-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaH3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
Methylation of histone H3 lysine 4 (H3K4me) is an evolutionarily conserved modification whose role in the regulation of gene expression has been extensively studied. In contrast, the function of H3K4 acetylation (H3K4ac) has received little attention because of a lack of tools to separate its function from that of H3K4me. Here we show that, in addition to being methylated, H3K4 is also acetylated in budding yeast. Genetic studies reveal that the histone acetyltransferases (HATs) Gcn5 and Rtt109 contribute to H3K4 acetylation in vivo. Whilst removal of H3K4ac from euchromatin mainly requires the histone deacetylase (HDAC) Hst1, Sir2 is needed for H3K4 deacetylation in heterochomatin. Using genome-wide chromatin immunoprecipitation (ChIP), we show that H3K4ac is enriched at promoters of actively transcribed genes and located just upstream of H3K4 tri-methylation (H3K4me3), a pattern that has been conserved in human cells. We find that the Set1-containing complex (COMPASS), which promotes H3K4me2 and -me3, also serves to limit the abundance of H3K4ac at gene promoters. In addition, we identify a group of genes that have high levels of H3K4ac in their promoters and are inadequately expressed in H3-K4R, but not in set1Δ mutant strains, suggesting that H3K4ac plays a positive role in transcription. Our results reveal a novel regulatory feature of promoter-proximal chromatin, involving mutually exclusive histone modifications of the same histone residue (H3K4ac and H3K4me).
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1001354
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001354Summary
Methylation of histone H3 lysine 4 (H3K4me) is an evolutionarily conserved modification whose role in the regulation of gene expression has been extensively studied. In contrast, the function of H3K4 acetylation (H3K4ac) has received little attention because of a lack of tools to separate its function from that of H3K4me. Here we show that, in addition to being methylated, H3K4 is also acetylated in budding yeast. Genetic studies reveal that the histone acetyltransferases (HATs) Gcn5 and Rtt109 contribute to H3K4 acetylation in vivo. Whilst removal of H3K4ac from euchromatin mainly requires the histone deacetylase (HDAC) Hst1, Sir2 is needed for H3K4 deacetylation in heterochomatin. Using genome-wide chromatin immunoprecipitation (ChIP), we show that H3K4ac is enriched at promoters of actively transcribed genes and located just upstream of H3K4 tri-methylation (H3K4me3), a pattern that has been conserved in human cells. We find that the Set1-containing complex (COMPASS), which promotes H3K4me2 and -me3, also serves to limit the abundance of H3K4ac at gene promoters. In addition, we identify a group of genes that have high levels of H3K4ac in their promoters and are inadequately expressed in H3-K4R, but not in set1Δ mutant strains, suggesting that H3K4ac plays a positive role in transcription. Our results reveal a novel regulatory feature of promoter-proximal chromatin, involving mutually exclusive histone modifications of the same histone residue (H3K4ac and H3K4me).
Introduction
Histones are covalently modified on many lysine residues. The outcome of these modifications is either to modify chromatin structure directly or provide docking sites for the binding of non-histone proteins [1]. The same chemical modification (e.g. methylation) on different residues can lead to distinct outcomes. Moreover, some histone modifications function in a combinatorial fashion to generate different functional outcomes [2]-[4]. This led to the notion that histone modifications may represent an epigenetic code that influences gene expression and serves as a “memory” of cell identity during development of cell lineages [5]-[6]. The recent development of high resolution mass spectrometry has enabled the identification of a great number of new histone modifications [7]-[9]. Elucidation of the functions of these new modifications is greatly facilitated in model organisms where, in contrast to vertebrate cells, histone gene mutations that abolish specific modifications can be readily introduced.
Histone H3 lysine 4 is a highly studied residue whose modification is important for many biological processes in a wide range of species [10]-[11]. The genomic localisation of H3K4 methylation (H3K4me) has been conserved through evolution. It is highly regulated and generally associated with transcriptionally active genes [12]-[18]. H3K4 tri-methylation (H3K4me3) is a hallmark of transcriptional start sites and is generally followed by H3K4me2 and H3K4me1 along gene coding regions [19]-[22]. The multiple functions of H3K4 are mediated by a number of chromatin-associated proteins that selectively bind to some of the four methylation states of H3K4: unmethylated, mono-, di - or trimethylated [23]-[37]. In yeast, H3K4 is methylated by Set1, a SET domain containing protein and a homolog of Trithorax. Set1 is part of a complex termed COMPASS (complex of proteins asociated with Set1) [38]-[42]. The regulation of the different forms of H3K4me is complex and requires not only the components of COMPASS, but also a trans-histone pathway which involves mono-ubiquitylation of H2B lysine 123 [43]-[44].
A close link exists between H3K4me2 and -me3 and acetylation of the H3 tail. H3K4me3 is often found together with acetylation of other residues (i.e. K9, K14, K18, K23 and K27) on the same H3 molecules [45]-[49]. In a subset of yeast genes, H3K4me3 directly binds to the PHD finger domain of Yng1, a subunit of the NuA3 histone acetyltransferase (HAT) complex that modifies H3K14, which couples the acetylation and methylation of H3 on different residues [33]. In contrast, through the recruitment of the SET3 complex, H3K4me2 in coding regions promotes deacetylation of H3 in the wake of RNA polymerase II (RNApol II) [50]. These results suggest that there is a highly dynamic and coordinated interplay between histone H3K4 methylation and the enzymes that control H3 acetylation during transcription.
Despite extensive studies of histone H3K4 methylation, the functional implications of other modifications that occur on the same residue have not been investigated. Here, we identified H3K4 acetylation (H3K4ac) in S. cerevisiae using mass spectrometry and a highly specific antibody that we developed. We found that GCN5 and, to a lesser extent, RTT109 are needed for both H3K4ac and H3K9ac in vivo. H3K4 deacetylation in euchromatin is mainly dependent upon HST1, and partly on SIR2. In contrast, removal of H3K4ac from heterochromatin requires SIR2 only. Genome-wide ChIP experiments revealed that H3K4ac is generally found upstream of H3K4me3 in active gene promoters, a pattern which has been conserved at many human CD4+ T-cell promoters [51]. We further demonstrate that H3K4me2 and –me3 mediated by the COMPASS complex limits global levels of H3K4ac at promoters and prevents it from spreading into the 5′-ends of coding regions. Using a genetic approach to separate the functions of H3K4ac and H3K4me, we identified a subset of S. cerevisiae genes whose expression depends upon H3K4ac, but not H3K4me. Altogether, our results strongly support a positive role for H3K4ac in gene transcription and identify a novel interplay between two modifications of the same histone residue.
Results
Histone H3 lysine 4 is acetylated in Saccharomyces cerevisiae
One of the most studied and conserved histone modifications is H3K4 methylation (H3K4me), which is tightly linked to transcription in many species [10]. Because of this, and the fact that methylation and acetylation of the same lysine residue are mutually exclusive, we investigated whether H3K4ac was present in S. cerevisiae, a powerful model organism for genetic analysis. For this purpose, we raised an antibody against H3K4ac and confirmed its specificity with peptide binding assays and immunoblotting competition assays (Figure 1A, 1B). Previous studies have detected the presence of H3K4ac in human cells and an antibody was made commercially available, however the specificity of this antibody was questioned as it showed cross-reactivity against the acetylated H4 N-terminal tail and H3K9ac peptides [51]-[52]. In contrast, our antibody does not show any strong cross-reactivity against H3K9ac or tetra-acetylated H4 peptides (Figure 1C).
Fig. 1. H3K4 Is Acetylated in S. cerevisiae. 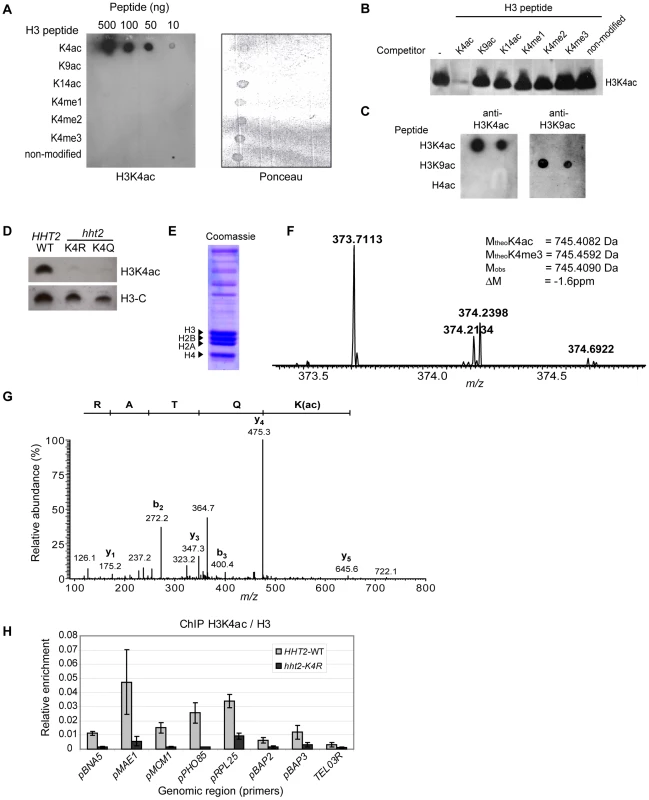
(A) Synthetic peptides derived from the histone H3 N-terminal tail (residues 1 to 10) with modifications at different residues were applied to a nitrocellulose membrane. The membrane was incubated with the H3K4ac antibody, a horseradish peroxidase-conjugated anti-rabbit IgG, and signals visualised by chemiluminescence (left). Ponceau S staining showing that all the peptides bound to nitrocellulose (right). (B) Immunoblots carried out with nuclear extracts from mouse thymocytes. The membrane was cut into strips and incubated with H3K4ac antibody that was pre-incubated with the indicated competitor peptides (1 µg/ml), resulting in a molar ratio of approximately 1000∶1 (peptide:antibody). (C) Dot blots of synthetic peptides (100ng each) containing H3K4ac, H3K9ac, and tetra-acetylated histone H4, (K5, 8, 12, 16)ac, were incubated with H3K4ac and H3K9ac antibodies. Antibody binding was detected as described above. (D) Whole-cell lysates from strains expressing either WT or mutant versions of H3 were analysed by immunoblotting for H3K4ac or a non-modified C-terminal peptide of H3. (E) Coomassie stained gel of histones purified from exponentially growing yeast cells. (F) Doubly charged precursor ion with the expected m/z ratio for tryptic peptide 3-TKacQTAR-8. The experimental and theoretical masses of the non-charged peptides are indicated in the inset along with the mass difference between the empirically determined and the predicted mass of the K4-acetylated peptide. (G) MS/MS spectrum of the doubly charged precursor ion with m/z 373.7113. The peptide sequence is shown from its C-terminus to its N-terminus above the spectrum. (H) Chromatin immunoprecipitation (ChIP) assays were analysed by quantitative PCR to determine the abundance of H3K4ac relative to non-modified H3 ChIP at different loci in WT and H3-K4R cells. We readily detected a single H3K4ac band in immunoblots of extracts prepared from wild-type (WT) S. cerevisiae cells (Figure 1D). Importantly, there was no H3K4ac signal in extracts from cells that express either H3-K4R or H3-K4Q as the only source of H3 (Figure 1D). We also confirmed the presence of H3K4ac by mass spectrometry (MS). Histones were isolated from exponentially growing yeast cells (Figure 1E). After trypsin digestion, histone peptides were analysed by nano-LC MS/MS. In histones purified from wild-type cells, we identified a doubly charged precursor ion with the expected mass-to-charge ratio (m/z) for the tryptic peptide 3-TKacQTAR-8. The m/z ratio for the monoisotopic form of this precursor ion was determined with high accuracy as 373.7113 (Figure 1F). The experimental mass of the non-protonated peptide corresponding to this precursor ion (745.4070 Da) was in excellent agreement (−1.6 ppm) with the theoretical mass expected for a K4-acetylated peptide (745.4082 Da) and clearly different from the predicted mass of a peptide containing trimethylated K4 (745.4592 Da). Fragmentation of this precursor peptide by collision-activated dissociation resulted in b - and y-type ion series (Figure 1G) with diagnostic fragment ions (e.g. the mass difference between the y3 and y5 fragments and the mass of the b2 fragment) that unambiguously proved the presence of an acetyl group on H3K4. However, technical limitations precluded us from determining the abundance of H3K4ac by MS. Using synthetic peptides, we found that the tryptic peptide containing H3K4ac was extremely poorly retained during reverse phase HPLC. Because of this we were not able to accurately assess the stoichiometry of H3K4 acetylation.
To investigate the presence of H3K4ac in chromatin, we performed chromatin immunoprecipitation (ChIP) in strains expressing either H3-WT or H3-K4R. Our initial screen of various genomic regions by qPCR revealed a strong enrichment of H3K4ac (relative to ChIP of non-modified H3) at some gene promoters (MAE1, PHO85, RPL25) a modest enrichment at other genes (BNA5, MCM1, BAP2, BAP3) and a very low enrichment at a region ∼1 Kb from telomere III-R (TEL03R) (Figure 1H). These results are consistent with our genome-wide ChIP-chip data showing an enrichment of H3K4ac in euchromatin at the promoters of highly transcribed genes (see below). We performed control experiments to confirm the target lysine specificity of our antibody in ChIP assays. The ChIP signal obtained with the H3K4ac antibody dropped significantly at all regions tested in the H3-K4R strain (Figure 1H). As an average for all the promoters tested, the signal obtained by ChIP with our H3K4ac antibody in the WT strain is about 7-fold higher than the non-specific signal (noise) derived from the H3-K4R strain. The average signal-to-noise ratio increases to 9-fold when considering only the four promoters where the signal is most abundant in WT cells. Certain promoters show a higher background, such as the RPL25 promoter (3-fold signal-to-noise), which may reflect some degree of non-specific binding of our H3K4ac to other acetylated lysine residues present in proteins in the vicinity of the RPL25 promoter. Nevertheless, our H3K4ac-specific enrichment values are comparable with previously published ChIP data using acetyl-lysine specific antibodies, where the signal-to-noise ratio ranged from 3 to 10-fold [53]. These results validate the specificity of our antibody in ChIP assays and confirm the presence of H3K4ac in gene promoters.
H3K4 acetylation depends upon GCN5 and RTT109
To screen for potential HATs that act on H3K4, we monitored H3K4ac by immunoblotting extracts derived from strains carrying gene disruptions of previously identified HATs. Using this approach, we found that GCN5 and RTT109 were required for H3K4ac in vivo (Figure 2A, lanes 4 and 7). Consistent with a recent report [54], gcn5Δ and rtt109Δ single mutants also exhibited lower levels of H3K9ac than WT cells (Figure 2A). A gcn5Δ rtt109Δ double mutant showed levels of H3K4ac comparable to the background observed in histones from H3-K4R cells (Figure 2B, 2C), suggesting that, as is the case for H3K9ac, GCN5 and RTT109 are responsible for essentially all H3K4ac in yeast.
Fig. 2. HATs and HDACs That Contribute to H3K4ac In Vivo. 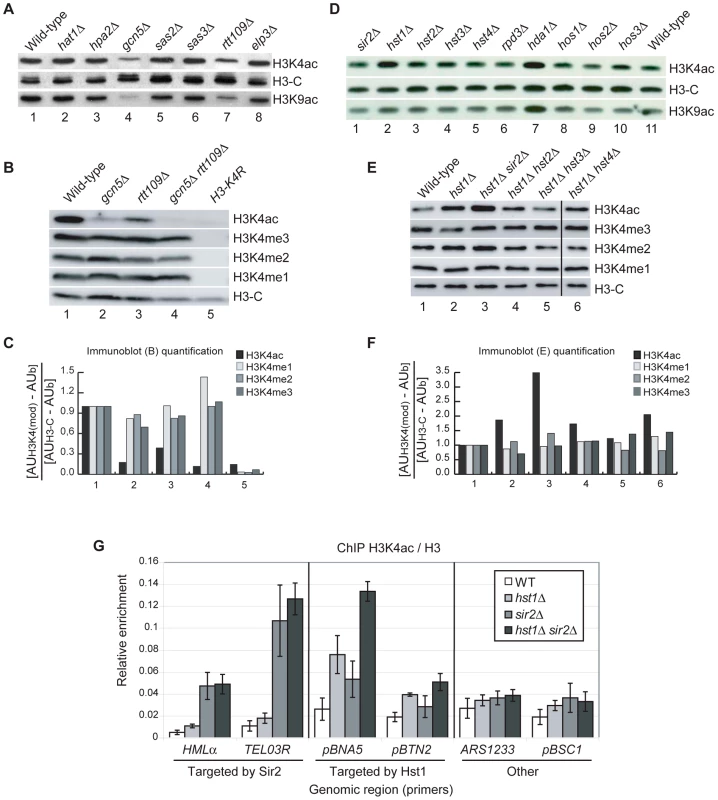
(A–B) Whole-cell lysates prepared from strains with deletions of known HAT encoding genes were analysed by immunoblotting with the indicated antibodies. (C) Quantification of the immunoblots shown in (B). Signals for each band were expressed in Arbitrary Units (AU) and, after background subtraction (AUb), normalised to the H3-C signal measured in each strain. (D–E) Whole-cell lysates obtained from strains with deletions of known HDAC encoding genes were analysed by immunoblotting with the indicated antibodies. (F) Quantification of the immunoblots shown in (E), calculated as described above. (G) Chromatin immunoprecipitation (ChIP) was performed by quantitative PCR to determine the abundance of H3K4ac at different loci in WT, hst1Δ, sir2Δ or hst1Δ sir2Δ strains. The six target regions were divided into three groups: regions targeted by Sir2, gene promoters targeted by Hst1 and other gene regions not targeted by Sir2 or Hst1. The TEL03R primers amplify a portion of ARS319, a unique sequence region approximately 1 Kb from the telomeric repeats on the right arm of chromosome III. Results are expressed as a ratio of ChIP signals obtained from the same extracts with antibodies against H3K4ac and a non-modified H3 C-terminal peptide. H3K4 deacetylation is dependent upon HST1 and SIR2
To identify potential HDACs responsible for H3K4 deacetylation we repeated our immunoblotting screen with extracts from different HDAC deletion mutants. This revealed that H3K4 deacetylation is, at least in part, dependent upon HST1 and HDA1 (Figure 2D, lanes 2 and 7). Deletion of HDA1 also led to an increase in H3K9ac, whereas the hst1Δ mutant showed an increase in H3K4ac but not H3K9ac (Figure 2D, bottom panel). Because of this, we focussed our experiments on Hst1. S. cerevisiae Hst1 is a member of a family of five NAD+-dependent HDACs, known as sirtuins because of their homology to Sir2. These enzymes are readily inhibited by nicotinamide, one of the products of the deacetylation reaction. Consistent with this, exposure of WT cells to nicotinamide caused an increase in H3K4ac (Figure S1). Treatment of hst1Δ cells with nicotinamide led to a further increase in H3K4ac (Figure S1), which suggested that other sirtuins also contribute to H3K4 deacetylation. We analysed H3K4ac in a collection of sirtuin double mutants and found that deletion of SIR2 in cells lacking Hst1 increased H3K4ac above the levels observed in WT cells and hst1Δ single mutants (Figure 2E, 2F, lanes 1-3). This suggests that SIR2 can compensate for the loss of HST1 for global deacetylation of H3K4. In S. cerevisiae, Sir2 promotes heterochromatin formation through histone deacetylation in specific chromosomal regions, namely the silent mating type loci (HMRa and HMLα ), sub-telomeric regions and a subset of ribosomal DNA (rDNA) repeats [55]. By immunoblotting whole-cell extracts, we found no global increase in H3K4ac in a sir2Δ single mutant (Figure 2D, lanes 1 and 11). We hypothesised that, in WT cells, Sir2 might deacetylate H3K4 mainly at heterochromatic loci, which represent a relatively small fraction of the genome. To test this hypothesis, we performed ChIP in WT, hst1Δ, sir2Δ and hst1Δ sir2Δ double mutant cells at different euchromatic and heterochromatic regions known to be deacetylated by Hst1 and Sir2 respectively: the promoters of BNA5 and BTN2, targeted by Hst1 [56], a sub-telomeric region (TEL03R) and the silent mating type locus HMLα, targeted by Sir2 [55]. We also analysed an origin of replication (ARS1233) and the BSC1 gene promoter, which are not known to be targeted by either Sir2 or Hst1. The sir2Δ single mutation elevated H3K4ac at the TEL03R and HMLα heterochromatic loci to levels comparable to those observed in the hst1Δ sir2Δ double mutant (Figure 2G). The deletion of HST1 had virtually no effect at these regions. We therefore conclude that H3K4 deacetylation in heterochromatic regions is solely dependent upon SIR2. Conversely, deletion of HST1 increased H3K4ac at the promoters of BNA5 and BTN2, but not at other euchromatic regions (Figure 2G). The SIR2 deletion affected H3K4ac only slightly at those loci whereas the hst1Δ sir2Δ double mutant showed higher levels of H3K4ac than in any of the single mutants (Figure 2G). This is consistent with a previous study [57] and suggests that Sir2 can also contribute to deacetylate H3K4 at Hst1-targeted loci.
Genome-wide presence of H3K4ac in euchromatin and active promoters
To localise H3K4ac throughout the genome, DNA recovered from ChIPs was analysed with high-resolution tiling arrays (Affymetrix 1.0R). Nucleosome density was controlled by normalising H3K4ac to the results of a ChIP experiment performed from the same cell lysates using an antibody against the non-modified C-terminal tail of H3 (H3-C).
The data showed that H3K4ac peaks are distributed along all chromosomes and relatively low levels of H3K4ac are observed at most sub-telomeric regions (Figure S2). This is most apparent on chromosome III, where H3K4ac is markedly depleted in large sub-telomeric regions that encompass HMLα and HMRa (Figure S2), where Sir2 was previously shown to promote histone deacetylation. Closer manual inspection of the results using the UCSC genome browser [58] indicated that the strongest peaks of H3K4ac are located at highly transcribed genes, such as ribosomal protein genes. Two examples (RPL37A and RPS31) are highlighted in Figure 3A. We performed a systematic analysis aimed at determining whether H3K4ac was generally enriched at promoters. For this purpose, we aligned H3K4ac data derived from genes with upstream intergenic regions (IGRs) that contain divergent promoters. Based on this criterion, these IGRs are devoid of DNA sequences corresponding to 3′-untranslated regions (3′-UTRs) and transcriptional termination sequences. This analysis revealed that H3K4ac peaks slightly upstream of the 5′-ends of genes (Figure 3B). Furthermore, the degree of H3K4ac enrichment correlated with transcriptional activity (as measured in [59]). For the most highly transcribed genes (50+ mRNA/hr), H3K4ac peaks upstream and appears to spread into coding regions (Figure 3B). In less frequently transcribed genes, H3K4ac also peaks just upstream of 5′ends, but sharply drops in coding regions (Figure 3B). We performed a similar analysis for genes that have converging 3′-ends. The corresponding IGRs contain 3′-UTRs and transcriptional termination sequences, but are devoid of promoters. These chromosomal regions revealed only a modest enrichment for H3K4ac in the most highly transcribed gene group (Figure 3C, coordinates 0.0 and beyond). This effect probably results from the aforementioned spreading of H3K4ac peaks over the whole body of highly transcribed genes (compare Figure 3B and 3C). From this data, it is clear that H3K4ac peaks at gene promoters and the degree of enrichment is proportional to the rate of transcription.
Fig. 3. H3K4ac Is Enriched at Promoters of Transcribed Genes. 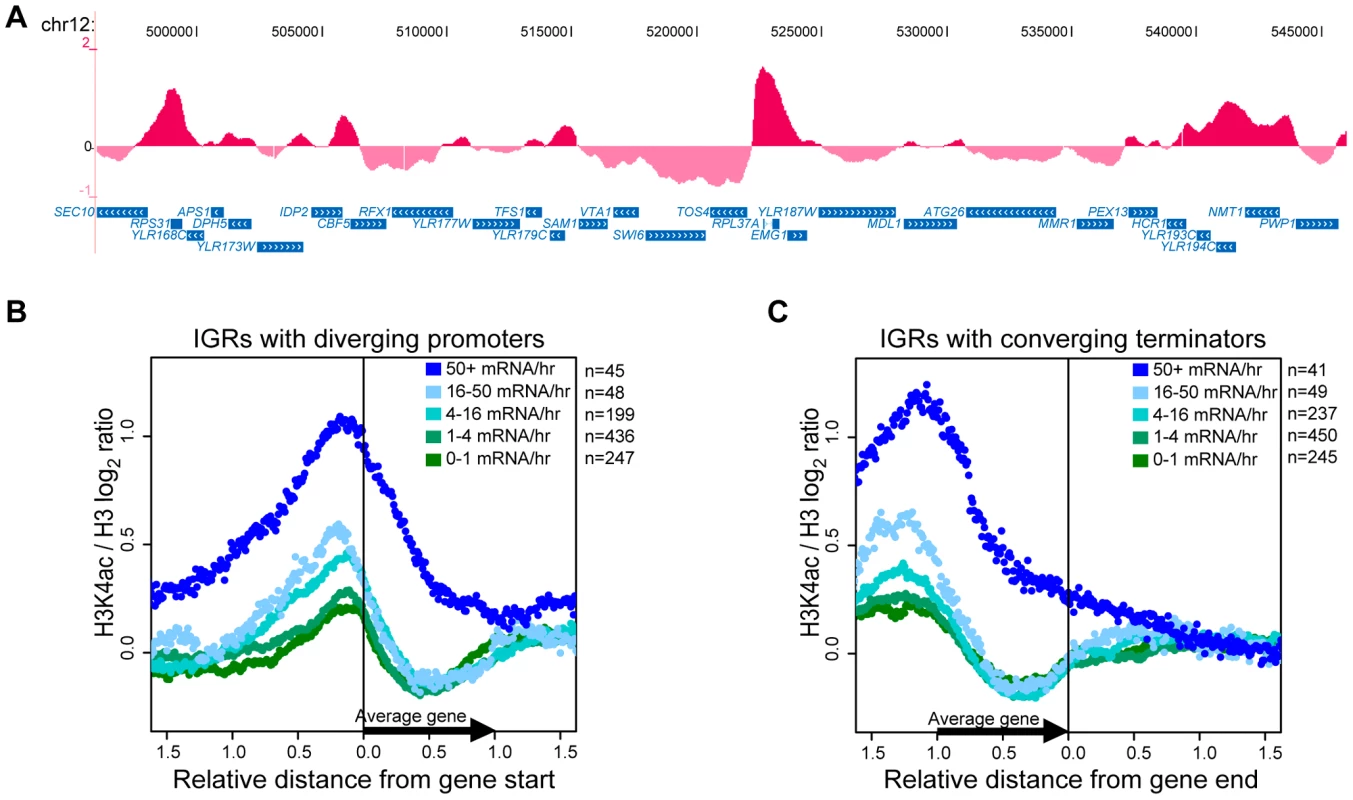
(A) Examples of the localisation pattern of H3K4ac along a 50 kb segment of chromosome 12, as displayed on the UCSC genome browser (http://genome.ucsc.edu/) [58]. The data (purple) are represented as a log2 ratio of H3K4ac over non-modified H3 C-terminus ChIP signals and represent the average of 3 biological replicates. The names, position and orientation of ORFs (S. cerevisiae assembly Oct. 2003) are shown (blue) at the bottom of each panel. (B–C) Systematic analyses of the genome-wide location of H3K4ac / H3-C ChIP signals at: (B) 5′-ends of ORFs with divergent promoter regions. (C) 3′-ends of ORFs with convergent terminator regions. The data was aligned to the closest 5′- or 3′-end of ORFs, normalised for gene length and sub-divided into 5 groups according to transcriptional activity (mRNA/hr) [59]. For simplicity, the relative position value for each data point was rounded to the nearest hundredth. The y-axes show the abundance of H3K4ac (average of three biological replicates) as a function of relative position along the nearest gene (x-axes). H3K4ac is upstream of H3K4me3 at gene promoters
We found that H3K4ac was enriched at transcriptionally active promoters (Figure 3B) and it has been reported that H3K4me3 is also present at this location in active genes [20]. To explore the relationship between H3K4ac and H3K4me, we compared our genome-wide H3K4ac localisation data with the previously published high-resolution data for H3K4me1, -me2 and -me3 [60]. Strong peaks of H3K4ac were generally located upstream of H3K4me3 in gene promoters, followed by the typical pattern of H3K4me2 and H3K4me1 enrichment along coding regions (Figure 4A and more examples in Figure S3). There is a considerable overlap between H3K4ac with H3K4me3, although their localisation patterns are not identical. We repeated the H3K4me3 ChIP-chip under our conditions (as described for H3K4ac) and our results (data not shown) produced an enrichment pattern nearly identical to the Kirmizis et al. dataset [60]. Systematic analysis showed that H3K4me3 spreads further into coding regions than H3K4ac in moderately and highly transcribed genes (from 4 to 50+ mRNA/hr) (Figure 4B). The peak intensity for the H3K4ac signals are slightly upstream of H3K4me3 (Figure 4B), which is consistent with the UCSC track observations (Figure 4A).
Fig. 4. H3K4ac Is Located Upstream of H3K4me3. 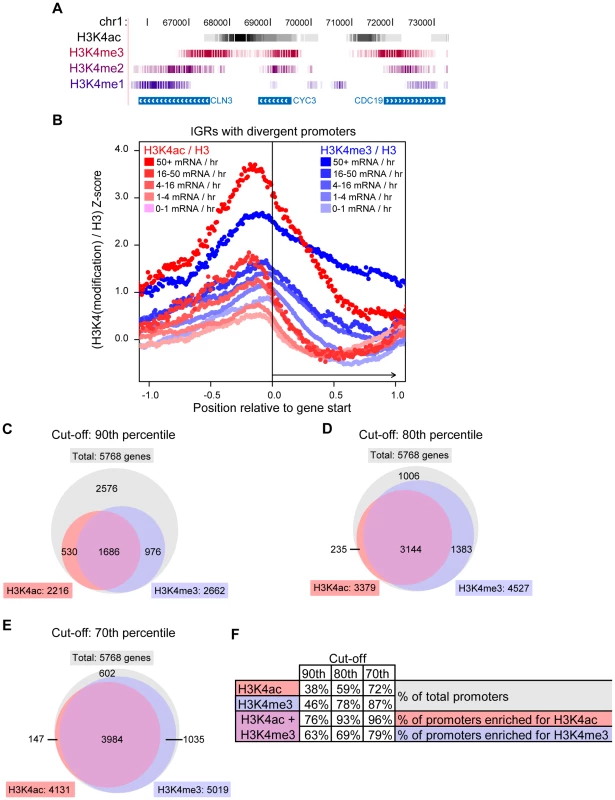
(A) The H3K4ac / H3 (black) ChIP-chip data were aligned with the H3K4me3 (burgundy), -me2 (purple) and -me1 (blue) data from [60] and displayed in the UCSC genome browser (http://genome.ucsc.edu/). Each vertical line represents one probe in the dataset and the intensity of the color is proportional to the enrichment of the modification. For clarity, only the probes with a log2 ratio above 0 are shown for each dataset. (B) Systematic alignement of H3K4ac / H3 (red shades) and H3K4me3 / H3 (blue shades) ChIP-chip data at intergenic regions (IGRs) with divergent promoters (as in Figure 3B). The raw ChIP-chip data were converted into Z-scores in order to be compared on the same scale (see Material and Methods). (C–E) Venn diagrams illustrating the overlapping number of promoters that are enriched for H3K4ac or H3K4me3. The cut-off indicates the H3K4(mod) / H3-C Z-score threshold at which a given promoter was judged to be enriched in either H3K4ac or H3K4me3. For example, with a cut-off at the 90th percentile (C), the red circle contains the number of promoters that have H3K4ac / H3 signal ratios in the top 10% of the genome. (F) A table displaying the data from the Venn diagrams as percentages. To investigate whether this pattern was evolutionarily conserved, we examined the localisation of H3K4 modifications in human CD4+ lymphocytes from previously published ChIP-seq data [19], [51]. Manual inspection of the data with the UCSC genome browser revealed H3K4ac and H3K4me3 overlapping at many transcriptional start sites, but H3K4ac was slightly upstream of H3K4me3 (representative examples are shown in Figure S4). Interestingly, H3K4ac peaks also overlap with RNApol II more closely than H3K4me3 at these genes (Figure S4), suggesting a function for H3K4ac in transcriptional regulation in human cells. From these data, we conclude that H3K4ac is present at transcribed gene promoters, and is adjacent to a unidirectional gradient of H3K4me3, -me2 and -me1 that proceeds from promoters into coding regions. This pattern of H3K4 modifications has been conserved during evolution.
Genome-wide overlap of H3K4ac and H3K4me3 at promoters
To investigate and compare the proportion of promoters that are enriched in H3K4ac or H3K4me3, we transformed the log-ratio data into Z-scores (see Materials and Methods). This was performed to normalise datasets with different scales, thus allowing a direct comparison of the H3K4ac and H3K4me3 datasets. We then established a threshold value above which a promoter was judged enriched in H3K4ac or H3K4me3 relative to H3. When the cut-off was set at the 90th percentile (only promoters with enrichment values scoring in the top 10% of all values), H3K4ac was found to be enriched at 38% of all gene promoters, whereas H3K4me3 was enriched at 46% of promoters (Figure 4C and 4F). Of the promoters enriched in H3K4ac, 74% were also enriched in H3K4me3. This value increased to 93% when the cut-off was set to the top 80th percentile (Figure 4D and 4F) and 96% at the top 70th percentile (Figure 4E and 4F), indicating that most promoters enriched in H3K4ac also have a significant enrichment in H3K4me3. However, a smaller proportion of H3K4me3-enriched promoters also contain H3K4ac: 63%, 69% and 79% for the top 90th, 80th and 70th percentile cut-offs respectively (Figure 4F). From this we conclude that, genome-wide, H3K4me3 is enriched at a greater number of promoters than H3K4ac, and that most promoters which harbour H3K4ac are also enriched in H3K4me3.
Set1-mediated H3K4me2 and me3 limit the accumulation of H3K4ac
Since H3K4ac and H3K4me3 overlap considerably at the promoters of many genes, we sought to determine if there is a competition for the lysine 4 substrate. Based on mass spectrometry, we found that a SET1 deletion caused a global increase in H3K4 acetylation (Figure 5A). This result was corroborated by immunoblotting data showing that H3K4ac, but not H3K9ac was elevated in set1Δ cells (Figure 5B, 5C). The immunoblotting result truly reflected an increase in H3K4ac in set1Δ cells, rather than cross-reaction to other sites of acetylation, because no signal was observed in set1Δ H3-K4R mutant cells (Figure 5D). In addition, an H3K4ac peptide, but not H3K9ac or non-modified K4 peptides, could compete the signal observed with the K4ac antibody in WT and set1Δ cells (Figure 5E). Set1 is part of COMPASS, a complex that has multiple subunits which contribute to the different degrees of H3K4 methylation (me1, me2 or me3) [61]-[63]. To determine which subunits of COMPASS were important to limit the accumulation of H3K4ac, we performed immunoblots using strains with single gene deletions of all the COMPASS subunits. Cells lacking SET1, BRE2, SDC1, SWD3 or SWD1 have in common a complete absence of H3K4me2 and me3, but mutations of these genes differentially affect H3K4me1 (Figure 5F). For instance, bre2Δ and sdc1Δ mutants have WT levels of H3K4me1 (Figure 5F, lanes 1, 3 and 5), whereas set1Δ, swd3Δ and swd1Δ mutants lack all forms of H3K4me (lanes 2, 6 and 8). Nonetheless, relative to WT cells, all these mutants increased H3K4ac to similar degrees (Figure 5F, compare lane 1 with lanes 2, 3, 5, 6 and 8). An spp1Δ mutant with nearly normal levels of H3K4me2, but reduced H3K4me3 showed a moderate increase of H3K4ac (Figure 5F, lanes 1 and 7). In striking contrast, an shg1Δ mutant that retained normal levels of H3K4me2 and H3K4me3 did not show a significant increase in H3K4ac compared with WT cells (Figure 5F, lanes 1 and 4). These results argue that the increase in H3K4ac observed in several mutants of the COMPASS complex occurs mainly because of the absence of H3K4me2 and me3, rather than a lack of H3K4me1.
Fig. 5. The COMPASS Complex Limits Global Levels of H3K4ac and Confines H3K4ac Localisation to Promoter Regions. 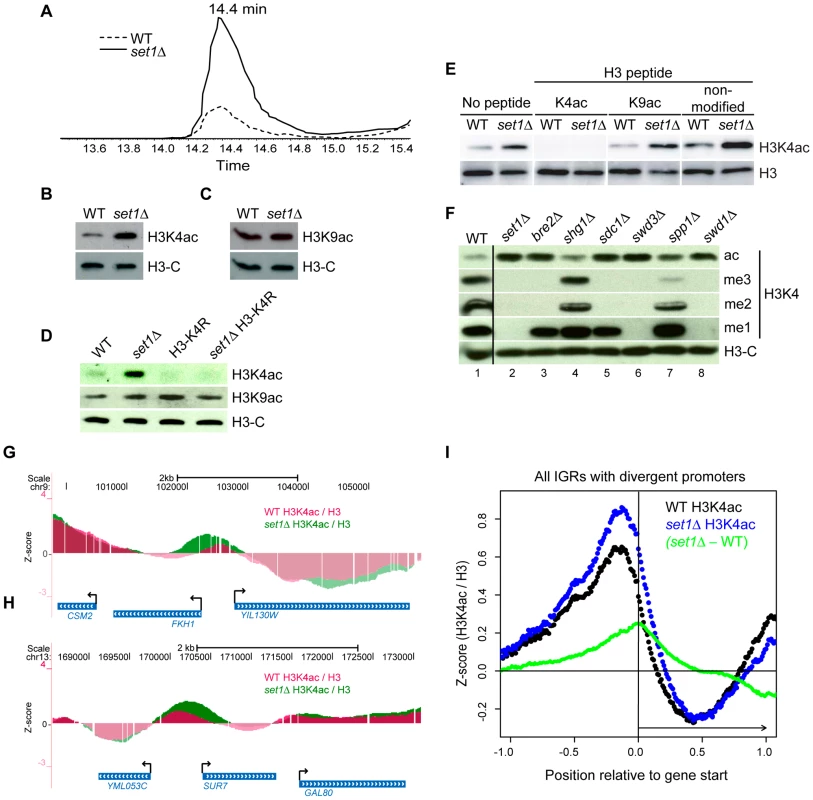
(A) Extracted ion chromatogram showing the retention time of peptide 3-TKacQTAR-8 during reverse phase HPLC and its relative abundance in WT and set1Δ cells. (B–F) Whole-cell lysates from the indicated mutants were analysed by immunoblotting. In (E), a peptide competition assay was performed as described in Figure 1B. (G–H) Examples of the localisation pattern of H3K4ac in either WT (purple) or set1Δ (green) strains at two genomic regions displayed using the UCSC genome browser (genome.ucsc.edu) [58]. The data represents the normalised (Z-score) log2 ratio of H3K4ac over H3-C (see Materials and Methods). (I) Systematic analyses of H3K4ac in WT versus set1Δ cells in all intergenic regions (IGRs) that contain divergent promoters. The raw H3K4ac ChIP-chip data in WT or set1Δ cells were normalised either against their respective H3-C ChIP (black: WT, blue: set1Δ), converted into Z-scores (see Material and Methods) and aligned relative to the 5′-end of the corresponding ORF (normalised as described in Figure 3). The difference between the set1Δ and WT datasets (set1Δ - WT, green) is also shown. Since a global reduction in H3K4me2 and me3 led to an increase in H3K4ac, we wished to investigate if the opposite was also true. Immunoblots of cell lysates prepared from gcn5Δ and gcn5Δ rtt109Δ strains, with strongly reduced H3K4ac, showed no significant increase in any form of H3K4me (Figure 2B, 2C). Conversely, we observed no apparent decrease in H3K4me1, me2 or me3 in cells with elevated levels of H3K4ac (hst1Δ or hst1Δ sir2Δ, (Figure 2E, 2F). Therefore, although H3K4ac increases in mutant cells lacking H3K4me2 and me3, changes in H3K4ac do not influence global levels of H3K4 methylation. These results suggest that the relative abundance of H3K4ac does not merely reflect a simple competition for the H3K4 substrate.
Set1 regulates H3K4ac at promoters and the 5′ end of coding regions
To see if the deletion of SET1 led to a change in the genome-wide distribution of H3K4ac, we performed ChIP-chip experiments to compare the localisation of H3K4ac in set1Δ and WT cells. Manual inspection of the data in the UCSC Genome Browser revealed changes in the distribution and the relative abundance of H3K4ac in set1Δ cells, at several gene promoters. For example, at the FKH1 and SUR7 genes, H3K4ac is increased at the promoter region and spreads towards the coding region in the set1Δ strain (Figure 5G, 5H). Systematic alignment of divergent promoter regions showed that the deletion of SET1 led to a global increase in the relative abundance of H3K4ac at gene promoters (Figure 5I, blue versus black dots). We also observed a slight shift of H3K4ac towards the 5′ ends of the coding region in set1Δ cells (Figure 5I). To better assess the changes in localisation of H3K4ac in the set1Δ strain, the H3K4ac ChIP-chip data from WT cells were subtracted from those of set1Δ cells. This analysis clearly showed that the strongest difference in H3K4ac occurs at position 0 relative to gene start (Figure 5I, green dots), which corresponds to the 5′ ends of coding regions. The fact that the shoulder of the H3K4ac peak in the set1Δ strain is skewed over downstream sequences argues that this increase is not merely due to increased peak signal in set1Δ cells, since this would lead to a symmetric increase in each shoulder. Instead, this shows that, on average, H3K4ac spreads further into the 5′ ends of coding regions in set1Δ cells. We also observed a global decrease in H3K4ac at the 3′ ends of coding regions in set1Δ mutants (Figure 5I). The significance of this decrease is unknown. Nevertheless, we can conclude that the global increase in H3K4ac observed in set1Δ mutant cells by MS and immunoblotting occurs more prominently at the promoters and 5′ ends of coding regions relative to other regions.
The global average increase of H3K4ac in set1Δ cells likely occurs at a subset of genes and not all genes. Manual inspection of the datasets clearly shows that the H3K4ac pattern remains unaffected at certain genes, for example GAL80 and YML053c (Figure 5H). To provide further evidence that the increase in H3K4ac observed in the set1Δ strain did not occur equally at all genes, we performed ChIP-qPCR experiments in WT and set1Δ strains at three genes: CPA2, ARG7 and FKH1. These genes were selected because they respectively showed little, moderate and large changes in H3K4ac in set1Δ strains according to our initial observations in the UCSC Genome Browser. Our ChIP-qPCR analyses confirmed the ChIP-chip results for H3K4ac in the set1Δ strain (Figure S5A-S5C). Therefore, although the absence of H3K4me2 and me3 leads to a global increase in H3K4ac (Figure 5A, 5B), ChIP assays demonstrate that the increase in H3K4ac is more prominent at some genes than others.
H3K4ac and gene expression
Since H3K4ac is enriched at highly transcribed gene promoters, we explored the possibility that H3K4ac might play a role in gene transcription. Genetic analysis of the function of H3K4ac is complicated by the fact that mutation of H3K4 abolishes both acetylation and methylation. To circumvent this problem, we compared the global mRNA profile of a set1Δ strain, which lacks H3K4me but retains H3K4ac, with that of an H3-K4R mutant where both H3K4 acetylation and methylation are lost. In order to get a global view of the expression profiles in both mutants, we ranked the genes by log2 fold change in mutant strains relative to WT cells (Figure 6A). At one end of the spectrum, we found many genes that are up-regulated in both the H3-K4R and the set1Δ mutant strains (Figure 6A, left part). Many of these genes are up-regulated only in H3-K4R mutant cells (Figure 6A, 6B). One possibility is that expression of some of these genes may be affected simply because of the lysine-to-arginine mutations, rather than a lack of modification. However, there is considerable overlap between the genes up-regulated in H3-K4R and set1Δ mutant strains (Figure 6B). We conclude that the repressed state of these genes in WT cells is likely to be dependent on H3K4me, rather than H3K4ac. This is consistent with previous reports showing a repressive function for SET1 [64]-[66].
Fig. 6. Global mRNA Expression Profiles of set1Δ and H3-K4R Mutant Strains. 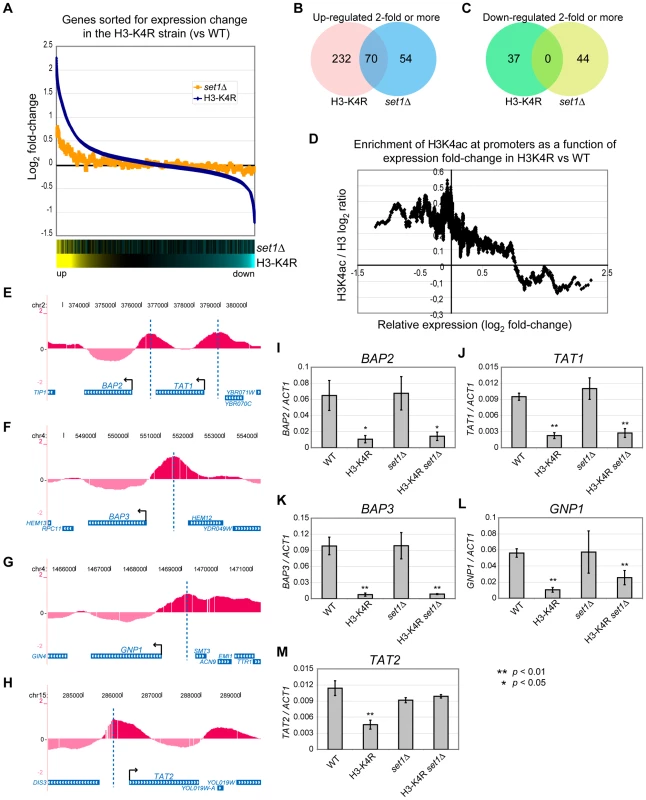
(A) Graph of mRNA expression fold-change relative to WT cells in the H3-K4R (blue) and set1Δ (yellow) strains for all the analysed genes. The genes were ranked according to log2 fold-change in the H3-K4R strain and a sliding average (window = 50, step = 1) was applied to the data on the y-axis. The bottom panels are heat map illustrations of the log2 fold-change, where yellow indicates an increase in mRNA abundance in the mutant strains relative to WT cells and blue indicates a decrease. (B–C) Venn diagrams showing the overlap between groups of genes up-regulated (B) or down-regulated (C) at least two-fold when either the H3-K4R or the set1Δ strain is compared with WT cells. (D) Graph of the H3K4ac / H3 ratio at the promoter regions of each gene (average for from −200 to +1bp relative to the start codon) derived from the ChIP-chip data as a function of their log2 fold-change in mRNA abundance in the H3-K4R mutant strain versus WT cells (derived from the mRNA expression microarray data). A sliding window average was applied to the data on both axes (window = 50, step = 1). (E–H) Localisation of H3K4ac near genes that belong to the ontology group of “amino acid transporters” and are poorly expressed in H3-K4R mutants compared with WT cells: (BAP2/YBR068C, TAT1/YBR069C, BAP3/YDR046C, GNP1/YDR508C, TAT2/YOL020W). The start sites of these genes are indicated by arrows and vertical dashed lines indicate the peaks of H3K4ac in their upstream regions. The display is from the UCSC genome browser (http://genome.ucsc.edu/). (I–M) RT-qPCR analyses of RNA extracted from the indicated strains (WT, H3-K4R, set1Δ, H3-K4R set1Δ). The values were normalised to ACT1. The data represent an average of three biological replicates. At the other end of the spectrum, we found a group of genes which were down-regulated in the H3-K4R strain, but were unaffected in the set1Δ strain (Figure 6A, right part), suggesting a positive role for H3K4ac in the expression of these genes. We identified 37 genes that were down-regulated 2-fold or more in the H3-K4R strain and 44 in the set1Δ strain. Strikingly, there was no overlap between these two groups of genes (Figure 6C), suggesting a function of H3K4 in gene expression that is independent of methylation. We next compared the H3K4ac abundance at promoter regions in WT (from our ChIP-chip dataset) with the expression change of the gene in the H3-K4R strain. The genes that are down-regulated in the H3-K4R strain have high levels of H3K4ac at their promoters in WT cells (Figure 6D), which is consistent with a direct role of H3K4ac in gene expression. Gene ontology term enrichment analysis of the genes down-regulated 2-fold or more in the H3-K4R strain, but whose expression was not affected in a set1Δ strain, identified genes encoding amino acid transporters (p = 1.42×10−5): BAP2/YBR068C, TAT1/YBR069C, BAP3/YDR046C, GNP1/YDR508C, TAT2/YOL020W. Inspection of our ChIP-chip data for these genes confirmed that they all had high levels of H3K4ac at their promoters in WT cells (Figure 6E-6H). We confirmed by RT-qPCR that these genes are down-regulated in an H3-K4R strain, but unaffected in a set1Δ strain (Figure 6I-6M). In contrast to the other RNAs that we monitored, the expression of the TAT2 RNA was unexpectedly restored in a set1Δ H3-K4R double mutant strain, suggesting an indirect effect of the SET1 deletion on this gene (Figure 6M). Taken together, our data suggest that H3K4ac has a positive and direct role in the transcription of a subset of yeast genes.
Discussion
Histone H3K4 methylation is an extensively studied modification in a wide range of organisms, and particularly in the budding yeast Saccharomyces cerevisiae. Using MS and highly specific antibodies we demonstrated that H3K4 is also acetylated in budding yeast. An earlier study of histone modifications had shown that both H3K4ac and H3K4me exist in human, mouse and ciliates [52] and, very recently, in Schizosaccharomyces pombe [67]. The versatility of genetic tools available in S. cerevisiae, and the ability to mutate histone H3K4, has enabled us to uncover a positive function for H3K4ac in the expression of a subset of genes that carry H3K4ac in their promoters. The presence of H3K4ac at active gene promoters has been conserved in human CD4+ cells [51], which suggests a similar role for H3K4ac in gene expression in mammals. We also discovered that the COMPASS complex and H3K4me2 and me3 appear to globally limit the abundance of H3K4ac at promoters and its spreading into the 5′-coding regions of many genes.
Acetylation of the H3 N-terminal tail by specific HATs
Our targeted screen of known HAT-encoding genes has allowed us to identify GCN5 and RTT109 as responsible for H3K4ac in vivo. Gcn5 is the catalytic subunit of the ADA, SAGA, SLIK/SALSA transcriptional co-activator complexes [68] and has been highly conserved during evolution [69]. Our results are consistent with the fact that Gcn5 has been shown to acetylate multiple lysine residues within the H3 N-terminal tail [70]-[71] and is localised to active gene promoters dispersed throughout the genome [56]. Interestingly, RTT109 is also important for the acetylation of H3K4. Furthermore, the gcn5Δ rtt109Δ double mutant has markedly less H3K4ac than the single mutants. Collectively, our results indicate that GCN5 and RTT109 promote H3K4ac via different pathways and/or in different contexts. If they were fully redundant, there should be no decrease in H3K4ac in the single mutants, which is not the case. However, we cannot exclude the possibility that they may also have partially redundant functions in mediating H3K4ac. RTT109 encodes a HAT that mediates H3K56ac [72]-[75], which is involved in nucleosome assembly during DNA replication and the DNA damage response [76]-[78]. In addition, Rtt109 has recently been implicated in acetylating H3K9 and H3K27 in vivo and in vitro [54], [79]-[80]. As is the case for H3K4ac, H3K9ac and H3K27ac are virtually undetectable by immunoblotting and mass spectrometry in gcn5Δ rtt109Δ cells [79]. Therefore, the same HATs are responsible for the bulk of both H3K4 and H3K9 acetylation.
H3 N-terminal tail acetylation and gene transcription
The genome-wide localisation pattern of H3K4ac showed strong peaks at the promoters of active genes. This enrichment has also been observed in the genome-wide patterns of other acetylated lysines in the H3 N-terminal tail, such as H3K9ac, H3K14ac and H3K18ac [20]-[21], suggesting that these modifications often occur together on chromatin. Our microarray analysis identified a relatively small group of genes (37 genes down-regulated by 2-fold or more, Figure 6C) that require H3K4ac, but not H3K4me, for normal expression. However, based on our ChIP-chip data, many other promoters contain high levels of H3K4ac. It is tempting to speculate that acetylation of H3K4 functions in a partially redundant manner with other sites of acetylation at many promoters to increase accessibility to the transcriptional initiation machinery and thereby facilitate gene expression. Interestingly, our gene ontology term analyses revealed a group of amino acid transporter genes (BAP2, BAP3, TAT1, TAT2, GNP1) that are co-regulated by levels of extracellular amino acids [81]-[84]. An interesting possibility is that the transcription factors involved in their expression require H3K4ac either alone or in combination with other acetylated residues to specifically promote initiation of these genes.
Regional and functional specificity of HST1 and SIR2
We identified a role for HST1 in global deacetylation of H3K4 in euchromatin. HST1 encodes a class III HDAC found in at least two complexes. Hst1 is part of a complex with Sum1 and Rfm1 [85]-[86]. Interestingly, the deletion of SUM1 did not result in a significant global increase in H3K4ac based on immunoblotting (data not shown). Hst1 has also been found as a sub-stoichiometric subunit of the SET3 complex (SET3C) [87], which represses middle-sporulation genes during mitosis [88]. However, neither deletion of HOS2 (encoding the main HDAC in the complex) (Figure 2C), nor that of any other components of SET3C led to a global increase in H3K4ac (data not shown). Our results suggest that Hst1 might act in a non-targeted manner to promote global deacetylation of H3K4. Alternatively, Hst1 might be part of another, as yet unidentified complex involved in H3K4 deacetylation.
Interestingly, we also observed some redundancy between HST1 and SIR2 for deacetylation of H3K4 in euchromatic regions. In specific conditions, Hst1 and Sir2 have been shown to act outside of the chromosomal regions where they “normally” exert their functions. For example, overexpression of HST1, but not HST2 or HST3, can specifically compensate for the loss of silencing at HMRa that occurs in a sir2Δ mutant [89]. Our results are also consistent with studies showing a strong connection between these two genes. Both genes share 71% identity in their sequence over 84% of the length of HST1 [90]. SIR2 was previously shown to affect the expression of euchromatic genes when HTZ1 and SET1 were deleted [91] and to interfere with the firing of a number of replication origins [92]. Moreover, studies of chimaeric proteins demonstrated that the N-terminal domains of Hst1 and Sir2 control the chromosomal location and function of both proteins. In contrast, other domains of Hst1 and Sir2 could be swapped, supporting the notion that these enzymes have similar substrate specificity [93]. We propose here that H3K4 is an important target for deacetylation by both Hst1 and Sir2. HST1 and SIR2 are both part of the same phylogenetic branch (subgroup Ia) of NAD+-dependent HDACs (class III) that also includes human SIRT1 and Drosophila Sir2 [94], which are, therefore likely candidates for deacetylation of H3K4 in these organisms.
Patterns of mutually exclusive histone modifications
The relationship between H3K4ac and H3K4me differs from that of other modifications that occur on the same lysine residue. The best studied case is that of H3K9, which is either acetylated or methylated. H3K9ac and H3K9me are present at different genomic regions [51] and have opposing roles in transcriptional regulation. H3K9ac is found in promoter regions and stimulates transcription through the recruitment of TFIID [95], whereas H3K9me2 and me3 contribute to gene repression and pericentric heterochromatin structure by binding HP1 proteins through their chromo-domains [96]-[97]. In this case, deacetylation of H3K9 is essential for the establishment of H3K9 methylation [98]-[100]. Another example is H3K36ac and H3K36me3, which are respectively present in promoters and coding regions of transcriptionally active genes [101]. In contrast to these modifications, H3K4ac overlaps considerably with H3K4me3 in promoter regions. Furthermore, H3K4me2/me3 globally limit the levels of H3K4ac, which suggests the existence of mechanisms to establish the correct patterns of localisation of these mutually exclusive modifications. For example, H3K4me2/me3 could prevent H3K4ac from occurring in gene coding regions by simple competition for the H3K4 substrate. Another possibility is that H3K4 methylation might be a prerequisite step in a negative feedback loop that enables Hst1 to fine tune the levels of H3K4 acetylation in promoters and 5′-coding regions. Mechanisms that confine H3K4ac to some promoters may contribute to prevent spurious transcriptional initiation events [102] or attenuation of transcriptional initiation [66]. The potential to restrict the spreading of histone H3K4ac suggests a novel function for H3K4 methylation and reveals a previously unrecognised layer of chromatin regulation linked to the regulation of transcription in vivo.
Materials and Methods
H3K4ac antibody production and validation
Immunisation procedures and animal handling were carried out by Eurogentec. Sera from ten animals were screened prior to immunisation. Rabbits were immunised with the following peptide: ART(acetyl-K)QTARKSC. The efficiency of antibody production was monitored using ELISA. The antibody was affinity-purified using the same peptide coupled to a Sulfolink column (Thermo) and its specificity tested by dot blots with synthetic peptides and experiments where 1 µg/ml of the peptides were incubated with the antibody prior to immunoblotting.
Yeast strains and media
For most experiments, yeast strains were grown at 30°C in YPD (1% yeast extract, 2% peptone, 2% dextrose). A complete list of yeast strains is available in Table S1.
Whole-cell lysates and immunoblots
Yeast whole-cell lysates were prepared using an alkaline lysis protocol [103]. Essentially, 5×107 cells were resuspended in 500 µl of distilled water and 500 µl of 0.2 M NaOH was added. After 5 min at room temperature, cells were spun down, and resuspended in 100 µl SDS-PAGE sample buffer, boiled for 3 min and spun down again. About 7 µl (equivalent to 3.5×106 cells) of supernatant was typically loaded per lane and, after electrophoresis through an SDS-15% polyacrylamide gel, proteins were transferred to PVDF membranes using a semi-dry apparatus with in 29 mM Tris, 193 mM glycine, 0.02% SDS, 5% methanol for 2 h at 1 mA /cm2 (set maximum:15 V). We used the following dilutions for antibody incubations: anti-H3K4ac: 1/500, anti-H3-C (AbCam ab1791 or home-made): 1/10 000, anti-H3K9ac (Millipore 07-539): 1/10 000, anti-H3K56ac (Millipore 07-677): 1/5000.
Histone sample preparation and mass spectrometry
Histones were acid extracted from set1Δ hst1Δ cells essentially as described previously [104], except that we added 100 mM sodium butyrate and 100 mM nicotinamide in the nuclear isolation buffer and wash buffers. H3 was purified and digested as previously described [105]. Intact core histones (approximately 10 µg of total protein acid extract) were separated using an Agilent 1200 HPLC system equipped with a fraction collector. Separations were performed using an ACE C8 column (5 µm, 300 Å), 150×4.6 mm i.d., with a solvent system consisting of 0.1% trifluoroacetic acid (TFA) in water (v/v) (A) and 0.1% TFA in acetonitrile (v/v) (B). Gradient elution was performed from 5–70% B in 60 minutes at 0.7 ml/min. Fractions were collected in conical tubes in 60 second time slices. Individual histone peaks that eluted over multiple fractions were pooled together. Histone fractions were evaporated in a Speed-Vac. Dried fractions were resuspended in 0.1 M ammonium bicarbonate (without pH adjustment) and digested overnight at 37°C using 1 µg of trypsin. Samples were acidified with 5% TFA in water (v/v) prior to LC-MS/MS analysis. Tryptic digests of histone extracts were injected onto a Thermo Electron LTQ-Orbitrap XL mass spectrometer equipped with an Eksigent nanoLC separation module. Samples were loaded onto a C18 trapping column at 10 µL/min using 0.2% formic acid (v/v) in water. Peptides were eluted onto a C18 analytical column (10 cm×150 µm i.d.) at 0.6 µl/min. Gradient elution was performed using a solvent system of 0.2% formic acid in water (A), and 0.2% formic acid in acetonitrile (B). Peptides were separated by gradient elution from 0 to 70% B in 60 minutes. For mass calibration, we used an internal lock mass [protonated (Si(CH3)2O))6; m/z 445.12057]. In each data collection cycle, one full MS scan (m/z 300–2000) was acquired in the Orbitrap (60000 resolution setting, automatic gain control (AGC) target of 106), followed by 3 data-dependent MS/MS scans in the LTQ (AGC target 10000; threshold 5000) for the 3 most abundant ions and collision induced dissociation (CID) for fragmentation.
Chromatin immunoprecipitation (ChIP)
Extracts containing fragments for chromatin imunoprecipitation (ChIP) were prepared as follows. 200 ml of exponentially growing cells (O.D.600 of 0.6 to 0.8) in YPD were treated with 1% formaldehyde at room temperature for 10 minutes. Formaldehyde was quenched with 0.125 M of glycine and cells were washed three times in cold water. Pellets (approximately 1.2×109 cells) were resuspended in 1.2 ml of lysis buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% Na-deoxycholate, 1 mM EDTA, 1 mM PMSF, 30 mM sodium butyrate, complete EDTA-free protease inhibitor cocktail, Roche), and split into three tubes. After adding one volume of glass beads to each sample, cells were disrupted using a bead beater (Disruptor Genie, Scientific Industries) at maximal speed for 2h at 4°C. After removing the glass beads, the lysates were transferred to fresh tubes and sonicated for 15 minutes (30 seconds ON, 30 seconds OFF) at high intensity in a Bioruptor (Diagenode) connected to a water cooler at 4°C. Lysates were clarified by centrifugation and pooled into a 2 ml tube. After addition of 800 µl of fresh lysis buffer and mixing, lysates were clarified again by centrifugation. 4 µl of lysate (1% of the whole cell extract, WCE) was kept aside as an input control. For immunoprecipitation, aliquots of 400 µl were prepared for each ChIP and mixed with the appropriate antibody: either 5 µl of anti-H3K4ac, 5 µl of anti-H3-C (Abcam ab1791), 3 µl of anti-H3K4me3 (Abcam ab8580) for at least 1h at 4°C. Before use, Dynabeads M-280 coupled to sheep anti-rabbit IgG (InVitrogen) (30 µl per ChIP) were washed in lysis buffer containing 5 mg/ml bovine serum albumin. The beads were added to each lysate and incubated at 4°C for at least 1 h. Beads were washed twice with lysis buffer, twice with lysis buffer containing 500 mM NaCl, twice in wash buffer (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 0.5% Nonidet-P40, 0.5% Na-deoxycholate, 1 mM EDTA) and once in TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA). To elute the DNA, 100 µl of a 10% Chelex 100 (Biorad) suspension in water was added to the beads (as well as to the 1% WCE) and incubated at 100°C for 10 minutes. Tubes were then cooled and treated with 0.2 µg/µl ribonuclease A at 50°C for 30 min, followed by 0.2 µg/µl proteinase K at 50°C for 30 min. Samples were incubated again at 100°C for 10 min to inactivate the proteinase K and then cooled down and spun down to get rid of debris. 80 µl of each supernatant was kept at −20°C for qPCR or ChIP-chip analysis.
ChIP-chip
All ChIP-on-chip datasets have been deposited in NCBI's Gene Expression Omnibus [106] and are accessible through GEO Series accession number GSE27307 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27307). Samples were processed according to the Affymetrix protocol for amplification, fragmentation and labelling of DNA for hybridisation to a GeneChip S. cerevisiae Tiling 1.0R Array (www.affymetrix.com). All ChIP-chips were performed in triplicate from independent colonies. Pre-processing of the data was carried out with the Affymetrix Tiling Analysis Software with default settings at 200bp bandwidth and expressed as a log2 ratio of the ChIP signal obtained for modified H3 (K4ac or K4me3) over that obtained from non-modified C-terminal H3 ChIP. The datasets used in Figure 4C and Figure 5F, 5G were normalised using the standard score (Z-score) equation: Z = (x−µ)/σ, where x is the [log2 (H3K4(mod) / H3-C)] raw value for each probe; µ is the mean raw value of all probes and σ is the standard deviation of the raw values for all probes. The resulting datasets have a normal distribution with a mean value of 0 and a standard deviation value of 1, and thus can be compared directly on the same scales. Data was displayed on the UCSC genome browser with the October 2003 S.cerevisiae genome assembly (http://genome.ucsc.edu/). Systematic alignment of ChIP-chip data to gene promoters and terminators were performed using the database management tools provided in the web application Galaxy (http://main.g2.bx.psu.edu/) and graphics were created with R (http://www.r-project.org/).
Promoter enrichment analyses and Venn diagrams
Gene promoters were defined as a genomic region −500 bp to +100 bp relative to gene start. Promoters containing Z-score values above the threshold value (90th percentile: Z>1.28 ; 80th percentile: Z>0.84 ; 70th percentile Z>0.52) were compared using the web application BioVenn (http://www.cmbi.ru.nl/cdd/biovenn/).
RNA extraction and expression analysis using microarrays
Total RNA was extracted from three independent colonies using the hot phenol method as described previously [107]. A total of 7 µg of RNA were processed for microarray analysis using the One-Cycle Target Labeling package from Affymetrix and hybridised to a GeneChip Yeast Genome 2.0 Array following the manufacturer's instructions (http://www.affymetrix.com/). Probe intensity data was pre-processed using the mas5 program of the R-Bioconductor software (http://www.bioconductor.org/index.html) [108]. The data from either H3-K4R or set1Δ mutants were normalised to their WT counterpart and the genes were sorted according to log2 fold change. The log2 fold change values for all genes are presented in Table S2 and represented graphically in Figure S6. For RT-qPCR analyses, 1 µg of total RNA was treated with DNase I, which was then inactivated following the manufacturer's instructions (DNA-free kit, Ambion). RT was performed using the First-Strand cDNA Synthesis Kit with M-MLV RT (Invitrogen) and oligo-dT primers, following the manufacturer's instructions.
Quantitative PCR
1 µl of the ChIP sample was mixed with 50 nM of region-specific primers and SYBR Green JumpStart Taq ReadyMix (Sigma) in a total volme of 20 µl and analysed on an Opticon real-time PCR machine (MJ Research/Bio-Rad). Relative binding values were extrapolated by subtracting the Ct of non-modified H3-C to that of modified H3 (K4ac or K4me3), and converting the ΔCt with the following formula: x = 2(−ΔCt), where x represents the relative binding value of modified H3 on non-modified H3-C.
Supporting Information
Zdroje
1. KouzaridesT
2007 Chromatin modifications and their function. Cell 128 693 705
2. SunZW
AllisCD
2002 Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418 104 108
3. DoverJ
SchneiderJ
Tawiah-BoatengMA
WoodA
DeanK
2002 Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277 28368 28371
4. FischleW
TsengBS
DormannHL
UeberheideBM
GarciaBA
2005 Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438 1116 1122
5. JenuweinT
AllisCD
2001 Translating the histone code. Science 293 1074 1080
6. TurnerBM
2002 Cellular memory and the histone code. Cell 111 285 291
7. TrelleMB
JensenON
2007 Functional proteomics in histone research and epigenetics. Expert Rev Proteomics 4 491 503
8. BrumbaughJ
PhanstielD
CoonJJ
2008 Unraveling the histone's potential: a proteomics perspective. Epigenetics 3 254 257
9. ZhangL
EugeniEE
ParthunMR
FreitasMA
2003 Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 112 77 86
10. ShilatifardA
2008 Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20 341 348
11. RuthenburgAJ
AllisCD
WysockaJ
2007 Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25 15 30
12. BernsteinBE
HumphreyEL
ErlichRL
SchneiderR
BoumanP
2002 Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A 99 8695 8700
13. StrahlBD
OhbaR
CookRG
AllisCD
1999 Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci U S A 96 14967 14972
14. SchneiderR
BannisterAJ
MyersFA
ThorneAW
Crane-RobinsonC
2003 Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol
15. SchubelerD
MacAlpineDM
ScalzoD
WirbelauerC
KooperbergC
2004 The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18 1263 1271
16. Santos-RosaH
SchneiderR
BannisterAJ
SherriffJ
BernsteinBE
2002 Active genes are tri-methylated at K4 of histone H3. Nature 419 407 411
17. GuentherMG
LevineSS
BoyerLA
JaenischR
YoungRA
2007 A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130 77 88
18. NgHH
RobertF
YoungRA
StruhlK
2003 Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11 709 719
19. BarskiA
CuddapahS
CuiK
RohTY
SchonesDE
2007 High-resolution profiling of histone methylations in the human genome. Cell 129 823 837
20. PokholokDK
HarbisonCT
LevineS
ColeM
HannettNM
2005 Genome-wide Map of Nucleosome Acetylation and Methylation in Yeast. Cell 122 517 527
21. LiuCL
KaplanT
KimM
BuratowskiS
SchreiberSL
2005 Single-Nucleosome Mapping of Histone Modifications in S. cerevisiae. PLoS Biol 3 e328 doi:10.1371/journal.pbio.0030328
22. ZhangX
BernatavichuteYV
CokusS
PellegriniM
JacobsenSE
2009 Genome-wide analysis of mono-, di - and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10 R62
23. FlanaganJF
MiLZ
ChruszczM
CymborowskiM
ClinesKL
2005 Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438 1181 1185
24. WysockaJ
SwigutT
XiaoH
MilneTA
KwonSY
2006 A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442 86 90
25. LiH
IlinS
WangW
DuncanEM
WysockaJ
2006 Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442 91 95
26. ShiX
HongT
WalterKL
EwaltM
MichishitaE
2006 ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442 96 99
27. MartinDG
BaetzK
ShiX
WalterKL
MacDonaldVE
2006 The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol 26 7871 7879
28. ShiX
KachirskaiaI
WalterKL
KuoJH
LakeA
2007 Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem 282 2450 2455
29. LanF
CollinsRE
De CegliR
AlpatovR
HortonJR
2007 Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448 718 722
30. MatthewsAG
KuoAJ
Ramon-MaiquesS
HanS
ChampagneKS
2007 RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450 1106 1110
31. VermeulenM
MulderKW
DenissovS
PijnappelWW
van SchaikFM
2007 Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131 58 69
32. SimsRJ3rd
MillhouseS
ChenCF
LewisBA
Erdjument-BromageH
2007 Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28 665 676
33. TavernaSD
IlinS
RogersRS
TannyJC
LavenderH
2006 Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell 24 785 796
34. OoiSK
QiuC
BernsteinE
LiK
JiaD
2007 DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448 714 717
35. HuangY
FangJ
BedfordMT
ZhangY
XuRM
2006 Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312 748 751
36. PenaPV
DavrazouF
ShiX
WalterKL
VerkhushaVV
2006 Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442 100 103
37. Santos-RosaH
SchneiderR
BernsteinBE
KarabetsouN
MorillonA
2003 Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell 12 1325 1332
38. RoguevA
SchaftD
ShevchenkoA
PijnappelWW
WilmM
2001 The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. Embo J 20 7137 7148
39. KroganNJ
DoverJ
KhorramiS
GreenblattJF
SchneiderJ
2002 COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277 10753 10755
40. MillerT
KroganNJ
DoverJ
Erdjument-BromageH
TempstP
2001 COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A 98 12902 12907
41. BriggsSD
BrykM
StrahlBD
CheungWL
DavieJK
2001 Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15 3286 3295
42. NagyPL
GriesenbeckJ
KornbergRD
ClearyML
2002 A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A 99 90 94
43. Vitaliano-PrunierA
MenantA
HobeikaM
GeliV
GwizdekC
2008 Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol 10 1365 1371
44. DehePM
GeliV
2006 The multiple faces of Set1. Biochem Cell Biol 84 536 548
45. JiangL
SmithJN
AndersonSL
MaP
MizzenCA
2007 Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. LYS4 trimethylation correlates with degree of acetylation on the same H3 tail. J Biol Chem 282 27923 27934
46. ZhangK
SiinoJS
JonesPR
YauPM
BradburyEM
2004 A mass spectrometric “Western blot” to evaluate the correlations between histone methylation and histone acetylation. Proteomics 4 3765 3775
47. NightingaleKP
GendreizigS
WhiteDA
BradburyC
HollfelderF
2007 Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J Biol Chem 282 4408 4416
48. YoungNL
DimaggioPA
Plazas-MayorcaMD
BalibanRC
FloudasCA
2009 High-throughput characterization of combinatorial histone codes. Mol Cell Proteomics
49. HazzalinCA
MahadevanLC
2005 Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol 3 e393 doi:10.1371/journal.pbio.0030393
50. KimT
BuratowskiS
2009 Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137 259 272
51. WangZ
ZangC
RosenfeldJA
SchonesDE
BarskiA
2008 Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40 897 903
52. GarciaBA
HakeSB
DiazRL
KauerM
MorrisSA
2007 Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem 282 7641 7655
53. SukaN
SukaY
CarmenAA
WuJ
GrunsteinM
2001 Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8 473 479
54. FillinghamJ
RechtJ
SilvaAC
SuterB
EmiliA
2008 Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol 28 4342 4353
55. RuscheLN
KirchmaierAL
RineJ
2003 The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72 481 516
56. RobertF
PokholokDK
HannettNM
RinaldiNJ
ChandyM
2004 Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell 16 199 209
57. HickmanMA
RuscheLN
2007 Substitution as a mechanism for genetic robustness: the duplicated deacetylases Hst1p and Sir2p in Saccharomyces cerevisiae. PLoS Genet 3 e126 doi:10.1371/journal.pgen.0030126
58. KarolchikD
BaertschR
DiekhansM
FureyTS
HinrichsA
2003 The UCSC Genome Browser Database. Nucleic Acids Res 31 51 54
59. HolstegeFC
JenningsEG
WyrickJJ
LeeTI
HengartnerCJ
1998 Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95 717 728
60. KirmizisA
Santos-RosaH
PenkettCJ
SingerMA
VermeulenM
2007 Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449 928 932
61. DehePM
DichtlB
SchaftD
RoguevA
PamblancoM
2006 Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem 281 35404 35412
62. SchneiderJ
WoodA
LeeJS
SchusterR
DuekerJ
2005 Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell 19 849 856
63. MorillonA
KarabetsouN
NairA
MellorJ
2005 Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell 18 723 734
64. DietvorstJ
BrandtA
2008 Flocculation in Saccharomyces cerevisiae is repressed by the COMPASS methylation complex during high-gravity fermentation. Yeast 25 891 901
65. CarvinCD
KladdeMP
2004 Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1-10. J Biol Chem 279 33057 33062
66. PinskayaM
GourvennecS
MorillonA
2009 H3 lysine 4 di - and tri-methylation deposited by cryptic transcription attenuates promoter activation. Embo J 28 1697 1707
67. XhemalceB
KouzaridesT
2010 A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev 24 647 652
68. BakerSP
GrantPA
2007 The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 26 5329 5340
69. NagyZ
ToraL
2007 Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26 5341 5357
70. VogelauerM
WuJ
SukaN
GrunsteinM
2000 Global histone acetylation and deacetylation in yeast. Nature 408 495 498
71. ZhangW
BoneJR
EdmondsonDG
TurnerBM
RothSY
1998 Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. Embo J 17 3155 3167
72. CollinsSR
MillerKM
MaasNL
RoguevA
FillinghamJ
2007 Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 806 810
73. HanJ
ZhouH
HorazdovskyB
ZhangK
XuRM
2007 Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315 653 655
74. DriscollR
HudsonA
JacksonSP
2007 Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315 649 652
75. SchneiderJ
BajwaP
JohnsonFC
BhaumikSR
ShilatifardA
2006 Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem 281 37270 37274
76. LiQ
ZhouH
WurteleH
DaviesB
HorazdovskyB
2008 Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134 244 255
77. ChenCC
CarsonJJ
FeserJ
TamburiniB
ZabaronickS
2008 Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134 231 243
78. MasumotoH
HawkeD
KobayashiR
VerreaultA
2005 A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436 294 298
79. TangY
HolbertMA
DelgoshaieN
WurteleH
GuillemetteB
2011 Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation. Structure 19 221 231
80. BurgessRJ
ZhouH
HanJ
ZhangZ
2010 A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell 37 469 480
81. DidionT
RegenbergB
JorgensenMU
Kielland-BrandtMC
AndersenHA
1998 The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol Microbiol 27 643 650
82. NielsenPS
van den HazelB
DidionT
de BoerM
JorgensenM
2001 Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol Gen Genet 264 613 622
83. SchmidtA
HallMN
KollerA
1994 Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol Cell Biol 14 6597 6606
84. ZhuX
GarrettJ
SchreveJ
MichaeliT
1996 GNP1, the high-affinity glutamine permease of S. cerevisiae. Curr Genet 30 107 114
85. RuscheLN
RineJ
2001 Conversion of a gene-specific repressor to a regional silencer. Genes Dev 15 955 967
86. McCordR
PierceM
XieJ
WonkatalS
MickelC
2003 Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol Cell Biol 23 2009 2016
87. PijnappelWW
SchaftD
RoguevA
ShevchenkoA
TekotteH
2001 The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev 15 2991 3004
88. XieJ
PierceM
Gailus-DurnerV
WagnerM
WinterE
1999 Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. Embo J 18 6448 6454
89. BrachmannCB
ShermanJM
DevineSE
CameronEE
PillusL
1995 The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev 9 2888 2902
90. DerbyshireMK
WeinstockKG
StrathernJN
1996 HST1, a new member of the SIR2 family of genes. Yeast 12 631 640
91. VenkatasubrahmanyamS
HwangWW
MeneghiniMD
TongAH
MadhaniHD
2007 Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci U S A 104 16609 16614
92. CramptonA
ChangF
PappasDLJr
FrischRL
WeinreichM
2008 An ARS element inhibits DNA replication through a SIR2-dependent mechanism. Mol Cell 30 156 166
93. MeadJ
McCordR
YoungsterL
SharmaM
GartenbergMR
2007 Swapping the gene-specific and regional silencing specificities of the Hst1 and Sir2 histone deacetylases. Mol Cell Biol 27 2466 2475
94. FryeRA
2000 Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273 793 798
95. AgaliotiT
ChenG
ThanosD
2002 Deciphering the transcriptional histone acetylation code for a human gene. Cell 111 381 392
96. LachnerM
O'CarrollD
ReaS
MechtlerK
JenuweinT
2001 Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410 116 120
97. BannisterAJ
ZegermanP
PartridgeJF
MiskaEA
ThomasJO
2001 Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410 120 124
98. ShankaranarayanaGD
MotamediMR
MoazedD
GrewalSI
2003 Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol 13 1240 1246
99. NakayamaJ
RiceJC
StrahlBD
AllisCD
GrewalSI
2001 Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292 110 113
100. ReaS
EisenhaberF
O'CarrollD
StrahlBD
SunZW
2000 Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406 593 599
101. MorrisSA
RaoB
GarciaBA
HakeSB
DiazRL
2007 Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J Biol Chem 282 7632 7640
102. PinskayaM
MorillonA
2009 Histone H3 lysine 4 di-methylation: A novel mark for transcriptional fidelity? Epigenetics 4
103. KushnirovVV
2000 Rapid and reliable protein extraction from yeast. Yeast 16 857 860
104. EdmondsonDG
SmithMM
RothSY
1996 Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev 10 1247 1259
105. DrogarisP
WurteleH
MasumotoH
VerreaultA
ThibaultP
2008 Comprehensive profiling of histone modifications using a label-free approach and its applications in determining structure-function relationships. Anal Chem 80 6698 6707
106. EdgarR
DomrachevM
LashAE
2002 Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 207 210
107. SchmittME
BrownTA
TrumpowerBL
1990 A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18 3091 3092
108. GentlemanRC
CareyVJ
BatesDM
BolstadB
DettlingM
2004 Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



