-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
The fitness effect of mutations can be influenced by their interactions with the environment, other mutations, or both. Previously, we constructed 32 ( = 25) genotypes that comprise all possible combinations of the first five beneficial mutations to fix in a laboratory-evolved population of Escherichia coli. We found that (i) all five mutations were beneficial for the background on which they occurred; (ii) interactions between mutations drove a diminishing returns type epistasis, whereby epistasis became increasingly antagonistic as the expected fitness of a genotype increased; and (iii) the adaptive landscape revealed by the mutation combinations was smooth, having a single global fitness peak. Here we examine how the environment influences epistasis by determining the interactions between the same mutations in two alternative environments, selected from among 1,920 screened environments, that produced the largest increase or decrease in fitness of the most derived genotype. Some general features of the interactions were consistent: mutations tended to remain beneficial and the overall pattern of epistasis was of diminishing returns. Other features depended on the environment; in particular, several mutations were deleterious when added to specific genotypes, indicating the presence of antagonistic interactions that were absent in the original selection environment. Antagonism was not caused by consistent pleiotropic effects of individual mutations but rather by changing interactions between mutations. Our results demonstrate that understanding adaptation in changing environments will require consideration of the combined effect of epistasis and pleiotropy across environments.
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003426
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003426Summary
The fitness effect of mutations can be influenced by their interactions with the environment, other mutations, or both. Previously, we constructed 32 ( = 25) genotypes that comprise all possible combinations of the first five beneficial mutations to fix in a laboratory-evolved population of Escherichia coli. We found that (i) all five mutations were beneficial for the background on which they occurred; (ii) interactions between mutations drove a diminishing returns type epistasis, whereby epistasis became increasingly antagonistic as the expected fitness of a genotype increased; and (iii) the adaptive landscape revealed by the mutation combinations was smooth, having a single global fitness peak. Here we examine how the environment influences epistasis by determining the interactions between the same mutations in two alternative environments, selected from among 1,920 screened environments, that produced the largest increase or decrease in fitness of the most derived genotype. Some general features of the interactions were consistent: mutations tended to remain beneficial and the overall pattern of epistasis was of diminishing returns. Other features depended on the environment; in particular, several mutations were deleterious when added to specific genotypes, indicating the presence of antagonistic interactions that were absent in the original selection environment. Antagonism was not caused by consistent pleiotropic effects of individual mutations but rather by changing interactions between mutations. Our results demonstrate that understanding adaptation in changing environments will require consideration of the combined effect of epistasis and pleiotropy across environments.
Introduction
The extent to which mutations interact with their genetic background (epistasis) and the role such interactions play in evolution is not well understood [1], [2]. Initial expectations were that epistatic interactions, defined as non-additive interactions among mutations, were common, causing fitness landscapes to be rugged and limiting the number of selectively accessible mutational paths [3]. Although early work revealed few such interactions (reviewed in [4]), more recent studies of defined combinations of mutations have revealed abundant epistasis in a range of systems [5]–[17]. Studies that focused on interactions among beneficial mutations have often found a tendency for antagonism [18]–[21], which is consistent with epistasis having a predictable influence on the curvature of fitness peaks in constant environments [22], [23].
In addition to interactions within a genome, mutations can also interact with the external environment [24]–[29]. Moreover, phenotypic plasticity and epistasis can combine so that the fate of a mutation depends on both the environment and its genetic background [27], [30]. This kind of dependence can have important evolutionary consequences. For example, Wright considered how fluctuating conditions — most often population size, but also the external environment — could change the sign of epistatic interactions and allow populations to evolve along otherwise maladaptive paths [31]–[35].
Relatively few studies have examined how epistasis and plasticity combine to influence mutational effects. One study that did found that all of 18 transposon insertion mutations were affected by either epistasis or plasticity, with half being affected by both [28]. It remains unclear, however, how interactions between beneficial mutations, which might be expected to depend strongly on a particular selective environment, will be affected by changes in the external environment. Such an understanding is vital for addressing questions concerning the course of adaptation in fluctuating conditions. For example: can the magnitude and sign of epistasis change with the external environment? If so, are there any overarching features of mutation interactions, e.g., a tendency towards antagonism, that nevertheless remain consistent? A few studies have begun to address these questions by examining how epistasis between pairs of mutations changes with genomic [36], [37] and environmental [38] contexts.
Here, we expand this investigation of how the external environment affects epistatic interactions between five beneficial mutations that fixed in one population of a long-term Escherichia coli evolution experiment [19]. We first screened the response of the ancestor and the evolved genotype having all five mutations over a total of 1,920 external environments. Next, we measured the fitness of a set of 32 ( = 25) strains comprising all mutation combinations in the two environments with the most extreme opposing plasticity. These measurements allowed us to isolate effects of epistasis (GxG), interactions between mutations and the environment (GxE), and interactions between epistasis and the environment (GxGxE) on the fitness of defined genotypes. More generally, we investigated how the adaptive landscape, and the indirect consequences of each mutational step, might change with the external environment.
Results
Interactions between beneficial mutations and their external environment
To determine how phenotypic plasticity changes following an adaptive walk, we used Biolog plates to compare the respiration (a measure of catabolic activity) of the ancestor and the strain containing five beneficial mutations (hereafter, rtsgp, where each letter indicates a mutation in a gene or gene region as follows: r = rbs, t = topA, s = spoT, g = glmUS and p = pykF) in 1,920 different environments. On average, rtsgp exhibited enhanced respiration over the ancestor (mean = 84.37±1.39 (SEM) compared to 72.45±1.14 (SEM); paired t-test, t1919 = 26.28, P = <0.0001) with significant differences in 203 environments (see Materials and Methods for criteria), involving 171 gains of function and 32 losses of function (Table S1). These environments contained 28 alternative carbon sources, 34 alternative nitrogen sources, five alternative phosphate sources, ten nutritional supplements, and 126 “stressors” including antibiotics and other potentially toxic chemicals. Providing a useful control, one of the carbon sources in which the rtsgp strain had decreased respiration was D-ribose, which was expected due to a large deletion of the rbs operon in this strain [39]. We confirmed that measured respiration changes reflected growth rate changes in eight of the environments (six gains of function and two losses of function) by direct growth comparisons (Table S2).
We focused on two environments that revealed large differences in respiration between rtsgp and the ancestor to examine how genotype and environment interact to affect fitness. The largest relative increase in respiration was in the presence of EGTA, a Ca++/Mg++chelator [40]. The largest decrease was in the presence of guanazole, a ribonucleotide DP reductase inhibitor [41]. In direct fitness competitions comparing rtsgp to its ancestor, rtsgp was significantly more fit in the environment containing the original selection medium (DM25, a minimal salts medium supplemented with glucose) supplemented with EGTA than in the environment containing the selection medium alone (DM25+EGTA: fitness = 1.497±0.068 (95% CI); DM25 : 1.299±0.061 (95% CI), t9 = 4.973, P = 0.0008). By contrast, rtsgp was less fit in DM25 supplemented with guanazole than in the selection environment (DM25+guanazole: fitness of 1.116±0.029 (95% CI); DM25 : 1.299±0.061 (95% CI), t9 = −5.779, P = 0.0003).
To examine the underlying genetic basis of this phenotypic plasticity, we quantified the fitness effect of each individual mutation in all three environments. The relative fitness of three of the five mutations significantly depended on the external environment (rbs: F2,11 = 1.100, P = 0.367; topA: F2,8 = 15.506, P = 0.002; spoT: F2,10 = 31.389, P<0.0001; glmUS: F2,10 = 3.513, P = 0.070; pykF: F2,10 = 149.730, P<0.0001) (Figure 1). In summary, three of the individual mutations present in the rtsgp genotype produced effects that differed significantly across environments.
Fig. 1. Effects of beneficial mutations in alternative environments. 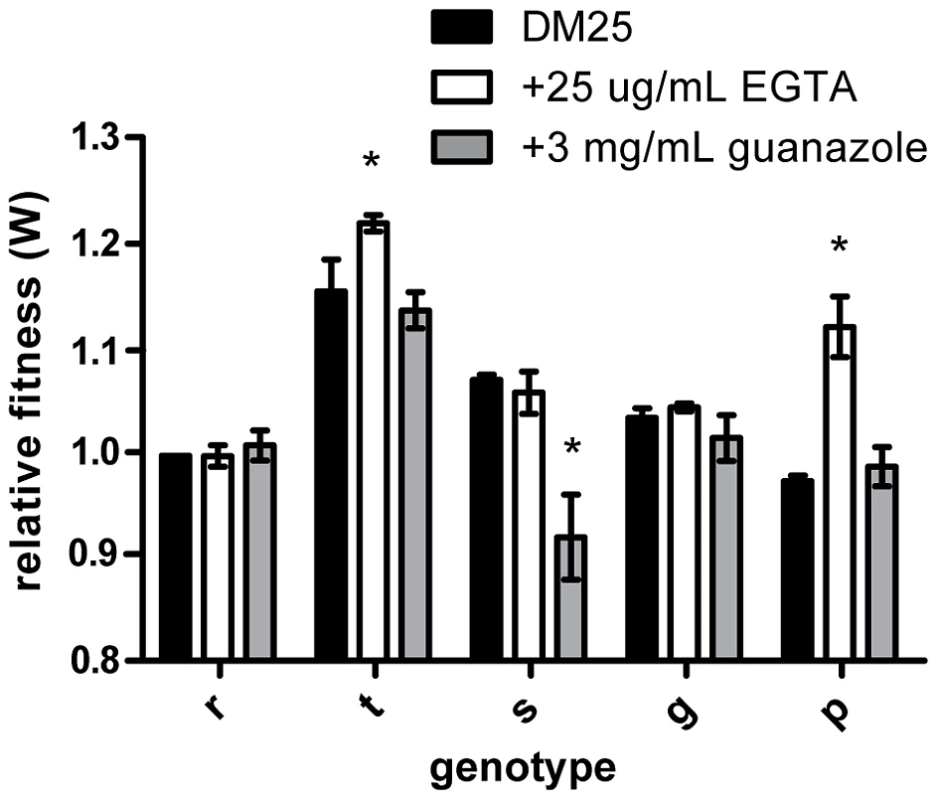
Genotypes are designated as single letters and define alleles: rbs (r), topA (t), spot (s), glmS (g), and pykF (p). Fill color defines the environment: black, DM25, white, EGTA, and grey, guanazole. Mutational effects were determined to depend on the environment using an ANOVA. Asterisks represent significance based on a P value < 0.05. Fitness varies across external environments due to plasticity and epistasis
The above results demonstrate that the effect of individual mutations depend on the environment (i.e., G×E). However, it is also possible that interactions between mutations depend on the environment (i.e., G×G×E), which would further influence the topology of the fitness landscape and make it much more difficult to predict the influence of environmental changes on evolutionary outcomes. To examine G×G×E we measured the fitness of all combinations of the five beneficial mutations in the two focal environments (i.e., the selection environment supplemented with EGTA and guanazole).
Fitness of each of the 32 genotypes comprising each mutation combination was quantified in both novel environments and compared with prior findings in the original selection environment [19] (Figure 2). To get some overall indication of the influence of environment on mutation effects we compare the number of “selectively accessible” mutational paths connecting the ancestor and rtsgp [42]. Although a different set of beneficial mutations would presumably be followed in guanazole and EGTA environments, considering a common set of genotypes allows a direct comparison of the effect of environment in altering selection pressures as a result of GxE and GxGxE. Of the 120 ( = 5!) paths connecting the ancestor and rtsgp, 86 had monotonically increasing fitness in the selection environment [19]. By contrast, only 43 paths in EGTA and 2 paths in guanazole are selectively accessible (Figure 2, Tables S3, S4). (The small number of selectively accessible paths in guanazole reflects, in large part, that rts and not rtsgp was the most fit genotype (Table S6).) In all, nine mutational steps became significantly deleterious, six in the EGTA environment and three in the guanazole environment (Tables S3 and S4), although only three of these steps in the EGTA environment remain significantly deleterious when we correct for multiple comparisons. Nevertheless, differences in the number of selectively accessible paths available in different environments clearly indicate that environment affects landscape topology and selective constraints.
Fig. 2. Fitness landscapes in the selection environment and in two novel environments. 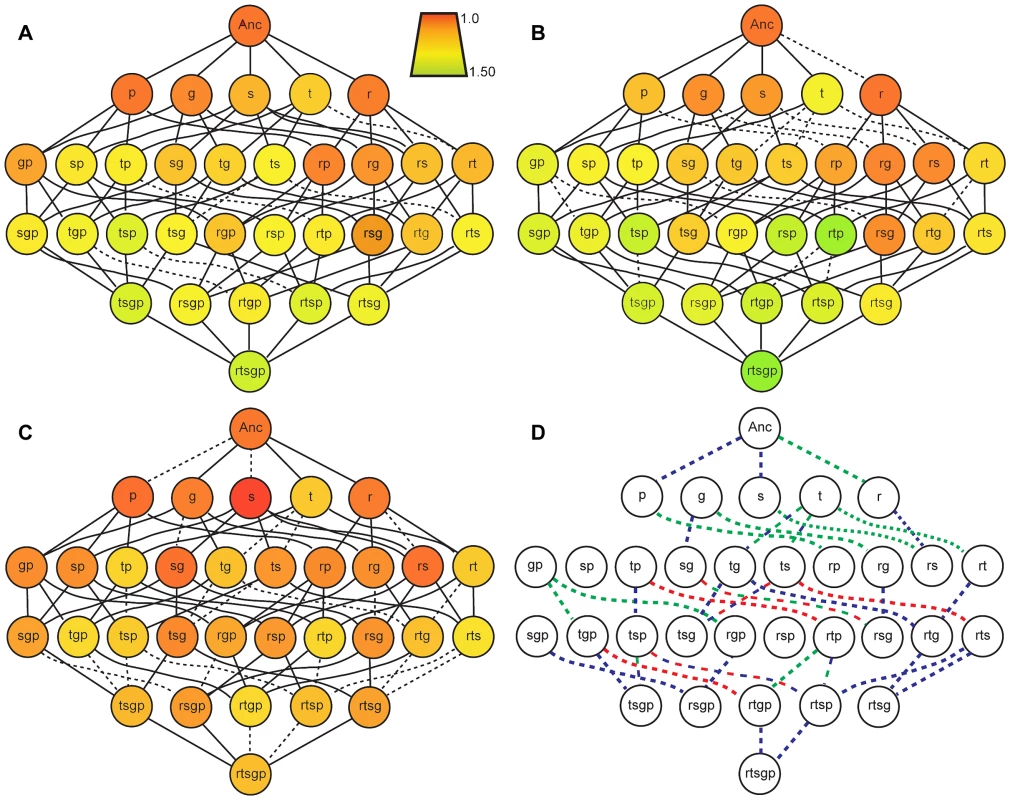
Genotypes are designated as single letters as defined previously. A, B, C: Node color indicates the fitness of a genotype relative to the ancestor in that environment. Solid lines indicate a selectively accessible step that increases genotype fitness and dashed lines indicate steps corresponding to fitness decreases. D: Colored dotted lines indicate selectively inaccessible mutational paths with a neutral or deleterious effects on fitness given mean relative fitness values (Tables S5 and S6); line color defines the environment: red, DM25; green, EGTA; blue, guanazole. DM25 data is adapted from Khan et al [19]. To further examine the patterns of epistasis in the novel environments we focused on the effect of epistasis in determining the fitness of individual genotypes (Tables S5, S6). In the EGTA environment mean epistasis was slightly, but not significantly, negative (mean absolute epistatic deviation, εm = −0.039±0.046 (95% C.I.), t25 = −1.740, P = 0.094) (Figure 3). In the guanazole environment mean epistasis was significantly positive (εm = 0.057±0.022 (95% C.I.), t25 = 5.303, P<0.0001) (Figure 3). In total, 16 and 5 genotypes exhibited significant epistasis in EGTA and guanazole, respectively (Tables S4, S5). Both environments also displayed markedly different effects of higher-order epistasis involving interactions between at least three mutations. In the EGTA environment, genotypes tended to be more fit than expected from the sum of the relevant lower-order interactions (mean higher-order epistatic deviation = 0.229±0.191 (95% C.I.), t15 = 2.556, P = 0.022). The opposite effect was seen in the guanazole environment (mean higher-order deviation = −0.247±0.196 (95% C.I.), t15 = 2.693, P = 0.017).
Fig. 3. Distributions of epistatic effects in two environments. 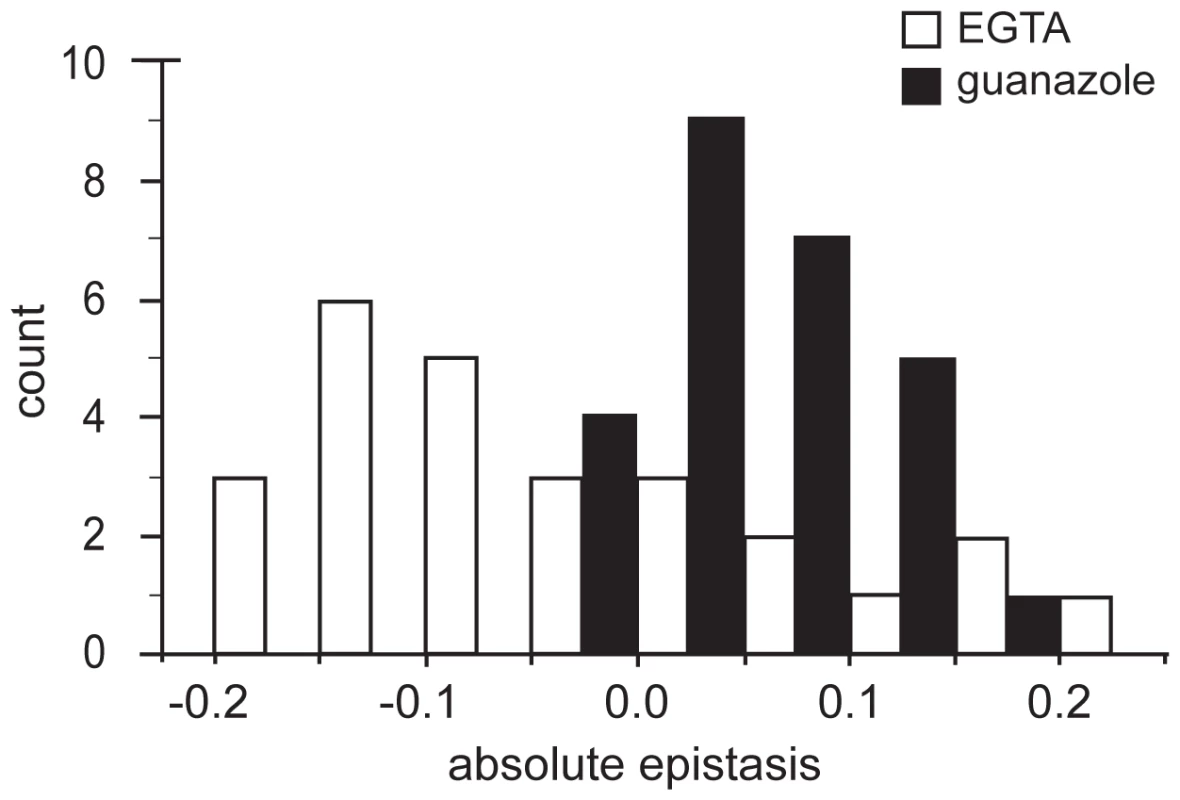
Observed and expected fitness were compared for 26 genotypes containing two or more mutations in two environments. Absolute epistasis was calculated as described in the text. Considering only the mean effect of epistasis can miss other underlying patterns. For example, we previously found that the strength of negative epistasis between the five beneficial mutations increased with the expected fitness of the genotype in the selection environment, despite a lack of any mean effect [19]. This pattern has been reported in several other studies [18], [20], [21] and is consistent with interactions between beneficial mutations acting to slow the rate of adaptation. In the guanazole environment we found the same negative correlation between epistasis and expected fitness that was seen in the selection environment (r = −0.748) (Figure 4, Figure S1, Table S5 and S6). The same correlation was only weakly negative in the EGTA environment (r = −0.281) (Figure 4 and Figure S2). We also evaluated whether interactions between mutations and the environment, either EGTA or guanazole, contributed significantly to the overall variation in fitness, and found significant interactions between mutations (GxG), mutations and the environment (GxE), and interactions of both (GxGxE) (overall model: F63,261 = 58.439, P<0.0001, Table S7, see Materials and Methods). Using variance partitioning, we determined that GxGxE interactions explained approximately 8% of the variance in fitness observed in our complete data set (Table S8).
