-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe G4 Genome
Recent experiments provide fascinating examples of how G4 DNA and G4 RNA structures—aka quadruplexes—may contribute to normal biology and to genomic pathologies. Quadruplexes are transient and therefore difficult to identify directly in living cells, which initially caused skepticism regarding not only their biological relevance but even their existence. There is now compelling evidence for functions of some G4 motifs and the corresponding quadruplexes in essential processes, including initiation of DNA replication, telomere maintenance, regulated recombination in immune evasion and the immune response, control of gene expression, and genetic and epigenetic instability. Recognition and resolution of quadruplex structures is therefore an essential component of genome biology. We propose that G4 motifs and structures that participate in key processes compose the G4 genome, analogous to the transcriptome, proteome, or metabolome. This is a new view of the genome, which sees DNA as not only a simple alphabet but also a more complex geography. The challenge for the future is to systematically identify the G4 motifs that form quadruplexes in living cells and the features that confer on specific G4 motifs the ability to function as structural elements.
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003468
Category: Review
doi: https://doi.org/10.1371/journal.pgen.1003468Summary
Recent experiments provide fascinating examples of how G4 DNA and G4 RNA structures—aka quadruplexes—may contribute to normal biology and to genomic pathologies. Quadruplexes are transient and therefore difficult to identify directly in living cells, which initially caused skepticism regarding not only their biological relevance but even their existence. There is now compelling evidence for functions of some G4 motifs and the corresponding quadruplexes in essential processes, including initiation of DNA replication, telomere maintenance, regulated recombination in immune evasion and the immune response, control of gene expression, and genetic and epigenetic instability. Recognition and resolution of quadruplex structures is therefore an essential component of genome biology. We propose that G4 motifs and structures that participate in key processes compose the G4 genome, analogous to the transcriptome, proteome, or metabolome. This is a new view of the genome, which sees DNA as not only a simple alphabet but also a more complex geography. The challenge for the future is to systematically identify the G4 motifs that form quadruplexes in living cells and the features that confer on specific G4 motifs the ability to function as structural elements.
G4 Motifs and G4 Structures
The sequence motif G≥3NxG≥3NxG≥3NxG≥3 confers the ability to form a four-stranded (“quadruplex”) structure in which interactions among strands are stabilized by G-quartets (Figure 1, see legend for details). G-quartets [1], G4 DNA [2], and G4 RNA [3] form readily in solution, creating structures of great variety [4], [5]. Strand orientation, conformation of glycosidic bonds of guanine bases in quartets, and sizes of and sequences of loops connecting the guanine runs contribute to structural intricacy. The diversity of G4 structures contrasts with the highly predictable B-form duplex. It has captivated chemists and fueled development of drugs that target quadruplexes and applications of quadruplexes to nanotechnology.
Fig. 1. Structural variety of G4 DNA. 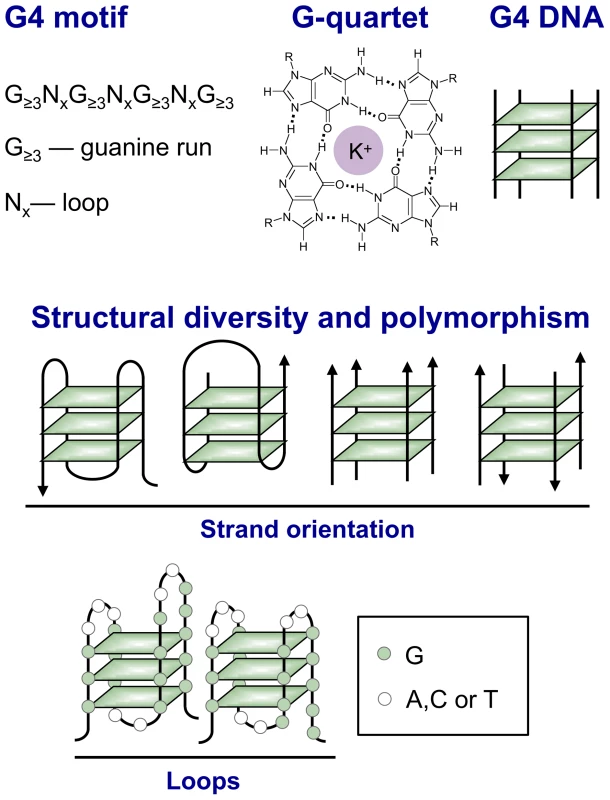
Top: A G4 motif consists of four runs of at least three guanines per run, separated by other bases (N). G4 motifs confer the ability to form a G4 DNA structure, also known as a “quadruplex” by analogy with the B-form DNA duplex. The essential unit of G4 DNA is the G-quartet, a planar array of guanines stabilized by Hoogsteen base pairing between the N7 group of one guanine and the extracyclic amino group of its neighbor. The guanines form a ring around a central channel that is occupied by a monovalent cation and associated water molecules (potassium is shown). G4 structures derive stability both from hydrogen bonding between guanines within G-quartets and from stacking of the planar, hydrophobic G-quartets. Middle: The length and sequence composition of the loops connecting planar arrays of G-quartets (left) and the parallel (near right) or antiparallel (far right) orientation of nucleic acid strands determine quadruplex topology. Bottom: There is considerable potential for structural polymorphism, as illustrated by the diagram of two possible conformations formed by the G4 motif G3N3G3N2G4N2G5. There are numerous G4 motifs in the human genome: over 350,000 allowing loops of 1–7 nt, and over 700,000 allowing loops up to 12 nt in length. G4 motifs are especially abundant in specific chromosomal domains, genomic regions, and genes. In human cells, the telomeres, rDNA, immunoglobulin switch regions (S regions), some variable number tandem repeats (VNTRs), and some single copy genes are all enriched for G4 motifs (“G4hi”). We do not yet know if every genomic G4 motif forms a quadruplex in a living cell. Nonetheless, G4 motifs provide a considerable potential repertoire for formation of diverse structures that may correlate with specific functions.
G4 Motifs in DNA Replication
DNA Replication Origins Coincide with G4 Motifs
Shared features of human DNA replication origins have been elusive. Origin identification by exhaustive high throughput sequencing of short nascent DNA strands has very recently revealed that the majority of the 250,000 human replication origins correspond to G4 motifs (67%, loop size 1–7 nt) [6]. This identifies half of the G4 motifs in the human genome as replication origins, and the fraction drops only slightly using more relaxed criteria for G4 motif identification. Origins proved to be highly conserved among four distinct human cell types: fibroblasts, embryonic stem cells, induced pluripotent stem cells, and HeLa cells. Quadruplexes are also implicated in recruitment of host factors for origin recognition by two human DNA viruses, SV40 [7] and Epstein-Barr virus [8]. It will be interesting to learn if components of the origin recognition complex recognize quadruplexes, and whether they discriminate among structures formed by distinct classes of G4 motifs.
Quadruplexes and Telomere Maintenance in Human Cells
Human telomeres provide an example of both the utility and potential hazards of G4 motifs in DNA replication. They consist of thousands of kilobases of duplex DNA repeats (TTAGGG) and a single-stranded G-rich 3′-tail. Human telomeric repeats form characteristic “beads on a string” quadruplex structures, with a propeller-like domain [9], [10]. The importance of telomeres in cancer and aging has made them a paradigm for the design of G4-targeting drugs [11].
Human telomeres are transcribed from subtelomeric promoters to generate long, noncoding TERRA RNAs consisting of UUAGGG repeats, which adopt a characteristic G4 RNA structure [12], [13]. Use of a novel dual-pyrene probe specific for TERRA RNA G-quadruplexes has enabled direct imaging of TERRA G4 RNA at the telomeres in human cells [14]. The telomere binding protein TRF2 interacts with TERRA RNA G-quadruplexes to promote telomere heterochromatinization [15], [16]. Thus, G4 RNA is a key participant in telomere biology and epigenetic regulation.
G4 structures can protect telomeres. Telomeric repeats are normally capped by a protein complex that identifies them as telomeres rather than damaged DNA and protects them from misguided cellular efforts at repair that are potentially destabilizing [17]–[19]. In Saccharomyces cerevisiae, depletion of Cdc13, a component of the telomere-capping complex, results in telomere instability that can be countered by drugs that stabilize G4 structures [20]. G4 DNA and RNA are very resistant to digestion by exonucleases, and this may confer stability to telomeres deprived of caps. G4 structures may also facilitate t-loop formation, to establish the lariat conformation adopted by telomere ends in living cells.
Replicative Instability at G4 Motifs
Quadruplexes may form spontaneously when DNA single strands are exposed during replication or transcription (Figure 2). These structures pose challenges to replication, and G4 helicases must recognize and unwind G4 DNA to maintain genetic stability. G4 helicases in human cells include BLM [21], WRN [22], FANCJ [23], [24], CHL1 [25], PIF1 [26], and quite probably RTEL1 [18], [27]. The RecQ family G4 helicases WRN [28] and BLM [29] and the iron-sulfur domain helicase RTEL1 [30], [31] are required to resolve telomeric quadruplexes. Deficiency in WRN helicase occurs in the human genetic disease Werner syndrome, the most salient feature of which is premature aging due to depletion of telomeric sequence. Deficiency in BLM helicase causes Bloom syndrome, characterized by immunodeficiency due to impaired recombination at the G4hi immunoglobulin S regions, as well as genomic instability and cancer predisposition. FANCJ deficiency is associated with instability at G4 motifs in human cells [23], [24], and causes an especially striking phenotype in Caenorhabditis elegans, evident as extended DNA deletions in which one end is bounded by a G4hi region [32].
Fig. 2. Structures form upon replication or transcription of regions bearing G4 motifs. 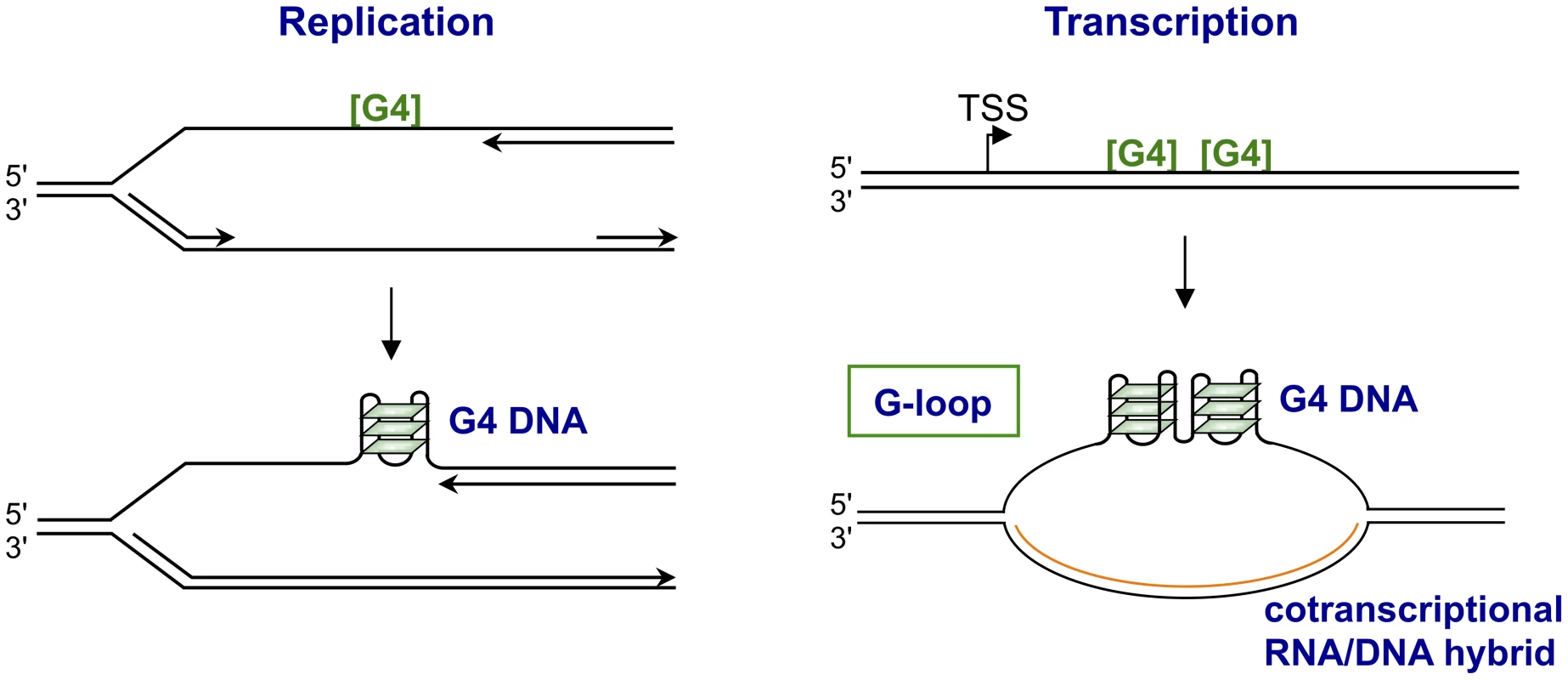
The figures illustrate how replication (left) or transcription (right) through G4 motifs ([G4]) may result in formation of structures. Replication is shown as arresting at a G4 motif in the leading DNA strand, as it does in Pif1-deficient yeast [33]. Transcription is shown as resulting in formation of a G-loop, which contains a persistent RNA/DNA on the template strand and G4 DNA interspersed with single-stranded regions on the nontemplate strand [39]. In S. cerevisiae, use of the G4hi human CEB1 VNTR (GGGGGGAGGGAGGGTGGCCTGCGGAGGTCCCTGGGCTGA) as a reporter identified Pif1 helicase as necessary for stability of G4 motifs [33], [34]. CEB1 instability was correlated with quadruplex structures by showing that a small molecule G4 ligand, PhenDC3, exacerbated instability, and that mutation that eliminated the potential to form quadruplexes conferred stability. Genomewide analysis of Pif1-deficient S. cerevisiae found endogenous G4 motifs enriched among sites of replication stalling [35]. Notably, instability associated with Pif1 deficiency exhibited an unexpected strand bias, and occurred if the G-rich strand was the template for leading (but not lagging) strand replication [34]. Helicases that unwind quadruplexes require an adjacent single-stranded region to load. The dependence of leading strand replication on Pif1, a 5′–3′ G4 helicase, may reflect the presence of exposed single-stranded region 5′ but not 3′ of a quadruplex structure on the leading strand, as new DNA synthesis may proceed to the very boundary of the quadruplex structure, thereby blocking the region where a 3′–5′ helicase would load [36].
G4 Motifs in Regulated Recombination: Immune Evasion and the Immune Response
The most detailed examination of the biological function of a specific G4 DNA structure has been carried out in the course of defining the mechanism of pathogenesis of Neisseria gonorrhoeae. This obligate pathogen evades the human immune response by varying expression of its cell surface pilin proteins. Antigen variation depends upon gene conversion at the active pilin expression locus pilE using a reservoir of silent pilS loci as sequence donors. A specific recombination activator element functions in cis to regulate gene conversion. This element is a G4 motif (G3TG3TTG3TG3), and the element must form G4 DNA to promote variation [37]. Quadruplex formation requires transcription of the activator element from a dedicated upstream promoter, which generates a noncoding transcript [38]. RecA recognition of the pilE quadruplex stimulates strand exchange, but other G4 motifs do not support regulated recombination at the pilE locus [39]. Thus, protein–quadruplex recognition can be highly selective.
There are clear mechanistic analogies between immune evasion by N. gonorrhoeae and immunoglobulin gene class switch recombination in the vertebrate immune response. Class switch recombination is targeted to S regions, 2 - to 8-kb repetitive G4hi motifs, and deletes a long region of genomic DNA to join a new constant region to the expressed variable region, thereby altering the mode of antigen clearance without affecting antigen recognition. Each S region has a dedicated promoter, and transcription through the S region is necessary to activate recombination and target it to specific S regions. Transcription of S regions (and other G4hi sequences) results in the formation of an unusual structure, a G-loop (Figure 2), containing a stable cotranscriptional RNA/DNA hybrid on the template strand and G4 DNA interspersed with single-stranded DNA on the nontemplate strand [40]–[42]. Quadruplexes formed in the transcribed S regions are the targets of factors that promote switch recombination, including BLM helicase, and MutSα (reviewed by [43]). MutSα also functions in telomere maintenance and in repair of DNA base mismatches and small loops. Human MutSα binds to quadruplexes in G-loops formed by transcribed S regions and can promote synapsis between distinct S regions in solution [44]. Escherichia coli MutS also binds quadruplexes [45], so quadruplex binding is a conserved property of this factor.
G4 Motifs in Genes and Transcripts
Characteristic Distribution of G4 Motifs within Genes
G4 motifs exhibit a characteristic distribution within human RefSeq genes (genomic sequences used as reference standards for well-characterized genes; http://www.ncbi.nlm.nih.gov/refseq/rsg/about/). Figure 3 diagrams a generic RefSeq gene, with the genomewide average of G4 motifs plotted relative to standard reference points including the transcription start site (TSS), 5′-UTR, exons, introns, and 3′-UTR. G4 motifs are enriched at the TSS, the 5′-UTR, and the 5′ end of the first intron, and depleted in coding regions. The coding regions of most genes are depleted for G4 motifs (G4lo), but some are enriched (G4hi) [46].
Fig. 3. G4 motif frequency in a generic human RefSeq gene. 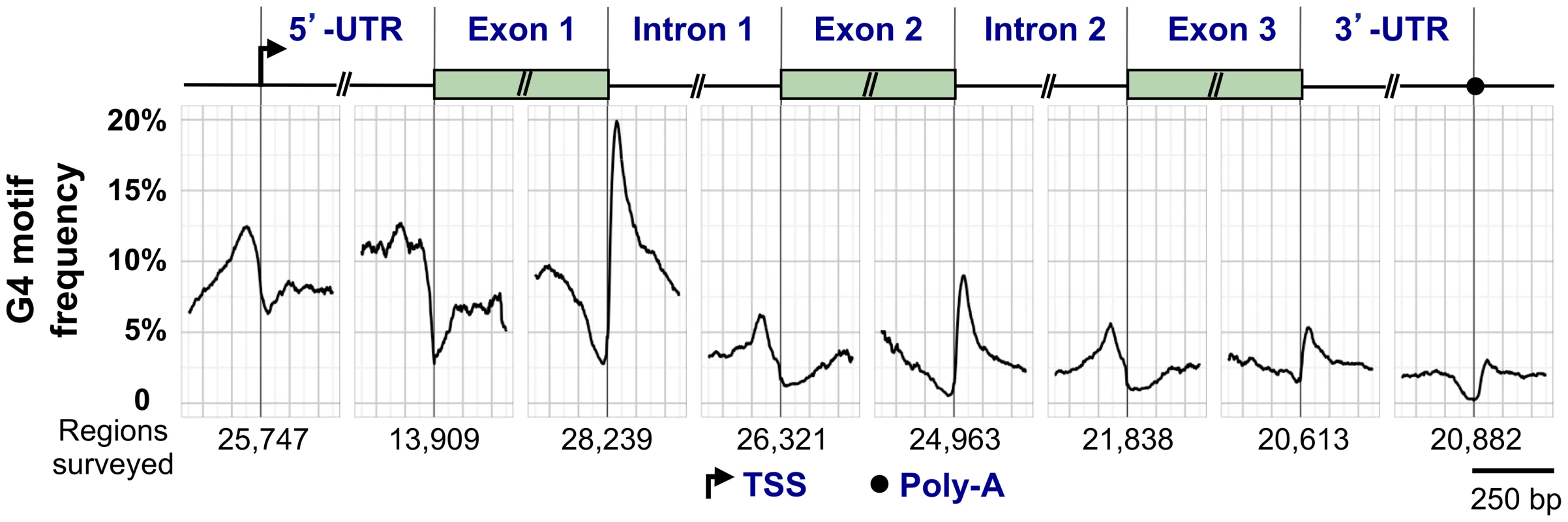
Above, key elements of a generic gene are shown, including the TSS, 5′-UTR, 5′ exons and introns, and the 3′-UTR and poly(A) signal. Below, the graph shows the frequency of G4 motifs in each region (loop size 1–12 nt). G4 motif frequency was calculated by counting the number of times G4 motifs overlapped each position, and dividing by the number of regions surveyed, which varied for each window. G4 locations at the 5′-UTR–exon 1 boundary were calculated only for genes with a 5′-UTR that did not span multiple exons. The regions flanking the TSS are G4hi [47]–[50]. The presence of G4 motifs in promoters of oncogenes, such as MYC and RAS, fueled efforts to develop small molecule ligands that would bind to a postulated quadruplex and downregulate gene expression [51]. However, it has proved challenging to develop a ligand specific for a single quadruplex in the genome of a human cell, and this drug development strategy has not yet been fruitful.
Current evidence for regulation by binding of factors to quadruplexes near the TSS is relatively limited. Analysis of genomewide associations by chromatin immunoprecipitation sequencing (ChIP-Seq) can establish protein enrichment at specific sequence motifs, and this has provided one line of evidence that promoter quadruplexes may regulate transcription. In those experiments, the ubiquitous transcription factor SP1, which binds a G-rich consensus motif GGGCGG in duplex DNA, was shown to bind G4 DNA in solution, and to preferentially associate with G4 motifs at promoter regions in living cells [52].
