-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
article has not abstract
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003480
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003480Summary
article has not abstract
Unexplained Cardiac Arrest
In this issue of PLOS Genetics, Nakano and colleagues from 13 centers in Japan report a candidate gene analysis in unrelated individuals with an unexplained cardiac arrest (UCA) [1]. Cardiac arrest is most often caused by a cardiac arrhythmia named ventricular fibrillation, which, if left untreated, will be lethal within minutes. Moreover, ventricular fibrillation is often the first expression of the disease, implying that patients die suddenly without any warning. It is thus understandable that cardiac arrest is an extremely difficult phenotype, as its victims will not be pre-symptomatically treated and the vast majority (∼85%) will not survive the event. In those few who do survive, the sequelae to cardiac ischemia in coronary artery disease appear to be the predominant cause of the cardiac arrest (i.e., the direct pro-arrhythmic effects of ischemia as well as the indirect pro-arrhythmic effects of successive cardiac remodeling and fibrosis). Several genetic variants that predispose to cardiac arrest in the setting of cardiac ischemia have already been documented [2], [3]. In the absence of coronary artery disease or other clear causes (e.g., myocarditis), structural heart disease (e.g., hypertrophic cardiomyopathy) and channelopathies (e.g., long QT syndrome, Brugada syndrome) are now recognized as the most prevalent causes of cardiac arrest, especially in younger individuals [4]. Nevertheless, in some individuals (<5%) the cause of the cardiac arrest remains unexplained: these unexplained cases comprise different, yet unidentified, pathologies (Figure 1).
Fig. 1. Illustration of the position of unexplained cardiac arrest among sudden cardiac arrest, focused on the occurrence of familial sudden cardiac arrest. 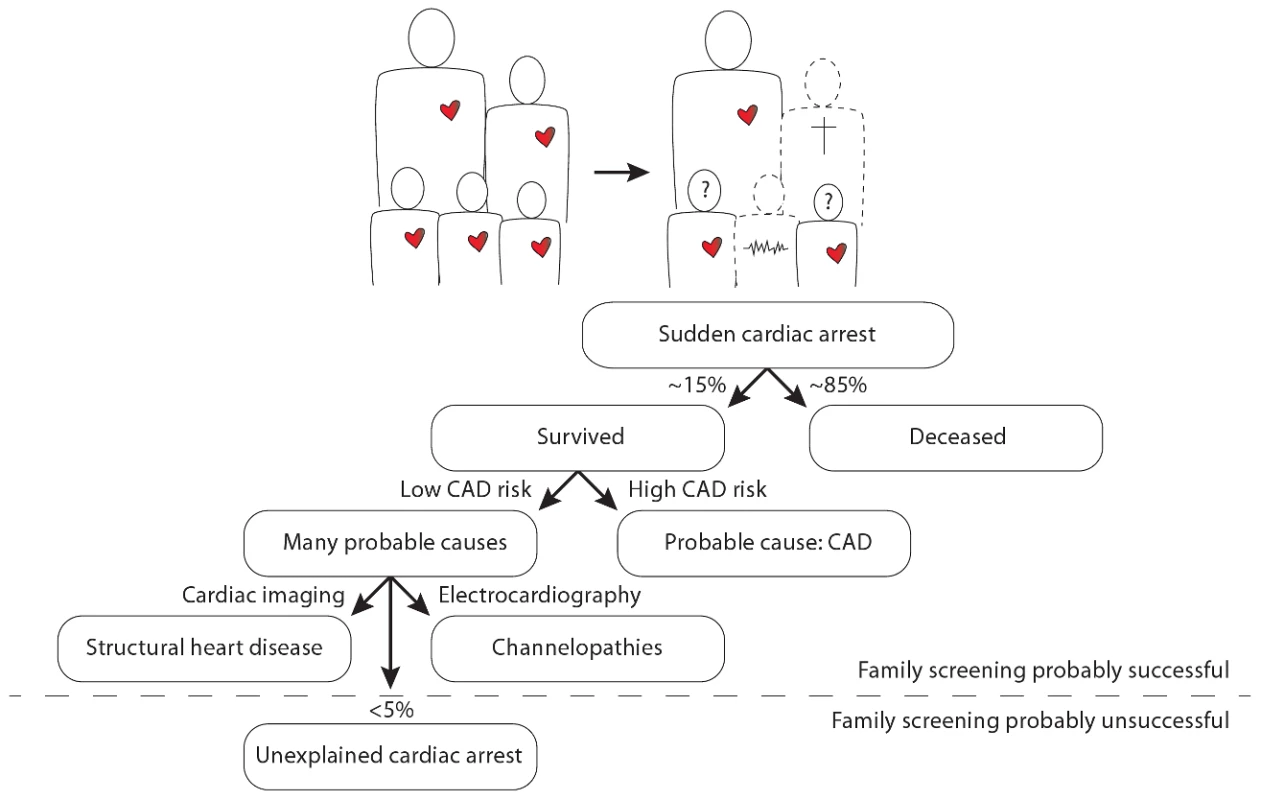
The question marks denote the uncertainty in the remaining siblings as to whether they too have an increased risk for sudden cardiac arrest and whether it is possible that they transmit this risk to their own offspring. CAD denotes coronary artery disease. Importantly, UCA appears to have a heritable component in several patients [5], which may thus set their whole family at risk for premature sudden death. A number of UCA cases with documented ventricular fibrillation can be defined as having idiopathic ventricular fibrillation (IVF). This more strict definition is reserved for those patients who survive ventricular fibrillation and in whom all extensive evaluations return normal. The first, and only, breakthrough in defining the genetic underpinnings of IVF was made in 2009 in The Netherlands [5]. Genome-wide haplotype analysis in several related IVF families revealed a conserved haplotype in the DPP6 gene, putatively involved in one of the main cardiac potassium currents: the transient outward current. This discovery has already led to the identification and pre-symptomatic treatment of dozens of patients at risk for IVF. However, mutations in the DPP6 gene have not yet been implicated in IVF/UCA outside The Netherlands.
This continuing lacuna hampers both treatment and cascade screening for the identification and treatment of affected family members and results in both under-treatment and over-treatment with substantial collateral damage (including mortality). Hence, UCA is both an uncommon and a difficult phenotype, and it represents a bothersome black box in current cardiology. Further efforts to delineate its underlying genetic profile will help us to understand this entity and, importantly, will hopefully guide us to the identification of as yet pre-symptomatic but similarly affected family members.
Semaphorin 3A and Unexplained Cardiac Arrest
Nakano and colleagues studied two populations with UCA and documented ventricular fibrillation. In east and west Japan, they retrospectively collected cases who survived UCA (n = 31 and n = 52, respectively) and controls (n = 912 and n = 2,046, respectively) from several cohorts. Guided by their previous results in mice studies [6], they focused on SEMA3A as a candidate gene. SEMA3A is vital for normal neuronal pattern development, and has been implicated in various disease conditions [7], [8]. Interestingly, murine cardiac-specific SEMA3A under-expression results in sinus bradycardia and SEMA3A over-expression in a susceptibility to ventricular tachycardia and sudden death, putatively due to differences in the pattern of cardiac innervation [6]. In their current study, through resequencing and SNP genotyping of SEMA3A, it was found that a nonsynonymous polymorphism (I334V, rs138694505A>G) in exon 10 of the SEMA3A gene was associated with UCA with an odds ratio of 3.1 (95%CI 1.7–5.7). UCA patients with SEMA3AI334V also displayed slightly different autonomic nervous system control, apparently as a result of a higher incidence of sinus bradycardia. Subsequent in vivo studies using cardiac biopsies revealed that UCA patients with SEMA3AI334V display aberrant sympathethic nerve fiber growth. Furthermore, in vitro studies using transfected HEK293T cells revealed that the I334V amino acid replacement in SEMA3A indeed disrupts the normal function of the protein in neural growth inhibition and control of cardiac innervation.
Strengths and Limitations
The strengths of the current study are several. The authors were able to unite data from the extremely rare but extremely challenging UCA cases who survived documented ventricular fibrillation from many different centers in Japan, and they replicated the association between UCA and SEMA3AI334V in two study groups (east and west Japan). Further clinical, in vitro, and in vivo studies substantiated their findings. These results, in combination with the previous studies in SEMA3A mice [6] and other studies, make a strong argument for a pivotal role of SEMA3A in cardiac innervation patterning and, if that goes wrong, UCA.
However, there are several limitations that should be acknowledged. Although it can be expected that cases with such a rare phenotype are difficult to assemble, the low numbers by current genetic association standards clearly prohibit definitive conclusions. Another important limitation is the lack of segregation data in the families of the victims. To be able to use SEMA3AI334V as a risk stratifier for sudden death in the remaining pre-symptomatic family members (which is the ultimate goal), one should have indisputable evidence that inheritance of the risk allele is the predominant cause of the increased risk for sudden death in these families. Only with this knowledge can one easily justify aggressive pre-symptomatic therapy such as implantation of a cardioverter defibrillator. However, as the family members of the studied cases apparently refused screening during this study, the clinical usefulness of SEMA3AI334V currently remains unknown.
Furthermore, this phenotype of UCA with documented ventricular fibrillation evidently has several different underlying causes, including IVF but also cardiomyopathies/channelopathies of unknown origin, as can be discerned from the electrocardiograms (Figure 2 from Nakano et al. [1]). It could be suggested that SEMA3A plays a role in all of these different underlying causes, but it is more likely that certain phenotypes will be more affected by SEMA3A dysregulation than others. It should be noted that 85% of SEMA3AI334V patients underwent a pilsicainide provocation test, which excluded Brugada syndrome in these patients (supplementary Table S2 from Nakano et al. [1]). This suggests that SEMA3AI334V does not result in a Brugada phenotype. However, it is believed that arrhythmias in Brugada syndrome are exacerbated by vagal stimulation, which might suggest SEMA3A as a candidate gene for future studies of Brugada syndrome.
The quest for the identification of genetic variants in UCA and IVF will certainly continue and meticulous phenotyping of the patients will remain an important issue. Genome-wide studies in unrelated individuals are unlikely to be revealing for the near future, but Nakano et al. demonstrate the ongoing value of candidate gene studies. More study populations are needed that are suitable for discerning phenotype–genotype relationships, which then can be further analyzed in translational studies.
Conclusions
Nakano and colleagues studied the rare and difficult but important phenotype of UCA, and revealed that SEMA3AI334V is associated with a higher incidence of cardiac arrest. These results were substantiated by in vitro and in vivo studies and point to a pivotal role for SEMA3A in cardiac innervation patterning and, if that goes wrong, a propensity for sudden cardiac arrest.
Zdroje
1. NakanoY, ChayamaK, OchiH, ToshishigeM, HayashidaY, et al. (2013) A nonsynonymous polymorphism in Semaphorin 3A as a risk factor for human unexplained cardiac arrest with documented ventricular fibrillation. PLoS Genet 9: e1003364 doi:10.1371/journal.pgen.1003364.
2. BezzinaCR, PazokiR, BardaiA, MarsmanRF, De JongJS, et al. (2010) Genome-wide association study identifies a susceptibility locus at 21q21 for ventricular fibrillation in acute myocardial infarction. Nat Genet 42 : 688–691.
3. ArkingDE, JunttilaMJ, GoyetteP, Huertas-VazquezA, EijgelsheimM, et al. (2011) Identification of a sudden cardiac death susceptibility locus at 2q24.2 through genome-wide association in European ancestry individuals. PLoS Genet 7: e1002158 doi:10.1371/journal.pgen.1002158.
4. Van der WerfC, Van LangenIM, WildeAA (2010) Sudden death in the young: what do we know about it and how to prevent? Circ Arrhythm Electrophysiol 3 : 96–104.
5. AldersM, KoopmannTT, ChristiaansI, PostemaPG, BeekmanL, et al. (2009) Haplotype-sharing analysis implicates chromosome 7q36 harboring DPP6 in familial idiopathic ventricular fibrillation. Am J Hum Genet 84 : 468–476.
6. IedaM, KanazawaH, KimuraK, HattoriF, IedaY, et al. (2007) Sema3a maintains normal heart rhythm through sympathetic innervation patterning. Nat Med 13 : 604–612.
7. KanekoS, IwanamiA, NakamuraM, KishinoA, KikuchiK, et al. (2006) A selective Sema3A inhibitor enhances regenerative responses and functional recovery of the injured spinal cord. Nat Med 12 : 1380–1389.
8. HanchateNK, GiacobiniP, LhuillierP, ParkashJ, EspyC, et al. (2012) SEMA3A, a gene involved in axonal pathfinding, is mutated in patients with Kallmann syndrome. PLoS Genet 8: e1002896 doi:10.1371/journal.pgen.1002896.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



