-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDisturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-ethylene Crosstalk in Seminal Roots
Observations gained from model organisms are essential, yet it remains unclear to which degree they are applicable to distant relatives. For example, in the dicotyledon Arabidopsis thaliana (Arabidopsis), auxin biosynthesis via indole-3-pyruvic acid (IPA) is essential for root development and requires redundant TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) and TAA1-RELATED (TAR) genes. A promoter T-DNA insertion in the monocotyledon Brachypodium distachyon (Brachypodium) TAR2-LIKE gene (BdTAR2L) severely down-regulates expression, suggesting reduced tryptophan aminotransferase activity in this mutant, which thus represents a hypomorphic Bdtar2l allele (Bdtar2lhypo). Counterintuitive however, Bdtar2lhypo mutants display dramatically elongated seminal roots because of enhanced cell elongation. This phenotype is also observed in another, stronger Bdtar2l allele and can be mimicked by treating wild type with L-kynerunine, a specific TAA1/TAR inhibitor. Surprisingly, L-kynerunine-treated as well as Bdtar2l roots display elevated rather than reduced auxin levels. This does not appear to result from compensation by alternative auxin biosynthesis pathways. Rather, expression of YUCCA genes, which are rate-limiting for conversion of IPA to auxin, is increased in Bdtar2l mutants. Consistent with suppression of Bdtar2lhypo root phenotypes upon application of the ethylene precursor 1-aminocyclopropane-1-carboxylic-acid (ACC), BdYUCCA genes are down-regulated upon ACC treatment. Moreover, they are up-regulated in a downstream ethylene-signaling component homolog mutant, Bd ethylene insensitive 2-like 1, which also displays a Bdtar2l root phenotype. In summary, Bdtar2l phenotypes contrast with gradually reduced root growth and auxin levels described for Arabidopsis taa1/tar mutants. This could be explained if in Brachypodium, ethylene inhibits the rate-limiting step of auxin biosynthesis in an IPA-dependent manner to confer auxin levels that are sub-optimal for root cell elongation, as suggested by our observations. Thus, our results reveal a delicate homeostasis of local auxin and ethylene activity to control cell elongation in Brachypodium roots and suggest alternative wiring of auxin-ethylene crosstalk as compared to Arabidopsis.
Published in the journal: . PLoS Genet 9(6): e32767. doi:10.1371/journal.pgen.1003564
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003564Summary
Observations gained from model organisms are essential, yet it remains unclear to which degree they are applicable to distant relatives. For example, in the dicotyledon Arabidopsis thaliana (Arabidopsis), auxin biosynthesis via indole-3-pyruvic acid (IPA) is essential for root development and requires redundant TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) and TAA1-RELATED (TAR) genes. A promoter T-DNA insertion in the monocotyledon Brachypodium distachyon (Brachypodium) TAR2-LIKE gene (BdTAR2L) severely down-regulates expression, suggesting reduced tryptophan aminotransferase activity in this mutant, which thus represents a hypomorphic Bdtar2l allele (Bdtar2lhypo). Counterintuitive however, Bdtar2lhypo mutants display dramatically elongated seminal roots because of enhanced cell elongation. This phenotype is also observed in another, stronger Bdtar2l allele and can be mimicked by treating wild type with L-kynerunine, a specific TAA1/TAR inhibitor. Surprisingly, L-kynerunine-treated as well as Bdtar2l roots display elevated rather than reduced auxin levels. This does not appear to result from compensation by alternative auxin biosynthesis pathways. Rather, expression of YUCCA genes, which are rate-limiting for conversion of IPA to auxin, is increased in Bdtar2l mutants. Consistent with suppression of Bdtar2lhypo root phenotypes upon application of the ethylene precursor 1-aminocyclopropane-1-carboxylic-acid (ACC), BdYUCCA genes are down-regulated upon ACC treatment. Moreover, they are up-regulated in a downstream ethylene-signaling component homolog mutant, Bd ethylene insensitive 2-like 1, which also displays a Bdtar2l root phenotype. In summary, Bdtar2l phenotypes contrast with gradually reduced root growth and auxin levels described for Arabidopsis taa1/tar mutants. This could be explained if in Brachypodium, ethylene inhibits the rate-limiting step of auxin biosynthesis in an IPA-dependent manner to confer auxin levels that are sub-optimal for root cell elongation, as suggested by our observations. Thus, our results reveal a delicate homeostasis of local auxin and ethylene activity to control cell elongation in Brachypodium roots and suggest alternative wiring of auxin-ethylene crosstalk as compared to Arabidopsis.
Introduction
The root system plays a fundamental role for plant growth and survival, not only by providing support, water and nutrients for the shoot, but also by participating in secondary functions, such as hormone biosynthesis or storage of photoassimilates [1], [2]. Root system architecture, that is the number and arrangement of different root types and their branching pattern, is highly plastic and determined by developmental and environmental factors that interact to optimize soil exploration. This is particularly important for the capture of growth limiting macronutrients, including nitrogen and phosphorus, whose edaphic distribution strongly influences post-embryonic root development and, therefore, root system architecture [2]–[4]. However, the root system can only respond to variation in such resources within its inherent developmental limits of growth rate and branching capacity, which are genetically determined. Optimization of root system architecture through breeding is therefore of particular interest in crops to increase root system plasticity with respect to biotic and abiotic stresses [5], [6].
Our knowledge about the molecular genetic control of root growth and branching has been largely obtained from analyses of the dicotyledon plant model system Arabidopsis thaliana (Arabidopsis) through mutagenesis approaches [2], [7]. The genes identified through these efforts have greatly benefitted the isolation of corresponding loci in monocotyledons, such as rice or maize [8]–[11]. Many of them encode proteins with regulatory functions, and among them components of plant hormone signaling pathways are particularly preeminent. For example, interference with the auxin-signaling pathway by mutation typically impairs primary root elongation or root branching, and in extreme cases even abolishes root formation [12]–[14]. The same is true for loss-of-function mutations in genes that encode enzymes involved in tryptophan-dependent auxin biosynthesis. In particular, auxin biosynthesis from tryptophan via indole-3-pyruvic acid (IPA) has been shown to be essential for root formation [15], [16]. Two enzyme classes define this pathway: the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) and TAA1-RELATED (TAR) proteins, which catalyze the conversion of tryptophan to IPA; and the family of YUCCA cytochrome P450s, which catalyze the conversion of IPA to indole-3-acetic acid (IAA), the major active form of auxin [15]–[18]. Whereas the YUCCA genes were originally identified through a gain-of-function approach that led to auxin over-accumulation [19], TAA1/TAR genes were identified through loss-of-function approaches [16], [20], [21]. For instance, one study isolated the taa1 mutant because of its root growth resistance to the application of 1-aminocyclopropane-1-carboxylic-acid (ACC), a rate-limiting precursor for the biosynthesis of another hormone, ethylene [16]. This phenotype arises as a consequence of reduced auxin biosynthesis, which is normally up-regulated by ethylene through induction of TAA1/TAR gene expression. This finding also illustrates the dosage-dependent action of auxin, because although auxin and its perception are essential for root formation and growth, excess auxin application, biosynthesis or signaling are eventually inhibitory [22]–[24]. Indeed, it has been suggested that depending on the species, auxin levels might be supra-optimal for root growth [25].
Phylogenetic analysis has identified bona fide TAA1/TAR homologous genes in monocotyledons, with varying degrees of redundancy. For instance, whereas maize contains five genes of this family, only two are found in both rice and the monocotyledon model system, Brachypodium distachyon (Brachypodium) [26]. So far, only one TAA1/TAR-related mutant has been identified in monocotyledons, the vanishing tassel 2 (vt2) mutant of maize [26]. Despite the presence of multiple TAA1/TAR homologs in maize, vt2 null mutants display rather severe shoot phenotypes, such as dwarfism, reduced axillary meristem formation and associated impaired inflorescence development. Free auxin levels are reduced to ca. one third of wild type levels in vt2 mutants, suggesting that VT2 encodes the predominant TAA1/TAR activity in maize. Here we report the isolation and characterization of a Brachypodium mutant in the TAR2-LIKE (BdTAR2L) gene. Unlike vt2, this Bdtar2l mutant displays only mild shoot phenotypes. However, we observed dramatic root phenotypes, which surprisingly appear to result from upwardly disturbed auxin homeostasis.
