-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
Vertebrate limb outgrowth is driven by a positive feedback loop that involves Sonic hedgehog (Shh) and Gremlin1 (Grem1) in the posterior limb bud mesenchyme and Fibroblast growth factors (Fgfs) in the overlying epithelium. Proper spatio-temporal control of these signaling activities is required to avoid limb malformations such as polydactyly. Here we show that, in Tbx2-deficient hindlimbs, Shh/Fgf4 signaling is prolonged, resulting in increased limb bud size and duplication of digit 4. In turn, limb-specific Tbx2 overexpression leads to premature termination of this signaling loop with smaller limbs and reduced digit number as phenotypic manifestation. We show that Tbx2 directly represses Grem1 in distal regions of the posterior limb mesenchyme allowing Bone morphogenetic protein (Bmp) signaling to abrogate Fgf4/9/17 expression in the overlying epithelium. Since Tbx2 itself is a target of Bmp signaling, our data identify a growth-inhibiting positive feedback loop (Bmp/Tbx2/Grem1). We propose that proliferative expansion of Tbx2-expressing cells mediates self-termination of limb bud outgrowth due to their refractoriness to Grem1 induction.
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003467
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003467Summary
Vertebrate limb outgrowth is driven by a positive feedback loop that involves Sonic hedgehog (Shh) and Gremlin1 (Grem1) in the posterior limb bud mesenchyme and Fibroblast growth factors (Fgfs) in the overlying epithelium. Proper spatio-temporal control of these signaling activities is required to avoid limb malformations such as polydactyly. Here we show that, in Tbx2-deficient hindlimbs, Shh/Fgf4 signaling is prolonged, resulting in increased limb bud size and duplication of digit 4. In turn, limb-specific Tbx2 overexpression leads to premature termination of this signaling loop with smaller limbs and reduced digit number as phenotypic manifestation. We show that Tbx2 directly represses Grem1 in distal regions of the posterior limb mesenchyme allowing Bone morphogenetic protein (Bmp) signaling to abrogate Fgf4/9/17 expression in the overlying epithelium. Since Tbx2 itself is a target of Bmp signaling, our data identify a growth-inhibiting positive feedback loop (Bmp/Tbx2/Grem1). We propose that proliferative expansion of Tbx2-expressing cells mediates self-termination of limb bud outgrowth due to their refractoriness to Grem1 induction.
Introduction
Polydactyly, the condition of having more than the normal number of toes and/or fingers is the most frequent form of limb malformation in humans with an incidence of 1∶500. It most commonly occurs as post-axial polydactyly (extra digit(s) towards 5th finger in limb), less common is pre-axial polydactyly (extra digit(s) towards thumb or toe), very rare is central (mesoaxial) polydactyly (with extra digits within the three middle digits). In all cases, the excess digits can be undeveloped and only attached by a little stalk mostly on the small finger side of the hand or fully formed and working. Polydactyly can occur by itself, or more commonly, as one feature of a syndrome of congenital anomalies as e.g. in Pallister-Hall syndrome, Smith-Lemli-Opitz syndrome or Bardet-Biedl syndrome (OMIM 146510, 270400, 209900).
Elucidation of the genetic, molecular and cellular changes that underlie polydactyly (as well as other limb defects) in humans has greatly benefitted from the analysis of normal limb development, and of the consequences of altered gene functions in suitable animal models such as the chicken and the mouse [1]. All of these studies unraveled that proper establishment and elaboration of the two main limb axes during development is crucial for setting up a correct number and identity of digits. In the limb primordium two signaling centers control the morphogenesis along these limb axes. The apical ectodermal ridge (AER), a distal thickening of the ectodermal jacket of the limb bud, controls proximal-distal (from shoulder to finger tips), whereas the zone of polarizing activity (ZPA) in the posterior region of the mesenchymal core mediates anterior-posterior (A-P, from thumb to the small finger) development. Fibroblast growth factors (Fgf4, Fgf8, Fgf9 and Fgf17) and Shh secreted from the AER and ZPA, respectively, specify distal and posterior positional values in the early limb bud mesenchyme (in the mouse until E9.5) [2], [3]. Removal of the signaling centers or the signals, results in time-dependent distal truncations [4] and loss of posterior limb positions (ulna and digits 2–5) [5], respectively. However, both AER-FGFs and ZPA-Shh do not only provide patterning functions, they also account for the massive outgrowth of the limb bud at subsequent stages (from E9.5 to 11.5 in the mouse) by promoting cell survival and proliferative expansion of the distal and posterior limb bud mesenchyme, respectively [6], [7]. During this phase the three primordia of the stylopod (the future upper arm/leg), the zeugopod (lower arm/leg) and the autopod (hand/foot) are laid down and expanded and digit formation is initiated. It has long been noted that AER and ZPA are mutually dependent on each other, thus linking the two signaling centers by an epithelial-mesenchymal (e-m) signaling loop [8], [9], [10]. Fgf signaling is likely to maintain Shh directly, whereas Shh signaling to the overlying AER is relayed in the posterior limb bud mesenchyme by Gremlin 1 (Grem1), a secreted antagonist of Bone morphogenetic protein (Bmp) signaling [11], [12], [13]. Inhibition of Bmp signaling allows Fgf4 expression in the posterior AER enabling the further propagation of the loop. Limb bud outgrowth comes to a halt (around E11.5 to E12.0 in the mouse) when Fgf4 and Shh expression is shut-off due to a rise of Bmp signaling, resulting in cell-cycle exit and initiation of chondrogenic differentiation [10], [14]. Although the precise mechanisms are unclear, refractoriness of ZPA descendants to induce Grem1 may be of critical importance to self-terminate the e-m signaling loop [15].
Tbx2 encodes a transcriptional repressor of the T-box gene family that has recently been implicated in digit development. In the mouse, Tbx2 is expressed in the anterior and posterior mesenchymal flanks of the early limb bud. From E11.5 on, the posterior domain of Tbx2 extends more distally and is then found in the interdigital mesenchyme (IDM), most prominently in IDM4, and at the distal tips of the digit condensates at E12.5 [16] (Figure S1). Limbs of Tbx2-deficient mice exhibit a hindlimb-specific duplication of digit 4 [17]. The spatially restricted nature of this phenotype may be due to redundancy with the closely related Tbx3 gene. While both genes are coexpressed in the proximal mesenchyme on either limb margin, Tbx3 is absent from the distal mesenychme of the posterior flank [16] (Figure S1). Exclusive expression of Tbx3 in the AER may relate to the variable distal truncations and oligodactyly observed in Tbx3−/− embryos [18]. Retroviral mis - and overexpression experiments in the chick model provided phenotypic outcomes that suggested additional or alternative functions for Tbx2 (and Tbx3) in regulating digit identity rather than digit number [19] and in anterior-posterior positioning of the limb bud [20].
Here, we set out to gain further insight into the role of Tbx2 in digit development by genetic loss - and gain-of-function experiments in the mouse. We show that increased digit number in Tbx2-mutant mice and oligodactyly in embryos overexpressing Tbx2 (in the limbs) relates to the maintenance of the e-m feedback loop in the posterior limb bud mesenchyme. Tbx2 terminates this loop by locally repressing Grem1. Our experiments identify a Bmp/Tbx2/Grem1 loop that counteracts the Shh/Grem1/Fgf4 loop to mediate self-termination of limb bud outgrowth.
Results
Etiology of digit 4 duplication in Tbx2-mutant hindlimbs
Mice homozygous for a Tbx2 null allele (Tbx2cre) maintained on a NMRI outbred background died shortly after birth due to craniofacial defects [21]. Mutant E18.5 embryos had normal forelimbs but hindlimbs displayed six instead of the normal five digits. Soft tissue webbing, i.e. the persistence of IDM tissue was not observed (Figure 1B). Hindlimb polydactyly showed 70% penetrance in homozygous embryos, heterozygous embryos were not affected. Skeletal preparations of hindlimbs of E18.5 Tbx2−/− embryos revealed that the proximal segment of digit 4 was broadened and split distally to connect to a duplicated pair of second and third phalangeal segments (Figure 1D). Analysis of chondrogenic elements at E15.5 and E13.5 showed that the duplication of skeletal elements of digit 4 was established shortly after onset of chondrogenesis in the mutant hindlimb (Figure 1H and 1L). Comparative expression analysis for the (pre-)chondrogenic marker gene Sox9 [22] and Raldh2, which marks the IDM [23], showed that the occurrence of duplicated distal cartilagenous segments of digit 4 was preceded by a posterior expansion of the prechondrogenic condensate at the expense of the adjacent IDM4 at E12.5 (Figure 1M–1P).
