-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSix Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
Adult skeletal muscles are classified into fast-type and slow-type, which display different resistance to muscle atrophy and metabolic protection against obesity. We identify in this manuscript a new mechanism controlling in vivo adult muscle fiber-type specification implicating a long intergenic non-coding RNA, linc-MYH. We demonstrate a three-element genetic partnership, where an enhancer under the control of the myogenic homeoprotein Six1 functions as a regulatory hub to control fibre phenotype. In this partnership, the enhancer controls positively the expression of both the adjacent fast myosin heavy chain (MYH) gene cluster and of linc-MYH. linc-MYH is present only in adult fast type skeletal myofibers and controls their phenotype by suppressing slow-type gene expression. The regulation of linc-MYH could provide a lead for new therapeutic approaches or drug development.
Published in the journal: . PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004386
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004386Summary
Adult skeletal muscles are classified into fast-type and slow-type, which display different resistance to muscle atrophy and metabolic protection against obesity. We identify in this manuscript a new mechanism controlling in vivo adult muscle fiber-type specification implicating a long intergenic non-coding RNA, linc-MYH. We demonstrate a three-element genetic partnership, where an enhancer under the control of the myogenic homeoprotein Six1 functions as a regulatory hub to control fibre phenotype. In this partnership, the enhancer controls positively the expression of both the adjacent fast myosin heavy chain (MYH) gene cluster and of linc-MYH. linc-MYH is present only in adult fast type skeletal myofibers and controls their phenotype by suppressing slow-type gene expression. The regulation of linc-MYH could provide a lead for new therapeutic approaches or drug development.
Introduction
Adult skeletal muscles are composed of slow and fast myofiber subtypes which selectively express the genes required for their specific contraction activity and metabolic properties [1]–[4]. These properties are acquired at the end of fetal development and during the neonatal period, when mixed skeletal myofibers expressing a panel of embryonic, fast and slow genes develop a specific slow or fast phenotype. The formation of efficient fast sarcomeric units, and Ca++ cycling and excitation/contraction/relaxation coupling in fast-myofibers, is achieved through the coordinate control of fast Myhs and associated fast sarcomeric genes (including Tnnt3, Tnni2, Tnnc2, Atp2a1 and Pvalb) [2],[4]. Myofibers can be classified by their MYH expression profile: slow-type myofibers in mice express MYH7 (also known as MYHCI, β or slow), and fast-myofibers express MYH2 (MYHCIIA), MYH1 (MYHCIIX) or MYH4 (MYHCIIB). Fast Myh genes found in developmental and adult stages (Myh3, Myh2, Myh1, Myh4, Myh8 and Myh13) are organized as a cluster within a 300 kb region on mouse chromosome 11 [5]. The spatio-temporal expression of one specific fast Myh gene at the Myh locus is reminiscent of the organization and expression of Globin genes at the beta-globin locus [6]. However, we are yet to investigate potential enhancers or the Myh locus control region (LCR) that could be responsible for sequential and specific Myh gene expression in myofibers. The coordination of fast-type and slow-type gene expression in fast myofibers is not currently understood. Distinct intramyofibrillary calcium transients, evoked by slow tonic motor neuron firing, induce a cascade of downstream signaling involving Calcineurin and CamK. This results in the activation of selective transcription activators and repressors in slow myofibres. However, the signaling pathways operating in distinct Myh2, Myh1 and Myh4 myofiber subtypes, which coordinate the activation of the other fast-type genes and the repression of slow-type genes, is less well understood [1]. Better knowledge of the mechanisms controlling muscle specialization and plasticity is important to enable the understanding and modulation of muscle adaptations in pathophysiological conditions.
Six homeoproteins are major myogenic transcription factors which directly bind to DNA sequences (called MEF3s) to control myogenesis [7],[8] and the genesis of fast-type myofibers during embryogenesis [9],[10]. In adult skeletal muscle, Six1 accumulates at a higher level in the nuclei of adult fast myofibers than in of slow myofibers. Forced expression of Six1 and its Eya1 cofactor in slow myofibers causes adult slow-twitch oxidative fibers toward a fast-twitch glycolytic phenotype [11]. Animals with a Six1 KO present severe muscle hypoplasia and die at birth [12]. This prevents the in vivo analysis of the adult phenotype and the ability to investigate the direct or indirect involvement of Six1 in the spatio-temporal control of the expression of genes in the fast Myh cluster.
The mammalian genome encodes thousands of long intergenic non-coding RNAs (lincRNAs) which have multiple functions [13],[14]. Some accumulate in the cytoplam as miRNAs decoys [15],[16]. Others accumulate in the nucleus and participate to gene regulation through chromatin remodeling and epigenetic modifications [14],[17],[18]. Here, they may act as cis [19] or trans [20] transcriptional activators, as transcriptional repressors [21],[22] or through DNA-RNA triplex formation [23],[24].
In this study we identify a new lincRNA, linc-MYH, and the mechanism of its control of adult muscle fast fiber-type specification in vivo. We demonstrate a three-element genetic partnership, where an enhancer element under the control of the myogenic homeoprotein Six1 functions as a regulatory hub to control fiber phenotype. In this partnership, the enhancer positively controls the expression of both the adjacent fast Myh gene cluster and linc-MYH, suppressing slow-type gene expression and facilitating fast fiber-type specialization.
Results
Six1 binds directly to a newly identified enhancer of the Myh genes cluster
Our previous studies suggested that Six1 could be directly involved in the control of the expression of fast Myh genes, since higher levels of this transcription factor accumulate in the nuclei of adult fast myofibers than in slow myofibers [11]. To investigate how Six1 could control the expression of fast Myh isoforms, we used computational analysis to locate MEF3 sites at the fast Myh locus (see Materials and Methods). Six clustered MEF3 sites are conserved across human, rat and mouse genomes in an intergenic region located 50 kb upstream of the Myh2 gene (Figures 1A and S1) and 4 kb upstream of a lincRNA (2310065F04Rik); we refer to this lincRNA as linc-MYH (Figures 1A and S2). Six1 binding at these MEF3 sites was demonstrated in vivo by ChIP (Chromatin Immunoprecipitation) experiments with Six1 antibodies on adult fast gastrocnemius plantaris (GP) and tibialis anterior (TA) muscles (Figure 1B) but not on adult slow Soleus (data not shown), and confirmed for five of these sites (sites 1, 2, 3, 4 and 6) by EMSA assays (Figure S3A). We asked whether this Myh intergenic region could constitute an enhancer element, controlling the spatio-temporal expression of Myh genes in this locus. A 2 kb DNA fragment of this region, including the six identified MEF3 sites and 1 kb of DNA fragments upstream of the transcription start site of fast-type Myh2, Myh1 and Myh4 genes, was isolated. The putative enhancer was ligated to each Myh promoter using luciferase pGL3 basic plasmids to generate pGL3-Enhancer-Myh2/1/4 constructs. To test the involvement of Six binding in enhancer activation of the Myh2, Myh1 and Myh4 promoters, we mutated all six MEF3 sites present in the enhancer, and named these reporters pGL3-mutEnhancer-Myh2/1/4. Luciferase activity was tested two weeks after the electroporation of reporter plasmids in adult TA muscles. The luciferase activity of pGL3-Enhancer-Myh2/1/4 was seven to twelve times higher for either of the promoters, than with pGL3-Myh2/1/4. Enhancer activity was not observed in plasmids with mutated MEF3 sites associated with either of the Myh promoters (Figure 1C). Enhancer activity was neither observed with the promoters of the slow Sln (Figure S3B) or Tnni1 genes, or with the promoter of the ubiquitous β-actin gene (Figure S3C). A weak enhancer activity was observed with Myh4 promoter in primary embryonic fibroblasts, in which Six1 is expressed (Figure S3D and data not shown). These data showed that high MYH enhancer activity was only reached in vivo and required specific interactions with MYH promoter elements. To determine in vivo interactions between the enhancer and each Myh gene, we performed chromatin conformation capture (3C) assays of adult fast EDL (Extensor digitorum longus) myofibers. These experiments revealed that the enhancer interacts with the promoter of Myh2/1/4 genes in native chromatin of EDL myonuclei (Figure 1D). The strongest interactions were observed with the Myh1 and Myh4 promoters, consistent with the expression profile of these two genes in EDL muscles. The data demonstrates that the identified conserved cis-element acts as an enhancer for the Myh locus and that MEF3 sites are essential for its enhancer activity in vivo.
Fig. 1. Six1 binds directly the enhancer of the fast Myh gene cluster. 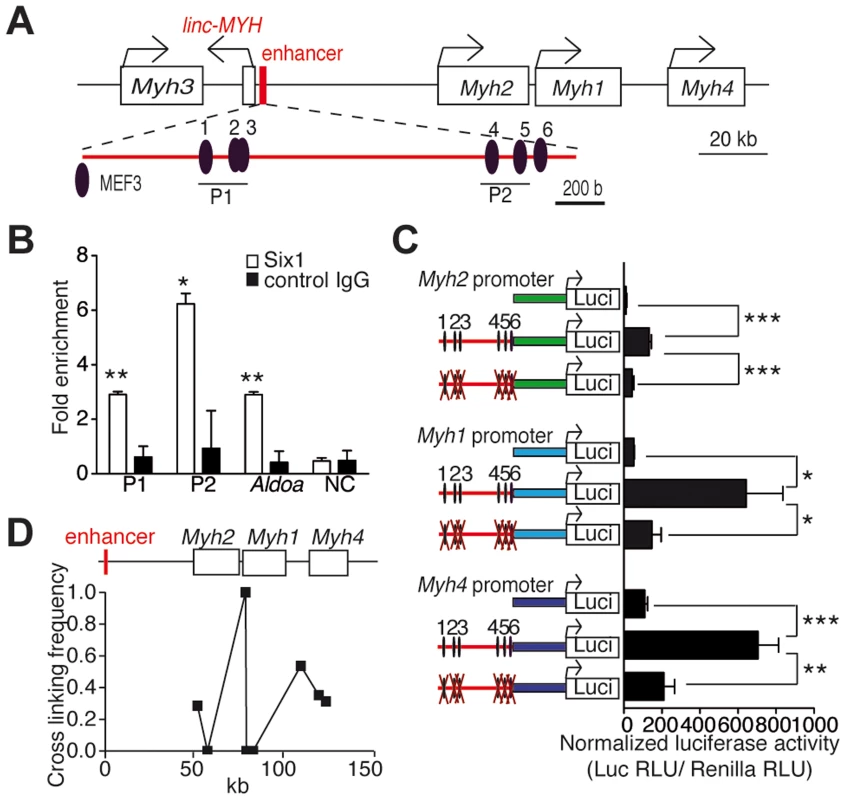
(A) Schematic representation of the fast Myh gene cluster. (B) qPCR values of ChIP experiments performed with Six1 antibodies, or IgG on GP and TA chromatin, showing Six1 binding to P1 and P2 regions of the fast Myh enhancer, to the muscle promoter of AldoA, and to an intergenic region located 86 kb 5′ upstream of Myh3 (NC) (n = 3). (C) Luciferase assays from adult TA muscles electroporated with luciferase vectors (indicated) and a TK-Rennila luciferase vector allowing normalization (n = 4). (D) qPCR experiments from 3C assays of wild type EDL muscle, showing the direct interactions of Myh2, Myh1 and Myh4 promoters with the fast Myh enhancer. *P<0.05, **P<0.01, ***P<0.001. Loss of Six1 impairs fast muscle genes and linc-MYH expression during post-natal development
To further characterize the role of Six1 in the control of fast Myh gene expression, we bred Six1flox/flox mice with transgenic mice expressing CRE recombinase under the control of the human skeletal actin (HSA) promoter and obtained Six1flox/flox;HSA-CRE conditional knockout mice (hereafter named cSix1 KO) [25],[26]. We analyzed the expression of fiber type specific genes in the back muscles of wild-type control mice and cSix1 KO mice at embryonic day 18.5 (E18.5) and at several post-natal stages (two weeks (P2W), four weeks (P4W) and eight weeks (P8W)) animals (Figure 2), as muscle fiber fast-subtype specialization takes place from the end of embryogenesis [9]. Six1 mRNA was not detectable in back muscles of cSix1 KO mice (Figure 2). The expression of fast-type genes (Myh4, Tnnt3, Tnni2, Tnnc2 and Pvalb) increased during postnatal development in control mice but that of slow-type genes (Myh7, Tnnt1, Tnni1, Tnnc1 and Sln) decreased. The linc-MYH RNA was detected after birth in muscle samples and its expression increased in line with that of Myh4 (Figure 2). The induction of fast-type genes and linc-MYH and the suppression of slow-type genes, were impaired in cSix1 KO mice. Expression of linc-MYH was reduced by three to five times in cSix1 KO mice during postnatal development (Figure 2). These results show that Six1 controls the induction of linc-MYH and fast-type genes during postnatal development, and is required for the downregulation of slow type genes.
Fig. 2. The expression of fast-type genes and linc-MYH is impaired in cSix1 KO mice during postnatal development. 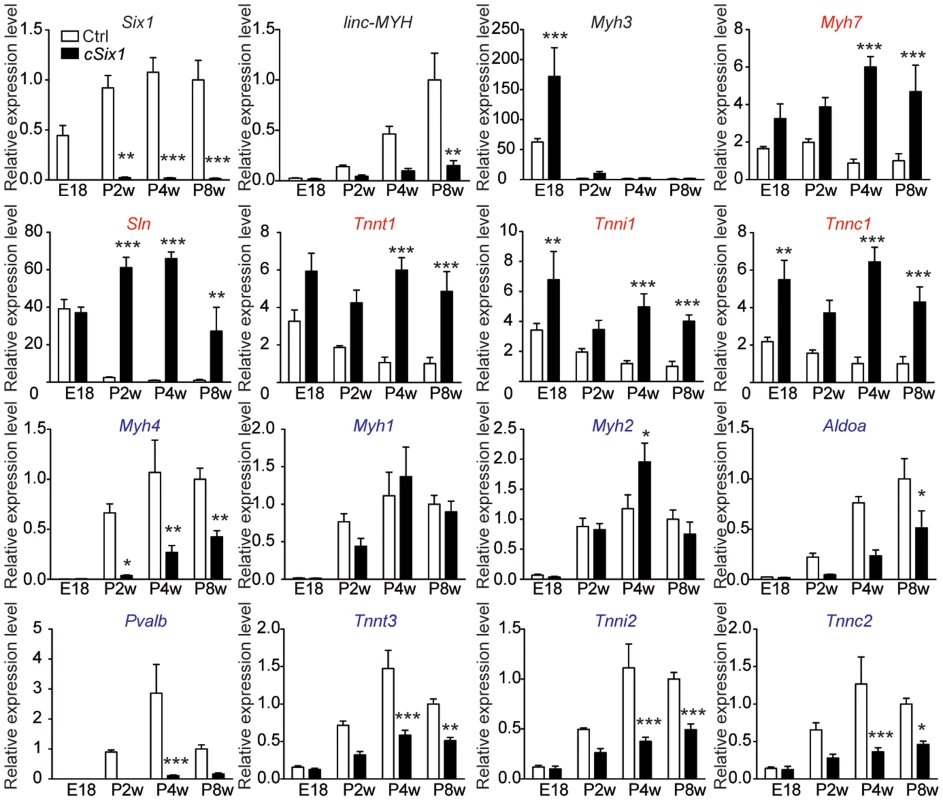
mRNA expression level of Six1, linc-MYH, Myh3, slow-type genes (red) and fast-type genes (blue) in back muscles of cSix1 KO mice at E18.5, P2W, P4W and P8W, as determined by qPCR experiments, (n = 3 to 6 for each point). *P<0.05, **P<0.01, ***P<0.001. Six1 deficiency impairs adult muscle fast phenotype
We next analyzed adult 12 week-old cSix1 KO mice to further characterize the role of Six1 in adult muscle. Six1 mRNA and protein were not detectable in GP enriched with fast-myofibers or soleus (SOL) muscle enriched with slow-myofibers (Figure 3A and B), and fatigue resistance of TA muscle was 35% higher (Figure 3C) in the cSix1 KO mice. We used immunohistochemistry to analyse the composition of MYH7, MYH2 and MYH4 in cSix1 mutant myofibers. Mutant TA muscles had a higher percentage of fibers containing MYH7 and MYH2, but a lower percentage of fibers containing MYH4 (Figures 3D and S4). We found consistent results during qPCR analysis of Myh mRNA i.e., higher levels of Myh7 and Myh2 mRNA and lower levels of Myh4 mRNA levels were observed in the fast TA muscles of cSix1 KO (Figure 3E). Expression levels of other specific fast and slow-type genes were also tested. We found in mutant TA muscles a downregulation of fast-type genes (Tnnt3, Tnni2, Tnnc2 and Pvalb) and a five and to 25 fold increase in the levels of slow-type genes (Myh7, Tnnt1, Tnni1, Tnnc1 and Sln) (Figure 3E). Nevertheless expression of slow Myh7 is increased more than ten fold at the mRNA level in cSix1 mutant TA myofibers, while by immunohistochemistry the number of MYH7 positive myofibers is increased less than two fold. This showed that there is no major phenotype switch in cSix1 mutant TA myofibers. This observation could be explained either by the higher amount of Myh7 mRNA accumulating in MYH7 positive fibers or by a general increase of Myh7 mRNA in TA myofibers, mRNAs that would not be translated efficiently and leading to the absence of increase of MYH7 positive fibers. The expression of linc-MYH expression was lower in the adult TA of cSix1 KO mice, than in control mice (Figure 3E). These results indicate that the Six1 homeoprotein can control the phenotype of fast skeletal myofibers in adult animals.
Fig. 3. Six1 deficiency impairs the adult phenotype of fast muscle. 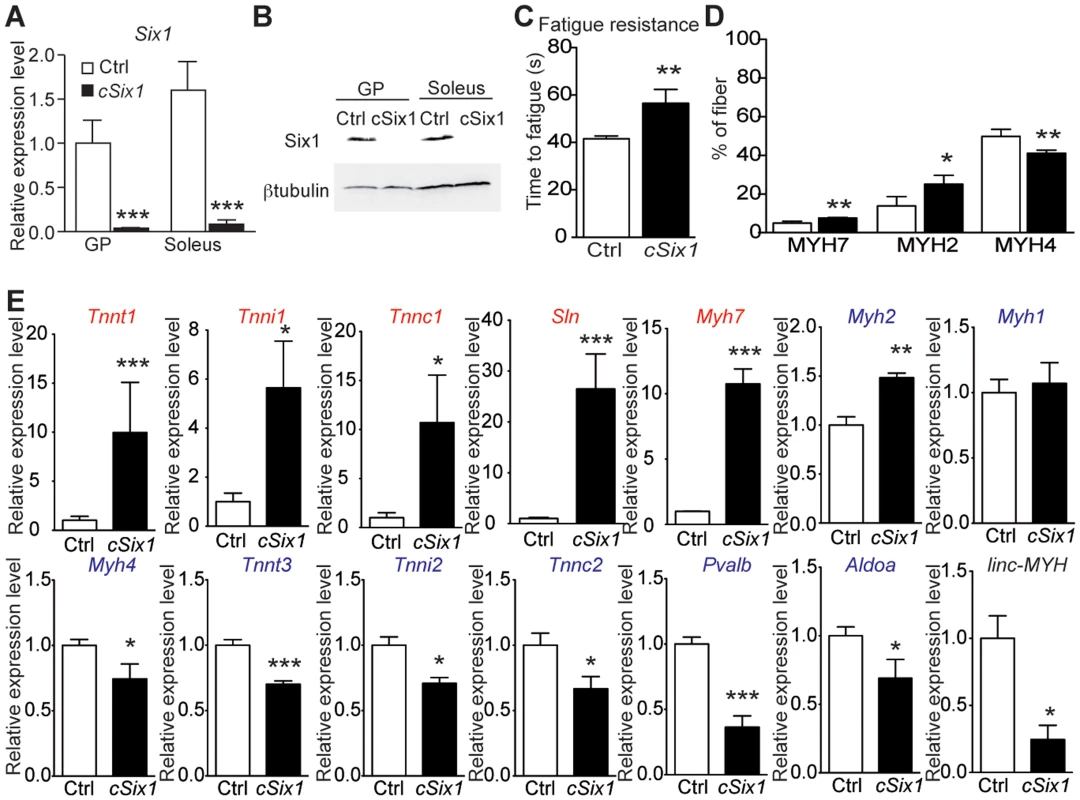
(A) Six1 mRNA expression levels in GP and Sol muscles of three month-old control (Ctrl, n = 4) and cSix1 KO (n = 3) mice. (B) Western blot analysis of Six1 and βtubulin expression in Sol and GP of Ctrl and cSix1 KO mice. (C) Time to fatigue ratio of TA muscles of Ctrl (n = 4) and cSix1 KO (n = 4) mice. (D) Percentage of myofibers expressing MYH7, MYH2 and MYH4 in TA muscles of three month-old Ctrl (n = 4) and cSix1 KO (n = 4) mice. (E) mRNA expression levels of slow-type genes (red), fast-type genes (blue) and linc-MYH in TA muscles of three month-old Ctrl (n = 4) and cSix1 KO (n = 4) mice. *P<0.05, **P<0.01, ***P<0.001. linc-MYH is expressed exclusively in adult fast-type muscles
We found that linc-MYH is expressed in fast-type skeletal muscles (GP, TA and EDL), but not in SOL, brain, kidney, heart or fat tissues, an expression pattern which parallels that of the fast-fiber Myh4 (Figure 4A). This suggested that linc-MYH is only expressed following robust nuclear accumulation of Six1, as takes place in the nuclei of MYH4 myofibers [11], and that the weaker nuclear accumulation of Six1 observed in SOL myonuclei does not allow efficient Six1 binding on the MYH enhancer and linc-MYH expression. We used luciferase reporter transfection assays (as described above) to test the requirement for Six binding on the MYH enhancer to activate linc-MYH expression. These transient transfection assays, performed in adult TA, show that the MYH enhancer activates linc-MYH expression in a Six-dependent manner, as measured two weeks after electroporation (Figure 4B). lincRNAs can localize in cytoplasm [16] or as a single focus [19] or multiple foci [20] in nuclei. To analyze linc-MYH localization in skeletal muscle fiber, we performed fluorescent in situ hybridization (FISH), using linc-MYH sense and antisense RNA on isolated myofibers from fast EDL. Intranuclear localization of linc-MYH was observed with the antisense linc-MYH RNA probe, with approximately 10 linc-MYH foci per nucleus (n = 10, Figure 4C), while the sense linc-MYH RNA probe gave no signal (data not shown). We concluded from these experiments that linc-Myh RNA accumulates at specific sites in the nucleus of fast myofibers.
Fig. 4. linc-MYH is expressed in adult fast-type skeletal muscles and accumulates in their nuclei. 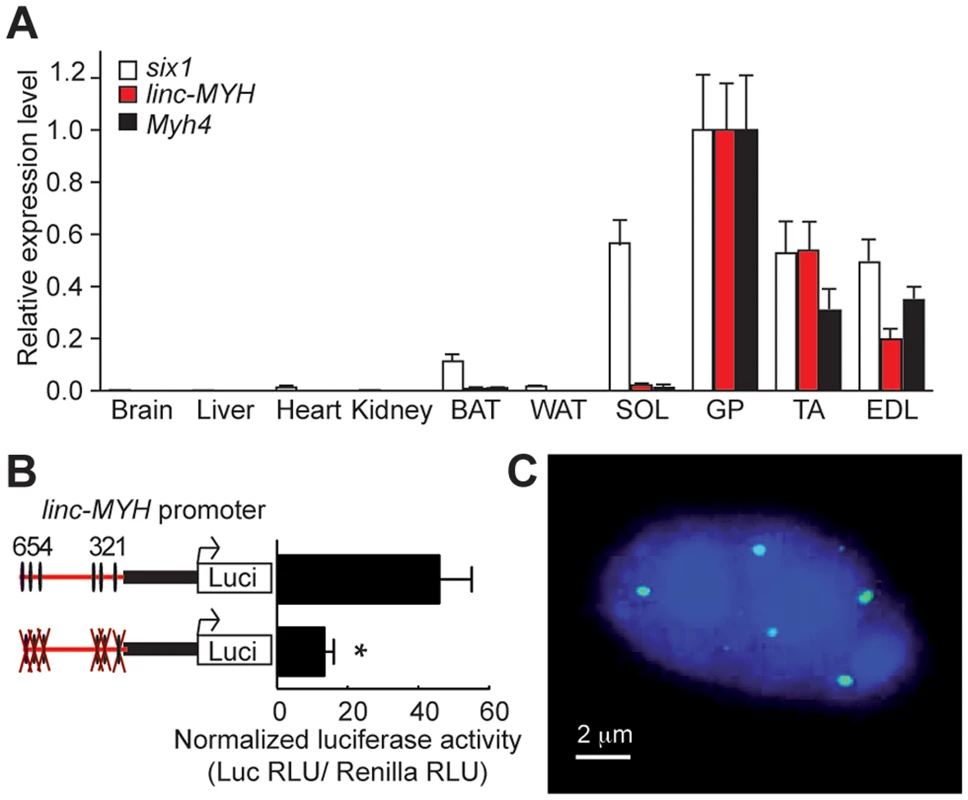
(A) Tissue distribution of Six1, Myh4 and linc-MYH RNAs. BAT, brown adipose tissue; WAT, white adipose tissue. (n = 4). (B) Luciferase assays of adult TA electroporated with linc-MYH promoter luciferase vectors (indicated) and a TK-Renilla luciferase vector (allowing normalization). *P<0.05. (C) FISH of isolated EDL myofiber with a linc-MYH antisense RNA fluorescent-labeled probe (green) and Dapi staining (blue). linc-MYH coordinates fiber-type gene expression
Due to the number of linc-MYH foci observed in fast type nuclei, we hypothesized that linc-MYH could act in trans [17] to control gene expression in fast myofibers. To test this theory, we used electroporation to introduce a shRNA against linc-MYH (shlinc-MYH) in TA muscle and analyzed the transfected samples after fourteen days. This method yielded the efficient knockdown of linc-MYH, with a 90% reduction of its expression (Figure 5A). To identify the consequences of linc-MYH knockdown and understand its mode of action, RNA samples from shlinc-MYH transfected adult TA were analyzed by Affymetrix microarrays (Figure 5D and Table S1), and validated by qPCR experiments (Figure 5A). The expression of linc-MYH was significantly lower in the absence of Six1, but Six1 expression was not affected by the absence of linc-MYH. Knockdown of linc-MYH led to robust gene expression modification; this knockdown strongly upregulated the expression of numerous slow genes (such as Sln, Tnni1, Tnnc1 and Tnnt1) and moderately downregulated the expression of several fast genes, (including Myh4, Tnnt3, Tnni2 and Pvalb) (Figure 5A). We noted that the expression of Sln and other slow genes remained far lower in linc-MYH knockdown TA than in wild type Soleus. This showed that linc-MYH knock down in TA is not sufficient to achieve the full transcription efficiency of these slow genes as is observed in SOL, where calcineurin and several kinases are required to achieve their efficient transcription [27]. In addition, and contrary to what was observed previously in the muscles of cSix1 KO mice, slow Myh7 expression level did not change in linc-MYH knock down TA. We next tested whether the transcription rate of Sln and Tnnt1 slow genes was modified in linc-MYH knock down TA by measuring their pre-mRNA expression level. As can be seen in Figure 5B we observed an upregulation of pre-mRNA of these slow genes after linc-MYH knock down, which is proportional to their respective mRNA accumulation, demonstrating that their transcription is increased after linc-MYH knock down in TA, and showing that linc-Myh can act in trans to decrease their transcription. Since we also observed a moderate down-regulation of the expression of fast genes in linc-MYH knock down TA, among which Myh4, we next tested whether the activity of the pGL3-Enhancer-Myh4 reporter could be modulated by the absence of linc-MYH. Two weeks after cotransfection of pGL3-Enhancer-Myh4 and shlinc-MYH in TA, we observed a statistically significant decrease of Luciferase activity, demonstrating the requirement of linc-MYH for efficient transcription of MYH4 (Figure 5C). Altogether these experiments show that accumulation of linc-Myh in the nuclei of fast myofibers participates in the transcriptional silencing of slow genes and is required for the full activation of fast genes.
Fig. 5. Slow-type gene expression is suppressed by linc-MYH. 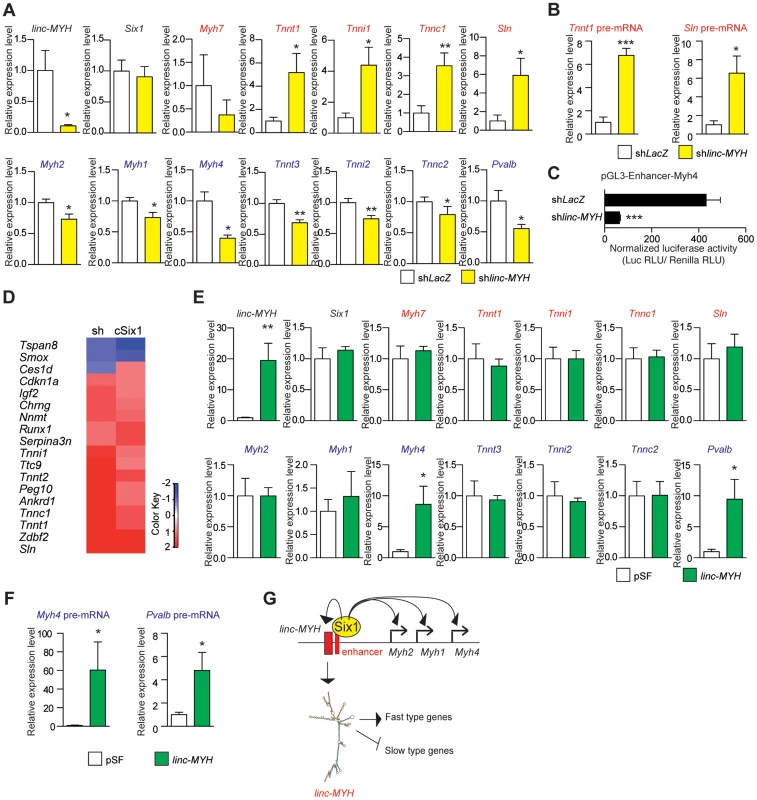
(A) qPCR experiments revealing mRNA expression levels of linc-MYH, Six1, slow-type genes (red) and fast-type genes (blue) in TA muscles expressing a shRNA directed against linc-MYH or LacZ (n = 5). (B) qPCR experiments revealing pre-mRNA expression of the Tnnt1 and Sln slow genes in TA muscles expressing a shRNA directed against linc-MYH or LacZ (n = 5) (C) Luciferase activity of pGL3-Enhancer-Myh4 in TA muscles expressing a shRNA directed against linc-MYH or LacZ (n = 4). (D) Microarray analysis of TA muscles transfected by shRNA against linc-MYH: a heat map of genes (red) upregulated to more than double the levels observed in cSix1KO. (E) qPCR experiments revealing mRNA expression levels of linc-MYH, Six1, slow-type genes (red) and fast-type genes (blue) in Soleus muscles expressing linc-Myh or the empty vector pSF (n = 4). (F) qPCR experiments revealing pre-mRNA expression of the Myh4 and Pvalb fast genes in Soleus muscles expressing linc-Myh or the empty vector pSF (n = 4) (G) A model of Six1 controlling the expression of the different MYH and of linc-MYH at the fast Myh locus in fast myofibers. Below, the hypothesis explaining the linc-MYH mode of action, as supported by the transcriptomic-wide analysis performed after linc-MYH knockdown in fast TA. *P<0.05, **P<0.01, ***P<0.001. Transcriptomic analysis of cSix1 and linc-MYH
We compared the networks of genes under the control of linc-MYH and of Six1 homeoprotein in adult muscles by the transcriptomic analysis of cSix1 and linc-MYH knockdown (Figure 5D and Table S2). We found that the six genes whose expression was the most increased in the linc-MYH knockdown were also significantly upregulated in cSix1 KO muscles (Figure S5). Besides slow muscle genes, two genes, Ankrd1 and Peg10, were more severely upregulated in the linc-MYH knockdown line (10 and 8 times, respectively) than in cSIX1 mutant myofibers (by 2.8 and 1.5 times, respectively). These non slow-type genes could be exclusively repressed by linc-MYH in adult fast myofibers since there is a stronger downregulation of linc-MYH accumulation after its knockdown than in cSix1 mutant myofibers. Transcriptomic analysis of adult myofibers deprived of either Six1 or of linc-Myh identified a strong qualitative and quantitative correlation in the expression of specific genes between linc-MYH knockdown and cSix1 adult mutant myofibers. The expression of slow muscle genes was 3 to 10 fold higher in linc-MYH knockdown samples, and 5 to 25 fold higher in cSix1KO samples, than in the wild type. We further showed that linc-MYH lies downstream of Six1 in the Six myogenic pathway and helps to repress slow muscle genes in fast myofibers. The downregulation of all fast-type genes (other than Myh4), and the upregulation of slow-type genes, was weaker in the linc-MYH knockdown than in the Six1cKO line. Six1 may control several inhibitory pathways, including the linc-MYH pathway, to prevent slow-type genes expression in adult fast myofibers. During fetal development, at a stage where linc-MYH expression is not yet activated, Six1/4 increases the nuclear accumulation of the slow muscle repressors Sox6 and HDAC4 to repress slow muscle gene expression [9],[28],[29]. In accordance with this, the expression of the slow genes Myh7, Sln, Tnni1 and Tnnt1 is upregulated in the muscle-specific cSox6 mutant [30]. This demonstrates that linc-MYH and Sox6, lying both downstream of Six1, directly participate in the downregulation of Sln, Tnni1 and Tnnt1 in fast myofibers. However, the repression of slow Myh7 in fast myofibers acts by a Six1-Sox6 dependent, but linc-MYH independent, repression mechanism. In this study, we observed that the levels of fast muscle gene expression decreased by 2–3 folds in the linc-MYH knockdown, with the highest decrease found for Myh4 expression. The expression of these genes decreased by a factor of 1.3 to 2.5 in cSix1KO, with the highest decrease found for Pvalb expression (Figures 3E and 5A). The presence of Six4 and Six5 proteins in adult myofibers [11], which have the same DNA binding specificity as Six1, could compensate its absence in cSix1 KO animals and enable the activation of downstream fast muscle targets. In this case, linc-MYH expression could be preferentially dependent upon Six1, rather than on Six4 or Six5. Altogether, these experiments suggest that the accumulation of linc-MYH transcripts in the nuclei of fast myofibers facilitates the regulation of a network of genes that drive myofiber specialization via the same pathway as Six1 and downstream of this transcription factor.
Forced expression of linc-MYH in adult slow Soleus
To test the possibility that linc-MYH could modulate the expression of specific muscle genes, we forced its expression in myogenic C2 cells and in primary myoblasts, where endogenous linc-MYH expression was faintly detectable even in myotubes four days after their differentiation (data not shown). Transfection of a 13 kb genomic fragment encompassing the whole linc-MYH gene lead to efficient linc-MYH RNA accumulation in myotubes, but after this forced expression, we were unable to detect any modification in the expression of slow or fast type genes (data not shown). These results suggest that specific cofactors of linc-MYH required for its appropriate functioning are lacking in cultured myotubes in culture, in agreement with the expression of linc-Myh only in adult fast type fibers. To circumvent the limitations of cultured cells, we turned to in vivo experiments in the Soleus in which linc-Myh is weakly expressed. Two weeks after linc-MYH gene transfection in the Soleus we observed that linc-MYH RNA accumulates up to approximately 80% of its expression level in TA (Figure 5E). We observed a selective upregulation of Myh4 and Pvalb mRNAs which were increased to approximately one-third their expression level observed in TA, while mRNA for slow genes remained unchanged (Figure 5E). To test whether the increase in Myh4 and Pvalb mRNA was due to an increased transcription of their genes, and potentially exclude a mechanism implicating mRNA stabilization, we measured pre-mRNA accumulation. As can be seen in Figure 5F, the transcription of these two fast genes is upregulated in the Soleus samples expressing linc-MYH proportionally to their mRNA accumulation, demonstrating that linc-Myh can work in trans and allow efficient activation of the transcription of specific fast genes. Absence of down regulation of the expression of slow genes in Soleus myofibers expressing linc-Myh suggests that, like in cultured myotubes, linc-Myh RNA needs specific protein-binding partners to achieve its function. Such specific protein-binding partners may be absent in Soleus as well as in cultured myotubes. Nuclear long non coding RNA are known to guide chromatin modifiers to specific gene loci, and by recruiting histone modifiers or DNA methyltransferase to modulate their transcription rate [31]. Potential linc-Myh protein partners expressed differentially in fast and slow adult myofibers may explain how linc-Myh efficiently represses the transcription of slow genes and activates the transcription of fast genes in fast myofibers, while in slow myofibers its forced expression is only able to activate the transcription of fast genes.
Discussion
The commitment and maintenance of muscle fiber fast sub type specialization relies on the specific expression of one of the fast Myosin heavy chain gene present at the fast Myh locus, and of specific isoforms of sarcomeric genes [1],[2],[4]. Myosin heavy chains are the primary determinant of the efficiency of muscle contraction. In this manuscript, we identified a novel mechanism for the specialization of the fast-myofiber subtype. We show that the long intergenic non-coding RNA linc-MYH and fast MYH genes, both of which are essential for myofiber specification, share a common enhancer which is regulated by Six1 homeoproteins. The linc-MYH specifically accumulates in nuclei of adult fast myofibers. Its function, as revealed here by in vivo knockdown and transcriptome-wide analysis, is to prevent slow-type muscle gene transcription and increase fast-type muscle gene expression in fast-type myofibers. We found linc-MYH downregulates the transcription of genes associated with slow muscle contractile properties like the slow genes Tnn and Sln (a known repressor of Serca1/Atp2a1 protein [32],[33] involved in Ca++ reuptake by the sarcoplasmic reticulum). These genes, which belong to the muscle contractile machinery and are repressed in adult fast myofiber, are positively controlled by Six1 in myogenic C2 cells [34], where linc-MYH expression is not detected. This suggests that their expression in adult fast myofiber may be restricted by an additional level of regulation involving the Six1-linc-MYH axis. As a result of our study we suggest that Six1 controls the acquisition of fast-type myofiber mechanical properties by binding to a single enhancer region of the fast Myh locus. It promotes the coordinated expression of fast Myhs and that of a strong repressor of genes controlling slow contractile properties. The modulation of Six activity (depending on fiber-type) facilitates changes in the expression levels of the fast genes Myh and Tnn; these changes are required for the formation of efficient sarcomeric units and the appropriate Ca++ cycling and excitation/contraction/relaxation coupling [1]–[4]. The Myh enhancer element therefore connects distinct regulatory hubs to achieve ultimate muscle fiber specialization. In this context, linc-MYH functions as an end-of-the-chain control element, conveying information on the state of fast Myh enhancer activity to repress slow-type specific genes and coordinates a finer level of regulation. This genomic organization at the fast Myh locus is reminiscent of the slow Myh7 locus where two microRNA miR-208b and miR-499 involved in fast myofiber program repression are co-regulated with Myh7 [35]. The precise molecular interactions between linc-Myh and higher order chromatin modifying complexes remains to be identified, to explain how linc-Myh coordinates the activation of target genes at specific sites in the nucleus, and the repression of others.
Materials and Methods
Mice, ethics statement
Animals were bred and handled as recommended by European Community guidelines. Experiments were performed in accordance with the guidelines of the French Veterinary Department. cSix1KO mice were obtained by breeding the Six1-LoxP mice [26] and transgenic mice expressing a CRE recombinase under the control of the human skeletal actin promoter (HSA) [25].
