-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Gut Odyssey: The Impact of the Microbiota on Spore Formation and Germination
article has not abstract
Published in the journal: . PLoS Pathog 11(10): e32767. doi:10.1371/journal.ppat.1005157
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1005157Summary
article has not abstract
Introduction
The Gram-positive, anaerobic, spore-forming bacterium Clostridium difficile is the leading cause of health care–associated infections and gastroenteritis-associated deaths in the United States [1]. C. difficile-associated disease is primarily toxin-mediated, although the organism’s natural antibiotic resistance and propensity to cause disease recurrence can lead to severe clinical complications, such as pseudomembranous colitis and toxic megacolon [2]. Antibiotic exposure potentiates C. difficile infections (CDI) by disrupting the colonization resistance conferred by the normal gut microbiota [3–5], while spore formation allows C. difficile to outlast antibiotic therapies and persist in the environment.
The remarkable success of fecal microbiota transplantation (FMT) in treating severe recurrent CDI provides the most direct evidence that our gut microbiota protects us from C. difficile invasion [4–6]. While the most effective antibiotic-based therapies lead to an ~20% CDI recurrence rate [1], FMT has an ~95% cure rate [6]. However, since FMT may cause unforeseen complications [4,7], there is obvious interest in determining the mechanisms that control colonization resistance in order to produce more targeted therapies. Several mechanisms have been suggested by which the microbiota antagonizes C. difficile, including increased competition for resources, inhibition of germination and/or vegetative growth, and enhancement of host defense mechanisms [3,5]. In this Pearl, we focus on how the microbiota alters the developmental life cycle of C. difficile during infection.
What Are the Dynamics of C. difficile’s Life Cycle during Infection?
As an obligate anaerobe, C. difficile uses its oxygen-tolerant spores to transmit infection [8]. Spores ingested from the environment germinate in the gut in response to specific bile salts and transform into toxin-secreting, vegetative cells [2]. While spore germination is often measured in colony-forming units, germination specifically refers to the events that cause loss of spore-specific properties, namely metabolic dormancy and resistance, while outgrowth refers to the conversion of germinated spores into vegetative cells [9]. In C. difficile, outgrowth takes approximately two hours to complete following germinant sensing [10]. When vegetative C. difficile cells start replicating in the gut, a subset of the population will initiate sporulation [11,12]. This developmental process generates the metabolically dormant, highly resistant spores that are essential for C. difficile to survive excretion from the host. Infected hosts shed large amounts of infectious spores, which serve as an environmental reservoir for C. difficile [2].
Koenigsknecht et al. recently described the spatiotemporal dynamics of CDI in mice [12]. After orally inoculating mice with C. difficile spores, they first detected vegetative cells in the colon at the six-hour time point, indicating that spore germination and outgrowth occurred within this timeframe. After 24 hours post-infection (hpi), vegetative C. difficile expanded by almost 5 logs and were most abundant in the cecum and colon (i.e., the large intestine, Fig 1). C. difficile spores were first detected in the large intestine at 24 hours; by 30–36 hpi, ~20% of the viable C. difficile in the large intestine were in the spore form. Notably, toxin levels were also highest in the large intestine at 24 hours, and disease symptoms were apparent within six hours after toxin detection.
