-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaIntegrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation
Elucidation of the epigenetic basis for cell-type specific gene regulation is key to gaining a full understanding of how the distinct phenotypes of differentiated cells are achieved and maintained. Here we examined how epigenetic changes are integrated with transcriptional activation to determine cell phenotype during differentiation. We performed epigenomic profiling in conjunction with transcriptomic profiling using in vitro differentiation of human primary alveolar epithelial cells (AEC). This model recapitulates an in vivo process in which AEC transition from one differentiated cell type to another during regeneration following lung injury. Interrogation of histone marks over time revealed enrichment of specific transcription factor binding motifs within regions of changing chromatin structure. Cross-referencing of these motifs with pathways showing transcriptional changes revealed known regulatory pathways of distal alveolar differentiation, such as the WNT and transforming growth factor beta (TGFB) pathways, and putative novel regulators of adult AEC differentiation including hepatocyte nuclear factor 4 alpha (HNF4A), and the retinoid X receptor (RXR) signaling pathways. Inhibition of the RXR pathway confirmed its functional relevance for alveolar differentiation. Our incorporation of epigenetic data allowed specific identification of transcription factors that are potential direct upstream regulators of the differentiation process, demonstrating the power of this approach. Integration of epigenomic data with transcriptomic profiling has broad application for the identification of regulatory pathways in other models of differentiation.
Published in the journal: . PLoS Genet 9(6): e32767. doi:10.1371/journal.pgen.1003513
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003513Summary
Elucidation of the epigenetic basis for cell-type specific gene regulation is key to gaining a full understanding of how the distinct phenotypes of differentiated cells are achieved and maintained. Here we examined how epigenetic changes are integrated with transcriptional activation to determine cell phenotype during differentiation. We performed epigenomic profiling in conjunction with transcriptomic profiling using in vitro differentiation of human primary alveolar epithelial cells (AEC). This model recapitulates an in vivo process in which AEC transition from one differentiated cell type to another during regeneration following lung injury. Interrogation of histone marks over time revealed enrichment of specific transcription factor binding motifs within regions of changing chromatin structure. Cross-referencing of these motifs with pathways showing transcriptional changes revealed known regulatory pathways of distal alveolar differentiation, such as the WNT and transforming growth factor beta (TGFB) pathways, and putative novel regulators of adult AEC differentiation including hepatocyte nuclear factor 4 alpha (HNF4A), and the retinoid X receptor (RXR) signaling pathways. Inhibition of the RXR pathway confirmed its functional relevance for alveolar differentiation. Our incorporation of epigenetic data allowed specific identification of transcription factors that are potential direct upstream regulators of the differentiation process, demonstrating the power of this approach. Integration of epigenomic data with transcriptomic profiling has broad application for the identification of regulatory pathways in other models of differentiation.
Introduction
Over the past two decades the relationship between gene expression and chromatin structure has been increasingly recognized [1]–[4]. Elucidation of the histone code and subsequent insights into the functional implications of post-translational modifications of histone tails have begun to provide a mechanistic understanding of the role that chromatin context plays in gene expression. One of the most widely studied histone marks of active gene transcription is acetylation of lysine residues in the N-terminal tail of histone H3. Acetylation of Lysines 9 and 14 (H3K9/14Ac), found at promoters and enhancers of actively transcribed genes [5]–[7], serves as a docking point for chromatin remodeling complexes that open chromatin, facilitating transcriptional activation [8]–[10]. In contrast, trimethylation of lysine 27 of histone H3 (H3K27me3) confers repression through binding of the polycomb repressive complex (PRC1/2) and chromatin compaction [11]–[13]. The H3K9/14Ac and H3K27me3 marks usually occur in distinct cell-type specific genomic regions. Many studies have examined the differentiation of stem cells into a variety of differentiated cell types, in processes that traverse large phenotypic (and presumably epigenetic) distances [14]. However, using stem cells to dissect the mechanism(s) by which the differentiated epigenotype is reached may be challenging due to the distant relationship between the starting and resulting cell populations. In contrast, examining epigenetic differences between two closely related yet phenotypically distinct cell types might offer more straightforward insights into the relationship between epigenetic changes and the establishment of new expression patterns. Isolated distal lung epithelial cells offer a compelling model system; primary human alveolar epithelial type 2 (AT2) cells can be purified in large numbers from remnant transplant lung, and can be differentiated in vitro in a manner closely mimicking both normal maintenance and regeneration following lung injury [15], [16].
The distal lung alveolar epithelium consists of two major cell types: cuboidal surfactant-producing AT2 cells and elongated type 1 (AT1) cells that facilitate gas exchange. AT2 cells are the implicated precursors of AT1 cells, and through differentiation restore function of damaged distal lung epithelium [17], [18]. This phenotypic transition can be recapitulated in vitro when purified primary AT2 cells are plated under defined cell culture conditions [19], [20]. Previous studies using purified rat AT2 cells showed that AT2 cell-specific markers, such as surfactant protein C and A (SFTPC, SFTPA1), decrease with time in culture, while AT1 cell markers, such as aquaporin 5 (AQP5 [21], [22]), caveolin 1 (CAV1, [23], [24]) and podoplanin (PDPN, [25], [26]) increase over time [27]. These expression changes are accompanied by profound changes in cellular morphology and function [28]. Recent advances have allowed for the isolation of human AT2 cells from remnant human transplant lungs [15], [29].
Differentiation of purified primary human AT2 cells into AT1-like cells represents a unique kinetic model system to study the process of epithelial cell differentiation without the caveats of cell line immortalization or mixed tissue analysis. Here, we simultaneously profiled the transcriptomic and epigenomic changes of differentiating human alveolar epithelial cells (AEC). Detailed integrated analysis facilitated identification of regulatory networks and participating transcription factors, pointing to roles for known and novel signaling pathways in distal lung epithelial differentiation. To our knowledge this is the first integrated analysis of human primary epithelial cell differentiation.
Results
Transcriptomic profiling of human alveolar epithelial type 2 cells during differentiation
AT2 cells were isolated from the lungs of three non-smoker donors (Figure S1A) and plated on collagen-coated polytetrafluoroethylene membranes. Purity was verified by immunostaining for AT2 cell-specific markers pro-SFTPC and transcription factor NK2 homeobox 1 (NKX2-1), as well as hematopoietic marker protein tyrosine phosphatase, receptor type, C (PTPRC, previously CD45) and mesenchymal marker vimentin (VIM) to check for contaminating cell types. Pro-SFTPC-positive cells averaged 86% purity (Figure S1A–1B). Over the course of 8 days the cells underwent differentiation into AT1-like cells, forming confluent monolayers with tight junctions expressing tight junction protein 1 (TJP1, previously ZO-1) and showing downregulation of SFTPC, upregulation of AQP5, and establishment of transepithelial resistance [27], [30], [31] (Figure S1C–1E).
Raw expression data across all samples had similar distributions (Figure S2), GEO accession #GSE38571. Thus, all samples, including a technical duplicate each of day 0 (D0) and day 4 (D4), were included in normalization (Figure S3). The relationship between gene expression profiles was examined by unsupervised hierarchical clustering using the top 5% of genes most variant across the dataset. Samples from different lungs clustered together based largely on the timing of differentiation, with the D0 and D2 samples each grouping together (Figure 1A). Importantly, the sample dendrogram indicates the major branch point is between D2 and D4, which has been observed previously as the point in time when the largest shift from AT2 to AT1 phenotype occurs in gene expression and morphology [32]. Clustering did not change using alternate cutoffs in the number of genes (top 2%, top 10%) (Figure S4). A two-dimensional principal component analysis plot showed that time in culture corresponded to PC1 (46% of total variation in the samples) (Figure 1B), suggesting that the differentiation process contributed most to inter-sample variation.