G4 motifs are enriched downstream of the TSS in the nontemplate DNA strand, where they may form quadruplexes in either the DNA or RNA [48]. Functions of these quadruplexes are potentially interesting, but need better definition. In 10%–15% of human genes, a G4 motif occurs in the region specifying the 5′-UTR of the encoded mRNA, leading to the suggestion that G4 RNA structures may promote translational repression (reviewed by [53]). At many human genes, RNA Pol2 initiates transcription but pauses near the 5′ end, and pausing correlates with enrichment of G4 motifs [50]. In nearly half of all human genes, one or more G4 motifs are present at the very 5′ end of the first intron, on the nontemplate strand, and will be transcribed into the encoded pre-mRNA [54]. The G4hi element in the first intron of the gene encoding the G4 helicase CHL1, has been shown to form a quadruplex structure not previously documented, and resembling the catalytic core of group I introns [55]. This RNA quadruplex may be the target of regulation, even by CHL1 itself, but this needs to be carefully studied.
Both quadruplexes and quadruplex binding proteins may function in 3′-end processing of mRNA transcripts. Cell stress regulates 3′-end processing of the P53 gene transcript, and altered 3′-end processing has been shown to depend upon recognition of a G4 RNA structure by hnRNP H/F [56]. More generally, RNA Pol2 must pause at a G4hi region just downstream of the poly(A) site to enable transcriptional termination at a subset of genes in normally proliferating cells, to enable 3′-end processing by XRN nuclease and the DNA/RNA helicase senataxin [57]. Deficiencies in senataxin are associated with two neurological diseases, ataxia oculomoter apraxia 2 and amyotrophic lateral sclerosis type 4.
Transcription-Induced Genomic Instability at R-Loops and G-Loops
Transcribed G4 motifs pose a special threat to genomic stability. Transcription of G-rich regions results in formation of R-loops, which are targets of transcription-associated genomic instability, as has been recently reviewed [58], [59]. Like G-loops (Figure 2), R-loops contain an RNA/DNA hybrid, but they do not necessarily contain the nontemplate strand G4 DNA that makes G-loops a target for some recombination factors [40]–[42]. Nonetheless, many of the regions that form cotranscriptional R-loops are G4hi, and the nontemplate strand is likely to contain quadruplexes that may contribute to biological function even if those structures are not explicitly acknowledged. For example, it has been reported that promoter regions that form R-loops are protected from de novo methylation at CpG dinucleotides [60]. The authors recognized that R-loop formation is a property of G-rich regions, but did not extend their analysis to include G4 motifs.
The contribution of transcription-induced G-quadruplexes with genomic instability has been established by genetic analyses, and by use of a G4 DNA ligand, pyridostatin [61]. Pyridostatin treatment of human cells resulted in transcription-dependent appearance of DNA damage markers, including γ-H2AX, and arrest in the G2 phase of the cell cycle. Genomewide analysis by ChIP-Seq showed that G4 motifs were enriched among sites of damage. Pyridostatin interacts with an exposed planar G-quartet rather than loop sequences in solution, suggesting that it might recognize a very broad spectrum of G4 structures in a living cell. However, damage induced by pyridostatin was restricted to a subset of G4hi genes—including the actively transcribed rDNA in the nucleolus, but not the telomeres—and the SRC (but not HRAS) oncogene [61]. This selectivity, which was not anticipated by biochemical characterization of pyridostatin, points to the importance of validating predicted cellular targets of reagents directed at quadruplexes.
Neurological Disease Associated with G4 Motif Repeat Expansions in Specific Genes
Expansions of G4 motifs in five different genes are associated with neurological disease (Table 1). Expansions in the FMR1 [62], C9orf72 [63]–[65], and NOP56 [66] genes produce pre-mRNAs that carry extended regions of quadruplex structures, which appear to titrate essential RNA binding proteins and impair mRNA processing. At the CTSB gene [67], the expanded CGCGGGGCGGGG repeat in the promoter is thought to promote excessive DNA methylation at CpG dinucleotides that downregulate gene expression.
Tab. 1. G4 motif expansions in neurological disease. 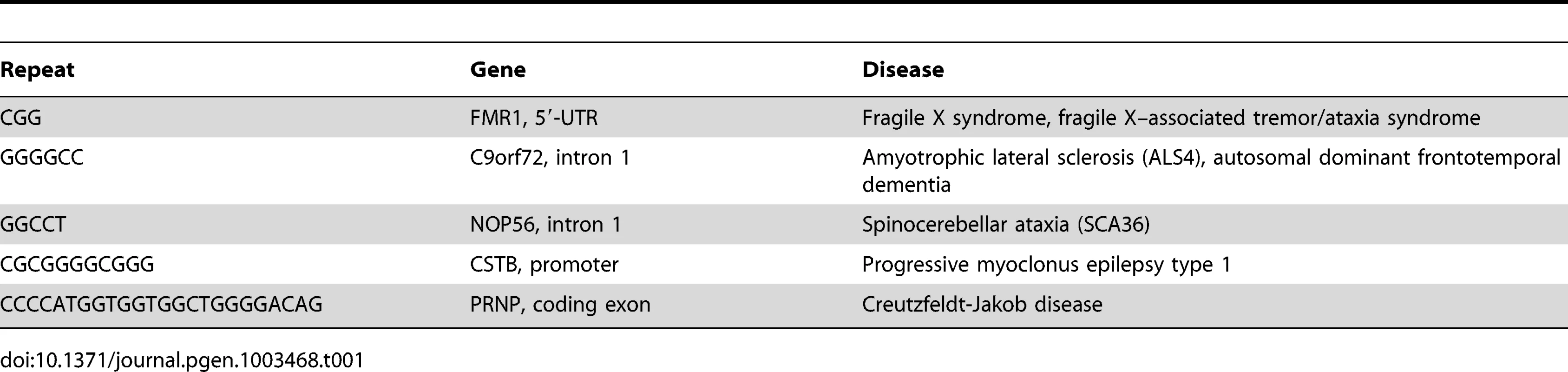
Especially intriguing is an expansion in the coding region of the PRNP gene (Table 1), which encodes the prion protein associated with Creutzfeldt-Jakob disease [68]. The normal prion protein contains five repeats of the sequence CCCCATGGTGGTGGCTGGGGACAG. Expansions, typically to 10–14 repeats, cause a dominant form of familial Creutzfeldt-Jakob disease exhibiting early onset and slow progression, which correlate with misfolding of the corresponding prion protein and formation of insoluble protein aggregates in solution [69].
The mechanisms that drive expansions of these G4 motifs have not been defined. Transcription may promote instability, as suggested by the inherent instability of regions prone to form persistent cotranscriptional RNA/DNA hybrids. Expansion may reflect mitotic instability, dramatically evident at G4hi VNTRs, which rank among the most unstable repeats in the human genome.
Epigenetic Instability at G4 Motifs
Unimpeded replication of G4 motifs is important not only in genetic stability but also in epigenetic stability. Deficiency in FANCJ, BLM, or WRN helicases can cause alterations of epigenetic modifications near G4 motifs, evident as increased expression of silenced genes or reduced expression of active genes [70], [71]. This may reflect replication slowdown at G4 motifs that prevents the local redeposition of marked histones necessary to maintain epigenetic status [72].
If G4 motifs do contribute to epigenetic regulation by enabling epigenetic marks to be reset upon replication, then genes that respond rapidly to external stimuli would be predicted to be G4hi. This is in fact the case. Genomewide analyses have shown that the G4hi or G4lo status of a gene extends throughout its length, including both exons and introns, and that it correlates with gene function [46]. G4hi genes include transcriptional activators, developmental regulators, and oncogenes, such as MYC, JUNB, FGF4, and TERT, which respond rapidly to developmental and environmental stimuli.
The possibility that G4 motifs may function as epigenetic regulatory elements is supported by analyses of ATRX, a SWI/SNF family member with robust G4 DNA binding activity [73]–[75]. ATRX is enriched at the G4hi telomeres and rDNA, and at G4 motifs elsewhere throughout the genome, including a polymorphic G4hi VNTR at the α-globin gene, CGCGGGGCGGGGG. Deficiencies in ATRX are associated with an X-linked genetic disease characterized by α-thalassemia and mental retardation. The severity of α-thalassemia resulting from ATRX deficiency correlates with α-globin VNTR length [75], recapitulating features of diseases associated with trinucleotide repeat instability [76].
Genomewide studies suggest that G4 motifs tend to be hypomethylated and depleted for nucleosomes, in normal cells and especially in human tumors, where hypomethylated G4 motifs predominate among sites of genomic instability leading to copy number variation [77]–[80]. The relatively relaxed state of hypomethylated chromatin may be conducive to quadruplex formation. Thus, epigenetic and genetic instability at G4 motifs may go hand in hand.
Future Challenges: Defining the G4 Genome
Which G4 Motifs Form Quadruplexes in Living Cells?
This question can be addressed by systematically defining genomewide targets of endogenous quadruplex binding proteins by ChIP-Seq, or by identifying targets of reagents with validated specificity for quadruplexes in living cells. Both approaches have been used in contexts noted above. For example, associations of SP1 in the human genome [52] and of Pif1 in the S. cerevisiae genome [35] have been mapped by ChIP-Seq, and the G4 ligands PhenDC3 and pyridostatin have been used to specifically exacerbate instability at quadruplexes in S. cerevisiae [33] and human cells [61]. Quite recently, a single chain antibody selective for quadruplexes in solution was shown to stain chromosomes in the nuclei of human cells [81]. If antibody specificity for quadruplexes in living cells can be validated (e.g., by ChIP-Seq), it will be a useful tool for studying the cell biology of quadruplexes.
How Are Specific Quadruplexes Recognized to Enlist Participation in Specific Pathways?
The diverse conformations of quadruplexes suggest that specific structural features may enable participation in specific pathways. To take one example, about half the G4 motifs in the human genome map to replication origins. What distinguishes those quadruplexes? Co-crystal or nuclear magnetic resonance structures of protein/quadruplex complexes are essential to identify the structural determinants that attract specific proteins to specific quadruplexes.
Zdroje
1. GellertM, LipsettMN, DaviesDR (1962) Helix formation by guanylic acid. Proc Natl Acad Sci U S A 48 : 2014–2018.
2. SenD, GilbertW (1988) Formation of parallel four-stranded complexes by guanine rich motifs in DNA and its implications for meiosis. Nature 334 : 364–366.