Fig. 1. Developmental life cycle of Clostridium difficile during infection. 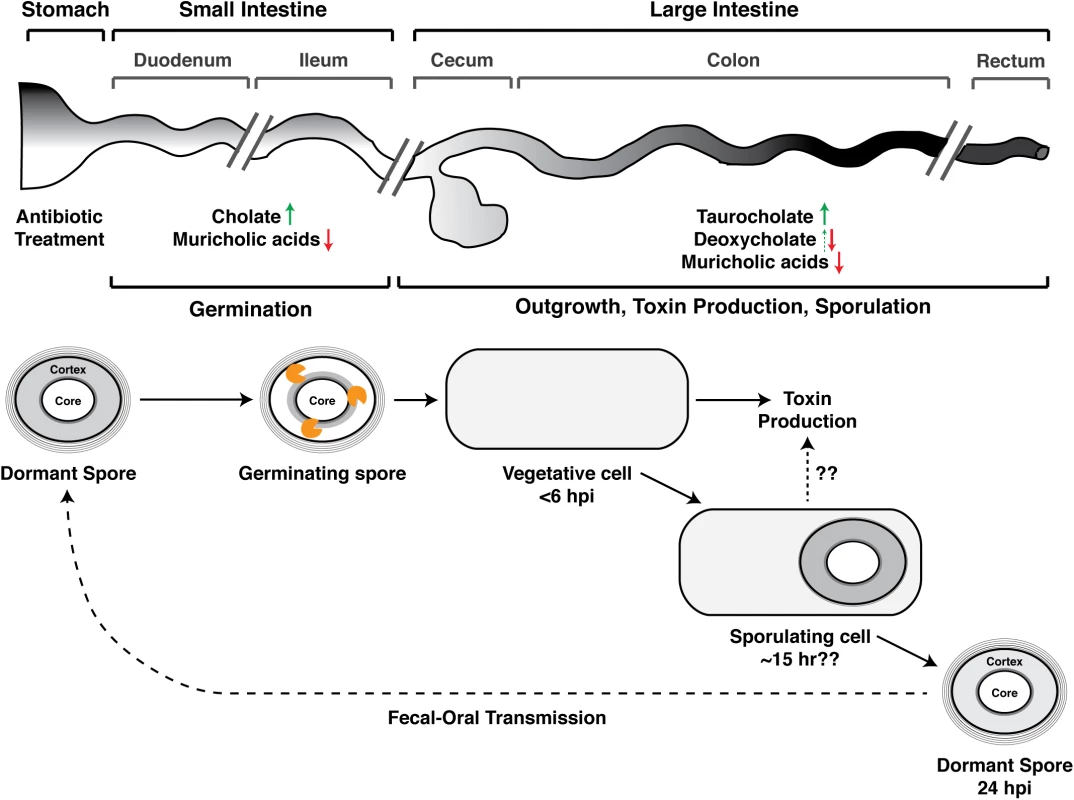
Spore germination occurs in the small intestine [13] and likely within the ileum [12,23]. Germinating spores are shown degrading the cortex layer (orange figure), a thick layer of modified peptidoglycan that maintains metabolic dormancy [19]. The times at which Koenigsknecht et al. detected the indicated developmental stages following C. difficile infection (CDI) are shown; the time at which sporulation is induced is unclear [12]. Koenigsknecht et al. propose a link between sporulation and toxin production, but whether sporulating cells themselves produce toxin during CDI requires further investigation. The effect of antibiotic exposure on bile acid composition of the indicated anatomical regions is summarized based on studies in mice [12,21,22], although it should be noted that antibiotics have differing effects on microbiota composition [21], and muricholic acids are murine-specific [20]. Weingarden et al. made similar observations in fecal extracts from patients with recurrent CDI [24]. Green arrows demarcate increases in germination-promoting bile acids, while red arrows indicate decreases in C. difficile growth-inhibitory bile acids. Deoxycholate promotes spore germination (dotted green arrow), but it also strongly inhibits C. difficile growth [14]. hpi designates hours post-infection. Where and How Do Spores Germinate in the Gut?
Over 30 years ago, Wilson et al. showed that C. difficile spores germinate within the small intestine, and identified the bile salt taurocholate as a potential in vivo germinant [13]. Almost two decades later, Sorg and Sonenshein demonstrated that cholate derivatives activate spore germination when combined with glycine or other amino acid co-germinants [14], whereas chenodeoxycholate derivatives competitively inhibited cholate-induced spore germination (Fig 2) [15]. While taurocholate was the most potent and rapid cholate-based germinant, the affinity of C. difficile spores was greater for chenodeoxycholate [15]. Giel et al. later showed that small intestinal extracts from mice induce C. difficile spore germination [16]. Since pre-treatment of these extracts with the bile salt sequestrant cholesterylamine abrogated their germination-stimulating activity [16], these studies suggested that bile salts signal to C. difficile spores that they have reached the gut. Although strain-to-strain variability in germinant responsiveness has been documented [17,18], all C. difficile strains likely sense bile acid germinants using the non-canonical germinant receptor, CspC, a subtilisin-like pseudoprotease that controls a unique signaling pathway reviewed elsewhere [19].
Fig. 2. Effect of bile acid metabolism on the developmental life cycle of C. difficile. 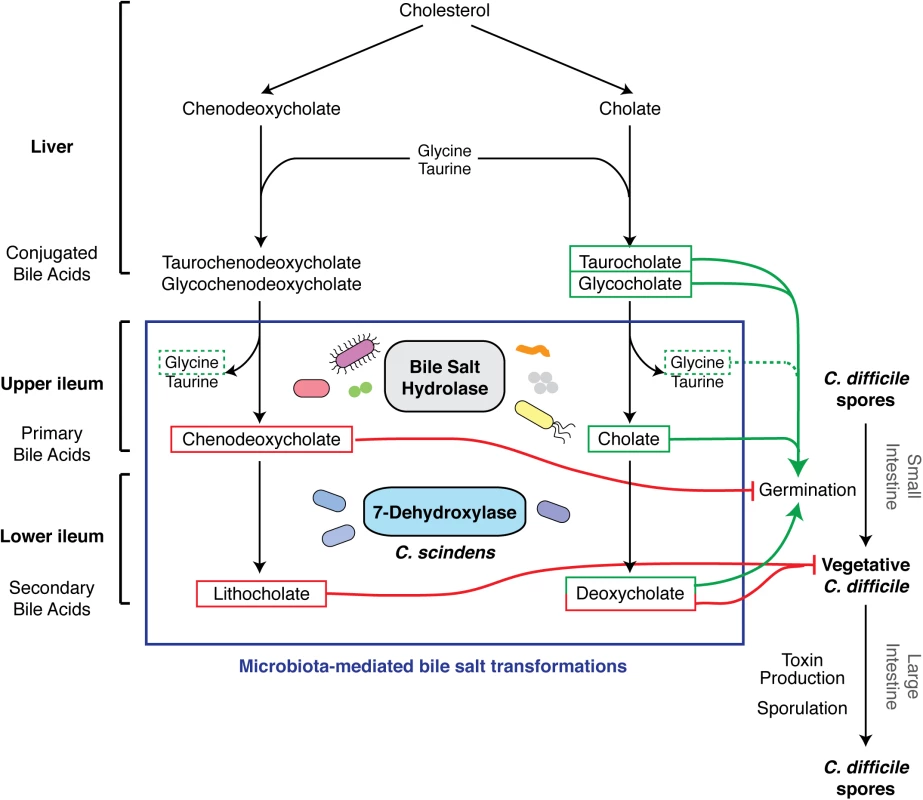
The figure represents an update of the schematic by Britton et al. [3]. The liver synthesizes bile acids from cholesterol and secretes them as conjugated bile acids. Ileum gut microbiota produces bile salt hydrolases that deconjugate bile acids to primary bile acids, chenodeoxycholate, and cholate [23]. Primary bile acids serve as substrates for 7-dehydroxylases, made by a small subset of clostridial organisms such as C. scindens, which generate secondary bile acids lithocholate and deoxycholate. Chenodeoxycholate and secondary bile acids are toxic to vegetative C. difficile (red lines) [20,21], while cholate derivatives promote germination when combined with amino acid co-germinants like glycine (dotted line) [14]. What Effect Does the Microbiota Have on Spore Germination and Vegetative Cell Growth?
Interestingly, although small intestinal extracts derived from untreated mice are capable of inducing germination, extracts prepared from antibiotic-treated mice are ~3–10-fold more potent at germinating C. difficile spores [12,16]. These observations imply that a healthy microbiota can dampen C. difficile germination in the small intestine. Consistent with this idea, antibiotic treatment of mice increases the ratio of the germinant cholate to the germination-inhibitory, murine-specific muricholic acids [20] in the small intestine (Fig 1) [12]. Furthermore, antibiotic treatment elevates the levels of the germinant taurocholate in the cecum [21,22] while reducing the levels of the cholate derivative deoxycholate (Fig 2) and muricholic acids [12]. As a result, cecal extracts from antibiotic-treated mice stimulate C. difficile spore germination by ~30-fold relative to those from untreated mice [16]. Although Koenigsknecht et al. did not observe this effect [12], deoxycholate and muricholic acids nevertheless inhibit vegetative C. difficile growth [15,20]. Cecal extracts from untreated mice decrease the viability of C. difficile cultures, whereas extracts from antibiotic-treated mice support their replication [21]. Furthermore, cholestyramine treatment of cecal extracts from untreated mice restores their ability to support C. difficile growth [21]. Collectively, these observations indicate that antibiotic treatment enhances C. difficile germination in the small intestine while simultaneously reducing the levels of growth-inhibitory secondary bile acids in the large intestine [14]. In addition, the elevated levels of taurocholate in the large intestine caused by antibiotic exposure may enhance CDI by preventing the normal microbiota from re-establishing itself, since taurocholate can inhibit the growth of some bacteria [23] but not C. difficile [14].
Notably, patients with recurrent CDI exhibit similar changes in bile acid composition: prior to FMT, they had elevated levels of primary bile acids and reduced levels of secondary bile acids relative to healthy individuals; after FMT, this bile acid imbalance was restored [24]. Although the bacterial species that confer protection against CDI by modulating bile acid composition are unknown, a recent study has identified a potential candidate. While many gut bacteria produce bile salt hydrolases that convert conjugated bile acids secreted by the liver into the primary bile acids, cholate and chenodeoxycholate (Fig 2) [23], only a small subset of gut bacteria encode the 7-dehydroxylases required to transform these primary bile acids into C. difficile-growth-inhibitory secondary bile acids, deoxycholate and lithocholate, respectively [23]. Buffie et al. recently showed that the 7-dehydroxylating bacterium Clostridium scindens can confer colonization resistance against CDI [21]. Using a systems-based approach to compare the microbiome and metabolome of allogeneic stem cell transplant patients who experienced CDI with those who did not, Buffie et al. correlated resistance to CDI with elevated levels of secondary bile acids and the presence of C. scindens. Adoptive transfer of C. scindens into mice infected with C. difficile provided partial protection against C. difficile-associated disease [21]. Remarkably, Sorg and Sonenshein previously predicted this result based on their observation that deoxycholate inhibits C. difficile growth [15] and 7-dehydroxylation activity decreases in the gut after antibiotic administration [25].
How and Where Is Sporulation Induced in the Gut?
While a fair amount is known about how C. difficile spores germinate in the gut, comparatively little is known about the reciprocal process of sporulation during infection.
Following oral challenge of mice with C. difficile spores, spores re-appear in the gut 24 hpi [12]. Since spore germination and outgrowth occur within 6 hpi, and spore formation takes approximately nine hours to complete in vitro [26], these observations suggest that sporulation is induced ~15 hpi. These kinetics are somewhat accelerated relative to in vivo transcriptomics analyses of CDI in gnotobiotic mice, which showed that sporulation genes are strongly induced 14 hours after gnotobiotic mice are infected with vegetative C. difficile [27]. The in vivo transcriptomics data [27], however, cannot be compared directly to the spatiotemporal analysis of CDI [12], since the former used vegetative cells to initiate infection while the latter used spores. Interestingly, C. difficile sporulation rates are reduced in mice colonized with human microbiota relative to monoxenic mice [27], suggesting that a healthy microbiota may dampen C. difficile sporulation and potentially reduce disease transmission and recurrence.