Fig. 1. Transcriptomic profiling of human AEC differentiation. 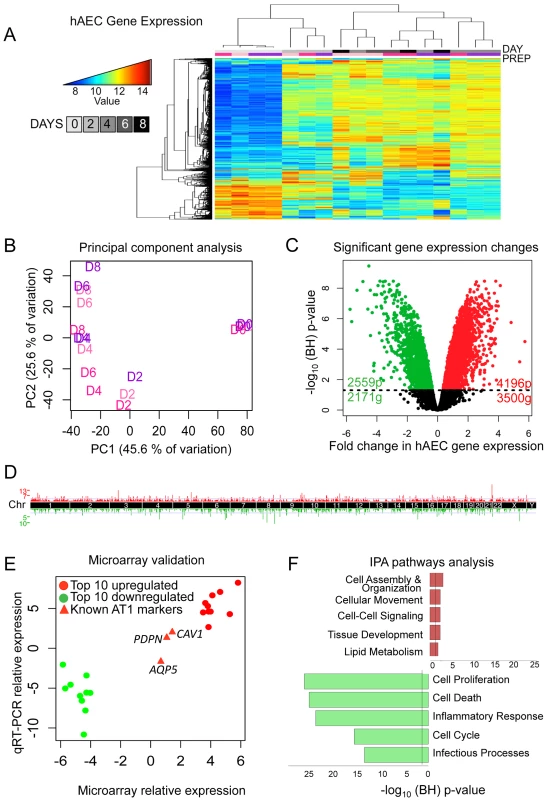
A) Heatmap of top 5% variant-VSN normalized gene expression probes. Blue = low expression, red = high expression. DAY = number of days AT2 cells were allowed to differentiate. “Prep” = donor lung origin by color (Figure S1). B) Principal component analysis of normalized hAEC samples. Samples color coded by donor lung as in (A). C) Significant changes in hAEC gene expression. Black line = BH-adjusted cutoff (FDR adjusted p≤0.05) calculated between D0 and D8. 20 genes show both significant up and downregulation for probes in different locations of the gene. D) Manhattan plot of differentially expressed genes. X-axis = chromosomal location, Y-axis = number of genes in each 2 MB region. E) qRT-PCR validation of microarray, data expressed in log2-fold change of differences between D0 and D8. Circles = top 10 up- and down-regulated genes, triangles = known AT1 cell differentiation markers (AQP5, PDPN, CAV1). F) IPA of significantly up- or down-regulated genes. Bars expressed as log10-BH corrected p-values of enrichment for pathway members in significant list against RefSeq db38 background. Whole figure: Red = upregulated, green = downregulated. A linear model was fit over the 5 time points (D0, D2, D4, D6, and D8) excluding the technical duplicates and a moderated t-test was used to determine significance of changes in gene expression from D0 (AT2 cells) to D8 (AT1-like cells). 6755 probes (5651 genes) showed statistically significant changes in gene expression; 4196 upregulated probes (3500 genes) and 2559 downregulated probes (2171 genes) (Figure 1C). These changes were distributed throughout the genome (Figure 1D). qRT-PCR of the top 10 up - and down-regulated genes showed a high degree of correlation with the microarray expression results (Figure 1E, Figure S5). Genes known to become activated during AT2 to AT1 cell differentiation (PDPN, CAV1, and AQP5) were also assessed using qRT-PCR and values were plotted for comparison alongside the top differentially expressed genes (Figure 1E, red triangles).
Ingenuity Pathways Analysis (IPA) revealed that the top upregulated pathways were cell assembly and organization, cell movement, cell-cell signaling, tissue development, and lipid metabolism, all of which are consistent with AT2 cells differentiating into larger, flat AT1 cells, while the most downregulated pathways pertained to cell proliferation, cell death, inflammatory response, cell cycle and infectious processes (Figure 1F). Signaling pathways in the upregulated networks included those of v-akt murine thymoma viral oncogene homolog (AKT), protein kinase C (PRKC), and the RAS family RAB genes, implicated in endocytosis, while FBJ murine osteosarcoma viral gene homolog (FOS) was the center of interconnectivity in the top downregulated network (Figure S6). Analysis using another pathway prediction program DAVID yielded results consistent with IPA (Figure S7).
Comparative transcriptomic profiling of human and rat alveolar epithelial cell differentiation
Differentiation of AT2 cells into AT1 cells has been extensively studied in the rat [33], [19]. To compare differentiation between rat AEC (rAEC) and human AEC (hAEC), purified rat AT2 cells were cultured under differentiation-permissive conditions and RNA was subjected to whole genome profiling. Technical variation within raw data was minimal (Figure S8). Data was preprocessed and clustered similarly to the human expression arrays (Figure S9). Purified AT2 cells clustered separately from AEC differentiating toward the AT1 cell phenotype (Figure 2A, Figure S10). We observed 4860 significantly changing probes corresponding to 4799 genes, with 2835 probes (2793 genes) upregulated and 1983 probes (1964 genes) downregulated (Figure 2B). For comparison, human and rat expression arrays were subset to include probes represented on both arrays; 13173 genes were represented in both species. This resulted in 3973 and 3662 statistically significant gene expression alterations in rat and human AEC respectively. Of these statistically significant gene sets derived separately from both species, 1514 genes were significantly differentially expressed during both human and rat AT2 to AT1 differentiation (Figure 2C–2D, p-value <2.2×10−16). Separating significantly up - or downregulated genes yielded similar degrees of overlap (Figure 2D). Therefore, differentiation of AT2 into AT1-like cells involves coordinated changes in thousands of genes, many of which occur in both human and rat. IPA analysis of genes concordantly changing in rat and human AT2 cell differentiation identified many similar altered pathways in both DAVID and IPA results (Figure 1E, Figure S11A). The most significant network from this joint analysis centered on genes involved in lipid metabolism (Figure S11B).
Fig. 2. Comparative transcriptomic profiling of human and rat AEC differentiation. 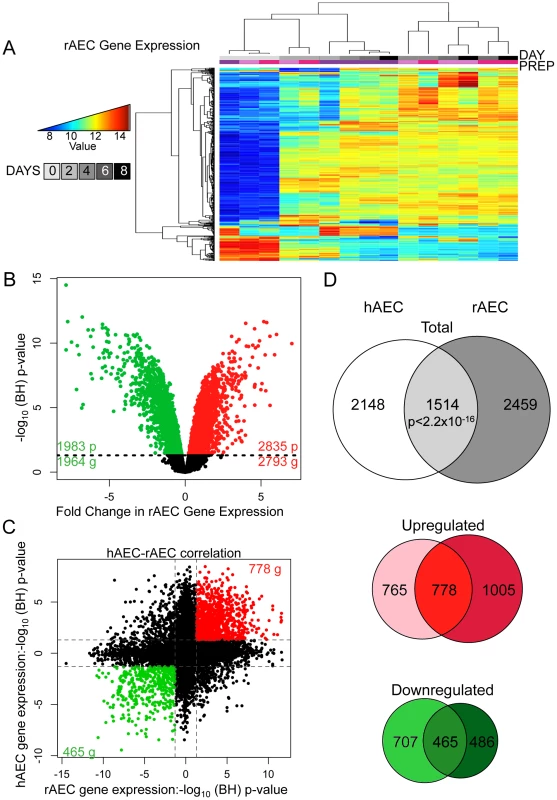
A) Heatmap of top 2% of variant-VSN normalized gene expression probes in rat AEC. Blue = low expression, red = high expression. Prep = separate rAEC purifications. B) Significant changes in rAEC gene expression: red = upregulated, green = downregulated, black line = BH-adjusted cutoff for significance (FDR adjusted p≤0.05) calculated between D0 and D8. C) Correlation between rAEC and hAEC statistically significant genes. Data points expressed as significance of change between D0 and D8. Direction of change derived from increase or decrease in gene expression. Red = statically significant upregulated genes in both hAEC and rAEC, green = statistically significant downregulated genes in both hAEC and rAEC. Dotted lines = BH-adjusted cutoff for significance (p.adjusted≤0.05) calculated between D0 and D8. D) Venn diagram of statistically significant gene overlap between hAEC and rAEC (Top), genes upregulated between hAEC and rAEC (Middle), and genes downregulated between hAEC and rAEC (Bottom). 271 genes were significant in both species but expression changed in opposite directions. In all three diagrams: pale color = hAEC-specific statistically significant gene expression changes, medium color = statistically significant overlap in both rAEC and hAEC, dark color = rAEC-specific statistically significant gene expression changes. The non-overlapping subsets of rAEC and hAEC genes were also of interest because they could indicate potential interspecies variation. While cell cycle control networks featured more prominently in the rat IPA networks, possibly reflecting species-specific differences, IPA of the hAEC and rAEC-specific gene sets revealed that 3 of the top 5 molecular signaling processes were identical (Figure S12, Figure S13), suggesting that different genes in a similar pathway were modulated in the two species to achieve similar effects. The top signaling network in hAEC-specific genes, cell growth and proliferation, centered on HNF4A and was the third most significant pathway in rAEC-specific alterations. TGFB featured prominently in rAEC-specific and in overall human IPA analysis.
