-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRecovery of Arrested Replication Forks by Homologous Recombination Is Error-Prone
Homologous recombination is a universal mechanism that allows repair of DNA and provides support for DNA replication. Homologous recombination is therefore a major pathway that suppresses non-homology-mediated genome instability. Here, we report that recovery of impeded replication forks by homologous recombination is error-prone. Using a fork-arrest-based assay in fission yeast, we demonstrate that a single collapsed fork can cause mutations and large-scale genomic changes, including deletions and translocations. Fork-arrest-induced gross chromosomal rearrangements are mediated by inappropriate ectopic recombination events at the site of collapsed forks. Inverted repeats near the site of fork collapse stimulate large-scale genomic changes up to 1,500 times over spontaneous events. We also show that the high accuracy of DNA replication during S-phase is impaired by impediments to fork progression, since fork-arrest-induced mutation is due to erroneous DNA synthesis during recovery of replication forks. The mutations caused are small insertions/duplications between short tandem repeats (micro-homology) indicative of replication slippage. Our data establish that collapsed forks, but not stalled forks, recovered by homologous recombination are prone to replication slippage. The inaccuracy of DNA synthesis does not rely on PCNA ubiquitination or trans-lesion-synthesis DNA polymerases, and it is not counteracted by mismatch repair. We propose that deletions/insertions, mediated by micro-homology, leading to copy number variations during replication stress may arise by progression of error-prone replication forks restarted by homologous recombination.
Published in the journal: . PLoS Genet 8(10): e32767. doi:10.1371/journal.pgen.1002976
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002976Summary
Homologous recombination is a universal mechanism that allows repair of DNA and provides support for DNA replication. Homologous recombination is therefore a major pathway that suppresses non-homology-mediated genome instability. Here, we report that recovery of impeded replication forks by homologous recombination is error-prone. Using a fork-arrest-based assay in fission yeast, we demonstrate that a single collapsed fork can cause mutations and large-scale genomic changes, including deletions and translocations. Fork-arrest-induced gross chromosomal rearrangements are mediated by inappropriate ectopic recombination events at the site of collapsed forks. Inverted repeats near the site of fork collapse stimulate large-scale genomic changes up to 1,500 times over spontaneous events. We also show that the high accuracy of DNA replication during S-phase is impaired by impediments to fork progression, since fork-arrest-induced mutation is due to erroneous DNA synthesis during recovery of replication forks. The mutations caused are small insertions/duplications between short tandem repeats (micro-homology) indicative of replication slippage. Our data establish that collapsed forks, but not stalled forks, recovered by homologous recombination are prone to replication slippage. The inaccuracy of DNA synthesis does not rely on PCNA ubiquitination or trans-lesion-synthesis DNA polymerases, and it is not counteracted by mismatch repair. We propose that deletions/insertions, mediated by micro-homology, leading to copy number variations during replication stress may arise by progression of error-prone replication forks restarted by homologous recombination.
Introduction
Maintenance of genome stability requires the faithful and accurate replication of the genetic material. Genome instability is a hallmark for most types of cancer and it is strongly associated with predisposition to cancer in many human syndromes (for a review, see [1], [2]). Genome instability is manifest at two levels: at the nucleotide level, resulting in base-substitutions, frame-shifts or in micro-insertions/deletions (referred to herein as mutations); and at the chromosomal level, resulting in duplications, deletions, inversions and translocations (referred to herein as gross chromosomal rearrangements or GCRs).
Genome instability during cancer development and in other human genomic disorders may be consequences of failures in chromosome replication (for a review, see [3], [4]). Origin spacing has recently been shown to cause chromosomal fragility at some human fragile sites [5], [6]. Impediments to replication fork movements per se may also cause genome instability [7]–[9]. Indeed, both slowing down and blockages to fork progression can lead to chromosomal fragilities or GCRs in human cells and yeast models [10]–[14]. However, how a blocked replication fork leads to genetic instability remains poorly understood.
In eukaryotes, DNA replication is initiated at numerous origins along linear chromosomes, and impediments to fork progression appear unavoidable during each S-phase (for a review, see [9], [15]). Impediments to fork progression can be caused by DNA lesions, by non-histone proteins tightly bound to DNA, by sequence-caused secondary structures such as cruciform structures and possibly G-quadruplexes, by nucleotide pool imbalance and by conflicts with transcription machinery (for a review, see [16], [17]). In case of failures in fork progression, DNA replication can be completed either by the recovery of the arrested fork by fork-restart mechanisms, or as a result of the progression of a converging fork which can be ensured by activation of dormant origins [7], [15], [18]. Fork restart is presumably essential in unidirectional replication regions, such as the rDNA locus, in regions of low densities of origins, such as some human fragile sites, and when two converging forks are both impeded [5], [19], [20].
To ensure faithful and complete DNA replication, cells coordinate DNA synthesis restart with specific pathways, including DNA replication checkpoint and homologous recombination mechanisms [17]. The integrity of replication forks is guaranteed by the DNA replication checkpoint that maintains the replisome in a replication-competent state to keep DNA polymerases at the site of nucleotide incorporation [21]. It remains unclear how the DNA replication checkpoint modulates replisome activities to maintain its function [21], [22]. The DNA replication checkpoint also regulates nuclease activities (e.g. Exo1 or Mus81) which contribute to preserving the integrity of stalled forks [23], [24]. If replisome function is lost or the replisome dissociates at broken replication forks, the resumption of DNA synthesis appears to require the replisome to be rebuilt. In E.coli, restart of a collapsed fork involves homologous recombination and the PriA helicase that allows replisome components to be loaded de novo on joint-molecule structures [25], [26]. In eukaryotes, the restart of collapsed or broken replication forks is dependent upon homologous recombination, but the mechanism of origin-independent loading of the replisome remains to be described [20], [27]–[30]. It has been proposed that the repair of a double-strand break (DSB) by recombination (break-induced replication, BIR) in budding yeast similarly involves the assembly of a replication fork (for a review, see [30]–[32]). When BIR occurs outside S-phase, recombination-dependent replication fork assembly can synthesise hundreds of kilobases (Kb). However, this DNA synthesis is highly inaccurate due to frequent template switching of nascent-strands and frame-shift mutations [33], [34].
We previously reported a system that displays replication fork arrest at a specific locus in the fission yeast S. pombe. The system is a polar replication fork barrier (RFB) regulated by the Rtf1 protein binding to its RTS1 binding site [35]. The RTS1-RFB causes fork arrest because of a non-histone protein complex binding to the DNA. As proposed for other polar RFBs, the RTS1-RFB is thought to block fork progression by directly (contact between proteins and the replisome) or indirectly (topological constraint) affecting the replicative helicase activity and thereby preventing DNA unwinding [36], [37]. Recovery of the arrested fork occurs by a DSB-independent mechanism and involves the recruitment of recombination proteins at the RTS1-RFB site. We proposed that recombination proteins associate with unwound nascent strands that then anneal with the initial template to allow DNA synthesis to restart [11], [20]. The causative protein barrier then has to be removed either by DNA helicase or by the recombination machinery itself to allow fork-progression to resume [38]–[40]. Occasionally, the unwound nascent strand can mistakenly anneal with a homologous template in the vicinity of the collapsed fork, resulting in the restart of DNA synthesis on non-contiguous template. This incorrect template switch of nascent strands results in inversions and iso-acentric and dicentric chromosomes in ∼2–3% of cells/generation [11], [20]. Error-free template switching between sister-chromatids provides an efficient mechanism for filling-in single-stranded gaps left behind damage-induced stalled forks [41]. Inverted chromosome fusions in yeast and rare-genome rearrangements in human genomic disorders, may both be consequences of template switching between ectopic repeats associated with impeded replication forks [8], [14].
Here, we used the RTS1-RFB to investigate the consequences of fork collapse on genome instability. We report that recovery from a collapsed fork is associated with a high frequency of instability, with a single fork arrest increasing the rates of mutation, deletion and translocation by 10, 40 and 5 fold, respectively. We show that genetic instability associated with fork arrest is dependent on homologous recombination. Fork-arrest-induced GCRs (deletion and translocation) result from inappropriate ectopic recombination at the site of the collapsed fork. We also demonstrate that restoration of fork progression by homologous recombination results in error-prone DNA synthesis due to frequent replication slippage between short tandem repeats. We investigated the molecular mechanisms of this replication slippage and found that post-replication repair, including ubiquitination of PCNA or trans-lesions-synthesis (TLS) DNA polymerases, is not involved in fork-arrest-induced replication slippage. Micro-deletions/insertions flanked by micro-homology associated with copy number variations (CNVs) in cancer cells or in response to replication stress may therefore be scars left following the restoration of forks progression by homologous recombination.
Results
The conditional replication fork barrier RTS1
We generated fork arrest constructs by manipulating the polar RTS1-RFB (Figure 1A). We introduced the RTS1 sequence on the centromere-proximal (cen-proximal) side of the ura4 locus, 5 kb away from the strong replication origin (ori) 3006/7 on chromosome III. This created the t-ura4<ori locus, in which “t” and “ori” refer to the telomere and the origin 3006/7, respectively; and “<” and“ >”refer to the RTS1-barrier and its polarity that is whether it blocks replication forks travelling from the ori 3006/7 towards the telomere or forks travelling from the telomere towards the ori 3006/7, respectively. We previously confirmed that forks moving from ori 3006/7 towards the telomere (tel) are efficiently blocked by the RTS1-RFB at the t-ura4<ori locus [35]. In this model system, fork arrest is activated by inducing the expression of rtf1+ gene that is under control of the thiamine repressible promoter nmt41. Thus, the RTS1-RFB is inactivated by adding thiamine to the media and it is activated in thiamine-free media. Efficient induction of Rtf1 expression requires incubation for 12–16 hours in thiamine-free media. Replication intermediates were analysed by native 2-dimensional gel electrophoresis (2DGE). In conditions of Rtf1 expression, more than 95% of replication forks were blocked by the RTS1-RFB at the t-ura4<ori locus (see black arrow on Figure 1B, t-ura4<ori ON). Arrested forks were not detected without Rtf1 induction (Figure 1B, t-ura4<ori OFF) [20].
