-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNovel Function of as a Gap Gene during Spider Segmentation
Despite many aspects of the regulation of segmentation being conserved among arthropods, the evolution of novel gene functions has played an important role in the evolution of developmental regulation and the emergence of new segmental structures. Moreover the study of such novel gene functions can be informative with respect to the patterns and direction of evolutionary changes in developmental programs. The homeobox gene Distal-less (Dll) is known for its conserved function in appendage development in metazoans. In arthropods, Dll is required for the specification of distal appendage structures. Here we describe a novel and unexpected role of Dll in the spider Achaearanea tepidariorum. We detect At-Dll transcripts not only in the appendages, but unexpectedly also in an anterior domain during early development, prior to the specification of the limb primordia. A similar early Dll domain is present in the distantly related spider Pholcus phalangioides. In A. tepidariorum this early At-Dll expression is required for head segmentation. RNA interference results in spiders that lack either the first or the first and the second walking leg segments. The early At-Dll expression is also required for the activation of the segment polarity genes engrailed and hedgehog in this region. Our work identifies the Distal-less gene as a novel factor in anterior spider segmentation with a gap gene-like function. This novel role of Dll is interesting because Dll expression is reduced in this region in crustaceans and the homologous insect segment, the mandible segment, does not express Dll and does not require this gene for patterning. We therefore discuss the possible implications of our results for understanding the evolution and diversification of the mandible segment.
Published in the journal: . PLoS Genet 7(10): e32767. doi:10.1371/journal.pgen.1002342
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002342Summary
Despite many aspects of the regulation of segmentation being conserved among arthropods, the evolution of novel gene functions has played an important role in the evolution of developmental regulation and the emergence of new segmental structures. Moreover the study of such novel gene functions can be informative with respect to the patterns and direction of evolutionary changes in developmental programs. The homeobox gene Distal-less (Dll) is known for its conserved function in appendage development in metazoans. In arthropods, Dll is required for the specification of distal appendage structures. Here we describe a novel and unexpected role of Dll in the spider Achaearanea tepidariorum. We detect At-Dll transcripts not only in the appendages, but unexpectedly also in an anterior domain during early development, prior to the specification of the limb primordia. A similar early Dll domain is present in the distantly related spider Pholcus phalangioides. In A. tepidariorum this early At-Dll expression is required for head segmentation. RNA interference results in spiders that lack either the first or the first and the second walking leg segments. The early At-Dll expression is also required for the activation of the segment polarity genes engrailed and hedgehog in this region. Our work identifies the Distal-less gene as a novel factor in anterior spider segmentation with a gap gene-like function. This novel role of Dll is interesting because Dll expression is reduced in this region in crustaceans and the homologous insect segment, the mandible segment, does not express Dll and does not require this gene for patterning. We therefore discuss the possible implications of our results for understanding the evolution and diversification of the mandible segment.
Introduction
The genetic regulation of head segmentation in Drosophila melanogaster is one of the best-studied examples of how tissues become specified and patterned during early embryonic development [1], [2]. Moreover, some features of the Drosophila segmentation gene cascade are evolutionarily conserved among other arthropods [3]. However, although the arthropod head itself is morphologically an ancient and evolutionary conserved structure, composed of a protocerebral (or ocular) region followed by a series of five homologous segments [4], [5], there is increasing evidence that the upstream mechanisms regulating head patterning vary among arthropods [3], [6].
The formation of the head in Drosophila melanogaster begins with a maternal and early zygotic gradient system involving the proteins encoded by the bicoid (bcd), hunchback (hb), and orthodenticle (otd) genes [2], [3], [7]–[9]. These gradients provide a coordinate system for a large number of subordinate genes that further subdivide the head epidermis. Thus, the initial gradient system controls the final location of the segmental borders and the correct number of head segments. Surprisingly, although all insect heads have the same segmental composition, neither the bcd gene, nor the role and regulation of hb and otd is conserved in other insect species [3], [6]. It appears, therefore, that the evolutionarily conserved architecture of insect heads is produced by an as yet comparatively unexplained diversity of developmental genetic mechanisms. This phenomenon also extends to other arthropods: recent work in the spider Achaearanea tepidariorum demonstrated novel head formation mechanisms involving dynamic gene expression [10]. Understanding head development in arthropods will thus shed light on the flexibility and evolvability of the gene network underlying the patterning of this ancient and evolutionarily conserved structure.
Head segmentation mechanisms not only differ between species, but between adjacent head segments within a species. Developmental studies in Drosophila have shown that the embryonic head consists of two parts that use different mechanisms for segmentation [1], [11]. The anterior head part (or procephalon) uses a set of head gap genes to establish the location of the segments [7]–[9]. The posterior head part (or gnathocephalon) uses the pair-rule genes for segmentation and thus closely resembles the establishment of the trunk segments [2], [12]. The segment that abuts this border between the two segmentation mechanisms is the mandible segment. It is the anterior-most gnathocephalic segment that (together with the posterior part of the intercalary segment) receives input from both the head gap genes and the pair-rule genes. This integration of two developmental mechanisms is mediated by the gene collier (col; also known as knot (kn)), which acts specifically at the interface of the pro - and gnathocephalic head segmentation mechanisms [13]. Recent studies in spiders have shown that the spider head is also divided into an anterior and a posterior part distinguished by different segmentation mechanisms [10], [14], [15], but these segmentation mechanisms differ substantially from those in Drosophila and other insects. First, the pair-rule gene ortholog hairy is involved in the segmentation of both parts of the spider head. Second, the anterior head region (procephalon) of the spider employs a dynamic wave of gene expression for segmentation [10], a mechanism that is not used for anterior patterning in Drosophila. Establishment of the posterior head region of the spider (gnathocephalon) depends on a gap gene mechanism, more similar to Drosophila [14]. Third, the col gene is not expressed at the interface of the pro - and gnathocephalon in spider embryos [16]. Despite these differences, the spider homolog of the insect mandible segment, the first walking leg segment (L1), also develops at the interface of the two different segmentation mechanisms in the spider head. Thus, although the pro - and gnathocephalic segmentation mechanisms have diverged during spider and insect evolution, the role of the mandible segment as the interface of the two regions of the embryo where these mechanisms act is evolutionarily ancient. How then is the L1 segment generated in spiders?
The homeobox gene Distal-less (Dll) is well known for its evolutionarily conserved role in distal appendage formation [17], [18]. However, we also find that the Dll gene of A. tepidariorum (At-Dll) is expressed unexpectedly early in the L1 region, before the formation of the germ band and the specification of the appendages, and concurrent with the dynamic gene expression in the procephalon. This early expression of Dll is also seen in the spider Pholcus phalangioides, which is only distantly related to A. tepidariorum. We demonstrate in A. tepidariorum that this early expression of Dll in the L1 region is necessary for the specification of the entire L1 segment. Our results show that loss of At-Dll function leads to the complete loss of the L1 segment in adult spiders, and the loss of this segment in the embryo is preceded by the loss of segmentation gene expression and increased cell death. Thus, At-Dll functions similarly to the head gap genes that pattern the procephalic head segments in Drosophila [7], [8], implicating At-Dll as a novel head gap gene in spider head segmentation. Our results suggest that in contrast to the procephalic segments of this spider, the L1 segment is specified in a manner mechanistically similar to procephalic segment specification in Drosophila (i.e. involving the action of statically expressed head gap genes). The gene expression data from the cellar spider P. phalangioides suggest that the head gap gene role is more widely conserved in spiders. It is interesting to note that in insects and other mandibulate arthropods the expression levels of Dll in the homologous segment are reduced [19], [20] and we discuss possible implications of the divergent roles of Dll in this segment for the evolution of the mandible segment in the arthropods.
Results
Unexpected early Dll expression in the spider head
The anterior (“head”) segments of the spider A. tepidariorum are specified very early in embryonic development, at the germ disc stage when the embryo still has radial symmetry, and well before the transition into the bilateral symmetric germ band [10], [14], [21], [22]. The Dll gene of A. tepidariorum is strongly expressed in a static ring in presumptive anterior cells of the L1 region already at early stage 5 (staging after [21]) (Figure 1A). This is surprising because this expression occurs very early in development, long before the specification of the limb primordia, and concurrent with the general anterior-posterior patterning of the head region [10], [14]. The ring of At-Dll expression persists through late stage 5 (Figure 1B) and opens during the transition from germ disc to germ band formation at stage 6 (Figure 1C). Subsequently, as the germ band forms during early stage 7, the ring of expression is transformed into a broad stripe in the L1 segment (Figure 1D, 1E). In addition, a weaker stripe of expression is detected in the second walking leg segment (L2). Expression at later stages is found predominantly in the limb buds and other domains known for Dll homologs in other arthropods, (e.g. [17], [18]) (Figure 1F–1H).
Fig. 1. Expression of Dll in Achaearanea tepidariorum embryos. 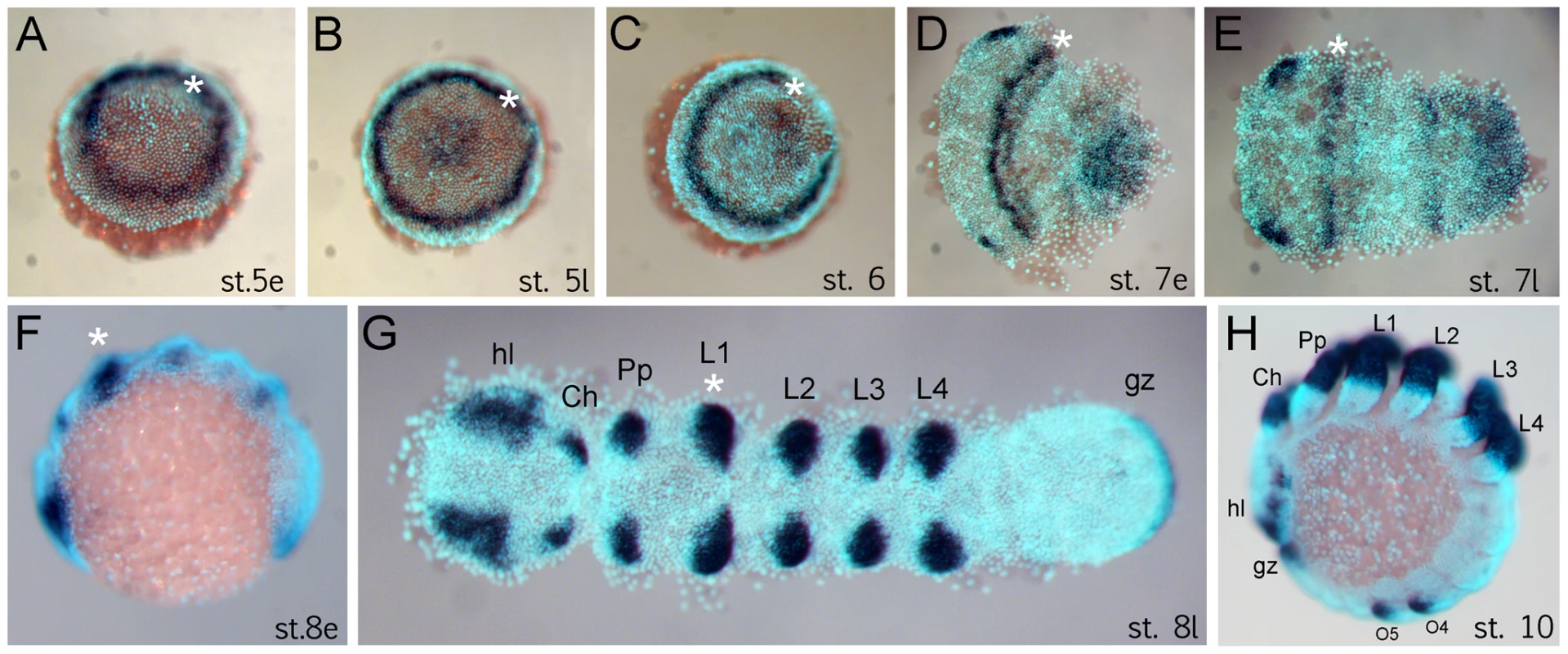
(A, B) Early ring of expression at early stage 5 (A) and late stage 5 (B). (C–E) Formation of the germ band. The Dll ring opens at stage 6 (C) and becomes an anterior stripe in the presumptive L1 region (D, E). (F–H) Late expression of Dll at early stage 8 (F), late stage 8 (G) and stage 10 (H) comprises additional domains in the appendages, head lobes, and growth zone. Abbreviations: Ch, chelicera; gz, growth zone; hl, head lobe; L, walking leg; O, opisthosomal segment; Pp, pedipalp. The asterisk marks the location of the early ring and its derivative head stripe in L1. Dll is required for the formation of a head segment
The unexpected early ring of At-Dll expression in the germ disc suggests that At-Dll plays a role in early head development in A. tepidariorum. We therefore tested the role of At-Dll in early head development using parental RNA interference (RNAi) (Table S1). RNAi led to a down-regulation of At-Dll transcripts in the cytoplasm in the early ring of anterior expression below the level of detection via in situ hybridisation (Figure S1A, S1B). In 64% of embryos, the RNAi effect was permanent, but in 19% the effect was no longer observed after stage 7 and thus led to a partial restoration of At-Dll transcripts in the subsequent stages of development (Figure S1D). In both cases, RNAi resulted in embryos lacking one entire leg-bearing segment, and in a few embryos with permanently down-regulated At-Dll expression, the loss of two entire leg-bearing segments was also observed. As expected, RNAi also affected appendage development resulting in severely truncated limbs in permanently affected embryos (Figure 2A–2C). In the other embryos, the development of the remaining legs was not affected owing to restoration of At-Dll expression in later stages. Importantly, this indicates that the patterning function of the early ring of At-Dll expression is required much earlier than the onset of appendage development and is necessary for segment formation.
Fig. 2. Phenotype of At-Dll RNAi animals. 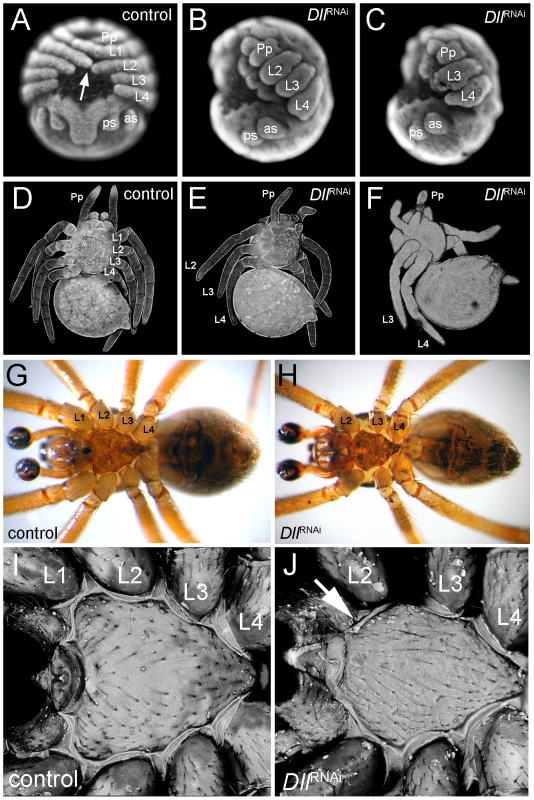
(A–C) Late inversion stage embryos of Achaearanea tepidariorum. In control embryos (A, embryo in a ventral view), the prosomal appendages are fully formed and the distal tips of L1 and L2 touch each other across the ventral midline (arrow). In strongly affected Dll RNAi embryos either L1 (B, embryo in lateral view) or L1 and L2 (C, embryo in lateral view) segments are missing and the remaining appendages are severely truncated. (D–F) Confocal scans of larvae. In control animals (D) all four walking leg segments are present. In Dll RNAi animals one (E) or two (F) leg segments are missing. (G, H) Ventral aspects of adult control male (G) and Dll RNAi male lacking the L1 segment (H). Note the fully sclerotized bulb of the pedipalps (dark colouring) indicating sexual maturity of the males. (I, J) Confocal scan of the sternum of adult control male (I) and Dll RNAi male (J). Note the missing anterior edge corresponding to L1 in the Dll RNAi animal (arrow in J). Abbreviations: L, walking leg; Pp, pedipalp; as, anterior spinneret; ps, posterior spinneret. While the permanently affected embryos did not survive, embryos where only the early At-Dll ring is down-regulated survived to reach the first larval instar. These juvenile spiders had only three pairs of walking legs (Figure 2E) or, in few cases, two pairs of walking legs (Figure 2F). The chelicerae, pedipalps, and the remaining legs were normal. The loss of some walking legs does not cause a discernible gap between the remaining appendages and this indicates that not only the legs, but also the corresponding segments are missing (see also below). The four-legged larvae did not survive, but the six-legged larvae survived and moulted normally and even reached adulthood (Figure 2G, 2H). Thus, under laboratory conditions the loss of a complete body segment does not appear to significantly compromise the viability and feeding behaviour of the animals (Video S1, Video S2).
The L1 segment, the mandible segment homolog, is missing in Dll RNAi animals
In order to establish the identity of the missing walking leg segments, we compared morphological and structural landmarks between wild type and the six-legged adult Dll RNAi spiders. Adult spiders have a thick cuticle plate (the sternum) on the ventral side of their anterior body part (prosoma). The sternum in the wildtype has a characteristic shape with three pointed edges, the anterior most edge being the most pronounced (Figure 2I). In A. tepidariorum, like in other spider species [23], each walking leg segment except for L4 is represented on the surface of the sternum by a characteristic slit sense organ (Figure S2A). The slit sense organ of L1 and L3 is divided into two separate slits, whereas the slit sense organ of L2 comprises a group of three slits. In At-Dll RNAi animals the anterior most edge of the sternum is missing (Figure 2J, Figure S3A, S3B, S3C) and the bipartite slit sensory organ of L1 is lacking (Figure S2B). We conclude that the L1 segment is missing in these animals.
As an additional means of confirming the identity of the missing walking leg segments, we have used the expression of the Hox gene Sex combs reduced (Scr) as a marker [14]. This gene is expressed differentially in the four walking leg segments (L1 to L4) (Figure 3A). It is not expressed in L1, weakly expressed in L2 and L4, and strongly expressed in L3. The expression level of Scr thus allows the four walking leg segments to be distinguished in the embryo. In six-legged At-Dll RNAi embryos, the weakly expressing L2 and L4, and the strongly expressing L3 remain, thus confirming that L1 is missing (Figure 3B). In four-legged At-Dll RNAi embryos only the strongly expressing L3 and the weakly expressing L4 remain, thus confirming that L1 and L2 are missing (Figure 3C). There is no indication of homeotic transformation of the identity of the segments, rather the L1 or L1 and L2 segments, respectively, are absent. These results suggest that the early expression of At-Dll in the germ disc is required for the formation of the L1 and L2 segments and that the lack of early At-Dll expression leads to a gap gene-like phenotype in the anterior body region of A. tepidariorum.
Fig. 3. Expression of Scr in At-Dll RNAi embryos. 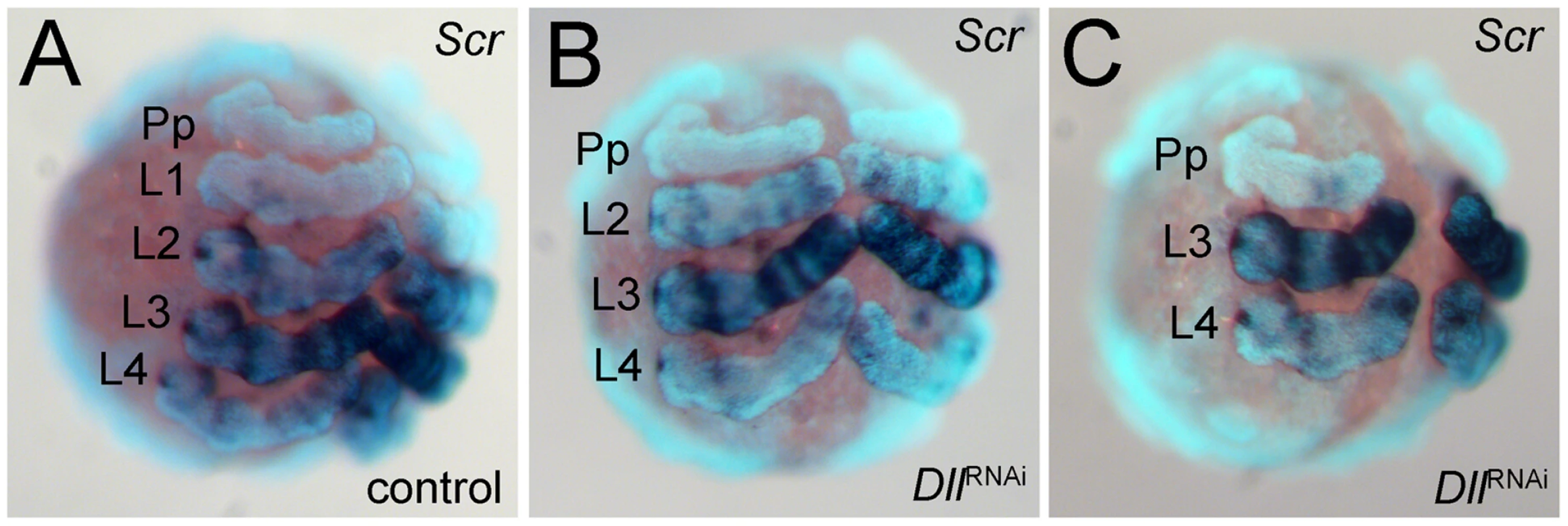
(A–C) Ventral views of stage 10 embryos. The expression levels distinguish the four walking leg segments in control embryos (A) and identify the identity of the remaining leg segments in six-legged Dll RNAi embryos (B) and four-legged Dll RNAi embryos (C). Abbreviations: L, walking leg segment; Pp pedipalpal segment. To understand the mode of segment loss after early At-Dll loss, we have studied the development of the presumptive L1 and L2 region in RNAi embryos. During early germ band stages, when the segments first become morphologically visible, in At-Dll RNAi embryos, the body portion between the pedipalpal and L3 segment is bulged and larger than a single normal segment and likely consists of the cells normally comprising both the L1 and L2 segments (Figure 4A–4C). Cell death (Figure 4A′–4D′), however, does not occur in this bulged segment before late stage 8 when many dead cells were then detected in this segment (Figure 4C′). These data show that the loss of early At-Dll function leads to a malformation of the L1 and L2 segments and that a large portion of the cells in this malformed segment is later removed by cell death. Our results suggest that in the majority of cases the presumptive L2 cells remain and can still form this segment later during development. However, occasionally cell death apparently removes more cells in this region, resulting in the animals lacking both L1 and L2. Thus, the loss of L2 may not be a specific effect of the loss of the early At-Dll ring, but rather a secondary effect of occasional excessive cell death.
Fig. 4. Effects of At-Dll RNAi. 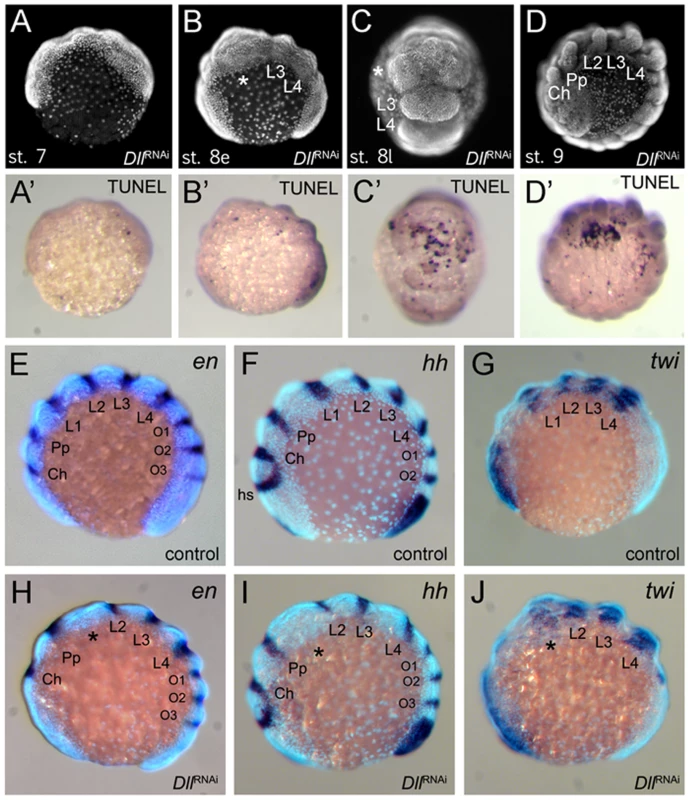
(A–D, A′–D′) Cell death during development of Dll RNAi embryos. (A–D) Morphology of the Dll RNAi embryos at stage 7 (A), early stage 8 (B), late stage 8 (C), and stage 9 (D) photographed under UV light. (A′–D′) These panels correspond to the panels A–D and show the same individuals as the panels A–D, except that the embryos were photographed under white light to document the TUNEL staining. (E–J) L1 specific elements of en and hh expression are deleted in At-Dll RNAi embryos. (E–G) Expression of en (E), hh (F), and twi (G) in control embryos. (H–J) Expression of en (H), hh (I), and twi (J) in Dll RNAi embryos. The asterisk denotes the malformed and bulged segment between pedipalpal and third walking leg segment. Abbreviations: Ch, cheliceral segment; L, walking leg segment; O, opisthosomal segment; Pp, pedipalpal segment. Dll acts as a head gap gene in the spider
The Dll RNAi phenotype in A. tepidariorum is reminiscent of the mutant phenotypes of the head gap genes in Drosophila (e.g. empty-spiracles (ems) and buttonhead (btd)) [7], which also show the loss of anterior segments. One hallmark of these head gap genes is that they are required for the activation of the segment polarity genes (e.g. engrailed (en), hedgehog (hh)) in the particular head segments they specify [7], [8], [24]. We therefore investigated the impact of At-Dll RNAi on segment polarity gene expression in A. tepidariorum. We examined the expression of hh and en, both of which are normally expressed in single stripes in the posterior of each segment (Figure 4E, 4F). In At-Dll RNAi animals the expression of these genes was affected in the bulged segment between the pedipalpal and the L3 segment. The stripes of hh and en normally found in the posterior portion of L2 were still present, but the L1 stripe of both genes was missing (Figure 4H, 4I). These data indicate that the activation of these segmentation genes in the L1 segment directly or indirectly requires At-Dll. The lack of the L1 en and hh stripes in the bulged segment is not due to the absence of the cells that normally express these genes, because cell death in this segment occurs later, as described above (late stage 8; see Figure 4C). Moreover, the absence of en and hh expression does not reflect a general lack of gene expression in this malformed segment because the gene twist (twi), which is a mesoderm marker and not related to ectodermal segmentation or patterning [25] (Figure 4G), is still expressed in this bulged segment (Figure 4J).
Recently, it was found that the gene hunchback (hb) is crucial for the development of anterior segments in A. tepidariorum and is required for the formation of the L1, L2 and L4 segments [14]. Since the phenotypic effect of At-hb RNAi and At-Dll RNAi are similar with respect to the L1 segment and in part to the L2 segment, we reasoned that there might be an interaction between At-hb and At-Dll in the formation of these segments. However, we could not detect any obvious differences in At-hb expression in At-Dll RNAi embryos at the germ disc stage (Figure S4A, S4B), and the early ring of At-Dll expression was also expressed normally in At-hb RNAi embryos at the germ disc stage (Figure S4C, S4D). We conclude from these data that there is no clear or obvious interaction between At-hb and At-Dll during L1 and L2 segment development.
Evidence for a head gap gene role of Dll in Pholcus phalangioides, a haplogyne spider
Our results show that Dll has a role in the formation and development of the L1 segment in A. tepidariorum, but is this a general mechanism for L1 formation in spiders or a novelty of this particular group of spiders like the role of bicoid in higher flies [26]? To answer this question, we studied Dll expression in the cellar spider Pholcus phalangioides (Figure S5A–S5E). The common house spider A. tepidariorum is a member of the Entelegynae that contain the vast majority of extant spiders. P. phalangioides, however, belongs to the Haplogynae, which is the sister group of the Entelegynae (Figure S6). Comparisons between A. tepidariorum and P. phalangioides can therefore reveal conserved features that trace from the common ancestor of entelegyne and haplogyne spiders.
The development of P. phalangioides differs somewhat from that of A. tepidariorum. The early stages of P. phalangioides also form a nearly round germ disc that undergoes a transition from a radial symmetric embryo to an axial symmetric embryo, which results in the formation of the germ band. In A. tepidariorum as well as in P. phalangioides the cumulus migrates from the centre of the germ disc to the rim of the disc, the future dorsal pole of the embryo [19], [27]. In A. tepidariorum the cumulus disappears after its migration, but in P. phalangioides the cumulus persists much longer and is still visible during early stages of germ band elongation (asterisk in Figure S5C). The earliest stages of P. phalangioides embryos that are accessible for in situ hybridisation are comparable to stage 6 or stage 7 embryos of A. tepidariorum. Pp-Dll in these embryos is expressed in an almost closed ring or stripe close to the anterior rim of the germ disc and forming germ band (Figure S5B, S5C). This expression is very similar to the early Dll ring that is observed in A. tepidariorum embryos (Figure S5B), and thus argues for the conservation of early Dll function in entelegyne and haplogyne spiders. The late pattern of Pp-Dll is also virtually identical to the pattern in A. tepidariorum (and other spiders) and includes strong expression in the cheliceres, pedipalps, and walking legs (Figure S5D, S5E).
Discussion
While the last common ancestor of all arthropods is generally accepted to have been segmented and, moreover, had a segmented head, the molecular and genetic mechanisms for trunk segmentation and head segmentation are diverse in extant arthropods [6]. Understanding the genetic basis of head development in arthropods thus provides us with insights into the diversification of the underlying gene regulatory networks that nonetheless specify an evolutionarily conserved structure. Here we describe a novel function of the highly conserved Dll gene in head segmentation in the spider Achaearanea tepidariorum and we present evidence that this function is conserved in a second distantly related spider species, the cellar spider Pholcus phalangioides.
A novel role for Dll as a head gap gene in Achaearanea
Previous results from the spider A. tepidariorum showed how positional values for procephalic segmentation genes are specified via the dynamic expression of the conserved head gene otd [10]. However, segmentation gene expression in the following walking leg segments (that constitute the gnathocephalon homolog of spiders) is static and does not depend on otd expression dynamics [10], [14]. The L1 segment has a special position in the spider head as it lies at the boundary between the procephalon and gnathocephalon that divides dynamic and static segmentation gene expression patterns. Our results identify a novel function of the appendage patterning gene Dll as a head gap gene in this particular head segment. An early static activation of At-Dll in L1 is necessary for the specification and formation of the L1 segment, and is required for the subsequent activation of the segmentation genes in this region. In these respects this early function of At-Dll is similar to the head gap genes that mediate procephalic segmentation in Drosophila, e.g. ems and btd [7], [8]. Remarkably, this head gap gene function of At-Dll represents a novel function of Dll and has not been observed in other arthropod groups. In addition, Dll function in A. tepidariorum is independent of the conserved early head factor hb and might act in parallel with this gene, whereas in Drosophila the head gap genes interact with hb [2], . Together, this indicates that the formation of the L1 segment in A. tepidariorum is mechanistically similar to procephalic segmentation in Drosophila (i.e. involving the action of statically expressed head gap genes), but employs some factors that are not used in segmentation in this insect. Indeed, it is currently unclear to what extent the mode of head development in Drosophila is representative of other arthropods. The specific functions of the maternal coordinate genes (e.g. bcd) are not conserved even within the higher flies (cyclorrhaphan dipterans) [26], [30], and in non-dipteran species the role of bcd appears to be assumed by different factors in different species [3], [6], [31]. The role of the head gap genes is only partially conserved in other insects, because recent results in the beetle Tribolium castaneum show that the btd gene does not have a role in head development [32], and the situation in non-insect arthropods is largely unknown. Thus, paradoxically, the evolutionarily conserved morphological structure of the arthropod head is produced by highly diverse molecular mechanisms. It is generally appreciated that the germ band stage in arthropod development represents the “phylotypic stage” for the phylum and functions as a bottleneck for the diverse developmental mechanism before and after that stage [33], [34]. Indeed, recent work has shown that the transcriptome that is active during the phylotypic stage is phylogenetically ancient and well-conserved [35], [36]. The morphology of the arthropod head is very similar at the germ band stage and thus is an integral part of the arthropod phylotypic stage. It thus appears that so long as the conserved transcriptome of the head is activated at the phylotypic stage, then what mechanisms actually activate this transcriptome does not really matter - hence the divergence in early head patterning mechanisms.
Implications for understanding the evolution of the mandibular segment
Our results also have additional evolutionary implications. As mentioned above, the spider L1 segment is homologous to the insect mandibular segment [4], [5]. This segment in insects is remarkable for its complete lack of Dll expression, but yet produces the insect typical gnathal appendage, the mandible [20]. In other mandibulate arthropods (crustaceans, myriapods) reduced Dll levels are expressed during mandibular development [17], [18], [20], [37]. In contrast, our results show that Dll is broadly expressed in the mandibular segment homolog of spiders and has an important role in the formation of this segment.
The function of Dll has been studied previously in another spider, the Central American wandering spider Cupiennius salei, but no role of Dll in L1 segment formation has been observed [38]. The lack of gap-like phenotypes in C. salei, however, may have resulted from methodological limitations in this species because parental RNAi is not applicable in C. salei [38], and thus an early function of Dll might have been missed in the embryonic RNAi experiments. Indeed, we have performed embryonic RNAi for Dll in A. tepidariorum embryos identical to the RNAi procedure in C. salei, and obtained similar phenotypes as in C.salei: appendage phenotypes, but no gap phenotypes (Figure S7A–S7C). This class of phenotype is absent in parental RNAi experiments in A. tepidariorum where the appendage phenotypes (if present) are always associated with gap phenotypes (Table S1). These data suggest that the RNAi mechanism requires some time to develop the full effect after the application of the dsRNA, and thus only the parental injections provide the effect early enough, whereas the embryonic injections fail to interfere with the early expression of Dll and thus only lead to appendage phenotypes.
Early embryonic stages of C. salei are not accessible for in situ hybridisation, therefore it is not possible to analyse the early expression of Dll in C. salei. However, we could demonstrate that in another spider, the cellar spider P. phalangioides, Dll is expressed in a similar early anterior ring like in A. tepidariorum. P. phalangioides belongs to the Haplogynae, the sister group of the Entelegynae, the group to which Achaearanea belongs and that contains the majority of all extant spider species. These data suggest that the early gap-like function of Dll has already been present in the common ancestor of the entelegyne and haplogyne spiders. The only non-spider chelicerate that has been studied for Dll function is the mite Tetranychus urticae, but no gap-like phenotypes were reported [39]. The results in T. urticae, however, might be explained by the fact that mites show a derived chelicerate body plan with a mite-specific tagmosis of their anterior segments (these are fused into a structure called capitulum) and might therefore also be derived in terms of their anterior developmental mechanisms.
In summary, we present evidence for a gap-like function of Dll in entelegyne and haplogyne spiders in addition to its widely conserved role as an appendage gene. We propose the hypothesis that this dual role of Dll in spiders might have prevented the reduction of Dll expression levels in the spider homolog of the mandibular segment, because this would have led to severe and possibly deleterious phenotypes. By contrast, we propose that in the mandibulates, where Dll has no dual role, the reduction of Dll levels in the mandible segment only led to a distal reduction of the mandibular appendages as can be seen in Crustacea [17], [18], [37]. This novel type of distally reduced appendage has then further diversified also leading to the fully gnathobasic (i.e. lacking all distal elements) insect mandible. In this way, diversification of Dll function may have driven the diversification of mandible segment morphology and facilitated the evolution of the mandible as a tool to tap into new food resources.
Whether the gap-like role of Dll in the L1/mandible segment is the ancestral state for the arthropods or has evolved later in the chelicerate lineage is difficult to be determined in the absence of functional data from outgroups (e.g. onychophorans (velvet worms), tardigrades (water bears)). However, the presence of early Dll expression in the anterior body region of a mollusc [40], a hemichordate [41] and a cnidarian [42] suggests that Dll is ancestrally involved in anterior development, and it has been proposed recently that the majority of the leg patterning genes (including Dll) originally were involved in setting up the anterior-posterior head axis in the bilaterian animals and were only later co-opted for proximal-distal axis formation of the appendages [43].
Materials and Methods
Animal culture and gene cloning
Embryos and adults of A. tepidariorum were obtained from our laboratory stocks in Göttingen. A partial sequence of the A. tepidariorum Dll gene was cloned previously [44]. Additional sequence was obtained by analysing the embryonic transcriptome (sequenced from RNA isolated from stages 1 to 10) and by RACE PCR using the SMART RACE cDNA Amplification Kit (Clontech Laboratories, Inc.) and the following primer sets:
At-Dll-5′RACE-1: GGCCGAGAGTTGTGGGCTGAGGCG;
At-Dll-5′RACE-2: CTGGGCATGGTGGGGCATACCCGC;
At-Dll-3′RACE-1: CTCATCCGTACCTGGGTTCGTATCCGG;
At-Dll-3′RACE-2: CCGGCTGTCCGCCATGTCCATCACC.
The full At-Dll gene sequence has been deposited with the EMBL sequence database under accession number FM876233.
Adult females of P. phalangioides were collected from several cellars in Göttingen, Frankfurt am Main and Munich. Embryos were fixed as described previously [45].
A fragment of the homeobox of the P. phalangioides Dll gene was isolated using the primers eDP-fw, eDP-bw, iDP-fw, and iDP-bw as described previously [46]. RACE-PCR to obtain the full Pp-Dll sequence was performed using the primers Pp-Dll-5′RACE (CCA GTG ATG CGG CGA GTT CGG CTC) and Pp-Dll-3′RACE (GGT TTC AAA GGA CGC AGT ACC TGG CGC). The full Pp-Dll sequence has been deposited with the EMBL sequence database under accession number HE585694.
RNA interference
Parental RNAi was performed as described previously [47] with minor modifications. To exclude off-target effects caused by the dsRNA injections, either the full length At-Dll fragment or a 1260 bp fragment, both including the homeobox, or two not overlapping fragments of 305 bp and 360 bp, both excluding the homeobox, were injected independently of each other into adult spider females. All injections led to identical phenotypes. For the knock-down of hunchback, dsRNA of a 1015 bp fragment was injected and led to the same phenotypes as described previously [14]. Control spiders were injected with water.
Embryonic RNAi was performed in A. tepidariorum by injections into the perivitelline space as described for Cupiennius salei [38]. A. tepidariorum embryos were injected at stage 4, being the earliest stage that has a perivitelline space that is wide enough for injections, thus sticking to the Cupiennius protocol [38] as closely as possible.
In situ hybridisation, DNA labeling, imaging
In situ hybridisation, TUNEL and nuclear staining were performed as described previously [44], [48], [49] with minor modifications. For the TUNEL experiments both negative and positive controls were performed as previously described [49] to confirm the specificity of the obtained stainings (Figure S8A–S8D). Images of spider embryos or adult spiders were either captured with a Zeiss Axioplan-2 microscope or with a Leica dissection microscope equipped with an Intas digital camera and UV-light. Confocal z-stacks of spider larvae and of the sternum of adult spiders were captured using a Zeiss LSM 510. Before scanning the animals were heat fixed, embedded in Voltalef H10S oil and covered with a cover slip.
Supporting Information
Zdroje
1. CohenSMJürgensG 1991 Drosophila headlines. Trends Genet 7 267 272
2. JürgensGHartensteinV 1993 The terminal regions of the body pattern. BateMMartinez AriasA The Development of Drosophila melanogaster Cold Spring Harbor Cold Spring Harbor Laboratory Press 687 746
3. RosenbergMILynchJADesplanC 2009 Heads and tails: Evolution of antero-posterior patterning in insects. Biochim Biophys Acta 1789 333 342
4. DamenWGMHausdorfMSeyfarthEATautzD 1998 A conserved mode of head segmentation in arthropods revealed by the expression pattern of Hox genes in a spider. Proc Natl Acad Sci USA 95 10665 10670
5. TelfordMJThomasRH 1998 Expression of homeobox genes shows chelicerate arthropods retain their deutocerebral segment. Proc Natl Acad Sci USA 95 10671 10675
6. LiuPZKaufmanTC 2005 Short and long germ segmentation: unanswered questions in the evolution of a developmental mode. Evol Dev 7 629 646
7. CohenSMJürgensG 1990 Mediation of Drosophila head development by gap-like segmentation genes. Nature 346 482 485
8. FinkelsteinRPerrimonN 1990 The orthodenticle gene is regulated by bicoid and torso and specifies Drosophila head development. Nature 346 485 488
9. DrieverW 1993 Maternal control of anterior development in the Drosophila embryo. BateMMartinez AriasA The Development of Drosophila melanogaster Cold Spring Harbor Cold Spring Harbor Laboratory Press 301 324
10. PechmannMMcGregorAPSchwagerEEFeitosaNMDamenWGM 2009 Dynamic gene expression is required for anterior regionalization in a spider. Proc Natl Acad Sci USA 106 1468 1472
11. VincentABlankenshipJTWieschausE 1997 Integration of the head and trunk segmentation systems controls cephalic furrow formation in Drosophila. Development 124 3747 3754
12. PankratzMJJäckleH 1993 Blastoderm segmentation. BateMMartinez AriasA The Development of Drosophila melanogaster Cold Spring Harbor Cold Spring Harbor Laboratory Press 467 516
13. CrozatierMValleDDuboisLIbnsoudaSVincentA 1999 Head versus trunk patterning in the Drosophila embryo; collier requirement for formation of the intercalary segment. Development 126 4385 4394
14. SchwagerEEPechmannMFeitosaNMMcGregorAPDamenWGM 2009 hunchback functions as a segmentation gene in the spider Achaearanea tepidariorum. Curr Biol 19 1333 1340
15. Akiyama-OdaYOdaH 2010 Cell migration that orients the dorsoventral axis is coordinated with anteroposterior patterning mediated by Hedgehog signaling in the early spider embryo. Development 137 1263 1273
16. SchaeperNDPechmannMDamenWGMPrpicNMWimmerEA 2010 Evolutionary plasticity of collier function in head development of diverse arthropods. Dev Biol 344 363 376
17. ScholtzGMittmannBGerberdingM 1998 The pattern of Distal-less expression in the mouthparts of crustaceans, myriapods and insects: new evidence for a gnathobasic mandible and the common origin of Mandibulata. Int J Dev Biol 42 801 810
18. PopadicAPanganibanGRuschDShearWAKaufmanTC 1998 Molecular evidence for the gnathobasic derivation of arthropod mandibles and for the appendicular origin of the labrum and other structures. Dev Genes Evol 208 142 150
19. PanganibanGNagyLCarrollSB 1994 The role of the Distal-less gene in the development and evolution of insect limbs. Curr Biol 4 671 675
20. PopadicARuschDPetersonMRogersBTKaufmanTC 1996 Origin of the arthropod mandible. Nature 380 395
21. Akiyama-OdaYOdaH 2003 Early patterning of the spider embryo: a cluster of mesenchymal cells at the cumulus produces Dpp signals received by germ disc epithelial cells. Development 130 1735 1747
22. OdaHAkiyama-OdaY 2008 Differing strategies for forming the arthropod body plan: Lessons from Dpp, Sog and Delta in the fly Drosophila and spider Achaearanea. Develop Growth Differ 50 203 214
23. BarthFGLiberaW 1970 Ein Atlas der Spaltsinnesorgane von Cupiennius salei Keys. Chelicerata (Araneae). Z Morph Tiere 68 343 369
24. MohlerJ 1995 Spatial regulation of segment polarity gene expression in the anterior terminal region of the Drosophila blastoderm embryo. Mech Dev 50 151 161
25. YamazakiKAkiyama-OdaYOdaH 2005 Expression patterns of a twist-related gene in embryos of the spider Achaearanea tepidariorum reveal divergent aspects of mesoderm development in the fly and spider. Zoolog Sci 22 177 185
26. McGregorAP 2005 How to get ahead: the origin, evolution and function of bicoid. BioEssays 27 904 913
27. McGregorAPHilbrantMPechmannMSchwagerEEPrpicNMDamenWGM 2008 Cupiennius salei and Achaearanea tepidariorum: spider models for investigating evolution and development. BioEssays 30 487 498
28. Simpson-BroseMTreismanJDesplanC 1994 Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell 78 855 865
29. WimmerEASimpson-BroseMCohenSMDesplanCJäckleH 1995 Trans - and cis-acting requirements for blastodermal expression of the head gap gene buttonhead. Mech Dev 53 235 245
30. LemkeSBuschSEAntonopoulosDAMeyerFDomanusMH 2010 Maternal activation of gap genes in the hover fly Episyrphus. Development 137 1709 1719
31. SchoppmeierMFischerSSchmitt-EngelCLöhrUKlinglerM 2009 An ancient anterior patterning system promotes caudal repression and head formation in ecdysozoa. Curr Biol 19 1811 1815
32. SchinkoJBKreuzerNOffenNPosnienNWimmerEA 2008 Divergent functions of orthodenticle, empty spiracles and buttonhead in early head patterning of the beetle Tribolium castaneum (Coleoptera). Dev Biol 317 600 613
33. SlackJMHollandPWGrahamCF 1993 The zootype and the phylotypic stage. Nature 361 490 492
34. TautzDSchmidKJ 1998 From genes to individuals: developmental genes and the generation of the phenotype. Philos Trans R Soc Lond B Biol Sci 353 231 240
35. KalinkaATVargaKMGerrardDTPreibischSCorcoranDL 2010 Gene expression divergence recapitulates the developmental hourglass model. Nature 468 811 814
36. Domazet-LosoTTautzD 2010 A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468 815 818
37. PanganibanGSebringANagyLCarrollS 1995 The development of crustacean limbs and the evolution of arthropods. Science 270 1363 1366
38. SchoppmeierMDamenWGM 2001 Double-stranded RNA interference in the spider Cupiennius salei: the role of Distal-less is evolutionarily conserved in arthropod appendage formation. Dev Genes Evol 211 76 82
39. KhilaAGrbicM 2007 Gene silencing in the spider mite Tetranychus urticae: ds RNA and siRNA parental silencing of the Distal-less gene. Dev Genes Evol 217 241 251
40. LeeSEJacobsDK 1999 Expression of Distal-less in molluscan eggs, embryos, and larvae. Evol Dev 1 172 179
41. LoweCJWuMSalicAEvansLLanderE 2003 Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell 113 853 865
42. RyanJFMazzaMEPangKMatusDQBaxevanisAD 2007 Pre-Bilaterian origins of the Hox cluster and the Hox code: evidence from the sea anemone, Nematostella vectensis. PLoS ONE 2 e153 doi:10.1371/journal.pone.0000153
43. LemonsDFritzenwankerJHGerhartJLoweCJMcGinnisW 2010 Co-option of an anteroposterior head axis patterning system for proximodistal patterning of appendages in early bilaterian evolution. Dev Biol 344 358 362
44. PechmannMPrpicNM 2009 Appendage patterning in the South American bird spider Acanthoscurria geniculata (Araneae: Mygalomorphae). Dev Genes Evol 219 189 198
45. PrpicNMSchoppmeierMDamenWGM 2008 Collection and fixation of spider embryos. CSH Protocols 3 930 932 doi:10.1101/pdb.prot5067
46. PrpicNMTautzD 2003 The expression of the proximodistal axis patterning genes Distal-less and dachshund in the appendages of Glomeris marginata (Myriapoda: Diplopoda) suggests a special role of these genes in patterning the head appendages. Dev Biol 260 97 112
47. Akiyama-OdaYOdaH 2006 Axis specification in the spider embryo: dpp is required for radial-to-axial symmetry transformation and sog for ventral patterning. Development 133 2347 2357
48. PrpicNMSchoppmeierMDamenWGM 2008 Whole-mount in situ hybridization of spider embryos. CSH Protocols 3 933 936 doi:10.1101/pdb.prot5068
49. PrpicNMDamenWGM 2005 Cell death during germ band inversion, dorsal closure and nervous system development in the spider Cupiennius salei. Dev Dyn 234 222 228
50. AbzhanovAKaufmanTC 2000 Homologs of Drosophila appendage genes in the patterning of arthropod limbs. Dev Biol 227 673 89
51. CoddingtonJALeviHW 1991 Systematics and evolution of spiders (Araneae). Ann Rev Ecol Syst 22 565 592
Štítky
Genetika Reprodukční medicína
Článek Macroautophagy Is Regulated by the UPR–Mediator CHOP and Accentuates the Phenotype of SBMA MiceČlánek Dynamic Replacement of Histone H3 Variants Reprograms Epigenetic Marks in Early Mouse EmbryosČlánek Mutations in a Guanylate Cyclase GCY-35/GCY-36 Modify Bardet-Biedl Syndrome–Associated Phenotypes in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 10
-
Všechny články tohoto čísla
- Transcriptional Robustness Complements Nonsense-Mediated Decay in Humans
- Identification, Replication, and Fine-Mapping of Loci Associated with Adult Height in Individuals of African Ancestry
- Genetic Determinants of Serum Testosterone Concentrations in Men
- A One Base Pair Deletion in the Canine Gene Causes Exon Skipping and Late-Onset Neuronal Ceroid Lipofuscinosis in the Tibetan Terrier
- Three Structure-Selective Endonucleases Are Essential in the Absence of BLM Helicase in
- Identification of Widespread Ultra-Edited Human RNAs
- Multiple Wnts Redundantly Control Polarity Orientation in Epithelial Stem Cells
- The Bicoid Stability Factor Controls Polyadenylation and Expression of Specific Mitochondrial mRNAs in
- Transcriptome-Wide Binding Sites for Components of the Non-Poly(A) Termination Pathway: Nrd1, Nab3, and Sen1
- Macroautophagy Is Regulated by the UPR–Mediator CHOP and Accentuates the Phenotype of SBMA Mice
- Genetic Rearrangements Can Modify Chromatin Features at Epialleles
- Novel Function of as a Gap Gene during Spider Segmentation
- A Genome-Wide Screen for Interactions Reveals a New Locus on 4p15 Modifying the Effect of Waist-to-Hip Ratio on Total Cholesterol
- Comparative Genomic Analysis of Human Fungal Pathogens Causing Paracoccidioidomycosis
- Genetic Diversity in Cytokines Associated with Immune Variation and Resistance to Multiple Pathogens in a Natural Rodent Population
- Mutator Suppression and Escape from Replication Error–Induced Extinction in Yeast
- Dynamic Replacement of Histone H3 Variants Reprograms Epigenetic Marks in Early Mouse Embryos
- A Barcode Screen for Epigenetic Regulators Reveals a Role for the NuB4/HAT-B Histone Acetyltransferase Complex in Histone Turnover
- HIF–VEGF Pathways Are Critical for Chronic Otitis Media in and Mouse Mutants
- A Conserved Developmental Patterning Network Produces Quantitatively Different Output in Multiple Species of Drosophila
- Role of Exonic Variation in Chemokine Receptor Genes on AIDS: Association with Pneumocystis Pneumonia
- Whole-Exome Sequencing Identifies Homozygous Mutations in a Spastic Ataxia-Neuropathy Syndrome Linked to Mitochondrial -AAA Proteases
- Von Hippel-Lindau () Inactivation in Sporadic Clear Cell Renal Cancer: Associations with Germline Polymorphisms and Etiologic Risk Factors
- A Systems Biology Approach Reveals the Role of a Novel Methyltransferase in Response to Chemical Stress and Lipid Homeostasis
- Identification of Genomic Regions Associated with Phenotypic Variation between Dog Breeds using Selection Mapping
- Global Mapping of Cell Type–Specific Open Chromatin by FAIRE-seq Reveals the Regulatory Role of the NFI Family in Adipocyte Differentiation
- Natural Selection Affects Multiple Aspects of Genetic Variation at Putatively Neutral Sites across the Human Genome
- MicroRNA Expression and Regulation in Human, Chimpanzee, and Macaque Brains
- An Adaptive Allelic Series Featuring Complex Gene Rearrangements
- Feed-Forward Microprocessing and Splicing Activities at a MicroRNA–Containing Intron
- Developmental Stability: A Major Role for in
- A Phenomics-Based Strategy Identifies Loci on , , and Associated with Metabolic Syndrome Phenotype Domains
- Association of , , , , and with Systemic Lupus Erythematosus
- Small RNAs Prevent Transcription-Coupled Loss of Histone H3 Lysine 9 Methylation in
- Successive Increases in the Resistance of to Viral Infection through a Transposon Insertion Followed by a Duplication
- Mutations in a Guanylate Cyclase GCY-35/GCY-36 Modify Bardet-Biedl Syndrome–Associated Phenotypes in
- The Glycobiome Reveals Mechanisms of Pentose and Hexose Co-Utilization in Bacteria
- Insights into Hox Protein Function from a Large Scale Combinatorial Analysis of Protein Domains
- Mutations Cause Seckel and Jawad Syndromes
- Zelda Binding in the Early Embryo Marks Regions Subsequently Activated at the Maternal-to-Zygotic Transition
- Temporal Coordination of Gene Networks by Zelda in the Early Embryo
- Genetic Interaction between MTMR2 and FIG4 Phospholipid Phosphatases Involved in Charcot-Marie-Tooth Neuropathies
- Oxr1 Is Essential for Protection against Oxidative Stress-Induced Neurodegeneration
- Transforming Growth Factor β Receptor Type 1 Is Essential for Female Reproductive Tract Integrity and Function
- Positional Cloning of a Type 2 Diabetes Quantitative Trait Locus; , a Negative Regulator of Insulin Secretion
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Glycobiome Reveals Mechanisms of Pentose and Hexose Co-Utilization in Bacteria
- Global Mapping of Cell Type–Specific Open Chromatin by FAIRE-seq Reveals the Regulatory Role of the NFI Family in Adipocyte Differentiation
- Genetic Determinants of Serum Testosterone Concentrations in Men
- MicroRNA Expression and Regulation in Human, Chimpanzee, and Macaque Brains
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