Results
Isolation of a T-DNA insertion mutant of BdTAR2L
In an effort to identify genetic factors that influence root system architecture in Brachypodium, we monitored seedlings from transgenic lines obtained in our lab through T-DNA transformation in tissue culture. One regenerated line stood out because of the occurrence of longer seminal (primary) roots (Fig. 1A–B), a phenotype that co-segregated recessively with the T-DNA insertion (χ2 test two-tailed p value = 0.7697). Isolation of the flanking genomic DNA by an inverse PCR strategy [27] revealed that this line contains only one T-DNA locus, whose integration site is located in the Bd2g04290 gene. Both the copy number and the insertion site were confirmed by whole genome sequencing of the homozygous mutant line (Fig. S1A). Bd2g04290 is one of the two TAA1/TAR homologs of Brachypodium, the other one being Bd2g34400 [26]. Based on their closest homologs in Arabidopsis, we named them Brachypodium distachyon TAR2-LIKE (BdTAR2L, Bd2g04290) and Brachypodium distachyon TAR1-LIKE (BdTAR1L, Bd2g34400), respectively. Quantitative RT-PCR (qPCR) to monitor expression of both genes in dissected seedling tissues indicated that BdTAR1L expression is dominant in the root meristem, whereas relative BdTAR2L expression increases strongly in the elongating and mature parts of the root, and in the shoot tissues (Fig. S1B). The T-DNA insertion in this Bdtar2l mutant is located 140 bp upstream of the ATG codon, thereby presumably disrupting the 5′ UTR, but not the coding sequence (Fig. 1C). To determine whether and to what extent the T-DNA insertion affects BdTAR2L expression, we quantified BdTAR2L mRNA levels by qPCR in 4-day-old seedlings. Indeed, expression was still detected both in shoot and root tissue, however at severely reduced levels of less than 20% and 5%, respectively, as compared to wild type or an unrelated transformant (the unrelated transformant line was included in all our assays to control for any tissue culture regeneration effects and contains a single copy T-DNA insert in a non-annotated, possibly repetitive region as determined by whole genome sequencing) (Fig. 1D–E). Therefore, it appears that our Bdtar2l mutation represents a hypomorphic allele, which we thus named Bdtar2lhypo. Interestingly, plants that are homozygous for the Bdtar2lhypo mutation display dramatically elongated roots (Fig. 1F–G), compared to which the shoot phenotype is rather mild. We could not detect any difference to wild type in the vegetative growth pattern, but observed a general decrease in overall leaf size to ca. 80% of wild type (Fig. 1H–M). Reproductive development in the mutant progresses normal without any apparent defects in inflorescence development, and plants are fully fertile.
Fig. 1. Isolation of the Bdtar2lhypo mutant and characterization of macroscopic phenotypes. 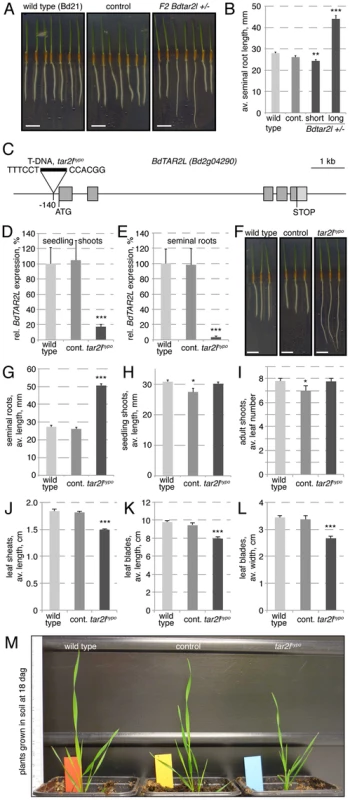
(A) Four-day-old tissue culture grown seedlings of wild type (accession Bd21), an unrelated control transformant (control) (the unrelated transformant line was included in all our assays to control for any tissue culture regeneration effects) and the transgenic line segregating the Bdtar2lhypo mutation. (B) Seminal root length quantification of the different genotypes at 4 days after germination (dag). (C) Schematic presentation of the BdTAR2L gene and the location of the T-DNA insertion in the Bdtar2lhypo mutant. (D–E) Relative expression level of BdTAR2L in different genotypes at 4 dag as determined by qPCR and normalized with respect to the housekeeping gene, BdUBC18. (F) Root elongation in wild type, control and homozygous Bdtar2lhypo mutants, assayed at 4 dag. (G–H) Quantification of seedling phenotypes at 4 dag. (I) Leaf number at 18 dag. (J–L) Different size parameters of the 5th leaf of plants, assayed at 18 dag. (M) Representative image of adult plants at 18 dag. Size bars are 1 cm; differences as compared to wild type are not significant unless indicated otherwise; error bars indicate standard error; * = p<0.05; ** = p<0.01; *** = p<0.001. Increased cellular anisotropy in Bdtar2lhypo roots
A closer look at the mutant roots revealed that their phenotype is principally due to increased cellular anisotropy, which is most apparent in the post-meristematic, differentiated region. For instance, mature cortical cell length in Bdtar2lhypo roots reaches typically ca. 150% of wild type control (Fig. 2A–B), which would largely account for the overall increase in root length. We did indeed not observe a difference in root meristem size, measured as the number of cells that constitute the division and transition zones of the meristem in the central metaxylem cell file (Fig. 2C–D). Also, metaxylem cell length at equal position in the meristem is similar in Bdtar2lhypo and wild type up to the elongation zone, from where on cells elongate dramatically faster in Bdtar2lhypo than in wild type (Fig. 2C). At the same time, the transverse total as well as stele area of mature roots is reduced in Bdtar2lhypo to ca. 85% of wild type, accompanied by a slight reduction in the number of cells along the circumference of the innermost cortex layer (Fig. 2E–H). A quantitative analysis of transverse sections using an automated segmentation pipeline indicated that the number of cells in the outer six cell layers is indeed slightly reduced in Bdtar2lhypo mutants (Fig. 2I–J). Moreover, except in the epidermal layer, transverse cell area is in tendency smaller in Bdtar2lhypo (Fig. 2K). Therefore, mature root cells are overall thinner and longer than in wild type. Interestingly, this change in cellular anisotropy also manifests in the morphology of the root hairs, which are extensions of the epidermal cells and shorter in Bdtar2lhypo (Fig. 2L–M). In summary, while the decreased diameter of Bdtar2lhypo roots can be explained by a combination of a slight decrease in cell proliferation and in expansion in the radial dimension, their increased length can be attributed to enhanced cell elongation. Thus, the Bdtar2lhypo root phenotype largely results from increased cellular anisotropy.
Fig. 2. Cellular root phenotypes of Bdtar2lhypo mutants. 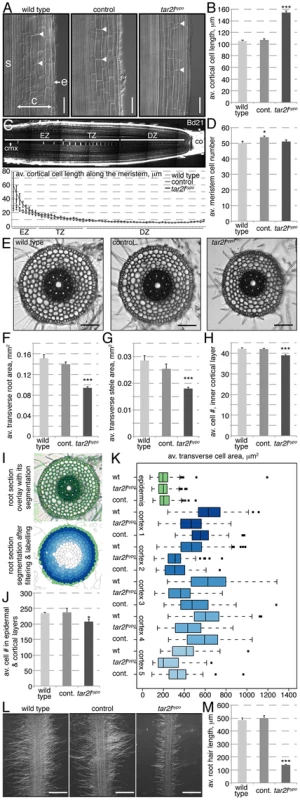
(A) Representative Nomarski optics images of mature root portions. s: stele; c: cortex layers; e: epidermis; arrowheads point out top and bottom of individual cells in the 3rd cortex layer; (B) Quantification of mature cortex cell length at 4 dag. (C) Confocal image of a 4-day-old Brachypodium wild type (Bd21) root meristem (top) and quantification of the progression of cell elongation per cell position (for the 60 cells above the stem cell niche) in the central metaxylem (cmx) (bottom). co: columella; DZ: division zone; TZ: transition zone; EZ: elongation zone; (D) Quantification of meristem size as number of metaxylem cells in the combined DZ and TZ. (E) Representative light microscopy images of transverse sections across the mature root. (F–H) Quantification of total area and stele area in root sections, as well as number of cells in the circumference of the innermost cortex layer. (I) Illustration of the segmentation process for quantification of root sections. Overlay of segmented cell shapes (green) on the section (top), labeling of cell layers after filtering (bottom). (J) Combined epidermal and cortical cell number in root sections. (K) Transverse cell area in root sections, for different cell layers from outside to inside. (L) Representative microscopy images of root hairs at 4 dag. (M) Quantification of root hair length in 4-day-old seedlings. Size bars are 100 µm; differences as compared to wild type or mock are not significant unless indicated otherwise; error bars indicate standard error; * = p<0.05; *** = p<0.001. Characterization of another loss-of-function allele of BdTAR2L
To independently corroborate the effects of reduced BdTAR2L expression, we obtained another mutant allele from the Brachypodium T-DNA collection in which the gene is disrupted by a T-DNA insertion in the second intron (Fig. S1C) [28]. In semi-quantitative RT-PCR, a cDNA fragment comprising the borders of exons 1 and 2 was nearly undetectable (Fig. S1D), and compared to the Bdtar2lhypo allele, BdTAR2L expression in the root as monitored by qPCR was even more severely reduced, to 1–2% of wild type levels (Fig. 3A). However, since we could not exclude production of some residual full-length transcript, we designated this allele a quasi-null mutant (Bdtar2lqnull). Compared to their wild type background, Bd21-3, Bdtar2lqnull mutants again display an elongated root phenotype, which is however not as drastic as in Bdtar2lhypo mutants (Fig. 3B–C). This could again be largely attributed to increased cell elongation, which reaches about 125% of wild type (Fig. 3D–E). Moreover, Bdtar2lqnull mutants also display shorter root hairs (Fig. 3F). At the same time, transverse root and stele area are reduced to about the levels observed in Bdtar2lhypo mutants, without a change in the number of cortical cell layers (Fig. 3G–I). Therefore, similar to Bdtar2lhypo, Bdtar2lqnull mutants display increased cell elongation and cellular anisotropy in the root. Unlike in Bdtar2lhypo mutants, however, exaggerated root growth is not sustained in Bdtar2lqnull mutants although the enhanced cell elongation is maintained (Fig. 3J). This is because of a gradual consumption of the root meristem as development proceeds (Fig. 3K). Compared to Bdtar2lhypo, Bdtar2lqnull mutants also display more severe shoot phenotypes, notably a clearly reduced shoot length in young seedlings (Fig. 3L) and a dwarf stature as an adult (Fig. 3M), which is accompanied by severely reduced fertility. Collectively, our mutant characterizations therefore suggest that the Bdtar2lhypo and Bdtar2lqnull mutants indeed represent an allelic series that displays the consequences of gradually reduced BdTAR2L dosage.
Fig. 3. Phenotypes of the Bdtar2lqnull mutant in comparison to its wild type background, Bd21-3. 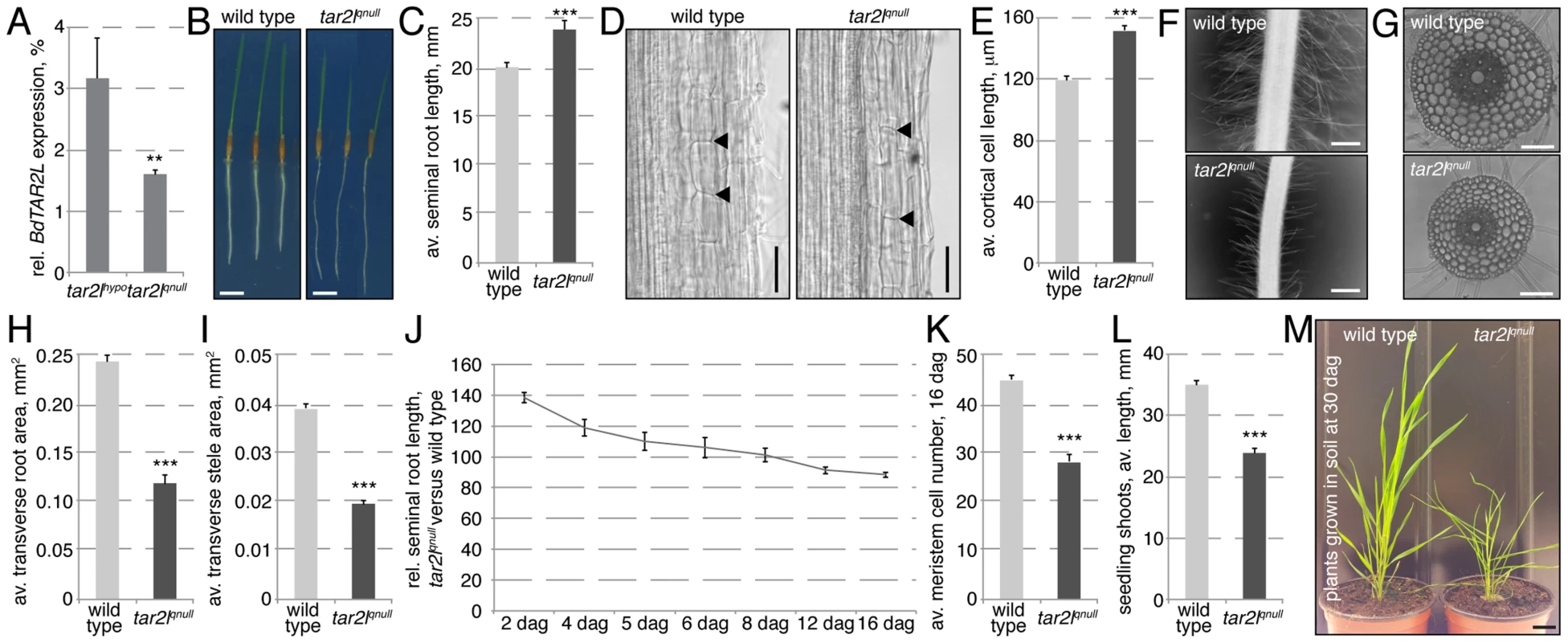
(A) Relative expression level of BdTAR2L in different genotypes at 4 dag as determined by qPCR and normalized with respect to the housekeeping gene, BdUBC18. (B–C) Root elongation in wild type and Bdtar2lqnull mutants, assayed at 4 dag. (D) Representative Nomarski optics images of mature root portions. Arrowheads point out top and bottom of individual cells in a cortex layer; (E) Quantification of mature cortex cell length at 4 dag. (F) Representative microscopy images of root hairs at 4 dag. (G) Representative light microscopy images of transverse sections across the mature root. (H–I) Quantification of total transverse area and stele area in sections from mature roots. (J) Relative seminal root length in Bdtar2lqnull mutants during root growth progression. (K) Progressive breakdown of root meristem as indicated by shrinkage of the meristematic zone. (L) Seedling shoot length at 4 dag. (M) Adult shoots. Size bars are 1 cm (B, M) or 100 µm (D,F,G); differences as compared to wild type or mock are not significant unless indicated otherwise; error bars indicate standard error; * = p<0.05; *** = p<0.001. Mimic of the Bdtar2lhypo phenotype by L-kynerunine treatment of wild type plants
The low BdTAR2L expression level in the mutants is not compensated by up-regulated BdTAR1L expression (Fig. S1E) and therefore should result in overall decreased tryptophan aminotransferase activity. However, the Bdtar2lhypo long root phenotype is counterintuitive in this respect, because progressive loss-of-function of TAA1/TAR activity in Arabidopsis leads to progressively impaired rather than enhanced root growth [16]. The same is true when Arabidopsis wild type plants are grown on a specific competitive inhibitor of TAA1/TAR enzymes, L-kynerunine [29]. To test whether L-kynerunine also inhibits root growth in Brachypodium, we transferred 2-day-old seedlings onto media with different L-kynerunine concentrations and assayed root growth two days later. Strikingly, root elongation was stimulated rather than inhibited already at concentrations as little as 1 µM (Fig. 4A–B). Higher concentrations, up to 100 µM, strongly promoted root elongation up to 150–200% of the mock controls. Moreover, we observed exaggerated cell elongation upon L-kynerunine treatment (Fig. 4C–D), which therefore mimics the Bdtar2lhypo root phenotype. Interestingly, unlike wild type, the Bdtar2lhypo mutant hardly responded to L-kynerunine treatment (Fig. 4A–D). Finally, similar to the Arabidopsis taa1 mutant [20], both Bdtar2l alleles were hypersensitive to the application of the toxic tryptophan analog, 5-methyl-tryptophan (Fig. 4E–F), which is an artificial substrate for TAA/TAR enzymes. 5-methyl-tryptophan can be detoxified by its conversion to IPA [20], and therefore 5-methyl-tryptophan hypersensitivity is indicative of reduced IPA production. Thus, the data are consistent with the idea that reduced TAA1/TAR activity in the Bdtar2l mutants is indeed responsible for the mutant phenotypes.
Fig. 4. Effect of L-kynerunine (L-kyn) treatment on root elongation of different genotypes. 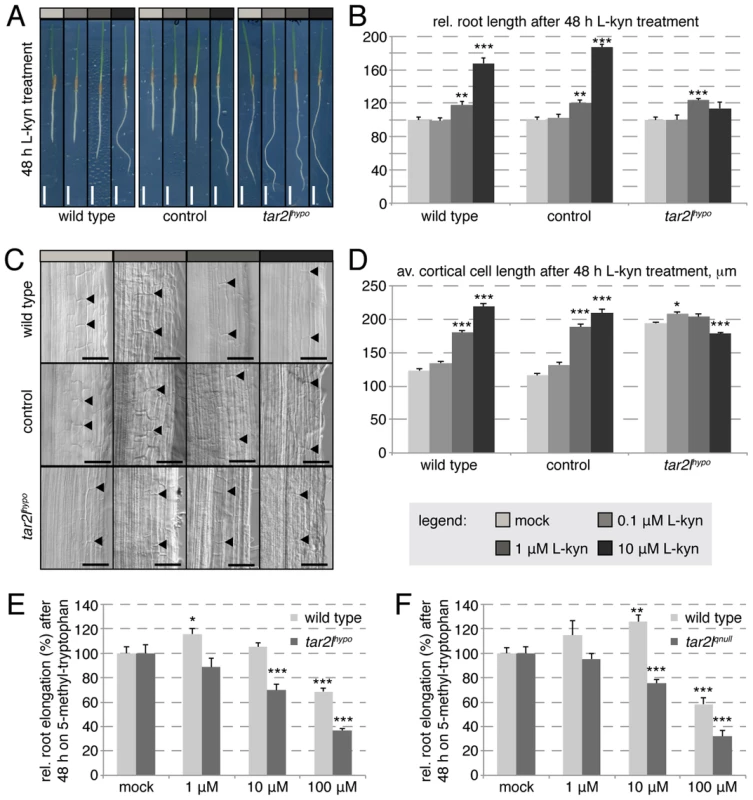
(A) Representative images of 4-day-old seedlings, transferred onto media with indicated L-kyn concentration at 2 dag. (B) Quantification of root length after 2 days of indicated L-kyn treatment. (C) Representative Nomarski optics images of mature root portions formed during indicated L-kyn treatment. Arrowheads point out top and bottom of individual cells in the 3rd cortex layer; (D) Quantification of mature cortex cell length after 2 days of indicated L-kyn treatment. (E–F) Relative root elongation of indicated mutants and their respective wild type backgrounds after 2 days of indicated 5-methyl-tryptophan treatment. Size bars are 1 cm (A) or 100 µm (C); differences as compared to wild type or mock are not significant unless indicated otherwise; error bars indicate standard error; * = p<0.05; ** = p<0.01; *** = p<0.001. Altered root branching patterns in the Bdtar2l mutants
While other auxin-dependent processes, such as gravitropism, appeared unaffected in Bdtar2l mutants (Fig. S1F), we also observed a root system branching phenotype. In Bdtar2lhypo mutants, coleoptile node root formation is slightly reduced (Fig. 5A), but unlike the seminal roots, coleoptile node roots elongate normally (Fig. 5B). Contrary to the coleoptile node root phenotype, the number of emerged lateral roots from the seminal root is increased in Bdtar2lhypo (Fig. 5C). This increase is also evident once lateral root number is normalized for total root length (Fig. 5D), even if the total number of lateral roots is small. Because it was difficult to follow this phenotype over a longer period in the tissue culture system (due to the limited growth space on our 20 cm dishes) [30], we employed an alternative assay, i.e. lateral root emergence that has been triggered by removal of the seminal root meristem. In this assay, Bdtar2lhypo mutants showed enhanced lateral root formation capacity (Fig. 5E), again also holding up once normalized for seminal root length (Fig. 5F). Again, this phenotype could be copied by L-kynerunine treatment of wild type (Fig. S1G). Considering that increased Bdtar2lhypo root length can be largely explained by cell elongation, it therefore appears that Bdtar2lhypo mutants have a genuinely higher capacity of seminal root branching.
Fig. 5. Root branching phenotypes of Bdtar2lhypo mutants. 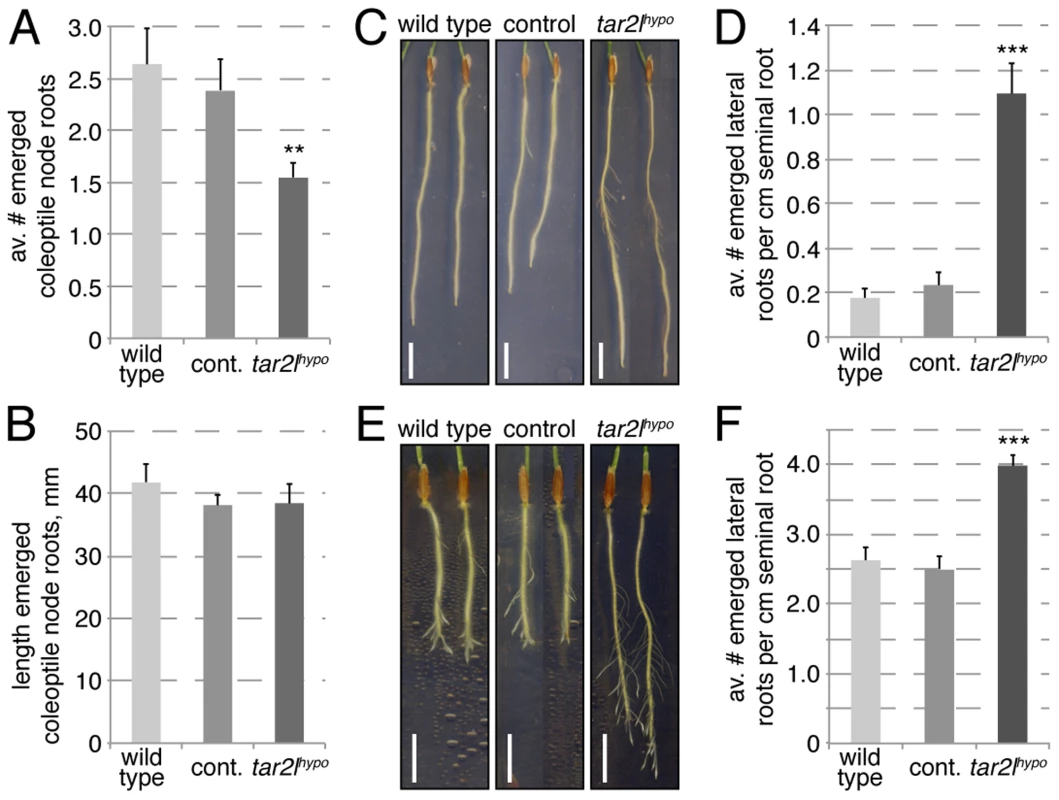
(A) Coleoptile node root formation in 25-day-old plants. (B) Quantification of coleoptile node root elongation. (C) Representative images of 8-day-old seminal roots, note emerged lateral roots. (D) Quantification of emerged lateral root number at 8 dag, normalized for seminal root length. (E) Representative images of 8-day-old seminal roots, 4 days after removal of the root tip. Note emerged lateral roots. (F) Quantification of emerged lateral root number after seminal root tip removal, normalized for seminal root length (per cm). Size bars are 1 cm; differences as compared to wild type are not significant unless indicated otherwise; error bars indicate standard error; ** = p<0.01; *** = p<0.001. Auxin levels are elevated rather than reduced in Bdtar2l roots
Collectively, our genetic as well as pharmacological analyses suggest that reduced tryptophan aminotransferase activity in Brachypodium results in increased root cell elongation and anisotropy. This contrasts with gradually reduced root growth in Arabidopsis taa1/tar single and double mutants. As expected, in Arabidopsis this root growth reduction is accompanied by gradually decreased free auxin levels [17]. Thus, the most parsimonious explanation for the Bdtar2l phenotype is that auxin levels might normally be supra-optimal for cell elongation in Brachypodium, similar to what has been proposed for rice [25]. To our surprise then, we found that free auxin levels are elevated rather than reduced in Bdtar2l seminal roots (Fig. 6A; Fig. S1H), in particular in the elongating and mature parts where the expression of BdTAR2L is relatively high as compared to BdTAR1L (Fig. S1A). Consistently, elevated auxin levels where also observed upon L-kynerunine treatment (Fig. S1I). To determine whether this could arise from compensatory up-regulation of proposed alternative auxin biosynthesis pathways [31], we checked the expression of various homologs of corresponding rate-limiting enzyme genes in Bdtar2l roots, i.e. AMIDASE-LIKE 1-LIKE (BdAMI1L), NITRILASE 1-LIKE (BdNIT1L), ALDEHYDE OXIDASE 1-LIKE (BdAO1L) and BdAO2L. However, with the exception of a slight increase in BdAO1L expression, no significant upward changes were detected (Fig. 6B). By contrast, the expression of four YUCCA homologs, selected for the reported root-specific expression of their respective counterparts in rice [32], is significantly up-regulated in Bdtar2l roots (Fig. 6C; Fig. S1J), amounting to more than triple in combined transcript levels in Bdtar2lhypo (Fig. 6C) and one-and-a-half in Bdtar2lqnull, corresponding with the respective auxin levels. The increased BdYUCCA expression could account for the increased auxin levels, because it has been determined that YUCCA gene expression is rate-limiting for auxin biosynthesis via the IPA pathway [17], [33].
Fig. 6. Auxin homeostasis in Bdtar2lhypo roots and its relation to the ethylene pathway. 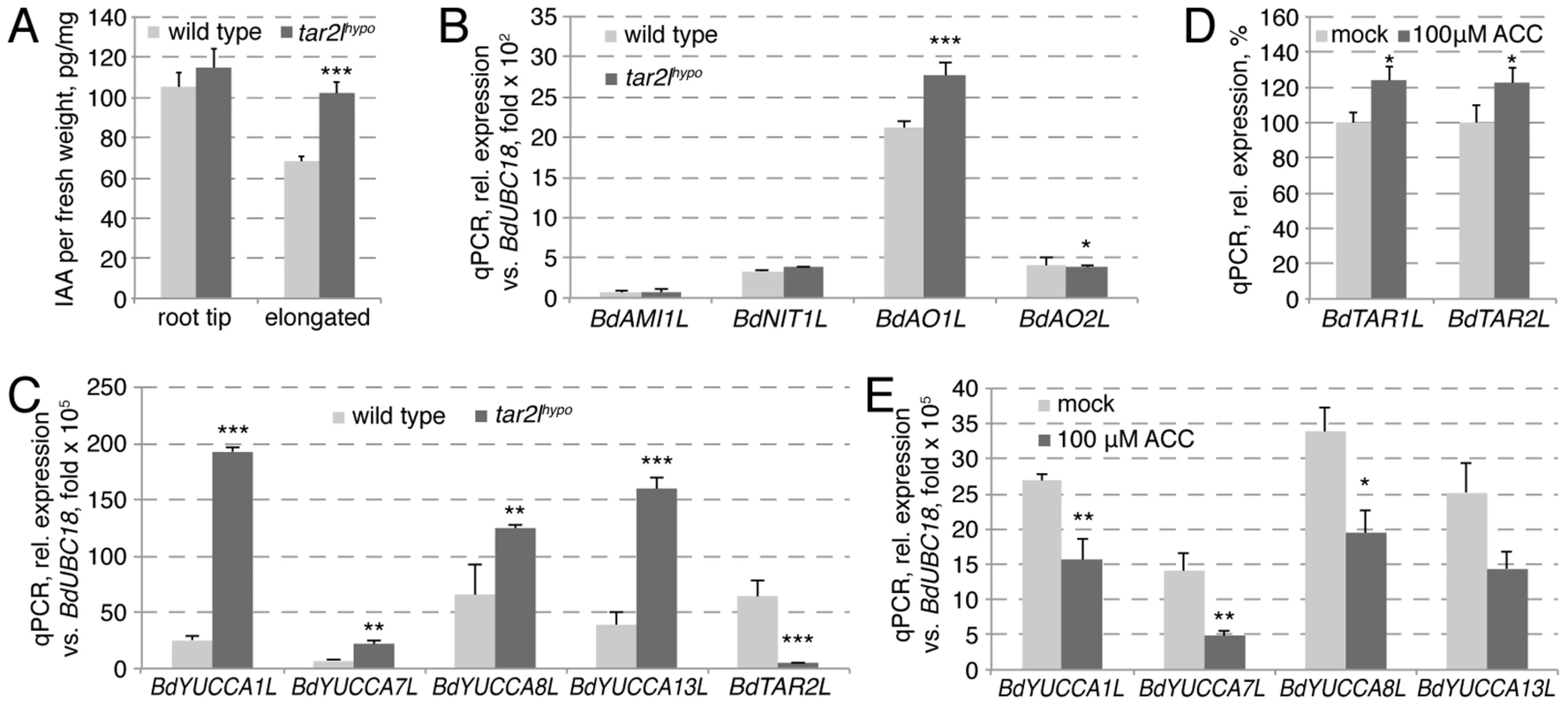
(A) Free auxin (IAA) content in wild type and Bdtar2lhypo root segments at 4 dag. The root tip comprised the terminal 8 mm of the roots, the elongated parts all above this. (B) Expression levels of the homologs of various genes encoding rate limiting enzymes in alternative auxin biosynthesis pathways in wild type and Bdtar2lhypo roots at 4 dag. (C) Expression levels of YUCCA homologs in wild type and Bdtar2lhypo roots at 4 dag. (D) Expression levels of BdTAR1L and BdTAR2L in wild type at 3 dag and after 3 h of ACC treatment. (E) Expression levels of YUCCA homologs in wild type at 3 dag and after 3 h of ACC treatment. All expression levels were determined by qPCR and normalized with respect to the housekeeping gene, BdUBC18; differences as compared to wild type or mock are not significant unless indicated otherwise; error bars indicate standard error; * = p<0.05; ** = p<0.01; *** = p<0.001. Bdtar2lhypo roots are restored to wild type by application of the ethylene precursor ACC
Root growth resistance to enhanced ethylene production, conferred by application of ACC, contributed to the isolation of the taa1/tar mutants in Arabidopsis, because ethylene promotes auxin biosynthesis via the IPA pathway through transcriptional regulation of TAA1/TAR and YUCCA genes [16], [34]. By contrast, we found that expression of BdTAR2L and BdTAR1L is only mildly ethylene-responsive (Fig. 6D). Moreover, the expression of the four BdYUCCA genes tested is negatively regulated by ACC application (Fig. 6E). Thus, in Brachypodium, the ethylene pathway might repress rather than promote auxin biosynthesis via the IPA pathway, mainly by down-regulating BdYUCCA expression. A prediction from this observation is that the Bdtar2l root phenotypes might be rescued by enhanced ethylene signaling. To test this notion, we transferred 2-day-old Bdtar2lhypo seedlings onto media containing increasing amounts of ACC and monitored root growth over the two days that followed. Indeed, ACC treatment strongly impaired Bdtar2lhypo root elongation and reduced growth to about the level of wild type mock controls (Fig. 7A–B). Moreover, ACC treatment restored cell elongation to wild type length in Bdtar2lhypo (Fig. 7E–F).
Fig. 7. Manipulation of the ethylene pathway and its impact on root growth. 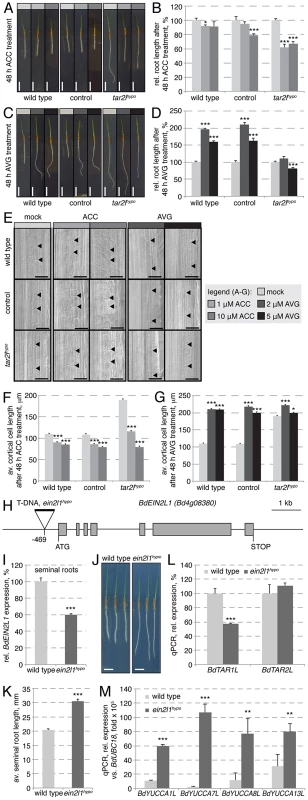
(A) Representative images of 4-day-old seedlings, transferred onto media with indicated ACC concentration at 2 dag. (B) Quantification of root length after 2 days of indicated ACC treatment. (C) Representative images of 4-day-old seedlings, transferred onto media with indicated AVG concentration at 2 dag. (D) Quantification of root length after 2 days of indicated AVG treatment. (E) Representative Nomarski optics images of mature root portions formed during indicated ACC or AVG treatment. Arrowheads point out top and bottom of individual cells in the 3rd cortex layer; (F–G) Quantification of mature cortex cell length after 2 days of indicated ACC or AVG treatment. (H) Schematic presentation of the BdEIN2L1 gene and the location of the T-DNA insertion in the Bdein2l1hypo mutant. (I) Relative expression level of BdEIN2L1 in the Bdein2l1hypo roots at 4 dag. (J) Representative seedlings of wild type and Bdein2l1hypo mutants at 4 dag. (K) Quantification of root length in wild type and Bdein2l1hypo mutants at 4 dag. (L) Expression levels of BdTAR1L and BdTAR2L in wild type and Bdein2l1hypo roots at 4 dag. (M) Expression levels of YUCCA homologs in wild type and Bdein2l1hypo roots at 4 dag. All expression levels were determined by qPCR and normalized with respect to the housekeeping gene, BdUBC18; size bars are 1 cm (A, C) or 100 µm (E); differences as compared to wild type or mock are not significant unless indicated otherwise; error bars indicate standard error; * = p<0.05; ** = p<0.01; *** = p<0.001. The Bdtar2lhypo root phenotype can be mimicked by reduced ethylene biosynthesis or signaling
The above results suggested that inhibition of ethylene biosynthesis or signaling in Brachypodium roots should mimic the Bdtar2lhypo root phenotype. We tested this notion by transferring 2-day-old seedlings onto media that contained aminoethoxyvinylglycine (AVG), an inhibitor of a rate-limiting enzyme in ethylene biosynthesis, ACC synthase [35]. Following root growth over the two days that followed revealed that Bdtar2lhypo roots are largely resistant to AVG, while wild type roots display a dramatic increase in elongation that approached the levels observed in Bdtar2lhypo (Fig. 7C–D). Investigation of cortical cells revealed that again this effect could be explained by increased cell elongation (Fig. 7E, G). Higher levels of AVG eventually slowed down elongation rate of Bdtar2lhypo roots, but still promoted root elongation in wild type. A cautionary note on AVG is that it not only inhibits ACC synthase, but also other enzymes that require pyridoxal 5′-phosphate (PLP) as a cofactor [36], [37]. Since the activity of TAA1/TAR enzymes is stimulated by PLP [16], it appears possible that AVG treatment impairs their function to some degree, mimicking L-kynerunine treatment. Thus, for independent confirmation we took advantage of a mutant from the Brachypodium T-DNA collection, in which a homolog of the Arabidopsis gene ETHYLENE INSENSITIVE 2 (EIN2), an essential positive regulator of ethylene signaling [38]–[40], carries a T-DNA insertion in the promoter, 469 bp upstream of the start codon (Fig. 7H). As a consequence, expression of this EIN2-LIKE (BdEIN2L1, Bd4g08380) gene is significantly down-regulated (Fig. 7I). Strikingly, this hypomorphic mutant (Bdein2l1hypo) displays a Bdtar2l root phenotype (Fig. 7J–K), and while this is not accompanied by up-regulation of BdTAR1L or BdTAR2L (Fig. 7L), it is accompanied by increased BdYUCCA expression (Fig. 7M). Finally, similar to Bdtar2l mutants, auxin levels are elevated in the elongating parts of Bdein2l1hypo roots (Fig. S1K), thereby corroborating our above findings.
Root elongation is only slightly stimulated by L-kynerunine or AVG treatment in Arabidopsis
The observed stimulatory effects of L-kynerunine and AVG treatment on root elongation have not been described for Arabidopsis. However, given the morphological differences between Arabidopsis and Brachypodium roots, in particular the more than three-fold difference in thickness, it is conceivable that the concentration of those substances required for root penetration and biological action might be different as well. The described largely inhibitory effect of those treatments on root elongation in Arabidopsis could therefore have resulted from application of too high concentrations. These considerations prompted us to revisit the response of Arabidopsis to an extended concentration range of both L-kynerunine and AVG. Interestingly, relatively low concentrations as compared to Brachypodium of both treatments indeed slightly promote root elongation (Fig. S1L–M), although by far not as strong as in Brachypodium.
Discussion
The root systems of dicotyledons and monocotyledons display some fundamental differences in their organization and ontogeny, as exemplified by the respective model systems, Arabidopsis and Brachypodium [30]. Despite these differences, the principal genes involved in root formation, growth vigor and branching are expected to be homologous in the two systems. This is based on experience in other species such as maize, where several causative mutations that affect root system development are in homologs of auxin signaling components [9], [41]. The effect of manipulating the IPA branch of auxin biosynthesis has been investigated in another monocotyledon crop, rice, through gain - and loss-of-function approaches. For instance, both over-expression and down-regulation of the YUCCA homolog OsYUCCA1 by transgenic means results in strongly reduced root growth [42], whereas a knockout in another YUCCA homolog, CONSTITUTIVELY WILTED 1, displays reduced root branching [43]. Compared to those mutants, the enhanced root elongation phenotype of Bdtar2l mutants is unusual. Our initial interpretation was therefore that auxin levels are supra-optimal for cell elongation in the Brachypodium seminal root, as has been suggested for seminal root growth in rice [25]. However, repeated independent measurements of multiple samples clearly indicated that auxin levels are increased rather than decreased in Bdtar2l mutant roots. This is particularly pronounced in the Bdtar2lhypo allele, and in tendency also observed in the Bdtar2lqnull allele, correlating with quantitatively corresponding BdYUCCA up-regulation. The comparatively severe shoot phenotypes of the Bdtar2lqnull allele, its less pronounced root cell elongation, and the observation that the root meristem gradually breaks down as development progresses indicate that compared to the Bdtar2lhypo allele, IPA levels are eventually limiting in Bdtar2lqnull mutants. This idea is supported by the dose-response curve of wild type to L-kynerunine, where increasing amounts promote cell elongation up to a certain threshold, beyond which root growth is inhibited. Further corroborating this idea, a threshold also exists for the Bdtar2lhypo mutant, which moreover is hypersensitive to L-kynerunine treatment as concentrations that still promote root elongation in wild type are inhibitory in Bdtar2lhypo.
A similar dose-response curve is observed for AVG treatment, which inhibits the rate-limiting step in ethylene biosynthesis, but might also impinge on TAA1/TAR activity because of its generic action on enzymes that use PLP as a co-factor [16], [36], [37]. Stimulation of root growth by AVG treatment has also been reported for rice [25], although the reported dosage response is quantitatively different from our assays with Brachypodium. For instance, while in rice 0.05 µM AVG promoted root growth and 1.0 µM was already inhibitory, in Brachypodium 5.0 µM was still stimulating. In part, this could be due to technical issues, for instance the concentration needed in the tissue culture media to reach the same tissue penetration in roots of different thickness or cell permeability. In light of our results, it appears possible that the response of rice to AVG treatment is similar to Brachypodium, i.e. that it could reflect a combined effect of reducing TAA1/TAR as well as ACC synthase activity, thereby boosting auxin levels by removing the inhibitory effect of ethylene on YUCCA expression as long as interference with TAA/TAR1 activity does not lead to limiting IPA levels. The finding that YUCCA expression is rate limiting for auxin biosynthesis in Arabidopsis [17], [33] supports this interpretation, suggesting that this is also likely the case in Brachypodium and/or rice.
Corroborating the effects of AVG application and circumventing its ambiguity, the root phenotype of the Bdein2l1hypo mutant confirms the involvement of the ethylene-signaling pathway in auxin homeostasis. However, based on the observed regulatory logic of this hormone crosstalk, a central finding of our study is that the regulation of the IPA branch of auxin biosynthesis through the ethylene pathway observed in Arabidopsis roots might not be conserved in Brachypodium. This idea is based on several convergent observations, for instance that unlike their Arabidopsis counterparts, expression of BdTAR2L as well as BdTAR1L is hardly ethylene-responsive or that BdYUCCAs are repressed upon ACC treatment and up-regulated in Bdein2l1hypo, consistent with the latter's Bdtar2l phenotype. Moreover, unlike Arabidopsis taa1/tar mutants, Bdtar2lhypo mutants are not ACC-resistant. Rather, ACC treatment essentially restores the Bdtar2lhypo phenotype to wild type. Thus, our data thus support a scenario in which the effects of auxin biosynthesis through the IPA branch on root cell elongation are mediated by the ethylene pathway rather than vice versa. Such an inversion of a regulatory relationship could alternatively reflect a shift in the key nodes of the regulatory network linking auxin and ethylene through feedback loops and is a simple way for evolutionary adaptation. Indeed, feedback of ethylene on auxin biosynthesis by repressing YUCCA expression, rather than promoting TAA1/TAR as well as YUCCA expression as in Arabidopsis, is a central feature of the mutant phenotypes described in our paper (Fig. 8). How this feedback is mediated remains unclear for the moment. The recent discovery of an enzymatic link between auxin and ethylene biosynthesis suggests that this crosstalk might very well respond directly to IPA levels [44]. Hypomorphic mutants, such as those employed in our study, might become a crucial tool in future efforts to elaborate such a scenario.
Fig. 8. A schematic overview of the regulation of tryptophan-dependent auxin (indole-3-acetic acid) biosynthesis via indole-3-pyruvic acid (IPA) by ethylene action in Arabidopsis (<i>A</i>) and Brachypodium (<i>B</i>). 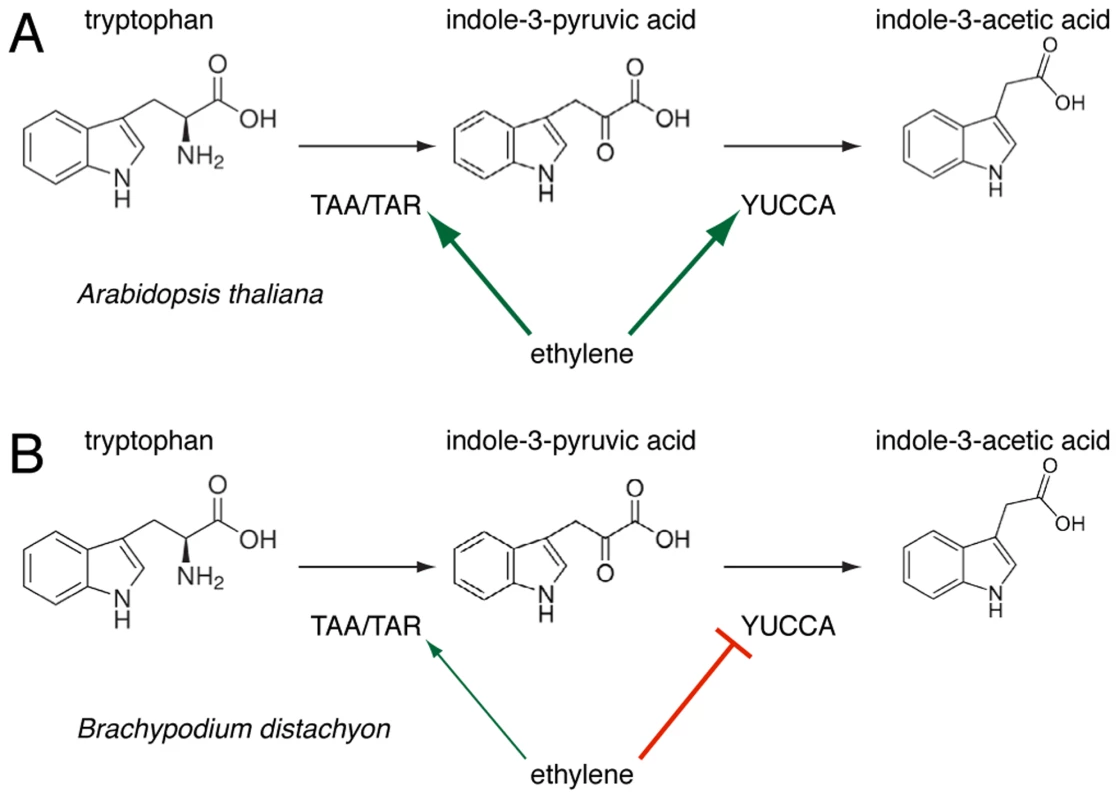
Materials and Methods
Molecular biology and genetics procedures, such as genomic DNA isolation, genotyping, sequencing or qPCR were performed according to standard procedures as described [45], [46].
Plant materials and growth conditions
The community standard diploid inbreed Brachypodium distachyon line Bd21 was used for transformation and as a control in all experiments [47], except for the Bdtar2lqnull mutant (stock id JJ9248.0) and the Bdein2l1hypo mutant (stock id JJ110.0), which together with their Bd21-3 wild type background line were obtained from a Brachypodium T-DNA collection (http://brachypodium.pw.usda.gov/TDNA/) [28]. Genotyping, for instance to establish homozygous mutant lines, was performed using oligonucleotides 5′-CGT GAG AGC TAG TGG GAT AG-3′ and 5′-ATG GGT GGC TGA TGG CGT AG-3′ (BdTAR2L wild type allele for Bdtar2lhypo), 5′-CGT GAG AGC TAG TGG GAT AG-3′ and 5′-TTG AAG GAG CCA CTC AGC CGC G-3′ (Bdtar2lhypo T-DNA insertion); 5′-GCG GTT CCC TGT TCA TCT TC-3′ and 5′-CAC AGC GAA ACA ACA CAC AG-3′ (BdTAR2L wild type allele control for Bdtar2lqnull), 5′-GCG GTT CCC TGT TCA TCT TC-3′ and 5′-TAC GAG CCG GAA GCA TA AAG-3′ (Bdtar2lqnull T-DNA insertion); 5′-GTA CCT TTC TCC GTC AAG AG-3′ and 5′-GAA GGA GGC ATC AGG ACA TG-3′ (BdEIN2L1 wild type allele), 5′ - GTA CCT TTC TCC GTC AAG AG -3′ and 5′-CTC CGC TCA TGA TCA GAT TG-3′ (Bdein2l1hypo T-DNA insertion); Arabidopsis thaliana experiments were performed with the standard Col-0 accession. For tissue culture growth, the lemma of mature seeds was carefully peeled off with forceps before seed sterilization in 1 ml of 70% ethanol per seed for 1 min. After ethanol removal, seeds were soaked in a solution of 1.3% sodium hypochlorite plus one drop of Tween-20 per 50 ml for 5 min. with gently rocking, then rinsed with sterile deionized water three times. The sterilized seeds were stratified for 2 days at 4°C to ensure synchronous germination on vertically oriented 10 or 24 cm square plates of half-strength Murashige-Skoog (MS) media (2.45 g/l MS salts with vitamins, 1% sucrose, 1% agar, pH 5.7) in a growth chamber under continuous light of 100–120 µE intensity at 22°C. To quantify leaf number, sheath/blade length and blade width, 2-day-old Brachypodium seedlings were transferred into pots with soil, watered every 2–3 days and incubated at 22°C under a 20 h photoperiod. Leaf features were measured 18 days after germination (dag), crown roots were counted 25 dag. Arabidopsis seedlings were grown as described [46].
Seminal root length and lateral root number quantification, and root gravitropism assays
To determine root length, seedlings growing in vertically oriented plates were either scanned or photographed with a digital camera to measure root length using the ImageJ software, version 1.47b. For lateral root quantification after seminal root meristem removal, 2 mm of the root tip were cut from the seminal root of 4-day-old plants with a scalpel. The number of visible lateral roots was then scored 4 days later. For gravitropism assays, Brachypodium seeds were germinated for 2 days in vertically oriented plates. To induce gravitropic response, plates were then rotated 90° and grown for another 24 hours. Plates were scanned on a flatbed scanner before and after gravitropic stimulation.
Transformation of Bd21 and FST-retrieval
Embryonic calli generation of Bd21 was performed according to [48], subsequent transformation with the pVec8GFP plasmid and plant regeneration according to [47], and retrieval and mapping of the region flanking the right border of the T-DNA insert in Bdtar2lhypo mutants according to [27]. A total of 48 transgenic lines were produced, among which the Bdtarlhypo mutant was a chance hit.
Whole genome sequencing and T-DNA insertion mapping
Whole genome sequencing of genomic DNA isolated from Brachypodium seedlings was performed on the Illumina HiSeq 2000 platform, generating more than 250 mio. paired-end reads of 100 bp length. The Bowtie 2 software [49] was used for the alignment on the Brachypodium distachyon reference genome (http://mips.helmholtz-muenchen.de/plant/brachypodium/download/index.jsp), revealing coverage of ca. 100 reads per bp. For detection of T-DNA insertions, reads that aligned on the T-DNA reference sequence were selected for alignment on the genome. This procedure confirmed the localization of the Bdtar2lhypo insert on chromosome 2 (position: 3,030,511). The precise position of the control line insert remains undetermined because it could not be mapped to a unique annotated region, however it is clear that it does not disrupt any annotated gene. Finally, coverage of the T-DNA reference sequence was similar to genome coverage, confirming the presence of a single insertion in both sequenced genomes.
Hormone and inhibitor treatments
The hormone and inhibitor treatments were done on plates, except in the case of qPCR, for which treatments were carried out in liquid media for 3 h. Briefly, Brachypodium seeds were germinated on standard plates as described above. At 2 days after germination, seedlings were then transferred to media containing the respective hormone or inhibitor, or mock. For Arabidopsis treatments, 4-day-old seedlings were transferred.
Auxin measurements
Auxin measurements were performed on eight independent samples of pooled roots per genotype excised from 4-day-old seedlings as described [50].
Microscopy
Seminal roots of 4-day-old seedlings were fixed in a solution of 1% glutaraldehyde, 4% formaldehyde and 50 mM sodium phosphate buffer (pH 7.2). Fixed roots were thoroughly rinsed four times with water. To determine transverse root and cell area, roots were cut into 0.5–1 cm pieces and embedded in 6% agarose. Sections of 75 µm were obtained approximately 2 cm from the root tip using a Leica-VT 1000S vibratome. Sections were stained with 0.1% toluidine blue solution for 30 s and washed. For quantification of cortical cell length, unstained roots were cleared with 10% potassium hydroxide solution at 95°C for 30 min. Roots were mounted on glass slides with 50% glycerol and photographed either in light field or differential interference contrast using a Leica DM5500B compound microscope. For visualization of meristem structure, seminal roots were stained following the mPS-PI procedure [45] before imaging with a Zeiss LSM 700 confocal microscope. Cortical cell length, root hair length, meristem size and central metaxylem cell length were quantified using the ImageJ software, version 1.47b.
qPCR and oligonucleotides
qPCR reactions were performed using a Stratagene MxPro 3005P Real-Time PCR System (Stratagene). Three technical replicates were analyzed for each sample. The specificity of each amplification reaction was verified by DNA melting curve analysis and gel electrophoresis of the amplified products. Not reverse transcribed samples and non-template controls were included in every assay to rule out genomic DNA contamination. The final threshold cycle (Ct), efficiency and initial fluorescence (R0) for every reaction were calculated with the Miner algorithm [51]. Relative expression levels were obtained from the ratio between R0 of the target gene and R0 of the reference gene, UBIQUITIN-CONJUGATING ENZYME 18 (BdUBC18). The following oligonucleotides were used: BdUBC18 (Bd4G00660), 5′-GGA GGC ACC TCA GGT CAT TT-3′ and 5′-ATA GCG GTC ATT GTC TTG CG-3′; BdTAR1L (Bd2G34400), 5′-GAA TCG GGA TGG TGG CCT CG-3′ and 5′-ATT GTC GGA TCG CCG TGA TC-3′; BdTAR2L (Bd2G04290), 5′-GGC TCC ATA CTA CTC TTC GTA TC-3′ and 5′-CAG TAG TAG GCC AGG TCG TG-3′; BdYUCCA1L (Bd1G28967), 5′-GCA ATG GCT CAA GGG AAG TG-3′ and 5′-TGT GGC AGT TTG ATG CTT CC-3′; BdYUCCA7L (Bd1G00587), 5′-GCA GTG GCT CAA GGG AAG C-3′ and 5′-TGT GGT ATG CTG TGG CGA TG-3′; BdYUCCA8L (Bd5G01327), 5′-CCC AGT TCA TCT CCT ACC TC-3′ and 5′-GGT ACT CGA CGG TGG ACT TC-3′; BdYUCCA13L (Bd2G10302), 5′-GTC GTC CGC AGC GAG CTT CA-3′ and 5′-GGG GGT TTG GAG CTT CAT GG-3′; BdAMI1L (Bd5G27490), 5′-CGA CTT CTC CCT CGG AAC TG-3′ and 5′-GTT GCT GAC GCG AGA CAA TG-3′; BdNIT1L (Bd3G49620), 5′-CCC CTG CCA CCA TTG ATA AAG-3′ and 5′-GTC TTC TTT TCC CTT GGC AG-3′; BdAO1L (Bd1G52740), 5′-GGC TGT GGC GAA GGT GGA TG-3′ and 5′-ACC CTC AGT GGT GAT AAC TG-3′; BdAO2L (Bd1G56667), 5′-GTG GAC CCA GTG CAA ATG TG-3′ and 5′-CAT ATA CAG CCT CCC CAG AAG-3′; BdEIN2L1 (Bd4G08380), 5′-AGA ATC TTG CCC AGA TTT GC-3′ and 5′-GCA AAC CAT ATG CCT GTG AG-3′;
Supporting Information
Zdroje
1. LynchJ (1995) Root Architecture and Plant Productivity. Plant Physiol 109 : 7–13.
2. OsmontKS, SiboutR, HardtkeCS (2007) Hidden branches: developments in root system architecture. Annu Rev Plant Biol 58 : 93–113.
3. FitterA, WilliamsonL, LinkohrB, LeyserO (2002) Root system architecture determines fitness in an Arabidopsis mutant in competition for immobile phosphate ions but not for nitrate ions. Proc Biol Sci 269 : 2017–2022.
4. LinkohrBI, WilliamsonLC, FitterAH, LeyserHM (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29 : 751–760.
5. AikenRM, SmuckerAJ (1996) Root system regulation of whole plant growth. Annu Rev Phytopathol 34 : 325–346.
6. de DorlodotS, ForsterB, PagesL, PriceA, TuberosaR, et al. (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 12 : 474–481.
7. BenfeyPN, BennettM, SchiefelbeinJ (2010) Getting to the root of plant biology: impact of the Arabidopsis genome sequence on root research. Plant J 61 : 992–1000.
8. HochholdingerF, TuberosaR (2009) Genetic and genomic dissection of maize root development and architecture. Curr Opin Plant Biol 12 : 172–177.
9. HochholdingerF, ZimmermannR (2008) Conserved and diverse mechanisms in root development. Curr Opin Plant Biol 11 : 70–74.
10. InukaiY, SakamotoT, Ueguchi-TanakaM, ShibataY, GomiK, et al. (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17 : 1387–1396.
11. LiuH, WangS, YuX, YuJ, HeX, et al. (2005) ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J 43 : 47–56.
12. HardtkeCS, BerlethT (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. Embo J 17 : 1405–1411.
13. HardtkeCS, CkurshumovaW, VidaurreDP, SinghSA, StamatiouG, et al. (2004) Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131 : 1089–1100.
14. DharmasiriN, DharmasiriS, WeijersD, LechnerE, YamadaM, et al. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9 : 109–119.
15. WonC, ShenX, MashiguchiK, ZhengZ, DaiX, et al. (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci U S A 108 : 18518–18523.
16. StepanovaAN, Robertson-HoytJ, YunJ, BenaventeLM, XieDY, et al. (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 : 177–191.
17. StepanovaAN, YunJ, RoblesLM, NovakO, HeW, et al. (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23 : 3961–3973.
18. ZhaoY, HullAK, GuptaNR, GossKA, AlonsoJ, et al. (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16 : 3100–3112.
19. ZhaoY, ChristensenSK, FankhauserC, CashmanJR, CohenJD, et al. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 : 306–309.
20. TaoY, FerrerJL, LjungK, PojerF, HongF, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133 : 164–176.
21. YamadaM, GreenhamK, PriggeMJ, JensenPJ, EstelleM (2009) The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 151 : 168–179.
22. GarrettJJ, MeentsMJ, BlackshawMT, BlackshawLC, HouH, et al. (2012) A novel, semi-dominant allele of MONOPTEROS provides insight into leaf initiation and vein pattern formation. Planta 236 : 297–312.
23. KroganNT, CkurshumovaW, MarcosD, CarageaAE, BerlethT (2012) Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol 194 : 391–401.
24. SiboutR, SukumarP, HettiarachchiC, HolmM, MudayGK, et al. (2006) Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet 2: e202.
25. YinC, WuQ, ZengH, XiaK, XuJ, et al. (2011) Endogenous Auxin is Required but Supraoptimal for Rapid Growth of Rice (Oryza sativa L.) Seminal Roots, and Auxin Inhibition of Rice Seminal Root Growth is Not Caused by Ethylene. Journal of Plant Growth Regulation 30 : 20–29.
26. PhillipsKA, SkirpanAL, LiuX, ChristensenA, SlewinskiTL, et al. (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23 : 550–566.
27. TholeV, AlvesSC, WorlandB, BevanMW, VainP (2009) A protocol for efficiently retrieving and characterizing flanking sequence tags (FSTs) in Brachypodium distachyon T-DNA insertional mutants. Nat Protoc 4 : 650–661.
28. BraggJN, WuJ, GordonSP, GuttmanME, ThilmonyR, et al. (2012) Generation and characterization of the Western Regional Research Center Brachypodium T-DNA insertional mutant collection. PLoS One 7: e41916.
29. HeW, BrumosJ, LiH, JiY, KeM, et al. (2011) A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23 : 3944–3960.
30. Pacheco-VillalobosD, HardtkeCS (2012) Natural genetic variation of root system architecture from Arabidopsis to Brachypodium: towards adaptive value. Philos Trans R Soc Lond B Biol Sci 367 : 1552–1558.
31. ManoY, NemotoK (2012) The pathway of auxin biosynthesis in plants. J Exp Bot 63 : 2853–2872.
32. TakehisaH, SatoY, IgarashiM, AbikoT, AntonioBA, et al. (2012) Genome-wide transcriptome dissection of the rice root system: implications for developmental and physiological functions. Plant J 69 : 126–140.
33. MashiguchiK, TanakaK, SakaiT, SugawaraS, KawaideH, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci U S A 108 : 18512–18517.
34. LiangX, WangH, MaoL, HuY, DongT, et al. (2012) Involvement of COP1 in ethylene - and light-regulated hypocotyl elongation. Planta 236 : 1791–1802.
35. AdamsDO, YangSF (1979) Ethylene biosynthesis: Identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc Natl Acad Sci U S A 76 : 170–174.
36. ClausenT, HuberR, MesserschmidtA, PohlenzHD, LaberB (1997) Slow-binding inhibition of Escherichia coli cystathionine beta-lyase by L-aminoethoxyvinylglycine: a kinetic and X-ray study. Biochemistry 36 : 12633–12643.
37. KrupkaHI, HuberR, HoltSC, ClausenT (2000) Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5′-phosphate-dependent protein acting as a haemolytic enzyme. Embo J 19 : 3168–3178.
38. AlonsoJM, HirayamaT, RomanG, NourizadehS, EckerJR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 : 2148–2152.
39. JuC, YoonGM, ShemanskyJM, LinDY, YingZI, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci U S A 109 : 19486–19491.
40. QiaoH, ShenZ, HuangSS, SchmitzRJ, UrichMA, et al. (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338 : 390–393.
41. HochholdingerF, ParkWJ, SauerM, WollK (2004) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9 : 42–48.
42. YamamotoY, KamiyaN, MorinakaY, MatsuokaM, SazukaT (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143 : 1362–1371.
43. WooYM, ParkHJ, Su'udiM, YangJI, ParkJJ, et al. (2007) Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol 65 : 125–136.
44. ZhengZ, GuoY, NovakO, DaiX, ZhaoY, et al. (2013) Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat Chem Biol 9 : 244–246.
45. ScacchiE, SalinasP, GujasB, SantuariL, KroganN, et al. (2010) Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proc Natl Acad Sci U S A 107 : 22734–22739.
46. SiboutR, PlantegenetS, HardtkeCS (2008) Flowering as a condition for xylem expansion in Arabidopsis hypocotyl and root. Curr Biol 18 : 458–463.
47. VogelJ, HillT (2008) High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep 27 : 471–478.
48. AlvesSC, WorlandB, TholeV, SnapeJW, BevanMW, et al. (2009) A protocol for Agrobacterium-mediated transformation of Brachypodium distachyon community standard line Bd21. Nat Protoc 4 : 638–649.
49. LangmeadB, SalzbergSL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9 : 357–359.
50. AndersenSU, BuechelS, ZhaoZ, LjungK, NovakO, et al. (2008) Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20 : 88–100.
51. ZhaoS, FernaldRD (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol 12 : 1047–1064.
Štítky
Genetika Reprodukční medicína
Článek PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant TransformationČlánek The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene ClusterČlánek Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish PathogenČlánek USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and TranscriptionČlánek Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA EditingČlánek Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 6
-
Všechny články tohoto čísla
- BMS1 Is Mutated in Aplasia Cutis Congenita
- High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants
- How Cool Is That: An Interview with Caroline Dean
- Genetic Architecture of Vitamin B and Folate Levels Uncovered Applying Deeply Sequenced Large Datasets
- Juvenile Hormone and Insulin Regulate Trehalose Homeostasis in the Red Flour Beetle,
- Meiosis-Specific Stable Binding of Augmin to Acentrosomal Spindle Poles Promotes Biased Microtubule Assembly in Oocytes
- Environmental Dependence of Genetic Constraint
- H3.3-H4 Tetramer Splitting Events Feature Cell-Type Specific Enhancers
- Network Topologies and Convergent Aetiologies Arising from Deletions and Duplications Observed in Individuals with Autism
- Effectively Identifying eQTLs from Multiple Tissues by Combining Mixed Model and Meta-analytic Approaches
- Altered Splicing of the BIN1 Muscle-Specific Exon in Humans and Dogs with Highly Progressive Centronuclear Myopathy
- The NADPH Metabolic Network Regulates Human Cardiomyopathy and Reductive Stress in
- Negative Regulation of Notch Signaling by Xylose
- A Genome-Wide, Fine-Scale Map of Natural Pigmentation Variation in
- Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals mC within Archaeal mRNAs
- Multiplexin Promotes Heart but Not Aorta Morphogenesis by Polarized Enhancement of Slit/Robo Activity at the Heart Lumen
- Latent Effects of Hsp90 Mutants Revealed at Reduced Expression Levels
- Impact of Natural Genetic Variation on Gene Expression Dynamics
- DeepSAGE Reveals Genetic Variants Associated with Alternative Polyadenylation and Expression of Coding and Non-coding Transcripts
- The Identification of -acting Factors That Regulate the Expression of via the Osteoarthritis Susceptibility SNP rs143383
- Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs
- The RNA Export Factor, Nxt1, Is Required for Tissue Specific Transcriptional Regulation
- Inferring Demographic History from a Spectrum of Shared Haplotype Lengths
- Histone Acetyl Transferase 1 Is Essential for Mammalian Development, Genome Stability, and the Processing of Newly Synthesized Histones H3 and H4
- PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant Transformation
- DNA Methylation Restricts Lineage-specific Functions of Transcription Factor Gata4 during Embryonic Stem Cell Differentiation
- The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Deregulation of the Protocadherin Gene Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy
- Evidence for Two Different Regulatory Mechanisms Linking Replication and Segregation of Chromosome II
- USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and Transcription
- Methylation of Histone H3 on Lysine 79 Associates with a Group of Replication Origins and Helps Limit DNA Replication Once per Cell Cycle
- A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue
- The Gene Desert Mammary Carcinoma Susceptibility Locus Regulates Modifying Mammary Epithelial Cell Differentiation and Proliferation
- Hooked and Cooked: A Fish Killer Genome Exposed
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
- Mutations in Cause Autosomal Recessive Congenital Ichthyosis in Humans
- Integrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation
- RSR-2, the Ortholog of Human Spliceosomal Component SRm300/SRRM2, Regulates Development by Influencing the Transcriptional Machinery
- Comparative Polygenic Analysis of Maximal Ethanol Accumulation Capacity and Tolerance to High Ethanol Levels of Cell Proliferation in Yeast
- SPO11-Independent DNA Repair Foci and Their Role in Meiotic Silencing
- Budding Yeast ATM/ATR Control Meiotic Double-Strand Break (DSB) Levels by Down-Regulating Rec114, an Essential Component of the DSB-machinery
- Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA Editing
- Functional Analysis of Neuronal MicroRNAs in Dauer Formation by Combinational Genetics and Neuronal miRISC Immunoprecipitation
- DNA Ligase IV Supports Imprecise End Joining Independently of Its Catalytic Activity
- Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
- Heritable Change Caused by Transient Transcription Errors
- From Many, One: Genetic Control of Prolificacy during Maize Domestication
- Neuronal Target Identification Requires AHA-1-Mediated Fine-Tuning of Wnt Signaling in
- Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish
- H-NS Can Facilitate Specific DNA-binding by RNA Polymerase in AT-rich Gene Regulatory Regions
- Prophage Dynamics and Contributions to Pathogenic Traits
- Global DNA Hypermethylation in Down Syndrome Placenta
- Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in
- Disturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-ethylene Crosstalk in Seminal Roots
- Causes and Consequences of Chromatin Variation between Inbred Mice
- Genome-scale Analysis of FNR Reveals Complex Features of Transcription Factor Binding
- Distinct and Atypical Intrinsic and Extrinsic Cell Death Pathways between Photoreceptor Cell Types upon Specific Ablation of in Cone Photoreceptors
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- BMS1 Is Mutated in Aplasia Cutis Congenita
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