Fig. 1. Conditional replication fork-arrest assays. 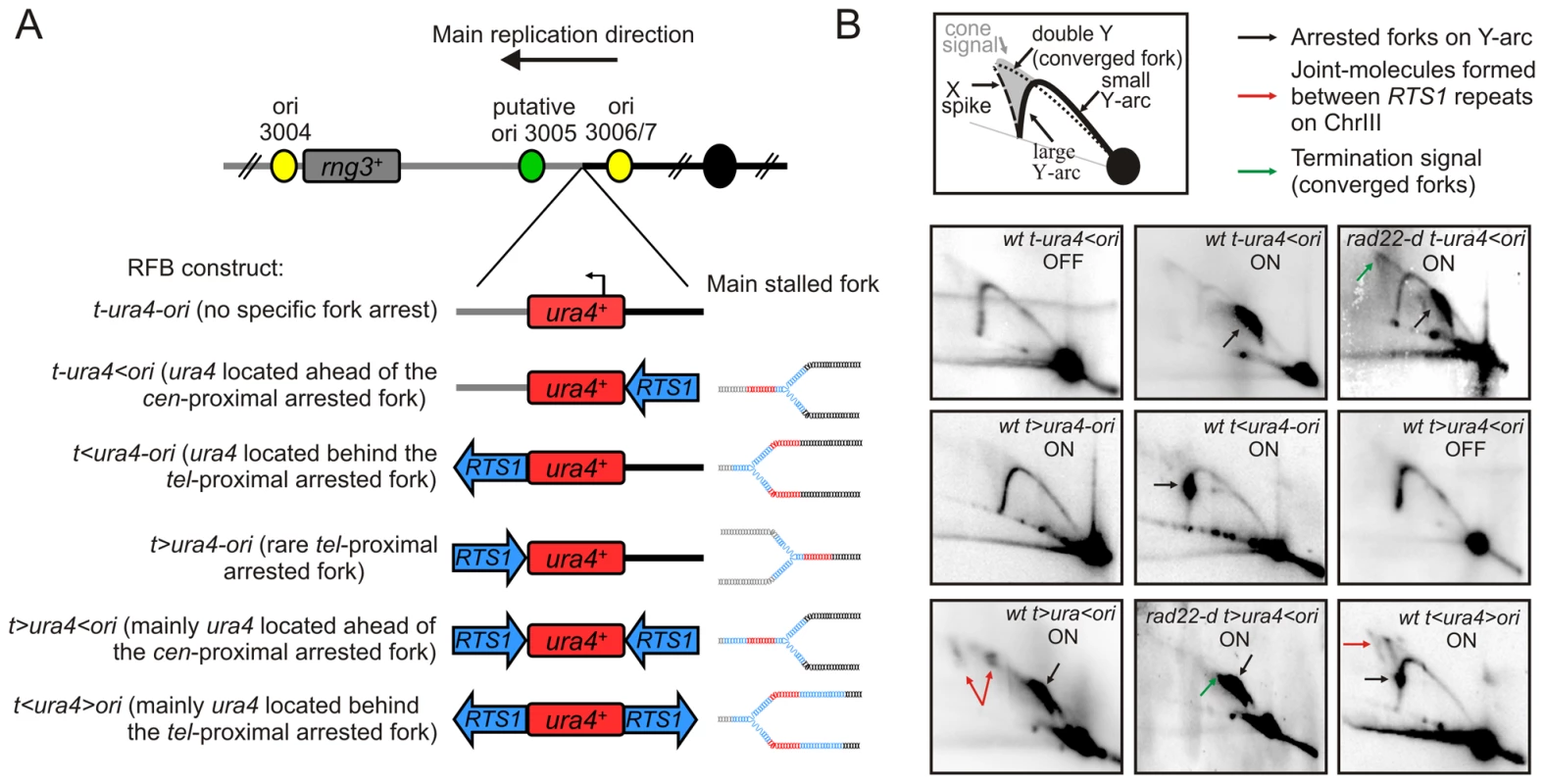
A. Diagrams of fork-arrest constructs. Centromere-proximal and telomere-proximal regions are represented in black and grey, respectively. Strong or putative replication origins (ori) and the centromere are indicated by yellow, green and black circles, respectively. Blues arrows indicate the polarity of the RTS1-RFB. The ura4+ gene is indicated in red and the arrow indicates its direction of transcription. Representations of the primary arrested fork structure are given for each construct. The name of each fork-arrest construct is given using the following nomenclature: “t” and “ori” refer to the telomere and the replication origin 3006/7, respectively; “<“ and ”>” indicate the RTS1-barrier and its polarity (< blocks replication forks moving from the ori3006/7 towards the telomere, and > blocks replication forks moving from the telomere towards the origin 3006/7. B. Diagrams of replication intermediates (RIs) within the AseI fragment analysed by 2DGE (top panel). Representative RIs analysed by 2DGE in indicated strains in OFF (Rtf1 being repressed) and ON (Rtf1 being expressed) conditions. Signal corresponding to arrested forks, joints-molecules (JMs) and termination structures are indicated by black, red and green arrows, respectively. Note that the t>ura4-ori construct does not result in a strong fork arrest as the RTS1-RFB is not orientated in the main direction of replication (see text for details). Conditional fork-arrest constructs to investigate fork-arrest-induced genome instability
The RTS1 sequence was inserted on the tel-proximal side of ura4 creating the t<ura4-ori locus. 2DGE analysis of this construct revealed a strong fork arrest signal on the descending large Y arc (Figure 1A and 1B, t<ura4-ori ON). The ura4+ gene, used in this system as a reporter to score genetic instability, is located behind the arrested fork when the RTS1-RFB is active at the t<ura4-ori locus and ahead of the arrested fork at the t-ura4<ori locus. This explains the distinct position of the arrested fork signal on the Y arc. Inversion of the RTS1 sequence at the tel-proximal side of ura4 created the t>ura4-ori locus and no fork arrest signal was detected for this construct by 2DGE when Rtf1 was expressed (Figure 1A and 1B, t>ura4-ori ON). Thus, RTS1 behaves as a polar RFB at the ura4 locus, and replication across this locus is strongly unidirectional due to the relative positions of the origins [42].
Introducing a second RTS1 sequence, such that the two RTS1 sequences are inverted repeats (IRs), created t>ura4<ori and t<ura4>ori loci (Figure 1A and 1B, t>ura4<ori and t<ura4>ori ON). Given the orientation of the polar RTS1-RFB in the t<ura4>ori strain, converging forks cannot be blocked. Whereas block of converging forks can virtually occur in the t>ura4<ori strain, 2DGE in this construct revealed that forks arrested on the cen-proximal side of ura4 were efficiently recovered by recombination before forks are arrested on the tel-proximal side. Indeed, joint-molecules (JMs) resulting from recombination between RTS1 repeats were detected by 2DGE (see red arrows on Figure 1B, t>ura4<ori and t<ura4>ori ON). Resolution of these JMs gives rise to chromosomal rearrangements [20]. In the absence of homologous recombination (i.e. in a rad22-d mutant), JMs were not detected and termination signals accumulated (see green arrow on Figure 1B, t>ura4<ori rad22-d strain). Similarly, termination signals accumulated in the rad22-d t-ura4<ori strain (see green arrow on Figure 1B, t-ura4<ori rad22-d), showing that, when arrested forks are not restarted by homologous recombination, the RTS1-RFB behaves as a hot spot for replication termination [20].
A single fork arrest induces genomic deletions
We investigated fork-arrest-induced genome instability by selecting for cell resistance to 5-FOAR, the result of loss of ura4+ function. Inducing fork-arrest at t-ura4<ori increased ura4 loss 3 fold (Table 1). Rtf1 expression in the t-ura4-ori and t>ura4-ori strains did not cause site-specific fork-arrest at ura4 as assessed by 2DGE and did not increase the rate of ura4 loss. Thus, ura4 loss results from the RTS1-RFB activity and not simply from the presence of RTS1 and/or Rtf1 expression (Table 1). To investigate the nature of this genetic instability, primers were designed to amplify the ura4 coding sequence and, as a control, the essential rng3 gene, mapping 30 kb tel-proximal to ura4, that should not be rearranged (Figure 2A and 2B) [35]. The absence of ura4 amplification was classified as a deletion event; sequencing of amplified ura4 sequence was used to identify point mutation events (Figure 2B).
Fig. 2. A single fork-arrest induces GCRs that are stimulated by inverted repeats near the site of fork arrest. 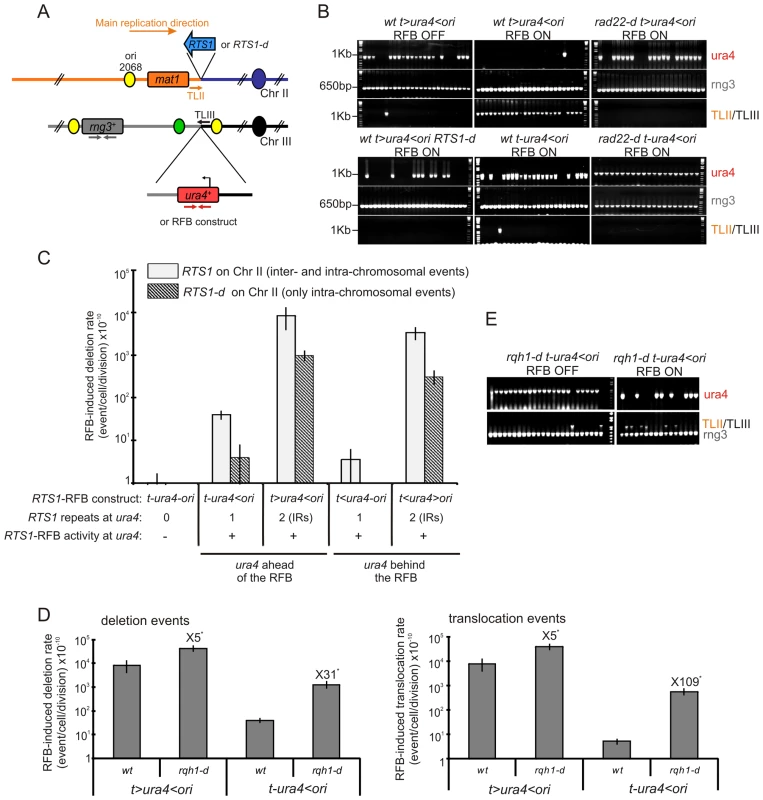
A. Diagrams of chromosome II containing or not the RTS1 sequence (blue arrow or RTS1-d) and of chromosome III containing ura4+ alone or associated with RTS1-RFB constructs. The RTS1 sequence maps near the mat1 locus where it helps to ensure unidirectional replication [62]. Primers used for amplifying the 1 Kb ura4 fragment or the 650 bp rng3 fragment are depicted in red and grey, respectively. Primers used to amplify the translocation junction (1.2 kb) are represented in orange on chromosome II (TLII) and in black on chromosome III (TLIII). B. Representative PCR-amplifications from 5-FOAR colonies of the indicated strains; ON and OFF refers to the RTS1-RFB being active or not, respectively. PCR products and their sizes are indicated on the figure. C. Effect of intra- and inter-chromosomal recombination between RTS1 repeats on fork-arrest-induced genomic deletion. RTS1-RFB activity and ura4 location with respect to the RFB are given for each construct. The % of deletion events, as determined by the PCR assay, was used to balance the rate of ura4 loss. Then, the RFB-induced deletion rate was calculated by subtracting the rate obtained in the presence of thiamine (Rtf1 being repressed) from the rate obtained in the absence of thiamine (Rtf1 being expressed). The values reported are means of at least 3 independent median rates. Error bars correspond to the standard error (SE). D. Effect of Rqh1 on RFB-induced deletions (left) and translocations (right), as described for panel C. Error bars indicate SE. Statistically significant fold differences between the rqh1-d and the wild-type strains are indicated with an *. E. Representative PCR amplifications from 5-FOAR colonies of the rqh1-d t-ura4<ori strain, as described for panel B. (Refer to Figure S1 for corresponding rates of deletion and translocation when Rtf1 is expressed or not). Tab. 1. Rates of ura4 loss (including genomic deletion, translocation, and mutation events), calculated using the method of the median. 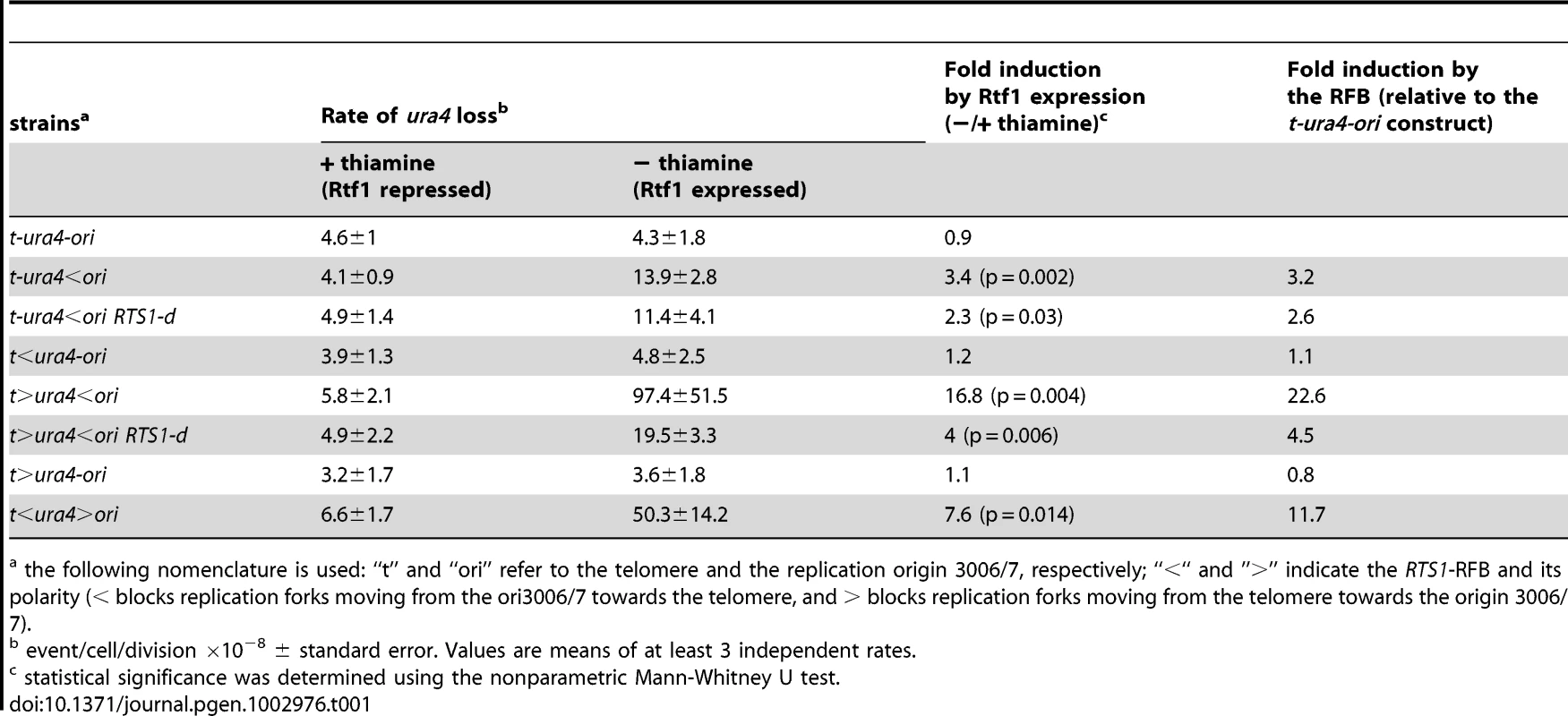
the following nomenclature is used: “t” and “ori” refer to the telomere and the replication origin 3006/7, respectively; “<“ and ”>” indicate the RTS1-RFB and its polarity (< blocks replication forks moving from the ori3006/7 towards the telomere, and > blocks replication forks moving from the telomere towards the origin 3006/7). A single arrested fork at the t-ura4<ori locus was sufficient to increase the rate of genomic deletion up to 40 times over spontaneous events (i.e. in the t-ura4-ori strain, p = 0.006) (Figure 2C and Figure S1A). Fork-arrest-induced deletion was recombination-dependent. Spontaneously (i.e. when the RTS1-barrier was inactivated), the rate of genomic deletion in rad22-d or rhp51-d strains was higher than that in the wild-type strain (Figure S1B). Nonetheless, no further increase in the rate of genomic deletion was observed in the surviving rad22-d or rhp51-d cells upon activation of the RTS1-barrier (Figure S1B, t-ura4<ori). Frequent spontaneous genomic deletion in the rad22-d or rhp51-d strains is consistent with previous reports showing that mutations in recombination genes are associated with an increase level of GCRs [14], [43], [44]. Deleting the natural RTS1 sequence from chromosome II abolished deletion events at collapsed forks, indicating that fork-arrest-induced deletion was also mediated by inter-chromosomal recombination (Figure 2C and t-ura4<ori RTS1-d on Figure S1A). Thus, these data are consistent with the view that homologous recombination makes a major contribution to suppressing genome instability, but can occasionally drive non allelic recombination events leading to GCRs [35], [45].
We detected no fork-arrest-induced deletion in the t<ura4-ori strain, in contrast to the t-ura4<ori strain (Figure S1A and Figure 2C). The ura4 marker is located behind and ahead of collapsed forks in the t<ura4-ori and t-ura4<ori strains, respectively (Figure 1A). Therefore, replicated regions, located behind collapsed forks, do not display instability, and fork-arrest-induced deletion occurs within unreplicated regions immediately in front of arrested forks. Overall, our data establish that genomic deletion at collapsed forks results from inappropriate recombination between ectopic sequences during the process of fork recovery by recombination proteins.
Inverted repeats stimulate fork-arrest-induced deletion by promoting inter - and intra-chromosomal recombination
Inverted repeats (IRs) are structural elements often associated with genome rearrangements [11], [14], [46], [47]. We investigated the effects of IRs in the vicinity of the RTS1-RFB on fork-arrest-induced genomic deletion. We first compared the t>ura4<ori strain (IRs flanking ura4) to the t-ura4<ori strain (no IRs near the RTS1-RFB). The rate of fork-arrest-induced genomic deletion was 200 times higher in the t>ura4<ori than that in the t-ura4<ori strain (p = 0.009, Figure 2C and Figure S1A). Thus, intra-chromosomal ectopic recombination permitted by the RTS1 sequence on the tel-proximal side of ura4 accounted for 99.5% of the genomic deletions observed in the t>ura4<ori strain (Figure 2C, compare with t-ura4>ori). Preventing inter-chromosomal recombination by deleting RTS1 from the chromosome II (t>ura4<ori RTS1-d) abolished 90% of deletion events (Figure 2C and Figure S1A). Thus, genomic deletions induced by fork-arrest near IRs are due to inter - and intra-chromosomal recombination events. In support of this, stimulation of fork-arrest-induced deletion by IRs is mediated by homologous recombination. Indeed, the rate of genomic deletion was not increased upon induction of the RTS1-RFB in the surviving population of t>ura4<ori rad22-d and rhp51-d strains (Figure S1B). These data indicate that IRs favour genomic deletion at collapsed forks by promoting inappropriate inter - and intra-chromosomal recombination during fork recovery by recombination proteins.
We verified that our data were not influenced by the orientation of IRs or by rare blocking of converging forks in the t>ura4<ori strain. We analysed the t<ura4>ori construct in which RTS1 repeats are in the opposite orientations relative to the t>ura4<ori construct, such that forks converging towards ura4 cannot be blocked (Figure 1). The rate of fork-arrest-induced genomic deletion was 1,000 times higher in the t<ura4>ori than that in the t<ura4-ori strain, that does not contain IRs near the RTS1-RFB (p = 0.008, Figure 2C and Figure S1A). Thus, intra-chromosomal recombination, permitted by the RTS1-RFB sequence on the cen-proximal side of ura4, accounted for nearly 100% of the genomic deletions observed in the t<ura4>ori strain (Figure 2C, compare with t<ura4-ori). Preventing inter-chromosomal recombination by deleting RTS1 from the chromosome II (t<ura4>ori RTS1-d) abolished 90% of deletion events (Figure 2C and Figure S1A). Importantly, the deletion rates for the t<ura4>ori and t>ura4<ori strains were not significantly different (Figure 2C), showing that IRs cause genomic deletion at collapsed forks irrespective of their orientations and independently of blockage of converging forks.
A single collapsed fork induces translocations that are stimulated by IRs
Fork-arrest at t>ura4<ori results in translocations between ectopic RTS1 repeats on chromosomes II and III [35]. We investigated the influence of IRs on fork-arrest-induced translocation. We designed primers to amplify the predicted translocation junction between chromosomes II and III (TLII and TLIII on Figure 2A and 2B). A single arrested fork at the t-ura4<ori locus was sufficient to increase the translocation rate to 5 times higher than the spontaneous rate (p = 0.002, Figure 2D and Figure S1C). The translocation rate for the t>ura4<ori construct (containing IRs) was 1,500 fold higher than that for the t-ura4<ori strain that does not contain IRs near the RTS1-RFB (p = 0.009, Figure 2D and Figure S1C). Thus, intra-chromosomal recombination accounted for nearly 99% of translocation events observed in the t>ura4<ori construct (Figure 2D and Figure S1C, compare with t-ura4<ori). No translocation events were detected when inter-chromosomal recombination was prevented by deleting RTS1 from the chromosome II (t>ura4<ori RTS1-d on Figure 2B). Therefore, as reported for genomic deletions, fork-arrest-induced translocation associated with IRs is due to inter - and intra-chromosomal ectopic recombination. No translocations were detected in the t<ura4>ori strain (data not shown), so we cannot formally exclude the possibility that fork-arrest-induced translocations in the t>ura4<ori strain was caused by blocking of converging forks. However, as no translocation event occurred in the absence of Rad22Rad52 or Rhp51Rad51, it is most likely that translocations occur during fork recovery by recombination (Figure 2B and [35]). Overall, our data indicate that recovery of a single collapsed fork causes translocations and IRs near the site of fork-arrest stimulate translocations by promoting inappropriate inter - and intra-chromosomal recombination.
Fork-arrest-induced GCRs are caused by inter - and intra-chromosomal recombination. We noticed a slightly greater contribution of intra - than inter-chromosomal recombination (Figure 2C). This is consistent with ectopic recombination preferentially occurring at the most proximal homologous sequence, as previously reported [48]. Nonetheless, the rate of fork-arrest-induced deletion in the t>ura4<ori strain (8.4 10−7) was not simply the sum of the rates of intra-chromosomal recombination events (9.9 10−8 in the t>ura4<ori RTS1-d strain) and inter-chromosomal recombination events (4 10−9 in the t-ura4>ori strain). Similar reasoning can be applied for the t<ura4>ori strain. Thus, independent intra - and inter-recombination events cannot themselves explain high rate of GCRs induced by fork arrest near IRs. Therefore, we infer that there is interplay between inter - and intra-chromosomal recombination such that fork-arrest-induced GCRs may involve recombination between three homologous sequences (tri-parental recombination).
The RecQ helicase Rqh1 prevents GCRs at collapsed forks
To confirm that fork-arrest-induced GCRs are the result of inappropriate ectopic recombination during fork recovery, we analysed the involvement of the RecQ helicase Rqh1. We previously reported that Rqh1 limits inappropriate template switching of stalled nascent strands without affecting the efficiency of fork restart [20]. In the t-ura4<ori construct (in which only inter-chromosomal recombination is possible), fork-arrest-induced deletion and translocation rates were 31 and 109 times higher in the rqh1-d strain than that in the wild-type control, respectively (p = 0.0003, Figure 2D–2E and Figure S1C). For the t>ura4<ori construct (containing IRs near fork-arrest), fork-arrest-induced deletion and translocation rates were 5 times higher in the rqh1-d than that in the wild-type control (p = 0.0007, Figure 2D–2E and Figure S1C). Thus, Rqh1 limits GCRs at collapsed forks by preventing inappropriate ectopic recombination during the process of fork recovery by recombination proteins.
A single fork arrest induces mutations
We analysed the effects of collapsed forks on the mutation rate. We sequenced the ura4 coding sequence from 5-FOAR isolated cells and identified base-substitutions, frame-shifts and small insertions and duplications between short tandem repeats (Table 2). A single collapsed fork in the t-ura4<ori strain increased the overall mutation rate up to 10 times over spontaneous events (Figure 3A, p = 0.003). Similar increases in the overall mutation rate were found for the strains with IRs near the arrested fork and those with RTS1 deleted from chromosome II (Figure 3A and Figure S2A). Thus, fork-arrest-induced mutation is not mediated by inappropriate ectopic recombination. Induction of the RTS1-RFB in the t<ura4-ori strain did not increase the mutation rate of the ura4 gene. Thus, as for GCRs, replicated regions behind arrested forks are not prone to mutation. This observation rules out the hypothesis that fork-arrest-induced mutation is a consequence of the accumulation of damaged single-stranded DNA behind collapsed forks (see discussion). Our data suggest that recovery from collapsed forks results in error-prone DNA-synthesis.
Fig. 3. Fork-arrest induces mutations. 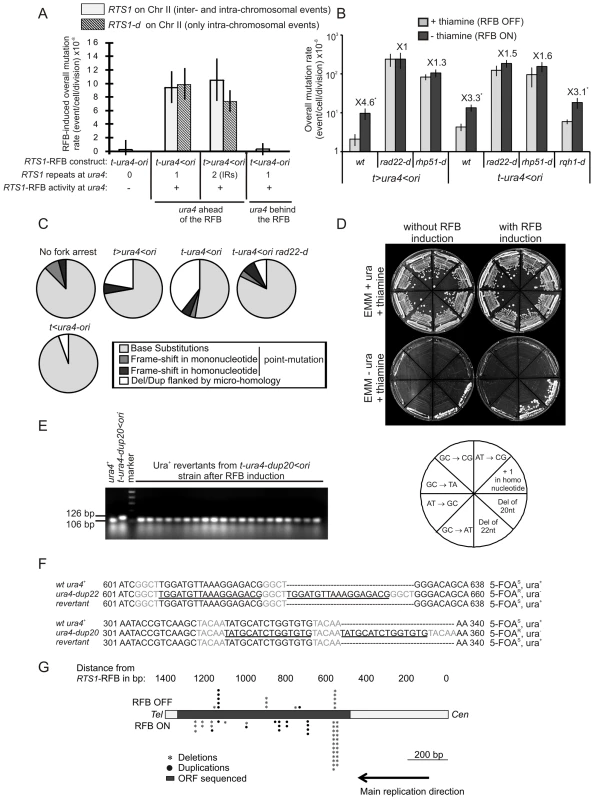
A. Effect of intra- and inter-chromosomal recombination between RTS1 repeats on fork-arrest-induced mutation rates (base-substitutions, frame-shifts and small insertions or deletions between short tandem repeats). RTS1-RFB activity and ura4 location with respect to the RFB are given for each construct. The % of mutation events, as determined by the PCR assay and sequencing, was used to balance the rate of ura4 loss. Then, the RFB-induced mutation rate was calculated by subtracting the rate obtained in the presence of thiamine (Rtf1 being repressed) from the rate obtained in the absence of thiamine (Rtf1 being expressed). The values reported are means of at least 3 independent median rates. Error bars correspond to SE. (Refer to Figure S2 for corresponding rates of mutation when Rtf1 is expressed or not). B. Rate of mutation for indicated strains; ON and OFF refers to the RTS1-RFB being active or not, respectively. The % of mutation events, as determined by the PCR assay and sequencing, was used to balance the rate of ura4 loss. The values reported are means of at least 3 independent median rates. Error bars correspond to SE. Statistically significant fold differences in mutation rates between the “OFF” and “ON” conditions are indicated with an *. C. Spectra of mutation events in indicated strains upon RFB induction (refer to Table 2 for exact numbers and to Figure S3 for mapping of deletions/duplications and their features). D. Strains harbouring the ura4 alleles with a single base-substitution or frame-shift or duplication of 20 or 22 nt, together with the RTS1-RFB in the t-ura4<ori configuration were streaked onto the indicated media after cell growth with (RFB “OFF”) or without (RFB “ON”) thiamine. The bottom diagram indicates strain positions and the mutation events required to obtain Ura+ revertants. E. PCR analysis of Ura+ revertants isolated from the t-ura4-dup20<ori strain (duplication of 20 nt in ura4) after RFB induction. With the primers used, a 106 bp fragment is amplified from the ura4+ strain and a 126 bp fragment is amplified from the t-ura4-dup20<ori strain. F. Sequence alignments of ura4-dup22, ura4-dup20, ura4+ alleles and corresponding Ura+ revertants. Micro-homologies are indicated in grey and duplicated sequences are underlined in black. The phenotype of each allele is indicated on the figure. G. Map of deletion and duplication events within the ura4 ORF. Tab. 2. Mutations spectra in the indicated strains. 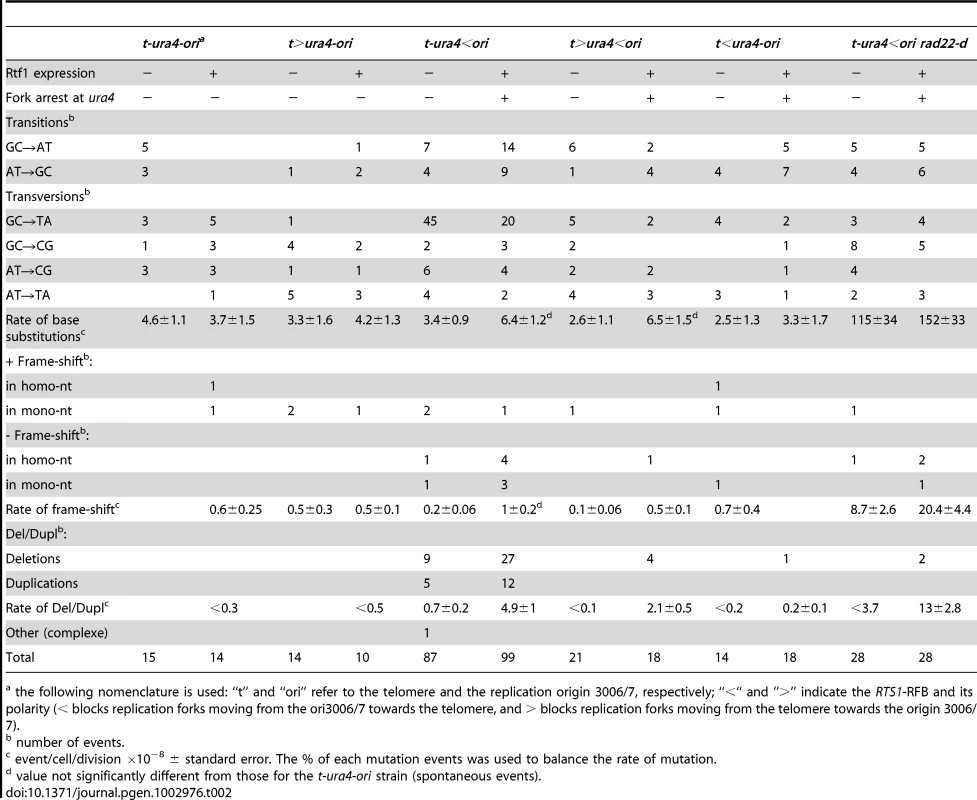
the following nomenclature is used: “t” and “ori” refer to the telomere and the replication origin 3006/7, respectively; “<“ and ”>” indicate the RTS1-RFB and its polarity (< blocks replication forks moving from the ori3006/7 towards the telomere, and > blocks replication forks moving from the telomere towards the origin 3006/7). Collapsed forks result specifically in replication slippage
We then analysed the spectra of mutations found in the ura4 ORF by sequencing the PCR products. The rates of base-substitutions and frame-shifts were not significantly increased by the RTS1-RFB activity over spontaneous events (i.e. compare to t-ura4-ori strain, Figure 3C and Table 2). In contrast, the rate of deletions and duplications (Del/Dup) flanked by short homology was increased by 7 times over spontaneous events in the t-ura4<ori strain, but not in the t<ura4-ori strain (Figure 3C and Table 1). These data further confirm that fork-arrest does not promote mutation events behind collapsed forks.
We used reversed mutation assays to test if fork-arrest at the RTS1-RFB specifically induced Del/Dup mutations. We made use of strains harbouring a single mutation within the ura4 ORF: either a single base-substitution or a −1 frame-shift in homo-nucleotide (Figure S2B). We also studied strains harbouring either a duplication of 20 or 22 nt flanked by 5 or 4 bp of micro-homology, respectively (defined as ura4-dup20 and ura4-dup22, Figure S2B). These non-functional ura4− alleles were inserted in front of the RTS1-RFB in the t-ura4<ori configuration and we then tested whether fork arrest could restore a functional ura4+ gene. Activation of the RTS1-RFB at ura4 increased the frequency of Ura+ revertants up to 15 and 7 times in strains harbouring ura4-dup22 and ura4-dup20, respectively (Figure 3D and Figure S2B). Thirty Ura+ colonies were studied by PCR and all gave a product of the same size as the wild-type ura4+ gene: they had therefore lost the duplication (Figure 3E and data not shown). Sequencing the full ura4 ORF confirmed that Ura+ revertants contained an intact ura4+ sequence, showing that the reversion of these alleles was due solely to the precise deletion of 20 or 22 nt (Figure 3F and data not shown). In contrast, activation of the RTS1-RFB did not increase the frequency of Ura+ revertants of strains harbouring ura4 alleles with a single base-substitution or a −1 frame-shift (Figure 3D and Figure S2B). Thus, collapsed forks tend to induce deletion events between short tandem repeats rather than base-substitution or frame-shift mutations.
Among Del/Dup events, deletions represented the two-third of events in the t-ura4<ori strain (Table 2). The median size of Del/Dup events was 24 and 22 nt respectively, and Del/Dup occurred between short direct repeats 1 to 10 nt long (Figure S3). Thus, the ura4-dup20 and ura4-dup22 alleles used in the reverse mutation assay were representative of the Del/Dup events observed. Del/Dup flanked by micro-homology result from intra-molecular template switching mechanisms in which nascent strands dissociate from the template and misalign with the template when restarting the elongation step. This leads to loop formation, either in the nascent strand or in the template, resulting in duplication or deletion events, respectively [49]. Consequently, we will hereafter refer to Del/Dup as replication slippage. Replication slippage was observed all along the ura4 ORF and up to 1.2 kb ahead of the arrested fork, even if a hot spot of deletion was present 500 bp away from the RTS1-RFB (Figure 3G and Figure S3B). Thus, our data suggest that the DNA synthesis is prone to replication slippage at least for the first 1,200 nt synthetized during the recovery of collapsed forks. Inaccuracy of DNA synthesis on further distances was not directly addressed.
Replication slippage results from error-prone DNA synthesis during fork recovery
To confirm that replication slippage occurs as forks recover, and not behind the fork in the DNA already replicated, we inserted the ura4-dup20 or the ura4-dup22 allele either behind (in the t<ura4-ori configuration) or in front of the RTS1-RFB (in the t-ura4<ori configuration) (Figure 4). This allows the analysis of the same event of replication slippage behind and ahead of collapsed forks. In the t-ura4<ori configuration, induction of the RTS1-RFB resulted in a 8 and 16 fold increases in the replication slippage frequency for the ura4-dup20 and ura4-dup22 alleles, respectively (Figure 4A and 4B). Similar increases in the rate of replication slippages were observed (Figure 4C). In contrast, in the t<ura4-ori background, the frequency of replication slippage was induced by only 2–3 fold by the RTS1-RFB (Figure 4B–4C). These data confirm that DNA located ahead of collapsed forks is more prone to replication slippage than replicated DNA adjacent to arrested forks, further evidence that replication slippage arises during fork recovery.
Fig. 4. Fork recovery by homologous recombination results in replication slippage. 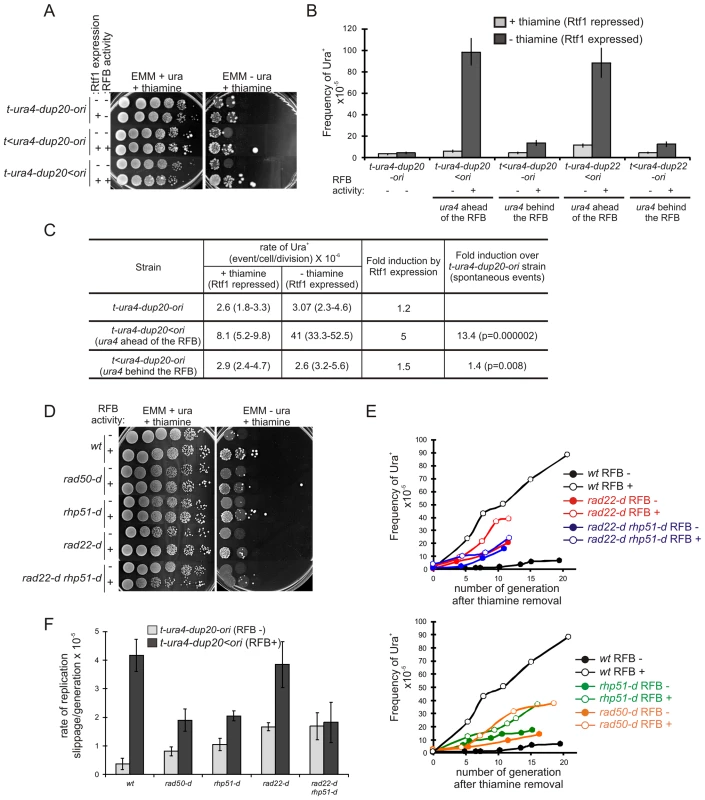
A. Serial tenfold-dilutions from indicated strains (t-ura4-dup20-ori associated or not with the RTS1-RFB) were spotted onto the indicated media after cell growth with (Rtf1 -, repressed) or without (Rtf1 +, expressed) thiamine. RTS1-RFB activity is given for each construct and condition. B. Frequency of Ura+ revertants in indicated strains after cell growth with (Rtf1 repressed) or without (Rtf1 expressed) thiamine. The RTS1-RFB activity is given for each construct and condition. Values correspond to the mean of at least three independent experiments and error bars correspond to the standard error of the mean (SEM). C. Rate of replication slippage in the indicated strains and conditions. The rate of Ura+ revertants was calculated using the method of the median from at least 11 independent cultures. Values in brackets indicate the 95% confidence interval. Statistical significance was detected using the nonparametric Mann-Whitney U test. D. Serial tenfold-dilutions from the strains indicated spotted onto the media indicated after cell growth without thiamine. RTS1-RFB activity “–” refers to the strain t-ura4-dup20-ori and “+” refers to the strain t-ura4-dup20<ori. E. Kinetics of Ura+ revertants frequency for the strains indicated as a function of the number of generations after thiamine removal. RTS1-RFB activity “–” refers to the strain t-ura4-dup20-ori and “+” refers to the strain t-ura4-dup20<ori. The values reported are the means of two experiments. F. The rate of replication slippage/generation for the strains indicated with (t-ura4-dup20<ori) or without (t-ura4-dup20-ori) the active RTS1-RFB. The rate was calculated from the slope of the curves presented in panel F. The values reported are means of three independent experiments and error bars correspond to SE. Replication slippage results from forks restarted by recombination
Replication slippage occurs in DNA in front of (and not behind) the arrested fork, this DNA being replicated only after restart of the fork. Thus, a defect preventing fork recovery would be expected to abolish the error-prone DNA synthesis during restart. We analyzed fork-arrest-induced mutation in recombination mutants in which collapsed forks at the RTS1-RFB cannot recover, resulting in cell death. Induction of the RTS1-RFB did not increase the overall mutation rate in the surviving populations of t>ura4<ori or t-ura4<ori rad22-d and rhp51-d strains (Figure 3B). In addition, only 7% of mutation events in the survivors of the rad22-d t-ura4<ori strain were Del/Dup mutations, compared to 40% in the wild-type strain (Figure 3C and Table 1). We currently cannot assess mutation events associated with defects in fork recovery because this appears to be lethal in the absence of recombination. Nevertheless, our data are consistent with fork-arrest-induced replication slippage being dependent on homologous recombination. The rad22-d and rhp51-d strains are themselves spontaneously mutagenic. Consequently, any small increase in the fork-arrest-induced mutation rate might be masked by the high frequency of spontaneous 5-FOAR cells in rad22-d and rhp51-d strains. We therefore used a more specific mutation assay, based on the ura4-dup20 allele, to determine the rate of replication slippage induced by the RTS1-RFB over spontaneous events.
Strains carrying mutations in recombination genes grow slowly, so replication slippage was scored as a function of the number of generations following thiamine removal (i.e. generations subject to fork arrest at ura4) (Figure 4D and 4E). In the wild-type strain, fork arrest at the RTS1-RFB resulted in a 10 fold-increase in the frequency of replication slippage, as expected. In recombination mutants (rad50-d, rhp51-d and rad22-d), fork-arrest at the RTS1-RFB increased the frequency of replication slippage by only 2 times over spontaneous events: therefore, replication slippage occurs less frequently in survivors from recombination mutants than those from the wild-type strain (Figure 4D–4F). Based on 2DGE analysis, fork-restart is severely impaired in the absence of Rad22Rad52 (Figure 1B and [20]), such that even the two-fold induction in replication slippage by fork arrest in the rad22-d strain was surprising. The rad22-d strain accumulates suppressors involving the Fbh1 helicase that limits Rhp51Rad51 - dependent recombination at blocked replication forks [50], [51]. Therefore, we analyzed replication slippage in the rad22-d rhp51-d double mutant in which no homologous recombination event occurs. In this background, there was no detectable fork-arrest-induced replication slippage (Figure 4D–4F). Thus, complete defect in fork restart results in a complete abolition of fork-arrest-induced replication slippage in the surviving population. Overall, our data establish that replication slippage results from inaccurate DNA synthesis during the restart of collapsed forks by recombination.
Replication stress leading to fork collapse induces replication slippage
We investigated the effects of replication stress, other than the replication block imposed by the RTS1-RFB, on replication slippage. Strains harbouring ura4− alleles (base-substitutions, −1 frame-shift, and ura4-dup20) were exposed to replication-blocking agents or UV-C-induced DNA damages and the frequency of Ura+ revertants was scored. Three hours of treatment with either the topoisomerase I inhibitor camptothecin (CPT) or mitomycin C (MMC), an inter-strand cross-linking agent (ICls), increased the frequency of Ura+ revertants by 3 to 4 fold in the ura4-dup20 strain (Figure 5A and 5B). At equivalent survival (70–90%), DNA-damages induced by a dose of 100 J/m2 of UV-C did not increase the frequency of Ura+ revertants in the ura4-dup20 strain. Increasing the UV-C dose (150 J/m2) resulted in an increased reversion effect. The other ura4 alleles exhibited an opposite behaviour pattern. As expected, UV-C-induced DNA damages, but not CPT or MMC treatment, increased the frequency of Ura+ revertants of the base-substitution and the −1 frame-shift mutants (Figure 5A). Thus, replication slippage, unlike other point mutations, appears to be a mutation event specifically induced by replication stress.
Fig. 5. Collapsed forks, but not stalled forks, induce replication slippage. 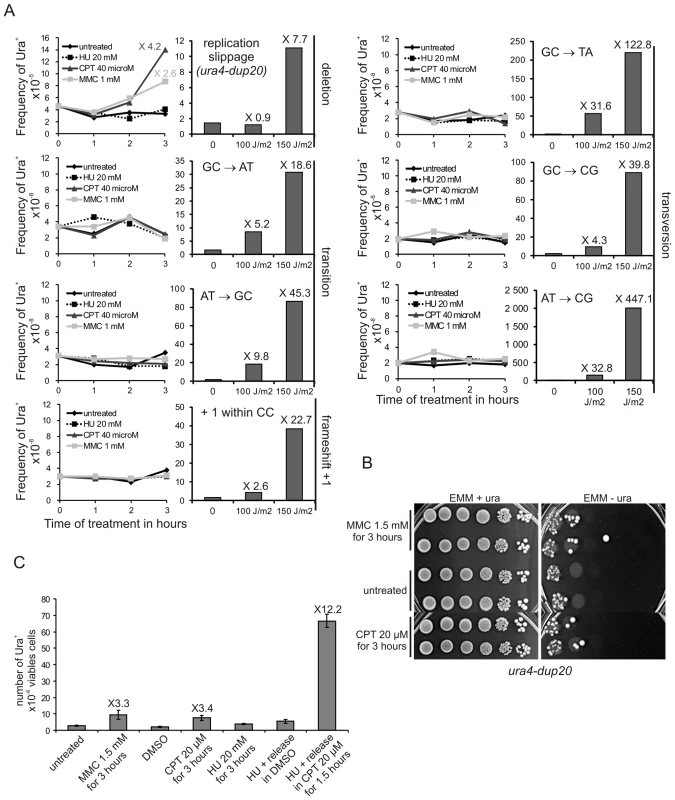
A. Left panel: the frequency of Ura+ revertants as a function of time-contact with indicated drugs for the indicated ura4 alleles (single base-substitution, frame-shift, duplication of 20 nt). Right panel: the frequency of Ura+ revertants in response to UV-C irradiation as a function of dose for the indicated ura4 alleles. The values reported are means of two independent experiments. Numbers indicate fold difference in the frequency of Ura+ revertants between the treated and untreated control samples. For ura4 alleles containing base-substitutions or frame-shifts, the mutation event required to obtain Ura+ revertants is indicated on the figure. B. Serial tenfold-dilutions from ura4-dup20 strain spotted onto the media indicated after treatment with MMC or CPT as indicated. C. Frequency of Ura+ revertants after the indicated treatments in the ura4-dup20 strain. DMSO (the vehicle) was used as control for CPT treatment. The values reported are means of at least three independent experiments. Error bars correspond to SEM. Hydroxyurea (HU) that prevents the bulk of dNTP synthesis during S-phase by inhibiting the ribonucleotide reductase, results in a slow-down of fork progression which did not induce replication slippage (Figure 5A). In contrast, CPT and MMC treatments that lead to replication stress by causing fork collapse induced replication slippage. Homologous recombination is repressed during HU treatment and recombination proteins are recruited to collapsed but not stalled forks [52]–[54]. Consistent with this, we found that the rad22-d mutant is highly sensitive to acute exposure to CPT, but not to HU (Figure S4). Thus, acute exposure to HU results in stalled forks that recover without recombination, whereas recombination may be required for restarting forks that have collapsed due to CPT or MMC treatment. We confirmed that CPT-induced replication slippage results from collapsed forks and was thus S-phase specific: the ura4-dup20 strain was synchronized in early S-phase by HU treatment and released into S-phase with or without CPT. HU-synchronization and release into DMSO (used as vehicle for CPT) did not induce replication slippage. In contrast, the release of cells into S-phase in the presence of CPT stimulated replication slippage up to 12 fold (Figure 5C). These data indicate that CPT-induced fork collapse results in error-prone DNA synthesis characterized by replication slippage. These experiments further support the view that replication slippage results from recovery of collapsed forks by recombination and point out that the RTS1-barrier is representative of collapsed forks restarted by homologous recombination.
TLS–DNA polymerases are not involved in fork-arrest-induced replication slippage
To investigate the inaccuracy of the DNA synthesis occurring immediately following the restart of collapsed forks, we analysed the involvement of TLS-DNA polymerases. In fission yeast, TLS pathways require either mono - or poly-ubiquitination of the clamp loader PCNA on lysine 164 [55]. We found that mutating this lysine to arginine residue did not affect replication slippage induced by the RTS1-RFB. None of Rev1, Rev3 or DinB DNA polymerases were required for fork-arrest-induced replication slippage (Figure S5). Therefore, the error-prone DNA synthesis associated with fork recovery by recombination does not rely on TLS DNA polymerases activity.
The mismatch repair pathway does not counteract fork-arrest-induced replication slippage
The mismatch repair (MMR) pathway is temporally coupled to DNA replication, and MMR components are associated with replication centres [56]. The heterodimer Msh2/Msh6 recognises mispaired DNA and Msh2/Msh3 recognises small DNA loops up to 31 bases long, arising from replication slippage [57]. The failure to repair small DNA loops results in more frequent insertions and deletions [58]. Therefore, MMR activity could potentially lead to an underestimation of the extent of fork-arrest-induced replication slippage. However, replication slippage induced by the RTS1-RFB activity was as frequent in msh2-d, msh6-d and msh3-d strains as in wild-type control. Also, spontaneous replication slippage at ura4 (without RTS1-RFB) was unaffected by the absence of MMR proteins (Figure S5). Therefore, there is no evidence that MMR repairs small DNA loops (20 nt) in fission yeast and fork-arrest-induced replication slippage is not counteracted by MMR in our model system.
Discussion
Using conditional fork arrest constructs, we studied the consequences for genome instability of impediments to replication forks progression. A single fork arrest results in large-scale genomic changes and mutations that occur during recombination-mediated fork recovery (Figure 6). Inappropriate ectopic recombination at arrested forks results in GCRs, whereas appropriate restarting of the fork on the initial template results in error-prone DNA synthesis. GCRs and mutations at collapsed forks are genetically separable: Rqh1 limits fork-arrest-induced GCRs but not mutations (Figure 2D and Figure 3B). We demonstrate here that collapsed forks whose progression resumes by recombination lose accuracy during DNA synthesis, resulting in frequent intra-template switches. Thus, homologous recombination contributes to completion of DNA replication when forks progression is impeded but also fuels genome modifications both at the chromosomal and nucleotide level.
Fig. 6. Model of replication-stress-induced genetic instability at collapsed forks. 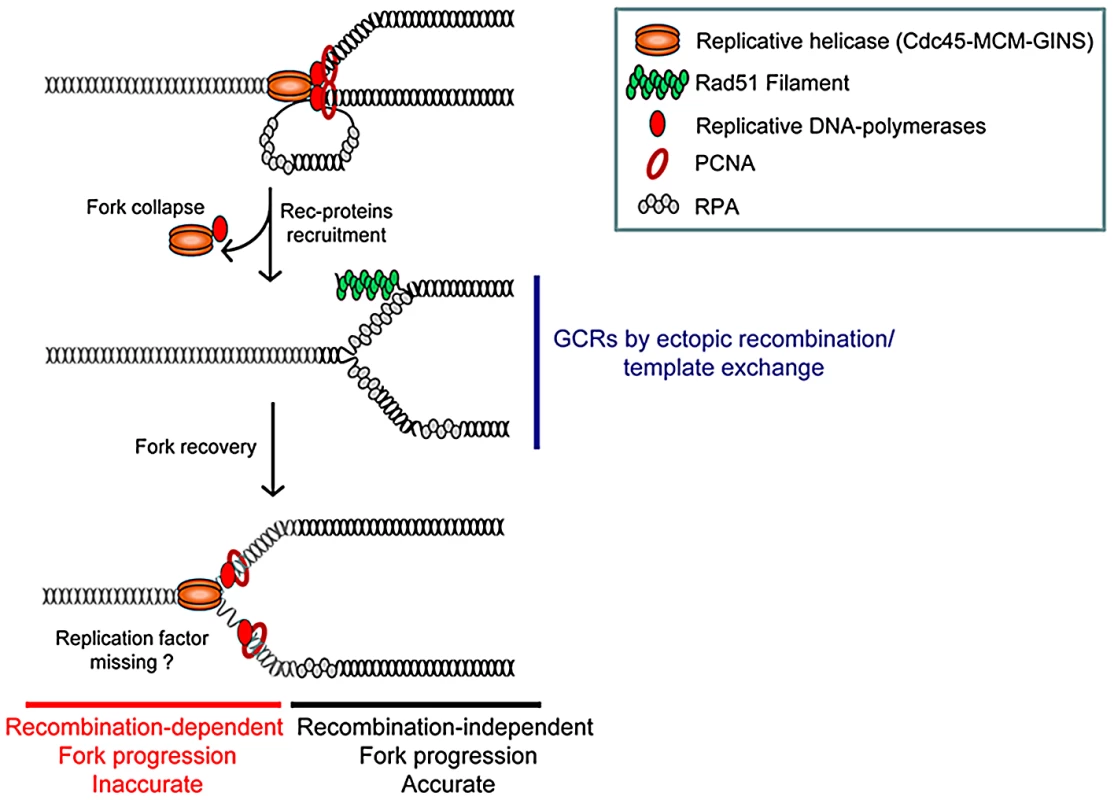
Collapsed forks might arise from torsional stress, fork breakage (i.e. at nick, ICLs), or proteins tightly-bound to DNA. Replisome disassembly at collapsed forks may favour the unwinding of the nascent strand on which Rad51 nucleates. At this initial stage of fork resumption by recombination, homology-driven template exchange can promote intra- or inter-recombination resulting in GCRs. Fork recovery by recombination overcomes the initial replication block and allows an inaccurate replisome to form (see text for details). Non allelic homologous recombination (NAHR) between low copy number repeats (LCR) contributes to recombination-mediated GCRs in mitosis and meiosis. NAHR is responsible for translocations, deletions, inversions and loss of heterozygosity [59]. Ou et al. predicted 1143 LCR pairs in the human genome liable to mediate recurrent translocations [60]. In budding yeast, a single DSB is sufficient to mediate recombination-dependent translocation [61]. Here, we report that a single collapsed fork increases the rate of genomic deletion 40 fold, and that of translocation 5 fold. Fork-arrest-induced GCRs are mediated by NAHR between heterologous chromosomes. It is not clear whether fork arrest on both homologous repeats contributes to fork-arrest-induced GCRs. We could not address this question in our model system, because the RTS1 sequence close to the mat1 locus on chromosome II has a low RFB activity [62]. Also, the recruitment of recombination proteins at the RTS1 sequence near the mat1 locus is not regulated by the level of Rtf1 expression, showing that regulating Rtf1 expression was in itself insufficient to regulate the RTS1-RFB activity at the mat1 locus [35].
Inverted repeats (IRs) are structural elements that contribute to genome instability. Impediments to replication forks progressing through IRs favour their fusion to generate acentric and dicentric inverted chromosomes [11], [14]. IRs in humans can also trigger the formation of inverted genomic segments and complex triplication rearrangements by a replication-based mechanism [47]. Here, we report that IRs near collapsed forks can increase the rate of GCRs by up to 1,500 fold. This high level of GCRs cannot be explained merely by the addition of independent inter - and intra-chromosomal recombination events. Rather, our analyses suggest that IRs may stimulate tri-parental recombination events induced by template switching of nascent strands at collapsed forks, such that three homologous sequences are involved. Similarly, recombination-dependent translocations induced by a single DSB in budding yeast is proposed to be the consequence of tri-parental recombination [63], [64]. One possible mechanism is that IRs-induced GCRs result from successive template switches initiated by nascent strands at collapsed forks, reminiscent of the multiple template switches during break-induced-replication (BIR) in budding yeast [33]. Interestingly, Rqh1 prevents fork-arrest-induced GCRs by limiting both inter - and intra-chromosomal recombination without affecting fork restart efficiency. Thus, tri-parental recombination might correspond to multiple and successive template switches between homologous repeats.
The high accuracy of DNA replication is compromised by impediments to fork progression, and recombination-mediated fork recovery results in decreased processivity of DNA synthesis (Figure 6). Recombination-induced mutations associated with DSBs or impeding DNA replication have been described previously. The formation of damaged single-stranded DNA during the resection of DSBs favours base-substitutions [65]. We detected the recruitment of the single-strand binding protein RPA up to 1.4 kb behind, but not ahead of the RTS1-arrested fork (data to be published), showing that fork-arrest-induced mutation is not correlated with damaged single-stranded DNA exposed behind collapsed forks. Nevertheless, there were rare replication slippage occurred behind RTS1-arrested forks (in the t<ura4-ori construct), suggesting that resumption of DNA synthesis can in some cases occur at a position behind the site of the collapsed fork. Recombination-dependent DNA synthesis occurring outside S-phase is also highly inaccurate during gene conversion and BIR, resulting in either template switches, base-substitutions or frame-shifts [33], [34], [66]. Elevated dNTP pools, due to activation of the DNA-damage checkpoint in G2 cells, contributes to the generation of mutations when hundreds of kbs are synthesised during BIR [34]. Here, we demonstrate that during normal S-phase progression, a single collapsed fork, restored by recombination, results in replication slippage up to 1.2 Kb away from the initial restarting point.
