-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaChiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
The chiasma is a structure that forms between a pair of homologous chromosomes by crossover recombination and physically links the homologous chromosomes during meiosis. Chiasmata are essential for the attachment of the homologous chromosomes to opposite spindle poles (bipolar attachment) and their subsequent segregation to the opposite poles during meiosis I. However, the overall function of chiasmata during meiosis is not fully understood. Here, we show that chiasmata also play a crucial role in the attachment of sister chromatids to the same spindle pole and in their co-segregation during meiosis I in fission yeast. Analysis of cells lacking chiasmata and the cohesin protector Sgo1 showed that loss of chiasmata causes frequent bipolar attachment of sister chromatids during anaphase. Furthermore, high time-resolution analysis of centromere dynamics in various types of chiasmate and achiasmate cells, including those lacking the DNA replication checkpoint factor Mrc1 or the meiotic centromere protein Moa1, showed the following three outcomes: (i) during the pre-anaphase stage, the bipolar attachment of sister chromatids occurs irrespective of chiasma formation; (ii) the chiasma contributes to the elimination of the pre-anaphase bipolar attachment; and (iii) when the bipolar attachment remains during anaphase, the chiasmata generate a bias toward the proper pole during poleward chromosome pulling that results in appropriate chromosome segregation. Based on these results, we propose that chiasmata play a pivotal role in the selection of proper attachments and provide a backup mechanism that promotes correct chromosome segregation when improper attachments remain during anaphase I.
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1001329
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001329Summary
The chiasma is a structure that forms between a pair of homologous chromosomes by crossover recombination and physically links the homologous chromosomes during meiosis. Chiasmata are essential for the attachment of the homologous chromosomes to opposite spindle poles (bipolar attachment) and their subsequent segregation to the opposite poles during meiosis I. However, the overall function of chiasmata during meiosis is not fully understood. Here, we show that chiasmata also play a crucial role in the attachment of sister chromatids to the same spindle pole and in their co-segregation during meiosis I in fission yeast. Analysis of cells lacking chiasmata and the cohesin protector Sgo1 showed that loss of chiasmata causes frequent bipolar attachment of sister chromatids during anaphase. Furthermore, high time-resolution analysis of centromere dynamics in various types of chiasmate and achiasmate cells, including those lacking the DNA replication checkpoint factor Mrc1 or the meiotic centromere protein Moa1, showed the following three outcomes: (i) during the pre-anaphase stage, the bipolar attachment of sister chromatids occurs irrespective of chiasma formation; (ii) the chiasma contributes to the elimination of the pre-anaphase bipolar attachment; and (iii) when the bipolar attachment remains during anaphase, the chiasmata generate a bias toward the proper pole during poleward chromosome pulling that results in appropriate chromosome segregation. Based on these results, we propose that chiasmata play a pivotal role in the selection of proper attachments and provide a backup mechanism that promotes correct chromosome segregation when improper attachments remain during anaphase I.
Introduction
During cell division, chromosomes that harbor genetic information are accurately segregated into daughter cells. Chromosome segregation depends on attachment of chromosomes to the spindle via chromosomal sites called kinetochores. The interaction between kinetochores and spindle microtubules, which extend from opposite spindle poles, generates pulling forces on the chromosomes from opposite directions, causing them to migrate toward opposite spindle poles. To understand the mechanisms underlying chromosome segregation, it is crucial to elucidate how chromosomes attach to the spindle.
In mitosis, sister chromatids are segregated to opposite poles (equational segregation; Figure 1). The sister chromatids are associated until anaphase via a protein complex called cohesin [1], [2], which is required for the back-to-back arrangement of the kinetochores that permits their attachment to opposite spindle poles [3]. In addition, when sister chromatids are pulled from opposite directions, the cohesion generates tension at the kinetochore that leads to stabilization of the kinetochore–microtubule interaction, probably via inactivation of aurora kinase [4]. When the cohesion is compromised, sister chromatids fail to attach to the spindle properly and are mis-segregated [5]–[8].
Fig. 1. Spindle attachment of chromosomes and their segregation during mitosis and meiosis I. 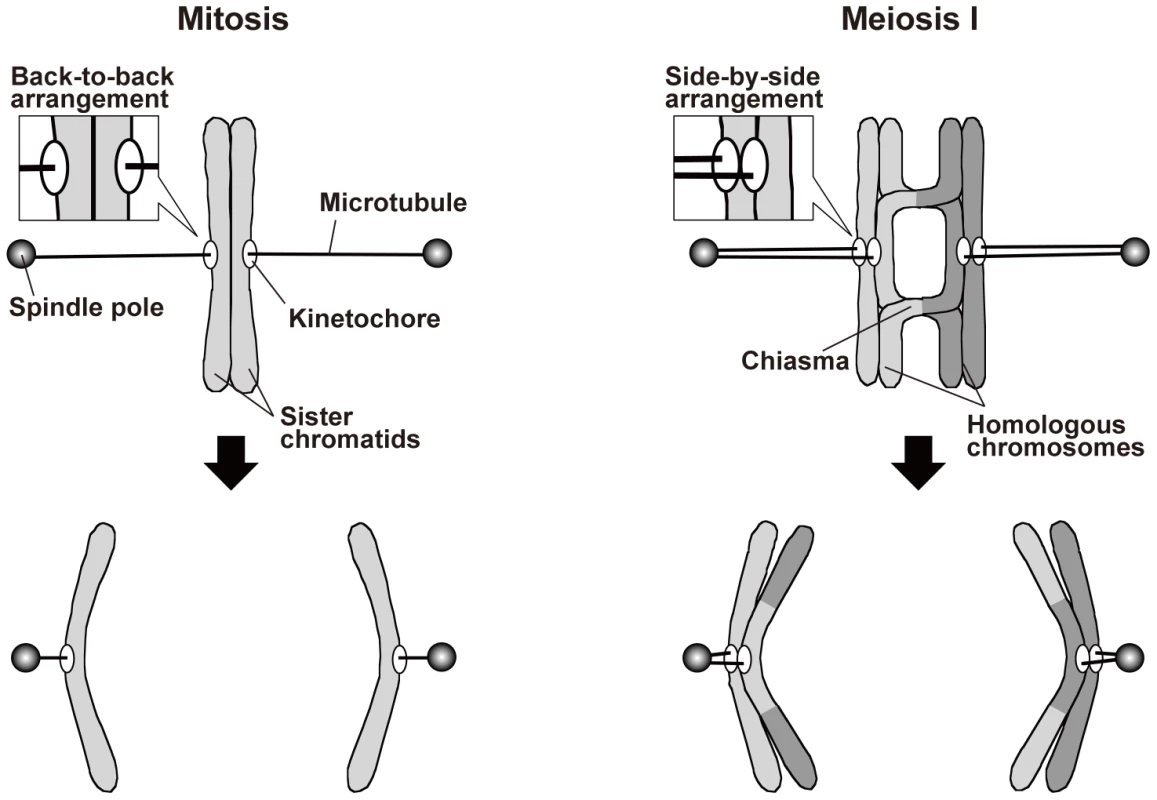
For simplicity, only a single kinetochore-interacting microtubule is shown for each kinetochore, and other microtubules are not shown. During meiosis, on the other hand, a physical association between homologous chromosomes additionally contributes to proper spindle attachment of chromosomes [3], [9], [10]. Meiosis occurs during gamete formation, and during meiosis, two rounds of chromosome segregation follow a single round of DNA replication, resulting in the production of gametes with half the original number of chromosomes. Chromosome segregation during meiosis I is specific to meiosis: Homologous chromosomes attach to opposite spindle poles, with each pair of sister chromatids attaching to the same pole (monopolar attachment), and are segregated to the opposite poles (reductional segregation; Figure 1). As in mitosis, sister chromatid cohesion is required for proper kinetochore arrangement during meiosis. However, a meiosis-specific type of cohesin mediates this cohesion [11]–[15], and sister kinetochores are arranged side by side facing the same direction so that they become attached to the same pole [16]. Furthermore, shugoshin proteins maintain centromeric cohesion during anaphase I [17]–[21]. These proteins inhibit the removal of centromeric cohesin and regulate centromeric aurora kinase [17]–[19], [21]–[24]. Elimination of both of these functions compromises sister chromatid segregation during meiosis I and II [17], [18], [22], [25]. Further, elimination of the cohesin-retention function alone causes sister chromatid separation after anaphase I but has little if any effect on sister chromatid segregation toward the same pole during anaphase I [17], [19]. Unlike the situation in mitosis, homologous chromosome association contributes to the generation of tension at the kinetochore in meiosis. Homologous chromosomes are physically associated with each other via the chiasmata that are formed by reciprocal recombination. When homologous chromosomes are pulled in opposite directions, the chiasmata generate tension at the kinetochore and stabilize the kinetochore–microtubule interaction. Elimination of chiasmata leads to non-disjunction of homologous chromosomes [26].
In addition to this widely accepted role, chiasmata appear to play additional roles in the attachment of chromosomes to the spindle. A lack of chiasmata results in the separation or fragmentation of sister chromatids during meiosis I in many species [27]–[29], suggesting that chiasmata prevent the bipolar attachment of sister chromatids. Furthermore, chiasmata greatly alter meiotic sister chromatid segregation patterns in several different types of fission yeast cells. Fission yeast cells normally undergo meiosis after responding to the mating pheromone [30], but meiosis can also be induced without mating pheromone response by inactivation of Pat1 kinase, a key negative regulator of meiosis [31], [32]. We previously reported that when haploid fission yeast cells lacking homologous chromosomes were forced to enter meiosis by Pat1 inactivation after a mating pheromone response, sister chromatids were primarily segregated to the same pole at meiosis I, as seen in normal diploid meiosis [33]. However, when they were induced to enter meiosis without a mating pheromone response, sister chromatids primarily underwent equational segregation. By contrast, when Pat1 inactivation forced diploid cells to enter meiosis without a mating pheromone response, the sister chromatids were primarily segregated to the same pole in a recombination-dependent manner. Similar recombination-dependent co-segregation of sister chromatids has been observed in several cohesin-related mutants of fission yeast [34]. These findings suggest that chiasmata promote the monopolar attachment of sister chromatids; however, because a loss of recombination causes only a negligible level of equational segregation during normal diploid meiosis in fission yeast cells, chiasmata have previously been thought to be dispensable for monopolar attachment of sister chromatids [35]. The contribution of chiasmata to the monopolar attachment during meiosis I, therefore, remains elusive.
To understand the mechanisms underlying meiotic chromosome segregation, we examined the functions of chiasmata in spindle attachment and segregation of sister chromatids during meiosis I in fission yeast. Our analysis of chromosome segregation and dynamics in several different types of achiasmate cells showed that in the absence of chiasmata, sister chromatids were frequently attached to opposite poles during anaphase I. High time-resolution analysis of centromere dynamics further showed that chiasmata contribute to the elimination of bipolar attachments during the pre-anaphase stage. Furthermore, when the bipolar attachments remain during anaphase I, chiasmata induce a bias toward the proper pole during poleward chromosome pulling from opposite directions that results in correct chromosome segregation. Based on our findings, we discuss how chiasmata contribute to spindle attachment and segregation of chromosomes and further extend our idea to include the general functions of chromosome association during mitotic and meiotic chromosome segregation.
Results
Sister centromeres frequently become dissociated and remain between the spindle poles during anaphase I in rec12 mutant cells
Elimination of chiasmata induced by depletion of Rec12, a recombination factor required for the formation of double-strand breaks [36], causes occasional equational segregation of sister chromatids [33] and frequent non-disjunction of homologous chromosomes [37]. As a first step toward understanding the role of chiasmata in the spindle attachment of sister chromatids, we re-examined chromosome segregation during meiosis I in more detail in rec12 mutant cells.
We examined chromosome segregation by visualizing centromere-linked loci of chromosome I (the lys1 locus: cen1) and chromosome II (the D107 locus: cen2) using green fluorescent protein (GFP) [33]. After the first division, homologous centromeres were partitioned into two nuclei and rarely into the same nucleus in wild-type cells (Figure 2A). In contrast, homologous centromeres were frequently partitioned into the same nucleus in rec12 mutant cells (Figure 2A). Furthermore, sister centromeres were partitioned into the same nucleus and rarely into two nuclei in wild-type cells but were occasionally partitioned into two nuclei in rec12 mutant cells (Figure 2B, +, rec+ and rec12Δ). The ∼4% of wild-type cells that showed a partition of cen1 into the distinct nuclei was most likely the result of recombination between the centromere and the lys1 locus used for this analysis. These results confirmed the mis-segregation of both homologous chromosomes and sister chromatids during meiosis I in rec12 mutant cells. The same mis-segregation phenotypes were also observed in cells lacking the Rec14 recombination factor, which functions together with Rec12 and the depletion of which eliminates recombination (Figure 2A and 2B) [38], [39].
Fig. 2. The effect of loss of chiasmata on chromosome segregation. 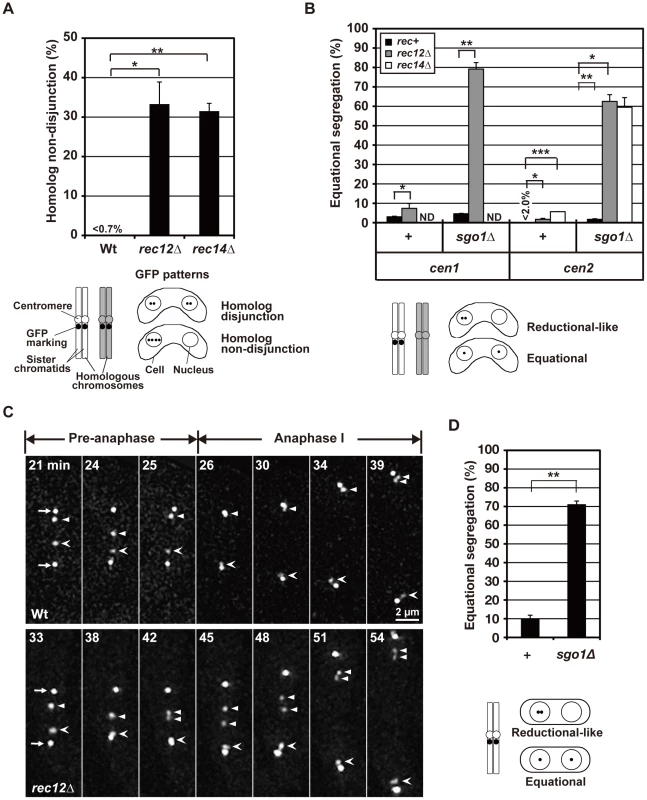
(A) The frequency of non-disjunction of homologous chromosomes during meiosis I was examined by GFP-visualized cen2. (B) Equational segregation of sister chromatids during meiosis I in various types of diploid cells. +: no sgo1 mutation; cen1: the lys1 locus; cen2: the D107 locus [33]; rec+: no rec12 or rec14 mutation. ND: not determined. (C) Centromere dynamics in wild-type and rec12 mutant cells during meiosis I. Arrows indicate the spindle pole bodies (SPBs). Arrowheads and barbed arrowheads show homologous centromeres (cen2). Pre-anaphase: the pre-anaphase stage, as determined by a constant pole-to-pole distance. Anaphase I: anaphase I, as determined by an increase in the pole-to-pole distance. Numbers indicate the time in minutes from the beginning of spindle formation. (D) Equational segregation of sister chromatids at meiosis I induced in haploid cells. Sister chromatid segregation was analyzed using GFP-visualized cen2. +: sgo1+ cells; sgo1Δ: sgo1 mutant cells. The lower illustrations in (A), (B), and (D) show how GFP was used to mark chromosomes (left) and the segregation patterns of the GFP signals after meiosis I (right). In all analyses in this study, with the exception of analyses in the supplementary results, sister chromatid segregation was analyzed in cells containing two DNA masses that underwent meiosis I. Each data point was obtained from two independent experiments, with the exception of the non-disjunction frequency of homologous chromosome in rec12 mutant cells, which was obtained from three independent experiments. More than 50 cells were examined in each experiment. Error bars indicate standard deviation. Asterisks indicate statistically significant differences and their associated p values, as determined by t-tests. *p<0.05; ** p<0.005; *** p<5×10−5. Segregation analysis showed that the overall mis-segregation frequency of sister chromatids in recombination-deficient, chiasmata-lacking cells (i.e., achiasmate cells) was small. However, live cell analysis of cen2 dynamics suggested that improper spindle attachment of sister centromeres occurs more frequently during anaphase I. Although the sister centromeres eventually moved to the pole in rec12 mutant cells, they frequently remained between the two spindle poles and were dissociated during anaphase I [observed for 7 out of 14 centromeres examined (50.0%); Figure 2C, rec12Δ]. These centromeres are called lagging centromeres, and they were not observed in wild-type cells [observed for 0 of 12 centromeres examined (0%); Figure 2C, Wt]. The chromosome lagging is most likely caused by a loss of chiasmata and not a loss of Rec12 function, because lagging chromosomes were also frequently observed when meiosis was induced in haploid cells [33], which do not form chiasmata due to their lack of homologous chromosomes (Figure S1). These results suggest that sister centromeres are frequently attached to both poles and are pulled from opposite directions during anaphase I in achiasmate cells.
Sgo1 depletion causes equational segregation of sister chromatids during meiosis I in achiasmate cells
To confirm the frequent bipolar attachment of sister chromatids in achiasmate cells, we depleted Sgo1, which inhibits the removal of centromeric cohesin during anaphase I [17], [19]. We hypothesized that although sister chromatids are frequently attached to both poles and are pulled from opposite directions, the centromere cohesion that persists until meiosis II should provide resistance against this force and prevent their separation during anaphase I in achiasmate cells. If so, depletion of Sgo1, which eliminates centromere cohesion during anaphase I, should lead to frequent equational segregation of sister chromatids.
Indeed, Sgo1 depletion led to a substantial increase in equational segregation in achiasmate cells. Equational segregation of sister centromeres was occasionally observed in sgo1 mutant cells but was more frequently observed in sgo1 rec12 and sgo1 rec14 double-mutant cells (Figure 2B, sgo1Δ). When meiosis I was induced in haploid cells, Sgo1 depletion similarly increased equational segregation (Figure 2D) irrespective of Rec12 depletion (data not shown). Therefore, the increased equational segregation is not specific to recombination-deficient cells but is common in achiasmate cells. These results confirm that the loss of chiasmata frequently leads to the bipolar attachment of sister chromatids during anaphase I.
Bipolar attachment of sister chromatids only partially depends on the spindle assembly checkpoint (SAC) in achiasmate cells
The SAC ensures faithful chromosome segregation by delaying anaphase initiation until all of the chromosomes become properly attached to the spindle [40], [41]. We previously reported that the SAC becomes activated to delay anaphase initiation at meiosis I in rec12 mutant cells, which is likely associated with improper spindle attachment of chromosomes [42]. Similarly, analysis of spindle length showed that anaphase initiation was substantially delayed in sgo1 rec12 double-mutant cells in a Mad2-dependent manner, as previously observed in rec12 mutant cells (Figure S2A, Text S1). Therefore, we next examined whether the SAC contributes to the bipolar attachment of sister chromatids in achiasmate cells by depleting the SAC factor Mad2.
Mad2 depletion led to decreased equational segregation of sister chromatids in rec12 mutant cells, but equational segregation was still observed at substantial levels in rec12 sgo1 double-mutant cells (Figure 3A). Likewise, Mad2 depletion decreased but did not abolish equational segregation during meiosis I in haploid cells (Figure 3B). These results showed that the SAC promotes the bipolar attachment of sister chromatids but is not essential for this process in achiasmate cells. Furthermore, as seen in rec12 mutant cells, sister centromeres frequently dissociated and failed to move to the pole during anaphase I in mad2 rec12 double-mutant cells [40.9% (22 centromeres); Figure 3C, mad2Δ rec12Δ]. However, lagging centromeres were rarely observed in mad2 mutant cells [0.1% (20 centromeres); Figure 3C, mad2Δ], although the timing of anaphase initiation was not much different between these mutants (Figure S2A). These results indicated that the lagging centromeres seen in achiasmate cells were not caused by SAC activation or delayed anaphase initiation. Thus, we conclude that the bipolar attachment of sister chromatids depends only partially on the SAC in achiasmate cells.
Fig. 3. A role for the SAC factor, Mad2, in chromosome segregation during meiosis I. 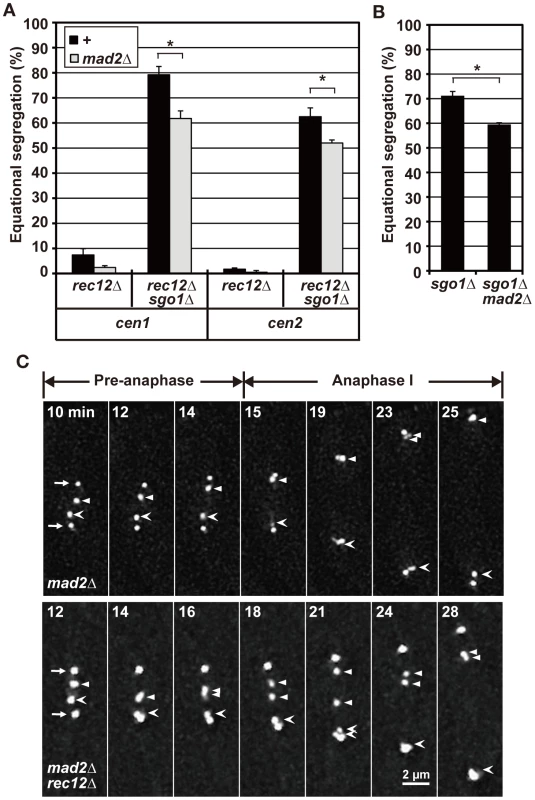
(A) Effects of Mad2 depletion on sister chromatid segregation during meiosis I in rec12 achiasmate cells. +: no mad2 mutation. (B) Effects of Mad2 depletion on sister chromatid segregation at meiosis I in haploid cells. Sister chromatid segregation was analyzed using GFP-visualized cen2. (C) Effects of Mad2 depletion on centromere dynamics during meiosis I. Arrows indicate the SPBs. Arrowheads and barbed arrowheads show each of the homologous centromeres (cen2). Numbers indicate the time in minutes from the beginning of spindle formation. Bar: 2 µm. In (A) and (B), each value was obtained from two independent experiments, with the exception of the equational frequencies for rec12 sgo1 mad2 cells, which were obtained from three independent experiments. Error bars indicate standard deviation. Asterisks show statistically significant differences (p<0.05). Bipolar attachment of sister chromatids occasionally occurs before anaphase irrespective of chiasma formation
Spindle attachment of chromosomes is established before anaphase, and the chiasma may prevent the bipolar attachment of sister chromatids from occurring during the pre-anaphase stage. To test this possibility, we examined the dynamics of sister centromeres before anaphase by time-lapse analysis with 10-s intervals. The time-lapse analysis of cen2 loci on both homologous chromosomes in wild-type and rec12 mutant cells confirmed our previous observations from time-lapse analyses with 1-min intervals, although they exhibited slight differences in dynamic parameters (Table S1) [42]. Homologous centromeres oscillated between the two spindle poles in a somewhat coordinated manner in wild-type cells; a pair of homologous centromeres often moved in the same direction (Figure 4A, Table S2). Accordingly, centromeres were mostly positioned around the middle point between the spindle pole and the spindle center with a tendency to be near the center (Figure 4B). These centromere dynamics presumably reflect the frequent bipolar attachment of homologous chromosomes that are linked by the chiasmata (Figure 4C). On the other hand, sister centromeres oscillated in an uncoordinated manner and tended to remain near the pole in rec12 mutant cells (Figure 4A), and centromere positioning was shifted toward the pole (Figure 4B). These centromere dynamics probably reflect the frequent attachment of each of the non-linked homologous chromosomes to one pole and the occasional switch in their attachment to the other pole (Figure 4C).
Fig. 4. Pre-anaphase centromere dynamics during meiosis I in wild-type and rec12 mutant cells. 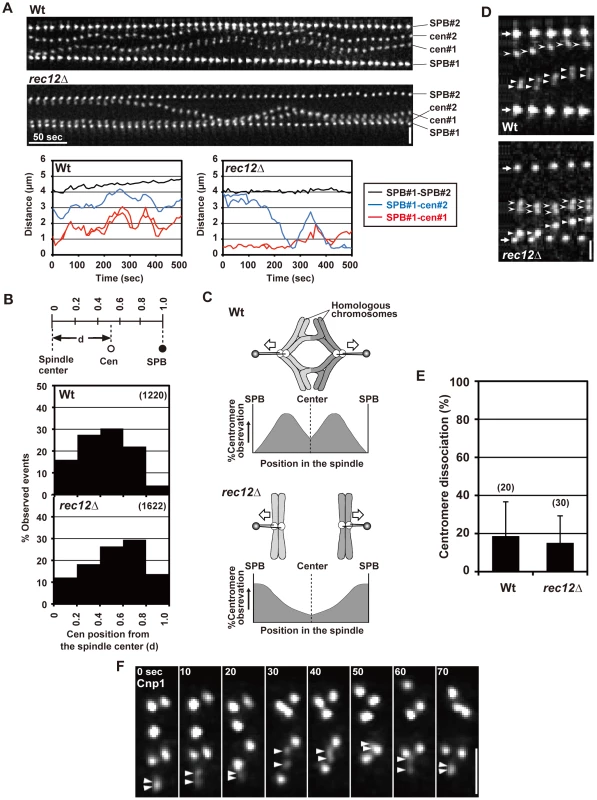
(A) Pre-anaphase dynamics of the spindle pole and centromere (cen2) during meiosis I. Photos were taken every 10 s and are shown in order from left to right. Horizontal bar: 50 s. Vertical bar: 2 µm. Graphs show changes in the distance between the SPB and each centromere (red, SPB#1-cen#1, and blue, SPB#1-cen#2) and between the two SPBs (black, SPB#1-SPB#2). (B) Observation frequencies of centromeres at distinct positions in the spindle during the pre-anaphase stage. The positions of centromeres are shown as relative distances from the spindle center (d), as determined in the upper illustration. Zero and 1.0 correspond to positions of the spindle center and the SPB, respectively. The number of examined positions is shown in parentheses. (C) Spindle attachment of chromosomes during meiosis I in wild-type (Wt) and rec12 mutants (rec12Δ) and expected observation frequencies of centromeres at distinct positions in the spindle. For rec12 mutant, only the attachment of homologous chromosomes to both poles is shown. (D) Dissociation of GFP-visualized sister centromeres (cen2). Arrows indicate the SPB. Arrowheads and barbed arrowheads show each of the homologous centromeres. Bar: 1 µm. (E) Average dissociation frequencies of sister centromeres in wild-type and rec12 mutant cells. The number of centromeres examined is shown in parentheses. (F) Dissociation of sister centromeres visualized by GFP-tagged Cnp1 during the pre-anaphase stage. The stage was determined based on centromere behavior and the distance between the spindle poles visualized using the DsRed-tagged SPB component Sad1 (not shown). Arrowheads indicate sister centromeres that underwent dissociation. Numbers at the top indicate the time in seconds. Bar: 1 µm. In analyses of centromere position and dissociation, 20 and 28 pairs of sister centromeres were examined for wild-type and rec12 mutant strains, respectively. More than 10 time points were examined for each centromere analysis. Error bars indicate standard deviation. Notably, we found that sister centromeres occasionally underwent a transient dissociation in both wild-type and rec12 mutant cells (Figure 4A and 4D, Table 1). In both types of cells, centromere dissociation was observed in ∼20% of events on average (Figure 4E). This dissociation was not the result of the integration into the chromosome of lacO repeats, which are used for visualization [33], or of the dissociation of only the visualized pericentromeric region; when all three homologous sets of sister centromeres were visualized by GFP tagging of the centromere-specific histone H3 variant Cnp1 [43], we observed more than six centromere signals together with a transient split of the signal into two (Figure 4F). These observations showed that bipolar attachment of sister chromatids occasionally occurs during the pre-anaphase stage, irrespective of chiasma formation. Similar centromere dynamics were also observed in cells lacking Sgo1. The occurrence of bipolar attachment in the presence of chiasmata is contradictory to the idea that chiasmata prevent the bipolar attachment of sister chromatids from occurring during the pre-anaphase stage.
Tab. 1. Centromere dissociation during the pre-anaphase stage of meiosis I in various fission yeast strains. 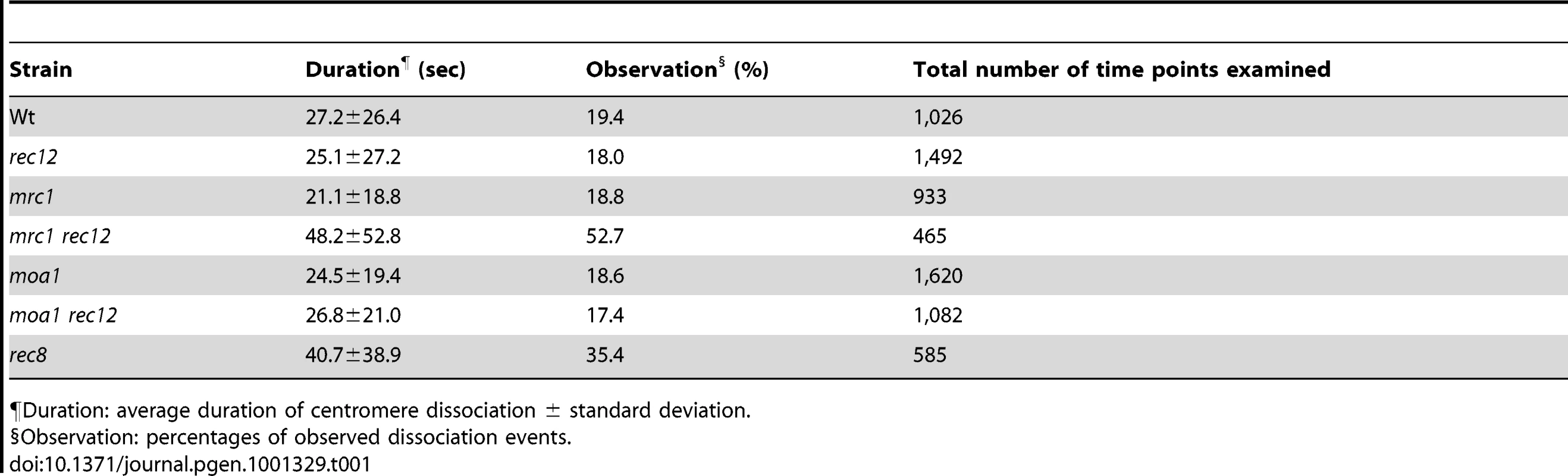
¶Duration: average duration of centromere dissociation ± standard deviation. Sister chromatids attach to both poles more frequently in mrc1 rec12 and moa1 rec12 double-mutant cells than in rec12 single-mutant cells
If chiasmata do not prevent the bipolar attachment of sister chromatids from occurring, they must contribute to the elimination of bipolar attachment of sister chromatids during the pre-anaphase stage. However, the overall frequency of centromere dissociation was not significantly different between wild-type and rec12 mutant cells (Figure 4E, Table 1), and chiasma-dependent elimination of the bipolar attachment was not evident. We hypothesized that if sister centromeres attach to both poles more frequently in the achiasmate background, the chiasma-dependent elimination of the bipolar attachment would be evident. Following this hypothesis, we examined mrc1 and moa1 mutant cells.
The mrc1 gene encodes a conserved DNA replication checkpoint factor, which delays cell cycle progression upon DNA replication stress, promotes proper fork progression, and contributes to sister chromatid cohesion in mitosis [44]–[50]. On the other hand, the moa1 gene encodes a meiosis-specific centromere protein that contributes to the proper centromere localization of the meiotic cohesin component Rec8 [34]. In both mrc1 and moa1 mutant cells, chromosome segregation as well as spindle dynamics, recombination, and spore formation are largely normal (Figures S2B and S3, Text S1) [34]. However, sister chromatids are primarily segregated equationally in a manner partly dependent on Mad2 when chiasmata are not formed (in the rec12Δ or the haploid background; Figure 5) [34]. Although these phenotypes are similar to the sgo1-mutant phenotypes, the equational segregation is primarily caused by defects in centromere features other than maintenance of centromere cohesion, because both mrc1 and moa1 mutant cells can maintain sister centromere cohesion until anaphase II if sister chromatids are not segregated equationally during meiosis I (Figure S3D, Text S1) [34]. Therefore, the equational segregation seen in the mrc1 rec12 and moa1 rec12 mutant cells is likely to be caused by frequent bipolar attachment of sister chromatids, and we expected that the chiasma effects would be more evident in the mrc1 and moa1 mutants.
Fig. 5. Sister chromatid segregation in mrc1 and moa1 mutants. 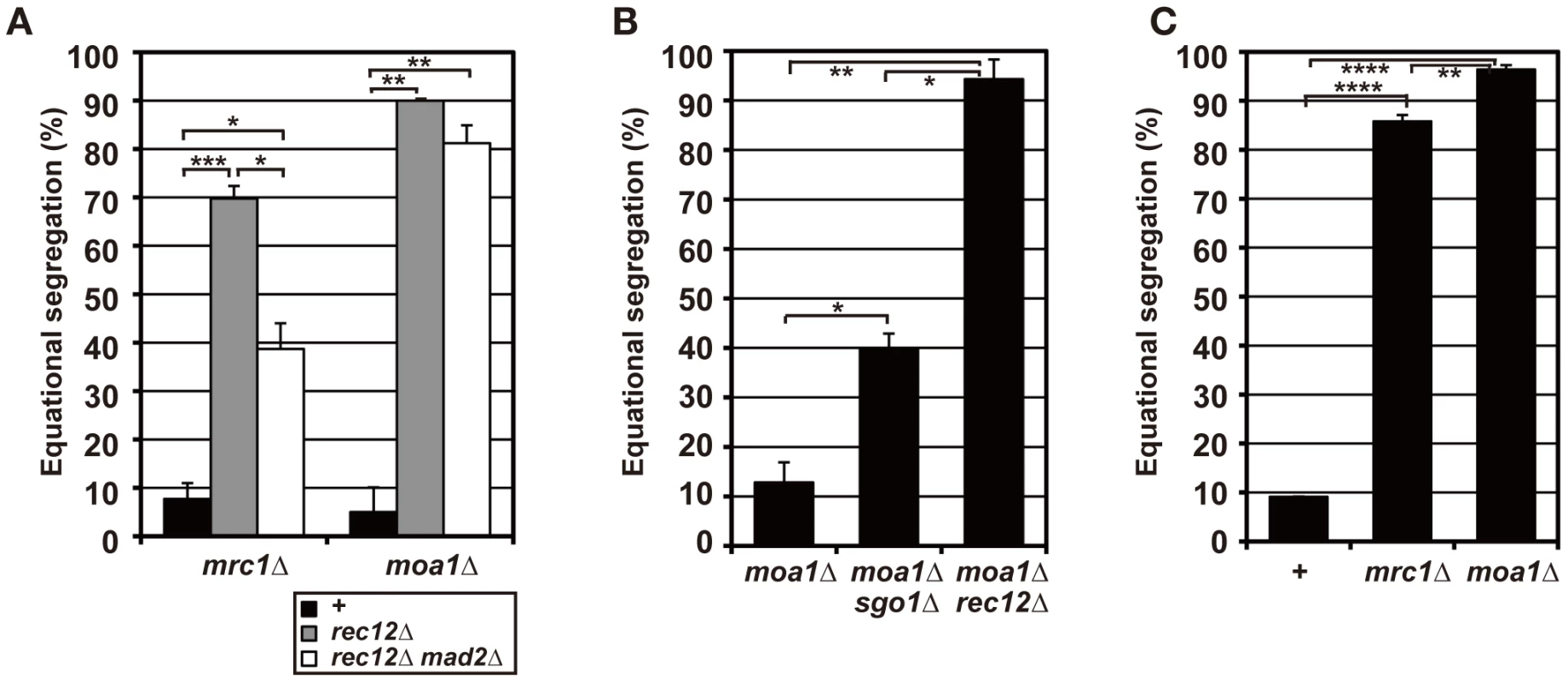
(A) Sister chromatid segregation in mrc1 and moa1 mutants and the effects of Rec12 or Mad2 depletion analyzed by the GFP-visualized cen2. +: no rec12 or mad2 mutation. (B) Sister chromatid segregation in moa1 mutant and the effects of Sgo1 or Rec12 depletion analyzed by the GFP-visualized cen1. (C) Effects of Mrc1 or Moa1 depletion on sister chromatid segregation at meiosis I in haploid cells. Sister chromatid segregation was analyzed by the GFP-visualized cen2. Data values in all graphs were obtained as described in Figure 2. Error bars indicate standard deviation. Asterisks show statistically significant differences and their associated p values. * p<0.05; ** p<0.005; *** p<0.0005; **** p<5×10−6. To evaluate chiasma effects in the mrc1 and moa1 mutants, we first examined the pre-anaphase centromere dynamics in the achiasmate mrc1 rec12 and moa1 rec12 double-mutant cells. In the mrc1 rec12 mutant cells, the sister centromeres dissociated more frequently (Figure 6A and 6B), with a significantly longer duration (Table 1), and were predominantly positioned around the spindle center, unlike those in the rec12 mutant cells (Figure 6C). In the moa1 rec12 mutant cells, the centromeres were also frequently positioned around the spindle center (Figure 6A and 6C), and in addition, the SAC was not activated as much as in rec12 mutant cells (Figure S2A, Text S1). These characteristics were expected to be associated with frequent bipolar attachment of sister chromatids (Figure 6D). Indeed, the frequent dissociation of the centromeres and their positioning around the spindle center together with the low level of SAC activation were observed during meiosis I in achiasmate rec8 mutant cells (Figure 6A–6C and Figure S2A, Table 1), in which sister chromatids efficiently attach to both poles to fully undergo equational segregation [12], [51]. They were also observed during mitotic division in wild-type diploid cells (Figure S4). These observations thus confirmed that sister centromeres attach to both poles more frequently in the mrc1 rec12 and moa1 rec12 double-mutant cells than in rec12 single-mutant cells. However, the centromere properties of the mrc1 and moa1 mutant cells differed from those of rec8 mutant or mitotic cells because the SAC substantially delayed anaphase initiation in mrc1 rec12 mutant cells (Figure S2A, Text S1), and centromere dissociation was not so frequent in moa1 rec12 mutant cells (Figure 6B).
Fig. 6. Pre-anaphase centromere dynamics during meiosis I in mrc1 and moa1 mutants. 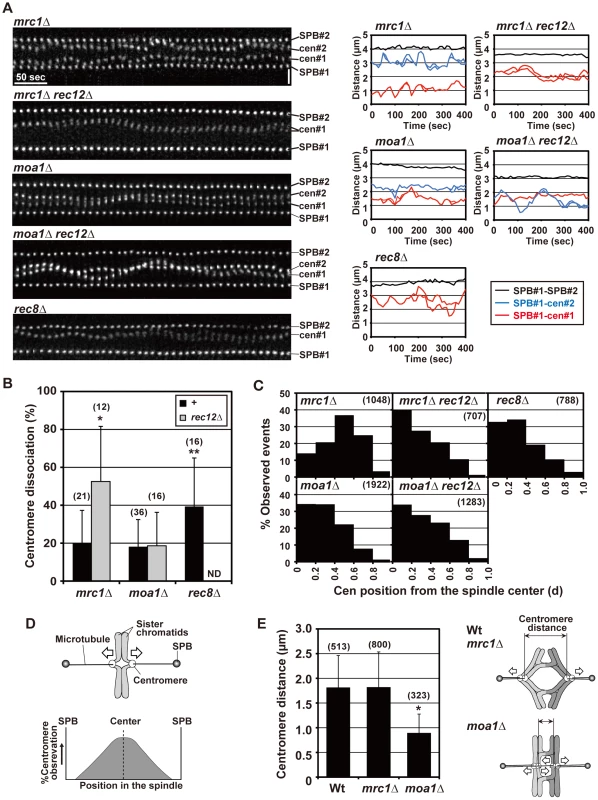
(A) Pre-anaphase dynamics of the spindle pole and centromere (cen2) at meiosis I, and changes in the distance between the spindle pole and the centromere and between the two spindle poles in mrc1, moa1, and rec8 mutants. Note that only one of the homologous centromeres is visualized in mrc1 rec12 and rec8 mutant cells. Horizontal bar: 50 s. Vertical bar: 2 µm. (B) Average centromere dissociation frequencies in mrc1, moa1, and rec8 mutant cells. The number of centromeres examined is shown in parentheses. +: no rec12 mutation. ND: not determined. Asterisks indicate dissociation frequencies that are statistically different from the frequency of wild type. * p<0.005; ** p<0.01. (C) Observation frequencies of centromeres at distinct positions in the spindle during the pre-anaphase stage. The positions of centromeres are shown based on their relative distance from the spindle center (d), as determined in Figure 4B. The number of examined positions is shown in parentheses. (D) Bipolar attachment of sister chromatids and expected observation frequencies of centromeres at distinct positions in the spindle. (E) Distance between homologous centromeres. The distance between homologous centromeres was measured at every time point in each strain, and an average distance is shown. When centromeres were dissociated, the distance between the nearest homologous pair of centromeres was measured. The asterisk indicates a distance statistically different from that of wild type (p<5×10−125). The number of distances examined is shown in parentheses. Right illustrations show models for spindle attachment of chromosomes and the resultant distance between the centromeres in wild-type, mrc1, and moa1 mutant cells. White arrows in all illustrations indicate forces exerted on chromosomes. Error bars in all graphs indicate standard deviations. Chiasmata prevent the bipolar attachment of sister chromatids in mrc1 mutant cells but not in moa1 mutant cells
We next examined the pre-anaphase centromere dynamics in the chiasmate mrc1 and moa1 single-mutant cells to evaluate chiasma effects. Remarkably, in mrc1 single-mutant cells, the level of centromere dissociation was almost identical to that in wild-type cells (Figure 6A and 6B, Table 1), indicating that bipolar attachment of sister chromatids was reduced to a wild-type level. Furthermore, centromere positioning and the distance between homologous centromeres were very similar to what was seen in wild-type cells (Figure 6C and 6E), indicating that homologous chromosomes attach to both poles as frequently as in wild-type cells. These results show that chiasmata eliminate the bipolar attachment of sister chromatids and promote the bipolar attachment of homologous chromosomes during the pre-anaphase stage in mrc1 mutant cells.
On the other hand, in moa1 mutant cells, centromere positioning and dissociation were not significantly different from those seen in achiasmate moa1 rec12 mutant cells (Figure 6A–6C, Table 1). Furthermore, homologous centromeres were not separated as widely as in wild-type cells (Figure 6E). These results indicate that sister chromatids still attach to both poles at a level similar to that in moa1 rec12 mutant cells and pulling forces are not properly exerted on homologous chromosomes in moa1 mutant cells (Figure 6E). Therefore, chiasmata fail to eliminate the bipolar attachment of sister chromatids during the pre-anaphase stage in moa1 mutant cells.
Chiasmata induce the preferential exertion of segregation forces on sister chromatids toward the proper pole during anaphase I in moa1 mutant cells
Because the bipolar attachment of sister centromeres did not appear to be eliminated during the pre-anaphase stage in chiasmate moa1 mutant cells, we examined whether their bipolar attachment is retained during anaphase by analyzing anaphase centromere dynamics. In wild-type cells, sister centromeres moved swiftly toward the poles (all 13 of the centromeres examined reached the poles within 130 s; Figure 7) and only occasionally dissociated during anaphase I [only three centromeres out of 13 (23.1%) were dissociated; Figure 7, Wt, lower panel]. The centromeres also moved swiftly to the pole and remained associated in mrc1 mutant cells (all 11 centromeres examined reached the pole within 80 s without dissociation; Figure 7). In contrast, in moa1 mutant cells, lagging and dissociation of centromeres were frequently observed during anaphase [10 out of 14 centromeres (71.4%) failed to reach the poles within 130 s, unlike wild-type centromeres, and 5 of them (35.7%) failed to reach the poles within 300 s; 6 centromeres (42.9%) were dissociated; Figure 7]. Furthermore, elimination of anaphase centromere cohesion by Sgo1 deletion substantially increased the equational segregation of sister chromatids (Figure 5B). These results showed that sister chromatids were frequently attached to both poles and pulled from opposite directions during anaphase I in moa1 mutant cells. Surprisingly, most of the lagging centromeres eventually moved to the proper pole (Figure 5A and 5B, Figure 7). This result indicates that although sister chromatids were pulled from opposite directions during anaphase, they were pulled toward the proper pole more strongly and/or continuously than they were pulled toward the improper pole in the chiasmate moa1 mutant cells. Therefore, the chiasma generates a bias toward the proper pole in poleward chromosome pulling from opposite directions that eventually results in proper chromosome segregation in moa1 mutant cells.
Fig. 7. Centromere dynamics during anaphase I in mrc1 and moa1 mutants. 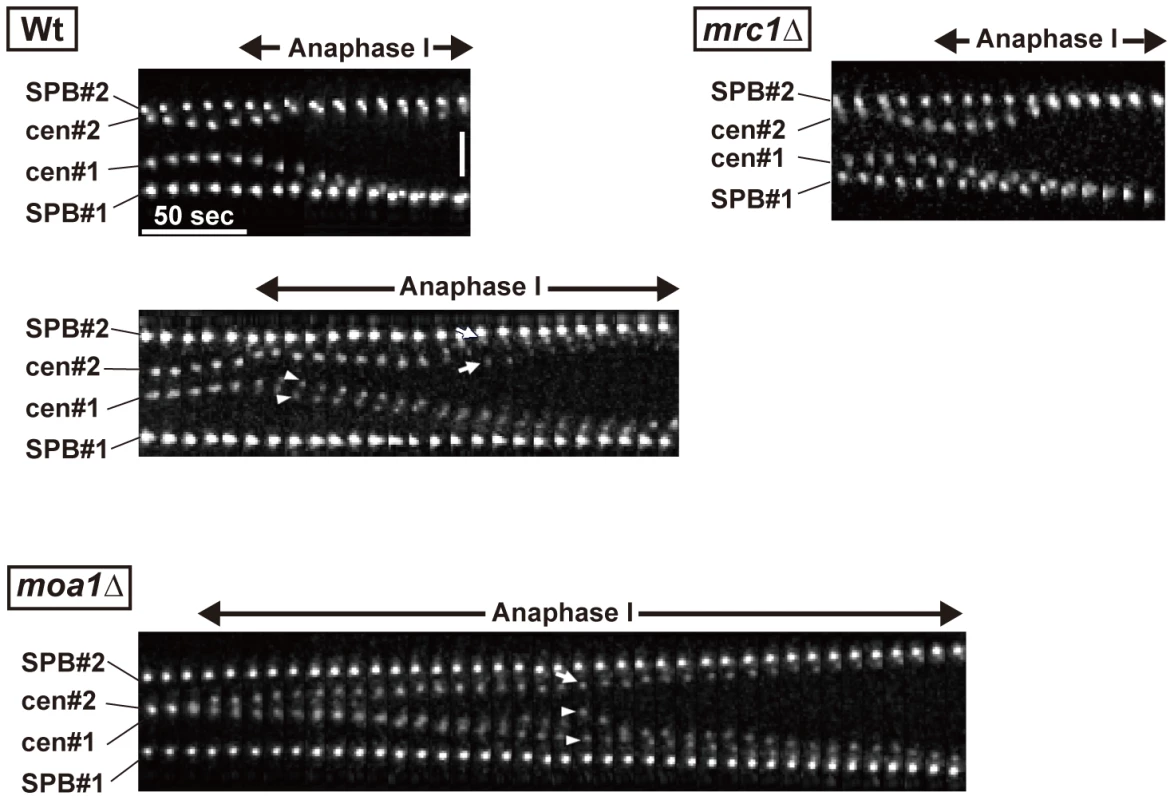
Arrows and arrowheads show each of the homologous centromeres (cen2), respectively, and the two arrowheads or arrows indicate dissociated sister centromeres. Horizontal bar: 50 s. Vertical bar: 2 µm. Discussion
Chiasmata play a crucial role in preventing the bipolar attachment of sister chromatids during anaphase I
In the current study, we examined the role of chiasmata by analyzing the segregation and dynamics of chromosomes during meiosis I induced in recombination-deficient diploid cells and in haploid cells. The analysis of these two distinct types of achiasmate cells provided two lines of evidence to show that sister chromatids frequently attach to both poles and experience pulling forces from opposite directions during anaphase I in achiasmate cells. First, sister centromeres frequently became transiently dissociated and/or failed to move to the pole during anaphase I (Figure 2C and Figure S1). Second, when sister centromere cohesion was resolved during anaphase by Sgo1 depletion, sister chromatids frequently underwent equational segregation during anaphase I (Figure 2B and 2D). Chiasmata therefore play a crucial role in preventing the bipolar attachment of sister chromatids during anaphase I. Because the bipolar attachment of sister chromatids has been observed during anaphase I in various achiasmate organisms [27]–[29], it is probably common among eukaryotes.
Two distinct tasks of chiasmata: elimination of the bipolar attachment of sister chromatids and induction of a bias in poleward chromosome pulling
We further examined how chiasmata prevent the bipolar attachment of sister chromatids. Loss of chiasmata causes activation of the SAC [42]. However, we showed that the bipolar attachment of sister chromatids depends only partially on the SAC in achiasmate cells. The reduction of the bipolar attachment that normally generates tension in the achiasmate background is consistent with the idea that the SAC promotes attachments that generate tension [40], [41].
We performed high time-resolution analysis of pre-anaphase centromere dynamics in several different types of chiasmate and achiasmate cells to understand how chiasmata contribute to the attachment. From this analysis, we have reached three conclusions. First, chiasmata cannot prevent occurrence of bipolar attachment of sister chromatids, based on the observation that the bipolar attachment occasionally occurred in chiasmate wild-type cells.
Second, analysis of mrc1 mutant cells showed that chiasmata contribute to the elimination of the bipolar attachment of sister chromatids during the pre-anaphase stage (Figure 8A). However, the elimination was not evident in wild-type cells in comparison with rec12 mutant cells. One possible explanation for this result is that the bipolar attachments occur more frequently in wild-type than in rec12 mutant cells because the centromere is positioned closer to the spindle center in wild-type cells (Figure 4B). Alternatively, chiasmata may eliminate bipolar attachments in mrc1 mutant cells but not in wild-type cells because of distinct centromere structures or functions. Furthermore, we cannot completely exclude the possibility that the chiasmata-dependent elimination depends in part on unknown Rec12 functions.
Fig. 8. Two major roles of chiasmata during meiosis I. 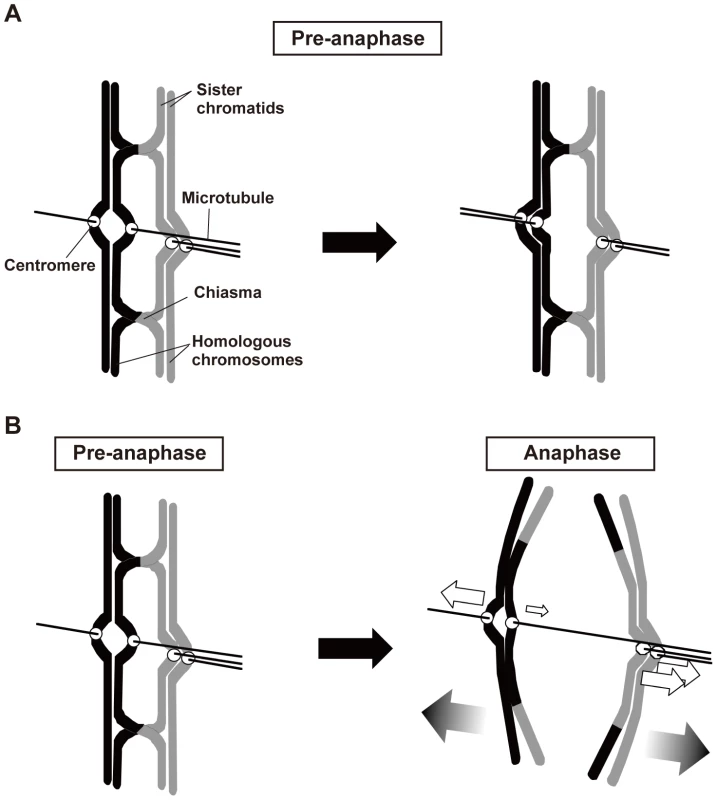
(A) Chiasmata eliminate the bipolar attachment of sister centromeres (centromeres on left sister chromatids) during the pre-anaphase stage of meiosis I. (B) When the bipolar attachment remains during anaphase, chiasmata generate bias in the poleward pulling forces to cause proper chromosome segregation. White arrows indicate the pulling forces exerted on chromosomes during anaphase I. A smaller arrow indicates a weaker or less continuously exerted force. For simplicity, only a single microtubule is shown to illustrate the spindle attachment of each kinetochore. Third, analysis of moa1 mutant cells showed that chiasmata induced a bias toward the proper pole in poleward chromosome pulling from opposite directions that resulted in proper chromosome segregation (Figure 8B). In moa1 mutant cells, sister centromeres were frequently pulled from opposite directions and dissociated during anaphase I, but they were pulled toward the proper pole more strongly and/or continuously than they were pulled toward the improper pole, and eventually moved to the appropriate pole. We also observed this chiasma effect, albeit occasionally, in wild-type cells (Figure 7, Wt, lower panel) and thereby speculate that the chiasma-induced bias is a backup mechanism that ensures proper meiotic chromosome segregation even when improper attachments remain.
Mechanism associated with chiasma-dependent elimination of bipolar attachments and biased chromosome pulling
How the chiasmata eliminate bipolar attachments and induce a bias in chromosome pulling remains elusive. Because chiasmata are essential for generating the tension that stabilizes kinetochore–microtubule interactions and increases kinetochore microtubules [9], [52], we speculate that chiasmata execute these different tasks via tension, as follows (see also Text S1). In wild-type cells, sister kinetochores occasionally attach to both poles (Figure S5A). In the presence of chiasmata, microtubules that attach to the proper poles generate sufficient tension, but those that attach to improper poles probably do not. As a result, improper attachments are eliminated while proper attachments are increased. Even when improper attachments are not eliminated, the increase in proper attachments presumably promotes the exertion of segregation forces in the appropriate direction (a similar scenario is shown in Figure S5A, rec12Δ). In contrast, improper attachments are not eliminated in rec12 mutant cells, possibly because the improper attachments also generate tension (Figure S5A).
In this model, chiasmata must prevent improper attachments from generating tension. During the pre-anaphase stage, chromosomes oscillate between the poles, and oscillation of the chiasma-linked chromosomes may reduce tension (Figure S5B). When a pair of sister chromatids follows the other homologous pair that is moving toward the spindle pole, the leading sister chromatid pair presumably exerts pulling forces on the chromosome arms of the following pair via chiasmata. These pulling forces are likely to reduce the tension that improper attachments generate but not those generated by proper attachments. As a result, only proper attachments (i.e., bipolar attachment of the homologous chromosomes) become stable and persist, whereas improper attachments (i.e., bipolar attachment of sister chromatids) do not. Alternatively, the chiasmata-dependent pulling may make the kinetochores on the following chromosomes face the side opposite the direction of chromosome movement to physically eliminate improper attachments. Although the above model can account for the observed chiasmata-dependent effects, we cannot completely rule out the possibility that chiasmata directly contribute to centromere function or structure to affect spindle attachment and segregation of chromosomes.
Effects of kinetochore arrangement on the chiasma-dependent elimination of improper attachments
Chiasmata eliminated bipolar attachment of sister chromatids in the mrc1 mutant but did not eliminate it in the moa1 mutant. Distinct kinetochore arrangements may account for this difference (Figure S5A, Text S1). Given the frequent monopolar attachment of sister chromatids in the chiasmate mrc1 single-mutant cells together with the substantial SAC activation in achiasmate mrc1 rec12 double-mutant cells, sister kinetochores probably face the same side in mrc1 mutants. However, the frequent bipolar attachment of sister chromatids seen in mrc1 rec12 mutant cells conversely implies that the kinetochores face opposite sides. This contradiction may be explained by the flexibility of the kinetochore arrangement (Figure S5A, Text S1). It is possible that in the mrc1 mutant cells, although sister kinetochores are initially arranged side by side, the kinetochores end up facing opposite sides when they are pulled from opposite directions, leading to the subsequent efficient bipolar attachment of sister centromeres.
On the other hand, in moa1 mutant cells, sister kinetochores perhaps face opposite sides to attach to both poles efficiently (Figure S5A, Text S1), as proposed previously [34]. Although kinetochore arrangement was previously proposed to be flexible in moa1 mutant cells [34], we speculate that the arrangement is conversely inflexible because of strong centromere cohesion, considering increased centromere accumulation of cohesin [34], infrequent sister centromere dissociation (Figure 6B), and a narrower dissociation distance (Figure S6). Bipolar attachment was not eliminated in moa1 single-mutant cells, perhaps because bipolar attachment is easily re-established due to the back-to-back kinetochore arrangement. An alternative possibility is that moa1 mutant cells are defective in destabilizing the kinetochore–microtubule interaction and fail to eliminate improper attachments efficiently.
Common mechanisms for chromosome segregation between mitosis and meiosis
Our findings have three important implications for understanding the mitotic chromosome segregation mechanism. First, the frequent bipolar attachment of sister chromatids seen in achiasmate cells indicates that kinetochore arrangement alone cannot prevent improper attachments and suggests that bipolar (merotelic) attachment of a single chromatid also occurs when sister chromatid cohesion is defective. Indeed, Courtheoux et al. recently reported that merotelic attachments occur during mitotic anaphase in rad21 fission yeast mutants defective in sister chromatid cohesion [53]. Furthermore, a lagging chromatid was frequently observed during anaphase II in sgo1 mutant of fission yeast, in which sister chromatids undergo precautious dissociation before anaphase II [17]. These observations may alter the interpretation of phenotypes associated with monopolin and heterochromatin mutants of fission yeast, which were proposed to be defective in the arrangement of microtubule-binding sites of kinetochores because these mutants frequently exhibited merotelic attachments during mitotic anaphase [54], [55]. However, defective sister centromere cohesion in the monopolin and heterochromatin mutants may have caused the merotelic attachments [56]–[58].
Second, the fact that sister chromatids, despite their bipolar attachment, move to the same pole in chiasmate cells indicates that monopolar attachment of sister chromatids is not a prerequisite for their proper segregation. This feature is probably common during mitotic chromosome segregation because the proper segregation of a single chromatid that is attached to both poles has also been observed in higher eukaryotes during mitosis [59]. Therefore, generation of bias in the segregation forces is probably a general mechanism that ensures correct chromosome segregation.
Finally, the chromosome oscillation-dependent model for the elimination of improper attachments may also account for the establishment of proper attachments during mitosis (Figure S5B). During mitosis, chromosomes oscillate during the establishment of their spindle attachment (Figure S4A) [60], [61], and merotelic attachment occurs in higher eukaryotes [62]. Furthermore, in fission yeast, the physical linkage between two kinetochores induces their bipolar attachment during mitosis [63]. These facts suggest that the oscillation of cohesin-linked sister chromatids destabilizes improper attachments and contributes to the selection of proper attachments during mitosis.
In summary, we have shown that chiasmata are essential for proper spindle attachment and segregation of sister chromatids during meiosis I. Based on our results, we propose that chiasmata play a pivotal role in the selection of proper attachments and establish a backup mechanism that promotes the appropriate segregation of chromosomes when improper attachments remain during anaphase I. Furthermore, we propose a model to explain how chromosome association contributes to correct spindle attachment of the chromosomes not only in meiosis but also in mitosis. Our findings increase understanding of the general mechanisms of chromosome segregation and contribute to knowledge about the mechanisms that underlie the chromosome mis-segregation associated with birth defects and/or tumorigenesis in humans.
Materials and Methods
Yeast strains and media
Table S3 lists the yeast strains used in this study, and strains used in figures are described in Text S1. Media used in this study have been described by Moreno et al. [64].
Analysis of chromosome segregation during meiosis I in diploid cells
Yeast strains were grown on solid YES medium at 30°C. For the segregation analyses of homologous chromosomes, two types of cells, both of which contained GFP-labeled centromeres (cen2 or lys1), were crossed on solid ME medium. For sister chromatid segregation analyses, cells containing GFP-labeled centromeres were crossed with cells lacking GFP-labeled centromeres. The resulting diploid cells were then induced to enter meiosis by incubation at 25°C for 16–18 h. Nuclear DNA in meiotic zygotes was stained with the DNA-specific dye, Hoechst 33342, as described [65]. GFP signal was examined in zygotes containing two round DNA masses that underwent meiosis I. Zygotes containing two DNA masses with a tear-drop shape and pointed ends facing each other were excluded because they were in the karyogamy stage.
Analysis of chromosome segregation during meiosis I in haploid cells
Haploid yeast cells were forced to enter meiosis by Pat1 inactivation following activation of the mating pheromone signaling pathway, as previously described [33]. Haploid pat1 temperature-sensitive mutant cells bearing the c-type mat gene of the opposite mating type, which is required for activation of the mating pheromone signaling pathway, were grown in YES-rich medium to a density of 3–5×106 cells/ml at 25°C. The cells were suspended in an equal volume of EMM2 medium lacking a source of nitrogen (EMM2-N) and incubated at 25°C for 14–16 h to synchronize the cells in G1 phase and activate the mating pheromone signaling pathway. The cells were resuspended in fresh EMM2-N medium and induced to enter meiosis by further incubation at 34°C. Meiotic progression was monitored by analysis of chromosomal DNA morphology at 1-h time intervals. Sister chromatid segregation was analyzed in cells containing two DNA masses that underwent meiosis I.
Live cell analysis of chromosome and spindle pole dynamics
The chromosome locus and spindle poles were visualized using the lacI/lacO recognition system and the GFP-tagged spindle pole component Sid4, respectively, as described previously [42]. Cells were grown on solid YES medium at 30°C and induced to undergo meiosis by incubation on solid ME medium at 25°C for 16–18 h. The cells were observed to determine the dynamics of the GFP-labeled spindle pole or chromosome locus at 25°C using a DeltaVision microscope system (Applied Precision Inc.) equipped with a 60X/1.42 numerical aperture Plan Apo oil-immersion objective lens (Olympus), as described previously [65]. The behavior of the GFP-labeled chromosome locus was observed every 1 min or 10 s. A set of images from six focal planes with 0.5-µm intervals or ten focal planes with 0.3-µm intervals was taken at each time point for 1-min or 10-s time-lapse analysis, respectively. Behavior of the GFP-tagged Cnp1 was observed in a manner similar to the 10-s time-lapse analysis of the GFP-labeled chromosome locus, except that a 100X/1.4 numerical aperture Plan Apo oil-immersion objective lens (Olympus) was used. All measurements were conducted in three dimensions.
Supporting Information
Zdroje
1. NasmythK
2002 Segregating sister genomes: the molecular biology of chromosome separation. Science 297 559 565
2. KoshlandDE
GuacciV
2000 Sister chromatid cohesion: the beginning of a long and beautiful relationship. Curr Opin Cell Biol 12 297 301
3. HaufS
WatanabeY
2004 Kinetochore orientation in mitosis and meiosis. Cell 119 317 327
4. KellyAE
FunabikiH
2009 Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol 21 51 58
5. GuacciV
KoshlandD
StrunnikovA
1997 A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91 47 57
6. MicchaelisC
CioskR
NasmythK
1977 Cohesins: chromosomal proteins that prevent premature separation of sister chroamtids. Cell 91 35 45
7. FuruyaK
TakahashiK
YanagidaM
1998 Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev 12 3408 3418
8. SonodaE
MatsusakaT
MorrisonC
VagnarelliP
HoshiO
2001 Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell 1 759 770
9. NicklasRB
1997 How cells get the right chromosomes. Science 275 632 637
10. PetronczkiM
SiomosMF
NasmythK
2003 Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112 423 440
11. KleinF
MahrP
GalovaM
BuonomoSBC
MichaelisC
1999 A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98 91 103
12. WatanabeY
NurseP
1999 Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400 461 464
13. KudoNR
AngerM
PetersAH
StemmannO
TheusslHC
2009 Role of cleavage by separase of the Rec8 kleisin subunit of cohesin during mammalian meiosis I. J Cell Sci 122 2686 2698
14. PasierbekP
JantschM
MelcherM
SchleifferA
SchweizerD
2001 A Caenorhabditis elegans cohesion protein with funcitons in meiotic chromosome pairing and disjunction. Genes Dev 15 1349 1360
15. BhattAM
ListerC
PageT
FranszP
FindlayK
1999 The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19 463 472
16. ParraMT
VieraA
GomezR
PageJ
BenaventeR
2004 Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J Cell Sci 117 1221 1234
17. RabitschKP
GreganJ
SchleifferA
JaverzatJP
EisenhaberF
2004 Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol 14 287 301
18. KatisVL
GalovaM
RabitschKP
GreganJ
NasmythK
2004 Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol 14 560 572
19. KitajimaTS
KawashimaSA
WatanabeY
2004 The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427 510 517
20. KerrebrockAW
MooreDP
WuJS
Orr-WeaverTL
1995 Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 83 247 256
21. MarstonAL
ThamWH
ShahH
AmonA
2004 A genome-wide screen identifies genes required for centromeric cohesion. Science 303 1367 1370
22. KawashimaSA
TsukaharaT
LangeggerM
HaufS
KitajimaTS
2007 Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev 21 420 435
23. VanoosthuyseV
PrykhozhijS
HardwickKG
2007 Shugoshin 2 regulates localization of the chromosomal passenger proteins in fission yeast mitosis. Mol Biol Cell 18 1657 1669
24. YuHG
KoshlandD
2007 The Aurora kinase Ipl1 maintains the centromeric localization of PP2A to protect cohesin during meiosis. J Cell Biol 176 911 918
25. HaufS
BiswasA
LangeggerM
KawashimaSA
TsukaharaT
2007 Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I. Embo J 26 4475 4486
26. HawleyRS
1988 Exchange and chromosomal segregation in eukaryotes.
KucherlapatiR
SmithGR
Genetic recombination Washington, DC American Society for Microbiology 497 527
27. HuntP
LeMaireR
EmburyP
SheeanL
MrozK
1995 Analysis of chromosome behavior in intact mammalian oocytes: monitoring the segregation of a univalent chromosome during female meiosis. Human Mol Genet 4 2007 2012
28. DarlingtonCD
1939 Misdivision and the genetics of the centromere. J Genet 37 341 363
29. MaguireMP
1987 Meiotic behavior of a tiny fragment chromosome that carries a transposed centromere. Genome 29 744 746
30. YamamotoM
ImaiY
WatanabeY
1997 Mating and sporulation in Schizosaccharomyces pombe.
PringleJR
BroachJR
JonesEW
The molecular and cellular biology of the yeast Saccharomyces; cell cycle and cell biology: Cold Spring Harbor Press 1037 1106
31. IinoY
YamamotoM
1985 Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet 198 416 421
32. McLeodM
BeachD
1988 A specific inhibitor of the ran1+ protein kinase regulates entry into meiosis in Schizosaccharomyces pombe. Nature 332 509 514
33. YamamotoA
HiraokaY
2003 Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J 22 2284 2296
34. YokobayashiS
WatanabeY
2005 The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123 803 817
35. KitajimaTS
YokobayashiS
YamamotoM
WatanabeY
2003 Distinct cohesin complexes organize meiotic chromosome domains. Science 300 1152 1155
36. CervantesMD
FarahJA
SmithGR
2000 Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell 5 883 888
37. DavisL
SmithGR
2003 Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics 163 857 874
38. EvansDH
LiYF
FoxME
SmithGR
1997 A WD repeat protein, Rec14, essential for meiotic recombination in Schizosaccharomyces pombe. Genetics 146 1253 1264
39. SteinerS
KohliJ
LudinK
2010 Functional interactions among members of the meiotic initiation complex in fission yeast. Curr Genet 56 237 249
40. MusacchioA
HardwickKG
2002 The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol 3 731 741
41. KaduraS
SazerS
2005 SAC-ing mitotic errors: how the spindle assembly checkpoint (SAC) plays defense against chromosome mis-segregation. Cell Motil Cytoskeleton 61 145 160
42. YamamotoA
KitamuraK
HiharaD
HiroseY
KatsuyamaS
2008 Spindle checkpoint activation at meiosis I advances anaphase II onset via meiosis-specific APC/C regulation. J Cell Biol 182 277 288
43. TakahashiK
ChenES
YanagidaM
2000 Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288 2215 2219
44. AlcasabasAA
OsbornAJ
BachantJ
HuF
WerlerPJ
2001 Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3 958 965
45. TanakaK
RussellP
2001 Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat Cell Biol 3 966 972
46. XuH
BooneC
KleinHL
2004 Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol Cell Biol 24 7082 7090
47. OsbornAJ
ElledgeSJ
2003 Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev 17 1755 1767
48. KatouY
KanohY
BandoM
NoguchiH
TanakaH
2003 S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424 1078 1083
49. TourriereH
VersiniG
Cordon-PreciadoV
AlabertC
PaseroP
2005 Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19 699 706
50. KumagaiA
DunphyWG
2000 Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell 6 839 849
51. YokobayashiS
YamamotoM
WatanabeY
2003 Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol 23 3965 3973
52. KingJM
NicklasRB
2000 Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J Cell Sci 113 Pt 21 3815 3823
53. CourtheouxT
GayG
GachetY
TournierS
2009 Ase1/Prc1-dependent spindle elongation corrects merotely during anaphase in fission yeast. J Cell Biol 187 399 412
54. RabitschKP
PetronczkiM
JaverzatJP
GenierS
ChwallaB
2003 Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell 4 535 548
55. GreganJ
RiedelCG
PidouxAL
KatouY
RumpfC
2007 The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol 17 1190 1200
56. NonakaN
KitajimaT
YokobayashiS
XiaoG
YamamotoM
2002 Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4 89 93
57. Monje-CasasF
PrabhuVR
LeeBH
BoselliM
AmonA
2007 Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 128 477 490
58. BernardP
MaureJF
PartridgeJF
GenierS
JaverzatJP
2001 Requirement of heterochromatin for cohesion at centromeres. Science 294 2539 2542
59. CiminiD
CameronLA
SalmonED
2004 Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol 14 2149 2155
60. InoueS
SalmonED
1995 Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell 6 1619 1640
61. TanakaTU
StarkMJ
TanakaK
2005 Kinetochore capture and bi-orientation on the mitotic spindle. Nat Rev Mol Cell Biol 6 929 942
62. CiminiD
MoreeB
CanmanJC
SalmonED
2003 Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci 116 4213 4225
63. SakunoT
TadaK
WatanabeY
2009 Kinetochore geometry defined by cohesion within the centromere. Nature 458 852 858
64. MorenoS
KlarA
NurseP
1991 Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194 793 823
65. YamamotoA
TsutsumiC
KojimaH
OiwaK
HiraokaY
2001 Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol Biol Cell 12 3933 3946
66. PetronczkiM
ChwallaB
SiomosMF
YokobayashiS
HelmhartW
2004 Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J Cell Sci 117 3547 3559
67. NabeshimaK
NakagawaT
StraightAF
MurrayA
ChikashigeY
1998 Dynamics of centromeres during metaphase-anaphase transition in fission yeast: dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell 9 3211 3225
68. TomlinGC
MorrellJL
GouldKL
2002 The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol Biol Cell 13 1203 1214
69. ChikashigeY
HiraokaY
2001 Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol 11 1618 1623
70. TakayamaY
SatoH
SaitohS
OgiyamaY
MasudaF
2008 Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol Biol Cell 19 682 690
71. LinY
SmithGR
1994 Transient, meiosis-specific expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics 136 769 779
72. KimSH
LinDP
MatsumotoS
KitazonoA
MatsumotoT
1998 Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science 279 1045 1047
73. MasudaH
MiyamotoR
HaraguchiT
HiraokaY
2006 The carboxy-terminus of Alp4 alters microtubule dynamics to induce oscillatory nuclear movement led by the spindle pole body in Schizosaccharomyces pombe. Genes Cells 11 337 352
74. MolnarM
BählerJ
SipiczkiM
KohliJ
1995 The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics 141 61 73
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and PhosphodiesterasesČlánek Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



