabilization of these SPA proteins in light-grown seedlings. Hence, a stabilization of SPA1, SPA3 and SPA4 in a phyA mutant background might lead to the complete rescue of the spa single mutant phenotypes.
The failure of other B receptors than phyA, such as cryptochromes, to cause rapid degradation of SPA2 in B is not due to a general lack of SPA2-cry interactions in vivo. However, our results demonstrate that SPA2 only associates with cry1 and not with cry2 in B-treated seedlings. Hence, the lack of a cry2-SPA2 interaction is likely in part responsible for the observed stability of SPA2 in B-treated phyA mutant seedlings. On the other hand, our results also show that SPA2 rapidly interacts with cry1 in B without causing rapid SPA2 degradation. Based on this finding we conclude that the failure of cry1 to cause rapid degradation of SPA2 is not due to a lack of a SPA2-cry1 interaction, especially since the SPA2-cry1 interaction is observed rapidly in vivo, i.e. within 5 min of B irradiation. Thus, cry1 interacting with SPA2 in B does not induce rapid degradation of SPA2; cry1 action thereby strongly differs from phytochrome actions on the SPA2 protein.
In contrast to SPA2 which only interacted with cry1 in our in vivo co-immunoprecipitation experiments, SPA1 interacted with both cryptochromes, as shown previously [46,47,50]. Hence, SPA1 and SPA2 clearly differ in their interaction capacity with cry2. cry2 was shown to interact with the N-terminal domain of SPA1 [50]. Though we do not know the cry2-interacting domain in SPA2, it is possible that the relatively high sequence divergence between the N-terminal domains of SPA1 and SPA2 might be the cause for their differential interaction capacities with cry2. cry1, in contrast, interacts with the WD-repeat domains of SPA1 and SPA2 [47], and this domain is highly conserved between SPA1 and SPA2 [55].
The mechanism of SPA2 degradation may essentially reflect ubiquitination by the COP1 (or COP1/SPA2) E3 ubiquitin ligase or the action of another E3 ligase. Recently, the COP1-interacting E3 ubiquitin ligase COP1 SUPPRESSOR1 (CSU1) was reported to de-stabilize COP1 and SPA1 in darkness, but not in the light. SPA2, SPA3 and SPA4 protein levels were not altered in csu1 mutants, neither in dark-grown nor in light-grown seedlings [56]. It is therefore unlikely that CSU1 is involved in the light-dependent degradation of SPA2. Indeed, light-induced SPA2 degradation was absent in a cop1-5 null mutant. Hence, ubiquitination of SPA2 by COP1 or the COP1/SPA2 ubiquitin ligase is the likely mechanism. This is supported by our finding that a ΔCC SPA2 deletion derivative which does not interact with COP1 in vivo is not degraded in the light. We therefore propose that light influences the E3 ligase activity of COP1/SPA2 in two ways: it inhibits COP1/SPA2 E3 ligase activity towards its substrate transcription factors, while it enhances COP1 (or COP1/SPA2) (auto)-ubiquitination activity towards SPA2 and, possibly, SPA1 as well (Fig 10). However, we cannot fully exclude the possibility that SPA2 is ubiquitinated by an indirect COP1-dependent mechanism. For example, COP1 might be a scaffolding protein required for SPA2 degradation or control the activity of another E3 ubiquitin ligase. Whether SPA3 and SPA4 protein stability is controlled by light remains to be determined. In humans, DNA damage increases COP1 autodegradation by ATM-mediated phosphorylation of COP1, followed by stabilization of the COP1 substrate p53 as a cell cycle check point [57]. Though the phosphorylated residue in human COP1 is not conserved neither in Arabidopsis COP1 nor in the SPA proteins, this finding shows that autodegradation of components of this E3 ligase is a regulatory mechanism used in both humans and plants.