3. KimJ, CheongC, MoorePB (1991) Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature 351 : 331–332.
4. PhanAT, KuryavyiV, PatelDJ (2006) DNA architecture: from G to Z. Curr Opin Struct Biol 16 : 288–298.
5. PatelDJ, PhanAT, KuryavyiV (2007) Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res 35 : 7429–7455.
6. BesnardE, BabledA, LapassetL, MilhavetO, ParrinelloH, et al. (2012) Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 19 : 837–844.
7. TuesuwanB, KernJT, ThomasPW, RodriguezM, LiJ, et al. (2008) Simian virus 40 large T-antigen G-quadruplex DNA helicase inhibition by G-quadruplex DNA-interactive agents. Biochemistry 47 : 1896–1909.
8. NorseenJ, JohnsonFB, LiebermanPM (2009) Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J Virol 83 : 10336–10346.
9. ParkinsonGN, LeeMP, NeidleS (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417 : 876–880.
10. YuHQ, MiyoshiD, SugimotoN (2006) Characterization of structure and stability of long telomeric DNA G-quadruplexes. J Am Chem Soc 128 : 15461–15468.
11. HaiderSM, NeidleS, ParkinsonGN (2011) A structural analysis of G-quadruplex/ligand interactions. Biochimie 93 : 1239–1251.
12. LukeB, LingnerJ (2009) TERRA: telomeric repeat-containing RNA. EMBO J 28 : 2503–2510.
13. MartadinataH, HeddiB, LimKW, PhanAT (2011) Structure of long human telomeric RNA (TERRA): G-quadruplexes formed by four and eight UUAGGG repeats are stable building blocks. Biochemistry 50 : 6455–6461.
14. XuY, SuzukiY, ItoK, KomiyamaM (2010) Telomeric repeat-containing RNA structure in living cells. Proc Natl Acad Sci U S A 107 : 14579–14584.
15. DengZ, NorseenJ, WiedmerA, RiethmanH, LiebermanPM (2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35 : 403–413.
16. BiffiG, TannahillD, BalasubramanianS (2012) An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J Am Chem Soc 134 : 11974–11976.
17. de LangeT (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19 : 2100–2110.
18. SfeirA, KosiyatrakulST, HockemeyerD, MacRaeSL, KarlsederJ, et al. (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138 : 90–103.
19. SfeirA, de LangeT (2012) Removal of shelterin reveals the telomere end-protection problem. Science 336 : 593–597.
20. SmithJS, ChenQ, YatsunykLA, NicoludisJM, GarciaMS, et al. (2011) Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nat Struct Mol Biol 18 : 478–485.
21. SunH, KarowJK, HicksonID, MaizelsN (1998) The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem 273 : 27587–27592.
22. FryM, LoebLA (1999) Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem 274 : 12797–12802.
23. LondonTB, BarberLJ, MosedaleG, KellyGP, BalasubramanianS, et al. (2008) FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem 283 : 36132–36139.
24. WuY, Shin-YaK, BroshRMJr (2008) FANCJ helicase defective in Fanconi anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol 28 : 4116–4128.
25. WuY, SommersJA, KhanI, de WinterJP, BroshRMJr (2012) Biochemical characterization of Warsaw breakage syndrome helicase. J Biol Chem 287 : 1007–1021.
26. SandersCM (2010) Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem J 430 : 119–128.
27. DingH, SchertzerM, WuX, GertsensteinM, SeligS, et al. (2004) Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell 117 : 873–886.
28. CrabbeL, VerdunRE, HaggblomCI, KarlsederJ (2004) Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306 : 1951–1953.
29. BarefieldC, KarlsederJ (2012) The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res 40 : 7358–7367.
30. UringaEJ, YoudsJL, LisaingoK, LansdorpPM, BoultonSJ (2011) RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res 39 : 1647–1655.
31. VannierJB, Pavicic-KaltenbrunnerV, PetalcorinMI, DingH, BoultonSJ (2012) RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149 : 795–806.
32. KruisselbrinkE, GuryevV, BrouwerK, PontierDB, CuppenE, et al. (2008) Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol 18 : 900–905.
33. PiazzaA, BouleJB, LopesJ, MingoK, LargyE, et al. (2010) Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res 38 : 4337–4348.
34. LopesJ, PiazzaA, BermejoR, KriegsmanB, ColosioA, et al. (2011) G-quadruplex-induced instability during leading-strand replication. EMBO J 30 : 4033–4046.
35. PaeschkeK, CapraJA, ZakianVA (2011) DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145 : 678–691.
36. DavisL, MaizelsN (2011) G4 DNA: at risk in the genome. EMBO J 30 : 3878–3879.
37. CahoonLA, SeifertHS (2009) An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 325 : 764–767.
38. CahoonLA, SeifertHS (2013) Transcription of a cis-acting, noncoding, small RNA is required for pilin antigenic variation in Neisseria gonorrhoeae. PLoS Pathog 9: e1003074 doi:10.1371/journal.ppat.1003074.
39. KuryavyiV, CahoonLA, SeifertHS, PatelDJ (2012) RecA-binding pilE G4 sequence essential for pilin antigenic variation forms monomeric and 5′ end-stacked dimeric parallel G-quadruplexes. Structure 20 : 2090–2102.
40. DuquetteML, HandaP, VincentJA, TaylorAF, MaizelsN (2004) Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev 18 : 1618–1629.
41. DuquetteML, PhamP, GoodmanMF, MaizelsN (2005) AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene 24 : 5791–5798.
42. DuquetteML, HuberMD, MaizelsN (2007) G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res 67 : 2586–2594.
43. MaizelsN (2006) Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol 13 : 1055–1059.
44. LarsonED, DuquetteML, CummingsWJ, StreiffRJ, MaizelsN (2005) MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr Biol 15 : 470–474.
45. EhratEA, JohnsonBR, WilliamsJD, BorchertGM, LarsonED (2012) G-quadruplex recognition activities of E. Coli MutS. BMC Mol Biol 13 : 23.
46. EddyJ, MaizelsN (2006) Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res 34 : 3887–3896.
47. HuppertJL, BalasubramanianS (2007) G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res 35 : 406–413.
48. EddyJ, MaizelsN (2009) Selection for the G4 DNA motif at the 5′ end of human genes. Mol Carcinog 48 : 319–325.
49. DuZ, ZhaoY, LiN (2009) Genome-wide colonization of gene regulatory elements by G4 DNA motifs. Nucleic Acids Res 37 : 6784–6798.
50. EddyJ, VallurAC, VarmaS, LiuH, ReinholdWC, et al. (2011) G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res 39 : 4975–4983.
51. BalasubramanianS, HurleyLH, NeidleS (2011) Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov 10 : 261–275.
52. RaiberEA, KranasterR, LamE, NikanM, BalasubramanianS (2012) A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res 40 : 1499–1508.
53. BugautA, BalasubramanianS (2012) 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res 40 : 4727–4741.
54. EddyJ, MaizelsN (2008) Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res 36 : 1321–1333.
55. KuryavyiV, PatelDJ (2010) Solution structure of a unique G-quadruplex scaffold adopted by a guanosine-rich human intronic sequence. Structure 18 : 73–82.
56. DecorsiereA, CayrelA, VagnerS, MillevoiS (2011) Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev 25 : 220–225.
57. Skourti-StathakiK, ProudfootNJ, GromakN (2011) Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42 : 794–805.
58. AguileraA, Garcia-MuseT (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46 : 115–124.
59. KimN, Jinks-RobertsonS (2012) Transcription as a source of genome instability. Nat Rev Genet 13 : 204–214.
60. GinnoPA, LottPL, ChristensenHC, KorfI, ChedinF (2012) R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45 : 814–825.
61. RodriguezR, MillerKM, FormentJV, BradshawCR, NikanM, et al. (2012) Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol 8 : 301–310.
62. SantoroMR, BraySM, WarrenST (2012) Molecular mechanisms of Fragile X syndrome: a twenty-year perspective. Annu Rev Pathol 7 : 219–245.
63. DeJesus-HernandezM, MackenzieIR, BoeveBF, BoxerAL, BakerM, et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72 : 245–256.
64. FrattaP, MizielinskaS, NicollAJ, ZlohM, FisherEM, et al. (2012) C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep 2 : 1016.
65. RentonAE, MajounieE, WaiteA, Simon-SanchezJ, RollinsonS, et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72 : 257–268.
66. KobayashiH, AbeK, MatsuuraT, IkedaY, HitomiT, et al. (2011) Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am J Hum Genet 89 : 121–130.
67. BorelC, MigliavaccaE, LetourneauA, GagnebinM, BenaF, et al. (2012) Tandem repeat sequence variation as causative cis-eQTLs for protein-coding gene expression variation: the case of CSTB. Hum Mutat 33 : 1302–1309.
68. MeadS, WebbTE, CampbellTA, BeckJ, LinehanJM, et al. (2007) Inherited prion disease with 5-OPRI: phenotype modification by repeat length and codon 129. Neurology 69 : 730–738.
69. LehmannS, HarrisDA (1996) Two mutant prion proteins expressed in cultured cells acquire biochemical properties reminiscent of the scrapie isoform. Proc Natl Acad Sci U S A 93 : 5610–5614.
70. SarkiesP, ReamsC, SimpsonLJ, SaleJE (2010) Epigenetic instability due to defective replication of structured DNA. Mol Cell 40 : 703–713.
71. SarkiesP, MuratP, PhillipsLG, PatelKJ, BalasubramanianS, et al. (2011) FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res 40 : 1485–1498.
72. SarkiesP, SaleJE (2011) Propagation of histone marks and epigenetic memory during normal and interrupted DNA replication. Cell Mol Life Sci 69 : 697–716.
73. GoldbergAD, BanaszynskiLA, NohKM, LewisPW, ElsaesserSJ, et al. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140 : 678–691.
74. WongLH, McGhieJD, SimM, AndersonMA, AhnS, et al. (2010) ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res 20 : 351–360.
75. LawMJ, LowerKM, VoonHP, HughesJR, GarrickD, et al. (2010) ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 143 : 367–378.
76. MirkinSM (2007) Expandable DNA repeats and human disease. Nature 447 : 932–940.
77. WongHM, HuppertJL (2009) Stable G-quadruplexes are found outside nucleosome-bound regions. Mol Biosyst 5 : 1713–1719.
78. HalderK, HalderR, ChowdhuryS (2009) Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol Biosyst 5 : 1703–1712.
79. HalderR, HalderK, SharmaP, GargG, SenguptaS, et al. (2010) Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol Biosyst 6 : 2439–2447.
80. DeS, MichorF (2011) DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol 18 : 950–955.
81. BiffiG, TannahillD, McCafferty, SubramanianS (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nature Chem 5 : 182–186.
Štítky
Genetika Reprodukční medicína
Článek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



