-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaActivation of Estrogen-Responsive Genes Does Not Require Their Nuclear Co-Localization
The spatial organization of the genome in the nucleus plays a role in the regulation of gene expression. Whether co-regulated genes are subject to coordinated repositioning to a shared nuclear space is a matter of considerable interest and debate. We investigated the nuclear organization of estrogen receptor alpha (ERα) target genes in human breast epithelial and cancer cell lines, before and after transcriptional activation induced with estradiol. We find that, contrary to another report, the ERα target genes TFF1 and GREB1 are distributed in the nucleoplasm with no particular relationship to each other. The nuclear separation between these genes, as well as between the ERα target genes PGR and CTSD, was unchanged by hormone addition and transcriptional activation with no evidence for co-localization between alleles. Similarly, while the volume occupied by the chromosomes increased, the relative nuclear position of the respective chromosome territories was unaffected by hormone addition. Our results demonstrate that estradiol-induced ERα target genes are not required to co-localize in the nucleus.
Published in the journal: . PLoS Genet 6(4): e32767. doi:10.1371/journal.pgen.1000922
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000922Summary
The spatial organization of the genome in the nucleus plays a role in the regulation of gene expression. Whether co-regulated genes are subject to coordinated repositioning to a shared nuclear space is a matter of considerable interest and debate. We investigated the nuclear organization of estrogen receptor alpha (ERα) target genes in human breast epithelial and cancer cell lines, before and after transcriptional activation induced with estradiol. We find that, contrary to another report, the ERα target genes TFF1 and GREB1 are distributed in the nucleoplasm with no particular relationship to each other. The nuclear separation between these genes, as well as between the ERα target genes PGR and CTSD, was unchanged by hormone addition and transcriptional activation with no evidence for co-localization between alleles. Similarly, while the volume occupied by the chromosomes increased, the relative nuclear position of the respective chromosome territories was unaffected by hormone addition. Our results demonstrate that estradiol-induced ERα target genes are not required to co-localize in the nucleus.
Introduction
Chromatin organisation in the vertebrate nucleus is non random: chromosomes adopt preferential positions with regard to the centre or edge of the nucleus and genes adopt preferential positions with regard to their own chromosome territory [1]. Moreover, preferential long-range associations have been found between loci, mainly in cis [2], [3] but also in trans [4]–[9]. Many of these associations have been suggested to be of functional significance for gene expression, either through the trans-interaction of genes and regulatory elements [4], [8], through the trans-sensing of homologous alleles prior to X chromosome inactivation [5], [9] or by the co-localisation of genes at the same transcription factory [7].
An instance of rapid and directed inter-chromosomal interactions has recently been reported for estrogen receptor α (ERα) target genes in primary human mammary epithelial cells (HMEC) and in a breast cancer cell line (MCF-7) [10]. ERα is a nuclear receptor that, in response to stimulation by 17β estradiol (E2), regulates gene expression by binding both promoters and more distal sites that may be long-range enhancers [2], [11]–[15]. E2 bound ERα accumulates in numerous nuclear foci [16], [17] which raises the possibility that there might be associations in the nucleus between multiple ERα binding sites, in cis and in trans. Activation of gene expression by ERα involves extensive chromatin remodelling mediated by the recruitment of histone modifying enzymes and nucleosome remodelling complexes [18]. Moreover, molecular motors such as dynein light chain (DLC1) have been reported to bind to ERα and to the promoters of ERα-responsive genes to potentiate their transcription [19], a dynactin component binds and modifies the function of ERα [20] and the microtubule network has also been implicated in ERα action [21]. These observations raise the possibility that directed long-range motion in the nucleus might be involved in ERα function.
Indeed, the rapid (within 1 hour) and directed long range movement of estrogen responsive genes reported after E2 exposure, was reported to be dependent on nuclear actin/myosin [10]. In particular, inter-chromosomal interactions detected by chromosome conformation capture (3C), and nuclear co-localisation revealed by fluorescence in situ hybridisation (FISH), were described between alleles of some estrogen inducible genes. More surprisingly, the movement was restricted to the gene loci concerned and involved rapid repositioning of the genes' chromosome territories within the nucleus. The estrogen-inducible genes that apparently showed this inter-chromosomal “kissing” [22] were TFF1 (also known as pS2) on chromosome 21 and GREB1 on human chromosome 2. Within 60 minutes of E2 addition to cells that had been grown in the absence of steroids, these genes were activated in ERα–positive MCF-7 cells and “monoallelic” and “biallelic” heterologous associations between GREB1 and TFF1 and between chromosomes paints for chromosomes 2 and 21 were reported, both in HMEC and MCF-7 cells [10].
Importantly, ERα activates the expression of these genes through de novo recruitment of RNA polymerase II (RNAPII), rather than, as is apparently the case for most ERα-responsive genes, through regulation of the phosphorylation state of RNAPII pre-loaded at the promoter [23]. Hence, it is possible that the reported nuclear co-localisation of these ERα-responsive genes represents their recruitment to a shared nuclear compartment that facilitates gene expression, such as transcription factories or splicing factor-enriched nuclear speckles [24].
Whilst there are other reported instances of rapid gene and locus motion within the nucleus [1], including an example where nuclear actin/myosin is involved [25], the view of rapid and extensive nuclear reorganisation induced by estrogen contrasts with other studies of the dynamics of specific loci or of whole chromosome territories. These have indicated that chromatin generally has limited mobility in mammalian cells. With the exception of the initial stages of G1 [26], chromatin motion appears to occur by constrained diffusion and is limited to a range of approximately 0.5 microns [27] over long periods (tens of minutes through to many hours) of interphase [27], [28]. Given this potential discrepancy, we sought to re-examine the nuclear organisation of TFF1 and GREB1 upon E2 stimulation in normal-like MCF10A and cancerous MCF-7 breast cancer cell lines and in primary HMECs. We found no evidence for nuclear co-localisation of TFF1 and GREB1 upon E2 stimulation in either situation and did not observe any directed, coordinated rearrangements of the chromosome 2 and 21 territories.
Results
Nuclear organisation of ERα–responsive genes in human mammary epithelial cells
The rapid inter-chromosomal co-localisation of estrogen responsive genes that are activated by addition of E2, and the nuclear repositioning of their chromosome territories, was reported in primary human mammary epithelial cells (HMECs) [10]. To reproduce this data, we prepared probes corresponding to the GREB1 and TFF1 loci and verified them, along with paints for chromosomes 2 and 21, by FISH to metaphase chromosomes from HT1080 and MCF10A cells, both of which have a near normal karyotype [29], [30]. The GREB1 and TFF1 probes mapped only to the expected positions at 2p25.1 and 21q22.3, respectively (Figure S1A and S1C), and each gave two distinct signals in the interphase nuclei of diploid cells (Figure S1B and S1D).
These probes were then used on nuclei from two independent cultures of HMECs grown either in charcoal-depleted stripped media, i.e. in the absence of E2 (-E2), or after 60 mins of stimulation by 100 nM E2 (+E2). Nuclear positions were analysed by both 2D and 3D FISH. 2D FISH affords faster image analysis and although it slightly exaggerates interphase distances compared to 3D [31] it gave remarkably similar results on TFF1-GREB1 distances compared to 3D analysis. Visual inspection of 3D FISH images revealed four distinct and separate hybridisation signals (two per gene) and so did not indicate any obvious co-localisation, either between homologous alleles of TFF1 or GREB1, or between heterologous alleles of these genes (Figure 1A). We measured the interphase distances between all combinations of the hybridisation signals, and we also normalised each inter-probe distance (d) by the radius (r) of a circle of equal area to that of the nucleus to account for any changes in nuclear size as a consequence of E2 addition (Figure 1B). In 3D analysis, there was no significant difference in the normalised inter-probe distances before and after addition of E2, either for homologous alleles p≥0.2, or for the heterologous TFF1-GREB1 probe pairs (p≥0.5). The mean separation between TFF1-GREB1 alleles after E2 addition was 11 µm, with only 0.5% of measurements ≤1 µm.
Fig. 1. Nuclear organisation of TFF1 and GREB1 in diploid primary and tumor epithelial cells. 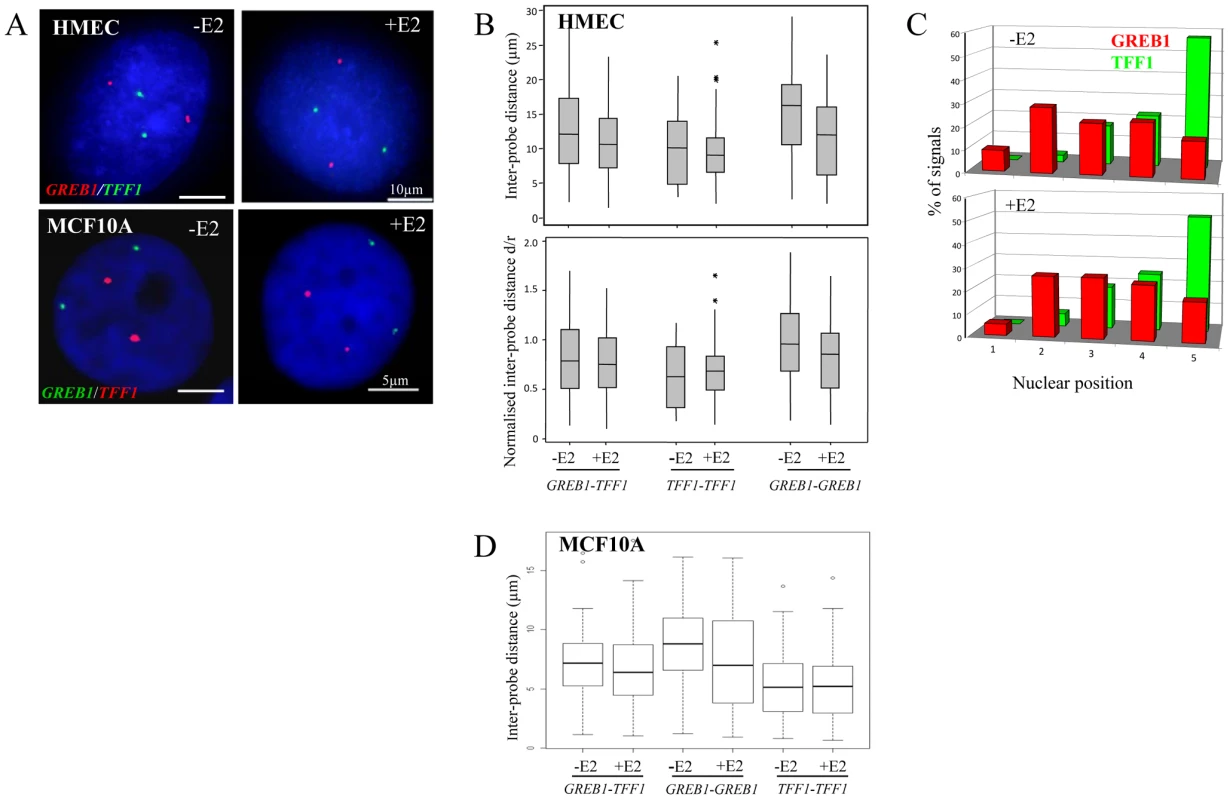
(A) Interphase FISH with probes (red or green) for TFF1 and GREB1 on nuclei from HMEC and MCF10A cells in the absence of estradiol (-E2), or after 60 mins of E2 addition (+E2). Nuclei were stained with DAPI (blue). Scale bars, 10 µm and 5 µm. (B) Box plots showing; top: inter-probe distances (d in µm) and bottom: inter-probe distances (d) normalized to nuclear radius (r) between homologous or heterologous TFF1 and GREB1 alleles, as measured either by 3D FISH in HMEC nuclei grown in the absence (-E2) or after 1 hr of 100 nM E2 addition. Shaded boxes show the mean and 25–75 percentiles of the data. Asterisks indicate data points beyond the 95th percentile. N = 50 cells. (C) Histograms showing the percentage of signals of the radial position of TFF1 and GREB1 alleles, before and after E2 addition, across 5 erosion shells placed between the edge (shell 1) and the centre (shell 5) of the nuclei. N = 50 cells. (D) Box plots showing inter-probe distances (µm) between homologous or heterologous TFF1 and GREB1 alleles, as measured by 3D FISH in MCF10A nuclei grown in the absence (-E2) or after 1 hr of 10 nM E2 addition. Asterisks indicate data points beyond the 95th percentile. N = 50 cells. The closest distances in HMEC nuclei were, in fact, recorded between the homologous TFF1 alleles in the absence of E2 (p≥0.004). We considered this likely due to the fact that TFF1 is located on the small acrocentric chromosomes 21, which are associated with the nucleolus and so constrained to a position within the small central volume of the nucleus. In contrast, chromosome 2, where GREB1 resides, has a more peripheral nuclear location, affording the possibility of much larger nuclear distances between the homologues [32]. Indeed, analysis of the radial nuclear position of these two genes confirmed the more central nuclear position of TFF1 alleles, with >50% of signals found in the innermost zone 5 of the nucleus (Figure 1C).
The absence of nuclear co-localisation of ERα-responsive genes after E2 addition to cultures of HMECs cells is not that surprising, since these cell types are generally considered to have low or undetectable levels of ERα [33], [34]. Indeed, immunohistochemical staining revealed the absence of detectable ERα in the nucleus of these cells (Figure 2B) and the absence of ERα in these cells was confirmed by western blot (Figure 2A). Similarly, MCF10A cells, which are spontaneously immortalized human breast epithelial cells [35], and which have a normal diploid complement of TFF1 and GREB1 alleles (Figure S1C), also have no detectable ERα protein levels (Figure 2A). As in HMECs, no co-localisation of homologous or heterologous TFF1 and GREB1 alleles was seen in these cells (Figure 1A). The mean inter-probe distances measured after 3D FISH between heterologous alleles were ∼7 µm, with no changes after E2 addition (less than 1% at ∼1 µm before and after E2 stimulation) (Figure 1D). Similarly, the smallest inter-probe distances were found in MCF10A cells between homologous TFF1 alleles, with ∼3% at less than 1 µm, in both the absence of E2 or after 1 h of E2 induction.
Fig. 2. Expression and genomic arrangement of TFF1 and GREB1 in breast epithelial cells and MCF-7 breast cancer cells. 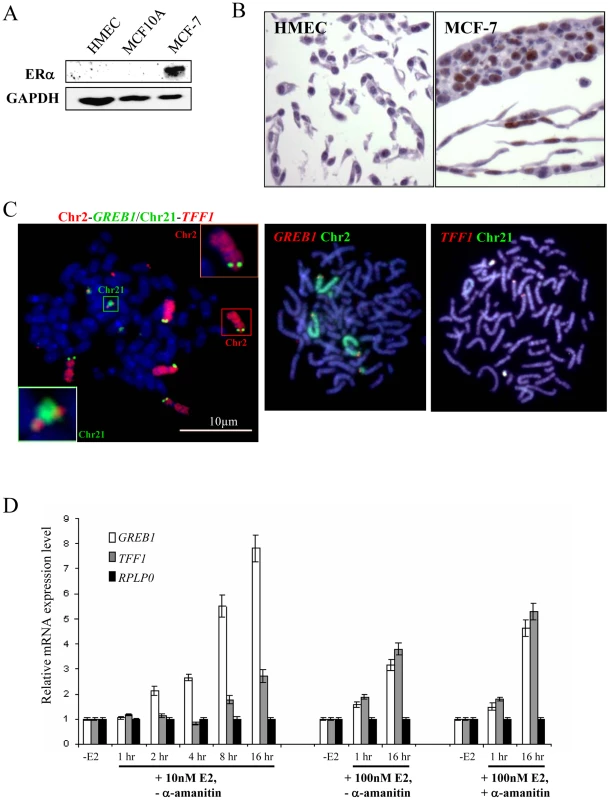
(A) Western blot to detect protein expression of ERα and GAPDH in total cell lysate prepared from HMEC, MCF10A and MCF-7 cells. (B) Immunohistochemical staining of HMEC and MCF-7 cells with antibody that detects ERα (brown). (C) FISH with probes for TFF1 and GREB1, and paints for chromosomes 2 and 21, on metaphase chromosomes prepared from MCF-7 cells. Bars, 10 µm. (D) Real-time qPCR analysis to detect the relative expression levels of GREB1, TFF1 and a RPLP0 control prepared from MCF-7 cells grown without E2 (-E2) and during 16 hr time courses in the presence of either 10 nM or 100 nM E2. Cells with (+) and without (−) pre-treatment with 2.5 nM α–amanitin were also examined. Chromosomal rearrangements of regions carrying ERα–responsive genes in MCF-7 cells
We therefore tested the same TFF1 and GREB1 probes on ERα–positive (Figure 2A) MCF-7 breast cancer cells, which have also been reported to demonstrate rapid nuclear colocalisation of TFF1 and GREB1 upon E2 addition [10]. In this cell line between 4–6 hybridisation signals were seen for each of the gene probes on both metaphase and interphase MCF-7 preparations (Figure 2C). Similarly, chromosome paints indicated extensive rearrangement and gain of material from chromosomes 2 and 21 in independent isolates of these cells. There appear to be two normal copies of chromosome 2 that carry GREB1 and one GREB1-carrying copy with additional material translocated onto the long arm of chromosome 2. A fourth copy of GREB1 is on a small portion of chromosome 2p translocated onto another chromosome. Similarly, two copies of TFF1 are on normal-looking chromosomes 21, with two or four additional copies translocated onto unidentified large chromosomes. Variable karyotypes have been described, by both cytogenetic and molecular methods, from MCF-7 cells grown in different labs at different times, but in all of them the cell line is highly aneuploid [36]–[41]. This genomic instability was recently confirmed by deep-sequencing [42] and our analysis is compatible with this.
Using quantitative RT-PCR (qRT-PCR) we confirmed that TFF1 and GREB1 expression was activated upon addition of E2 to MCF-7 cells. Activation of E2-responsive genes has been reported after addition of both 10nM [17], [18] and 100 nM [10], [23], [43] E2. Indeed, relative TFF1 and GREB1 mRNA levels in our MCF-7 cells increased 3 and 8 fold, respectively, after 16 h exposure to 10 nM E2 and 4 and 3 fold, respectively, using 100 nM E2 (Figure 2D). Moreover, steady-state levels of GREB1 and TFF1 mRNA increased 2 fold within 1 hr of 100 nM E2 addition, the time period during which co-localisation of these gene loci has been reported [10]. Several previous studies have also used a pre-treatment with the RNA polymerase II inhibitor α-amanitin before the addition of E2, in order to remove any active ongoing transcription from genes before their induction [10], [18]. In our analysis, α-amanitin did not change the initial response of cells to E2, but steady state levels of TFF1 and GREB1 mRNAs after 16 hrs E2 exposure were higher in the α-amanitin pre-treated cells, compared to untreated cells (Figure 2D).
Absence of nuclear co-localisation of ERα–responsive genes in MCF-7 cells
We did not observe co-localisation of GREB1-TFF1 alleles in untreated or E2 treated MCF-7 cells (Figure 3A). We analysed the inter-probe distances (Figure 3B) and the normalised inter-probe distances (Figure S2A) between all possible homologous or heterologous pairs of gene signals in nuclei from MCF-7 cells grown in steroid-free media in the absence of E2, and then 1 and 16 hrs after the addition of either 10 nM or 100 nM E2. We scored >750 inter-probe distances of TFF1-GREB1 alleles and found on average 5%<2 µm (2–11 measured distances per experiment with or without E2) and 2.5%<1 µm (1–3 distances per experiment). We never observed a single nucleus in which all possible TFF1-GREB1 distances were <1 or 2 µm. Overall in ∼10% of nuclei one or two inter-probe distances <2 µm were observed. This proportion was highly variable (3–18%) from experiment to experiment since there were so few small distances. Such close proximity between two genes may thus be a transient and randomly occurring situation. In addition we also analysed the nuclear organisation of these genes in response to 100 nM E2 addition in cells which had been pre-treated with α-amanitin, the experimental conditions under which nuclear colocalisation of TFF1 and GREB1 has previously been reported 1 hr after E2 addition [10]. There was also no difference in the TFF1-GREB1 distances before and after E2 addition (p>0.2). The average inter-probe distance in 100 nM E2 treated cells was 6 µm, and only 3% and 2% of distances between heterologous alleles were ≤1 µm in untreated or α-amanitin pre-treated MCF-7 cells, respectively. We did also not observe any changes in distances between GREB1-GREB1 alleles (p>0.2) or TFF1-TFF1 alleles (p>0.4) upon E2 stimulation.
Fig. 3. TFF1 and GREB1 nuclear organisation in ERα–positive MCF-7 breast cancer cells. 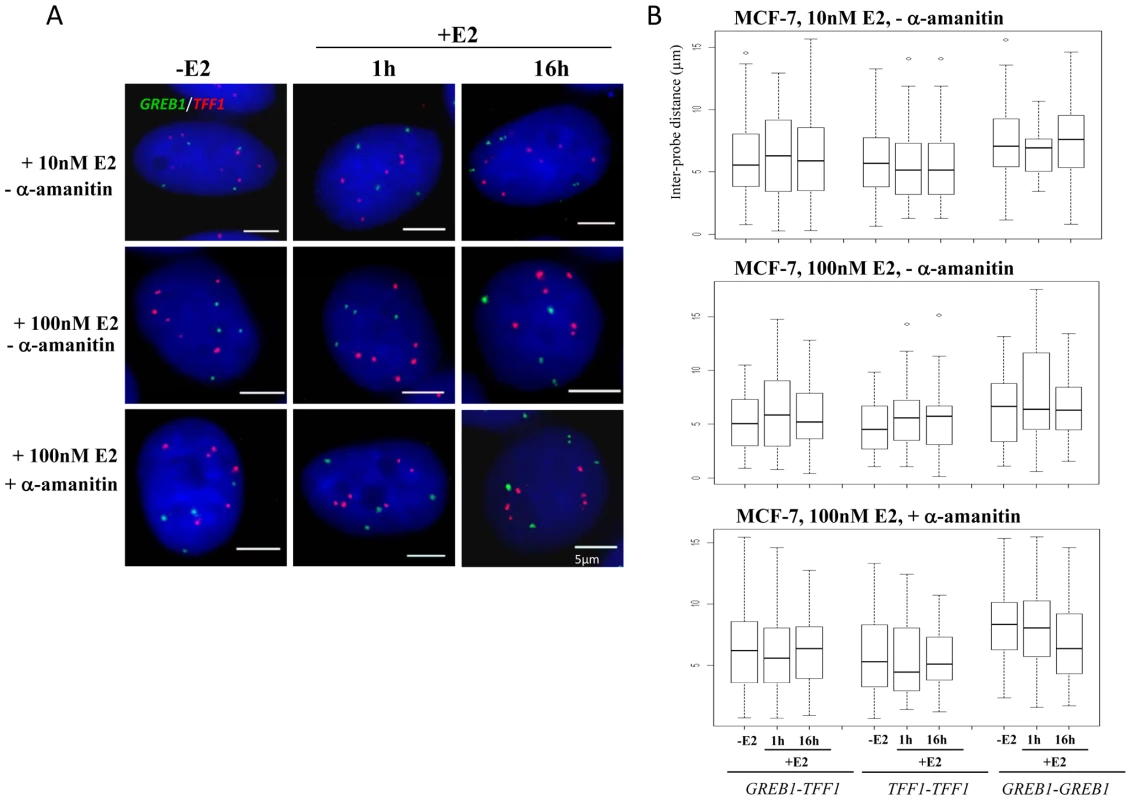
(A) 3D Interphase FISH with probes for TFF1 (red) and GREB1(green) on nuclei of MCF-7 cells grown either in the absence of estrogen (-E2), or 1 and 16 hrs after the addition of 10 nM (top row) or 100 nM (middle row) E2 (+E2). Bottom row: 1 h and 16 hrs of 100 nM E2 addition following pre-treament with 2.5 nM α-amanitin. Nuclei were stained with DAPI (blue). Scale bars = 5 µm. (B) Box plots show inter-probe distances (µm) between homologous or heterologous TFF1 and GREB1 alleles, as measured either by 3D FISH in nuclei of MCF-7 cells grown in the absence of E2 (-E2) or 1 and 16 hr after the addition of 10 nM (top) or 100 nM (middle) E2. Bottom row: 1 h and 16 hrs of 100 nM E2 addition following pre-treament with 2.5 nM α–amanitin. Asterisks indicate data points beyond the 95th percentile. N = 40 cells. To determine whether the absence of nuclear colocalisation between TFF1 and GREB1 was an exception or a more general feature of estrogen induced transcription, we analysed the positioning of the progesterone receptor (PGR) and Cathepsin-D (CTSD) genes, both located on different regions of chromosome 11. Metaphase FISH indicated that both alleles of these genes reside on normal-looking chromosomes 11 in MCF-7 cells (Figure 4A). There are four additional translocation fragments that contain material from the centromere of chromosome 11 as reported by Bautista et al. [44], but these contain no additional copies of our genes of interest. Using qRT-PCR, we confirmed that PGR and CTSD expression was activated upon addition of E2 to MCF-7 cells (Figure 4B) with relative mRNA levels increasing 3 and 8 fold, respectively, after 16 h exposure to 10 nM E2.
Fig. 4. Localisation, expression, and nuclear organisation of PGR and CTSD in MCF-7 cells. 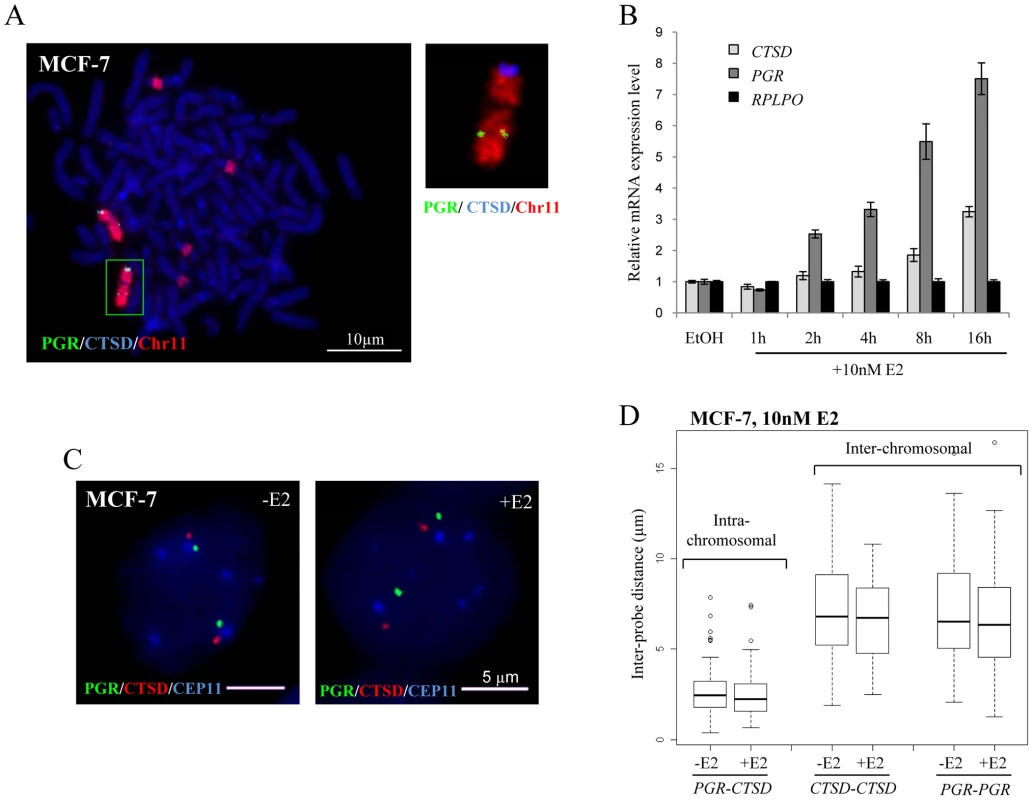
(A) FISH with probes for CTSD (blue) and PGR (green) and paint for chromosome 11 (red) on metaphase chromosome spreads of MCF-7 cells. Scale bars = 10 µm. (B) Real-time qPCR analysis to detect the relative expression levels of PGR, CTSD and a RPLP0 control prepared from MCF-7 cells grown without E2 (-E2) and during 16 hr time courses in the presence of 10 nM E2. (C) 3D Interphase FISH with probes for CTSD(red), PGR(green) and the centromere of chromosome 11 CEP11 (blue) on nuclei of MCF-7 cells grown in the absence of estrogen (-E2) and 3 hrs after the addition of 10 nM E2 (+E2). Nuclei were counterstained with DAPI (blue). Scale bars = 5 µm. (D) Box plots show inter- and intra-chromosomal distances (µm) between PGR and CTSD alleles, as measured by 3D FISH in nuclei of MCF-7 cells grown in the absence of E2 (-E2) and 3 hrs after the addition of 10 nM E2. Asterisks indicate data points beyond the 95th percentile. N = 50 cells. As for the GREB1-TFF1 alleles, we did not observe any change in nuclear separation for the PGR-CTSD alleles in untreated versus E2 treated MCF-7 cells (Figure 4C). Indeed, there was no difference in intra-chromosomal PGR-CTSD, or inter-chromosomal PGR-PGR or CTSD-CTSD distances before and after E2 (10 nM, 3 hrs) addition (p>0.5) (Figure 4D). As expected from the existence of chromosome territories, the average intra-chromosomal inter-probe distances were less than the inter-chromosomal ones, both before and after E2 addition (PGR-CTSD, 2.5 µm; PGR-PGR, 6.8 µm; CTSD-CTSD, 6.6 µm). Less than 1% of the inter-chromosomal and 10% of the intra-chromosomal probe distances were ≤1 µm. We did not observe any nuclei in which probe signals overlapped (<250 nm separation).
Absence of nuclear co-localisation of ERα–responsive genes in cells with elevated TFF1
In addition, we investigated the position of GREB1 and TFF1 genes in LCC1 and LCC9 cells which are clonal derivatives of the MCF-7 cell line [45], [46]. LCC1 is hormone independent for growth but still responsive to estrogen and anti-estrogens. LCC9 cells are resistant to anti-estrogens, fulvestrant and tamoxifen. Compared to MCF-7 cells ESR1 mRNA levels are tripled in LCC1 cells and reduced to about 30% in LCC9 cells, yet both cell lines express very high baseline levels of TFF1 (Figure 5A).
Fig. 5. TFF1 and GREB1 nuclear organisation in LCC1 and LCC9 cells. 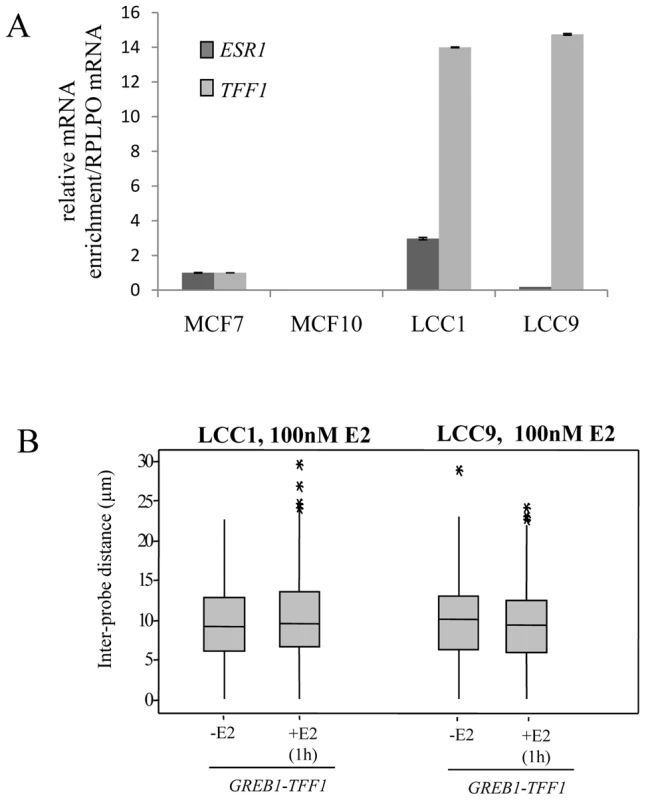
(A) Real-time qPCR analysis to detect the relative expression levels of ERα (ESR1) and TFF1 prepared from MCF-7, MCF10A, LCC1 and LCC9 cells grown in phenol red containing DMEM. (B) Box plots showing inter-probe distances (d) between heterologous TFF1 and GREB1 alleles, as measured by 2D FISH in nuclei of LCC1 or LCC9 cells grown in the absence of E2 (-E2) or after 1 hr in the presence of 100 nM E2. Asterisks indicate data points beyond the 95th percentile. N = 50 cells. As for the parental MCF-7 cells, we found no evidence for nuclear co-localisation of TFF1 and GREB1 before or after E2 addition in either LCC1 or LCC9 cell lines (Figure 5B and Figure S2B). These results suggest that increased transcription rates of the TFF1 gene do not promote interaction with another ERα-regulated gene.
Absence of nuclear re-organisation of chromosome territories
Finally, we examined the relative nuclear position of the territories of chromosomes 2 and 21 by 3D FISH. In MCF10A cells, the two territories of chromosome 2 (Chr2) are frequently near the nuclear periphery, while, as expected, Chr21 localised near the nucleoli in a more central nuclear position (Figure 6A) [32]. In 40–50% of the untreated cells, Chr2 and Chr21 are adjacent to each other yet without significant overlap. Upon 1 h exposure to 10 nM E2, the relative positions of Chr2 and Chr21 and their general localisation in the nucleus did not vary from that observed in untreated cells. Notably, we did not observe an increase in cells in which Chr2 and Chr21 co-localised. We analysed the radial nuclear position of Chr2 and Chr21 and we confirmed more than 40% of Chr21 in the central nuclear space (shell 5) compare to the edge (shell 1) of the nuclei (Figure 6B). These observations are in good correlation with the results for TFF1 genes (Figure 1C) that are located on Chr21.
Fig. 6. Chromosome territory organisation in MCF10A and MCF-7 cells. 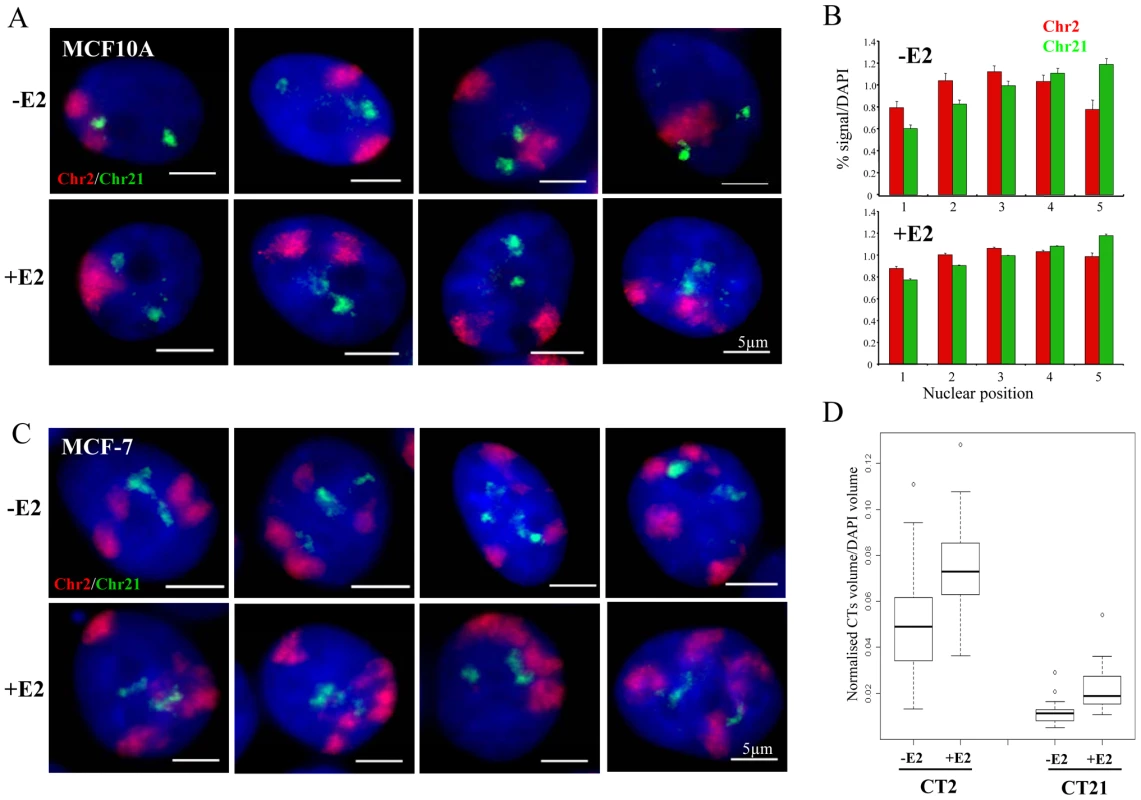
(A) 3D interphase FISH with chromosome paint to chromosome territory 2 (red) and 21 (green) on nuclei from MCF10A cells imaged in the absence of estrogen (-E2), or after 60 mins of 10 nM E2 addition (+E2). Nuclei were stained with DAPI (blue). Scale bars = 5 µm. (B) Histograms showing the percentage of chromosome paint hybridization signal normalized to the percentage of DAPI signal, before and after E2 addition, across 5 erosion shells placed between the edge (shell 1) and the centre (shell 5) of the nuclei. N = 25−30 cells. (C) 3D interphase FISH with chromosome paint to chromosome territory 2 (red) and 21 (green) on nuclei from MCF-7 cells imaged in the absence of estrogen (-E2), or after 60 mins of 100 nM E2 addition (+E2). Nuclei were stained with DAPI (blue). Scale bars = 5 µm. (D) Box plots show the percentage of chromosome paint hybridization signal (CT2 and CT21) normalized to the percentage of DAPI signal, before and after E2 addition. Asterisks indicate data points beyond the 95th percentile. N = 30 cells. We also examined the position of Chr2 and Chr21 in MCF-7 cells, where inter-chromosomal associations between the territories of these two heterologous chromosomes have been reported to be induced by E2 [10]. We did not observe any obvious significant overlap between Chr2 and Chr21 (Figure 6C, representative pictures are shown) upon E2 stimulation. However, the highly aneuploid and highly rearranged nature of these chromosomes in this breast cancer cell line (Figure 2C) makes it impossible to unequivocally differentiate the large number of chromosome territories and to objectively quantify their spatial relationships. Interestingly, we observed that after estradiol treatment both chromosome territories expanded. We examined the volume of chromosome territories and we detected ∼1.5 fold increase after E2 stimulation in MCF-7 cells (Figure 6D). We note that Chr2 and Chr21 are frequently neighboring each other, but this relative position did not change upon E2 addition and did not lead to colocalisation of TFF1 and/or GREB1 genes (Figure 3A).
Discussion
The regulation of gene expression by elements located in cis, even when >1 Mb from the gene, is well accepted and increasingly well understood. In one case, this has been shown to be accompanied by co-localisation in the nucleus between the distant enhancer and the gene [47]. Because there is a degree of intermingling between chromosome territories [48] and because individual loci can loop out of their own chromosome territory [49], it is conceivable that a regulatory element could interact with a gene locus located on another chromosome. Such an interaction might be between homologous chromosomes when there is a need to establish differential expression states on these chromosome pairs, such as in X-chromosome inactivation (XCI) or imprinting. Whereas the transient pairing between X chromosomes during the establishment of XCI has been confirmed by two independent groups [5], [9], nuclear co-localisation between imprinted regions of autosomes has not been substantiated [50]–[52].
In vertebrate cells, even if reports of nuclear co-localisation between genes and regulatory elements in trans were to be verified [4], [8], [10], their functional significance remains unclear in the absence of direct genetic evidence for specific regulation in trans between heterologous chromosomes [53]. Such a deterministic view of nuclear organisation is also at odds with the non-heritable relative position of heterologous chromosomes. Although the disposition of chromosome territories in the nucleus is non-random, with a gene-density related radial organisation well described, it is also probabilistic in that the precise neighbourhoods of any particular chromosome change from one cell cycle to the next [28], [54]. This seems too loose an arrangement of chromosomes to ensure the dependable spatial juxtaposition of genes and regulatory elements in trans.
A less rigid spatial arrangement of genes in trans may come about through the sharing of multiple, but limited, nuclear compartments by genes that are active or inducible in a particular cell type. Such compartments might be rather general, for example transcription factories, or splicing factor enriched speckles [24], [55], [56], indeed it is suggested that the co-localised alleles of TFF1 and GREB1 in E2-treated breast epithelial cells are associated with the latter nuclear compartment [10]. Alternatively, such compartments may be specific to a particular pathway of regulated gene expression [57]. Nuclear hormone receptors have been reported to concentrate in discrete foci in the nucleus. In the case of the progesterone receptor, these foci do appear to correspond to sites of RNA synthesis, raising the possibility that they represent the specific nuclear sites of transcription of genes regulated by this hormone receptor [58], however, foci of the glucocorticoid receptor do not seem to correspond to sites where genes are expressed [59]. For ERα, the presence of the agonist ligand E2 results in rapid redistribution of the protein to discrete nuclear foci [16], [17]. Although these foci do correspond to a less mobile form of ERα - indicating binding, and they are the sites of recruitment of co-activators such as SRC-1, they do not significantly overlap with sites of transcription, calling into question whether they are the actual nuclear compartments for the activation of ERα-induced genes. Nor do these foci overlap with splicing factor enriched nuclear speckles [17], which are the nuclear structures where ERα-induced gene loci have been reported to co-localise [10].
E2-induced nuclear foci are numerous – there appear to be many 100 s or 1000 s of them [17] and their number is not dissimilar to the estimated range of E2-upregulated genes (∼100–700) or ERα binding sites (∼1000–3500) in the genome of MCF-7 cells [12], [14]. Therefore, it is not clear why up to 60% [10] of any two particular heterologous E2-induced genes (e.g. TFF1-GREB1) should co-localise at just one nuclear site, rather than each gene localizing at any of the many other ERα sites or splicing factor enriched speckles, perhaps with any of the other 100 s of E2-induced alleles. If this were a reflection of a pre-existing spatial proximity of the two genes in the nucleus, then one might rather expect to see preferential co-localisation of the two homologous alleles of TFF1, since we have shown (Figure 1B) that these alleles have a closer spatial proximity to each other, than to the GREB1 alleles. We suggest that this reflects the location of TFF1 on the small acrocentric chromosomes 21 that are constrained, through their nucleolar association, to the small central volume of the nucleus (Figure 1C). Indeed, a recent re-evaluation of reported pairing between imprinted alleles on human chromosomes 15, suggested that this was a secondary effect due to the convergence of the acrocentric chromosome 15 s at the nucleolus [52].
In normal and cancer breast epithelial cells, we can find no evidence for E2-induced nuclear co-localisation between the heterologous or homologous alleles of TFF1 and GREB1 (Figure 1, Figure 2, Figure 3, and Figure 5) or indeed for any allele combination of another pair of ERα-regulated genes, PGR and CTSD located in cis on the same chromosome as each other (Figure 4). Given the existence of chromosome territories as a major principle of nuclear organisation, this might have afforded increased opportunity for gene co-localisation in cis, albeit at long range. We cannot exclude that “chromosome kissing” between these genes is very transient and that we have just missed a critical time-window in our analysis, either before 1 hour of induction or between 1 and 16 hours, or that the increased steady state levels after the addition of E2 is the result of transcription from a very small proportion of alleles at any one moment in time [60]. However, TFF1-GREB1 co-localisation has been reported as soon as 15 minutes after E2 addition and until 60 minutes after hormone addition, and precedes gene activation itself [10]. Similarly, ERα and its co-activators are bound to promoters of responsive genes within 30 minutes of E2 addition and are still there at 60 minutes [18], [43], [61]. Even at very long time periods (16 hours) after E2 addition, when we see maximal expression of PGR, CTSD, TFF1 and GREB1 in MCF-7 cells (Figure 2D and Figure 4B), we find no evidence for nuclear co-localisation of these genes (Figure 3B, Figure 4C). Moreover, our demonstration that HMECs do not express ERα (Figure 2A and 2B) and that MCF-7 cells are karyotypically abnormal and aneuploid for GREB1/Chr 2 and TFF1/Chr21 (Figure 2C, Figure 3A, and Figure 6C) precludes these cellular systems as having being appropriate models in which to record bi-allelic “kissing” of these loci and chromosomes in response to stimulation by estrogen.
In agreement with the notion that chromosome order established at the exit of mitosis remains stable throughout interphase [28], the relative nuclear position of the analysed chromosome territories was also unaffected by hormone addition in normal and cancerous cells. We noticed, however, that upon E2 treatment the nuclear volume occupied by the chromosomes increased about 1.5 fold. The association of agonist bound ERα with ∼100 s of target genes at numerous intranuclear sites causes decondensation [62] of chromatin simultaneously at all sites of activated transcription. This general chromatin decompaction thus supports our view that hormone stimulated gene activation occurs at multiple sites throughout the nucleoplasm.
Consistent with our findings, a recent genome-wide analysis, using a new CHIA-PET method, of long-range ERα-bound chromatin interactions in E2 stimulated MCF-7 cells [63] detected 689 intra-chromosomal interactions, but no validated inter-chromosomal interactions were found. Even if there were limitations in the coverage or interpretation of this data set, this experiment indicates that any specific ERα-mediated interactions between TFF1-GREB1 [10], or indeed between any other functionally relevant combination of ERα targets on different chromosomes, occurs at a low frequency compared with intra-chromosomal interactions. Moreover, the intra-chromosomal interactions detected were mostly within a 100 kb size range, including those for TFF1, GREB1, PGR and CTSD, <1% encompassed distances of >1 Mb [63]. We therefore conclude that ERα does not generally mediate rapid interactions between distant target genes, either located >1 MB in cis (PGR-CTSD) or in trans (TFF1-GREB1), but that changes in chromatin and nuclear structure mediated by ERα are relatively local (generally within a 100 kb size range).
Materials and Methods
Cell culture and cell lines
Primary HMEC cells of luminal origin were isolated from normal breast tissue as previously described [64] and were maintained in CnT22 medium (CellnTEC) supplemented with 10% heat-inactivated fetal calf serum (FCS). The normal-like diploid human breast epithelial cell line MCF10A (ATCC purchase at July 2008 as a passage No. 97 and we used up to 10th passages of MCF10A in all experiments) was maintained in Dulbecco's modified Eagle's medium F-12 (DMEM F-12) with Glutamax containing Mammary Epithelial Growth Supplement (MEGS), 10 ng/ml hEGF and 100 ng/ml cholera toxin.
Experiments on the human ERα–positive breast cancer cell line MCF-7 were conducted independently in both Toulouse and Edinburgh. In Toulouse, the cells were from ATCC, purchased at February 2008 as a passage No. 146 and used up to 10th passages, maintained in DMEM F-12 with Glutamax containing 50 µg/ml gentamicin, 1 mM sodium pyruvate and 10% FCS. In Edinburgh MCF-7, as well as LCC1 and LCC9 cells were a gift of Bob Clarke (Georgetown University School of Medicine, Washington DC). These cells were maintained in DMEM without phenol red and supplemented with 5% L-Glutamine, 5% Penicillin/Streptomycin and 10% FCS. All cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
To study the effects of 17β estradiol (E2) on ERα-target gene dynamics, cells were grown for 3 days in medium containing phenol red-free media supplemented with 5–10% charcoal-stripped FCS (-E2) and subsequently treated with 10 nM or 100 nM E2 (Sigma) for the indicated times. The cells synchronized by α-amanitin were pre-treated for 2 h with 2.5 nM α-amanitin, followed by washing and recovering for 1 h in normal steroid and phenol-red free media and then stimulated with 100 nM E2 for 1 h and 16 h.
IHC
HMECs and MCF-7 cells were harvested by cell scraping and the cell pellets were mixed in 2% agarose/PBS. The pellet mix was left to cool and then paraffin-embedded using a Leica ASP300S automatic processor. Standard immunohistochemistry protocol was performed on 3 micron sections using the REAL EnVision mouse/rabbit kit (Dako), according to manufacturer's instructions. Antigen retrieval for ERα was performed using sodium citrate buffer (18 µM Citric Acid, 82 µM sodium citrate, pH 6.0). Anti-human ERα antibody (Vector Labs, VP-E613) was used at a dilution of 1∶50 for 1 h in room temperature.
FISH
2D FISH on metaphase and interphase cells was as previously described [65]. In Toulouse, 3D FISH experiments were adapted from a previously protocol [66]. Cells were grown for 3 days on coverslips in DMEM without phenol red, containing 5% charcoal-stripped FCS, before addition of 10 nM or 100 nM E2 for the indicated times. Coverslips were washed twice with PBS, fixed in 4% paraformaldehyde (pFA)/PBS for 10 mins at room temperature and during the last minute 200 µl of 0.5% Triton X-100/PBS were added into 500 µl of 4% pFA. After fixation, the cells were washed three-times for 3 mins in 0.01% Triton X-100/PBS, incubated in 0.5% Triton X-100/PBS for 10 mins at room temperature and then incubated with 0.1 mg/ml RNase in 2xSSC for 1 hour at 37°C. After 3×10 mins washes in PBS, cells were incubated in 0.1 M HCl for 5 mins at room temperature, twice in 2xSSC for 3 mins and then equilibrated overnight in 50% formamide/2xSSC (pH = 7.2). MCF10A cells, with a large cytoplasm, underwent optional treatment with pepsin. In brief, equilibrated slides (kept in 50% formamide/2xSSC) were incubated in 2xSSC for 2 mins at room temperature, equilibrated in PBS for 3 mins at room temperature, incubated in 0.005% pepsin/0.01 M HCl pre-warmed to 37°C for 5 mins, incubated twice in 50 mM MgCl2 for 5 mins at room temperature, post-fixed in 1% pFA/PBS for 10 mins at room temperature, washed in PBS for 5 mins at room temperature, washed twice in 2xSSC for 5 mins and then returned into 50% formamide/2xSSC (pH = 7.2) for at least 1 hour before hybridization. Denaturation of the probes and target DNA is performed simultaneously at 85°C for 2 minutes.
In Edinburgh, cells were grown on Superfrost slides in the appropriate media, washed 3x in PBS before fixation in 4%pFa/PBS for 10 mins. After 3 further washes in PBS the cells were permeabilised in 0.5% Triton X-100/PBS for 10 mins and then washed 3x in PBS again. The slides were then incubated in 0.1 mg/ml RNase in 2xSSC for 1 hour at 37°C, washed again and then put through an ethanol series (70%, 90%, 100%) for 2 minutes each. The slides were warmed to 70°C in an oven for 5 minutes prior to denaturation in 70% deionised formamide, 2xSSC, pH 7.0 for 15 minutes at 80°C.
DNA from BAC clones (Toulouse) which include TFF1 (RPCIB753I15619Q), GREB1 (RZPDB737C102019D), PGR (RP11-788M5) or CTSD (RZPDB737F022085D) were directly labeled using nick translation (BioPrime DNA Labeling System, Invitrogen) by incorporation of fluorochrome-conjugated nucleotides Atto647N-dUTP-NT (Jena Bioscience) or ChromaTide AlexaFluor 488-5-dUTP (Molecular Probes). DNA from fosmid clones (Edinburgh) which encompass TFF1 (G248P89501F6) or GREB1 (G248P80076843) were nick-translated with biotin-16-dUTP or digoxigenin-11-dUTP. Chromosome paints were obtained from Genetix Ltd., UK (Toulouse) or from Cambio (Edinburgh). We used 100 ng labeled DNA probe or 100 ng chromosome paint together with 7 µg Cot-1 DNA and 5 µg sonicated salmon sperm DNA per slide. CEP11 (chromosome 11 alpha-satellite probe D11Z1) was obtained from MP Biomedicals.
Microscopy and quantification
In Toulouse cells were examined by fluorescence microscopy using an Olympus IX-81 microscope, equipped with a CoolSNAPHQ camera (Roper Scientific) and imaged through an Olympus oil-immersion objective 100x PLANAPO NA1.4. Subnuclear position was captured on 21-image (200 nm step size) stacks and analysis of inter-probe distances between the centroid of each signal was performed using Metamorph software (Universal Imaging). Images were processed using Adobe Photoshop 9.0.2.
In Edinburgh, 2D specimens were examined with a Zeiss Axioplan II microscope fitted with Plan-neofluar oil-immersion objectives, a 100 W Hg source and Chroma #8300 triple band pass filter set. Image capture and analysis of nuclear size and distance between the centroids of the hybridization signals was performed with scripts written for IPLab Spectrum (Scanalytics Copr, Fairfax, VA) as previously described [67]. Three-dimensional images were captured at 200 nm intervals in the z axis, using an objective fitted with a Pifoc motor.
Images from 30–50 nuclei were analysed in each experiment and the significance of any difference in the data distributions was assessed using the non-parametric Mann-Whitney U test. A p-value ≤0.05 was considered statistically significant.
Western blotting
The cells were washed with ice-cold PBS and total cell lysates were prepared by resuspending the cells in lysis buffer. The samples were boiled for 20 min at 95°C and cleared by centrifugation at 12 000×g for 10 mins. Next, the samples were subjected to SDS-PAGE and the proteins transferred onto nitrocellulose membrane. Western blot analysis was performed using ERα (HC-20, Santa Cruz Biotechnology, Inc.) and GAPDH (MAB374, Chemicon International) antibodies and processed using the MultiGauge Software from FUJI.
qRT–PCR
Total RNAs were extracted using TRIzol Reagent (Invitrogen). 1–5 µg of total RNA was reverse transcribed in a final volume of 20 µl using SuperScriptr III Reverse Transcriptase. cDNA was stored at −80°C. All target transcripts were detected using quantitative RT-PCR (SYBRGreen) assays on a Mastercycler Realplex device using RPLP0 as endogenous control for normalization of the data. The following primer pairs were used for amplification:
RPLP0: (Fwd) 5′-TGGCAGCATCTACAACCCTGAA -3′
(Rev) 5′-ACACTGGCAACATTGCGGACA -3′
GREB1: (Fwd) 5′-GTGGTAGCCGAGTGGACAAT-3′
(Rev) 5′-AAACCCGTCTGTGGTACAGC-3′
TFF1: (Fwd) 5′-CCCCTGGTGCTTCTATCCTAAT-3′
(Rev) 5′-CAGATCCCTGCAGAAGTGTCTA-3′
PGR: (Fwd) 5′-CTTAATCAACTAGGCGAGAG-3′
(Rev) 5′-AAGCTCATCCAAGAATACTG-3′
CTSD: (Fwd) 5′-GCGAGTACATGATCCCCTGT-3′
(Rev) 5′-CTCTGGGGACAGCTTGTAGC-3′
The thermal cycling condition comprised 2 mins at 50°C and 2 mins at 95°C followed by 40 PCR cycles (95°C for 15 sec, 58°C for 30 sec, 72°C for 20 sec). Melting curves were recorded from 60°C to 95°C and all PCR products revealed single bands. The results were analyzed using Mastercycler Realplex and qBASE software.
Supporting Information
Zdroje
1. FraserP
BickmoreW
2007 Nuclear organization of the genome and the potential for gene regulation. Nature 447 413 417
2. FullwoodMJ
RuanY
2009 ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem 107 30 39
3. SimonisM
KlousP
SplinterE
MoshkinY
WillemsenR
2006 Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38 1348 1354
4. ApostolouE
ThanosD
2008 Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134 85 96
5. BacherCP
GuggiariM
BrorsB
AuguiS
ClercP
2006 Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol 8 293 299
6. LomvardasS
BarneaG
PisapiaDJ
MendelsohnM
KirklandJ
2006 Interchromosomal interactions and olfactory receptor choice. Cell 126 403 413
7. OsborneCS
ChakalovaL
BrownKE
CarterD
HortonA
2004 Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36 1065 1071
8. SpilianakisCG
LaliotiMD
TownT
LeeGR
FlavellRA
2005 Interchromosomal associations between alternatively expressed loci. Nature 435 637 645
9. XuN
TsaiCL
LeeJT
2006 Transient homologous chromosome pairing marks the onset of X inactivation. Science 311 1149 1152
10. HuQ
KwonYS
NunezE
CardamoneMD
HuttKR
2008 Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A 105 19199 19204
11. CarrollJS
MeyerCA
SongJ
LiW
GeistlingerTR
2006 Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38 1289 1297
12. KininisM
ChenBS
DiehlAG
IsaacsGD
ZhangT
2007 Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol 27 5090 5104
13. LaganiereJ
DebloisG
LefebvreC
BatailleAR
RobertF
2005 From the Cover: Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A 102 11651 11656
14. LinCY
VegaVB
ThomsenJS
ZhangT
KongSL
2007 Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet 3 e87 doi:10.1371/journal.pgen.0030087
15. KwonYS
Garcia-BassetsI
HuttKR
ChengCS
JinM
2007 Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci U S A 104 4852 4857
16. HtunH
HolthLT
WalkerD
DavieJR
HagerGL
1999 Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor. Mol Biol Cell 10 471 486
17. StenoienDL
ManciniMG
PatelK
AllegrettoEA
SmithCL
2000 Subnuclear trafficking of estrogen receptor-alpha and steroid receptor coactivator-1. Mol Endocrinol 14 518 534
18. MetivierR
PenotG
HubnerMR
ReidG
BrandH
2003 Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115 751 763
19. RayalaSK
den HollanderP
BalasenthilS
YangZ
BroaddusRR
2005 Functional regulation of oestrogen receptor pathway by the dynein light chain 1. EMBO Rep 6 538 544
20. LeeSJ
ChaeC
WangMM
2009 p150/glued modifies nuclear estrogen receptor function. Mol Endocrinol 23 620 629
21. ManavathiB
AcconciaF
RayalaSK
KumarR
2006 An inherent role of microtubule network in the action of nuclear receptor. Proc Natl Acad Sci U S A 103 15981 15986
22. CavalliG
2007 Chromosome kissing. Curr Opin Genet Dev 17 443 450
23. KininisM
IsaacsGD
CoreLJ
HahN
KrausWL
2009 Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol Cell Biol 29 1123 1133
24. SutherlandH
BickmoreWA
2009 Transcription factories: gene expression in unions? Nat Rev Genet 10 457 466
25. ChuangCH
CarpenterAE
FuchsovaB
JohnsonT
de LanerolleP
2006 Long-range directional movement of an interphase chromosome site. Curr Biol 16 825 831
26. ThomsonI
GilchristS
BickmoreWA
ChubbJR
2004 The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol 14 166 172
27. ChubbJR
BoyleS
PerryP
BickmoreWA
2002 Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 12 439 445
28. WalterJ
SchermellehL
CremerM
TashiroS
CremerT
2003 Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol 160 685 697
29. CowellJK
LaDucaJ
RossiMR
BurkhardtT
NowakNJ
2005 Molecular characterization of the t(3;9) associated with immortalization in the MCF10A cell line. Cancer Genet Cytogenet 163 23 29
30. KrystosekA
1998 Repositioning of human interphase chromosomes by nucleolar dynamics in the reverse transformation of HT1080 fibrosarcoma cells. Exp Cell Res 241 202 209
31. MoreyC
Da SilvaNR
PerryP
BickmoreWA
2007 Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development 134 909 919
32. BoyleS
GilchristS
BridgerJM
MahyNL
EllisJA
2001 The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 10 211 219
33. BowieML
DietzeEC
DelrowJ
BeanGR
TrochMM
2004 Interferon-regulatory factor-1 is critical for tamoxifen-mediated apoptosis in human mammary epithelial cells. Oncogene 23 8743 8755
34. SenguptaK
BanerjeeS
SaxenaNK
JonathanNB
CampbellDR
2004 Differential expression of VEGF-A mRNA by 17beta-estradiol in breast tumor cells lacking classical ER-alpha may be mediated through a variant form of ER-alpha. Mol Cell Biochem 262 215 224
35. SouleHD
MaloneyTM
WolmanSR
PetersonWDJr
BrenzR
1990 Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res 50 6075 6086
36. JonesC
PayneJ
WellsD
DelhantyJD
LakhaniSR
2000 Comparative genomic hybridization reveals extensive variation among different MCF-7 cell stocks. Cancer Genet Cytogenet 117 153 158
37. JonssonG
StaafJ
OlssonE
HeidenbladM
Vallon-ChristerssonJ
2007 High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes Cancer 46 543 558
38. KytolaS
RummukainenJ
NordgrenA
KarhuR
FarneboF
2000 Chromosomal alterations in 15 breast cancer cell lines by comparative genomic hybridization and spectral karyotyping. Genes Chromosomes Cancer 28 308 317
39. NeveRM
ChinK
FridlyandJ
YehJ
BaehnerFL
2006 A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10 515 527
40. OsborneCK
HobbsK
TrentJM
1987 Biological differences among MCF-7 human breast cancer cell lines from different laboratories. Breast Cancer Res Treat 9 111 121
41. ShadeoA
LamWL
2006 Comprehensive copy number profiles of breast cancer cell model genomes. Breast Cancer Res 8 R9
42. HamptonOA
Den HollanderP
MillerCA
DelgadoDA
LiJ
2009 A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res 19 167 177
43. ShangY
HuX
DiRenzoJ
LazarMA
BrownM
2000 Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103 843 852
44. BautistaS
TheilletC
1998 CCND1 and FGFR1 coamplification results in the colocalization of 11q13 and 8p12 sequences in breast tumor nuclei. Genes Chromosomes Cancer 22 268 277
45. BrunnerN
BoulayV
FojoA
FreterCE
LippmanME
1993 Acquisition of hormone-independent growth in MCF-7 cells is accompanied by increased expression of estrogen-regulated genes but without detectable DNA amplifications. Cancer Res 53 283 290
46. BrunnerN
BoysenB
JirusS
SkaarTC
Holst-HansenC
1997 MCF7/LCC9: an antiestrogen-resistant MCF-7 variant in which acquired resistance to the steroidal antiestrogen ICI 182,780 confers an early cross-resistance to the nonsteroidal antiestrogen tamoxifen. Cancer Res 57 3486 3493
47. AmanoT
SagaiT
TanabeH
MizushinaY
NakazawaH
2009 Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell 16 47 57
48. BrancoMR
PomboA
2006 Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol 4 e138 doi:10.1371/journal.pbio.0040138
49. HeardE
BickmoreW
2007 The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol 19 311 316
50. LaSalleJM
LalandeM
1996 Homologous association of oppositely imprinted chromosomal domains. Science 272 725 728
51. NogamiM
KohdaA
TaguchiH
NakaoM
IkemuraT
2000 Relative locations of the centromere and imprinted SNRPN gene within chromosome 15 territories during the cell cycle in HL60 cells. J Cell Sci 113 (Pt 12) 2157 2165
52. TellerK
SoloveiI
BuitingK
HorsthemkeB
CremerT
2007 Maintenance of imprinting and nuclear architecture in cycling cells. Proc Natl Acad Sci U S A 104 14970 14975
53. FussSH
OmuraM
MombaertsP
2007 Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell 130 373 384
54. CvackovaZ
MasataM
StanekD
FidlerovaH
RaskaI
2009 Chromatin position in human HepG2 cells: although being non-random, significantly changed in daughter cells. J Struct Biol 165 107 117
55. BrownJM
GreenJ
das NevesRP
WallaceHA
SmithAJ
2008 Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol 182 1083 1097
56. OsborneCS
ChakalovaL
MitchellJA
HortonA
WoodAL
2007 Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol 5 e192 doi:10.1371/journal.pbio.0050192
57. XuM
CookPR
2008 Similar active genes cluster in specialized transcription factories. J Cell Biol 181 615 623
58. Arnett-MansfieldRL
GrahamJD
HansonAR
MotePA
GompelA
2007 Focal subnuclear distribution of progesterone receptor is ligand dependent and associated with transcriptional activity. Mol Endocrinol 21 14 29
59. GrandeMA
van der KraanI
de JongL
van DrielR
1997 Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci 110 (Pt 15) 1781 1791
60. LarsonDR
SingerRH
ZenklusenD
2009 A single molecule view of gene expression. Trends Cell Biol 19 630 637
61. FleuryL
GerusM
LavigneAC
Richard-FoyH
BystrickyK
2008 Eliminating epigenetic barriers induces transient hormone-regulated gene expression in estrogen receptor negative breast cancer cells. Oncogene 27 4075 4085
62. NyeAC
RajendranRR
StenoienDL
ManciniMA
KatzenellenbogenBS
2002 Alteration of large-scale chromatin structure by estrogen receptor. Mol Cell Biol 22 3437 3449
63. FullwoodMJ
LiuMH
PanYF
LiuJ
XuH
2009 An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462 58 64
64. HollidayDL
BrouiletteKT
MarkertA
GordonLA
JonesJL
2009 Novel multicellular organotypic models of normal and malignant breast: tools for dissecting the role of the microenvironment in breast cancer progression. Breast Cancer Res 11 R3
65. FantesJA
BickmoreWA
FletcherJM
BallestaF
HansonIM
1992 Submicroscopic deletions at the WAGR locus, revealed by nonradioactive in situ hybridization. Am J Hum Genet 51 1286 1294
66. CremerM
GrasserF
LanctotC
MullerS
NeusserM
2008 Multicolor 3D Fluorescence In Situ Hybridization for Imaging Interphase Chromosomes. Methods Mol Biol 463 205 239
67. ChambeyronS
BickmoreWA
2004 Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18 1119 1130
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 4
-
Všechny články tohoto čísla
- Whole-Genome SNP Association in the Horse: Identification of a Deletion in Myosin Va Responsible for Lavender Foal Syndrome
- Human Telomeres Are Hypersensitive to UV-Induced DNA Damage and Refractory to Repair
- Fragilities Caused by Dosage Imbalance in Regulation of the Budding Yeast Cell Cycle
- Admixture Mapping Scans Identify a Locus Affecting Retinal Vascular Caliber in Hypertensive African Americans: the Atherosclerosis Risk in Communities (ARIC) Study
- Activation of Estrogen-Responsive Genes Does Not Require Their Nuclear Co-Localization
- Genetic Tests for Ecological and Allopatric Speciation in Anoles on an Island Archipelago
- A Pax3/Dmrt2/Myf5 Regulatory Cascade Functions at the Onset of Myogenesis
- Two New Loci for Body-Weight Regulation Identified in a Joint Analysis of Genome-Wide Association Studies for Early-Onset Extreme Obesity in French and German Study Groups
- Hypomethylation of a LINE-1 Promoter Activates an Alternate Transcript of the MET Oncogene in Bladders with Cancer
- Candidate Causal Regulatory Effects by Integration of Expression QTLs with Complex Trait Genetic Associations
- Combined Inactivation of pRB and Hippo Pathways Induces Dedifferentiation in the Retina
- Allele-Specific Virulence Attenuation of the HopZ1a Type III Effector via the ZAR1 Resistance Protein
- Down-Regulation of Honey Bee Gene Biases Behavior toward Food Rich in Protein
- A Microarray-Based Genetic Screen for Yeast Chronological Aging Factors
- Actin-Related Protein Arp6 Influences H2A.Z-Dependent and -Independent Gene Expression and Links Ribosomal Protein Genes to Nuclear Pores
- Phosphorylation of the Conserved Transcription Factor ATF-7 by PMK-1 p38 MAPK Regulates Innate Immunity in
- A -Regulatory Signature for Chordate Anterior Neuroectodermal Genes
- Genetic Analysis of Fin Development in Zebrafish Identifies Furin and Hemicentin1 as Potential Novel Fraser Syndrome Disease Genes
- Assembly of a 40 Mb Eukaryotic Genome from Short Sequence Reads: , a Model Organism for Fungal Morphogenesis
- Trait-Associated SNPs Are More Likely to Be eQTLs: Annotation to Enhance Discovery from GWAS
- Absence of Evidence for MHC–Dependent Mate Selection within HapMap Populations
- The TALE Class Homeobox Gene Defines the Anterior Compartment for Head Regeneration
- Cyclic Expression of Lhx2 Regulates Hair Formation
- Genetic Evidence for Hybrid Trait Speciation in Butterflies
- Epigenetic Regulation of a Murine Retrotransposon by a Dual Histone Modification Mark
- Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with Expression
- S Phase Progression in Human Cells Is Dictated by the Genetic Continuity of DNA Foci
- The Next Generation Becomes the Now Generation
- Acts as a Tumor Suppressor in a Murine Retinoblastoma Model by Facilitating Tumor Cell Death
- Genome-Wide Association Study of Lp-PLA Activity and Mass in the Framingham Heart Study
- The Five Zinc Transporters Undergo Different Evolutionary Fates towards Adaptive Evolution to Zinc Tolerance in
- MicroRNA–Directed siRNA Biogenesis in
- Deletion of the WD40 Domain of LRRK2 in Zebrafish Causes Parkinsonism-Like Loss of Neurons and Locomotive Defect
- Incipient Balancing Selection through Adaptive Loss of Aquaporins in Natural Populations
- GTPase Activity Plays a Key Role in the Pathobiology of LRRK2
- Natural Single-Nucleosome Epi-Polymorphisms in Yeast
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Genome SNP Association in the Horse: Identification of a Deletion in Myosin Va Responsible for Lavender Foal Syndrome
- Admixture Mapping Scans Identify a Locus Affecting Retinal Vascular Caliber in Hypertensive African Americans: the Atherosclerosis Risk in Communities (ARIC) Study
- Genetic Tests for Ecological and Allopatric Speciation in Anoles on an Island Archipelago
- Human Telomeres Are Hypersensitive to UV-Induced DNA Damage and Refractory to Repair
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



