-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster
The ascomycete fungus Tolypocladium inflatum, a pathogen of beetle larvae, is best known as the producer of the immunosuppressant drug cyclosporin. The draft genome of T. inflatum strain NRRL 8044 (ATCC 34921), the isolate from which cyclosporin was first isolated, is presented along with comparative analyses of the biosynthesis of cyclosporin and other secondary metabolites in T. inflatum and related taxa. Phylogenomic analyses reveal previously undetected and complex patterns of homology between the nonribosomal peptide synthetase (NRPS) that encodes for cyclosporin synthetase (simA) and those of other secondary metabolites with activities against insects (e.g., beauvericin, destruxins, etc.), and demonstrate the roles of module duplication and gene fusion in diversification of NRPSs. The secondary metabolite gene cluster responsible for cyclosporin biosynthesis is described. In addition to genes necessary for cyclosporin biosynthesis, it harbors a gene for a cyclophilin, which is a member of a family of immunophilins known to bind cyclosporin. Comparative analyses support a lineage specific origin of the cyclosporin gene cluster rather than horizontal gene transfer from bacteria or other fungi. RNA-Seq transcriptome analyses in a cyclosporin-inducing medium delineate the boundaries of the cyclosporin cluster and reveal high levels of expression of the gene cluster cyclophilin. In medium containing insect hemolymph, weaker but significant upregulation of several genes within the cyclosporin cluster, including the highly expressed cyclophilin gene, was observed. T. inflatum also represents the first reference draft genome of Ophiocordycipitaceae, a third family of insect pathogenic fungi within the fungal order Hypocreales, and supports parallel and qualitatively distinct radiations of insect pathogens. The T. inflatum genome provides additional insight into the evolution and biosynthesis of cyclosporin and lays a foundation for further investigations of the role of secondary metabolite gene clusters and their metabolites in fungal biology.
Published in the journal: . PLoS Genet 9(6): e32767. doi:10.1371/journal.pgen.1003496
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003496Summary
The ascomycete fungus Tolypocladium inflatum, a pathogen of beetle larvae, is best known as the producer of the immunosuppressant drug cyclosporin. The draft genome of T. inflatum strain NRRL 8044 (ATCC 34921), the isolate from which cyclosporin was first isolated, is presented along with comparative analyses of the biosynthesis of cyclosporin and other secondary metabolites in T. inflatum and related taxa. Phylogenomic analyses reveal previously undetected and complex patterns of homology between the nonribosomal peptide synthetase (NRPS) that encodes for cyclosporin synthetase (simA) and those of other secondary metabolites with activities against insects (e.g., beauvericin, destruxins, etc.), and demonstrate the roles of module duplication and gene fusion in diversification of NRPSs. The secondary metabolite gene cluster responsible for cyclosporin biosynthesis is described. In addition to genes necessary for cyclosporin biosynthesis, it harbors a gene for a cyclophilin, which is a member of a family of immunophilins known to bind cyclosporin. Comparative analyses support a lineage specific origin of the cyclosporin gene cluster rather than horizontal gene transfer from bacteria or other fungi. RNA-Seq transcriptome analyses in a cyclosporin-inducing medium delineate the boundaries of the cyclosporin cluster and reveal high levels of expression of the gene cluster cyclophilin. In medium containing insect hemolymph, weaker but significant upregulation of several genes within the cyclosporin cluster, including the highly expressed cyclophilin gene, was observed. T. inflatum also represents the first reference draft genome of Ophiocordycipitaceae, a third family of insect pathogenic fungi within the fungal order Hypocreales, and supports parallel and qualitatively distinct radiations of insect pathogens. The T. inflatum genome provides additional insight into the evolution and biosynthesis of cyclosporin and lays a foundation for further investigations of the role of secondary metabolite gene clusters and their metabolites in fungal biology.
Introduction
Fungi are prolific producers of secondary metabolites and an important source of novel and commercially important pharmaceuticals, mycoinsecticides, and antibiotics. Cyclosporin A (CsA; CAS ID: 59865-13-3), the well-known immunosuppressant drug which revolutionized organ transplantation from an experimental to a relatively routine lifesaving procedure [1], was first discovered in the insect pathogenic and ubiquitous soil fungus, Tolypocladium inflatum. CsA targets and binds with high affinity human cyclophilin A (hCypA, peptidylprolyl isomerase A, EC: 5.2.1.8), a conserved immunophilin found across eukaryotes [2], [3]. The CsA-hCypA complex suppresses the vertebrate immune system by binding to and inhibiting calcineurin, a conserved calcium-calmodulin activated serine/threonine-specific protein phosphatase (EC: 3.1.3.16) [4], [5]. Inhibition of calcineurin blocks activity of NF-AT (nuclear factor of activated T-cells), a regulator of transcription of Interleukin 2 in T-lymphocytes [6]. CsA also impairs the immune response in insects and shows both antifungal [7] and antiviral activity [8]. CsA and related cyclosporins (B-Z and isoforms) form a family of cyclic undecapeptides produced by nonribosomal peptide synthetases (NRPSs), a class of large multimodular enzymes that produce peptides via a nonribosomal mechanism [9]. While the 45.8 kb locus encoding the NRPS synthetase (simA) responsible for biosynthesis of cyclosporin was cloned in 1994 [10], the complete biosynthetic cluster remains uncharacterized.
T. inflatum belongs to the fungal order Hypocreales, containing fungi known to produce a high diversity of bioactive secondary metabolites [11]. In addition to cyclosporins, T. inflatum synthesizes a number of other products via both NRPSs and polyketide synthetases (PKSs), another class of multimodular enzyme involved in secondary metabolite production in bacteria and fungi [12]. Other hypocrealean fungi are known to produce NRPS or PKS products with activity against insects, such as destruxins (Metarhizium robertsii) [13], efrapeptins (Tolypocladium spp.) [14], and ergot alkaloids (clavicipitalean endophytes of grasses including Claviceps and Epichloë spp.) [15]. Many of these compounds also have pharmaceutical applications and/or roles in antibiosis, pathogenesis, and competitive interactions between organisms [13], [14]. The genome sequence of T. inflatum thus provides an opportunity to characterize the secondary metabolite arsenal of an insect-pathogenic fungus with potential for both elucidation of the biosynthetic cluster of the immunosuppressant drug cyclosporin and discovery of novel gene clusters and metabolites with applications in medicine and agriculture.
Hypocrealean fungi display considerable flexibility of lifestyles. They include plant-pathogens, plant-saprobes, plant-endophytes, mycoparasites, and pathogens of insects, spiders, rotifers, and nematodes. Transitions between different lifestyles have occurred multiple times in the evolutionary history of the order [16], [17]. T. inflatum is a pathogen of beetle larvae [18], but is also able to live saprotrophically in soil during the asexual phase of its lifecycle (Figure 1). It is one of the few insect pathogenic (entomopathogenic) fungi sequenced to date, although Hypocreales contains three families (Clavicipitaceae, Cordycipitaceae, and Ophiocordycipitaceae) that are particularly rich in entomopathogenic species. The genomes of M. robertsii and M. acridum (Clavicipitaceae) have provided insights into expansions of gene families, especially those for secreted proteins, with roles in insect pathogenesis [19]. Other studies have shown changes in the profiles of carbohydrate active enzymes (CAZymes), cytochrome P450s, and proteases in insect pathogens when compared to closely related plant pathogens [20]. The genome of Cordyceps militaris (Cordycipitaceae), a common pathogen of moth pupae used in traditional Chinese Medicine, revealed aspects of the mating systems of entomopathogenic fungi [21]. These taxa belong to three separate families of Hypocreales that represent parallel diversifications of entomopathogenic fungi [16].
Fig. 1. Lifecycle of T. inflatum. 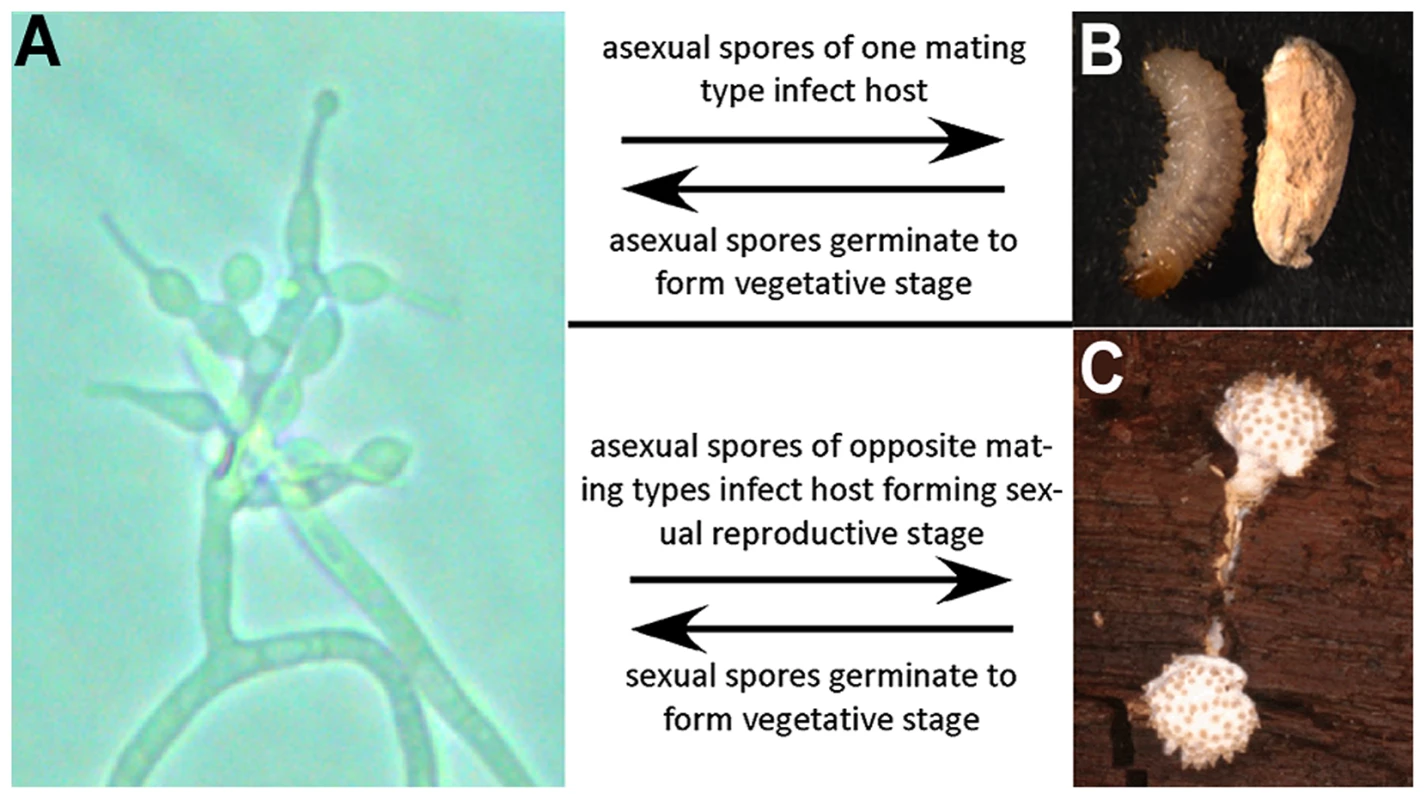
The lifecycle of T. inflatum comprises both an asexual stage (primarily saprotrophic growth in soil) and a sexual stage (only occurs on an insect host). A) Asexual reproductive stage growing on cornmeal agar produces asexual spores on phialides with diagnostic inflated bases. B) Uninfected beetle larva (left) and beetle larva infected with asexual spores (right). C) Sexual reproductive stage growing off of an infected beetle host buried in wood. The sexual stage only occurs when two spores of opposite mating type infect the same insect host. Here we present the results from whole genome sequencing and RNA-Seq analyses of T. inflatum (Ophiocordycipitaceae), which represents the first draft genome from the third entomopathogenic family within Hypocreales. Through phylogenomic and comparative genomic analyses we demonstrate that the NRPS responsible for cyclosporin biosynthesis (simA) exhibits complex patterns of homology with other NRPSs and that the cyclosporin gene cluster is unique to Tolypocladium with no evidence for horizontal transfer of a complete cluster from other fungi or bacteria. RNA-Seq analyses in a cyclosporin-inducing medium clearly delineate a secondary metabolite cluster responsible for cyclosporin biosynthesis. RNA-Seq analyses in media simulating insect pathogenesis indicate that several genes within the cluster, including the homolog of the cyclosporin binding protein cyclophilin, are also upregulated in response to insect hemolymph, supporting a role for both cyclosporin and the cyclophilin gene in pathogenesis of insects.
Results/Discussion
General Genome Features
A karyotype study of the sequenced strain T. inflatum NRRL 8044 (ATCC 34921) indicated that T. inflatum has 6 chromosomes ranging in size from 3.8 to 6.6 Mb and a mini supernumerary chromosome of 1 Mb with a total genome size of approximately 30.45 Mb [22]. The total size of the assembly (30.348 Mb) closely matched this estimate and contained 194 contigs in 101 scaffolds with an N50 of 1.5 Mb and an Nmax of 3.56 Mb. The MAKER 2.0 annotation pipeline [23] predicted 9,998 protein coding genes (loci tagged as TINF) with greater than 90% having support from either protein (Fusarium graminearum, Nectria haematococca, Trichoderma reesei, Tr. virens, and M. robertsii) or EST data (C. militaris, Beauveria bassiana, M. robertsii, Tr. reesei, and assembled T. inflatum RNA-Seq reads). Analysis using the Core Eukaryotic Genes Mapping Approach (CEGMA) pipeline [24] estimated that the annotations represent >98% of coding regions based on completeness of a conserved set of eukaryotic proteins. The average gene length (1.67 kb), exon length (570 bp) and intron length (77.5 bp) were similar to estimates from other Ascomycota (Table 1) [25]. However, T. inflatum has a higher average GC content (58%) and a more compact genome with higher gene density (329 genes/Mb) than closely related filamentous ascomycetes (Table 1). Only the MAT1-2 mating type locus was detected in the sequenced strain, indicating that T. inflatum is likely heterothallic (Figure S1).