Recombination-induced replication slippage has been reported previously.
In fission yeast, a defect in pol alpha (swi7-H4 mutant) is associated with a recombination-mediated mutator phenotype characterized by an increased frequency of base-substitutions and Del/Dup between short direct repeats [67]. DNA-polymerase kappa (DinB) and zeta (Rev3) are responsible for the increased base-substitution rate, but the DNA-polymerases involved in Del/Dup mutations were not identified [68]. In budding yeast, a defect in polymerase delta (pol3-t mutant) results in an increased level of replication slippage, mediated by homologous recombination [49]. In contrast, the increased rate of replication slippage in the absence of the accessory subunit of polymerase delta, Pol32, does not depend on a functional recombination pathway [69]. Here, we report that recovery of collapsed forks by recombination is specifically associated with replication slippage. Nonetheless, spontaneous replication slippage events are also increased in strains mutated for recombination genes (Figure 4D–4F). Pol32 is required for BIR and replication-induced template switches leading to segmental duplication [70], [71]. Recombination is responsible for only half of these segmental duplications. Thus, it is possible that fork-restart mechanisms dependent on Pol32 and homologous recombination are prone to replication slippage and that in the absence of these pathways alternative micro-homology mediated mechanisms are revealed.
We suggest that at least two steps of the recombination-based fork recovery mechanism can compromise genome stability (Figure 6). At an initial stage, recruitment of recombination proteins on stalled nascent strands favours both fork recovery and ectopic template switches leading to GCRs. At a later stage, once the replisome has been reconstituted and the fork has resumed its progression, the nascent strands are prone to intra-template switching leading to replication slippage. The reasons for the inaccuracy of DNA synthesis associated with restarted forks during scheduled DNA replication (i.e. in S-phase) remain to be determined. One possibility is that one or more factors are missing in the rebuilt replisome during recovery by recombination. Oncogene-induced replication stress results from unbalanced DNA replication that contributes to genome instability in precancerous cells [12]. Completion of DNA replication in such stress conditions is likely to rely on recombination-mediated fork recovery that in turn generates genome instability. Insertions/deletions flanked by micro-homology, responsible for copy number variations (CNVs), have been identified both in cancer cells and also in response to replication inhibition [72], [73]; their reported sizes are between 1 Kb and several tens of mega bases, but the analysis of these features has been limited by the resolution of array-based genomics approaches. Sub-microscopic insertions/deletions flanked by micro-homology have been also described at loci in which replication origins are scarce, including the human fragile site FRA3B, the instability of which is a consequence of replication stress [74]. Interestingly, homologous recombination contributes to the stability of fragile sites by facilitating complete replication or by repairing gaps and breaks at these sites. Thus, we propose that micro-homology-mediated CNVs could be viewed as scars left by error-prone replication forks restarted by recombination.
Materials and Methods
Standard genetic and molecular biology
Strains used were constructed by standard genetic techniques and are listed in Table S1. 2DGE was performed as previously described [20]. To create ura4-dup20 and ura4-dup22 alleles associated or not with the RTS1-RFB, genomic DNA was isolated from selected 5-FOAR cells containing duplications identified by sequencing. A PCR fragment containing duplications within the ura4 ORF was amplified using the following primers: TTCTGTTCCAACA-CCAATGTTT and TCACGTTTATTTTCAAACATCCA. The PCR products were purified and used to transform strains SL206 (ura4+), SL350 (t-ura4<ori) and SL504 (t<ura4-ori). Transformants were selected on 5-FOA-containing plates. Appropriate replacement of ura4+ by ura4-dup20 or ura4-dup22 was verified by PCR and sequencing.
ura4 loss assay
A minimum of 11 independent single colonies from appropriate strains growing with or without thiamine were inoculated in 10 ml of non-selective media (with or without thiamine) and grown to stationary phase. Appropriate dilutions were plated on supplemented YEA to determine plating efficiency and on 5-FOA-containing plates. Colonies were counted after 5–7 days of incubation at 30°C. The rate of ura4 loss was determined with the method of the median and data are presented on Table 1. Statistical significance was detected using the nonparametric Mann-Whitney U test.
PCR assays and sequencing to determine the rates of genomic deletion, translocation, and mutation
At least 200 5-FOAR colonies per strain and condition were subjected to PCR analysis with the following primers: AAAACAAACGCAAACAAGGC and GTTTAACTATGCTTCGTCGG to amplify ura4 ORF, TGAATCCTCCGTTCAGTAGG and AAGGACTGCGTTCTTCTAGC to amplify rng3 and TTTCCTTTCACGGCTAACCC (TLII) and TGTACCCATGAGCAAACTGC (TLIII) to amplify the translocation junction. The amplified ura4 fragments were then sequenced on both strands, with the primers used to amplify the ura4 ORF. Only mutations present on both strands were considered to determine mutation spectra. Deletions, mutations and translocations were scored as percentages of all events and these values were used to balance the rates of ura4 loss to determine the respective rates of deletion, mutation and translocation (see Figure S1 for deletion and translocation rates and Figure S2 for mutation rates).
The fork-arrest-induced deletion, translocation and mutation rates (Figure 2C–2E and Figure 3A) were calculated by subtracting the rate obtained in presence of thiamine (Rtf1 being repressed, OFF) from the rate obtained in the absence of thiamine (Rtf1 being expressed, ON). This method allows the spontaneous instability of IRs and the leakiness of the RTS1-RFB activity to be disregarded to determine strictly the rate of events induced by fork-arrest. The nonparametric Mann-Whitney U test was used to test for statistically significant differences.
Reverse Mutation Assay
Exponentially growing cells were treated with 20 mM of HU, 40 µM of CPT or 1 mM of MMC. At indicated times, samples were taken and appropriate dilutions were plated on supplemented minimal media to determine plating efficiency and on uracil-free plates. Colonies were counted after incubation at 30°C for 5–7 days and the frequency of Ura+ colonies was determined.
Replication Slippage Assay Using Ura4-Dup20 And Ura4-Dup22 Strains
For strains showing a slow growth phenotype (recombination mutants), the frequency of Ura+ revertants was determined as a function of the number of generations experiencing fork arrest at ura4. Exponentially growing 5-FOAR cells were washed twice in water and used to inoculate uracil-containing media without thiamine. Every 24 hours, cells were counted to determine the number of generations, and appropriate dilutions were plated on supplemented minimal media and on uracil-free plates. Colonies were counted after incubation at 30°C for 5–7 days and the frequency of Ura+ colonies was determined. The slope of the curves presented on Figure 4F corresponds to the rate of replication slippage/generation. For strains showing similar growth to wild-type cells, a single 5-FOAR colony was grown on uracil-containing plates with or without thiamine for 2–3 days, and then grown in uracil-containing media with or without thiamine for 2 days at 30°C. Appropriate dilutions were plated on supplemented minimal media and on uracil-free plates. Colonies were counted after incubation at 30°C for 5–7 days and the frequency of Ura+ colonies was determined.
Supporting Information
Zdroje
1. AguileraA, Gomez-GonzalezB (2008) Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9 : 204–217.
2. BranzeiD, FoianiM (2010) Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11 : 208–219.
3. HalazonetisTD, GorgoulisVG, BartekJ (2008) An oncogene-induced DNA damage model for cancer development. Science 319 : 1352–1355.
4. ZhangF, CarvalhoCM, LupskiJR (2009) Complex human chromosomal and genomic rearrangements. Trends Genet 25 : 298–307.
5. LetessierA, MillotGA, KoundrioukoffS, LachagesAM, VogtN, et al. (2011) Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470 : 120–123.
6. Le TallecB, DutrillauxB, LachagesAM, MillotGA, BrisonO, et al. (2011) Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol 18 : 1421–1423.
7. PetermannE, HelledayT (2010) Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11 : 683–687.
8. HastingsPJ, IraG, LupskiJR (2009) A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327 doi:10.1371/journal.pgen.1000327
9. WeinertT, KaocharS, JonesH, PaekA, ClarkAJ (2009) The replication fork's five degrees of freedom, their failure and genome rearrangements. Curr Opin Cell Biol 21 : 778–784.
10. LemoineFJ, DegtyarevaNP, LobachevK, PetesTD (2005) Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120 : 587–598.
11. MizunoK, LambertS, BaldacciG, MurrayJM, CarrAM (2009) Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev 23 : 2876–2886.
12. BesterAC, RonigerM, OrenYS, ImMM, SarniD, et al. (2011) Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145 : 435–446.
13. Ozeri-GalaiE, LebofskyR, RahatA, BesterAC, BensimonA, et al. (2011) Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell 43 : 122–131.
14. PaekAL, KaocharS, JonesH, ElezabyA, ShanksL, et al. (2009) Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev 23 : 2861–2875.
15. BlowJJ, GeXQ, JacksonDA (2011) How dormant origins promote complete genome replication. Trends Biochem Sci 36 : 405–414.
16. MirkinEV, MirkinSM (2007) Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 71 : 13–35.
17. LambertS, FrogetB, CarrAM (2007) Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst) 6 : 1042–1061.
18. KawabataT, LuebbenSW, YamaguchiS, IlvesI, MatiseI, et al. (2011) Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell 41 : 543–553.
19. MurrayJM, CarrAM (2008) Smc5/6: a link between DNA repair and unidirectional replication? Nat Rev Mol Cell Biol 9 : 177–182.
20. LambertS, MizunoK, BlaisonneauJ, MartineauS, ChanetR, et al. (2010) Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol cell 39 : 346–359.
21. KatouY, KanohY, BandoM, NoguchiH, TanakaH, et al. (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424 : 1078–1083.
22. De PiccoliG, KatouY, ItohT, NakatoR, ShirahigeK, et al. (2012) Replisome stability at defective DNA replication forks is independent of s phase checkpoint kinases. Mol Cell 45 : 696–704.
23. FrogetB, BlaisonneauJ, LambertS, BaldacciG (2008) Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Mol Biol Cell 19 : 445–456.
24. Cotta-RamusinoC, FachinettiD, LuccaC, DoksaniY, LopesM, et al. (2005) Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17 : 153–159.
25. HellerRC, MariansKJ (2006) Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol 7 : 932–943.
26. MichelB, BoubakriH, BaharogluZ, LeMassonM, LestiniR (2007) Recombination proteins and rescue of arrested replication forks. DNA Repair (Amst) 6 : 967–980.
27. HashimotoY, PudduF, CostanzoV (2011) RAD51 - and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol 19 : 17–24.
28. RoseaulinL, YamadaY, TsutsuiY, RussellP, IwasakiH, et al. (2008) Mus81 is essential for sister chromatid recombination at broken replication forks. Embo J 27 : 1378–1387.
29. Moriel-CarreteroM, AguileraA (2010) A postincision-deficient TFIIH causes replication fork breakage and uncovers alternative Rad51 - or Pol32-mediated restart mechanisms. Mol Cell 37 : 690–701.
30. LydeardJR, Lipkin-MooreZ, SheuYJ, StillmanB, BurgersPM, et al. (2010) Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev 24 : 1133–1144.
31. LlorenteB, SmithCE, SymingtonLS (2008) Break-induced replication: what is it and what is it for? Cell Cycle 7 : 859–864.
32. McEachernMJ, HaberJE (2006) Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75 : 111–135.
33. SmithCE, LlorenteB, SymingtonLS (2007) Template switching during break-induced replication. Nature 447 : 102–105.
34. DeemA, KeszthelyiA, BlackgroveT, VaylA, CoffeyB, et al. (2011) Break-induced replication is highly inaccurate. PLoS Biol 9: e1000594 doi:10.1371/journal.pbio.1000594
35. LambertS, WatsonA, SheedyDM, MartinB, CarrAM (2005) Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121 : 689–702.
36. EydmannT, SommarivaE, InagawaT, MianS, KlarAJ, et al. (2008) Rtf1-mediated eukaryotic site-specific replication termination. Genetics 180 : 27–39.
37. KaplanDL, BastiaD (2009) Mechanisms of polar arrest of a replication fork. Mol Microbiol 72 : 279–285.
38. McInerneyP, O'DonnellM (2007) Replisome fate upon encountering a leading strand block and clearance from DNA by recombination proteins. J Biol Chem 282 : 25903–25916.
39. SabouriN, McDonaldKR, WebbCJ, CristeaIM, ZakianVA (2012) DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev 26 : 581–593.
40. SteinacherR, OsmanF, DalgaardJZ, LorenzA, WhitbyMC (2012) The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev 26 : 594–602.
41. BranzeiD, VanoliF, FoianiM (2008) SUMOylation regulates Rad18-mediated template switch. Nature 456 : 915–920.
42. MiyabeI, KunkelTA, CarrAM (2011) The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet 7: e1002407 doi:10.1371/journal.pgen.1002407
43. MyungK, ChenC, KolodnerRD (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411 : 1073–1076.
44. ChenC, KolodnerRD (1999) Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 23 : 81–85.
45. PutnamCD, HayesTK, KolodnerRD (2009) Specific pathways prevent duplication-mediated genome rearrangements. Nature 460 : 984–989.
46. VoineaguI, NarayananV, LobachevKS, MirkinSM (2008) Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci U S A 105 : 9936–9941.
47. CarvalhoCM, RamockiMB, PehlivanD, FrancoLM, Gonzaga-JaureguiC, et al. (2011) Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet 43 : 1074–1081.
48. LichtenM, HaberJE (1989) Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics 123 : 261–268.
49. TranHT, DegtyarevaNP, KolotevaNN, SuginoA, MasumotoH, et al. (1995) Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol Cell Biol 15 : 5607–5617.
50. LorenzA, OsmanF, FolkyteV, SofuevaS, WhitbyMC (2009) Fbh1 limits Rad51-dependent recombination at blocked replication forks. Mol Cell Biol 29 : 4742–4756.
51. OsmanF, DixonJ, BarrAR, WhitbyMC (2005) The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol 25 : 8084–8096.
52. AlabertC, BiancoJN, PaseroP (2009) Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. Embo J 28 : 1131–1141.
53. LisbyM, BarlowJH, BurgessRC, RothsteinR (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118 : 699–713.
54. MeisterP, TaddeiA, VernisL, PoidevinM, GasserSM, et al. (2005) Temporal separation of replication and recombination requires the intra-S checkpoint. J Cell Biol 168 : 537–544.
55. CoulonS, RamasubramanyanS, AliesC, PhilippinG, LehmannA, et al. (2010) Rad8Rad5/Mms2-Ubc13 ubiquitin ligase complex controls translesion synthesis in fission yeast. Embo J 29 : 2048–2058.
56. HombauerH, CampbellCS, SmithCE, DesaiA, KolodnerRD (2011) Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell 147 : 1040–1053.
57. TranHT, GordeninDA, ResnickMA (1996) The prevention of repeat-associated deletions in Saccharomyces cerevisiae by mismatch repair depends on size and origin of deletions. Genetics 143 : 1579–1587.
58. SiaEA, Jinks-RobertsonS, PetesTD (1997) Genetic control of microsatellite stability. Mutat Res 383 : 61–70.
59. LiuP, CarvalhoCM, HastingsP, LupskiJR (2012) Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev
60. OuZ, StankiewiczP, XiaZ, BremanAM, DawsonB, et al. (2010) Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes. Genome Res 21 : 33–46.
61. BoscoG, HaberJE (1998) Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150 : 1037–1047.
62. DalgaardJZ, KlarAJ (2001) A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev 15 : 2060–2068.
63. RuizJF, Gomez-GonzalezB, AguileraA (2009) Chromosomal translocations caused by either pol32-dependent or pol32-independent triparental break-induced replication. Mol Cell Biol 29 : 5441–5454.
64. SchmidtKH, WuJ, KolodnerRD (2006) Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae homolog of the Bloom's syndrome protein. Mol Cell Biol 26 : 5406–5420.
65. YangY, SterlingJ, StoriciF, ResnickMA, GordeninDA (2008) Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet 4: e1000264 doi:10.1371/journal.pgen.1000264
66. HicksWM, KimM, HaberJE (2010) Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329 : 82–85.
67. KaiM, BoddyMN, RussellP, WangTS (2005) Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev 19 : 919–932.
68. KaiM, WangTS (2003) Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev 17 : 64–76.
69. HuangME, de CalignonA, NicolasA, GalibertF (2000) POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr Genet 38 : 178–187.
70. PayenC, KoszulR, DujonB, FischerG (2008) Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet 4: e1000175 doi:10.1371/journal.pgen.1000175
71. LydeardJR, JainS, YamaguchiM, HaberJE (2007) Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448 : 820–823.
72. ColnaghiR, CarpenterG, VolkerM, O'DriscollM (2011) The consequences of structural genomic alterations in humans: Genomic Disorders, genomic instability and cancer. Semin Cell Dev Biol 22 : 875–885.
73. ArltMF, WilsonTE, GloverTW (2012) Replication stress and mechanisms of CNV formation. Curr Opin Genet Dev
74. DurkinSG, RaglandRL, ArltMF, MulleJG, WarrenST, et al. (2008) Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc Natl Acad Sci U S A 105 : 246–251.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 10
-
Všechny články tohoto čísla
- The Germline Genome Provides a Niche for Intragenic Parasitic DNA: Evolutionary Dynamics of Internal Eliminated Sequences
- Classical Genetics Meets Next-Generation Sequencing: Uncovering a Genome-Wide Recombination Map in
- Calpain-5 Mutations Cause Autoimmune Uveitis, Retinal Neovascularization, and Photoreceptor Degeneration
- Cofilin-1: A Modulator of Anxiety in Mice
- The Date of Interbreeding between Neandertals and Modern Humans
- Embryos of Robertsonian Translocation Carriers Exhibit a Mitotic Interchromosomal Effect That Enhances Genetic Instability during Early Development
- Viral Evasion of a Bacterial Suicide System by RNA–Based Molecular Mimicry Enables Infectious Altruism
- Phosphatase-Dead Myotubularin Ameliorates X-Linked Centronuclear Myopathy Phenotypes in Mice
- Full-Length Synaptonemal Complex Grows Continuously during Meiotic Prophase in Budding Yeast
- MOV10 RNA Helicase Is a Potent Inhibitor of Retrotransposition in Cells
- A Likelihood-Based Framework for Variant Calling and Mutation Detection in Families
- The Contribution of RNA Decay Quantitative Trait Loci to Inter-Individual Variation in Steady-State Gene Expression Levels
- New Partners in Regulation of Gene Expression: The Enhancer of Trithorax and Polycomb Corto Interacts with Methylated Ribosomal Protein L12 Its Chromodomain
- Mining the Unknown: A Systems Approach to Metabolite Identification Combining Genetic and Metabolic Information
- Mutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
- The Many Landscapes of Recombination in
- Faster-X Evolution of Gene Expression in
- Loss of Slc4a1b Chloride/Bicarbonate Exchanger Function Protects Mechanosensory Hair Cells from Aminoglycoside Damage in the Zebrafish Mutant
- Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection
- and the BTB Adaptor Are Key Regulators of Sleep Homeostasis and a Dopamine Arousal Pathway in Drosophila
- Mutation and Fetal Ethanol Exposure Synergize to Produce Midline Signaling Defects and Holoprosencephaly Spectrum Disorders in Mice
- Specific Missense Alleles of the Arabidopsis Jasmonic Acid Co-Receptor COI1 Regulate Innate Immune Receptor Accumulation and Function
- Deep Genome-Wide Measurement of Meiotic Gene Conversion Using Tetrad Analysis in
- Mismatch Repair Balances Leading and Lagging Strand DNA Replication Fidelity
- Distinguishing between Selective Sweeps from Standing Variation and from a Mutation
- Cytokinesis-Based Constraints on Polarized Cell Growth in Fission Yeast
- Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes
- Functional Antagonism between Sas3 and Gcn5 Acetyltransferases and ISWI Chromatin Remodelers
- The SET-Domain Protein SUVR5 Mediates H3K9me2 Deposition and Silencing at Stimulus Response Genes in a DNA Methylation–Independent Manner
- Morphogenesis and Cell Fate Determination within the Adaxial Cell Equivalence Group of the Zebrafish Myotome
- Muscle-Specific Splicing Factors ASD-2 and SUP-12 Cooperatively Switch Alternative Pre-mRNA Processing Patterns of the ADF/Cofilin Gene in
- Maize Is Required for Maintaining Silencing Associated with Paramutation at the and Loci
- Increasing Signal Specificity of the TOL Network of mt-2 by Rewiring the Connectivity of the Master Regulator XylR
- Use of Pleiotropy to Model Genetic Interactions in a Population
- RAB-Like 2 Has an Essential Role in Male Fertility, Sperm Intra-Flagellar Transport, and Tail Assembly
- Variants Affecting Exon Skipping Contribute to Complex Traits
- Topoisomerase II– and Condensin-Dependent Breakage of -Sensitive Fragile Sites Occurs Independently of Spindle Tension, Anaphase, or Cytokinesis
- Comparison of Family History and SNPs for Predicting Risk of Complex Disease
- Recovery of Arrested Replication Forks by Homologous Recombination Is Error-Prone
- A Mutation in the Gene Causes Alternative Splicing Defects and Deafness in the Bronx Waltzer Mouse
- Comparative Genomics Suggests an Independent Origin of Cytoplasmic Incompatibility in
- It Was Heaven: An Interview with Evelyn Witkin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Mutation in the Gene Causes Alternative Splicing Defects and Deafness in the Bronx Waltzer Mouse
- Mutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
- Classical Genetics Meets Next-Generation Sequencing: Uncovering a Genome-Wide Recombination Map in
- Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