Fig. 10. Model for the effect of light on the ubiquitination activities of the COP1/SPA2 E3 ubiquitin ligase. 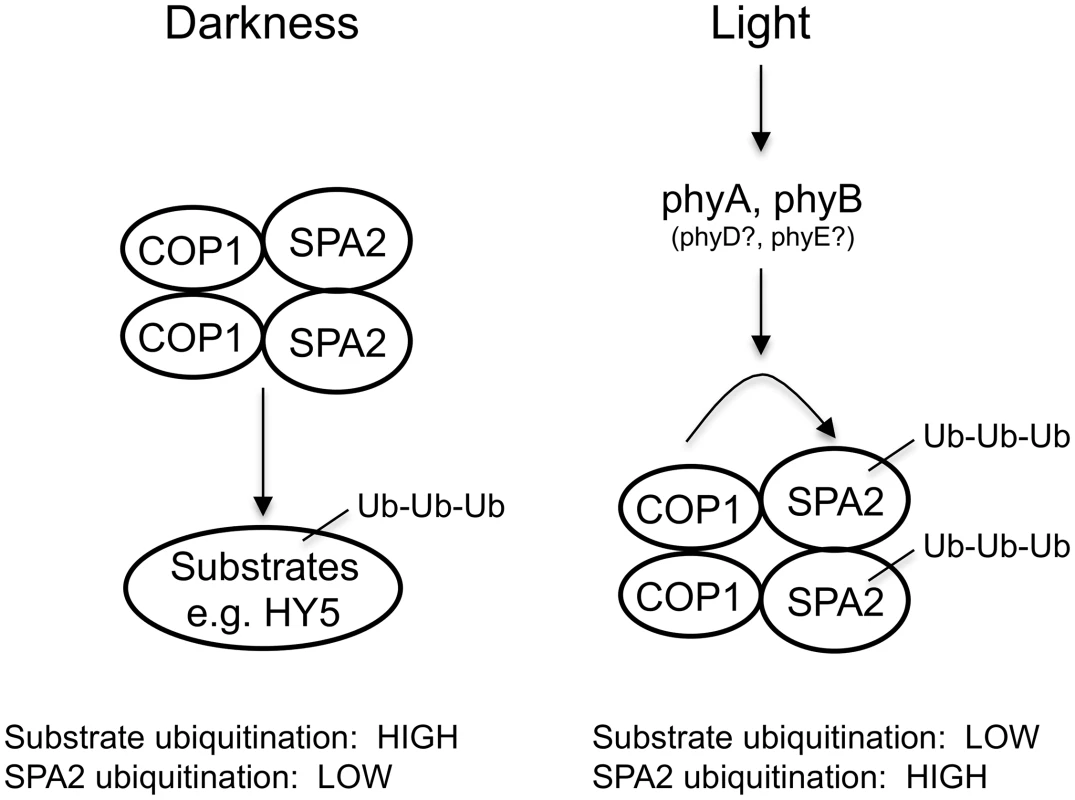
Materials and Methods
Plant materials, light sources and growth conditions
Wild-type Arabidopsis thaliana accessions Col-0, RLD and Ler were used in this study. Photoreceptor mutants phyA-211 (Col-0) [58], phyA-101 (RLD) [59], phyB-1 (introgressed into RLD) [60,61], phyA-101 phyB-1 (RLD), phyA-201 (Ler) [58], cry1 cry2 (Ler) and cry1 cry2 phyA-201 (Ler) [62] were described previously. The transgenic lines with constitutively active photoreceptors expressed the phytochromes AY242H and BY276H [7], PHYA406-YFP-DD/NLS [10], CRY1G380R [8] or GUS-CCT1 [9]. The transgenic lines SPA2::SPA1-HA 28, SPA2::SPA1-HA 70, SPA1::SPA2-HA 64 and SPA2::SPA2-HA 32 were described previously [42]. The mutants spa1-7 spa2-1 spa3-1 and spa1-7 spa3-1 spa4-1 [51] were used whenever no allele information is provided. spa1-100 spa3-1 spa4-3 [23], cop1-4 [63] and cop1-5 [64] were described. The spa1-7 spa3-1 spa4-1 phyA-211 quadruple mutant was generated by crossing the spa1-7 spa3-1 spa4-1 triple mutant with the phyA-211 single mutant and was confirmed in the F2 and F3 progenies by the phyA phenotype and a genotypic analysis using molecular markers that can distinguish between mutant and wild-type spa alleles.
To obtain SPA2::SPA2-HA cop1-5 (-/-) seed, the transgenic line SPA2::SPA2-HA 32 was crossed with cop1-5 (+/-). Transgenic homozygous cop1-5 seeds were selected in a segregating F4 population based on their black seed phenotype which was scored using a stereo microscope. Seeds with normal seed color served as a control that is homo - or heterozygous for the wild-type COP1 allele.
LED light sources and seedling growth conditions were as described previously [22,54]. Growth conditions for the SPA2::SPA2-HA cop1-5 (-/-) experiment were as follows: after stratification of imbibed seeds for 3 days at 4°C, seeds were irradiated with white light for 3 h to break the dormancy and were subsequently kept in darkness for another 21 h. Seeds were then transferred from darkness to Rc (40 μmol m–2 s–1) for 6 h.
Generation of transgenic plants expressing ΔCC SPA2-HA
SPA2::ΔCC SPA2-HA lines express a deletion derivative lacking the amino acids 580–702 in the SPA2 protein. To generate the construct, two PCR fragments were amplified from the full-length SPA2 ORF lacking the stop codon using the primer pairs SC_SPA2deltaCC_ApaI_F1 and SC_SPA2deltaCC_R1 or SC_SPA2deltaCC_F2 and SPA2deltaN-NotI-R. Both PCR products were purified, combined and subsequently used as templates for amplifying the ΔCC SPA2 sequence using the primers SC_SPA2deltaCC_ApaI_F1 and SPA2 delta N NotI R, thereby also introducing a 5’ ApaI restriction site and a 3’ NotI restriction site. After PCR-amplification of ΔCC SPA2, the resulting fragment was introduced into the pJET1.2 vector (Thermo Scientific). After sequencing of the insert to confirm the correct sequence, the deletion construct was digested with ApaI and NotI and ligated into the ApaI and NotI sites of the pBS vector carrying the SPA2 5’ and 3’ regulatory sequences as described previously [42], resulting in the SPA2::ΔCC SPA2 construct in pBS. The 3xHA tag with stop codon was subsequently cloned into the NotI site and the complete insert was cloned into the pJHA212 binary vector [65] as described in [42] to generate SPA2::ΔCC SPA2-HA. This construct was transformed into spa1-7 spa2-1 spa3-1 mutant plants by floral dipping. T2 plants were used for analysis.
Isolation of nuclear protein fractions
In order to detect the native SPA2 protein using an α-SPA2 antibody [42], nuclear proteins were enriched from seedlings as described previously [66].
Isolation of total proteins
Approximately 200 mg of seedlings or, for cop1-5 related experiments, approximately 20 μl volume-equivalents of imbibed seeds were homogenized to a fine powder using liquid nitrogen. Lysis buffer [50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Triton X-100, 5 mM DTT, 1% protease inhibitor cocktail (Sigma-Aldrich), 10 μM MG132] was added to the ground tissue at a ratio of 150 μl per 100 mg tissue. The mixtures were thawed on ice and centrifuged at 20.000 g at 4°C for 12 min. 5x Laemmli buffer was added to the supernatant to a final concentration of 1x before heating at 96°C for 5 min. Protein concentrations were determined by Bradford assay (Bio-Rad).
Immunoblot analysis
For separating nuclear-enriched protein extracts by SDS-PAGE, equal volumes of nuclear-enriched extracts were loaded. To separate total protein extracts, equal amounts of protein were resolved by SDS-PAGE. Protein samples were subsequently blotted onto PVDF membranes. After blotting, membranes were blocked with Rotiblock (Roth) reagent and incubated with the respective primary antibody followed by a horseradish peroxidase (HRP)-conjugated secondary antibody. HRP activity was detected using the SuperSignal West Femto Maximum Sensitivity kit (Thermo Scientific) and visualized by a LAS-4000 Mini bioimager (GE Healthcare Life Sciences). Signal intensities were quantified using Multi-Gauge software (GE Healthcare Life Sciences). Commercial antibodies used were HRP-conjugated α-HA (Roche), α-Histone H3 (Abcam), α-HSC70 (Stressgen), α-α-Tubulin (Sigma-Aldrich), α-rabbit IgG-HRP (Sigma-Aldrich) and α-mouse IgG-HRP (Sigma-Aldrich). α-SPA2 and α-COP1 antibodies were described previously in [42]. α-cry1 [67] and α-cry2 [68] antibodies were used to detect cry1 and cry2, respectively.
Co-immunoprecipitations
Co-immunoprecipitation experiments were performed using μMACS Anti-HA Starting Kits (Miltenyi Biotec) according to the manufacturer’s protocol with minor modification. Total proteins were extracted as described above. Protein lysates were incubated with 10 μl μMACS Anti-HA MicroBeads. After incubation on ice for 30 min, the mixture was applied onto prepared μ Columns which were placed in the magnetic field of μMACS Separator attached to a MACS MultiStand. The columns were washed four times with lysis buffer and once with Wash Buffer 2 provided by the kit. Elution was performed at 95°C with Elution Buffer according to the manufacturer’s manual. For cry1 and cry2 pull-down experiments, seedlings were pre-infiltrated with 100 μM MG132 and 10 μM clasto-Lactacystin β-lactone twice, 15 min each, before light treatment. Furthermore, five times more protein extract was used for the SPA2-HA immunoprecipitation than for the SPA1-HA immunoprecipitation.
Hypocotyl length measurement
Seedlings were flattened on the surface of solid MS plates and photographed with a Nikon D5000 digital camera. Images were analyzed by ImageJ 1.43u (Wayne Rasband, National Institutes of Health) to obtain hypocotyl lengths.
Transcript level analysis
Total RNA isolation, DNase I treatment, first-strand cDNA synthesis and qRT-PCR were performed as described in [42]. Primers used to amplify HA-tag and UBQ10 were previously described [42]. Two biological replicates were included. Relative transcript levels were calculated using the ΔΔCt method with UBQ10 as a normalization transcript.
Accession numbers
COP1 (At2g32950), SPA1 (At2g46340), SPA2 (At4g11110), SPA3 (At3g15354), SPA4 (At1g53090), cry1 (AT4G08920), cry2 (AT1G04400), phyA (AT1G09570), phyB (AT2G18790).
Supporting Information
Zdroje
1. Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91 : 29–66. doi: 10.1016/S0070-2153(10)91002-8 20705178
2. Casal JJ (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64 : 403–427. doi: 10.1146/annurev-arplant-050312-120221 23373700
3. Casal JJ, Candia AN, Sellaro R (2014) Light perception and signalling by phytochrome A. J Exp Bot 65 : 2835–2845. doi: 10.1093/jxb/ert379 24220656
4. Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61 : 11–24. doi: 10.1093/jxb/erp304 19815685
5. Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, et al. (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62 : 335–364. doi: 10.1146/annurev-arplant-042110-103759 21526969
6. Liu H, Liu B, Zhao C, Pepper M, Lin C (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci 16 : 684–691. doi: 10.1016/j.tplants.2011.09.002 21983106
7. Su YS, Lagarias JC (2007) Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19 : 2124–2139. 17660358
8. Gu NN, Zhang YC, Yang HQ (2012) Substitution of a conserved glycine in the PHR domain of Arabidopsis cryptochrome 1 confers a constitutive light response. Mol Plant 5 : 85–97. doi: 10.1093/mp/ssr052 21765176
9. Yang HQ, Wu YJ, Tang RH, Liu DM, Liu Y, et al. (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103 : 815–827. 11114337
10. Viczian A, Adam E, Wolf I, Bindics J, Kircher S, et al. (2012) A short amino-terminal part of Arabidopsis phytochrome A induces constitutive photomorphogenic response. Mol Plant 5 : 629–641. doi: 10.1093/mp/sss035 22498774
11. Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, et al. (2013) The UVR8 UV-B photoreceptor: perception, signaling and response. Arabidopsis Book 11: e0164. doi: 10.1199/tab.0164 23864838
12. Jenkins GI (2014) The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26 : 21–37. doi: 10.1105/tpc.113.119446 24481075
13. Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1 : 20 years later. Trends Plant Sci 17 : 584–593. doi: 10.1016/j.tplants.2012.05.004 22705257