Fig. 1. Duplication of digit 4 is preceded by reduced apoptosis and expanded chondrogenesis in the posterior limb mesenchyme. 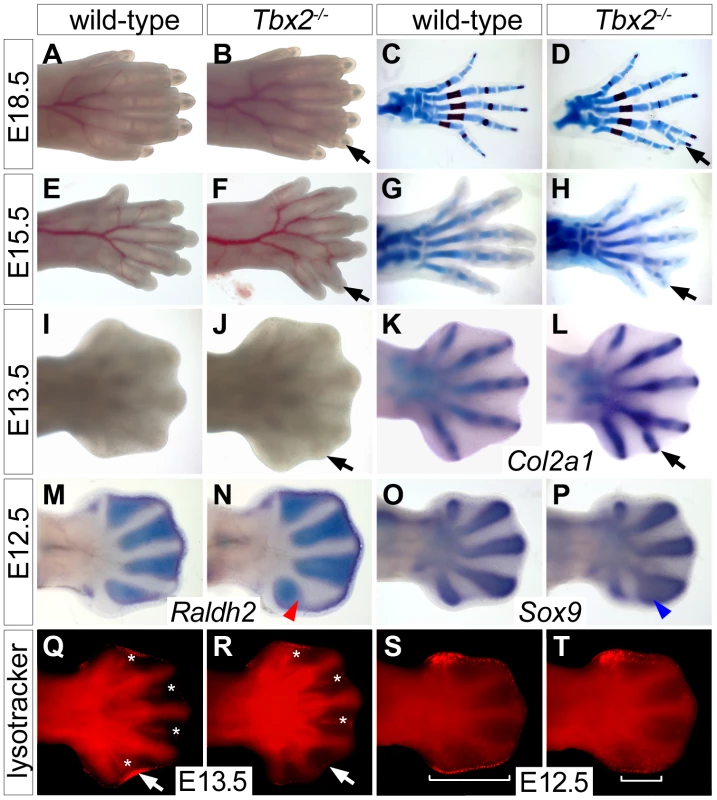
(A–L) Morphology (A, B, E, F, I, J), Alcian blue/Alizarin red skeletal preparations (C, D, G, H) and Col2a1 in situ hybridization (K, L) in wild-type and Tbx2−/− hindlimbs at different stages. Black arrows point to the duplicated digit 4. (M–P) Whole mount in situ hybridization analysis for Raldh2 and Sox9 expression in E12.5 wild-type and Tbx2−/− hindlimbs. An expansion of the digit 4 condensation (blue arrowhead) and a reduction of IDM4 (red arrowhead) are observed. (Q–T) Detection of programmed cell death (bright red signals) by Lysotracker Red staining. (Q, R) Asterisks mark interdigital, arrows sub-AER apoptosis. (S, T) Brackets indicate the spatial extent of apoptosis at the posterior limb bud margin. Since Tbx2 expression in the developing limb is confined to flank and ID mesenchyme that are removed by programmed cell death [24], we determined the distribution of apoptotic cells by Lysotracker Red staining. We noted a specific reduction of apoptotic cells in IDM4 of Tbx2−/− hindlimbs at E13.5, whereas other domains of programmed cell death (IDM1-3) were unchanged (Figure 1Q and 1R). In E12.5 wild-type embryos, apoptosis was confined to the mesenchyme underlying the AER at the anterior and posterior margins of the hindlimb (Figure 1S). Programmed cell death appeared unaffected in the anterior region whereas it was largely reduced in the posterior mesenchymal region of mutant hindlimbs at this stage (Figure 1T). The latter may explain the increased size and the characteristic outward curvature of the posterior edge of the mutant hindlimb. Together with the strong expression of Tbx2 in the distal region of the posterior flank mesenchyme at E11.5 and in IDM4 at E12.5 (Figure S1), this suggests that Tbx2 functions after E11.5 in the development of the posterior autopod region to maintain IDM4 fate and to restrict the expansion of chondrogenic material at the posterior limit of the digit 4 condensation.
The signaling loop between the AER and the underlying mesenchyme is disturbed in the posterior region of Tbx2-deficient hindlimbs
Proliferative expansion and apoptotic removal of the limb bud mesenchyme is governed by reciprocal signaling with the overlying AER [1]. Given the increased posterior size and the decreased apoptosis of E12.5 Tbx2-deficient hindlimbs, we studied the expression of factors that are involved in e-m signaling before the manifestation of morphological defects. In E10.5 hindlimb buds expression of Fgf4, Fgf8, Fgf9, Fgf17 and Shh was unaffected (Figure 2A–2E). At E11.5, however, we found increased expression of Fgf4 (in 7/8 embryos), Fgf9 (in 3/3 embryos) and Fgf17 (in 2/3 embryos) in the posterior AER of Tbx2−/− hindlimbs (Figure 2F–2H). At E12.5, Fgf4 expression was no longer detected in wild-type hindlimbs, but residual AER expression of Fgf4 was observed in 3/8 mutant embryos (Figure 2K). Shh was unchanged at E11.5 (Figure 2I), but expression was aberrantly maintained at E12.5 (in 5/5 embryos) (Figure 2N). Fgf8, which is continuously expressed in the entire AER, was unaffected in Tbx2-deficient hindlimbs at all analyzed stages (Figure 2E, 2J and 2O, and data not shown). Together we conclude that Tbx2 is required to assure correct termination of the Fgf4/9/17-Shh signaling loop in the posterior mesenchyme of the hindlimb bud.
