-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAn In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
Abnormal signaling by the epidermal growth factor receptor (EGFR) contributes to the development of various human diseases, including different cancer types. One important mechanism that controls intracellular signal transduction is by regulation of the subcellular receptor localization in the signal-receiving cell. We are investigating the regulation of the EGFR homolog LET-23 in the Nematode C. elegans by observing the localization of the EGFR in the epithelial cells of live animals. This approach has allowed us to study the dynamics of receptor trafficking in cells embedded in their natural environment and receiving physiological concentrations of various extracellular signals. In a systematic RNA interference screen, we have identified 81 genes controlling EGFR localization and signaling in different subcellular compartments. One new regulator of EGFR signaling identified in this screen encodes the Ezrin Homolog ERM-1. We show genetic and biochemical evidence indicating that ERM-1 is part of a buffering mechanism to maintain a pool of immobile EGFR in the basolateral membrane compartment of the epithelial cells. This mechanism permits the generation of a long-lasting EGFR signal during multiple rounds of cell divisions. The control of receptor localization is thus necessary for the precise temporal regulation of signal transduction during animal development.
Published in the journal: . PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004341
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004341Summary
Abnormal signaling by the epidermal growth factor receptor (EGFR) contributes to the development of various human diseases, including different cancer types. One important mechanism that controls intracellular signal transduction is by regulation of the subcellular receptor localization in the signal-receiving cell. We are investigating the regulation of the EGFR homolog LET-23 in the Nematode C. elegans by observing the localization of the EGFR in the epithelial cells of live animals. This approach has allowed us to study the dynamics of receptor trafficking in cells embedded in their natural environment and receiving physiological concentrations of various extracellular signals. In a systematic RNA interference screen, we have identified 81 genes controlling EGFR localization and signaling in different subcellular compartments. One new regulator of EGFR signaling identified in this screen encodes the Ezrin Homolog ERM-1. We show genetic and biochemical evidence indicating that ERM-1 is part of a buffering mechanism to maintain a pool of immobile EGFR in the basolateral membrane compartment of the epithelial cells. This mechanism permits the generation of a long-lasting EGFR signal during multiple rounds of cell divisions. The control of receptor localization is thus necessary for the precise temporal regulation of signal transduction during animal development.
Introduction
The formation of epithelial tissues involves the polarized distribution of growth factor receptors that determine cell proliferation and differentiation. Notably, changes in EGFR localization have a major impact on signaling and organogenesis [1]–[3].
In C. elegans, the let-23 gene encodes the sole member of the EGFR/ErbB family of receptor tyrosine kinases. let-23 is involved in a variety of developmental processes including the induction of the hermaphrodite vulva [4]. In early second stage (L2) larvae, LET-23 is expressed at equal levels in the six equivalent vulval precursor cells (VPCs) (P3.p through P8.p) (Figure 1A) [5], [6]. Beginning in the L2 stage, the gonadal anchor cell (AC) secretes the EGF ortholog LIN-3, which binds to LET-23 on the basolateral plasma membrane of the VPCs to activate the LET-60 RAS/MPK-1 MAPK signaling pathway [4] (Figure 1B). In order to reach high levels of receptor activity, LET-23 must be retained on the basolateral membrane of the VPCs by a ternary protein complex consisting of the PDZ-domain proteins LIN-2 CASK, LIN-10 MINT and LIN-7 VELIS. LIN-7 directly binds to the C-terminal PDZ binding motif of LET-23 [5]. The VPC that is nearest to the AC, P6.p, receives most of the inductive LIN-3 signal and hence adopts the primary (1°) cell fate. P6.p then produces several DELTA ligands, which induce via the NOTCH pathway the secondary (2°) cell fate in the neighboring VPCs P5.p and P7.p [7], [8] (Figure 1B). NOTCH signaling blocks RAS/MAPK signaling and results in the endocytosis and degradation of LET-23 in the 2° VPCs [9]–[11]. The distal VPCs P3.p, P4.p and P8.p, which receive only little inductive signal, down-regulate LET-23 expression and adopt the tertiary (3°), uninduced cell fate. As the pathway components are conserved, the study of vulval induction of the worm can be used to find new core components or those required for fine-tune the signaling output. For this purpose, we performed the first systematic in vivo screen for regulators of LET-23 EGFR localization and expression in live C. elegans larvae.
Fig. 1. Identification of genes regulating LET-23 EGFR localization and signaling. 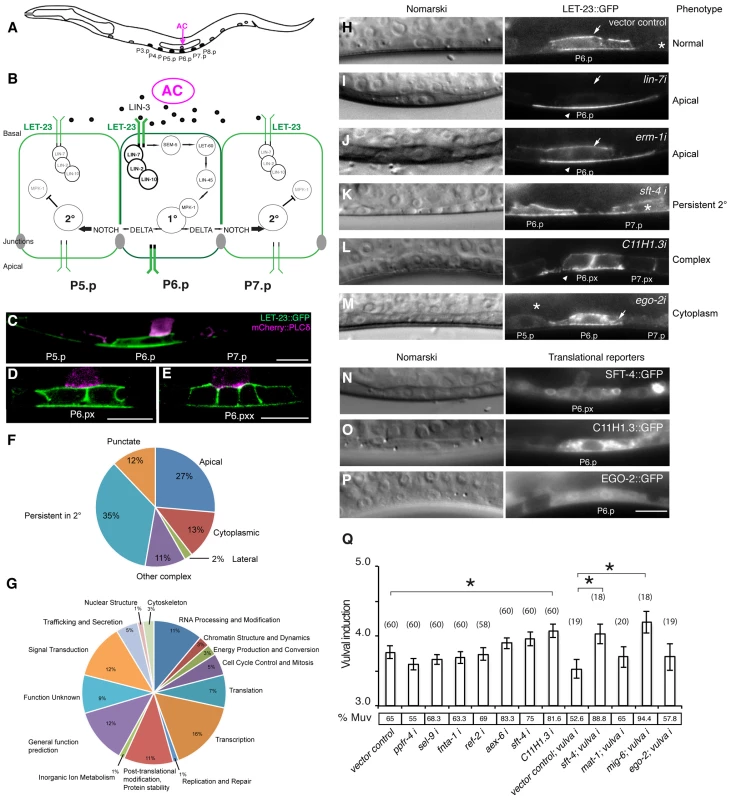
(A) Schematic drawing of an L2 larva with the location of the VPCs and AC. P5.p, P6.p and P7.p get induced to form the mature vulva. P3.p, P4.p and P8.p divide once and fuse to the hypodermis. (B) Overview of the LET-23 EGFR and NOTCH signaling network controlling 1° and 2° vulval fate specification. (C) LET-23::GFP expression (green) in P6.p of a late L2 larva during vulval induction. The AC is labeled with an mCherry::plcδPH reporter (magenta) [35]. Note the low LET-23::GFP levels in the 2° P5.p and P7.p. (D) Expression of the LET-23::GFP reporter in the 1° lineage at the Pn.px and (E) Pn.pxx stage. (F) Pie charts indicating the frequencies of the different classes of mislocalization phenotypes observed after RNAi and (G) the Clusters of Orthologous Groups (KOGs) of the 81 genes identified in the screen (H–M). Examples of different genes identified in the LET-23 localization screen. Left panels show the corresponding Nomarski images and right panels LET-23::GFP expression in the 1° cells and their neighbors (asterisks). (H) The negative empty vector control and (I) lin-7 RNAi as positive control. (J) erm-1 RNAi as an example for reduced basolateral (arrow) and increased apical localization (arrow head), (K) sft-4 RNAi with normal localization in P6.p but persistent expression in P7.p (asterisk), and (L) C11H1.3 RNAi (Pn.px stage) with punctate apical accumulation (arrow head). (M) ego-2 RNAi with cytoplasmic accumulation of LET-23::GFP in P6.p (arrow head) and P5.p (asterisk). (N) perinuclear localization of SFT-4::GFP in the vulval cells and the AC and (O) intracellular punctate expression of C11H1.3::GFP in P6.p. (P) Cytoplasmic and nuclear expression of EGO-2::GFP in P6.p. (Q) Vulval induction in let-60(n1046gf) larvae treated with different RNAi clones. Vulval induction (VI) indicates the average number of induced VPCs per animal. “vulva i” indicates Pn.p cell-specific RNAi in the rde-1(lf);let-60(n1046gf); [Plin-31::rde-1] background. %Muv (Multivulva) indicates the fraction of animals with VI>3. The numbers of animals scored are indicated in brackets. * Indicates p<0.05 as determined in a two tailed student's t-test - two-sample unequal variance. t-test values in RNAi: C11H1.3 (0.003), sft-4 (0.013), mig-6 (0.002). Error bars represent the standard error of the mean. The scale bars are 10 µm. Through this approach, we have identified 81 genes causing a variety of LET-23::GFP mislocalization phenotypes upon RNAi knock-down. A subset of these genes also controls the strength of the LET-23 EGFR signal produced in the VPCs. We have identified ERM-1, the homologue of mammalian Ezrin, Radixin and Moesin proteins, as a temporal regulator of LET-23 EGFR signaling. Based on our genetic and biochemical data, we propose that ERM-1 binds to and sequesters the LET-23 EGFR in an inactive compartment at or close to the basolateral membrane of the VPCs. In this manner, ERM-1 competes with the activating LET-23/LIN-2/LIN-7/LIN-10 complex [5]. ERM-1 may act as a buffer that prevents the immediate activation of the entire pool of basolateral LET-23 EGFR at vulval induction and thus allows the generation of a prolonged signal.
Results
An In Vivo Screen Identifies Novel Regulators of LET-23 EGFR Localization and Expression
We performed RNAi knock-down of all genes (705 clones) reported to exhibit a protruding vulva (Pvl) phenotype, which is indicative of a defect in vulval fate specification or execution (Table S1) and examined LET-23 localization and expression in the vulval epithelium of live L3 larvae expressing a functional LET-23::GFP reporter (Figure 1C–E). The LET-23::GFP reporter used showed the same vulval expression pattern as endogenous LET-23 detected by antibody staining [5], and LET-23::GFP protein levels in total extracts were comparable to endogenous LET-23 levels (Figure S1A). Moreover, let-23::gfp efficiently rescued the let-23(sy1) vulvaless (Vul) phenotype (Figure S1B), and RNAi against lin-7 or a mutation in lin-2 caused an apical mislocalization of LET-23::GFP, as shown previously for endogenous LET-23 by antibody staining [5] (Figure 1I, and Figure S1D). In total, we identified 81 candidates that change different aspects of LET-23::GFP expression or localization (Table 1). We further classified these genes according to the specific mislocalization phenotypes observed (Figure 1F): Apical enrichment (24 genes, Figure 1I,J), accumulation in intracellular punctae or uniform cytoplasmic distribution (23 genes, Figure 1M and Figure S2,C,D), persisting expression in the 2° cells (31 genes, Figure 1K and Figure S2E), enrichment on the lateral membrane (2 genes, Figure S2F) and complex mislocalization phenotypes (10 genes, Figure 1L). Grouping the 81 genes into Clusters of Orthologous Groups (KOGs) indicated that a variety of processes are involved in regulating LET-23 localization (Figure 1G) [12]. In particular, genes involved in transcription, intracellular trafficking, signal transduction and protein stability and posttranslational modification were slightly overrepresented, while genes involved in chromatin modification, DNA replication and repair were underrepresented when compared to the distribution of the KOGs among the genes causing a Pvl phenotype that were screened.
Tab. 1. Genes that control the localization or expression of LET-23::GFP. 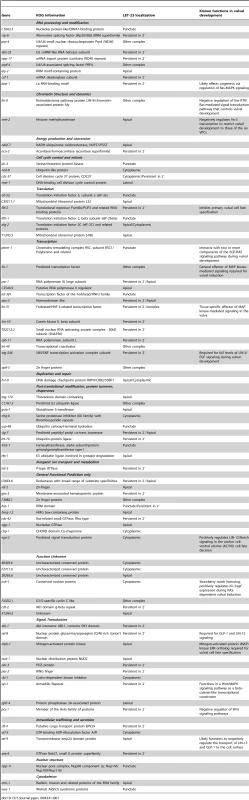
Changes in LET-23 EGFR Localization Alter RAS-Mediated Signaling
For a subset of the candidates with predicted roles in signaling or trafficking, we examined whether inhibition of these genes altered the activity of the RAS/MAPK pathway. This was tested by performing RNAi in the sensitized let-60 ras(n1046) gain-of-function background in which more than 3 VPCs are induced [13] and scoring the average number of induced VPCs per animal (Figure 1Q). It should be noted that the VPCs in the let-60 ras(n1046) gain-of-function background are still sensitive to the AC signal [13]. In those cases where RNAi caused a penetrant embryonic or larval lethal phenotype, we performed Pn.p cell-specific RNAi using an rde-1(ne219lf); let-60(n1046gf) RNAi resistant background expressing rde-1(wt) from the Pn.p cell-specific lin-31 promoter [14]. For example, RNAi against sft-4 or against the small GTPase aex-6 caused persistent LET-23::GFP expression in 2° VPCs (Figure 1K and Table 1). Moreover, Pn.p cell-specific sft-4 RNAi significantly enhanced vulval induction in the let-60(gf) background (Figure 1Q). The yeast sft-4 homolog ERV29 encodes a SURF protein with a putative di-lysine endoplasmic reticulum (ER) localization signal that sorts secretory cargo proteins in the ER into COPII vesicles [15]. An sft-4::gfp translational reporter showed expression in the VPCs in perinuclear structures that resemble the ER (Figure 1N). Thus, ER to Golgi transport might be involved in controlling LET-23 turnover in the VPCs.
RNAi of C11H1.3 caused a complex mislocalization phenotype with a moderate apical enrichment, punctate LET-23::GFP accumulation at or close to the apical membrane (Figure 1L) and an increase in vulval induction in the let-60(n1046gf) background (Figure 1Q). C11H1.3 encodes a predicted E3 ubiquitin ligase that is expressed in the VPCs in intracellular vesicles (Figure 1O). Therefore, C11H1.3 may control LET-23 localization and stability through ubiquitination of the receptor itself or of an associated factor. A penetrant mislocalization phenotype with punctate cytoplasmic accumulation of LET-23::GFP was observed in ego-2 RNAi treated animals (Figure 1M), and a translational ego-2::gfp reporter was expressed in the cytoplasm and nuclei of all the VPCs (Figure 1P). ego-2 encodes a BRO1 domain protein that is related to mammalian PTPN23, which regulates the transport of ubiquitinated EGFR through the ESCRT III complex to the intralumenal vesicles of multivesicular bodies [16]. Interestingly, ego-2 has also been reported to regulate GLP-1 NOTCH signaling during germ cell development and embryogenesis as well as LIN-12 NOTCH signaling during somatic gonad development [17]. Therefore, ego-2 might be a general regulator of LET-23 EGFR and LIN-12/GLP-1 NOTCH via control of their endocytic transport.
ERM-1 Controls LET-23 EGFR Trafficking
One of the strongest apical enrichment mislocalization phenotypes was observed in erm-1 RNAi treated animals (Figure 1J), prompting us to analyze the role of ERM-1 in LET-23 localization and signaling in more detail. erm-1 encodes the sole C. elegans member of the Ezrin, Radixin and Moesin (ERM) protein family. ERM proteins link the cortical actin cytoskeleton and the plasma membrane and recruit transmembrane proteins to specific membrane compartment [18]. In addition, C. elegans ERM-1 is required for apical lumen morphogenesis in the intestine [19] [20]. In contrast to the apical localization observed in the intestine, an ERM-1::mCherry reporter showed basolateral and junctional localization and a partial overlap with LET-23::GFP in the VPCs and their descendants (Figure 2A-A″). Only after vulval invagination (at the Pn.pxxx stage), ERM-1 relocalized to the apical, luminal plasma membrane of the vulval toroids (data not shown). To confirm the RNAi phenotype, we examined LET-23::GFP expression in erm-1(tm677) null mutants. Homozygous erm-1(tm677) larvae showed decreased basolateral and increased apical membrane localization of LET-23::GFP in the VPCs and their descendants, resulting in a significantly increased ratio of apical to basolateral LET-23::GFP signal intensity when compared to heterozygous erm-1(tm677)/+ controls (Figure 2B–D). The localization of the apical junction marker DLG-1::RFP [21] or the plasma membrane marker CED-10::GFP [22] were not changed, indicating that overall polarity of the VPCs was not altered in erm-1(tm677) mutants (data not shown and Figure S3). However, we detected a reduced basolateral staining of the F-actin reporter lifeAct::GFP [23] in the VPCs of erm-1(tm677) mutants, which is consistent with the role of ERM proteins as membrane linkers for cortical F-actin (Figure 2D–F′).
Fig. 2. ERM-1 controls LET-23::GFP localization at the basolateral membrane of the vulval cells. 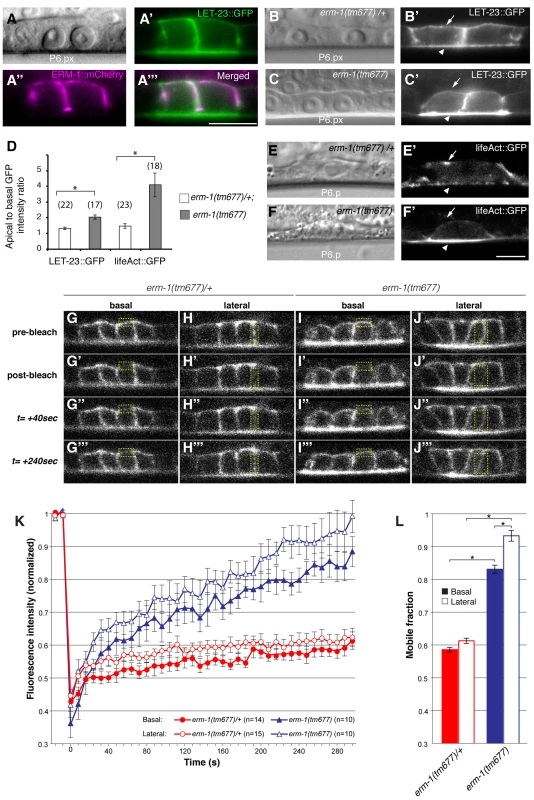
(A) Nomarski image, (A′) LET-23::GFP (green) and (A″) ERM-1::mCherry (magenta) expression at the Pn.px stage. (A′″) shows a merged image of (A′) and (A″) indicating partial co-localization at the basolateral membrane. (B) Nomarski and (B′) LET-23::GFP expression in a heterozygous erm-1(tm677)/+ and (C, C′) a homozygous erm-1(tm677) larva at the Pn.px stage. Arrows indicate the basal and arrowheads the apical membrane domains. (D) Apical to basal LET-23::GFP and lifeAct::GFP intensity ratios in P6.p in erm-1(tm677)/+ versus erm-1(tm677). The numbers of animals analyzed are indicated in brackets. Error bars represent the standard error of the mean. (E) Nomarski and (E′) lifeAct::GFP expression in P6.p of a heterozygous erm-1(tm677)/+ and (F, F′) a homozygous erm-1(tm677) larva. (G–J′″) Example images of the FRAP experiment at the time points indicated to the left of panels G-G′″. (G-G′″ and I-I′″) Basal and (H-H′″ and J-J′″) lateral membrane regions outlined with the dotted yellow boxes were photobleached in heterozygous erm-1(tm677)/+ and homozygous erm-1(tm677) larvae, respectively, at the Pn.pxxx stage. (K) Quantification of the FRAP experiments. The y axis indicates LET-23::GFP intensity normalized to the signal intensity measured before bleaching inside the bleached areas and to the total signal intensity in the cell, and the x-axis the time after photo-bleaching. The numbers of animals analyzed are shown in brackets. (L) Quantification of the mobile fraction from the FRAP curves. *Indicates p<0.05, as determined in a two tailed student's t-test - two-sample unequal variance. The scale bars are 10 µm. To test whether the reduced basolateral expression of LET-23::GFP is due to decreased basolateral secretion or to an increased membrane mobility and recycling rate of LET-23, we performed Fluorescence Recovery After Photobleaching (FRAP) experiments on the basal and lateral membranes of the vulval cells at the Pn.pxx stage and calculated the mobile fraction and half time of recovery (t1/2) of LET-23::GFP (Figure 2G–L) (see materials and methods). In erm-1(tm677) larvae, the total mobile fraction of LET-23::GFP was significantly higher than in heterozygous controls in both the basal and lateral compartments, while the t1/2 was not significantly changed (Figure 2L, t1/2 = 76 sec in heterozygous erm-1(tm677)/+ vs. 81 sec in homozygous erm-1(tm677) mutants). Thus, erm-1(tm677) mutants exhibit an increased mobility of LET-23::GFP on the basolateral plasma membrane, rather than a decreased rate of basolateral secretion or retention.
ERM-1 Inhibits Ligand-Dependent Internalization of LET-23 EGFR
Changes in the ligand concentrations could alter the steady-state levels of LET-23 EGFR on the basolateral membrane. For example, reducing the dose of LIN-3 EGF may decrease receptor endocytosis and thus diminish the ratio of apical to basal EGFR, while increasing the dose of LIN-3 may promote receptor endocytosis on the basolateral membrane and therefore increase the apical to basal ratio. On the other hand, mutations in components of the LIN-2/LIN-7/LIN-10 complex that is necessary to retain the EGFR on the basolateral membrane also cause a strong reduction in basolateral EGFR localization, yet they result in reduced receptor activation [5]. To distinguish between these different scenarios, we tested if the increased apical LET-23::GFP localization in erm-1(tm677) mutants could be due to a higher rate of LET-23 endocytosis after binding to LIN-3 EGF secreted from the AC. In lin-3(e1417) mutants, in which LIN-3 activity in the AC is strongly reduced [24], apical LET-23::GFP localization was nearly two-fold reduced (Figure 3A,C). However, in erm-1(tm677); lin-3(e1417) double mutants the apical to basal LET-23::GFP ratio was lower than in erm-1(tm677) but higher than in lin-3(e1417) single mutants (Figure 3B,C). Since the viable lin-3(e1417) allele used does not eliminate all LIN-3 activity, we conclude that the apical accumulation of LET-23::GFP in the absence of ERM-1 is at least in part ligand-dependent. On the other hand, a pulse of ectopic LIN-3 ubiquitously expressed under control of the heat-shock promoter hs::lin-3 [25] caused the almost complete disappearance of LET-23::GFP from the basolateral membrane and accumulation on the apical membrane within 230 minutes (Figure 3D,F). In homozygous erm-1(tm677) mutants, however, a LIN-3 pulse caused a smaller increase in the apical LET-23::GFP pool and persisting receptor expression on the basolateral membrane (Figure 3E,F). Thus, not only LET-23 endocytosis but also basolateral recycling are increased in erm-1(tm677) mutants, which is consistent with the increased mobile fraction of LET-23::GFP observed in the FRAP experiments (Figure 2L). By contrast, activation of the EGFR signaling pathway downstream of the receptor using for example the let-60(gf) mutation did not change LET-23::GFP localization (data not shown). Thus, the LIN-3 ligand stimulates and ERM-1 inhibits internalization and recycling of LET-23 on the basolateral membrane.
Fig. 3. The LIN-3 EGF ligand stimulates and ERM-1 inhibits internalization and recycling of LET-23 on the basolateral membrane. 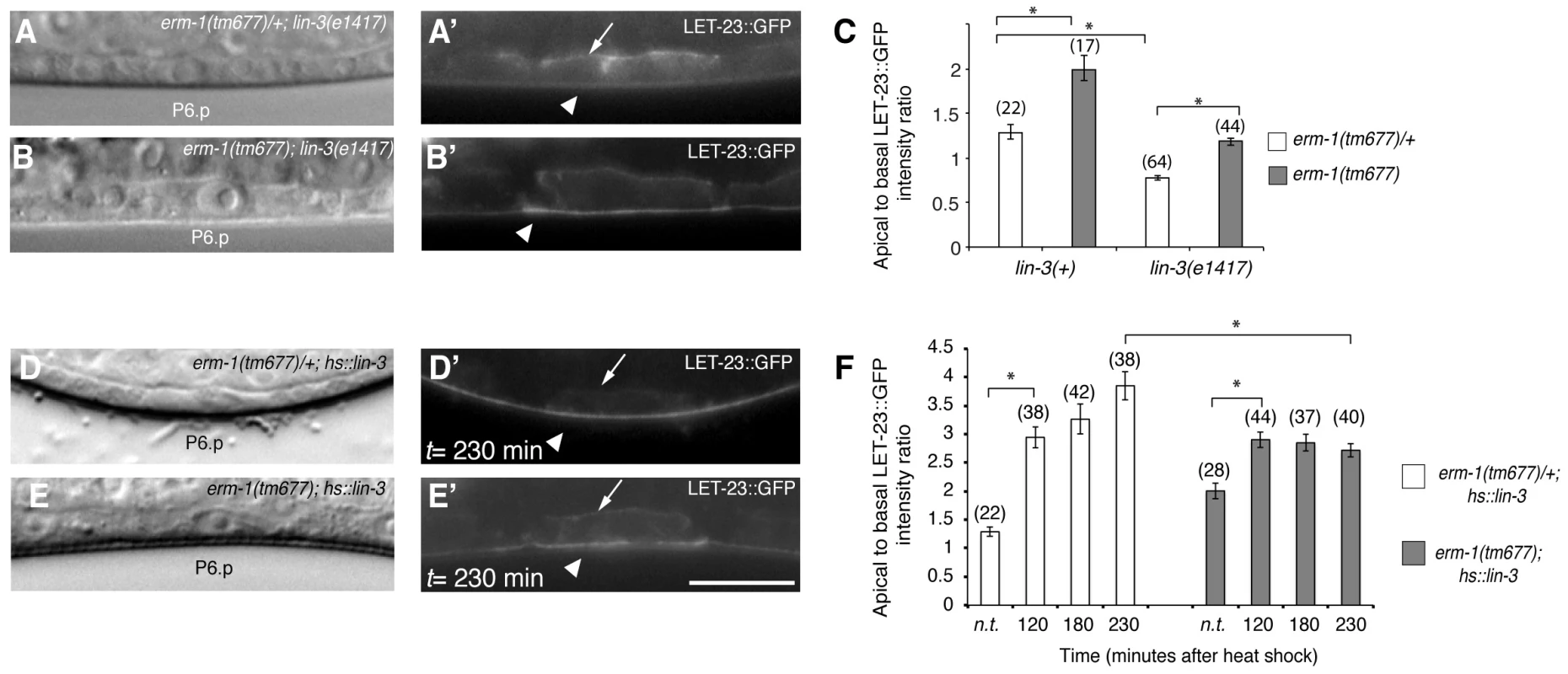
(A) Nomarski image and (A′) LET-23::GFP expression in erm-1(tm677)/+; lin-3(e1417) and (B, B′) erm-1(tm677); lin-3(e1417) double mutants at the Pn.p stage. Arrows indicate the basal and arrowheads the apical membrane domains. (C) Apical to basal LET-23::GFP intensity ratios in P6.p in erm-1(tm677)/+ versus erm-1(tm677) single and in erm-1(tm677)/+; lin-3(e1417) versus erm-1(tm677); lin-3(e1417) double mutants. (D) Nomarski image and (D′) LET-23::GFP expression in a heterozygous erm-1(tm677)/+ and (E,E′) a homozygous erm-1(tm677) mutant 230 minutes after heat-shock induction of LIN-3. (F) Apical to basal LET-23::GFP intensity ratios at different time points after heat-shock. n.t. indicates animals of the same genotype that were not subjected to a heat-shock. *Indicates p<0.001 as determined in a two tailed student's t-test - two-sample unequal variance. The scale bars are 10 µm. ERM-1 Acts as a Negative Regulator of the EGFR/RAS/MAPK Pathway
Enhanced receptor endocytosis could result in the attenuation of LET-23 signaling, while faster recycling to the plasma membrane could promote signaling [1]. To determine how the altered LET-23 dynamics in erm-1 mutants affects signaling, we performed epistasis analysis by combining erm-1(tm677) with mutations in different components of the EGFR/RAS/MAPK pathway [4] and quantifying vulval induction. In erm-1(tm677) single mutants, the three proximal VPCs were always induced as in the wild-type (Figure 4A). However, in let-60(gf); erm-1(tm677) double mutants, the average number of induced VPCs was significantly increased compared to let-60(gf); erm-1(tm677)/+ controls, resulting in an enhanced Multivulva (Muv) phenotype (Figure 4A). Thus, ERM-1 negatively regulates RAS/MAPK signaling during vulval induction. Mutations in the lin-2/lin-7/lin-10 receptor localization complex or in the PDZ binding motif in let-23(sy1) cause a penetrant Vulvaless (Vul) phenotype because LET-23 mislocalized to the apical membrane cannot bind to LIN-3 [5]. Interestingly, erm-1(tm677) partially suppressed the lin-2(n397), lin-7(e1413), lin-10(e1439) and let-23(sy1) Vul phenotypes (Figure 4A), indicating that ERM-1 inhibits vulval induction independently of the LIN-2/LIN-7/LIN-10 receptor localization complex. However, the suppression of the lin-2(n397) Vul phenotype was not accompanied by a visible re-localization of LET-23::GFP to the basolateral membrane (data not shown). In contrast, erm-1(tm677) did not suppress the lin-3(e1417) Vul phenotype, suggesting that vulval induction in erm-1(tm677) mutants still depends on the AC signal (Figure 4A).
Fig. 4. ERM-1 negatively regulates vulval induction and binds to LET-23. 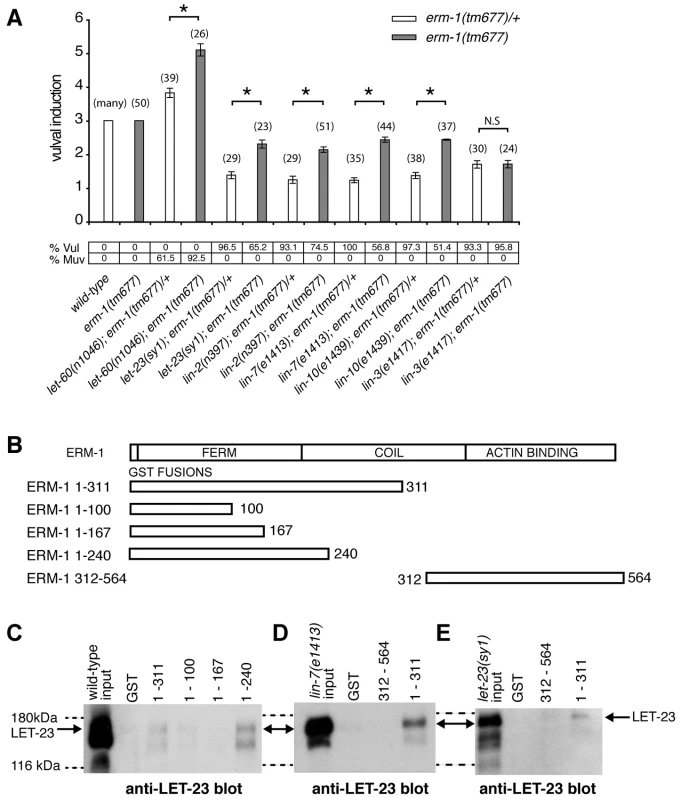
(A) Genetic epistasis analysis between erm-1 and components of the egfr/ras/mapk pathway. Vulval induction (VI) indicates the average numbers of induced VPCs in different double mutant combinations scored in erm-1(tm677) heterozygous (white bars) versus homozygous (gray bars) animals. %Vul indicates the fraction of animals with VI<3 and % Muv the fraction of animals with VI>3. The numbers of animals scored for each genotype are indicated in brackets. N.S: no significant change. *Indicates p<0.05 as determined in a two tailed student's t-test - two-sample unequal variance. (B) Structures of the GST::ERM-1 fusion proteins tested for LET-23 binding. (C) Interaction of LET-23 from wild-type extracts with different GST::ERM-1 fusion proteins detected on an anti-LET-23 Western blot. (D) Binding of LET-23 extracted from lin-7(e1413) and (E) from let-23(sy1) mutants to GST::ERM-1 proteins. The dashed lines indicate the approximate positions of the 180 kDa and 116 kDA protein standards. ERM-1 Forms a Complex with LET-23 EGFR Independently of the LIN-2/LIN-7/LIN-10 Complex
ERM proteins are composed of an N-terminal FERM domain and a C-terminal actin-binding domain [26]. They can switch from a closed, inactive conformation in the cytoplasm to an open, active conformation at the plasma membrane [27]. The FERM domain in the open conformation interacts with plasma membrane proteins either directly or indirectly through adaptor proteins [26], while binding of the actin cytoskeleton to the C-terminus of ERM proteins regulates the activity of the entire complex [28]. Since our genetic analysis indicated that ERM-1 controls LET-23 signaling independently of the LIN-2/LIN-7/LIN-10 complex, we tested if ERM-1 and LET-23 might exist in an alternate complex. For this purpose, different portions of purified recombinant ERM-1 tagged at the N-terminus with glutathione S-transferase (GST) were incubated with total worm lysates, and bound LET-23 was detected on Western-blots. LET-23 from wild-type worm extracts bound to N-terminal fragments containing the entire FERM domain (GST::ERM-11–240 and GST::ERM-11–311), while a C-terminal fragment (GST::ERM-1312–564) or truncated FERM domains (GST::ERM-11–100 or GST::ERM-11–167) did not bind LET-23 (Figure 4B,C). Moreover, LET-23 extracted from sy1 mutants, in which LET-23 lacks the PDZ binding motif, or from lin-7(e1413) mutants still bound to the ERM-1 FERM domain (Figure 4D,E). Thus, LET-23 and ERM-1 exist in a complex that is distinct from the LIN-2/LIN-7/LIN-10 localization complex.
ERM-1 Is a Temporal Regulator of EGFR Signaling
The increased basolateral LET-23 mobility in erm-1(tm677) mutants may result in an overall elevated activity of the RAS/MAPK pathway, as more LET-23 molecules are available to interact with LIN-3. The co-localization of LET-23 and ERM-1 together with the in vitro protein interaction experiments suggested that both proteins form a complex at the basolateral membrane of the VPCs. We thus hypothesized that ERM-1 may prevent the instant activation of the entire basolateral pool of LET-23 once the AC begins to secrete LIN-3 at the mid L2 stage, allowing the cells to maintain a high LET-23 activity after vulval induction. To test this model, we quantified the expression levels of the RAS/MAPK target EGL-17::CFP [29] in the descendants of the VPCs. In wild-type mid L3 larvae, we observed a peak of EGL-17::CFP expression after vulval induction in the 1° descendants of P6.p (Figure 5A–C′,G). By contrast, erm-1(tm677) mutants showed a gradual decrease rather than an increase in EGL-17::CFP expression after vulval induction (Figure 5D–G). Thus, ERM-1 is required for the generation of a long-lasting RAS/MAPK signal in the 1° vulval cells after fate specification has occurred.
Fig. 5. Temporal regulation of LET-23 EGFR signaling by ERM-1. 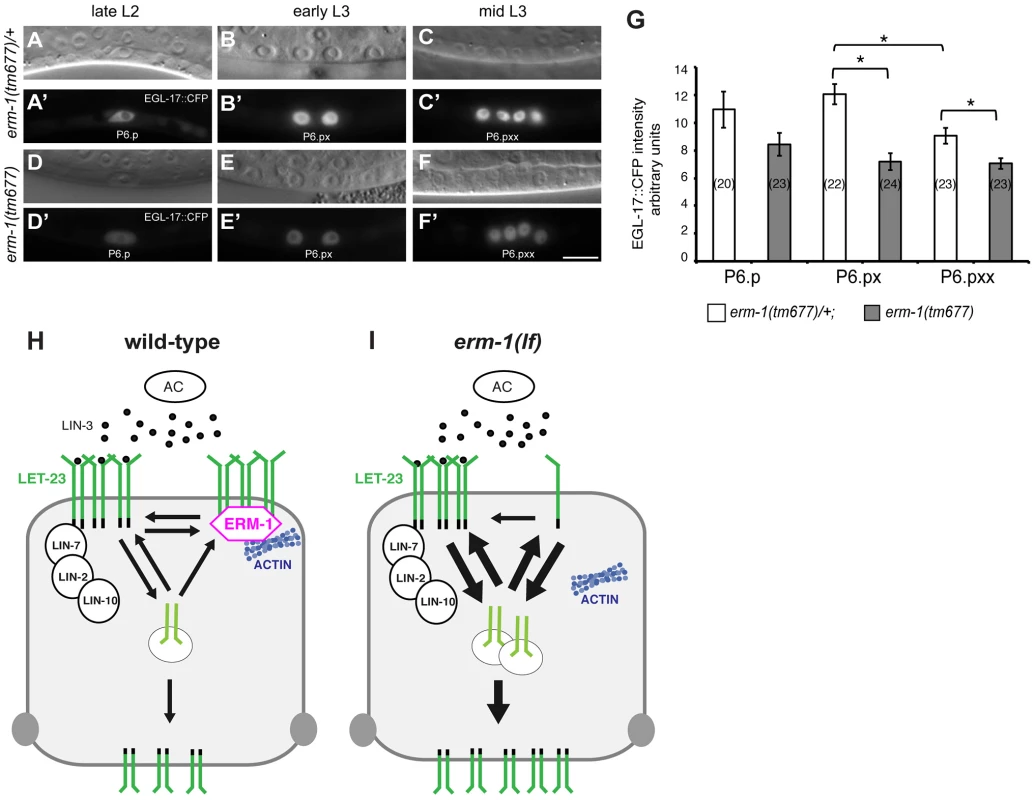
(A, B, C) Nomarski images and (A′, B′, C′) EGL-17::CFP expression in erm-1(tm677)/+ controls versus (D through F′) homozygous erm-1(tm677) mutants at the Pn.p (A′, D′), Pn.px (B′, E′) and Pn.pxx (C′, F′) stages. The scale bar is 10 µm. (G) Quantification of nuclear EGL-17::CFP intensities in P6.p and its descendants. The numbers of animals analyzed are indicated in brackets. *Indicates p<0.01 as determined in a two tailed student's t-test - two-sample unequal variance. Error bars represent the standard error of the mean. (H) Two antagonistic complexes control LET-23 localization at the basolateral membrane of the VPCs. The ternary LIN-2/LIN-7/LIN-10 complex promotes receptor activation, while the ERM-1 complex (magenta) prevents LET-23 endocytosis and signaling from basolateral membrane. ERM-1 may sequester LET-23 in an actin-rich membrane compartment to prevent receptor activation. (I) In the absence of ERM-1, more LET-23 can be activated via the LIN-2/LIN-7/LIN-10 complex, resulting in enhanced receptor endocytosis and recycling, and ultimately an increased accumulation of LET-23 on the apical plasma membrane. Discussion
Regulation of EGFR Localization Allows Pattern Formation
In order to systematically search for regulators of LET-23 EGFR trafficking and signaling, we performed an in vivo receptor localization screen in C. elegans larvae. There do exist certain limitations of this system, such as the inability to isolate individual cell for biochemical studies. However, an important advantage of our approach over previous studies performed with cultured epithelial cells [1] is the ability to observe the dynamics of receptor trafficking under normal conditions, in epithelial cells embedded in their natural environment and receiving physiological concentrations of various extracellular signals. The different regulators of LET-23 EGFR localization and signaling identified in our screen point at a complex network controlling LET-23 EGFR trafficking and signaling in different sub-cellular compartments. In a system, such as the VPCs, where ligand availability is limiting [8], [30], these additional control mechanisms at the level of the receptor are necessary to prevent too many cells from engaging in signaling at the same time and to focus the inductive signal on a single cell (P6.p). A perturbation of LET-23 EGFR trafficking can lead to a multivulva phenotype because decreased ligand sequestering by the 1° VPC P6.p results in increased LET-23 EGFR activation in the distal VPCs [30]. The down regulation of the LET-23 EGFR in all but the 1° VPC is therefore an important mechanism to break the symmetry of the initially equivalent VPCs and select a single cell for the 1° fate. The most frequent phenotype we observed in our screen (31 genes) was persisting LET-23 EGFR expression in 2° VPCs, and for those genes that had a significant effect on signaling we found increased rather than decreased vulval induction in the let-60 background. This suggests that a relatively large number of negative regulators of EGFR signaling is required to generate the invariant pattern of vulval cell fates with a single 1° cell flanked by two 2° cells.
Genes Regulating LET-23 EGFR Trafficking Are Conserved
A recent study has indicated that around 38% of all predicted protein coding genes in C. elegans possess at least one human homolog [31]. However, we found for 91% of the genes identified in our screen (74 of 81) at least one human homolog in the OrthoList, suggesting that the mechanisms regulating EGFR trafficking are strongly conserved. Further studies of these mammalian homologs may provide new means of interfering with deregulated EGFR signaling in human cells.
ERM-1 Is a Temporal Regulator of EGFR Signaling
We describe a new function of the C. elegans Ezrin homolog ERM-1 in regulating EGFR signaling on the basolateral membrane of the vulval cells. Based on the subcellular localization and dynamics and on genetic and biochemical data, we propose that ERM-1 forms a complex with the LET-23 EGFR at the basolateral plasma membrane to recruit the receptor into an actin-rich inactive membrane compartment and limit receptor activation (Figure 5H,I). A similar function has been proposed for mammalian NF2 Merlin, which shares similarity to Ezrin/Radixin/Moesin proteins. In confluent cultured epithelial cells, Merlin coordinates adherens junction stabilization with EGFR signaling by recruiting the EGFR into an apical membrane compartment, where the receptor cannot be activated [32]. In analogy, ERM-1 may link a fraction of the LET-23 EGFR pool at the basolateral membrane to cortical F-actin and restrict the access of the receptor to the activating LIN-2/LIN-7/LIN-10 complex [5]. In the absence of the tripartite LIN-2/LIN-7/LIN-10 complex, most of the residual basolateral LET-23 EGFR is probably bound and inactivated by ERM-1. The inhibitory ERM-1 complex thus antagonizes the activating LIN-2/LIN-7/LIN-10 complex to prevent the instant activation and subsequent degradation of the entire basolateral pool of LET-23 EGFR once the AC begins to secrete LIN-3 in the mid-L2 stage. This mechanism allows the vulval cells to maintain high LET-23 EGFR activity at later time points after vulval induction. LET-23 EGFR may be released from the ERM-1 complex when the vulval lumen is formed and ERM-1 relocalizes to the apical plasma membrane of the toroids. Such a buffering mechanism may be important, as sustained RAS/MAPK signaling is required during the subsequent phase of vulval morphogenesis when RAS/MAPK activity induces the expression of genes required for the execution of the vulval fates [4], [33]. Thus, the strength and duration of EGFR activation during development must be precisely controlled to achieve the correct levels of RAS/MAPK activity required for organogenesis.
Materials and Methods
Strains and General Methods
C. elegans strains were maintained at 20°C on standard nematode growth media [34] and the reference wild-type strain of C. elegans used was Bristol N2. Mutant strains used: LGI: erm-1(tm677)/hT2[bli-4(e937) let(q782) qIs48] (I;III), rde1(ne219), lin-10(e1339). LGII: let-23(sy1), lin-7(e1413), syIs12[hs::lin-3EGF][25]. LGIII: unc-119(ed3), unc-119(e2498). LGIV: lin-3(e1417), let-60(n1046). LGX: lin-2(n397). Integrated and extra-chromosomal arrays: qyIs23[Pcdh-3::mCherry::plcδPH; unc-119(+)] II [35], zhIs038[let-23::gfp, unc-119(+)] IV, zhEx484[C11H1.3::gfp; Pmyo-2::mCherry], zhIs396[Pdlg-1::lifeact::gfp::unc-54 3′utr, Plin-48::gfp] [23], zhEx486[sft-4::gfp; Pmyo-2::mCherry], zhEx487[ego-2::gfp; Pmyo-2::mCherry], zhEx519 [erm-1::mCherry; unc-119(+);Pmyo-2::mCherry]; zhEx418[Plin-31::rde-1; Pmyo-2::mcherry].
The construction of the translational reporter constructs used in this study is described in the Text S1 and Table S2 (see also Figure S4).
Extra-chromosomal arrays were obtained by microinjection of plasmids at 20 to 50 ng/µl along with the coinjection marker Pmyo2::mCherry or unc-119(+) at 2 to 10 ng/µl and pBluescript to a final concentration of 150 to 200 ng/µl as described [36]. zhIs038 was obtained by bombardment of unc-119 mutants with plasmid coated gold particles as described [37]. Primers used and details on the construction of plasmids can be found in the Supplementary Material.
GST Pull-down and Western Blots
GST fusion proteins were purified from E. coli BL-21 with glutathione-sepharose beads, incubated with 500 µg total protein worm extracts, and bound LET-23 was detected on Western blots with affinity-purified rabbit anti-LET-23 antibodies (1∶1000 dilution) raised against the C-terminal 196 amino acids [5].
RNAi Screen and Microscopy
RNAi was performed by bacterial feeding as described [38]. LET-23::GFP localization was scored in L3 larvae of the F1 generation mounted on 3% agarose pads supplemented with 5 mM tetramisol. For each RNAi clone, the vulval cells in 30 to 50 animals were observed at 40 to 63-fold magnification with a Leica DMRA wide-field microscope. Positive RNAi clones from the rescreen were verified by DNA sequencing. Images were recorded with a Hamamatsu ORCA-ER CCD camera controlled by the Openlab 5 software package (Improvision). Confocal images were recorded with a Olympus FV-1000 or a Zeiss LSM710 confocal microscope and analyzed with ImageJ [39]. Apical to basolateral intensity ratios were determined in mid-sagittal frames taken with the same illumination and same exposure settings by manually selecting the basal and apical membrane compartments and measuring total fluorescence intensities.
Fluorescence Recovery after Photobleaching
Larvae at the Pn.pxx stage were imaged at 20°C using a 63×/1.4 NA oil lens on an Zeiss LSM710 confocal microscope equipped with 458/488/514 nm argon and 405 nm diode lasers. A selected area of the basal or lateral membrane was bleached using the 488 nm argon laser at 85% power setting for 886.6 µsec to bleach around 70% of the signal, and fluorescence recovery was monitored over the following 296 seconds taking a frame every 8 seconds with a 488 nm laser excitation at 20% power intensity, a pinhole equivalent to 2.12 Airey units, a frame size of 256×256 dpi, and a pixel size of 0.53 µm. Data were analyzed in ImageJ by first registering the images with the StackReg plugin and then using the FRAP Norm plugin by Joris Meys [40] to measure recovery. Normalized curves were fitted to the formula I(t) = A·(1-e−kt)+C using the solver function in MS Excel to calculate the total mobile fraction A and half time as t1/2 = 1/k.
Heat Shock Treatment
Synchronized L3 larvae were heat-treated at 33°C for 30 minutes in a water bath, transferred to 20°C, and imaged at 120, 180, 230 minute after induction under the same illumination and exposure conditions. Quantification of the apical to basal LET-23::GFP intensity ratio with and without heat-shock treatment as described in the results.
Supporting Information
Zdroje
1. SorkinA, GohLK (2009) Endocytosis and intracellular trafficking of ErbBs. Experimental Cell Research 315 : 683–696.
2. ShtiegmanK, KochupurakkalBS, ZwangY, PinesG, StarrA, et al. (2007) Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene 26 : 6968–6978 doi:10.1038/sj.onc.1210503
3. SweeneyWE, ChenY, NakanishiK, FrostP, AvnerED (2000) Treatment of polycystic kidney disease with a novel tyrosine kinase inhibitor. Kidney Int 57 : 33–40 doi:10.1046/j.1523-1755.2000.00829.x
4. SternbergPW (2005) Vulval development. Wormbook 1–28 doi:10.1895/wormbook.1.6.1
5. KaechSM, WhitfieldCW, KimSK (1998) The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell 94 : 761–771.
6. WhitfieldCW, BénardC, BarnesT, HekimiS, KimSK (1999) Basolateral localization of the Caenorhabditis elegans epidermal growth factor receptor in epithelial cells by the PDZ protein LIN-10. Mol Biol Cell 10 : 2087–2100.
7. ChenN, GreenwaldI (2004) The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev Cell 6 : 183–192.
8. SimskeJS, KimSK (1995) Sequential signalling during Caenorhabditis elegans vulval induction. Nature 375 : 142–146 doi:10.1038/375142a0
9. BersetT, HoierEF, BattuG, CanevasciniS, HajnalA (2001) Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291 : 5 doi:10.1126/science.1055642
10. StetakA, HoierEF, CroceA, CassataG, Di FiorePP, et al. (2006) Cell fate-specific regulation of EGF receptor trafficking during Caenorhabditis elegans vulval development. EMBO J 25 : 11 doi:10.1038/sj.emboj.7601137
11. YooAS, BaisC, GreenwaldI (2004) crosstalk between the egfr and lin-12/notch pathways in C. elegans vulval development. Science 303 : 5 doi:10.1126/science.1091639
12. TatusovRL, FedorovaND, JacksonJD, JacobsAR, KiryutinB, et al. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4 : 41 doi:10.1186/1471-2105-4-41
13. BeitelGJ, ClarkSG, HorvitzHR (1990) Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348 : 503–509 doi:10.1038/348503a0
14. QadotaH, InoueM, HikitaT, KöppenM, HardinJD, et al. (2007) Establishment of a tissue-specific RNAi system in C. elegans. Gene 400 : 8 doi:10.1016/j.gene.2007.06.020
15. FoleyDA, SharpeHJ, OtteS (2007) Membrane topology of the endoplasmic reticulum to Golgi transport factor Erv29p. Mol Membr Biol 24 : 259–268 doi:10.1080/09687860601178518
16. AliN, ZhangL, TaylorS, MironovA, UrbéS, et al. (2013) Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr Biol 23 : 453–461 doi:10.1016/j.cub.2013.02.033
17. LiuY, MaineEM (2007) The Bro1-domain protein, EGO-2, promotes Notch signaling in Caenorhabditis elegans. Genetics 176 : 2265–2277 doi:10.1534/genetics.107.071225
18. AlgrainM, TurunenO, VaheriA, LouvardD, ArpinM (1993) Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. The Journal of Cell Biology 120 : 129–139.
19. Van FürdenD, JohnsonK, SegbertC, BossingerO (2004) The C. elegans ezrin-radixin-moesin protein ERM-1 is necessary for apical junction remodelling and tubulogenesis in the intestine. Developmental Biology 272 : 262–276 doi:10.1016/j.ydbio.2004.05.012
20. GöbelV, BarrettPL, HallDH, FlemingJT (2004) Lumen morphogenesis in C. elegans requires the membrane-cytoskeleton linker erm-1. Dev Cell 6 : 865–873 doi:10.1016/j.devcel.2004.05.018
21. DiogonM, WisslerF, QuintinS, NagamatsuY, SookhareeaS, et al. (2007) The RhoGAP RGA-2 and LET-502/ROCK achieve a balance of actomyosin-dependent forces in C. elegans epidermis to control morphogenesis. Development 134 : 2469–2479 doi:10.1242/dev.005074
22. LundquistE, ReddienP, HartwiegE, HorvitzHR, BargmannC (2001) Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128 : 14.
23. FarooquiS, PellegrinoMW, RimannI, MorfMK, MüllerL, et al. (2012) Coordinated lumen contraction and expansion during vulval tube morphogenesis in Caenorhabditis elegans. Dev Cell 23 : 494–506 doi:10.1016/j.devcel.2012.06.019
24. HwangBJ (2004) A cell-specific enhancer that specifies lin-3 expression in the C. elegans anchor cell for vulval development. Development 131 : 143–151 doi:10.1242/dev.00924
25. KatzWS, HillRJ, ClandininTR, SternbergPW (1995) Different levels of the C. elegans growth factor LIN-3 promote distinct vulval precursor fates. Cell 82 : 11.
26. HughesSC, FehonRG (2007) Understanding ERM proteins–the awesome power of genetics finally brought to bear. Current Opinion in Cell Biology 19 : 6 doi:10.1016/j.ceb.2006.12.004
27. NakamuraF, AmievaMR, FurthmayrH (1995) Phosphorylation of threonine 558 in the carboxyl-terminal actin-binding domain of moesin by thrombin activation of human platelets. J Biol Chem 270 : 31377–31385.
28. TurunenO, WahlströmT, VaheriA (1994) Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. The Journal of Cell Biology 126 : 1445–1453.
29. BurdineRD, BrandaCS, SternMJ (1998) EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development 125 : 1083–1093.
30. HajnalA, WhitfieldCW, KimSK (1997) Inhibition of Caenorhabditis elegans vulval induction by gap-1 and by let-23 receptor tyrosine kinase. Gene 11 : 2715–2728 doi:10.1101/gad.11.20.2715
31. ShayeDD, GreenwaldI (2011) OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE 6: e20085 doi:10.1371/journal.pone.0020085
32. CurtoM, ColeBK, LallemandD, LiuC-H, McClatcheyAI (2007) Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. The Journal of Cell Biology 177 : 893–903 doi:10.1083/jcb.200703010
33. PellegrinoMW, FarooquiS, FröhliE, RehrauerH, Kaeser-PebernardS, et al. (2011) LIN-39 and the EGFR/RAS/MAPK pathway regulate C. elegans vulval morphogenesis via the VAB-23 zinc finger protein. Development 138 : 4649–4660 doi:10.1242/dev.071951
34. BrennerS (1974) The genetics of Caenorhabditis elegans.. Genetics 77 : 24.
35. ZielJW, HagedornEJ, AudhyaA, SherwoodDR (2009) UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nature Cell Biology 11 : 183–189 doi:10.1038/ncb1825
36. MelloCC, KramerJM, StinchcombD (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10 : 3959–3970.
37. PraitisV, CaseyE, CollarD, AustinJ (2001) Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157 : 1217–1226.
38. KamathRS, FraserAG, DongY, PoulinG, GottaM, et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 : 231–237.
39. SchneiderCA, RasbandWS, EliceiriKW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9 : 671–675.
40. PhairRD, ScaffidiP, ElbiC, VecerováJ, DeyA, et al. (2004) Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Molecular and Cellular Biology 24 : 6393–6402 doi:10.1128/MCB.24.14.6393-6402.2004
Štítky
Genetika Reprodukční medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 5
-
Všechny články tohoto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