Tab. 1. Comparison of genome features with closely related ascomycetes. 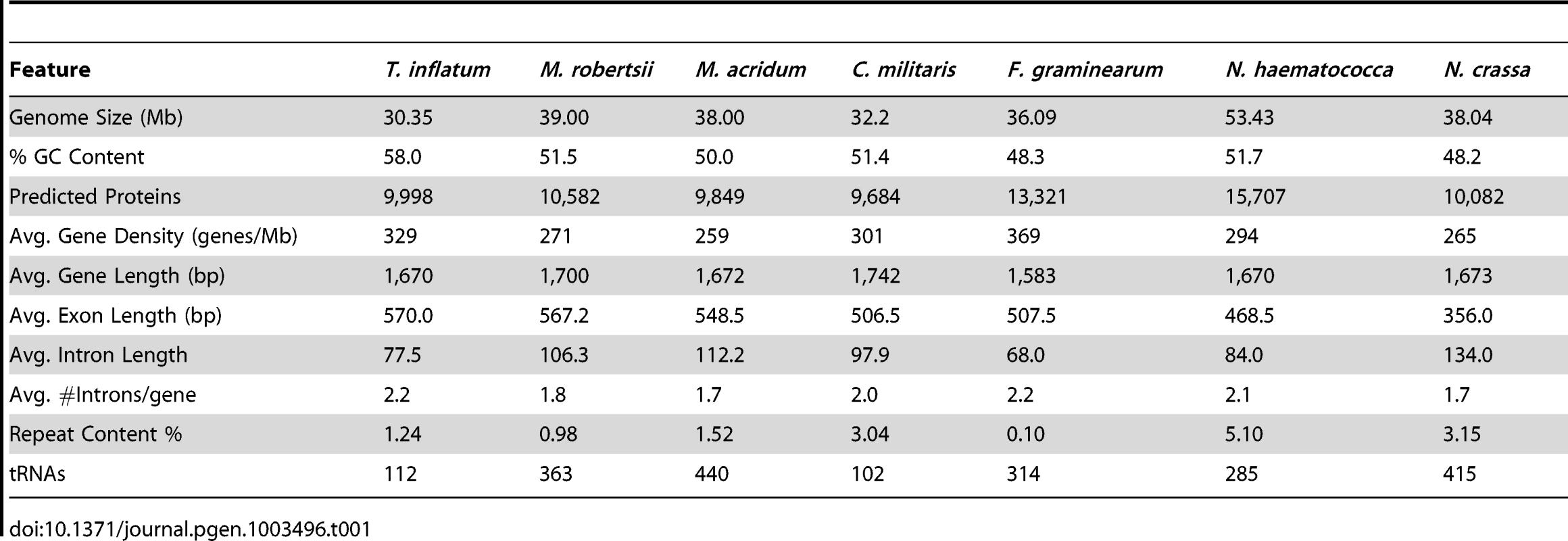
Repeat Elements
The estimated proportion of repeat sequence (1.24%), which agrees well with previous experimental estimates (1%) [26], is relatively low compared to other filamentous ascomycetes (Table 1). In total, T. inflatum contained a slightly larger number of retrotransposons compared to DNA transposons (Table S1). Retrotransposons were dominated by two classes, LINE elements and the Gypsy family of LTRs, while DNA transposons were mostly comprised of the hAT family (Table S1). Several novel repeat elements, including the CPA (cyclosporin production associated) element [26] and the first characterized fungal hAT transposon (Restless) [27], were previously identified in the sequenced strain (NRRL 8044). The CPA element, which shows greatest similarity to a RecQ DNA helicase in M. robertsii [28], was named based on the observation that multiple copies were found only in cyclosporin producing strains of T. inflatum while only a single copy was present in other strains of T. inflatum and related Tolypocladium and Beauveria species. We identified 12 copies of the CPA element in T. inflatum NRRL 8044 that were found dispersed across eleven scaffolds, but none of them associated with the scaffold containing the cyclosporin biosynthetic cluster. A single partial copy was also found in C. militaris and Tr. virens and multiple copies were present in Tr. reesei, Metarhizium spp., and especially N. haematococca, in which the transposon has undergone expansion to 27 copies (Table S1). We conclude that presence and expansion of CPA elements is not unique to T. inflatum and that it is not associated with the evolution or production of cyclosporin. Similarly, copies of Restless were also found in other hypocrealean taxa and were particularly expanded in F. oxysporum, which harbored over double (55) the number of elements in T. inflatum (26) (Table S1). Restless generates partial deletion copies of the transposon and several deletion variants (ΔRst 1-6) have been found in T. inflatum, Neurospora crassa, and Penicillium chrysogenum [29]. With the exception of Tr. atroviride, all hypocrealean taxa contained either an intact or a deletion variant of Restless, indicating the transposon was present in the ancestor of Hypocreales. However, of the genomes analyzed, deletion variants were most abundant and diverse in T. inflatum (Table S1), suggesting it has been particularly active in T. inflatum.
Phylogenomic Relationships and Orthologous Gene Clusters
T. inflatum is a member of the class Sordariomycetes and is related to the widely studied filamentous ascomycete N. crassa, which together with the wilt pathogens Verticillium dahliae and Verticillium albo-atrum, served as an outgroup to the order Hypocreales in our phylogenomic analyses (Figure 2A, node 1). Orthologous clustering of proteins using MCL [30] identified a total of 36,532 orthologous clusters of proteins across the 14 taxa analyzed (N. crassa, V. dahliae, V. albo-atrum, N. haematococca, F. graminearum, F. oxysporum, F. verticillioides, Tr. virens, Tr. atroviride, Tr. reesei, C. militaris, M. robertsii, M. acridum, and T. inflatum). Using the phylogenomic pipeline Hal [31], these 36,532 clusters were filtered to identify 2,769 clusters containing only single-copy orthologous proteins. A concatenated alignment was built from this subset of clusters and used to construct a maximum likelihood phylogeny.
Fig. 2. Phylogenetic relationships and orthologous gene clusters. 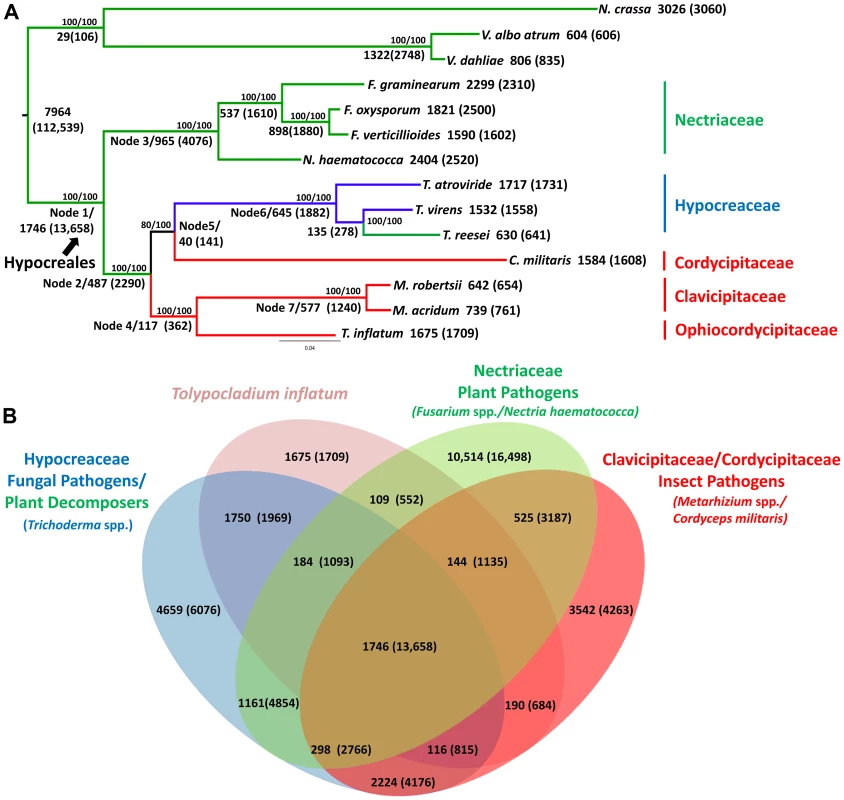
A) Maximum likelihood phylogeny created from a concatenated alignment of 2769 groups of single copy orthologs identified by the Hal pipeline. Phylogeny constructed using RAxML with best models for each cluster partition identified using ProtTest. Bootstrap values for analyses with the original alignment (top)/the alignment with fast-evolving sites removed (bottom) are shown above nodes. Larger numbers beneath or adjacent to nodes and terminal taxa indicate the number of clusters and genes (in parentheses) within those clusters that map to each node in the phylogeny or are unique to a species. Color coding corresponds to fungal host: green = plant associated, blue = fungal associated, red = animal or insect associated. Hypocreales is delineated at node 1. A major shift from early diverging taxa that have primarily plant-associated hosts to either animal/insect or fungal hosts occurs at node 2. B) The number of both clusters and number of genes (in parentheses) in those clusters that are shared by T. inflatum with each of the major families and associated ecologies within Hypocreales: green = Nectriaceae, primarily plant associated including F. graminearum, F. oxysporum, F. verticillioides, and N. haematococca; blue = Hypocreaceae, primarily fungal associated (Tr. atroviride and Tr. virens) or plant saprobic (Tr. reesei); red = Clavicipitaceae/Cordycipitaceae, primarily animal or insect associated including C. militaris, M. robertsii, M. acridum; and pink = T. inflatum. The inferred phylogeny recovered a topology consistent with the four major families of Hypocreales (Figure 2A). The earliest diverging group, Nectriaceae, comprises primarily plant pathogenic species including the wheat head blight fungus F. graminearum and related species F. oxysporum, F. verticillioides, and N. haematococca (Figure 2A, node 3, green). Sequenced representatives of Hypocreaceae include members of the genus Trichoderma (Tr. atroviride, Tr. reesei, Tr.virens). Although the ability of Trichoderma spp. to grow on and digest plant-based compounds is well-documented, recent comparative genomic studies support an evolutionary history characterized by mycoparasitism [32](Figure 2A, node 6, blue). Hypocreaceae forms a sister group (node 5) to the primarily insect pathogenic Cordycipitaceae, which includes the moth pathogen and traditional Chinese medicinal fungus C. militaris. The remaining two families, Ophiocordycipitaceae, of which T. inflatum is the first sequenced representative, and Clavicipitaceae (Figure 2A, node 7, red), which includes the two insect pathogenic biocontrol species M. robertsii and M. acridum, comprise, with the exception of the clavicipitaceous endophytes, primarily insect pathogens. This topology is consistent with standard multigene (e.g., five to seven loci) phylogenetic analyses that have sampled an order of magnitude more species of Hypocreales and ancestral character state reconstructions from previously published multigene datasets [16], [17], which support a major transition within the order from plant hosts/substrates in early diverging lineages (Nectriaceae) to primarily insect (Clavicipitaceae, Cordycipitaceae, Ophiocordycipitaceae) or fungal (Hypocreaceae) hosts (Figure 2A, node 2). However, the placement of Cordycipitaceae has been controversial. The removal of fast evolving sites [33] in these genome-scale analyses provides stronger bootstrap support for the placement of C. militaris (Cordycipitaceae) as monophyletic with Trichoderma (Hypocreaceae) and not with the other insect pathogens of Metarhizium (Clavicipitaceae) and Tolypocladium (Ophiocordycipitaceae). These results provide additional support for polyphyletic origins and parallel diversifications of insect pathogenic fungi in three separate families in Hypocreales [34].
Out of the total 36,532 orthologous clusters, those shared by one or more descendants of each node were mapped to the phylogeny to produce a phylogenetic profile of orthologous clusters (Figure 2A). A total of 7,964 clusters containing 112,539 proteins from both within Hypocreales and from outgroup taxa (N. crassa, V. dahliae and V. albo atrum), while 1,746 clusters containing 13,658 proteins mapped uniquely to the node representing the origin of Hypocreales (Figure 2A, node 1). Within Hypocreales, plant associated species in the Nectriaceae had the largest number of unique clusters although these genomes also contained a larger number of protein coding genes per genome (Table 1). T. inflatum had 1,675 species-unique clusters containing 1,709 proteins (out of a total of 9998). T. inflatum shared a larger number of clusters with fungal pathogens in Hypocreaceae (1750) than with other insect pathogens in Clavicipitaceae and Cordycipitaceae (190) or with plant pathogens in Nectriaceae (109) (Figure 2B).
Gene Content and Evolution
All hypocrealean taxa, including T. inflatum, shared a similar profile of Gene Ontology (GO) Slim categories (Figures 3A, S2). However, GO Slim profiles of genes found in orthologous clusters unique to each of the insect pathogens C. militaris (Cordycipitaceae), M. robertsii and M. acridum (Clavicipitaceae), and T. inflatum (Ophiocordycipitaceae) showed lineage specific differences (Figure 3B). Metarhizium spp. (Clavicipitaceae) had a larger proportion of species-unique genes associated with GO molecular functions of protein binding (22–26% vs 13–16%), oxidoreductase activity (20% vs 8–11%), and peptidase activity (7–9% vs 2–3%) relative to either T. inflatum or C. militaris (Figures 3B, S2B). These results are consistent with expansions of both proteases (peptidase activity) and P450s (oxidative activity) in Metarhizium spp., Tr. virens, and all plant pathogenic species in Nectriaceae, but not in the other insect pathogens T. inflatum or C. militaris (Table S2). In fact, M. acridum and particularly M. robertsii, which is known to live in association with the plant rhizosphere [35], showed overall profiles of CAZymes, proteases, and P450s more similar to plant pathogens than to the other insect pathogens (Table S2). T. inflatum contained a larger proportion of species-unique genes associated with the GO molecular function of transporter activity (7% vs 2%) than other insect pathogens, while C. militaris had a larger proportion of species-unique genes associated with transferase activity (39% vs 13–22%) (Figures 3B, S2B). For GO biological process categories, T. inflatum also contained a larger proportion of species-unique genes related to transport (11% vs 3–4%) but a smaller proportion of genes involved in DNA-dependent transcription (7% vs 18–23%) relative to other insect pathogens (Figure S2D). A large proportion of species-unique genes in all insect pathogens (31–74%) were associated with membranes, particularly endomembrane systems (Figure S2F), consistent with the importance of secreted proteins in these fungi [19]. These differences in gene content and ontology between the three insect pathogenic lineages corroborate phylogenetic evidence for parallel and qualitatively distinct evolutionary radiations of insect pathogens in Hypocreales that reflect adaptations to distinct ecologies.
Fig. 3. Functional classification of gene content. 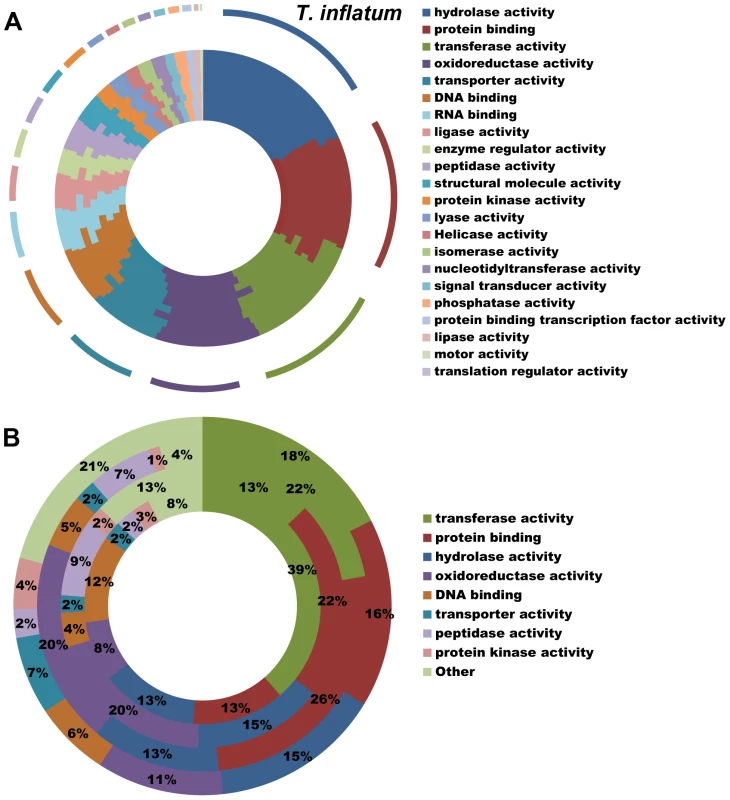
A) Profiles of GO Slim (Aspergillus GO Slim) molecular function categories for hypocrealean taxa. Taxa from inside of circle to outside of circle are F. oxysporum, F. verticillioides, F. graminearum, N. haematococca, Tr. atroviride, Tr. reesei, Tr. virens, C. militaris, M. acridum, M. robertsii, and T. inflatum. B) Profiles of GO Slim molecular function categories for genes in species-unique orthologous clusters (Figure 3A) showing percentage of genes in each category out of total annotated genes. Taxa from inside to outside are C. militaris, M. robertsii, M. acridum, and T. inflatum. Overview of Secondary Metabolite Gene Clusters
While T. inflatum is best known as the original source of CsA [36], it is also known to produce other bioactive secondary metabolites including insecticidal compounds such as efrapeptins [37] and tolypin [38], diketopiperazines [39], and the carboxysterol antibiotic ergokonin-C [39]. In addition to well-known core enzymes involved in producing fungal secondary metabolites (NRPSs, PKSs, prenyltransferases, and terpene cyclases (TC)), a large number of modifying enzymes such as racemases, methyltransferases, acetyl transferases, prenyltransferases, cytochrome P450 monooxygenases (P450s) and oxidoreductases are often required for synthesis of the final bioactive products. In fungi, these are often found clustered with the core enzymes to form secondary metabolite biosynthetic gene clusters [40], which some hypothesize may facilitate or be driven by horizontal transfer [41]. Others suggest clustering may minimize the number of coordinated interactions between regulatory elements [42]. We identified a total of 14 NRPSs, 20 PKSs, 4 Hybrid PKS/NRPSs, 11 putative NRPS-like enzymes, 5 putative PKS-like enzymes, and one dimethylallyl-tryptophan synthase (DMATS) in the T. inflatum genome (Table S3), indicating that T. inflatum has a large potential for secondary metabolite production. The majority of these core enzymes fell within one of the 36 secondary metabolite clusters identified by SMURF [43] or an additional 2 clusters (38 total) identified by antiSMASH [44].
In addition to the NPRS in the cyclosporin cluster, phylogenomic analyses of T. inflatum NRPS adenylation domains (Figure S3, Table S4) identified homologs of a number of functionally characterized NRPSs from fungi, including three peptaibol synthetases (TINF05969, TINF07827, TINF07876), both intracellular (ChNPS2 - TINF08996) and extracellular (ChNPS6 - TINF01764 and TINF06175) siderophore synthetases, and a homolog of conserved NRPS-like proteins involved in morphological development (ChNPS10 TINF09755) (Figure S3, Table S4).
We also identified an NRPS (TINF02556) whose A-domains group with those of the ergot alkaloid synthetases (cpps1-4) from the grass endophyte Claviceps purpurea (Figures S3, S4A). The ergot alkaloid cluster in C. purpurea contains two trimodular NRPSs (cpps1, cpps4), two monomodular NRPSs (cpps2, cpps3), and a DMATS [45], [46]. The alkaloid secondary metabolite clusters recently discovered in the insect pathogens M. robertsii and M. acridum [19] contain homologs of the two monomodular NRPSs (cpps2, cpps3), the DMAT synthase, and the majority of modifying enzymes found on the 5′ end of the C. purpurea cluster. In contrast, the antiSMASH predicted cluster in T. inflatum lacks homologs of these monomodular NRPSs and other ergot alkaloid biosynthetic genes but contains a four modular NRPS (TINF02556) with A-domains that show closest similarity to the trimodular NRPSs (cpps1 and cpps4) from C. purpurea, as well as other genes involved in secondary metabolism (Figure S4B). We also identified an additional cluster in Metarhizium spp. which contains a 7 modular NRPS (MAA_06559, MAC_08899) with A-domains that also show homology to cpps1 and cpps4, but which also lacks other genes from the ergot alkaloid cluster (Figure S4B). T. inflatum does contain one DMAT synthase. However, it is located on a different scaffold in a distinct secondary metabolite cluster predicted by antiSMASH to be involved in terpene biosynthesis (Figure S4C). While further chemical data is needed, we conclude it is unlikely that the T. inflatum cluster produces an ergot alkaloid compound similar to those produced by C. purpurea.
Evolution of Cyclosporin Synthetase (simA)
Cyclosporins are cyclic depsipeptides belonging to a class of cyclic undecapeptides [10], [47], and T. inflatum is known to produce 25 different analogs of cyclosporin (cyclosporins A-I and K-Z) [48], [49]. CsA is composed of 11 substrate molecules produced by an NRPS encoded by the single 45.8 kb simA locus, and like the products of many NRPSs, CsA contains several non-proteinogenic substrates including 2-aminobutyric acid, D-alanine, and (4R)-4-[(E)-2-butenyl]-4-methyl-threonine (Bmt) [10]. The simA gene displays a modular structure typical of NRPSs, consisting of 11 modules comprised of three core catalytic domains: adenylation (A), which binds and activates the substrate, thiolation (T), which attaches substrates to the NRPS, and condensation (C), which forms a peptide bond between adjacent substrates. Each module activates one of the eleven substrates and additional methylation (M) domains are present which methylate substrates 2 (Leu), 3, (Leu), 4 (Val), 5 (Bmt), 8 (Leu) and 10 (Leu). Various other fungi within Hypocreales (Acremonium, Chaunopycnis, Fusarium, Isaria, Nectria, Neocosmospora, Trichoderma and Verticillium) have been reported to synthesize a common profile of cyclosporins A-D and E-F [49], [50]. Only a few fungi outside of Hypocreales (Aspergillus terreus) [51] have been reported to produce cyclosporin A, while others (Leptostroma, Cylindrotrichum, Stachybotrys) produce a single and often novel cyclosporin-related compound [52], [53].
A previous phylogenomic study of NRPSs from 37 complete fungal genomes found that simA grouped sister to a clade of bacterial NRPSs but found no complete (11 modular) homolog of simA in either bacteria or other fungi [54]. Similarly, we employed BLAST searches of the NCBI database and HMMER searches across closely related hypocrealean fungi and found no complete homologs of simA (Figures 4, S3, S5). The phylogenetic tree constructed from A-domain sequences of the top 50 BLAST hits to simA from the NCBI nr database (Figure 4, S5) showed that no bacterial sequences group within the simA clade. This suggests that simA likely evolved by duplication of modules within fungi rather than through recent horizontal transfer from bacteria. Individual adenylation domains from several other fungal NRPSs group within the simA clade with 100% bootstrap support (Figures 4B, S5). These include NRPSs synthesizing several other known fungal cyclic depsipeptides such as enniatin synthetase (esyn1) (Fusarium equiseti) [55], beauvericin (bbBeas) and bassianolide (bbBsls) synthetases (Beauveria bassiana), the NRPS responsible for biosynthesis of the antifungal compound aureobasidin A (aba1) (Aureobasidium pullulans), and two modules of destruxin synthetase (dtxS1) (M. robertsii) (Figures 4B, S5). These compounds share similar functions, having either anti-insect (beauvericin, bassianolide, destruxin, and cyclosporin A) [13], [56]–[58] and/or antifungal properties (aureobasidin A [59] and cyclosporin A [60]).
Fig. 4. Phylogeny of cyclosporin synthetase (simA). 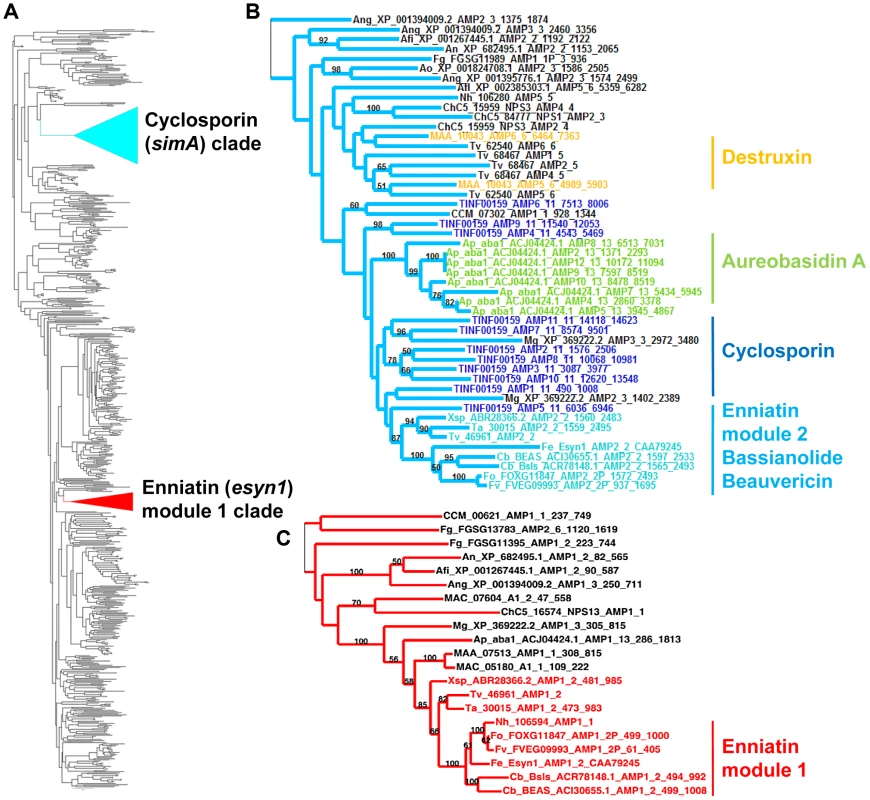
A) Locations of the cyclosporin (simA) and enniatin synthetase (esyn1) module1 clade showing their disparate locations in the larger NRPS A-domain phylogeny. B) Expanded view of the simA clade, showing modules from six other fungal NRPSs producing cyclic depsipeptides and containing A-domains in the simA clade: module 2 of enniatin synthetase (esyn1), module 2 of beauvericin synthetase (bbBeas), module 2 of bassianolide synthetase (bbBsls), aureobasidin A synthetase (aba1), and modules 5 and 6 of destruxin A synthetase (dtxS1). C) Expanded view of the enniatin synthetase (esyn1) module 1 clade containing module 1 of esyn1, bbBeas, and bbBsls. Fungal NRPSs containing adenylation (A) domains found in the simA clade, share a complex history of evolution through duplication and fusion of modules [61]. For example, the A-domain of module 1 of the NRPSs synthesizing enniatin (esyn1), beauvericin (bbBeas), and bassianolide (bbBsls) synthetases all code for an identical non-amino acid substrate, D-2-hydroxyisovaleric acid (Hiv) and group together phylogenetically in a clade distinct from the simA clade (Figures 4A, C, 5, S5 [enniatin module 1 clade]). In contrast, the A-domain from module 2 of enniatin and the C-terminal modules of all of these genes fall within the simA clade (Figures 4A, B, 5, S5 [enniatin module 2 clade]), suggesting fusion of modules. Other fungal NRPSs from Magnaporthe grisea (XP_369222.2) and Aspergillus species (XP_001394009.2, XP 682495.1, XP 001267445.1) display a similar pattern. Others, such as NPS1 and NPS3 from Cochliobolus heterostrophus and destruxin synthetase from M. robertsii, contain two A-domains grouping in the simA clade, but remaining A-domains group with the Epichloë festucae NRPS perA, outside both the simA and the enniatin module 1 clades (Figures 5, S3).
Fig. 5. Modular domain structure and A-domain specificities of NRPSs grouping within the simA clade. 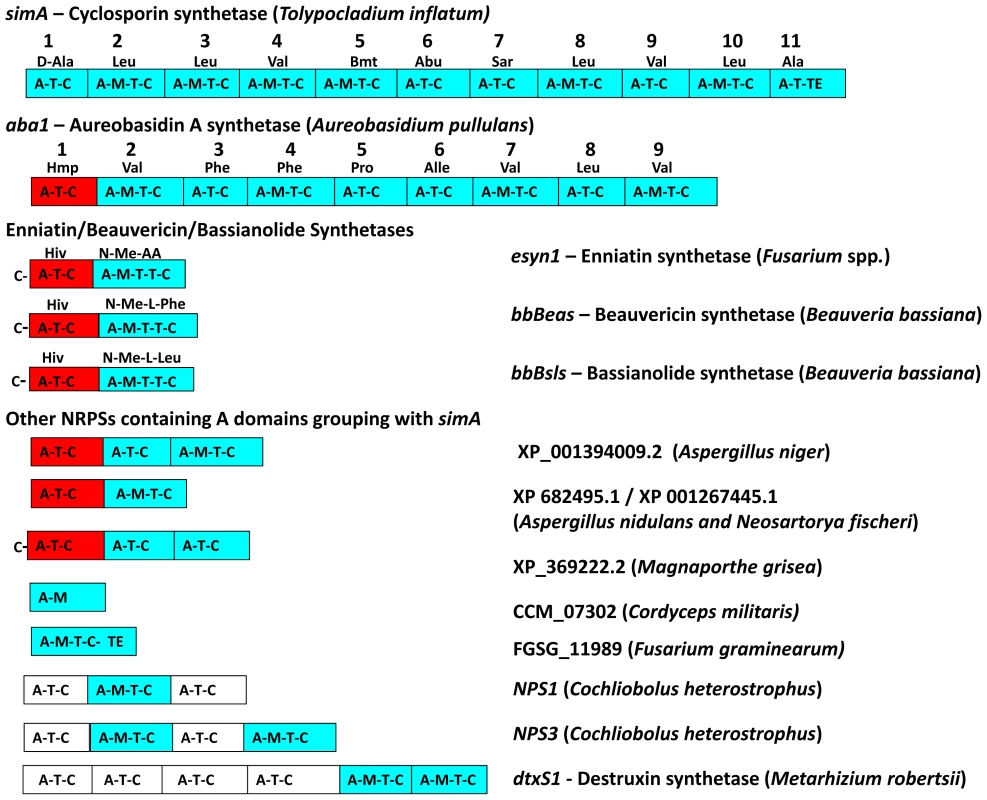
Color coding of NRPS modules denotes clade assignment of A-domains in phylogeny (Figure 4, Figure S3): light blue = groups within cyclosporin (simA) clade, red = groups within enniatin (esyn1) module 1 clade, white = groups with perA-like outside both simA and enniatin module 1 clade (Figure S3). Abbreviations for unusual amino acid substrates: Bmt = (4R)-4-[(E)-2-butenyl]-4-methyl-threonine, Abu = Aminobutyric acid, Hiv = D-2-hydroxyvaleric acid, Hmp = D-Hmp, D-2-hydroxy-3-methylpentanoic acid. The C-terminal modules of these same NRPSs, containing A-domains in the simA clade, show evidence of duplication of NRPS modules followed by divergence of A-domain substrate specificities. Enniatins, for example, are known to vary in the N-Me-amino acid incorporated by the second A-domain [62]. The C-terminal modules (2–8) of aureobasidin A synthetase (aba1) provide the clearest example of extensive module duplication and divergence within a single species as all A-domains group as a single monophyletic group with 100% bootstrap support and many share over 95% sequence similarity (Figure 4B) [63]. While the sequence of duplications within simA is more complex, A-domains of simA that group together with greater than 50% bootstrap support encode for identical substrate amino acid specificities ([A2, A3, A8, A10; bs = 78%) for Leu and (A4 and A9; bs = 98%) for Val]), suggesting these domains represent more recent duplications that have not diverged in specificity encoding regions (Figures 4B, 5). The average pairwise dN/dS ratio across all A-domains was low (ώ = 0.182), indicating that most sites within A domains are under purifying selection. However, the branch-site REL method of the HYPHY package [64] detected significant evidence of episodic positive selection (p<0.0001) on the branch separating the clade coding for Leu (A2, A3, A8, A10) from other A-domains. These results are consistent with a process in which duplication followed by lineage specific changes at a few amino-acid positions contributed to the evolution of the species-unique cyclosporin metabolite.
Computational and Transcriptional Identification of the simA Biosynthetic Cluster
The two computational methods used for defining secondary metabolite clusters, SMURF and antiSMASH, identified slightly different boundaries to the simA cluster (Figures 6, 7). The secondary metabolite gene cluster surrounding simA predicted by antiSMASH contains 22 genes and spans over 112 kb, while SMURF delineated a slightly smaller cluster of 10 genes spanning approximately 93 kb (Figures 6, 7).
Fig. 6. Computational and transcriptional identification of the cyclosporin metabolite cluster. 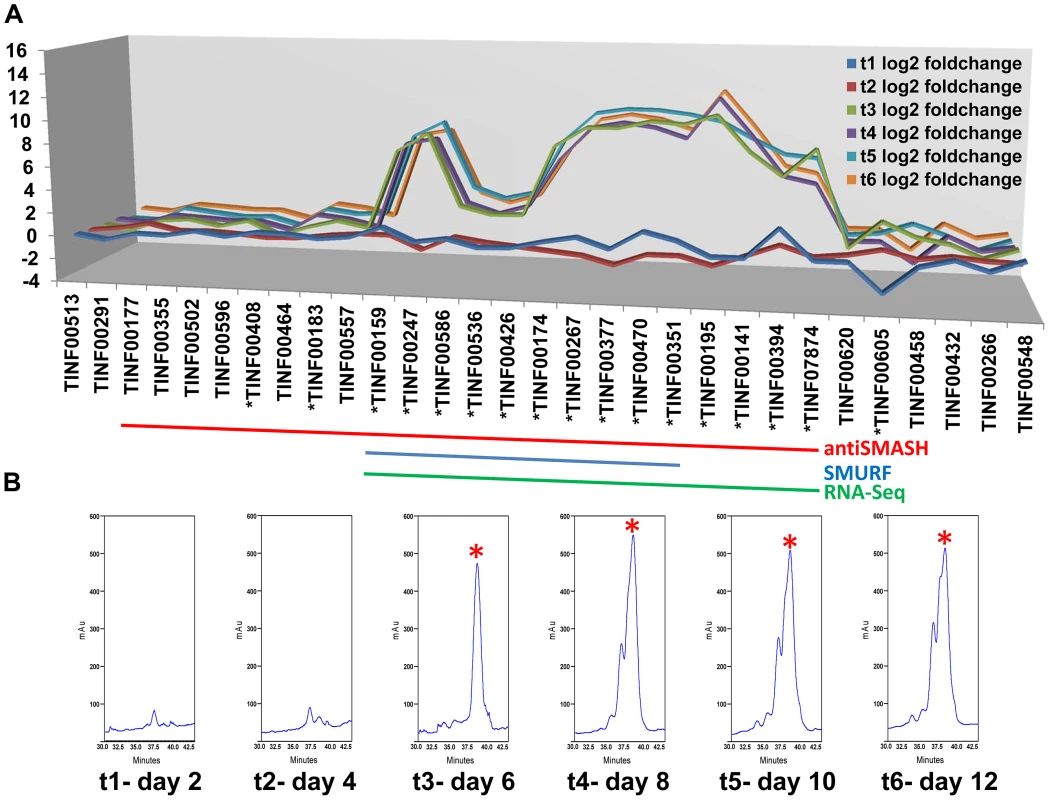
A) Fold changes (log2 transformed) of gene expression levels from SDB to SM media at time points 1–6 (days 2, 4, 6, 8, 10, 12). Strong upregulation of gene expression occurs after time point 3 (day 6). All genes marked below with an * are upregulated with q-value<0.001 for at least one time point. The boundaries to the cyclosporin simA cluster predicted by antiSMASH (red), SMURF (blue), and RNA-Seq data (green) are indicated by bars below. B) Partial HPLC traces showing the major cyclosporin A peak at 38 min. (marked with a red asterisk) in SM medium for each harvest time point. Trace amounts of cyclosporin A are found at time points 1 and 2, but production spikes at time point 3 (day 6) and peaks at time point 4 (day 8). Additional peaks surrounding the 38 min. major peak are observed after time point 4, consistent with depletion of substrates in the culture media leading to relaxed specificity of NRPS A-domains and production of additional cyclosporin analogs. Fig. 7. Organization of the simA biosynthetic cluster. 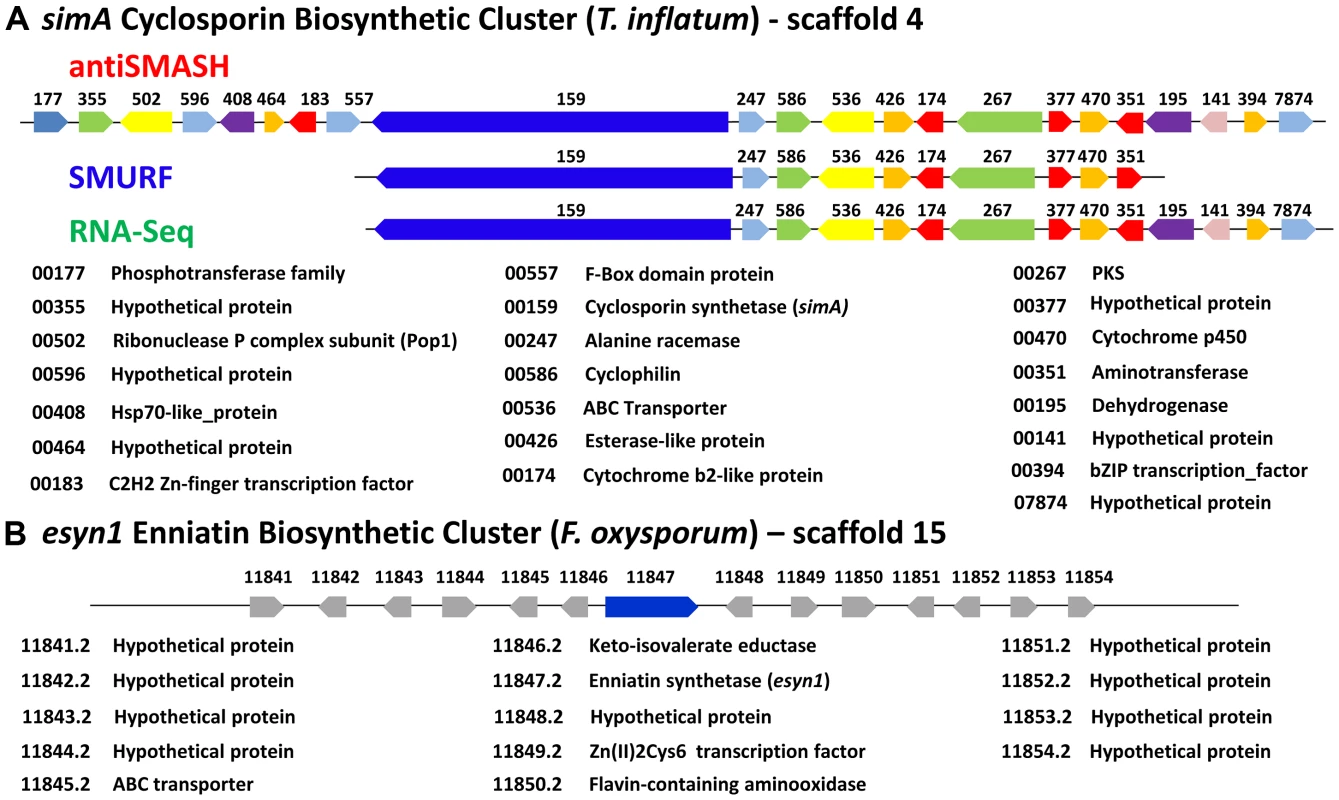
A) The secondary metabolite gene cluster responsible for cyclosporin biosynthesis as identified by antiSMASH (red), SMURF (blue), and RNA-Seq (green) with predicted protein functions or families listed below. B) The enniatin biosynthetic cluster of F. oxysporum showing genes orthologous to those in the simA cluster in blue [only enniatin synthetase (esyn1)] and genes without orthologs in the simA cluster in grey. In order to utilize transcriptional data to define the cyclosporin metabolite cluster, an RNA-Seq time course experiment in a cyclosporin-inducing medium (SM medium) [65] was conducted. Three biological replicates each were grown in the inducing (SM) and a rich control medium, Sabouraud Dextrose Broth (SDB), and were sampled at two-day intervals for a total of six time points (days 2, 4, 6, 8, 10, and 12). Total RNA isolated from these samples was prepared for RNA-Seq and remaining mycelia and culture filtrate was harvested and analyzed by LS-MS in order to correlate gene expression patterns with production of cyclosporin metabolites.
Expression profiling under cyclosporin-inducing conditions (SM medium) clearly identified a cluster of genes surrounding simA that were significantly (q-value<0.01) upregulated (Table S5) during production of cyclosporins as detected by an HPLC peak (Figures 6, S6) that included analogs with nominal molecular masses of 1188, 1202 (Cyclosporin A), 1216, and 1218 Da (data not shown). Upregulation of genes within the cluster became highly significant at time point 3 (day 6) which corresponded with the first detection of large quantities of cyclosporin in the culture filtrate by LC-MS and were consistently and strongly upregulated at time points 4, 5, and 6 that also showed an HPLC peak for CsA (Figure 6, S6, Table S5). The 5′ boundary of this cluster corresponded to the SMURF computational prediction beginning at simA (TINF00159), while the 3′ edge of this cluster corresponded to the 3′ boundary of the antiSMASH cluster prediction (TINF07874) (Figures 6, 7, Table S5).
LC-MS profiles for the cyclosporin-containing fraction were generated consistently from culture filtrate extracts obtained for each time point. Under the HPLC protocol used, analogs of cyclosporins eluted predominantly between 32–40 min, with the peak maximum for cyclosporin A (the major product, molecular mass 1202 Da) at 38 min. The overall production of cyclosporins peaked at time point 4 (day 8) in SM medium (Figure 6, Figure S6). Beginning at time point 4, an increase in the number of overlapping peaks for closely-eluting analogs of cyclosporin A is consistent with depletion of specific amino acids in the culture media leading to relaxed substrate specificity of the cyclosporin NRPS A-domains and production of distinct cyclosporin analogs (Figure 6, Figure S6). However, the existence of more than one isoform with the same molecular mass prevents the rigorous assignment of these metabolites (molecular masses 1188, 1202, 1216, and 1218 Da) by mass spectrometry alone.
This combination of computational and experimental approaches provided a more robust method for characterization of secondary metabolite clusters and demonstrates the utility of transcriptional data in confirming cluster boundaries.
Organization and Components of the simA Cluster
Although NRPSs such as simA and esyn1 contain A-domains with shared ancestry (Figure 4, S3, S5), the metabolite clusters containing these core metabolite genes do not share other homologous genes (Figure 7). Components of secondary metabolite clusters other than the core backbone enzymes function in synthesis of precursors, mediation of intermediate steps, transport and delivery, and modifications of the final metabolite [66]. While further functional studies (e.g. gene knockouts) are needed, the simA cluster contains genes which likely function in both synthesizing substrates for the NRPS and modification or activation of the cyclosporin product. The unusual non-proteinogenic amino acid substrate D-alanine must be supplied by an independent alanine racemase [67] as simA itself does not contain racemase activity. Similarly, it was shown that one of the unusual amino acid substrates of cyclosporin, (4R)-4-[(E)-2-butenyl]-4-methyl-threonine (Bmt), is synthesized by a polyketide biosynthetic mechanism [68]. As a cyclosporin mutant (Cyb56) was shown to accumulate Bmt [69], this substrate is likely synthesized by T. inflatum. The discovery of both a D-alanine racemase (TINF00247) and a PKS gene (TINF00267) within the cluster strongly suggests their involvement in production of these two unusual substrate molecules (Figure 7). The cluster also contains an aminotransferase (TINF00351), an enzyme involved in the synthesis of branched chain amino acids such as the Leu and Val residues found in cyclosporin. Several genes in the cluster belong to gene families commonly found in fungal secondary metabolite clusters, including a cytochrome P450 (TINF00470) and a dehydrogenase (TINF00195). Two transcription factors, a C2H2 zinc-finger transcription factor (TINF00183) on the 5′-edge of the cluster and a putative basic leucine zipper (bZIP) transcription factor (TINF0394) on the 3′-end of the cluster, are candidates for a cluster-specific transcriptional regulator (Figure 7).
Adjacent to the alanine racemase (TINF0247) is a gene (TINF00586) belonging to the cyclophilin family of peptidylprolyl isomerases (IPR002130) (Figure 7). The first isolated cyclophilin, human cyclophilin A (hCypA), was identified almost thirty years ago as the cellular target of cyclosporin [3]. Binding to hCypA is a prerequisite for the immunosuppressive activity of CsA as it causes CsA to undergo a conformational change to an entirely trans peptide conformation, which puts the calcineurin-binding motif of CsA in the reverse orientation compared to the crystal structure of CsA bound to a tetrapeptide substrate [70], [71] and primes CsA to create a better fit to its cellular target calcineurin. Cyclophilins have since been identified in nearly all kingdoms of life, including animals, plants, insects, fungi, protists, and bacteria, and they are classified based on their cellular location, domain organization and function [72], [73]. While different cyclophilins vary in their binding affinity for CsA, all exhibit petidyl isomerase activity that facilitates conformational changes from cis to trans at peptide bonds preceding prolines (peptidyl-prolyl bonds), and thus may function as general molecular chaperones in protein folding [74]. They are also implicated in diverse cellular processes including cell signaling, cell cycle control, intracellular transport, stress response, and virulence in both plant [75] and animal pathogens [76].
Using an HMM model to the conserved cyclophilin-like domain (CLD) of cyclophilins (Pfam: PF00160.16), we identified ten proteins, including TINF00586, containing the conserved CLD domain (PF00160.16) in the T. inflatum genome (Figure 8 A, B). The simA cluster cyclophilin (TINF00586) has highest and equally scoring BLAST hits to two S. cerevisiae proteins, Cpr1 (YDR155C) (e−41), the yeast homolog of hCypA, and the yeast mitochondrial cyclophilin Cpr3 (YML078W) (e-41), and it contains an N-terminal signal peptide with a localization signal to mitochondria (Figure 8B) [77]. In a phylogeny including the conserved CLD domains of major cyclophilins from other fungi, animals, bacteria, and protists, the T. inflatum cyclophilins group in diverse locations in the phylogeny, suggesting that T. inflatum, like other eukaryotes, contains a full suite of cyclophilins (Figures 8A, S7).
Fig. 8. Cyclophilins in T. inflatum genome. 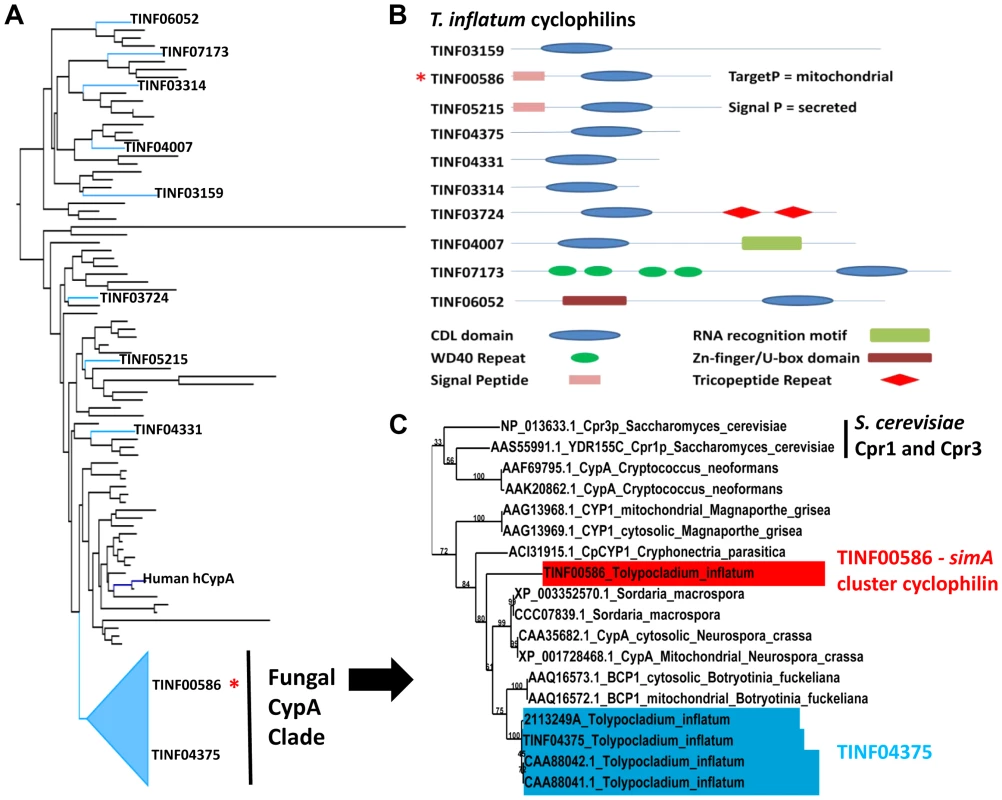
A) Backbone of maximum likelihood phylogeny of major cyclophilins from T. inflatum, H. sapiens, C. elegans, D. melanogaster, and characterized cyclophilins from other fungi, bacteria, and protists (Figure S7) showing phylogenetic positions of the ten T. inflatum cyclophilins and the simA cluster cyclophilin (red asterisk) within the fungal CypA clade. B) Domain organization of cyclophilins identified in the T. inflatum genome. The cyclosporin cluster cyclophilin (TINF00586) is indicated with a red asterisk and contains a mitochondrial localization signal. C) Expanded view of the fungal CypA clade showing S. cerevisiae Cpr1 and Cpr3, the simA cluster cyclophilin (TINF00586) in red, and TINF04375 in blue. Note that all of the products in blue are likely produced by alternative splicing of the single gene TINF04375. The simA cluster cyclophilin is similar in domain structure to hCypA and other cyclophilin A homologs. It groups phylogenetically with greater than 70% bootstrap support in a clade with another T. inflatum cyclophilin gene (TINF04375) and a number of fungal cyclophilins with roles in morphological development and pathogenesis in either plant (BCP1 (Botrytis cinerea) [78], CpCYP1 (Cryphonectria parasiticus) [75], and CYP1 (Magnaporthe grisea) [79]) or animal (Cpa1 and Cpa2 (Cryptococcus neoformans) systems [80] (Figures 8A, C [Fungal CypA Clade], S7). Several putative cyclophilins previously cloned from T. inflatum, including two that are hypothesized to be alternately spliced products of a single gene targeted to the cytosol and mitochondria, respectively [81], and another gene coding for an approximately 19.5 kDa protein (cptA) [82], are nearly identical in sequence and all group closely with TINF04375 (Figure 8C, S7). Alternative splicing of this single gene is consistent with the finding that other cyclophilin genes in this clade CypA (N. crassa) [83] and BCP1 (B. cinerea) [78]), also produce alternately spliced mitochondrial and cytosolic isoforms (Figure 8C, S7). The simA cluster cyclophilin (TINF00586) is distinct in sequence from TINF04375 and groups at the base of this clade with the CpCYP1 of C. parasiticus (Figure 8C, S7).
While the direct mechanism of toxicity of CsA in insects remains unknown, histopathological changes consistent with Mitochondrial Pore Transition Permeability (MPT), such as swollen, electron-dense, and occasionally lysed mitochondria, have been observed in several insect species treated with CsA [84]. We hypothesize that the simA cluster cyclophilin may be involved in targeting CsA to the insect mitochondria. Other possible functions include a role in folding of the cyclosporin peptide during export, creation of a pre-activated CsA-CYPA cocktail prior to delivery to the host, protection of CsA from proteolysis by endopeptidases [85], binding to detoxifying proteins in hemolymph [86], or auto protection for T. inflatum against CsA toxicity.
Transcriptional Responses in the simA Gene Cluster in Relation to Insect Pathogenesis
Toxins and other secondary metabolites are suspected to function in insect pathogenesis but their expression patterns and modes of action remain poorly characterized. Previous studies suggest that many secondary metabolites are expressed at very low levels under most experimental conditions [87], and their expression is elicited only in response to specific stimuli. Like many insect pathogenic fungi, T. inflatum exhibits a complex lifecycle encompassing a saprobic growth phase in soil and a pathogenic growth phase on and within the insect host (Figure 1). The pathogenic phase initiates with an infection phase that involves growth on, and penetration of, the insect cuticle. This infection phase is followed by a colonization phase, which initially involves a yeast-like (hyphal body) growth phase within insect hemolymph, and ultimately switches to a filamentous growth form that colonizes the insect to form an endosclerotium. Previous studies have shown that cyclosporin has immunosuppressive functions in insects [58], as well as in humans, suggesting a role for and expression of cyclosporin inside the insect. In order to evaluate the expression of the simA cluster in relation to insect pathogenesis, RNA-Seq was carried out on fungal cultures grown on (1) minimal medium supplemented with insect cuticle (infection stage), and (2) Grace's insect medium supplemented with insect hemolymph (colonization stage). Each of these treatment samples (cuticle and hemolymph) were compared to a control grown on SDB. Transcriptional responses in these media were compared qualitatively to responses in the cyclosporin-inducing (SM) medium.
In the strongly inducing SM medium, nearly all genes within the cluster were upregulated with extremely high significance (q-value<0.0005) (Figure 6, Table S5), but their relative expression levels varied widely (Figures 9A, S6, Table S5). The most highly expressed gene in the cluster was the cyclophilin gene (TINF00586). However, this gene also had relatively high levels of constitutive expression in the control SDB medium and underwent only a 3.18× log2 fold increase in expression during time point 5 (Figure 6, Table S5). In contrast, most other genes within the cluster, with the exception of TINF00536 and TINF00426, had very low levels of constitutive expression in control SDB medium, but were more highly upregulated in SM medium (Figures 6, 9A, S6, Table S5). For example, the PKS (TINF00267), the cytochrome P450 (TINF00470), the D-alanine racemase (TINF00247), the dehydrogenase (TINF00195), a hypothetical protein (TINF00377), and the aminotransferase (TINF00351) had over a 9× log2 fold increase in expression in SM medium compared to the control (Figure 6, Table S5). Several other genes, including simA (TINF00159), a cytochrome b-2-like protein (TINF0174), two hypothetical proteins (TINF00141 and TINF07874), and the bZIP transcription factor (TINF00394) experienced between 5–9× log2 fold increases in expression (Figure 6, Table S5). The C2H2 transcription factor on the 5′ end of the cluster (TINF00183) experienced less than 1× log2 fold increases in expression, suggesting that the bZIP transcription factor (TINF001374) is more likely the cluster-specific transcriptional regulator (Figure 6, Table S5).
Fig. 9. Expression profiling of genes in the simA cluster in relation to insect pathogenesis. 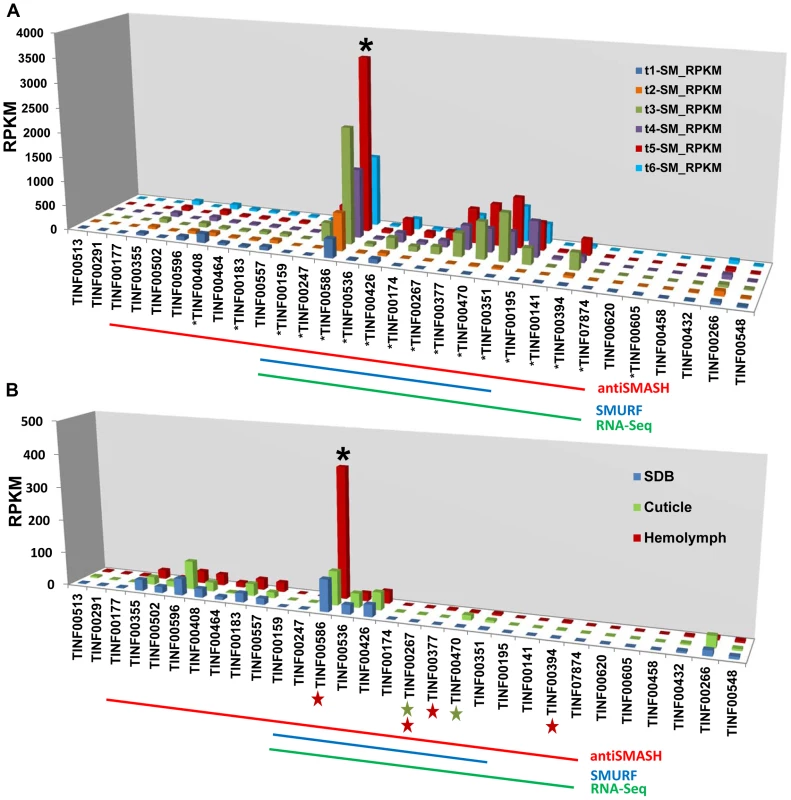
A) Relative expression levels in Reads Per Kilobase per million mapped reads (RPKM) of genes in the simA cluster in SM medium. Significantly (p<.01) upregulated genes are shown with an asterisk below gene. B) Relative expression levels (RPKM) of genes in the simA cluster in Sabouraud Dextrose Broth (SDB), cuticle, and hemolymph media. Genes that are significantly (p<.05) upregulated are shown with either a green star (cuticle media) and/or red star (hemolymph media) below gene. In both A and B, the Y-axis is RPKM, the cluster cyclophilin (TINF00586) is indicated with an asterisk above the histogram bar and the antiSMASH (red), SMURF (blue), and RNA-Seq (green) predicted clusters are indicated by lines below. While the simA NRPS itself was not significantly upregulated in media simulating stages of insect pathogenesis (cuticle and hemolymph media), several cluster genes showed significant upregulation (q-value<0.05) (Figure 9B, Table S6). In cuticle medium, the PKS gene (TINF00267) and the cytochrome P450 (TINF00470) were significantly upregulated (Figure 9B, Table S6). In hemolymph medium, a greater number of cluster genes including the PKS gene (TINF00267), the bZIP transcription factor (TINF00394), the cyclophilin homolog (TINF00586), and a hypothetical protein containing a thioester/thiol ester dehydrase-isomerase domain (TINF00377) were significantly upregulated (Figure 9B, Table S6). The P450 (TINF00470) was the most highly upregulated gene in the cuticle medium (3.25× log2 fold), while the bZIP transcription factor (TINF00394) was the most highly upregulated gene in hemolymph medium (3.58× log2 fold) (Table S6). Importantly, the cyclophilin gene (TINF00586) showed significant upregulation (1.82× log2 fold) only in hemolymph media and had the highest relative expression level of all cluster genes in hemolymph media (Figure 9B, Table S6).
However, culture media conditions can only simulate conditions of insect pathogenesis, and differential expression cannot be solely attributed to the added insect components as differences in the composition and pH of the basal media used may also influence expression patterns. RNA samples were also harvested 24 hours after transfer from a rich SDB medium to hemolymph medium. Given that full induction of cyclosporin production took nearly 6 days in a strongly inducing medium, it is possible that the weaker response observed in media containing insect components reflects an early stage of cyclosporin induction. The larger number of significantly upregulated genes and the strong upregulation of the bZIP transcription factor (TINF00394) only in hemolymph media, however, suggests a possible role for cyclosporin and the cyclophilin homolog during the colonization phase inside an insect host (Figure 9B, Table S6).
Evolution and Synteny of the simA Cluster
The disjunct distribution of cyclosporin biosynthesis across fungal taxa poses interesting questions about the origins and evolution of the cyclosporin biosynthetic cluster. In order to search for homologs of cyclosporin cluster genes in other fungi, the top 25 BLAST hits to the antiSMASH predicted simA cluster genes plus 10 flanking genes on either side in the NCBI nr database were aligned and phylogenies constructed using maximum likelihood (Figure S8). Pairwise BLASTP searches among the set of the fourteen hypocrealean fungi analyzed for phylogenomic analyses were also performed to identify reciprocal best-pair hits between these genomes and these best-pair BLASTP hits were considered as orthologs. These analyses revealed that only one gene (TINF00195) between the C2H2 transcription factor (TINF00183), adjacent to the 5′ end of the RNA-Seq defined cluster (Figure 10, red line), and the 3′-end of the RNA-Seq defined cluster (Figure 10, blue line, at TINF07874) had hits above e−05 to bacterial genes (Figure S8). Similarly, only a few genes (TINF00557, TINF00586, TINF00426, TINF00174, TINF00267, TIN00470, and TINF00394) had orthologs in other sequenced hypocrealean taxa (Figures 10, S8). In contrast, most genes flanking this region on both sides, as well as the 8 genes at the 5′-end of the antiSMASH predicted cluster (TINF00177 to TINF00557) contained single-copy orthologs in nearly all other hypocrealean taxa that showed relatively conserved synteny with those in T. inflatum (Figures 10, S8). The 8 genes on the 5′ end of the antiSMASH predicted cluster that had homologs in other Hypocreales were excluded from the cluster by both SMURF and the RNA-Seq predicted cluster (Figures 6, 7). Additionally, the few homologs of genes within the RNA-Seq defined cluster in other hypocrealean taxa were scattered elsewhere in these genomes and not located between these conserved flanking regions (Figure 10, S8). In most species, no additional genes were found between the 5′ flank (TINF00183) and the 3′ flank (TINF07874) of the RNA-Seq defined cluster and the intervening region between these boundaries was less than 5 kb (Figure 10). F. oxysporum contained a single additional gene in this region, while C. militaris underwent an inversion that added additional genes, none of which were orthologs of the simA cluster genes (Figure 10). These results suggest one of two hypotheses regarding the origin of the nearly 100 kb simA cluster region in T. inflatum that is missing in other Hypocreales: 1) the cluster has been horizontally transferred from another fungal species into this site in T. inflatum, or 2) this region has evolved by recruitment of genes from other regions of the T. inflatum genome.
Fig. 10. Synteny of regions flanking the cyclosporin cluster in other hypocrealean taxa. 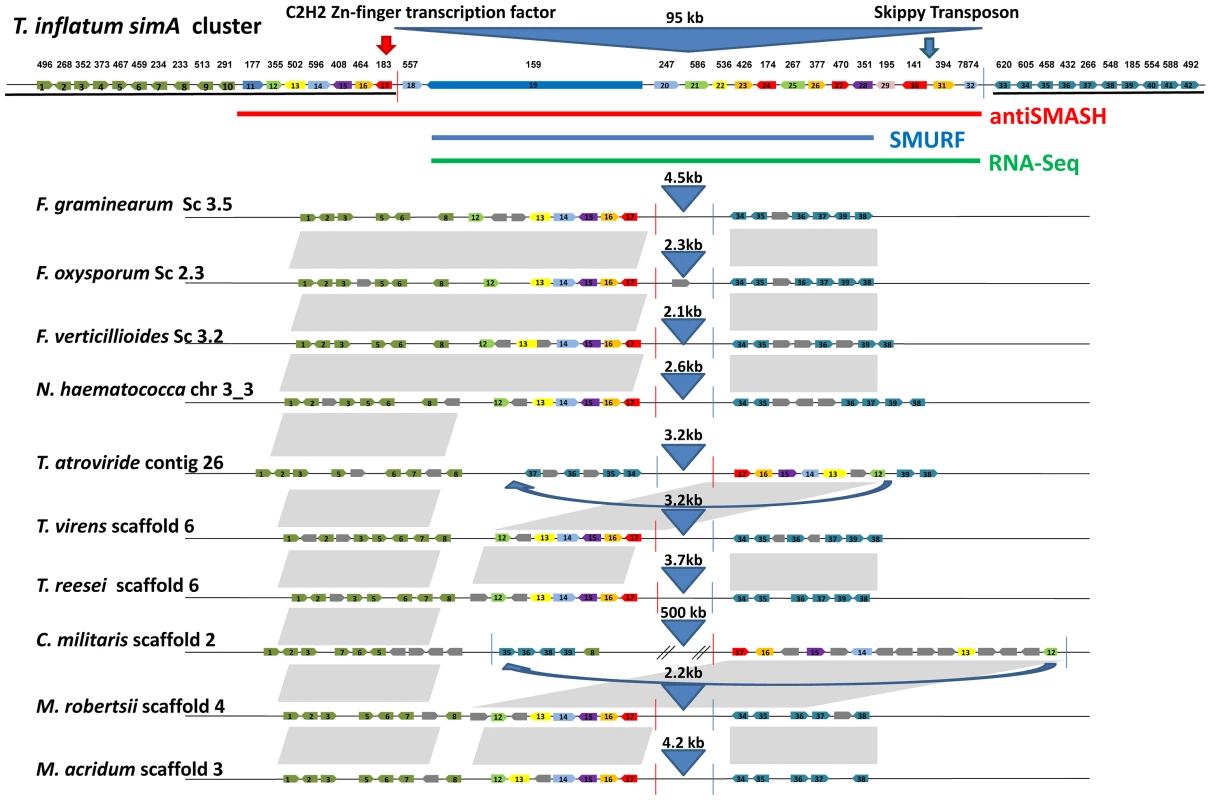
Genes within the cyclosporin biosynthetic cluster as delineated by antiSMASH (red), SMURF (blue), and RNA-Seq (green) plus ten genes on the 5′ (green) and 3′ (blue) flanks of the antiSMASH predicted cluster are shown at top and numbered from left (5′) to right (3′) (1–42). Orthologous genes in other hypocrealean taxa identified by best-pairwise BLAST searches are shown below for each species. Grey genes indicate additional genes present in other species while grey shaded areas show regions of synteny between genomes. Genes in T. inflatum in the region between the C2H2 Zn-finger transcription factor (TINF00183) on the 5′end (red line) and the RNA-Seq predicted 3′- end of the simA cluster (at TINF007874) (blue line) mostly lack orthologs in other hypocrealean genomes. The few best-pair orthologs identified in other Hypocreales were found elsewhere in these genomes. Numbers above blue triangles show length of intervening sequence between the 5′ (red line) and 3′ (blue line) flanks of the cluster (approximately 95 kb in T. inflatum) which is less than 5 kb in all other hypocrealean taxa except C. militaris and Tr. atroviride. Blue arrows show regions inverted in Tr. atroviride and C. militaris. In Tr. atroviride, an inversion has occurred but the region between adjacent genes is still <5 kb and contains no additional genes. In C. militaris, the inversion has added nearly 500 kb of sequence containing additional genes, none of which were found to have orthologs in the simA cluster or to belong to other secondary metabolite clusters. Horizontal transfer of complete secondary metabolite clusters between heterologous bacterial species by transposition has been demonstrated [88], and evidence exists for horizontal transfer of large secondary metabolite clusters among fungi [89], [90]. However, BLAST searches did not detect a homologous cluster in other fungal taxa. To further test for horizontal transfer, we utilized the customized pipeline (CRAP) [89] to scan for syntenic homologs of simA cluster genes in 195 sequenced fungal genomes. We did not detect a cluster containing both the NRPS and PKS genes and a majority of accessory genes from the simA cluster. While these results do not preclude the existence of a nearly complete cluster in a yet to be sequenced fungus, available evidence suggests that the cluster more likely evolved by a process of recruitment of genes into the cluster from elsewhere within the T. inflatum genome.
The fact that simA and esyn1 synthetases clearly share related A-domains while the clusters containing these NRPSs lack other shared genes leads us to hypothesize that these clusters have evolved by recruitment of distinct modifying enzymes into regions surrounding these core NRPSs through rearrangement and transposition. Transposons have been found adjacent to other secondary metabolite gene clusters in fungi, such as the gliotoxin cluster in Aspergillus fumigatus [91]. While their role in shaping the evolution of secondary metabolite clusters remains speculative, they represent a potential mechanism for gene recruitment. A gypsy LTR retrotransposon, related to F. oxysporum SKIPPY [92], was found on the 3′ end of the simA cluster (Figure 10), and several lines of evidence suggest that the region surrounding the simA cluster may be prone to rearrangement in other taxa. Both Tr. atroviride and C. militaris show evidence of an inversion or transposition in this region of the genome. In Tr. atroviride, this is simply an inversion which did not add additional genes. In C. militaris, an additional 0.5 Mb of sequence distinct from the T. inflatum sequence is found between the two rearranged flanking regions, but this additional sequence does not contain any homologs of simA cluster genes nor is it predicted by SMURF or antiSMASH to contain other secondary metabolite clusters (Figure 10).
Conclusions
Although the simA clade itself was shown previously to group sister to a clade of bacterial NRPSs [54], no bacterial homolog of simA were found within the simA clade. Thus, we conclude that simA evolved through duplication and divergence of fungal NRPS modules within T. inflatum rather than by recent horizontal transfer from bacteria (Figures 4, S5, S8). A number of related NRPSs that share homologous A-domains with simA, including enniatin, beauvericin, bassianolide, and aureobasidin A synthetases, evolved through a similar process of module duplication, but also fusion of distantly related NRPS modules (Figures 4, 5, S5). Interestingly, all of these metabolites possess insecticidal or fungicidal properties. Other genes within the secondary metabolite gene clusters containing these NRPSs, however, do not show homology with those in the simA cluster and are not syntenic with the simA cluster. Regions syntenic with the simA cluster in other hypocrealean fungi instead lack the nearly 100 kb of the simA cluster present in T. inflatum. However, searches for orthologs of the simA cluster genes in other fungi using BLAST searches of the NCBI nr database and the CRAP pipeline found no clusters containing more than a few genes showing similarity to those in the simA cluster. While horizontal transfer from a yet un-sampled fungus cannot be ruled out, we suggest that the simA cluster instead had a lineage specific origin, having evolved through recruitment of genes from other locations in the T. inflatum genome.
The discovery of a homolog of hCypA, the cellular target of cyclosporin in mammalian systems, within the simA cluster of T. inflatum is novel. This is the first report of a cyclophilin gene located within a secondary metabolite gene cluster, although several genes shown to be dependent on the activity of calcineurin, the target of CsA, are suspected of being located within a secondary metabolite biosynthetic cluster in B. cinerea [78]. The up-regulation of the cluster cyclophilin in hemolymph and high expression levels under both inducing conditions and in insect hemolymph medium suggests a role for this gene in mediating the activity of cyclosporin in vivo. Elucidation of the simA gene cluster through a combination of computational and transcriptional approaches opens the door for functional studies and chemical analyses to address mechanisms of action in nature and potential novel applications of these compounds in pathogenicity and medicine.
Materials and Methods
Strains and Culture Conditions
Tolypocladium inflatum NRRL 8044, the original strain from which cyclosporin was isolated, was obtained from NRRL. Cultures were grown for 2.5 weeks on cornmeal agar to induce sporulation. For DNA extractions, conidia from agar plates were used to inoculate potato dextrose broth cultures, which were grown for 3 days before harvesting.
DNA Isolation and Sequencing
Lyophilized mycelia were ground in liquid nitrogen and genomic DNA was isolated using the Qiagen genomic tip 500 following the manufacturer-supplied protocol for isolation of genomic DNA from plants and filamentous fungi (Qiagen). A 350 bp insert Illumina library was prepared by shearing two samples of approximately 5 µg of genomic DNA in a Biorupter XL sonicator for 20 min with a cycle of 30 sec on and 30 sec off. These samples were pooled and prepared following the Illumina protocol for paired-end sequencing. Illumina sequencing was performed on the Illumina GAII machine at the Center for Genome Research and Biocomputing (CGRB) at Oregon State University. For 454 sequencing, a shotgun library and a 3-kb paired end 454 library were each prepared and sequenced on a full plate using titanium chemistry at the Duke IGSP Sequencing Core Facility at Duke University.
Genome Assembly
454 reads were assembled using the Newbler Assembler version 2.3 (454 Life Sciences) using a combined shotgun and 3 kb paired end library assembly. Out of a total of 4,260,863 input 454 reads (1,516,236 single end shotgun reads, 1,067,664 mate pair reads with both pairs, and 1,676,963 singleton mate pair reads), 4,077,744 (95.70%) were assembled into the Newbler Assembly. Illumina reads were first trimmed to 50 bp, and sequences containing adaptors, N's, or greater than 2 bp with a quality score below 20 were filtered out of the dataset. A total of 37,643,435 quality filtered reads, containing 33,586,130 paired end reads, were submitted to SOAP Gapcloser [93] to fill gaps in the Newbler scaffolds. Finally, a mapping assembly was performed in MIRA to map the quality filtered Illumina reads to the SOAP Gapcloser assembly to correct for homopolymer base pair errors.
Gene Predictions
Genome annotations were created in MAKER 2.00 [23] using three ab initio gene prediction models: an AUGUSTUS model trained for F. graminearum, a GeneMark model trained for T. inflatum via self-training, and a SNAP model trained for F. graminearum. Protein datasets from F. graminearum, Nectria haematococca, Tr. reesei, Tr. virens, and M. robertsii were submitted to MAKER as protein evidence. ESTs from C. militaris, B. bassiana, M. robertsii, and Tr. reesei downloaded from GenBank were included as EST evidence. Illumina PE RNA-Seq reads from T. inflatum PDB grown cultures (see RNA isolation and sequencing) were trimmed to 70 bp and filtered to remove sequences containing adaptors, N's, or greater than 2 bp with a quality score below 20, and assembled into transcripts using OASES with a coverage cutoff of 3 [94]. Assembled transcripts were input into MAKER as EST evidence. Transfer RNAs (tRNAs) were identified with tRNA scan-SE [95].
Repeat Elements
Repeat elements were identified with Repeat Masker (Smit, AFA, Hubley, R & Green, P. RepeatMasker Open-3.2.8 1996–2010 http://www.repeatmasker.org) using the fungal transposon species library (database version 20090604) as input and crossmatch version .990329. The number of CPA (NCBI Accession AM990997.1) and Restless (NCBI Accession Z69893.1) elements was identified in hypocrealean taxa by supplying these sequences as a library to Repeat Masker using the same settings.
Phylogenetic Relationships and Orthologous Clusters
Proteins for the set of hypocrealean taxa utilized for functional annotations, as well as three Sordariomycete outgroups (N. crassa, V. dahliae, and V. albo-atrum), were clustered using MCL [30] at inflation parameter 3 within the Hal pipeline [31] and then filtered to identify single-copy orthologs. Single copy orthologs were aligned separately using MUSCLE [96] and then concatenated. The concatenated alignment was used to infer a phylogeny using RAxML with the best fit model of amino acid substitution for each gene partition estimated by ProtTest [97] and branch support estimated from 1000 bootstrap replicates. In a second set of analyses, amino acid rate categories were estimated across eight categories using PAML [98] and the effect of amino acid positions with high rates of substitution was determined by repeating the same RAxML analyses without the 6th, 7th, and 8th rate categories. The ultrametric tree used for CAFÉ analyses was computed in r8s [99] from the phylogeny inferred by the first RAxML analysis using previously estimated divergence dates for Hypocreales [16] (Table S2). The number of clusters and proteins within clusters were mapped to nodes using a custom perl script.
Functional Annotation
A functional annotation pipeline was developed for the hypocrealean fungi F. graminearum, F. oxysporum, F. verticillioides, N. haematococca, Tr. reesei, Tr. virens, Tr. atroviride, M. robertsii, M. acridum, C. militaris, and T. inflatum. Functional annotations were classified with InterproScan (http://www.ebi.ac.uk/interpro) using hmmpfam, PatternScan, ProfileScan and blastprodom databases and converted to GO with the Interpro2GO mapping (version 11/05/2011). Orthologous groups of proteins for these eleven fungi as well as the model fungi S. cerevisiae, Schizosaccharomyces pombe, and N. crassa were determined with Inparanoid [100], [101]. GO enrichments were performed by transferring additional GO annotations associated with proteins from model fungi to uncharacterized proteins within the same orthologous cluster. Protein localization signals excluding the plastid location were identified using TargetP [102] and Predotar [103] while transmembrane proteins were characterized using TMHMM [104]. Carbohydrate active enzymes (CAZymes) were determined by BLAST searches against the CAZymes database with a cutoff of <e−30. Proteases were identified by BLASTP searches of the MEROPS [105] database (http://merops.sanger.ac.uk/) with default settings (cutoff of e−05), and P450 enzymes were identified as BLAST hits to entries in the Nelson P450 [106] database below a cutoff of <e−20. The program CAFÉ, using the default settings, was used to analyze gene families (CAZymes, P450s, and proteases) expanded or contracted in taxa within Hypocreales.
Secondary Metabolite Characterization
NRPS and PKS secondary metabolite genes and their domain structures were characterized using three methods: 1) HMMER searches using models build for Adenylation (A), Thiolation (T), and Condensation (C) domains of NRPSs [54], 2) SMURF [43], and 3) antiSMASH [44]. Adenylation (A) domains from NRPSs and ketosynthase (KS) domains from PKSs were extracted from the same eleven hypocrealean taxa included in the functional annotation pipeline and aligned together with known NRPSs from fungi and several outgroup A-domains (Acetyl CoA Synthetases, AcylCoA ligases, Ochratoxin, and CPS1) using MAFFT [107]. The alignment was examined and manually edited to remove regions of ambiguous alignment and a maximum likelihood phylogeny constructed in RAxML [108] using 1000 bootstrap replicates and the best fit model identified by ProtTest [97] (REVF plus gamma for A domains). The simA NRPS and the PKS gene found within the cyclosporin cluster were also subjected to BLAST against the NCBI nr database and A-domains from the top 50 hits were extracted, aligned, and analyzed as described above to identify any putative bacterial homologs of simA. The identified secondary metabolite genes were placed into secondary metabolite gene clusters using SMURF and antiSMASH.
Synteny of the Cyclosporin Cluster
Protein sequences from the T. inflatum cyclosporin cluster plus ten flanking genes were used to search the genomes of related hypocrealean fungi using TBLASTN and BLASTP searches. The TBLASTN searches identified genes flanking the cyclosporin cluster that localized to the same contig or scaffold in other species. Genome-wide best-pair BLASTP searches were used to refine this search and identify those genes in the simA cluster with best pairwise BLASTP hits in other genomes and these were considered putative orthologs. The genomic sequence was also aligned with MAUVE at the DNA level to confirm synteny relationships (data not shown) [109]. To identify potential homologs in fungal species other than hypocrealean taxa and to address phylogenetic evidence for horizontal transfer, all T. inflatum cyclosporin cluster genes plus ten flanking genes of the antiSMASH predicted cluster were subjected to BLASTP against the nr database and the top 25 hits were extracted and aligned with MAFFT [107]. These alignments were filtered to remove regions of poor alignment using Gblocks (with relaxed setting), and ProtTest [97] was used to identify best-fit models for each gene alignment. Maximum likelihood phylogenies were constructed using RAxML [108] with the corresponding best-fit protein model and 100 bootstrap replicates. The simA cluster genes were also run through a customized pipeline (CRAP) [89] to search 195 sequenced fungal genomes and 1150 bacterial genomes for syntenic homologs of the simA cluster.
RNA Isolation and Sequencing
For initial coverage of the transcriptome, conidia from 2 1/2 week-old cultures grown on cornmeal agar were adjusted to a concentration of 1×107 spores/mL and 1 mL was used to inoculate a 100 mL liquid culture of potato dextrose broth. Mycelia were harvested after three days, flash frozen and ground in liquid nitrogen. RNA was extracted using the Qiagen RNAeasy kit. cDNA was prepared using the Mint cDNA Kit (Evrogen), sheared by nebulization for 6 min at 34 psi, prepared using the Illumina protocol for PE sequencing, and sequenced in one lane of a paired end 80 bp run on the Illumina GAII. For the RNA-Seq time course experiment in cyclosporin inducing (SM medium), two 200 mL flasks containing 100 mL of YM medium (4 g yeast extract, 20 g malt extract in 1L ddH20) were each inoculated with approximately 1 mL of 1×107 conidia/mL. After two days growth in YM medium, 10 mL of the YM culture was transferred to 125 mL of SM production media or Sabouraud Dextrose Broth (SDB) in 200 mL flasks. All cultures were grown at 21°C. Three flasks (3 biological replicates) for each treatment (SM or SDB) were harvested every two days for a total of six time points (days 2, 4, 6, 8, 10, 12). Tissue was flash frozen and ground in liquid nitrogen and total RNA extracted in TRIzol®. Remaining tissue and culture filtrate was frozen at −20°C until chemical extraction for HPLC analyses. The RNA samples were prepared using the Illumina TruSeq RNA sequencing kit, randomized, and sequenced across three lanes of a 50 bp SE run on the Illumina HiSeq2000. For RNA-Seq expression studies in insect media, cultures were grown under three conditions: 1) a rich medium of Sabouraud dextrose broth (SDB), 2) a minimal salts medium [110] supplemented with 10% (w/v) black vine weevil (Otiorhynchus sulcatus, Coleoptera) insect cuticle cleaned from soft tissue with sodium tetraborate [111] to simulate the infection stage, and 3) Grace's insect medium (Gibco, unsupplemented) with 10% (v/v) filter sterilized black vine weevil hemolymph added to simulate the colonization stage. The minimal salts medium utilized for cuticle media contained 0.02% KH2PO4, 0.01% MgSO4, 0.2 p.p.m. FeSO4, 1.0 p.p.m. ZnSO4, 0.02 p.p.m. NaMoO4, 0.02 p.p.m. CuSO4, 0.02 p.p.m. MnCl2 adjusted to pH 6.5 These cultures were grown in a 2-stage fermentation that has proven a reproducible method for eliciting expression of proteins in response to specific elicitors in insect pathogenic fungi [112]. Conidia from cultures grown on cornmeal agar for 2.5 weeks were used to inoculate a 100 mL culture of rich media (SDB) at a final concentration of approximately 1×106 conidia/mL. These cultures were grown on a shaking incubator for 48 hours, washed in sterile water, and approximately 500 mg wet weight of mycelia transferred to three replicates of 2 mL cultures for each media condition. Mycelia were grown for an additional 24 hours before harvesting. All cultures were grown at 21°C. RNA was extracted with TriZol® according the manufacturers protocol (Invitrogen), polyA RNA isolated using the Ambion PolyA Purist kit, and cDNA prepared using the Superscript III kit and random hexamer primers (Invitrogen). A 450 bp insert library was prepared for each biological replicate according to the Illumina PE protocol and all samples were multiplexed in each of three lanes of a SE 40 bp run.
RNA-Seq Analyses
Barcodes were trimmed from 40 bp Illumina reads to a length of 36 bp for the insect pathogenesis experiment and the first and last nine bases were trimmed from the 50 bp reads based on quality score profiles to a length of 40 bp for the cyclosporin inducing experiment. Reads were mapped to gene models using GENE-Counter [113]. Differential expression was analyzed in GENE-Counter, which utilizes a negative binomial model and the NBP-Seq R package to model differential gene expression [114]. For the time course experiment, the three biological replicates in SM media were compared with the three biological replicates in the SDB media at each time point separately. For the insect assays, pairwise comparisons of SDB vs cuticle medium and SDB vs hemolymph medium were performed in GENE-Counter. Q-values and fold changes (log2 transformed) were calculated using the normalized expression values from NBP-Seq. Relative expression levels were calculated as Reads Per Kilobase of transcript per Million mapped reads (RPKM) [115].
Chemical Extraction and HPLC/LC-MS Profiling
Culture filtrates from three biological replicates at each time point were pooled for chemical extraction. Each culture filtrate was applied to a glass column containing Diaion® HP20 resin (20 g, Supelco), which had been sonicated in MeOH (to de-gas) and then pre-washed with H2O (200 mL). The column loaded with sample was then eluted sequentially with H2O (200 mL, to desalt the sample), MeOH (100 mL) and acetone (100 mL). The latter two organic solvent eluents were combined and concentrated to provide an organic extract from each culture filtrate. In each case, the organic extract was applied to a C18 reversed-phase solid phase extraction (RP18 SPE) cartridge (10 g), which had been primed in 100% methanol, and then equilibrated in 70% methanol in water. The SPE cartridge was then eluted sequentially with 70% and 100% methanol in H2O before being washed with dichloromethane. The cyclosporin-containing SPE fractions (100% methanol, determined by direct injection MS) from each SM culture filtrate extract were selected for comparative HPLC (used to establish protocols for peak collection) and also LC-MS profiling, alongside the corresponding control SDB medium extracts. HPLC of each sample (50 µg per 5 injection) was performed using a linear gradient from 60–100% methanol in H2O over 40 min followed by isocratic 100% methanol for 20 min (column: Synergi Hydro-RP, 4.6×250 mm, 0.6 mL/min). LC-MS of 5 µg-containing aliquots was performed under identical HPLC solvent conditions using a Synergi Hydro-RP, 2×100 mm column with a flow rate of 0.2 mL/min.
HPLC was performed on a Shimadzu HPLC system comprising a SIL-20AC autosampler, dual LC-20AD solvent pumps and a SPD-M20A UV/VIS photodiode array detector. LC-ESI(+) MS data were obtained using an AB SCIEX QTrap 3200 mass spectrometer interfaced with a Shimadzu Prominence HPLC system. HPLC-grade solvents were used for all chemical extraction and fractionation.
Accession Numbers
The Whole Genome Shotgun projects have been deposited at DDBJ/EMBL/GenBank under the accession number AOHE00000000.
Supporting Information
Zdroje
1. BorelJF (1997) Cyclosporin in immunology: Past, present and future. Biodrugs 8 : 1–3.
2. WangP, HeitmanJ (2005) The cyclophilins. Genome Biology 6 : 226.
3. HandschumacherRE, HardingMW, RiceJ, DruggeRJ (1984) Cyclophilin - A specific cytosolic binding-protein for Cyclosporin-A. Science 226 : 544–547.
4. LiuJ, FarmerJD, LaneWS, FriedmanJ, WeissmanI, et al. (1991) Calcineurin is a common target of cyclophilin-Cyclosporine-A and FKBP-FK506 complexes. Cell 66 : 807–815.
5. JorgensenKA, Koefoed-NielsenPB, KaramperisN (2003) Calcineurin phosphatase activity and immunosuppression. A review on the role of calcineurin phosphatase activity and the immunosuppressive effect of cyclosporin A and tacrolimus. Scandinavian Journal of Immunology 57 : 93–98.
6. OkeefeSJ, TamuraJ, KincaidRL, TocciMJ, OneillEA (1992) FK-506-sensitive and CsA-sensitive activation of the Interleukin-2 promotor by calcineurin. Nature 357 : 692–694.
7. CruzMC, Del PoetaM, WangP, WengerR, ZenkeG, et al. (2000) Immunosuppressive and nonimmunosuppressive cyclosporine analogs are toxic to the opportunistic fungal pathogen Cryptococcus neoformans via cyclophilin-dependent inhibition of calcineurin. Antimicrobial Agents and Chemotherapy 44 : 143–149.
8. NakagawaM, SakamotoN, EnomotoN, TanabeY, KanazawaN, et al. (2004) Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochemical and Biophysical Research Communications 313 : 42–47.
9. MarahielMA, StachelhausT, MootzHD (1997) Modular peptide synthetases involved in nonribosomal peptide synthesis. Chemical Reviews 97 : 2651–2673.
10. WeberG, SchorgendorferK, SchneiderscherzerE, LeitnerE (1994) The peptide synthetase catalyzing cyclosporine production in Tolypocladium-niveum is encoded by a giant 45.8-kilobase open reading frame. Current Genetics 26 : 120–125.
11. Isaka M, Kittakoop P (2003) Secondary Metabolites of Clavicipitalean Fungi. In: White JF, Bacon CW, Hywel-Jones NL, Spatafora JW, editors. Clavicipitalean Fungi: Evolutionary Biology, Chemistry, Biocontrol, and Cultural Impacts. New York, NY: Marcel Dekker Inc. pp 355–397.
12. CaneDE, WalshCT (1999) The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chemistry & Biology 6: R319–R325.
13. PalS, LegerRJS, WuLP (2007) Fungal peptide destruxin a plays a specific role in suppressing the innate immune response in Drosophila melanogaster. Journal of Biological Chemistry 282 : 8969–8977.
14. BandaniAR, KhambayBPS, FaullJL, NewtonR, DeadmanM, et al. (2000) Production of efrapeptins by Tolypocladium species and evaluation of their insecticidal and antimicrobial properties. Mycological Research 104 : 537–544.
15. TorresMS, SinghAP, VorsaN, WhiteJFJr (2008) An analysis of ergot alkaloids in the Clavicipitaceae (Hypocreales, Ascomycota) and ecological implications. Symbiosis 46 : 11–19.
16. SungGH, PoinarGO, SpataforaJW (2008) The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal-arthropod symbioses. Molecular Phylogenetics and Evolution 49 : 495–502.
17. SpataforaJW, SungGH, SungJM, Hywel-JonesNL, WhiteJF (2007) Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Molecular Ecology 16 : 1701–1711.
18. HodgeK, KrasnoffS, HumberRA (1996) Tolypocladium inflatum is the anamorph of Cordyceps subsessilis. Mycologia 88 : 715–719.
19. GaoQA, JinK, YingSH, ZhangYJ, XiaoGH, et al. (2011) Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genetics 7: e1001264.
20. BidochkaMJ, St LegerRJ, StuartA, GowanlockK (1999) Nuclear rDNA phylogeny in the fungal genus Verticillium and its relationship to insect and plant virulence, extracellular proteases and carbohydrases. Microbiology-UK 145 : 955–963.
21. ZhengPeng, XiaYongliang, XiaoGuohua, Chenghui Xiong, HuXiao, et al. (2011) Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biology 12: R116.
22. StimbergN, WalzM, SchorgendorferK, KuckU (1992) Electrophoretic karyotyping from Tolypocladium-inflatum and 6 related strains allows differentiation of morphologically similar species. Applied Microbiology and Biotechnology 37 : 485–489.
23. HoltC, YandellM (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12 : 491.
24. ParraG, BradnamK, KorfI (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23 : 1061–1067.
25. StajichJE, DeietrichFS, RoySW (2007) Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biology 8: R223.
26. KempkenF, SchreinerC, SchorgendorferK, KuchU (1995) A unique repeated DNA sequence in the cyclosporin-producing strain of Tolypocladium inflatum (ATCC 34921). Experimental Mycology 19 : 305–313.
27. KempkenF, KuckU (1996) restless, an active Ac-like transposon from the fungus Tolypocladium inflatum: Structure, expression, and alternative RNA splicing. Molecular and Cellular Biology 16 : 6563–6572.
28. KempkenF (2008) The Tolypocladium inflatum CPA element encodes a RecQ helicase-like gene. Journal of Basic Microbiology 48 : 496–499.
29. WindhoferF, HauckK, CatchesideDEA, KuckU, KempkenF (2002) Ds-like Restless deletion derivatives occur in Tolypocladium inflatum and two foreign hosts, Neurospora crassa and Penicillium chrysogenum. Fungal Genetics and Biology 35 : 171–182.
30. Van DongenS (2008) Graph clustering via a discrete uncoupling process. Siam Journal on Matrix Analysis and Applications 30 : 121–141.
31. RobbertseB, YoderR, BoydA, ReevesJ, SpataforaJ (2011) Hal: an automated pipeline for phylogenetic analyses of genomic data. PLoS Curr 3: RRN1213.
32. KubicekCP, Herrera-EstrellaA, Seidl-SeibothV, MartinezDA, DruzhininaIS, et al. (2011) Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biology 12: R40.
33. Rodriguez-EzpeletaN, BrinkmannH, RoureB, LartillotN, LangBF, et al. (2007) Detecting and overcoming systematic errors in genome-scale phylogenies. Systematic Biology 56 : 389–399.
34. XiaoG, YingS-H, ZhengP, WangZ-L, ZhangS, et al. (2012) Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Scientific Reports 2 : 483.
35. Pava-RipollM, AngeliniC, FangWG, WangSB, PosadaFJ, et al. (2011) The rhizosphere-competent entomopathogen Metarhizium anisopliae expresses a specific subset of genes in plant root exudate. Microbiology-SGM 157 : 47–55.
36. DreyfussM, HarriE, HofmannH, KobelH, PacheW, et al. (1976) Cyclosporin-A and C new metabolites from Trichoderma-polysporum (Link Ex Pers) rifai. European Journal of Applied Microbiology 3 : 125–133.
37. KrasnoffSB, GuptaS (1992) Efrapeptin production by Tolypocladium fungi (Deuteromycotina, Hyphomycetes) - Intraspecific and interspecific Variation. Journal of Chemical Ecology 18 : 1727–1741.
38. WeiserJ, MathaV (1988) Tolypin, a new insecticidal metabolite of fungi of the genus Tolypocladium. Journal of Invertebrate Pathology 51 : 94–96.
39. ChuM, MierzwaR, TruumeesI, GentileF, PatelM, et al. (1993) 2 novel diketopiperazines isolated from the fungus Tolypocladium sp. Tetrahedron Letters 34 : 7537–7540.
40. KellerNP, HohnTM (1997) Metabolic pathway gene clusters in filamentous fungi. Fungal Genetics and Biology 21 : 17–29.
41. WaltonJD (2000) Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: An hypothesis. Fungal Genetics and Biology 30 : 167–171.
42. GacekA, StraussJ (2012) The chromatin code of fungal secondary metabolite gene clusters. Applied Microbiology and Biotechnology 95 : 1389–1404.
43. KhaldiN, SeifuddinFT, TurnerG, HaftD, NiermanWC, et al. (2010) SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genetics and Biology 47 : 736–741.
44. MedemaMH, BlinK, CimermancicP, de JagerV, ZakrzewskiP, et al. (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Research 39: W339–W346.
45. PanaccioneDG (2005) Origins and significance of ergot alkaloid diversity in fungi. FEMS Microbiology Letters 251 : 9–17.
46. TudzynskiP, HoelterK, CorreiaT, ArntzC, GrammelN, et al. (1999) Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Molecular and General Genetics 261 : 133–141.
47. HoffmeisterD, KellerNP (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Natural Product Reports 24 : 393–416.
48. VonwartburgA, TraberR (1986) Chemistry of the natural cyclosporine metabolites. Progress in Allergy 38 : 28–45.
49. TraberR, DreyfussMM (1996) Occurrence of cyclosporins and cyclosporin-like peptolides in fungi. Journal of Industrial Microbiology & Biotechnology 17 : 397–401.
50. NakajimaH, HamasakiT, TanakaK, KimuraY, UdagawaS, et al. (1989) Production of cyclosporin by fungi belonging to the genus Neocosmospora. Agric Biol Chem 53 : 2291–2292.
51. SallamLAR, El-RefaiAMH, HamdyAHA, El-MinofiHA, Abdel-SalamIS (2003) Role of some fermentation parameters on cyclosporin A production by a new isolate of Aspergillus terreus. Journal of General and Applied Microbiology 49 : 321–328.
52. SakamotoK, TsujiiE, MiyauchiM, NakanishiT, YamashitaM, et al. (1993) FR901459, a novel immunosuppressant isolated from Stachybotrys chartarum No. 19392. J Antibiotics 46 : 1788–1798.
53. DreyfussMM (1986) Neue Erkenntnisse aus einem pharmakologischen Screening. Sydowia 39 : 22–36.
54. BushleyKE, TurgeonBG (2010) Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evolutionary Biology 10 : 26.
55. HaeseA, SchubertM, HerrmannM, ZocherR (1993) Molecular characterization of the Enniatin synthetase gene encoding a multifunctional enzyme catalyzing N-methyldepsipeptide formation in Fusarium-scirpi. Molecular Microbiology 7 : 905–914.
56. XuYQ, OrozcoR, WijeratneEMK, GunatilakaAAL, StockSP, et al. (2008) Biosynthesis of the cyclooligomer depsipeptide Beauvericin, a virulence factor of the entomopathogenic fungus Beauveria bassiana. Chemistry & Biology 15 : 898–907.
57. XuYQ, RozcoR, WijeratneEMK, Espinosa-ArtilesP, GunatilakaAAL, et al. (2009) Biosynthesis of the cyclooligomer depsipeptide bassianolide, an insecticidal virulence factor of Beauveria bassiana. Fungal Genetics and Biology 46 : 353–364.
58. FiolkaM (2008) Immunosuppressive effect of cyclosporin A on insect humoral immune response. Journal of Invertebrate Pathology 98 : 287–292.
59. EndoM, TakesakoK, KatoI, YamaguchiH (1997) Fungicidal action of aureobasidin A, a cyclic depsipeptide antifungal antibiotic, against Saccharomyces cerevisiae. Antimicrobial Agents and Chemotherapy 41 : 672–676.
60. CardenasME, CruzMC, Del PoetaM, ChungNJ, PerfectJR, et al. (1999) Antifungal activities of antineoplastic agents Saccharomyces cerevisiae as a model system to study drug action. Clinical Microbiology Reviews 12 : 583–611.
61. BushleyKE, RipollDR, TurgeonBG (2008) Module evolution and substrate specificity of fungal nonribosomal peptide synthetases involved in siderophore biosynthesis. BMC Evolutionary Biology 8 : 328.
62. PieperR, KleinkaufH, ZocherR (1992) Enniatin synthetases from different Fusaria exhibiting distinct amino-acid specificities. Journal of Antibiotics 45 : 1273–1277.
63. SlightomJL, MetzgerBR, LuuHT, ElhammerAP (2009) Cloning and molecular characterization of the gene encoding the Aureobasidin A biosynthesis complex in Aureobasidium pullulans BP-1938. Gene 431 : 67–79.
64. PondSLK, MurrellB, FourmentM, FrostSDW, DelportW, et al. (2011) A Random Effects Branch-Site Model for Detecting Episodic Diversifying Selection. Molecular Biology and Evolution 28 : 3033–3043.
65. LeeMJ, LeeHN, HanK, KimES (2008) Spore inoculum optimization to maximize cyclosporin a production in Tolypocladium niveum. Journal of Microbiology and Biotechnology 18 : 913–917.
66. SamelSA, MarahielMA, EssenLO (2008) How to tailor non-ribosomal peptide products - new clues about the structures and mechanisms of modifying enzymes. Molecular Biosystems 4 : 387–393.
67. HoffmannK, SchneiderscherzerE, KleinkaufH, ZocherR (1994) Purification and characterization of eukaryotic alanine racemase acting as key enzyme in cyclosporine biosynthesis. Journal of Biological Chemistry 269 : 12710–12714.
68. OffenzellerM, SanterG, TotschnigK, SuZ, MoserH, et al. (1996) Biosynthesis of the unusual amino acid (4R)-4-[(E)-2-butenyl]-4-methyl-L-threonine of cyclosporin A: Enzymatic analysis of the reaction sequence including identification of the methylation precursor in a polyketide pathway. Biochemistry 35 : 8401–8412.
69. SanglierJJ, TraberR, BuckRH, HofmannH, KobelH (1990) Isolation of (4-R)-R-(E)-2-Butenyl-4-Methyl-L-Threonine, the characteristic structural element of cyclosporins, rrom a blocked mutant of Tolypocladium-inflatum. Journal of Antibiotics 43 : 707–714.
70. ThériaultYves, LoganTimothy M, MeadowsRobert, YuLiping, OlejniczakEdward T, et al. (1993) Solution structure of the cyclosporin A/cyclophilin complex by NMR. Nature 361 : 88–90.
71. PflüglGaston, KallenJörg, SchirmerTilman, JansoniusJohan N, MauroGM, et al. (1993) X-ray structure of a decameric cyclophilin-cyclosporin crystal complex. Nature 361 : 91–93.
72. GalatA (1999) Variations of sequences and amino acid compositions of proteins that sustain their biological functions: An analysis of the cyclophilin family of proteins. Archives of Biochemistry and Biophysics 371 : 149–162.
73. GalatA (2003) Peptidylprolyl cis/trans isomerases (immunophilins): Biological diversity targets - Functions. Current Topics in Medicinal Chemistry 3 : 1315–1347.
74. GothelSF, MarahielMA (1999) Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cellular and Molecular Life Sciences 55 : 423–436.
75. ChenMM, JiangMG, ShangJJ, LanXW, YangF, et al. (2011) CYP1, a hypovirus-regulated cyclophilin, is required for virulence in the chestnut blight fungus. Molecular Plant Pathology 12 : 239–246.
76. HermansPWM, AdrianPV, AlbertC, EstevaoS, HoogenboezemT, et al. (2006) The streptococcal lipoprotein rotamase A (SlrA) is a functional peptidyl-prolyl isomerase involved in pneumococcal colonization. Journal of Biological Chemistry 281 : 968–976.
77. EmanuelssonO, NielsenH, BrunakS, von HeijneG (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology 300 : 1005–1016.
78. ViaudM, Brunet-SimonA, BrygooY, PradierJM, LevisC (2003) Cyclophilin A and calcineurin functions investigated by gene inactivation, cyclosporin A inhibition and cDNA arrays approaches in the phytopathogenic fungus Botrytis cinerea. Molecular Microbiology 50 : 1451–1465.
79. ViaudMC, BalhaderePV, TalbotNJ (2002) A Magnaporthe grisea cyclophilin acts as a virulence determinant during plant infection. Plant Cell 14 : 917–930.
80. WangP, CardenasME, CoxCM, PerfectJR, HeitmanJ (2001) Two cyclophilin A homologs with shared and distinct functions important for growth and virulence of Cryptococcus neoformans. Embo Reports 2 : 511–518.
81. HornbogenT, ZocherR (1995) Cloning and sequencing of a cyclophilin gene from the cyclosporine producer Tolypocladium-niveum. Biochemistry and Molecular Biology International 36 : 169–176.
82. WeberG, LeitnerE (1994) Disruption of the cyclosporine synthetase gene of Tolypocladium-niveum. Current Genetics 26 : 461–467.
83. TropschugM, NicholsonDW, HartlFU, KohlerH, PfannerN, et al. (1988) Cyclosporin A-binding protein (Cyclophilin) of Neurospora crassa - One gene codes for both the cytosolic and mitochondrial Forms. Journal of Biological Chemistry 263 : 14433–14440.
84. DumasC, RavallecM, MathaV, VeyA (1996) Comparative study of the cytological aspects of the mode of action of destruxins and other peptidic fungal metabolites on target epithelial cells. Journal of Invertebrate Pathology 67 : 137–146.
85. HornbogenT, PieperR, HoffmannK, KleinkaufH, ZocherR (1992) 2 New cyclophilins from Fusarium sambucinum and Aspergillus-niger - Resistance of cyclophilin Cyclosporine A complexes against proteolysis. Biochemical and Biophysical Research Communications 187 : 791–796.
86. VilcinskasA, KopacekP, JegorovA, VeyA, MathaV (1997) Detection of lipophorin as the major cyclosporin-binding protein in the hemolymph of the greater wax moth Galleria mellonella. Comparative Biochemistry and Physiology C-Pharmacology Toxicology & Endocrinology 117 : 41–45.
87. LeeBN, KrokenS, ChouDYT, RobbertseB, YoderOC, et al. (2005) Functional analysis of all nonribosomal peptide synthetases in Cochliobolus heterostrophus reveals a factor, NPS6, involved in virulence and resistance to oxidative stress. Eukaryotic Cell 4 : 545–555.
88. FuJ, WenzelSC, PerlovaO, WangJP, GrossF, et al. (2008) Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucleic Acids Research 36: e113.
89. SlotJC, RokasA (2011) Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Current Biology 21 : 134–139.
90. KhaldiN, CollemareJ, LebrunMH, WolfeKH (2008) Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biology 9: R18.
91. GardinerDM, HowlettBJ (2005) Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiology Letters 248 : 241–248.
92. AnayaN, RonceroMIG (1995) skippy, a retrotransposon from the fungal plant pathogen Fusarium oxysporum. Molecular & General Genetics 249 : 637–647.
93. LeaRuiqiang (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24 : 713–714.
94. SchulzMH, ZerbinoDR, VingronM, BirneyE (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28 : 1086–1092.
95. LoweTM, EddySR (1997) tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research 25 : 955–964.
96. EdgarRC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32 : 1792–1797.
97. AbascalF, ZardoyaR, PosadaD (2005) ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21 : 2104–2105.
98. YangZH (2007) PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24 : 1586–1591.
99. SandersonMJ (2003) r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19 : 301–302.
100. O'BrienKP, RemmM, SonnhammerELL (2005) Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Research 33: D476–D480.
101. BerglundAC, SjolundE, OstlundG, SonnhammerELL (2008) InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Research 36: D263–D266.
102. EmanuelssonO, BrunakS, von HeijneG, NielsenH (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2 : 953–971.
103. SmallI, PeetersN, LegeaiF, LurinC (2004) Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4 : 1581–1590.
104. KroghA, LarssonB, von HeijneG, SonnhammerELL (2001) Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. Journal of Molecular Biology 305 : 567–580.
105. RawlingsND, BarrettAJ, BatemanA (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research 40: D343–D350.
106. NelsonDR (2002) Mining databases for cytochrome P450 genes. Methods in Enzymology: Cytochrome P450, Part C 357 : 3–15.
107. KatohM, KumaMiyata (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res 30 : 3059–3066.
108. StamatakisA, HooverP, RougemontJ (2008) A rapid bootstrap algorithm for the RAxML web-servers. Systematic Biology 75 : 758–771.
109. DarlingAE, MauB, PernaNT (2010) progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. Plos One 5: e11147.
110. St LegerRJ, NelsonJO, ScreenSE (1999) The entomopathogenic fungus Metarhizium anisopliae alters ambient pH, allowing extracellular protease production and activity. Microbiology-UK 145 : 2691–2699.
111. St LegerRJ, BidochkaMJ, RobertsDW (1994) Isoforms of the cuticle degrading Pr1 protease and production of a metalloproteinase by Metarhizium anisopliae. Arch Biochem Biophysical Journal 313 : 1–7.
112. St. LegerRJ, BidochkaMJ, RobertsDW (1994) Characterization of a novel carboxypeptidase produced by the entomopathogenic fungus Metarhizium anisopliae. Arch Biochem Biophys 314 : 392–398.
113. CumbieJS, KimbrelJA, DiYM, SchaferDW, WilhelmLJ, et al. (2011) GENE-Counter: A Computational Pipeline for the Analysis of RNA-Seq Data for Gene Expression Differences. PloS ONE 6: e25279.
114. DiYM, SchaferDW, CumbieJS, ChangJH (2011) The NBP Negative Binomial Model for Assessing Differential Gene Expression from RNA-Seq. Statistical Applications in Genetics and Molecular Biology 10 doi: 10.2202/1544-6115.1637
115. MortazaviA, WilliamsBA, McCueK, SchaefferL, WoldB (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5 : 621–628.
Štítky
Genetika Reprodukční medicína
Článek PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant TransformationČlánek Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish PathogenČlánek USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and TranscriptionČlánek Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA EditingČlánek Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 6
-
Všechny články tohoto čísla
- BMS1 Is Mutated in Aplasia Cutis Congenita
- High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants
- How Cool Is That: An Interview with Caroline Dean
- Genetic Architecture of Vitamin B and Folate Levels Uncovered Applying Deeply Sequenced Large Datasets
- Juvenile Hormone and Insulin Regulate Trehalose Homeostasis in the Red Flour Beetle,
- Meiosis-Specific Stable Binding of Augmin to Acentrosomal Spindle Poles Promotes Biased Microtubule Assembly in Oocytes
- Environmental Dependence of Genetic Constraint
- H3.3-H4 Tetramer Splitting Events Feature Cell-Type Specific Enhancers
- Network Topologies and Convergent Aetiologies Arising from Deletions and Duplications Observed in Individuals with Autism
- Effectively Identifying eQTLs from Multiple Tissues by Combining Mixed Model and Meta-analytic Approaches
- Altered Splicing of the BIN1 Muscle-Specific Exon in Humans and Dogs with Highly Progressive Centronuclear Myopathy
- The NADPH Metabolic Network Regulates Human Cardiomyopathy and Reductive Stress in
- Negative Regulation of Notch Signaling by Xylose
- A Genome-Wide, Fine-Scale Map of Natural Pigmentation Variation in
- Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals mC within Archaeal mRNAs
- Multiplexin Promotes Heart but Not Aorta Morphogenesis by Polarized Enhancement of Slit/Robo Activity at the Heart Lumen
- Latent Effects of Hsp90 Mutants Revealed at Reduced Expression Levels
- Impact of Natural Genetic Variation on Gene Expression Dynamics
- DeepSAGE Reveals Genetic Variants Associated with Alternative Polyadenylation and Expression of Coding and Non-coding Transcripts
- The Identification of -acting Factors That Regulate the Expression of via the Osteoarthritis Susceptibility SNP rs143383
- Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs
- The RNA Export Factor, Nxt1, Is Required for Tissue Specific Transcriptional Regulation
- Inferring Demographic History from a Spectrum of Shared Haplotype Lengths
- Histone Acetyl Transferase 1 Is Essential for Mammalian Development, Genome Stability, and the Processing of Newly Synthesized Histones H3 and H4
- PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant Transformation
- DNA Methylation Restricts Lineage-specific Functions of Transcription Factor Gata4 during Embryonic Stem Cell Differentiation
- The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Deregulation of the Protocadherin Gene Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy
- Evidence for Two Different Regulatory Mechanisms Linking Replication and Segregation of Chromosome II
- USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and Transcription
- Methylation of Histone H3 on Lysine 79 Associates with a Group of Replication Origins and Helps Limit DNA Replication Once per Cell Cycle
- A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue
- The Gene Desert Mammary Carcinoma Susceptibility Locus Regulates Modifying Mammary Epithelial Cell Differentiation and Proliferation
- Hooked and Cooked: A Fish Killer Genome Exposed
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
- Mutations in Cause Autosomal Recessive Congenital Ichthyosis in Humans
- Integrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation
- RSR-2, the Ortholog of Human Spliceosomal Component SRm300/SRRM2, Regulates Development by Influencing the Transcriptional Machinery
- Comparative Polygenic Analysis of Maximal Ethanol Accumulation Capacity and Tolerance to High Ethanol Levels of Cell Proliferation in Yeast
- SPO11-Independent DNA Repair Foci and Their Role in Meiotic Silencing
- Budding Yeast ATM/ATR Control Meiotic Double-Strand Break (DSB) Levels by Down-Regulating Rec114, an Essential Component of the DSB-machinery
- Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA Editing
- Functional Analysis of Neuronal MicroRNAs in Dauer Formation by Combinational Genetics and Neuronal miRISC Immunoprecipitation
- DNA Ligase IV Supports Imprecise End Joining Independently of Its Catalytic Activity
- Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
- Heritable Change Caused by Transient Transcription Errors
- From Many, One: Genetic Control of Prolificacy during Maize Domestication
- Neuronal Target Identification Requires AHA-1-Mediated Fine-Tuning of Wnt Signaling in
- Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish
- H-NS Can Facilitate Specific DNA-binding by RNA Polymerase in AT-rich Gene Regulatory Regions
- Prophage Dynamics and Contributions to Pathogenic Traits
- Global DNA Hypermethylation in Down Syndrome Placenta
- Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in
- Disturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-ethylene Crosstalk in Seminal Roots
- Causes and Consequences of Chromatin Variation between Inbred Mice
- Genome-scale Analysis of FNR Reveals Complex Features of Transcription Factor Binding
- Distinct and Atypical Intrinsic and Extrinsic Cell Death Pathways between Photoreceptor Cell Types upon Specific Ablation of in Cone Photoreceptors
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- BMS1 Is Mutated in Aplasia Cutis Congenita
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