Fig. 4. Relationship between relative epistasis and expected fitness assuming no epistasis in each foreign environment. 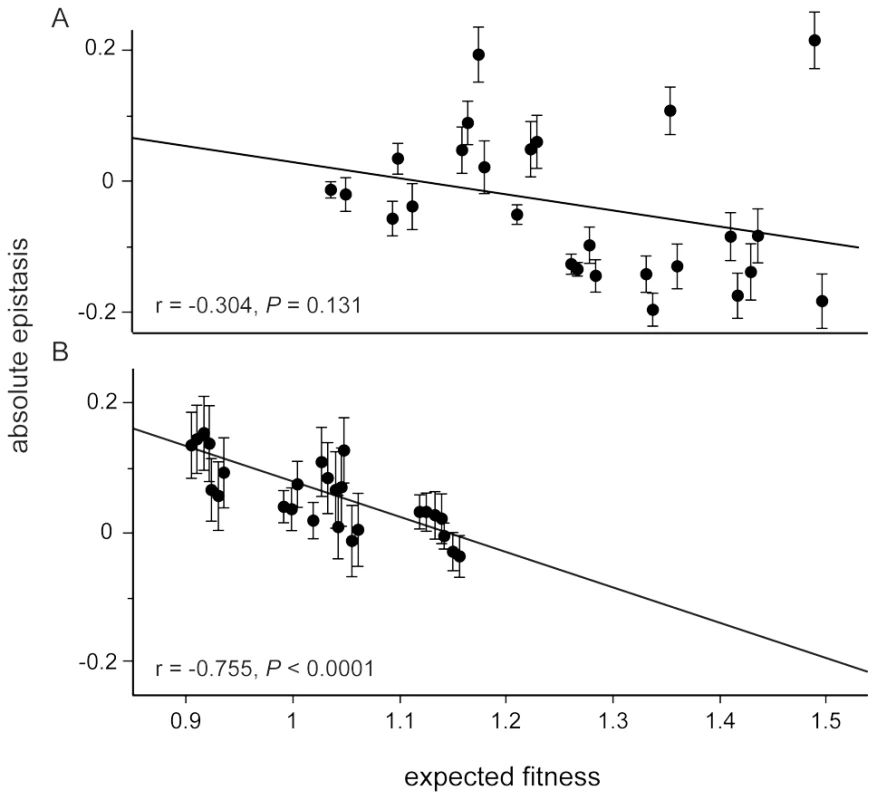
Each point refers to one of the 32 genotypes assayed for fitness in both environments (a, EGTA, b, guanazole). Error bars represent the standard deviation approximated through the method of error propagation. The solid lines are the best linear fit with the text below reporting the correlation (r) and significance (P values). Correlations between epistasis and expected fitness could reflect a general trend but could also be leveraged by outlying fitness or epistatic effects of an individual mutation. To distinguish between these possibilities we performed a series of ANCOVA analyses to test whether the presence or absence of each focal mutation influenced the overall relationship between epistasis and expected fitness (Figure S3 and S4). Only the pykF mutation explained a significant portion of the variation in the relationship between epistasis and expected fitness in the guanazole environment (Figure S3). Genotypes with this mutation tended to be more fit while the negative correlation, consistent with diminishing returns epistasis, with or without this mutation was retained.
In the EGTA environment, considering genotypes distinguished by the presence or absence of either topA or pykF mutations revealed their significant contributions to the overall pattern of epistasis (Figure S4). The topA mutation tended to effect epistasis so as to decrease fitness (mean epistasis of genotypes with topA = −0.088 compared to mean epistasis of genotypes without = 0.042, t24 = 3.272, P = 0.003) whereas pykF altered epistasis to generally increase genotype fitness (mean epistasis of genotypes with pykF = 0.007 compared to mean epistasis of genotypes without = −0.087, t24 = −2.156, P = 0.041). Genotypes lacking the rbs mutation again displayed a strong negative correlation between epistasis and expected fitness (r = −0.783), but adding the rbs mutation weakened the negative association between epistasis and expected genotype fitness without changing mean epistasis among these genotypes (Figure S4, genotypes with rbs, r = 0.089, compared to without P = 0.033; mean epistasis with rbs = 0.0001 compared to mean epistasis of genotypes without = −0.078, t24 = −1.720, P = 0.098).
Epistasis is common in a variety of external environments
We used a higher-throughput approach using overall population growth (AUC, see Materials and Methods) as a proxy for fitness to assay for epistasis in nine additional environments. Seven of these environments were not expected to interact with the five mutations based on the initial Biolog screen comparing the rtsgp and ancestral strains (Table S1, Materials and Methods). In each environment, the growth of each single mutant was compared with the ancestor, rtsgp, and a randomly selected double-mutant, gp (Figure 5). In seven of nine environments, growth of either rtsgp or gp differed significantly from additive expectations assuming no epistasis (Figure 5, Tables S9, S10). The nature of these interactions also changed with the environment. For example, gp was significantly less fit than expected in two environments and significantly more fit than expected in four environments (Table S9). In summary, the sign and magnitude of epistasis among generally beneficial mutations may vary widely even with relatively small changes in the external environment.