Profiling of altered chromatin states during AT2 to AT1 cell differentiation
Using subsets of the same batches of human AT2 cells (D0) and fully differentiated AT1-like cells (D8) used for expression analyses, we assessed alterations to the chromatin environment by chromatin immunoprecipitation followed by sequencing (ChIP-seq). Specifically, H3K9/14Ac was used to interrogate active promoter and enhancer elements, and H3K27me3 was used to assess the repressed chromatin state. Peaks were called using both the Spatial clustering approach for the Identification of ChIP Enriched Regions (SICER) and the Model-based Analysis of ChIP-Seq (MACS) methods. Each analysis revealed thousands of enriched regions, with a significant fraction (20–40%) shared between D0 and D8 and the remainder of them specific to D0 or D8 (Figure S14). Changes in chromatin were distributed throughout the genome (Figure 3A). The transcription factor binding site (TFBS) predictor program Hypergeometric Optimization of Motif EnRichment (HOMER) was used to identify conserved sequence motifs enriched within day-specific chromatin marks [34]. For each of the 135 TFBSs in HOMER, the strongest association with either D0 or D8 H3K9/14Ac from SICER-called peaks was determined, and likewise for D0 and D8 H3K27me3 (Table S1). We then plotted the strongest H3K9/14Ac significance level versus the strongest H3K27me3 significance level for all TFBSs (with negative significance values representing D0 and positive D8) (Figure 3B), revealing a striking correlation – those motifs associated with regions losing H3K9/14Ac from D0 to D8 were consistently associated with regions gaining H3K27me3 from D0 to D8. Conversely, motifs associated with regions gaining H3K9/14Ac were consistently associated with regions losing H3K27me3. While this basic trend was expected based on the antagonism between these two marks, the identification of so many transcription factors apparently involved in regulation of both these marks was surprising. Some of the stronger associations with activating chromatin changes included motifs for zinc finger protein 711 (ZNF711), transcription factor 3 (TCF3 or E2A), TCF4 (a WNT signaling target), and RXR. TFBSs strongly associated with repressive chromatin changes included those for forkhead box (FOX) proteins FOXA1 and FOXA2, TATA-box binding protein TBP, and CCAAT/enhancer-binding protein CEBP. Two example loci illustrate the location of particular motifs within differentially marked chromatin regions – an upregulated gene, frizzled family receptor 2 (FZD2), which shows spreading acetylation and loss of H3K27me3, is predicted to have an RXR site within activating chromatin marks (Figure 3C), while a downregulated gene, progastricin (PGC), shows loss of acetylation but no gain of H3K27me3 and is predicted to have numerous FOXA1 sites within silenced regions (Figure 3D). These two examples illustrate that different combinations of marks might be found on activated and repressed genes. To further investigate this, we examined the association between transcriptomic and epigenetic changes by analyzing the relationship between changes in gene expression and all possible combinations of H3K9/14Ac and H3K27me3 from D0 to D8.
Fig. 3. Chromatin changes during AEC differentiation. 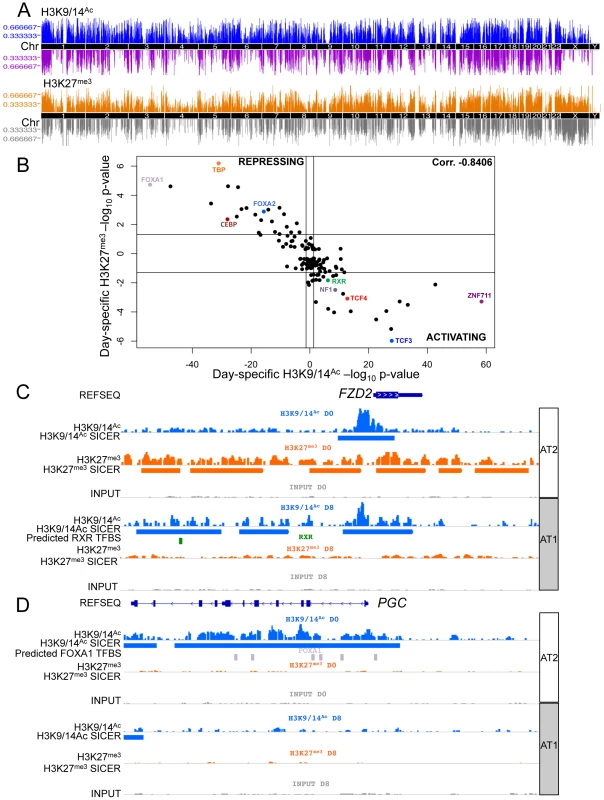
A) Manhattan plot of differential chromatin changes. X-axis = chromosomal location, Y-axis = number of cell type-specific chromatin changes within 2 MB region. Upper panel = H3K9/14Ac changes, blue = AT2 cell-specific acetylation, purple = AT1 cell-specific acetylation. Lower panel = H3K27me3 changes, orange = AT2 cell-specific methylation, grey = AT1 cell-specific methylation. B) 135 TFBS enrichment in domains of chromatin change from HOMER. X-axis = H3K9/14Ac, Y-axis = H3K27me3 enrichment. AT2 enrichment is shown as the log10 TFBS p-value, AT1 enrichment is shown as the −log10 TFBS p-value. C) Example of chromatin changes at an upregulated gene, FZD2, using IGV to visualize chromatin tracks. Blue = H3K9/14Ac raw reads and SICER peaks called, green = predicted RXR binding site from HOMER analysis. D) Example of downregulated gene expression at the PGC gene locus. Lavender = predicted FOXA1 binding sites from HOMER analysis. AT2 = AEC chromatin signature (D0), AT1 = AEC chromatin signature (D8). Integration of gene expression data with epigenetic alterations
Activating chromatin changes were strongly associated with upregulated gene expression, while repressive chromatin changes were loosely associated with downregulated gene expression (Figure 4A). 3011 (53%) of the genes showing altered expression were associated with at least one mark, and the largest single category was that of genes showing only acetylation at D0 (Figure 4B). Since we sampled just two of several dozen known histone tail post-translational modifications, the statistically significantly activated or repressed genes that were not associated with either of these marks may be regulated by epigenetic events and chromatin marks not evaluated in the current study.
Fig. 4. Integration of gene expression data with epigenetic alterations. 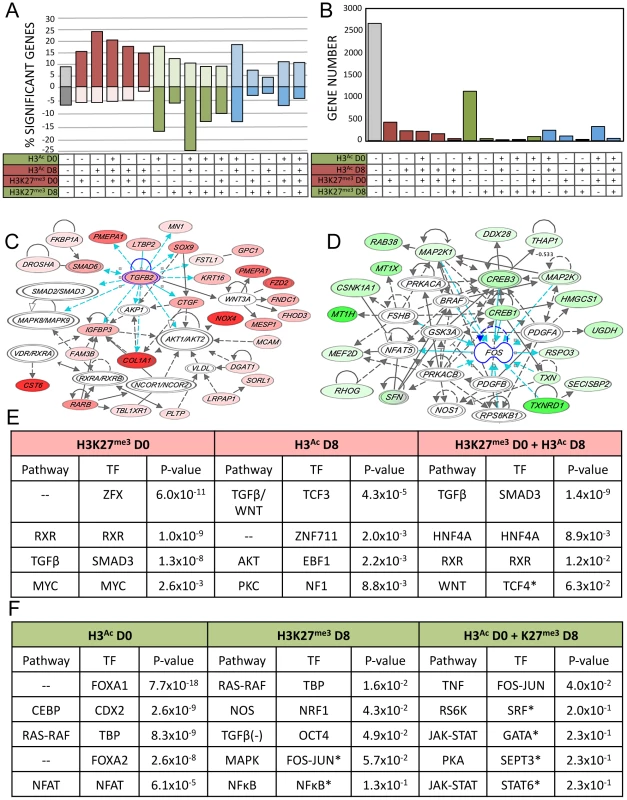
A–B–C) Relationship between all 16 possible combinations of chromatin changes and gene expression. Grey = unassociated with H3K9/14Ac or H3K27me3 changes, red = potentially activating chromatin changes, green = potentially repressive chromatin changes, blue = mixture of both. A) Significant expression changes in genes as a percentage of all genes associated with each histone mark for each of the possible 16 combinations of chromatin marks. Upregulated = above x-axis, downregulated = below x-axis. B) Total number of genes with significant gene expression changes associated with each chromatin combination. C) Representative IPA network of upregulated genes with both H3K9/14Ac gain and H3K27me3 loss. D) Representative IPA network of downregulated genes with H3K9/14Ac loss. E and F) IPA ranked networks of genes subset by chromatin context. Corresponding TFBS present in subset chromatin and enrichment p-value from HOMER analysis, for each chromatin-associated gene subset. Red = upregulated gene expression and activating chromatin changes, green = downregulated gene expression and deactivating chromatin changes. (*) Indicates below significance threshold in HOMER but still present in IPA. We performed IPA analysis on sets of genes associated with genomic regions carrying combinations of active or repressive chromatin marks that were associated with positive and negative expression changes respectively (Figure 4C). We simultaneously performed TFBS motif analysis on those genomic regions. For each class, we investigated the most significant TFBS matches, which were then matched to a transcription factor identified in the corresponding IPA gene networks enriched in the same combined chromatin/expression class (Figure 4C–D). Upregulated pathways included TGFB, WNT, HNF4A, RXR, and AKT (Figure 4E), while downregulated pathways included CEBP, RAS-RAF, and tumor necrosis factor (TNF) as well as FOXA1 and FOXA2 (Figure 4F). This three-way integration of whole-genome gene expression data, chromatin data, and TFBS identified several pathways already implicated in AEC differentiation, such as the WNT and TGFB signaling pathways [35], [36], along with transcription factors previously implicated in lung development such as FOXA1 and FOXA2 [7], [36]. Data integration allowed us to distinguish those genes (and regulatory sequences) likely to be direct transcriptional targets for each pathway, such as PGC, encoding an AT2-specific protease [37] as a potential direct target of FOXA1 (Figure 3D). Our analysis identified several pathways not previously implicated in lung regeneration, such as RXR, HNF4A, and TNF. The chromatin data enabled us to focus on those genes likely to be directly targeted by these novel pathways, such as FZD2, encoding a WNT receptor and candidate target of RXR (Figure 3C).
Validation of predicted transcription factor signaling pathways
As noted, one signaling pathway previously reported to be involved in AEC differentiation [35], [38], [39] and confirmed through our integrated analysis is the WNT signaling pathway. Specific examination of the microarray data for genes in the WNT signaling pathway showed altered expression of many genes, verified through qRT-PCR (Figure S15) and enrichment of the WNT signaling target transcription factor TCF4 in activated chromatin regions (Figure 3B, Figure 4E).
Of the newly implicated pathways, we chose the RXR pathway for validation. Retinoid X receptors can homodimerize or heterodimerize with a large variety of proteins, including retinoic acid receptors (RARs), thyroid hormone receptor (THR), the vitamin D receptor (VDR), farnesoid X receptor (NR1H4) and other nuclear receptors (NRs), as well as the family of peroxisome proliferator-activated receptors (PPARs) [40]. While retinoic acid and its receptor has been implicated in lung development and injury [41]–[48] the precise role of RXR remains to be clarified. Because of their many potential binding partners, the function of RXRs can be complex; certain NR heterodimers (e.g. NR1H4:RXR, PPAR:RXR) are permissive, responding to an RXR ligand (“rexinoid”) or the corresponding NR ligand, while other complexes are non-permissive, responding only to rexinoids in the presence of ligands for the NR partner (such as RXR:RAR, RXR:VDR, RXR:THR) [49]. Our integrated data suggested RXR-based activation of certain genes that lose the H3K27me3 mark (Figure 4E). To test a functional role for RXR pathway activation in AEC differentiation, it was important to target these receptors specifically. Thus, freshly isolated rAT2 cells were plated in the presence or absence of UVI-3003 (7.5 µM), a selective RXR inhibitor that blocks transactivation by RXR agonists but has been previously demonstrated to have minimal binding to structurally similar RAR family members [50]. Treatment with UVI-3003 markedly reduced AT1 cell marker induction (AQP5) while delaying downregulation of AT2 cell marker pro-SFTPC (Figure 5A) and significantly reducing transepithelial resistance (p<0.0001) (Figure 5B). Rescue of UVI-3003 treatment by drug removal on D4 of differentiation restored tight junctions. Analysis of the regulatory region of rat Aqp5 (a well-established AT1-specific gene) revealed 34 predicted PPARA:RXR heterodimer binding sites (Figure 5C) and an average of 9.6 predicted PPARA:RXR sites per kb across 4 kb upstream regions of the rat, mouse, and human AQP5 promoters. A similar phenomenon was seen with other AT1-specific genes (Pdpn, Cav1) (Figure 5C). Luciferase assays using the 4.3-kb upstream region of the rat Aqp5 gene transfected into mouse lung epithelial (MLE-15) cells revealed that UVI-3003 inhibited ∼50% of Aqp5 promoter activity (Figure 5D). ChIP assays of cultured rAEC revealed little detectable binding of RXR to the Aqp5 promoter at D0, but a marked increase in precipitation of a site about 4 kb upstream of the transcription start site was seen at D8, when the cells had achieved their AT1 cell-like phenotype (Figure 5E). Taken together, these analyses support a function for RXR signaling in AEC differentiation, and illustrate the utility of an integrated transcriptomic/epigenomic approach to identify new pathways involved in differentiation.
Fig. 5. Functional validation of a transcription factor signaling pathway predicted from bioinformatics analysis. 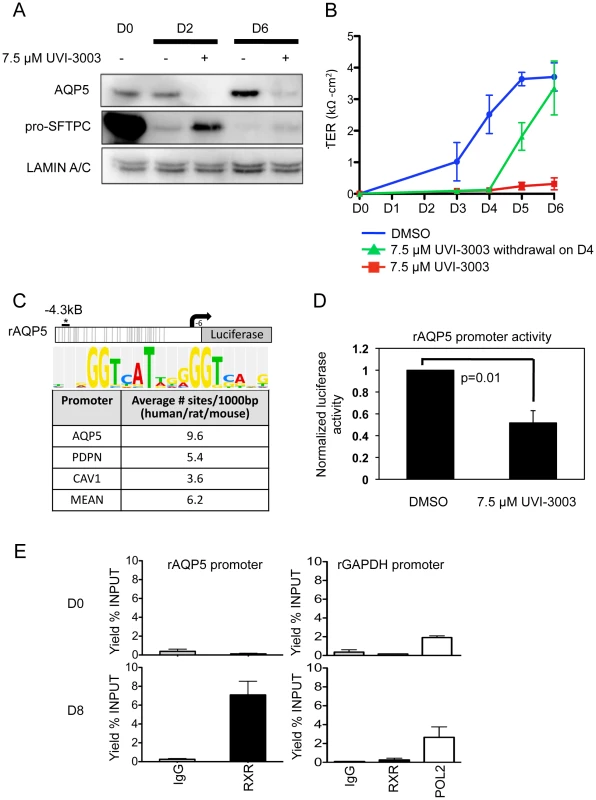
A) Western blots examining AT2 and AT1 cell markers during differentiation in the presence or absence of RXR antagonist UVI-3003. LAMIN A/C is the loading control. B) Transepithelial resistance as measured in kΩ-cm2 over the course of differentiation. Error bars represent technical duplicates for each plating. C) Rat Aqp5-luciferase 4.3 kb promoter construct. Grey lines = 34 putative PPARA:RXR binding sites (Explain3.0). No sites were predicted from −900 to +6 bp due to lack of rat sequence information in the Explain v3.0 database. The asterisk marks the approximate location in the promoter of the ChIPed RXR site in E, below. The average number of PPARA:RXR sites per kilobase in the listed human/rat/mouse promoters is given in the table, with consensus site listed at the top. D) MLE-15 cells were transiently transfected with the Aqp5-luciferase construct and treated for 48 hours with vehicle (DMSO) or 7.5 µM UVI-3003. UV1-3003 treatment reduced Aqp5-luc activity by 48%±0.06. Values were normalized to vehicle control and represent the mean, error bars represent SEM, N = 3. All experiments represent 3 biological replicates. E) ChIP was performed on primary cultured rat AEC at day 0 (AT2, D0, n = 2) and day 8 (AT1-like, D8, n = 3). A region ∼4 kb upstream of the transcription start site specifically precipitated with RXR in day 8 samples. ChIP of GAPDH with RXR was performed as a control, and POL2 (POLR2A) binding to the GAPDH promoter was included as a positive control for the quality of day 0 DNA. Discussion
AT1-like cells, differentiated in vitro from AT2 cells, exhibit many properties of AT1 cells in vivo, including morphology and expression of known phenotypic markers. Direct isolation of fragile AT1 cells from human lung is very challenging [51], due in part to the fact that strong cell-specific markers remain to be identified. However, the ability to differentiate AT2 cells in vitro into AT1-like cells offers a tractable model system to study not only the transcriptomic and epigenomic differences between these two cell types, but also the kinetic mechanisms controlling epithelial cell differentiation. Transcriptomic analysis of differentiating primary human and rat AEC identified thousands of genes undergoing significantly altered expression, a large number of which overlap between the two species. ChIP-seq identified D0 - and D8-specific acetylation (H3K9/14Ac) and methylation events (H3K27me3) and allowed identification of corresponding TFBSs enriched within AT2 - or AT1-specific chromatin patterns. Interestingly, almost all of the TFBS enriched within D8-specific H3K9/14Ac regions were also enriched within D0-specific H3K27me3 regions, and conversely for D0-specific H3K9/14Ac and D8-specific H3K27me3. This near perfect concordance is unexpected since H3K27me3 is thought to represent only one of several silencing mechanisms active in development [3].
Integrated analysis showed that upregulation of gene expression was associated with individual or combined gain of H3K9/14Ac and loss of H3K27me3, while downregulation was primarily associated with loss of H3K9/14Ac. Approximately half of the genes showing altered expression were not associated with either chromatin mark. This could be because marks were present but distant from the gene, or because other chromatin marks or regulatory mechanisms were involved in their up or downregulation. With application of ChIP-seq to the examination of other chromatin marks, remodeling complexes, and transcription factors, and the addition of information on nucleosome positioning, non-coding RNAs, microRNAs, chromatin conformation capture technologies, and DNA methylation, the current model system will lend itself well to a detailed understanding of the epigenomic basis for the differentiation of adult epithelial cells.
This study demonstrates how expression datasets and chromatin mapping are a potent combination to obtain an integrated picture of signaling pathway activity, transcription factors and their genomic targets. Transcriptional profiling can identify altered gene expression and corresponding regulatory pathways, but identifying transcription factors is difficult without knowing which genomic regions are implicated; epigenomic profiling can pinpoint the specific genomic regions where transcription factors and other regulatory proteins are likely to bind. Our approach revealed known regulators, such as TGFB, WNT, FOXA1 and FOXA2 [52]–[55], [7] as well as new potential regulators of AEC differentiation, including AKT, RXR and HNF4A.
As proof-of-principle, we investigated whether RXR signaling was required for the differentiation process of adult alveolar epithelium. Use of an RXR-specific inhibitor delayed differentiation as measured by inhibition of AT1-specific AQP5 expression and delayed TER. A role for RXR in normal AQP5 expression in AT1 cells is further supported by the inhibition of the Aqp5 promoter by UVI-3003, as well as the specific detection of RXR on the Aqp5 promoter in rAT1-like cells. Retinoic acid receptors (RARs) are one of the many potential binding partners with RXRs and numerous reports have implicated retinoic acid and/or RARs in lung development and injury [40]–[49]. Thus, it is possible that RXR effects are mediated through interactions with RAR. However, evaluation of stringently predicted RXR binding sites in the AQP5 promoter in human and rat shows that the presence of adjacent predicted RAR binding sites is rare or absent, while adjacent predicted VDR sites are more common, and predicted estrogen and glucocorticoid receptor sites also abut RXR binding site [56]. Given the multitude of interacting partners of RXRs (over 19 described [40]), dissecting the mechanism of RXR action on the promoter of AQP5 and other genes will require a very detailed examination. While the molecular mechanisms of altered RXR signaling remain to be further defined, inhibition of RXR in vitro as well as ChIP data showing increased promoter occupancy in AT1-like cells on day 8 support its functional role and illustrate the utility of our system and the potential of epigenomic/transcriptomic data integration to reveal novel regulators of biologic processes.
In summary, our analysis enabled identification of known and novel signaling pathways, gene regulatory networks and associated TFBS implicated in morphologic and phenotypic changes that occur during AEC differentiation. Full characterization of normal differentiation is critical to determine the precise mechanisms that are perturbed in disease. In the distal lung, this might shed light on the molecular basis of chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. The analysis presented here shows that purified human primary epithelial cells undergoing in vitro differentiation can serve as a powerful tool for the mechanistic investigation of normal and aberrant epithelial cell differentiation.
Materials and Methods
Ethics statement
Remnant human transplant lungs were obtained in compliance with Institutional Review Board-approved protocols for the use of human source material in research (HS-07-00660) and processed within 3 days of death. Rat AT2 cells were isolated in compliance with IACUC protocol #11360.
Isolation and culture of human and rat alveolar epithelial cells
Human lung tissue was processed as previously described [29] with inclusion of anti-EpCAM conjugated beads to select for epithelial cells. Cells were plated in 50∶50 [DMEM High glucose media (GIBCO 21063): DMEM-F12 (Sigma D6421)]. Differentiation into AT1-like cells was verified by measuring SFTPC and AQP5 expression using RNA extracted with the Illustra TriplePrep Kit (GE LifeSciences, Piscataway, NJ), and by measuring transepithelial resistance. Rat AT2 cells were isolated as previously described [57], [19]. Total RNA, DNA, and protein were simultaneously isolated from AT2 cells (D0), intermediate cell phenotypes (D2-6) and AT1-like cells (D8). Chromatin was isolated in tandem at D0 and D8.
Immunofluorescence
Freshly isolated hAT2 cells were fixed with 4% paraformadehyde for 10 min at room temperature (RT), permeabilized with 0.3% Triton, and blocked with CAS blocking reagent (Invitrogen Cat #00-8020, Camarillo, CA) for 30 min at RT. Slides were incubated with rabbit anti-pro-SFTPC (Seven Hills #WRAB-SPC serum) or anti-PTPRC (Santa Cruz sc-25590) antibodies and diluted in CAS-block at 4°C overnight. Slides were washed in Tris-Buffered Saline & Tween 20 (TBST) and incubated with anti-rabbit-FITC fluorescent secondary antibody in CAS-block for 1 hr at RT. For vimentin staining, mouse anti-VIM (Sigma V2258) and biotinylated anti-mouse IgM (Vector # BA-2020) antibodies were used. Sections were viewed with a NIKON Eclipse microscope equipped with a QImaging Retica 200R charge-coupled-device camera (QImaging, Surrey, BC, Canada). Images were processed with the NIS-Elements BR program (NIKON).
Gene expression analysis by Illumina HT-12v4 and RatRef-12 arrays
1 µg of RNA was converted into cRNA using Illumina TotalPrep RNA amplification kit, (Life Technologies, USA) and used for Illumina HT-12v4 or RatRef-12 expression analysis at the Southern California Genotyping Consortium, University of California Los Angeles. BeadStudio was used to convert images to raw signal data. Using R (version 2.11.1), Variant Stabilization and Normalization (VSN) was performed using LUMI [58] to allow for a large number of differentially expressed genes. Statistical analyses were performed using LIMMA [59]. A linear regression model was fitted over the time-course of differentiation, technical replicates were removed, and t-tests performed between D0 and D8. False-discovery rate was controlled using the Benjamini-Hochberg (BH) correction [60]. R was used for principal component analysis and heatmap generation. Heatmaps were generated by selecting the top 5% of probes most variant across the whole dataset and clustering with Ward's method. Pathways analysis was performed using IPA (Ingenuity Systems, www.ingenuity.com) or DAVID [61], [62]. Correlation of human and rat gene expression was performed using Entrez identifiers and the Mouse Genome Informatics (MGI) Web database [63].
Quantitative polymerase chain reactions
RNA was reverse transcribed using random hexamers and M-MLV reverse transcriptase per manufacturer's guidelines (Invitrogen), followed by qRT-PCR using SYBR green (BioRad, Hercules, CA) with primers listed in Table S2. qRT-PCR reactions were performed using a DNA engine Opticon (MJ Research, Waltham, MA) and normalized to 18S rRNA.
ChIP and ChIP-seq on primary human and rat epithelial cells
Chromatin immunoprecipitations for H3K9/14Ac (Millipore, #06-599), H3K27me3 (Millipore, #07-449), POL2 (POLR2A, Santa Cruz Biotechnology, sc-899X), and RXR (Santa Cruz Biotechnology, sc-774X) were performed as previously described [64]. Primer sequences are noted in Table S3. For rat ChIP at the Gapdh promoter, POL2 was used as a positive control. For human ChIP-seq, positive control loci were the promoter regions of GAPDH and MUC4 for H3K9/14Ac and H3K27me3 respectively; ≥10-fold enrichment was considered successful. ChIP-seq libraries were constructed using the New England Biolabs library prep kit (NEB Cat#E6200, Ipswich, MA). ChIP products underwent Illumina GAII single-end sequencing; reads were aligned to the hg18 human genome build using the MAQ 0.7.1 aligner. All datasets are deposited in the public GEO database (GEO# GSE38571).
Data access
All datasets are deposited in the public GEO database (GEO# GSE38571).
Peak calling
Integrative Genomics Viewer v1.5 (The Broad Institute) was used to visually inspect peak quality. SICER [65] peak calling was performed using a window and gap size of 600 bp. Input DNA was used for background normalization. The second peak-calling algorithm, MACS, (http://cistrome.org/ap/) was performed using default parameters and input DNA for background normalization. SICER - and MACS-called peaks had a large degree of overlap, as measured by the correlation coefficient (Figure S16A), calculated using Genome Graphs (http:/genome.ucsc.edu). A lack of correlation was observed between H3K9/14Ac and H3K27me3 (Figure S16B) [66]–[70]. ChIP-seq reads from both AT2 and AT1 cells were observed at or near read saturation (Figure S17). Differential peak occupancy was determined using the UCSC table browser. Manhattan plots were generated using Genome Graphs.
Motif analysis & data integration
Peaks were annotated to the nearest transcription start site (TSS) and motif analysis was performed with HOMER [34]. The opposite AEC cell type was used as background. Gene expression data was merged with annotated chromatin peaks using Entrez ID. For the AQP5 TFBS analysis PPAR:RXR predicted binding sites were evaluated using P-match algorithms within ExPlain 3.0 across human, mouse and rat species. ExPlain motif V$PPARA_02 with a high probability (>87%) were counted and averaged in the region 4.3 kb upstream of the promoter for the three species. Similar results were obtained with the Match program in Biobase [56], which was used to examine the frequency and nature of binding site adjacent to predicted RXR targets.
RXR inhibitor studies
1×106 rat AT2 (≥95% purity) cells were plated on 1.1-cm2 polycarbonate filters (Corning Costar #3401) and treated with either 7.5 µM UVI-3003 (Tocris Biosciences, Ellisville Missouri) or DMSO vehicle control from the time of plating (D0) through completion of the study. Whole cell lysates were extracted in 2% SDS lysis buffer (62.5 mM Tris pH 6.8, 2% SDS, 10% glycerol, and protease inhibitor cocktail III (Calbiochem #539134)).
Western blot analysis
Western blots were performed as previously described [16]. Primary antibodies (all rabbit) were anti-AQP5 (Alomone Labs AQP-005), anti-CAV1 (Abcam ab2910), anti-pro-SFTPC (Millipore AB3786) and anti-LAMIN A/C (sc-20681, Santa Cruz Biotechnology). Blots were analyzed by chemiluminescence and visualized by West Fempto Super Sensitivity Kit (Thermo Scientific) with a FluorChem 8900 Imaging System (Alpha Innotech).
Luciferase assays
Mouse lung epithelial cells (MLE-15) were plated at a density of 4×104 cells per well on 24-well plates 1.5 days prior to transfection. Duplicate wells were transiently transfected with 500 ng rAqp5-luc containing the 4.3 kb rat Aqp5 promoter or pGL2 empty vector using Superfect (Qiagen). Three hours post-transfection, media was changed to contain 7.5 µM UVI-3003 or DMSO vehicle control. 48 hours post transfection, luciferase assays were performed as previously described [33]. Raw luciferase values were normalized to protein concentration and then to pGL2 empty vector controls. Significance was measured using the student's t-test.
Supporting Information
Zdroje
1. SharmaS, KellyTK, JonesPA (2010) Epigenetics in cancer. Carcinogenesis 31 : 27–36.
2. VasanthiD, MishraRK (2008) Epigenetic regulation of genes during development: a conserved theme from flies to mammals. J Genet Genomics 35 : 413–429.
3. BernsteinBE, MeissnerA, LanderES (2007) The mammalian epigenome. Cell 128 : 669–681.
4. HirschhornJN, BrownSA, ClarkCD, WinstonF (1992) Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev 6 : 2288–2298.
5. GuentherMG, LevineSS, BoyerLA, JaenischR, YoungRA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130 : 77–88.
6. JiaL, ShenHC, WantrobaM, KhalidO, WangQ, et al. (2006) Locus-wide chromatin remodeling and enhanced androgen receptor-mediated transcription in recurrent prostate tumor cells. Mol Cell Bio 26 : 7331–7341.
7. WanH, DingleS, XuY, BesnardV, KaestnerKH, et al. (2005) Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem 280 : 13809–13816.
8. MaiA, MassaS, RotiliD, CerbaraI, ValenteS, et al. (2005) Histone deacetylation in epigenetics: an attractive target for anticancer therapy. Med Res Rev 25 : 261–309.
9. SaraivaNZ, OliveiraCS, GarciaJM (2010) Histone acetylation and its role in embryonic stem cell differentiation. World J Stem Cells 2 : 121–126.
10. BoegerH, GriesenbeckJ, StrattanJS, KornbergRD (2003) Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11 : 1587–1598.
11. SparmannA, van LohuizenM (2006) Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6 : 846–856.
12. MüllerJ, HartCM, FrancisNJ, VargasML, SenguptaA, et al. (2002) Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell 111 : 197–208.
13. SchwartzYB, PirrottaV (2008) Polycomb complexes and epigenetic states. Curr Opin Cell Biol 20 : 266–273.
14. MurryCE, KellerG (2008) Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132 : 661–680.
15. FuchsS, HollinsAJ, LaueM, SchaeferUF, RoemerK, et al. (2003) Differentiation of human alveolar epithelial cells in primary culture: morphological characterization and synthesis of caveolin-1 and surfactant protein-C. Cell Tissue Res 311 : 31–45.
16. ZhouB, AnnDK, FlodbyP, MinooP, LieblerJM, et al. (2008) Rat aquaporin-5 4.3-kb 5′-flanking region differentially regulates expression in salivary gland and lung in vivo. Am J Physiol Cell Physiol 295: C111–C120.
17. FehrenbachH (2001) Alveolar epithelial type II cell: defender of the alveolus revisited. Respir Res 2 : 33–46.
18. ChapmanHA, LiX, AlexanderJP, BrumwellA, LorizioW, et al. (2011) Integrin α6β4 identifies an adult distal epithelial population with regenerative potential in mice. J Clin Invest 121 : 2855–2862.
19. BorokZ, DantoSI, LubmanRL, CaoY, WilliamsMC, et al. (1998) Modulation of t1α expression with alveolar epithelial cell phenotype in vitro. Am J Physiol Lung Cell Mol Physiol 275: L155–164.
20. FangX, SongY, ZemansR, HirschJ, MatthayMA (2004) Fluid transport across cultured rat alveolar epithelial cells: a novel in vitro system. Am J Physiol Lung Cell Mol Physiol 287: L104–10.
21. FlodbyP, BorokZ, BanfalviA, ZhouB, GaoD, et al. (2010) Directed expression of Cre in alveolar epithelial type 1 cells. Am J Respir Cell Mol Biol 43 : 173–178.
22. NielsenS, KingLS, ChristensenBM, AgreP (1997) Aquaporins in complex tissues. II. Subcellular distribution in respiratory and glandular tissues of rat. Am J Physiol Cell Physiol 273: C1549–C1561.
23. CampbellL, HollinsAJ, Al-EidA, NewmanGR, von RuhlandC, et al. (1999) Caveolin-1 expression and caveolae biogenesis during cell transdifferentiation in lung alveolar epithelial primary cultures. Biochem Biophys Res Commun 262 : 744–751.
24. NewmanDR, ZhangH, BortoffK, BonnerJC, SannesPL (2010) Alveolar epithelial differentiation during repair involves FoxA1, Wnt7A, and TGF-β. Proc Am Thorac Soc 7 : 155–156.
25. RamirezMI, MillienG, HindsA, CaoY, SeldinDC, et al. (2003) T1alpha, a lung type I cell differentiation gene, is required for normal lung cell proliferation and alveolus formation at birth. Dev Biol 256 : 61–72.
26. MillienG, SpiraA, HindsA, WangJ, WilliamsMC, et al. (2006) Alterations in gene expression in T1α null lung: a model of deficient alveolar sac development. BMC Dev Biol 6 : 35.
27. MasonRJ, WilliamsMC, WiddicombeJH, SandersMJ, MisfeldtDS, et al. (1982) Transepithelial transport by pulmonary alveolar type II cells in primary culture. Proc Natl Acad Sci U S A 79 : 6033–6037.
28. QiaoR, ZhouB, LieblerJM, LiX, CrandallED, et al. (2003) Identification of three genes of known function expressed by alveolar epithelial type I cells. Am J Respir Cell Mol Biol 29 : 98–105.
29. BallardPL, LeeJW, FangX, ChapinC, AllenL, et al. (2010) Regulated gene expression in cultured type II cells of adult human lung. Am J Physiol Lung Cell Mol Physiol 299: L36–50.
30. CheekJM, EvansMJ, CrandallED (1989) Type I cell-like morphology in tight alveolar epithelial monolayers. Exp Cell Res 184 : 375–387.
31. CheekJM, KimKJ, CrandallED (1989) Tight monolayers of rat alveolar epithelial cells: bioelectric properties and active sodium transport. Am J Physiol 256: C688–93.
32. WangJ, EdeenK, ManzerR, ChangY, WangS, et al. (2007) Differentiated human alveolar epithelial cells and reversibility of their phenotype in vitro. Am J Respir Cell Mol Biol 36 : 661–668.
33. ZhouB, FrancisTA, YangH, TsengW, ZhongQ, et al. (2008) GATA-6 mediates transcriptional activation of aquaporin-5 through interactions with Sp1. Am J Physiol Cell Physiol 295: C1141–C1150.
34. HeinzS, BennerC, SpannN, BertolinoE, LinYC, et al. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38 : 576–589.
35. FlozakAS, LamAP, RussellS, JainM, PeledON, et al. (2010) Beta-catenin/T cell factor signaling is activated during lung injury and promotes the survival and migration of alveolar epithelial cells. J Biol Chem 285 : 3157–3167.
36. NewmanGR, CampbellL, von RuhlandC, JasaniB, GumbletonM (1999) Caveolin and its cellular and subcellular immunolocalisation in lung alveolar epithelium: implications for alveolar epithelial type I cell function. Cell Tissue Res 295 : 111–120.
37. FosterC, AktarA, KopfD, ZhangP, GuttentagS (2004) Pepsinogen C: a type 2 cell-specific protease. Am J Physiol Lung Cell Mol Physiol 286: L382–L387.
38. MucenskiML, WertSE, NationJM, LoudyDE, HuelskenJ, et al. (2003) β-catenin is required for specification of proximal/distal cell fate during lung morphogenesis. J Biol Chem 278 : 40231–40238.
39. TebarM, DestreeO, de VreeWJ, Ten Have-OpbroekAA (2001) Expression of Tcf/Lef and sFrp and localization of beta-catenin in the developing mouse lung. Mech Dev 109 : 437–440.
40. LefebvreP, BenomarY, StaelsB (2010) Retinoid X receptors: common heterodimerization partners with distinct functions. Trends Endocrinol Metabol 21 : 676–683.
41. McGowanS, JacksonSK, Jenkins-MooreM, DaiHH, ChambonP, et al. (2000) Mice bearing deletions of retinoic acid receptors demonstrate reduced lung elastin and alveolar numbers. Am J Respir Cell Mol Biol 23 : 162–167.
42. SimonDM, MarianiTJ (2007) Role of PPARs and retinoid X receptors in the regulation of lung maturation and development. PPAR Res 2007 : 91240.
43. ZhangH, GarberSJ, CuiZ, FoleyJP, MohanGS, et al. (2009) The angiogenic factor midkine is regulated by dexamethasome and retinoic acid during alveolarization and in alveolar epithelial cells. Respir Res 10 : 77.
44. KimuraY, SuzukiT, KanekoC, DarnelAD, MoriyaT, et al. (2002) Retinoid receptors in the developing human lung. Clin Sci 103 : 613–621.
45. HindM, MadenM (2004) Retinoic acid induces alveolar regeneration in the adult mouse lung. Eur Respir J 23 : 20–27.
46. SugimotoK, TakayasuH, NakazawaN, MontedonicoS, PuriP (2008) Prenatal treatment with retinoic acid accelerates type 1 alveolar cell proliferation of the hypoplastic lung in the nitrofen model of congenital diaphragmatic hernia. J Pediatr Surg 43 : 367–372.
47. MontedocinoS, SugimotoK, FelleP, BanniganJ, PuriP (2008) Prenatal treatment with retinoic acid promotes pulmonary alveologenesis in the nitrofen model of congenital diaphragmatic hernia. J Pediatr Surg 43 : 500–507.
48. BaybuttRC, SmithBW, DonskayaEV, HuL, LiT, et al. (2010) The proliferative effects of retinoic acid on primary cultures of adult rat type II pneumocytes depend upon cell density. In Vitro Cell Dev Biol Anim 46 : 20–27.
49. PerezE, BourguetW, GronemeyerH, de LeraAR (2012) Modulation of RXR function through ligand design. Biochim Biophys Acta 1821 : 57–69.
50. NahoumV, PérezE, GermainP, Rodríquez-BarriosF, ManzoF, et al. (2007) Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function. Proc Natl Acad Sci U S A 104 : 17323–17328.
51. FujinoN, KuboH, OtaC, SuzukiT, SuzukiS, et al. (2012) A novel method for isolating individual cellular components from the adult human distal lung. Am J Respir Cell Mol Biol 46 : 442–430.
52. SugaharaK, SadoharaT, SugitaM, IyamaK, TakiguchiM (1999) Differential expression of CCAAT enhancer binding protein family in rat alveolar epithelial cell proliferation and in acute lung injury. Cell Tissue Res 297 : 261–270.
53. BalciO, OzdemirS, MahmoudAS, AcarA, ColakogluMC (2010) The effect of antenatal steroids on fetal lung maturation between the 34th and 36th week of pregnancy. Gynecol Obstet Invest 70 : 95–99.
54. OikonomouN, HarokoposV, ZalevskyJ, ValavanisC, KotanidouA, et al. (2006) Soluble TNF mediates the transition from pulmonary inflammation to fibrosis. PLoS One 1: e108.
55. BesnardV, WertSE, KaestnerKH, WhitsettJA (2005) Stage-specific regulation of respiratory epithelial cell differentiation by Foxa1. Am J Physiol Lung Cell Mol Physiol 289: L750–L759.
56. KelAE, GösslingE, ReuterI, CheremushkinE, Kel-MargoulisOV, et al. (2003) MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 31 : 3576–3579.
57. ZhouB, ZhongQ, MinooP, LiC, AnnDK, et al. (2008) Foxp2 inhibits Nkx2.1-mediated transcription of SP-C via interactions with the Nkx2.1 homeodomain. Am J Respir Cell Mol Biol 38 : 750–758.
58. DuP, KibbeWA, LinSM (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics 24 : 1547–1548.
59. SmythGK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 Article 3.
60. BenjaminiY, HochbergY (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57 : 289–300.
61. HuangDW, ShermanBT, LempickiRA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc 4 : 44–57.
62. HuangDW, ShermanBT, LempickiRA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37 : 1–13.
63. Mouse Genome Informatics (MGI) Web, The Jackson Laboratory, Bar Harbor, Maine. (URL: http://www.informatics.jax.org). [retrieved 2/2011].
64. JiaL, KimJ, ShenH, ClarkPE, TilleyWD, et al. (2003) Androgen receptor activity at the prostate specific antigen locus: steroidal and non-steroidal mechanisms. Mol Cancer Res 1 : 385–392.
65. ZangC, SchonesDE, ZengC, CuiK, ZhaoK, et al. (2009) A clustering approach for identification of enriched domains from histone modification ChIP-seq data. Bioinformatics 25 : 1952–1958.
66. HeintzmanND, StuartRK, HonG, FuY, ChingCW, et al. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39 : 311–318.
67. BarskiA, CuddapahS, CuiK, RohTY, SchonesDE, et al. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129 : 823–837.
68. MikkelsenTS, KuM, JaffeDB, IssacB, LiebermanE, et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448 : 553–560.
69. PanG, TianS, NieJ, YangC, RuottiV, et al. (2007) Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1 : 299–312.
70. ZhaoXD, HanX, ChewJL, LiuJ, ChiuKP, et al. (2007) Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1 : 286–298.
Štítky
Genetika Reprodukční medicína
Článek PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant TransformationČlánek The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene ClusterČlánek Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish PathogenČlánek USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and TranscriptionČlánek Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA EditingČlánek Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 6
-
Všechny články tohoto čísla
- BMS1 Is Mutated in Aplasia Cutis Congenita
- High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants
- How Cool Is That: An Interview with Caroline Dean
- Genetic Architecture of Vitamin B and Folate Levels Uncovered Applying Deeply Sequenced Large Datasets
- Juvenile Hormone and Insulin Regulate Trehalose Homeostasis in the Red Flour Beetle,
- Meiosis-Specific Stable Binding of Augmin to Acentrosomal Spindle Poles Promotes Biased Microtubule Assembly in Oocytes
- Environmental Dependence of Genetic Constraint
- H3.3-H4 Tetramer Splitting Events Feature Cell-Type Specific Enhancers
- Network Topologies and Convergent Aetiologies Arising from Deletions and Duplications Observed in Individuals with Autism
- Effectively Identifying eQTLs from Multiple Tissues by Combining Mixed Model and Meta-analytic Approaches
- Altered Splicing of the BIN1 Muscle-Specific Exon in Humans and Dogs with Highly Progressive Centronuclear Myopathy
- The NADPH Metabolic Network Regulates Human Cardiomyopathy and Reductive Stress in
- Negative Regulation of Notch Signaling by Xylose
- A Genome-Wide, Fine-Scale Map of Natural Pigmentation Variation in
- Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals mC within Archaeal mRNAs
- Multiplexin Promotes Heart but Not Aorta Morphogenesis by Polarized Enhancement of Slit/Robo Activity at the Heart Lumen
- Latent Effects of Hsp90 Mutants Revealed at Reduced Expression Levels
- Impact of Natural Genetic Variation on Gene Expression Dynamics
- DeepSAGE Reveals Genetic Variants Associated with Alternative Polyadenylation and Expression of Coding and Non-coding Transcripts
- The Identification of -acting Factors That Regulate the Expression of via the Osteoarthritis Susceptibility SNP rs143383
- Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs
- The RNA Export Factor, Nxt1, Is Required for Tissue Specific Transcriptional Regulation
- Inferring Demographic History from a Spectrum of Shared Haplotype Lengths
- Histone Acetyl Transferase 1 Is Essential for Mammalian Development, Genome Stability, and the Processing of Newly Synthesized Histones H3 and H4
- PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant Transformation
- DNA Methylation Restricts Lineage-specific Functions of Transcription Factor Gata4 during Embryonic Stem Cell Differentiation
- The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Deregulation of the Protocadherin Gene Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy
- Evidence for Two Different Regulatory Mechanisms Linking Replication and Segregation of Chromosome II
- USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and Transcription
- Methylation of Histone H3 on Lysine 79 Associates with a Group of Replication Origins and Helps Limit DNA Replication Once per Cell Cycle
- A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue
- The Gene Desert Mammary Carcinoma Susceptibility Locus Regulates Modifying Mammary Epithelial Cell Differentiation and Proliferation
- Hooked and Cooked: A Fish Killer Genome Exposed
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
- Mutations in Cause Autosomal Recessive Congenital Ichthyosis in Humans
- Integrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation
- RSR-2, the Ortholog of Human Spliceosomal Component SRm300/SRRM2, Regulates Development by Influencing the Transcriptional Machinery
- Comparative Polygenic Analysis of Maximal Ethanol Accumulation Capacity and Tolerance to High Ethanol Levels of Cell Proliferation in Yeast
- SPO11-Independent DNA Repair Foci and Their Role in Meiotic Silencing
- Budding Yeast ATM/ATR Control Meiotic Double-Strand Break (DSB) Levels by Down-Regulating Rec114, an Essential Component of the DSB-machinery
- Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA Editing
- Functional Analysis of Neuronal MicroRNAs in Dauer Formation by Combinational Genetics and Neuronal miRISC Immunoprecipitation
- DNA Ligase IV Supports Imprecise End Joining Independently of Its Catalytic Activity
- Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
- Heritable Change Caused by Transient Transcription Errors
- From Many, One: Genetic Control of Prolificacy during Maize Domestication
- Neuronal Target Identification Requires AHA-1-Mediated Fine-Tuning of Wnt Signaling in
- Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish
- H-NS Can Facilitate Specific DNA-binding by RNA Polymerase in AT-rich Gene Regulatory Regions
- Prophage Dynamics and Contributions to Pathogenic Traits
- Global DNA Hypermethylation in Down Syndrome Placenta
- Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in
- Disturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-ethylene Crosstalk in Seminal Roots
- Causes and Consequences of Chromatin Variation between Inbred Mice
- Genome-scale Analysis of FNR Reveals Complex Features of Transcription Factor Binding
- Distinct and Atypical Intrinsic and Extrinsic Cell Death Pathways between Photoreceptor Cell Types upon Specific Ablation of in Cone Photoreceptors
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- BMS1 Is Mutated in Aplasia Cutis Congenita
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