ChIP experiments
GP and TA muscles of 2 months old mice were minced with scissors just after sampling and fixed in 1% formaldehyde for 10 minutes. Formaldehyde was quenched by addition of 0.125 M glycine, and muscles were washed twice in PBS. The muscles were incubated on ice in lysis buffer (10 mM Tris-HCl pH 7.9, 85 mM KCl, 0.5% NP40, protease inhibitors (cOmplete, Roche)) for 10 minutes and homogenized with a mortar and, subsequently with a Dounce homogenizer. The nuclei were obtained by centrifugation, incubated in SDS lysis buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 1% SDS, protease inhibitors) for 10 minutes, and sonicated in a Bioruptor apparatus (Diagenode). The debris were removed by centrifugation. The sonicated DNA was incubated with 1 µg of Six1 antibodies (HPA001893, Sigma) under agitation at +4°C overnight. 20 µl of Dynabeads protein G (Invitrogen) were added to the samples and incubated under rotation at +4°C for 1 hour. The beads were washed with low-salt buffer (2 mM EDTA, 20 mM Tris-HCl pH 8, 150 mM NaCl, 1% TritonX-100, 0.1% SDS), high salt buffer (2 mM EDTA, 20 mM Tris-HCl pH 8, 0.5M NaCl, 1% TritonX-100, 0.1% SDS), LiCl buffer (1 mM EDTA, 10 mM Tris-HCl pH 8, 0.25M LiCl, 1% NP40, 1% deoxycholate) and TE buffer (1 mM EDTA, 10 mM Tris-HCl pH 8). The DNA was eluted with elution buffer (1% SDS, 0.1 M NaHCO3) containing 0.1 mg/ml proteinase K (Invitrogen) at 62°C for 2 hours, and, proteinase K was inactivated by incubation at 95°C for 10 minutes. The DNA was finally purified with MinElute PCR purification kit (Qiagen). The amount of specific amplified DNA is normalized by beta-Actin promoter amplification. The sequences of the oligonucleotides used in this study are as follows. Enh 1F, 5′-ATC TCC ACC TCC CTC CAA CT; Enh 1R, 5′-ACC CCC TAG CTT TGA CAG GT; Enh 2F, 5′-AAT CTG ACG ACA GGG TGA GC; Enh 2R, 5′-GGT CGC CTG ACC TGA TAG AG; AldoaF, 5′-CTC TCA AGG CAA ACC AAA GC; AldoaR, 5′-CCA GTG TCC CAG ACC TTC TC; ActbF, 5′-TGT TAC CAA CTG GGA CGA CA; ActbR, 5′-ACC TGG GTC ATC TTT TCA CG, NCF, 5′-ATC CTG CCC CAC TGT GTT AG; NCR, 5′-GCC AGC AAT TTG GTT TGA AT.
3C experiments
3C experiments were performed as described [36] with few modifications. Single myofibers were obtained from adult EDL muscles as previously described [26], cross-linked in 2% formaldehyde, 10 mM Tris-HCl pH 7.9, 85 mM KCl, 0.5% NP40 for 10 min at room temperature. Crosslinking reaction was quenched by 1 M glycine. Cross-linked myofibers were lysed for 10 minutes with lysis buffer (10 mM Tris-HCl pH 7.9, 85 mM KCl, 0.5% NP40) on ice, and the nuclei were harvested. Nuclei were resuspended in appropriate restriction enzyme buffer, 0.3% SDS and incubated for 1 hour at 37°C with shaking. Triton X-100 was added to 2%, and samples were incubated for 1 hour at 37°C. Samples were digested with Hind III overnight at 37°C. DNA ligation was performed for 4 hours at 16°C and for 30 minutes at room temperature. Cross-links were reversed, and DNA was then purified by phenol extraction and ethanol precipitation. To correct for the PCR amplification efficiency of different primer sets, a BAC clone containing the mouse Myh locus (RP23-61C14) was digested, ligated and used as control templates. Quantification of the data was performed by quantitative real-time PCR using the Lightcycler 480 probe master (Roche Diagnostic). The sequences of the oligonucleotides used in this study are given in Table S3.
RNA preparation
TA, back, soleus and GP muscles were collected from cSix1 KO and control mice. Total RNAs were extracted by Trizol Reagent (Invitrogen) according to manufacturer's instruction.
cDNA synthesis and qPCR
RNAs were treated with DNase I (Turbo DNA-free, Invitrogen) and were reverse transcribed with Superscript III kit (Invitrogen) according to manufacture's instruction. Reverse transcription was performed with 1 µg of total RNA. Quantitative real time PCR (Light Cycler 480, Roche) was performed using Light Cycler 480 SYBR Green I Master Kit (Roche) according to the manufacturer's protocols. PCR was performed for 40 cycles of 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 15 seconds. Genes expression level was normalized by the expression level of the housekeeping gene Actb. The sequences of the oligonucleotides used in this study are given in Table S4. Pre-mRNA qPCR experiments to measure RNA transcription rate were performed in the same conditions. Reverse oligonucleotides were complementary to intronic sequences, while forward oligonucleotides were complementary to exonic sequences. Samples without reverse transcription were used as controls, and signal due to contaminating DNA was subtracted to the values obtained with cDNA. We noticed that genomic DNA contamination was very low (less than hundred fold level of qPCR value observed with cDNA).
Muscle contraction test
Skeletal muscle function was evaluated by measuring in situ muscle contraction, as described previously [37]. 12 week-old male mice were anesthetized (intraperitoneal injection of pentobarbital sodium, 50 mg/kg). Body temperature was maintained at 37°C using radiant heat. The distal tendon of the TA muscle was attached to an isometric transducer (Harvard Bioscience) using a silk ligature. The sciatic nerves were proximally crushed and distally stimulated by a bipolar silver electrode using supramaximal square wave pulses of 0.1 ms duration. Responses to tetanic stimulation (pulse frequency 50–143 Hz) were successively recorded. Maximal forces were determined at optimal length (length at which maximal force was obtained during the tetanus). Fatigue resistance was then determined after a 5-minutes rest period. The muscle was continuously stimulated at 50 Hz for 2 minutes (sub-maximal continuous tetanus), and the duration corresponding to a 20% decrease in force was recorded.
RNA-FISH
Fluorescent-labeled antisense linc-MYH probes were synthesized according to manufacturer's instruction (FISH Tag RNA kit, Invitrogen). FISH experiments were performed on isolated EDL myofibres and images acquired on a Leica SP2 confocal microscope.
Generation of shRNA against mouse linc-MYH
Five distinct shRNAs targeting mouse linc-MYH were designed, called shlincMYH, and inserted into the psiSTRIKE hMGFP system (Promega). The efficiency of each shRNA was established by determination of linc-MYH transcript levels in TA muscles transfected by each shlincMYH. The shRNA against 5′-TTC TGC TCA CCA CCT ACA ATT-3′ sequence was selected for the knockdown experiment. For knock down experiments using shlincMYH, a plasmid coding for shLacZ was electroporated in the contra-lateral TA as a negative control.
Electroporation
In vivo transfections were also carried out on 10-weeks old C57Bl6 mice. For each experimental conditions, three to five Tibialis anterior (TA) or Soleus (Sol) muscles belonging to different mice were used. Under isoflurane anesthesia, legs were shaved and muscles were pre-treated by injection of a sterile 0.9% NaCl solution containing 0.4 U of bovine hyaluronidase/µl two hours before plasmid injection. Ten µg of shRNA-expressing vector were introduced into TA muscles of 8 week-old mice by electroporation as previously described [11]. Two weeks following electroporation, TA myofibers expressing GFP were dissected under a Nikon SMZ1500 stereo microscope and frozen in liquid nitrogen before processing for Luciferase assays or RNA purification.
Immunohistochemistry
TA, soleus and gastrocnemius muscles were embedded in cryomatrix and quickly frozen in isopentane cooled with liquid nitrogen. Cryostat sections (10 µm) were fixed in 4% PFA, washed in PBS, permeabilized with 0.1% Triton X-100 and left for 1 hour in blocking solution (1× PBS, 1.5% goat serum, 0.1% Triton X-100). Rabbit poly-clonal antibodies directed against Laminin (Z0097, Dako) (1/100 dilution), and monoclonal antibodies against MYH7 (NOQ7.5.4D, Sigma) (1/1000 dilution), MYH2 (SC-71, Developmental Studies Hybridoma Bank) (1/20 dilution) and against MYH4 (BF-F3, Developmental Studies Hybridoma Bank) (1/20 dilution) were applied overnight at 4°C to the treated sections. The next day, after three washes with 1× PBS containing 0.05% Tween-20, cryosections were incubated for 1 h with appropriate fluorescent secondary antibodies (Alexa Fluor 488 goat anti-rabbit IgG 1/1000 dilution, Alexa Fluor 594 goat anti-mouse IgG 1/1000 dilution, Invitrogen). After three washes with 1× PBS containing 0.05% Tween 20, samples were mounted in Vectashield mounting medium.
Microarray
After validation of RNA quality with the Bioanalyzer 2100 (using Agilent RNA6000 nano chip kit), 50 ng of total RNA were reverse transcribed following the Ovation PicoSL WTA System (Nugen). Briefly, the resulting double-strand cDNA was used for amplification based on SPIA technology. After purification according to Nugen protocol, 5 µg of single strand DNA was used for generation of Sens Target DNA using Ovation Exon Module kit (Nugen). 2.5 µg of Sens Target DNA were fragmented and labelled with biotin using Encore Biotin Module kit (Nugen). After control of fragmentation using Bioanalyzer 2100, the cDNA was then hybridized to GeneChip Mouse Gene 1.0 ST (Affymetrix) at 45°C for 17 hours. After overnight hybridization, the ChIPs were washed using the fluidic station FS450 following specific protocols (Affymetrix) and scanned using the GCS3000 7G. The scanned images were then analyzed with Expression Console software (Affymetrix) to obtain raw data (cel files) and metrics for Quality Controls. The analysis of some of these metrics and the study of the distribution of raw data show no outlier experiment. RMA normalization was performed using R and normalized data was subjected to statistical tests.
EMSA
EMSA was carried out with Six1 full-length mouse cDNA cloned into the pCR3 vector (Clontech) as previously described [38]. Recombinant mouse Six1 protein was obtained with a T7 transcription/translation kit (Promega). The oligonucleotide containing double-stranded myogenin MEF3 site was incubated with recombinant proteins. Competition experiments were performed in the presence of a ten-fold and hundred-fold molar excess of unlabeled identified Myh enhancer MEF3 sites (Enh1 to Enh6) or mutated Myh MEF3 sites (mtEnh1 to mtEnh6), or Myogenin promoter NFI or MEF3 sites. The sequences of the oligonucleotides used are as follows, the MEF3 consensus sequence is underlined; Enh1F 5′-CTC TTG GGT AAC TGG AGC CCC TC-3′. Enh1R 5′-GAG GGG CTC CAG TTA CCC AAG AG-3′. Enh2R 5′-GGT TGA CTT AGA TTT CCT TAT GA-3′. Enh2F 5′-TCA TAA GGA AAT CTA AGT CAA CC-3′. Enh3F 5′-TGT AAG AGA AAC TGA AAT AAA AT-3′. Enh3R 5′-ATT TTA TTT CAG TTT CTC TTA CA-3′. Enh4F 5′-GGG GTA AGA AAT CTG ACG ACA GG-3′. Enh4R 5′-CCT GTC GTC AGA TTT CTT ACC CC-3′. Enh5F 5′-CTA TCA GGT CAG GCG ACC TCA GT-3′. Enh5R 5′-ACT GAG GTC GCC TGA CCT GAT AG-3′. Enh6F 5′-CGT CAA GGA AAC CTT ATT CCA TC-3′. Enh6R 5′-GAT GGA ATA AGG TTT CCT TGA CG-3′. MyogF 5′-TGG GGG GGC TCA GGT TTC TGT GGC GT-3′. MyogR 5′-ACG CCA CAG AAA CCT GAG CCC CCC CA-3′. NF1F 5′-TAT CTC TGG GTT CAT GCC AGC AGG G-3′. NF1R 5′-CCC TGC TGG CAT GAA CCC AGA GAT A-3′. mtEnh1F 5′-CTC TTG GGT AGG ATC CGC CCC TC-3′. mtEnh1R 5′-GAG GGG CGG ATC CTA CCC AAG AG-3′. mtEnh2F 5′-GGT TGA CGA ATT CTT GCT TAT GA-3′. mtEnh2R 5′-TCA TAA GCA AGA ATT CGT CAA CC-3′. mtEnh3F 5′-TGT AAG ACC AAC TGA AAT AAA AT-3′. mtEnh3R 5′-ATT TTA TTT CAG TTG GTC TTA CA-3′. mtEnh4F 5′-GGG GTA AGA AGG ATC CCG ACA GG-3′. mtEnh4R 5′-CCT GTC GGG ATC CTT CTT ACC CC-3′. mtEnh5F 5′-CTA TCA GGT CGG ATC CCC TCA GT-3′. mtEnh5R 5′-ACT GAG GGG ATC CGA CCT GAT AG-3′. mtEnh6F 5′-CGT CAA GGA AGG ATC CTT CCA TC-3′. mtEnh6R 5′-GAT GGA AGG ATC CTT CCT TGA CG-3′.
Western blot
Western blots were performed with protein extracts of GP and soleus muscles from cSix1KO mice and control mice as previously described [9]. 1∶1000 dilutions of anti-Six1 antibodies (HPA001893, Sigma) or anti-β−tubulin antibodies (2128, Cell Signaling) were used.
Statistical analysis
All graphs represent mean values ± SEM. Significant differences between mean values were evaluated using two-tailed, unpaired Student's t test (when two groups were analyzed) or one-way ANOVA followed by Student Newman-Keuls test (for three or more groups).
Plasmids construction
For the construction of the pGL3-Myh2/1/4, c57bl6N mouse DNA was first used as a template to clone 1.1 kbp promoters of Myh2/1/4 with forward KpnI/SacI 5′-TTC AGA AAC TGC ATC ACT TAA A-3′ and reverse MluI, 5′-GCA GCT CGG GCA GTG GCC AGT GT-3′, forward KpnI/SacI 5′ - CAT ATC TGC ATC TCT AGA TAC C-3′ and reverse MluI, 5′ - GGC AGC AGC AGC CAG GAT GTG T-3′, forward KpnI/SacI 5′ - ACC GCT AGC CTT GAG CCT TTG-3′ and reverse MluI, 5′ - ATA GCG AGA GCC CTT TGT TCT C-3′, respectively. Myh2/1/4 promoter fragments were subsequently inserted into an KpnI-MluI digested pGL3 basic plasmid. For the construction of the pGL3-Enhancer-Myh2/1/4, mouse DNA was first used as a template to clone the enhancer with forward KpnI 5′ - GCG TTT CTA ATT CGG CTT GAA C-3′ and reverse SacI, 5′ - CAT TTC CTT CCT CTA AAG GCT CTT TATT C-3′. This enhancer fragment was subsequently inserted into KpnI-SacI digested pGL3-Myh2/1/4 plasmids. For the construction of the pGL3-mutEnhancer-Myh2/1/4, the six MEF3 sites of the enhancer were mutated as follows; MEF3 1 : 5′-GTAACTGGA to 5′-GTAGGATCC; MEF3 2 : 5′-TTAGATTTC to 5′-GAATTCTTG; MEF3 3 : 5′-GAAACTGAA to 5′-CCAACTGAA; MEF3 4 : 5′-GAAATCTGA to 5′-GAAGGATCC; MEF3 5 : 5′-GTCAGGCGA to 5′-GTCGGATCC; MEF3 6 : 5′-GAAACCTTA to 5′-GAAGGATCC. All plasmids sequence was confirmed by sequencing.
For the construction of the pSF-pA-CMVe-linc-MYH, genomic DNA fragment containing linc-MYH was obtained from digestion of a BAC clone containing (RP23-61C14) by BsaWI and AvrII. The 13.3 kbp DNA fragment was subsequently inserted into a SpeI-XmaI digested pSF-pA-CMVe plasmid. For linc-Myh gain of function experiments, the empty pSF-pA-CMVe plasmid was electroporated in the contra-lateral Soleus as a negative control. For the construction of the pGL3-Actb, pGL3-Sln and pGL3-Tnni1, mouse DNA was first used as a template to clone the promoters of Actb, Sln, and Tnni1 with forward 5′ - TTT CTC TAT CGA TAG GTA CCT TTG AGC TCC TGA CCC CGT GTG TAG CTC T-3′ and reverse 5′ - GAG CCC GGG CTA GCA CGC GTA AGG AGC TGC AAA GAA GCT G-3′, forward 5′ - TTT CTC TAT CGA TAG GTA CCT TTG AGC TCT ACC GAC TAT CAT GCC CAC A-3′ and reverse MluI, 5′ - GAG CCC GGG CTA GCA CGC GTC AGG CTA CCA AGG ACC TCA G-3′, forward 5′ - TTT CTC TAT CGA TAG GTA CCT TTG AGC TCC TGG GAT TTG AAC CCA TGA C-3′ and reverse 5′ - GAG CCC GGG CTA GCA CGC GTC CTC ACC ACA GAC TGC AGA G-3′, respectively. Actb, Sln, and Tnni1 promoter fragments were subsequently inserted into a KpnI-MluI site of pGL3 basic plasmid by GeneArt kit (Life Technologies). For the construction of the pGL3-Enhancer-Actb, pGL3-Enhancer-Sln and pGL3-Enhancer-Tnni1, the enhancer fragment was subsequently inserted into an KpnI-SacI site of pGL3-Actb, pGL3-Sln and pGL3-Tnni1plasmids. All plasmids sequence were confirmed by sequencing.