Fig. 2. Prolonged posterior e-m signaling in Tbx2-deficient hindlimbs. 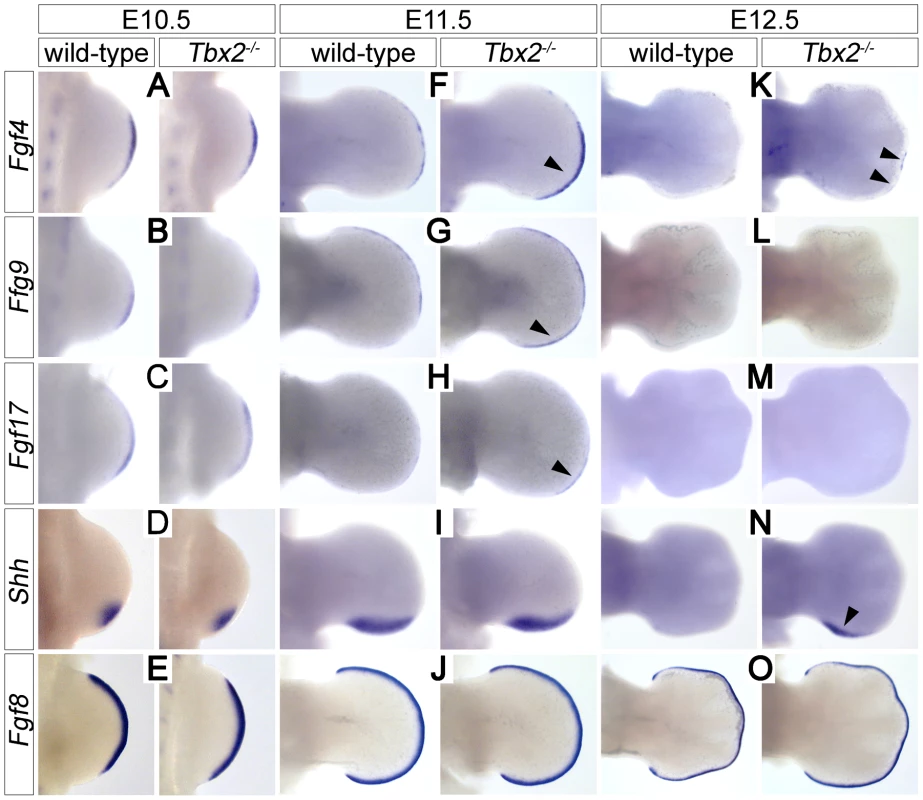
Whole-mount in situ hybridization analysis of Fgf4, Fgf9, Fgf17, Shh and Fgf8 expression in E10.5 (A–E), E11.5 (F–J) and E12.5 (K–O) wild-type and Tbx2−/− hindlimbs. (F, G, H) Arrowheads show prolonged AER expression of Fgf4/9/17 in the posterior limb bud at E11.5. (K, N) Arrowheads show maintenance of Fgf4 and Shh expression at E12.5 in Tbx2−/− hindlimbs. Fgf8 expression is unchanged in Tbx2-mutant limb buds. Polydactyly in Tbx2-deficient mice is ameliorated by reduced mesenchymal Fgf-signaling
To explore if a gain in e-m signaling indeed causes polydactyly in Tbx2-deficient hindlimbs, we genetically reduced the level of mesenchymal Fgf-signaling. We took advantage of cre recombinase driven from our Tbx2 mutant allele to delete a floxed allele of the Fgf-receptor 1 gene in the posterior limb bud mesenchyme. Double heterozygous animals (Tbx2cre/+;Fgfr1fl/+) were intercrossed and the limb skeleton of compound mutants was analyzed. Homozygous loss of Fgfr1 (Tbx2cre/+;Fgfr1fl/fl) caused a reduced number of 4 digits in fore - and hindlimbs (Figure 3F). This result is consistent with previous experiments using the Shh-cre line that recombines in a domain very similar to that of Tbx2cre [7], [25] (Figure S2A–S2D). At E16.5 Tbx2cre/cre;Fgfr1fl/+ embryos were underrepresented and Tbx2cre/cre;Fgfr1fl/fl embryos were absent, most likely due to a synthetic lethal cardiac defect. Double heterozygous Tbx2cre/+;Fgfr1fl/+ embryos exhibited a normal limb skeleton (Figure 3B) in agreement with previous results using the Prrx1-cre line that deletes in the entire limb bud mesenchyme [25], [26]. In Tbx2-deficient embryos, however, dose-reduction of Fgfr1 strongly reduced the penetrance of polydactyly (3/7 compared to 7/10 in Tbx2cre/cre;Fgfr1wt/wt embryos) or even caused oligodactyly (4 digits in 3/7 embryos analyzed) (Figure 3D and 3E). In oligodactic limbs the characteristic shape of the proximal end of the posterior metacarpal indicated a digit 5 identity (white arrows). Absence of Eomes expression in E12.5 Tbx2cre/cre;Fgfr1fl/fl hindlimbs argued for a specific loss of digit 4 (Figure S2E, S2F) [27]. Our results suggest that polydactyly in Tbx2-deficient mice critically depends on increased activity of the e-m signaling loop.
Fig. 3. Polydactyly in Tbx2-deficient limbs depends on the Fgf signaling level. 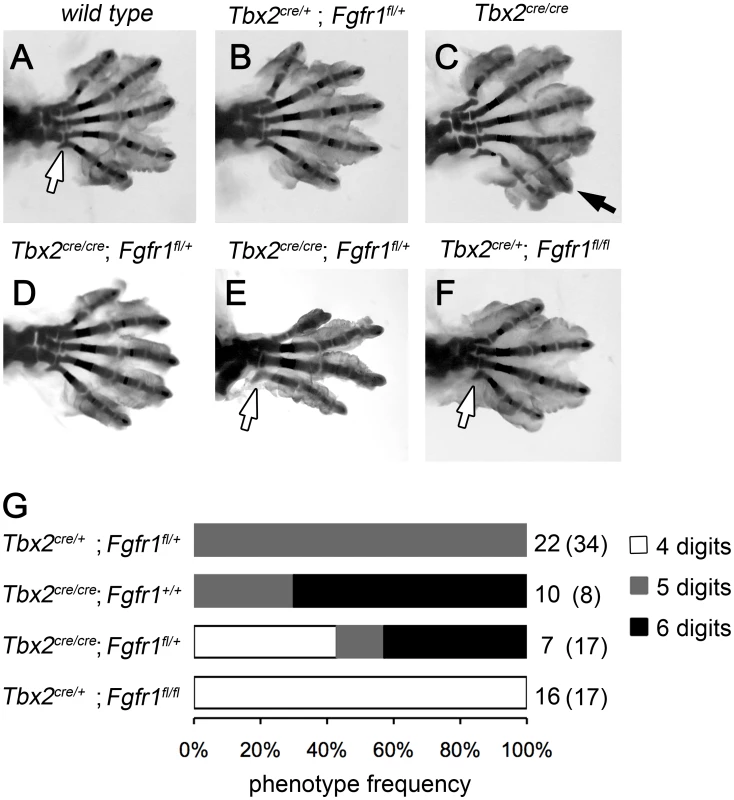
(A–F) Skeletal analysis of E18.5 hindlimbs. Normal digit development in the wild-type (A) and after heterozygous loss of Fgfr1 (Tbx2cre/+;Fgfr1fl/+, B). Duplicated digit 4 in Tbx2-deficient limbs (C, black arrow). Loss of one Fgfr1 allele on the Tbx2-mutant background (Tbx2cre/cre;Fgfr1fl/+) rescues polydactyly (D), or even causes oligodactyly (E), as observed in homozygous Fgfr1-mutant embryos (Tbx2cre/+;Fgfr1fl/fl, F). Morphology of the proximal part of the most posterior metacarpal suggests normal formation of digit 5 (white arrows). (G) Phenotype frequencies in Tbx2cre/Fgfr1fl compound mutant limbs (oligodactyly – 4 digits; normal hindlimb – 5 digits; digit 4 duplication – 6 digits). A total of 135 embryos was analyzed. All phenotypes were found in a bilateral symmetric fashion. Numbers on the right side indicate observed and expected counts (in brackets) of embryos for each genotype. Double mutant embryos (Tbx2cre/cre;Fgfr1fl/fl) were not obtained at E18.5. Tbx2 directly represses Grem1 in the posterior mesenchyme of hindlimb buds
Since Tbx2 encodes a nuclear transcriptional repressor [28], we sought to identify targets that may help to explain the observed molecular changes. We judged it unlikely, that Shh itself is a target of Tbx2 repression since Tbx2 and Shh are broadly coexpressed in the posterior limb bud mesenchyme. Furthermore, unchanged expression of Shh in E11.5 mutant hindlimbs cannot explain up-regulation of Fgf4 in the AER at this stage. Because AER expression of Fgf4 is mediated by inhibition of Bmp signaling by the secreted Bmp antagonist Grem1 [12], [13], we explored the possibility of Grem1 regulation by Tbx2. In E12.0 and E11.5 wild-type embryos, the expression of Grem1 in two crescent-shaped domains at the dorsal and ventral surface of the limb mesenchyme was sharply excluded from the Tbx2-positive posterior hindlimb mesenchyme (Figure 4A and 4B, 4D and 4E). In Tbx2−/− hindlimbs, the expression of Grem1 was unaffected at E10.5 and E13.0 (data not shown), but appeared specifically up-regulated at E12.0 and E11.5 in the posterior region (Figure 4C and 4F; arrows). On adjacent sagittal sections of E11.5 Tbx2cre/+ hindlimbs, cre and Grem1 were expressed in neighboring, non-overlapping domains (Figure 4G–4I). In Tbx2cre/cre - embryos, however, Grem1 expression was shifted posteriorly and overlapped with the domain of cre expression that itself was unchanged. Consistent with local expansion of the Bmp-antagonist Grem1 the Bmp target gene Dkk1 [29] was absent in the mesenchyme underlying the posterior AER in Tbx2−/− hindlimbs at E11.5 (Figure S3A). Expression of Bmp targets Msx1 and Msx2 [30] was unaffected (Figure S3B and S3C), suggesting that the level of Bmps was still sufficient to activate these genes.
Fig. 4. Tbx2 directly represses Grem1 expression in the posterior hindlimb mesenchyme. 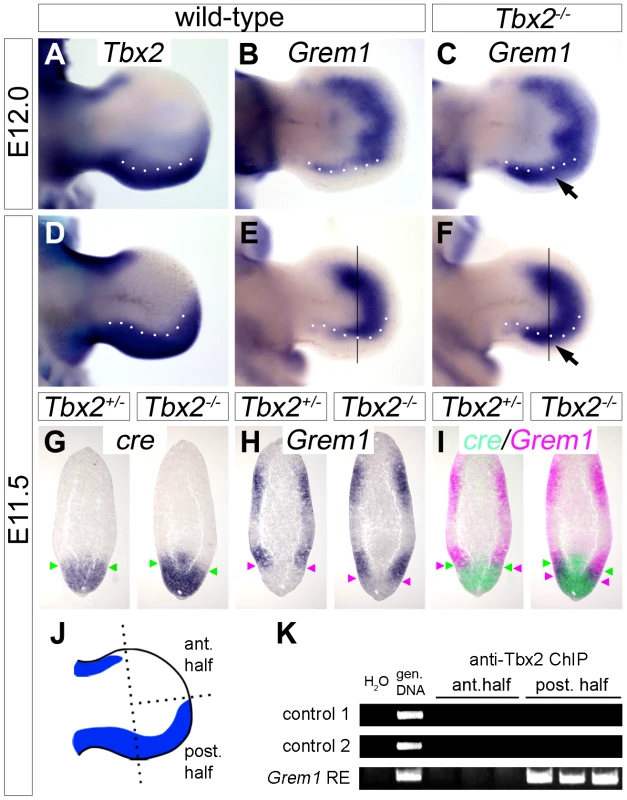
(A–F) Whole mount in situ hybridization analysis of Tbx2 and Grem1 expression in E12.0 and E11.5 hindlimbs of wild-type and Tbx2−/− embryos. Arrows (C, F) point to posterior expansion of Grem1 expression in Tbx2−/− limbs. White dotted lines demarcate the anterior limit of Tbx2 expression. (G–I) Comparative expression analysis of cre, driven from the mutant Tbx2-locus, and Grem1 by in situ hybridization on adjacent sagital hindlimb sections of E11.5 Tbx2cre/+ (control) and Tbx2cre/cre (mutant) embryos. Section planes are indicated by black lines (E, F). Arrowheads mark the anterior limit of cre expression (in green), and the posterior limit of Grem1 expression (in pink). False-colored overlays in (I) show posterior expansion of Grem1 in Tbx2−/− hindlimbs. (J–K) ChIP analysis using chromatin from anterior or posterior halves of E11.5 wild-type hindlimbs. Scheme (J) shows Tbx2 expression (blue) exclusively in the posterior limb half. (K) PCR detection of Tbx2-bound DNA regions. Total genomic DNA was used as positive control. In posterior limb halves Tbx2 binds to the reported Grem1 limb regulatory element but not to genomic control regions. ChIP experiments were performed in triplicates. To explore if Tbx2 mediates Grem1 repression directly, we performed chromatin immunoprecipitation experiments (ChIP) using anti-Tbx2 IgG and E11.5 wild-type hindlimb tissue (Figure 4J and 4K). We found that in posterior limb-bud halves Tbx2 protein specifically interacted with the known Grem1 limb bud enhancer [31] but not with control genomic regions. No binding was observed to chromatin prepared from anterior limb bud halves that lack Tbx2 expression, demonstrating specificity of this assay. Together our results strongly suggest that Tbx2 terminates Fgf4/Shh signaling in the posterior limb bud by direct repression of Grem1.
Tbx2 is a target of Bmp signaling
Bmp ligands are expressed in the AER and at the margins of the limb mesenchyme [13] and represent good candidates as activators of Tbx2 expression, similar to other developmental contexts [32], [33], [34], [35]. To explore this possibility, we analyzed the effect of Bmp on Tbx2 expression by bead implantation experiments. We found that Bmp4-soaked beads implanted into the central mesenchyme of E10.5 forelimb buds caused upregulation of Tbx2 after 16 hours of culture (Figure 5A). This effect was further increased after ectoderm removal (Figure 5B). We established micromass cultures of E10.5 limb bud mesenchyme and studied Tbx2 expression by quantitative RT-PCR 2 hours after addition of Bmp4 to the medium. A dose-dependent increase of Tbx2 mRNA was observed that closely resembled induction of the known Bmp targets Msx1 and Msx2 (Figure 5C). To study Bmp requirement of Tbx2 expression, we added Dorsomorphin, a selective inhibitor of bone morphogenetic protein (BMP) type I receptors [36], during the last two hours of culture. We observed a twofold reduction of basal Tbx2 expression in the presence of 1 µM Dorsomorphin that again resembled the response of Msx1 and Msx2 (Figure 5D). Thus, Bmp signaling is necessary and sufficient to induce Tbx2 expression in the limb. Tbx2 – via repression of Grem1 – therefore operates in a positive feedback loop with Bmp(s) at the posterior margin of the limb.
Fig. 5. Bmp signaling induces Tbx2 expression in the limb bud mesenchyme. 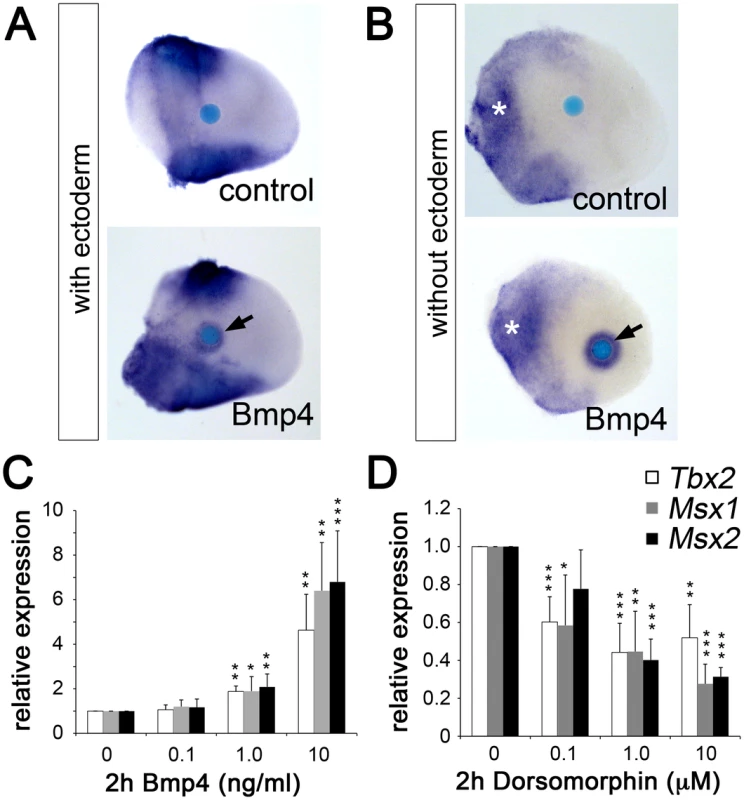
(A, B) Whole mount in situ hybridization analysis demonstrates that Bmp4-soaked beads (100 µg/ml) but not BSA-soaked beads (control) induce Tbx2 expression in the central limb bud mesenchyme after 18 h of culture (black arrows). White asterisks show an expansion of Tbx2 expression into the proximal-central region following ectoderm removal. (C, D) Influence of Bmp signaling on Tbx2 expression in micromass cultures of dissociated E10.5 limb bud mesenchyme. Bmp4 (C) or the Bmp-inhibitor Dorsomorphin (D) was added during the last 2 hours of in total 18 hours of culture. The relative expression levels of Tbx2 and of the known Bmp targets Msx1 and Msx2 were analyzed by qPCR analysis. Means ± SD of four independent cDNAs. Significant changes compared to control are labeled with asterisks (*: p<0.05; **: p<0.01; ***: p<0.001, as determined by t-test). Ectopic Tbx2 abrogates Grem1 expression in the early limb bud mesenchyme
Next we used a cre/loxP-based misexpression approach to analyze if expression of Tbx2 in the entire limb bud would interfere with e-m signaling and digit development. In HprtTBX2 mice expression of the human TBX2 cDNA (introduced into the X-chromosomal Hypoxanthine guanine phosphoribosyl transferase) locus, is inducible by cre-mediated recombination [37]. We employed the Prrx1-cre mouse line to drive transgene expression in the entire limb mesenchyme [38]. X-chromosome inactivation in females causes mosaicism, we therefore only analyzed hemizygous Prrx1-cre/+;HprtTBX2/Y male embryos that express the transgene in a uniform manner (abbreviated as Prrx1-TBX2). By Western blot analysis and immunostainings we confirmed ubiquitous expression of TBX2 in the E10.5 limb mesenchyme at levels comparable to the endogenous Tbx2 protein (Figure S4A–S4D). At E18.5, transgenic embryos exhibited oligodactyly with a dramatic reduction of the limb length (Figure 6A–6D). Forelimbs (3 digits in 9; 4 digits in 1 out of 10 embryos) were stronger affected than hindlimbs (4 digits in 5; 5 digits in 4 and 6 digits in 1 out of 10 embryos), most likely reflecting the relatively delayed onset of Prrx1-cre mediated recombination in hindlimbs [38]. We invariantly observed a single zeugopodial element and a reduction of pectoral and pelvic girdles. Forelimb stylo - and zeugopod elements were fused, in the hindlimb the femur was strongly reduced.
Fig. 6. Ectopic Tbx2 represses Grem1 and disrupts e-m signaling in the limb. 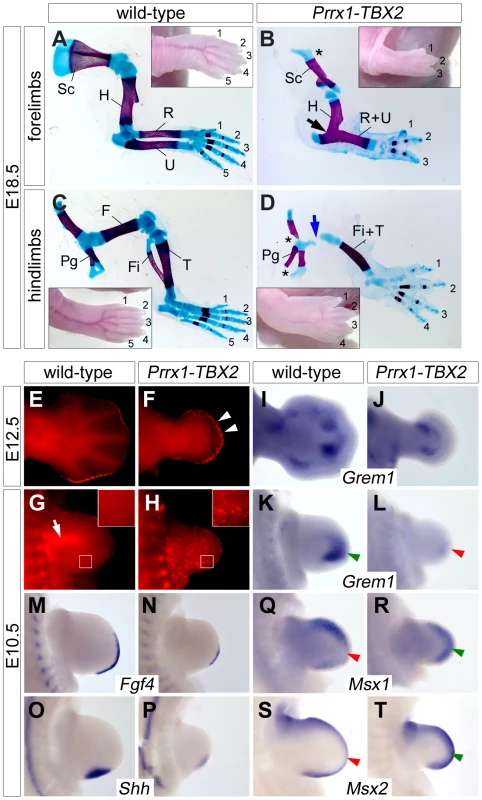
(A–D) Skeletal preparations of E18.5 fore- and hindlimbs from wild-type and Prrx1-TBX2 (Prrx1-cre/+;HprtTBX2/Y) embryos. Insets show morphology of the autopod. Asterisks indicate reduced scapula in (B), and pelvic girdle in (D). Blue arrow in (D) shows the reduced femur. Abbreviations are: F, femur; Fi, fibula; H, humerus; Pg, pelvic gridle; R, radius; U, ulna; S, scapula; Ti, tibia. (E–H) Detection of apoptosis by Lysotracker Red staining in wild-type and Prrx1-TBX2 forelimbs. White arrowheads in (F) indicate increased mesenchymal apoptosis beneath the AER. White arrow in (G) shows apoptosis in the proximal mesenchyme of wild-type limbs. White boxes indicate magnified regions. (I–T) Whole-mount in situ hybridization analysis of Grem1, Fgf4, Shh, Msx1 and Msx2 expression in E12.5 and E10.5 wild-type and Prrx1-TBX2 forelimb buds. Green and red arrowheads highlight complementary expression of Grem1 and Msx1/2. The delayed onset of recombination in the hindlimbs led us to study the early consequences of TBX2 misexpression in forelimbs that demonstrated a complete lack of autopod outgrowth at E12.5. In sharp contrast to the Tbx2 loss-of-function situation, Lysotracker Red staining showed apoptotic mesenchymal cells underneath the entire AER at this stage in Prrx1-TBX2 embryos (Figure 6E and 6F). In E10.5 wild-type forelimbs, apoptosis was confined to the proximal mesenchyme as previously reported [6] (Figure 6G). Age matched Prrx1-TBX2 embryos exhibited strongly reduced forelimb buds and widespread apoptosis throughout the mesenchyme as observed by LysoTracker Red and by TUNEL staining (Figure 6H and Figure S4F).
A highly similar phenotypic spectrum of limb defects including reduced limb length and autopod outgrowth, oligodactyly, fusion of zeugopodial elements, as well as early and widespread mesenchymal apoptosis was reported in Grem1−/− mutants [12], [13], suggesting that loss of Grem1 expression may account for the observed effects in Prrx1-TBX2 embryos. Indeed, we found strong reduction of Grem1 expression in E10.5 transgenic forelimb buds (Figure 6K and 6L) and residual expression at E12.5 at the base of the autopod (Figure 6I and 6J). Thus, Tbx2 represses Grem1 expression distally, whereas the more proximal expression domain at E12.5 might be controlled by an independent mechanism. Next, we analyzed the effects of TBX2 misexpression on known downstream effectors of Grem1. At E10.5, expression of both Fgf4 and Shh was strongly reduced (Figure 6M–6P), again closely resembling the situation in Grem1−/− limbs. Reduction of the target genes Spry4 and Ptch1 [39], [40] confirmed a decrease in Fgf4 and Shh signaling (Figure S5). Fgf9 and Fgf17 expression was reduced but expression of Fgf8 and of the more broadly expressed Fgf target Etv4 (Pea3) [41] was unaffected, demonstrating that Tbx2 acts specifically on posterior e-m signaling (Figure S5C and S5D). To study if reduction of Grem1 is associated with increased Bmp signaling we analyzed Msx1 and Msx2 expression, and found that both genes were upregulated in the distal limb bud (Figure 5Q–5T), i.e. in regions normally devoid of Msx1/2 expression due to Grem1-mediated Bmp-antagonism (compare Figure 5K) [12], [13].
Notably, we did neither observe transformations of digit identity nor changes in limb positioning (data not shown), as reported for Tbx2 and Tbx3 misexpression in the chick model [19], [20]. Hence, control of digit formation by local repression of Grem1 is the primary function of Tbx2 in the mouse.
Discussion
Precise termination of the e-m signaling loop involving Shh, Grem1 and Fgfs is crucial to restrict limb bud size and to assure a normal digit number. Studies in chick and mouse have indicated that downregulation of Grem1 drives termination of this loop but suggested two different molecular mechanisms: The one is based on the observation that Shh expressing cells as well as their descendants are unable to express Grem1 [15], [42]. Proliferative expansion of ZPA-derived cells [7] would thereby displace the source of Grem1 secretion from the AER to a point where the distal range of Grem1 diffusion is eventually exceeded. As a consequence, Bmp signaling increases and suppresses AER-Fgfs, followed by termination of Shh expression and proliferative expansion. Although this model is supported by the sequence of signal terminations in the chick, the factor responsible for the cell-autonomous repression of Grem1 in the Shh lineage cells has remained enigmatic (Scherz et al., 2004). As an alternative mechanism, recent loss-of-function experiments in the mouse have supported the existence of an inhibitory Fgf/Grem1 signaling loop that becomes progressively activated once the positive Shh/Grem1/Fgf loop has induced sufficiently high levels of Fgfs [43]. This model can elegantly rationalize the regulation of limb bud size via interconnected, self-terminating signaling loops. However, it fails to explain the selective absence of Grem1 from the posterior limb bud mesenchyme.
The parallel upregulation and prolonged expression of Grem1, Fgf4/9/17 and Shh in Tbx2-deficient hindlimbs and their coordinated downregulation upon limb-specific TBX2 overexpression identifies Tbx2 as an essential factor for the termination of the e-m signaling loop (summarized in Figure 7). Given the virtual overlap of Tbx2 and Shh cell lineages (see Figure S2A–S2D and [7]), we propose that Tbx2 renders Shh-descendant cells unable to induce Grem1. Consistently we found that the transcriptional repressor Tbx2 binds to the Grem1 limb enhancer in vivo. This 437 bp element has been identified previously by genome-wide, limb-specific ChIP analysis of Gli3, the transcription factor that mediates Shh-dependent gene transcription in the limb bud [31]. In transgenic mice the element largely recapitulates the complex Grem1 limb expression pattern, which argues for an integration of both activating (Gli3) as well as repressive modules (Tbx2). Absence of Tbx2 expression from the anterior limb margin can explain earlier observations that Shh loaded beads are sufficient to induce Grem1 in this region but not in the posterior limb mesenchyme [15]. Importantly, we noted that in Tbx2-deficient hindlimbs the posterior mesenchyme directly underneath the AER remained Grem1 negative (Figure 4F). This suggests that the negative Fgf-Grem1 signaling loop [43] stays active in Tbx2−/− embryos and argues that both termination mechanisms (see above) operate in parallel in adjacent domains of the limb mesenchyme to achieve spatio-temporal control of Grem1 expression.
Fig. 7. Model of Tbx2 function in the posterior hindlimb mesenchyme. 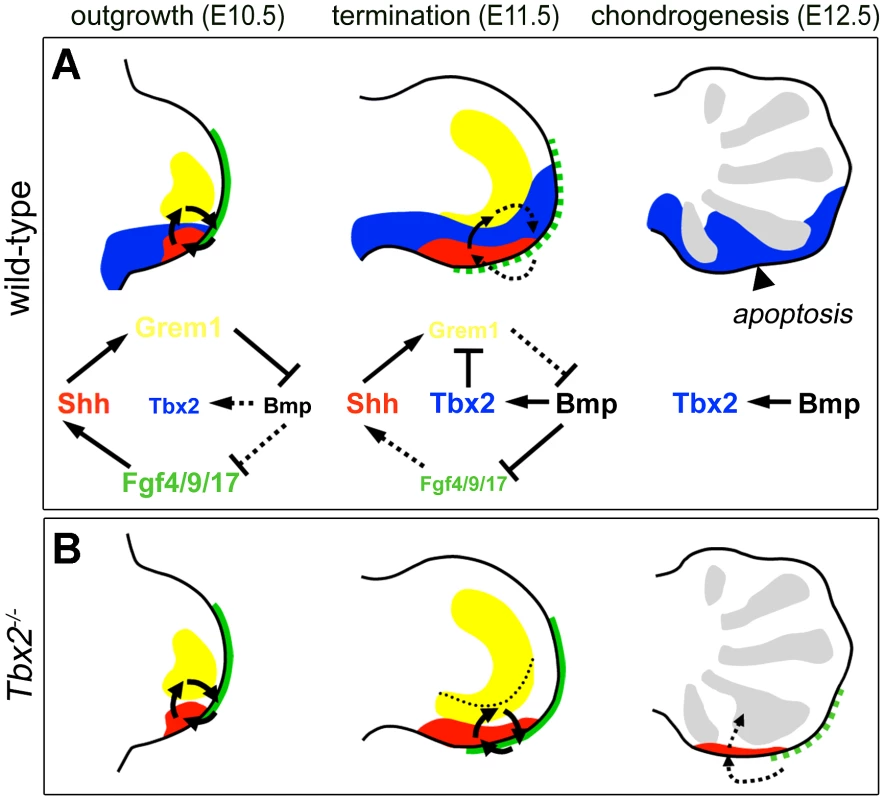
(A) During normal limb bud outgrowth (E10.5) the expression domains of Shh (red), Grem1 (yellow) and Fgf4/9/17 (green) are in close proximity allowing propagation of the e-m signaling loop. Following proliferative expansion of ZPA-derived cells, a broadened Tbx2 expression (blue) causes Grem1 repression. Progressive displacement of Grem1-secreting cells from the AER terminates e-m signaling (E11.5, dotted arrows). The diagrams illustrate these dynamic signaling activities. (B) Failure of Grem1 repression in Tbx2-deficient hindlimbs causes prolonged e-m signaling. The dotted line shows the posterior limit of Grem1 expression in the wild-type. Impaired termination of outgrowth reduces apoptosis and causes expansion of the digit4 condensation (E12.5, grey regions). The hindlimb-specific requirement for Tbx2 cannot easily be explained at this point. It may result from differences in the proliferative expansion of fore - and hindlimbs, differential Shh/Fgf signaling activities, or the existence of additional repressors that might operate redundantly with Tbx2 in forelimbs. Tbx3 is unlikely to compensate for the loss of Tbx2 given the absence of Tbx3 expression at the distal margin of forelimb buds (Figure S1). However, we can clearly exclude a function of mouse Tbx2 in specification of digit identity, as previously suggested from experiments in chicken embryos [19]. Although some of the observed differences may relate to species-specific variations in the underlying molecular programs, experimental caveats due to unphysiological levels of protein obtained after retroviral mis - and overexpression cannot be excluded at this point.
We have shown that Bmp signaling activates Tbx2 expression in the limb mesenchyme. This notion is supported by reduction of Tbx2 expression in embryos with reduced levels of Bmp signaling in the limb [44]. Since Tbx2 expression is normal in Prrx1-cre;Bmp2fl/fl;Bmp4fl/fl or Prrx1-cre;Bmp2fl/fl;Bmp7−/− mice [45], several Bmp ligands are likely to operate redundantly. Recent transgenic analysis of the Tbx2 promoter led to the identification of Smad binding sites [34], [35] that mediate Tbx2 expression in the limb (supporting material in [35]) further stressing the relevance of Bmp signaling for Tbx2 expression in this organ. In chick embryos the non-AER marginal ectoderm activates Tbx2 expression, whereas transplantation of the AER or Fgf-soaked beads repress Tbx2 expression [45]. In contrast to chick embryos, however, murine Tbx2 expression is not excluded from mesenchymal regions underling the AER and extends more distally, demonstrating species-specific differences in Tbx2 regulation. Abrogation of Shh signaling by cyclopamine treatment has shown that Shh is dispensable for posterior Tbx2 expression [45]. The fact that implantation of Shh-soaked beads can induce Tbx2 expression in the anterior mesenchyme [46] may therefore be secondary to induction of Bmp expression in these regions [42]. Thus, Bmp signaling constitutes a major positive input for Tbx2 limb expression but additional factors may feed-in to fine-tune its expression. However, the mutual relationship: Bmp-induction of Tbx2 and Tbx2 repression of Grem1-mediated Bmp-antagonism, constitute a positive feedback loop that acts locally to diminish and terminate Shh/Fgf4 signaling (Figure 7). Postaxial polydactyly, expansion of Sox9 expression and reduced apoptosis in Bmp pathway mutants [44], [47] represent phenotypic similarities that support engagement in a common pathway.
While loss of Grem1 (following TBX2 misexpression) causes a general increase in Bmp signaling, as indicated by induction of distal Msx1/2 expression, both genes were unaffected in Tbx2−/− limbs. Here, absence of the Bmp target Dkk1 was observed in the sub-AER mesenchyme, indicating that Dkk1 expression requires higher levels of Bmp signaling. In fact, the loss of Dkk1 expression might explain reduced apoptosis in the Tbx2−/− sub-AER mesenchyme as ectopic Dkk1 induces programmed cell death [29]. Moreover, the postaxial polydactyly in Dkk1 mutants [48] is compatible with a role downstream of Tbx2.
Mesoaxial polydactyly has been suggested as a characteristic feature of Oral-Facial-Digital syndrome (OFDS) type IV (OMIM 258860) [49]. Interestingly, a variant case of OFDS, that partially resembles OFDS type IV and type II (also known as Mohr Syndrome, OMIM 252100) shows a malformation spectrum including endocardial cushion defects, cleft palate and central polydactyly with bifurcated Y-shaped metacarpals of the forth digit [50] phenocopying Tbx2 loss-of-function in the mouse [17], [21], [51].
Together, our data allow the integration and refinement of existing models for termination of distal limb outgrowth, and emphasizes how local differences of signaling activities are translated into the architecture of the adult skeleton, i.e. the number or digits. They show that central polydactyly like preaxial and postaxial variants arise from perturbation in components of the signaling loops including Shh, Grem, Bmp and Fgf signaling.
Materials and Methods
Ethics statement
All animal work conducted for this study was approved by H. Hedrich, state head of the animal facility at Medizinische Hochschule Hannover and performed according to German legislation.
Mice and genonotyping
Mice carrying a null allele of Tbx2 (Tbx2tm1.1(cre)Vmc, synonyms: Tbx2−, Tbx2cre) [52], a floxed allele of Fgfr1 [53], the transgenic lines Tg(Prrx1-cre)1Cjt/J) (synonym: Prx1-Cre) [38] and the reporter lines R26lacZ (synonym: R26R) [54] and Tg(CAG-Bgeo/GFP)21Lbe) (synonym: Z/EG) [55] and mice with integration of the human TBX2 gene in the Hprt locus (Hprttm2(CAG-TBX2,-EGFP)Akis, synonym: HprtTBX2) [51] were maintained on an outbred (NMRI) background. For timed pregnancies, vaginal plugs were checked in the morning after mating; noon was taken as embryonic day (E) 0.5. Pregnant females were sacrificed by cervical dislocation; embryos were harvested in phosphate-buffered saline, decapitated, fixed in 4% paraformaldehyde overnight, and stored in 100% methanol at −20°C before further use. Genomic DNA prepared from yolk sacs or tail biopsies was used for genotyping by polymerase chain reaction (PCR).
Limb culture experiments
Limb buds from E10.5 wild-type NMRI embryos were dissected in PBS and placed on Nucleopore filters (Whatman, pore size 1.0 µm) on top of a stainless steel mesh at the air-liquid interface in 3.5 cm cell culture dishes. The surgical removal of the ectoderm was performed with forceps in DMEM/10% FCS, after incubation of limb buds in 2% Trypsin/PBS (w/v) for 20 min at 4°C. Affi-Gel blue beads (100–200 µm diameter, Bio-Rad) were rinsed in PBS and incubated at room temperature for 1 h in either recombinant human BMP4 (100 µg/ml, AbD Serotech) or in 1 mg/ml BSA (control). Beads were rinsed in PBS before implantation into the limb mesenchyme. The culture was performed at 37°C and 5% CO2 in organ culture medium (DMEM/10% FCS, 1× solutions of Penicillin/Streptomycin, Glutamax, sodium pyruvate, and non-essential amino acids [Gibco]).
Limb micromass cultures
Micromass cultures were established by dissociation of E10.5 fore - and hindlimb buds in DMEM/10% FCS, after incubation in 2% Trypsin/PBS (w/v) for 5 min at 37°C. A single cell suspension was obtained by gentle pipetting; clumps of ectoderm were removed after sedimentation. Cells were adjusted to 1.5×107 cells/ml in organ culture medium (as above), before 10 µl spots were placed on 24 well plates. Cells were incubated for 1 hour at 37°C to allow adherence, before the wells were filled with medium. Recombinant BMP4 (as above) or Dorsomorphin (Sigma) were added to the medium after 16 h of culture, 2 hours before RNA isolation. Total RNA was extracted from single micromass cultures with PeqGOLD reagent (Peqlab). RNA (500 ng) was reverse transcribed using oligo dT primer and RevertAid M-MuLV Reverse Transcriptase (Fermentas) following the manufacturer's recommendations. Relative gene expression was measured using iQ−SYBR Green reagent (Biorad) and calculated using the DDCT method by normalization to Hprt expression. The error bars show standard deviation from 4 independent experiments.
In situ hybridization, skeletal Preparations, β-galactosidase and apoptosis assays, and immunostainings
Skeletal preparations with Alcian blue and Alizarin red, β-galactosidase stainings, detection of apoptosis by the TUNEL assay, and in situ hybridization analyses on whole embryos and on 10 µm paraffin sections were performed as previously described [56], [57], [58]. All experiments were performed on at least three independent embryos. For experiments that showed variable results, numbers of used specimens are mentioned in the text. For detection of apoptotic cells, embryos were collected in PBS and incubated for 30 min at 37°C in prewarmed PBS containing 2.5 µM LysoTracker Red (Sigma), followed by several washes in PBS. Tbx2 antibody (#07-318, Millipore) was used 1∶100 for immunostainings on 5 µm paraffin sections. Signals were amplified by Tyramide Signal Amplification (PerkinElmer).
Chromatin immunoprecipitation
Distal pieces of E11.5 wild-type hindlimb buds were separated in anterior and posterior halves (as indicated in Figure 4J) and collected in 3 separate pools of 8 embryos each and treated with 4% paraformaldehyde overnight. ChIP experiments using an anti-Tbx2 antibody were performed essentially as previously described [59]. Primer sequences for the Grem1 enhancer were TTCCCCTCCTCTTCCACAGTAGG and GGCCAAATAACCACACAGGAAAC, corresponding to a 447 bp fragment previously tested in transgenic animals [31]. Primer pairs of control regions were TGAAAACCCCAAGGAGTCTG, CATGGGCAGGATACTACGCT (193 bp product, 25.5 kbp distal) and AGCCTGACTCTCCCATCTCA, GGCACTGGATAAAACTCCCA (273 bp product, 24.3 kbp distal from the Grem1 limb enhancer).
Image analysis
Whole-mount specimens were photographed on Leica M420 with Fujix digital camera HC-300Z. Whole-mount GFP-epifluorescence was documented on a Leica MZFLIII macroscope equipped with a Leica DFC300 camera. Sections of in situ hybridizations were photographed using a Leica DM5000 microscope with a Leica DFC300FX camera. All images were processed in Adobe Photoshop CS.
Supporting Information
Zdroje
1. ZellerR, Lopez-RiosJ, ZunigaA (2009) Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet 10 : 845–858.
2. MarianiFV, AhnCP, MartinGR (2008) Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature 453 : 401–405.
3. ZhuJ, NakamuraE, NguyenMT, BaoX, AkiyamaH, et al. (2008) Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell 14 : 624–632.
4. SummerbellD (1974) A quantitative analysis of the effect of excision of the AER from the chick limb-bud. J Embryol Exp Morphol 32 : 651–660.
5. ChiangC, LitingtungY, HarrisMP, SimandlBK, LiY, et al. (2001) Manifestation of the limb prepattern: limb development in the absence of sonic hedgehog function. Dev Biol 236 : 421–435.
6. SunX, MarianiFV, MartinGR (2002) Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature 418 : 501–508.
7. HarfeBD, ScherzPJ, NissimS, TianH, McMahonAP, et al. (2004) Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118 : 517–528.
8. LauferE, NelsonCE, JohnsonRL, MorganBA, TabinC (1994) Sonic hedgehog and Fgf-4 act through a signaling cascade and feedback loop to integrate growth and patterning of the developing limb bud. Cell 79 : 993–1003.
9. NiswanderL, JeffreyS, MartinGR, TickleC (1994) A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature 371 : 609–612.
10. BenazetJD, BischofbergerM, TieckeE, GoncalvesA, MartinJF, et al. (2009) A self-regulatory system of interlinked signaling feedback loops controls mouse limb patterning. Science 323 : 1050–1053.
11. ZunigaA, HaramisAP, McMahonAP, ZellerR (1999) Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature 401 : 598–602.
12. KhokhaMK, HsuD, BrunetLJ, DionneMS, HarlandRM (2003) Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat Genet 34 : 303–307.
13. MichosO, PanmanL, VinterstenK, BeierK, ZellerR, et al. (2004) Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development 131 : 3401–3410.
14. Lopez-RiosJ, SpezialeD, RobayD, ScottiM, OsterwalderM, et al. (2012) GLI3 constrains digit number by controlling both progenitor proliferation and BMP-dependent exit to chondrogenesis. Dev Cell 22 : 837–848.
15. ScherzPJ, HarfeBD, McMahonAP, TabinCJ (2004) The limb bud Shh-Fgf feedback loop is terminated by expansion of former ZPA cells. Science 305 : 396–399.
16. ChapmanDL, GarveyN, HancockS, AlexiouM, AgulnikSI, et al. (1996) Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn 206 : 379–390.
17. HarrelsonZ, KellyRG, GoldinSN, Gibson-BrownJJ, BollagRJ, et al. (2004) Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development 131 : 5041–5052.
18. DavenportTG, Jerome-MajewskaLA, PapaioannouVE (2003) Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development 130 : 2263–2273.
19. SuzukiT, TakeuchiJ, Koshiba-TakeuchiK, OguraT (2004) Tbx Genes Specify Posterior Digit Identity through Shh and BMP Signaling. Dev Cell 6 : 43–53.
20. RallisC, Del BuonoJ, LoganMP (2005) Tbx3 can alter limb position along the rostrocaudal axis of the developing embryo. Development 132 : 1961–1970.
21. ZirzowS, LudtkeTH, BronsJF, PetryM, ChristoffelsVM, et al. (2009) Expression and requirement of T-box transcription factors Tbx2 and Tbx3 during secondary palate development in the mouse. Dev Biol 336 : 145–155.
22. WrightE, HargraveMR, ChristiansenJ, CooperL, KunJ, et al. (1995) The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet 9 : 15–20.
23. NiederreitherK, McCafferyP, DragerUC, ChambonP, DolleP (1997) Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev 62 : 67–78.
24. Salas-VidalE, ValenciaC, CovarrubiasL (2001) Differential tissue growth and patterns of cell death in mouse limb autopod morphogenesis. Dev Dyn 220 : 295–306.
25. VerheydenJM, LewandoskiM, DengC, HarfeBD, SunX (2005) Conditional inactivation of Fgfr1 in mouse defines its role in limb bud establishment, outgrowth and digit patterning. Development 132 : 4235–4245.
26. MaoJ, McGlinnE, HuangP, TabinCJ, McMahonAP (2009) Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Dev Cell 16 : 600–606.
27. HancockSN, AgulnikSI, SilverLM, PapaioannouVE (1999) Mapping and expression analysis of the mouse ortholog of Xenopus Eomesodermin. Mech Dev 81 : 205–208.
28. CarreiraS, DexterTJ, YavuzerU, EastyDJ, GodingCR (1998) Brachyury-related transcription factor Tbx2 and repression of the melanocyte-specific TRP-1 promoter. Mol Cell Biol 18 : 5099–5108.
29. GrotewoldL, RutherU (2002) The Wnt antagonist Dickkopf-1 is regulated by Bmp signaling and c-Jun and modulates programmed cell death. EMBO J 21 : 966–975.
30. PizetteS, Abate-ShenC, NiswanderL (2001) BMP controls proximodistal outgrowth, via induction of the apical ectodermal ridge, and dorsoventral patterning in the vertebrate limb. Development 128 : 4463–4474.
31. VokesSA, JiH, WongWH, McMahonAP (2008) A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev 22 : 2651–2663.
32. MaL, LuMF, SchwartzRJ, MartinJF (2005) Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development 132 : 5601–5611.
33. BehestiH, HoltJK, SowdenJC (2006) The level of BMP4 signaling is critical for the regulation of distinct T-box gene expression domains and growth along the dorso-ventral axis of the optic cup. BMC Dev Biol 6 : 62.
34. SinghR, HorsthuisT, FarinHF, GrieskampT, NordenJ, et al. (2009) Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ Res 105 : 442–452.
35. ShiraiM, Imanaka-YoshidaK, SchneiderMD, SchwartzRJ, MorisakiT (2009) T-box 2, a mediator of Bmp-Smad signaling, induced hyaluronan synthase 2 and Tgfbeta2 expression and endocardial cushion formation. Proc Natl Acad Sci U S A 106 : 18604–18609.
36. YuPB, HongCC, SachidanandanC, BabittJL, DengDY, et al. (2008) Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol 4 : 33–41.
37. SinghR, HoogaarsWM, BarnettP, GrieskampT, RanaMS, et al. (2011) Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell Mol Life Sci 69 : 1377–1389.
38. LoganM, MartinJF, NagyA, LobeC, OlsonEN, et al. (2002) Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33 : 77–80.
39. MinowadaG, JarvisLA, ChiCL, NeubuserA, SunX, et al. (1999) Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development 126 : 4465–4475.
40. GoodrichLV, JohnsonRL, MilenkovicL, McMahonJA, ScottMP (1996) Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev 10 : 301–312.
41. RoehlH, Nusslein-VolhardC (2001) Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr Biol 11 : 503–507.
42. NissimS, HassoSM, FallonJF, TabinCJ (2006) Regulation of Gremlin expression in the posterior limb bud. Dev Biol 299 : 12–21.
43. VerheydenJM, SunX (2008) An Fgf/Gremlin inhibitory feedback loop triggers termination of limb bud outgrowth. Nature 454 : 638–641.
44. SeleverJ, LiuW, LuMF, BehringerRR, MartinJF (2004) Bmp4 in limb bud mesoderm regulates digit pattern by controlling AER development. Dev Biol 276 : 268–279.
45. NissimS, AllardP, BandyopadhyayA, HarfeBD, TabinCJ (2007) Characterization of a novel ectodermal signaling center regulating Tbx2 and Shh in the vertebrate limb. Dev Biol 304 : 9–21.
46. Gibson-BrownJJ, AgulnikSI, SilverLM, NiswanderL, PapaioannouVE (1998) Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development 125 : 2499–2509.
47. BandyopadhyayA, TsujiK, CoxK, HarfeBD, RosenV, et al. (2006) Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet 2: e216 doi:10.1371/journal.pgen.0020216.
48. MukhopadhyayM, ShtromS, Rodriguez-EstebanC, ChenL, TsukuiT, et al. (2001) Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev Cell 1 : 423–434.
49. PorettiA, VitielloG, HennekamRC, ArrigoniF, BertiniE, et al. (2012) Delineation and diagnostic criteria of Oral-Facial-Digital Syndrome type VI. Orphanet J Rare Dis 7 : 4.
50. HsiehYC, HouJW (1999) Oral-facial-digital syndrome with Y-shaped fourth metacarpals and endocardial cushion defect. Am J Med Genet 86 : 278–281.
51. SinghR, HoogaarsWM, BarnettP, GrieskampT, RanaMS, et al. (2012) Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell Mol Life Sci 69 : 1377–1389.
52. AanhaanenWT, BronsJF, DominguezJN, RanaMS, NordenJ, et al. (2009) The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res 104 : 1267–1274.
53. HochRV, SorianoP (2006) Context-specific requirements for Fgfr1 signaling through Frs2 and Frs3 during mouse development. Development 133 : 663–673.
54. SorianoP (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21 : 70–71.
55. NovakA, GuoC, YangW, NagyA, LobeCG (2000) Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis 28 : 147–155.
56. WilkinsonDG, NietoMA (1993) Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol 225 : 361–373.
57. MoormanAF, HouwelingAC, de BoerPA, ChristoffelsVM (2001) Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J Histochem Cytochem 49 : 1–8.
58. BussenM, PetryM, Schuster-GosslerK, LeitgesM, GosslerA, et al. (2004) The T-box transcription factor Tbx18 maintains the separation of anterior and posterior somite compartments. Genes Dev 18 : 1209–1221.
59. LüdtkeTH-W, FarinHF, RudatC, Schuster-GosslerK, PetryM, et al. (2012) Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes Cdkn1a and Cdkn1b. PLoS Genet 9: e1003189 doi:10.1371/journal.pgen.1003189.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



