-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAdaptive Evolution of the Lactose Utilization Network in Experimentally Evolved Populations of
Adaptation to novel environments is often associated with changes in gene regulation. Nevertheless, few studies have been able both to identify the genetic basis of changes in regulation and to demonstrate why these changes are beneficial. To this end, we have focused on understanding both how and why the lactose utilization network has evolved in replicate populations of Escherichia coli. We found that lac operon regulation became strikingly variable, including changes in the mode of environmental response (bimodal, graded, and constitutive), sensitivity to inducer concentration, and maximum expression level. In addition, some classes of regulatory change were enriched in specific selective environments. Sequencing of evolved clones, combined with reconstruction of individual mutations in the ancestral background, identified mutations within the lac operon that recapitulate many of the evolved regulatory changes. These mutations conferred fitness benefits in environments containing lactose, indicating that the regulatory changes are adaptive. The same mutations conferred different fitness effects when present in an evolved clone, indicating that interactions between the lac operon and other evolved mutations also contribute to fitness. Similarly, changes in lac regulation not explained by lac operon mutations also point to important interactions with other evolved mutations. Together these results underline how dynamic regulatory interactions can be, in this case evolving through mutations both within and external to the canonical lactose utilization network.
Published in the journal: . PLoS Genet 8(1): e32767. doi:10.1371/journal.pgen.1002444
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002444Summary
Adaptation to novel environments is often associated with changes in gene regulation. Nevertheless, few studies have been able both to identify the genetic basis of changes in regulation and to demonstrate why these changes are beneficial. To this end, we have focused on understanding both how and why the lactose utilization network has evolved in replicate populations of Escherichia coli. We found that lac operon regulation became strikingly variable, including changes in the mode of environmental response (bimodal, graded, and constitutive), sensitivity to inducer concentration, and maximum expression level. In addition, some classes of regulatory change were enriched in specific selective environments. Sequencing of evolved clones, combined with reconstruction of individual mutations in the ancestral background, identified mutations within the lac operon that recapitulate many of the evolved regulatory changes. These mutations conferred fitness benefits in environments containing lactose, indicating that the regulatory changes are adaptive. The same mutations conferred different fitness effects when present in an evolved clone, indicating that interactions between the lac operon and other evolved mutations also contribute to fitness. Similarly, changes in lac regulation not explained by lac operon mutations also point to important interactions with other evolved mutations. Together these results underline how dynamic regulatory interactions can be, in this case evolving through mutations both within and external to the canonical lactose utilization network.
Introduction
Changes in gene regulation are an important and common cause of adaptation. Support for this comes from bioinformatic evidence that changes in regulatory elements are associated with presumably adaptive phenotypic changes (reviewed in [1], [2]), comparative experimental studies [3] and from experimental evolution studies, which often find regulatory changes occurring during adaptation to novel environments [4]–[12]. Indeed, in some of these cases direct links have been established between regulatory changes and adaptation [6]–[8], [10]. These experiments directly demonstrate that small and local regulatory changes can significantly contribute to adaptation. In most cases, however, the physiological basis for selection of regulatory changes is unknown.
Previously, we described the evolution of populations of Escherichia coli in defined environments that differed only in the number and presentation of the limiting resource [13]. Populations were evolved in environments supplemented with a single limiting resource or combinations of two limiting resources either presented together or fluctuating daily. These populations adapted to the environments in which they were evolved and this adaptation was, at least to some extent, environment-specific [13]. Here, we focus on a subset of 24 populations that evolved in environments supplemented with glucose and/or lactose and examine changes in the regulation of the lac operon in these populations.
Several attributes make the lac operon a good candidate in which to observe regulatory changes and relate them to their physiological and fitness effects. First, the costs and benefits of lac operon expression are environmentally dependent. Expression of the lac operon is necessary for utilization of lactose for growth, but expression in the absence of lactose can impose a significant cost [14]–[16]. Second, the molecular components of the lactose utilization network are well characterized and their activity can be measured in vivo. The ability to assay changes in the lac regulatory network ‘output’ provides a means to identify and test activity of evolved regulatory changes. Third, the lac operon has been the subject of much theoretical work, leading to the development of mathematical models to explain important features of lac operon physiology [14], [17]–[22] and evolution [16], [23], [24]. Fourth, the utility of the lac operon for examining evolution of regulatory changes has been demonstrated experimentally. For example, lac operon constitutive [25], [26], loss of function [14] and duplication [11] mutants have been recovered following growth in different selective environments, demonstrating that lac operon regulation is evolutionarily flexible and can be a target of selection. It has even been possible to predict the evolution of regulatory changes on the basis of their expected fitness effects. Dekel and Alon (2005) used a cost-benefit analysis to predict the optimum expression level of the lac operon in different inducer concentration environments. They found that populations selected in environments containing a high level of gratuitous inducer, but various concentrations of lactose, generally evolved to regulate expression of the lac operon to the predicted level.
In this study, we examine changes in lac operon regulation associated with selection in environments differing in the presentation of its natural substrate and inducer, lactose, and repressor, glucose. In addition to quantitatively characterizing the changes that have occurred, we examine the genetic and demographic basis for selection of different modes of lac regulation. We find that regulatory changes in the lac operon evolved in many replicate populations selected in environments containing lactose. Much, but not all, of these changes were due to mutations in the LacI repressor or its major operator binding site within the lac promoter. By themselves, these mutations conferred significant fitness benefits in all of the evolution environments that contained lactose. We also present evidence for interactions between lac mutations and mutations in genes outside of the canonical lac utilization network and show that these interactions impact lac operon regulation and fitness. Finally, operator and repressor mutations fixed at different frequencies in different selective environments, although the selective basis of this is currently not known.
Results
Qualitative characterization of evolved changes to lac operon regulation
Previously we reported propagation of 24 replicate populations of E. coli B REL606 for 2000 generations in one of four environments that differed only in the concentration and/or presentation of glucose and lactose [13]. Environments used were glucose (Glu), lactose (Lac), glucose and lactose presented simultaneously (G+L) or alternating daily between glucose and lactose (G/L). To determine whether regulation of the lac operon had changed during the evolution of our experimental populations, we screened ≥1000 clones from each population on LacZ indicator plates (see Materials and Methods). All six Glu populations had LacZ phenotypes that were indistinguishable from the ancestor. By contrast, in all other evolution environments at least some replicate populations showed clear changes in LacZ activity (Lac, 3 of 6; G+L, 6 of 6; G/L, 6 of 6) (Table 1, Figure 1A). Some of these populations also had within-population variation in LacZ activity and colony morphology. To facilitate molecular and physiological studies of these changes we identified and isolated 46 clones (at least one from each evolved population) that encompassed the range of LacZ activity and morphology types present across all populations (Table 1). These clones were used in all subsequent analyses.
Fig. 1. Characterization of lac operon regulation. 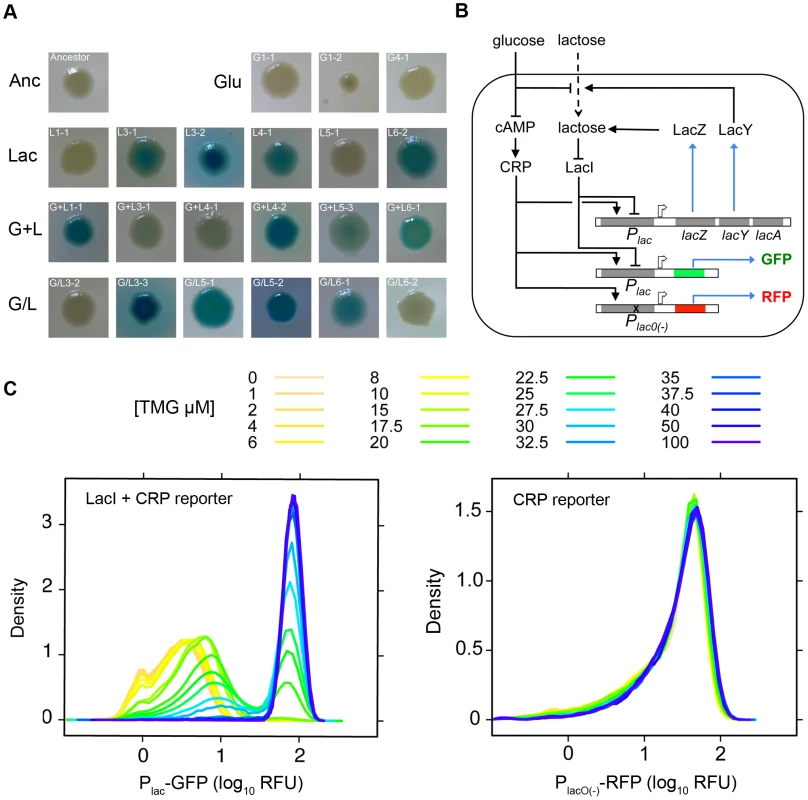
A) Examples of the range of evolved LacZ activity phenotypes present in the four evolution environments. Degree of blue coloration on TGX plates gives a qualitative measure of LacZ activity. B) Schematic of lac operon and reporters used to measure lac operon regulation. lacZ encodes the β-galactosidase responsible for lactose catabolism and lacY encodes a lactose permease. The expression of lacZYA is directly controlled by LacI and CRP. LacI is a negative regulator, binding to operator sites within the lacZYA promoter (Plac). LacI binding is inhibited by lactose and gratuitous inducers, such as TMG. CRP is a positive regulator, activating lacZYA expression when cAMP levels are elevated in response to low glucose concentrations. High levels of glucose also repress lac expression by inhibiting import of lactose through LacY. Two reporters were designed to measure LacI and CRP inputs into lac operon regulation. The native lac promoter drives expression of GFP and is subject to regulation by both LacI and CRP. A second reporter utilizes a mutant lac promoter that cannot bind LacI to drive expression of DsRedExpress2. This reporter is only subject to regulation by CRP. Solid lines indicate positive (arrows) and negative (blunt arrow) regulatory interactions; dotted lines indicate the transfer of metabolites; blue lines indicate the production of proteins; open arrows indicate expression start sites. Figure adapted from Ozbudak et al. 2004. C) Ancestral inducer response profile. Shown are flow cytometry histograms for the ancestor grown in a range of TMG concentrations. Plac-GFP and Plac(O-)-RFP measurements were taken simultaneously from the same cultures. Tab. 1. Characterization of evolved clones. 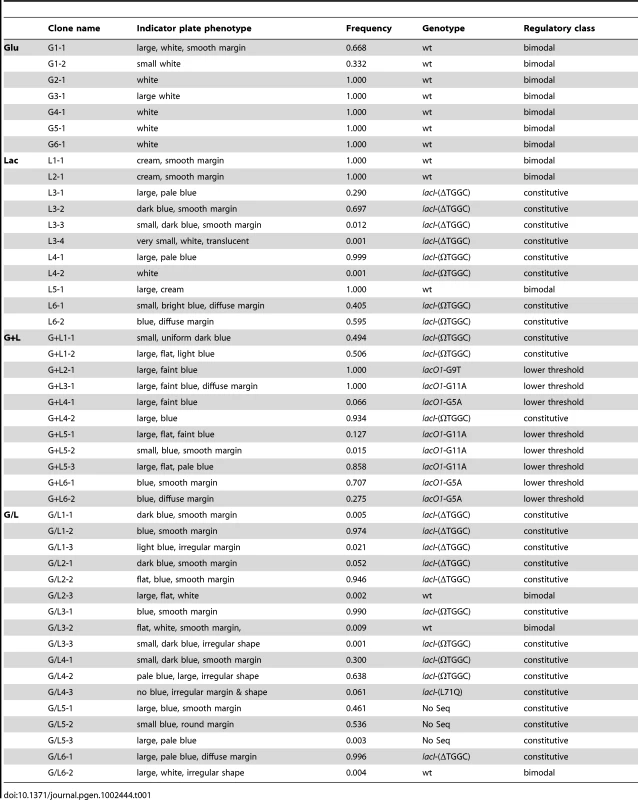
Quantitative analysis of evolved changes to lac operon regulation
To characterize evolved changes in lac operon regulation, we used a dual fluorescence reporter system to independently quantify with single cell resolution the activity of the major transcriptional regulators of the lac operon, LacI and CRP. LacI and CRP bind the lac promoter at different locations, repressing and activating transcription of the lac operon, respectively (Figure 1B). The ancestor and each of the 46 evolved clones were transformed with Plac-GFP (LacI and CRP reporter) and PlacO(-)-RFP (CRP reporter) constructs. For each strain we used flow cytometry to measure the population distribution of steady-state GFP and RFP expression over a range of thiomethyl-galactoside (TMG) concentrations (TMG is a non-metabolizable inducer of the lac operon). The Plac-GFP reporter captured several key features of lac regulation. The inducer response of the ancestor is ultra-sensitive, showing a sharp transition from low to high expression states as a function of TMG concentration and is bimodal, with populations showing a mix of non-induced and fully induced cells at intermediate levels of TMG [19], [27], [28] (Figure 1C, Figure 2). The CRP-only reporter (PlacO(-)-RFP) shows a unimodal distribution with a constant mean over the range of TMG concentrations, confirming that CRP activity of the ancestor is independent of LacI activity and lac operon expression state (Figure 1C). To facilitate comparisons between the ancestral and evolved genotypes, we quantified three aspects of lac regulation: the TMG concentration required for half maximal population expression (TMG½ Max), the range of TMG concentrations causing a bimodal population response and the fully induced (maximum) steady state level of lac expression. For the ancestor, TMG½ Max is 25 µM, the range of bimodality is between 15–30 µM TMG, and the level of Plac-GFP at full induction (100 µM TMG) is ∼84 RFU (Figure 2).
Fig. 2. Inducer response profiles of evolved clones. 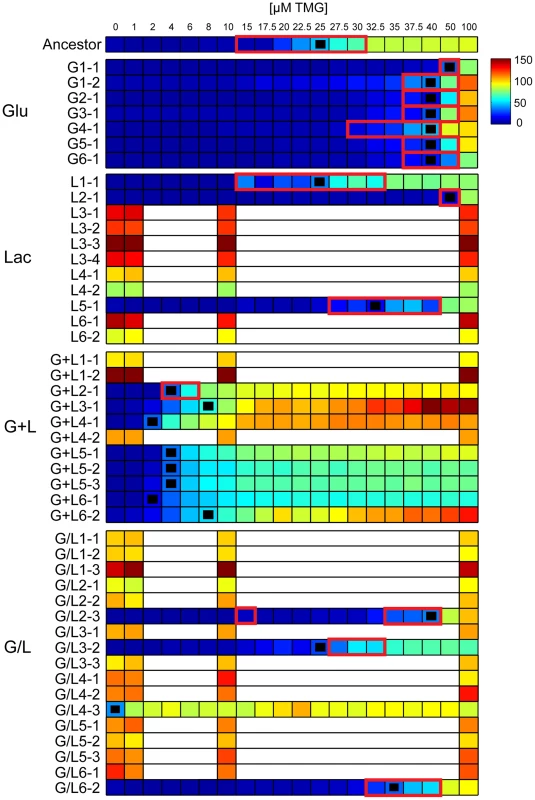
Heat maps show the mean response of the Plac-GFP reporter (RFU) for evolved clones at different concentrations of inducer (TMG). Black squares indicate the concentration of TMG that gave half maximal expression (TMG½Max) and red outlines indicate the presence of bimodal expression within the population. Inducer responses for clones with constitutive lac operons were only generated with four TMG concentrations because lac expression was independent of inducer concentration such that finer resolution measurements were redundant. Evolved changes in the inducer response profiles were common and fell into three broad classes (Figure 2). 1) Constitutive: operationally defined as mean Plac-GFP expression varying by less than 2-fold across the range of tested TMG concentrations. Constitutive clones were observed in at least some populations of each of the treatments containing lactose (Lac, G+L and G/L), but were not observed in any of the Glu evolved populations. 2) Lower threshold/graded response: increased sensitivity to inducer with TMG½ Max values ranging from 2 µM to 8 µM. In addition, all but one clone with a lower induction threshold (G+L2-1) also evolved a graded response, with a unimodal distribution of Plac-GFP expression that increased continuously as a function of TMG concentration. 3) Bimodal: bimodal induction response differing from the ancestor by generally being less sensitive to the inducer, with TMG½ Max values ranging from 25 µM to 50 µM TMG (Figure 2). Higher inducer thresholds were observed for all clones evolved in Glu as well as some clones evolved in Lac and G/L environments. Representative inducer response curves of each class are shown in Figure 3A and the distribution of response types across environments is shown in Figure 3B. Interestingly, clones with a lower threshold response were found exclusively in populations evolved in the G+L environment, fixing in four of the six populations. This pattern is unlikely to occur by chance (Fisher's exact test omitting the polymorphic G+L population, P = 0.002). By contrast, populations evolved in the G/L environment were almost exclusively composed of clones with constitutive lac expression and populations evolved in Lac had either bimodal or constitutive regulation. All clones evolved in Glu showed a bimodal response type (Figure 2, Figure 3B).
Fig. 3. Inducer response classes and association with evolution environment. 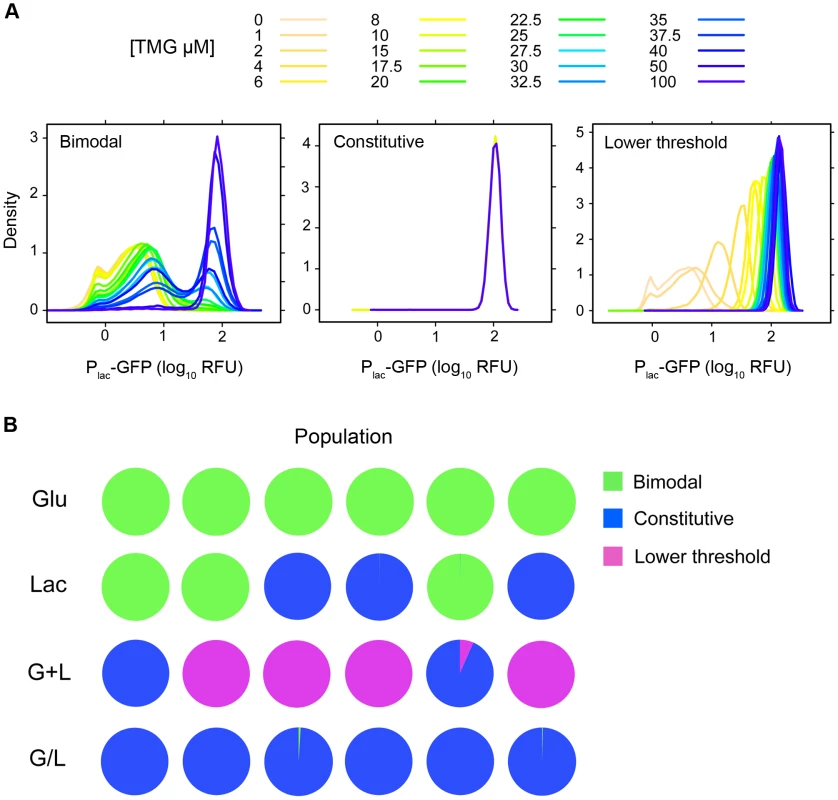
A) Inducer response classes for evolved clones. 1) Bimodal with a higher threshold for lac induction, 2) Constitutive with expression levels that are independent of inducer, 3) Lower threshold for lac induction with graded response to inducer. Inducer response histograms are shown for clones representative of each class (Bimodal, L5-1; Constitutive, G/L5-1; Lower threshold, G+L3-1). B) Distribution of inducer response types by population and evolution environment. Pie charts show the fraction of clones with each inducer response type (Bimodal, Constitutive, Lower threshold) within a given population. The level of Plac-GFP expression at maximum induction (100 µM TMG) was higher than the ancestor in 36 of the 46 evolved clones (Figure 2). To examine this observation in more detail, we repeated our measurements of Plac-GFP expression in each evolved clone, but this time only in the presence of 100 µM TMG, which enabled us to include all clones in duplicate in the same experimental block (Figure 4A). We again saw a strong trend toward an increase in lac expression with 40 of 46 clones having a mean reporter expression level greater than the ancestor. Furthermore, the mean change in expression level of clones evolved in lactose containing environments was significantly higher than the ancestor (two tailed t-test with unequal variance: Lac, P = 0.008; G+L, P = 0.004; G/L, P<0.001) but not significantly different for clones evolved in the Glu environment (P = 0.242). To verify that changes in the level of our GFP reporter accurately reflected changes in the expression level of the native lac operon, we used a direct assay to measure the lac promoter activity of a focal evolved clone G+L3-1, which shows an approximate 2-fold increase in maximal Plac-GFP expression [29]. We found that expression from the lac promoter was significantly higher in the G+L3-1 clone than the ancestor and this increase agreed with our estimate based on flow cytometry (Figure 4B). Measurements taken at two time-points during exponential growth gave similar promoter activity estimates for all strains tested, indicating that promoter activities are representative of the lac system at steady state.
Fig. 4. Changes in maximal lac expression for evolved clones. 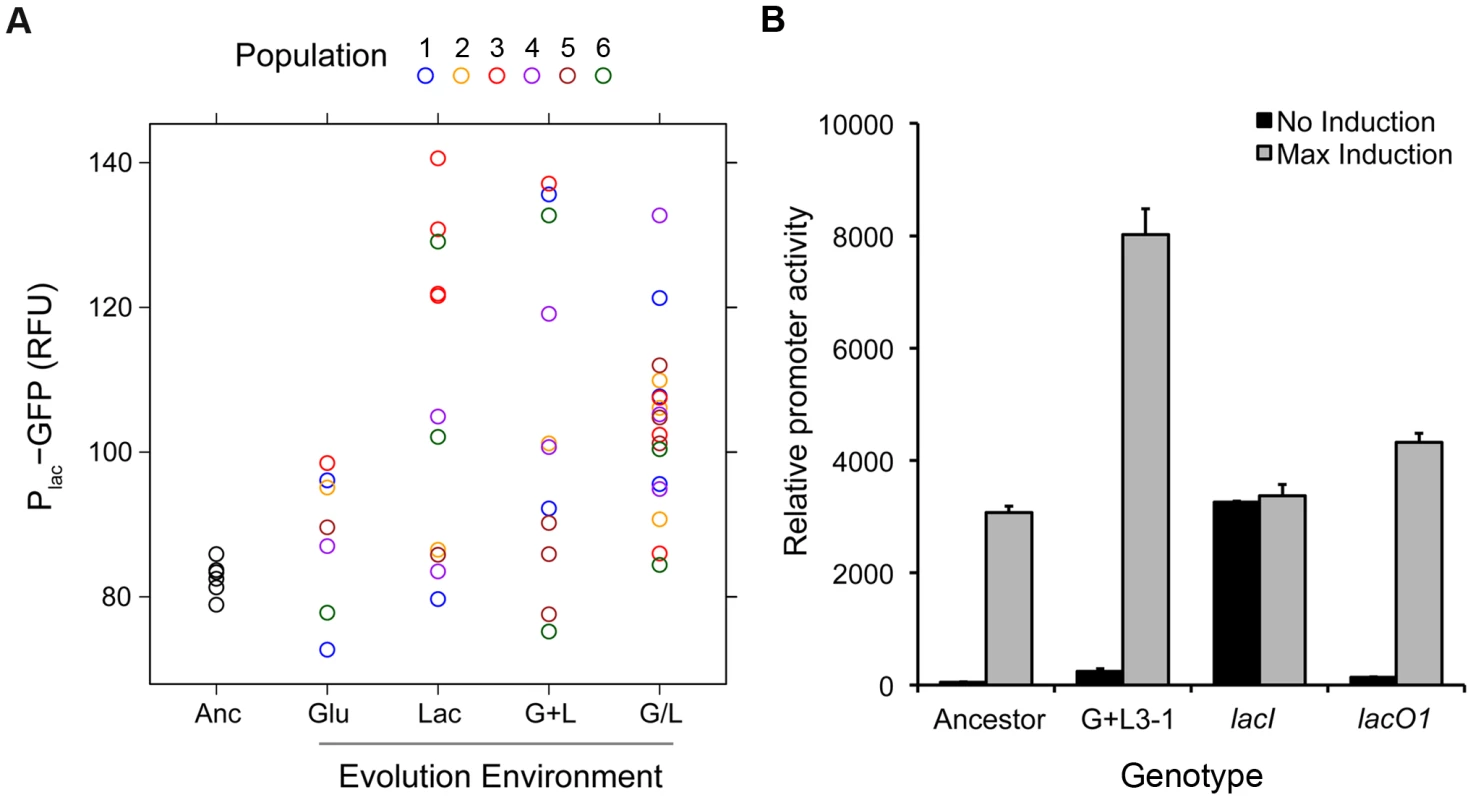
A) Steady state Plac-GFP levels (RFU) are shown for evolved clones grown in the presence of 100 µM TMG. Clones are divided into categories by evolution environment: Glu, Lac, G+L and G/L. Markers of the same color denote clones recovered from the same evolved population. Data points are the average of two independent replicates. As a reference, data for six independent ancestral (Anc) samples is also shown. B) Native lac promoter activity for the ancestor, evolved clone G+L3-1 and the ancestor with lacI and lacO1 mutations. Basal promoter activity was measured for strains grown in the absence of inducer, and maximum promoter activity was measured in the presence of saturating levels of inducer. Lastly, we measured the expression level of the CRP activity reporter (PlacO(-)-RFP) as a function of inducer concentration. All evolved clones showed a unimodal distribution with a mean response that was independent of inducer concentration, suggesting that, similar to the ancestor, evolved clones maintained predominantly non-cooperative interactions between the LacI and CRP regulators and that CRP activity is independent of lac expression level (Figure S1).
Genetic basis of evolved changes in lac regulation
To determine the genetic basis of changes to lac regulation we first sequenced the main lac regulatory regions, lacI and Plac, of each evolved clone (excluding G/L5 clones whose lac regulatory region could not be amplified by PCR). We found that all but one clone classified as constitutive had either a deletion or insertion of a 4 bp sequence within the lacI gene, which results in a frame shift (Figure 5). This region of lacI has three 4 bp direct repeats and is known to be a mutational hotspot, accounting for ∼75% of all spontaneous lacI null mutations [30], [31]. Constitutive clone G/L4-3 had a nonsynonymous mutation in lacI conferring a leucine to glutamine change at residue 71. This mutation is predicted to cause a severe defect in the ability of LacI to repress lac expression [32]. In addition, we found that all clones that evolved a lower induction threshold contained a single base pair substitution in the primary LacI repressor binding site of the lac promoter (lacO1). We identified three unique lacO1 mutations, two of which occurred twice in independent G+L populations (Figure 5). Previous work has demonstrated that all three lacO1 mutations can reduce the binding efficiency of LacI to the operator, thereby reducing repression of the lac operon in the absence of a specific inducer [33]–[36]. Lastly, populations in the bimodal inducer response class did not have any mutations within lacI or the lac promoter region, even though other aspects of their lac regulation, such as the region of bimodalilty and TMG½Max values, were altered (Figure 2).
Fig. 5. Identification lac mutations in evolved clones. 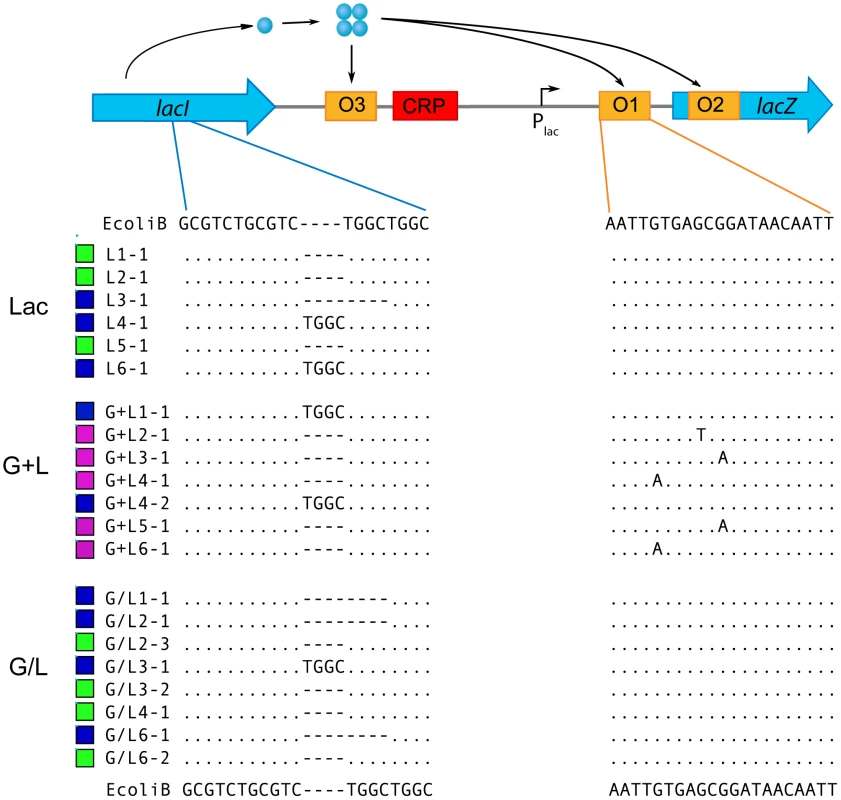
Sequence alignments for evolved clones relative to the ancestor are shown for the lacI mutation hotspot region (left) and the primary lacO1 operator (right). Clones were chosen that represent the diversity in lac genotypes within each population. Colored boxes by sequences indicate the inducer response type for each clone. Bimodal is shown as green, constitutive is shown as blue and lower threshold response is shown as purple. No sequence was obtained for the G/L5 clones. Sequence alignments are not shown for Glu clones since all have wt lac regulatory sequences. Regulation of evolved promoter reporters
The inducer response profiles presented above used a GFP reporter controlled by the ancestral Plac promoter. It is possible that this promoter does not accurately reflect the activity of the mutant promoters that evolved in the G+L isolated strains. To investigate the effect of this difference we constructed a version of the Plac-GFP reporter with the lacO1G11A mutation found in the G+L3 and G+L5 populations (PlacO1-GFP) and used it to generate inducer response profiles in the G+L3-1 clone (Figure S2A). Inducer response profiles for G+L3-1 with PlacO1-GFP are qualitatively similar to those obtained with the Plac-GFP reporter, showing a graded inducer response and a lower induction threshold. These characteristics were confirmed with β-Gal enzymatic assays that directly examined mean LacZ activity (Figure S2B).
Reconstruction of lac mutations and inducer response profiles
The lacI insertion/deletion and lacO1 mutations are clearly good candidates to explain the evolved changes in lac operon regulation, but additional mutations may also be influential. To test the effect of the identified mutations on lac expression, we added the evolved lacI 4 bp deletion allele and the lacO1G11A mutation individually into the ancestral reporter strain. If these mutations play a major role in determining the evolved regulatory change, we expect these constructed strains to have inducer profiles similar to those of the evolved strains from which the mutations were isolated. Indeed, the lacI deletion recapitulated the inducer response profiles found in all constitutive clones, causing Plac-GFP expression levels to become independent of TMG concentration (Figure 6A, lacIΔTGGC versus Figure 3A, Constitutive). Similarly, adding the lacO1G11A mutation into the ancestral background recreated the lower threshold/graded inducer response associated with evolved clones harboring mutations in lacO1 (Figure 6A, lacO1G11A versus Figure 3A, Lower threshold). The TMG½Max value for the lacO1G11A reconstructed strain was 4 µM, which is similar to the TMG½Max of evolved clones with this mutation (G+L3-1, 8 µM; G+L5-1, 4 µM), demonstrating that the lacO1 mutation is the primary cause of evolved changes in inducer sensitivity.
Fig. 6. Contribution of lac mutations to evolved inducer responses. 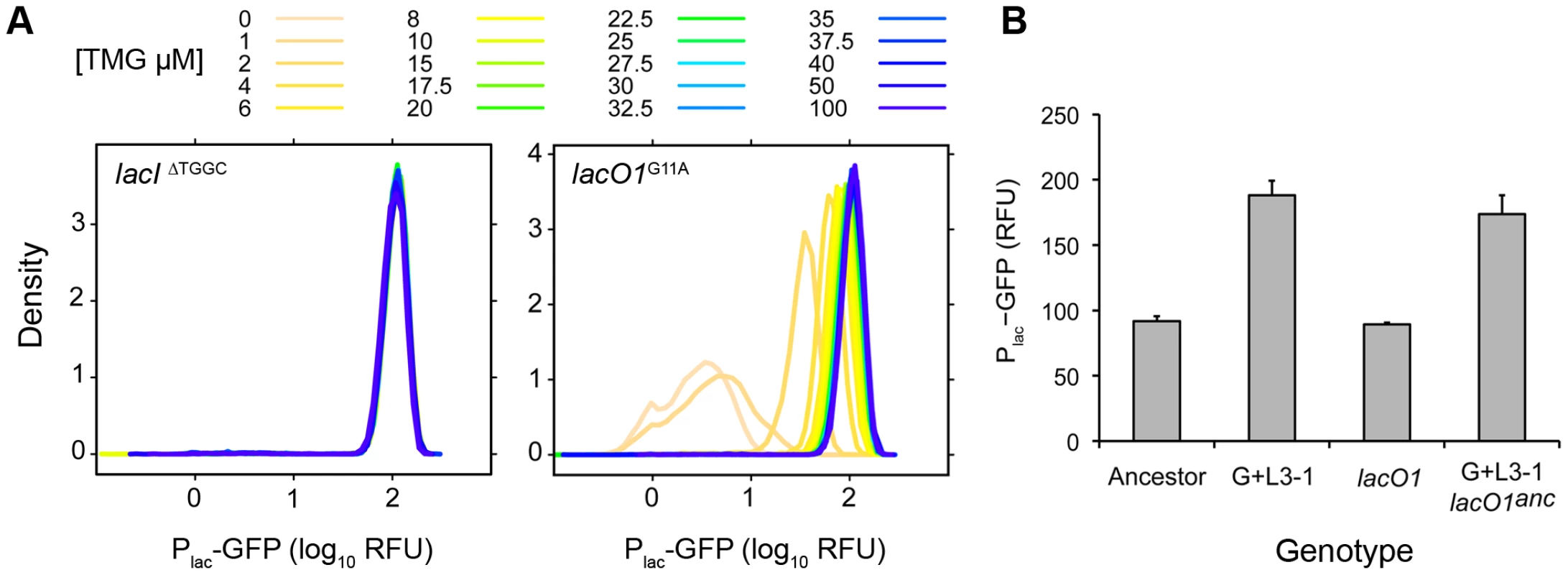
A) Inducer response histograms for reconstructed lacIΔTGGC and lacO1G11A mutants. The lacI mutation confers constitutive lac expression, whereas the lacO1 mutation confers a lower induction threshold and a graded response to inducer. B) Effect of the lacO1 mutation on the maximum lac expression level. Mean Plac-GFP expression levels during growth in saturating levels of inducer (100 µM TMG) are shown for the ancestor, evolved clone G+L3-1, lacO1G11A single mutant and the G+L3-1 clone with lacO1 reverted to the ancestral sequence (G+L3-1 lacO1anc). Standard error is shown, n = 4. The lacI and lacO1 mutations can explain many, but not all, of the lac regulation changes seen in the evolved strains. Specifically, neither mutation confers the increase in maximal expression that was seen in most evolved clones (Figure 6B, Figure 4B). To conclusively establish that additional evolved mutations impacted lac regulation in the G+L3-1 evolved clone, we replaced the lacO1 mutation with the ancestral operator sequence. This strain maintained the ∼2-fold increase in maximum Plac-GFP expression level relative to the ancestor, indicating that mutations outside the canonical lac operon regulatory network contribute to evolved changes in lac regulation (Figure 6B). Whole genome sequencing of G+L3-1 found 6 additional mutations (in the genes or gene regions rbsDACB, ECB_00822, fabF, sapF, mreB and malT), none of which are in genes previously characterized as affecting lac operon expression.
Fitness analysis of lacI and lacO1 mutants
That multiple lacO1 and lacI mutations arose independently in replicate populations and affect a trait of relevance in the evolution environments suggests that they confer a selective advantage. Further, the presence of lacO1 mutations exclusively in the G+L evolved populations suggests that they confer a greater advantage in this environment than do lacI mutations. To test these predictions we introduced the lacI and lacO1 mutants individually into the ancestor and measured the fitness of these constructed strains relative to the ancestor in each of the four evolution environments. We found that lacO1 and lacI mutations conferred a fitness benefit in all environments containing lactose (mean relative fitness effect and 2-tailed t-test: Lac environment. lacI: 8.9%, P<0.001; lacO1: 7.9%, P<0.001. G+L environment. lacI: 8.4%, P<0.001; lacO1: 8.0%, P = 0.001. G/L environment. lacI: 6.6%, P<0.001; lacO1: 2.2%, P = 0.02). By contrast, both mutations imposed a small fitness cost in the glucose environment, consistent with them not being observed in Glu populations (Glu environment: lacI: −4.0%, P<0.001; lacO1: −1.8%, P = 0.16) (Figure 7). Intriguingly, despite the lacO1 mutations being significantly overrepresented among G+L populations, they did not confer a greater fitness advantage in this environment. Similarly, lacI mutations did not confer a greater advantage in Lac or G/L environments, where they were dominant. To address the possibility that some non-transitive interaction could complicate our indirect comparison of the relative fitness benefits of the two mutations, we also performed direct competitions between the two constructed strains. Again, we found that the fitness of the lacI mutant was not significantly different from that of the lacO1 mutant in any environment (fitness of lacI relative to lacO1, 2-tailed t-test: Glu environment: 0.5%, P = 0.69; Lac environment: 0.4%, P = 0.60; G+L environment: −0.5%, P = 0.39; G/L environment: 2.1%, P = 0.05) (Figure 7).
Fig. 7. Effect of lac mutations on fitness. 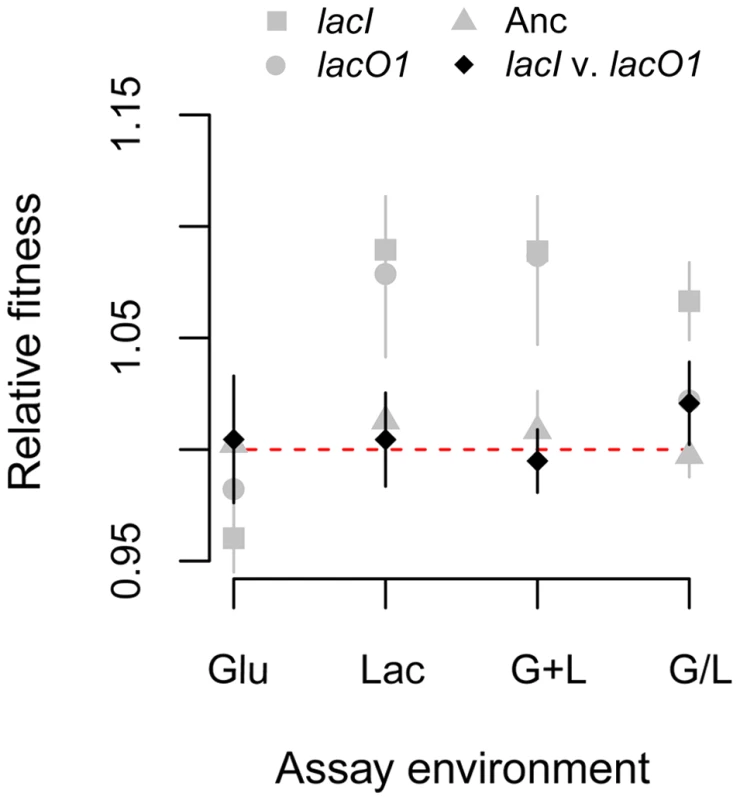
A) Fitness of lacI and lacO1 mutants in the four evolution environments. Competitions were performed for lacI versus the ancestor, lacO1 versus the ancestor and lacI versus lacO1. As a control, we included the ancestor competed against itself. The red dotted line indicates a relative fitness of 1 (no fitness difference). 95% confidence intervals are shown for each competition (n≥8). Physiological basis of the fitness advantage of lacI and lacO1 mutants
To examine the basis of the fitness effects conferred by the lacI and lacO1 mutations, we quantified their effect on population growth dynamics, focusing on the transitions between glucose and lactose utilization that are encountered in the G+L and G/L environments (environments in which lacO1 and lacI mutants predominated). We found that the lacI and lacO1 mutations significantly decreased the lag phase, relative to the corresponding ancestral alleles, following a shift from growth on glucose to growth on lactose (a part of the G/L environment) (Figure 8A, Table 2). Furthermore, both lacI and lacO1 mutations eliminated the diauxic lag phase measured for the ancestor when switching from glucose to lactose utilization in the G+L environment. Neither mutation had a significant effect on lag time following a shift from growth in lactose to glucose, indicating that the change in lag time was specific to lactose utilization. The maximum growth rate constant (μMax) for lacI and lacO1 mutants was not significantly different from that of the ancestor, except during growth on lactose in the G+L environment (Figure 8A, Table 2). In this case, both lacI and lacO1 mutants had significantly higher growth rates than the ancestor. In agreement with fitness measurements, strains containing the lacI and lacO1 mutations show no significant differences in the length of lag phases or maximum growth rates in any of the environments tested. To decrease experimental noise in these experiments, we used higher sugar concentrations than present in the evolution experiment. Analysis of growth dynamics using the exact evolution environments yielded qualitatively similar results (Figure S3).
Fig. 8. Physiological characterization of lac mutants. 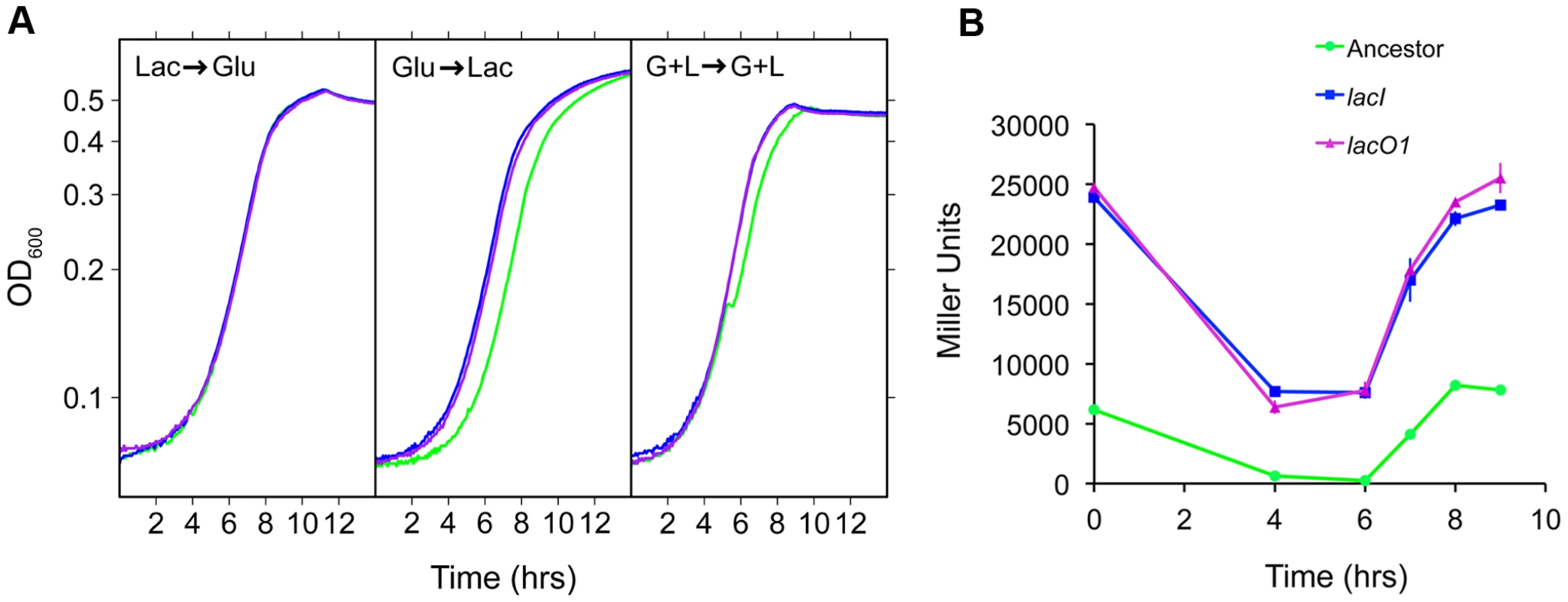
A) Growth curves for the ancestor (green), and reconstructed lacI (blue) and lacO1 (purple) mutants. Conditions used were: Lac→Glu, Glu→Lac and G+L→G+L, where the sugars indicate pre-conditioning and measurement environments, respectively. These transitions correspond to those present in the G/L and G+L evolution environments. OD values are plotted on a log10 scaled axis. B) LacZ expression time course for the ancestor and the ancestor with lacI and lacO1 mutations during growth in the G+L evolution environment. Values are the average of two independent replicates with standard deviation shown. Tab. 2. Growth parameters for ancestor, lacI and lacO1 mutants. 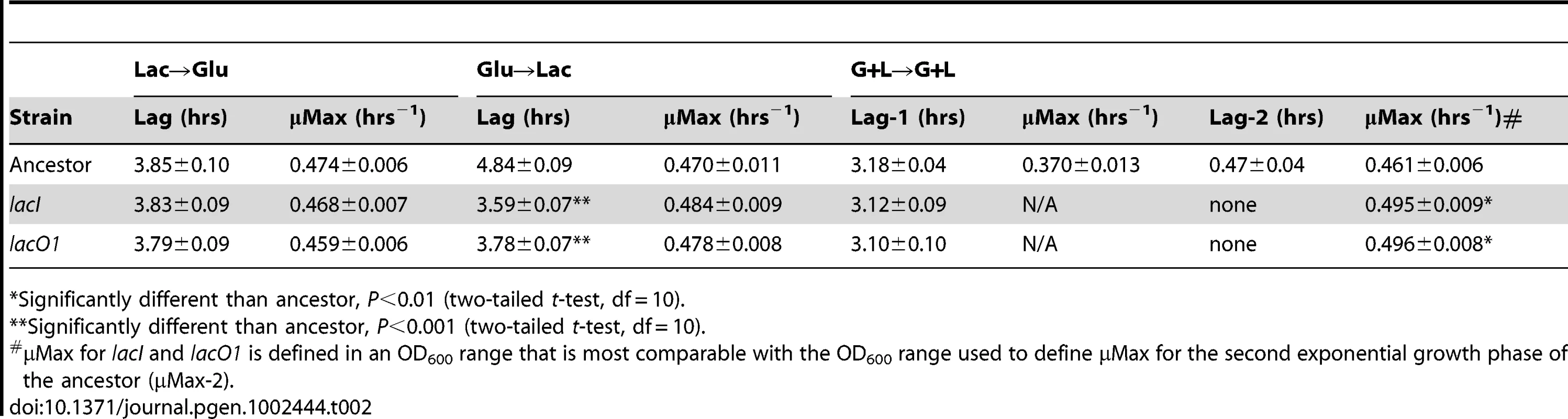
*Significantly different than ancestor, P<0.01 (two-tailed t-test, df = 10). In contrast to the lacI mutant, the lacO1 mutant can repress lac expression to some degree (Figure 2, Figure 3A). To examine how this difference in regulation translates to the evolution environment, we measured LacZ activity of the ancestor and the lacI and lacO1 mutants in the G+L environment (Figure 8B). The ancestor shows the anticipated LacZ expression profile, with activity decreasing 97% during growth on glucose and increasing back up to the initial level of activity after switching from growth on glucose to lactose. Interestingly, lacI and lacO1 mutants showed indistinguishable LacZ activity profiles, with LacZ activity much higher than the ancestor at all time points during growth in G+L medium. These results suggest that the relatively low levels of lactose present in the G+L environment induce the lacO1 mutant lac operon, even in the presence of glucose concentrations sufficient to prevent induction of the ancestral lac operon. Analysis of the steady state levels of lac expression during growth in DM+Glu (2 mg/mL) with and without lactose (87.5 µg/mL) supports this conclusion, with repression of lac expression only occurring in the absence of lactose (Figure S4). In summary, in the ancestral background, lacO1 and lacI mutations are indistinguishable with respect to their effect on fitness, growth dynamics and lac expression dynamics in the G+L environment.
Genetic interactions with lac mutations and their effect on fitness
A possible explanation for the success of lacO1 mutations in the G+L populations despite them not conferring any advantage relative to more frequent lacI mutations is that they interact synergistically with other mutations that fixed during the evolution of these populations [37]. To test this, we compared the fitness advantage conferred by lacO1 and lacI alleles in the ancestral background relative to the advantage they confer in the genetic background of evolved clone G+L3-1, which substituted a mutation in lacO1 during evolution in the G+L environment (Figure 5). If epistasis was important in selection of lacO1 alleles in the G+L environment, we predicted that the fitness advantage conferred by the lacO1 mutation would be significantly larger in the background of this evolved clone than in the ancestral background, and that this positive effect will be less pronounced for the lacI mutation. The lacO1 mutation did confer a bigger benefit in the evolved background (two tailed t-test, fitness in evolved background minus fitness in ancestral background = 4.4%, P = 0.03) (Figure 9). However, a similar effect was seen for the lacI mutation (two tailed t-test, fitness in evolved minus fitness in ancestral background = 4.8%, P<0.001) and there was no significant difference in the fitness conferred by the lacI and lacO1 mutations in the evolved background when they were directly competed against one another (lacI versus lacO1 in evolved background, relative fitness difference = 1.3%, P = 0.07). Therefore, epistatic interactions increase the fitness effect of both the lacI and lacO1 mutations in the evolved background, but do not explain the enrichment of lacO1 mutations in populations evolved in the G+L environment.
Fig. 9. Fitness effect of lacI and lacO1 mutations depend on genetic background. 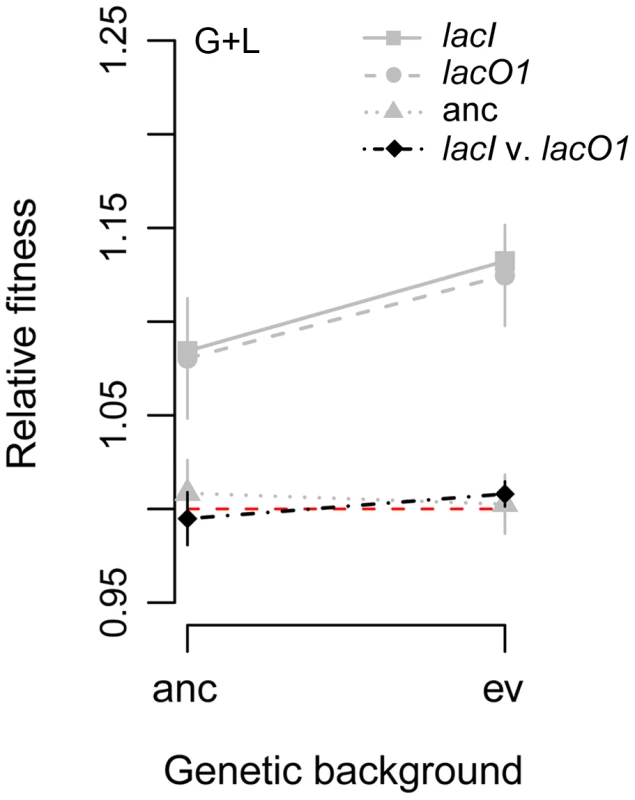
The fitness effect of lacO1 and lacI mutations were measured in the ancestor and the G+L3-1 evolved genetic backgrounds when competed in the G+L environment. Gray points indicate fitness effect in competitions against the corresponding progenitor strains that do not have the added lac mutation; black points indicate fitness of lacI and lacO1 mutations competed directly against each other. Lines connect competitions of the same type but in different genetic backgrounds. Alternative explanations of the occurrence of lacO1 mutations
In the absence of a measurable difference in the fitness conferred by lacI and lacO1 mutations, what could explain our finding that lacOI mutations only occurred in populations evolved in the G+L environment? If the lacI and lacO1 mutations occurred with equal probability, the distribution of mutation types over selection environments we observed is unlikely to have occurred by chance (Fisher's exact test omitting the polymorphic G+L population, P = 0.002). In fact, our observations are even more unlikely than this test implies because the lacO1 mutation will almost certainly occur at a much lower rate than the lacI mutation. The frequency of lacI null mutations in the E. coli strain used in this experiment is ∼3×10−6 per generation (Hana Noh and TFC unpub. obs.), which is in good agreement with a previous measurement for E. coli K12 [31]. By contrast, we conservatively estimate the lacO1 mutation frequency to be <3×10−9 per generation (Materials and Methods). Without some unique advantage, it is difficult to see how lacO1 mutations could reach high frequency in any population, let alone predominate as in the G+L populations. The ∼1000 fold difference in mutation frequency does, however, provide an explanation for the absence of lacO1 mutations in Lac or G/L environments.
We considered the possibility that cross-contamination could be responsible for the occurrence of identical lacO1 mutations in two of the G+L populations, which would reduce the number of independent lacO1 populations to three. This is unlikely since replicate populations were not propagated in adjacent wells, and no evidence for cross contamination was found during the course of the experiment (see Materials and Methods). Furthermore, the environmental association remains significant even if only unique lacO1 mutations are considered (Fisher's exact test omitting one polymorphic population and populations with non-unique lacO1 mutations, P = 0.006).
Finally, it is possible that the statistical association between the lacO1 mutation and the G+L evolution environment, despite its high significance, is nevertheless spurious. In this case, repeat evolutionary experiments of the ancestor in the same G+L and G/L environments (where selection of lacI mutations was most consistent) would not be expected to lead to significant mutation-environment association. To test this, we began 12 ‘replay’ populations in each of the G+L and G/L environments, founding each population with the same ancestor as used in the original experiment. Every 100 generations, we examined the frequency of lacI and lacO1 mutations in each population using LacZ indicator plates and by sequencing select clones. Although lacO1 mutants were detected in the majority of G+L replay populations, their frequencies never rose above 4% in any one population (Table S1). In contrast, lacI mutants rose to high frequency, accounting for >98% of clones in all populations by 400 generations. Replay experiments in the G/L environment followed a similar trend, although lacO1 mutations were detected in only 2 of the 12 populations over the course of the experiment (Table S2). In summary, despite being highly improbable, the failure of lacO1 mutations to establish in our replay experiments suggests that their enrichment over competing lacI mutations in the original G+L populations may have occurred by chance.
Discussion
We sought to test whether evolution in environments that differed only in the availability and presentation of lactose selected for changes in the regulation of the lac utilization network. Our analysis of inducer response profiles found three broad classes of lac regulation change among evolved clones. Two of these classes, constitutive expression and a lower threshold/graded inducer response, represent substantial changes from ancestral regulation and were observed only in populations evolved in environments containing lactose. Sequencing of lac regulatory regions in evolved clones uncovered mutations in the lac repressor (lacI) and the primary lac operator (lacO1) that correlated with the constitutive and the lower threshold/graded inducer response, respectively. Addition of these mutations to the ancestor demonstrated that they explained many, but not all, of the broad scale changes in regulation we observed and that, by themselves, they can confer fitness benefits in environments containing lactose. These fitness benefits were relatively large, representing 20%, 27% and 28% of the total mean fitness improvement in the Lac (lacI mutation), G+L (lacO1 mutation) and G/L (lacI mutation) evolved populations, respectively [13]. Together these results indicate that regulatory changes were common, complex — occurring both within and outside of the recognized lac regulatory elements — and adaptive.
Extensive previous study of lac operon regulation offers the opportunity to connect the genetic and phenotypic changes we observed. Twenty-one of the 22 lacI mutants we identified mapped to a mutational hotspot within lacI [30], [31]. These mutations generate a frameshift in the coding sequence that results in expression of a nonfunctional repressor, which provides a good explanation for the complete loss of negative regulation we observed in lacI mutants. Similarly, the three lacO1 mutations we identified in the G+L evolved populations have been reported as reducing the binding affinity of LacI for this binding site [33]–[35]. Consistent with the mutations reducing, but not completely preventing, LacI binding, strains containing lacO1 mutations are able to repress the lac operon, but are induced at much lower TMG concentrations than the ancestor. lacO1 mutations also conferred a graded response to increasing inducer concentration, which contrasted with the canonical bimodal response of the ancestor. The same regulatory outcome was demonstrated by Ozbudak et al. (2004), who found that decreasing the effective concentration of LacI by providing extra copies of lacO1 binding sites resulted in a graded unimodal induction of the lac operon [19]. The similarity in regulatory changes suggests that the graded induction we observe is a consequence of decreased LacI-lacO1 affinity, reducing the effective concentration of LacI repressor. More generally, our results support the concept that small changes in the activity of cis-regulators have the potential to transform the output of a regulatory network between binary and graded responses [38].
Both lacI and lacO1 mutations were shown to confer significant fitness benefits in the three lactose containing evolution environments (Lac, G+L and G/L). Analysis of the growth dynamics of strains containing only these mutations indicated that a large part of this benefit is due to a reduction in lag phase when switching from glucose to lactose utilization. Interestingly, when added to the ancestor, both lacO1 and lacI mutations abolished the diauxic lag that separates glucose and lactose growth phases during growth in the G+L environment. This phenomenon is well documented for lacI mutants [39], but to the best of our knowledge has not been demonstrated for lacO1 mutants. In the case of lacI mutants, constitutive expression of the lac operon primes the cell for utilization of lactose as soon as glucose resources are exhausted. In contrast to lacI mutants, lacO1 mutants are still capable of repressing lac expression in the absence of inducer. However, when grown in media containing both glucose and lactose the lacO1 mutation essentially phenocopies a lacI mutant, causing constitutive lac expression. Evidently glucose-mediated blockage of lactose import through LacY (inducer exclusion) is insufficient to prevent lactose accumulating in cells to a concentration sufficient to allow lac operon induction in lacO1 mutants [39], [40].
The loss of lac repression in lactose (3/6), but not glucose (0/6), evolved populations is consistent with the ‘use it or lose it’ hypothesis [23], [24]. This hypothesis proposes that negative regulation will be maintained during evolution in environments in which gene products, in this case the LacI repressor, are used because mutants that lose the repressor will needlessly express the lac operon and be selected against. If a repressor is seldom used, as in the lactose evolution environment, loss of function mutations will not be effectively selected against and can fix through genetic drift. However, in its simplest form, this mutation accumulation mechanism does not capture the dynamics of the regulatory changes we see. First, loss of the lacI repressor actually confers a benefit during growth on lactose, so that underlying mutations will increase in frequency faster than expected if they were neutral. Second, repressor mutations also occurred in environments where glucose was just as common as lactose. Analysis of growth curves suggest a mechanism for this; lac repressor mutants were able to quickly begin growth following a switch from glucose to lactose. Fitness measurements indicated that this advantage outweighed the cost of unnecessary lac expression during growth in glucose.
Given the large benefit conferred by lacI mutations in the Lac environment, it is interesting that only three of the six Lac populations were enriched for lacI mutations. We identify two possible explanations for this observation. First, clonal interference may have resulted in lacI mutations being outcompeted by higher effect beneficial mutations. Second, populations that did not enrich lacI mutations may have fixed alternative mutations that genetically interact with lacI mutations to reduce their fitness benefit. To distinguish between these possibilities, we are continuing the evolution experiment and tracking the frequency of lacI mutations in the Lac populations. In addition, we are examining the fitness benefit conferred by lacI mutations when introduced into clones from Lac evolved populations that did not fix lacI.
We can explain why lac mutations occurred only in lactose containing selective environments. A second layer of environment specificity is less clear; why do lacO1 mutations only reach high frequency in the G+L environment? The lacI and lacO1 mutations had no differential effect on fitness in either the ancestor or an evolved background and conferred indistinguishable growth dynamics in all evolution environments. Without a selective advantage over lacI mutations it is difficult to understand how lacO1 mutants were fixed in 4 of 6 G+L populations, especially considering that the rate of lacI mutations is likely on the order of 1000-fold greater than for lacO1 mutations. In the absence of a plausible mechanism to explain enrichment of lacO1 mutations in the G+L environment, we investigated whether environment-specific selection of lacO1 mutants was reproducible. This was not the case, with all 12 of the independent replay populations selected in G+L eventually fixing (>98%) lacI mutations. It remains formally possible that subtle differences in media or experimental conditions during competition assays or the replay evolution experiments could affect the fitness advantage experienced by lacO1 mutants in focal G+L evolved populations. However, taken at face value, the different outcome between replay and primary populations suggests that, notwithstanding mutation rate differences and the strong statistical association between environment and mutation type, the enrichment of lacO1 mutations over lacI mutations in the G+L environment might have occurred by chance.
Models incorporating interactions between key regulatory elements can successfully predict aspects of lac operon regulation [18], [19]. Nevertheless, recent studies demonstrate that regulation of the lac operon is evolutionarily plastic, such that interactions can arise or be altered to fine tune regulation and better fit E. coli to its environment [11], [14], [41]. By characterizing changes in regulation without a priori assumptions as to the nature of regulatory changes or the mutations causing them, we were able to identify evolved clones with changes in lac regulation that are likely due to novel interactions with mutations in genes outside of the canonical lac operon. Two results support this conclusion. First, we identified numerous clones with maximal steady state expression levels of the lac operon that were higher than the ancestor. Further examination of evolved clone G+L3-1 indicated that the increase in maximal lac expression level could not be explained by the lacO1 mutation present in this clone. Second, the fitness benefit conferred by the lacO1 mutation in this same evolved clone was significantly greater than in the ancestor, indicating that one or more evolved mutations interact with the lacO1 mutation to determine its effect on fitness. Whole genome sequencing of G+L3-1 identified six additional mutations, however, none of these mutations mapped to the lac operon or genes known to directly impact CRP-cAMP activity. It seems likely, therefore, that one or more mutations in the G+L3-1 clone have directly or indirectly led to new regulatory control of the lac operon.
Is there an optimal level of lac expression in each of the three lactose environments? Dekkel and Alon (2005) found that, in the presence of a gratuitous inducer, lac operon expression evolved to a level predicted on the basis of a cost-benefit analysis, dependent on the concentration of lactose in the selection environment [14]. Our results support the idea that maximal expression level is a plastic feature of the lac operon and can be tuned to best fit the environment. At this time, however, we do not know the genetic or molecular basis for the widespread increase in maximum lac expression observed in many evolved clones. Possible ‘local’ explanations include: increases in the level of cAMP, mutations in the lacZYA genes that affect mRNA stability, or changes in DNA supercoiling that increase lac operon transcription. It is also possible that changes in lac maximum expression reflect an alteration in some global process. For example, changes in the function or concentration of ribosomes could affect expression of all genes. In future work we aim to identify the evolved mutations that are responsible for changes in maximum lac expression and then construct strains that will allow us to test the adaptive value of different expression levels as well as probe the underlying molecular mechanisms.
An additional widespread change in lac regulation was that evolved clones displaying bimodal inducer responses tended to also have higher induction thresholds (TMG½Max) than the ancestor. This trend was not environment specific, occurring in clones isolated from Glu, Lac and G/L evolved populations. However, the parallel and large-scale increases in induction threshold observed for Glu-evolved clones suggests that this change in lac regulation is a direct or correlated response to adaptation. The mechanistic bases of increases in induction threshold are currently not understood, but could be the result of both direct and indirect mechanisms. For example, reduction in the activity and/or concentration of the permease LacY could increase the concentration of extracellular inducer required to achieve intracellular levels of inducer sufficient to inactivate LacI. In glucose evolved populations, changes in LacY activity may result from mutations in the PTS system that optimize glucose transport but lead to elevated levels of unphosphorylated EIIAglc, which is a known inhibitor of LacY activity [40]. Alternatively, higher growth rates of evolved strains will also tend to decrease the steady state intracellular concentration of inducer thereby increasing the external concentration required for induction of the lac operon. Further study will be required to discern between these and other hypotheses.
In summary, we have identified and characterized widespread changes in lac operon regulation that occurred during selection of replicate populations in different lactose containing environments. In our view, the most important aspects of our findings are how common these changes were and that they likely involve mutations both within and outside of the set of genes that are recognized as regulating the lac operon. Identification of these changes will provide a rare insight into how regulatory networks can be rewired in response to an environmental change.
Materials and Methods
Strains and growth conditions
The ancestral strains used for experimental evolution studies were E. coli B REL606 (ara−) and an otherwise isogenic ara+ derivative, REL607 [42]. For routine culturing, cells were grown in lysogeny broth (LB) medium [43]. Davis minimal (DM) medium was used for experimental evolution and subsequent analysis of evolved clones [42]. Sugars were added to base DM medium at the following concentrations to make single and mixed resource environments: glucose (Glu) 175 µg/mL, lactose (Lac) 210 µg/mL, and Glucose+Lactose (Glu+Lac) 87.5 µg/mL and 105 µg/mL, respectively. These concentrations were chosen to ensure that each environment supports approximately the same stationary phase density of bacteria (∼3.5×108 cfu/mL) [13]. Strains were grown at 37°C unless otherwise stated. T medium contains 1% Bacto tryptone, 0.1% yeast extract and 0.5% sodium chloride. Antibiotics were used at the following concentrations: chloramphenicol (Cm), 20 µg/mL; kanamycin (Km), 35 µg/mL; streptomycin (Sm), 100 µg/mL.
Qualitative characterization of lac operon regulation within evolved populations
Evolved populations were screened for qualitative changes to lac operon regulation using TGX medium, which consisted of T agar plates supplemented with 0.5% glucose and 30 µg/mL of the colorimetric LacZ substrate 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-Gal). On this medium, the degree of blue coloration correlates with the degree of LacZ activity and, therefore, lac operon expression. Clones representing the diversity in LacZ activity and colony morphology within each evolved population were recovered, scored and stored for future analysis. TGX plates enabled us to distinguish between the ancestor and clones with lacO1 and lacI mutations (mutations that we subsequently identified in evolved populations), which appear white, faint blue and dark blue, respectively.
Quantitative tools for analysis of changes to lac operon regulation
We developed a dual fluorescent reporter system that enabled us to independently monitor LacI and CRP activity with single cell resolution. In this system the native lac promoter (containing LacI binding sites O1 and O3), coupled with an optimized ribosomal binding site, drives expression of the fast maturing GFP derivative, GFPmut3.1 [44]. This Plac-GFP module was cloned into a mini-Tn7 delivery vector, to make pTn7-Plac-GFP, and integrated into the chromosome in a site-specific manner [45].
Expression of GFP from the Plac-GFP reporter depends on both LacI and CRP activity. To isolate these effects we developed a second reporter to monitor the contribution of CRP to lac operon expression independent of LacI. To do this, we constructed a version of the lac promoter (PlacO(-)) with defined mutations in the O1 and O3 operators which have been shown to prevent binding of the repressor LacI while maintaining the integrity of the major binding site for CRP [46]. This synthetic promoter was used to drive expression of the red shifted fluorescent protein DsRed express2 [47]. This reporter was cloned into a stable low copy plasmid (∼5 copies per cell) to generate pRM102-3. Control experiments confirmed that the introduced mutations abolished LacI binding while retaining promoter response to cAMP dependent activation of CRP (Figure S5). This reporter system differs from a previously published system in three important ways [19]. First, the fluorescent signal is bright enough to allow analysis by flow cytometry. Second, the CRP reporter is a derivative of the lac promoter that has the LacI binding sites deleted, rather than an unrelated reporter subject to regulation by CRP. Third, DsRed express2 shows low cytotoxicity relative to other commonly used RFP's [47].
Transformation of evolved clones with LacI and CRP reporters
The Tn7-Plac-GFP reporter was integrated into the chromosome of focal evolved clones by tri-parental mating with the MFDpir (pTn7-Plac-GFP) donor strain and the MFDpir (pTSN2) helper strain [48]. Strains were grown overnight in LB and washed once in LB before being resuspended in 1/10 volume LB. Recipient, donor and helper strains were mixed at a 4∶1∶1 ratio and incubated on an LB plate for 3 hrs. Transconjugants were selected on LB plates supplemented with Km and Sm. A PCR assay was used to confirm that the mini-Tn7 had inserted into the single characterized chromosomal integration site [49]. Finally, all reporter strains were screened to ensure that they retained the LacZ phenotype that formed the basis of their initial selection from evolved populations.
Inducer response profiles
Strains were inoculated from glycerol stocks into 500 µL LB media in 2 mL deep-well plates (Phenix Research) and grown overnight at 37°C on a microplate shaker at 750 rpm (Heidolph Titramax 1000). After overnight growth, cultures were diluted 1∶1000 into 500 µL DM+0.4% glycerol and incubated for a further 24 hrs at 37°C. To determine the inducer response of each strain, overnight cultures were diluted 1∶1000 into separate wells of a 96-well plate containing DM+0.4% glycerol supplemented with TMG at concentrations ranging from 0 to 100 µM and incubated at 37°C for a further 15–18 hrs. This time period encompassed approximate steady state reporter expression in ancestral and evolved clones (Figure S6). TMG induces the lac operon by binding and inactivating the LacI repressor. Unlike the natural inducer, allolactose, TMG is metabolically stable, which is advantageous for quantitative studies because it allows accurate control of TMG concentrations through the course of the experiment. Importantly, import of TMG into the cell is dependent on the lactose permease LacY, which is not true of other commonly used synthetic inducers such as Isopropyl β-D-1-thiogalactopyranoside (IPTG).
Flow cytometry was performed with a FACScalibur (BD Biosciences) equipped with a high throughput sampler. PMT voltages for the flow cytometer were set as follows: SSC-H - E02, FSC-H 580 V, FL1-H 800 V and FL2-H 800 V. The threshold was set at 250 on the SSC-H channel. For each expression assay, a total of 25,000 events were captured at a rate of 1000–3000 events/s. Data was acquired in log mode with no hardware compensation. We examined day-to-day reproducibility by measuring ancestral inducer response curves on 5 separate days using independent cultures (Figure S7). The level and distribution of Plac-GFP expression in response to TMG was similar between replicates, indicating that our protocol for measuring inducer response profiles was robust.
Manipulation and analysis of flow cytometry data
Routine analysis of the flow cytometry data and plotting of inducer response profiles was performed in R (version 2.12) using the Bioconductor packages FlowCore and FlowViz [50]–[52]. To control for cross talk between GFP and RFP reporter detection, a compensation matrix was calculated using the appropriate single reporter control strains and used to correct flow cytometry data post acquisition. To minimize detection noise and enrich for cells of similar size, all data was filtered with FlowCore's norm2Filter on the FSC-H and SSC-H channels using the default settings. This typically resulted in retention of 50–60% of all collected events.
For quantitative analysis of inducer responses, flow cytometry data were processed using Matlab (Mathworks, Inc.). An elliptical gate corresponding to a Mahalanobis distance of 0.5, centered at peak cell density in the FSC-SSC coordinates, was used to minimize the effects of varying cell size and granularity on the resulting fluorescence histograms. A custom bimodality detection algorithm (to be described elsewhere) was applied to determine the region of bimodality for each histogram. Inducer sensitivity was determined as the point where the population-mean of GFP expression was halfway between baseline and saturation.
β-Gal assays
Assays were carried out as described by Zhang and Bremer (1995) with modifications [53]. Specifically, 1–2 mL of cell culture was pelleted and resuspended in 250 µL unsupplemented DM medium to remove any remaining lactose. Cell concentration was estimated by measuring absorbance at OD600 with a microplate reader (Tecan). Cells were permeabilized by mixing 20 µL of cell suspension with 80 µL of permeabilization solution (100 mM Na2HPO4, 20 mM KCl, 2 mM MgSO4, 0.8 mg/mL cetyl-trimethylammonium bromide (CTAB), 0.4 mg/mL sodium deoxycholate, 5 µL/mL β-mercaptoethonal). After incubation for 10 minutes at room temperature, measurement of LacZ activity was initiated by adding 150 µL of o-nitrophenyl-β-D-galactoside (ONPG, 4 mg/mL) and mixing. Yellow color development was stopped by addition of 250 µL 1 M sodium carbonate and the reaction time recorded. Samples were centrifuged to remove cell debris and absorbance at OD420 was measured for 200 µL of the supernatant. LacZ activity (Miller units) was calculated as (1000×OD420)/(volume (mL)×OD600×reaction time (min)).
Growth analysis
Strains were inoculated into LB medium from freezer stocks, incubated overnight at 37°C and then diluted 1∶100 into DM medium supplemented with Glu, Lac or G+L at the concentrations used in the original evolution experiment. Strains were grown overnight and diluted 1∶100 into fresh DM media and incubated for a further 24 hrs to allow them to become physiologically adapted to their resource environment. To measure growth dynamics, a 1∶100 dilution of pre-conditioned culture was inoculated into 200 µL of DM supplemented with Glu, Lac or G+L in a clear 96-well plate. Concentration of sugars was either the same as in the evolution environments or to facilitate higher cell densities and correspondingly more precise OD measurements, were as follows: Glu, 0.2 mg/mL; Lac, 1.8 mg/mL; G+L, 0.2 mg/mL & 1.5 mg/mL, respectively. Incubation and optical density measurements were performed with a Bioscreen C plate reader (Oy Growth curves AB Ltd) at 37°C with continuous shaking and OD600 measured at 5 min intervals. The maximal growth rate constant (μMax) of each strain was calculated by linear regression of the plot of ln(OD600) versus time (hrs) using a sliding window of 10 data points. The steepest of these slopes was used to calculate μMax with units hrs−1. Lag time was calculated by extrapolating the μMax regression line to its intersection with OD600 = 0.06. Extrapolating to the initial density of individual growth curves would have been preferable, however, we found that these measurements were quite variable. To account for this we adopted the approach described by Friesen et al. (2004) where a constant reference density is used, assuming that starting biomass is similar for all strains [54]. For strains showing diauxic growth, we analyzed both growth phases separately to derive μMax-1 and μMax-2. The diauxic lag phase (lag-2) was calculated by determining the difference between the times when the regression lines for μMax-1 and μMax-2 intersect with the OD600 value coinciding with the end of the first growth phase.
Promoter activity assays
Assays were performed as described by Kuhlman et al. (2007) [29]. Briefly, steady state levels of LacZ activity were measured for strains grown in DM+0.2% glucose supplemented with a saturating concentration of the gratuitous inducer IPTG (1 mM). β-Gal assays were performed as described above except that color development was followed over time by measuring absorbance at OD420 and linear regression used to fit a line of best slope to the plot of OD420 vs time (min). LacZ activity (Miller units) was calculated as (1000×slope)/(assay volume (mL)×OD600). Doubling rate was measured for each strain in the experimental conditions by linear regression of log2(OD600) plotted against time, with the steepest of these slopes designated as the maximum doubling rate (doublings/hr). Promoter activity was calculated as the product of LacZ activity (Miller units) and doubling rate (hrs−1). To confirm that the inducer concentration we used was sufficient to completely inactivate LacI, we also measured expression from a lacI null mutant. This mutation caused a similar expression increase as induction with 1 mM IPTG, indicating that this level of inducer was sufficient to fully induce the lac operon.
Allele exchange and strain construction
Constructs and approaches used for the manipulation of each mutation were as follows. The PlacO1 and araA - mutations were introduced using a suicide plasmid approach that has been described previously [6]. Briefly, PCR products containing the relevant evolved alleles were separately cloned into pDS132 [55]. Resulting plasmids were introduced into recipients by conjugation and CmR cells (formed by chromosomal integration of the plasmid) were selected. Resistant clones were streaked onto LB+sucrose agar to select cells that lost the plasmid (which carries the sacB gene conferring susceptibility to killing by sucrose). These cells were then screened for the presence of the evolved alleles by a PCR-RFLP approach using the enzyme HaeIII (araA-) or on LacZ indicator medium (PlacO1). Putative allelic replacements of the evolved ara - and lacO1 alleles were confirmed by sequencing.
The lacI(-) mutation was obtained by isolating spontaneous mutants of relevant strains that could grow on minimal media supplemented with P-Gal as the only carbon source and confirmed by sequencing [56]. We isolated independent lacI(-) mutants that had either insertion or deletion mutations in a previously identified mutational hotspot [57]. Preliminary experiments indicated that these mutant types had identical fitness in each of the environments used here. For this reason, we used only the deletion mutant in the experiments reported in Results.
Fitness assays and analysis
The fitness of constructed strains was measured relative to the ancestor strain used to found the evolution experiment or directly to each other. Competing strains contained opposite Ara marker or lac regulation types, which allowed them to be distinguished on tetrazolium arabinose (TA) [42] or LB+X-Gal indicator medium, respectively. Before each fitness assay, competing strains were grown separately for one complete propagation cycle in the environment to be used in the assay so that they reached comparable cell densities and physiological states. Following this step, competitors were each diluted 1∶200 into the assay environment. A sample was taken immediately and plated on indicator plates to estimate the initial densities of the competing strains. At the end of the competition a further sample was plated to obtain the final density of each competitor. The fitness of the test strain relative to the reference strain was calculated as ln(NT2/NT0)/ln(NR2/NR0), where NT0 and NR0 represent the initial densities of the test and reference strains, respectively, and NT2 and NR2 represent their corresponding densities at the end of the competition with correction for the number of transfer cycles the competition occurred over. Competitions were generally carried out over two transfer cycles. All assays were carried out with at least four-fold replication unless reported otherwise in Results.
Estimation of lacO1 mutation frequency
To estimate the per generation frequency of loss of function lacO1 mutations, we assume that mutation of the lacO1 region is random and equally likely for each of the 21 bp that define lacO1. Using a mutation rate of 5×10−10 bp/generation [58] the probability of generating a single substitution in lacO1 is ∼1×10−8 per generation. However, only a subset of these mutations will severely compromise LacI binding. A survey of the literature indicates that approximately 16 single base pair substitutions within lacO1 have been reported to reduce the affinity of LacI by >90% and/or reduce the effective repression of LacZ expression >90% [33], [34]. Based on this, we conservatively estimate that a third of the possible 63 single base pair substitutions will severely compromise LacI binding, giving a mutation frequency of ∼3×10−9 per generation. This is likely an overestimate since only three lacO1 alleles were selected during evolution, two of which were selected twice in independent populations, indicating that relatively few of the possible lacO1 mutations may actually confer an advantage in the evolution environments. In addition, the lacO1 alleles selected during evolution have been reported to reduce the affinity for LacI by >98%, further reinforcing the stringency of our operational criteria for estimating the number of possible lacO1 mutations [33], [36].
Supporting Information
Zdroje
1. CarrollSB 2005 Evolution at two levels: on genes and form. PLoS Biol 3 e245 doi:10.1371/journal.pbio.0030245
2. BornemanARGianoulisTAZhangZDYuHRozowskyJ 2007 Divergence of transcription factor binding sites across related yeast species. Science 317 815 819 doi:10.1126/science.1140748
3. WittkoppPJHaerumBKClarkAG 2008 Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40 346 350 doi:10.1038/ng.77
4. ZambranoMMSiegeleDAAlmirónMTormoAKolterR 1993 Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259 1757 1760
5. Notley-McRobbLFerenciT 1999 The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ Microbiol 1 45 52
6. CooperTFRozenDELenskiRE 2003 Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci USA 100 1072 1077 doi:10.1073/pnas.0334340100
7. ChouH-HBerthetJMarxCJ 2009 Fast growth increases the selective advantage of a mutation arising recurrently during evolution under metal limitation. PLoS Genet 5 e1000652 doi:10.1371/journal.pgen.1000652
8. McDonaldMJGehrigSMMeintjesPLZhangX-XRaineyPB 2009 Adaptive divergence in experimental populations of Pseudomonas fluorescens. IV. Genetic constraints guide evolutionary trajectories in a parallel adaptive radiation. Genetics 183 1041 1053 doi:10.1534/genetics.109.107110
9. WangLSpiraBZhouZFengLMaharjanRP 2010 Divergence involving global regulatory gene mutations in an Escherichia coli population evolving under phosphate limitation. Genome Biol Evol 2 478 487 doi:10.1093/gbe/evq035
10. StoebelDMDormanCJ 2010 The effect of mobile element IS10 on experimental regulatory evolution in Escherichia coli. Mol Biol Evol 27 2105 2112 doi:10.1093/molbev/msq101
11. ZhongSKhodurskyADykhuizenDEDeanAM 2004 Evolutionary genomics of ecological specialization. Proc Natl Acad Sci USA 101 11719 11724 doi:10.1073/pnas.0404397101
12. SpencerCCBertrandMTravisanoMDoebeliM 2007 Adaptive diversification in genes that regulate resource use in Escherichia coli. PLoS Genet 3 e15 doi:10.1371/journal.pgen.0030015
13. CooperTFLenskiRE 2010 Experimental evolution with E. coli in diverse resource environments. I. Fluctuating environments promote divergence of replicate populations. BMC Evol Biol 10 11 doi:10.1186/1471-2148-10-11
14. DekelEAlonU 2005 Optimality and evolutionary tuning of the expression level of a protein. Nature 436 588 592 doi:10.1038/nature03842
15. StoebelDMDeanAMDykhuizenDE 2008 The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics 178 1653 1660 doi:10.1534/genetics.107.085399
16. PerfeitoLGhozziSBergJSchnetzKLässigM 2011 Nonlinear fitness landscape of a molecular pathway. PLoS Genet 7 e1002160 doi:10.1371/journal.pgen.1002160
17. DykhuizenDEDeanAMHartlDL 1987 Metabolic flux and fitness. Genetics 115 25 31
18. SettyYmayoAESuretteMGAlonU 2003 Detailed map of a cis-regulatory input function. Proc Natl Acad Sci USA 100 7702 7707 doi:10.1073/pnas.1230759100
19. OzbudakEMThattaiMLimHNShraimanBIVan OudenaardenA 2004 Multistability in the lactose utilization network of Escherichia coli. Nature 427 737 740 doi:10.1038/nature02298
20. van HoekMHogewegP 2007 The effect of stochasticity on the lac operon: an evolutionary perspective. PLoS Comput Biol 3 e111 doi:10.1371/journal.pcbi.0030111
21. SantillánMMackeyMCZeronES 2007 Origin of bistability in the lac Operon. Biophys J 92 3830 3842 doi:10.1529/biophysj.106.101717
22. RobertLPaulGChenYTaddeiFBaiglD 2010 Pre-dispositions and epigenetic inheritance in the Escherichia coli lactose operon bistable switch. Mol Syst Biol 6 doi:10.1038/msb.2010.12
23. SavageauMA 1998 Demand theory of gene regulation. II. Quantitative application to the lactose and maltose operons of Escherichia coli. Genetics 149 1677 1691
24. GerlandUHwaT 2009 Evolutionary selection between alternative modes of gene regulation. Proc Natl Acad Sci USA 106 8841 8846 doi:10.1073/pnas.0808500106
25. SilverRSMatelesRI 1969 Control of mixed-substrate utilization in continuous cultures of Escherichia coli. J Bacteriol 97 535 543
26. DykhuizenDDaviesM 1980 An experimental model: bacterial specialists and generalists competing in chemostats. Ecology 61 1213 1227
27. NovickAWeinerM 1957 Enzyme Induction as an all-or-none phenomenon. Proc Natl Acad Sci USA 43 553 566
28. ChoiPJCaiLFriedaKXieXS 2008 A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science 322 442 446 doi:10.1126/science.1161427
29. KuhlmanTZhangZSaierMHHwaT 2007 Combinatorial transcriptional control of the lactose operon of Escherichia coli. Proc Natl Acad Sci USA 104 6043 6048 doi:10.1073/pnas.0606717104
30. CoulondreCMillerJHFarabaughPJGilbertW 1978 Molecular basis of base substitution hotspots in Escherichia coli. Nature 274 775 780
31. FarabaughPJSchmeissnerUHoferMMillerJH 1978 Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol 126 847 857
32. MarkiewiczPKleinaLGCruzCEhretSMillerJH 1994 Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as “spacers” which do not require a specific sequence. J Mol Biol 240 421 433 doi:10.1006/jmbi.1994.1458
33. BetzJLSasmorHMBuckFInsleyMYCaruthersMH 1986 Base substitution mutants of the lac operator: in vivo and in vitro affinities for lac repressor. Gene 50 123 132
34. MaquatLThorntonK 1980 lac promoter mutations located downstream from the transcription start site. J Mol Biol 139 537 549
35. MossingMCRecordMT 1985 Thermodynamic origins of specificity in the lac repressor-operator interaction. Adaptability in the recognition of mutant operator sites. J Mol Biol 186 295 305
36. FalconCMMatthewsKS 2000 Operator DNA sequence variation enhances high affinity binding by hinge helix mutants of lactose repressor protein. Biochemistry 39 11074 11083 doi:10.1021/bi000924z
37. KhanAIDinhDMSchneiderDLenskiRECooperTF 2011 Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332 1193 1196 doi:10.1126/science.1203801
38. BiggarSRCrabtreeGR 2001 Cell signaling can direct either binary or graded transcriptional responses. EMBO J 20 3167 3176 doi:10.1093/emboj/20.12.3167
39. InadaTKimataKAibaH 1996 Mechanism responsible for glucose-lactose diauxie in Escherichia coli: challenge to the cAMP model. Genes Cells 1 293 301
40. GörkeBStülkeJ 2008 Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Micro 6 613 624 doi:10.1038/nrmicro1932
41. MitchellARomanoGHGroismanBYonaADekelE 2009 Adaptive prediction of environmental changes by microorganisms. Nature 460 220 224 doi:10.1038/nature08112
42. LenskiRRoseMSimpsonS 1991 Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138 1315 1341
43. BertaniG 2004 Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol 186 595 600
44. ZaslaverABrenARonenMItzkovitzSKikoinI 2006 A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Meth 3 623 628 doi:10.1038/nmeth895
45. ChoiK-HGaynorJBWhiteKGLopezCBosioCM 2005 A Tn7-based broad-range bacterial cloning and expression system. Nat Meth 2 443 448 doi:10.1038/nmeth765
46. OehlerSEismannERKrämerHMüller-HillB 1990 The three operators of the lac operon cooperate in repression. EMBO J 9 973 979
47. StrackRLStronginDEBhattacharyyaDTaoWBermanA 2008 A noncytotoxic DsRed variant for whole-cell labeling. Nat Meth 5 955 957 doi:10.1038/nmeth.1264
48. FerrièresLHémeryGNhamTGuéroutA-MMazelD 2010 Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol 192 6418 6427 doi:10.1128/JB.00621-10
49. MondsRDNewellPDGrossRHO'TooleGA 2007 Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol 63 656 679 doi:10.1111/j.1365-2958.2006.05539.x
50. GentlemanRCCareyVJBatesDMBolstadBDettlingM 2004 Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80 doi:10.1186/gb-2004-5-10-r80
51. HahneFLeMeurNBrinkmanRREllisBHaalandP 2009 flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics 10 106 doi:10.1186/1471-2105-10-106
52. SarkarDLe MeurNGentlemanR 2008 Using flowViz to visualize flow cytometry data. Bioinformatics 24 878 879 doi:10.1093/bioinformatics/btn021
53. ZhangXBremerH 1995 Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J Biol Chem 270 11181 11189
54. FriesenMLSaxerGTravisanoMDoebeliM 2004 Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58 245 260
55. PhilippeNAlcarazJ-PCoursangeEGeiselmannJSchneiderD 2004 Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51 246 255 doi:10.1016/j.plasmid.2004.02.003
56. NghiemYCabreraMCupplesCGMillerJH 1988 The mutY gene: a mutator locus in Escherichia coli that generates GC to TA transversions. Proc Natl Acad Sci USA 85 2709 2713
57. SchaaperRMDanforthBNGlickmanBW 1986 Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol 189 273 284
58. DrakeJW 1991 A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA 88 7160 7164
Štítky
Genetika Reprodukční medicína
Článek USF-1 Is Critical for Maintaining Genome Integrity in Response to UV-Induced DNA PhotolesionsČlánek Checkpoints in a Yeast Differentiation Pathway Coordinate Signaling during Hyperosmotic StressČlánek Two-Component Elements Mediate Interactions between Cytokinin and Salicylic Acid in Plant Immunity
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 1
-
Všechny články tohoto čísla
- DNA Methylation and Gene Expression Changes in Monozygotic Twins Discordant for Psoriasis: Identification of Epigenetically Dysregulated Genes
- An siRNA Screen in Pancreatic Beta Cells Reveals a Role for in Insulin Production
- Parallel Mapping and Simultaneous Sequencing Reveals Deletions in and Associated with Discrete Inherited Disorders in a Domestic Dog Breed
- Cytoplasmic Polyadenylation Element Binding Protein Deficiency Stimulates PTEN and Stat3 mRNA Translation and Induces Hepatic Insulin Resistance
- Nucleolar Association and Transcriptional Inhibition through 5S rDNA in Mammals
- USF-1 Is Critical for Maintaining Genome Integrity in Response to UV-Induced DNA Photolesions
- Heterochromatin Formation Promotes Longevity and Represses Ribosomal RNA Synthesis
- Genetic Evidence for an Indispensable Role of Somatic Embryogenesis Receptor Kinases in Brassinosteroid Signaling
- Poly(ADP-Ribose) Polymerase 1 (PARP-1) Regulates Ribosomal Biogenesis in Nucleoli
- Genome Engineering in : A Feasible Approach to Address Biological Issues
- RIC-7 Promotes Neuropeptide Secretion
- Adaptation and Preadaptation of to Bile
- Checkpoints in a Yeast Differentiation Pathway Coordinate Signaling during Hyperosmotic Stress
- Progressive Polycomb Assembly on H3K27me3 Compartments Generates Polycomb Bodies with Developmentally Regulated Motion
- A High Density SNP Array for the Domestic Horse and Extant Perissodactyla: Utility for Association Mapping, Genetic Diversity, and Phylogeny Studies
- Two-Component Elements Mediate Interactions between Cytokinin and Salicylic Acid in Plant Immunity
- Cdc5-Dependent Asymmetric Localization of Bfa1 Fine-Tunes Timely Mitotic Exit
- A Genome-Wide Analysis of Promoter-Mediated Phenotypic Noise in
- Contribution of Intragenic DNA Methylation in Mouse Gametic DNA Methylomes to Establish Oocyte-Specific Heritable Marks
- Adaptive Evolution of the Lactose Utilization Network in Experimentally Evolved Populations of
- Microenvironmental Regulation by Fibrillin-1
- Unraveling the Regulatory Mechanisms Underlying Tissue-Dependent Genetic Variation of Gene Expression
- A Half-Century of Inspiration: An Interview with Hamilton Smith
- Reduced Lentivirus Susceptibility in Sheep with Mutations
- High-Density SNP Mapping of the HLA Region Identifies Multiple Independent Susceptibility Loci Associated with Selective IgA Deficiency
- Calpains Mediate Integrin Attachment Complex Maintenance of Adult Muscle in
- Genomic Ancestry of North Africans Supports Back-to-Africa Migrations
- Functional Specialization of the Plant miR396 Regulatory Network through Distinct MicroRNA–Target Interactions
- The Seminal Fluid Protease “Seminase” Regulates Proteolytic and Post-Mating Reproductive Processes
- Insulin Signaling Regulates Fatty Acid Catabolism at the Level of CoA Activation
- A Genome-Wide Association Study Identified as a Susceptibility Locus for Systemic Lupus Eyrthematosus in Japanese
- A Spontaneous Mutation of the Rat Gene Leads to Impaired Function of Regulatory T Cells Linked to Inflammatory Bowel Disease
- The Yeast Complex I Equivalent NADH Dehydrogenase Rescues Mutants
- A Flexible Bayesian Model for Studying Gene–Environment Interaction
- Sex Pheromone Evolution Is Associated with Differential Regulation of the Same Desaturase Gene in Two Genera of Leafroller Moths
- Genome-Wide Assessment of AU-Rich Elements by the ARE Algorithm
- Inference of Population Structure using Dense Haplotype Data
- A Gene Regulatory Network for Root Epidermis Cell Differentiation in Arabidopsis
- Sequencing of Pooled DNA Samples (Pool-Seq) Uncovers Complex Dynamics of Transposable Element Insertions in
- A Genome-Wide Association Scan on the Levels of Markers of Inflammation in Sardinians Reveals Associations That Underpin Its Complex Regulation
- Tempo and Mode in Evolution of Transcriptional Regulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Poly(ADP-Ribose) Polymerase 1 (PARP-1) Regulates Ribosomal Biogenesis in Nucleoli
- Microenvironmental Regulation by Fibrillin-1
- Parallel Mapping and Simultaneous Sequencing Reveals Deletions in and Associated with Discrete Inherited Disorders in a Domestic Dog Breed
- Two-Component Elements Mediate Interactions between Cytokinin and Salicylic Acid in Plant Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



