-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAssociation of , , , , and with Systemic Lupus Erythematosus
Systemic lupus erythematosus (SLE) is a complex trait characterised by the production of a range of auto-antibodies and a diverse set of clinical phenotypes. Currently, ∼8% of the genetic contribution to SLE in Europeans is known, following publication of several moderate-sized genome-wide (GW) association studies, which identified loci with a strong effect (OR>1.3). In order to identify additional genes contributing to SLE susceptibility, we conducted a replication study in a UK dataset (870 cases, 5,551 controls) of 23 variants that showed moderate-risk for lupus in previous studies. Association analysis in the UK dataset and subsequent meta-analysis with the published data identified five SLE susceptibility genes reaching genome-wide levels of significance (Pcomb<5×10−8): NCF2 (Pcomb = 2.87×10−11), IKZF1 (Pcomb = 2.33×10−9), IRF8 (Pcomb = 1.24×10−8), IFIH1 (Pcomb = 1.63×10−8), and TYK2 (Pcomb = 3.88×10−8). Each of the five new loci identified here can be mapped into interferon signalling pathways, which are known to play a key role in the pathogenesis of SLE. These results increase the number of established susceptibility genes for lupus to ∼30 and validate the importance of using large datasets to confirm associations of loci which moderately increase the risk for disease.
Published in the journal: . PLoS Genet 7(10): e32767. doi:10.1371/journal.pgen.1002341
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002341Summary
Systemic lupus erythematosus (SLE) is a complex trait characterised by the production of a range of auto-antibodies and a diverse set of clinical phenotypes. Currently, ∼8% of the genetic contribution to SLE in Europeans is known, following publication of several moderate-sized genome-wide (GW) association studies, which identified loci with a strong effect (OR>1.3). In order to identify additional genes contributing to SLE susceptibility, we conducted a replication study in a UK dataset (870 cases, 5,551 controls) of 23 variants that showed moderate-risk for lupus in previous studies. Association analysis in the UK dataset and subsequent meta-analysis with the published data identified five SLE susceptibility genes reaching genome-wide levels of significance (Pcomb<5×10−8): NCF2 (Pcomb = 2.87×10−11), IKZF1 (Pcomb = 2.33×10−9), IRF8 (Pcomb = 1.24×10−8), IFIH1 (Pcomb = 1.63×10−8), and TYK2 (Pcomb = 3.88×10−8). Each of the five new loci identified here can be mapped into interferon signalling pathways, which are known to play a key role in the pathogenesis of SLE. These results increase the number of established susceptibility genes for lupus to ∼30 and validate the importance of using large datasets to confirm associations of loci which moderately increase the risk for disease.
Introduction
Systemic lupus erythematosus (SLE) is a relapsing-remitting complex trait which most commonly affects women of child-bearing age, with a ratio of 9∶1 in female to males. The disease prevalence varies with ethnicity, being more prevalent in non-European populations (approximately 1∶500 in populations with African ancestry and 1∶2500 in Northern Europeans) [1]. The condition is characterised by the production of a diverse range of auto-antibodies against serological, intra-cellular, nucleic acid and cell surface antigens [2]. The wide-ranging clinical phenotypes include skin rash, neuropsychiatric and musculosketal symptoms and lupus nephritis, which may be partially mediated by the extensive deposition of immune complexes. Today, thanks to improved treatments, the 10-year survival rate after diagnosis has increased to 90%, with lower survival rates being related to disease severity or complications from treatment [3]. Increased understanding of the underlying genetic basis for lupus is of key importance in improving the prognosis for lupus patients.
Until recently, the genetic basis of lupus remained largely undetermined, with only about ∼8% of the genetic contribution known [4]. However, within the last three years, tremendous progress has been made in defining novel loci, through three moderate-sized genome-wide association studies in European American cohorts and a replication study in a US-Swedish cohort [5]–[7]. The loci previously identified for SLE include genes involved in the innate immune response (eg. IRF5), T and B cell signalling (eg. STAT4, TNFSF4 and BLK), autophagy/apoptosis (eg. ATG5), ubiquitinylation (UBE2L3, TNAIP3, TNIP1) and phagocytosis (ITGAM, FCGR3A and FCGR3B). All of these pathways are of potential importance in lupus pathogenesis [8]–[10].
To date, a total of 1729 independent SLE cases have been subjected to genome-wide association genotyping using three genotyping platforms: Illumina 317 K BeadChip [5], Illumina 550 K BeadChip [6] and Affymetrix 500 K array [7]. There is currently no published meta-analysis of these datasets.
The aim of the current work was to perform a replication study using our UK SLE cohort on loci that showed some evidence for association in previous studies in order to extend the list of confirmed susceptibility genes for lupus.
Results
To identify additional susceptibility loci for SLE, we first identified the independent genetic variants that showed moderate risk (5×10−3<P>5×10−8) in a combined US-Swedish dataset comprising 3273 SLE cases and 12188 controls [4]. We then genotyped 27 independent SNPs in a replication cohort of 905 UK SLE cases and 5551 UK control samples (Table 1), that included both British 1958 Birth Cohort samples and additional controls from the WTCCC2 project.
Tab. 1. SLE Case-Control Study Cohorts used in the study. 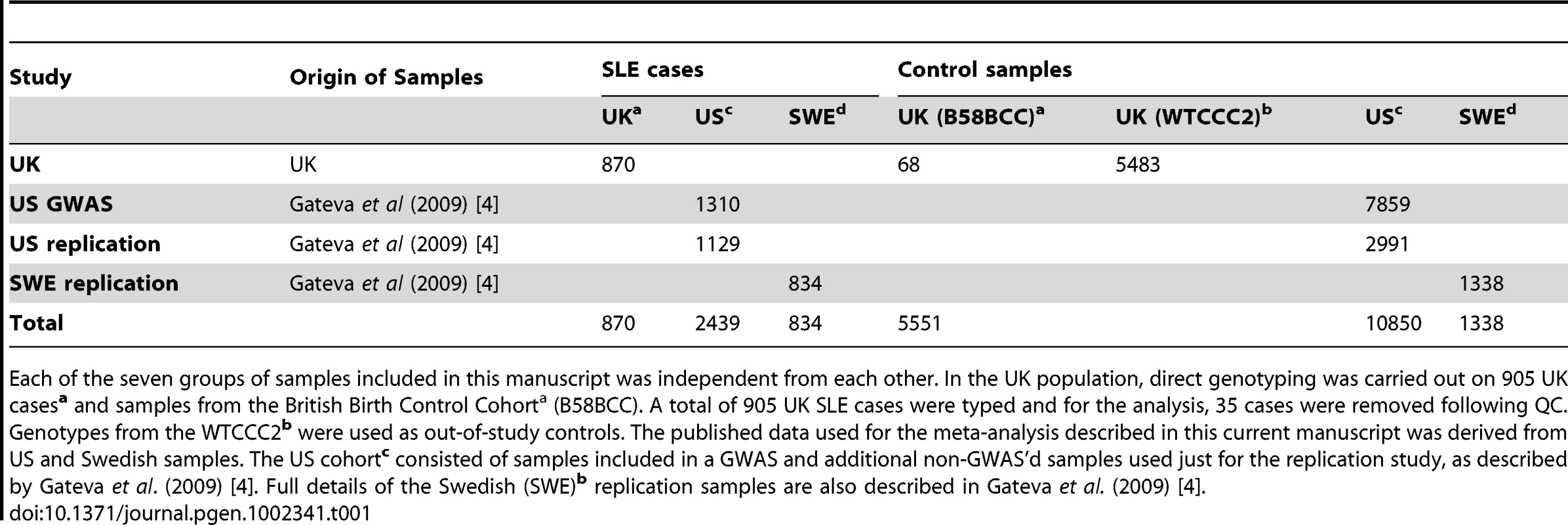
Each of the seven groups of samples included in this manuscript was independent from each other. In the UK population, direct genotyping was carried out on 905 UK casesa and samples from the British Birth Control Cohorta (B58BCC). A total of 905 UK SLE cases were typed and for the analysis, 35 cases were removed following QC. Genotypes from the WTCCC2b were used as out-of-study controls. The published data used for the meta-analysis described in this current manuscript was derived from US and Swedish samples. The US cohortc consisted of samples included in a GWAS and additional non-GWAS'd samples used just for the replication study, as described by Gateva et al. (2009) [4]. Full details of the Swedish (SWE)b replication samples are also described in Gateva et al. (2009) [4]. For the 27 genotyped SNPs, 10 variants which had not been genotyped by the WTCCC2 project, were imputed using IMPUTE2 [11]. This imputation was performed using CEPH HapMap samples as the phased reference sequence and the boundary of the surrounding haplotype blocks used to demarcate the imputation interval. The subsequent association analysis excluded two of these ten imputed SNPs because they had less than 95% certainty for the imputation (Table S2). In the US/SWE dataset, imputation of selected SNPs not genotyped previously [4] was performed using IMPUTE1 for HapMap. Phase II CEU sample haplotypes were used as reference with subsequent association analysis performed using SNPTEST and a genomic control factor (lambda-GC) values of: 1.05 (US dataset) and 1.10 (SWE dataset) after correction for population stratification.
In the UK replication sample by performing allelic association analysis using PLINK for the 23 SNPs passing QC (Tables S2 and S3), we demonstrated moderate association (P≤0.05) for twelve variants - with a lambda-GC of 1.01 following ancestry correction (see Table 2 and Table 3). Under the null hypothesis, only 1 of the 23 loci would be expected to have P≤0.05. The observed enrichment of associated SLE genes in the UK dataset suggested that many of these loci were likely to be true-positive associations.
Tab. 2. Novel SNPs showing genome-wide significance (P = 5×10−8) in SLE following meta-analysis of UK, US, and Swedish cohorts. 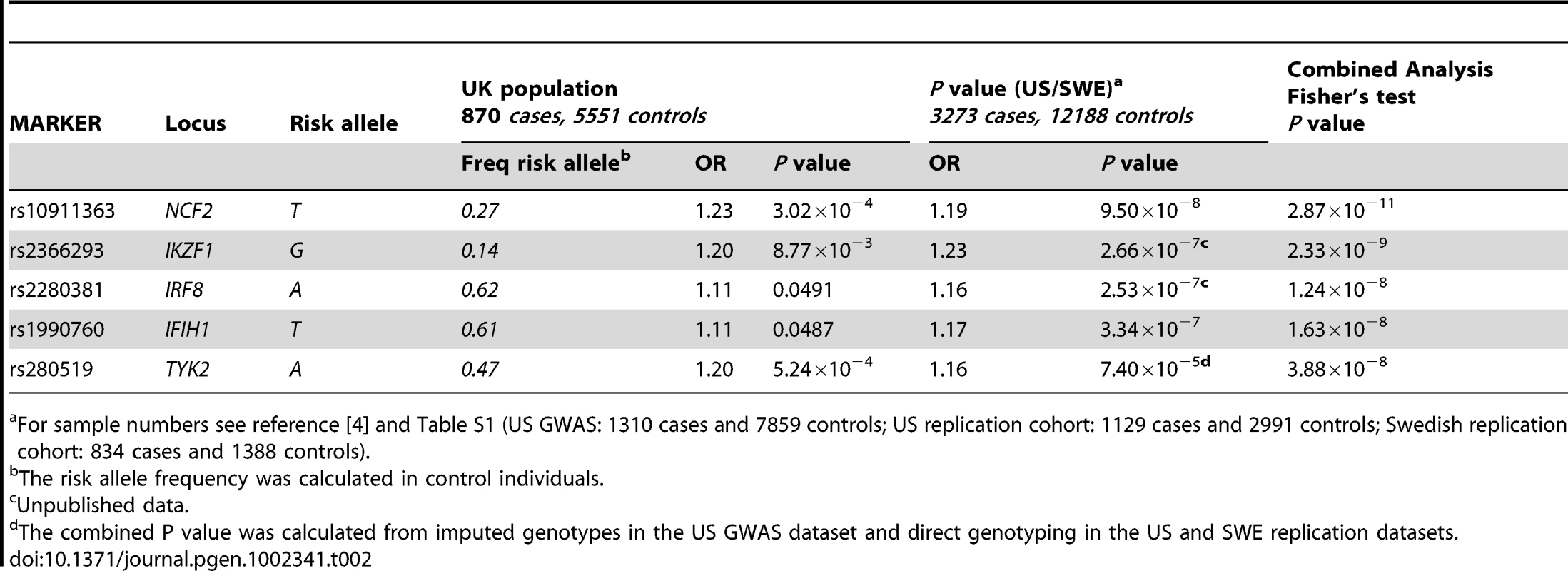
For sample numbers see reference [4] and Table S1 (US GWAS: 1310 cases and 7859 controls; US replication cohort: 1129 cases and 2991 controls; Swedish replication cohort: 834 cases and 1388 controls). Tab. 3. Additional SNPs showing association with SLE in the UK, US, and Swedish cohorts. 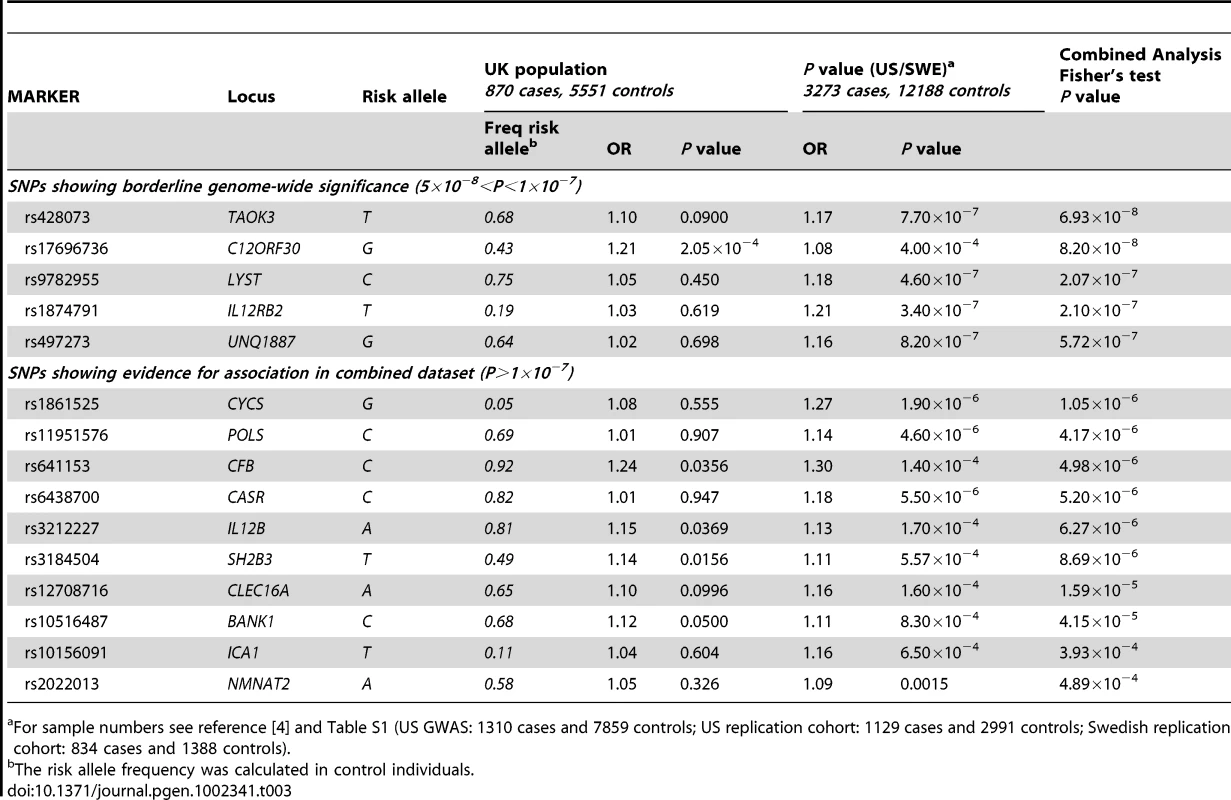
For sample numbers see reference [4] and Table S1 (US GWAS: 1310 cases and 7859 controls; US replication cohort: 1129 cases and 2991 controls; Swedish replication cohort: 834 cases and 1388 controls). We confirmed the similarity of odds-ratios (Het P value) and direction of the effect between the UK and US-SWE datasets (Table S4) and then performed a meta-analysis using Fisher's combined P-value (see Materials and Methods). This meta-analysis revealed five novel associated loci with P<5×10−8 (Table 2): NCF2 (neutrophil cytosolic factor 2) (rs10911363, Pcomb = 2.87×10−11, ORcomb = 1.19); IKZF1 (Ikaros family zinc-finger 1) (rs2366293, Pcomb = 2.33×10−9, ORcomb = 1.24); IRF8 (interferon regulatory factor 8) (rs2280381, Pcomb = 1.24×10−8, ORcomb = 1.16); IFIH1 (interferon-induced helicase C domain-containing protein 1) (rs1990760, Pcomb = 1.63×10−8, ORcomb = 1.15) and TYK2 (tyrosine kinase 2) (rs280519, Pcomb = 3.88×10−8, ORcomb = 1.17)(Table 1). The strength of these associations was similar to those found from a weighted meta-analysis, using the METAL programme (Table S4). A case-only analysis using PLINK in the combined UK/US/SWE dataset revealed no non-additive interactions between the five newly associated variants (P>0.05). These new SLE loci are discussed in more detail below and with additional information in Text S1.
Three of the SNPs tested were for loci that had shown genome-wide levels of significance in other SLE GWAS studies (Table S5). In the UK cohort we found further support for the association at JAZF1 (rs849142 PUK = 0.0243, ORUK = 1.13) and identified a third associated variant in the first intron of TNIP1 (rs6889239 PUK = 9.06×10−6, ORUK = 1.30), which is in strong LD (r2 = 0.895) with both the previous report in Europeans [4] and in perfect LD with a third SNP (rs10036748), first reported in a Chinese GWAS [12]. All three variants in TNIP1 are located within a 661 bp region of intron 1. We did not replicate the previous association with IL10 (rs3024505, PUK = 0.209 ORUK = 1.09) (Table S5).
These analyses increased the evidence of association for a number of additional loci that had shown borderline significance in the original US/SWE GWAS (Table 3), including CFB, C12ORF30, SH2B3, and IL12B. Genotyping of additional samples will be required to determine if the association signals shown in Table 3 represent confirmed genetic loci for SLE.
Discussion
The work presented here confirms five new susceptibility loci for SLE at the level of genome-wide significance (P<5×10−8). Each of the associated variants lie within, or close to, the coding sequence for genes with known roles in immune regulation: NCF2, IKZF1, IRF8, IFIH1 and TYK2. Interestingly, each of these genes has been implicated in interferon signalling. While the interferons have classically been defined as anti-viral cytokines, recent studies have suggested an important role for interferon in the pathophysiology of SLE [13]. While most evidence points to the role of type I interferon in SLE [14] there is substantial data suggesting that type II interferon (IFNγ) is also involved in SLE pathogenesis [15].
NCF2 (neutrophil cytosolic factor 2) (1q25), is induced by IFNγ and specifically expressed in a number of immune-cell types, including B-cells. Our data suggest that the NCF2 association is independent from the previously reported signal in the neighbouring locus NMNAT2, [5] because we found no evidence of strong LD between the genotyped SNP within NMNAT2 (rs2022013) and that in NCF2 (rs10911363) (r2 = 0.136). Logistic regression in the UK replication cohort confirmed that NMNAT2 did not contribute to the association at NCF2 (P = 0.777).
NCF2, as a cytosolic subunit of NADPH-oxidase, may have a role in the increased production of the free radicals characterising B-cell activation [16] (Figure 1) which increases auto-antibody levels and may suggest a mechanism for the involvement of NCF2 as a susceptibility gene for SLE.
Fig. 1. Pathways of immuno-dysregulation by five new susceptibility genes for SLE. 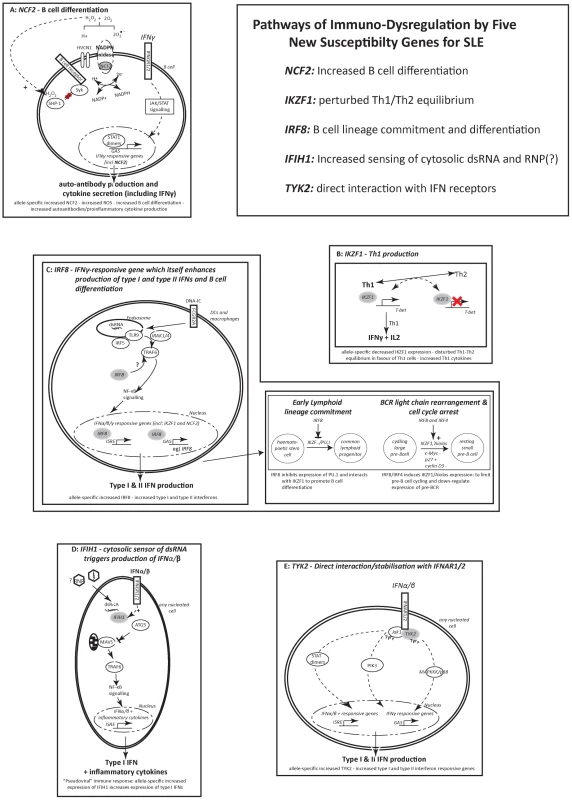
The figure shows the interferon-related pathways involving NCF2, IKZF1, IRF8, IFIH1 and TYK2 and and the ways in which these pathways may contribute to lupus susceptibility. In SLE these five genes contribute to increasing the levels of type-I and –II interferons, imbalances in Th1/Th2 related to disease severity, perturbations in B cell physiology and production of a diverse set of auto-antibodies. There are allele-specific significant expression differences for rs10911363, following a recessive model of basal expression for the risk T allele of rs10911363 in CEPH individuals but not in YRI and ASN (CHB+JPT) HapMap cohorts (PCEPH = 0.03) (Figure 2A). There is also a significant difference in gene expression for a variant (rs3845466) located 2 kb away from rs10911363 in intron 2 of NCF2 (Figure S2A), using lymphoblastoid cell lines (LCLs) from umbilical cords of 75 individuals which were taken from the GENEVAR collection (P = 0.0228). The population-specific nature of this correlation could be because of local differences in the pattern of LD within NCF2 between the CEU, YRI and ASN (CHB+JPT) HapMap cohorts. These population specific differences in LD may be between the genotyped SNP and an unknown causal allele(s) responsible for an expression difference seen in multiple ethnic backgrounds or between the genotyped marker and an unknown causal allele(s) exhibiting population-specific differences in gene expression itself. However, it will be necessary to confirm these findings in primary cells and tissues, because the EBV-transformed B cells model system may not entirely reflect the physiological conditions in peripheral B cells. Indeed a recent report showed that there may be systematic changes in gene expression within EBV-transformed B cells [17]. Nevertheless, with this caveat in mind, and taking each locus on a case-by-case basis, the model-based approach can provide important insights into measurement of transcript levels in ex vivo cells. For example, the increases in transcript levels that we initially observed in EBV-LCLs for OX40L, were also confirmed in peripheral blood B cells [18].
Fig. 2. Expression pattern in EBV-transformed lymphoblastoid cell lines. 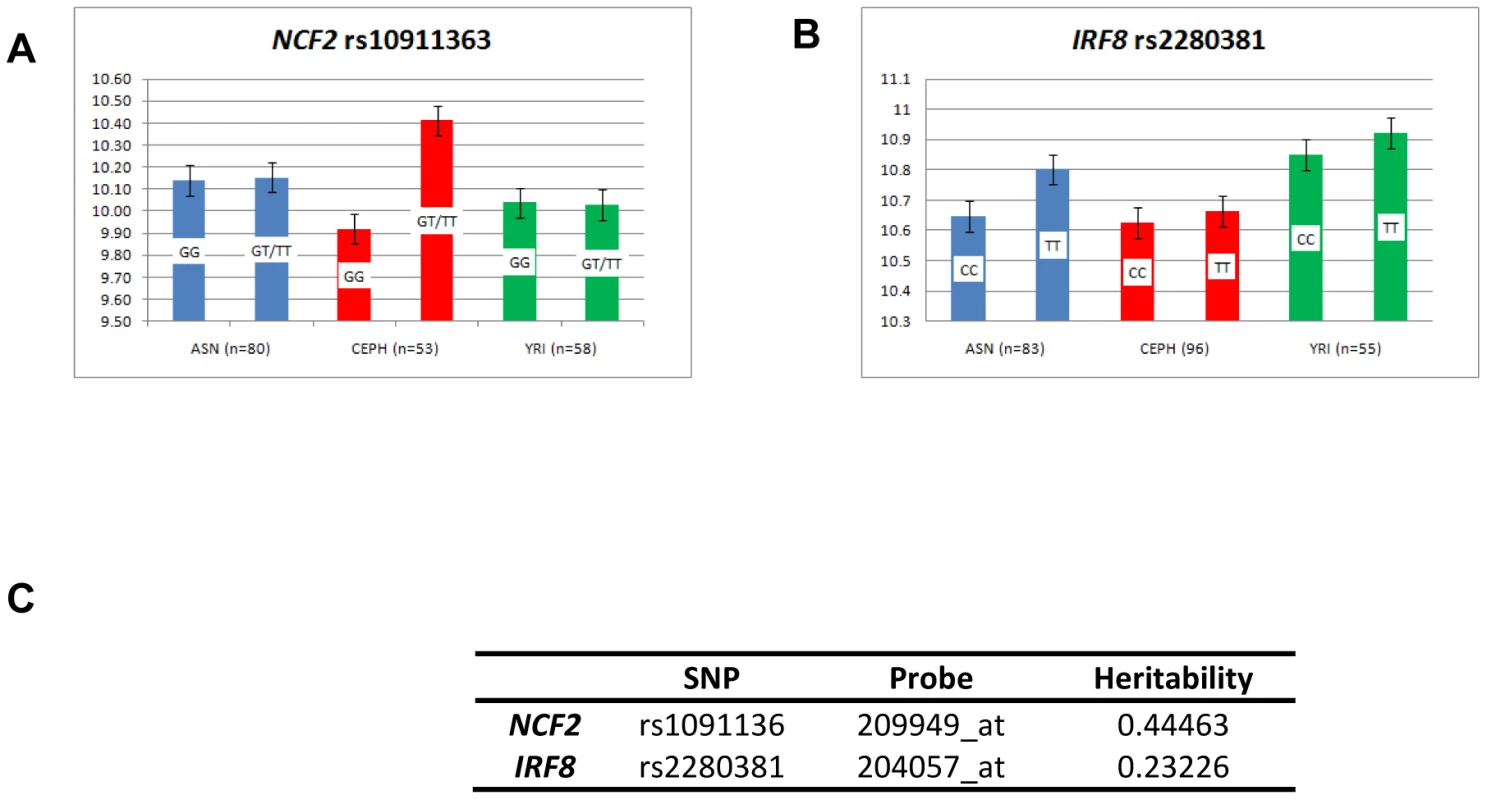
Regression analysis, as described in the materials and methods, was performed on publically available genotype data from EBV-transformed B cells which were part of the HAPMAP collection and expression data on the same individuals taken from the GEO database. Four populations were used: CEPH, YRI and CHB/JPT (ASN) [36]. The GEO dataset was GSE12526 and the expression probes were: A) NCF2 (209949_at), B) IRF8 (204057_at). For each graph, the mean expression per risk (R) allele and that for the non-risk (r) allele was plotted for each population. The alleles are listed on each bar and for each SNP, the total number of individuals for which there was both genotype and expression data are quoted for the three populations analysed. C) Heritability estimates for each locus were taken from the mRNA by SNP browser (http://www.sph.umich.edu/csg/liang/asthma/). IKZF1 (Ikaros family zinc-finger 1) (17p14.3) is a transcription factor essential for dendritic cell and lymphocyte development. The association with rs2366293 is supported by a report of a second associated variant, rs921916 (Pcomb = 2.0×10−6) [4], found 860 bp away from rs2362293, which is in strong LD with rs2366293 (r2 = −0.746, D′ = 0.925) (Figure S2B). A third SNP, rs4917014, located ∼200 kb upstream of IKZF1, showed association with SLE in a Chinese GWAS (PGWAS = 2.93×10−06), but it was a separate signal from the European SNPs (r2<0.0002) [9], [12]. IKZF1 has a role in the production of IFNγ, by blocking the production of the Th1 master-regulator T-bet (Figure 1). The shifted Th1/Th2 equilibrium (in favour of Th1 cells) increases the levels of IFNγ directly [19] rather than indirectly as a result of cross-talk between the type-I and type-II IFN signalling pathways eg) via type-I interferon mediated activation of STAT1 homodimers, which are the primary means of signalling from IFNγ [20] and have recently been shown to be associated with SLE in a Swedish cohort [21].
The transcription factor IRF8 (interferon regulatory factor 8) (16q24.1), shows immune-cell restricted expression. rs2280381 is found 64 kb downstream of IRF8, and is in LD with the coding region (Figure S2C), but independent from a susceptibility allele for multiple sclerosis (rs17445836), 1 kb away [22]. The lupus variant influences IRF8 gene expression, since LCLs from three HapMap cohorts, showed a significant increase in IRF8 transcript levels in homozygotes for the risk allele (TT) compared to homozygotes for the non-risk allele (CC) (P = 0.045) (Figure 2A). IRF8 also has a key role in regulating the differentiation of myeloid and B-cells and in mice, IRF8 restricts myeloid cell differentiation but promotes B-cell differentiation [23](Figure 1).
IFIH1 (interferon-induced helicase C domain-containing protein 1) (2q24.3) is an ubiquitiously expressed, cytoplasmic sensor of dsRNA. The SLE risk allele for rs1990760 (Table 1) is identical to that previously reported in two organ-specific autoimmune diseases: T1D [24] and Graves' Disease [25]. Regression analysis using publically available genotype data from HapMap and expression data from GEO dataset GSE12526 revealed that individuals who were homozygous for the common risk T allele of rs1990760 had significantly higher IFIH1 transcript levels compared to individuals who were homozygous for the non-risk allele (P = 0.8.19×10−5) (Figure S3B). Furthermore, a recent paper showed that the presence of the risk T allele of rs1990760 was correlated with increased levels of IFN-induced gene expression, in lupus patients who were positive for anti-dsDNA antibodies [26]. Another report demonstrated that IFIHI was rapidly up-regulated by type-I IFNs (Figure 1), and that IFIH1 signalled downstream through NF-κB, to further increase IFN-α production [27].
TYK2 (tyrosine kinase 2) (19p13.2) phosphorylates the receptor subunits of cytokine receptors, including type-I IFN receptors which are found on all nucleated cells, leading to increased production of type I interferon responsive genes (Figure 1). The significant association in intron 11 TYK2 for rs280519 in our UK cohort (P = 5.24×10−4) crossed the threshold for genome-wide significance when combined with the US/Swedish cohort. The association for rs280519 increases the genetic evidence for the involvement of TYK2 reported in a smaller UK family-based SLE cohort [28]. There was an earlier report, using a Swedish/Finnish population, of association in TYK2. This Swedish/Finnish study showed association for a missense mutation in exon 8 (rs2304256) (Pcomb = 5.60×10−5, PSwe = 9.60×10−5) [29]. The Swedish individuals used in the earlier analysis are a subset of the Swedish individuals analysed for this current manuscript and rs2304526 is in moderate LD with the TYK2 SNP that we typed in this current study - rs280519 (r2CEPH-HapMap = 0.373). The association for rs2304256 was replicated in a second moderate sized European study [30], but not in the GWAS from the SLEGEN consortium [5]. In preliminary analysis in UK cases and controls, there are data to support the fact that rs280519 is enriched in SLE cases (n = 345) with renal disease compared to healthy controls (n = 5551) (P = 0.033).
There were variants in several loci for which we have found evidence of association (P<0.05) in our UK cohort, but which did not reach genome-wide significance in the combined analysis. One of these variants was rs17696736, located in intron 15 of C12ORF30 (MDM20). This protein is a subunit of N-acetyltransferase complex B (NatB), and may promote apoptosis by reducing cell cycle progression [31]. In the joint cohort, rs17696736 was in LD (r2 = 0.625) with a second variant on chromosome 12q24, a missense W262R allele (rs3184504) in the lymphocyte adaptor protein SH2B3. SH2B3 facilitates T-cell activation by mediating the interaction between the T-cell receptor and T cell signalling molecules [32]. Both MDM20 and SH2B3 are also associated with T1D [33], and SH2B3 is additionally associated with celiac disease [34] and both myocardial infarction and asthma [35]. The associated variant within IL12B, rs3212227, is located in the 3′ UTR region, and the SLE risk allele is the same as previously reported for psoriasis [36]. IL12B encodes for the larger subunit (p40) of two cytokines, IL12 and IL23, and thereby contributes to both Th1 [37] and Th17 [38] immune responses.
In summary, we have identified five new genes contributing to SLE risk: NCF2, IKZF1, IRF8, IFIH1 and TYK2. Dense fine-mapping and/or genomic re-sequencing of each locus will be required to reveal the functional alleles for each gene with respect to immune dysregulation in lupus. Taken together, these findings further support an important role of interferon pathway dysregulation in lupus pathogenesis.
Materials and Methods
Ethics statement
The ethical approval for the study was obtained from the London Multi-Centre Research Ethics Committee (London MREC).
Details of the UK SLE samples in the study cohort
All of the 905 UK SLE cases conformed to the ACR criteria for SLE [39] with a diagnosis of SLE being established by telephone interview, health questionnaire and details from clinical notes. Written consent was obtained from all participants. Genomic DNA from the UK samples was isolated from anti-coagulated whole blood by a standard phenol-chloroform extraction.
Genotyping methodology
Each of the 27 SNPs were genotyped on a custom Illumina chip, using the BeadXpress platform at the Oklahoma Medical Research Foundation (OMRF), Oklahoma. The panel of ancestry informative markers was typed independently on an Illumina platform at Gen-Probe, Livingstone.
Power calculations
Power calculations were performed in the UK case-control dataset for each of the markers tested, using the algorithm described by Purcell et al [40]. Taking into account varying minor allele frequencies for the risk alleles and the differences in effect size (OR), and by employing a population prevalence of 0.002 and D′ of 1, with an type I error rates, alpha = 0.05, each of the SNPs showing novel genome-wide significance in the meta-analysis showed a power of >48% (2) to detect an association in our cohort.
Quality control of genotyping
Markers were excluded from the analysis if they showed a genotyping success rate of less than 95% or had a Hardy-Weinberg P value in the B58BCC control samples of less than P = 0.001. A total of 21 cases were removed from the final analysis due to low percentage genotyping (<95%). All samples were filtered for cryptic relatedness and duplication using an identity by state test in PLINK (PI_HAT score >0.1). The full list of genotyped variants and the results of the QC analysis are shown in (Table S3).
Correction for ancestry
A total of 35887 markers, distributed across each autosome, were selected for ancestry correction in the UK case-control cohort, these markers had all been typed as part of the HapMap project and on the WTCCC2 samples. The 35887 SNPs were chosen from a set of Illumina 317 K markers pruned for LD (r2<0.25) after removing regions of known extended LD, including the extended MHC and the region covering the inverted repeat on chromosome 8 (pers commun. David Morris, King's College and Kim Taylor, UCSF). This list of AIMs is available directly from the corresponding author, Professor Timothy Vyse.
The EIGENSTRAT PCA analysis was performed on the UK cases and also the control samples, both from the genotyped B58BCC and the WTCCC2 out-of-study controls. The eleven populations from HapMap3 were used as external references. Each SNP included in the PCA analysis showed >95% genotyping in the each dataset. Following EIGENSTRAT analysis, a graph was plotted of PC1 against PC2 for all the cases and controls in the UK study cohort (Figure S1). Individuals were only retained for association analysis if the values for their first two principal components fell within 6 SD of the mean for the CEPH HapMap samples. The genomic inflation factor (lambda-GC) for each population was calculated using PLINK.
Statistical analysis
All sample genotype and phenotype data was managed by, and analysis files generated with BC/SNPmax and BC/CLIN software (Biocomputing Platforms Ltd, Finland).
The imputation intervals for each imputed variant, defined as the bounds of the haplotype blocks, calculated using the Gabriel algorithm in Haploview, (for details of the intervals see Table S2). For SNPs which were not genotyped as part of the WTCCC2 project, we performed imputation using a method described by Marchini et al [11] to generate the missing genotypes for case-control association analysis. Each un-typed variant from our list of tested SNPs, was imputed in the WTCCC2 samples, using HAPMAP as the phased reference sequence. The LD pattern around each un-typed variant was examined using the CEPH cohort from HapMap. The boundaries of the haplotype blocks were determined using the default settings for the Gabriel et al algorithm in Haploview. For each imputed variant, these haplotype boundaries were used to define the boundaries of the imputation interval (Table S2). Only SNPs with greater than a 95% certainty in imputation, assessed using the quality score from the IMPUTE2 output file, were used for subsequent analysis.
Allelic association testing, using UK SLE cases with either genotyped control samples or imputed genotypes, was carried out using PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/).
Prior to performing the meta-analysis, the heterogeneity of odds ratios was tested using METAL and the Cochran-Mantel-Haenszel test (Table S4). SNPs with P value<0.001 between the two studies were discarded. Combined analysis of P values generated in the UK samples with those from the US/SWE cohort in published data [4] was conducted using Fisher's combined P value and with a meta-analysis using the programme METAL, which weighted the effect size, based on the inverse of the standard error.
To determine whether there was any allele-specific effect on the level of gene expression, we used publically available genotype data on unrelated EBV-transformed B cells (CEU, YRI and CHB/JPT individuals which were part of the HapMap project) and expression data from the same individuals (GSE12526, GEO database) [41]. For each locus, which reached genome-wide significance by meta-analysis, we categorised the expression data based on the SNP genotype for the respective associated variant (homozygote risk allele, heterozygote and homozygous non-risk allele). The significance of the correlation between genotype and expression level was then calculated using logistic regression analysis in SNPTEST, using gender as a covariate.
Interactions between the five SNPs reaching genome-wide significance following meta-analysis, were assessed using the epistatic option in PLINK. To maximize the power of this test, we restricted our analysis to the SLE affected individuals from the combined US/SWE/UK cohort.
Supporting Information
Zdroje
1. JohnsonAEGordonCPalmerRGBaconPA 1995 The prevalence and incidence of systemic lupus erythematosus in Birmingham, England. Relationship to ethnicity and country of birth. Arthritis Rheum 38 551 558
2. IppolitoAWallaceDJGladmanDFortinPRUrowitzM 2011 Autoantibodies in systemic lupus erythematosus: comparison of historical and current assessment of seropositivity. Lupus 20 250 255
3. DoriaAIaccarinoLGhirardelloAZampieriSArientiS 2006 Long-term prognosis and causes of death in systemic lupus erythematosus. Am J Med 119 700 706
4. GatevaVSandlingJKHomGTaylorKEChungSA 2009 A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. NatGenet 41 1228 1233
5. HarleyJBAlarcon-RiquelmeMECriswellLAJacobCOKimberlyRP 2008 Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 40 204 210
6. HomGGrahamRRModrekBTaylorKEOrtmannW 2008 Association of Systemic Lupus Erythematosus with C8orf13 - BLK and ITGAM - ITGAX. NEnglJMed 358 956 961
7. GrahamRRCotsapasCDaviesLHackettRLessardCJ 2008 A genome-wide association scan identifies Tumour Necrosis Factor Alpha Inducible Protein 3 (TNFAIP3/A20) as a susceptibility locus for Systemic Lupus Erythematosus. Nat Genet 40 1059 1061
8. RhodesBVyseTJ 2008 The genetics of SLE: an update in the light of genome-wide association studies. Rheumatology (Oxford) 47 1603 1611
9. MoserKLKellyJALessardCJHarleyJB 2009 Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun 10 373 379
10. HarleyITKaufmanKMLangefeldCDHarleyJBKellyJA 2009 Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet 10 285 290
11. MarchiniJHowieBMyersSMcVeanGDonnellyP 2007 A new multipoint method for genome-wide association studies by imputation of genotypes. NatGenet 39 906 913
12. HanJWZhengHFCuiYSunLDYeDQ 2009 Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 41 1234 1237
13. RonnblomLAlmGVElorantaML 2009 Type I interferon and lupus. Curr Opin Rheumatol 21 471 477
14. RonnblomLAlmGVElorantaML 2011 The type I interferon system in the development of lupus. Semin Immunol 23 113 121
15. TheofilopoulosANKoundourisSKonoDHLawsonBR 2001 The role of IFN-gamma in systemic lupus erythematosus: a challenge to the Th1/Th2 paradigm in autoimmunity. Arthritis Res 3 136 141
16. VeneRDelfinoLCastellaniPBalzaEBertolottiM 2010 Redox remodeling allows and controls B-cell activation and differentiation. Antioxid Redox Signal 13 1145 1155
17. CaliskanMCusanovichDAOberCGiladY 2011 The effects of EBV transformation on gene expression levels and methylation profiles. Human molecular genetics 20 1643 1652
18. Cunninghame GrahamDSGrahamRRMankuHWongAKWhittakerJC 2008 Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. NatGenet 40 83 89
19. ThomasRMChenCChunderNMaLTaylorJ 2010 Ikaros silences T-bet expression and interferon-gamma production during T helper 2 differentiation. J Biol Chem 285 2545 2553
20. BironCA 2001 Interferons alpha and beta as immune regulators–a new look. Immunity 14 661 664
21. SandlingJKGarnierSSigurdssonSWangCNordmarkG 2011 A candidate gene study of the type I interferon pathway implicates IKBKE and IL8 as risk loci for SLE. Eur J Hum Genet 19 479 484
22. De JagerPLJiaXWangJde BakkerPIOttoboniL 2009 Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet 41 776 782
23. WangHMorseHC3rd 2009 IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol Res 43 109 117
24. SmythDJCooperJDBaileyRFieldSBurrenO 2006 A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 38 617 619
25. SutherlandADaviesJOwenCJVaikkakaraSWalkerC 2007 Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to Graves' disease susceptibility. J Clin Endocrinol Metab 92 3338 3341
26. RobinsonTKariukiSNFranekBSKumabeMKumarAA 2011 Autoimmune Disease Risk Variant of IFIH1 Is Associated with Increased Sensitivity to IFN-{alpha} and Serologic Autoimmunity in Lupus Patients. Journal of immunology
27. YoneyamaMKikuchiMMatsumotoKImaizumiTMiyagishiM 2005 Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175 2851 2858
28. Cunninghame GrahamDSAkilMVyseTJ 2007 Association of polymorphisms across the tyrosine kinase gene, TYK2 in UK SLE families. Rheumatology (Oxford) 46 927 930
29. SigurdssonSNordmarkGGoringHHLindroosKWimanAC 2005 Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. AmJHumGenet 76 528 537
30. Saurez-GestalMCalazaMPullmannRRosJOSebastianiGD 2009 Replication of recently identified Systemic Lupus Erythematosus genetic factors: a case control study. Arthritis Res Ther
31. StarheimKKArnesenTGromykoDRyningenAVarhaugJE 2008 Identification of the human N(alpha)-acetyltransferase complex B (hNatB): a complex important for cell-cycle progression. Biochem J 415 325 331
32. TakakiSWattsJDForbushKANguyenNTHayashiJ 1997 Characterization of Lnk. An adaptor protein expressed in lymphocytes. J Biol Chem 272 14562 14570
33. ToddJAWalkerNMCooperJDSmythDJDownesK 2007 Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 39 857 864
34. HuntKAZhernakovaATurnerGHeapGAFrankeL 2008 Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet 40 395 402
35. GudbjartssonDFBjornsdottirUSHalapiEHelgadottirASulemP 2009 Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet 41 342 347
36. HuffmeierULascorzJBohmBLohmannJWendlerJ 2009 Genetic variants of the IL-23R pathway: association with psoriatic arthritis and psoriasis vulgaris, but no specific risk factor for arthritis. J Invest Dermatol 129 355 358
37. LiYBegovichAB 2009 Unraveling the genetics of complex diseases: susceptibility genes for rheumatoid arthritis and psoriasis. Semin Immunol 21 318 327
38. YangJChuYYangXGaoDZhuL 2009 Th17 and natural Treg cell population dynamics in systemic lupus erythematosus. Arthritis Rheum 60 1472 1483
39. TanEMCohenASFriesJFMasiATMcShaneDJ 1982 The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 25 1271 1277
40. PurcellSChernySSShamPC 2003 Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19 149 150
41. NayakRRKearnsMSpielmanRSCheungVG 2009 Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. Genome Res 19 1953 1962
Štítky
Genetika Reprodukční medicína
Článek Macroautophagy Is Regulated by the UPR–Mediator CHOP and Accentuates the Phenotype of SBMA MiceČlánek Dynamic Replacement of Histone H3 Variants Reprograms Epigenetic Marks in Early Mouse EmbryosČlánek Mutations in a Guanylate Cyclase GCY-35/GCY-36 Modify Bardet-Biedl Syndrome–Associated Phenotypes in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 10
-
Všechny články tohoto čísla
- Transcriptional Robustness Complements Nonsense-Mediated Decay in Humans
- Identification, Replication, and Fine-Mapping of Loci Associated with Adult Height in Individuals of African Ancestry
- Genetic Determinants of Serum Testosterone Concentrations in Men
- A One Base Pair Deletion in the Canine Gene Causes Exon Skipping and Late-Onset Neuronal Ceroid Lipofuscinosis in the Tibetan Terrier
- Three Structure-Selective Endonucleases Are Essential in the Absence of BLM Helicase in
- Identification of Widespread Ultra-Edited Human RNAs
- Multiple Wnts Redundantly Control Polarity Orientation in Epithelial Stem Cells
- The Bicoid Stability Factor Controls Polyadenylation and Expression of Specific Mitochondrial mRNAs in
- Transcriptome-Wide Binding Sites for Components of the Non-Poly(A) Termination Pathway: Nrd1, Nab3, and Sen1
- Macroautophagy Is Regulated by the UPR–Mediator CHOP and Accentuates the Phenotype of SBMA Mice
- Genetic Rearrangements Can Modify Chromatin Features at Epialleles
- Novel Function of as a Gap Gene during Spider Segmentation
- A Genome-Wide Screen for Interactions Reveals a New Locus on 4p15 Modifying the Effect of Waist-to-Hip Ratio on Total Cholesterol
- Comparative Genomic Analysis of Human Fungal Pathogens Causing Paracoccidioidomycosis
- Genetic Diversity in Cytokines Associated with Immune Variation and Resistance to Multiple Pathogens in a Natural Rodent Population
- Mutator Suppression and Escape from Replication Error–Induced Extinction in Yeast
- Dynamic Replacement of Histone H3 Variants Reprograms Epigenetic Marks in Early Mouse Embryos
- A Barcode Screen for Epigenetic Regulators Reveals a Role for the NuB4/HAT-B Histone Acetyltransferase Complex in Histone Turnover
- HIF–VEGF Pathways Are Critical for Chronic Otitis Media in and Mouse Mutants
- A Conserved Developmental Patterning Network Produces Quantitatively Different Output in Multiple Species of Drosophila
- Role of Exonic Variation in Chemokine Receptor Genes on AIDS: Association with Pneumocystis Pneumonia
- Whole-Exome Sequencing Identifies Homozygous Mutations in a Spastic Ataxia-Neuropathy Syndrome Linked to Mitochondrial -AAA Proteases
- Von Hippel-Lindau () Inactivation in Sporadic Clear Cell Renal Cancer: Associations with Germline Polymorphisms and Etiologic Risk Factors
- A Systems Biology Approach Reveals the Role of a Novel Methyltransferase in Response to Chemical Stress and Lipid Homeostasis
- Identification of Genomic Regions Associated with Phenotypic Variation between Dog Breeds using Selection Mapping
- Global Mapping of Cell Type–Specific Open Chromatin by FAIRE-seq Reveals the Regulatory Role of the NFI Family in Adipocyte Differentiation
- Natural Selection Affects Multiple Aspects of Genetic Variation at Putatively Neutral Sites across the Human Genome
- MicroRNA Expression and Regulation in Human, Chimpanzee, and Macaque Brains
- An Adaptive Allelic Series Featuring Complex Gene Rearrangements
- Feed-Forward Microprocessing and Splicing Activities at a MicroRNA–Containing Intron
- Developmental Stability: A Major Role for in
- A Phenomics-Based Strategy Identifies Loci on , , and Associated with Metabolic Syndrome Phenotype Domains
- Association of , , , , and with Systemic Lupus Erythematosus
- Small RNAs Prevent Transcription-Coupled Loss of Histone H3 Lysine 9 Methylation in
- Successive Increases in the Resistance of to Viral Infection through a Transposon Insertion Followed by a Duplication
- Mutations in a Guanylate Cyclase GCY-35/GCY-36 Modify Bardet-Biedl Syndrome–Associated Phenotypes in
- The Glycobiome Reveals Mechanisms of Pentose and Hexose Co-Utilization in Bacteria
- Insights into Hox Protein Function from a Large Scale Combinatorial Analysis of Protein Domains
- Mutations Cause Seckel and Jawad Syndromes
- Zelda Binding in the Early Embryo Marks Regions Subsequently Activated at the Maternal-to-Zygotic Transition
- Temporal Coordination of Gene Networks by Zelda in the Early Embryo
- Genetic Interaction between MTMR2 and FIG4 Phospholipid Phosphatases Involved in Charcot-Marie-Tooth Neuropathies
- Oxr1 Is Essential for Protection against Oxidative Stress-Induced Neurodegeneration
- Transforming Growth Factor β Receptor Type 1 Is Essential for Female Reproductive Tract Integrity and Function
- Positional Cloning of a Type 2 Diabetes Quantitative Trait Locus; , a Negative Regulator of Insulin Secretion
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Glycobiome Reveals Mechanisms of Pentose and Hexose Co-Utilization in Bacteria
- Global Mapping of Cell Type–Specific Open Chromatin by FAIRE-seq Reveals the Regulatory Role of the NFI Family in Adipocyte Differentiation
- Genetic Determinants of Serum Testosterone Concentrations in Men
- MicroRNA Expression and Regulation in Human, Chimpanzee, and Macaque Brains
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