Luciferase assays
Two µg of Luciferase-expressing vector and one hundred ng of pRL-TK vector (Promega) were introduced into TA muscles of 8 week-old mice by electroporation as previously described [11]. Two weeks following electroporation, the TA muscles were dissected and frozen in liquid nitrogen before processing. The TA muscles were homogenized in Passive Lysis Buffer (Dual-Luciferase Reporter Assay System, Promega) and rotated for 15 minutes. The homogenate were centrifuged to remove debris, and the supernatant was used for measurement of Luciferase activity according to manufacture's instruction (Dual-Luciferase Reporter Assay System, Promega).
Computational analysis
In order to computationally identify MEF3 binding sites, we built a PWM (position-specific weight matrix) for MEF3, starting from a list of 15 binding sites (see Table S5) that were previously tested by Electrophoretic Mobility Shift Assay on the basis of their proximity to the MEF3 Myogenin consensus GAAACCTGA [10]. The PWM (shown in Figure S6 and Table S6) was generated by running the de novo motif finder Imogene [39] on small DNA fragments (see Table S5) that contained these binding sites. Imogene used phylogeny to enrich mouse set of DNA fragments with orthologs in 11 other mammalian sequenced genomes and to produce a refined PWM. The information content of the PWM (the genmot Sg parameter of Imogene) was set to 8.7 bits. Binding sites were predicted using a prediction threshold (the scangen Ss parameter of Imogene) of 9 bits and requiring conservation, as explained in [39].
Supporting Information
Zdroje
1. GundersenK (2011) Excitation-transcription coupling in skeletal muscle: the molecular pathways of exercise. Biol Rev Camb Philos Soc 86 : 564–600.
2. SchiaffinoS, ReggianiC (2011) Fiber types in mammalian skeletal muscles. Physiol Rev 91 : 1447–1531.
3. BraunT, GautelM (2011) Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol 12 : 349–361.
4. GreisingSM, GranseeHM, MantillaCB, SieckGC (2012) Systems biology of skeletal muscle: fiber type as an organizing principle. Wiley Interdiscip Rev Syst Biol Med 4 : 457–473.
5. ShragerJB, DesjardinsPR, BurkmanJM, KonigSK, StewartSK, et al. (2000) Human skeletal myosin heavy chain genes are tightly linked in the order embryonic-IIa-IId/x-ILb-perinatal-extraocular. J Muscle Res Cell Motil 21 : 345–355.
6. PalstraR-J, de LaatW, GrosveldF (2008) Beta-globin regulation and long-range interactions. Adv Genet 61 : 107–142.
7. GrifoneR, DemignonJ, HoubronC, SouilE, NiroC, et al. (2005) Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development 132 : 2235–2249.
8. RelaixF, DemignonJ, LaclefC, PujolJ, SantoliniM, et al. (2013) Six Homeoproteins Directly Activate Myod Expression in the Gene Regulatory Networks That Control Early Myogenesis. PLoS Genet 9: e1003425.
9. RichardA-F, DemignonJ, SakakibaraI, PujolJ, FavierM, et al. (2011) Genesis of muscle fiber-type diversity during mouse embryogenesis relies on Six1 and Six4 gene expression. Dev Biol 359 : 303–320.
10. NiroC, DemignonJ, VincentS, LiuY, GiordaniJ, et al. (2010) Six1 and Six4 gene expression is necessary to activate the fast-type muscle gene program in the mouse primary myotome. Dev Biol 338 : 168–182.
11. GrifoneR, LaclefC, LopezS, DemignonJ, GuidottiJ, et al. (2004) Six1 and Eya1 Expression Can Reprogram Adult Muscle from the Slow-Twitch Phenotype into the Fast-Twitch Phenotype. Mol Cell Biol 24 : 6253–6267.
12. LaclefC, HamardG, DemignonJ, SouilE, HoubronC, et al. (2003) Altered myogenesis in Six1-deficient mice. Development 130 : 2239–2252.
13. MercerTR, MattickJS (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20 : 300–307.
14. LeeJT (2012) Epigenetic Regulation by Long Noncoding RNAs. Science (80-) 338 : 1435–1439.
15. GuttmanM, RinnJL (2012) Modular regulatory principles of large non-coding RNAs. Nature 482 : 339–346.
16. CesanaM, CacchiarelliD, LegniniI, SantiniT, SthandierO, et al. (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147 : 358–369.
17. RinnJL, ChangHY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81 : 145–166.
18. CabiancaDS, CasaV, BodegaB, XynosA, GinelliE, et al. (2012) A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149 : 819–831.
19. WangKC, YangYW, LiuB, SanyalA, Corces-ZimmermanR, et al. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472 : 120–124.
20. YangL, LinC, LiuW, ZhangJ, Ohgi Ka, et al. (2011) ncRNA - and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147 : 773–788.
21. TsaiM-C, ManorO, WanY, MosammaparastN, WangJK, et al. (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329 : 689–693.
22. RinnJL, KerteszM, WangJK, SquazzoSL, XuX, et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129 : 1311–1323.
23. MartianovI, RamadassA, Serra BarrosA, ChowN, AkoulitchevA (2007) Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445 : 666–670.
24. SchmitzK-M, MayerC, PostepskaA, GrummtI (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24 : 2264–2269.
25. MiniouP, TizianoD, FrugierT, RoblotN, Le MeurM, et al. (1999) Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res 27: e27.
26. Le GrandF, GrifoneR, MourikisP, HoubronC, GigaudC, et al. (2012) Six1 regulates stem cell repair potential and self-renewal during skeletal muscle regeneration. J Cell Biol 198 : 815–832.
27. Bassel-DubyR, OlsonEN (2006) Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem 75 : 19–37.
28. AnC-I, DongY, HagiwaraN (2011) Genome-wide mapping of Sox6 binding sites in skeletal muscle reveals both direct and indirect regulation of muscle terminal differentiation by Sox6. BMC Dev Biol 11 : 59.
29. PotthoffMJ, WuH, ArnoldMA, SheltonJM, BacksJ, et al. (2007) Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest 117 : 2459–2467.
30. QuiatD, Voelker Ka, PeiJ, Grishin NV, GrangeRW, et al. (2011) Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc Natl Acad Sci U S A 108 : 10196–10201.
31. FaticaA, BozzoniI (2014) Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15 : 7–21.
32. ToyoshimaC, IwasawaS, OgawaH, HirataA, TsuedaJ, et al. (2013) Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nature 495 : 260–264.
33. WintherA-ML, BublitzM, KarlsenJL, MøllerJV, HansenJB, et al. (2013) The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm. Nature 495 : 265–269.
34. LiuY, ChuA, ChakrounI, IslamU, BlaisA (2010) Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res 38 : 6857–6871.
35. Van RooijE, QuiatD, JohnsonBa, SutherlandLB, QiX, et al. (2009) A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 17 : 662–673.
36. HagègeH, KlousP, BraemC, SplinterE, DekkerJ, et al. (2007) Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2 : 1722–1733.
37. JoanneP, HourdéC, OchalaJ, CaudéranY, MedjaF, et al. (2012) Impaired adaptive response to mechanical overloading in dystrophic skeletal muscle. PLoS One 7: e35346.
38. GiordaniJ, BajardL, DemignonJ, DaubasP, BuckinghamM, et al. (2007) Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proc Natl Acad Sci U S A 104 : 11310–11315.
39. RouaultH, SantoliniM, SchweisguthF, HakimV (2014) Imogene: identification of motifs and cis-regulatory modules underlying gene co-regulation. Nucleic Acids Res doi: 10.1093/nar/gku209
40. GruberAR, LorenzR, BernhartSH, NeuböckR, HofackerIL (2008) The Vienna RNA websuite. Nucleic Acids Res 36: W70–4.
Štítky
Genetika Reprodukční medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 5
-
Všechny články tohoto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