14. Huang X, Ouyang X, Deng XW (2014) Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr Opin Plant Biol 21C: 96–103.
15. Weidler G, Zur Oven-Krockhaus S, Heunemann M, Orth C, Schleifenbaum F, et al. (2012) Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome A. Plant Cell 24 : 2610–2623. doi: 10.1105/tpc.112.098210 22739826
16. Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18 : 617–622. 15031264
17. Shalitin D, Yang HY, Mockler TC, Maymon M, Guo HW, et al. (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417 : 763–767. 12066190
18. Debrieux D, Trevisan M, Fankhauser C (2013) Conditional involvement of CONSTITUTIVE PHOTOMORPHOGENIC1 in the degradation of phytochrome A. Plant Physiol 161 : 2136–2145. doi: 10.1104/pp.112.213280 23391578
19. Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, et al. (2008) Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20 : 2307–2323. doi: 10.1105/tpc.107.056580 18812498
20. Ranjan A, Dickopf S, Ullrich KK, Rensing SA, Hoecker U (2014) Functional analysis of COP1 and SPA orthologs from Physcomitrella and rice during photomorphogenesis of transgenic Arabidopsis reveals distinct evolutionary conservation. BMC Plant Biol 14 : 178. doi: 10.1186/1471-2229-14-178 24985152
21. Deng X-W, Caspar T, Quail PH (1991) cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev 5 : 1172–1182. 2065972
22. Laubinger S, Fittinghoff K, Hoecker U (2004) The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16 : 2293–2306. 15308756
23. Ordonez-Herrera N, Fackendahl P, Yu X, Schaefer S, Koncz C, et al. (2015) A cop1 spa mutant deficient in COP1 and SPA proteins reveals partial co-action of COP1 and SPA during Arabidopsis post-embryonic development and photomorphogenesis. Mol Plant 8 : 479–481. doi: 10.1016/j.molp.2014.11.026 25667004
24. Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, et al. (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133 : 3213–3222. 16854975
25. Rolauffs S, Fackendahl P, Sahm J, Fiene G, Hoecker U (2012) Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low red light to far-red light ratio. Plant Physiol 160 : 2015–2027. doi: 10.1104/pp.112.207233 23093358
26. Maier A, Schrader A, Kokkelink L, Falke C, Welter B, et al. (2013) Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J 74 : 638–651. doi: 10.1111/tpj.12153 23425305
27. Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, et al. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. Embo J 27 : 1277–1288. doi: 10.1038/emboj.2008.68 18388858
28. Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, et al. (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20 : 292–306. doi: 10.1105/tpc.107.057281 18296627
29. Wang CQ, Sarmast MK, Jiang J, Dehesh K (2015) The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell: 27 : 1128–1139. doi: 10.1105/tpc.15.00044 25841036
30. Ranjan A, Fiene G, Fackendahl P, Hoecker U (2011) The Arabidopsis repressor of light signaling SPA1 acts in the phloem to regulate seedling de-etiolation, leaf expansion and flowering time. Development 138 : 1851–1862. doi: 10.1242/dev.061036 21447551
31. Jang IC, Yang JY, Seo HS, Chua NH (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19 : 593–602. 15741320
32. Yang J, Lin R, Hoecker U, Liu B, Xu L, et al. (2005) Repression of light signaling by Arabidopsis SPA1 involves post-translational regulation of HFR1 protein accumulation. Plant J 43 : 131–141. 15960622
33. Hoecker U, Xu Y, Quail PH (1998) SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10 : 19–33. 9477570
34. Jackson S, Xiong Y (2009) CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci 34 : 562–570. doi: 10.1016/j.tibs.2009.07.002 19818632
35. Biedermann S, Hellmann H (2011) WD40 and CUL4-based E3 ligases: lubricating all aspects of life. Trends Plant Sci 16 : 38–46. doi: 10.1016/j.tplants.2010.09.007 20965772
36. Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, et al. (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22 : 108–123. doi: 10.1105/tpc.109.065490 20061554
37. Hoecker U, Quail PH (2001) The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem 276 : 38173–38178. 11461903
38. Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, et al. (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17 : 2642–2647. 14597662
39. Holm M, Hardtke CS, Gaudet R, Deng XW (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20 : 118–127. 11226162
40. Hoecker U, Tepperman JM, Quail PH (1999) SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284 : 496–499. 10205059
41. Deng XW, Matsui M, Wei N, Wagner D, Chu AM, et al. (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71 : 791–801. 1423630
42. Balcerowicz M, Fittinghoff K, Wirthmueller L, Maier A, Fackendahl P, et al. (2011) Light exposure of Arabidopsis seedlings causes rapid de-stabilization as well as selective post-translational inactivation of the repressor of photomorphogenesis SPA2. Plant J 65 : 712–723. doi: 10.1111/j.1365-313X.2010.04456.x 21235648
43. Pacin M, Legris M, Casal JJ (2014) Rapid decline in nuclear CONSTITUTIVE PHOTOMORPHOGENESIS1 abundance anticipates the stabilization of its target ELONGATED HY5 in the light. Plant Physiol 164 : 1134–1138. doi: 10.1104/pp.113.234245 24434030
44. von Arnim AG, Deng X-W (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79 : 1035–1045. 8001131
45. Fankhauser C, Ulm R (2011) Light-regulated interactions with SPA proteins underlie cryptochrome-mediated gene expression. Genes Dev 25 : 1004–1009. doi: 10.1101/gad.2053911 21576261
46. Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25 : 1029–1034. doi: 10.1101/gad.2025011 21511871
47. Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, et al. (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25 : 1023–1028. doi: 10.1101/gad.2025111 21511872
48. Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, et al. (2015) Red light-dependent interaction of phyB with SPA1 promotes COP1–SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8 : 467–478. doi: 10.1016/j.molp.2014.11.025 25744387
49. Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, et al. (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27 : 189–201. doi: 10.1105/tpc.114.134775 25627066
50. Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21 : 841–847. doi: 10.1016/j.cub.2011.03.048 21514160
51. Fittinghoff K, Laubinger S, Nixdorf M, Fackendahl P, Baumgardt RL, et al. (2006) Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J 47 : 577–590. 16813571
52. Osterlund MT, Deng XW (1998) Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J 16 : 201–208. 9839465
53. Osterlund MT, Wei N, Deng XW (2000) The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of arabidopsis seedling development. Plant Physiol 124 : 1520–1524. 11115869
54. Baumgardt RL, Oliverio KA, Casal JJ, Hoecker U (2002) SPA1, a component of phytochrome A signal transduction, regulates the light signaling current. Planta 215 : 745–753. 12244439
55. Laubinger S, Hoecker U (2003) The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J 35 : 373–385. 12887588
56. Xu D, Lin F, Jiang Y, Huang X, Li J, et al. (2014) The RING-finger E3 ubiquitin ligase COP1 SUPPRESSOR1 negatively regulates COP1 abundance in maintaining COP1 homeostasis in dark-grown Arabidopsis seedlings. Plant Cell 26 : 1981–1991. 24838976
57. Dornan D, Shimizu H, Mah A, Dudhela T, Eby M, et al. (2006) ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 313 : 1122–1126. 16931761
58. Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and Phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiology 104 : 1139–1149. 12232154
59. Parks BM, Quail PH (1993) Hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell 5 : 39–48. 8439743
60. Quail PH, Briggs WR, Chory J, Hangarter RP, Harberd NP, et al. (1994) Spotlight on phytochrome nomenclature. Plant Cell 6 : 468–471. 12244245
61. Smith H, Xu Y, Quail PH (1997) Antagonistic but complementary actions of phytochromes A and B allow seedling de-etiolation. Plant Physiol 114 : 637–641. 9193095
62. Mazzella MA, Cerdan PD, Staneloni RJ, Casal JJ (2001) Hierarchical coupling of phytochromes and cryptochromes reconciles stability and light modulation of Arabidopsis development. Development 128 : 2291–2299. 11493548
63. Deng XW, Quail PH (1992) Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J 2 : 83–95.
64. McNellis TW, Von Arnim AG, Araki T, Komeda Y, Miséra S, et al. (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6 : 487–500. 8205001
65. Yoo SY, Bomblies K, Yoo SK, Yang JW, Choi MS, et al. (2005) The 35S promoter used in a selectable marker gene of a plant transformation vector affects the expression of the transgene. Planta 221 : 523–530. 15682278
66. Xia YJ, Nikolau BJ, Schnable PS (1997) Developmental and hormonal regulation of the Arabidopsis CER2 gene that codes for a nuclear-localized protein required for the normal accumulation of cuticular waxes. Plant Physiology 115 : 925–937. 9390429
67. Lin C, Ahmad M, Gordon D, Cashmore AR (1995) Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue, UV-a, and green light. Proc NatAcad Sci USA 92 : 8423–8427.
68. Ahmad M, Jarillo JA, Cashmore AR (1998) Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10 : 197–207. 9490743
Štítky
Genetika Reprodukční medicína
Článek The Chromatin Protein DUET/MMD1 Controls Expression of the Meiotic Gene during Male Meiosis inČlánek Tissue-Specific Gain of RTK Signalling Uncovers Selective Cell Vulnerability during Embryogenesis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 9
-
Všechny články tohoto čísla
- Retraction: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Signaling from Within: Endocytic Trafficking of the Robo Receptor Is Required for Midline Axon Repulsion
- A Splice Region Variant in Lowers Non-high Density Lipoprotein Cholesterol and Protects against Coronary Artery Disease
- The Chromatin Protein DUET/MMD1 Controls Expression of the Meiotic Gene during Male Meiosis in
- A NIMA-Related Kinase Suppresses the Flagellar Instability Associated with the Loss of Multiple Axonemal Structures
- Slit-Dependent Endocytic Trafficking of the Robo Receptor Is Required for Son of Sevenless Recruitment and Midline Axon Repulsion
- Expression of Concern: Protein Under-Wrapping Causes Dosage Sensitivity and Decreases Gene Duplicability
- Mutagenesis by AID: Being in the Right Place at the Right Time
- Identification of as a Genetic Modifier That Regulates the Global Orientation of Mammalian Hair Follicles
- Bridges Meristem and Organ Primordia Boundaries through , , and during Flower Development in
- Evaluating the Performance of Fine-Mapping Strategies at Common Variant GWAS Loci
- KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome
- Differential Expression of Ecdysone Receptor Leads to Variation in Phenotypic Plasticity across Serial Homologs
- Receptor Polymorphism and Genomic Structure Interact to Shape Bitter Taste Perception
- Cognitive Function Related to the Gene Acquired from an LTR Retrotransposon in Eutherians
- Critical Function of γH2A in S-Phase
- Arabidopsis AtPLC2 Is a Primary Phosphoinositide-Specific Phospholipase C in Phosphoinositide Metabolism and the Endoplasmic Reticulum Stress Response
- XBP1-Independent UPR Pathways Suppress C/EBP-β Mediated Chondrocyte Differentiation in ER-Stress Related Skeletal Disease
- Integration of Genome-Wide SNP Data and Gene-Expression Profiles Reveals Six Novel Loci and Regulatory Mechanisms for Amino Acids and Acylcarnitines in Whole Blood
- A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism
- Cell Cycle Regulates Nuclear Stability of AID and Determines the Cellular Response to AID
- A Genome-Wide Association Analysis Reveals Epistatic Cancellation of Additive Genetic Variance for Root Length in
- Tissue-Specific Gain of RTK Signalling Uncovers Selective Cell Vulnerability during Embryogenesis
- RAB-10-Dependent Membrane Transport Is Required for Dendrite Arborization
- Basolateral Endocytic Recycling Requires RAB-10 and AMPH-1 Mediated Recruitment of RAB-5 GAP TBC-2 to Endosomes
- Dynamic Contacts of U2, RES, Cwc25, Prp8 and Prp45 Proteins with the Pre-mRNA Branch-Site and 3' Splice Site during Catalytic Activation and Step 1 Catalysis in Yeast Spliceosomes
- ARID1A Is Essential for Endometrial Function during Early Pregnancy
- Predicting Carriers of Ongoing Selective Sweeps without Knowledge of the Favored Allele
- An Interaction between RRP6 and SU(VAR)3-9 Targets RRP6 to Heterochromatin and Contributes to Heterochromatin Maintenance in
- Photoreceptor Specificity in the Light-Induced and COP1-Mediated Rapid Degradation of the Repressor of Photomorphogenesis SPA2 in Arabidopsis
- Autophosphorylation of the Bacterial Tyrosine-Kinase CpsD Connects Capsule Synthesis with the Cell Cycle in
- Multimer Formation Explains Allelic Suppression of PRDM9 Recombination Hotspots
- Rescheduling Behavioral Subunits of a Fixed Action Pattern by Genetic Manipulation of Peptidergic Signaling
- A Gene Regulatory Program for Meiotic Prophase in the Fetal Ovary
- Cell-Autonomous Gβ Signaling Defines Neuron-Specific Steady State Serotonin Synthesis in
- Discovering Genetic Interactions in Large-Scale Association Studies by Stage-wise Likelihood Ratio Tests
- The RCC1 Family Protein TCF1 Regulates Freezing Tolerance and Cold Acclimation through Modulating Lignin Biosynthesis
- The AMPK, Snf1, Negatively Regulates the Hog1 MAPK Pathway in ER Stress Response
- The Parkinson’s Disease-Associated Protein Kinase LRRK2 Modulates Notch Signaling through the Endosomal Pathway
- Multicopy Single-Stranded DNA Directs Intestinal Colonization of Enteric Pathogens
- Recurrent Domestication by Lepidoptera of Genes from Their Parasites Mediated by Bracoviruses
- Three Different Pathways Prevent Chromosome Segregation in the Presence of DNA Damage or Replication Stress in Budding Yeast
- Identification of Four Mouse Diabetes Candidate Genes Altering β-Cell Proliferation
- The Intolerance of Regulatory Sequence to Genetic Variation Predicts Gene Dosage Sensitivity
- Synergistic and Dose-Controlled Regulation of Cellulase Gene Expression in
- Genome Sequence and Transcriptome Analyses of : Metabolic Tools for Enhanced Algal Fitness in the Prominent Order Prymnesiales (Haptophyceae)
- Ty3 Retrotransposon Hijacks Mating Yeast RNA Processing Bodies to Infect New Genomes
- FUS Interacts with HSP60 to Promote Mitochondrial Damage
- Point Mutations in Centromeric Histone Induce Post-zygotic Incompatibility and Uniparental Inheritance
- Genome-Wide Association Study with Targeted and Non-targeted NMR Metabolomics Identifies 15 Novel Loci of Urinary Human Metabolic Individuality
- Outer Hair Cell Lateral Wall Structure Constrains the Mobility of Plasma Membrane Proteins
- A Large-Scale Functional Analysis of Putative Target Genes of Mating-Type Loci Provides Insight into the Regulation of Sexual Development of the Cereal Pathogen
- A Genetic Selection for Mutants Reveals an Interaction between DNA Polymerase IV and the Replicative Polymerase That Is Required for Translesion Synthesis
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Arabidopsis AtPLC2 Is a Primary Phosphoinositide-Specific Phospholipase C in Phosphoinositide Metabolism and the Endoplasmic Reticulum Stress Response
- Bridges Meristem and Organ Primordia Boundaries through , , and during Flower Development in
- KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome
- XBP1-Independent UPR Pathways Suppress C/EBP-β Mediated Chondrocyte Differentiation in ER-Stress Related Skeletal Disease
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



