-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaIL-4 Induced Innate CD8 T Cells Control Persistent Viral Infection
Over the course of viral infection there may be a limited time period during which the host system can eliminate the virus. When viruses are not eliminated within this period of time, virus can establish persistent infection. Here, we show that IL-4-induced innate CD8+ T cells are able to effectively control chronic virus infection. Innate T cells are heterogeneous population of T cells that acquire effector/memory phenotype as a result of their maturation process in thymus, unlike conventional T cells that differentiate into memory cells after antigen encounter in periphery. Previous data suggest that innate T cells might serve as a first-line of defense against certain bacterial pathogens. IL-4-induced innate CD8+ T cells are a unique subset of innate T cells that were recently identified in both mouse and human. We found that IL-4-induced innate CD8+ T cells immediately accumulated after viral infection and produced a robust amount of effector cytokines. Thereby, IL-4-induced innate CD8+ T cells provide an effective barrier to the establishment of persistent infection via effective virus control during the early phase of viral infection. Collectively our data show that IL-4-induced innate CD8+ T cells works as an early defense mechanism against chronic viral infection.
Published in the journal: . PLoS Pathog 11(10): e32767. doi:10.1371/journal.ppat.1005193
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005193Summary
Over the course of viral infection there may be a limited time period during which the host system can eliminate the virus. When viruses are not eliminated within this period of time, virus can establish persistent infection. Here, we show that IL-4-induced innate CD8+ T cells are able to effectively control chronic virus infection. Innate T cells are heterogeneous population of T cells that acquire effector/memory phenotype as a result of their maturation process in thymus, unlike conventional T cells that differentiate into memory cells after antigen encounter in periphery. Previous data suggest that innate T cells might serve as a first-line of defense against certain bacterial pathogens. IL-4-induced innate CD8+ T cells are a unique subset of innate T cells that were recently identified in both mouse and human. We found that IL-4-induced innate CD8+ T cells immediately accumulated after viral infection and produced a robust amount of effector cytokines. Thereby, IL-4-induced innate CD8+ T cells provide an effective barrier to the establishment of persistent infection via effective virus control during the early phase of viral infection. Collectively our data show that IL-4-induced innate CD8+ T cells works as an early defense mechanism against chronic viral infection.
Introduction
Conventional T cells take on naive phenotypes when they emigrate out from the thymus, whereas innate T cells from the thymus are phenotypically of the effector/memory form [1]. Compared with conventional T cells, these innate T cells, such as natural killer T (NKT) cells, mucosal-associated invariant T (MAIT) cells and H2-M3-specific T cells, are selected by interaction with hematopoietic cells rather than thymic epithelial cells, and their development is dependent on IL–15 and the SAP (SLAM-associated protein) signaling pathway [1]. Moreover, most innate T cells express T cell receptors (TCRs) specific for MHC class Ib molecules [1,2].
Memory-like CD8+ T cells expressing eomesodermin (Eomes) are another subset of innate T cells [3]. Although this type of cells is not abundant in wild type C57BL/6 mice, they initially described in Tec-kinase-deficient mice [4,5] and subsequently found in the thymus of a variety of mice in which T-cell-associated genes are deficient [6–13] or CIITA-transgenic (CIITATg) mice in which MHC class II molecules are expressed in thymocytes [14]. Recently, a substantial number of these innate CD8+ T cells was also identified in wild-type BALB/c mice [6] and in human [14]. Eomes+ CD8+ T cells from both mice and human thymus exhibit immediate effector function upon TCR stimulation [6,14]; however, this type of CD8+ T cells has unique characteristics that make them different from MHC class Ib-restricted innate T cells. Firstly, common gamma chain cytokines, particularly IL–4 in this case, drive the expression of Eomes during the intrathymic developmental process [6,14]. Promyelocytic leukemia zinc finger protein (PLZF)+ NKT cells are the major source of IL–4 in wild-type BALB/c and Klf2-deficient mice [6], whereas in CIITATg mice PLZF+ T-T CD4+ T cells are responsible for the production of IL–4 [14,15]. In humans, IL–4 would be produced by both PLZF+ T-T CD4+ T and NKT cells [14]. Secondly, MHC class Ib-restricted innate T cells have a highly restricted TCR repertoire [16], whereas IL-4-induced Eomes+ innate CD8+ T cells from CIITATg mice have a diverse TCR repertoire very much like conventional T cells [14]. This difference in TCR repertoire suggests that they are selected by diverse self-peptides presented by classical MHC class I molecules and raises the possibility that IL-4-induced innate CD8+ T cells perform some functions distinct from those of MHC class Ib-restricted innate T cells during a variety of immune responses. However, the biological relevance of IL-4-induced innate CD8+ T cells has not been elucidated.
Although CD8+ T cells are crucial for the control or elimination of various viral infections, many viruses are able to establish a chronic infection by escaping virus-specific CD8+ T cell responses. The functional inactivation of antigen-specific CD8+ T cells through the triggering of co-inhibitory receptors such as programmed death–1 (PD–1) and cytotoxic T-lymphocyte antigen–4 (CTLA–4) is currently considered to be a conserved mechanism for not only maintaining viral persistence, but also for limiting immunopathology [17,18]. Moreover, an increase in frequency of virus-specific naïve CD8+ T-cell precursors was reported to help control initial viremia but cause differing outcomes with either clearance of wild-type chronic virus or emergence of a T-cell epitope escape mutant virus [19]. However, little is known regarding the impact that the type of CD8+ T cells present during infection has on protection against viral persistence.
In the present study, we investigated the in vivo role of IL-4-induced innate CD8+ T cells in controlling initial viremia using the lymphocytic choriomeningitis virus (LCMV) clone 13 (CL–13) chronic virus infection model. One of the most notable findings from this experiment is that IL-4-induced innate CD8+ T cells produce a robust amount of cytokines such as IFN-γ and TNF-α upon LCMV infection, resulting in the efficient control of viruses from the body and providing an effective barrier to the establishment of viral persistence.
Results
The anti-viral CD8+ T-cell response is enhanced in CIITATg mice infected with LCMV CL–13
To explore the in vivo function of IL-4-induced Eomes+ CD8+ T cells, we used CIITATg mice in which thymocytes express MHC class II molecules. As reported previously [14], thymus of CIITATg mice contain high numbers of Eomes+ CD8+ T cells, whereas wild-type C57BL/6 mice have only a small number of these cells (Fig 1A). These Eomes+ CD8+ T cells exhibited a phenotype similar to that of Eomes+ memory-like CD8+ T cells identified in other types of gene-manipulated mice [3,6] in that they highly express CXCR3, CD124 (IL-4Rα), CD122 (IL-2Rβ) and CD44, and exhibit low expression of CD24 (Fig 1B). We initially infected both CIITATg and wild-type mice with a conventional dose (2 x 106 PFU/mouse) of LCMV CL–13 and found that CIITATg mice succumbed to early death, whereas wild-type mice did not (Fig 1C). Histopathological analysis of LCMV CL-13-infected CIITATg mice showed edematous lungs where most of the alveolar spaces were filled with transudate (Fig 1D), suggesting that the mice died due to immunopathologic tissue damage.
Fig. 1. Clone 13 infection caused the immunopathology in CIITATg mice with abundant IL-4-induced innate CD8+ T cells. 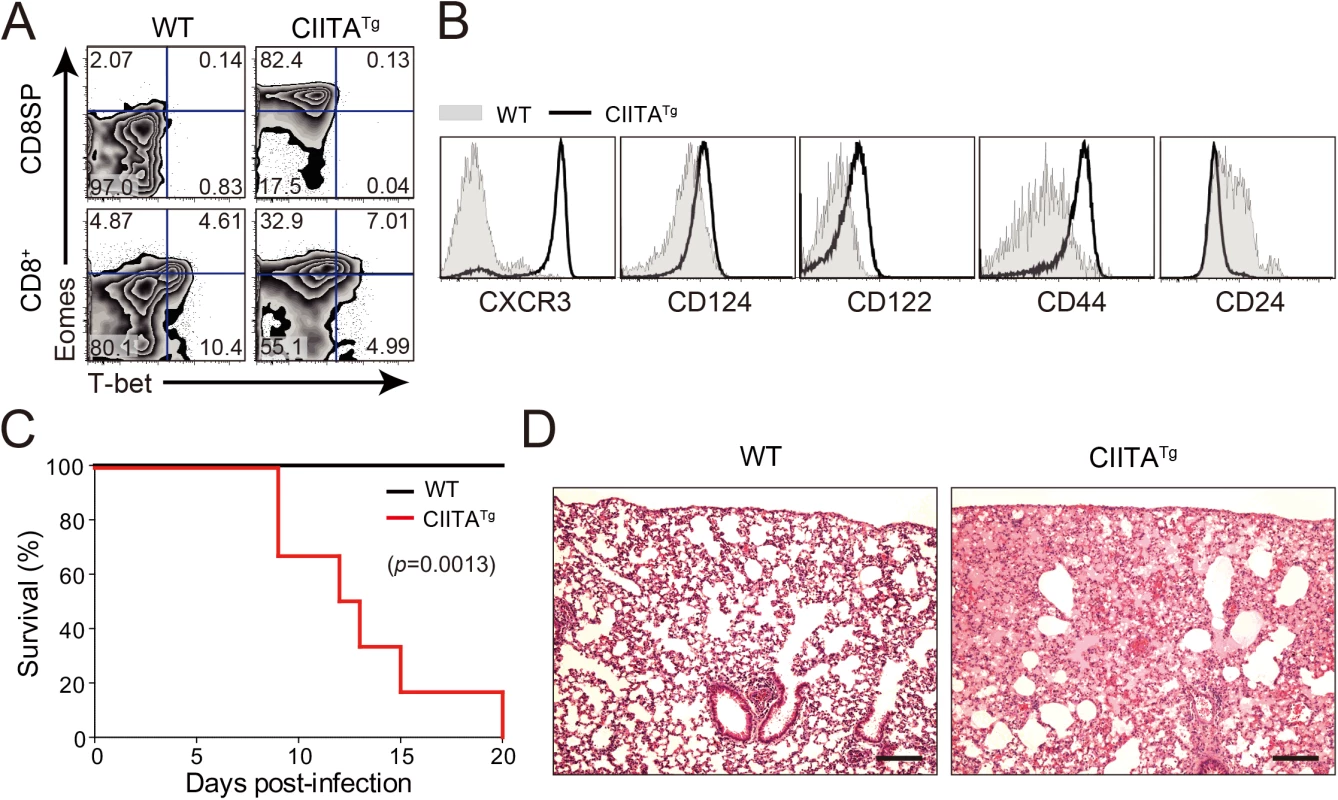
(A) Cells were isolated from the thymus and spleen of wild-type (WT) and CIITATg mice and expression of Eomes and T-bet on CD8 single positive (CD8SP) thymocytes and splenic CD8+ T cells were analyzed by flow cytometry. Numbers in the plots indicate the percentages of cells in each quadrant. (B) CXCR3, CD124, CD122, CD44, and CD24 expression levels were compared on CD8SP thymocytes of wild-type and CIITATg mice. (C) Wild-type and CIITATg mice were infected with 2 x 106 PFU of LCMV CL–13. Survival of wild-type and CIITATg mice were monitored at indicated time points after chronic infection with LCMV CL–13. P value = 0.0013, wild-type versus CIITATg mice upon conventional dose of LCMV CL–13 infection (log-rank (Mantel-Cox) test). (D) Lung pathology in wild-type and CIITATg mice at 8 days post-infection (DPI). Data are pooled from two experiments or are representative of at least two experiments (n≥3 per group in each experiment). Original magnification x100, bar = 200 μm. We next infected mice with a diverse range of viral doses to determine the minimal infectious dose capable of causing chronic infection in wild-type C57BL/6 mice. We found that inoculation of LCMV CL–13 into wild-type mice established chronic virus infection with sustained expression of PD–1 molecules on CD8+ T cells (S1A Fig) and virus persistency in serum (S1B Fig). Based on this, we challenged CIITATg and wild-type mice with this dose of LCMV CL–13 for subsequent studies and monitored the frequency of CD8+ T cells and viral titer in the blood for 28 DPI. Notably, the frequency and number of endogenous CD8+ T cells specific for the LCMV GP33-41 epitope (GP33) were greatly enhanced in CIITATg mice when compared with those in wild-type mice (Fig 2A and 2B). In addition, sustained expression of PD–1 on GP33-specific CD8+ T cells was not induced in CIITATg mice upon LCMV CL–13 infection, although wild-type mice exhibited strongly induced PD–1 expression patterns (Fig 2C and 2D). In the CIITATg mice, the expression of CD127, which are known to be downregulated on exhausted virus-specific CD8+ T cells [19,20] was significantly increased (Fig 2E). This data coincides with a rapid drop in virus titer in blood of CIITATg mice, but not in that of wild-type mice (Fig 2F).
Fig. 2. Accelerated viral control in CIITATg mice during LCMV clone 13 infection. 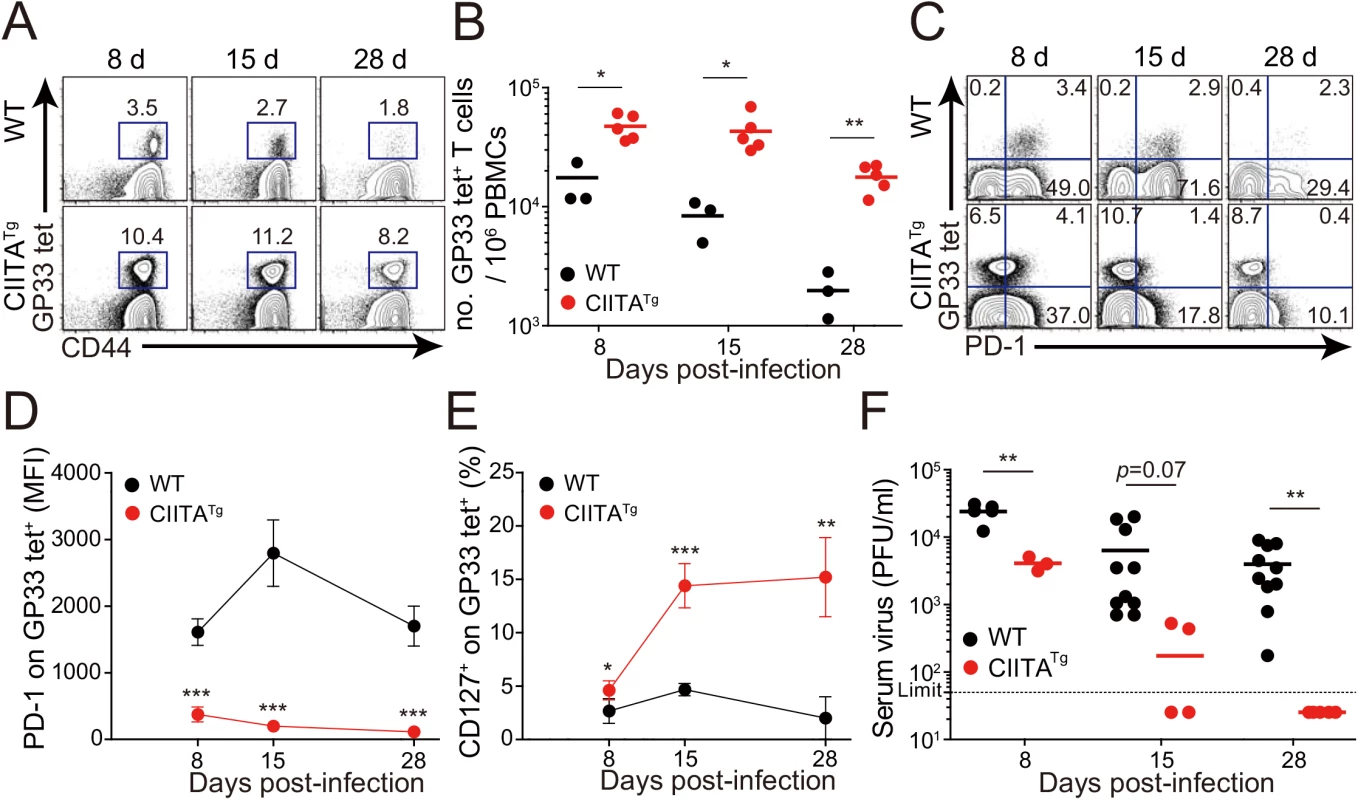
(A-E) Wild-type (WT) and CIITATg mice were infected with 5 x 105 PFU of LCMV CL–13 per mouse. Peripheral blood mononuclear cells (PBMCs) were collected at indicated days post infection (DPI). Frequency of GP33-41 (GP33) tetramer-positive cells among CD8+ T cells were analyzed by flow cytometry (A) and the numbers of GP33 tetramer-positive CD8+ T cells per 106 PBMCs during the course of LCMV CL–13 infection are represented (B). Numbers in plots indicate percentage of the tetramer-positive cells among CD8+ T cells. PD–1 (C and D) and CD127 (E) expression by GP33 tetramer-positive cells after gating on CD8+ T cells were also analyzed, and summary showing the PD–1 expression level on GP33 tetramer-positive CD8+ T cells is represented by mean fluorescence intensity (MFI) (D). The percentage of CD127+ cells among GP33 tetramer-positive CD8+ T cells is also summarized (E). Line graph shows mean ± SD. Data are representatives of four independent experiments (n≥3 per group in each experiment). (F) Serum were collected at indicated DPI from the infected wild-type and CIITATg mice, and viral titer were checked. Dashed line indicates the virus detection limit. Undetectable samples were given a half of detection limit. Line graph shows mean ± SD. Data of (A-E) are representative of three independent experiments, and data of (F) is pooled from two independent experiments (n≥3 per group in each experiment).*P<0.05; **P<0.01; ***P<0.001. We next asked whether the enhanced CD8+ T-cell response was also present in peripheral tissues, particularly during the late phase of viral infection. To this end, we dissected the CD8+ T-cell response with respect to their number and cytokine responses in the spleen and lungs at 31 DPI. In this experiment, endogenous CD8+ T cells specific for the LCMV GP276-286 epitope (GP276) as well as for LCMV GP33 were analyzed phenotypically and functionally. As expected, the CIITATg mice contained higher numbers of GP33 - or GP276-specific CD8+ T cells in the spleen (Fig 3A) and these cells exhibited much lower levels of PD–1 expression on their surface (Fig 3B) compared with wild-type mice. As was the case in peripheral blood, a substantial population of CD127hi virus-specific memory CD8+ T cells was detected in the spleen and lung of CIITATg mice (Fig 3C). A particularly important point in this experiment is the fact that CD8+ T cells from CIITATg mice showed very strong cytokine responses compared with T cells from wild-type mice, in this case IFN-γ and TNF-α release upon ex vivo restimulation with GP33, GP276, or pooled peptides (Fig 3D and 3E). Like that of peripheral blood, this enhanced function of virus-specific CD8+ T cells was associated with decreased viral titers in the spleen and especially in the kidney, which is well known to be a life-long reservoir of chronic LCMV, of CIITATg mice compared with wild-type mice (Fig 3F). Taken together with data from peripheral blood, this strongly suggests that enhanced virus-specific CD8+ T-cell responses both quantitatively and qualitatively contribute to the accelerated control of viremia in CIITATg mice.
Fig. 3. Phenotype and function of virus-specific CD8+ T cells in tissues of LCMV infected CIITATg mice. 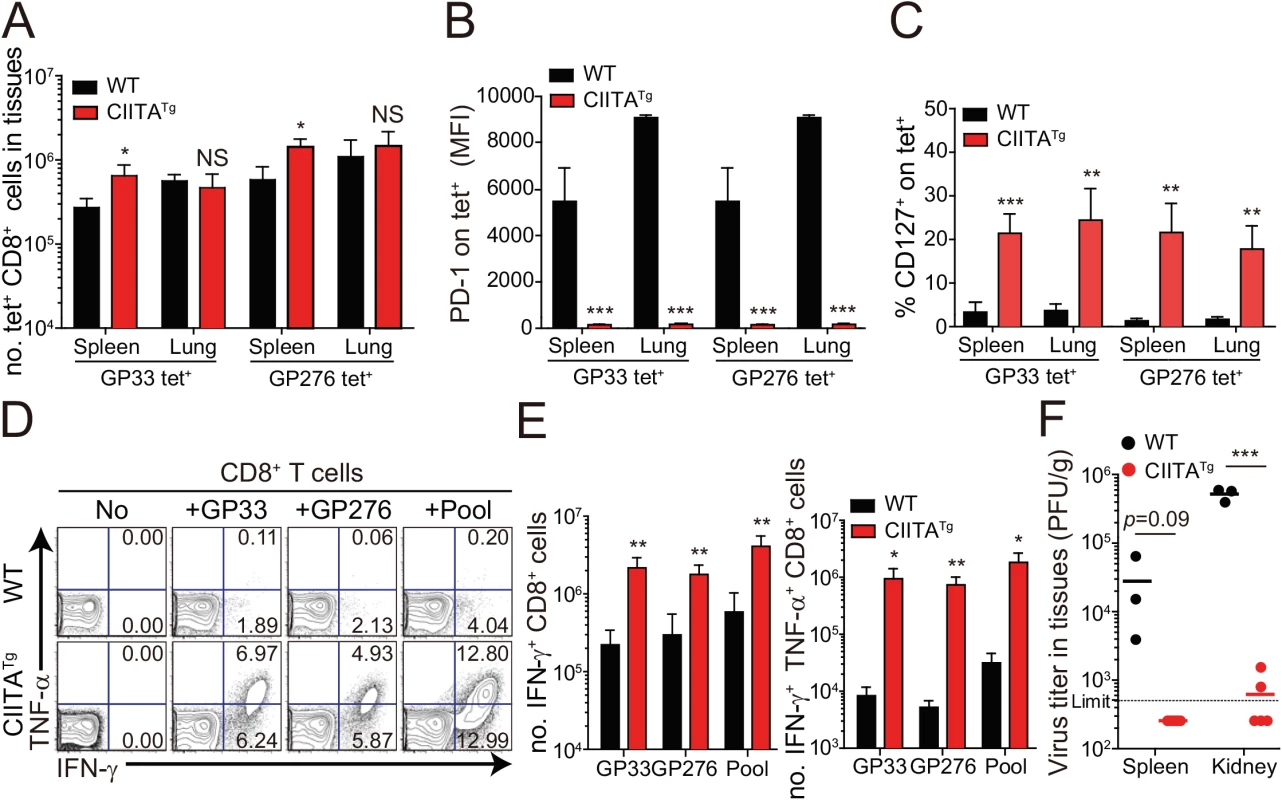
(A-C) Lymphocytes were isolated from the spleen and lungs of LCMV CL–13 (5 x 105 PFU per mouse) infected wild-type and CIITATg mice at 31 DPI and analyzed by flow cytometry. Total numbers of GP33 and GP276-286 (GP276) tetramer-positive CD8+ T cells (A) and PD–1 expression level on tetramer-specific CD8+ T cells (B) in the spleen and lungs of wild-type and CIITATg mice are represented. Percentages of CD127+ cells among virus-specific CD8+ T cells in the indicated tissues are also summarized in the bar graph (C). (D and E) Splenocytes were restimulated in vitro with GP33, GP276, or LCMV peptide pool (GP33, GP276, GP118-125, GP92-101, and GP70-77) and stained for CD8+ T cells. Frequency of IFN-γ- and TNF-α-producing CD8+ T cells were analyzed by flow cytometry (D), and absolute numbers of CD8+ T cells producing IFN-γ and producing both IFN-γ and TNF-α in the spleen are summarized, respectively (E). Numbers in the plots indicate the percentage of TNF-α+ and TNF-α- CD8+ T cells producing IFN-γ, respectively. (F) Viral titers were checked in the spleen and kidney extracted from LCMV CL-13-infected mice at 31 DPI. Dashed line indicates the virus detection limit. Undetectable samples were given a half of detection limit. Bar graphs show mean + SD. Data are representative of three independent experiments (n≥3 per group in each experiment). NS, not significant; *P<0.05; **P<0.01; ***P<0.001. Mice deficient for IL-4-induced innate CD8+ T cells fail to control LCMV CL–13 infection
The development of IL-4-induced innate CD8+ T cells is dependent on PLZF+ T cells, such as PLZF+ innate CD4+ T cells [14] and NKT cells, as a source of IL–4 [6]. Consistent with the previous report [14], the frequency of innate CD8+ T cells co-expressing CD44 and CXCR3 in the thymus was significantly lower in CIITATgIL-4KO mice than in CIITATg mice (Fig 4A). Thus, to determine whether the enhanced anti-viral CD8+ T-cell response in CIITATg mice is dependent on IL-4-induced innate CD8+ T cells, we used CIITATgIL-4KO mice. When these mice were infected with CL–13 and virus-specific CD8+ T-cell numbers in peripheral blood were monitored until 31 DPI, the overall magnitude of these cells in CIITATgIL-4KO mice was found to be as low as that in control IL-4KO mice infected with CL–13 (Fig 4B). Functionally, virus-specific CD8+ T cells in both CIITATgIL-4KO and IL-4KO mice exhibited an exhausted phenotype in terms of sustained PD–1 expression (Fig 4C and 4D). In addition, the robust cytokine production exhibited in cells from CL-13-infected CIITATg mice upon ex vivo restimulation with epitope peptides was not present in CIITATgIL-4KO mice (Fig 4E). This defect in CD8+ T-cell function suggests a failure of virus control in CIITATgIL-4KO mice. Indeed, virus was not cleared in these mice (Fig 4F). Thus, these results imply that the enhanced CD8+ T-cell response in CIITATg mice infected with CL–13 is dependent on the existence of IL-4-induced innate CD8+ T cells.
Fig. 4. CD8+ T cells responses in IL-4-deficient CIITATg and control mice during LCMV clone 13 infection. 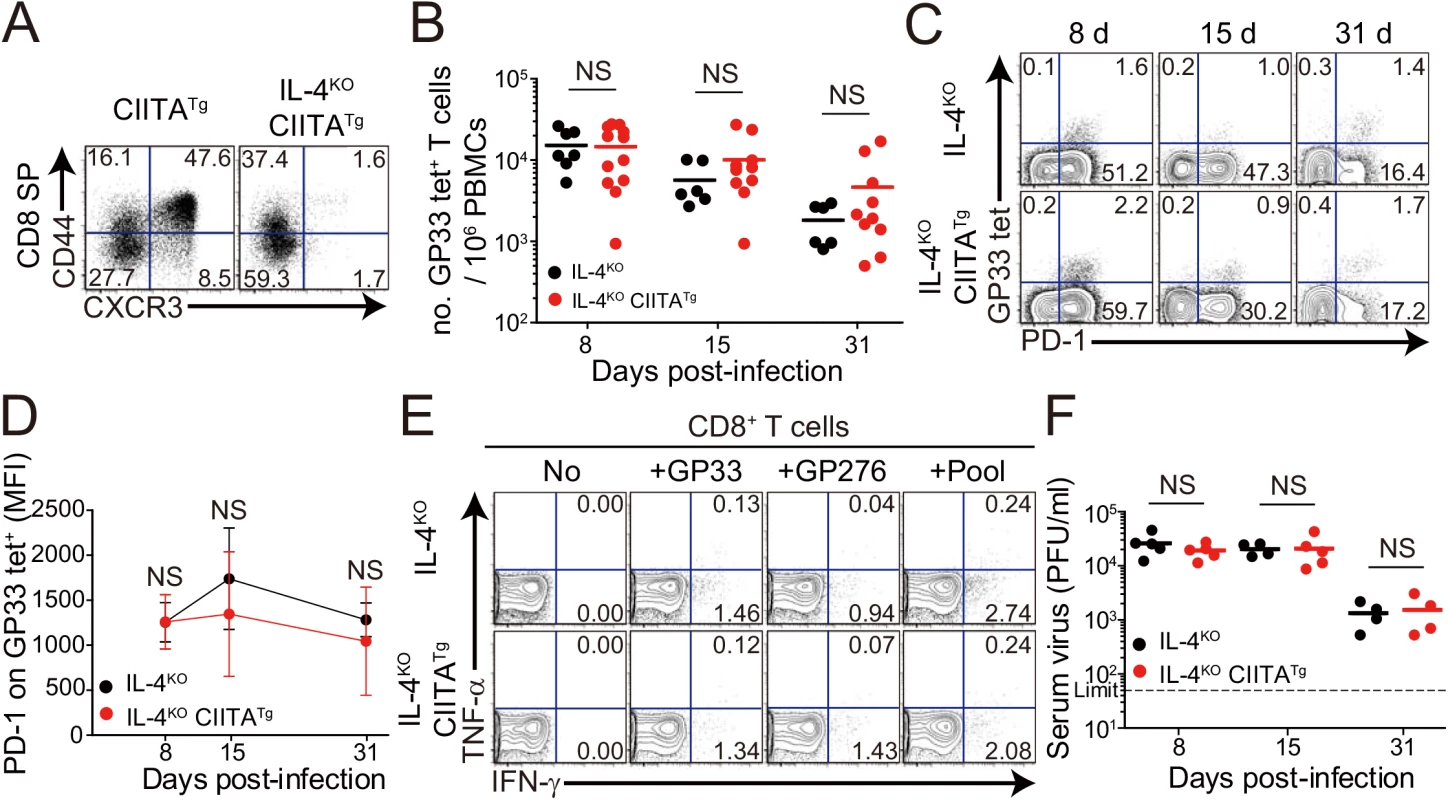
(A) Cells were isolated from the thymus of CIITATg and IL-4KOCIITATg mice and expression of CXCR3 and CD44 on CD8 single positive (CD8SP) thymocytes were analyzed by flow cytometry. Numbers in the plots indicate the percentages of cells in each quadrant. (B-F) IL-4KO and IL-4KOCIITATg mice were infected with 5 x 105 PFU of LCMV CL–13 per mouse. PBMCs were collected at the indicated DPI for analysis. The numbers of GP33 tetramer-positive CD8+ T cells per 106 PBMCs (B) and frequency of GP33 tetramer-specific CD8+ T cells and their PD–1 expression among CD8+ T cells (C) were assessed during the course of LCMV CL–13 infection by flow cytometry. Numbers in the plots indicate percentage of PD–1+ or PD–1- GP33 tetramer-positive cells among CD8+ T cells. PD–1 expression level on GP33 tetramer-positive CD8+ T cells in PBMCs represented by MFI value (D). (E) Lymphocytes isolated from the spleen of mice at 31 DPI were restimulated in vitro with GP33, GP276, or LCMV peptide pool for CD8+ T cells. Frequency of IFN-γ- and TNF-α-producing CD8+ T cells were analyzed by flow cytometry. (F) Serum viral titer in IL-4KO and IL-4KOCIITATg mice at the indicated DPI. Dashed line indicates the virus detection limit. Line graph shows mean ± SD. Data of (B-D) are data pooled from two independent experiments. Data of (E and F) are representative of four independent experiments (n≥3 per group in each experiment). NS, not significant. It is possible that IL–4 deficiency probably has effects not only on the loss of the IL-4-induced innate CD8+ T cells but also on the development of antibody responses, an important component in determining the outcome of virus infection. To exclude the possibility, we examined cytotoxic T lymphocyte (CTL) and antibody response between wild-type and IL-4KO mice during the course of CL–13 infection. The number of virus-specific CD8+ T cells and their PD–1 expression were not different between wild-type and IL-4KO mice (S2A and S2B Fig). When serum level of LCMV-specific IgG was measured, the level was also similar between these mice (S2C Fig). Accordingly, absolute numbers of follicular helper T (TFH) cells and germinal center (GC) B cells was not different between IL-4KO and wild-type mice (S2D and S2E Fig), which both could not control viruses for a certain period (S2F and S2G Fig). These data suggest that IL–4 itself does not affect the control of chronic viruses.
IL-4-induced innate CD8+ T cells mediate early control of CL–13 infection
Next, to confirm the anti-viral function of IL-4-induced innate CD8+ T cells, we generated these cells in P14 TCR transgenic mice, which express a TCR recognizing LCMV GP33 peptide presented by H-2Db molecules. To produce P14 Eomes+ CD8+ T cells, we injected a mixture of BM cells isolated from CIITATg and Thy1.1 P14 mice into irradiated CIITATgPIVKO mice (Fig 5A, T-T P14). In CIITA promoter type IV null (PIVKO) mice, MHC class II molecules are not expressed only on cortical thymic epithelial cells [21], therefore most of CD4+ T cells are selected by MHC class II+ thymocyte-thymocyte interaction pathway in CIITATgPIVKO mice [15]. As a result, CIITATgPIVKO mice generate much higher number of PLZF+ CD4+ T cells as compared to CIITATg mice, so that these mice are able to get much higher amount of IL–4 in thymic environment. As expected, this mixed chimerism allowed most of the P14 CD8+ T cells to express Eomes (Fig 5B). Consistent with a previous report [14], Eomes+ P14 cells developed with a CD127hiCD44hi memory phenotype (Fig 5C). In contrast, wild-type C57BL/6 mice that received Thy1.1 P14 BM cells contained only Eomes- naïve P14 cells (Fig 5A–5C, T-E P14). Eomes+ P14 cells also exhibited enhanced ability for effector cytokine production and cytotoxicity compared with the Eomes- counterpart: a higher fraction of Eomes+ cells produced IFN-γ and expressed a marker of degranulation, CD107a, upon in vitro stimulation with GP33 peptide (Fig 5D).
Fig. 5. Generation and characterization of virus-specific IL–4 induced innate CD8+ T cells. 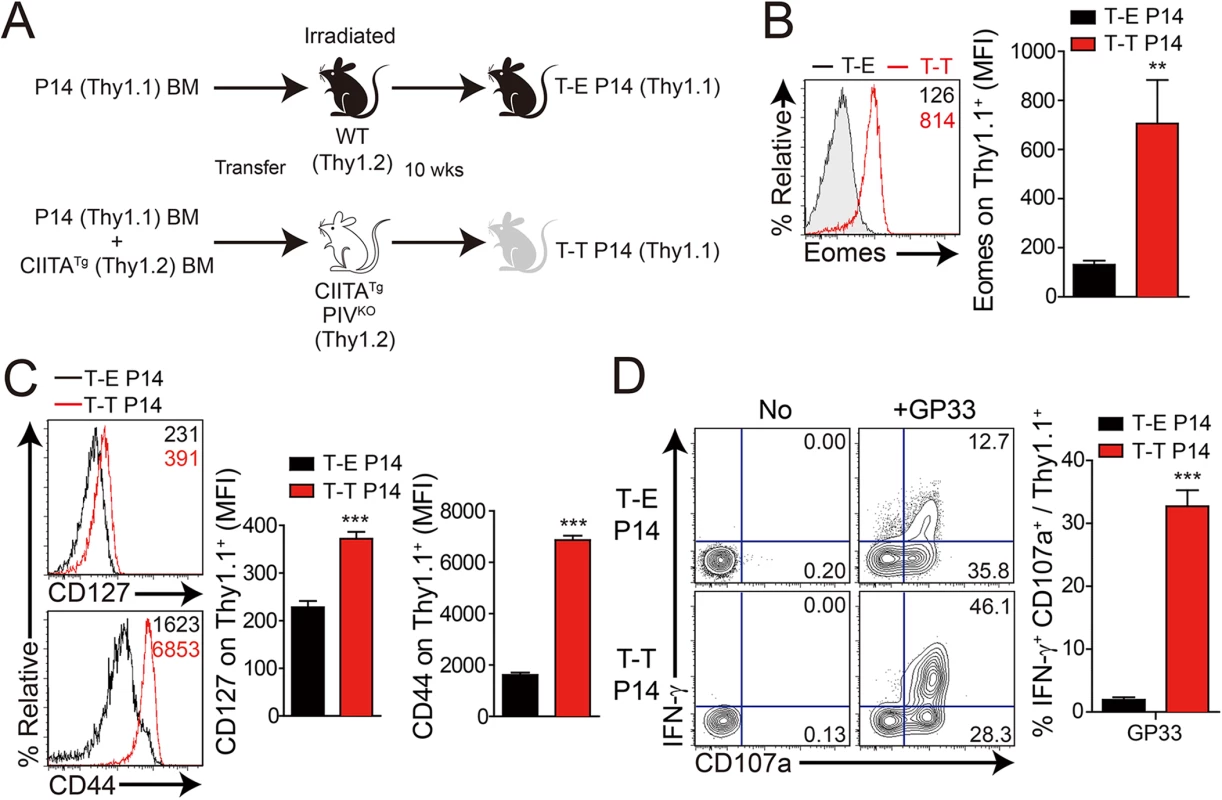
(A) Wild-type (WT) and CIITATgPIVKO host mice (Thy1.2+) were sublethally irradiated. Irradiated wild-type and CIITATgPIVKO mice were reconstituted for 10 weeks with P14 (Thy1.1+) BM (T-E P14) and a 1:1 mixture of P14 (Thy1.1+) and CIITATg (Thy1.2+) BM (T-T P14), respectively. (B and C) Lymphocytes were isolated from the spleen of T-E and T-T P14 mice. Eomes expression on P14 cells were analyzed by flow cytometry (B). Numbers in the plots indicate MFI values of Eomes staining on T-E and T-T P14 cells. MFI values are summarized in the bar graphs. CD127 and CD44 expression level on P14 cells of the spleen in T-E and T-T P14 mice were also analyzed (C). Numbers in the plots indicate CD127 and CD44 MFI value on T-E and T-T P14 cells. MFI values of CD127 and CD44 are also summarized in the bar graph. (D) Splenocytes from the BM chimeric mice were stimulated in vitro with GP33 for P14 cells for 5 hours, and stained with anti-Thy1.1, CD107a, and IFN-γ. Numbers in quadrants indicate the percentage of CD107a+ IFN-γ-producing or nonproducing P14 cells. Data were representative from more than two independent experiments and summarized data are shown; bars indicate the mean + SD. **P<0.01; ***P<0.001. After generation of virus-specific IL-4-induced innate CD8+ T cells via mixed BM chimerism, we evaluated the anti-viral function of these innate CD8+ T cells in vivo. For this, we transferred Eomes+ or Eomes- P14 cells into congenic hosts, which were then infected with CL–13 (Fig 6A). This strategy allowed us to track the response of IL-4-induced and Eomes- conventional CD8+ T cells after CL–13 infection and to judge their individual contributions to the protection against viral persistence. To examine the proliferation capability of Eomes+ or Eomes- P14 cells after CL–13 infection, each of the cells were labelled and transferred into naïve congenic mice. After 2.5 DPI, Eomes+ P14 cells showed a more division and a higher expression of CD44 compared to Eomes- P14 cells (Fig 6B). In the spleen, a 10-fold higher number of P14 cells accumulated at an early time point (5 DPI) in the mice that received Eomes+ P14 cells than in those that received Eomes- cells, although these cells were detected with similar abundance at a later time point (18 DPI) (Fig 6C). In the mice that received Eomes+ P14 cells, PD–1 was only transiently expressed on P14 cells at 5 DPI and was then later downregulated (Fig 6C). In contrast, the high level of PD–1 expression was sustained on Eomes- P14 cells. Functionally, P14 cells from mice received Eomes+ P14 cells were superior to those from recipients of Eomes- cell, with a higher fraction of these cells capable of producing both IFN-γ and TNF-α (Fig 6D). To test whether the increased CD8+ T-cell activity corresponded to better viral control we measured viral titer in the serum. Indeed, Eomes+ P14 cells also had an effect on reducing viral load during CL–13 infection (Fig 6E). These results indicate that Eomes+ P14 cells provide superior support for anti-viral activity compared to Eomes- P14 cells.
Interestingly, most of the responding CD8+ T cells of recipients were PD-1-negative in the mice that received Eomes+ P14 cells (Fig 6C) compared to those that received Eomes- P14 cells at 18 DPI. Thus, we examined the PD–1 expression on endogenous virus-specific CD8+ T cells, which are initially Eomes-negative before infection, and their cytokine production. PD–1 expression on endogenous GP276 tetramer+ CD8+ T cells was comparable to the donor Eomes+ P14 cells (Fig 6F). In addition, endogenous virus-specific CD8+ T cells in the mice that received Eomes+ P14 cells were not exhausted (Fig 6G). These data suggest that PD–1 expression in both donor Eomes+ P14 cells and endogenous virus-specific CD8+ T cells during the course of CL–13 infection presumably depend on antigen levels rather than intrinsic property of the Eomes+ or Eomes- responding cells.
Fig. 6. Control of chronic LCMV infection by innate CD8+ T cells. 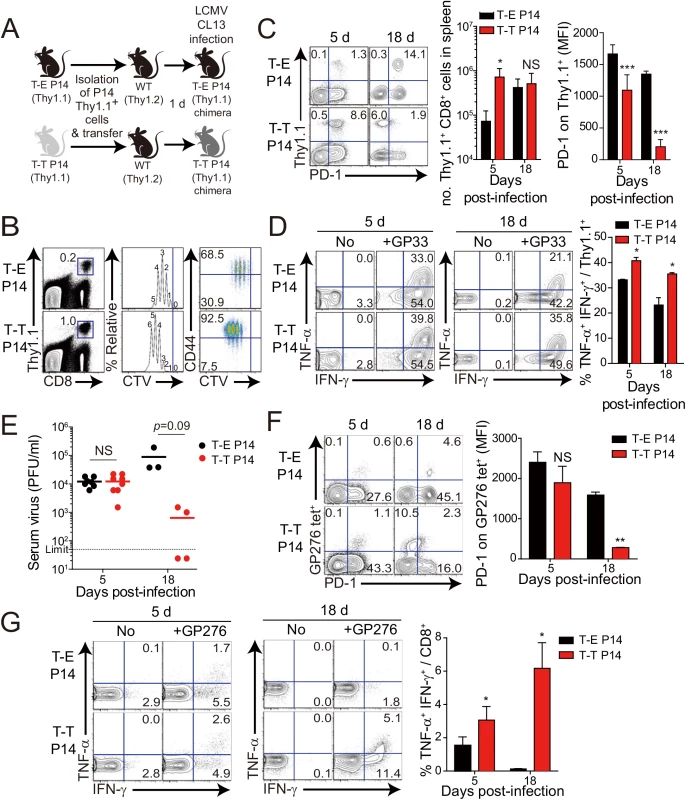
(A) T-E and T-T P14 cells were isolated from the spleens of each BM chimera as shown in Fig 5 and transferred into Thy1.2+ congenic C57BL/6 mice. One day after transfer, the mice were infected with 5 x 105 PFU of LCMV CL–13 per mouse. (B-D) Thy1.1+ CD8+ donor P14 cells were gated for following analysis. (B) CellTrace Violet (CTV)-labelled donor T-E and T-T P14 cells were analyzed at 2.5 DPI by flow cytometry. Frequency of Thy1.1+ CD8+ P14 cells among splenocytes are depicted. The gated donor Thy1.1+ CD8+ P14 cells are examined for CTV dilution along with CD44 expression. Division time and frequency of CD44hi cell population are indicated in histogram plot. (C) Lymphocytes were isolated from the spleens of T-E and T-T P14 chimeric mice at 5 and 18 DPI and analyzed by flow cytometry. The number of Thy1.1+ P14 cells in the spleen and PD–1 expression among CD8+ T cells were analyzed at indicated DPI. Numbers in the plots indicate PD–1+ or PD–1- Thy1.1+ transferred cells. PD–1 expression level (MFI) on Thy1.1+ transferred cells is summarized in the graph. (D) Splenocytes of T-E and T-T P14 chimeric mice were restimulated in vitro with GP33 for P14 cells, and frequency of IFN-γ- and TNF-α-producing Thy1.1+ transferred cells were analyzed. Numbers in the plots indicate the percentage of TNF-α+ and TNF-α- CD8+ T cells producing IFN-γ, respectively. Frequency of Thy1.1+ transferred cells producing both IFN-γ and TNF-α was summarized in the graph. (E) Serum viral titers were checked in T-E and T-T P14 chimeras at the indicated DPI. Dashed line indicates the virus detection limit. Undetectable samples were given a half of detection limit. (F-G) CD8+ T cells were gated for following analysis. (F) PD–1 expression on GP276 tetramer-positive CD8+ T cells was analyzed at indicated DPI. Numbers in the plots indicate PD–1+ or PD–1- GP276 tetramer-positive cells among CD8+ T cells. PD–1 expression level (MFI) on GP276 tetramer-positive CD8+ T cells is summarized in the graph. (G) Splenocytes of T-E and T-T P14 chimeric mice were restimulated in vitro with GP276 peptide for endogenous CD8+ T cells, and frequency of IFN-γ- and TNF-α-producing CD8+ T cells was analyzed. Numbers in the plots indicate the percentage of TNF-α+ and TNF-α- CD8+ T cells producing IFN-γ, respectively. Frequency of CD8+ T cells producing both IFN-γ and TNF-α was summarized in the graph. Bar graphs show mean + SD. Data are representative of more than two independent experiments (n≥3 per group in each experiment). NS, not significant; *P<0.05; **P<0.01; ***P<0.001. Our observation that IL-4-induced Eomes+ innate CD8+ T cells are more effective at controlling the CL–13 infection than Eomes- conventional CD8+ T cells (Fig 6) raises a question regarding the underlying mechanism. As shown in Fig 5D, Eomes+ P14 cells were able to produce more effector cytokine per cell level upon antigen stimulation than Eomes- P14 cells. In addition to an elevated effector function, Eomes+ P14 cells also showed better proliferative capability (Fig 6B), resulting in the higher frequency of these cells in the mice that received Eomes+ P14 than Eomes- P14 cells (Fig 6C). Therefore, an enhanced quantity and quality of Eomes+ P14 cells compared to Eomes- P14 cells could contribute to the accelerated control of CL–13 infection. However, it is possible that functional and numerical differences in the two populations depend on viral titers. To address this issue, we co-transferred both Eomes+ and Eomes- P14 cells into same mice (Fig 7A). At 5 DPI, we found that the frequency of Eomes+ P14 cells was significantly higher than that of Eomes- P14 cells (Fig 7B). In addition, the ability to produce effector cytokines, IFN-γ and TNF-α, was better in Eomes+ P14 cells than in Eomes- P14 cells (Fig 7C). These data indicate that functionality and proliferative capability of Eomes+ and Eomes- P14 cells upon antigen stimulation is intrinsically different independently of viral titer.
Fig. 7. Co-adoptive transfers of T-E and T-T P14 cells. 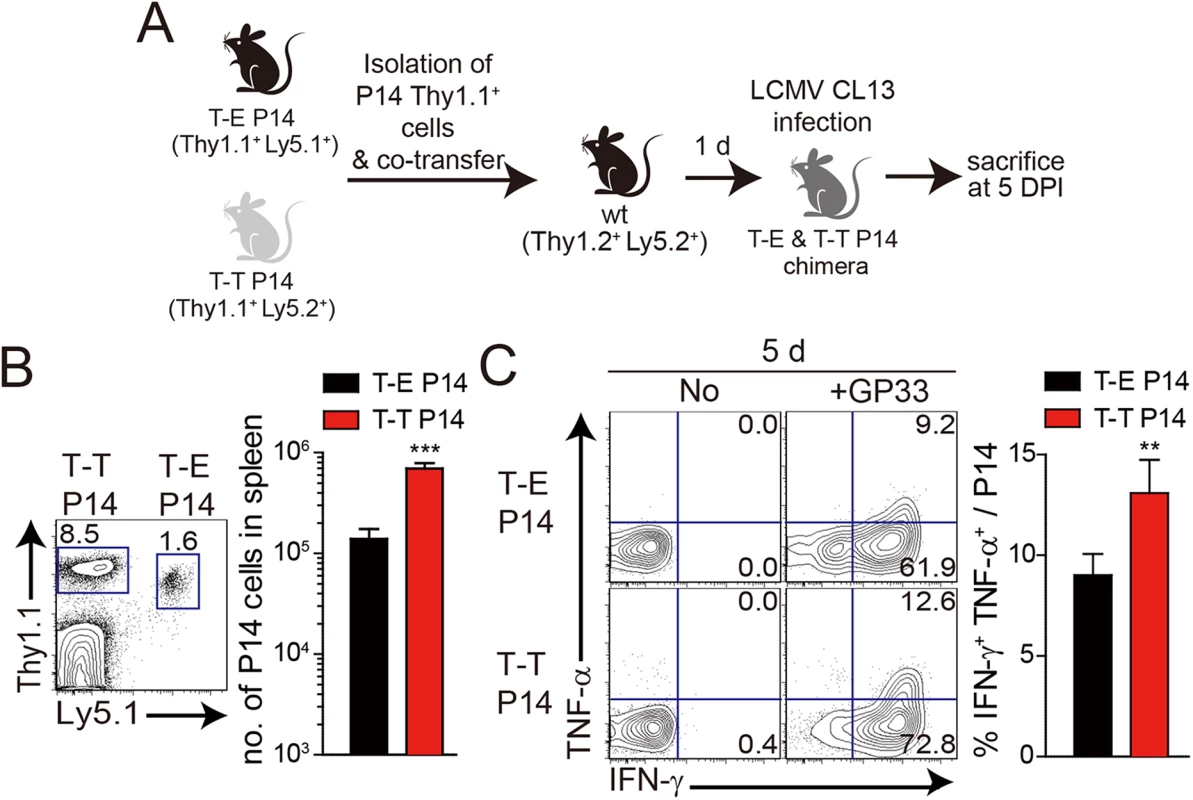
(A) T-E P14 (Thy1.1+Ly5.1+) and T-T P14 (Thy1.1+Ly5.2+) cells were isolated from the spleens of each BM chimera as shown in Fig 5 and transferred into Thy1.2+Ly5.2+ congenic C57BL/6 mice. One day after transfer, the mice were infected with LCMV CL–13. (B) Lymphocytes were isolated from the spleens of recipient mice at 5 DPI and analyzed by flow cytometry. Numbers in the plot indicates the frequency of transferred T-E or T-T P14 cells among CD8+ T cells in the spleen and absolute numbers of Thy1.1+Ly5.1+ and Thy1.1+Ly5.2+ cells in the spleen are summarized in the graph. (C) Splenocytes of T-E and T-T P14 chimeric mice were restimulated in vitro with GP33 for P14 cells, and frequency of IFN-γ- and TNF-α-producing P14 cells were analyzed. Numbers in the plots indicate the percentage of TNF-α+ and TNF-α- P14 cells producing IFN-γ, respectively. Frequency of P14 cells producing both IFN-γ and TNF-α was summarized in the graph. Bar graphs show mean + SD. n≥4 per group in each experiment. **P<0.01; ***P<0.001. Lack of IL-4-induced innate CD8+ T cells in CD1dKO BALB/c mice is associated with susceptibility to chronic viral infection
Next, we wanted to determine whether the anti-viral effect of IL-4-induced innate CD8+ T cells also occur in a wild-type host. Unlike C57BL/6 mice, BALB/c mice have an abundant population of IL-4-induced innate CD8+ T cells, whose generation is supported by IL–4 produced by intrathymic PLZF+ NKT cells [6]. Prior to investigating anti-viral CTL responses in BALB/c strain, we compared IL–4 induced innate CD8+ T-cell phenotypes on CD8+ T cells in uninfected wild-type BALB/c and CD1dKO BALB/c, in which IL-4-induced innate CD8+ T-cell generation is defective due to the absence of NKT cells. The number of virus-specific CD8+ T cells, NP118-126 (NP118) tetramer+ CD8+ T cells, was not different between wild-type and CD1dKO BALB/c mice (Fig 8A). However, the NP118 tetramer+ CD8+ T cells displayed a slightly higher Eomes expression level and the population co-expressing CD44 and CXCR3 among the NP118 tetramer+ CD8+ T cells was significantly increased in wild-type BALB/c mice than in CD1dKO BALB/c mice (Fig 8B). Similarly, the other CD8+ T cells than NP118 tetramer+ CD8+ T cells also contained the higher population of CD44hiCXCR3+ cells in wild-type BALB/c mice than in CD1dKO BALB/c mice (Fig 8C), indicating that the frequency of pre-existing innate CD8+ T cells is higher in wild-type BALB/c than in CD1dKO BALB/c prior to infection, irrespectively of antigen-specificity of CD8+ T cells.
Fig. 8. Accelerated viral control in BALB/c wild-type mice compared to BALB/c-CD1dKO mice. 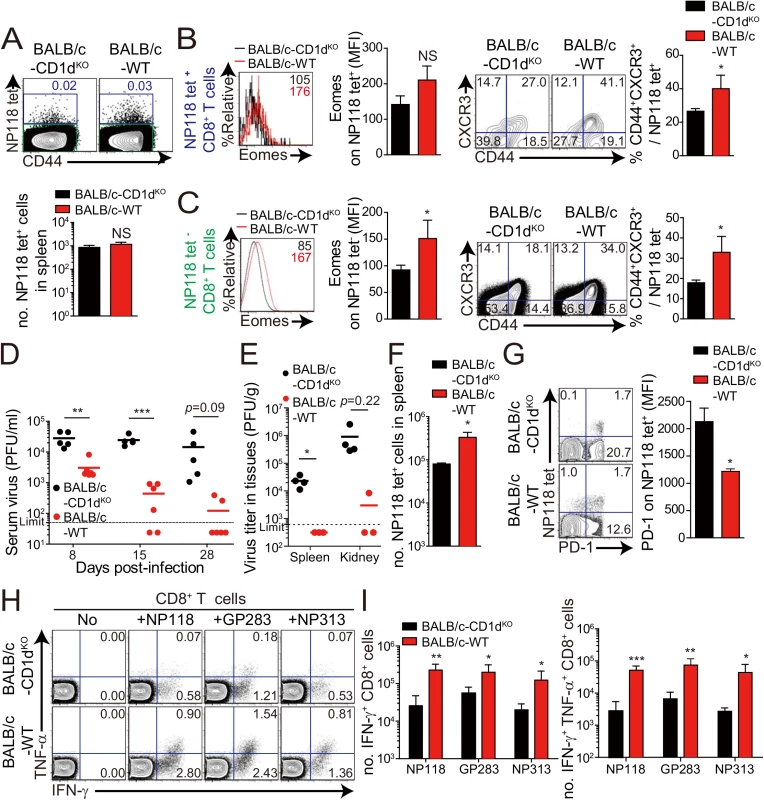
(A-C) CD8+ T cells were enriched from the spleens of BALB/c-CD1dKO and BALB/c wild-type by using magnetic sorting. NP118 tetramer+ or tetramer- CD8+ T cells were analyzed by flow cytometry. (A) NP118 tetramer+ cells are gated in blue and NP118 tetramer- cells are gated in green. The numbers in the plots indicated the frequency of NP118 tetramer+ cells among CD8+ T cells. Absolute number of NP118 tetramer+ cells in the spleen is represented in the bar graph. (B and C) Eomes expression on NP118 tetramer+ or tetramer- cells in BALB/c-CD1dKO and BALB/c wild-type. Numbers in the histogram plots indicate MFI values of Eomes stained on NP118 tetramer+ or tetramer- cells. MFI value is summarized in the bar graph. Numbers in the contour plots indicate the frequency of each population in the quadrants depending on CD44 and CXCR3 expression. The frequency of CD44hiCXCR3+ cells are summarized in the bar graph. (D-I) BALB/c-CD1dKO and BALB/c wild-type mice were infected with 2 x 105 PFU per mouse of LCMV CL–13. (D and E) Virus titers were checked in serum at the indicated DPI and in spleen and kidney extracted at 29 DPI. Dashed line indicates the virus detection limit. Undetectable samples were given a half of detection limit. (F) Absolute numbers of NP118 tetramer-positive CD8+ T cells in the spleen and lung of CL–13 infected mice were analyzed at 29 DPI. (G) Numbers in plots indicate percentage of PD–1+ or PD–1- CD8+ T cells. PD–1 expression by NP118 tetramer-positive cells after gating on CD8+ T cells were also analyzed, and summary showing the PD–1 expression level on NP118 tetramer-positive CD8+ T cells is represented by mean fluorescence intensity (MFI). (H and I) Splenocytes were restimulated in vitro with NP118, GP283-291 (GP283), or NP313-322 (NP313), and frequency of IFN-γ- and TNF-α-producing CD8+ T cells were analyzed by flow cytometry (H). Numbers in the plots indicate the percentage of TNF-α+ and TNF-α- CD8+ T cells producing IFN-γ, respectively. Absolute numbers of CD8+ T cells producing IFN-γ or both IFN-γ and TNF-α in the spleen are summarized (I). Bar graphs show mean + SD. Data are representative of two independent experiments (n≥3 per group in each experiment). *P<0.05; **P<0.01; ***P<0.001. Next, to compare anti-viral CD8+ T-cell responses in wild-type BALB/c mice with those of CD1dKO BALB/c mice, we challenged wild-type BALB/c mice and CD1dKO BALB/c mice with CL–13 (2 x 105 PFU/mouse). The viral load in peripheral blood of wild-type BALB/c mice was significantly decreased compared with that of CD1dKO hosts (Fig 8D). Moreover, wild-type BALB/c mice exhibited decreased viral titer in spleen and kidney and more abundant virus-specific CD8+ T cells in spleen and lungs than CD1dKO mice (Fig 8E and 8F). As was the case in CIITATg mice, PD–1 expression on virus-specific CD8+ T cells in wild-type BALB/c mice was not sustained at a later time point after viral infection in wild-type BALB/c mice (Fig 8G). Furthermore, we observed that CD8+ T cells from wild-type BALB/c mice produced higher amounts of effector cytokines such as IFN-γ and TNF-α than did those from CD1dKO mice (Fig 8H and 8I). These data suggest that IL-4-induced innate CD8+ T cells in a wild-type BALB/c host also contributed to reduce CL–13 viral load compared with CD1dKO mice.
Discussion
Over the course of viral infection there may be a limited time period during which the host system can eliminate the virus [22]. When viruses are not eliminated within this period of time, virus-specific CD8+ T cells are exhausted via PD–1 (programmed death 1) and its ligand, PD-L1 interaction, resulting in a chronic infection [23]. In this study we demonstrated that IL-4-induced innate CD8+ T cells are able to effectively control the chronic viral infection. For this, we first compared T-cell responses to chronic viral infection induced by LCMV CL–13 in CIITATg and wild-type C57BL/6 mice. The immune system of the CIITATg mouse resembles that of humans with respect to MHC class II expression in both thymic epithelial cells and thymocytes making it a suitable model [24–27]. Thus, PLZF+ T-T CD4+ T cells are generated in response to TCR signals from the MHC class II/peptide complex expressed on thymocytes [15,27,28] and provide IL–4 for the development of IL-4-induced innate CD8+ T cells [14]. When mice were infected with CL–13, CIITATg mice were able to control viral titers below detection levels in selected tissues such as the spleen and serum within a month, whereas wild-type mice succumbed to persistent infection. Furthermore, the ability of CIITATg mice to control the virus was dependent on IL-4-induced innate CD8+ T cells as CIITATg and control mice did not show a difference in serum viral titers on the IL–4 deficient background, which causes a lack of intrathymic generation of IL-4-induced innate CD8+ T cells. Moreover, adoptive transfer of LCMV-specific IL-4-induced innate CD8+ T cells into wild-type hosts further confirmed the crucial role of these innate T cells as the primary effector mechanism for viral control. We also demonstrated that wild-type BALB/c mice, which have abundant IL-4-induced innate CD8+ T cells, exhibit notably enhanced anti-viral CTL responses compared with CD1dKO BALB/c mice, which only possess a very small fraction of these cells. As expected, expression level of Eomes and innate CD8+ T cell marker (CD44 and CXCR3) was higher in virus-specific CD8+ T cells wild-type mice, as compared to those of CD1dKO BALB/c mouse (Fig 8B), while we could not found any difference in LCMV NP118-specific CD8+ T cell numbers in these mice (Fig 8A). Considering the results from P14 cell adoptive transfer and CL–13 infection (Fig 6), these data favor the idea that IL-4-induced innate CD8+ T cells in a wild-type BALB/c host contributed to reduce CL–13 viral load compared with CD1dKO mice, although the genetic perturbation in CD1dKO mice is not specific for the innate CD8+ T cell population.
Eomes is a key transcription factor in the cytotoxic T-cell lineage [29]. During the activation and differentiation of mature CD8+ T cells Eomes induces effector function and cooperates with T-bet to sustain memory CD8+ T-cell homeostasis [30]. In particular, the central memory population is diminished in CD8+ T cells lacking Eomes [29,30]. Moreover, in the thymus Eomes also seems to confer effector function and memory phenotypes to innate CD8+ T cells as IL-4-induced innate CD8+ T cells acquire a CD44hiCD62Lhi central memory cell-like phenotype [14]. In addition to the previous reports, our comparison data of phenotype and function in between IL-4-induced innate CD8+ T cells and memory CD8+ T cells showed that the expression levels of Eomes and CXCR3 were similar but those of CD44, CD124, CD24, and NKG2D were different (S3A Fig). IFN-γ production and degranulation ability upon antigen stimulation were also different (S3B Fig). Although these IL-4-induced innate CD8+ T cells are less functional than memory CD8+ T cells, their function is evidently better than those of naïve CD8+ T cells. These data demonstrate that IL-4-induced innate CD8+ T cells are phenotypically and functionally different from conventional memory CD8+ T cells as well as naïve CD8+ T cells.
On the other hand, the expression of Eomes mRNA and protein are markedly elevated in exhausted CD8+ T cells during chronic virus infection compared to that in effector or memory CD8+ T cells [20,31]. These finding suggest that Eomes alone is not sufficient to stimulate the effector function of exhausted CD8+ T cells under the conditions of established chronic virus infection. This is despite the fact that upregulation of Eomes initially triggers the effector function of CD8+ T cells upon TCR stimulation and contributes to preserve the functionality of memory CD8+ T cells. In the present study, when CD8+ T cells already express a significantly high level of Eomes, these cells acquire obviously enhanced ability to produce effector cytokines such as IFN-γ and TNF-α upon viral antigen challenge. Taken together, these data suggest that a high level of Eomes expression allows IL-4-induced innate CD8+ T cells to exhibit their prompt and significant effector function, thereby controlling viremia during the early phase of virus infection.
Many researchers have attempted to control chronic virus infection using immunotherapeutic interventions such as blockade of the inhibitory receptors PD–1 and CTLA–4, administration of type I IFN, and regulation of microRNAs [32] [33,34]. Additionally, for chronic hepatitis B virus infection treatment, adoptive T-cell therapy using either in vitro expanded hepatitis B virus antigen-specific T cells or grafting T cells with recombinant TCR has been investigated as an approach [35]. Based on our data, virus-specific IL-4-induced innate CD8+ T cells have the potential to be used in adoptive T-cell therapy. Interestingly, when we adoptively transferred Eomes+ LCMV-specific innate CD8+ T cells into CL-13-infected mice, the CTL response was slightly increased even though the transferred cells were seemed still exhausted (S4 Fig). From this experiment, we hypothesize that combination therapy with adoptive transfer of IL-4-induced innate CD8+ T cells for prompt control of virus and PD–1 blockade for rejuvenating CTL function could be effective. Further experiments will be required to test this theory.
X-linked lymph proliferative disease (XLP) is a human immunodeficiency caused by germ-line mutations in SH2D1A gene and characterized by an inability to respond appropriately to infections such as Epstein-Barr virus [36]. The SH2D1A gene encodes the SAP molecule that is a component of the SLAM (signaling lymphocytic activation molecule) signaling pathway. Signaling through the SLAM family of receptors is crucial for the development of NKT cells, and thus the absence of the SAP causes an arrest in NKT cell development [37] [38,39]). In this context, the deficiency of NKT cells has been considered as one of the cellular bases of XLP [38]. However, considering the crucial role of SAP for T-T CD4+ T-cell development [40], the development and function of T-T CD4+ T cells would also be expected to be defective in XLP patients [27]. Both NKT and T-T CD4+ T cells are engaged in the generation of innate CD8+ T cells via IL–4 production and thus, the development of IL-4-induced innate CD8+ T cells is also dependent on the adaptor SAP [41]. In the present study, we demonstrated that IL-4-induced innate CD8+ T cells are able to rapidly proliferate, secrete cytokines, and decrease viral load after LCMV CL–13 infection. Taken together, it is necessary to consider the possibility that defective development of IL-4-induced innate CD8+ T cells causes the heighten susceptibility to Epstein-Barr virus infection in XLP patients.
Materials and Methods
Ethics statement
Animals were maintained and procedures were performed with approval of the IACUCs of Seoul National University (permit number: SNUIBC-R100524-1) and Yonsei University (permit number: 2013–0115) in accordance to LABORATORY ANIMAL ACT of Korean Ministry of Food and Drug Safety for enhancing the ethics and reliability on animal testing through appropriate administration of laboratory animals and animal testing.
Mice, infection, and titration
C57BL/6, IL-4KO, BALB/c and BALB/c-CD1dKO mice were purchased from the Jackson Laboratory. The CIITATg mice were previously generated at Seoul National University [26] and CIITATg mice were bred to IL-4KO and PIVKO mice in our laboratory to generate CIITATgIL-4KO and CIITATgPIVKO. LCMV epitope-specific TCR transgenic P14 Thy1.1+ Ly5.2+ mice were obtained from the Emory Vaccine Center, USA and P14 Thy1.1+ Ly5.1+ mice were obtained from POSTECH, Korea. All mice were maintained in the specific pathogen-free facility of the Yonsei Laboratory Animal Research Center at Yonsei University and the Center for Animal Resource Development at Seoul National University College of Medicine (Seoul, Korea). LCMV CL–13, a variant derived from an LCMV ARM CA1371 carrier mouse [42], was obtained from Rafi Ahmed (Emory Vaccine Center, Atlanta). Six - to ten-week old mice were infected with 1 x 105 to 2 x 106 PFU of LCMV CL–13 diluted in serum-free RPMI medium per 20 g of mouse body weight by intravenous infection or with 2 x 105 PFU of LCMV Armstrong diluted in serum-free RPMI medium per 20 g of mouse body weight by intraperitoneal infection. For serum virus titration, three to four drops of blood were individually collected by microcapillary tube at the indicated time points post infection, and the serum was directly stored at -70°C. For tissue titration, small pieces of the spleen and kidney were put in DMEM containing 1% FBS (HyClone) and stored at -70°C. The tissues were later homogenized completely using a homogenizer (Kinematica) before titration. Viral titers from sera or homogenized samples were determined by plaque assay on Vero cells as previously described [43]. Undetectable samples were given a half of each detection limit.
Cell isolation, antibodies, and staining
Peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood using density gradient centrifugation underlaid with Histopaque–1077 (Sigma-Aldrich). Lymphocytes from the thymus, spleen and lung were isolated as previously described [23]. For phenotypic analysis of lymphocytes, single-cell suspensions were stained with the following antibodies; fluorochrome-conjugated antibodies against CD4 (RM4-5), CD127 (A7R34), and PD–1 (RMP1-13) were from BioLegend, antibodies against CD8 (53–6.7), CD24 (30-F1), CD44 (IM7), CXCR3 (CXCR3-173), and CD19 (eBio1D3) were from eBioscience, and antibodies against CD44 (IM7), CD90.1 (Thy1.1; OX–1), CD122 (IL-2Rβ; TM-b1), CD124 (IL-4Rα; mIL4R-M1), CXCR5 (2G8), CD45R/B220 (RA3-6B2), CD95 (Fas; Jo2), T - and B-cell activation antigen (GL7), and NKG2D (CX5) were from BD Biosciences. H-2Db tetramers bound to GP33-41 or GP276-286 peptide and H-2Ld tetramer bound to NP118-126 peptide were generated and used as previously described [44]. To detect cytokine production by virus-specific CD8+ T cells, splenocytes from C57BL/6 mice were restimulated in vitro with 0.2 μg/ml of LCMV GP33-41, GP276-286, or peptide pool including GP33-41, GP276-286, GP70-77, GP92-101, NP166-175, NP205-212, NP235-249, and NP396-404 in the presence of Golgi plug/Golgi stop (BD Biosciences) and anti-CD107a (1D4B, BD Biosciences) Ab for 5 hours followed by intracellular cytokine staining using anti-IFN-γ (XMG1.2, BD Biosciences) and anti-TNF-α (MP6-XT22, BioLegend) antibodies. In case of in vitro restimulation of splenocytes from BALB/c mice, LCMV NP118-126, GP283-291, or NP313-322 peptide was used. For IL-4-induced innate CD8+ T-cell staining, lymphocytes were fixed and permeabilized with Fixation/Permeabilization solution (eBioscience) and stained with anti-Eomes (Dan11mag, eBioscience) Ab. The Live/Dead fixable Stain Kit (Invitrogen) was used to remove the dead cell population in most staining procedures. All stained samples were read by FACS CANTO II or LSR II (BD Biosciences), and analyzed by FlowJo software (Tree Star).
Histopathology
Lungs were fixed in 10% neutral buffered formalin (Sigma-Aldrich), and paraffin-embedded tissues were sectioned to a thickness of 4 μm and stained with hematoxylin and eosin. Microscopic observations were performed with an ECLIPSE 80i Bright-Field Microscope Set (Nikon) equipped with CFI 10×/22 eyepiece, Plan Fluor objectives (with 4×, 10×, 20×, and 100× objectives) and DS-Fi1 camera. We used NIS-Elements BR 3.1 software (Nikon) for image acquisition.
ELISA (enzyme-linked immunosorbent assay)
LCMV-specific antibodies were measured by ELISA. LCMV CL-13-infected BHK–21 cell lysate was used as capture antigen. Ninety-six-well Polysorp plates (Nunc) were coated with sonicated lysate for 3 days before performing the ELISA. Three fold serial dilutions of serum samples were incubated and detected with IgG-specific horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin (Southern Biotech). TMB (Sigma-Aldrich) was used as a substrate, and the reaction was stopped by sulfuric acid and read at 450nm.
BM chimeras
BM cells were isolated from femurs and tibias of donor mice and T cells were depleted through magnetic sorting using a mixture of CD4 and CD8 depleting microbeads (Miltenyi Biotech, Auburn, CA). Then, 3 x 106 T cell-depleted BM cells were intravenous injected into each recipient mouse preconditioned with 13,000 rad irradiation (two doses of 650 rad applied 4 h apart) from a 137Cs source 1 day prior.
Cell purification and adoptive transfer
For CD8+ T cell enrichment, CD8+ T cells were isolated from the spleen of wild-type BALB/c and CD1dKO BALB/c mice after negative selection using a CD8+ T-cell isolation kit (Miltenyi Biotech).
For adoptive transfer of P14 Thy1.1+ Ly5.2+ CD8+ T cells and P14 Thy1.1+ Ly5.1+ CD8+ T cells, the cells were isolated from the spleen of BMT chimera mice after negative selection using a CD8+ T-cell isolation kit and subsequent positive selection using Thy1.1+ cell microbeads (Miltenyi Biotech). After isolation, 5 x 103 purified P14 Thy1.1+ CD8+ T cells were transferred alone or with P14 Ly5.1+ CD8+ T cells into wild-type C57BL/6 mice via tail vein. Mice were infected with LCMV CL–13 at 1d after the adoptive transfer.
In vivo proliferation assay
LCMV GP33-41-specific P14 CD8+ T cells were isolated from P14 transgenic mice using CD8+ isolation kit (Myltenyi Biotec). Purified P14 CD8+ T cells were labelled with CellTrace Violet (CTV) proliferation kit at concentration of 10uM (Invitrogen). Labelled P14 CD8+ T cells (1 x 106 cells) were adoptively transferred into naïve mice. Mice were infected with LCMV CL–13 at 1d after the adoptive transfer.
Therapeutic adoptive transfer
Thy1.2+ congenic C57BL/6 mice were infected with 2 x 106 PFU of LCMV CL–13. At 3 weeks post infection, P14 Thy1.1+ CD8+ T cells were isolated from the spleen of BMT chimera mice for adoptive transfer after negative selection using a CD8+ T-cell isolation kit and subsequent positive selection using Thy1.1+ cell microbeads (Miltenyi Biotech). After isolation, 2 x 106 purified P14 Thy1.1+ CD8+ T cells were transferred into CL-13-infected C57BL/6 mice via tail vein.
Statistical analysis
Survival curve was evaluated by log-rank (Mantel-Cox) test and other data were analyzed by two-tailed unpaired Student’s t-test using GraphPad Prism software. A p value less than 0.05 was considered statistically significant.
Supporting Information
Zdroje
1. Berg LJ (2007) Signalling through TEC kinases regulates conventional versus innate CD8+ T-cell development. Nat Rev Immunol 7 : 479–485. 17479128
2. Veillette A, Dong Z, Latour S (2007) Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes. Immunity 27 : 698–710. 18031694
3. Lee YJ, Jameson SC, Hogquist KA (2011) Alternative memory in the CD8 T cell lineage. Trends Immunol 32 : 50–56. doi: 10.1016/j.it.2010.12.004 21288770
4. Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ (2006) The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity 25 : 79–91. 16860759
5. Broussard C, Fleischacker C, Horai R, Chetana M, Venegas AM, Sharp LL, et al. (2006) Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity 25 : 93–104. 16860760
6. Weinreich MA, Odumade OA, Jameson SC, Hogquist KA (2010) T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol 11 : 709–716. doi: 10.1038/ni.1898 20601952
7. Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA (2009) KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity 31 : 122–130. doi: 10.1016/j.immuni.2009.05.011 19592277
8. Fukuyama T, Kasper LH, Boussouar F, Jeevan T, van Deursen J, Brindle PK (2009) Histone acetyltransferase CBP is vital to demarcate conventional and innate CD8+ T-cell development. Mol Cell Biol 29 : 3894–3904. doi: 10.1128/MCB.01598-08 19433445
9. Jordan MS, Smith JE, Burns JC, Austin JE, Nichols KE, Aschenbrenner AC, et al. (2008) Complementation in trans of altered thymocyte development in mice expressing mutant forms of the adaptor molecule SLP76. Immunity 28 : 359–369. doi: 10.1016/j.immuni.2008.01.010 18342008
10. Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, et al. (2010) Development of promyelocytic zinc finger and ThPOK-expressing innate γδ T cells is controlled by strength of TCR signaling and Id3. J Immunol 184 : 1268–1279. doi: 10.4049/jimmunol.0903218 20038637
11. Verykokakis M, Boos MD, Bendelac A, Kee BL (2010) SAP protein-dependent natural killer T-like cells regulate the development of CD8+ T cells with innate lymphocyte characteristics. Immunity 33 : 203–215. doi: 10.1016/j.immuni.2010.07.013 20674402
12. Ueda-Hayakawa I, Mahlios J, Zhuang Y (2009) Id3 restricts the developmental potential of γδ lineage during thymopoiesis. J Immunol 182 : 5306–5316. doi: 10.4049/jimmunol.0804249 19380777
13. Verykokakis M, Boos MD, Bendelac A, Adams EJ, Pereira P, Kee BL (2010) Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent "innate" γδ T cells. PLoS One 5: e9303. doi: 10.1371/journal.pone.0009303 20174563
14. Min HS, Lee YJ, Jeon YK, Kim EJ, Kang BH, Jung KC, et al. (2011) MHC class II-restricted interaction between thymocytes plays an essential role in the production of innate CD8+ T cells. J Immunol 186 : 5749–5757. doi: 10.4049/jimmunol.1002825 21478404
15. Lee YJ, Jeon YK, Kang BH, Chung DH, Park CG, Shin HY, et al. (2010) Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. J Exp Med 207 : 237–246. doi: 10.1084/jem.20091519 20038602
16. Treiner E, Lantz O (2006) CD1d - and MR1-restricted invariant T cells: of mice and men. Curr Opin Immunol 18 : 519–526. 16870416
17. Virgin HW, Wherry EJ, Ahmed R (2009) Redefining chronic viral infection. Cell 138 : 30–50. doi: 10.1016/j.cell.2009.06.036 19596234
18. West EE, Youngblood B, Tan WG, Jin HT, Araki K, Alexe G, et al. (2011) Tight regulation of memory CD8+ T cells limits their effectiveness during sustained high viral load. Immunity 35 : 285–298. doi: 10.1016/j.immuni.2011.05.017 21856186
19. Blattman JN, Wherry EJ, Ha SJ, van der Most RG, Ahmed R (2009) Impact of epitope escape on PD–1 expression and CD8 T-cell exhaustion during chronic infection. J Virol 83 : 4386–4394. doi: 10.1128/JVI.02524-08 19211743
20. Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. (2007) Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27 : 670–684. 17950003
21. Waldburger JM, Suter T, Fontana A, Acha-Orbea H, Reith W (2001) Selective abrogation of major histocompatibility complex class II expression on extrahematopoietic cells in mice lacking promoter IV of the class II transactivator gene. J Exp Med 194 : 393–406. 11514597
22. Okazaki T, Honjo T (2006) Rejuvenating exhausted T cells during chronic viral infection. Cell 124 : 459–461. 16469690
23. Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, et al. (2006) Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439 : 682–687. 16382236
24. Park SH, Bae YM, Kim TJ, Ha IS, Kim S, Chi JG, et al. (1992) HLA-DR expression in human fetal thymocytes. Hum Immunol 33 : 294–298. 1639632
25. Li W, Kim MG, Gourley TS, McCarthy BP, Sant'Angelo DB, Chang CH (2005) An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity 23 : 375–386. 16226503
26. Choi EY, Jung KC, Park HJ, Chung DH, Song JS, Yang SD, et al. (2005) Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity 23 : 387–396. 16226504
27. Lee YJ, Jung KC, Park SH (2009) MHC class II-dependent T-T interactions create a diverse, functional and immunoregulatory reaction circle. Immunol Cell Biol 87 : 65–71. doi: 10.1038/icb.2008.85 19030015
28. Qiao Y, Zhu L, Sofi H, Lapinski PE, Horai R, Mueller K, et al. (2012) Development of promyelocytic leukemia zinc finger-expressing innate CD4 T cells requires stronger T-cell receptor signals than conventional CD4 T cells. Proc Natl Acad Sci U S A 109 : 16264–16269. doi: 10.1073/pnas.1207528109 22988097
29. Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, et al. (2010) Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol 185 : 4988–4992. doi: 10.4049/jimmunol.1002042 20935204
30. Kaech SM, Cui W (2012) Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12 : 749–761. doi: 10.1038/nri3307 23080391
31. Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, et al. (2012) Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338 : 1220–1225. doi: 10.1126/science.1229620 23197535
32. Ha SJ, Mueller SN, Wherry EJ, Barber DL, Aubert RD, Sharpe AH, et al. (2008) Enhancing therapeutic vaccination by blocking PD-1-mediated inhibitory signals during chronic infection. J Exp Med 205 : 543–555. doi: 10.1084/jem.20071949 18332181
33. Wijesundara DK, Xi Y, Ranasinghe C (2014) Unraveling the convoluted biological roles of type I interferons in infection and immunity: a way forward for therapeutics and vaccine design. Front Immunol 5 : 412. doi: 10.3389/fimmu.2014.00412 25221557
34. van der Ree MH, van der Meer AJ, de Bruijne J, Maan R, van Vliet A, Welzel TM, et al. (2014) Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res 111 : 53–59. doi: 10.1016/j.antiviral.2014.08.015 25218783
35. Bohne F, Protzer U (2007) Adoptive T-cell therapy as a therapeutic option for chronic hepatitis B. J Viral Hepat 14 Suppl 1 : 45–50. 17958642
36. MacDonald HR, Schumann J (2005) The need for natural killer T cells. Nat Med 11 : 256–257. 15746936
37. Godfrey DI, Berzins SP (2007) Control points in NKT-cell development. Nat Rev Immunol 7 : 505–518. 17589542
38. Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, et al. (2005) Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med 11 : 340–345. 15711562
39. Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, et al. (2007) Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 27 : 751–762. 18031695
40. Li W, Sofi MH, Rietdijk S, Wang N, Terhorst C, Chang CH (2007) The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity 27 : 763–774. 18031696
41. Horai R, Mueller KL, Handon RA, Cannons JL, Anderson SM, Kirby MR, et al. (2007) Requirements for selection of conventional and innate T lymphocyte lineages. Immunity 27 : 775–785. 18031697
42. Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB (1984) Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med 160 : 521–540. 6332167
43. Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R (2003) Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 77 : 4911–4927. 12663797
44. Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, et al. (1998) Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8 : 177–187. 9491999
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized VirusČlánek Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell ProliferationČlánek Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite InterfaceČlánek Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 10- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Expression of Concern: Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Preparing for the Next Epidemic with Basic Virology
- Effectively Communicating the Uncertainties Surrounding Ebola Virus Transmission
- Translating Basic Research into Clinical Applications: Malaria Research at an NIH Lab
- A Gut Odyssey: The Impact of the Microbiota on Spore Formation and Germination
- Papillomavirus E6 Oncoproteins Take Common Structural Approaches to Solve Different Biological Problems
- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens
- Addressing the Complications of Ebola and Other Viral Hemorrhagic Fever Infections: Using Insights from Bacterial and Fungal Sepsis
- Time for Chocolate: Current Understanding and New Perspectives on Cacao Witches’ Broom Disease Research
- Ganglioside and Non-ganglioside Mediated Host Responses to the Mouse Polyomavirus
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Structure Elucidation of Coxsackievirus A16 in Complex with GPP3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses
- CD39 Expression Identifies Terminally Exhausted CD8 T Cells
- Abiotic Stresses Antagonize the Rice Defence Pathway through the Tyrosine-Dephosphorylation of OsMPK6
- Dissociation of Tissue Destruction and Bacterial Expansion during Bubonic Plague
- Interferon-γ: The Jekyll and Hyde of Malaria
- CCR2 Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8 T Cell Response to the Protective YopE Epitope during Infection
- A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1
- The Suramin Derivative NF449 Interacts with the 5-fold Vertex of the Enterovirus A71 Capsid to Prevent Virus Attachment to PSGL-1 and Heparan Sulfate
- Trans-generational Immune Priming Protects the Eggs Only against Gram-Positive Bacteria in the Mealworm Beetle
- Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection
- Respiratory Syncytial Virus Disease Is Mediated by Age-Variable IL-33
- TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized Virus
- Modeling the Effects of Vorinostat Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells
- Identification of a Novel Lipoprotein Regulator of Spore Germination
- Calcium Regulation of Hemorrhagic Fever Virus Budding: Mechanistic Implications for Host-Oriented Therapeutic Intervention
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
- Comparative Life Cycle Transcriptomics Revises Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates
- The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy
- Carcinogenic Parasite Secretes Growth Factor That Accelerates Wound Healing and Potentially Promotes Neoplasia
- Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell Proliferation
- Dengue Virus Infection of Requires a Putative Cysteine Rich Venom Protein
- Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma
- Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite Interface
- Phenotypic and Functional Alterations in Circulating Memory CD8 T Cells with Time after Primary Infection
- Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems
- Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses
- Myeloid Cell Arg1 Inhibits Control of Arthritogenic Alphavirus Infection by Suppressing Antiviral T Cells
- The White-Nose Syndrome Transcriptome: Activation of Anti-fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis
- Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles
- Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine
- Differentiation-Dependent KLF4 Expression Promotes Lytic Epstein-Barr Virus Infection in Epithelial Cells
- The Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
- C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in
- Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
- Crystal Structure of the Human Cytomegalovirus Glycoprotein B
- Depletion of . GlmU from Infected Murine Lungs Effects the Clearance of the Pathogen
- Immunologic Control of Papillomavirus Type 1
- Requires Host Rab1b for Survival in Macrophages
- Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi
- PD-L1 Expression on Retrovirus-Infected Cells Mediates Immune Escape from CD8 T Cell Killing
- Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction
- IL-4 Induced Innate CD8 T Cells Control Persistent Viral Infection
- Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
- BCG Skin Infection Triggers IL-1R-MyD88-Dependent Migration of EpCAM CD11b Skin Dendritic cells to Draining Lymph Node During CD4+ T-Cell Priming
- Antigenic Characterization of the HCMV gH/gL/gO and Pentamer Cell Entry Complexes Reveals Binding Sites for Potently Neutralizing Human Antibodies
- Rescue of a Plant Negative-Strand RNA Virus from Cloned cDNA: Insights into Enveloped Plant Virus Movement and Morphogenesis
- Geminivirus Activates to Accelerate Cytoplasmic DCP2-Mediated mRNA Turnover and Weakens RNA Silencing in
- Disruption of Sphingolipid Biosynthesis Blocks Phagocytosis of
- The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps
- The Timing of Stimulation and IL-2 Signaling Regulate Secondary CD8 T Cell Responses
- Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity
- The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Interferon-γ: The Jekyll and Hyde of Malaria
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