Fig. 5. The magnitude and direction of epistatic effects on growth vary with external environment. 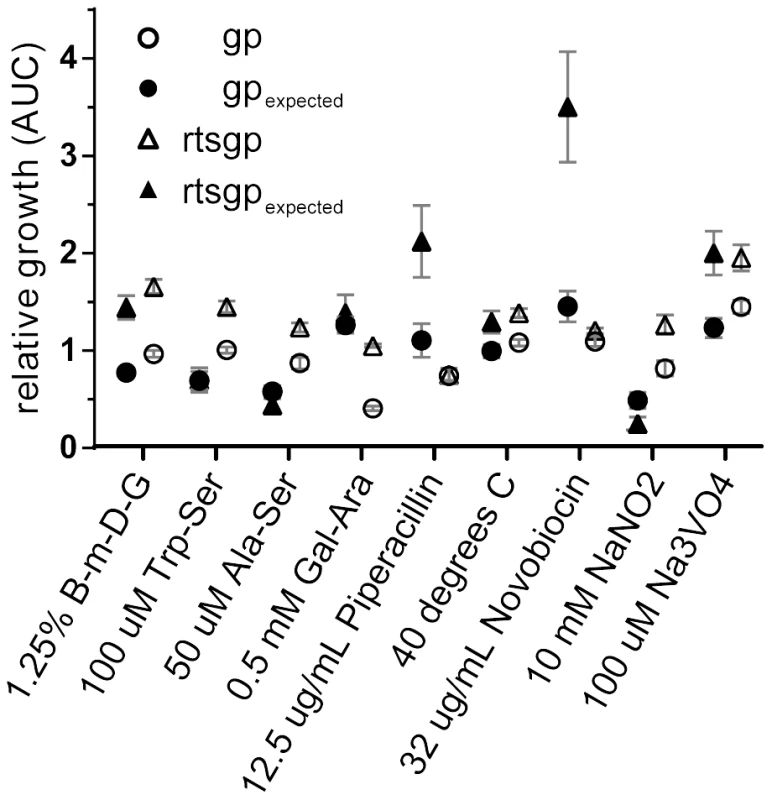
Symbols represent growth (AUC) of a particular genotype relative to the ancestor (triangles = gp, circles = rtsgp). Filled symbols represent expected relative growth and open symbols represent observed relative growth based on a multiplicative model assuming no epistatic interactions. Differences between observed and expected values were determined using the t statistic (Table S9 and S10). Discussion
Recent theoretical work has applied population genetic models to empirically constructed fitness landscapes to make basic predictions about the likelihood of particular evolutionary outcomes [8], [14]. These outcomes depend crucially on the shape of the fitness landscape, which is determined by the form and extent of epistatic interactions between mutations. How sensitive these interactions, and therefore the repeatability of evolutionary outcomes, are to environmental change remains uncertain. To address this point experimentally we analyzed a set of strains including all combinations of the first five beneficial mutations that fixed during the adaptation of a population of E. coli to a constant laboratory environment (Table 1, [19]). By measuring the fitness of these strains in contrasting environments we generated two new empirical fitness landscapes that reveal how epistasis may change with the environmental context. Comparing these landscapes to the one determined in the original selection environment, we found interactions between mutations and their environment to be both common and complex.
Tab. 1. The five mutations in the order in that they arose and fixed in a population of E. coli from Lenski et al. 2002. 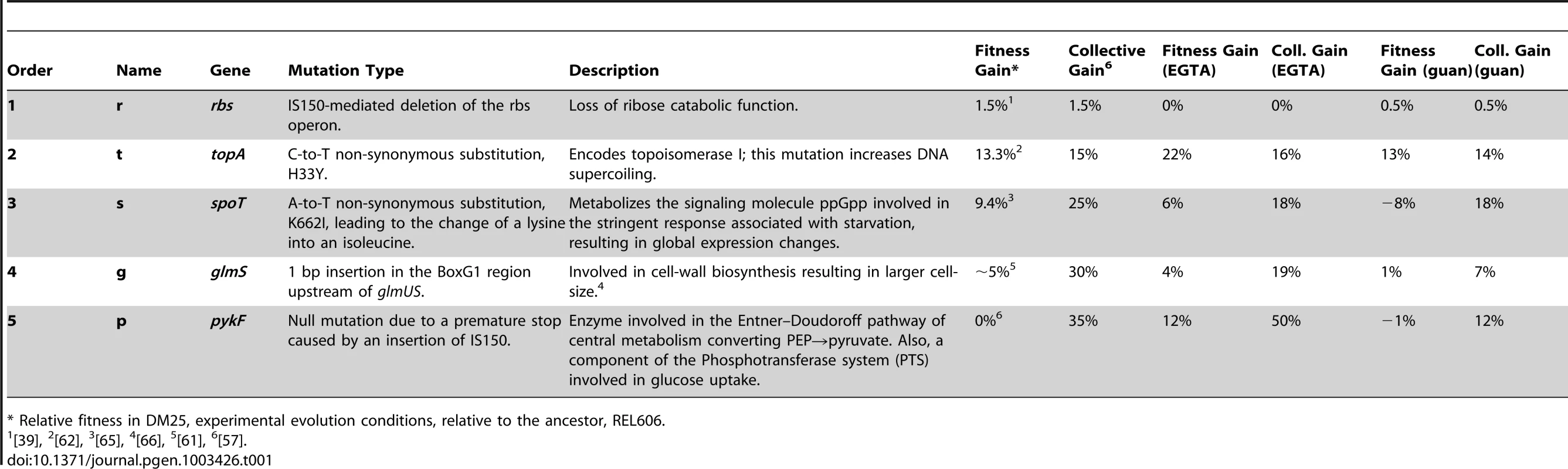
Relative fitness in DM25, experimental evolution conditions, relative to the ancestor, REL606. Previous work has shown that the diet breadth of 12 E. coli populations, including the population that was the source of the mutations used in our experiments, declined substantially during long-term evolution in a constant environment with a single carbon source [39], [43], [44]. However, it is difficult to distinguish if this trend was caused by few mutations of strong pleiotropic effect or if the beneficial substitutions display antagonistic pleiotropy in general. In an effort to distinguish these explanations, one study specifically focused on pleiotropic effects of beneficial mutations in five different environments. Mutations that were beneficial in the selected environment tended to be beneficial in others, and although there were exceptions, limited antagonistic pleiotropy was observed [45], [46]. Here, we also report limited antagonistic pleiotropy with five beneficial mutations with an increased sample size of 1,920 environments from our initial Biolog screen (Table S1). This result supports the inference that antagonistic effects may be limited to a subset of beneficial variation. Since both studies focused on a collection of beneficial mutations contributing to initial adaptation to a minimal glucose environment, we speculate that early adaptation may be characterized by niche expansion with limited cost [47].
Epistasis was frequent in all environments and generally followed a pattern of diminishing returns. Nevertheless, both the individual effects of mutations and their interactions were environmentally dependent, in several cases resulting in mutations changing from being beneficial to deleterious or neutral (Figure 1, Figure 4, Figure S2 and S3). Perhaps most strikingly, different numbers of paths to the rtsgp genotype were found in each environment, one of which featured a different global peak. Our results also suggest that selective constraints in fluctuating environments may depend on how the environment influences epistasis between contending adaptive alleles, and not just the pleiotropic effects of individual mutations alone (Table S5). For example, the topA and glmS mutations were more beneficial in the EGTA environment alone (positive pleiotropy) (Figure 1), but in combination the tg genotype was much less fit than expected (Table S4). Since the fitness of this genotype did not significantly deviate from expectations in the guanazole environment (Table S5), environmental effects on epistasis (GxGxE) were not predicted by GxE interactions. More broadly, these results indicate that variable fitness of a genotype under different conditions can arise from altered interactions among the alleles comprising that genotype and not from any single mutation. This conclusion is robust to fitness measurements in environments not found to effect rtsgp respiration, suggesting that it is not dependent on our initial focus on the two environments in which GxE was most extreme (Figure 5).