In order to test this hypothesis, a clearer understanding of the signaling pathways that trigger sporulation, as well as the environmental cues that they respond to, is necessary. Similar to other spore-forming organisms [28], C. difficile entry into sporulation depends upon a classical two-component signaling pathway involving the phosphorylation of the master transcriptional regulator Spo0A [8]. Although two histidine kinases have been implicated in phosphorylating Spo0A [29], the regulatory inputs controlling their activity are unknown. Two peptide transporters, Opp and App, were recently identified as negative regulators of C. difficile sporulation [30]. In contrast, opp and app mutants in the model spore-former Bacillus subtilis are positive regulators of sporulation, indicating that distinct mechanisms control sporulation induction between spore-formers [29]. The signaling pathway controlling C. difficile Spo0A activation appears to be considerably simpler than B. subtilis, which uses a highly regulated phosphorelay consisting of five histidine kinases to tightly control entry into sporulation under conditions of nutrient limitation [29]. Interestingly, a gut-adapted B. subtilis strain has a simplified pathway for regulating Spo0A, which allows it to induce sporulation more rapidly than the well-studied, lab-adapted strain [31]. Since the regulatory pathway of gut-adapted B. subtilis resembles C. difficile, and some intestinal symbionts divide exclusively using sporulation [28], gut-adapted organisms may use fewer checkpoints to initiate sporulation.
Future Directions
Clearly, a great deal remains to be discovered regarding how the microbiota confers colonization resistance against CDI. Addressing the following questions will provide important insight into CDI and may identify new avenues for therapeutic intervention.
What environmental cues induce sporulation in the gut, and how do they activate the signaling pathway?
When does C. difficile induce sporulation in the gut?
Does the microbiota affect the induction of C. difficile sporulation?
Is there a link between sporulation and toxin production, as suggested by [12]?
Does loss of 7-dehydroxylase activity in the gut confer sensitivity to CDI; i.e., does a C. scindens 7-dehydroxylase mutant lose its ability to confer resistance against CDI?
Zdroje
1. Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, et al. (2015) Burden of Clostridium difficile infection in the United States. N Engl J Med 372 : 825–834. doi: 10.1056/NEJMoa1408913 25714160
2. Carroll K, Bartlett J (2011) Biology of Clostridium difficile: implications for epidemiology and diagnosis. Annual review of microbiology 65 : 501–521. doi: 10.1146/annurev-micro-090110-102824 21682645
3. Britton RA, Young VB (2012) Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol 20 : 313–319. doi: 10.1016/j.tim.2012.04.001 22595318
4. Britton RA, Young VB (2014) Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 146 : 1547–1553. doi: 10.1053/j.gastro.2014.01.059 24503131
5. Yurist-Doutsch S, Arrieta MC, Vogt SL, Finlay BB (2014) Gastrointestinal microbiota-mediated control of enteric pathogens. Annu Rev Genet 48 : 361–382. doi: 10.1146/annurev-genet-120213-092421 25251855
6. van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, et al. (2013) Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368 : 407–415. doi: 10.1056/NEJMoa1205037 23323867
7. Alang N, Kelly CR (2015) Weight gain after fecal microbiota transplantation. Open Forum Infect Dis 2: ofv004. doi: 10.1093/ofid/ofv004 26034755
8. Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, et al. (2012) The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80 : 2704–2711. doi: 10.1128/IAI.00147-12 22615253
9. Setlow P (2003) Spore germination. Curr Opin Microbiol 6 : 550–556. 14662349
10. Dembek M, Stabler RA, Witney AA, Wren BW, Fairweather NF (2013) Transcriptional analysis of temporal gene expression in germinating Clostridium difficile 630 endospores. PLoS One 8: e64011. doi: 10.1371/journal.pone.0064011 23691138
11. Howerton A, Patra M, Abel-Santos E (2013) Fate of ingested Clostridium difficile spores in mice. PLoS One 8: e72620. doi: 10.1371/journal.pone.0072620 24023628
12. Koenigsknecht MJ, Theriot CM, Bergin IL, Schumacher CA, Schloss PD, et al. (2015) Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun 83 : 934–941. doi: 10.1128/IAI.02768-14 25534943
13. Wilson KH, Sheagren JN, Freter R (1985) Population dynamics of ingested Clostridium difficile in the gastrointestinal tract of the Syrian hamster. J Infect Dis 151 : 355–361. 3968453
14. Sorg JA, Sonenshein AL (2008) Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190 : 2505–2512. doi: 10.1128/JB.01765-07 18245298
15. Sorg JA, Sonenshein AL (2010) Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192 : 4983–4990. doi: 10.1128/JB.00610-10 20675492
16. Giel J, Sorg J, Sonenshein A, Zhu J (2010) Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS One 5: e8740. doi: 10.1371/journal.pone.0008740 20090901
17. Carlson PE Jr., Kaiser AM, McColm SA, Bauer JM, Young VB, et al. (2015) Variation in germination of Clostridium difficile clinical isolates correlates to disease severity. Anaerobe 33 : 64–70. doi: 10.1016/j.anaerobe.2015.02.003 25681667
18. Heeg D, Burns DA, Cartman ST, Minton NP (2012) Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS One 7: e32381. doi: 10.1371/journal.pone.0032381 22384234
19. Paredes-Sabja D, Shen A, Sorg JA (2014) Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol 22 : 406–416. doi: 10.1016/j.tim.2014.04.003 24814671
20. Francis MB, Allen CA, Sorg JA (2013) Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS One 8: e73653. doi: 10.1371/journal.pone.0073653 24040011
21. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, et al. (2015) Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517 : 205–208. doi: 10.1038/nature13828 25337874
22. Theriot CM, Koenigsknecht MJ, Carlson PE Jr., Hatton GE, Nelson AM, et al. (2014) Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5 : 3114. doi: 10.1038/ncomms4114 24445449
23. Ridlon JM, Kang DJ, Hylemon PB (2006) Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47 : 241–259. 16299351
24. Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, et al. (2014) Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol 306: G310–319. doi: 10.1152/ajpgi.00282.2013 24284963
25. Northfield TC, McColl I (1973) Postprandial concentrations of free and conjugated bile acids down the length of the normal human small intestine. Gut 14 : 513–518. 4729918
26. Pereira FC, Saujet L, Tome AR, Serrano M, Monot M, et al. (2013) The Spore Differentiation Pathway in the Enteric Pathogen Clostridium difficile. PLoS Genet 9: e1003782. doi: 10.1371/journal.pgen.1003782 24098139
27. Janoir C, Deneve C, Bouttier S, Barbut F, Hoys S, et al. (2013) Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun 81 : 3757–3769. doi: 10.1128/IAI.00515-13 23897605
28. Hutchison EA, Miller DA, Angert ER (2014) Sporulation in Bacteria: Beyond the Standard Model. Microbiol Spectr 2.
29. Edwards AN, McBride SM (2014) Initiation of sporulation in Clostridium difficile: a twist on the classic model. FEMS Microbiol Lett 358 : 110–118. doi: 10.1111/1574-6968.12499 24910370
30. Edwards AN, Nawrocki KL, McBride SM (2014) Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect Immun 82 : 4276–4291. doi: 10.1128/IAI.02323-14 25069979
31. Serra CR, Earl AM, Barbosa TM, Kolter R, Henriques AO (2014) Sporulation during growth in a gut isolate of Bacillus subtilis. J Bacteriol 196 : 4184–4196. doi: 10.1128/JB.01993-14 25225273
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized VirusČlánek Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell ProliferationČlánek Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite InterfaceČlánek Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 10- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Expression of Concern: Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Preparing for the Next Epidemic with Basic Virology
- Effectively Communicating the Uncertainties Surrounding Ebola Virus Transmission
- Translating Basic Research into Clinical Applications: Malaria Research at an NIH Lab
- A Gut Odyssey: The Impact of the Microbiota on Spore Formation and Germination
- Papillomavirus E6 Oncoproteins Take Common Structural Approaches to Solve Different Biological Problems
- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens
- Addressing the Complications of Ebola and Other Viral Hemorrhagic Fever Infections: Using Insights from Bacterial and Fungal Sepsis
- Time for Chocolate: Current Understanding and New Perspectives on Cacao Witches’ Broom Disease Research
- Ganglioside and Non-ganglioside Mediated Host Responses to the Mouse Polyomavirus
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Structure Elucidation of Coxsackievirus A16 in Complex with GPP3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses
- CD39 Expression Identifies Terminally Exhausted CD8 T Cells
- Abiotic Stresses Antagonize the Rice Defence Pathway through the Tyrosine-Dephosphorylation of OsMPK6
- Dissociation of Tissue Destruction and Bacterial Expansion during Bubonic Plague
- Interferon-γ: The Jekyll and Hyde of Malaria
- CCR2 Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8 T Cell Response to the Protective YopE Epitope during Infection
- A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1
- The Suramin Derivative NF449 Interacts with the 5-fold Vertex of the Enterovirus A71 Capsid to Prevent Virus Attachment to PSGL-1 and Heparan Sulfate
- Trans-generational Immune Priming Protects the Eggs Only against Gram-Positive Bacteria in the Mealworm Beetle
- Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection
- Respiratory Syncytial Virus Disease Is Mediated by Age-Variable IL-33
- TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized Virus
- Modeling the Effects of Vorinostat Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells
- Identification of a Novel Lipoprotein Regulator of Spore Germination
- Calcium Regulation of Hemorrhagic Fever Virus Budding: Mechanistic Implications for Host-Oriented Therapeutic Intervention
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
- Comparative Life Cycle Transcriptomics Revises Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates
- The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy
- Carcinogenic Parasite Secretes Growth Factor That Accelerates Wound Healing and Potentially Promotes Neoplasia
- Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell Proliferation
- Dengue Virus Infection of Requires a Putative Cysteine Rich Venom Protein
- Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma
- Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite Interface
- Phenotypic and Functional Alterations in Circulating Memory CD8 T Cells with Time after Primary Infection
- Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems
- Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses
- Myeloid Cell Arg1 Inhibits Control of Arthritogenic Alphavirus Infection by Suppressing Antiviral T Cells
- The White-Nose Syndrome Transcriptome: Activation of Anti-fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis
- Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles
- Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine
- Differentiation-Dependent KLF4 Expression Promotes Lytic Epstein-Barr Virus Infection in Epithelial Cells
- The Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
- C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in
- Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
- Crystal Structure of the Human Cytomegalovirus Glycoprotein B
- Depletion of . GlmU from Infected Murine Lungs Effects the Clearance of the Pathogen
- Immunologic Control of Papillomavirus Type 1
- Requires Host Rab1b for Survival in Macrophages
- Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi
- PD-L1 Expression on Retrovirus-Infected Cells Mediates Immune Escape from CD8 T Cell Killing
- Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction
- IL-4 Induced Innate CD8 T Cells Control Persistent Viral Infection
- Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
- BCG Skin Infection Triggers IL-1R-MyD88-Dependent Migration of EpCAM CD11b Skin Dendritic cells to Draining Lymph Node During CD4+ T-Cell Priming
- Antigenic Characterization of the HCMV gH/gL/gO and Pentamer Cell Entry Complexes Reveals Binding Sites for Potently Neutralizing Human Antibodies
- Rescue of a Plant Negative-Strand RNA Virus from Cloned cDNA: Insights into Enveloped Plant Virus Movement and Morphogenesis
- Geminivirus Activates to Accelerate Cytoplasmic DCP2-Mediated mRNA Turnover and Weakens RNA Silencing in
- Disruption of Sphingolipid Biosynthesis Blocks Phagocytosis of
- The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps
- The Timing of Stimulation and IL-2 Signaling Regulate Secondary CD8 T Cell Responses
- Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity
- The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Interferon-γ: The Jekyll and Hyde of Malaria
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



