-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaIdentification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
Asymmetrical segregation of differentiated sister chromatids is thought to be important for cellular differentiation in higher eukaryotes. Similarly, in fission yeast, cellular differentiation involves the asymmetrical segregation of a chromosomal imprint. This imprint has been shown to consist of two ribonucleotides that are incorporated into the DNA during lagging-strand synthesis in response to a replication pause, but the underlying mechanism remains unknown. Here we present key novel discoveries important for unravelling this process. Our data show that cis-acting sequences within the mat1 cassette mediate pausing of replication forks at the proximity of the imprinting site, and the results suggest that this pause dictates specific priming at the position of imprinting in a sequence-independent manner. Also, we identify a novel type of cis-acting spacer region important for the imprinting process that affects where subsequent primers are put down after the replication fork is released from the pause. Thus, our data suggest that the imprint is formed by ligation of a not-fully-processed Okazaki fragment to the subsequent fragment. The presented work addresses how differentiated sister chromatids are established during DNA replication through the involvement of replication barriers.
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1001328
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001328Summary
Asymmetrical segregation of differentiated sister chromatids is thought to be important for cellular differentiation in higher eukaryotes. Similarly, in fission yeast, cellular differentiation involves the asymmetrical segregation of a chromosomal imprint. This imprint has been shown to consist of two ribonucleotides that are incorporated into the DNA during lagging-strand synthesis in response to a replication pause, but the underlying mechanism remains unknown. Here we present key novel discoveries important for unravelling this process. Our data show that cis-acting sequences within the mat1 cassette mediate pausing of replication forks at the proximity of the imprinting site, and the results suggest that this pause dictates specific priming at the position of imprinting in a sequence-independent manner. Also, we identify a novel type of cis-acting spacer region important for the imprinting process that affects where subsequent primers are put down after the replication fork is released from the pause. Thus, our data suggest that the imprint is formed by ligation of a not-fully-processed Okazaki fragment to the subsequent fragment. The presented work addresses how differentiated sister chromatids are established during DNA replication through the involvement of replication barriers.
Introduction
Differentiated sister chromatids, coupled with non-random segregation, have been proposed to control cell fate during the development and differentiation of multicellular organisms [1]. Similarly, cellular differentiation in the fission yeast Schizosaccharomyces pombe involves the establishment of differentiated chromatids and their asymmetrical segregation. The nature of this epigenetic mark or imprint that acts to discriminate the chromatids has been defined, however, the mechanism that leads to its introduction remains unknown. Fission yeast cells exhibit two different mating-types denoted as M and P. Homothallic strains are able to switch between mating-types in a pattern where only one granddaughter of a newly switched cell will have switched mating-type (Figure 1A) [2]–[6]. The mating-type of a cell is determined by the transcriptionally expressed mat1 locus located on chromosome II, which encodes either an M- or a P-specific gene cassette (Figure 1B) [7], [8]. Switching between the two mating-types occurs through a recombination event that replaces the cassette present at mat1 with that of the opposite mating-type, using one of the two transcriptionally silenced loci, mat2P and mat3M, as a donor of the information.
Fig. 1. Mating-type switching in S. pombe. 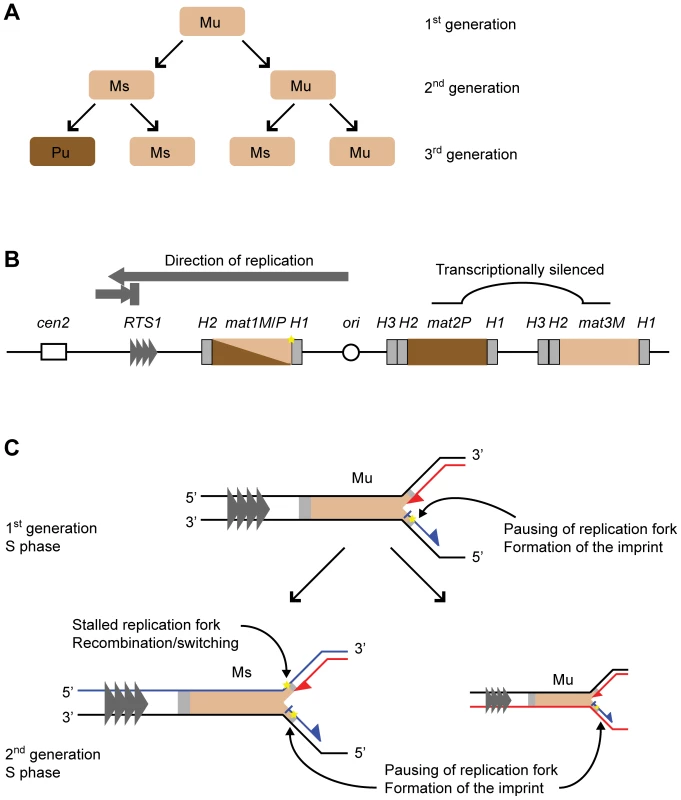
(A) Switching pedigree. The cells of minus (M) and plus (P) mating type are represented with light and dark brown colours, respectively. Division of an unswitchable (u) cell leads to the formation of an unswitchable and a switchable (s) cell of the same mating type. Division of the switchable cell gives rise to a switchable cell of the same mating type and an unswitchable cell of the opposite mating type. (B) The mating-type region on chromosome II. The transcriptionally active mat1 locus and the silenced donor loci, mat2P and mat3M, are shown. The M and P information is indicated with light and dark brown colours, respectively. The homology domains H1, H2 and H3 flanking the loci are represented by grey boxes. The centromere (cen2) and the origin of replication located centromere-distal to mat1 are shown with a hollow box and a circle, respectively. The replication termination element RTS1 is indicated with grey triangles, and direction of fork movement through mat1 is shown above with grey arrows, representing unidirectional replication. The imprint is indicated by a yellow star. (C) The replication of the mat1 locus in unswitchable and switchable cells. The nascent leading and lagging strands are represented by red and blue lines, respectively. In the second generation, the inherited template strands are also shown in red or blue, depending on which strand they were replicated as during the S phase of the parent. The polarities of DNA strands are indicated with 3′ and 5′ symbols. The S phases of both cell types involves pausing of the replication fork at mat1 and the introduction of the imprint in the newly synthesized upper strand. In addition, during the S phase of a switchable cell, the imprint on the upper template strand acts as a lesion to block leading-strand synthesis. The stalled leading-strand replication complex induces homologous recombination between mat1 and the donor loci leading to mating-type switching. The molecular mechanism underlying mating-type switching is tightly linked to DNA replication, and the asymmetrical pattern of switching reflects the asymmetry of the replication process. The mat1 locus is replicated unidirectionally in the centromere-proximal direction (see below). An imprint, which marks the mat1 locus of switchable cells (Figure 1B, 1C), is introduced during lagging-strand synthesis of the mat1 DNA in only one of the two sister chromatids formed in S phase [9], [10]. This imprint is maintained for one cell cycle, then during the next S phase, the imprint acts as a barrier for leading-strand replication, resulting in the replication-coupled recombination event that underlies mating-type switching [11]–[13]. The recombination event progresses by the “synthesis dependent single-strand annealing mechanism” where only one DNA strand at the donor locus provides the genetic information that is transferred to mat1 [14]. The homology needed for both the initiation and resolution of this recombination event is provided by short homology domains, named H1 and H2, that flank both the mat1 locus and each of the two donor loci, mat2P and mat3M (Figure 1B) [8], [15]. As a result of this recombination event, the mating-type cassette of only one of the two sister chromatids is replaced by a cassette containing the opposite mating-type information, such that the subsequent cell division gives rise to two cells with opposite mating-types.
The imprint was first identified genetically as a strand-specific, chromosomally inherited mark that is linked to the mat1 locus and is required for the induction of switching [4]–[6], [16]. Subsequently it was shown that the imprint consists of two ribonucleotides that are incorporated into the DNA duplex in a strand - and site-specific manner ([12] and reviewed in [17]). These two ribonucleotides are located at the junction between the mating-type cassette and the homology domain H1 (Figure 1B) [18]. The bonds of the RNA-DNA hybrid at the imprint site appear to be efficiently hydrolyzed by cellular enzymes, making purification of intact imprinted DNA difficult. During standard purification methods, the imprint is converted into a double-stranded break that can be detected by Southern analysis [10], [19]. However, we have managed to purify the intact DNA, and have shown that subsequent hydrolysis of the imprint can be achieved using RNAse T2 or alkali treatment [12]. In addition, we have shown that when DNA from a wild-type strain is purified under conditions that are known to hydrolyze RNA, a one-nucleotide gap can be detected in the imprinted strand [17]. This study also detected the presence of a 3′-terminal ribonucleotide, suggesting that the wild-type imprint consists of two ribonucleotides. A separate study using a mutant strain where a PstI restriction site has been introduced at the site of the imprint detected a nick with a 5′ hydroxyl at the site of the imprint [20]. Importantly, the presence of a 5′ hydroxyl is expected from the hydrolysis of a DNA-RNA-DNA hybrid molecule.
Several trans-acting factors have been identified that are required for the introduction of the imprint [21]. These include the checkpoint proteins Swi1 and Swi3 that travel with the replication fork and DNA polymerase alpha (Swi7), which is predominantly involved in lagging-strand replication [9], [22], [23]. Imprinting also correlates with a replication pause occurring at the site named MPS1, which can be observed both in P and M cells in the proximity of the imprint and is dependent on the trans-acting factors Swi1 and Swi3, but not on Swi7 [12], [24]. Replication pausing and imprinting are dependent on the direction of replication at the mat1 locus, which is tightly regulated by a site-specific polar replication termination site named RTS1 [25]. RTS1 is located at the centromere-proximal side of mat1 (Figure 1B), such that mat1 is only replicated by forks originating from the centomere-distal direction. Changing this direction of replication through mat1, by introducing replication origins on the centromere-proximal side, by inverting the mat1 locus relative to the RTS1 element, or by transposing the RTS1 element to the centromere-distal side of mat1 in the inverted orientation, leads to a reduction or an abolishment of imprinting [10], [25]. Inversion of the mat1 locus relative to the RTS1 element also leads to abolishment of replication pausing at MPS1, showing that the signal for pausing only acts on replication forks moving in one direction [24]. Such polarity is observed for all known site-specific replication barriers (reviewed in [26]).
Various cis-acting sequences involved in pausing at MPS1 and imprinting have been identified. These include the smt0 deletion that removes 263 base pairs (bp) starting at a position 33 bp centromere-distal to the mat1 cassette within the H1 homology domain and extending distally [27]. The smt0 mutation leads to the abolishment of imprinting but does not affect pausing at MPS1 [24], [27]. A smaller deletion named Δ17 that only removes sequences centromere-distal to the H1 domain also abolishes imprinting [28]. Experiments suggest that the essential factor Sap1 binds within this region, to the site named SAS1, and deletion of this site affects Sap1 binding in vitro, as well as imprinting in vivo. In addition, a linker-scanning mutagenesis study of the H1 domain identified a 6-bp substitution within the H1 domain called mut3 that reduces pausing at MPS1 [13]. Two others substitutions located within the part of the H1 domain that is covered by the smt0 deletion, named mut5 and mut7, also reduce imprinting. Characterization of the mut7 substitution suggested that it might affect the maintenance of the imprint throughout the cell cycle. During an earlier study done to characterize replication intermediates at the mat1 locus, replication initiation point (RIP) mapping was used to identify priming sites in the region. This method utilizes the fact that RNA primers on Okazaki fragments protect the fragments from degradation by λ-exonuclease, allowing their selective purification and use for primer extension [29]. Using this method, we detected a strong priming site within the mat1M cassette, approximately 350 bp centromere-proximal to the imprint, but not at the site of the imprint itself [12]. A weak leading-strand replication pause site was also observed in this region. Here we identify and analyze the cis-acting sequences required for replication pausing and imprinting. Our results obtained by characterization of replication intermediates allow us to propose a mechanism for imprinting in fission yeast.
Results
Identification of two regions encoded by the M cassette that are required for imprinting and pausing
In order to analyze the functional importance of the sequences within the mating-type cassette DNA, we utilized a strain that carried M information at the mat1 locus and was deleted for the two donor loci mat2P and mat3M. In this genetic background, MPS1 pausing and imprinting still occur, but the replication forks stalled at the imprint are resolved by sister chromatid recombination instead of recombination with the donor loci [30]. Thus, the M-cassette DNA is maintained at mat1, allowing mutations to be stably introduced at the locus. In an initial attempt to investigate the importance of the M-cassette sequence in pausing and imprinting, we deleted a 406-bp region starting just proximal to, but not including, where the two imprinted nucleotides are located and extending into the cassette DNA such that it ended within the Mc transcript encoded by the cassette (Figure 2A, strain SS4). Interestingly, analysis of this strain showed that the deletion abolished both imprinting, as detected by standard Southern blot analysis, and replication pausing at MPS1, as measured by 2D-gel electrophoresis (Figure 2B, 2C). Since this deletion covers a large region, it could potentially include several cis-acting sequences required for replication pausing and imprinting. Therefore, several shorter deletions within this region of interest were constructed (Figure 2). The 60-bp deletion in strain SS15, which included the previously mapped lagging-strand priming site (see Introduction), did not have an effect on pausing and only slightly reduced the level of imprinting. This deletion gave rise to 75.7% and 102% of wild-type levels of imprinting and pausing, respectively. Extending this deletion a further 60 bp centromere-proximal into the M cassette (strain SS14) also had little effect on pausing and imprinting, implying that this region is not required for these processes. However, deletion of the remaining centromere-distal 316-bp region (strain SS3) abolished both imprinting and replication pausing, establishing that cis-acting sequences required for both these processes are located within this region. To further define the location of these cis-acting sequences, the 316-bp region was divided into two, deleting either the 205-bp centromere-proximal segment or the 111-bp centromere-distal segment separately (Figure 2, strains SS13 & SS2, respectively). Surprisingly, deletion of the centromere-proximal segment completely abolished imprinting but only slightly affected replication pausing. Deletion of the centromere-distal part however, completely abolished both imprinting and replication pausing. Therefore, this initial analysis identified two regions within the M cassette that are of importance for the imprinting process in different ways.
Fig. 2. Identification of two regions required for imprinting and pausing within the mat1M cassette. 
(A) Line drawing on the left, a simple representation of the intact mat1M cassette flanked with the homology domains is shown. The site of the imprint is indicated by a vertical arrow. The nucleotide length of the region is given. The positions of the two encoded transcripts, Mi and Mc, are shown by horizontal arrows. The full-length wild-type DNA is represented by an intact line, whereas deletions are shown as gapped lines and the nucleotide positions of the start and the end of each deletion are given below the lines. The position of the specific lagging-strand priming site described in 12] is marked by a vertical line. The text box on right shows the names of the strains used, followed by quantification of the double-strand break (DSB) products (including standard error) and of the replication pause signal observed on 2D gels. The results are given as percentages of the wild-type values (100%). For DSB products, the values lower or equal to the negative control threshold are indicated as “below detection” (B.D.), whilst the absence of a pause signal is indicated by a dash. (B) Southern analysis of the strains shown in panel A. Genomic DNA was digested with HindIII and probed with the mat1P HindIII fragment. During DNA isolation, the imprint is hydrolyzed to a DSB and the resulting break products migrate as two bands. The strain names are given on top, while bands corresponding to the full-length mat1 fragment and the fragments generated by the DSB are given to the left. (C) 2D-gel analysis of the strains shown in panel A. Genomic DNA from log-phase cells was digested with NdeI and subjected to 2D-gel electrophoresis. A line drawing of the analyzed fragment and the positions of the restriction enzyme sites and the probe is shown below. The site of the imprint is indicated with a circle. The obtained blots were probed with a mat1M-specific probe. The strain names are given above each 2D gel. Identification of a region required for imprinting that acts as a spacer
To characterize the region defined by the deletion in strain SS13 in more detail, we constructed two sets of nested deletions, starting from both ends of the region (Figure 3). Deletion of 31 bp, 111 bp or 150 bp starting from the centromere-proximal side (strains SS6, SS5 and SS12, respectively) led to a gradual reduction in levels of imprinting, from 87.7% to 70.7% and ending with 46.6% of the wild-type level, suggesting that each deleted segment of this region partly contributes to the effectiveness of the imprinting process (Figure 3A, 3B). Similarly, introducing nested deletions of 55 bp, 94 bp of 174 bp in length starting from the centromere-distal side (strains SS22, SS23 and SS24, respectively), led to reduced imprinting to 85.5%, 55.5% and 16.9% of the wild-type level (Figure 3A, 3B). Again, this suggested that each of the deleted segments of this region makes a contribution to the imprinting process. All the deletions had a similar effect on replication pausing at MPS1, leading to an approximately 50% reduction in the intensity of the pause signal (Figure 3A, 3C). There are two possible explanations for the observed effect on imprinting; either specific sequences within this region contribute separately to the efficiency of the imprinting process or the 205-bp region acts as a spacer to separate regions of importance on either side. In the latter case, the observed gradual reduction in imprinting would be due to the gradual decrease in the distance between these flanking regions. If this region indeed acts as a spacer, reinstating the distance with an unrelated DNA sequence should restore imprinting. When we performed this experiment, the level of imprinting was indeed restored to 72.3% of the wild-type level and the level of pausing was restored to 80% (Figure 3, strain SS25). This result confirms the main role of this 205-bp region as a spacer separating flanking regions and the importance of this function for imprinting. The requirement of this 205-bp region for the imprinting process was also confirmed by deleting the equivalent sequences from the mat3M donor locus of a wild-type strain (Figure 3D). Imprinting in this strain was undetectable (Figure 3F) and colonies only displayed 1% sporulation efficiency compared to 72% observed for the wild-type colonies (Figure 3E). These results are consistent with the transposition of the mutant cassette sequence from the mat3M donor locus to the expressed mat1 locus, where the cis-acting deletion prevents subsequent imprinting events, and therefore any further mating-type switching.
Fig. 3. Identification of a cis-acting “spacer” element in mat1M. 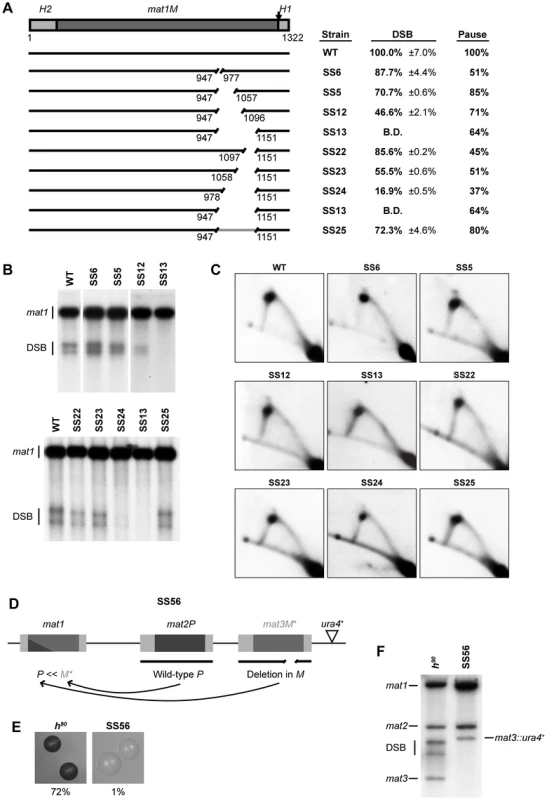
(A) Left, line drawing representation of the strains used is given. The grey line segment indicates the fragment that is substituted by a random stretch of DNA. Right, the names of the strains and quantification of the DSB products (including standard error) and of the pause signals are given. (B) Southern analysis of the strains shown in panel A. See Figure 2 legend for method. (C) 2D-gel analysis of the strains shown in panel A. See Figure 2 legend for method. (D) Schematic representation of the mating-type region of the strain used for analyzing the spacer deletion in vivo. The largest spacer deletion (same as in SS13, see A) was introduced into the donor locus mat3M to yield a mutant form of the donor M information (denoted as M*). The ura4+ gene linked to the mat3M locus, which was used during strain construction, is marked by a triangle. (E) Sporulation phenotype caused by the spacer deletion. The h90 wild-type strain and the mutant described in panel D (SS56) were allowed to form single colonies on sporulation media (PMA+), which were subsequently stained by iodine vapours. Iodine reacts with the starch found in spores to yield dark-coloured colonies. (F) Southern analysis of the strain SS56. Genomic DNA from h90 wild type and SS56 strains was digested with HindIII and probed for the mat1 locus. Because of the presence of homology, the donor loci are also visualized. The positions of the bands corresponding to mat1, mat2P, mat3M, DSB products, and mat3M linked to the ura4+ gene (mat3::ura4+) are indicated. Transcription initiated from the Mc gene does not interfere with imprinting
One possible function of the spacer region is that it separates the transcriptional unit Mc, which is encoded by the gene cassette, from the region of imprinting. Mc transcription is initiated from a promoter located within the cassette and terminates just prior to the spacer region where transcription termination and poly-adenylation signals are located. (Figure 4B). Thus, deletion of the spacer region brings these signals closer to the site of the imprint, where they could interfere with the imprinting process. It has been shown that insertion of the repressible nmt1 promoter at a position centromere-distal to the H1 domain such that it transcribes into the imprinting region leads to transcription-dependent inhibition of replication pausing as well as imprinting [11]. To test if Mc transcription interferes with imprinting as well when the spacer region is deleted, we used two approaches. Mc transcription has been shown to be repressed in rich media [8], therefore the levels of imprinting in the spacer region deletion strains were investigated under different growth conditions (Figure 4A). Quantification of the imprinting levels showed only minor differences between strains grown in poor (PMA+) and rich (YEA) media, suggesting that Mc transcription did not interfere with the imprinting process. To exclude the possibility that repressed levels of Mc transcription were still sufficient to inhibit imprinting, we deleted the promoter region of the transcriptional unit (Figure 4B–4D, strain SS66). Abolishment of the Mc transcript was confirmed by northern analysis, and this deletion produced only a slight reduction in the level of imprinting (90.6% of the wild-type). Importantly, the combination of the promoter deletion with the spacer deletion (strain SS63) did not lead to a restoration of imprinting (Figure 4B–4D). Thus, the reduction in imprinting observed when the spacer region is deleted is not due to inhibitory effects of Mc transcription.
Fig. 4. Transcription of the mating-type-specific gene Mc does not affect imprinting. 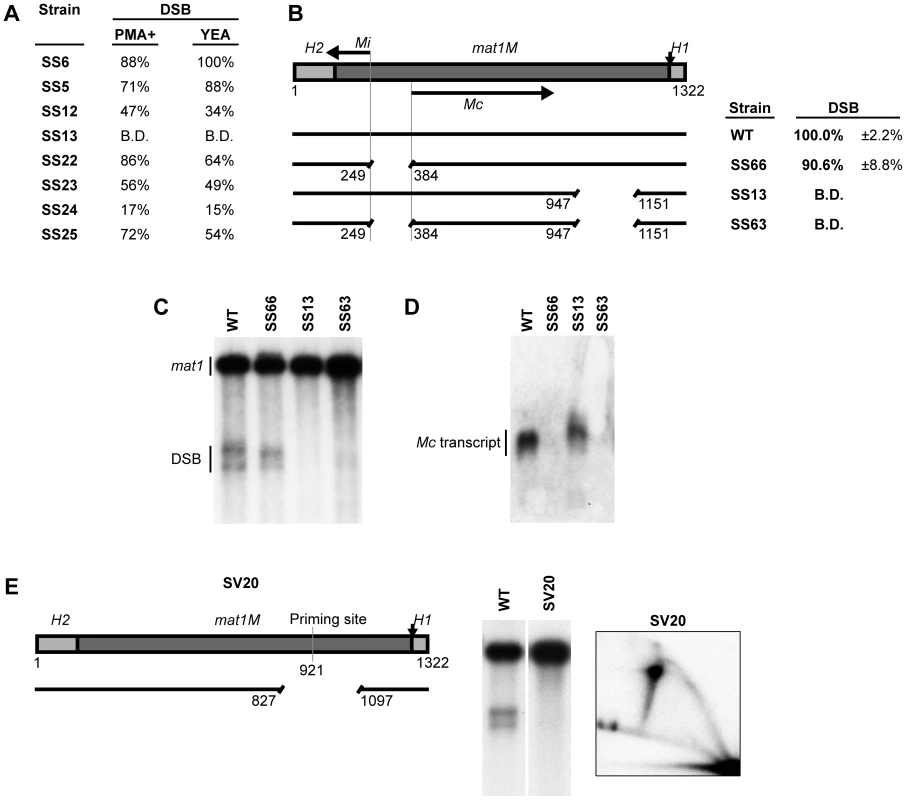
(A) The names of the strains and quantification of the DSB products generated during growth in either low-nitrogen sporulation (PMA+) or rich (YEA) media are given. The levels of imprinting, as assessed by DSB products, are similar for both growth conditions. (B) The effect of abolishing Mc transcription on the spacer deletion phenotype. Left, schematic representation of the mat1M cassette and the strains used is given. The positions of the Mi and Mc transcripts are shown by horizontal arrows. Deletion of the promoter region between the two transcripts (in strains SS66 and SS63) was performed, thus abolishing transcription. Right, the names of the strains and quantification of the DSB products (including standard error) are given. (C) Southern analysis of the strains shown in panel B. See Figure 2 legend for method. (D) Northern analysis of total RNA from the strains shown in panel B. The blot was probed for Mc using a double-stranded fragment generated by PCR. (E) Characterization of the strain (SV20) carrying a deletion of most of the spacer region and the region containing the previously mapped priming site. See Figure 2 legend for method. The position of the deletion is shown as a line drawing. Please note that fork-regression at MPS1 is observed in the 2D-gel displayed, in the absence of the imprint. This observation is fully analyzed in a related study (Vengrova and Dalgaard, in preparation). The region just centromere-proximal to the spacer region contains the strong priming site that we determined previously by RIP mapping [12], in addition to the putative Mc transcription termination and poly-adenylation signals. Therefore, the spacer deletions could be causing an inhibitory effect by bringing this region closer to the site of imprinting. To investigate this possibility, a 271-bp deletion was introduced that covered most of the spacer region and the potentially inhibitory region containing the priming site and the putative transcription signals (Figure 4E, strain SV20). This deletion had no effect on replication pausing, however, imprinting was still abolished in this strain. Therefore, the reduced levels of imprinting when the spacer region is shortened or fully removed are not due to a flanking inhibitory region brought closer to the site of the imprint.
The spacer region influences the position of lagging-strand priming sites
The spacer deletion could affect the replication process by changing where RNA primers are synthesized relative to the imprint site during lagging-strand replication. Therefore, high-resolution Southern blot analysis of replication intermediates was performed, using a single-stranded probe specific to the nascent lagging-strand (Figure 5A–5D). This method is the most direct way to analyze replication intermediates and avoids the problem of numerous unspecific bands observed when using the RIP mapping analysis. Two strong bands were detected in the imprinting-proficient wild-type strain (WT) and strain SS25, where the spacer region has been replaced by a scrambled sequence, the migrations of which are consistent with that predicted for fragments generated by hydrolysis of the imprinted parental upper strand (Figure 5B). Additionally, two weaker signals approximately 30 bp and 350 bp into the cassette were detected in these strains, which could be putative priming sites (indicated by arrowheads in Figure 5B). The signal 350 bp into the cassette corresponds to the position of the strong priming site that we had detected previously using RIP mapping [12]. Interestingly, both of these signals were absent in the lane containing replication intermediates from strain SS13, which carries the spacer deletion, suggesting that the deletion affects where primers are put down during lagging-strand replication of the region. Surprisingly, a signal in the proximity of the imprinting site was detected in this strain (Figure 5B). This signal is not due to hydrolysis of imprinted DNA, as we did not observe a signal where the centromere-proximal fragment would migrate. Instead, it suggests that a priming event is taking place at the site of imprinting, although this was undetected by our RIP mapping analysis [12]. To test if we could also observe leading-strand pausing at this position, we analyzed the leading-strand replication intermediates from the same strains (Figure 5C). A signal was detected at the site of the imprint in both the wild-type strain and strain SS25, which corresponds to previously observed stalling of leading-strand replication one nucleotide before the imprint [12], [13]. However, in the strain carrying the spacer deletion (SS13) and therefore lacking the imprint, a weaker signal can also be observed at this position. This suggests that leading-strand replication pausing at the MPS1 site occurs in the proximity of the imprinting site.
Fig. 5. Southern analysis of mat1 replication intermediates. 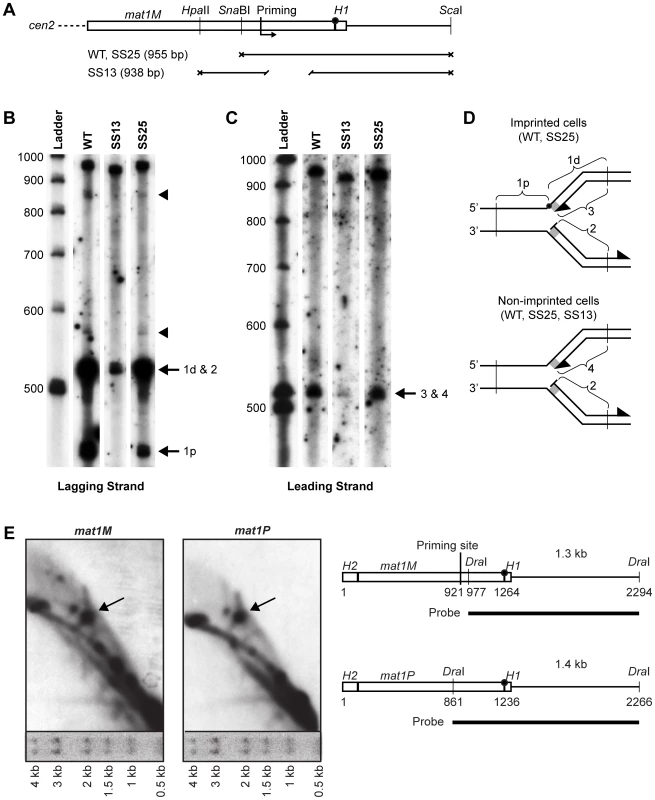
(A) Line drawing of the fragments generated for different strains for denaturing polyacrylamide gel electrophoresis (PAGE) analysis of replicating mat1M DNA shown in panels B and C. The positions of the restriction enzyme sites and the mapped priming site, and the lengths of the fragments are given. The site of the imprint is indicated with a circle. (B) Denaturing PAGE analysis of the mat1M lagging strand. Replicating DNA from log-phase cells was isolated as for 2D-gel analysis, digested with restriction enzymes given in panel A, then was subjected to denaturing PAGE analysis. After blotting, the membrane was probed with a strand-specific probe that hybridizes to the nascent lagging strand and the parental upper strand. Denatured and labelled DNA ladder is given to the left with size markers. The strains that exhibit imprinting (WT and SS25) produced two strong bands that correspond to the centromere-distal and proximal DSB products created by hydrolysis of the imprint (1d and 1p, respectively). Specific priming products at the site of the imprint (2) migrate to the same position as the distal DSB product observed for the WT and SS25 strains (1d). See panel D. Two fainter putative priming products that are shared between WT and SS25 are marked by arrowheads. (C) Denaturing PAGE analysis of the mat1M leading strand. The nascent leading strand and the parental lower strand were detected by the strand-specific probe. The co-migrating signals from the leading strand intermediates stalled at the imprint in WT and SS25 strains (3) and at MPS1 in the strain SS13 (4) are indicated. See panel D. Denatured and labelled DNA ladder is given to the left with size markers. (D) Line drawings of the replication intermediates observed in panel B and C are given. Fragments that produce bands in PAGE analysis are marked. The H1 domain is represented as a grey rectangle. The imprint is shown as a black circle. The positions of the restriction enzyme sites used are indicated by vertical lines. (E) 2D-gel analysis of the mat1P and mat1M imprint regions contained in DraI fragments. Migration of size markers after the first-dimension electrophoresis step is shown below each 2D gel. A line drawing of the region is given on right. The positions of the mapped priming site and the probes are shown. The site of the imprint is indicated with a circle. These results makes us reconsider our data presented earlier where we had mapped the paused replication fork at MPS1 to a position approximately 350 bp centromere-proximal to the imprinting site [12]. In order to discriminate between these two models, we performed a 2D-gel analysis of a small DraI fragment, which included the region of imprinting but not the region where we had previously proposed that stalling of the replication fork occurred (Figure 5E, left panel). The presence or absence of the pause signal on the Y-arc would determine which model is correct. The analysis clearly detected a strong pause site at a position that corresponds well to the site of the imprint. A similar analysis of the P cassette gave the same result (Figure 5E, right panel). Thus, the data suggest that the MPS1 signal stalls the replication fork at the region of imprinting and not within the cassette.
Definition of the MPS1 signal sequences
To define the sequences required for the MPS1 signal, we analyzed the 111-bp region deleted in the strain SS2 (Figure 2) by dividing it into three shorter segments, named a, b and c (Figure 6A). Deletion of each segment individually led to a complete abolishment of imprinting and a dramatic reduction in replication pausing (10%, 9% and 23% residual pausing when region a, b and c was deleted, respectively; Figure 6). Substitution of regions a and b with random sequences had similar effects to deleting them, resulting in abolished imprinting and reduced replication pausing to about 10% of wild-type levels (Figure 6). This demonstrates that these sequences are required for pausing and imprinting. Substitution of region c, which almost entirely consists of thymidines, with a random sequence caused only a reduction in imprinting to 28.4% of wild-type levels and a 47% reduction in pausing (Figure 6). Therefore, although this region clearly plays a role in both imprinting and pausing processes, there is not an absolute requirement for a specific sequence. The observation that deletion of region c abolishes imprinting, whilst replacement of it leads to only a reduction suggests that region c partly acts as a spacer.
Fig. 6. Characterization of a cis-acting region named abc that is required for replication pausing and imprinting. 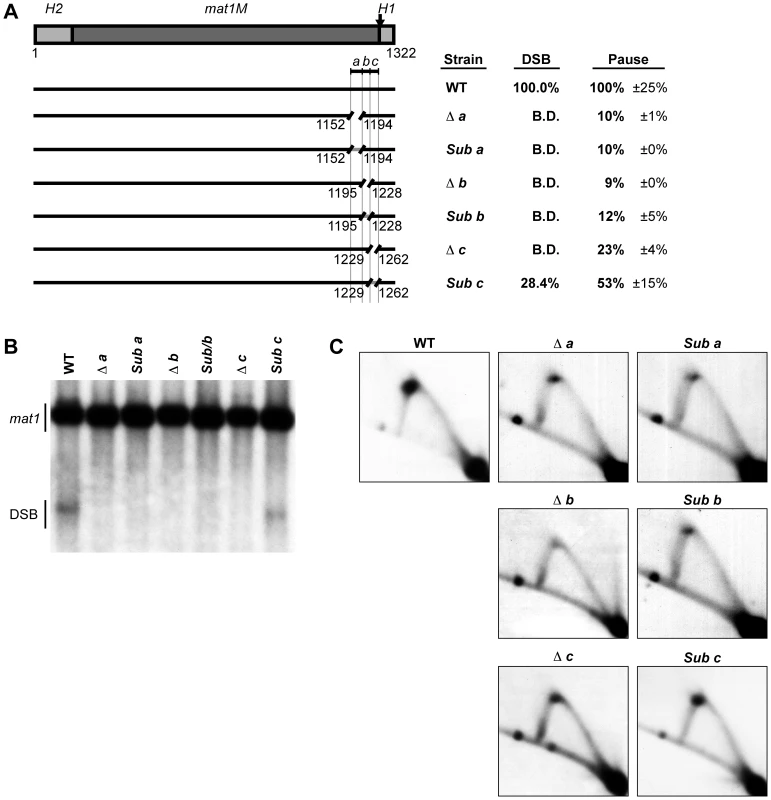
(A) Left, line drawing representation of the strains used. A gap represents a deletion and a gray line segment represents a substitution of a DNA sequence. The defined abc region is shown and the borders of the element are indicated by vertical lines. Right, names of the strains and quantification of DSB products and pause signals on 2D gels (including standard error) are given. (B) Southern analysis of the strains shown in panel A. See Figure 2 legend for method. (C) 2D-gel analysis of the strains shown in panel A. See Figure 2 legend for method. To investigate if the abc region could act as a replication barrier when transposed to a different locus and to define the sequences sufficient for barrier activity, we inserted the region into the plasmid pREP81K, next to the origin of replication [31]. Initially the transposed sequences included the abc region and the first 32 nucleotides of the H1 domain (Figure 7B, line drawing), as an earlier study had showed that a six-bp substitution within this region, called mut3, significantly reduced replication pausing [13]. Also, the first 32 nucleotides is what is left of the H1 domain in the strain carrying the cis-acting 263-bp smt0 deletion that abolishes imprinting, but does not affect replication pausing at MPS1 (see Introduction). The experiments were performed in a genetic background where the mat1 locus and the mat2P and mat3M donor loci had been deleted to avoid interference due to sequence homology (strain SS61). 2D-gel analysis of the plasmid detected replication pausing at a position on the Y-arc that corresponded to the position of the inserted sequences, whilst no barrier activity could be observed for the empty plasmid (Figure 7B, left panels). This plasmid-based system was used to further define which sequences within the abc region were required for replication pausing. Ten 11–12 bp substitutions were introduced, covering the entire abc region (Figure 7A). The substitutions Sub1, Sub2, Sub9 and Sub10 had little effect on pausing, however, the six substitutions within the middle of the abc region all reduced pausing to a significant degree (Figure 7B). Sub4, Sub5, Sub6 and Sub8 displayed about 20% residual pausing, while slightly more pausing could be observed for Sub3 and Sub7 plasmids (39% and 49% of the wild-type, respectively). Efficient pausing at MPS1 therefore requires at least the middle 66 bp of the abc region. The abc region alone and two shorter fragments comprising of the regions covered by the Sub4 to Sub8 and Sub4 to Sub9 substitutions (M abc, M R48 and M R49, respectively) were analyzed to determine if they were sufficient for pausing without the additional sequences of the H1 domain (Figure 7C; left panels). Omitting the H1 sequences had no effect on pausing, indicating that the region defined by the mut3 substitution is not important in this system. Significant pause signals could be observed for both the Sub4 to Sub8 and Sub4 to Sub9 fragments, the latter being of a similar intensity to the full abc region. Thus, the data suggest that the 77-bp Sub3-Sub8 region located 11 bp centromere-proximal to the imprinted nucleotides at mat1 is required and sufficient for maximum replication pausing in this assay.
Fig. 7. Characterization of the cis-acting region required for replication pausing. 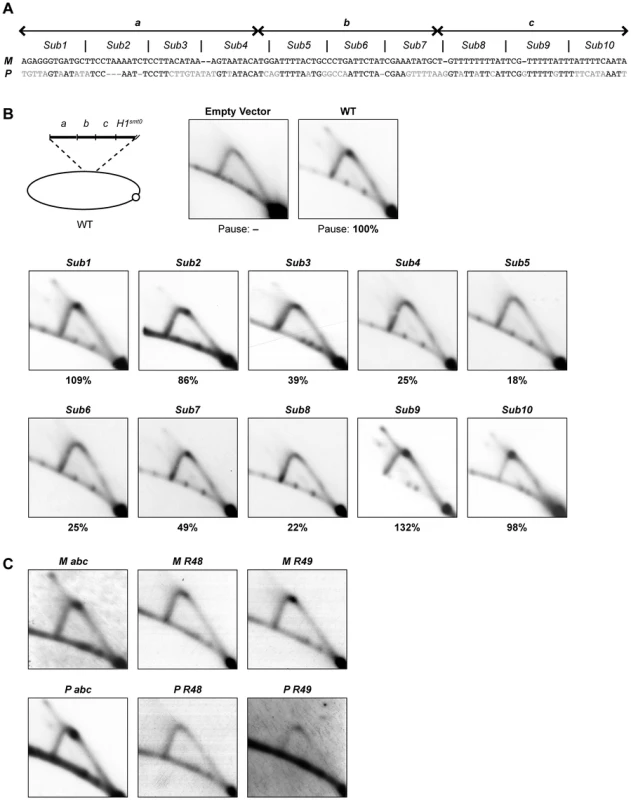
(A) DNA sequence of the abc region, aligned to the corresponding sequence in the P cassette, is given. The borders of a, b, and c regions are indicated by double arrows above the sequence. The sequence was divided into 10 segments and each segment was substituted by a random DNA sequence and denominated as substitution 1 to 10 (Sub1-10). (B) 2D-gel analysis of plasmids that carry different substitutions within the abc region. Top left, line drawing of the parent plasmid. The analyzed fragment contains the abc region and the first 32 bp of the H1 domain. The plasmids were digested with MpsA1I and AfeI, which produce a 2.5-kb fragment for the empty vector. The membranes were probed with same fragment of the parental plasmid. Relative intensities of the pause signals observed in plasmids carrying the substitutions are given as percentages of the wild-type value below each 2D gel. (C) 2D-gel analysis of replication of plasmids with different DNA fragments inserted. This requirement for specific sequences within the mat1M cassette was very surprising because replication pausing is observed both when mat1 contains P - and M-cassette DNA, which generally share no sequence similarity. We therefore analyzed the sequence corresponding to the M abc region in the P cassette, to see if replication pausing was also induced by this fragment. Indeed, a strong pause site could be observed by 2D-gel analysis (Figure 7C, P abc). Interestingly, an alignment of the two regions shows that there is a low level of sequence similarity between these M - and P-cassette segments, which is greatest in the region covered by substitutions Sub4 to Sub9 (Figure 7A). However, 2D-gel analysis of shorter segments of the P cassette, which correspond to the Sub4 to Sub8 and Sub4 to Sub9 regions of M abc, did not detect any pause signals (Figure 7C, P R48 & P R49). Thus, while there is low similarity between the P and M abc sequences, the size of the region required for pausing differs.
Does a trans-acting factor(s) interact with the abc region?
The replication pause signal observed in our plasmid assay is not as intense as that at the chromosomal mat1 locus (compare WT signals in Figure 2C and Figure 7B). This is partly due to the fact that even though the abc region is inserted next to the origin in the plasmid, it is replicated in both directions [32], and that only the forks moving in the same direction as at the genomic mat1 locus give rise to a barrier signal. However, we think that the weaker signal cannot be fully explained by this observation. Another possibility was that sequences outside the abc region contributed to pausing in the chromosomal context. However, inclusion of a 323-bp sequence centromere-proximal to abc or the entire centromere-distal H1 domain did not increase the intensity of the pause signal in our plasmid assay (data not shown). Thus, flanking sequences are not required for more efficient pausing. Alternatively, the plasmid used is present in multiple copies and therefore there could be an out-titration of a putative abc-interacting trans-acting factor(s) required for pausing. To test this possibility, we introduced plasmids containing the M abc or P abc region into h90 wild-type background to examine if an inhibitory effect could be observed on imprinting and switching, which both depend on MPS1 pausing. No effect was observed. We then constructed plasmids with arrays of the M or P abc region to increase the copy number. Each array contained approximately 10 tandem copies. Introduction of two different plasmids containing M abc arrays led to a reduction in sporulation to 57% of the wild-type levels and a 11.8% decrease in imprinting (Figure 8A, 8B). A similar effect was observed when two plasmids containing P abc arrays were introduced (71% of wild-type sporulation and a 11.1% decrease in imprinting; Figure 8A, 8B). However, we were unable to detect a corresponding reduction in the intensity of the MPS1 signal, as a value of 11–12% is too small to be statistically significant when quantifying signals on 2D gels. Importantly, while these data indicate that a trans-acting factor(s) binds to the M and P abc regions, and is required for imprinting, sporulation and potentially pausing, they also show that out-titration of this factor is not the cause of the weaker pause signal observed in our plasmid assay compared to that at the chromosomal locus. We therefore considered whether chromatin structure of a locus might affect the efficiency of replication pausing. In this context, it is interesting that the P and M abc regions are present at mat2P and mat3M donor loci that are transcriptionally silenced due to heterochromatin, yet no imprinting occurs at these loci. We therefore analyzed these loci by 2D-gel electrophoresis and could not observe pausing (Figure 8C). It should be noted that the K-region that separates the two loci replicates early [33], so the loci are replicated in opposite directions, with only mat2P being replicated in the direction where pausing would be expected. However, the data clearly show that the chromatin of a region can affect whether a cis-acting sequence mediates replication barrier activity. Potentially, heterochromatin could be preventing the binding of a trans-acting factor(s) required for replication pausing. Thus, although we do not directly show it, the variance in the intensities of the pause signals observed for the genomic and plasmid M abc could at least be partly due to the differences in chromatin of the regions analyzed.
Fig. 8. Out-competition of the putative trans-acting factor(s) that interacts with the M abc and P abc regions. 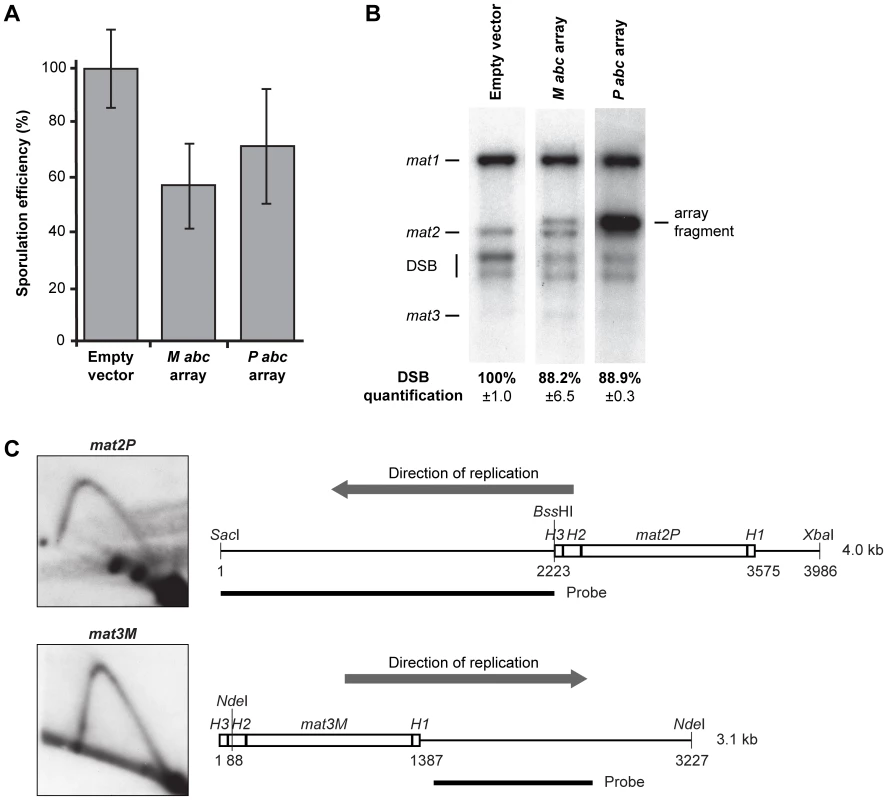
(A) The effect of plasmids that carry arrays of the abc region on sporulation efficiency. Two different plasmids (constructed using pREP3X or pREP4X parental vectors) that each carries an array of approximately 10 copies of either the M abc or P abc region were introduced in the h90 wild-type background (strain JZ1), such that two plasmid-borne arrays of either M or P abc were present. The sporulation efficiency relative to that observed for a strain carrying empty vectors (100%) are given as a graph. The deviation is shown as error bars. (B) Quantification of mat1 imprinting in strains used in panel A. HindIII fragments were run and probed with the mat1P HindIII fragment. The intensity of the signal from the P abc array is higher due to greater sequence homology. Quantification of intensities of the DSB products relative to the strain that carries the empty vectors (100%) is given below. (C) Neither P abc nor M abc induces replication pausing at the transcriptionally silenced donor loci. Left, 2D-gel analyses of mat2P and mat3M replication are shown. Right, line drawings of the fragments analyzed are given. The direction of replication through each locus is indicated with grey arrows. The positions of the restriction enzyme sites and the probes used are shown. The sizes of the fragments are given. There are no sequence requirements to the imprinted nucleotides or the surrounding region
One question that remained was whether there was some kind of sequence recognition at the site of imprinting. In both P and M cells, the imprint is introduced at the junction of the H1 domain within a region predominantly comprising of thymidines (The M sequence is shown in Figure 9A). A published study has shown that a 6-bp PstI restriction site introduced by substitution at the site of imprinting did not affect the overall level of imprinting [13]. However, characterization of the strain detected a nick with a 5′ hydroxyl, a result that is consistent with the hydrolysis of a single ribonucleotide present in the DNA. Separate studies showed that when wild-type DNA was purified under conditions that are known to hydrolyze RNA, a one-nucleotide gap was observed at the site of the imprint [17], [20], [34]. Since the introduction of the PstI site only led to the mutation of one of the two thymidine residues the imprint consists of, the possibility was raised that there was a sequence specificity to the introduction the ribonucleotides into the DNA. To test this, we substituted the two imprinted thymidine residues with either two adenosines, two guanosines or two cytidines (Figure 9A). None of these substitutions affected the level of imprinting and precise mapping of the position of the imprint in hydrolyzed DNA showed that the mutations did not affect where the imprint was introduced (Figure 9B, 9C). Therefore imprinting process is unaffected by the identity of the imprinted nucleotides.
Fig. 9. Substitutions in the region around and of the imprinted nucleotides. 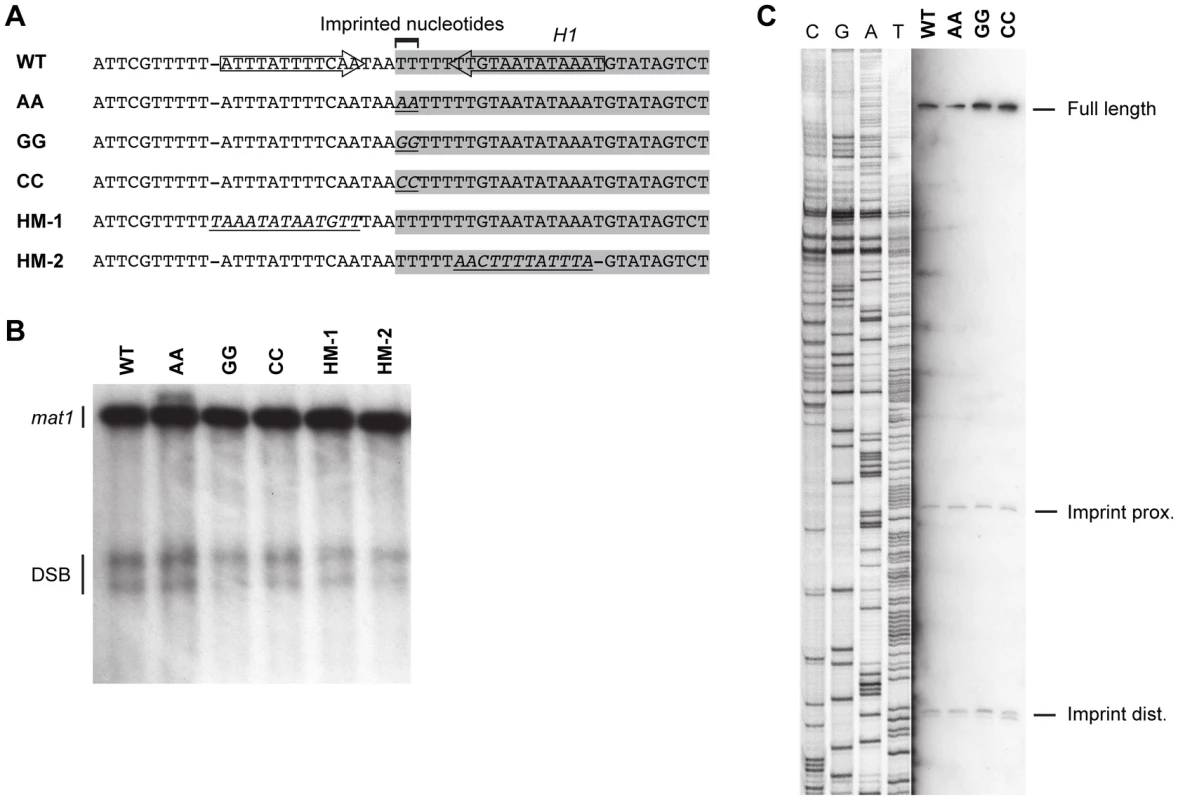
(A) The wild-type sequence is given on top. The H1 domain sequences are highlighted with a grey shadow. The position of the inverted repeat that flank the imprint is shown with arrows. Below, the mutant sequences introduced are given. Substituted nucleotides are shown in italics and are underlined. (B) Southern analysis of the strains given in panel A. See Figure 2 legend for method. The position of the DSB products is shown. (C) High-resolution Southern blot of the wild-type and mutant strains carrying substitutions at the imprinted nucleotides. A strand-specific probe that hybridizes to the imprinted strand was used. For detailed explanation of the method, see the Materials and Methods section. The positions of the fragments generated by hydrolysis of the imprint are shown. Potentially the sequence of the region surrounding the imprint site may be important for the imprinting process. Substitution of the c section of the abc region, which is immediately centromere-proximal to imprint site, reduced the level of imprinting to approximately 50% (Figure 6). However, the substituted region contained the segment defined by Sub8, which is required for efficient pausing in our plasmid assay (Figure 7), therefore the reduction in imprinting is likely to result from the reduction in the replication pausing. Additionally, an inverted repeat in this region that flanks the site of the imprint may affect the imprinting process [18]. Therefore, the effects of a 12-bp substitution of a region located three bp centromere-proximal to the imprinting site, which substitutes one side of the inverted repeat present in the sequence, were investigated (Figure 9, HM-1). This substitution had no effect on the level of imprinting. Similarly, substitution of the 13-bp region located three bp centromere-distal to the imprinted nucleotides, which substitutes the other side of the inverted repeat, had no effect on imprinting (Figure 9, HM-2). Thus, the DNA sequence of neither the imprinted nucleotides nor the regions flanking them is of importance for the imprinting process.
Discussion
Replication barriers have been identified in the rDNA, at tRNA genes, and telomeric and centromeric sequences in several eukaryotic organisms, and at the RTS1 and MPS1 elements of the mating-type region in S. pombe (reviewed in [26]). The MPS1 site is particularly interesting as the replication pause leads to the introduction of the imprint marking the mat1 cassette in switchable cells of S. pombe, thereby establishing a functional link between DNA replication and cellular differentiation. Surprisingly, our analysis shows that cis-acting sequences required for MPS1 activity are located within the mating-type cassette DNA that is swapped during the switching process, such that the observed pause signal is constituted by forks stalled at not one, but two different sequences. Our results contrast a previously published study that showed that a 6-bp substitution, mut3, within the H1 domain flanking the mating-type cassette caused a significant reduction in imprinting and pausing [13]. This region is not required for pausing in our plasmid assay (Figure 7). Instead, by characterization of different deletions and substitutions, we were able to identify a 77-bp M-cassette region located 11 bp centromere-proximal to the site of the imprint that is required for both MPS1 activity and imprinting (Figure 6 and Figure 7). Importantly, a 2D-gel analysis of the corresponding region from the P cassette also detected a pause signal, despite the limited sequence identity between the two regions (Figure 7A, 7C). How these two cis-acting sequences induce replication pausing remains unknown, but most likely a trans-acting factor(s) mediates the activity (Figure 10, upper line drawing): Firstly, we have been unable to identify any secondary structure in the regions that could act as a barrier for the progressing replication fork. Secondly, MPS1 pausing is dependent on the trans-acting factors Swi1 and Swi3 that travel with the replication fork [24], [35]. These factors are also required for replication pausing and termination at the S. pombe RTS1 element and at rDNA arrays where barrier activities are mediated by trans-acting factors [26]. Thirdly, we observed reduced mat1 imprinting and sporulation efficiency when plasmids containing arrays of M or P abc regions were introduced in otherwise wild-type background (Figure 8A, 8B). These data are consistent with the model that a trans-acting factor(s) required for imprinting, and by deduction replication pausing, is binding to the mat1 abc region and that out-titration occurs by competitive binding to the multiple plasmid copies present. Lastly, we were unable to detect any barrier activity at the donor loci mat2P and mat3M (Figure 8C). The K-region that separates the two donor loci has been shown to replicate early [33], suggesting that mat2P, but not mat3M, is replicated in the direction of replication where the barrier is active. Also, this observation does not support the possibility that the secondary structure of the mat1 abc region mediates replication pausing, as the same structures would be expected to arise at the mat2P locus. Instead, it suggests that heterochromatin at the mat2P locus prevents the interaction of a trans-acting factor(s). The observation that the chromatin state of a cis-acting sequence affects its barrier activity is important as it establishes a mechanism, in addition to control of direction of replication, by which replication barriers could be regulated in other systems. Importantly, while we think a trans-acting factor(s) is required for pausing at both the mat1 cassettes, we are unable to predict, due to the limited similarity between the P abc and M abc sequences, whether it is the same factor(s) that is acting at the two sites.
Fig. 10. Working model for MPS1 replication pausing and mat1 imprinting. 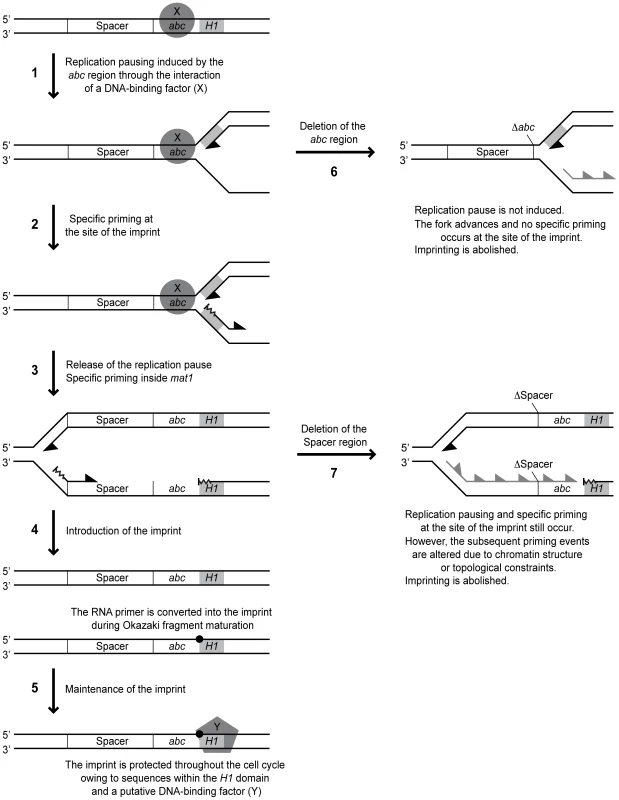
The mat1 locus and the events that occur during replication of it are depicted with line drawings. Left, molecular events that take place in the wild-type strain. Right, events occurring when cis-acting mutations are introduced. Positions of the spacer and abc regions and the H1 domain (grey boxes) are shown. The orientations of the DNA strands are indicated. The putative proteins mediating MPS1 activity and maintenance of the imprint are displayed as a circle and a pentagon, respectively. The imprint is marked with a black spot. Leading- and lagging-strand replication is represented with black half arrowheads. Specific priming is depicted as wavy lines, while loss of it is indicated with grey half arrowheads. Left, positions of the cis-acting deletions are given. See main text for description of each step. One interesting question is where the replication fork pauses in response to the MPS1 signal. Two lines of evidence suggest that the replication pausing takes place in the proximity of the imprint, and not further into the cassette as suggested earlier (Figure 10, arrow 1) [12]: i) We observe a leading-strand pause signal in the proximity of the imprint in the spacer deletion strain (SS13) where imprinting is abolished, but MPS1 pausing still occurs (Figure 5C). Importantly, it is not possible to distinguish a corresponding signal in the wild-type strain due to the leading-strand intermediates from stalled forks at the imprint that migrate to the same position [12], [13]. ii) 2D-gel analysis detected a strong pause signal during the replication of a fragment containing only the imprint region and not the position where our previous analysis suggested MPS1 pausing occurred (Figure 5E). Instead, the weak leading-strand pause signal that we observed earlier in the vicinity of the strong priming site located approximately 350 bp into the cassette might reflect a minor pause of the replication fork while the priming event takes place. In this context, it is noteworthy that during bacteriophage replication, priming has been shown to pause leading-strand progression [36].
All the available data suggest that stalling of the replication fork at MPS1 is a prerequisite for imprint formation. However, the observation of pausing at MPS1 without subsequent imprinting indicates that pausing is only an initial step of the process by which the imprint is introduced. Our analysis identified a novel type of cis-acting element that is required for imprinting but not pausing. Deletion of a 205-bp region located on the centromere-proximal side of the pause signal abolished imprinting, but only slightly reduced pausing at MPS1 (Figure 2). Substitution of this region with a random sequence however, restored imprinting to 72% of wild-type levels, suggesting that this region mainly acts as a spacer to separate important flanking sequences. To our knowledge, this is the first description of such a genetic element. The important flanking sequences on one side of the spacer are defined by the abc region, while the flanking sequences of importance on the other side are unknown. However, the deletion of the 205-bp region leads to a complete abolishment of imprinting, therefore preventing us from using this assay to determine if the spacer extends further into the cassette. Also, we do not know whether extending the length of the spacer would affect imprinting; if this region is necessary for separation of two flanking sites, it is possible that enlarging it would prevent their concerted action. Alternatively, the wild-type spacer region could represent the minimum size and there will still be appropriate imprinting when it is lengthened.
The reduction in imprinting caused by the spacer deletion appears not to be due to an inhibitory effect of transcription of the Mc gene, which is brought into close proximity of the imprinting site by the deletion, since neither repression of Mc transcription by growth in rich media nor deletion of the Mc promoter region leads to a restoration of imprinting (Figure 4). Reduced imprinting is also not due to a repressive effect of the strong priming site that we have mapped previously, which is located in the region flanking the spacer on the centromere-proximal side, as extension of the deleted region to include these sequences does not restore imprinting (Figure 4E). This priming site is also not directly required for imprinting or pausing, as deletion of it only causes minor reductions in the efficiencies of these processes (Figure 2).
An alternative explanation is that it is not the specific sequence, but the chromatin structure of the transcribed Mc region that inhibits imprinting in the absence of the spacer. One possibility is that there is a requirement for a non-transcribed spacer, as for example the nucleosome arrangement in such a region might affect where primers are synthesized during lagging-strand replication. Using high-resolution Southern analysis of replication intermediates, we attempted to map the priming sites used in the region of the wild-type, spacer-deleted and spacer-substituted strains. In the imprinting-proficient wild-type and spacer-substituted stains, we detected a putative priming signal within the mating-type cassette, approximately 350 bp from the site of the imprint, at a position that corresponds to where we earlier detected a strong priming site using the RIP mapping method (Figure 5B) [12]. Another putative priming signal was observed approximately 30 bp centromere-proximal from the imprinting site, which was undetected by our earlier RIP mapping analysis. Both these signals were absent in replication intermediates from the strain carrying the spacer deletion, suggesting that the non-transcribed spacer region indeed influences the sites of primer synthesis after the replication fork is released from pausing at MPS1 (Figure 10, arrows 3 & 7). If the imprint is introduced by ligation of a not fully processed Okazaki fragment to the subsequent fragment, the length and position of these could be crucial, maybe due to topological constrains during the replication process.
Interestingly, a putative priming signal is detected at the site of imprinting in the spacer deletion strain (Figure 5B and Figure 10, arrow 2). Because of the strong signals created by hydrolysis of the imprinted DNA, it is impossible to determine whether such a priming event also occurs at this position in the wild-type strain. This priming site was not observed for the wild-type strain in our previous RIP mapping analysis [12]. However, it is possible that priming at this site is special, involving a shorter RNA primer that does not protect the fragment from λ-exonuclease digestion used for RIP mapping [29]. Also, the high-resolution Southern analysis is a more direct method for analysis of replication intermediates than RIP mapping. The detection of specific priming at the imprinting site does provide strong support for a model proposing that the imprint is introduced by a mechanism involving the fusion of a not fully processed Okazaki fragment to the subsequent fragment (Figure 10, arrow 4). In support of this possibility, DNA polymerase alpha (Swi7), which initiates lagging-strand replication together with primase, is involved in imprinting [9].
Another observation that supports a model for the imprinting process that involves a priming event is that we were not able to detect any sequence requirements for either the two imprinted nucleotides or the 12-bp and 13-bp regions flanking the site of the imprint (Figure 9). This observation is consistent with another study, in which substitution of five of the first six bp of H1, including one of the imprinted nucleotides, as well as the 9th to the 14th bp, had no effect on imprinting levels [13]. Similarly, we observed that whilst deletion of the last 34 bp of the M cassette that flank the imprinted nucleotides (region c) abolished imprinting, substitution of this region only caused a reduction in imprinting levels (Figure 6). Therefore there appears to be no absolute sequence requirement at or around the site of the imprint for the imprinting process to take place. Additionally, region c may partially act as a spacer element, again highlighting the potential importance of the position of priming events in the imprinting process. Thus, our data suggest that the imprint is introduced at a certain distance from the replication pause site in a sequence-independent manner. The position of this priming event might not only affect whether the imprint is formed, but also whether it is maintained. Here, the distance to the sequences within the H1 domain, which has already been implicated in the maintenance of the imprint, might be important (Figure 10, arrow 5) [13].
Our data presented here suggest that ribonucleotide imprints might be introduced at other replication barriers, but not maintained. Such temporary marks could be important for replication-associated processes such as DNA damage repair or the establishment of certain chromatin. Also, the production of epigenetic marks during the replication process and subsequent asymmetric segregation of the differentiated sister chromatids may be a more general mechanism for cellular differentiation and development in eukaryotes. Understanding the process of imprinting in fission yeast and identifying the cis - and trans-acting factors involved will help to establish whether this specific type of epigenetic mark also plays a role in cellular differentiation of higher eukaryotes. Finally, unravelling the molecular processes at the replication fork barriers involved in imprinting may also increase our understanding of the events that occur at unintentional replication barriers and how they lead to genome instability.
Materials and Methods
Construction of S. pombe strains
Deletions and substitutions were introduced into the M cassette using inverse PCR on plasmids containing mat1M DNA. Deletions and substitutions were verified by sequencing. The mutated fragments were subsequently transformed into the strain EG571, which carries the ura4+ gene inserted at the H1 domain and is deleted for the donor loci mat2P and mat3M (gift from Olaf Nielsen). 5-fluoroorotic acid-resistant colonies were selected, and the replacement of the parental ura4+-containing allele of mat1 with the mutated allele was verified by PCR, followed by sequencing and Southern analysis. The sequences of the substituted segments were: Strain SS25, AATTGAACGTTGTTTTTCCATTTTGTATAAGCCGTCGCCGGTT TAGAAGCGTTGAATGTCAGGTATTATGAATAGTAAAATCTTATTTATCTATTTTGAAGGTCAATTGTTATCATTTCTTAAAGTGTGATTGTTATATATCAGTAGGTTTAAATATATTAACACTAGATAAATTCTTGTCTCATTATTTATTGATTCCACATAA; Sub a, CTGTGTCTGTCGAGTTGCCCATCAGTACGGAGCTAAAACGAAT; Sub b, GTAA-GTCACCGATAAACGCGCCTGGAATGTTTCT; Sub c, TTTGATCGCCTGGA-ACCATGAACGACCTAAGGTC; Sub 1-10, TCTCTGCAGGCT, CACTGCAGCGT, CGCTGCAGATC, TTCTGCAGGCC, CCCTGCAGGAA, GTCTGCAGGTA, TTCTGCAGCTC, TCCTGCAGGTC, GGCTGCAGCGC, GCCTGCAGGAT, respectively. The recipes of media and the method for the transformation procedure are given in [37].
Quantification of imprinting levels by Southern blot analysis
Unless otherwise stated, the chromosomal DNA was purified from cells grown in PMA+ media as described in [10]. The DNA samples were digested with the HindIII restriction enzyme and separated on a 0.6% agarose gel. Southern analysis was performed with a probe specific to the 10.6-kilo base pairs (kb) mat1P HindIII fragment. The level of imprinting was determined by the amount of the mat1 fragments that were broken during the purification process. The intensity of signals was measured using a BioRad Phosphorimager. Two independent measurements were performed for each strain and the average was given in the figures.
2D-gel analysis of replication intermediates
2D gels were performed as described in [38], except the first - and second-dimension gels contained 0.5% and 1.2% agarose, respectively. The replication intermediates were digested with the restriction enzymes given in Figure 2C, Figure 5E, Figure 7, and Figure 8C. The intensity of the replication signals was determined using a BioRad Phosphorimager. The pause signal level was given relative to the intensity of the ascending Y-arc, as well as to the intensity of the wild-type pause signal. The average measurement from two gels was given for each strain.
High-resolution Southern blot analysis of replication intermediates
Yeast chromosomal DNA was purified as described in [39]. The replication intermediates were digested with the restriction enzymes given in Figure 5A. Replication intermediates were separated from non-replicating DNA by BND-cellulose chromatography [40] and separated on a 4% denaturing polyacrylamide gel. After electrophoresis, the DNA was transferred to a GeneScreen membrane (NEN) by electro-blotting, and probed with strand-specific probes that had been generated as described in [41].
Precise mapping of the position of the imprint
Chromosomal DNA was purified as described before [10]. The DNA were digested with SspI and precipitated overnight with isopropanol, redissolved in loading buffer, and loaded on a 6% denaturing polyacrylamide gel. Sequencing reactions, performed using the primer mat1M-SspI-R (ATTAGTGAGTATATTATGGTAGGGAGTGC) and a bacterial plasmid containing the EcoRI fragment of mat1M (pBZ85), were run in parallel as a marker. The DNA was electro-blotted onto a GeneScreen membrane (NEN). The strand-specific probe to detect the imprinted mat1 strand was synthesized by linear primer extension, using the primer mat1M-SspI-F (ATTTTGTGTACCCCATTTGCGTTGAG) and a 440-bp mat1M SspI fragment as template [41]. Chain-termination sequencing reactions were performed following the manufacturer's instructions.
Northern analysis
Total RNA was purified from logarithmically growing cells using the method described in [42]. 10 µg of total RNA was analyzed using a 1% denaturing agarose gel containing 2.2% formaldehyde [43]. A double-stranded PCR product of the Mc transcribed region was used for generation of the probe.
Zdroje
1. TajbakhshS
2008 Stem cell identity and template DNA strand segregation. Curr Opin Cell Biol 20 716 722
2. MiyataH
MiyataM
1981 Mode of conjugation in homotallic cells of Schizosaccharomyces pombe. J Gen Appl Microbiol 27 365 371
3. EgelR
EieB
1987 Cell lineage asymmetry for Schizosaccharomyces pombe: Unilateral transmission of a high-frequency state of mating-type switching in diploid pedigrees. Curr Genet 12 429 433
4. EgelR
1984 The pedigree pattern of mating-type switching in Schizosaccharomyces pombe. Curr Genet 8 205 210
5. KlarAJ
1987 Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature 326 466 470
6. KlarAJ
1990 The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands. EMBO J 9 1407 1415
7. BeachDH
1983 Cell type switching by DNA transposition in fission yeast. Nature 305 682 687
8. KellyM
BurkeJ
SmithM
KlarA
BeachD
1988 Four mating-type genes control sexual differentiation in the fission yeast. EMBO J 7 1537 1547
9. SinghJ
KlarAJ
1993 DNA polymerase-alpha is essential for mating-type switching in fission yeast. Nature 361 271 273
10. DalgaardJZ
KlarAJ
1999 Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400 181 184
11. HolmesAM
KaykovA
ArcangioliB
2005 Molecular and cellular dissection of mating-type switching steps in Schizosaccharomyces pombe. Mol Cell Biol 25 303 311
12. VengrovaS
DalgaardJZ
2004 RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev 18 794 804
13. KaykovA
HolmesAM
ArcangioliB
2004 Formation, maintenance and consequences of the imprint at the mating-type locus in fission yeast. EMBO J 23 930 938
14. Yamada-InagawaT
KlarAJ
DalgaardJZ
2007 Schizosaccharomyces pombe switches mating type by the synthesis-dependent strand-annealing mechanism. Genetics 177 255 265
15. ArcangioliB
de LahondesR
2000 Fission yeast switches mating type by a replication-recombination coupled process. EMBO J 19 1389 1396
16. KlarAJ
BonaduceMJ
1993 The mechanism of fission yeast mating-type interconversion: evidence for two types of epigenetically inherited chromosomal imprinted events. Cold Spring Harb Symp Quant Biol 58 457 465
17. VengrovaS
DalgaardJZ
2006 The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep 7 59 65
18. NielsenO
EgelR
1989 Mapping the double-strand breaks at the mating-type locus in fission yeast by genomic sequencing. EMBO J 8 269 276
19. ArcangioliB
1998 A site - and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J 17 4503 4510
20. KaykovA
ArcangioliB
2004 A programmed strand-specific and modified nick in S. pombe constitutes a novel type of chromosomal imprint. Curr Biol 14 1924 1928
21. EgelR
BeachDH
KlarAJ
1984 Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc Natl Acad Sci U S A 81 3481 3485
22. SommarivaE
PellnyTK
KarahanN
KumarS
HubermanJA
2005 Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol Cell Biol 25 2770 2784
23. NoguchiE
NoguchiC
DuLL
RussellP
2003 Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol 23 7861 7874
24. DalgaardJZ
KlarAJ
2000 swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102 745 751
25. DalgaardJZ
KlarAJ
2001 A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev 15 2060 2068
26. DalgaardJZ
EydmannT
KoulintchenkoM
SayracS
VengrovaS
2009 Random and site-specific replication termination. Methods Mol Biol 521 35 53
27. StyrkarsdottirU
EgelR
NielsenO
1993 The smt-0 mutation which abolishes mating-type switching in fission yeast is a deletion. Curr Genet 23 184 186
28. ArcangioliB
CopelandTD
KlarAJ
1994 Sap1, a protein that binds to sequences required for mating-type switching, is essential for viability in Schizosaccharomyces pombe. Mol Cell Biol 14 2058 2065
29. GerbiSA
BielinskyAK
1997 Replication initiation point mapping. Methods 13 271 280
30. RoseaulinL
YamadaY
TsutsuiY
RussellP
IwasakiH
2008 Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J 27 1378 1387
31. CravenRA
GriffithsDJ
SheldrickKS
RandallRE
HaganIM
1998 Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221 59 68
32. EydmannT
SommarivaE
InagawaT
MianS
KlarAJ
2008 Rtf1-mediated eukaryotic site-specific replication termination. Genetics 180 27 39
33. KimSM
DubeyDD
HubermanJA
2003 Early-replicating heterochromatin. Genes Dev 17 330 335
34. VengrovaS
DalgaardJZ
2005 The Schizosaccharomyces pombe imprint—nick or ribonucleotide(s)? Curr Biol : - 15 R326 327; author reply R327
35. NoguchiE
NoguchiC
McDonaldWH
YatesJR3rd
RussellP
2004 Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol 24 8342 8355
36. LeeJB
HiteRK
HamdanSM
XieXS
RichardsonCC
2006 DNA primase acts as a molecular brake in DNA replication. Nature 439 621 624
37. MorenoS
KlarA
NurseP
1991 Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194 795 823
38. BrewerBJ
FangmanWL
1987 The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51 463 471
39. HubermanJA
SpotilaLD
NawotkaKA
el-AssouliSM
DavisLR
1987 The in vivo replication origin of the yeast 2 microns plasmid. Cell 51 473 481
40. KigerJAJr
SinsheimerRL
1969 Vegetative lambda DNA. IV. Fractionation of replicating lambda DNA on benzoylated-naphthoylated DEAE cellulose. J Mol Biol 40 467 490
41. RuvenHJ
SeelenCM
LohmanPH
MullendersLH
van ZeelandAA
1994 Efficient synthesis of 32P-labeled single-stranded DNA probes using linear PCR; application of the method for analysis of strand-specific DNA repair. Mutat Res 315 189 195
42. SchmittME
BrownTA
TrumpowerBL
1990 A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18 3091 3092
43. RosenKM
LampertiED
Villa-KomaroffL
1990 Optimizing the northern blot procedure. Biotechniques 8 398 403
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and PhosphodiesterasesČlánek Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



