-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaInsights into Hox Protein Function from a Large Scale Combinatorial Analysis of Protein Domains
Protein function is encoded within protein sequence and protein domains. However, how protein domains cooperate within a protein to modulate overall activity and how this impacts functional diversification at the molecular and organism levels remains largely unaddressed. Focusing on three domains of the central class Drosophila Hox transcription factor AbdominalA (AbdA), we used combinatorial domain mutations and most known AbdA developmental functions as biological readouts to investigate how protein domains collectively shape protein activity. The results uncover redundancy, interactivity, and multifunctionality of protein domains as salient features underlying overall AbdA protein activity, providing means to apprehend functional diversity and accounting for the robustness of Hox-controlled developmental programs. Importantly, the results highlight context-dependency in protein domain usage and interaction, allowing major modifications in domains to be tolerated without general functional loss. The non-pleoitropic effect of domain mutation suggests that protein modification may contribute more broadly to molecular changes underlying morphological diversification during evolution, so far thought to rely largely on modification in gene cis-regulatory sequences.
Published in the journal: . PLoS Genet 7(10): e32767. doi:10.1371/journal.pgen.1002302
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002302Summary
Protein function is encoded within protein sequence and protein domains. However, how protein domains cooperate within a protein to modulate overall activity and how this impacts functional diversification at the molecular and organism levels remains largely unaddressed. Focusing on three domains of the central class Drosophila Hox transcription factor AbdominalA (AbdA), we used combinatorial domain mutations and most known AbdA developmental functions as biological readouts to investigate how protein domains collectively shape protein activity. The results uncover redundancy, interactivity, and multifunctionality of protein domains as salient features underlying overall AbdA protein activity, providing means to apprehend functional diversity and accounting for the robustness of Hox-controlled developmental programs. Importantly, the results highlight context-dependency in protein domain usage and interaction, allowing major modifications in domains to be tolerated without general functional loss. The non-pleoitropic effect of domain mutation suggests that protein modification may contribute more broadly to molecular changes underlying morphological diversification during evolution, so far thought to rely largely on modification in gene cis-regulatory sequences.
Introduction
How the diversity of animal body plans is established remains a central question in developmental and evolutionary biology [1], [2]. A key step towards understanding the molecular basis underlying diversity is to decipher mechanisms controlling proper genome expression, and how variations in these mechanisms have been at the origin of developmental and evolutionary diversity. While a large number of studies have focused on the impact of cis-regulatory sequences organization (reviewed in [3]), deciphering the intrinsic functional organization of trans-acting transcription factors remains largely unaddressed. Studies have identified functional domains ([4]–[9] and [7], [10], [11] for reviews), but how different protein domains jointly and collectively act for defining the overall activity has been poorly assessed. Yet, a recent study highlights that the synthetic shuffling of protein domains within proteins of the yeast-mating signaling pathway results in the diversification of the mating behavior, demonstrating the importance of protein domain interactions for functional diversification [12].
Hox genes, which encode homeodomain (HD)-containing transcription factors, provide a suitable paradigm to decipher how function is encoded within protein sequence, and how associated changes may constitute the origin of functional specification and diversification. Hox genes have arisen from duplication events of ancestral genes, followed by sequence divergence that promoted the emergence of up to 14 paralogous groups in vertebrates. Hox paralogue proteins display distinct regulatory functions, promoting axial morphological diversification in all bilaterian animals [13]–[17]. Previous work has established that sequence changes in the HD, the DNA binding domain, and a few additional protein domains, have played a major role in the diversification of Hox protein function [4]–[9], [18]–[21]. However, how protein domains functionally interact to shape overall protein activity remains elusive.
We focused on three protein domains from the Drosophila central Hox paralogue protein Abdominal (AbdA, Figure 1). These domains are related by their demonstrated or potential involvement in the recruitment of the Extradenticle (Exd) cofactor, homologous to vertebrate PBX proteins, known to have key roles in establishing Hox functional specificity. The first domain, known as hexapeptide (HX) or PID (Pbx Interacting Domain), with a core YPWM sequence, is found in all Hox paralogue groups, with the exception of some posterior Hox proteins. Biochemical, structural and functional studies have shown that this motif mediates interaction with the Exd/PBX class of Hox cofactors (collectively referred as PBC). The second domain, termed UbdA (UA) is specifically found in the central Hox proteins AbdA and Ultrabithorax (Ubx). This paralogue-specific domain was recently shown to be required for Exd recruitment in the repression of the limb-promoting gene Distalless (Dll) [8], [22]. The third domain (TD), similar in sequence (TDWM) to the YPWM motif, is also paralogue-specific. The TD motif retains the W that provides strong contact with the PBC class proteins, and matches the sequence of the HX motif in some Hox proteins (eg., Hoxa1). Evidence for an Exd recruiting role of the TD domain in AbdA however remains to be demonstrated.
Fig. 1. Combinatorial analyses of AbdA protein domains using a variety of biological readouts. 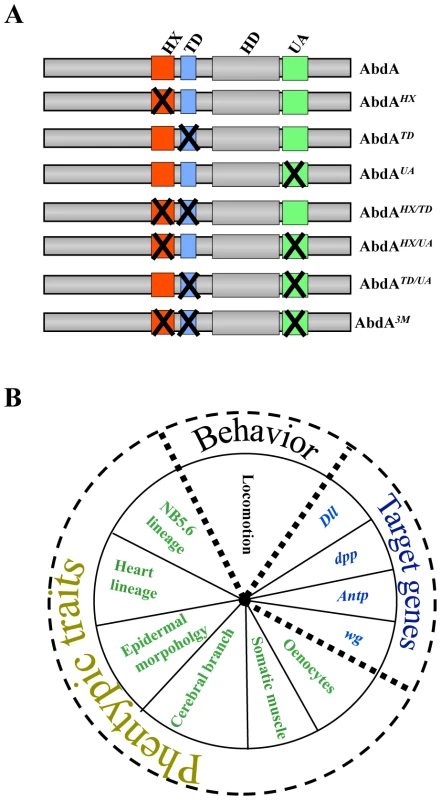
A. AbdA protein variants generated. Crosses indicate domain mutations. B. Biological readouts used in this study. To start unraveling how protein domains collectively shape Hox protein activity, the effect of single, combined double or triple domain mutations were analyzed using most known AbdA functions as biological readouts. The large functional window covered by the study allows identifying functional attributes of protein domains taken in isolation and collectively, and a quantitative analysis by hierarchical clustering highlights the functional organization of the Hox protein AbdA. Given the phylogeny of the studied protein domains, the work has also implication regarding the mechanisms underlying the evolution of AbdA protein function.
Results
Expression of AbdA variants and biological readouts
AbdA variants bearing single or all possible combinations of protein domain mutations (Figure 1A) were ectopically expressed through the binary UAS-Gal4 expression system [23]. Protein levels following induced expression were quantified and experimental conditions ensuring levels close to that of endogenous AbdA were selected (see Materials and Methods). Impact of AbdA variants on target gene control, phenotypic traits and locomotion behavior (Figure 1B), covering AbdA functions of increasing complexity in different tissues, were evaluated in the anterior region where the endogenous AbdA protein is absent. Quantified results (see Text S1) are presented as loss (and in few cases as gain) of regulatory potential.
Eleven functional assays were used to assess domain requirements for AbdA activity (Figure 1B). Four assays rely on the regulation of AbdA target genes, for which evidence of a direct regulation has been previously reported, including the regulation of Distalless (Dll) [8], [24], [25] and Antennapedia (Antp) [26] in the epidermis, and the regulation of wingless (wg) [27] and decapentaplegic (dpp) [28], [29] in the visceral mesoderm. Six assays rely on analysis of phenotypic traits. One of these phenotypic trait, oenocyte specification, results from the regulation of a single target gene [30]. Others, cerebral branch [31], somatic muscles [32], A2 epidermal morphology [33], [34], neuroblast [35], [36] and heart cell lineage specification [37] likely depend of the coordinated regulation of several target genes. Finally, we also used a behavioral trait, larval locomotion, thought to rely on integrated AbdA function in two distinct tissues, the somatic musculature and the nervous system [38].
Dispensability of the HX, TD, and UA protein domains for somatic muscle specification
In the somatic musculature, the abdominal specific pattern is characterized by the presence of muscle located ventrally and absent in thoracic segments, a feature that can be visualized by the expression of nautilus (nau) [32]. This distinction was previously shown to result, at least in part, from the activity of AbdA [32]. Accordingly, anterior ectopic expression of AbdA using the mesodermal driver (24B-Gal4) results in ectopic ventral expression of Nau in anterior segments (Figure 2). We found however that none of the AbdA protein domains under study, alone or in combination, was required to specify the abdominal specific features of the somatic musculature (Figure 2 and Figure S1). In the same conditions, a point mutation at position 50 of the homeodomain that impairs AbdA binding to DNA resulted in the loss of Nau inducing capacity (Figure 2 and Figure S1). The dispensability of the HX, TD and UA domains for specifying abdominal features of somatic muscle pattern is consistent with the fact that nau activation by AbdA is not dependent upon Exd activity [32], although results below argue that these domains assume other functions than Exd recruitment.
Fig. 2. Dispensability of HX, TD, and UA protein domains for somatic muscle specification. 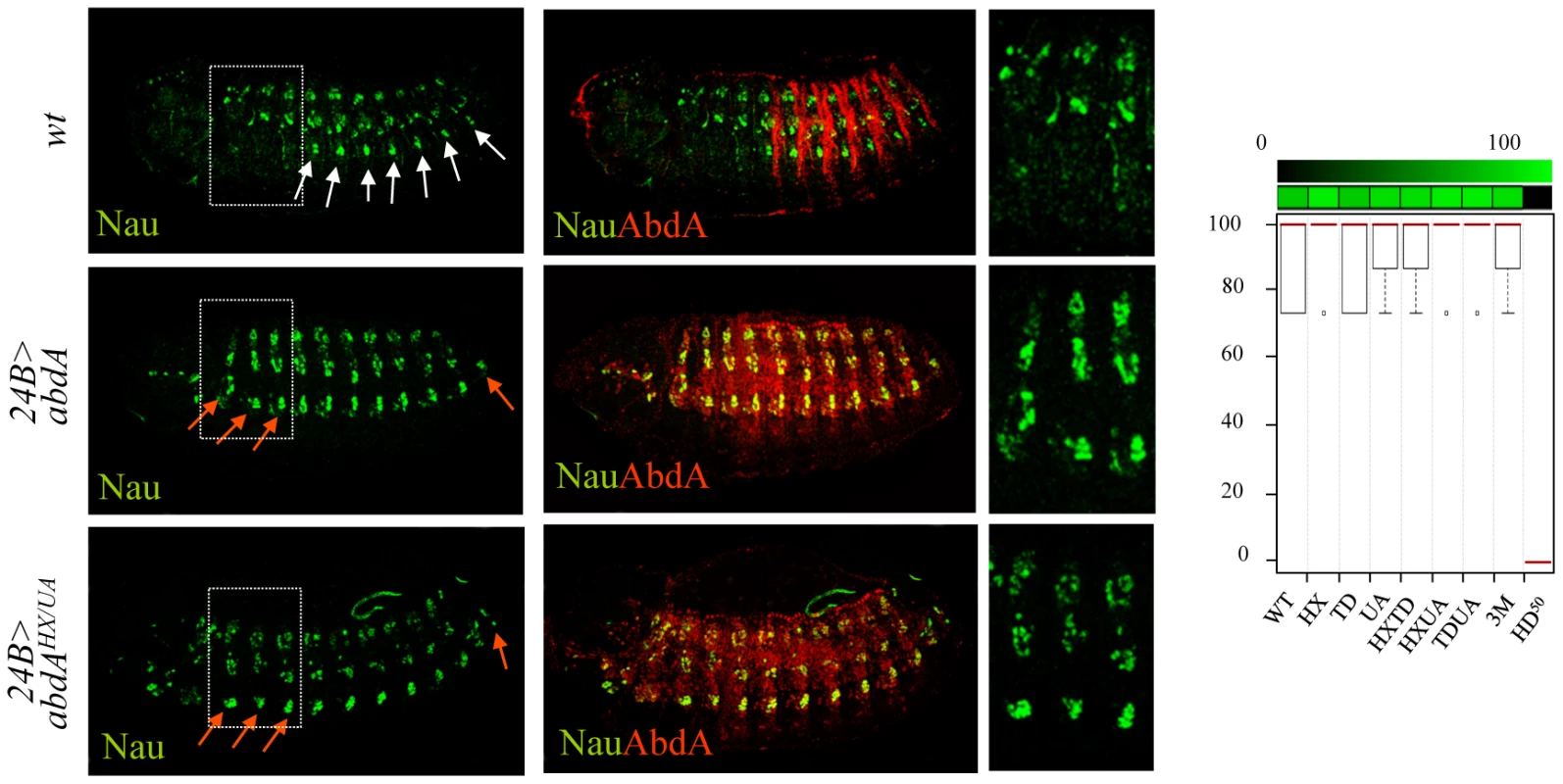
Somatic muscles are visualized by Nautilus (Nau, green) expression (upper panels). Expression of AbdA (red) in the thorax using the 24B-Gal4 driver induces abdominal specific muscle pattern (white arrows) in thoracic segments (red arrows) (middle panels). The effect of the AbdAHX,UA variant is illustrated (lower panels). Right panels are magnifications of boxed areas. Graphs (% of remaining activities compared to the wild type AbdA protein (WT) following domain mutations) using the boxplot representation on the right summarize quantitative analyses (see Text S1 and Figure S1 for full illustration). A graded color-coded bar above the graphs illustrates the level of protein activity, ranging from light green (full activity) to black (no activity). Single protein domain requirement for NB5–6 CNS lineage specification
In the embryonic central nervous system, a subset of 30 neuroblasts (NB's) found in each hemisegment, including the NB5–6, generate a larger lineage in the thorax than in the abdomen. Recent studies demonstrated that posterior Hox genes, such as abdA, impose in the abdomen a smaller NB5–6 lineage by triggering an early cell cycle exit [39]. Misexpression of AbdA within NB5–6 in the thorax using ladybird(K)-Gal4 result in an early lineage truncation, mimicking the situation that normally occurs in the abdomen, ultimately leading to a smaller thoracic NB5–6 lineage size (Figure 3). Average number of NB5–6 cells in wild type thoracic and abdominal segments was previously estimated at 16 and 6 cells respectively: these values were considered as references for full (100%) or complete loss (0%) of repressive activities of AbdA variants on NB5–6 lineage. Intermediate repressive levels upon ectopic expression with ladybird(K)-Gal4 were deduced from the quantification of NB5–6 lineage cell numbers in thoracic segments T2/3 (see methods). Results obtained indicate that lineage truncation triggered by AbdA is similarly affected following UA, HX/UA, TD/UA and HX/TD/UA mutations (Figure 3), which can be best explained by a unique requirement of the UA domain for AbdA function.
Fig. 3. Single protein domain requirement for NB5–6 lineage truncation. 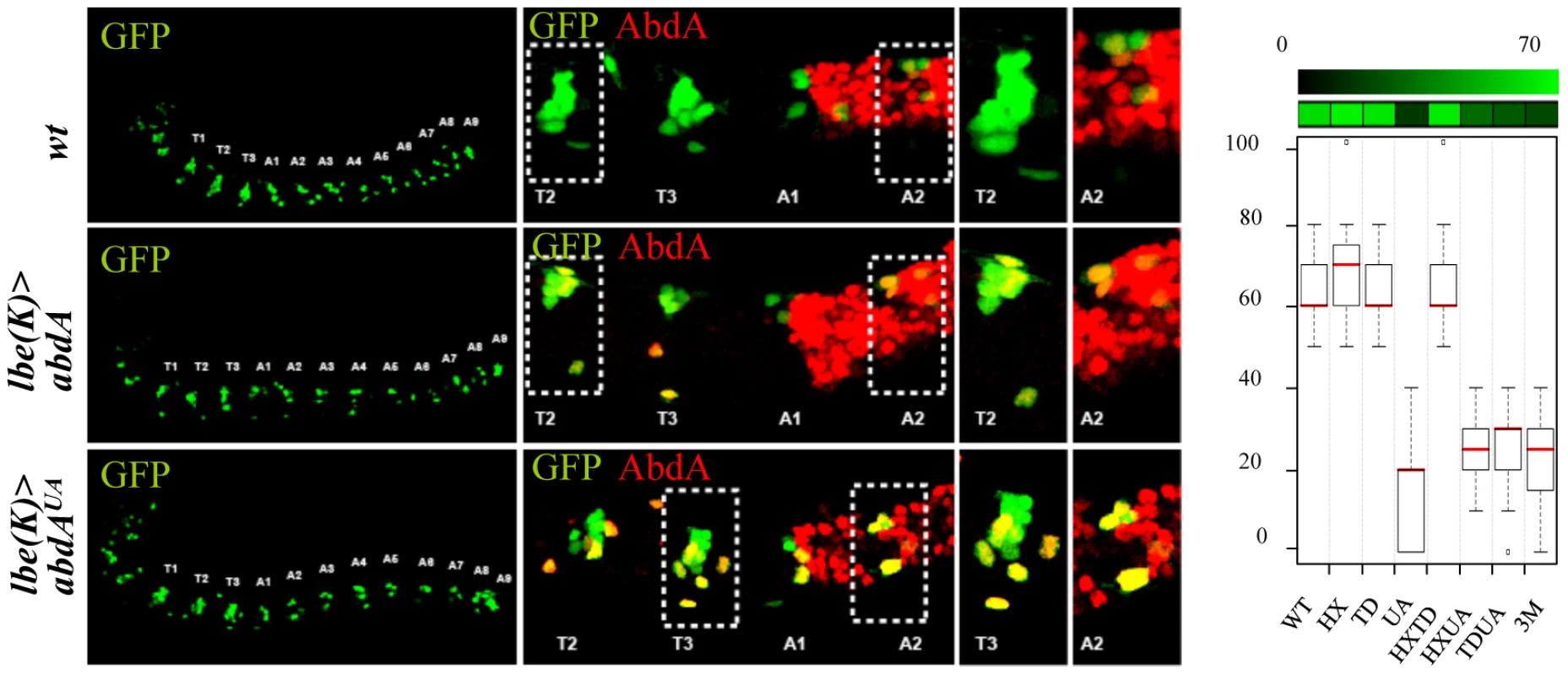
Neuroblast 5–6 are visualized using the ladybird early lbe(K)-Gal4 driver expressing nuclear GFP (green; upper panels). Dotted rectangles highlight differences in cell number of the T2/3 thoracic NB5–6 lineage. Expression of AbdA (red) in NB5–6, through the lbe(K)-Gal4 driver, leads to thoracic lineage truncation (middle panels). The effect of AbdAUA is illustrated in the lower panel. Graphs (% of remaining activities compared to the wild type AbdA protein (WT) following domain mutations) using the boxplot representation on the right summarize quantitative analysis (see Text S1). A graded color-coded bar above the graphs illustrates the level of protein activity, ranging from light green (full activity) to black (no activity). Protein domain mutations induces neomorphic activity in the regulation of the dpp and wg target genes
In the visceral mesoderm, AbdA is expressed in parasegment (PS)8–12. The target genes wg and dpp are respectively activated (in PS8) and repressed (in PS8–12) by AbdA in the visceral mesoderm. Restricted (PS8) activation of wg by AbdA results from the action of the Dpp signal, locally produced by PS7 cells under the control of the Ubx protein [40]. Accordingly, anterior ectopic expression of AbdA only results in a mild activation of wg, as activation only occurs in cells experiencing partial repression of dpp [27]. Previous work has shown that the HX mutation results in a protein that activates dpp instead of repressing it, and consequently more efficiently activates wg [41].
AbdA variants were expressed with the 24B-Gal4 driver. Levels of regulatory activities were deduced following fluorescent in situ hybridization against dpp or wg in the visceral mesoderm of stage 14 embryos in PS1–PS7, ie anterior to endogenous AbdA expressing cells (PS8–12; Figure 4 and Figures S2 and S3). Arbitrary values have been assigned to regulatory activities of AbdA variants. For dpp (Figure 4A and Figure S2), no effect on dpp expression was scored by 0, normal repression of dpp expression in PS7 by 100 (partial repression was never observed) and ectopic activation (instead of repression) of dpp was scored by negative values (depending of the number of ectopic sites (see Text S1). For wg, in a manner similar to dpp, no effect was scored by 0, and positive and negative values were respectively assigned to normal (activation) or abnormal (repression) activities on wg expression (Figure 4B and Figure S3; see Text S1).
Fig. 4. Protein domain mutations inducing neomorphic activities. 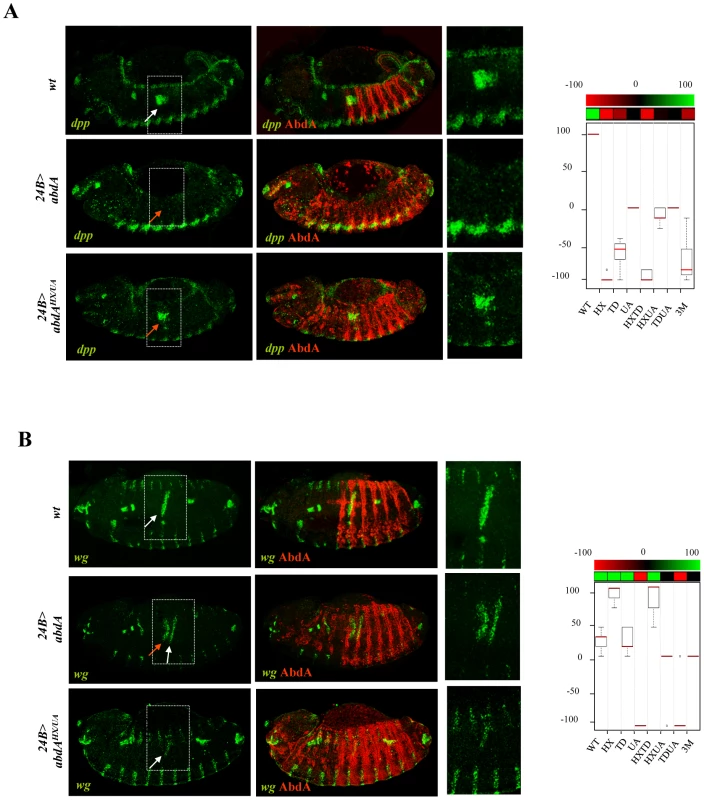
A. Localized PS7 dpp expression (green) in the visceral mesoderm (in situ hybridization, white arrow) relies on posterior repression by AbdA (upper panels). Ubiquitous mesodermal (24B-Gal4 driven) expression of AbdA (red) represses dpp expression in PS7 (red arrow; middle panels) B. Localized PS8 expression of wg (green) in the visceral mesoderm (in situ hybridization, white arrow) relies on activation by AbdA. Ubiquitous AbdA in the mesoderm (24B-Gal4) induces anterior ectopic wg expression (red arrow; middle panels). In A and B, the effect of the AbdAHX,UA variant on dpp (A) or wg (B) expression is illustrated (lower panels). Right panels are magnifications of the boxed areas. Graphs (% of remaining activities compared to the wild type AbdA protein (WT) following domain mutations) using the boxplot representation on the right summarize quantitative analyses (see Text S1 and Figure S2 (dpp) and S3 (wg) for full illustration). A graded color-coded bar above the graphs illustrates the level of protein activity, ranging from light green (full activity) to black (no activity). Neomorphic activities, ie qualitative changes in protein activity, are depicted in red. Results obtained allow two conclusions. First, single domain mutations result in strong modification of AbdA activity. Second, domain mutations often result not only in a quantitative, but also in a qualitative (neomorphic) modification of activity, changing AbdA from an activator to a repressor, or reversely from a repressor to an activator.
Additive contribution of protein domains for oenocyte specification
Oenocytes form under AbdA control in segments A1–A7. This occurs through AbdA-dependent activation of Rhomboid (Rho) in a chordotonal organ precursor cell called C1. Expression of Rho then enables the secretion of the EGF ligand Spitz that will instruct neighboring epidermal cells to differentiate into oenocytes [30]. In absence of AbdA, the EGF pathway is not locally activated and oenocytes are not specified [30]. Reversely, ectopic expression of AbdA induces oenocytes in thoracic segments.
AbdA variants were ubiquitously expressed with the armadillo (arm)-Gal4 driver. Oenocyte inducing potential of AbdA variants, visualised with the seven-up (svp)-lacZ enhancer trap reporter construct, was deduced from the number of thoracic segments that contain ectopic oenocytes (see Text S1). This inductive potential is reduced following single mutations of the UA domain and combined mutation of the HX/TD or TD/UA domains, and is abolished following HX/UA and HX/TD/UA mutations (Figure 5 and Figure S4). These observations suggest an additive contribution of the HX, TD and UA protein domains for oenocyte induction by AbdA, consistent with protein domains acting independently of each other, and contributing uniquely through additive contribution to protein activity.
Fig. 5. Additive contribution of protein domains. 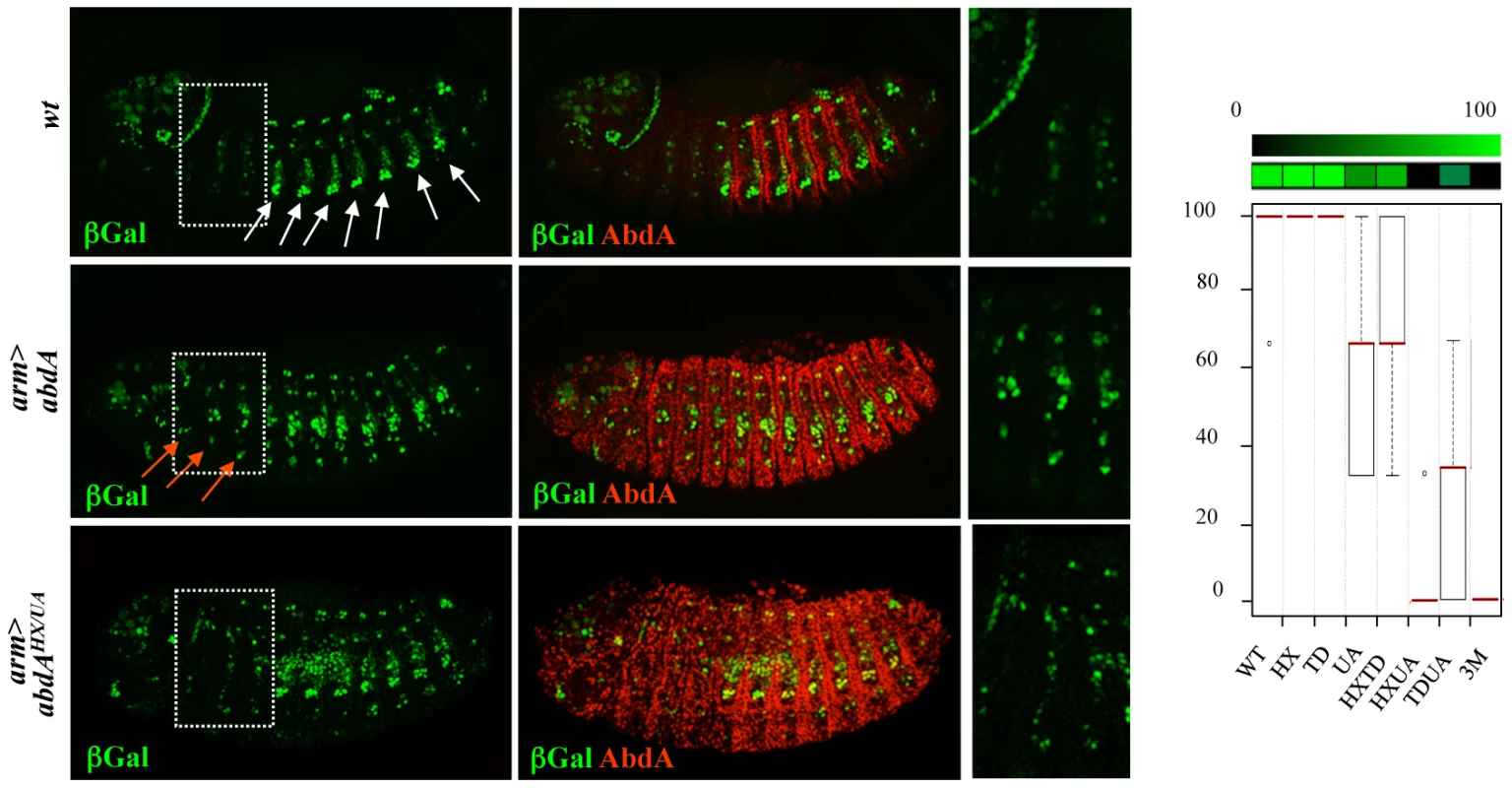
Oenocytes ventrally located in the abdomen are visualized by β-gal (green) driven by the seven-up (svp) promoter (white arrows, upper panel). Expression of AbdA (red) in the thorax through the arm-Gal4 driver induces ectopic oenocytes in the thorax (red arrows, middle panels). The effect of the AbdAHX/UA variants is illustrated (lower panels). Boxed areas highlight thoracic segments. Right panels are magnifications of the boxed areas. Graphs (% of remaining activities compared to the wild type AbdA protein (WT) following domain mutations) using the boxplot representation on the right summarize quantitative analyses (see Text S1 and Figure S4 for full illustration). A graded color-coded bar above the graphs illustrates the level of protein activity, ranging from light green (full activity) to black (no activity). Functional redundancy in protein domain usage for trachea and heart lineage specification
The tracheal cerebral branch forms dorsally exclusively in the second thoracic segment T2, in response to repressive activities of Bithorax Hox proteins in T3-A8 segments [42]. This phenotypic trait can be followed by a breathless (btl) driven GFP reporter that extends posteriorly in the absence of Bithorax complex genes, and that is suppressed in T2 following Btl-driven expression of AbdA in the tracheal system (Figure 6A). Only full repression of cerebral branches was considered and repressive activities of AbdA variants thus correspond to either 0% (no repression) or 100% (full repression) (see Text S1). We found that the repression of the cerebral branch by AbdA is impaired following TD/UA and HX/TD/UA but not HX/UA or HX/TD mutations, revealing a functional redundancy between the TD and UA domains (Figure 6A, and Figure S5).
Fig. 6. Functional redundancy of the HX, TD, and UA protein domains. 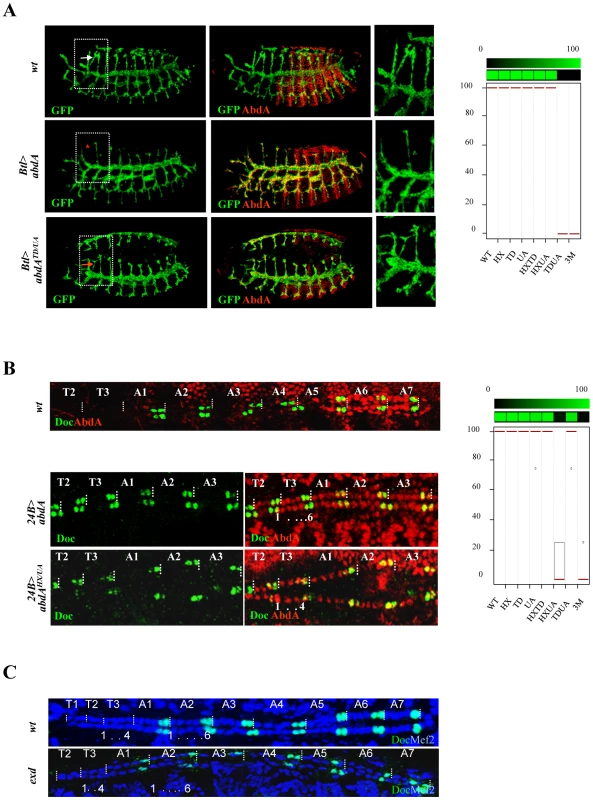
A. Tracheal branches are visualized by GFP-driven through the breathless btl-Gal4 driver (green). The cerebral branch (white arrowhead) forms only in T2 as a result of abdominal repression mediated by AbdA (upper panels). Expression of AbdA (red) in thoracic segments through the btl-Gal4 driver suppresses cerebral branch formation (red star, middle panels). The effect of the AbdATD/UA variants is illustrated (arrow, lower panels). Right panels are magnifications of boxed thoracic areas. B. Double immunostaining for AbdA (red) and Doc1 (green) in wild type embryo (upper panel). Magnifications of thoracic segments T2/T3 and abdominal segments A1–3 (middle and lower panels). Expression of AbdA in thoracic segments through the 24B-Gal4 driver promotes a six cell lineage state, with the two anterior most cells expressing Doc1 (middle panels). The effect of the AbdAHX/UA variants is illustrated (lower panels). Graphs in A and B (% of remaining activities compared to the wild type AbdA protein (WT) following domain mutations) using the boxplot representation summarize quantitative analyses (see Text S1 and Figure S5 (cerebral branch) and S6 (heart lineage) for full illustration). A graded color-coded bar above the graphs illustrates the level of protein activity, ranging from light green (full activity) to black (no activity). C. Abdominal hemi-segments in the cardiac tube are composed of six cardiac cells, labeled in blue by Mef2. The two most posterior cells express Doc1 (green). The thoracic hemi-segments lack the anterior Doc1 positive cells. This distinction between thoracic and abdominal segments is not affected following maternal and zygotic loss of exd. In the embryonic heart, abdominal segments are made of six pairs of cells, instead of four in thoracic segments [37]. This difference was shown to result from AbdA (and Ubx) promoting the six cell lineage in the abdomen [37], and in the thorax following AbdA ubiquitous expression in the mesoderm driven by the 24B-Gal4 driver ([37], Figure 6B). The visualization of the lineage is facilitated by a Dorsocross (Doc) staining, that labels two cells in each hemisegment, allowing to unambiguously identify each hemisegment. Effects of AbdA variants in cardiac cells specification were visualized by double fluorescent immunostaining against AbdA and Dorsocross (Doc). The six cell lineage inductive capacity of AbdA was scored by counting the number of cardiac cells in the T2 and T3 segments (see Text S1). Results showed that the six cell lineage inductive ability of AbdA is lost following HX/UA and HX/TD/UA mutations (Figure 6B and Figure S6). These observations again highlight functional redundancy, but between the UA and HX domains, instead of TD and UA domain as observed in cerebral branch specification. Additional examples of functional redundancy, yet in more complex pattern of interactions between protein domains were found in the biological contexts described below.
Mutually suppressive interaction of protein domains in the regulation of the Dll and Antp direct target genes, the specification of epidermal morphology, and larval locomotion
The limb-promoting gene Distalles (Dll) and Hox gene Antennapedia (Antp) are direct targets of AbdA [26], [43]. The ability of AbdA variants, following ubiquitous expression through the arm-Gal4 driver, to repress Dll (Figure 7A and Figure S7) and Antp (Figure 7B and Figure S8) was evaluated by examining the activity of a Hox responsive Dll enhancer (DME, [44]) and the expression of the Antp protein, respectively (see Text S1). Single domain mutations do not strongly affect repressive activities of AbdA on Dll and Antp, leading to a mean loss of 40%, with the exception of the TD mutation, which affects more (60%) the repressive activities on Antp. Combining domain mutations leads to stronger effects: in the case of Dll, simultaneous mutation of the HX and UA domains almost completely abolishes AbdA repressive activities, while in the case of Antp simultaneous mutation of the HX and UA domains or TD and UA domains results in a loss of 70% of AbdA repressive activity. More surprisingly, simultaneous mutation of the HX, TD and UA domains does not compromise further AbdA activity but instead restores a significant level of repressive activity, comparable to that of single domain mutated AbdA variants. This indicates that the three protein domains do not provide independent regulatory input, but likely act in interactive and mutually inhibitory ways.
Fig. 7. Mutually suppressive interaction of protein domains. 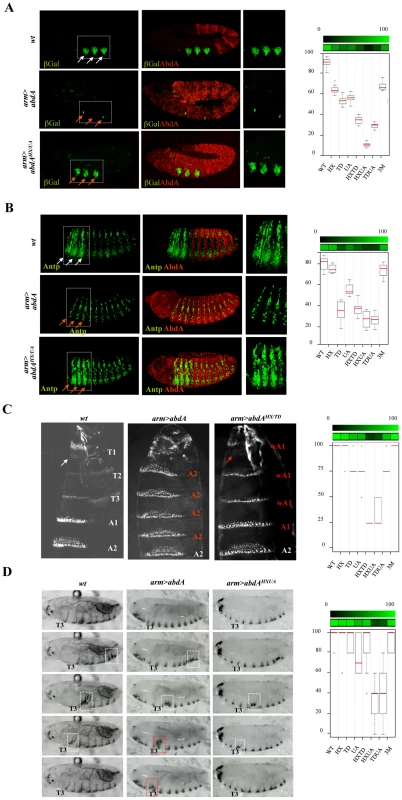
A. Thoracic restricted expression of Dll (white arrows) followed by Dll enhancer driven β-gal (green) results from repression by AbdA (red) in the abdomen (upper panel). Ubiquitous AbdA expression driven by arm-Gal4 represses Dll thoracic expression (red arrows, middle panels). The effect of the AbdAHX/UA variants is illustrated (lower panels). Right panels are magnification of boxed thoracic areas. B. Increased thoracic Antp expression (green, white arrows) results from AbdA (red) repression in the abdomen (upper panels). Ubiquitous AbdA expression driven by arm-Gal4 represses Antp expression in the thorax (red arrows, middle panels). The effect of the AbdAHX/UA variants is illustrated (lower panels). Right panels are magnification of boxed thoracic areas. C. Abdominal segments are characterized by refringent denticles organized in a trapezoidal shape in segments A2 but not A1, while T2/T3 thoracic segments harbors thinner denticles (left panel). Upon AbdA thoracic expression driven by arm-Gal4, the first abdominal segment A1 and thoracic segments acquire abdominal features, including abdominal type of denticles, trapezoidal organization of denticles and suppression of a T1 specific feature (white arrow), the “beard” (middle panel). Full or intermediate transformations were observed for AbdA variants (see Text S1 for quantifying criteria). The effect of the AbdAHX/TD variants is illustrated (right panel). Weak A1 (wA1) stands for a transformation of thoracic denticles toward abdominal type of denticles, with an organization typical of A1, but with only a partial suppression of the beard in T1 (arrow). D. Snapshots from movies illustrating locomotion in wild type larvae (left panels), or in larvae expressing ubiquitously AbdA (middle panels) or AbdAHX/UA variant (right panels) driven by the arm-Gal4 driver. White boxed areas show the progression of a peristaltic waves in the abdomen. The red boxed area shows an ectopic peristaltic wave in the thorax following ectopic AbdA expression in the thorax. Graphs in A–D (% of remaining activities compared to the wild type AbdA protein (WT) following domain mutations) using the boxplot representation summarize quantitative analyses (see Text S1 and Figure S7 (Dll), S8 (Antp), and S9 (A2 epidermal morphology) for full illustration, and Figure S10 for data on larval locomotion experiments. A graded color-coded bar above the graphs illustrates the level of protein activity, ranging from light green (full activity) to black (no activity). A similar yet more complex pattern of domain interactions was observed in the specification of A2 epidermal morphology. In this tissue, AbdA promotes the formation of a stereotyped trapezoidal arrangement of denticle belts (Figure 7C). The potential of AbdA variants to specify A2 epidermal morphology was assessed following arm-Gal4 driven expression by scoring the denticle belts morphology and organisation in transformed A1 and thoracic segments (Figure 7C and Figure S9). Epidermal specification was not impaired by HX and slightly reduced by UA or TD mutations. Simultaneous mutation in two domains suggests functional redundancy between HX and TD, UA and HX but not between UA and TD domains. As noticed previously for the regulation of Dll and Antp in the epidermis, mutating the three domains simultaneously restores the activity, generating a protein that displays an activity close to the wild type protein.
In many animals including vertebrates, locomotion results from the coordinated action of regionally distinct sets of movements. Drosophila larvae crawl by means of three region specific movements [38]. The locomotion cycle starts by a contraction of the most posterior abdominal segments (A8/A9), followed by a wave of peristaltic movement in A1–A7, where each segment is transiently lifted up (D/V movement), pulled forward and lowered, starting from A7. When the wave reaches A1, the thoracic and head segments start moving by a telescopic type of movement (A/P movement), occurring through contraction of anterior segments [38]. It was established that AbdA is necessary and sufficient to specify the abdominal type of movement, namely abdominal peristalsis [38]. The potential of wild type and AbdA variants to promote abdominal peristalsis was evaluated following arm-Gal4 driven expression (Figure 7B), by scoring in the T3 thoracic segment D/V movements (see Text S1). Single domain mutations do not significantly alter promotion of abdominal peristalsis (Figure 7D and Figure S10). Again, two types of functional redundancy were observed: between the TD and UA domains, and to a lesser extent between the HX and UA domains. As in the case of Dll and Antp regulation and A2 epidermal morphology specification, triple domain mutation corrected the effects of double mutations, with a protein promoting abdominal peristalsis as efficiently as the wild type protein, providing an additional example of mutually suppressive activity of protein domains.
Multifunctionality of protein domains revealed by Exd-dependency
Previous studies have established that Exd is required for Dll [25] and wg [45] regulation, oenocytes [30] and epidermal morphology specification [46], and neuroblast lineage commitment [37], while dispensable for Antp [46] and dpp [47] regulation. In the case of cerebral branch specification, no conclusion could be reached since loss of Exd results in the absence of cerebral branch formation in the T2 segment [48]: this positive input of Exd hinders the assessment of a possible contribution for AbdA mediated cerebral branch repression in abdominal segments.
The potential implication of Exd in AbdA-mediated heart lineage commitment and larval locomotion is not known. Staining for Doc1 in embryos deprived for maternal and zygotic Exd showed that the abdominal hemi segments adopt the AbdA-dependent six cell lineage, showing the dispensability of Exd for this AbdA function (Figure 6C). The requirement of Exd for larval locomotion has been examined in homothorax (hth) mutant that impairs Exd nuclear transport and mimics exd maternal and zygotic loss [49]. The absence of peristaltic waves in this genetic context indicates a strict requirement of Exd for abdominal peristalsis (Figure S10).
Taken together with the protein domain requirement results, the exd dependency indicates that the HX, UA and TD domains, known (HX and UA) or candidate (TD) Exd recruiting domains, are also required for Exd-independent function. This is supported by the HX/UA requirement for heart lineage specification, by the HX and UA requirement for proper regulation of the dpp target gene, the HX/TD requirement for Antp repression and the requirement of TD for dpp target regulation. Collectively, this highlights that the HX and UA (and likely TD) protein domains are multifunctional, serving in some biological context Exd interaction function, while in others, they are used differently, for a molecular activity that still remains to be defined.
Hierarchical clustering reveals two functional modules and a predominant role for the UA domain
The complete set of quantitative data was analyzed using a hierarchical clustering method (Figure 8; see Materials and Methods). Clustering according to biological readouts does not reveal any clear grouping, regarding for instance developmental stage or tissue type, suggesting that the forces that govern domain usage and interaction between protein domains mostly reside in the regulated target gene. By contrast, clustering according to protein domains clearly reveals a hierarchical requirement of the domains for the various AbdA functions analyzed here. A bipartition of AbdA variants is observed, with the mutants for the HX, the TD and HX/TD domains on the one hand, and variants mutant for the UA domain, alone or in combination, on the other hand. Such bipartition suggests the existence of two functional modules that can be distinguished based on UA domain requirement. The first module, which relies mostly on the HX and TD domains, is used for a small subset of AbdA functions only. The second module relies on the activity of the HX, TD and UA domains, yet the requirements of the HX and TD domains are revealed only in UA deficient context. Thus, the driving force in this second functional module is the UA domain, as its mutation unmasks the requirement for the HX and TD domains, which is not revealed by their single or combined mutations. These results identify a prominent role of the UA domain in AbdA function.
Fig. 8. Hierarchical clustering of AbdA domain requirements. 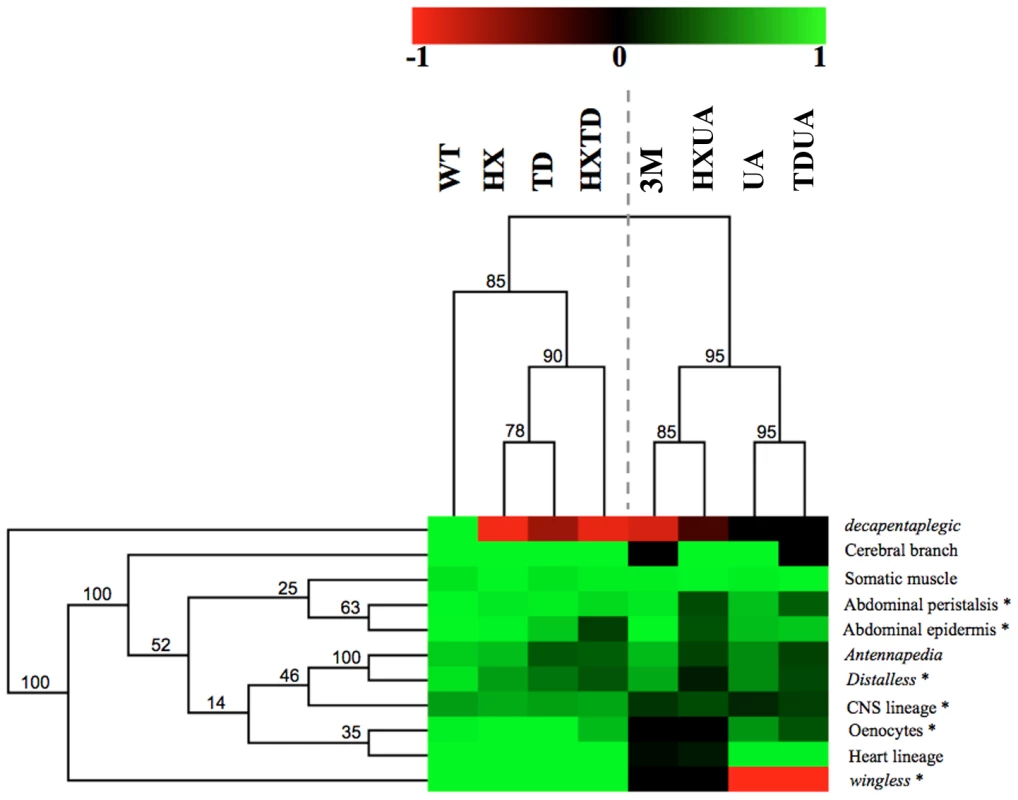
Hierarchical clustering of domain contribution reveals two functional modules and a predominant role for the UA domain. A graded color-coded bar above the graphs illustrates the level of protein activity, ranging from light green (full activity) to black (no activity). Neomorphic activity is depicted in red. Asterisks indicate Exd dependent biological readouts. Jacknife values are indicated for each node. Discussion
A different complementary approach to Hox protein function
Studies towards deciphering the mode of action of Hox proteins have so far essentially concentrated on how individual protein domains contribute to protein function. These focused approaches allowed in depth analyses, unraveling the intimate molecular and sometimes structural details of how protein domains contribute to protein function, providing decisive insights into how Hox proteins reach specificity. This work provides a different complementary approach towards deciphering the mode of action of Hox proteins. First it aims at studying protein domains in combinations, using combined and not only single protein domain mutations, considering that the overall protein activity is likely not a sum of the activity of individual protein domains, and that novel properties may emerge from interactions between protein domains. Second, it uses extensive in vivo biological readout, (most of the known AbdA functions), instead of a single or a few functions. While impairing the in depth analyses of previous focused approaches, the large functional window covered by this study allows the identification of features underlying the intrinsic functional organization of the Hox protein AbdA.
Although the approach taken relies on a gain of function strategy, special care was taken to select experimental conditions where proteins were expressed closed to physiological levels of expression. Biological readouts considered are functions that AbdA can sustain in ectopic places, suggesting that availability of AbdA protein partners is not a limitation of the experimental strategy chosen. Finally, the effects of expressing the AbdA variants (in all eleven biological readouts) were scored in regions anterior to the endogenous AbdA expression domain (ie in cells where the endogenous wild type gene product is not present), avoiding any further complexity that may result from competition with the endogenous AbdA protein.
Below, we summarize how results obtained shed light on the mode of action of the Hox protein AbdA and discuss the evolutionary implications.
Functional attributes and mode of protein domain usage: Implication for robustness and diversity
This study identifies salient features underlying the intrinsic functional organization of the AbdA Hox transcription factor. Protein domains often display functional redundancy, with strong effects in most cases requiring simultaneous mutations of two or three domains. Redundancy was frequently observed between the HX and UA domains, or between the TD and UA domains, while redundancy between the HX and TD domains is less frequent (Figure 8). This indicates that redundancy does not necessarily rely on functional compensation through structurally related domains, since the HX and TD are closely related domains, while the UA domain is completely unrelated. Thus, functional redundancy rather reflects the potential to perform similar activities through distinct molecular strategies. This property likely confers robustness to Hox protein activity, accommodating mutations in protein domains without generally impacting on regulatory activities.
Protein domains within AbdA also generally do not act as independent functional modules, but instead display a high degree of interactivity, as demonstrated by the non-additive effects of domain mutations in the majority of the biological readouts studied. In addition, protein domains are often multifunctional, in the sense that they serve different molecular functions. This is illustrated by the fact that the HX and UA domains, previously described to mediate Exd recruitment, are also required for Exd-independent processes. Thus domain interactivity and multifunctionality are hallmarks of AbdA regulatory activity. These properties provide means to apprehend the bases underlying Hox functional diversity with a restricted number of functional modules, and therefore may account for the variety of Hox-controlled biological functions.
Protein domain usage and interaction between protein domains in AbdA strongly depends on the biological readout, suggesting that domain usage largely depends on the regulated target gene, and hence on the identity of the gene cis regulatory sequences. Recent reports support that DNA sequences impact on Hox protein activity: Hox binding site neighboring sequences are important for proper regulation of the reaper downstream target [50]; Sex combs reduced changes its conformation and activity depending on the cognate sequence [51]. Of note, a role for the target sequence in controlling the structure and activity of the glucocorticoid receptor has also been recently reported [52], indicating that this may generally apply for many DNA binding transcription factors.
Mechanisms underlying the evolution of protein function
Our results also have implication on how modifications in protein sequences are translated into changes in protein function during evolution. The HX domain, common to all Hox proteins, is ancient and found in all bilaterians, and provides a generic mode of PBC interaction (Figure 9). The UA domain, specific to some central Hox proteins (AbdA and Ubx in Drosophila), was acquired later, at the time of protostome/deuterostome radiation. It provides a distinct yet to be characterised PBC interaction mode, specific to some Hox paralogues only, allowing fine-tuning of Hox protein activity [22]. TD is found only in insect AbdA and not in Ubx proteins, suggesting that it arose after the duplication that generated Ubx and AbdA in the common ancestor of insects (Figure 9). Remarkably, within AbdA arthropod proteins, the HX domain has significantly diverged in some lineages like anopheles, while the TD domain has been strictly conserved.
Fig. 9. Phylogeny of the HX, TD, and UbdA protein domains. 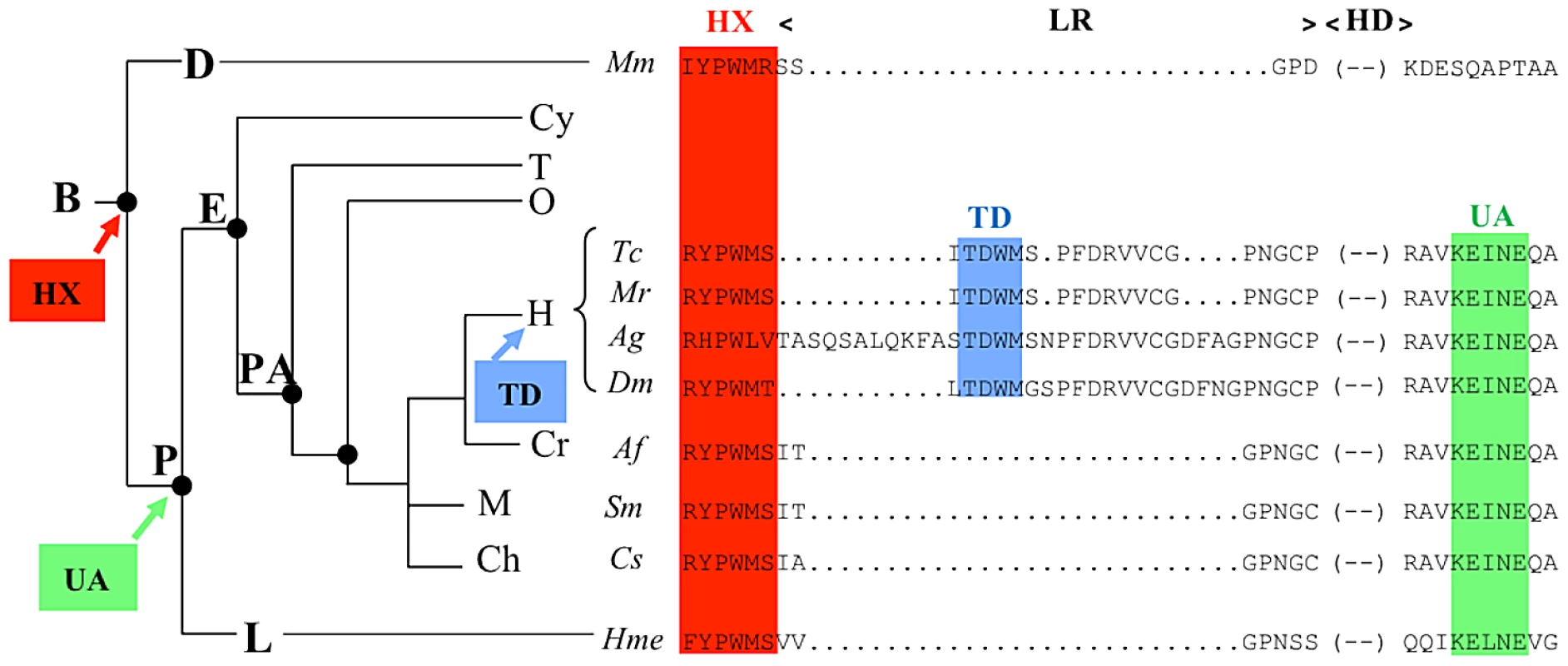
Abbreviations are as follows: B: bilaterians; P: protostomes; D: deuterostomes; L: lophotrochozoa; E:ecdysozoa; Cy, cycloneuralia. PA: pan-arthopods; O: onycophores; T: tardigrades; Ch: chelicerates; M; myriapods; Cr: crustaceans; H:hexapods. Sequence alignment spanning the HX, TD and UA domains are shown for representatives of the four main arthropod branches (Tc: Tribolium castaneum; Mr: Myrmica rubra; Ag: Anophela gambiae; Dm: Drosophila melanogaster) and for a representative of deuterostomes (Mus musculus, Mm) and lophotrochozoa (Hirudo medicinalis, Hme). Conceptually, two non-exclusive models could account for the evolution of protein function following the acquisition of a novel protein domain. In the first one, the acquisition provides a novel molecular and functional property, which adds to pre-existing ones. This is for example the case for the acquisition of the QA domain that confers repressive function to Ubx [6], and the acquisition/loss of HX or LRALLT domains by Futzitarazu (Ftz) from distinct insect species, which provides Ftz with the capacity to recruit either Exd or FtzF1 cofactors and switches its activity from a Hox to a segmentation protein [53]. In the second model, the acquisition of a novel protein domain interferes with the activity of pre existing domains, reorganizing the intrinsic functional organization of the protein. This view is supported by the predominant role of the UA domain and the widespread domain interactivity seen in this study.
More room for changes in protein activity during morphological evolution
Evolutionary changes in animal morphology is thought to mostly rely on changes in cis-regulatory sequences [1]. This is conceptually supported by the modular organization of cis-regulatory sequences, allowing subtle and cell specific changes in gene expression not deleterious for the animal. Experimentally, it is largely supported by the correlation between expression of key developmental regulatory genes and morphological changes (for example see [54]), and by changes in cis-regulatory sequences that impact on morphological traits [55]–[59]. Changes in animal morphology could also result from changes in protein sequence and function, as shown for Hox proteins in the morphological diversification in arthropods [4], [6]. However, changes in protein function are not believed to broadly contribute to morphological diversification during animal evolution, based on the assumption that changes in protein sequences are expected to have pleiotropic effects, which as such, do not provide a mean to convey subtle and viable evolutionary changes.
Our work grasps redundancy and selectivity in protein domain usage and as salient features of AbdA transcription factor intrinsic regulatory logic: even the HX domain, evolutionarily conserved in all Hox proteins, is essential for only one AbdA function, and often acts in a redundant way with the TD or the UA protein domains. Selective use of protein domains is also supported by findings of a few smaller scale studies of three other Drosophila Hox proteins: viable missense or small deletion mutations within the Scr protein coding sequences falls in different allelic series when examined for three distinct biological readouts [60]; deletion of C-terminal sequences of the Ubx protein, starting from an insect specific QA protein domain preferentially affects a subset of Ubx function [61]; dispensability of the HX was reported for the leg inducing capabilities of the Antp Hox protein, while required for other Antp functions [62]. This context dependent selective mode of protein domain usage, or differential pleiotropy, may be essential for the evolution of Hox protein functions, as it ensures developmental robustness of a Hox-controlled program while being permissive to evolutionary changes endowing novel functions to preexisting protein domains. In addition, our work also establishes that interactivity between protein domains is highly context dependent, suggesting that Hox protein function not only relies on selective mode of protein domain usage but also on selective mode of protein domain interactivity. Altogether, these observations challenge the view that changes in protein sequences necessarily have pleiotropic effects, giving more room for protein changes in the evolution of animal body plans.
Materials and Methods
Flies, egg collections, cuticle preparations, in situ hybridization, and immunostaining
24B-Gal4 and arm-Gal4 were used as embryonic mesodermal and ubiquitous drivers, respectively. Btl-Gal4 and lbe(K)-Gal4 for specific expression in the tracheal system and NB5–6 neuroblasts, respectively. The DME-lacZ and svp-lacZ lines are respectively from R. Mann (Columbia Univ., NY, USA) and S. Zaffran (IBDML, Marseille, France). exdXP11 and hthP2 alleles were used. Embryo collections, cuticle preparations, in situ hybridizations, and immunodetections were performed according to standard procedures. Digoxigenin RNA-labelled probes were generated according to the manufacturer's protocol (Boehringer Mannheim, Gaithersburg, MD) from wg and dpp cDNAs cloned in Bluescript. Primary antibodies used are: anti-Antp (4C3, dilution 1/100, Developmental Studies Hybridoma Bank (DSHB)); rabbit anti-AbdA (1/1000); guinea-pig anti-Doc2+3 (1/400) and rabbit anti-Dmef2 (1/2000) from L. Perrin (IBDML, Marseille, France); rabbit anti-Exd (1/1000) from R. Mann; rabbit anti-Nau (1/100) from BM Paterson (University of Texas Southwestern Medical Center, Dallas, TX); rabbit (1/500) or mouse (1/200) anti-GFP (1/500) from Molecular Probes; chicken anti-GFP (1/1000) from Aves labs; mouse anti-β-galactosidase (1/1000) from Promega; rabbit anti-β-galactosidase (1/1000) from MP Biomedical; anti-digoxigenin coupled to biotin (1/500) from Jackson. Secondary antibodies coupled to Alexa 488, Alexa 555 (Molecular Probes) or to biotin (Jackson) were used at a 1/500 dilution.
Constructs, transgenic lines, biological readouts, and quantification procedures
AbdA variant were generated by PCR. Domain mutations were YPWM→AAAA; TDWM→AVAI; KEINE→KAAAA. The homeodomain point mutation alleviating DNA binding is a mutation of position 50 (Q→K; [47]). Constructs were cloned in pUAST or pUASTattB vectors for transgenic line establishment. Lines were crossed with the appropriate driver, and collected embryos were stained with anti-AbdA to select the conditions (line and temperature) that result in expression levels similar (+/−15%) to AbdA wild type levels in A2 (see [22] for a detailed description of the procedure). Procedures used for quantification of biological readouts using at least 10 embryos of each genotype are provided in Text S1.
Hierarchical clustering of domain requirements
A matrix containing the values corresponding to the readout was built. The extreme values were given to the total loss of activity (value 0), and to the wild type activity (value 1 for 100% of activity). A hierarchical clustering algorithm (with Euclidian distance and average linking) was applied to the matrix using the MeV software suite [63]. The jacknife method was used for re-sampling the data and provides a statistical support for each tree node.
Boxplot data representation
Boxplots drawn using the R-Software. Boxplot depicts the value distribution obtained for each tested genotype. Black points correspond to individual counts.
Supporting Information
Zdroje
1. CarrollSB 2008 Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134 25 36
2. PearsonJCLemonsDMcGinnisW 2005 Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6 893 904
3. Prud'hommeBGompelNCarrollSB 2007 Emerging principles of regulatory evolution. Proc Natl Acad Sci U S A 104 Suppl 1 8605 8612
4. RonshaugenMMcGinnisNMcGinnisW 2002 Hox protein mutation and macroevolution of the insect body plan. Nature 415 914 917
5. TourEHittingerCTMcGinnisW 2005 Evolutionarily conserved domains required for activation and repression functions of the Drosophila Hox protein Ultrabithorax. Development 132 5271 5281
6. GalantRCarrollSB 2002 Evolution of a transcriptional repression domain in an insect Hox protein. Nature 415 910 913
7. MerabetSHudryBSaadaouiMGrabaY 2009 Classification of sequence signatures: a guide to Hox protein function. Bioessays 31 500 511
8. MerabetSSaadaouiMSambraniNHudryBPradelJ 2007 A unique Extradenticle recruitment mode in the Drosophila Hox protein Ultrabithorax. Proc Natl Acad Sci U S A 104 16946 16951
9. ChanS-KMannRS 1993 The segment identity functions of Ultrabithorax are contained within its homeo domain and carboxy-terminal sequences. Genes Dev 7 796 811
10. MannRSLelliKMJoshiR 2009 Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol 88 63 101
11. Merabet sSNPradelJGrabaY 2009 Regulation of Hox activity: insights from protein motifs. In Hox genes, studies form the 20th to 21st century Landes Bioscience
12. PeisajovichSGGarbarinoJEWeiPLimWA 2010 Rapid diversification of cell signaling phenotypes by modular domain recombination. Science 328 368 372
13. HueberSDLohmannI 2008 Shaping segments: Hox gene function in the genomic age. Bioessays 30 965 979
14. GehringWJKloterUSugaH 2009 Evolution of the Hox gene complex from an evolutionary ground state. Curr Top Dev Biol 88 35 61
15. WellikDM 2009 Hox genes and vertebrate axial pattern. Curr Top Dev Biol 88 257 278
16. TumpelSWiedemannLMKrumlaufR 2009 Hox genes and segmentation of the vertebrate hindbrain. Curr Top Dev Biol 88 103 137
17. KrumlaufR 2009 Hox Genes and Patterning in Mammals. Annu Rev Cell Dev Biol
18. GibsonGSchierALeMottePGehringWJ 1990 The specificities of Sex combs reduced and Antennapedia are defined by a distinct portion of each protein that includes the homeodomain. Cell 62 1087 1103
19. KuzioraMAMcGinnisW 1989 A homeodomain substitution changes the regulatory specificity of the Deformed protein in Drosophila embryo. Cell 59 563 571
20. ZengWAndrewDJMathiesLDHornerMAScottMP 1993 Ectopic expression and function of the Antp and Scr homeotic genes: The N terminus of the homeodomain is critical to functional specificity. Development 118 339 352
21. ChauvetSMerabetSBilderDScottMPPradelJ 2000 Distinct hox protein sequences determine specificity in different tissues. Proc Natl Acad Sci U S A 97 4064 4069
22. SaadaouiMMerabetSLitim-MecheriIArbeilleESambraniN 2011 Selection of distinct Hox-Extradenticle interaction modes fine-tunes Hox protein activity. Proc Natl Acad Sci U S A 108 2276 2281
23. BrandAPerrimonN 1993 Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401 415
24. GebeleinBCuliJRyooHDZhangWMannRS 2002 Specificity of distalless repression and limb primordia development by abdominal hox proteins. Dev Cell 3 487 498
25. GebeleinBMcKayDJMannRS 2004 Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature 431 653 659
26. AppelBSakonjuS 1993 Cell-type-specific mechanisms of transcriptional repression by the homeotic gene products UBX and ABD-A in Drosophila embryos. EMBO J 12 1099 1109
27. GrienenbergerAMerabetSManakJIltisIFabreA 2003 Tgf{beta} signaling acts on a Hox response element to confer specificity and diversity to Hox protein function. Development 130 5445 5455
28. ManakJRMathiesLCScottMP 1994 Regulation of a decapentaplegic midgut enhancer by homeotic proteins. Development 120 3605 3619
29. CapovillaMBrandtMBotasJ 1994 Direct regulation of decapentaplegic by Ultrabithorax and its role in midugt morphogenesis. Cell 76 461 475
30. BroduVElstobPRGouldAP 2002 abdominal A specifies one cell type in Drosophila by regulating one principal target gene. Development 129 2957 2963
31. ChiangCYoungKEBeachyPA 1995 Control of Drosophila tracheal branching by the novel homeodomain gene unplugged, a regulatory target for genes of the bithorax complex. Development 121 3901 3912
32. MichelsonAM 1994 Muscle pattern diversification in Drosophila is determined by the autonomous function of homeotic genes in the embryonic mesoderm. Development 120 755 768
33. LewisEB 1978 A gene complex controlling segmentation in Drosophila. Nature 276 565 570
34. Sanchez-HerreroEVernosIMarcoRMorataG 1985 Genetic organization of Drosophila bithorax complex. Nature 313 108 113
35. BelloBCHirthFGouldAP 2003 A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron 37 209 219
36. BergerCPallaviSKPrasadMShashidharaLSTechnauGM 2005 Cyclin E acts under the control of Hox-genes as a cell fate determinant in the developing central nervous system. Cell Cycle 4 422 425
37. PerrinLMonierBPonzielliRAstierMSemerivaM 2004 Drosophila cardiac tube organogenesis requires multiple phases of Hox activity. Dev Biol 272 419 431
38. DixitRVijayraghavanKBateM 2008 Hox genes and the regulation of movement in Drosophila. Dev Neurobiol 68 309 316
39. KarlssonDBaumgardtMThorS 2010 Segment-specific neuronal subtype specification by the integration of anteroposterior and temporal cues. PLoS Biol 8 e1000368 doi:10.1371/journal.pbio.1000368
40. BilderDGrabaYScottMP 1998 Wnt and TGFbeta signals subdivide the AbdA Hox domain during Drosophila mesoderm patterning. Development 125 1781 1790
41. MerabetSKambrisZCapovillaMBerengerHPradelJ 2003 The Hexapeptide and Linker Regions of the AbdA Hox Protein Regulate Its Activating and Repressive Functions. Dev Cell 4 761 768
42. ChiangCYoungKEBeachyPA 1995 Control of Drosophila tracheal branching by the novel homeodomain gene unplugged, a regulatory target for genes of the bithorax complex. Development 121 3901 3912
43. VachonGCohenBPfeifleCMcGuffinMEBotasJ 1992 Homeotic genes of the bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell 71 437 450
44. GebeleinBCuliJRyooHDZhangWMannRS 2002 Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev Cell 3 487 498
45. RauskolbCWieschausE 1994 Coordinate regulation of downstream genes by extradenticle and the homeotic selector proteins. Embo J 13 3561 3569
46. PeiferMWieschausE 1990 Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev 4 1209 1223
47. CapovillaMBotasJ 1998 Functional dominance among Hox genes: repression dominates activation in the regulation of Dpp. Development 125 4949 4957
48. MerabetSEbnerAAffolterM 2005 The Drosophila Extradenticle and Homothorax selector proteins control branchless/FGF expression in mesodermal bridge-cells. EMBO Rep 6 762 768
49. RieckhofGECasaresFRyooHDAbu-ShaarMMannRS 1997 Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91 171 183
50. StobePSteinMAHabring-MullerABezdanDFuchsAL 2009 Multifactorial regulation of a hox target gene. PLoS Genet 5 e1000412 doi:10.1371/journal.pgen.1000412
51. JoshiRPassnerJMRohsRJainRSosinskyA 2007 Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131 530 543
52. MeijsingSHPufallMASoAYBatesDLChenL 2009 DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324 407 410
53. LohrUPickL 2005 Cofactor-interaction motifs and the cooption of a homeotic Hox protein into the segmentation pathway of Drosophila melanogaster. Curr Biol 15 643 649
54. BurkeACNelsonCEMorganBATabinC 1995 Hox genes and the evolution of vertebrate axial morphology. Development in press
55. Prud'hommeBGompelNRokasAKassnerVAWilliamsTM 2006 Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440 1050 1053
56. GompelNPrud'hommeBWittkoppPJKassnerVACarrollSB 2005 Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433 481 487
57. JeongSRebeizMAndolfattoPWernerTTrueJ 2008 The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 132 783 793
58. JeongSRokasACarrollSB 2006 Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125 1387 1399
59. WilliamsTMSelegueJEWernerTGompelNKoppA 2008 The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134 610 623
60. SivanantharajahLPercival-SmithA 2009 Analysis of the sequence and phenotype of Drosophila Sex combs reduced alleles reveals potential functions of conserved protein motifs of the Sex combs reduced protein. Genetics 182 191 203
61. HittingerCTSternDLCarrollSB 2005 Pleiotropic functions of a conserved insect-specific Hox peptide motif. Development 132 5261 5270
62. PrinceFKatsuyamaTOshimaYPlazaSResendez-PerezD 2008 The YPWM motif links Antennapedia to the basal transcriptional machinery. Development 135 1669 1679
63. SaeedAIBhagabatiNKBraistedJCLiangWSharovV 2006 TM4 microarray software suite. Methods Enzymol 411 134 193
Štítky
Genetika Reprodukční medicína
Článek Macroautophagy Is Regulated by the UPR–Mediator CHOP and Accentuates the Phenotype of SBMA MiceČlánek Dynamic Replacement of Histone H3 Variants Reprograms Epigenetic Marks in Early Mouse Embryos
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 10
-
Všechny články tohoto čísla
- Transcriptional Robustness Complements Nonsense-Mediated Decay in Humans
- Identification, Replication, and Fine-Mapping of Loci Associated with Adult Height in Individuals of African Ancestry
- Genetic Determinants of Serum Testosterone Concentrations in Men
- A One Base Pair Deletion in the Canine Gene Causes Exon Skipping and Late-Onset Neuronal Ceroid Lipofuscinosis in the Tibetan Terrier
- Three Structure-Selective Endonucleases Are Essential in the Absence of BLM Helicase in
- Identification of Widespread Ultra-Edited Human RNAs
- Multiple Wnts Redundantly Control Polarity Orientation in Epithelial Stem Cells
- The Bicoid Stability Factor Controls Polyadenylation and Expression of Specific Mitochondrial mRNAs in
- Transcriptome-Wide Binding Sites for Components of the Non-Poly(A) Termination Pathway: Nrd1, Nab3, and Sen1
- Macroautophagy Is Regulated by the UPR–Mediator CHOP and Accentuates the Phenotype of SBMA Mice
- Genetic Rearrangements Can Modify Chromatin Features at Epialleles
- Novel Function of as a Gap Gene during Spider Segmentation
- A Genome-Wide Screen for Interactions Reveals a New Locus on 4p15 Modifying the Effect of Waist-to-Hip Ratio on Total Cholesterol
- Comparative Genomic Analysis of Human Fungal Pathogens Causing Paracoccidioidomycosis
- Genetic Diversity in Cytokines Associated with Immune Variation and Resistance to Multiple Pathogens in a Natural Rodent Population
- Mutator Suppression and Escape from Replication Error–Induced Extinction in Yeast
- Dynamic Replacement of Histone H3 Variants Reprograms Epigenetic Marks in Early Mouse Embryos
- A Barcode Screen for Epigenetic Regulators Reveals a Role for the NuB4/HAT-B Histone Acetyltransferase Complex in Histone Turnover
- HIF–VEGF Pathways Are Critical for Chronic Otitis Media in and Mouse Mutants
- A Conserved Developmental Patterning Network Produces Quantitatively Different Output in Multiple Species of Drosophila
- Role of Exonic Variation in Chemokine Receptor Genes on AIDS: Association with Pneumocystis Pneumonia
- Whole-Exome Sequencing Identifies Homozygous Mutations in a Spastic Ataxia-Neuropathy Syndrome Linked to Mitochondrial -AAA Proteases
- Von Hippel-Lindau () Inactivation in Sporadic Clear Cell Renal Cancer: Associations with Germline Polymorphisms and Etiologic Risk Factors
- A Systems Biology Approach Reveals the Role of a Novel Methyltransferase in Response to Chemical Stress and Lipid Homeostasis
- Identification of Genomic Regions Associated with Phenotypic Variation between Dog Breeds using Selection Mapping
- Global Mapping of Cell Type–Specific Open Chromatin by FAIRE-seq Reveals the Regulatory Role of the NFI Family in Adipocyte Differentiation
- Natural Selection Affects Multiple Aspects of Genetic Variation at Putatively Neutral Sites across the Human Genome
- MicroRNA Expression and Regulation in Human, Chimpanzee, and Macaque Brains
- An Adaptive Allelic Series Featuring Complex Gene Rearrangements
- Feed-Forward Microprocessing and Splicing Activities at a MicroRNA–Containing Intron
- Developmental Stability: A Major Role for in
- A Phenomics-Based Strategy Identifies Loci on , , and Associated with Metabolic Syndrome Phenotype Domains
- Association of , , , , and with Systemic Lupus Erythematosus
- Small RNAs Prevent Transcription-Coupled Loss of Histone H3 Lysine 9 Methylation in
- Successive Increases in the Resistance of to Viral Infection through a Transposon Insertion Followed by a Duplication
- Mutations in a Guanylate Cyclase GCY-35/GCY-36 Modify Bardet-Biedl Syndrome–Associated Phenotypes in
- The Glycobiome Reveals Mechanisms of Pentose and Hexose Co-Utilization in Bacteria
- Insights into Hox Protein Function from a Large Scale Combinatorial Analysis of Protein Domains
- Mutations Cause Seckel and Jawad Syndromes
- Zelda Binding in the Early Embryo Marks Regions Subsequently Activated at the Maternal-to-Zygotic Transition
- Temporal Coordination of Gene Networks by Zelda in the Early Embryo
- Genetic Interaction between MTMR2 and FIG4 Phospholipid Phosphatases Involved in Charcot-Marie-Tooth Neuropathies
- Oxr1 Is Essential for Protection against Oxidative Stress-Induced Neurodegeneration
- Transforming Growth Factor β Receptor Type 1 Is Essential for Female Reproductive Tract Integrity and Function
- Positional Cloning of a Type 2 Diabetes Quantitative Trait Locus; , a Negative Regulator of Insulin Secretion
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Glycobiome Reveals Mechanisms of Pentose and Hexose Co-Utilization in Bacteria
- Global Mapping of Cell Type–Specific Open Chromatin by FAIRE-seq Reveals the Regulatory Role of the NFI Family in Adipocyte Differentiation
- Genetic Determinants of Serum Testosterone Concentrations in Men
- MicroRNA Expression and Regulation in Human, Chimpanzee, and Macaque Brains
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



