-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Genetic and Structural Study of Genome Rearrangements Mediated by High Copy Repeat Ty1 Elements
Ty elements are high copy number, dispersed repeated sequences in the Saccharomyces cerevisiae genome known to mediate gross chromosomal rearrangements (GCRs). Here we found that introduction of Ty912, a previously identified Ty1 element, onto the non-essential terminal region of the left arm of chromosome V led to a 380-fold increase in the rate of accumulating GCRs in a wild-type strain. A survey of 48 different mutations identified those that either increased or decreased the rate of Ty-mediated GCRs and demonstrated that suppression of Ty-mediated GCRs differs from that of both low copy repeat sequence - and single copy sequence-mediated GCRs. The majority of the Ty912-mediated GCRs observed were monocentric nonreciprocal translocations mediated by RAD52-dependent homologous recombination (HR) between Ty912 and a Ty element on another chromosome arm. The remaining Ty912-mediated GCRs appeared to involve Ty912-mediated formation of unstable dicentric translocation chromosomes that were resolved by one or more Ty-mediated breakage-fusion-bridge cycles. Overall, the results demonstrate that the Ty912-mediated GCR assay is an excellent model for understanding mechanisms and pathways that suppress genome rearrangements mediated by high copy number repeat sequences, as well as the mechanisms by which such rearrangements occur.
Published in the journal: . PLoS Genet 7(5): e32767. doi:10.1371/journal.pgen.1002089
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002089Summary
Ty elements are high copy number, dispersed repeated sequences in the Saccharomyces cerevisiae genome known to mediate gross chromosomal rearrangements (GCRs). Here we found that introduction of Ty912, a previously identified Ty1 element, onto the non-essential terminal region of the left arm of chromosome V led to a 380-fold increase in the rate of accumulating GCRs in a wild-type strain. A survey of 48 different mutations identified those that either increased or decreased the rate of Ty-mediated GCRs and demonstrated that suppression of Ty-mediated GCRs differs from that of both low copy repeat sequence - and single copy sequence-mediated GCRs. The majority of the Ty912-mediated GCRs observed were monocentric nonreciprocal translocations mediated by RAD52-dependent homologous recombination (HR) between Ty912 and a Ty element on another chromosome arm. The remaining Ty912-mediated GCRs appeared to involve Ty912-mediated formation of unstable dicentric translocation chromosomes that were resolved by one or more Ty-mediated breakage-fusion-bridge cycles. Overall, the results demonstrate that the Ty912-mediated GCR assay is an excellent model for understanding mechanisms and pathways that suppress genome rearrangements mediated by high copy number repeat sequences, as well as the mechanisms by which such rearrangements occur.
Introduction
Gross chromosomal rearrangements (GCRs) are associated with many different diseases. Disease-causing GCRs include translocations, deletions, and inversions that can inactivate genes, form chimeric genes encoding proteins with altered activity, or change gene copy numbers or gene expression. The human genome contains many highly duplicated elements, such as Alu and LINE elements, which collectively comprise nearly 40–50% of the human genome [1]–[3]. Non-allelic homologous recombination (HR) between repeated sequences can mediate rearrangements leading to segmental duplications [4], numerous human genetic diseases [5] including certain sex disorders thought to be due to HR between palindromic regions on the Y chromosome [6], and many of the GCRs present in adult solid tumors [7]. Despite the importance of suppressing non-allelic recombination between highly duplicated repeats to maintain genome stability, little is known about the genetic factors that suppress these types of rearrangements.
Studies in the yeast Saccharomyces cerevisiae have contributed greatly to our general understanding of the suppression and formation of GCRs mediated by both single-copy sequence and, more recently, low copy number segmental duplications [8]–[11]. These studies, however, have not generally addressed the roles of highly repeated genomic elements. The Ty1 family of retrotransposons is the most common class of retrotransposons in S. cerevisiae [12]. A full length Ty1 element is ∼5.9 kb long and consists of ∼5.2 kb of unique sequence (known as the epsilon sequence) flanked by one copy of a ∼332 bp Long Terminal Repeat (LTR) sequence (also known as a delta sequence) at each end. The LTR sequences are both oriented in the same direction and homologous recombination between them results in the deletion of the internal Ty1 epsilon sequence and one copy of the delta sequences, giving rise to a “solo delta” element [13]. The reference S288c S. cerevisiae genome sequence contains at least 32 full length Ty1s and at least 217 solo delta sequences, comprising at least 2.1% of the genome [12]. Because Ty1-related sequences are the most repetitive components of the S. cerevisiae genome, they are the best S. cerevisiae analog to the highly repetitive human Alu sequences, which are smaller than Ty elements, and LINE sequences, which are similar in size to Ty elements.
Like highly repetitive human elements, Ty1s appear to mediate many types of chromosomal rearrangements, including inversions, deletions, and both reciprocal and non-reciprocal translocations [14]–[18]. Such events are believed to result from the repair of DNA double strand breaks (DSBs) at or near Ty1 sequences and indeed induction of such DSBs through fragile sites, unstable inverted repeats, ionizing radiation and formation of unstable dicentric chromosomes stimulates Ty-mediated GCRs [18]–[27]. A number of mechanisms have been proposed to account for these Ty-mediated GCRs, including Break Induced Replication (BIR) between a Ty element on a broken chromosome and a Ty element at another site on either a broken or intact chromosome, and crossing over between two Ty elements potentially mediated by single strand annealing (SSA) and other HR mechanisms [17], [22], [25], [27]–[30]. Ty1 sequences are also a target of GCR-causing rearrangements involving non-repetitive sequences [31]. Because GCRs mediated by highly repetitive genome sequences underlie a number of human diseases and because there is currently a dearth of information about which pathways prevent such GCRs, we developed a quantitative genetic assay that measures the rate of Ty1-mediated GCRs. Our results demonstrate that repetitive sequences greatly contribute to genomic instability and we identify genes and pathways that suppress and promote these Ty1-mediated GCRs. In addition, we characterized 88 Ty1-mediated GCRs at varying levels of detail and demonstrated that the most common Ty1-mediated GCRs appear to involve non-reciprocal HR between ectopic Ty sequences that often results in the duplication of stretches of sequences bounded by a target Ty element at one end and a telomere at the other end. In a small number of cases, we observed complex rearrangements consistent with multiple exchanges between target sequences, as well as rearrangements consistent with the formation and resolution of dicentric chromosomes initially formed by HR between Ty elements.
Results
A Ty912 element increases GCR rates
To identify how Ty elements influence GCRs, we placed Ty912, a Ty1 retrotransposon originally isolated during a screen for spontaneous histidine auxotrophic mutants [32], in a nonessential region of the left arm of chromosome V between the NPR2 and CIN8 genes (Figure 1a). This site is between the most telomeric essential gene on the left arm of chromosome V (PCM1) and two counter-selectable genes (CAN1 and hxt13::URA3) used in the original GCR assay [9]. We chose this integration site for the Ty912 because it allows direct analysis of the effect of a Ty element on GCRs mediated by a well characterized single copy sequence GCR breakpoint region. We determined the rate of accumulating GCRs by measuring the rate of simultaneous loss of CAN1 and URA3 by fluctuation analysis. The presence of Ty912 on chromosome V (hereafter referred to as +Ty912) in a wild-type strain resulted in a 380-fold increase in the rate of accumulating Canr 5FOAr progeny compared to an isogenic wild-type strain without the Ty912 insertion (hereafter referred to as −Ty) (Table 1). As will be demonstrated below, the Canr 5FOAr progeny that accumulated in the +Ty912 strain were the result of Ty1-mediated translocations.
Fig. 1. Assay model and a summary of the aCGH data. 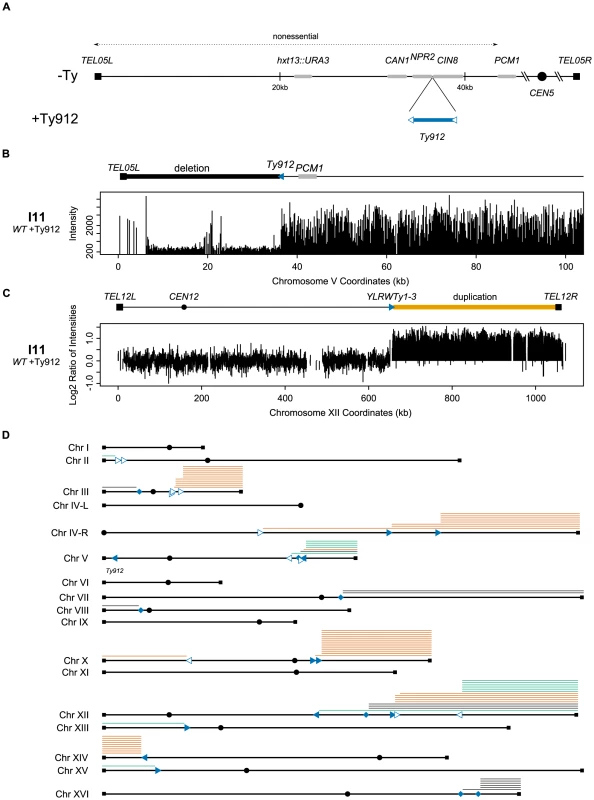
A. Schematic of the −Ty and +Ty912 GCR assays on their respective chromosome Vs. Genes and Ty912 are not drawn to scale. B. Example of the Ty912-TEL05L deletion. Absolute intensities of the probes in the deletion region are noticeably lower than other regions where DNA is present. Signal spikes in the deletion represent redundant sequences (telomeric sequence on the left and DSF1 and HXT13 sequences in the middle). C. Example of a Class II duplication. The log2 ratio of intensities indicates a doubling of the genomic content from YLRWTy1-3 to TEL12R. D. Overview of the aCGH data. Filled squares represent telomeres, filled circles centromeres. Solid triangles represent full-length Ty1 elements. Hollow triangles represent solo deltas. The orientations of the triangles reflect the transcriptional orientation of the elements. Filled diamonds represent loci containing multiple Ty1 elements transcriptionally oriented in opposite directions. Lines above the chromosome arms represent duplicated regions. Orange lines represent duplications of genomic DNA between a telomere and a telomere oriented Ty element. Green lines represent duplications of genomic DNA between a telomere and a centromere oriented Ty element. Black lines represent duplications oriented between a telomere and a mixed orientation multiple Ty loci site. Ty1 elements targeted by independent translocations 3 or more times include YCRWdelta10, YCRWdelta11, YDRWTy1-5, YERWdelta21, YJRWTy1-2, YLRWTy1-3, YLRCdelta21, the YLRCdelta9/YLRWTy1-2/YLRCdelta12 multiple Ty loci, YNLCTy1-1, and YPRWTy1-3. Tab. 1. Wild-type and mutant GCR rates. 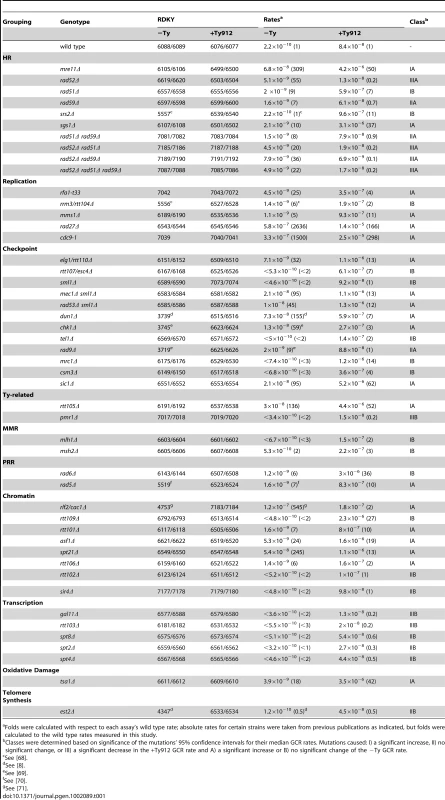
Folds were calculated with respect to each assay's wild type rate; absolute rates for certain strains were taken from previous publications as indicated, but folds were calculated to the wild type rates measured in this study. Diverse genes and pathways suppress Ty912-mediated GCRs
We next surveyed a series of mutations for their effects on GCR rates in both the −Ty and +Ty912 strain backgrounds (Table 1; Table S1). The selected mutations affected many pathways, including HR, DNA replication, checkpoints, Ty transposition, mismatch repair, post replication repair, chromatin structure and assembly, transcription, accumulation of oxidative DNA damage, and telomere synthesis. To simplify the analysis of the mutations in this survey, we divided the mutations into three classes, Class I, II, and III, that caused +Ty912 GCR rates that were higher than, the same as, or lower than the wild-type +Ty912 GCR rate, respectively. Each class was then divided into two subclasses, A and B, depending on whether or not the −Ty GCR rate was greater than, or the same or lower than, the wild-type −Ty GCR rate.
Class IA mutations increased the GCR rate in both assays and comprised the largest proportion of the mutations tested (20 of 48; 42%). Of these, 4 caused a similar fold increase in both the −Ty and +Ty912 rates, 13 caused a greater fold increase in the −Ty rate, and 3 (sgs1Δ, mms1Δ, and tsa1Δ) caused a greater fold increase in the +Ty912 rate. Both sgs1Δ and tsa1Δ were previously identified as mutations that significantly increased the GCR rate in the segmental duplication-mediated GCR assay relative to assays that detected single copy sequence-mediated GCRs (Table S1) [11], [33]. There were a smaller number of Class IB mutations that increased the +Ty912 GCR rate and had no effect on the −Ty GCR rate. Mutations in many of the Class IB genes, including SRS2, RRM3, MRC1, MSH2 MLH1 and RTT109 increased GCRs mediated by segmental duplications (Table S1) [11], which suggests that the Class IB genes play general roles in suppressing various aspects of non-allelic HR. In addition, this class of mutations also included csm3Δ and rtt107Δ, which have not been tested in the segmental duplication assay. Intriguingly, RTT107/ESC4 and RTT109 are both genes that affect the rate of Ty transposition [34], modulate chromatin structure [35], [36], and play roles in processing stalled replication forks [37].
Three Class IIA mutations (rad9Δ and rad59Δ single mutations, and the rad51Δ rad59Δ double mutation) did not significantly increase the +Ty912 GCR rate but did increase the −Ty GCR rate. In contrast, there were 8 Class IIB mutations that caused little to no effect in either GCR assay. These Class IIB mutations represented individual deletions of a number of genes, including SPT2, SPT4, SPT8, RTT102, and SIR4, involved in suppressing Ty1 transposition and/or transcription [34], [38]–[40].
The Class IIIA mutations that decreased the +Ty912 GCR rate and increased the −Ty GCR rate were restricted to a small number of mutations in mutant backgrounds containing a deletion of RAD52 (rad52Δ, rad52Δ rad51Δ, rad52Δ rad59Δ, and rad52Δ rad51Δ rad59Δ). There were also 3 Class IIIB mutations that did not appear to affect the −Ty GCR rate, but did decrease the +Ty912 GCR rate. Each of these mutations have been previously identified as affecting Ty1 biology; deletions of PMR1 and RTT103 alter the rate of Ty1 transposition [34], [41] and deletion of GAL11 affects Ty1 transcription [42].
Ty912-mediated GCRs in wild-type strains involve homologous recombination
Mutations affecting genes encoding core HR proteins belonged to several different mutation classes (IA, IIA, and IIIA), indicating a range of effects on the +Ty912 GCR assay. Deletion of RAD52, which eliminates most if not all HR in S. cerevisiae [43], suppressed the +Ty912 GCR rate (Table 1) to a level not significantly different from that caused by a RAD52 deletion in the −Ty background (unpaired Wilcoxon rank sum test; p = 0.067). This suggested that Ty1 sequence-specific GCRs are RAD52-dependent. RAD52 is involved in two HR subpathways mediated by RAD51 and RAD59. In contrast to a deletion of RAD52, the rad51Δ mutation increased the GCR rate in the +Ty912 GCR assay by 7-fold (unpaired Wilcoxon rank sum test; p = 4.89×10−7), suggesting RAD51-dependent repair events normally suppress Ty1-mediated GCRs. This is somewhat consistent with a previous report, which found that deletion of RAD51 caused an increase in the rate of deletions mediated by direct repeat recombination between Ty1 LTRs but caused a decrease in recombination of Ty1s resulting in conversion events [44]. In contrast, deletion of RAD59 alone had no significant effect on the GCR rate in the +Ty912 GCR assay (unpaired Wilcoxon rank sum test; p = 0.34). However, deletion of RAD59 suppressed the increased GCR rate of a rad51Δ mutant and led to a GCR rate that was not statistically different from that of a wild-type strain (unpaired Wilcoxon rank sum test; p = 0.89); this suggests that RAD59 is responsible for mediating the formation of many of the GCRs that occur in a rad51Δ mutant, consistent with previous reports of RAD59-dependent, RAD51-independent recombination events [28], [45]–[48]. Furthermore, the GCR rate of the rad51Δ rad59Δ double mutant in the +Ty912 GCR assay was significantly higher than the GCR rate caused by a rad52Δ mutation in the +Ty912 GCR assay (unpaired Wilcoxon rank sum test; p = 5.59×10−4). This is consistent with the existence of an inefficient RAD52-dependent, RAD51 - and RAD59-independent HR pathway [11], [46]; accordingly, the rad52Δ rad51Δ, rad52Δ rad59Δ, and rad52Δ rad51Δ rad59Δ double and triple mutants had GCR rates in the +Ty912 GCR assay that were not statistically different than the GCR rate seen in a rad52Δ mutant with the +Ty912 GCR assay (unpaired Wilcoxon rank sum tests; p = 0.21, 0.10, 0.95, respectively).
Mutations affecting Ty biology have variable effects on the +Ty912 GCR assay
Among the mutations selected for testing were a number originally isolated as altering either transcription [38], [39], [42] or transposition [34] of Ty1 elements, many of which had not previously been tested for their effects on GCR rates. Mutations affecting the transcription of Ty1 elements had no (spt2Δ, spt4Δ, and spt8Δ), suppressive (gal11Δ), or stimulating (spt21Δ) effects on the +Ty912 GCR rate. Of the mutations affecting Ty1 transcription, only spt21Δ significantly increased the GCR rate in the −Ty assay; it was not possible to determine if the gal11Δ mutant had reduced GCR rates in the −Ty assay because the −Ty GCR rate of this mutant was too low to measure. Mutations known to affect the transposition of Ty1 elements also had variable effects on the +Ty912 GCR rate, with some mutations causing no change (rtt102Δ and rtt106Δ), an increased rate (rtt101Δ, rrm3Δ, rtt105Δ, rtt107Δ, rtt109Δ, and elg1Δ), or a decreased rate (rtt103Δ and pmr1Δ). Half of the mutations causing increased +Ty912 GCR rates (rtt101Δ, rtt105Δ, and elg1Δ), also increased the GCR rate in the −Ty assay; rrm3Δ and rtt109Δ are also known to increase the rate of GCRs mediated by segmental duplications (Table S1) [11]. It was not possible to determine if the rtt103Δ and pmr1Δ mutants had reduced GCR rates in the −Ty assay because the −Ty GCR rate of these mutants were too low to measure. Thus, many of the mutations that affect both the +Ty912 GCR rate and either Ty1 transcription or transposition that could be evaluated in the −Ty GCR assay also similarly affect non-Ty1 mediated genome rearrangements. Taken together, these data are consistent with a view in which Ty-mediated genome instability is generally independent of mechanisms of Ty propagation and Ty-mediated gene silencing and suggest results from the +Ty912 assay will be broadly applicable to the study of genome instability mediated by highly repetitive sequences.
Non Ty-mediated events lose portions of the left arm of chromosome V
To understand how Ty912 increases GCR rates, we first used array Comparative Genomic Hybridization (aCGH) to analyze 7 independent GCR-containing strains isolated from the wild-type −Ty strain (I1–I7; Table S2). All 7 GCR-containing strains had terminal deletions of the left arm of chromosome V starting at positions between the PCM1 and CAN1 genes and extending to the left telomere TEL05L (Figure S1a). These strains contained no additional copy number changes of other chromosomal regions. The aCGH results were consistent with the smaller chromosome Vs identified by a combination of pulse-field gel electrophoresis (PFGE) followed by Southern blot analysis using a probe to MCM3, an essential gene on the left arm of chromosome V (Figure S1b). The data suggest that GCRs from all 7 isolates from the −Ty background were formed by breakage of the left arm of chromosome V followed by healing of the chromosome end by de novo telomere addition, similar to GCRs formed in other wild-type strains lacking repetitive elements in their breakpoint regions [9], [49].
Most GCRs isolated in the +Ty912 assay duplicated large chromosomal regions bordered by Tys and telomeres
We then performed aCGH analysis on 10 GCR-containing strains isolated from the wild-type +Ty912 strain, and 78 GCR-containing strains isolated from 11 different mutant +Ty912 strains (I8–I94, Table S2). Unlike the wild-type −Ty GCR-containing strains, only 5 of 88 GCR-containing strains isolated from the +Ty912 GCR assay had aCGH patterns consistent with terminal deletions associated with de novo telomere additions (Class I GCRs; Table 2; Figure S2a–g). Two of these events also contained putative copy number changes of small internal regions on the terminally deleted chromosome V with no duplications or deletions on other chromosomes. Strikingly, the majority (83 of 88) of GCR-containing strains isolated using the +Ty912 GCR assay and analyzed by aCGH had a deletion from Ty912 to TEL05L (Ty912-TEL05L deletion) and had one or more duplicated regions that were bounded at least on one side by a full-length Ty1 or a solo Ty1 or Ty2 delta element in the reference genome. None of these duplicated regions spanned centromeres. In addition, 5 of the 83 cases (4 occurring in the rtt109Δ mutant and 1 occurring in the rad53Δ sml1Δ mutant) appeared to also be disomic for at least one chromosome (I53, I55, I56, I58, and I67; Table S2). Based on the aCGH analysis of these 83 isolates, we observed four additional classes of rearrangements (Class II to V GCRs) containing the chromosome V Ty912-TEL05L deletion and additional copy number changes bounded by Ty-related sequences that are described below.
Tab. 2. Summary of GCR events determined by aCGH data. 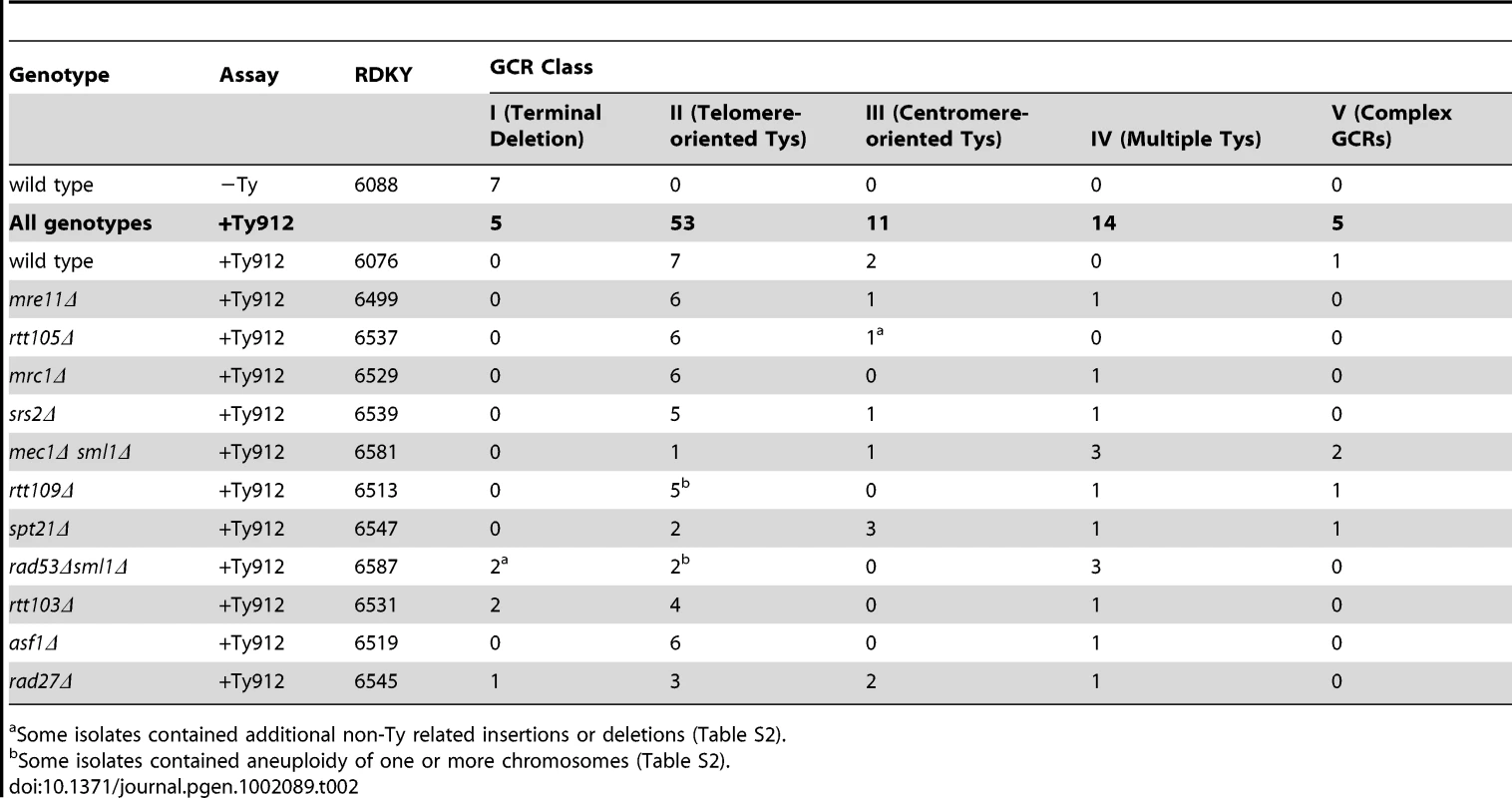
Some isolates contained additional non-Ty related insertions or deletions (Table S2). Class II GCRs contained duplications bordered by a telomere and one or more telomere-oriented Ty1s and were fused to chromosome V at Ty912
The 53 Class II GCR-containing strains (60.2% of the total surveyed) contained both the chromosome V Ty912-TEL05L deletion (Figure 1b) and a single duplicated region of another chromosome arm extending from a Ty1 element or solo Ty1 delta element to a telomere. In this class, the Ty1 elements were transcriptionally oriented towards the telomere and away from the centromere (Figure 1c; Table 2); this is the same orientation as the Ty912 element. In spite of the fact that 121 full-length Ty1, solo Ty1 delta, and solo Ty2 delta elements are oriented appropriately to generate Class II GCRs, only 16 such elements in the S288c genome reference sequence were involved as targets in the observed Class II rearrangements as determined by the aCGH data, suggesting the distribution of Ty elements mediating the rearrangements is non-random (Figure 1d).
Several lines of evidence support the idea that a non-reciprocal HR-mediated process, such as BIR or half-crossovers [29], [43], [50]–[55], occurred between Ty912 and the Ty target to generate the observed products. First, the orientations of the Ty1 sequences at the boundaries of the duplications relative to the duplicated regions are consistent with a homology-driven translocation process. Second, PFGE and Southern blotting with a probe to MCM3 revealed that the seven analyzed GCR-containing strains in this class (I8, I10, I11, I13, I15, I16, and I17) had a single abnormally-sized chromosome V that was consistent with loss of 36 kb from the left arm of chromosome V (from the Ty912-TEL05L deletion) and gain of sequence equal to the length of the duplicated region from the target chromosome (Figure 2a; Table 3). Third, the predicted junctions from these seven isolates could be amplified by PCR using primers in unique sequences flanking the Ty-mediated junction or verified by cloning the junction and sequencing of the resulting plasmid; the sequences of these amplified junction had SNPs in the 5′ ends of the junctions that generally corresponded to the Ty912 sequence from chromosome V and had SNPs in the 3′ ends of the junctions that generally corresponded to the target Ty1 sequences (Figure 2b; Table 4). Fourth, the pattern of SNPs observed across the Ty fusion junction indicated that breakpoints occurred throughout the region of homology between Ty912 and the target Ty sequences and were not restricted to terminal delta elements.
Fig. 2. Ty912 mediates GCRs. 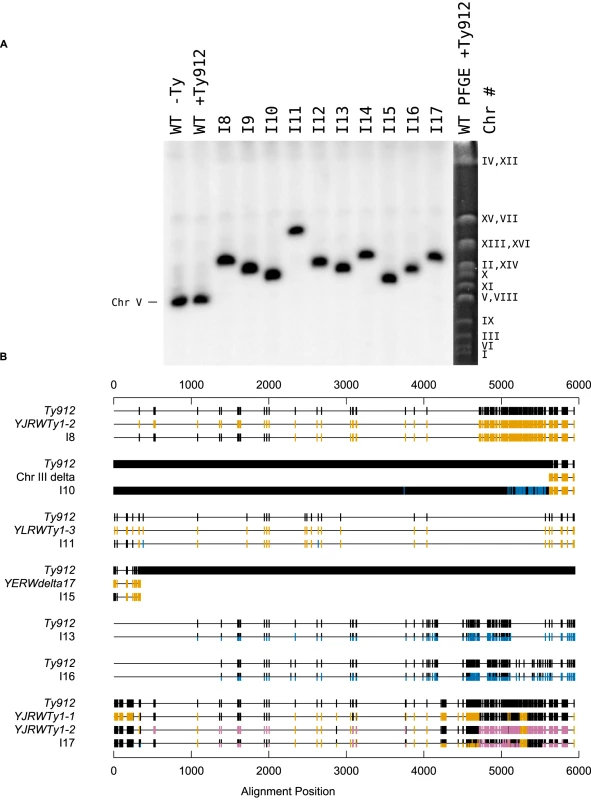
A. Southern blot of isolates I8–I17 using a probe specific to MCM3, an essential gene on chromosome V. The MCM3 probe was amplified using JCP447 & JCP 448. A PFGE reference column is included. B. An analysis of sequenced fusion Tys mediating the translocations. SNPs are attributable to either Ty912 (black), the target Ty(s) bordering the duplication (gold or magenta), or neither (blue). For isolate I17, the analysis was performed in three steps; SNPs consistent with YJRWTy1-1 were first colored gold, then SNPs consistent with YJRWTy1-2 were colored magenta, and finally SNPs belonging to either Ty912 or none of the Tys above were colored black and blue, respectively. Tab. 3. Observed and predicted Chr Vs from a Southern blot of isolates I8–I17. 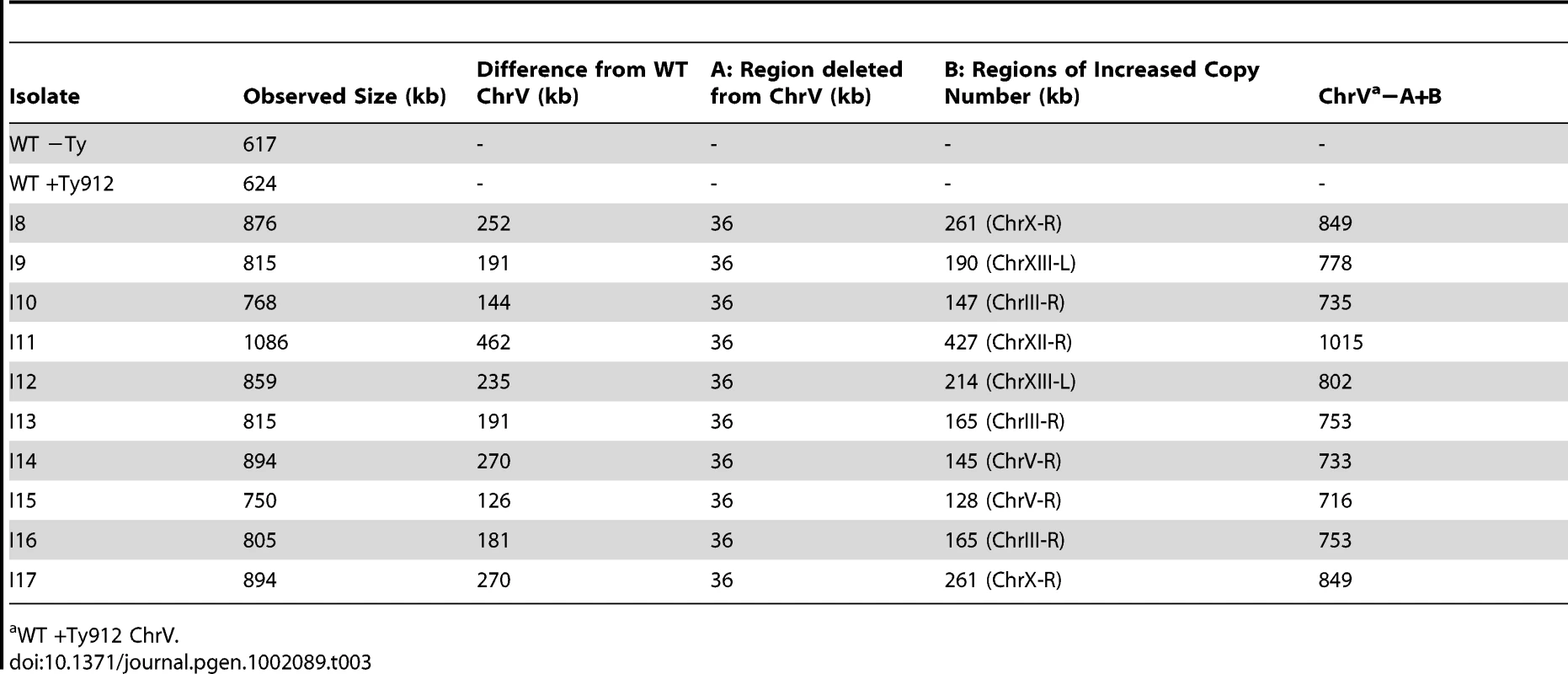
WT +Ty912 ChrV. Tab. 4. Analysis of SNPs in Ty fusion junctions. 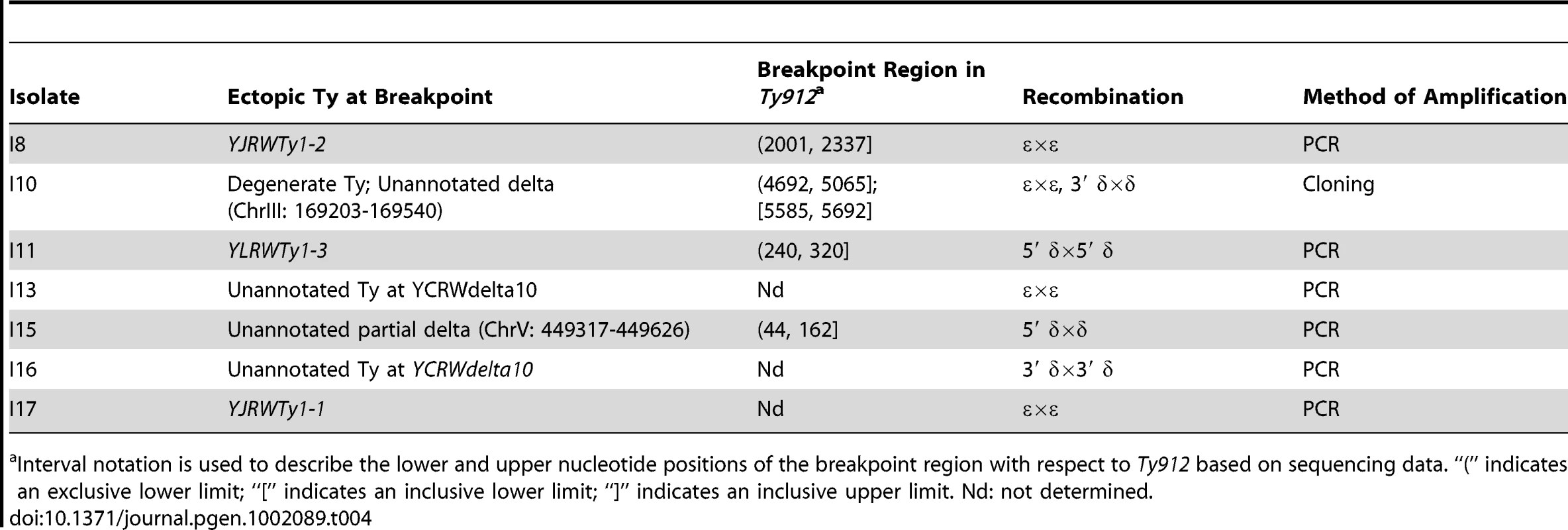
Interval notation is used to describe the lower and upper nucleotide positions of the breakpoint region with respect to Ty912 based on sequencing data. “(” indicates an exclusive lower limit; “[” indicates an inclusive lower limit; “]” indicates an inclusive upper limit. Nd: not determined. Although all of the 7 Class II GCR fusion junctions that were amplified and analyzed featured a pattern of 5′ SNPs consistent with Ty912 sequence and a pattern of 3′ SNPs consistent with the target Ty1 sequence bordering the duplication, the fusion junctions from I10, I13, I16, and I17 contained other notable features (Figure 2b). The junction sequence from isolate I10 featured an almost continuous block of SNPs at the 3′ end of its epsilon sequence that could not be attributed to the Ty912 or to an unannotated solo delta near YCRWdelta10 that bordered the duplicated region. However, this sequence region had 100% sequence identity to other Ty1s elsewhere in the genome (YHRCTy1-1, YMLWTy1-1, YPRWTy1-3, and YDRWTy1-4). This SNP pattern was consistent with a tripartite fusion in which Ty912 first recombined with one of the 4 ectopic Ty1 elements (YHRCTy1-1, YMLWTy1-1, YPRWTy1-3, or YDRWTy1-4) followed by a second recombination event between the 3′ delta sequence of the target Ty1 and the unannotated delta sequence next to YCRWdelta10 on chromosome III. In contrast, the amplified junction regions from I13, I16, and I17 were approximately twice the size of a full-length Ty1 element, and sequencing of the regions with primers internal to Ty1 elements revealed heterozygous SNPs consistent with the fusion junction containing more than one Ty element. In the case of I17, the chromosome X target consisted of two tandem Ty1s (YJRWTy1-1 and YJRWTy1-2). The size of the amplified region and the fact that the observed SNPs included Ty912 SNPs, YJRWTy1-1 SNPs, and a mixture of homozygous and heterozygous SNPs from YJRWTy1-1 and YJRWTy1-2, is consistent with a Ty912 fusion to YJRWTy1-1 resulting in a junction containing two Ty1 elements. The features of I13 and I16 suggest a similar rearrangement structure as I17; however, because the sequences at the target junctions were annotated in the reference sequence as solo deltas (an unannotated delta near YCRWdelta10 and YCRWdelta10 itself), we lacked sufficient sequence information to perform full SNP analyses of the multiple Ty1 elements found in the fusion junctions.
Class III GCRs containing duplications from telomeres to centromere-oriented Ty1s were generated by complex rearrangements
The 11 Class III GCR-containing strains (12.5%) resembled the more common Class II GCRs, except that the duplicated region on the target chromosomes were bounded by Ty sequences that the reference genome suggested were transcriptionally oriented towards the centromere rather than towards the telomere (Table 2). In each of these 11 cases, homology-driven rearrangements between the telomere-oriented Ty912 and the centromere-oriented target would be expected to duplicate a region on the centromeric side of the target Ty element in contrast to duplication on the telomeric side, as was observed in all 11 cases. Such events have been previously observed [17], [30], but their structure has not been investigated in detail. Eight of the 133 centromere-oriented Ty sequences present in the reference sequence bordered regions that were duplicated in the 11 Class III GCR-containing strains (Figure 1d). To understand the nature of this unexpected class of GCRs, we first examined 6 of these 8 native Ty loci; the remaining 2 loci were located in the repetitive regions of chromosome XII and could not be definitively analyzed. Southern analyses of the six target Ty loci in the wild-type parental strain (RDKY6076) revealed that only YMLWTy1-1 (targeted in isolate I9) had increased size compared to the reference sequence (Table 5). The change in size of the YMLWTy1-1 locus was consistent with the presence of an additional full-length Ty1 element and subsequent analysis showed that our strains contained 2 tandem Ty1s (termed YMLWTy1-1A and YMLWTy1-1B below) at the YMLWTy1-1 locus that were oriented towards the centromere. We chose two isolates of this class (I9 and I14) to analyze further. Both isolates had a single abnormally-sized chromosome V consistent with fusion of Ty912 to the duplicated region (Figure 2a; Table 3). Details of the structure of each isolate are described below.
Tab. 5. Analysis of centromere-oriented Ty target loci. 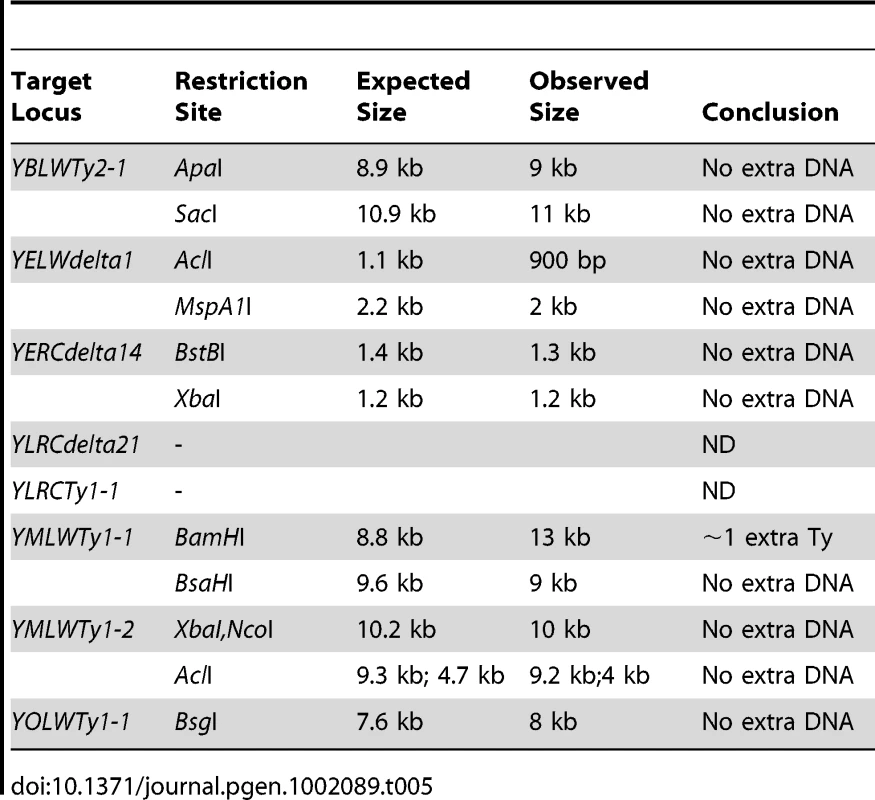
Isolate I9, derived from a wild-type strain, contained a GCR associated with the chromosome V Ty912-TEL05L deletion (Figure 3a) and a 184 kb chromosome XIII duplication from TEL13L to the tandem centromere-oriented Ty1s, YMLWTy1-1A and YMLWTy1-1B (Figure 3b); no other region of the genome was observed to be duplicated. To confirm that chromosome V was indeed fused to a copy of the left arm of chromosome XIII, we cloned the fusion junction by: 1) integrating plasmid pRDK1564 into the region adjacent to the junction on chromosome V, 2) isolating genomic DNA, 3) cutting the genomic DNA with the restriction enzyme Xba I that cut once within the plasmid but not within Ty-related sequences, 4) circularizing the resulting fragments, and 5) recovering the plasmid by transformation into E. coli (Figure 3c). Partial sequencing of the cloned junction confirmed that the I9 GCR contained the duplicated region of chromosome XIII at the YMLWTy1-1A/B locus fused to chromosome V at the Ty912 locus. This analysis also verified the orientations of YMLWTy1-1A/B and Ty912. Restriction mapping of the recovered plasmid indicated that the cloned Ty fusion junction was unexpectedly large (∼25 kb) and consistent with the size of approximately four Ty1 elements. Sequencing of the junction with internal Ty1-specific primers resulted in a highly heterozygous sequencing read consistent with the simultaneous sequencing of 2 or more unique Ty1 sequences (Figure 3d). The large size of the junction, combined with the sequencing data and the fact that only chromosome XIII sequences were observed to be duplicated, suggests that the extra Ty1 sequences in the fusion junction arose as a result of a complex rearrangement mechanism, such as a breakage-fusion-bridge event driven by the formation of an initial dicentric chromosome that amplified the Ty1 sequences present on the 2 partner chromosomes (Figure 3e). Ty-mediated resolution of dicentric translocation chromosomes have been previously observed during both an analysis of the structure of dicentric GCRs [25] and an analysis of GCRs derived from endogenous inverted Ty1 repeats [27].
Fig. 3. Analysis of isolate I9 reveals a structure consistent with breakage-fusion-bridge. 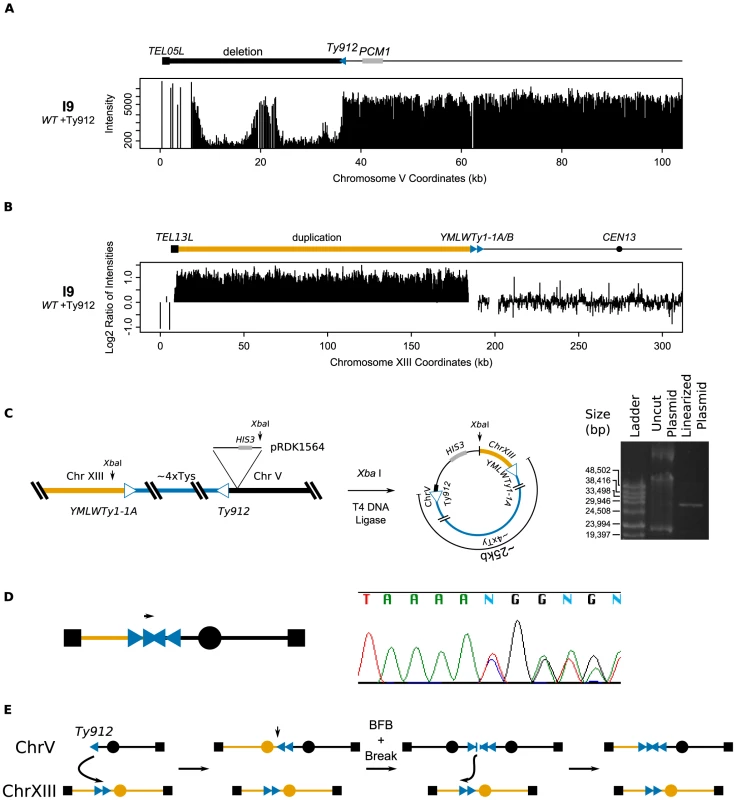
A. Deletion of the region between TEL05L and Ty912 in isolate I9. B. Duplication of the genomic region between TEL13L and YMLWTy1-1A/B. The duplicated region is located upstream of the 5′ end of YMLWTy1-1A/B as indicated. C. Schematic of the integration of pRDK1564 near the fusion junction and the subsequent steps utilized to create a vector cloning the fusion junction. The linearized form of the vector measures approximately 30 kb; 25 kb of the cloned fusion vector is predicted to be Ty DNA. D. Internal primer hybridizing within the epsilon sequence of the Ty reveals a heterogeneous chromatogram, indicating presence of at least two unique Ty sequences. E. A proposed mechanism for the creation of the observed structure. Exposed Ty912 sequence invades YMLWTy1-1A, resulting in a dicentric chromosome. Formation of the dicentric results in one round of breakage-fusion-bridge (with the break at the vertical arrow) followed by a subsequent break due to the formation of a second dicentric. The new break invades a wild type copy of chromosome XIII at YMLWTy1-1A/B resulting in the observed duplication and correctly sized fusion junction. We also investigated isolate I14, which contained both the chromosome V Ty912-TEL05L deletion and a 145 kb duplication of the right arm of chromosome V between YERCdelta14 and TEL05R (Figure 4a, 4b). Based on the aCGH data, we originally predicted that Ty912 had fused to YERCdelta14. However, after cloning and sequencing the fusion junction containing Ty912, we found that Ty912 underwent a HR-mediated fusion with YERCTy1-1, a centromere-oriented Ty1 located approximately 17.5 kb telomeric to YERCdelta14 (Figure 4c). The size of the cloned junction was consistent with a single Ty1 element fused to the unique chromosome V sequence downstream of YERCTy1-1. This cloned Ty912/YERCTy1-1 fusion indicated the initial rearrangement could have resulted in either a dicentric chromosome if two copies of chromosome V were involved (Figure 4d) or a ring chromosome if only one copy of chromosome V was involved. The data strongly suggest that I14 initially formed a dicentric chromosome that later rearranged rather than a ring chromosome because (1) chromosome V from I14 was not trapped in a well during PFGE (Figure 2a), (2) the region telomeric to YERCTy1-1 was duplicated, (3) the region centromeric to YERCdelta14 was not duplicated, and (4) dicentric chromosomes, but not ring chromosomes, are known to be unstable [25]. The aCGH data are consistent with the initial dicentric chromosome breaking at or near the centromere-oriented delta sequence YERCdelta14 allowing the chromosome end to then invade the telomere-oriented YERWdelta17 or an unannotated telomere-oriented delta we call YERWdelta20B (ChrV: 449316-449625; Figure 4c) and copy to the telomere, possibly by BIR (Figure 4d). Regardless of the final fusion event, it is clear that the GCR present in isolate I14, like the GCR present in isolate I9, was the product of multiple rearrangements.
Fig. 4. The dicentric structure of isolate I14 is resolved by Ty-mediated recombination. 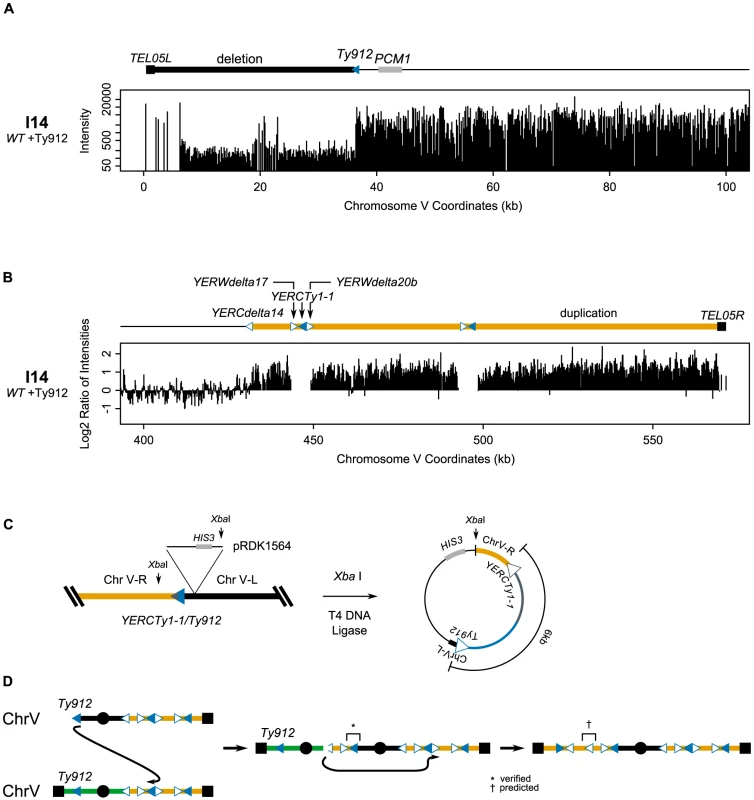
A. Deletion of the region between TEL05L and Ty912 in isolate I14. B. Duplication of the region between YERCdelta14 and TEL05R. C. Schematic of the integration of pRDK1564 near the fusion junction and subsequent steps utilized to create a vector cloning the fusion junction. The Ty fusion junction measures approximately 6 kb, the size of one full length Ty1 element. D. Exposed Ty912 DNA invades YERCTy1-1, copying through the centromere and creating a dicentric chromosome. The unstable dicentric chromosome breaks at YERCdelta14 and invades YERWdelta20b, copying the rest of the duplicated region to TEL05R. * is a verified junction; † a predicted junction. Class IV GCRs contain duplications from telomeres to clusters of Tys in both telomeric and centromeric orientations
The 14 Class IV GCR-containing strains each had both the chromosome V Ty912-TEL05L deletion and a single duplicated region of another chromosome extending from a telomere to a region of DNA containing a cluster of Ty-related elements in both telomeric and centromeric orientations (Figure 1d; Table 2; Figure S3a–S3c). No other duplicated or deleted regions were identified. Based on the hypothesis that these rearrangements arose by the same mechanisms that gave rise to Class II and III GCRs, we successfully amplified fusion junctions for isolates I23, I65, I76, I58, and I48, indicating the telomere-oriented Ty1 loci YLRWTy1-2, YLRWTy1-2, YLRWTy1-2, YHLCdelta1, and YGRWTy1-1 mediated the respective fusion junctions (Figure S3d) and that these 5 GCRs were all Class II GCRs. Since Class IV rearrangements were unlikely to be mechanistically distinct from Class II and III GCRs and since Class IV GCRs had similar aCGH patterns to those analyzed above, we did not further analyze other GCRs of this class.
Class V GCRs contain multiple duplicated regions
A complex GCR containing a duplicated and triplicated region
Isolate I12, originally obtained from a wild-type strain, contained the chromosome V Ty912-TEL05L deletion (Figure 5a), a 6.2 kb triplicated region of chromosome XIII bordered by the centromere-oriented YMLWTy1-1A/B at one end and the centromere-oriented YMLWTy1-2 at the other end, and a 184 kb duplicated region of chromosome XIII bordered by YMLWTy1-1A/B and TEL13L (Figure 5b). The copy numbers of the amplified chromosome XIII regions were confirmed by qPCR (data not presented). PFGE analysis of this isolate revealed an abnormal chromosome V, consistent with the size of a copy of the terminally deleted chromosome V joined to 2 copies of the 6.2 kb triplicated segment and 1 copy of the 184 kb duplicated segment (Figure 2c; Table 3). All of the other chromosomes present in this isolate appeared to be of wild-type size. We therefore cloned and sequenced the fusion junction containing Ty912 and found that Ty912 was fused to the centromere-oriented YMLWTy1-1A; moreover, the size of the cloned fusion junction was ∼12 kb, consistent with the presence of both a hybrid Ty912/YMLWTy1-1A Ty element and the YMLWTy1-1B element at the junction (Figure 5c). Southern blots of Acl I-digested genomic DNA using probes flanking Ty912 and YMLWTy1-1A/B were consistent with a hybrid Ty912/YMLWTy1-1A junction and further indicated the presence of a wild-type copy of chromosome XIII (Figure 5d), consistent with PFGE results (data not presented). Cloning and sequencing of the second fusion junction involving YMLWTy1-2 revealed that each side of the fusion junction contained a copy of the triplicated region adjacent to YMLWTy1-2, demonstrating that the fusion junction contained two copies of YMLWTy1-2 in an inverted orientation (Figure 5e). Consistent with this, restriction mapping indicated that the fusion junction was ∼11.4 kb in length, slightly shorter than the length of two full-length Ty1s. This inverted duplication is consistent with a product from breakage-fusion-bridge cycles involving a dicentric chromosome generated by an initial Ty912/YMLWTy1-1A fusion (Figure 5f). Moreover, given the fact that a Southern blot with a probe hybridizing to the triplicated region flanking YMLWTy1-1A/B revealed only two discrete bands and a Southern blot with a probe hybridizing to the duplicated region flanking YMLWTy1-1A/B revealed only one discrete band instead of the three and two bands suggested by the triplication and duplication of the regions, it is likely the inverted triplicated DNA is fused to a wild-type configuration of the YMLWTy1-1A/B locus. This is consistent with a breakage-fusion-bridge mechanism that would have fused together two inverted copies of YMLWTy1-2, as the dicentric chromosome created from the breakage-fusion-bridge cycle would have broken again and, by HR, copied from the triplicated region to the left telomere of chromosome XIII. Thus, the chromosome V-chromosome XIII translocation predicted by both junctions accounts for the duplicated and triplicated regions observed in the aCGH data.
Fig. 5. I12's tripartite recombination involves breakage-fusion-bridge. 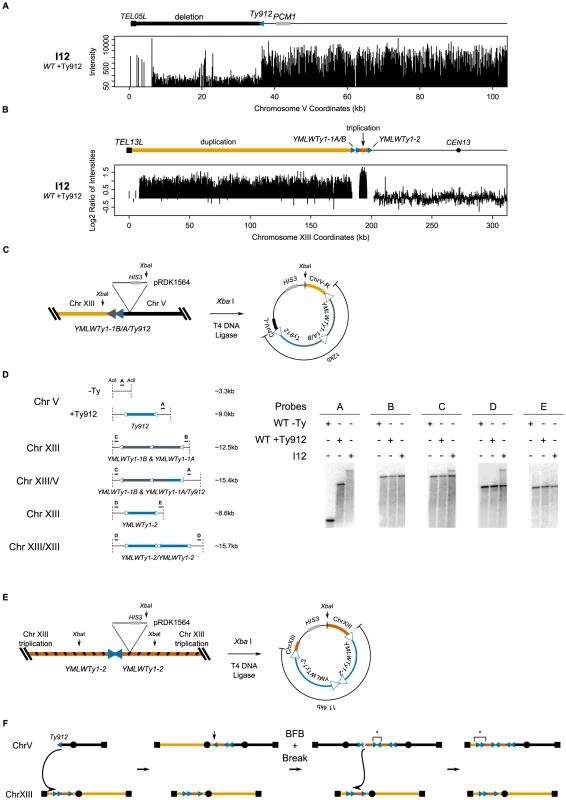
A. Deletion of the region between TEL05L and Ty912 in isolate I12. B. Duplication of the region between TEL13L and YMLWTy1-1A/B. Triplication of the region between YMLWTy1-1A/B and YMLWTy1-2. C. Schematic of the integration of pRDK1564 near the fusion junction on chromosome V and subsequent steps utilized to create a vector cloning the fusion junction. The Ty fusion vector measures approximately 12 kb, as would be expected if Ty912 recombined with YMLWTy1-1A. D. Map of Southern probes and sizes of the expected fragments. DNA from the wild-type −Ty (WT −Ty), wild-type +Ty912 (WT +Ty912), and I12 were cut with restriction enzyme Acl I and subsequently probed with the indicated Southern probes. Bands within a dotted box represent equivalently sized bands. E. Schematic of the integration of pRDK1564 near a copy of YMLWTy1-2 and subsequent steps utilized to create a vector cloning the fusion junction. Results indicated two copies of the triplicated region fused together in an inverted fashion with the Ty fusion measuring approximately 11.4 kb, as would be expected if YMLWTy1-2 underwent a break-fusion-bridge cycle. F. Exposed Ty912 DNA invades YMLWTy1-1A on chromosome XIII. This leads to the formation of a dicentric that then leads to a breakage fusion bridge event at the vertical arrow followed by another break; the broken chromosome is repaired by mediating a translocation to homologous DNA on the wild type chromosome XIII. * indicates verified junctions. GCRs associated with multiple duplications bordered by Ty elements
Among the GCRs studied were 3 that contained aCGH patterns indicative of the chromosome V Ty912-TEL05L deletion and duplication of two different chromosomal segments from one or two different chromosomes. In each isolate, one duplicated region was bounded on both sides by Ty sequences and the other duplicated region was bounded by a Ty sequence and a telomere. Each isolate appears to have undergone two rounds of Ty-mediated translocations. Isolate I46 was obtained from a mec1Δ sml1Δ mutant and contained a GCR that was associated with the chromosome V Ty912-TEL05L deletion, a ∼377 kb duplicated region of the right arm of chromosome XII (centromeric border YLRCdelta9, YLRWTy1-2, and YLRWdelta12; telomeric border YLRCTy2-2), and a ∼91 kb duplicated region of chromosome XVI (centromeric border YPRWTy1-3, YPRCdelta22, YPRCTy1-4; telomeric border TEL16R) (Figure S4a–S4c). This isolate can be explained by two simple translocations between two different groups of Ty loci: one between Ty912 on chromosome V and YLRWTy1-2 on chromosome XII and another translocation between YLRCTy2-2 on chromosome XII and YPRCTy1-4 on chromosome XVI (Figure S4d). Similarly, isolate I47, also obtained from a mec1Δ sml1Δ mutant, contained a GCR that was associated with the chromosome V Ty912-TEL05L deletion, a ∼174 kb duplication of chromosome IV (centromeric border YDRWdelta11 and YDR170W-A; telomeric border YDRWTy2-3 and YDRCTy1-3), and another ∼179 kb duplication of chromosome IV (centromeric border YDRWdelta29 and YDRWdelta30; telomeric border TEL04R) (Figure S4e, S4f). This isolate can also be explained by two simple translocations between two groups of Ty elements: an initial translocation between Ty912 on chromosome V and YDRWdelta11 on chromosome IV and a second between YDRWTy2-3 and YDRWdelta29 (Figure S4g). The final isolate in this class, isolate I54, was obtained from an rtt109Δ mutant and contained a GCR associated with the chromosome V Ty912-TEL05L deletion, a ∼105 kb duplication of the right arm of chromosome IV (centromeric border YDRWTy1-4; telomeric border YDRWTy1-5), and a ∼262 kb duplication of chromosome X (centromeric border YJRWTy1-1 and YJRWTy1-2; telomeric border TEL10R) (Figure S4h–S4j). Like the previous two isolates, this isolate can be explained by two Ty-mediated translocations: the first between Ty912 on chromosome V and YDRWTy1-4 on chromosome IV, and the second between YDRWTy1-5 on chromosome IV and YJRWTy-1-1/2 on chromosome X. In all cases, the aCGH data were consistent with the GCR-containing strains also containing a complete copy of the chromosomes that were the source of the duplicated regions. We were unable to amplify the predicted translocation breakpoint junctions in these three isolates by PCR. While it was not possible to completely determine the structure of each GCR, the available data were consistent with the hypothesis that each of these 3 GCRs involved non-reciprocal translocations that joined 3 translocated segments. In the case of I47 and I54, the orientations of the Ty elements at the breakpoint junctions were only consistent with mechanisms involving template switching between 3 templates and were inconsistent with the formation of an intermediate dicentric chromosome. In contrast, in the case of I46, the orientations of the Ty elements at the breakpoint junctions were consistent with mechanisms involving switching between 3 templates but could also have been created by the formation of an intermediate dicentric translocation chromosome formed between chromosome V and chromosome XII.
A complex translocation containing a microhomology-mediated fusion
Isolate I66 was obtained from a spt21Δ mutant and resembled isolates I46, I47, and I54 described above except that one of the junctions involved a fusion mediated by a microhomology breakpoint. I66 contained the chromosome V Ty912-TEL05L deletion, a ∼100 kb duplication of chromosome X (centromeric border YJRWTy1-1 and YJRWTy1-2; telomeric border part of the CSN12 gene), and a ∼32 kb duplication of chromosome XVI (centromeric border part of the SKI3 gene; telomeric border TEL16R) (Figure S5a–S5c). We confirmed the fusion junction between Ty912 and YJRWTy1-1 by PCR (Figure S5d). Amplification of CSN12 and SKI3 by PCR revealed wild-type copies of both genes and PCR amplification and sequencing confirmed the presence of a CSN12-SKI3 fusion gene mediated by a region of microhomology (5′-CTTC-3′) between the two genes (Figure S5e). The data were most consistent with the presence of a tripartite translocation consisting of chromosome V at Ty912 joined to a copy of a fragment of chromosome X at YJRWTy1-1 that was then joined at CSN12 to a copy of a fragment of chromosome XVI at SKI3 extending to TEL16R (Figure S5f). This rearrangement was likely a product of a non-reciprocal translocation as the aCGH data were consistent with the presence of a complete copy of both chromosomes X and XVI. Furthermore, the orientations of the sequences at the breakpoint junctions were only consistent with mechanisms involving switching between the chromosome X and XVI templates and were inconsistent with the formation of an intermediate dicentric chromosome.
Discussion
In the present study, we describe the development of a quantitative genetic assay that allows for the assessment of the impact of genetic defects on the rate of Ty1-mediated GCRs and facilitates analyses of the structures of the resulting Ty1-mediated GCRs. The results described here demonstrate that presence of a telomere-oriented Ty912 on a nonessential terminal arm of chromosome V greatly increases the spontaneous rate of loss of that chromosome arm. Furthermore, this loss appears to be driven primarily by non-reciprocal translocations between Ty912 and other Ty-related elements in the genome, resulting in a broken chromosome V joined to a fragment of another chromosome that terminates with a telomere. The observed rearrangement products are consistent with HR-mediated processes, such as BIR and half crossovers [29], [43], [50]–[55], which result in translocation breakpoints occurring at regions of homology mediated by RAD52-dependent HR.
The majority of the Ty1-mediated GCRs observed (∼60.2%) were simple non-reciprocal translocations likely mediated by HR between the telomere-oriented Ty912 on chromosome V and a single telomere-oriented Ty elements located on another chromosome arm. These results are consistent with a simple model in which a resection of a spontaneous DSB on chromosome V exposes single stranded Ty912 DNA that then invades a telomere oriented Ty element on another chromosome arm and leads to the replication of DNA from this Ty element to the telomere by BIR [17], [22], [24], [25], [28], [56]. The results are also consistent with another model in which spontaneous DSBs form on both chromosome V and another target chromosome during the G2 phase of the cell cycle. These DSBs are followed by HR between Ty912 on chromosome V and a Ty element on the broken target chromosome. Mitosis then occurs, and the cell containing the remaining intact copy of chromosome V is selected against in the assay selection system; such HR events could be mediated by SSA as well as other HR mechanisms [27], [29], [30], [55], [57]. Other GCRs identified, such as those that involved the duplication of telomeric regions adjacent to centromere-oriented Ty elements or those that involved the duplication of multiple chromosomal regions, appear to be the products of more complicated mechanisms, ranging from the formation and resolution of unstable dicentric translocation chromosomes [25], [27] to sequential linked monocentric translocations consistent with template switching during BIR [58], [59]. In all of the events observed, it is possible that the initiating DSBs occurred at the site of participating Ty elements. However, it is more likely that the initiating DSBs occurred randomly and were resected to the participating Ty elements; the selection of Ty-mediated GCR events was due to the fact that the rates of HR mediated GCRs are much higher than those of single copy sequence mediated GCRs (Table 1; [11]). Surprisingly, most of the genetic defects that increased the rate of Ty912-mediated GCRs did not appear to significantly alter the types of GCRs recovered (Table S2) in spite of the fact that Ty elements targeted in rearrangements had an apparently nonrandom distribution (Figure 1d). This observation raises the possibility that Ty-mediated repair of DNA damage may be biased to target specific locations or Ty elements in the genome. However, because our data were pooled from the results of analyses of GCRs isolated in multiple mutant backgrounds, and because we currently lack a rapid, cost efficient method to identify very large numbers of chromosome arm duplications, we were unable to determine whether this result was due to the analysis of a biased set of GCRs or was a manifestation of true repair target bias.
To gain insights into whether the pathways affecting Ty1-mediated GCRs were similar to those affecting other types of GCRs, we surveyed mutations previously demonstrated to affect GCR rates as well as mutations known to affect Ty metabolism. Most of the mutations that increased rates of single copy sequence-mediated GCRs also increased GCR rates in the +Ty912 GCR assay, as well as in the segmental duplication GCR assay [11], suggesting that these increases are likely due to elevated levels of DNA damage leading to aberrant repair. In addition, all of the mutations tested that specifically increase GCRs mediated by segmental duplications (mrc1Δ, sgs1Δ, srs2Δ, rrm3Δ, rtt109Δ, and rad6Δ) increased the rate of Ty912-mediated GCRs, indicating an overlap between the pathways that suppress segmental duplication-mediated GCRs and the Ty912-mediated GCRs. These mutations potentially cause defects in pathways that specifically suppress non-allelic HR. Interestingly, the wild-type strain containing the Ty912 on chromosome V had a higher GCR rate than that of the wild-type strain containing the segmental duplication-mediated GCR assay. This is likely due at least partially to both the larger size of the Ty912 element and the larger number of potential alternative repair templates available in the genome. In contrast, most of the gene defects affecting Ty1 transcription and transposition seemed to have little or no specific effects on the rate of Ty912-mediated GCRs, as many of the mutations that increased the +Ty912 GCR rate also increased the GCR rates in other GCR assays that did not involve Ty1 elements. This suggests that the +Ty912 GCR assay is an excellent model for understanding mechanisms suppressing rearrangements between high copy number repeats.
Some of the mutations tested had distinct effects in the Ty1-mediated GCR assay that were surprisingly different than their effect in other GCR assays, including the segmental duplication assay. First, a rad52Δ that decreases HR increases the rate of single copy sequence-mediated GCRs [8] and decreases the rate of Ty1 - and other duplication-mediated GCRs [11]; this is expected as HR is thought to play a central role in the formation of duplication mediated GCRs, but promotes the correct repair of DNA damage that would otherwise lead to single copy sequence-mediated GCRs. Second, deletion of rad51Δ increased both single copy-mediated and Ty-mediated GCRs (Table 1), but had relatively little effect on low-copy segmental duplication-mediated GCRs. This was surprising, especially given the fact that products of Ty-mediated HR in a wild-type strain were most consistent with the products of BIR, a process that is highly dependent on RAD51 [50]. A previous report found that a rad51Δdeletion caused an increased Ty1 recombination rate that led to a rise in the formation of solo LTRs but suppression of Ty conversion events [44]. Our study also revealed an increase in Ty-mediated GCRs upon deletion of RAD51 and suggested that another HR mechanism, such as a RAD59-dependent RAD51-independent single-stranded annealing event followed by a half-crossover [29], [55], could be responsible for the formation of Ty-mediated GCRs and that furthermore, the mechanism of repair was mutagenic and suppressed by RAD51. Deletion of both RAD51 and RAD59 resulted in a GCR rate equivalent to that of a wild-type strain in the +Ty912 GCR assay (Table 1), which is consistent with a view that RAD59 promotes the mutagenic repair of DNA in the presence of the Ty912 and absence of RAD51. Third, the rad51Δ rad59Δ double deletion resulted in a GCR rate that was higher than that of a rad52Δ strain (unpaired Wilcoxon rank sum test; p = 5.59×10−4), a pattern which was not observed in the segmental duplication assay [11], but has been noted in a previous assay [46]. This difference suggests that other RAD52-dependent factors besides RAD51 and RAD59 play a larger role in the formation of Ty1-mediated GCRs than in the formation of lower copy segmental duplication-mediated GCRs. Fourth, we identified mutations that significantly reduced the +Ty912 GCR rate, but did not affect the rate of single copy sequence-mediated GCRs. These mutations include pmr1Δ, rtt103Δ and gal11Δ, all of which alter Ty metabolism [34], [41], [42]. This suggests that some aspects of normal Ty metabolism may impact the formation of GCRs. Overall, the genetic analysis performed as part of the present study indicates that the +Ty912 GCR assay is a high sensitivity assay suitable for the analysis of pathways that affect the rate of GCRs mediated by repetitive DNA and provides a means to detect common pathways that suppress genome instability, novel pathways that affect repetitive DNA, and different aspects of known GCR suppression pathways not previously studied.
The work presented here indicates that dispersed repetitive elements in S. cerevisiae DNA, like the Ty elements that are analogous to human LINE and Alu elements in abundance, are chromosomal features that result in increased genomic instability. Analysis of this genomic instability has provided insights into both the HR-based mechanisms that yield Ty-mediated GCRs and the pathways that normally act to prevent such GCRs. In humans, suppression of non-allelic HR is likely important for preventing GCRs from occurring due to the large numbers of dispersed repetitive sequences in the human genome, particularly because such GCRs have been seen to underlie genetic diseases and are found among the genome rearrangements in many cancers. Our observations on the pathways that preferentially suppress Ty-mediated GCRs and on the mechanisms that produce such GCRs suggest that the structures of GCRs observed in disease situations will provide signatures diagnostic for particular genome instability-causing genetic defects.
Methods
General methods
PCR
All PCR reactions used a mixture of Pfu from Stratagene and KlenTaq from Ab Peptides. A master mix of 16∶1 KlenTaq (25 U/ul) to Pfu (2.5 U/ul) by volume was made for a total unit ratio of 160 U KlenTaq per 10 U Pfu per microliter.
DNA isolation
In general, the Gentra Puregene Kit (Qiagen) was used to isolate S. cerevisiae DNA for the microarray hybridizations and PCRs as described by the manufacturer. We modified a previous protocol [60] to isolate DNA for use in Southern blots, cloning, and amplification of Ty fusion junctions. Modifications included the use of 5 ml cultures instead of 10 ml cultures, an extra chloroform extraction step after the phenol∶chloroform∶isoamyl-alcohol (25∶24∶1) extraction step, incubation at 37°C for 10 minutes instead of 5 minutes after addition of RNase A, use of 5 mg/ml RNase A instead of 1 mg/ml RNase A, and an additional phenol∶chloroform∶isoamyl-alcohol and chloroform extraction after incubation of the DNA with RNase A.
Statistical tools
R (version≥2.9.2) was used to calculate p-values for Wilcoxon rank-sum tests. Ninety-five percent confidence intervals for the median were calculated using a two-sided nonparametric test (http://www.math.unb.ca/~knight/utility/MedInt95.htm).
Ty912 cassette construction
We constructed the plasmid pRDK1251 containing Ty912 [32] surrounded by chromosome V targeting sequence to integrate the Ty1 element into the NPR2-CIN8 intergenic region. The flanking targeting sequence from CIN8 (ChrV: 39724-36426) was amplified by PCR from S. cerevisiae genomic DNA with primers JCP41 and JCP42 (Table S3), which introduced flanking Sac I and Sma I restriction sites. The flanking targeting sequence from NPR2 (ChrV: 36340-33913) was also amplified with primers JCP43 and JCP44, which introduced flanking BamH I and Xba I restriction sites. Ty912 was amplified from plasmid B155 (FB118), a generous gift of Dr. Fred Winston (Harvard Medical School), with JCP39 and JCP40, which introduced flanking Sma I and BamH I restriction sites. The pRDK1251 plasmid was generated by sequentially subcloning the Sac I-CIN8-Sma I, Sma I-Ty912-Xba I, and BamH I-NPR2-Xba I fragments into pUC19 [61], such that the NPR2 fragment was joined to the Ty912-XbaI site at the Xba I site present at genomic coordinate 35,997.
Construction of assay-containing strains
We sequentially transformed URA3 and HIS3 markers into the intergenic region between NPR2 and CIN8 in RDKY6078 (MATa lys2-10A, hom3-10, ura3Δ0, leu2Δ0, trp1Δ63, his3Δ200) to generate strain RDKY6081. We released the Ty912 cassette from pRDK1251 by Xba I digestion and then transformed the Ty912 cassette (∼10 ug) into RDKY6081 using standard lithium acetate transformation protocols and plated the putative transformed colonies onto YPD plates. After one day of growth at 30°C, this YPD plate was replica-plated to a uracil dropout plate containing 1 g/L 5-fluoroorotic Acid (5FOA). After ∼2 days of growth at 30°C, the resulting colonies were replica-plated from the dropout plate containing 5FOA to a separate histidine dropout plate. Colonies were screened for growth on the uracil dropout plate containing 5FOA and for non-growth on the histidine dropout plate. We verified the insertion of Ty912 between NPR2 and CIN8 on the Crick strand of RDKY6082 by PCR (primer pair JCP44 & JCP85; primer pair JCP125 & JCP349 (Table S3)). A hxt13::URA3 KO cassette was amplified from RDKY3615 (MATa, ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2-Bgl, hom3-10, ade2Δ1, ade8, hxt13::URA3) by PCR (primer pair JCP28 & JCP29 (Table S3)) and transformed into RDKY6082 to create RDKY6084. RDKY6084 was then backcrossed to RDKY6079 (MATalpha lys2-10A, hom3-10, ura3Δ0, leu2Δ0, trp1Δ63, his3Δ200) and our +Ty912 wild type haploids RDKY6076/6077 (MATa lys2-10A, hom3-10, ura3Δ0, leu2Δ0, trp1Δ63, his3Δ200 iYEL062w::Ty912 hxt13::URA3) were isolated. The −Ty wild-type strains RDKY6088/6089 (MATa lys2-10A, hom3-10, ura3Δ0, leu2Δ0, trp1Δ63, his3Δ200 hxt13::URA3) were isolated by first transforming the previously isolated hxt13::URA3 cassette into RDKY6078, backcrossing the resulting strain to RDKY6079, and isolating haploids. All strains used in the experiments were isogenic to either RDKY6076 (+Ty912) or RDKY6088 (−Ty), which differ only by the presence of the Ty912 insertion (Table S4).
Construction of mutant strains
Strains with kanMX4 marked deletions of interest were created using kanMX4 cassettes amplified from the systematic S. cerevisiae knockout library [62]. Strains with deletions marked with TRP1 and HIS3 cassettes were created by amplifying the cassettes from the pRS series of plasmids [63] with PCR primers that added 50 bases of the target homology of interest. These cassettes were then transformed into strains of interest using standard lithium acetate transformation protocols followed by verification of the correct insertion by PCR with flanking and internal primers. All strains used in the experiments are available upon request (Table S4), as are the primer sequences used in their construction.
GCR rate calculations
General methods, including use of YPD and synthetic dropout medias, have been described previously [9]. For each strain, we used 14 or more independent cultures in our fluctuation analyses [64] to calculate the median rates [65].
Array comparative genomic hybridization
For each strain of interest, 1 µg of DNA was labeled with either Cy5 or Cy3 and applied to one of four wells of a Nimblegen 4-plex microarray. The GeneChip Microarray Core (UC San Diego School of Medicine) performed the hybridization and scanning. Probes on the array had a median base pair spacing of ∼200 bp between probes. DNA for seven independent GCR isolates and one −Ty wild type strain (RDKY6088) were applied to each 4-plex microarray. Each microarray thus contained either a GCR isolate hybridized along with the −Ty wild-type DNA or with DNA from another GCR isolate (Table S5). The R package Ringo (> = 1.8.0) [66], an add-on to the Bioconductor suite (> = 2.4.1) [67], was used in combination with the SignalMap software (> = 1.9) from Nimblegen to visualize the aCGH data. Increased copy numbers of probes were revealed by identifying continuous regions whose log ratios deviated from 0. Deletions were identified by analyses of the raw intensity data and identification of regions with continuously low regional intensity.
Ty fusion junction amplification
PCR
Ty fusion junctions were amplified using the following protocol: 5 min 95°C initial denaturing step; 25 cycles of 12 or 20 sec 95°C denaturation, 30 sec 63.8°C annealing, 7 min 68 or 72°C extension; final 7 minute 68 or 72°C extension. The resulting amplicons were gel-purified and sequenced using an ABI3730 sequencer and standard protocols.
Construction and insertion of a plasmid for subcloning of GCR junctions
Plasmid pRDK1564 was constructed by cloning a HIS3 marker flanked by Sac I and Sma I restriction sites (amplified using PCR primers JCP404 and JCP405 (Table S3)) into the multiple cloning site of pUC19. Primers with homology centromeric to Ty912 (primer pair JCP445 and JCP446 (Table S3)) were used to amplify and linearize the plasmid for insertion into the intergenic region between Ty912 and CIN8 in GCR isolates. The amplicon was transformed into the GCR isolates using the standard lithium acetate transformation protocol.
Subcloning of chromosomal GCR breakpoints
One µg of genomic DNA containing the integrated plasmid construct was digested with Xba I. The DNA was diluted to 2.5 ng/ul and ligated overnight at 16°C with 1,600 U of T4 ligase. The plasmid was then recovered by transforming the DNA into SURE electrocompetent cells (e14-(McrA-) Δ(mcrCB-hsdSMR-mrr)171 endA1 gyrA96 thi-1 supE44 relA1 lac recB recJ sbcC umuC::Tn5 (Kanr) uvrC [F′ proAB lacIqZΔM15 Tn10 (Tetr)]) from Stratagene. The resulting plasmids were then characterized by restriction and PCR mapping, and sequencing using standard methods as described under individual experiments.
Supporting Information
Zdroje
1. VenterJCAdamsMDMyersEWLiPWMuralRJ 2001 The sequence of the human genome. Science 291 1304 1351
2. LanderESLintonLMBirrenBNusbaumCZodyMC 2001 Initial sequencing and analysis of the human genome. Nature 409 860 921
3. LevySSuttonGNgPCFeukLHalpernAL 2007 The diploid genome sequence of an individual human. PLoS Biol 5 e254 doi:10.1371/journal.pbio.0050254
4. BaileyJALiuGEichlerEE 2003 An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet 73 823 834
5. DeiningerPLBatzerMA 1999 Alu repeats and human disease. Mol Genet Metab 67 183 193
6. LangeJSkaletskyHvan DaalenSKEmbrySLKorverCM 2009 Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell 138 855 869
7. LengauerCKinzlerKWVogelsteinB 1998 Genetic instabilities in human cancers. Nature 396 643 649
8. MyungKChenCKolodnerRD 2001 Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411 1073 1076
9. ChenCKolodnerRD 1999 Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 23 81 85
10. HuangMEKolodnerRD 2005 A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol Cell 17 709 720
11. PutnamCDHayesTKKolodnerRD 2009 Specific pathways prevent duplication-mediated genome rearrangements. Nature 460 984 989
12. KimJMVanguriSBoekeJDGabrielAVoytasDF 1998 Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 8 464 478
13. RoederGSFinkGR 1980 DNA rearrangements associated with a transposable element in yeast. Cell 21 239 249
14. ChaleffDTFinkGR 1980 Genetic events associated with an insertion mutation in yeast. Cell 21 227 237
15. RoederGSFinkGR 1982 Movement of yeast transposable elements by gene conversion. Proc Natl Acad Sci U S A 79 5621 5625
16. DunhamMJBadraneHFereaTAdamsJBrownPO 2002 Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 99 16144 16149
17. UmezuKHiraokaMMoriMMakiH 2002 Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160 97 110
18. SuroskyRTTyeBK 1985 Resolution of dicentric chromosomes by Ty-mediated recombination in yeast. Genetics 110 397 419
19. KupiecMPetesTD 1988 Meiotic recombination between repeated transposable elements in Saccharomyces cerevisiae. Mol Cell Biol 8 2942 2954
20. KupiecMPetesTD 1988 Allelic and ectopic recombination between Ty elements in yeast. Genetics 119 549 559
21. VincentAPetesTD 1989 Mitotic and meiotic gene conversion of Ty elements and other insertions in Saccharomyces cerevisiae. Genetics 122 759 772
22. ArguesoJLWestmorelandJMieczkowskiPAGawelMPetesTD 2008 Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci U S A 105 11845 11850
23. HoangMLTanFJLaiDCCelnikerSEHoskinsRA 2010 Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet 6 e1001228 doi:10.1371/journal.pgen.1001228
24. LemoineFJDegtyarevaNPLobachevKPetesTD 2005 Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120 587 598
25. PennaneachVKolodnerRD 2009 Stabilization of dicentric translocations through secondary rearrangements mediated by multiple mechanisms in S. cerevisiae. PLoS ONE 4 e6389 doi:10.1371/journal.pone.0006389
26. NarayananVMieczkowskiPAKimHMPetesTDLobachevKS 2006 The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125 1283 1296
27. DowningBMorganRVanHulleKDeemAMalkovaA 2008 Large inverted repeats in the vicinity of a single double-strand break strongly affect repair in yeast diploids lacking Rad51. Mutat Res 645 9 18
28. VanHulleKLemoineFJNarayananVDowningBHullK 2007 Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol 27 2601 2614
29. DeemABarkerKVanhulleKDowningBVaylA 2008 Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 179 1845 1860
30. CasperAMGreenwellPWTangWPetesTD 2009 Chromosome aberrations resulting from double-strand DNA breaks at a naturally occurring yeast fragile site composed of inverted ty elements are independent of Mre11p and Sae2p. Genetics 183 423 439 421SI–426SI
31. KolodnerRDPutnamCDMyungK 2002 Maintenance of genome stability in Saccharomyces cerevisiae. Science 297 552 557
32. HenrySADonahueTFCulbertsonMR 1975 Selection of spontaneous mutants by inositol starvation in yeast. Mol Gen Genet 143 5 11
33. PutnamCDHayesTKKolodnerRD 2010 Post-replication repair suppresses duplication-mediated genome instability. PLoS Genet 6 e1000933 doi:10.1371/journal.pgen.1000933
34. ScholesDTBanerjeeMBowenBCurcioMJ 2001 Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159 1449 1465
35. ChinJKBashkirovVIHeyerWDRomesbergFE 2006 Esc4/Rtt107 and the control of recombination during replication. DNA Repair (Amst) 5 618 628
36. HanJZhouHHorazdovskyBZhangKXuRM 2007 Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315 653 655
37. RobertsTMZaidiIWVaisicaJAPeterMBrownGW 2008 Regulation of rtt107 recruitment to stalled DNA replication forks by the cullin rtt101 and the rtt109 acetyltransferase. Mol Biol Cell 19 171 180
38. WinstonFChaleffDTValentBFinkGR 1984 Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107 179 197
39. WinstonFDollardCMaloneEAClareJKapakosJG 1987 Three genes are required for trans-activation of Ty transcription in yeast. Genetics 115 649 656
40. RadfordSJBoyleMLSheelyCJGrahamJHaeusserDP 2004 Increase in Ty1 cDNA recombination in yeast sir4 mutant strains at high temperature. Genetics 168 89 101
41. BoltonECMildvanASBoekeJD 2002 Inhibition of reverse transcription in vivo by elevated manganese ion concentration. Mol Cell 9 879 889
42. FasslerJSWinstonF 1988 Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae. Genetics 118 203 212
43. KroghBOSymingtonLS 2004 Recombination proteins in yeast. Annu Rev Genet 38 233 271
44. LiefshitzBParketAMayaRKupiecM 1995 The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics 140 1199 1211
45. RattrayAJSymingtonLS 1994 Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics 138 587 595
46. BaiYSymingtonLS 1996 A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev 10 2025 2037
47. SignonLMalkovaANaylorMLKleinHHaberJE 2001 Genetic requirements for RAD51 - and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol 21 2048 2056
48. ChenQIjpmaAGreiderCW 2001 Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol 21 1819 1827
49. PutnamCDPennaneachVKolodnerRD 2004 Chromosome healing through terminal deletions generated by de novo telomere additions in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 101 13262 13267
50. DavisAPSymingtonLS 2004 RAD51-dependent break-induced replication in yeast. Mol Cell Biol 24 2344 2351
51. Voelkel-MeimanKRoederGS 1990 Gene conversion tracts stimulated by HOT1-promoted transcription are long and continuous. Genetics 126 851 867
52. MorrowDMConnellyCHieterP 1997 “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147 371 382
53. BoscoGHaberJE 1998 Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150 1037 1047
54. MalkovaAIvanovELHaberJE 1996 Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci U S A 93 7131 7136
55. SmithCELamAFSymingtonLS 2009 Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol Cell Biol 29 1432 1441
56. LemoineFJDegtyarevaNPKokoskaRJPetesTD 2008 Reduced levels of DNA polymerase delta induce chromosome fragile site instability in yeast. Mol Cell Biol 28 5359 5368
57. HaberJEHearnM 1985 Rad52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics 111 7 22
58. SmithCELlorenteBSymingtonLS 2007 Template switching during break-induced replication. Nature 447 102 105
59. SchmidtKHWuJKolodnerRD 2006 Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae homolog of the Bloom's syndrome protein. Mol Cell Biol 26 5406 5420
60. AmbergDCBurkeDJStrathernJN 2006 Isolation of Yeast Genomic DNA for Southern Blot Analysis. Cold Spring Harbor Protocols 2006 pdb.prot4149-
61. Yanisch-PerronCVieiraJMessingJ 1985 Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33 103 119
62. WinzelerEAShoemakerDDAstromoffALiangHAndersonK 1999 Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901 906
63. SikorskiRSHieterP 1989 A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19 27
64. LuriaSEDelbruckM 1943 Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics 28 491 511
65. LeaDECoulsonCA 1949 The distribution of the numbers of mutants in bacterial populations. Journal of Genetics 49 264 285
66. ToedlingJSkylarOKruegerTFischerJJSperlingS 2007 Ringo–an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 8 221
67. GentlemanRCCareyVJBatesDMBolstadBDettlingM 2004 Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80
68. SchmidtKHKolodnerRD 2006 Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. Proc Natl Acad Sci U S A 103 18196 18201
69. MyungKDattaAKolodnerRD 2001 Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104 397 408
70. KatsESEnserinkJMMartinezSKolodnerRD 2009 The Saccharomyces cerevisiae Rad6 postreplication repair and Siz1/Srs2 homologous recombination-inhibiting pathways process DNA damage that arises in asf1 mutants. Mol Cell Biol 29 5226 5237
71. MyungKPennaneachVKatsESKolodnerRD 2003 Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci U S A 100 6640 6645
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 5
-
Všechny články tohoto čísla
- Structural and Functional Differences in the Long Non-Coding RNA in Mouse and Human
- Identification, Replication, and Functional Fine-Mapping of Expression Quantitative Trait Loci in Primary Human Liver Tissue
- A −436C>A Polymorphism in the Human Gene Promoter Associated with Severe Childhood Malaria
- A Decline in p38 MAPK Signaling Underlies Immunosenescence in
- The Operon Balances the Requirements for Vegetative Stability and Conjugative Transfer of Plasmid R388
- Novel and Conserved Protein Macoilin Is Required for Diverse Neuronal Functions in
- Ixr1 Is Required for the Expression of the Ribonucleotide Reductase Rnr1 and Maintenance of dNTP Pools
- Genome of Strain SmR1, a Specialized Diazotrophic Endophyte of Tropical Grasses
- A Deficiency of Ceramide Biosynthesis Causes Cerebellar Purkinje Cell Neurodegeneration and Lipofuscin Accumulation
- A Latent Pro-Survival Function for the Mir-290-295 Cluster in Mouse Embryonic Stem Cells
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time
- Prion Formation and Polyglutamine Aggregation Are Controlled by Two Classes of Genes
- Integrated Genome-Scale Prediction of Detrimental Mutations in Transcription Networks
- Post-Embryonic Nerve-Associated Precursors to Adult Pigment Cells: Genetic Requirements and Dynamics of Morphogenesis and Differentiation
- A Novel Mouse Synaptonemal Complex Protein Is Essential for Loading of Central Element Proteins, Recombination, and Fertility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
- A Genetic and Structural Study of Genome Rearrangements Mediated by High Copy Repeat Ty1 Elements
- A Missense Mutation in Causes a Major QTL Effect on Ear Size in Pigs
- A Flurry of Folding Problems: An Interview with Susan Lindquist
- Meiotic Recombination Intermediates Are Resolved with Minimal Crossover Formation during Return-to-Growth, an Analogue of the Mitotic Cell Cycle
- A Nervous Origin for Fish Stripes
- The ISWI Chromatin Remodeler Organizes the hsrω ncRNA–Containing Omega Speckle Nuclear Compartments
- The Telomerase Subunit Est3 Binds Telomeres in a Cell Cycle– and Est1–Dependent Manner and Interacts Directly with Est1
- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Characterizing Genetic Risk at Known Prostate Cancer Susceptibility Loci in African Americans
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



