-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDNA Methylation and Gene Expression Changes in Monozygotic Twins Discordant for Psoriasis: Identification of Epigenetically Dysregulated Genes
Monozygotic (MZ) twins do not show complete concordance for many complex diseases; for example, discordance rates for autoimmune diseases are 20%–80%. MZ discordance indicates a role for epigenetic or environmental factors in disease. We used MZ twins discordant for psoriasis to search for genome-wide differences in DNA methylation and gene expression in CD4+ and CD8+ cells using Illumina's HumanMethylation27 and HT-12 expression assays, respectively. Analysis of these data revealed no differentially methylated or expressed genes between co-twins when analyzed separately, although we observed a substantial amount of small differences. However, combined analysis of DNA methylation and gene expression identified genes where differences in DNA methylation between unaffected and affected twins were correlated with differences in gene expression. Several of the top-ranked genes according to significance of the correlation in CD4+ cells are known to be associated with psoriasis. Further, gene ontology (GO) analysis revealed enrichment of biological processes associated with the immune response and clustering of genes in a biological pathway comprising cytokines and chemokines. These data suggest that DNA methylation is involved in an epigenetic dysregulation of biological pathways involved in the pathogenesis of psoriasis. This is the first study based on data from MZ twins discordant for psoriasis to detect epigenetic alterations that potentially contribute to development of the disease.
Published in the journal: . PLoS Genet 8(1): e32767. doi:10.1371/journal.pgen.1002454
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002454Summary
Monozygotic (MZ) twins do not show complete concordance for many complex diseases; for example, discordance rates for autoimmune diseases are 20%–80%. MZ discordance indicates a role for epigenetic or environmental factors in disease. We used MZ twins discordant for psoriasis to search for genome-wide differences in DNA methylation and gene expression in CD4+ and CD8+ cells using Illumina's HumanMethylation27 and HT-12 expression assays, respectively. Analysis of these data revealed no differentially methylated or expressed genes between co-twins when analyzed separately, although we observed a substantial amount of small differences. However, combined analysis of DNA methylation and gene expression identified genes where differences in DNA methylation between unaffected and affected twins were correlated with differences in gene expression. Several of the top-ranked genes according to significance of the correlation in CD4+ cells are known to be associated with psoriasis. Further, gene ontology (GO) analysis revealed enrichment of biological processes associated with the immune response and clustering of genes in a biological pathway comprising cytokines and chemokines. These data suggest that DNA methylation is involved in an epigenetic dysregulation of biological pathways involved in the pathogenesis of psoriasis. This is the first study based on data from MZ twins discordant for psoriasis to detect epigenetic alterations that potentially contribute to development of the disease.
Introduction
Psoriasis is a common chronic inflammatory disease, which affects mainly the skin, but also the joints. The worldwide prevalence is reported to range between 1–11.8% depending on ethnicity, geographical area and method of assessment [1]. Psoriasis is known to have a strong genetic component with an estimated heritability of 66% [2]. Linkage peaks [3], copy number variations (CNVs) [4]–[6] and genes identified by GWAS [7]–[13] are associated with psoriasis. However, the combined effect of these loci does not account for the genetic variation underlying the observed susceptibility to psoriasis, and indicates the involvement of additional genetic, epigenetic or environmental factors [7], [14]–[15]. Further evidence for the role of epigenetic or environmental factors comes from the fact that the concordance rate among MZ twins is only 35–72% [2], [14], [16].
Phenotypic differences are a result of genetic and epigenetic variation, as well as environmental influences. The study of discordant MZ twins provides an attractive model to investigate epigenetic mechanisms in disease. Environmentally driven epigenetic changes are thought to contribute to development of autoimmune diseases through alteration of epigenetic profiles, but exactly how the environment modulates epigenetic states is not well understood. Besides twin discordance, accumulating evidence supports the contribution of epigenetics in the development of autoimmune diseases through dysregulation of the immune system [17]–[20]. Psoriasis is considered a T cell-mediated autoimmune disease, and T cell activation is a key event in the pathogenesis [21]. Antigen binding results in a complex downstream signaling pathway in which epigenetic mechanisms have an important role [21], [22]. A likely scenario involves an abnormal activation and migration of T cells into the skin, followed by a cascade of events, which ultimately results in aggregation of the inflammatory cells and development of psoriasis. The development of psoriatic plaques is a result of an activation of cells in focal skin regions, which is mediated by CD4+ (helper) and CD8+ (cytotoxic) cells [23].
The aim of this study was to identify epigenetically dysregulated genes, which contribute to development of psoriasis. To do this, we assessed the extent of epigenetic and transcriptomic differences in CD4+ and CD8+ cells isolated from MZ twins discordant for psoriasis. This study design enabled identification of cell-type specific DNA methylation differences which correlate with gene expression, and thereby identification of genes where DNA methylation may have a functional role in development of psoriasis.
Results/Discussion
Since genome-wide patterns of DNA methylation are known to differ between cell-types [24]–[26], and different cell-types in the immune system are implicated in the pathogenesis of psoriasis, we isolated and studied CD4+ and CD8+ cells to overcome the issue of epigenetic heterogeneity in whole blood. Both cell-types are relatively abundant in blood and have important functions in the immune system, thus they are good targets for epigenetic alterations which might contribute to the development of psoriasis. Comparisons of CD4+ and CD8+ cells revealed many significant differences for both DNA methylation (1,288 of the 26,690 CpG sites, 4,8%, n = 12, Table S1) and gene expression (2,126 of the 37,846 transcripts, 5,6%, n = 13, Table S2) in unaffected individual twins (Figure 1), which clearly demonstrate the importance of isolating specific cell-types.
Fig. 1. Volcano plots of cell-type specific differences in DNA methylation and gene expression. 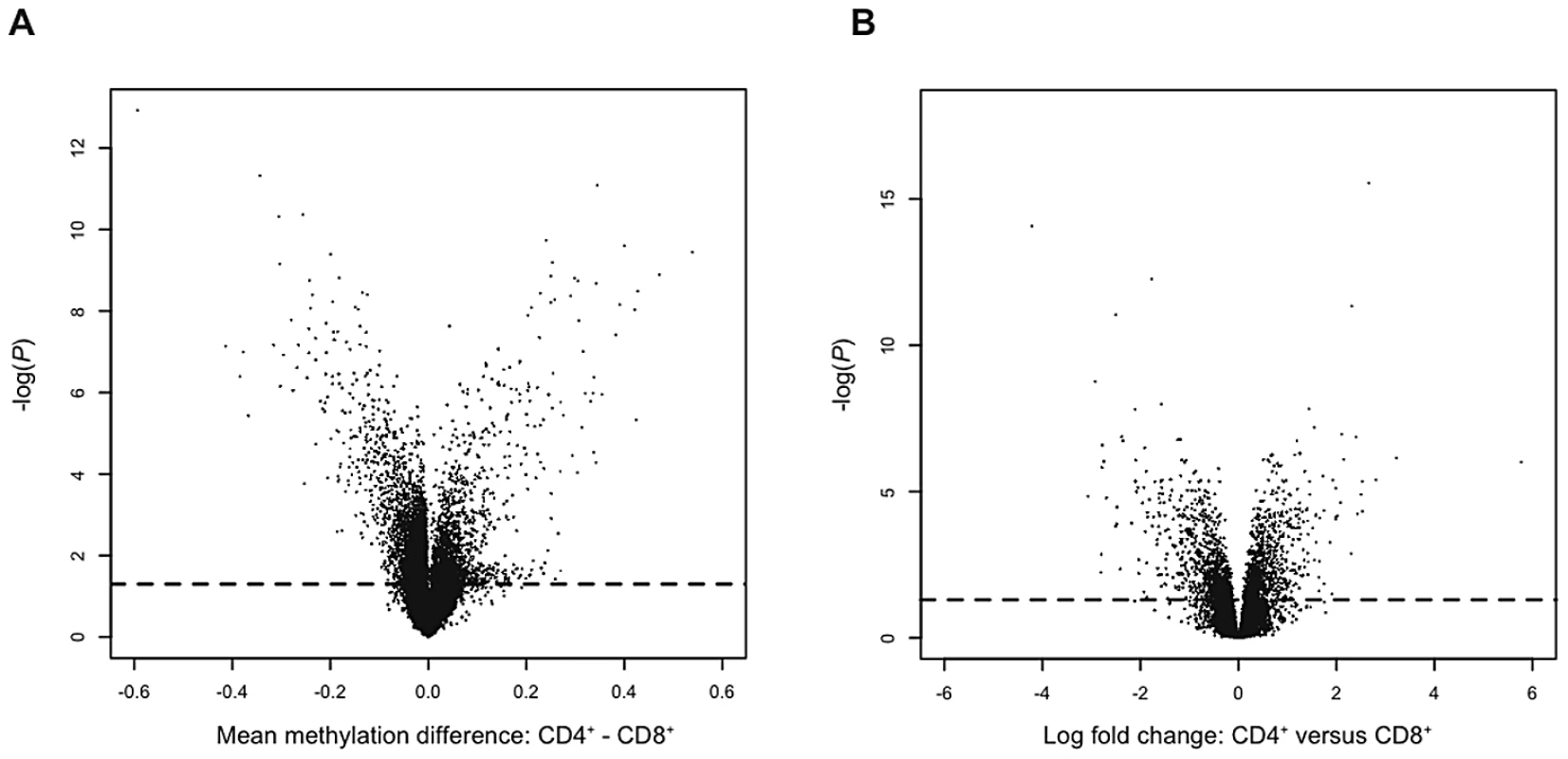
(A) Differences in DNA methylation between CD4+ and CD8+ cells. Each point represents a CpG site, with mean β-value difference across 12 unaffected individuals along the x-axis and −log10 of a corrected P-value from a paired t-test along the y-axis. (B) Differences in gene expression between CD4+ and CD8+ cells. Each point represents a gene, with mean log2 fold change across 13 unaffected individuals along the x-axis and −log10 of a corrected P-value from a paired t-test along the y-axis. Dashed lines represent the FDR of 5%. MZ co-twins are highly correlated for DNA methylation in both CD4+ cells (n = 17 pairs, mean ρ = 0.98, range = 0.96–0.99) and CD8+ cells (n = 13 pairs, mean ρ = 0.97, range = 0.95–0.98) (Figure 2A and 2B). Both analyses of individual CpG sites and mean methylation per gene did not reveal any differentially methylated sites between unaffected and affected co-twins. Individual scatter plots of DNA methylation clearly demonstrate greater similarity among the MZ twins than among unrelated individuals (Figure S1). To ensure that the observed differences in DNA methylation between co-twins were genuine rather than technical artifacts, we ran internal replicates on a subset of the twins. Specifically, we replicated 7 pairs and calculated technical and biological differences between replicated samples (self-self comparisons) and between co-twins, respectively. The overall distributions of the technical differences in DNA methylation per CpG site were significantly smaller than the biological differences (Kolmogorov-Smirnov test, two-sided, P-value<2.2×10−16, Figure S2). This clearly shows that the observed biological differences between unaffected and affected co-twins are genuine, although they are small. Similarly, differences in gene expression between MZ co-twins were small in both CD4+ cells (n = 17 pairs, mean r = 0.99, range = 0.97–0.99) and CD8+ cells (n = 14 pairs, mean r = 0.99, range = 0.98–0.99). Although there are many small differences, we did not detect any genome-wide significant differences in DNA methylation or gene expression between co-twins discordant for psoriasis when analyzed separately (Figure 2C and 2D).
Fig. 2. Volcano plots of differences in DNA methylation and gene expression in discordant MZ twins. 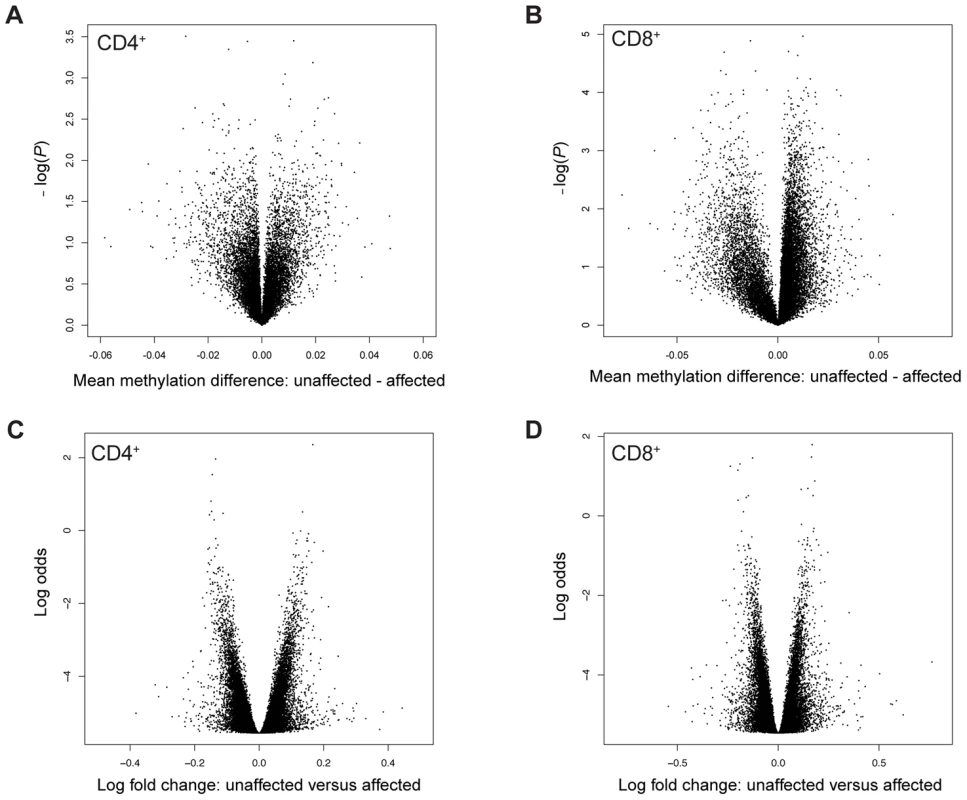
(A–B) Differences in DNA methylation in CD4+ (n = 17 pairs) and CD8+ cells (n = 13 pairs), respectively. Each point represents a gene, with mean co-twin β-value difference along the x-axis and −log10 of the uncorrected P-value from a paired t-test along the y-axis. (C–D) Differences in gene expression in CD4+ cells (n = 17 pairs) and CD8+ cells (n = 14 pairs), respectively. Each point represents a gene, with mean log2 fold change along the x-axis and log odds along the y-axis. DNA methylation is essential for the regulation of gene expression. We reasoned that a combined analysis of DNA methylation and gene expression could select functional methylation sites involved in regulating gene expression. We therefore investigated if co-twin differences in DNA methylation and gene expression were correlated. To do this, we compared the differences (between co-twins) in mean β-values for the CpGs associated with each gene, with the log fold changes of the gene expression. Using Spearman's rho as a measure for correlation, we then ranked the genes according to the significance of the correlation coefficients. This combined analysis of DNA methylation and gene expression revealed cell-type specific differences, identifying genes known to be involved in immune response and associated with psoriasis, only in CD4+ cells. Table 1 shows the top 50 genes ranked according to the significance of the correlation in CD4+ cells. Genes associated with psoriasis (shown in bold) are overrepresented in this list (Fisher's exact test, P = 3.3×10−5, Table S3). IL13 have been identified in GWAS [8], [10], [11], [27], and ALOX5AP [28], PTHLH [29] and TNFSF11 [30] have all been linked to different aspects of the disease. The entire list of the 11,933 genes studied ranked according to the significance of the correlation in CD4+ and CD8+ can be found as Table S4 in supporting information. Scatter plots depicting the relationship of MZ co-twin differences in DNA methylation and gene expression of TNFSF11 demonstrate the strong correlation in CD4+ cells compared to a non-significant correlation in CD8+ cells (Figure 3).
Fig. 3. Scatter plots of differences in DNA methylation against the differences in gene expression of TNFSF11. 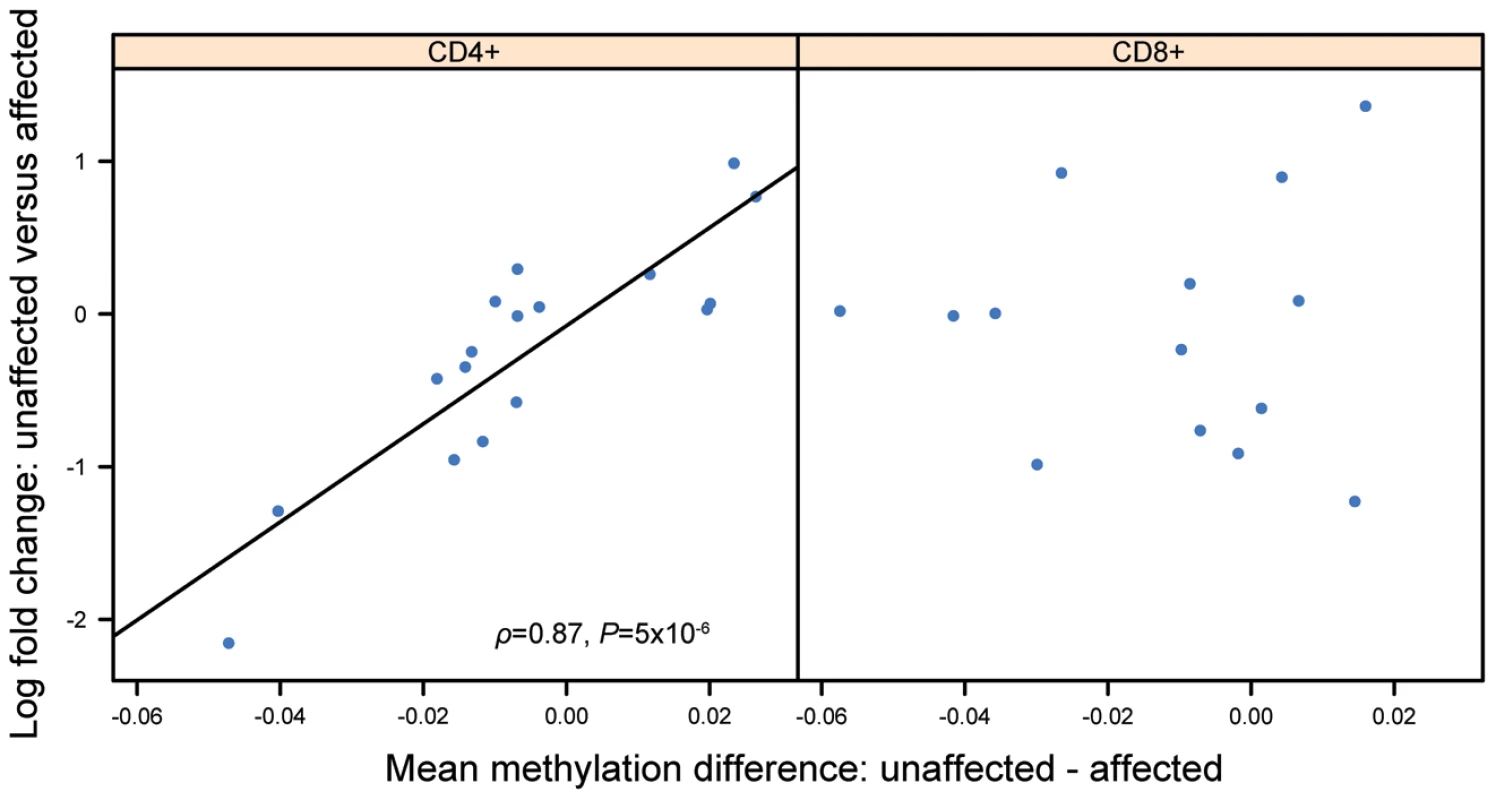
The plots show a correlation of DNA methylation differences and gene expression differences in MZ co-twins. Each point represents a twin pair, with the mean difference in DNA methylation β-value (of 2 CpG sites) along the x-axis, and log2 fold change along the y-axis. A correlation of 0.87 was calculated in CD4+ cells. Tab. 1. Subset of genes with a correlated difference in DNA methylation and gene expression. 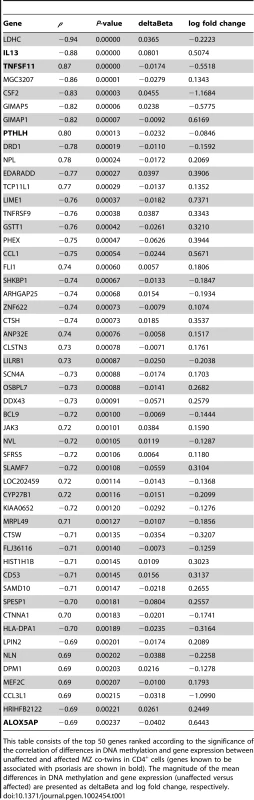
This table consists of the top 50 genes ranked according to the significance of the correlation of differences in DNA methylation and gene expression between unaffected and affected MZ co-twins in CD4+ cells (genes known to be associated with psoriasis are shown in bold). The magnitude of the mean differences in DNA methylation and gene expression (unaffected versus affected) are presented as deltaBeta and log fold change, respectively. We used DAVID [31], [32] to explore the potential of shared biological pathways among the genes in the list we generated from the combined analysis of DNA methylation and gene expression in CD4+ and CD8+ cells (Table S4). GO analyses identified significant enrichment of GO terms in CD4+ cells (Table 2), whereas the analysis did not detect any enriched terms in CD8+ cells. A significant portion of the top 1% of the genes ranked at the top of the list were found to be involved in the immune response (GO: 0006955, 12.5%, P = 0.042), positive regulation of response to stimulus (GO: 0048584, 7.5%, P = 0.037), immune system process (GO: 0002376, 15%, P = 0.043) and regulation of response to stimulus (GO: 0048583, 10.8%, P = 0.043). All of these categories contain genes involved in the immune response, which are potentially important in autoimmune diseases. Interestingly, a subset of the genes identified in the GO analysis (IL13, IL23R, CCL1, CCL5, CSF2, TNFSF11, LTB and SF9) comprises part of the cytokine-cytokine receptor interaction pathway, which is essential in communication between cells in the immune system. Skewed cytokine levels of pro-inflammatory and anti-inflammatory cytokines characterize the pathogenesis of psoriasis. Cytokines and chemokines are essential in the communication between cells in the immune system. Whereas cytokines generally influence proliferation, differentiation and secretion of pro - or anti-inflammatory factors, chemokines primarily have an effect on the movement of cells [33]. Thus, pathways like the cytokine-cytokine receptor interaction are indeed relevant in the etiology of psoriasis. In this context, our results suggest that DNA methylation is important in regulation of the cytokine cascade and signaling pathways involved in psoriasis.
Tab. 2. Gene ontology results of significantly enriched GO terms identified in a combined analysis of DNA methylation and gene expression in CD4+ cells. 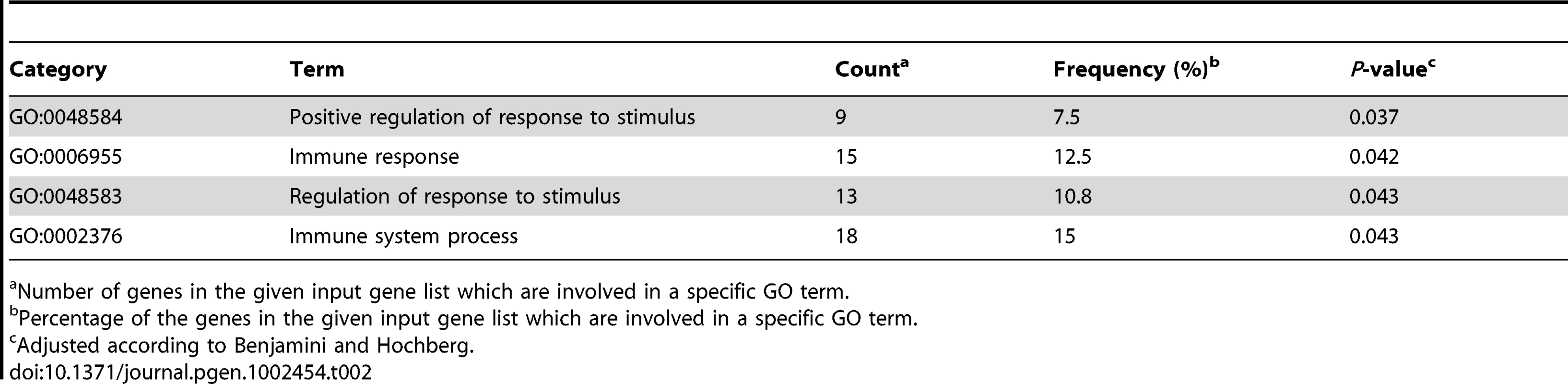
Number of genes in the given input gene list which are involved in a specific GO term. Both CD4+ and CD8+ cells are known to be present in psoriatic plaques, and current evidence indicates that CD4+ cells play a more critical role than CD8+ cells [34]. Our data strongly suggest that CD4+ cells are important in the pathogenesis of psoriasis. However, in this context it is important to recognize the complexity of CD4+ cells, which consists of several subpopulations with specific roles in the immune system (i.e. upon activation, naïve CD4+ cells develop into several lineages; Th1 cells, Th2 cells, Th22 and T regulatory cells). Recently, much attention has been drawn towards Th17 cells and the role in the pathogenesis of psoriasis [35]. It has also been speculated that CD4+ cells at different differentiation states may be present, which complicates the picture even further [36]. Distinctive compositions of these subpopulations can potentially contribute to the observed intra-pair differences. In addition, the complexity of the CD4+ and CD8+ cells could explain the small intra-pair differences by averaging out the level of DNA methylation and gene expression. Thus, a disease-associated change in DNA methylation and gene expression in a subset of cells can ultimately appear as an overall small difference.
Recently, several genes and pathways associated with psoriasis have been identified in GWAS [7]–[13]. Many of these have an essential role in the immune system and this clearly demonstrates the importance of immune response regulation in the disease. The molecular mechanisms driving the inflammation in skin causing psoriasis are complex. Our findings identify new potential susceptibility genes and point to different plausible biological pathways in psoriasis that are under epigenetic regulation and suggest an epigenetic dysregulation of biological pathways implicated in immune function. It will be important to expand on these findings in larger twin and other non-twin cohorts.
Materials and Methods
Twin Collection
The twins were recruited pair-wise from the Norwegian Twin Registry (NTR). Altogether 105 pairs were invited to participate, 60 pairs were invited from the cohorts born 1967–1979 [37], [38] and 45 pairs were invited from the cohorts born 1924–1960 [39]. The selection of discordant MZ pairs was based on a two-step procedure. Initial screening was conducted using self-reported data collected via questionnaires in earlier studies [37]–[39]. Pairs for which both twins consented to participate were called in to a clinical dermatology interview and skin examination at Oslo University Hospital, where additional information was also collected. Among the 105 pairs invited through the initial screening phase, 35 pairs consented and 27 pairs clinically evaluated to be discordant for psoriasis participated. The affected twins generally presented with a mild form of psoriasis, mainly affecting the scalp, knees and elbows. Scores for body surface area affected (BSA) were generally low and less than 10% for all the affected. The study was approved by the regional ethical committee and written informed consents were obtained.
Cell Separation
Lymphocyte subpopulations were isolated in a semi-automated way. Briefly, 60 ml EDTA-blood was diluted 1∶1 with RPMI, split into three aliquots and layered over Lymphoprep solution (Axis-Shield) in 50 ml centrifugation tubes. After centrifugation, PBMCs formed a distinct band that was harvested and washed twice to remove contaminating platelets. CD4+ cells and CD8+ cells were then sequentially isolated using positive and negative isolation kits from Miltenyi on an autoMACS Pro separator (Miltenyi). CD8+ cells were positively isolated using CD8+ MicroBeads and the Possel program. CD4+ cells were then separated from the negative fraction by negative isolation (i.e. by labeling all other cells but the CD4+ cells) using CD4+ T Cell Isolation kit II and the Deplete program.
Zygosity Testing
The zygosity for all twin pairs was determined based on 13 microsatellites on chromosomes 13, 18, 21, X and Y.
Cell Culturing
CD4+ cells were cultured for 7 days in X-VIVO (Lonza), exogenous rIL-2 (10 ng/µl) and Dynabeads CD3/CD28 T Cell Expander (Invitrogen) using ½ bead per cell. CD8+ cells were cultured for 14 days under the same conditions.
DNA Isolation and RNA Isolation
DNA was isolated from cultured cells on a Gentra autopure LS (Qiagen) using the 2–5×107 protocol. This resulted in high yields of pure DNA with an A260-A280 between 1.7 and 1.9. Total RNA was prepared manually from cultured CD4+ and CD8+ lymphocytes using RNAqueous Small Scale phenol-Free Total RNA Isolation Kit (Ambion) according to manufacturers instructions. RNA quality was checked using an Agilent 2100 Bioanalyser (Agilent Technologies).
DNA Methylation and Gene Expression Analysis
DNA methylation status was assessed using the Infinium HumanMethylation27 BeadChip, according to manufactures instructions (Illumina). These arrays enabled detection of the methylation status of 27,578 individual CpGs predominantly distributed in the promoters of 14,475 coding genes throughout the genome. The fluorescence data were analyzed in BeadStudio (Illumina) to determine the β-values (quantitative measurement of the methylation) for each CpG and normalized in Bioconductor lumi package [40]. We selected 26,690 probes that unambiguously mapped to the genome (hg18) with up to 2 mismatches as was done in Bell et al. [41]. In DNA methylation analysis, we excluded 1 out of 18 pairs in CD4+ cells from all analysis due to low bisulfite conversion.
Gene expression profiling was performed using the HumanHT-12 v3 Expression BeadChip, targeting >25,000 annotated genes, according to manufactures instructions (Illumina). The data were quartile normalized in BeadStudio (Illumina).
Statistical Analysis
All statistical tests were conducted in R (http://www.r-project.org/). The significance of the differences in DNA methylation between CD4+ and CD8+ cells were calculated based on a paired t-test in unaffected twins with overlapping data from both cell-types. Correlation of DNA methylation in discordant MZ co-twins was computed based on a nonparametric Spearman rank correlation. To search for differentially methylated genes between unaffected and affected twins we used a paired t-test on the mean β-value on all CpG sites associated with each gene. In addition, we also searched for differentially methylated CpG sites based on the β-value per CpG separately. The FDR significance thresholds were calculated using stats (R package), after raw P-values of a paired t-test had been computed. Nonparametric permutation tests were also performed, where P-values were calculated by comparing the t-statistic of the true data set with the t-statistics resulting from permutations of the affection status of the twins in all possible combinations. The results from these permutation tests produced similar results.
To search for differentially expressed genes between CD4+ and CD8+ cells and between unaffected and affected twins, the data was first log2 transformed and an empirical Bayes moderated t-test was then applied using the Limma package [42]. Correlation of gene expression between MZ co-twins was computed based on the parametric Pearson correlation. All statistical tests were done two-tailed and a false discovery rate (FDR) below 5% was considered significant.
Of the genes represented on the HumanMethylation27 BeadChip, 11,933 were also present on the HumanHT-12 Expression BeadChip, which enabled integrated analysis of the methylation status and gene expression. In the combined analysis of DNA methylation and gene expression, we compared the differences (between co-twins) in mean β-values for the CpGs associated with each gene, with the log fold changes of the gene expression. Using Spearman's rho as a measure for correlation, we then ranked the genes according to the significance of the correlation coefficients.
GO Analysis
GO analysis was conducted using the DAVID functional annotation tool [31], [32] with the 120 most significantly correlated genes as input (1%) and the 11,933 genes that was used in the combined analysis of DNA methylation and gene expression as background. Analysis was done with default parameters and results corrected for multiple testing by the method of Benjamini and Hochberg [43].
Supporting Information
Zdroje
1. ChandranVRaychaudhuriSP 2010 Geoepidemiology and environmental factors of psoriasis and psoriatic arthritis. J Autoimmun 34 J314 J321
2. GrjibovskiAMOlsenAOMagnusPHarrisJR 2007 Psoriasis in Norwegian twins: contribution of genetic and environmental effects. J Eur Acad Dermatol Venereol 21 1337 1343
3. BowcockAMKruegerJG 2005 Getting under the skin: the immunogenetics of psoriasis. Nat Rev Immunol 5 699 711
4. HolloxEJHuffmeierUZeeuwenPLJMPallaRLascorzJ 2008 Psoriasis is associated with increased β-defensin genomic copy number. Nat Genet 40 23 25
5. ZhangXJHuangWYangSSunLDZhangFY 2009 Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet 41 205 210
6. de CidRRiveira-MunozEZeeuwenPLJMRobargeJLiaoW 2009 Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet 41 211 215
7. LiuYHelmsCLiaoWZabaLCDuanS 2008 A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet 4 e1000041 doi:10.1371/journal.pgen.1000041
8. NairRPDuffinKCHelmsCDingJStuartPE 2009 Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat Genet 41 199 204
9. Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2 2010 A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet 42 985 990
10. SunLDChengHWangZXZhangAPWangPG 2010 Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet 42 1005 1009
11. EllinghausEEllinghausDStuartPENairRPDebrusS 2010 Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat Genet 42 991 995
12. HuffmeierUUebeSEkiciABBowesJGiardinaE 2010 Common variants at TRAF3IP2 are associated with susceptibility to psoriatic arthritis and psoriasis. Nat Genet 42 996 999
13. StuartPENairRPEllinghausEDingJTejasviT 2010 Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat Genet 42 1000 1004
14. BowcockAM 2005 THE GENETICS OF PSORIASIS AND AUTOIMMUNITY. Annu Rev Genomics Hum Genet 6 93 122
15. FengBJSunLDSoltani-ArabshahiRBowcockAMNairRP 2009 Multiple Loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet 5 e1000606 doi:10.1371/journal.pgen.1000606
16. BallestarE 2009 Epigenetics Lessons from Twins: Prospects for Autoimmune Disease. Clin Rev Allergy Immunol 39 30 41
17. JavierreBMFernandezAFRichterJAl-ShahrourFMartin-SuberoJI 2010 Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res 20 170 179
18. GaraudSLe DantecCJousse-JoulinSHanrotel-SaliouCSarauxA 2009 IL-6 Modulates CD5 Expression in B Cells from Patients with Lupus by Regulating DNA Methylation. J Immunol 182 5623 5632
19. LeiWLuoYLeiWLuoYYanK 2009 Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand J Rheumatol 38 369 374
20. StricklandFMRichardsonBC 2008 Epigenetics in human autoimmunity. Autoimmunity 41 278 286
21. KruegerJGBowcockA 2005 Psoriasis pathophysiology: current concepts of pathogenesis. Ann Rheum Dis 64 Suppl 2 ii30 ii36
22. CuddapahSBarskiAZhaoK 2010 Epigenomics of T cell activation, differentiation, and memory. Curr Opin Immunol 22 341 347
23. LowesMABowcockAMKruegerJG 2007 Pathogenesis and therapy of psoriasis. Nature 445 866 873
24. EckhardtFLewinJCorteseRRakyanVKAttwoodJ 2006 DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 38 1378 1385
25. RakyanVKDownTAThorneNPFlicekPKuleshaE 2008 An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res 18 1518 1529
26. BibikovaMChudinEWuBZhouLGarciaEW 2006 Human embryonic stem cells have a unique epigenetic signature. Genome Res 16 1075 1083
27. ChangMLiYYanCCallis-DuffinKPMatsunamiN 2007 Variants in the 5q31 cytokine gene cluster are associated with psoriasis. Genes Immun 9 176 181
28. DixonRAFDiehlREOpasERandsEVickersPJ 1990 Requirement of a 5-lipoxygenase-activating protein for leukotriene synthesis. Nature 343 282 284
29. TaylorJMStreetTLHaoLCopleyRTaylorMS 2009 Dynamic and Physical Clustering of Gene Expression during Epidermal Barrier Formation in Differentiating Keratinocytes. PLoS ONE 4 e7651 doi:10.1371/journal.pone.0007651
30. HolickMFChimehFNRayS 2003 Topical PTH (1–34) is a novel, safe and effective treatment for psoriasis: a randomized self-controlled trial and an open trial. Br J Dermatol 149 370 376
31. DennisGShermanBHosackDYangJGaoW 2003 DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4 P3
32. HuangDWShermanBTLempickiRA 2008 Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols 4 44 57
33. NickoloffBJXinHNestleFOQinJZ 2007 The cytokine and chemokine network in psoriasis. Clin Dermatol 25 568 573
34. GhoreschiKWeigertCRöckenM 2011 Immunopathogenesis and role of T cells in psoriasis. Clin Dermatol 25 574 580
35. WilsonNJBonifaceKChanJRMcKenzieBSBlumenscheinWM 2007 Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol 8 950 957
36. O'SheaJJPaulWE 2010 Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327 1098 1102
37. HarrisJRMagnusPTambsK 2002 The Norwegian Institute of Public Health Twin Panel: a description of the sample and program of research. Twin Res 5 415 423
38. HarrisJRMagnusPTambsK 2006 The Norwegian Institute of Public Health twin program of research: an update. Twin Res Hum Genet 9 858 864
39. BergemAL 2002 Norwegian Twin Registers and Norwegian twin studies–an overview. Twin Res 5 407 414
40. DuPKibbeWALinSM 2008 lumi: a pipeline for processing Illumina microarray. Bioinformatics 24 1547 1548
41. BellJTPaiAAPickrellJKGaffneyDJPique-RegiR 2011 DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol 12 R10
42. SmythGK 2004 Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 Article3
43. BenjaminiYHochbergY 1995 Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Stat Methodol 57 289 300
Štítky
Genetika Reprodukční medicína
Článek USF-1 Is Critical for Maintaining Genome Integrity in Response to UV-Induced DNA PhotolesionsČlánek Checkpoints in a Yeast Differentiation Pathway Coordinate Signaling during Hyperosmotic StressČlánek Two-Component Elements Mediate Interactions between Cytokinin and Salicylic Acid in Plant ImmunityČlánek Adaptive Evolution of the Lactose Utilization Network in Experimentally Evolved Populations of
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 1
-
Všechny články tohoto čísla
- DNA Methylation and Gene Expression Changes in Monozygotic Twins Discordant for Psoriasis: Identification of Epigenetically Dysregulated Genes
- An siRNA Screen in Pancreatic Beta Cells Reveals a Role for in Insulin Production
- Parallel Mapping and Simultaneous Sequencing Reveals Deletions in and Associated with Discrete Inherited Disorders in a Domestic Dog Breed
- Cytoplasmic Polyadenylation Element Binding Protein Deficiency Stimulates PTEN and Stat3 mRNA Translation and Induces Hepatic Insulin Resistance
- Nucleolar Association and Transcriptional Inhibition through 5S rDNA in Mammals
- USF-1 Is Critical for Maintaining Genome Integrity in Response to UV-Induced DNA Photolesions
- Heterochromatin Formation Promotes Longevity and Represses Ribosomal RNA Synthesis
- Genetic Evidence for an Indispensable Role of Somatic Embryogenesis Receptor Kinases in Brassinosteroid Signaling
- Poly(ADP-Ribose) Polymerase 1 (PARP-1) Regulates Ribosomal Biogenesis in Nucleoli
- Genome Engineering in : A Feasible Approach to Address Biological Issues
- RIC-7 Promotes Neuropeptide Secretion
- Adaptation and Preadaptation of to Bile
- Checkpoints in a Yeast Differentiation Pathway Coordinate Signaling during Hyperosmotic Stress
- Progressive Polycomb Assembly on H3K27me3 Compartments Generates Polycomb Bodies with Developmentally Regulated Motion
- A High Density SNP Array for the Domestic Horse and Extant Perissodactyla: Utility for Association Mapping, Genetic Diversity, and Phylogeny Studies
- Two-Component Elements Mediate Interactions between Cytokinin and Salicylic Acid in Plant Immunity
- Cdc5-Dependent Asymmetric Localization of Bfa1 Fine-Tunes Timely Mitotic Exit
- A Genome-Wide Analysis of Promoter-Mediated Phenotypic Noise in
- Contribution of Intragenic DNA Methylation in Mouse Gametic DNA Methylomes to Establish Oocyte-Specific Heritable Marks
- Adaptive Evolution of the Lactose Utilization Network in Experimentally Evolved Populations of
- Microenvironmental Regulation by Fibrillin-1
- Unraveling the Regulatory Mechanisms Underlying Tissue-Dependent Genetic Variation of Gene Expression
- A Half-Century of Inspiration: An Interview with Hamilton Smith
- Reduced Lentivirus Susceptibility in Sheep with Mutations
- High-Density SNP Mapping of the HLA Region Identifies Multiple Independent Susceptibility Loci Associated with Selective IgA Deficiency
- Calpains Mediate Integrin Attachment Complex Maintenance of Adult Muscle in
- Genomic Ancestry of North Africans Supports Back-to-Africa Migrations
- Functional Specialization of the Plant miR396 Regulatory Network through Distinct MicroRNA–Target Interactions
- The Seminal Fluid Protease “Seminase” Regulates Proteolytic and Post-Mating Reproductive Processes
- Insulin Signaling Regulates Fatty Acid Catabolism at the Level of CoA Activation
- A Genome-Wide Association Study Identified as a Susceptibility Locus for Systemic Lupus Eyrthematosus in Japanese
- A Spontaneous Mutation of the Rat Gene Leads to Impaired Function of Regulatory T Cells Linked to Inflammatory Bowel Disease
- The Yeast Complex I Equivalent NADH Dehydrogenase Rescues Mutants
- A Flexible Bayesian Model for Studying Gene–Environment Interaction
- Sex Pheromone Evolution Is Associated with Differential Regulation of the Same Desaturase Gene in Two Genera of Leafroller Moths
- Genome-Wide Assessment of AU-Rich Elements by the ARE Algorithm
- Inference of Population Structure using Dense Haplotype Data
- A Gene Regulatory Network for Root Epidermis Cell Differentiation in Arabidopsis
- Sequencing of Pooled DNA Samples (Pool-Seq) Uncovers Complex Dynamics of Transposable Element Insertions in
- A Genome-Wide Association Scan on the Levels of Markers of Inflammation in Sardinians Reveals Associations That Underpin Its Complex Regulation
- Tempo and Mode in Evolution of Transcriptional Regulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Poly(ADP-Ribose) Polymerase 1 (PARP-1) Regulates Ribosomal Biogenesis in Nucleoli
- Microenvironmental Regulation by Fibrillin-1
- Parallel Mapping and Simultaneous Sequencing Reveals Deletions in and Associated with Discrete Inherited Disorders in a Domestic Dog Breed
- Two-Component Elements Mediate Interactions between Cytokinin and Salicylic Acid in Plant Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