These interactions may be especially important in determining evolutionary outcomes given initially rugged fitness landscapes [14], [48], [49] or in naturally variable environments. In one study [17], a more beneficial allele was eventually outcompeted by a less fit allele because of epistatic limits to the adaptive path of the former allele. However, a different outcome may have occurred in a fluctuating or seasonal external environment. Given prevalent genotype-by-environment interactions, epistatic interactions producing low fitness intermediates could be alleviated in alternative environments and allow new combinations of alleles to overcome evolutionary dead-ends and rise to fixation. This process could represent a mechanism for maintaining conditional, yet beneficial, variation in the population [50]–[52]. As evidence, the three significantly maladaptive steps in the EGTA environment are alleviated by a shift to either the guanazole or the original selection environment (Figure 2). Fluctuating environments may therefore provide a solution to evolutionary dead-ends in an inherently rugged fitness landscape.
In summary, the combination of phenotypic plasticity and epistasis can strongly influence how an organism adapts to a new environment. Although the five mutations examined here would not likely be the same favored in these new environments, our results demonstrate that epistatic interactions are not static and can determine which trajectories are selectively accessible during an adaptive walk in a fluctuating environment. As a result, the fate of a mutation depends on its individual effect, epistasis with preexisting mutations and on interactions with the prevailing environment. With growing opportunities to survey dynamics of many genotypes within evolving populations, studies of both inherent properties of individual alleles and effects of their interactions in multiple conditions would address how frequently pleiotropy and epistasis guide adaptive evolution.
Materials and Methods
Bacterial strains and growth conditions
Twelve populations of E. coli have been propagated for more than 50,000 generations in Davis Minimal (DM) medium supplemented with 25 µg/ml glucose (DM25) in a long-term evolution experiment studying the dynamics and genetic basis of adaptation [43], [53]–[59]. Mutations identified in one of these populations, as described previously, are studied here [39], [57], [60]–[62]. Other types of media used in this study include Tryptic soy (Tsoy) broth, tetrazolium-arabinose (TA) agar plates, and DM media supplemented with sugars other than glucose or with glucose and additional compounds. E. coli strains were grown in rich Tsoy liquid media overnight from −80°C freezer stocks. Aliquots of overnight culture were transferred to 10 mL DM25 media to precondition the cultures for 24 hours prior to growth curves or fitness assays.
Biolog phenotypic microarrays
To identify external environments that interact with the five mutations (Table 1), the respiration of the rtsgp genotype was compared to the ancestor of the long-term evolution experiment, REL606, using Biolog's Phenotypic Microarray Services in duplicate (Biolog, Hayward CA). This method utilizes a high-throughput approach to compare respiration of two strains in 1,920 different environments, consisting of a variety of carbon, nitrogen, phosphorous and sulfur sources, differences in pH, and an assortment of chemical agents that target a variety of cellular processes. This approach uses the reduction of a tetrazolium dye as a terminal electron acceptor to assess respiratory activity. The amount of respiration was quantified by the extent of color production taking readings every 15 mins and graphed as a kinetic response curve. Incubation, recording and quality control analysis of PM plates 1–20 were performed by Biolog staff using an OmniLog instrument. Relative respiration in each environment was compared using the average height of the kinetic response curves (h). The two strains were considered to have differential growth in an environment if h differed by more than 3 standard deviations of the means of h for both strains.
Since differences in respiration do not necessarily reflect differences in growth or fitness, growth rates and in some cases relative fitness (see below) of the rtsgp strain was compared with the ancestral strain in a variety of these external environments to confirm that respiration was representative of growth or fitness. These follow-up growth rate assays were confirmatory and qualitative, not quantitative (Table S2).
Fitness assays
The fitness of each constructed strain was determined relative to the ancestor by direct competitions as described previously [53]. Briefly, competitions were typically carried out at 37°C in 10 mL of DM25, the same medium used in the original long-term evolution experiment, in 50 mL flasks with 10 mL beakers as covers. For some competitions glucose was replaced with another carbon source (β-methyl-D-glucoside) or supplemented with another compound at various concentrations (all others). These compounds and concentrations were as follows: 1.25% β-methyl-D-glucoside, 0.5 mM 3-0-β-D-galactopyranosyl-D-arabinose, 50 µM Ara-Ser, 3 mg/mL guanazole, 25 µg/ml EGTA, 100 µM Trp-Ser, 12 µg/mL piperacillin, 100 µM sodium orthovanadate, 32 µg/mL novobiocin, and 10 mM sodium nitrite. The constructed strains were competed against a marked Ara+ ancestral strain (REL607) that is able to utilize the sugar arabinose. The arabinose utilization phenotype was found to be neutral in each of these competitions but allowed for the two different cell types to be easily distinguishable on TA agar plates.
Competitors were pre-conditioned in the medium used for the competition for 24 hours prior to all competitions. Each competitor was then standardized based on OD600 values and added to the competition environment. Competitions were typically carried out for three days with a 1∶100 mixture transferred to fresh media every 24 hours. Since the fitness effect of some mutations was small, multiday fitness assays were used to amplify subtle advantages. Mixtures of competing strains were plated on TA agar at the start and end of each competition to determine fitness. Relative fitness (w) was calculated as the ratio of natural logarithms of realized growth by each competitor over three days of competition. Assays were typically carried out with five-fold replication and no less than three-fold replication.
The fitness values of genotypes in the selective environment assayed in our lab were generally lower than previously reported in a study carried out at the University of Houston with these strains [19]. We do not know the reason for this discrepancy, though lab-specific differences in fitness effects, for example due to differences in water source, have been seen previously [63]. We also tested whether different preconditioning methods influenced the outcome of these fitness assays (that is, preconditioning cultures in the original evolution environment (DM25) or under competition conditions). We found no significant difference in the fitness of two genotypes, tp and sgp, when competed against the ancestor under either preconditioning method (tp, F2,6 = 0.667, P = 0.244; sgp, F2,6 = 0.047, P = 0.258). Notwithstanding the difference, relative features of the fitness landscape do not seem to have changed (all five beneficial mutations remained beneficial in the selective environment (DM25) (Figure 1)). We note also that the analyses reported in this work generally consider the fitness effects of genotypes within a single environment or across two novel environments (DM25 glucose supplemented with EGTA or guanazole) used in the experiments carried out at the University of New Hampshire. Importantly, the key result observed in the dataset reported by Khan et al. [19], that epistasis was negatively correlated with expected fitness, is also seen in the work presented here.
Fitness epistasis
Relative fitness, w, was calculated as described above based on the change in the relative density of strains in direct competition with one another. The terms that we use to describe and quantify epistasis were adopted from da Silva et al. [15]. The effect of the interactions among adaptive mutations on relative fitness was calculated as absolute epistasis:
where is the set of mutations, is the fitness of the genotype with the entire set of mutations, and is the relative fitness of a mutant with mutation from that set. The null model assumes no interactions and under this model the fitness of a combination of beneficial mutations is equal to the product of the fitness of those mutations individually. We refer to this null hypothesis as the expected fitness of any combination of mutations. Any significant difference between the observed and expected fitness of a genotype indicates the presence of epistatic interactions. Moreover, the sign of the absolute epistasis is important, suggesting either a negative or positive interaction on the fitness of the genotype.Genotypes consisting of more than two adaptive mutations were further analyzed for net higher-order epistatic interactions, defined as epistasis that occurs between three or more mutations that cannot be explained as the result of constituent lower-order interactions. As a result, net higher-order epistasis was calculated by subtracting the effect of lower-order interactions as shown in equation 2,
where represents the number of mutations present and represents the fitness of a subset of the mutations present. We used this combination of methods to determine what types of interactions are most important in producing the observed phenotypes.Given the error inherent to calculations of expected fitness and hence , we used the method of error propagation to approximate the error of both parameters [64]. Since expected fitness of a particular genotype is equal to the product of the fitness of those mutations individually, the error () is calculated from the sum of the relative errors of the individual mutations as shown in equation 3,
where is the standard deviation of single-mutation fitnesses present. Since the uncertainty of ε depends on both and , the error of is the summation of the uncertainty of both as shown in equation 4, Epistasis was considered significant using a t-test with the t-statistic calculated as the ratio of the mean relative fitness to its standard deviation and the degrees of freedom based on the number of replicate assays to determine significance (Table S5 and S6).Growth curves
To identify potential epistatic interactions among the five beneficial mutations in different environments, growth over 24 hours was quantified for the constructed strains containing only one of the five beneficial mutations and compared to both the ancestral strain and the constructed strain containing all five mutations, rtsgp. Cells were grown in 200 µL of DM25 media in 96-well plates with 12 replicates per strain. Relative growth was quantified as AUC based on OD600 measured every 15 minutes for 24 hours, compared to the ancestor, REL606, and averaged across replicates. Average relative growth of genotypes containing only a single mutation were then used to calculate an expected additive value for gp and rtsgp assuming no epistatic interactions between mutations. The error for expected values was approximated using the method of error propagation described above. Observed and expected relative growth for both gp and rtsgp was compared in each environment using a t-test with the t-statistic calculated as the ratio of the mean relative growth to its standard deviation and the degrees of freedom based on the number of replicate assays to determine significance (Table S9, S10).
Supporting Information
Zdroje
1. de VisserJAGM, CooperTF, ElenaSF (2011) The causes of epistasis. Proceedings of the Royal Society B: Biological Sciences 278 : 3617–3624.
2. ØstmanB, HintzeA, AdamiC (2012) Impact of epistasis and pleiotropy on evolutionary adaptation. Proceedings of the Royal Society B: Biological Sciences 279 : 247–256.
3. WrightS (1930) Review of fisher (1930). Journal of Heredity 21 : 349–356.
4. HillWG, GoddardME, VisscherPM (2008) Data and Theory Point to Mainly Additive Genetic Variance for Complex Traits. PLoS Genet 4: e1000008 doi:10.1371/journal.pgen.1000008.
5. PoonA, ChaoL (2005) The rate of compensatory mutation in the DNA bacteriophage phiX174. Genetics 170 : 989–999.
6. LeeYH, LMDS, FoxGE (1997) Equally parsimonious pathways through an RNA sequence space are not equally likely. J Mol Evol 45 : 278–284.
7. LozovskyER, ChookajornT, BrownKM, ImwongM, ShawPJ, et al. (2009) Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc Natl Acad Sci U S A 106 : 12025–12030.
8. de VisserJA, ParkSC, KrugJ (2009) Exploring the effect of sex on empirical fitness landscapes. Am Nat 174 Suppl 1: S15–30.
9. SanjuanR, CuevasJM, MoyaA, ElenaSF (2005) Epistasis and the adaptability of an RNA virus. Genetics 170 : 1001–1008.
10. Maisnier-PatinS, AnderssonDI (2004) Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res Microbiol 155 : 360–369.
11. TongAH, LesageG, BaderGD, DingH, XuH, et al. (2004) Global mapping of the yeast genetic interaction network. Science 303 : 808–813.
12. SegreD, DelunaA, ChurchGM, KishonyR (2005) Modular epistasis in yeast metabolism. Nat Genet 37 : 77–83.
13. Van DriesscheN, DemsarJ, BoothEO, HillP, JuvanP, et al. (2005) Epistasis analysis with global transcriptional phenotypes. Nat Genet 37 : 471–477.
14. WeinreichDM, DelaneyNF, DepristoMA, HartlDL (2006) Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312 : 111–114.
15. da SilvaJ, CoetzerM, NedellecR, PastoreC, MosierDE (2009) Fitness epistasis and constraints on adaptation in a human immunodeficiency virus type 1 protein region. Genetics 185 : 293–303.
16. LunzerM, GoldingGB, DeanAM (2010) Pervasive cryptic epistasis in molecular evolution. PLoS Genet 6: e1001162 doi:10.1371/journal.pgen.1001162.
17. WoodsRJ, BarrickJE, CooperTF, ShresthaU, KauthMR, et al. (2011) Second-order selection for evolvability in a large Escherichia coli population. Science 331 : 1433–1436.
18. ChouHH, ChiuHC, DelaneyNF, SegreD, MarxCJ (2011) Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332 : 1190–1192.
19. KhanAI, DinhDM, SchneiderD, LenskiRE, CooperTF (2011) Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332 : 1193–1196.
20. MacLeanRC, PerronGG, GardnerA (2010) Diminishing returns from beneficial mutations and pervasive epistasis shape the fitness landscape for rifampicin resistance in Pseudomonas aeruginosa. Genetics 186 : 1345–1354.
21. RokytaDR, JoyceP, CaudleSB, MillerC, BeiselCJ, et al. (2011) Epistasis between beneficial mutations and the phenotype-to-fitness Map for a ssDNA virus. PLoS Genet 7: e1002075 doi:10.1371/journal.pgen.1002075.
22. KryazhimskiyS, TkacikG, PlotkinJB (2009) The dynamics of adaptation on correlated fitness landscapes. Proc Natl Acad Sci U S A 106 : 18638–18643.
23. MartinG, ElenaSF, LenormandT (2007) Distributions of epistasis in microbes fit predictions from a fitness landscape model. Nat Genet 39 : 555–560.
24. AlizonS, van BaalenM (2005) Emergence of a convex trade-off between transmission and virulence. Am Nat 165: E155–167.
25. FrankSA (1996) Models of parasite virulence. Q Rev Biol 71 : 37–78.
26. KimSC, RiesebergLH (2001) The contribution of epistasis to species differences in annual sunflowers. Mol Ecol 10 : 683–690.
27. KishonyR, LeiblerS (2003) Environmental stresses can alleviate the average deleterious effect of mutations. J Biol 2 : 14.
28. RemoldSK, LenskiRE (2004) Pervasive joint influence of epistasis and plasticity on mutational effects in Escherichia coli. Nat Genet 36 : 423–426.
29. Wagner A (2005) Robustness and evolvability in living systems. Princeton University Press
30. CooperTF, LenskiRE, ElenaSF (2005) Parasites and mutational load: an experimental test of a pluralistic theory for the evolution of sex. Proc R Soc Lond B Biol Sci 272 : 311–317.
31. CoyneJA, BartonNH, TurelliM (1997) Perspective: A critique of Sewall Wright's shifting balance theory of evolution. Evolution 51 : 643–671.
32. WadeMJ, GoodnightCJ (1998) ‘Perspective: The Theories of Fisher and Wright in the Context of Metapopulations: When Nature Does Many Small Experiments’. Evolution 52 : 1537–1548.
33. CoyneJA, BartonNH, TurelliM (2000) Is Wright's shifting balance process important in evolution? Evolution 54 : 306–317.
34. GoodnightCJ, WadeMJ (2000) The ongoing synthesis: a reply to Coyne, Barton, and Turelli. Evolution 54 : 317–324.
35. WrightS (1988) Surfaces of Selective Value Revisited. The American Naturalist 131 : 115–123.
36. WangY, Díaz ArenasC, StoebelDM, CooperTF (2012) Genetic background affects epistatic interactions between two beneficial mutations. Biology Letters
37. PearsonVM, MillerCR, RokytaDR (2012) The Consistency of Beneficial Fitness Effects of Mutations across Diverse Genetic Backgrounds. PLoS ONE 7: e43864 doi:10.1371/journal.pone.0043864.
38. LalicJ, ElenaSF (2013) Epistasis between mutations is host-dependent for an RNA virus. Biol Lett 9 : 20120396.
39. CooperVS, SchneiderD, BlotM, LenskiRE (2001) Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. Journal of Bacteriology 183 : 2834–2841.
40. RileyWWJr, PfeifferDR (1986) Rapid and extensive release of Ca2+ from energized mitochondria induced by EGTA. J Biol Chem 261 : 28–31.
41. BrockmanRW, ShaddixS, LasterWRJr, SchabelFMJr (1970) Inhibition of ribonucleotide reductase, DNA synthesis, and L1210 leukemia by guanazole. Cancer Res 30 : 2358–2368.
42. WeinreichDM (2005) The rank ordering of genotypic fitness values predicts genetic constraint on natural selection on landscapes lacking sign epistasis. Genetics 171 : 1397–1405.
43. CooperVS, LenskiRE (2000) The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407 : 736–739.
44. CooperVS (2002) Long-term experimental evolution in Escherichia coli. X. Quantifying the fundamental and realized niche. BMC Evol Biol 2 : 12.
45. OstrowskiEA, RozenDE, LenskiRE (2005) Pleiotropic effects of beneficial mutations in Escherichia coli. Evolution 59 : 2343–2352.
46. OstrowskiEA, WoodsRJ, LenskiRE (2008) The genetic basis of parallel and divergent phenotypic responses in evolving populations of Escherichia coli. Proc Biol Sci 275 : 277–284.
47. DuffyS, TurnerPE, BurchCL (2006) Pleiotropic costs of niche expansion in the RNA bacteriophage phi 6. Genetics 172 : 751–757.
48. WhitlockMC (1995) Multiple fitness peaks and epistasis. Annual Review of Ecology and Systematics 26 : 601–629.
49. RozenDE, HabetsMG, HandelA, de VisserJA (2008) Heterogeneous adaptive trajectories of small populations on complex fitness landscapes. PLoS ONE 3: e1715 doi:10.1371/journal.pone.0001715.
50. ElenaSFL, R.E (1997) Long-term experimental evolution in Escherichia coli. VII. Mechanisms maintaining genetic variability within populations. Evolution 51 : 1058–1067.
51. RaineyPB, TravisanoM (1998) Adaptive radiation in a heterogeneous environment. Nature 394 : 69–72.
52. HedrickPW (1999) Perspective: Highly Variable Loci and Their Interpretation in Evolution and Conservation. Evolution 53 : 313–318.
53. LenskiRE (1991) Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138 : 1315–1341.
54. LenskiRE, TravisanoM (1994) Dynamics of adaptation and diversification: A 10,000-generation experiment with bacterial populations. Proc Natl Acad Sci USA 91 : 6808–6814.
55. LenskiRE (2004) Phenotypic and genomic evolution during a 20,000 generation experiment with the bacterium Escherichia coli. Plant Breeding Reviews 24 : 225–265.
56. PapadopoulosD, SchneiderD, Meier-EissJ, ArberW, LenskiRE, et al. (1999) Genomic evolution during a 10,000-generation experiment with bacteria. Proc Natl Acad Sci USA 96 : 3807–3812.
57. SchneiderD, DuperchyE, CoursangeE, LenskiRE, BlotM (2000) Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156 : 477–488.
58. SniegowskiPD, GerrishPJ, LenskiRE (1997) Evolution of high mutation rates in experimental populations of E. coli. Nature 387 : 703–705.
59. TravisanoM, VasiF, LenskiRE (1995) Long-term experimental evolution in Escherichia coli. III. Variation among replicate populations in correlated responses to novel environments. Evolution 49 : 189–200.
60. CooperTF, RozenDE, LenskiRE (2003) Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci U S A 100 : 1072–1077.
61. StanekMT, CooperTF, LenskiRE (2009) Identification and dynamics of a beneficial mutation in a long-term evolution experiment with Escherichia coli. BMC Evol Biol 9 : 302.
62. CrozatE, PhilippeN, LenskiRE, GeiselmannJ, SchneiderD (2005) Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics 169 : 523–532.
63. O'KeefeKJ, MoralesNM, ErnstbergerH, BenoitG, TurnerPE (2006) Laboratory-dependent bacterial ecology: a cautionary tale. Appl Environ Microbiol 72 : 3032–3035.
64. KuH (1966) Notes on the use of propagation of error formulas. J Research of National Bureau of Standards-C Engineering and Instrumentation 70C: 263–273.
65. PelosiL, KuhnL, GuettaD, GarinJ, GeiselmannJ, et al. (2006) Parallel changes in global protein profiles during long-term experimental evolution in Escherichia coli. Genetics 173 : 1851–1869.
66. LenskiRE (2000) Cell size, shape, and fitness in evolving populations of bacteria. Scaling in Biology 221–235.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání







