-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
Holoprosencephaly (HPE) is a failure of the forebrain to bifurcate and is the most common structural malformation of the embryonic brain. Mutations in SHH underlie most familial (17%) cases of HPE; and, consistent with this, Shh is expressed in midline embryonic cells and tissues and their derivatives that are affected in HPE. It has long been recognized that a graded series of facial anomalies occurs within the clinical spectrum of HPE, as HPE is often found in patients together with other malformations such as acrania, anencephaly, and agnathia. However, it is not known if these phenotypes arise through a common etiology and pathogenesis. Here we demonstrate for the first time using mouse models that Hedgehog acyltransferase (Hhat) loss-of-function leads to holoprosencephaly together with acrania and agnathia, which mimics the severe condition observed in humans. Hhat is required for post-translational palmitoylation of Hedgehog (Hh) proteins; and, in the absence of Hhat, Hh secretion from producing cells is diminished. We show through downregulation of the Hh receptor Ptch1 that loss of Hhat perturbs long-range Hh signaling, which in turn disrupts Fgf, Bmp and Erk signaling. Collectively, this leads to abnormal patterning and extensive apoptosis within the craniofacial primordial, together with defects in cartilage and bone differentiation. Therefore our work shows that Hhat loss-of-function underscrores HPE; but more importantly it provides a mechanism for the co-occurrence of acrania, holoprosencephaly, and agnathia. Future genetic studies should include HHAT as a potential candidate in the etiology and pathogenesis of HPE and its associated disorders.
Published in the journal: . PLoS Genet 8(10): e32767. doi:10.1371/journal.pgen.1002927
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002927Summary
Holoprosencephaly (HPE) is a failure of the forebrain to bifurcate and is the most common structural malformation of the embryonic brain. Mutations in SHH underlie most familial (17%) cases of HPE; and, consistent with this, Shh is expressed in midline embryonic cells and tissues and their derivatives that are affected in HPE. It has long been recognized that a graded series of facial anomalies occurs within the clinical spectrum of HPE, as HPE is often found in patients together with other malformations such as acrania, anencephaly, and agnathia. However, it is not known if these phenotypes arise through a common etiology and pathogenesis. Here we demonstrate for the first time using mouse models that Hedgehog acyltransferase (Hhat) loss-of-function leads to holoprosencephaly together with acrania and agnathia, which mimics the severe condition observed in humans. Hhat is required for post-translational palmitoylation of Hedgehog (Hh) proteins; and, in the absence of Hhat, Hh secretion from producing cells is diminished. We show through downregulation of the Hh receptor Ptch1 that loss of Hhat perturbs long-range Hh signaling, which in turn disrupts Fgf, Bmp and Erk signaling. Collectively, this leads to abnormal patterning and extensive apoptosis within the craniofacial primordial, together with defects in cartilage and bone differentiation. Therefore our work shows that Hhat loss-of-function underscrores HPE; but more importantly it provides a mechanism for the co-occurrence of acrania, holoprosencephaly, and agnathia. Future genetic studies should include HHAT as a potential candidate in the etiology and pathogenesis of HPE and its associated disorders.
Introduction
Holoprosencephaly (HPE) is a congenital malformation resulting from the failure of the forebrain to divide into left and right hemispheres [1], [2]. HPE occurs at a frequency of 1 in 10,000–20,000 live births, although affected embryonic individuals are thought to be as high as 1 in 250 pregnancies, making it the most common brain anomaly in humans [3], [4]. Within the clinical spectrum of HPE, a broad range of brain malformations are observed. Alobar holoprosencephaly is the most severe form, and in this case hemisphere bifurcation completely fails to occur resulting in the forebrain developing as a single holosphere together with a single cyclopic eye [5]. In milder instances such as lobar holoprosencephaly, near complete hemisphere separation occurs but cortical structures are hypoplastic and specific brain nuclei remain congruous [5]. It has long been recognized that the face predicts the brain [6] and HPE manifests with a range of craniofacial anomalies in humans. In extreme cases of HPE, cyclopia and a nasal proboscis can occur [3], [7]. In very mild forms, such as cebocephaly, only a single nostril or incisor might be present and the philtral ridges may be absent [8]. Thus phenotypically, HPE is a heterogeneous disorder and this is also true etiologically.
Exposure to environmental teratogens such as alcohol [9], [10] and retinoids [9] can result in HPE phenotypes. Gestational diabetes is also a factor as 1–2% of newborn infants of diabetic mothers exhibit HPE [11]. Genetically, HPE is similarly heterogeneous and is currently associated with mutations in at least 12 different loci encompassing multiple signaling pathways such as BMP, NODAL, ZIC, SIX, and SHH [4]. What is common amongst many of the loci and signaling pathways is that they play important roles in the development of the ventral brain and midline structures of the embryo. This is particularly true for Sonic Hedgehog (SHH) signaling.
Shh is a member of the Hedgehog (Hh) family of signaling molecules that also includes Indian (Ihh) and Desert Hedgehog (Dhh). Each Hh is a secreted glycoprotein that undergoes autoproteolytic cleavage and dual lipid post-translational modification to generate its proper active form. Autoproteolytic cleavage of Hh precursor molecules generates an N-terminal fragment (Hh-N) referred to as the mature form. Hh-N is then modified via the addition of a cholesterol moiety to its C-terminus [12], [13], followed by addition of a palmitoyl moiety to its N-terminus [14]. These lipid modifications are required for Hh protein multimerisation, distribution and activity.
To date, mutational analyses of human HPE, together with naturally occurring and engineered mouse mutants, have identified the genetic lesions responsible for only about 20% of individuals with HPE. Hence it is critical to identify additional candidate genes for the majority of patients whose genetic lesions remain unknown. Furthermore, HPE is a heterogeneous disorder and often found in patients together with other malformations such as acrania, anencephaly and agnathia and it is not known if these phenotypes arise through a common etiology and pathogenesis. Here we describe the AP2-Cre (“Creface”) mouse [15] as an insertional mutation in Hhat, and define Hhat as a novel HPE associated gene which can mechanistically explain the co-occurrence of HPE together with acrania and agnathia.
Results
“Creface” mice exhibit acrania-holoprosencephaly-agnathia
“Creface” (AP2-Cre) is a transgenic line of mice [15] in which nuclear localizing Cre recombinase is driven by a specific TFAP2A enhancer element [16]. We discovered that interbreeding heterozygous Creface+/T mice failed to generate any post-natal viable homozygous CrefaceT/T animals. Therefore we investigated the etiology and pathogenesis of the CrefaceT/T mutant phenotype during embryogenesis. Morphological abnormalities in CrefaceT/T embryos are readily identifiable as early as E9.5. In contrast to control littermates, CrefaceT/T embryos exhibited smaller telencephalic hemispheres together with diencephalic and mesencephalic hypoplasia (Figure 1A, 1B). Pax6 expression demarcates the telencephalon and prosomere (P) territories 1 and 2 of the diencephalon and in situ hybridization analyses with Pax6 revealed the specific absence of P2 as well as abnormal neural morphology in E9.5 CrefaceT/T embryos (Figure 1A–1F). Pax6 also labels the optic placode and interestingly, although present, the optic vesicles are displaced ventrally and medially in CrefaceT/T embryos (Figure 1C, 1D, 1G, 1H). At later stages of gestation, the forebrain in CrefaceT/T embryos often lacked a ventricular canal and instead persisted as a single–lobed or incompletely bifurcated neuroepithelium. In contrast, control littermates, displayed bifurcated hemispheres surrounding the forebrain ventricle (Figure 1I, 1J). Ocular anomalies in CrefaceT/T embryos manifested as microphthalmia but in addition, the eye often remained embedded in grossly disorganized brain tissue and the lack of contact with the surface ectoderm resulted in a failure to form tissues such as the cornea (Figure 1K, 1L).
Fig. 1. CrefaceT/T embryos exhibit brain anomalies. 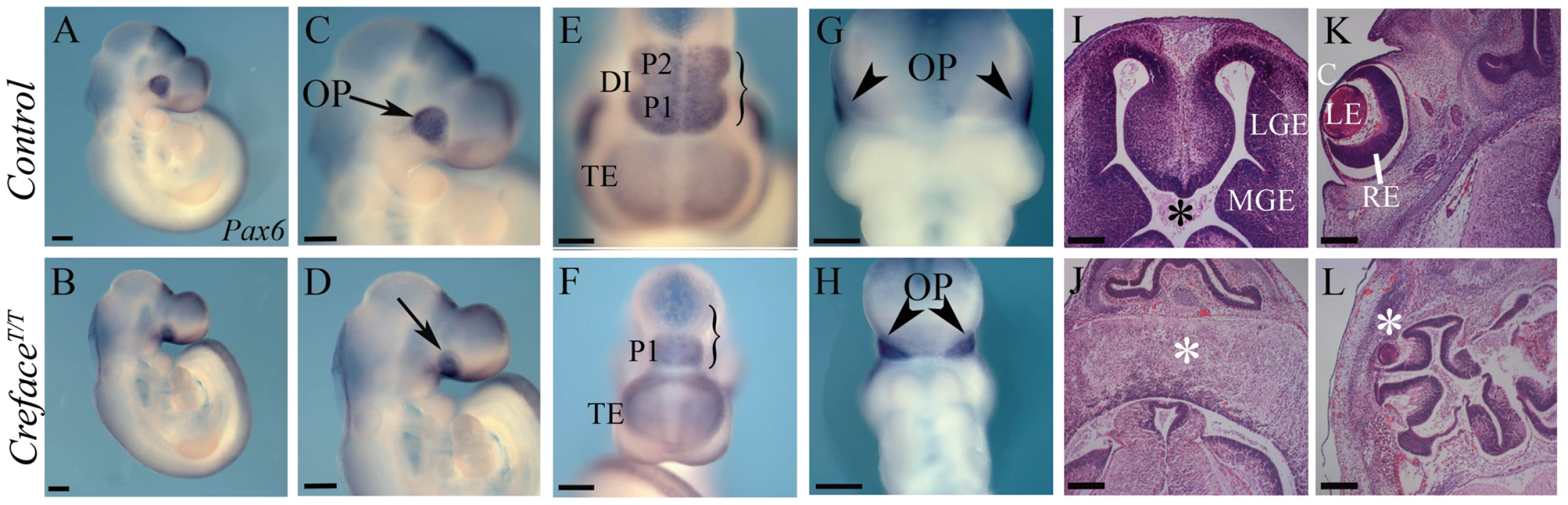
Pax6 whole embryo in situ hybridization in E9.5 control (A, C, E, G) and CrefaceT/T (B, D, F, H) embryos demonstrating ventrally displaced optic cup (arrows, arrowheads) and agenesis of prosomere 2 in the diencephalon (parentheses) of mutant embryos. Hematoxylin and eosin stained transverse sections through the forebrain and eye regions of control (I, K) and CrefaceT/T (J, L) embryos reveal absence of the ventricle due to bifurcation failure (asterisk in I, J) and agenesis of the cornea in association with microphthalmia (asterisk in L) in mutants. Abbreviations: C, cornea; DI, diencephalon; LE, lens; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; OP, optic placode; P, prosomere; RE, retina; TE, telencephalon. Scale bars: A–H 500 uM; I–L 200 uM. E10.5 CrefaceT/T mutant embryos are noticeably smaller in size than control littermates and exhibit more prominent craniofacial abnormalities (Figure 2A, 2B). In particular, the frontonasal region of the embryo as defined by the medial nasal prominences and spacing between the bilateral nasal slits is dramatically reduced in size to the extent that only a single slit is present in CrefaceT/T mutant embryos (Figure 2C, 2D). Craniofacial anomalies in CrefaceT/T embryos are not limited to the brain and frontonasal region as the maxillary and mandibular components of the first pharyngeal arch are also hypoplastic at E9.5–10.5 (Figure 2E, 2F). This manifests in E14.5 mutant embryos as a narrow protruding midface, together with more severely pronounced maxillary and mandibular hypoplasia (Figure 2G, 2H). In addition to craniofacial defects, mutant embryos exhibited limb defects including oligodactyly (Figure 2G, 2H and data not shown). E14.5 CrefaceT/T embryos displayed considerable edema with the outer layer of skin being displaced from the body cavity, most likely due to defects in lymphatic development (Figure 2G, 2H). Large areas of blood pooling were also often observed in the anterior region of the embryos, which may be indicative of more general vascular anomalies. These lymphatic and vascular anomalies progressively worsened coinciding with embryonic lethality prior to birth.
Fig. 2. CrefaceT/T embryos exhibit general facial prominence hypoplasia. 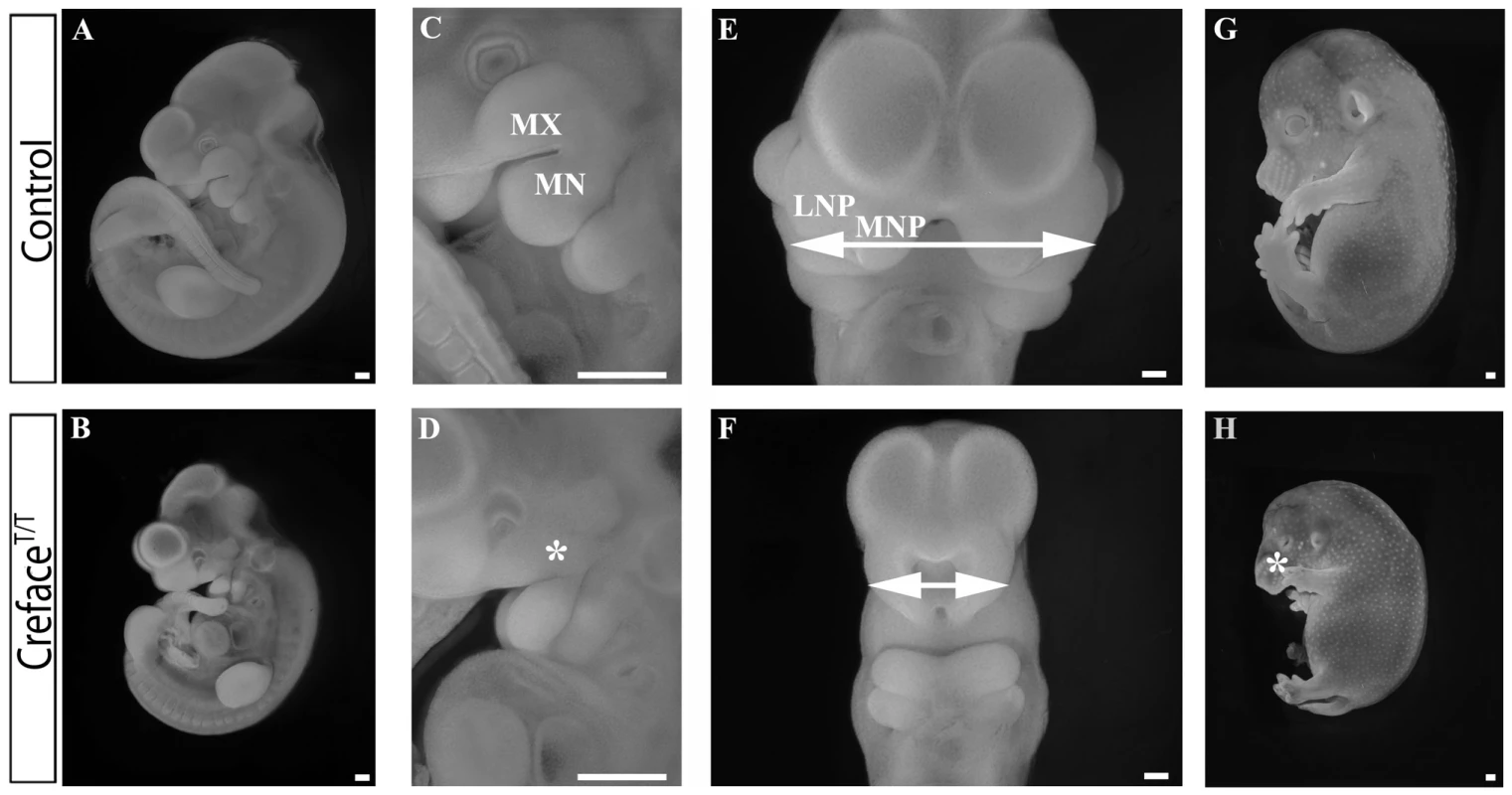
Fluorescent images of whole DAPI stained E10.5 control (A, C, E) and CrefaceT/T (B, D, F) embryos highlighting the pharyngeal (asterisk) and midfacial hypoplasia evident in the form of a single nasal slit (double arrows) in mutant embryos. Bright field images of E14.5 control (G) and CrefaceT/T (H) embryos illustrating the severe frontonasal defects in mutant embryos (asterisk). Abbreviations: LNP, lateral nasal prominence; MN, mandibular prominence; MNP, medial nasal prominence; MX, maxillary prominence. Scale bars: 200 uM. In the current analysis we concentrated on the molecular and structural changes associated with the CrefaceT/T craniofacial defects. In control embryos Eya2 bilaterally demarcates the nasal placode ectoderm (Figure 3A, 3C, 3E) while Six2 is expressed bilaterally in the mesenchyme of each medial nasal prominence (Figure 3G, 3I). In E9.5–10.5 CrefaceT/T mutants, there is a single continuous central domain of placodal Eya2 activity (Figure 3B, 3D, 3F), while Six2 expression is absent from midline tissues (Figure 3H, 3J). This is consistent with frontonasal agenesis, nasal placode fusion and a single nasal pit/slit in CrefaceT/T mutant embryos (Figure 2C, 2D). We next examined the signaling molecules Fgf8 and Bmp4, which are known to regulate craniofacial development [17]. At E10.5, Fgf8 normally labels the epithelium flanking the nasal pits almost uniformly (Figure 4A, 4C), while Bmp4 marks only specific ventral domains of the nasal prominences (Figure 4E, 4G). However, CrefaceT/T mutants display single continuous domains of epithelial Fgf8 and Bmp4 expression (Figure 4B, 4D, 4F, 4H). These findings are also consistent with frontonasal agenesis, nasal placode fusion and a single nasal pit/slit in CrefaceT/T mutant embryos.
Fig. 3. CrefaceT/T embryos exhibit frontonasal placode patterning defects. 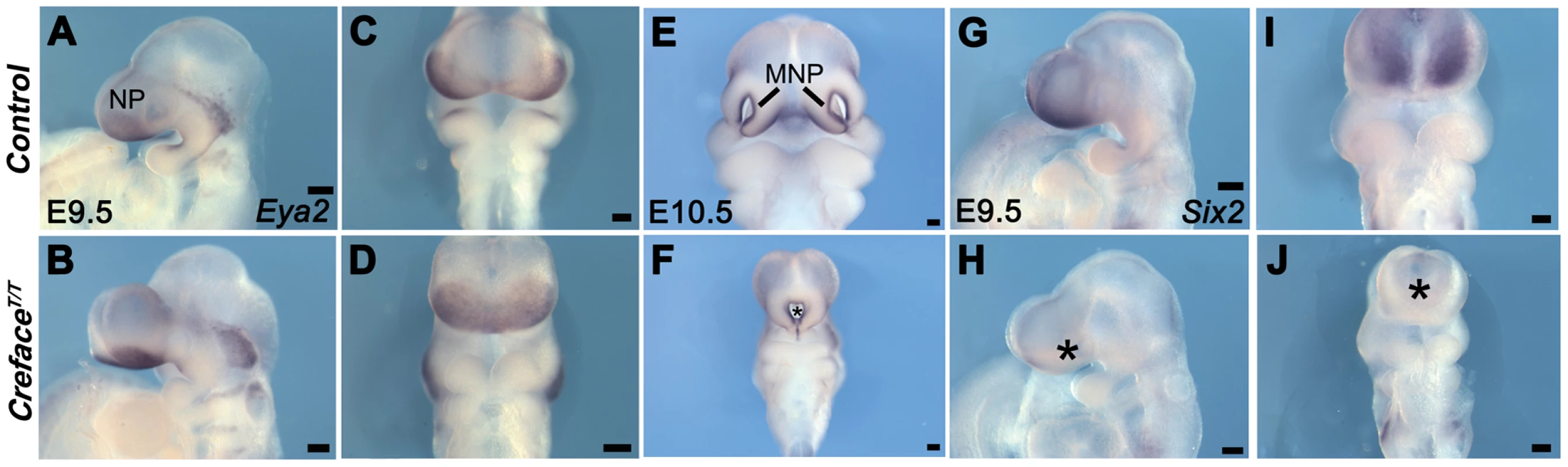
Eya2 (A–F) and Six2 (G–J) whole embryo in situ hybridization in E9.5 and E10.5 control (A, C, E, G, I) and CrefaceT/T (B, D, F, H, J) embryos showing agenesis of frontonasal mesenchyme (I, J), nasal placode fusion (C, D) and formation of a single nasal slit in mutant embryos (asterisks). Abbreviations: NP, nasal placode. Scale bars: A and G 500 uM; all others 200 uM. Fig. 4. CrefaceT/T embryos exhibit frontonasal and pharyngeal epithelial patterning defects. 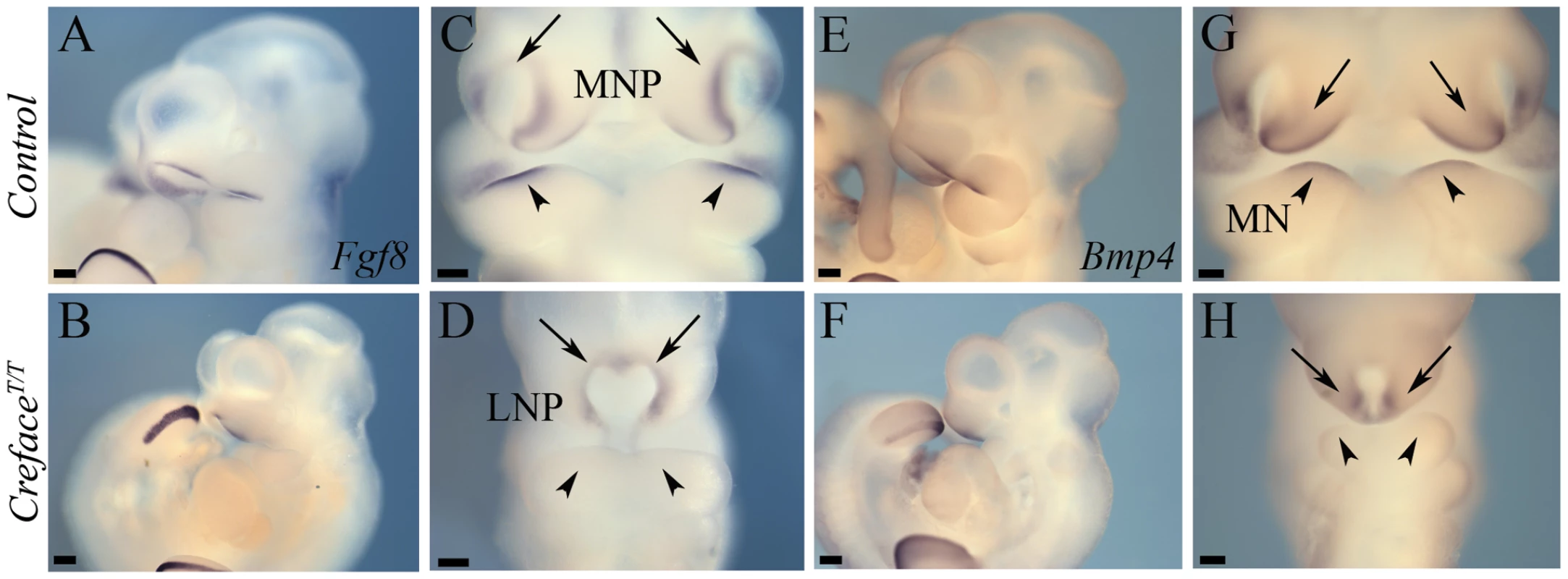
Fgf8 (A–D) and Bmp4 (E–H) whole embryo in situ hybridization in E10.5 control (A, C, E, G) and CrefaceT/T (B, D, F, H) embryos highlighting the midfacial hypoplasia, single nasal slit (arrows) and abnormal proximodistal pharyngeal arch patterning (arrowheads) defects in mutant embryos. Scale bars: 200 uM. To characterize any defects in skeletogenesis in CrefaceT/T mutants, we stained embryos at E15.5 and 17.5 (Figure 5) with Alcian blue and Alizarin red to label cartilage and bone, respectively. At E15.5 control embryos exhibited considerable domains of differentiated cartilage within the head, particularly within the developing cranium, ear and nasal regions as evidenced by extensive blue tissue staining (Figure 5A). Ossification was clearly evident in multiple elements including the frontal and parietal bones but was particularly well advanced in components of the viscerocranium including the premaxillary, maxillary and dentary bones. In marked contrast, E15.5 CrefaceT/T littermates demonstrated a complete absence of Alizarin red stained ossified bone within the skull and jaw (Figure 5B). Mutant embryos also displayed considerably diminished chondrogenesis of calvarial, nasal and otic mesenchyme (Figure 5B). Not only were cranial elements such as the nasal cartilage hypoplastic or missing, but staining of cartilage in the vertebral column was also absent. Section histology revealed that compared to control littermates, which have bifurcated continuous nasal cavities separated by a nasal septum, E14.5 CrefaceT/T embryos lacked a nasal septum and possessed only a single yet discontinuous nasal cavity (Figure 5E, 5F). In addition, although the palatal shelves were readily identifiable adjacent to the tongue in control embryos, this was not the case in CrefaceT/T littermates, and is indicative of a defect in vertical extension and medial growth of the palatal shelves toward the midline (Figure 5G, 5H). The oral cavity was considerably narrower in mutant embryos and the tongue was hypoplastic.
Fig. 5. CrefaceT/T embryos exhibit major cranioskeletal malformations. 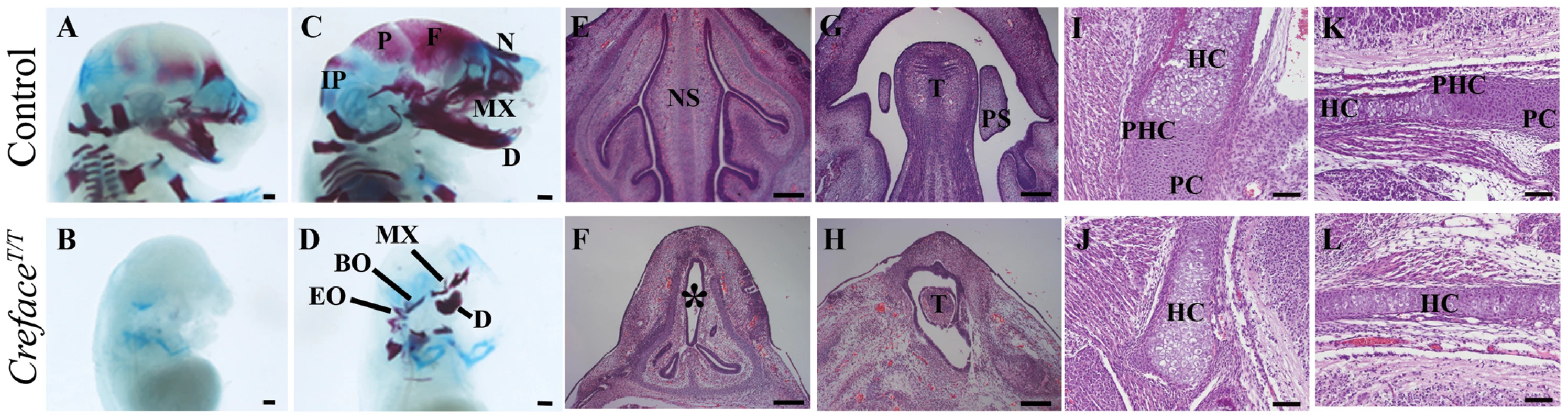
Alcian blue carilage and Alizarin red bone staining of E15.5 (A, B) and E17.5 (C, D) control (A, C) and CrefaceT/T (B, D) embryos highlighting acrania, agnathia and general cranioskeletal agenesis in mutant embryos. Hematoxylin and eosin stained sections through the nasal septum region (E, F), palatal mesenchyme (G, H) exoccipital (I, J) and basioccipital (K, L) bones of E14.5 control (E, G) and CrefaceT/T (F, H) and E17.5 control (I, K) and CrefaceT/T (J, L) embryos illustrating tissue agenesis (asterisk) and hypoplasia as well as defects in endochondral ossification in mutant embryos. Abbreviations: BO, basioccipital; D, dentary; EO, exoccipital; F, frontal; HC, hypertrophic chondrocytes; IP, interparietal; MX, maxilla; N, nasal; NS, nasal septum; P, parietal, PC, proliferating chondrocytes; PHC, prehypotrophic chondrocytes; PS, palatal shelves; T, tongue. Scale bars: A–D 500 uM; I–L 100 uM; E–H 200 uM. By E17.5, cranioskeletal mineralization was almost complete in control embryos. The frontal, parietal, interparietal, supraoccipital, exoccipital and temporal bones that contribute to the neurocranium are nearly fully ossified as were the nasal, premaxilla, maxilla, jugal and dentary bones within the viscerocranium (Figure 5C). In contrast CrefaceT/T littermate embryos exhibited acrania as the frontal parietal, interparietal and supraoccipital, bones were all completely absent (Figure 5D). The nasal cartilage that was present was a tube-shaped structure that was devoid of ossification. The basioccipital and exoccipital bones that constitute the base of the skull were present, but were dramatically reduced in size and abnormally shaped. E17.5 CrefaceT/T embryos also display agnathia as we observed the presence of only rudimentary premaxilla, maxilla and dentary bones. The dentary bones lacked all processes including the condyle, angular and coronoid processes. Furthermore CrefaceT/T embryos also exhibited disrupted tooth development as incisor and alveolar molar tooth primordia which were readily identifiable in control embryos, were absent from mutant littermates (data not shown).
Given the extensive nature of cranioskeletal anomalies in CrefaceT/T embryos, we characterised their specific pathogenesis in more detail. During endochondral ossification proliferating chondrocytes at distal ends of a skeletal element undergo hypertrophy and dramatically increase in size. As development proceeds, hypertrophic chondrocytes undergo apoptosis and are replaced by invading osteoblasts (reviewed in [18]). Sagittal sections through the exoccipital and basioccipital bones of E15.5 Creface+/T embryos revealed the presence of proliferating chondrocytes at the distal end of the element (Figure 5I, 5K). These cells were located adjacent to a domain of prehypertrophic chondrocytes which were in turn were flanked by noticeably larger hypertrophic chondrocytes positioned centrally within each element. In contrast, sections of CrefaceT/T mutants revealed a significant perturbation in chondrocyte development. The skeletal elements were reduced in size and this appeared to be associated with a diminished zone of proliferating chondrocytes (Figure 5J, 5L).
Hedgehog acyltransferase (Hhat) is disrupted in CrefaceT/T mutants
The constellation of craniofacial abnomalities observed in CrefaceT/T embryos is consistent with acrania-holoprosencephaly-agnathia. Holoprosencephaly and limb anomalies including digit oligodactyly are classic features of perturbed Hh signaling during early embryogenesis. We therefore hypothesized that the AP2-Cre transgene used to generate Creface mice, inserted into or interfered with a locus important for Hh signaling. However, to rule out the possibility that the insertion occurred in Hh signaling genes previously associated with HPE, we initially performed complementation tests by intercrossing Creface+/T mice with Shh+/− [19] and Ptch1+/− [20] mice. None of the Creface+/T;Shh+/−, or Creface+/T;Ptch1-LacZ+ embryos harvested at E17.5–18.5 displayed craniofacial anomalies consistent with HPE, acrania or agnathia (data not shown). This indicated that the CrefaceT/T phenotype was not the result of transgene integration into Shh, or Ptch1 genomic loci.
Given that the genetic lesions responsible for HPE have only been identified in about 20% of affected individuals, there is considerable interest in discovering new genes that contribute to its etiology and pathogenesis. This is particularly true where HPE is a subcomponent of more complex syndromes. Therefore, using a PCR-based method for DNA walking and mapping known as Vectorette, we determined that the Creface transgene had integrated into intron 9 of Hedgehog acyltransferase (Hhat) at nucleotide position 22949 (Figure S1A). To further verify that Hhat was indeed the gene disrupted, we genotyped E10.5 embryos obtained from intercrosses of Creface+/T mice using control and transgene-specific reactions. Genotyping of the endogenous Hhat intron 9 region produced a 1.4 kb DNA band from both wild-type and heterozygous (Creface+/T) embryos, but did not produce a band from mutant (CrefaceT/T) embryos as expected (Figure S1B). Furthermore, using a primer designed to recognize the Creface:Hhat fusion, this enabled amplification of an integration specific 250 bp fragment from Creface+/T heterozygous and CrefaceT/T mutant embryos but not from wild-type embryos (Figure S1B). Collectively, these results confirmed that the Creface transgene integrated within intron 9 of Hhat. It is well recognized that transgenes integrate into host genomes as concatamers and can disrupt endogenous gene expression [21]–[23]. Consistent with this we were unable to RT-PCR amplify full length Hhat using RNA obtained from CrefaceT/T mutant embryos, in contrast to wild-type and Creface+/T heterozygous embryos (Figure S1C).
To examine the expression of Hhat in control and mutant embryos, we generated a riboprobe corresponding specifically to exons 8–10 of Hhat (Figure 6A). Through in situ hybdrization of E9.5–10.5 embryos followed by sectioning, we observed that Hhat is expressed in and around the notochord ventral to the neural tube in wild-type embryos. In contrast, the level of Hhat expression was considerably diminished or absent in CrefaceT/T mutant littermate embryos (Figure 6B, 6C). This indicates that Hhat activity is disrupted through Creface transgene insertion. These data clearly validate that integration of the Creface transgene physically disrupts Hhat rendering the gene non-functional, which results in the morphological defects characteristic of CrefaceT/T mutant mice. Creface+/T heterozygous and CrefaceT/T homozygous mice will herein be referred to as Hhat+/Creface and HhatCreface/Creface respectively.
Fig. 6. Hhat loss-of-function perturbs gene expression and palmitoylation of Shh. 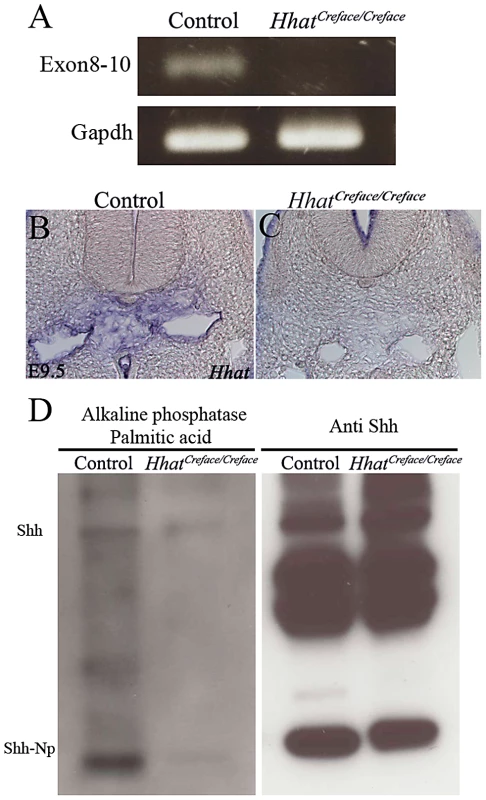
Hhat gene expression analysis by RT-PCR (A) and in situ hybridization (B, C) showing diminished or absent activity in HhatCreface/Creface mutant embryos compared to controls, particularly with respect to expression in the mesenchymal cells surrounding the notochord. (D) Palmitoylation assay performed using MEFs from control and HhatCreface/Creface mice. HhatCreface/Creface MEFs exhibited a considerable reduction in palmitoylated Shh both in unprocessed (Shh) and processed (Shh-Np) protein forms (left panel) despite western blotting highlighting equivalent amounts of Shh protein in mutant compared to control mice (right panel). The expression of Hhat mRNA according to the mutation site was absent in HhatCreface/Creface mutant (B). In situ hybridization revealed that Hhat mRNA level of HhatCreface/Creface mice (C, right panel) are impaired at the mesenchyme cells around the notochord compared with control mice (C, left panel). Loss of Hhat function disrupts Shh signaling
Hhat encodes an acyltransferase that palmitoylates Hh proteins [14]. This fatty acid modification increases the potency of Hh signaling in vitro [24] and is also required for long-range Hh signaling in vivo [25]. Consistent with this, Drosophila mutants lacking Hh palmitoylation exhibit patterning defects in Hh-responsive cells [26]–[29]. We therefore hypothesized that Hhat loss-of-function should disrupt the palmitoylation of Shh and perturb its signaling effects in HhatCreface/Creface embryos. To test this, we generated mouse embryonic fibroblast (MEFs) from control and HhatCreface/Creface embryos, transfected each with a Shh expression vector, and incubated them with palmitic acid for 12 hours. Incorporated palmitic acid was then tagged with biotin. Protein extraction, followed by western blotting with an anti Shh antibody and alkaline phosphatase streptavidin staining, confirmed that despite the presence of similar levels of total Shh protein, the degree of palmitoylation in HhatCreface/Creface MEFs was considerably diminished compared to controls (Figure 6D). This clearly demonstrates that the functionality of Hhat is compromised in HhatCreface/Creface embryos.
Our expectation from Hhat loss-of-function in concert with the holoprosencephaly phenotype was that the lack of palmitoylation should perturb the spatiotemporal activity of Shh signaling throughout HhatCreface/Creface embryos. Hence we initially examined the expression of Shh via in situ hybridization in control and mutant embryos. Compared to control littermates, Shh was expressed in the notochord in E8.5 and E9.5 HhatCreface/Creface embryos albeit at slightly reduced levels but was absent from the ventral telencephalon and floor plate (Figure 7A–7F, 7I–7L). Shh expression was also absent from the branchial arch (BA) ectoderm and pharyngeal endoderm of E10.5–11.5 HhatCreface/Creface embryos (Figure 7G, 7H). Shh activity in tissues such as the ventral forebrain and branchial arch ectoderm is induced by and dependent upon prior expression of Shh in the notochord and endoderm respectively [30], [31]. Thus the alterations to Shh expression in HhatCreface/Creface embryos were indicative of specific disruptions to Shh signaling gradients as a consequence of Hhat loss-of-function and lack of Shh palmitoylation.
Fig. 7. Shh expression is altered in HhatCreface/Creface embryos. 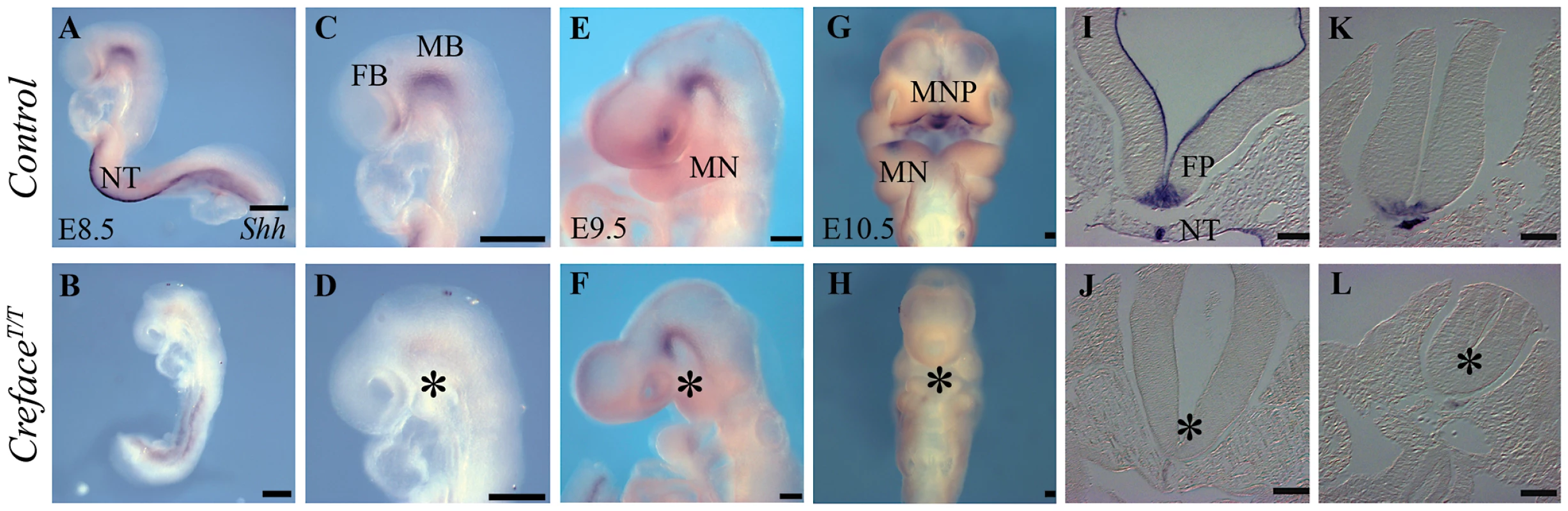
Shh in situ hybridization in whole E8.5–10.5 control (A, C, E, G) and HhatCreface/Creface (B, D, F, H) embryos illustrating the downregulation or loss of Shh expression in mutant embryos. Sections through the neural tube and notochord of E9.5 control (I, K) and HhatCreface/Creface (J, L) highlighting the absence of Shh expression in the floor plate of mutant embryos (asterisk). Abbreviations: FB, forebrain; FP, floor plate; MB, midbrain; NT, notochord. Scale bars: A–D 200 uM; E 500 uM; F–H 200 uM; I–L 50 uM. To confirm this, we characterized the distribution of Shh protein in E9.5 embryos (Figure 8). Control embryos exhibited Shh activity in the ventral telencephalon, branchial arch ectoderm, pharyngeal endoderm, notochord and floor plate (Figure 8A, 8C, 8E, 8G). However, in HhatCreface/Creface embryos, Shh protein was absent from all these sites with the exception of the notochord (Figure 8B, 8D, 8F, 8H). These findings are consistent with our observations of Shh expression via in situ hybridization (Figure 7) and correlate with the localization of Hhat activity. Thus our results indicate that localized tissue specific production of Shh can occur in HhatCreface/Creface embryos, however the failure to induce secondary domains of activity suggests a disruption in long-range Shh signaling gradients as a function of perturbed palmitoylation.
Fig. 8. Shh activity is disrupted in HhatCreface/Creface embryos. 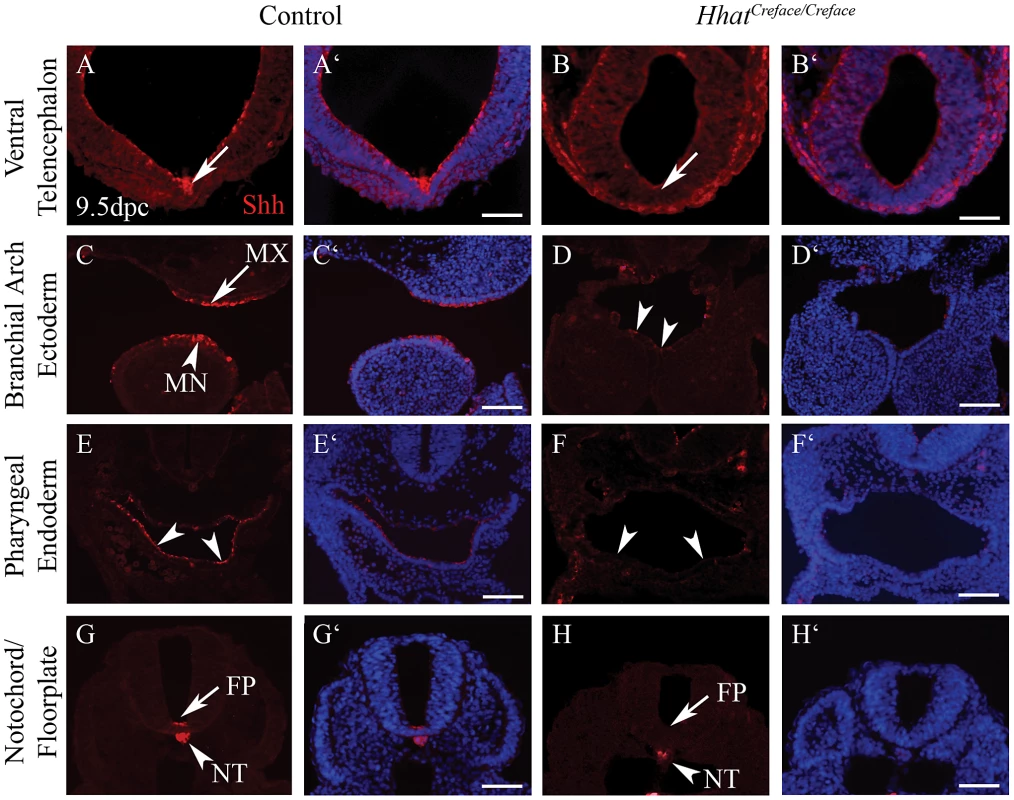
Shh immunostaining (red) and DAPI nuclei staining (blue) of transverse sections of E10.5 control and HhatCreface/Creface embryos demonstrating the absence or reduction of Shh activity in the ventral telencephalon (A–B′; arrows), branchial arch ectoderm (C, C′ arrows; D, D′ arrowheads), pharyngeal endoderm (E–F′, arrowheads) and floor plate (G–H′, arrows) of mutant embryos. Scale bars: 200 uM. HhatCreface/Creface embryos have decreased Patched1 activity
Ptch1 is a receptor for Hh ligands and its activity is widely used as a read out of the gradient and range of Hh signaling. Therefore to better document the degree of spatial perturbation of Hh signaling in HhatCreface/Creface embryos, we outcrossed Hhat+/Creface mice to Ptch1-LacZ mice [20] and used the LacZ reporter as a measure of Ptch1 activity. E9.5–11.5 Hhat+/Creface;Ptch1-LacZ+ embryos exhibit intense LacZ expression in the ventral telencephalon (arrows), pharyngeal endoderm, notochord (arrowheads) and surrounding mesenchyme as well as in the floorplate (Figure 9A, 9C, 9E, 9G, 9I, 9K). Often, a gradient of Ptch1 activity is present within or adjacent to these sites of Shh synthesis, which reflects the extensive range of Hh signaling in each of these regions (Figure 9I, 9K). In contrast, Ptch1-LacZ activity is considerably reduced in HhatCreface/Creface embryos (Figure 9B, 9D). More specifically these mutant embryos lack LacZ expression in the ventral telencephalon, branchial arch ectoderm, pharyngeal endoderm and floor plate (Figure 9F, 9H, 9J, 9L). LacZ activity in the mesenchyme surrounding the notochord is present in HhatCreface/Creface mutants, but is considerably reduced compared to control littermates (Figure 9I–9L). These results specifically highlight the spatiotemporal diminishment of the gradient and range of Shh activity in HhatCreface/Creface embryos.
Fig. 9. Patched (Ptch1) activation is diminished in HhatCreface/Creface embryos. 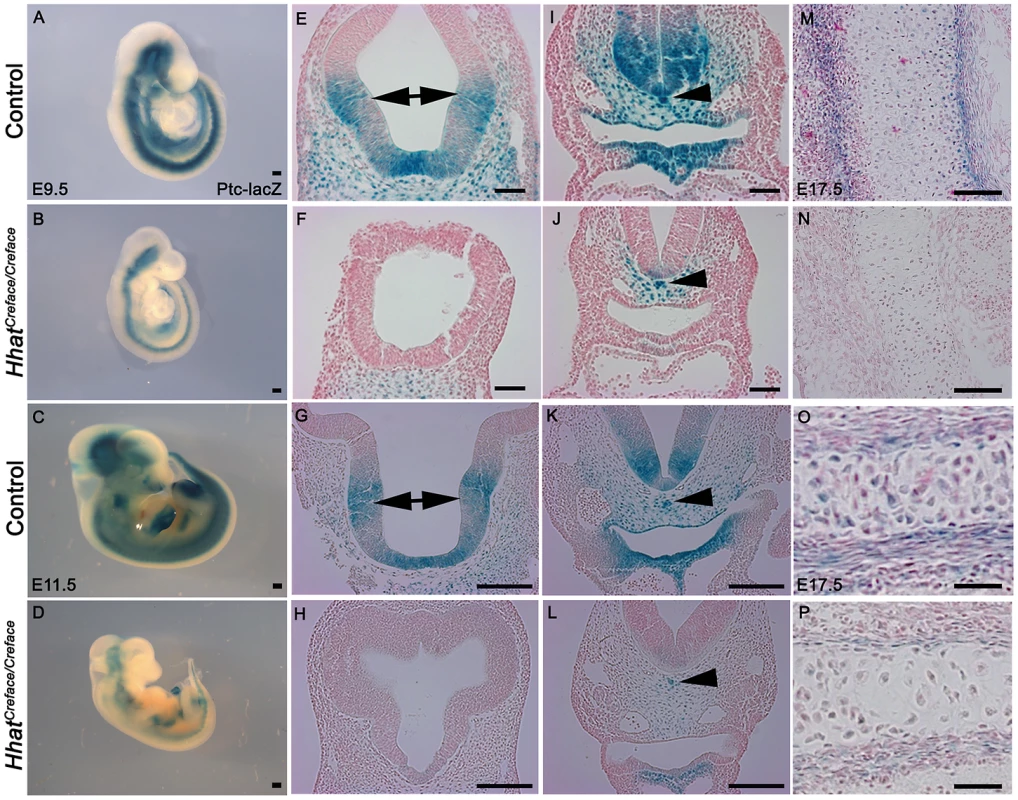
Ptch1-LacZ reporter staining in whole and transverse sectioned E9.5 and 11.5 control (A, C, E, G, I, K) and HhatCreface/Creface (B, D, F, H, J, L) embryos revealed a reduction in Shh signaling gradients and range throughout the telencephalon (E–H; double arrows), spinal cord, peri-notochord mesenchyme (arrowhead) and pharyngeal tissues (I–L) in mutant embryos. Ptch1-LacZ reporter staining of cranial sagittal sections through the exoccipital and basioccipital bones from E17.5 control (M, O) and HhatCreface/Creface (N, P) embryos illustrate the reduction of Shh signaling during endochondral ossification of cranioskeletal elements. Scale bars: A and B 200 uM; C and D 500 uM; E,F,I,J 50 uM; G,H,K,L 100 uM; M,N 100 uM; O,P 200 um. Hhat is required for the palmitoylation of all Hh proteins, not just Shh. Furthermore the cartilage and bones which are disrupted in HhatCreface/Creface embryos are dependent upon the actions of both Shh [19] and Indian hedgehog (Ihh) [32]. Consistent with this Shh−/− mutant embryos exhibit agenesis of the calvarial bones, while loss of Ihh results in reduction of cranial bone size and all markers of osteodifferentiation during endochondral as well as membranous ossification [33]. Therefore we also documented the activity of Ptch1 during ossification to determine if global Hh signaling was impaired during cranioskeletal differentiation and if this also contributed to the pathogenesis of craniofacial anomalies in HhatCreface/Creface embryos. In sections of E17.5–18.5 control embryos, strong Ptch1-LacZ activity was observed in the perichondrium and chondrocytes within cranioskeletal elements such as the exoccipital and basioccipital bones (Figure 9M, 9O). In contrast, Ptch1-LacZ activity was apparently absent from the exoccipital bone in HhatCreface/Creface;Ptch1-LacZ+ mutants (Figure 9N) and was considerably reduced in the basioccipital bone (Figure 9P). The disruptions in Ptch1 activity are indicative of a global diminishment of Hh signaling in HhatCreface/Creface embryos. Our results therefore demonstrate a direct in vivo requirement for Hhat in establishing Hh signaling gradients during early and late stages of embryogenesis. Thus perturbation of Hh signaling directly underpins the etiology of cranioskeletal defects in HhatCreface/Creface embryos.
Neural crest lineages are altered in HhatCreface/Creface mutants
HhatCreface/Creface embryos display decreased Shh signaling in the ventral telencephalon and branchial arches from E8.5–10.5. Temporally, this coincides with cranial neural crest cell migration and colonisation of the frontonasal, maxillary and mandibular facial prominences. Neural crest cells generate most of the cartilage, bone and connective tissue in the head and face [34], [35] and Shh is known to be required for their proper migration and survival [36]–[39]. We hypothesized that defects in neural crest cell development could therefore be a major contributing factor to the pathogenesis of cranioskeletal anomalies in HhatCreface/Creface embryos. Hence, we compared the formation, migration and survival of neural crest cells in control and HhatCreface/Creface embryos. No obvious defects in the initial specification of cranial neural crest cells were observed from in situ hybridization analyses with general neural crest cell markers such as Snail1 and Crabp1 (Figure S2A–S2H), or more lineage specific markers such as Sox9 and Sox10 (Figure S2I–S2P). Additionally, we performed lineage tracing analyses using Wnt1-Cre together with Rosa26R mice to permanently label neural crest cells. In E9.5–10.5 control embryos, streams of LacZ positive neural crest cells migrated into and colonized the developing frontonasal prominences as well as the pharyngeal arches (Figure S3A, S3B). In HhatCreface/Creface;Wnt1-Cre+;R26R+ embryos LacZ positive neural crest cells were present in similar patterns to control littermates, further demonstrating that there was no significant defect in neural crest cell formation, migration or colonization of the facial prominences and pharyngeal arches in HhatCreface/Creface embryos (Figure S3C, S3D). However, the patterns of LacZ staining highlighted the hypoplasia of medial, lateral, maxillary and mandibular facial prominences in HhatCreface/Creface embryos, and also revealed gross anomalies in development of the cranial ganglia as the trigeminal appeared to be hypoplastic, and the hypoglossal and vagal ganglia were fused (Figure S3C, S3D).
HhatCreface/Creface embryos exhibit cranial ganglia defects
Cranial ganglia form part of the peripheral nervous system and are derived from both neural crest and sensory placode cells [40]. The gross cranial ganglia defects observed via lacZ staining in E10.5 HhatCreface/Creface;Wnt1-Cre+;R26R+ embryos were confirmed through anti-neurofilament (2H3) immunostaining (Figure S4). Not only were the cranial ganglia and in particular the trigeminal hypoplastic, but interestingly, immunostaining more discretely revealed that the maxillary branch of the trigeminal was consistently narrower in HhatCreface/Creface embryos compared to control littermates (Figure S4A, S4D). This correlates with maxillary hypoplasia in mutant embryos (Figure 2A–2D). In addition, the hypoglossal and vagal ganglia are aberrantly fused and the oculomotor nerve which is readily identifiable in control embryos is missing in HhatCreface/Creface littermates.
The dual origin of cranial ganglia from neural crest and ectodermal placode cells prompted us to examine whether defects in development of either progenitor population underpinned the ganglia defects in HhatCreface/Creface mutants. E10.5 embryos were processed via in situ hybridization for Sox10 and Eya2, which demarcate neurogenic neural crest and sensory placode cells respectively. Although the spatiotemporal patterns of Sox10 and Eya2 expression were quite similar between control and mutant embryos, the domains of activity highlighted the hypoplasia of the trigeminal ganglia and fusion of the hypoglossal and vagal ganglia in HhatCreface/Creface embryos (Figure S4B, S4C, S4E, S4F). Collectively, these results indicate that Hhat loss-of-function and disrupted Hh signaling does not globally affect the induction or migration of neural crest and sensory placode cells. However, hypoplasia and aberrant fusion of the cranial ganglia as evidenced by both neural crest (Sox10) and sensory placode (Eya2) markers suggested that Hh signaling plays a regionalized role in the development and patterning of both progenitor cell populations.
HhatCreface/Creface embryos exhibit increased apoptosis
Hypoplasia of the facial prominences together with the trigeminal ganglia, suggested that Hhat - through its effects on mediating the gradient and range of Hh signaling - may play a critical role in progenitor cell survival. Consistent with this idea, Shh has been shown to be critical for neural crest cell survival during craniofacial morphogenesis [36], [37], [41]. Therefore we examined whether there were any alterations in progenitor cell survival in HhatCreface/Creface embryos by characterizing the spatiotemporal differences in cell death in control versus mutant embryos. Using the apoptotic marker cleaved-Caspase3, we observed a considerable increase in apoptotic cells in the neural crest cell derived frontonasal, maxillary, mandibular and prospective palatal mesenchyme of E9.5–10.5 HhatCreface/Creface embryos compared to control littermates (Figure 10A–10H). In contrast, we did not observe any significant differences in the number of proliferative cells using the marker phospho-Histone 3 (data not shown). Our data demonstrates that Hhat loss-of-function perturbs Hh signaling during early embryogenesis resulting in increased apoptosis in neural crest cell derived craniofacial mesenchyme. This underscores hypoplasia of the facial prominences during early embryogenesis and together with defects in differentiation, subsequently contributes to the characteristic acrania-holoprosencephaly-agnathia malformations observed in HhatCreface/Creface embryos.
Fig. 10. HhatCreface/Creface embryos exhibit increased cell death in craniofacial mesenchyme. 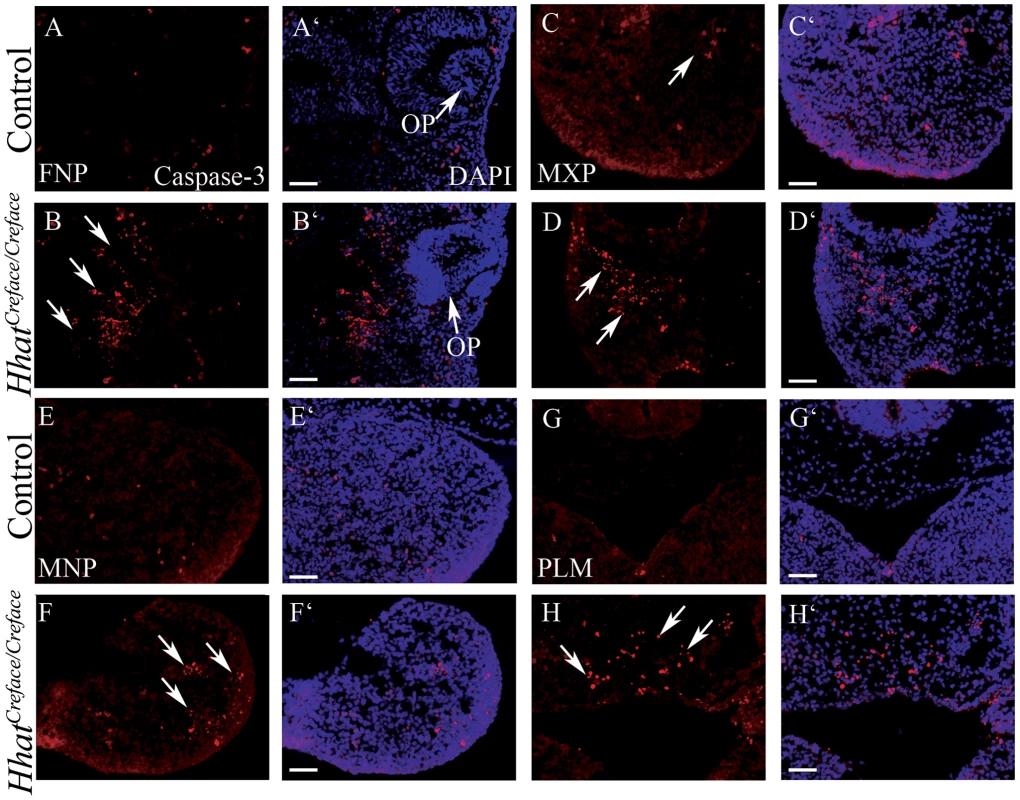
Caspase3 immunostaining (red) and DAPI (blue) nuclear staining of transverse sectioned E10.5 control (A, A′, C, C′, E, E′, G, G′) and HhatCreface/Creface (B, B′, D, D′, F, F′, H, H′) embryos revealed elevated levels of apoptotic cells (arrows) in the frontonasal (A–B′), maxillary (C–D′) and mandibular (E–F′) prominences as well as in the palatal mesenchyme (G–H′) of mutant embryos. Abbreviations: FNP, frontonasal process; PLM, palatal mesenchyme. Scale bars: 200 uM. HhatCreface/Creface embryos exhibit altered pharyngeal arch patterning
In addition to regulating cell survival, Hh signaling also plays important roles in governing branchial arch patterning. In fact, branchial arch ectoderm-derived Shh signaling, regulates downstream targets such as Fgf8 and Bmp4, which respectively influence proximal versus distal pharyngeal arch and jaw patterning. Fgf8 is normally expressed in the proximal maxillo-mandibular ectoderm of E9.5–10.5 embryos and is flanked (Figure 4A, 4C) distally within the mandible by an adjacent ectoderm domain of Bmp4 (Figure 4E, 4G). Compared to control littermates, Fgf8 (Figure 4B, 4D) and Bmp4 (Figure 4F, 4H) expression were absent from the first pharyngeal arch ectoderm of E10.5 HhatCreface/Creface embryos demonstrating their dependence on Hh signaling. Both Bmp and Fgf signaling are known to play critical temporal and tissue context dependent roles in cell proliferation and survival. Hence we examined the extent of their respective roles in the pathogenesis of craniofacial anomalies in HhatCreface/Creface mutants during early embryogenesis.
Bmps transduce signals by binding to complexes of type I and II serine/threonine kinase receptors which then activate canonical signaling via receptor Smads (R-Smads) 1, 5 and 8. Immunostaining with phospho-SMAD1/5/8 revealed no obvious alterations in SMAD signaling in the frontonasal, maxillary or mandibular mesenchyme of E10.5 HhatCreface/Creface embryos compared to control littermates (data not shown). The lack of any significant change in SMAD signaling is likely due to redundancy with other Bmp signals such as Bmp7 and furthermore is consistent with previous analyses that conditionally deleted Bmp4 from the branchial arch ectoderm [42].
In contrast, FGF signaling functions through membrane bound receptors which activate downstream effectors, such has mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK). We observed a considerable decrease in both the domain and level of phosphorylated Erk1/2 staining within the frontonasal, maxillary and mandibular mesenchyme of HhatCreface/Creface mutant embryos compared to control littermates (Figure S5A, S5B). This indicates that diminished Erk1/2 signaling impacts upon neural crest cell survival and contributes to hypoplasia of the facial prominences underpinning the pathogenesis of craniofacial anomalies in HhatCreface/Creface embryos. Consistent with this, conditional deletion of Erk1/2 in neural crest cells results in extensive agenesis of the neural crest cell derived craniofacial skeleton (P.A.T. unpublished). Furthermore, activation and repression of Shh signaling has been shown to respectively increase and decrease the levels of phosphorylated Erk1/2 in vitro [43]. Taken together with our in vivo data, this collectively highlights the vital role played by Hh-Fgf-Erk signaling in promoting neural crest cell survival and preventing apoptosis during normal craniofacial morphogenesis and in the pathogenesis of craniofacial anomalies in HhatCreface/Creface embryos.
Discussion
HhatCreface/Creface: a model of acrania-holoprosencephaly-agnathia
SHH was the first locus mapped in association with HPE in affected patients [1], [2] and knockouts of Shh in mice recapitulate the characteristics of HPE observed in human individuals. Mutations in Hh signaling pathway members, such as Patched1 (Ptch1) and Smoothened (Smo) also result in HPE [4]. Collectively, this illustrates the importance of Hh signaling in embryonic craniofacial development and in the etiology of HPE. To date, mutational analyses of human HPE together with naturally occurring and engineered mouse mutants have identified the genetic lesions responsible for only about 20% of the cases of HPE. Hence it is critical to identify additional candidate genes for the majority of patients whose genetic lesions remain unknown.
Here we describe the AP2-Cre (“Creface”) mouse [15] as an insertion mutation in Hhat. HhatCreface/Creface embryos exhibit holoprosencephaly; extensive apoptosis in the craniofacial mesenchyme; frontonasal, mediolateral, maxillary and mandibular prominence hypoplasia; patterning defects in the facial prominences and cranial ganglia; and diminished chondrogenesis and osteogenesis. Collectively these anomalies contribute to midfacial hypoplasia and agenesis of the jaw. In addition to holoprosencephaly and agnathia, HhatCreface/Creface mutant embryos lack all but the ventral bones of the skull. We observed complete agenesis of the nasal, frontal, parietal, and interparietal bones amongst others, while the basioccipital, exoccipital, and basisphenoid bones were all severely hypoplastic. Thus we have identified HhatCreface/Creface as a novel mouse model of acrania-holoprosencephaly-agnathia.
These phenotypes are consistent with perturbation of Hh signaling. Hh proteins are secreted glycoproteins that undergo autoproteolytic cleavage to generate their active forms. However dual cholesterol and palmitoyl post-translational lipid modification are also critical for proper Hh protein multimerization, distribution and activity [12]–[14]. Hhat encodes an acyltransferase and our results demonstrate that Hhat loss-of-function results in diminished palmitoylation of Shh. In support of our observations, Skinny hedgehog (Skn) which is also known as Hhat, was recently shown to catalyse the palmitoylation of all Hh proteins in invertebrates such as Drosophila [27], and mammals such as mice [25]–[29]. Furthermore, Drosophila mutants lacking Hh palmitoylation, exhibit patterning defects in Hh-responsive cells during wing and eye development, while limb development and spinal cord neurogenesis are disrupted in Skn mutant mice [25]–[29]. In agreement with this, HhatCreface/Creface mutants exhibit a diminished gradient and range of Hh signaling as early as E9.5 as evidenced by the specific spatiotemporal loss of Hh production and downregulation of Ptch1. Neither Shh protein nor Ptch1-lacZ reporter activity could be detected in the ventral forebrain, floor plate or pharyngeal arch ectoderm of HhatCreface/Creface embryos. During normal embryogenesis, Shh expression and subsequent Ptch1 activity in these tissues is induced by or dependent upon gradients of Shh emanating from other tissues such as the notochord and pharyngeal endoderm [30], [31],[38], [44]. Although some localized and possibly short-range Hh signaling remained intact, our data indicates that long-range Hh signaling gradients fail to be properly established during mammalian embryogenesis in the absence of Hhat. Thus the absence of palmitoylation prevents the formation of Hh multimeric complexes, subsequently limiting establishment of a Hh gradient. Therefore our analyses of HhatCreface/Creface mouse embryos highlight the conserved requirement for Hhat in establishing Hh signaling gradients in vivo during mammalian embryogenesis and in particular during craniofacial development. Thus Hhat plays a critical role in the regulation of Hh signaling during embryogenesis.
Patterning and cell survival are disrupted in HhatCreface/Creface embryos
Hh signaling performs a number of key roles during craniofacial development. Hh is required to maintain the viability of neural crest cells [37] during their colonization of the facial prominences and also regulates patterning of the frontonasal region and pharyngeal arches [45], [46]. Interestingly, Fgf8 is expressed in the proximal pharyngeal arch ectoderm [47], while Bmp4 is expressed in the distal ectoderm and underlying mesenchyme [48]. Fgf8 and Bmp4 are critical regulators of jaw development as the loss of either gene results in specific jaw defects. Conditional deletion of Fgf8 from the pharyngeal ectoderm results in the absence of molar teeth and most of the chondrocranial and dermatocranial elements that form the proximal jaw [49], [50]. In contrast, conditional inactivation of Bmp4 in the mandibular ectoderm results in agensis of incisor teeth together with severe distal truncations of the jaw [42]. Thus, the jaw is patterned in a proximo-distal manner through Fgf and Bmp activity respectively [42], [50].
HhatCreface/Creface embryos exhibit a complete absence of Fgf8 and Bmp4 expression in the pharyngeal arch ectoderm which demonstrates these genes are downstream targets of Hh signaling. Consistent with this idea, Fgf8 and Bmp4 expression is similarly diminished in Shh loss-of-function embryos [39], [51] and also as a consequence of pharyngeal endoderm ablation in chick embryos [44]. Taken together, this data indicates that Shh signaling from the pharyngeal endoderm is required for activation of Shh in the pharyngeal ectoderm and that this domain of activity in turn is essential for ectodermal Fgf8 and Bmp4 signaling during jaw development. Therefore, the presence of agnathia and anodontia observed in HhatCreface/Creface embryos correlates with the loss of Fgf8 and Bmp4 activity in the pharyngeal arch ectoderm.
Fgf8 and Bmp4, both play critical roles in neural crest cell and craniofacial mesenchyme survival [42], [49] and examination of Erk1/2 and Smad1/5/8 activity as downstream readouts of Fgf and Bmp signaling respectively, indicated that Fgf signaling was primarily affected in HhatCreface/Creface embryos. Interestingly, Shh is known to be required for the viability of neural crest cells [37] and activation and repression of Hh signaling in vitro increases and decreases Erk1/2 activity respectively [43]. Our analyses therefore suggest that Hh signaling acts through Fgf-Erk to promote neural crest cell viability and facial prominence outgrowth during craniofacial development. Consistent with this, Hhat is expressed at low but consistent levels throughout the facial prominences between E10.5–12.5 [52].
Shh loss-of-function embryos exhibit a reduction in the overall size of the brain, a single optic vesicle with agenesis of external eye structures, nasal proboscis and malformations of the craniofacial skeleton. Shh mutants also exhibit agenesis of all the calvarial bones irrespective of their neural crest or mesoderm origins [19]. The similarity in phenotypes between Shh−/− embryos and HhatCreface/Creface mutants suggests that loss of Shh signaling in HhatCreface mutants may also primarily account for the extensive defects in cranial vault. However, Ihh also plays an important role in craniofacial development during late gestation. Loss of Ihh results in reduced chondrocyte proliferation, an absence of bone calcification, hypoplasia of the cranial bones and diminished markers of osteodifferentiation during endochondral as well as membranous ossification [32]. [33]. Although there are three members of the Hh gene family, neither Ihh nor Dhh is appreciably expressed in craniofacial tissues prior to E12.5 [36], [53], [54]. Thus, whereas early neural crest cell survival and craniofacial morphogenesis depends primarily upon Shh signaling, later craniofacial differentiation and maturation requires both Shh and Ihh. Therefore during early embryogenesis, perturbation of Shh signaling is likely to be primarily responsible for apoptosis in the craniofacial mesenchyme and defects in facial prominence and pharyngeal arch patterning and outgrowth in HhatCreface/Creface mutants. Consistent with this idea, perturbation of Hh signaling in neural crest cells through conditional deletion of Smo has no effect on neural crest migration but elicits a profound effect on the formation of skeletal and non-skeletal elements of the head [36].
Thus the combination of defects in patterning of the facial prominences, extensive apoptosis in the neural crest cell derived craniofacial mesenchyme and altered skeletogenic differentiation, underpins the craniofacial malformations in HhatCreface/Creface embryos. In summary, we have identified HhatCreface/Creface mice as a novel model of acrania-holoprosencephal-agnathia. Hhat palmitoylates Hh proteins and is critical for establishing proper gradients of Hh signaling during embryogenesis. Our work therefore highlights the importance of long-range Hh signaling in craniofacial development and suggests HHAT is a potential candidate in the etiology and pathogenesis of HPE and its associated congenital human disorders.
Materials and Methods
Mouse lines and maintenance
Mice were housed in the Laboratory Animal Services Facility at the Stowers Institute for Medical Research according to IACUC animal welfare guidelines. Hhat+/Creface were maintained on a CD1 background; genotyping of the Hhat+/Creface mice was performed as described below. The Patched1(Ptch1)-LacZ [20], Rosa 26 Reporter (R26R) [55], Shh [19], and Wnt1-Cre [56], [57] mice were maintained as previously reported. For embryo collection, the day of plug was noted as embryonic (E) day 0 and control embryos described in the analyses were either wild-type or heterozygous littermates. A minimum of 4 embryos were used for each assay performed unless otherwise stated. Embryo morphology was visualized by staining whole embryos with DAPI to label all cell nuclei and imaging fluorescent signal. Embryos were fixed in 4% paraformaldehyde overnight at 4°C then stained with 2 ug/ml DAPI in PBS and imaged by confocal microscopy. Scale bars designate magnification of embryos however in some cases may be a close approximation.
Identification of the Creface transgene insertion site
The insertion location of the Creface transgene was identified using the Vectorette kit (Sigma, St. Louis, MO). A Creface;Vectorette DNA library was created using the EcoRI-Vectorette unit according to manufacturer's instructions. To amplify clone DNA containing the transgene and surrounding genomic region, the primer (5′-ACA TCT GGG GTG AAG GGA ATT AGG GAG TTG-3′) was used in addition to the Vectorette-specific primer to amplify DNA bands containing the integration site. A step-down PCR was used according to manufacturer's instructions; a 5 kb band, clone E1N3, was gel-purified (Gel Extraction Kit, Qiagen, Valencia, CA) and sequenced using the Vectorette sequencing primer. The blasted sequence aligned to intron 9–10 of the Hhat gene, indicating Hhat as the gene disrupted in Creface mutants.
Verification of transgene insertion site was performed using PCR in wild-type, heterozygous and mutant genomic DNA from embryos at E10.5. Amplification of the region spanning the insertion site used the transgene-specific primer (5′-TGG TTA CCT TCC TCC AGA TAG TATG-3′) and Hhat-specific primer (5′-CAC TTG CTA ACT AGA AGG AAC TTCC-3′) produced a 250 bp band; wild-type primers (5′-CCT GGG AAG GAA AAA CCA ATA TGTA-3′) and (5′-GGT CCT ATC ATG CTA CCA AGA AA-3′) amplified a 1.4 kb band. Samples were denatured at 94°C for 30 sec, annealed at 57°C for 30 sec, and extended at 72°C for 30 sec for 30 cycles for both reactions; PCR bands for the wildtype and mutant reactions were gel purified and sequenced, confirming Hhat as the gene disrupted by the Creface transgene.
To confirm the absence of Hhat mRNA in HhatCreface/Creface mutants, RNA was isolated from wild-type and mutant embryos (RNeasy kit, Qiagen, Valencia, CA) and cDNA libraries were created using the Superscript first strand kit (Invitrogen, Carlsbad, CA). Primers (5′-AGG TTC TGG TGG GAC CCT GTGT-3′) and (5′-AGA AAG CAG TGT CCC CAA CAGG-3′) were used to amplify the full length Hhat mRNA in wild-type, heterozygous and mutant embryos. Primers to glyceraldehyde 3-phosphate dehydrogenase (Gapdh) (5′-AGC CTC GTC CCG TAG ACA AAAT-3′) and (5′-ACC AGG AAA TGA GCT TGA CAAA-3′) were used as an internal positive control.
In situ hybridization
Embryos were collected as described above, fixed overnight in 4% paraformaldehyde (PFA) at 4°C, dehydrated in methanol (MeOH), and stored at −20°C until used in the staining protocol. In situ hybridizations were performed following the standard protocol described by [58]. Anti-sense digoxigenin-labeled mRNA riboprobes were synthesized for Bmp4 (R. Arkell), Crabp1 (S. Schneider-Maunoury), Eya2 (P. Trainor), Fgf8 (I. Mason), Shh (A. McMahon), Six2 (P. Trainor), Snail (A. Nieto), Sox9 (R. Krumlauf), and Sox10 (M. Gassmann). Hhat forward 5′-CTG CGT GAG CAC CAT GTT CA-3′ and Hhat reverse 5′-TCT CCA CAG TGA CTC CCA GC-3′ primers were used to generate an exon 8–10 specific probe and whole embryos which was stained with Hhat antisense probe were mounted in Tissue Tek OCT (VWR, West Chester, PA) and sectioned at 20 µm thick to observe spatiotemporal activity.
β-galactosidase staining
Hhat+/Creface mice were mated to Rosa 26-lacZ reporter (R26R) mice and embryos were collected at E9.5–11.5 as described above. Embryos were stained using the β-galacatosidase staining solution kit (Chemicon/Millipore, Billerica, MA) according to manufacturer's instructions. Briefly, embryos were fixed on wet ice for 20–30 min in the Tissue Fixative Solution, washed in Tissue Rinse Solution A for 30 min and Tissue Rinse Solution B for 5 min at RT. Embryos were incubated O/N at 37°C in Tissue Stain Solution with X-gal (40 mg/mL in DMF) (Invitrogen, Carlsbad, CA). After incubation, embryos were washed PBS and re-fixed overnight. Embryos were processed in paraffin, cut in 10micron sections, and counterstained in Nuclear Fast Red (NFR) (Sigma, St. Louis, MO).
For β-galactosidase staining on sections, E15.5 embryos were harvested and immersed in LacZ fixative (0.25%glutaraldehyde, 10% 0.5 M EGTA, and 10% 1 M MgCl2 in PBS) for 2.5 hrs at 4°C. Embryos were processed through a sucrose gradient (15% then 30% sucrose in PBS) and snap frozen in OCT. Eight micron sections were cut and re-fixed for 10 min at room temperature. Sections were then washed in LacZ wash buffer (0.2% MgCl2, 0.01% NaDOC, 0.02% NP40 in PBS) three times for 5 min. β-galactosidase staining was developed using a LacZ stain solution (0.002% Potassium ferrocyanide, 0.01% Potassium ferricyanide, 0.04% X-gal (25 mg/mL in DMF) in LacZ wash buffer) to the desired intensity. The sections were then rinsed three times in LacZ wash buffer for 5 min, counterstained with nuclear fast red and rinsed in distilled water for 1–2 min. Sections were mounted and photographed using a Ziess Axioplan microscope and processed using Photoshop CS2 (Adobe, San Jose, CA).
Immunohistochemistry
For whole-mount staining using a neurofilament antibody, embryos were fixed in 4% PFA overnight at 4°C and dehydrated in a graded MeOH series. Dehydrated embryos were bleached in methanol∶DMSO∶30% H2O2 (4∶1∶1) for 5 hrs at room temperature and rehydrated in 50% methanol∶PBS and 15% methanol∶PBS, PBS for 30 minutes each. Embryos were blocked in PBSMT (2% milk powder, 0.1% Triton X-100 in PBS) twice for 1 hour at room temperature. Embryos were incubated in primary antibody to Neurofilament (2H3) diluted at 1∶250 O/N at 4°C in PBSMT. Embryos were rinsed in PBSMT and incubated in secondary antibody using a 1∶200 dilution in PBSMT of HRP-coupled goat anti-rabbit IgG (Jackson Immunoresearch, West Grove, PA). For color development, embryos were rinsed in PBSMT and PBT (0.2% BSA, Sigma, 0.1% Triton X-100) three times, 20 minutes each at room temperature and washed in 3,3-Diaminobenzidine (0.3 mg/mL) (Sigma, St. Louis, MO) in PBT for 20 minutes; 0.03% H2O2 was added and color was developed to the desired intensity.
For section immunohistochemistry, embryos were fixed overnight in 1% PFA at 4°C. Embryos were processed through a sucrose gradient (15%, then 30% sucrose in PBS), mounted in Tissue Tek O.C.T. (VWR, West Chester, PA) and sectioned at 10microns. Sections were rinsed three times in PBT (PBS with 0.1% Triton X-100) for 5 min and blocked in 10% goat serum (Invitrogen, Carlsbad, CA) in PBT for 1 hr at RT. Slides were incubated in primary antibody diluted in 10% goat serum/PBT overnight at 4°C. The mouse monoclonal antibodies to cleaved-Caspase-3 and pErk1/2 (Cell Signaling Technologies, Danvers, MA), phospho-Histone-3 (Upstate/Millipore, Billerica, MA) and Shh (5e1) were used at 1∶500 (Caspase-3, anti-pH3), 1∶250 (pErk1/2) and 1∶25 (5e1) respectively.
Slides were rinsed three times in PBT for 10 min each at room temperature and incubated in a goat anti-mouse Alexa 488 secondary antibody at 1∶250 (Molecular probes/Invitrogen, Carlsbad, CA) for 2 hours at 4°C. Sections were counterstained with a 1/1000 dilution of 2 mg/ml DAPI (Sigma, St. Louis, MO) in PBS for 5 minutes, followed by multiple rinses in PBS. Slides were then mounted with fluorescent mounting medium (DakoCytomation, Carpinteria, CA). All images were collected using a Ziess Axioplan microscope and processed using Photoshop CS2 (Adobe, San Jose, CA). Antibodies to Shh (5e1) and Neurofilament (2H3) were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences (Iowa City, IA 52242).
Cartilage and bone staining
Embryos were collected at E15.5 and 17.5 and fixed in 95% ethanol (EtOH) overnight. Embryos were washed in a stain solution containing 0.5% Alizarin red (Sigma, St. Louis, MO) and 0.4% Alcian blue 8× (Sigma, St. Louis, MO) in 60% EtOH overnight at room temperature. For E15.5dpc embryos, soft tissue was dissolved in 2% KOH for three hours and transferred to 0.25% KOH for 30 min. For E17.5 staining, the embryos were anesthetized in PBS for 1 hr at 4°C, fixed overnight in 95% EtOH, skinned and eviscerated prior to staining; all remaining soft tissue was dissolved in 2% KOH for six hours and transferred to 0.25% KOH for 30 min. Embryos were cleared in glycerol∶KOH (20%∶0.25%; 33%∶0.25%; 50%∶0.25%). Embryos were stored in 50% glycerol∶0.25% KOH until photographed.
Palmitoylation assay
Immortalized mouse embryonic fibroblast (MEFs) derived from control and HhatCreface/Creface mouse embryos were transfected Shh expression vector (gift from Dr. Pao-Tien Chuang) using Amaxa Nucleofactor (Amaxa, Germany) according to manufactures protocol. Twenty-four hours after transfection, cells were incubated with 50 µM Click-It palmitic acid (Invitrogen, Carlsbad,CA). After 12 hrs incubation, cells were washed three times with cold PBS and whole cell lysate was harvested using RIPA lyses buffer. Protein extracts were subjected to Click labeling reaction using Click-It protein reaction buffer kit (Invitrogen, Carlsbad,CA) in order to attach biotin to incorporated palmitic acid. Precipitated samples were again dissolved into RIPA buffer and mixed with mouse anti-Shh antibody (5e1) overnight at 4°C with rocking, and subsequently immunoprecipitated with protein G beads (Thermo scientific, Rockford, IL) with rocking for 2 hrs at room temperature. The beads were resuspended in SDS sample buffer and separated by SDS-PAGE and transferred onto a PVDF membrane (GE healthcare life sciences, Buckinghamshire, UK). The membrane was incubated with alkaline phosphatase streptavidin (Vector laboratories, Burlingame, CA) and treated with fluorescent substrate for alkaline phosphatase (Vector laboratories, Burlingame, CA), exposed to X-ray film to detect the palmitoylated Shh. Same membrane was incubated with anti Shh antibody (H-160; Santa Cruz Biotechnology Inc, Santa Cruz, CA) and subsequently HRP conjugated secondary antibody (Sigma, St.Louis, MO) in order to confirm the amount of Shh protein in each samples. The signals were detected with ECL plus western blotting detection system (GE healthcare life sciences, Buckinghamshire, UK) and exposed to X-ray film.
Supporting Information
Zdroje
1. BelloniE, MuenkeM, RoesslerE, TraversoG, Siegel-BarteltJ, et al. (1996) Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nature Genetics 14 : 353–356.
2. RoesslerE, BelloniE, GaudenzK, JayP, BertaP, et al. (1996) Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet 14 : 357–360.
3. MuenkeM, BeachyPA (2000) Genetics of ventral forebrain development and holoprosencephaly. Curr Opin Genet Dev 10 : 262–269.
4. WallisD, MuenkeM (2000) Mutations in holoprosencephaly. Hum Mutat 16 : 99–108.
5. DeMyer ME (1977) Holoprosencephaly (cyclopia-arhinencephaly); Vinken PJ, Bruyn GW, editors. Amerstam: North-Hollan.
6. DemyerW, ZemanW, PalmerCG (1964) The Face Predicts the Brain: Diagnostic Significance of Median Facial Anomalies for Holoprosencephaly (Arhinencephaly). Pediatrics 34 : 256–263.
7. DemyerW, ZemanW (1963) Alobar holoprosencephaly (arhinencephaly) with median cleft lip and palate: clinical, electroencephalographic and nosologic considerations. Confin Neurol 23 : 1–36.
8. MartinRA, JonesKL, BenirschkeK (1996) Absence of the lateral philtral ridges: a clue to the structural basis of the philtrum. Am J Med Genet 65 : 117–123.
9. CohenMMJr, ShiotaK (2002) Teratogenesis of holoprosencephaly. Am J Med Genet 109 : 1–15.
10. AotoK, YayoiS, HigashiyamaD, ShiotaK, MotoyamaJ (2008) Fetal ethanol exposure activates protein kinase a and impairs Shh expression in prechordal mesendoderm cells in the pathogenesis of holoprosencephaly. Birth Defects Res A Clin Mol Teratol
11. BarrMJr, HansonJW, CurreyK, SharpS, TorielloH, et al. (1983) Holoprosencephaly in infants of diabetic mothers. J Pediatr 102 : 565–568.
12. PorterJA, EkkerSC, ParkWJ, von KesslerDP, YoungKE, et al. (1996) Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 86 : 21–34.
13. PorterJA, YoungKE, BeachyPA (1996) Cholesterol modification of hedgehog signaling proteins in animal development. Science 274 : 255–259.
14. PepinskyRB, ZengC, WenD, RayhornP, BakerD, et al. (1998) Identification of a palmitic acid-modified form of human Sonic hedgehog. Journal of Biological Chemistry 273 : 14037–14045.
15. NelsonDK, WilliamsT (2004) Frontonasal process-specific disruption of AP-2alpha results in postnatal midfacial hypoplasia, vascular anomalies, and nasal cavity defects. Dev Biol 267 : 72–92.
16. ZhangJ, WilliamsT (2003) Identification and regulation of tissue-specific cis-acting elements associated with the human AP-2alpha gene. Dev Dyn 228 : 194–207.
17. ChaiY, MaxsonREJr (2006) Recent advances in craniofacial morphogenesis. Dev Dyn 235 : 2353–2375.
18. KronenbergHM (2003) Developmental regulation of the growth plate. Nature 423 : 332–336.
19. ChiangC, LitingtungY, LeeE, YoungKE, CordenJL, et al. (1996) Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383 : 407–413.
20. GoodrichLV, MilenkovicL, HigginsKM, ScottMP (1997) Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277 : 1109–1113.
21. CostantiniF, LacyE (1981) Introduction of a rabbit beta-globin gene into the mouse germ line. Nature 294 : 92–94.
22. JaenischR (1988) Transgenic animals. Science 240 : 1468–1474.
23. HeaneyJD, BronsonSK (2006) Artificial chromosome-based transgenes in the study of genome function. Mamm Genome 17 : 791–807.
24. TaylorFR, WenD, GarberEA, CarmilloAN, BakerDP, et al. (2001) Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry 40 : 4359–4371.
25. ChenMH, LiYJ, KawakamiT, XuSM, ChuangPT (2004) Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev 18 : 641–659.
26. AmanaiK, JiangJ (2001) Distinct roles of Central missing and Dispatched in sending the Hedgehog signal. Development 128 : 5119–5127.
27. ChamounZ, MannRK, NellenD, von KesslerDP, BellottoM, et al. (2001) Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 293 : 2080–2084.
28. LeeJD, TreismanJE (2001) Sightless has homology to transmembrane acyltransferases and is required to generate active Hedgehog protein. Curr Biol 11 : 1147–1152.
29. MicchelliCA, TheI, SelvaE, MogilaV, PerrimonN (2002) Rasp, a putative transmembrane acyltransferase, is required for Hedgehog signaling. Development 129 : 843–851.
30. MacdonaldR, BarthKA, XuQ, HolderN, MikkolaI, et al. (1995) Midline signalling is required for Pax gene regulation and patterning of the eyes. Development 121 : 3267–3278.
31. DaleJ, VesqueC, LintsT, SampathK, FurleyA, et al. (1997) Cooperation of BMP7 and SHH in the induction of forebrain ventral midline cells by prechordal mesoderm. Cell 90 : 257–269.
32. St-JacquesB, HammerschmidtM, McMahonAP (1999) Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev 13 : 2072–2086.
33. LentonK, JamesAW, ManuA, BrugmannSA, BirkerD, et al. (2011) Indian hedgehog positively regulates calvarial ossification and modulates bone morphogenetic protein signaling. Genesis 49 : 784–796.
34. Le Douarin N, Kalcheim C (1999) The Neural Crest; Bard J, Barlow P, Kirk D, editors: Cambridge Univesity Press.
35. NodenD (1978) The control of avian cephalic neural crest cytodifferentiation I. skeletal and connective tissues. Devl Biol 67 : 296–312.
36. JeongJ, MaoJ, TenzenT, KottmannAH, McMahonAP (2004) Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev 18 : 937–951.
37. AhlgrenSC, Bronner-FraserM (1999) Inhibition of sonic hedgehog signaling in vivo results in craniofacial neural crest cell death. Curr Biol 9 : 1304–1314.
38. Moore-ScottBA, ManleyNR (2005) Differential expression of Sonic hedgehog along the anterior-posterior axis regulates patterning of pharyngeal pouch endoderm and pharyngeal endoderm-derived organs. Dev Biol 278 : 323–335.
39. YamagishiC, YamagishiH, MaedaJ, TsuchihashiT, IveyK, et al. (2006) Sonic hedgehog is essential for first pharyngeal arch development. Pediatr Res 59 : 349–354.
40. Larson WJ (1993) Essentials of Human Embryology: Churchill Livingstone Inc.
41. AhlgrenSC, ThakurV, Bronner-FraserM (2002) Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proc Natl Acad Sci U S A 99 : 10476–10481.
42. LiuW, SeleverJ, MuraliD, SunX, BruggerSM, et al. (2005) Threshold-specific requirements for Bmp4 in mandibular development. Dev Biol 283 : 282–293.
43. ChangH, LiQ, MoraesRC, LewisMT, HamelPA (2010) Activation of Erk by sonic hedgehog independent of canonical hedgehog signalling. Int J Biochem Cell Biol 42 : 1462–1471.
44. BritoJM, TeilletMA, Le DouarinNM (2006) An early role for sonic hedgehog from foregut endoderm in jaw development: ensuring neural crest cell survival. Proc Natl Acad Sci U S A 103 : 11607–11612.
45. HuD, MarcucioRS (2009) A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development 136 : 107–116.
46. MarcucioRS, CorderoDR, HuD, HelmsJA (2005) Molecular interactions coordinating the development of the forebrain and face. Dev Biol 284 : 48–61.
47. CrossleyPH, MartinGR (1995) The mouse FGF-8 gene encodes a family of polypeptides expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121 : 439–451.
48. BennettJH, HuntP, ThorogoodP (1995) Bone morphogenetic protein-2 and -4 expression during murine orofacial development. Arch Oral Biol 40 : 847–854.
49. TrumppA, DepewMJ, RubensteinJL, BishopJM, MartinGR (1999) Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev 13 : 3136–3148.
50. TuckerAS, YamadaG, GrigoriouM, PachnisV, SharpePT (1999) Fgf-8 determines rostral-caudal polarity in the first branchial arch. Development 126 : 51–61.
51. HaworthKE, WilsonJM, GrevellecA, CobourneMT, HealyC, et al. (2007) Sonic hedgehog in the pharyngeal endoderm controls arch pattern via regulation of Fgf8 in head ectoderm. Dev Biol 303 : 244–258.
52. FengW, LeachSM, TipneyH, PhangT, GeraciM, et al. (2009) Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PLoS ONE 4: e8066 doi:10.1371/journal.pone.0008066.
53. BitgoodMJ, McMahonAP (1995) Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol 172 : 126–138.
54. KronmillerJE, NguyenT (1996) Spatial and temporal distribution of Indian hedgehog mRNA in the embryonic mouse mandible. Arch Oral Biol 41 : 577–583.
55. SorianoP (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21 : 70–71.
56. JiangX, RowitchDH, SorianoP, McMahonAP, SucovHM (2000) Fate of the mammalian cardiac neural crest. Development 127 : 1607–1616.
57. ChaiY, JiangX, ItoY, BringasPJr, HanJ, et al. (2000) Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127 : 1671–1679.
58. Nagy A, Gertsenstein M, Vintersten K, Behringer RR (2003) Manipulating the mouse embryo. Cold Spring Harbor: Cold Spring Harbor Laboratory.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 10
-
Všechny články tohoto čísla
- The Germline Genome Provides a Niche for Intragenic Parasitic DNA: Evolutionary Dynamics of Internal Eliminated Sequences
- Classical Genetics Meets Next-Generation Sequencing: Uncovering a Genome-Wide Recombination Map in
- Calpain-5 Mutations Cause Autoimmune Uveitis, Retinal Neovascularization, and Photoreceptor Degeneration
- Cofilin-1: A Modulator of Anxiety in Mice
- The Date of Interbreeding between Neandertals and Modern Humans
- Embryos of Robertsonian Translocation Carriers Exhibit a Mitotic Interchromosomal Effect That Enhances Genetic Instability during Early Development
- Viral Evasion of a Bacterial Suicide System by RNA–Based Molecular Mimicry Enables Infectious Altruism
- Phosphatase-Dead Myotubularin Ameliorates X-Linked Centronuclear Myopathy Phenotypes in Mice
- Full-Length Synaptonemal Complex Grows Continuously during Meiotic Prophase in Budding Yeast
- MOV10 RNA Helicase Is a Potent Inhibitor of Retrotransposition in Cells
- A Likelihood-Based Framework for Variant Calling and Mutation Detection in Families
- The Contribution of RNA Decay Quantitative Trait Loci to Inter-Individual Variation in Steady-State Gene Expression Levels
- New Partners in Regulation of Gene Expression: The Enhancer of Trithorax and Polycomb Corto Interacts with Methylated Ribosomal Protein L12 Its Chromodomain
- Mining the Unknown: A Systems Approach to Metabolite Identification Combining Genetic and Metabolic Information
- Mutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
- The Many Landscapes of Recombination in
- Faster-X Evolution of Gene Expression in
- Loss of Slc4a1b Chloride/Bicarbonate Exchanger Function Protects Mechanosensory Hair Cells from Aminoglycoside Damage in the Zebrafish Mutant
- Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection
- and the BTB Adaptor Are Key Regulators of Sleep Homeostasis and a Dopamine Arousal Pathway in Drosophila
- Mutation and Fetal Ethanol Exposure Synergize to Produce Midline Signaling Defects and Holoprosencephaly Spectrum Disorders in Mice
- Specific Missense Alleles of the Arabidopsis Jasmonic Acid Co-Receptor COI1 Regulate Innate Immune Receptor Accumulation and Function
- Deep Genome-Wide Measurement of Meiotic Gene Conversion Using Tetrad Analysis in
- Mismatch Repair Balances Leading and Lagging Strand DNA Replication Fidelity
- Distinguishing between Selective Sweeps from Standing Variation and from a Mutation
- Cytokinesis-Based Constraints on Polarized Cell Growth in Fission Yeast
- Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes
- Functional Antagonism between Sas3 and Gcn5 Acetyltransferases and ISWI Chromatin Remodelers
- The SET-Domain Protein SUVR5 Mediates H3K9me2 Deposition and Silencing at Stimulus Response Genes in a DNA Methylation–Independent Manner
- Morphogenesis and Cell Fate Determination within the Adaxial Cell Equivalence Group of the Zebrafish Myotome
- Muscle-Specific Splicing Factors ASD-2 and SUP-12 Cooperatively Switch Alternative Pre-mRNA Processing Patterns of the ADF/Cofilin Gene in
- Maize Is Required for Maintaining Silencing Associated with Paramutation at the and Loci
- Increasing Signal Specificity of the TOL Network of mt-2 by Rewiring the Connectivity of the Master Regulator XylR
- Use of Pleiotropy to Model Genetic Interactions in a Population
- RAB-Like 2 Has an Essential Role in Male Fertility, Sperm Intra-Flagellar Transport, and Tail Assembly
- Variants Affecting Exon Skipping Contribute to Complex Traits
- Topoisomerase II– and Condensin-Dependent Breakage of -Sensitive Fragile Sites Occurs Independently of Spindle Tension, Anaphase, or Cytokinesis
- Comparison of Family History and SNPs for Predicting Risk of Complex Disease
- Recovery of Arrested Replication Forks by Homologous Recombination Is Error-Prone
- A Mutation in the Gene Causes Alternative Splicing Defects and Deafness in the Bronx Waltzer Mouse
- Comparative Genomics Suggests an Independent Origin of Cytoplasmic Incompatibility in
- It Was Heaven: An Interview with Evelyn Witkin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Mutation in the Gene Causes Alternative Splicing Defects and Deafness in the Bronx Waltzer Mouse
- Mutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
- Classical Genetics Meets Next-Generation Sequencing: Uncovering a Genome-Wide Recombination Map in
- Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



