-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
Reklamaand the BTB Adaptor Are Key Regulators of Sleep Homeostasis and a Dopamine Arousal Pathway in Drosophila
Sleep is homeostatically regulated, such that sleep drive reflects the duration of prior wakefulness. However, despite the discovery of genes important for sleep, a coherent molecular model for sleep homeostasis has yet to emerge. To better understand the function and regulation of sleep, we employed a reverse-genetics approach in Drosophila. An insertion in the BTB domain protein CG32810/insomniac (inc) exhibited one of the strongest baseline sleep phenotypes thus far observed, a ∼10 h sleep reduction. Importantly, this is coupled to a reduced homeostatic response to sleep deprivation, consistent with a disrupted sleep homeostat. Knockdown of the INC-interacting protein, the E3 ubiquitin ligase Cul3, results in reduced sleep duration, consolidation, and homeostasis, suggesting an important role for protein turnover in mediating INC effects. Interestingly, inc and Cul3 expression in post-mitotic neurons during development contributes to their adult sleep functions. Similar to flies with increased dopaminergic signaling, loss of inc and Cul3 result in hyper-arousability to a mechanical stimulus in adult flies. Furthermore, the inc sleep duration phenotype can be rescued by pharmacological inhibition of tyrosine hydroxylase, the rate-limiting enzyme for dopamine biosynthesis. Taken together, these results establish inc and Cul3 as important new players in setting the sleep homeostat and a dopaminergic arousal pathway in Drosophila.
Published in the journal: . PLoS Genet 8(10): e32767. doi:10.1371/journal.pgen.1003003
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003003Summary
Sleep is homeostatically regulated, such that sleep drive reflects the duration of prior wakefulness. However, despite the discovery of genes important for sleep, a coherent molecular model for sleep homeostasis has yet to emerge. To better understand the function and regulation of sleep, we employed a reverse-genetics approach in Drosophila. An insertion in the BTB domain protein CG32810/insomniac (inc) exhibited one of the strongest baseline sleep phenotypes thus far observed, a ∼10 h sleep reduction. Importantly, this is coupled to a reduced homeostatic response to sleep deprivation, consistent with a disrupted sleep homeostat. Knockdown of the INC-interacting protein, the E3 ubiquitin ligase Cul3, results in reduced sleep duration, consolidation, and homeostasis, suggesting an important role for protein turnover in mediating INC effects. Interestingly, inc and Cul3 expression in post-mitotic neurons during development contributes to their adult sleep functions. Similar to flies with increased dopaminergic signaling, loss of inc and Cul3 result in hyper-arousability to a mechanical stimulus in adult flies. Furthermore, the inc sleep duration phenotype can be rescued by pharmacological inhibition of tyrosine hydroxylase, the rate-limiting enzyme for dopamine biosynthesis. Taken together, these results establish inc and Cul3 as important new players in setting the sleep homeostat and a dopaminergic arousal pathway in Drosophila.
Introduction
Sleep is a homeostatically regulated process, consuming roughly one-third of our lives, yet its function remains a mystery. To identify novel pathways governing sleep, we and others have employed a genetic approach in Drosophila. The fruit fly shares several core features of sleep with its mammalian counterparts, including behavioral quiescence, reduced responsiveness to sensory stimuli, and homeostatic responses to sleep deprivation [1], [2]. To date, several forward-genetics screens have been performed, successfully identifying mutants that increase or decrease sleep duration to varying degrees, highlighting the roles of (1) membrane excitability via the Shaker potassium channel [3]–[6], (2) neurotransmitters such as dopamine [7]–[10], (3) growth factors such as epidermal growth factor [11], and (4) signal transduction pathways among others [12]–[14]. Of these mutations, those affecting Shaker or dopamine yield the most robust phenotypes [4]–[6], [9], [15]. Yet how these key pathways regulate sleep homeostasis remains unclear.
Here we report the result of a reverse-genetics approach aimed at identifying regulators of sleep and arousal in Drosophila. We focused on the gene with the most robust phenotype, insomniac (inc), a target identifier for the E3 ubiquitin ligase Cullin-3 (Cul3) [16]. We find that flies lacking inc or Cul3 exhibit strikingly reduced and poorly consolidated sleep. Developmental expression of inc and Cul3 in post-mitotic neurons contributes to these adult sleep phenotypes. In addition to their baseline sleep phenotypes, both inc and Cul3 also exhibit reduced homeostatic responses to sleep deprivation as well as hyper-arousability to mechanical stimuli. Baseline sleep in flies deficient for inc or Cul3 can be rescued by pharmacological inhibition of dopamine synthesis, but are behaviorally resistant to pharmacologically increased dopamine synthesis, consistent with the hypothesis that these genes operate in a dopamine arousal pathway. Taken together, our data indicate a central role for inc and Cul3 in sleep homeostasis and dopamine-mediated arousal.
Results
A reverse-genetics screen for sleep genes
To identify novel sleep genes, we performed a reverse-genetics screen, focusing on genes previously reported to have sleep/wake-dependent expression [3], [17], circadian expression [3], [18], Clk-target genes [19], kinases/phosphatases, GTPase-activating proteins, guanine nucleotide exchange factors, G-protein coupled receptors, ion channels, and synaptic components (Flybase). Of the initial 2203 genes, we were able to analyze 1015 with potential loss-of-function alleles (Figure 1A). To compensate for potential differences in genetic background, previously existing alleles were tested over a deficiency (Df) from the isogenic DrosDel collection [20], and allele/Df combinations shifted at least 2 standard deviations from the population mean for sleep duration and/or average sleep bout length (ABL) in males in 3 separate behavior experiments were considered hits. In the case of X-linked genes, allele virgins were crossed to X-linked deficiency males and the F1 allele/Y males were tested.
Fig. 1. A reverse-genetics screen identifies novel sleep genes. 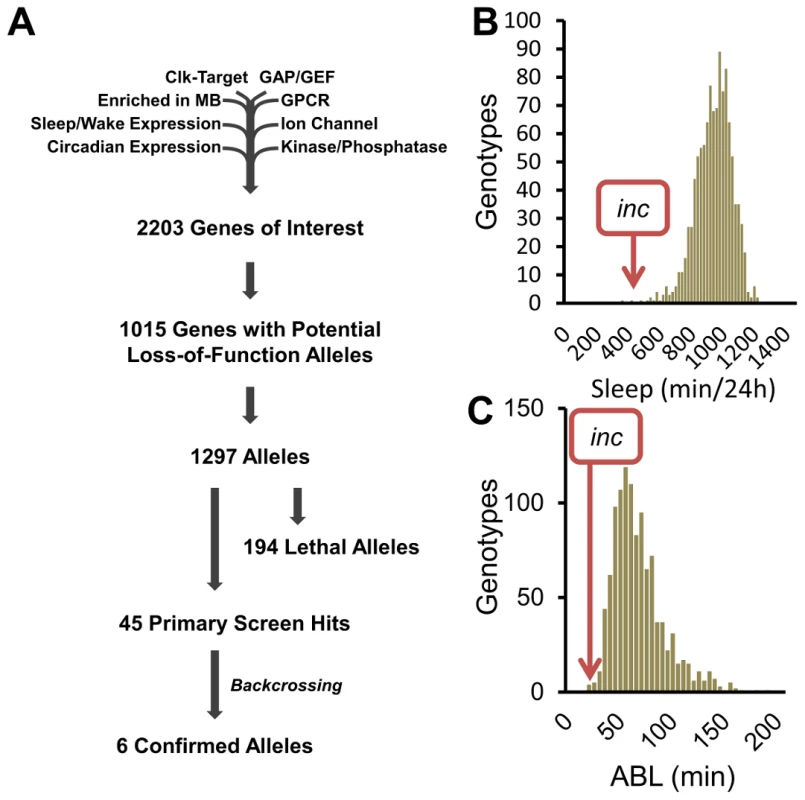
(A) Flow chart shows gene selection at each stage of the screen. (B) The sleep duration and (C) average sleep bout length (ABL) distributions from the reverse-genetics screen. At the conclusion of the primary screen we identified 45 alleles with reproducible sleep duration or bout length phenotypes (Figure 1A–1C). To determine the influence of genetic background on these phenotypes we backcrossed the 45 allele hits for 5 generations into the isogenic iso31 background developed by DrosDel [20]. Surprisingly, despite outcrossing the alleles to isogenic Df lines in the primary screen, only 6 of the hits retained their sleep phenotypes after backcrossing (Figure 1A). For example, in the primary screen we identified the following insertion alleles as having a striking effect on sleep behavior: (1) mXrDG17503 exhibited increased sleep duration, (2) CG9135f03307 had increased ABL, and (3) RhoGDIEY02738 resulted in reduced sleep (Figure S1A–S1C). However, after backcrossing into the iso31 background the sleep phenotypes are no longer observable (Figure S1A–S1C). To distinguish between a potential suppressor in the iso31 background and a flanking sleep mutant in the original RhoGDIEY02738 background, we analyzed sleep in precise excisions of the EY02738 transposon. Importantly, we found that the RhoGDIEY02738 short-sleep phenotype persists after precise excision of the P-element, suggesting that a distinct mutation in this background is responsible for the phenotype. Taken together, these observations highlight the important modulatory effect genetic background has on sleep. Furthermore, these results make clear that simply outcrossing an allele to a deficiency line is insufficient to rule out genetic background as a primary cause of phenotype. Importantly, these results do not exclude a role for sleep regulation for the 39 primary screen hits that do not retain a sleep phenotype after backcrossing, as either the iso31 or the original background may have a modifier that enhances or suppresses the sleep phenotype. Future work will be required to confirm a sleep regulatory role for these alleles.
The homeostatic regulation of sleep is disrupted in insomniac mutants
Despite the influence of genetic background, we were able to identify one allele with a robust and reproducible sleep reduction even after backcrossing: f00285, a piggyBac insertion in the 5′ untranslated region of insomniac (inc, CG32810) (Figure 1B–1C, Figure S2A), a gene selected for its BTB protein-protein interaction domain and recently linked to sleep regulation [21]. To complement this allele we created a second allele by knocking in a miniwhite gene just upstream of the inc stop codon (Figure S2A, incmw). After backcrossing, expression of inc transcript in incf00285 flies was nearly undetectable (Figure 2A); furthermore, although we do not expect incmw to affect transcript levels given the insertion location, we observed that INC protein was undetectable in both incf00285 and incmw (Figure 2B) indicating that the insertions strongly disrupt inc function. We found that these backcrossed inc alleles sleep greater than 600 minutes less than their isogenic control flies (Figure 2C–2D).
Fig. 2. Sleep is reduced throughout the day in inc mutants. 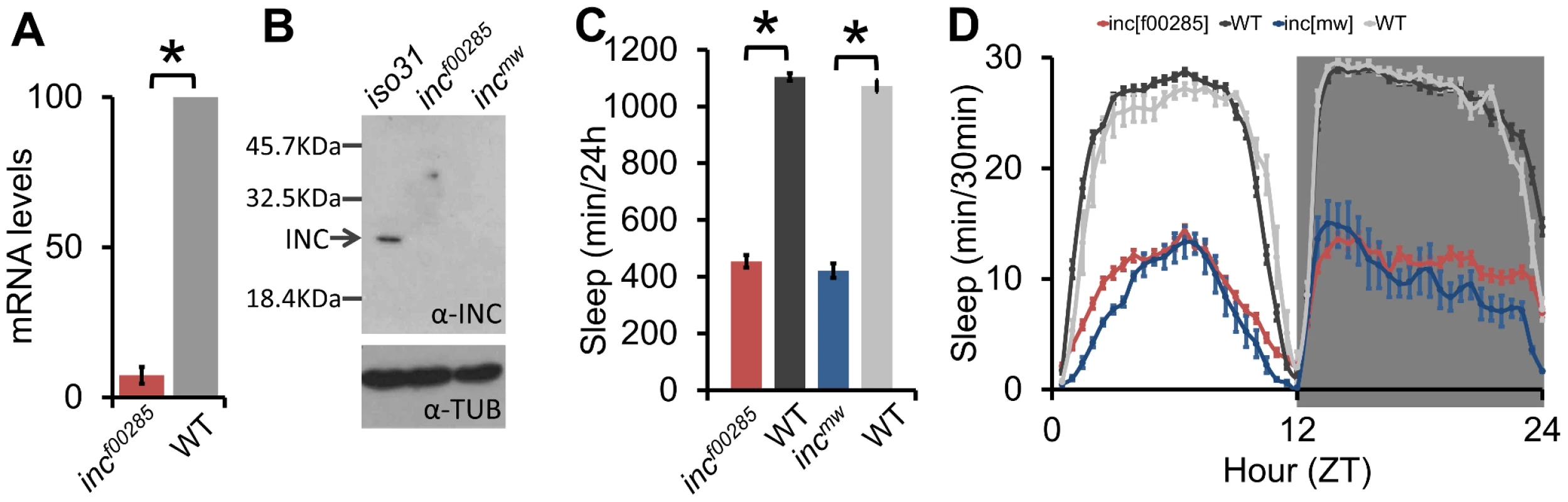
(A) inc mRNA transcript levels are reduced in incf00285 (n = 4×20 male heads/genotype). (B) Western blot shows INC protein is detectable in wild type (iso31), but not incf00285 or incmw male heads (n = 30 male heads/genotype). (C) Sleep duration in incf00285 (red, n = 44 males), incf00285 isogenic control (WT, dark grey, n = 47 males), incmw (blue, n = 72 males), and incmw isogenic control (WT, light grey, n = 32 males). (D) Sleep duration per 30 min bins. Data is shown as 30 min periods averaged from 4 days 12 h∶12 h light∶dark (ZT0 is lights-on, ZT12 is lights-off) in incf00285 (red, n = 88 males), incf00285 isogenic control (WT, dark grey, n = 47 males), incmw (blue, n = 72 males), and incmw isogenic control (WT, light grey, n = 32 males), showing that inc mutant flies exhibit reduced sleep throughout a 24 h day and anticipatory locomotor activity before lights-on and –off. Error bars are SEM. * p<0.005 with Student's t test. To further show the sleep relevant phenotypes were due to a disruption of inc, we demonstrated that sleep duration and bout length phenotypes could be rescued with Y-linked genomic duplications that included the inc genomic region, but not with a Y-linked duplication from the same collection that did not contain inc (Figure S2A–S2D). Furthermore, we found that a deletion that removes inc, as well as inc transheterozygotes, failed to complement the recessive inc phenotype (Figure S3). inc mutants displayed reduced sleep during both light and dark periods with increases in locomotor activity; nonetheless, their sleep levels dropped in anticipation of light-dark and dark-light transitions, consistent with intact circadian clock function (Figure 2D). These effects on sleep duration are comparable to those observed for the Shmns allele [15] in the iso31 background under our conditions (data not shown) and represent one of the strongest sleep phenotypes thus far observed in Drosophila or any animal model.
The sleep reduction is associated with specific changes in sleep architecture. First, inc flies displayed a decrease in sleep bout length (Figure 3A) that was accompanied by an increase in sleep bout number (Figure 3B), suggesting that flies were repeatedly attempting to initiate sleep but were unable to maintain it. Flies, like humans, typically fall asleep rapidly after the lights turn-off. However, inc files exhibited an increased latency to sleep after lights-off (Figure 3C). Despite the dramatic reduction in sleep levels, inc flies were not hyperactive, instead displaying a modest reduction in activity during wakefulness, suggesting a primary effect on sleep rather than activity (Figure 3D).
Fig. 3. Sleep architecture is altered in inc mutants. 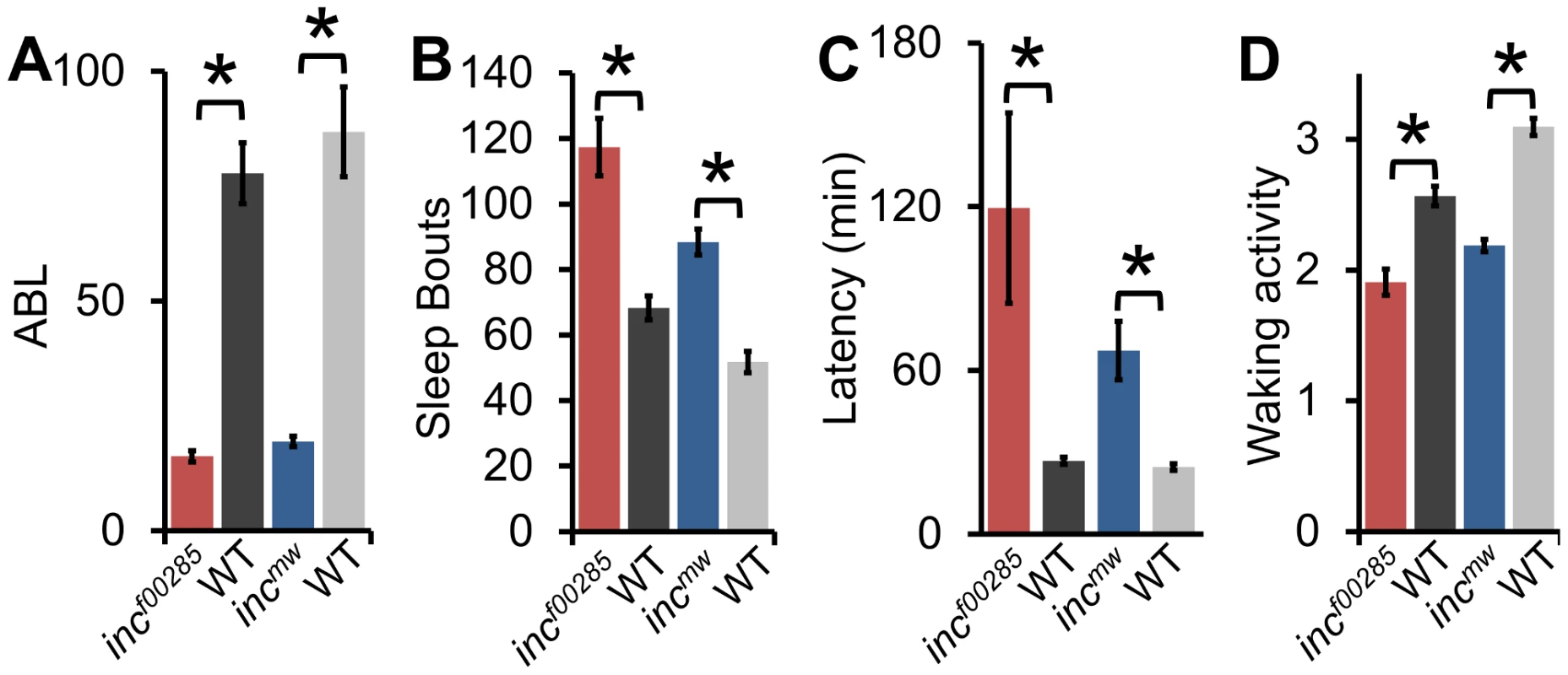
(A) Average sleep bout length (ABL), (B) total number of sleep bouts, (C) latency to sleep after lights-off, (D) and waking activity in incf00285 (red, n = 44 males), incf00285 isogenic control (WT, dark grey, n = 47 males), incmw (blue, n = 72 males), and incmw isogenic control (WT, light grey, n = 32 males). Error bars are SEM. * p<0.005 with Student's t test for all data except sleep bout length. Sleep bout length is not normally distributed; therefore, Mann-Whitney U Test was used. Although sleep duration and consolidation are important behavioral characteristics for determining the role a gene plays in sleep homeostasis, the gold standard is to analyze the homeostatic response to sleep loss, thus determining the role a gene plays in the sleep homeostat. Interestingly, whereas isogenic control flies had a significant increase in sleep after 12 hours of sleep deprivation by mechanical stimulation, we found that inc flies did not exhibit a detectable sleep rebound (Figure 4). Importantly, increasing the length of deprivation to 24 hours, which equalized the magnitude sleep loss in inc mutant flies to iso31 12 h sleep deprivation, still did not reveal a significant rebound in inc mutants (Figure S4).
Fig. 4. Flies lacking inc exhibit reduced behavioral sleep homeostasis. 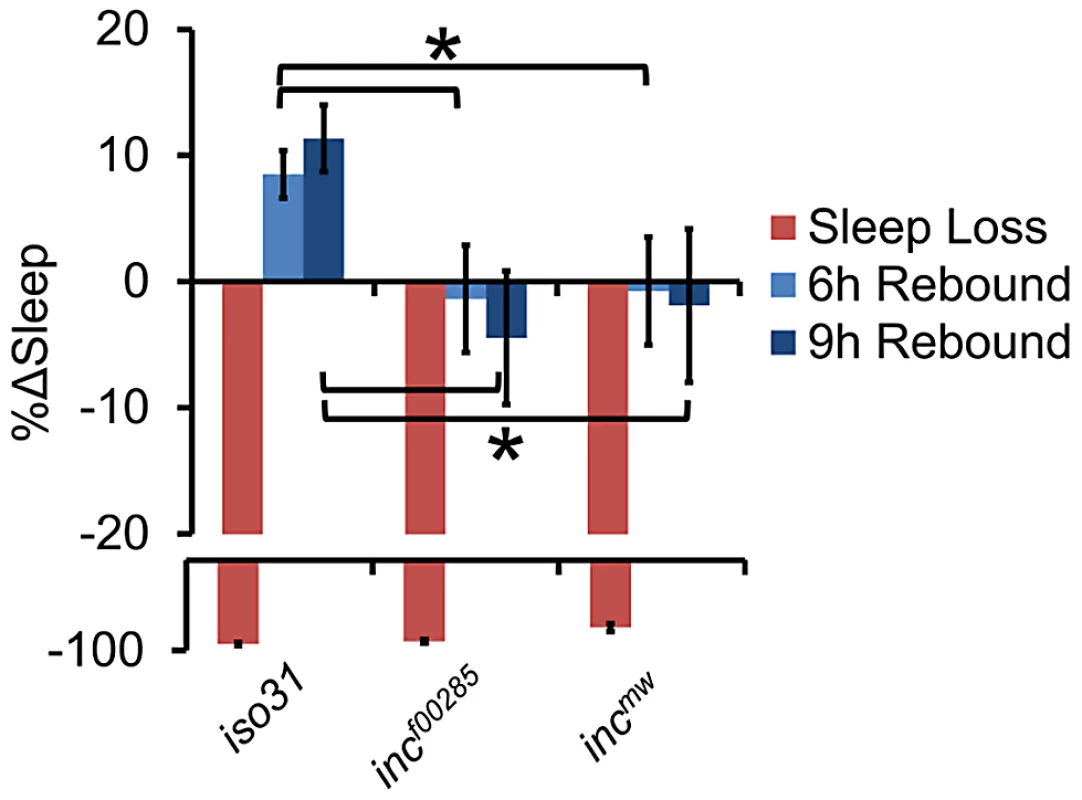
The graph shows the percent change in sleep (%Δsleep) after 12 h of mechanical sleep deprivation (red), 6 h of sleep recovery (light blue), and 9 h of sleep recovery (dark blue) in incf00285 (n = 45 females), incmw (n = 68 females), and iso31 (n = 41 females) showing that the homeostatic response to sleep loss is disrupted in inc mutants. Error bars are SEM. * p<0.05 with Student's t test. Cholinergic neurons likely mediate the effects of INC on sleep homeostasis
To test whether inc is required in neurons for proper sleep/wake regulation, we employed the Gal4/UAS system to knock down inc in all post-mitotic neurons with elav-Gal4. Using two distinct UAS-inc-RNAi transgenes targeting different parts of the inc transcript (Figure S2A) in concert with UAS-dcr2 to enhance RNAi effects [22] we found that RNAi phenocopies the inc mutant sleep duration and consolidation phenotypes (Figure 5A–5B). We were unable to identify more-refined Gal4 drivers that phenocopy the inc mutant sleep phenotype in combination with UAS-inc-RNAi alone.
Fig. 5. inc functional neuroanatomy. 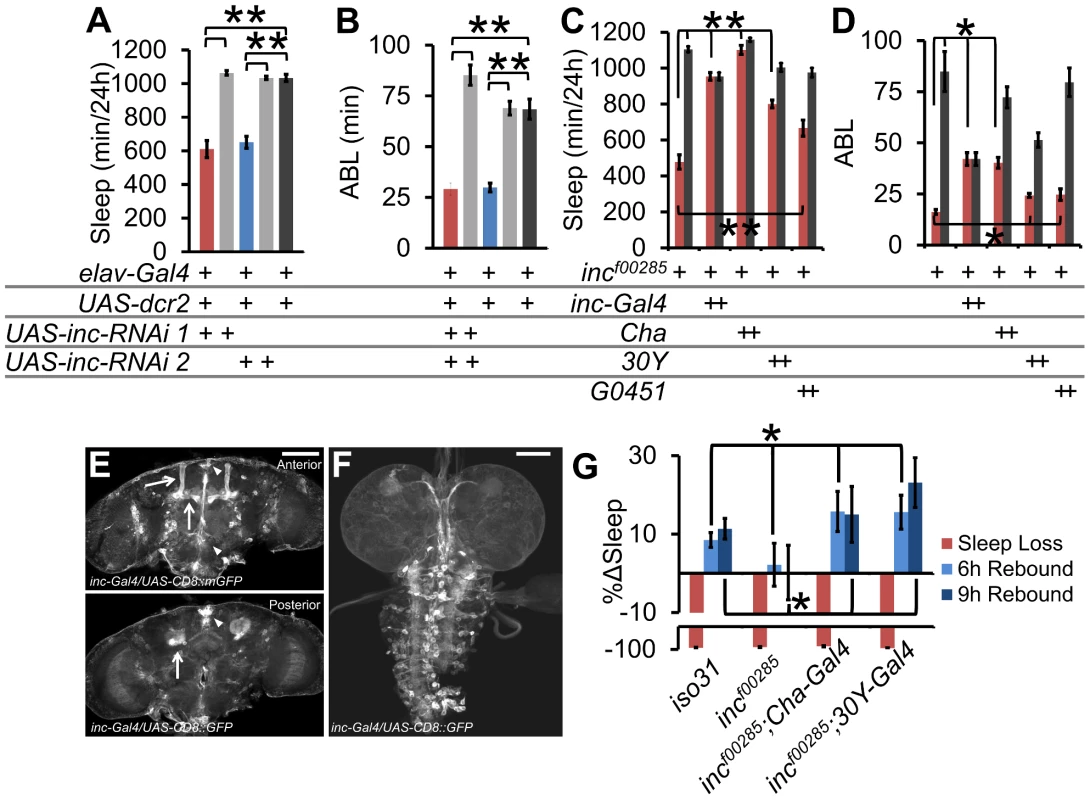
(A) Sleep duration and (B) average sleep bout length (ABL) in pan-neuronal knockdown of inc with 2 distinct UAS-inc-RNAi lines. (C) Sleep duration and (D) ABL in rescue of incf00285 (red) or wild type isogenic control (grey) with inc-Gal4, Cha-Gal4, 30Y-Gal4, and G0451-Gal4 (incf00285 is a piggybac insertion that contains a UAS element, n>30 male flies for all conditions). (E) inc-Gal4 driving UAS-CD8::GFP in adult brain and (F) L3 wandering larval brain. Arrows identify the mushroom bodies (MB), triangle identifies the pars intercerebralis (PI). (G) Rescue of homeostatic sleep regulation in incf00285 (n = 45 females) by Cha-Gal4 (n = 34 females), and 30Y-Gal4 (n = 36 females), as compared to iso31 (n = 41 females). Graph shows the percent change in sleep (%Δsleep) after 12 h of mechanical sleep deprivation (red), 6 h of sleep recovery (light blue), and 9 h of sleep recovery (dark blue). Error bars are SEM. ** p<<0.001, * p<0.05 with Student's t test. Scale bars are 100 µm. Next we took advantage of the UAS element present in the piggyBac element in incf00285 (Figure S2A) to rescue inc phenotypes. We screened Gal4s with expression patterns in known sleep-regulatory regions of the brain, including the mushroom bodies (MB) (247, 30Y, c309, G0451, c305a), the pars intercerebralis (PI; 50Y, c767, dilp2), the ellipsoid bodies (EB; c547, c305a), circadian cells (pdf, tim), glia (repo), and a number of functional neuronal groups, such as dopaminergic neurons (Figure S5). In addition, we generated and tested flies in which the putative inc promoter (−2550…+340 bp relative to the transcription start site) drives Gal4 expression. We find that both the cholinergic driver Cha-Gal4 and one of the inc-Gal4 lines provided rescue of the sleep duration phenotypes (Figure 5C–5D, Figure S5). Notably, the SH regulator sleepless (sss) also functions in Cha-Gal4 neurons to regulate sleep [5]. Furthermore, the rescuing inc-Gal4 line drove expression in known sleep regulatory loci including the MBs [23], [24], PI [11], [25], and fan-shaped body [26] as well as in the larval ventral nerve cord (Figure 5E–5F). Consistent with this pattern, we observed that 3 Gal4 drivers that overlap in the MB gave partial rescue (Figure 5C–5D, Figures S5 and S6; 30Y, c309, G0451); however, the more highly restricted MB driver 247-Gal4 did not rescue suggesting that additional neural loci and/or broader MB expression may be required.
We next sought to determine if Gal4s that rescue baseline sleep also rescue sleep homeostasis in inc mutants. We focused on Cha-Gal4 and 30Y-Gal4 given their robust rescue and functional or regional expression specificity (Figure S6). We found that expression of inc with either Cha-Gal4 or 30Y-Gal4 was sufficient to rescue sleep homeostasis defects (Figure 5G).
CUL3 interacts with INC and regulates sleep homeostasis
The highly conserved mammalian homolog of INC, KCTD5, physically interacts with the E3 ubiquitin ligase CULLIN-3 (CUL3) and ubiquitin, consistent with a role as a substrate recognition adaptor for targeting ubiquitin-dependent degradation [16], [27]. In agreement with similar studies in Drosophila [21], we verified these interactions by co-immunoprecipitation of epitope-tagged CUL3 and INC proteins in S2 cells (Figure 6A). Furthermore, we found that INC could physically interact with itself, similar to findings reported for the mammalian homologue [16] (Figure 6A). To test if inc regulates sleep through Cul3, we analyzed pan-neuronal RNAi knockdown of Cul3 (Cul3 is an essential gene, and therefore there were no viable loss-of-function alleles to analyze). We found that 2 distinct insertions of an RNAi construct against Cul3, when driven in post-mitotic neurons, phenocopied the inc mutant phenotype, exhibiting reduced, poorly consolidated sleep, and increased latency to sleep. Furthermore, we found that the baseline sleep phenotypes could be partially rescued by co-expression of wild-type Cul3 using UAS-Cul3 (Figure 6B–6D, Figure S7A–S7C). Importantly, we observed reduced Cul3 transcript levels with pan-neuronal RNAi knockdown (Figure 6E, Figure S7D).
Fig. 6. The INC-interactor CUL3 is necessary for proper sleep behavior. 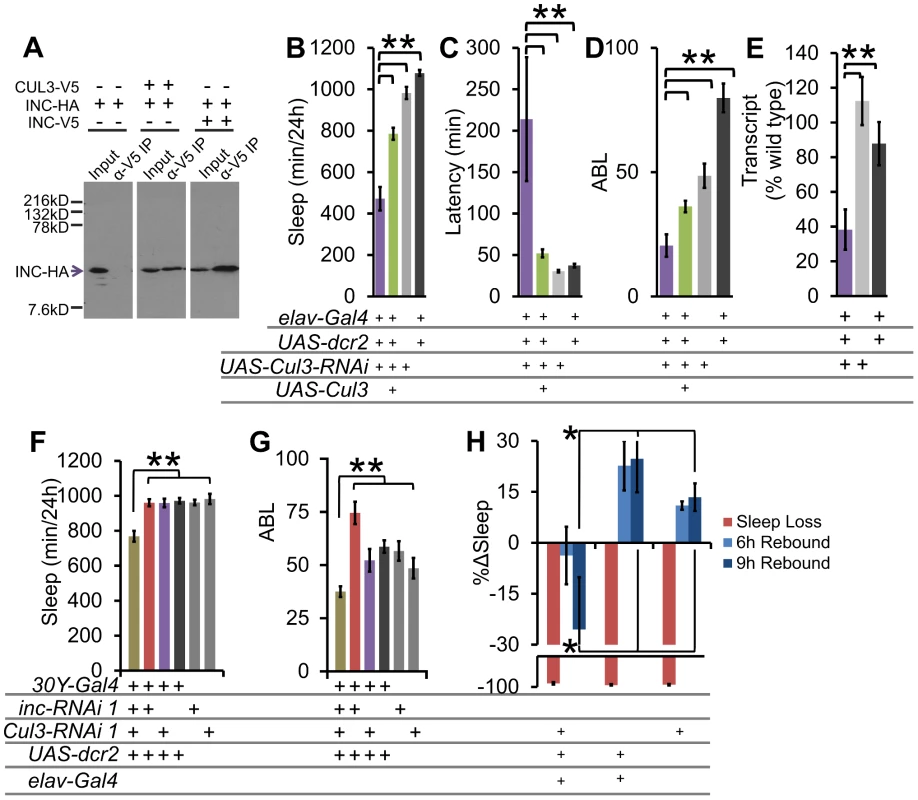
(A) Western blot shows results of an α-V5 immunoprecipitation from S2 cell supernatant transfected with Cul3-V5, inc-HA, and/or inc-V5, and blotted with α-HA to detect INC-HA. INC interacts with both CUL3 and itself under these conditions. Sleep duration (B), latency to sleep after lights-off (C), and average sleep bout length (ABL) (D) with Cul3-RNAi knockdown and UAS-Cul3 rescue in post-mitotic neurons in males (n>26 male flies for all conditions). (E) Pan-neuronal knockdown with Cul3-RNAi results in reduced Cul3 transcript levels as compared with heterozygous controls (n = 3×20 male heads/genotype). (F) Sleep duration and (G) ABL in 30Y-Gal4 knockdown of inc and Cul3. (H) Graph shows the percent change in sleep (%Δsleep) after 12 h of mechanical sleep deprivation (red), 6 h of sleep recovery (light blue), and 9 h of sleep recovery (dark blue) in elav-Gal4;;UAS-dcr2/Cul3-RNAi (n = 76 females), elav-Gal4;;UAS-dcr2/+ (n = 31 females), and Cul3-RNAi/+ (n = 54 females) showing that the homeostatic response to sleep loss is disrupted in flies lacking Cul3. Error bars are SEM. ** p<0.01, * p<0.05 with Student's t test. To examine genetic interactions between Cul3 and inc, we employed the 30Y-Gal4 driver in combination with RNAi. We found that whereas 30Y-Gal4 driven RNAi knockdown of either inc or Cul3 alone was insufficient to affect sleep, knockdown of both simultaneously results in a significant synthetic decrease in sleep duration and consolidation (Figure 6F–6G), consistent with the hypothesis that each partially impairs the same pathway.
To determine if Cul3-RNAi displays a similar sleep homeostasis phenotype as inc, we examined the behavioral response of Cul3-RNAi flies to mechanically induced sleep deprivation. We did not detect any significant rebound in these Cul3-RNAi knockdown flies (Figure 6H). These results further support a role for the INC/CUL3 complex in sleep homeostasis.
Developmental expression of inc and Cul3 may contribute to adult sleep behavior
With few exceptions, and largely limited to overexpression [11], [14], prior discoveries of sleep mutants have not typically been accompanied by a direct assay to establish if effects are due to their function in development or in adulthood. To determine if inc and Cul3 expression must be initiated developmentally or acutely in the adult to regulate sleep we employed the RU486-inducible pan-neuronal Gal4 driver elavGeneSwitch [28]. To drive adult expression only, adult flies were placed on RU486-laced behavior food starting 48 h prior to monitoring sleep behavior. To initiate developmental expression, parent flies were mated on RU486-laced food, after which eclosed F1 progeny were removed to drug-free food for 5 days prior to monitoring behavior (Figure 7A). We find that incf00285;elavGeneSwitch flies fed RU486 - or vehicle-laced food after eclosion are indistinguishable for sleep (Figure 7B), whereas flies exposed to RU486, but not vehicle alone, during development exhibit rescue of sleep behavior (Figure 7C). Likewise, the short-sleep phenotypes observed with inc- and Cul3-RNAi knockdown were only present when driven during development, but not in adult flies (Figure 7D–7G). We next tested the effectiveness of elavGeneSwitch-driven rescue of inc by asking (1) is INC protein detectable in adult heads after adult only rescue and (2) is cessation of RU486 exposure for 5 d sufficient to remove residual INC protein? We found that incf00285;elavGeneSwitch flies fed RU486-laced food as adults exhibit wild-type INC protein levels in their heads (Figure 7H). However, incf00285;elavGeneSwitch flies exposed to RU486-laced food prior to eclosion retained some INC even after 5 d on RU486-free food (Figure 7H). These results suggest that INC has a long half-life, and raise the possibility that developmental transcription may contribute to adult protein levels. Taken together, these data demonstrate that inc and Cul3 expression during development in post-mitotic neurons may contribute to adult sleep.
Fig. 7. Developmental expression of inc and Cul3 contributes to adult sleep behavior. 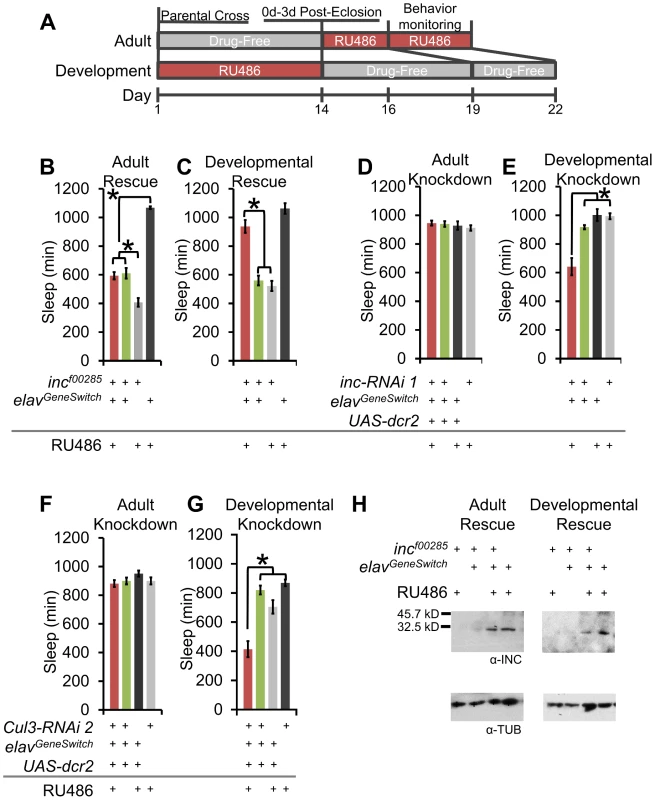
(A) The schematic shows the time-course for administering RU486 for adult and developmental expression using elavGeneSwitch. (B–C) Graphs show sleep duration of incf00285 flies after pan-neuronal rescue starting 48 h prior to behavior (Adult Rescue) or from conception to eclosion ending 5 d before behavior (Developmental Rescue) with elavGeneSwitch-Gal4 (n>37 male flies for all conditions). (D–E) Graphs show sleep duration after pan-neuronal RNAi knockdown of inc starting 48 h prior to behavior (Adult Knockdown) or from conception to eclosion ending 5 d before behavior (Developmental Knockdown) with elavGeneSwitch-Gal4 (n>31 male flies for all conditions). (F–G) Graphs show sleep duration after pan-neuronal RNAi knockdown of Cul3 starting 48 h prior to behavior (Adult Knockdown) or from conception to eclosion ending 5 d before behavior (Developmental Knockdown) with elavGeneSwitch-Gal4 (n>36 male flies for all conditions). (H) Western blots show head INC levels for elavGeneSwitch driven rescue of incf00285 after adult RU486 exposure (3 d on RU486 or vehicle-laced after eclosion) or developmental RU486 exposure (5 d on drug-free food after eclosion) (INC levels with Adult v Dev rescue). α-TUBULIN (α-TUB) was used as a load control. RU486-induced elavGeneSwitch expression is stronger during development than during adult, to counteract this, developmental expression used 50 µM RU486 and adult expression used 500 µM. Error bars are SEM. * p<0.001 with Student's t test. If inc and Cul3 have developmental functions, we would predict morphological phenotypes, especially in sleep-regulatory regions. Whereas we observed no gross defects in the PI, clock neurons, or dopaminergic neurons (Figure S8), we did observe a low penetrant stochastic branching defect in the MB (Figure S9A–S9C). We labeled the MB with α-FASII immunofluorescence and 247dsRed to visualize the α/β, α′/β′, and γ lobes. Whereas all MB lobes were observable in 15 of 15 wild-type brains, a subset of inc mutant flies lacked either an α - or β-lobe (incf00285: 10/50; incmw: 12/38 brains; Figure S9B–S9C). These observed defects were non-symmetrical, such that only a single lobe was missing from a brain (i.e., the same lobe was present in the other hemisphere). Notably, Cul3 has also been reported to play a role in MB branching [29]. We found that Shmns and DATfmn flies had normal MB morphology, arguing that reduced sleep or increased dopaminergic signaling alone is not the underlying cause of the morphological phenotype (Figure S9D–S9E). To determine if it was possible for these defects to be the underlying cause of the inc sleep phenotype we compared the morphological penetrance to the sleep behavior penetrance. Whereas 20–32% of inc mutants exhibited the MB morphological defect, >90% of inc mutant flies slept less than the shortest sleeping iso31 fly (Figure S9F), and almost 75% of inc mutant flies exhibited less consolidated sleep than the most extreme iso31 example (Figure S9G). Based on these findings the MB branching defect cannot be the sole cause of the sleep phenotypes.
inc and Cul3 regulate arousability
To further elucidate the underlying mechanism of the inc/Cul3 phenotype, we next asked whether inc flies had difficulty maintaining and initiating sleep because they were hyper-arousable. To test arousability, we used a mechanical apparatus to rotate the DAM monitors, and therefore the glass capillary tubes that housed the flies, off horizontal at ZT16 and examined the waking response of flies that were asleep prior to the stimulus (Figure 8A, see methods). Controlling for flies that spontaneously awoke in the absence of a stimulus, we found that whereas roughly 25% of sleeping wild-type flies woke up in response to this rotational stimulus, >85% of incf00285 and incmw flies woke up, arguing that inc mutants indeed are hyper-arousable (Figure 8B). In addition, we also examined arousability in flies expressing Cul3-RNAi pan-neuronally and observed similar results with >80% of flies responding to the stimulus (Figure 8C). This makes inc mutants and Cul3-RNAi flies distinct from Shmns and CnA knockdown flies, which exhibit short-sleep phenotypes in the absence of hyper-arousability [15], [30].
Fig. 8. Flies lacking inc or Cul3 are hyper-arousable. 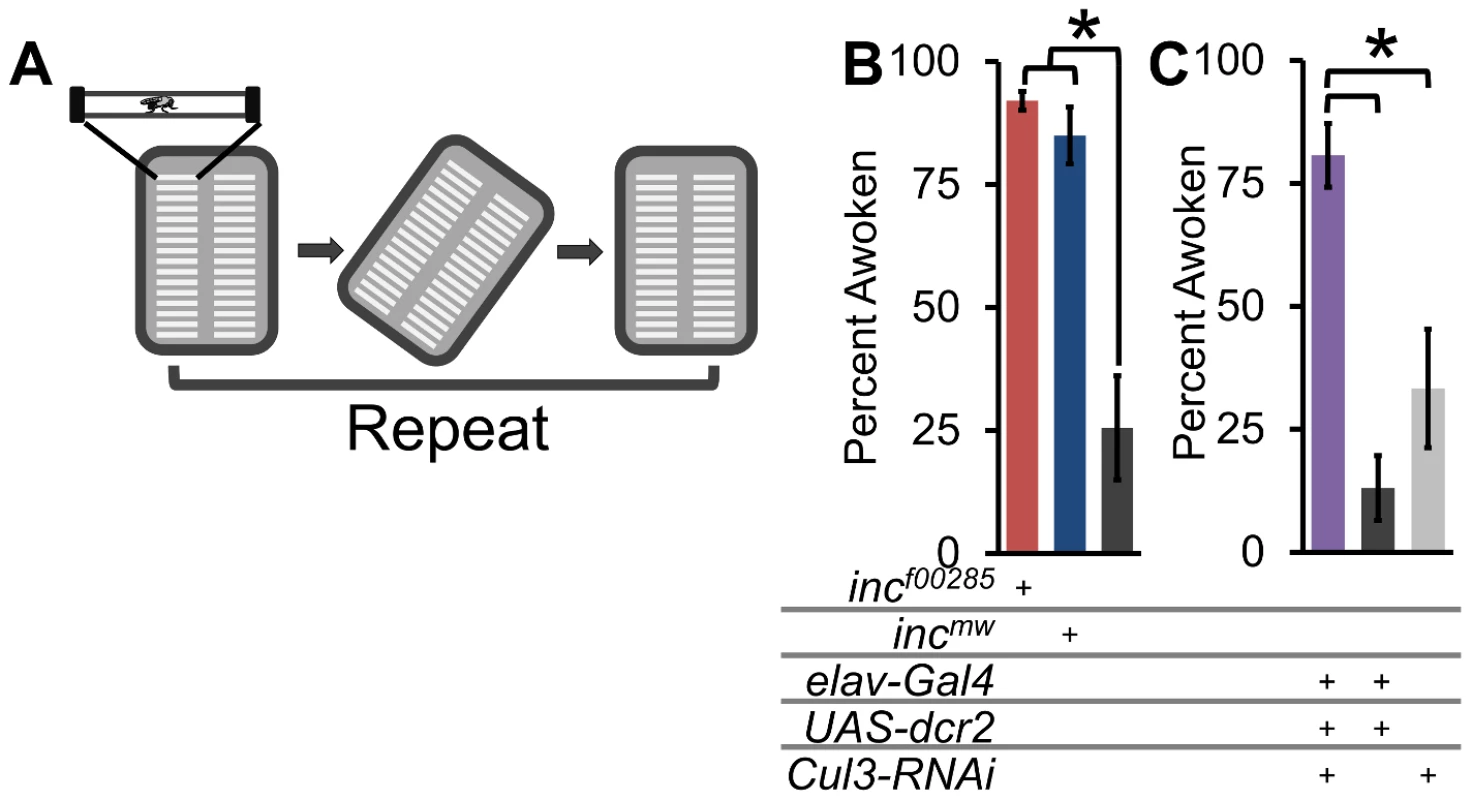
(A) Schematic shows arousal threshold paradigm. At ZT16, flies were rotated off horizontal and back, and the number of sleeping flies to wake up within 5 min were counted (see methods). (B) Graph shows arousal threshold (percent flies awoken by a rotational stimulus 2 h after lights-off) in incf00285 (red), incmw (dark blue), and iso31 (grey). (C) Graph shows arousal threshold in elav-Gal4;Cul3-RNAi (purple), elav-Gal4 heterozygotes (dark grey), and Cul3-RNAi heterozygotes (light grey). n>55 sleeping male flies for all genotypes. Error bars are SEM. * p<0.01. inc effects on sleep require dopamine
Dopaminergic signaling is a key regulator of arousal in both flies and mammals [7]–[10], [31], [32]. To determine if inc functions in a dopaminergic arousal pathway we first took a genetic approach and asked if the sleep duration phenotype in inc mutants was additive with the dopamine transporter mutant DATfmn. We found that incf00285;DATfmn and incmw;DATfmn flies did not sleep significantly less than single mutants (Figure 9A), in support of our hypothesis that inc and DAT operate in the same arousal pathway.
Fig. 9. inc effects on sleep require a dopamine sleep-regulatory pathway. 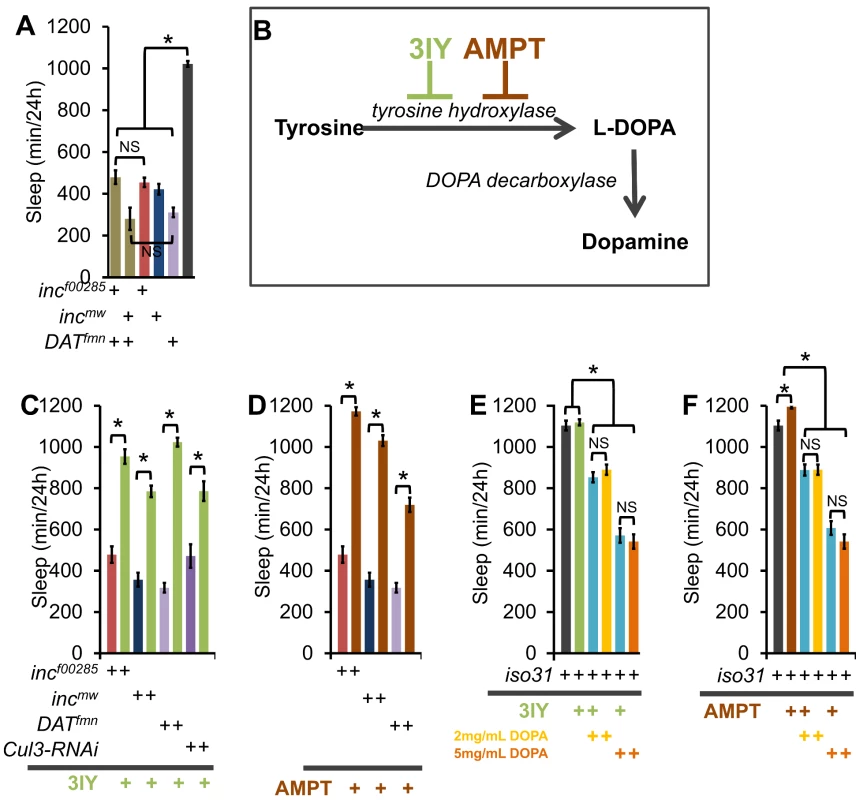
(A) Graph shows sleep duration in incf00285;DATfmn (tan), incmw;DATfmn (tan), incf00285 (red), incmw (dark blue), DATfmn (light purple), and iso31 (grey) males showing a non-additive effect on sleep duration between inc and DATfmn (n>38 males for all genotypes). (B) Schematic shows the dopamine synthesis pathway. 3-iodo-tyrosine (3IY) and α-methyl-p-tyrosine methyl ester (AMPT) are inhibitors of tyrosine hydroxylase (TH). (C–D) Graphs show sleep duration in incf00285 (red), incmw (dark blue), DATfmn (light purple), and elav-Gal4;UAS-Cul3-RNAi/UAS-dcr2 (Cul3-RNAi, dark purple) after consumption of food laced with TH antagonists 2 mg/mL 3IY (C; green) or 1 mg/mL AMPT (D; brown; n≥31 males for all conditions). (E–F) Graphs show effects of 2 mg/mL 3IY (green), 1 mg/mL AMPT (brown), 2 mg/mL L-DOPA (yellow), and 5 mg/mL L-DOPA (orange) on sleep duration in iso31 flies. L-DOPA is the product of TH, and the precursor to dopamine. A non-significant effect of 3IY or AMPT on the reduction in sleep caused by L-DOPA (blue) demonstrates pathway specificity for these TH inhibitors (n≥31 males for all conditions). Error bars are SEM. * p<0.001 with Student's t test. To test the specificity of inc function we took a pharmacological approach to modulate two known arousal pathways. Flies were fed (1) L-DOPA to increase dopaminergic arousal or (2) carbamazepine (CBZ), an antagonist of the GABA receptor Rdl previously demonstrated to increase arousal by modulation of the PDF cells [33]–[35]. We found that, whereas wild-type iso31 and inc mutants exhibit robust reductions in sleep with CBZ (Figure S10A), inc mutants were resistant to the sleep effects of L-DOPA (Figure S10B) arguing that the inc effects are specific to a dopaminergic, but not the Rdl, arousal pathway. The CBZ data further indicate that there is not a “floor” effect preventing further reductions in inc sleep.
We next pharmacologically examined the dopamine-dependence of inc and Cul3 sleep phenotypes. Flies were fed one of two inhibitors of the rate limiting step in dopamine synthesis, tyrosine hydroxylase (TH): (1) 3IY (3-iodo-tyrosine) [36], or (2) AMPT (α-methyl-p-tyrosine methyl ester, Figure 9B) [37]. Inhibition of dopamine synthesis with either drug suppressed the inc mutant and Cul3-RNAi short-sleep phenotypes, as well as that of DATfmn, which is thought to increase arousal through increased dopaminergic signaling [9] (Figure 9C–9D). One possibility is that 3IY and AMPT act non-specifically to increase sleep; therefore, we next sought to determine the specificity of these drugs by restoring L-DOPA, the enzymatic product of TH (Figure 9B). We found that wild-type iso31 flies fed L-DOPA alone exhibited a dose-dependent decrease in sleep; furthermore, 3IY and AMPT did not suppress the L-DOPA effect, as expected if they operate upstream of L-DOPA (Figure 9E–9F). Importantly, we also observed decreased head dopamine levels after 3IY consumption, and increased levels after L-DOPA consumption, biochemically verifying drug mechanism/efficacy (Figure S11). Interestingly, inc phenotypes were not rescued by expression in dopaminergic neurons using a tyrosine hydroxylase-Gal4 (TH-Gal4) or a Dopa decarboxylase-Gal4 (Ddc-Gal4) (Figure S5); furthermore, TH protein levels were not altered in inc mutant brains (Figure S11C), suggesting that inc does not control sleep via its function in dopaminergic neurons.
Given that Cul3 and inc affect sleep homeostasis and dopaminergic arousal pathways, we next sought to determine if these phenotypes are linked. We found that wild-type flies fed 3IY displayed intact sleep homeostasis (Figure 10). On the other hand, 3IY consumption restored sleep homeostasis in Cul3-RNAi, indicating the dopamine dependence of the homeostatic defect in these flies. We also found that 3IY could restore rebound sleep in incf00285 but not in incmw (Figure 10). While the molecular lesion differs between the two inc alleles, we are not aware of how this might explain the different drug responses between these two mutants. Although we did test a 5-fold higher concentration of 3IY than that sufficient to rescue Cul3-RNAi and incf00285 and found that it was still insufficient to rescue incmw (Figure 10), we cannot rule out a trivial explanation for the negative results in incmw such as insufficient drug uptake/dopamine suppression. Nonetheless, the positive results with Cul3 and incf00285 support a model in which the homeostatic defect in these flies depends on dopamine. We are not aware of another example of pharmacological rescue of sleep rebound, at least in Drosophila. Taken as a whole, these results suggest that inc functions in a group of cholinergic neurons, as defined by Cha-Gal4 and 30Y-Gal4, and that in its absence excess dopaminergic signaling underlies the resulting sleep phenotype.
Fig. 10. 3IY rescues sleep homeostasis defects in Cul3-RNAi and incf00285 but not incmw. 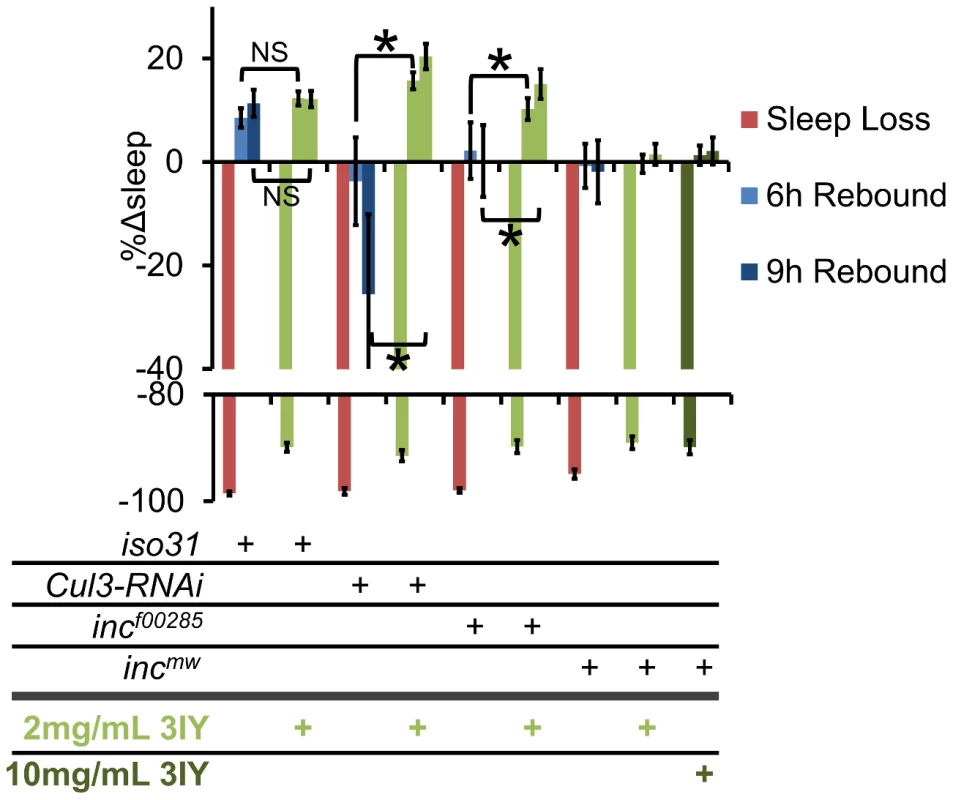
The graph shows the percent change in sleep (%Δsleep) after 12 h of mechanical sleep deprivation (red), 6 h of sleep recovery (light blue), and 9 h of sleep recovery (dark blue) in iso31, elav-Gal4;Cul3-RNAi/UAS-dcr2 (Cul3-RNAi), incf00285, and incmw with 2 mg/mL 3IY (light green), 10 mg/mL 3IY (dark green) or no 3IY. n≥41 females for all conditions. * p<0.05. Error bars are SEM. Discussion
Our detailed phenotypic analyses of inc and Cul3 reveal novel insights into the molecular basis of sleep homeostasis and dopaminergic arousal pathways. inc mutants display one of the strongest baseline and homeostatic sleep phenotypes thus far observed. Furthermore, we find that the reduced sleep phenotype depends largely on a single neurotransmitter, dopamine, establishing a transmitter basis to inc function.
Widespread modulatory effects of genetic background on sleep
Using genetic backcrossing to an isogenic (iso31) strain, we made the striking observation that the vast majority of the mutants (39/45, nearly 90%) identified in our primary genetic screen did not have a significant phenotype after backcrossing, indicating a remarkably pervasive role for genetic background in mediating sleep phenotypes in a variety of mutant strains. There are two main possibilities for how genetic background influences sleep phenotypes: (1) the tested allele indeed affects sleep; however, there are suppressors of this phenotype present in the iso31 background but absent from the original background. (2) The sleep phenotype is not due to the transposon insertion but instead is caused by one or more flanking mutations present in the original mutant background but absent from the iso31 background. Isolated examples of (1) have been observed in the case of Sh and mutants of the Sh regulatory subunit Hyperkinetic [6], [15] as well as Crc and Sema-5c effects on olfactory, startle, and sleep behavior [38] and of (2) in the discovery of DATfmn mutants in the background of a timeless mutant strain [9]. These are consistent with observations in C. elegans indicating the limitations of backcrossing for removing flanking mutations [39] and in Drosophila indicating the widespread presence of background mutations that can suppress mutant-induced behavioral phenotypes [38]. Our experience with RhoGDIEY02738 suggests that scenario 2 may be more common than previously thought (Figure S1C). Nonetheless, the sheer number of examples observed here indicates that the presence of genetic variation at sleep regulatory loci among laboratory stocks is both prevalent and perhaps even sufficiently important to mask or induce significant sleep phenotypes. Moreover, in the case of Sh a single outcross was sufficient to unmask the short sleep phenotype. Indeed, we assumed that if we outcrossed mutant alleles to a deficiency strain this would effectively remove the influence of accumulated recessive mutations that flank the allele. However, our backcrossing data indicates that this strategy did not remove those concerns, suggesting that background variants may exert dominant effects (see also [38]). Practically, our experience suggests that outcrossing to deletion stocks alone may not be sufficient to verify the function of a genetic locus in sleep. Overall, this observation has important implications for the role of genetic modifiers in sleep, the conduct and design of sleep genetic screens, and for the interpretation of sleep and other behavioral mutant phenotypes in general. While backcrossing can remove flanking genetic variants that may contribute to an observed phenotype, alone it is not sufficient to definitively establish genotype-phenotype causation.
inc and Cul3 are key regulators of sleep homeostasis in Drosophila
Despite the large modulatory effect of genetic background, we were able to observe persistent phenotypes with inc, which showed the most robust and reproducible sleep phenotypes, in particular demonstrating an important role in the homeostatic regulation of sleep. Several independent lines of evidence support the role of inc in sleep homeostasis. First, 2 inc alleles (incf00285 and incmw) were backcrossed for 5 generations into an isogenic background and retained their short sleep and suppressed sleep homeostasis phenotypes, each among the strongest observed, as compared to isogenic control lines. Second, we rescued incf00285 in 2 distinct ways: (1) with genomic duplications encompassing the gene but not those that do not include the gene, and (2) using the GAL4/UAS system, the latter rescuing both baseline and homeostatic phenotypes. Third, we demonstrated failure to complement with a deletion removing the inc genomic locus, or inc transheterozygotes. Fourth, we demonstrated that two independent RNAi lines that target two different regions of inc phenocopy the inc mutant phenotype.
In addition, we provided evidence that the INC-interacting protein, the E3 ubiquitin ligase CUL3, functions to regulate sleep levels suggesting that inc links protein turnover to sleep homeostasis. Two independent inserts of a Cul3-RNAi line that effectively suppress Cul3 mRNA levels resulted in reduced sleep, and induction of a wild-type Cul3 transgene could rescue these phenotypes. We verified the CUL3/INC interaction in S2 cells and observed synthetic genetic interactions between Cul3 and inc using RNAi, consistent with the model that they operate together to affect sleep.
A core concept in understanding sleep behavior is its homeostatic regulation, i.e., the observation that the drive to sleep reflects the duration of prior wakefulness. Sleep homeostasis typically is measured by enforcing wakefulness/depriving sleep for a defined period and assaying the increase in subsequent rebound sleep. Importantly, we demonstrated that both inc and Cul3 have robust effects on sleep homeostasis where reduced inc or Cul3 was accompanied by suppressed or absent sleep rebound under our conditions. These results suggest that inc and Cul3, and by extension, protein degradation, are important for the accumulation of sleep need during wake and/or dissipation of sleep need after deprivation.
Expression of inc and Cul3 during development in post-mitotic neurons may contribute to adult sleep
For the large majority of sleep mutants that have been described, assessment of developmental and adult contributions has not formally been addressed, raising questions regarding their precise function in sleep. Here we provided evidence that inc induction or Cul3-RNAi knockdown during development, but not exclusively during adulthood, could rescue (in the case of induction) or phenocopy (in the case of knockdown) their respective mutant/RNAi phenotypes. The Cul3 results are consistent with an established role for Cul3 in dendritic and axonal arborization, in which dendritic and axonal arborization are reduced in Cul3 mutants [29], [40]. Our data also revealed a stochastic branching defect in MB neurons in 26% of inc mutants, in which they lack a single α - or β-lobe. Based on the incomplete penetrance of this morphological defect, it cannot explain the sleep behavior phenotype; however, it may be reflective of other morphological phenotypes that are causative for behavior. Alternatively, the necessity for developmental expression of Cul3 and inc may be for the appropriate processing, maturation and/or localization of these proteins in the adult. The apparent long half-life/persistence of this protein after induction only during development is consistent with the possibility that developmentally expressed transcription is important for adult protein expression and function. Regardless, it will be of interest to examine the relative adult and developmental requirements of other sleep mutants.
inc and Cul3 function in a dopaminergic arousal pathway
We found that the reduced sleep phenotype depends largely on a single neurotransmitter, dopamine, establishing a transmitter basis to inc/Cul3 function. Dopaminergic signaling is a key regulator of sleep/wake behavior. In humans, sleep deprivation has been associated with increased brain levels of dopamine [41]. Treatment of Parkinson's disease with L-DOPA can alleviate daytime sleepiness, or in the extreme result in insomnia [42]–[45]. In Drosophila, genetic loss or pharmacological inhibition of tyrosine hydroxylase increases sleep [46]–[48]. Furthermore, flies that lack a functional copy of DopR exhibit increased sleep and general arousal defects, including reduced arousing effects of caffeine [8], [49]. Conversely, in DATfmn flies, or flies fed dopamine-enhancing methamphetamine, sleep levels are severely reduced [9], [48]. Dopamine arousal effects are modulated by light [50]. Moreover, sleep deprivation induced reductions in learning can be suppressed by enhancing dopaminergic signaling [51]. Other than dopamine receptors and DAT, members of the dopaminergic arousal pathway remain largely unknown.
We report here that inc and Cul3 function in the dopaminergic arousal pathway. First, inc mutants, Cul3-RNAi, and DATfmn all showed robust sleep duration and consolidation phenotypes. Second, all three groups were hyper-arousable to mechanical stimuli. Third, disruption of inc, Cul3, and DAT all exhibited suppressed or absent homeostatic responses to sleep deprivation. Fourth, the short-sleep phenotypes of inc and DATfmn were non-additive in double mutants. Fifth, while wild-type flies exhibited reduced sleep when fed the dopamine precursor L-DOPA, inc mutants were resistant to these effects, but not the arousing effects of the Rdl antagonist CBZ. Finally, the sleep duration phenotypes in flies with disrupted inc, Cul3, and DAT could be suppressed by pharmacologically inhibiting dopamine synthesis with 3IY or AMPT, linking short sleep to excess dopamine function. Importantly, we demonstrated that inhibition of dopamine synthesis via tyrosine hydroxylase inhibition does not affect L-DOPA-induced sleep reductions. We also observed that 3IY could restore sleep homeostasis to Cul3-RNAi. Similar 3IY effects on homeostasis were only observed in one of the two inc alleles. Nonetheless, these studies do further link dopamine signaling to sleep homeostasis. To our knowledge inc and Cul3 are the first genes that are not known dopamine receptors reported to function in the dopaminergic arousal pathway, further reinforcing the pivotal role of dopamine in sleep homeostasis.
Our data suggests Cul3/inc function to regulate dopaminergic signaling downstream of dopamine. inc phenotypes did not map to dopaminergic neurons nor were we able to identify consistent changes in global dopamine levels among Cul3-RNAi and inc mutants (data not shown). Thus, Cul3/inc may be involved in active turnover of dopamine receptors or their effectors in neurons defined by Cha-GAL4 and 30Y-GAL4. We examined double mutants of inc and a major dopamine receptor involved in arousal in Drosophila, DopR, and failed to observe suppression of inc baseline phenotypes; moreover, we found that DopR mutant flies were responsive to 3IY consumption (i.e. exhibit increased sleep; data not shown), suggesting that additional dopamine receptors function in Cul3/inc-based dopamine arousal. Drosophila has 2 other dopamine receptors and we have observed partial suppression of inc with DopR and DopR2 RNAi (data not shown), suggesting that multiple dopamine receptors may contribute to these effects. Alternatively, Cul3/inc may be important for protein turnover of other homeostatically regulated components. For example, extensive and dose-dependent changes in synaptic protein expression throughout the brain with sleep deprivation and recovery [52]–[54] may depend on Cul3/inc-dependent turnover of these proteins during sleep.
Interestingly, Cul3 has also been linked to sleep behavior via a candidate gene for Restless Leg Syndrome (RLS) and BTB gene, BTBD9 [55]. Unlike our studies, disruption of the Drosophila BTBD9 is not associated with reduced sleep, a reduced level of waking activity, nor elevated dopaminergic signaling. In addition, the phenotypes map in part to dopaminergic neurons in the case of BTBD9 rather than cholinergic neurons for inc. Thus, Cul3/inc likely represents a distinct pathway regulating sleep. Nonetheless, these studies further highlight the importance of Cul3/BTB adaptor pathways in sleep regulation in both Drosophila and humans. Future work will be required to identify the dopamine and sleep-relevant ubiquitination target(s) of inc and Cul3.
Materials and Methods
Flies
Flies were raised on cornmeal-yeast-agar food at 25°C, 12 h∶12 h Light∶Dark. Alleles and deficiencies for the reverse-genetics screen were acquired from the Drosophila Stock Centers based in Bloomington, Kyoto, Harvard, and Szeged. The deficiencies were from the isogenic collection created by DrosDel. Stocks of particular interest: inc-spanning duplications (Bloomington #6021 [Dp(1;Y)Sz280], 33872 [Dp(1;Y)BSC308], 33871 [Dp(1;Y)BSC307]), non-inc-spanning (#33875 [Dp(1;Y)BSC311]), inc-spanning deficiency (Bloomington #934 [Df(1)S39]), inc-RNAi (Vienna Drosophila RNAi Center #18226, #108816), Cul3-RNAi (National Institute of Genetics – Kyoto #11861R-1, #11861R-2), UAS-Cul3 (Bloomington #9936), Cdk4-spanning deficiency (Bloomington #9213 [Df(2R)ED3181]), mXr-spanning deficiency (Bloomington #9276 [Df(2R)ED1742]), CG9135-spanning deficiency (Bloomington #9186 [Df(2L)ED353]).
The following Gal4s were used: 50Y, c929, 5HT7, 5HT1a, ple, Ddc, DopR, DopR2, Hdc, Tdc, vGlut, repo, Cha, elav (Bloomington #30820, 25373, 23066, 27807, 27820, 8848, 7010, 24743, 19491, 25260, 9313, 26160, 7415, 6793, 8765), 247, 30Y, c309, c767, c547, c305a [24], pdf [34], tim [56], Trh [57], G0451 [58], dilp2 [59], Gad [60], elavGeneSwitch [28].
inc-Gal4 was created using 3 Kb upstream of the inc ATG start site (−2550–+340 bp relative to the transcription start site) from an inc genomic BAC (CHORI: CH223-4018). The promoter region was inserted into pPTG4, and the inc-Gal4 vector was injected into embryos by BestGene Inc.
incmw was created by cloning 4197 bp upstream of the inc stop codon (left arm) and 4052 bp downstream and including the stop codon (right arm) into pw25 (DGRC 1166), flanking a miniwhite gene. Constructs were injected into embryos by BestGene Inc. To knock in the miniwhite, the fragment with inc flanking regions was mobilized by hsFLP, digested in vivo with Sce-I (Bloomington #6934), and candidate knock-in flies were screened behaviorally and molecularly by PCR.
Conditional expression
For adult-specific expression with elavGeneSwitch, flies within 3 d after eclosion were placed on behavior food laced with 500 µM RU486 (Sigma) or vehicle alone (4% ethanol final concentration) for 48 h prior to behavior monitoring, and then monitored for 3 d in behavior. For developmental expression, parental flies were crossed on normal cornmeal-based food laced with 50 µM RU486 or vehicle alone (0.4% ethanol final concentration). Within 3 d after eclosion, F1 progeny were moved to drug-free food for 5 d prior to behavior monitoring, and then monitored for 3 d in behavior.
Genetic screen
To identify new genetic regulators of sleep we screened genes with sleep/wake-regulated expression [3], [17], genes with circadian expression patterns [3], [17]–[19], enriched in the MB [61], and genes involved in neuronal and intracellular signaling (flybase.com). We screened 1297 alleles covering 1015 genes. In each case a previously existing allele was tested over a deficiency (Df) from the isogenic DrosDel collection [20], and allele/Df combinations shifted ≥2SD from the population mean for sleep duration and/or average sleep bout length in males were considered hits. In the case of X-linked genes, allele virgins were crossed to X-linked deficiency males and the F1 allele/Y males were tested. Whenever possible we tested proven loss-of-function alleles, followed by mutations that affect in descending order of preference: exonic regions, 5′untranslated region, 3′untranslated region, intronic regions, promoter regions.
Quantitative PCR
Flies 4–8d post-eclosion were frozen on dry ice, heads were removed by dry ice cold vortexing and isolated on frozen sieves. RNA was isolated from 20 heads/sample with Trizol (Invitrogen). qPCR was performed using a QuantiTect SYBR Green PCR Kit (Qiagen).
Behavior
Sleep behavior was analyzed using the Drosophila Activity Monitor system (Trikinetics), and processed with a custom written Excel macro [62]. Flies 2–5d post-eclosion were individually loaded into 5×65 mm glass capillary tubes with a 5% sucrose 2% agar food source and analyzed for 5 d 25°C 12 h∶12 h Light∶Dark. Sleep was defined as ≥5 min inactivity (zero infrared beam crossings).
To determine lifespan, flies were maintained in DAM monitors as described above and transferred to fresh behavior tubes every 7 d.
Sleep deprivation
Sleep deprivation was performed as described previously [24]. Briefly, activity monitors were placed in an apparatus that rotates and jostles the flies at varying intervals. Flies age-matched within 24 h were loaded into behavior 2 d after eclosion and allowed at least 36 h to acclimate followed by 24 h without sleep deprivation to determine 24 h baseline sleep. For behavioral analyses flies were deprived ZT11–ZT23 for 12 h deprivation and ZT0-ZT0 for 24 h deprivation. Non-deprived controls were handled similarly to deprived flies, in a separate incubator from the sleep deprivation apparatus. We confirmed behaviorally that the flies lost ≥90% of their sleep with this protocol. To determine Δsleep, baseline sleep was subtracted from sleep obtained during the recovery period for individual flies from both sleep-deprived and non-deprived populations then non-deprived Δsleep was subtracted from sleep-deprived Δsleep.
Arousability
Flies were loaded into the same apparatus as for sleep deprivation (see above Methods section), and given 1 day to acclimate. On the second night flies were stimulated by mechanically rotating the DAM monitors 36° off horizontal and back 10 times for inc experiments and 18° 2 times for Cul3 experiments at ZT16, and the number of sleeping flies to wake up within 5 min of the stimulus was determined. We took into account the number of flies that would wake spontaneously by determining the number of flies to wake up at ZT15 : 50 and normalizing the percent awoken with the following formula:
Dopamine drug treatment
Flies were raised on cornmeal-yeast-agar food and presented with drug-labeled food throughout the behavior experiment. 3IY, AMPT, and L-DOPA were dissolved in 5% sucrose 2% agar behavior food as the sole food-source during the behavior experiment. For 3IY - and AMPT-only experiments, flies were presented with drug-laced food for 12–16 h before monitoring behavior for 2 d. For L-DOPA experiments, flies were presented with drug-laced food for 12–16 h before monitoring behavior for 1 d (after 48 h on L-DOPA the flies become unhealthy).
Immunostaining and quantification
The following antibodies were used in this study: ms-α-PDF (DSHB; 1∶100), rab-α-tyrosine hydroxylase [63] (1∶1000), ms-α-FASII (DSHB: 1∶50), gt-α-ms-Alexa488, gt-α-rab-Alexa488 (Invitrogen; 1∶500). Flies were dissected in 1% TritonX-100, 3.7% formaldehyde in PBS, fixed for 1 h post-dissection in 3.7% formaldehyde in PBS, and permeablized in 0.3% TritonX-100 in PBS overnight. Brains were incubated with all antibodies in 0.3% TritonX-100, 7% goat normal serum in PBS overnight. Wash steps were with 0.3% TritonX-100 in PBS.
Coimmunoprecipitations and Western blotting
Expression constructs were made using inc cDNA (DGRC #GM03763) and Cul3 cDNA (DGRC #LD10516). V5-tagged constructs were made by cloning the coding region into pAc5.1-V5/His (Invitrogen). HA-tagged constructs were made using a modified version of pAc5.1-V5/His, in which the coding region for V5/His was replaced with HA. Transfections were done on Drosophila S2 cells using Effectene reagent (Qiagen). 24 h after transfection, cells were lysed with T150 lysis buffer (25 mM Tris-Cl pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.5% NP-40, 1 mM PMSF), supernatant was mixed with α-V5 agarose beads (Sigma) for 1.5 h at 4°C, after washing beads were boiled with SDS loading buffer, and eluted sample was run on 10% acrylamide gels. Hybond membranes (GE Lifesciences) were blotted with α-HA (1∶2500, Roche), and developed with ECL Plus (GE Lifesciences).
For INC head western blots, 30 flies/sample were flash frozen on dry ice at ZT6, and heads were homogenized in 30 µL lysis buffer (20 mM HEPES [pH 7.5], 100 mM KCl, 10 mM EDTA, 50 mM NaCl, 0.1% Triton X-100, 10% glycerol). Samples were run on 15% acrylamide gels. Hybond membranes (GE Lifesciences) were blotted with α-INC (1∶2000 [21]), and developed with ECL Prime (GE Lifesciences).
Dopamine levels
20 age-matched male flies were presented with drug-free behavior food, or food laced with 2 mg/mL 3IY, 2 mg/mL L-DOPA, or 5 mg/mL L-DOPA for 2 d under 12 h light∶12 h dark conditions. They were then frozen on dry ice at ZT6, heads were removed by dry ice cold vortexing and isolated on frozen sieves. Dopamine levels were determined by HPLC by Dr. Raymond F Johnson at the Vanderbilt University Neurochemistry Core Lab.
Statistics
To compare quantifiable groups with normal distributions (as determined by the Shapiro-Wilk Test) we used the two-tailed Student's t test. To compare sleep bout lengths, which are not normally distributed, we used the Mann-Whitney U Test. p<0.05 was considered statistically significant.
Supporting Information
Zdroje
1. HendricksJC, FinnSM, PanckeriKA, ChavkinJ, WilliamsJA, et al. (2000) Rest in Drosophila is a sleep-like state. Neuron 25 : 129–138.
2. ShawPJ, CirelliC, GreenspanRJ, TononiG (2000) Correlates of sleep and waking in Drosophila melanogaster. Science 287 : 1834–1837.
3. CirelliC, LaVauteTM, TononiG (2005) Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem 94 : 1411–1419.
4. KohK, JoinerWJ, WuMN, YueZ, SmithCJ, et al. (2008) Identification of SLEEPLESS, a sleep-promoting factor. Science 321 : 372–376.
5. WuMN, JoinerWJ, DeanT, YueZ, SmithCJ, et al. (2010) SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci 13 : 69–75.
6. BusheyD, HuberR, TononiG, CirelliC (2007) Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J Neurosci 27 : 5384–5393.
7. ShangY, HaynesP, PirezN, HarringtonKI, GuoF, et al. (2011) Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci 14 : 889–895.
8. LebestkyT, ChangJS, DankertH, ZelnikL, KimYC, et al. (2009) Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron 64 : 522–536.
9. KumeK, KumeS, ParkSK, HirshJ, JacksonFR (2005) Dopamine is a regulator of arousal in the fruit fly. J Neurosci 25 : 7377–7384.
10. AndreticR, van SwinderenB, GreenspanRJ (2005) Dopaminergic modulation of arousal in Drosophila. Curr Biol 15 : 1165–1175.
11. FoltenyiK, GreenspanRJ, NewportJW (2007) Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci 10 : 1160–1167.
12. HendricksJC, WilliamsJA, PanckeriK, KirkD, TelloM, et al. (2001) A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci 4 : 1108–1115.
13. NakaiY, HoriuchiJ, TsudaM, TakeoS, AkahoriS, et al. (2011) Calcineurin and Its Regulator Sra/DSCR1 Are Essential for Sleep in Drosophila. J Neurosci 31 : 12759–12766.
14. SeugnetL, SuzukiY, MerlinG, GottschalkL, DuntleySP, et al. (2011) Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr Biol 21 : 835–840.
15. CirelliC, BusheyD, HillS, HuberR, KreberR, et al. (2005) Reduced sleep in Drosophila Shaker mutants. Nature 434 : 1087–1092.
16. BayonY, TrinidadAG, de la PuertaML, Del Carmen RodriguezM, BogetzJ, et al. (2008) KCTD5, a putative substrate adaptor for cullin3 ubiquitin ligases. Febs J 275 : 3900–3910.
17. ZimmermanJE, RizzoW, ShockleyKR, RaizenDM, NaidooN, et al. (2006) Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol Genomics 27 : 337–350.
18. KeeganKP, PradhanS, WangJP, AlladaR (2007) Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol 3: e208 doi:10.1371/journal.pcbi.0030208.
19. KadenerS, StoleruD, McDonaldM, NawatheanP, RosbashM (2007) Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev 21 : 1675–1686.
20. RyderE, BlowsF, AshburnerM, Bautista-LlacerR, CoulsonD, et al. (2004) The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167 : 797–813.
21. StavropoulosN, YoungM (2011) insomniac and Cullin-3 Regulate Sleep and Wakefulness in Drosophila. Neuron 72 : 964–1040.
22. DietzlG, ChenD, SchnorrerF, SuKC, BarinovaY, et al. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 : 151–156.
23. JoinerWJ, CrockerA, WhiteBH, SehgalA (2006) Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441 : 757–760.
24. PitmanJL, McGillJJ, KeeganKP, AlladaR (2006) A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441 : 753–756.
25. CrockerA, ShahidullahM, LevitanIB, SehgalA (2010) Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron 65 : 670–681.
26. DonleaJM, ThimganMS, SuzukiY, GottschalkL, ShawPJ (2011) Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332 : 1571–1576.
27. DementievaIS, TereshkoV, McCrossanZA, SolomahaE, ArakiD, et al. (2009) Pentameric assembly of potassium channel tetramerization domain-containing protein 5. J Mol Biol 387 : 175–191.
28. OsterwalderT, YoonKS, WhiteBH, KeshishianH (2001) A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A 98 : 12596–12601.
29. ZhuS, PerezR, PanM, LeeT (2005) Requirement of Cul3 for axonal arborization and dendritic elaboration in Drosophila mushroom body neurons. J Neurosci 25 : 4189–4197.
30. TomitaJ, MitsuyoshiM, UenoT, AsoY, TanimotoH, et al. (2011) Pan-neuronal knockdown of calcineurin reduces sleep in the fruit fly, Drosophila melanogaster. The Journal of neuroscience : the official journal of the Society for Neuroscience 31 : 13137–13183.
31. DzirasaK, RibeiroS, CostaR, SantosLM, LinSC, et al. (2006) Dopaminergic control of sleep-wake states. J Neurosci 26 : 10577–10589.
32. LalouxC, DerambureP, HoudayerE, JacquessonJM, BordetR, et al. (2008) Effect of dopaminergic substances on sleep/wakefulness in saline - and MPTP-treated mice. J Sleep Res 17 : 101–110.
33. AgostoJ, ChoiJC, PariskyKM, StilwellG, RosbashM, et al. (2008) Modulation of GABA(A) receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci 11 : 354–359.
34. ChungBY, KilmanVL, KeathJR, PitmanJL, AlladaR (2009) The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol 19 : 386–390.
35. PariskyKM, AgostoJ, PulverSR, ShangY, KuklinE, et al. (2008) PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60 : 672–682.
36. NeckameyerWS (1998) Dopamine and mushroom bodies in Drosophila: experience-dependent and -independent aspects of sexual behavior. Learning and Memory 5 : 157–165.
37. BangS, HyunS, HongS-T, KangJ, JeongK, et al. (2011) Dopamine signalling in mushroom bodies regulates temperature-preference behaviour in Drosophila. PLoS Genet 7: e1001346 doi:10.1371/journal.pgen.1001346.
38. SwarupS, HarbisonS, HahnL, MorozovaT, YamamotoA, et al. (2012) Extensive epistasis for olfactory behaviour, sleep and waking activity in Drosophila melanogaster. Genetics research 94 : 9–29.
39. SarinS, BertrandV, BigelowH, BoyanovA, DoitsidouM, et al. (2010) Analysis of multiple ethyl methanesulfonate-mutagenized Caenorhabditis elegans strains by whole-genome sequencing. Genetics 185 : 417–447.
40. DjagaevaI, DoronkinS (2009) COP9 limits dendritic branching via Cullin3-dependent degradation of the actin-crosslinking BTB-domain protein Kelch. PLoS ONE 4: e7598 doi:10.1371/journal.pone.0007598.
41. VolkowND, WangGJ, TelangF, FowlerJS, LoganJ, et al. (2008) Sleep deprivation decreases binding of [11C]raclopride to dopamine D2/D3 receptors in the human brain. J Neurosci 28 : 8454–8461.
42. TandbergE, LarsenJP, KarlsenK (1999) Excessive daytime sleepiness and sleep benefit in Parkinson's disease: a community-based study. Mov Disord 14 : 922–927.
43. OndoWG, Dat VuongK, KhanH, AtassiF, KwakC, et al. (2001) Daytime sleepiness and other sleep disorders in Parkinson's disease. Neurology 57 : 1392–1396.
44. ClarenbachP (2000) Parkinson's disease and sleep. J Neurol 247 Suppl 4: IV/20–23.
45. ArnulfI, KonofalE, Merino-AndreuM, HouetoJL, MesnageV, et al. (2002) Parkinson's disease and sleepiness: an integral part of PD. Neurology 58 : 1019–1024.
46. RiemenspergerT, IsabelG, CoulomH, NeuserK, SeugnetL, et al. (2011) Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc Natl Acad Sci U S A 108 : 834–839.
47. CattersonJH, Knowles-BarleyS, JamesK, HeckMM, HarmarAJ, et al. (2010) Dietary modulation of Drosophila sleep-wake behaviour. PLoS ONE 5: e12062 doi:10.1371/journal.pone.0012062.
48. AndreticR, van SwinderenB, GreenspanR (2005) Dopaminergic modulation of arousal in Drosophila. Current biology : CB 15 : 1165–1240.
49. AndreticR, KimYC, JonesFS, HanKA, GreenspanRJ (2008) Drosophila D1 dopamine receptor mediates caffeine-induced arousal. Proc Natl Acad Sci U S A 105 : 20392–20397.
50. ShangY, HaynesP, PírezN, HarringtonK, GuoF, et al. (2011) Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nature Neuroscience 14 : 889–984.
51. SeugnetL, SuzukiY, VineL, GottschalkL, ShawPJ (2008) D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr Biol 18 : 1110–1117.
52. BusheyD, TononiG, CirelliC (2011) Sleep and synaptic homeostasis: structural evidence in Drosophila. Science 332 : 1576–1581.
53. GilestroGF, TononiG, CirelliC (2009) Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324 : 109–112.
54. DonleaJM, RamananN, ShawPJ (2009) Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324 : 105–108.
55. FreemanA, PranskiE, MillerR, RadmardS, BernhardD, et al. (2012) Sleep fragmentation and motor restlessness in a Drosophila model of restless legs syndrome. Current biology : CB 22 : 1142–1150.
56. AlladaR, KadenerS, NandakumarN, RosbashM (2003) A recessive mutant of Drosophila Clock reveals a role in circadian rhythm amplitude. Embo J 22 : 3367–3375.
57. ChenJ, CondronBG (2008) Branch architecture of the fly larval abdominal serotonergic neurons. Dev Biol 320 : 30–38.
58. LinHH, LaiJS, ChinAL, ChenYC, ChiangAS (2007) A map of olfactory representation in the Drosophila mushroom body. Cell 128 : 1205–1217.
59. RulifsonEJ, KimSK, NusseR (2002) Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296 : 1118–1120.
60. NgM, RoordaRD, LimaSQ, ZemelmanBV, MorcilloP, et al. (2002) Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron 36 : 463–474.
61. KobayashiM, MichautL, InoA, HonjoK, NakajimaT, et al. (2006) Differential microarray analysis of Drosophila mushroom body transcripts using chemical ablation. Proc Natl Acad Sci U S A 103 : 14417–14422.
62. PfeiffenbergerC, LearBC, KeeganKP, AlladaR (2010) Processing sleep data created with the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc 2010: pdb prot5520.
63. NeckameyerWS, WoodromeS, HoltB, MayerA (2000) Dopamine and senescence in Drosophila melanogaster. Neurobiol Aging 21 : 145–152.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 10
-
Všechny články tohoto čísla
- The Germline Genome Provides a Niche for Intragenic Parasitic DNA: Evolutionary Dynamics of Internal Eliminated Sequences
- Classical Genetics Meets Next-Generation Sequencing: Uncovering a Genome-Wide Recombination Map in
- Calpain-5 Mutations Cause Autoimmune Uveitis, Retinal Neovascularization, and Photoreceptor Degeneration
- Cofilin-1: A Modulator of Anxiety in Mice
- The Date of Interbreeding between Neandertals and Modern Humans
- Embryos of Robertsonian Translocation Carriers Exhibit a Mitotic Interchromosomal Effect That Enhances Genetic Instability during Early Development
- Viral Evasion of a Bacterial Suicide System by RNA–Based Molecular Mimicry Enables Infectious Altruism
- Phosphatase-Dead Myotubularin Ameliorates X-Linked Centronuclear Myopathy Phenotypes in Mice
- Full-Length Synaptonemal Complex Grows Continuously during Meiotic Prophase in Budding Yeast
- MOV10 RNA Helicase Is a Potent Inhibitor of Retrotransposition in Cells
- A Likelihood-Based Framework for Variant Calling and Mutation Detection in Families
- The Contribution of RNA Decay Quantitative Trait Loci to Inter-Individual Variation in Steady-State Gene Expression Levels
- New Partners in Regulation of Gene Expression: The Enhancer of Trithorax and Polycomb Corto Interacts with Methylated Ribosomal Protein L12 Its Chromodomain
- Mining the Unknown: A Systems Approach to Metabolite Identification Combining Genetic and Metabolic Information
- Mutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
- The Many Landscapes of Recombination in
- Faster-X Evolution of Gene Expression in
- Loss of Slc4a1b Chloride/Bicarbonate Exchanger Function Protects Mechanosensory Hair Cells from Aminoglycoside Damage in the Zebrafish Mutant
- Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection
- and the BTB Adaptor Are Key Regulators of Sleep Homeostasis and a Dopamine Arousal Pathway in Drosophila
- Mutation and Fetal Ethanol Exposure Synergize to Produce Midline Signaling Defects and Holoprosencephaly Spectrum Disorders in Mice
- Specific Missense Alleles of the Arabidopsis Jasmonic Acid Co-Receptor COI1 Regulate Innate Immune Receptor Accumulation and Function
- Deep Genome-Wide Measurement of Meiotic Gene Conversion Using Tetrad Analysis in
- Mismatch Repair Balances Leading and Lagging Strand DNA Replication Fidelity
- Distinguishing between Selective Sweeps from Standing Variation and from a Mutation
- Cytokinesis-Based Constraints on Polarized Cell Growth in Fission Yeast
- Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes
- Functional Antagonism between Sas3 and Gcn5 Acetyltransferases and ISWI Chromatin Remodelers
- The SET-Domain Protein SUVR5 Mediates H3K9me2 Deposition and Silencing at Stimulus Response Genes in a DNA Methylation–Independent Manner
- Morphogenesis and Cell Fate Determination within the Adaxial Cell Equivalence Group of the Zebrafish Myotome
- Muscle-Specific Splicing Factors ASD-2 and SUP-12 Cooperatively Switch Alternative Pre-mRNA Processing Patterns of the ADF/Cofilin Gene in
- Maize Is Required for Maintaining Silencing Associated with Paramutation at the and Loci
- Increasing Signal Specificity of the TOL Network of mt-2 by Rewiring the Connectivity of the Master Regulator XylR
- Use of Pleiotropy to Model Genetic Interactions in a Population
- RAB-Like 2 Has an Essential Role in Male Fertility, Sperm Intra-Flagellar Transport, and Tail Assembly
- Variants Affecting Exon Skipping Contribute to Complex Traits
- Topoisomerase II– and Condensin-Dependent Breakage of -Sensitive Fragile Sites Occurs Independently of Spindle Tension, Anaphase, or Cytokinesis
- Comparison of Family History and SNPs for Predicting Risk of Complex Disease
- Recovery of Arrested Replication Forks by Homologous Recombination Is Error-Prone
- A Mutation in the Gene Causes Alternative Splicing Defects and Deafness in the Bronx Waltzer Mouse
- Comparative Genomics Suggests an Independent Origin of Cytoplasmic Incompatibility in
- It Was Heaven: An Interview with Evelyn Witkin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Mutation in the Gene Causes Alternative Splicing Defects and Deafness in the Bronx Waltzer Mouse
- Mutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
- Classical Genetics Meets Next-Generation Sequencing: Uncovering a Genome-Wide Recombination Map in
- Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání







