-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Tradeoff Drives the Evolution of Reduced Metal Resistance in
Natural Populations of Yeast
Various types of genetic modification and selective forces have been implicated
in the process of adaptation to novel or adverse environments. However, the
underlying molecular mechanisms are not well understood in most natural
populations. Here we report that a set of yeast strains collected from Evolution
Canyon (EC), Israel, exhibit an extremely high tolerance to the heavy metal
cadmium. We found that cadmium resistance is primarily caused by an enhanced
function of a metal efflux pump, PCA1. Molecular analyses
demonstrate that this enhancement can be largely attributed to mutations in the
promoter sequence, while mutations in the coding region have a minor effect.
Reconstruction experiments show that three single nucleotide substitutions in
the PCA1 promoter quantitatively increase its activity and thus
enhance the cells' cadmium resistance. Comparison among different yeast
species shows that the critical nucleotides found in EC strains are conserved
and functionally important for cadmium resistance in other species, suggesting
that they represent an ancestral type. However, these nucleotides had diverged
in most Saccharomyces cerevisiae populations, which gave cells
growth advantages under conditions where cadmium is low or absent. Our results
provide a rare example of a selective sweep in yeast populations driven by a
tradeoff in metal resistance.
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1002034
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002034Summary
Various types of genetic modification and selective forces have been implicated
in the process of adaptation to novel or adverse environments. However, the
underlying molecular mechanisms are not well understood in most natural
populations. Here we report that a set of yeast strains collected from Evolution
Canyon (EC), Israel, exhibit an extremely high tolerance to the heavy metal
cadmium. We found that cadmium resistance is primarily caused by an enhanced
function of a metal efflux pump, PCA1. Molecular analyses
demonstrate that this enhancement can be largely attributed to mutations in the
promoter sequence, while mutations in the coding region have a minor effect.
Reconstruction experiments show that three single nucleotide substitutions in
the PCA1 promoter quantitatively increase its activity and thus
enhance the cells' cadmium resistance. Comparison among different yeast
species shows that the critical nucleotides found in EC strains are conserved
and functionally important for cadmium resistance in other species, suggesting
that they represent an ancestral type. However, these nucleotides had diverged
in most Saccharomyces cerevisiae populations, which gave cells
growth advantages under conditions where cadmium is low or absent. Our results
provide a rare example of a selective sweep in yeast populations driven by a
tradeoff in metal resistance.Introduction
Unicellular microorganisms are often challenged by fluctuating environmental conditions. Especially for those organisms having limited mobility, adaptation to such environmental stresses is critical for survival of their populations. However, mutations beneficial for survival in one environment may impose a cost under other conditions [1], [2]. Cells need to fine-tune the evolved gene function or regulation in order to maintain an optimal physiology under a range of conditions. It is important to understand how cells adapt to novel or adverse environments. Such information may not only allow us to dissect the factors affecting evolution of organisms, but may also provide us some insights into pathway or functional network flexibility (or evolvability) of the cell. To address this issue, identifying the mutations responsible for the adaptive phenotypes is the most direct approach, and yet it remains challenging even in simple organisms such as E. coli. Moreover, even if a mutation is identified, detailed population and phylogeny data are required in order to deduce the evolutionary trajectory of adaptive traits.
Experimental evolution represents a simplified approach since it allows scientists to follow the evolutionary history of populations exposed to known selective pressures. Several adaptive mutations in microorganisms have been discovered and characterized at the molecular level from laboratory experimental evolution, adding greatly to our understanding of adaptive evolution [1], [3]–[9]. On the other hand, studies related to natural adaptation are more complicated. Although the mechanistic basis or phylogeny of adaptive traits have been revealed in several previous studies [10]–[15], systematic approaches dealing with both aspects are still rare [16]–[18].
Metal ions such as copper, iron, zinc, potassium and sodium are essential nutrients involved in a broad range of biological processes [19], [20]. These essential metals function as catalysts for biochemical reactions, stabilizers of protein structures or cell walls, or regulators of intracellular osmotic balance. Despite their importance, unbalanced metal concentrations can cause deleterious effects, sometimes leading to programmed cell death [21], and thus represent a double-edged sword. It is important for cells to tightly regulate homeostasis of these metal ions.
In natural environments, cells often encounter other nonessential metal ions. Some of them such as cadmium, lead and arsenic are highly toxic to cells. Toxicity often occurs through the displacement of essential metals from their native binding sites or through ligand interactions, resulting in altered structural conformations or interference with biochemical reactions [22]. These metal ions can induce the generation of reactive oxygen species and cause damages to various cellular components [23]–[25]. Organisms have evolved several different mechanisms to cope with metal induced stresses, including specific metal transporters, metal sequestration proteins or compartments, and different detoxification enzymes [12], [22], [26], [27]. These various systems often cooperate with each other to quickly respond to variations in environmental metal concentrations, indicating the importance of metal ion balance to cells.
In this study, we observed that a subset of diploid yeast strains collected from different locations of the EC could tolerate a heavy metal, cadmium, to a level unseen in most known yeast strains. We found that the cadmium-resistant phenotype is primarily caused by regulatory changes in the PCA1 gene, which encodes a P-type ATPase required for cadmium efflux [28], [29]. By performing functional assays and phylogenetic analyses, we show that PCA1 has experienced several rounds of selective adaptation during yeast evolution. More strikingly, we observe that a weak PCA1 allele spread to most S. cerevisiae populations, probably due to a tradeoff between metal resistance and fitness under low cadmium conditions.
Results/Discussion
One subset of EC yeast strains is highly resistant to cadmium
Evolution Canyon is an east-west-oriented canyon at Lower Nahal Oren, Israel. It originated 3–5 million years ago and is believed to have experienced minimal human disturbance [30], [31]. In contrast to other wild yeast, the strains collected from EC are often polyploid and most of them are heterothallic [32],[33]. Previous studies have revealed high allelic diversity among EC yeast strains [32], [33]. To assess whether these strains also carry specific adaptive phenotypes, we performed a panel of phenotypic assays including cell growth under several stress conditions. Only diploid strains were included in this study since triploid and tetraploid strains are less amenable to further genetic analyses. The results showed that a subset of EC strains (EC9, 10, 35, 36, 39, 57, and 58) was resistant to a very high concentration of cadmium (0.8 mM CdCl2), while all other strains analyzed were unable to grow on plates containing 0.2 mM CdCl2 (Figure S1A).
Because chromosomal rearrangement has been suggested to be involved in adaptive evolution [17], [34],[35], we first examined the karyotype of 14 diploid EC strains. Pulsed-field gel electrophoresis (PFGE) analysis revealed that these EC strains comprised three major karyotypes, EC-C1, EC-C2 and EC-C3 (with some minor deviations)(Figure S1B). Interestingly, all cadmium-resistant strains belong to EC-C1, suggesting that the metal-resistant phenotypes have already evolved before the EC-C1 populations split. Therefore, we chose to use mainly one strain (EC9) from EC-C1 for subsequent genetic analyses.
The cadmium-resistant phenotype is caused by a mutant allele of PCA1 (PCA1-C1)
We performed a genetic analysis to assess how many genetic loci are involved in the cadmium-resistant phenotype. A haploid Cd-resistant clone (EC9-8) was crossed with two Cd-sensitive strains (EC13 from EC-C2 and lab strain S288C). Both hybrid diploids were Cd resistant, indicating that the Cd-resistant phenotype was dominant. The hybrid diploids were then induced to sporulate, and their haploid segregants were examined for their cadmium tolerance. These segregrants showed a 2∶2 (resistant to sensitive) segregation pattern, indicating that the Cd-resistant phenotype was primarily controlled by a single genetic locus.
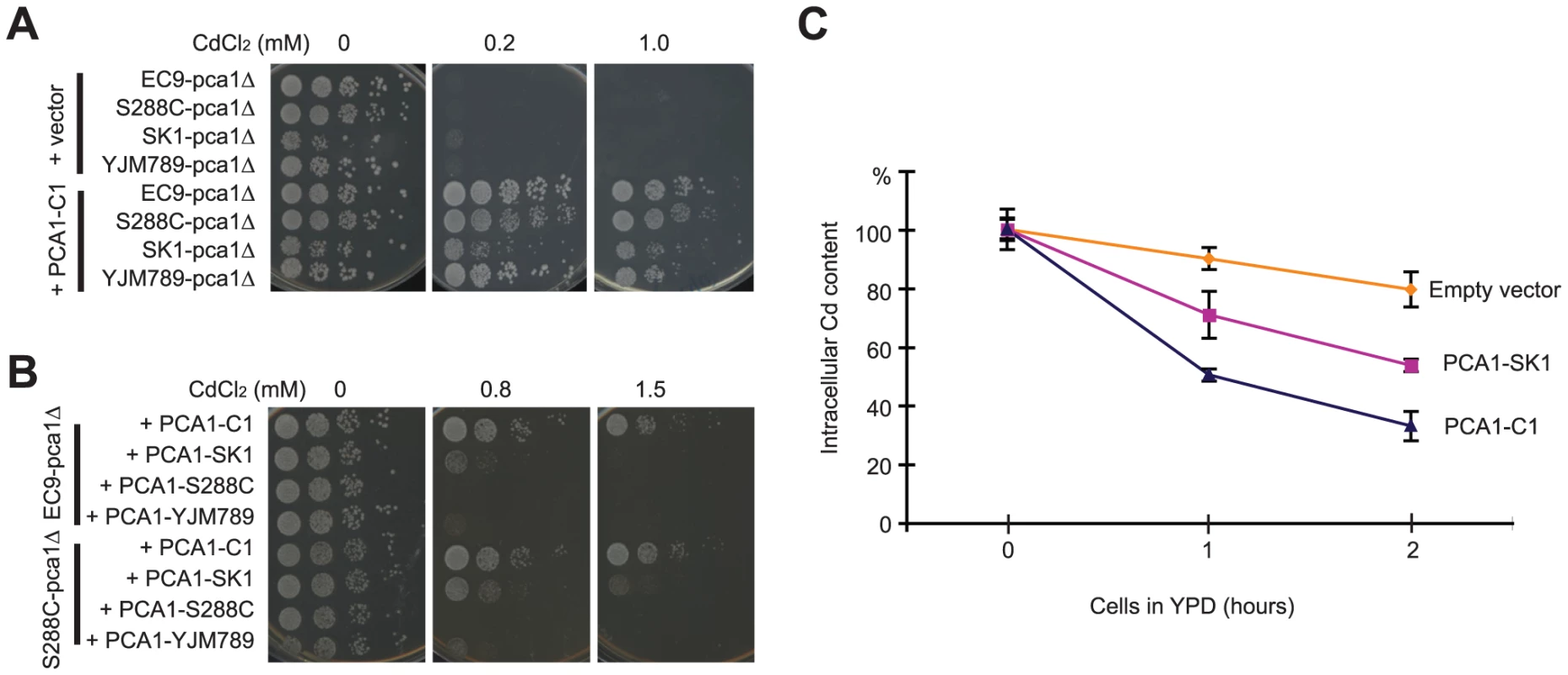
To screen for the gene responsible for cadmium resistance, a genomic DNA library constructed from EC9 genomic DNA was transformed into Cd-sensitive cells. All Cd-resistant colonies carried plasmids containing PCA1. Sequencing the PCA1 allele (PCA1-C1) of the EC-C1 strains revealed many mutations in both promoter and protein-coding regions as compared with the PCA1 sequences from other strains (Table S1). Next, we directly tested whether PCA1-C1 alone is able to improve the cadmium tolerance of cells. Plasmids carrying PCA1-C1 were transformed into three Cd-sensitive strains (S288C, SK1, and YJM789) in which the PCA1 gene had been deleted. As shown in Figure 1A, the different strains carrying PCA1-C1 all exhibited a level of cadmium resistance close to the level of EC-C1. By contrast, when the PCA1 alleles from Cd-sensitive strains were transformed into an EC9 pca1Δ mutant, the transformants remained cadmium sensitive (Figure 1B). Finally, we sequenced the PCA1 alleles of the segregants obtained from the previous genetic analysis and confirmed that all Cd-resistant segregants carry the PCA1-C1 allele.
A previous study by Adle and co-workers has shown that the PCA1-dependent cadmium resistance is mainly a consequence of active cadmium export (efflux) [29]. To determine whether cadmium efflux is higher in cells containing PCA1-C1, cells carrying PCA1-C1 or PCA1-SK1 were pretreated with cadmium, washed to remove extracellular cadmium, resuspended in fresh media, and collected at different time points to measure the cellular cadmium content using inductively coupled plasma-atomic emission spectroscopy (ICP-AES). The PCA1-SK1 allele from the SK1 strain was chosen for comparison because this allele is phylogenetically more related to PCA1-C1 based on an analysis of the corresponding ORF sequences (Figure S2D) and because it is a cadmium-sensitive allele lacking the G970R mutation (which abolishes the activity of Pca1) present in other laboratory strains. Indeed, cells carrying PCA1-C1 could reduce the intracellular cadmium concentration more quickly than cells carrying PCA1-SK1. These data indicate that cells containing PCA1-C1 have a very efficient cadmium efflux (Figure 1C).
Mutations in both the promoter and coding regions of PCA1-C1 contribute to cadmium resistance
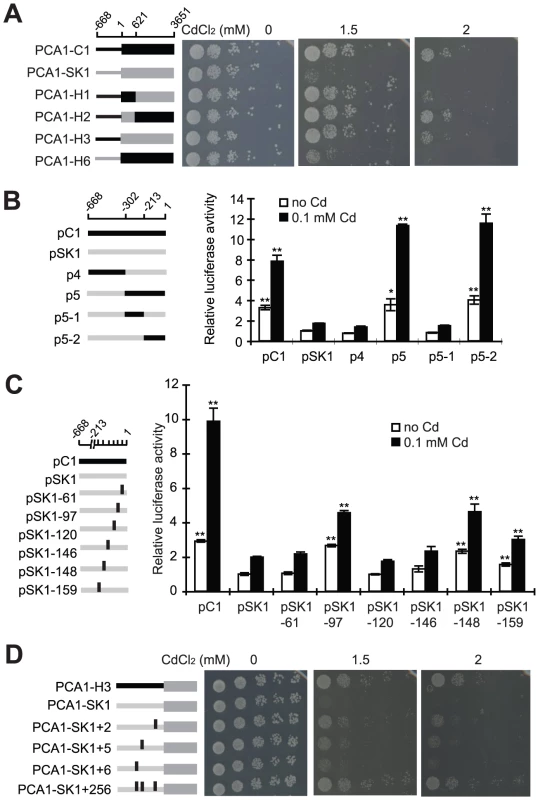
In order to understand how PCA1-C1 has evolved a high cadmium resistance, chimeric proteins with regions from PCA1-C1 and PCA1-SK1 were constructed and assayed for their ability to complement cadmium sensitivity of the pca1Δ mutant. We found that swapping the promoters drastically affected cadmium resistance (Figure 2A, compare C1 with H6 and SK1 with H3), whereas swapping the region between amino acids 207 and 1216 had a mild effect (C1 vs. H1 and H2 vs. H3). Four nonsynonymous mutations are present in this coding region: N223, T358, T363, and G365. Previous studies have identified a few domains important for the stability or function of Pca1 [29], [36]. However, none of these four mutations are located within these functional domains.
To assess whether the different levels of cadmium resistance resulted from differences in promoter strength, we fused the PCA1-C1 or PCA1-SK1 promoters to a luciferase reporter and assayed the luciferase activity of these constructs. The expression driven by the PCA1-C1 promoter was about four-fold higher than that driven by the PCA1-SK1 promoter in the absence and presence of cadmium treatments, suggesting that mutations in the PCA1-C1 promoter increased the degree of cadmium resistance by increasing the PCA1 gene expression without destroying its regulation (Figure 2B). We also performed quantitative PCR to determine the level of PCA1 mRNA in EC9 and SK1 strains. The data were consistent with the results from the luciferase reporter gene assay.
The increase in PCA1-C1 expression is mainly caused by three point mutations in the promoter region
By comparing the promoter sequences (including 600 bp upstream of the initiation codon) of PCA1-C1 and PCA1-SK1, we observed 18 single nucleotide polymorphisms (17 single nucleotide substitutions and one 1-bp deletion) and one 10-bp insertion (Table S1A). Because altered PCA1-C1 expression plays a key role in enhancing cadmium resistance, we sought to understand how this gene evolved and the degree to which changes in its promoter contribute to expression differences. To address the latter issue, we fused chimeric promoters with regions from PCA1-C1 and PCA1-SK1 promoters to a luciferase reporter and assayed their expression levels. We found that only the region immediately upstream of the initiation codon (−213 to −1) contributed significantly to the enhanced gene expression (Figure 2B, p5-2). Only 6 of the 18 single nucleotide polymorphisms are present in this region. To determine which mutations led to the enhanced promoter activity, we introduced the PCA1-C1 version of each of these sites into the PCA1-SK1 promoter and assayed the reporter gene expression. Only mutations in three nucleotides (PCA1-SK1 to PCA1-C1: −97T > C, −148T > G, and −159G > T) had obvious effects (Figure 2C; Table S1A).
To confirm that the changed expression is important for the cadmium resistance, we introduced the same mutations into the PCA1-SK1 allele and then measured the cadmium resistance of cells carrying these mutant alleles. The expression level of the PCA1-SK1 mutants was indeed correlated with the cadmium resistance (Figure 2D). When all three mutations were combined together into a single mutant clone (PCA1-SK1+2/5/6), cells carrying this clone were as resistant to cadmium as the cells carrying PCA1-H3, in which the PCA1-C1 promoter is fused to the PCA1-SK1 ORF. This result demonstrated that the enhanced cadmium resistance of EC-C1 strains is mainly caused by three single nucleotide substitutions in the promoter of PCA1-C1. Currently, it is still unclear how PCA1 transcription is regulated. Although we could not identify any transcription factor binding motif in the sequences where the critical mutations (−97C, −148G and −159T) are located, it is quite possible that these regions contain some of the regulatory elements of PCA1.
Cadmium resistance is an ancestral phenotype
To determine whether the Cd-resistant phenotype is specific to EC-C1 strains, we examined the cadmium sensitivity of two closely related species, S. paradoxus and S. mikatae. Two S. mikatae strains and 28 S. paradoxus strains isolated from different niches were tested [37]. Although both species could not tolerate, unlike EC-C1, a high level of cadmium (2.0 mM CdCl2), they exhibited much higher Cd resistance (0.4–0.8 mM CdCl2) than the other S. cerevisiae strains (Figure S3). To confirm that the medium level of cadmium resistance in S. paradoxus was also mediated through PCA1, we cloned the S. paradoxus PCA1 gene (Sp-PCA1) into a plasmid, transformed the resulting vector into S. cerevisiae pca1Δ mutant cells, and then assayed the transformants for cadmium sensitivity. The result showed that Sp-PCA1 was able to generate a medium level of cadmium resistance in S. cerevisiae pca1Δ mutants (Figure S3B). Consistent with these results, deletions of PCA1 in S. paradoxus strains resulted in a Cd-sensitive phenotype (Figure S3C).
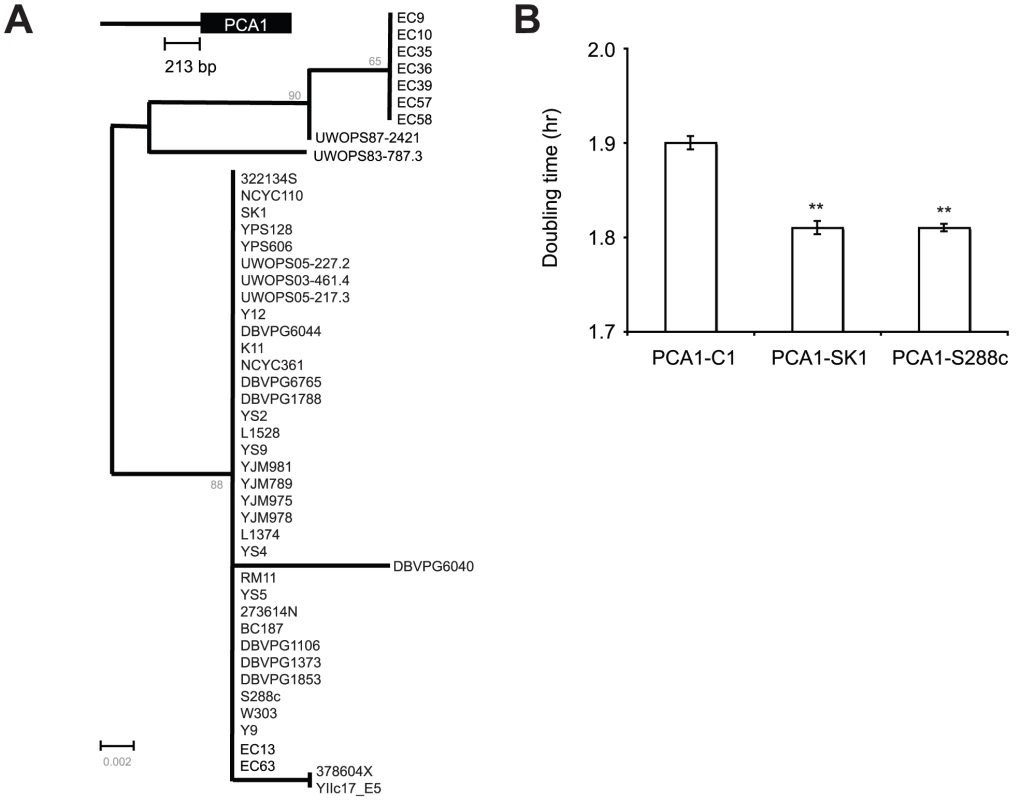
Interestingly, comparison between S. cerevisiae-, S. paradoxus-, and S. mikatae-PCA1 promoter sequences revealed that those critical residues (−97C, −148G and −159T) identified in the previous experiment are conserved between EC-C1 strains, S. paradoxus (28 strains) and S. mikatae (2 strains)(Figure 3). When we mutated these nucleotides of Sp-PCA1 (−100C, −149G and −162T) to non-EC-C1 Sc-PCA1 sequences, the mutant allele became cadmium sensitive (Figure S3D), indicating that these nucleotides were also critical for the function of Sp-PCA1. Since both S. paradoxus and S. mikatae can tolerate a medium level of cadmium, it is likely that this phenotype represents an original phenotype of the common ancestor of S. cerevisiae, S. paradoxus, and S. mikatae, which has been lost in most S. cerevisiae populations (Figure S4).
Loss of cadmium resistance provides a fitness advantage under cadmium-free conditions in S. cerevisiae
If the common ancestor of S. cerevisiae and S. paradoxus was cadmium resistant, why did most S. cerevisiae populations become cadmium sensitive? By comparing the promoter sequences of PCA1 from EC-C1, EC-C2, EC-C3, and 38 other S. cerevisiae strains (collected from various habitats on different continents; see [37]), a striking pattern was revealed: most S. cerevisiae strains carry a weak PCA1 promoter similar to the one in the SK1 strain (Figure 4A). Hence, reduced promoter strength accounts for the cadmium-sensitive phenotype observed in these strains (Figure S4). The only exceptions are EC-C1, UWOPS87_2421, and UWOPS83_787_3. Interestingly, the PCA1 promoters of UWOPS87_2421 and UWOPS83_787_3 also contain the mutations critical for cadmium resistance (−97C and −159T in UWOPS87_2421 and −97C in UWOPS83_787_3), and both strains show cadmium-resistant phenotypes (Table S1).
A previous study showed that cells overexpressing PCA1 in a medium without cadmium suffered reduced fitness [38]. To determine whether expression of PCA1-C1 also imposes a high fitness cost on cells, we conducted a competition assay to measure the fitness of cells containing PCA1-C1, PCA1-SK1 or PCA1-S288C. Plasmids carrying either PCA1-C1, PCA1-SK1 or PCA1-S288C were transformed into S288C pca1Δ mutants. The resulting transformants were then mixed with a reference strain carrying a green fluorescent protein-tagged Pgk1 protein and grown in a medium without cadmium. The results showed that cells containing PCA1-C1 had a lower fitness than cells containing PCA1-SK1 or PCA1-S288C (p<0.001, two-tailed t-test), suggesting a tradeoff between high Cd resistance and the fitness of cells under Cd-free conditions (Figure 4B). It is possible that Sc-PCA1 was selected to reduce the fitness cost, thus resulting in lower Cd resistance if most S. cerevisiae cells were constantly living in environments containing low levels of cadmium. Alternatively, Sc-PCA1 might have been selected, at a cost of reduced Cd resistance, to enhance other activities. Previous studies have suggested that Pca1 is also involved in copper resistance [38]. However, we found that cells carrying PCA1-C1 or PCA1-SK1 showed a similar level of copper resistance, indicating that the mutations in PCA1-C1 are specific to cadmium resistance (Figure S5).
It is unclear why cells carrying PCA1-C1 have a lower fitness under Cd-free conditions. Since Pca1 is not a highly abundant protein, the fitness reduction is unlikely due to the energy cost for producing extra amounts of the Pca1 protein. PCA1 belongs to a P-type ATPase family whose members have been shown to transport metal ions such as cadmium, copper, zinc, cobalt, and lead [39], [40]. Hence, the fitness cost of PCA1-C1 under non-cadmium conditions may result from a depletion of essential metal ions caused by enhanced PCA1 expression. In S. cerevisiae, it has been shown that Pca1 exports cadmium, not copper [28]. However, a study by Adle et al. has also demonstrated that Pca1 affects copper balance by chelating copper ions in a manner analogous to metallothionine [29]. Thus, it is possible that the high expression of PCA1-C1 depletes copper or other unidentified vital metal ions by metal sequestration or by metal exportation.
In nature, S. paradoxus and S. cerevisiae were found to occasionally coexist in the same ecological niches [41]. We have shown that most S. cerevisiae strains have lost the ancestral cadmium-resistant phenotype probably due to its fitness cost. Why do S. paradoxus populations still maintain this phenotype? Using the aforementioned competitive fitness assay, we found that S. paradoxus cells carrying either wild-type or low-expression alleles of Sp-PCA1 exhibit similar fitness under Cd-free conditions (Figure S6). These data suggest that S. paradoxus has evolved other mechanisms to offset the fitness cost of high PCA1 expression.
Evolution of PCA1 in S. cerevisiae populations
In EC-C1 cells, the increase in the expression of PCA1-C1 was caused by three nucleotide substitutions in the promoter that were also shared by the S. paradoxus and S. mikatae PCA1 genes. Horizontal gene transfer between different species of yeast has been observed in previous studies [42], [43]. One possible explanation for the high Cd resistance of EC-C1 strains is that the ancestor of EC-C1 strains acquired a S. paradoxus PCA1 allele through a horizontal gene transfer event, and that the transferred Sp-PCA1 function was reinforced later on by natural selection in EC-C1 strains. If that was the case, we would expect to see that the sequence of PCA1-C1 is more similar to that of Sp-PCA1 than to sequences of PCA1 alleles from other S. cerevisiae strains. Phylogenetic analyses using the PCA1 coding or promoter sequences, however, showed that the distance between Sp-PCA1 and PCA1-C1 is farther than that between Sp-PCA1 and other S. cerevisiae PCA1 alleles, suggesting that PCA1-C1 was not derived from Sp-PCA1 (Figure 3 and Figure S7). Moreover, we can rule out the possibility that gene conversion of a small region between Sc-PCA1 and Sp-PCA1 has occurred in the ancestor of EC-C1 strains. At alignment of PCA1 promoter sequences, we found that even in the region containing the critical nucleotides several nucleotides (−99, −112, −115, −119, −124, and −137) were shared by all S. cerevisiae strains, but did not exist in S. paradoxus strains (Figure 3); it should be noted, however, that these sequences were also conserved in all 28 S. paradoxus strains.
We have shown that, unlike S. paradoxus and S. mikatae, most S. cerevisiae strains except for EC-C1 and two other strains (UWOPS87_2421 and UWOPS83_787_3) are highly sensitive to cadmium. Intriguingly, we found that sequences of the PCA1 promoter in Cd-sensitive strains showed high identity (Table S1A). When population data were analyzed, a dramatic decrease in the frequency of DNA polymorphisms was observed in this region, inconsistent with the phylogenetic relationship observed in both upstream and downstream regions (Figure 4A and Figure S2; Tajima's D = −1.94, p<0.05). This result suggests that the cadmium-sensitive phenotype did not evolve independently in different strains. Instead, it was caused by a selective sweep of a weak PCA1 promoter in S. cerevisiae populations. A selective sweep of nonfunctional aquaporin alleles in S. cerevisiae populations has been reported recently [44]. However, unlike the aquaporin case, the selective sweep of S. cerevisiae PCA1 is mainly caused by a single allele and covers a wider range of populations. In addition, the PCA1 allele involved in the sweep is still functional. In S. cerevisiae populations, only the PCA1 alleles from S288C and W303 have lost the function completely due to a mutation (R970G) in the catalytic domain [28], [29]. Expression of PCA1-SK1 is upregulated when cells sense environmental cadmium and deletion of PCA1 in other Cd-sensitive S. cerevisiae strains makes mutant cells at least 20-fold more sensitive to cadmium.
The reduced cadmium resistance in most S. cerevisiae strains is probably a result of regulatory fine-tuning that allows cells to maintain a certain level of cadmium efflux activity, without compromising their fitness, under normal conditions. Such an ‘optimized’ PCA1 might explain why this weak allele spread so efficiently to different S. cerevisae populations. On the other hand, the EC-C1 strains maintained and even reinforced the ancestral Pca1 activity, probably due to constant selection in their living environments. We measured the soil cadmium concentrations at the collection sites of Evolution Canyon using inductively coupled plasma-atomic emission spectroscopy (see Materials and Methods) and found that they ranged from 2.5 to 4.2 ppm, which is about 17–28-fold higher the median soil cadmium concentration in Europe [45]. It is intriguing that some cadmium-sensitive strains (EC-C2 and EC-C3) were isolated from the same areas as the EC-C1 strains. We found that the PCA1 sequences of EC-C2 and EC-C3 strains are more closely related to those in the European isolates than to those in EC-C1 strains (Figure 4A and Figure S2). One possible explanation is that these cadmium-sensitive strains arrived in Evolution Canyon more recently and have not yet adapted to the environment. An in-depth study combining genomics and population distribution of the EC strains will help us address this issue.
Phenotypic studies in budding yeast have suggested that resistance to various metal ions in different yeast strains is quite diverse [16], [37], [46]. A recent genomic analysis of both promoter and coding regions of three S. cerevisiae strains also indicates that metal ion transporter genes are significantly enriched in the gene group showing signatures of positive selection [47]. Our data with PCA1 provide a clear example how a metal transporter gene evolves after experiencing various types of selection that occurred at both inter - and intra-species levels.
In the present study, we found that mutations in the regulatory and coding regions both contribute to the adaptive phenotype. However, the mutations in the regulatory region have a more profound effect as compared to those in the coding region. Recent studies in a variety of organisms have suggested that regulatory changes are critical for adaptive evolution [46], [48]–[53]. It is possible that the promoter is more flexible to accommodate functional changes since it has less structural constraints than the coding region [54]. Our results showed that, by fine-tuning the PCA1 gene expression, Cd tolerance and cell growth could be dramatically affected in natural yeast isolates, further emphasizing the importance of regulatory changes in evolution.
Materials and Methods
Strains and genetic procedures
EC diploid strains consist of S. cerevisiae collected from an east-west-oriented canyon (Evolution Canyon) at Lower Nahal Oren, Israel [32], [55]. Our strain numbers are the same as the numbers shown in Figure 2 of reference 26. In brief, EC3, 5, 23, 33, 34, 35 and 36 were isolated from the south-facing slope (SFS), EC7, 9, 10, 39, 40, 45 and 48 were isolated from the valley bottom (VB), and EC13, 14, 57, 58, 59, 60 and 63 were isolated from the north-facing slope (NFS). S paradoxus, S. mikatae and other S. cerevisiae strains were obtained from the collections of Dr. Duncan Greig (University College London, UK) and Dr. Edward Louis (University of Nottingham) [37]. Substitutive and integrative transformations were carried out by the lithium acetate procedure [56]. Media, microbial, and genetic techniques were as described [57].
Karyotyping of EC strains
A total amount of ∼2×108 yeast cells was used for plug preparation. Cells were washed with 1 ml EDTA/Tris (50 mM EDTA, 10 mM Tris-HCl, pH 7.5) and transferred into EDTA/Tris containing 0.13 mg/ml zymolyase (Seikagaku America Inc., St. Petersburg, FL). The cell mixtures were incubated for 30 s at 42°C and then embedded in low melting point agarose (Sigma-Aldrich, St. Louis, MO). The agarose plugs were then incubated at 37°C overnight for zymolyase digestion. After digestion, the agarose plugs were placed in LET solution (0.5 M EDTA, 10 mM Tris-HCl (pH 7.5), 2 mg/ml protease K, and 1% N-lauroylsarcosine) at 50°C overnight. This step was repeated three times. The plugs were transferred to EDTA/Tris solution and dialyzed four times for 1 h at 37°C. Yeast chromosomes were separated in 0.7% agarose gels by pulsed-field gel electrophoresis (PFGE) using a Rotaphor Type V apparatus (Biometra, Göttingen, Germany). Electrophoresis was performed for 48 h at 13°C in 0.5× TBE buffer at a fixed voltage of 120 V and an angle of 115°, with pulse time intervals of 30 s.
Genomic DNA library screening
To construct an EC9 genomic DNA library, yeast genomic DNA was extracted using the Qiagen Genomic-Tip 100/G kit (Qiagen, Valencia, CA), digested with restriction enzymes, and ligated into a yeast vector pRS416 as described [57]. To screen for Cd-resistant genes, a Cd-sensitive lab strain (S288C) was transformed with the EC9 genomic DNA library and plated on cadmium-containing plates (0.4 mM CdCl2). Plasmids isolated from the Cd-resistant colonies were sequenced to identify the insert-containing genes.
Functional assay
To measure the intracellular cadmium concentration, log-phase cells carrying different alleles of PCA1 were pretreated with 0.4 mM CdCl2 for 1 h, washed with PBS containing 10 mM EDTA, resuspended in rich medium without cadmium, and then collected at indicated time points. Collected cells were immediately washed with PBS containing 10 mM EDTA. Total intracellular cadmium levels were measured by inductively coupled plasma-atomic emission spectroscopy (ICP-AES).
Quantitative PCR
After cadmium treatment (0.1 mM CdCl2) for 2 h at 28°C, total RNA was isolated from cells using the Qiagen RNeasy Midi Kit (Qiagen, Valencia, CA). First-strand cDNA was synthesized for 2 h at 37°C using the High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems, Foster City, CA). A 20-fold dilution of the reaction products was then subjected to real-time quantitative PCR using gene-specific primers, the SYBR Green PCR master mix, and an ABI-7000 sequence detection system (Applied Biosystems). Data were analyzed using the built-in analysis program.
Reporter assay
To construct the luciferase reporter plasmids, different promoter regions (up to 668 bp upstream of the start codon) of PCA1-C1 and PCA1-SK1 were amplified by PCR. The luciferase coding region (from Renilla reniforms) was also amplified by PCR. The PCR fragments were co-transformed with pRS416 digested with XhoI and SacI into the lab strain S288C. The genomic DNA of Ura+ colonies was isolated and transformed into component E. coli cells. The plasmids from ampicillin-resistant clones were isolated and sequenced. The constructed luciferase reporter plasmids were transformed into an EC9 pca1Δ mutant.
Yeast cells carrying different luciferase reporter plasmids were treated with 0.1 mM of CdCl2 for 2 h. After the treatment, 0.5×107 cells were harvested for detection of the luciferase activity on a luminometer (PE Victor3 luminometer plus autojector, Perkin Elmer, Waltham, MA). To the test samples, 100 µl of 1 µM substrate (coelenterazine) was added. Following a 5 second equilibration time, luminescence was measured with a 10 second integration time.
Competitive relative fitness assay
We measured the fitness of the testing strains by competing them against a reference strain expressing PGK1::GFP in CSM-URA media at 28°C. The testing cells and reference cells were inoculated in the CSM-URA medium individually and acclimated for 24 h. The cells were subsequently diluted and refreshed in new media for another 4 h. The reference and testing cells were then mixed at a 1∶1 ratio, diluted into fresh medium at a final cell concentration of 5×103 cells/ml, and allowed to compete for 21 h, which represents about 12 generations of growth. The ratio of the two competitors was quantified at the initial and final time points using a fluorescence activated cell sorter (FACSCalibur, Becton Dickinson, Franklin Lakes, NJ). Five independent replicates for each fitness measurement were performed.
Phylogenetic tree construction
The evolutionary history of the PCA1 ORF (3651 bps), SUL1 ORF (2580 bps), PCA1 promoter (213 bps) and PCA1-SUL1 intergenic region (820 bps) was inferred using the Neighbor-Joining method [58]. Sequences were obtained from previously released data [37]. The percentage of replicate trees in which the associated taxa were clustered together in the bootstrap test (500 replicates) is shown next to the branches, analogous to a previous study [59]. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method [60] and are expressed as number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). Phylogenetic analyses were conducted in MEGA4 [61]. Tajima's D of the PCA1 promoter (213 bp) was calculated by DnaSP V5 [62].
Measurement of soil cadmium concentrations
Soil samples were collected at 7 locations of Evolution Canyon corresponding to the collection sites of the EC yeast strains (3 at the SFS, one at the VB, and 3 at the NFS). Soil cadmium levels were measured by inductively coupled plasma-atomic emission spectroscopy (ICP-AES) using at least 200 g of individual samples.
Supporting Information
Zdroje
1. NilssonAIZorzetAKanthADahlstromSBergOG
2006
Reducing the fitness cost of antibiotic resistance by
amplification of initiator tRNA genes.
Proc Natl Acad Sci U S A
103
6976
6981
2. TodescoMBalasubramanianSHuTTTrawMBHortonM
2010
Natural allelic variation underlying a major fitness trade-off in
Arabidopsis thaliana.
Nature
465
632
636
3. CowenLESanglardDCalabreseDSirjusinghCAndersonJB
2000
Evolution of drug resistance in experimental populations of
Candida albicans.
J Bacteriol
182
1515
1522
4. SegreAVMurrayAWLeuJY
2006
High-resolution mutation mapping reveals parallel experimental
evolution in yeast.
PLoS Biol
4
e256
doi:10.1371/journal.pbio.0040256
5. BeaumontHJGallieJKostCFergusonGCRaineyPB
2009
Experimental evolution of bet hedging.
Nature
462
90
93
6. BarrickJEYuDSYoonSHJeongHOhTK
2009
Genome evolution and adaptation in a long-term experiment with
Escherichia coli.
Nature
461
1243
1247
7. CooperTFRozenDELenskiRE
2003
Parallel changes in gene expression after 20,000 generations of
evolution in Escherichiacoli.
Proc Natl Acad Sci U S A
100
1072
1077
8. ChouHHBerthetJMarxCJ
2009
Fast growth increases the selective advantage of a mutation
arising recurrently during evolution under metal limitation.
PLoS Genet
5
e1000652
doi:10.1371/journal.pgen.1000652
9. VelicerGJYuYT
2003
Evolution of novel cooperative swarming in the bacterium
Myxococcus xanthus.
Nature
425
75
78
10. GuptaAMatsuiKLoJFSilverS
1999
Molecular basis for resistance to silver cations in
Salmonella.
Nat Med
5
183
188
11. AndersonJB
2005
Evolution of antifungal-drug resistance: mechanisms and pathogen
fitness.
Nat Rev Microbiol
3
547
556
12. HaferburgGKotheE
2007
Microbes and metals: interactions in the
environment.
J Basic Microbiol
47
453
467
13. DeutschbauerAMDavisRW
2005
Quantitative trait loci mapped to single-nucleotide resolution in
yeast.
Nat Genet
37
1333
1340
14. FidalgoMBarralesRRIbeasJIJimenezJ
2006
Adaptive evolution by mutations in the FLO11
gene.
Proc Natl Acad Sci U S A
103
11228
11233
15. SteinmetzLMSinhaHRichardsDRSpiegelmanJIOefnerPJ
2002
Dissecting the architecture of a quantitative trait locus in
yeast.
Nature
416
326
330
16. FayJCMcCulloughHLSniegowskiPDEisenMB
2004
Population genetic variation in gene expression is associated
with phenotypic variation in Saccharomyces cerevisiae.
Genome Biol
5
R26
17. Perez-OrtinJEQuerolAPuigSBarrioE
2002
Molecular characterization of a chromosomal rearrangement
involved in the adaptive evolution of yeast strains.
Genome Res
12
1533
1539
18. GerkeJLorenzKCohenB
2009
Genetic interactions between transcription factors cause natural
variation in yeast.
Science
323
498
501
19. WilsonCJApiyoDWittung-StafshedeP
2004
Role of cofactors in metalloprotein folding.
Q Rev Biophys
37
285
314
20. PierrelFCobinePAWingeDR
2007
Metal Ion availability in mitochondria.
Biometals
20
675
682
21. LiangQZhouB
2007
Copper and manganese induce yeast apoptosis via different
pathways.
Mol Biol Cell
18
4741
4749
22. BruinsMRKapilSOehmeFW
2000
Microbial resistance to metals in the
environment.
Ecotoxicol Environ Saf
45
198
207
23. ValkoMMorrisHCroninMT
2005
Metals, toxicity and oxidative stress.
Curr Med Chem
12
1161
1208
24. ThorsenMPerroneGGKristianssonETrainiMYeT
2009
Genetic basis of arsenite and cadmium tolerance in Saccharomyces
cerevisiae.
BMC Genomics
10
105
25. ShiHShiXLiuKJ
2004
Oxidative mechanism of arsenic toxicity and
carcinogenesis.
Mol Cell Biochem
255
67
78
26. RosenBP
2002
Transport and detoxification systems for transition metals, heavy
metals and metalloids in eukaryotic and prokaryotic
microbes.
Comp Biochem Physiol A Mol Integr Physiol
133
689
693
27. SilverSPhung leT
2005
A bacterial view of the periodic table: genes and proteins for
toxic inorganic ions.
J Ind Microbiol Biotechnol
32
587
605
28. ShiraishiEInouheMJohoMTohoyamaH
2000
The cadmium-resistant gene, CAD2, which is a mutated putative
copper-transporter gene (PCA1), controls the intracellular cadmium-level in
the yeast S. cerevisiae.
Curr Genet
37
79
86
29. AdleDJSinaniDKimHLeeJ
2007
A cadmium-transporting P1B-type ATPase in yeast Saccharomyces
cerevisiae.
J Biol Chem
282
947
955
30. NevoE
1995
Asian, African and European Biota Meet at Evolution-Canyon Israel
- Local Tests of Global Biodiversity and Genetic Diversity
Patterns.
Proceedings of the Royal Society of London Series B-Biological
Sciences
262
149
155
31. GrishkanINevoEWasserSPBeharavA
2003
Adaptive spatiotemporal distribution of soil microfungi in
‘Evolution canyon’ II, Lower Nahal Keziv, western Upper Galilee,
Israel.
Biological Journal of the Linnean Society
78
527
539
32. EzovTKBoger-NadjarEFrenkelZKatsperovskiIKemenyS
2006
Molecular-genetic biodiversity in a natural population of the
yeast Saccharomyces cerevisiae from “Evolution Canyon”:
microsatellite polymorphism, ploidy and controversial sexual
status.
Genetics
174
1455
1468
33. Katz EzovTChangSLFrenkelZSegreAVBahalulM
2010
Heterothallism in Saccharomyces cerevisiae isolates from nature:
effect of HO locus on the mode of reproduction.
Mol Ecol
19
121
131
34. DunhamMJBadraneHFereaTAdamsJBrownPO
2002
Characteristic genome rearrangements in experimental evolution of
Saccharomyces cerevisiae.
Proc Natl Acad Sci U S A
99
16144
16149
35. InfanteJJDombekKMRebordinosLCantoralJMYoungET
2003
Genome-wide amplifications caused by chromosomal rearrangements
play a major role in the adaptive evolution of natural
yeast.
Genetics
165
1745
1759
36. AdleDJLeeJ
2008
Expressional control of a cadmium-transporting P1B-type ATPase by
a metal sensing degradation signal.
J Biol Chem
283
31460
31468
37. LitiGCarterDMMosesAMWarringerJPartsL
2009
Population genomics of domestic and wild yeasts.
Nature
458
337
341
38. RadMRKirchrathLHollenbergCP
1994
A putative P-type Cu(2+)-transporting ATPase gene on
chromosome II of Saccharomyces cerevisiae.
Yeast
10
1217
1225
39. WilliamsLEMillsRF
2005
P(1B)-ATPases–an ancient family of transition metal pumps
with diverse functions in plants.
Trends Plant Sci
10
491
502
40. ArguelloJMErenEGonzalez-GuerreroM
2007
The structure and function of heavy metal transport
P1B-ATPases.
Biometals
20
233
248
41. SniegowskiPDDombrowskiPGFingermanE
2002
Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a
natural woodland site in North America and display different levels of
reproductive isolation from European conspecifics.
FEMS Yeast Res
1
299
306
42. LitiGPeruffoAJamesSARobertsINLouisEJ
2005
Inferences of evolutionary relationships from a population survey
of LTR-retrotransposons and telomeric-associated sequences in the
Saccharomyces sensu stricto complex.
Yeast
22
177
192
43. NovoMBigeyFBeyneEGaleoteVGavoryF
2009
Eukaryote-to-eukaryote gene transfer events revealed by the
genome sequence of the wine yeast Saccharomyces cerevisiae
EC1118.
Proc Natl Acad Sci U S A
106
16333
16338
44. WillJLKimHSClarkeJPainterJCFayJC
2010
Incipient balancing selection through adaptive loss of aquaporins
in natural Saccharomyces cerevisiae populations.
PLoS Genet
6
e1000893
doi:10.1371/journal.pgen.1000893
45. PanJPlantJAVoulvoulisNOatesCJIhlenfeldC
2010
Cadmium levels in Europe: implications for human
health.
Environ Geochem Health
32
1
12
46. KvitekDJWillJLGaschAP
2008
Variations in stress sensitivity and genomic expression in
diverse S. cerevisiae isolates.
PLoS Genet
4
e1000223
doi:10.1371/journal.pgen.1000223
47. LiYDLiangHGuZLinZGuanW
2009
Detecting positive selection in the budding yeast
genome.
J Evol Biol
22
2430
2437
48. FraserHBMosesAMSchadtEE
2010
Evidence for widespread adaptive evolution of gene expression in
budding yeast.
Proc Natl Acad Sci U S A
107
2977
2982
49. FayJCWittkoppPJ
2008
Evaluating the role of natural selection in the evolution of gene
regulation.
Heredity
100
191
199
50. WrayGA
2007
The evolutionary significance of cis-regulatory
mutations.
Nat Rev Genet
8
206
216
51. ChanYFMarksMEJonesFCVillarrealGJrShapiroMD
2010
Adaptive evolution of pelvic reduction in sticklebacks by
recurrent deletion of a Pitx1 enhancer.
Science
327
302
305
52. GompelNPrud'hommeBWittkoppPJKassnerVACarrollSB
2005
Chance caught on the wing: cis-regulatory evolution and the
origin of pigment patterns in Drosophila.
Nature
433
481
487
53. TurnerTLBourneECVon WettbergEJHuTTNuzhdinSV
2010
Population resequencing reveals local adaptation of Arabidopsis
lyrata to serpentine soils.
Nat Genet
42
260
263
54. TiroshIBarkaiNVerstrepenKJ
2009
Promoter architecture and the evolvability of gene
expression.
J Biol
8
95
55. NagornayaSSBabichTVPodgorskyVSBeharavANevoE
2003
Yeast interslope divergence in soils and plants of
“Evolution canyon”, Lower Nahal Oren, Mount Carmel,
Israel.
Israel Journal of Plant Sciences
51
55
57
56. ItoHFukudaYMurataKKimuraA
1983
Transformation of intact yeast cells treated with alkali
cations.
J Bacteriol
153
163
168
57. GuthrieCFinkG
2004
Guide to yeast genetics and molecular and cell biology
San Diego
Elsevier Academic Press
58. SaitouNNeiM
1987
The neighbor-joining method: a new method for reconstructing
phylogenetic trees.
Mol Biol Evol
4
406
425
59. FelsensteinJ
1985
Confidence-Limits on Phylogenies - an Approach Using the
Bootstrap.
Evolution
39
783
791
60. TamuraKNeiMKumarS
2004
Prospects for inferring very large phylogenies by using the
neighbor-joining method.
Proc Natl Acad Sci U S A
101
11030
11035
61. TamuraKDudleyJNeiMKumarS
2007
MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software
version 4.0.
Mol Biol Evol
24
1596
1599
62. LibradoPRozasJ
2009
DnaSP v5: a software for comprehensive analysis of DNA
polymorphism data.
Bioinformatics
25
1451
1452
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and PhosphodiesterasesČlánek Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání