-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
Plant immunity must be tightly regulated, as over - or constitutive activation of plant defenses can cause detrimental effects, such as dwarf stature and enhanced cell death. EDR1, a Raf-like mitogen-activated protein kinase (MAPK) kinase kinase, negatively regulates defenses in Arabidopsis. The highly conserved MAPK cascades modulate diverse biological processes, including plant immunity. However, whether EDR1 affects the regulation of one of the MAPK pathways was not previously known. Here, we show that EDR1 physically associates with MKK4 and MKK5, two MAP kinase kinases, and negatively regulates the protein levels of MKK4, MKK5, MPK3 and MPK6. We further show that edr1-mediated disease resistance requires MKK4, MKK5 and MPK3 function. Over-expression of MKK4 or MKK5 in wild-type increased resistance to powdery mildew and caused mildew-induced cell death. Our study suggests that EDR1 negatively regulates defenses and directly modulates the MKK4/MKK5-MPK3/MPK6 cascade to fine-tune plant immunity.
Published in the journal: . PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004389
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004389Summary
Plant immunity must be tightly regulated, as over - or constitutive activation of plant defenses can cause detrimental effects, such as dwarf stature and enhanced cell death. EDR1, a Raf-like mitogen-activated protein kinase (MAPK) kinase kinase, negatively regulates defenses in Arabidopsis. The highly conserved MAPK cascades modulate diverse biological processes, including plant immunity. However, whether EDR1 affects the regulation of one of the MAPK pathways was not previously known. Here, we show that EDR1 physically associates with MKK4 and MKK5, two MAP kinase kinases, and negatively regulates the protein levels of MKK4, MKK5, MPK3 and MPK6. We further show that edr1-mediated disease resistance requires MKK4, MKK5 and MPK3 function. Over-expression of MKK4 or MKK5 in wild-type increased resistance to powdery mildew and caused mildew-induced cell death. Our study suggests that EDR1 negatively regulates defenses and directly modulates the MKK4/MKK5-MPK3/MPK6 cascade to fine-tune plant immunity.
Introduction
Mitogen-activated protein kinase (MAPK) cascades are highly conserved signaling modules that control diverse signal transduction pathways in eukaryotes, including defenses against infection [1]. Activation of MAPK cascades is thought to be one of the earliest events in plant immunity [1]–[3]. For instance, treatment of Arabidopsis with a conserved 22-amino acid peptide fragment of bacterial flagellin, a pathogen-associated molecular pattern (PAMP), specifically recognized by the pattern recognition receptor FLS2, can trigger activation of MKK4/MKK5 and MPK3/MPK6 [4], [5], which subsequently promotes expression of the downstream target gene FRK1 and activates plant defenses [5]. Other PAMPs, such as EF-Tu, chitin, harpin, oligogalacturonides and xylanase, also trigger the activation of MPK3 and/or MPK6 [6]–[9]. In addition, MPK3 and MPK6 regulate phytoalexin biosynthesis by activating the transcription factor WRKY33, which is required for resistance to necrotrophic fungal pathogens [10]–[12]; also, the full priming of stress responses in Arabidopsis requires MPK3 and MPK6 [13]. MKK4 and MKK5 function redundantly and act upstream of MPK3 and MPK6. Constitutive activation of MKK4 and MKK5 in Arabidopsis leads to HR-like cell death associated with the generation of reactive oxygen species [14]. Plants expressing active forms of MKK4 and MKK5 have enhanced resistance to Pseudomonas syringae and Botrytis cinerea [5].
Plant basal defenses require the MKK4/MKK5-MPK3/MPK6 kinase cascade. To effectively invade plants, bacterial pathogens block PAMP-induced defenses with effectors, such as AvrPto, AvrPtoB, HopAI1, and HopF2, that directly repress MKK4/MKK5 and MPK3/MPK6 activities or inhibit the components that act upstream of MAP kinase cascades [15]–[20]. In Arabidopsis, five phosphatases have been reported that regulate MPK3 and MPK6 kinase activity by dephosphorylation. For instance, the PP2C-type phosphatase AP2C1 inactivates stress-induced kinase activity of MPK4 and MPK6 [21]; PP2C5 also regulates ABA-mediated activation of MPK3 and MPK6 [22]. Also, MKP1 and PTP1 phosphatases repress salicylic acid biosynthesis and SNC1-mediated responses by inactivating MPK3 and MPK6 [23]. Also, MKP2 interacts with and dephosphorylates MPK3 and MPK6 to regulate oxidative stress and plant defense responses [24], [25]. However, the mechanisms of the negative regulation of MKK4/MKK5 in Arabidopsis remain unclear.
The Raf-like MAPK kinase kinase (MAPKKK) EDR1 functions as a negative regulator of plant defense. For example, edr1 mutants have enhanced resistance to pathogens including powdery mildew fungus, bacteria and oomycetes [26], [27]. The edr1 mutants also show enhanced ethylene-induced senescence [28]. EDR1 protein consists of an N-terminal functionally unknown domain and a C-terminal kinase domain. The kinase activity of EDR1 has been demonstrated in vitro [29]. KEEP ON GOING (KEG) encodes a protein containing RING E3 ligase domain, kinase domain, ankyrin repeats and HERC2-like repeats [30] and the recessive missense mutant keg-4 suppresses the phenotype of edr1 mutants. KEG could directly interact with EDR1 and recruit EDR1 to the trans-Golgi network/early endosome [31].
In Arabidopsis, the EDR1 homolog CTR1 (Constitutive Triple Response 1), a Raf-like MAPKKK [29], plays an essential role in the negative regulation of ethylene signaling [32]. CTR1 and the MKK9-MPK3/MPK6 cascade antagonistically regulate ethylene responses [33]. However, whether EDR1 affects the regulation of one specific MAPK cascade pathway remains unknown. The edr1 mutants are constitutively primed for salicylic acid-inducible defenses and enhanced callose deposition, which may be mediated by the regulation of MPK3 and MPK6 in Arabidopsis [13], [34]. However, the molecular mechanisms leading to enhanced resistance and cell death in edr1 are still not well understood. Here, we report that EDR1 negatively regulates the MKK4/MKK5-MPK3/MPK6 kinase cascade pathway through direct interaction with MKK4 and MKK5 to modulate plant defense and cell death.
Results
EDR1 negatively affects MPK3 and MPK6 protein levels and kinase activity
EDR1 belongs to the MAPKKK family, but a mechanistic link to MAP kinase cascades has remained elusive. After treatment with benzothiadiazole (BTH), the activity of MPK3 and MPK6 was higher in edr1 mutants than in Col-0 [13]. In addition, large-scale co-expression data analysis [35], [36] showed that MKP1, which is involved in the negative regulation of MPK3/MPK6 by dephosphorylation in Arabidopsis [23], is one of the top-ranking genes co-expressed with EDR1 (Figure S1). Based on these results, we hypothesized that EDR1 may function as a negative regulator of the MPK3/MPK6 kinase cascade pathway in pathogen responses. Consistent with this notion, the accumulation of FRK1 transcript, a target of the MPK3/MPK6 cascade [37], was significantly higher in edr1 compared to wild-type upon infection with powdery mildew Golovinomyces cichoracearum or Pseudomonas syringae pv. tomato (Pto) DC3000 (Figure 1A and 1B).
Fig. 1. EDR1 negatively regulates the kinase activity of MPK3 and MPK6. 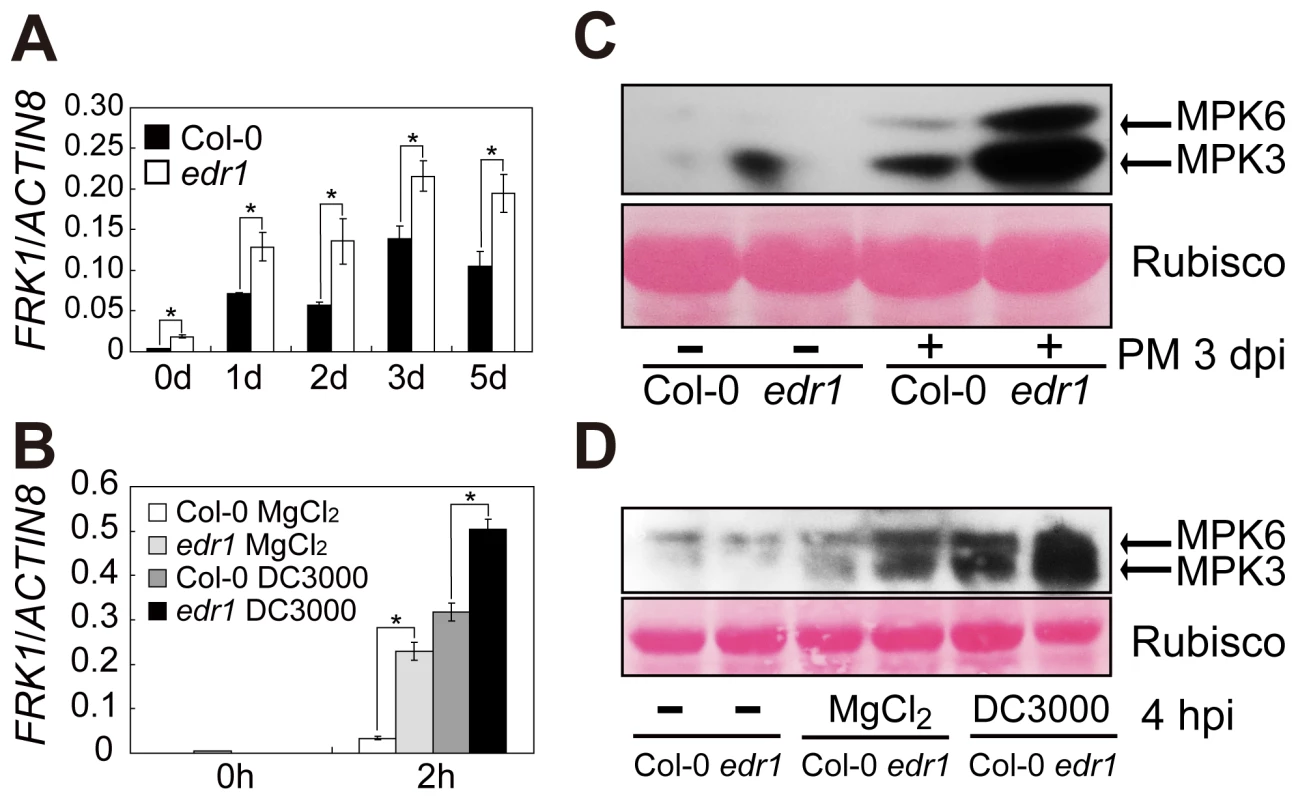
(A–B) The transcript accumulation of FRK1 was measured by quantitative real-time RT-PCR. Leaves were collected for RNA isolation at different time points after infection with G. cichoracearum (A) or Pto DC3000 (in 10 mM MgCl2) (B). Error bars represent the standard deviation of three biological replicates. Asterisks indicate statistically significant differences (P<0.05, Student's t-test). (C–D) The plants were infected with G. cichoracearum (C) and Pto DC3000 (D), respectively. Immunoblotting was performed using an anti-phospho-p44/42 MAPK (Thr202/Tyr204) (anti-pTEpY) antibody. The large subunit of Rubisco is shown as a protein loading control. The experiment was repeated at least three times with similar results. PM: powdery mildew infection. Along with up-regulation of the FRK1 gene, MPK3 and MPK6 kinase activation also increased in edr1 compared to wild-type after infection by powdery mildew or Pto DC3000 (Figure 1C and 1D). These data indicate that EDR1 negatively affects the MPK3/MPK6 cascade.
To further examine the role of EDR1 in the MPK3/MPK6 pathway, we over-expressed EDR1 in Arabidopsis and investigated whether over-expression of EDR1 reduces MPK3 and MPK6 kinase activity. Previously, an attempt to over-express EDR1 with cauliflower mosaic virus (CaMV) 35S promoter driven EDR1 coding sequence (CDS) was not successful [29]. Therefore, we generated over-expression lines by introducing GFP-tagged EDR1 genomic sequence under its native promoter into the edr1 mutant. This construct complemented all edr1 mutant phenotypes, including edr1-mediated enhanced resistance to powdery mildew and enhanced cell death (Figure S2A, S2B and S2C). For further analysis, we selected one transgenic line with three-fold up-regulation of EDR1 and with properly expressed EDR1-GFP protein, as demonstrated by immunoblotting assays (Figure 2A and 2B). The EDR1 over-expressing plants showed enhanced susceptibility to powdery mildew, as significantly more spores of G. cichoracearum were produced after 5 days inoculation compared to wild-type (Figure S2D). However, the growth of Hyaloperonospora arabidopsidis Noco2 on the EDR1 over-expressing plants and wild-type was not significantly different (Figure S2E). The EDR1 over-expressing plants showed delayed ethylene-induced senescence compared to wild-type (Figure S2F and S2G). Furthermore, the EDR1 over-expressing plants showed lower levels of PR-1 and FRK1 expression and lower MPK3 and MPK6 kinase activity than wild-type upon infection by powdery mildew (Figure 2C, 2D and 2E).
Fig. 2. Over-expression of EDR1 reduced the kinase activity and protein levels of MPK3 and MPK6. 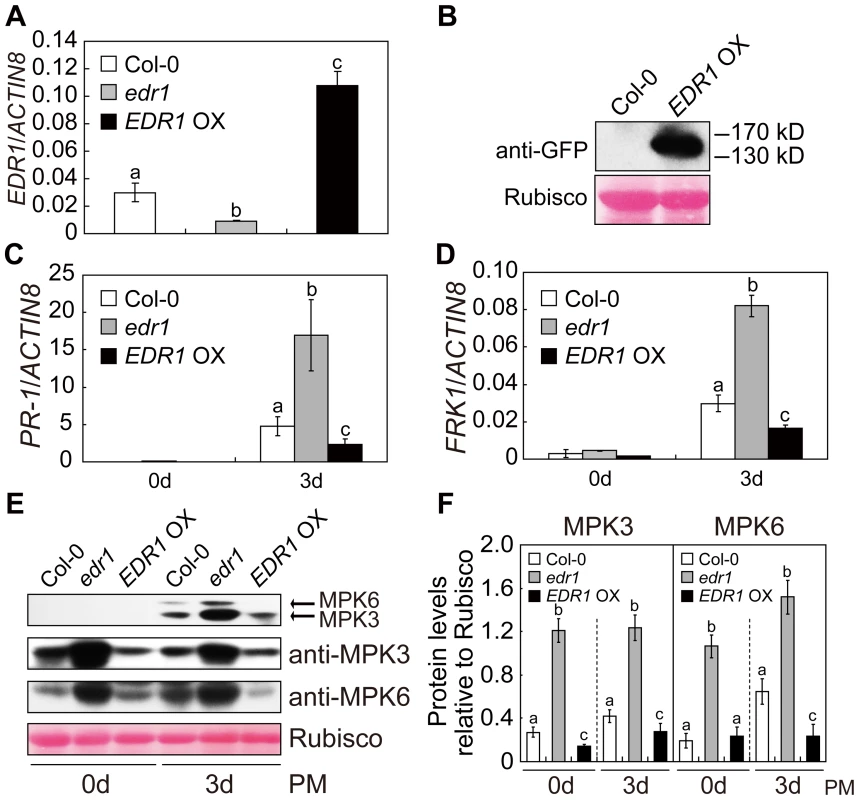
(A) The transcript accumulation of EDR1 was examined by quantitative real-time RT-PCR for wild-type Col-0, edr1 and EDR1 over-expressing plants. ACTIN8 was used as an internal control. Error bars represent the standard deviation of three biological replicates. Different letters represent statistically significant differences (P<0.05, one-way ANOVA). (B) EDR1-GFP is properly expressed in EDR1 over-expressing plants. Immunoblotting was performed using anti-GFP antibody in four-week-old plants. The large subunit of Rubisco is shown as a protein loading control. (C–D) The transcript accumulation of PR-1 (C) or FRK1 (D) was examined by quantitative real-time RT-PCR. Leaves from Col-0, edr1 and EDR1 over-expressing plants after infection by powdery mildew for 0 d and 3 d were collected for RNA isolation. Error bars represent the standard deviation of three biological replicates. Different letters represent statistically significant differences (P<0.05, one-way ANOVA). (E) Col-0, edr1 and EDR1 over-expressing plants were infected by G. cichoracearum. Immunoblot was performed using anti-pTEpY, anti-MPK3 and anti-MPK6 antibodies, as indicated. The large subunit of Rubisco is shown as a protein loading control. The experiment was repeated three times with similar results. PM: powdery mildew infection. (F) The protein bands of MPK3 and MPK6, as well as Rubisco, were quantified with ImageJ. The protein levels of MPK3 and MPK6 in each sample were evaluated by comparing to Rubisco. The error bars represent the standard deviation of three biological replicates. Different letters represent statistically significant differences (P<0.05, one-way ANOVA). PM: powdery mildew infection. We then examined the accumulation of MPK3 and MPK6 proteins in edr1 and EDR1 over-expressing plants using specific anti-MPK3 and anti-MPK6 antibodies. Compared to wild-type, the protein levels of MPK3 and MPK6 were significantly higher in the edr1 mutants, but lower in EDR1 over-expressing plants, even in normal conditions without infection by pathogens (Figure 2E and 2F). To further demonstrate that EDR1 negatively regulates the protein level of MPK3, we transiently expressed MPK3 alone or with EDR1 in Nicotiana benthamiana. The level of MPK3 was significantly lower when we co-expressed MPK3 with EDR1 (Figure S3A). Transient expression in N. benthamiana further showed that expression of the N-terminal presumptive regulatory domain of EDR1 (1–657 aa), but not the C-terminal kinase domain (658–933 aa), was sufficient to suppress the accumulation of MPK3 (Figure S3B and S3C). In contrast, the accumulation of MPK3 and MPK6 transcripts was not significantly affected in the edr1 mutants infected with powdery mildew (Figure S4A and S4B), indicating that the regulation of MPK3 and MPK6 by EDR1 mainly functions at the protein level, not the mRNA level.
As EDR1 plays a negative role in plant defense, we also examined abundance of EDR1 protein during pathogen infection. Leaves of EDR1-Flag transgenic plants were infected by G. cichoracearum, and the proteins were extracted 0 d, 2 d, and 5 d after inoculation, respectively. We found that levels of EDR1 protein significantly declined upon powdery mildew infection (Figure S5), consistent with the negative role of EDR1 in defense responses.
A missense mutation (keg-4) in the E3 ubiquitin ligase KEG suppresses all edr1-associated phenotypes [30]. To examine whether the keg-4 mutation also counteracts the elevated activation of the MPK3 kinase cascade pathway, we examined the protein level of MPK3 in the edr1 keg-4 double mutant before and after infection with Pto DC3000. In the edr1 keg-4 mutant, the MPK3 protein level was significantly lower than in the edr1 mutants, indicating that the keg-4 mutation also suppressed increased accumulation of MPK3 protein in the edr1 mutants (Figure S6).
Loss of MPK3 suppresses the edr1 phenotype
To further examine the role of MPK3/MPK6 in edr1-mediated defense, we conducted crosses to make double mutants of edr1 and mpk3-1 or mpk6-3 (Figure 3A and S7A). The mpk3-1 mutation suppressed the early senescence and spontaneous cell death associated with edr1 mutants (Figure 3B); it also partially suppressed the resistance of edr1 to powdery mildew and H. a. Noco2. In addition, the mpk3-1 mutation counteracted edr1-mediated enhanced ethylene-induced senescence, restoring it to wild-type levels (Figure 3C–3H). In contrast, mpk6-3 did not suppress these edr1 phenotypes (). These observations indicate that edr1-mediated resistance to pathogens and cell death requires MPK3, but not MPK6.
Fig. 3. The mpk3-1 mutation suppressed the edr1 phenotype. 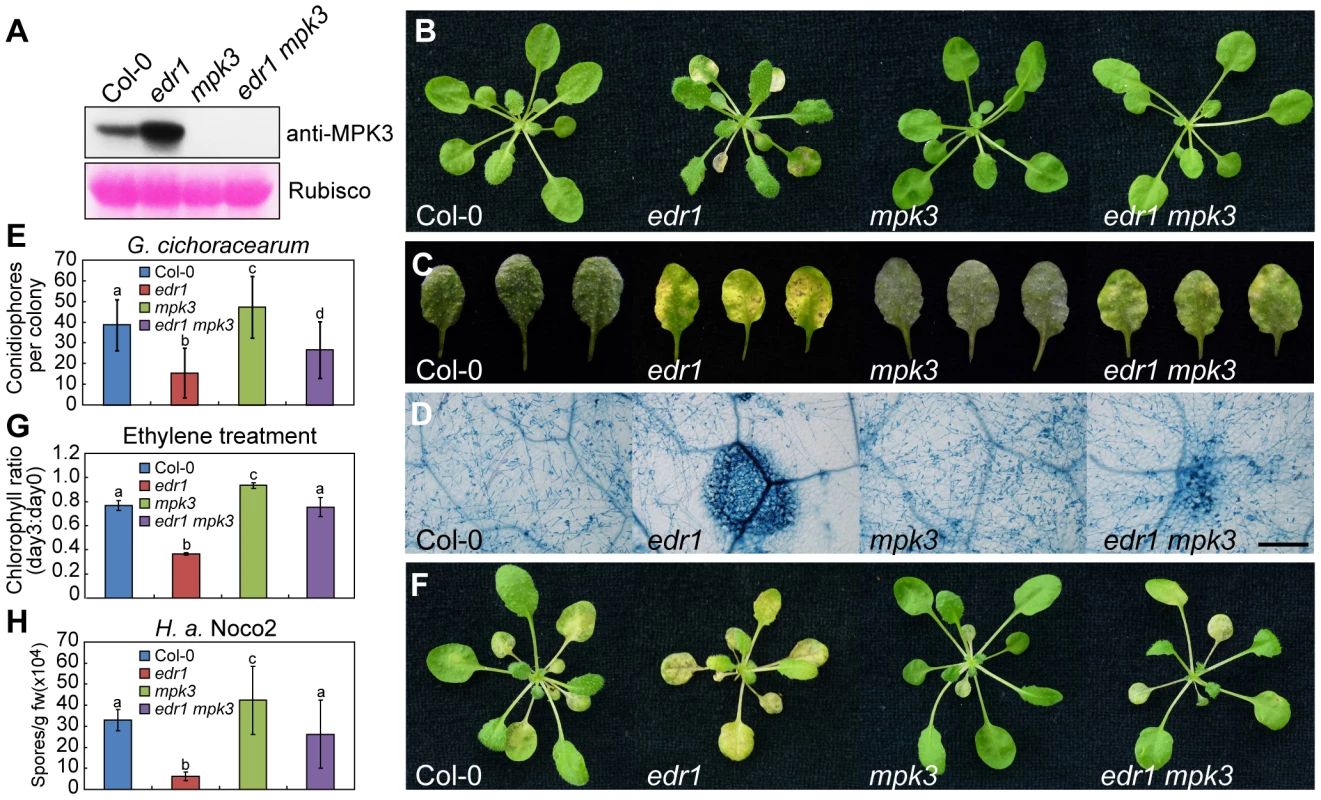
(A) Immunoblot for Col-0, edr1, mpk3-1 and edr1 mpk3-1 was performed using specific anti-MPK3 antibody. The large subunit of Rubisco is shown as a protein loading control. (B) Col-0, edr1, mpk3-1 and edr1 mpk3-1 were grown in the greenhouse at 22°C and a 9 h light/15 h dark cycle. Pictures were taken after 5 weeks growth. (C) Plants were infected with G. cichoracearum. Pictures were taken at 7 dpi. (D) Powdery mildew infected leaves at 7 dpi were stained by trypan blue. Bar = 0.3 mm. (E) Quantification of fungal growth by counting the number of conidiophores per colony at 5 dpi. At least 30 colonies were counted for each sample. Error bars represent the standard deviation. Different letters represent statistically significant differences (P<0.05, one-way ANOVA). (F) Four-week-old plants of Col-0, edr1, mpk3-1 and edr1 mpk3-1 were treated with ethylene (100 µL/L) for three days in a sealed chamber. Pictures were taken after 3 days. (G) Chlorophyll content was measured in wild-type Col-0, edr1, mpk3-1 and edr1 mpk3-1 before and after treatment of ethylene (3 days). The ratio of chlorophyll content at day 3 to day 0 was calculated for each sample. Error bars represent the standard deviation of ten plants. Different letters represent statistically significant differences (P<0.05, one-way ANOVA). (H) Three-week-old Col-0, edr1, mpk3-1 and edr1 mpk3-1 plants were infected by H. a. Noco2. Spores were counted at 7 dpi. Error bars represent the standard deviation of three biological replicates. Different letters represent statistically significant differences (P<0.05, one-way ANOVA). The above experiments were repeated three times with similar results. To further examine the functions of MPK3 and MPK6 in plant defense, we transformed wild-type plants with MPK3 and MPK6 expressed under the control of the 35S promoter. Although we obtained a number of MPK3 transgenic lines, none of them showed higher MPK3 expression, suggesting that Arabidopsis may not tolerate the over-expression of MPK3. We did obtain transgenic plants with higher expression levels of MPK6 (Figure S8A and S8B), but these MPK6 over-expressing plants showed wild-type-like responses to powdery mildew (Figure S8C). This suggests that the high level of MPK6 in edr1 mutants does not contribute to edr1-mediated resistance to powdery mildew, consistent with the observation that the mpk6 mutation did not affect edr1 phenotypes.
EDR1 interacts with MKK4 and MKK5
To examine whether EDR1 directly regulates the MPK3/6 kinase cascade, we used a yeast two-hybrid assay to test for potential interactions of EDR1 with MPK3 or MPK6, or the upstream MAP kinases MKK4 and MKK5 [5], [38]. We found that EDR1 interacts with MKK4 and MKK5 (Figure 4A), but not with MPK3 or MPK6. As a control, we also assayed the interaction of EDR1 with the well-studied MAP kinase kinases MKK1 and MKK2. EDR1 did not interact with either MKK1 or MKK2 (Figure 4A), indicating that EDR1 interacts specifically with MKK4 and MKK5. To determine which domain of EDR1 is responsible for the interaction with MKK4 and MKK5, we tested the EDR1 N-terminal domain and C-terminal kinase domain and found that the EDR1 N-terminal domain is responsible for the interaction with MKK4 and MKK5 (Figure 4A).
Fig. 4. EDR1 interacts with MKK4 and MKK5. 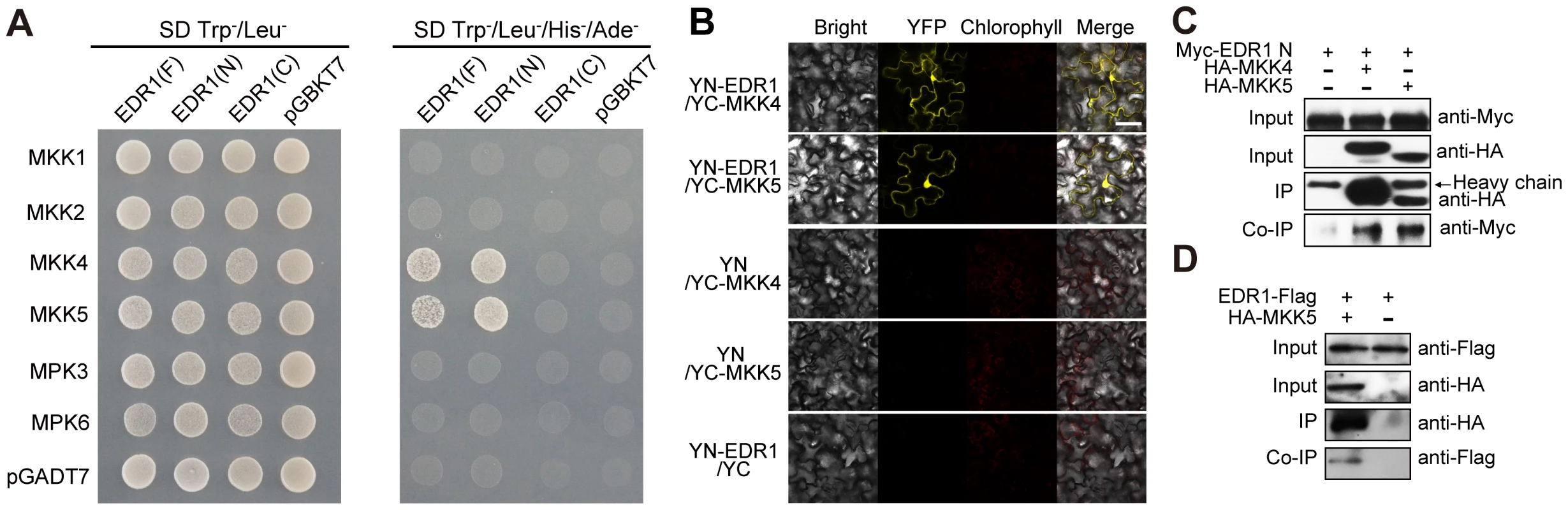
(A) EDR1 full length (F), EDR1 N-terminal domain (N) and EDR1 C-terminal domain (C) were fused to the Gal4 DNA binding domain (BD). MKK1, MKK2, MKK4, MKK5, MPK3 and MPK6 were fused to the Gal4 transactivation domain (AD). Different pairs of constructs were cotransformed into yeast isolate AH109 to test the interaction. 10 µL suspension (OD600 = 0.5) of each cotransformant was dropped on the synthetic dropout (SD) medium lacking Leu and Trp (left) and SD medium lacking Ade, His, Leu and Trp (right), respectively. Pictures were taken after 2 days incubation. (B) YFPYN-fused EDR1 and YFPYC-fused MKK4/MKK5 were co-expressed in N. benthamiana. YFP fluorescence was detected by confocal microscopy. Cotransformants of YFPYN-EDR1 and YFPYC, YFPYN and YFPYC-MKK4, or YFPYN and YFPYC-MKK5 were used as controls. Bar = 50 µm. (C) EDR1 N-terminal domain was expressed alone or co-expressed with MKK4 and MKK5 in N. benthamiana. Proteins were extracted after 48 h, and subjected to immunoprecipitation by anti-HA antibody, followed by immunoblotting using anti-Myc and anti-HA antibodies, respectively. (D) EDR1-Flag transgenic plants and EDR1-Flag/HA-MKK5 double transgenic plants were used for co-IP. The proteins were analyzed by immunoblotting using anti-Flag or anti-HA antibody, respectively. The above experiments were repeated three times with similar results. We used several complementary approaches to examine whether EDR1 also interacts with MKK4 and MKK5 in vivo. First, we performed bimolecular fluorescence complementation (BiFC) assays by transiently co-expressing YFPYN-fused EDR1 and YFPYC-fused MKK4 or MKK5, in N. benthamiana. We detected YFP fluorescence only in cells co-expressing YFPYN-EDR1 with YFPYC-MKK4 or YFPYN-EDR1 with YFPYC-MKK5, but not in the negative controls (Figure 4B). Second, to confirm the association of EDR1 and MKK4/MKK5, we performed co-immunoprecipitation (co-IP) assays and found that MKK4 and MKK5 immunoprecipitated the EDR1 N-terminal domain upon transient expression in N. benthamiana (Figure 4C). Furthermore, MKK5 and EDR1 could also be precipitated from stable transgenic Arabidopsis plants expressing both EDR1-Flag and MKK5-HA (Figure 4D). Third and finally, we examined whether EDR1 and MKK4/MKK5 co-localize in Arabidopsis. We crossed plants harboring GFP-tagged MKK4 or MKK5 transgenes with plants harboring a Cherry-tagged EDR1 transgene, and imaged GFP and Cherry fluorescence by confocal microscopy in the F1 generation. We found that EDR1 and MKK4/MKK5 co-localize in the cytoplasm and, partially, in the nucleus (Figure S9A and S9B).
EDR1 negatively regulates the levels of MKK4 and MKK5
Since EDR1 physically associates with MKK4 and MKK5, we reasoned that EDR1 may also affect the protein levels of MKK4 and MKK5. As MKK4 and MKK5 specific antibodies are not available, we used the same transgenic lines described above to examine the accumulation of GFP-tagged MKK4 or MKK5 in plants with higher or lower levels of EDR1. Analyzing more than 30 individuals for each transgene combination by confocal microscopy, we found that plants expressing both MKK4 g-GFP and EDR1 g-Cherry, or both MKK5 g-GFP and EDR1 g-Cherry showed less-intense GFP fluorescence than transgenic plants expressing MKK4 g-GFP or MKK5 g-GFP alone (Figure 5A and 5B). In contrast, the intensity of Cherry fluorescence was not affected. Immunoblotting assays confirmed that protein levels of MKK4 and MKK5 were significantly lower in the presence of an EDR1 transgene (Figure 5C). Furthermore, we combined the MKK5 g-GFP transgene with the edr1 mutation; as expected, MKK5-GFP accumulated to higher levels in the edr1 background compared to wild-type on the basis of both GFP fluorescence intensity and immunoblotting assays (Figure 5D and 5E). These data indicate that EDR1 negatively affects the protein levels of MKK4 and MKK5.
Fig. 5. EDR1 regulates the protein levels of MKK4 and MKK5. 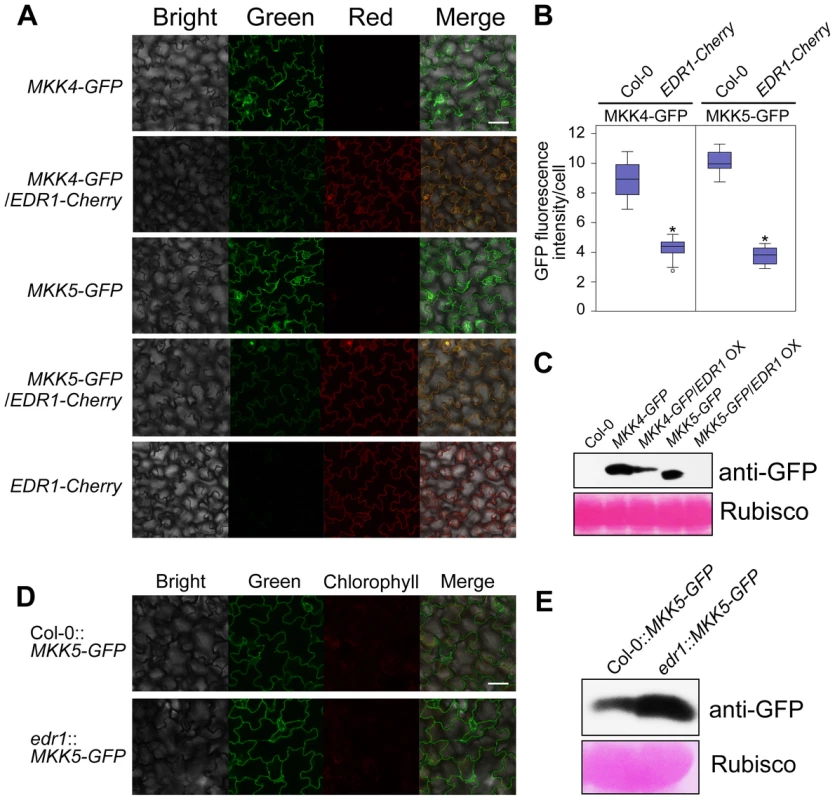
(A) GFP and Cherry fluorescence of seedlings of transgenic plants that express MKK4-GFP or MKK5-GFP alone or with EDR1-Cherry, was detected by confocal microscopy using the same parameters. Bar = 50 µm. (B) The GFP fluorescence intensity was quantified by using ImageJ software. 30 cells from 10 independent leaves of each transgenic plant were used for the quantification of the intensity of GFP fluorescence. The results are shown as a box plot graph. Asterisks represent statistically significant differences (P<0.05, Student's t-test). (C) Immunoblot was performed for each sample using anti-GFP antibody. The large subunit of Rubisco is shown as a protein loading control. (D) GFP fluorescence of seedlings of transgenic plants Col-0::MKK5-GFP and edr1::MKK5-GFP was detected by confocal microscopy using the same parameters. Bar = 50 µm. (E) Immunoblot was performed for Col-0::MKK5-GFP and edr1::MKK5-GFP using anti-GFP antibody. The large subunit of Rubisco is shown as a protein loading control. edr1-mediated resistance to powdery mildew requires MKK4 and MKK5
To investigate whether the enhanced powdery mildew resistance of edr1 mutants is due to the elevated protein levels of MKK4 and MKK5, we generated double mutant combinations of edr1 and loss-of-function alleles of MKK4 or MKK5. The mkk4–18 mutation results in the substitution of proline-240, a conserved position in the catalytic site, to serine. yda-2, a strong loss-of-function allele of the MAPKKK YDA, harbors the same exchange in the homologous position [39], suggesting that this amino acid is important for protein function. The mutation in mkk5–18 leads to premature termination of translation (R72stop). Both the mkk4 and mkk5 mutation suppressed edr1-mediated early senescence and cell death after 5 weeks growth (Figure 6A). Following inoculation with G. cichoracearum, both the mkk4 and mkk5 mutation counteracted the resistance conferred by edr1, such that susceptibility of double mutants was close to wild-type (Figure 6B, 6C and 6D). In addition, the leaves of mkk4 and mkk5 single mutants produced more spores of powdery mildew than wild-type leaves, indicating that both mutants show higher susceptibility to powdery mildew. This enhanced susceptibility phenotype could be complemented by introducing a genomic DNA fragment spanning the MKK4 or MKK5 locus, respectively (Figure S10), indicating that MKK4 and MKK5 are involved in disease resistance to powdery mildew. Finally, the activity of the MPK3/MPK6 kinases was lower in edr1 mkk4 and edr1 mkk5 than in edr1 after infection by powdery mildew (Figure 6E).
Fig. 6. mkk4 and mkk5 suppress edr1-mediated resistance to powdery mildew and cell death. 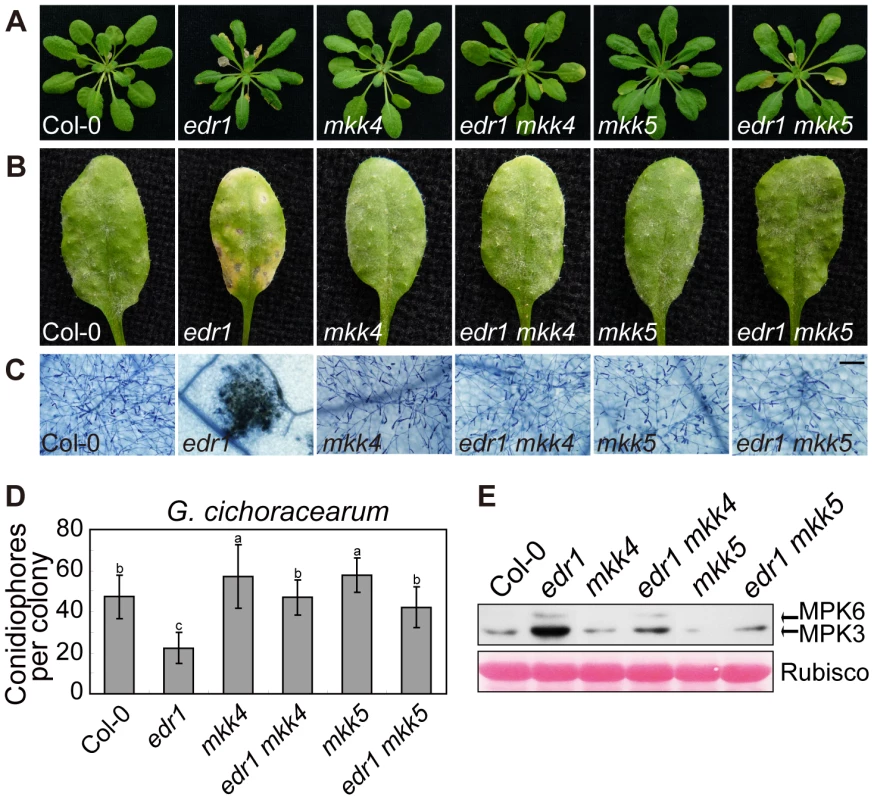
(A) Col-0, edr1, mkk4, edr1 mkk4, mkk5 and edr1 mkk5 were grown in the greenhouse at 22°C and a 9 h light/15 h dark regime. Pictures were taken after 5 weeks of growth. (B) Plants were infected by G. cichoracearum. Pictures were taken at 7 dpi. (C) Powdery mildew infected leaves at 7 dpi were stained by trypan blue. Bar = 0.1 mm. (D) Fungal growth was assessed by counting the number of conidiophores at 5 dpi. At least 30 colonies were counted for each sample. Error bars represent the standard deviation. Different letters represent statistically significant differences (P<0.05, one-way ANOVA). (E) Plants were infected by G. cichoracearum for 3 days. Immunoblot was performed using anti-pTEpY antibody. The large subunit of Rubisco is shown as a protein loading control. The experiment was repeated three times with similar results. To further study the functions of MKK4 and MKK5 in plant defense, we overexpressed MKK4 and MKK5 in wild-type (Figure S11A, S11B and S11C); plants over-expressing MKK4 or MKK5 showed enhanced resistance to powdery mildew and displayed edr1-like spontaneous cell death (Figure S11D and S11E). In summary, these results indicate that MKK4 and MKK5 play a positive role in powdery mildew resistance, and implicate the elevated protein levels of MKK4 and MKK5 as the main mechanism leading to pathogen resistance in the edr1 mutant.
Discussion
The essential MKK4/MKK5-MPK3/MPK6 kinase cascade transduces extracellular stimuli in many different response pathways. Activation of this cascade must be tightly controlled, as inappropriate activation of MAPKs can inhibit growth or even cause lethality. For instance, in Arabidopsis, constitutively activated MKK4 and MKK5 cause an accelerated cell death phenotype [14]. Over-expression of MPK3 may be lethal for the plant, as we (and others) have found it difficult to obtain MPK3 over-expressing plants ([13], [40] and this study). Therefore, tight regulation of MKK4/MKK5 and MPK3/MPK6 kinase activity and protein accumulation seems essential for survival and adaptation to environmental challenges. The kinase activity of MPK3/MPK6 can be repressed through dephosphorylation mediated by several different phosphatases [21]–[25]. Modulation of MKK4 and MKK5 protein levels by EDR1 could be another mechanism for fine-tuning the activity of the MKK4/MKK5-MPK3/MPK6 kinase cascade in response to pathogen attack.
How EDR1 affects accumulation of MKK4 and MKK5 protein remains unresolved. Our study showed that the N-terminal regulatory domain of EDR1 interacts with MKK4 and MKK5, and negatively affects MKK4 and MKK5 accumulation. One possible mechanism for this effect may be that EDR1 acts as a scaffold protein that keeps MKK4 and MKK5 in a catalytically inactive state. While MKK4 and MKK5 are not active, the plants would deploy a feedback regulation mechanism to keep MKK4 and MKK5 protein levels low in order to avoid detrimental spontaneous activation of defense responses. Low levels of MKK4 and MKK5 protein in turn would lead to low accumulation of MPK3 and MPK6 protein. An alternative explanation for our finding is that EDR1 affects the stability of MKK4 and MKK5 directly, perhaps through interactions with the 26S proteasome degradation machinery. Consistent with this possibility, EDR1 has been reported to associate with the E3 ubiquitin ligase KEG [30], [31], a component of the SCF ligase complex, and that mutations in KEG suppress edr1-mediated resistance. Furthermore, we recently showed that edr1-mediated defense responses require RPN1a, a subunit of the 26S proteasome [41], suggesting that 26S proteasome degradation machinery is involved in the EDR1 pathway.
Although MKK4 and MKK5 are commonly considered, by implication, to be important components of plant immunity pathways, little genetic support for this notion has been reported. Here, we identified mkk4 and mkk5 loss-of-function mutants, and showed that they display enhanced susceptibility to powdery mildew. We also showed that edr1-mediated resistance requires MKK4 and MKK5 function, providing direct genetic evidence that MKK4 and MKK5 are key players in plant immunity responses. The mkk4 and mkk5 alleles described in this study will be valuable tools for dissecting the role of the MKK4/5 mediated MAPK cascade in biotic and abiotic stresses.
The MKK4 and MKK5 proteins are considered to be functionally equivalent [1], [42]. Interestingly, our study revealed that single mutation of either MKK4 or MKK5 significantly affect edr1-mediated defense. Consistent with this observation, both mkk4 and mkk5 single mutant showed enhanced susceptibility to powdery mildew, indicating that loss-of-function of either one of these two genes causes a defect in immune responses. Dosage effects may be a possible explanation for these observations. In this scenario, the accumulation of MKK4 and MKK5 protein is limiting, such that the activity of both genes is required for normal immune responses. This view is supported by our finding that over-expression of either MKK4 or MKK5 enhanced resistance to powdery mildew and resulted in edr1-like cell death. However, the possibility that MKK4 and MKK5 have non-identical roles in plant immunity has not been rigorously tested and cannot be dismissed at this time.
Arabidopsis has more than sixty MAPKKKs, which can be divided into two subfamilies, including 12 MEKK1-like kinases and approximately 50 Raf-like kinases [43]. Several members of the MEKK1 family have been shown to function as MAPKKKs upstream of MAPKKs in MAP kinase cascades [1]; in contrast, no evidence has implicated any of the Raf-like kinases as canonical MAPKKKs. EDR1 and CTR1 are the only two well-characterized Raf-like kinases in plants. CTR1 directly regulates EIN2 by phosphorylation; EIN2, in turn, inactivates EIN3/EIL1-dependent ethylene responses. This indicates that CTR1, at least in this context, does not function as a MAPKKK [44], [45]. Here, we show that the N-terminus of EDR1 associates with MKK4/MKK5 and negatively affects the accumulation of MKK4/5 protein, suggesting that EDR1 does not act as a MAPKKK either. Instead, EDR1 may fine-tune responses to biotic and abiotic stresses by controlling this MAP kinase cascade.
Several proteins that were originally identified as negative regulators of plant immunity, such as LSD1 and ACD11, have on closer analysis turned out not be true negative regulators, as loss-of-function alleles lead to inappropriate activation of NBS-LRR proteins, suggesting that LSD1 and ACD11 may be guarded pathogen targets [46], [47]. However, EDR1 appears to be different from those proteins, as over-expression of EDR1 leads to enhanced susceptibility and lower activation of MAPK cascade, indicating that EDR1 could serve as a negative regulator of defense by repressing the MAPK pathway. In the absence of pathogen, EDR1 inactivates the MAPK pathways; however, upon pathogen infection, plants quickly activate defenses, possibly by de-repression of the inhibition of MAPK pathways by EDR1.
In conclusion, we show that EDR1 directly associates with MKK4 and MKK5, and negatively affects protein levels of MKK4, MKK5, MPK3 and MPK6, which may represent an important mechanism that fine-tunes plant defense responses.
Materials and Methods
Plant materials and growth conditions
The Arabidopsis thaliana mpk3-1 (SALK_151594) and mpk6-3 (SALK_127507) mutants were obtained from the Arabidopsis Stock Center (ABRC; Ohio State University; Columbus, OH). The homozygous T-DNA insertion mutants were confirmed by PCR. The edr1 mutant was described previously [26]. Mutations in MKK4 and MKK5 were isolated by the Arabidopsis TILLING facility from EMS-mutagenized Col er-105 plants [48]. The mkk4–18 allele has a substitution at a conserved position of the catalytic domain (proline-240 to serine; CCT to TCT). The mkk5–18 allele has a premature stop codon (arginine-72 to opal; CGA to TGA). The er-105 mutation was removed by crossing with Col-0. Plants were grown in the growth room at 20–22°C as described previously [49]. For molecular complementation of mkk4 and mkk5 mutants, genomic DNA fragments spanning both loci (MKK4 : 2.3 kb total, including approximately 0.6 kb upstream and downstream of the coding sequence; MKK5 : 2.5 kb total, including approximately 0.6 kb upstream and 0.9 kb downstream of the coding sequence) were introduced into mkk4 or mkk5 mutants, respectively. Three independent lines from each transformation were selected for further analyses.
Pathogen infection and ethylene assay
For powdery mildew infection, four-week-old plants were inoculated with G. cichoracearum strain UCSC1 as described previously [50]. To quantify the resistance, the conidiophores per colony were counted at 5 dpi. P. syringae pv. tomato (Pto) DC3000 infection assay and ethylene treatment assays were performed as described previously [51]. For H. a. Noco2 infection assay, three-week-old plants were used for infection [52].
Construction of plasmids
To generate EDR1 genomic-GFP and EDR1 genomic-Cherry constructs, the EDR1 genomic sequence including 1077 bp upstream of the ATG start codon was amplified (TOYOBO), and cloned into the pDONR207 ENTRY vector, and then into pMDC107 and pMDC163-Cherry destination vectors, respectively, using the Gateway cloning system (Invitrogen). The MKK4 and MKK5 genomic-GFP constructs were constructed using a similar strategy. For HA tagged MKK4, MKK5, MPK3 and MPK6 constructs, the coding sequence (CDS) of each gene was amplified and cloned into the pDNOR207 ENTRY vector and then into the pEarleyGate 201 destination vector.
For the yeast two-hybrid assay, the CDS sequences of full-length EDR1, EDR1 N-terminal domain and EDR1 C-terminal domain were amplified and ligated into vector pGBKT-7, and the CDS sequences of MKK1, MKK2, MKK4, MKK5, MPK3 and MPK6 were amplified and ligated into vector pGADT-7. For the BiFC assay, 35S-YN-EDR1, 35S-YC-MKK4 and 35S-YC-MKK5 were constructed according to the procedure described previously [53].
Immunoblotting and co-immunoprecipitation analysis
For protein extraction, leaves collected from Nicotiana benthamiana or Arabidopsis were ground in liquid nitrogen and the proteins were extracted using native extraction buffer (50 mM Tris-MES pH 8.0, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT and protease inhibitor cocktail S8830 (Sigma)). The total extraction was mixed well and centrifuged at 12000 rpm and 4°C for 30 min. The suspension was transferred to a new tube for further analysis. For immunoblotting, proteins were separated by SDS-PAGE (10% acrylamide gel) and transferred to PVDF membrane (Millipore) by electro-transfer at 80 V for 90 min. The membrane was blocked in 1× TBS buffer containing 5% skim milk powder and further incubated with primary antibody and secondary antibody. Finally the bands were detected using chemiluminescent HRP substrate (Millipore). For the co-IP assay, 1 ml protein extraction was incubated with 3 µL HA antibody for 4 hrs and then 40 µL Protein G (50% slurry, Millipore) was added to the cell lysates for another 2 h to capture the immunocomplex. The mixture was washed 3 times with cold PBS buffer containing 0.1% IGEPAL CA-630 (Sigma) and once with cold PBS buffer. Finally the agarose beads were resuspended in 50 µL 2× Laemmli sample buffer and 20 µL of supernatant was used for immunoblot. Antibodies used for immunoblotting were as follows: anti-HA antibody (1∶3000, H3663, Sigma), anti-GFP antibody (1∶2000, 632569, Clontech), anti-Myc antibody (1∶2000, M20002M, Abmart), anti-Flag antibody (1∶2000, F1804, Sigma), anti-MPK3 antibody (1∶10000, Sigma), anti-MPK6 antibody (1∶10000, Sigma) and anti–Phospho-p44/p42 MAPK (anti-pTEpY) (1∶2000, Cell Signaling Technology).
Yeast two-hybrid assay
For yeast two-hybrid assays, two constructs (pGBKT-7 and pGADT-7 containing the respective genes) were cotransformed into yeast AH109 strain (Clontech). The transformants were plated on synthetic dropout (SD) agar plates containing adenine and histidine for selection. A single colony for each transformant was incubated in the SD liquid medium containing adenine and histidine for 2 days. Subsequently, the concentration of each suspension was measured and diluted to OD 0.5, and 10 µL of each dilution was dropped on the selection medium SD for 2 days incubation.
Fluorescence assay
For BiFC assay, YFPN and YFPC fused proteins or YFPN and YFPC empty vector were co-transformed into N. benthamiana by Agrobacterium-mediated transformation. 48 hours later, the YFP fluorescence was detected by confocal microscopy.
For the colocalization assay, EDR1 pro-EDR1 genomic sequence-Cherry construct (EDR1 g-Cherry), MKK4 pro-MKK4 genomic sequence-GFP construct (MKK4 g-GFP) and MKK5 pro-MKK5 genomic sequence-GFP construct (MKK5 g-GFP) were transformed into wild-type, respectively. Then the copy number of the MKK4 g-GFP, MKK5 g-GFP and EDR1 g-Cherry expression constructs was determined, and the plants contained a single locus of the transgene were selected. And then, MKK4 g-GFP, MKK5 g-GFP transgenic plants were crossed with EDR1 g-Cherry transgenic plants, respectively, to generate double transgenic plants. The fluorescence of GFP and Cherry was observed by confocal microscopy.
For the comparison of the intensity of fluorescence, MKK4 g-GFP/EDR1 g-Cherry or MKK5 g-GFP/EDR1 g-Cherry double transgenic plants mentioned above were selected to compare with the corresponding MKK4 g-GFP or MKK5 g-GFP transgenic line. The transgenes are all homozygous, so the plants have the same genetic background, except for the specific transgene of interest. Identical parameters, including same laser strength and same pinhole, were applied for all samples. ImageJ software (http://rsb.info.nih.gov/ij) was used to quantify the intensity of GFP fluorescence.
Supporting Information
Zdroje
1. MengX, ZhangS (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51 : 245–266.
2. InnesRW (2001) Mapping out the roles of MAP Kinases in plant defense. Trends Plant Sci 6 : 392–395.
3. RodriguezMC, PetersenM, MundyJ (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61 : 621–649.
4. AlbrechtC, BoutrotF, SegonzacC, SchwessingerB, Gimenez-IbanezS, et al. (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci U S A 109 : 303–308.
5. AsaiT, TenaG, PlotnikovaJ, WillmannMR, ChiuWL, et al. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 : 977–983.
6. RouxM, SchwessingerB, AlbrechtC, ChinchillaD, JonesA, et al. (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23 : 2440–2455.
7. DesikanR, HancockJT, IchimuraK, ShinozakiK, NeillSJ (2001) Harpin induces activation of the Arabidopsis mitogen-activated protein kinases AtMPK4 and AtMPK6. Plant Physiol 126 : 1579–1587.
8. NuhseTS, PeckSC, HirtH, BollerT (2000) Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J Biol Chem 275 : 7521–7526.
9. MiyaA, AlbertP, ShinyaT, DesakiY, IchimuraK, et al. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A 104 : 19613–19618.
10. ZhengZ, QamarSA, ChenZ, MengisteT (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48 : 592–605.
11. RenD, LiuY, YangKY, HanL, MaoG, et al. (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci U S A 105 : 5638–5643.
12. MaoG, MengX, LiuY, ZhengZ, ChenZ, et al. (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23 : 1639–1653.
13. BeckersGJ, JaskiewiczM, LiuY, UnderwoodWR, HeSY, et al. (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21 : 944–953.
14. RenD, YangH, ZhangS (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem 277 : 559–565.
15. ZhangJ, ShaoF, LiY, CuiH, ChenL, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1 : 175–185.
16. HeP, ShanL, LinNC, MartinGB, KemmerlingB, et al. (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125 : 563–575.
17. XiangT, ZongN, ZouY, WuY, ZhangJ, et al. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18 : 74–80.
18. ShanL, HeP, LiJ, HeeseA, PeckSC, et al. (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4 : 17–27.
19. WangY, LiJ, HouS, WangX, LiY, et al. (2010) A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22 : 2033–2044.
20. GoehreV, SpallekT, HaewekerH, MersmannS, MentzelT, et al. (2008) Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Current Biology 18 : 1824–1832.
21. SchweighoferA, KazanaviciuteV, ScheiklE, TeigeM, DocziR, et al. (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19 : 2213–2224.
22. BrockAK, WillmannR, KolbD, GrefenL, LajunenHM, et al. (2010) The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol 153 : 1098–1111.
23. BartelsS, AndersonJC, Gonzalez BesteiroMA, CarreriA, HirtH, et al. (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21 : 2884–2897.
24. LeeJS, EllisBE (2007) Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J Biol Chem 282 : 25020–25029.
25. LumbrerasV, VilelaB, IrarS, SoleM, CapelladesM, et al. (2010) MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J 63 : 1017–1030.
26. FryeCA, InnesRW (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10 : 947–956.
27. PanH, LiuS, TangD (2011) HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J 69 : 831–843.
28. FryeCA, TangD, InnesRW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci U S A 98 : 373–378.
29. TangD, InnesRW (2002) Overexpression of a kinase-deficient form of the EDR1 gene enhances powdery mildew resistance and ethylene-induced senescence in Arabidopsis. Plant J 32 : 975–983.
30. WawrzynskaA, ChristiansenKM, LanY, RodibaughNL, InnesRW (2008) Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol 148 : 1510–1522.
31. GuY, InnesRW (2011) The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-golgi network/early endosome vesicles. Plant Physiology 155 : 1827–1838.
32. KieberJJ, RothenbergM, RomanG, FeldmannKA, EckerJR (1993) CTR1, a negative regulator of the ethylene response pathway in arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72 : 427–441.
33. YooSD, ChoYH, TenaG, XiongY, SheenJ (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451 : 789–795.
34. van HultenM, PelserM, van LoonLC, PieterseCM, TonJ (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci U S A 103 : 5602–5607.
35. ObayashiT, HayashiS, SaekiM, OhtaH, KinoshitaK (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 37: D987–991.
36. ManfieldIW, JenCH, PinneyJW, MichalopoulosI, BradfordJR, et al. (2006) Arabidopsis Co-expression Tool (ACT): web server tools for microarray-based gene expression analysis. Nucleic Acids Res 34: W504–509.
37. BoudsocqM, WillmannMR, McCormackM, LeeH, ShanL, et al. (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464 : 418–422.
38. MengX, WangH, HeY, LiuY, WalkerJC, et al. (2012) A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24 : 4948–4960.
39. LukowitzW, RoederA, ParmenterD, SomervilleC (2004) A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116 : 109–119.
40. LuC, HanMH, Guevara-GarciaA, FedoroffNV (2002) Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci U S A 99 : 15812–15817.
41. YaoC, WuY, NieH, TangD (2012) RPN1a, a 26S proteasome subunit, is required for innate immunity in Arabidopsis. Plant J 71 : 1015–1028.
42. WangH, NgwenyamaN, LiuY, WalkerJC, ZhangS (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19 : 63–73.
43. IchimuraK, ShinozakiK, TenaG, SheenJ, HenryY, et al. (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7 : 301–308.
44. JuC, YoonGM, ShemanskyJM, LinDY, YingZI, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci U S A 109 : 19486–19491.
45. QiaoH, ShenZ, HuangSS, SchmitzRJ, UrichMA, et al. (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338 : 390–393.
46. BonardiV, TangS, StallmannA, RobertsM, CherkisK, et al. (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci U S A 108 : 16463–16468.
47. PalmaK, ThorgrimsenS, MalinovskyFG, FiilBK, NielsenHB, et al. (2010) Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog 6: e1001137.
48. TillBJ, ReynoldsSH, GreeneEA, CodomoCA, EnnsLC, et al. (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res 13 : 524–530.
49. ShiH, ShenQ, QiY, YanH, NieH, et al. (2013) BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25 : 1143–1157.
50. WangY, NishimuraMT, ZhaoT, TangD (2011) ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J 68 : 74–87.
51. NieH, ZhaoC, WuG, WuY, ChenY, et al. (2012) SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol 158 : 1847–1859.
52. LiX, ClarkeJD, ZhangY, DongX (2001) Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact 14 : 1131–1139.
53. Bracha-DroriK, ShichrurK, KatzA, OlivaM, AngeloviciR, et al. (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40 : 419–427.
Štítky
Genetika Reprodukční medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 5
-
Všechny články tohoto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



